m6A Target Gene Information
General Information of the m6A Target Gene (ID: M6ATAR00298)
Full List of m6A Methylation Regulator of This Target Gene and Corresponding Disease/Drug Response(s)
ITGB1
can be regulated by the following regulator(s), and cause disease/drug response(s). You can browse detail information of regulator(s) or disease/drug response(s).
Browse Regulator
Browse Disease
Fat mass and obesity-associated protein (FTO) [ERASER]
| Representative RNA-seq result indicating the expression of this target gene regulated by FTO | ||
| Cell Line | 253J cell line | Homo sapiens |
|
Treatment: siFTO 253J cells
Control: 253J cells
|
GSE150239 | |
| Regulation |
  |
logFC: 6.90E-01 p-value: 1.74E-07 |
| More Results | Click to View More RNA-seq Results | |
| In total 1 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [1] | |||
| Response Summary | FTO was an independent risk factor for overall survival (OS) of GC patients and FTO could promote GC metastasis by upregulating the expression of Integrin beta-1 (ITGB1) via decreasing its m6A level. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Gastric cancer | ICD-11: 2B72 | ||
| In-vitro Model | SNU-216 | Gastric tubular adenocarcinoma | Homo sapiens | CVCL_3946 |
| MKN7 | Gastric tubular adenocarcinoma | Homo sapiens | CVCL_1417 | |
| MGC-803 | Gastric mucinous adenocarcinoma | Homo sapiens | CVCL_5334 | |
| HGC-27 | Gastric carcinoma | Homo sapiens | CVCL_1279 | |
| BGC-823 | Gastric carcinoma | Homo sapiens | CVCL_3360 | |
| AGS | Gastric adenocarcinoma | Homo sapiens | CVCL_0139 | |
Methyltransferase-like 3 (METTL3) [WRITER]
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | LX2 cell line | Homo sapiens |
|
Treatment: shMETTL3 LX2 cells
Control: shLuc LX2 cells
|
GSE207909 | |
| Regulation |
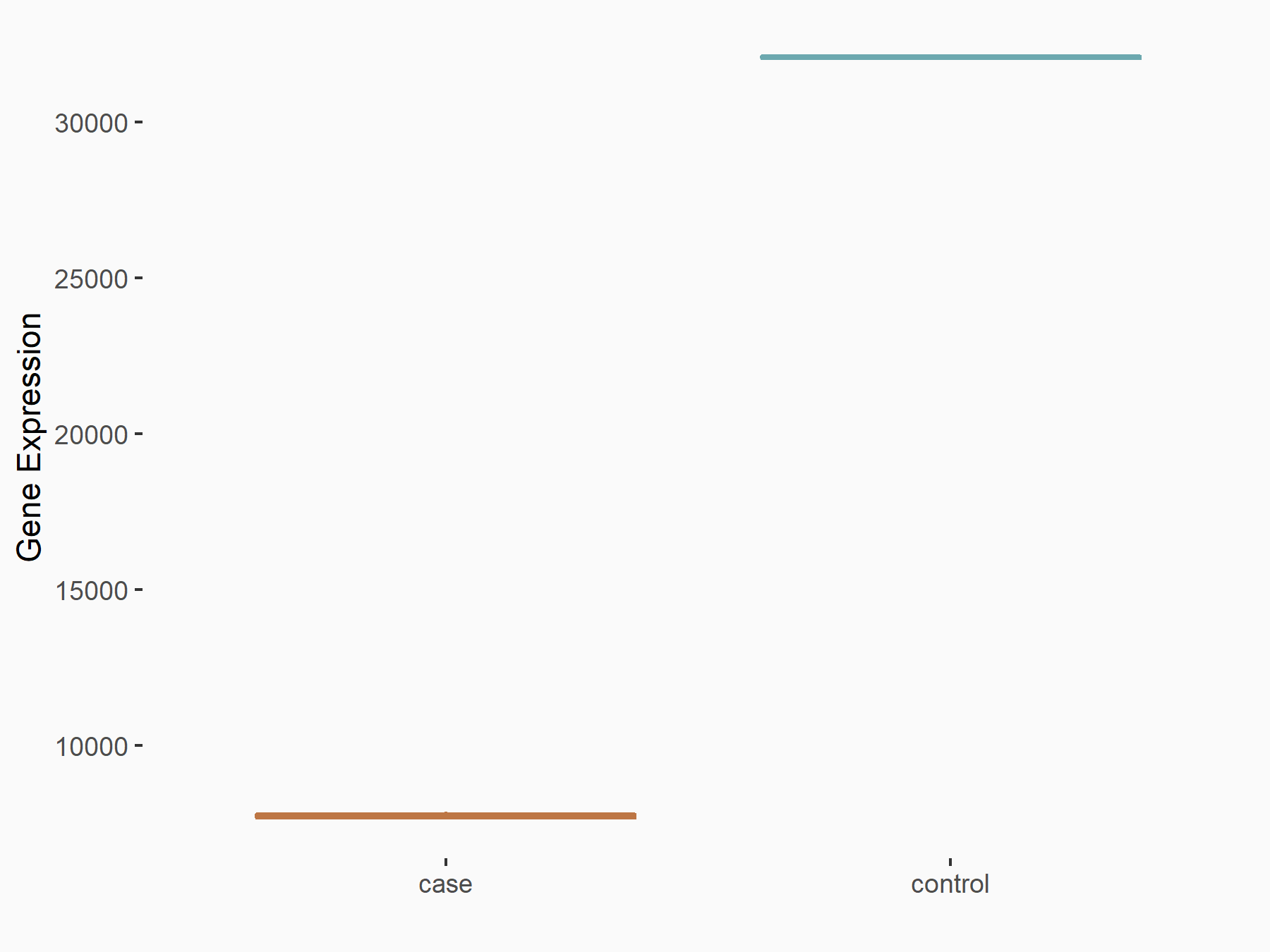 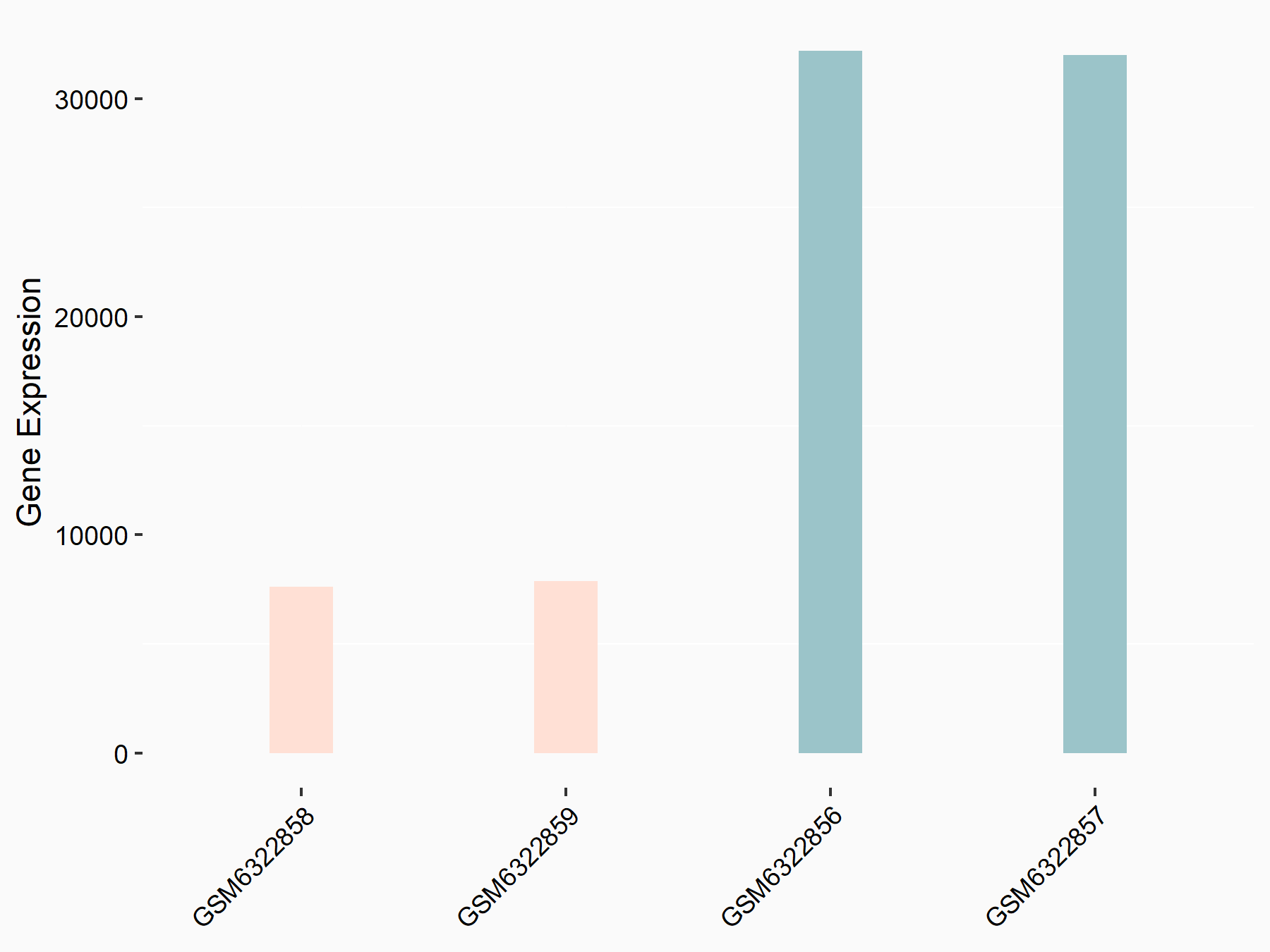 |
logFC: -2.05E+00 p-value: 1.83E-215 |
| More Results | Click to View More RNA-seq Results | |
| In total 1 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [2] | |||
| Response Summary | METTL3 regulates the expression of Integrin beta-1 (ITGB1) through m6A-HuR-dependent mechanism, which affects the binding of ITGB1 to Collagen I and tumor cell motility, so as to promote the bone metastasis of prostate cancer. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Prostate cancer | ICD-11: 2C82 | ||
| Pathway Response | Cell adhesion molecules | hsa04514 | ||
| Cell Process | Cell adhesion | |||
| In-vitro Model | LNCaP | Prostate carcinoma | Homo sapiens | CVCL_0395 |
| PC-3 | Prostate carcinoma | Homo sapiens | CVCL_0035 | |
| In-vivo Model | All the mice were randomly divided into five groups of four mice each, including PC3-shNC, PC3-shMETTL3-1/2, LNCaP-vector and LNCaP-METTL3 group. | |||
YTH domain-containing family protein 2 (YTHDF2) [READER]
| In total 1 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [5] | |||
| Response Summary | KAT1 triggers YTHDF2-mediated Integrin beta-1 (ITGB1) mRNA instability to alleviate the progression of DR. | |||
| Target Regulation | Down regulation | |||
| Responsed Disease | Diabetic retinopathy | ICD-11: 9B71.0 | ||
| Pathway Response | PI3K-Akt signaling pathway | hsa04151 | ||
| Cell Process | RNA stability | |||
| In-vitro Model | RMECs (Retinal microvascular endothelial cells (RMECs) purchased from Olaf (Worcester, MA, USA)) | |||
| rMCs (Retinal Muller cells (rMCs) from Kerafast Inc. (Boston, MA)) | ||||
| In-vivo Model | Male mice (8-10 weeks old, SLAC Laboratory Animal Co., Ltd., Shanghai, China) were administrated with STZ through intraperitoneal injection (I.P) for continuous 5 days (d). | |||
Gastric cancer [ICD-11: 2B72]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [1] | |||
| Response Summary | FTO was an independent risk factor for overall survival (OS) of GC patients and FTO could promote GC metastasis by upregulating the expression of Integrin beta-1 (ITGB1) via decreasing its m6A level. | |||
| Responsed Disease | Gastric cancer [ICD-11: 2B72] | |||
| Target Regulator | Fat mass and obesity-associated protein (FTO) | ERASER | ||
| Target Regulation | Up regulation | |||
| In-vitro Model | SNU-216 | Gastric tubular adenocarcinoma | Homo sapiens | CVCL_3946 |
| MKN7 | Gastric tubular adenocarcinoma | Homo sapiens | CVCL_1417 | |
| MGC-803 | Gastric mucinous adenocarcinoma | Homo sapiens | CVCL_5334 | |
| HGC-27 | Gastric carcinoma | Homo sapiens | CVCL_1279 | |
| BGC-823 | Gastric carcinoma | Homo sapiens | CVCL_3360 | |
| AGS | Gastric adenocarcinoma | Homo sapiens | CVCL_0139 | |
Lung cancer [ICD-11: 2C25]
Prostate cancer [ICD-11: 2C82]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [2] | |||
| Response Summary | METTL3 regulates the expression of Integrin beta-1 (ITGB1) through m6A-HuR-dependent mechanism, which affects the binding of ITGB1 to Collagen I and tumor cell motility, so as to promote the bone metastasis of prostate cancer. | |||
| Responsed Disease | Prostate cancer [ICD-11: 2C82] | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Cell adhesion molecules | hsa04514 | ||
| Cell Process | Cell adhesion | |||
| In-vitro Model | LNCaP | Prostate carcinoma | Homo sapiens | CVCL_0395 |
| PC-3 | Prostate carcinoma | Homo sapiens | CVCL_0035 | |
| In-vivo Model | All the mice were randomly divided into five groups of four mice each, including PC3-shNC, PC3-shMETTL3-1/2, LNCaP-vector and LNCaP-METTL3 group. | |||
Retinopathy [ICD-11: 9B71]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [5] | |||
| Response Summary | KAT1 triggers YTHDF2-mediated Integrin beta-1 (ITGB1) mRNA instability to alleviate the progression of DR. | |||
| Responsed Disease | Diabetic retinopathy [ICD-11: 9B71.0] | |||
| Target Regulator | YTH domain-containing family protein 2 (YTHDF2) | READER | ||
| Target Regulation | Down regulation | |||
| Pathway Response | PI3K-Akt signaling pathway | hsa04151 | ||
| Cell Process | RNA stability | |||
| In-vitro Model | RMECs (Retinal microvascular endothelial cells (RMECs) purchased from Olaf (Worcester, MA, USA)) | |||
| rMCs (Retinal Muller cells (rMCs) from Kerafast Inc. (Boston, MA)) | ||||
| In-vivo Model | Male mice (8-10 weeks old, SLAC Laboratory Animal Co., Ltd., Shanghai, China) were administrated with STZ through intraperitoneal injection (I.P) for continuous 5 days (d). | |||
Full List of Crosstalk(s) between m6A Modification and Epigenetic Regulation Related to This Regulator
DNA modification
m6A Regulator: Methyltransferase-like 3 (METTL3)
| In total 2 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT02124 | ||
| Epigenetic Regulator | DNA (cytosine-5)-methyltransferase 1 (DNMT1) | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Pancreatic cancer | |
| Crosstalk ID: M6ACROT02133 | ||
| Epigenetic Regulator | Cysteine methyltransferase DNMT3A (DNMT3A) | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Pancreatic cancer | |
Histone modification
m6A Regulator: YTH domain-containing family protein 2 (YTHDF2)
| In total 1 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT03215 | ||
| Epigenetic Regulator | Histone acetyltransferase type B catalytic subunit (HAT1) | |
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Diabetic retinopathy | |
RNA Modification Sequencing Data Associated with the Target (ID: M6ATAR00298)
| In total 1 m6A sequence/site(s) in this target gene | |||
| mod ID: AC4SITE000140 | Click to Show/Hide the Full List | ||
| mod site | chr10:32920290-32920291:- | [6] | |
| Sequence | CAAGAACGGGGTGAATGGAACAGGGGAAAATGGAAGAAAAT | ||
| Cell/Tissue List | H1 | ||
| Seq Type List | ac4C-seq | ||
| Transcript ID List | ENST00000396033.6; ENST00000423113.5; ENST00000302278.8 | ||
| External Link | RMBase: ac4C_site_203 | ||
5-methylcytidine (m5C)
| In total 2 m6A sequence/site(s) in this target gene | |||
| mod ID: M5CSITE004690 | Click to Show/Hide the Full List | ||
| mod site | chr10:32912126-32912127:- | [7] | |
| Sequence | GCAATTTTGGCATGTTTCTTCAGGTGCAATGAAGGGCGTGT | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000423113.5; ENST00000302278.8; ENST00000396033.6 | ||
| External Link | RMBase: m5C_site_5206 | ||
| mod ID: M5CSITE004691 | Click to Show/Hide the Full List | ||
| mod site | chr10:32954260-32954261:- | ||
| Sequence | AAGCTGGGGGTGCTGGTTGGCTGGAGCAGAGGGGAAATGAT | ||
| Cell/Tissue List | T24 | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000417122.6; ENST00000396033.6; ENST00000475184.5; ENST00000534049.5; ENST00000528877.5; ENST00000488494.5; ENST00000302278.8; ENST00000414670.2; ENST00000474568.5; ENST00000484088.5; ENST00000480226.5; ENST00000437302.5 | ||
| External Link | RMBase: m5C_site_5207 | ||
N6-methyladenosine (m6A)
| In total 85 m6A sequence/site(s) in this target gene | |||
| mod ID: M6ASITE096216 | Click to Show/Hide the Full List | ||
| mod site | chr10:32900498-32900499:- | [8] | |
| Sequence | TGAGTTGCCCATCTTGTTTCACACTAGTCACATTCTTGTTT | ||
| Motif Score | 2.047297619 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000396033.6; ENST00000302278.8; ENST00000423113.5 | ||
| External Link | RMBase: m6A_site_97504 | ||
| mod ID: M6ASITE096217 | Click to Show/Hide the Full List | ||
| mod site | chr10:32900549-32900550:- | [9] | |
| Sequence | TACTTTGAGTTAGTGCCATAACAGACCACTGTATGTTTACT | ||
| Motif Score | 2.168095238 | ||
| Cell/Tissue List | liver | ||
| Seq Type List | m6A-REF-seq | ||
| Transcript ID List | ENST00000396033.6; ENST00000423113.5; ENST00000302278.8 | ||
| External Link | RMBase: m6A_site_97505 | ||
| mod ID: M6ASITE096218 | Click to Show/Hide the Full List | ||
| mod site | chr10:32900620-32900621:- | [8] | |
| Sequence | TCAATTTGCCTTTTTAATGAACATGTGAAGTTATACTGTGG | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000396033.6; ENST00000302278.8; ENST00000423113.5 | ||
| External Link | RMBase: m6A_site_97506 | ||
| mod ID: M6ASITE096219 | Click to Show/Hide the Full List | ||
| mod site | chr10:32900786-32900787:- | [10] | |
| Sequence | AAAGTATTTGCTGAATGGGGACCTTTTGAGTTGAATTTATT | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; Huh7 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000302278.8; ENST00000423113.5; ENST00000396033.6 | ||
| External Link | RMBase: m6A_site_97507 | ||
| mod ID: M6ASITE096220 | Click to Show/Hide the Full List | ||
| mod site | chr10:32900808-32900809:- | [8] | |
| Sequence | TTACATACAGGCCATACTTTACAAAGTATTTGCTGAATGGG | ||
| Motif Score | 2.07285119 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000396033.6; ENST00000423113.5; ENST00000302278.8 | ||
| External Link | RMBase: m6A_site_97508 | ||
| mod ID: M6ASITE096221 | Click to Show/Hide the Full List | ||
| mod site | chr10:32900826-32900827:- | [8] | |
| Sequence | TCCTTGATTTAGCACTATTTACATACAGGCCATACTTTACA | ||
| Motif Score | 2.07285119 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000302278.8; ENST00000396033.6; ENST00000423113.5 | ||
| External Link | RMBase: m6A_site_97509 | ||
| mod ID: M6ASITE096222 | Click to Show/Hide the Full List | ||
| mod site | chr10:32900870-32900871:- | [10] | |
| Sequence | TTCAAGCCTTAGATAAAAGAACCGAGCAATTTTCTGCTAAA | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | HeLa; HEK293T; Huh7 | ||
| Seq Type List | m6A-seq; DART-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000396033.6; ENST00000302278.8; ENST00000423113.5 | ||
| External Link | RMBase: m6A_site_97510 | ||
| mod ID: M6ASITE096223 | Click to Show/Hide the Full List | ||
| mod site | chr10:32900895-32900896:- | [8] | |
| Sequence | AACTTTTATGCCTTCACTTTACAAATTCAAGCCTTAGATAA | ||
| Motif Score | 2.07285119 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000396033.6; ENST00000302278.8; ENST00000423113.5 | ||
| External Link | RMBase: m6A_site_97511 | ||
| mod ID: M6ASITE096224 | Click to Show/Hide the Full List | ||
| mod site | chr10:32901008-32901009:- | [8] | |
| Sequence | ACGTTTGAAATAGTTGATCTACAAAGGCCATGGGAAAAATT | ||
| Motif Score | 2.078666667 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000302278.8; ENST00000423113.5; ENST00000396033.6; ENST00000488427.1 | ||
| External Link | RMBase: m6A_site_97512 | ||
| mod ID: M6ASITE096225 | Click to Show/Hide the Full List | ||
| mod site | chr10:32901059-32901060:- | [8] | |
| Sequence | ACTGATGACATATCTGAAAGACAAGTATGTTGAGAGTTGCT | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000396033.6; ENST00000423113.5; ENST00000488427.1; ENST00000302278.8 | ||
| External Link | RMBase: m6A_site_97513 | ||
| mod ID: M6ASITE096226 | Click to Show/Hide the Full List | ||
| mod site | chr10:32901084-32901085:- | [8] | |
| Sequence | TTTTGCTATTTGCCTGTTAGACATGACTGATGACATATCTG | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000488427.1; ENST00000396033.6; ENST00000423113.5; ENST00000302278.8 | ||
| External Link | RMBase: m6A_site_97514 | ||
| mod ID: M6ASITE096227 | Click to Show/Hide the Full List | ||
| mod site | chr10:32901184-32901185:- | [8] | |
| Sequence | TAAAACTGATTTTTAGCTTTACAAATATGTCAGTTTGCAGT | ||
| Motif Score | 2.07285119 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000423113.5; ENST00000488427.1; ENST00000396033.6; ENST00000302278.8 | ||
| External Link | RMBase: m6A_site_97515 | ||
| mod ID: M6ASITE096228 | Click to Show/Hide the Full List | ||
| mod site | chr10:32901200-32901201:- | [10] | |
| Sequence | TCTGTTAATCATGTATTAAAACTGATTTTTAGCTTTACAAA | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000488427.1; ENST00000302278.8; ENST00000423113.5; ENST00000396033.6 | ||
| External Link | RMBase: m6A_site_97516 | ||
| mod ID: M6ASITE096229 | Click to Show/Hide the Full List | ||
| mod site | chr10:32901307-32901308:- | [10] | |
| Sequence | TTTTGACCTTTTCTTCCTGGACTATTGAAATCAAGCTTATT | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa; HEK293T; HepG2; fibroblasts; A549 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000423113.5; ENST00000396033.6; ENST00000488427.1; ENST00000302278.8 | ||
| External Link | RMBase: m6A_site_97517 | ||
| mod ID: M6ASITE096230 | Click to Show/Hide the Full List | ||
| mod site | chr10:32901338-32901339:- | [8] | |
| Sequence | TACTCTTGTCAGCTAAGGTCACATTGTGCCTTTTTGACCTT | ||
| Motif Score | 2.047297619 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000302278.8; ENST00000396033.6; ENST00000488427.1; ENST00000423113.5 | ||
| External Link | RMBase: m6A_site_97518 | ||
| mod ID: M6ASITE096231 | Click to Show/Hide the Full List | ||
| mod site | chr10:32901368-32901369:- | [10] | |
| Sequence | GGACTGACAAAAGACTTGAGACAGGATGGTTACTCTTGTCA | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000423113.5; ENST00000396033.6; ENST00000302278.8; ENST00000488427.1 | ||
| External Link | RMBase: m6A_site_97519 | ||
| mod ID: M6ASITE096232 | Click to Show/Hide the Full List | ||
| mod site | chr10:32901375-32901376:- | [10] | |
| Sequence | ATGCCAGGGACTGACAAAAGACTTGAGACAGGATGGTTACT | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000423113.5; ENST00000302278.8; ENST00000488427.1; ENST00000396033.6 | ||
| External Link | RMBase: m6A_site_97520 | ||
| mod ID: M6ASITE096233 | Click to Show/Hide the Full List | ||
| mod site | chr10:32901382-32901383:- | [8] | |
| Sequence | GTAATTCATGCCAGGGACTGACAAAAGACTTGAGACAGGAT | ||
| Motif Score | 2.859755952 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000488427.1; ENST00000302278.8; ENST00000396033.6; ENST00000423113.5 | ||
| External Link | RMBase: m6A_site_97521 | ||
| mod ID: M6ASITE096234 | Click to Show/Hide the Full List | ||
| mod site | chr10:32901386-32901387:- | [10] | |
| Sequence | TGTTGTAATTCATGCCAGGGACTGACAAAAGACTTGAGACA | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000488427.1; ENST00000423113.5; ENST00000396033.6; ENST00000302278.8 | ||
| External Link | RMBase: m6A_site_97522 | ||
| mod ID: M6ASITE096235 | Click to Show/Hide the Full List | ||
| mod site | chr10:32901450-32901451:- | [11] | |
| Sequence | TGTGCAGGTTTTGAAAATGTACAATATGTATAATTTTTAAA | ||
| Motif Score | 2.856142857 | ||
| Cell/Tissue List | HEK293T; hESC-HEK293T | ||
| Seq Type List | DART-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000396033.6; ENST00000423113.5; ENST00000488427.1; ENST00000302278.8 | ||
| External Link | RMBase: m6A_site_97523 | ||
| mod ID: M6ASITE096236 | Click to Show/Hide the Full List | ||
| mod site | chr10:32901475-32901476:- | [11] | |
| Sequence | GGCAATATTGCCATGGTTTTACTCATGTGCAGGTTTTGAAA | ||
| Motif Score | 2.494845238 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000423113.5; ENST00000302278.8; ENST00000396033.6; ENST00000488427.1 | ||
| External Link | RMBase: m6A_site_97524 | ||
| mod ID: M6ASITE096237 | Click to Show/Hide the Full List | ||
| mod site | chr10:32901513-32901514:- | [9] | |
| Sequence | AAGTAGCAATTTCCATAGTCACAGTTAGGTAGCTTTAGGGC | ||
| Motif Score | 2.047297619 | ||
| Cell/Tissue List | kidney | ||
| Seq Type List | m6A-REF-seq | ||
| Transcript ID List | ENST00000488427.1; ENST00000396033.6; ENST00000302278.8; ENST00000423113.5 | ||
| External Link | RMBase: m6A_site_97525 | ||
| mod ID: M6ASITE096238 | Click to Show/Hide the Full List | ||
| mod site | chr10:32901548-32901549:- | [8] | |
| Sequence | GTACTGCCCGTGCAAATCCCACAACACTGAATGCAAAGTAG | ||
| Motif Score | 2.053113095 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000423113.5; ENST00000488427.1; ENST00000396033.6; ENST00000302278.8 | ||
| External Link | RMBase: m6A_site_97526 | ||
| mod ID: M6ASITE096239 | Click to Show/Hide the Full List | ||
| mod site | chr10:32901604-32901605:- | [8] | |
| Sequence | CTATTTATAAGAGTGCCGTAACAACTGTGGTCAATCCGAAG | ||
| Motif Score | 2.168095238 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000494395.1; ENST00000488427.1; ENST00000423113.5; ENST00000396033.6; ENST00000302278.8 | ||
| External Link | RMBase: m6A_site_97527 | ||
| mod ID: M6ASITE096240 | Click to Show/Hide the Full List | ||
| mod site | chr10:32907093-32907094:- | [10] | |
| Sequence | TAATAATTTCAAGAATCCAAACTACGGACGTAAAGCTGGTC | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa; hNPCs | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000396033.6; ENST00000494395.1; ENST00000423113.5; ENST00000302278.8; ENST00000488427.1 | ||
| External Link | RMBase: m6A_site_97528 | ||
| mod ID: M6ASITE096241 | Click to Show/Hide the Full List | ||
| mod site | chr10:32908371-32908372:- | [10] | |
| Sequence | GAAAATGAATGCCAAATGGGACACGGTAAGTTACAAAACAT | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; hESC-HEK293T | ||
| Seq Type List | m6A-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000423113.5; ENST00000396033.6; ENST00000302278.8; ENST00000494395.1; ENST00000488427.1 | ||
| External Link | RMBase: m6A_site_97529 | ||
| mod ID: M6ASITE096242 | Click to Show/Hide the Full List | ||
| mod site | chr10:32908515-32908516:- | [8] | |
| Sequence | AGAGTGTCCCACTGGTCCAGACATCATTCCAATTGTAGCTG | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000302278.8; ENST00000494395.1; ENST00000396033.6; ENST00000423113.5 | ||
| External Link | RMBase: m6A_site_97530 | ||
| mod ID: M6ASITE096243 | Click to Show/Hide the Full List | ||
| mod site | chr10:32910257-32910258:- | [8] | |
| Sequence | TACGTATTCAGTGAATGGGAACAACGAGGTCATGGTTCATG | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000396033.6; ENST00000302278.8; ENST00000423113.5; ENST00000494395.1 | ||
| External Link | RMBase: m6A_site_97531 | ||
| mod ID: M6ASITE096244 | Click to Show/Hide the Full List | ||
| mod site | chr10:32910356-32910357:- | [8] | |
| Sequence | TACCAAGGTAGAAAGTCGGGACAAATTACCCCAGCCGGTCC | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000302278.8; ENST00000423113.5; ENST00000396033.6; ENST00000494395.1 | ||
| External Link | RMBase: m6A_site_97532 | ||
| mod ID: M6ASITE096245 | Click to Show/Hide the Full List | ||
| mod site | chr10:32910410-32910411:- | [8] | |
| Sequence | CAATAAAGGAGAAAAGAAAGACACATGCACACAGGAATGTT | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000302278.8; ENST00000423113.5; ENST00000494395.1; ENST00000396033.6 | ||
| External Link | RMBase: m6A_site_97533 | ||
| mod ID: M6ASITE096246 | Click to Show/Hide the Full List | ||
| mod site | chr10:32911475-32911476:- | [10] | |
| Sequence | AAACGTGTGAGATGTGTCAGACCTGCCTTGGTGTCTGTGCT | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000396033.6; ENST00000302278.8; ENST00000423113.5; ENST00000494395.1 | ||
| External Link | RMBase: m6A_site_97534 | ||
| mod ID: M6ASITE096247 | Click to Show/Hide the Full List | ||
| mod site | chr10:32911566-32911567:- | [10] | |
| Sequence | ACTTGTGAAGCCAGCAACGGACAGATCTGCAATGGCCGGGG | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; HepG2; HEC-1-A | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000302278.8; ENST00000396033.6; ENST00000423113.5; ENST00000494395.1 | ||
| External Link | RMBase: m6A_site_97535 | ||
| mod ID: M6ASITE096248 | Click to Show/Hide the Full List | ||
| mod site | chr10:32911624-32911625:- | [8] | |
| Sequence | GTGTGAGTGCAACCCCAACTACACTGGCAGTGCATGTGACT | ||
| Motif Score | 2.078666667 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000494395.1; ENST00000423113.5; ENST00000302278.8; ENST00000396033.6 | ||
| External Link | RMBase: m6A_site_97536 | ||
| mod ID: M6ASITE096249 | Click to Show/Hide the Full List | ||
| mod site | chr10:32911917-32911918:- | [11] | |
| Sequence | CTGCGAGTGTGATAATTTCAACTGTGATAGATCCAATGGCT | ||
| Motif Score | 2.595904762 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000423113.5; ENST00000396033.6; ENST00000302278.8; ENST00000494395.1 | ||
| External Link | RMBase: m6A_site_97537 | ||
| mod ID: M6ASITE096250 | Click to Show/Hide the Full List | ||
| mod site | chr10:32911963-32911964:- | [11] | |
| Sequence | TTTGTAGGAAGAGGGATAATACAAATGAAATTTATTCTGGC | ||
| Motif Score | 2.110482143 | ||
| Cell/Tissue List | HEK293T; hESC-HEK293T | ||
| Seq Type List | DART-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000494395.1; ENST00000302278.8; ENST00000396033.6; ENST00000423113.5 | ||
| External Link | RMBase: m6A_site_97538 | ||
| mod ID: M6ASITE096251 | Click to Show/Hide the Full List | ||
| mod site | chr10:32911991-32911992:- | [10] | |
| Sequence | AATGGAGAGTGCGTCTGCGGACAGTGTGTTTGTAGGAAGAG | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; HepG2; HEC-1-A | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000494395.1; ENST00000423113.5; ENST00000302278.8; ENST00000396033.6 | ||
| External Link | RMBase: m6A_site_97539 | ||
| mod ID: M6ASITE096252 | Click to Show/Hide the Full List | ||
| mod site | chr10:32912034-32912035:- | [10] | |
| Sequence | TGCTTACTGCAGGAAAGAAAACAGTTCAGAAATCTGCAGTA | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa; HepG2; endometrial; HEC-1-A | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000494395.1; ENST00000302278.8; ENST00000396033.6; ENST00000423113.5 | ||
| External Link | RMBase: m6A_site_97540 | ||
| mod ID: M6ASITE096253 | Click to Show/Hide the Full List | ||
| mod site | chr10:32912061-32912062:- | [10] | |
| Sequence | AGATGAAGTTAACAGTGAAGACATGGATGCTTACTGCAGGA | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa; HepG2; endometrial; HEC-1-A | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000302278.8; ENST00000494395.1; ENST00000396033.6; ENST00000423113.5 | ||
| External Link | RMBase: m6A_site_97541 | ||
| mod ID: M6ASITE096254 | Click to Show/Hide the Full List | ||
| mod site | chr10:32912099-32912100:- | [10] | |
| Sequence | AATGAAGGGCGTGTTGGTAGACATTGTGAATGCAGCACAGA | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa; hESC-HEK293T; HepG2; endometrial; HEC-1-A | ||
| Seq Type List | m6A-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000396033.6; ENST00000302278.8; ENST00000423113.5 | ||
| External Link | RMBase: m6A_site_97542 | ||
| mod ID: M6ASITE096255 | Click to Show/Hide the Full List | ||
| mod site | chr10:32919906-32919907:- | [10] | |
| Sequence | AGTGTCATGAAGGAAATGGGACATTTGAGTGTGGCGCGTGC | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; hESC-HEK293T; HepG2; endometrial; HEC-1-A | ||
| Seq Type List | m6A-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000423113.5; ENST00000302278.8; ENST00000396033.6 | ||
| External Link | RMBase: m6A_site_97543 | ||
| mod ID: M6ASITE096256 | Click to Show/Hide the Full List | ||
| mod site | chr10:32919974-32919975:- | [8] | |
| Sequence | AGTAGAGGTTATTCTTCAGTACATCTGTGAATGTGAATGCC | ||
| Motif Score | 2.856142857 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000396033.6; ENST00000302278.8; ENST00000423113.5 | ||
| External Link | RMBase: m6A_site_97544 | ||
| mod ID: M6ASITE096257 | Click to Show/Hide the Full List | ||
| mod site | chr10:32920291-32920292:- | [10] | |
| Sequence | GCAAGAACGGGGTGAATGGAACAGGGGAAAATGGAAGAAAA | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000396033.6; ENST00000423113.5; ENST00000302278.8 | ||
| External Link | RMBase: m6A_site_97545 | ||
| mod ID: M6ASITE096258 | Click to Show/Hide the Full List | ||
| mod site | chr10:32920323-32920324:- | [9] | |
| Sequence | AGAAGGCGTAACAATAAGTTACAAATCTTACTGCAAGAACG | ||
| Motif Score | 2.07285119 | ||
| Cell/Tissue List | kidney; liver | ||
| Seq Type List | m6A-REF-seq | ||
| Transcript ID List | ENST00000302278.8; ENST00000396033.6; ENST00000423113.5 | ||
| External Link | RMBase: m6A_site_97546 | ||
| mod ID: M6ASITE096259 | Click to Show/Hide the Full List | ||
| mod site | chr10:32920333-32920334:- | [8] | |
| Sequence | GCAAATTGTCAGAAGGCGTAACAATAAGTTACAAATCTTAC | ||
| Motif Score | 2.168095238 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000302278.8; ENST00000423113.5; ENST00000396033.6 | ||
| External Link | RMBase: m6A_site_97547 | ||
| mod ID: M6ASITE096260 | Click to Show/Hide the Full List | ||
| mod site | chr10:32922260-32922261:- | [8] | |
| Sequence | TCAGTTGATCATTGATGCATACAATGTGAGTACATCTTAAA | ||
| Motif Score | 2.110482143 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000302278.8; ENST00000423113.5; ENST00000396033.6 | ||
| External Link | RMBase: m6A_site_97548 | ||
| mod ID: M6ASITE096261 | Click to Show/Hide the Full List | ||
| mod site | chr10:32922309-32922310:- | [10] | |
| Sequence | TCCCTAAGTCAGCAGTAGGAACATTATCTGCAAATTCTAGC | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa; hESC-HEK293T; HepG2 | ||
| Seq Type List | m6A-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000396033.6; ENST00000302278.8; ENST00000423113.5 | ||
| External Link | RMBase: m6A_site_97549 | ||
| mod ID: M6ASITE096262 | Click to Show/Hide the Full List | ||
| mod site | chr10:32922335-32922336:- | [10] | |
| Sequence | TTAATTGCAGGAGCTGAAAAACTTGATCCCTAAGTCAGCAG | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa; HepG2 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000302278.8; ENST00000423113.5; ENST00000396033.6 | ||
| External Link | RMBase: m6A_site_97550 | ||
| mod ID: M6ASITE096263 | Click to Show/Hide the Full List | ||
| mod site | chr10:32922643-32922644:- | [8] | |
| Sequence | TGAAGAATTTCAGCCTGTTTACAAGGTAAATGTGTGAGATA | ||
| Motif Score | 2.07285119 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000302278.8; ENST00000396033.6; ENST00000423113.5 | ||
| External Link | RMBase: m6A_site_97551 | ||
| mod ID: M6ASITE096264 | Click to Show/Hide the Full List | ||
| mod site | chr10:32922680-32922681:- | [10] | |
| Sequence | TGAGTGAAAATAATATTCAGACAATTTTTGCAGTTACTGAA | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa; hESC-HEK293T | ||
| Seq Type List | m6A-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000302278.8; ENST00000423113.5; ENST00000396033.6 | ||
| External Link | RMBase: m6A_site_97552 | ||
| mod ID: M6ASITE096265 | Click to Show/Hide the Full List | ||
| mod site | chr10:32922702-32922703:- | [10] | |
| Sequence | ATTGCTCACCTTGTCCAGAAACTGAGTGAAAATAATATTCA | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa; HEK293T | ||
| Seq Type List | m6A-seq; DART-seq | ||
| Transcript ID List | ENST00000423113.5; ENST00000302278.8; ENST00000396033.6 | ||
| External Link | RMBase: m6A_site_97553 | ||
| mod ID: M6ASITE096266 | Click to Show/Hide the Full List | ||
| mod site | chr10:32923629-32923630:- | [10] | |
| Sequence | ATTGTTTTACCAAATGATGGACAATGTCACCTGGAAAATAA | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; hESC-HEK293T | ||
| Seq Type List | m6A-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000423113.5; ENST00000302278.8; ENST00000396033.6 | ||
| External Link | RMBase: m6A_site_97554 | ||
| mod ID: M6ASITE096267 | Click to Show/Hide the Full List | ||
| mod site | chr10:32923659-32923660:- | [10] | |
| Sequence | CACTTTGCTGGAGATGGGAAACTTGGTGGCATTGTTTTACC | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000302278.8; ENST00000396033.6; ENST00000423113.5 | ||
| External Link | RMBase: m6A_site_97555 | ||
| mod ID: M6ASITE096268 | Click to Show/Hide the Full List | ||
| mod site | chr10:32923694-32923695:- | [9] | |
| Sequence | CACGGCTGCTGGTGTTTTCCACAGATGCCGGGTTTCACTTT | ||
| Motif Score | 2.053113095 | ||
| Cell/Tissue List | liver | ||
| Seq Type List | m6A-REF-seq | ||
| Transcript ID List | ENST00000396033.6; ENST00000302278.8; ENST00000423113.5 | ||
| External Link | RMBase: m6A_site_97556 | ||
| mod ID: M6ASITE096269 | Click to Show/Hide the Full List | ||
| mod site | chr10:32923715-32923716:- | [8] | |
| Sequence | TGATTGGCTGGAGGAATGTTACACGGCTGCTGGTGTTTTCC | ||
| Motif Score | 2.07285119 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000302278.8; ENST00000396033.6; ENST00000423113.5 | ||
| External Link | RMBase: m6A_site_97557 | ||
| mod ID: M6ASITE096270 | Click to Show/Hide the Full List | ||
| mod site | chr10:32925942-32925943:- | [10] | |
| Sequence | TTTAATGAACTTGTTGGAAAACAGCGCATATCTGGAAATTT | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa; HepG2 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000396033.6; ENST00000302278.8; ENST00000423113.5 | ||
| External Link | RMBase: m6A_site_97558 | ||
| mod ID: M6ASITE096271 | Click to Show/Hide the Full List | ||
| mod site | chr10:32925954-32925955:- | [10] | |
| Sequence | AAAGGAGAAGTATTTAATGAACTTGTTGGAAAACAGCGCAT | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa; HepG2; HEC-1-A; AML | ||
| Seq Type List | m6A-seq; miCLIP | ||
| Transcript ID List | ENST00000423113.5; ENST00000396033.6; ENST00000302278.8 | ||
| External Link | RMBase: m6A_site_97559 | ||
| mod ID: M6ASITE096272 | Click to Show/Hide the Full List | ||
| mod site | chr10:32925980-32925981:- | [11] | |
| Sequence | ACAAAAATGTGCTCAGTCTTACTAATAAAGGAGAAGTATTT | ||
| Motif Score | 2.494845238 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000423113.5; ENST00000484088.5; ENST00000302278.8; ENST00000396033.6 | ||
| External Link | RMBase: m6A_site_97560 | ||
| mod ID: M6ASITE096273 | Click to Show/Hide the Full List | ||
| mod site | chr10:32926000-32926001:- | [8] | |
| Sequence | CTGCACCAGCCCATTTAGCTACAAAAATGTGCTCAGTCTTA | ||
| Motif Score | 2.078666667 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000396033.6; ENST00000484088.5; ENST00000423113.5; ENST00000302278.8 | ||
| External Link | RMBase: m6A_site_97561 | ||
| mod ID: M6ASITE096274 | Click to Show/Hide the Full List | ||
| mod site | chr10:32926021-32926022:- | [10] | |
| Sequence | CCCTTGCACAAGTGAACAGAACTGCACCAGCCCATTTAGCT | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; HEC-1-A | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000396033.6; ENST00000484088.5; ENST00000302278.8; ENST00000423113.5 | ||
| External Link | RMBase: m6A_site_97562 | ||
| mod ID: M6ASITE096275 | Click to Show/Hide the Full List | ||
| mod site | chr10:32926026-32926027:- | [10] | |
| Sequence | AGGAACCCTTGCACAAGTGAACAGAACTGCACCAGCCCATT | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; HEC-1-A | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000484088.5; ENST00000396033.6; ENST00000423113.5; ENST00000302278.8 | ||
| External Link | RMBase: m6A_site_97563 | ||
| mod ID: M6ASITE096276 | Click to Show/Hide the Full List | ||
| mod site | chr10:32926042-32926043:- | [10] | |
| Sequence | AACACCAGCTAAGCTCAGGAACCCTTGCACAAGTGAACAGA | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; HEC-1-A | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000484088.5; ENST00000396033.6; ENST00000423113.5; ENST00000302278.8 | ||
| External Link | RMBase: m6A_site_97564 | ||
| mod ID: M6ASITE096277 | Click to Show/Hide the Full List | ||
| mod site | chr10:32926072-32926073:- | [8] | |
| Sequence | GGAAAAGACTGTGATGCCTTACATTAGCACAACACCAGCTA | ||
| Motif Score | 2.07285119 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000423113.5; ENST00000484088.5; ENST00000396033.6; ENST00000302278.8 | ||
| External Link | RMBase: m6A_site_97565 | ||
| mod ID: M6ASITE096278 | Click to Show/Hide the Full List | ||
| mod site | chr10:32926085-32926086:- | [10] | |
| Sequence | TTGGCTCATTTGTGGAAAAGACTGTGATGCCTTACATTAGC | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; HEC-1-A | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000484088.5; ENST00000302278.8; ENST00000423113.5; ENST00000396033.6 | ||
| External Link | RMBase: m6A_site_97566 | ||
| mod ID: M6ASITE096279 | Click to Show/Hide the Full List | ||
| mod site | chr10:32928104-32928105:- | [10] | |
| Sequence | AATGAGGAGGATTACTTCGGACTTCAGAATTGGTAGGAATG | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; HEC-1-A | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000484088.5; ENST00000396033.6; ENST00000423113.5; ENST00000302278.8 | ||
| External Link | RMBase: m6A_site_97567 | ||
| mod ID: M6ASITE096280 | Click to Show/Hide the Full List | ||
| mod site | chr10:32928141-32928142:- | [10] | |
| Sequence | AGAATGTAAAAAGTCTTGGAACAGATCTGATGAATGAAATG | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa; HEC-1-A | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000302278.8; ENST00000484088.5; ENST00000423113.5; ENST00000396033.6 | ||
| External Link | RMBase: m6A_site_97568 | ||
| mod ID: M6ASITE096281 | Click to Show/Hide the Full List | ||
| mod site | chr10:32928191-32928192:- | [10] | |
| Sequence | TGACCTCTACTACCTTATGGACCTGTCTTACTCAATGAAAG | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; BGC823; H1299; HEC-1-A | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000484088.5; ENST00000396033.6; ENST00000474568.5; ENST00000480226.5; ENST00000423113.5; ENST00000302278.8 | ||
| External Link | RMBase: m6A_site_97569 | ||
| mod ID: M6ASITE096282 | Click to Show/Hide the Full List | ||
| mod site | chr10:32928221-32928222:- | [10] | |
| Sequence | AAAATTCAAGAGAGCTGAAGACTATCCCATTGACCTCTACT | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa; BGC823; H1299; HEC-1-A | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000480226.5; ENST00000484088.5; ENST00000302278.8; ENST00000474568.5; ENST00000423113.5; ENST00000396033.6; ENST00000488494.5; ENST00000534049.5 | ||
| External Link | RMBase: m6A_site_97570 | ||
| mod ID: M6ASITE096283 | Click to Show/Hide the Full List | ||
| mod site | chr10:32928246-32928247:- | [8] | |
| Sequence | TAGGGGAGCCACAGACATTTACATTAAAATTCAAGAGAGCT | ||
| Motif Score | 2.07285119 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000474568.5; ENST00000480226.5; ENST00000302278.8; ENST00000437302.5; ENST00000396033.6; ENST00000534049.5; ENST00000484088.5; ENST00000488494.5; ENST00000423113.5 | ||
| External Link | RMBase: m6A_site_97571 | ||
| mod ID: M6ASITE096284 | Click to Show/Hide the Full List | ||
| mod site | chr10:32928252-32928253:- | [10] | |
| Sequence | TCTAATTAGGGGAGCCACAGACATTTACATTAAAATTCAAG | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa; BGC823; hNPCs; H1299; HEC-1-A | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000302278.8; ENST00000423113.5; ENST00000480226.5; ENST00000534049.5; ENST00000396033.6; ENST00000474568.5; ENST00000484088.5; ENST00000437302.5; ENST00000488494.5 | ||
| External Link | RMBase: m6A_site_97572 | ||
| mod ID: M6ASITE096285 | Click to Show/Hide the Full List | ||
| mod site | chr10:32929849-32929850:- | [11] | |
| Sequence | GATATTACTCAGATCCAACCACAGCAGTTGGTTTTGCGATT | ||
| Motif Score | 2.053113095 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000484088.5; ENST00000437302.5; ENST00000423113.5; ENST00000474568.5; ENST00000475184.5; ENST00000488494.5; ENST00000480226.5; ENST00000302278.8; ENST00000396033.6; ENST00000534049.5 | ||
| External Link | RMBase: m6A_site_97573 | ||
| mod ID: M6ASITE096286 | Click to Show/Hide the Full List | ||
| mod site | chr10:32929893-32929894:- | [10] | |
| Sequence | TAACCAACCGTAGCAAAGGAACAGCAGAGAAGCTCAAGCCA | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa; BGC823; hNPCs; fibroblasts; H1299; endometrial; HEC-1-A | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000528877.5; ENST00000475184.5; ENST00000423113.5; ENST00000302278.8; ENST00000396033.6; ENST00000488494.5; ENST00000484088.5; ENST00000480226.5; ENST00000534049.5; ENST00000437302.5; ENST00000474568.5 | ||
| External Link | RMBase: m6A_site_97574 | ||
| mod ID: M6ASITE096287 | Click to Show/Hide the Full List | ||
| mod site | chr10:32929961-32929962:- | [11] | |
| Sequence | GAAGGGTTGCCCTCCAGATGACATAGAAAATCCCAGAGGCT | ||
| Motif Score | 2.859755952 | ||
| Cell/Tissue List | HEK293T; hESC-HEK293T | ||
| Seq Type List | DART-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000534049.5; ENST00000488494.5; ENST00000417122.6; ENST00000493758.5; ENST00000302278.8; ENST00000474568.5; ENST00000437302.5; ENST00000484088.5; ENST00000396033.6; ENST00000480226.5; ENST00000423113.5; ENST00000464001.1; ENST00000475184.5; ENST00000528877.5 | ||
| External Link | RMBase: m6A_site_97575 | ||
| mod ID: M6ASITE096288 | Click to Show/Hide the Full List | ||
| mod site | chr10:32932522-32932523:- | [11] | |
| Sequence | GGCCAAATTGTGGGTGGTGCACAAATTCAGTAAGTAAGGTT | ||
| Motif Score | 2.830589286 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000488494.5; ENST00000464001.1; ENST00000437302.5; ENST00000534049.5; ENST00000484088.5; ENST00000396033.6; ENST00000480226.5; ENST00000474568.5; ENST00000528877.5; ENST00000472147.1; ENST00000493758.5; ENST00000475184.5; ENST00000414670.2; ENST00000302278.8; ENST00000423113.5; ENST00000417122.6 | ||
| External Link | RMBase: m6A_site_97576 | ||
| mod ID: M6ASITE096289 | Click to Show/Hide the Full List | ||
| mod site | chr10:32932550-32932551:- | [8] | |
| Sequence | AAATCATGTGGAGAATGTATACAAGCAGGGCCAAATTGTGG | ||
| Motif Score | 2.110482143 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000484088.5; ENST00000437302.5; ENST00000474568.5; ENST00000423113.5; ENST00000488494.5; ENST00000464001.1; ENST00000302278.8; ENST00000480226.5; ENST00000414670.2; ENST00000534049.5; ENST00000528877.5; ENST00000472147.1; ENST00000396033.6; ENST00000475184.5; ENST00000493758.5; ENST00000417122.6 | ||
| External Link | RMBase: m6A_site_97577 | ||
| mod ID: M6ASITE096290 | Click to Show/Hide the Full List | ||
| mod site | chr10:32935494-32935495:- | [10] | |
| Sequence | TTTGCTGTGTGTTTGCTCAAACAGGTAAAAACAAAAAATTT | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa; A549; fibroblasts; H1299; HEK293A-TOA; iSLK; MSC; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000472147.1; ENST00000437302.5; ENST00000488494.5; ENST00000396033.6; ENST00000302278.8; ENST00000417122.6; ENST00000484088.5; ENST00000475184.5; ENST00000528877.5; ENST00000423113.5; ENST00000534049.5; ENST00000414670.2; ENST00000480226.5; ENST00000493758.5; ENST00000474568.5 | ||
| External Link | RMBase: m6A_site_97578 | ||
| mod ID: M6ASITE096291 | Click to Show/Hide the Full List | ||
| mod site | chr10:32935528-32935529:- | [10] | |
| Sequence | CAACCAATTTTCTGGATTGGACTGATCAGTTCAGTTTGCTG | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa; A549; fibroblasts; H1299; HEK293A-TOA; iSLK; MSC; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000488494.5; ENST00000396033.6; ENST00000423113.5; ENST00000302278.8; ENST00000484088.5; ENST00000480226.5; ENST00000474568.5; ENST00000528877.5; ENST00000414670.2; ENST00000534049.5; ENST00000417122.6; ENST00000493758.5; ENST00000472147.1; ENST00000437302.5; ENST00000475184.5 | ||
| External Link | RMBase: m6A_site_97579 | ||
| mod ID: M6ASITE096292 | Click to Show/Hide the Full List | ||
| mod site | chr10:32935549-32935550:- | [8] | |
| Sequence | TTTACATTTCAGATGAATTTACAACCAATTTTCTGGATTGG | ||
| Motif Score | 2.07285119 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000493758.5; ENST00000302278.8; ENST00000396033.6; ENST00000475184.5; ENST00000472147.1; ENST00000484088.5; ENST00000417122.6; ENST00000414670.2; ENST00000437302.5; ENST00000480226.5; ENST00000534049.5; ENST00000474568.5; ENST00000528877.5; ENST00000423113.5; ENST00000488494.5 | ||
| External Link | RMBase: m6A_site_97580 | ||
| mod ID: M6ASITE096293 | Click to Show/Hide the Full List | ||
| mod site | chr10:32935900-32935901:- | [10] | |
| Sequence | TAGAGATTCTATGGGCTGAGACCATTGGTAGGTTTTATGGT | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa; A549; fibroblasts; H1299; HEK293A-TOA; iSLK; MSC; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000474568.5; ENST00000534049.5; ENST00000396033.6; ENST00000302278.8; ENST00000475184.5; ENST00000528877.5; ENST00000472147.1; ENST00000437302.5; ENST00000488494.5; ENST00000484088.5; ENST00000417122.6; ENST00000480226.5; ENST00000414670.2 | ||
| External Link | RMBase: m6A_site_97581 | ||
| mod ID: M6ASITE096294 | Click to Show/Hide the Full List | ||
| mod site | chr10:32955646-32955647:- | [12] | |
| Sequence | TAATAATGAAAAGTGGTCAAACAACTTAACATGGTGCCAGT | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa; A549; U2OS; HEK293T; iSLK | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000528877.5; ENST00000417122.6; ENST00000302278.8; ENST00000437302.5; ENST00000474568.5; ENST00000475184.5; ENST00000480226.5; ENST00000534049.5; ENST00000396033.6; ENST00000484088.5; ENST00000488494.5; ENST00000414670.2 | ||
| External Link | RMBase: m6A_site_97582 | ||
| mod ID: M6ASITE096295 | Click to Show/Hide the Full List | ||
| mod site | chr10:32957814-32957815:- | [13] | |
| Sequence | GGCTGCCGGAGCTGCGGGGGACCGGGCCCGAACGGCCCCTG | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HepG2; HeLa; A549; U2OS; GSC-11; HEK293T; HEK293A-TOA; iSLK; TIME | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000484088.5; ENST00000414670.2; ENST00000417122.6; ENST00000302278.8; ENST00000396033.6; ENST00000474568.5; ENST00000488494.5; ENST00000475184.5; ENST00000528877.5; ENST00000534049.5 | ||
| External Link | RMBase: m6A_site_97583 | ||
| mod ID: M6ASITE096296 | Click to Show/Hide the Full List | ||
| mod site | chr10:32958193-32958194:- | [10] | |
| Sequence | GGCCCAGCGGGAGTCGCGGAACAGCAGGCCCGAGCCCACCG | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa; HepG2; A549; U2OS; H1A; H1B; fibroblasts; Jurkat; CD4T; GSC-11; HEK293T; HEK293A-TOA; iSLK; MSC; TIME; HEC-1-A; GSCs | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000414670.2; ENST00000484088.5; ENST00000534049.5; ENST00000417122.6; ENST00000475184.5; ENST00000488494.5; ENST00000302278.8; ENST00000528877.5; ENST00000474568.5 | ||
| External Link | RMBase: m6A_site_97584 | ||
| mod ID: M6ASITE096297 | Click to Show/Hide the Full List | ||
| mod site | chr10:32958568-32958569:- | [10] | |
| Sequence | CGCCCTTCGCAGAGGAGGAAACTGAGGCACGGCGACTGTCT | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa; A549; U2OS; GSC-11; HEK293T; HEK293A-TOA; GSCs | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000534049.5; ENST00000475184.5; ENST00000528877.5; ENST00000439974.3 | ||
| External Link | RMBase: m6A_site_97585 | ||
| mod ID: M6ASITE096298 | Click to Show/Hide the Full List | ||
| mod site | chr10:32958826-32958827:- | [10] | |
| Sequence | GCCAGTTCCCTTCCAGAAGGACCACGTGGTTTTTGGCCCCA | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; HEK293A-TOA; TREX | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000528877.5; ENST00000534049.5; ENST00000439974.3; ENST00000475184.5 | ||
| External Link | RMBase: m6A_site_97586 | ||
| mod ID: M6ASITE096299 | Click to Show/Hide the Full List | ||
| mod site | chr10:32960298-32960299:- | [10] | |
| Sequence | CACCCCCTGGTTGACCTTAGACATGGAAAGGTGAGAGCTAT | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000528877.5; ENST00000439974.3; ENST00000475184.5; ENST00000534049.5 | ||
| External Link | RMBase: m6A_site_97588 | ||
| mod ID: M6ASITE096300 | Click to Show/Hide the Full List | ||
| mod site | chr10:32960333-32960334:- | [10] | |
| Sequence | AGCCATCTGGTCGACCTTAGACACGGGAAGGCAAGCACCCC | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000439974.3; ENST00000528877.5; ENST00000475184.5; ENST00000534049.5 | ||
| External Link | RMBase: m6A_site_97589 | ||
2'-O-Methylation (2'-O-Me)
| In total 3 m6A sequence/site(s) in this target gene | |||
| mod ID: 2OMSITE000550 | Click to Show/Hide the Full List | ||
| mod site | chr10:32920025-32920026:- | [14] | |
| Sequence | AAAAAAGGATTCTGACAGCTTTAAAATTAGGCCTCTGGGCT | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | Nm-seq | ||
| Transcript ID List | ENST00000423113.5; ENST00000396033.6; ENST00000302278.8 | ||
| External Link | RMBase: Nm_site_701 | ||
| mod ID: 2OMSITE000551 | Click to Show/Hide the Full List | ||
| mod site | chr10:32922329-32922330:- | [14] | |
| Sequence | GCAGGAGCTGAAAAACTTGATCCCTAAGTCAGCAGTAGGAA | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | Nm-seq | ||
| Transcript ID List | ENST00000302278.8; ENST00000396033.6; ENST00000423113.5 | ||
| External Link | RMBase: Nm_site_702 | ||
| mod ID: 2OMSITE000552 | Click to Show/Hide the Full List | ||
| mod site | chr10:32935509-32935510:- | [14] | |
| Sequence | GACTGATCAGTTCAGTTTGCTGTGTGTTTGCTCAAACAGGT | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | Nm-seq | ||
| Transcript ID List | ENST00000534049.5; ENST00000472147.1; ENST00000302278.8; ENST00000488494.5; ENST00000493758.5; ENST00000414670.2; ENST00000474568.5; ENST00000396033.6; ENST00000437302.5; ENST00000528877.5; ENST00000480226.5; ENST00000417122.6; ENST00000475184.5; ENST00000484088.5; ENST00000423113.5 | ||
| External Link | RMBase: Nm_site_703 | ||
References