m6A Target Gene Information
General Information of the m6A Target Gene (ID: M6ATAR00171)
Full List of m6A Methylation Regulator of This Target Gene and Corresponding Disease/Drug Response(s)
ADAM19
can be regulated by the following regulator(s), and cause disease/drug response(s). You can browse detail information of regulator(s) or disease/drug response(s).
Browse Regulator
Browse Disease
Browse Drug
Methyltransferase-like 14 (METTL14) [WRITER]
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL14 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
|
Treatment: siMETTL14 MDA-MB-231 cells
Control: MDA-MB-231 cells
|
GSE81164 | |
| Regulation |
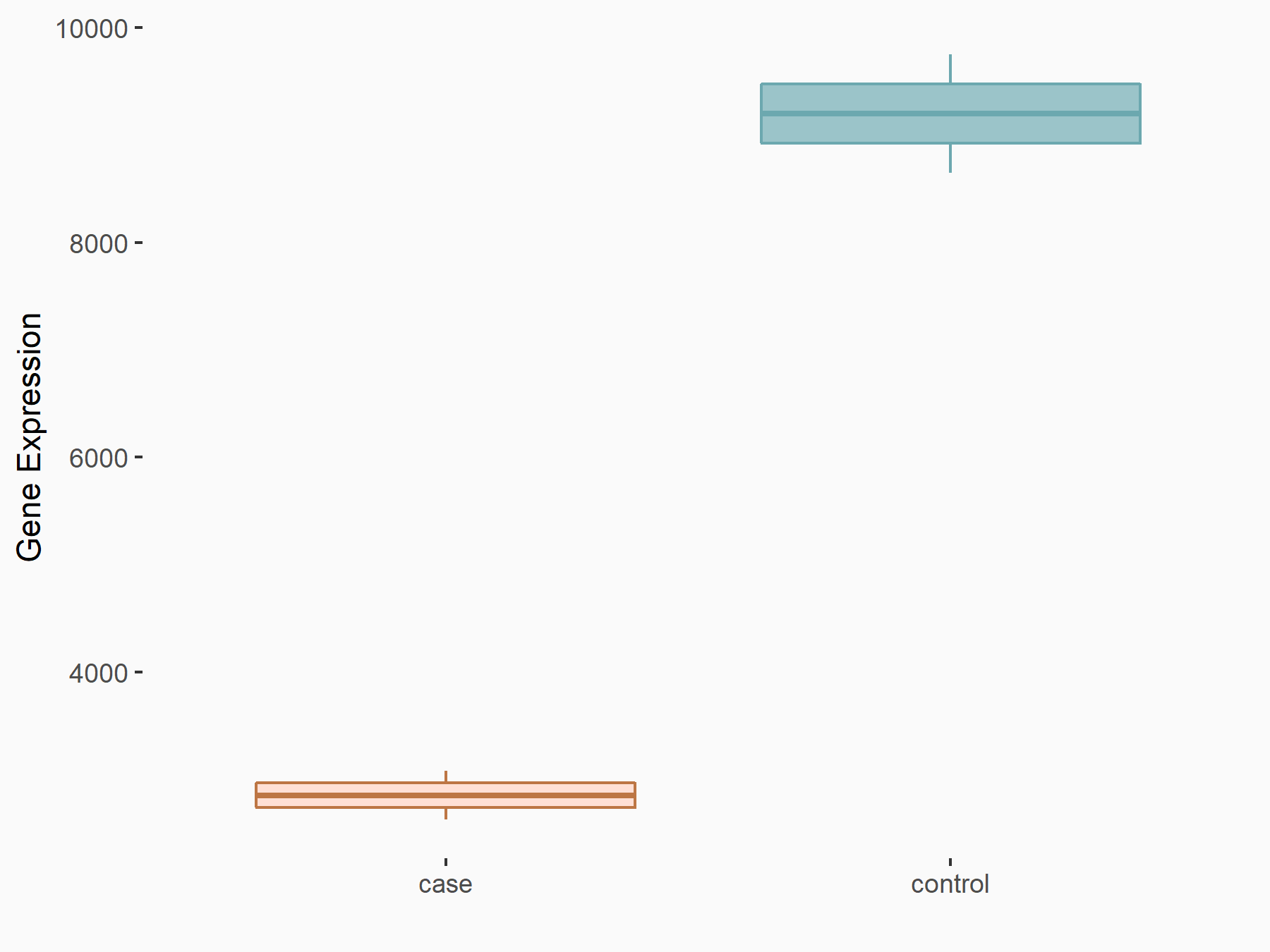  |
logFC: -1.69E+00 p-value: 2.38E-31 |
| More Results | Click to View More RNA-seq Results | |
| In total 1 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [1] | |||
| Response Summary | Knockdown of METTL3 or METTL14 induced changes in mRNA m6A enrichment and altered mRNA expression of genes (e.g., Meltrin-beta (ADAM19)) with critical biological functions in GSCs. Treatment with MA2, a chemical inhibitor of FTO, dramatically suppressed GSC-induced tumorigenesis and prolonged lifespan in GSC-grafted animals. | |||
| Target Regulation | Down regulation | |||
| Responsed Disease | Glioblastoma | ICD-11: 2A00.00 | ||
| Responsed Drug | Ethyl ester form of meclofenamic acid | Approved | ||
| Cell Process | Cells growth | |||
| Cells self-renewal | ||||
| Tumorigenesis | ||||
| MicroRNAs in cancer (hsa05206) | ||||
| In-vitro Model | GSC | Glioma | Epinephelus akaara | CVCL_M752 |
| In-vivo Model | 2 × 105 dissociated cells in 2 uL PBS were injected into the following site (anteroposterior [AP] +0.6 mm, mediolateral [ML] +1.6 mm, and dorsoventricular [DV] 2.6 mm) with a rate of 1 uL/min. | |||
Methyltransferase-like 3 (METTL3) [WRITER]
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | HUVEC cell line | Homo sapiens |
|
Treatment: shMETTL3 HUVEC cells
Control: shScramble HUVEC cells
|
GSE157544 | |
| Regulation |
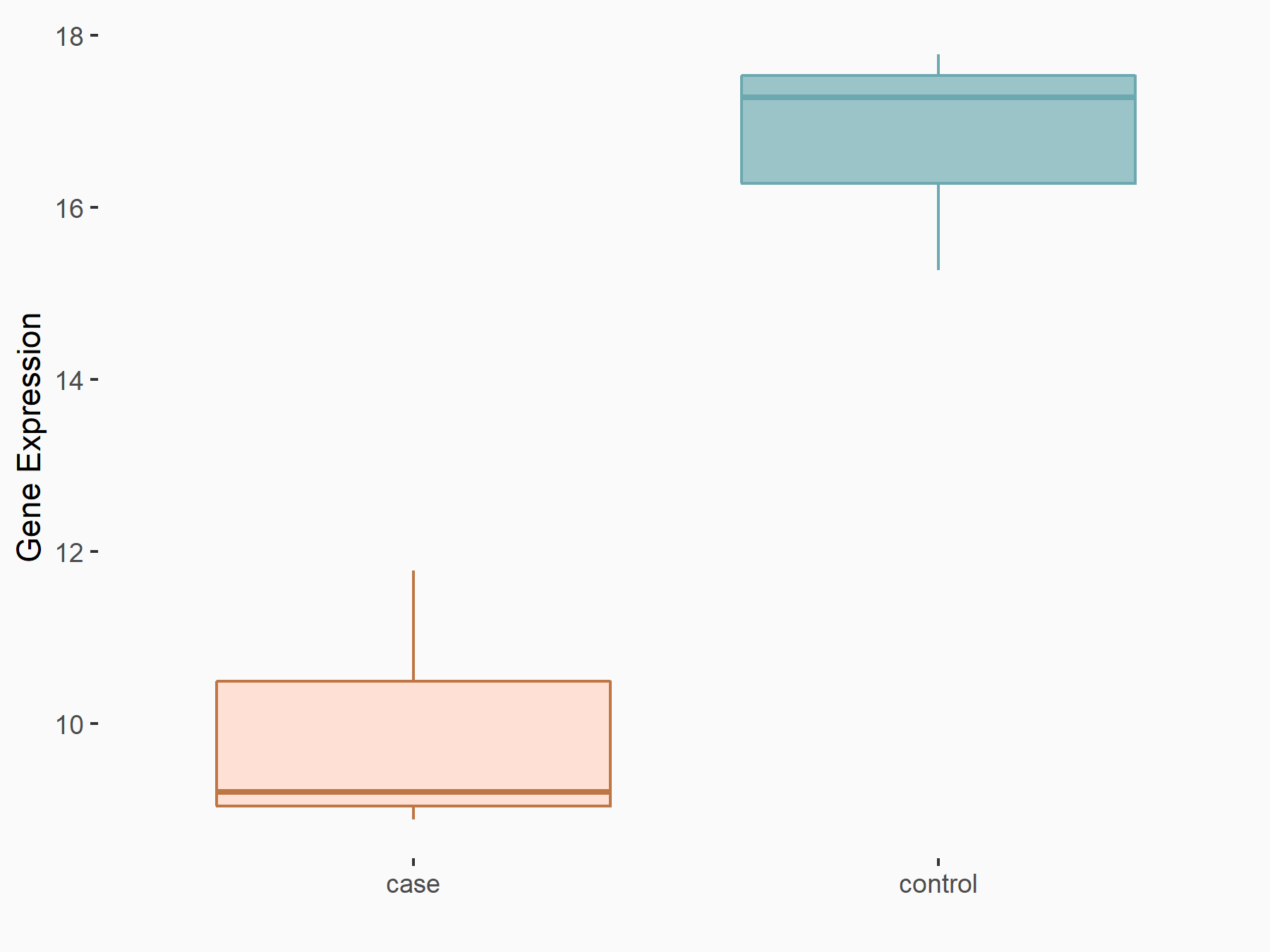  |
logFC: -7.06E-01 p-value: 4.35E-03 |
| More Results | Click to View More RNA-seq Results | |
| In total 1 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [1] | |||
| Response Summary | Knockdown of METTL3 or METTL14 induced changes in mRNA m6A enrichment and altered mRNA expression of genes (e.g., Meltrin-beta (ADAM19)) with critical biological functions in GSCs. Treatment with MA2, a chemical inhibitor of FTO, dramatically suppressed GSC-induced tumorigenesis and prolonged lifespan in GSC-grafted animals. | |||
| Target Regulation | Down regulation | |||
| Responsed Disease | Glioblastoma | ICD-11: 2A00.00 | ||
| Responsed Drug | Ethyl ester form of meclofenamic acid | Approved | ||
| Cell Process | Cells growth | |||
| Cells self-renewal | ||||
| Tumorigenesis | ||||
| MicroRNAs in cancer (hsa05206) | ||||
| In-vitro Model | GSC | Glioma | Epinephelus akaara | CVCL_M752 |
| In-vivo Model | 2 × 105 dissociated cells in 2 uL PBS were injected into the following site (anteroposterior [AP] +0.6 mm, mediolateral [ML] +1.6 mm, and dorsoventricular [DV] 2.6 mm) with a rate of 1 uL/min. | |||
Brain cancer [ICD-11: 2A00]
| In total 2 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [1] | |||
| Response Summary | Knockdown of METTL3 or METTL14 induced changes in mRNA m6A enrichment and altered mRNA expression of genes (e.g., Meltrin-beta (ADAM19)) with critical biological functions in GSCs. Treatment with MA2, a chemical inhibitor of FTO, dramatically suppressed GSC-induced tumorigenesis and prolonged lifespan in GSC-grafted animals. | |||
| Responsed Disease | Glioblastoma [ICD-11: 2A00.00] | |||
| Target Regulator | Methyltransferase-like 14 (METTL14) | WRITER | ||
| Target Regulation | Down regulation | |||
| Responsed Drug | Ethyl ester form of meclofenamic acid | Approved | ||
| Cell Process | Cells growth | |||
| Cells self-renewal | ||||
| Tumorigenesis | ||||
| MicroRNAs in cancer (hsa05206) | ||||
| In-vitro Model | GSC | Glioma | Epinephelus akaara | CVCL_M752 |
| In-vivo Model | 2 × 105 dissociated cells in 2 uL PBS were injected into the following site (anteroposterior [AP] +0.6 mm, mediolateral [ML] +1.6 mm, and dorsoventricular [DV] 2.6 mm) with a rate of 1 uL/min. | |||
| Experiment 2 Reporting the m6A-centered Disease Response | [1] | |||
| Response Summary | Knockdown of METTL3 or METTL14 induced changes in mRNA m6A enrichment and altered mRNA expression of genes (e.g., Meltrin-beta (ADAM19)) with critical biological functions in GSCs. Treatment with MA2, a chemical inhibitor of FTO, dramatically suppressed GSC-induced tumorigenesis and prolonged lifespan in GSC-grafted animals. | |||
| Responsed Disease | Glioblastoma [ICD-11: 2A00.00] | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Target Regulation | Down regulation | |||
| Responsed Drug | Ethyl ester form of meclofenamic acid | Approved | ||
| Cell Process | Cells growth | |||
| Cells self-renewal | ||||
| Tumorigenesis | ||||
| MicroRNAs in cancer (hsa05206) | ||||
| In-vitro Model | GSC | Glioma | Epinephelus akaara | CVCL_M752 |
| In-vivo Model | 2 × 105 dissociated cells in 2 uL PBS were injected into the following site (anteroposterior [AP] +0.6 mm, mediolateral [ML] +1.6 mm, and dorsoventricular [DV] 2.6 mm) with a rate of 1 uL/min. | |||
Solid tumour/cancer [ICD-11: 2A00-2F9Z]
Ethyl ester form of meclofenamic acid
[Approved]
| In total 2 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response | [1] | |||
| Response Summary | Knockdown of METTL3 or METTL14 induced changes in mRNA m6A enrichment and altered mRNA expression of genes (e.g., Meltrin-beta (ADAM19)) with critical biological functions in GSCs. Treatment with MA2, a chemical inhibitor of FTO, dramatically suppressed GSC-induced tumorigenesis and prolonged lifespan in GSC-grafted animals. | |||
| Target Regulator | Methyltransferase-like 14 (METTL14) | WRITER | ||
| Target Regulation | Down regulation | |||
| Responsed Disease | Glioblastoma | ICD-11: 2A00.00 | ||
| Cell Process | Cells growth | |||
| Cells self-renewal | ||||
| Tumorigenesis | ||||
| MicroRNAs in cancer (hsa05206) | ||||
| In-vitro Model | GSC | Glioma | Epinephelus akaara | CVCL_M752 |
| In-vivo Model | 2 × 105 dissociated cells in 2 uL PBS were injected into the following site (anteroposterior [AP] +0.6 mm, mediolateral [ML] +1.6 mm, and dorsoventricular [DV] 2.6 mm) with a rate of 1 uL/min. | |||
| Experiment 2 Reporting the m6A-centered Drug Response | [1] | |||
| Response Summary | Knockdown of METTL3 or METTL14 induced changes in mRNA m6A enrichment and altered mRNA expression of genes (e.g., Meltrin-beta (ADAM19)) with critical biological functions in GSCs. Treatment with MA2, a chemical inhibitor of FTO, dramatically suppressed GSC-induced tumorigenesis and prolonged lifespan in GSC-grafted animals. | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Target Regulation | Down regulation | |||
| Responsed Disease | Glioblastoma | ICD-11: 2A00.00 | ||
| Cell Process | Cells growth | |||
| Cells self-renewal | ||||
| Tumorigenesis | ||||
| MicroRNAs in cancer (hsa05206) | ||||
| In-vitro Model | GSC | Glioma | Epinephelus akaara | CVCL_M752 |
| In-vivo Model | 2 × 105 dissociated cells in 2 uL PBS were injected into the following site (anteroposterior [AP] +0.6 mm, mediolateral [ML] +1.6 mm, and dorsoventricular [DV] 2.6 mm) with a rate of 1 uL/min. | |||
Full List of Crosstalk(s) between m6A Modification and Epigenetic Regulation Related to This Regulator
Histone modification
m6A Regulator: Methyltransferase-like 3 (METTL3)
| In total 1 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT03299 | ||
| Epigenetic Regulator | Histone-lysine N-methyltransferase EZH2 (EZH2) | |
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Brain cancer | |
| Drug | Ethyl ester form of meclofenamic acid | |
RNA Modification Sequencing Data Associated with the Target (ID: M6ATAR00171)
| In total 41 m6A sequence/site(s) in this target gene | |||
| mod ID: A2ISITE001259 | Click to Show/Hide the Full List | ||
| mod site | chr5:157477749-157477750:- | [2] | |
| Sequence | CCCTGCCTACTTCCTAATTTATTCTTATTTGACAGAGGGTA | ||
| Transcript ID List | ENST00000257527.8; ENST00000517951.5; rmsk_1799868; ENST00000517374.5 | ||
| External Link | RMBase: RNA-editing_site_113303 | ||
| mod ID: A2ISITE001260 | Click to Show/Hide the Full List | ||
| mod site | chr5:157477776-157477777:- | [3] | |
| Sequence | CCTTTCCAATCTTTATTGGAAGTCTCTCCCTGCCTACTTCC | ||
| Transcript ID List | ENST00000257527.8; ENST00000517374.5; rmsk_1799868; ENST00000517951.5 | ||
| External Link | RMBase: RNA-editing_site_113304 | ||
| mod ID: A2ISITE001261 | Click to Show/Hide the Full List | ||
| mod site | chr5:157477822-157477823:- | [3] | |
| Sequence | TCCCTTCCCCCTGAAGCAATAGCCCCTCCCCACCTCCTGCA | ||
| Transcript ID List | ENST00000517951.5; rmsk_1799868; ENST00000257527.8; ENST00000517374.5 | ||
| External Link | RMBase: RNA-editing_site_113305 | ||
| mod ID: A2ISITE001262 | Click to Show/Hide the Full List | ||
| mod site | chr5:157477824-157477825:- | [3] | |
| Sequence | AGTCCCTTCCCCCTGAAGCAATAGCCCCTCCCCACCTCCTG | ||
| Transcript ID List | ENST00000517951.5; rmsk_1799868; ENST00000517374.5; ENST00000257527.8 | ||
| External Link | RMBase: RNA-editing_site_113306 | ||
| mod ID: A2ISITE001263 | Click to Show/Hide the Full List | ||
| mod site | chr5:157477844-157477845:- | [3] | |
| Sequence | GTGTTTTTTTGTATTGCCATAGTCCCTTCCCCCTGAAGCAA | ||
| Transcript ID List | ENST00000517951.5; ENST00000517374.5; rmsk_1799868; ENST00000257527.8 | ||
| External Link | RMBase: RNA-editing_site_113307 | ||
| mod ID: A2ISITE001264 | Click to Show/Hide the Full List | ||
| mod site | chr5:157477909-157477910:- | [2] | |
| Sequence | TTCTGCCCCAGCTCTGAGACACTTGCAGATCTTAAGGTCTG | ||
| Transcript ID List | ENST00000257527.8; ENST00000517374.5; ENST00000517951.5; rmsk_1799868 | ||
| External Link | RMBase: RNA-editing_site_113308 | ||
| mod ID: A2ISITE001265 | Click to Show/Hide the Full List | ||
| mod site | chr5:157477913-157477914:- | [3] | |
| Sequence | CTTTTTCTGCCCCAGCTCTGAGACACTTGCAGATCTTAAGG | ||
| Transcript ID List | ENST00000517951.5; rmsk_1799868; ENST00000257527.8; ENST00000517374.5 | ||
| External Link | RMBase: RNA-editing_site_113309 | ||
| mod ID: A2ISITE001266 | Click to Show/Hide the Full List | ||
| mod site | chr5:157477920-157477921:- | [2] | |
| Sequence | GTAAAGCCTTTTTCTGCCCCAGCTCTGAGACACTTGCAGAT | ||
| Transcript ID List | ENST00000257527.8; ENST00000517374.5; rmsk_1799868; ENST00000517951.5 | ||
| External Link | RMBase: RNA-editing_site_113310 | ||
| mod ID: A2ISITE001267 | Click to Show/Hide the Full List | ||
| mod site | chr5:157477936-157477937:- | [2] | |
| Sequence | ACTTCTCCCTATAAAAGTAAAGCCTTTTTCTGCCCCAGCTC | ||
| Transcript ID List | ENST00000257527.8; rmsk_1799868; ENST00000517374.5; ENST00000517951.5 | ||
| External Link | RMBase: RNA-editing_site_113311 | ||
| mod ID: A2ISITE001268 | Click to Show/Hide the Full List | ||
| mod site | chr5:157477938-157477939:- | [2] | |
| Sequence | TGACTTCTCCCTATAAAAGTAAAGCCTTTTTCTGCCCCAGC | ||
| Transcript ID List | ENST00000257527.8; rmsk_1799868; ENST00000517374.5; ENST00000517951.5 | ||
| External Link | RMBase: RNA-editing_site_113312 | ||
| mod ID: A2ISITE001269 | Click to Show/Hide the Full List | ||
| mod site | chr5:157477941-157477942:- | [3] | |
| Sequence | TGCTGACTTCTCCCTATAAAAGTAAAGCCTTTTTCTGCCCC | ||
| Transcript ID List | rmsk_1799868; ENST00000517951.5; ENST00000517374.5; ENST00000257527.8 | ||
| External Link | RMBase: RNA-editing_site_113313 | ||
| mod ID: A2ISITE001270 | Click to Show/Hide the Full List | ||
| mod site | chr5:157477942-157477943:- | [2] | |
| Sequence | TTGCTGACTTCTCCCTATAAAAGTAAAGCCTTTTTCTGCCC | ||
| Transcript ID List | ENST00000517374.5; ENST00000257527.8; ENST00000517951.5; rmsk_1799868 | ||
| External Link | RMBase: RNA-editing_site_113314 | ||
| mod ID: A2ISITE001271 | Click to Show/Hide the Full List | ||
| mod site | chr5:157477943-157477944:- | [2] | |
| Sequence | CTTGCTGACTTCTCCCTATAAAAGTAAAGCCTTTTTCTGCC | ||
| Transcript ID List | ENST00000517374.5; ENST00000257527.8; ENST00000517951.5; rmsk_1799868 | ||
| External Link | RMBase: RNA-editing_site_113315 | ||
| mod ID: A2ISITE001272 | Click to Show/Hide the Full List | ||
| mod site | chr5:157477946-157477947:- | [2] | |
| Sequence | AGCCTTGCTGACTTCTCCCTATAAAAGTAAAGCCTTTTTCT | ||
| Transcript ID List | ENST00000257527.8; rmsk_1799868; ENST00000517374.5; ENST00000517951.5 | ||
| External Link | RMBase: RNA-editing_site_113316 | ||
| mod ID: A2ISITE001273 | Click to Show/Hide the Full List | ||
| mod site | chr5:157477956-157477957:- | [3] | |
| Sequence | GATTCCTTTCAGCCTTGCTGACTTCTCCCTATAAAAGTAAA | ||
| Transcript ID List | rmsk_1799868; ENST00000517951.5; ENST00000257527.8; ENST00000517374.5 | ||
| External Link | RMBase: RNA-editing_site_113317 | ||
| mod ID: A2ISITE001274 | Click to Show/Hide the Full List | ||
| mod site | chr5:157477966-157477967:- | [3] | |
| Sequence | GCTAGCCATAGATTCCTTTCAGCCTTGCTGACTTCTCCCTA | ||
| Transcript ID List | ENST00000257527.8; rmsk_1799868; ENST00000517374.5; ENST00000517951.5 | ||
| External Link | RMBase: RNA-editing_site_113318 | ||
| mod ID: A2ISITE001275 | Click to Show/Hide the Full List | ||
| mod site | chr5:157478364-157478365:- | [2] | |
| Sequence | ACTTTTCTTTTATTGGAAGAAGTAAACATGGCTAGAAAGAA | ||
| Transcript ID List | ENST00000257527.8; ENST00000517374.5; ENST00000517951.5 | ||
| External Link | RMBase: RNA-editing_site_113319 | ||
| mod ID: A2ISITE001276 | Click to Show/Hide the Full List | ||
| mod site | chr5:157478373-157478374:- | [3] | |
| Sequence | CAGAAAAGGACTTTTCTTTTATTGGAAGAAGTAAACATGGC | ||
| Transcript ID List | ENST00000517374.5; ENST00000257527.8; ENST00000517951.5 | ||
| External Link | RMBase: RNA-editing_site_113320 | ||
| mod ID: A2ISITE001277 | Click to Show/Hide the Full List | ||
| mod site | chr5:157478387-157478388:- | [3] | |
| Sequence | TCTCAAAGCCCAGGCAGAAAAGGACTTTTCTTTTATTGGAA | ||
| Transcript ID List | ENST00000517374.5; ENST00000517951.5; ENST00000257527.8 | ||
| External Link | RMBase: RNA-editing_site_113321 | ||
| mod ID: A2ISITE001278 | Click to Show/Hide the Full List | ||
| mod site | chr5:157478388-157478389:- | [3] | |
| Sequence | GTCTCAAAGCCCAGGCAGAAAAGGACTTTTCTTTTATTGGA | ||
| Transcript ID List | ENST00000257527.8; ENST00000517951.5; ENST00000517374.5 | ||
| External Link | RMBase: RNA-editing_site_113322 | ||
| mod ID: A2ISITE001279 | Click to Show/Hide the Full List | ||
| mod site | chr5:157478389-157478390:- | [3] | |
| Sequence | CGTCTCAAAGCCCAGGCAGAAAAGGACTTTTCTTTTATTGG | ||
| Transcript ID List | ENST00000257527.8; ENST00000517374.5; ENST00000517951.5 | ||
| External Link | RMBase: RNA-editing_site_113323 | ||
| mod ID: A2ISITE001280 | Click to Show/Hide the Full List | ||
| mod site | chr5:157478396-157478397:- | [3] | |
| Sequence | CTGCAAGCGTCTCAAAGCCCAGGCAGAAAAGGACTTTTCTT | ||
| Transcript ID List | ENST00000517951.5; ENST00000517374.5; ENST00000257527.8 | ||
| External Link | RMBase: RNA-editing_site_113324 | ||
| mod ID: A2ISITE001281 | Click to Show/Hide the Full List | ||
| mod site | chr5:157478402-157478403:- | [2] | |
| Sequence | CAAGATCTGCAAGCGTCTCAAAGCCCAGGCAGAAAAGGACT | ||
| Transcript ID List | ENST00000257527.8; ENST00000517374.5; ENST00000517951.5 | ||
| External Link | RMBase: RNA-editing_site_113325 | ||
| mod ID: A2ISITE001282 | Click to Show/Hide the Full List | ||
| mod site | chr5:157478411-157478412:- | [2] | |
| Sequence | GACCGATGGCAAGATCTGCAAGCGTCTCAAAGCCCAGGCAG | ||
| Transcript ID List | ENST00000517951.5; ENST00000517374.5; ENST00000257527.8 | ||
| External Link | RMBase: RNA-editing_site_113326 | ||
| mod ID: A2ISITE001283 | Click to Show/Hide the Full List | ||
| mod site | chr5:157478420-157478421:- | [2] | |
| Sequence | GCTCAGACCGACCGATGGCAAGATCTGCAAGCGTCTCAAAG | ||
| Transcript ID List | ENST00000257527.8; ENST00000517374.5; ENST00000517951.5 | ||
| External Link | RMBase: RNA-editing_site_113327 | ||
| mod ID: A2ISITE001284 | Click to Show/Hide the Full List | ||
| mod site | chr5:157478436-157478437:- | [2] | |
| Sequence | TGGGAATAGTGAAAAGGCTCAGACCGACCGATGGCAAGATC | ||
| Transcript ID List | ENST00000517951.5; ENST00000517374.5; ENST00000257527.8 | ||
| External Link | RMBase: RNA-editing_site_113328 | ||
| mod ID: A2ISITE001285 | Click to Show/Hide the Full List | ||
| mod site | chr5:157478484-157478485:- | [2] | |
| Sequence | TGCAGAAGGGGAAAGGGAATATTGCAAAAGGGAGAGGAAGG | ||
| Transcript ID List | ENST00000257527.8; ENST00000517374.5; ENST00000517951.5 | ||
| External Link | RMBase: RNA-editing_site_113329 | ||
| mod ID: A2ISITE001286 | Click to Show/Hide the Full List | ||
| mod site | chr5:157478491-157478492:- | [2] | |
| Sequence | GCACGATTGCAGAAGGGGAAAGGGAATATTGCAAAAGGGAG | ||
| Transcript ID List | ENST00000257527.8; ENST00000517374.5; ENST00000517951.5 | ||
| External Link | RMBase: RNA-editing_site_113330 | ||
| mod ID: A2ISITE001287 | Click to Show/Hide the Full List | ||
| mod site | chr5:157478546-157478547:- | [2] | |
| Sequence | GAGGTGTCAAATACAAATAAATCTGGGCTTAGGGAAGGAGA | ||
| Transcript ID List | ENST00000257527.8; ENST00000517374.5; ENST00000517951.5 | ||
| External Link | RMBase: RNA-editing_site_113331 | ||
| mod ID: A2ISITE001288 | Click to Show/Hide the Full List | ||
| mod site | chr5:157478547-157478548:- | [3] | |
| Sequence | TGAGGTGTCAAATACAAATAAATCTGGGCTTAGGGAAGGAG | ||
| Transcript ID List | ENST00000257527.8; ENST00000517374.5; ENST00000517951.5 | ||
| External Link | RMBase: RNA-editing_site_113332 | ||
| mod ID: A2ISITE001289 | Click to Show/Hide the Full List | ||
| mod site | chr5:157478548-157478549:- | [4] | |
| Sequence | TTGAGGTGTCAAATACAAATAAATCTGGGCTTAGGGAAGGA | ||
| Transcript ID List | ENST00000517374.5; ENST00000257527.8; ENST00000517951.5 | ||
| External Link | RMBase: RNA-editing_site_113333 | ||
| mod ID: A2ISITE001290 | Click to Show/Hide the Full List | ||
| mod site | chr5:157478550-157478551:- | [2] | |
| Sequence | TTTTGAGGTGTCAAATACAAATAAATCTGGGCTTAGGGAAG | ||
| Transcript ID List | ENST00000257527.8; ENST00000517951.5; ENST00000517374.5 | ||
| External Link | RMBase: RNA-editing_site_113334 | ||
| mod ID: A2ISITE001291 | Click to Show/Hide the Full List | ||
| mod site | chr5:157478551-157478552:- | [3] | |
| Sequence | GTTTTGAGGTGTCAAATACAAATAAATCTGGGCTTAGGGAA | ||
| Transcript ID List | ENST00000517951.5; ENST00000257527.8; ENST00000517374.5 | ||
| External Link | RMBase: RNA-editing_site_113335 | ||
| mod ID: A2ISITE001292 | Click to Show/Hide the Full List | ||
| mod site | chr5:157478552-157478553:- | [4] | |
| Sequence | TGTTTTGAGGTGTCAAATACAAATAAATCTGGGCTTAGGGA | ||
| Transcript ID List | ENST00000517374.5; ENST00000517951.5; ENST00000257527.8 | ||
| External Link | RMBase: RNA-editing_site_113336 | ||
| mod ID: A2ISITE001293 | Click to Show/Hide the Full List | ||
| mod site | chr5:157478556-157478557:- | [4] | |
| Sequence | ATGCTGTTTTGAGGTGTCAAATACAAATAAATCTGGGCTTA | ||
| Transcript ID List | ENST00000257527.8; ENST00000517374.5; ENST00000517951.5 | ||
| External Link | RMBase: RNA-editing_site_113337 | ||
| mod ID: A2ISITE001294 | Click to Show/Hide the Full List | ||
| mod site | chr5:157478557-157478558:- | [3] | |
| Sequence | TATGCTGTTTTGAGGTGTCAAATACAAATAAATCTGGGCTT | ||
| Transcript ID List | ENST00000517951.5; ENST00000517374.5; ENST00000257527.8 | ||
| External Link | RMBase: RNA-editing_site_113338 | ||
| mod ID: A2ISITE001295 | Click to Show/Hide the Full List | ||
| mod site | chr5:157478558-157478559:- | [4] | |
| Sequence | GTATGCTGTTTTGAGGTGTCAAATACAAATAAATCTGGGCT | ||
| Transcript ID List | ENST00000517951.5; ENST00000257527.8; ENST00000517374.5 | ||
| External Link | RMBase: RNA-editing_site_113339 | ||
| mod ID: A2ISITE001296 | Click to Show/Hide the Full List | ||
| mod site | chr5:157504655-157504656:- | [2] | |
| Sequence | AGGGCCTGTTTATGATTCTTAGAGAATGGTGTTTCAATGTC | ||
| Transcript ID List | ENST00000517951.5; ENST00000257527.8; ENST00000517905.1 | ||
| External Link | RMBase: RNA-editing_site_113340 | ||
| mod ID: A2ISITE001297 | Click to Show/Hide the Full List | ||
| mod site | chr5:157505006-157505007:- | [2] | |
| Sequence | TCTGCCTTAGCCTCCCGAGTAGCTGGGAATACAGGCGCCTG | ||
| Transcript ID List | ENST00000517905.1; ENST00000257527.8; ENST00000517951.5 | ||
| External Link | RMBase: RNA-editing_site_113341 | ||
| mod ID: A2ISITE001298 | Click to Show/Hide the Full List | ||
| mod site | chr5:157510539-157510540:- | [2] | |
| Sequence | CAATAGACATGAGGGATCTCACCGATGTGATGAAAATATTC | ||
| Transcript ID List | ENST00000517951.5; rmsk_1799935; ENST00000517905.1; ENST00000257527.8 | ||
| External Link | RMBase: RNA-editing_site_113342 | ||
| mod ID: A2ISITE001299 | Click to Show/Hide the Full List | ||
| mod site | chr5:157556397-157556398:- | [2] | |
| Sequence | GGTGAAACCTGGTCTCTACTAAAAATACAAAAATTAGCTGG | ||
| Transcript ID List | ENST00000517951.5; ENST00000517905.1; rmsk_1800051; ENST00000257527.8 | ||
| External Link | RMBase: RNA-editing_site_113343 | ||
5-methylcytidine (m5C)
N6-methyladenosine (m6A)
| In total 21 m6A sequence/site(s) in this target gene | |||
| mod ID: M6ASITE072576 | Click to Show/Hide the Full List | ||
| mod site | chr5:157477449-157477450:- | [5] | |
| Sequence | TTTTATTTGGGAGATTGTTCACGGAAAACAGATCTTCTTCT | ||
| Motif Score | 2.021232143 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000257527.8; ENST00000517951.5; ENST00000517374.5 | ||
| External Link | RMBase: m6A_site_695525 | ||
| mod ID: M6ASITE072577 | Click to Show/Hide the Full List | ||
| mod site | chr5:157480659-157480660:- | [6] | |
| Sequence | ACTGGCAGCTTCTCCTCCAGACTCCTAGCCCCGAGGTGCTG | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | CD4T; HEK293A-TOA; iSLK; MSC; endometrial | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000257527.8; ENST00000517951.5; ENST00000517374.5 | ||
| External Link | RMBase: m6A_site_695526 | ||
| mod ID: M6ASITE072578 | Click to Show/Hide the Full List | ||
| mod site | chr5:157480758-157480759:- | [7] | |
| Sequence | TTCCTTGGAAGCCCAGAGGGACTATGATCTGATGGCCTCTA | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | H1A; H1B; fibroblasts; A549; GM12878; CD4T; GSC-11; HEK293A-TOA; iSLK; TIME; MSC; endometrial | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000257527.8; ENST00000517951.5; ENST00000517374.5 | ||
| External Link | RMBase: m6A_site_695527 | ||
| mod ID: M6ASITE072579 | Click to Show/Hide the Full List | ||
| mod site | chr5:157480826-157480827:- | [7] | |
| Sequence | CCTCCCACCCTCCTCCAGGAACCTCCACATCTCCAAAAATC | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | H1B; H1A; fibroblasts; A549; GM12878; CD4T; GSC-11; HEK293T; HEK293A-TOA; iSLK; MSC; endometrial | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000517374.5; ENST00000257527.8; ENST00000517951.5 | ||
| External Link | RMBase: m6A_site_695528 | ||
| mod ID: M6ASITE072580 | Click to Show/Hide the Full List | ||
| mod site | chr5:157480880-157480881:- | [7] | |
| Sequence | AGAGGACCCATGGCCATGGAACCCTGAAGAAGCATGTCTGG | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | H1A; H1B; fibroblasts; A549; GM12878; CD4T; GSC-11; HEK293T; HEK293A-TOA; iSLK; endometrial | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000257527.8; ENST00000517951.5; ENST00000517374.5 | ||
| External Link | RMBase: m6A_site_695529 | ||
| mod ID: M6ASITE072581 | Click to Show/Hide the Full List | ||
| mod site | chr5:157480895-157480896:- | [7] | |
| Sequence | CTCTCTGGACACTGCAGAGGACCCATGGCCATGGAACCCTG | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | H1A; H1B; fibroblasts; A549; GM12878; CD4T; GSC-11; HEK293T; HEK293A-TOA; iSLK; endometrial | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000517951.5; ENST00000257527.8; ENST00000517374.5 | ||
| External Link | RMBase: m6A_site_695530 | ||
| mod ID: M6ASITE072582 | Click to Show/Hide the Full List | ||
| mod site | chr5:157480907-157480908:- | [7] | |
| Sequence | CCTTTCCTTGAGCTCTCTGGACACTGCAGAGGACCCATGGC | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | H1A; H1B; fibroblasts; A549; GM12878; CD4T; GSC-11; HEK293T; HEK293A-TOA; iSLK; endometrial | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000517951.5; ENST00000257527.8; ENST00000517374.5 | ||
| External Link | RMBase: m6A_site_695531 | ||
| mod ID: M6ASITE072583 | Click to Show/Hide the Full List | ||
| mod site | chr5:157480947-157480948:- | [7] | |
| Sequence | TGATTAGCTCGAAAATCTAGACCTGTCCAAGGGGCTTCTCC | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | H1B; fibroblasts; A549; GM12878; endometrial | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000257527.8; ENST00000517951.5; ENST00000517374.5 | ||
| External Link | RMBase: m6A_site_695532 | ||
| mod ID: M6ASITE072584 | Click to Show/Hide the Full List | ||
| mod site | chr5:157491703-157491704:- | [8] | |
| Sequence | GGTCTGTAACAACAACCAGAACTGCCACTGCCTGCCGGGCT | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | endometrial | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000517905.1; ENST00000517951.5; ENST00000517374.5; ENST00000257527.8 | ||
| External Link | RMBase: m6A_site_695533 | ||
| mod ID: M6ASITE072585 | Click to Show/Hide the Full List | ||
| mod site | chr5:157491869-157491870:- | [8] | |
| Sequence | GGAACACCTCCTTCTTTGAAACTGAAGGCTGTGGGAAGAAG | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | endometrial | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000517905.1; ENST00000517951.5; ENST00000517374.5; ENST00000257527.8 | ||
| External Link | RMBase: m6A_site_695534 | ||
| mod ID: M6ASITE072586 | Click to Show/Hide the Full List | ||
| mod site | chr5:157491886-157491887:- | [8] | |
| Sequence | CTTTGAGGGGCAGTGCAGGAACACCTCCTTCTTTGAAACTG | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | endometrial | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000257527.8; ENST00000517951.5; ENST00000517374.5; ENST00000517905.1 | ||
| External Link | RMBase: m6A_site_695535 | ||
| mod ID: M6ASITE072587 | Click to Show/Hide the Full List | ||
| mod site | chr5:157492992-157492993:- | [8] | |
| Sequence | CAGGGCTGGTGATGACTGGAACCAAGTGTGGCTACAACCAT | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | endometrial | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000517374.5; ENST00000517951.5; ENST00000517905.1; ENST00000257527.8 | ||
| External Link | RMBase: m6A_site_695536 | ||
| mod ID: M6ASITE072588 | Click to Show/Hide the Full List | ||
| mod site | chr5:157493015-157493016:- | [8] | |
| Sequence | GGAGGAGGGTGACATGCTGGACCCAGGGCTGGTGATGACTG | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | endometrial | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000257527.8; ENST00000517951.5; ENST00000517374.5; ENST00000517905.1 | ||
| External Link | RMBase: m6A_site_695537 | ||
| mod ID: M6ASITE072589 | Click to Show/Hide the Full List | ||
| mod site | chr5:157494706-157494707:- | [8] | |
| Sequence | GGAAAGGACATGAATGGTGAACACAGGAAGTGCAACATGAG | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | endometrial | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000257527.8; ENST00000517905.1; ENST00000517951.5; ENST00000517374.5 | ||
| External Link | RMBase: m6A_site_695538 | ||
| mod ID: M6ASITE072590 | Click to Show/Hide the Full List | ||
| mod site | chr5:157494719-157494720:- | [8] | |
| Sequence | CTTTGGAAACTGTGGAAAGGACATGAATGGTGAACACAGGA | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | endometrial | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000257527.8; ENST00000517951.5; ENST00000517374.5; ENST00000517905.1 | ||
| External Link | RMBase: m6A_site_695539 | ||
| mod ID: M6ASITE072591 | Click to Show/Hide the Full List | ||
| mod site | chr5:157494731-157494732:- | [8] | |
| Sequence | GGCAGGAGACACCTTTGGAAACTGTGGAAAGGACATGAATG | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | endometrial | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000517951.5; ENST00000517374.5; ENST00000517905.1; ENST00000257527.8 | ||
| External Link | RMBase: m6A_site_695540 | ||
| mod ID: M6ASITE072592 | Click to Show/Hide the Full List | ||
| mod site | chr5:157570879-157570880:- | [9] | |
| Sequence | GAAAAGGTAAGACTCAAAAGACCTCATTTACACTGGCAGGA | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | GSC-11; iSLK | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000517951.5; ENST00000517905.1; ENST00000257527.8; ENST00000519752.1 | ||
| External Link | RMBase: m6A_site_695541 | ||
| mod ID: M6ASITE072593 | Click to Show/Hide the Full List | ||
| mod site | chr5:157570888-157570889:- | [10] | |
| Sequence | CCCGTGAGAGAAAAGGTAAGACTCAAAAGACCTCATTTACA | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | U2OS; GSC-11; iSLK | ||
| Seq Type List | MeRIP-seq; m6A-seq | ||
| Transcript ID List | ENST00000517951.5; ENST00000257527.8; ENST00000519752.1; ENST00000517905.1 | ||
| External Link | RMBase: m6A_site_695542 | ||
| mod ID: M6ASITE072594 | Click to Show/Hide the Full List | ||
| mod site | chr5:157570920-157570921:- | [10] | |
| Sequence | TTATCATACCTCAGTGGAAGACTTCAGAAAGCCCCGTGAGA | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | U2OS; GSC-11; HEK293T; iSLK; MSC; GSCs | ||
| Seq Type List | MeRIP-seq; m6A-seq | ||
| Transcript ID List | ENST00000257527.8; ENST00000519752.1; ENST00000517951.5; ENST00000517905.1 | ||
| External Link | RMBase: m6A_site_695543 | ||
| mod ID: M6ASITE072595 | Click to Show/Hide the Full List | ||
| mod site | chr5:157570942-157570943:- | [10] | |
| Sequence | AGCCCCAAGCTGCAGCATGAACTTATCATACCTCAGTGGAA | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | U2OS; GSC-11; HEK293T; iSLK; MSC; GSCs | ||
| Seq Type List | MeRIP-seq; m6A-seq | ||
| Transcript ID List | ENST00000517951.5; ENST00000519752.1; ENST00000517905.1; ENST00000257527.8 | ||
| External Link | RMBase: m6A_site_695544 | ||
| mod ID: M6ASITE072596 | Click to Show/Hide the Full List | ||
| mod site | chr5:157575605-157575606:- | [10] | |
| Sequence | CGGCGCGGGAGCCTGGATGGACAAGTAAGTGGCCAGGCGCG | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | U2OS; GSC-11; HEK293T; HEK293A-TOA; iSLK; MSC; GSCs | ||
| Seq Type List | MeRIP-seq; m6A-seq | ||
| Transcript ID List | ENST00000257527.8; ENST00000517905.1; ENST00000517951.5; ENST00000519752.1 | ||
| External Link | RMBase: m6A_site_695545 | ||
References