m6A-centered Crosstalk Information
Mechanism of Crosstalk between m6A Modification and Epigenetic Regulation
| Crosstalk ID |
M6ACROT05999
|
[1] | |||
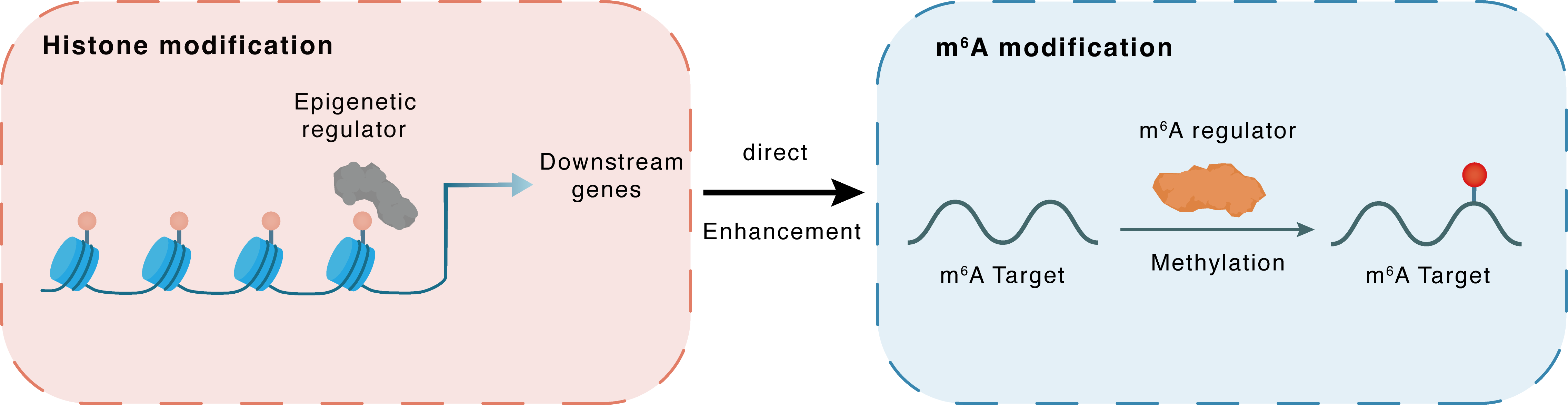 Histone modification
H3K27me3
KDM6B
IL12B
Direct
Enhancement
m6A modification
IL12B
IL12B
METTL3
Methylation
Histone modification
H3K27me3
KDM6B
IL12B
Direct
Enhancement
m6A modification
IL12B
IL12B
METTL3
Methylation
 : m6A sites : m6A sites
|
|||||
| m6A Modification: | |||||
|---|---|---|---|---|---|
| m6A Regulator | Methyltransferase-like 3 (METTL3) | WRITER | |||
| m6A Target | Interleukin-12 subunit beta (IL12B) | ||||
| Epigenetic Regulation that have Cross-talk with This m6A Modification: | |||||
| Epigenetic Regulation Type | Histone modification (HistMod) | ||||
| Epigenetic Regulator | Lysine-specific demethylase 6B (KDM6B) | ERASER | View Details | ||
| Regulated Target | Histone H3 lysine 27 trimethylation (H3K27me3) | View Details | |||
| Downstream Gene | IL12B | View Details | |||
| Crosstalk Relationship | Histone modification → m6A | Enhancement | |||
| Crosstalk Mechanism | Histone modification directly impacts m6A modification through recruiting m6A regulator | ||||
| Crosstalk Summary | Knockout (KO) of YTHDF2, an m6A reader, markedly enhanced demethylation of Histone H3 lysine 27 trimethylation (H3K27me3) on the promoters of proinflammatory cytokines (e.g., IL-6 and Interleukin-12 subunit beta (IL12B)). Furthermore, we identified H3K27me3 as a barrier for m6A modification during transcription. KDM6B recruits the m6A methyltransferase complex (METTL3/METTL14/WTAP) to facilitate the methylation of m6A in transcribing mRNA by removing adjacent H3K27me3 barriers. These results revealed cross-talk between m6A and H3K27me3 during bacterial infection, which has broader implications for deciphering epitranscriptomics in immune homeostasis. | ||||
| Responsed Disease | Inflammatory response | ICD-11: MG46 | |||
| Cell Process | RNA decay | ||||
In-vitro Model |
THP-1 | Childhood acute monocytic leukemia | Homo sapiens | CVCL_0006 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | ||
Full List of Potential Compound(s) Related to This m6A-centered Crosstalk
| Lysine-specific demethylase 6B (KDM6B) | 2 Compound(s) Regulating the Target | Click to Show/Hide the Full List | ||
| GSK-J1 | Investigative | [2] | ||
| Synonyms |
GSK J1; 1373422-53-7; 3-{[2-(pyridin-2-yl)-6-(2,3,4,5-tetrahydro-1H-3-benzazepin-3-yl)pyrimidin-4-yl]amino}propanoic acid; 3-((6-(4,5-Dihydro-1H-benzo[d]azepin-3(2H)-yl)-2-(pyridin-2-yl)pyrimidin-4-yl)amino)propanoic acid; 3-[[2-Pyridin-2-Yl-6-(1,2,4,5-Tetrahydro-3-Benzazepin-3-Yl)pyrimidin-4-Yl]amino]propanoic Acid; GSKJ1; MLS006010249; GTPL7027; SCHEMBL10157115; CHEMBL3188597; BDBM60875; EX-A571; AOB3940; CHEBI:131152; MolPort-023-278-906; EX-A1744; BCP08262; ZINC95616592; s7581; 2442AH; AKOS024458240
Click to Show/Hide
|
|||
| MOA | Inhibitor | |||
| Activity | IC50 = 16 nM | |||
| External Link | ||||
| IOX1 | Investigative | [3] | ||
| Synonyms |
5852-78-8; 8-Hydroxyquinoline-5-Carboxylic Acid; 8-Hydroxy-5-quinolinecarboxylic acid; 5-Carboxy-8-hydroxyquinoline; IOX 1; UNII-JM015YQC1C; IOX-1; 5-carboxy-8HQ; 5-Quinolinecarboxylic acid, 8-hydroxy-; JM015YQC1C; CHEMBL1230640; 4bio; 4jht; 8XQ; 4ie4; AC1LA0UV; MLS002729056; GTPL8230; SCHEMBL6068195; KS-00000PPH; CHEBI:93239; CTK1E0142; DTXSID20207236; AOB6499; JGRPKOGHYBAVMW-UHFFFAOYSA-N; MolPort-006-673-354; HMS3653E21; ZINC5933707; BCP16996; s7234; BDBM50396018; 2184AH; IOX1, > AKOS016371793
Click to Show/Hide
|
|||
| MOA | Inhibitor | |||
| Activity | IC50 = 100 nM | |||
| External Link | ||||
References