m6A Target Gene Information
General Information of the m6A Target Gene (ID: M6ATAR00341)
Full List of m6A Methylation Regulator of This Target Gene and Corresponding Disease/Drug Response(s)
MYC
can be regulated by the following regulator(s), and cause disease/drug response(s). You can browse detail information of regulator(s) or disease/drug response(s).
Browse Regulator
Browse Disease
Browse Drug
Methyltransferase-like 3 (METTL3) [WRITER]
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | LNCaP cell line | Homo sapiens |
|
Treatment: shMETTL3 LNCaP cells
Control: shControl LNCaP cells
|
GSE147884 | |
| Regulation |
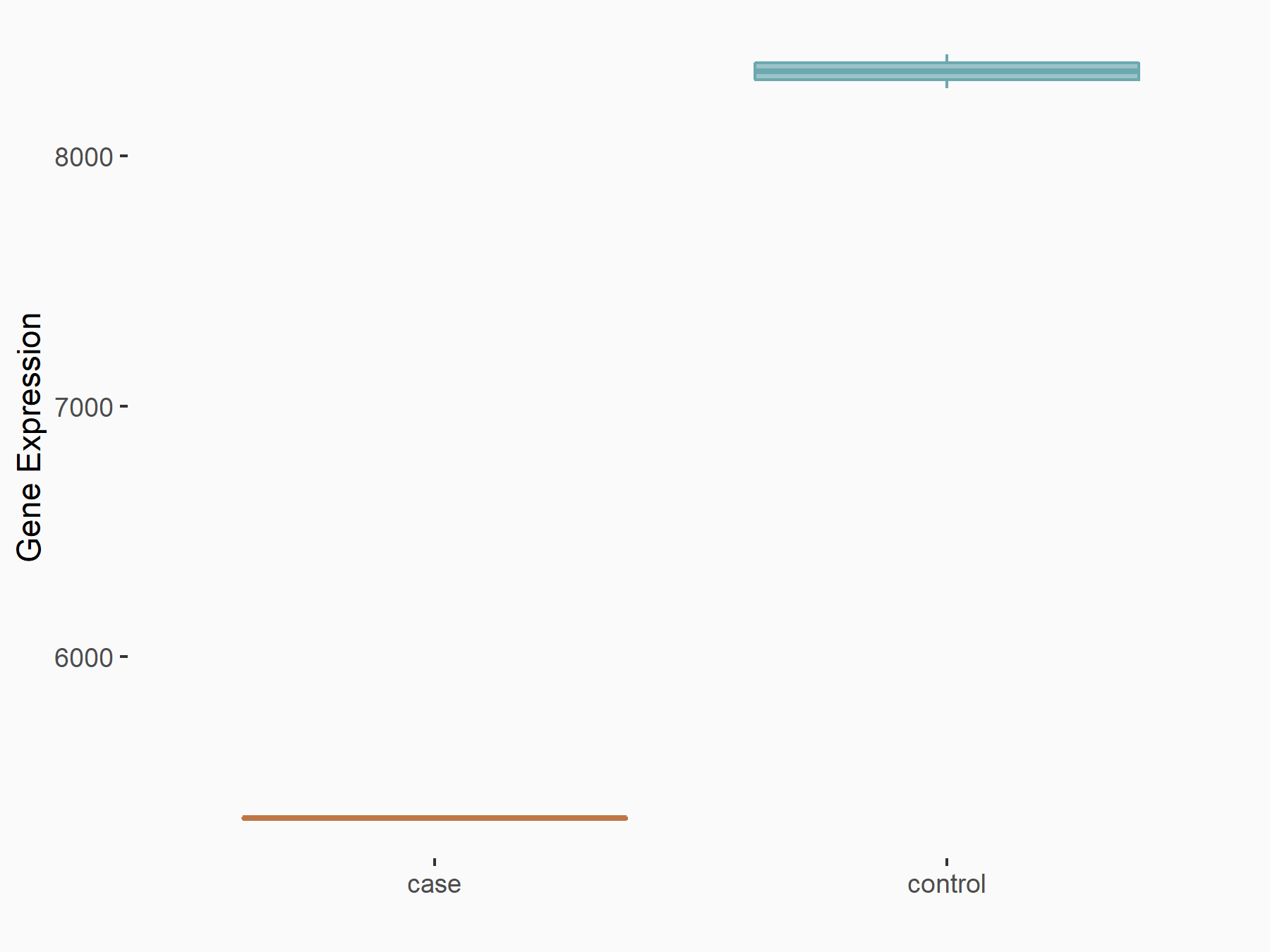 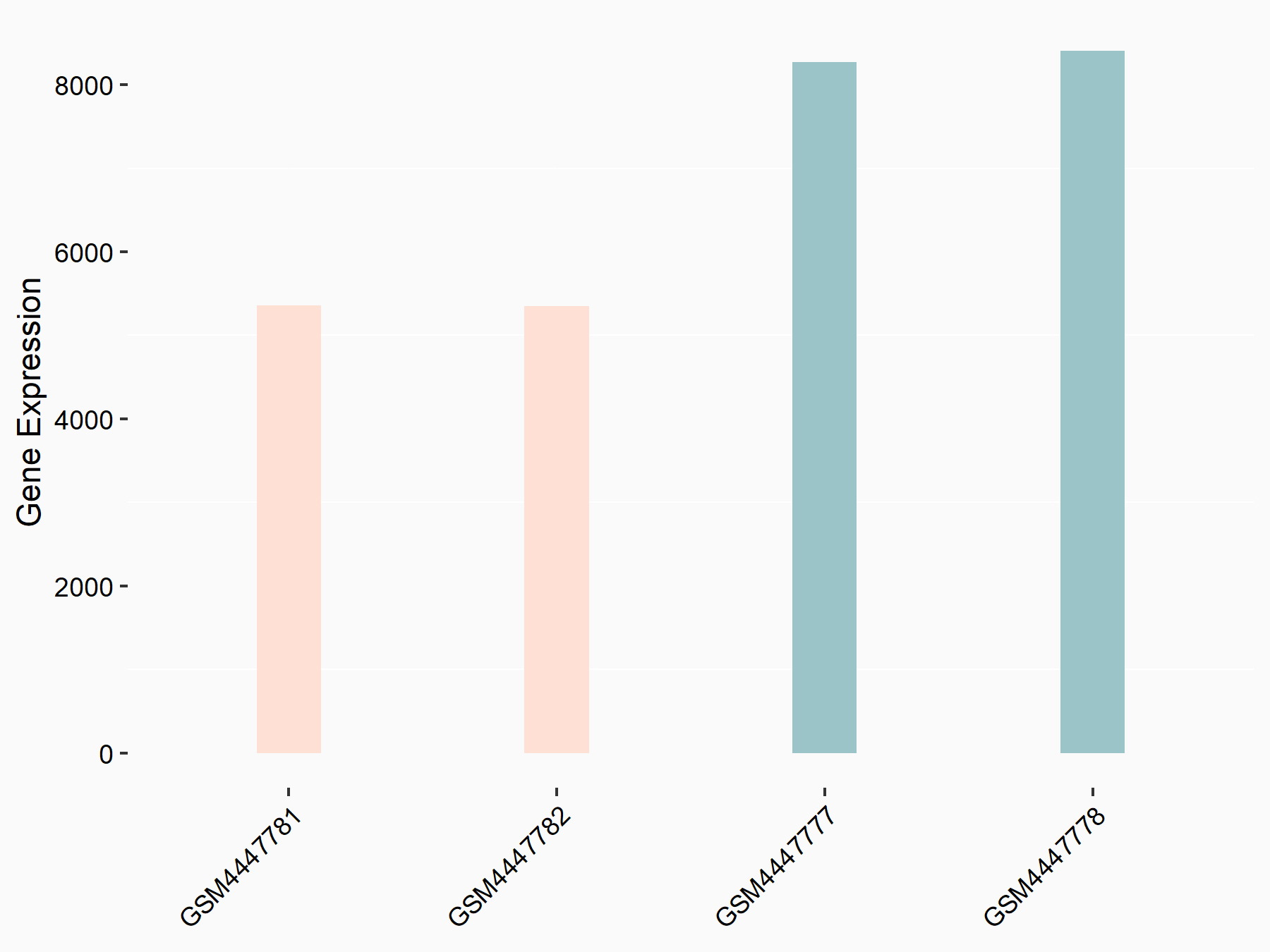 |
logFC: -6.39E-01 p-value: 4.30E-103 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between MYC and the regulator | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 1.10E+00 | GSE60213 |
| In total 12 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [1] | |||
| Response Summary | METTL3 depletion in human myeloid leukemia cell lines induces cell differentiation and apoptosis and delays leukemia progression in recipient mice in vivo. Single-nucleotide-resolution mapping of m6A coupled with ribosome profiling reveals that m6A promotes the translation of Myc proto-oncogene protein (MYC), BCL2 and PTEN mRNAs in the human acute myeloid leukemia MOLM-13 cell line. Moreover, loss of METTL3 leads to increased levels of phosphorylated AKT. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Acute myeloid leukaemia | ICD-11: 2A60 | ||
| Cell Process | Cell differentiation and apoptosis | |||
| Apoptosis (hsa04210) | ||||
| In-vitro Model | HSPC (Human hematopoietic stem cell) | |||
| In-vivo Model | 500,000 selected cells were injected via tail vein or retro-orbital route into female NSG (6-8 week old) recipient mice that had been sublethally irradiated with 475 cGy one day before transplantation. | |||
| Experiment 2 Reporting the m6A Methylation Regulator of This Target Gene | [2] | |||
| Response Summary | METTL3 level was slightly increased in AML-M5 patients,and its expression was significantly higher in immature cells than in mature monocytes.METTL3 acts as an oncogene in MOLM13 cells by upregulating Myc proto-oncogene protein (MYC) expression. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Acute myeloid leukaemia | ICD-11: 2A60 | ||
| Cell Process | Cell proliferation | |||
| In-vitro Model | MOLM-13 | Adult acute myeloid leukemia | Homo sapiens | CVCL_2119 |
| Experiment 3 Reporting the m6A Methylation Regulator of This Target Gene | [3] | |||
| Response Summary | In oral squamous cell carcinoma, YTH N6-methyladenosine RNA binding protein 1 (YTH domain family, member 1 [YTHDF1]) mediated the m6A-increased stability of Myc proto-oncogene protein (MYC) mRNA catalyzed by METTL3. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Oral squamous cell carcinoma | ICD-11: 2B6E.0 | ||
| Pathway Response | RNA degradation | hsa03018 | ||
| Cell Process | RNA stability | |||
| In-vitro Model | CAL-27 | Tongue squamous cell carcinoma | Homo sapiens | CVCL_1107 |
| NHOK (Normal oral keratinocytes) | ||||
| SCC-15 | Tongue squamous cell carcinoma | Homo sapiens | CVCL_1681 | |
| SCC-25 | Tongue squamous cell carcinoma | Homo sapiens | CVCL_1682 | |
| TSCCa | Endocervical adenocarcinoma | Homo sapiens | CVCL_VL15 | |
| In-vivo Model | The stable transfection of SCC25 cells (1 × 107 cells in 0.1 mL) with lenti-sh-METTL3 or blank vectors was injected subcutaneously into BALB/c nude mice. | |||
| Experiment 4 Reporting the m6A Methylation Regulator of This Target Gene | [4] | |||
| Response Summary | In gastric cancer, several component molecules (e.g., MCM5, MCM6, etc.) of Myc proto-oncogene protein (MYC) target genes were mediated by METTL3 via altered m6A modification. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Gastric cancer | ICD-11: 2B72 | ||
| Cell Process | Cell proliferation | |||
| Cell migration | ||||
| Cell invasion | ||||
| In-vitro Model | GES-1 | Normal | Homo sapiens | CVCL_EQ22 |
| HGC-27 | Gastric carcinoma | Homo sapiens | CVCL_1279 | |
| MGC-803 | Gastric mucinous adenocarcinoma | Homo sapiens | CVCL_5334 | |
| MKN45 | Gastric adenocarcinoma | Homo sapiens | CVCL_0434 | |
| MKN74 | Gastric tubular adenocarcinoma | Homo sapiens | CVCL_2791 | |
| pGCC (Primary GC cells) | ||||
| SGC-7901 | Gastric carcinoma | Homo sapiens | CVCL_0520 | |
| In-vivo Model | A total of 2 × 106 GC cells were injected into the flank of nude mice in a 1:1 suspension of BD Matrigel (BD Biosciences) in phosphate-buffered saline (PBS) solution. | |||
| Experiment 5 Reporting the m6A Methylation Regulator of This Target Gene | [5] | |||
| Response Summary | METTL3 enhanced Myc proto-oncogene protein (MYC) m6A methylation and increased MYC translation, which could potentiate the proliferation, migration and invasion of gastric cancer cells. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Gastric cancer | ICD-11: 2B72 | ||
| Cell Process | Cell proliferation | |||
| Cell migration | ||||
| Cell invasion | ||||
| Cell apoptosis | ||||
| In-vitro Model | AGS | Gastric adenocarcinoma | Homo sapiens | CVCL_0139 |
| AZ-521 | Duodenal adenocarcinoma | Homo sapiens | CVCL_2862 | |
| GES-1 | Normal | Homo sapiens | CVCL_EQ22 | |
| MKN45 | Gastric adenocarcinoma | Homo sapiens | CVCL_0434 | |
| SGC-7901 | Gastric carcinoma | Homo sapiens | CVCL_0520 | |
| In-vivo Model | The GC cell line MKN-45 stably infected with lentivirus expressing sh-HBXIP was prepared into 5 × 107 cells/mL cell suspension. | |||
| Experiment 6 Reporting the m6A Methylation Regulator of This Target Gene | [5] | |||
| Response Summary | Expressions of HBXIP, METTL3 and Myc proto-oncogene protein (MYC) were all determined to be upregulated in both GC tissues and cells. HBXIP plays an oncogenic role in GC via METTL3-mediated MYC mRNA m6A modification. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Gastric cancer | ICD-11: 2B72 | ||
| Cell Process | Cell proliferation | |||
| Cell migration | ||||
| Cell invasion | ||||
| Cell apoptosis | ||||
| In-vitro Model | SGC-7901 | Gastric carcinoma | Homo sapiens | CVCL_0520 |
| MKN45 | Gastric adenocarcinoma | Homo sapiens | CVCL_0434 | |
| GES-1 | Normal | Homo sapiens | CVCL_EQ22 | |
| AZ-521 | Duodenal adenocarcinoma | Homo sapiens | CVCL_2862 | |
| AGS | Gastric adenocarcinoma | Homo sapiens | CVCL_0139 | |
| In-vivo Model | The GC cell line MKN-45 stably infected with lentivirus expressing sh-HBXIP was prepared into 5 × 107 cells/mL cell suspension. The cell suspension was injected into the left axilla of nude mice using a 1 mL syringe as the sh-HBXIP group (n = 6). The GC cell line MKN-45 infected with the lentivirus expressing sh-NC was dispersed into the cell suspension, which was injected into nude mice as the sh-NC group (n = 6). Tumor growth was observed and data were recorded after inoculation. On the 26th day, all nude mice were euthanized by cervical dislocation and the tumors were resected and weighed. | |||
| Experiment 7 Reporting the m6A Methylation Regulator of This Target Gene | [6] | |||
| Response Summary | METTL3 exerted its function through enhancing Myc proto-oncogene protein (MYC) expression, at least partially in an m6A-IGF2BP1-dependent manner. Knockdown of METTL3 suppressed colorectal cancer cell proliferation in vitro and in vivo. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Colorectal cancer | ICD-11: 2B91 | ||
| Cell Process | Cell proliferation | |||
| In-vitro Model | HEK293T | Normal | Homo sapiens | CVCL_0063 |
| Caco-2 | Colon adenocarcinoma | Homo sapiens | CVCL_0025 | |
| HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 | |
| HT29 | Colon cancer | Mus musculus | CVCL_A8EZ | |
| LoVo | Colon adenocarcinoma | Homo sapiens | CVCL_0399 | |
| SW480 | Colon adenocarcinoma | Homo sapiens | CVCL_0546 | |
| SW620 | Colon adenocarcinoma | Homo sapiens | CVCL_0547 | |
| In-vivo Model | METTL3 stable knockdown or overexpression HCT116 cells were collected and resuspended at a density of 5 × 106 or 3 × 106 cells per 150 uL PBS. | |||
| Experiment 8 Reporting the m6A Methylation Regulator of This Target Gene | [7] | |||
| Response Summary | LCAT3 upregulation is attributable to N6-methyladenosine (m6A) modification mediated by methyltransferase like 3 (METTL3), leading to LCAT3 stabilization. LCAT3 as a novel oncogenic lncRNA in the lung, and validated the LCAT3-FUBP1-Myc proto-oncogene protein (MYC) axis as a potential therapeutic target for lung adenocarcinomas. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Lung adenocarcinoma | ICD-11: 2C25.0 | ||
| In-vitro Model | A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 |
| Calu-1 | Lung squamous cell carcinoma | Homo sapiens | CVCL_0608 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| HOP-62 | Lung adenocarcinoma | Homo sapiens | CVCL_1285 | |
| In-vivo Model | For the in vivo tumorigenicity assay, female BALB/c nude mice (ages 4-5 weeks) were randomly divided into two groups (n = 6/group). Calu1 cells (4 × 106) that had been stably transfected with sh-LCAT3 or scramble were implanted subcutaneously into the nude mice. Tumor growth was measured after one week, and tumor volumes were calculated with the following formula: Volume (cm3) = (length × width2)/2. After four weeks, the mice were euthanized, and the tumors were collected and weighed. For the in vivo tumor invasion assay, 1.2 × 106 scramble or shLCAT3 cells were injected intravenously into the tail vein of nude mice (n = 6/group). 1.5 mg luciferin (Gold Biotech, St Louis, MO, USA) was administered once a week for 4 weeks, to monitor metastases using an IVIS@ Lumina II system (Caliper Life Sciences, Hopkinton, MA, USA). | |||
| Experiment 9 Reporting the m6A Methylation Regulator of This Target Gene | [8] | |||
| Response Summary | This study revealed that m6A methylation is closely related to the poor prognosis of non-small cell lung cancer patients via interference with the TIME, which suggests that m6A plays a role in optimizing individualized immunotherapy management and improving prognosis. The expression levels of METTL3, FTO and YTHDF1 in non-small cell lung cancer were changed. Patients in Cluster 1 had lower immunoscores, higher programmed death-ligand 1 (PD-L1) expression, and shorter overall survival compared to patients in Cluster 2. The Myc proto-oncogene protein (MYC) targets, E2 transcription Factor (E2F) targets were significantly enriched. | |||
| Responsed Disease | Non-small-cell lung carcinoma | ICD-11: 2C25.Y | ||
| Pathway Response | p53 signaling pathway | hsa04115 | ||
| Central carbon metabolism in cancer | hsa05230 | |||
| PD-L1 expression and PD-1 checkpoint pathway in cancer | hsa05235 | |||
| Experiment 10 Reporting the m6A Methylation Regulator of This Target Gene | [9] | |||
| Response Summary | METTL3 enhanced Myc proto-oncogene protein (MYC) expression by increasing m6A levels of MYC mRNA transcript, leading to oncogenic functions in prostate cancer. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Prostate cancer | ICD-11: 2C82 | ||
| Cell Process | Cell proliferation | |||
| Cell migration | ||||
| Cell invasion | ||||
| In-vitro Model | LNCaP C4-2 | Prostate carcinoma | Homo sapiens | CVCL_4782 |
| DU145 | Prostate carcinoma | Homo sapiens | CVCL_0105 | |
| LNCaP | Prostate carcinoma | Homo sapiens | CVCL_0395 | |
| PC-3 | Prostate carcinoma | Homo sapiens | CVCL_0035 | |
| Experiment 11 Reporting the m6A Methylation Regulator of This Target Gene | [10] | |||
| Response Summary | AF4/FMR2 family member 4 (AFF4), two key regulators of NF-Kappa-B pathway (IKBKB and RELA) and Myc proto-oncogene protein (MYC) were further identified as direct targets of METTL3-mediated m6A modification.overexpression of METTL3 significantly promoted Bladder cancer cell growth and invasion. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Bladder cancer | ICD-11: 2C94 | ||
| Cell Process | Glucose metabolism | |||
| Experiment 12 Reporting the m6A Methylation Regulator of This Target Gene | [11] | |||
| Response Summary | Mettl3 activates the cyst-promoting c-Myc and cAMP pathways through enhanced Myc proto-oncogene protein (MYC) and Avpr2 mRNA m6A modification and translation. Thus, Mettl3 promotes Autosomal dominant polycystic kidney disease and links methionine utilization to epitranscriptomic activation of proliferation and cyst growth. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Polycystic kidney disease | ICD-11: GB81 | ||
| In-vitro Model | mIMCD-3 | Normal | Mus musculus | CVCL_0429 |
| In-vivo Model | The clone, with one wild-type Mettl3 allele and one L1L2_Bact_P cassette inserted allele, was injected into C57BL/6 blastocysts. Mettl3-targeted mouse line was established from a germline-transmitting chimera. The chimeric mouse was crossed to C57BL/6 Flp mice to excise the neomycin resistance system. | |||
YTH domain-containing family protein 1 (YTHDF1) [READER]
| Representative RNA-seq result indicating the expression of this target gene regulated by YTHDF1 | ||
| Cell Line | Embryonic stem cells | Mus musculus |
|
Treatment: YTHDF1-/- mESCs
Control: Wild type ESCs
|
GSE147849 | |
| Regulation |
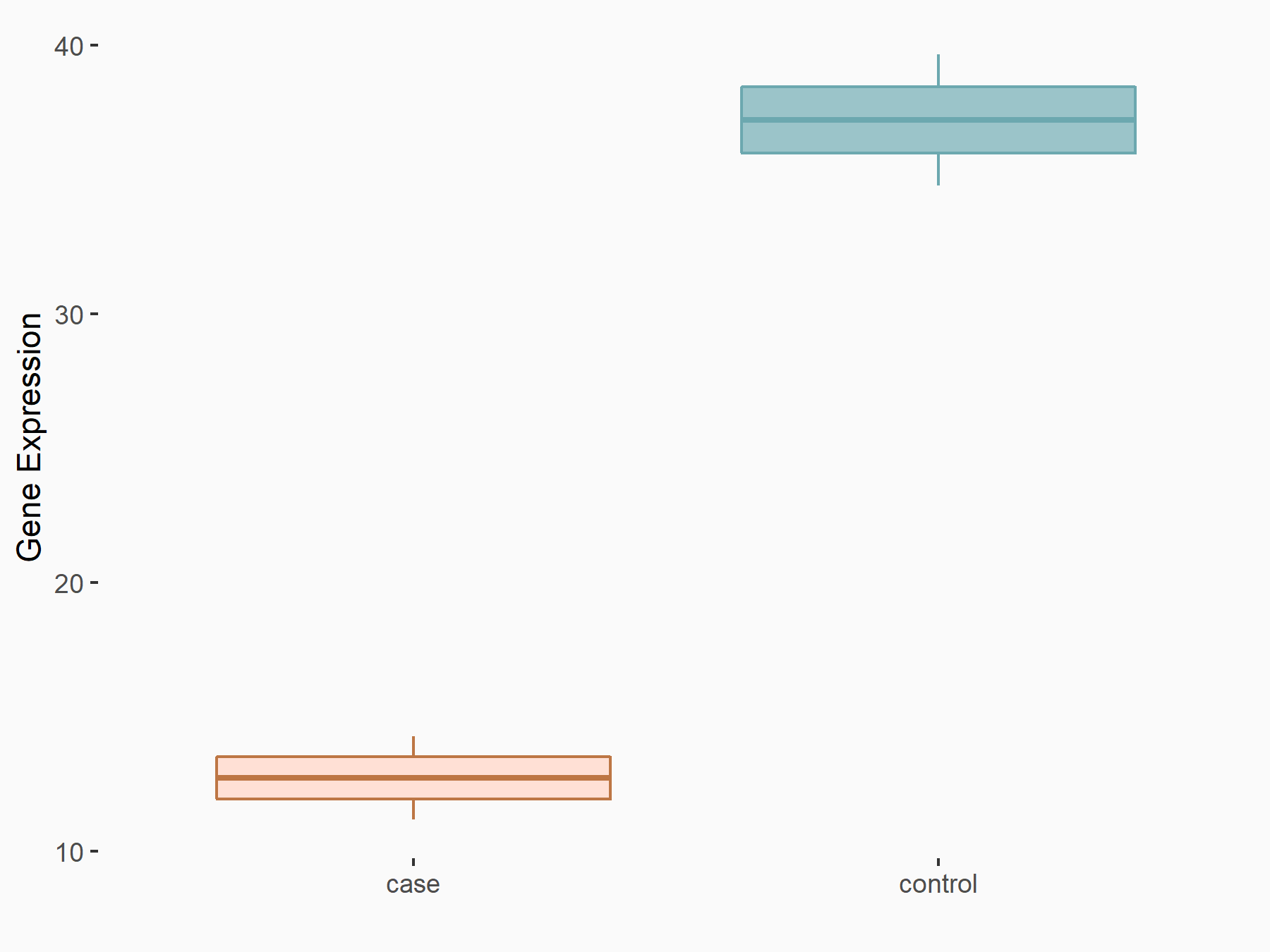 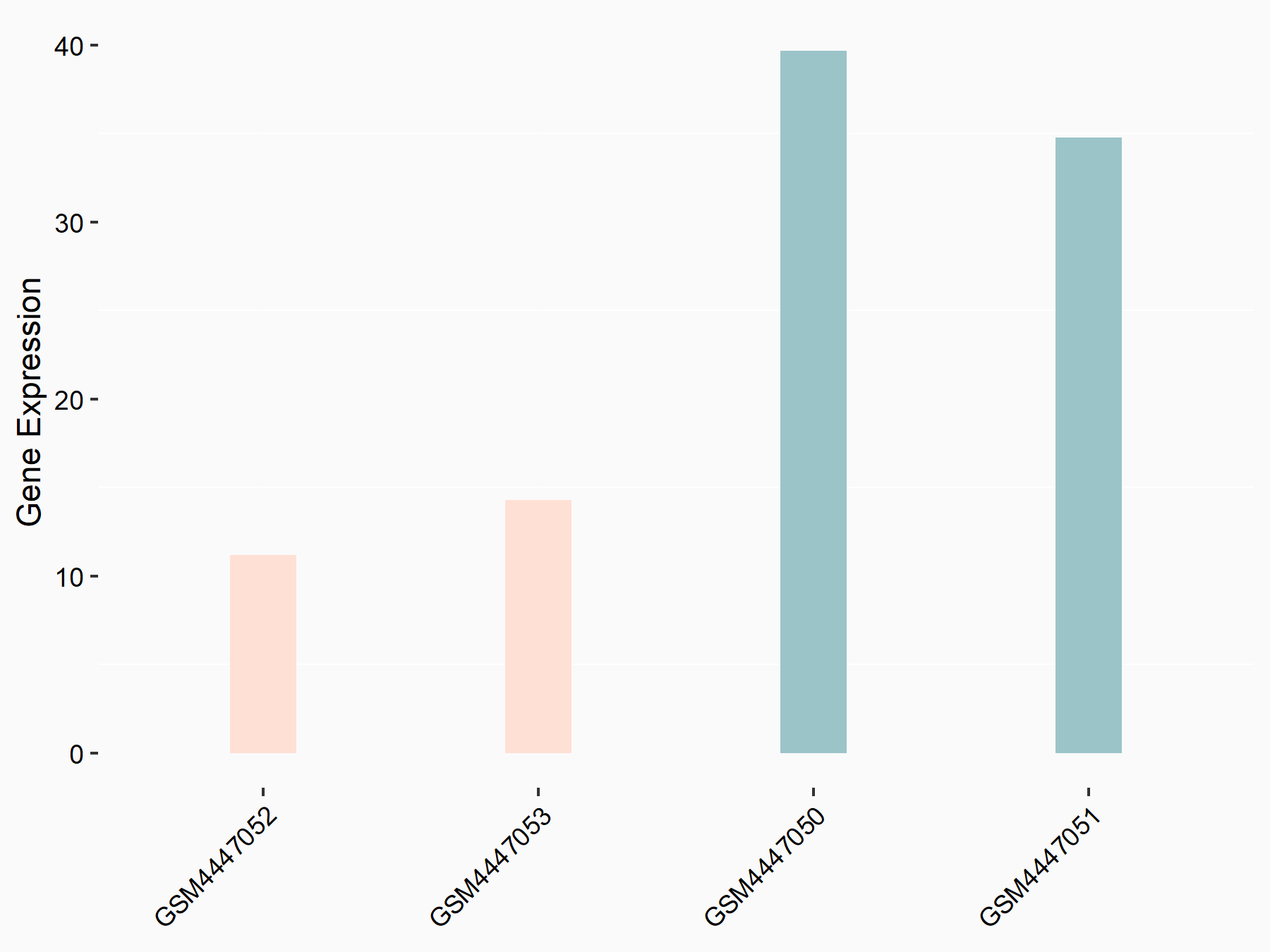 |
logFC: -1.55E+00 p-value: 2.76E-03 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between MYC and the regulator | ||
| Cell Line | Hela | Homo sapiens |
| Regulation | logFC: 1.21E+00 | GSE63591 |
| In total 2 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [3] | |||
| Response Summary | In oral squamous cell carcinoma, YTH N6-methyladenosine RNA binding protein 1 (YTH domain family, member 1 [YTHDF1]) mediated the m6A-increased stability of Myc proto-oncogene protein (MYC) mRNA catalyzed by METTL3. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Oral squamous cell carcinoma | ICD-11: 2B6E.0 | ||
| Pathway Response | RNA degradation | hsa03018 | ||
| Cell Process | RNA stability | |||
| In-vitro Model | CAL-27 | Tongue squamous cell carcinoma | Homo sapiens | CVCL_1107 |
| NHOK (Normal oral keratinocytes) | ||||
| SCC-15 | Tongue squamous cell carcinoma | Homo sapiens | CVCL_1681 | |
| SCC-25 | Tongue squamous cell carcinoma | Homo sapiens | CVCL_1682 | |
| TSCCa | Endocervical adenocarcinoma | Homo sapiens | CVCL_VL15 | |
| In-vivo Model | The stable transfection of SCC25 cells (1 × 107 cells in 0.1 mL) with lenti-sh-METTL3 or blank vectors was injected subcutaneously into BALB/c nude mice. | |||
| Experiment 2 Reporting the m6A Methylation Regulator of This Target Gene | [8] | |||
| Response Summary | This study revealed that m6A methylation is closely related to the poor prognosis of non-small cell lung cancer patients via interference with the TIME, which suggests that m6A plays a role in optimizing individualized immunotherapy management and improving prognosis. The expression levels of METTL3, FTO and YTHDF1 in non-small cell lung cancer were changed. Patients in Cluster 1 had lower immunoscores, higher programmed death-ligand 1 (PD-L1) expression, and shorter overall survival compared to patients in Cluster 2. The Myc proto-oncogene protein (MYC) targets, E2 transcription Factor (E2F) targets were significantly enriched. | |||
| Responsed Disease | Non-small-cell lung carcinoma | ICD-11: 2C25.Y | ||
| Pathway Response | p53 signaling pathway | hsa04115 | ||
| Central carbon metabolism in cancer | hsa05230 | |||
| PD-L1 expression and PD-1 checkpoint pathway in cancer | hsa05235 | |||
YTH domain-containing family protein 2 (YTHDF2) [READER]
| Representative RNA-seq result indicating the expression of this target gene regulated by YTHDF2 | ||
| Cell Line | GSC11 cell line | Homo sapiens |
|
Treatment: siYTHDF2 GSC11 cells
Control: siControl GSC11 cells
|
GSE142825 | |
| Regulation |
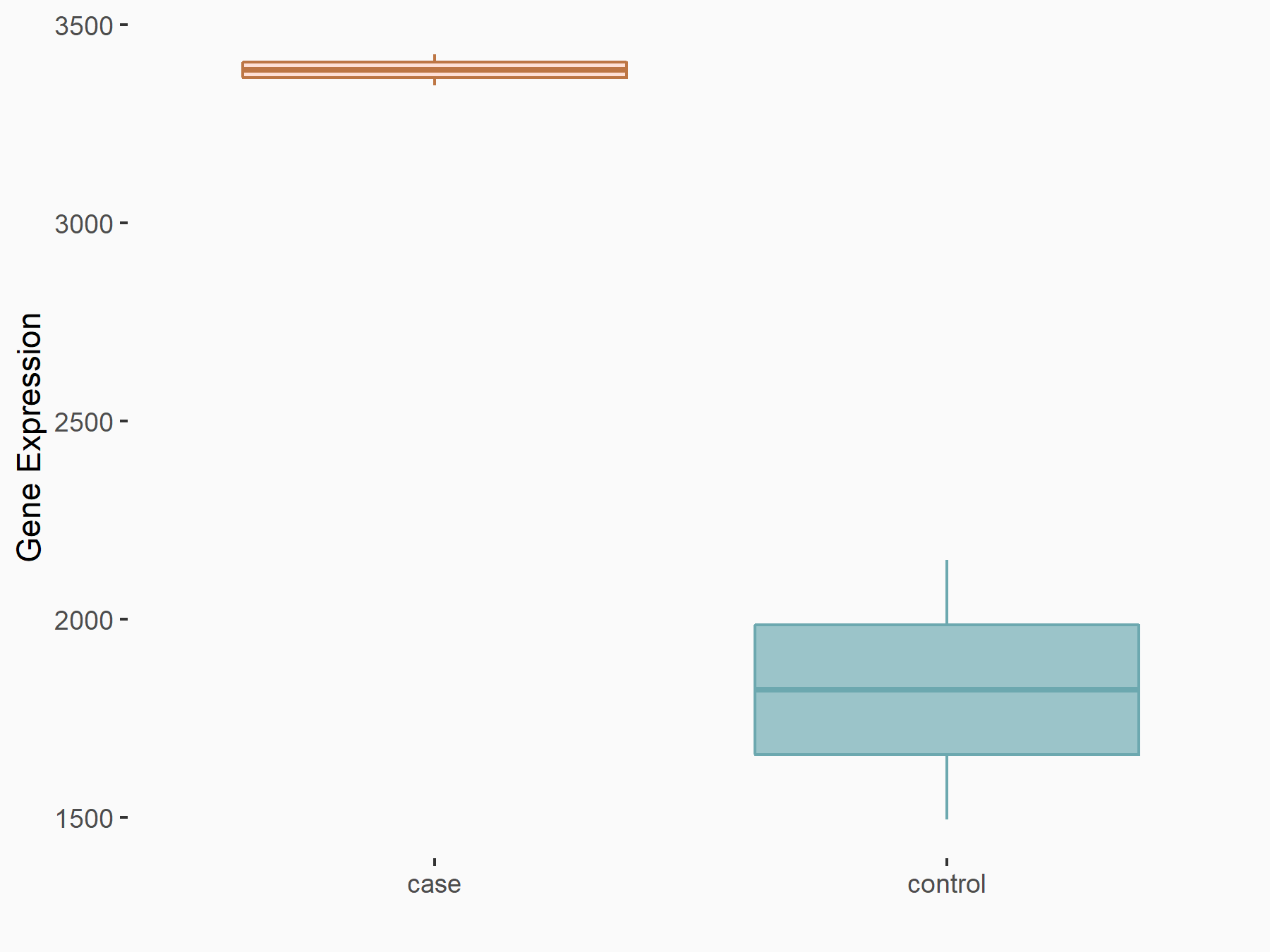  |
logFC: 8.94E-01 p-value: 8.57E-07 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between MYC and the regulator | ||
| Cell Line | Hela | Homo sapiens |
| Regulation | logFC: 1.40E+00 | GSE49339 |
| In total 3 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [12] | |||
| Response Summary | The m6A reader YTHDF2 stabilized Myc proto-oncogene protein (MYC) mRNA specifically in cancer stem cells. Given the challenge of targeting MYC, YTHDF2 presents a therapeutic target to perturb MYC signaling in glioblastoma. The IGF1/IGF1R inhibitor linsitinib preferentially targeted YTHDF2-expressing cells, inhibiting GSC viability without affecting NSCs and impairing in vivo glioblastoma growth. YTHDF2 links RNA epitranscriptomic modifications and GSC growth, laying the foundation for the YTHDF2-MYC-IGFBP3 axis as a specific and novel therapeutic target in glioblastoma. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Glioma | ICD-11: 2A00.0 | ||
| Responsed Drug | Linsitinib | Phase 3 | ||
| Pathway Response | RNA degradation | hsa03018 | ||
| Cell Process | RNA stability | |||
| In-vitro Model | () | |||
| HNP1 (A human neural progenitor cell) | ||||
| NHA (Normal human astrocytes) | ||||
| NSC11 (Pluripotent derived neural progenitor cell) | ||||
| In-vivo Model | For in vivo drug treatment studies, intracranial xenografts were generated by implanting 5000 patient-derived GSCs (387 and 4121) into the right cerebral cortex of NSG mice as described above. | |||
| Experiment 2 Reporting the m6A Methylation Regulator of This Target Gene | [12] | |||
| Response Summary | The IGF1/IGF1R inhibitor, linsitinib for further investigation based upon the role of the IGF pathway member, IGFBP3, as a downstream effector of YTHDF2-Myc proto-oncogene protein (MYC) axis in GSCs. Inhibiting glioblastoma stem cells viability without affecting NSCs and impairing in vivo glioblastoma growth. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Glioblastoma | ICD-11: 2A00.00 | ||
| Responsed Drug | Linsitinib | Phase 3 | ||
| In-vitro Model | NSC11 (Pluripotent derived neural progenitor cell) | |||
| NHA (Normal human astrocytes) | ||||
| HNP1 (A human neural progenitor cell) | ||||
| ENSA (A human embryonic stem derived neural progenitor cell) | ||||
| In-vivo Model | Implanting 5000 human derived GSCs into the right cerebral cortex of NSG mice at a depth of 3.5 mm under a University of California, San Diego Institutional Animal Care and Use Committee (IACUC) approved protocol. Brains were harvested and fixed in 4% formaldehyde, cryopreserved in 30% sucrose, and then cryosectioned. Hematoxylin and eosin (H&E) staining was performed on sections for histological analysis. In parallel survival experiments, mice were observed until the development of neurological signs. For in vivo drug treatment studies, intracranial xenografts were generated by implanting 5000 patient-derived GSCs (387 and 4121) into the right cerebral cortex of NSG mice as described above. Mice recovered for 7 days were randomly assigned into drug vs. treatment group by a blinded investigator. Mice were then treated daily with either vehicle (25 mM Tartaric acid) or 50 mg/kg linsitinib by oral gavage. | |||
| Experiment 3 Reporting the m6A Methylation Regulator of This Target Gene | [13] | |||
| Response Summary | LCAT3 upregulation is attributable to m6A modification mediated by METTL3, leading to LCAT3 stabilization. Treated cells with tamoxifen to induce MYC activity. Highlights the therapeutic potential of RBPs by uncovering a critical role for YTHDF2 in counteracting the global increase of mRNA synthesis in Myc proto-oncogene protein (MYC)-driven breast cancers. | |||
| Responsed Disease | Breast cancer | ICD-11: 2C60 | ||
| Responsed Drug | Tamoxifen | Approved | ||
| Pathway Response | MAPK signaling pathway | hsa04010 | ||
| Cell Process | Epithelial-to-mesenchymal transition | |||
| Cell apoptosis | ||||
| In-vitro Model | HEK293T | Normal | Homo sapiens | CVCL_0063 |
| MCF-7 | Invasive breast carcinoma | Homo sapiens | CVCL_0031 | |
| MDA-MB-231 | Breast adenocarcinoma | Homo sapiens | CVCL_0062 | |
| MBA-MD-231 (Human breast cancer cell) | ||||
| MYC-ER HMEC (Human mammary epithelial cells expressing a MYC estrogen receptor fusion) | ||||
| SK-BR-3 | Breast adenocarcinoma | Homo sapiens | CVCL_0033 | |
| In-vivo Model | To induce recombination at 8 weeks of age both CAG-CreERT;Ythdf2fl/fl and Ythdf2fl/fl littermates were injected with 75mg/kg body weight tamoxifen dissolved in corn oil daily for 5 days. | |||
Fat mass and obesity-associated protein (FTO) [ERASER]
| Representative RNA-seq result indicating the expression of this target gene regulated by FTO | ||
| Cell Line | NB4 cell line | Homo sapiens |
|
Treatment: shFTO NB4 cells
Control: shNS NB4 cells
|
GSE103494 | |
| Regulation |
  |
logFC: -6.81E-01 p-value: 3.45E-03 |
| More Results | Click to View More RNA-seq Results | |
| In total 8 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [14] | |||
| Response Summary | This work demonstrates anti-tumor effects of 2HG in inhibiting proliferation/survival of FTO-high cancer cells via targeting FTO/m6A/Myc proto-oncogene protein (MYC)/CEBPA signaling.High levels of FTO sensitize leukemia cells to R-2HG, whereas hyperactivation of MYC signaling confers resistance that can be reversed by the inhibition of MYC signaling. R-2HG also displays anti-tumor activity in glioma. High levels of FTO sensitize leukemic cells to R-2HG, whereas hyperactivation of MYC signaling confers resistance that can be reversed by the inhibition of MYC signaling. | |||
| Target Regulation | Down regulation | |||
| Responsed Disease | Glioma | ICD-11: 2A00.0 | ||
| Responsed Drug | R-2HG | Investigative | ||
| Cell Process | Glutamine metabolism | |||
| Cell apoptosis | ||||
| In-vitro Model | 8-MG-BA | Glioblastoma | Homo sapiens | CVCL_1052 |
| A-172 | Glioblastoma | Homo sapiens | CVCL_0131 | |
| DK-MG | Glioblastoma | Homo sapiens | CVCL_1173 | |
| GaMG | Glioblastoma | Homo sapiens | CVCL_1226 | |
| HEL | Erythroleukemia | Homo sapiens | CVCL_0001 | |
| Jurkat | T acute lymphoblastic leukemia | Homo sapiens | CVCL_0065 | |
| KOCL-45 | B acute lymphoblastic leukemia | Homo sapiens | CVCL_3993 | |
| KOCL-48 | Childhood acute monocytic leukemia | Homo sapiens | CVCL_6867 | |
| KOCL-50 | B acute lymphoblastic leukemia | Homo sapiens | CVCL_6866 | |
| KOCL-51 | B acute lymphoblastic leukemia | Homo sapiens | CVCL_6865 | |
| KOCL-69 | B acute lymphoblastic leukemia | Homo sapiens | CVCL_3995 | |
| KOPN-1 | B acute lymphoblastic leukemia | Homo sapiens | CVCL_3937 | |
| LN-18 | Glioblastoma | Homo sapiens | CVCL_0392 | |
| LN-229 | Glioblastoma | Homo sapiens | CVCL_0393 | |
| MA9.3 (MA9.3) | ||||
| MA9.6ITD (MLL-AF9 plus FLT3-ITD) | ||||
| MA9.6RAS (MLL-AF9 plus NRasG12D) | ||||
| MA9.6 (MLL-AF9) | ||||
| MA9.6ITD (MLL-AF9 plus FLT3-ITD) | ||||
| MA9.6RAS (MLL-AF9 plus NRasG12D) | ||||
| ME-1 [Human leukemia] | Adult acute myeloid leukemia | Homo sapiens | CVCL_2110 | |
| ML-2 | Adult acute myeloid leukemia | Homo sapiens | CVCL_1418 | |
| MV4-11 | Childhood acute monocytic leukemia | Homo sapiens | CVCL_0064 | |
| NB4 | Acute promyelocytic leukemia | Homo sapiens | CVCL_0005 | |
| NOMO-1 | Adult acute monocytic leukemia | Homo sapiens | CVCL_1609 | |
| PL21 | Familial adenomatous polyposis | Homo sapiens | CVCL_JM48 | |
| T98G | Glioblastoma | Homo sapiens | CVCL_0556 | |
| THP-1 | Childhood acute monocytic leukemia | Homo sapiens | CVCL_0006 | |
| U-87MG ATCC | Glioblastoma | Homo sapiens | CVCL_0022 | |
| U-937 | Adult acute monocytic leukemia | Homo sapiens | CVCL_0007 | |
| In-vivo Model | For R-2HG injection mouse models, sensitive (NOMO-1 and MA9.3ITD) or resistant (MA9.3RAS) cells were injected into NSGS or NRGS intravenously, and then R-2HG (6mg/kg body weight) or PBS were injected once daily through tail vein for 12 consecutive days starting from day 11 post xeno-transplantation. | |||
| Experiment 2 Reporting the m6A Methylation Regulator of This Target Gene | [15] | |||
| Response Summary | Wnt/Beta-catenin-mediated FTO downregulation and underscored the role of m6A modifications of Myc proto-oncogene protein (MYC) mRNA in regulating tumor cell glycolysis and growth. | |||
| Target Regulation | Down regulation | |||
| Responsed Disease | Solid tumour/cancer | ICD-11: 2A00-2F9Z | ||
| Pathway Response | Wnt signaling pathway | hsa04310 | ||
| Central carbon metabolism in cancer | hsa05230 | |||
| Glycolysis / Gluconeogenesis | hsa00010 | |||
| Cell Process | Glycolysis | |||
| In-vitro Model | NCI-H322 | Minimally invasive lung adenocarcinoma | Homo sapiens | CVCL_1556 |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| In-vivo Model | Mice were randomized into several groups. For the subcutaneous implantation model, 1 × 106 cells were injected subcutaneously into the flank regions of female BALB/c nude mice (4-5 weeks). For lung colonization assays, 1 × 106 cells were injected into the tail vein of female NOD/SCID mice (6-7 weeks), and 6 weeks later the lung was removed and fixed with 10% formalin. | |||
| Experiment 3 Reporting the m6A Methylation Regulator of This Target Gene | [14] | |||
| Response Summary | This work demonstrates anti-tumor effects of 2HG in inhibiting proliferation/survival of FTO-high cancer cells via targeting FTO/m6A/Myc proto-oncogene protein (MYC)/CEBPA signaling.High levels of FTO sensitize leukemia cells to R-2HG, whereas hyperactivation of MYC signaling confers resistance that can be reversed by the inhibition of MYC signaling. R-2HG also displays anti-tumor activity in glioma. High levels of FTO sensitize leukemic cells to R-2HG, whereas hyperactivation of MYC signaling confers resistance that can be reversed by the inhibition of MYC signaling. | |||
| Target Regulation | Down regulation | |||
| Responsed Disease | Leukaemia | ICD-11: 2B33.4 | ||
| Responsed Drug | R-2HG | Investigative | ||
| Cell Process | Glutamine metabolism | |||
| Cell apoptosis | ||||
| In-vitro Model | 8-MG-BA | Glioblastoma | Homo sapiens | CVCL_1052 |
| A-172 | Glioblastoma | Homo sapiens | CVCL_0131 | |
| DK-MG | Glioblastoma | Homo sapiens | CVCL_1173 | |
| GaMG | Glioblastoma | Homo sapiens | CVCL_1226 | |
| HEL | Erythroleukemia | Homo sapiens | CVCL_0001 | |
| Jurkat | T acute lymphoblastic leukemia | Homo sapiens | CVCL_0065 | |
| KOCL-45 | B acute lymphoblastic leukemia | Homo sapiens | CVCL_3993 | |
| KOCL-48 | Childhood acute monocytic leukemia | Homo sapiens | CVCL_6867 | |
| KOCL-50 | B acute lymphoblastic leukemia | Homo sapiens | CVCL_6866 | |
| KOCL-51 | B acute lymphoblastic leukemia | Homo sapiens | CVCL_6865 | |
| KOCL-69 | B acute lymphoblastic leukemia | Homo sapiens | CVCL_3995 | |
| KOPN-1 | B acute lymphoblastic leukemia | Homo sapiens | CVCL_3937 | |
| LN-18 | Glioblastoma | Homo sapiens | CVCL_0392 | |
| LN-229 | Glioblastoma | Homo sapiens | CVCL_0393 | |
| MA9.3 (MA9.3) | ||||
| MA9.6ITD (MLL-AF9 plus FLT3-ITD) | ||||
| MA9.6RAS (MLL-AF9 plus NRasG12D) | ||||
| MA9.6 (MLL-AF9) | ||||
| MA9.6ITD (MLL-AF9 plus FLT3-ITD) | ||||
| MA9.6RAS (MLL-AF9 plus NRasG12D) | ||||
| ME-1 [Human leukemia] | Adult acute myeloid leukemia | Homo sapiens | CVCL_2110 | |
| ML-2 | Adult acute myeloid leukemia | Homo sapiens | CVCL_1418 | |
| MV4-11 | Childhood acute monocytic leukemia | Homo sapiens | CVCL_0064 | |
| NB4 | Acute promyelocytic leukemia | Homo sapiens | CVCL_0005 | |
| NOMO-1 | Adult acute monocytic leukemia | Homo sapiens | CVCL_1609 | |
| PL21 | Familial adenomatous polyposis | Homo sapiens | CVCL_JM48 | |
| T98G | Glioblastoma | Homo sapiens | CVCL_0556 | |
| THP-1 | Childhood acute monocytic leukemia | Homo sapiens | CVCL_0006 | |
| U-87MG ATCC | Glioblastoma | Homo sapiens | CVCL_0022 | |
| U-937 | Adult acute monocytic leukemia | Homo sapiens | CVCL_0007 | |
| In-vivo Model | For R-2HG injection mouse models, sensitive (NOMO-1 and MA9.3ITD) or resistant (MA9.3RAS) cells were injected into NSGS or NRGS intravenously, and then R-2HG (6mg/kg body weight) or PBS were injected once daily through tail vein for 12 consecutive days starting from day 11 post xeno-transplantation. | |||
| Experiment 4 Reporting the m6A Methylation Regulator of This Target Gene | [16] | |||
| Response Summary | MiR-96 antagomir could potentially retard the cancerogenesis in colorectal cancer via AMPK-alpha-2-dependent inhibition of FTO and blocking FTO-mediated m6A modification of Myc proto-oncogene protein (MYC). | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Colorectal cancer | ICD-11: 2B91 | ||
| Cell Process | Cell proliferation | |||
| Cell apoptosis | ||||
| In-vitro Model | HCT 8 | Colon adenocarcinoma | Homo sapiens | CVCL_2478 |
| SW480 | Colon adenocarcinoma | Homo sapiens | CVCL_0546 | |
| SW620 | Colon adenocarcinoma | Homo sapiens | CVCL_0547 | |
| In-vivo Model | RC cells SW480 at logarithmic growth phase were prepared into cell suspension with a concentration of about 1 × 107/100 L, which was then injected into the left axilla of nude mice with a 1 ml syringe to establish a subcutaneous mouse xenograft model. Once the tumor volume reached about 50 mm3, the nude mice were injected with miR-96 antagomir or NC antagomir (10 nmol once every 5 days for 5 weeks). After 5 weeks, the mice were euthanized, after which the subcutaneous transplanted tumor was removed, and weighed. | |||
| Experiment 5 Reporting the m6A Methylation Regulator of This Target Gene | [17] | |||
| Response Summary | GSK3beta inhibited MZF1 expression by mediating FTO-regulated m6A modification of MZF1 and then decreased the proto-oncogene Myc proto-oncogene protein (MYC) expression, thus hampering CRC cell proliferation. | |||
| Target Regulation | Down regulation | |||
| Responsed Disease | Colorectal cancer | ICD-11: 2B91 | ||
| Pathway Response | Ubiquitin mediated proteolysis | hsa04120 | ||
| Cell Process | Cell proliferation | |||
| In-vitro Model | SW620 | Colon adenocarcinoma | Homo sapiens | CVCL_0547 |
| SW480 | Colon adenocarcinoma | Homo sapiens | CVCL_0546 | |
| HCT 8 | Colon adenocarcinoma | Homo sapiens | CVCL_2478 | |
| In-vivo Model | Twenty-four specific pathogen free female BALB/c nude mice (age: 6 weeks, weight: 15 ~ 18 g) were purchased from Slac Laboratory Animal Co., Ltd., and subcutaneously injected with SW620 cells stably transfected with oe-NC, oe-GSK3-Beta + oe-NC, or oe-GSK3-Beta + oe-c-Myc to establish a subcutaneous xenograft tumour model in nude mice. | |||
| Experiment 6 Reporting the m6A Methylation Regulator of This Target Gene | [18] | |||
| Response Summary | FTO has been indicated to interact with Myc proto-oncogene protein (MYC) proto-oncogene, bHLH transcription factor and to enhance its stability by decreasing its m6A level.the aforementioned observations indicate a novel mechanism for the regulation of pancreatic cancer cells by FTO. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Pancreatic cancer | ICD-11: 2C10 | ||
| Cell Process | Cell proliferation | |||
| In-vitro Model | BxPC-3 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0186 |
| HPDE | Normal | Homo sapiens | CVCL_4376 | |
| PANC-1 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0480 | |
| SW1990 | Pancreatic adenocarcinoma | Homo sapiens | CVCL_1723 | |
| Experiment 7 Reporting the m6A Methylation Regulator of This Target Gene | [8] | |||
| Response Summary | This study revealed that m6A methylation is closely related to the poor prognosis of non-small cell lung cancer patients via interference with the TIME, which suggests that m6A plays a role in optimizing individualized immunotherapy management and improving prognosis. The expression levels of METTL3, FTO and YTHDF1 in non-small cell lung cancer were changed. Patients in Cluster 1 had lower immunoscores, higher programmed death-ligand 1 (PD-L1) expression, and shorter overall survival compared to patients in Cluster 2. The Myc proto-oncogene protein (MYC) targets, E2 transcription Factor (E2F) targets were significantly enriched. | |||
| Responsed Disease | Non-small-cell lung carcinoma | ICD-11: 2C25.Y | ||
| Pathway Response | p53 signaling pathway | hsa04115 | ||
| Central carbon metabolism in cancer | hsa05230 | |||
| PD-L1 expression and PD-1 checkpoint pathway in cancer | hsa05235 | |||
| Experiment 8 Reporting the m6A Methylation Regulator of This Target Gene | [19] | |||
| Response Summary | FTO interacts with transcripts of E2F1 and Myc proto-oncogene protein (MYC), inhibition of FTO significantly impairs the translation efficiency of E2F1 and Myc.FTO plays important oncogenic role in regulating cervical cancer cells' proliferation. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Cervical cancer | ICD-11: 2C77 | ||
| Cell Process | Cell proliferation and migration | |||
| In-vitro Model | HeLa | Endocervical adenocarcinoma | Homo sapiens | CVCL_0030 |
| SiHa | Cervical squamous cell carcinoma | Homo sapiens | CVCL_0032 | |
Insulin-like growth factor 2 mRNA-binding protein 1 (IGF2BP1) [READER]
| Representative RNA-seq result indicating the expression of this target gene regulated by IGF2BP1 | ||
| Cell Line | HepG2 cell line | Homo sapiens |
|
Treatment: siIGF2BP1 HepG2 cells
Control: siControl HepG2 cells
|
GSE161086 | |
| Regulation |
  |
logFC: -9.94E-01 p-value: 1.72E-04 |
| More Results | Click to View More RNA-seq Results | |
| In total 4 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [20] | |||
| Response Summary | YBX1 selectively functions in regulating survival of myeloid leukemia cells. YBX1 interacts with insulin-like growth factor 2 messenger RNA (mRNA)-binding proteins (IGF2BPs) and stabilizes m6A-tagged RNA. YBX1 deficiency dysregulates the expression of apoptosis-related genes and promotes mRNA decay of Myc proto-oncogene protein (MYC) and BCL2 in an m6A-dependent manner, which contributes to the defective survival that results from deletion of YBX1. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Myeloid leukaemia | ICD-11: 2B33.1 | ||
| Cell Process | Cell apoptosis | |||
| In-vitro Model | Leukemia stem cell line (Leukemia stem cell line) | |||
| Kasumi-1 | Myeloid leukemia with maturation | Homo sapiens | CVCL_0589 | |
| MOLM-13 | Adult acute myeloid leukemia | Homo sapiens | CVCL_2119 | |
| THP-1 | Childhood acute monocytic leukemia | Homo sapiens | CVCL_0006 | |
| MV4-11 | Childhood acute monocytic leukemia | Homo sapiens | CVCL_0064 | |
| BV-173 | Chronic myelogenous leukemia | Homo sapiens | CVCL_0181 | |
| NOMO-1 | Adult acute monocytic leukemia | Homo sapiens | CVCL_1609 | |
| K-562 | Chronic myelogenous leukemia | Homo sapiens | CVCL_0004 | |
| KG-1a | Adult acute myeloid leukemia | Homo sapiens | CVCL_1824 | |
| Experiment 2 Reporting the m6A Methylation Regulator of This Target Gene | [21] | |||
| Response Summary | IGF2BP1 upregulated in GC tissue and acted as a predictor of poor prognosis for GC patients. IGF2BP1 directly interacted with Myc proto-oncogene protein (MYC) mRNA via m6A-dependent manner to by stabilize its stability. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Gastric cancer | ICD-11: 2B72 | ||
| Pathway Response | Central carbon metabolism in cancer | hsa05230 | ||
| Glycolysis / Gluconeogenesis | hsa00010 | |||
| Cell Process | Aerobic glycolysis | |||
| In-vitro Model | SNU-216 | Gastric tubular adenocarcinoma | Homo sapiens | CVCL_3946 |
| MKN45 | Gastric adenocarcinoma | Homo sapiens | CVCL_0434 | |
| HGC-27 | Gastric carcinoma | Homo sapiens | CVCL_1279 | |
| GES-1 | Normal | Homo sapiens | CVCL_EQ22 | |
| AGS | Gastric adenocarcinoma | Homo sapiens | CVCL_0139 | |
| In-vivo Model | A total of 5 × 106 transfected MKN-45 cells, stably transfected with sh-IGF2BP1 vector or empty vector were subcutaneously injected into the flank of the mice. Tumor growth was measured every three days, and calculated using the following equation = a × b2/2 (a for longitudinal diameter; and b for latitudinal diameter). Three weeks after injection, mice were sacrificed. | |||
| Experiment 3 Reporting the m6A Methylation Regulator of This Target Gene | [22] | |||
| Response Summary | In contrast to the mRNA-decay-promoting function of YTH domain-containing family protein 2, IGF2BPs promote the stability and storage of their target mRNAs (for example, MYC) in an m6A-dependent manner under normal and stress conditions and therefore affect gene expression output. Four representative high confidence targets, including Myc proto-oncogene protein (MYC), FSCN1, TK1, and MARCKSL1, exhibit strong binding with IGF2BPs around their m6A motifs in control cells. Knocking down of each individual IGF2BPs in Hela (cervical cancer) and HepG2 (liver cancer) cells significantly repressed MYC expression. | |||
| Responsed Disease | Hepatocellular carcinoma | ICD-11: 2C12.02 | ||
| Cell Process | RNA decay | |||
| In-vitro Model | Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| HeLa | Endocervical adenocarcinoma | Homo sapiens | CVCL_0030 | |
| Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 | |
| Experiment 4 Reporting the m6A Methylation Regulator of This Target Gene | [23] | |||
| Response Summary | Hypoxia-induced lncRNA KB-1980E6.3 is involved in the self-renewal and stemness maintenance of breast cancer stem cells by recruiting IGF2BP1 to regulate Myc proto-oncogene protein (MYC) mRNA stability. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Breast cancer | ICD-11: 2C60 | ||
| Pathway Response | Signaling pathways regulating pluripotency of stem cells | hsa04550 | ||
| In-vitro Model | BT-474 | Invasive breast carcinoma | Homo sapiens | CVCL_0179 |
| BT-549 | Invasive breast carcinoma | Homo sapiens | CVCL_1092 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| Hs 578T | Invasive breast carcinoma | Homo sapiens | CVCL_0332 | |
| MCF-7 | Invasive breast carcinoma | Homo sapiens | CVCL_0031 | |
| MDA-MB-231 | Breast adenocarcinoma | Homo sapiens | CVCL_0062 | |
| MDA-MB-453 | Breast adenocarcinoma | Homo sapiens | CVCL_0418 | |
| MDA-MB-468 | Breast adenocarcinoma | Homo sapiens | CVCL_0419 | |
| T-47D | Invasive breast carcinoma | Homo sapiens | CVCL_0553 | |
| In-vivo Model | The enriched mammosphere cells derived from engineered BT549 and Hs578T with silenced lncRNA KB-1980E6.3 (shKB/vector), BT549, and Hs578T with lncRNA KB-1980E6.3 knockdown combined with ectopic c-Myc (shKB/c-Myc), BT549, and Hs578T with silenced IGF2BP1 (shIGF2BP1/vector), BT549, and Hs578T with knocked down IGF2BP1 combined with ectopic c-Myc (shIGF2BP1/c-Myc), and BT549, and Hs578T/shNC/vector control cells were used in Xenograft experiments. Three doses (1 × 105, 1 × 104 and 1 × 103) of spheres derived from the engineered Hs578T and 1 × 105 of spheres derived from the engineered BT549 were subcutaneously inoculated into 4- to 6-week-old female nude mice (n = 5 per group). Mice were then treated with either bevacizumab (10 mg/kg every 3 days) to form a hypoxic tumor microenvironment or vehicle PBS to form a non-hypoxic condition. | |||
Methyltransferase-like 14 (METTL14) [WRITER]
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL14 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
|
Treatment: siMETTL14 MDA-MB-231 cells
Control: MDA-MB-231 cells
|
GSE81164 | |
| Regulation |
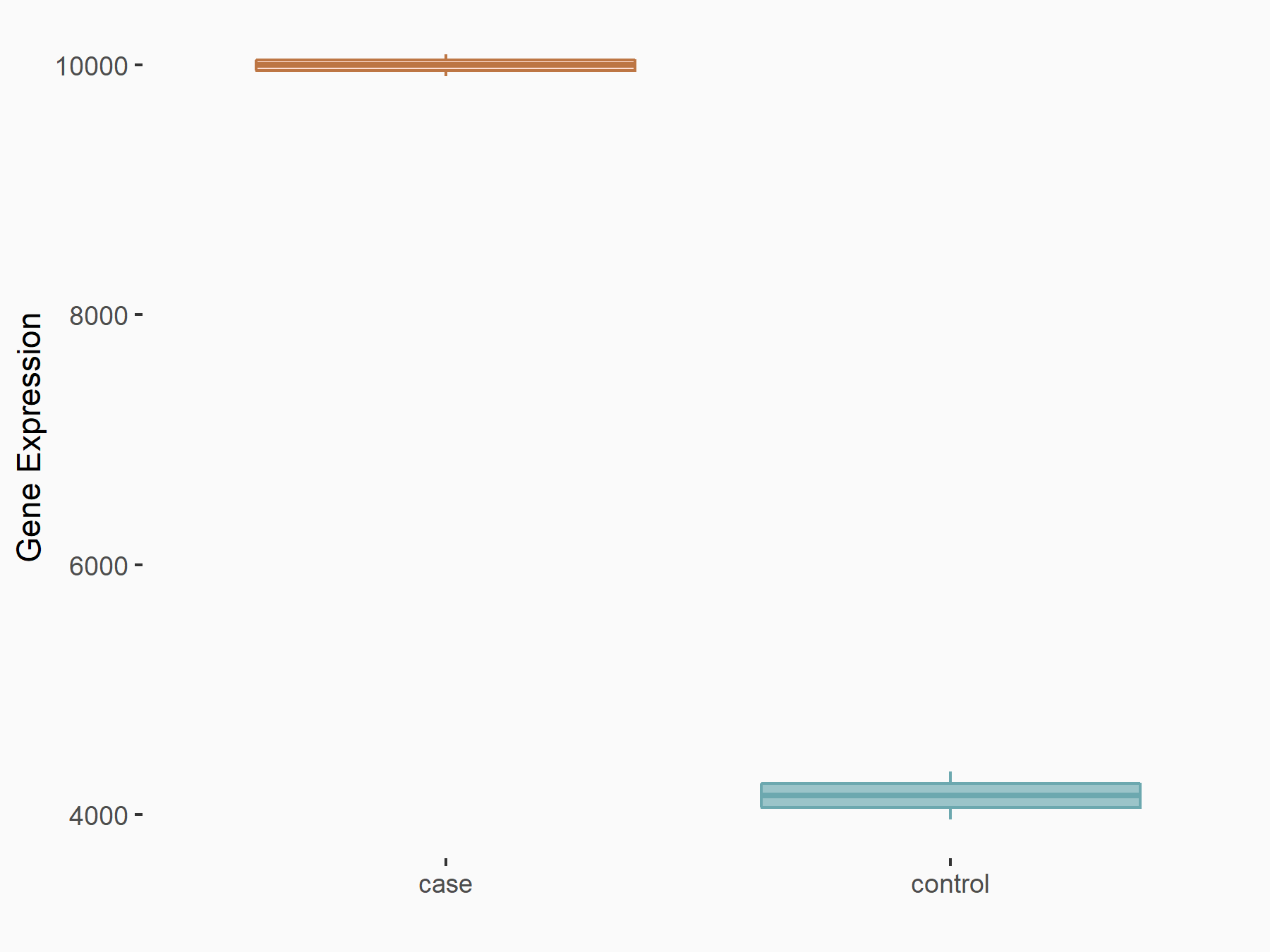  |
logFC: 1.27E+00 p-value: 6.21E-24 |
| More Results | Click to View More RNA-seq Results | |
| In total 1 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [24] | |||
| Response Summary | METTL14 in normal myelopoiesis and AML pathogenesis, as featured by blocking myeloid differentiation and promoting self-renewal of normal HSPCs and LSCs/LICs. METTL14 exerts its oncogenic role by regulating its mRNA targets (e.g., MYB and Myc proto-oncogene protein (MYC)) through m6A modification, while the protein itself is negatively regulated by SPI1. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Acute myeloid leukaemia | ICD-11: 2A60 | ||
| Cell Process | Cell survival/proliferation | |||
| In-vitro Model | HEK293T | Normal | Homo sapiens | CVCL_0063 |
| HSPC (Human hematopoietic stem cell) | ||||
| MNC (Cord blood pluripotent stem cells) | ||||
| OP9 | Normal | Mus musculus | CVCL_4398 | |
| U-937 | Adult acute monocytic leukemia | Homo sapiens | CVCL_0007 | |
| In-vivo Model | Lin- HSPCs were purified from BM of wildtype mice and 0.1×106 cells were seeded in 2 mL OP9 medium onto the OP9 cells with the addition of 10 ng/mL mouse IL-3, 10 ng/mL human IL-6, 10 ng/mL mouse IL-7, 10 ng/mL mouse Flt-3L, and 50 ng/mL mouse stem cell factor (SCF). | |||
Methyltransferase-like 5 (METTL5) [WRITER]
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL5 | ||
| Cell Line | Mouse liver cells | Mus musculus |
|
Treatment: METTL5 knockout liver cells
Control: WT liver cells
|
GSE174418 | |
| Regulation |
  |
logFC: 2.11E+00 p-value: 1.69E-05 |
| More Results | Click to View More RNA-seq Results | |
| In total 1 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [25] | |||
| Response Summary | The study revealed important roles for METTL5 in the development of pancreatic cancer and present the METTL5/Myc proto-oncogene protein (MYC) axis as a novel therapeutic strategy for treatment. | |||
| Responsed Disease | Pancreatic cancer | ICD-11: 2C10 | ||
| Cell Process | Cell proliferation | |||
| Cell migration | ||||
| Cell invasion | ||||
Wilms tumor 1-associating protein (WTAP) [WRITER]
| Representative RNA-seq result indicating the expression of this target gene regulated by WTAP | ||
| Cell Line | mice hepatocyte | Mus musculus |
|
Treatment: Wtap Hknockout mice hepatocyte
Control: Wtap flox/flox mice hepatocyte
|
GSE168850 | |
| Regulation |
  |
logFC: 2.89E+00 p-value: 1.82E-04 |
| More Results | Click to View More RNA-seq Results | |
| In total 1 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [26] | |||
| Response Summary | WTAP made acute myeloid leukemia cells resistant to daunorubicin. In further investigations, m6A methylation level was downregulated when knocking down WTAP, and Myc proto-oncogene protein (MYC) was upregulated due to the decreased m6A methylation of MYC mRNA. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Acute myeloid leukaemia | ICD-11: 2A60 | ||
| Responsed Drug | Daunorubicin | Approved | ||
| Cell Process | Cell cycle | |||
| Cell proliferation | ||||
| Cell apoptosis | ||||
| In-vitro Model | K-562 | Chronic myelogenous leukemia | Homo sapiens | CVCL_0004 |
| MV4-11 | Childhood acute monocytic leukemia | Homo sapiens | CVCL_0064 | |
Insulin-like growth factor 2 mRNA-binding protein 2 (IGF2BP2) [READER]
| In total 4 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [20] | |||
| Response Summary | YBX1 selectively functions in regulating survival of myeloid leukemia cells. YBX1 interacts with insulin-like growth factor 2 messenger RNA (mRNA)-binding proteins (IGF2BPs) and stabilizes m6A-tagged RNA. YBX1 deficiency dysregulates the expression of apoptosis-related genes and promotes mRNA decay of Myc proto-oncogene protein (MYC) and BCL2 in an m6A-dependent manner, which contributes to the defective survival that results from deletion of YBX1. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Myeloid leukaemia | ICD-11: 2B33.1 | ||
| Cell Process | Cell apoptosis | |||
| In-vitro Model | Leukemia stem cell line (Leukemia stem cell line) | |||
| Kasumi-1 | Myeloid leukemia with maturation | Homo sapiens | CVCL_0589 | |
| MOLM-13 | Adult acute myeloid leukemia | Homo sapiens | CVCL_2119 | |
| THP-1 | Childhood acute monocytic leukemia | Homo sapiens | CVCL_0006 | |
| MV4-11 | Childhood acute monocytic leukemia | Homo sapiens | CVCL_0064 | |
| BV-173 | Chronic myelogenous leukemia | Homo sapiens | CVCL_0181 | |
| NOMO-1 | Adult acute monocytic leukemia | Homo sapiens | CVCL_1609 | |
| K-562 | Chronic myelogenous leukemia | Homo sapiens | CVCL_0004 | |
| KG-1a | Adult acute myeloid leukemia | Homo sapiens | CVCL_1824 | |
| Experiment 2 Reporting the m6A Methylation Regulator of This Target Gene | [54] | |||
| Response Summary | LINRIS blocked K139 ubiquitination of IGF2BP2, maintaining its stability. This process prevented the degradation of IGF2BP2 through the autophagy-lysosome pathway (ALP). The LINRIS-IGF2BP2-Myc proto-oncogene protein (MYC) axis promotes the progression of Colorectal cancer and is a promising therapeutic target. MYC-mediated glycolysis was influenced by the interaction between LINRIS and IGF2BP2. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Colorectal cancer | ICD-11: 2B91 | ||
| Pathway Response | Ubiquitin mediated proteolysis | hsa04120 | ||
| Glycolysis / Gluconeogenesis | hsa00010 | |||
| Cell Process | Autophagy-lysosome pathway | |||
| Ubiquitination | ||||
| Glycolysis | ||||
| In-vitro Model | DLD-1 | Colon adenocarcinoma | Homo sapiens | CVCL_0248 |
| HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 | |
| In-vivo Model | For the orthotopic models, 2 × 106 cells with negative control (NC, sh-NC), sh-1 or sh-2 in 0.5 mL of PBS were subcutaneously injected into the dorsal flank of 2 mice respectively. Then 15 mice were separated into 3 groups (sh-NC, sh-1 and sh-2), of which the tumor pieces were tied to the base of the ceca. The growth of the tumors was monitored every 2 weeks after intraperitoneal injection of D-luciferin with a Xenogen IVIS 100 Bioluminescent Imaging System. | |||
| Experiment 3 Reporting the m6A Methylation Regulator of This Target Gene | [55] | |||
| Response Summary | LncRNA-PACERR which bound to IGF2BP2 acts as an m6A-dependent manner to enhance the stability of KLF12 and Myc proto-oncogene protein (MYC) in cytoplasm. This study found that LncRNA-PACERR functions as key regulator of TAMs in PDAC microenvironment and revealed the novel mechanisms in cytoplasm and in nucleus. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Pancreatic ductal adenocarcinoma | ICD-11: 2C10.0 | ||
| Pathway Response | mRNA surveillance pathway | hsa03015 | ||
| RNA degradation | hsa03018 | |||
| Cell Process | RNA stability | |||
| In-vitro Model | THP-1 | Childhood acute monocytic leukemia | Homo sapiens | CVCL_0006 |
| PATU-8988 (Human pancreatic adenocarcinoma cell) | ||||
| PANC-1 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0480 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| 37 (Pancreatic cancer cell) | ||||
| In-vivo Model | BALB/c nude mice which were co-injected with THP-1 cells and PATU-8988 cells subcutaneously. | |||
| Experiment 4 Reporting the m6A Methylation Regulator of This Target Gene | [22] | |||
| Response Summary | In contrast to the mRNA-decay-promoting function of YTH domain-containing family protein 2, IGF2BPs promote the stability and storage of their target mRNAs (for example, MYC) in an m6A-dependent manner under normal and stress conditions and therefore affect gene expression output. Four representative high confidence targets, including Myc proto-oncogene protein (MYC), FSCN1, TK1, and MARCKSL1, exhibit strong binding with IGF2BPs around their m6A motifs in control cells. Knocking down of each individual IGF2BPs in Hela (cervical cancer) and HepG2 (liver cancer) cells significantly repressed MYC expression. | |||
| Responsed Disease | Hepatocellular carcinoma | ICD-11: 2C12.02 | ||
| Cell Process | RNA decay | |||
| In-vitro Model | Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| HeLa | Endocervical adenocarcinoma | Homo sapiens | CVCL_0030 | |
| Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 | |
Insulin-like growth factor 2 mRNA-binding protein 3 (IGF2BP3) [READER]
| In total 3 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [12] | |||
| Response Summary | The m6A reader YTHDF2 stabilized Myc proto-oncogene protein (MYC) mRNA specifically in cancer stem cells. Given the challenge of targeting MYC, YTHDF2 presents a therapeutic target to perturb MYC signaling in glioblastoma. The IGF1/IGF1R inhibitor linsitinib preferentially targeted YTHDF2-expressing cells, inhibiting GSC viability without affecting NSCs and impairing in vivo glioblastoma growth. YTHDF2 links RNA epitranscriptomic modifications and GSC growth, laying the foundation for the YTHDF2-MYC-IGFBP3 axis as a specific and novel therapeutic target in glioblastoma. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Glioma | ICD-11: 2A00.0 | ||
| Responsed Drug | Linsitinib | Phase 3 | ||
| Pathway Response | RNA degradation | hsa03018 | ||
| Cell Process | RNA stability | |||
| In-vitro Model | () | |||
| HNP1 (A human neural progenitor cell) | ||||
| NHA (Normal human astrocytes) | ||||
| NSC11 (Pluripotent derived neural progenitor cell) | ||||
| In-vivo Model | For in vivo drug treatment studies, intracranial xenografts were generated by implanting 5000 patient-derived GSCs (387 and 4121) into the right cerebral cortex of NSG mice as described above. | |||
| Experiment 2 Reporting the m6A Methylation Regulator of This Target Gene | [20] | |||
| Response Summary | YBX1 selectively functions in regulating survival of myeloid leukemia cells. YBX1 interacts with insulin-like growth factor 2 messenger RNA (mRNA)-binding proteins (IGF2BPs) and stabilizes m6A-tagged RNA. YBX1 deficiency dysregulates the expression of apoptosis-related genes and promotes mRNA decay of Myc proto-oncogene protein (MYC) and BCL2 in an m6A-dependent manner, which contributes to the defective survival that results from deletion of YBX1. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Myeloid leukaemia | ICD-11: 2B33.1 | ||
| Cell Process | Cell apoptosis | |||
| In-vitro Model | Leukemia stem cell line (Leukemia stem cell line) | |||
| Kasumi-1 | Myeloid leukemia with maturation | Homo sapiens | CVCL_0589 | |
| MOLM-13 | Adult acute myeloid leukemia | Homo sapiens | CVCL_2119 | |
| THP-1 | Childhood acute monocytic leukemia | Homo sapiens | CVCL_0006 | |
| MV4-11 | Childhood acute monocytic leukemia | Homo sapiens | CVCL_0064 | |
| BV-173 | Chronic myelogenous leukemia | Homo sapiens | CVCL_0181 | |
| NOMO-1 | Adult acute monocytic leukemia | Homo sapiens | CVCL_1609 | |
| K-562 | Chronic myelogenous leukemia | Homo sapiens | CVCL_0004 | |
| KG-1a | Adult acute myeloid leukemia | Homo sapiens | CVCL_1824 | |
| Experiment 3 Reporting the m6A Methylation Regulator of This Target Gene | [22] | |||
| Response Summary | In contrast to the mRNA-decay-promoting function of YTH domain-containing family protein 2, IGF2BPs promote the stability and storage of their target mRNAs (for example, MYC) in an m6A-dependent manner under normal and stress conditions and therefore affect gene expression output. Four representative high confidence targets, including Myc proto-oncogene protein (MYC), FSCN1, TK1, and MARCKSL1, exhibit strong binding with IGF2BPs around their m6A motifs in control cells. Knocking down of each individual IGF2BPs in Hela (cervical cancer) and HepG2 (liver cancer) cells significantly repressed MYC expression. | |||
| Responsed Disease | Hepatocellular carcinoma | ICD-11: 2C12.02 | ||
| Cell Process | RNA decay | |||
| In-vitro Model | Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| HeLa | Endocervical adenocarcinoma | Homo sapiens | CVCL_0030 | |
| Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 | |
Insulin-like growth factor-binding protein 3 (IGFBP3) [READER]
| In total 1 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [12] | |||
| Response Summary | YTHDF2 depletion downregulated IGFBP3 mRNA and protein levels, without affecting its mRNA stability. YTHDF2 regulated IGFBP3 levels via Myc proto-oncogene protein (MYC) in glioblastoma stem cells. | |||
| Responsed Disease | Glioblastoma | ICD-11: 2A00.00 | ||
| In-vitro Model | ENSA (A human embryonic stem derived neural progenitor cell) | |||
| HNP1 (A human neural progenitor cell) | ||||
| NSC11 (Pluripotent derived neural progenitor cell) | ||||
| NHA (Normal human astrocytes) | ||||
| In-vivo Model | Implanting 5000 human derived GSCs into the right cerebral cortex of NSG mice at a depth of 3.5 mm under a University of California, San Diego Institutional Animal Care and Use Committee (IACUC) approved protocol. Brains were harvested and fixed in 4% formaldehyde, cryopreserved in 30% sucrose, and then cryosectioned. Hematoxylin and eosin (H&E) staining was performed on sections for histological analysis. In parallel survival experiments, mice were observed until the development of neurological signs. For in vivo drug treatment studies, intracranial xenografts were generated by implanting 5000 patient-derived GSCs (387 and 4121) into the right cerebral cortex of NSG mice as described above. Mice recovered for 7 days were randomly assigned into drug vs. treatment group by a blinded investigator. Mice were then treated daily with either vehicle (25 mM Tartaric acid) or 50 mg/kg linsitinib by oral gavage. | |||
Brain cancer [ICD-11: 2A00]
| In total 5 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [14] | |||
| Response Summary | This work demonstrates anti-tumor effects of 2HG in inhibiting proliferation/survival of FTO-high cancer cells via targeting FTO/m6A/Myc proto-oncogene protein (MYC)/CEBPA signaling.High levels of FTO sensitize leukemia cells to R-2HG, whereas hyperactivation of MYC signaling confers resistance that can be reversed by the inhibition of MYC signaling. R-2HG also displays anti-tumor activity in glioma. High levels of FTO sensitize leukemic cells to R-2HG, whereas hyperactivation of MYC signaling confers resistance that can be reversed by the inhibition of MYC signaling. | |||
| Responsed Disease | Glioma [ICD-11: 2A00.0] | |||
| Target Regulator | Fat mass and obesity-associated protein (FTO) | ERASER | ||
| Target Regulation | Down regulation | |||
| Responsed Drug | R-2HG | Investigative | ||
| Cell Process | Glutamine metabolism | |||
| Cell apoptosis | ||||
| In-vitro Model | 8-MG-BA | Glioblastoma | Homo sapiens | CVCL_1052 |
| A-172 | Glioblastoma | Homo sapiens | CVCL_0131 | |
| DK-MG | Glioblastoma | Homo sapiens | CVCL_1173 | |
| GaMG | Glioblastoma | Homo sapiens | CVCL_1226 | |
| HEL | Erythroleukemia | Homo sapiens | CVCL_0001 | |
| Jurkat | T acute lymphoblastic leukemia | Homo sapiens | CVCL_0065 | |
| KOCL-45 | B acute lymphoblastic leukemia | Homo sapiens | CVCL_3993 | |
| KOCL-48 | Childhood acute monocytic leukemia | Homo sapiens | CVCL_6867 | |
| KOCL-50 | B acute lymphoblastic leukemia | Homo sapiens | CVCL_6866 | |
| KOCL-51 | B acute lymphoblastic leukemia | Homo sapiens | CVCL_6865 | |
| KOCL-69 | B acute lymphoblastic leukemia | Homo sapiens | CVCL_3995 | |
| KOPN-1 | B acute lymphoblastic leukemia | Homo sapiens | CVCL_3937 | |
| LN-18 | Glioblastoma | Homo sapiens | CVCL_0392 | |
| LN-229 | Glioblastoma | Homo sapiens | CVCL_0393 | |
| MA9.3 (MA9.3) | ||||
| MA9.6ITD (MLL-AF9 plus FLT3-ITD) | ||||
| MA9.6RAS (MLL-AF9 plus NRasG12D) | ||||
| MA9.6 (MLL-AF9) | ||||
| MA9.6ITD (MLL-AF9 plus FLT3-ITD) | ||||
| MA9.6RAS (MLL-AF9 plus NRasG12D) | ||||
| ME-1 [Human leukemia] | Adult acute myeloid leukemia | Homo sapiens | CVCL_2110 | |
| ML-2 | Adult acute myeloid leukemia | Homo sapiens | CVCL_1418 | |
| MV4-11 | Childhood acute monocytic leukemia | Homo sapiens | CVCL_0064 | |
| NB4 | Acute promyelocytic leukemia | Homo sapiens | CVCL_0005 | |
| NOMO-1 | Adult acute monocytic leukemia | Homo sapiens | CVCL_1609 | |
| PL21 | Familial adenomatous polyposis | Homo sapiens | CVCL_JM48 | |
| T98G | Glioblastoma | Homo sapiens | CVCL_0556 | |
| THP-1 | Childhood acute monocytic leukemia | Homo sapiens | CVCL_0006 | |
| U-87MG ATCC | Glioblastoma | Homo sapiens | CVCL_0022 | |
| U-937 | Adult acute monocytic leukemia | Homo sapiens | CVCL_0007 | |
| In-vivo Model | For R-2HG injection mouse models, sensitive (NOMO-1 and MA9.3ITD) or resistant (MA9.3RAS) cells were injected into NSGS or NRGS intravenously, and then R-2HG (6mg/kg body weight) or PBS were injected once daily through tail vein for 12 consecutive days starting from day 11 post xeno-transplantation. | |||
| Experiment 2 Reporting the m6A-centered Disease Response | [12] | |||
| Response Summary | The m6A reader YTHDF2 stabilized Myc proto-oncogene protein (MYC) mRNA specifically in cancer stem cells. Given the challenge of targeting MYC, YTHDF2 presents a therapeutic target to perturb MYC signaling in glioblastoma. The IGF1/IGF1R inhibitor linsitinib preferentially targeted YTHDF2-expressing cells, inhibiting GSC viability without affecting NSCs and impairing in vivo glioblastoma growth. YTHDF2 links RNA epitranscriptomic modifications and GSC growth, laying the foundation for the YTHDF2-MYC-IGFBP3 axis as a specific and novel therapeutic target in glioblastoma. | |||
| Responsed Disease | Glioma [ICD-11: 2A00.0] | |||
| Target Regulator | Insulin-like growth factor 2 mRNA-binding protein 3 (IGF2BP3) | READER | ||
| Target Regulation | Up regulation | |||
| Responsed Drug | Linsitinib | Phase 3 | ||
| Pathway Response | RNA degradation | hsa03018 | ||
| Cell Process | RNA stability | |||
| In-vitro Model | () | |||
| HNP1 (A human neural progenitor cell) | ||||
| NHA (Normal human astrocytes) | ||||
| NSC11 (Pluripotent derived neural progenitor cell) | ||||
| In-vivo Model | For in vivo drug treatment studies, intracranial xenografts were generated by implanting 5000 patient-derived GSCs (387 and 4121) into the right cerebral cortex of NSG mice as described above. | |||
| Experiment 3 Reporting the m6A-centered Disease Response | [12] | |||
| Response Summary | YTHDF2 depletion downregulated IGFBP3 mRNA and protein levels, without affecting its mRNA stability. YTHDF2 regulated IGFBP3 levels via Myc proto-oncogene protein (MYC) in glioblastoma stem cells. | |||
| Responsed Disease | Glioblastoma [ICD-11: 2A00.00] | |||
| Target Regulator | Insulin-like growth factor-binding protein 3 (IGFBP3) | READER | ||
| In-vitro Model | ENSA (A human embryonic stem derived neural progenitor cell) | |||
| HNP1 (A human neural progenitor cell) | ||||
| NSC11 (Pluripotent derived neural progenitor cell) | ||||
| NHA (Normal human astrocytes) | ||||
| In-vivo Model | Implanting 5000 human derived GSCs into the right cerebral cortex of NSG mice at a depth of 3.5 mm under a University of California, San Diego Institutional Animal Care and Use Committee (IACUC) approved protocol. Brains were harvested and fixed in 4% formaldehyde, cryopreserved in 30% sucrose, and then cryosectioned. Hematoxylin and eosin (H&E) staining was performed on sections for histological analysis. In parallel survival experiments, mice were observed until the development of neurological signs. For in vivo drug treatment studies, intracranial xenografts were generated by implanting 5000 patient-derived GSCs (387 and 4121) into the right cerebral cortex of NSG mice as described above. Mice recovered for 7 days were randomly assigned into drug vs. treatment group by a blinded investigator. Mice were then treated daily with either vehicle (25 mM Tartaric acid) or 50 mg/kg linsitinib by oral gavage. | |||
| Experiment 4 Reporting the m6A-centered Disease Response | [12] | |||
| Response Summary | The m6A reader YTHDF2 stabilized Myc proto-oncogene protein (MYC) mRNA specifically in cancer stem cells. Given the challenge of targeting MYC, YTHDF2 presents a therapeutic target to perturb MYC signaling in glioblastoma. The IGF1/IGF1R inhibitor linsitinib preferentially targeted YTHDF2-expressing cells, inhibiting GSC viability without affecting NSCs and impairing in vivo glioblastoma growth. YTHDF2 links RNA epitranscriptomic modifications and GSC growth, laying the foundation for the YTHDF2-MYC-IGFBP3 axis as a specific and novel therapeutic target in glioblastoma. | |||
| Responsed Disease | Glioma [ICD-11: 2A00.0] | |||
| Target Regulator | YTH domain-containing family protein 2 (YTHDF2) | READER | ||
| Target Regulation | Up regulation | |||
| Responsed Drug | Linsitinib | Phase 3 | ||
| Pathway Response | RNA degradation | hsa03018 | ||
| Cell Process | RNA stability | |||
| In-vitro Model | () | |||
| HNP1 (A human neural progenitor cell) | ||||
| NHA (Normal human astrocytes) | ||||
| NSC11 (Pluripotent derived neural progenitor cell) | ||||
| In-vivo Model | For in vivo drug treatment studies, intracranial xenografts were generated by implanting 5000 patient-derived GSCs (387 and 4121) into the right cerebral cortex of NSG mice as described above. | |||
| Experiment 5 Reporting the m6A-centered Disease Response | [12] | |||
| Response Summary | The IGF1/IGF1R inhibitor, linsitinib for further investigation based upon the role of the IGF pathway member, IGFBP3, as a downstream effector of YTHDF2-Myc proto-oncogene protein (MYC) axis in GSCs. Inhibiting glioblastoma stem cells viability without affecting NSCs and impairing in vivo glioblastoma growth. | |||
| Responsed Disease | Glioblastoma [ICD-11: 2A00.00] | |||
| Target Regulator | YTH domain-containing family protein 2 (YTHDF2) | READER | ||
| Target Regulation | Up regulation | |||
| Responsed Drug | Linsitinib | Phase 3 | ||
| In-vitro Model | NSC11 (Pluripotent derived neural progenitor cell) | |||
| NHA (Normal human astrocytes) | ||||
| HNP1 (A human neural progenitor cell) | ||||
| ENSA (A human embryonic stem derived neural progenitor cell) | ||||
| In-vivo Model | Implanting 5000 human derived GSCs into the right cerebral cortex of NSG mice at a depth of 3.5 mm under a University of California, San Diego Institutional Animal Care and Use Committee (IACUC) approved protocol. Brains were harvested and fixed in 4% formaldehyde, cryopreserved in 30% sucrose, and then cryosectioned. Hematoxylin and eosin (H&E) staining was performed on sections for histological analysis. In parallel survival experiments, mice were observed until the development of neurological signs. For in vivo drug treatment studies, intracranial xenografts were generated by implanting 5000 patient-derived GSCs (387 and 4121) into the right cerebral cortex of NSG mice as described above. Mice recovered for 7 days were randomly assigned into drug vs. treatment group by a blinded investigator. Mice were then treated daily with either vehicle (25 mM Tartaric acid) or 50 mg/kg linsitinib by oral gavage. | |||
Solid tumour/cancer [ICD-11: 2A00-2F9Z]
| In total 2 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [] | |||
| Response Summary | In this review, we discuss the specific roles of m6A "writers", "erasers", and "readers" in normal physiology and how their altered expression promotes tumorigenesis. We also describe the potential of exploiting the aberrant expression of these enzymes for cancer diagnosis, prognosis, and the development of novel therapies. The abnormal expression of m6A regulatory enzymes affects m6A abundance and consequently dysregulates the expression of tumor suppressor genes and oncogenes, including Myc proto-oncogene protein (MYC), SOCS2, ADAM19, and PTEN. | |||
| Responsed Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | |||
| Experiment 2 Reporting the m6A-centered Disease Response | [15] | |||
| Response Summary | Wnt/Beta-catenin-mediated FTO downregulation and underscored the role of m6A modifications of Myc proto-oncogene protein (MYC) mRNA in regulating tumor cell glycolysis and growth. | |||
| Responsed Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | |||
| Target Regulator | Fat mass and obesity-associated protein (FTO) | ERASER | ||
| Target Regulation | Down regulation | |||
| Pathway Response | Wnt signaling pathway | hsa04310 | ||
| Central carbon metabolism in cancer | hsa05230 | |||
| Glycolysis / Gluconeogenesis | hsa00010 | |||
| Cell Process | Glycolysis | |||
| In-vitro Model | NCI-H322 | Minimally invasive lung adenocarcinoma | Homo sapiens | CVCL_1556 |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| In-vivo Model | Mice were randomized into several groups. For the subcutaneous implantation model, 1 × 106 cells were injected subcutaneously into the flank regions of female BALB/c nude mice (4-5 weeks). For lung colonization assays, 1 × 106 cells were injected into the tail vein of female NOD/SCID mice (6-7 weeks), and 6 weeks later the lung was removed and fixed with 10% formalin. | |||
Acute myeloid leukaemia [ICD-11: 2A60]
| In total 4 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [24] | |||
| Response Summary | METTL14 in normal myelopoiesis and AML pathogenesis, as featured by blocking myeloid differentiation and promoting self-renewal of normal HSPCs and LSCs/LICs. METTL14 exerts its oncogenic role by regulating its mRNA targets (e.g., MYB and Myc proto-oncogene protein (MYC)) through m6A modification, while the protein itself is negatively regulated by SPI1. | |||
| Responsed Disease | Acute myeloid leukaemia [ICD-11: 2A60] | |||
| Target Regulator | Methyltransferase-like 14 (METTL14) | WRITER | ||
| Target Regulation | Up regulation | |||
| Cell Process | Cell survival/proliferation | |||
| In-vitro Model | HEK293T | Normal | Homo sapiens | CVCL_0063 |
| HSPC (Human hematopoietic stem cell) | ||||
| MNC (Cord blood pluripotent stem cells) | ||||
| OP9 | Normal | Mus musculus | CVCL_4398 | |
| U-937 | Adult acute monocytic leukemia | Homo sapiens | CVCL_0007 | |
| In-vivo Model | Lin- HSPCs were purified from BM of wildtype mice and 0.1×106 cells were seeded in 2 mL OP9 medium onto the OP9 cells with the addition of 10 ng/mL mouse IL-3, 10 ng/mL human IL-6, 10 ng/mL mouse IL-7, 10 ng/mL mouse Flt-3L, and 50 ng/mL mouse stem cell factor (SCF). | |||
| Experiment 2 Reporting the m6A-centered Disease Response | [1] | |||
| Response Summary | METTL3 depletion in human myeloid leukemia cell lines induces cell differentiation and apoptosis and delays leukemia progression in recipient mice in vivo. Single-nucleotide-resolution mapping of m6A coupled with ribosome profiling reveals that m6A promotes the translation of Myc proto-oncogene protein (MYC), BCL2 and PTEN mRNAs in the human acute myeloid leukemia MOLM-13 cell line. Moreover, loss of METTL3 leads to increased levels of phosphorylated AKT. | |||
| Responsed Disease | Acute myeloid leukaemia [ICD-11: 2A60] | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Target Regulation | Up regulation | |||
| Cell Process | Cell differentiation and apoptosis | |||
| Apoptosis (hsa04210) | ||||
| In-vitro Model | HSPC (Human hematopoietic stem cell) | |||
| In-vivo Model | 500,000 selected cells were injected via tail vein or retro-orbital route into female NSG (6-8 week old) recipient mice that had been sublethally irradiated with 475 cGy one day before transplantation. | |||
| Experiment 3 Reporting the m6A-centered Disease Response | [2] | |||
| Response Summary | METTL3 level was slightly increased in AML-M5 patients,and its expression was significantly higher in immature cells than in mature monocytes.METTL3 acts as an oncogene in MOLM13 cells by upregulating Myc proto-oncogene protein (MYC) expression. | |||
| Responsed Disease | Acute myeloid leukaemia [ICD-11: 2A60] | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Target Regulation | Up regulation | |||
| Cell Process | Cell proliferation | |||
| In-vitro Model | MOLM-13 | Adult acute myeloid leukemia | Homo sapiens | CVCL_2119 |
| Experiment 4 Reporting the m6A-centered Disease Response | [26] | |||
| Response Summary | WTAP made acute myeloid leukemia cells resistant to daunorubicin. In further investigations, m6A methylation level was downregulated when knocking down WTAP, and Myc proto-oncogene protein (MYC) was upregulated due to the decreased m6A methylation of MYC mRNA. | |||
| Responsed Disease | Acute myeloid leukaemia [ICD-11: 2A60] | |||
| Target Regulator | Wilms tumor 1-associating protein (WTAP) | WRITER | ||
| Target Regulation | Up regulation | |||
| Responsed Drug | Daunorubicin | Approved | ||
| Cell Process | Cell cycle | |||
| Cell proliferation | ||||
| Cell apoptosis | ||||
| In-vitro Model | K-562 | Chronic myelogenous leukemia | Homo sapiens | CVCL_0004 |
| MV4-11 | Childhood acute monocytic leukemia | Homo sapiens | CVCL_0064 | |
Malignant haematopoietic neoplasm [ICD-11: 2B33]
| In total 2 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [14] | |||
| Response Summary | This work demonstrates anti-tumor effects of 2HG in inhibiting proliferation/survival of FTO-high cancer cells via targeting FTO/m6A/Myc proto-oncogene protein (MYC)/CEBPA signaling.High levels of FTO sensitize leukemia cells to R-2HG, whereas hyperactivation of MYC signaling confers resistance that can be reversed by the inhibition of MYC signaling. R-2HG also displays anti-tumor activity in glioma. High levels of FTO sensitize leukemic cells to R-2HG, whereas hyperactivation of MYC signaling confers resistance that can be reversed by the inhibition of MYC signaling. | |||
| Responsed Disease | Leukaemia [ICD-11: 2B33.4] | |||
| Target Regulator | Fat mass and obesity-associated protein (FTO) | ERASER | ||
| Target Regulation | Down regulation | |||
| Responsed Drug | R-2HG | Investigative | ||
| Cell Process | Glutamine metabolism | |||
| Cell apoptosis | ||||
| In-vitro Model | 8-MG-BA | Glioblastoma | Homo sapiens | CVCL_1052 |
| A-172 | Glioblastoma | Homo sapiens | CVCL_0131 | |
| DK-MG | Glioblastoma | Homo sapiens | CVCL_1173 | |
| GaMG | Glioblastoma | Homo sapiens | CVCL_1226 | |
| HEL | Erythroleukemia | Homo sapiens | CVCL_0001 | |
| Jurkat | T acute lymphoblastic leukemia | Homo sapiens | CVCL_0065 | |
| KOCL-45 | B acute lymphoblastic leukemia | Homo sapiens | CVCL_3993 | |
| KOCL-48 | Childhood acute monocytic leukemia | Homo sapiens | CVCL_6867 | |
| KOCL-50 | B acute lymphoblastic leukemia | Homo sapiens | CVCL_6866 | |
| KOCL-51 | B acute lymphoblastic leukemia | Homo sapiens | CVCL_6865 | |
| KOCL-69 | B acute lymphoblastic leukemia | Homo sapiens | CVCL_3995 | |
| KOPN-1 | B acute lymphoblastic leukemia | Homo sapiens | CVCL_3937 | |
| LN-18 | Glioblastoma | Homo sapiens | CVCL_0392 | |
| LN-229 | Glioblastoma | Homo sapiens | CVCL_0393 | |
| MA9.3 (MA9.3) | ||||
| MA9.6ITD (MLL-AF9 plus FLT3-ITD) | ||||
| MA9.6RAS (MLL-AF9 plus NRasG12D) | ||||
| MA9.6 (MLL-AF9) | ||||
| MA9.6ITD (MLL-AF9 plus FLT3-ITD) | ||||
| MA9.6RAS (MLL-AF9 plus NRasG12D) | ||||
| ME-1 [Human leukemia] | Adult acute myeloid leukemia | Homo sapiens | CVCL_2110 | |
| ML-2 | Adult acute myeloid leukemia | Homo sapiens | CVCL_1418 | |
| MV4-11 | Childhood acute monocytic leukemia | Homo sapiens | CVCL_0064 | |
| NB4 | Acute promyelocytic leukemia | Homo sapiens | CVCL_0005 | |
| NOMO-1 | Adult acute monocytic leukemia | Homo sapiens | CVCL_1609 | |
| PL21 | Familial adenomatous polyposis | Homo sapiens | CVCL_JM48 | |
| T98G | Glioblastoma | Homo sapiens | CVCL_0556 | |
| THP-1 | Childhood acute monocytic leukemia | Homo sapiens | CVCL_0006 | |
| U-87MG ATCC | Glioblastoma | Homo sapiens | CVCL_0022 | |
| U-937 | Adult acute monocytic leukemia | Homo sapiens | CVCL_0007 | |
| In-vivo Model | For R-2HG injection mouse models, sensitive (NOMO-1 and MA9.3ITD) or resistant (MA9.3RAS) cells were injected into NSGS or NRGS intravenously, and then R-2HG (6mg/kg body weight) or PBS were injected once daily through tail vein for 12 consecutive days starting from day 11 post xeno-transplantation. | |||
| Experiment 2 Reporting the m6A-centered Disease Response | [20] | |||
| Response Summary | YBX1 selectively functions in regulating survival of myeloid leukemia cells. YBX1 interacts with insulin-like growth factor 2 messenger RNA (mRNA)-binding proteins (IGF2BPs) and stabilizes m6A-tagged RNA. YBX1 deficiency dysregulates the expression of apoptosis-related genes and promotes mRNA decay of Myc proto-oncogene protein (MYC) and BCL2 in an m6A-dependent manner, which contributes to the defective survival that results from deletion of YBX1. | |||
| Responsed Disease | Myeloid leukaemia [ICD-11: 2B33.1] | |||
| Target Regulator | Insulin-like growth factor 2 mRNA-binding protein 1 (IGF2BP1) | READER | ||
| Target Regulation | Up regulation | |||
| Cell Process | Cell apoptosis | |||
| In-vitro Model | Leukemia stem cell line (Leukemia stem cell line) | |||
| Kasumi-1 | Myeloid leukemia with maturation | Homo sapiens | CVCL_0589 | |
| MOLM-13 | Adult acute myeloid leukemia | Homo sapiens | CVCL_2119 | |
| THP-1 | Childhood acute monocytic leukemia | Homo sapiens | CVCL_0006 | |
| MV4-11 | Childhood acute monocytic leukemia | Homo sapiens | CVCL_0064 | |
| BV-173 | Chronic myelogenous leukemia | Homo sapiens | CVCL_0181 | |
| NOMO-1 | Adult acute monocytic leukemia | Homo sapiens | CVCL_1609 | |
| K-562 | Chronic myelogenous leukemia | Homo sapiens | CVCL_0004 | |
| KG-1a | Adult acute myeloid leukemia | Homo sapiens | CVCL_1824 | |
Head and neck squamous carcinoma [ICD-11: 2B6E]
| In total 2 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [3] | |||
| Response Summary | In oral squamous cell carcinoma, YTH N6-methyladenosine RNA binding protein 1 (YTH domain family, member 1 [YTHDF1]) mediated the m6A-increased stability of Myc proto-oncogene protein (MYC) mRNA catalyzed by METTL3. | |||
| Responsed Disease | Oral squamous cell carcinoma [ICD-11: 2B6E.0] | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Target Regulation | Up regulation | |||
| Pathway Response | RNA degradation | hsa03018 | ||
| Cell Process | RNA stability | |||
| In-vitro Model | CAL-27 | Tongue squamous cell carcinoma | Homo sapiens | CVCL_1107 |
| NHOK (Normal oral keratinocytes) | ||||
| SCC-15 | Tongue squamous cell carcinoma | Homo sapiens | CVCL_1681 | |
| SCC-25 | Tongue squamous cell carcinoma | Homo sapiens | CVCL_1682 | |
| TSCCa | Endocervical adenocarcinoma | Homo sapiens | CVCL_VL15 | |
| In-vivo Model | The stable transfection of SCC25 cells (1 × 107 cells in 0.1 mL) with lenti-sh-METTL3 or blank vectors was injected subcutaneously into BALB/c nude mice. | |||
| Experiment 2 Reporting the m6A-centered Disease Response | [3] | |||
| Response Summary | In oral squamous cell carcinoma, YTH N6-methyladenosine RNA binding protein 1 (YTH domain family, member 1 [YTHDF1]) mediated the m6A-increased stability of Myc proto-oncogene protein (MYC) mRNA catalyzed by METTL3. | |||
| Responsed Disease | Oral squamous cell carcinoma [ICD-11: 2B6E.0] | |||
| Target Regulator | YTH domain-containing family protein 1 (YTHDF1) | READER | ||
| Target Regulation | Up regulation | |||
| Pathway Response | RNA degradation | hsa03018 | ||
| Cell Process | RNA stability | |||
| In-vitro Model | CAL-27 | Tongue squamous cell carcinoma | Homo sapiens | CVCL_1107 |
| NHOK (Normal oral keratinocytes) | ||||
| SCC-15 | Tongue squamous cell carcinoma | Homo sapiens | CVCL_1681 | |
| SCC-25 | Tongue squamous cell carcinoma | Homo sapiens | CVCL_1682 | |
| TSCCa | Endocervical adenocarcinoma | Homo sapiens | CVCL_VL15 | |
| In-vivo Model | The stable transfection of SCC25 cells (1 × 107 cells in 0.1 mL) with lenti-sh-METTL3 or blank vectors was injected subcutaneously into BALB/c nude mice. | |||
Gastric cancer [ICD-11: 2B72]
| In total 4 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [21] | |||
| Response Summary | IGF2BP1 upregulated in GC tissue and acted as a predictor of poor prognosis for GC patients. IGF2BP1 directly interacted with Myc proto-oncogene protein (MYC) mRNA via m6A-dependent manner to by stabilize its stability. | |||
| Responsed Disease | Gastric cancer [ICD-11: 2B72] | |||
| Target Regulator | Insulin-like growth factor 2 mRNA-binding protein 1 (IGF2BP1) | READER | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Central carbon metabolism in cancer | hsa05230 | ||
| Glycolysis / Gluconeogenesis | hsa00010 | |||
| Cell Process | Aerobic glycolysis | |||
| In-vitro Model | SNU-216 | Gastric tubular adenocarcinoma | Homo sapiens | CVCL_3946 |
| MKN45 | Gastric adenocarcinoma | Homo sapiens | CVCL_0434 | |
| HGC-27 | Gastric carcinoma | Homo sapiens | CVCL_1279 | |
| GES-1 | Normal | Homo sapiens | CVCL_EQ22 | |
| AGS | Gastric adenocarcinoma | Homo sapiens | CVCL_0139 | |
| In-vivo Model | A total of 5 × 106 transfected MKN-45 cells, stably transfected with sh-IGF2BP1 vector or empty vector were subcutaneously injected into the flank of the mice. Tumor growth was measured every three days, and calculated using the following equation = a × b2/2 (a for longitudinal diameter; and b for latitudinal diameter). Three weeks after injection, mice were sacrificed. | |||
| Experiment 2 Reporting the m6A-centered Disease Response | [4] | |||
| Response Summary | In gastric cancer, several component molecules (e.g., MCM5, MCM6, etc.) of Myc proto-oncogene protein (MYC) target genes were mediated by METTL3 via altered m6A modification. | |||
| Responsed Disease | Gastric cancer [ICD-11: 2B72] | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Target Regulation | Up regulation | |||
| Cell Process | Cell proliferation | |||
| Cell migration | ||||
| Cell invasion | ||||
| In-vitro Model | GES-1 | Normal | Homo sapiens | CVCL_EQ22 |
| HGC-27 | Gastric carcinoma | Homo sapiens | CVCL_1279 | |
| MGC-803 | Gastric mucinous adenocarcinoma | Homo sapiens | CVCL_5334 | |
| MKN45 | Gastric adenocarcinoma | Homo sapiens | CVCL_0434 | |
| MKN74 | Gastric tubular adenocarcinoma | Homo sapiens | CVCL_2791 | |
| pGCC (Primary GC cells) | ||||
| SGC-7901 | Gastric carcinoma | Homo sapiens | CVCL_0520 | |
| In-vivo Model | A total of 2 × 106 GC cells were injected into the flank of nude mice in a 1:1 suspension of BD Matrigel (BD Biosciences) in phosphate-buffered saline (PBS) solution. | |||
| Experiment 3 Reporting the m6A-centered Disease Response | [5] | |||
| Response Summary | METTL3 enhanced Myc proto-oncogene protein (MYC) m6A methylation and increased MYC translation, which could potentiate the proliferation, migration and invasion of gastric cancer cells. | |||
| Responsed Disease | Gastric cancer [ICD-11: 2B72] | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Target Regulation | Up regulation | |||
| Cell Process | Cell proliferation | |||
| Cell migration | ||||
| Cell invasion | ||||
| Cell apoptosis | ||||
| In-vitro Model | AGS | Gastric adenocarcinoma | Homo sapiens | CVCL_0139 |
| AZ-521 | Duodenal adenocarcinoma | Homo sapiens | CVCL_2862 | |
| GES-1 | Normal | Homo sapiens | CVCL_EQ22 | |
| MKN45 | Gastric adenocarcinoma | Homo sapiens | CVCL_0434 | |
| SGC-7901 | Gastric carcinoma | Homo sapiens | CVCL_0520 | |
| In-vivo Model | The GC cell line MKN-45 stably infected with lentivirus expressing sh-HBXIP was prepared into 5 × 107 cells/mL cell suspension. | |||
| Experiment 4 Reporting the m6A-centered Disease Response | [5] | |||
| Response Summary | Expressions of HBXIP, METTL3 and Myc proto-oncogene protein (MYC) were all determined to be upregulated in both GC tissues and cells. HBXIP plays an oncogenic role in GC via METTL3-mediated MYC mRNA m6A modification. | |||
| Responsed Disease | Gastric cancer [ICD-11: 2B72] | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Target Regulation | Up regulation | |||
| Cell Process | Cell proliferation | |||
| Cell migration | ||||
| Cell invasion | ||||
| Cell apoptosis | ||||
| In-vitro Model | SGC-7901 | Gastric carcinoma | Homo sapiens | CVCL_0520 |
| MKN45 | Gastric adenocarcinoma | Homo sapiens | CVCL_0434 | |
| GES-1 | Normal | Homo sapiens | CVCL_EQ22 | |
| AZ-521 | Duodenal adenocarcinoma | Homo sapiens | CVCL_2862 | |
| AGS | Gastric adenocarcinoma | Homo sapiens | CVCL_0139 | |
| In-vivo Model | The GC cell line MKN-45 stably infected with lentivirus expressing sh-HBXIP was prepared into 5 × 107 cells/mL cell suspension. The cell suspension was injected into the left axilla of nude mice using a 1 mL syringe as the sh-HBXIP group (n = 6). The GC cell line MKN-45 infected with the lentivirus expressing sh-NC was dispersed into the cell suspension, which was injected into nude mice as the sh-NC group (n = 6). Tumor growth was observed and data were recorded after inoculation. On the 26th day, all nude mice were euthanized by cervical dislocation and the tumors were resected and weighed. | |||
Colorectal cancer [ICD-11: 2B91]
| In total 4 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [16] | |||
| Response Summary | MiR-96 antagomir could potentially retard the cancerogenesis in colorectal cancer via AMPK-alpha-2-dependent inhibition of FTO and blocking FTO-mediated m6A modification of Myc proto-oncogene protein (MYC). | |||
| Responsed Disease | Colorectal cancer [ICD-11: 2B91] | |||
| Target Regulator | Fat mass and obesity-associated protein (FTO) | ERASER | ||
| Target Regulation | Up regulation | |||
| Cell Process | Cell proliferation | |||
| Cell apoptosis | ||||
| In-vitro Model | HCT 8 | Colon adenocarcinoma | Homo sapiens | CVCL_2478 |
| SW480 | Colon adenocarcinoma | Homo sapiens | CVCL_0546 | |
| SW620 | Colon adenocarcinoma | Homo sapiens | CVCL_0547 | |
| In-vivo Model | RC cells SW480 at logarithmic growth phase were prepared into cell suspension with a concentration of about 1 × 107/100 L, which was then injected into the left axilla of nude mice with a 1 ml syringe to establish a subcutaneous mouse xenograft model. Once the tumor volume reached about 50 mm3, the nude mice were injected with miR-96 antagomir or NC antagomir (10 nmol once every 5 days for 5 weeks). After 5 weeks, the mice were euthanized, after which the subcutaneous transplanted tumor was removed, and weighed. | |||
| Experiment 2 Reporting the m6A-centered Disease Response | [17] | |||
| Response Summary | GSK3beta inhibited MZF1 expression by mediating FTO-regulated m6A modification of MZF1 and then decreased the proto-oncogene Myc proto-oncogene protein (MYC) expression, thus hampering CRC cell proliferation. | |||
| Responsed Disease | Colorectal cancer [ICD-11: 2B91] | |||
| Target Regulator | Fat mass and obesity-associated protein (FTO) | ERASER | ||
| Target Regulation | Down regulation | |||
| Pathway Response | Ubiquitin mediated proteolysis | hsa04120 | ||
| Cell Process | Cell proliferation | |||
| In-vitro Model | SW620 | Colon adenocarcinoma | Homo sapiens | CVCL_0547 |
| SW480 | Colon adenocarcinoma | Homo sapiens | CVCL_0546 | |
| HCT 8 | Colon adenocarcinoma | Homo sapiens | CVCL_2478 | |
| In-vivo Model | Twenty-four specific pathogen free female BALB/c nude mice (age: 6 weeks, weight: 15 ~ 18 g) were purchased from Slac Laboratory Animal Co., Ltd., and subcutaneously injected with SW620 cells stably transfected with oe-NC, oe-GSK3-Beta + oe-NC, or oe-GSK3-Beta + oe-c-Myc to establish a subcutaneous xenograft tumour model in nude mice. | |||
| Experiment 3 Reporting the m6A-centered Disease Response | [54] | |||
| Response Summary | LINRIS blocked K139 ubiquitination of IGF2BP2, maintaining its stability. This process prevented the degradation of IGF2BP2 through the autophagy-lysosome pathway (ALP). The LINRIS-IGF2BP2-Myc proto-oncogene protein (MYC) axis promotes the progression of Colorectal cancer and is a promising therapeutic target. MYC-mediated glycolysis was influenced by the interaction between LINRIS and IGF2BP2. | |||
| Responsed Disease | Colorectal cancer [ICD-11: 2B91] | |||
| Target Regulator | Insulin-like growth factor 2 mRNA-binding protein 2 (IGF2BP2) | READER | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Ubiquitin mediated proteolysis | hsa04120 | ||
| Glycolysis / Gluconeogenesis | hsa00010 | |||
| Cell Process | Autophagy-lysosome pathway | |||
| Ubiquitination | ||||
| Glycolysis | ||||
| In-vitro Model | DLD-1 | Colon adenocarcinoma | Homo sapiens | CVCL_0248 |
| HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 | |
| In-vivo Model | For the orthotopic models, 2 × 106 cells with negative control (NC, sh-NC), sh-1 or sh-2 in 0.5 mL of PBS were subcutaneously injected into the dorsal flank of 2 mice respectively. Then 15 mice were separated into 3 groups (sh-NC, sh-1 and sh-2), of which the tumor pieces were tied to the base of the ceca. The growth of the tumors was monitored every 2 weeks after intraperitoneal injection of D-luciferin with a Xenogen IVIS 100 Bioluminescent Imaging System. | |||
| Experiment 4 Reporting the m6A-centered Disease Response | [6] | |||
| Response Summary | METTL3 exerted its function through enhancing Myc proto-oncogene protein (MYC) expression, at least partially in an m6A-IGF2BP1-dependent manner. Knockdown of METTL3 suppressed colorectal cancer cell proliferation in vitro and in vivo. | |||
| Responsed Disease | Colorectal cancer [ICD-11: 2B91] | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Target Regulation | Up regulation | |||
| Cell Process | Cell proliferation | |||
| In-vitro Model | HEK293T | Normal | Homo sapiens | CVCL_0063 |
| Caco-2 | Colon adenocarcinoma | Homo sapiens | CVCL_0025 | |
| HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 | |
| HT29 | Colon cancer | Mus musculus | CVCL_A8EZ | |
| LoVo | Colon adenocarcinoma | Homo sapiens | CVCL_0399 | |
| SW480 | Colon adenocarcinoma | Homo sapiens | CVCL_0546 | |
| SW620 | Colon adenocarcinoma | Homo sapiens | CVCL_0547 | |
| In-vivo Model | METTL3 stable knockdown or overexpression HCT116 cells were collected and resuspended at a density of 5 × 106 or 3 × 106 cells per 150 uL PBS. | |||
Pancreatic cancer [ICD-11: 2C10]
| In total 3 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [18] | |||
| Response Summary | FTO has been indicated to interact with Myc proto-oncogene protein (MYC) proto-oncogene, bHLH transcription factor and to enhance its stability by decreasing its m6A level.the aforementioned observations indicate a novel mechanism for the regulation of pancreatic cancer cells by FTO. | |||
| Responsed Disease | Pancreatic cancer [ICD-11: 2C10] | |||
| Target Regulator | Fat mass and obesity-associated protein (FTO) | ERASER | ||
| Target Regulation | Up regulation | |||
| Cell Process | Cell proliferation | |||
| In-vitro Model | BxPC-3 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0186 |
| HPDE | Normal | Homo sapiens | CVCL_4376 | |
| PANC-1 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0480 | |
| SW1990 | Pancreatic adenocarcinoma | Homo sapiens | CVCL_1723 | |
| Experiment 2 Reporting the m6A-centered Disease Response | [55] | |||
| Response Summary | LncRNA-PACERR which bound to IGF2BP2 acts as an m6A-dependent manner to enhance the stability of KLF12 and Myc proto-oncogene protein (MYC) in cytoplasm. This study found that LncRNA-PACERR functions as key regulator of TAMs in PDAC microenvironment and revealed the novel mechanisms in cytoplasm and in nucleus. | |||
| Responsed Disease | Pancreatic ductal adenocarcinoma [ICD-11: 2C10.0] | |||
| Target Regulator | Insulin-like growth factor 2 mRNA-binding protein 2 (IGF2BP2) | READER | ||
| Target Regulation | Up regulation | |||
| Pathway Response | mRNA surveillance pathway | hsa03015 | ||
| RNA degradation | hsa03018 | |||
| Cell Process | RNA stability | |||
| In-vitro Model | THP-1 | Childhood acute monocytic leukemia | Homo sapiens | CVCL_0006 |
| PATU-8988 (Human pancreatic adenocarcinoma cell) | ||||
| PANC-1 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0480 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| 37 (Pancreatic cancer cell) | ||||
| In-vivo Model | BALB/c nude mice which were co-injected with THP-1 cells and PATU-8988 cells subcutaneously. | |||
| Experiment 3 Reporting the m6A-centered Disease Response | [25] | |||
| Response Summary | The study revealed important roles for METTL5 in the development of pancreatic cancer and present the METTL5/Myc proto-oncogene protein (MYC) axis as a novel therapeutic strategy for treatment. | |||
| Responsed Disease | Pancreatic cancer [ICD-11: 2C10] | |||
| Target Regulator | Methyltransferase-like 5 (METTL5) | WRITER | ||
| Cell Process | Cell proliferation | |||
| Cell migration | ||||
| Cell invasion | ||||
Liver cancer [ICD-11: 2C12]
| In total 2 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [] | |||
| Response Summary | m6A-related genes have a prognostic value in liver cancer, and the constructed riskscore can identify patients who are high risk and can enable individualized therapy. Gene set enrichment analysis showed that tumorigenic markers, including DNA repair, E2F targets, G2M checkpoint, and Myc proto-oncogene protein (MYC) targets V1, were enriched in Cluster2. | |||
| Responsed Disease | Liver cancer [ICD-11: 2C12] | |||
| Experiment 2 Reporting the m6A-centered Disease Response | [22] | |||
| Response Summary | In contrast to the mRNA-decay-promoting function of YTH domain-containing family protein 2, IGF2BPs promote the stability and storage of their target mRNAs (for example, MYC) in an m6A-dependent manner under normal and stress conditions and therefore affect gene expression output. Four representative high confidence targets, including Myc proto-oncogene protein (MYC), FSCN1, TK1, and MARCKSL1, exhibit strong binding with IGF2BPs around their m6A motifs in control cells. Knocking down of each individual IGF2BPs in Hela (cervical cancer) and HepG2 (liver cancer) cells significantly repressed MYC expression. | |||
| Responsed Disease | Hepatocellular carcinoma [ICD-11: 2C12.02] | |||
| Target Regulator | Insulin-like growth factor 2 mRNA-binding protein 1 (IGF2BP1) | READER | ||
| Cell Process | RNA decay | |||
| In-vitro Model | Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| HeLa | Endocervical adenocarcinoma | Homo sapiens | CVCL_0030 | |
| Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 | |
Lung cancer [ICD-11: 2C25]
| In total 4 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [8] | |||
| Response Summary | This study revealed that m6A methylation is closely related to the poor prognosis of non-small cell lung cancer patients via interference with the TIME, which suggests that m6A plays a role in optimizing individualized immunotherapy management and improving prognosis. The expression levels of METTL3, FTO and YTHDF1 in non-small cell lung cancer were changed. Patients in Cluster 1 had lower immunoscores, higher programmed death-ligand 1 (PD-L1) expression, and shorter overall survival compared to patients in Cluster 2. The Myc proto-oncogene protein (MYC) targets, E2 transcription Factor (E2F) targets were significantly enriched. | |||
| Responsed Disease | Non-small-cell lung carcinoma [ICD-11: 2C25.Y] | |||
| Target Regulator | Fat mass and obesity-associated protein (FTO) | ERASER | ||
| Pathway Response | p53 signaling pathway | hsa04115 | ||
| Central carbon metabolism in cancer | hsa05230 | |||
| PD-L1 expression and PD-1 checkpoint pathway in cancer | hsa05235 | |||
| Experiment 2 Reporting the m6A-centered Disease Response | [7] | |||
| Response Summary | LCAT3 upregulation is attributable to N6-methyladenosine (m6A) modification mediated by methyltransferase like 3 (METTL3), leading to LCAT3 stabilization. LCAT3 as a novel oncogenic lncRNA in the lung, and validated the LCAT3-FUBP1-Myc proto-oncogene protein (MYC) axis as a potential therapeutic target for lung adenocarcinomas. | |||
| Responsed Disease | Lung adenocarcinoma [ICD-11: 2C25.0] | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Target Regulation | Up regulation | |||
| In-vitro Model | A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 |
| Calu-1 | Lung squamous cell carcinoma | Homo sapiens | CVCL_0608 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| HOP-62 | Lung adenocarcinoma | Homo sapiens | CVCL_1285 | |
| In-vivo Model | For the in vivo tumorigenicity assay, female BALB/c nude mice (ages 4-5 weeks) were randomly divided into two groups (n = 6/group). Calu1 cells (4 × 106) that had been stably transfected with sh-LCAT3 or scramble were implanted subcutaneously into the nude mice. Tumor growth was measured after one week, and tumor volumes were calculated with the following formula: Volume (cm3) = (length × width2)/2. After four weeks, the mice were euthanized, and the tumors were collected and weighed. For the in vivo tumor invasion assay, 1.2 × 106 scramble or shLCAT3 cells were injected intravenously into the tail vein of nude mice (n = 6/group). 1.5 mg luciferin (Gold Biotech, St Louis, MO, USA) was administered once a week for 4 weeks, to monitor metastases using an IVIS@ Lumina II system (Caliper Life Sciences, Hopkinton, MA, USA). | |||
| Experiment 3 Reporting the m6A-centered Disease Response | [8] | |||
| Response Summary | This study revealed that m6A methylation is closely related to the poor prognosis of non-small cell lung cancer patients via interference with the TIME, which suggests that m6A plays a role in optimizing individualized immunotherapy management and improving prognosis. The expression levels of METTL3, FTO and YTHDF1 in non-small cell lung cancer were changed. Patients in Cluster 1 had lower immunoscores, higher programmed death-ligand 1 (PD-L1) expression, and shorter overall survival compared to patients in Cluster 2. The Myc proto-oncogene protein (MYC) targets, E2 transcription Factor (E2F) targets were significantly enriched. | |||
| Responsed Disease | Non-small-cell lung carcinoma [ICD-11: 2C25.Y] | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Pathway Response | p53 signaling pathway | hsa04115 | ||
| Central carbon metabolism in cancer | hsa05230 | |||
| PD-L1 expression and PD-1 checkpoint pathway in cancer | hsa05235 | |||
| Experiment 4 Reporting the m6A-centered Disease Response | [8] | |||
| Response Summary | This study revealed that m6A methylation is closely related to the poor prognosis of non-small cell lung cancer patients via interference with the TIME, which suggests that m6A plays a role in optimizing individualized immunotherapy management and improving prognosis. The expression levels of METTL3, FTO and YTHDF1 in non-small cell lung cancer were changed. Patients in Cluster 1 had lower immunoscores, higher programmed death-ligand 1 (PD-L1) expression, and shorter overall survival compared to patients in Cluster 2. The Myc proto-oncogene protein (MYC) targets, E2 transcription Factor (E2F) targets were significantly enriched. | |||
| Responsed Disease | Non-small-cell lung carcinoma [ICD-11: 2C25.Y] | |||
| Target Regulator | YTH domain-containing family protein 1 (YTHDF1) | READER | ||
| Pathway Response | p53 signaling pathway | hsa04115 | ||
| Central carbon metabolism in cancer | hsa05230 | |||
| PD-L1 expression and PD-1 checkpoint pathway in cancer | hsa05235 | |||
Breast cancer [ICD-11: 2C60]
| In total 2 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [23] | |||
| Response Summary | Hypoxia-induced lncRNA KB-1980E6.3 is involved in the self-renewal and stemness maintenance of breast cancer stem cells by recruiting IGF2BP1 to regulate Myc proto-oncogene protein (MYC) mRNA stability. | |||
| Responsed Disease | Breast cancer [ICD-11: 2C60] | |||
| Target Regulator | Insulin-like growth factor 2 mRNA-binding protein 1 (IGF2BP1) | READER | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Signaling pathways regulating pluripotency of stem cells | hsa04550 | ||
| In-vitro Model | BT-474 | Invasive breast carcinoma | Homo sapiens | CVCL_0179 |
| BT-549 | Invasive breast carcinoma | Homo sapiens | CVCL_1092 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| Hs 578T | Invasive breast carcinoma | Homo sapiens | CVCL_0332 | |
| MCF-7 | Invasive breast carcinoma | Homo sapiens | CVCL_0031 | |
| MDA-MB-231 | Breast adenocarcinoma | Homo sapiens | CVCL_0062 | |
| MDA-MB-453 | Breast adenocarcinoma | Homo sapiens | CVCL_0418 | |
| MDA-MB-468 | Breast adenocarcinoma | Homo sapiens | CVCL_0419 | |
| T-47D | Invasive breast carcinoma | Homo sapiens | CVCL_0553 | |
| In-vivo Model | The enriched mammosphere cells derived from engineered BT549 and Hs578T with silenced lncRNA KB-1980E6.3 (shKB/vector), BT549, and Hs578T with lncRNA KB-1980E6.3 knockdown combined with ectopic c-Myc (shKB/c-Myc), BT549, and Hs578T with silenced IGF2BP1 (shIGF2BP1/vector), BT549, and Hs578T with knocked down IGF2BP1 combined with ectopic c-Myc (shIGF2BP1/c-Myc), and BT549, and Hs578T/shNC/vector control cells were used in Xenograft experiments. Three doses (1 × 105, 1 × 104 and 1 × 103) of spheres derived from the engineered Hs578T and 1 × 105 of spheres derived from the engineered BT549 were subcutaneously inoculated into 4- to 6-week-old female nude mice (n = 5 per group). Mice were then treated with either bevacizumab (10 mg/kg every 3 days) to form a hypoxic tumor microenvironment or vehicle PBS to form a non-hypoxic condition. | |||
| Experiment 2 Reporting the m6A-centered Disease Response | [13] | |||
| Response Summary | LCAT3 upregulation is attributable to m6A modification mediated by METTL3, leading to LCAT3 stabilization. Treated cells with tamoxifen to induce MYC activity. Highlights the therapeutic potential of RBPs by uncovering a critical role for YTHDF2 in counteracting the global increase of mRNA synthesis in Myc proto-oncogene protein (MYC)-driven breast cancers. | |||
| Responsed Disease | Breast cancer [ICD-11: 2C60] | |||
| Target Regulator | YTH domain-containing family protein 2 (YTHDF2) | READER | ||
| Responsed Drug | Tamoxifen | Approved | ||
| Pathway Response | MAPK signaling pathway | hsa04010 | ||
| Cell Process | Epithelial-to-mesenchymal transition | |||
| Cell apoptosis | ||||
| In-vitro Model | HEK293T | Normal | Homo sapiens | CVCL_0063 |
| MCF-7 | Invasive breast carcinoma | Homo sapiens | CVCL_0031 | |
| MDA-MB-231 | Breast adenocarcinoma | Homo sapiens | CVCL_0062 | |
| MBA-MD-231 (Human breast cancer cell) | ||||
| MYC-ER HMEC (Human mammary epithelial cells expressing a MYC estrogen receptor fusion) | ||||
| SK-BR-3 | Breast adenocarcinoma | Homo sapiens | CVCL_0033 | |
| In-vivo Model | To induce recombination at 8 weeks of age both CAG-CreERT;Ythdf2fl/fl and Ythdf2fl/fl littermates were injected with 75mg/kg body weight tamoxifen dissolved in corn oil daily for 5 days. | |||
Cervical cancer [ICD-11: 2C77]
| In total 2 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [19] | |||
| Response Summary | FTO interacts with transcripts of E2F1 and Myc proto-oncogene protein (MYC), inhibition of FTO significantly impairs the translation efficiency of E2F1 and Myc.FTO plays important oncogenic role in regulating cervical cancer cells' proliferation. | |||
| Responsed Disease | Cervical cancer [ICD-11: 2C77] | |||
| Target Regulator | Fat mass and obesity-associated protein (FTO) | ERASER | ||
| Target Regulation | Up regulation | |||
| Cell Process | Cell proliferation and migration | |||
| In-vitro Model | HeLa | Endocervical adenocarcinoma | Homo sapiens | CVCL_0030 |
| SiHa | Cervical squamous cell carcinoma | Homo sapiens | CVCL_0032 | |
| Experiment 2 Reporting the m6A-centered Disease Response | [22] | |||
| Response Summary | In contrast to the mRNA-decay-promoting function of YTH domain-containing family protein 2, IGF2BPs promote the stability and storage of their target mRNAs (for example, MYC) in an m6A-dependent manner under normal and stress conditions and therefore affect gene expression output. Four representative high confidence targets, including Myc proto-oncogene protein (MYC), FSCN1, TK1, and MARCKSL1, exhibit strong binding with IGF2BPs around their m6A motifs in control cells. Knocking down of each individual IGF2BPs in Hela (cervical cancer) and HepG2 (liver cancer) cells significantly repressed MYC expression. | |||
| Responsed Disease | Cervical cancer [ICD-11: 2C77] | |||
| Target Regulator | Insulin-like growth factor 2 mRNA-binding protein 1 (IGF2BP1) | READER | ||
| Cell Process | RNA decay | |||
| In-vitro Model | Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| HeLa | Endocervical adenocarcinoma | Homo sapiens | CVCL_0030 | |
| Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 | |
Prostate cancer [ICD-11: 2C82]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [9] | |||
| Response Summary | METTL3 enhanced Myc proto-oncogene protein (MYC) expression by increasing m6A levels of MYC mRNA transcript, leading to oncogenic functions in prostate cancer. | |||
| Responsed Disease | Prostate cancer [ICD-11: 2C82] | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Target Regulation | Up regulation | |||
| Cell Process | Cell proliferation | |||
| Cell migration | ||||
| Cell invasion | ||||
| In-vitro Model | LNCaP C4-2 | Prostate carcinoma | Homo sapiens | CVCL_4782 |
| DU145 | Prostate carcinoma | Homo sapiens | CVCL_0105 | |
| LNCaP | Prostate carcinoma | Homo sapiens | CVCL_0395 | |
| PC-3 | Prostate carcinoma | Homo sapiens | CVCL_0035 | |
Bladder cancer [ICD-11: 2C94]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [10] | |||
| Response Summary | AF4/FMR2 family member 4 (AFF4), two key regulators of NF-Kappa-B pathway (IKBKB and RELA) and Myc proto-oncogene protein (MYC) were further identified as direct targets of METTL3-mediated m6A modification.overexpression of METTL3 significantly promoted Bladder cancer cell growth and invasion. | |||
| Responsed Disease | Bladder cancer [ICD-11: 2C94] | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Target Regulation | Up regulation | |||
| Cell Process | Glucose metabolism | |||
Polycystic kidney disease [ICD-11: GB81]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [11] | |||
| Response Summary | Mettl3 activates the cyst-promoting c-Myc and cAMP pathways through enhanced Myc proto-oncogene protein (MYC) and Avpr2 mRNA m6A modification and translation. Thus, Mettl3 promotes Autosomal dominant polycystic kidney disease and links methionine utilization to epitranscriptomic activation of proliferation and cyst growth. | |||
| Responsed Disease | Polycystic kidney disease [ICD-11: GB81] | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Target Regulation | Up regulation | |||
| In-vitro Model | mIMCD-3 | Normal | Mus musculus | CVCL_0429 |
| In-vivo Model | The clone, with one wild-type Mettl3 allele and one L1L2_Bact_P cassette inserted allele, was injected into C57BL/6 blastocysts. Mettl3-targeted mouse line was established from a germline-transmitting chimera. The chimeric mouse was crossed to C57BL/6 Flp mice to excise the neomycin resistance system. | |||
Daunorubicin
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response | [26] | |||
| Response Summary | WTAP made acute myeloid leukemia cells resistant to daunorubicin. In further investigations, m6A methylation level was downregulated when knocking down WTAP, and Myc proto-oncogene protein (MYC) was upregulated due to the decreased m6A methylation of MYC mRNA. | |||
| Target Regulator | Wilms tumor 1-associating protein (WTAP) | WRITER | ||
| Target Regulation | Up regulation | |||
| Responsed Disease | Acute myeloid leukaemia | ICD-11: 2A60 | ||
| Cell Process | Cell cycle | |||
| Cell proliferation | ||||
| Cell apoptosis | ||||
| In-vitro Model | K-562 | Chronic myelogenous leukemia | Homo sapiens | CVCL_0004 |
| MV4-11 | Childhood acute monocytic leukemia | Homo sapiens | CVCL_0064 | |
Tamoxifen
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response | [13] | |||
| Response Summary | LCAT3 upregulation is attributable to m6A modification mediated by METTL3, leading to LCAT3 stabilization. Treated cells with tamoxifen to induce MYC activity. Highlights the therapeutic potential of RBPs by uncovering a critical role for YTHDF2 in counteracting the global increase of mRNA synthesis in Myc proto-oncogene protein (MYC)-driven breast cancers. | |||
| Target Regulator | YTH domain-containing family protein 2 (YTHDF2) | READER | ||
| Responsed Disease | Breast cancer | ICD-11: 2C60 | ||
| Pathway Response | MAPK signaling pathway | hsa04010 | ||
| Cell Process | Epithelial-to-mesenchymal transition | |||
| Cell apoptosis | ||||
| In-vitro Model | HEK293T | Normal | Homo sapiens | CVCL_0063 |
| MCF-7 | Invasive breast carcinoma | Homo sapiens | CVCL_0031 | |
| MDA-MB-231 | Breast adenocarcinoma | Homo sapiens | CVCL_0062 | |
| MBA-MD-231 (Human breast cancer cell) | ||||
| MYC-ER HMEC (Human mammary epithelial cells expressing a MYC estrogen receptor fusion) | ||||
| SK-BR-3 | Breast adenocarcinoma | Homo sapiens | CVCL_0033 | |
| In-vivo Model | To induce recombination at 8 weeks of age both CAG-CreERT;Ythdf2fl/fl and Ythdf2fl/fl littermates were injected with 75mg/kg body weight tamoxifen dissolved in corn oil daily for 5 days. | |||
Linsitinib
[Phase 3]
| In total 3 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response | [12] | |||
| Response Summary | The m6A reader YTHDF2 stabilized Myc proto-oncogene protein (MYC) mRNA specifically in cancer stem cells. Given the challenge of targeting MYC, YTHDF2 presents a therapeutic target to perturb MYC signaling in glioblastoma. The IGF1/IGF1R inhibitor linsitinib preferentially targeted YTHDF2-expressing cells, inhibiting GSC viability without affecting NSCs and impairing in vivo glioblastoma growth. YTHDF2 links RNA epitranscriptomic modifications and GSC growth, laying the foundation for the YTHDF2-MYC-IGFBP3 axis as a specific and novel therapeutic target in glioblastoma. | |||
| Target Regulator | Insulin-like growth factor 2 mRNA-binding protein 3 (IGF2BP3) | READER | ||
| Target Regulation | Up regulation | |||
| Responsed Disease | Glioma | ICD-11: 2A00.0 | ||
| Pathway Response | RNA degradation | hsa03018 | ||
| Cell Process | RNA stability | |||
| In-vitro Model | () | |||
| HNP1 (A human neural progenitor cell) | ||||
| NHA (Normal human astrocytes) | ||||
| NSC11 (Pluripotent derived neural progenitor cell) | ||||
| In-vivo Model | For in vivo drug treatment studies, intracranial xenografts were generated by implanting 5000 patient-derived GSCs (387 and 4121) into the right cerebral cortex of NSG mice as described above. | |||
| Experiment 2 Reporting the m6A-centered Drug Response | [12] | |||
| Response Summary | The m6A reader YTHDF2 stabilized Myc proto-oncogene protein (MYC) mRNA specifically in cancer stem cells. Given the challenge of targeting MYC, YTHDF2 presents a therapeutic target to perturb MYC signaling in glioblastoma. The IGF1/IGF1R inhibitor linsitinib preferentially targeted YTHDF2-expressing cells, inhibiting GSC viability without affecting NSCs and impairing in vivo glioblastoma growth. YTHDF2 links RNA epitranscriptomic modifications and GSC growth, laying the foundation for the YTHDF2-MYC-IGFBP3 axis as a specific and novel therapeutic target in glioblastoma. | |||
| Target Regulator | YTH domain-containing family protein 2 (YTHDF2) | READER | ||
| Target Regulation | Up regulation | |||
| Responsed Disease | Glioma | ICD-11: 2A00.0 | ||
| Pathway Response | RNA degradation | hsa03018 | ||
| Cell Process | RNA stability | |||
| In-vitro Model | () | |||
| HNP1 (A human neural progenitor cell) | ||||
| NHA (Normal human astrocytes) | ||||
| NSC11 (Pluripotent derived neural progenitor cell) | ||||
| In-vivo Model | For in vivo drug treatment studies, intracranial xenografts were generated by implanting 5000 patient-derived GSCs (387 and 4121) into the right cerebral cortex of NSG mice as described above. | |||
| Experiment 3 Reporting the m6A-centered Drug Response | [12] | |||
| Response Summary | The IGF1/IGF1R inhibitor, linsitinib for further investigation based upon the role of the IGF pathway member, IGFBP3, as a downstream effector of YTHDF2-Myc proto-oncogene protein (MYC) axis in GSCs. Inhibiting glioblastoma stem cells viability without affecting NSCs and impairing in vivo glioblastoma growth. | |||
| Target Regulator | YTH domain-containing family protein 2 (YTHDF2) | READER | ||
| Target Regulation | Up regulation | |||
| Responsed Disease | Glioblastoma | ICD-11: 2A00.00 | ||
| In-vitro Model | NSC11 (Pluripotent derived neural progenitor cell) | |||
| NHA (Normal human astrocytes) | ||||
| HNP1 (A human neural progenitor cell) | ||||
| ENSA (A human embryonic stem derived neural progenitor cell) | ||||
| In-vivo Model | Implanting 5000 human derived GSCs into the right cerebral cortex of NSG mice at a depth of 3.5 mm under a University of California, San Diego Institutional Animal Care and Use Committee (IACUC) approved protocol. Brains were harvested and fixed in 4% formaldehyde, cryopreserved in 30% sucrose, and then cryosectioned. Hematoxylin and eosin (H&E) staining was performed on sections for histological analysis. In parallel survival experiments, mice were observed until the development of neurological signs. For in vivo drug treatment studies, intracranial xenografts were generated by implanting 5000 patient-derived GSCs (387 and 4121) into the right cerebral cortex of NSG mice as described above. Mice recovered for 7 days were randomly assigned into drug vs. treatment group by a blinded investigator. Mice were then treated daily with either vehicle (25 mM Tartaric acid) or 50 mg/kg linsitinib by oral gavage. | |||
R-2HG
[Investigative]
| In total 2 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response | [14] | |||
| Response Summary | This work demonstrates anti-tumor effects of 2HG in inhibiting proliferation/survival of FTO-high cancer cells via targeting FTO/m6A/Myc proto-oncogene protein (MYC)/CEBPA signaling.High levels of FTO sensitize leukemia cells to R-2HG, whereas hyperactivation of MYC signaling confers resistance that can be reversed by the inhibition of MYC signaling. R-2HG also displays anti-tumor activity in glioma. High levels of FTO sensitize leukemic cells to R-2HG, whereas hyperactivation of MYC signaling confers resistance that can be reversed by the inhibition of MYC signaling. | |||
| Target Regulator | Fat mass and obesity-associated protein (FTO) | ERASER | ||
| Target Regulation | Down regulation | |||
| Responsed Disease | Glioma | ICD-11: 2A00.0 | ||
| Cell Process | Glutamine metabolism | |||
| Cell apoptosis | ||||
| In-vitro Model | 8-MG-BA | Glioblastoma | Homo sapiens | CVCL_1052 |
| A-172 | Glioblastoma | Homo sapiens | CVCL_0131 | |
| DK-MG | Glioblastoma | Homo sapiens | CVCL_1173 | |
| GaMG | Glioblastoma | Homo sapiens | CVCL_1226 | |
| HEL | Erythroleukemia | Homo sapiens | CVCL_0001 | |
| Jurkat | T acute lymphoblastic leukemia | Homo sapiens | CVCL_0065 | |
| KOCL-45 | B acute lymphoblastic leukemia | Homo sapiens | CVCL_3993 | |
| KOCL-48 | Childhood acute monocytic leukemia | Homo sapiens | CVCL_6867 | |
| KOCL-50 | B acute lymphoblastic leukemia | Homo sapiens | CVCL_6866 | |
| KOCL-51 | B acute lymphoblastic leukemia | Homo sapiens | CVCL_6865 | |
| KOCL-69 | B acute lymphoblastic leukemia | Homo sapiens | CVCL_3995 | |
| KOPN-1 | B acute lymphoblastic leukemia | Homo sapiens | CVCL_3937 | |
| LN-18 | Glioblastoma | Homo sapiens | CVCL_0392 | |
| LN-229 | Glioblastoma | Homo sapiens | CVCL_0393 | |
| MA9.3 (MA9.3) | ||||
| MA9.6ITD (MLL-AF9 plus FLT3-ITD) | ||||
| MA9.6RAS (MLL-AF9 plus NRasG12D) | ||||
| MA9.6 (MLL-AF9) | ||||
| MA9.6ITD (MLL-AF9 plus FLT3-ITD) | ||||
| MA9.6RAS (MLL-AF9 plus NRasG12D) | ||||
| ME-1 [Human leukemia] | Adult acute myeloid leukemia | Homo sapiens | CVCL_2110 | |
| ML-2 | Adult acute myeloid leukemia | Homo sapiens | CVCL_1418 | |
| MV4-11 | Childhood acute monocytic leukemia | Homo sapiens | CVCL_0064 | |
| NB4 | Acute promyelocytic leukemia | Homo sapiens | CVCL_0005 | |
| NOMO-1 | Adult acute monocytic leukemia | Homo sapiens | CVCL_1609 | |
| PL21 | Familial adenomatous polyposis | Homo sapiens | CVCL_JM48 | |
| T98G | Glioblastoma | Homo sapiens | CVCL_0556 | |
| THP-1 | Childhood acute monocytic leukemia | Homo sapiens | CVCL_0006 | |
| U-87MG ATCC | Glioblastoma | Homo sapiens | CVCL_0022 | |
| U-937 | Adult acute monocytic leukemia | Homo sapiens | CVCL_0007 | |
| In-vivo Model | For R-2HG injection mouse models, sensitive (NOMO-1 and MA9.3ITD) or resistant (MA9.3RAS) cells were injected into NSGS or NRGS intravenously, and then R-2HG (6mg/kg body weight) or PBS were injected once daily through tail vein for 12 consecutive days starting from day 11 post xeno-transplantation. | |||
| Experiment 2 Reporting the m6A-centered Drug Response | [14] | |||
| Response Summary | This work demonstrates anti-tumor effects of 2HG in inhibiting proliferation/survival of FTO-high cancer cells via targeting FTO/m6A/Myc proto-oncogene protein (MYC)/CEBPA signaling.High levels of FTO sensitize leukemia cells to R-2HG, whereas hyperactivation of MYC signaling confers resistance that can be reversed by the inhibition of MYC signaling. R-2HG also displays anti-tumor activity in glioma. High levels of FTO sensitize leukemic cells to R-2HG, whereas hyperactivation of MYC signaling confers resistance that can be reversed by the inhibition of MYC signaling. | |||
| Target Regulator | Fat mass and obesity-associated protein (FTO) | ERASER | ||
| Target Regulation | Down regulation | |||
| Responsed Disease | Leukaemia | ICD-11: 2B33.4 | ||
| Cell Process | Glutamine metabolism | |||
| Cell apoptosis | ||||
| In-vitro Model | 8-MG-BA | Glioblastoma | Homo sapiens | CVCL_1052 |
| A-172 | Glioblastoma | Homo sapiens | CVCL_0131 | |
| DK-MG | Glioblastoma | Homo sapiens | CVCL_1173 | |
| GaMG | Glioblastoma | Homo sapiens | CVCL_1226 | |
| HEL | Erythroleukemia | Homo sapiens | CVCL_0001 | |
| Jurkat | T acute lymphoblastic leukemia | Homo sapiens | CVCL_0065 | |
| KOCL-45 | B acute lymphoblastic leukemia | Homo sapiens | CVCL_3993 | |
| KOCL-48 | Childhood acute monocytic leukemia | Homo sapiens | CVCL_6867 | |
| KOCL-50 | B acute lymphoblastic leukemia | Homo sapiens | CVCL_6866 | |
| KOCL-51 | B acute lymphoblastic leukemia | Homo sapiens | CVCL_6865 | |
| KOCL-69 | B acute lymphoblastic leukemia | Homo sapiens | CVCL_3995 | |
| KOPN-1 | B acute lymphoblastic leukemia | Homo sapiens | CVCL_3937 | |
| LN-18 | Glioblastoma | Homo sapiens | CVCL_0392 | |
| LN-229 | Glioblastoma | Homo sapiens | CVCL_0393 | |
| MA9.3 (MA9.3) | ||||
| MA9.6ITD (MLL-AF9 plus FLT3-ITD) | ||||
| MA9.6RAS (MLL-AF9 plus NRasG12D) | ||||
| MA9.6 (MLL-AF9) | ||||
| MA9.6ITD (MLL-AF9 plus FLT3-ITD) | ||||
| MA9.6RAS (MLL-AF9 plus NRasG12D) | ||||
| ME-1 [Human leukemia] | Adult acute myeloid leukemia | Homo sapiens | CVCL_2110 | |
| ML-2 | Adult acute myeloid leukemia | Homo sapiens | CVCL_1418 | |
| MV4-11 | Childhood acute monocytic leukemia | Homo sapiens | CVCL_0064 | |
| NB4 | Acute promyelocytic leukemia | Homo sapiens | CVCL_0005 | |
| NOMO-1 | Adult acute monocytic leukemia | Homo sapiens | CVCL_1609 | |
| PL21 | Familial adenomatous polyposis | Homo sapiens | CVCL_JM48 | |
| T98G | Glioblastoma | Homo sapiens | CVCL_0556 | |
| THP-1 | Childhood acute monocytic leukemia | Homo sapiens | CVCL_0006 | |
| U-87MG ATCC | Glioblastoma | Homo sapiens | CVCL_0022 | |
| U-937 | Adult acute monocytic leukemia | Homo sapiens | CVCL_0007 | |
| In-vivo Model | For R-2HG injection mouse models, sensitive (NOMO-1 and MA9.3ITD) or resistant (MA9.3RAS) cells were injected into NSGS or NRGS intravenously, and then R-2HG (6mg/kg body weight) or PBS were injected once daily through tail vein for 12 consecutive days starting from day 11 post xeno-transplantation. | |||
Full List of Crosstalk(s) between m6A Modification and Epigenetic Regulation Related to This Regulator
RNA modification
m6A Regulator: Fat mass and obesity-associated protein (FTO)
| In total 4 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT00027 | ||
| Epigenetic Regulator | tRNA methyltransferase 10 homolog A (TRMT10A) | |
| Regulated Target | tRNA-Gln (anticodon TTG) 1-1 (TRQ-TTG1-1) | |
| Crosstalk relationship | m1G → m6A | |
| Drug | AVI-5126 | |
| Crosstalk ID: M6ACROT00029 | ||
| Epigenetic Regulator | tRNA methyltransferase 10 homolog A (TRMT10A) | |
| Regulated Target | tRNA-Arg (anticodon CCG) 1-1 (TRR-CCG1-1) | |
| Crosstalk relationship | m1G → m6A | |
| Drug | AVI-5126 | |
| Crosstalk ID: M6ACROT00031 | ||
| Epigenetic Regulator | tRNA methyltransferase 10 homolog A (TRMT10A) | |
| Regulated Target | tRNA-Thr (anticodon CGT) 1-1 (TRT-CGT1-1) | |
| Crosstalk relationship | m1G → m6A | |
| Drug | AVI-5126 | |
| Crosstalk ID: M6ACROT00045 | ||
| Epigenetic Regulator | tRNA methyltransferase 10 homolog A (TRMT10A) | |
| Regulated Target | Myc proto-oncogene protein (MYC) | |
| Crosstalk relationship | m1G → m6A | |
| Drug | AVI-5126 | |
m6A Regulator: YTH domain-containing family protein 2 (YTHDF2)
| In total 3 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT00028 | ||
| Epigenetic Regulator | tRNA methyltransferase 10 homolog A (TRMT10A) | |
| Regulated Target | tRNA-Gln (anticodon TTG) 1-1 (TRQ-TTG1-1) | |
| Crosstalk relationship | m1G → m6A | |
| Drug | AVI-5126 | |
| Crosstalk ID: M6ACROT00030 | ||
| Epigenetic Regulator | tRNA methyltransferase 10 homolog A (TRMT10A) | |
| Regulated Target | tRNA-Arg (anticodon CCG) 1-1 (TRR-CCG1-1) | |
| Crosstalk relationship | m1G → m6A | |
| Drug | AVI-5126 | |
| Crosstalk ID: M6ACROT00032 | ||
| Epigenetic Regulator | tRNA methyltransferase 10 homolog A (TRMT10A) | |
| Regulated Target | tRNA-Thr (anticodon CGT) 1-1 (TRT-CGT1-1) | |
| Crosstalk relationship | m1G → m6A | |
| Drug | AVI-5126 | |
m6A Regulator: Insulin-like growth factor 2 mRNA-binding protein 1 (IGF2BP1)
| In total 1 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT00228 | ||
| Epigenetic Regulator | Y-box-binding protein 1 (YBX1) | |
| Regulated Target | Myc proto-oncogene protein (MYC) | |
| Crosstalk relationship | m6A → m5C | |
| Drug | AVI-5126 | |
DNA modification
m6A Regulator: RNA demethylase ALKBH5 (ALKBH5)
| In total 1 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT02036 | ||
| Epigenetic Regulator | DNA (cytosine-5)-methyltransferase 1 (DNMT1) | |
| Regulated Target | RNA demethylase ALKBH5 (ALKBH5) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Lung cancer | |
m6A Regulator: YTH domain-containing family protein 2 (YTHDF2)
| In total 1 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT02039 | ||
| Epigenetic Regulator | DNA (cytosine-5)-methyltransferase 1 (DNMT1) | |
| Regulated Target | RNA demethylase ALKBH5 (ALKBH5) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Lung cancer | |
m6A Regulator: Insulin-like growth factor 2 mRNA-binding protein 1 (IGF2BP1)
| In total 5 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT02075 | ||
| Epigenetic Regulator | Cysteine methyltransferase DNMT3A (DNMT3A) | |
| Regulated Target | Protein tyrosine phosphatase non-receptor type 13 (PTPN13) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Liver cancer | |
| Crosstalk ID: M6ACROT02194 | ||
| Epigenetic Regulator | Cysteine methyltransferase DNMT3A (DNMT3A) | |
| Regulated Target | Protein tyrosine phosphatase non-receptor type 13 (PTPN13) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Liver cancer | |
| Crosstalk ID: M6ACROT05864 | ||
| Epigenetic Regulator | Cysteine methyltransferase DNMT3A (DNMT3A) | |
| Regulated Target | Long intergenic non-protein coding RNA 261 (LINC00261) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Pancreatic cancer | |
| Crosstalk ID: M6ACROT05865 | ||
| Epigenetic Regulator | DNA (cytosine-5)-methyltransferase 3B (DNMT3B) | |
| Regulated Target | Long intergenic non-protein coding RNA 261 (LINC00261) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Pancreatic cancer | |
| Crosstalk ID: M6ACROT05866 | ||
| Epigenetic Regulator | DNA (cytosine-5)-methyltransferase 1 (DNMT1) | |
| Regulated Target | Long intergenic non-protein coding RNA 261 (LINC00261) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Pancreatic cancer | |
Histone modification
m6A Regulator: Methyltransferase-like 3 (METTL3)
| In total 6 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT03106 | ||
| Regulated Target | Histone H3 lysine 18 lactylation (H3K18la) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Esophageal Squamous Cell Carcinoma | |
| Crosstalk ID: M6ACROT03550 | ||
| Epigenetic Regulator | Histone acetyltransferase p300 (P300) | |
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
| Crosstalk ID: M6ACROT03595 | ||
| Epigenetic Regulator | N-lysine methyltransferase SMYD2 (SMYD2) | |
| Regulated Target | Histone H3 lysine 4 trimethylation (H3K4me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
| Crosstalk ID: M6ACROT03648 | ||
| Epigenetic Regulator | Histone-lysine N-methyltransferase EZH2 (EZH2) | |
| Regulated Target | Histone H3 lysine 27 trimethylation (H3K27me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Gastric cancer | |
| Crosstalk ID: M6ACROT03651 | ||
| Epigenetic Regulator | Histone-lysine N-methyltransferase EZH2 (EZH2) | |
| Regulated Target | Histone H3 lysine 27 trimethylation (H3K27me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Gastric cancer | |
| Crosstalk ID: M6ACROT03660 | ||
| Epigenetic Regulator | Histone-lysine N-methyltransferase EZH2 (EZH2) | |
| Regulated Target | Histone H3 lysine 27 trimethylation (H3K27me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Gastric cancer | |
m6A Regulator: Insulin-like growth factor 2 mRNA-binding protein 2 (IGF2BP2)
| In total 4 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT03133 | ||
| Epigenetic Regulator | Histone acetyltransferase p300 (P300) | |
| Regulated Target | Histone H3 lysine 18 lactylation (H3K18la) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Pancreatic cancer | |
| Crosstalk ID: M6ACROT03476 | ||
| Epigenetic Regulator | Histone acetyltransferase p300 (P300) | |
| Regulated Target | Histone H3 lysine 18 lactylation (H3K18la) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Pancreatic cancer | |
| Crosstalk ID: M6ACROT03477 | ||
| Epigenetic Regulator | Histone acetyltransferase p300 (P300) | |
| Regulated Target | Histone H3 lysine 18 lactylation (H3K18la) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Pancreatic cancer | |
| Crosstalk ID: M6ACROT06042 | ||
| Epigenetic Regulator | Histone acetyltransferase p300 (P300) | |
| Regulated Target | Histone H3 lysine 18 lactylation (H3K18la) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Pancreatic cancer | |
| Drug | Gemcitabine | |
m6A Regulator: Insulin-like growth factor 2 mRNA-binding protein 1 (IGF2BP1)
| In total 2 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT03155 | ||
| Epigenetic Regulator | Histone acetyltransferase p300 (P300) | |
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Intrahepatic cholangiocarcinoma | |
| Crosstalk ID: M6ACROT05863 | ||
| Epigenetic Regulator | Histone-lysine N-methyltransferase EZH2 (EZH2) | |
| Regulated Target | Histone H3 lysine 27 trimethylation (H3K27me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Pancreatic cancer | |
m6A Regulator: YTH domain-containing family protein 2 (YTHDF2)
| In total 3 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT03419 | ||
| Regulated Target | Histone H3 lysine 4 trimethylation (H3K4me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Liver cancer | |
| Crosstalk ID: M6ACROT03430 | ||
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Liver cancer | |
| Crosstalk ID: M6ACROT03539 | ||
| Epigenetic Regulator | Histone-lysine N-methyltransferase EZH2 (EZH2) | |
| Regulated Target | Histone H3 lysine 27 trimethylation (H3K27me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Brain cancer | |
m6A Regulator: Fat mass and obesity-associated protein (FTO)
| In total 2 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT03431 | ||
| Epigenetic Regulator | Histone deacetylase 1 (HDAC1) | |
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
| Crosstalk ID: M6ACROT03447 | ||
| Epigenetic Regulator | Histone deacetylase 1 (HDAC1) | |
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
Non-coding RNA
m6A Regulator: Insulin-like growth factor 2 mRNA-binding protein 1 (IGF2BP1)
| In total 7 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT05004 | ||
| Epigenetic Regulator | Long intergenic non-protein coding RNA 261 (LINC00261) | |
| Regulated Target | Insulin like growth factor 2 mRNA binding protein 1 (IGF2BP1) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Pancreatic cancer | |
| Crosstalk ID: M6ACROT05008 | ||
| Epigenetic Regulator | Neuroblastoma associated transcript 1 (NBAT1) | |
| Regulated Target | Insulin like growth factor 2 mRNA binding protein 1 (IGF2BP1) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Liver cancer | |
| Crosstalk ID: M6ACROT05058 | ||
| Epigenetic Regulator | Septin 14 pseudogene 20 (SEPTIN14P20/LINC00266-1) | |
| Regulated Target | Insulin like growth factor 2 mRNA binding protein 1 (IGF2BP1) | |
| Crosstalk relationship | ncRNA → m6A | |
| Crosstalk ID: M6ACROT05172 | ||
| Epigenetic Regulator | FERM, ARH/RhoGEF and pleckstrin domain protein 1 (FARP1) | |
| Regulated Target | Insulin like growth factor 2 mRNA binding protein 1 (IGF2BP1) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Acute myeloid leukaemia | |
| Drug | Cytarabine | |
| Crosstalk ID: M6ACROT05366 | ||
| Epigenetic Regulator | KB-1980E6.3 | |
| Regulated Target | Insulin like growth factor 2 mRNA binding protein 1 (IGF2BP1) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Breast cancer | |
| Crosstalk ID: M6ACROT05867 | ||
| Epigenetic Regulator | Long intergenic non-protein coding RNA 261 (LINC00261) | |
| Regulated Target | Insulin like growth factor 2 mRNA binding protein 1 (IGF2BP1) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Pancreatic cancer | |
| Crosstalk ID: M6ACROT05891 | ||
| Epigenetic Regulator | Testis associated oncogenic lncRNA (THORLNC) | |
| Regulated Target | Insulin like growth factor 2 mRNA binding protein 1 (IGF2BP1) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Nasopharyngeal carcinoma | |
| Drug | Triptonide | |
m6A Regulator: Insulin-like growth factor 2 mRNA-binding protein 3 (IGF2BP3)
| In total 2 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT05012 | ||
| Epigenetic Regulator | DARS1 antisense RNA 1 (DARS1-AS1) | |
| Regulated Target | Insulin like growth factor 2 mRNA binding protein 3 (IGF2BP3) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Cervical cancer | |
| Crosstalk ID: M6ACROT05216 | ||
| Epigenetic Regulator | LOC101929709 | |
| Regulated Target | Insulin like growth factor 2 mRNA binding protein 3 (IGF2BP3) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Gastric cancer | |
m6A Regulator: YTH domain-containing family protein 2 (YTHDF2)
| In total 1 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT05047 | ||
| Epigenetic Regulator | MicroRNA 145 (MIR145) | |
| Regulated Target | YTH domain-containing family protein 2 (YTHDF2) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Liver cancer | |
m6A Regulator: Insulin-like growth factor 2 mRNA-binding protein 2 (IGF2BP2)
| In total 3 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT05055 | ||
| Epigenetic Regulator | Long intergenic non-protein coding RNA 920 (LINC00920) | |
| Regulated Target | Insulin like growth factor 2 mRNA binding protein 2 (IGF2BP2) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Colorectal cancer | |
| Crosstalk ID: M6ACROT05319 | ||
| Epigenetic Regulator | PTGS2 antisense RNA 1 (PACERR) | |
| Regulated Target | Insulin like growth factor 2 mRNA binding protein 2 (IGF2BP2) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Pancreatic ductal adenocarcinoma | |
| Crosstalk ID: M6ACROT05950 | ||
| Epigenetic Regulator | Long intergenic non-protein coding RNA 901 (LINC00901) | |
| Regulated Target | Insulin like growth factor 2 mRNA binding protein 2 (IGF2BP2) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | pancreatic cancer | |
m6A Regulator: Methyltransferase-like 3 (METTL3)
| In total 3 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT05103 | ||
| Epigenetic Regulator | hsa-miR-493-5p | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Acute myeloid leukaemia | |
| Crosstalk ID: M6ACROT05104 | ||
| Epigenetic Regulator | Maternally expressed 3 (MEG3) | |
| Regulated Target | hsa-miR-493-5p | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Acute myeloid leukaemia | |
| Crosstalk ID: M6ACROT05327 | ||
| Epigenetic Regulator | hsa-miR-338-5p | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Lung cancer | |
m6A Regulator: RNA-binding protein Musashi homolog 2 (MSI2)
| In total 1 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT05107 | ||
| Epigenetic Regulator | Long intergenic non-protein coding RNA 942 (LINC00942) | |
| Regulated Target | Musashi RNA binding protein 2 (MSI2) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Gastric cancer | |
| Drug | Cisplatin | |
m6A Regulator: YTH domain-containing family protein 1 (YTHDF1)
| In total 1 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT05115 | ||
| Epigenetic Regulator | DLGAP1 antisense RNA 2 (DLGAP1-AS2) | |
| Regulated Target | YTH domain-containing family protein 1 (YTHDF1) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Non-small cell lung cancer | |
m6A Regulator: Fat mass and obesity-associated protein (FTO)
| In total 10 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT05127 | ||
| Epigenetic Regulator | hsa-let-7b-5p | |
| Regulated Target | FTO alpha-ketoglutarate dependent dioxygenase (FTO) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Acute myeloid leukaemia | |
| Crosstalk ID: M6ACROT05163 | ||
| Epigenetic Regulator | Long intergenic non-protein coding RNA 858 (LINC00858) | |
| Regulated Target | Zinc finger protein 184 (ZNF184) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Esophageal Squamous Cell Carcinoma | |
| Crosstalk ID: M6ACROT05181 | ||
| Epigenetic Regulator | hsa-miR-218-5p | |
| Regulated Target | FTO alpha-ketoglutarate dependent dioxygenase (FTO) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Colorectal cancer | |
| Crosstalk ID: M6ACROT05182 | ||
| Epigenetic Regulator | hsa_circ_0001739 (Circ_ZNF277) | |
| Regulated Target | hsa-miR-218-5p | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Colorectal cancer | |
| Crosstalk ID: M6ACROT05183 | ||
| Epigenetic Regulator | AC159540.1 | |
| Regulated Target | hsa-miR-218-5p | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Colorectal cancer | |
| Crosstalk ID: M6ACROT05237 | ||
| Epigenetic Regulator | hsa-miR-607 | |
| Regulated Target | FTO alpha-ketoglutarate dependent dioxygenase (FTO) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Non-small cell lung cancer | |
| Crosstalk ID: M6ACROT05251 | ||
| Epigenetic Regulator | hsa_circ_0072309 (Circ_LIFR) | |
| Regulated Target | hsa-miR-607 | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Non-small cell lung cancer | |
| Crosstalk ID: M6ACROT05320 | ||
| Epigenetic Regulator | hsa-miR-96 | |
| Regulated Target | Protein kinase AMP-activated catalytic subunit alpha 2 (PRKAA2) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Colorectal cancer | |
| Crosstalk ID: M6ACROT05437 | ||
| Epigenetic Regulator | MicroRNA 23a (MIR23A) | |
| Regulated Target | Max-interacting protein 1 (MXI1) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Brain cancer | |
| Drug | Temozolomide | |
| Crosstalk ID: M6ACROT05438 | ||
| Epigenetic Regulator | MicroRNA 155 (MIR155) | |
| Regulated Target | Max-interacting protein 1 (MXI1) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Brain cancer | |
| Drug | Temozolomide | |
m6A Regulator: Protein lin-28 homolog B (LIN28B)
| In total 1 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT05217 | ||
| Epigenetic Regulator | LOC101929709 | |
| Regulated Target | Protein lin-28 homolog B (LIN28B) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Gastric cancer | |
RNA Modification Sequencing Data Associated with the Target (ID: M6ATAR00341)
| In total 5 m6A sequence/site(s) in this target gene | |||
| mod ID: 2OMSITE000491 | Click to Show/Hide the Full List | ||
| mod site | chr8:127741082-127741083:+ | [73] | |
| Sequence | GTCTTGAGACTGAAAGATTTAGCCATAATGTAAACTGCCTC | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | Nm-seq | ||
| Transcript ID List | ENST00000651626.1; ENST00000377970.6; ENST00000524013.1; ENST00000621592.6; ENST00000652288.1 | ||
| External Link | RMBase: Nm_site_6512 | ||
| mod ID: 2OMSITE000488 | Click to Show/Hide the Full List | ||
| mod site | chr8:127736579-127736580:+ | [73] | |
| Sequence | TCTCTGAAAGGCTCTCCTTGCAGCTGCTTAGACGCTGGATT | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | Nm-seq | ||
| Transcript ID List | ENST00000517291.1; ENST00000651626.1; ENST00000520751.1; ENST00000377970.6; ENST00000524013.1; ENST00000641036.1; ENST00000621592.6; ENST00000259523.10 | ||
| External Link | RMBase: Nm_site_6509 | ||
| mod ID: 2OMSITE000487 | Click to Show/Hide the Full List | ||
| mod site | chr8:127736571-127736572:+ | [73] | |
| Sequence | GACCCGCTTCTCTGAAAGGCTCTCCTTGCAGCTGCTTAGAC | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | Nm-seq | ||
| Transcript ID List | ENST00000641036.1; ENST00000651626.1; ENST00000524013.1; ENST00000621592.6; ENST00000520751.1; ENST00000517291.1; ENST00000259523.10; ENST00000377970.6 | ||
| External Link | RMBase: Nm_site_6508 | ||
| mod ID: 2OMSITE000489 | Click to Show/Hide the Full List | ||
| mod site | chr8:127740592-127740593:+ | [73] | |
| Sequence | CTCCCTCCACTCGGAAGGACTATCCTGCTGCCAAGAGGGTC | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | Nm-seq | ||
| Transcript ID List | ENST00000651626.1; ENST00000524013.1; ENST00000621592.6; ENST00000652288.1; ENST00000613283.2; ENST00000652131.1; ENST00000377970.6 | ||
| External Link | RMBase: Nm_site_6510 | ||
| mod ID: 2OMSITE000490 | Click to Show/Hide the Full List | ||
| mod site | chr8:127741079-127741080:+ | [73] | |
| Sequence | TGAGTCTTGAGACTGAAAGATTTAGCCATAATGTAAACTGC | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | Nm-seq | ||
| Transcript ID List | ENST00000652288.1; ENST00000377970.6; ENST00000621592.6; ENST00000524013.1; ENST00000651626.1 | ||
| External Link | RMBase: Nm_site_6511 | ||
5-methylcytidine (m5C)
| In total 11 m6A sequence/site(s) in this target gene | |||
| mod ID: M5CSITE004339 | Click to Show/Hide the Full List | ||
| mod site | chr8:127739465-127739466:+ | ||
| Sequence | CTATCTACAAAAATGAGGGGCTGTGTTTAGAGGCTAGGCAG | ||
| Cell/Tissue List | T24 | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000377970.6; ENST00000259523.10; ENST00000613283.2; ENST00000524013.1; ENST00000652288.1; ENST00000651626.1; ENST00000652131.1; ENST00000621592.6 | ||
| External Link | RMBase: m5C_site_41754 | ||
| mod ID: M5CSITE004340 | Click to Show/Hide the Full List | ||
| mod site | chr8:127740154-127740155:+ | ||
| Sequence | TTTTAGTAGAGATGGGGTTTCATCGTGTTGGCCAGGATGGT | ||
| Cell/Tissue List | T24 | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000651626.1; ENST00000652288.1; ENST00000621592.6; ENST00000524013.1; ENST00000613283.2; ENST00000652131.1; ENST00000259523.10; ENST00000377970.6 | ||
| External Link | RMBase: m5C_site_41755 | ||
| mod ID: M5CSITE004341 | Click to Show/Hide the Full List | ||
| mod site | chr8:127740157-127740158:+ | ||
| Sequence | TAGTAGAGATGGGGTTTCATCGTGTTGGCCAGGATGGTCTC | ||
| Cell/Tissue List | T24 | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000652288.1; ENST00000259523.10; ENST00000621592.6; ENST00000377970.6; ENST00000524013.1; ENST00000651626.1; ENST00000613283.2; ENST00000652131.1 | ||
| External Link | RMBase: m5C_site_41756 | ||
| mod ID: M5CSITE004342 | Click to Show/Hide the Full List | ||
| mod site | chr8:127740165-127740166:+ | ||
| Sequence | ATGGGGTTTCATCGTGTTGGCCAGGATGGTCTCTCCTGACC | ||
| Cell/Tissue List | T24 | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000377970.6; ENST00000652288.1; ENST00000259523.10; ENST00000652131.1; ENST00000621592.6; ENST00000524013.1; ENST00000651626.1; ENST00000613283.2 | ||
| External Link | RMBase: m5C_site_41757 | ||
| mod ID: M5CSITE004343 | Click to Show/Hide the Full List | ||
| mod site | chr8:127740166-127740167:+ | ||
| Sequence | TGGGGTTTCATCGTGTTGGCCAGGATGGTCTCTCCTGACCT | ||
| Cell/Tissue List | T24 | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000377970.6; ENST00000259523.10; ENST00000652131.1; ENST00000613283.2; ENST00000652288.1; ENST00000621592.6; ENST00000651626.1; ENST00000524013.1 | ||
| External Link | RMBase: m5C_site_41758 | ||
| mod ID: M5CSITE004344 | Click to Show/Hide the Full List | ||
| mod site | chr8:127740206-127740207:+ | ||
| Sequence | TCACGATCCGCCCACCTCGGCCTCCCAAAGTGCTGGGATTA | ||
| Cell/Tissue List | T24 | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000259523.10; ENST00000377970.6; ENST00000651626.1; ENST00000524013.1; ENST00000621592.6; ENST00000652288.1; ENST00000652131.1; ENST00000613283.2 | ||
| External Link | RMBase: m5C_site_41759 | ||
| mod ID: M5CSITE004345 | Click to Show/Hide the Full List | ||
| mod site | chr8:127740207-127740208:+ | ||
| Sequence | CACGATCCGCCCACCTCGGCCTCCCAAAGTGCTGGGATTAC | ||
| Cell/Tissue List | T24 | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000621592.6; ENST00000652131.1; ENST00000377970.6; ENST00000651626.1; ENST00000652288.1; ENST00000524013.1; ENST00000613283.2; ENST00000259523.10 | ||
| External Link | RMBase: m5C_site_41760 | ||
| mod ID: M5CSITE004346 | Click to Show/Hide the Full List | ||
| mod site | chr8:127740209-127740210:+ | ||
| Sequence | CGATCCGCCCACCTCGGCCTCCCAAAGTGCTGGGATTACAG | ||
| Cell/Tissue List | T24 | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000621592.6; ENST00000613283.2; ENST00000524013.1; ENST00000259523.10; ENST00000377970.6; ENST00000651626.1; ENST00000652131.1; ENST00000652288.1 | ||
| External Link | RMBase: m5C_site_41761 | ||
| mod ID: M5CSITE004347 | Click to Show/Hide the Full List | ||
| mod site | chr8:127740210-127740211:+ | ||
| Sequence | GATCCGCCCACCTCGGCCTCCCAAAGTGCTGGGATTACAGG | ||
| Cell/Tissue List | T24 | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000613283.2; ENST00000621592.6; ENST00000652288.1; ENST00000259523.10; ENST00000652131.1; ENST00000524013.1; ENST00000651626.1; ENST00000377970.6 | ||
| External Link | RMBase: m5C_site_41762 | ||
| mod ID: M5CSITE004348 | Click to Show/Hide the Full List | ||
| mod site | chr8:127740218-127740219:+ | ||
| Sequence | CACCTCGGCCTCCCAAAGTGCTGGGATTACAGGTGTGAGCC | ||
| Cell/Tissue List | T24 | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000621592.6; ENST00000259523.10; ENST00000652288.1; ENST00000651626.1; ENST00000524013.1; ENST00000377970.6; ENST00000652131.1; ENST00000613283.2 | ||
| External Link | RMBase: m5C_site_41763 | ||
| mod ID: M5CSITE004349 | Click to Show/Hide the Full List | ||
| mod site | chr8:127740974-127740975:+ | [74] | |
| Sequence | GTAAGGAAAAGTAAGGAAAACGATTCCTTCTAACAGAAATG | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000377970.6; ENST00000524013.1; ENST00000651626.1; ENST00000652288.1; ENST00000621592.6 | ||
| External Link | RMBase: m5C_site_41764 | ||
N6-methyladenosine (m6A)
| In total 74 m6A sequence/site(s) in this target gene | |||
| mod ID: M6ASITE086875 | Click to Show/Hide the Full List | ||
| mod site | chr8:127735610-127735611:+ | [75] | |
| Sequence | TTTCCTCCACTCTCCCTGGGACTCTTGATCAAAGCGCGGCC | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa; MSC | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000517291.1; ENST00000641252.1; ENST00000259523.10 | ||
| External Link | RMBase: m6A_site_809494 | ||
| mod ID: M6ASITE086876 | Click to Show/Hide the Full List | ||
| mod site | chr8:127735752-127735753:+ | [76] | |
| Sequence | CGATGATTTATACTCACAGGACAAGGATGCGGTTTGTCAAA | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; MT4; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000517291.1; ENST00000651626.1; ENST00000259523.10; ENST00000641252.1 | ||
| External Link | RMBase: m6A_site_809495 | ||
| mod ID: M6ASITE086877 | Click to Show/Hide the Full List | ||
| mod site | chr8:127735772-127735773:+ | [76] | |
| Sequence | ACAAGGATGCGGTTTGTCAAACAGTACTGCTACGGAGGAGC | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa; MT4; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000259523.10; ENST00000641252.1; ENST00000517291.1; ENST00000651626.1 | ||
| External Link | RMBase: m6A_site_809496 | ||
| mod ID: M6ASITE086878 | Click to Show/Hide the Full List | ||
| mod site | chr8:127736069-127736070:+ | [76] | |
| Sequence | GAGGGTCTGGACGGCTGAGGACCCCCGAGCTGTGCTGCTCG | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; HepG2; A549; MT4; CD4T; peripheral-blood; HEK293T; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000517291.1; ENST00000651626.1; ENST00000641036.1; ENST00000621592.6; ENST00000259523.10 | ||
| External Link | RMBase: m6A_site_809497 | ||
| mod ID: M6ASITE086879 | Click to Show/Hide the Full List | ||
| mod site | chr8:127736427-127736428:+ | [76] | |
| Sequence | ACCCTTGCCGCATCCACGAAACTTTGCCCATAGCAGCGGGC | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa; HepG2; A549; MM6; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293T; iSLK; TIME; TREX; MSC; endometrial; HEC-1-A; GSCs; NB4; AML | ||
| Seq Type List | m6A-seq; MeRIP-seq; m6A-CLIP/IP; miCLIP | ||
| Transcript ID List | ENST00000641036.1; ENST00000524013.1; ENST00000621592.6; ENST00000517291.1; ENST00000259523.10; ENST00000651626.1; ENST00000520751.1; ENST00000377970.6 | ||
| External Link | RMBase: m6A_site_809498 | ||
| mod ID: M6ASITE086880 | Click to Show/Hide the Full List | ||
| mod site | chr8:127736465-127736466:+ | [76] | |
| Sequence | GGCGGGCACTTTGCACTGGAACTTACAACACCCGAGCAAGG | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa; HepG2; A549; CD8T; MM6; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293T; TIME; TREX; MSC; endometrial; HEC-1-A; GSCs; NB4; AML | ||
| Seq Type List | m6A-seq; MeRIP-seq; m6A-CLIP/IP; miCLIP | ||
| Transcript ID List | ENST00000520751.1; ENST00000377970.6; ENST00000641036.1; ENST00000517291.1; ENST00000524013.1; ENST00000651626.1; ENST00000621592.6; ENST00000259523.10 | ||
| External Link | RMBase: m6A_site_809499 | ||
| mod ID: M6ASITE086882 | Click to Show/Hide the Full List | ||
| mod site | chr8:127736469-127736470:+ | [77] | |
| Sequence | GGCACTTTGCACTGGAACTTACAACACCCGAGCAAGGACGC | ||
| Motif Score | 2.07285119 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000651626.1; ENST00000377970.6; ENST00000641036.1; ENST00000524013.1; ENST00000259523.10; ENST00000520751.1; ENST00000517291.1; ENST00000621592.6 | ||
| External Link | RMBase: m6A_site_809500 | ||
| mod ID: M6ASITE086883 | Click to Show/Hide the Full List | ||
| mod site | chr8:127736472-127736473:+ | [77] | |
| Sequence | ACTTTGCACTGGAACTTACAACACCCGAGCAAGGACGCGAC | ||
| Motif Score | 2.173910714 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000259523.10; ENST00000377970.6; ENST00000517291.1; ENST00000641036.1; ENST00000520751.1; ENST00000621592.6; ENST00000651626.1; ENST00000524013.1 | ||
| External Link | RMBase: m6A_site_809501 | ||
| mod ID: M6ASITE086884 | Click to Show/Hide the Full List | ||
| mod site | chr8:127736530-127736531:+ | [76] | |
| Sequence | GCTATTCTGCCCATTTGGGGACACTTCCCCGCCGCTGCCAG | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; CD34; HepG2; HEK293; A549; hESC-HEK293T; MM6; Jurkat; CD4T; peripheral-blood; GSC-11; MSC; endometrial; HEC-1-A; GSCs; NB4; AML | ||
| Seq Type List | m6A-seq; m6A-REF-seq; MeRIP-seq; MAZTER-seq; miCLIP | ||
| Transcript ID List | ENST00000377970.6; ENST00000259523.10; ENST00000621592.6; ENST00000641036.1; ENST00000524013.1; ENST00000520751.1; ENST00000517291.1; ENST00000651626.1 | ||
| External Link | RMBase: m6A_site_809502 | ||
| mod ID: M6ASITE086885 | Click to Show/Hide the Full List | ||
| mod site | chr8:127736552-127736553:+ | [76] | |
| Sequence | ACTTCCCCGCCGCTGCCAGGACCCGCTTCTCTGAAAGGCTC | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; CD34; HepG2; A549; MM6; Huh7; Jurkat; CD4T; peripheral-blood; endometrial; HEC-1-A; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000651626.1; ENST00000259523.10; ENST00000524013.1; ENST00000621592.6; ENST00000517291.1; ENST00000377970.6; ENST00000520751.1; ENST00000641036.1 | ||
| External Link | RMBase: m6A_site_809503 | ||
| mod ID: M6ASITE086886 | Click to Show/Hide the Full List | ||
| mod site | chr8:127736618-127736619:+ | [76] | |
| Sequence | TTTTTTTCGGGTAGTGGAAAACCAGGTAAGCACCGAAGTCC | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | HeLa; HepG2; A549; fibroblasts; Huh7; peripheral-blood; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000520751.1; ENST00000621592.6; ENST00000377970.6; ENST00000259523.10; ENST00000524013.1; ENST00000517291.1; ENST00000613283.2; ENST00000651626.1; ENST00000652131.1 | ||
| External Link | RMBase: m6A_site_809504 | ||
| mod ID: M6ASITE086887 | Click to Show/Hide the Full List | ||
| mod site | chr8:127738293-127738294:+ | [76] | |
| Sequence | CGTTAGCTTCACCAACAGGAACTATGACCTCGACTACGACT | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa; HepG2; fibroblasts; Huh7 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000517291.1; ENST00000613283.2; ENST00000524013.1; ENST00000652288.1; ENST00000652131.1; ENST00000259523.10; ENST00000520751.1; ENST00000377970.6; ENST00000621592.6; ENST00000651626.1 | ||
| External Link | RMBase: m6A_site_809505 | ||
| mod ID: M6ASITE086888 | Click to Show/Hide the Full List | ||
| mod site | chr8:127738350-127738351:+ | [76] | |
| Sequence | CTACTGCGACGAGGAGGAGAACTTCTACCAGCAGCAGCAGC | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa; HepG2; fibroblasts; MT4; MM6; Huh7; peripheral-blood; HEK293T; TIME; endometrial; AML | ||
| Seq Type List | m6A-seq; MeRIP-seq; miCLIP | ||
| Transcript ID List | ENST00000377970.6; ENST00000524013.1; ENST00000520751.1; ENST00000259523.10; ENST00000652288.1; ENST00000621592.6; ENST00000517291.1; ENST00000613283.2; ENST00000651626.1; ENST00000652131.1 | ||
| External Link | RMBase: m6A_site_809506 | ||
| mod ID: M6ASITE086889 | Click to Show/Hide the Full List | ||
| mod site | chr8:127738493-127738494:+ | [77] | |
| Sequence | CGCCCTCCTACGTTGCGGTCACACCCTTCTCCCTTCGGGGA | ||
| Motif Score | 2.047297619 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000621592.6; ENST00000613283.2; ENST00000524013.1; ENST00000517291.1; ENST00000259523.10; ENST00000652131.1; ENST00000651626.1; ENST00000377970.6; ENST00000652288.1 | ||
| External Link | RMBase: m6A_site_809507 | ||
| mod ID: M6ASITE086890 | Click to Show/Hide the Full List | ||
| mod site | chr8:127738515-127738516:+ | [76] | |
| Sequence | ACCCTTCTCCCTTCGGGGAGACAACGACGGCGGTGGCGGGA | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa; CD34; HepG2; HEK293T; hESC-HEK293T; MT4; H1299; MM6; Huh7; peripheral-blood; MSC; TIME; iSLK; endometrial; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000377970.6; ENST00000517291.1; ENST00000524013.1; ENST00000652131.1; ENST00000652288.1; ENST00000613283.2; ENST00000259523.10; ENST00000621592.6; ENST00000651626.1 | ||
| External Link | RMBase: m6A_site_809508 | ||
| mod ID: M6ASITE086891 | Click to Show/Hide the Full List | ||
| mod site | chr8:127738587-127738588:+ | [76] | |
| Sequence | GACCGAGCTGCTGGGAGGAGACATGGTGAACCAGAGTTTCA | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa; CD34; HepG2; HEK293T; A549; hESC-HEK293T; H1A; H1B; fibroblasts; LCLs; MT4; H1299; MM6; Huh7; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; TREX; endometrial; HEC-1-A; GSCs; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000524013.1; ENST00000652288.1; ENST00000621592.6; ENST00000259523.10; ENST00000652131.1; ENST00000613283.2; ENST00000517291.1; ENST00000377970.6; ENST00000651626.1 | ||
| External Link | RMBase: m6A_site_809509 | ||
| mod ID: M6ASITE086892 | Click to Show/Hide the Full List | ||
| mod site | chr8:127738596-127738597:+ | [76] | |
| Sequence | GCTGGGAGGAGACATGGTGAACCAGAGTTTCATCTGCGACC | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | HeLa; CD34; HepG2; HEK293T; A549; H1A; H1B; fibroblasts; LCLs; MT4; H1299; MM6; Huh7; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; TREX; endometrial; HEC-1-A; GSCs; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000377970.6; ENST00000621592.6; ENST00000524013.1; ENST00000613283.2; ENST00000517291.1; ENST00000652288.1; ENST00000259523.10; ENST00000651626.1; ENST00000652131.1 | ||
| External Link | RMBase: m6A_site_809510 | ||
| mod ID: M6ASITE086893 | Click to Show/Hide the Full List | ||
| mod site | chr8:127738620-127738621:+ | [78] | |
| Sequence | GAGTTTCATCTGCGACCCGGACGACGAGACCTTCATCAAAA | ||
| Motif Score | 3.616982143 | ||
| Cell/Tissue List | CD8T; A549 | ||
| Seq Type List | m6A-CLIP/IP | ||
| Transcript ID List | ENST00000621592.6; ENST00000259523.10; ENST00000652288.1; ENST00000524013.1; ENST00000613283.2; ENST00000517291.1; ENST00000377970.6; ENST00000652131.1; ENST00000651626.1 | ||
| External Link | RMBase: m6A_site_809511 | ||
| mod ID: M6ASITE086894 | Click to Show/Hide the Full List | ||
| mod site | chr8:127738628-127738629:+ | [76] | |
| Sequence | TCTGCGACCCGGACGACGAGACCTTCATCAAAAACATCATC | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa; CD34; HepG2; HEK293T; A549; H1A; H1B; hESCs; fibroblasts; LCLs; CD8T; MT4; H1299; MM6; Huh7; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; TREX; endometrial; HEC-1-A; GSCs; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq; m6A-CLIP/IP | ||
| Transcript ID List | ENST00000621592.6; ENST00000651626.1; ENST00000259523.10; ENST00000517291.1; ENST00000652288.1; ENST00000613283.2; ENST00000377970.6; ENST00000652131.1; ENST00000524013.1 | ||
| External Link | RMBase: m6A_site_809512 | ||
| mod ID: M6ASITE086895 | Click to Show/Hide the Full List | ||
| mod site | chr8:127738641-127738642:+ | [76] | |
| Sequence | CGACGAGACCTTCATCAAAAACATCATCATCCAGGACTGTA | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; A549; hESC-HEK293T; H1A; H1B; hESCs; fibroblasts; LCLs; MT4; H1299; MM6; Huh7; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; TREX; endometrial; HEC-1-A; GSCs; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000652131.1; ENST00000259523.10; ENST00000517291.1; ENST00000651626.1; ENST00000613283.2; ENST00000377970.6; ENST00000621592.6; ENST00000652288.1; ENST00000524013.1 | ||
| External Link | RMBase: m6A_site_809513 | ||
| mod ID: M6ASITE086896 | Click to Show/Hide the Full List | ||
| mod site | chr8:127738656-127738657:+ | [76] | |
| Sequence | CAAAAACATCATCATCCAGGACTGTATGTGGAGCGGCTTCT | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; A549; H1A; H1B; hESCs; fibroblasts; LCLs; CD8T; MT4; H1299; MM6; Huh7; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; TREX; endometrial; HEC-1-A; GSCs; NB4; AML | ||
| Seq Type List | m6A-seq; MeRIP-seq; m6A-CLIP/IP; miCLIP | ||
| Transcript ID List | ENST00000621592.6; ENST00000377970.6; ENST00000651626.1; ENST00000652288.1; ENST00000517291.1; ENST00000259523.10; ENST00000613283.2; ENST00000652131.1; ENST00000524013.1 | ||
| External Link | RMBase: m6A_site_809514 | ||
| mod ID: M6ASITE086897 | Click to Show/Hide the Full List | ||
| mod site | chr8:127738734-127738735:+ | [76] | |
| Sequence | CTACCAGGCTGCGCGCAAAGACAGCGGCAGCCCGAACCCCG | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; A549; hESC-HEK293T; H1B; H1A; hESCs; fibroblasts; LCLs; H1299; MM6; Huh7; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; TREX; endometrial; HEC-1-A; NB4; AML | ||
| Seq Type List | m6A-seq; MeRIP-seq; DART-seq; MAZTER-seq; miCLIP | ||
| Transcript ID List | ENST00000517291.1; ENST00000524013.1; ENST00000259523.10; ENST00000651626.1; ENST00000377970.6; ENST00000652288.1; ENST00000621592.6; ENST00000613283.2; ENST00000652131.1 | ||
| External Link | RMBase: m6A_site_809515 | ||
| mod ID: M6ASITE086898 | Click to Show/Hide the Full List | ||
| mod site | chr8:127738749-127738750:+ | [76] | |
| Sequence | CAAAGACAGCGGCAGCCCGAACCCCGCCCGCGGCCACAGCG | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; A549; H1B; H1A; hESCs; fibroblasts; LCLs; H1299; MM6; Huh7; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; TREX; endometrial; HEC-1-A; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000517291.1; ENST00000613283.2; ENST00000524013.1; ENST00000651626.1; ENST00000652131.1; ENST00000259523.10; ENST00000652288.1; ENST00000621592.6; ENST00000377970.6 | ||
| External Link | RMBase: m6A_site_809516 | ||
| mod ID: M6ASITE086899 | Click to Show/Hide the Full List | ||
| mod site | chr8:127738896-127738897:+ | [76] | |
| Sequence | CAAGTCCTGCGCCTCGCAAGACTCCAGCGCCTTCTCTCCGT | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa; CD34; HepG2; A549; MM6; Huh7; peripheral-blood; HEK293T; MSC; TIME; iSLK; endometrial; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000652131.1; ENST00000613283.2; ENST00000652288.1; ENST00000651626.1; ENST00000259523.10; ENST00000621592.6; ENST00000524013.1; ENST00000377970.6 | ||
| External Link | RMBase: m6A_site_809517 | ||
| mod ID: M6ASITE086900 | Click to Show/Hide the Full List | ||
| mod site | chr8:127738991-127738992:+ | [76] | |
| Sequence | CCCTGGTGCTCCATGAGGAGACACCGCCCACCACCAGCAGC | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa; CD34; HepG2; hESC-HEK293T; MM6; Huh7; peripheral-blood; iSLK; MSC; endometrial; NB4; AML | ||
| Seq Type List | m6A-seq; MAZTER-seq; MeRIP-seq; miCLIP | ||
| Transcript ID List | ENST00000621592.6; ENST00000259523.10; ENST00000524013.1; ENST00000613283.2; ENST00000377970.6; ENST00000651626.1; ENST00000652288.1; ENST00000652131.1 | ||
| External Link | RMBase: m6A_site_809518 | ||
| mod ID: M6ASITE086901 | Click to Show/Hide the Full List | ||
| mod site | chr8:127740402-127740403:+ | [76] | |
| Sequence | TCCTTTCTTAAAGAGGAGGAACAAGAAGATGAGGAAGAAAT | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa; CD34; HepG2; HEK293; hESC-HEK293T; MM6; Huh7; peripheral-blood; endometrial; HEC-1-A; NB4 | ||
| Seq Type List | m6A-seq; m6A-REF-seq; MAZTER-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000613283.2; rmsk_2646882; ENST00000524013.1; ENST00000652288.1; ENST00000652131.1; ENST00000259523.10; ENST00000377970.6; ENST00000621592.6; ENST00000651626.1 | ||
| External Link | RMBase: m6A_site_809519 | ||
| mod ID: M6ASITE086902 | Click to Show/Hide the Full List | ||
| mod site | chr8:127740504-127740505:+ | [76] | |
| Sequence | TCTGCTGGAGGCCACAGCAAACCTCCTCACAGCCCACTGGT | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | HeLa; CD34; HepG2; HEK293T; U2OS; MT4; A549; H1299; MM6; Huh7; CD4T; peripheral-blood; GSC-11; MSC; endometrial; HEC-1-A; NB4; AML | ||
| Seq Type List | m6A-seq; MeRIP-seq; miCLIP | ||
| Transcript ID List | ENST00000621592.6; ENST00000377970.6; ENST00000613283.2; ENST00000651626.1; ENST00000652131.1; ENST00000652288.1; ENST00000524013.1 | ||
| External Link | RMBase: m6A_site_809520 | ||
| mod ID: M6ASITE086903 | Click to Show/Hide the Full List | ||
| mod site | chr8:127740557-127740558:+ | [77] | |
| Sequence | CCACGTCTCCACACATCAGCACAACTACGCAGCGCCTCCCT | ||
| Motif Score | 2.830589286 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000651626.1; ENST00000652131.1; ENST00000621592.6; ENST00000613283.2; ENST00000377970.6; ENST00000652288.1; ENST00000524013.1 | ||
| External Link | RMBase: m6A_site_809521 | ||
| mod ID: M6ASITE086904 | Click to Show/Hide the Full List | ||
| mod site | chr8:127740590-127740591:+ | [76] | |
| Sequence | GCCTCCCTCCACTCGGAAGGACTATCCTGCTGCCAAGAGGG | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa; CD34; HepG2; HEK293T; A549; U2OS; H1A; hESCs; fibroblasts; GM12878; LCLs; CD8T; MT4; H1299; MM6; Huh7; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; TREX; endometrial; HEC-1-A; NB4; AML | ||
| Seq Type List | m6A-seq; MeRIP-seq; m6A-CLIP/IP; miCLIP | ||
| Transcript ID List | ENST00000613283.2; ENST00000652288.1; ENST00000651626.1; ENST00000621592.6; ENST00000377970.6; ENST00000652131.1; ENST00000524013.1 | ||
| External Link | RMBase: m6A_site_809522 | ||
| mod ID: M6ASITE086905 | Click to Show/Hide the Full List | ||
| mod site | chr8:127740620-127740621:+ | [76] | |
| Sequence | TGCCAAGAGGGTCAAGTTGGACAGTGTCAGAGTCCTGAGAC | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; CD34; HepG2; HEK293T; A549; hESC-HEK293T; U2OS; H1A; H1B; hESCs; fibroblasts; GM12878; LCLs; CD8T; MT4; H1299; MM6; Huh7; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; TREX; endometrial; HEC-1-A; NB4; AML | ||
| Seq Type List | m6A-seq; MeRIP-seq; MAZTER-seq; m6A-CLIP/IP; miCLIP | ||
| Transcript ID List | ENST00000652288.1; ENST00000613283.2; ENST00000621592.6; ENST00000651626.1; ENST00000652131.1; ENST00000377970.6; ENST00000524013.1 | ||
| External Link | RMBase: m6A_site_809523 | ||
| mod ID: M6ASITE086906 | Click to Show/Hide the Full List | ||
| mod site | chr8:127740639-127740640:+ | [76] | |
| Sequence | GACAGTGTCAGAGTCCTGAGACAGATCAGCAACAACCGAAA | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa; CD34; HepG2; HEK293T; A549; hESC-HEK293T; U2OS; H1A; H1B; hESCs; fibroblasts; GM12878; LCLs; MT4; H1299; MM6; Huh7; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; TREX; endometrial; HEC-1-A; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq; m6A-REF-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000524013.1; ENST00000651626.1; ENST00000377970.6; ENST00000621592.6; ENST00000652288.1; ENST00000613283.2; ENST00000652131.1 | ||
| External Link | RMBase: m6A_site_809524 | ||
| mod ID: M6ASITE086907 | Click to Show/Hide the Full List | ||
| mod site | chr8:127740650-127740651:+ | [77] | |
| Sequence | AGTCCTGAGACAGATCAGCAACAACCGAAAATGCACCAGCC | ||
| Motif Score | 2.173910714 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000652131.1; ENST00000377970.6; ENST00000524013.1; ENST00000651626.1; ENST00000621592.6; ENST00000652288.1; ENST00000613283.2 | ||
| External Link | RMBase: m6A_site_809525 | ||
| mod ID: M6ASITE086908 | Click to Show/Hide the Full List | ||
| mod site | chr8:127740683-127740684:+ | [76] | |
| Sequence | CACCAGCCCCAGGTCCTCGGACACCGAGGAGAATGTCAAGA | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; CD34; HepG2; HEK293T; A549; hESC-HEK293T; U2OS; H1A; H1B; hESCs; fibroblasts; GM12878; LCLs; MT4; H1299; MM6; Huh7; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; TREX; endometrial; HEC-1-A; GSCs; NB4; AML | ||
| Seq Type List | m6A-seq; MeRIP-seq; m6A-REF-seq; MAZTER-seq; miCLIP | ||
| Transcript ID List | ENST00000652288.1; ENST00000651626.1; ENST00000613283.2; ENST00000524013.1; ENST00000652131.1; ENST00000621592.6; ENST00000377970.6 | ||
| External Link | RMBase: m6A_site_809526 | ||
| mod ID: M6ASITE086909 | Click to Show/Hide the Full List | ||
| mod site | chr8:127740709-127740710:+ | [76] | |
| Sequence | AGGAGAATGTCAAGAGGCGAACACACAACGTCTTGGAGCGC | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa; CD34; HepG2; HEK293T; A549; U2OS; H1A; H1B; hESCs; fibroblasts; GM12878; LCLs; MT4; H1299; MM6; Huh7; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; TREX; endometrial; HEC-1-A; GSCs; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000652288.1; ENST00000377970.6; ENST00000524013.1; ENST00000621592.6; ENST00000651626.1; ENST00000613283.2; ENST00000652131.1 | ||
| External Link | RMBase: m6A_site_809527 | ||
| mod ID: M6ASITE086910 | Click to Show/Hide the Full List | ||
| mod site | chr8:127740713-127740714:+ | [77] | |
| Sequence | GAATGTCAAGAGGCGAACACACAACGTCTTGGAGCGCCAGA | ||
| Motif Score | 2.084928571 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000652288.1; ENST00000377970.6; ENST00000652131.1; ENST00000524013.1; ENST00000613283.2; ENST00000621592.6; ENST00000651626.1 | ||
| External Link | RMBase: m6A_site_809528 | ||
| mod ID: M6ASITE086911 | Click to Show/Hide the Full List | ||
| mod site | chr8:127740794-127740795:+ | [76] | |
| Sequence | CCAGATCCCGGAGTTGGAAAACAATGAAAAGGCCCCCAAGG | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa; CD34; HepG2; HEK293T; A549; hESC-HEK293T; U2OS; H1A; H1B; hESCs; fibroblasts; GM12878; LCLs; MT4; H1299; MM6; Huh7; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; TREX; endometrial; HEC-1-A; GSCs; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000621592.6; ENST00000652131.1; ENST00000524013.1; ENST00000377970.6; ENST00000613283.2; ENST00000651626.1; ENST00000652288.1 | ||
| External Link | RMBase: m6A_site_809529 | ||
| mod ID: M6ASITE086912 | Click to Show/Hide the Full List | ||
| mod site | chr8:127740842-127740843:+ | [77] | |
| Sequence | CCTTAAAAAAGCCACAGCATACATCCTGTCCGTCCAAGCAG | ||
| Motif Score | 2.110482143 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000377970.6; ENST00000613283.2; ENST00000524013.1; ENST00000621592.6; ENST00000652131.1; ENST00000651626.1; ENST00000652288.1 | ||
| External Link | RMBase: m6A_site_809530 | ||
| mod ID: M6ASITE086913 | Click to Show/Hide the Full List | ||
| mod site | chr8:127740890-127740891:+ | [76] | |
| Sequence | AAAGCTCATTTCTGAAGAGGACTTGTTGCGGAAACGACGAG | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; A549; BGC823; U2OS; H1A; H1B; hNPCs; hESCs; fibroblasts; GM12878; LCLs; CD8T; H1299; MM6; Huh7; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; TREX; endometrial; HEC-1-A; GSCs; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq; m6A-CLIP/IP | ||
| Transcript ID List | ENST00000621592.6; ENST00000524013.1; ENST00000652288.1; ENST00000651626.1; ENST00000652131.1; ENST00000377970.6; ENST00000613283.2 | ||
| External Link | RMBase: m6A_site_809531 | ||
| mod ID: M6ASITE086914 | Click to Show/Hide the Full List | ||
| mod site | chr8:127740912-127740913:+ | [76] | |
| Sequence | TTGTTGCGGAAACGACGAGAACAGTTGAAACACAAACTTGA | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; A549; hESC-HEK293T; BGC823; U2OS; H1A; H1B; hNPCs; hESCs; fibroblasts; GM12878; LCLs; CD8T; H1299; MM6; Huh7; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; TREX; endometrial; HEC-1-A; GSCs; NB4; AML | ||
| Seq Type List | m6A-seq; MeRIP-seq; MAZTER-seq; m6A-CLIP/IP; miCLIP | ||
| Transcript ID List | ENST00000651626.1; ENST00000377970.6; ENST00000524013.1; ENST00000613283.2; ENST00000652288.1; ENST00000621592.6; ENST00000652131.1 | ||
| External Link | RMBase: m6A_site_809532 | ||
| mod ID: M6ASITE086915 | Click to Show/Hide the Full List | ||
| mod site | chr8:127740921-127740922:+ | [76] | |
| Sequence | AAACGACGAGAACAGTTGAAACACAAACTTGAACAGCTACG | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; A549; hESC-HEK293T; BGC823; U2OS; H1A; H1B; hNPCs; hESCs; fibroblasts; GM12878; LCLs; CD8T; H1299; MM6; Huh7; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; TREX; endometrial; HEC-1-A; GSCs; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq; DART-seq; MAZTER-seq; m6A-CLIP/IP | ||
| Transcript ID List | ENST00000651626.1; ENST00000377970.6; ENST00000621592.6; ENST00000613283.2; ENST00000652131.1; ENST00000524013.1; ENST00000652288.1 | ||
| External Link | RMBase: m6A_site_809533 | ||
| mod ID: M6ASITE086916 | Click to Show/Hide the Full List | ||
| mod site | chr8:127740927-127740928:+ | [76] | |
| Sequence | CGAGAACAGTTGAAACACAAACTTGAACAGCTACGGAACTC | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; A549; BGC823; U2OS; H1A; H1B; hNPCs; hESCs; fibroblasts; GM12878; LCLs; CD8T; H1299; MM6; Huh7; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; TREX; endometrial; HEC-1-A; GSCs; NB4; AML | ||
| Seq Type List | m6A-seq; MeRIP-seq; m6A-CLIP/IP; miCLIP | ||
| Transcript ID List | ENST00000377970.6; ENST00000621592.6; ENST00000652131.1; ENST00000613283.2; ENST00000651626.1; ENST00000652288.1; ENST00000524013.1 | ||
| External Link | RMBase: m6A_site_809534 | ||
| mod ID: M6ASITE086917 | Click to Show/Hide the Full List | ||
| mod site | chr8:127740933-127740934:+ | [76] | |
| Sequence | CAGTTGAAACACAAACTTGAACAGCTACGGAACTCTTGTGC | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; A549; hESC-HEK293T; BGC823; U2OS; H1A; H1B; hNPCs; hESCs; fibroblasts; GM12878; LCLs; H1299; MM6; Huh7; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; TREX; endometrial; HEC-1-A; GSCs; NB4; AML | ||
| Seq Type List | m6A-seq; MeRIP-seq; MAZTER-seq; m6A-CLIP/IP; miCLIP | ||
| Transcript ID List | ENST00000651626.1; ENST00000524013.1; ENST00000613283.2; ENST00000652288.1; ENST00000621592.6; ENST00000377970.6; ENST00000652131.1 | ||
| External Link | RMBase: m6A_site_809535 | ||
| mod ID: M6ASITE086918 | Click to Show/Hide the Full List | ||
| mod site | chr8:127740944-127740945:+ | [76] | |
| Sequence | CAAACTTGAACAGCTACGGAACTCTTGTGCGTAAGGAAAAG | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; A549; BGC823; U2OS; H1A; H1B; hNPCs; hESCs; fibroblasts; GM12878; LCLs; H1299; MM6; Huh7; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; TREX; endometrial; HEC-1-A; GSCs; NB4; AML | ||
| Seq Type List | m6A-seq; MeRIP-seq; m6A-CLIP/IP; miCLIP | ||
| Transcript ID List | ENST00000377970.6; ENST00000651626.1; ENST00000621592.6; ENST00000652288.1; ENST00000652131.1; ENST00000524013.1; ENST00000613283.2 | ||
| External Link | RMBase: m6A_site_809536 | ||
| mod ID: M6ASITE086919 | Click to Show/Hide the Full List | ||
| mod site | chr8:127740973-127740974:+ | [79] | |
| Sequence | CGTAAGGAAAAGTAAGGAAAACGATTCCTTCTAACAGAAAT | ||
| Motif Score | 2.179660714 | ||
| Cell/Tissue List | HEK293T; CD8T | ||
| Seq Type List | DART-seq; m6A-CLIP/IP | ||
| Transcript ID List | ENST00000524013.1; ENST00000621592.6; ENST00000377970.6; ENST00000652288.1; ENST00000651626.1 | ||
| External Link | RMBase: m6A_site_809537 | ||
| mod ID: M6ASITE086920 | Click to Show/Hide the Full List | ||
| mod site | chr8:127740986-127740987:+ | [80] | |
| Sequence | AAGGAAAACGATTCCTTCTAACAGAAATGTCCTGAGCAATC | ||
| Motif Score | 2.168095238 | ||
| Cell/Tissue List | HEK293; hESC-HEK293T; CD8T | ||
| Seq Type List | m6A-REF-seq; MAZTER-seq; m6A-CLIP/IP | ||
| Transcript ID List | ENST00000651626.1; ENST00000621592.6; ENST00000377970.6; ENST00000652288.1; ENST00000524013.1 | ||
| External Link | RMBase: m6A_site_809538 | ||
| mod ID: M6ASITE086921 | Click to Show/Hide the Full List | ||
| mod site | chr8:127741015-127741016:+ | [76] | |
| Sequence | TCCTGAGCAATCACCTATGAACTTGTTTCAAATGCATGATC | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; A549; BGC823; U2OS; H1A; H1B; hNPCs; hESCs; fibroblasts; GM12878; LCLs; CD8T; H1299; MM6; Huh7; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; TREX; endometrial; HEC-1-A; GSCs; NB4; AML | ||
| Seq Type List | m6A-seq; MeRIP-seq; m6A-CLIP/IP; miCLIP | ||
| Transcript ID List | ENST00000621592.6; ENST00000377970.6; ENST00000524013.1; ENST00000651626.1; ENST00000652288.1 | ||
| External Link | RMBase: m6A_site_809539 | ||
| mod ID: M6ASITE086922 | Click to Show/Hide the Full List | ||
| mod site | chr8:127741043-127741044:+ | [78] | |
| Sequence | CAAATGCATGATCAAATGCAACCTCACAACCTTGGCTGAGT | ||
| Motif Score | 2.153267857 | ||
| Cell/Tissue List | CD8T | ||
| Seq Type List | m6A-CLIP/IP | ||
| Transcript ID List | ENST00000524013.1; ENST00000377970.6; ENST00000651626.1; ENST00000621592.6; ENST00000652288.1 | ||
| External Link | RMBase: m6A_site_809540 | ||
| mod ID: M6ASITE086923 | Click to Show/Hide the Full List | ||
| mod site | chr8:127741048-127741049:+ | [79] | |
| Sequence | GCATGATCAAATGCAACCTCACAACCTTGGCTGAGTCTTGA | ||
| Motif Score | 2.047297619 | ||
| Cell/Tissue List | HEK293T; hESC-HEK293T | ||
| Seq Type List | DART-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000524013.1; ENST00000651626.1; ENST00000377970.6; ENST00000652288.1; ENST00000621592.6 | ||
| External Link | RMBase: m6A_site_809541 | ||
| mod ID: M6ASITE086924 | Click to Show/Hide the Full List | ||
| mod site | chr8:127741051-127741052:+ | [78] | |
| Sequence | TGATCAAATGCAACCTCACAACCTTGGCTGAGTCTTGAGAC | ||
| Motif Score | 2.153267857 | ||
| Cell/Tissue List | CD8T | ||
| Seq Type List | m6A-CLIP/IP | ||
| Transcript ID List | ENST00000621592.6; ENST00000651626.1; ENST00000377970.6; ENST00000524013.1; ENST00000652288.1 | ||
| External Link | RMBase: m6A_site_809542 | ||
| mod ID: M6ASITE086925 | Click to Show/Hide the Full List | ||
| mod site | chr8:127741070-127741071:+ | [76] | |
| Sequence | AACCTTGGCTGAGTCTTGAGACTGAAAGATTTAGCCATAAT | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; A549; U2OS; H1A; H1B; hNPCs; hESCs; fibroblasts; GM12878; LCLs; CD8T; H1299; MM6; Huh7; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; TREX; endometrial; HEC-1-A; GSCs; NB4; AML | ||
| Seq Type List | m6A-seq; MeRIP-seq; DART-seq; m6A-CLIP/IP; miCLIP | ||
| Transcript ID List | ENST00000377970.6; ENST00000652288.1; ENST00000621592.6; ENST00000651626.1; ENST00000524013.1 | ||
| External Link | RMBase: m6A_site_809543 | ||
| mod ID: M6ASITE086926 | Click to Show/Hide the Full List | ||
| mod site | chr8:127741095-127741096:+ | [76] | |
| Sequence | AAGATTTAGCCATAATGTAAACTGCCTCAAATTGGACTTTG | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; A549; U2OS; H1A; H1B; hESCs; fibroblasts; GM12878; LCLs; H1299; MM6; Huh7; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; TREX; HEC-1-A; GSCs; NB4; AML | ||
| Seq Type List | m6A-seq; MeRIP-seq; m6A-CLIP/IP; miCLIP | ||
| Transcript ID List | ENST00000651626.1; ENST00000524013.1; ENST00000377970.6; ENST00000621592.6; ENST00000652288.1 | ||
| External Link | RMBase: m6A_site_809544 | ||
| mod ID: M6ASITE086927 | Click to Show/Hide the Full List | ||
| mod site | chr8:127741110-127741111:+ | [76] | |
| Sequence | TGTAAACTGCCTCAAATTGGACTTTGGGCATAAAAGAACTT | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; A549; U2OS; H1A; H1B; hESCs; fibroblasts; GM12878; LCLs; CD8T; H1299; MM6; Huh7; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; TREX; HEC-1-A; GSCs; NB4; AML | ||
| Seq Type List | m6A-seq; MeRIP-seq; m6A-CLIP/IP; miCLIP | ||
| Transcript ID List | ENST00000652288.1; ENST00000524013.1; ENST00000651626.1; ENST00000377970.6; ENST00000621592.6 | ||
| External Link | RMBase: m6A_site_809545 | ||
| mod ID: M6ASITE086928 | Click to Show/Hide the Full List | ||
| mod site | chr8:127741127-127741128:+ | [76] | |
| Sequence | TGGACTTTGGGCATAAAAGAACTTTTTTATGCTTACCATCT | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; A549; U2OS; hESCs; fibroblasts; GM12878; LCLs; CD8T; H1299; MM6; Huh7; peripheral-blood; HEK293A-TOA; iSLK; MSC; TIME; TREX; HEC-1-A; AML | ||
| Seq Type List | m6A-seq; MeRIP-seq; m6A-CLIP/IP; miCLIP | ||
| Transcript ID List | ENST00000524013.1; ENST00000621592.6; ENST00000652288.1; ENST00000377970.6; ENST00000651626.1 | ||
| External Link | RMBase: m6A_site_809546 | ||
| mod ID: M6ASITE086929 | Click to Show/Hide the Full List | ||
| mod site | chr8:127741206-127741207:+ | [79] | |
| Sequence | TTTAAAAAATTTTAAGATTTACACAATGTTTCTCTGTAAAT | ||
| Motif Score | 2.07285119 | ||
| Cell/Tissue List | HEK293T; hESC-HEK293T | ||
| Seq Type List | DART-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000524013.1; ENST00000651626.1; ENST00000377970.6; ENST00000652288.1; ENST00000621592.6 | ||
| External Link | RMBase: m6A_site_809547 | ||
| mod ID: M6ASITE086930 | Click to Show/Hide the Full List | ||
| mod site | chr8:127741258-127741259:+ | [79] | |
| Sequence | ATGTAAATAACTTTAATAAAACGTTTATAGCAGTTACACAG | ||
| Motif Score | 2.179660714 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000524013.1; ENST00000651626.1; ENST00000652288.1; ENST00000621592.6; ENST00000377970.6 | ||
| External Link | RMBase: m6A_site_809548 | ||
| mod ID: M6ASITE086931 | Click to Show/Hide the Full List | ||
| mod site | chr8:127741273-127741274:+ | [77] | |
| Sequence | ATAAAACGTTTATAGCAGTTACACAGAATTTCAATCCTAGT | ||
| Motif Score | 2.07285119 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000652288.1; ENST00000524013.1; ENST00000651626.1; ENST00000621592.6; ENST00000377970.6 | ||
| External Link | RMBase: m6A_site_809549 | ||
| mod ID: M6ASITE086932 | Click to Show/Hide the Full List | ||
| mod site | chr8:127741324-127741325:+ | [81] | |
| Sequence | TAGTATTATAGGTACTATAAACCCTAATTTTTTTTATTTAA | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | HEK293T; HepG2; GM12878; Huh7; iSLK | ||
| Seq Type List | MeRIP-seq; m6A-seq | ||
| Transcript ID List | ENST00000524013.1; ENST00000652288.1; ENST00000651626.1; ENST00000621592.6; ENST00000377970.6 | ||
| External Link | RMBase: m6A_site_809550 | ||
| mod ID: M6ASITE086933 | Click to Show/Hide the Full List | ||
| mod site | chr8:127741347-127741348:+ | [79] | |
| Sequence | CTAATTTTTTTTATTTAAGTACATTTTGCTTTTTAAAGTTG | ||
| Motif Score | 2.856142857 | ||
| Cell/Tissue List | HEK293T; hESC-HEK293T | ||
| Seq Type List | DART-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000652288.1; ENST00000621592.6; ENST00000651626.1; ENST00000377970.6; ENST00000524013.1 | ||
| External Link | RMBase: m6A_site_809551 | ||
| mod ID: M6ASITE086934 | Click to Show/Hide the Full List | ||
| mod site | chr8:127741611-127741612:+ | [76] | |
| Sequence | TTCTAAGATGCTTCCTGGAGACTATGATAACAGCCAGAGTT | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000621592.6 | ||
| External Link | RMBase: m6A_site_809552 | ||
| mod ID: M6ASITE086935 | Click to Show/Hide the Full List | ||
| mod site | chr8:127741735-127741736:+ | [76] | |
| Sequence | GTACACAAAGAGGCATAAGGACTGGGGAGTTGGGAGGAAGG | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000621592.6 | ||
| External Link | RMBase: m6A_site_809553 | ||
| mod ID: M6ASITE086936 | Click to Show/Hide the Full List | ||
| mod site | chr8:127741766-127741767:+ | [76] | |
| Sequence | GGGAGGAAGGTGAGGAAGAAACTCCTGTTACTTTAGTTAAC | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000621592.6 | ||
| External Link | RMBase: m6A_site_809554 | ||
| mod ID: M6ASITE086937 | Click to Show/Hide the Full List | ||
| mod site | chr8:127741852-127741853:+ | [76] | |
| Sequence | TTGATGGGGACACGGTGGGAACCAGCTTCTGCTGCCTTCAC | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000621592.6 | ||
| External Link | RMBase: m6A_site_809555 | ||
| mod ID: M6ASITE086938 | Click to Show/Hide the Full List | ||
| mod site | chr8:127741910-127741911:+ | [76] | |
| Sequence | GTCCATGGGTTATCTCGCAAACCCCAGAGGATCTCTGGGAG | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000621592.6 | ||
| External Link | RMBase: m6A_site_809556 | ||
| mod ID: M6ASITE086939 | Click to Show/Hide the Full List | ||
| mod site | chr8:127741959-127741960:+ | [76] | |
| Sequence | CTATTAACCCTATTTCACAAACAAGGAAATAGAAGAGCTCA | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000621592.6 | ||
| External Link | RMBase: m6A_site_809557 | ||
| mod ID: M6ASITE086940 | Click to Show/Hide the Full List | ||
| mod site | chr8:127742035-127742036:+ | [82] | |
| Sequence | AATACAAAGCAGCAATCTGGACCCATTCTGTTCAAAACACT | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | MeRIP-seq | ||
| Transcript ID List | ENST00000621592.6 | ||
| External Link | RMBase: m6A_site_809558 | ||
| mod ID: M6ASITE086941 | Click to Show/Hide the Full List | ||
| mod site | chr8:127742051-127742052:+ | [82] | |
| Sequence | CTGGACCCATTCTGTTCAAAACACTTAACCCTTCGCTATCA | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | MeRIP-seq | ||
| Transcript ID List | ENST00000621592.6 | ||
| External Link | RMBase: m6A_site_809559 | ||
| mod ID: M6ASITE086942 | Click to Show/Hide the Full List | ||
| mod site | chr8:127742120-127742121:+ | [82] | |
| Sequence | TGAGATCAAGAAGGTTTAGGACCTAATGGACAGACTCAAGT | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | MeRIP-seq | ||
| Transcript ID List | ENST00000621592.6 | ||
| External Link | RMBase: m6A_site_809560 | ||
| mod ID: M6ASITE086943 | Click to Show/Hide the Full List | ||
| mod site | chr8:127742129-127742130:+ | [82] | |
| Sequence | GAAGGTTTAGGACCTAATGGACAGACTCAAGTCATAACAAT | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | MeRIP-seq | ||
| Transcript ID List | ENST00000621592.6 | ||
| External Link | RMBase: m6A_site_809561 | ||
| mod ID: M6ASITE086944 | Click to Show/Hide the Full List | ||
| mod site | chr8:127742276-127742277:+ | [82] | |
| Sequence | GGAAGCTGAGGCACACAAAGACTCATCCACATGCCCAAGAT | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | MeRIP-seq | ||
| Transcript ID List | ENST00000621592.6 | ||
| External Link | RMBase: m6A_site_809562 | ||
| mod ID: M6ASITE086945 | Click to Show/Hide the Full List | ||
| mod site | chr8:127742330-127742331:+ | [82] | |
| Sequence | AAAGTGGAAGCGAGATTTGAACCCAGGCTGTTTACTCCTAA | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | MeRIP-seq | ||
| Transcript ID List | ENST00000621592.6 | ||
| External Link | RMBase: m6A_site_809563 | ||
| mod ID: M6ASITE086946 | Click to Show/Hide the Full List | ||
| mod site | chr8:127742586-127742587:+ | [82] | |
| Sequence | TTAGCCATTTGTTGAATCAGACTCATGACGGCTCCTGGGAA | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | MeRIP-seq | ||
| Transcript ID List | ENST00000621592.6 | ||
| External Link | RMBase: m6A_site_809564 | ||
| mod ID: M6ASITE086947 | Click to Show/Hide the Full List | ||
| mod site | chr8:127742633-127742634:+ | [82] | |
| Sequence | AGTTCAGATCATAAAATAAAACATATTTATTCTTTGTCATG | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | MeRIP-seq | ||
| Transcript ID List | ENST00000621592.6 | ||
| External Link | RMBase: m6A_site_809565 | ||
| mod ID: M6ASITE086948 | Click to Show/Hide the Full List | ||
| mod site | chr8:127742672-127742673:+ | [82] | |
| Sequence | TGGGAGTCATTATTTTAGAAACTACAAACTCTCCTTGCTTC | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | MeRIP-seq | ||
| Transcript ID List | ENST00000621592.6 | ||
| External Link | RMBase: m6A_site_809566 | ||
| mod ID: M6ASITE086949 | Click to Show/Hide the Full List | ||
| mod site | chr8:127742679-127742680:+ | [82] | |
| Sequence | CATTATTTTAGAAACTACAAACTCTCCTTGCTTCCATCCTT | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | MeRIP-seq | ||
| Transcript ID List | ENST00000621592.6 | ||
| External Link | RMBase: m6A_site_809567 | ||
Pseudouridine (Pseudo)
| In total 1 m6A sequence/site(s) in this target gene | |||
| mod ID: PSESITE000266 | Click to Show/Hide the Full List | ||
| mod site | chr8:127736350-127736351:+ | [83] | |
| Sequence | GAAGGGAGATCCGGAGCGAATAGGGGGCTTCGCCTCTGGCC | ||
| Transcript ID List | ENST00000377970.6; ENST00000641036.1; ENST00000259523.10; ENST00000520751.1; ENST00000517291.1; ENST00000524013.1; ENST00000621592.6; ENST00000651626.1 | ||
| External Link | RMBase: Pseudo_site_4664 | ||
References