m6A Target Gene Information
General Information of the m6A Target Gene (ID: M6ATAR00339)
Full List of m6A Methylation Regulator of This Target Gene and Corresponding Disease/Drug Response(s)
MTOR
can be regulated by the following regulator(s), and cause disease/drug response(s). You can browse detail information of regulator(s) or disease/drug response(s).
Browse Regulator
Browse Disease
Browse Drug
Methyltransferase-like 3 (METTL3) [WRITER]
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | Liver | Mus musculus |
|
Treatment: Mettl3 knockout liver
Control: Wild type liver cells
|
GSE198512 | |
| Regulation |
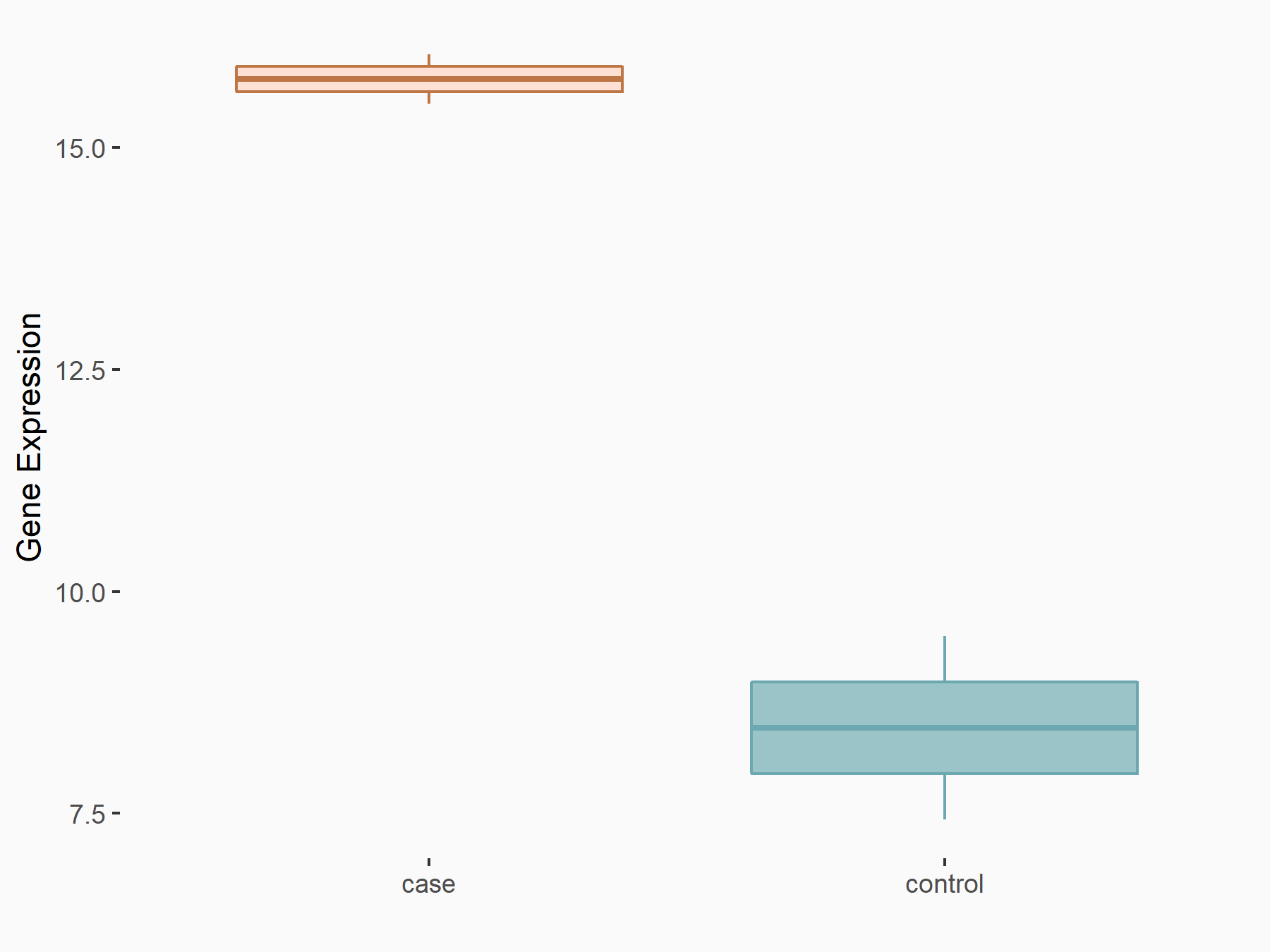 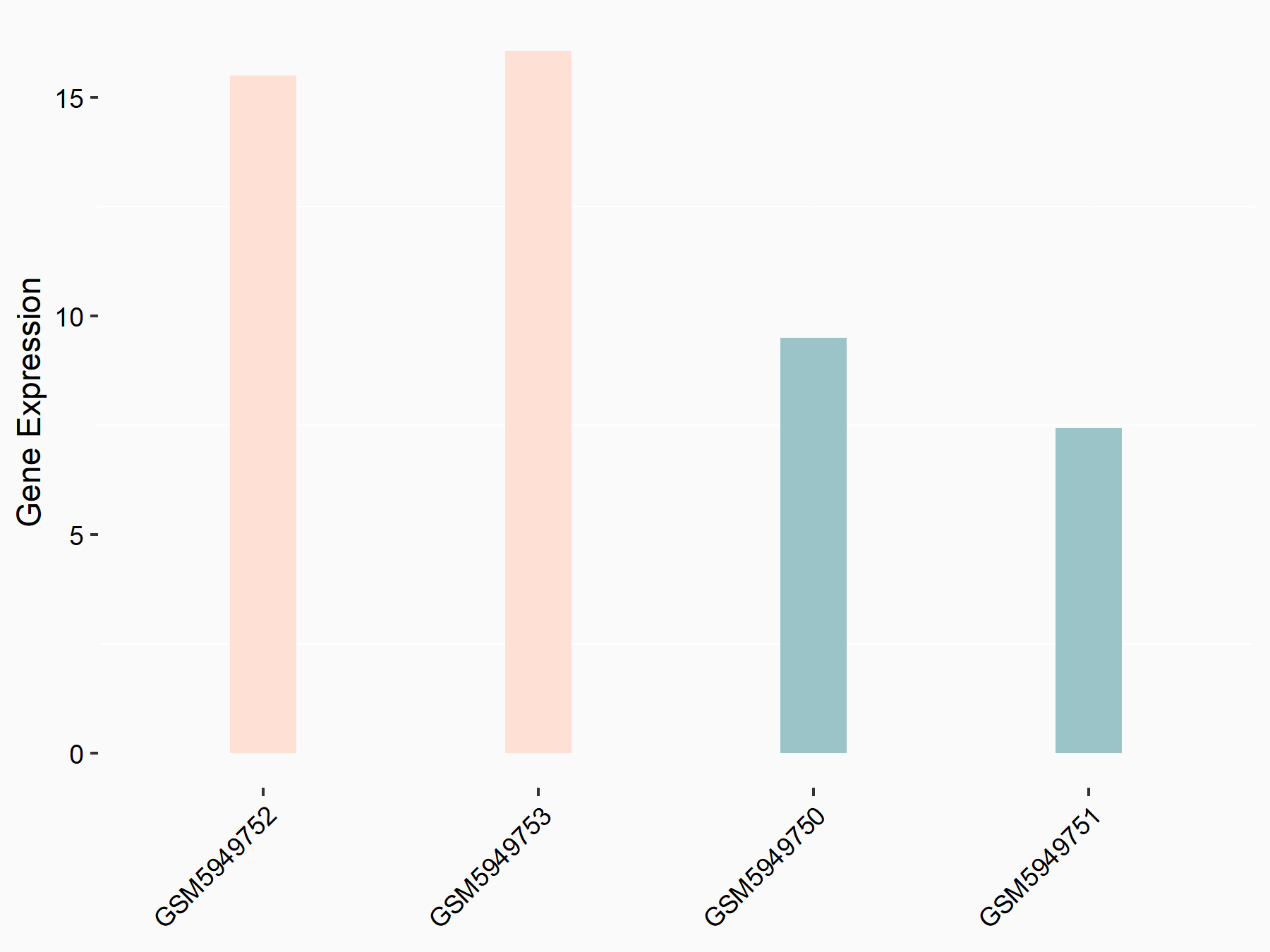 |
logFC: 8.34E-01 p-value: 1.58E-02 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between MTOR and the regulator | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 1.47E+00 | GSE60213 |
| In total 4 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [1] | |||
| Response Summary | METTL3-mediated m6 A methylation promotes lung cancer progression via activating PI3K/AKT/Serine/threonine-protein kinase mTOR (MTOR) pathway. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Lung cancer | ICD-11: 2C25 | ||
| Pathway Response | mTOR signaling pathway | hsa04150 | ||
| PI3K-Akt signaling pathway | hsa04151 | |||
| In-vitro Model | A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 |
| In-vivo Model | 5 × 106 A549 cells overexpressing METTL3 (Lv-METTL3) or control (Lv-Ctrl) were suspended in 100 uL phosphate-buffered saline (PBS), and were subcutaneously injected into mouse lower right flank. Drug treatment started in the Lv-METTL3 group when the tumour volume reached around 100 mm3. Mice were randomly divided into three groups to receive vehicle, GSK2536771 (30 mg/kg) or rapamycin (1 mg/kg). Drugs were administrated daily through intraperitoneal injection for 18 days. Treatment conditions were chosen as previously reported. | |||
| Experiment 2 Reporting the m6A Methylation Regulator of This Target Gene | [2] | |||
| Response Summary | Knockdown of METTL3 could obviously promote cell proliferation, migration and invasion function, and induce G0/G1 arrest,METTL3 acts as a novel marker for tumorigenesis, development and survival of RCC. Knockdown of METTL3 promoted changes in pI3K/AKT/Serine/threonine-protein kinase mTOR (MTOR) markers' expression with a gain in p-PI3k, p-AKT, p-mTOR and p-p70, and a loss of p-4EBP1. | |||
| Target Regulation | Down regulation | |||
| Responsed Disease | Renal cell carcinoma | ICD-11: 2C90 | ||
| Cell Process | Epithelial-to-mesenchymal transition | |||
| Arrest cell cycle at G0/G1 phase | ||||
| In-vitro Model | ACHN | Papillary renal cell carcinoma | Homo sapiens | CVCL_1067 |
| Caki-1 | Clear cell renal cell carcinoma | Homo sapiens | CVCL_0234 | |
| Caki-2 | Papillary renal cell carcinoma | Homo sapiens | CVCL_0235 | |
| HK2 | Normal | Acipenser baerii | CVCL_YE28 | |
| In-vivo Model | Cells (5×106 cells in 200 uL) were suspended with 100 uL PBS and 100 uL Matrigel Matrix, and injected subcutaneously into the left armpit of each mouse. | |||
| Experiment 3 Reporting the m6A Methylation Regulator of This Target Gene | [3] | |||
| Response Summary | METTL3 promotes the progression of retinoblastoma through PI3K/AKT/Serine/threonine-protein kinase mTOR (MTOR) pathways in vitro and in vivo. METTL3 has an impact on the PI3K-AKT-mTOR-P70S6K/4EBP1 pathway. The cell proliferation results show that the stimulatory function of METTL3 is lost after rapamycin treatment. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Retinoblastoma | ICD-11: 2D02.2 | ||
| Responsed Drug | Rapamycin | Approved | ||
| Pathway Response | PI3K-Akt signaling pathway | hsa04151 | ||
| mTOR signaling pathway | hsa04150 | |||
| Cell Process | Cell proliferation | |||
| Cell migration | ||||
| Cell invasion | ||||
| Cell apoptosis | ||||
| In-vitro Model | WERI-Rb-1 | Retinoblastoma | Homo sapiens | CVCL_1792 |
| Y-79 | Retinoblastoma | Homo sapiens | CVCL_1893 | |
| In-vivo Model | To establish a subcutaneous tumour model in nude mice, 2 × 107 Y79 cells (METTL3 knockdown group: shNC, shRNA1 and shRNA2; METTL3 up-regulated group: NC and METLL3) were resuspended in 1 mL of pre-cooled PBS, and 200 uL of the cell suspension was injected subcutaneously into the left side of the armpit to investigate tumour growth (4 × 106 per mouse). | |||
| Experiment 4 Reporting the m6A Methylation Regulator of This Target Gene | [4] | |||
| Response Summary | The contribution of METTL3-mediated m6A modification of Ddit4 mRNA to macrophage metabolic reprogramming in non-alcoholic fatty liver disease and obesity. In METTL3-deficient macrophages, there is a significant downregulation of Serine/threonine-protein kinase mTOR (MTOR) and nuclear factor Kappa-B (NF-Kappa-B) pathway activity in response to cellular stress and cytokine stimulation, which can be restored by knockdown of DDIT4. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Non-alcoholic fatty liver disease | ICD-11: DB92 | ||
| Pathway Response | mTOR signaling pathway | hsa04150 | ||
| HIF-1 signaling pathway | hsa04066 | |||
| In-vivo Model | The 8-10 weeks old mice were fed either a high fat diet or HF-CDAA , ad lib for 6-12 weeks. Chow diet was used as control for HFD.The mouse liver was perfused with PBS through portal vein, and liver tissue was cut into small pieces by a scissor. The single cell was made using syringe plunger to mull the tissue, and passed through a 40 uM cell strainer. | |||
YTH domain-containing family protein 1 (YTHDF1) [READER]
| Representative RIP-seq result supporting the interaction between MTOR and the regulator | ||
| Cell Line | Hela | Homo sapiens |
| Regulation | logFC: 1.73E+00 | GSE63591 |
| In total 1 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [5] | |||
| Response Summary | YTHDF1 contributes to the progression of HCC by activating PI3K/AKT/Serine/threonine-protein kinase mTOR (MTOR) signaling pathway and inducing EMT. | |||
| Target Regulation | Down regulation | |||
| Responsed Disease | Hepatocellular carcinoma | ICD-11: 2C12.02 | ||
| Pathway Response | PI3K-Akt signaling pathway | hsa04151), mTOR signaling pathway | ||
| Cell Process | Epithelial-mesenchymal transition | |||
| Cell migration | ||||
| Cell invasion | ||||
| Cell proliferation | ||||
| In-vitro Model | SNU-398 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_0077 |
| SK-HEP-1 | Liver and intrahepatic bile duct epithelial neoplasm | Homo sapiens | CVCL_0525 | |
| PLC/PRF/5 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_0485 | |
| L-02 | Endocervical adenocarcinoma | Homo sapiens | CVCL_6926 | |
| Huh-7 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_0336 | |
| Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 | |
| Hep 3B2.1-7 | Childhood hepatocellular carcinoma | Homo sapiens | CVCL_0326 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| HCCLM3 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_6832 | |
| In-vivo Model | Ten four-week-old BALB/c male nude mice (GemPharmatech, Jiangsu, China) were subcutaneously injected with control Huh7 cells 2 × 106 (left-back) and stable knockdown of YTHDF1 Huh7 cells 2 × 106 (right-back). These cells were respectively premixed with 50 ul Matrigel (Corning, 354,234) in 100 ul PBS. | |||
Fat mass and obesity-associated protein (FTO) [ERASER]
| In total 4 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [6] | |||
| Response Summary | In colorectal cancer, Glutaminolysis inhibition upregulated ATF4 expression in an m6A-dependent manner to activate pro-survival autophagy through transcriptional activation of the mTOR inhibitor DDIT4. Determined the relationship between FTO alpha-ketoglutarate dependent dioxygenase (FTO), YTH N6-methyladenosine RNA binding protein 2 (YTHDF2), and ATF4. ATF4 transcriptionally upregulated DDIT4 to suppress Serine/threonine-protein kinase mTOR (MTOR), which induced pro-survival autophagy during glutaminolysis inhibition. | |||
| Responsed Disease | Colorectal cancer | ICD-11: 2B91 | ||
| Responsed Drug | Asparagine inhibitor | Approved | ||
| Pathway Response | mTOR signaling pathway | hsa04150 | ||
| Autophagy | hsa04140 | |||
| Cell Process | RNA decay | |||
| Cell growth and death | ||||
| Cell autophagy | ||||
| In-vitro Model | HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 |
| SW480 | Colon adenocarcinoma | Homo sapiens | CVCL_0546 | |
| Experiment 2 Reporting the m6A Methylation Regulator of This Target Gene | [6] | |||
| Response Summary | In colorectal cancer, Glutaminolysis inhibition upregulated ATF4 expression in an m6A-dependent manner to activate pro-survival autophagy through transcriptional activation of the mTOR inhibitor DDIT4. Determined the relationship between FTO alpha-ketoglutarate dependent dioxygenase (FTO), YTH N6-methyladenosine RNA binding protein 2 (YTHDF2), and ATF4. ATF4 transcriptionally upregulated DDIT4 to suppress Serine/threonine-protein kinase mTOR (MTOR), which induced pro-survival autophagy during glutaminolysis inhibition. | |||
| Responsed Disease | Colorectal cancer | ICD-11: 2B91 | ||
| Responsed Drug | Chloroquine | Approved | ||
| Pathway Response | mTOR signaling pathway | hsa04150 | ||
| Autophagy | hsa04140 | |||
| Cell Process | RNA decay | |||
| Cell growth and death | ||||
| Cell autophagy | ||||
| In-vitro Model | HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 |
| SW480 | Colon adenocarcinoma | Homo sapiens | CVCL_0546 | |
| Experiment 3 Reporting the m6A Methylation Regulator of This Target Gene | [6] | |||
| Response Summary | In colorectal cancer, Glutaminolysis inhibition upregulated ATF4 expression in an m6A-dependent manner to activate pro-survival autophagy through transcriptional activation of the mTOR inhibitor DDIT4. Determined the relationship between FTO alpha-ketoglutarate dependent dioxygenase (FTO), YTH N6-methyladenosine RNA binding protein 2 (YTHDF2), and ATF4. Serine/threonine-protein kinase mTOR (MTOR) transcriptionally upregulated DDIT4 to suppress mTOR, which induced pro-survival autophagy during glutaminolysis inhibition. | |||
| Responsed Disease | Colorectal cancer | ICD-11: 2B91 | ||
| Responsed Drug | Meclofenamate sodium | Approved | ||
| Pathway Response | mTOR signaling pathway | hsa04150 | ||
| Autophagy | hsa04140 | |||
| Cell Process | RNA decay | |||
| Cell growth and death | ||||
| Cell autophagy | ||||
| In-vitro Model | HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 |
| SW480 | Colon adenocarcinoma | Homo sapiens | CVCL_0546 | |
| Experiment 4 Reporting the m6A Methylation Regulator of This Target Gene | [6] | |||
| Response Summary | In colorectal cancer, Glutaminolysis inhibition upregulated ATF4 expression in an m6A-dependent manner to activate pro-survival autophagy through transcriptional activation of the mTOR inhibitor DDIT4. Determined the relationship between FTO alpha-ketoglutarate dependent dioxygenase (FTO), YTH N6-methyladenosine RNA binding protein 2 (YTHDF2), and ATF4. Serine/threonine-protein kinase mTOR (MTOR) transcriptionally upregulated DDIT4 to suppress mTOR, which induced pro-survival autophagy during glutaminolysis inhibition. | |||
| Responsed Disease | Colorectal cancer | ICD-11: 2B91 | ||
| Responsed Drug | CB-839 | Phase 2 | ||
| Pathway Response | mTOR signaling pathway | hsa04150 | ||
| Autophagy | hsa04140 | |||
| Cell Process | RNA decay | |||
| Cell growth and death | ||||
| Cell autophagy | ||||
| In-vitro Model | HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 |
| SW480 | Colon adenocarcinoma | Homo sapiens | CVCL_0546 | |
Methyltransferase-like 14 (METTL14) [WRITER]
| In total 1 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [7] | |||
| Response Summary | The m6A modification level was decreased in GC and METTL14 was a key regulator resulting in m6A disorder in GC. METTL14 overexpression suppressed GC cell proliferation and aggression by deactivating the PI3K/AKT/Serine/threonine-protein kinase mTOR (MTOR) pathway and the EMT pathway, respectively. | |||
| Target Regulation | Down regulation | |||
| Responsed Disease | Gastric cancer | ICD-11: 2B72 | ||
| Pathway Response | PI3K-Akt signaling pathway | hsa04151 | ||
| mTOR signaling pathway | hsa04150 | |||
| In-vitro Model | SGC-7901 | Gastric carcinoma | Homo sapiens | CVCL_0520 |
| MGC-803 | Gastric mucinous adenocarcinoma | Homo sapiens | CVCL_5334 | |
| GES-1 | Normal | Homo sapiens | CVCL_EQ22 | |
YTH domain-containing family protein 2 (YTHDF2) [READER]
| In total 4 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [6] | |||
| Response Summary | In colorectal cancer, Glutaminolysis inhibition upregulated ATF4 expression in an m6A-dependent manner to activate pro-survival autophagy through transcriptional activation of the mTOR inhibitor DDIT4. Determined the relationship between FTO alpha-ketoglutarate dependent dioxygenase (FTO), YTH N6-methyladenosine RNA binding protein 2 (YTHDF2), and ATF4. ATF4 transcriptionally upregulated DDIT4 to suppress Serine/threonine-protein kinase mTOR (MTOR), which induced pro-survival autophagy during glutaminolysis inhibition. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Colorectal cancer | ICD-11: 2B91 | ||
| Responsed Drug | Asparagine inhibitor | Approved | ||
| Pathway Response | mTOR signaling pathway | hsa04150 | ||
| Autophagy | hsa04140 | |||
| Cell Process | RNA decay | |||
| Cell growth and death | ||||
| Cell autophagy | ||||
| In-vitro Model | HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 |
| SW480 | Colon adenocarcinoma | Homo sapiens | CVCL_0546 | |
| Experiment 2 Reporting the m6A Methylation Regulator of This Target Gene | [6] | |||
| Response Summary | In colorectal cancer, Glutaminolysis inhibition upregulated ATF4 expression in an m6A-dependent manner to activate pro-survival autophagy through transcriptional activation of the mTOR inhibitor DDIT4. Determined the relationship between FTO alpha-ketoglutarate dependent dioxygenase (FTO), YTH N6-methyladenosine RNA binding protein 2 (YTHDF2), and ATF4. Serine/threonine-protein kinase mTOR (MTOR) transcriptionally upregulated DDIT4 to suppress mTOR, which induced pro-survival autophagy during glutaminolysis inhibition. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Colorectal cancer | ICD-11: 2B91 | ||
| Responsed Drug | Chloroquine | Approved | ||
| Pathway Response | mTOR signaling pathway | hsa04150 | ||
| Autophagy | hsa04140 | |||
| Cell Process | RNA decay | |||
| Cell growth and death | ||||
| Cell autophagy | ||||
| In-vitro Model | HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 |
| SW480 | Colon adenocarcinoma | Homo sapiens | CVCL_0546 | |
| Experiment 3 Reporting the m6A Methylation Regulator of This Target Gene | [6] | |||
| Response Summary | In colorectal cancer, Glutaminolysis inhibition upregulated ATF4 expression in an m6A-dependent manner to activate pro-survival autophagy through transcriptional activation of the mTOR inhibitor DDIT4. Determined the relationship between FTO alpha-ketoglutarate dependent dioxygenase (FTO), YTH N6-methyladenosine RNA binding protein 2 (YTHDF2), and ATF4. Serine/threonine-protein kinase mTOR (MTOR) transcriptionally upregulated DDIT4 to suppress mTOR, which induced pro-survival autophagy during glutaminolysis inhibition. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Colorectal cancer | ICD-11: 2B91 | ||
| Responsed Drug | Meclofenamate sodium | Approved | ||
| Pathway Response | mTOR signaling pathway | hsa04150 | ||
| Autophagy | hsa04140 | |||
| Cell Process | RNA decay | |||
| Cell growth and death | ||||
| Cell autophagy | ||||
| In-vitro Model | HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 |
| SW480 | Colon adenocarcinoma | Homo sapiens | CVCL_0546 | |
| Experiment 4 Reporting the m6A Methylation Regulator of This Target Gene | [6] | |||
| Response Summary | In colorectal cancer, Glutaminolysis inhibition upregulated ATF4 expression in an m6A-dependent manner to activate pro-survival autophagy through transcriptional activation of the mTOR inhibitor DDIT4. Determined the relationship between FTO alpha-ketoglutarate dependent dioxygenase (FTO), YTH N6-methyladenosine RNA binding protein 2 (YTHDF2), and ATF4. ATF4 transcriptionally upregulated DDIT4 to suppress Serine/threonine-protein kinase mTOR (MTOR), which induced pro-survival autophagy during glutaminolysis inhibition. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Colorectal cancer | ICD-11: 2B91 | ||
| Responsed Drug | CB-839 | Phase 2 | ||
| Pathway Response | mTOR signaling pathway | hsa04150 | ||
| Autophagy | hsa04140 | |||
| Cell Process | RNA decay | |||
| Cell growth and death | ||||
| Cell autophagy | ||||
| In-vitro Model | HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 |
| SW480 | Colon adenocarcinoma | Homo sapiens | CVCL_0546 | |
Gastric cancer [ICD-11: 2B72]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [7] | |||
| Response Summary | The m6A modification level was decreased in GC and METTL14 was a key regulator resulting in m6A disorder in GC. METTL14 overexpression suppressed GC cell proliferation and aggression by deactivating the PI3K/AKT/Serine/threonine-protein kinase mTOR (MTOR) pathway and the EMT pathway, respectively. | |||
| Responsed Disease | Gastric cancer [ICD-11: 2B72] | |||
| Target Regulator | Methyltransferase-like 14 (METTL14) | WRITER | ||
| Target Regulation | Down regulation | |||
| Pathway Response | PI3K-Akt signaling pathway | hsa04151 | ||
| mTOR signaling pathway | hsa04150 | |||
| In-vitro Model | SGC-7901 | Gastric carcinoma | Homo sapiens | CVCL_0520 |
| MGC-803 | Gastric mucinous adenocarcinoma | Homo sapiens | CVCL_5334 | |
| GES-1 | Normal | Homo sapiens | CVCL_EQ22 | |
Colorectal cancer [ICD-11: 2B91]
| In total 5 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [] | |||
| Response Summary | WM_Score correlated highly with the regulation of transcription and post-transcriptional events contributing to the development of colorectal cancer. In response to anti-cancer drugs, WM_Score highly negatively correlated (drug sensitive) with drugs which targeted oncogenic related pathways, such as MAPK, EGFR, and Serine/threonine-protein kinase mTOR (MTOR) signaling pathways, positively correlated (drug resistance) with drugs which targeted in apoptosis and cell cycle. Importantly, the WM_Score was associated with the therapeutic efficacy of PD-L1 blockade, suggesting that the development of potential drugs targeting these "writers" to aid the clinical benefits of immunotherapy. | |||
| Responsed Disease | Colorectal cancer [ICD-11: 2B91] | |||
| Pathway Response | MAPK signaling pathway | hsa04010 | ||
| VEGF signaling pathway | hsa04370 | |||
| mTOR signaling pathway | hsa04150 | |||
| PD-L1 expression and PD-1 checkpoint pathway in cancer | hsa05235 | |||
| Cell Process | Cell apoptosis | |||
| Experiment 2 Reporting the m6A-centered Disease Response | [6] | |||
| Response Summary | In colorectal cancer, Glutaminolysis inhibition upregulated ATF4 expression in an m6A-dependent manner to activate pro-survival autophagy through transcriptional activation of the mTOR inhibitor DDIT4. Determined the relationship between FTO alpha-ketoglutarate dependent dioxygenase (FTO), YTH N6-methyladenosine RNA binding protein 2 (YTHDF2), and ATF4. ATF4 transcriptionally upregulated DDIT4 to suppress Serine/threonine-protein kinase mTOR (MTOR), which induced pro-survival autophagy during glutaminolysis inhibition. | |||
| Responsed Disease | Colorectal cancer [ICD-11: 2B91] | |||
| Target Regulator | Fat mass and obesity-associated protein (FTO) | ERASER | ||
| Responsed Drug | Asparagine inhibitor | Approved | ||
| Pathway Response | mTOR signaling pathway | hsa04150 | ||
| Autophagy | hsa04140 | |||
| Cell Process | RNA decay | |||
| Cell growth and death | ||||
| Cell autophagy | ||||
| In-vitro Model | HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 |
| SW480 | Colon adenocarcinoma | Homo sapiens | CVCL_0546 | |
| Experiment 3 Reporting the m6A-centered Disease Response | [6] | |||
| Response Summary | In colorectal cancer, Glutaminolysis inhibition upregulated ATF4 expression in an m6A-dependent manner to activate pro-survival autophagy through transcriptional activation of the mTOR inhibitor DDIT4. Determined the relationship between FTO alpha-ketoglutarate dependent dioxygenase (FTO), YTH N6-methyladenosine RNA binding protein 2 (YTHDF2), and ATF4. ATF4 transcriptionally upregulated DDIT4 to suppress Serine/threonine-protein kinase mTOR (MTOR), which induced pro-survival autophagy during glutaminolysis inhibition. | |||
| Responsed Disease | Colorectal cancer [ICD-11: 2B91] | |||
| Target Regulator | Fat mass and obesity-associated protein (FTO) | ERASER | ||
| Responsed Drug | Chloroquine | Approved | ||
| Pathway Response | mTOR signaling pathway | hsa04150 | ||
| Autophagy | hsa04140 | |||
| Cell Process | RNA decay | |||
| Cell growth and death | ||||
| Cell autophagy | ||||
| In-vitro Model | HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 |
| SW480 | Colon adenocarcinoma | Homo sapiens | CVCL_0546 | |
| Experiment 4 Reporting the m6A-centered Disease Response | [6] | |||
| Response Summary | In colorectal cancer, Glutaminolysis inhibition upregulated ATF4 expression in an m6A-dependent manner to activate pro-survival autophagy through transcriptional activation of the mTOR inhibitor DDIT4. Determined the relationship between FTO alpha-ketoglutarate dependent dioxygenase (FTO), YTH N6-methyladenosine RNA binding protein 2 (YTHDF2), and ATF4. Serine/threonine-protein kinase mTOR (MTOR) transcriptionally upregulated DDIT4 to suppress mTOR, which induced pro-survival autophagy during glutaminolysis inhibition. | |||
| Responsed Disease | Colorectal cancer [ICD-11: 2B91] | |||
| Target Regulator | Fat mass and obesity-associated protein (FTO) | ERASER | ||
| Responsed Drug | Meclofenamate sodium | Approved | ||
| Pathway Response | mTOR signaling pathway | hsa04150 | ||
| Autophagy | hsa04140 | |||
| Cell Process | RNA decay | |||
| Cell growth and death | ||||
| Cell autophagy | ||||
| In-vitro Model | HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 |
| SW480 | Colon adenocarcinoma | Homo sapiens | CVCL_0546 | |
| Experiment 5 Reporting the m6A-centered Disease Response | [6] | |||
| Response Summary | In colorectal cancer, Glutaminolysis inhibition upregulated ATF4 expression in an m6A-dependent manner to activate pro-survival autophagy through transcriptional activation of the mTOR inhibitor DDIT4. Determined the relationship between FTO alpha-ketoglutarate dependent dioxygenase (FTO), YTH N6-methyladenosine RNA binding protein 2 (YTHDF2), and ATF4. Serine/threonine-protein kinase mTOR (MTOR) transcriptionally upregulated DDIT4 to suppress mTOR, which induced pro-survival autophagy during glutaminolysis inhibition. | |||
| Responsed Disease | Colorectal cancer [ICD-11: 2B91] | |||
| Target Regulator | Fat mass and obesity-associated protein (FTO) | ERASER | ||
| Responsed Drug | CB-839 | Phase 2 | ||
| Pathway Response | mTOR signaling pathway | hsa04150 | ||
| Autophagy | hsa04140 | |||
| Cell Process | RNA decay | |||
| Cell growth and death | ||||
| Cell autophagy | ||||
| In-vitro Model | HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 |
| SW480 | Colon adenocarcinoma | Homo sapiens | CVCL_0546 | |
Gastrointestinal cancer [ICD-11: 2C11]
Liver cancer [ICD-11: 2C12]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [5] | |||
| Response Summary | YTHDF1 contributes to the progression of HCC by activating PI3K/AKT/Serine/threonine-protein kinase mTOR (MTOR) signaling pathway and inducing EMT. | |||
| Responsed Disease | Hepatocellular carcinoma [ICD-11: 2C12.02] | |||
| Target Regulator | YTH domain-containing family protein 1 (YTHDF1) | READER | ||
| Target Regulation | Down regulation | |||
| Pathway Response | PI3K-Akt signaling pathway | hsa04151), mTOR signaling pathway | ||
| Cell Process | Epithelial-mesenchymal transition | |||
| Cell migration | ||||
| Cell invasion | ||||
| Cell proliferation | ||||
| In-vitro Model | SNU-398 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_0077 |
| SK-HEP-1 | Liver and intrahepatic bile duct epithelial neoplasm | Homo sapiens | CVCL_0525 | |
| PLC/PRF/5 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_0485 | |
| L-02 | Endocervical adenocarcinoma | Homo sapiens | CVCL_6926 | |
| Huh-7 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_0336 | |
| Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 | |
| Hep 3B2.1-7 | Childhood hepatocellular carcinoma | Homo sapiens | CVCL_0326 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| HCCLM3 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_6832 | |
| In-vivo Model | Ten four-week-old BALB/c male nude mice (GemPharmatech, Jiangsu, China) were subcutaneously injected with control Huh7 cells 2 × 106 (left-back) and stable knockdown of YTHDF1 Huh7 cells 2 × 106 (right-back). These cells were respectively premixed with 50 ul Matrigel (Corning, 354,234) in 100 ul PBS. | |||
Lung cancer [ICD-11: 2C25]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [1] | |||
| Response Summary | METTL3-mediated m6 A methylation promotes lung cancer progression via activating PI3K/AKT/Serine/threonine-protein kinase mTOR (MTOR) pathway. | |||
| Responsed Disease | Lung cancer [ICD-11: 2C25] | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Target Regulation | Up regulation | |||
| Pathway Response | mTOR signaling pathway | hsa04150 | ||
| PI3K-Akt signaling pathway | hsa04151 | |||
| In-vitro Model | A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 |
| In-vivo Model | 5 × 106 A549 cells overexpressing METTL3 (Lv-METTL3) or control (Lv-Ctrl) were suspended in 100 uL phosphate-buffered saline (PBS), and were subcutaneously injected into mouse lower right flank. Drug treatment started in the Lv-METTL3 group when the tumour volume reached around 100 mm3. Mice were randomly divided into three groups to receive vehicle, GSK2536771 (30 mg/kg) or rapamycin (1 mg/kg). Drugs were administrated daily through intraperitoneal injection for 18 days. Treatment conditions were chosen as previously reported. | |||
Renal cell carcinoma [ICD-11: 2C90]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [2] | |||
| Response Summary | Knockdown of METTL3 could obviously promote cell proliferation, migration and invasion function, and induce G0/G1 arrest,METTL3 acts as a novel marker for tumorigenesis, development and survival of RCC. Knockdown of METTL3 promoted changes in pI3K/AKT/Serine/threonine-protein kinase mTOR (MTOR) markers' expression with a gain in p-PI3k, p-AKT, p-mTOR and p-p70, and a loss of p-4EBP1. | |||
| Responsed Disease | Renal cell carcinoma [ICD-11: 2C90] | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Target Regulation | Down regulation | |||
| Cell Process | Epithelial-to-mesenchymal transition | |||
| Arrest cell cycle at G0/G1 phase | ||||
| In-vitro Model | ACHN | Papillary renal cell carcinoma | Homo sapiens | CVCL_1067 |
| Caki-1 | Clear cell renal cell carcinoma | Homo sapiens | CVCL_0234 | |
| Caki-2 | Papillary renal cell carcinoma | Homo sapiens | CVCL_0235 | |
| HK2 | Normal | Acipenser baerii | CVCL_YE28 | |
| In-vivo Model | Cells (5×106 cells in 200 uL) were suspended with 100 uL PBS and 100 uL Matrigel Matrix, and injected subcutaneously into the left armpit of each mouse. | |||
Retina cancer [ICD-11: 2D02]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [3] | |||
| Response Summary | METTL3 promotes the progression of retinoblastoma through PI3K/AKT/Serine/threonine-protein kinase mTOR (MTOR) pathways in vitro and in vivo. METTL3 has an impact on the PI3K-AKT-mTOR-P70S6K/4EBP1 pathway. The cell proliferation results show that the stimulatory function of METTL3 is lost after rapamycin treatment. | |||
| Responsed Disease | Retinoblastoma [ICD-11: 2D02.2] | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Target Regulation | Up regulation | |||
| Responsed Drug | Rapamycin | Approved | ||
| Pathway Response | PI3K-Akt signaling pathway | hsa04151 | ||
| mTOR signaling pathway | hsa04150 | |||
| Cell Process | Cell proliferation | |||
| Cell migration | ||||
| Cell invasion | ||||
| Cell apoptosis | ||||
| In-vitro Model | WERI-Rb-1 | Retinoblastoma | Homo sapiens | CVCL_1792 |
| Y-79 | Retinoblastoma | Homo sapiens | CVCL_1893 | |
| In-vivo Model | To establish a subcutaneous tumour model in nude mice, 2 × 107 Y79 cells (METTL3 knockdown group: shNC, shRNA1 and shRNA2; METTL3 up-regulated group: NC and METLL3) were resuspended in 1 mL of pre-cooled PBS, and 200 uL of the cell suspension was injected subcutaneously into the left side of the armpit to investigate tumour growth (4 × 106 per mouse). | |||
Non-alcoholic fatty liver disease [ICD-11: DB92]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [4] | |||
| Response Summary | The contribution of METTL3-mediated m6A modification of Ddit4 mRNA to macrophage metabolic reprogramming in non-alcoholic fatty liver disease and obesity. In METTL3-deficient macrophages, there is a significant downregulation of Serine/threonine-protein kinase mTOR (MTOR) and nuclear factor Kappa-B (NF-Kappa-B) pathway activity in response to cellular stress and cytokine stimulation, which can be restored by knockdown of DDIT4. | |||
| Responsed Disease | Non-alcoholic fatty liver disease [ICD-11: DB92] | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Target Regulation | Up regulation | |||
| Pathway Response | mTOR signaling pathway | hsa04150 | ||
| HIF-1 signaling pathway | hsa04066 | |||
| In-vivo Model | The 8-10 weeks old mice were fed either a high fat diet or HF-CDAA , ad lib for 6-12 weeks. Chow diet was used as control for HFD.The mouse liver was perfused with PBS through portal vein, and liver tissue was cut into small pieces by a scissor. The single cell was made using syringe plunger to mull the tissue, and passed through a 40 uM cell strainer. | |||
Asparagine inhibitor
[Approved]
| In total 2 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response | [6] | |||
| Response Summary | In colorectal cancer, Glutaminolysis inhibition upregulated ATF4 expression in an m6A-dependent manner to activate pro-survival autophagy through transcriptional activation of the mTOR inhibitor DDIT4. Determined the relationship between FTO alpha-ketoglutarate dependent dioxygenase (FTO), YTH N6-methyladenosine RNA binding protein 2 (YTHDF2), and ATF4. ATF4 transcriptionally upregulated DDIT4 to suppress Serine/threonine-protein kinase mTOR (MTOR), which induced pro-survival autophagy during glutaminolysis inhibition. | |||
| Target Regulator | Fat mass and obesity-associated protein (FTO) | ERASER | ||
| Responsed Disease | Colorectal cancer | ICD-11: 2B91 | ||
| Pathway Response | mTOR signaling pathway | hsa04150 | ||
| Autophagy | hsa04140 | |||
| Cell Process | RNA decay | |||
| Cell growth and death | ||||
| Cell autophagy | ||||
| In-vitro Model | HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 |
| SW480 | Colon adenocarcinoma | Homo sapiens | CVCL_0546 | |
| Experiment 2 Reporting the m6A-centered Drug Response | [6] | |||
| Response Summary | In colorectal cancer, Glutaminolysis inhibition upregulated ATF4 expression in an m6A-dependent manner to activate pro-survival autophagy through transcriptional activation of the mTOR inhibitor DDIT4. Determined the relationship between FTO alpha-ketoglutarate dependent dioxygenase (FTO), YTH N6-methyladenosine RNA binding protein 2 (YTHDF2), and ATF4. ATF4 transcriptionally upregulated DDIT4 to suppress Serine/threonine-protein kinase mTOR (MTOR), which induced pro-survival autophagy during glutaminolysis inhibition. | |||
| Target Regulator | YTH domain-containing family protein 2 (YTHDF2) | READER | ||
| Target Regulation | Up regulation | |||
| Responsed Disease | Colorectal cancer | ICD-11: 2B91 | ||
| Pathway Response | mTOR signaling pathway | hsa04150 | ||
| Autophagy | hsa04140 | |||
| Cell Process | RNA decay | |||
| Cell growth and death | ||||
| Cell autophagy | ||||
| In-vitro Model | HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 |
| SW480 | Colon adenocarcinoma | Homo sapiens | CVCL_0546 | |
Chloroquine
[Approved]
| In total 2 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response | [6] | |||
| Response Summary | In colorectal cancer, Glutaminolysis inhibition upregulated ATF4 expression in an m6A-dependent manner to activate pro-survival autophagy through transcriptional activation of the mTOR inhibitor DDIT4. Determined the relationship between FTO alpha-ketoglutarate dependent dioxygenase (FTO), YTH N6-methyladenosine RNA binding protein 2 (YTHDF2), and ATF4. ATF4 transcriptionally upregulated DDIT4 to suppress Serine/threonine-protein kinase mTOR (MTOR), which induced pro-survival autophagy during glutaminolysis inhibition. | |||
| Target Regulator | Fat mass and obesity-associated protein (FTO) | ERASER | ||
| Responsed Disease | Colorectal cancer | ICD-11: 2B91 | ||
| Pathway Response | mTOR signaling pathway | hsa04150 | ||
| Autophagy | hsa04140 | |||
| Cell Process | RNA decay | |||
| Cell growth and death | ||||
| Cell autophagy | ||||
| In-vitro Model | HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 |
| SW480 | Colon adenocarcinoma | Homo sapiens | CVCL_0546 | |
| Experiment 2 Reporting the m6A-centered Drug Response | [6] | |||
| Response Summary | In colorectal cancer, Glutaminolysis inhibition upregulated ATF4 expression in an m6A-dependent manner to activate pro-survival autophagy through transcriptional activation of the mTOR inhibitor DDIT4. Determined the relationship between FTO alpha-ketoglutarate dependent dioxygenase (FTO), YTH N6-methyladenosine RNA binding protein 2 (YTHDF2), and ATF4. Serine/threonine-protein kinase mTOR (MTOR) transcriptionally upregulated DDIT4 to suppress mTOR, which induced pro-survival autophagy during glutaminolysis inhibition. | |||
| Target Regulator | YTH domain-containing family protein 2 (YTHDF2) | READER | ||
| Target Regulation | Up regulation | |||
| Responsed Disease | Colorectal cancer | ICD-11: 2B91 | ||
| Pathway Response | mTOR signaling pathway | hsa04150 | ||
| Autophagy | hsa04140 | |||
| Cell Process | RNA decay | |||
| Cell growth and death | ||||
| Cell autophagy | ||||
| In-vitro Model | HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 |
| SW480 | Colon adenocarcinoma | Homo sapiens | CVCL_0546 | |
Meclofenamate sodium
[Approved]
| In total 2 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response | [6] | |||
| Response Summary | In colorectal cancer, Glutaminolysis inhibition upregulated ATF4 expression in an m6A-dependent manner to activate pro-survival autophagy through transcriptional activation of the mTOR inhibitor DDIT4. Determined the relationship between FTO alpha-ketoglutarate dependent dioxygenase (FTO), YTH N6-methyladenosine RNA binding protein 2 (YTHDF2), and ATF4. Serine/threonine-protein kinase mTOR (MTOR) transcriptionally upregulated DDIT4 to suppress mTOR, which induced pro-survival autophagy during glutaminolysis inhibition. | |||
| Target Regulator | Fat mass and obesity-associated protein (FTO) | ERASER | ||
| Responsed Disease | Colorectal cancer | ICD-11: 2B91 | ||
| Pathway Response | mTOR signaling pathway | hsa04150 | ||
| Autophagy | hsa04140 | |||
| Cell Process | RNA decay | |||
| Cell growth and death | ||||
| Cell autophagy | ||||
| In-vitro Model | HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 |
| SW480 | Colon adenocarcinoma | Homo sapiens | CVCL_0546 | |
| Experiment 2 Reporting the m6A-centered Drug Response | [6] | |||
| Response Summary | In colorectal cancer, Glutaminolysis inhibition upregulated ATF4 expression in an m6A-dependent manner to activate pro-survival autophagy through transcriptional activation of the mTOR inhibitor DDIT4. Determined the relationship between FTO alpha-ketoglutarate dependent dioxygenase (FTO), YTH N6-methyladenosine RNA binding protein 2 (YTHDF2), and ATF4. Serine/threonine-protein kinase mTOR (MTOR) transcriptionally upregulated DDIT4 to suppress mTOR, which induced pro-survival autophagy during glutaminolysis inhibition. | |||
| Target Regulator | YTH domain-containing family protein 2 (YTHDF2) | READER | ||
| Target Regulation | Up regulation | |||
| Responsed Disease | Colorectal cancer | ICD-11: 2B91 | ||
| Pathway Response | mTOR signaling pathway | hsa04150 | ||
| Autophagy | hsa04140 | |||
| Cell Process | RNA decay | |||
| Cell growth and death | ||||
| Cell autophagy | ||||
| In-vitro Model | HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 |
| SW480 | Colon adenocarcinoma | Homo sapiens | CVCL_0546 | |
Rapamycin
[Approved]
| In total 3 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response | [6] | |||
| Response Summary | In colorectal cancer, Glutaminolysis inhibition upregulated ATF4 expression in an m6A-dependent manner to activate pro-survival autophagy through transcriptional activation of the mTOR inhibitor DDIT4. Determined the relationship between FTO alpha-ketoglutarate dependent dioxygenase (FTO), YTH N6-methyladenosine RNA binding protein 2 (YTHDF2), and ATF4. Serine/threonine-protein kinase mTOR (MTOR) transcriptionally upregulated DDIT4 to suppress mTOR, which induced pro-survival autophagy during glutaminolysis inhibition. | |||
| Target Regulator | Fat mass and obesity-associated protein (FTO) | ERASER | ||
| Responsed Disease | Colorectal cancer | ICD-11: 2B91 | ||
| Pathway Response | mTOR signaling pathway | hsa04150 | ||
| Autophagy | hsa04140 | |||
| Cell Process | RNA decay | |||
| Cell growth and death | ||||
| Cell autophagy | ||||
| In-vitro Model | HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 |
| SW480 | Colon adenocarcinoma | Homo sapiens | CVCL_0546 | |
| Experiment 2 Reporting the m6A-centered Drug Response | [3] | |||
| Response Summary | METTL3 promotes the progression of retinoblastoma through PI3K/AKT/Serine/threonine-protein kinase mTOR (MTOR) pathways in vitro and in vivo. METTL3 has an impact on the PI3K-AKT-mTOR-P70S6K/4EBP1 pathway. The cell proliferation results show that the stimulatory function of METTL3 is lost after rapamycin treatment. | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Target Regulation | Up regulation | |||
| Responsed Disease | Retinoblastoma | ICD-11: 2D02.2 | ||
| Pathway Response | PI3K-Akt signaling pathway | hsa04151 | ||
| mTOR signaling pathway | hsa04150 | |||
| Cell Process | Cell proliferation | |||
| Cell migration | ||||
| Cell invasion | ||||
| Cell apoptosis | ||||
| In-vitro Model | WERI-Rb-1 | Retinoblastoma | Homo sapiens | CVCL_1792 |
| Y-79 | Retinoblastoma | Homo sapiens | CVCL_1893 | |
| In-vivo Model | To establish a subcutaneous tumour model in nude mice, 2 × 107 Y79 cells (METTL3 knockdown group: shNC, shRNA1 and shRNA2; METTL3 up-regulated group: NC and METLL3) were resuspended in 1 mL of pre-cooled PBS, and 200 uL of the cell suspension was injected subcutaneously into the left side of the armpit to investigate tumour growth (4 × 106 per mouse). | |||
| Experiment 3 Reporting the m6A-centered Drug Response | [6] | |||
| Response Summary | In colorectal cancer, Glutaminolysis inhibition upregulated ATF4 expression in an m6A-dependent manner to activate pro-survival autophagy through transcriptional activation of the mTOR inhibitor DDIT4. Determined the relationship between FTO alpha-ketoglutarate dependent dioxygenase (FTO), YTH N6-methyladenosine RNA binding protein 2 (YTHDF2), and ATF4. Serine/threonine-protein kinase mTOR (MTOR) transcriptionally upregulated DDIT4 to suppress mTOR, which induced pro-survival autophagy during glutaminolysis inhibition. | |||
| Target Regulator | YTH domain-containing family protein 2 (YTHDF2) | READER | ||
| Target Regulation | Up regulation | |||
| Responsed Disease | Colorectal cancer | ICD-11: 2B91 | ||
| Pathway Response | mTOR signaling pathway | hsa04150 | ||
| Autophagy | hsa04140 | |||
| Cell Process | RNA decay | |||
| Cell growth and death | ||||
| Cell autophagy | ||||
| In-vitro Model | HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 |
| SW480 | Colon adenocarcinoma | Homo sapiens | CVCL_0546 | |
CB-839
[Phase 2]
| In total 2 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response | [6] | |||
| Response Summary | In colorectal cancer, Glutaminolysis inhibition upregulated ATF4 expression in an m6A-dependent manner to activate pro-survival autophagy through transcriptional activation of the mTOR inhibitor DDIT4. Determined the relationship between FTO alpha-ketoglutarate dependent dioxygenase (FTO), YTH N6-methyladenosine RNA binding protein 2 (YTHDF2), and ATF4. Serine/threonine-protein kinase mTOR (MTOR) transcriptionally upregulated DDIT4 to suppress mTOR, which induced pro-survival autophagy during glutaminolysis inhibition. | |||
| Target Regulator | Fat mass and obesity-associated protein (FTO) | ERASER | ||
| Responsed Disease | Colorectal cancer | ICD-11: 2B91 | ||
| Pathway Response | mTOR signaling pathway | hsa04150 | ||
| Autophagy | hsa04140 | |||
| Cell Process | RNA decay | |||
| Cell growth and death | ||||
| Cell autophagy | ||||
| In-vitro Model | HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 |
| SW480 | Colon adenocarcinoma | Homo sapiens | CVCL_0546 | |
| Experiment 2 Reporting the m6A-centered Drug Response | [6] | |||
| Response Summary | In colorectal cancer, Glutaminolysis inhibition upregulated ATF4 expression in an m6A-dependent manner to activate pro-survival autophagy through transcriptional activation of the mTOR inhibitor DDIT4. Determined the relationship between FTO alpha-ketoglutarate dependent dioxygenase (FTO), YTH N6-methyladenosine RNA binding protein 2 (YTHDF2), and ATF4. ATF4 transcriptionally upregulated DDIT4 to suppress Serine/threonine-protein kinase mTOR (MTOR), which induced pro-survival autophagy during glutaminolysis inhibition. | |||
| Target Regulator | YTH domain-containing family protein 2 (YTHDF2) | READER | ||
| Target Regulation | Up regulation | |||
| Responsed Disease | Colorectal cancer | ICD-11: 2B91 | ||
| Pathway Response | mTOR signaling pathway | hsa04150 | ||
| Autophagy | hsa04140 | |||
| Cell Process | RNA decay | |||
| Cell growth and death | ||||
| Cell autophagy | ||||
| In-vitro Model | HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 |
| SW480 | Colon adenocarcinoma | Homo sapiens | CVCL_0546 | |
GLS-IN-968
[Investigative]
| In total 2 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response | [6] | |||
| Response Summary | In colorectal cancer, Glutaminolysis inhibition upregulated ATF4 expression in an m6A-dependent manner to activate pro-survival autophagy through transcriptional activation of the mTOR inhibitor DDIT4. Determined the relationship between FTO alpha-ketoglutarate dependent dioxygenase (FTO), YTH N6-methyladenosine RNA binding protein 2 (YTHDF2), and ATF4. ATF4 transcriptionally upregulated DDIT4 to suppress Serine/threonine-protein kinase mTOR (MTOR), which induced pro-survival autophagy during glutaminolysis inhibition. | |||
| Target Regulator | Fat mass and obesity-associated protein (FTO) | ERASER | ||
| Responsed Disease | Colorectal cancer | ICD-11: 2B91 | ||
| Pathway Response | mTOR signaling pathway | hsa04150 | ||
| Autophagy | hsa04140 | |||
| Cell Process | RNA decay | |||
| Cell growth and death | ||||
| Cell autophagy | ||||
| In-vitro Model | HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 |
| SW480 | Colon adenocarcinoma | Homo sapiens | CVCL_0546 | |
| Experiment 2 Reporting the m6A-centered Drug Response | [6] | |||
| Response Summary | In colorectal cancer, Glutaminolysis inhibition upregulated ATF4 expression in an m6A-dependent manner to activate pro-survival autophagy through transcriptional activation of the mTOR inhibitor DDIT4. Determined the relationship between FTO alpha-ketoglutarate dependent dioxygenase (FTO), YTH N6-methyladenosine RNA binding protein 2 (YTHDF2), and ATF4. ATF4 transcriptionally upregulated DDIT4 to suppress Serine/threonine-protein kinase mTOR (MTOR), which induced pro-survival autophagy during glutaminolysis inhibition. | |||
| Target Regulator | YTH domain-containing family protein 2 (YTHDF2) | READER | ||
| Target Regulation | Up regulation | |||
| Responsed Disease | Colorectal cancer | ICD-11: 2B91 | ||
| Pathway Response | mTOR signaling pathway | hsa04150 | ||
| Autophagy | hsa04140 | |||
| Cell Process | RNA decay | |||
| Cell growth and death | ||||
| Cell autophagy | ||||
| In-vitro Model | HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 |
| SW480 | Colon adenocarcinoma | Homo sapiens | CVCL_0546 | |
Full List of Crosstalk(s) between m6A Modification and Epigenetic Regulation Related to This Regulator
DNA modification
m6A Regulator: Methyltransferase-like 3 (METTL3)
| In total 4 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT02151 | ||
| Epigenetic Regulator | Cysteine methyltransferase DNMT3A (DNMT3A) | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Lung cancer | |
| Drug | Metformin | |
| Crosstalk ID: M6ACROT02164 | ||
| Epigenetic Regulator | DNA (cytosine-5)-methyltransferase 3B (DNMT3B) | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Lung cancer | |
| Drug | Metformin | |
| Crosstalk ID: M6ACROT02177 | ||
| Epigenetic Regulator | Cysteine methyltransferase DNMT3A (DNMT3A) | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Lung cancer | |
| Drug | Osimertinib | |
| Crosstalk ID: M6ACROT02190 | ||
| Epigenetic Regulator | DNA (cytosine-5)-methyltransferase 3B (DNMT3B) | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Lung cancer | |
| Drug | Osimertinib | |
Histone modification
m6A Regulator: YTH domain-containing family protein 2 (YTHDF2)
| In total 6 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT03340 | ||
| Epigenetic Regulator | Histone acetyltransferase p300 (P300) | |
| Regulated Target | Histone H3 lysine 18 lactylation (H3K18la) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
| Drug | Rapamycin | |
| Crosstalk ID: M6ACROT03341 | ||
| Epigenetic Regulator | Histone acetyltransferase p300 (P300) | |
| Regulated Target | Histone H3 lysine 18 lactylation (H3K18la) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
| Drug | GLS-IN-968 | |
| Crosstalk ID: M6ACROT03342 | ||
| Epigenetic Regulator | Histone acetyltransferase p300 (P300) | |
| Regulated Target | Histone H3 lysine 18 lactylation (H3K18la) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
| Drug | CB-839 | |
| Crosstalk ID: M6ACROT03343 | ||
| Epigenetic Regulator | Histone acetyltransferase p300 (P300) | |
| Regulated Target | Histone H3 lysine 18 lactylation (H3K18la) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
| Drug | Meclofenamate sodium | |
| Crosstalk ID: M6ACROT03344 | ||
| Epigenetic Regulator | Histone acetyltransferase p300 (P300) | |
| Regulated Target | Histone H3 lysine 18 lactylation (H3K18la) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
| Drug | Chloroquine | |
| Crosstalk ID: M6ACROT03345 | ||
| Epigenetic Regulator | Histone acetyltransferase p300 (P300) | |
| Regulated Target | Histone H3 lysine 18 lactylation (H3K18la) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
| Drug | Asparagine inhibitor | |
m6A Regulator: Methyltransferase-like 3 (METTL3)
| In total 2 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT03387 | ||
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Lung cancer | |
| Drug | Gefitinib | |
| Crosstalk ID: M6ACROT03400 | ||
| Regulated Target | Histone H3 lysine 4 monomethylation (H3K4me1) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Lung cancer | |
| Drug | Gefitinib | |
m6A Regulator: Fat mass and obesity-associated protein (FTO)
| In total 6 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT03438 | ||
| Epigenetic Regulator | Histone deacetylase 1 (HDAC1) | |
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
| Drug | Rapamycin | |
| Crosstalk ID: M6ACROT03439 | ||
| Epigenetic Regulator | Histone deacetylase 1 (HDAC1) | |
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
| Drug | GLS-IN-968 | |
| Crosstalk ID: M6ACROT03440 | ||
| Epigenetic Regulator | Histone deacetylase 1 (HDAC1) | |
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
| Drug | CB-839 | |
| Crosstalk ID: M6ACROT03441 | ||
| Epigenetic Regulator | Histone deacetylase 1 (HDAC1) | |
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
| Drug | Meclofenamate sodium | |
| Crosstalk ID: M6ACROT03442 | ||
| Epigenetic Regulator | Histone deacetylase 1 (HDAC1) | |
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
| Drug | Chloroquine | |
| Crosstalk ID: M6ACROT03443 | ||
| Epigenetic Regulator | Histone deacetylase 1 (HDAC1) | |
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
| Drug | Asparagine inhibitor | |
m6A Regulator: Methyltransferase-like 14 (METTL14)
| In total 1 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT03500 | ||
| Regulated Target | Histone H3 lysine 18 lactylation (H3K18la) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Gastric cancer | |
RNA Modification Sequencing Data Associated with the Target (ID: M6ATAR00339)
| In total 4 m6A sequence/site(s) in this target gene | |||
| mod ID: A2ISITE002482 | Click to Show/Hide the Full List | ||
| mod site | chr1:11120405-11120406:- | [8] | |
| Sequence | AGCTGGGATTACAGGCACGTACCACCATGTGGGTTAATTTT | ||
| Transcript ID List | ENST00000376838.5; ENST00000361445.8 | ||
| External Link | RMBase: RNA-editing_site_1299 | ||
| mod ID: A2ISITE002511 | Click to Show/Hide the Full List | ||
| mod site | chr1:11142255-11142256:- | [8] | |
| Sequence | TCCCAAAGTTCTGGGATTACAGGCATGAGCCACCATGCCTG | ||
| Transcript ID List | ENST00000495435.1; ENST00000361445.8 | ||
| External Link | RMBase: RNA-editing_site_1300 | ||
| mod ID: A2ISITE002515 | Click to Show/Hide the Full List | ||
| mod site | chr1:11235859-11235860:- | [8] | |
| Sequence | CCTCCTGAGTAGCTGGGACTACAGGCGCCCGCCACCACGCC | ||
| Transcript ID List | ENST00000361445.8 | ||
| External Link | RMBase: RNA-editing_site_1301 | ||
| mod ID: A2ISITE002516 | Click to Show/Hide the Full List | ||
| mod site | chr1:11236316-11236317:- | [8] | |
| Sequence | ATGACAAACCACGTCTCTACAAAATATACGAAAGTTAGCTG | ||
| Transcript ID List | ENST00000361445.8; rmsk_21463 | ||
| External Link | RMBase: RNA-editing_site_1302 | ||
2'-O-Methylation (2'-O-Me)
| In total 1 m6A sequence/site(s) in this target gene | |||
| mod ID: 2OMSITE000219 | Click to Show/Hide the Full List | ||
| mod site | chr1:11247410-11247411:- | [9] | |
| Sequence | ACAGAGAGGCAATTCTATGGCCAAGCAAATTTTCATGTATG | ||
| Cell/Tissue List | HEK293 | ||
| Seq Type List | RiboMeth-seq | ||
| Transcript ID List | ENST00000361445.8 | ||
| External Link | RMBase: Nm_site_30 | ||
5-methylcytidine (m5C)
N6-methyladenosine (m6A)
| In total 205 m6A sequence/site(s) in this target gene | |||
| mod ID: M6ASITE074317 | Click to Show/Hide the Full List | ||
| mod site | chr1:11106538-11106539:- | [10] | |
| Sequence | AAATGACATCAGAATTTTAAACATATGTATATGAGTGGCGT | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa; HEK293T; Huh7 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000376838.5; ENST00000361445.8 | ||
| External Link | RMBase: m6A_site_7120 | ||
| mod ID: M6ASITE074346 | Click to Show/Hide the Full List | ||
| mod site | chr1:11106553-11106554:- | [11] | |
| Sequence | AATTTTTGTGCCAATAAATGACATCAGAATTTTAAACATAT | ||
| Motif Score | 2.859755952 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000376838.5; ENST00000361445.8 | ||
| External Link | RMBase: m6A_site_7121 | ||
| mod ID: M6ASITE074431 | Click to Show/Hide the Full List | ||
| mod site | chr1:11106593-11106594:- | [10] | |
| Sequence | TAAAGTGTAGCCATGTCTAGACACCATGTTGTATCAGAATA | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa; HEK293T; Huh7 | ||
| Seq Type List | m6A-seq; MeRIP-seq; DART-seq | ||
| Transcript ID List | ENST00000361445.8; ENST00000376838.5 | ||
| External Link | RMBase: m6A_site_7122 | ||
| mod ID: M6ASITE074432 | Click to Show/Hide the Full List | ||
| mod site | chr1:11106635-11106636:- | [11] | |
| Sequence | AAATTATGAGCAGAACAAATACTCAACTAAATGCACAAAGT | ||
| Motif Score | 2.53247619 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000376838.5; ENST00000361445.8 | ||
| External Link | RMBase: m6A_site_7123 | ||
| mod ID: M6ASITE074499 | Click to Show/Hide the Full List | ||
| mod site | chr1:11106641-11106642:- | [10] | |
| Sequence | TATTGGAAATTATGAGCAGAACAAATACTCAACTAAATGCA | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa; HEK293T; hESCs; Huh7 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000376838.5; ENST00000361445.8 | ||
| External Link | RMBase: m6A_site_7124 | ||
| mod ID: M6ASITE074528 | Click to Show/Hide the Full List | ||
| mod site | chr1:11106682-11106683:- | [10] | |
| Sequence | TACTGTCATGGAGGTGCTGAACACAGGGAAGGTCTGGTACA | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa; HEK293T; hESC-HEK293T; hESCs; Huh7 | ||
| Seq Type List | m6A-seq; MeRIP-seq; DART-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000376838.5; ENST00000361445.8 | ||
| External Link | RMBase: m6A_site_7125 | ||
| mod ID: M6ASITE074529 | Click to Show/Hide the Full List | ||
| mod site | chr1:11106709-11106710:- | [10] | |
| Sequence | TTAGAGCAAGGGCTCAGAAAACAGAAATACTGTCATGGAGG | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa; HEK293T; hESCs; A549; Huh7 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000376838.5; ENST00000361445.8 | ||
| External Link | RMBase: m6A_site_7126 | ||
| mod ID: M6ASITE074534 | Click to Show/Hide the Full List | ||
| mod site | chr1:11106792-11106793:- | [10] | |
| Sequence | GGGGATCACTGTGCAGTGGGACCACCCTCACTGGCCTTCTG | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; HEK293T; HepG2; Huh7 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000361445.8; ENST00000376838.5 | ||
| External Link | RMBase: m6A_site_7127 | ||
| mod ID: M6ASITE074535 | Click to Show/Hide the Full List | ||
| mod site | chr1:11106876-11106877:- | [10] | |
| Sequence | CTCACACGCTTCAATTCAAGACCTGACCGCTAGTAGGGAGG | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; Huh7; peripheral-blood; HEK293A-TOA; iSLK; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000376838.5; ENST00000361445.8 | ||
| External Link | RMBase: m6A_site_7128 | ||
| mod ID: M6ASITE074623 | Click to Show/Hide the Full List | ||
| mod site | chr1:11106893-11106894:- | [12] | |
| Sequence | ACCCAGTGATGCTGCGACTCACACGCTTCAATTCAAGACCT | ||
| Motif Score | 2.047297619 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000361445.8; ENST00000376838.5 | ||
| External Link | RMBase: m6A_site_7129 | ||
| mod ID: M6ASITE074624 | Click to Show/Hide the Full List | ||
| mod site | chr1:11106913-11106914:- | [10] | |
| Sequence | AAAGGTCTGTCTTCCATCAGACCCAGTGATGCTGCGACTCA | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; A549; Huh7; Jurkat; peripheral-blood; GSC-11; HEK293A-TOA; iSLK; NB4; MM6 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000361445.8; ENST00000376838.5 | ||
| External Link | RMBase: m6A_site_7130 | ||
| mod ID: M6ASITE074625 | Click to Show/Hide the Full List | ||
| mod site | chr1:11106936-11106937:- | [11] | |
| Sequence | CAAGATAGGCCAAAATGAGTACAAAAGGTCTGTCTTCCATC | ||
| Motif Score | 2.856142857 | ||
| Cell/Tissue List | HEK293T; hESC-HEK293T | ||
| Seq Type List | DART-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000376838.5; ENST00000361445.8 | ||
| External Link | RMBase: m6A_site_7131 | ||
| mod ID: M6ASITE074650 | Click to Show/Hide the Full List | ||
| mod site | chr1:11106975-11106976:- | [10] | |
| Sequence | AAGACCTCACTGGTCTGTGGACAGCAGCAGAAATGTTTGCA | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; A549; hESC-HEK293T; H1A; H1B; hNPCs; hESCs; fibroblasts; GM12878; H1299; MM6; Huh7; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; TREX; endometrial; HEC-1-A; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000376838.5; ENST00000361445.8 | ||
| External Link | RMBase: m6A_site_7132 | ||
| mod ID: M6ASITE074651 | Click to Show/Hide the Full List | ||
| mod site | chr1:11106992-11106993:- | [10] | |
| Sequence | CCCCTGCCACCTATCCCAAGACCTCACTGGTCTGTGGACAG | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; A549; U2OS; H1A; H1B; hNPCs; hESCs; fibroblasts; GM12878; H1299; MM6; Huh7; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; TREX; endometrial; HEC-1-A; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000361445.8; ENST00000376838.5 | ||
| External Link | RMBase: m6A_site_7133 | ||
| mod ID: M6ASITE074652 | Click to Show/Hide the Full List | ||
| mod site | chr1:11107031-11107032:- | [11] | |
| Sequence | GGATCCTAGACTGTAAAGACACAGAAGATGCTGACCTCACC | ||
| Motif Score | 2.084928571 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000361445.8; ENST00000376838.5 | ||
| External Link | RMBase: m6A_site_7134 | ||
| mod ID: M6ASITE074655 | Click to Show/Hide the Full List | ||
| mod site | chr1:11107033-11107034:- | [10] | |
| Sequence | TAGGATCCTAGACTGTAAAGACACAGAAGATGCTGACCTCA | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; A549; hESC-HEK293T; U2OS; H1A; H1B; hNPCs; hESCs; fibroblasts; GM12878; CD8T; H1299; MM6; Huh7; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; TREX; endometrial; HEC-1-A; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq; DART-seq; MAZTER-seq; m6A-CLIP/IP | ||
| Transcript ID List | ENST00000376838.5; ENST00000361445.8 | ||
| External Link | RMBase: m6A_site_7135 | ||
| mod ID: M6ASITE074689 | Click to Show/Hide the Full List | ||
| mod site | chr1:11107042-11107043:- | [10] | |
| Sequence | AACATGGATTAGGATCCTAGACTGTAAAGACACAGAAGATG | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; A549; U2OS; H1A; H1B; hNPCs; hESCs; fibroblasts; GM12878; H1299; MM6; Huh7; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; TREX; endometrial; HEC-1-A; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq; DART-seq; m6A-CLIP/IP | ||
| Transcript ID List | ENST00000376838.5; ENST00000361445.8 | ||
| External Link | RMBase: m6A_site_7136 | ||
| mod ID: M6ASITE074690 | Click to Show/Hide the Full List | ||
| mod site | chr1:11107061-11107062:- | [10] | |
| Sequence | GAAGCCTCAGGTCGTGGAGAACATGGATTAGGATCCTAGAC | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; A549; hESC-HEK293T; U2OS; H1A; H1B; hNPCs; hESCs; fibroblasts; GM12878; H1299; MM6; Huh7; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; TREX; endometrial; HEC-1-A; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000361445.8; ENST00000376838.5 | ||
| External Link | RMBase: m6A_site_7137 | ||
| mod ID: M6ASITE074691 | Click to Show/Hide the Full List | ||
| mod site | chr1:11107089-11107090:- | [13] | |
| Sequence | ATGGGTACAGCAAACTCAGCACAGCCAAGAAGCCTCAGGTC | ||
| Motif Score | 2.830589286 | ||
| Cell/Tissue List | HEK293; HEK293T | ||
| Seq Type List | m6A-REF-seq; DART-seq | ||
| Transcript ID List | ENST00000376838.5; ENST00000361445.8 | ||
| External Link | RMBase: m6A_site_7138 | ||
| mod ID: M6ASITE074692 | Click to Show/Hide the Full List | ||
| mod site | chr1:11107096-11107097:- | [10] | |
| Sequence | TCTCAACATGGGTACAGCAAACTCAGCACAGCCAAGAAGCC | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; A549; U2OS; H1A; H1B; hNPCs; hESCs; fibroblasts; GM12878; H1299; MM6; Huh7; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; TREX; endometrial; HEC-1-A; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000376838.5; ENST00000361445.8 | ||
| External Link | RMBase: m6A_site_7139 | ||
| mod ID: M6ASITE074693 | Click to Show/Hide the Full List | ||
| mod site | chr1:11107111-11107112:- | [13] | |
| Sequence | TGACCTAGTTGCTCCTCTCAACATGGGTACAGCAAACTCAG | ||
| Motif Score | 2.173910714 | ||
| Cell/Tissue List | brain; liver; hESC-HEK293T | ||
| Seq Type List | m6A-REF-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000361445.8; ENST00000376838.5 | ||
| External Link | RMBase: m6A_site_7140 | ||
| mod ID: M6ASITE074694 | Click to Show/Hide the Full List | ||
| mod site | chr1:11107129-11107130:- | [11] | |
| Sequence | TACTTGCTGATGGAAGAATGACCTAGTTGCTCCTCTCAACA | ||
| Motif Score | 2.839113095 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000376838.5; ENST00000361445.8 | ||
| External Link | RMBase: m6A_site_7141 | ||
| mod ID: M6ASITE074695 | Click to Show/Hide the Full List | ||
| mod site | chr1:11107161-11107162:- | [10] | |
| Sequence | TTGACTTAACTCACAAGAGAACTCATCATAAGTACTTGCTG | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa; CD34; HepG2; HEK293T; A549; U2OS; hNPCs; hESCs; fibroblasts; GM12878; MM6; Huh7; HEK293A-TOA; iSLK; MSC; TIME; TREX | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000361445.8; ENST00000376838.5 | ||
| External Link | RMBase: m6A_site_7142 | ||
| mod ID: M6ASITE074696 | Click to Show/Hide the Full List | ||
| mod site | chr1:11107169-11107170:- | [11] | |
| Sequence | TACGGGTTTTGACTTAACTCACAAGAGAACTCATCATAAGT | ||
| Motif Score | 2.047297619 | ||
| Cell/Tissue List | HEK293T; hESC-HEK293T | ||
| Seq Type List | DART-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000361445.8; ENST00000376838.5 | ||
| External Link | RMBase: m6A_site_7143 | ||
| mod ID: M6ASITE074697 | Click to Show/Hide the Full List | ||
| mod site | chr1:11107173-11107174:- | [11] | |
| Sequence | GAAATACGGGTTTTGACTTAACTCACAAGAGAACTCATCAT | ||
| Motif Score | 2.590089286 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000376838.5; ENST00000361445.8 | ||
| External Link | RMBase: m6A_site_7144 | ||
| mod ID: M6ASITE074698 | Click to Show/Hide the Full List | ||
| mod site | chr1:11107178-11107179:- | [11] | |
| Sequence | CTTTAGAAATACGGGTTTTGACTTAACTCACAAGAGAACTC | ||
| Motif Score | 3.28175 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000376838.5; ENST00000361445.8 | ||
| External Link | RMBase: m6A_site_7145 | ||
| mod ID: M6ASITE074699 | Click to Show/Hide the Full List | ||
| mod site | chr1:11107199-11107200:- | [11] | |
| Sequence | TGGGGAACAGAAGATCCATAACTTTAGAAATACGGGTTTTG | ||
| Motif Score | 2.590089286 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000361445.8; ENST00000376838.5 | ||
| External Link | RMBase: m6A_site_7146 | ||
| mod ID: M6ASITE074733 | Click to Show/Hide the Full List | ||
| mod site | chr1:11107213-11107214:- | [10] | |
| Sequence | CAAGCCATTCATTTTGGGGAACAGAAGATCCATAACTTTAG | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; U2OS; hNPCs; hESCs; fibroblasts; A549; GM12878; MM6; Huh7; peripheral-blood; HEK293A-TOA; iSLK; MSC; TIME; TREX | ||
| Seq Type List | m6A-seq; MeRIP-seq; DART-seq | ||
| Transcript ID List | ENST00000361445.8; ENST00000376838.5 | ||
| External Link | RMBase: m6A_site_7147 | ||
| mod ID: M6ASITE074802 | Click to Show/Hide the Full List | ||
| mod site | chr1:11107241-11107242:- | [11] | |
| Sequence | CAGTGAAACATAGTAATACCACGTAAATCAAGCCATTCATT | ||
| Motif Score | 2.027047619 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000376838.5; ENST00000361445.8 | ||
| External Link | RMBase: m6A_site_7148 | ||
| mod ID: M6ASITE074803 | Click to Show/Hide the Full List | ||
| mod site | chr1:11107254-11107255:- | [10] | |
| Sequence | TTGGTTCCCAGGACAGTGAAACATAGTAATACCACGTAAAT | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; hESC-HEK293T; U2OS; hNPCs; hESCs; fibroblasts; A549; GM12878; MM6; Huh7; peripheral-blood; HEK293A-TOA; iSLK; MSC; TIME; TREX | ||
| Seq Type List | m6A-seq; MeRIP-seq; DART-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000376838.5; ENST00000361445.8 | ||
| External Link | RMBase: m6A_site_7149 | ||
| mod ID: M6ASITE074804 | Click to Show/Hide the Full List | ||
| mod site | chr1:11107262-11107263:- | [10] | |
| Sequence | GGCTTGATTTGGTTCCCAGGACAGTGAAACATAGTAATACC | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; hESC-HEK293T; U2OS; hNPCs; hESCs; fibroblasts; A549; GM12878; MM6; Huh7; peripheral-blood; HEK293A-TOA; iSLK; MSC; TIME; TREX | ||
| Seq Type List | m6A-seq; MeRIP-seq; MAZTER-seq; m6A-CLIP/IP | ||
| Transcript ID List | ENST00000376838.5; ENST00000361445.8 | ||
| External Link | RMBase: m6A_site_7150 | ||
| mod ID: M6ASITE074833 | Click to Show/Hide the Full List | ||
| mod site | chr1:11107286-11107287:- | [11] | |
| Sequence | AAGAATATATTGTCAGAAACACAAGGCTTGATTTGGTTCCC | ||
| Motif Score | 2.084928571 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000361445.8; ENST00000376838.5; ENST00000473471.5 | ||
| External Link | RMBase: m6A_site_7151 | ||
| mod ID: M6ASITE074909 | Click to Show/Hide the Full List | ||
| mod site | chr1:11107288-11107289:- | [10] | |
| Sequence | TGAAGAATATATTGTCAGAAACACAAGGCTTGATTTGGTTC | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; hESC-HEK293T; hNPCs; hESCs; fibroblasts; A549; GM12878; MM6; Huh7; peripheral-blood; HEK293A-TOA; iSLK; MSC; TIME; TREX | ||
| Seq Type List | m6A-seq; MeRIP-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000361445.8; ENST00000376838.5; ENST00000473471.5 | ||
| External Link | RMBase: m6A_site_7152 | ||
| mod ID: M6ASITE074926 | Click to Show/Hide the Full List | ||
| mod site | chr1:11107328-11107329:- | [10] | |
| Sequence | TATATTAAAAGTTGGTTTGAACCAACTTTCTAGCTGCTGTT | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | HeLa; HEK293T; HepG2; hNPCs; hESCs; fibroblasts; A549; GM12878; MM6; Huh7; HEK293A-TOA; iSLK; MSC; TIME | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000490931.1; ENST00000473471.5; ENST00000376838.5; ENST00000361445.8 | ||
| External Link | RMBase: m6A_site_7153 | ||
| mod ID: M6ASITE074927 | Click to Show/Hide the Full List | ||
| mod site | chr1:11107355-11107356:- | [10] | |
| Sequence | TTGAAATGTAAATGAAAAGAACTACTGTATATTAAAAGTTG | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa; HEK293T; HepG2; hNPCs; fibroblasts; A549; GM12878; CD8T; Huh7; HEK293A-TOA | ||
| Seq Type List | m6A-seq; MeRIP-seq; m6A-CLIP/IP | ||
| Transcript ID List | ENST00000361445.8; ENST00000473471.5; ENST00000376838.5; ENST00000490931.1 | ||
| External Link | RMBase: m6A_site_7154 | ||
| mod ID: M6ASITE074944 | Click to Show/Hide the Full List | ||
| mod site | chr1:11107390-11107391:- | [11] | |
| Sequence | AACCATGGTGAGAAAGTTTGACTTTGTTAAATATTTTGAAA | ||
| Motif Score | 3.28175 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000361445.8; ENST00000473471.5; ENST00000376838.5; ENST00000490931.1 | ||
| External Link | RMBase: m6A_site_7155 | ||
| mod ID: M6ASITE074962 | Click to Show/Hide the Full List | ||
| mod site | chr1:11107409-11107410:- | [11] | |
| Sequence | AATGCTTCCACTAAACTGAAACCATGGTGAGAAAGTTTGAC | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | HEK293T; HepG2; HeLa; A549 | ||
| Seq Type List | MeRIP-seq; DART-seq; m6A-seq | ||
| Transcript ID List | ENST00000490931.1; ENST00000376838.5; ENST00000473471.5; ENST00000361445.8 | ||
| External Link | RMBase: m6A_site_7156 | ||
| mod ID: M6ASITE074963 | Click to Show/Hide the Full List | ||
| mod site | chr1:11107415-11107416:- | [14] | |
| Sequence | TTAGTAAATGCTTCCACTAAACTGAAACCATGGTGAGAAAG | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HEK293T; HepG2; HeLa; A549 | ||
| Seq Type List | MeRIP-seq; m6A-seq | ||
| Transcript ID List | ENST00000361445.8; ENST00000490931.1; ENST00000376838.5; ENST00000473471.5 | ||
| External Link | RMBase: m6A_site_7157 | ||
| mod ID: M6ASITE074964 | Click to Show/Hide the Full List | ||
| mod site | chr1:11107438-11107439:- | [11] | |
| Sequence | GTTTTTTCTGAGGCTTTTGTACTTTAGTAAATGCTTCCACT | ||
| Motif Score | 3.278136905 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000376838.5; ENST00000490931.1; ENST00000361445.8; ENST00000473471.5 | ||
| External Link | RMBase: m6A_site_7158 | ||
| mod ID: M6ASITE074965 | Click to Show/Hide the Full List | ||
| mod site | chr1:11108264-11108265:- | [11] | |
| Sequence | TCGGGACTTCTCTCATGATGACACTTTGGATGTTCCAACGC | ||
| Motif Score | 2.859755952 | ||
| Cell/Tissue List | HEK293T; hESC-HEK293T | ||
| Seq Type List | DART-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000490931.1; ENST00000376838.5; ENST00000361445.8; ENST00000473471.5 | ||
| External Link | RMBase: m6A_site_7159 | ||
| mod ID: M6ASITE074966 | Click to Show/Hide the Full List | ||
| mod site | chr1:11111575-11111576:- | [15] | |
| Sequence | AAAAGGGGACTTCACTGTGAACCGTAGCTGCGAAGAGAGCA | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | Huh7 | ||
| Seq Type List | MeRIP-seq | ||
| Transcript ID List | ENST00000361445.8; ENST00000490931.1; ENST00000473471.5; ENST00000455339.1; ENST00000376838.5 | ||
| External Link | RMBase: m6A_site_7160 | ||
| mod ID: M6ASITE074967 | Click to Show/Hide the Full List | ||
| mod site | chr1:11111587-11111588:- | [15] | |
| Sequence | GTGGAGATTCAGAAAAGGGGACTTCACTGTGAACCGTAGCT | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | Huh7 | ||
| Seq Type List | MeRIP-seq | ||
| Transcript ID List | ENST00000455339.1; ENST00000376838.5; ENST00000473471.5; ENST00000490931.1; ENST00000361445.8 | ||
| External Link | RMBase: m6A_site_7161 | ||
| mod ID: M6ASITE074968 | Click to Show/Hide the Full List | ||
| mod site | chr1:11112901-11112902:- | [12] | |
| Sequence | CCTAGCAAATACCAAAGGCAACAAGCGATCCCGAACGAGGA | ||
| Motif Score | 2.173910714 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000473471.5; ENST00000490931.1; ENST00000455339.1; ENST00000361445.8; ENST00000376838.5 | ||
| External Link | RMBase: m6A_site_7162 | ||
| mod ID: M6ASITE075011 | Click to Show/Hide the Full List | ||
| mod site | chr1:11114319-11114320:- | [12] | |
| Sequence | GCTGAACTGGAGGCTGATGGACAGTGAGTATCATCAAGTGC | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000473471.5; ENST00000361445.8; ENST00000455339.1; ENST00000376838.5; ENST00000490931.1 | ||
| External Link | RMBase: m6A_site_7163 | ||
| mod ID: M6ASITE075049 | Click to Show/Hide the Full List | ||
| mod site | chr1:11114334-11114335:- | [11] | |
| Sequence | TGTCTATGACCCCTTGCTGAACTGGAGGCTGATGGACAGTG | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000361445.8; ENST00000455339.1; ENST00000490931.1; ENST00000376838.5; ENST00000473471.5 | ||
| External Link | RMBase: m6A_site_7164 | ||
| mod ID: M6ASITE075050 | Click to Show/Hide the Full List | ||
| mod site | chr1:11114838-11114839:- | [12] | |
| Sequence | AGAAGATTCCATTTAGACTAACAAGAATGTTGACCAATGCT | ||
| Motif Score | 2.168095238 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000376838.5; ENST00000455339.1; ENST00000361445.8; ENST00000490931.1 | ||
| External Link | RMBase: m6A_site_7165 | ||
| mod ID: M6ASITE075075 | Click to Show/Hide the Full List | ||
| mod site | chr1:11115414-11115415:- | [11] | |
| Sequence | TGGGAAGATCCTGCACATTGACTTTGGGGACTGCTTTGAGG | ||
| Motif Score | 3.28175 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000361445.8; ENST00000490931.1; ENST00000455339.1; ENST00000376838.5 | ||
| External Link | RMBase: m6A_site_7166 | ||
| mod ID: M6ASITE075076 | Click to Show/Hide the Full List | ||
| mod site | chr1:11115420-11115421:- | [12] | |
| Sequence | TCTGAGTGGGAAGATCCTGCACATTGACTTTGGGGACTGCT | ||
| Motif Score | 2.830589286 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000455339.1; ENST00000361445.8; ENST00000376838.5; ENST00000490931.1 | ||
| External Link | RMBase: m6A_site_7167 | ||
| mod ID: M6ASITE075077 | Click to Show/Hide the Full List | ||
| mod site | chr1:11115467-11115468:- | [12] | |
| Sequence | CTGTCTTCTGTTTCTCAAAGACACCCATCCAACCTGATGCT | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000361445.8; ENST00000455339.1; ENST00000490931.1; ENST00000376838.5 | ||
| External Link | RMBase: m6A_site_7168 | ||
| mod ID: M6ASITE075092 | Click to Show/Hide the Full List | ||
| mod site | chr1:11121357-11121358:- | [10] | |
| Sequence | CCTGATGCAGATGGCTCCGGACTATGACCACTTGACTCTGA | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000361445.8; ENST00000376838.5 | ||
| External Link | RMBase: m6A_site_7169 | ||
| mod ID: M6ASITE075140 | Click to Show/Hide the Full List | ||
| mod site | chr1:11122003-11122004:- | [12] | |
| Sequence | GAAGAAGAAGATCCTTCTCAACATCGAGCATCGCATCATGT | ||
| Motif Score | 2.173910714 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000361445.8; ENST00000376838.5 | ||
| External Link | RMBase: m6A_site_7170 | ||
| mod ID: M6ASITE075141 | Click to Show/Hide the Full List | ||
| mod site | chr1:11122033-11122034:- | [10] | |
| Sequence | ACTGCACGCCCTCATCCGGGACTACAGGGAGAAGAAGAAGA | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000376838.5; ENST00000361445.8 | ||
| External Link | RMBase: m6A_site_7171 | ||
| mod ID: M6ASITE075142 | Click to Show/Hide the Full List | ||
| mod site | chr1:11122057-11122058:- | [12] | |
| Sequence | TGGCTGGGTTCCCCACTGTGACACACTGCACGCCCTCATCC | ||
| Motif Score | 2.859755952 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000361445.8; ENST00000376838.5 | ||
| External Link | RMBase: m6A_site_7172 | ||
| mod ID: M6ASITE075143 | Click to Show/Hide the Full List | ||
| mod site | chr1:11124503-11124504:- | [10] | |
| Sequence | CCCAACATCTCTTCGGAAAAACCTCAGGTATTCAGAAACCC | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000361445.8; ENST00000376838.5 | ||
| External Link | RMBase: m6A_site_7173 | ||
| mod ID: M6ASITE075144 | Click to Show/Hide the Full List | ||
| mod site | chr1:11124519-11124520:- | [12] | |
| Sequence | CCCTTCTGGCCAATGACCCAACATCTCTTCGGAAAAACCTC | ||
| Motif Score | 2.173910714 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000361445.8; ENST00000376838.5 | ||
| External Link | RMBase: m6A_site_7174 | ||
| mod ID: M6ASITE075145 | Click to Show/Hide the Full List | ||
| mod site | chr1:11124542-11124543:- | [12] | |
| Sequence | GCAGCTCTTCGGCCTGGTTAACACCCTTCTGGCCAATGACC | ||
| Motif Score | 2.168095238 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000361445.8; ENST00000376838.5 | ||
| External Link | RMBase: m6A_site_7175 | ||
| mod ID: M6ASITE075146 | Click to Show/Hide the Full List | ||
| mod site | chr1:11124622-11124623:- | [10] | |
| Sequence | TTCCTTACAGGCAGCAACGGACATGAGTTTGTTTTCCTTCT | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; hESC-HEK293T | ||
| Seq Type List | m6A-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000361445.8; ENST00000376838.5 | ||
| External Link | RMBase: m6A_site_7176 | ||
| mod ID: M6ASITE075147 | Click to Show/Hide the Full List | ||
| mod site | chr1:11126657-11126658:- | [12] | |
| Sequence | CACCGTCTTTGCAAGTCATCACATCCAAGCAGAGGCCCCGG | ||
| Motif Score | 2.047297619 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000376838.5; ENST00000361445.8 | ||
| External Link | RMBase: m6A_site_7177 | ||
| mod ID: M6ASITE075148 | Click to Show/Hide the Full List | ||
| mod site | chr1:11126720-11126721:- | [10] | |
| Sequence | TTGAATTGGCTGTGCCAGGAACATATGACCCCAACCAGCCA | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa; hESC-HEK293T | ||
| Seq Type List | m6A-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000376838.5; ENST00000361445.8 | ||
| External Link | RMBase: m6A_site_7178 | ||
| mod ID: M6ASITE075149 | Click to Show/Hide the Full List | ||
| mod site | chr1:11126743-11126744:- | [10] | |
| Sequence | AAAACTTCTGATGTGCCGGGACCTTGAATTGGCTGTGCCAG | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000361445.8; ENST00000376838.5 | ||
| External Link | RMBase: m6A_site_7179 | ||
| mod ID: M6ASITE075175 | Click to Show/Hide the Full List | ||
| mod site | chr1:11126760-11126761:- | [10] | |
| Sequence | CTGCAATATGTTTCCCCAAAACTTCTGATGTGCCGGGACCT | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000376838.5; ENST00000361445.8 | ||
| External Link | RMBase: m6A_site_7180 | ||
| mod ID: M6ASITE075209 | Click to Show/Hide the Full List | ||
| mod site | chr1:11126792-11126793:- | [12] | |
| Sequence | TTGGTAATTATTTACAGCTCACATCCTTAGAGCTGCAATAT | ||
| Motif Score | 2.047297619 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000361445.8; ENST00000376838.5 | ||
| External Link | RMBase: m6A_site_7181 | ||
| mod ID: M6ASITE075210 | Click to Show/Hide the Full List | ||
| mod site | chr1:11127055-11127056:- | [10] | |
| Sequence | GGACCTCACCCAAGCCTGGGACCTCTATTATCATGTGTTCC | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000376838.5; ENST00000361445.8 | ||
| External Link | RMBase: m6A_site_7182 | ||
| mod ID: M6ASITE075211 | Click to Show/Hide the Full List | ||
| mod site | chr1:11127073-11127074:- | [10] | |
| Sequence | GAAATCAGGGAATGTCAAGGACCTCACCCAAGCCTGGGACC | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000361445.8; ENST00000376838.5 | ||
| External Link | RMBase: m6A_site_7183 | ||
| mod ID: M6ASITE075263 | Click to Show/Hide the Full List | ||
| mod site | chr1:11127097-11127098:- | [12] | |
| Sequence | CCAAGAGTGGTGCAGGAAGTACATGAAATCAGGGAATGTCA | ||
| Motif Score | 2.856142857 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000361445.8; ENST00000376838.5 | ||
| External Link | RMBase: m6A_site_7184 | ||
| mod ID: M6ASITE075264 | Click to Show/Hide the Full List | ||
| mod site | chr1:11127637-11127638:- | [10] | |
| Sequence | GCCCCCAGACTCTGAAGGAAACATCCTTTAATCAGGTATGG | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa; hESC-HEK293T | ||
| Seq Type List | m6A-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000361445.8; ENST00000376838.5 | ||
| External Link | RMBase: m6A_site_7185 | ||
| mod ID: M6ASITE075265 | Click to Show/Hide the Full List | ||
| mod site | chr1:11127649-11127650:- | [10] | |
| Sequence | TGATGGAACGGGGCCCCCAGACTCTGAAGGAAACATCCTTT | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000376838.5; ENST00000361445.8 | ||
| External Link | RMBase: m6A_site_7186 | ||
| mod ID: M6ASITE075266 | Click to Show/Hide the Full List | ||
| mod site | chr1:11128028-11128029:- | [12] | |
| Sequence | GAACATGTGTGAGCACAGCAACACCCTGGTCCAGCAGGCCA | ||
| Motif Score | 2.173910714 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000376838.5; ENST00000361445.8 | ||
| External Link | RMBase: m6A_site_7187 | ||
| mod ID: M6ASITE075267 | Click to Show/Hide the Full List | ||
| mod site | chr1:11128046-11128047:- | [10] | |
| Sequence | AGCCAACAAGATTCTGAAGAACATGTGTGAGCACAGCAACA | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa; Jurkat | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000361445.8; ENST00000376838.5 | ||
| External Link | RMBase: m6A_site_7188 | ||
| mod ID: M6ASITE075306 | Click to Show/Hide the Full List | ||
| mod site | chr1:11128475-11128476:- | [10] | |
| Sequence | CATTCACCAGCTTCTCACAGACATTGGTCGGTACCACCCCC | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa; hESC-HEK293T | ||
| Seq Type List | m6A-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000361445.8; ENST00000376838.5 | ||
| External Link | RMBase: m6A_site_7189 | ||
| mod ID: M6ASITE075384 | Click to Show/Hide the Full List | ||
| mod site | chr1:11128513-11128514:- | [10] | |
| Sequence | GCAAGAATTGATACGCCCAGACCCTTGGTGGGACGTCTCAT | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000376838.5; ENST00000361445.8 | ||
| External Link | RMBase: m6A_site_7190 | ||
| mod ID: M6ASITE075385 | Click to Show/Hide the Full List | ||
| mod site | chr1:11129744-11129745:- | [12] | |
| Sequence | GAGGCAACAACCTCCAGGATACACTCAGGTATCAGAGAAGG | ||
| Motif Score | 2.110482143 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000376838.5; ENST00000361445.8 | ||
| External Link | RMBase: m6A_site_7191 | ||
| mod ID: M6ASITE075386 | Click to Show/Hide the Full List | ||
| mod site | chr1:11129758-11129759:- | [12] | |
| Sequence | CATCTCCTTGTCACGAGGCAACAACCTCCAGGATACACTCA | ||
| Motif Score | 2.173910714 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000376838.5; ENST00000361445.8 | ||
| External Link | RMBase: m6A_site_7192 | ||
| mod ID: M6ASITE075387 | Click to Show/Hide the Full List | ||
| mod site | chr1:11129812-11129813:- | [12] | |
| Sequence | GTCCAAAACCCTCCTGATGTACACGGTGCCTGCCGTCCAGG | ||
| Motif Score | 2.856142857 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000361445.8; ENST00000376838.5 | ||
| External Link | RMBase: m6A_site_7193 | ||
| mod ID: M6ASITE075388 | Click to Show/Hide the Full List | ||
| mod site | chr1:11129825-11129826:- | [10] | |
| Sequence | TCCAACAGGATCTGTCCAAAACCCTCCTGATGTACACGGTG | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | HeLa; HepG2; CD4T; HEK293T; HEC-1-A; MM6 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000376838.5; ENST00000361445.8 | ||
| External Link | RMBase: m6A_site_7194 | ||
| mod ID: M6ASITE075389 | Click to Show/Hide the Full List | ||
| mod site | chr1:11130568-11130569:- | [10] | |
| Sequence | CGAGGCCGAGAGCACCGAGAACAGCCCCACCCCATCGCCGC | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa; HepG2; MT4; Jurkat; CD4T; HEK293T; endometrial; HEC-1-A; NB4; MM6 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000376838.5; ENST00000361445.8 | ||
| External Link | RMBase: m6A_site_7195 | ||
| mod ID: M6ASITE075390 | Click to Show/Hide the Full List | ||
| mod site | chr1:11130670-11130671:- | [12] | |
| Sequence | GCGTCATGCCAGCGGGGCCAACATCACCAACGCCACCACTG | ||
| Motif Score | 2.173910714 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000361445.8; ENST00000376838.5 | ||
| External Link | RMBase: m6A_site_7196 | ||
| mod ID: M6ASITE075391 | Click to Show/Hide the Full List | ||
| mod site | chr1:11130693-11130694:- | [10] | |
| Sequence | GCCCGCGATGAGAAGAAGAAACTGCGTCATGCCAGCGGGGC | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa; MT4; Huh7; Jurkat; HEK293T; endometrial; MM6 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000361445.8; ENST00000376838.5 | ||
| External Link | RMBase: m6A_site_7197 | ||
| mod ID: M6ASITE075392 | Click to Show/Hide the Full List | ||
| mod site | chr1:11130718-11130719:- | [10] | |
| Sequence | GCTACACTACAAACATCAGAACCAAGCCCGCGATGAGAAGA | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | HeLa; MT4; Huh7; endometrial | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000361445.8; ENST00000376838.5 | ||
| External Link | RMBase: m6A_site_7198 | ||
| mod ID: M6ASITE075393 | Click to Show/Hide the Full List | ||
| mod site | chr1:11130726-11130727:- | [10] | |
| Sequence | GAAGCTGTGCTACACTACAAACATCAGAACCAAGCCCGCGA | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa; hESC-HEK293T; MT4; Huh7; endometrial | ||
| Seq Type List | m6A-seq; MAZTER-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000361445.8; ENST00000376838.5 | ||
| External Link | RMBase: m6A_site_7199 | ||
| mod ID: M6ASITE075394 | Click to Show/Hide the Full List | ||
| mod site | chr1:11130735-11130736:- | [12] | |
| Sequence | ATGAACTTCGAAGCTGTGCTACACTACAAACATCAGAACCA | ||
| Motif Score | 2.078666667 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000361445.8; ENST00000376838.5 | ||
| External Link | RMBase: m6A_site_7200 | ||
| mod ID: M6ASITE075395 | Click to Show/Hide the Full List | ||
| mod site | chr1:11130751-11130752:- | [10] | |
| Sequence | GCATGCGTGGGCAGTGATGAACTTCGAAGCTGTGCTACACT | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa; Huh7; endometrial | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000376838.5; ENST00000361445.8 | ||
| External Link | RMBase: m6A_site_7201 | ||
| mod ID: M6ASITE075396 | Click to Show/Hide the Full List | ||
| mod site | chr1:11130877-11130878:- | [10] | |
| Sequence | TTGCTTTATGGACACACAGAACAGGCAGGAGCAGCTGTTCC | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000376838.5; ENST00000361445.8 | ||
| External Link | RMBase: m6A_site_7202 | ||
| mod ID: M6ASITE075424 | Click to Show/Hide the Full List | ||
| mod site | chr1:11130886-11130887:- | [10] | |
| Sequence | TTGAAGTGTTTGCTTTATGGACACACAGAACAGGCAGGAGC | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000361445.8; ENST00000376838.5 | ||
| External Link | RMBase: m6A_site_7203 | ||
| mod ID: M6ASITE075455 | Click to Show/Hide the Full List | ||
| mod site | chr1:11131492-11131493:- | [10] | |
| Sequence | CTATCTGGTGACCTGTCAAAACTTTGCTCCCTTGAAATATT | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000361445.8; ENST00000376838.5 | ||
| External Link | RMBase: m6A_site_7204 | ||
| mod ID: M6ASITE075519 | Click to Show/Hide the Full List | ||
| mod site | chr1:11133083-11133084:- | [12] | |
| Sequence | AGAGCACGACCGCAGCTGGTACAAGGTGATCAGAAGCAGGC | ||
| Motif Score | 2.856142857 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000361445.8; ENST00000495435.1 | ||
| External Link | RMBase: m6A_site_7205 | ||
| mod ID: M6ASITE075523 | Click to Show/Hide the Full List | ||
| mod site | chr1:11133184-11133185:- | [10] | |
| Sequence | GCTTCCAGATGCTTCCTGAAACTTGGAGAGTGGCAGCTGAA | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa; Huh7; endometrial | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000495435.1; ENST00000361445.8 | ||
| External Link | RMBase: m6A_site_7206 | ||
| mod ID: M6ASITE075529 | Click to Show/Hide the Full List | ||
| mod site | chr1:11134365-11134366:- | [12] | |
| Sequence | GCAGCATAAGCAGGAACTGCACAAGCTCATGGCCCGGTGAG | ||
| Motif Score | 2.830589286 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000361445.8; ENST00000495435.1 | ||
| External Link | RMBase: m6A_site_7207 | ||
| mod ID: M6ASITE075530 | Click to Show/Hide the Full List | ||
| mod site | chr1:11134370-11134371:- | [10] | |
| Sequence | GACCAGCAGCATAAGCAGGAACTGCACAAGCTCATGGCCCG | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa; Huh7 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000361445.8; ENST00000495435.1 | ||
| External Link | RMBase: m6A_site_7208 | ||
| mod ID: M6ASITE075531 | Click to Show/Hide the Full List | ||
| mod site | chr1:11134389-11134390:- | [10] | |
| Sequence | GCATGCCATCGCTACTGAGGACCAGCAGCATAAGCAGGAAC | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; Huh7 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000361445.8; ENST00000495435.1 | ||
| External Link | RMBase: m6A_site_7209 | ||
| mod ID: M6ASITE075532 | Click to Show/Hide the Full List | ||
| mod site | chr1:11134429-11134430:- | [10] | |
| Sequence | ACATGCAGCATTTTGTCCAGACCATGCAGCAACAGGCCCAG | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa; Huh7 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000495435.1; ENST00000361445.8 | ||
| External Link | RMBase: m6A_site_7210 | ||
| mod ID: M6ASITE075533 | Click to Show/Hide the Full List | ||
| mod site | chr1:11138957-11138958:- | [10] | |
| Sequence | TTGTAAATACACAGACTAGAACTTTCACAAGACCAATTTGG | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa; Huh7 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000361445.8; ENST00000476768.1; ENST00000495435.1 | ||
| External Link | RMBase: m6A_site_7211 | ||
| mod ID: M6ASITE075534 | Click to Show/Hide the Full List | ||
| mod site | chr1:11138963-11138964:- | [10] | |
| Sequence | GCAGATTTGTAAATACACAGACTAGAACTTTCACAAGACCA | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa; Huh7 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000495435.1; ENST00000361445.8; ENST00000476768.1 | ||
| External Link | RMBase: m6A_site_7212 | ||
| mod ID: M6ASITE075535 | Click to Show/Hide the Full List | ||
| mod site | chr1:11139003-11139004:- | [10] | |
| Sequence | CTACCCCTAAGAGAAGATGGACTGTTTGGTACAAAAAAGAG | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000476768.1; ENST00000361445.8; ENST00000495435.1 | ||
| External Link | RMBase: m6A_site_7213 | ||
| mod ID: M6ASITE075536 | Click to Show/Hide the Full List | ||
| mod site | chr1:11139043-11139044:- | [10] | |
| Sequence | TGTATCTAGAAAGTGCTGGGACATAGAAAGGGCACACTTTC | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000361445.8; ENST00000495435.1; ENST00000476768.1 | ||
| External Link | RMBase: m6A_site_7214 | ||
| mod ID: M6ASITE075537 | Click to Show/Hide the Full List | ||
| mod site | chr1:11139094-11139095:- | [10] | |
| Sequence | GTGTGTATACCATTGATCAAACCAATGAATGTGATCACAGT | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000361445.8; ENST00000476768.1; ENST00000495435.1 | ||
| External Link | RMBase: m6A_site_7215 | ||
| mod ID: M6ASITE075538 | Click to Show/Hide the Full List | ||
| mod site | chr1:11139362-11139363:- | [13] | |
| Sequence | AACTTGACCATCCTCTGCCAACAGTTCACCCTCAGGTGACC | ||
| Motif Score | 2.173910714 | ||
| Cell/Tissue List | kidney | ||
| Seq Type List | m6A-REF-seq | ||
| Transcript ID List | ENST00000476768.1; ENST00000361445.8; ENST00000495435.1 | ||
| External Link | RMBase: m6A_site_7216 | ||
| mod ID: M6ASITE075539 | Click to Show/Hide the Full List | ||
| mod site | chr1:11139419-11139420:- | [10] | |
| Sequence | TTAAGGCTCTTGCTCATAAAACTTTAGTGTTGCTCCTGGGA | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000476768.1; ENST00000361445.8; ENST00000495435.1 | ||
| External Link | RMBase: m6A_site_7217 | ||
| mod ID: M6ASITE075540 | Click to Show/Hide the Full List | ||
| mod site | chr1:11139576-11139577:- | [10] | |
| Sequence | GCCCTCATGAAGACATGAGAACCTGGCTCAAGTATGCAAGC | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000476768.1; ENST00000495435.1; ENST00000361445.8 | ||
| External Link | RMBase: m6A_site_7218 | ||
| mod ID: M6ASITE075541 | Click to Show/Hide the Full List | ||
| mod site | chr1:11139584-11139585:- | [10] | |
| Sequence | TGTGGTCAGCCCTCATGAAGACATGAGAACCTGGCTCAAGT | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa; hESC-HEK293T | ||
| Seq Type List | m6A-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000476768.1; ENST00000495435.1; ENST00000361445.8 | ||
| External Link | RMBase: m6A_site_7219 | ||
| mod ID: M6ASITE075546 | Click to Show/Hide the Full List | ||
| mod site | chr1:11139635-11139636:- | [10] | |
| Sequence | CTGCCAGCGTATCGTAGAGGACTGGCAGAAAATCCTTATGG | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000361445.8; ENST00000495435.1; ENST00000476768.1 | ||
| External Link | RMBase: m6A_site_7220 | ||
| mod ID: M6ASITE075581 | Click to Show/Hide the Full List | ||
| mod site | chr1:11144653-11144654:- | [10] | |
| Sequence | CGCCAGATCTGGTGGGAGAGACTGCAGGTGAGCTTGGTAGA | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000361445.8; ENST00000495435.1 | ||
| External Link | RMBase: m6A_site_7221 | ||
| mod ID: M6ASITE075582 | Click to Show/Hide the Full List | ||
| mod site | chr1:11144701-11144702:- | [10] | |
| Sequence | GAGGAGGTTATCCAGTACAAACTTGTCCCCGAGCGACGAGA | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000361445.8; ENST00000495435.1 | ||
| External Link | RMBase: m6A_site_7223 | ||
| mod ID: M6ASITE075583 | Click to Show/Hide the Full List | ||
| mod site | chr1:11144705-11144706:- | [12] | |
| Sequence | GCTGGAGGAGGTTATCCAGTACAAACTTGTCCCCGAGCGAC | ||
| Motif Score | 2.856142857 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000361445.8; ENST00000495435.1 | ||
| External Link | RMBase: m6A_site_7224 | ||
| mod ID: M6ASITE075584 | Click to Show/Hide the Full List | ||
| mod site | chr1:11145025-11145026:- | [10] | |
| Sequence | GTGCATTGACAAGGCCAGGGACCTGCTGGATGCTGAATTAA | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000361445.8 | ||
| External Link | RMBase: m6A_site_7231 | ||
| mod ID: M6ASITE075585 | Click to Show/Hide the Full List | ||
| mod site | chr1:11145037-11145038:- | [12] | |
| Sequence | GTTTCTCTTCTAGTGCATTGACAAGGCCAGGGACCTGCTGG | ||
| Motif Score | 2.859755952 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000361445.8 | ||
| External Link | RMBase: m6A_site_7232 | ||
| mod ID: M6ASITE075586 | Click to Show/Hide the Full List | ||
| mod site | chr1:11146681-11146682:- | [12] | |
| Sequence | CAGGACCTCTTCTCCTTGGCACAACAGGTAAAAACCTACAT | ||
| Motif Score | 2.830589286 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000361445.8 | ||
| External Link | RMBase: m6A_site_7233 | ||
| mod ID: M6ASITE075591 | Click to Show/Hide the Full List | ||
| mod site | chr1:11146697-11146698:- | [10] | |
| Sequence | TGTGCTGGCACTGCATCAGGACCTCTTCTCCTTGGCACAAC | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000361445.8 | ||
| External Link | RMBase: m6A_site_7234 | ||
| mod ID: M6ASITE075692 | Click to Show/Hide the Full List | ||
| mod site | chr1:11146745-11146746:- | [10] | |
| Sequence | CACCTGTATGATCCCTCGGGACACCCATGATGGGGCATTTT | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; hESC-HEK293T | ||
| Seq Type List | m6A-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000361445.8 | ||
| External Link | RMBase: m6A_site_7235 | ||
| mod ID: M6ASITE075792 | Click to Show/Hide the Full List | ||
| mod site | chr1:11146781-11146782:- | [10] | |
| Sequence | TGTGTGTATAGGTCAGTGGGACAGCATGGAAGAATACACCT | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000361445.8 | ||
| External Link | RMBase: m6A_site_7236 | ||
| mod ID: M6ASITE075799 | Click to Show/Hide the Full List | ||
| mod site | chr1:11150173-11150174:- | [10] | |
| Sequence | GGACCCTGGTTAATGATGAGACCCAAGCCAAGATGGCCCGG | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000361445.8 | ||
| External Link | RMBase: m6A_site_7237 | ||
| mod ID: M6ASITE075807 | Click to Show/Hide the Full List | ||
| mod site | chr1:11150191-11150192:- | [10] | |
| Sequence | AGCAGTGCTGTGAAAAGTGGACCCTGGTTAATGATGAGACC | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; Jurkat | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000361445.8 | ||
| External Link | RMBase: m6A_site_7238 | ||
| mod ID: M6ASITE075818 | Click to Show/Hide the Full List | ||
| mod site | chr1:11157211-11157212:- | [12] | |
| Sequence | TGACAAGAAAATGGACACCAACAAGGACGACCCAGAGCTGA | ||
| Motif Score | 2.173910714 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000361445.8 | ||
| External Link | RMBase: m6A_site_7239 | ||
| mod ID: M6ASITE075829 | Click to Show/Hide the Full List | ||
| mod site | chr1:11157217-11157218:- | [10] | |
| Sequence | GGCCTATGACAAGAAAATGGACACCAACAAGGACGACCCAG | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000361445.8 | ||
| External Link | RMBase: m6A_site_7240 | ||
| mod ID: M6ASITE075840 | Click to Show/Hide the Full List | ||
| mod site | chr1:11157229-11157230:- | [12] | |
| Sequence | GGATGCCCTTGTGGCCTATGACAAGAAAATGGACACCAACA | ||
| Motif Score | 2.859755952 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000361445.8 | ||
| External Link | RMBase: m6A_site_7241 | ||
| mod ID: M6ASITE075851 | Click to Show/Hide the Full List | ||
| mod site | chr1:11157264-11157265:- | [10] | |
| Sequence | CAGGCTACCTGGTATGAGAAACTGCACGAGTGGGAGGATGC | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000361445.8 | ||
| External Link | RMBase: m6A_site_7242 | ||
| mod ID: M6ASITE075862 | Click to Show/Hide the Full List | ||
| mod site | chr1:11167456-11167457:- | [10] | |
| Sequence | GTGTTAGAATATGCCATGAAACACTTTGGAGAGCTGGTAAG | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa; hESC-HEK293T | ||
| Seq Type List | m6A-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000361445.8 | ||
| External Link | RMBase: m6A_site_7243 | ||
| mod ID: M6ASITE075873 | Click to Show/Hide the Full List | ||
| mod site | chr1:11199307-11199308:- | [10] | |
| Sequence | AAAGCACTACACTACAAAGAACTGGAGTTCCAGAAAGGCCC | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000361445.8 | ||
| External Link | RMBase: m6A_site_7244 | ||
| mod ID: M6ASITE075876 | Click to Show/Hide the Full List | ||
| mod site | chr1:11199314-11199315:- | [12] | |
| Sequence | ATATGCCAAAGCACTACACTACAAAGAACTGGAGTTCCAGA | ||
| Motif Score | 2.078666667 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000361445.8 | ||
| External Link | RMBase: m6A_site_7245 | ||
| mod ID: M6ASITE075877 | Click to Show/Hide the Full List | ||
| mod site | chr1:11199319-11199320:- | [12] | |
| Sequence | CGAGCATATGCCAAAGCACTACACTACAAAGAACTGGAGTT | ||
| Motif Score | 2.078666667 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000361445.8 | ||
| External Link | RMBase: m6A_site_7246 | ||
| mod ID: M6ASITE075878 | Click to Show/Hide the Full List | ||
| mod site | chr1:11199380-11199381:- | [12] | |
| Sequence | CCCCCTGCCACTGAGAGATGACAATGGCATTGTTCTGCTGG | ||
| Motif Score | 2.859755952 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000361445.8 | ||
| External Link | RMBase: m6A_site_7247 | ||
| mod ID: M6ASITE075879 | Click to Show/Hide the Full List | ||
| mod site | chr1:11199552-11199553:- | [10] | |
| Sequence | AACTTGGCTGAATTCATGGAACACAGTGACAAGGTGAGACT | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000361445.8 | ||
| External Link | RMBase: m6A_site_7248 | ||
| mod ID: M6ASITE075880 | Click to Show/Hide the Full List | ||
| mod site | chr1:11199571-11199572:- | [10] | |
| Sequence | AGTCACACAGACCCTCTTAAACTTGGCTGAATTCATGGAAC | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000361445.8 | ||
| External Link | RMBase: m6A_site_7249 | ||
| mod ID: M6ASITE075881 | Click to Show/Hide the Full List | ||
| mod site | chr1:11199581-11199582:- | [10] | |
| Sequence | ACATCGCTGAAGTCACACAGACCCTCTTAAACTTGGCTGAA | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000361445.8 | ||
| External Link | RMBase: m6A_site_7250 | ||
| mod ID: M6ASITE075937 | Click to Show/Hide the Full List | ||
| mod site | chr1:11199601-11199602:- | [10] | |
| Sequence | GTTGGCCCTCACCTCACAAGACATCGCTGAAGTCACACAGA | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000361445.8 | ||
| External Link | RMBase: m6A_site_7251 | ||
| mod ID: M6ASITE075943 | Click to Show/Hide the Full List | ||
| mod site | chr1:11199606-11199607:- | [12] | |
| Sequence | ATCGAGTTGGCCCTCACCTCACAAGACATCGCTGAAGTCAC | ||
| Motif Score | 2.047297619 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000361445.8 | ||
| External Link | RMBase: m6A_site_7252 | ||
| mod ID: M6ASITE075944 | Click to Show/Hide the Full List | ||
| mod site | chr1:11199663-11199664:- | [10] | |
| Sequence | TTTGTGTCCTGCTGGTCTGAACTGAATGAAGATCAACAGGA | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000361445.8 | ||
| External Link | RMBase: m6A_site_7253 | ||
| mod ID: M6ASITE075945 | Click to Show/Hide the Full List | ||
| mod site | chr1:11204575-11204576:- | [12] | |
| Sequence | CTGGGCCCTGGCACAGGCCTACAACCCGATGGCCAGGTATA | ||
| Motif Score | 2.078666667 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000361445.8 | ||
| External Link | RMBase: m6A_site_7254 | ||
| mod ID: M6ASITE075959 | Click to Show/Hide the Full List | ||
| mod site | chr1:11204623-11204624:- | [10] | |
| Sequence | GAGCCTGGAGCTGCTGAAGGACTCATCATCGCCCTCCCTGC | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa; endometrial | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000361445.8 | ||
| External Link | RMBase: m6A_site_7255 | ||
| mod ID: M6ASITE076060 | Click to Show/Hide the Full List | ||
| mod site | chr1:11209341-11209342:- | [10] | |
| Sequence | GAAACAGGACCCATGAAGAAACTGCACGTCAGCACCATCAA | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000361445.8 | ||
| External Link | RMBase: m6A_site_7256 | ||
| mod ID: M6ASITE076118 | Click to Show/Hide the Full List | ||
| mod site | chr1:11209353-11209354:- | [10] | |
| Sequence | AGTGGACCAGTGGAAACAGGACCCATGAAGAAACTGCACGT | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000361445.8 | ||
| External Link | RMBase: m6A_site_7257 | ||
| mod ID: M6ASITE076119 | Click to Show/Hide the Full List | ||
| mod site | chr1:11209358-11209359:- | [10] | |
| Sequence | TGGCTAGTGGACCAGTGGAAACAGGACCCATGAAGAAACTG | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000361445.8 | ||
| External Link | RMBase: m6A_site_7258 | ||
| mod ID: M6ASITE076120 | Click to Show/Hide the Full List | ||
| mod site | chr1:11209368-11209369:- | [10] | |
| Sequence | GGGGATGCATTGGCTAGTGGACCAGTGGAAACAGGACCCAT | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000361445.8 | ||
| External Link | RMBase: m6A_site_7259 | ||
| mod ID: M6ASITE076121 | Click to Show/Hide the Full List | ||
| mod site | chr1:11209453-11209454:- | [12] | |
| Sequence | AAATCTTTGGTTGTAGGGATACACACTTGCTGATGAAGAGG | ||
| Motif Score | 2.110482143 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000361445.8 | ||
| External Link | RMBase: m6A_site_7260 | ||
| mod ID: M6ASITE076122 | Click to Show/Hide the Full List | ||
| mod site | chr1:11210864-11210865:- | [12] | |
| Sequence | GTGAATAAAGTTCTGGTGCGACACCGAATCAATCATCAGCG | ||
| Motif Score | 2.865571429 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000361445.8 | ||
| External Link | RMBase: m6A_site_7261 | ||
| mod ID: M6ASITE076123 | Click to Show/Hide the Full List | ||
| mod site | chr1:11212348-11212349:- | [10] | |
| Sequence | ACTGCGCTCCACAGCCATGGACACGCTGTCTTCACTTGTTT | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; hESC-HEK293T | ||
| Seq Type List | m6A-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000361445.8 | ||
| External Link | RMBase: m6A_site_7262 | ||
| mod ID: M6ASITE076124 | Click to Show/Hide the Full List | ||
| mod site | chr1:11212368-11212369:- | [10] | |
| Sequence | ACACTGGACCAGAGCCCAGAACTGCGCTCCACAGCCATGGA | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa; Huh7 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000361445.8 | ||
| External Link | RMBase: m6A_site_7263 | ||
| mod ID: M6ASITE076125 | Click to Show/Hide the Full List | ||
| mod site | chr1:11212381-11212382:- | [10] | |
| Sequence | CCCTATTGTTCGAACACTGGACCAGAGCCCAGAACTGCGCT | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; Huh7 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000361445.8 | ||
| External Link | RMBase: m6A_site_7264 | ||
| mod ID: M6ASITE076126 | Click to Show/Hide the Full List | ||
| mod site | chr1:11212388-11212389:- | [10] | |
| Sequence | TCATTCACCCTATTGTTCGAACACTGGACCAGAGCCCAGAA | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa; Huh7 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000361445.8 | ||
| External Link | RMBase: m6A_site_7265 | ||
| mod ID: M6ASITE076127 | Click to Show/Hide the Full List | ||
| mod site | chr1:11212453-11212454:- | [10] | |
| Sequence | GGCAGCGCTAGAGACTGTGGACCGCCTGACGGAGTCCCTGG | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; Huh7 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000361445.8 | ||
| External Link | RMBase: m6A_site_7266 | ||
| mod ID: M6ASITE076128 | Click to Show/Hide the Full List | ||
| mod site | chr1:11212460-11212461:- | [10] | |
| Sequence | GTTGTAGGGCAGCGCTAGAGACTGTGGACCGCCTGACGGAG | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa; Huh7 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000361445.8 | ||
| External Link | RMBase: m6A_site_7267 | ||
| mod ID: M6ASITE076129 | Click to Show/Hide the Full List | ||
| mod site | chr1:11213455-11213456:- | [12] | |
| Sequence | TACCTGCCCCAGCTGATCCCACACATGCTGCGTGTCTTCAT | ||
| Motif Score | 2.053113095 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000361445.8 | ||
| External Link | RMBase: m6A_site_7268 | ||
| mod ID: M6ASITE076130 | Click to Show/Hide the Full List | ||
| mod site | chr1:11213549-11213550:- | [10] | |
| Sequence | GTAGGAATTCTGGGTCATGAACACCTCAATTCAGAGCACGA | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa; hESC-HEK293T | ||
| Seq Type List | m6A-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000361445.8 | ||
| External Link | RMBase: m6A_site_7269 | ||
| mod ID: M6ASITE076131 | Click to Show/Hide the Full List | ||
| mod site | chr1:11216180-11216181:- | [10] | |
| Sequence | TTTGTGAAGAGCCACATCAGACCTTATATGGATGAAATAGT | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000361445.8 | ||
| External Link | RMBase: m6A_site_7270 | ||
| mod ID: M6ASITE076132 | Click to Show/Hide the Full List | ||
| mod site | chr1:11216187-11216188:- | [12] | |
| Sequence | GGTGTCCTTTGTGAAGAGCCACATCAGACCTTATATGGATG | ||
| Motif Score | 2.053113095 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000361445.8 | ||
| External Link | RMBase: m6A_site_7271 | ||
| mod ID: M6ASITE076133 | Click to Show/Hide the Full List | ||
| mod site | chr1:11228748-11228749:- | [10] | |
| Sequence | TTCATCTTCAAGTCCCTGGGACTCAAATGTGTGCAGTTCCT | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000361445.8 | ||
| External Link | RMBase: m6A_site_7272 | ||
| mod ID: M6ASITE076134 | Click to Show/Hide the Full List | ||
| mod site | chr1:11228794-11228795:- | [12] | |
| Sequence | CCAGTCACTCTCTCATCATCACACCATGGTTGTCCAGGCCA | ||
| Motif Score | 2.047297619 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000361445.8 | ||
| External Link | RMBase: m6A_site_7273 | ||
| mod ID: M6ASITE076135 | Click to Show/Hide the Full List | ||
| mod site | chr1:11228815-11228816:- | [10] | |
| Sequence | CCTGATGCGGATCTTCCGAGACCAGTCACTCTCTCATCATC | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa; Huh7 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000361445.8 | ||
| External Link | RMBase: m6A_site_7274 | ||
| mod ID: M6ASITE076136 | Click to Show/Hide the Full List | ||
| mod site | chr1:11228878-11228879:- | [10] | |
| Sequence | AATGCTGGTCAACATGGGAAACTTGCCTCTGGATGAGTTCT | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000361445.8 | ||
| External Link | RMBase: m6A_site_7275 | ||
| mod ID: M6ASITE076137 | Click to Show/Hide the Full List | ||
| mod site | chr1:11228887-11228888:- | [12] | |
| Sequence | CACTAGTGAAATGCTGGTCAACATGGGAAACTTGCCTCTGG | ||
| Motif Score | 2.173910714 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000361445.8 | ||
| External Link | RMBase: m6A_site_7276 | ||
| mod ID: M6ASITE076147 | Click to Show/Hide the Full List | ||
| mod site | chr1:11230983-11230984:- | [10] | |
| Sequence | AGTGAACATTGGCATGATAGACCAGTCCCGGGATGCCTCTG | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000361445.8 | ||
| External Link | RMBase: m6A_site_7277 | ||
| mod ID: M6ASITE076158 | Click to Show/Hide the Full List | ||
| mod site | chr1:11230998-11230999:- | [10] | |
| Sequence | TCCTTACAAGCACAAAGTGAACATTGGCATGATAGACCAGT | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa; hESC-HEK293T | ||
| Seq Type List | m6A-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000361445.8 | ||
| External Link | RMBase: m6A_site_7278 | ||
| mod ID: M6ASITE076169 | Click to Show/Hide the Full List | ||
| mod site | chr1:11231013-11231014:- | [12] | |
| Sequence | TTTAGGGGCTTTGGATCCTTACAAGCACAAAGTGAACATTG | ||
| Motif Score | 2.07285119 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000361445.8 | ||
| External Link | RMBase: m6A_site_7279 | ||
| mod ID: M6ASITE076180 | Click to Show/Hide the Full List | ||
| mod site | chr1:11231310-11231311:- | [12] | |
| Sequence | AGACTGAGCAGAACCAGGGTACACGCAGAGAGGTAGGGGAC | ||
| Motif Score | 2.856142857 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000361445.8 | ||
| External Link | RMBase: m6A_site_7280 | ||
| mod ID: M6ASITE076191 | Click to Show/Hide the Full List | ||
| mod site | chr1:11231318-11231319:- | [10] | |
| Sequence | TTTTCTGAAGACTGAGCAGAACCAGGGTACACGCAGAGAGG | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000361445.8 | ||
| External Link | RMBase: m6A_site_7281 | ||
| mod ID: M6ASITE076202 | Click to Show/Hide the Full List | ||
| mod site | chr1:11231328-11231329:- | [10] | |
| Sequence | TGCTACTGAATTTTCTGAAGACTGAGCAGAACCAGGGTACA | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000361445.8 | ||
| External Link | RMBase: m6A_site_7282 | ||
| mod ID: M6ASITE076213 | Click to Show/Hide the Full List | ||
| mod site | chr1:11231375-11231376:- | [13] | |
| Sequence | TGGCTATGTAGTAGAGCCCTACAGGAAGTACCCTACTTTGC | ||
| Motif Score | 2.078666667 | ||
| Cell/Tissue List | HEK293 | ||
| Seq Type List | m6A-REF-seq | ||
| Transcript ID List | ENST00000361445.8 | ||
| External Link | RMBase: m6A_site_7283 | ||
| mod ID: M6ASITE076224 | Click to Show/Hide the Full List | ||
| mod site | chr1:11231413-11231414:- | [10] | |
| Sequence | GTGGCTCTGTGGACCCTGGGACAGTTGGTGGCCAGCACTGG | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000361445.8 | ||
| External Link | RMBase: m6A_site_7284 | ||
| mod ID: M6ASITE076235 | Click to Show/Hide the Full List | ||
| mod site | chr1:11231421-11231422:- | [10] | |
| Sequence | CTATCCAGGTGGCTCTGTGGACCCTGGGACAGTTGGTGGCC | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000361445.8 | ||
| External Link | RMBase: m6A_site_7285 | ||
| mod ID: M6ASITE076258 | Click to Show/Hide the Full List | ||
| mod site | chr1:11232472-11232473:- | [10] | |
| Sequence | ACTTTTTATTATCATCATGGACATGCTCCAGGATTCCTCTT | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; hESC-HEK293T | ||
| Seq Type List | m6A-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000361445.8 | ||
| External Link | RMBase: m6A_site_7286 | ||
| mod ID: M6ASITE076323 | Click to Show/Hide the Full List | ||
| mod site | chr1:11232492-11232493:- | [10] | |
| Sequence | ATGAGGAAATGGGTTGATGAACTTTTTATTATCATCATGGA | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000361445.8 | ||
| External Link | RMBase: m6A_site_7287 | ||
| mod ID: M6ASITE076345 | Click to Show/Hide the Full List | ||
| mod site | chr1:11233417-11233418:- | [12] | |
| Sequence | TGATCAATAATGTCCTGGCAACAATAGGAGAATTGGCACAG | ||
| Motif Score | 2.173910714 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000361445.8 | ||
| External Link | RMBase: m6A_site_7288 | ||
| mod ID: M6ASITE076381 | Click to Show/Hide the Full List | ||
| mod site | chr1:11233446-11233447:- | [10] | |
| Sequence | AGATCCAGACCCTGATCCAAACCCAGGTGTGATCAATAATG | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000361445.8 | ||
| External Link | RMBase: m6A_site_7289 | ||
| mod ID: M6ASITE076423 | Click to Show/Hide the Full List | ||
| mod site | chr1:11233458-11233459:- | [10] | |
| Sequence | TTTGAAACTGAAAGATCCAGACCCTGATCCAAACCCAGGTG | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000361445.8 | ||
| External Link | RMBase: m6A_site_7290 | ||
| mod ID: M6ASITE076438 | Click to Show/Hide the Full List | ||
| mod site | chr1:11233472-11233473:- | [10] | |
| Sequence | CTCTAGGCATTAATTTTGAAACTGAAAGATCCAGACCCTGA | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000361445.8 | ||
| External Link | RMBase: m6A_site_7291 | ||
| mod ID: M6ASITE076468 | Click to Show/Hide the Full List | ||
| mod site | chr1:11234161-11234162:- | [12] | |
| Sequence | CCCCCGACTCATCCGCCCCTACATGGAGCCTATTCTGAAGG | ||
| Motif Score | 2.078666667 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000361445.8 | ||
| External Link | RMBase: m6A_site_7292 | ||
| mod ID: M6ASITE076477 | Click to Show/Hide the Full List | ||
| mod site | chr1:11237885-11237886:- | [10] | |
| Sequence | GGGCCGACTCAGTAGCATGAACCCTGCCTTTGTCATGCCTT | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000361445.8 | ||
| External Link | RMBase: m6A_site_7293 | ||
| mod ID: M6ASITE076478 | Click to Show/Hide the Full List | ||
| mod site | chr1:11237975-11237976:- | [10] | |
| Sequence | ACACCTGGCCCAGGCGGAGAACTTGCAGGCCTTGTTTGTGG | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000361445.8 | ||
| External Link | RMBase: m6A_site_7294 | ||
| mod ID: M6ASITE076479 | Click to Show/Hide the Full List | ||
| mod site | chr1:11237995-11237996:- | [12] | |
| Sequence | CTGGACGAGCGCTTTGATGCACACCTGGCCCAGGCGGAGAA | ||
| Motif Score | 2.830589286 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000361445.8 | ||
| External Link | RMBase: m6A_site_7295 | ||
| mod ID: M6ASITE076506 | Click to Show/Hide the Full List | ||
| mod site | chr1:11238041-11238042:- | [12] | |
| Sequence | CATTGCTCTTGCAGACCCTGACATTCGCTACTGTGTCTTGG | ||
| Motif Score | 2.859755952 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000361445.8 | ||
| External Link | RMBase: m6A_site_7296 | ||
| mod ID: M6ASITE076591 | Click to Show/Hide the Full List | ||
| mod site | chr1:11238429-11238430:- | [10] | |
| Sequence | GTGGCAGATGTGCTTAGCAAACTGCTCGTAGTTGGGATAAC | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000361445.8 | ||
| External Link | RMBase: m6A_site_7297 | ||
| mod ID: M6ASITE076633 | Click to Show/Hide the Full List | ||
| mod site | chr1:11238464-11238465:- | [10] | |
| Sequence | ATGCTCATGTGGTTAGCCAGACCGCAGTGCAAGTGGTGGCA | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000361445.8 | ||
| External Link | RMBase: m6A_site_7298 | ||
| mod ID: M6ASITE076634 | Click to Show/Hide the Full List | ||
| mod site | chr1:11238512-11238513:- | [12] | |
| Sequence | GCACCTGCTCCCGCCTGCTCACACCCTCCATCCACCTCATC | ||
| Motif Score | 2.047297619 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000361445.8 | ||
| External Link | RMBase: m6A_site_7299 | ||
| mod ID: M6ASITE076635 | Click to Show/Hide the Full List | ||
| mod site | chr1:11238559-11238560:- | [13] | |
| Sequence | TCATTTCCTGAACAGTGAGCACAAGGAGATCCGCATGGAGG | ||
| Motif Score | 2.830589286 | ||
| Cell/Tissue List | brain; hESC-HEK293T | ||
| Seq Type List | m6A-REF-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000361445.8 | ||
| External Link | RMBase: m6A_site_7300 | ||
| mod ID: M6ASITE076636 | Click to Show/Hide the Full List | ||
| mod site | chr1:11238568-11238569:- | [10] | |
| Sequence | CTGTGCGGATCATTTCCTGAACAGTGAGCACAAGGAGATCC | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa; brain | ||
| Seq Type List | m6A-seq; m6A-REF-seq | ||
| Transcript ID List | ENST00000361445.8 | ||
| External Link | RMBase: m6A_site_7301 | ||
| mod ID: M6ASITE076637 | Click to Show/Hide the Full List | ||
| mod site | chr1:11240438-11240439:- | [10] | |
| Sequence | TCCCTGGTCCTTATGCACAAACCCCTTCGCCACCCAGGCAT | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000361445.8 | ||
| External Link | RMBase: m6A_site_7302 | ||
| mod ID: M6ASITE076638 | Click to Show/Hide the Full List | ||
| mod site | chr1:11240442-11240443:- | [12] | |
| Sequence | GCTGTCCCTGGTCCTTATGCACAAACCCCTTCGCCACCCAG | ||
| Motif Score | 2.830589286 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000361445.8 | ||
| External Link | RMBase: m6A_site_7303 | ||
| mod ID: M6ASITE076707 | Click to Show/Hide the Full List | ||
| mod site | chr1:11240487-11240488:- | [10] | |
| Sequence | GATTCCACAGCTAAAGAAGGACATTCAAGATGGGCTACTGA | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; hESC-HEK293T; endometrial | ||
| Seq Type List | m6A-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000361445.8 | ||
| External Link | RMBase: m6A_site_7304 | ||
| mod ID: M6ASITE076708 | Click to Show/Hide the Full List | ||
| mod site | chr1:11241557-11241558:- | [10] | |
| Sequence | GAGCCCATGCTGGCAGTGGGACTAAGGTGGGTGTCAGAAAC | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa; endometrial | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000361445.8 | ||
| External Link | RMBase: m6A_site_7305 | ||
| mod ID: M6ASITE076709 | Click to Show/Hide the Full List | ||
| mod site | chr1:11243125-11243126:- | [10] | |
| Sequence | AGCGGCCCTGCCCCCAAAGGACTTCGCCCATAAGTAAGCAC | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000361445.8 | ||
| External Link | RMBase: m6A_site_7306 | ||
| mod ID: M6ASITE076710 | Click to Show/Hide the Full List | ||
| mod site | chr1:11243155-11243156:- | [10] | |
| Sequence | CTATTTGCCTCGCGTGCTGGACATCATCCGAGCGGCCCTGC | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; hESC-HEK293T | ||
| Seq Type List | m6A-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000361445.8 | ||
| External Link | RMBase: m6A_site_7307 | ||
| mod ID: M6ASITE076711 | Click to Show/Hide the Full List | ||
| mod site | chr1:11243272-11243273:- | [10] | |
| Sequence | GTATCTCCAAGATACCATGAACCATGTCCTAAGCTGTGTCA | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000361445.8 | ||
| External Link | RMBase: m6A_site_7308 | ||
| mod ID: M6ASITE076720 | Click to Show/Hide the Full List | ||
| mod site | chr1:11247678-11247679:- | [11] | |
| Sequence | AGAACTCGCTGATCCAAATGACAATCCTTAATTTGTTGCCC | ||
| Motif Score | 2.859755952 | ||
| Cell/Tissue List | HEK293T; hESC-HEK293T | ||
| Seq Type List | DART-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000361445.8 | ||
| External Link | RMBase: m6A_site_7309 | ||
| mod ID: M6ASITE076784 | Click to Show/Hide the Full List | ||
| mod site | chr1:11247695-11247696:- | [10] | |
| Sequence | GAAATGCAGGAATAGCAAGAACTCGCTGATCCAAATGACAA | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000361445.8 | ||
| External Link | RMBase: m6A_site_7310 | ||
| mod ID: M6ASITE076817 | Click to Show/Hide the Full List | ||
| mod site | chr1:11247843-11247844:- | [10] | |
| Sequence | GGAGAGCCGGTGTTGCAGAGACTTGATGGAGGAGAAATTTG | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000361445.8 | ||
| External Link | RMBase: m6A_site_7311 | ||
| mod ID: M6ASITE076818 | Click to Show/Hide the Full List | ||
| mod site | chr1:11247895-11247896:- | [10] | |
| Sequence | AAGGCCTCATGGGATTTGGGACCTCCCCCAGTCCAGCTAAG | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000361445.8 | ||
| External Link | RMBase: m6A_site_7312 | ||
| mod ID: M6ASITE076860 | Click to Show/Hide the Full List | ||
| mod site | chr1:11248007-11248008:- | [10] | |
| Sequence | CTCATGGGCTTCGGAACAAAACCTCGTCACATTACCCCCTT | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | HeLa; Huh7 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000361445.8 | ||
| External Link | RMBase: m6A_site_7313 | ||
| mod ID: M6ASITE076884 | Click to Show/Hide the Full List | ||
| mod site | chr1:11248012-11248013:- | [10] | |
| Sequence | AAGATCTCATGGGCTTCGGAACAAAACCTCGTCACATTACC | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa; Huh7 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000361445.8 | ||
| External Link | RMBase: m6A_site_7314 | ||
| mod ID: M6ASITE076885 | Click to Show/Hide the Full List | ||
| mod site | chr1:11248066-11248067:- | [12] | |
| Sequence | GAGAAGAAATGGAAGAAATCACACAGCAGCAGCTGGTACAC | ||
| Motif Score | 2.047297619 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000361445.8 | ||
| External Link | RMBase: m6A_site_7315 | ||
| mod ID: M6ASITE076886 | Click to Show/Hide the Full List | ||
| mod site | chr1:11253936-11253937:- | [10] | |
| Sequence | CAGAGAAGGGATTTGATGAGACCTTGGCCAAAGAGAAGGGC | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa; CD34; Huh7 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000361445.8 | ||
| External Link | RMBase: m6A_site_7316 | ||
| mod ID: M6ASITE076887 | Click to Show/Hide the Full List | ||
| mod site | chr1:11253971-11253972:- | [12] | |
| Sequence | ACCCTGTCTCTCTACATAGCACACATTTGAAGAAGCAGAGA | ||
| Motif Score | 2.830589286 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000361445.8 | ||
| External Link | RMBase: m6A_site_7317 | ||
| mod ID: M6ASITE076888 | Click to Show/Hide the Full List | ||
| mod site | chr1:11256038-11256039:- | [12] | |
| Sequence | TTCGTGCCTGTCTGATTCTCACAACCCAGCGTGAGCCGAAG | ||
| Motif Score | 2.047297619 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000361445.8 | ||
| External Link | RMBase: m6A_site_7318 | ||
| mod ID: M6ASITE076889 | Click to Show/Hide the Full List | ||
| mod site | chr1:11256090-11256091:- | [10] | |
| Sequence | GTGGCCGTGTGGGACCCCAAACAGGCCATCCGTGAGGGAGC | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa; CD34 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000361445.8 | ||
| External Link | RMBase: m6A_site_7319 | ||
| mod ID: M6ASITE076890 | Click to Show/Hide the Full List | ||
| mod site | chr1:11256097-11256098:- | [10] | |
| Sequence | CATTTTTGTGGCCGTGTGGGACCCCAAACAGGCCATCCGTG | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; CD34 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000361445.8 | ||
| External Link | RMBase: m6A_site_7320 | ||
| mod ID: M6ASITE076891 | Click to Show/Hide the Full List | ||
| mod site | chr1:11256121-11256122:- | [12] | |
| Sequence | GCAAGTGCAACCCTTCTTTGACAACATTTTTGTGGCCGTGT | ||
| Motif Score | 2.859755952 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000361445.8 | ||
| External Link | RMBase: m6A_site_7321 | ||
| mod ID: M6ASITE076892 | Click to Show/Hide the Full List | ||
| mod site | chr1:11256941-11256942:- | [10] | |
| Sequence | GACCGCAATGAGGGCCGGAGACATGCAGCTGTGAGTGTCTA | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa; hESC-HEK293T | ||
| Seq Type List | m6A-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000361445.8 | ||
| External Link | RMBase: m6A_site_7322 | ||
| mod ID: M6ASITE076893 | Click to Show/Hide the Full List | ||
| mod site | chr1:11257023-11257024:- | [10] | |
| Sequence | CCGTCTTGCCATGGCAGGGGACACTTTTACCGCTGAGTACG | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; hESC-HEK293T | ||
| Seq Type List | m6A-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000361445.8 | ||
| External Link | RMBase: m6A_site_7323 | ||
| mod ID: M6ASITE076894 | Click to Show/Hide the Full List | ||
| mod site | chr1:11257098-11257099:- | [10] | |
| Sequence | ATTTGCCAACTATCTTCGGAACCTCCTCCCCTCCAATGACC | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000361445.8 | ||
| External Link | RMBase: m6A_site_7324 | ||
| mod ID: M6ASITE076895 | Click to Show/Hide the Full List | ||
| mod site | chr1:11258546-11258547:- | [12] | |
| Sequence | CTATGACCAACTGAACCATCACATTTTTGAATTGGTTTCCA | ||
| Motif Score | 2.047297619 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000361445.8 | ||
| External Link | RMBase: m6A_site_7325 | ||
| mod ID: M6ASITE076961 | Click to Show/Hide the Full List | ||
| mod site | chr1:11258552-11258553:- | [10] | |
| Sequence | TCGCTTCTATGACCAACTGAACCATCACATTTTTGAATTGG | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000361445.8 | ||
| External Link | RMBase: m6A_site_7326 | ||
| mod ID: M6ASITE076962 | Click to Show/Hide the Full List | ||
| mod site | chr1:11259256-11259257:- | [10] | |
| Sequence | CAGCACTATGTCACCATGGAACTCCGAGAGGTGCTTCTGGG | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa; HepG2; Jurkat | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000361445.8 | ||
| External Link | RMBase: m6A_site_7327 | ||
| mod ID: M6ASITE076965 | Click to Show/Hide the Full List | ||
| mod site | chr1:11259303-11259304:- | [10] | |
| Sequence | TAAAGAGCCGGAATGAGGAAACCAGGGCCAAAGCCGCCAAG | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | HeLa; HepG2; A549; Jurkat; CD4T; GSC-11; HEK293T | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000361445.8 | ||
| External Link | RMBase: m6A_site_7328 | ||
| mod ID: M6ASITE076976 | Click to Show/Hide the Full List | ||
| mod site | chr1:11259366-11259367:- | [12] | |
| Sequence | CCGCCACCACCGCTGCCACCACATCTAGCAATGTGAGCGTC | ||
| Motif Score | 2.053113095 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000361445.8 | ||
| External Link | RMBase: m6A_site_7329 | ||
| mod ID: M6ASITE076984 | Click to Show/Hide the Full List | ||
| mod site | chr1:11259394-11259395:- | [10] | |
| Sequence | GGCAAGATGCTTGGAACCGGACCTGCCGCCGCCACCACCGC | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; HepG2; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293T; HEK293A-TOA; MSC; TREX; iSLK; HEC-1-A; GSCs; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000361445.8 | ||
| External Link | RMBase: m6A_site_7330 | ||
| mod ID: M6ASITE076995 | Click to Show/Hide the Full List | ||
| mod site | chr1:11259399-11259400:- | [10] | |
| Sequence | CTCAGGGCAAGATGCTTGGAACCGGACCTGCCGCCGCCACC | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | HeLa; HepG2; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293T; HEK293A-TOA; MSC; TREX; iSLK; HEC-1-A; GSCs; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000361445.8 | ||
| External Link | RMBase: m6A_site_7331 | ||
Pseudouridine (Pseudo)
| In total 1 m6A sequence/site(s) in this target gene | |||
| mod ID: PSESITE000163 | Click to Show/Hide the Full List | ||
| mod site | chr1:11127723-11127724:- | [16] | |
| Sequence | AGAGGCATCTCGTTTGTACTTTGGGGAAAGGAACGTGAAAG | ||
| Transcript ID List | ENST00000361445.8; ENST00000376838.5 | ||
| External Link | RMBase: Pseudo_site_32 | ||
References