m6A Regulator Information
General Information of the m6A Regulator (ID: REG00008)
| Regulator Name | YTH domain-containing family protein 2 (YTHDF2) | ||||
|---|---|---|---|---|---|
| Synonyms |
DF2; CLL-associated antigen KW-14; High-glucose-regulated protein 8; Renal carcinoma antigen NY-REN-2; HGRG8
Click to Show/Hide
|
||||
| Gene Name | YTHDF2 | ||||
| Sequence |
MSASSLLEQRPKGQGNKVQNGSVHQKDGLNDDDFEPYLSPQARPNNAYTAMSDSYLPSYY
SPSIGFSYSLGEAAWSTGGDTAMPYLTSYGQLSNGEPHFLPDAMFGQPGALGSTPFLGQH GFNFFPSGIDFSAWGNNSSQGQSTQSSGYSSNYAYAPSSLGGAMIDGQSAFANETLNKAP GMNTIDQGMAALKLGSTEVASNVPKVVGSAVGSGSITSNIVASNSLPPATIAPPKPASWA DIASKPAKQQPKLKTKNGIAGSSLPPPPIKHNMDIGTWDNKGPVAKAPSQALVQNIGQPT QGSPQPVGQQANNSPPVAQASVGQQTQPLPPPPPQPAQLSVQQQAAQPTRWVAPRNRGSG FGHNGVDGNGVGQSQAGSGSTPSEPHPVLEKLRSINNYNPKDFDWNLKHGRVFIIKSYSE DDIHRSIKYNIWCSTEHGNKRLDAAYRSMNGKGPVYLLFSVNGSGHFCGVAEMKSAVDYN TCAGVWSQDKWKGRFDVRWIFVKDVPNSQLRHIRLENNENKPVTNSRDTQEVPLEKAKQV LKIIASYKHTTSIFDDFSHYEKRQEEEESVKKERQGRGK Click to Show/Hide
|
||||
| Family | YTHDF family; YTHDF2 subfamily | ||||
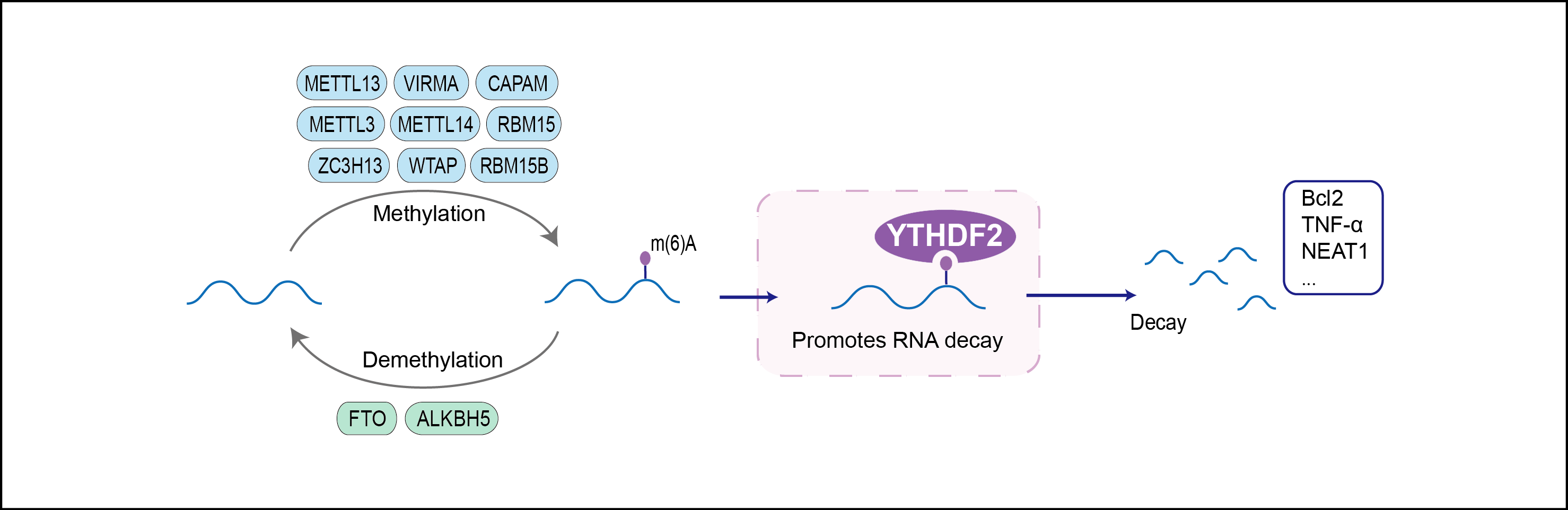
| Function |
Specifically recognizes and binds N6-methyladenosine (m6A)-containing RNAs, and regulates their stability. M6A is a modification present at internal sites of mRNAs and some non-coding RNAs and plays a role in mRNA stability and processing. Acts as a regulator of mRNA stability by promoting degradation of m6A-containing mRNAs via interaction with the CCR4-NOT and ribonuclease P/MRP complexes, depending on the context. The YTHDF paralogs (YTHDF1, YTHDF2 and YTHDF3) share m6A-containing mRNAs targets and act redundantly to mediate mRNA degradation and cellular differentiation. M6A-containing mRNAs containing a binding site for RIDA/HRSP12 (5'-GGUUC-3') are preferentially degraded by endoribonucleolytic cleavage: cooperative binding of RIDA/HRSP12 and YTHDF2 to transcripts leads to recruitment of the ribonuclease P/MRP complex. Other m6A-containing mRNAs undergo deadenylation via direct interaction between YTHDF2 and CNOT1, leading to recruitment of the CCR4-NOT and subsequent deadenylation of m6A-containing mRNAs. Required maternally to regulate oocyte maturation: probably acts by binding to m6A-containing mRNAs, thereby regulating maternal transcript dosage during oocyte maturation, which is essential for the competence of oocytes to sustain early zygotic development (By similarity). Also required during spermatogenesis: regulates spermagonial adhesion by promoting degradation of m6A-containing transcripts coding for matrix metallopeptidases (By similarity). Also involved in hematopoietic stem cells specification by binding to m6A-containing mRNAs, leading to promote their degradation. Also acts as a regulator of neural development by promoting m6A-dependent degradation of neural development-related mRNA targets (By similarity). Inhibits neural specification of induced pluripotent stem cells by binding to methylated neural-specific mRNAs and promoting their degradation, thereby restraining neural differentiation. Regulates circadian regulation of hepatic lipid metabolism: acts by promoting m6A-dependent degradation of PPARA transcripts. Regulates the innate immune response to infection by inhibiting the type I interferon response: acts by binding to m6A-containing IFNB transcripts and promoting their degradation. May also act as a promoter of cap-independent mRNA translation following heat shock stress: upon stress, relocalizes to the nucleus and specifically binds mRNAs with some m6A methylation mark at their 5'-UTR, protecting demethylation of mRNAs by FTO, thereby promoting cap-independent mRNA translation. Regulates mitotic entry by promoting the phase-specific m6A-dependent degradation of WEE1 transcripts . Promotes formation of phase-separated membraneless compartments, such as P-bodies or stress granules, by undergoing liquid-liquid phase separation upon binding to mRNAs containing multiple m6A-modified residues: polymethylated mRNAs act as a multivalent scaffold for the binding of YTHDF proteins, juxtaposing their disordered regions and thereby leading to phase separation . The resulting mRNA-YTHDF complexes then partition into different endogenous phase-separated membraneless compartments, such as P-bodies, stress granules or neuronal RNA granules. May also recognize and bind RNAs modified by C5-methylcytosine (m5C) and act as a regulator of rRNA processing; (Microbial infection) Promotes viral gene expression and virion production of kaposis sarcoma-associated herpesvirus (KSHV) at some stage of the KSHV life cycle (in iSLK.219 and iSLK.BAC16 cells). Acts by binding to N6-methyladenosine (m6A)-containing viral RNAs.
Click to Show/Hide
|
||||
| Gene ID | 51441 | ||||
| Uniprot ID | |||||
| Regulator Type | WRITER ERASER READER | ||||
| Mechanism Diagram | Click to View the Original Diagram | ||||
|
|||||
| Target Genes | Click to View Potential Target Genes of This Regulator | ||||
Full List of Target Gene(s) of This m6A Regulator and Corresponding Disease/Drug Response(s)
YTHDF2 can regulate the m6A methylation of following target genes, and result in corresponding disease/drug response(s). You can browse corresponding disease or drug response(s) resulted from the regulation of certain target gene.
Browse Target Gene related Disease
Browse Target Gene related Drug
Arrestin domain-containing protein 4 (ARRDC4)
| Representative RNA-seq result indicating the expression of this target gene regulated by YTHDF2 | ||
| Cell Line | Human umbilical cord blood CD34+ cells | Homo sapiens |
|
Treatment: YTHDF2 knockdown UCB CD34+ cells
Control: Wild type UCB CD34+ cells
|
GSE107956 | |
| Regulation |
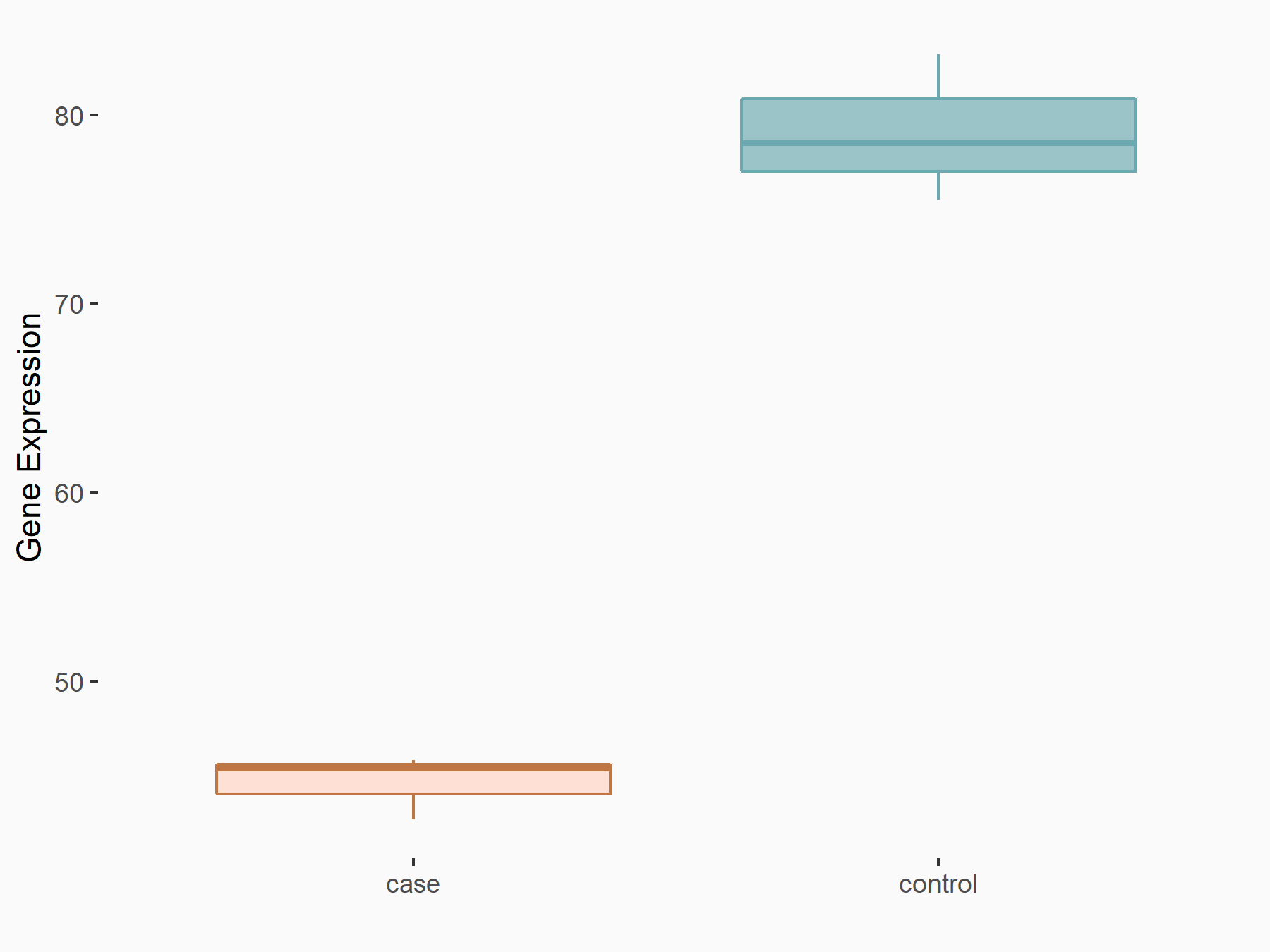  |
logFC: -8.12E-01 p-value: 3.74E-05 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and YTHDF2 | ||
| Cell Line | Hela | Homo sapiens |
| Regulation | logFC: 1.87E+00 | GSE49339 |
Colorectal cancer [ICD-11: 2B91]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [1] | |||
| Responsed Disease | Colorectal cancer [ICD-11: 2B91] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | mRNA surveillance pathway | hsa03015), RNA degradation | ||
| Cell Process | RNA stability | |||
In-vitro Model |
SW620 | Colon adenocarcinoma | Homo sapiens | CVCL_0547 |
| SW480 | Colon adenocarcinoma | Homo sapiens | CVCL_0546 | |
| RKO | Colon carcinoma | Homo sapiens | CVCL_0504 | |
| NCM460 | Normal | Homo sapiens | CVCL_0460 | |
| LoVo | Colon adenocarcinoma | Homo sapiens | CVCL_0399 | |
| HT29 | Colon cancer | Mus musculus | CVCL_A8EZ | |
| HCT 8 | Colon adenocarcinoma | Homo sapiens | CVCL_2478 | |
| HCT 15 | Colon adenocarcinoma | Homo sapiens | CVCL_0292 | |
| HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 | |
| In-vivo Model | Equal amount of HCT116 cells (2 × 106) stably expression of relevant plasmids was injected into the right flank of mice, tumor bulks was monitored once a week after injection and volumes were counted as 0.5 × a2 × b (a and b respectively indicated short and long diameter of tumor). | |||
| Response Summary | Knockdown of METTL14 significantly enhanced Arrestin domain-containing protein 4 (ARRDC4) mRNA stability relying on the "reader" protein YTHDF2 dependent manner in colorectal cancer. | |||
Bcl-2-modifying factor (BMF)
| Representative RNA-seq result indicating the expression of this target gene regulated by YTHDF2 | ||
| Cell Line | Human umbilical cord blood CD34+ cells | Homo sapiens |
|
Treatment: YTHDF2 knockdown UCB CD34+ cells
Control: Wild type UCB CD34+ cells
|
GSE107956 | |
| Regulation |
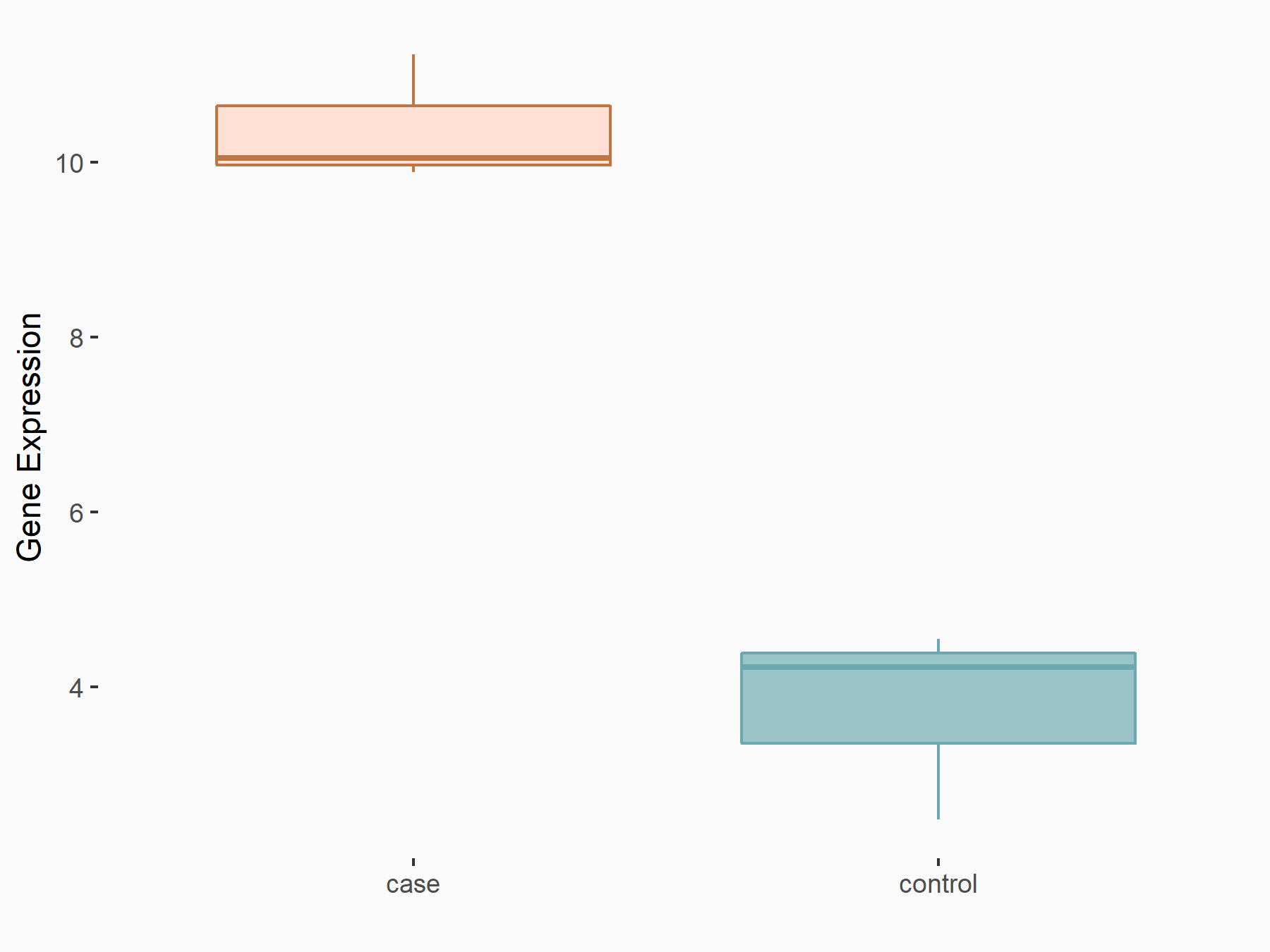 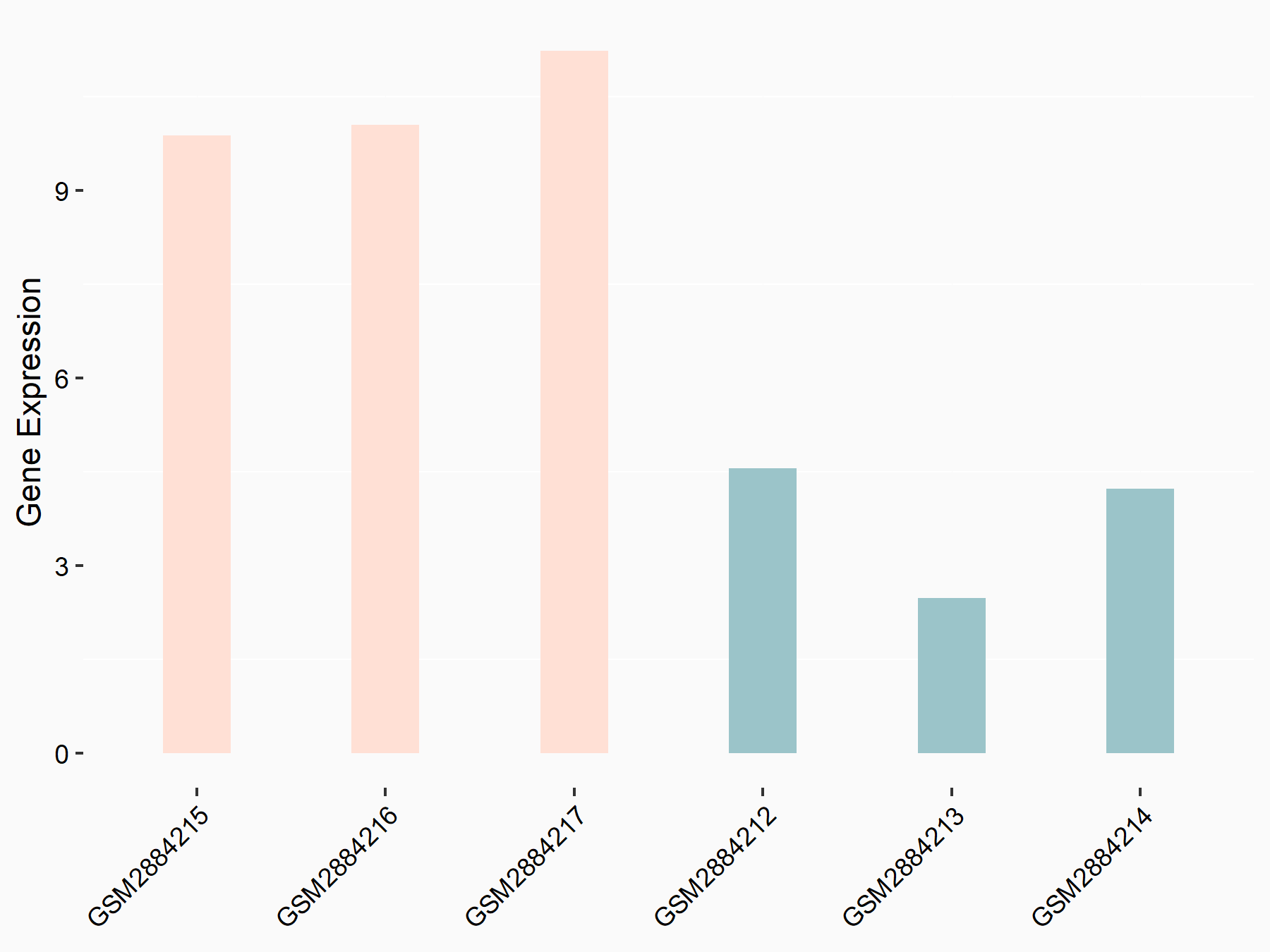 |
logFC: 1.29E+00 p-value: 2.46E-03 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and YTHDF2 | ||
| Cell Line | Hela | Homo sapiens |
| Regulation | logFC: 2.27E+00 | GSE49339 |
Ovarian cancer [ICD-11: 2C73]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [2] | |||
| Responsed Disease | Ovarian cancer [ICD-11: 2C73] | |||
| Target Regulation | Down regulation | |||
| Cell Process | RNA stability | |||
| Cell apoptosis | ||||
In-vitro Model |
SK-OV-3 | Ovarian serous cystadenocarcinoma | Homo sapiens | CVCL_0532 |
| OVCAR-8 | High grade ovarian serous adenocarcinoma | Homo sapiens | CVCL_1629 | |
| OVCA429 | Ovarian cystadenocarcinoma | Homo sapiens | CVCL_3936 | |
| OVCA420 | Ovarian serous adenocarcinoma | Homo sapiens | CVCL_3935 | |
| In-vivo Model | 5 × 106 cells were suspended in 100 uL PBS and then were inoculated subcutaneously. | |||
| Response Summary | FBW7 suppresses tumor growth and progression via antagonizing YTHDF2-mediated Bcl-2-modifying factor (BMF) mRNA decay in ovarian cancer. | |||
C-X-C chemokine receptor type 4 (CXCR4)
| Representative RNA-seq result indicating the expression of this target gene regulated by YTHDF2 | ||
| Cell Line | Testis | Mus musculus |
|
Treatment: YTHDF2 knockout mice testis
Control: Mice testis
|
GSE147574 | |
| Regulation |
  |
logFC: 7.39E-01 p-value: 4.60E-02 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and YTHDF2 | ||
| Cell Line | Hela | Homo sapiens |
| Regulation | logFC: 2.20E+00 | GSE49339 |
Melanoma [ICD-11: 2C30]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [3] | |||
| Responsed Disease | Melanoma [ICD-11: 2C30] | |||
| Responsed Drug | PMID31239444-anti-PD1 antibody | Investigative | ||
| Target Regulation | Down regulation | |||
| Pathway Response | PD-L1 expression and PD-1 checkpoint pathway in cancer | hsa05235 | ||
| Cell Process | mRNA decay | |||
In-vitro Model |
B16-F10 | Mouse melanoma | Mus musculus | CVCL_0159 |
| CHL-1 | Melanoma | Homo sapiens | CVCL_1122 | |
| 624-mel | Melanoma | Homo sapiens | CVCL_8054 | |
| NHEM (Normal Human Epidermal Melanocytes) | ||||
| SK-MEL-30 | Cutaneous melanoma | Homo sapiens | CVCL_0039 | |
| WM115 | Melanoma | Homo sapiens | CVCL_0040 | |
| WM35 | Melanoma | Homo sapiens | CVCL_0580 | |
| WM3670 | Melanoma | Homo sapiens | CVCL_6799 | |
| WM793 | Melanoma | Homo sapiens | CVCL_8787 | |
| In-vivo Model | When the tumors reached a volume of 80-100 mm3, mice were treated with anti-PD-1 or isotype control antibody (200 ug/mouse) by i.p. injection, every other day for three times. For IFNγ blockade treatment, C57BL/6 mice were treated with anti-IFNγ antibody or isotype control IgG (250 ug/mouse) every other day after tumor cell inoculation. | |||
| Response Summary | These findings demonstrate a crucial role of FTO as an m6A demethylase in promoting melanoma tumorigenesis and anti-PD-1 resistance, and suggest that the combination of FTO inhibition with anti-PD-1 blockade reduces the resistance to immunotherapy in melanoma. Knockdown of FTO increases m6A methylation in the critical protumorigenic melanoma cell-intrinsic genes including PD-1 (PDCD1), C-X-C chemokine receptor type 4 (CXCR4), and SOX10, leading to increased RNA decay through the m6A reader YTHDF2. | |||
Ephrin type-B receptor 3 (EPHB3)
| Representative RNA-seq result indicating the expression of this target gene regulated by YTHDF2 | ||
| Cell Line | Mouse-cerebellum granule cell | Mus musculus |
|
Treatment: YTHDF2 knockdown mouse-cerebellum granule cell
Control: Wild type mouse-cerebellum granule cell
|
GSE153688 | |
| Regulation |
 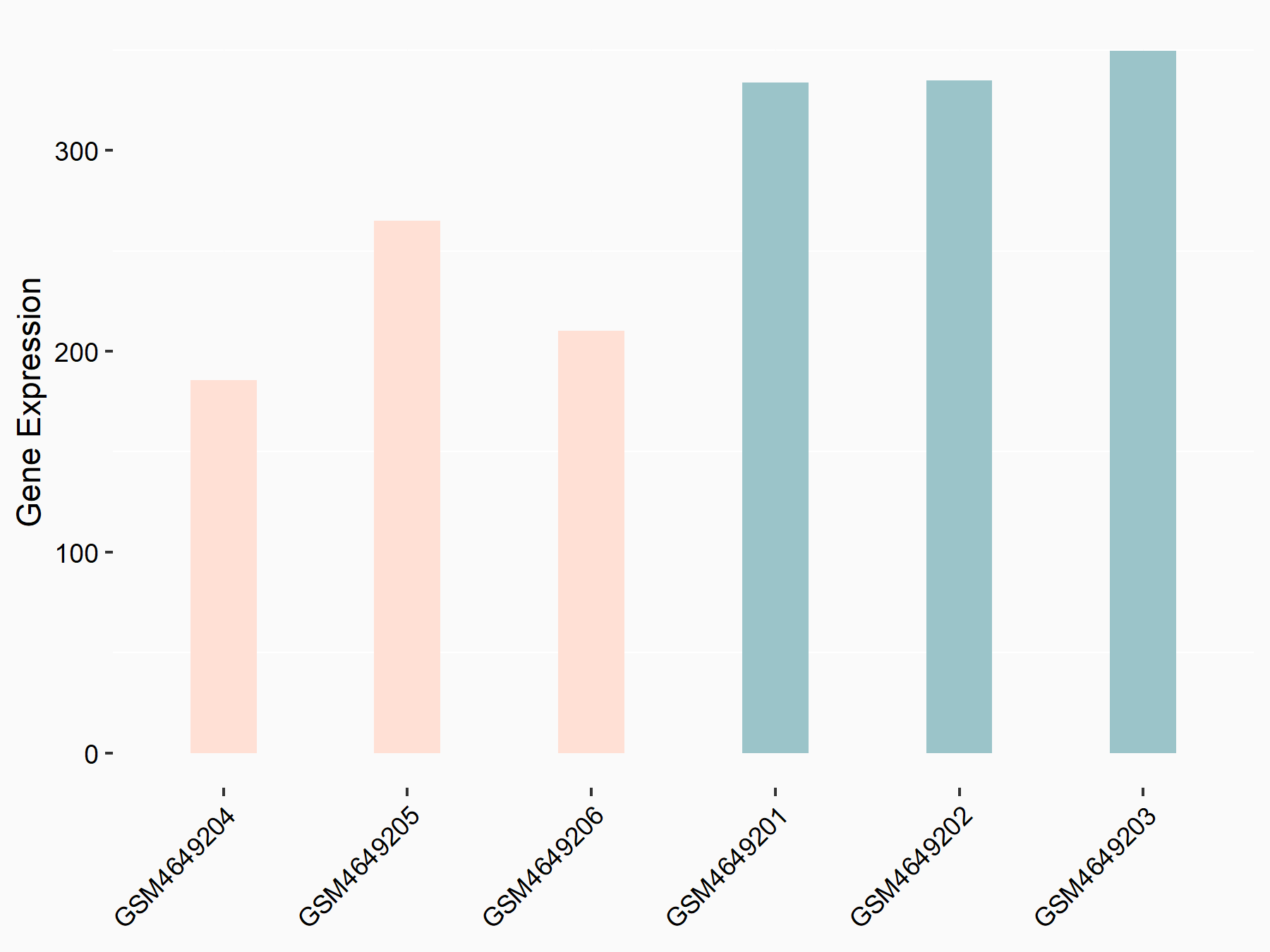 |
logFC: -6.25E-01 p-value: 1.90E-03 |
| More Results | Click to View More RNA-seq Results | |
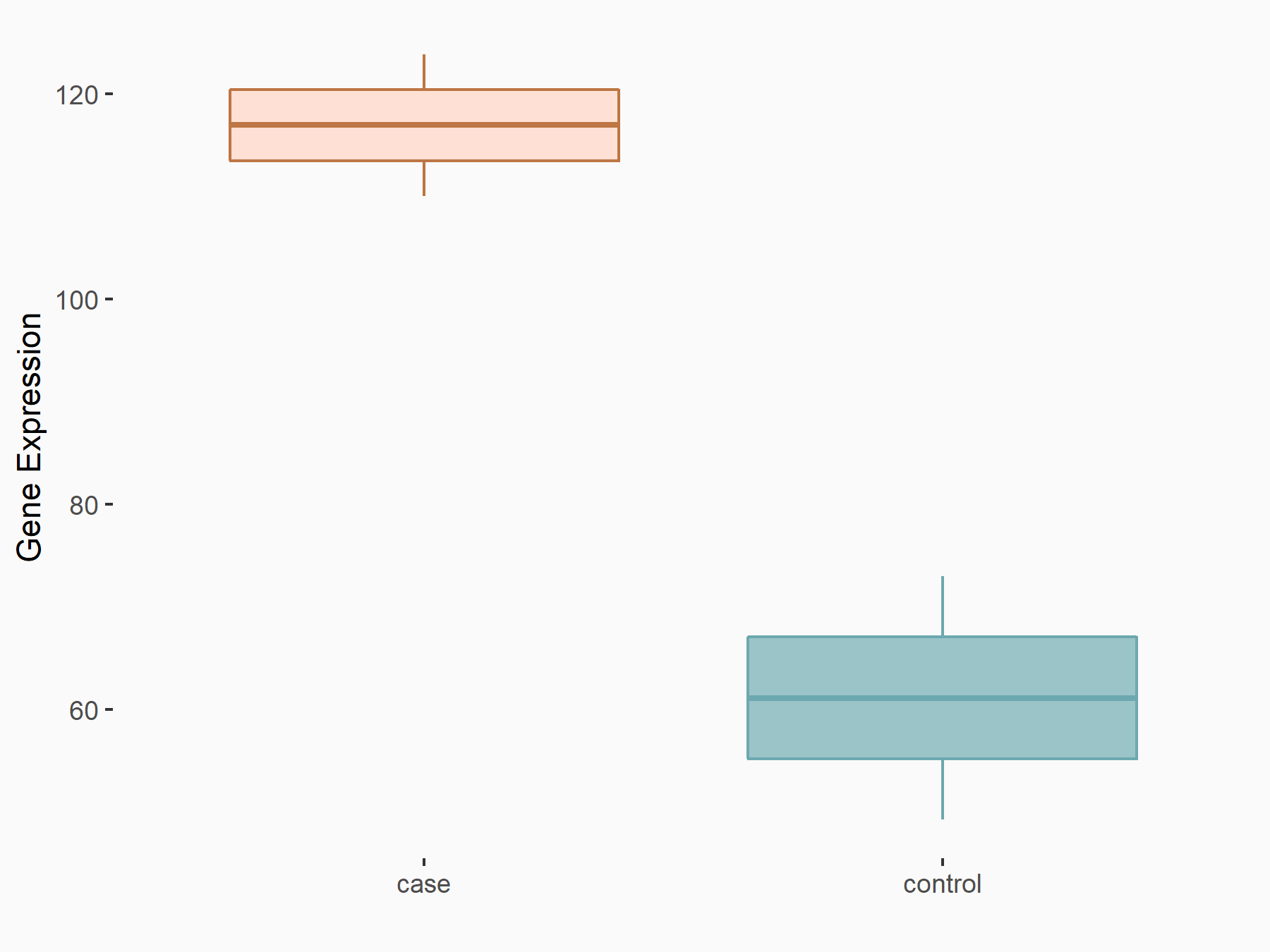
| Representative RIP-seq result supporting the interaction between the target gene and YTHDF2 | ||
| Cell Line | Hela | Homo sapiens |
| Regulation | logFC: 1.21E+00 | GSE49339 |
Brain cancer [ICD-11: 2A00]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [4] | |||
| Responsed Disease | Glioblastoma [ICD-11: 2A00.00] | |||
| Responsed Drug | Temozolomide | Approved | ||
| Target Regulation | Down regulation | |||
| Pathway Response | PI3K-Akt signaling pathway | hsa04151 | ||
| Cell Process | RNA stability | |||
In-vitro Model |
T98G | Glioblastoma | Homo sapiens | CVCL_0556 |
| LN-229 | Glioblastoma | Homo sapiens | CVCL_0393 | |
| In-vivo Model | 5 × 106 infected T98G cells (LV-NC or LV-YTHDF2) were injected into the flanks of mice through subcutaneous. | |||
| Response Summary | YTHDF2 enhanced TMZ resistance in GBM by activation of the PI3K/Akt and NF-Kappa-B signalling pathways via inhibition of Ephrin type-B receptor 3 (EPHB3) and TNFAIP3. | |||
Homeobox protein Nkx-3.1 (NKX3-1)
| Representative RNA-seq result indicating the expression of this target gene regulated by YTHDF2 | ||
| Cell Line | GSC11 cell line | Homo sapiens |
|
Treatment: siYTHDF2 GSC11 cells
Control: siControl GSC11 cells
|
GSE142825 | |
| Regulation |
 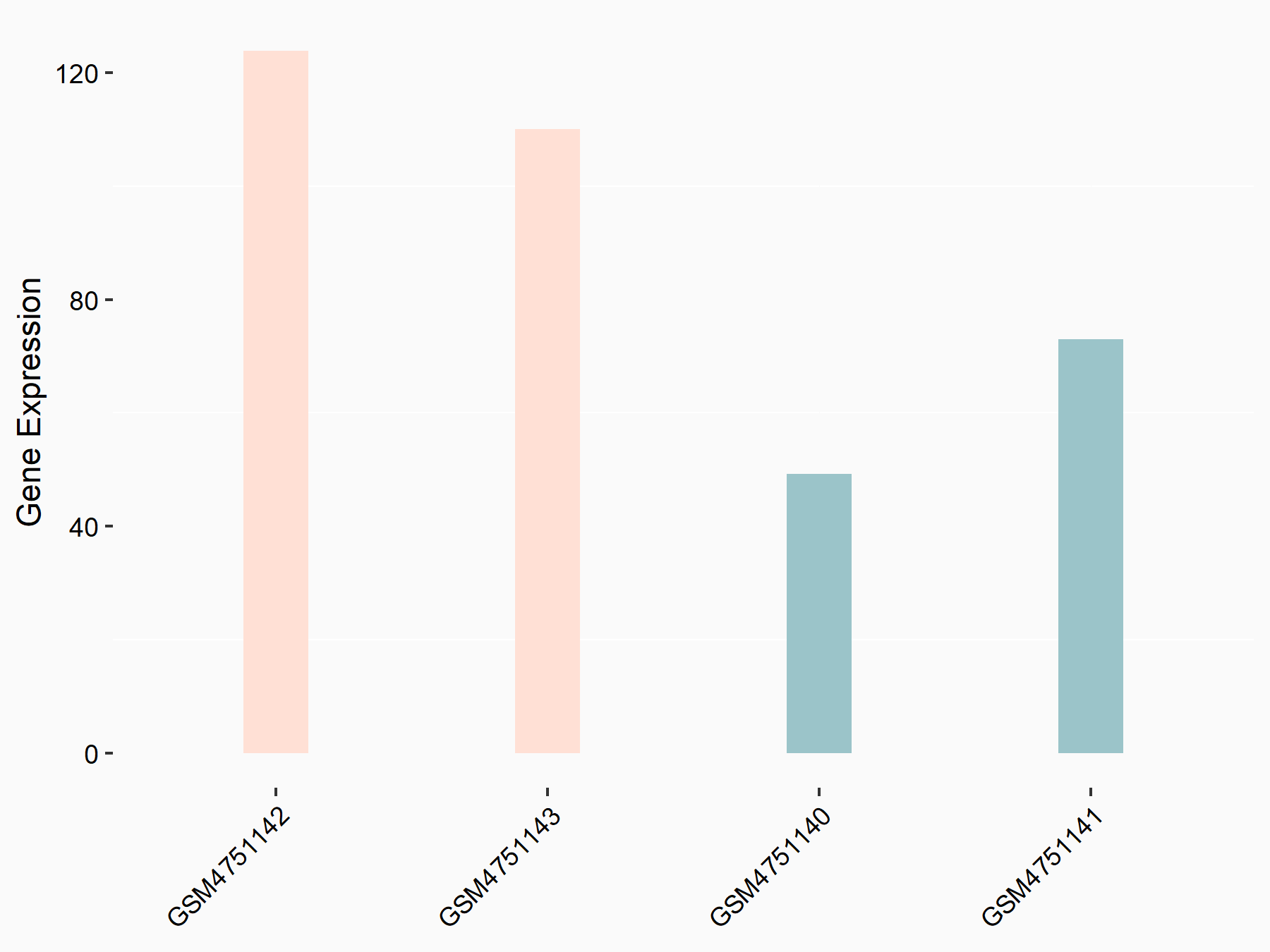 |
logFC: 9.36E-01 p-value: 4.17E-02 |
| More Results | Click to View More RNA-seq Results | |
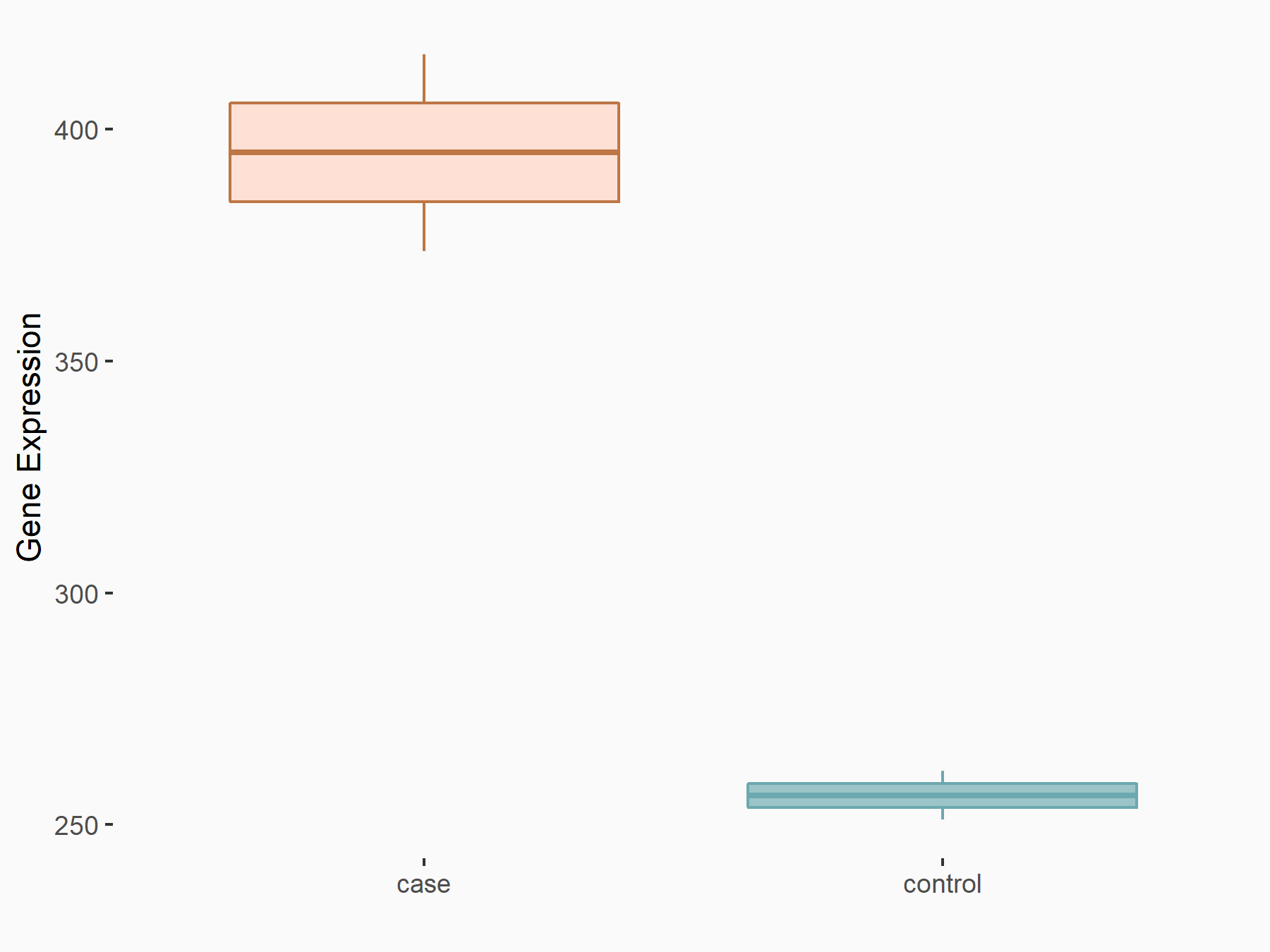
| Representative RIP-seq result supporting the interaction between the target gene and YTHDF2 | ||
| Cell Line | Hela | Homo sapiens |
| Regulation | logFC: 1.20E+00 | GSE49339 |
Prostate cancer [ICD-11: 2C82]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [5] | |||
| Responsed Disease | Prostate cancer [ICD-11: 2C82] | |||
| Target Regulation | Down regulation | |||
| Pathway Response | Oxidative phosphorylation | hsa00190 | ||
In-vitro Model |
VCaP | Prostate carcinoma | Homo sapiens | CVCL_2235 |
| RWPE-1 | Normal | Homo sapiens | CVCL_3791 | |
| PC-3 | Prostate carcinoma | Homo sapiens | CVCL_0035 | |
| DU145 | Prostate carcinoma | Homo sapiens | CVCL_0105 | |
| 22Rv1 | Prostate carcinoma | Homo sapiens | CVCL_1045 | |
| In-vivo Model | Approximately 2 × 106 PCa cells (PC-3 shNC, shYTHDF2, shMETTL3 cell lines) per mouse suspended in 100 uL PBS were injected in the flank of male BALB/c nude mice (4 weeks old). During the 40-day observation, the tumor size (V = (width2×length ×0.52)) was measured with vernier caliper. Approximately 1.5 × 106 PCa cells suspended in 100 uL of PBS (PC-3 shNC, shYTHDF2, and shMETTL3 cell lines) per mouse were injected into the tail vein of male BALB/c nude mice (4 weeks old). The IVIS Spectrum animal imaging system (PerkinElmer) was used to evaluate the tumor growth (40 days) and whole metastasis conditions (4 weeks and 6 weeks) with 100 uL XenoLight D-luciferin Potassium Salt (15 mg/ml, Perkin Elmer) per mouse. Mice were anesthetized and then sacrificed for tumors and metastases which were sent for further organ-localized imaging as above, IHC staining and hematoxylin-eosin (H&E) staining. | |||
| Response Summary | Knock-down of YTHDF2 or METTL3 significantly induced the expression of LHPP and Homeobox protein Nkx-3.1 (NKX3-1) at both mRNA and protein level with inhibited phosphorylated AKT. YTHDF2 mediates the mRNA degradation of the tumor suppressors LHPP and NKX3-1 in m6A-dependent way to regulate AKT phosphorylation-induced tumor progression in prostate cancer. | |||
Krueppel-like factor 4 (KLF4)
| Representative RNA-seq result indicating the expression of this target gene regulated by YTHDF2 | ||
| Cell Line | GSC11 cell line | Homo sapiens |
|
Treatment: siYTHDF2 GSC11 cells
Control: siControl GSC11 cells
|
GSE142825 | |
| Regulation |
 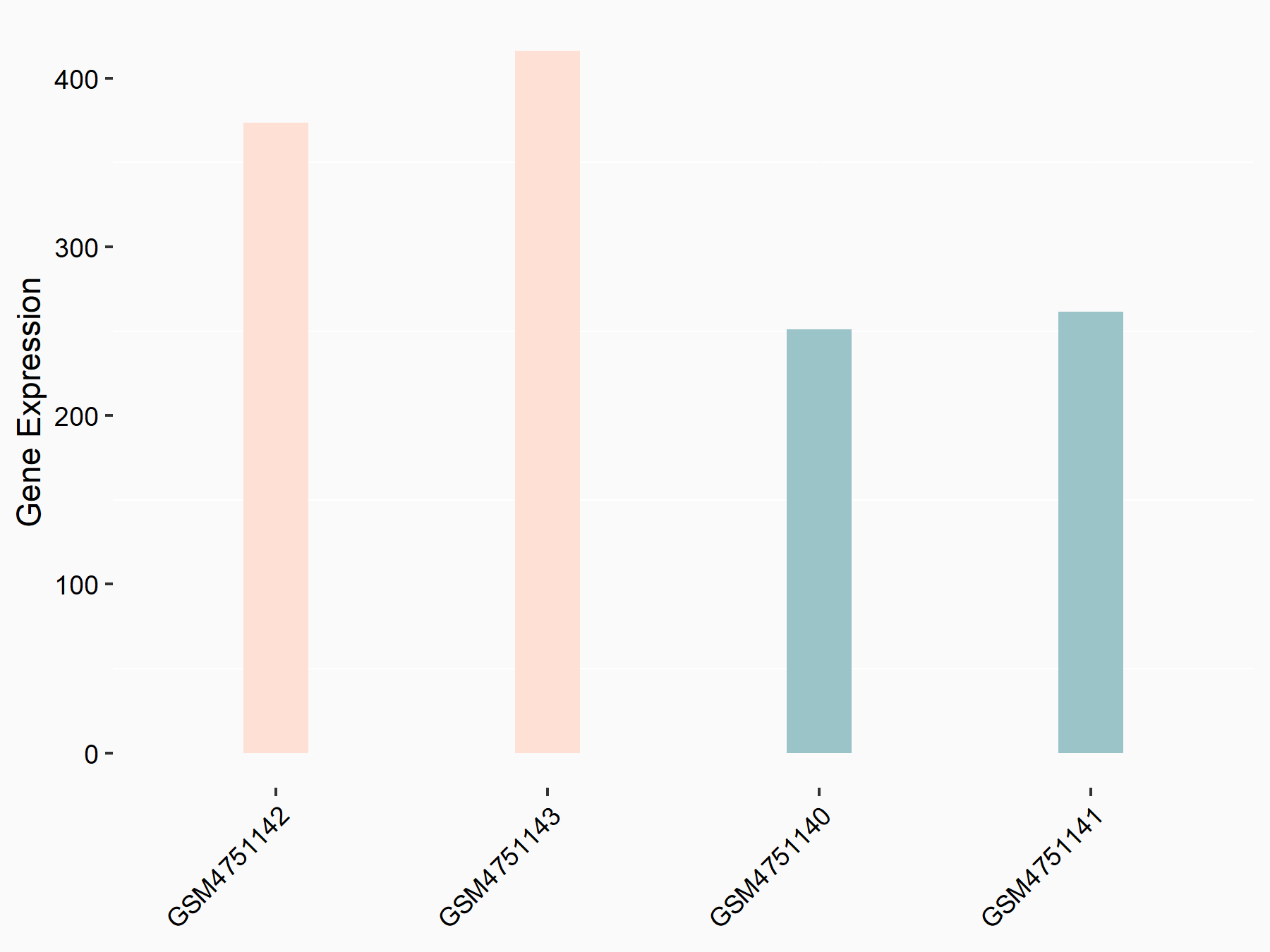 |
logFC: 6.24E-01 p-value: 8.92E-03 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and YTHDF2 | ||
| Cell Line | Hela | Homo sapiens |
| Regulation | logFC: 2.55E+00 | GSE49339 |
Lung cancer [ICD-11: 2C25]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [6] | |||
| Responsed Disease | Non-small-cell lung carcinoma [ICD-11: 2C25.Y] | |||
| Target Regulation | Down regulation | |||
| Pathway Response | TNF signaling pathway | hsa04668 | ||
| Cell Process | Cell migration | |||
| Cell invasion | ||||
| Epithelial-mesenchymal transition | ||||
In-vitro Model |
NCI-H1975 | Lung adenocarcinoma | Homo sapiens | CVCL_1511 |
| A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 | |
| Response Summary | MiR-1915-3p expression was regulated by METTL3/YTHDF2 m6A axis through transcription factor Krueppel-like factor 4 (KLF4). miR-1915-3p function as a tumor suppressor by targeting SET and has an anti-metastatic therapeutic potential for lung cancer treatment. | |||
Bladder cancer [ICD-11: 2C94]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [7] | |||
| Responsed Disease | Bladder cancer [ICD-11: 2C94] | |||
| Target Regulation | Down regulation | |||
| Cell Process | Cancer proliferation | |||
| Cancer metastasis | ||||
In-vitro Model |
SV-HUC-1 | Normal | Homo sapiens | CVCL_3798 |
| T24 | Bladder carcinoma | Homo sapiens | CVCL_0554 | |
| UM-UC-3 | Bladder carcinoma | Homo sapiens | CVCL_1783 | |
| In-vivo Model | For the subcutaneous implantation model, UM-UC-3 cells (2 × 106 cells per mouse) stably METTL3 knocked down (shMETTL3-1, shMETTL3-2) were injected into the flanks of mice. | |||
| Response Summary | METTL3/YTHDF2/SETD7/Krueppel-like factor 4 (KLF4) m6 A axis provide the insight into the underlying mechanism of carcinogenesis and highlight potential therapeutic targets for bladder cancer. | |||
MOB kinase activator 3B (MOB3B)
| Representative RNA-seq result indicating the expression of this target gene regulated by YTHDF2 | ||
| Cell Line | GSC11 cell line | Homo sapiens |
|
Treatment: siYTHDF2 GSC11 cells
Control: siControl GSC11 cells
|
GSE142825 | |
| Regulation |
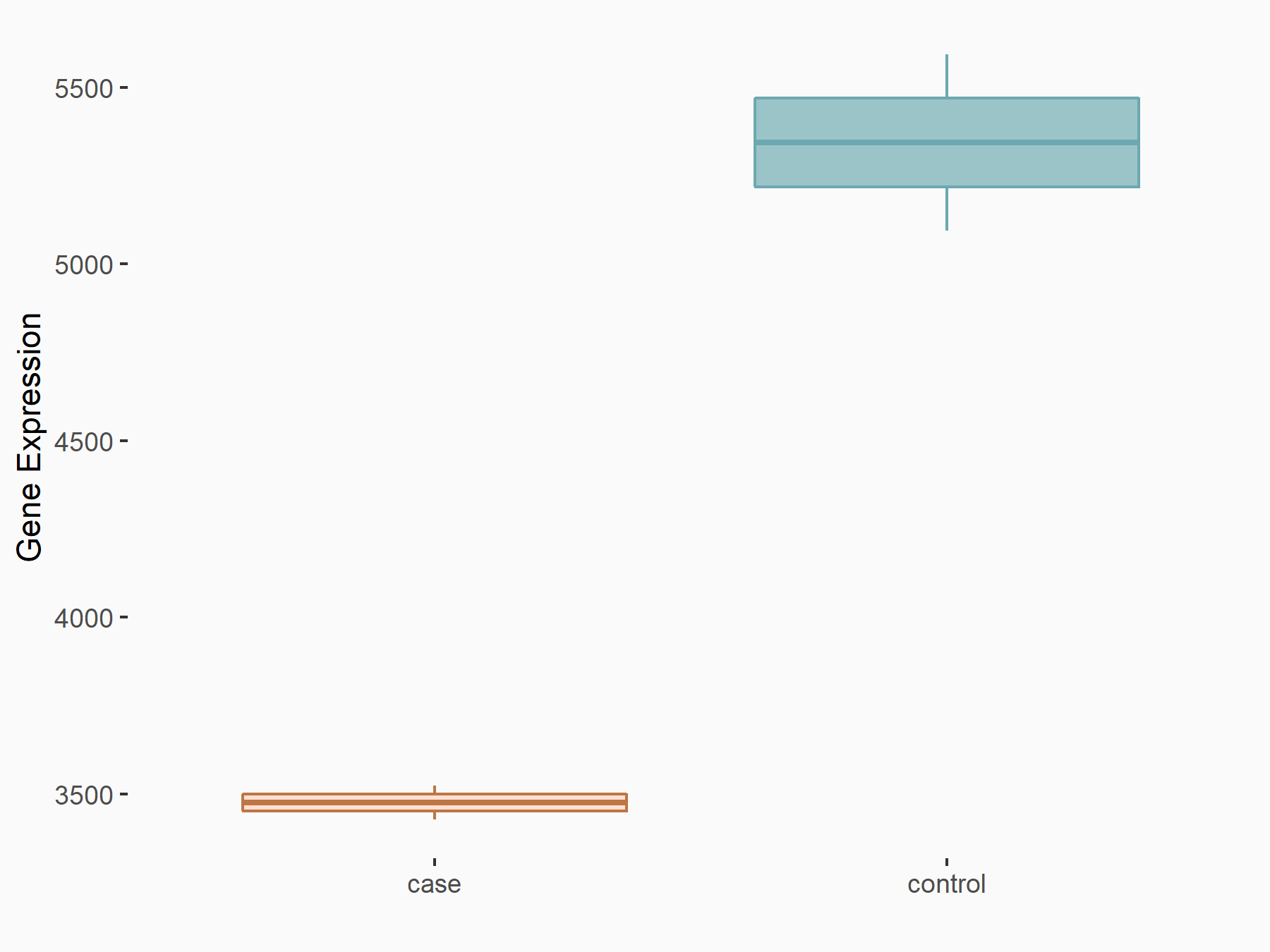 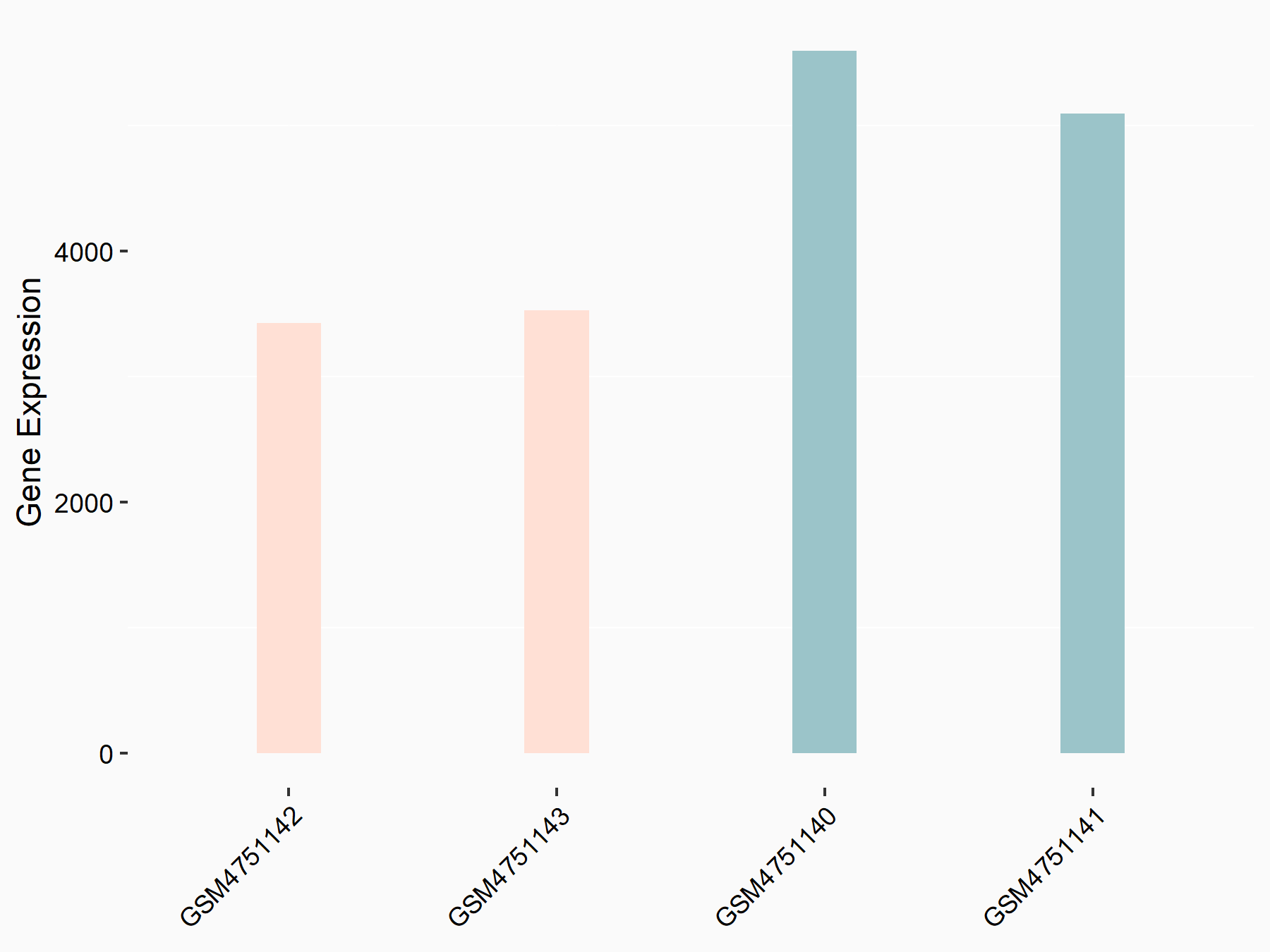 |
logFC: -6.21E-01 p-value: 1.40E-08 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and YTHDF2 | ||
| Cell Line | Hela | Homo sapiens |
| Regulation | logFC: 1.42E+00 | GSE49339 |
Prostate cancer [ICD-11: 2C82]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [8] | |||
| Responsed Disease | Prostate cancer [ICD-11: 2C82] | |||
| Target Regulation | Down regulation | |||
| Cell Process | Cell proliferation | |||
| Cell migration | ||||
| Cell invasion | ||||
| Cell apoptosis | ||||
In-vitro Model |
DU145 | Prostate carcinoma | Homo sapiens | CVCL_0105 |
| LNCaP C4-2 | Prostate carcinoma | Homo sapiens | CVCL_4782 | |
| PC-3 | Prostate carcinoma | Homo sapiens | CVCL_0035 | |
| In-vivo Model | Under anesthesia with ether, the nude mice were disinfected and subcutaneously inoculated with cells transfected with oe-NC, oe-KDM5A + oe-NC and oe-KDM5A + oe-MOB3B at a density of 1 × 106 cells/mouse (200 uL) at the back of the right hind leg. | |||
| Response Summary | Activation of the KDM5A/miRNA-495/YTHDF2/m6A-MOB3B axis facilitates prostate cancer progression. YTHDF2 could inhibit MOB kinase activator 3B (MOB3B) expression by recognizing m6A modification of MOB3B mRNA and inducing mRNA degradation. | |||
Mothers against decapentaplegic homolog 3 (SMAD3)
| Representative RNA-seq result indicating the expression of this target gene regulated by YTHDF2 | ||
| Cell Line | mouse embryonic stem cells | Mus musculus |
|
Treatment: shYthdf2 embryonic stem cells
Control: shLuc embryonic stem cells
|
GSE156437 | |
| Regulation |
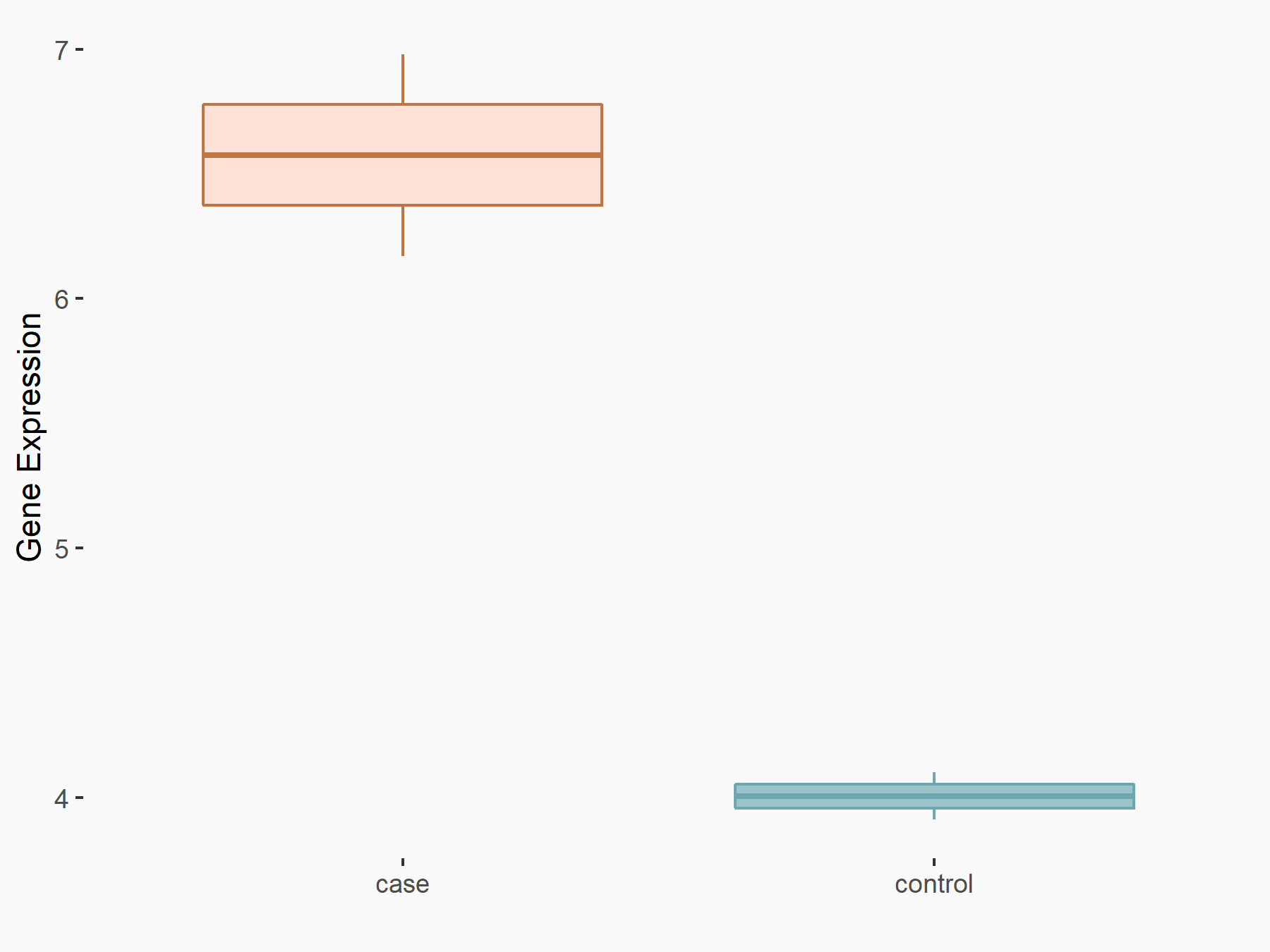 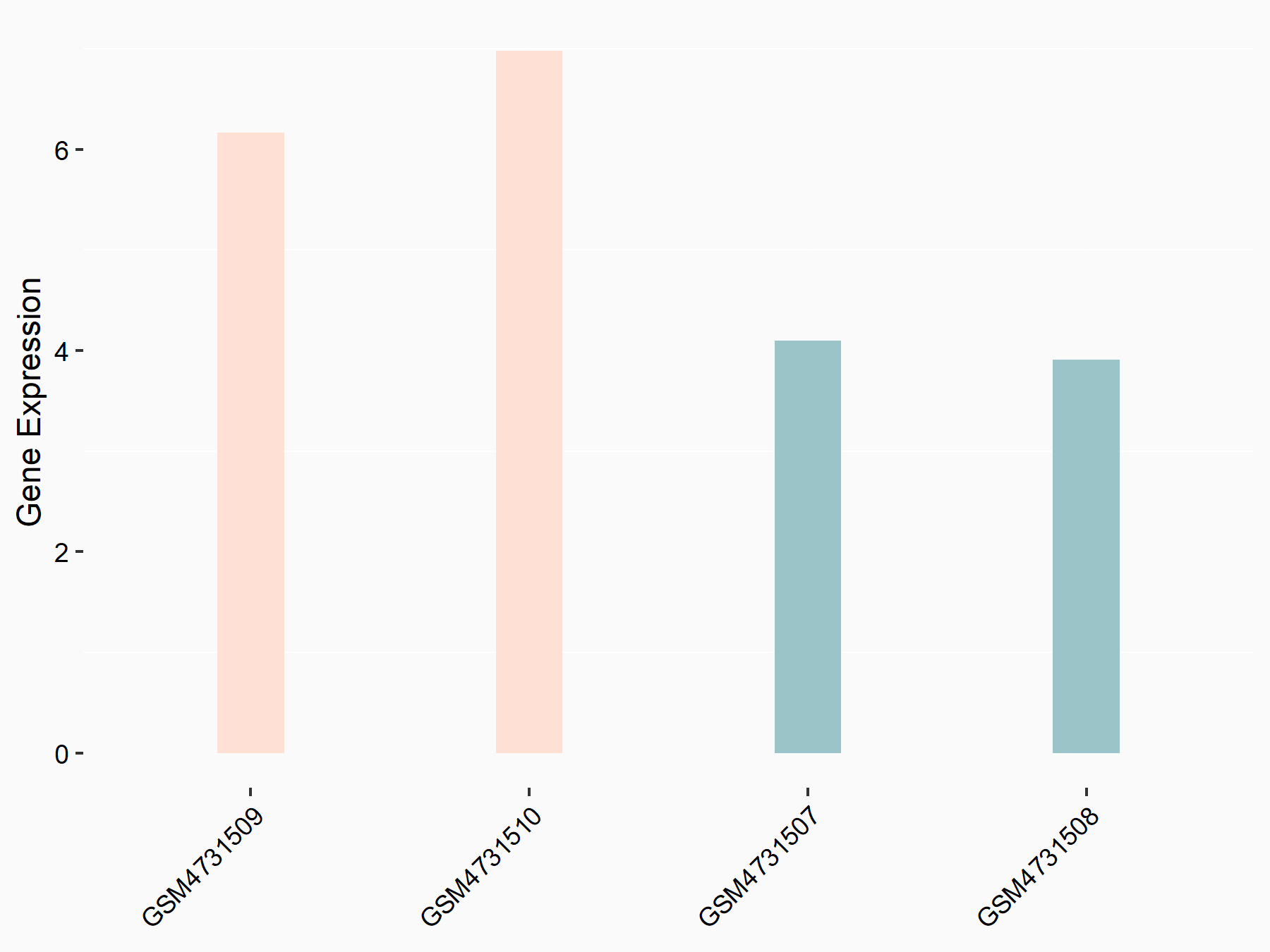 |
logFC: 5.96E-01 p-value: 6.99E-03 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and YTHDF2 | ||
| Cell Line | Hela | Homo sapiens |
| Regulation | logFC: 1.21E+00 | GSE49339 |
Lung cancer [ICD-11: 2C25]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [9] | |||
| Responsed Disease | Lung cancer [ICD-11: 2C25] | |||
| Target Regulation | Down regulation | |||
| Pathway Response | mRNA surveillance pathway | hsa03015 | ||
| Cell Process | Epithelial-mesenchymal transition | |||
In-vitro Model |
NCI-H1299 | Lung large cell carcinoma | Homo sapiens | CVCL_0060 |
| A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 | |
| Response Summary | YTHDF2 inhibits the migration and invasion of lung adenocarcinoma cells by regulating the FAM83D-TGFbeta1-Mothers against decapentaplegic homolog 3 (SMAD3) pathway, which will play an important role in lung cancer metastasis. | |||
Mothers against decapentaplegic homolog 7 (SMAD7)
| Representative RNA-seq result indicating the expression of this target gene regulated by YTHDF2 | ||
| Cell Line | Human umbilical cord blood CD34+ cells | Homo sapiens |
|
Treatment: YTHDF2 knockdown UCB CD34+ cells
Control: Wild type UCB CD34+ cells
|
GSE107956 | |
| Regulation |
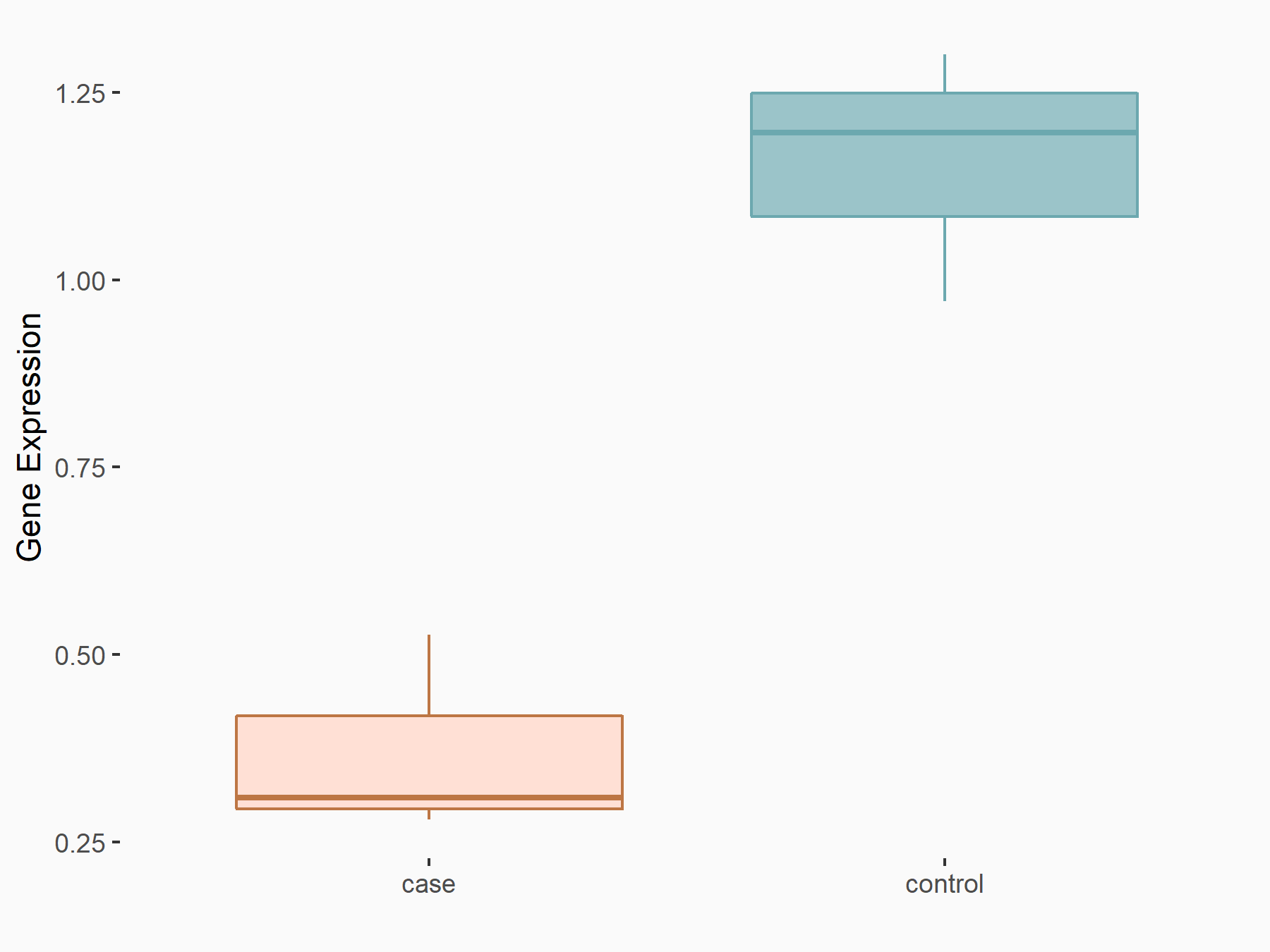 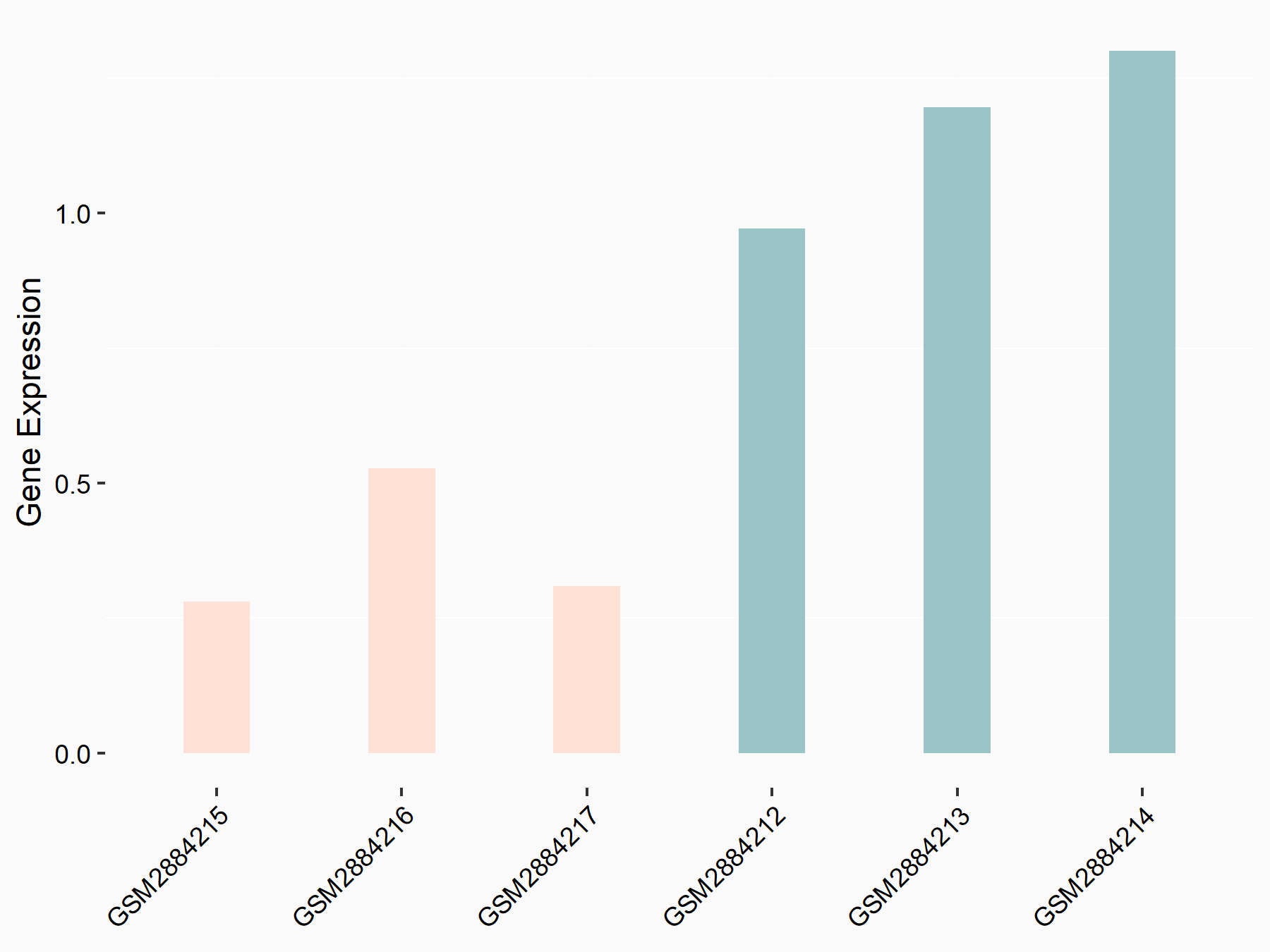 |
logFC: -6.53E-01 p-value: 1.90E-03 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and YTHDF2 | ||
| Cell Line | Hela | Homo sapiens |
| Regulation | logFC: 1.67E+00 | GSE49339 |
Inflammatory response [ICD-11: MG46]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [10] | |||
| Responsed Disease | Inflammatory response [ICD-11: MG46] | |||
| Target Regulation | Down regulation | |||
| Pathway Response | MAPK signaling pathway | hsa04010 | ||
| Cell Process | RNA stability | |||
In-vitro Model |
MC3T3-E1 | Normal | Mus musculus | CVCL_0409 |
| Response Summary | METTL3 knockdown inhibits osteoblast differentiation and Smad-dependent signaling by stabilizing Mothers against decapentaplegic homolog 7 (SMAD7) and Smurf1 mRNA transcripts via YTHDF2 involvement and activates the inflammatory response by regulating MAPK signaling in LPS-induced inflammation. | |||
Lung cancer [ICD-11: 2C25]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [85] | |||
| Responsed Disease | Lung cancer [ICD-11: 2C25] | |||
| Target Regulation | Down regulation | |||
In-vitro Model |
MRC-9 | Normal | Homo sapiens | CVCL_2629 |
| NCI-H1299 | Lung large cell carcinoma | Homo sapiens | CVCL_0060 | |
| NCI-H1650 | Minimally invasive lung adenocarcinoma | Homo sapiens | CVCL_1483 | |
| NCI-H1703 | Lung squamous cell carcinoma | Homo sapiens | CVCL_1490 | |
| H1795 (Lung cancer H1795 cell lines were purchased from ATCC, USA) | ||||
| NCI-H1792 | Lung adenocarcinoma | Homo sapiens | CVCL_1495 | |
| A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 | |
| DLD-1 | Colon adenocarcinoma | Homo sapiens | CVCL_0248 | |
| SW480 | Colon adenocarcinoma | Homo sapiens | CVCL_0546 | |
| PANC-1 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0480 | |
| MIA PaCa-2 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0428 | |
Myc proto-oncogene protein (MYC)
| Representative RNA-seq result indicating the expression of this target gene regulated by YTHDF2 | ||
| Cell Line | GSC11 cell line | Homo sapiens |
|
Treatment: siYTHDF2 GSC11 cells
Control: siControl GSC11 cells
|
GSE142825 | |
| Regulation |
 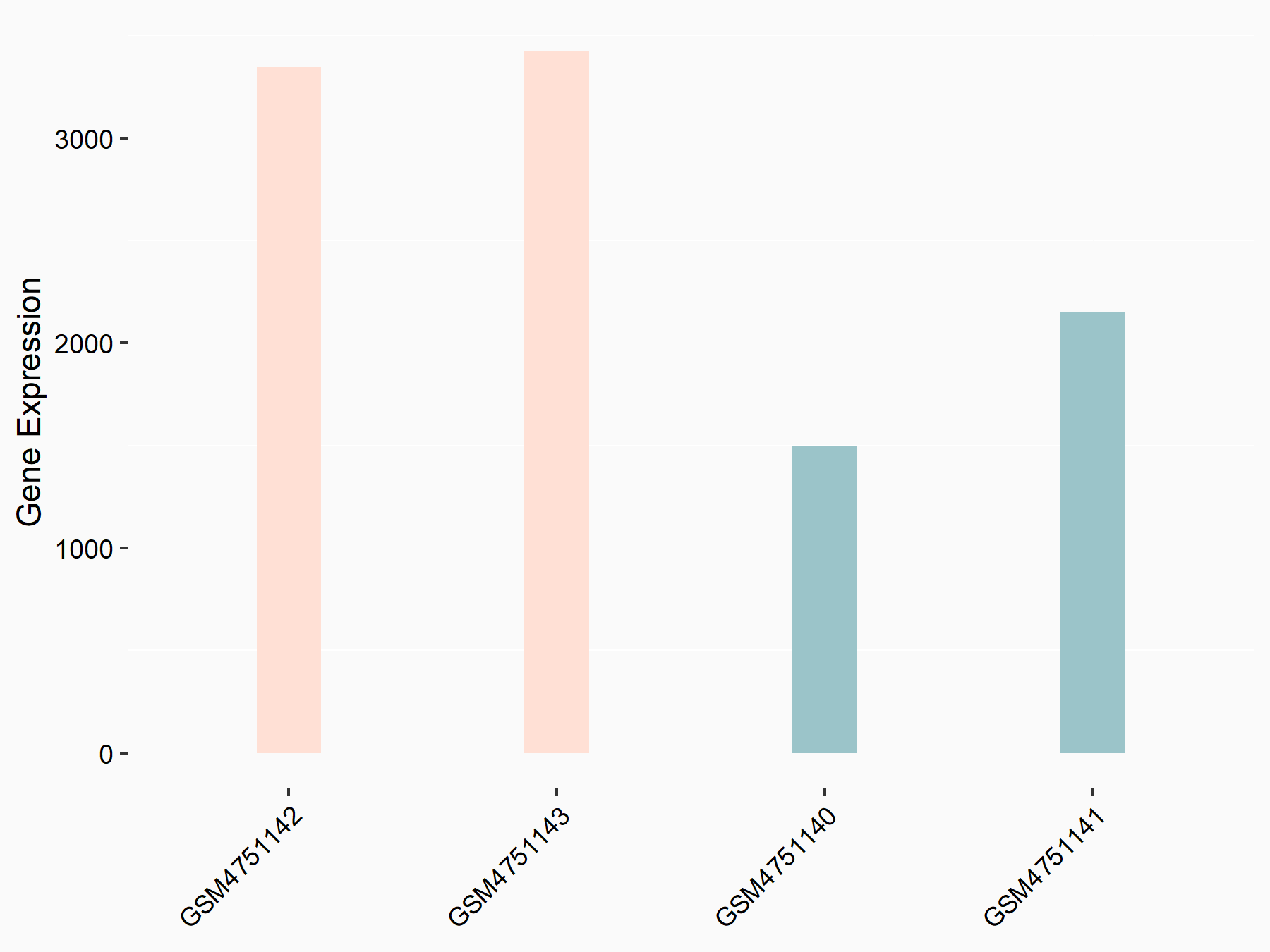 |
logFC: 8.94E-01 p-value: 8.57E-07 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and YTHDF2 | ||
| Cell Line | Hela | Homo sapiens |
| Regulation | logFC: 1.40E+00 | GSE49339 |
Brain cancer [ICD-11: 2A00]
| In total 2 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [11] | |||
| Responsed Disease | Glioma [ICD-11: 2A00.0] | |||
| Responsed Drug | Linsitinib | Phase 3 | ||
| Target Regulation | Up regulation | |||
| Pathway Response | RNA degradation | hsa03018 | ||
| Cell Process | RNA stability | |||
In-vitro Model |
() | |||
| HNP1 (A human neural progenitor cell) | ||||
| NHA (Normal human astrocytes) | ||||
| NSC11 (Pluripotent derived neural progenitor cell) | ||||
| In-vivo Model | For in vivo drug treatment studies, intracranial xenografts were generated by implanting 5000 patient-derived GSCs (387 and 4121) into the right cerebral cortex of NSG mice as described above. | |||
| Response Summary | The m6A reader YTHDF2 stabilized Myc proto-oncogene protein (MYC) mRNA specifically in cancer stem cells. Given the challenge of targeting MYC, YTHDF2 presents a therapeutic target to perturb MYC signaling in glioblastoma. The IGF1/IGF1R inhibitor linsitinib preferentially targeted YTHDF2-expressing cells, inhibiting GSC viability without affecting NSCs and impairing in vivo glioblastoma growth. YTHDF2 links RNA epitranscriptomic modifications and GSC growth, laying the foundation for the YTHDF2-MYC-IGFBP3 axis as a specific and novel therapeutic target in glioblastoma. | |||
| Experiment 2 Reporting the m6A-centered Disease Response of This Target Gene | [86] | |||
| Responsed Disease | Brain cancer [ICD-11: 2A00] | |||
| Target Regulation | Up regulation | |||
Breast cancer [ICD-11: 2C60]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [12] | |||
| Responsed Disease | Breast cancer [ICD-11: 2C60] | |||
| Responsed Drug | Tamoxifen | Approved | ||
| Pathway Response | MAPK signaling pathway | hsa04010 | ||
| Cell Process | Epithelial-to-mesenchymal transition | |||
| Cell apoptosis | ||||
In-vitro Model |
HEK293T | Normal | Homo sapiens | CVCL_0063 |
| MCF-7 | Invasive breast carcinoma | Homo sapiens | CVCL_0031 | |
| MDA-MB-231 | Breast adenocarcinoma | Homo sapiens | CVCL_0062 | |
| MBA-MD-231 (Human breast cancer cell) | ||||
| MYC-ER HMEC (Human mammary epithelial cells expressing a MYC estrogen receptor fusion) | ||||
| SK-BR-3 | Breast adenocarcinoma | Homo sapiens | CVCL_0033 | |
| In-vivo Model | To induce recombination at 8 weeks of age both CAG-CreERT;Ythdf2fl/fl and Ythdf2fl/fl littermates were injected with 75mg/kg body weight tamoxifen dissolved in corn oil daily for 5 days. | |||
| Response Summary | LCAT3 upregulation is attributable to m6A modification mediated by METTL3, leading to LCAT3 stabilization. Treated cells with tamoxifen to induce MYC activity. Highlights the therapeutic potential of RBPs by uncovering a critical role for YTHDF2 in counteracting the global increase of mRNA synthesis in Myc proto-oncogene protein (MYC)-driven breast cancers. | |||
Liver cancer [ICD-11: 2C12]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [75] | |||
| Responsed Disease | Liver hepatocellular carcinoma [ICD-11: 2C12.02] | |||
| Target Regulation | Down regulation | |||
| In-vivo Model | Male BALB/c nude mice (aged 4-6 weeks; n = 5/group) were obtained from Vital River Laboratory Animal Technology (Beijing, China). MHCC97H or Huh7 cells (2 × 106 cells/mouse) stably transfected with lentivirus containing different plasmids in 100 μL DMEM were subcutaneously or orthotopically implanted into the nude mice. | |||
Lung cancer [ICD-11: 2C25]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [85] | |||
| Responsed Disease | Lung cancer [ICD-11: 2C25] | |||
| Target Regulation | Down regulation | |||
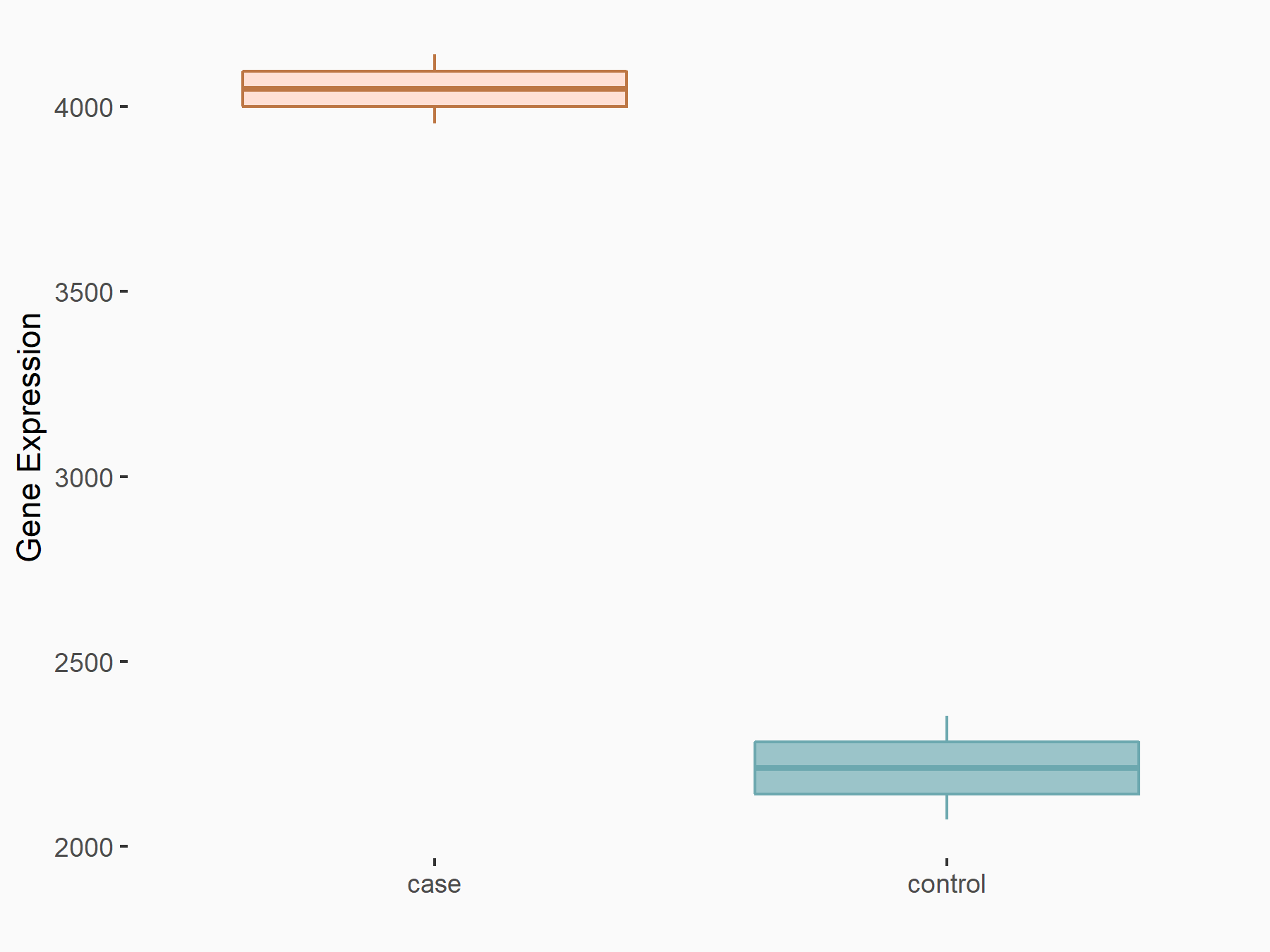
Peroxisome proliferator-activated receptor alpha (PPARalpha/PPARA)
| Representative RNA-seq result indicating the expression of this target gene regulated by YTHDF2 | ||
| Cell Line | GSC11 cell line | Homo sapiens |
|
Treatment: siYTHDF2 GSC11 cells
Control: siControl GSC11 cells
|
GSE142825 | |
| Regulation |
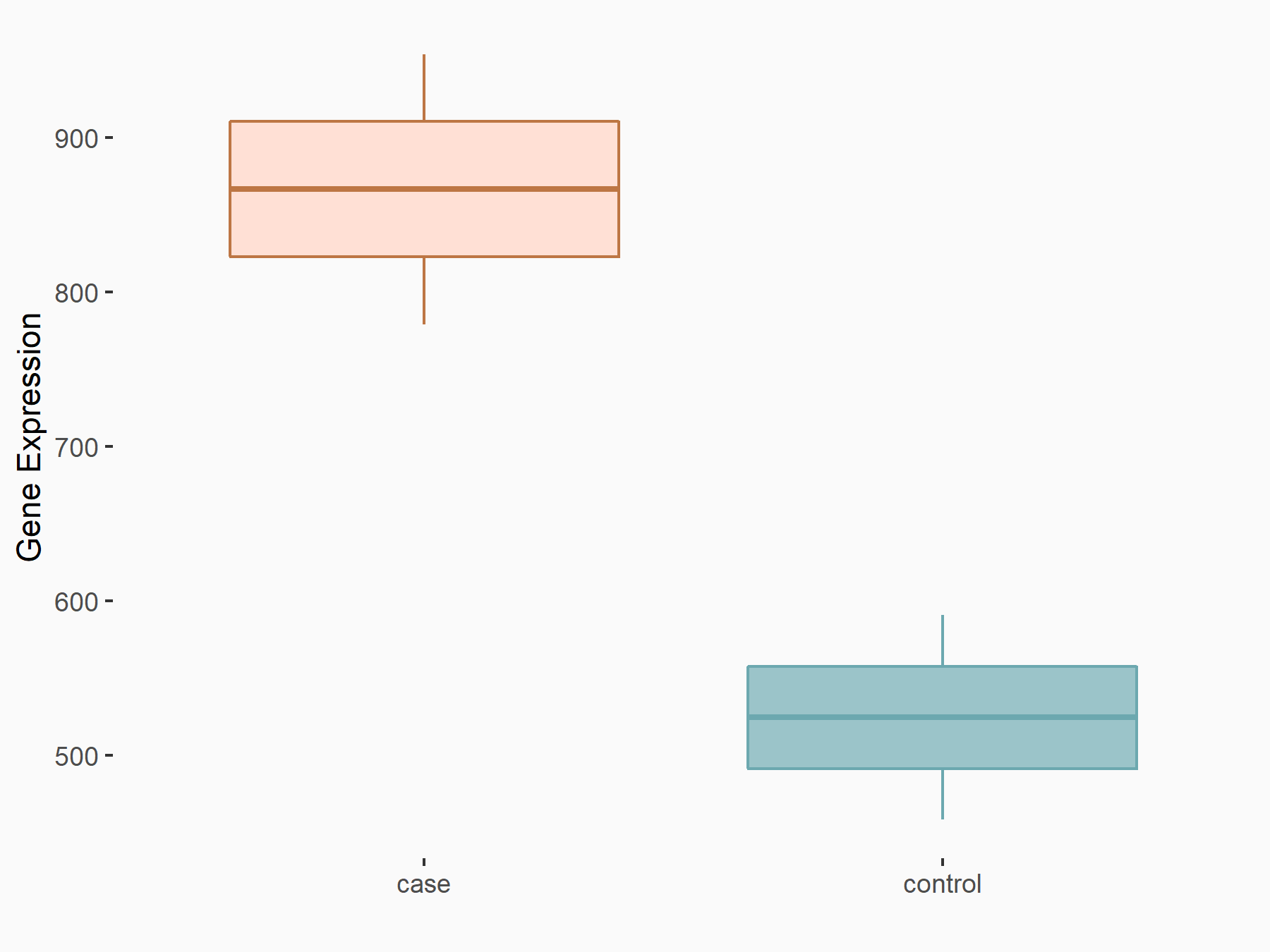 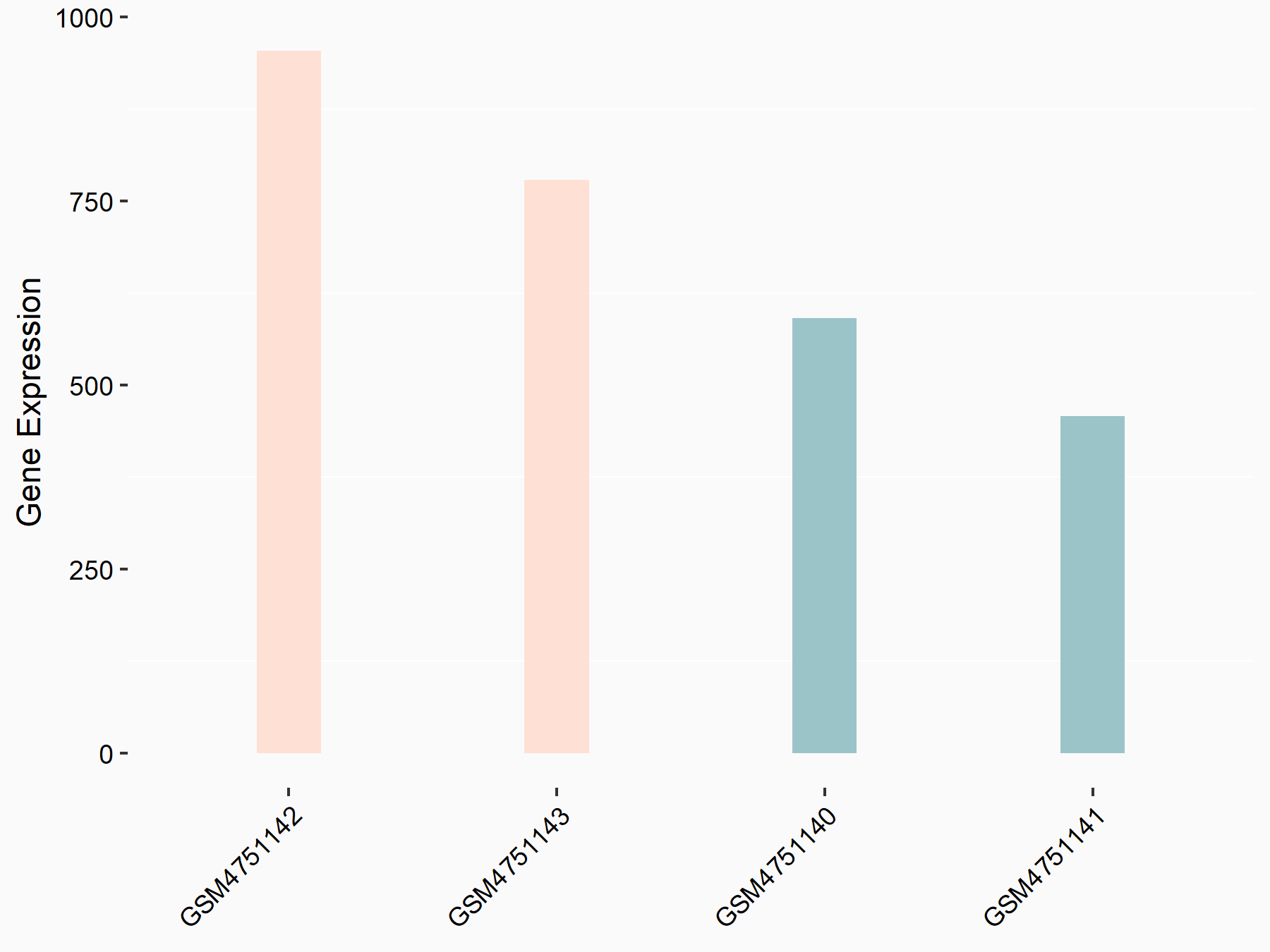 |
logFC: 7.24E-01 p-value: 8.81E-04 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and YTHDF2 | ||
| Cell Line | Hela | Homo sapiens |
| Regulation | logFC: 1.45E+00 | GSE49339 |
Metabolic disorders [ICD-11: 5D2Z]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [13] | |||
| Responsed Disease | Metabolic disorders [ICD-11: 5D2Z] | |||
| Target Regulation | Down regulation | |||
| Pathway Response | PPAR signaling pathway | hsa03320 | ||
| Adipocytokine signaling pathway | hsa04920 | |||
| Cell Process | Llipid metabolism | |||
In-vitro Model |
Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 |
| Hepa 1-6 | Hepatocellular carcinoma of the mouse | Mus musculus | CVCL_0327 | |
| In-vivo Model | Liver-specific Bmal1f/f-AlbCre-knockout mice were purchased from Jackson Laboratory. C57BI/6J or Bmal1f/f-AlbCre-knockout male mice were maintained under a 12 hr light/12 hr dark (LD) cycle (ZT0 = 6 AM) and fed ad libitum with normal rodent chow (2018 Global 18% Protein diet, Envigo) and water. At 10-14 weeks of age, 10 male mice per group were sacrificed via CO2 asphyxiation at Zeitgeber Time (ZT) 0,2,6,10,12,14,18,22. In order to induce high levels of ROS in the liver, WT male mice were fasted 12 h and followed by intraperitoneal injection with 300 mg/kg APAP dissolved in PBS and re-fed. | |||
| Response Summary | PPaRalpha to mediate its mRNA stability to regulate lipid metabolism. Hepatic deletion of Bmal1 increases m6A mRNA methylation, particularly of Peroxisome proliferator-activated receptor alpha (PPARalpha/PPARA). Inhibition of m6A methylation via knockdown of m6A methyltransferase METTL3 decreases PPaR-Alpha m6A abundance and increases PPaRalpha mRNA lifetime and expression, reducing lipid accumulation in cells in vitro. YTHDF2 binds to PPaRalpha to mediate its mRNA stability to regulate lipid metabolism. Transcriptional regulation of circadian rhythms is essential for lipid metabolic homeostasis, disruptions of which can lead to metabolic diseases. | |||
Platelet-derived growth factor C (PDGFC)
| Representative RNA-seq result indicating the expression of this target gene regulated by YTHDF2 | ||
| Cell Line | GSC11 cell line | Homo sapiens |
|
Treatment: siYTHDF2 GSC11 cells
Control: siControl GSC11 cells
|
GSE142825 | |
| Regulation |
 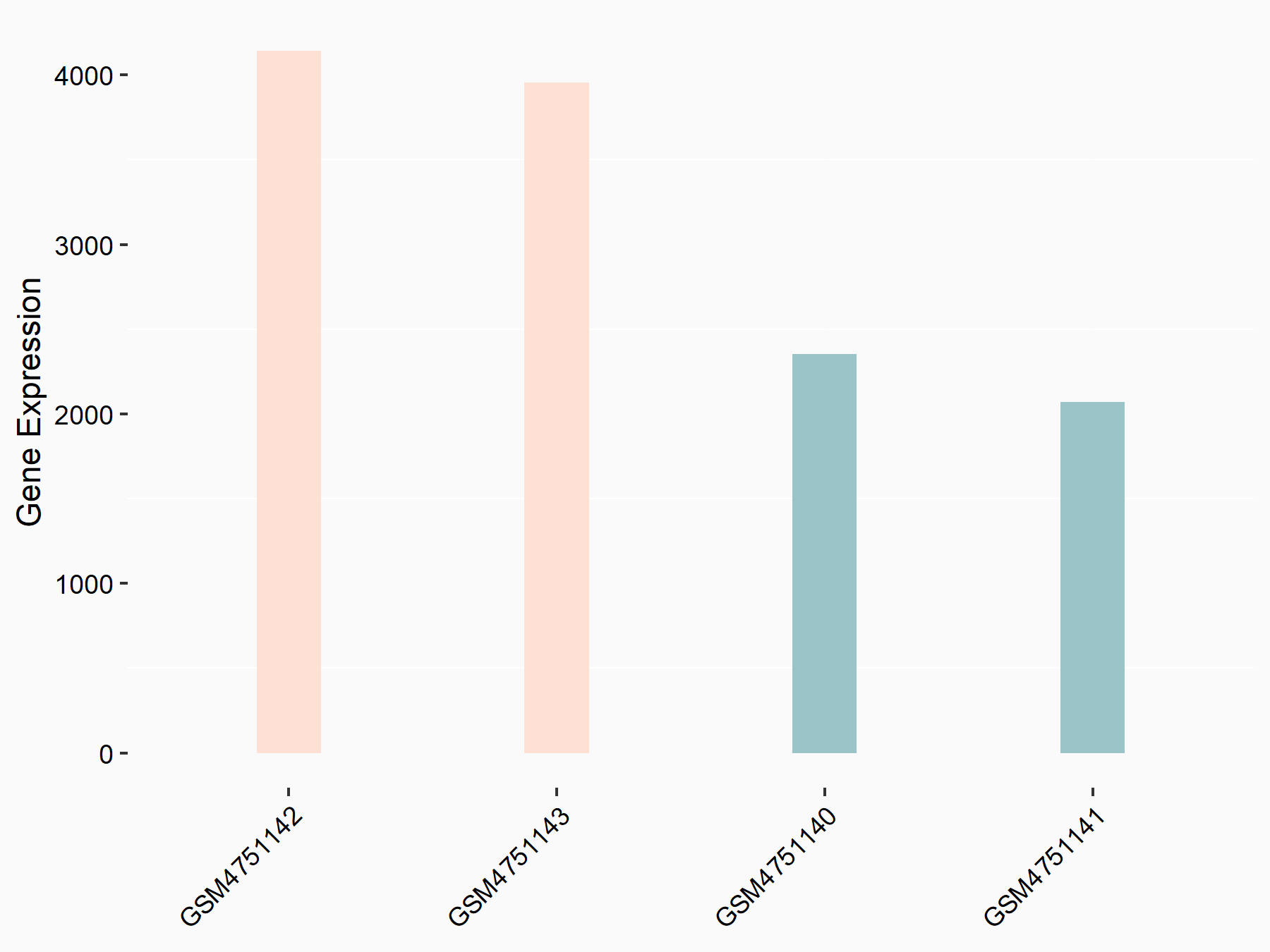 |
logFC: 8.71E-01 p-value: 1.24E-12 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and YTHDF2 | ||
| Cell Line | Hela | Homo sapiens |
| Regulation | logFC: 1.41E+00 | GSE49339 |
Pancreatic cancer [ICD-11: 2C10]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [14] | |||
| Responsed Disease | Pancreatic cancer [ICD-11: 2C10] | |||
| Target Regulation | Down regulation | |||
| Pathway Response | PI3K-Akt signaling pathway | hsa04151 | ||
In-vitro Model |
SW1990 | Pancreatic adenocarcinoma | Homo sapiens | CVCL_1723 |
| PANC-1 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0480 | |
| MIA PaCa-2 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0428 | |
| HPDE | Normal | Homo sapiens | CVCL_4376 | |
| CFPAC-1 | Cystic fibrosis | Homo sapiens | CVCL_1119 | |
| Capan-1 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0237 | |
| BxPC-3 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0186 | |
| AsPC-1 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0152 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| In-vivo Model | The right flanks of mice were injected subcutaneously with 2 × 106 MiaPaCa-2 cells stably expressing shFTO and a scrambled shRNA in 100 uL PBS. Tumors were measured using an external caliper once per week, and tumor volume was calculated with the formula: (length × width2)/2. | |||
| Response Summary | FTO downregulation leads to increased m6A modifications in the 3' UTR of Platelet-derived growth factor C (PDGFC) and then modulates the degradation of its transcriptional level in an m6A-YTHDF2-dependent manner, highlighting a potential therapeutic target for PDAC treatment and prognostic prediction. | |||
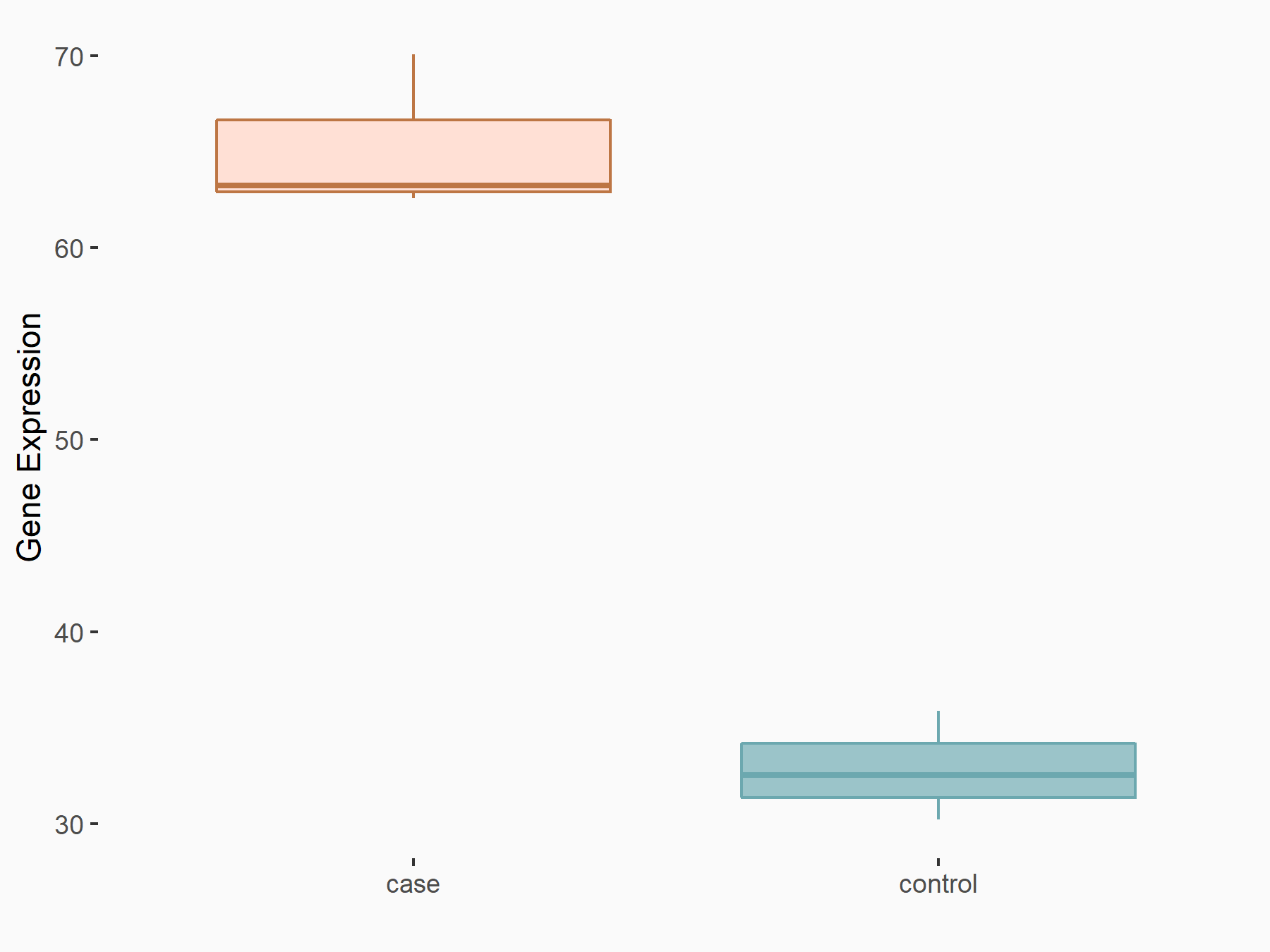
Protein FAM83D (FAM83D)
| Representative RNA-seq result indicating the expression of this target gene regulated by YTHDF2 | ||
| Cell Line | Human umbilical cord blood CD34+ cells | Homo sapiens |
|
Treatment: YTHDF2 knockdown UCB CD34+ cells
Control: Wild type UCB CD34+ cells
|
GSE107956 | |
| Regulation |
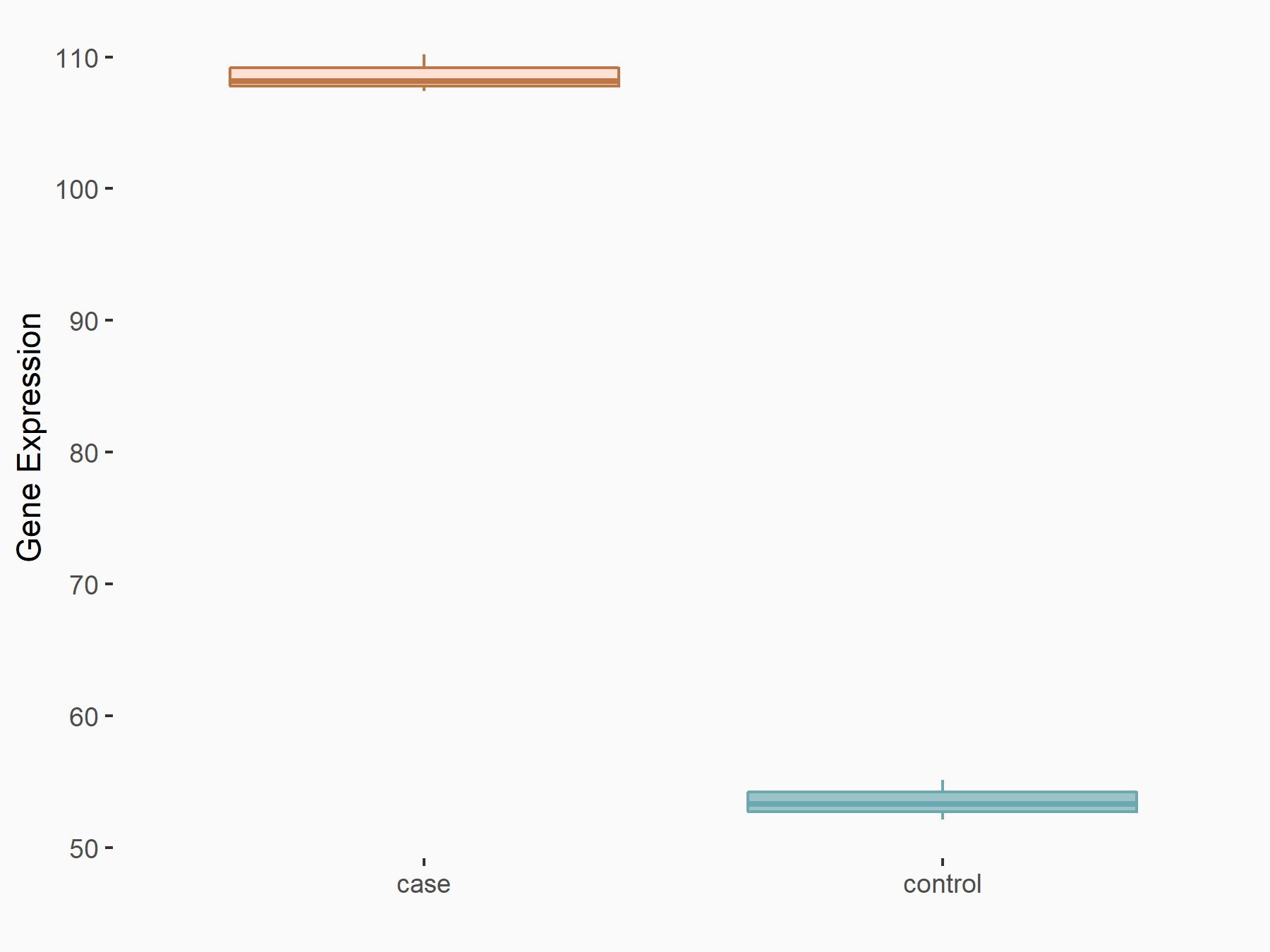 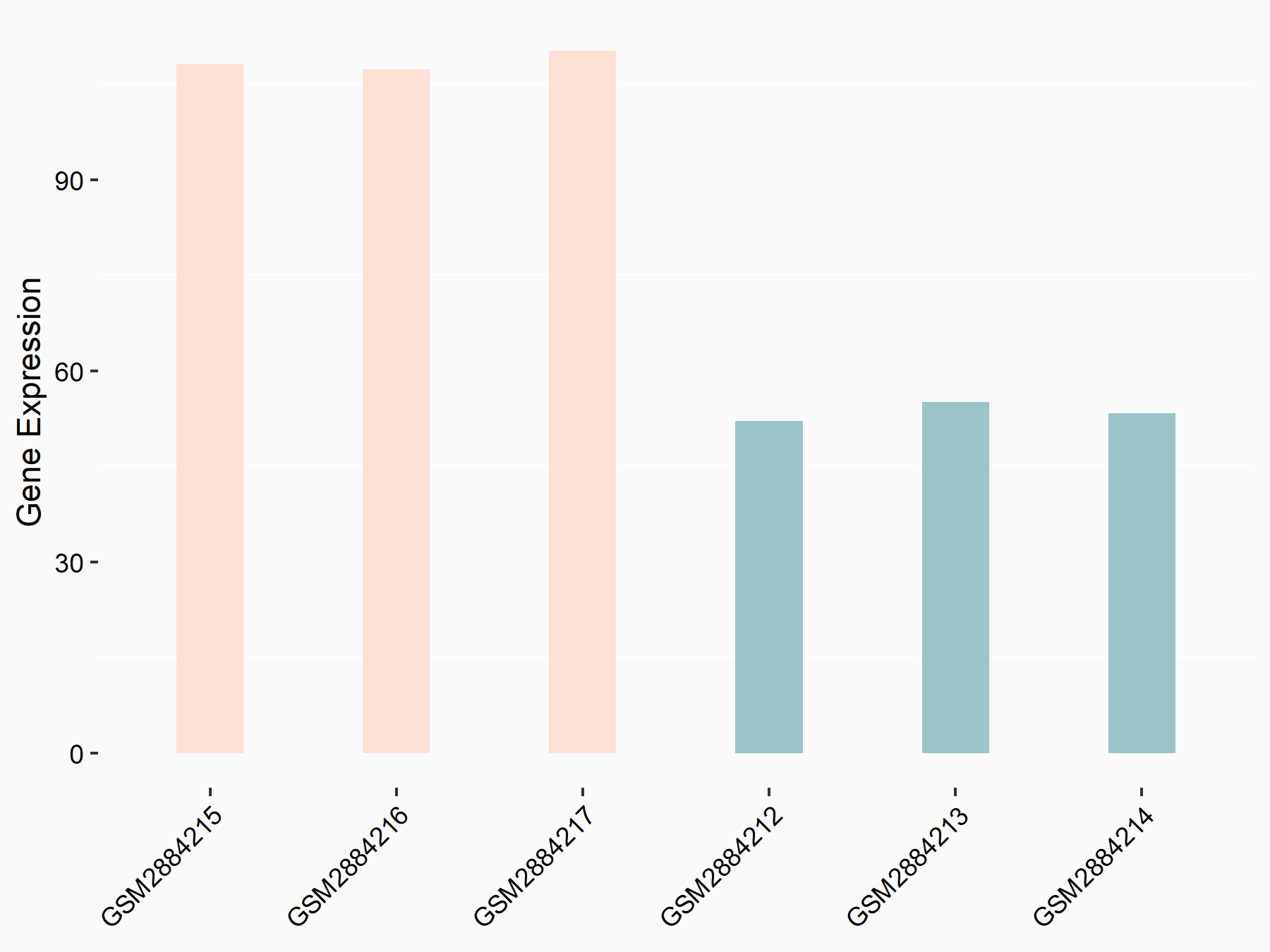 |
logFC: 1.01E+00 p-value: 7.31E-07 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and YTHDF2 | ||
| Cell Line | Hela | Homo sapiens |
| Regulation | logFC: 1.69E+00 | GSE49339 |
Lung cancer [ICD-11: 2C25]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [9] | |||
| Responsed Disease | Lung cancer [ICD-11: 2C25] | |||
| Target Regulation | Down regulation | |||
| Pathway Response | mRNA surveillance pathway | hsa03015 | ||
| Cell Process | Epithelial-mesenchymal transition | |||
In-vitro Model |
NCI-H1299 | Lung large cell carcinoma | Homo sapiens | CVCL_0060 |
| A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 | |
| Response Summary | YTHDF2 inhibits the migration and invasion of lung adenocarcinoma cells by regulating the Protein FAM83D (FAM83D)-TGFbeta1-pSMAD2/3 pathway, which will play an important role in lung cancer metastasis. | |||
Suppressor of cytokine signaling 2 (SOCS2)
| Representative RNA-seq result indicating the expression of this target gene regulated by YTHDF2 | ||
| Cell Line | Human umbilical cord blood CD34+ cells | Homo sapiens |
|
Treatment: YTHDF2 knockdown UCB CD34+ cells
Control: Wild type UCB CD34+ cells
|
GSE107956 | |
| Regulation |
 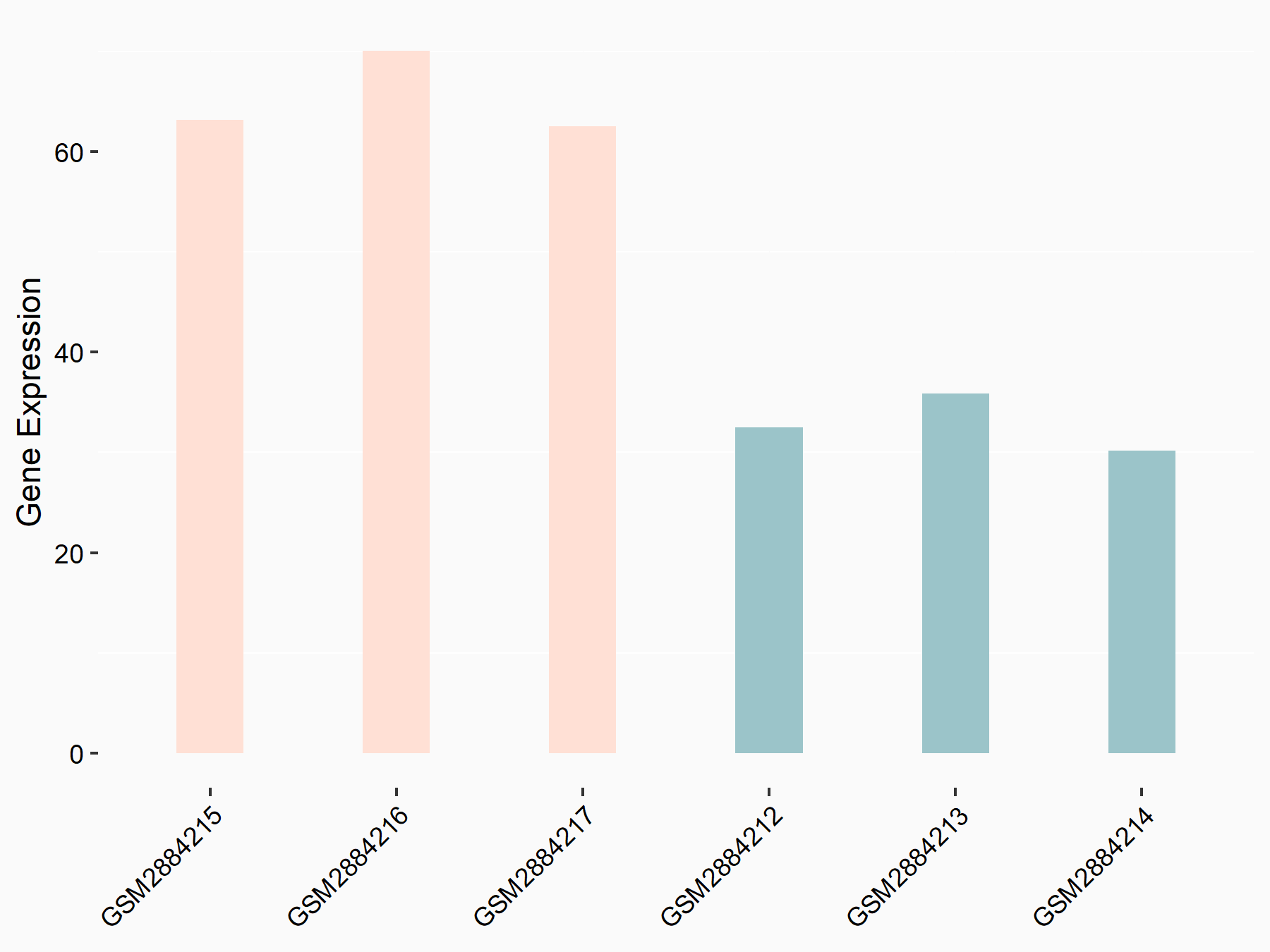 |
logFC: 9.71E-01 p-value: 1.71E-04 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and YTHDF2 | ||
| Cell Line | Hela | Homo sapiens |
| Regulation | logFC: 1.81E+00 | GSE49339 |
Liver cancer [ICD-11: 2C12]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [15] | |||
| Responsed Disease | Liver cancer [ICD-11: 2C12] | |||
| Target Regulation | Down regulation | |||
| Cell Process | Cells proliferation | |||
| Cells migration | ||||
| Cells invasion | ||||
| RNA degradation (hsa03018) | ||||
In-vitro Model |
Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 |
| Huh-7 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_0336 | |
| MHCC97-L | Adult hepatocellular carcinoma | Homo sapiens | CVCL_4973 | |
| In-vivo Model | For the subcutaneous implantation model, 2 × 106 METTL3 stable knockdown Huh-7 cells or METTL3 overexpression MHCC97L cells were injected subcutaneously into BABL/cAnN-nude mice. For orthotopic implantation, wild-type and METTL3 knockout Huh-7 cells were luciferase labelled, and 2 × 106 cells were then injected orthotopically into the left liver lobe of nude mice. | |||
| Response Summary | METTL3 is frequently up-regulated in human HCC and contributes to HCC progression. METTL3 represses Suppressor of cytokine signaling 2 (SOCS2) expression in HCC through an m6A-YTHDF2-dependent mechanism. | |||
Suppressor of cytokine signaling 3 (SOCS3)
| Representative RNA-seq result indicating the expression of this target gene regulated by YTHDF2 | ||
| Cell Line | mouse embryonic stem cells | Mus musculus |
|
Treatment: shYthdf2 embryonic stem cells
Control: shLuc embryonic stem cells
|
GSE156437 | |
| Regulation |
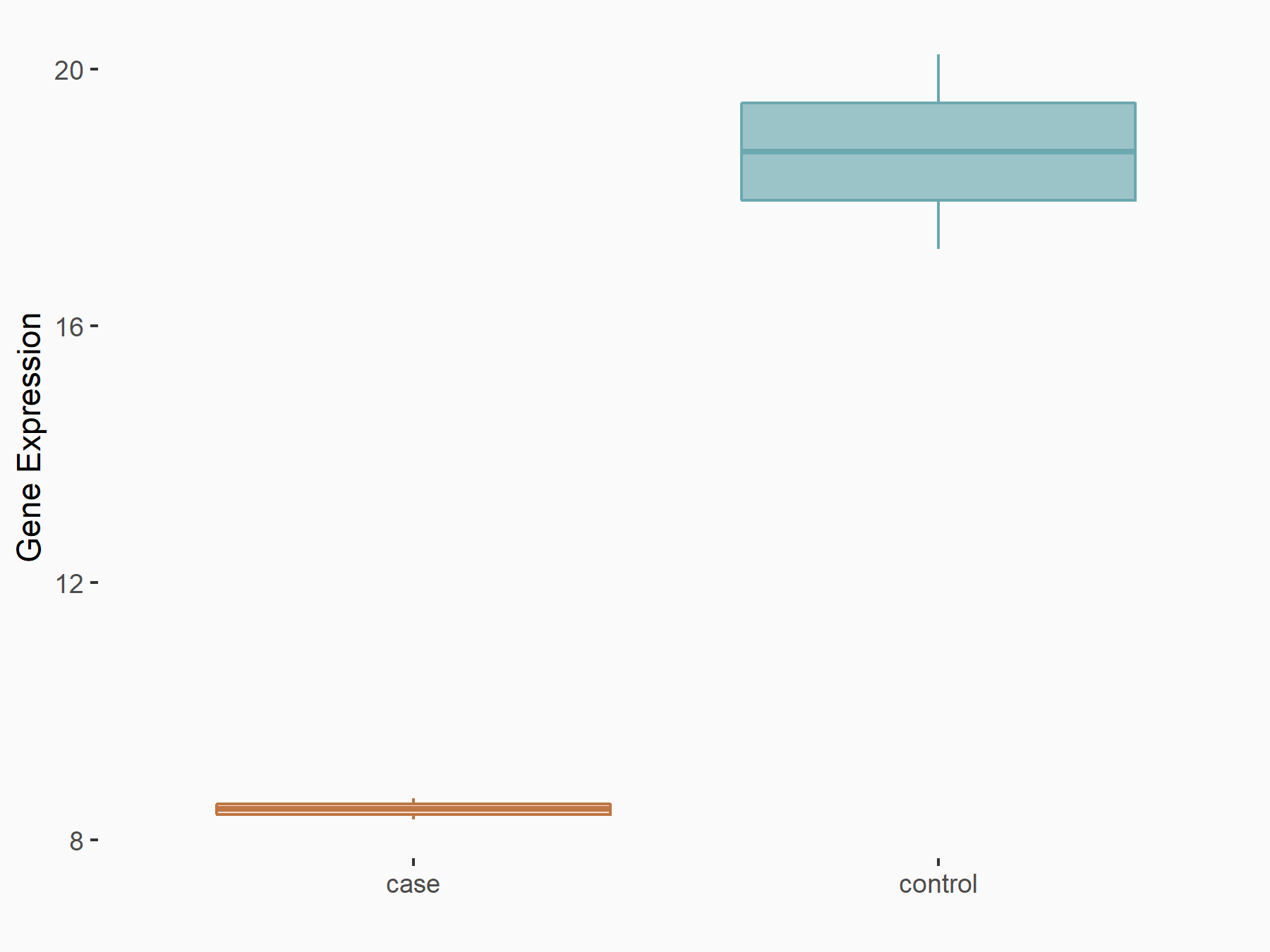  |
logFC: -1.05E+00 p-value: 3.86E-03 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and YTHDF2 | ||
| Cell Line | Hela | Homo sapiens |
| Regulation | logFC: 1.46E+00 | GSE49339 |
Osteosarcoma [ICD-11: 2B51]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [16] | |||
| Responsed Disease | Osteosarcoma [ICD-11: 2B51] | |||
| Target Regulation | Down regulation | |||
| Pathway Response | JAK-STAT signaling pathway | hsa04630 | ||
In-vitro Model |
U2OS | Osteosarcoma | Homo sapiens | CVCL_0042 |
| OS3 [Human osteosarcoma] | Osteosarcoma | Homo sapiens | CVCL_F866 | |
| OS2 [Human osteosarcoma] | Osteosarcoma | Homo sapiens | CVCL_F865 | |
| OS1 [Human osteosarcoma] | Osteosarcoma | Homo sapiens | CVCL_F864 | |
| KHOS/NP | Osteosarcoma | Homo sapiens | CVCL_2546 | |
| Response Summary | ALKBH5 inactivated STAT3 pathway by increasing Suppressor of cytokine signaling 3 (SOCS3) expression via an m6A-YTHDF2-dependent manner.Reducing m6A mRNA levels in human osteosarcoma cells through ALKBH5 up-regulation lead to cell proliferation inhibition, cell apoptosis and cycle arrest. | |||
Human skin lesions [ICD-11: ME60]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [110] | |||
| Responsed Disease | Human skin lesions [ICD-11: ME60] | |||
| Responsed Drug | Arsenite | Phase 2 | ||
| Target Regulation | Down regulation | |||
| Pathway Response | JAK-STAT signaling pathway | hsa04630 | ||
In-vitro Model |
HaCaT | Normal | Homo sapiens | CVCL_0038 |
Thrombospondin-1 (THBS1)
| Representative RNA-seq result indicating the expression of this target gene regulated by YTHDF2 | ||
| Cell Line | Human umbilical cord blood CD34+ cells | Homo sapiens |
|
Treatment: YTHDF2 knockdown UCB CD34+ cells
Control: Wild type UCB CD34+ cells
|
GSE107956 | |
| Regulation |
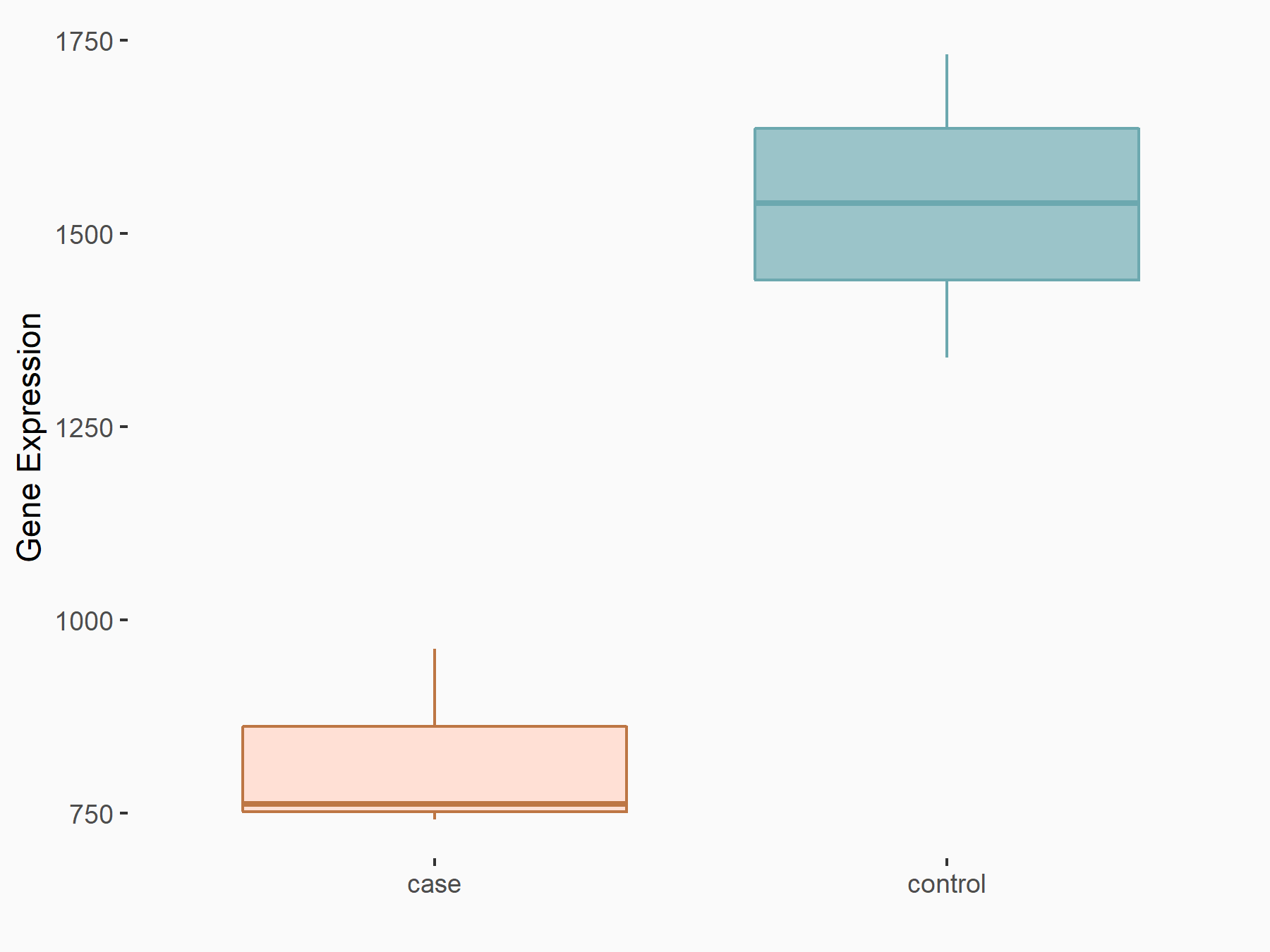 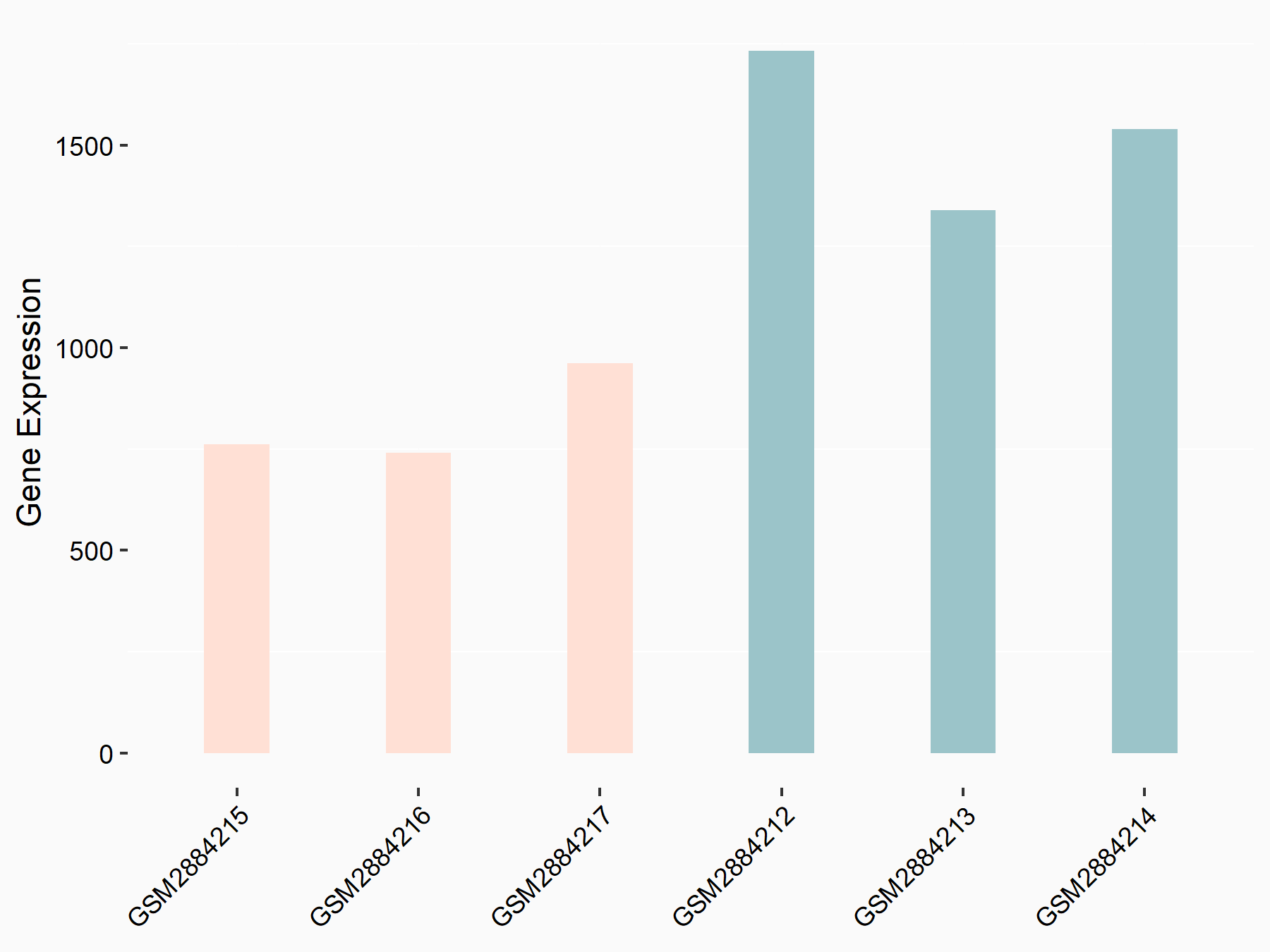 |
logFC: -9.06E-01 p-value: 2.97E-03 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and YTHDF2 | ||
| Cell Line | Hela | Homo sapiens |
| Regulation | logFC: 2.48E+00 | GSE49339 |
Prostate cancer [ICD-11: 2C82]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [17] | |||
| Responsed Disease | Prostate cancer [ICD-11: 2C82] | |||
| Target Regulation | Down regulation | |||
| Pathway Response | RNA degradation | hsa03018 | ||
| Cell Process | Cell proliferation | |||
In-vitro Model |
PC-3 | Prostate carcinoma | Homo sapiens | CVCL_0035 |
| DU145 | Prostate carcinoma | Homo sapiens | CVCL_0105 | |
| In-vivo Model | Stably transfected shMETTL14 and shNC DU145 cells (5×106 cells) suspended in a mixture of 100uL PBS were subcutaneously injected into the right flank of male nude BALB/C mice (6-8 weeks old) to induce tumor formation. | |||
| Response Summary | In prostate cancer, METTL14 downregulated Thrombospondin-1 (THBS1) expression in an m6A-dependent manner, which resulted in the recruitment of YTHDF2 to recognize and degrade Thrombospondin 1 (THBS1) mRNA. | |||
Transcription factor HIVEP2 (HIVEP2)
| Representative RNA-seq result indicating the expression of this target gene regulated by YTHDF2 | ||
| Cell Line | GSC11 cell line | Homo sapiens |
|
Treatment: siYTHDF2 GSC11 cells
Control: siControl GSC11 cells
|
GSE142825 | |
| Regulation |
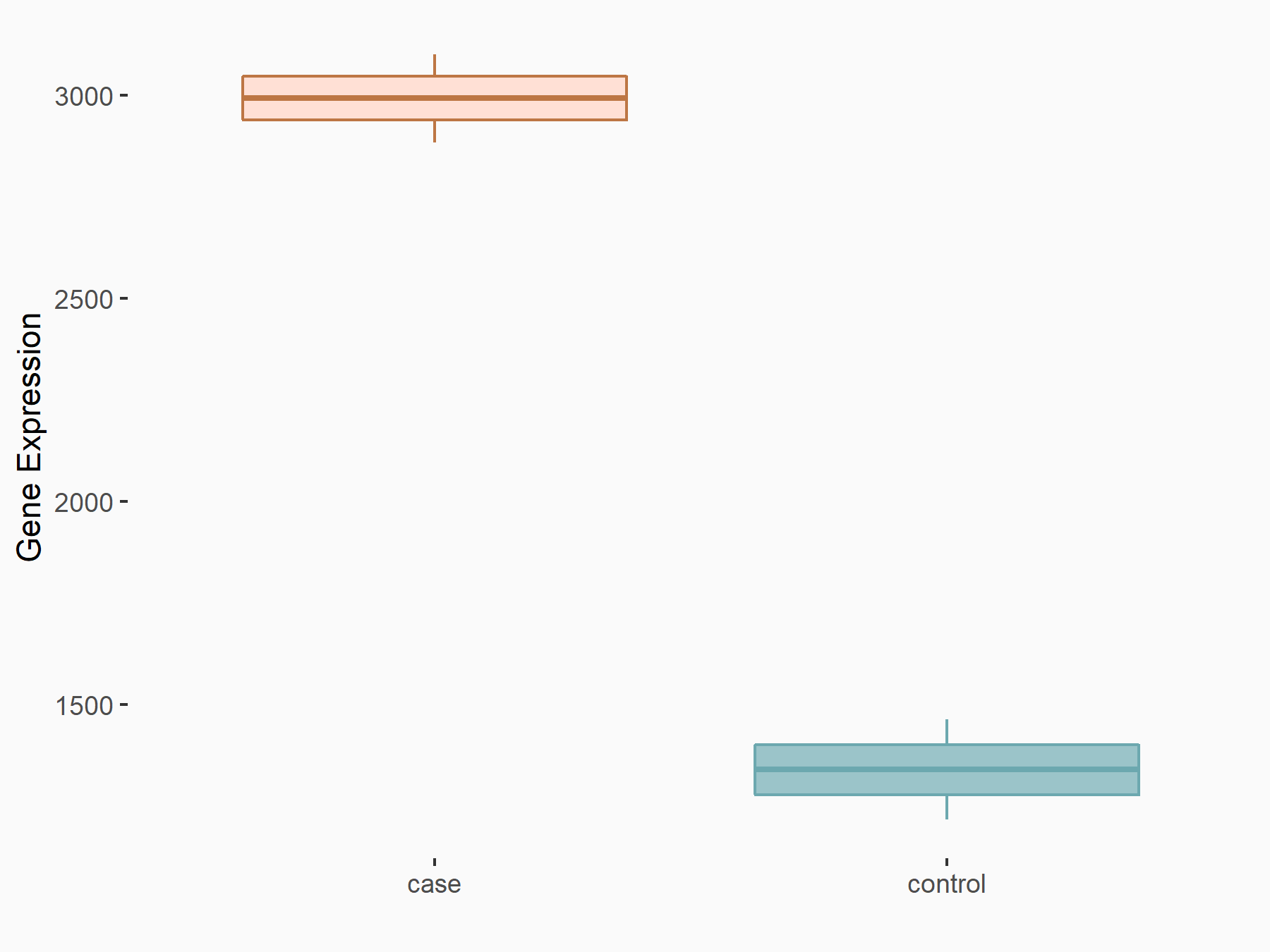  |
logFC: 1.16E+00 p-value: 1.15E-15 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and YTHDF2 | ||
| Cell Line | Hela | Homo sapiens |
| Regulation | logFC: 1.70E+00 | GSE49339 |
Brain cancer [ICD-11: 2A00]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [18] | |||
| Responsed Disease | Glioblastoma [ICD-11: 2A00.00] | |||
| Target Regulation | Down regulation | |||
| Pathway Response | mRNA surveillance pathway | hsa03015), RNA degradation | ||
| Cell Process | RNA stability | |||
In-vitro Model |
U-87MG ATCC | Glioblastoma | Homo sapiens | CVCL_0022 |
| U-251MG | Astrocytoma | Homo sapiens | CVCL_0021 | |
| T98G | Glioblastoma | Homo sapiens | CVCL_0556 | |
| SW1783 | Anaplastic astrocytoma | Homo sapiens | CVCL_1722 | |
| LN-229 | Glioblastoma | Homo sapiens | CVCL_0393 | |
| Hs 683 | Oligodendroglioma | Homo sapiens | CVCL_0844 | |
| GSC7-2 (GSC7-2 were obtained from fresh surgical specimens of human primary and recurrent glioma) | ||||
| GSC6-27 (GSC6-27 were obtained from fresh surgical specimens of human primary and recurrent glioma) | ||||
| GSC23 | Glioblastoma | Homo sapiens | CVCL_DR59 | |
| GSC20 (GSC20 were obtained from fresh surgical specimens of human primary and recurrent glioma) | ||||
| GSC17 | Glioblastoma | Homo sapiens | CVCL_DR57 | |
| GSC11 | Glioblastoma | Homo sapiens | CVCL_DR55 | |
| In-vivo Model | For the studies of investigating mice survival, mice were intracranially injected with 10,000 GSC11, 10,000 GSC7-2, or 500,000 LN229 cells. | |||
| Response Summary | YTHDF2 facilitates m6A-dependent mRNA decay of LXRA and Transcription factor HIVEP2 (HIVEP2), which impacts the glioma patient survival. YTHDF2 promotes tumorigenesis of GBM cells, largely through the downregulation of LXRA and HIVEP2. | |||
Transcription factor SOX-4 (SOX4)
| Representative RNA-seq result indicating the expression of this target gene regulated by YTHDF2 | ||
| Cell Line | GSC11 cell line | Homo sapiens |
|
Treatment: siYTHDF2 GSC11 cells
Control: siControl GSC11 cells
|
GSE142825 | |
| Regulation |
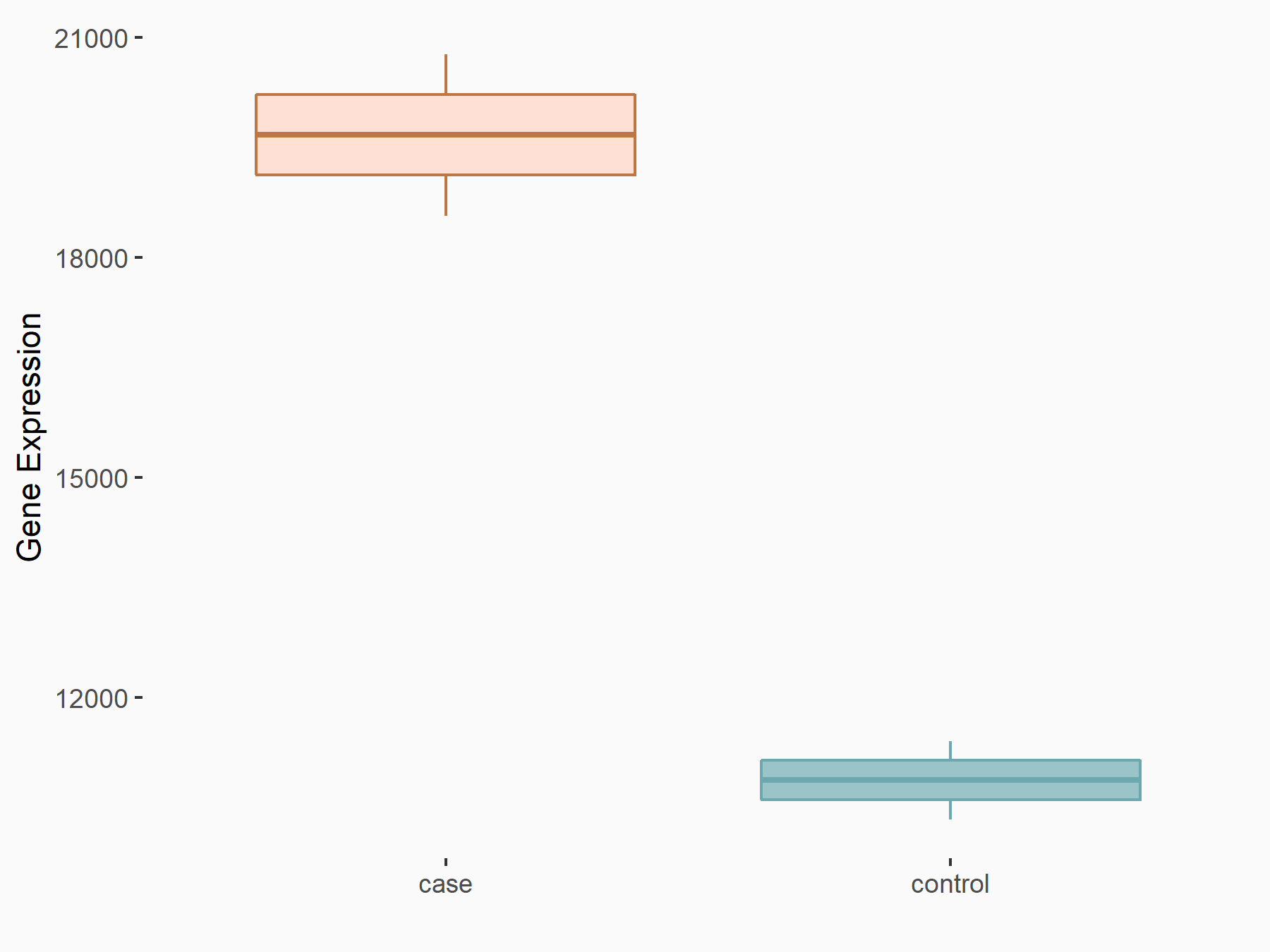  |
logFC: 8.56E-01 p-value: 4.96E-15 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and YTHDF2 | ||
| Cell Line | Hela | Homo sapiens |
| Regulation | logFC: 1.82E+00 | GSE49339 |
Endometrial cancer [ICD-11: 2C76]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [19] | |||
| Responsed Disease | Endometrial cancer [ICD-11: 2C76] | |||
| Target Regulation | Up regulation | |||
| Cell Process | Cell proliferation | |||
| Cell apoptosis | ||||
In-vitro Model |
HEC-1-B | Endometrial adenocarcinoma | Homo sapiens | CVCL_0294 |
| Response Summary | YTHDF2-mediated LncRNA FENDRR degradation promotes cell proliferation by elevating Transcription factor SOX-4 (SOX4) expression in endometrioid endometrial carcinoma. | |||
Colorectal cancer [ICD-11: 2B91]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [113] | |||
| Responsed Disease | Colorectal cancer [ICD-11: 2B91] | |||
| Target Regulation | Down regulation | |||
In-vitro Model |
NCM460 | Normal | Homo sapiens | CVCL_0460 |
| HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 | |
| HCT 8 | Colon adenocarcinoma | Homo sapiens | CVCL_2478 | |
| SW620 | Colon adenocarcinoma | Homo sapiens | CVCL_0547 | |
| SW480 | Colon adenocarcinoma | Homo sapiens | CVCL_0546 | |
| HT29 | Colon cancer | Mus musculus | CVCL_A8EZ | |
| DLD-1 | Colon adenocarcinoma | Homo sapiens | CVCL_0248 | |
| In-vivo Model | All animal experiments were approved by the animal care Committee of Nanjing First Hospital, Nanjing Medial University (acceptance No. SYXK 20160006). 2 × 106 transfected HCT116 cells in 0.2 ml PBS were injected into the tail vein of nude mice which were randomly divided into nine groups (eight mice per group). After 3 months of injection, mice were sacrificed, and their lungs were removed and stained by Hematoxylin and Eosin (HE) Staining. | |||
Transcriptional coactivator YAP1 (YAP1)
| Representative RNA-seq result indicating the expression of this target gene regulated by YTHDF2 | ||
| Cell Line | GSC11 cell line | Homo sapiens |
|
Treatment: siYTHDF2 GSC11 cells
Control: siControl GSC11 cells
|
GSE142825 | |
| Regulation |
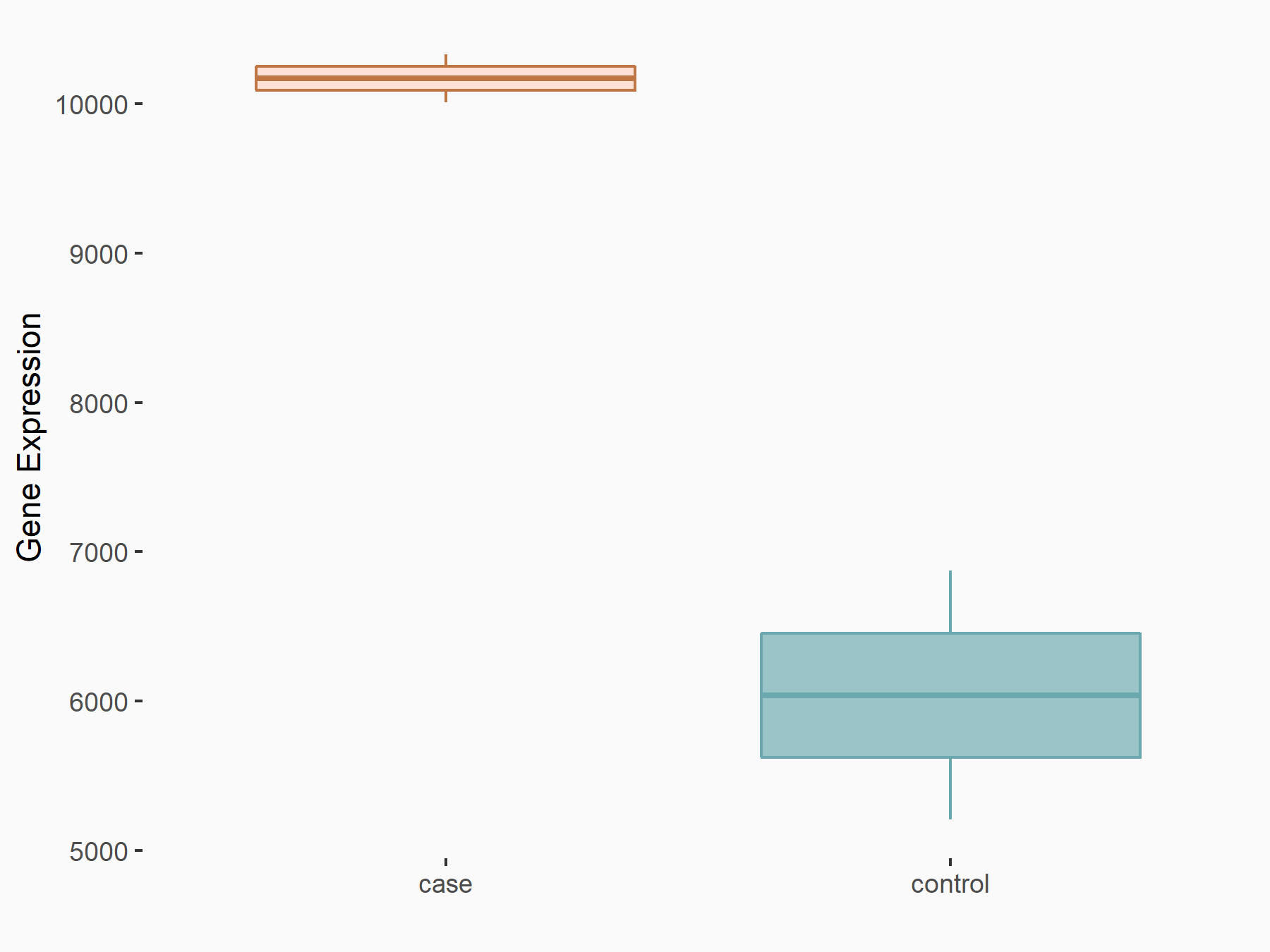  |
logFC: 7.52E-01 p-value: 2.51E-07 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and YTHDF2 | ||
| Cell Line | Hela | Homo sapiens |
| Regulation | logFC: 1.34E+00 | GSE49339 |
Osteosarcoma [ICD-11: 2B51]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [20] | |||
| Responsed Disease | Osteosarcoma [ICD-11: 2B51] | |||
| Cell Process | Cell growth | |||
| Cell migration | ||||
| Cell invasion | ||||
| Cell apoptosis | ||||
In-vitro Model |
U2OS | Osteosarcoma | Homo sapiens | CVCL_0042 |
| In-vivo Model | Three-week-old BABL/c female nude mice were randomized into three groups. 5 × 106 143B cells were subcutaneously injected in mice, and the tumor volume was assessed every 2 weeks. Eight weeks after injection, the animals were killed. The xenograft tumors were harvested and the tumor volumes were calculated by the standard formula: length × width2/2. | |||
| Response Summary | ALKBH5 is an anti-tumor factor or a pro-apoptotic factor, acting at least partially by suppressing Transcriptional coactivator YAP1 (YAP1) expression through dual mechanisms with direct m6A methylation of YAP and indirect downregulation of YAP level due to methylation of pre-miR-181b-1. Further results revealed that m6A methylated pre-miR-181b-1 was subsequently recognized by m6A-binding protein YTHDF2 to mediate RNA degradation. However, methylated YAP transcripts were recognized by YTHDF1 to promote its translation. ALKBH5 overexpression was considered a new approach of replacement therapy for osteosarcoma treatment. | |||
Pancreatic cancer [ICD-11: 2C10]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [21] | |||
| Responsed Disease | Pancreatic cancer [ICD-11: 2C10] | |||
| Target Regulation | Down regulation | |||
| Pathway Response | Hippo signaling pathway | hsa04390 | ||
| Cell Process | Cells proliferation | |||
| Cells migration | ||||
| Cells invasion | ||||
| Epithelial-mesenchymal transition | ||||
In-vitro Model |
BxPC-3 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0186 |
| PaTu 8988s | Pancreatic adenocarcinoma | Homo sapiens | CVCL_1846 | |
| SW1990 | Pancreatic adenocarcinoma | Homo sapiens | CVCL_1723 | |
| Response Summary | YTHDF2 knockdown significantly increases the total YAP expression, but inhibits TGF-beta/Smad signaling, indicating that YTHDF2 regulates EMT probably via Transcriptional coactivator YAP1 (YAP1) signaling. YTHDF2 is a new predictive biomarker of development of pancreatic cancer. | |||
Triple-negative breast cancer [ICD-11: 2C6Z]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [114] | |||
| Responsed Disease | Triple-negative breast cancer [ICD-11: 2C6Z] | |||
| Target Regulation | Down regulation | |||
Brain and muscle ARNT-like 1 (Bmal1/ARNTL)
| Representative RNA-seq result indicating the expression of this target gene regulated by YTHDF2 | ||
| Cell Line | B18-hi B cell line | Mus musculus |
|
Treatment: YTHDF2 knockout B18-hi B cells
Control: Wild type B18-hi B cells
|
GSE189819 | |
| Regulation |
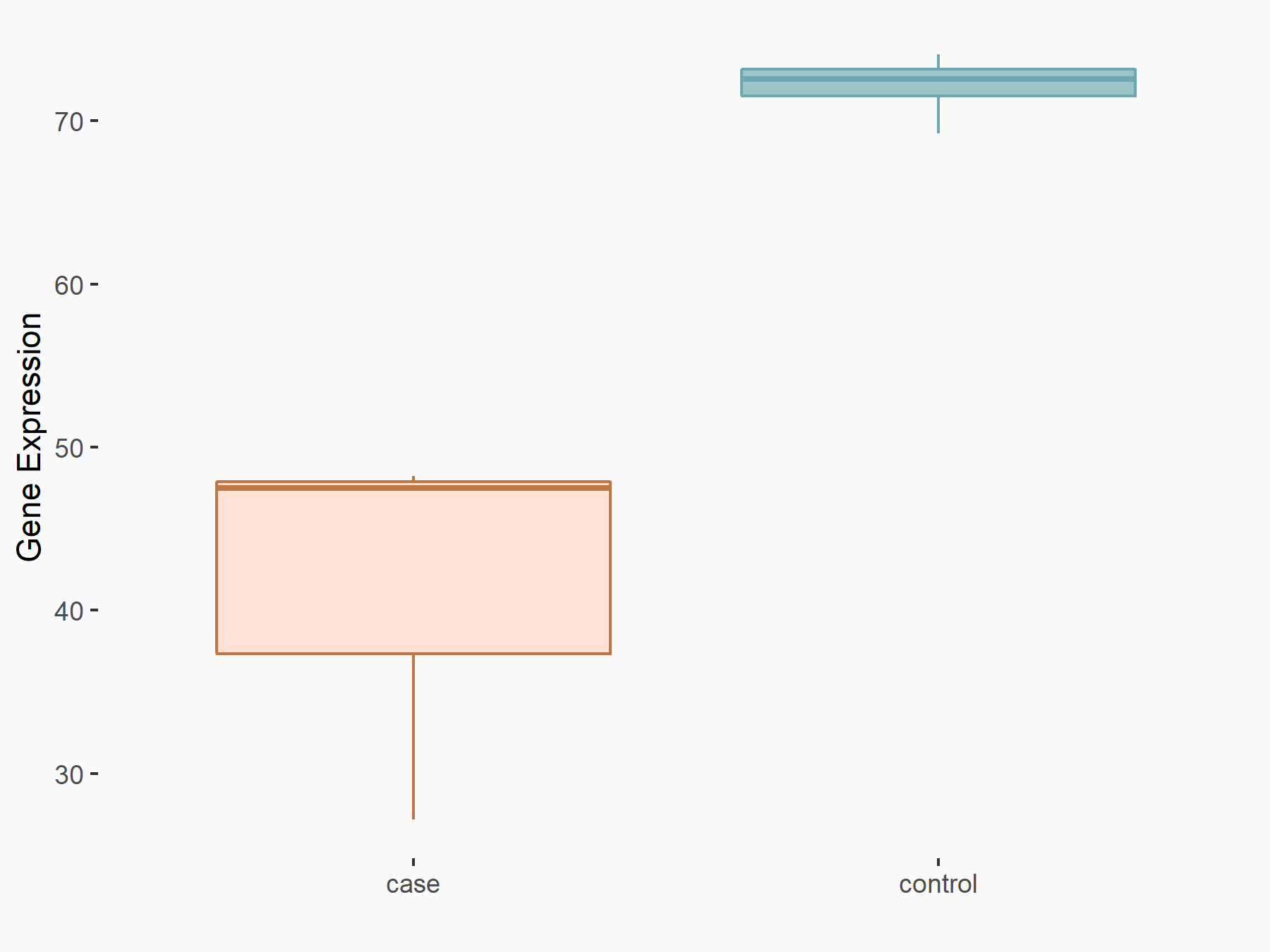  |
logFC: -7.97E-01 p-value: 5.55E-03 |
| More Results | Click to View More RNA-seq Results | |
Metabolic disorders [ICD-11: 5D2Z]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [13] | |||
| Responsed Disease | Metabolic disorders [ICD-11: 5D2Z] | |||
| Target Regulation | Down regulation | |||
| Pathway Response | PPAR signaling pathway | hsa03320 | ||
| Adipocytokine signaling pathway | hsa04920 | |||
| Cell Process | Llipid metabolism | |||
In-vitro Model |
Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 |
| Hepa 1-6 | Hepatocellular carcinoma of the mouse | Mus musculus | CVCL_0327 | |
| In-vivo Model | Liver-specific Bmal1f/f-AlbCre-knockout mice were purchased from Jackson Laboratory. C57BI/6J or Bmal1f/f-AlbCre-knockout male mice were maintained under a 12 hr light/12 hr dark (LD) cycle (ZT0 = 6 AM) and fed ad libitum with normal rodent chow (2018 Global 18% Protein diet, Envigo) and water. At 10-14 weeks of age, 10 male mice per group were sacrificed via CO2 asphyxiation at Zeitgeber Time (ZT) 0,2,6,10,12,14,18,22. In order to induce high levels of ROS in the liver, WT male mice were fasted 12 h and followed by intraperitoneal injection with 300 mg/kg APAP dissolved in PBS and re-fed. | |||
| Response Summary | PPaRalpha to mediate its mRNA stability to regulate lipid metabolism. Hepatic deletion of Brain and muscle ARNT-like 1 (Bmal1/ARNTL) increases m6A mRNA methylation, particularly of PPaRalpha. Inhibition of m6A methylation via knockdown of m6A methyltransferase METTL3 decreases PPaR-Alpha m6A abundance and increases PPaRalpha mRNA lifetime and expression, reducing lipid accumulation in cells in vitro. YTHDF2 binds to PPaRalpha to mediate its mRNA stability to regulate lipid metabolism. Transcriptional regulation of circadian rhythms is essential for lipid metabolic homeostasis, disruptions of which can lead to metabolic diseases. | |||
Death-associated protein kinase 2 (DAPK2)
| Representative RNA-seq result indicating the expression of this target gene regulated by YTHDF2 | ||
| Cell Line | Human umbilical cord blood CD34+ cells | Homo sapiens |
|
Treatment: YTHDF2 knockdown UCB CD34+ cells
Control: Wild type UCB CD34+ cells
|
GSE107956 | |
| Regulation |
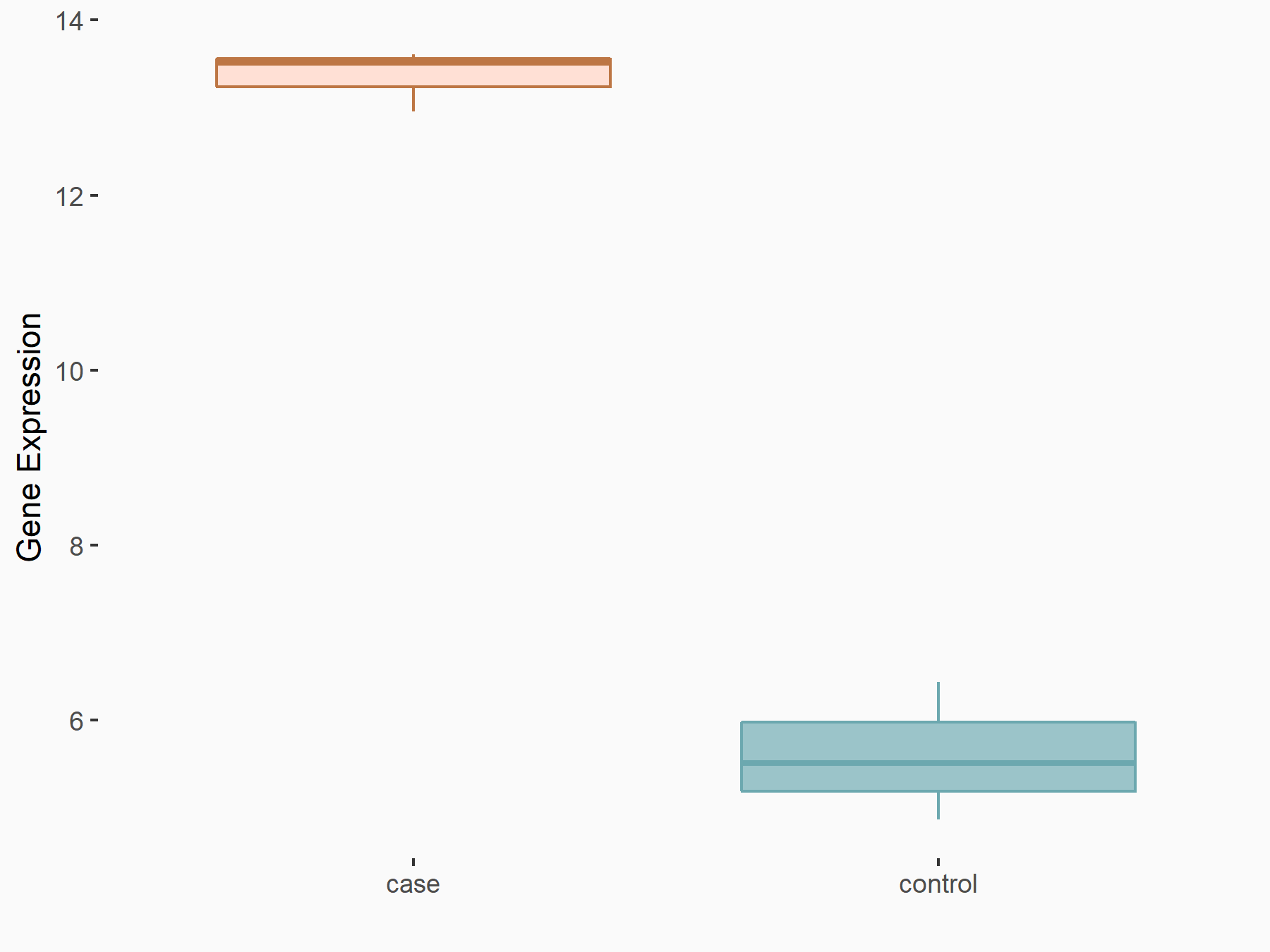 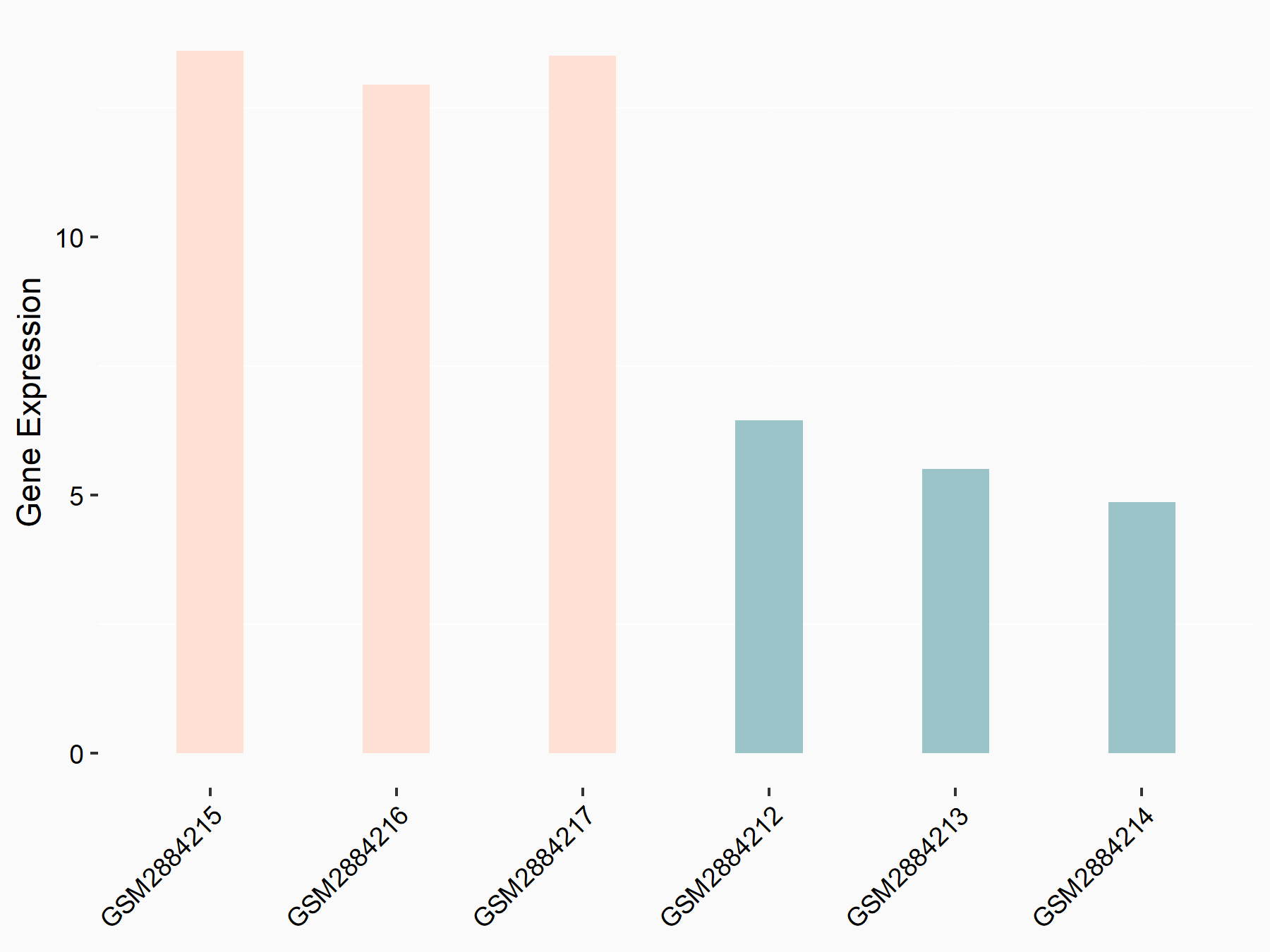 |
logFC: 1.13E+00 p-value: 1.77E-04 |
| More Results | Click to View More RNA-seq Results | |
Lung cancer [ICD-11: 2C25]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [22] | |||
| Responsed Disease | Non-small-cell lung carcinoma [ICD-11: 2C25.Y] | |||
| Target Regulation | Down regulation | |||
In-vitro Model |
NCI-H838 | Lung adenocarcinoma | Homo sapiens | CVCL_1594 |
| A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 | |
| In-vivo Model | The nude mice were maintained under pathogen-free conditions and kept under timed lighting conditions mandated by the committee with food and water provided ad libitum. For xenograft experiments, nude mice were injected subcutaneously with 5 × 106 cells resuspended in 0.1 mL PBS. When a tumor was palpable, it was measured every 3 days. | |||
| Response Summary | Cigarette smoking induced aberrant N6-methyladenosine modification of Death-associated protein kinase 2 (DAPK2), which resulted in decreased DAPK2 mRNA stability and expression of its mRNA and protein. This modification was mediated by the m6A "writer" METTL3 and the m6A "reader" YTHDF2. BAY 11-7085, a NF-Kappa-B signaling selective inhibitor, was shown to efficiently suppressed downregulation of DAPK2-induced oncogenic phenotypes of NSCLC cells. | |||
E3 ubiquitin-protein ligase TRIM7 (TRIM7)
| Representative RNA-seq result indicating the expression of this target gene regulated by YTHDF2 | ||
| Cell Line | Mouse-cerebellum granule cell | Mus musculus |
|
Treatment: YTHDF2 knockdown mouse-cerebellum granule cell
Control: Wild type mouse-cerebellum granule cell
|
GSE153688 | |
| Regulation |
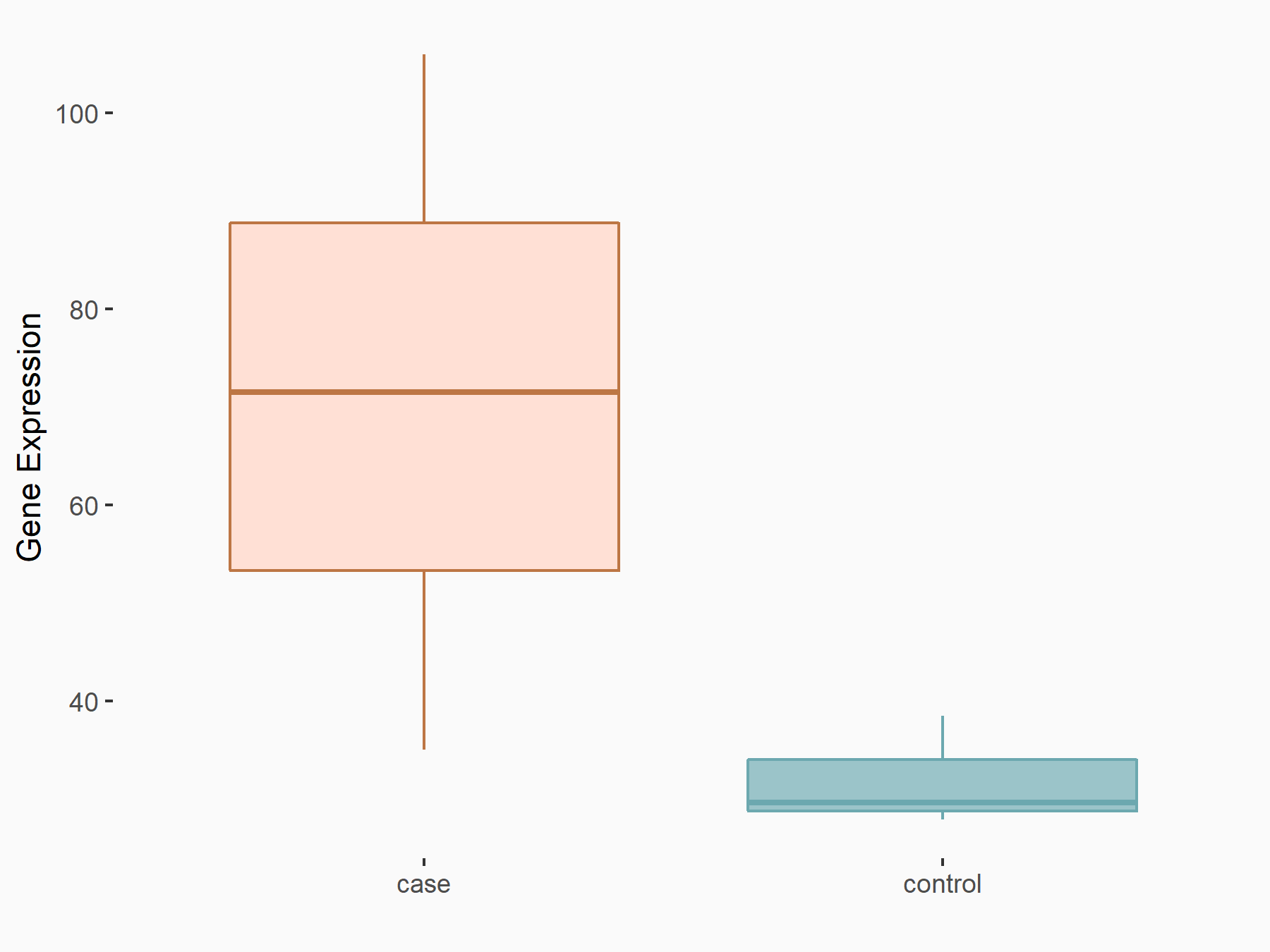 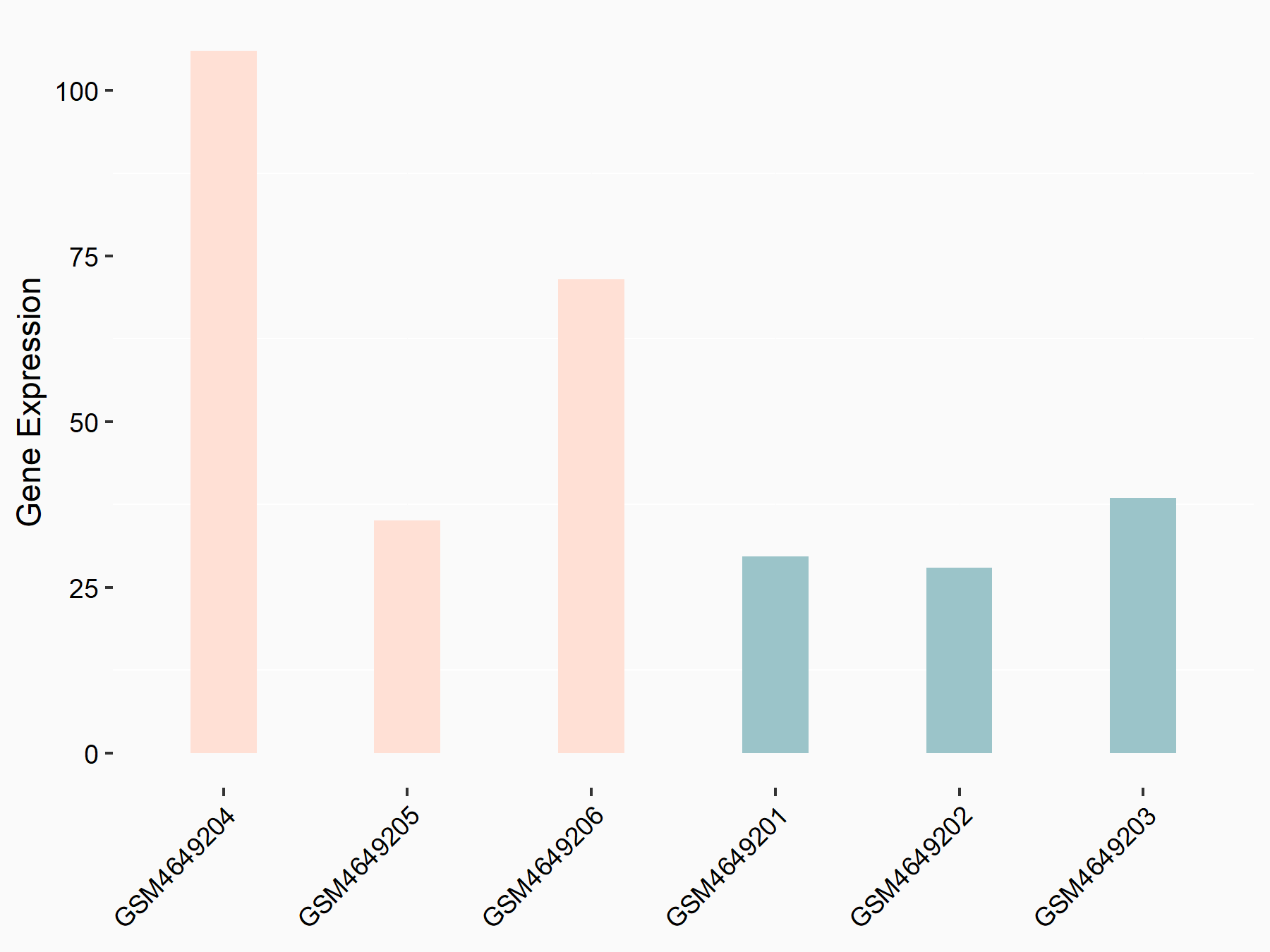 |
logFC: 1.15E+00 p-value: 8.92E-03 |
| More Results | Click to View More RNA-seq Results | |
Osteosarcoma [ICD-11: 2B51]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [23] | |||
| Responsed Disease | Osteosarcoma [ICD-11: 2B51] | |||
| Target Regulation | Down regulation | |||
| Pathway Response | Ubiquitin mediated proteolysis | hsa04120 | ||
| Cell Process | Proteasome pathway degradation | |||
In-vitro Model |
U2OS | Osteosarcoma | Homo sapiens | CVCL_0042 |
| SaOS-2 | Osteosarcoma | Homo sapiens | CVCL_0548 | |
| MG-63 | Osteosarcoma | Homo sapiens | CVCL_0426 | |
| HOS | Osteosarcoma | Homo sapiens | CVCL_0312 | |
| hFOB 1.19 | Normal | Homo sapiens | CVCL_3708 | |
| In-vivo Model | MG63 cells transduced with lentivirus expressing shTRIM7 or shNC, and SAOS2 cells transduced with lentivirus expressing TRIM7, BRMS1, TRIM7 plus BRMS1 or control vector, were injected via the tail vein into the nude mice (1 × 106 cells/mouse) (n = 11 per group). | |||
| Response Summary | E3 ubiquitin-protein ligase TRIM7 (TRIM7) mRNA stability was regulated by the METTL3/14-YTHDF2-mRNA in a decay-dependent manner. TRIM7 plays a key role in regulating metastasis and chemoresistance in osteosarcoma through ubiquitination of BRMS1. | |||
Endoplasmic reticulum chaperone BiP (Grp78)
| Representative RNA-seq result indicating the expression of this target gene regulated by YTHDF2 | ||
| Cell Line | B18-hi B cell line | Mus musculus |
|
Treatment: YTHDF2 knockout B18-hi B cells
Control: Wild type B18-hi B cells
|
GSE189819 | |
| Regulation |
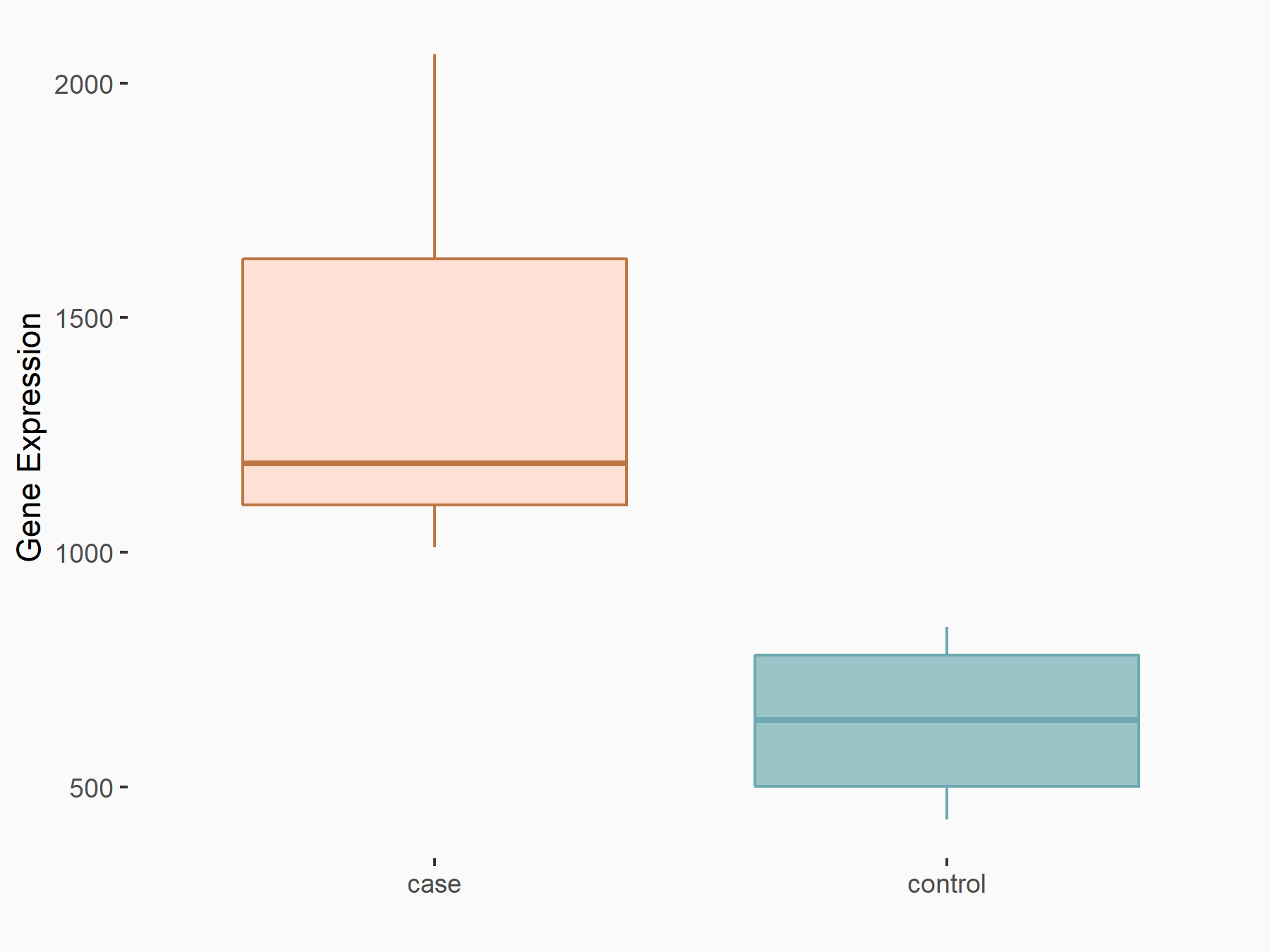 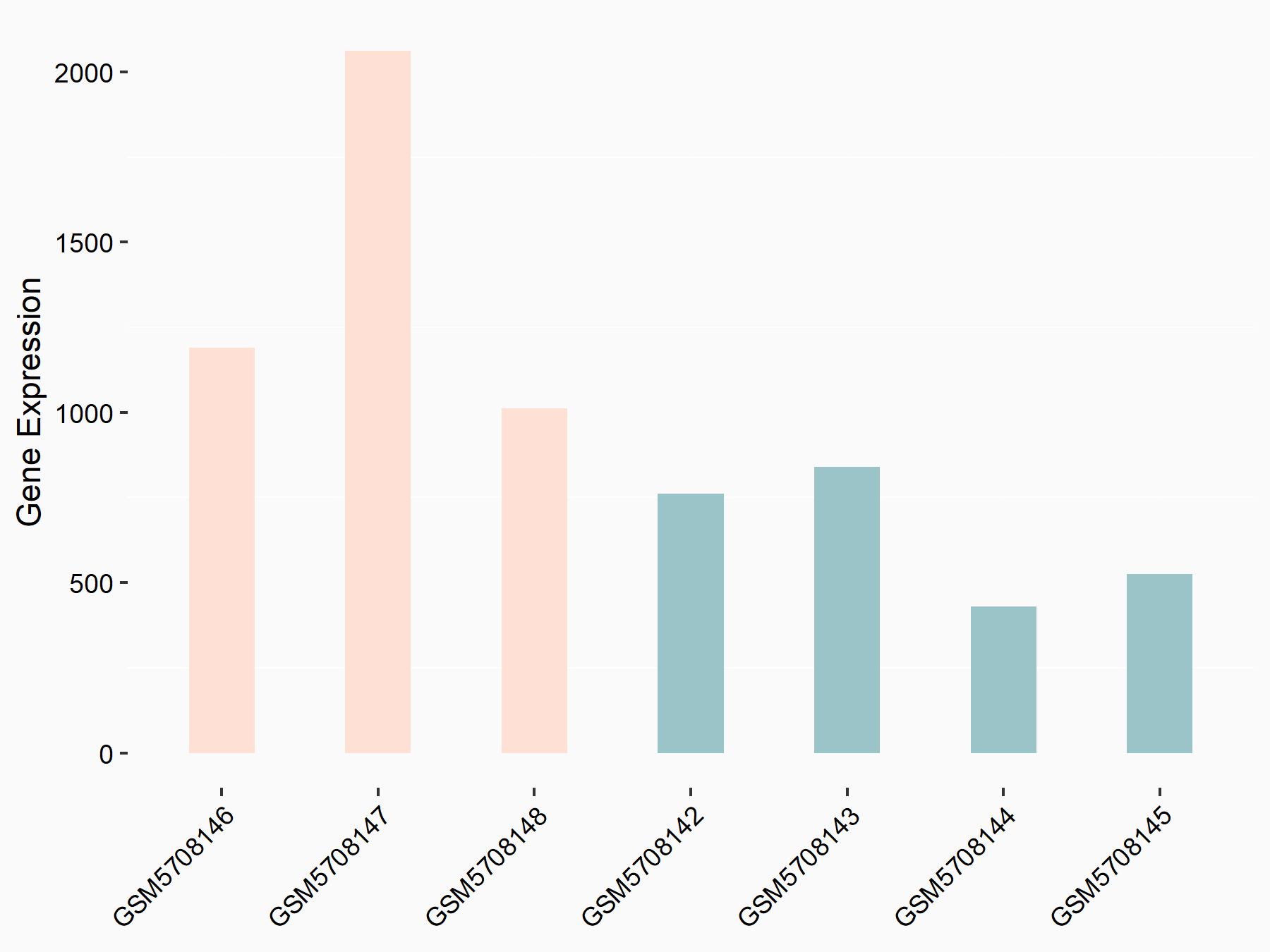 |
logFC: 1.16E+00 p-value: 1.91E-04 |
| More Results | Click to View More RNA-seq Results | |
Diseases of the musculoskeletal system [ICD-11: FC0Z]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [24] | |||
| Responsed Disease | Diseases of the musculoskeletal system [ICD-11: FC0Z] | |||
| Target Regulation | Down regulation | |||
| Pathway Response | RNA degradation | hsa03018 | ||
| Cell Process | RNA stability | |||
In-vitro Model |
MC3T3-E1 | Normal | Mus musculus | CVCL_0409 |
| Response Summary | METTL3 knockdown enhanced Endoplasmic reticulum chaperone BiP (Grp78) expression through YTHDF2-mediated RNA degradation, which elicited ER stress, thereby promoting osteoblast apoptosis and inhibiting cell proliferation and differentiation under LPS-induced inflammatory condition. | |||
G1/S-specific cyclin-D1 (CCND1)
| Representative RNA-seq result indicating the expression of this target gene regulated by YTHDF2 | ||
| Cell Line | GSC11 cell line | Homo sapiens |
|
Treatment: siYTHDF2 GSC11 cells
Control: siControl GSC11 cells
|
GSE142825 | |
| Regulation |
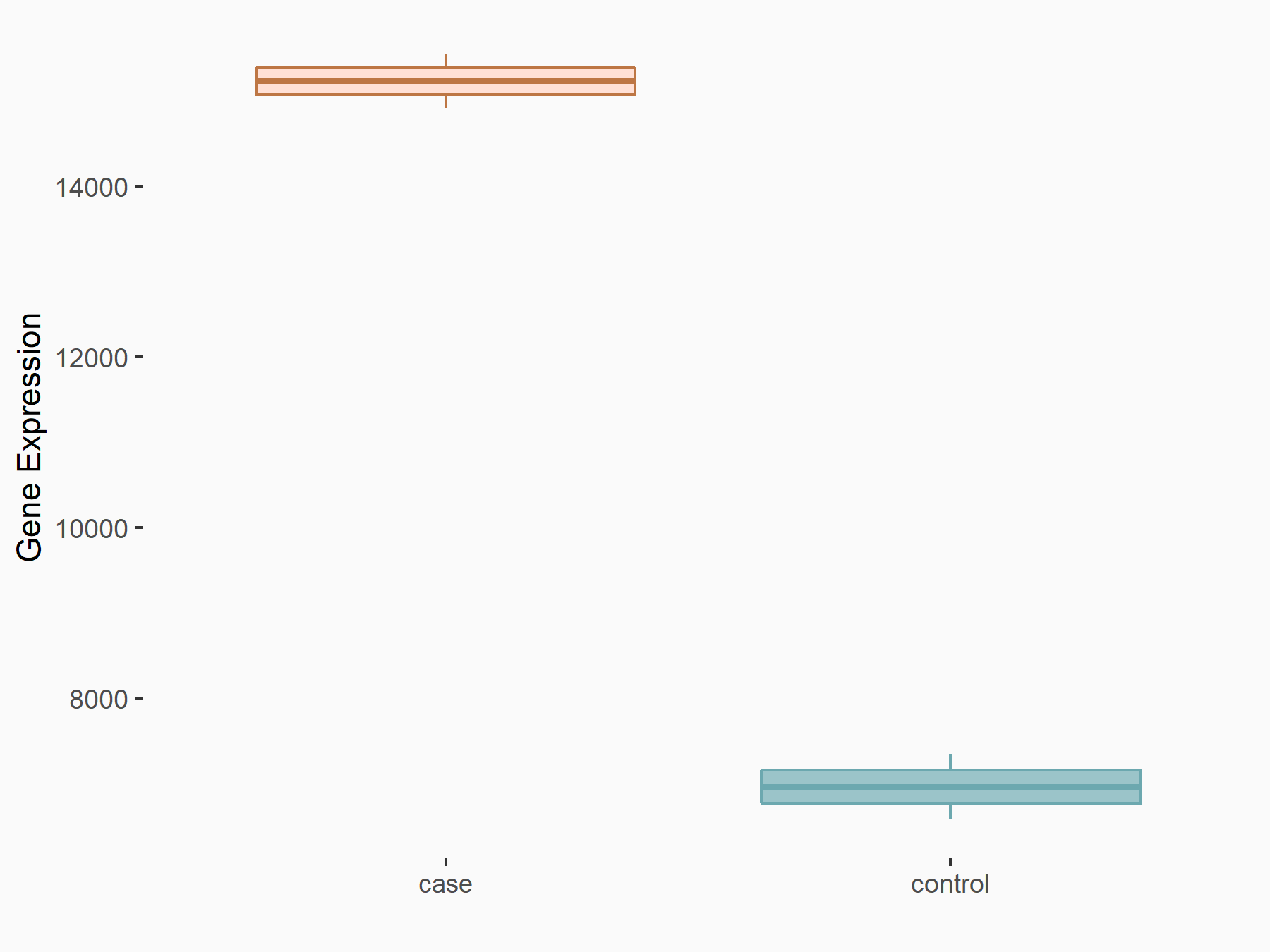 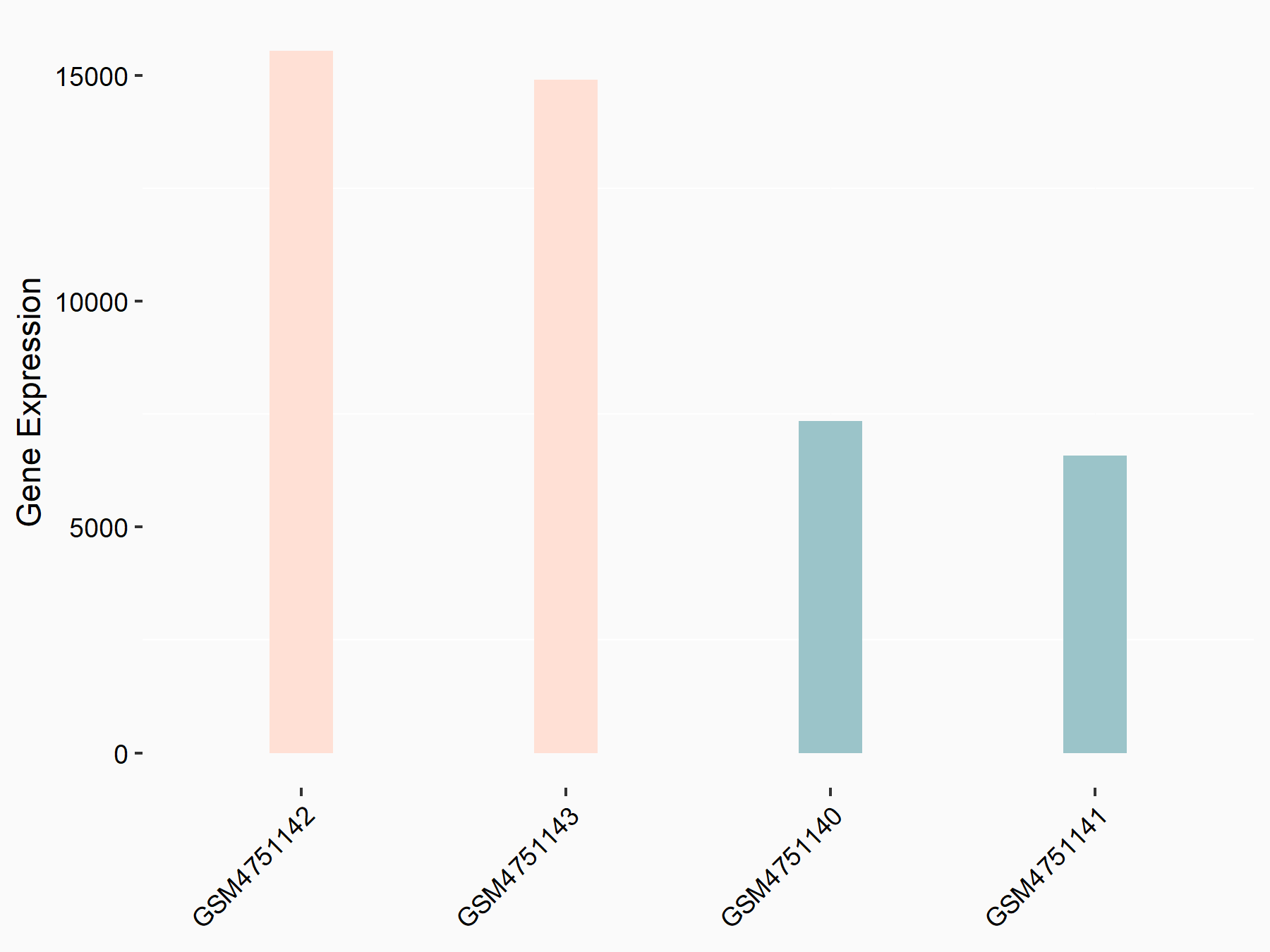 |
logFC: 1.13E+00 p-value: 1.41E-27 |
| More Results | Click to View More RNA-seq Results | |
Obesity [ICD-11: 5B81]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [25] | |||
| Responsed Disease | Obesity [ICD-11: 5B81] | |||
| Target Regulation | Down regulation | |||
| Pathway Response | Cell cycle | hsa04110 | ||
| Cell Process | Mitotic clonal | |||
| Prolonged G1/S transition | ||||
In-vitro Model |
3T3-L1 | Normal | Mus musculus | CVCL_0123 |
| Response Summary | Obesity is becoming a global problem. ZFP217 knockdown-induced adipogenesis inhibition was caused by G1/S-specific cyclin-D1 (CCND1), which was mediated by METTL3 and YTHDF2 in an m6A-dependent manner. | |||
Glia-derived nexin (SERPINE2)
| Representative RNA-seq result indicating the expression of this target gene regulated by YTHDF2 | ||
| Cell Line | Human umbilical cord blood CD34+ cells | Homo sapiens |
|
Treatment: YTHDF2 knockdown UCB CD34+ cells
Control: Wild type UCB CD34+ cells
|
GSE107956 | |
| Regulation |
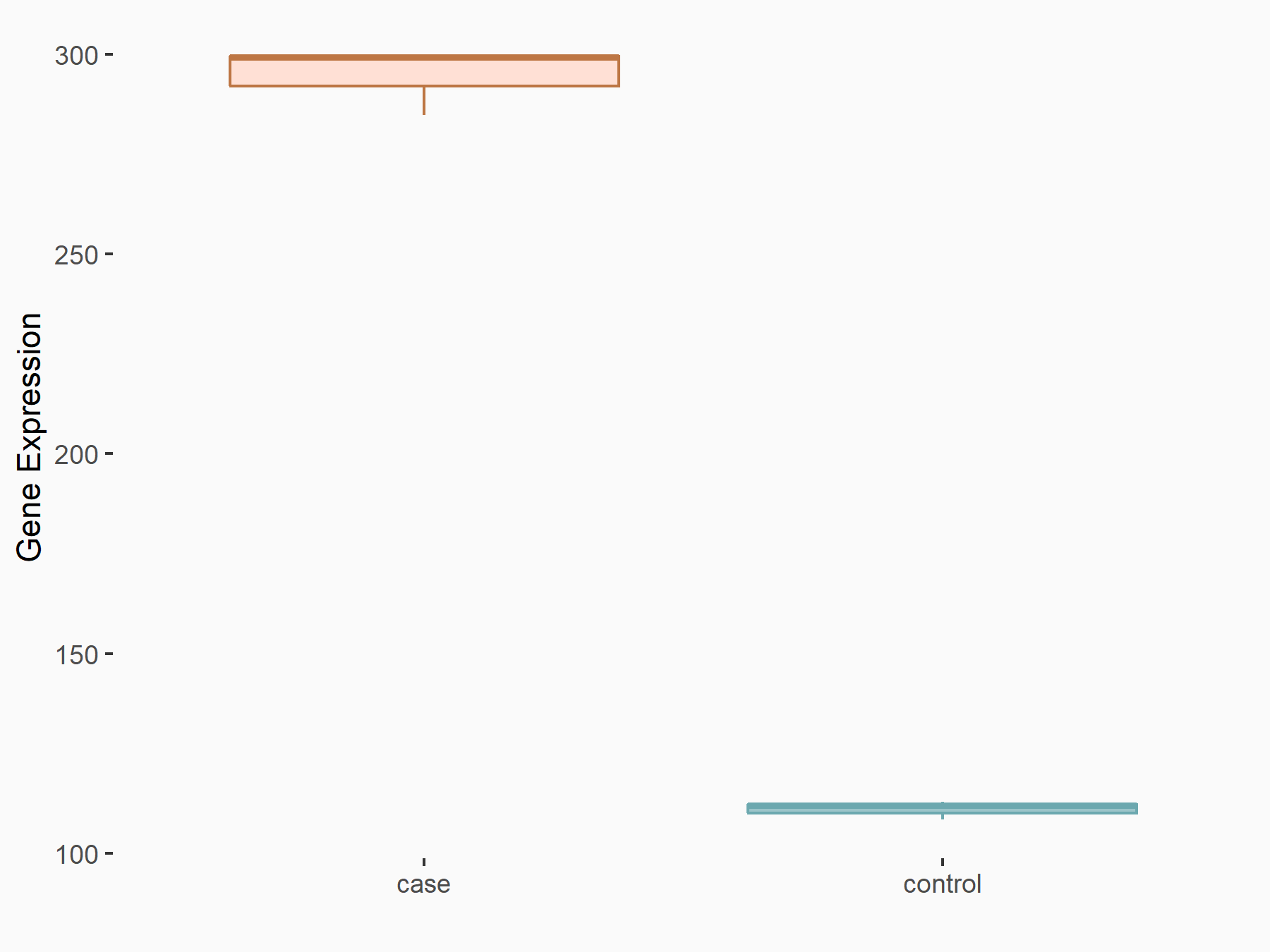 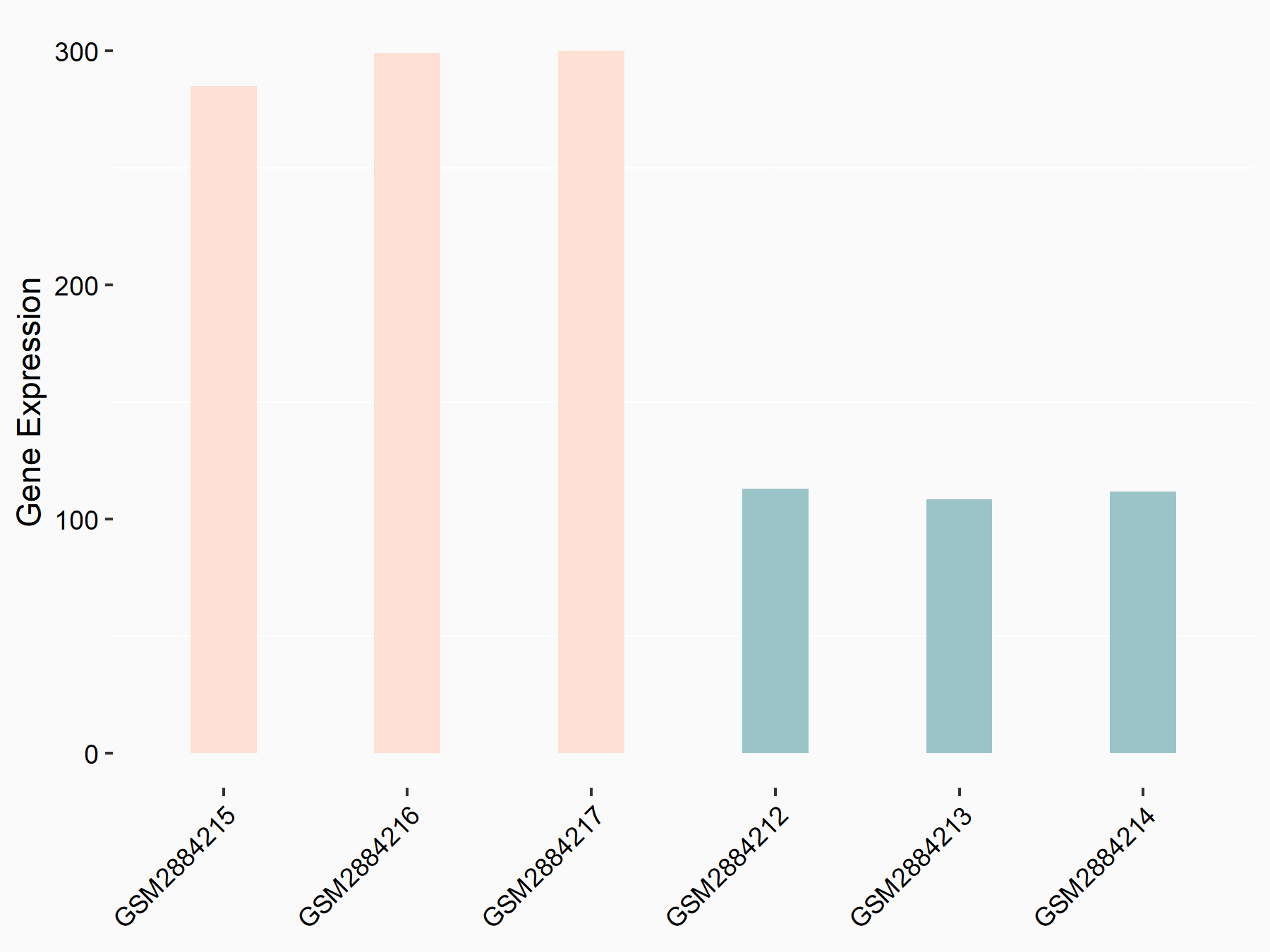 |
logFC: 1.40E+00 p-value: 3.23E-07 |
| More Results | Click to View More RNA-seq Results | |
Liver cancer [ICD-11: 2C12]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [27] | |||
| Responsed Disease | Hepatocellular carcinoma [ICD-11: 2C12.02] | |||
| Target Regulation | Down regulation | |||
| Pathway Response | HIF-1 signaling pathway | hsa04066 | ||
| Cell Process | Biological regulation | |||
In-vitro Model |
MHCC97-H | Adult hepatocellular carcinoma | Homo sapiens | CVCL_4972 |
| SMMC-7721 | Endocervical adenocarcinoma | Homo sapiens | CVCL_0534 | |
| In-vivo Model | A number of 5 × 106 SMMC7721 or MHCC97H cells re-suspended in 100 uL of PBS were subcutaneously injected into the right flank of 6-week old male NCG mice. | |||
| Response Summary | YTHDF2 processed the decay of m6A-containing interleukin 11 (IL11) and Glia-derived nexin (SERPINE2) mRNAs. YTHDF2 transcription succumbed to hypoxia-inducible factor-2-alpha (HIF-2-alpha). Administration of a HIF-2-alpha antagonist (PT2385) restored YTHDF2-programed epigenetic machinery and repressed liver cancer. | |||
Histone-lysine N-methyltransferase SETD7 (SETD7)
| Representative RNA-seq result indicating the expression of this target gene regulated by YTHDF2 | ||
| Cell Line | Human umbilical cord blood CD34+ cells | Homo sapiens |
|
Treatment: YTHDF2 knockdown UCB CD34+ cells
Control: Wild type UCB CD34+ cells
|
GSE107956 | |
| Regulation |
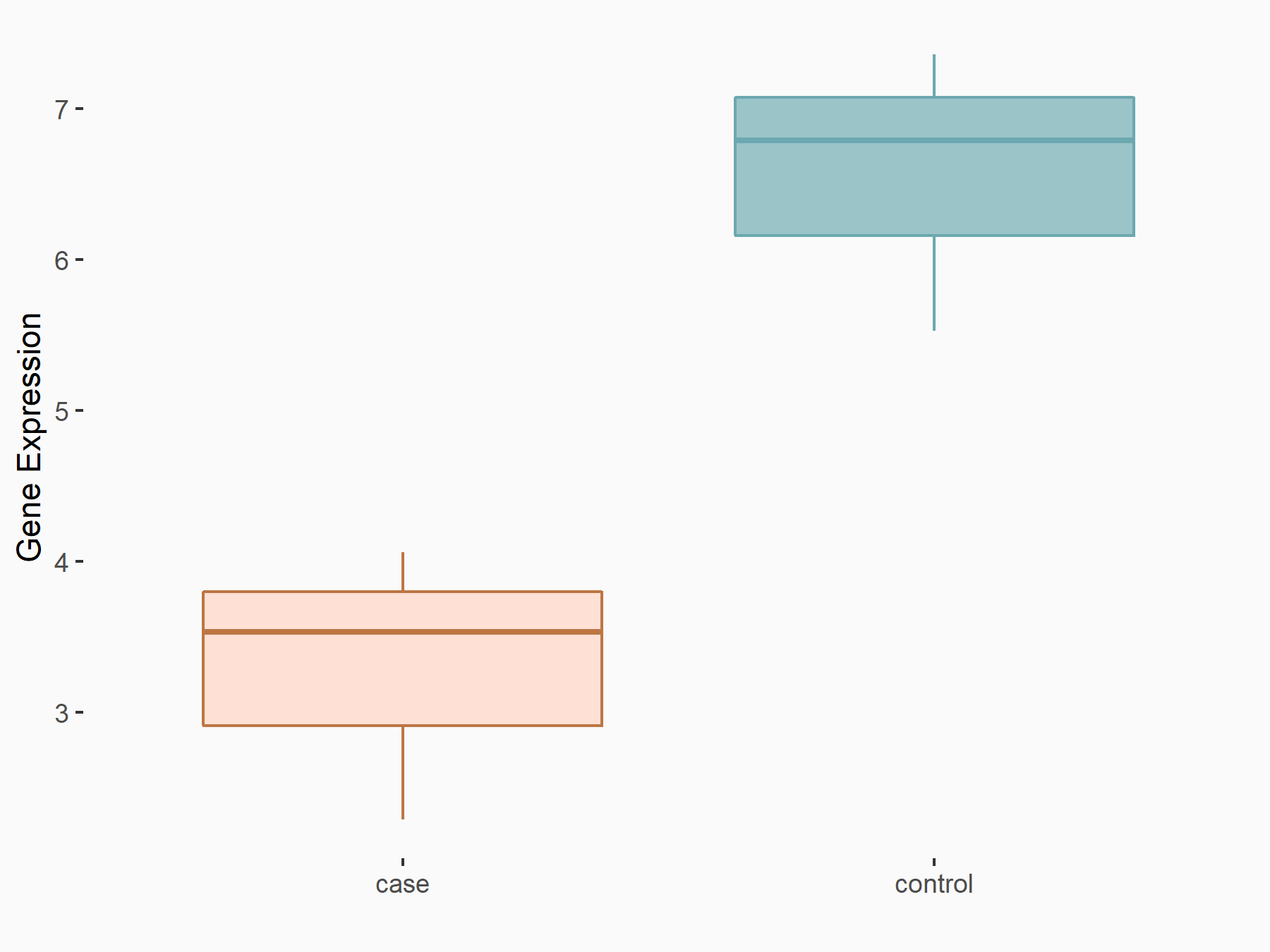 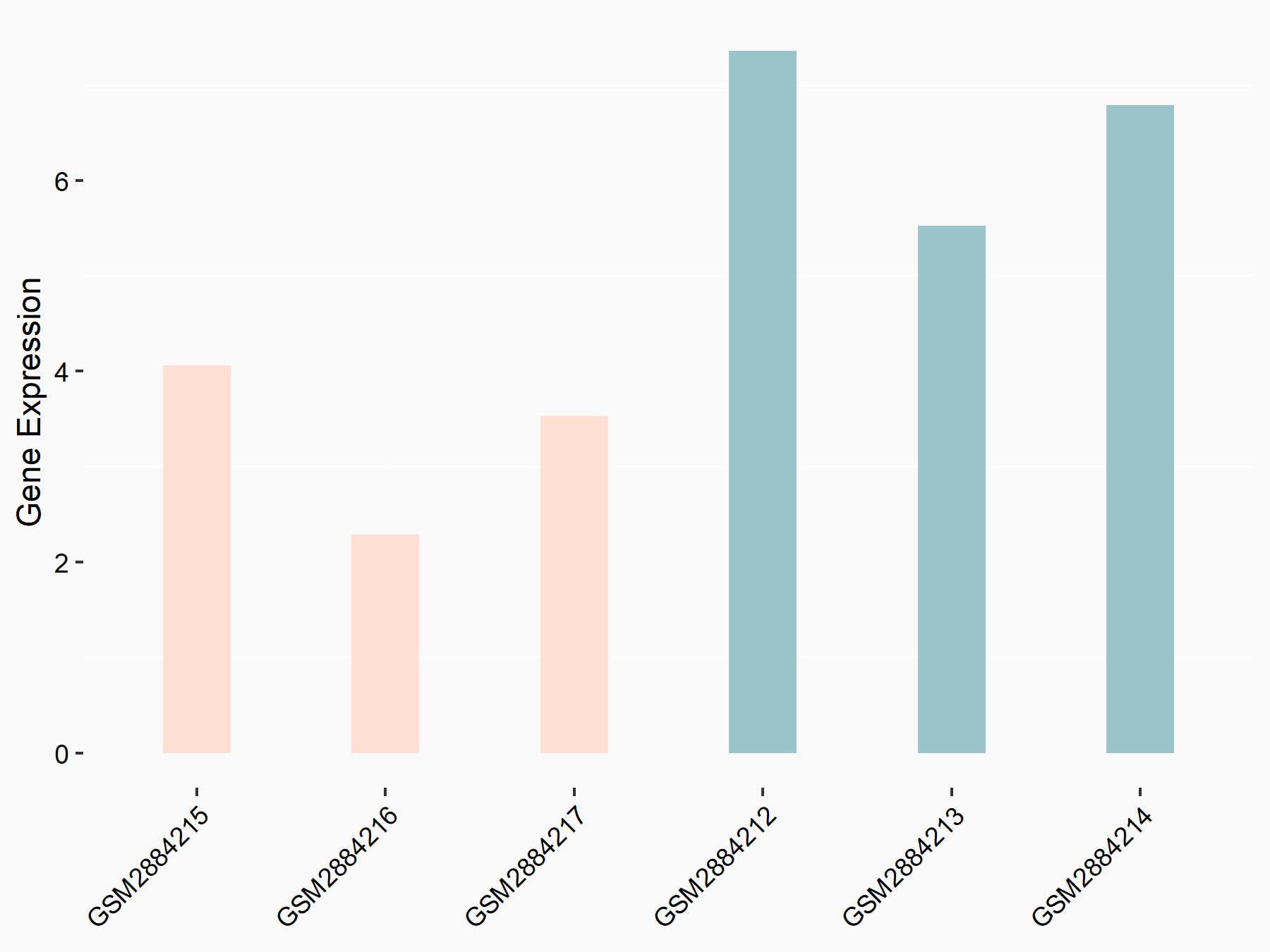 |
logFC: -8.31E-01 p-value: 1.26E-02 |
| More Results | Click to View More RNA-seq Results | |
Bladder cancer [ICD-11: 2C94]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [7] | |||
| Responsed Disease | Bladder cancer [ICD-11: 2C94] | |||
| Target Regulation | Down regulation | |||
| Cell Process | Cancer proliferation | |||
| Cancer metastasis | ||||
In-vitro Model |
SV-HUC-1 | Normal | Homo sapiens | CVCL_3798 |
| T24 | Bladder carcinoma | Homo sapiens | CVCL_0554 | |
| UM-UC-3 | Bladder carcinoma | Homo sapiens | CVCL_1783 | |
| In-vivo Model | For the subcutaneous implantation model, UM-UC-3 cells (2 × 106 cells per mouse) stably METTL3 knocked down (shMETTL3-1, shMETTL3-2) were injected into the flanks of mice. | |||
| Response Summary | METTL3/YTHDF2/Histone-lysine N-methyltransferase SETD7 (SETD7)/KLF4 m6 A axis provide the insight into the underlying mechanism of carcinogenesis and highlight potential therapeutic targets for bladder cancer. | |||
Methylcytosine dioxygenase TET1 (TET1)
| Representative RNA-seq result indicating the expression of this target gene regulated by YTHDF2 | ||
| Cell Line | Mouse-cerebellum granule cell | Mus musculus |
|
Treatment: YTHDF2 knockdown mouse-cerebellum granule cell
Control: Wild type mouse-cerebellum granule cell
|
GSE153688 | |
| Regulation |
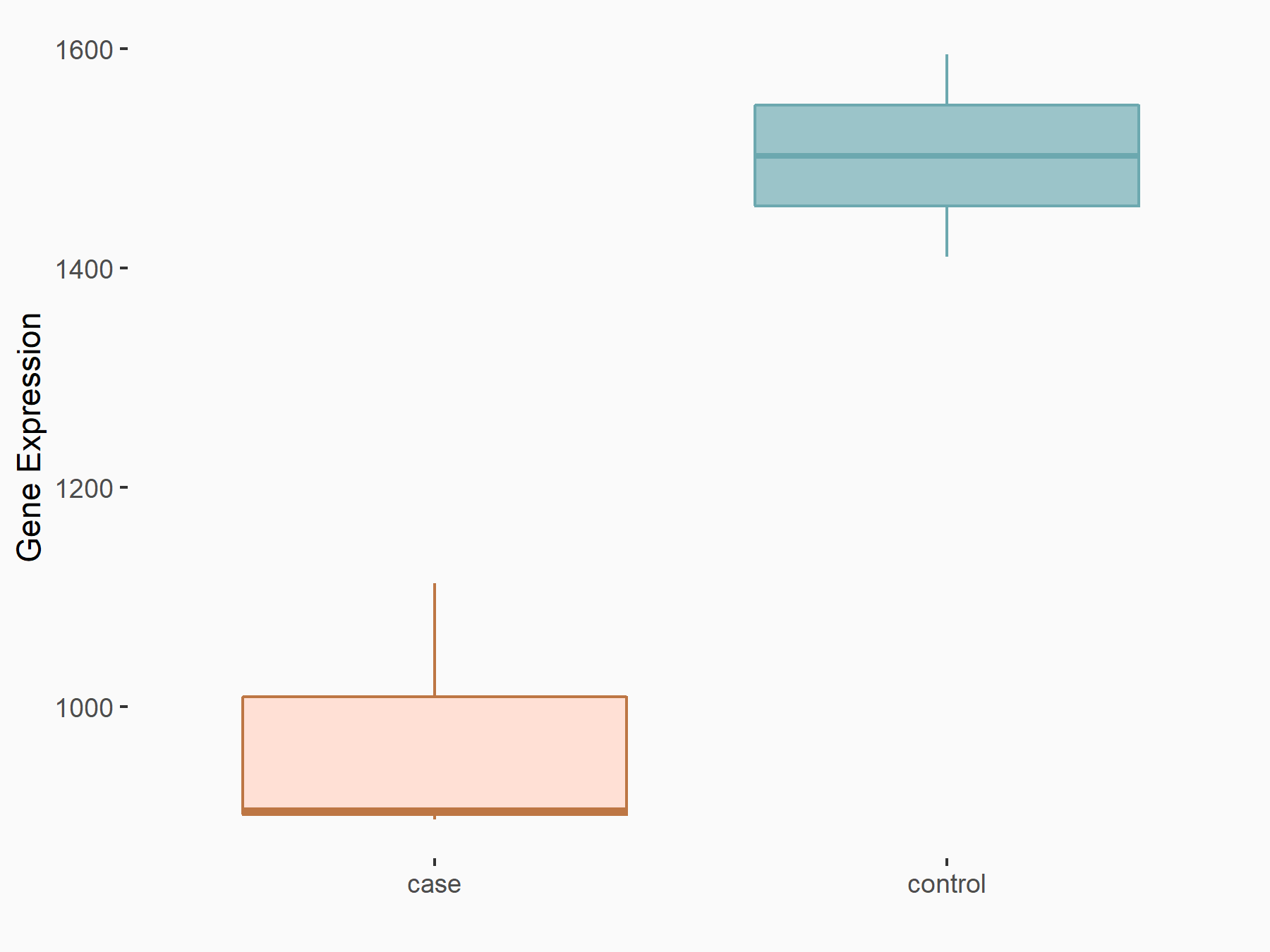  |
logFC: -6.29E-01 p-value: 8.41E-05 |
| More Results | Click to View More RNA-seq Results | |
Chronic pain [ICD-11: MG30]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [28] | |||
| Responsed Disease | Chronic pain [ICD-11: MG30] | |||
| Target Regulation | Down regulation | |||
| Response Summary | Downregulated spinal cord METTL3 coordinating with YTHDF2 contributes to the modulation of inflammatory pain through stabilizing upregulation of Methylcytosine dioxygenase TET1 (TET1) in spinal neurons. | |||
Myosin-7 (Myh7)
| Representative RNA-seq result indicating the expression of this target gene regulated by YTHDF2 | ||
| Cell Line | GSC11 cell line | Homo sapiens |
|
Treatment: siYTHDF2 GSC11 cells
Control: siControl GSC11 cells
|
GSE142825 | |
| Regulation |
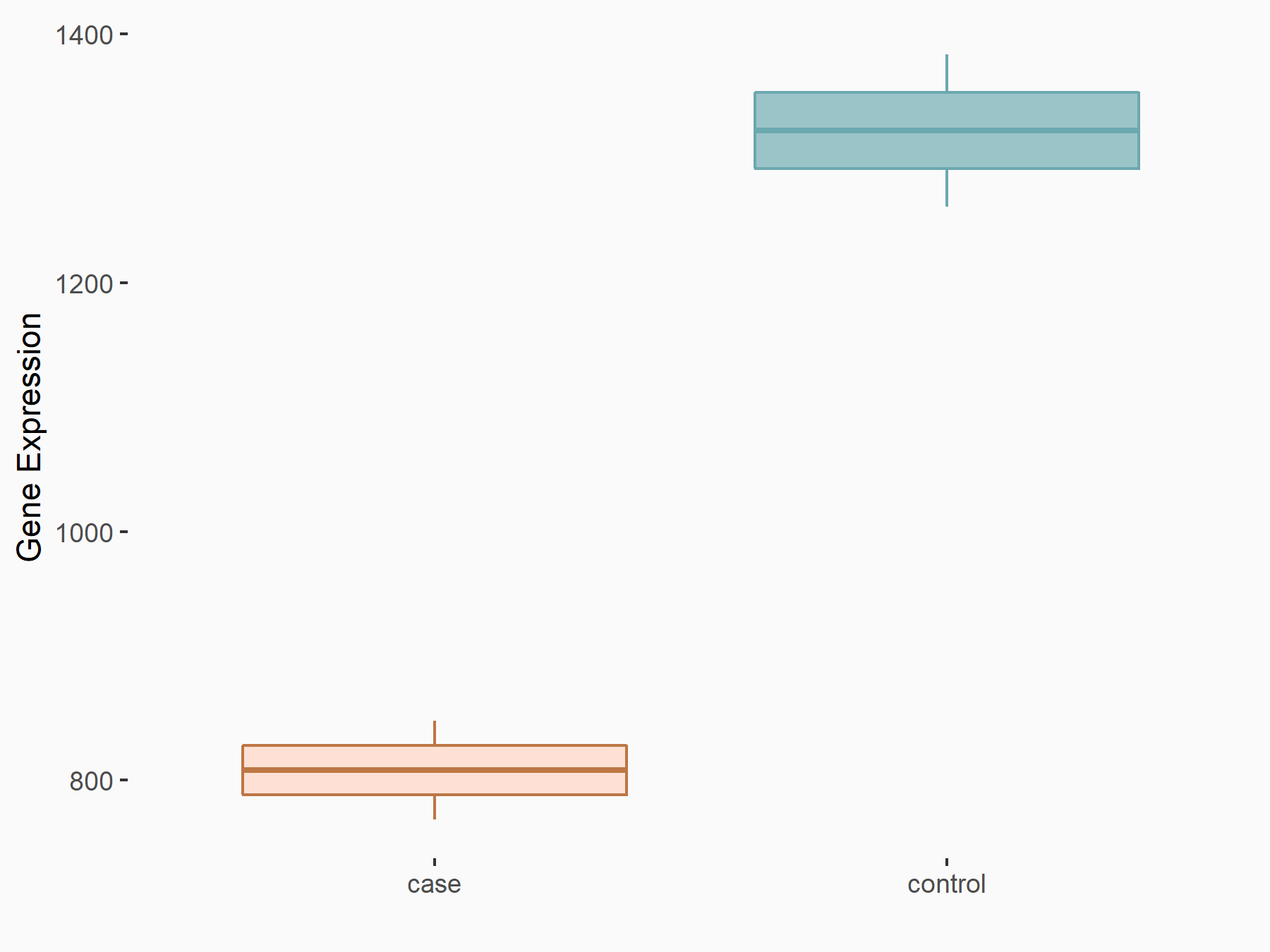  |
logFC: -7.12E-01 p-value: 5.98E-06 |
| More Results | Click to View More RNA-seq Results | |
Congestive heart failure [ICD-11: BD10]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [29] | |||
| Responsed Disease | Congestive heart failure [ICD-11: BD10] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
Neonatal rat ventricular cardiomyocytes (Primary myocyte cells) | |||
| In-vivo Model | Mice were anesthetized with 0.3% sodium pentobarbital (75 mg×kg-1) intraperitoneally, and the aortic arch was tied with a 6-0 nylon suture between the brachiocephalic and left common artery with a homemade L-shaped 26G cushion needle. After ligation, the needle was quickly removed, and the skin was closed. The sham operation was identical, except that the thread was not ligated. Moreover, mice were injected with rAAV9 (4 × 1011 vector genomes (vg)/mouse) carrying an empty vector, YTHDF2 or YTH-del via the tail vein. | |||
| Response Summary | Pathological cardiac hypertrophy is a major contributor of heart failure (HF), the m6A Reader YTHDF2 suppresses cardiac hypertrophy via Myosin-7 (Myh7) mRNA decoy in an m6A-dependent manner. | |||
Nucleobindin-1 (NUCB1)
| Representative RNA-seq result indicating the expression of this target gene regulated by YTHDF2 | ||
| Cell Line | B18-hi B cell line | Mus musculus |
|
Treatment: YTHDF2 knockout B18-hi B cells
Control: Wild type B18-hi B cells
|
GSE189819 | |
| Regulation |
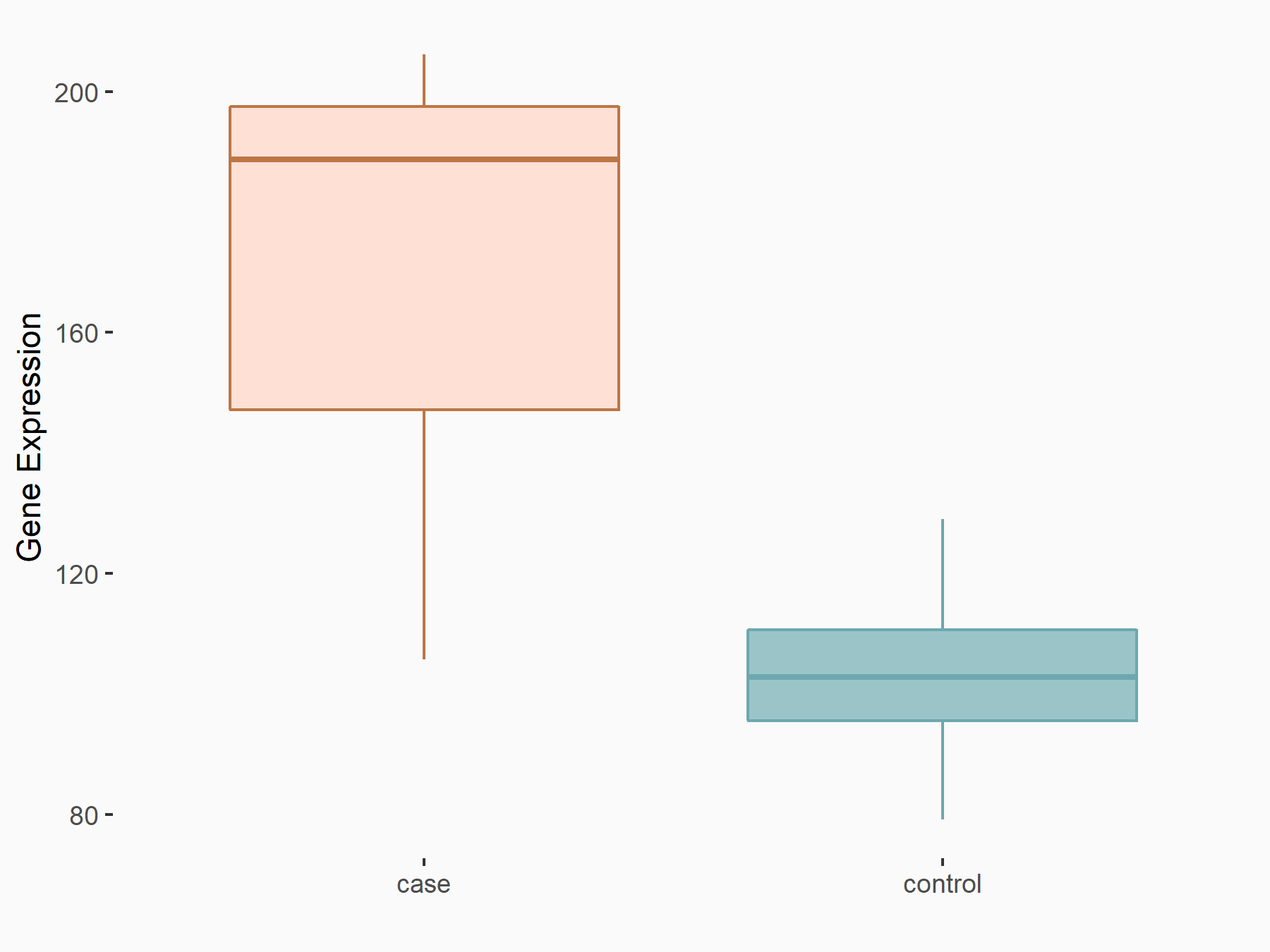  |
logFC: 6.95E-01 p-value: 1.62E-02 |
| More Results | Click to View More RNA-seq Results | |
Pancreatic cancer [ICD-11: 2C10]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [30] | |||
| Responsed Disease | Pancreatic ductal adenocarcinoma [ICD-11: 2C10.0] | |||
| Responsed Drug | Gemcitabine | Approved | ||
| Target Regulation | Down regulation | |||
| Pathway Response | Autophagy | hsa04140 | ||
| Cell Process | Cell proliferation | |||
| Cell autophagy | ||||
In-vitro Model |
SW1990 | Pancreatic adenocarcinoma | Homo sapiens | CVCL_1723 |
| PANC-1 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0480 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| CFPAC-1 | Cystic fibrosis | Homo sapiens | CVCL_1119 | |
| BxPC-3 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0186 | |
| AsPC-1 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0152 | |
| In-vivo Model | 5 × 106 SW1990 cells expressing NUCB1 (oeNUCB1) or control vector (oeNC) were injected subcutaneously. | |||
| Response Summary | METTL3-mediated m6A modification on Nucleobindin-1 (NUCB1) 5'UTR via the reader YTHDF2 as a mechanism for NUCB1 downregulation in PDAC. This study revealed crucial functions of NUCB1 in suppressing proliferation and enhancing the effects of gemcitabine in pancreatic cancer cells. | |||
Oxysterols receptor LXR-alpha (LXRA)
| Representative RNA-seq result indicating the expression of this target gene regulated by YTHDF2 | ||
| Cell Line | GSC11 cell line | Homo sapiens |
|
Treatment: siYTHDF2 GSC11 cells
Control: siControl GSC11 cells
|
GSE142825 | |
| Regulation |
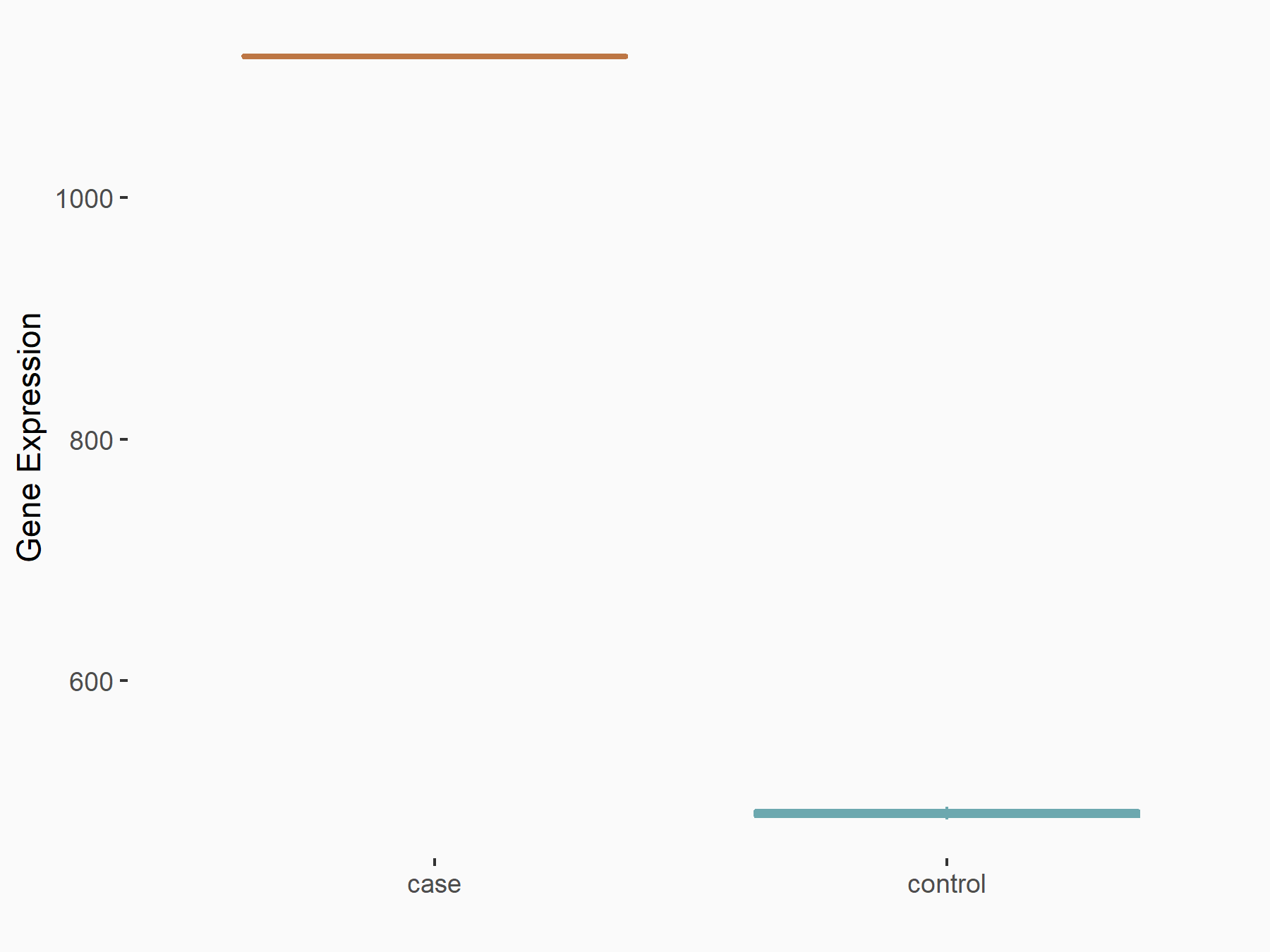  |
logFC: 1.19E+00 p-value: 2.27E-13 |
| More Results | Click to View More RNA-seq Results | |
Brain cancer [ICD-11: 2A00]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [18] | |||
| Responsed Disease | Glioblastoma [ICD-11: 2A00.00] | |||
| Target Regulation | Down regulation | |||
| Pathway Response | mRNA surveillance pathway | hsa03015), RNA degradation | ||
| Cell Process | RNA stability | |||
In-vitro Model |
U-87MG ATCC | Glioblastoma | Homo sapiens | CVCL_0022 |
| U-251MG | Astrocytoma | Homo sapiens | CVCL_0021 | |
| T98G | Glioblastoma | Homo sapiens | CVCL_0556 | |
| SW1783 | Anaplastic astrocytoma | Homo sapiens | CVCL_1722 | |
| LN-229 | Glioblastoma | Homo sapiens | CVCL_0393 | |
| Hs 683 | Oligodendroglioma | Homo sapiens | CVCL_0844 | |
| GSC7-2 (GSC7-2 were obtained from fresh surgical specimens of human primary and recurrent glioma) | ||||
| GSC6-27 (GSC6-27 were obtained from fresh surgical specimens of human primary and recurrent glioma) | ||||
| GSC23 | Glioblastoma | Homo sapiens | CVCL_DR59 | |
| GSC20 (GSC20 were obtained from fresh surgical specimens of human primary and recurrent glioma) | ||||
| GSC17 | Glioblastoma | Homo sapiens | CVCL_DR57 | |
| GSC11 | Glioblastoma | Homo sapiens | CVCL_DR55 | |
| In-vivo Model | For the studies of investigating mice survival, mice were intracranially injected with 10,000 GSC11, 10,000 GSC7-2, or 500,000 LN229 cells. | |||
| Response Summary | YTHDF2 facilitates m6A-dependent mRNA decay of Oxysterols receptor LXR-alpha (LXRA) and HIVEP2, which impacts the glioma patient survival. YTHDF2 promotes tumorigenesis of GBM cells, largely through the downregulation of LXRA and HIVEP2. | |||
Diseases of the urinary system [ICD-11: GC2Z]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [97] | |||
| Responsed Disease | Diseases of the urinary system [ICD-11: GC2Z] | |||
In-vitro Model |
MCP-5 [Mouse]
|
N.A. | Mus musculus | CVCL_R863 |
Period circadian protein homolog 1 (PER1)
| Representative RNA-seq result indicating the expression of this target gene regulated by YTHDF2 | ||
| Cell Line | Mouse-cerebellum granule cell | Mus musculus |
|
Treatment: YTHDF2 knockdown mouse-cerebellum granule cell
Control: Wild type mouse-cerebellum granule cell
|
GSE153688 | |
| Regulation |
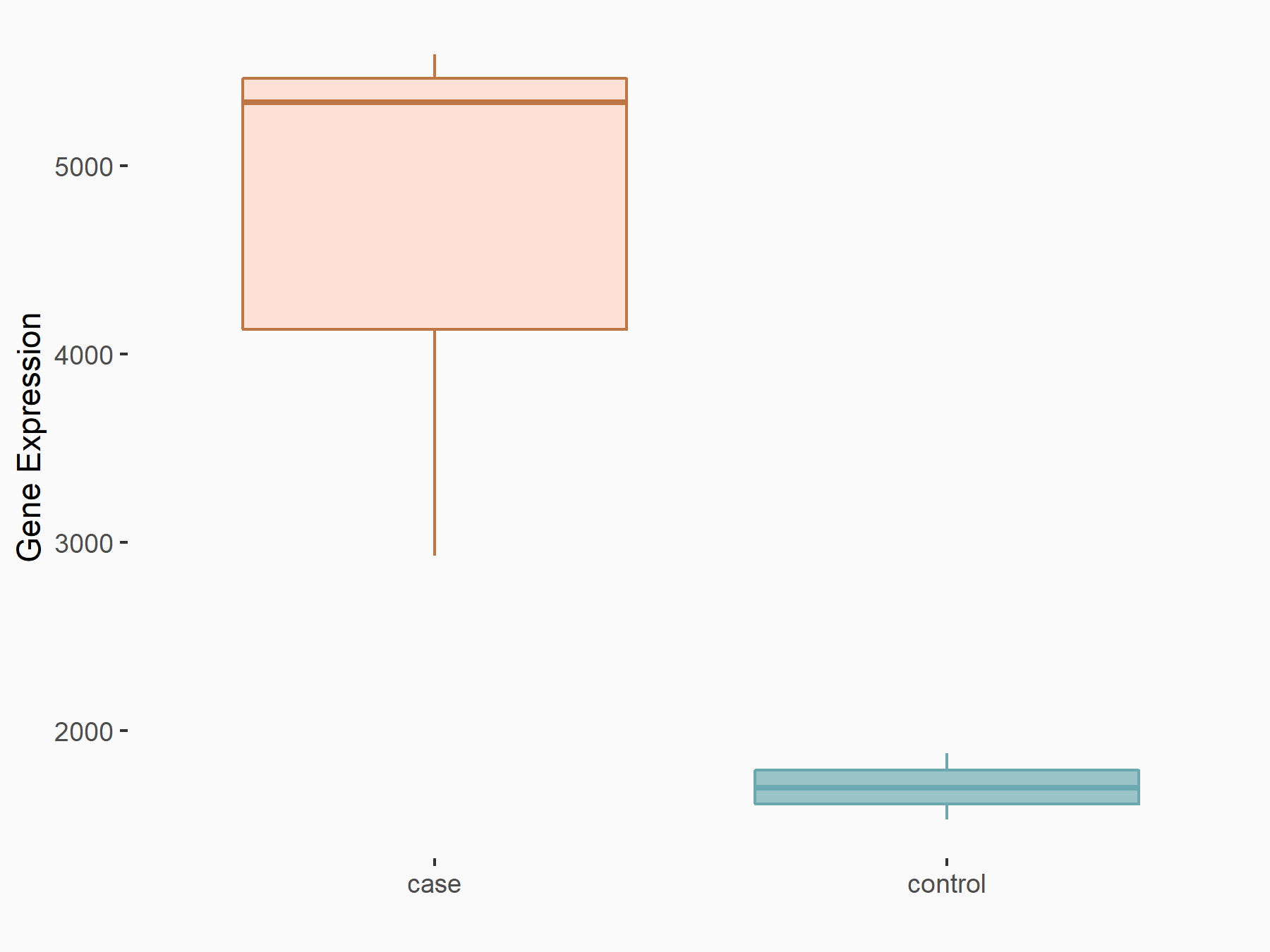  |
logFC: 1.44E+00 p-value: 9.54E-09 |
| More Results | Click to View More RNA-seq Results | |
Pancreatic cancer [ICD-11: 2C10]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [31] | |||
| Responsed Disease | Pancreatic cancer [ICD-11: 2C10] | |||
| Target Regulation | Down regulation | |||
| Pathway Response | p53 signaling pathway | hsa04115 | ||
| Cell Process | Cell proliferation | |||
| Cell migration | ||||
| Cell invasion | ||||
In-vitro Model |
AsPC-1 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0152 |
| BxPC-3 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0186 | |
| CFPAC-1 | Cystic fibrosis | Homo sapiens | CVCL_1119 | |
| HPDE6c7 | Normal | Homo sapiens | CVCL_0P38 | |
| PANC-1 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0480 | |
| SW1990 | Pancreatic adenocarcinoma | Homo sapiens | CVCL_1723 | |
| Response Summary | ALKBH5 serves as a pancreatic cancer suppressor by regulating the posttranscriptional activation of Period circadian protein homolog 1 (PER1) through m6A abolishment, which highlights a demethylation-based approach for PC diagnosis and therapy. ALKBH5 loss downregulated PER1 mRNA levels in an m6A-YTHDF2-dependent manner. | |||
Melanoma of uvea [ICD-11: 2D0Y]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [55] | |||
| Responsed Disease | Melanoma of uvea [ICD-11: 2D0Y] | |||
In-vitro Model |
OCM-1 | Amelanotic melanoma | Homo sapiens | CVCL_6934 |
| CRMM-1 | Conjunctival melanoma | Homo sapiens | CVCL_M593 | |
Phospholysine phosphohistidine inorganic pyrophosphate phosphatase (LHPP)
| Representative RNA-seq result indicating the expression of this target gene regulated by YTHDF2 | ||
| Cell Line | B18-hi B cell line | Mus musculus |
|
Treatment: YTHDF2 knockout B18-hi B cells
Control: Wild type B18-hi B cells
|
GSE189819 | |
| Regulation |
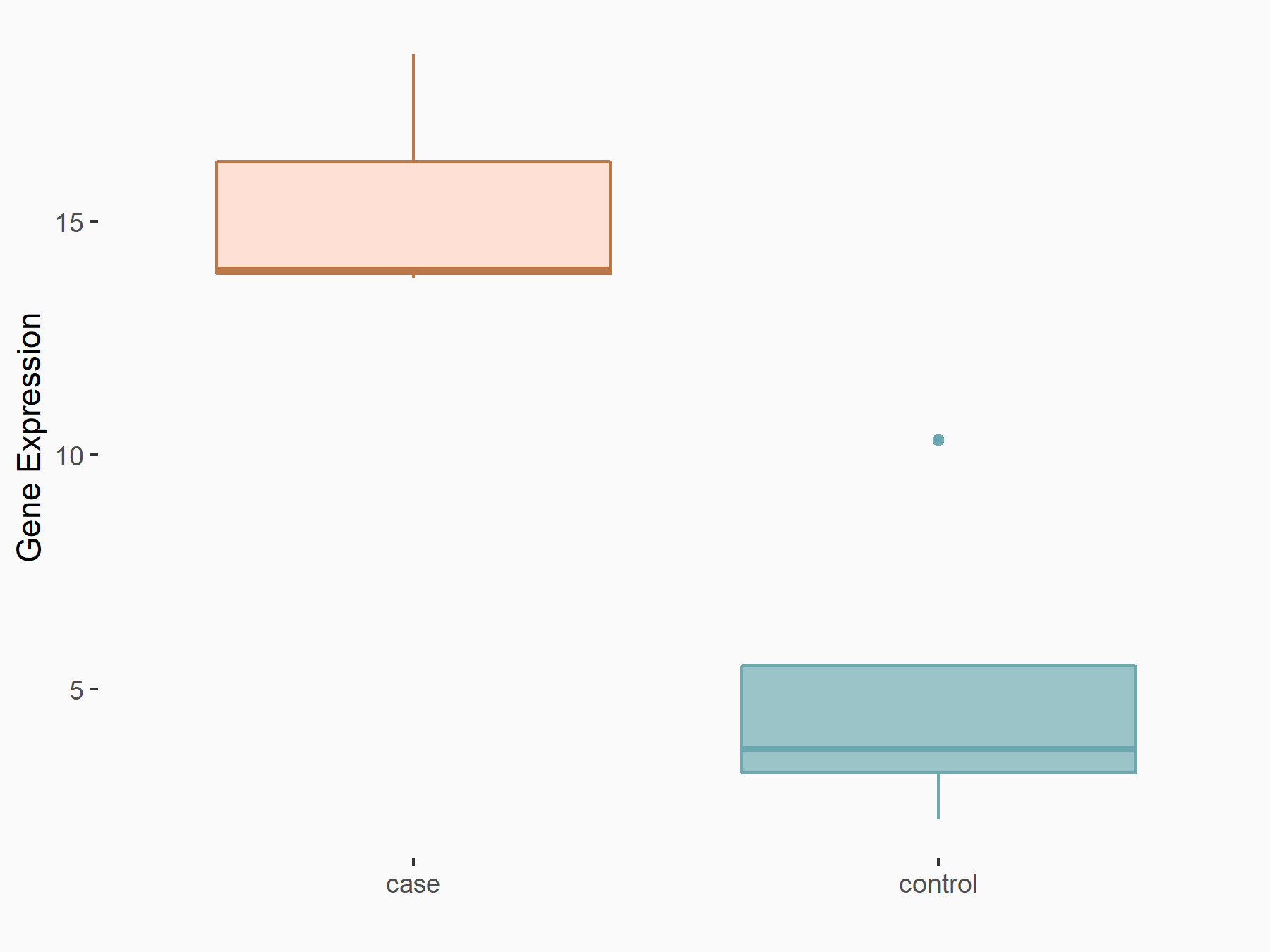  |
logFC: 1.54E+00 p-value: 1.87E-02 |
| More Results | Click to View More RNA-seq Results | |
Prostate cancer [ICD-11: 2C82]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [5] | |||
| Responsed Disease | Prostate cancer [ICD-11: 2C82] | |||
| Target Regulation | Down regulation | |||
| Pathway Response | Oxidative phosphorylation | hsa00190 | ||
In-vitro Model |
VCaP | Prostate carcinoma | Homo sapiens | CVCL_2235 |
| RWPE-1 | Normal | Homo sapiens | CVCL_3791 | |
| PC-3 | Prostate carcinoma | Homo sapiens | CVCL_0035 | |
| DU145 | Prostate carcinoma | Homo sapiens | CVCL_0105 | |
| 22Rv1 | Prostate carcinoma | Homo sapiens | CVCL_1045 | |
| In-vivo Model | Approximately 2 × 106 PCa cells (PC-3 shNC, shYTHDF2, shMETTL3 cell lines) per mouse suspended in 100 uL PBS were injected in the flank of male BALB/c nude mice (4 weeks old). During the 40-day observation, the tumor size (V = (width2×length ×0.52)) was measured with vernier caliper. Approximately 1.5 × 106 PCa cells suspended in 100 uL of PBS (PC-3 shNC, shYTHDF2, and shMETTL3 cell lines) per mouse were injected into the tail vein of male BALB/c nude mice (4 weeks old). The IVIS Spectrum animal imaging system (PerkinElmer) was used to evaluate the tumor growth (40 days) and whole metastasis conditions (4 weeks and 6 weeks) with 100 uL XenoLight D-luciferin Potassium Salt (15 mg/ml, Perkin Elmer) per mouse. Mice were anesthetized and then sacrificed for tumors and metastases which were sent for further organ-localized imaging as above, IHC staining and hematoxylin-eosin (H&E) staining. | |||
| Response Summary | Knock-down of YTHDF2 or METTL3 significantly induced the expression of Phospholysine phosphohistidine inorganic pyrophosphate phosphatase (LHPP) and NKX3-1 at both mRNA and protein level with inhibited phosphorylated AKT. YTHDF2 mediates the mRNA degradation of the tumor suppressors LHPP and NKX3-1 in m6A-dependent way to regulate AKT phosphorylation-induced tumor progression in prostate cancer. | |||
Protein SET (SET)
| Representative RNA-seq result indicating the expression of this target gene regulated by YTHDF2 | ||
| Cell Line | GSC11 cell line | Homo sapiens |
|
Treatment: siYTHDF2 GSC11 cells
Control: siControl GSC11 cells
|
GSE142825 | |
| Regulation |
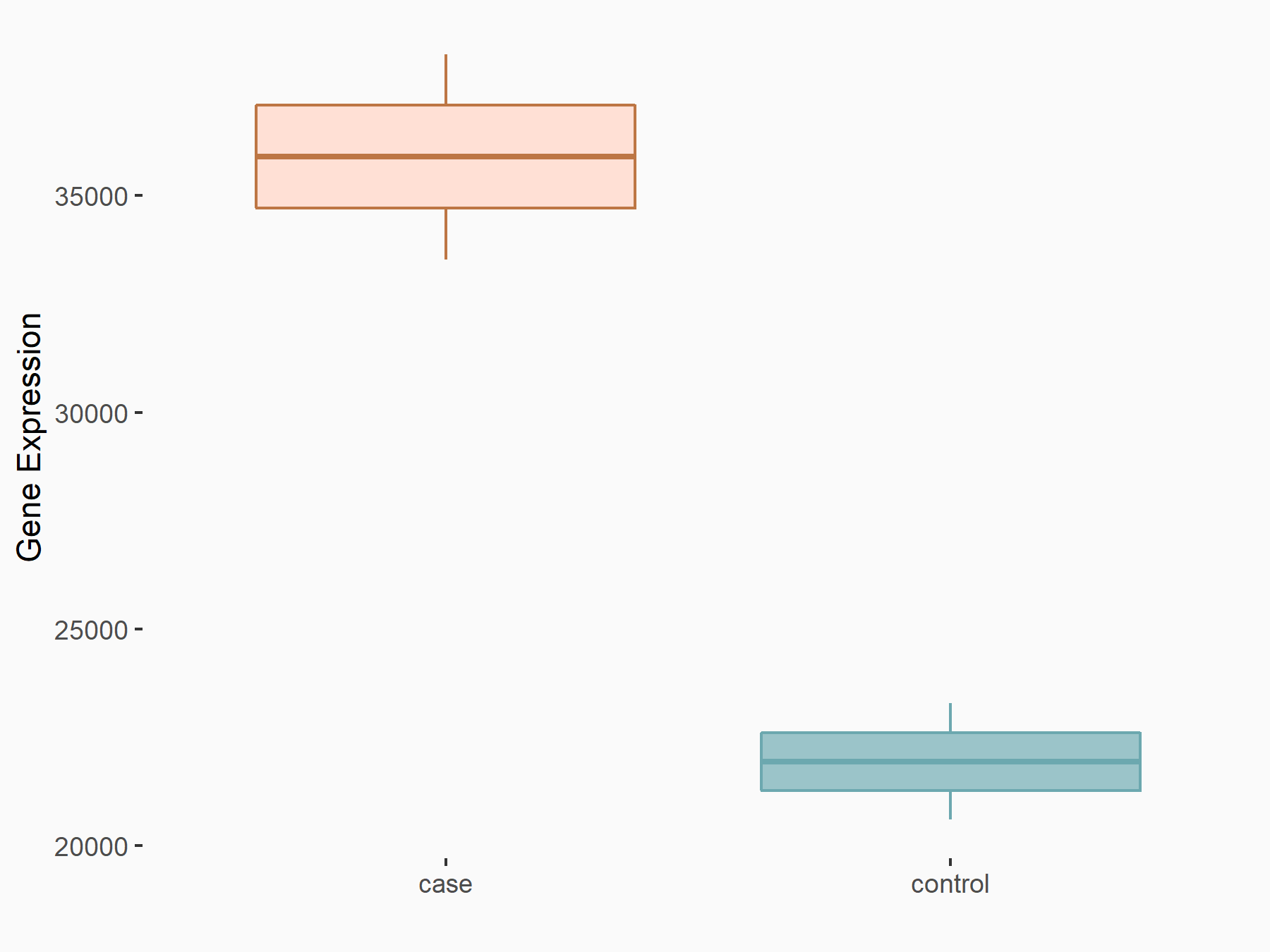 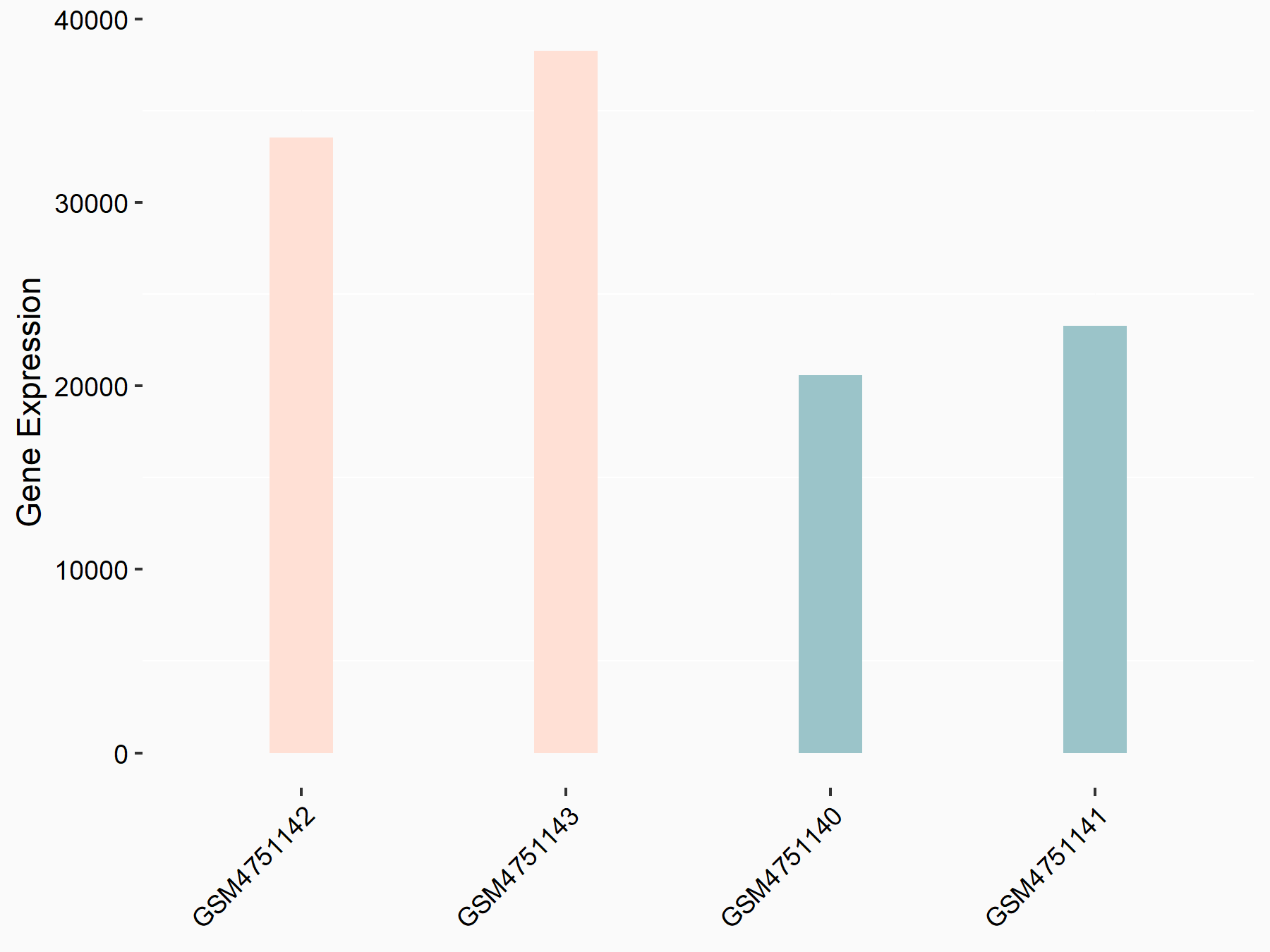 |
logFC: 7.10E-01 p-value: 6.25E-10 |
| More Results | Click to View More RNA-seq Results | |
Lung cancer [ICD-11: 2C25]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [6] | |||
| Responsed Disease | Non-small-cell lung carcinoma [ICD-11: 2C25.Y] | |||
| Target Regulation | Down regulation | |||
| Pathway Response | TNF signaling pathway | hsa04668 | ||
| Cell Process | Cell migration | |||
| Cell invasion | ||||
| Epithelial-mesenchymal transition | ||||
In-vitro Model |
NCI-H1975 | Lung adenocarcinoma | Homo sapiens | CVCL_1511 |
| A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 | |
| Response Summary | MiR-1915-3p expression was regulated by METTL3/YTHDF2 m6A axis through transcription factor KLF4. miR-1915-3p function as a tumor suppressor by targeting Protein SET (SET) and has an anti-metastatic therapeutic potential for lung cancer treatment. | |||
Protein yippee-like 5 (YPEL5)
| Representative RNA-seq result indicating the expression of this target gene regulated by YTHDF2 | ||
| Cell Line | Human umbilical cord blood CD34+ cells | Homo sapiens |
|
Treatment: YTHDF2 knockdown UCB CD34+ cells
Control: Wild type UCB CD34+ cells
|
GSE107956 | |
| Regulation |
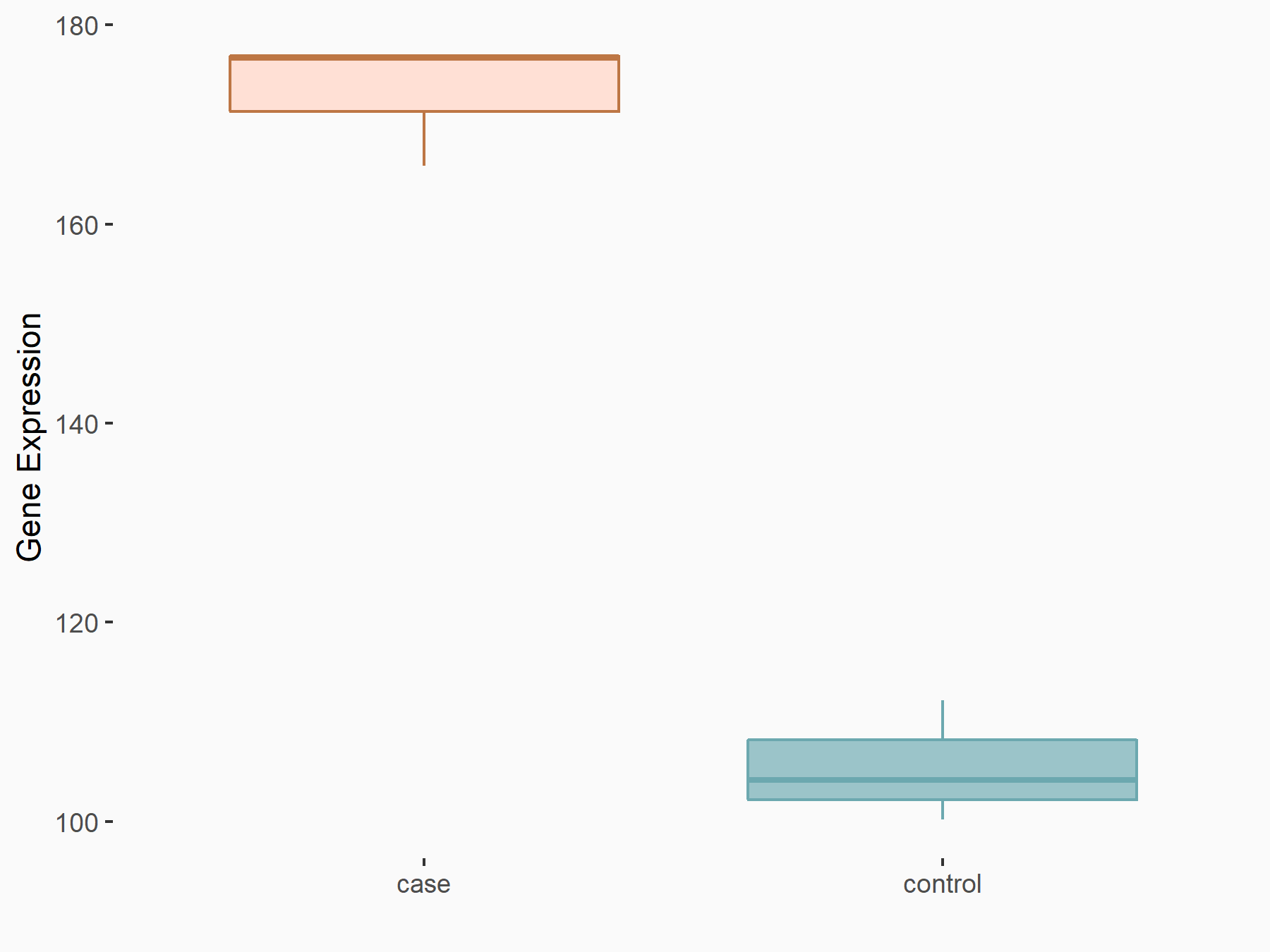 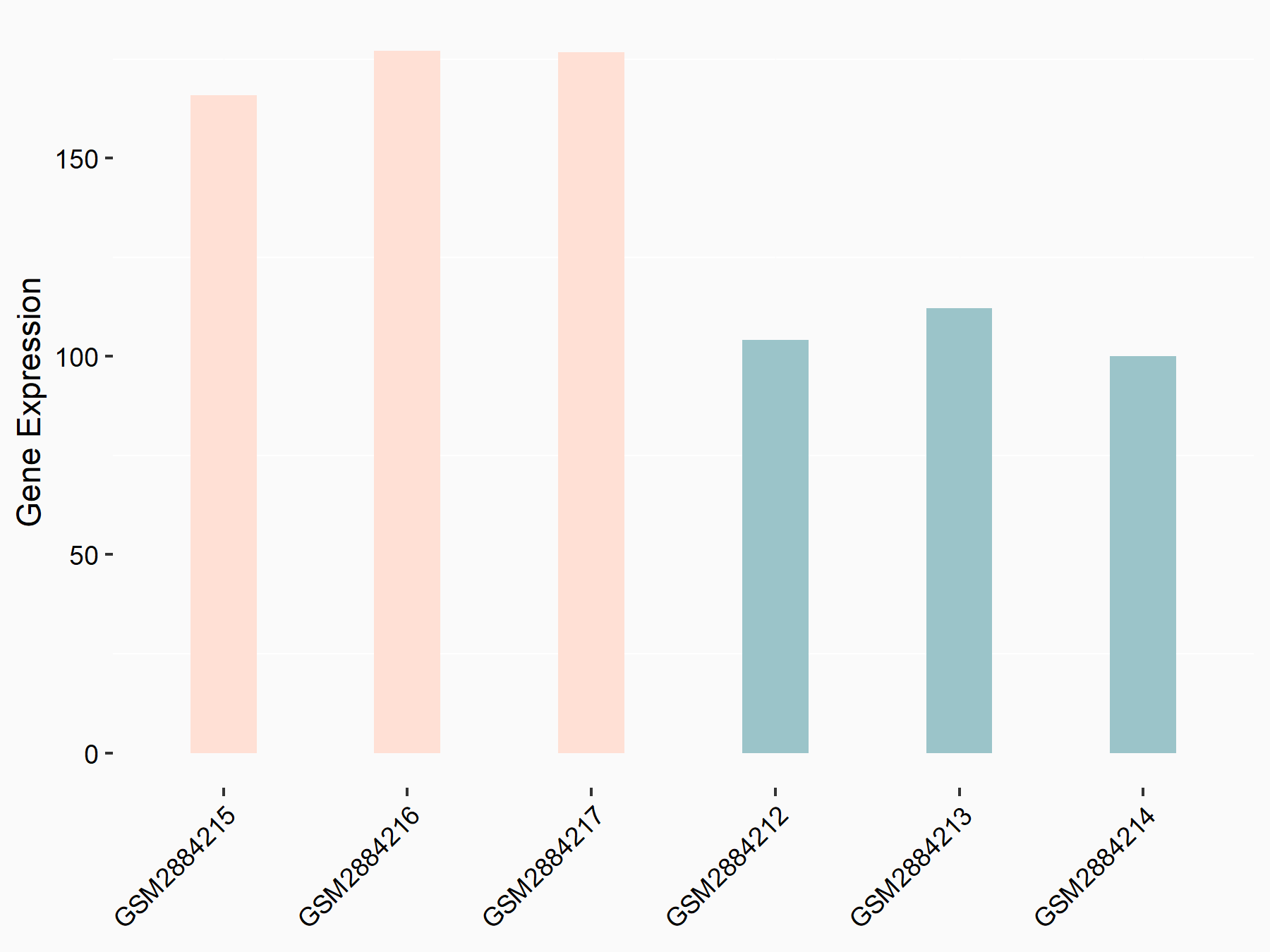 |
logFC: 7.11E-01 p-value: 1.04E-04 |
| More Results | Click to View More RNA-seq Results | |
Colorectal cancer [ICD-11: 2B91]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [32] | |||
| Responsed Disease | Colorectal cancer [ICD-11: 2B91] | |||
| Target Regulation | Down regulation | |||
In-vitro Model |
SW620 | Colon adenocarcinoma | Homo sapiens | CVCL_0547 |
| SW480 | Colon adenocarcinoma | Homo sapiens | CVCL_0546 | |
| NCM460 | Normal | Homo sapiens | CVCL_0460 | |
| HT29 | Colon cancer | Mus musculus | CVCL_A8EZ | |
| HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 | |
| In-vivo Model | For the xenograft model, METTL3 stable overexpressed SW620 cells (1 × 107) or control cells were subcutaneously injected into the right axilla of the female anesthetized BALB/C nude mice (4-6 weeks old, 18-20 g, four mice per group), respectively. The body weight and tumor volumes (length × width2 × 0.5) were measured twice a week. After 21 days, all mice were sacrificed and tumors were surgically removed for hematoxylin-eosin (H&E) staining.For the metastasis model, MTTL3 stable overexpressed SW620 cells (1 × 106) or control cells were injected into the exposed spleen of the anesthetized BALB/C nude mice, respectively. After 21 days, liver metastases were carefully detected using a fluorescent stereoscope and embedded for H&E staining. | |||
| Response Summary | METTL3-catalyzed m6A modification in CRC tumorigenesis, wherein it facilitates CRC tumor growth and metastasis through suppressing Protein yippee-like 5 (YPEL5) expression in an m6A-YTHDF2-dependent manner. | |||
Proto-oncogene c-Rel (c-Rel)
| Representative RNA-seq result indicating the expression of this target gene regulated by YTHDF2 | ||
| Cell Line | Mouse-cerebellum granule cell | Mus musculus |
|
Treatment: YTHDF2 knockdown mouse-cerebellum granule cell
Control: Wild type mouse-cerebellum granule cell
|
GSE153688 | |
| Regulation |
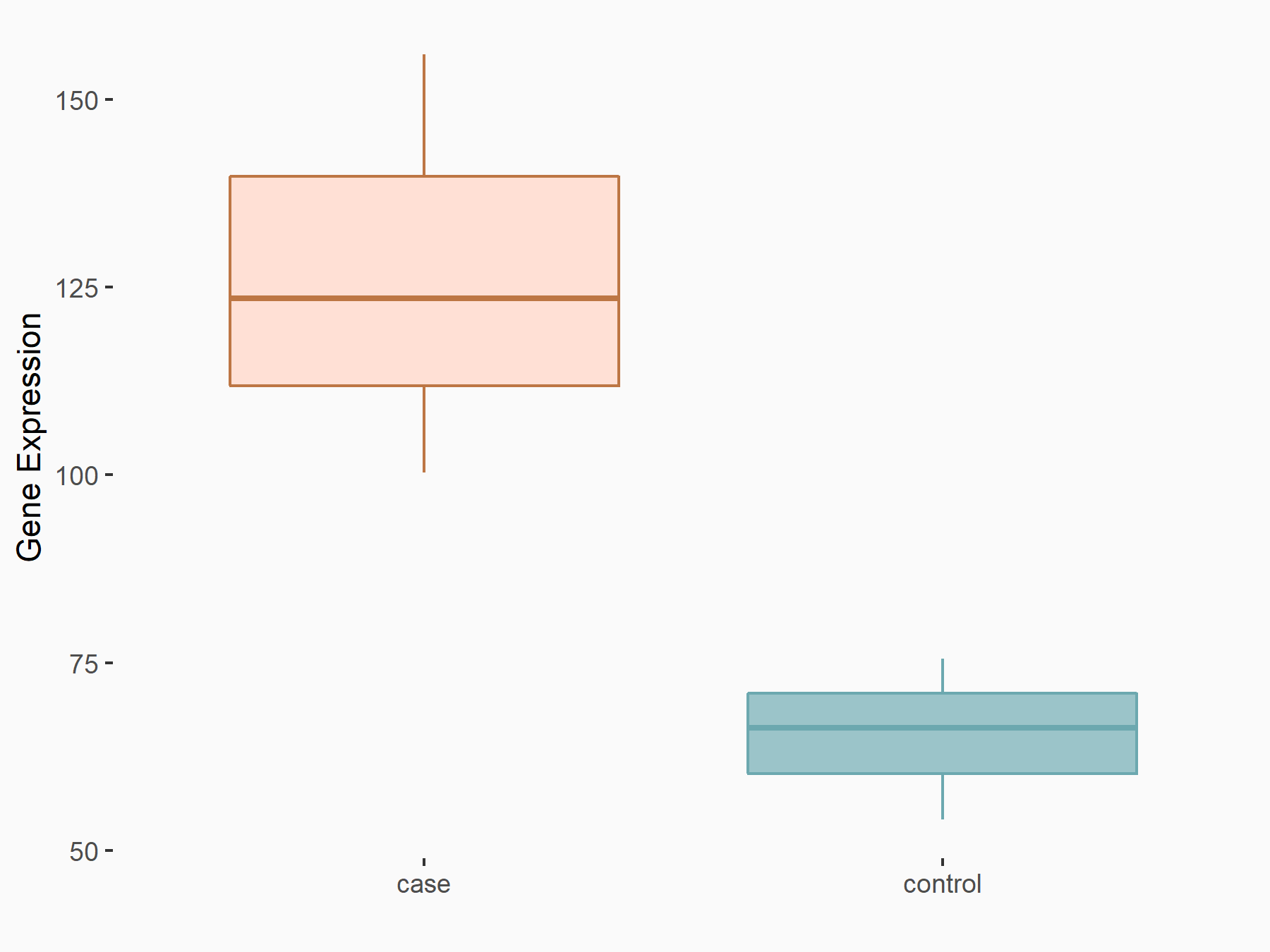  |
logFC: 9.51E-01 p-value: 8.19E-04 |
| More Results | Click to View More RNA-seq Results | |
Thyroid Cancer [ICD-11: 2D10]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [33] | |||
| Responsed Disease | Papillary thyroid cancer [ICD-11: 2D10.1] | |||
| Target Regulation | Down regulation | |||
| Pathway Response | NF-kappa B signaling pathway | hsa04064 | ||
In-vitro Model |
TPC-1 | Thyroid gland papillary carcinoma | Homo sapiens | CVCL_6298 |
| Nthy-ori 3-1 | Normal | Homo sapiens | CVCL_2659 | |
| KTC-1 | Thyroid carcinoma | Homo sapiens | CVCL_6300 | |
| B-CPAP | Thyroid gland carcinoma | Homo sapiens | CVCL_0153 | |
| In-vivo Model | For xenograft models, 5 × 106 BCPAP or KTC-1 cells from each group were injected subcutaneously into the flanks of female BALB/c nude mice (4-6 weeks old, Shanghai SLAC Laboratory Animal, China, n = 5 per group) in a volume of 150 uL PBS. Tumor growth was measured with a digital caliper every 4 days and calculated using the following formula: (length × width2)/2. To study the effect of IL-8 on tumor growth in vivo, scramble or shMETTL3 BCPAP cells were implanted hypodermically into BALB/c nude mice (2 × 106 cells in 150 uL PBS, n = 10 per group). When palpable tumors formed on day 14, mice were treated with DMSO or the IL-8 inhibitor SB225002 (10 mg/kg) by intraperitoneal injection 3 times per week for 3 weeks. Six weeks post-injection, the mice were sacrificed, and the tumors were collected to analyze the frequency of TANs by flow cytometry. For the lung metastasis model, BCPAP and KTC-1 cells (2 × 106 cells in 100 uL PBS) with the corresponding vectors were injected into the tail veins of BALB/c nude mice. Eight weeks after injection, the mice were euthanized, and metastatic lung nodules were analyzed (n = 5 for each group). | |||
| Response Summary | METTL3 played a pivotal tumor-suppressor role in papillary thyroid cancer carcinogenesis through Proto-oncogene c-Rel (c-Rel) and RelA inactivation of the nuclear factor Kappa-B (NF-Kappa-B) pathway by cooperating with YTHDF2 and altered TAN infiltration to regulate tumor growth. | |||
Signal transducer and activator of transcription 3 (STAT3)
| Representative RNA-seq result indicating the expression of this target gene regulated by YTHDF2 | ||
| Cell Line | GSC11 cell line | Homo sapiens |
|
Treatment: siYTHDF2 GSC11 cells
Control: siControl GSC11 cells
|
GSE142825 | |
| Regulation |
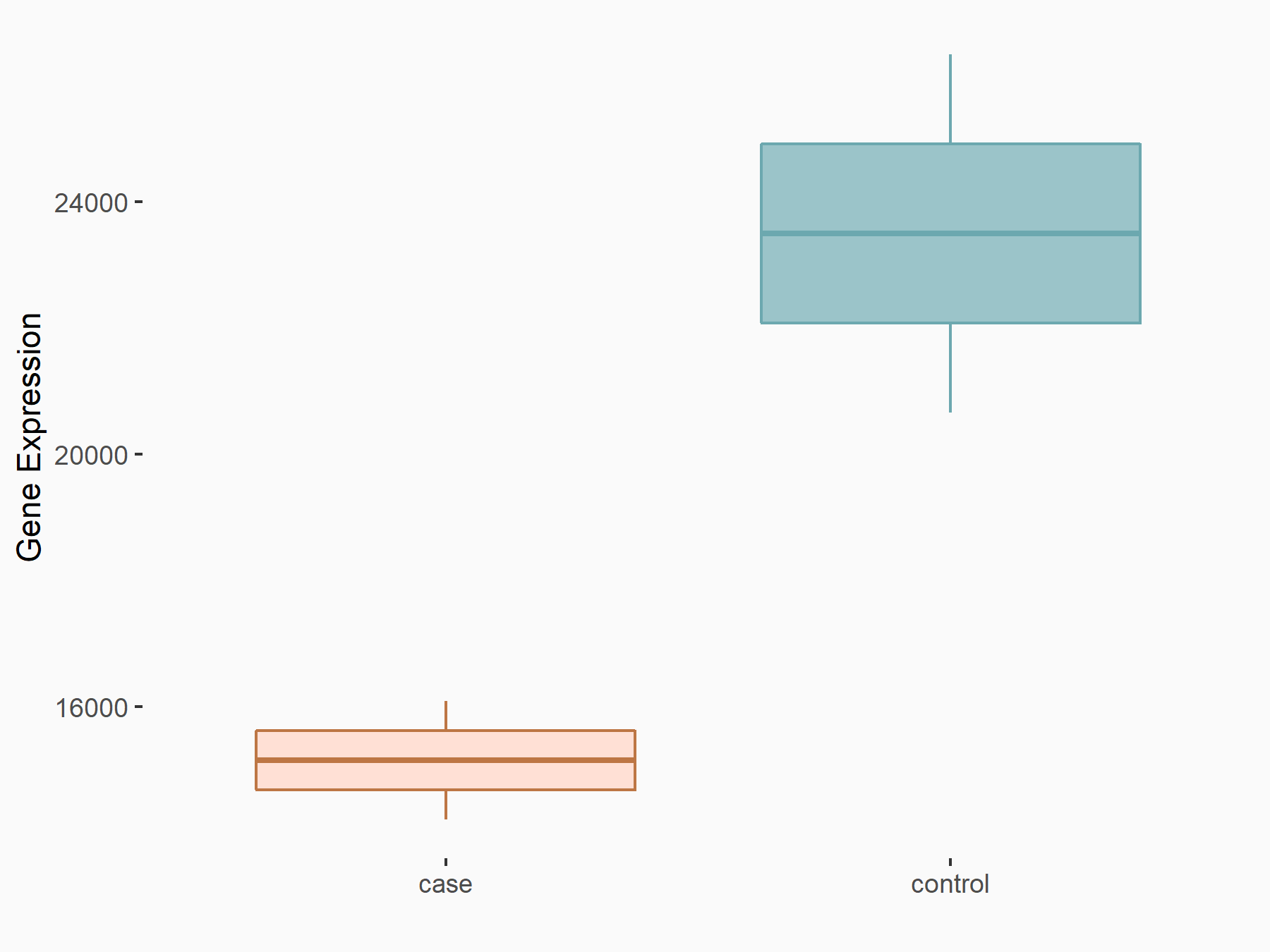  |
logFC: -6.33E-01 p-value: 6.04E-06 |
| More Results | Click to View More RNA-seq Results | |
Osteosarcoma [ICD-11: 2B51]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [16] | |||
| Responsed Disease | Osteosarcoma [ICD-11: 2B51] | |||
| Target Regulation | Down regulation | |||
| Pathway Response | JAK-STAT signaling pathway | hsa04630 | ||
In-vitro Model |
U2OS | Osteosarcoma | Homo sapiens | CVCL_0042 |
| OS3 [Human osteosarcoma] | Osteosarcoma | Homo sapiens | CVCL_F866 | |
| OS2 [Human osteosarcoma] | Osteosarcoma | Homo sapiens | CVCL_F865 | |
| OS1 [Human osteosarcoma] | Osteosarcoma | Homo sapiens | CVCL_F864 | |
| KHOS/NP | Osteosarcoma | Homo sapiens | CVCL_2546 | |
| Response Summary | ALKBH5 inactivated Signal transducer and activator of transcription 3 (STAT3) pathway by increasing SOCS3 expression via an m6A-YTHDF2-dependent manner. Reducing m6A mRNA levels in human osteosarcoma cells through ALKBH5 up-regulation lead to cell proliferation inhibition, cell apoptosis and cycle arrest. | |||
Transcription factor p65 (RELA)
| Representative RNA-seq result indicating the expression of this target gene regulated by YTHDF2 | ||
| Cell Line | GSC11 cell line | Homo sapiens |
|
Treatment: siYTHDF2 GSC11 cells
Control: siControl GSC11 cells
|
GSE142825 | |
| Regulation |
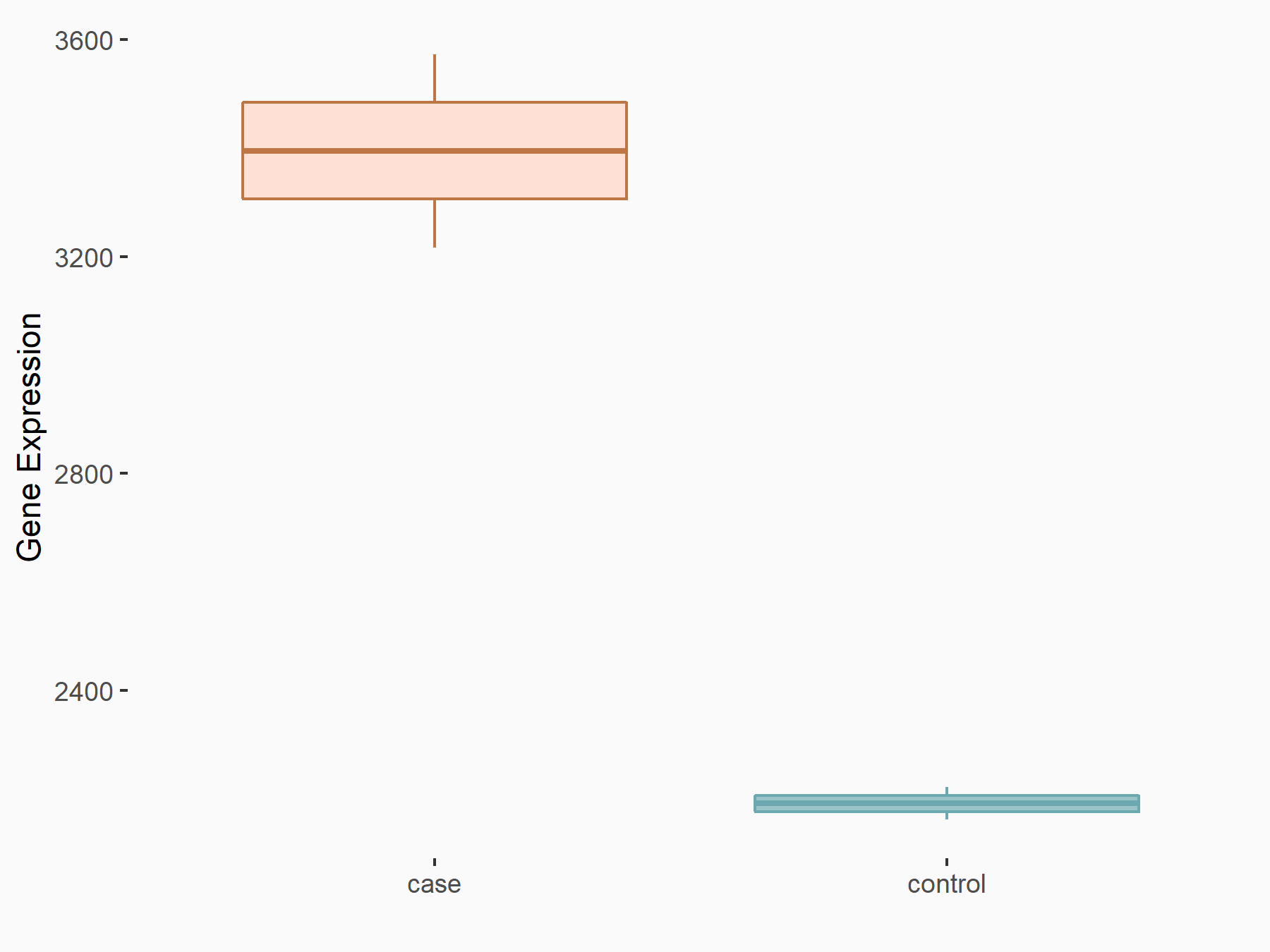 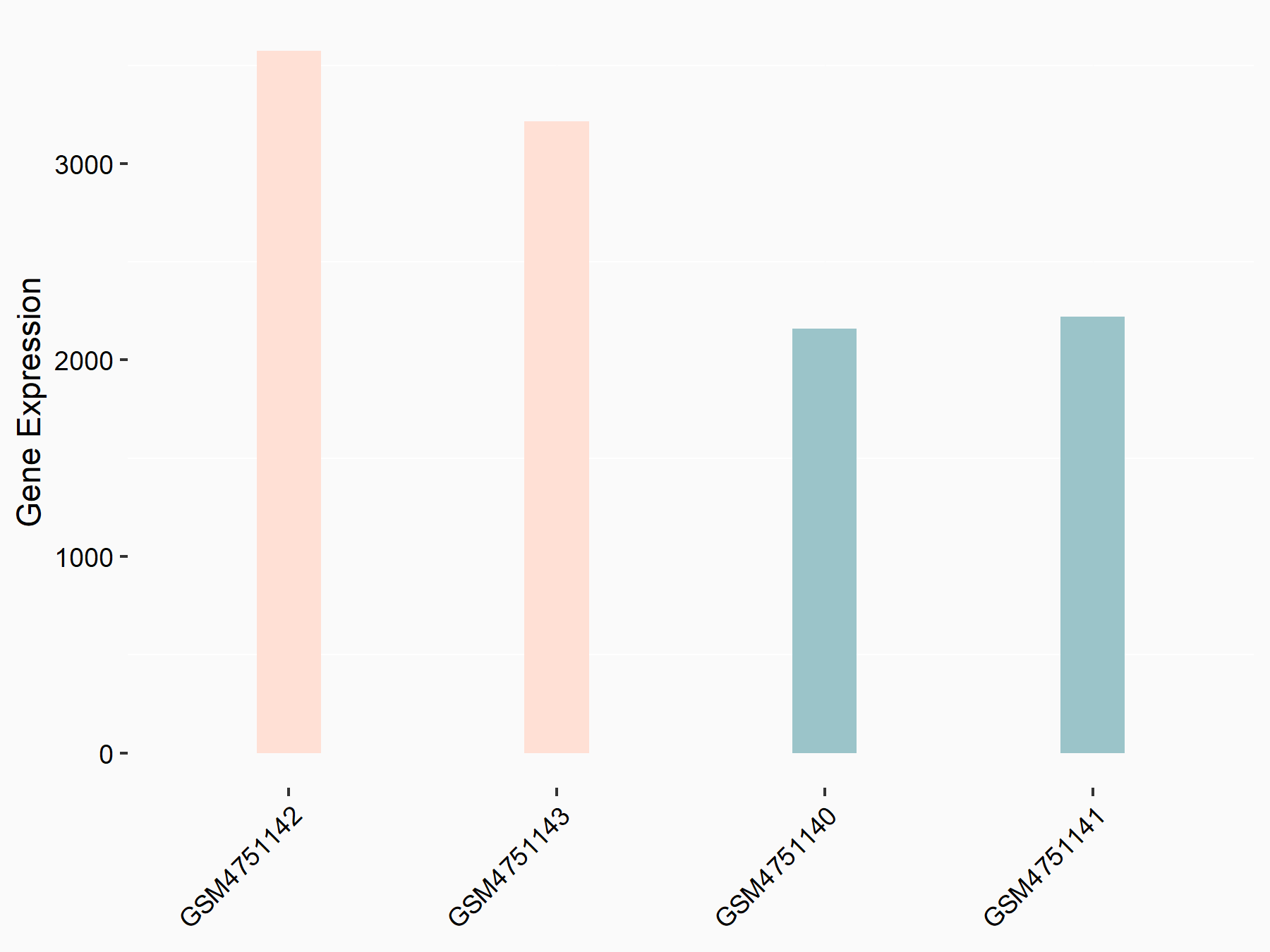 |
logFC: 6.31E-01 p-value: 1.53E-07 |
| More Results | Click to View More RNA-seq Results | |
Thyroid Cancer [ICD-11: 2D10]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [33] | |||
| Responsed Disease | Papillary thyroid cancer [ICD-11: 2D10.1] | |||
| Target Regulation | Down regulation | |||
| Pathway Response | NF-kappa B signaling pathway | hsa04064 | ||
In-vitro Model |
TPC-1 | Thyroid gland papillary carcinoma | Homo sapiens | CVCL_6298 |
| Nthy-ori 3-1 | Normal | Homo sapiens | CVCL_2659 | |
| KTC-1 | Thyroid carcinoma | Homo sapiens | CVCL_6300 | |
| B-CPAP | Thyroid gland carcinoma | Homo sapiens | CVCL_0153 | |
| In-vivo Model | For xenograft models, 5 × 106 BCPAP or KTC-1 cells from each group were injected subcutaneously into the flanks of female BALB/c nude mice (4-6 weeks old, Shanghai SLAC Laboratory Animal, China, n = 5 per group) in a volume of 150 uL PBS. Tumor growth was measured with a digital caliper every 4 days and calculated using the following formula: (length × width2)/2. To study the effect of IL-8 on tumor growth in vivo, scramble or shMETTL3 BCPAP cells were implanted hypodermically into BALB/c nude mice (2 × 106 cells in 150 uL PBS, n = 10 per group). When palpable tumors formed on day 14, mice were treated with DMSO or the IL-8 inhibitor SB225002 (10 mg/kg) by intraperitoneal injection 3 times per week for 3 weeks. Six weeks post-injection, the mice were sacrificed, and the tumors were collected to analyze the frequency of TANs by flow cytometry. For the lung metastasis model, BCPAP and KTC-1 cells (2 × 106 cells in 100 uL PBS) with the corresponding vectors were injected into the tail veins of BALB/c nude mice. Eight weeks after injection, the mice were euthanized, and metastatic lung nodules were analyzed (n = 5 for each group). | |||
| Response Summary | METTL3 played a pivotal tumor-suppressor role in papillary thyroid cancer carcinogenesis through c-Rel and Transcription factor p65 (RELA) inactivation of the nuclear factor Kappa-B (NF-Kappa-B) pathway by cooperating with YTHDF2 and altered TAN infiltration to regulate tumor growth. | |||
Deleted in lymphocytic leukemia 2 (DLEU2/LINC00022)
| Representative RNA-seq result indicating the expression of this target gene regulated by YTHDF2 | ||
| Cell Line | Human umbilical cord blood CD34+ cells | Homo sapiens |
|
Treatment: YTHDF2 knockdown UCB CD34+ cells
Control: Wild type UCB CD34+ cells
|
GSE107956 | |
| Regulation |
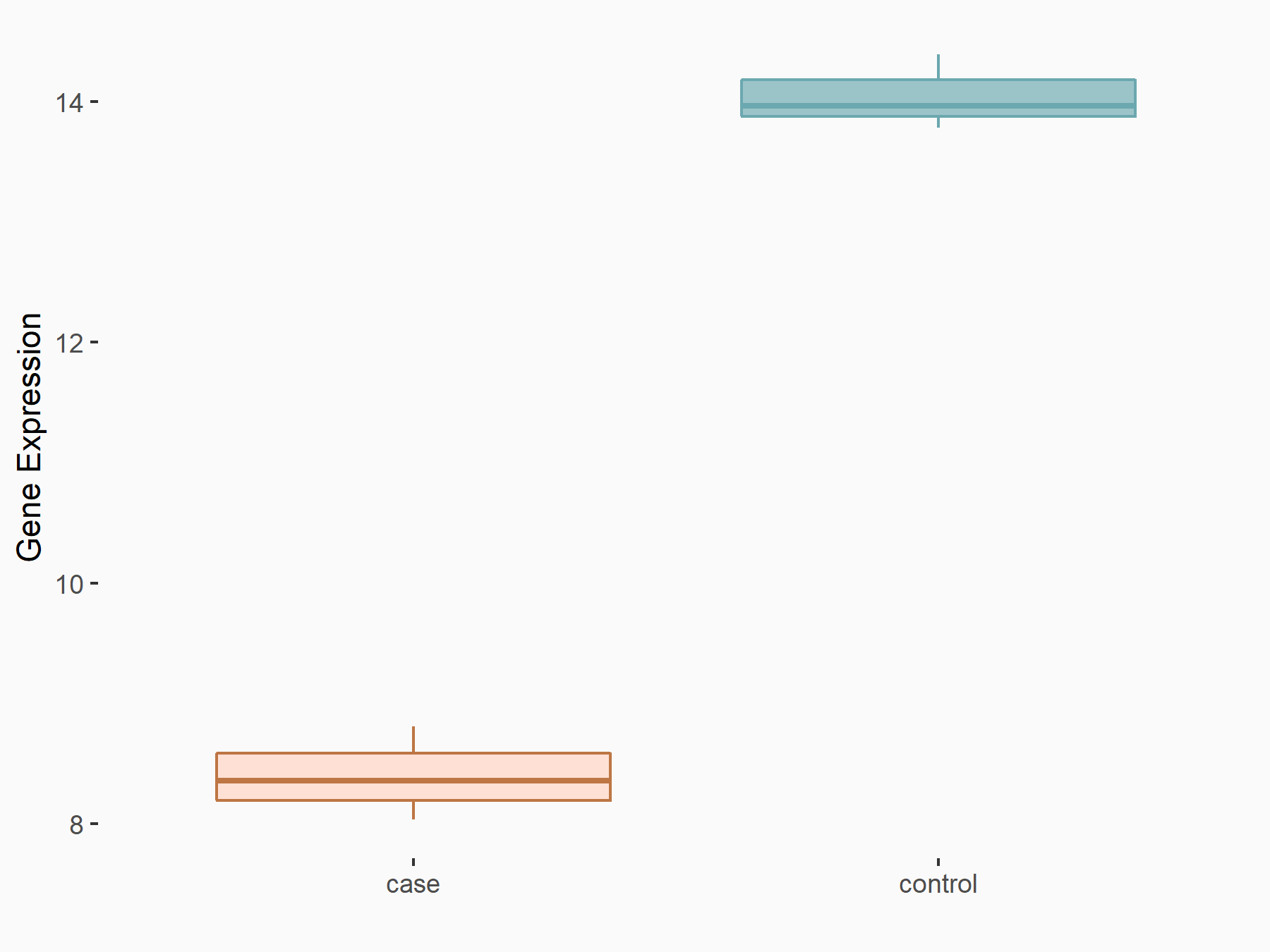 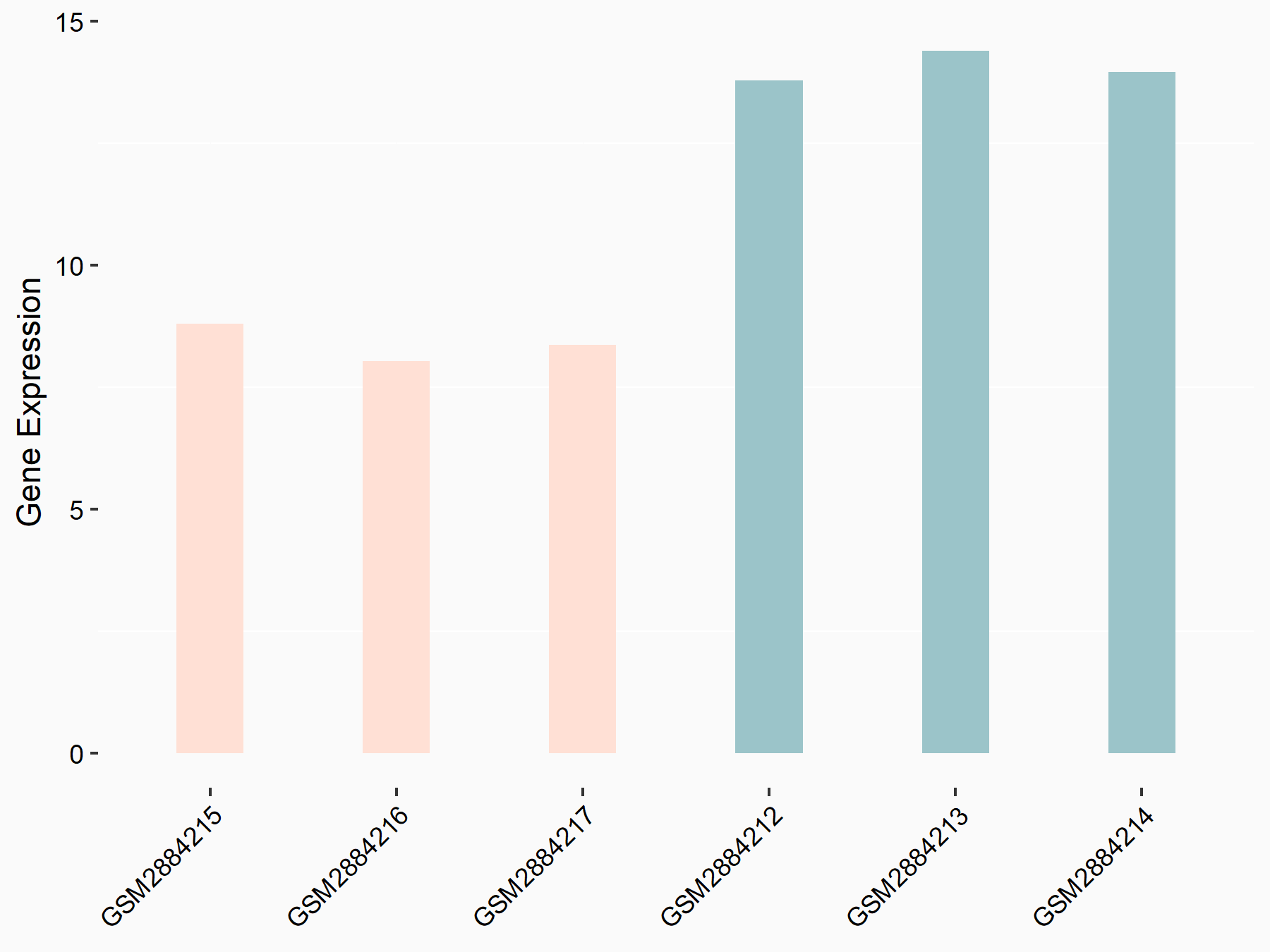 |
logFC: -6.80E-01 p-value: 2.37E-05 |
| More Results | Click to View More RNA-seq Results | |
Esophageal cancer [ICD-11: 2B70]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [34] | |||
| Responsed Disease | Esophageal squamous cell carcinoma [ICD-11: 2B70.1] | |||
| Target Regulation | Down regulation | |||
| Pathway Response | Cell cycle | hsa04110 | ||
| Ubiquitin mediated proteolysis | hsa04120 | |||
| Cell Process | Ubiquitination degradation | |||
| Cell apoptosis | ||||
| Decreased G0/G1 phase | ||||
In-vitro Model |
HET-1A | Normal | Homo sapiens | CVCL_3702 |
| KYSE-150 | Esophageal squamous cell carcinoma | Homo sapiens | CVCL_1348 | |
| KYSE-450 | Esophageal squamous cell carcinoma | Homo sapiens | CVCL_1353 | |
| KYSE-70 | Esophageal squamous cell carcinoma | Homo sapiens | CVCL_1356 | |
| TE-1 | Esophageal squamous cell carcinoma | Homo sapiens | CVCL_1759 | |
| In-vivo Model | The number of cells inoculated in each mouse was 4 × 106, 1 × 106, 2 × 106 and 1 × 106, respectively. | |||
| Response Summary | The elevated FTO in esophageal squamous cell carcinoma decreased m6A methylation of LINC00022 transcript, leading to the inhibition of Deleted in lymphocytic leukemia 2 (DLEU2/LINC00022) decay via the m6A reader YTHDF2. | |||
KCNQ1 opposite strand/antisense transcript 1 (KCNQ1OT1)
| Representative RNA-seq result indicating the expression of this target gene regulated by YTHDF2 | ||
| Cell Line | Human umbilical cord blood CD34+ cells | Homo sapiens |
|
Treatment: YTHDF2 knockdown UCB CD34+ cells
Control: Wild type UCB CD34+ cells
|
GSE107956 | |
| Regulation |
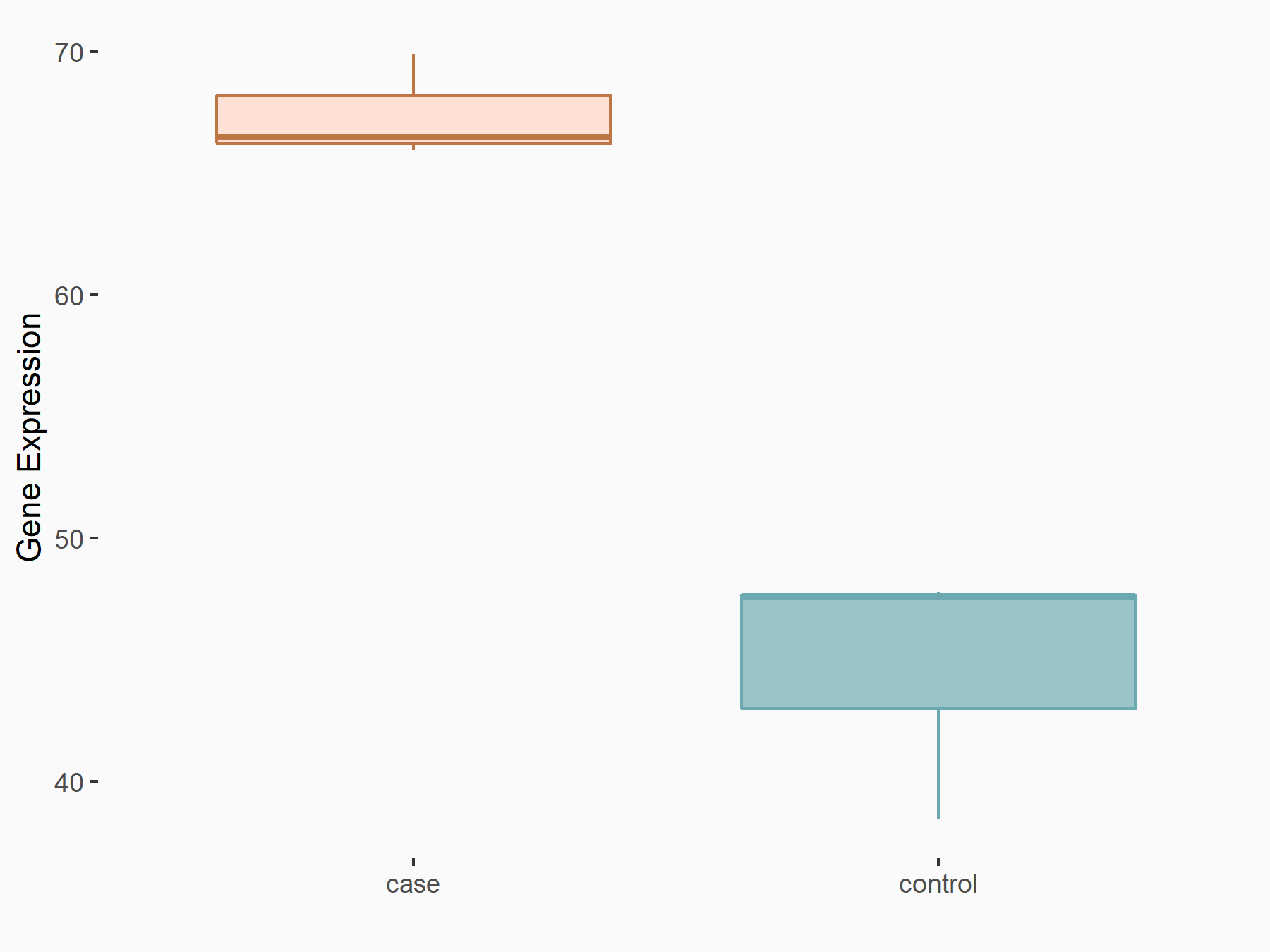 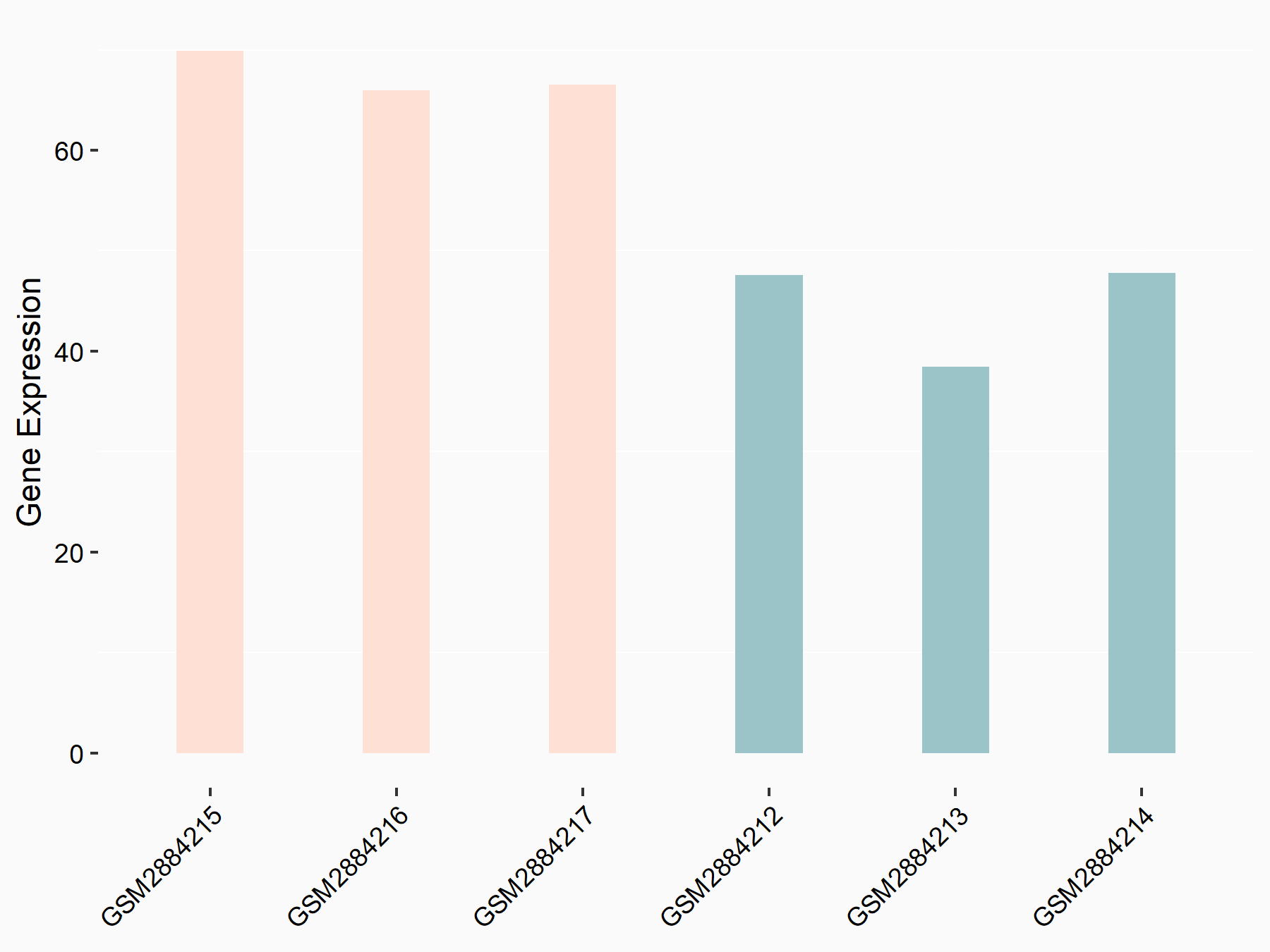 |
logFC: 5.92E-01 p-value: 2.94E-03 |
| More Results | Click to View More RNA-seq Results | |
Laryngeal cancer [ICD-11: 2C23]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [35] | |||
| Responsed Disease | Laryngeal cancer [ICD-11: 2C23] | |||
| Target Regulation | Up regulation | |||
| Cell Process | Cell proliferation and metastasis | |||
In-vitro Model |
AMC-HN-8 | Laryngeal squamous cell carcinoma | Homo sapiens | CVCL_5966 |
| HOK | Normal | Hexagrammos otakii | CVCL_YE19 | |
| Tu 212 | Head and neck squamous cell carcinoma | Homo sapiens | CVCL_4915 | |
| In-vivo Model | 1 × 106 (100 ul) cells of infected and uninfected by lentiviral were, respectively, injected subcutaneously into nude mice which divided randomly into scramble group and shKCNQ1OT1-1 group. | |||
| Response Summary | ALKBH5 mediates KCNQ1 opposite strand/antisense transcript 1 (KCNQ1OT1) expression via an m6A-YTHDF2-dependent manner and KCNQ1OT1 could directly bind to HOXA9 to further regulate the proliferation, invasion and metastasis of laryngeal squamous cell carcinoma cells. | |||
Pvt1 oncogene (PVT1)
| Representative RNA-seq result indicating the expression of this target gene regulated by YTHDF2 | ||
| Cell Line | Human umbilical cord blood CD34+ cells | Homo sapiens |
|
Treatment: YTHDF2 knockdown UCB CD34+ cells
Control: Wild type UCB CD34+ cells
|
GSE107956 | |
| Regulation |
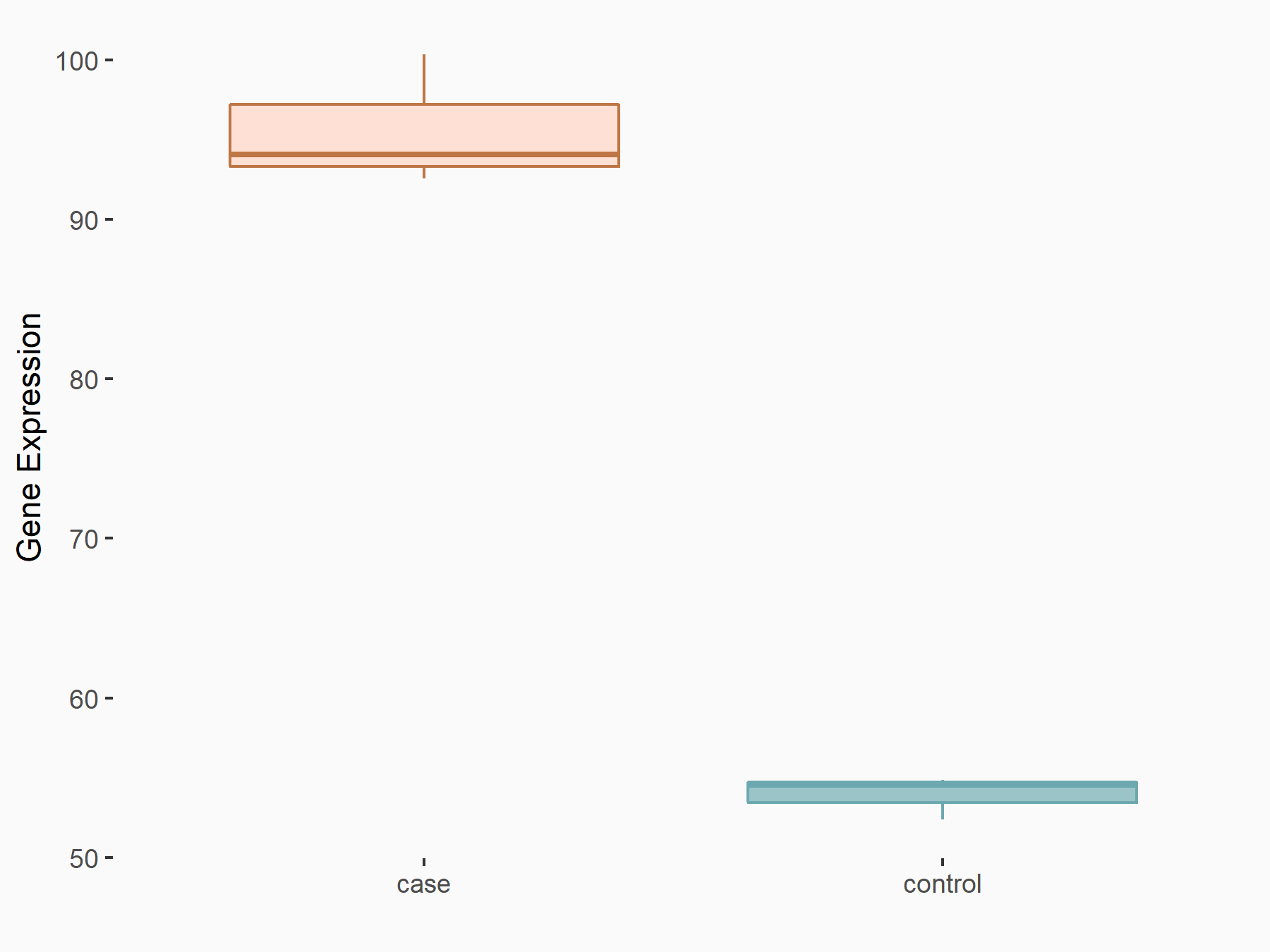 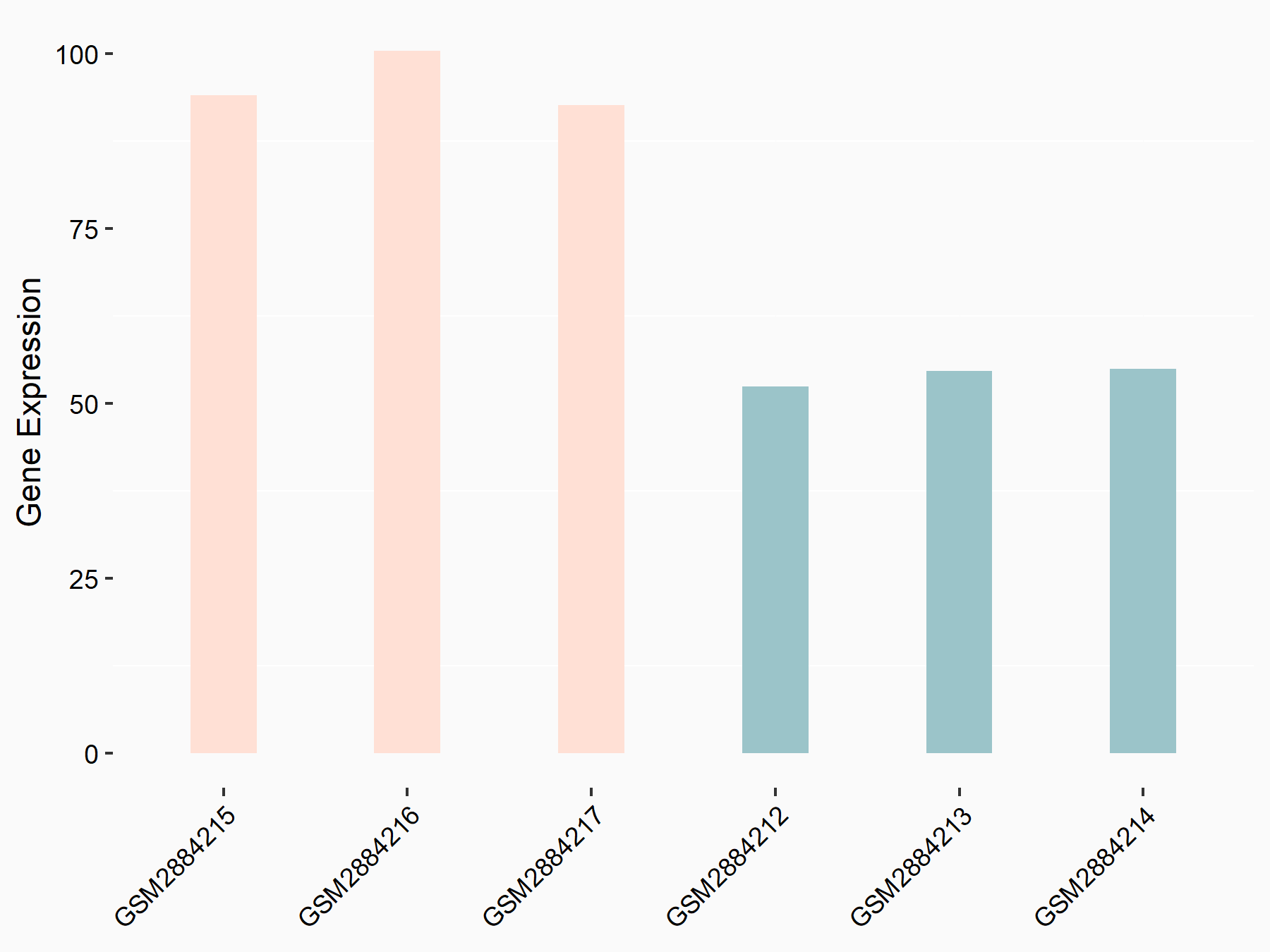 |
logFC: 8.15E-01 p-value: 1.40E-05 |
| More Results | Click to View More RNA-seq Results | |
Osteosarcoma [ICD-11: 2B51]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [36] | |||
| Responsed Disease | Osteosarcoma [ICD-11: 2B51] | |||
| Target Regulation | Up regulation | |||
| Cell Process | Cell proliferation | |||
| Cell migration | ||||
| Cell invasion | ||||
In-vitro Model |
143B | Osteosarcoma | Homo sapiens | CVCL_2270 |
| HOS | Osteosarcoma | Homo sapiens | CVCL_0312 | |
| SaOS-LM7 | Osteosarcoma | Homo sapiens | CVCL_0515 | |
| MG-63 | Osteosarcoma | Homo sapiens | CVCL_0426 | |
| NHOst (Normal human osteoblast cells) | ||||
| SaOS-2 | Osteosarcoma | Homo sapiens | CVCL_0548 | |
| U2OS | Osteosarcoma | Homo sapiens | CVCL_0042 | |
| In-vivo Model | Indicated stable 143B cells were subcutaneously injected into nude mice. | |||
| Response Summary | ALKBH5 decreased the m6A modification of Pvt1 oncogene (PVT1), thus inhibiting the binding of reader protein YTHDF2 in PVT1. ALKBH5-mediated PVT1 upregulation promoted the osteosarcoma cell proliferation in vitro and tumor growth in vivo. | |||
Diabetic encephalopathy [ICD-11: 8E47]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [49] | |||
| Responsed Disease | Diabetic encephalopathy [ICD-11: 8E47] | |||
Autophagy protein 5 (ATG5)
| Representative RIP-seq result supporting the interaction between the target gene and YTHDF2 | ||
| Cell Line | Hela | Homo sapiens |
| Regulation | logFC: 2.24E+00 | GSE49339 |
Obesity [ICD-11: 5B81]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [37] | |||
| Responsed Disease | Obesity [ICD-11: 5B81] | |||
| Target Regulation | Down regulation | |||
| Pathway Response | Autophagy | hsa04140 | ||
| Cell Process | Autophagy | |||
| Adipogenesis regulation | ||||
In-vitro Model |
3T3-L1 | Normal | Mus musculus | CVCL_0123 |
| Pig primary preadipocytes (Isolated from cervical subcutaneous adipose tissue of piglets) | ||||
| In-vivo Model | Mice were maintained at 22 ± 2 ℃ with a humidity of 35 ± 5% under a 12 h light and 12 h dark cycle, with free access to water and food. For the HFD experiment, female control (Ftoflox/flox) and adipose-selective fto knockout (Fabp4-Cre Ftoflox/flox, fto-AKO) mice were fed with high-fat diet (60% fat in calories; Research Diets, D12492) for the desired periods of time, and food intake and body weight were measured every week after weaning (at 3 weeks of age). | |||
| Response Summary | Autophagy protein 5 (ATG5) and Atg7 were the targets of YTHDF2 (YTH N6-methyladenosine RNA binding protein 2). Upon FTO silencing, Atg5 and Atg7 transcripts with higher m6A levels were captured by YTHDF2, which resulted in mRNA degradation and reduction of protein expression, thus alleviating autophagy and adipogenesis. | |||
Osteoarthritis [ICD-11: FA05]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [47] | |||
| Responsed Disease | Osteoarthritis [ICD-11: FA05] | |||
Epidermal growth factor receptor (EGFR)
| Representative RIP-seq result supporting the interaction between the target gene and YTHDF2 | ||
| Cell Line | Hela | Homo sapiens |
| Regulation | logFC: 1.61E+00 | GSE49339 |
Liver cancer [ICD-11: 2C12]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [38] | |||
| Responsed Disease | Hepatocellular carcinoma [ICD-11: 2C12.02] | |||
| Target Regulation | Down regulation | |||
| Pathway Response | MAPK signaling pathway | hsa04010 | ||
| Cell Process | Glucose metabolism | |||
In-vitro Model |
BEL-7402 | Endocervical adenocarcinoma | Homo sapiens | CVCL_5492 |
| Hep 3B2.1-7 | Childhood hepatocellular carcinoma | Homo sapiens | CVCL_0326 | |
| QGY-7703 | Endocervical adenocarcinoma | Homo sapiens | CVCL_6715 | |
| SMMC-7721 | Endocervical adenocarcinoma | Homo sapiens | CVCL_0534 | |
| In-vivo Model | 5 × 106 of HEP3B and SMMC7721 stable cells were resuspended in 0.1 ml of PBS and subcutaneously injected into the flank of mice. | |||
| Response Summary | YTHDF2 acts as a tumor suppressor to repress cell proliferation and growth via destabilizing the Epidermal growth factor receptor (EGFR) mRNA in HCC. | |||
Insulin receptor substrate 1 (IRS1)
| Representative RIP-seq result supporting the interaction between the target gene and YTHDF2 | ||
| Cell Line | Hela | Homo sapiens |
| Regulation | logFC: 1.86E+00 | GSE49339 |
Endometrial cancer [ICD-11: 2C76]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [39] | |||
| Responsed Disease | Endometrial cancer [ICD-11: 2C76] | |||
| Target Regulation | Down regulation | |||
| Pathway Response | PI3K-Akt signaling pathway | hsa04151 | ||
In-vitro Model |
T HESCs | Normal | Homo sapiens | CVCL_C464 |
| RL95-2 | Endometrial adenosquamous carcinoma | Homo sapiens | CVCL_0505 | |
| HEC-1-A | Endometrial adenocarcinoma | Homo sapiens | CVCL_0293 | |
| Response Summary | YTHDF2 inhibited the proliferation and invasion of endometrial cancer(EC) via inhibiting Insulin receptor substrate 1 (IRS1) expression in m6A epigenetic way, which suggests a potential therapeutic target for EC. | |||
MAP kinase kinase 4 (MAP2K4)
| Representative RIP-seq result supporting the interaction between the target gene and YTHDF2 | ||
| Cell Line | Hela | Homo sapiens |
| Regulation | logFC: 2.75E+00 | GSE49339 |
Diseases of the circulatory system [ICD-11: BE2Z]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [40] | |||
| Responsed Disease | Diseases of the circulatory system [ICD-11: BE2Z] | |||
| Response Summary | YTHDF2 regulates the stability of MAP kinase kinase 4 (MAP2K4) and MAP4K4 mRNAs.This study identified that dasatinib and quercetin alleviate LPS-induced senescence in HUVECs via the TRAF6-MAPK-NF-Kappa-B axis in a YTHDF2-dependent manner, providing novel ideas for clinical treatment of age-related cardiovascular diseases. | |||
Inflammatory response [ICD-11: MG46]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [41] | |||
| Responsed Disease | Inflammatory response [ICD-11: MG46] | |||
| Target Regulation | Down regulation | |||
| Pathway Response | MAPK signaling pathway | hsa04010 | ||
In-vitro Model |
RAW 264.7 | Mouse leukemia | Mus musculus | CVCL_0493 |
| Response Summary | YTHDF2 knockdown increases mRNA expression levels of MAP kinase kinase 4 (MAP2K4) and MAP4K4 via stabilizing the mRNA transcripts, which activate MAPK and NF-Kappa-B signaling pathways, which promote the expression of proinflammatory cytokines and aggravate the inflammatory response in LPS-stimulated RAW 264.7 cells. | |||
PI3-kinase subunit beta (PIK3CB)
| Representative RIP-seq result supporting the interaction between the target gene and YTHDF2 | ||
| Cell Line | Hela | Homo sapiens |
| Regulation | logFC: 1.28E+00 | GSE49339 |
Pancreatic cancer [ICD-11: 2C10]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [42] | |||
| Responsed Disease | Pancreatic cancer [ICD-11: 2C10] | |||
| Responsed Drug | AZD6482 | Terminated | ||
| Target Regulation | Down regulation | |||
| Pathway Response | PI3K-Akt signaling pathway | hsa04151 | ||
| Glycolysis / Gluconeogenesis | hsa00010 | |||
| Cell Process | Glucose metabolism | |||
In-vitro Model |
BxPC-3 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0186 |
| PANC-1 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0480 | |
| In-vivo Model | Established cohorts of mice bearing tumour xenografts driven by PTEN-deficient BxPC-3 and PANC-1 cells with PIK3CB overexpression. When tumours grew to ~300 mm3, mice were grouped and administered with vehicle (DMSO) or KIN-193 via intraperitoneal injection (20 mg/kg) once daily. | |||
| Response Summary | N6-methyladenosine mRNA methylation of PIK3CB regulates AKT signalling to promote PTEN-deficient pancreatic cancer progression. Rs142933486 is significantly associated with the overall survival of PDAC by reducing the PIK3CB m6A level, which facilitated its mRNA and protein expression levels mediated by the m6A 'writer' complex (METTL13/METTL14/WTAP) and the m6A 'reader' YTHDF2. KIN-193, a PI3-kinase subunit beta (PIK3CB)-selective inhibitor, is shown to serve as an effective anticancer agent for blocking PTEN-deficient PDAC. | |||
T-cell acute lymphocytic leukemia protein 1 (TAL1)
| Representative RIP-seq result supporting the interaction between the target gene and YTHDF2 | ||
| Cell Line | Hela | Homo sapiens |
| Regulation | logFC: 2.17E+00 | GSE49339 |
Hematological disorders [ICD-11: 3C0Z]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [43] | |||
| Responsed Disease | Hematological disorders [ICD-11: 3C0Z] | |||
| Target Regulation | Down regulation | |||
| Cell Process | RNA stability | |||
In-vitro Model |
Hematopoietic stem cells (Hematopoietic stem cells) | |||
| In-vivo Model | For the rescue experiment, LSK cells were sorted from wt and Ythdf2 KO mouse BM and cultured overnight in StemSpan SFEM medium (Stem Cell Technologies) supplemented with 10 ug/mL heparin (Sigma), 0.5 × penicillin/streptomycin (Sigma), 10 ng/mL recombinant mouse (rm) stem cell factor (SCF), and 20 ng/mL Tpo 75 at 37 ℃ in a 5% CO2 5% O2 atmosphere. | |||
| Response Summary | Knocking down one of YTHDF2's key targets, T-cell acute lymphocytic leukemia protein 1 (TAL1) mRNA, partially rescued the phenotype.the function of YTHDF2 in adult stem cell maintenance and identifies its important role in regulating HSC ex vivo expansion. | |||
TNF alpha-induced protein 3 (TNFAIP3)
| Representative RIP-seq result supporting the interaction between the target gene and YTHDF2 | ||
| Cell Line | Hela | Homo sapiens |
| Regulation | logFC: 1.94E+00 | GSE49339 |
Brain cancer [ICD-11: 2A00]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [4] | |||
| Responsed Disease | Glioblastoma [ICD-11: 2A00.00] | |||
| Responsed Drug | Temozolomide | Approved | ||
| Target Regulation | Down regulation | |||
| Pathway Response | PI3K-Akt signaling pathway | hsa04151 | ||
| Cell Process | RNA stability | |||
In-vitro Model |
T98G | Glioblastoma | Homo sapiens | CVCL_0556 |
| LN-229 | Glioblastoma | Homo sapiens | CVCL_0393 | |
| In-vivo Model | 5 × 106 infected T98G cells (LV-NC or LV-YTHDF2) were injected into the flanks of mice through subcutaneous. | |||
| Response Summary | YTHDF2 enhanced TMZ resistance in GBM by activation of the PI3K/Akt and NF-Kappa-B signalling pathways via inhibition of EPHB3 and TNF alpha-induced protein 3 (TNFAIP3). | |||
Lupus erythematosus [ICD-11: 4A40]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [44] | |||
| Responsed Disease | Lupus erythematosus [ICD-11: 4A40] | |||
| Target Regulation | Up regulation | |||
| Cell Process | Immunity | |||
In-vitro Model |
PBMCs (Human peripheral blood mononuclear cells (PBMCs) are isolated from peripheral blood and identified as any blood cell with a round nucleus) | |||
| Response Summary | MiR-19a regulated TNF alpha-induced protein 3 (TNFAIP3) degradation by downregulating the expression of YTH N6-methyladenosine RNA-binding protein 2 (YTHDF2). The circGARS sponges miR-19a to regulate YTHDF2 expression to promote SLE progression through the A20/NF-Kappa-B axis and acts as an independent biomarker to help the treatment of SLE patients. | |||
FOXF1 adjacent non-coding developmental regulatory RNA (FENDRR)
| Representative RIP-seq result supporting the interaction between the target gene and YTHDF2 | ||
| Cell Line | Hela | Homo sapiens |
| Regulation | logFC: 1.55E+00 | GSE49339 |
Endometrial cancer [ICD-11: 2C76]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [19] | |||
| Responsed Disease | Endometrial cancer [ICD-11: 2C76] | |||
| Target Regulation | Down regulation | |||
| Cell Process | Cell proliferation | |||
| Cell apoptosis | ||||
In-vitro Model |
HEC-1-B | Endometrial adenocarcinoma | Homo sapiens | CVCL_0294 |
| Response Summary | YTHDF2-mediated LncRNA FOXF1 adjacent non-coding developmental regulatory RNA (FENDRR) degradation promotes cell proliferation by elevating SOX4 expression in endometrioid endometrial carcinoma. | |||
6-phosphogluconate dehydrogenase, decarboxylating (6PGD/PGD)
Lung cancer [ICD-11: 2C25]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [45] | |||
| Responsed Disease | Lung cancer [ICD-11: 2C25] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | Pentose phosphate pathway | hsa00030 | ||
| Cell Process | Cell growth | |||
In-vitro Model |
A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 |
| NCI-H1299 | Lung large cell carcinoma | Homo sapiens | CVCL_0060 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| Response Summary | YTHDF2 directly binds to the m6A modification site of 6-phosphogluconate dehydrogenase, decarboxylating (6PGD/PGD) three prime untranslated region (3'-UTR) to promote 6PGD mRNA translation in lung cancer cells. | |||
ATP-binding cassette sub-family C member 10 (ABCC10)
Lung cancer [ICD-11: 2C25]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [46] | |||
| Responsed Disease | Non-small-cell lung carcinoma [ICD-11: 2C25.Y] | |||
| Responsed Drug | Gefitinib | Approved | ||
| Target Regulation | Down regulation | |||
| Pathway Response | ABC transporters | hsa02010 | ||
In-vitro Model |
PC-9 | Lung adenocarcinoma | Homo sapiens | CVCL_B260 |
| NCI-H1975 | Lung adenocarcinoma | Homo sapiens | CVCL_1511 | |
| In-vivo Model | Mice were randomized into three groups (n = 7/group), 1 × 107 PC9 cells absorbed exosomes were subcutaneously injected into the Bilateral groin of mice. Treatment began 1 week following injection, the mice were intraperitoneally injected with gefitinib (30 mg/kg/day). | |||
| Response Summary | Not only FTO knockdown enhanced the gefitinib sensitivity of GR cells but also FTO reduction in donor exosomes alleviated the acquired resistance of recipient non-small cell lung cancer PC9 cells. FTO/YTHDF2/ATP-binding cassette sub-family C member 10 (ABCC10) axis played a role in intercellular transmission of GR cell-derived exosome-mediated gefitinib resistance. | |||
Axin-1 (AXIN1)
Cervical cancer [ICD-11: 2C77]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [48] | |||
| Responsed Disease | Cervical cancer [ICD-11: 2C77] | |||
| Responsed Drug | Cisplatin | Approved | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Wnt signaling pathway | hsa04310 | ||
| Cell Process | Epithelial-mesenchymal transition | |||
In-vitro Model |
SiHa | Cervical squamous cell carcinoma | Homo sapiens | CVCL_0032 |
| HeLa | Endocervical adenocarcinoma | Homo sapiens | CVCL_0030 | |
| Ect1/E6E7 | Normal | Homo sapiens | CVCL_3679 | |
| Ca Ski | Cervical squamous cell carcinoma | Homo sapiens | CVCL_1100 | |
| C-33 A | Cervical squamous cell carcinoma | Homo sapiens | CVCL_1094 | |
| Response Summary | YTHDF2 interference could suppress the EMT of cervical cancer cells and enhance cisplatin chemosensitivity by regulating Axin-1 (AXIN1). | |||
Parkinson disease [ICD-11: 8A00]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [49] | |||
| Responsed Disease | Parkinson disease [ICD-11: 8A00] | |||
Bax inhibitor 1 (TMBIM6)
Pre-eclampsia [ICD-11: JA24]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [50] | |||
| Responsed Disease | Pre-eclampsia [ICD-11: JA24] | |||
| Target Regulation | Down regulation | |||
In-vitro Model |
HTR-8/SVneo | Normal | Homo sapiens | CVCL_7162 |
BCL2/adenovirus E1B 19 kDa protein-interacting protein 3 (BNIP3)
Sepsis [ICD-11: 1G40]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [52] | |||
| Responsed Disease | Sepsis [ICD-11: 1G40] | |||
| In-vivo Model | To assess mortality rates, mice were intraperitoneally administered LPS (30 mg/kg), with mdivi (3 mg/kg) given intraperitoneally 1 h before LPS challenge and then continued for 3 consecutive days. The 72-h mortality was subsequently recorded. For evaluating heart injury, mice received an injection of LPS (10 mg/kg), with songorine (10 or 50 mg/kg) administered 1 h before and 12 h after LPS treatment. Mice were euthanized 24 h later for heart collection. | |||
Osteoarthritis [ICD-11: FA05]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [47] | |||
| Responsed Disease | Osteoarthritis [ICD-11: FA05] | |||
Injury of heart [ICD-11: NB31]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [53] | |||
| Responsed Disease | Myocardial injury [ICD-11: NB31.Z] | |||
| Target Regulation | Down regulation | |||
In-vitro Model |
H9c2(2-1) | Normal | Rattus norvegicus | CVCL_0286 |
| In-vivo Model | All rats were arranged into 4 groups, with 10 rats in each group: sham, IR, IR + adeno-associated virus-mediated YTHDF2 overexpression vector (AAV-YTHDF2), and IR + negative control of AAV-YTHDF2 (AAV-NC) groups. Rats in the sham group underwent the same procedure without ligation. Rats in the IR group experienced 30-min ischemia and then 24-h reperfusion. | |||
Calcium/calmodulin-dependent protein kinase kinase 2 (CAMKK2)
Male infertility [ICD-11: GB04]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [54] | |||
| Responsed Disease | Azoospermia [ICD-11: GB04.0] | |||
| Target Regulation | Down regulation | |||
| Pathway Response | Autophagy | hsa04140 | ||
| Cell Process | RNA stability | |||
| Cell autophagy | ||||
In-vitro Model |
TM3 | Normal | Mus musculus | CVCL_4326 |
| In-vivo Model | Male SPF BALB/c mice (qls02-0202) were purchased from Qinglongshan animal breeding farm. Mice were sacrificed by CO2 asphyxiation and testes were obtained for following histopathological analyses. | |||
| Response Summary | m6A modification promoted translation of PPM1A (protein phosphatase 1A, magnesium dependent, alpha isoform), a negative AMP-activated protein kinase (AMPK) regulator, but decreased expression of Calcium/calmodulin-dependent protein kinase kinase 2 (CAMKK2) (calcium/calmodulin-dependent protein kinase kinase 2, beta), a positive AMPK regulator, by reducing its RNA stability. Similar regulation of METTL14, ALKBH5, and m6A was also observed in LCs upon treatment with human chorionic gonadotropin (HsCG). Knock down of YTHDF1 failed to change the expression of CAMKK2 Providing insight into novel therapeutic strategies by exploiting m6A RNA methylation as targets for treating azoospermatism and oligospermatism patients with reduction in serum testosterone. knock down of YTHDF2 increased expression of Camkk2 at both mRNA and protein levels to a similar extent as knock down of METTL14 in both TM3 cells and primary LCs. | |||
Cellular tumor antigen p53 (TP53/p53)
Melanoma of uvea [ICD-11: 2D0Y]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [55] | |||
| Responsed Disease | Melanoma of uvea [ICD-11: 2D0Y] | |||
In-vitro Model |
OCM-1 | Amelanotic melanoma | Homo sapiens | CVCL_6934 |
| CRMM-1 | Conjunctival melanoma | Homo sapiens | CVCL_M593 | |
Cyclic AMP-dependent transcription factor ATF-4 (ATF4)
Colorectal cancer [ICD-11: 2B91]
| In total 6 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [56] | |||
| Responsed Disease | Colorectal cancer [ICD-11: 2B91] | |||
| Responsed Drug | Asparagine inhibitor | Approved | ||
| Target Regulation | Down regulation | |||
| Pathway Response | mTOR signaling pathway | hsa04150 | ||
| Cell Process | RNA decay | |||
| Cell growth and death | ||||
| Cell autophagy | ||||
In-vitro Model |
HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 |
| SW480 | Colon adenocarcinoma | Homo sapiens | CVCL_0546 | |
| Response Summary | In colorectal cancer, Glutaminolysis inhibition upregulated ATF4 expression in an m6A-dependent manner to activate pro-survival autophagy through transcriptional activation of the mTOR inhibitor DDIT4. Determined the relationship between FTO alpha-ketoglutarate dependent dioxygenase (FTO), YTH N6-methyladenosine RNA binding protein 2 (YTHDF2), and ATF4. ATF4 transcriptionally upregulated DDIT4 to suppress Cyclic AMP-dependent transcription factor ATF-4 (ATF4), which induced pro-survival autophagy during glutaminolysis inhibition. | |||
| Experiment 2 Reporting the m6A-centered Disease Response of This Target Gene | [56] | |||
| Responsed Disease | Colorectal cancer [ICD-11: 2B91] | |||
| Responsed Drug | Chloroquine | Approved | ||
| Target Regulation | Down regulation | |||
| Pathway Response | mTOR signaling pathway | hsa04150 | ||
| Cell Process | RNA decay | |||
| Cell growth and death | ||||
| Cell autophagy | ||||
In-vitro Model |
HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 |
| SW480 | Colon adenocarcinoma | Homo sapiens | CVCL_0546 | |
| Response Summary | In colorectal cancer, Glutaminolysis inhibition upregulated ATF4 expression in an m6A-dependent manner to activate pro-survival autophagy through transcriptional activation of the mTOR inhibitor DDIT4. Determined the relationship between FTO alpha-ketoglutarate dependent dioxygenase (FTO), YTH N6-methyladenosine RNA binding protein 2 (YTHDF2), and ATF4. Cyclic AMP-dependent transcription factor ATF-4 (ATF4) transcriptionally upregulated DDIT4 to suppress mTOR, which induced pro-survival autophagy during glutaminolysis inhibition. | |||
| Experiment 3 Reporting the m6A-centered Disease Response of This Target Gene | [56] | |||
| Responsed Disease | Colorectal cancer [ICD-11: 2B91] | |||
| Responsed Drug | Meclofenamate sodium | Approved | ||
| Target Regulation | Down regulation | |||
| Pathway Response | mTOR signaling pathway | hsa04150 | ||
| Cell Process | RNA decay | |||
| Cell growth and death | ||||
| Cell autophagy | ||||
In-vitro Model |
HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 |
| SW480 | Colon adenocarcinoma | Homo sapiens | CVCL_0546 | |
| Response Summary | In colorectal cancer, Glutaminolysis inhibition upregulated ATF4 expression in an m6A-dependent manner to activate pro-survival autophagy through transcriptional activation of the mTOR inhibitor DDIT4. Determined the relationship between FTO alpha-ketoglutarate dependent dioxygenase (FTO), YTH N6-methyladenosine RNA binding protein 2 (YTHDF2), and ATF4. Cyclic AMP-dependent transcription factor ATF-4 (ATF4) transcriptionally upregulated DDIT4 to suppress mTOR, which induced pro-survival autophagy during glutaminolysis inhibition. | |||
| Experiment 4 Reporting the m6A-centered Disease Response of This Target Gene | [56] | |||
| Responsed Disease | Colorectal cancer [ICD-11: 2B91] | |||
| Responsed Drug | Rapamycin | Approved | ||
| Target Regulation | Down regulation | |||
| Pathway Response | mTOR signaling pathway | hsa04150 | ||
| Cell Process | RNA decay | |||
| Cell growth and death | ||||
| Cell autophagy | ||||
In-vitro Model |
HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 |
| SW480 | Colon adenocarcinoma | Homo sapiens | CVCL_0546 | |
| Response Summary | In colorectal cancer, Glutaminolysis inhibition upregulated ATF4 expression in an m6A-dependent manner to activate pro-survival autophagy through transcriptional activation of the mTOR inhibitor DDIT4. Determined the relationship between FTO alpha-ketoglutarate dependent dioxygenase (FTO), YTH N6-methyladenosine RNA binding protein 2 (YTHDF2), and ATF4. Cyclic AMP-dependent transcription factor ATF-4 (ATF4) transcriptionally upregulated DDIT4 to suppress mTOR, which induced pro-survival autophagy during glutaminolysis inhibition. | |||
| Experiment 5 Reporting the m6A-centered Disease Response of This Target Gene | [56] | |||
| Responsed Disease | Colorectal cancer [ICD-11: 2B91] | |||
| Responsed Drug | CB-839 | Phase 2 | ||
| Target Regulation | Down regulation | |||
| Pathway Response | mTOR signaling pathway | hsa04150 | ||
| Cell Process | RNA decay | |||
| Cell growth and death | ||||
| Cell autophagy | ||||
In-vitro Model |
HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 |
| SW480 | Colon adenocarcinoma | Homo sapiens | CVCL_0546 | |
| Response Summary | In colorectal cancer, Glutaminolysis inhibition upregulated ATF4 expression in an m6A-dependent manner to activate pro-survival autophagy through transcriptional activation of the mTOR inhibitor DDIT4. Determined the relationship between FTO alpha-ketoglutarate dependent dioxygenase (FTO), YTH N6-methyladenosine RNA binding protein 2 (YTHDF2), and ATF4. ATF4 transcriptionally upregulated DDIT4 to suppress Cyclic AMP-dependent transcription factor ATF-4 (ATF4), which induced pro-survival autophagy during glutaminolysis inhibition. | |||
| Experiment 6 Reporting the m6A-centered Disease Response of This Target Gene | [56] | |||
| Responsed Disease | Colorectal cancer [ICD-11: 2B91] | |||
| Responsed Drug | GLS-IN-968 | Investigative | ||
| Target Regulation | Down regulation | |||
| Pathway Response | mTOR signaling pathway | hsa04150 | ||
| Cell Process | RNA decay | |||
| Cell growth and death | ||||
| Cell autophagy | ||||
In-vitro Model |
HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 |
| SW480 | Colon adenocarcinoma | Homo sapiens | CVCL_0546 | |
| Response Summary | In colorectal cancer, Glutaminolysis inhibition upregulated ATF4 expression in an m6A-dependent manner to activate pro-survival autophagy through transcriptional activation of the mTOR inhibitor DDIT4. Determined the relationship between FTO alpha-ketoglutarate dependent dioxygenase (FTO), YTH N6-methyladenosine RNA binding protein 2 (YTHDF2), and ATF4. ATF4 transcriptionally upregulated DDIT4 to suppress Cyclic AMP-dependent transcription factor ATF-4 (ATF4), which induced pro-survival autophagy during glutaminolysis inhibition. | |||
Cyclic-AMP-dependent transcription factor ATF-3 (ATF3)
Breast cancer [ICD-11: 2C60]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [57] | |||
| Responsed Disease | Breast cancer [ICD-11: 2C60] | |||
| Target Regulation | Down regulation | |||
In-vitro Model |
MCF-7 | Invasive breast carcinoma | Homo sapiens | CVCL_0031 |
| Response Summary | The increased expression of Cyclic-AMP-dependent transcription factor ATF-3 (ATF3) in tamoxifen-resistant cells arises from the decreased expression of the m6A reader protein YTHDF2 and the ensuing elevated stability of ATF3 mRNA, which ultimately promotes the translation of ATF3. ATF3 as a candidate therapeutic target for mitigating drug resistance in breast cancer cells. | |||
Cyclin-A2 (CCNA2)
Obesity [ICD-11: 5B81]
| In total 2 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [58] | |||
| Responsed Disease | Obesity [ICD-11: 5B81] | |||
| Responsed Drug | Epigallocatechin gallate | Phase 3 | ||
| Target Regulation | Down regulation | |||
| Cell Process | Adipogenesis | |||
In-vitro Model |
3T3-L1 | Normal | Mus musculus | CVCL_0123 |
| Response Summary | m6A-dependent Cyclin-A2 (CCNA2) and CDK2 expressions mediated by FTO and YTHDF2 contributed to EGCG-induced adipogenesis inhibition. | |||
| Experiment 2 Reporting the m6A-centered Disease Response of This Target Gene | [59] | |||
| Responsed Disease | Obesity [ICD-11: 5B81] | |||
| Target Regulation | Down regulation | |||
| Pathway Response | Cell cycle | hsa04110 | ||
| Cell Process | Adipogenesis | |||
| Arrest cell cycle at S phase | ||||
In-vitro Model |
3T3-L1 | Normal | Mus musculus | CVCL_0123 |
| Response Summary | FTO knockdown markedly decreased the expression of Cyclin-A2 (CCNA2) and CDK2, crucial cell cycle regulators, leading to delayed entry of MDI-induced cells into G2 phase. m6A-binding protein YTHDF2 recognized and decayed methylated mRNAs of CCNA2 and CDK2, leading to decreased protein expression, thereby prolonging cell cycle progression and suppressing adipogenesis. The adipocyte life cycle, including proliferation and adipogenesis, has become a potential target for many bioactive compounds and drugs for the prevention and treatment of obesity. | |||
Cyclin-dependent kinase 2 (CDK2)
Obesity [ICD-11: 5B81]
| In total 2 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [58] | |||
| Responsed Disease | Obesity [ICD-11: 5B81] | |||
| Responsed Drug | Epigallocatechin gallate | Phase 3 | ||
| Target Regulation | Down regulation | |||
| Cell Process | Adipogenesis | |||
In-vitro Model |
3T3-L1 | Normal | Mus musculus | CVCL_0123 |
| Response Summary | m6A-dependent CCNA2 and Cyclin-dependent kinase 2 (CDK2) expressions mediated by FTO and YTHDF2 contributed to EGCG-induced adipogenesis inhibition. | |||
| Experiment 2 Reporting the m6A-centered Disease Response of This Target Gene | [59] | |||
| Responsed Disease | Obesity [ICD-11: 5B81] | |||
| Target Regulation | Down regulation | |||
| Pathway Response | Cell cycle | hsa04110 | ||
| Cell Process | Adipogenesis | |||
| Arrest cell cycle at S phase | ||||
In-vitro Model |
3T3-L1 | Normal | Mus musculus | CVCL_0123 |
| Response Summary | FTO knockdown markedly decreased the expression of CCNA2 and Cyclin-dependent kinase 2 (CDK2), crucial cell cycle regulators, leading to delayed entry of MDI-induced cells into G2 phase. m6A-binding protein YTHDF2 recognized and decayed methylated mRNAs of CCNA2 and CDK2, leading to decreased protein expression, thereby prolonging cell cycle progression and suppressing adipogenesis. The adipocyte life cycle, including proliferation and adipogenesis, has become a potential target for many bioactive compounds and drugs for the prevention and treatment of obesity. | |||
Retinopathy [ICD-11: 9B71]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [61] | |||
| Responsed Disease | Diabetic retinopathy [ICD-11: 9B71.0] | |||
| Responsed Drug | FB23-2 | Investigative | ||
| Target Regulation | Down regulation | |||
Cyclin-dependent kinase inhibitor 1B (CDKN1B/p27)
Liver cancer [ICD-11: 2C12]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [62] | |||
| Responsed Disease | Intrahepatic cholangiocarcinoma [ICD-11: 2C12.10] | |||
| Responsed Drug | Cisplatin | Approved | ||
| Target Regulation | Down regulation | |||
| Pathway Response | Cell cycle | hsa04110 | ||
| Cell Process | Cell proliferation | |||
| Arrest cell cycle at G0/G1 phase | ||||
In-vitro Model |
HuCC-T1 | Intrahepatic cholangiocarcinoma | Homo sapiens | CVCL_0324 |
| RBE | Intrahepatic cholangiocarcinoma | Homo sapiens | CVCL_4896 | |
| HCCC-9810 (The intrahepatic cholangiocarcinoma cell lines (HCCC-9810) were purchased from Cellcook Co., Ltd. (Guangzhou, China).) | ||||
| HIBEC (The normal intrahepatic bile duct cell line (HIBEC) were purchased from Cellcook Co., Ltd. (Guangzhou, China).) | ||||
| In-vivo Model | For tumour xenograft models, 1 × 107 HuCC-T1 cells in knockdown group or control group were implanted into the right flank of 5-week-old female nude mice. The volumes of tumour were recorded every 4 days by calliper. The volumes were calculated as length × width2/2. For patient-derived xenograft (PDX) model (PDX0075), ICC tissues from a patient, who relapsed in 6 months after R0 resection and subsequent chemotherapy with cisplatin and gemcitabine, were diced into 3 mm3 pieces and transplanted subcutaneously into the right flank of 5-week-old female B-NDG mice. | |||
| Response Summary | The role of YTHDF2 in tumourigenesis and cisplatin-desensitising function by promoting the degradation of Cyclin-dependent kinase inhibitor 1B (CDKN1B/p27) mRNA in an m6 A-dependent manner. YTHDF2 exhibits tumour oncogenic and cisplatin-desensitising properties, which offer insight into the development of novel combination therapeutic strategies for intrahepatic cholangiocarcinoma. | |||
Cyclin-dependent kinase inhibitor 2A (CDKN2A)
Renal cell carcinoma [ICD-11: 2C90]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [63] | |||
| Responsed Disease | Renal cell carcinoma [ICD-11: 2C90] | |||
| Target Regulation | Down regulation | |||
| Cell Process | RNA stability | |||
In-vitro Model |
UOK120 | Papillary renal cell carcinoma | Homo sapiens | CVCL_B099 |
| UOK109 | Renal cell carcinoma | Homo sapiens | CVCL_B087 | |
| HK-2 [Human kidney] | Normal | Homo sapiens | CVCL_0302 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| ACHN | Papillary renal cell carcinoma | Homo sapiens | CVCL_1067 | |
| 786-O | Renal cell carcinoma | Homo sapiens | CVCL_1051 | |
| In-vivo Model | The mice were subcutaneously inoculated with 786-O cells stably transfected with lentiviruses carrying sh-NC/sh-circMET, respectively (5 × 106, 200 uL). | |||
| Response Summary | CircMET enhances mRNA decay of Cyclin-dependent kinase inhibitor 2A (CDKN2A) by direct interaction and recruitment of YTHDF2. CircMET promotes the development of NONO-TFE3 tRCC, and the regulation to both CDKN2A and SMAD3 of circMET was revealed. | |||
Cystathionine beta-synthase (CBS)
Gastric cancer [ICD-11: 2B72]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [64] | |||
| Responsed Disease | Gastric cancer [ICD-11: 2B72] | |||
| Target Regulation | Down regulation | |||
| Pathway Response | Ubiquitin mediated proteolysis | hsa04120 | ||
| Cell Process | Proteasome pathway degradation | |||
In-vitro Model |
SGC-7901 | Gastric carcinoma | Homo sapiens | CVCL_0520 |
| MKN45 | Gastric adenocarcinoma | Homo sapiens | CVCL_0434 | |
| MKN28 | Gastric tubular adenocarcinoma | Homo sapiens | CVCL_1416 | |
| MGC-803 | Gastric mucinous adenocarcinoma | Homo sapiens | CVCL_5334 | |
| GSE-1 (Gse-1 is a human gastric epithelial cell line) | ||||
| BGC-823 | Gastric carcinoma | Homo sapiens | CVCL_3360 | |
| AGS | Gastric adenocarcinoma | Homo sapiens | CVCL_0139 | |
| Response Summary | CBSLR interacted with YTHDF2 to form a CBSLR/YTHDF2/CBS signaling axis that decreased the stability of Cystathionine beta-synthase (CBS) mRNA by enhancing the binding of YTHDF2 with the m6A-modified coding sequence (CDS) of CBS mRNA. Reveal a novel mechanism in how HIF1-Alpha/CBSLR modulates ferroptosis/chemoresistance in GC, illuminating potential therapeutic targets for refractory hypoxic tumors. | |||
Cystine/glutamate transporter (SLC7A11)
Atherosclerosis [ICD-11: BD40]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [65] | |||
| Responsed Disease | Atherosclerosis [ICD-11: BD40.Z] | |||
| Target Regulation | Down regulation | |||
In-vitro Model |
HUVEC-C | Normal | Homo sapiens | CVCL_2959 |
Injury of heart [ICD-11: NB31]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [66] | |||
| Responsed Disease | Myocardial injury [ICD-11: NB31.Z] | |||
| Target Regulation | Down regulation | |||
In-vitro Model |
H9c2(2-1) | Normal | Rattus norvegicus | CVCL_0286 |
Death-associated protein kinase 3 (DAPK3)
Gallbladder cancer [ICD-11: 2C13]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [67] | |||
| Responsed Disease | Gallbladder cancer [ICD-11: 2C13] | |||
| Responsed Drug | Gemcitabine | Approved | ||
| Target Regulation | Down regulation | |||
| In-vivo Model | Male BALB/c nude mice (6 weeks old) were used for xenograft experiments. A 150 μL-cell suspension containing 5 × 106 cells was injected s.c. into each nude mouse. Tumor volumes were measured every 4 days for 1 month. To evaluate chemosensitivity to gemcitabine, mice were i.p. injected with gemcitabine (50 mg/kg) every 4 days when the tumor volume reached 100 mm3.Male NOD/SCID mice (6 weeks old) were used for lung metastasis experiments. A 150 μL-cell suspension containing 5 × 105 cells was injected i.v. into the tail vein of each mouse. After 1 month, the lungs were harvested and photographed, and the number of metastatic lesions was calculated. | |||
DNA (cytosine-5)-methyltransferase 3B (DNMT3B)
Intervertebral disc degeneration [ICD-11: FA80]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [68] | |||
| Responsed Disease | Intervertebral disc degeneration [ICD-11: FA80] | |||
| Target Regulation | Down regulation | |||
DNA replication licensing factor MCM5 (MCM5)
Liver cancer [ICD-11: 2C12]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [69] | |||
| Responsed Disease | Liver hepatocellular carcinoma [ICD-11: 2C12.02] | |||
| Responsed Drug | OSMI-1 | Investigative | ||
| Target Regulation | Up regulation | |||
In-vitro Model |
HepAD38 | Hepatoblastoma | Homo sapiens | CVCL_M177 |
| Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 | |
| PLC/PRF/5 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_0485 | |
| MHCC97-H | Adult hepatocellular carcinoma | Homo sapiens | CVCL_4972 | |
| Huh-7 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_0336 | |
| HEK293 | Normal | Homo sapiens | CVCL_0045 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| In-vivo Model | For the xenograft implantation model, 2 × 106 MHCC-97H cells were subcutaneously injected into the flank of nude mice. | |||
DNA-binding protein inhibitor ID-2 (ID2)
Pancreatic cancer [ICD-11: 2C10]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [70] | |||
| Responsed Disease | Pancreatic cancer [ICD-11: 2C10] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | PI3K-Akt signaling pathway | hsa04151 | ||
In-vitro Model |
PANC-1 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0480 |
| MIA PaCa-2 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0428 | |
| SW1990 | Pancreatic adenocarcinoma | Homo sapiens | CVCL_1723 | |
| BxPC-3 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0186 | |
| CFPAC-1 | Cystic fibrosis | Homo sapiens | CVCL_1119 | |
| HPDE6c7 | Normal | Homo sapiens | CVCL_0P38 | |
| Panc02 | Mouse pancreatic ductal adenocarcinoma | Mus musculus | CVCL_D627 | |
| In-vivo Model | For the subcutaneous tumor model, cells from the treatment group and the control group (1 × 107) were subcutaneously injected into the right groin of BALB/c nude mice (4-6 weeks old, 18-20 g, 4 mice each). Both the treatment group and control group had 5 BALB/c nude mice each. After 21 days, all mice were sacrificed and tumor volume and weight were measured. For the pancreatic cancer in situ tumor model, mice were anesthetized with Avertin at a dose of 0.2 ml/10 g body weight, and 5 × 105 panc2 treatment group cells and control group cells were injected into the pancreas of mice respectively, and the abdomen was closed for disinfection. Both the treatment group and control group had 5 C57 mice each. After 21 days, the mice were sacrificed, the in-situ tumor was removed, and the volume and weight were measured for subsequent experiments. | |||
E3 ubiquitin-protein ligase SMURF1 (SMURF1)
Inflammatory response [ICD-11: MG46]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [10] | |||
| Responsed Disease | Inflammatory response [ICD-11: MG46] | |||
| Target Regulation | Down regulation | |||
| Pathway Response | MAPK signaling pathway | hsa04010 | ||
| Cell Process | RNA stability | |||
In-vitro Model |
MC3T3-E1 | Normal | Mus musculus | CVCL_0409 |
| Response Summary | METTL3 knockdown inhibits osteoblast differentiation and Smad-dependent signaling by stabilizing Smad7 and E3 ubiquitin-protein ligase SMURF1 (SMURF1) mRNA transcripts via YTHDF2 involvement and activates the inflammatory response by regulating MAPK signaling in LPS-induced inflammation. | |||
ELAV-like protein 1 (HuR/ELAVL1)
Prostate cancer [ICD-11: 2C82]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [71] | |||
| Responsed Disease | Prostate cancer [ICD-11: 2C82] | |||
| Target Regulation | Down regulation | |||
In-vitro Model |
PC-3 | Prostate carcinoma | Homo sapiens | CVCL_0035 |
| LNCaP | Prostate carcinoma | Homo sapiens | CVCL_0395 | |
| DU145 | Prostate carcinoma | Homo sapiens | CVCL_0105 | |
| In-vivo Model | A total of 1 × 106 PC3 cells or DU145 cells suspended in a mixture of 100 uL PBS and Matrigel were subcutaneously injected into BALB/c nude mice. Tumor weight were measured 2 months after the engraftment. To evaluate the role of METTL3 in tumor metastasis, PC3 cells with or without knockdown of METTL3 were injected into SCID mice through the tail vein (1 × 106 cells per mouse). After eight weeks, mice were sacrificed and their lung tissues were collected for subsequent analyses. | |||
| Response Summary | m6A modification levels were markedly upregulated in human PCa tissues due to increased expression of METTL3. METTL3 mediates m6A modification of USP4 mRNA at A2696, and m6A reader protein YTHDF2 binds to and induces degradation of USP4 mRNA by recruiting RNA-binding protein HNRNPD to the mRNA. Decrease of USP4 fails to remove the ubiquitin group from ELAV-like protein 1 (HuR/ELAVL1) protein, resulting in a reduction of ELAVL1 protein. Lastly, downregulation of ELAVL1 in turn increases ARHGDIA expression, promoting migration and invasion of PCa cells. | |||
Eukaryotic translation initiation factor 4 gamma 1 (EIF4G1)
Head and neck squamous carcinoma [ICD-11: 2B6E]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [72] | |||
| Responsed Disease | Oral squamous cell carcinoma [ICD-11: 2B6E.0] | |||
| Responsed Drug | Rapamycin | Approved | ||
| Target Regulation | Down regulation | |||
| Pathway Response | Autophagy | hsa04140 | ||
| Cell Process | Cell autophagy | |||
| Response Summary | Rapamycin inhibited FTO activity, and directly targeted Eukaryotic translation initiation factor 4 gamma 1 (EIF4G1) transcripts and mediated their expression in an m6A-dependent manner in oral squamous cell carcinoma. After FTO silencing, YTHDF2 captured eIF4G1 transcripts containing m6A, resulting in mRNA degradation and decreased expression of eIF4G1 protein, thereby promoting autophagy and reducing tumor occurrence. | |||
Forkhead box protein O1 (FOXO1)
Female infertility [ICD-11: GA31]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [73] | |||
| Responsed Disease | Female infertility [ICD-11: GA31] | |||
| Target Regulation | Down regulation | |||
In-vitro Model |
T HESCs | Normal | Homo sapiens | CVCL_C464 |
| HTR-8/SVneo | Normal | Homo sapiens | CVCL_7162 | |
| HTR-8 | Normal | Homo sapiens | CVCL_D728 | |
| In-vivo Model | Five female mice were subjected to a normal pregnancy assay. Uterine tissues of 14 female donor mice were cut up and injected into the abdominal cavity of 28 female recipient mice. After 21 d, 10 male C57 mice were mated with the recipient mice, and the next day, when vaginal plugs were observed, was regarded as day 1. Eight recipient mice were euthanized by cervical dislocation after deep pentobarbital anesthesia on day 8, and the number of blastocysts in the uterus was counted. At the night of day 3, 10 μL of STM2457, a METTL3 inhibitor (Sellcek, S9870, 10 μM) and 10 μL dimethyl sulfoxide (DMSO) were individually injected into the uterine horns of 20 recipient mice. The mice were euthanized on day 8, and the number of blastocysts in the uterus was counted. The uterine tissues were collected and fixed in 4% (w/v) paraformaldehyde for histological and IHC analyses. | |||
Forkhead box protein O3 (FOXO3)
Intervertebral disc degeneration [ICD-11: FA80]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [74] | |||
| Responsed Disease | Intervertebral disc degeneration [ICD-11: FA80] | |||
| Target Regulation | Down regulation | |||
Glucose transporter type 1 (GLUT1)
Liver cancer [ICD-11: 2C12]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [75] | |||
| Responsed Disease | Liver hepatocellular carcinoma [ICD-11: 2C12.02] | |||
| Target Regulation | Down regulation | |||
| In-vivo Model | Male BALB/c nude mice (aged 4-6 weeks; n = 5/group) were obtained from Vital River Laboratory Animal Technology (Beijing, China). MHCC97H or Huh7 cells (2 × 106 cells/mouse) stably transfected with lentivirus containing different plasmids in 100 μL DMEM were subcutaneously or orthotopically implanted into the nude mice. | |||
Glycogen synthase kinase-3 beta (GSK3Beta/GSK3B)
Colorectal cancer [ICD-11: 2B91]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [76] | |||
| Responsed Disease | Colorectal cancer [ICD-11: 2B91] | |||
In-vitro Model |
SW480 | Colon adenocarcinoma | Homo sapiens | CVCL_0546 |
| RKO | Colon carcinoma | Homo sapiens | CVCL_0504 | |
Histone deacetylase 4 (HDAC4)
Pancreatic cancer [ICD-11: 2C10]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [77] | |||
| Responsed Disease | Pancreatic cancer [ICD-11: 2C10] | |||
| Target Regulation | Up regulation | |||
Histone-lysine N-methyltransferase EHMT2 (G9a)
Pain disorders [ICD-11: 8E43]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [78] | |||
| Responsed Disease | Neuropathic pain [ICD-11: 8E43.0] | |||
| Target Regulation | Down regulation | |||
Histone-lysine N-methyltransferase EZH2 (EZH2)
Endometriosis [ICD-11: GA10]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [79] | |||
| Responsed Disease | Endometriosis [ICD-11: GA10] | |||
| Target Regulation | Down regulation | |||
HPK/GCK-like kinase HGK (MAP4K4)
Diseases of the circulatory system [ICD-11: BE2Z]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [40] | |||
| Responsed Disease | Diseases of the circulatory system [ICD-11: BE2Z] | |||
| Response Summary | YTHDF2 regulates the stability of MAP2K4 and HPK/GCK-like kinase HGK (MAP4K4) mRNAs.This study identified that dasatinib and quercetin alleviate LPS-induced senescence in HUVECs via the TRAF6-MAPK-NF-Kappa-B axis in a YTHDF2-dependent manner, providing novel ideas for clinical treatment of age-related cardiovascular diseases. | |||
Inflammatory response [ICD-11: MG46]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [41] | |||
| Responsed Disease | Inflammatory response [ICD-11: MG46] | |||
| Target Regulation | Down regulation | |||
| Pathway Response | MAPK signaling pathway | hsa04010 | ||
In-vitro Model |
RAW 264.7 | Mouse leukemia | Mus musculus | CVCL_0493 |
| Response Summary | YTHDF2 knockdown increases mRNA expression levels of MAP2K4 and HPK/GCK-like kinase HGK (MAP4K4) via stabilizing the mRNA transcripts, which activate MAPK and NF-Kappa-B signaling pathways, which promote the expression of proinflammatory cytokines and aggravate the inflammatory response in LPS-stimulated RAW 264.7 cells. | |||
Integrin beta-1 (ITGB1)
Malignant mixed epithelial mesenchymal tumour [ICD-11: 2B5D]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [80] | |||
| Responsed Disease | Malignant mixed epithelial mesenchymal tumour of ovary [ICD-11: 2B5D.0] | |||
| Target Regulation | Down regulation | |||
In-vitro Model |
HO-8910 | Endocervical adenocarcinoma | Homo sapiens | CVCL_6868 |
| A2780 | Ovarian endometrioid adenocarcinoma | Homo sapiens | CVCL_0134 | |
| In-vivo Model | We injected 8 × 105 HO8910 cells into the footpads of mice. The mice were randomized two weeks after tumor cell injection to receive either an ITGB1 blocking antibody (Santa, CA, USA) or an IgG isotype antibody twice per week for four weeks 22,23. Y15 (30 mg/kg) against p-FAK (Tyr397) and PBS as a control were intraperitoneally injected twice per week for four weeks in the FAK-treatment assays. The main tumors and popliteal LNs were harvested and fixed in paraformaldehyde after six weeks of tumor cell inoculation. LN volumes were calculated as follows: LN volume (mm3) = (length [mm]) × (width [mm])2 × 0.52. The formalin-fixed, paraffin-embedded samples were analysed using immunohistochemistry and haematoxylin and eosin (H&E) staining. | |||
Retinopathy [ICD-11: 9B71]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [81] | |||
| Responsed Disease | Diabetic retinopathy [ICD-11: 9B71.0] | |||
| Target Regulation | Down regulation | |||
| Pathway Response | PI3K-Akt signaling pathway | hsa04151 | ||
| Cell Process | RNA stability | |||
In-vitro Model |
RMECs (Retinal microvascular endothelial cells (RMECs) purchased from Olaf (Worcester, MA, USA)) | |||
| rMCs (Retinal Muller cells (rMCs) from Kerafast Inc. (Boston, MA)) | ||||
| In-vivo Model | Male mice (8-10 weeks old, SLAC Laboratory Animal Co., Ltd., Shanghai, China) were administrated with STZ through intraperitoneal injection (I.P) for continuous 5 days (d). | |||
| Response Summary | KAT1 triggers YTHDF2-mediated Integrin beta-1 (ITGB1) mRNA instability to alleviate the progression of DR. | |||
Interferon regulatory factor 1 (Irf1)
Colorectal cancer [ICD-11: 2B91]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [82] | |||
| Responsed Disease | Colorectal cancer [ICD-11: 2B91] | |||
| Target Regulation | Down regulation | |||
| Pathway Response | PD-L1 expression and PD-1 checkpoint pathway in cancer | hsa05235 | ||
| Cell Process | Immunity | |||
In-vitro Model |
CT26 | Mouse colon adenocarcinoma | Mus musculus | CVCL_7254 |
| B16-GM-CSF (B16-GM-CSF cell line was a kind gift from Drs. Glenn Dranoff and Michael Dougan (Dana-Farber/Harvard Cancer Center)) | ||||
| B16-F10 | Mouse melanoma | Mus musculus | CVCL_0159 | |
| In-vivo Model | 2 × 106 CT26 cells with knockout of Mettl3, Mettl14, Mettl3/Stat1, Mettl3/Irf1, Mettl14/Stat1, or Mettl14/Irf1 and control were suspended in 200 uL of PBS/Matrigel (Corning) (1:1) and then subcutaneously inoculated into flank of each mouse. | |||
| Response Summary | In colorectal cancer, Mettl3- or Mettl14-deficient tumors increased cytotoxic tumor-infiltrating CD8+ T cells and elevated secretion of IFN-gamma, Cxcl9, and Cxcl10 in tumor microenvironment in vivo. Mechanistically, Mettl3 or Mettl14 loss promoted IFN-gamma-Stat1-Irf1 signaling through stabilizing the Stat1 and Interferon regulatory factor 1 (Irf1) mRNA via Ythdf2. | |||
Interferon regulatory factor 3 (IRF3)
Picornavirus infections presenting in the skin or mucous membranes [ICD-11: 1F05]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [83] | |||
| Responsed Disease | Picornavirus infections presenting in the skin or mucous membranes [ICD-11: 1F05.3] | |||
In-vitro Model |
PK-15
|
N.A. | Sus scrofa | CVCL_2160 |
| HT-29 | Colon adenocarcinoma | Homo sapiens | CVCL_0320 | |
|
IB-RS-2
|
N.A. | Sus scrofa | CVCL_4528 | |
|
CPTC-SVIL-1
|
N.A. | Oryctolagus cuniculus | CVCL_C2LN | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
|
BHK-21
|
N.A. | Mesocricetus auratus | CVCL_1914 | |
| RD | Embryonal rhabdomyosarcoma | Homo sapiens | CVCL_1649 | |
Interleukin-11 (IL11)
Liver cancer [ICD-11: 2C12]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [27] | |||
| Responsed Disease | Hepatocellular carcinoma [ICD-11: 2C12.02] | |||
| Target Regulation | Down regulation | |||
| Pathway Response | HIF-1 signaling pathway | hsa04066 | ||
| Cell Process | Biological regulation | |||
In-vitro Model |
MHCC97-H | Adult hepatocellular carcinoma | Homo sapiens | CVCL_4972 |
| SMMC-7721 | Endocervical adenocarcinoma | Homo sapiens | CVCL_0534 | |
| In-vivo Model | A number of 5 × 106 SMMC7721 or MHCC97H cells re-suspended in 100 uL of PBS were subcutaneously injected into the right flank of 6-week old male NCG mice. | |||
| Response Summary | YTHDF2 processed the decay of m6A-containing Interleukin-11 (IL11) and serpin family E member 2 (SERPINE2) mRNAs. YTHDF2 transcription succumbed to hypoxia-inducible factor-2-alpha (HIF-2-alpha). Administration of a HIF-2-alpha antagonist (PT2385) restored YTHDF2-programed epigenetic machinery and repressed liver cancer. | |||
Metalloproteinase inhibitor 3 (TIMP3)
Intervertebral disc degeneration [ICD-11: FA80]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [84] | |||
| Responsed Disease | Intervertebral disc degeneration [ICD-11: FA80] | |||
In-vitro Model |
Pten-/- P2 | Carcinoma of the mouse prostate gland | Mus musculus | CVCL_VQ83 |
| In-vivo Model | We also used surgery to cause spine instability in rats to observe IVD changes under abnormal stress (Figure 1f). X-ray scans 6 months after surgery showed that the lumbar vertebral sequence line was continuous, with no stenosis in various intervertebral foramen in the ctrl group. | |||
Mothers against decapentaplegic homolog 2 (SMAD2)
Lung cancer [ICD-11: 2C25]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [9] | |||
| Responsed Disease | Lung cancer [ICD-11: 2C25] | |||
| Target Regulation | Down regulation | |||
| Pathway Response | mRNA surveillance pathway | hsa03015 | ||
| Cell Process | Epithelial-mesenchymal transition | |||
In-vitro Model |
NCI-H1299 | Lung large cell carcinoma | Homo sapiens | CVCL_0060 |
| A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 | |
| Response Summary | YTHDF2 inhibits the migration and invasion of lung adenocarcinoma cells by regulating the FAM83D-TGFbeta1-Mothers against decapentaplegic homolog 2 (SMAD2) pathway, which will play an important role in lung cancer metastasis. | |||
Mutated in multiple advanced cancers 1 (PTEN)
Brain cancer [ICD-11: 2A00]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [86] | |||
| Responsed Disease | Brain cancer [ICD-11: 2A00] | |||
| Target Regulation | Down regulation | |||
Gastric cancer [ICD-11: 2B72]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [87] | |||
| Responsed Disease | Gastric cancer [ICD-11: 2B72] | |||
| Target Regulation | Down regulation | |||
Pulmonary hypertension [ICD-11: BB01]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [88] | |||
| Responsed Disease | Pulmonary hypertension due to lung disease or hypoxia [ICD-11: BB01.2] | |||
| Target Regulation | Down regulation | |||
| Pathway Response | PI3K-Akt signaling pathway | hsa04151 | ||
| Cell Process | Cell apoptosis | |||
In-vitro Model |
PASMC cell line (Pulmonary artery smooth muscle cell) | |||
| In-vivo Model | 10 rats were divided into control and HPH group. In detail, 5 rats of the hypoxia groups were exposed to hypoxia (10%O2) chamber (AiPu XBS-02B, China) for 4 weeks. In addition, 5 rats of control group were kept under normoxic conditions (21% O2) for 4 weeks. Rats were housed in standard polypropylene cages under controlled photocycle (12 h light/12 h dark) under 22-25 ℃ temperature. | |||
| Response Summary | METTL3/YTHDF2/Mutated in multiple advanced cancers 1 (PTEN) axis exerts a significant role in hypoxia induced PASMCs proliferation, providing a novel therapeutic target for hypoxic pulmonary hypertension. | |||
NACHT, LRR and PYD domains-containing protein 3 (NLRP3)
Lung cancer [ICD-11: 2C25]
| In total 2 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [89] | |||
| Responsed Disease | Lung cancer [ICD-11: 2C25] | |||
| Responsed Drug | Gefitinib | Approved | ||
In-vitro Model |
PC-9 | Lung adenocarcinoma | Homo sapiens | CVCL_B260 |
| HCC827 | Lung adenocarcinoma | Homo sapiens | CVCL_2063 | |
| A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 | |
| SPC-A1 | Endocervical adenocarcinoma | Homo sapiens | CVCL_6955 | |
| PC-9/GR2 | Lung adenocarcinoma | Homo sapiens | CVCL_DI29 | |
|
HCC827/GR
|
N.A. | Homo sapiens | CVCL_E7R9 | |
| In-vivo Model | To establish xenograft models, five-week-old male mice were orthotopically injected with the same number of the PC9/GR cells. Tumours had developed after 4 days, at which time the xenografted mice were randomly divided into the following two experimental groups (each group with 6 mice): (1) the control group and (2) the gefitinib group. | |||
| Experiment 2 Reporting the m6A-centered Disease Response of This Target Gene | [89] | |||
| Responsed Disease | Lung cancer [ICD-11: 2C25] | |||
| Target Regulation | Up regulation | |||
Rheumatoid arthritis [ICD-11: FA20]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [90] | |||
| Responsed Disease | Rheumatoid arthritis [ICD-11: FA20] | |||
| Target Regulation | Down regulation | |||
NAD-dependent protein deacetylase sirtuin-1 (SIRT1)
Endometriosis [ICD-11: GA10]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [91] | |||
| Responsed Disease | Endometriosis [ICD-11: GA10] | |||
| Target Regulation | Down regulation | |||
| In-vivo Model | For in vivo studies, the shMETTL3 sequence was subcloned into adeno-associated virus (AAV) construct 9 (AAV9). Recombinant AAV9 was manufactured by GenePharma (Shanghai, China). The ectopic endometrium was obtained from a 28-year-old woman who had undergone an operation for ovarian endometriosis. The tissue sample was washed twice with PBS and cut into three 3- to 5-mm pieces, and suspended by PBS.he animals were administered intraperitoneal injections of 30 mg/kg 17-oestradiol every three days after the endometrial injections. Xenografts were generated under bilateral axillae after 14 days with tumour volumes of approximately 10 mm3. | |||
Neurogenic locus notch homolog protein 1 (NOTCH1)
Aortic aneurysm or dissection [ICD-11: BD50]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [92] | |||
| Responsed Disease | Thoracic aortic dissection [ICD-11: BD50.3] | |||
| Target Regulation | Down regulation | |||
Nuclear factor erythroid 2-related factor 2 (NFE2L2)
Epilepsy due to structural or metabolic conditions or diseases [ICD-11: 8A60]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [93] | |||
| Responsed Disease | Epilepsy due to structural or metabolic conditions or diseases [ICD-11: 8A60.5] | |||
| Target Regulation | Down regulation | |||
Acute ischemic stroke [ICD-11: 8B11]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [94] | |||
| Responsed Disease | Acute ischemic stroke [ICD-11: 8B11] | |||
| Target Regulation | Down regulation | |||
In-vitro Model |
SH-SY5Y | Neuroblastoma | Homo sapiens | CVCL_0019 |
| In-vivo Model | Cerebral I/R injury was induced by MCAO/R surgery following previous method [10]. Rats were anesthetized by intraperitoneal injection of 2% sodium pentobarbital. After cutting the cervical skin of the rats with surgical scissors, the internal, external and common carotid arteries were separated. Then the proximal ends of the external and the common carotid arteries were ligated and the distal internal carotid artery was clipped. After cutting an incision 1 cm away from the bifurcation of the internal carotid artery with a surgical scissors, the left common carotid artery was slowly inserted into the internal carotid artery of the rats with sutures coated with silicone (head diameter 0.36±0.02 mm, Beijing Sunbio Biotech Co., Ltd., China) until sutures reached the bifurcation of the middle cerebral artery, and the wound was sutured after the sutures were fixed. After blocking blood flow for 1 h, the plug was removed to restore blood flow and reperfusion was performed. Other rats were selected for the Sham group, and the operation procedure was the same as above except for no ligation and insertion. | |||
Reproductive System disorders [ICD-11: SM9Z-SN0Z]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [95] | |||
| Responsed Disease | Reproductive System disorders [ICD-11: SM9Z-SN0Z] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
TM3 | Normal | Mus musculus | CVCL_4326 |
Nuclear factor NF-kappa-B p105 subunit (NF-Kappa-B/NFKB1)
Brain cancer [ICD-11: 2A00]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [96] | |||
| Responsed Disease | Glioma [ICD-11: 2A00.0] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | NF-kappa B signaling pathway | hsa04064 | ||
In-vitro Model |
U87 (A primary glioblastoma cell line) | |||
| N33 (The GBM patient-derived cell line) | ||||
| LN-229 | Glioblastoma | Homo sapiens | CVCL_0393 | |
| H4 | Astrocytoma | Homo sapiens | CVCL_1239 | |
| In-vivo Model | Five-week-old female BALB/c nude mice (Charles Rivers, Beijing, China) were selected for the experiments. U87 cells (5 × 105) transfected with an empty vector, YTHDF2 overexpression, or METTL3 overexpression vectors were suspended in PBS and injected into the right frontal node of nude mice. The inoculation position was 2 mm lateral and 2 mm posterior to the anterior fontanel. Tumor size was estimated from luciferase volume measurements and MRI. The mice were sacrificed when they exhibited disturbed activity or convulsion. The brain was then harvested and embedded in paraffin. | |||
| Response Summary | YTHDF2 accelerated UBXN1 mRNA degradation via METTL3-mediated m6A, which, in turn, promoted Nuclear factor NF-kappa-B p105 subunit (NF-Kappa-B/NFKB1) activation. YTHDF2 promotes the malignant progression of gliomas and revealed important insight into the upstream regulatory mechanism of NF-Kappa-B activation via UBXN1 with a primary focus on m6A modification. | |||
POU domain, class 5, transcription factor 1 (POU5F1)
Liver cancer [ICD-11: 2C12]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [98] | |||
| Responsed Disease | Hepatocellular carcinoma [ICD-11: 2C12.02] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | RNA degradation | hsa03018 | ||
| Cell Process | Cancer metastasis | |||
In-vitro Model |
Hep 3B2.1-7 | Childhood hepatocellular carcinoma | Homo sapiens | CVCL_0326 |
| Huh-7 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_0336 | |
| Response Summary | YTHDF2 promotes the CSC liver phenotype and cancer metastasis by modulating the m6A methylation of POU domain, class 5, transcription factor 1 (POU5F1) mRNA. YTHDF2 expression is positively correlated with OCT4 expression and m6A levels in the 5'-UTR of OCT4 mRNA in clinical hepatocellular carcinoma specimens. | |||
PPAR-gamma coactivator 1-alpha (PGC-1a/PPARGC1A)
Insulin resistance [ICD-11: 5A44]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [99] | |||
| Responsed Disease | Insulin resistance [ICD-11: 5A44] | |||
| Responsed Drug | Arsenite | Phase 2 | ||
In-vitro Model |
L-02 | Endocervical adenocarcinoma | Homo sapiens | CVCL_6926 |
Programmed cell death 1 (PD-1)
Melanoma [ICD-11: 2C30]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [3] | |||
| Responsed Disease | Melanoma [ICD-11: 2C30] | |||
| Responsed Drug | PMID31239444-anti-PD1 antibody | Investigative | ||
| Target Regulation | Down regulation | |||
| Pathway Response | PD-L1 expression and PD-1 checkpoint pathway in cancer | hsa05235 | ||
| Cell Process | mRNA decay | |||
In-vitro Model |
B16-F10 | Mouse melanoma | Mus musculus | CVCL_0159 |
| CHL-1 | Melanoma | Homo sapiens | CVCL_1122 | |
| 624-mel | Melanoma | Homo sapiens | CVCL_8054 | |
| NHEM (Normal Human Epidermal Melanocytes) | ||||
| SK-MEL-30 | Cutaneous melanoma | Homo sapiens | CVCL_0039 | |
| WM115 | Melanoma | Homo sapiens | CVCL_0040 | |
| WM35 | Melanoma | Homo sapiens | CVCL_0580 | |
| WM3670 | Melanoma | Homo sapiens | CVCL_6799 | |
| WM793 | Melanoma | Homo sapiens | CVCL_8787 | |
| In-vivo Model | When the tumors reached a volume of 80-100 mm3, mice were treated with anti-PD-1 or isotype control antibody (200 ug/mouse) by i.p. injection, every other day for three times. For IFNγ blockade treatment, C57BL/6 mice were treated with anti-IFNγ antibody or isotype control IgG (250 ug/mouse) every other day after tumor cell inoculation. | |||
| Response Summary | These findings demonstrate a crucial role of FTO as an m6A demethylase in promoting melanoma tumorigenesis and anti-PD-1 resistance, and suggest that the combination of FTO inhibition with anti-PD-1 blockade reduces the resistance to immunotherapy in melanoma. Knockdown of FTO increases m6A methylation in the critical protumorigenic melanoma cell-intrinsic genes including Programmed cell death 1 (PD-1) (PDCD1), CXCR4, and SOX10, leading to increased RNA decay through the m6A reader YTHDF2. | |||
Proliferation-associated protein 2G4 (PA2G4)
Liver cancer [ICD-11: 2C12]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [100] | |||
| Responsed Disease | Hepatocellular carcinoma [ICD-11: 2C12.02] | |||
| Target Regulation | Up regulation | |||
| Cell Process | Cell metastatic | |||
In-vitro Model |
PLC/PRF/5 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_0485 |
| MHCC97-H | Adult hepatocellular carcinoma | Homo sapiens | CVCL_4972 | |
| Huh-7 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_0336 | |
| HLF | Adult hepatocellular carcinoma | Homo sapiens | CVCL_2947 | |
| Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 | |
| Hep 3B2.1-7 | Childhood hepatocellular carcinoma | Homo sapiens | CVCL_0326 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| HCCLM3 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_6832 | |
| In-vivo Model | For lung metastasis model, 1 × 106 HCC cells suspended in 100 ul serum free DMEM were injected via the tail vein of nude mice. | |||
| Response Summary | Proliferation-associated protein 2G4 (PA2G4) plays a pro-metastatic role by increasing FYN expression through binding with YTHDF2 in HCC. PA2G4 becomes a reliable prognostic marker or therapeutic target for HCC patients. | |||
Pyruvate kinase PKM (PKM2/PKM)
Liver cancer [ICD-11: 2C12]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [75] | |||
| Responsed Disease | Liver hepatocellular carcinoma [ICD-11: 2C12.02] | |||
| Target Regulation | Down regulation | |||
| In-vivo Model | Male BALB/c nude mice (aged 4-6 weeks; n = 5/group) were obtained from Vital River Laboratory Animal Technology (Beijing, China). MHCC97H or Huh7 cells (2 × 106 cells/mouse) stably transfected with lentivirus containing different plasmids in 100 μL DMEM were subcutaneously or orthotopically implanted into the nude mice. | |||
RAC-alpha serine/threonine-protein kinase (AKT1)
Prostate cancer [ICD-11: 2C82]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [5] | |||
| Responsed Disease | Prostate cancer [ICD-11: 2C82] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | Oxidative phosphorylation | hsa00190 | ||
In-vitro Model |
VCaP | Prostate carcinoma | Homo sapiens | CVCL_2235 |
| RWPE-1 | Normal | Homo sapiens | CVCL_3791 | |
| PC-3 | Prostate carcinoma | Homo sapiens | CVCL_0035 | |
| DU145 | Prostate carcinoma | Homo sapiens | CVCL_0105 | |
| 22Rv1 | Prostate carcinoma | Homo sapiens | CVCL_1045 | |
| Response Summary | Knock-down of YTHDF2 or METTL3 significantly induced the expression of LHPP and NKX3-1 at both mRNA and protein level with inhibited phosphorylated RAC-alpha serine/threonine-protein kinase (AKT1). YTHDF2 mediates the mRNA degradation of the tumor suppressors LHPP and NKX3-1 in m6A-dependent way to regulate AKT phosphorylation-induced tumor progression in prostate cancer. | |||
RB1-inducible coiled-coil protein 1 (RB1CC1/FIP200)
Brain cancer [ICD-11: 2A00]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [49] | |||
| Responsed Disease | Glioblastoma [ICD-11: 2A00.00] | |||
RIG-I-like receptor 1 (RIG-I)
Bladder cancer [ICD-11: 2C94]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [101] | |||
| Responsed Disease | Bladder cancer [ICD-11: 2C94] | |||
| Responsed Drug | Bacillus Calmette | Investigative | ||
| Target Regulation | Down regulation | |||
In-vitro Model |
MGH-U3 | Bladder carcinoma | Homo sapiens | CVCL_9827 |
| SW780 | Bladder carcinoma | Homo sapiens | CVCL_1728 | |
| 5637 | Bladder carcinoma | Homo sapiens | CVCL_0126 | |
| T24 | Bladder carcinoma | Homo sapiens | CVCL_0554 | |
| UM-UC-3 | Bladder carcinoma | Homo sapiens | CVCL_1783 | |
| In-vivo Model | For xenograft studies, female mice between 4 and 6 weeks of age were used. Monoclonal cell lines showing a nearly complete absence of Ythdf2 and Ddx58 were used for in vivo experimentation. Mice were implanted with 2 × 106 MGHU3 cells resuspended in 100 μL of Matrigel Matrix High Concentration (354248) and PBS mixture subcutaneously into the right flanks. Subcutaneous tumor growth was measured weekly by calipers, and the volume was calculated using the formula: volume = (length × width2)/2. | |||
Serine/threonine-protein kinase mTOR (MTOR)
Colorectal cancer [ICD-11: 2B91]
| In total 6 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [56] | |||
| Responsed Disease | Colorectal cancer [ICD-11: 2B91] | |||
| Responsed Drug | Asparagine inhibitor | Approved | ||
| Target Regulation | Up regulation | |||
| Pathway Response | mTOR signaling pathway | hsa04150 | ||
| Autophagy | hsa04140 | |||
| Cell Process | RNA decay | |||
| Cell growth and death | ||||
| Cell autophagy | ||||
In-vitro Model |
HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 |
| SW480 | Colon adenocarcinoma | Homo sapiens | CVCL_0546 | |
| Response Summary | In colorectal cancer, Glutaminolysis inhibition upregulated ATF4 expression in an m6A-dependent manner to activate pro-survival autophagy through transcriptional activation of the mTOR inhibitor DDIT4. Determined the relationship between FTO alpha-ketoglutarate dependent dioxygenase (FTO), YTH N6-methyladenosine RNA binding protein 2 (YTHDF2), and ATF4. ATF4 transcriptionally upregulated DDIT4 to suppress Serine/threonine-protein kinase mTOR (MTOR), which induced pro-survival autophagy during glutaminolysis inhibition. | |||
| Experiment 2 Reporting the m6A-centered Disease Response of This Target Gene | [56] | |||
| Responsed Disease | Colorectal cancer [ICD-11: 2B91] | |||
| Responsed Drug | Chloroquine | Approved | ||
| Target Regulation | Up regulation | |||
| Pathway Response | mTOR signaling pathway | hsa04150 | ||
| Autophagy | hsa04140 | |||
| Cell Process | RNA decay | |||
| Cell growth and death | ||||
| Cell autophagy | ||||
In-vitro Model |
HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 |
| SW480 | Colon adenocarcinoma | Homo sapiens | CVCL_0546 | |
| Response Summary | In colorectal cancer, Glutaminolysis inhibition upregulated ATF4 expression in an m6A-dependent manner to activate pro-survival autophagy through transcriptional activation of the mTOR inhibitor DDIT4. Determined the relationship between FTO alpha-ketoglutarate dependent dioxygenase (FTO), YTH N6-methyladenosine RNA binding protein 2 (YTHDF2), and ATF4. Serine/threonine-protein kinase mTOR (MTOR) transcriptionally upregulated DDIT4 to suppress mTOR, which induced pro-survival autophagy during glutaminolysis inhibition. | |||
| Experiment 3 Reporting the m6A-centered Disease Response of This Target Gene | [56] | |||
| Responsed Disease | Colorectal cancer [ICD-11: 2B91] | |||
| Responsed Drug | Meclofenamate sodium | Approved | ||
| Target Regulation | Up regulation | |||
| Pathway Response | mTOR signaling pathway | hsa04150 | ||
| Autophagy | hsa04140 | |||
| Cell Process | RNA decay | |||
| Cell growth and death | ||||
| Cell autophagy | ||||
In-vitro Model |
HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 |
| SW480 | Colon adenocarcinoma | Homo sapiens | CVCL_0546 | |
| Response Summary | In colorectal cancer, Glutaminolysis inhibition upregulated ATF4 expression in an m6A-dependent manner to activate pro-survival autophagy through transcriptional activation of the mTOR inhibitor DDIT4. Determined the relationship between FTO alpha-ketoglutarate dependent dioxygenase (FTO), YTH N6-methyladenosine RNA binding protein 2 (YTHDF2), and ATF4. Serine/threonine-protein kinase mTOR (MTOR) transcriptionally upregulated DDIT4 to suppress mTOR, which induced pro-survival autophagy during glutaminolysis inhibition. | |||
| Experiment 4 Reporting the m6A-centered Disease Response of This Target Gene | [56] | |||
| Responsed Disease | Colorectal cancer [ICD-11: 2B91] | |||
| Responsed Drug | Rapamycin | Approved | ||
| Target Regulation | Up regulation | |||
| Pathway Response | mTOR signaling pathway | hsa04150 | ||
| Autophagy | hsa04140 | |||
| Cell Process | RNA decay | |||
| Cell growth and death | ||||
| Cell autophagy | ||||
In-vitro Model |
HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 |
| SW480 | Colon adenocarcinoma | Homo sapiens | CVCL_0546 | |
| Response Summary | In colorectal cancer, Glutaminolysis inhibition upregulated ATF4 expression in an m6A-dependent manner to activate pro-survival autophagy through transcriptional activation of the mTOR inhibitor DDIT4. Determined the relationship between FTO alpha-ketoglutarate dependent dioxygenase (FTO), YTH N6-methyladenosine RNA binding protein 2 (YTHDF2), and ATF4. Serine/threonine-protein kinase mTOR (MTOR) transcriptionally upregulated DDIT4 to suppress mTOR, which induced pro-survival autophagy during glutaminolysis inhibition. | |||
| Experiment 5 Reporting the m6A-centered Disease Response of This Target Gene | [56] | |||
| Responsed Disease | Colorectal cancer [ICD-11: 2B91] | |||
| Responsed Drug | CB-839 | Phase 2 | ||
| Target Regulation | Up regulation | |||
| Pathway Response | mTOR signaling pathway | hsa04150 | ||
| Autophagy | hsa04140 | |||
| Cell Process | RNA decay | |||
| Cell growth and death | ||||
| Cell autophagy | ||||
In-vitro Model |
HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 |
| SW480 | Colon adenocarcinoma | Homo sapiens | CVCL_0546 | |
| Response Summary | In colorectal cancer, Glutaminolysis inhibition upregulated ATF4 expression in an m6A-dependent manner to activate pro-survival autophagy through transcriptional activation of the mTOR inhibitor DDIT4. Determined the relationship between FTO alpha-ketoglutarate dependent dioxygenase (FTO), YTH N6-methyladenosine RNA binding protein 2 (YTHDF2), and ATF4. ATF4 transcriptionally upregulated DDIT4 to suppress Serine/threonine-protein kinase mTOR (MTOR), which induced pro-survival autophagy during glutaminolysis inhibition. | |||
| Experiment 6 Reporting the m6A-centered Disease Response of This Target Gene | [56] | |||
| Responsed Disease | Colorectal cancer [ICD-11: 2B91] | |||
| Responsed Drug | GLS-IN-968 | Investigative | ||
| Target Regulation | Up regulation | |||
| Pathway Response | mTOR signaling pathway | hsa04150 | ||
| Autophagy | hsa04140 | |||
| Cell Process | RNA decay | |||
| Cell growth and death | ||||
| Cell autophagy | ||||
In-vitro Model |
HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 |
| SW480 | Colon adenocarcinoma | Homo sapiens | CVCL_0546 | |
| Response Summary | In colorectal cancer, Glutaminolysis inhibition upregulated ATF4 expression in an m6A-dependent manner to activate pro-survival autophagy through transcriptional activation of the mTOR inhibitor DDIT4. Determined the relationship between FTO alpha-ketoglutarate dependent dioxygenase (FTO), YTH N6-methyladenosine RNA binding protein 2 (YTHDF2), and ATF4. ATF4 transcriptionally upregulated DDIT4 to suppress Serine/threonine-protein kinase mTOR (MTOR), which induced pro-survival autophagy during glutaminolysis inhibition. | |||
Serine/threonine-protein kinase PINK1, mitochondrial (PINK1)
Chronic kidney disease [ICD-11: GB61]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [102] | |||
| Responsed Disease | Diabetic nephropathy [ICD-11: GB61.Z] | |||
| Target Regulation | Down regulation | |||
In-vitro Model |
HK-2 [Human kidney] | Normal | Homo sapiens | CVCL_0302 |
| In-vivo Model | Five week old male C57bl/6 mice (20 ± 2 g) were divided into the DKD model group and the control group. The fasting blood glucose of all the mice was measured before the experiment, and was required to be less than 7 mmol/L. The mice in the DKD model group were fed with a high-fat and high-sugar diet for 8 weeks. Then, 1 day after restoring the high-fat and high-sugar diet, the mice in the DKD model group were fasted for 16 h and injected with STZ at 50 mg/kg per day for 5 days. The fasting blood glucose was measured 2 weeks after the last injection. If the fasting blood glucose was greater than 16 mmol/L, the medium-term diabetes model had been established successfully. Then, the rat urine was collected and the ratio of urinary albumin to urinary muscle intoxication (AIb/Cr) was measured. The AIb/Cr was greater than 30 mg/g, indicating a diagnosis of DKD. The mice in the control group received standard chow and were injected with the same amount of normal saline. Subsequently, all DKD mice were randomly divided into the sh-NC group and the sh-METTL3 group. After establishment of the DKD model, the lentiviruses carrying sh-NC or sh-METTL3 (MOI = 50) were injected into the caudal vein at a dose of 1 μg/g according to the weight of mice, once a week for 8 weeks. | |||
Serum response factor (SRF)
Brain cancer [ICD-11: 2A00]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [104] | |||
| Responsed Disease | Glioblastoma [ICD-11: 2A00.00] | |||
| Responsed Drug | Tislelizumab | Approved | ||
In-vitro Model |
U-87MG ATCC | Glioblastoma | Homo sapiens | CVCL_0022 |
Signal transducer and activator of transcription 5A (STAT5A)
Multiple myeloma [ICD-11: 2A83]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [105] | |||
| Responsed Disease | Multiple myeloma [ICD-11: 2A83.1] | |||
| Target Regulation | Down regulation | |||
| Pathway Response | MAPK signaling pathway | hsa04010 | ||
| Response Summary | The analyses of m6A-RIP-seq and RIP-PCR indicated that Signal transducer and activator of transcription 5A (STAT5A) was the downstream target of YTHDF2, which was binding to the m6A modification site of STAT5A to promote its mRNA degradation. ChIP-seq and PCR assays revealed that STAT5A suppressed multiple myelomacell proliferation by occupying the transcription site of MAP2K2 to decrease ERK phosphorylation. | |||
Splicing factor 3A subunit 3 (SF3A3)
Liver cancer [ICD-11: 2C12]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [106] | |||
| Responsed Disease | Hepatocellular carcinoma [ICD-11: 2C12.02] | |||
| Target Regulation | Up regulation | |||
| Cell Process | Immunity | |||
| Response Summary | YTHDF2 expression was associated positively with Splicing factor 3A subunit 3 (SF3A3) expression, which implied that they cooperate in LIHC progression. | |||
Stearoyl-CoA desaturase (SCD)
Colon cancer [ICD-11: 2B90]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [107] | |||
| Responsed Disease | Colon cancer [ICD-11: 2B90] | |||
In-vitro Model |
SW480 | Colon adenocarcinoma | Homo sapiens | CVCL_0546 |
| SW620 | Colon adenocarcinoma | Homo sapiens | CVCL_0547 | |
Stimulator of interferon genes protein (STING1)
Picornavirus infections presenting in the skin or mucous membranes [ICD-11: 1F05]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [109] | |||
| Responsed Disease | Picornavirus infections presenting in the skin or mucous membranes [ICD-11: 1F05.3] | |||
Thioredoxin domain-containing protein 5 (TXNDC5)
Cervical cancer [ICD-11: 2C77]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [111] | |||
| Responsed Disease | Cervical cancer [ICD-11: 2C77] | |||
Transcription factor E2F3 (E2F3)
Liver cancer [ICD-11: 2C12]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [112] | |||
| Responsed Disease | Liver cancer [ICD-11: 2C12] | |||
| Target Regulation | Down regulation | |||
Transcription factor ISGF-3 components p91/p84 (Stat1)
Colorectal cancer [ICD-11: 2B91]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [82] | |||
| Responsed Disease | Colorectal cancer [ICD-11: 2B91] | |||
| Target Regulation | Down regulation | |||
| Pathway Response | PD-L1 expression and PD-1 checkpoint pathway in cancer | hsa05235 | ||
| Cell Process | Immunity | |||
In-vitro Model |
CT26 | Mouse colon adenocarcinoma | Mus musculus | CVCL_7254 |
| B16-GM-CSF (B16-GM-CSF cell line was a kind gift from Drs. Glenn Dranoff and Michael Dougan (Dana-Farber/Harvard Cancer Center)) | ||||
| B16-F10 | Mouse melanoma | Mus musculus | CVCL_0159 | |
| In-vivo Model | 2 × 106 CT26 cells with knockout of Mettl3, Mettl14, Mettl3/Stat1, Mettl3/Irf1, Mettl14/Stat1, or Mettl14/Irf1 and control were suspended in 200 uL of PBS/Matrigel (Corning) (1:1) and then subcutaneously inoculated into flank of each mouse. | |||
| Response Summary | In colorectal cancer, Mettl3- or Mettl14-deficient tumors increased cytotoxic tumor-infiltrating CD8+ T cells and elevated secretion of IFN-gamma, Cxcl9, and Cxcl10 in tumor microenvironment in vivo. Mechanistically, Mettl3 or Mettl14 loss promoted IFN-gamma-Stat1-Irf1 signaling through stabilizing the Transcription factor ISGF-3 components p91/p84 (Stat1) and Irf1 mRNA via Ythdf2. | |||
Transcription factor SOX-10 (SOX10)
Melanoma [ICD-11: 2C30]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [3] | |||
| Responsed Disease | Melanoma [ICD-11: 2C30] | |||
| Responsed Drug | PMID31239444-anti-PD1 antibody | Investigative | ||
| Target Regulation | Down regulation | |||
| Pathway Response | PD-L1 expression and PD-1 checkpoint pathway in cancer | hsa05235 | ||
| Cell Process | mRNA decay | |||
In-vitro Model |
B16-F10 | Mouse melanoma | Mus musculus | CVCL_0159 |
| CHL-1 | Melanoma | Homo sapiens | CVCL_1122 | |
| 624-mel | Melanoma | Homo sapiens | CVCL_8054 | |
| NHEM (Normal Human Epidermal Melanocytes) | ||||
| SK-MEL-30 | Cutaneous melanoma | Homo sapiens | CVCL_0039 | |
| WM115 | Melanoma | Homo sapiens | CVCL_0040 | |
| WM35 | Melanoma | Homo sapiens | CVCL_0580 | |
| WM3670 | Melanoma | Homo sapiens | CVCL_6799 | |
| WM793 | Melanoma | Homo sapiens | CVCL_8787 | |
| In-vivo Model | When the tumors reached a volume of 80-100 mm3, mice were treated with anti-PD-1 or isotype control antibody (200 ug/mouse) by i.p. injection, every other day for three times. For IFNγ blockade treatment, C57BL/6 mice were treated with anti-IFNγ antibody or isotype control IgG (250 ug/mouse) every other day after tumor cell inoculation. | |||
| Response Summary | These findings demonstrate a crucial role of FTO as an m6A demethylase in promoting melanoma tumorigenesis and anti-PD-1 resistance, and suggest that the combination of FTO inhibition with anti-PD-1 blockade reduces the resistance to immunotherapy in melanoma. Knockdown of FTO increases m6A methylation in the critical protumorigenic melanoma cell-intrinsic genes including PD-1 (PDCD1), CXCR4, and Transcription factor SOX-10 (SOX10), leading to increased RNA decay through the m6A reader YTHDF2. | |||
Transcription factor SOX-2 (SOX2)
Lung cancer [ICD-11: 2C25]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [85] | |||
| Responsed Disease | Lung cancer [ICD-11: 2C25] | |||
| Target Regulation | Down regulation | |||
In-vitro Model |
MRC-9 | Normal | Homo sapiens | CVCL_2629 |
| NCI-H1299 | Lung large cell carcinoma | Homo sapiens | CVCL_0060 | |
| NCI-H1650 | Minimally invasive lung adenocarcinoma | Homo sapiens | CVCL_1483 | |
| NCI-H1703 | Lung squamous cell carcinoma | Homo sapiens | CVCL_1490 | |
| H1795 (Lung cancer H1795 cell lines were purchased from ATCC, USA) | ||||
| NCI-H1792 | Lung adenocarcinoma | Homo sapiens | CVCL_1495 | |
| A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 | |
| DLD-1 | Colon adenocarcinoma | Homo sapiens | CVCL_0248 | |
| SW480 | Colon adenocarcinoma | Homo sapiens | CVCL_0546 | |
| PANC-1 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0480 | |
| MIA PaCa-2 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0428 | |
Transforming growth factor beta-1 proprotein (TGFB1)
Lung cancer [ICD-11: 2C25]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [9] | |||
| Responsed Disease | Lung cancer [ICD-11: 2C25] | |||
| Target Regulation | Down regulation | |||
| Pathway Response | mRNA surveillance pathway | hsa03015 | ||
| Cell Process | Epithelial-mesenchymal transition | |||
In-vitro Model |
NCI-H1299 | Lung large cell carcinoma | Homo sapiens | CVCL_0060 |
| A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 | |
| Response Summary | YTHDF2 inhibits the migration and invasion of lung adenocarcinoma cells by regulating the FAM83D-Transforming growth factor beta-1 proprotein (TGFB1)-pSMAD2/3 pathway, which will play an important role in lung cancer metastasis. | |||
Tyrosine-protein kinase Fyn (FYN)
Liver cancer [ICD-11: 2C12]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [100] | |||
| Responsed Disease | Hepatocellular carcinoma [ICD-11: 2C12.02] | |||
| Target Regulation | Up regulation | |||
| Cell Process | Cell metastatic | |||
In-vitro Model |
PLC/PRF/5 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_0485 |
| MHCC97-H | Adult hepatocellular carcinoma | Homo sapiens | CVCL_4972 | |
| Huh-7 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_0336 | |
| HLF | Adult hepatocellular carcinoma | Homo sapiens | CVCL_2947 | |
| Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 | |
| Hep 3B2.1-7 | Childhood hepatocellular carcinoma | Homo sapiens | CVCL_0326 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| HCCLM3 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_6832 | |
| In-vivo Model | For lung metastasis model, 1 × 106 HCC cells suspended in 100 ul serum free DMEM were injected via the tail vein of nude mice. | |||
| Response Summary | PA2G4 plays a pro-metastatic role by increasing Tyrosine-protein kinase Fyn (FYN) expression through binding with YTHDF2 in HCC. PA2G4 becomes a reliable prognostic marker or therapeutic target for HCC patients. | |||
Ubiquitin carboxyl-terminal hydrolase 4 (USP4)
Prostate cancer [ICD-11: 2C82]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [71] | |||
| Responsed Disease | Prostate cancer [ICD-11: 2C82] | |||
| Target Regulation | Down regulation | |||
In-vitro Model |
PC-3 | Prostate carcinoma | Homo sapiens | CVCL_0035 |
| LNCaP | Prostate carcinoma | Homo sapiens | CVCL_0395 | |
| DU145 | Prostate carcinoma | Homo sapiens | CVCL_0105 | |
| In-vivo Model | A total of 1 × 106 PC3 cells or DU145 cells suspended in a mixture of 100 uL PBS and Matrigel were subcutaneously injected into BALB/c nude mice. Tumor weight were measured 2 months after the engraftment. To evaluate the role of METTL3 in tumor metastasis, PC3 cells with or without knockdown of METTL3 were injected into SCID mice through the tail vein (1 × 106 cells per mouse). After eight weeks, mice were sacrificed and their lung tissues were collected for subsequent analyses. | |||
| Response Summary | m6A modification levels were markedly upregulated in human PCa tissues due to increased expression of METTL3. METTL3 mediates m6A modification of Ubiquitin carboxyl-terminal hydrolase 4 (USP4) mRNA at A2696, and m6A reader protein YTHDF2 binds to and induces degradation of USP4 mRNA by recruiting RNA-binding protein HNRNPD to the mRNA. Decrease of USP4 fails to remove the ubiquitin group from ELAVL1 protein, resulting in a reduction of ELAVL1 protein. Lastly, downregulation of ELAVL1 in turn increases ARHGDIA expression, promoting migration and invasion of PCa cells. | |||
Ubiquitin-like modifier-activating enzyme ATG7 (ATG7)
Obesity [ICD-11: 5B81]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [37] | |||
| Responsed Disease | Obesity [ICD-11: 5B81] | |||
| Target Regulation | Down regulation | |||
| Pathway Response | Autophagy | hsa04140 | ||
| Cell Process | Autophagy | |||
| Adipogenesis regulation | ||||
In-vitro Model |
3T3-L1 | Normal | Mus musculus | CVCL_0123 |
| Pig primary preadipocytes (Isolated from cervical subcutaneous adipose tissue of piglets) | ||||
| In-vivo Model | Mice were maintained at 22 ± 2 ℃ with a humidity of 35 ± 5% under a 12 h light and 12 h dark cycle, with free access to water and food. For the HFD experiment, female control (Ftoflox/flox) and adipose-selective fto knockout (Fabp4-Cre Ftoflox/flox, fto-AKO) mice were fed with high-fat diet (60% fat in calories; Research Diets, D12492) for the desired periods of time, and food intake and body weight were measured every week after weaning (at 3 weeks of age). | |||
| Response Summary | Atg5 and Ubiquitin-like modifier-activating enzyme ATG7 (ATG7) were the targets of YTHDF2 (YTH N6-methyladenosine RNA binding protein 2). Upon FTO silencing, Atg5 and Atg7 transcripts with higher m6A levels were captured by YTHDF2, which resulted in mRNA degradation and reduction of protein expression, thus alleviating autophagy and adipogenesis. | |||
Osteoarthritis [ICD-11: FA05]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [47] | |||
| Responsed Disease | Osteoarthritis [ICD-11: FA05] | |||
UBX domain-containing protein 1 (UBXN1)
Brain cancer [ICD-11: 2A00]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [96] | |||
| Responsed Disease | Glioma [ICD-11: 2A00.0] | |||
| Target Regulation | Down regulation | |||
| Pathway Response | NF-kappa B signaling pathway | hsa04064 | ||
In-vitro Model |
U87 (A primary glioblastoma cell line) | |||
| N33 (The GBM patient-derived cell line) | ||||
| LN-229 | Glioblastoma | Homo sapiens | CVCL_0393 | |
| H4 | Astrocytoma | Homo sapiens | CVCL_1239 | |
| In-vivo Model | Five-week-old female BALB/c nude mice (Charles Rivers, Beijing, China) were selected for the experiments. U87 cells (5 × 105) transfected with an empty vector, YTHDF2 overexpression, or METTL3 overexpression vectors were suspended in PBS and injected into the right frontal node of nude mice. The inoculation position was 2 mm lateral and 2 mm posterior to the anterior fontanel. Tumor size was estimated from luciferase volume measurements and MRI. The mice were sacrificed when they exhibited disturbed activity or convulsion. The brain was then harvested and embedded in paraffin. | |||
| Response Summary | YTHDF2 accelerated UBX domain-containing protein 1 (UBXN1) mRNA degradation via METTL3-mediated m6A, which, in turn, promoted NF-Kappa-B activation. YTHDF2 promotes the malignant progression of gliomas and revealed important insight into the upstream regulatory mechanism of NF-Kappa-B activation via UBXN1 with a primary focus on m6A modification. | |||
Growth arrest specific 5 (GAS5)
Cervical cancer [ICD-11: 2C77]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [115] | |||
| Responsed Disease | Cervical cancer [ICD-11: 2C77] | |||
| Target Regulation | Up regulation | |||
| Cell Process | Cell proliferation and metastasis | |||
In-vitro Model |
C-33 A | Cervical squamous cell carcinoma | Homo sapiens | CVCL_1094 |
| Ca Ski | Cervical squamous cell carcinoma | Homo sapiens | CVCL_1100 | |
| HeLa | Endocervical adenocarcinoma | Homo sapiens | CVCL_0030 | |
| SiHa | Cervical squamous cell carcinoma | Homo sapiens | CVCL_0032 | |
| Normal cervical epithelium cell line (HCvEpC) (Isolated from cervical tissue) | ||||
| In-vivo Model | 200 uL PBS containing 1×107 cells of stable cells were subcutaneously injected into male BALB/c athymic nude mice (6-week old, 18-20 g). | |||
| Response Summary | The GAS5-AS1 expression in cervical cancer tissues was markedly decreased when compared with that in the adjacent normal tissues. GAS5-AS1 interacted with the tumor suppressor Growth arrest specific 5 (GAS5), and increased its stability by interacting with RNA demethylase ALKBH5 and decreasing GAS5 N6-methyladenosine (m6A) modification. m6A-mediated GAS5 RNA degradation relied on the m6A reader protein YTHDF2-dependent pathway. | |||
Vascular disorders of the liver [ICD-11: DB98]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [116] | |||
| Responsed Disease | Vascular disorders of the liver [ICD-11: DB98.8] | |||
| Target Regulation | Down regulation | |||
| In-vivo Model | The in vivo assay was approved by the Animal Care and Use Committee of Anhui Medical University. And all experimental procedures and animal care were in accordance with the institutional ethics guidelines for animal experiments. The C57BL/6 mice (male, 8-10 weeks old) were housed (six per cage) in a specific and opportunistic pathogen-free facility and maintained on a 12-hlight-dark cycle with casually access to food and water. Detailed descriptions of the cell culture, cardiac fibrosis model, and treatment with lentiviral, were given in the Supplementary methods online. | |||
LINC00902 (TUSC7)
Lung cancer [ICD-11: 2C25]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [117] | |||
| Responsed Disease | Lung adenocarcinoma [ICD-11: 2C25.0] | |||
| Responsed Drug | Erlotinib | Approved | ||
| Target Regulation | Down regulation | |||
| Pathway Response | Notch signaling pathway | hsa04330), EGFR tyrosine kinase inhibitor resistance | ||
In-vitro Model |
PC-9 | Lung adenocarcinoma | Homo sapiens | CVCL_B260 |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| HCC827 | Lung adenocarcinoma | Homo sapiens | CVCL_2063 | |
| In-vivo Model | Control vector, TUSC7 knockout, FLI-06 treated H1975 cells (1*107) cells were suspended in 100 uL of serum-free DMEM medium (Hyclone, USA), mixed with matrix gel (Corning, USA), and then were injected subcutaneously. The changes in the tumor size were recorded every 3 or 5 days. | |||
| Response Summary | In lung adenocarcinoma, The miR-146a/Notch signaling was sustained highly activated in a m6A dependent manner, and the m6A regulator of YTHDF2 suppressed LINC00902 (TUSC7), both of which contributed to the resistant features. Functionally, the sponge type of TUSC7 regulation of miR-146a inhibited Notch signaling functions, and affected the cancer progression and stem cells' renewal in Erlotinib resistant PC9 cells (PC9ER) and Erlotinib resistant HCC827 cells (HCC827ER) cells. | |||
Long intergenic non-protein coding RNA 1273 (LINC01273)
Liver cancer [ICD-11: 2C12]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [118] | |||
| Responsed Disease | Hepatocellular carcinoma [ICD-11: 2C12.02] | |||
| Responsed Drug | Sorafenib | Approved | ||
| Target Regulation | Down regulation | |||
In-vitro Model |
SMMC-7721 | Endocervical adenocarcinoma | Homo sapiens | CVCL_0534 |
| Huh-7 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_0336 | |
| Response Summary | Long intergenic non-protein coding RNA 1273 (LINC01273) was modified with m6A, METTL3 increased LINC01273 m6A modification, followed by LINC01273 decay in the presence of YTHDF2, a m6A 'reader'. And LINC01273 plays a key role in sorafenib resistant HCC cells. | |||
Long intergenic non-protein coding RNA 470 (LINC00470)
Gastric cancer [ICD-11: 2B72]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [87] | |||
| Responsed Disease | Gastric cancer [ICD-11: 2B72] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | Gastric cancer | hsa05226 | ||
| Cell Process | Cell proliferation | |||
| Cell migration | ||||
| Cell invasion | ||||
In-vitro Model |
AGS | Gastric adenocarcinoma | Homo sapiens | CVCL_0139 |
| BGC-823 | Gastric carcinoma | Homo sapiens | CVCL_3360 | |
| GES-1 | Normal | Homo sapiens | CVCL_EQ22 | |
| HGC-27 | Gastric carcinoma | Homo sapiens | CVCL_1279 | |
| MGC-803 | Gastric mucinous adenocarcinoma | Homo sapiens | CVCL_5334 | |
| MKN45 | Gastric adenocarcinoma | Homo sapiens | CVCL_0434 | |
| Response Summary | Long intergenic non-protein coding RNA 470 (LINC00470)-METTL3-mediated PTEN mRNA degradation relied on the m6A reader protein YTHDF2-dependent pathway. LINC00470 served as a therapeutic target for Gastric cancer patients. | |||
Nuclear paraspeckle assembly transcript 1 (NEAT1)
Renal cell carcinoma [ICD-11: 2C90]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [119] | |||
| Responsed Disease | Renal cell carcinoma [ICD-11: 2C90] | |||
| Target Regulation | Down regulation | |||
| Cell Process | Cell proliferation and metastasis | |||
In-vitro Model |
769-P | Renal cell carcinoma | Homo sapiens | CVCL_1050 |
| 786-O | Renal cell carcinoma | Homo sapiens | CVCL_1051 | |
| HK2 | Normal | Acipenser baerii | CVCL_YE28 | |
| In-vivo Model | Mouse subcutaneous xenograft and lung metastasis experiments were carried out with six 4-week-old male BALB/c nude mice. | |||
| Response Summary | In renal cell carcinoma, YTHDF2 accelerated the degradation of Nuclear paraspeckle assembly transcript 1 (NEAT1)_1 by selectively recognizing METTL14-mediated m6A marks on Nuclear paraspeckle assembly transcript 1 (NEAT1)_1. | |||
X inactive specific transcript (XIST)
Colorectal cancer [ICD-11: 2B91]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [120] | |||
| Responsed Disease | Colorectal cancer [ICD-11: 2B91] | |||
| Target Regulation | Down regulation | |||
| Cell Process | Tumorigenicity and metastasis | |||
In-vitro Model |
HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 |
| HT29 | Colon cancer | Mus musculus | CVCL_A8EZ | |
| LoVo | Colon adenocarcinoma | Homo sapiens | CVCL_0399 | |
| NCM460 | Normal | Homo sapiens | CVCL_0460 | |
| SW480 | Colon adenocarcinoma | Homo sapiens | CVCL_0546 | |
| SW620 | Colon adenocarcinoma | Homo sapiens | CVCL_0547 | |
| In-vivo Model | For liver metastasis model, mice were anaesthetized and an incision was made through the skin and peritoneum to expose the spleen. 1 × 106 HCT116 cells were injected into the spleen (n = 4 each group). | |||
| Response Summary | In colorectal cancer, knockdown of METTL14 substantially abolished m6A level of X inactive specific transcript (XIST) and augmented XIST expression. m6A-methylated XIST was recognized by YTHDF2, a m6A reader protein, to mediate the degradation of XIST. | |||
microRNA 126 (MIR126)
Acute myeloid leukaemia [ICD-11: 2A60]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [121] | |||
| Responsed Disease | Acute myeloid leukaemia [ICD-11: 2A60] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
Mono-Mac-6 | Adult acute monocytic leukemia | Homo sapiens | CVCL_1426 |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| In-vivo Model | Cells were collected from the CD45.2 Ythdf2fl/flMx1-Cre mice, enriched for lin-population, and transduced with retroviral MA9 alone or retroviral MA9 + MSCV-pre-miR-126/MSCV-pre-miR-Control by spinoculation as described before. Clonal cells from CFA assays were washed and suspended together with healthy mouse BM cells (0.5 million CFA cells + 1 million healthy BM per mouse) with ice-cold PBS, and subsequently injected via the tail vein into lethally (900 rads) irradiated 8 to 10-week-old B6.SJL (CD45.1) recipient mice. Ten days after the injection, knockout was induced by i.p. delivering of poly I:C (428750 R&D Systems) at 300 μg each time per mice every other day for five times. Mice were observed regularly after cell injection and samples were collected at the end point. | |||
microRNA 145 (MIR145)
Epilepsy due to structural or metabolic conditions or diseases [ICD-11: 8A60]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [49] | |||
| Responsed Disease | Epilepsy due to structural or metabolic conditions or diseases [ICD-11: 8A60.5] | |||
hsa-mir-181b-1
Osteosarcoma [ICD-11: 2B51]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [20] | |||
| Responsed Disease | Osteosarcoma [ICD-11: 2B51] | |||
| Target Regulation | Down regulation | |||
| Cell Process | Cell growth | |||
| Cell migration | ||||
| Cell invasion | ||||
| Cell apoptosis | ||||
In-vitro Model |
U2OS | Osteosarcoma | Homo sapiens | CVCL_0042 |
| In-vivo Model | Three-week-old BABL/c female nude mice were randomized into three groups. 5 × 106 143B cells were subcutaneously injected in mice, and the tumor volume was assessed every 2 weeks. Eight weeks after injection, the animals were killed. The xenograft tumors were harvested and the tumor volumes were calculated by the standard formula: length × width2/2. | |||
| Response Summary | ALKBH5 is an anti-tumor factor or a pro-apoptotic factor, acting at least partially by suppressing YAP expression through dual mechanisms with direct m6A methylation of YAP and indirect downregulation of YAP level due to methylation of hsa-mir-181b-1. Further results revealed that m6A methylated pre-miR-181b-1 was subsequently recognized by m6A-binding protein YTHDF2 to mediate RNA degradation. However, methylated YAP transcripts were recognized by YTHDF1 to promote its translation. ALKBH5 overexpression was considered a new approach of replacement therapy for osteosarcoma treatment. | |||
hsa-miR-1915-3p
Lung cancer [ICD-11: 2C25]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [6] | |||
| Responsed Disease | Non-small-cell lung carcinoma [ICD-11: 2C25.Y] | |||
| Target Regulation | Down regulation | |||
| Pathway Response | TNF signaling pathway | hsa04668 | ||
| Cell Process | Cell migration | |||
| Cell invasion | ||||
| Epithelial-mesenchymal transition | ||||
In-vitro Model |
NCI-H1975 | Lung adenocarcinoma | Homo sapiens | CVCL_1511 |
| A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 | |
| Response Summary | hsa-miR-1915-3p expression was regulated by METTL3/YTHDF2 m6A axis through transcription factor KLF4. miR-1915-3p function as a tumor suppressor by targeting SET and has an anti-metastatic therapeutic potential for lung cancer treatment. | |||
5-hydroxytryptamine receptor 3A (HTR3A)
Pain disorders [ICD-11: 8E43]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [122] | |||
| Responsed Disease | Neuropathic pain [ICD-11: 8E43.0] | |||
| Target Regulation | Down regulation | |||
Alpha-synuclein (SNCA)
Parkinson disease [ICD-11: 8A00]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [123] | |||
| Responsed Disease | Parkinson disease [ICD-11: 8A00] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
SN4741
|
N.A. | Mus musculus | CVCL_S466 |
| In-vivo Model | Male nude mice (6 weeks old) were purchased from the Shanghai Laboratory Animal Central (Shanghai, China). 95D cells (1 × 107) transfected with sh-HNRNPA2B1 or sh-NC lentiviruses were injected subcutaneously into the right flanks of mice. After 8 weeks, the mice were sacrificed, and the xenografted tumors were collected for hematoxylin-eosin (HE) staining and IHC analysis. | |||
Apoptosis regulatory protein Siva (SIVA1)
Colorectal cancer [ICD-11: 2B91]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [124] | |||
| Responsed Disease | Colorectal cancer [ICD-11: 2B91] | |||
| Responsed Drug | Fluorouracil | Approved | ||
| Target Regulation | Down regulation | |||
In-vitro Model |
HCT 8 | Colon adenocarcinoma | Homo sapiens | CVCL_2478 |
| HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| MC-38 | Mouse colon adenocarcinoma | Mus musculus | CVCL_B288 | |
| In-vivo Model | For the tumor metastasis mouse model, 5-week-old C57BL/6 mice were randomly grouped and injected with 5 × 105 Control or shFTO stable MC38 cells via tail vein. Drug administration was adopted after 48 h. Drug administration (intraperitoneally): DMSO, 5-FU (50 mg/kg every 2 days) or Rhein (10 mg/kg every 2 days). To detect lung metastasis, mice were killed 2 weeks after tumor cell injection. Lung tissues were harvested and fixed with 4% PFA for paraffin-embedded section and lung metastases were detected with Nikon microscopy. For tumor intraperitoneal mouse model, 2 × 106 Dox-shCtrl, Dox-shFTO stable HCT8/5-FU cells were injected into 5-week-old male BALB/C nude mice. Drug administration was adopted after 48 h. Drug administration (intraperitoneally): DMSO, 5-FU (50 mg/kg every 2 days) or FB23-2 (10 mg/kg every 2 days). For tumor intraperitoneal mouse model, 5 × 105 shCtrl, shFTO-1, shFTO-2 stable MC38 cells were injected into 5-week-old C57BL/6 mice. Drug administration was adopted after 48 h. Drug administration (intraperitoneally): DMSO, 5-FU (50 mg/kg every 2 days) or Rhein (10 mg/kg every 2 days). | |||
Aryl hydrocarbon receptor (AHR)
Brain cancer [ICD-11: 2A00]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [104] | |||
| Responsed Disease | Glioblastoma [ICD-11: 2A00.00] | |||
| Responsed Drug | Tislelizumab | Approved | ||
In-vitro Model |
U-87MG ATCC | Glioblastoma | Homo sapiens | CVCL_0022 |
Carbohydrate sulfotransferase 11 (CHST11)
Diffuse large B-cell lymphomas [ICD-11: 2A81]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [125] | |||
| Responsed Disease | Diffuse large B-cell lymphomas [ICD-11: 2A81] | |||
| Target Regulation | Down regulation | |||
In-vitro Model |
OCI-Ly1 | Diffuse large B-cell lymphoma | Homo sapiens | CVCL_1879 |
| OCI-Ly8 | Diffuse large B-cell lymphoma germinal center B-cell type | Homo sapiens | CVCL_8803 | |
| OCI-Ly3 | Diffuse large B-cell lymphoma activated B-cell type | Homo sapiens | CVCL_8800 | |
| VAL | Diffuse large B-cell lymphoma | Homo sapiens | CVCL_1819 | |
| U-2932 | Diffuse large B-cell lymphoma | Homo sapiens | CVCL_1896 | |
| In-vivo Model | A total of 1 × 107 KIAA1429 stable knockdown OCI-LY1 cells or CHST11 stable knockdown OCI-LY1 cells were injected subcutaneously into the right armpit of mice. Two investigators who were blinded to the mice allocation observed the general condition of mice and tumor growth every 2 days, measuring tumor size with a vernier caliper upon the tumor size was higher than the skin surface and recording it. | |||
Cardiac mesoderm enhancer-associated non-coding RNA (CARMN)
Cervical cancer [ICD-11: 2C77]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [126] | |||
| Responsed Disease | Cervical cancer [ICD-11: 2C77] | |||
| Target Regulation | Down regulation | |||
In-vitro Model |
HeLa | Endocervical adenocarcinoma | Homo sapiens | CVCL_0030 |
| SiHa | Cervical squamous cell carcinoma | Homo sapiens | CVCL_0032 | |
| In-vivo Model | After approval by Ethics Committee of School of Public Health, Southeast University, BALB/c nude mice (4-5 weeks old) were purchased and raised in SPF animal house. 1 × 107 HeLa cells or NC cells with stable overexpression of CARMN were prepared in each nude mouse. The mice were killed 3 weeks later, and tumor volume was measured. | |||
Carnitine O-palmitoyltransferase 1, liver isoform (CPT1A)
Heart failure [ICD-11: BD1Z]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [127] | |||
| Responsed Disease | Heart failure [ICD-11: BD1Z] | |||
| Target Regulation | Down regulation | |||
CBSLR
Gastric cancer [ICD-11: 2B72]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [64] | |||
| Responsed Disease | Gastric cancer [ICD-11: 2B72] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | Ubiquitin mediated proteolysis | hsa04120 | ||
| Cell Process | Proteasome pathway degradation | |||
In-vitro Model |
SGC-7901 | Gastric carcinoma | Homo sapiens | CVCL_0520 |
| MKN45 | Gastric adenocarcinoma | Homo sapiens | CVCL_0434 | |
| MKN28 | Gastric tubular adenocarcinoma | Homo sapiens | CVCL_1416 | |
| MGC-803 | Gastric mucinous adenocarcinoma | Homo sapiens | CVCL_5334 | |
| GSE-1 (Gse-1 is a human gastric epithelial cell line) | ||||
| BGC-823 | Gastric carcinoma | Homo sapiens | CVCL_3360 | |
| AGS | Gastric adenocarcinoma | Homo sapiens | CVCL_0139 | |
| Response Summary | CBSLR interacted with YTHDF2 to form a CBSLR/YTHDF2/CBS signaling axis that decreased the stability of CBS mRNA by enhancing the binding of YTHDF2 with the m6A-modified coding sequence (CDS) of CBS mRNA. Reveal a novel mechanism in how HIF1-Alpha/CBSLR modulates ferroptosis/chemoresistance in GC, illuminating potential therapeutic targets for refractory hypoxic tumors. | |||
CD40 ligand (CD40L)
Immune-related diseases [ICD-11: 4B4Z]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [128] | |||
| Responsed Disease | Immune-related diseases [ICD-11: 4B4Z] | |||
| Target Regulation | Down regulation | |||
CD70 antigen (CD70)
Thyroid Cancer [ICD-11: 2D10]
| In total 2 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [129] | |||
| Responsed Disease | Papillary thyroid cancer [ICD-11: 2D10.1] | |||
| Responsed Drug | JNJ-74494550 | Phase 2 | ||
| Target Regulation | Down regulation | |||
In-vitro Model |
Nthy-ori 3-1 | Normal | Homo sapiens | CVCL_2659 |
| B-CPAP | Thyroid gland carcinoma | Homo sapiens | CVCL_0153 | |
| K1 | Thyroid gland papillary carcinoma | Homo sapiens | CVCL_2537 | |
| C-643 | Thyroid gland anaplastic carcinoma | Homo sapiens | CVCL_5969 | |
| BHT-101 | Thyroid gland anaplastic carcinoma | Homo sapiens | CVCL_1085 | |
| THP-1 | Childhood acute monocytic leukemia | Homo sapiens | CVCL_0006 | |
| In-vivo Model | NCG mice (female, 4-5 weeks old, purchased from Jiangsu GemPharmatech) were acclimated for 1 week and were selected randomly to subcutaneously (s.c.) injected with 100 μl suspensions of 1 × 106 thyroid cancer cell lines. All mice were housed in animal facility under specific pathogen-free conditions. After 7 day, 5 × 106 huPBMCs were intravenously (i.v.) inoculated into mice. Mice were treated intravenously with anti-PD-1 mAb (nivolumab, 200 μg per injection), Combination (nivolumab + anti-CD70 mAb cusatuzumab, 200 μg per injection each), or PBS once a week. The treatment with M2-TAMs EVs or PBS continued in mice at 2ug EVs per injection at the tumor site twice a week [24]. The body weight of mice and the length (mm) and width (mm) of tumors were monitored every 3 or 4 days. Tumor volume (mm3) was calculated as Length × Width2 × 1/2. Tumor growth analyses were limited to 3 to 4 weeks because following this period mice started to show signs of xenograft-versus-host disease (xGVHD). Therefore, after 14 days or 21 days of PBMCs implantation, the mice were euthanized. | |||
| Experiment 2 Reporting the m6A-centered Disease Response of This Target Gene | [129] | |||
| Responsed Disease | Papillary thyroid cancer [ICD-11: 2D10.1] | |||
| Target Regulation | Down regulation | |||
In-vitro Model |
Nthy-ori 3-1 | Normal | Homo sapiens | CVCL_2659 |
| B-CPAP | Thyroid gland carcinoma | Homo sapiens | CVCL_0153 | |
| K1 | Thyroid gland papillary carcinoma | Homo sapiens | CVCL_2537 | |
| C-643 | Thyroid gland anaplastic carcinoma | Homo sapiens | CVCL_5969 | |
| BHT-101 | Thyroid gland anaplastic carcinoma | Homo sapiens | CVCL_1085 | |
| THP-1 | Childhood acute monocytic leukemia | Homo sapiens | CVCL_0006 | |
| In-vivo Model | NCG mice (female, 4-5 weeks old, purchased from Jiangsu GemPharmatech) were acclimated for 1 week and were selected randomly to subcutaneously (s.c.) injected with 100 μl suspensions of 1 × 106 thyroid cancer cell lines. All mice were housed in animal facility under specific pathogen-free conditions. After 7 day, 5 × 106 huPBMCs were intravenously (i.v.) inoculated into mice. Mice were treated intravenously with anti-PD-1 mAb (nivolumab, 200 μg per injection), Combination (nivolumab + anti-CD70 mAb cusatuzumab, 200 μg per injection each), or PBS once a week. The treatment with M2-TAMs EVs or PBS continued in mice at 2ug EVs per injection at the tumor site twice a week [24]. The body weight of mice and the length (mm) and width (mm) of tumors were monitored every 3 or 4 days. Tumor volume (mm3) was calculated as Length × Width2 × 1/2. Tumor growth analyses were limited to 3 to 4 weeks because following this period mice started to show signs of xenograft-versus-host disease (xGVHD). Therefore, after 14 days or 21 days of PBMCs implantation, the mice were euthanized. | |||
Circ_AFF2
Colorectal cancer [ICD-11: 2B91]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [130] | |||
| Responsed Disease | Colorectal cancer [ICD-11: 2B91] | |||
| Target Regulation | Down regulation | |||
In-vitro Model |
RKO | Colon carcinoma | Homo sapiens | CVCL_0504 |
| SW480 | Colon adenocarcinoma | Homo sapiens | CVCL_0546 | |
| HT-29 | Colon adenocarcinoma | Homo sapiens | CVCL_0320 | |
| LoVo | Colon adenocarcinoma | Homo sapiens | CVCL_0399 | |
| HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 | |
| SW620 | Colon adenocarcinoma | Homo sapiens | CVCL_0547 | |
| NCM460 | Normal | Homo sapiens | CVCL_0460 | |
Circ_ASK1
Lung cancer [ICD-11: 2C25]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [131] | |||
| Responsed Disease | Lung adenocarcinoma [ICD-11: 2C25.0] | |||
| Responsed Drug | Gefitinib | Approved | ||
| Target Regulation | Down regulation | |||
| Cell Process | Cell apoptosis | |||
In-vitro Model |
SPC-A1 | Endocervical adenocarcinoma | Homo sapiens | CVCL_6955 |
| SK-LU-1 | Lung adenocarcinoma | Homo sapiens | CVCL_0629 | |
| NCI-H1993 | Lung adenocarcinoma | Homo sapiens | CVCL_1512 | |
| NCI-H1975 | Lung adenocarcinoma | Homo sapiens | CVCL_1511 | |
| NCI-H1650 | Minimally invasive lung adenocarcinoma | Homo sapiens | CVCL_1483 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| HCC827 | Lung adenocarcinoma | Homo sapiens | CVCL_2063 | |
| BEAS-2B | Normal | Homo sapiens | CVCL_0168 | |
| A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 | |
| 16HBE14o- | Normal | Homo sapiens | CVCL_0112 | |
| In-vivo Model | Established a xenograft model in BALB/c nude mice by inoculating HCC827-GR cells transfected with the constructs for circASK1 silencing, ASK1-272a.a overexpression and ASK1-272a.a overexpression/circASK1 knockdown. | |||
| Response Summary | Increased YTHDF2-mediated endoribonucleolytic cleavage of m6A-modified Circ_ASK1 accounts for its downregulation in gefitinib-resistant cells. Either METTL3 silencing or YTHDF2 silencing suppressed the decay of circASK1 in HCC827-GR cells. This study provides a novel therapeutic target to overcome gefitinib resistance in LUAD patients. | |||
Circ_FUT8
Lung cancer [ICD-11: 2C25]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [132] | |||
| Responsed Disease | Lung adenocarcinoma [ICD-11: 2C25.0] | |||
In-vitro Model |
A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 |
| PC-9 | Lung adenocarcinoma | Homo sapiens | CVCL_B260 | |
| NCI-H1299 | Lung large cell carcinoma | Homo sapiens | CVCL_0060 | |
| SPC-A1 | Endocervical adenocarcinoma | Homo sapiens | CVCL_6955 | |
| NCI-H1975 | Lung adenocarcinoma | Homo sapiens | CVCL_1511 | |
| In-vivo Model | Thirty female BALB/c nude mice weighing 18-22 g were randomly assigned to six groups. PC9-Mock, PC9-circFUT8, A549-sh-scramble, and A549-sh-circFUT8 cells were prepared as a suspension of 4 × 105 cells in 200 μL saline, respectively, and injected into the tail vein. Mice were sacrificed at 6 weeks post injection and examined microscopically by hematoxylin and eosin (H&E) staining for the development of lung metastases. | |||
Circ_GPATCH2L
Spinal pain [ICD-11: ME84]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [133] | |||
| Responsed Disease | Spinal pain [ICD-11: ME84.2] | |||
Circ_IRF2
Hepatic fibrosis/cirrhosis [ICD-11: DB93]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [134] | |||
| Responsed Disease | Hepatic fibrosis/cirrhosis [ICD-11: DB93] | |||
| Target Regulation | Down regulation | |||
In-vitro Model |
LX-2
|
N.A. | Homo sapiens | CVCL_5792 |
| In-vivo Model | For the TAA (Sigma-Aldrich, USA) induced mouse liver fibrosis model, briefly, TAA (200 mg/kg, diluted in saline) was injected 3 times weekly for 8 weeks. Mice were sacrificed 24 h after the last administration. | |||
Circ_RERE
Osteoarthritis [ICD-11: FA05]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [135] | |||
| Responsed Disease | Osteoarthritis [ICD-11: FA05] | |||
| Responsed Drug | ICG-001 | Investigative | ||
| Target Regulation | Down regulation | |||
Circ_RNF13
Cervical cancer [ICD-11: 2C77]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [136] | |||
| Responsed Disease | Cervical cancer [ICD-11: 2C77] | |||
| Target Regulation | Down regulation | |||
In-vitro Model |
SiHa | Cervical squamous cell carcinoma | Homo sapiens | CVCL_0032 |
| C-4-I | Cervical squamous cell carcinoma | Homo sapiens | CVCL_2253 | |
| HeLa | Endocervical adenocarcinoma | Homo sapiens | CVCL_0030 | |
| C-33 A | Cervical squamous cell carcinoma | Homo sapiens | CVCL_1094 | |
| In-vivo Model | Stably transfected cell lines were created by silencing circRNF13 in CC SiHa cells. Once xenografts were established, the tumors reached an approximate volume of 200 mm3. A single dose of 15 Gy irradiation was administered to female BALB/c nude mice (4-5 weeks old) in the murine model. The tumor volume was measured and recorded using vernier calipers every five days after irradiation. After 30 days, the mice were euthanized under anesthesia, and tumor tissue was collected for further investigations. | |||
Circ_SLC9A6
Non-alcoholic fatty liver disease [ICD-11: DB92]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [137] | |||
| Responsed Disease | Nonalcoholic fatty liver disease [ICD-11: DB92.Z] | |||
| Target Regulation | Down regulation | |||
Complement C1q subcomponent subunit A (C1QA)
Diffuse large B-cell lymphomas [ICD-11: 2A81]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [138] | |||
| Responsed Disease | Diffuse large B-cell lymphomas [ICD-11: 2A81] | |||
| Responsed Drug | Rituximab | Approved | ||
| Target Regulation | Down regulation | |||
| In-vivo Model | Approximately 2 × 106 Farage/R or Farage/S cells stably transfected with shNC, shC1qA or shMETTL3 were subcutaneously injected into the left flank of each mouse. When the tumor volume reached ~100 mm3, each mouse received an intraperitoneal injection of Rituximab (20 mg/kg) every 4 days for a total of 5 injections. The diameter of each tumor was examined every 4 days using a caliper, and tumor volume was calculated as follows: (length × width2)/2. At 28 days after xenograft, the mice were sacrificed and the tumors were weighed and collected. | |||
Cysteine methyltransferase DNMT3A (DNMT3A)
Chronic obstructive pulmonary disease [ICD-11: CA22]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [140] | |||
| Responsed Disease | Chronic obstructive pulmonary disease [ICD-11: CA22] | |||
| Target Regulation | Down regulation | |||
| In-vivo Model | Male BALB/c mice (SJA Laboratory Animal Company, Hunan, China) with age of 6-8 weeks were used in this study to establish COPD model. Mice were housed in individually ventilated cages under a pathogen-free condition, with ad libitum access to food and water. Animal welfare was monitored daily, and all efforts were made to minimize suffering. All animal procedures were conducted in accordance with the guidelines for use of laboratory animals, with approval from the Institutional Animal Care and Use Committee at Jiangxi Provincial People's Hospital (The First Affiliated Hospital of Nanchang Medical College). | |||
Cysteine protease ATG4A (ATG4A)
Intervertebral disc degeneration [ICD-11: FA80]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [141] | |||
| Responsed Disease | Intervertebral disc degeneration [ICD-11: FA80] | |||
| Target Regulation | Down regulation | |||
D26496
Injury of nerves at hip or thigh level [ICD-11: NC74]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [142] | |||
| Responsed Disease | Injury of nerves at hip or thigh level [ICD-11: NC74.0] | |||
| Target Regulation | Down regulation | |||
In-vitro Model |
Schwann cells (A type of glial cell that surrounds neurons) | |||
| In-vivo Model | Rats were randomly divided into 4 groups: sham group (6 rats), SNI model group (24 rats), SNI + vector group (6 rats), and SNI + pcDNA-D26496 group (6 rats). D26496 overexpressed Adeno associated virus (AAVs, virus titer: 1.88E + 13) and vector (OBiO Technology Corp., Shanghai, China) were injected into the epineurium of sciatic nerve of rats (5 μL for each) in the SNI + vector group and SNI + pcDNA-D26496 group, respectively. | |||
DNA replication licensing factor MCM7 (MCM2)
Liver cancer [ICD-11: 2C12]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [69] | |||
| Responsed Disease | Liver hepatocellular carcinoma [ICD-11: 2C12.02] | |||
| Responsed Drug | OSMI-1 | Investigative | ||
| Target Regulation | Up regulation | |||
In-vitro Model |
HepAD38 | Hepatoblastoma | Homo sapiens | CVCL_M177 |
| Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 | |
| PLC/PRF/5 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_0485 | |
| MHCC97-H | Adult hepatocellular carcinoma | Homo sapiens | CVCL_4972 | |
| Huh-7 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_0336 | |
| HEK293 | Normal | Homo sapiens | CVCL_0045 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| In-vivo Model | For the xenograft implantation model, 2 × 106 MHCC-97H cells were subcutaneously injected into the flank of nude mice. | |||
Dual specificity protein phosphatase 1 (DUSP1)
Bacterial infection [ICD-11: 1C41]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [143] | |||
| Responsed Disease | Bacterial infection [ICD-11: 1C41] | |||
| Target Regulation | Down regulation | |||
In-vitro Model |
RAW 264.7 | Mouse leukemia | Mus musculus | CVCL_0493 |
E3 ubiquitin-protein ligase pellino homolog 2 (PELI2)
Unspecific body region injury [ICD-11: ND56]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [144] | |||
| Responsed Disease | Cutaneous wound [ICD-11: ND56.0] | |||
| Target Regulation | Down regulation | |||
In-vitro Model |
HaCaT | Normal | Homo sapiens | CVCL_0038 |
Elongation factor 2 (EEF2)
Heart failure [ICD-11: BD1Z]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [145] | |||
| Responsed Disease | Heart failure [ICD-11: BD1Z] | |||
| In-vivo Model | All experiments were performed in 10-wk-old male mice unless otherwise indicated. Mice carrying a floxed Ythdf2 allele [24] and the alpha-MHC Cre Ribo-tag mouse [21] were crossed to obtain a cardiomyocyte specific Ythdf2 KO mouse expressing a HA-tagged RPL22. The mice were housed in a temperature- and humidity-controlled facility with a 12-h light-dark cycle. At 10 wk. of age, male mice underwent TAC (27 gauge needle) or sham operation, as previously described [25]. For echocardiography, the mice were anesthetized with 2% isoflurane and scanned using a Vevo2100 imaging system (Visual Sonics) as previously described [46]. Institutional Animal Care and Use Committee approval was obtained for all animal studies. | |||
ETS translocation variant 5 (ETV5)
Liver cancer [ICD-11: 2C12]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [146] | |||
| Responsed Disease | Liver cancer [ICD-11: 2C12] | |||
| Target Regulation | Up regulation | |||
Fibulin-1 (FBLN1)
Maxillofacial bone defect is a critical obstacle for maxillofacial tumors and periodontal diseases. [ICD-11: NA02]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [147] | |||
| Responsed Disease | Maxillofacial bone defect is a critical obstacle for maxillofacial tumors and periodontal diseases. [ICD-11: NA02] | |||
| Target Regulation | Down regulation | |||
In-vitro Model |
HEK293T | Normal | Homo sapiens | CVCL_0063 |
Glutamate--cysteine ligase regulatory subunit (GCLM)
Brain cancer [ICD-11: 2A00]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [148] | |||
| Responsed Disease | Glioblastoma [ICD-11: 2A00.00] | |||
In-vitro Model |
HEK293T | Normal | Homo sapiens | CVCL_0063 |
| ENSA (A human embryonic stem derived neural progenitor cell) | ||||
| NSC11 (Pluripotent derived neural progenitor cell) | ||||
| In-vivo Model | Intracranial xenografts were generated by implanting 20,000 patient derived GSCs into the right cerebral cortex of mice at a depth of 3.5 mm. Housing conditions and animal status were supervised by a veterinarian. Euthanasia was taken until neurologic symptoms included hunched posture, gait changes, or lethargy were observed, at which point they were sacrificed. Brains were harvested and frozen at -80 °C with O.C.T. compound (4583, Tissue-Tek) directly or fixed in 4 % formaldehyde for 48 hours then stored in 70 % ethanol, and sectioned. | |||
GTP-binding protein 4 (GTPBP4)
Picornavirus infections presenting in the skin or mucous membranes [ICD-11: 1F05]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [83] | |||
| Responsed Disease | Picornavirus infections presenting in the skin or mucous membranes [ICD-11: 1F05.3] | |||
In-vitro Model |
PK-15
|
N.A. | Sus scrofa | CVCL_2160 |
| HT-29 | Colon adenocarcinoma | Homo sapiens | CVCL_0320 | |
|
IB-RS-2
|
N.A. | Sus scrofa | CVCL_4528 | |
|
CPTC-SVIL-1
|
N.A. | Oryctolagus cuniculus | CVCL_C2LN | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
|
BHK-21
|
N.A. | Mesocricetus auratus | CVCL_1914 | |
| RD | Embryonal rhabdomyosarcoma | Homo sapiens | CVCL_1649 | |
Heat shock 70 kDa protein 6 (HSPA6)
Endometrial cancer [ICD-11: 2C76]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [149] | |||
| Responsed Disease | Endometrial cancer [ICD-11: 2C76] | |||
In-vitro Model |
Ishikawa | Endometrial adenocarcinoma | Homo sapiens | CVCL_2529 |
| HEK293 | Normal | Homo sapiens | CVCL_0045 | |
| HeLa | Endocervical adenocarcinoma | Homo sapiens | CVCL_0030 | |
Heme oxygenase 1 (HMOX1)
Pulmonary hypertension [ICD-11: BB01]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [150] | |||
| Responsed Disease | Pulmonary hypertension [ICD-11: BB01] | |||
| Target Regulation | Down regulation | |||
In-vitro Model |
MH-S
|
N.A. | Mus musculus | CVCL_3855 |
Diseases of the female genital system [ICD-11: GA6Z]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [151] | |||
| Responsed Disease | Diseases of the female genital system [ICD-11: GA6Z] | |||
| Target Regulation | Down regulation | |||
| In-vivo Model | Daily vaginal smears were collected between 4 and 6 PM, and rats showing four consecutive 4-day estrus cycles were chosen for constructing the endometrial injury model. Anesthesia was induced by injecting 3% mebumalnatrium (dose: 1.4 mL/kg) into the lumbar region, followed by a low midline abdominal incision for uterus exposure. | |||
Histone acetyltransferase KAT2A (KAT2A)
Cardiomyopathy [ICD-11: BC43]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [152] | |||
| Responsed Disease | Diabetic cardiomyopathy [ICD-11: BC43.7] | |||
| Target Regulation | Up regulation | |||
Insulin-like growth factor-binding protein 3 (IGFBP3)
Alzheimer disease [ICD-11: 8A20]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [49] | |||
| Responsed Disease | Alzheimer disease [ICD-11: 8A20] | |||
Interleukin-6 receptor subunit alpha (IL6R)
Sepsis [ICD-11: 1G40]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [153] | |||
| Responsed Disease | Sepsis [ICD-11: 1G40] | |||
| Target Regulation | Down regulation | |||
In-vitro Model |
RAW 264.7 | Mouse leukemia | Mus musculus | CVCL_0493 |
long intergenic non-protein coding RNA 115 (LINC00115)
Triple-negative breast cancer [ICD-11: 2C6Z]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [154] | |||
| Responsed Disease | Triple-negative breast cancer [ICD-11: 2C6Z] | |||
| Responsed Drug | Acetaminophen | Approved | ||
| Target Regulation | Down regulation | |||
Lysine-specific demethylase 6B (KDM6B)
Inflammatory response [ICD-11: MG46]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [155] | |||
| Responsed Disease | Inflammatory response [ICD-11: MG46] | |||
In-vitro Model |
THP-1 | Childhood acute monocytic leukemia | Homo sapiens | CVCL_0006 |
Metalloproteinase inhibitor 4 (TIMP4)
Osteoarthritis [ICD-11: FA05]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [156] | |||
| Responsed Disease | Osteoarthritis [ICD-11: FA05] | |||
| Target Regulation | Up regulation | |||
| In-vivo Model | The male C57BL/6 mice (SPF, Eight-week-old) were randomly divided into four groups: sham (n = 6), model (n = 6), model + adeno-associated virus-negative control (AVV-shNC) (n = 6), and model + AVV-shWTAP groups (n = 6). After acclimating for one week, mice in other groups were treated with DMM surgery to induce OA as previously described except for the sham groups [19]. The mice in sham group were underwent only the skin of the right knee joint incision. The AAV-shNC and AAV-shWTAP were constructed by HANBIO (Shanghai, China). The AAV-shNC and AAV-shWTAP groups were intra-articularly injected with 10 μL of AAV-shNC and AAV-shWTAP (1 × 1015 vg/ml) through the medial parapatellar area at two weeks after the DMM operation, respectively. | |||
Metalloreductase STEAP3 (STEAP3)
Colorectal cancer [ICD-11: 2B91]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [157] | |||
| Responsed Disease | Colorectal cancer [ICD-11: 2B91] | |||
| Target Regulation | Down regulation | |||
In-vitro Model |
HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 |
| RKO | Colon carcinoma | Homo sapiens | CVCL_0504 | |
| DLD-1 | Colon adenocarcinoma | Homo sapiens | CVCL_0248 | |
| LoVo | Colon adenocarcinoma | Homo sapiens | CVCL_0399 | |
| SW480 | Colon adenocarcinoma | Homo sapiens | CVCL_0546 | |
| SW620 | Colon adenocarcinoma | Homo sapiens | CVCL_0547 | |
| HT29 | Colon cancer | Mus musculus | CVCL_A8EZ | |
| NCM460 | Normal | Homo sapiens | CVCL_0460 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| In-vivo Model | Six to eight weeks old male BALB/c nu/nu mice were used. For orthotopic implantation, 1 × 107 PBS suspended cells was injected subcutaneously. For the spleen injection model, 5 × 105 cells were injected into the spleen through an incision on the left side of abdomen. | |||
Mothers against decapentaplegic homolog 4 (SMAD4)
Hematological disorders [ICD-11: 3C0Z]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [43] | |||
| Responsed Disease | Hematological disorders [ICD-11: 3C0Z] | |||
| Target Regulation | Down regulation | |||
Myocardial zonula adherens protein (MYZAP)
Cardiomyopathy [ICD-11: BC43]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [159] | |||
| Responsed Disease | Cardiomyopathy [ICD-11: BC43] | |||
| Target Regulation | Up regulation | |||
Nuclear factor of activated T-cells, cytoplasmic 1 (NFATC1)
Spinal deformities [ICD-11: FA70]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [160] | |||
| Responsed Disease | Spinal deformities [ICD-11: FA70.1] | |||
| Target Regulation | Down regulation | |||
In-vitro Model |
C2C12 | Normal | Mus musculus | CVCL_0188 |
Paired box protein Pax-6 (PAX6)
Acute ischemic stroke [ICD-11: 8B11]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [161] | |||
| Responsed Disease | Acute ischemic stroke [ICD-11: 8B11] | |||
In-vitro Model |
BV-2 | Normal | Mus musculus | CVCL_0182 |
| In-vivo Model | C57BL/6 mice were anesthetized intraperitoneally with 1% sodium pentobarbital (100 mg/kg). The mouse was placed on a thermostatic blanket to maintain the rectal temperature at 37.0°C± 0.5°C during surgery. The left common carotid artery (CCA), external carotid artery (ECA), and internal carotid artery (ICA) were exposed through a midline incision in the cervico abdominal region. The CCA was ligated distally with surgical nylon monofilament. | |||
Potassium channel subfamily K member 6 (KCNK6)
Colorectal cancer [ICD-11: 2B91]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [163] | |||
| Responsed Disease | Colorectal cancer [ICD-11: 2B91] | |||
| Target Regulation | Up regulation | |||
Prolyl endopeptidase FAP (FAP)
Lung cancer [ICD-11: 2C25]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [164] | |||
| Responsed Disease | Non-small cell lung cancer [ICD-11: 2C25.Y] | |||
| Responsed Drug | VS-6063 | Phase 2 | ||
In-vitro Model |
NCI-H1299 | Lung large cell carcinoma | Homo sapiens | CVCL_0060 |
| A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 | |
| NCI-H1650 | Minimally invasive lung adenocarcinoma | Homo sapiens | CVCL_1483 | |
| NCI-H460 | Lung large cell carcinoma | Homo sapiens | CVCL_0459 | |
| HCC827 | Lung adenocarcinoma | Homo sapiens | CVCL_2063 | |
| PC-9 | Lung adenocarcinoma | Homo sapiens | CVCL_B260 | |
| SK-MES-1 | Lung squamous cell carcinoma | Homo sapiens | CVCL_0630 | |
| BEAS-2B | Normal | Homo sapiens | CVCL_0168 | |
| In-vivo Model | The Laboratory Animal Center of Soochow University provided female BALB/c nude mice (5 weeks old). Mice were kept under specific pathogen-free conditions. They were injected intravenously (i.v.) with FTO-overexpressing and control A549 cells (1.8 × 106 cells/mouse) to establish the in vivo model of NSCLC metastasis, and were then gavaged with dimethyl sulfoxide (DMSO) (25 mg/kg, daily) or the FAK inhibitor defactinib (VS6063) (Cat#S7654, Selleck, USA) (25 mg/kg, daily) beginning in the fifth week after injection. The mice were euthanized eight weeks after being inoculated, and their lungs were taken out and preserved in Bouin's solution for macroscopic investigation of metastatic nodules. Hematoxylin and eosin (H&E) staining of lung tissues was used to look for micrometastatic foci. | |||
Protein flightless-1 homolog (FLII)
Prostate cancer [ICD-11: 2C82]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [165] | |||
| Responsed Disease | Prostate adenocarcinoma [ICD-11: 2C82.0] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
RWPE-1 | Normal | Homo sapiens | CVCL_3791 |
| In-vivo Model | For tumor growth measurement, 5 × 106 stably transfected 22RV1 cells were subcutaneously injected into mice at the flank site. Then, the volume (V) of xenograft tumors was examined once per week as follows: V = length × width2/2. After 4 weeks, the mice were euthanized via intraperitoneal injection of 150 mg/kg pentobarbital sodium, and the tumors were taken out and weighed, and collected for immunohistochemistry (IHC) to examine the expression of the proliferation marker Ki67 (1:200, ab16667, Abcam) in xenograft tumors. For tumor metastasis measurement, 5 × 106 stably transfected 22RV1 cells were injected into mice through tail vein. | |||
Protein NLRC5
Endometrial cancer [ICD-11: 2C76]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [166] | |||
| Responsed Disease | Endometrial cancer [ICD-11: 2C76] | |||
| Target Regulation | Up regulation | |||
Protein wntless homolog (WLS)
Pain disorders [ICD-11: 8E43]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [167] | |||
| Responsed Disease | Neuropathic pain [ICD-11: 8E43.0] | |||
| Target Regulation | Down regulation | |||
In-vitro Model |
PC-12 | Lung papillary adenocarcinoma | Homo sapiens | CVCL_S979 |
| In-vivo Model | Rats were anesthetized with isoflurane (1.5%-2.5%), and a lateral thigh incision was made. The left sciatic nerve and its three peripheral branches were isolated after sciatic nerve resection. A 5-0 suture was used to ligate the common peroneal and tibial nerves. Then, distal transverse incisions were made at the ligation. All surgical procedures were completed within 20 min, with special care taken to avoid damaging the sural nerve. The rats in the sham group underwent the same sciatic nerve exposure operation as the SNI group except for nerve ligation. | |||
Protein-tyrosine kinase 2-beta (PYK2)
Inflammatory response [ICD-11: MG46]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [168] | |||
| Responsed Disease | Inflammatory response [ICD-11: MG46] | |||
In-vitro Model |
RAW 264.7 | Mouse leukemia | Mus musculus | CVCL_0493 |
Pseudorabies Virus (PRV)
Rabies [ICD-11: 1C82]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [169] | |||
| Responsed Disease | Rabies [ICD-11: 1C82] | |||
| Responsed Drug | 3-deazidenosine | Investigative | ||
| Target Regulation | Up regulation | |||
In-vitro Model |
PK-15
|
N.A. | Sus scrofa | CVCL_2160 |
Ras-related protein R-Ras (RRAS)
Bladder cancer [ICD-11: 2C94]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [170] | |||
| Responsed Disease | Bladder cancer [ICD-11: 2C94] | |||
| Target Regulation | Down regulation | |||
In-vitro Model |
SV-HUC-1 | Normal | Homo sapiens | CVCL_3798 |
| UM-UC-3 | Bladder carcinoma | Homo sapiens | CVCL_1783 | |
| T24 | Bladder carcinoma | Homo sapiens | CVCL_0554 | |
| J82 | Bladder carcinoma | Homo sapiens | CVCL_0359 | |
| TCCSUP | Bladder carcinoma | Homo sapiens | CVCL_1738 | |
| 5637 | Bladder carcinoma | Homo sapiens | CVCL_0126 | |
| In-vivo Model | A total of 16 male, 4- to 6-week-old BALB/c nude mice weighing 18-26 g, were purchased from the Institute of Model Animals, Nanjing University and raised in a specific pathogen-free (SPF) environment for the xenograft model. The mice were raised in animal individually ventilated cage (IVC cages) with a room temperature of 24°C and relative humidity of 70% and the air exchange rate was 15 times/h. The mice could drink filtered tap water and commercial feed ad libitum under a strict 12-h light/dark cycle. The animal laboratory was cleaned twice one day and sterilized with ultraviolet light for 1 h each week. Each mouse was injected with 100 μl PBS containing 1×106 T24 cell lines expressing either LV-NC or LV-shMETTL3 subcutaneously under the right armpit. Tumor volumes were recorded on day 7 after the injection and then measured each week. At day 28, the mice were sacrificed by cervical dislocation following the inhalation of 3% isoflurane anesthesia and the death of the mice was confirmed by respiratory arrest. Tumor weights were also compared. The data from each group of mice are expressed as the mean ± standard deviation (SD). | |||
Ras-related protein Rab-5A (RAB5A)
Colorectal cancer [ICD-11: 2B91]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [171] | |||
| Responsed Disease | Colorectal cancer [ICD-11: 2B91] | |||
| Target Regulation | Down regulation | |||
| In-vivo Model | Briefly, 4 × 106 transfected HCT116 cells or 8 × 106 transfected SW480 cells in 0.1 mL PBS were injected into mice subcutaneously, and these mice were randomly divided into the experimental and control group. | |||
Retinoic acid receptor RXR-alpha (RXRA)
Autoimmune liver disease [ICD-11: DB96]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [172] | |||
| Responsed Disease | Autoimmune liver disease [ICD-11: DB96.0] | |||
| Target Regulation | Down regulation | |||
RNA demethylase ALKBH5 (ALKBH5)
Diabetic encephalopathy [ICD-11: 8E47]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [49] | |||
| Responsed Disease | Diabetic encephalopathy [ICD-11: 8E47] | |||
RNA-binding protein Nova-2 (NOVA2)
Hepatic fibrosis/cirrhosis [ICD-11: DB93]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [173] | |||
| Responsed Disease | Hepatic fibrosis/cirrhosis [ICD-11: DB93] | |||
| Target Regulation | Down regulation | |||
Runt-related transcription factor 1 (RUNX1)
Hematological disorders [ICD-11: 3C0Z]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [43] | |||
| Responsed Disease | Hematological disorders [ICD-11: 3C0Z] | |||
| Target Regulation | Down regulation | |||
Semaphorin-3F (SEMA3F)
Prostate cancer [ICD-11: 2C82]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [174] | |||
| Responsed Disease | Prostate cancer [ICD-11: 2C82] | |||
In-vitro Model |
RWPE-1 | Normal | Homo sapiens | CVCL_3791 |
| DU145 | Prostate carcinoma | Homo sapiens | CVCL_0105 | |
| PC-3 | Prostate carcinoma | Homo sapiens | CVCL_0035 | |
| 22Rv1 | Prostate carcinoma | Homo sapiens | CVCL_1045 | |
| LNCaP | Prostate carcinoma | Homo sapiens | CVCL_0395 | |
| LNCaP C4-2B | Prostate carcinoma | Homo sapiens | CVCL_4784 | |
| VCaP | Prostate carcinoma | Homo sapiens | CVCL_2235 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| In-vivo Model | All the animal studies and protocols followed the institutional guidelines of the First Affiliated Hospital, School of Medicine, Zhejiang University. | |||
Serine/threonine-protein kinase LATS1 (LATS1)
Breast cancer [ICD-11: 2C60]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [175] | |||
| Responsed Disease | Breast cancer [ICD-11: 2C60] | |||
| Target Regulation | Down regulation | |||
In-vitro Model |
SK-BR-3 | Breast adenocarcinoma | Homo sapiens | CVCL_0033 |
| MCF-7 | Invasive breast carcinoma | Homo sapiens | CVCL_0031 | |
| T-47D | Invasive breast carcinoma | Homo sapiens | CVCL_0553 | |
| BT-474 | Invasive breast carcinoma | Homo sapiens | CVCL_0179 | |
| MDA-MB-468 | Breast adenocarcinoma | Homo sapiens | CVCL_0419 | |
| MDA-MB-231 | Breast adenocarcinoma | Homo sapiens | CVCL_0062 | |
| In-vivo Model | All animal experiments were approved by the Institutional Animal Care and Use Committee of Southern Medical University. Breast cancer cells (5 × 106) were implanted into the subcutaneous axilla of the forelimb of 3-4-week-old BALB/c nude mice. Seven days after transplantation, the diameter of the tumors was measured, and the tumors were removed after three weeks. | |||
Serine/threonine-protein kinase PAK 6 (PAK5)
Cervical cancer [ICD-11: 2C77]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [176] | |||
| Responsed Disease | Cervical cancer [ICD-11: 2C77] | |||
| Target Regulation | Down regulation | |||
In-vitro Model |
C-33 A | Cervical squamous cell carcinoma | Homo sapiens | CVCL_1094 |
| HeLa | Endocervical adenocarcinoma | Homo sapiens | CVCL_0030 | |
| SiHa | Cervical squamous cell carcinoma | Homo sapiens | CVCL_0032 | |
| Ca Ski | Cervical squamous cell carcinoma | Homo sapiens | CVCL_1100 | |
| In-vivo Model | Tumor xenograft models were established, with each group consisting of 5 mice (n = 5 per group), including ALKBH5, ALKBH5/shPAK5, NC, shCon, shPAK5, shALKBH5, and shALKBH5. Subcutaneous injections of stably transfected HeLa cells (2.0 × 106 cells) were administered into the armpit region of the nude mice. The tumor volume was regularly monitored using the formula: volume = 0.5 × length × width2. After 5 weeks, the mice were euthanized and the weight of tumor was measured. In the lung metastasis models, which were conducted in nude mice, the groups consisted of 5 mice per group, similar to the tumor xenograft models. In these models, HeLa cells (2.0 × 106) that had been stably transfected were injected into the mice through the tail vein. After 5 weeks, the mice were euthanized. The lung tissues were excised, photographed, and fixed in 4% paraformaldehyde for H&E staining. | |||
Serine/threonine-protein phosphatase 2A catalytic subunit alpha isoform (PPP2CA)
Gastric cancer [ICD-11: 2B72]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [177] | |||
| Responsed Disease | Gastric cancer [ICD-11: 2B72] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
HGC-27 | Gastric carcinoma | Homo sapiens | CVCL_1279 |
| MGC-803 | Gastric mucinous adenocarcinoma | Homo sapiens | CVCL_5334 | |
| SGC-7901 | Gastric carcinoma | Homo sapiens | CVCL_0520 | |
| BGC-823 | Gastric carcinoma | Homo sapiens | CVCL_3360 | |
| In-vivo Model | We injected 2 × 106 stably infected GC cells subcutaneously into each dorsal flank of mice (n = 6). After 4 weeks, each histologically intact tumor was removed, weighed and photographed. To construct the model of mice pulmonary metastasis, we injected 2 × 106 stably infected GC cells into the tail vein of the mice (n = 5). | |||
Taurine up-regulated 1 protein (TUG1)
Acute myeloid leukaemia [ICD-11: 2A60]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [178] | |||
| Responsed Disease | Acute myeloid leukaemia [ICD-11: 2A60] | |||
| Target Regulation | Down regulation | |||
testis expressed 41 (TEX41)
Renal cell carcinoma [ICD-11: 2C90]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [179] | |||
| Responsed Disease | Renal cell carcinoma [ICD-11: 2C90] | |||
| Target Regulation | Up regulation | |||
Tnfrsf2
Acute myeloid leukaemia [ICD-11: 2A60]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [180] | |||
| Responsed Disease | Acute myeloid leukaemia [ICD-11: 2A60] | |||
| Target Regulation | Down regulation | |||
| Pathway Response | Signaling pathways regulating pluripotency of stem cells | hsa04550 | ||
| Cell Process | mRNA decay | |||
In-vitro Model |
THP-1 | Childhood acute monocytic leukemia | Homo sapiens | CVCL_0006 |
| In-vivo Model | THP-1 cells transduced with CTL or KD lentiviruses were tail vein injected into non-irradiated 12 week-old female non-obese diabetic (NOD)/LtSz-severe combined immune-deficiency (SCID) IL-2Rγcnull (NSG) mice (1x106 cells per 200 uL per mouse). | |||
| Response Summary | Acute myeloid leukemia (AML) is an aggressive clonal disorder of hematopoietic stem cells (HSCs) and primitive progenitors that blocks their myeloid differentiation, generating self-renewing leukemic stem cells (LSCs). YTHDF2 decreases the half-life of diverse m6A transcripts that contribute to the overall integrity of LSC function, including the tumor necrosis factor receptor Tnfrsf2, whose upregulation in Ythdf2-deficient LSCs primes cells for apoptosis. | |||
Transcription factor E2F6 (E2F6)
Liver cancer [ICD-11: 2C12]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [112] | |||
| Responsed Disease | Liver cancer [ICD-11: 2C12] | |||
| Target Regulation | Down regulation | |||
Transcription factor GATA-6 (GATA6)
Injury of other or unspecified intrathoracic organs [ICD-11: NB32]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [181] | |||
| Responsed Disease | Injury of other or unspecified intrathoracic organs [ICD-11: NB32.3] | |||
| Target Regulation | Down regulation | |||
In-vitro Model |
BEAS-2B | Normal | Homo sapiens | CVCL_0168 |
| In-vivo Model | To perform in vivo plasmid transfection, the researchers mixed the pcDNA3.1-EGFP-METTL3 plasmid with in vivo jetPEI (Polyplus-transfection; Illkirch) per the manufacturer's guidelines. They mixed 40 μg of plasmid nucleic acid in 5% glucose (25 μl) with 6.4 μl of jetPEI solution in 5% glucose (25 μl) to obtain a final nitrogen and phosphorus ratio of 8. The mixture was incubated at room temperature for 30 minutes to form stable complexes before tracheal intubation of mice. | |||
Transcription factor SOX-9 (SOX9)
Rheumatoid arthritis [ICD-11: FA20]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [182] | |||
| Responsed Disease | Rheumatoid arthritis [ICD-11: FA20] | |||
Tripartite motif-containing protein 72 (TRIM72)
Preeclampsia [ICD-11: JA23]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [183] | |||
| Responsed Disease | Preeclampsia [ICD-11: JA23] | |||
| Target Regulation | Down regulation | |||
Tyrosine-protein kinase JAK1 (JAK1)
Gastric cancer [ICD-11: 2B72]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [184] | |||
| Responsed Disease | Gastric cancer [ICD-11: 2B72] | |||
| Target Regulation | Down regulation | |||
In-vitro Model |
BGC-823 | Gastric carcinoma | Homo sapiens | CVCL_3360 |
| In-vivo Model | BGC-823 (3 × 106) cells that stably expressed or silenced ALKBH5 and LINC00659 and their paired control cells were injected into the left side of each mouse. For cell metastasis experiments in vivo, BGC-823 (3 × 106) cells that stably overexpressed or silenced ALKBH5 and LINC00659 and their paired control cells were injected through tail vein. | |||
Ubiquitin carboxyl-terminal hydrolase 38 (USP38)
Bladder cancer [ICD-11: 2C94]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [185] | |||
| Responsed Disease | Bladder cancer [ICD-11: 2C94] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
SV-HUC-1 | Normal | Homo sapiens | CVCL_3798 |
| In-vivo Model | BALB/c nude mice were purchased from the Slac Laboratories (Shanghai, China) and randomly divided into two groups. The xenograft mouse model was generated via tail-vein injection of METTL14-expressing T24 cells (5 × 105 cells per mouse) into experimental mice while those injected with pcDNA3.1 empty vector transfected cells as control. | |||
Voltage-dependent L-type calcium channel subunit alpha-1C (CACNA1C)
Supraventricular tachyarrhythmia [ICD-11: BC81]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [186] | |||
| Responsed Disease | Supraventricular tachyarrhythmia [ICD-11: BC81.3] | |||
| In-vivo Model | Under isoflurane anesthesia, bipolar leads were inserted into the right atrial (RA) appendage and connected to pacemakers. Following a 24-h recovery, the atrial pacemaker was programmed to pace the RA appendage at atrial tachycardia pacing for 2 weeks. | |||
Unspecific Target Gene
Solid tumour/cancer [ICD-11: 2A00-2F9Z]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [187] | |||
| Responsed Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | |||
In-vitro Model |
HeLa | Endocervical adenocarcinoma | Homo sapiens | CVCL_0030 |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| NCI-H1299 | Lung large cell carcinoma | Homo sapiens | CVCL_0060 | |
| HEK293-FT | Normal | Homo sapiens | CVCL_6911 | |
| In-vivo Model | Stable H1299 cell lines diluted into 100uL were injected subcutaneously into 5-week-old nude mice (n = 5) at the final concentration of 3 × 106 cells. Mice were sacrificed 4 weeks later, and tumors were weighed and photographed. | |||
| Response Summary | SUMOylation of YTHDF2 has little impact on its ubiquitination and localization, but significantly increases its binding affinity of m6A-modified mRNAs and subsequently results in deregulated gene expressions which accounts for cancer progression. | |||
Gastric cancer [ICD-11: 2B72]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [188] | |||
| Responsed Disease | Gastric cancer [ICD-11: 2B72] | |||
| Target Regulation | Down regulation | |||
| Cell Process | Cell proliferation | |||
| Cell apoptosis | ||||
In-vitro Model |
MGC-803 | Gastric mucinous adenocarcinoma | Homo sapiens | CVCL_5334 |
| Response Summary | YTHDF2 mRNA in gastric cancer was significantly higher than that in the normal tissues.Knockdown of YTHDF2 in MGC-803 cells inhibits cell proliferation and promotes apoptosis. | |||
Liver cancer [ICD-11: 2C12]
| In total 2 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [189] | |||
| Responsed Disease | Hepatocellular carcinoma [ICD-11: 2C12.02] | |||
| Responsed Drug | Teniposide | Approved | ||
| Pathway Response | Nucleotide excision repair | hsa03420 | ||
| mTOR signaling pathway | hsa04150 | |||
| Cell Process | DNA repair | |||
In-vitro Model |
Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 |
| Huh-7 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_0336 | |
| In-vivo Model | To establish a tumour model, C57BL/6 mice were intraperitoneal injected with 25 mg/kg diethylnitrosamine at 2 weeks of age. | |||
| Response Summary | The m6A model includes LRPPRC, YTHDF2, KIAA14219, and RBM15B, classified A-hepatocellular carcinoma patients into high/low-risk subtypes. The expression of Immunosuppressive cytokines DNMT1/EZH2 was up-regulated in A-hepatocellular carcinoma patients, and teniposide can be a potential therapeutic drug for A-hepatocellular carcinoma. | |||
| Experiment 2 Reporting the m6A-centered Disease Response of This Target Gene | [190] | |||
| Responsed Disease | Hepatocellular carcinoma [ICD-11: 2C12.02] | |||
In-vitro Model |
Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 |
| Response Summary | MIR145 modulates m6A levels by targeting the 3'-UTR of YTHDF2 mRNA in hepatocellular carcinoma cells. | |||
Lung cancer [ICD-11: 2C25]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [192] | |||
| Responsed Disease | Lung squamous cell carcinoma [ICD-11: 2C25.2] | |||
| Pathway Response | mTOR signaling pathway | hsa04150 | ||
| PI3K-Akt signaling pathway | hsa04151 | |||
| Cell Process | Cell proliferation | |||
| Cell invasion | ||||
In-vitro Model |
SK-MES-1 | Lung squamous cell carcinoma | Homo sapiens | CVCL_0630 |
| NCI-H226 | Pleural epithelioid mesothelioma | Homo sapiens | CVCL_1544 | |
| In-vivo Model | The mice were randomly divided into two groups which were inoculated with stable YTHDF2-expressing LUSC cells and the vector LUSC cells. A total of 5 × 106 of the cells were suspended in 0.1 ml of PBS and then injected subcutaneously into the flanks of mice. | |||
| Response Summary | High-YTHDF2 expression predicted a worse prognosis of LUSC, while hypoxia-mediated YTHDF2 overexpression promotes lung squamous cell carcinoma progression by activation of the mTOR/AKT signaling pathway. | |||
Ovarian cancer [ICD-11: 2C73]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [193] | |||
| Responsed Disease | Ovarian cancer [ICD-11: 2C73] | |||
| Cell Process | Cell proliferation and migration | |||
| Cell apoptosis | ||||
In-vitro Model |
SK-OV-3 | Ovarian serous cystadenocarcinoma | Homo sapiens | CVCL_0532 |
| Response Summary | YTHDF2 and microRNA 145, as two crucial m6A regulators, were involved in the progression of epithelial ovarian cancer by indirectly modulating m6A levels. | |||
Endometrial cancer [ICD-11: 2C76]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [194] | |||
| Responsed Disease | Endometrial cancer [ICD-11: 2C76] | |||
In-vitro Model |
RL95-2 | Endometrial adenosquamous carcinoma | Homo sapiens | CVCL_0505 |
| Ishikawa | Endometrial adenocarcinoma | Homo sapiens | CVCL_2529 | |
| HHUA | Endometrial adenocarcinoma | Homo sapiens | CVCL_3866 | |
| HEC-1-A | Endometrial adenocarcinoma | Homo sapiens | CVCL_0293 | |
| Response Summary | YTHDF2 expression can accurately assess the depth of myometrial invasion (DMI) in EMAC when EMAC coexists with adenomyosis. | |||
Prostate cancer [ICD-11: 2C82]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [195] | |||
| Responsed Disease | Prostate cancer [ICD-11: 2C82] | |||
| Cell Process | Cell proliferation | |||
| Cell migration | ||||
In-vitro Model |
DU145 | Prostate carcinoma | Homo sapiens | CVCL_0105 |
| PC-3 | Prostate carcinoma | Homo sapiens | CVCL_0035 | |
| Response Summary | YTHDF2 and miR-493-3p provide new insights into the carcinogenesis and new potential therapeutic targets for PCa. | |||
Hematological disorders [ICD-11: 3C0Z]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [196] | |||
| Responsed Disease | Hematological disorders [ICD-11: 3C0Z] | |||
| Cell Process | RNA stability | |||
In-vitro Model |
Hematopoietic stem cells (Hematopoietic stem cells) | |||
| In-vivo Model | Utilized three Cre transgenic mouse lines (Mx1-Cre, Ert-Cre and Vav-Cre) to cross with Ythdf2fl/fl mice to deplete the expression of YTHDF2 specifically in adult HSCs of mouse bone marrow (BM), respectively. | |||
| Response Summary | YTHDF2 deletion uniquely affects the homeostasis of HSPCs. | |||
Lupus erythematosus [ICD-11: 4A40]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [197] | |||
| Responsed Disease | Lupus erythematosus [ICD-11: 4A40] | |||
| Response Summary | These findings suggested decreased YTHDF2 that was associated with disease activity play an important role in the pathogenesis of SLE, METTL14 and ALKBH5 were concomitantly decreased. | |||
Liver disease [ICD-11: DB9Z]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [198] | |||
| Responsed Disease | Liver disease [ICD-11: DB9Z] | |||
| Pathway Response | PPAR signaling pathway | hsa03320 | ||
| Cell Process | Fatty degeneration | |||
| In-vivo Model | A total of 24 male mice were randomly allocated to LFD (low-fat diet), LFDR (low-fat diet + resveratrol), HFD (high-fat diet), and HFDR (high-fat diet + resveratrol) groups for 12 weeks (n = 6/group). | |||
| Response Summary | The beneficial effect of resveratrol on lipid metabolism disorder under HFD is due to a decrease of m6A RNA methylation and an increase of PPARalpha mRNA, providing mechanistic insights into the function of resveratrol in alleviating the disturbance of lipid metabolism in mice. The resveratrol in HFD increased the transcript levels of methyltransferase like 3 (METTL3), alkB homolog 5 (ALKBH5), fat mass and obesity associated protein (FTO), and YTH domain family 2 (YTHDF2), whereas it decreased the level of YTH domain family 3 (YTHDF3) and m6A abundance in mice liver. | |||
Chondropathies [ICD-11: FB82]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [199] | |||
| Responsed Disease | Steroid-induced necrosis of the femoral head [ICD-11: FB82.1] | |||
Male infertility [ICD-11: GB04]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [200] | |||
| Responsed Disease | Male infertility [ICD-11: GB04] | |||
| Cell Process | Apoptosis in germ cells | |||
In-vitro Model |
Spermatogenic cells (Prepared from undifferentiated mouse spermatogonial cells) | |||
| In-vivo Model | The knockout first allele was converted into a conditional allele by crossing Ythdf2 knockout first mice with Flp deleter mice. The resulting floxed Ythdf2 mice were crossed with Vasa-GFPCre knock-in mice to allow specific knockout of Ythdf2 in germ cells. | |||
| Response Summary | Germ cell-specific Ythdf2 mutants (Ythdf2-vKO) at a C57BL/6J background and demonstrated that YTHDF2 is essential for mouse spermatogenesis and fertility. Demonstrates the fundamental role of YTHDF2 during mouse spermatogenesis and provides a potential candidate for the diagnosis of male infertility with the oligoasthenoteratozoospermia syndrome. | |||
C-X-C chemokine receptor type 4 (CXCR4)
| Representative RNA-seq result indicating the expression of this target gene regulated by YTHDF2 | ||
| Cell Line | Testis | Mus musculus |
|
Treatment: YTHDF2 knockout mice testis
Control: Mice testis
|
GSE147574 | |
| Regulation |
 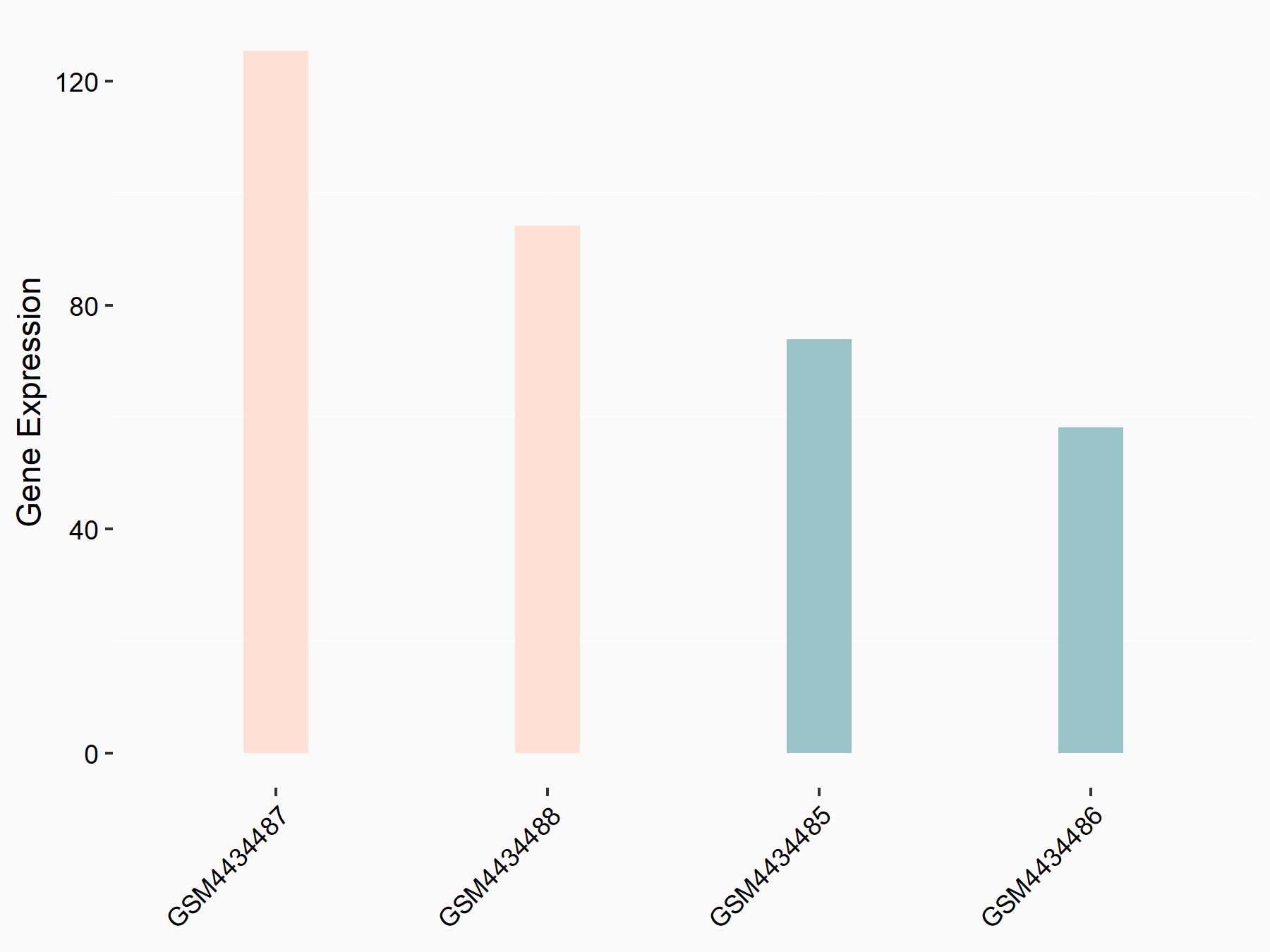 |
logFC: 7.39E-01 p-value: 4.60E-02 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and YTHDF2 | ||
| Cell Line | Hela | Homo sapiens |
| Regulation | logFC: 2.20E+00 | GSE49339 |
PMID31239444-anti-PD1 antibody
[Investigative]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [3] | |||
| Responsed Disease | Melanoma | ICD-11: 2C30 | ||
| Target Regulation | Down regulation | |||
| Pathway Response | PD-L1 expression and PD-1 checkpoint pathway in cancer | hsa05235 | ||
| Cell Process | mRNA decay | |||
| In-vitro Model | B16-F10 | Mouse melanoma | Mus musculus | CVCL_0159 |
| CHL-1 | Melanoma | Homo sapiens | CVCL_1122 | |
| 624-mel | Melanoma | Homo sapiens | CVCL_8054 | |
| NHEM (Normal Human Epidermal Melanocytes) | ||||
| SK-MEL-30 | Cutaneous melanoma | Homo sapiens | CVCL_0039 | |
| WM115 | Melanoma | Homo sapiens | CVCL_0040 | |
| WM35 | Melanoma | Homo sapiens | CVCL_0580 | |
| WM3670 | Melanoma | Homo sapiens | CVCL_6799 | |
| WM793 | Melanoma | Homo sapiens | CVCL_8787 | |
| In-vivo Model | When the tumors reached a volume of 80-100 mm3, mice were treated with anti-PD-1 or isotype control antibody (200 ug/mouse) by i.p. injection, every other day for three times. For IFNγ blockade treatment, C57BL/6 mice were treated with anti-IFNγ antibody or isotype control IgG (250 ug/mouse) every other day after tumor cell inoculation. | |||
| Response Summary | These findings demonstrate a crucial role of FTO as an m6A demethylase in promoting melanoma tumorigenesis and anti-PD-1 resistance, and suggest that the combination of FTO inhibition with anti-PD-1 blockade reduces the resistance to immunotherapy in melanoma. Knockdown of FTO increases m6A methylation in the critical protumorigenic melanoma cell-intrinsic genes including PD-1 (PDCD1), C-X-C chemokine receptor type 4 (CXCR4), and SOX10, leading to increased RNA decay through the m6A reader YTHDF2. | |||
Ephrin type-B receptor 3 (EPHB3)
| Representative RNA-seq result indicating the expression of this target gene regulated by YTHDF2 | ||
| Cell Line | Mouse-cerebellum granule cell | Mus musculus |
|
Treatment: YTHDF2 knockdown mouse-cerebellum granule cell
Control: Wild type mouse-cerebellum granule cell
|
GSE153688 | |
| Regulation |
 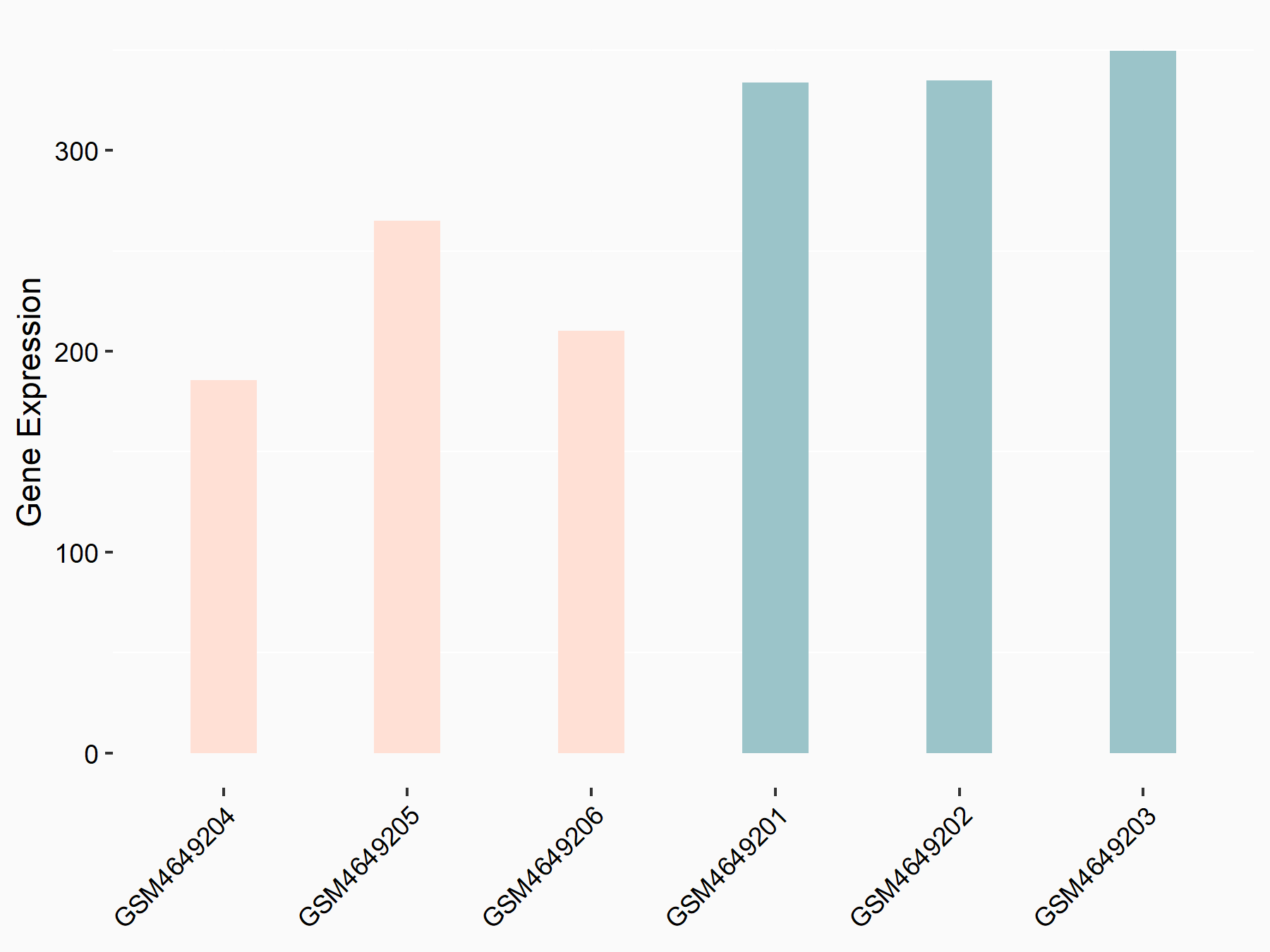 |
logFC: -6.25E-01 p-value: 1.90E-03 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and YTHDF2 | ||
| Cell Line | Hela | Homo sapiens |
| Regulation | logFC: 1.21E+00 | GSE49339 |
Temozolomide
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [4] | |||
| Responsed Disease | Glioblastoma | ICD-11: 2A00.00 | ||
| Target Regulation | Down regulation | |||
| Pathway Response | PI3K-Akt signaling pathway | hsa04151 | ||
| Cell Process | RNA stability | |||
| In-vitro Model | T98G | Glioblastoma | Homo sapiens | CVCL_0556 |
| LN-229 | Glioblastoma | Homo sapiens | CVCL_0393 | |
| In-vivo Model | 5 × 106 infected T98G cells (LV-NC or LV-YTHDF2) were injected into the flanks of mice through subcutaneous. | |||
| Response Summary | YTHDF2 enhanced TMZ resistance in GBM by activation of the PI3K/Akt and NF-Kappa-B signalling pathways via inhibition of Ephrin type-B receptor 3 (EPHB3) and TNFAIP3. | |||
Myc proto-oncogene protein (MYC)
| Representative RNA-seq result indicating the expression of this target gene regulated by YTHDF2 | ||
| Cell Line | GSC11 cell line | Homo sapiens |
|
Treatment: siYTHDF2 GSC11 cells
Control: siControl GSC11 cells
|
GSE142825 | |
| Regulation |
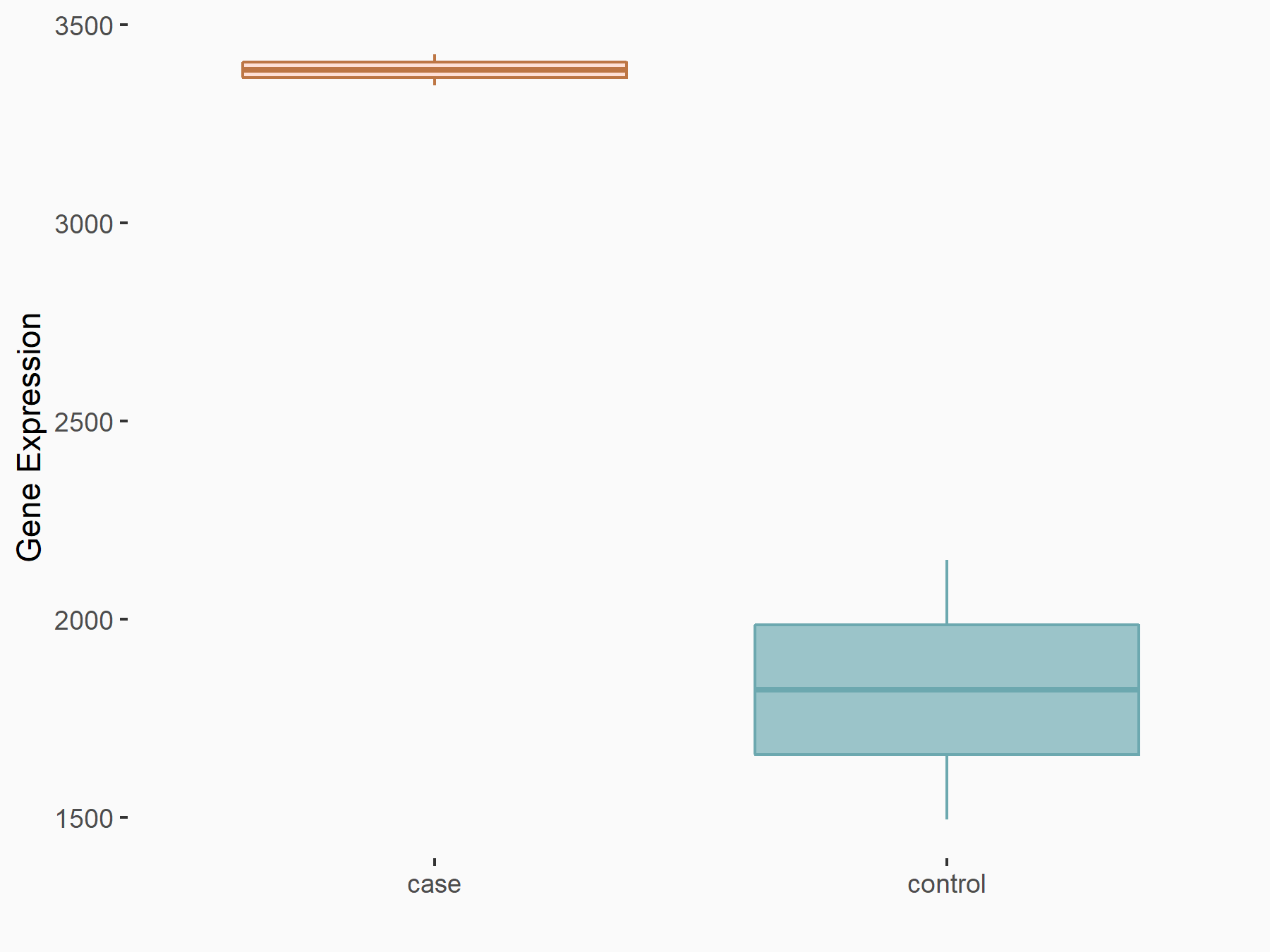 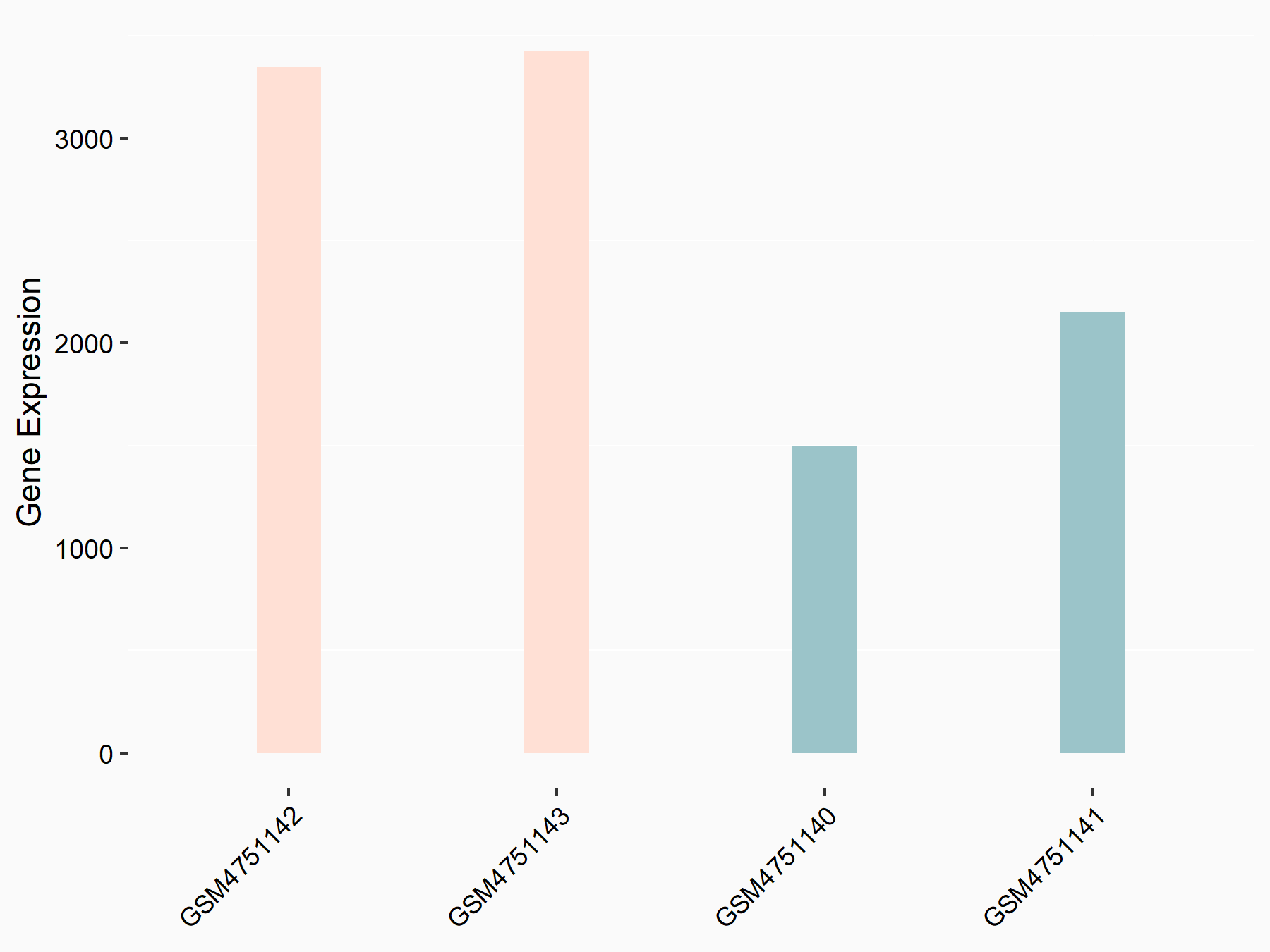 |
logFC: 8.94E-01 p-value: 8.57E-07 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and YTHDF2 | ||
| Cell Line | Hela | Homo sapiens |
| Regulation | logFC: 1.40E+00 | GSE49339 |
Linsitinib
[Phase 3]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [11] | |||
| Responsed Disease | Glioma | ICD-11: 2A00.0 | ||
| Target Regulation | Up regulation | |||
| Pathway Response | RNA degradation | hsa03018 | ||
| Cell Process | RNA stability | |||
| In-vitro Model | () | |||
| HNP1 (A human neural progenitor cell) | ||||
| NHA (Normal human astrocytes) | ||||
| NSC11 (Pluripotent derived neural progenitor cell) | ||||
| In-vivo Model | For in vivo drug treatment studies, intracranial xenografts were generated by implanting 5000 patient-derived GSCs (387 and 4121) into the right cerebral cortex of NSG mice as described above. | |||
| Response Summary | The m6A reader YTHDF2 stabilized Myc proto-oncogene protein (MYC) mRNA specifically in cancer stem cells. Given the challenge of targeting MYC, YTHDF2 presents a therapeutic target to perturb MYC signaling in glioblastoma. The IGF1/IGF1R inhibitor linsitinib preferentially targeted YTHDF2-expressing cells, inhibiting GSC viability without affecting NSCs and impairing in vivo glioblastoma growth. YTHDF2 links RNA epitranscriptomic modifications and GSC growth, laying the foundation for the YTHDF2-MYC-IGFBP3 axis as a specific and novel therapeutic target in glioblastoma. | |||
Tamoxifen
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [12] | |||
| Responsed Disease | Breast cancer | ICD-11: 2C60 | ||
| Pathway Response | MAPK signaling pathway | hsa04010 | ||
| Cell Process | Epithelial-to-mesenchymal transition | |||
| Cell apoptosis | ||||
| In-vitro Model | HEK293T | Normal | Homo sapiens | CVCL_0063 |
| MCF-7 | Invasive breast carcinoma | Homo sapiens | CVCL_0031 | |
| MDA-MB-231 | Breast adenocarcinoma | Homo sapiens | CVCL_0062 | |
| MBA-MD-231 (Human breast cancer cell) | ||||
| MYC-ER HMEC (Human mammary epithelial cells expressing a MYC estrogen receptor fusion) | ||||
| SK-BR-3 | Breast adenocarcinoma | Homo sapiens | CVCL_0033 | |
| In-vivo Model | To induce recombination at 8 weeks of age both CAG-CreERT;Ythdf2fl/fl and Ythdf2fl/fl littermates were injected with 75mg/kg body weight tamoxifen dissolved in corn oil daily for 5 days. | |||
| Response Summary | LCAT3 upregulation is attributable to m6A modification mediated by METTL3, leading to LCAT3 stabilization. Treated cells with tamoxifen to induce MYC activity. Highlights the therapeutic potential of RBPs by uncovering a critical role for YTHDF2 in counteracting the global increase of mRNA synthesis in Myc proto-oncogene protein (MYC)-driven breast cancers. | |||
Nucleobindin-1 (NUCB1)
| Representative RNA-seq result indicating the expression of this target gene regulated by YTHDF2 | ||
| Cell Line | B18-hi B cell line | Mus musculus |
|
Treatment: YTHDF2 knockout B18-hi B cells
Control: Wild type B18-hi B cells
|
GSE189819 | |
| Regulation |
 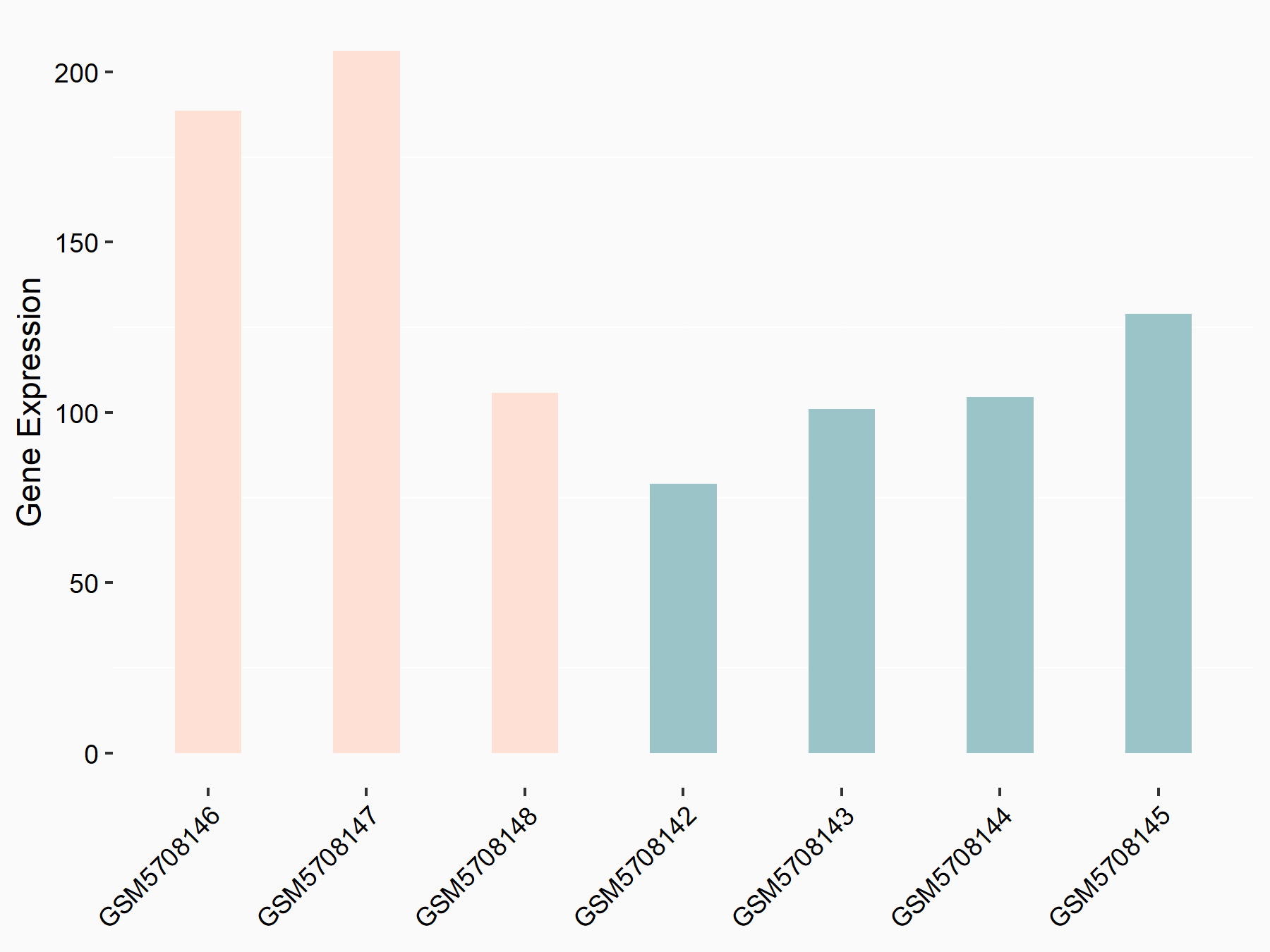 |
logFC: 6.95E-01 p-value: 1.62E-02 |
| More Results | Click to View More RNA-seq Results | |
Gemcitabine
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [30] | |||
| Responsed Disease | Pancreatic ductal adenocarcinoma | ICD-11: 2C10.0 | ||
| Target Regulation | Down regulation | |||
| Pathway Response | Autophagy | hsa04140 | ||
| Cell Process | Cell proliferation | |||
| Cell autophagy | ||||
| In-vitro Model | SW1990 | Pancreatic adenocarcinoma | Homo sapiens | CVCL_1723 |
| PANC-1 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0480 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| CFPAC-1 | Cystic fibrosis | Homo sapiens | CVCL_1119 | |
| BxPC-3 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0186 | |
| AsPC-1 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0152 | |
| In-vivo Model | 5 × 106 SW1990 cells expressing NUCB1 (oeNUCB1) or control vector (oeNC) were injected subcutaneously. | |||
| Response Summary | METTL3-mediated m6A modification on Nucleobindin-1 (NUCB1) 5'UTR via the reader YTHDF2 as a mechanism for NUCB1 downregulation in PDAC. This study revealed crucial functions of NUCB1 in suppressing proliferation and enhancing the effects of gemcitabine in pancreatic cancer cells. | |||
PI3-kinase subunit beta (PIK3CB)
| Representative RIP-seq result supporting the interaction between the target gene and YTHDF2 | ||
| Cell Line | Hela | Homo sapiens |
| Regulation | logFC: 1.28E+00 | GSE49339 |
AZD6482
[Terminated]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [42] | |||
| Responsed Disease | Pancreatic cancer | ICD-11: 2C10 | ||
| Target Regulation | Down regulation | |||
| Pathway Response | PI3K-Akt signaling pathway | hsa04151 | ||
| Glycolysis / Gluconeogenesis | hsa00010 | |||
| Cell Process | Glucose metabolism | |||
| In-vitro Model | BxPC-3 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0186 |
| PANC-1 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0480 | |
| In-vivo Model | Established cohorts of mice bearing tumour xenografts driven by PTEN-deficient BxPC-3 and PANC-1 cells with PIK3CB overexpression. When tumours grew to ~300 mm3, mice were grouped and administered with vehicle (DMSO) or KIN-193 via intraperitoneal injection (20 mg/kg) once daily. | |||
| Response Summary | N6-methyladenosine mRNA methylation of PIK3CB regulates AKT signalling to promote PTEN-deficient pancreatic cancer progression. Rs142933486 is significantly associated with the overall survival of PDAC by reducing the PIK3CB m6A level, which facilitated its mRNA and protein expression levels mediated by the m6A 'writer' complex (METTL13/METTL14/WTAP) and the m6A 'reader' YTHDF2. KIN-193, a PI3-kinase subunit beta (PIK3CB)-selective inhibitor, is shown to serve as an effective anticancer agent for blocking PTEN-deficient PDAC. | |||
TNF alpha-induced protein 3 (TNFAIP3)
| Representative RIP-seq result supporting the interaction between the target gene and YTHDF2 | ||
| Cell Line | Hela | Homo sapiens |
| Regulation | logFC: 1.94E+00 | GSE49339 |
Temozolomide
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [4] | |||
| Responsed Disease | Glioblastoma | ICD-11: 2A00.00 | ||
| Target Regulation | Down regulation | |||
| Pathway Response | PI3K-Akt signaling pathway | hsa04151 | ||
| Cell Process | RNA stability | |||
| In-vitro Model | T98G | Glioblastoma | Homo sapiens | CVCL_0556 |
| LN-229 | Glioblastoma | Homo sapiens | CVCL_0393 | |
| In-vivo Model | 5 × 106 infected T98G cells (LV-NC or LV-YTHDF2) were injected into the flanks of mice through subcutaneous. | |||
| Response Summary | YTHDF2 enhanced TMZ resistance in GBM by activation of the PI3K/Akt and NF-Kappa-B signalling pathways via inhibition of EPHB3 and TNF alpha-induced protein 3 (TNFAIP3). | |||
ATP-binding cassette sub-family C member 10 (ABCC10)
Gefitinib
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [46] | |||
| Responsed Disease | Non-small-cell lung carcinoma | ICD-11: 2C25.Y | ||
| Target Regulation | Down regulation | |||
| Pathway Response | ABC transporters | hsa02010 | ||
| In-vitro Model | PC-9 | Lung adenocarcinoma | Homo sapiens | CVCL_B260 |
| NCI-H1975 | Lung adenocarcinoma | Homo sapiens | CVCL_1511 | |
| In-vivo Model | Mice were randomized into three groups (n = 7/group), 1 × 107 PC9 cells absorbed exosomes were subcutaneously injected into the Bilateral groin of mice. Treatment began 1 week following injection, the mice were intraperitoneally injected with gefitinib (30 mg/kg/day). | |||
| Response Summary | Not only FTO knockdown enhanced the gefitinib sensitivity of GR cells but also FTO reduction in donor exosomes alleviated the acquired resistance of recipient non-small cell lung cancer PC9 cells. FTO/YTHDF2/ATP-binding cassette sub-family C member 10 (ABCC10) axis played a role in intercellular transmission of GR cell-derived exosome-mediated gefitinib resistance. | |||
Axin-1 (AXIN1)
Cisplatin
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [48] | |||
| Responsed Disease | Cervical cancer | ICD-11: 2C77 | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Wnt signaling pathway | hsa04310 | ||
| Cell Process | Epithelial-mesenchymal transition | |||
| In-vitro Model | SiHa | Cervical squamous cell carcinoma | Homo sapiens | CVCL_0032 |
| HeLa | Endocervical adenocarcinoma | Homo sapiens | CVCL_0030 | |
| Ect1/E6E7 | Normal | Homo sapiens | CVCL_3679 | |
| Ca Ski | Cervical squamous cell carcinoma | Homo sapiens | CVCL_1100 | |
| C-33 A | Cervical squamous cell carcinoma | Homo sapiens | CVCL_1094 | |
| Response Summary | YTHDF2 interference could suppress the EMT of cervical cancer cells and enhance cisplatin chemosensitivity by regulating Axin-1 (AXIN1). | |||
BCL2/adenovirus E1B 19 kDa protein-interacting protein 3 (BNIP3)
Chromium
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [51] | |||
| Target Regulation | Down regulation | |||
Cyclic AMP-dependent transcription factor ATF-4 (ATF4)
Asparagine inhibitor
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [56] | |||
| Responsed Disease | Colorectal cancer | ICD-11: 2B91 | ||
| Target Regulation | Down regulation | |||
| Pathway Response | mTOR signaling pathway | hsa04150 | ||
| Cell Process | RNA decay | |||
| Cell growth and death | ||||
| Cell autophagy | ||||
| In-vitro Model | HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 |
| SW480 | Colon adenocarcinoma | Homo sapiens | CVCL_0546 | |
| Response Summary | In colorectal cancer, Glutaminolysis inhibition upregulated ATF4 expression in an m6A-dependent manner to activate pro-survival autophagy through transcriptional activation of the mTOR inhibitor DDIT4. Determined the relationship between FTO alpha-ketoglutarate dependent dioxygenase (FTO), YTH N6-methyladenosine RNA binding protein 2 (YTHDF2), and ATF4. ATF4 transcriptionally upregulated DDIT4 to suppress Cyclic AMP-dependent transcription factor ATF-4 (ATF4), which induced pro-survival autophagy during glutaminolysis inhibition. | |||
Chloroquine
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [56] | |||
| Responsed Disease | Colorectal cancer | ICD-11: 2B91 | ||
| Target Regulation | Down regulation | |||
| Pathway Response | mTOR signaling pathway | hsa04150 | ||
| Cell Process | RNA decay | |||
| Cell growth and death | ||||
| Cell autophagy | ||||
| In-vitro Model | HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 |
| SW480 | Colon adenocarcinoma | Homo sapiens | CVCL_0546 | |
| Response Summary | In colorectal cancer, Glutaminolysis inhibition upregulated ATF4 expression in an m6A-dependent manner to activate pro-survival autophagy through transcriptional activation of the mTOR inhibitor DDIT4. Determined the relationship between FTO alpha-ketoglutarate dependent dioxygenase (FTO), YTH N6-methyladenosine RNA binding protein 2 (YTHDF2), and ATF4. Cyclic AMP-dependent transcription factor ATF-4 (ATF4) transcriptionally upregulated DDIT4 to suppress mTOR, which induced pro-survival autophagy during glutaminolysis inhibition. | |||
Meclofenamate sodium
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [56] | |||
| Responsed Disease | Colorectal cancer | ICD-11: 2B91 | ||
| Target Regulation | Down regulation | |||
| Pathway Response | mTOR signaling pathway | hsa04150 | ||
| Cell Process | RNA decay | |||
| Cell growth and death | ||||
| Cell autophagy | ||||
| In-vitro Model | HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 |
| SW480 | Colon adenocarcinoma | Homo sapiens | CVCL_0546 | |
| Response Summary | In colorectal cancer, Glutaminolysis inhibition upregulated ATF4 expression in an m6A-dependent manner to activate pro-survival autophagy through transcriptional activation of the mTOR inhibitor DDIT4. Determined the relationship between FTO alpha-ketoglutarate dependent dioxygenase (FTO), YTH N6-methyladenosine RNA binding protein 2 (YTHDF2), and ATF4. Cyclic AMP-dependent transcription factor ATF-4 (ATF4) transcriptionally upregulated DDIT4 to suppress mTOR, which induced pro-survival autophagy during glutaminolysis inhibition. | |||
Rapamycin
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [56] | |||
| Responsed Disease | Colorectal cancer | ICD-11: 2B91 | ||
| Target Regulation | Down regulation | |||
| Pathway Response | mTOR signaling pathway | hsa04150 | ||
| Cell Process | RNA decay | |||
| Cell growth and death | ||||
| Cell autophagy | ||||
| In-vitro Model | HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 |
| SW480 | Colon adenocarcinoma | Homo sapiens | CVCL_0546 | |
| Response Summary | In colorectal cancer, Glutaminolysis inhibition upregulated ATF4 expression in an m6A-dependent manner to activate pro-survival autophagy through transcriptional activation of the mTOR inhibitor DDIT4. Determined the relationship between FTO alpha-ketoglutarate dependent dioxygenase (FTO), YTH N6-methyladenosine RNA binding protein 2 (YTHDF2), and ATF4. Cyclic AMP-dependent transcription factor ATF-4 (ATF4) transcriptionally upregulated DDIT4 to suppress mTOR, which induced pro-survival autophagy during glutaminolysis inhibition. | |||
CB-839
[Phase 2]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [56] | |||
| Responsed Disease | Colorectal cancer | ICD-11: 2B91 | ||
| Target Regulation | Down regulation | |||
| Pathway Response | mTOR signaling pathway | hsa04150 | ||
| Cell Process | RNA decay | |||
| Cell growth and death | ||||
| Cell autophagy | ||||
| In-vitro Model | HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 |
| SW480 | Colon adenocarcinoma | Homo sapiens | CVCL_0546 | |
| Response Summary | In colorectal cancer, Glutaminolysis inhibition upregulated ATF4 expression in an m6A-dependent manner to activate pro-survival autophagy through transcriptional activation of the mTOR inhibitor DDIT4. Determined the relationship between FTO alpha-ketoglutarate dependent dioxygenase (FTO), YTH N6-methyladenosine RNA binding protein 2 (YTHDF2), and ATF4. ATF4 transcriptionally upregulated DDIT4 to suppress Cyclic AMP-dependent transcription factor ATF-4 (ATF4), which induced pro-survival autophagy during glutaminolysis inhibition. | |||
GLS-IN-968
[Investigative]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [56] | |||
| Responsed Disease | Colorectal cancer | ICD-11: 2B91 | ||
| Target Regulation | Down regulation | |||
| Pathway Response | mTOR signaling pathway | hsa04150 | ||
| Cell Process | RNA decay | |||
| Cell growth and death | ||||
| Cell autophagy | ||||
| In-vitro Model | HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 |
| SW480 | Colon adenocarcinoma | Homo sapiens | CVCL_0546 | |
| Response Summary | In colorectal cancer, Glutaminolysis inhibition upregulated ATF4 expression in an m6A-dependent manner to activate pro-survival autophagy through transcriptional activation of the mTOR inhibitor DDIT4. Determined the relationship between FTO alpha-ketoglutarate dependent dioxygenase (FTO), YTH N6-methyladenosine RNA binding protein 2 (YTHDF2), and ATF4. ATF4 transcriptionally upregulated DDIT4 to suppress Cyclic AMP-dependent transcription factor ATF-4 (ATF4), which induced pro-survival autophagy during glutaminolysis inhibition. | |||
Cyclin-A2 (CCNA2)
Epigallocatechin gallate
[Phase 3]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [58] | |||
| Responsed Disease | Obesity | ICD-11: 5B81 | ||
| Target Regulation | Down regulation | |||
| Cell Process | Adipogenesis | |||
| In-vitro Model | 3T3-L1 | Normal | Mus musculus | CVCL_0123 |
| Response Summary | m6A-dependent Cyclin-A2 (CCNA2) and CDK2 expressions mediated by FTO and YTHDF2 contributed to EGCG-induced adipogenesis inhibition. | |||
Cyclin-dependent kinase 2 (CDK2)
Epigallocatechin gallate
[Phase 3]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [58] | |||
| Responsed Disease | Obesity | ICD-11: 5B81 | ||
| Target Regulation | Down regulation | |||
| Cell Process | Adipogenesis | |||
| In-vitro Model | 3T3-L1 | Normal | Mus musculus | CVCL_0123 |
| Response Summary | m6A-dependent CCNA2 and Cyclin-dependent kinase 2 (CDK2) expressions mediated by FTO and YTHDF2 contributed to EGCG-induced adipogenesis inhibition. | |||
FB23-2
[Investigative]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [61] | |||
| Responsed Disease | Diabetic retinopathy | ICD-11: 9B71.0 | ||
| Target Regulation | Down regulation | |||
Cyclin-dependent kinase inhibitor 1B (CDKN1B/p27)
Cisplatin
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [62] | |||
| Responsed Disease | Intrahepatic cholangiocarcinoma | ICD-11: 2C12.10 | ||
| Target Regulation | Down regulation | |||
| Pathway Response | Cell cycle | hsa04110 | ||
| Cell Process | Cell proliferation | |||
| Arrest cell cycle at G0/G1 phase | ||||
| In-vitro Model | HuCC-T1 | Intrahepatic cholangiocarcinoma | Homo sapiens | CVCL_0324 |
| RBE | Intrahepatic cholangiocarcinoma | Homo sapiens | CVCL_4896 | |
| HCCC-9810 (The intrahepatic cholangiocarcinoma cell lines (HCCC-9810) were purchased from Cellcook Co., Ltd. (Guangzhou, China).) | ||||
| HIBEC (The normal intrahepatic bile duct cell line (HIBEC) were purchased from Cellcook Co., Ltd. (Guangzhou, China).) | ||||
| In-vivo Model | For tumour xenograft models, 1 × 107 HuCC-T1 cells in knockdown group or control group were implanted into the right flank of 5-week-old female nude mice. The volumes of tumour were recorded every 4 days by calliper. The volumes were calculated as length × width2/2. For patient-derived xenograft (PDX) model (PDX0075), ICC tissues from a patient, who relapsed in 6 months after R0 resection and subsequent chemotherapy with cisplatin and gemcitabine, were diced into 3 mm3 pieces and transplanted subcutaneously into the right flank of 5-week-old female B-NDG mice. | |||
| Response Summary | The role of YTHDF2 in tumourigenesis and cisplatin-desensitising function by promoting the degradation of Cyclin-dependent kinase inhibitor 1B (CDKN1B/p27) mRNA in an m6 A-dependent manner. YTHDF2 exhibits tumour oncogenic and cisplatin-desensitising properties, which offer insight into the development of novel combination therapeutic strategies for intrahepatic cholangiocarcinoma. | |||
Death-associated protein kinase 3 (DAPK3)
Gemcitabine
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [67] | |||
| Responsed Disease | Gallbladder cancer | ICD-11: 2C13 | ||
| Target Regulation | Down regulation | |||
| In-vivo Model | Male BALB/c nude mice (6 weeks old) were used for xenograft experiments. A 150 μL-cell suspension containing 5 × 106 cells was injected s.c. into each nude mouse. Tumor volumes were measured every 4 days for 1 month. To evaluate chemosensitivity to gemcitabine, mice were i.p. injected with gemcitabine (50 mg/kg) every 4 days when the tumor volume reached 100 mm3.Male NOD/SCID mice (6 weeks old) were used for lung metastasis experiments. A 150 μL-cell suspension containing 5 × 105 cells was injected i.v. into the tail vein of each mouse. After 1 month, the lungs were harvested and photographed, and the number of metastatic lesions was calculated. | |||
DNA replication licensing factor MCM5 (MCM5)
OSMI-1
[Investigative]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [69] | |||
| Responsed Disease | Liver hepatocellular carcinoma | ICD-11: 2C12.02 | ||
| Target Regulation | Up regulation | |||
| In-vitro Model | HepAD38 | Hepatoblastoma | Homo sapiens | CVCL_M177 |
| Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 | |
| PLC/PRF/5 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_0485 | |
| MHCC97-H | Adult hepatocellular carcinoma | Homo sapiens | CVCL_4972 | |
| Huh-7 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_0336 | |
| HEK293 | Normal | Homo sapiens | CVCL_0045 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| In-vivo Model | For the xenograft implantation model, 2 × 106 MHCC-97H cells were subcutaneously injected into the flank of nude mice. | |||
Eukaryotic translation initiation factor 4 gamma 1 (EIF4G1)
Rapamycin
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [72] | |||
| Responsed Disease | Oral squamous cell carcinoma | ICD-11: 2B6E.0 | ||
| Target Regulation | Down regulation | |||
| Pathway Response | Autophagy | hsa04140 | ||
| Cell Process | Cell autophagy | |||
| Response Summary | Rapamycin inhibited FTO activity, and directly targeted Eukaryotic translation initiation factor 4 gamma 1 (EIF4G1) transcripts and mediated their expression in an m6A-dependent manner in oral squamous cell carcinoma. After FTO silencing, YTHDF2 captured eIF4G1 transcripts containing m6A, resulting in mRNA degradation and decreased expression of eIF4G1 protein, thereby promoting autophagy and reducing tumor occurrence. | |||
Hypoxia-inducible factor 1-alpha (HIF-1-Alpha/HIF1A)
Chromium
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [51] | |||
| Target Regulation | Down regulation | |||
NACHT, LRR and PYD domains-containing protein 3 (NLRP3)
Gefitinib
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [89] | |||
| Responsed Disease | Lung cancer | ICD-11: 2C25 | ||
| In-vitro Model | PC-9 | Lung adenocarcinoma | Homo sapiens | CVCL_B260 |
| HCC827 | Lung adenocarcinoma | Homo sapiens | CVCL_2063 | |
| A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 | |
| SPC-A1 | Endocervical adenocarcinoma | Homo sapiens | CVCL_6955 | |
| PC-9/GR2 | Lung adenocarcinoma | Homo sapiens | CVCL_DI29 | |
|
HCC827/GR
|
N.A. | Homo sapiens | CVCL_E7R9 | |
| In-vivo Model | To establish xenograft models, five-week-old male mice were orthotopically injected with the same number of the PC9/GR cells. Tumours had developed after 4 days, at which time the xenografted mice were randomly divided into the following two experimental groups (each group with 6 mice): (1) the control group and (2) the gefitinib group. | |||
PPAR-gamma coactivator 1-alpha (PGC-1a/PPARGC1A)
Arsenite
[Phase 2]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [99] | |||
| Responsed Disease | Insulin resistance | ICD-11: 5A44 | ||
| In-vitro Model | L-02 | Endocervical adenocarcinoma | Homo sapiens | CVCL_6926 |
Programmed cell death 1 (PD-1)
PMID31239444-anti-PD1 antibody
[Investigative]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [3] | |||
| Responsed Disease | Melanoma | ICD-11: 2C30 | ||
| Target Regulation | Down regulation | |||
| Pathway Response | PD-L1 expression and PD-1 checkpoint pathway in cancer | hsa05235 | ||
| Cell Process | mRNA decay | |||
| In-vitro Model | B16-F10 | Mouse melanoma | Mus musculus | CVCL_0159 |
| CHL-1 | Melanoma | Homo sapiens | CVCL_1122 | |
| 624-mel | Melanoma | Homo sapiens | CVCL_8054 | |
| NHEM (Normal Human Epidermal Melanocytes) | ||||
| SK-MEL-30 | Cutaneous melanoma | Homo sapiens | CVCL_0039 | |
| WM115 | Melanoma | Homo sapiens | CVCL_0040 | |
| WM35 | Melanoma | Homo sapiens | CVCL_0580 | |
| WM3670 | Melanoma | Homo sapiens | CVCL_6799 | |
| WM793 | Melanoma | Homo sapiens | CVCL_8787 | |
| In-vivo Model | When the tumors reached a volume of 80-100 mm3, mice were treated with anti-PD-1 or isotype control antibody (200 ug/mouse) by i.p. injection, every other day for three times. For IFNγ blockade treatment, C57BL/6 mice were treated with anti-IFNγ antibody or isotype control IgG (250 ug/mouse) every other day after tumor cell inoculation. | |||
| Response Summary | These findings demonstrate a crucial role of FTO as an m6A demethylase in promoting melanoma tumorigenesis and anti-PD-1 resistance, and suggest that the combination of FTO inhibition with anti-PD-1 blockade reduces the resistance to immunotherapy in melanoma. Knockdown of FTO increases m6A methylation in the critical protumorigenic melanoma cell-intrinsic genes including Programmed cell death 1 (PD-1) (PDCD1), CXCR4, and SOX10, leading to increased RNA decay through the m6A reader YTHDF2. | |||
RIG-I-like receptor 1 (RIG-I)
Bacillus Calmette
[Investigative]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [101] | |||
| Responsed Disease | Bladder cancer | ICD-11: 2C94 | ||
| Target Regulation | Down regulation | |||
| In-vitro Model | MGH-U3 | Bladder carcinoma | Homo sapiens | CVCL_9827 |
| SW780 | Bladder carcinoma | Homo sapiens | CVCL_1728 | |
| 5637 | Bladder carcinoma | Homo sapiens | CVCL_0126 | |
| T24 | Bladder carcinoma | Homo sapiens | CVCL_0554 | |
| UM-UC-3 | Bladder carcinoma | Homo sapiens | CVCL_1783 | |
| In-vivo Model | For xenograft studies, female mice between 4 and 6 weeks of age were used. Monoclonal cell lines showing a nearly complete absence of Ythdf2 and Ddx58 were used for in vivo experimentation. Mice were implanted with 2 × 106 MGHU3 cells resuspended in 100 μL of Matrigel Matrix High Concentration (354248) and PBS mixture subcutaneously into the right flanks. Subcutaneous tumor growth was measured weekly by calipers, and the volume was calculated using the formula: volume = (length × width2)/2. | |||
Serine/threonine-protein kinase mTOR (MTOR)
Asparagine inhibitor
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [56] | |||
| Responsed Disease | Colorectal cancer | ICD-11: 2B91 | ||
| Target Regulation | Up regulation | |||
| Pathway Response | mTOR signaling pathway | hsa04150 | ||
| Autophagy | hsa04140 | |||
| Cell Process | RNA decay | |||
| Cell growth and death | ||||
| Cell autophagy | ||||
| In-vitro Model | HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 |
| SW480 | Colon adenocarcinoma | Homo sapiens | CVCL_0546 | |
| Response Summary | In colorectal cancer, Glutaminolysis inhibition upregulated ATF4 expression in an m6A-dependent manner to activate pro-survival autophagy through transcriptional activation of the mTOR inhibitor DDIT4. Determined the relationship between FTO alpha-ketoglutarate dependent dioxygenase (FTO), YTH N6-methyladenosine RNA binding protein 2 (YTHDF2), and ATF4. ATF4 transcriptionally upregulated DDIT4 to suppress Serine/threonine-protein kinase mTOR (MTOR), which induced pro-survival autophagy during glutaminolysis inhibition. | |||
Chloroquine
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [56] | |||
| Responsed Disease | Colorectal cancer | ICD-11: 2B91 | ||
| Target Regulation | Up regulation | |||
| Pathway Response | mTOR signaling pathway | hsa04150 | ||
| Autophagy | hsa04140 | |||
| Cell Process | RNA decay | |||
| Cell growth and death | ||||
| Cell autophagy | ||||
| In-vitro Model | HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 |
| SW480 | Colon adenocarcinoma | Homo sapiens | CVCL_0546 | |
| Response Summary | In colorectal cancer, Glutaminolysis inhibition upregulated ATF4 expression in an m6A-dependent manner to activate pro-survival autophagy through transcriptional activation of the mTOR inhibitor DDIT4. Determined the relationship between FTO alpha-ketoglutarate dependent dioxygenase (FTO), YTH N6-methyladenosine RNA binding protein 2 (YTHDF2), and ATF4. Serine/threonine-protein kinase mTOR (MTOR) transcriptionally upregulated DDIT4 to suppress mTOR, which induced pro-survival autophagy during glutaminolysis inhibition. | |||
Meclofenamate sodium
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [56] | |||
| Responsed Disease | Colorectal cancer | ICD-11: 2B91 | ||
| Target Regulation | Up regulation | |||
| Pathway Response | mTOR signaling pathway | hsa04150 | ||
| Autophagy | hsa04140 | |||
| Cell Process | RNA decay | |||
| Cell growth and death | ||||
| Cell autophagy | ||||
| In-vitro Model | HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 |
| SW480 | Colon adenocarcinoma | Homo sapiens | CVCL_0546 | |
| Response Summary | In colorectal cancer, Glutaminolysis inhibition upregulated ATF4 expression in an m6A-dependent manner to activate pro-survival autophagy through transcriptional activation of the mTOR inhibitor DDIT4. Determined the relationship between FTO alpha-ketoglutarate dependent dioxygenase (FTO), YTH N6-methyladenosine RNA binding protein 2 (YTHDF2), and ATF4. Serine/threonine-protein kinase mTOR (MTOR) transcriptionally upregulated DDIT4 to suppress mTOR, which induced pro-survival autophagy during glutaminolysis inhibition. | |||
Rapamycin
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [56] | |||
| Responsed Disease | Colorectal cancer | ICD-11: 2B91 | ||
| Target Regulation | Up regulation | |||
| Pathway Response | mTOR signaling pathway | hsa04150 | ||
| Autophagy | hsa04140 | |||
| Cell Process | RNA decay | |||
| Cell growth and death | ||||
| Cell autophagy | ||||
| In-vitro Model | HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 |
| SW480 | Colon adenocarcinoma | Homo sapiens | CVCL_0546 | |
| Response Summary | In colorectal cancer, Glutaminolysis inhibition upregulated ATF4 expression in an m6A-dependent manner to activate pro-survival autophagy through transcriptional activation of the mTOR inhibitor DDIT4. Determined the relationship between FTO alpha-ketoglutarate dependent dioxygenase (FTO), YTH N6-methyladenosine RNA binding protein 2 (YTHDF2), and ATF4. Serine/threonine-protein kinase mTOR (MTOR) transcriptionally upregulated DDIT4 to suppress mTOR, which induced pro-survival autophagy during glutaminolysis inhibition. | |||
CB-839
[Phase 2]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [56] | |||
| Responsed Disease | Colorectal cancer | ICD-11: 2B91 | ||
| Target Regulation | Up regulation | |||
| Pathway Response | mTOR signaling pathway | hsa04150 | ||
| Autophagy | hsa04140 | |||
| Cell Process | RNA decay | |||
| Cell growth and death | ||||
| Cell autophagy | ||||
| In-vitro Model | HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 |
| SW480 | Colon adenocarcinoma | Homo sapiens | CVCL_0546 | |
| Response Summary | In colorectal cancer, Glutaminolysis inhibition upregulated ATF4 expression in an m6A-dependent manner to activate pro-survival autophagy through transcriptional activation of the mTOR inhibitor DDIT4. Determined the relationship between FTO alpha-ketoglutarate dependent dioxygenase (FTO), YTH N6-methyladenosine RNA binding protein 2 (YTHDF2), and ATF4. ATF4 transcriptionally upregulated DDIT4 to suppress Serine/threonine-protein kinase mTOR (MTOR), which induced pro-survival autophagy during glutaminolysis inhibition. | |||
GLS-IN-968
[Investigative]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [56] | |||
| Responsed Disease | Colorectal cancer | ICD-11: 2B91 | ||
| Target Regulation | Up regulation | |||
| Pathway Response | mTOR signaling pathway | hsa04150 | ||
| Autophagy | hsa04140 | |||
| Cell Process | RNA decay | |||
| Cell growth and death | ||||
| Cell autophagy | ||||
| In-vitro Model | HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 |
| SW480 | Colon adenocarcinoma | Homo sapiens | CVCL_0546 | |
| Response Summary | In colorectal cancer, Glutaminolysis inhibition upregulated ATF4 expression in an m6A-dependent manner to activate pro-survival autophagy through transcriptional activation of the mTOR inhibitor DDIT4. Determined the relationship between FTO alpha-ketoglutarate dependent dioxygenase (FTO), YTH N6-methyladenosine RNA binding protein 2 (YTHDF2), and ATF4. ATF4 transcriptionally upregulated DDIT4 to suppress Serine/threonine-protein kinase mTOR (MTOR), which induced pro-survival autophagy during glutaminolysis inhibition. | |||
Serine/threonine-protein kinase ULK1 (ULK1/ATG1)
Rapamycin
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [103] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | Autophagy | hsa04140 | ||
| Cell Process | RNA stability | |||
| Cell autophagy | ||||
| In-vitro Model | HEK293T | Normal | Homo sapiens | CVCL_0063 |
| HeLa | Endocervical adenocarcinoma | Homo sapiens | CVCL_0030 | |
| Response Summary | The m6A changes caused by FTO influence the stability of ULK1 transcripts, likely through a YTHDF2-dependent manner.Under both basal and rapamycin-induced autophagy conditions, depletion of FTO significantly reduced the formation of GFP-LC3B puncta. The level of p62/SQSTM1 (an autophagy substrate) was higher in FTO-knockdown cells than that in control cells. FTO specifically upregulates the Serine/threonine-protein kinase ULK1 (ULK1) protein abundance. ULK1 mRNA undergoes m6A modification in the 3'-UTR and the m6A-marked ULK1 transcripts can further be targeted for degradation by YTHDF2. | |||
Serum response factor (SRF)
Tislelizumab
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [104] | |||
| Responsed Disease | Glioblastoma | ICD-11: 2A00.00 | ||
| In-vitro Model | U-87MG ATCC | Glioblastoma | Homo sapiens | CVCL_0022 |
Stimulator of interferon genes protein (STING1)
Fluorene-9-bisphenol
[Investigative]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [108] | |||
| Target Regulation | Up regulation | |||
Suppressor of cytokine signaling 3 (SOCS3)
Arsenite
[Phase 2]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [110] | |||
| Responsed Disease | Human skin lesions | ICD-11: ME60 | ||
| Target Regulation | Down regulation | |||
| Pathway Response | JAK-STAT signaling pathway | hsa04630 | ||
| In-vitro Model | HaCaT | Normal | Homo sapiens | CVCL_0038 |
Transcription factor SOX-10 (SOX10)
PMID31239444-anti-PD1 antibody
[Investigative]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [3] | |||
| Responsed Disease | Melanoma | ICD-11: 2C30 | ||
| Target Regulation | Down regulation | |||
| Pathway Response | PD-L1 expression and PD-1 checkpoint pathway in cancer | hsa05235 | ||
| Cell Process | mRNA decay | |||
| In-vitro Model | B16-F10 | Mouse melanoma | Mus musculus | CVCL_0159 |
| CHL-1 | Melanoma | Homo sapiens | CVCL_1122 | |
| 624-mel | Melanoma | Homo sapiens | CVCL_8054 | |
| NHEM (Normal Human Epidermal Melanocytes) | ||||
| SK-MEL-30 | Cutaneous melanoma | Homo sapiens | CVCL_0039 | |
| WM115 | Melanoma | Homo sapiens | CVCL_0040 | |
| WM35 | Melanoma | Homo sapiens | CVCL_0580 | |
| WM3670 | Melanoma | Homo sapiens | CVCL_6799 | |
| WM793 | Melanoma | Homo sapiens | CVCL_8787 | |
| In-vivo Model | When the tumors reached a volume of 80-100 mm3, mice were treated with anti-PD-1 or isotype control antibody (200 ug/mouse) by i.p. injection, every other day for three times. For IFNγ blockade treatment, C57BL/6 mice were treated with anti-IFNγ antibody or isotype control IgG (250 ug/mouse) every other day after tumor cell inoculation. | |||
| Response Summary | These findings demonstrate a crucial role of FTO as an m6A demethylase in promoting melanoma tumorigenesis and anti-PD-1 resistance, and suggest that the combination of FTO inhibition with anti-PD-1 blockade reduces the resistance to immunotherapy in melanoma. Knockdown of FTO increases m6A methylation in the critical protumorigenic melanoma cell-intrinsic genes including PD-1 (PDCD1), CXCR4, and Transcription factor SOX-10 (SOX10), leading to increased RNA decay through the m6A reader YTHDF2. | |||
LINC00902 (TUSC7)
Erlotinib
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [117] | |||
| Responsed Disease | Lung adenocarcinoma | ICD-11: 2C25.0 | ||
| Target Regulation | Down regulation | |||
| Pathway Response | Notch signaling pathway | hsa04330), EGFR tyrosine kinase inhibitor resistance | ||
| In-vitro Model | PC-9 | Lung adenocarcinoma | Homo sapiens | CVCL_B260 |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| HCC827 | Lung adenocarcinoma | Homo sapiens | CVCL_2063 | |
| In-vivo Model | Control vector, TUSC7 knockout, FLI-06 treated H1975 cells (1*107) cells were suspended in 100 uL of serum-free DMEM medium (Hyclone, USA), mixed with matrix gel (Corning, USA), and then were injected subcutaneously. The changes in the tumor size were recorded every 3 or 5 days. | |||
| Response Summary | In lung adenocarcinoma, The miR-146a/Notch signaling was sustained highly activated in a m6A dependent manner, and the m6A regulator of YTHDF2 suppressed LINC00902 (TUSC7), both of which contributed to the resistant features. Functionally, the sponge type of TUSC7 regulation of miR-146a inhibited Notch signaling functions, and affected the cancer progression and stem cells' renewal in Erlotinib resistant PC9 cells (PC9ER) and Erlotinib resistant HCC827 cells (HCC827ER) cells. | |||
Long intergenic non-protein coding RNA 1273 (LINC01273)
Sorafenib
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [118] | |||
| Responsed Disease | Hepatocellular carcinoma | ICD-11: 2C12.02 | ||
| Target Regulation | Down regulation | |||
| In-vitro Model | SMMC-7721 | Endocervical adenocarcinoma | Homo sapiens | CVCL_0534 |
| Huh-7 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_0336 | |
| Response Summary | Long intergenic non-protein coding RNA 1273 (LINC01273) was modified with m6A, METTL3 increased LINC01273 m6A modification, followed by LINC01273 decay in the presence of YTHDF2, a m6A 'reader'. And LINC01273 plays a key role in sorafenib resistant HCC cells. | |||
Apoptosis regulatory protein Siva (SIVA1)
Fluorouracil
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [124] | |||
| Responsed Disease | Colorectal cancer | ICD-11: 2B91 | ||
| Target Regulation | Down regulation | |||
| In-vitro Model | HCT 8 | Colon adenocarcinoma | Homo sapiens | CVCL_2478 |
| HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| MC-38 | Mouse colon adenocarcinoma | Mus musculus | CVCL_B288 | |
| In-vivo Model | For the tumor metastasis mouse model, 5-week-old C57BL/6 mice were randomly grouped and injected with 5 × 105 Control or shFTO stable MC38 cells via tail vein. Drug administration was adopted after 48 h. Drug administration (intraperitoneally): DMSO, 5-FU (50 mg/kg every 2 days) or Rhein (10 mg/kg every 2 days). To detect lung metastasis, mice were killed 2 weeks after tumor cell injection. Lung tissues were harvested and fixed with 4% PFA for paraffin-embedded section and lung metastases were detected with Nikon microscopy. For tumor intraperitoneal mouse model, 2 × 106 Dox-shCtrl, Dox-shFTO stable HCT8/5-FU cells were injected into 5-week-old male BALB/C nude mice. Drug administration was adopted after 48 h. Drug administration (intraperitoneally): DMSO, 5-FU (50 mg/kg every 2 days) or FB23-2 (10 mg/kg every 2 days). For tumor intraperitoneal mouse model, 5 × 105 shCtrl, shFTO-1, shFTO-2 stable MC38 cells were injected into 5-week-old C57BL/6 mice. Drug administration was adopted after 48 h. Drug administration (intraperitoneally): DMSO, 5-FU (50 mg/kg every 2 days) or Rhein (10 mg/kg every 2 days). | |||
Aryl hydrocarbon receptor (AHR)
Tislelizumab
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [104] | |||
| Responsed Disease | Glioblastoma | ICD-11: 2A00.00 | ||
| In-vitro Model | U-87MG ATCC | Glioblastoma | Homo sapiens | CVCL_0022 |
CD70 antigen (CD70)
JNJ-74494550
[Phase 2]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [129] | |||
| Responsed Disease | Papillary thyroid cancer | ICD-11: 2D10.1 | ||
| Target Regulation | Down regulation | |||
| In-vitro Model | Nthy-ori 3-1 | Normal | Homo sapiens | CVCL_2659 |
| B-CPAP | Thyroid gland carcinoma | Homo sapiens | CVCL_0153 | |
| K1 | Thyroid gland papillary carcinoma | Homo sapiens | CVCL_2537 | |
| C-643 | Thyroid gland anaplastic carcinoma | Homo sapiens | CVCL_5969 | |
| BHT-101 | Thyroid gland anaplastic carcinoma | Homo sapiens | CVCL_1085 | |
| THP-1 | Childhood acute monocytic leukemia | Homo sapiens | CVCL_0006 | |
| In-vivo Model | NCG mice (female, 4-5 weeks old, purchased from Jiangsu GemPharmatech) were acclimated for 1 week and were selected randomly to subcutaneously (s.c.) injected with 100 μl suspensions of 1 × 106 thyroid cancer cell lines. All mice were housed in animal facility under specific pathogen-free conditions. After 7 day, 5 × 106 huPBMCs were intravenously (i.v.) inoculated into mice. Mice were treated intravenously with anti-PD-1 mAb (nivolumab, 200 μg per injection), Combination (nivolumab + anti-CD70 mAb cusatuzumab, 200 μg per injection each), or PBS once a week. The treatment with M2-TAMs EVs or PBS continued in mice at 2ug EVs per injection at the tumor site twice a week [24]. The body weight of mice and the length (mm) and width (mm) of tumors were monitored every 3 or 4 days. Tumor volume (mm3) was calculated as Length × Width2 × 1/2. Tumor growth analyses were limited to 3 to 4 weeks because following this period mice started to show signs of xenograft-versus-host disease (xGVHD). Therefore, after 14 days or 21 days of PBMCs implantation, the mice were euthanized. | |||
Circ_ASK1
Gefitinib
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [131] | |||
| Responsed Disease | Lung adenocarcinoma | ICD-11: 2C25.0 | ||
| Target Regulation | Down regulation | |||
| Cell Process | Cell apoptosis | |||
| In-vitro Model | SPC-A1 | Endocervical adenocarcinoma | Homo sapiens | CVCL_6955 |
| SK-LU-1 | Lung adenocarcinoma | Homo sapiens | CVCL_0629 | |
| NCI-H1993 | Lung adenocarcinoma | Homo sapiens | CVCL_1512 | |
| NCI-H1975 | Lung adenocarcinoma | Homo sapiens | CVCL_1511 | |
| NCI-H1650 | Minimally invasive lung adenocarcinoma | Homo sapiens | CVCL_1483 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| HCC827 | Lung adenocarcinoma | Homo sapiens | CVCL_2063 | |
| BEAS-2B | Normal | Homo sapiens | CVCL_0168 | |
| A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 | |
| 16HBE14o- | Normal | Homo sapiens | CVCL_0112 | |
| In-vivo Model | Established a xenograft model in BALB/c nude mice by inoculating HCC827-GR cells transfected with the constructs for circASK1 silencing, ASK1-272a.a overexpression and ASK1-272a.a overexpression/circASK1 knockdown. | |||
| Response Summary | Increased YTHDF2-mediated endoribonucleolytic cleavage of m6A-modified Circ_ASK1 accounts for its downregulation in gefitinib-resistant cells. Either METTL3 silencing or YTHDF2 silencing suppressed the decay of circASK1 in HCC827-GR cells. This study provides a novel therapeutic target to overcome gefitinib resistance in LUAD patients. | |||
Circ_RERE
ICG-001
[Investigative]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [135] | |||
| Responsed Disease | Osteoarthritis | ICD-11: FA05 | ||
| Target Regulation | Down regulation | |||
Complement C1q subcomponent subunit A (C1QA)
Rituximab
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [138] | |||
| Responsed Disease | Diffuse large B-cell lymphomas | ICD-11: 2A81 | ||
| Target Regulation | Down regulation | |||
| In-vivo Model | Approximately 2 × 106 Farage/R or Farage/S cells stably transfected with shNC, shC1qA or shMETTL3 were subcutaneously injected into the left flank of each mouse. When the tumor volume reached ~100 mm3, each mouse received an intraperitoneal injection of Rituximab (20 mg/kg) every 4 days for a total of 5 injections. The diameter of each tumor was examined every 4 days using a caliper, and tumor volume was calculated as follows: (length × width2)/2. At 28 days after xenograft, the mice were sacrificed and the tumors were weighed and collected. | |||
CREB-binding protein (CREBBP)
PT2385
[Investigative]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [139] | |||
| Target Regulation | Up regulation | |||
DNA replication licensing factor MCM7 (MCM2)
OSMI-1
[Investigative]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [69] | |||
| Responsed Disease | Liver hepatocellular carcinoma | ICD-11: 2C12.02 | ||
| Target Regulation | Up regulation | |||
| In-vitro Model | HepAD38 | Hepatoblastoma | Homo sapiens | CVCL_M177 |
| Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 | |
| PLC/PRF/5 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_0485 | |
| MHCC97-H | Adult hepatocellular carcinoma | Homo sapiens | CVCL_4972 | |
| Huh-7 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_0336 | |
| HEK293 | Normal | Homo sapiens | CVCL_0045 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| In-vivo Model | For the xenograft implantation model, 2 × 106 MHCC-97H cells were subcutaneously injected into the flank of nude mice. | |||
GTP cyclohydrolase 1 (GCH1)
Fluorene-9-bisphenol
[Investigative]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [108] | |||
| Target Regulation | Up regulation | |||
long intergenic non-protein coding RNA 115 (LINC00115)
Acetaminophen
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [154] | |||
| Responsed Disease | Triple-negative breast cancer | ICD-11: 2C6Z | ||
| Target Regulation | Down regulation | |||
Mitogen-activated protein kinase 13 (MAPK13)
Rapamycin
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [158] | |||
| Target Regulation | Down regulation | |||
| In-vitro Model | 621-101 | Lung lymphangioleiomyomatosis | Homo sapiens | CVCL_S879 |
|
HEK293-EBNA
|
N.A. | Homo sapiens | CVCL_6974 | |
| MCF-7 | Invasive breast carcinoma | Homo sapiens | CVCL_0031 | |
| BT-549 | Invasive breast carcinoma | Homo sapiens | CVCL_1092 | |
| UMB1949 | Kidney angiomyolipoma | Homo sapiens | CVCL_C471 | |
Peroxiredoxin-5, mitochondrial (PRDX5)
FB23-2
[Investigative]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [162] | |||
| Target Regulation | Up regulation | |||
| In-vitro Model | HaCaT | Normal | Homo sapiens | CVCL_0038 |
Prolyl endopeptidase FAP (FAP)
VS-6063
[Phase 2]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [164] | |||
| Responsed Disease | Non-small cell lung cancer | ICD-11: 2C25.Y | ||
| In-vitro Model | NCI-H1299 | Lung large cell carcinoma | Homo sapiens | CVCL_0060 |
| A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 | |
| NCI-H1650 | Minimally invasive lung adenocarcinoma | Homo sapiens | CVCL_1483 | |
| NCI-H460 | Lung large cell carcinoma | Homo sapiens | CVCL_0459 | |
| HCC827 | Lung adenocarcinoma | Homo sapiens | CVCL_2063 | |
| PC-9 | Lung adenocarcinoma | Homo sapiens | CVCL_B260 | |
| SK-MES-1 | Lung squamous cell carcinoma | Homo sapiens | CVCL_0630 | |
| BEAS-2B | Normal | Homo sapiens | CVCL_0168 | |
| In-vivo Model | The Laboratory Animal Center of Soochow University provided female BALB/c nude mice (5 weeks old). Mice were kept under specific pathogen-free conditions. They were injected intravenously (i.v.) with FTO-overexpressing and control A549 cells (1.8 × 106 cells/mouse) to establish the in vivo model of NSCLC metastasis, and were then gavaged with dimethyl sulfoxide (DMSO) (25 mg/kg, daily) or the FAK inhibitor defactinib (VS6063) (Cat#S7654, Selleck, USA) (25 mg/kg, daily) beginning in the fifth week after injection. The mice were euthanized eight weeks after being inoculated, and their lungs were taken out and preserved in Bouin's solution for macroscopic investigation of metastatic nodules. Hematoxylin and eosin (H&E) staining of lung tissues was used to look for micrometastatic foci. | |||
Pseudorabies Virus (PRV)
3-deazidenosine
[Investigative]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [169] | |||
| Responsed Disease | Rabies | ICD-11: 1C82 | ||
| Target Regulation | Up regulation | |||
| In-vitro Model |
PK-15
|
N.A. | Sus scrofa | CVCL_2160 |
Unspecific Target Gene
Teniposide
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [189] | |||
| Responsed Disease | Hepatocellular carcinoma | ICD-11: 2C12.02 | ||
| Pathway Response | Nucleotide excision repair | hsa03420 | ||
| mTOR signaling pathway | hsa04150 | |||
| Cell Process | DNA repair | |||
| In-vitro Model | Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 |
| Huh-7 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_0336 | |
| In-vivo Model | To establish a tumour model, C57BL/6 mice were intraperitoneal injected with 25 mg/kg diethylnitrosamine at 2 weeks of age. | |||
| Response Summary | The m6A model includes LRPPRC, YTHDF2, KIAA14219, and RBM15B, classified A-hepatocellular carcinoma patients into high/low-risk subtypes. The expression of Immunosuppressive cytokines DNMT1/EZH2 was up-regulated in A-hepatocellular carcinoma patients, and teniposide can be a potential therapeutic drug for A-hepatocellular carcinoma. | |||
Full List of Crosstalk(s) between m6A Modification and Epigenetic Regulation Related to This Regulator
RNA modification
m6A Target: Ubiquitin domain-containing protein TINC (TINCR)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT00017 | ||
| Epigenetic Regulator | tRNA (adenine(58)-N(1))-methyltransferase non-catalytic subunit TRM6 (TRMT6) | |
| Regulated Target | TINCR ubiquitin domain containing (TINCR) | |
| Crosstalk relationship | m1A → m6A | |
| Crosstalk ID: M6ACROT00018 | ||
| Epigenetic Regulator | tRNA (adenine(58)-N(1))-methyltransferase catalytic subunit TRMT61A (TRMT61A) | |
| Regulated Target | TINCR ubiquitin domain containing (TINCR) | |
| Crosstalk relationship | m1A → m6A | |
m6A Target: Protein SON
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT00019 | ||
| Epigenetic Regulator | tRNA (adenine(58)-N(1))-methyltransferase non-catalytic subunit TRM6 (TRMT6) | |
| Regulated Target | Protein SON | |
| Crosstalk relationship | m1A → m6A | |
| Crosstalk ID: M6ACROT00020 | ||
| Epigenetic Regulator | tRNA (adenine(58)-N(1))-methyltransferase catalytic subunit TRMT61A (TRMT61A) | |
| Regulated Target | Protein SON | |
| Crosstalk relationship | m1A → m6A | |
m6A Target: Circ_MATR3
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT00021 | ||
| Epigenetic Regulator | tRNA (adenine(58)-N(1))-methyltransferase non-catalytic subunit TRM6 (TRMT6) | |
| Regulated Target | Circ_MATR3 | |
| Crosstalk relationship | m1A → m6A | |
| Crosstalk ID: M6ACROT00022 | ||
| Epigenetic Regulator | tRNA (adenine(58)-N(1))-methyltransferase catalytic subunit TRMT61A (TRMT61A) | |
| Regulated Target | Circ_MATR3 | |
| Crosstalk relationship | m1A → m6A | |
m6A Target: Circ_FAM20B
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT00023 | ||
| Epigenetic Regulator | tRNA (adenine(58)-N(1))-methyltransferase non-catalytic subunit TRM6 (TRMT6) | |
| Regulated Target | Circ_FAM20B | |
| Crosstalk relationship | m1A → m6A | |
| Crosstalk ID: M6ACROT00024 | ||
| Epigenetic Regulator | tRNA (adenine(58)-N(1))-methyltransferase catalytic subunit TRMT61A (TRMT61A) | |
| Regulated Target | Circ_FAM20B | |
| Crosstalk relationship | m1A → m6A | |
m6A Target: Circ_SLC45A4
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT00025 | ||
| Epigenetic Regulator | tRNA (adenine(58)-N(1))-methyltransferase non-catalytic subunit TRM6 (TRMT6) | |
| Regulated Target | Circ_SLC45A4 | |
| Crosstalk relationship | m1A → m6A | |
| Crosstalk ID: M6ACROT00026 | ||
| Epigenetic Regulator | tRNA (adenine(58)-N(1))-methyltransferase catalytic subunit TRMT61A (TRMT61A) | |
| Regulated Target | Circ_SLC45A4 | |
| Crosstalk relationship | m1A → m6A | |
m6A Target: Myc proto-oncogene protein (MYC)
| In total 3 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT00028 | ||
| Epigenetic Regulator | tRNA methyltransferase 10 homolog A (TRMT10A) | |
| Regulated Target | tRNA-Gln (anticodon TTG) 1-1 (TRQ-TTG1-1) | |
| Crosstalk relationship | m1G → m6A | |
| Drug | AVI-5126 | |
| Crosstalk ID: M6ACROT00030 | ||
| Epigenetic Regulator | tRNA methyltransferase 10 homolog A (TRMT10A) | |
| Regulated Target | tRNA-Arg (anticodon CCG) 1-1 (TRR-CCG1-1) | |
| Crosstalk relationship | m1G → m6A | |
| Drug | AVI-5126 | |
| Crosstalk ID: M6ACROT00032 | ||
| Epigenetic Regulator | tRNA methyltransferase 10 homolog A (TRMT10A) | |
| Regulated Target | tRNA-Thr (anticodon CGT) 1-1 (TRT-CGT1-1) | |
| Crosstalk relationship | m1G → m6A | |
| Drug | AVI-5126 | |
m6A Target: Forkhead box protein D1 (FOXD1)
| In total 3 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT00034 | ||
| Epigenetic Regulator | tRNA methyltransferase 10 homolog A (TRMT10A) | |
| Regulated Target | tRNA-Gln (anticodon TTG) 1-1 (TRQ-TTG1-1) | |
| Crosstalk relationship | m1G → m6A | |
| Crosstalk ID: M6ACROT00036 | ||
| Epigenetic Regulator | tRNA methyltransferase 10 homolog A (TRMT10A) | |
| Regulated Target | tRNA-Arg (anticodon CCG) 1-1 (TRR-CCG1-1) | |
| Crosstalk relationship | m1G → m6A | |
| Crosstalk ID: M6ACROT00038 | ||
| Epigenetic Regulator | tRNA methyltransferase 10 homolog A (TRMT10A) | |
| Regulated Target | tRNA-Thr (anticodon CGT) 1-1 (TRT-CGT1-1) | |
| Crosstalk relationship | m1G → m6A | |
m6A Target: Small ribosomal subunit protein mS38 (AURKAIP1)
| In total 3 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT00040 | ||
| Epigenetic Regulator | tRNA methyltransferase 10 homolog A (TRMT10A) | |
| Regulated Target | tRNA-Gln (anticodon TTG) 1-1 (TRQ-TTG1-1) | |
| Crosstalk relationship | m1G → m6A | |
| Crosstalk ID: M6ACROT00042 | ||
| Epigenetic Regulator | tRNA methyltransferase 10 homolog A (TRMT10A) | |
| Regulated Target | tRNA-Arg (anticodon CCG) 1-1 (TRR-CCG1-1) | |
| Crosstalk relationship | m1G → m6A | |
| Crosstalk ID: M6ACROT00044 | ||
| Epigenetic Regulator | tRNA methyltransferase 10 homolog A (TRMT10A) | |
| Regulated Target | tRNA-Thr (anticodon CGT) 1-1 (TRT-CGT1-1) | |
| Crosstalk relationship | m1G → m6A | |
m6A Target: Transcription factor SOX-9 (SOX9)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT00068 | ||
| Epigenetic Regulator | 5-methylcytosine rRNA methyltransferase NSUN4 (NSUN4) | |
| Regulated Target | Transcription factor SOX-9 (SOX9) | |
| Crosstalk relationship | m6A → m5C | |
m6A Target: Double-stranded RNA-specific editase 1 (ADARB1)
| In total 27 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT00082 | ||
| Epigenetic Regulator | Double-stranded RNA-specific editase 1 (ADARB1) | |
| Regulated Target | Filamin-A (FLNA) | |
| Crosstalk relationship | m6A → A-to-I | |
| Drug | PTI-125 | |
| Crosstalk ID: M6ACROT00084 | ||
| Epigenetic Regulator | Double-stranded RNA-specific editase 1 (ADARB1) | |
| Regulated Target | Glutamate ionotropic receptor AMPA type subunit 2 (GRIA2) | |
| Crosstalk relationship | m6A → A-to-I | |
| Drug | E-2007 | |
| Crosstalk ID: M6ACROT00086 | ||
| Epigenetic Regulator | Double-stranded RNA-specific editase 1 (ADARB1) | |
| Regulated Target | Cytoplasmic FMR1 interacting protein 2 (CYFIP2) | |
| Crosstalk relationship | m6A → A-to-I | |
| Crosstalk ID: M6ACROT00090 | ||
| Epigenetic Regulator | Double-stranded RNA-specific editase 1 (ADARB1) | |
| Regulated Target | Cell division cycle 14B (CDC14B) | |
| Crosstalk relationship | m6A → A-to-I | |
| Crosstalk ID: M6ACROT00094 | ||
| Epigenetic Regulator | Double-stranded RNA-specific editase 1 (ADARB1) | |
| Regulated Target | Zinc finger protein GLI1 (GLI1) | |
| Crosstalk relationship | m6A → A-to-I | |
| Drug | PMID26666870-Compound-16 | |
| Crosstalk ID: M6ACROT00096 | ||
| Epigenetic Regulator | Double-stranded RNA-specific editase 1 (ADARB1) | |
| Regulated Target | microRNA 16-1 (MIR16-1) | |
| Crosstalk relationship | m6A → A-to-I | |
| Crosstalk ID: M6ACROT00099 | ||
| Epigenetic Regulator | Double-stranded RNA-specific editase 1 (ADARB1) | |
| Regulated Target | microRNA 122 (MIR122) | |
| Crosstalk relationship | m6A → A-to-I | |
| Crosstalk ID: M6ACROT00101 | ||
| Epigenetic Regulator | Double-stranded RNA-specific editase 1 (ADARB1) | |
| Regulated Target | microRNA 214 (MIR214) | |
| Crosstalk relationship | m6A → A-to-I | |
| Crosstalk ID: M6ACROT00103 | ||
| Epigenetic Regulator | Double-stranded RNA-specific editase 1 (ADARB1) | |
| Regulated Target | Oligophrenin 1 (OPHN1) | |
| Crosstalk relationship | m6A → A-to-I | |
| Crosstalk ID: M6ACROT00105 | ||
| Epigenetic Regulator | Double-stranded RNA-specific editase 1 (ADARB1) | |
| Regulated Target | microRNA 21 (MIR21) | |
| Crosstalk relationship | m6A → A-to-I | |
| Crosstalk ID: M6ACROT00107 | ||
| Epigenetic Regulator | Double-stranded RNA-specific editase 1 (ADARB1) | |
| Regulated Target | microRNA 221 (MIR221) | |
| Crosstalk relationship | m6A → A-to-I | |
| Crosstalk ID: M6ACROT00109 | ||
| Epigenetic Regulator | Double-stranded RNA-specific editase 1 (ADARB1) | |
| Regulated Target | microRNA 222 (MIR222) | |
| Crosstalk relationship | m6A → A-to-I | |
| Crosstalk ID: M6ACROT00119 | ||
| Epigenetic Regulator | Double-stranded RNA-specific editase 1 (ADARB1) | |
| Regulated Target | Podocalyxin like (PODXL) | |
| Crosstalk relationship | m6A → A-to-I | |
| Crosstalk ID: M6ACROT00122 | ||
| Epigenetic Regulator | Double-stranded RNA-specific editase 1 (ADARB1) | |
| Regulated Target | Insulin-like growth factor-binding protein 7 (IGFBP7) | |
| Crosstalk relationship | m6A → A-to-I | |
| Crosstalk ID: M6ACROT00134 | ||
| Epigenetic Regulator | Double-stranded RNA-specific editase 1 (ADARB1) | |
| Regulated Target | Thiol methyltransferase 1A (TMT1A) | |
| Crosstalk relationship | m6A → A-to-I | |
| Crosstalk ID: M6ACROT00140 | ||
| Epigenetic Regulator | Double-stranded RNA-specific editase 1 (ADARB1) | |
| Regulated Target | Pellino E3 ubiquitin protein ligase 1 (PELI1) | |
| Crosstalk relationship | m6A → A-to-I | |
| Crosstalk ID: M6ACROT00142 | ||
| Epigenetic Regulator | Double-stranded RNA-specific editase 1 (ADARB1) | |
| Regulated Target | Protein sprouty homolog 2 (SPRY2) | |
| Crosstalk relationship | m6A → A-to-I | |
| Crosstalk ID: M6ACROT00145 | ||
| Epigenetic Regulator | Double-stranded RNA-specific editase 1 (ADARB1) | |
| Regulated Target | Heterogeneous nuclear ribonucleoprotein L like (HNRNPLL) | |
| Crosstalk relationship | m6A → A-to-I | |
| Crosstalk ID: M6ACROT00148 | ||
| Epigenetic Regulator | Double-stranded RNA-specific editase 1 (ADARB1) | |
| Regulated Target | microRNA 142 (MIR142) | |
| Crosstalk relationship | m6A → A-to-I | |
| Crosstalk ID: M6ACROT00151 | ||
| Epigenetic Regulator | Double-stranded RNA-specific editase 1 (ADARB1) | |
| Regulated Target | Protein SON | |
| Crosstalk relationship | m6A → A-to-I | |
| Crosstalk ID: M6ACROT00153 | ||
| Epigenetic Regulator | Double-stranded RNA-specific editase 1 (ADARB1) | |
| Regulated Target | Glutamate ionotropic receptor kainate type subunit 2 (GRIK2) | |
| Crosstalk relationship | m6A → A-to-I | |
| Drug | 2,4-epi-neodysiherbaine | |
| Crosstalk ID: M6ACROT00163 | ||
| Epigenetic Regulator | Double-stranded RNA-specific editase 1 (ADARB1) | |
| Regulated Target | Cytochrome P450 2B6 (CYP2B6) | |
| Crosstalk relationship | m6A → A-to-I | |
| Crosstalk ID: M6ACROT00165 | ||
| Epigenetic Regulator | Double-stranded RNA-specific editase 1 (ADARB1) | |
| Regulated Target | Cytochrome P450 2C8 (CYP2C8) | |
| Crosstalk relationship | m6A → A-to-I | |
| Crosstalk ID: M6ACROT00167 | ||
| Epigenetic Regulator | Double-stranded RNA-specific editase 1 (ADARB1) | |
| Regulated Target | Hepatocyte nuclear factor 4 alpha (HNF4A) | |
| Crosstalk relationship | m6A → A-to-I | |
| Drug | BI6015 | |
| Crosstalk ID: M6ACROT00170 | ||
| Epigenetic Regulator | Double-stranded RNA-specific editase 1 (ADARB1) | |
| Regulated Target | CXADR cell adhesion molecule (CXADR) | |
| Crosstalk relationship | m6A → A-to-I | |
| Crosstalk ID: M6ACROT00185 | ||
| Epigenetic Regulator | Double-stranded RNA-specific editase 1 (ADARB1) | |
| Regulated Target | Farnesyltransferase, CAAX box, subunit alpha (FNTA) | |
| Crosstalk relationship | m6A → A-to-I | |
| Crosstalk ID: M6ACROT00202 | ||
| Epigenetic Regulator | Double-stranded RNA-specific editase 1 (ADARB1) | |
| Regulated Target | Nuclear receptor subfamily 1 group I member 2 (NR1I2) | |
| Crosstalk relationship | m6A → A-to-I | |
| Drug | Pregnenolone-16alpha-carbonitrile | |
m6A Target: Runt-related transcription factor 1 (RUNX1)
| In total 3 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT00419 | ||
| Epigenetic Regulator | Double-stranded RNA-specific editase 1 (ADARB1) | |
| Regulated Target | MicroRNA 21 (MIR21) | |
| Crosstalk relationship | A-to-I → m6A | |
| Crosstalk ID: M6ACROT00420 | ||
| Epigenetic Regulator | Interferon-inducible protein 4 (ADAR1) | |
| Regulated Target | MicroRNA 21 (MIR21) | |
| Crosstalk relationship | A-to-I → m6A | |
| Crosstalk ID: M6ACROT00514 | ||
| Epigenetic Regulator | Interferon-inducible protein 4 (ADAR1) | |
| Regulated Target | hsa-mir-18a | |
| Crosstalk relationship | A-to-I → m6A | |
m6A Target: X inactive specific transcript (XIST)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT00423 | ||
| Epigenetic Regulator | Double-stranded RNA-specific editase 1 (ADARB1) | |
| Regulated Target | MicroRNA 21 (MIR21) | |
| Crosstalk relationship | m6A → A-to-I | |
| Crosstalk ID: M6ACROT00424 | ||
| Epigenetic Regulator | Interferon-inducible protein 4 (ADAR1) | |
| Regulated Target | MicroRNA 21 (MIR21) | |
| Crosstalk relationship | m6A → A-to-I | |
m6A Target: Cellular tumor antigen p53 (TP53/p53)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT00433 | ||
| Epigenetic Regulator | Methyltransferase-like protein 1 (METTL1) | |
| Regulated Target | MicroRNA 125a (MIR125A) | |
| Crosstalk relationship | m7G → m6A | |
| Crosstalk ID: M6ACROT00438 | ||
| Epigenetic Regulator | Double-stranded RNA-specific editase 1 (ADARB1) | |
| Regulated Target | MicroRNA 214 (MIR214) | |
| Crosstalk relationship | A-to-I → m6A | |
m6A Target: Epidermal growth factor receptor (EGFR)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT00442 | ||
| Epigenetic Regulator | Double-stranded RNA-specific editase 1 (ADARB1) | |
| Regulated Target | MicroRNA 214 (MIR214) | |
| Crosstalk relationship | A-to-I → m6A | |
m6A Target: Pvt1 oncogene (PVT1)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT00451 | ||
| Epigenetic Regulator | Double-stranded RNA-specific editase 1 (ADARB1) | |
| Regulated Target | MicroRNA 214 (MIR214) | |
| Crosstalk relationship | m6A → A-to-I | |
m6A Target: Zinc finger protein SNAI1 (SNAI1)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT00485 | ||
| Epigenetic Regulator | Fat mass and obesity-associated protein (FTO) | |
| Regulated Target | hsa-mir-22 | |
| Crosstalk relationship | m6Am → m6A | |
m6A Target: Cyclic-AMP-dependent transcription factor ATF-3 (ATF3)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT00507 | ||
| Epigenetic Regulator | Double-stranded RNA-specific editase 1 (ADARB1) | |
| Regulated Target | MicroRNA 222 (MIR222) | |
| Crosstalk relationship | A-to-I → m6A | |
| Crosstalk ID: M6ACROT00508 | ||
| Epigenetic Regulator | Interferon-inducible protein 4 (ADAR1) | |
| Regulated Target | MicroRNA 222 (MIR222) | |
| Crosstalk relationship | A-to-I → m6A | |
m6A Target: Forkhead box protein O3 (FOXO3)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT00520 | ||
| Epigenetic Regulator | Double-stranded RNA-specific editase 1 (ADARB1) | |
| Regulated Target | MicroRNA 221 (MIR221) | |
| Crosstalk relationship | A-to-I → m6A | |
| Crosstalk ID: M6ACROT00555 | ||
| Epigenetic Regulator | Y-box-binding protein 1 (YBX1) | |
| Regulated Target | Growth arrest specific 5 (GAS5) | |
| Crosstalk relationship | m5C → m6A | |
m6A Target: Mothers against decapentaplegic homolog 4 (SMAD4)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT00550 | ||
| Epigenetic Regulator | Interferon-inducible protein 4 (ADAR1) | |
| Regulated Target | MicroRNA 210 (MIR210) | |
| Crosstalk relationship | A-to-I → m6A | |
DNA modification
m6A Target: Methylcytosine dioxygenase TET1 (TET1)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT02011 | ||
| Epigenetic Regulator | Methylcytosine dioxygenase TET1 (TET1) | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | m6A → DNA modification | |
| Crosstalk ID: M6ACROT02095 | ||
| Epigenetic Regulator | Methylcytosine dioxygenase TET1 (TET1) | |
| Regulated Target | Signal transducer and activator of transcription 3 (STAT3) | |
| Crosstalk relationship | m6A → DNA modification | |
| Disease | Inflammatory pain | |
m6A Target: Semaphorin-3F (SEMA3F)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT02018 | ||
| Epigenetic Regulator | DNA (cytosine-5)-methyltransferase 1 (DNMT1) | |
| Regulated Target | Semaphorin-3F (SEMA3F) | |
| Crosstalk relationship | m6A → DNA modification | |
| Disease | Prostate cancer | |
| Drug | Docetaxel | |
m6A Target: Peroxisome proliferator-activated receptor alpha (PPARalpha/PPARA)
| In total 3 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT02022 | ||
| Epigenetic Regulator | DNA (cytosine-5)-methyltransferase 1 (DNMT1) | |
| Regulated Target | FTO alpha-ketoglutarate dependent dioxygenase (FTO) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Renal ischemia-reperfusion injury | |
| Drug | 5-azacytidine | |
| Crosstalk ID: M6ACROT02023 | ||
| Epigenetic Regulator | Cysteine methyltransferase DNMT3A (DNMT3A) | |
| Regulated Target | FTO alpha-ketoglutarate dependent dioxygenase (FTO) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Renal ischemia-reperfusion injury | |
| Drug | 5-azacytidine | |
| Crosstalk ID: M6ACROT02024 | ||
| Epigenetic Regulator | DNA (cytosine-5)-methyltransferase 3B (DNMT3B) | |
| Regulated Target | FTO alpha-ketoglutarate dependent dioxygenase (FTO) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Renal ischemia-reperfusion injury | |
| Drug | 5-azacytidine | |
m6A Target: DNA (cytosine-5)-methyltransferase 3B (DNMT3B)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT02026 | ||
| Epigenetic Regulator | DNA (cytosine-5)-methyltransferase 3B (DNMT3B) | |
| Regulated Target | Transcription factor E4F1 (E4F1) | |
| Crosstalk relationship | m6A → DNA modification | |
| Disease | Intervertebral disc degeneration | |
m6A Target: Transcription factor SOX-2 (SOX2)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT02037 | ||
| Epigenetic Regulator | DNA (cytosine-5)-methyltransferase 1 (DNMT1) | |
| Regulated Target | RNA demethylase ALKBH5 (ALKBH5) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Lung cancer | |
m6A Target: Mothers against decapentaplegic homolog 7 (SMAD7)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT02038 | ||
| Epigenetic Regulator | DNA (cytosine-5)-methyltransferase 1 (DNMT1) | |
| Regulated Target | RNA demethylase ALKBH5 (ALKBH5) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Lung cancer | |
m6A Target: Myc proto-oncogene protein (MYC)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT02039 | ||
| Epigenetic Regulator | DNA (cytosine-5)-methyltransferase 1 (DNMT1) | |
| Regulated Target | RNA demethylase ALKBH5 (ALKBH5) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Lung cancer | |
m6A Target: Cysteine methyltransferase DNMT3A (DNMT3A)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT02071 | ||
| Epigenetic Regulator | Cysteine methyltransferase DNMT3A (DNMT3A) | |
| Regulated Target | Dachshund family transcription factor 1 (DACH1) | |
| Crosstalk relationship | m6A → DNA modification | |
| Disease | Chronic obstructive pulmonary disease | |
m6A Target: 6-phosphogluconate dehydrogenase, decarboxylating (6PGD/PGD)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT02136 | ||
| Epigenetic Regulator | DNA (cytosine-5)-methyltransferase 1 (DNMT1) | |
| Regulated Target | RNA demethylase ALKBH5 (ALKBH5) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Lung cancer | |
m6A Target: Protein FAM83D (FAM83D)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT02137 | ||
| Epigenetic Regulator | DNA (cytosine-5)-methyltransferase 1 (DNMT1) | |
| Regulated Target | RNA demethylase ALKBH5 (ALKBH5) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Lung cancer | |
m6A Target: Transforming growth factor beta-1 proprotein (TGFB1)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT02138 | ||
| Epigenetic Regulator | DNA (cytosine-5)-methyltransferase 1 (DNMT1) | |
| Regulated Target | RNA demethylase ALKBH5 (ALKBH5) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Lung cancer | |
m6A Target: Mothers against decapentaplegic homolog 2 (SMAD2)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT02139 | ||
| Epigenetic Regulator | DNA (cytosine-5)-methyltransferase 1 (DNMT1) | |
| Regulated Target | RNA demethylase ALKBH5 (ALKBH5) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Lung cancer | |
m6A Target: Mothers against decapentaplegic homolog 3 (SMAD3)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT02140 | ||
| Epigenetic Regulator | DNA (cytosine-5)-methyltransferase 1 (DNMT1) | |
| Regulated Target | RNA demethylase ALKBH5 (ALKBH5) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Lung cancer | |
Histone modification
m6A Target: DNA (cytosine-5)-methyltransferase 3B (DNMT3B)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03006 | ||
| Epigenetic Regulator | Lysine-specific demethylase 4A (KDM4A) | |
| Regulated Target | Histone H3 lysine 9 trimethylation (H3K9me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Intervertebral disc degeneration | |
m6A Target: Lysine-specific demethylase 6B (KDM6B)
| In total 3 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03014 | ||
| Epigenetic Regulator | Lysine-specific demethylase 6B (KDM6B) | |
| Regulated Target | Histone H3 lysine 27 trimethylation (H3K27me3) | |
| Crosstalk relationship | m6A → Histone modification | |
| Disease | Inflammatory response | |
| Crosstalk ID: M6ACROT03240 | ||
| Epigenetic Regulator | Lysine-specific demethylase 6B (KDM6B) | |
| Regulated Target | Histone H3 lysine 27 trimethylation (H3K27me3) | |
| Crosstalk relationship | m6A → Histone modification | |
| Crosstalk ID: M6ACROT05995 | ||
| Epigenetic Regulator | Lysine-specific demethylase 6B (KDM6B) | |
| Regulated Target | Histone H3 lysine 27 trimethylation (H3K27me3) | |
| Crosstalk relationship | m6A → Histone modification | |
| Disease | Inflammatory response | |
m6A Target: Transcription factor SOX-4 (SOX4)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03037 | ||
| Epigenetic Regulator | Lysine-specific demethylase 5C (KDM5C) | |
| Regulated Target | Histone H3 lysine 4 trimethylation (H3K4me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
m6A Target: Period circadian protein homolog 1 (PER1)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03047 | ||
| Epigenetic Regulator | Lactate dehydrogenase A (LDHA) | |
| Regulated Target | Histone H3 lysine 18 lactylation (H3K18la) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Melanoma of uvea | |
| Crosstalk ID: M6ACROT03048 | ||
| Epigenetic Regulator | L-lactate dehydrogenase B chain (LDHB) | |
| Regulated Target | Histone H3 lysine 18 lactylation (H3K18la) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Melanoma of uvea | |
m6A Target: Cellular tumor antigen p53 (TP53/p53)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03049 | ||
| Epigenetic Regulator | Lactate dehydrogenase A (LDHA) | |
| Regulated Target | Histone H3 lysine 18 lactylation (H3K18la) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Melanoma of uvea | |
| Crosstalk ID: M6ACROT03050 | ||
| Epigenetic Regulator | L-lactate dehydrogenase B chain (LDHB) | |
| Regulated Target | Histone H3 lysine 18 lactylation (H3K18la) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Melanoma of uvea | |
m6A Target: MOB kinase activator 3B (MOB3B)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03073 | ||
| Epigenetic Regulator | Lysine-specific demethylase 5A (KDM5A) | |
| Regulated Target | Histone H3 lysine 4 trimethylation (H3K4me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Prostate cancer | |
m6A Target: Histone-lysine N-methyltransferase EZH2 (EZH2)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03082 | ||
| Epigenetic Regulator | Histone-lysine N-methyltransferase EZH2 (EZH2) | |
| Regulated Target | Histone H3 lysine 27 trimethylation (H3K27me3) | |
| Crosstalk relationship | m6A → Histone modification | |
| Disease | Endometriosis | |
m6A Target: Histone deacetylase 4 (HDAC4)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03083 | ||
| Epigenetic Regulator | Histone deacetylase 4 (HDAC4) | |
| Regulated Target | Hypoxia-inducible factor 1-alpha (HIF-1-Alpha/HIF1A) | |
| Crosstalk relationship | m6A → Histone modification | |
| Disease | Pancreatic cancer | |
m6A Target: Potassium channel subfamily K member 6 (KCNK6)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03085 | ||
| Epigenetic Regulator | Histone acetyltransferase p300 (P300) | |
| Regulated Target | Histone H3 lysine 18 lactylation (H3K18la) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
m6A Target: Cyclin-dependent kinase 2 (CDK2)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03087 | ||
| Epigenetic Regulator | Histone acetyltransferase p300 (P300) | |
| Regulated Target | Histone H3 lysine 18 lactylation (H3K18la) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Diabetic retinopathy | |
| Drug | FB23-2 | |
m6A Target: Histone acetyltransferase KAT2A (KAT2A)
| In total 4 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03089 | ||
| Epigenetic Regulator | Histone acetyltransferase KAT2A (KAT2A) | |
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | m6A → Histone modification | |
| Disease | Diabetic cardiomyopathy | |
| Crosstalk ID: M6ACROT03091 | ||
| Epigenetic Regulator | Histone acetyltransferase KAT2A (KAT2A) | |
| Regulated Target | Histone H3 lysine 9 acetylation (H3K9Ac) | |
| Crosstalk relationship | m6A → Histone modification | |
| Disease | Diabetic cardiomyopathy | |
| Crosstalk ID: M6ACROT05845 | ||
| Epigenetic Regulator | Histone acetyltransferase KAT2A (KAT2A) | |
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | m6A → Histone modification | |
| Disease | Diabetic cardiomyopathy | |
| Crosstalk ID: M6ACROT05847 | ||
| Epigenetic Regulator | Histone acetyltransferase KAT2A (KAT2A) | |
| Regulated Target | Histone H3 lysine 9 acetylation (H3K9ac) | |
| Crosstalk relationship | m6A → Histone modification | |
| Disease | Diabetic cardiomyopathy | |
m6A Target: long intergenic non-protein coding RNA 115 (LINC00115)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03105 | ||
| Epigenetic Regulator | Histone-lysine N-methyltransferase SETDB1 (SETDB1) | |
| Regulated Target | Polo like kinase 3 (PLK3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Triple-negative breast cancer | |
| Drug | Acetaminophen | |
m6A Target: NACHT, LRR and PYD domains-containing protein 3 (NLRP3)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03108 | ||
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Lung cancer | |
| Drug | Gefitinib | |
| Crosstalk ID: M6ACROT03110 | ||
| Regulated Target | Histone H3 lysine 4 monomethylation (H3K4me1) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Lung cancer | |
| Drug | Gefitinib | |
m6A Target: Serine/threonine-protein kinase PAK 6 (PAK5)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03118 | ||
| Epigenetic Regulator | Histone acetyltransferase p300 (P300) | |
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Cervical cancer | |
| Crosstalk ID: M6ACROT03120 | ||
| Epigenetic Regulator | WD repeat-containing protein 5 (WDR5) | |
| Regulated Target | Histone H3 lysine 4 trimethylation (H3K4me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Cervical cancer | |
m6A Target: ETS translocation variant 5 (ETV5)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03122 | ||
| Regulated Target | Histone H3 lysine 4 trimethylation (H3K4me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Liver cancer | |
| Crosstalk ID: M6ACROT03123 | ||
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Liver cancer | |
m6A Target: 5-hydroxytryptamine receptor 3A (HTR3A)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03130 | ||
| Epigenetic Regulator | Histone deacetylase 11 (HDAC11) | |
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Neuropathic pain | |
m6A Target: Transcriptional coactivator YAP1 (YAP1)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03140 | ||
| Epigenetic Regulator | Lysine-specific histone demethylase 1A (KDM1A) | |
| Regulated Target | Histone H3 lysine 4 dimethylation (H3K4me2) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Triple-negative breast cancer | |
m6A Target: Cysteine protease ATG4A (ATG4A)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03161 | ||
| Epigenetic Regulator | Histone-lysine N-methyltransferase 2A (KMT2A) | |
| Regulated Target | Histone H3 lysine 4 trimethylation (H3K4me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Intervertebral disc degeneration | |
m6A Target: Circ_SLC9A6
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03164 | ||
| Epigenetic Regulator | Histone acetyltransferase KAT8 (KAT8) | |
| Regulated Target | Histone H4 lysine 16 acetylation (H4K16ac) | |
| Crosstalk relationship | m6A → Histone modification | |
| Disease | Nonalcoholic fatty liver disease | |
m6A Target: Thioredoxin domain-containing protein 5 (TXNDC5)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03171 | ||
| Epigenetic Regulator | Histone acetyltransferase p300 (P300) | |
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Cervical cancer | |
| Crosstalk ID: M6ACROT03172 | ||
| Epigenetic Regulator | WD repeat-containing protein 5 (WDR5) | |
| Regulated Target | Histone H3 lysine 4 trimethylation (H3K4me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Cervical cancer | |
m6A Target: Methylcytosine dioxygenase TET1 (TET1)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03184 | ||
| Epigenetic Regulator | Methylcytosine dioxygenase TET1 (TET1) | |
| Regulated Target | Histone H3 lysine 4 trimethylation (H3K4me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Crosstalk ID: M6ACROT03185 | ||
| Epigenetic Regulator | Methylcytosine dioxygenase TET1 (TET1) | |
| Regulated Target | Histone H3 lysine 27 trimethylation (H3K27me3) | |
| Crosstalk relationship | Histone modification → m6A | |
m6A Target: testis expressed 41 (TEX41)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03197 | ||
| Epigenetic Regulator | Polycomb protein SUZ12 (SUZ12) | |
| Regulated Target | Histone H3 lysine 27 trimethylation (H3K27me3) | |
| Crosstalk relationship | m6A → Histone modification | |
| Disease | Renal cell carcinoma | |
m6A Target: Taurine up-regulated 1 protein (TUG1)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03203 | ||
| Epigenetic Regulator | Histone-lysine N-methyltransferase EHMT2 (EHMT2) | |
| Regulated Target | Histone H3 lysine 9 dimethylation (H3K9me2) | |
| Crosstalk relationship | m6A → Histone modification | |
| Disease | Acute myeloid leukaemia | |
| Drug | Adriamycin | |
m6A Target: Integrin beta-1 (ITGB1)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03215 | ||
| Epigenetic Regulator | Histone acetyltransferase type B catalytic subunit (HAT1) | |
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Diabetic retinopathy | |
m6A Target: Tripartite motif-containing protein 72 (TRIM72)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03217 | ||
| Epigenetic Regulator | Histone-lysine N-methyltransferase 2A (KMT2A) | |
| Regulated Target | Histone H3 lysine 4 trimethylation (H3K4me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Preeclampsia | |
| Drug | Actinomycin D | |
m6A Target: CREB-binding protein (CREBBP)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03219 | ||
| Epigenetic Regulator | CREB-binding protein (CREBBP) | |
| Crosstalk relationship | m6A → Histone modification | |
| Drug | BHBA | |
m6A Target: Mutated in multiple advanced cancers 1 (PTEN)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03221 | ||
| Epigenetic Regulator | Histone-lysine N-methyltransferase EZH2 (EZH2) | |
| Regulated Target | Histone H3 lysine 27 trimethylation (H3K27me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Brain cancer | |
m6A Target: Histone-lysine N-methyltransferase EHMT2 (G9a)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03232 | ||
| Epigenetic Regulator | Histone-lysine N-methyltransferase EHMT2 (EHMT2) | |
| Regulated Target | Histone H3 lysine 9 dimethylation (H3K9me2) | |
| Crosstalk relationship | m6A → Histone modification | |
| Disease | Neuropathic pain | |
m6A Target: Forkhead box protein O3 (FOXO3)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03238 | ||
| Epigenetic Regulator | CREB-binding protein (CREBBP) | |
| Crosstalk relationship | m6A → Histone modification | |
| Disease | Intervertebral disc degeneration | |
m6A Target: X inactive specific transcript (XIST)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03289 | ||
| Epigenetic Regulator | Lysine-specific demethylase 5C (KDM5C) | |
| Regulated Target | Histone H3 lysine 4 trimethylation (H3K4me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
| Crosstalk ID: M6ACROT03333 | ||
| Epigenetic Regulator | Histone acetyltransferase p300 (P300) | |
| Regulated Target | Histone H3 lysine 18 lactylation (H3K18la) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
m6A Target: Transcription factor ISGF-3 components p91/p84 (Stat1)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03290 | ||
| Epigenetic Regulator | Lysine-specific demethylase 5C (KDM5C) | |
| Regulated Target | Histone H3 lysine 4 trimethylation (H3K4me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
| Crosstalk ID: M6ACROT03346 | ||
| Epigenetic Regulator | Histone acetyltransferase p300 (P300) | |
| Regulated Target | Histone H3 lysine 18 lactylation (H3K18la) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
m6A Target: Interferon regulatory factor 1 (Irf1)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03291 | ||
| Epigenetic Regulator | Lysine-specific demethylase 5C (KDM5C) | |
| Regulated Target | Histone H3 lysine 4 trimethylation (H3K4me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
| Crosstalk ID: M6ACROT03347 | ||
| Epigenetic Regulator | Histone acetyltransferase p300 (P300) | |
| Regulated Target | Histone H3 lysine 18 lactylation (H3K18la) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
m6A Target: Arrestin domain-containing protein 4 (ARRDC4)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03292 | ||
| Epigenetic Regulator | Lysine-specific demethylase 5C (KDM5C) | |
| Regulated Target | Histone H3 lysine 4 trimethylation (H3K4me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
| Crosstalk ID: M6ACROT03348 | ||
| Epigenetic Regulator | Histone acetyltransferase p300 (P300) | |
| Regulated Target | Histone H3 lysine 18 lactylation (H3K18la) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
m6A Target: Thrombospondin-1 (THBS1)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03327 | ||
| Epigenetic Regulator | Lysine-specific demethylase 5A (KDM5A) | |
| Regulated Target | Histone H3 lysine 4 trimethylation (H3K4me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Prostate cancer | |
m6A Target: RAC-alpha serine/threonine-protein kinase (AKT1)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03328 | ||
| Epigenetic Regulator | Lysine-specific demethylase 5A (KDM5A) | |
| Regulated Target | Histone H3 lysine 4 trimethylation (H3K4me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Prostate cancer | |
m6A Target: Phospholysine phosphohistidine inorganic pyrophosphate phosphatase (LHPP)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03329 | ||
| Epigenetic Regulator | Lysine-specific demethylase 5A (KDM5A) | |
| Regulated Target | Histone H3 lysine 4 trimethylation (H3K4me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Prostate cancer | |
m6A Target: Homeobox protein Nkx-3.1 (NKX3-1)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03330 | ||
| Epigenetic Regulator | Lysine-specific demethylase 5A (KDM5A) | |
| Regulated Target | Histone H3 lysine 4 trimethylation (H3K4me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Prostate cancer | |
m6A Target: Ubiquitin carboxyl-terminal hydrolase 4 (USP4)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03331 | ||
| Epigenetic Regulator | Lysine-specific demethylase 5A (KDM5A) | |
| Regulated Target | Histone H3 lysine 4 trimethylation (H3K4me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Prostate cancer | |
m6A Target: ELAV-like protein 1 (HuR/ELAVL1)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03332 | ||
| Epigenetic Regulator | Lysine-specific demethylase 5A (KDM5A) | |
| Regulated Target | Histone H3 lysine 4 trimethylation (H3K4me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Prostate cancer | |
m6A Target: Cyclic AMP-dependent transcription factor ATF-4 (ATF4)
| In total 6 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03334 | ||
| Epigenetic Regulator | Histone acetyltransferase p300 (P300) | |
| Regulated Target | Histone H3 lysine 18 lactylation (H3K18la) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
| Drug | Rapamycin | |
| Crosstalk ID: M6ACROT03335 | ||
| Epigenetic Regulator | Histone acetyltransferase p300 (P300) | |
| Regulated Target | Histone H3 lysine 18 lactylation (H3K18la) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
| Drug | GLS-IN-968 | |
| Crosstalk ID: M6ACROT03336 | ||
| Epigenetic Regulator | Histone acetyltransferase p300 (P300) | |
| Regulated Target | Histone H3 lysine 18 lactylation (H3K18la) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
| Drug | CB-839 | |
| Crosstalk ID: M6ACROT03337 | ||
| Epigenetic Regulator | Histone acetyltransferase p300 (P300) | |
| Regulated Target | Histone H3 lysine 18 lactylation (H3K18la) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
| Drug | Meclofenamate sodium | |
| Crosstalk ID: M6ACROT03338 | ||
| Epigenetic Regulator | Histone acetyltransferase p300 (P300) | |
| Regulated Target | Histone H3 lysine 18 lactylation (H3K18la) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
| Drug | Chloroquine | |
| Crosstalk ID: M6ACROT03339 | ||
| Epigenetic Regulator | Histone acetyltransferase p300 (P300) | |
| Regulated Target | Histone H3 lysine 18 lactylation (H3K18la) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
| Drug | Asparagine inhibitor | |
m6A Target: Serine/threonine-protein kinase mTOR (MTOR)
| In total 6 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03340 | ||
| Epigenetic Regulator | Histone acetyltransferase p300 (P300) | |
| Regulated Target | Histone H3 lysine 18 lactylation (H3K18la) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
| Drug | Rapamycin | |
| Crosstalk ID: M6ACROT03341 | ||
| Epigenetic Regulator | Histone acetyltransferase p300 (P300) | |
| Regulated Target | Histone H3 lysine 18 lactylation (H3K18la) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
| Drug | GLS-IN-968 | |
| Crosstalk ID: M6ACROT03342 | ||
| Epigenetic Regulator | Histone acetyltransferase p300 (P300) | |
| Regulated Target | Histone H3 lysine 18 lactylation (H3K18la) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
| Drug | CB-839 | |
| Crosstalk ID: M6ACROT03343 | ||
| Epigenetic Regulator | Histone acetyltransferase p300 (P300) | |
| Regulated Target | Histone H3 lysine 18 lactylation (H3K18la) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
| Drug | Meclofenamate sodium | |
| Crosstalk ID: M6ACROT03344 | ||
| Epigenetic Regulator | Histone acetyltransferase p300 (P300) | |
| Regulated Target | Histone H3 lysine 18 lactylation (H3K18la) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
| Drug | Chloroquine | |
| Crosstalk ID: M6ACROT03345 | ||
| Epigenetic Regulator | Histone acetyltransferase p300 (P300) | |
| Regulated Target | Histone H3 lysine 18 lactylation (H3K18la) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
| Drug | Asparagine inhibitor | |
m6A Target: Protein yippee-like 5 (YPEL5)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03349 | ||
| Epigenetic Regulator | Histone acetyltransferase p300 (P300) | |
| Regulated Target | Histone H3 lysine 18 lactylation (H3K18la) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
m6A Target: Circ_AFF2
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03350 | ||
| Epigenetic Regulator | Histone acetyltransferase p300 (P300) | |
| Regulated Target | Histone H3 lysine 18 lactylation (H3K18la) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
m6A Target: Apoptosis regulatory protein Siva (SIVA1)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03351 | ||
| Epigenetic Regulator | Histone acetyltransferase p300 (P300) | |
| Regulated Target | Histone H3 lysine 18 lactylation (H3K18la) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
| Drug | Fluorouracil | |
m6A Target: Ras-related protein Rab-5A (RAB5A)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03352 | ||
| Epigenetic Regulator | Histone acetyltransferase p300 (P300) | |
| Regulated Target | Histone H3 lysine 18 lactylation (H3K18la) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
m6A Target: Growth arrest specific 5 (GAS5)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03405 | ||
| Epigenetic Regulator | Histone acetyltransferase p300 (P300) | |
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Cervical cancer | |
| Crosstalk ID: M6ACROT03408 | ||
| Epigenetic Regulator | WD repeat-containing protein 5 (WDR5) | |
| Regulated Target | Histone H3 lysine 4 trimethylation (H3K4me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Cervical cancer | |
m6A Target: Epidermal growth factor receptor (EGFR)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03409 | ||
| Regulated Target | Histone H3 lysine 4 trimethylation (H3K4me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Liver cancer | |
| Crosstalk ID: M6ACROT03420 | ||
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Liver cancer | |
m6A Target: POU domain, class 5, transcription factor 1 (POU5F1)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03410 | ||
| Regulated Target | Histone H3 lysine 4 trimethylation (H3K4me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Liver cancer | |
| Crosstalk ID: M6ACROT03421 | ||
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Liver cancer | |
m6A Target: Interleukin-11 (IL11)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03411 | ||
| Regulated Target | Histone H3 lysine 4 trimethylation (H3K4me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Liver cancer | |
| Drug | PT2385* | |
| Crosstalk ID: M6ACROT03422 | ||
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Liver cancer | |
| Drug | PT2385* | |
m6A Target: Glia-derived nexin (SERPINE2)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03412 | ||
| Regulated Target | Histone H3 lysine 4 trimethylation (H3K4me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Liver cancer | |
| Drug | PT2385* | |
| Crosstalk ID: M6ACROT03423 | ||
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Liver cancer | |
| Drug | PT2385* | |
m6A Target: Splicing factor 3A subunit 3 (SF3A3)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03413 | ||
| Regulated Target | Histone H3 lysine 4 trimethylation (H3K4me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Liver cancer | |
| Crosstalk ID: M6ACROT03424 | ||
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Liver cancer | |
m6A Target: Tyrosine-protein kinase Fyn (FYN)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03414 | ||
| Regulated Target | Histone H3 lysine 4 trimethylation (H3K4me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Liver cancer | |
| Crosstalk ID: M6ACROT03425 | ||
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Liver cancer | |
m6A Target: Proliferation-associated protein 2G4 (PA2G4)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03415 | ||
| Regulated Target | Histone H3 lysine 4 trimethylation (H3K4me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Liver cancer | |
| Crosstalk ID: M6ACROT03426 | ||
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Liver cancer | |
m6A Target: Long intergenic non-protein coding RNA 1273 (LINC01273)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03416 | ||
| Regulated Target | Histone H3 lysine 4 trimethylation (H3K4me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Liver cancer | |
| Drug | Sorafenib | |
| Crosstalk ID: M6ACROT03427 | ||
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Liver cancer | |
| Drug | Sorafenib | |
m6A Target: Glucose transporter type 1 (GLUT1)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03417 | ||
| Regulated Target | Histone H3 lysine 4 trimethylation (H3K4me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Liver cancer | |
| Crosstalk ID: M6ACROT03428 | ||
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Liver cancer | |
m6A Target: Pyruvate kinase PKM (PKM2/PKM)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03418 | ||
| Regulated Target | Histone H3 lysine 4 trimethylation (H3K4me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Liver cancer | |
| Crosstalk ID: M6ACROT03429 | ||
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Liver cancer | |
m6A Target: Myc proto-oncogene protein (MYC)
| In total 3 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03419 | ||
| Regulated Target | Histone H3 lysine 4 trimethylation (H3K4me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Liver cancer | |
| Crosstalk ID: M6ACROT03430 | ||
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Liver cancer | |
| Crosstalk ID: M6ACROT03539 | ||
| Epigenetic Regulator | Histone-lysine N-methyltransferase EZH2 (EZH2) | |
| Regulated Target | Histone H3 lysine 27 trimethylation (H3K27me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Brain cancer | |
m6A Target: UBX domain-containing protein 1 (UBXN1)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03540 | ||
| Epigenetic Regulator | Histone-lysine N-methyltransferase EZH2 (EZH2) | |
| Regulated Target | Histone H3 lysine 27 trimethylation (H3K27me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Brain cancer | |
m6A Target: Nuclear factor NF-kappa-B p105 subunit (NF-Kappa-B/NFKB1)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03541 | ||
| Epigenetic Regulator | Histone-lysine N-methyltransferase EZH2 (EZH2) | |
| Regulated Target | Histone H3 lysine 27 trimethylation (H3K27me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Brain cancer | |
m6A Target: Long intergenic non-protein coding RNA 657 (NORAD)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05974 | ||
| Epigenetic Regulator | Lysine-specific demethylase 5A (KDM5A) | |
| Regulated Target | Histone H3 lysine 4 trimethylation (H3K4me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Intervertebral disc degeneration | |
Non-coding RNA
m6A Target: Epidermal growth factor receptor (EGFR)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05037 | ||
| Epigenetic Regulator | MicroRNA 145 (MIR145) | |
| Regulated Target | YTH domain-containing family protein 2 (YTHDF2) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Liver cancer | |
m6A Target: POU domain, class 5, transcription factor 1 (POU5F1)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05038 | ||
| Epigenetic Regulator | MicroRNA 145 (MIR145) | |
| Regulated Target | YTH domain-containing family protein 2 (YTHDF2) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Liver cancer | |
m6A Target: Interleukin-11 (IL11)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05039 | ||
| Epigenetic Regulator | MicroRNA 145 (MIR145) | |
| Regulated Target | YTH domain-containing family protein 2 (YTHDF2) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Liver cancer | |
m6A Target: Glia-derived nexin (SERPINE2)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05040 | ||
| Epigenetic Regulator | MicroRNA 145 (MIR145) | |
| Regulated Target | YTH domain-containing family protein 2 (YTHDF2) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Liver cancer | |
m6A Target: Splicing factor 3A subunit 3 (SF3A3)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05041 | ||
| Epigenetic Regulator | MicroRNA 145 (MIR145) | |
| Regulated Target | YTH domain-containing family protein 2 (YTHDF2) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Liver cancer | |
m6A Target: Tyrosine-protein kinase Fyn (FYN)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05042 | ||
| Epigenetic Regulator | MicroRNA 145 (MIR145) | |
| Regulated Target | YTH domain-containing family protein 2 (YTHDF2) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Liver cancer | |
m6A Target: Proliferation-associated protein 2G4 (PA2G4)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05043 | ||
| Epigenetic Regulator | MicroRNA 145 (MIR145) | |
| Regulated Target | YTH domain-containing family protein 2 (YTHDF2) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Liver cancer | |
m6A Target: Long intergenic non-protein coding RNA 1273 (LINC01273)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05044 | ||
| Epigenetic Regulator | MicroRNA 145 (MIR145) | |
| Regulated Target | YTH domain-containing family protein 2 (YTHDF2) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Liver cancer | |
| Crosstalk ID: M6ACROT05612 | ||
| Epigenetic Regulator | Long intergenic non-protein coding RNA 1273 (LINC01273) | |
| Regulated Target | hsa-miR-600 | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Liver cancer | |
| Drug | Sorafenib | |
m6A Target: Glucose transporter type 1 (SLC2A1)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05045 | ||
| Epigenetic Regulator | MicroRNA 145 (MIR145) | |
| Regulated Target | YTH domain-containing family protein 2 (YTHDF2) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Liver cancer | |
| Crosstalk ID: M6ACROT05317 | ||
| Epigenetic Regulator | FTO intronic transcript 1 (FTO-IT1) | |
| Regulated Target | FTO alpha-ketoglutarate dependent dioxygenase (FTO) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Liver cancer | |
m6A Target: Pyruvate kinase PKM (PKM2/PKM)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05046 | ||
| Epigenetic Regulator | MicroRNA 145 (MIR145) | |
| Regulated Target | YTH domain-containing family protein 2 (YTHDF2) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Liver cancer | |
| Crosstalk ID: M6ACROT05316 | ||
| Epigenetic Regulator | FTO intronic transcript 1 (FTO-IT1) | |
| Regulated Target | FTO alpha-ketoglutarate dependent dioxygenase (FTO) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Liver cancer | |
m6A Target: Myc proto-oncogene protein (MYC)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05047 | ||
| Epigenetic Regulator | MicroRNA 145 (MIR145) | |
| Regulated Target | YTH domain-containing family protein 2 (YTHDF2) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Liver cancer | |
m6A Target: Growth arrest specific 5 (GAS5)
| In total 3 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05050 | ||
| Epigenetic Regulator | GAS5 antisense RNA 1 (GAS5-AS1) | |
| Regulated Target | RNA demethylase ALKBH5 (ALKBH5) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Cervical cancer | |
| Crosstalk ID: M6ACROT05398 | ||
| Epigenetic Regulator | Growth arrest specific 5 (GAS5) | |
| Regulated Target | GAS5 antisense RNA 1 (GAS5-AS1) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Cervical cancer | |
| Crosstalk ID: M6ACROT05718 | ||
| Epigenetic Regulator | Growth arrest specific 5 (GAS5) | |
| Regulated Target | Dynamin-1-like protein (DRP1) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Vascular disorders of the liver | |
m6A Target: Fibulin-1 (FBLN1)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05061 | ||
| Epigenetic Regulator | hsa-miR-615-3p | |
| Regulated Target | YTH domain-containing family protein 2 (YTHDF2) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Maxillofacial bone defect is a critical obstacle for maxillofacial tumors and periodontal diseases. | |
m6A Target: Transcription factor E2F6 (E2F6)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05068 | ||
| Epigenetic Regulator | Read-through Circular RNA E2F | |
| Regulated Target | Insulin like growth factor 2 mRNA binding protein 2 (IGF2BP2) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Liver cancer | |
m6A Target: Transcription factor E2F3 (E2F3)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05069 | ||
| Epigenetic Regulator | Read-through Circular RNA E2F | |
| Regulated Target | Insulin like growth factor 2 mRNA binding protein 2 (IGF2BP2) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Liver cancer | |
m6A Target: Ubiquitin carboxyl-terminal hydrolase 38 (USP38)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05175 | ||
| Epigenetic Regulator | hsa-miR-3165 | |
| Regulated Target | Methyltransferase-like protein 14 (METTL14) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Bladder cancer | |
m6A Target: Tyrosine-protein kinase JAK1 (JAK1)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05209 | ||
| Epigenetic Regulator | Long intergenic non-protein coding RNA 659 (LINC00659) | |
| Regulated Target | RNA demethylase ALKBH5 (ALKBH5) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Gastric cancer | |
m6A Target: Metalloreductase STEAP3 (STEAP3)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05211 | ||
| Epigenetic Regulator | STEAP3 antisense RNA 1 (STEAP3-AS1) | |
| Regulated Target | YTH domain-containing family protein 2 (YTHDF2) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Colorectal cancer | |
m6A Target: NACHT, LRR and PYD domains-containing protein 3 (NLRP3)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05224 | ||
| Epigenetic Regulator | MIR570 host gene (MIR570HG) | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Lung cancer | |
| Drug | Gefitinib | |
m6A Target: Glycogen synthase kinase-3 beta (GSK3Beta/GSK3B)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05278 | ||
| Epigenetic Regulator | hsa-miR-6125 | |
| Regulated Target | YTH domain-containing family protein 2 (YTHDF2) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Colorectal cancer | |
m6A Target: Heme oxygenase 1 (HMOX1)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05280 | ||
| Epigenetic Regulator | hsa-miR-340-3p | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Diseases of the female genital system | |
m6A Target: Mutated in multiple advanced cancers 1 (PTEN)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05288 | ||
| Epigenetic Regulator | Long intergenic non-protein coding RNA 470 (LINC00470) | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Gastric cancer | |
m6A Target: MOB kinase activator 3B (MOB3B)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05297 | ||
| Epigenetic Regulator | hsa-miR-495 | |
| Regulated Target | YTH domain-containing family protein 2 (YTHDF2) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Prostate cancer | |
m6A Target: Bcl-2-modifying factor (BMF)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05308 | ||
| Epigenetic Regulator | MicroRNA 145 (MIR145) | |
| Regulated Target | YTH domain-containing family protein 2 (YTHDF2) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Ovarian cancer | |
m6A Target: Cystathionine beta-synthase (CBS)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05350 | ||
| Epigenetic Regulator | CBS mRNA stabilizing lncRNA (CBSLR) | |
| Regulated Target | YTH domain-containing family protein 2 (YTHDF2) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Gastric cancer | |
m6A Target: Transcription factor SOX-9 (SOX9)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05371 | ||
| Epigenetic Regulator | Circ_FTO | |
| Regulated Target | Methyltransferase-like protein 14 (METTL14) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Rheumatoid arthritis | |
m6A Target: Carnitine O-palmitoyltransferase 1, liver isoform (CPT1A)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05373 | ||
| Epigenetic Regulator | Myocardial infarction associated transcript (MIAT) | |
| Regulated Target | YTH domain-containing family protein 2 (YTHDF2) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Heart failure | |
m6A Target: Long intergenic non-protein coding RNA 470 (LINC00470)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05401 | ||
| Epigenetic Regulator | Long intergenic non-protein coding RNA 470 (LINC00470) | |
| Regulated Target | Mutated in multiple advanced cancers 1 (PTEN) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Gastric cancer | |
m6A Target: Pvt1 oncogene (PVT1)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05418 | ||
| Epigenetic Regulator | Pvt1 oncogene (PVT1) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Osteosarcoma | |
| Crosstalk ID: M6ACROT05818 | ||
| Epigenetic Regulator | Pvt1 oncogene (PVT1) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Diabetic encephalopathy | |
m6A Target: X inactive specific transcript (XIST)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05420 | ||
| Epigenetic Regulator | X inactive specific transcript (XIST) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Colorectal cancer | |
m6A Target: Testis associated oncogenic lncRNA (THORLNC)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05442 | ||
| Epigenetic Regulator | Testis associated oncogenic lncRNA (THORLNC) | |
| Crosstalk relationship | m6A → ncRNA | |
m6A Target: hsa-mir-181b-1
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05467 | ||
| Epigenetic Regulator | hsa-mir-181b-1 | |
| Regulated Target | Transcriptional coactivator YAP1 (YAP1) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Osteosarcoma | |
m6A Target: FOXF1 adjacent non-coding developmental regulatory RNA (FENDRR)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05483 | ||
| Epigenetic Regulator | FOXF1 adjacent non-coding developmental regulatory RNA (FENDRR) | |
| Regulated Target | Transcription factor SOX-4 (SOX4) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Endometrial cancer | |
m6A Target: Deleted in lymphocytic leukemia 2 (DLEU2/LINC00022)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05518 | ||
| Epigenetic Regulator | Deleted in lymphocytic leukemia 2 (DLEU2/LINC00022) | |
| Regulated Target | Cyclin-dependent kinase inhibitor 1 (CDKN1A) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Esophageal Squamous Cell Carcinoma | |
m6A Target: Nuclear paraspeckle assembly transcript 1 (NEAT1)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05539 | ||
| Epigenetic Regulator | Nuclear paraspeckle assembly transcript 1 (NEAT1) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Renal cell carcinoma | |
m6A Target: KCNQ1 opposite strand/antisense transcript 1 (KCNQ1OT1)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05541 | ||
| Epigenetic Regulator | KCNQ1 opposite strand/antisense transcript 1 (KCNQ1OT1) | |
| Regulated Target | Homeobox A9 (HOXA9) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Laryngeal cancer | |
m6A Target: Circ_ASK1
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05585 | ||
| Epigenetic Regulator | Circ_ASK1 | |
| Regulated Target | Mitogen-activated protein kinase kinase kinase 5 (MAP3K5) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Lung cancer | |
| Drug | Gefitinib | |
m6A Target: hsa-miR-1915-3p
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05609 | ||
| Epigenetic Regulator | hsa-miR-1915-3p | |
| Regulated Target | Protein SET (SET) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Non-small cell lung cancer | |
m6A Target: LINC00902 (TUSC7)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05633 | ||
| Epigenetic Regulator | LINC00902 (TUSC7) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Lung cancer | |
| Drug | Erlotinib | |
m6A Target: Circ_RERE
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05642 | ||
| Epigenetic Regulator | Circ_RERE | |
| Regulated Target | hsa-miR-195-5p | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Osteoarthritis | |
| Drug | ICG-001 | |
m6A Target: Circ_IRF2
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05650 | ||
| Epigenetic Regulator | Circ_IRF2 | |
| Regulated Target | hsa-miR-29b-1-5p | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Hepatic fibrosis/cirrhosis | |
m6A Target: Cardiac mesoderm enhancer-associated non-coding RNA (CARMN)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05667 | ||
| Epigenetic Regulator | Cardiac mesoderm enhancer-associated non-coding RNA (CARMN) | |
| Regulated Target | hsa-miR-21-5p | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Cervical cancer | |
m6A Target: microRNA 126 (MIR126)
| In total 7 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05682 | ||
| Epigenetic Regulator | MicroRNA 126 (MIR126) | |
| Regulated Target | ADAM metallopeptidase domain 9 (ADAM9) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Acute myeloid leukaemia | |
| Crosstalk ID: M6ACROT05925 | ||
| Epigenetic Regulator | MicroRNA 126 (MIR126) | |
| Regulated Target | Scaffold protein ILK (ILK) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Acute myeloid leukaemia | |
| Crosstalk ID: M6ACROT05927 | ||
| Epigenetic Regulator | MicroRNA 126 (MIR126) | |
| Regulated Target | Cyclin-dependent kinase 3 (CDK3) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Acute myeloid leukaemia | |
| Crosstalk ID: M6ACROT05933 | ||
| Epigenetic Regulator | MicroRNA 126 (MIR126) | |
| Regulated Target | Target of Myb1 membrane trafficking protein (TOM1) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Acute myeloid leukaemia | |
| Crosstalk ID: M6ACROT05935 | ||
| Epigenetic Regulator | MicroRNA 126 (MIR126) | |
| Regulated Target | High affinity immunoglobulin gamma Fc receptor I (FCGR1A) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Acute myeloid leukaemia | |
| Crosstalk ID: M6ACROT05937 | ||
| Epigenetic Regulator | MicroRNA 126 (MIR126) | |
| Regulated Target | Ficolin-1 (FCN1) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Acute myeloid leukaemia | |
| Crosstalk ID: M6ACROT05939 | ||
| Epigenetic Regulator | MicroRNA 126 (MIR126) | |
| Regulated Target | Leucine-rich repeat and calponin homology domain-containing protein 4 (LRCH4) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Acute myeloid leukaemia | |
m6A Target: Circ_AFF2
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05683 | ||
| Epigenetic Regulator | Circ_AFF2 | |
| Regulated Target | Cullin associated and neddylation dissociated 1 (CAND1) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Colorectal cancer | |
m6A Target: Circ_FUT8
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05692 | ||
| Epigenetic Regulator | Circ_FUT8 | |
| Regulated Target | Fucosyltransferase 8 (FUT8) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Lung cancer | |
m6A Target: Circ_RNF13
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05707 | ||
| Epigenetic Regulator | Circ_RNF13 | |
| Regulated Target | C-X-C motif chemokine ligand 1 (CXCL1) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Cervical cancer | |
m6A Target: Circ_GPATCH2L
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05710 | ||
| Epigenetic Regulator | Circ_GPATCH2L | |
| Regulated Target | Tripartite motif containing 28 (TRIM28) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Spinal pain | |
m6A Target: microRNA 145 (MIR145)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05819 | ||
| Epigenetic Regulator | MicroRNA 145 (MIR145) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Epilepsy due to structural or metabolic conditions or diseases | |
m6A Target: Long intergenic non-protein coding RNA 657 (NORAD)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05970 | ||
| Epigenetic Regulator | Long intergenic non-protein coding RNA 657 (NORAD) | |
| Regulated Target | Pumilio homolog 1 (PUM1) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Intervertebral disc degeneration | |
| Crosstalk ID: M6ACROT05972 | ||
| Epigenetic Regulator | Long intergenic non-protein coding RNA 657 (NORAD) | |
| Regulated Target | Pumilio homolog 2 (PUM2) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Intervertebral disc degeneration | |
Xenobiotics Compound(s) Regulating the m6A Methylation Regulator
| Compound Name | Rapamycin | Approved |
|---|---|---|
| Synonyms |
Rapamune; Rapamycin (Sirolimus); AY-22989; Rapammune; sirolimusum; WY-090217; RAPA; Antibiotic AY 22989; AY 22989; UNII-W36ZG6FT64; CCRIS 9024; CHEBI:9168; SILA 9268A; W36ZG6FT64; HSDB 7284; C51H79NO13; NSC 226080; DE-109; NCGC00021305-05; DSSTox_CID_3582; DSSTox_RID_77091; DSSTox_GSID_23582; Cypher; Supralimus; Wy 090217; Perceiva; RAP; RPM; Rapamycin from Streptomyces hygroscopicus; SIIA 9268A; LCP-Siro; MS-R001; Rapamune (TN); Rapamycin (TN); Sirolimus (RAPAMUNE); Rapamycin C-7, analog 4; Sirolimus (USAN/INN); Sirolimus [USAN:BAN:INN]; Sirolimus, Rapamune,Rapamycin; Heptadecahydro-9,27-dihydroxy-3-[(1R)-2-[(1S,3R,4R)-4-hydroxy; 23,27-Epoxy-3H-pyrido(2,1-c)(1,4)oxaazacyclohentriacontine; 23,27-Epoxy-3H-pyrido[2,1-c][1,4]oxaazacyclohentriacontine; 23,27-epoxy-3H-pyrido[2,1-c][1,4]oxaazacyclohentriacontine-1,5,11,28,29; 3H-pyrido(2,1-c)(1,4)oxaazacyclohentriacontine-1,5,11,28,29(4H,6H,31H)-pentone; Sirolimus (MTOR inhibitor)
Click to Show/Hide
|
|
| External link | ||
| Description |
Rapamycin inhibited FTO activity, and directly targetedeIF4G1 transcripts and mediated their expression in an m6A-dependent manner inoral squamous cell carcinoma. After FTO silencing, YTHDF2 captured eIF4G1 transcripts containing m6A, resulting in mRNA degradation and decreased expression of eIF4G1 protein, thereby promoting autophagy and reducing tumor occurrence.
|
[72] |
| Compound Name | Linsitinib | Phase 3 |
| Synonyms |
Linsitinib; 867160-71-2; OSI-906; Linsitinib(OSI-906); OSI906; OSI 906; OSI-906AA; OSI-906 (Linsitinib); UNII-15A52GPT8T; Kinome_3532; ASP-7487; 3-[8-amino-1-(2-phenylquinolin-7-yl)imidazo[1,5-a]pyrazin-3-yl]-1-methylcyclobutan-1-ol; 15A52GPT8T; CHEMBL1091644; MMV676605; cis-3-[8-Amino-1-(2-phenyl-7-quinolinyl)imidazo[1,5-a]pyrazin-3-yl]-1-methylcyclobutanol; C26H23N5O; cis-3-(8-amino-1-(2-phenyl-7-quinolinyl)imidazo(1,5-a)pyrazin-3-yl)-1-methylcyclobutanol; Linsitinib [USAN:INN]; OSI906/Linsitinib/; Linsitinib; OSI-906
Click to Show/Hide
|
|
| External link | ||
| Description |
The m6A reader YTHDF2 stabilizedMYC mRNA specifically in cancer stem cells. Given the challenge of targeting MYC, YTHDF2 presents a therapeutic target to perturb MYC signaling in glioblastoma. The IGF1/IGF1R inhibitor linsitinib preferentially targeted YTHDF2-expressing cells, inhibiting GSC viability without affecting NSCs and impairing in vivoglioblastoma growth. YTHDF2 links RNA epitranscriptomic modifications and GSC growth, laying the foundation for the YTHDF2-MYC-IGFBP3 axis as a specific and novel therapeutic target in glioblastoma.
|
[11] |
| Compound Name | PT2385 | Phase 2 |
| Synonyms |
ONBSHRSJOPSEGS-INIZCTEOSA-N; PT-2385; UNII-6O16716DXP; 1672665-49-4; 6O16716DXP; SCHEMBL16555810; ZINC230453533; AKOS030526641; HY-12867; PT2385,1672665-49-4, PT 2385,PT-2385; Benzonitrile, 3-(((1S)-2,2-difluoro-2,3-dihydro-1-hydroxy-7-(methylsulfonyl)-1H-inden-4-yl)oxy)-5-fluoro-; 3-{[(1s)-2,2-Difluoro-1-Hydroxy-7-(Methylsulfonyl)-2,3-Dihydro-1h-Inden-4-Yl]oxy}-5-Fluorobenzonitrile; 3-(((1S)-2,2-Difluoro-1-hydroxy-7-methanesulfonyl-2,3-dihydro-1hinden-4-yl)oxy)-5-fluorobenzonitrile; 79A
Click to Show/Hide
|
|
| External link | ||
| Description |
YTHDF2 processed the decay of m6A-containinginterleukin 11 (IL11) and serpin family E member 2 (SERPINE2) mRNAs. YTHDF2 transcription succumbed to hypoxia-inducible factor-2-alpha (HIF-2-alpha). Administration of a HIF-2-alpha antagonist (PT2385) restored YTHDF2-programed epigenetic machinery and repressedliver cancer.
|
[27] |
| Compound Name | TL13-68 | Investigative |
| Synonyms |
TL13-68; CHEMBL4129274; BDBM50269622; HY-136849; CS-0134015
Click to Show/Hide
|
|
| External link | ||
| Activity |
IC50=33 nM
|
[201] |
References