m6A Target Gene Information
General Information of the m6A Target Gene (ID: M6ATAR00190)
Full List of m6A Methylation Regulator of This Target Gene and Corresponding Disease/Drug Response(s)
ATG7
can be regulated by the following regulator(s), and cause disease/drug response(s). You can browse detail information of regulator(s) or disease/drug response(s).
Browse Regulator
Browse Disease
Browse Drug
Methyltransferase-like 3 (METTL3) [WRITER]
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | LX2 cell line | Homo sapiens |
|
Treatment: shMETTL3 LX2 cells
Control: shLuc LX2 cells
|
GSE207909 | |
| Regulation |
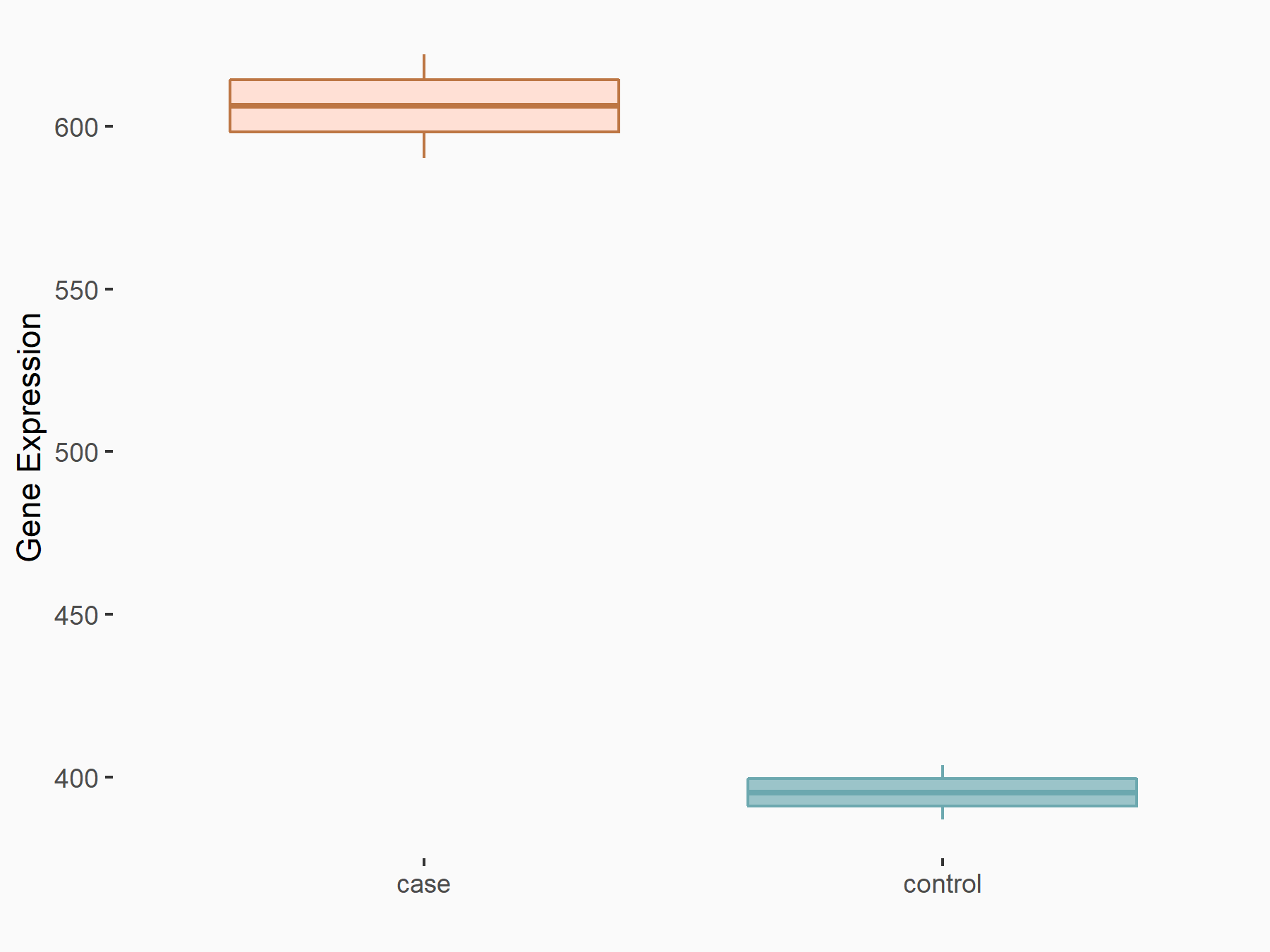  |
logFC: 6.17E-01 p-value: 3.67E-05 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between ATG7 and the regulator | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 4.77E+00 | GSE60213 |
| In total 6 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [1] | |||
| Response Summary | Knocking down METTL3 prevented Enterovirus 71-induced cell death and suppressed Enterovirus 71-induced expression of Bax while rescuing Bcl-2 expression after Enterovirus 71 infection. Knocking down METTL3 inhibited Enterovirus 71-induced expression of Atg5, Ubiquitin-like modifier-activating enzyme ATG7 (ATG7) and LC3 II. Knocking down METTL3 inhibited Enterovirus 71-induced apoptosis and autophagy. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Enterovirus | ICD-11: 1A2Y | ||
| Pathway Response | Autophagy | hsa04140 | ||
| Cell Process | Cell proliferation and metastasis | |||
| Cell apoptosis | ||||
| Cell autophagy | ||||
| In-vitro Model | Schwann cells (A type of glial cell that surrounds neurons) | |||
| Experiment 2 Reporting the m6A Methylation Regulator of This Target Gene | [2] | |||
| Response Summary | METTL3 can sensitise hepatocellular carcinoma cells to sorafenib through stabilising forkhead box class O3 (FOXO3) in an m6A-dependent manner and translated by YTHDF1, thereby inhibiting the transcription of autophagy-related genes, including ATG3, ATG5, Ubiquitin-like modifier-activating enzyme ATG7 (ATG7), ATG12, and ATG16L1. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Hepatocellular carcinoma | ICD-11: 2C12.02 | ||
| Responsed Drug | Sorafenib | Approved | ||
| Pathway Response | FoxO signaling pathway | hsa04068 | ||
| Autophagy | hsa04140 | |||
| Cell Process | Cell autophagy | |||
| Experiment 3 Reporting the m6A Methylation Regulator of This Target Gene | [3] | |||
| Response Summary | METTL3 could positively regulate the autophagy by targeting the autophagy-related genes such as ATG5, Ubiquitin-like modifier-activating enzyme ATG7 (ATG7), LC3B, and SQSTM1. beta-elemene inhibited the autophagy flux by preventing autophagic lysosome acidification, resulting in increasing expression of SQSTM1 and LC3B-II. beta-elemene could reverse gefitinib resistance in non-small cell lung cancer cells by inhibiting cell autophagy process in a manner of chloroquine. METTL3-mediated autophagy in reversing gefitinib resistance of NSCLC cells by beta-elemene, which shed light on providing potential molecular-therapy target and clinical-treatment method in NSCLC patients with gefitinib resistance. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Non-small-cell lung carcinoma | ICD-11: 2C25.Y | ||
| Responsed Drug | Chloroquine | Approved | ||
| Pathway Response | Autophagy | hsa04140 | ||
| Cell Process | Autophagic lysosome acidification | |||
| In-vitro Model | Gefitinib-resistant cell line HCC827GR (Gefitinib-resistant HCC827 cell line) | |||
| Gefitinib-resistant cell line PC9GR (Gefitinib-resistant PC9 cell line) | ||||
| HCC827 | Lung adenocarcinoma | Homo sapiens | CVCL_2063 | |
| PC-9 | Lung adenocarcinoma | Homo sapiens | CVCL_B260 | |
| In-vivo Model | NSCLC gefitinib-resistant cells (5 × 106 cells in 100 uL PBS) were injected subcutaneously into the lateral surface of the left abdomen of 6-week-old female BALB/c nude mice (at least five mice per group to ensure accuracy). | |||
| Experiment 4 Reporting the m6A Methylation Regulator of This Target Gene | [3] | |||
| Response Summary | METTL3 could positively regulate the autophagy by targeting the autophagy-related genes such as ATG5, Ubiquitin-like modifier-activating enzyme ATG7 (ATG7), LC3B, and SQSTM1. beta-elemene inhibited the autophagy flux by preventing autophagic lysosome acidification, resulting in increasing expression of SQSTM1 and LC3B-II. beta-elemene could reverse gefitinib resistance in non-small cell lung cancer cells by inhibiting cell autophagy process in a manner of chloroquine. METTL3-mediated autophagy in reversing gefitinib resistance of NSCLC cells by beta-elemene, which shed light on providing potential molecular-therapy target and clinical-treatment method in NSCLC patients with gefitinib resistance. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Non-small-cell lung carcinoma | ICD-11: 2C25.Y | ||
| Responsed Drug | Gefitinib | Approved | ||
| Pathway Response | Autophagy | hsa04140 | ||
| Cell Process | Autophagic lysosome acidification | |||
| In-vitro Model | Gefitinib-resistant cell line HCC827GR (Gefitinib-resistant HCC827 cell line) | |||
| Gefitinib-resistant cell line PC9GR (Gefitinib-resistant PC9 cell line) | ||||
| HCC827 | Lung adenocarcinoma | Homo sapiens | CVCL_2063 | |
| PC-9 | Lung adenocarcinoma | Homo sapiens | CVCL_B260 | |
| In-vivo Model | NSCLC gefitinib-resistant cells (5 × 106 cells in 100 uL PBS) were injected subcutaneously into the lateral surface of the left abdomen of 6-week-old female BALB/c nude mice (at least five mice per group to ensure accuracy). | |||
| Experiment 5 Reporting the m6A Methylation Regulator of This Target Gene | [3] | |||
| Response Summary | METTL3 could positively regulate the autophagy by targeting the autophagy-related genes such as ATG5, Ubiquitin-like modifier-activating enzyme ATG7 (ATG7), LC3B, and SQSTM1. beta-elemene inhibited the autophagy flux by preventing autophagic lysosome acidification, resulting in increasing expression of SQSTM1 and LC3B-II. beta-elemene could reverse gefitinib resistance in non-small cell lung cancer cells by inhibiting cell autophagy process in a manner of chloroquine. METTL3-mediated autophagy in reversing gefitinib resistance of NSCLC cells by beta-elemene, which shed light on providing potential molecular-therapy target and clinical-treatment method in NSCLC patients with gefitinib resistance. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Non-small-cell lung carcinoma | ICD-11: 2C25.Y | ||
| Responsed Drug | Beta-Elemen | Phase 3 | ||
| Pathway Response | Autophagy | hsa04140 | ||
| Cell Process | Autophagic lysosome acidification | |||
| In-vitro Model | Gefitinib-resistant cell line HCC827GR (Gefitinib-resistant HCC827 cell line) | |||
| Gefitinib-resistant cell line PC9GR (Gefitinib-resistant PC9 cell line) | ||||
| HCC827 | Lung adenocarcinoma | Homo sapiens | CVCL_2063 | |
| PC-9 | Lung adenocarcinoma | Homo sapiens | CVCL_B260 | |
| In-vivo Model | NSCLC gefitinib-resistant cells (5 × 106 cells in 100 uL PBS) were injected subcutaneously into the lateral surface of the left abdomen of 6-week-old female BALB/c nude mice (at least five mice per group to ensure accuracy). | |||
| Experiment 6 Reporting the m6A Methylation Regulator of This Target Gene | [4] | |||
| Response Summary | In osteoarthritis METTL3-mediated m6A modification decreased the expression of autophagy-related 7, an E-1 enzyme crucial for the formation of autophagosomes, by attenuating its RNA stability. Silencing METTL3 enhanced autophagic flux and inhibited Ubiquitin-like modifier-activating enzyme ATG7 (ATG7) expression in OA-FLS. | |||
| Target Regulation | Down regulation | |||
| Responsed Disease | Osteoarthritis | ICD-11: FA05 | ||
| Pathway Response | Autophagy | hsa04140 | ||
| Cell Process | Cellular senescence | |||
| Cell autophagy | ||||
| In-vitro Model | C-28/I2 | Normal | Homo sapiens | CVCL_0187 |
| FLS (Rat fibroblast synovial cell line) | ||||
| In-vivo Model | Mice were anaesthetized with isoflurane supplied in a mouse anaesthesia apparatus, followed with joint surgery on the right joint by sectioning the medial meniscotibial ligament. | |||
Fat mass and obesity-associated protein (FTO) [ERASER]
| Representative RNA-seq result indicating the expression of this target gene regulated by FTO | ||
| Cell Line | NB4 cell line | Homo sapiens |
|
Treatment: shFTO NB4 cells
Control: shNS NB4 cells
|
GSE103494 | |
| Regulation |
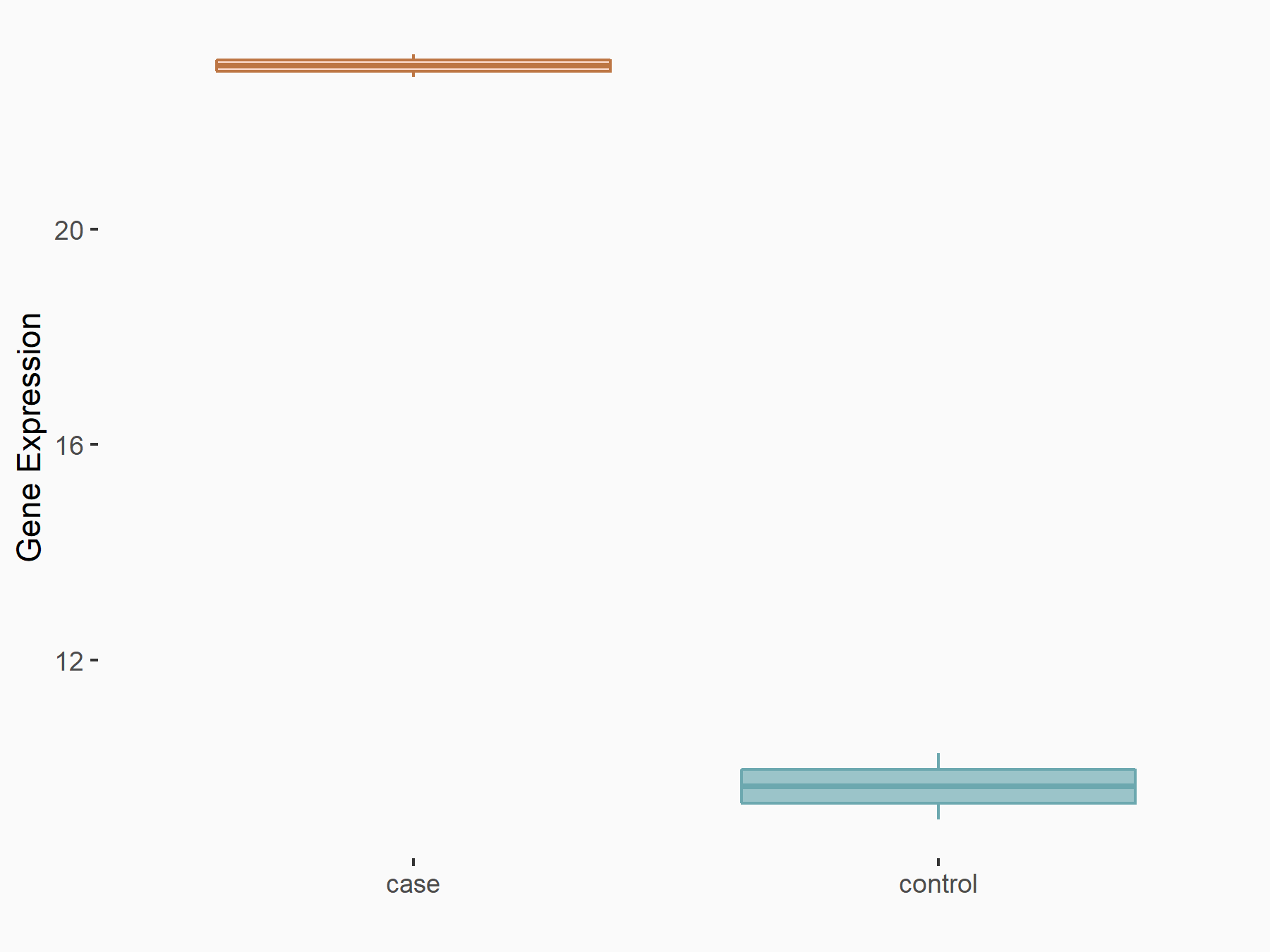  |
logFC: 1.18E+00 p-value: 1.81E-03 |
| More Results | Click to View More RNA-seq Results | |
| In total 2 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [5] | |||
| Response Summary | CircRAB11FIP1 promoted autophagy flux of ovarian cancer through DSC1 and miR-129. CircRAB11FIP1 can serve as the possible marker for EOC diagnosis and treatment. CircRAB11FIP1 regulated the mechanism of autophagy through m6A modification and direct binding to mRNA. CircRAB11FIP1 bound to the mRNA of FTO and promoted its expression. CircRAB11FIP1 directly bound to miR-129 and regulated its targets Ubiquitin-like modifier-activating enzyme ATG7 (ATG7) and ATG14. CircRAB11FIP1 bound to desmocollin 1to facilitate its interaction with ATG101. CircRAB11FIP1 mediated mRNA expression levels of ATG5 and ATG7 depending on m6A. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Malignant mixed epithelial mesenchymal tumour of ovary | ICD-11: 2B5D.0 | ||
| Pathway Response | Autophagy | hsa04140 | ||
| Cell Process | Cell autophagy | |||
| In-vitro Model | SK-OV-3 | Ovarian serous cystadenocarcinoma | Homo sapiens | CVCL_0532 |
| In-vivo Model | The SKOV3 ovarian cancer cell line was transfected with LV2-1 or LV2-NC. Thereafter, BALB/c nude mice (6-week old) were intraperitoneally injected with SKOV3 cells. The mice were killed after 5 weeks, and the number of ascites was determined. | |||
| Experiment 2 Reporting the m6A Methylation Regulator of This Target Gene | [6] | |||
| Response Summary | Atg5 and Ubiquitin-like modifier-activating enzyme ATG7 (ATG7) were the targets of YTHDF2 (YTH N6-methyladenosine RNA binding protein 2). Upon FTO silencing, Atg5 and Atg7 transcripts with higher m6A levels were captured by YTHDF2, which resulted in mRNA degradation and reduction of protein expression, thus alleviating autophagy and adipogenesis. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Obesity | ICD-11: 5B81 | ||
| Pathway Response | Autophagy | hsa04140 | ||
| Cell Process | Autophagy | |||
| Adipogenesis regulation | ||||
| In-vitro Model | 3T3-L1 | Normal | Mus musculus | CVCL_0123 |
| Pig primary preadipocytes (Isolated from cervical subcutaneous adipose tissue of piglets) | ||||
| In-vivo Model | Mice were maintained at 22 ± 2 ℃ with a humidity of 35 ± 5% under a 12 h light and 12 h dark cycle, with free access to water and food. For the HFD experiment, female control (Ftoflox/flox) and adipose-selective fto knockout (Fabp4-Cre Ftoflox/flox, fto-AKO) mice were fed with high-fat diet (60% fat in calories; Research Diets, D12492) for the desired periods of time, and food intake and body weight were measured every week after weaning (at 3 weeks of age). | |||
YTH domain-containing family protein 1 (YTHDF1) [READER]
| In total 1 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [2] | |||
| Response Summary | METTL3 can sensitise hepatocellular carcinoma cells to sorafenib through stabilising forkhead box class O3 (FOXO3) in an m6A-dependent manner and translated by YTHDF1, thereby inhibiting the transcription of autophagy-related genes, including ATG3, ATG5, Ubiquitin-like modifier-activating enzyme ATG7 (ATG7), ATG12, and ATG16L1. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Hepatocellular carcinoma | ICD-11: 2C12.02 | ||
| Responsed Drug | Sorafenib | Approved | ||
| Pathway Response | FoxO signaling pathway | hsa04068 | ||
| Autophagy | hsa04140 | |||
| Cell Process | Cell autophagy | |||
YTH domain-containing family protein 2 (YTHDF2) [READER]
| In total 1 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [6] | |||
| Response Summary | Atg5 and Ubiquitin-like modifier-activating enzyme ATG7 (ATG7) were the targets of YTHDF2 (YTH N6-methyladenosine RNA binding protein 2). Upon FTO silencing, Atg5 and Atg7 transcripts with higher m6A levels were captured by YTHDF2, which resulted in mRNA degradation and reduction of protein expression, thus alleviating autophagy and adipogenesis. | |||
| Target Regulation | Down regulation | |||
| Responsed Disease | Obesity | ICD-11: 5B81 | ||
| Pathway Response | Autophagy | hsa04140 | ||
| Cell Process | Autophagy | |||
| Adipogenesis regulation | ||||
| In-vitro Model | 3T3-L1 | Normal | Mus musculus | CVCL_0123 |
| Pig primary preadipocytes (Isolated from cervical subcutaneous adipose tissue of piglets) | ||||
| In-vivo Model | Mice were maintained at 22 ± 2 ℃ with a humidity of 35 ± 5% under a 12 h light and 12 h dark cycle, with free access to water and food. For the HFD experiment, female control (Ftoflox/flox) and adipose-selective fto knockout (Fabp4-Cre Ftoflox/flox, fto-AKO) mice were fed with high-fat diet (60% fat in calories; Research Diets, D12492) for the desired periods of time, and food intake and body weight were measured every week after weaning (at 3 weeks of age). | |||
Enterovirus [ICD-11: 1A2Y]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [1] | |||
| Response Summary | Knocking down METTL3 prevented Enterovirus 71-induced cell death and suppressed Enterovirus 71-induced expression of Bax while rescuing Bcl-2 expression after Enterovirus 71 infection. Knocking down METTL3 inhibited Enterovirus 71-induced expression of Atg5, Ubiquitin-like modifier-activating enzyme ATG7 (ATG7) and LC3 II. Knocking down METTL3 inhibited Enterovirus 71-induced apoptosis and autophagy. | |||
| Responsed Disease | Enterovirus [ICD-11: 1A2Y] | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Autophagy | hsa04140 | ||
| Cell Process | Cell proliferation and metastasis | |||
| Cell apoptosis | ||||
| Cell autophagy | ||||
| In-vitro Model | Schwann cells (A type of glial cell that surrounds neurons) | |||
Malignant mixed epithelial mesenchymal tumour [ICD-11: 2B5D]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [5] | |||
| Response Summary | CircRAB11FIP1 promoted autophagy flux of ovarian cancer through DSC1 and miR-129. CircRAB11FIP1 can serve as the possible marker for EOC diagnosis and treatment. CircRAB11FIP1 regulated the mechanism of autophagy through m6A modification and direct binding to mRNA. CircRAB11FIP1 bound to the mRNA of FTO and promoted its expression. CircRAB11FIP1 directly bound to miR-129 and regulated its targets Ubiquitin-like modifier-activating enzyme ATG7 (ATG7) and ATG14. CircRAB11FIP1 bound to desmocollin 1to facilitate its interaction with ATG101. CircRAB11FIP1 mediated mRNA expression levels of ATG5 and ATG7 depending on m6A. | |||
| Responsed Disease | Malignant mixed epithelial mesenchymal tumour of ovary [ICD-11: 2B5D.0] | |||
| Target Regulator | Fat mass and obesity-associated protein (FTO) | ERASER | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Autophagy | hsa04140 | ||
| Cell Process | Cell autophagy | |||
| In-vitro Model | SK-OV-3 | Ovarian serous cystadenocarcinoma | Homo sapiens | CVCL_0532 |
| In-vivo Model | The SKOV3 ovarian cancer cell line was transfected with LV2-1 or LV2-NC. Thereafter, BALB/c nude mice (6-week old) were intraperitoneally injected with SKOV3 cells. The mice were killed after 5 weeks, and the number of ascites was determined. | |||
Liver cancer [ICD-11: 2C12]
| In total 2 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [2] | |||
| Response Summary | METTL3 can sensitise hepatocellular carcinoma cells to sorafenib through stabilising forkhead box class O3 (FOXO3) in an m6A-dependent manner and translated by YTHDF1, thereby inhibiting the transcription of autophagy-related genes, including ATG3, ATG5, Ubiquitin-like modifier-activating enzyme ATG7 (ATG7), ATG12, and ATG16L1. | |||
| Responsed Disease | Hepatocellular carcinoma [ICD-11: 2C12.02] | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Target Regulation | Up regulation | |||
| Responsed Drug | Sorafenib | Approved | ||
| Pathway Response | FoxO signaling pathway | hsa04068 | ||
| Autophagy | hsa04140 | |||
| Cell Process | Cell autophagy | |||
| Experiment 2 Reporting the m6A-centered Disease Response | [2] | |||
| Response Summary | METTL3 can sensitise hepatocellular carcinoma cells to sorafenib through stabilising forkhead box class O3 (FOXO3) in an m6A-dependent manner and translated by YTHDF1, thereby inhibiting the transcription of autophagy-related genes, including ATG3, ATG5, Ubiquitin-like modifier-activating enzyme ATG7 (ATG7), ATG12, and ATG16L1. | |||
| Responsed Disease | Hepatocellular carcinoma [ICD-11: 2C12.02] | |||
| Target Regulator | YTH domain-containing family protein 1 (YTHDF1) | READER | ||
| Target Regulation | Up regulation | |||
| Responsed Drug | Sorafenib | Approved | ||
| Pathway Response | FoxO signaling pathway | hsa04068 | ||
| Autophagy | hsa04140 | |||
| Cell Process | Cell autophagy | |||
Lung cancer [ICD-11: 2C25]
| In total 3 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [3] | |||
| Response Summary | METTL3 could positively regulate the autophagy by targeting the autophagy-related genes such as ATG5, Ubiquitin-like modifier-activating enzyme ATG7 (ATG7), LC3B, and SQSTM1. beta-elemene inhibited the autophagy flux by preventing autophagic lysosome acidification, resulting in increasing expression of SQSTM1 and LC3B-II. beta-elemene could reverse gefitinib resistance in non-small cell lung cancer cells by inhibiting cell autophagy process in a manner of chloroquine. METTL3-mediated autophagy in reversing gefitinib resistance of NSCLC cells by beta-elemene, which shed light on providing potential molecular-therapy target and clinical-treatment method in NSCLC patients with gefitinib resistance. | |||
| Responsed Disease | Non-small-cell lung carcinoma [ICD-11: 2C25.Y] | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Target Regulation | Up regulation | |||
| Responsed Drug | Chloroquine | Approved | ||
| Pathway Response | Autophagy | hsa04140 | ||
| Cell Process | Autophagic lysosome acidification | |||
| In-vitro Model | Gefitinib-resistant cell line HCC827GR (Gefitinib-resistant HCC827 cell line) | |||
| Gefitinib-resistant cell line PC9GR (Gefitinib-resistant PC9 cell line) | ||||
| HCC827 | Lung adenocarcinoma | Homo sapiens | CVCL_2063 | |
| PC-9 | Lung adenocarcinoma | Homo sapiens | CVCL_B260 | |
| In-vivo Model | NSCLC gefitinib-resistant cells (5 × 106 cells in 100 uL PBS) were injected subcutaneously into the lateral surface of the left abdomen of 6-week-old female BALB/c nude mice (at least five mice per group to ensure accuracy). | |||
| Experiment 2 Reporting the m6A-centered Disease Response | [3] | |||
| Response Summary | METTL3 could positively regulate the autophagy by targeting the autophagy-related genes such as ATG5, Ubiquitin-like modifier-activating enzyme ATG7 (ATG7), LC3B, and SQSTM1. beta-elemene inhibited the autophagy flux by preventing autophagic lysosome acidification, resulting in increasing expression of SQSTM1 and LC3B-II. beta-elemene could reverse gefitinib resistance in non-small cell lung cancer cells by inhibiting cell autophagy process in a manner of chloroquine. METTL3-mediated autophagy in reversing gefitinib resistance of NSCLC cells by beta-elemene, which shed light on providing potential molecular-therapy target and clinical-treatment method in NSCLC patients with gefitinib resistance. | |||
| Responsed Disease | Non-small-cell lung carcinoma [ICD-11: 2C25.Y] | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Target Regulation | Up regulation | |||
| Responsed Drug | Gefitinib | Approved | ||
| Pathway Response | Autophagy | hsa04140 | ||
| Cell Process | Autophagic lysosome acidification | |||
| In-vitro Model | Gefitinib-resistant cell line HCC827GR (Gefitinib-resistant HCC827 cell line) | |||
| Gefitinib-resistant cell line PC9GR (Gefitinib-resistant PC9 cell line) | ||||
| HCC827 | Lung adenocarcinoma | Homo sapiens | CVCL_2063 | |
| PC-9 | Lung adenocarcinoma | Homo sapiens | CVCL_B260 | |
| In-vivo Model | NSCLC gefitinib-resistant cells (5 × 106 cells in 100 uL PBS) were injected subcutaneously into the lateral surface of the left abdomen of 6-week-old female BALB/c nude mice (at least five mice per group to ensure accuracy). | |||
| Experiment 3 Reporting the m6A-centered Disease Response | [3] | |||
| Response Summary | METTL3 could positively regulate the autophagy by targeting the autophagy-related genes such as ATG5, Ubiquitin-like modifier-activating enzyme ATG7 (ATG7), LC3B, and SQSTM1. beta-elemene inhibited the autophagy flux by preventing autophagic lysosome acidification, resulting in increasing expression of SQSTM1 and LC3B-II. beta-elemene could reverse gefitinib resistance in non-small cell lung cancer cells by inhibiting cell autophagy process in a manner of chloroquine. METTL3-mediated autophagy in reversing gefitinib resistance of NSCLC cells by beta-elemene, which shed light on providing potential molecular-therapy target and clinical-treatment method in NSCLC patients with gefitinib resistance. | |||
| Responsed Disease | Non-small-cell lung carcinoma [ICD-11: 2C25.Y] | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Target Regulation | Up regulation | |||
| Responsed Drug | Beta-Elemen | Phase 3 | ||
| Pathway Response | Autophagy | hsa04140 | ||
| Cell Process | Autophagic lysosome acidification | |||
| In-vitro Model | Gefitinib-resistant cell line HCC827GR (Gefitinib-resistant HCC827 cell line) | |||
| Gefitinib-resistant cell line PC9GR (Gefitinib-resistant PC9 cell line) | ||||
| HCC827 | Lung adenocarcinoma | Homo sapiens | CVCL_2063 | |
| PC-9 | Lung adenocarcinoma | Homo sapiens | CVCL_B260 | |
| In-vivo Model | NSCLC gefitinib-resistant cells (5 × 106 cells in 100 uL PBS) were injected subcutaneously into the lateral surface of the left abdomen of 6-week-old female BALB/c nude mice (at least five mice per group to ensure accuracy). | |||
Obesity [ICD-11: 5B81]
| In total 2 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [6] | |||
| Response Summary | Atg5 and Ubiquitin-like modifier-activating enzyme ATG7 (ATG7) were the targets of YTHDF2 (YTH N6-methyladenosine RNA binding protein 2). Upon FTO silencing, Atg5 and Atg7 transcripts with higher m6A levels were captured by YTHDF2, which resulted in mRNA degradation and reduction of protein expression, thus alleviating autophagy and adipogenesis. | |||
| Responsed Disease | Obesity [ICD-11: 5B81] | |||
| Target Regulator | Fat mass and obesity-associated protein (FTO) | ERASER | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Autophagy | hsa04140 | ||
| Cell Process | Autophagy | |||
| Adipogenesis regulation | ||||
| In-vitro Model | 3T3-L1 | Normal | Mus musculus | CVCL_0123 |
| Pig primary preadipocytes (Isolated from cervical subcutaneous adipose tissue of piglets) | ||||
| In-vivo Model | Mice were maintained at 22 ± 2 ℃ with a humidity of 35 ± 5% under a 12 h light and 12 h dark cycle, with free access to water and food. For the HFD experiment, female control (Ftoflox/flox) and adipose-selective fto knockout (Fabp4-Cre Ftoflox/flox, fto-AKO) mice were fed with high-fat diet (60% fat in calories; Research Diets, D12492) for the desired periods of time, and food intake and body weight were measured every week after weaning (at 3 weeks of age). | |||
| Experiment 2 Reporting the m6A-centered Disease Response | [6] | |||
| Response Summary | Atg5 and Ubiquitin-like modifier-activating enzyme ATG7 (ATG7) were the targets of YTHDF2 (YTH N6-methyladenosine RNA binding protein 2). Upon FTO silencing, Atg5 and Atg7 transcripts with higher m6A levels were captured by YTHDF2, which resulted in mRNA degradation and reduction of protein expression, thus alleviating autophagy and adipogenesis. | |||
| Responsed Disease | Obesity [ICD-11: 5B81] | |||
| Target Regulator | YTH domain-containing family protein 2 (YTHDF2) | READER | ||
| Target Regulation | Down regulation | |||
| Pathway Response | Autophagy | hsa04140 | ||
| Cell Process | Autophagy | |||
| Adipogenesis regulation | ||||
| In-vitro Model | 3T3-L1 | Normal | Mus musculus | CVCL_0123 |
| Pig primary preadipocytes (Isolated from cervical subcutaneous adipose tissue of piglets) | ||||
| In-vivo Model | Mice were maintained at 22 ± 2 ℃ with a humidity of 35 ± 5% under a 12 h light and 12 h dark cycle, with free access to water and food. For the HFD experiment, female control (Ftoflox/flox) and adipose-selective fto knockout (Fabp4-Cre Ftoflox/flox, fto-AKO) mice were fed with high-fat diet (60% fat in calories; Research Diets, D12492) for the desired periods of time, and food intake and body weight were measured every week after weaning (at 3 weeks of age). | |||
Osteoarthritis [ICD-11: FA05]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [4] | |||
| Response Summary | In osteoarthritis METTL3-mediated m6A modification decreased the expression of autophagy-related 7, an E-1 enzyme crucial for the formation of autophagosomes, by attenuating its RNA stability. Silencing METTL3 enhanced autophagic flux and inhibited Ubiquitin-like modifier-activating enzyme ATG7 (ATG7) expression in OA-FLS. | |||
| Responsed Disease | Osteoarthritis [ICD-11: FA05] | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Target Regulation | Down regulation | |||
| Pathway Response | Autophagy | hsa04140 | ||
| Cell Process | Cellular senescence | |||
| Cell autophagy | ||||
| In-vitro Model | C-28/I2 | Normal | Homo sapiens | CVCL_0187 |
| FLS (Rat fibroblast synovial cell line) | ||||
| In-vivo Model | Mice were anaesthetized with isoflurane supplied in a mouse anaesthesia apparatus, followed with joint surgery on the right joint by sectioning the medial meniscotibial ligament. | |||
Chloroquine
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response | [3] | |||
| Response Summary | METTL3 could positively regulate the autophagy by targeting the autophagy-related genes such as ATG5, Ubiquitin-like modifier-activating enzyme ATG7 (ATG7), LC3B, and SQSTM1. beta-elemene inhibited the autophagy flux by preventing autophagic lysosome acidification, resulting in increasing expression of SQSTM1 and LC3B-II. beta-elemene could reverse gefitinib resistance in non-small cell lung cancer cells by inhibiting cell autophagy process in a manner of chloroquine. METTL3-mediated autophagy in reversing gefitinib resistance of NSCLC cells by beta-elemene, which shed light on providing potential molecular-therapy target and clinical-treatment method in NSCLC patients with gefitinib resistance. | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Target Regulation | Up regulation | |||
| Responsed Disease | Non-small-cell lung carcinoma | ICD-11: 2C25.Y | ||
| Pathway Response | Autophagy | hsa04140 | ||
| Cell Process | Autophagic lysosome acidification | |||
| In-vitro Model | Gefitinib-resistant cell line HCC827GR (Gefitinib-resistant HCC827 cell line) | |||
| Gefitinib-resistant cell line PC9GR (Gefitinib-resistant PC9 cell line) | ||||
| HCC827 | Lung adenocarcinoma | Homo sapiens | CVCL_2063 | |
| PC-9 | Lung adenocarcinoma | Homo sapiens | CVCL_B260 | |
| In-vivo Model | NSCLC gefitinib-resistant cells (5 × 106 cells in 100 uL PBS) were injected subcutaneously into the lateral surface of the left abdomen of 6-week-old female BALB/c nude mice (at least five mice per group to ensure accuracy). | |||
Gefitinib
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response | [3] | |||
| Response Summary | METTL3 could positively regulate the autophagy by targeting the autophagy-related genes such as ATG5, Ubiquitin-like modifier-activating enzyme ATG7 (ATG7), LC3B, and SQSTM1. beta-elemene inhibited the autophagy flux by preventing autophagic lysosome acidification, resulting in increasing expression of SQSTM1 and LC3B-II. beta-elemene could reverse gefitinib resistance in non-small cell lung cancer cells by inhibiting cell autophagy process in a manner of chloroquine. METTL3-mediated autophagy in reversing gefitinib resistance of NSCLC cells by beta-elemene, which shed light on providing potential molecular-therapy target and clinical-treatment method in NSCLC patients with gefitinib resistance. | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Target Regulation | Up regulation | |||
| Responsed Disease | Non-small-cell lung carcinoma | ICD-11: 2C25.Y | ||
| Pathway Response | Autophagy | hsa04140 | ||
| Cell Process | Autophagic lysosome acidification | |||
| In-vitro Model | Gefitinib-resistant cell line HCC827GR (Gefitinib-resistant HCC827 cell line) | |||
| Gefitinib-resistant cell line PC9GR (Gefitinib-resistant PC9 cell line) | ||||
| HCC827 | Lung adenocarcinoma | Homo sapiens | CVCL_2063 | |
| PC-9 | Lung adenocarcinoma | Homo sapiens | CVCL_B260 | |
| In-vivo Model | NSCLC gefitinib-resistant cells (5 × 106 cells in 100 uL PBS) were injected subcutaneously into the lateral surface of the left abdomen of 6-week-old female BALB/c nude mice (at least five mice per group to ensure accuracy). | |||
Sorafenib
[Approved]
| In total 2 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response | [2] | |||
| Response Summary | METTL3 can sensitise hepatocellular carcinoma cells to sorafenib through stabilising forkhead box class O3 (FOXO3) in an m6A-dependent manner and translated by YTHDF1, thereby inhibiting the transcription of autophagy-related genes, including ATG3, ATG5, Ubiquitin-like modifier-activating enzyme ATG7 (ATG7), ATG12, and ATG16L1. | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Target Regulation | Up regulation | |||
| Responsed Disease | Hepatocellular carcinoma | ICD-11: 2C12.02 | ||
| Pathway Response | FoxO signaling pathway | hsa04068 | ||
| Autophagy | hsa04140 | |||
| Cell Process | Cell autophagy | |||
| Experiment 2 Reporting the m6A-centered Drug Response | [2] | |||
| Response Summary | METTL3 can sensitise hepatocellular carcinoma cells to sorafenib through stabilising forkhead box class O3 (FOXO3) in an m6A-dependent manner and translated by YTHDF1, thereby inhibiting the transcription of autophagy-related genes, including ATG3, ATG5, Ubiquitin-like modifier-activating enzyme ATG7 (ATG7), ATG12, and ATG16L1. | |||
| Target Regulator | YTH domain-containing family protein 1 (YTHDF1) | READER | ||
| Target Regulation | Up regulation | |||
| Responsed Disease | Hepatocellular carcinoma | ICD-11: 2C12.02 | ||
| Pathway Response | FoxO signaling pathway | hsa04068 | ||
| Autophagy | hsa04140 | |||
| Cell Process | Cell autophagy | |||
Beta-Elemen
[Phase 3]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response | [3] | |||
| Response Summary | METTL3 could positively regulate the autophagy by targeting the autophagy-related genes such as ATG5, Ubiquitin-like modifier-activating enzyme ATG7 (ATG7), LC3B, and SQSTM1. beta-elemene inhibited the autophagy flux by preventing autophagic lysosome acidification, resulting in increasing expression of SQSTM1 and LC3B-II. beta-elemene could reverse gefitinib resistance in non-small cell lung cancer cells by inhibiting cell autophagy process in a manner of chloroquine. METTL3-mediated autophagy in reversing gefitinib resistance of NSCLC cells by beta-elemene, which shed light on providing potential molecular-therapy target and clinical-treatment method in NSCLC patients with gefitinib resistance. | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Target Regulation | Up regulation | |||
| Responsed Disease | Non-small-cell lung carcinoma | ICD-11: 2C25.Y | ||
| Pathway Response | Autophagy | hsa04140 | ||
| Cell Process | Autophagic lysosome acidification | |||
| In-vitro Model | Gefitinib-resistant cell line HCC827GR (Gefitinib-resistant HCC827 cell line) | |||
| Gefitinib-resistant cell line PC9GR (Gefitinib-resistant PC9 cell line) | ||||
| HCC827 | Lung adenocarcinoma | Homo sapiens | CVCL_2063 | |
| PC-9 | Lung adenocarcinoma | Homo sapiens | CVCL_B260 | |
| In-vivo Model | NSCLC gefitinib-resistant cells (5 × 106 cells in 100 uL PBS) were injected subcutaneously into the lateral surface of the left abdomen of 6-week-old female BALB/c nude mice (at least five mice per group to ensure accuracy). | |||
Full List of Crosstalk(s) between m6A Modification and Epigenetic Regulation Related to This Regulator
Non-coding RNA
m6A Regulator: Fat mass and obesity-associated protein (FTO)
| In total 1 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT05873 | ||
| Epigenetic Regulator | Circ_RAB11FIP1 | |
| Regulated Target | FTO alpha-ketoglutarate dependent dioxygenase (FTO) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Ovarian cancer | |
RNA Modification Sequencing Data Associated with the Target (ID: M6ATAR00190)
| In total 28 m6A sequence/site(s) in this target gene | |||
| mod ID: A2ISITE011496 | Click to Show/Hide the Full List | ||
| mod site | chr3:11283916-11283917:+ | [9] | |
| Sequence | ACTCCAGCCTGGACGACAGAACGAGACTCTGTCTCAAAGAA | ||
| Transcript ID List | ENST00000419112.5; ENST00000451830.5; ENST00000354956.9; rmsk_887454; ENST00000444619.5; ENST00000460444.5; ENST00000451513.5; ENST00000354449.7; ENST00000418682.5; ENST00000469654.2; ENST00000423116.1; ENST00000446450.6; ENST00000470474.1; ENST00000435760.5 | ||
| External Link | RMBase: RNA-editing_site_94731 | ||
| mod ID: A2ISITE011497 | Click to Show/Hide the Full List | ||
| mod site | chr3:11314635-11314636:+ | [9] | |
| Sequence | CATTGCATTATGTACTTTAAATGGGTGAATTGGGCCAGGCA | ||
| Transcript ID List | rmsk_887527; ENST00000451830.5; ENST00000354956.9; ENST00000354449.7; ENST00000418682.5; ENST00000488924.5; ENST00000424071.5; ENST00000434066.6; ENST00000446450.6 | ||
| External Link | RMBase: RNA-editing_site_94732 | ||
| mod ID: A2ISITE011498 | Click to Show/Hide the Full List | ||
| mod site | chr3:11334837-11334838:+ | [9] | |
| Sequence | TGTGGTGGCGGGCGTCTGTAATCCCAGCTCTCAGGAGGCTG | ||
| Transcript ID List | ENST00000461278.1; ENST00000424071.5; ENST00000434066.6; ENST00000478638.5; ENST00000354956.9; ENST00000354449.7; rmsk_887581; ENST00000446450.6 | ||
| External Link | RMBase: RNA-editing_site_94733 | ||
| mod ID: A2ISITE011499 | Click to Show/Hide the Full List | ||
| mod site | chr3:11399226-11399227:+ | [9] | |
| Sequence | GTATGGTGGCCTGTACATATAGTCCCAGCTACTCAGGAGGC | ||
| Transcript ID List | ENST00000446450.6; ENST00000427759.5; rmsk_887721; ENST00000354449.7; ENST00000354956.9; ENST00000414717.5; ENST00000446110.1 | ||
| External Link | RMBase: RNA-editing_site_94734 | ||
| mod ID: A2ISITE011500 | Click to Show/Hide the Full List | ||
| mod site | chr3:11428091-11428092:+ | [9] | |
| Sequence | ACTTGCCATTAGTCCTTTTAAGTTGCAATTCTTCCCACCAG | ||
| Transcript ID List | ENST00000354956.9; ENST00000446110.1; ENST00000446450.6; ENST00000427759.5; ENST00000414717.5; ENST00000354449.7 | ||
| External Link | RMBase: RNA-editing_site_94735 | ||
| mod ID: A2ISITE011501 | Click to Show/Hide the Full List | ||
| mod site | chr3:11428129-11428130:+ | [9] | |
| Sequence | CAGCTAAAGAGAAATATATAAATTATTTCTAGCTAGGTGGC | ||
| Transcript ID List | ENST00000354449.7; ENST00000427759.5; ENST00000354956.9; ENST00000446110.1; ENST00000414717.5; ENST00000446450.6 | ||
| External Link | RMBase: RNA-editing_site_94736 | ||
| mod ID: A2ISITE011502 | Click to Show/Hide the Full List | ||
| mod site | chr3:11428143-11428144:+ | [10] | |
| Sequence | TATATAAATTATTTCTAGCTAGGTGGCAAAACTCCCCCAGC | ||
| Transcript ID List | ENST00000446110.1; ENST00000354956.9; ENST00000427759.5; ENST00000446450.6; ENST00000354449.7; ENST00000414717.5 | ||
| External Link | RMBase: RNA-editing_site_94737 | ||
| mod ID: A2ISITE011503 | Click to Show/Hide the Full List | ||
| mod site | chr3:11428193-11428194:+ | [9] | |
| Sequence | AAATGATTGCATGGGTGCAAAGATCTATAAATTCCTCCATA | ||
| Transcript ID List | ENST00000427759.5; ENST00000446450.6; ENST00000354449.7; ENST00000354956.9; ENST00000446110.1; ENST00000414717.5 | ||
| External Link | RMBase: RNA-editing_site_94738 | ||
| mod ID: A2ISITE011504 | Click to Show/Hide the Full List | ||
| mod site | chr3:11428714-11428715:+ | [11] | |
| Sequence | GTACTGCCTGATGGTATGGAAGAGGTAACATAGATGTTTGT | ||
| Transcript ID List | ENST00000354449.7; ENST00000414717.5; ENST00000446450.6; ENST00000446110.1; ENST00000354956.9; ENST00000427759.5 | ||
| External Link | RMBase: RNA-editing_site_94739 | ||
| mod ID: A2ISITE011505 | Click to Show/Hide the Full List | ||
| mod site | chr3:11428725-11428726:+ | [9] | |
| Sequence | TGGTATGGAAGAGGTAACATAGATGTTTGTAAACATACAGT | ||
| Transcript ID List | ENST00000446450.6; ENST00000427759.5; ENST00000354956.9; ENST00000414717.5; ENST00000446110.1; ENST00000354449.7 | ||
| External Link | RMBase: RNA-editing_site_94740 | ||
| mod ID: A2ISITE011506 | Click to Show/Hide the Full List | ||
| mod site | chr3:11428741-11428742:+ | [12] | |
| Sequence | ACATAGATGTTTGTAAACATACAGTCATTTGATGGCTAAGG | ||
| Transcript ID List | ENST00000427759.5; ENST00000354449.7; ENST00000354956.9; ENST00000446450.6; ENST00000414717.5; ENST00000446110.1 | ||
| External Link | RMBase: RNA-editing_site_94741 | ||
| mod ID: A2ISITE011507 | Click to Show/Hide the Full List | ||
| mod site | chr3:11428758-11428759:+ | [9] | |
| Sequence | CATACAGTCATTTGATGGCTAAGGAAGTTTTGCCACCCAGC | ||
| Transcript ID List | ENST00000446450.6; ENST00000446110.1; ENST00000354956.9; ENST00000414717.5; ENST00000427759.5; ENST00000354449.7 | ||
| External Link | RMBase: RNA-editing_site_94742 | ||
| mod ID: A2ISITE011508 | Click to Show/Hide the Full List | ||
| mod site | chr3:11428759-11428760:+ | [12] | |
| Sequence | ATACAGTCATTTGATGGCTAAGGAAGTTTTGCCACCCAGCT | ||
| Transcript ID List | ENST00000354449.7; ENST00000446450.6; ENST00000354956.9; ENST00000414717.5; ENST00000446110.1; ENST00000427759.5 | ||
| External Link | RMBase: RNA-editing_site_94743 | ||
| mod ID: A2ISITE011509 | Click to Show/Hide the Full List | ||
| mod site | chr3:11431604-11431605:+ | [9] | |
| Sequence | TAATCTTCTTTCTGTCTGTAAGGATTTTTCTATTCTGGACA | ||
| Transcript ID List | ENST00000354449.7; ENST00000414717.5; ENST00000446450.6; ENST00000354956.9; ENST00000446110.1; ENST00000427759.5 | ||
| External Link | RMBase: RNA-editing_site_94744 | ||
| mod ID: A2ISITE011510 | Click to Show/Hide the Full List | ||
| mod site | chr3:11436783-11436784:+ | [12] | |
| Sequence | GGATGAACATTGAAAATGTTACACTAATTGAAGAAGCCAGT | ||
| Transcript ID List | ENST00000354449.7; rmsk_887804; ENST00000354956.9; ENST00000446110.1; ENST00000446450.6; ENST00000427759.5; ENST00000414717.5 | ||
| External Link | RMBase: RNA-editing_site_94745 | ||
| mod ID: A2ISITE011511 | Click to Show/Hide the Full List | ||
| mod site | chr3:11442809-11442810:+ | [9] | |
| Sequence | ACATGCCTGTAGTCCCAGCTACTTGGGAGGCTGAGATGGGA | ||
| Transcript ID List | ENST00000446450.6; ENST00000354956.9; ENST00000354449.7; ENST00000414717.5; ENST00000446110.1; ENST00000427759.5; rmsk_887820 | ||
| External Link | RMBase: RNA-editing_site_94746 | ||
| mod ID: A2ISITE011512 | Click to Show/Hide the Full List | ||
| mod site | chr3:11446769-11446770:+ | [9] | |
| Sequence | CTGGAACCAAGGCACAGTCAAGGTGAACCAGCTATGCTCCA | ||
| Transcript ID List | ENST00000354956.9; ENST00000427759.5; ENST00000414717.5; ENST00000354449.7; ENST00000446110.1; ENST00000446450.6 | ||
| External Link | RMBase: RNA-editing_site_94747 | ||
| mod ID: A2ISITE011513 | Click to Show/Hide the Full List | ||
| mod site | chr3:11467505-11467506:+ | [12] | |
| Sequence | TCTGCCTCAGCCTCCCGAGTAGCTGGATTATAGGCATCCAC | ||
| Transcript ID List | ENST00000354956.9; ENST00000427759.5; ENST00000354449.7; ENST00000446110.1; ENST00000446450.6 | ||
| External Link | RMBase: RNA-editing_site_94748 | ||
| mod ID: A2ISITE011514 | Click to Show/Hide the Full List | ||
| mod site | chr3:11500665-11500666:+ | [12] | |
| Sequence | TTTTTTTGTTTGTTTGTCTGAGACGGAGTTTTGCTCTTGTT | ||
| Transcript ID List | ENST00000446450.6; ENST00000446110.1; ENST00000354449.7; ENST00000354956.9 | ||
| External Link | RMBase: RNA-editing_site_94749 | ||
| mod ID: A2ISITE011515 | Click to Show/Hide the Full List | ||
| mod site | chr3:11506540-11506541:+ | [13] | |
| Sequence | CAGCCTGGTCAACATGGTGAAACCCCATCTCTACAAAAAAA | ||
| Transcript ID List | ENST00000354956.9; ENST00000446110.1; ENST00000354449.7; rmsk_887944; ENST00000446450.6 | ||
| External Link | RMBase: RNA-editing_site_94750 | ||
| mod ID: A2ISITE011516 | Click to Show/Hide the Full List | ||
| mod site | chr3:11506546-11506547:+ | [13] | |
| Sequence | GGTCAACATGGTGAAACCCCATCTCTACAAAAAAAAAAAAA | ||
| Transcript ID List | rmsk_887944; ENST00000354449.7; ENST00000446450.6; ENST00000354956.9; ENST00000446110.1 | ||
| External Link | RMBase: RNA-editing_site_94751 | ||
| mod ID: A2ISITE011517 | Click to Show/Hide the Full List | ||
| mod site | chr3:11506630-11506631:+ | [13] | |
| Sequence | AGGTGCCTGTAATCCCAGCTACTCAGGAGGCTGAGGCAGGA | ||
| Transcript ID List | rmsk_887944; ENST00000446450.6; ENST00000446110.1; ENST00000354956.9; ENST00000354449.7 | ||
| External Link | RMBase: RNA-editing_site_94752 | ||
| mod ID: A2ISITE011518 | Click to Show/Hide the Full List | ||
| mod site | chr3:11506643-11506644:+ | [13] | |
| Sequence | CCCAGCTACTCAGGAGGCTGAGGCAGGACAATCACTTGAAC | ||
| Transcript ID List | ENST00000354449.7; ENST00000446110.1; ENST00000354956.9; rmsk_887944; ENST00000446450.6 | ||
| External Link | RMBase: RNA-editing_site_94753 | ||
| mod ID: A2ISITE011519 | Click to Show/Hide the Full List | ||
| mod site | chr3:11506662-11506663:+ | [13] | |
| Sequence | GAGGCAGGACAATCACTTGAACCCAGGAGGCAGAGGTTGTG | ||
| Transcript ID List | rmsk_887944; ENST00000446110.1; ENST00000354449.7; ENST00000354956.9; ENST00000446450.6 | ||
| External Link | RMBase: RNA-editing_site_94754 | ||
| mod ID: A2ISITE011520 | Click to Show/Hide the Full List | ||
| mod site | chr3:11506911-11506912:+ | [9] | |
| Sequence | TTCCCTAGGTAAGGATTCATAGCATTTTTTGTGCCATGGAT | ||
| Transcript ID List | ENST00000446110.1; ENST00000446450.6; ENST00000354449.7; ENST00000354956.9 | ||
| External Link | RMBase: RNA-editing_site_94755 | ||
| mod ID: A2ISITE011521 | Click to Show/Hide the Full List | ||
| mod site | chr3:11521570-11521571:+ | [9] | |
| Sequence | TTTTTTGTTTTTTTTTTTTTAAGACGGAGTCTCGCTCTGTC | ||
| Transcript ID List | ENST00000354956.9; ENST00000354449.7; ENST00000446450.6 | ||
| External Link | RMBase: RNA-editing_site_94756 | ||
| mod ID: A2ISITE011522 | Click to Show/Hide the Full List | ||
| mod site | chr3:11524529-11524530:+ | [9] | |
| Sequence | GCACTTTGAGAGGCCTAGGCAGGAGGATCACTTGAGCCCAG | ||
| Transcript ID List | ENST00000354956.9; ENST00000354449.7; ENST00000446450.6; rmsk_887982 | ||
| External Link | RMBase: RNA-editing_site_94757 | ||
| mod ID: A2ISITE011523 | Click to Show/Hide the Full List | ||
| mod site | chr3:11526366-11526367:+ | [9] | |
| Sequence | ACTCCACTGCACTCCAGCCTAGGTGGCAGAGTGAGACTCAA | ||
| Transcript ID List | ENST00000354449.7; ENST00000354956.9; rmsk_887988; ENST00000446450.6 | ||
| External Link | RMBase: RNA-editing_site_94758 | ||
5-methylcytidine (m5C)
N6-methyladenosine (m6A)
| In total 62 m6A sequence/site(s) in this target gene | |||
| mod ID: M6ASITE059491 | Click to Show/Hide the Full List | ||
| mod site | chr3:11272587-11272588:+ | [14] | |
| Sequence | TGGTCCTTCCTCCTCCAGGGACTCACTGATTCCGGCTGGCG | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | peripheral-blood | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000435760.5; ENST00000451513.5; ENST00000418682.5; ENST00000469654.2; ENST00000446450.6; ENST00000354956.9; ENST00000460444.5; ENST00000470474.1; ENST00000419112.5; ENST00000444619.5; ENST00000451830.5; ENST00000354449.7 | ||
| External Link | RMBase: m6A_site_577081 | ||
| mod ID: M6ASITE059492 | Click to Show/Hide the Full List | ||
| mod site | chr3:11272636-11272637:+ | [14] | |
| Sequence | CTGTAGCCGCGTCCCCTCAGACTGGTTCAGTCCGGGGTCTT | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | peripheral-blood | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000470474.1; ENST00000435760.5; ENST00000460444.5; ENST00000419112.5; ENST00000469654.2; ENST00000354449.7; ENST00000444619.5; ENST00000418682.5; ENST00000446450.6; ENST00000354956.9; ENST00000451513.5; ENST00000451830.5 | ||
| External Link | RMBase: m6A_site_577082 | ||
| mod ID: M6ASITE059493 | Click to Show/Hide the Full List | ||
| mod site | chr3:11282240-11282241:+ | [15] | |
| Sequence | CAGTGCATCCGTTCTCAAAAACTGAAGCTGATGGACACCAG | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | iSLK | ||
| Seq Type List | MeRIP-seq | ||
| Transcript ID List | ENST00000354449.7; ENST00000451830.5; ENST00000469654.2; ENST00000444619.5; ENST00000451513.5; ENST00000435760.5; ENST00000446450.6; ENST00000419112.5; ENST00000354956.9; ENST00000418682.5; ENST00000470474.1; ENST00000423116.1; ENST00000460444.5 | ||
| External Link | RMBase: m6A_site_577083 | ||
| mod ID: M6ASITE059494 | Click to Show/Hide the Full List | ||
| mod site | chr3:11282254-11282255:+ | [15] | |
| Sequence | TCAAAAACTGAAGCTGATGGACACCAGATTGCATGTGAAGG | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | iSLK | ||
| Seq Type List | MeRIP-seq | ||
| Transcript ID List | ENST00000460444.5; ENST00000354449.7; ENST00000354956.9; ENST00000451513.5; ENST00000469654.2; ENST00000451830.5; ENST00000418682.5; ENST00000446450.6; ENST00000419112.5; ENST00000470474.1; ENST00000444619.5; ENST00000435760.5; ENST00000423116.1 | ||
| External Link | RMBase: m6A_site_577084 | ||
| mod ID: M6ASITE059495 | Click to Show/Hide the Full List | ||
| mod site | chr3:11282299-11282300:+ | [16] | |
| Sequence | GGATATTATAGCAGAAGGAAACCAAAATAAAAGAAAAACAG | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | HeLa; HEK293T; iSLK; MSC | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000435760.5; ENST00000444619.5; ENST00000470474.1; ENST00000460444.5; ENST00000451830.5; ENST00000423116.1; ENST00000354449.7; ENST00000354956.9; ENST00000418682.5; ENST00000419112.5; ENST00000446450.6; ENST00000451513.5; ENST00000469654.2 | ||
| External Link | RMBase: m6A_site_577085 | ||
| mod ID: M6ASITE059496 | Click to Show/Hide the Full List | ||
| mod site | chr3:11282316-11282317:+ | [16] | |
| Sequence | GAAACCAAAATAAAAGAAAAACAGCTCCTGGAAGTAACCAA | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa; HEK293T; iSLK; MSC | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000354449.7; ENST00000423116.1; ENST00000418682.5; ENST00000470474.1; ENST00000446450.6; ENST00000460444.5; ENST00000444619.5; ENST00000419112.5; ENST00000451830.5; ENST00000469654.2; ENST00000354956.9; ENST00000435760.5; ENST00000451513.5 | ||
| External Link | RMBase: m6A_site_577086 | ||
| mod ID: M6ASITE059497 | Click to Show/Hide the Full List | ||
| mod site | chr3:11282345-11282346:+ | [16] | |
| Sequence | GGAAGTAACCAATGAGAAGGACAGATAAGAGTTTGAAGACT | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; HEK293T; Huh7; iSLK; MSC | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000354956.9; ENST00000460444.5; ENST00000419112.5; ENST00000354449.7; ENST00000418682.5; ENST00000444619.5; ENST00000451830.5; ENST00000469654.2; ENST00000446450.6; ENST00000451513.5; ENST00000470474.1; ENST00000423116.1; ENST00000435760.5 | ||
| External Link | RMBase: m6A_site_577087 | ||
| mod ID: M6ASITE059498 | Click to Show/Hide the Full List | ||
| mod site | chr3:11282363-11282364:+ | [16] | |
| Sequence | GGACAGATAAGAGTTTGAAGACTGTGGATGTGACTGCTTGT | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa; HEK293T; Huh7; iSLK | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000451830.5; ENST00000354956.9; ENST00000469654.2; ENST00000418682.5; ENST00000354449.7; ENST00000444619.5; ENST00000451513.5; ENST00000446450.6; ENST00000419112.5; ENST00000460444.5; ENST00000423116.1; ENST00000435760.5; ENST00000470474.1 | ||
| External Link | RMBase: m6A_site_577088 | ||
| mod ID: M6ASITE059499 | Click to Show/Hide the Full List | ||
| mod site | chr3:11288704-11288705:+ | [16] | |
| Sequence | TGGGGATTTGATCTTAAAAAACAAAAGATTTAGATCCATGT | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa; iSLK | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000451830.5; ENST00000444619.5; ENST00000451513.5; ENST00000469654.2; ENST00000354449.7; ENST00000423116.1; ENST00000446450.6; ENST00000470474.1; ENST00000418682.5; ENST00000354956.9; ENST00000419112.5; ENST00000435760.5; ENST00000460444.5 | ||
| External Link | RMBase: m6A_site_577089 | ||
| mod ID: M6ASITE059500 | Click to Show/Hide the Full List | ||
| mod site | chr3:11288727-11288728:+ | [16] | |
| Sequence | AAAGATTTAGATCCATGTGGACAGAAACTTTTGGTAAGTAC | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; iSLK | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000460444.5; ENST00000444619.5; ENST00000446450.6; ENST00000469654.2; ENST00000418682.5; ENST00000435760.5; ENST00000419112.5; ENST00000470474.1; ENST00000451830.5; ENST00000354449.7; ENST00000451513.5; ENST00000354956.9; ENST00000423116.1 | ||
| External Link | RMBase: m6A_site_577090 | ||
| mod ID: M6ASITE059501 | Click to Show/Hide the Full List | ||
| mod site | chr3:11288733-11288734:+ | [16] | |
| Sequence | TTAGATCCATGTGGACAGAAACTTTTGGTAAGTACTCATTC | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa; iSLK | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000354956.9; ENST00000419112.5; ENST00000435760.5; ENST00000354449.7; ENST00000460444.5; ENST00000423116.1; ENST00000470474.1; ENST00000446450.6; ENST00000444619.5; ENST00000451513.5; ENST00000469654.2; ENST00000418682.5; ENST00000451830.5 | ||
| External Link | RMBase: m6A_site_577091 | ||
| mod ID: M6ASITE059502 | Click to Show/Hide the Full List | ||
| mod site | chr3:11298721-11298722:+ | [17] | |
| Sequence | GCAGCTACGGGGGATCCTGGACTCTCTAAACTGCAGTTTGC | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | A549; iSLK; TIME; MSC | ||
| Seq Type List | MeRIP-seq | ||
| Transcript ID List | ENST00000435760.5; ENST00000444619.5; ENST00000419112.5; ENST00000354449.7; ENST00000418682.5; ENST00000460444.5; ENST00000470474.1; ENST00000451830.5; ENST00000451513.5; ENST00000446450.6; ENST00000423116.1; ENST00000469654.2; ENST00000354956.9 | ||
| External Link | RMBase: m6A_site_577092 | ||
| mod ID: M6ASITE059503 | Click to Show/Hide the Full List | ||
| mod site | chr3:11298730-11298731:+ | [17] | |
| Sequence | GGGGATCCTGGACTCTCTAAACTGCAGTTTGCCCCTTTTAG | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | A549; iSLK; TIME; MSC | ||
| Seq Type List | MeRIP-seq | ||
| Transcript ID List | ENST00000354956.9; ENST00000470474.1; ENST00000451830.5; ENST00000451513.5; ENST00000418682.5; ENST00000460444.5; ENST00000423116.1; ENST00000435760.5; ENST00000446450.6; ENST00000419112.5; ENST00000469654.2; ENST00000354449.7; ENST00000444619.5 | ||
| External Link | RMBase: m6A_site_577093 | ||
| mod ID: M6ASITE059504 | Click to Show/Hide the Full List | ||
| mod site | chr3:11298831-11298832:+ | [16] | |
| Sequence | GCTGGATGAAGCTCCCAAGGACATTAAGGGTTATTACTACA | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; hESC-HEK293T; MSC | ||
| Seq Type List | m6A-seq; MAZTER-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000460444.5; ENST00000446450.6; ENST00000469654.2; ENST00000451513.5; ENST00000451830.5; ENST00000354956.9; ENST00000418682.5; ENST00000470474.1; ENST00000423116.1; ENST00000419112.5; ENST00000435760.5; ENST00000354449.7; ENST00000444619.5 | ||
| External Link | RMBase: m6A_site_577094 | ||
| mod ID: M6ASITE059505 | Click to Show/Hide the Full List | ||
| mod site | chr3:11299412-11299413:+ | [18] | |
| Sequence | ATTGGAGTTCAGTGCTTTTGACATGTGAGTATTTATTTGTT | ||
| Motif Score | 2.859755952 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000451830.5; ENST00000469654.2; ENST00000423116.1; ENST00000418682.5; ENST00000354449.7; ENST00000444619.5; ENST00000419112.5; ENST00000354956.9; ENST00000451513.5; ENST00000446450.6; ENST00000460444.5 | ||
| External Link | RMBase: m6A_site_577095 | ||
| mod ID: M6ASITE059506 | Click to Show/Hide the Full List | ||
| mod site | chr3:11306982-11306983:+ | [18] | |
| Sequence | GTTGCTGCCCAGCTATTGGAACACTGTATAACACCAACACA | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000418682.5; ENST00000423116.1; ENST00000451513.5; ENST00000419112.5; ENST00000451830.5; ENST00000354956.9; ENST00000444619.5; ENST00000354449.7; ENST00000446450.6 | ||
| External Link | RMBase: m6A_site_577096 | ||
| mod ID: M6ASITE059507 | Click to Show/Hide the Full List | ||
| mod site | chr3:11307018-11307019:+ | [16] | |
| Sequence | ACACACTCGAGTCTTTCAAGACTGCAGATAAGAAGCTCCTT | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000354449.7; ENST00000423116.1; ENST00000446450.6; ENST00000451830.5; ENST00000354956.9; ENST00000418682.5; ENST00000451513.5 | ||
| External Link | RMBase: m6A_site_577097 | ||
| mod ID: M6ASITE059508 | Click to Show/Hide the Full List | ||
| mod site | chr3:11307044-11307045:+ | [16] | |
| Sequence | GATAAGAAGCTCCTTTTGGAACAAGCAGCAAATGAGGTTAG | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa; hESC-HEK293T | ||
| Seq Type List | m6A-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000451513.5; ENST00000423116.1; ENST00000354956.9; ENST00000451830.5; ENST00000418682.5; ENST00000354449.7; ENST00000446450.6 | ||
| External Link | RMBase: m6A_site_577098 | ||
| mod ID: M6ASITE059509 | Click to Show/Hide the Full List | ||
| mod site | chr3:11309035-11309036:+ | [18] | |
| Sequence | TGAAAACCCTGTACTCCTCAACAAGTTCCTCCTCTTGACAT | ||
| Motif Score | 2.173910714 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000451513.5; ENST00000418682.5; ENST00000451830.5; ENST00000446450.6; ENST00000424071.5; ENST00000354449.7; ENST00000488924.5; ENST00000464282.1; ENST00000354956.9 | ||
| External Link | RMBase: m6A_site_577099 | ||
| mod ID: M6ASITE059510 | Click to Show/Hide the Full List | ||
| mod site | chr3:11309052-11309053:+ | [19] | |
| Sequence | TCAACAAGTTCCTCCTCTTGACATTTGCAGTAAGTAAATGG | ||
| Motif Score | 2.859755952 | ||
| Cell/Tissue List | HEK293T; hESC-HEK293T | ||
| Seq Type List | DART-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000354956.9; ENST00000446450.6; ENST00000464282.1; ENST00000354449.7; ENST00000451513.5; ENST00000451830.5; ENST00000488924.5; ENST00000424071.5; ENST00000418682.5 | ||
| External Link | RMBase: m6A_site_577100 | ||
| mod ID: M6ASITE059511 | Click to Show/Hide the Full List | ||
| mod site | chr3:11315431-11315432:+ | [16] | |
| Sequence | CTTAATCAAGTATGATGAGAACATGGTGCTGGTTTCCTTGC | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000446450.6; ENST00000488924.5; ENST00000434066.6; ENST00000354956.9; ENST00000354449.7; ENST00000418682.5; ENST00000424071.5 | ||
| External Link | RMBase: m6A_site_577101 | ||
| mod ID: M6ASITE059512 | Click to Show/Hide the Full List | ||
| mod site | chr3:11315456-11315457:+ | [16] | |
| Sequence | GTGCTGGTTTCCTTGCTTAAACACTACAGTGATTTCTTCCA | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000354956.9; ENST00000424071.5; ENST00000488924.5; ENST00000434066.6; ENST00000446450.6; ENST00000418682.5; ENST00000354449.7 | ||
| External Link | RMBase: m6A_site_577102 | ||
| mod ID: M6ASITE059513 | Click to Show/Hide the Full List | ||
| mod site | chr3:11331342-11331343:+ | [18] | |
| Sequence | GTCTTTTTTGTTCACAGATAACAATTGGTGTATATGATCCC | ||
| Motif Score | 2.168095238 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000446450.6; ENST00000354956.9; ENST00000488924.5; ENST00000434066.6; ENST00000424071.5; ENST00000354449.7 | ||
| External Link | RMBase: m6A_site_577103 | ||
| mod ID: M6ASITE059514 | Click to Show/Hide the Full List | ||
| mod site | chr3:11340674-11340675:+ | [16] | |
| Sequence | AGCAGTTGGATGGGAAAAGAACCAGAAAGGAGGCATGGGAC | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000446450.6; ENST00000434066.6; ENST00000478638.5; ENST00000461278.1; ENST00000354956.9; ENST00000424071.5; ENST00000354449.7 | ||
| External Link | RMBase: m6A_site_577104 | ||
| mod ID: M6ASITE059515 | Click to Show/Hide the Full List | ||
| mod site | chr3:11340693-11340694:+ | [16] | |
| Sequence | AACCAGAAAGGAGGCATGGGACCAAGGATGGTGAACCTCAG | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000354956.9; ENST00000354449.7; ENST00000478638.5; ENST00000461278.1; ENST00000424071.5; ENST00000446450.6; ENST00000434066.6 | ||
| External Link | RMBase: m6A_site_577105 | ||
| mod ID: M6ASITE059516 | Click to Show/Hide the Full List | ||
| mod site | chr3:11340707-11340708:+ | [16] | |
| Sequence | CATGGGACCAAGGATGGTGAACCTCAGTGAATGTATGGACC | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000446450.6; ENST00000424071.5; ENST00000434066.6; ENST00000354956.9; ENST00000461278.1; ENST00000478638.5; ENST00000354449.7 | ||
| External Link | RMBase: m6A_site_577106 | ||
| mod ID: M6ASITE059517 | Click to Show/Hide the Full List | ||
| mod site | chr3:11340725-11340726:+ | [16] | |
| Sequence | GAACCTCAGTGAATGTATGGACCCTAAAAGGTATATTTGGG | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000446450.6; ENST00000354956.9; ENST00000424071.5; ENST00000478638.5; ENST00000434066.6; ENST00000354449.7; ENST00000461278.1 | ||
| External Link | RMBase: m6A_site_577107 | ||
| mod ID: M6ASITE059518 | Click to Show/Hide the Full List | ||
| mod site | chr3:11342199-11342200:+ | [16] | |
| Sequence | GAGATTGGTTCCTACTTTAGACTTGGACAAGGTTGTGTCTG | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000424071.5; ENST00000354956.9; ENST00000446450.6; ENST00000354449.7; ENST00000460291.1; ENST00000461278.1; ENST00000478638.5; ENST00000434066.6 | ||
| External Link | RMBase: m6A_site_577108 | ||
| mod ID: M6ASITE059519 | Click to Show/Hide the Full List | ||
| mod site | chr3:11342205-11342206:+ | [16] | |
| Sequence | GGTTCCTACTTTAGACTTGGACAAGGTTGTGTCTGTCAAAT | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; hESC-HEK293T | ||
| Seq Type List | m6A-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000354449.7; ENST00000461278.1; ENST00000434066.6; ENST00000354956.9; ENST00000478638.5; ENST00000424071.5; ENST00000446450.6; ENST00000460291.1 | ||
| External Link | RMBase: m6A_site_577109 | ||
| mod ID: M6ASITE059521 | Click to Show/Hide the Full List | ||
| mod site | chr3:11346538-11346539:+ | [16] | |
| Sequence | TTTGCAGATTAGCCATTTAAACTGTGTAGCATTCCAGGAAA | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000354449.7; ENST00000424071.5; ENST00000478638.5; ENST00000460291.1; ENST00000354956.9; ENST00000446450.6 | ||
| External Link | RMBase: m6A_site_577110 | ||
| mod ID: M6ASITE059522 | Click to Show/Hide the Full List | ||
| mod site | chr3:11346558-11346559:+ | [16] | |
| Sequence | ACTGTGTAGCATTCCAGGAAACCAAAAGTTAAGTGTGTGCA | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000424071.5; ENST00000460291.1; ENST00000354449.7; ENST00000446450.6; ENST00000354956.9; ENST00000478638.5 | ||
| External Link | RMBase: m6A_site_577111 | ||
| mod ID: M6ASITE059523 | Click to Show/Hide the Full List | ||
| mod site | chr3:11347890-11347891:+ | [16] | |
| Sequence | ACACAGGGTTGGGGCGTGAGACACATCACATTTGTGGACAA | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa; hESC-HEK293T | ||
| Seq Type List | m6A-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000446450.6; ENST00000354449.7; ENST00000424071.5; ENST00000460291.1; ENST00000354956.9; ENST00000478638.5 | ||
| External Link | RMBase: m6A_site_577112 | ||
| mod ID: M6ASITE059524 | Click to Show/Hide the Full List | ||
| mod site | chr3:11347907-11347908:+ | [18] | |
| Sequence | GAGACACATCACATTTGTGGACAATGCCAAGATCTCCTACT | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000478638.5; ENST00000354956.9; ENST00000460291.1; ENST00000424071.5; ENST00000446450.6; ENST00000354449.7 | ||
| External Link | RMBase: m6A_site_577113 | ||
| mod ID: M6ASITE059525 | Click to Show/Hide the Full List | ||
| mod site | chr3:11348006-11348007:+ | [16] | |
| Sequence | CAAGGCTCTGGCAGCAGCGGACCGGCTCCAGAAAATATTCC | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000354449.7; ENST00000354956.9; ENST00000446450.6; ENST00000460291.1; ENST00000424071.5 | ||
| External Link | RMBase: m6A_site_577114 | ||
| mod ID: M6ASITE059526 | Click to Show/Hide the Full List | ||
| mod site | chr3:11358433-11358434:+ | [18] | |
| Sequence | CTAGAATGCCAGAGGATTCAACATGAGCATACCTATGCCTG | ||
| Motif Score | 2.173910714 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000446450.6; ENST00000354956.9; ENST00000354449.7; ENST00000424071.5 | ||
| External Link | RMBase: m6A_site_577115 | ||
| mod ID: M6ASITE059527 | Click to Show/Hide the Full List | ||
| mod site | chr3:11358559-11358560:+ | [18] | |
| Sequence | TGTCGTCTTCCTATTGATGGACACCAGGGAGAGCCGGTGGC | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000446450.6; ENST00000424071.5; ENST00000354449.7; ENST00000354956.9 | ||
| External Link | RMBase: m6A_site_577116 | ||
| mod ID: M6ASITE059528 | Click to Show/Hide the Full List | ||
| mod site | chr3:11360627-11360628:+ | [18] | |
| Sequence | GACACATTTGTTGTCATGAGACATGGTCTGAAGAAACCAAA | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000467121.1; ENST00000354956.9; ENST00000446450.6; ENST00000354449.7 | ||
| External Link | RMBase: m6A_site_577117 | ||
| mod ID: M6ASITE059529 | Click to Show/Hide the Full List | ||
| mod site | chr3:11360725-11360726:+ | [18] | |
| Sequence | GGGCTCATCGCTTTTTGCCAACATCCCTGGTTACAAGCTTG | ||
| Motif Score | 2.173910714 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000467121.1; ENST00000354956.9; ENST00000354449.7; ENST00000446450.6 | ||
| External Link | RMBase: m6A_site_577118 | ||
| mod ID: M6ASITE059530 | Click to Show/Hide the Full List | ||
| mod site | chr3:11380026-11380027:+ | [19] | |
| Sequence | TCCCGTCAGCCTGGCATTTGACAAATGTACAGCTTGTTCTT | ||
| Motif Score | 2.859755952 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000354449.7; ENST00000414717.5; ENST00000446450.6; ENST00000427759.5; ENST00000354956.9; ENST00000446110.1 | ||
| External Link | RMBase: m6A_site_577119 | ||
| mod ID: M6ASITE059532 | Click to Show/Hide the Full List | ||
| mod site | chr3:11426862-11426863:+ | [18] | |
| Sequence | GCCAAGGTGTTTAATTCTTCACATTCCTTCTTAGAAGACTT | ||
| Motif Score | 2.047297619 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000446450.6; ENST00000446110.1; ENST00000427759.5; ENST00000354956.9; ENST00000354449.7; ENST00000414717.5 | ||
| External Link | RMBase: m6A_site_577120 | ||
| mod ID: M6ASITE059533 | Click to Show/Hide the Full List | ||
| mod site | chr3:11426911-11426912:+ | [16] | |
| Sequence | TTACATTGCTGCATCAAGAAACCCAAGCTGCTGAGGTAAGA | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000414717.5; ENST00000446450.6; ENST00000354449.7; ENST00000354956.9; ENST00000446110.1; ENST00000427759.5 | ||
| External Link | RMBase: m6A_site_577121 | ||
| mod ID: M6ASITE059534 | Click to Show/Hide the Full List | ||
| mod site | chr3:11446549-11446550:+ | [16] | |
| Sequence | TTGATACCTACTGTGACAAAACACACATCCAATGACAGTGT | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000414717.5; ENST00000446110.1; ENST00000427759.5; ENST00000354449.7; ENST00000446450.6; ENST00000354956.9 | ||
| External Link | RMBase: m6A_site_577122 | ||
| mod ID: M6ASITE059535 | Click to Show/Hide the Full List | ||
| mod site | chr3:11510298-11510299:+ | [16] | |
| Sequence | TCTACCAGCTCTCAAGCAGAACAATTCCTTGCATTCATACT | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000354956.9; ENST00000354449.7; ENST00000446110.1; ENST00000446450.6 | ||
| External Link | RMBase: m6A_site_577123 | ||
| mod ID: M6ASITE059536 | Click to Show/Hide the Full List | ||
| mod site | chr3:11554817-11554818:+ | [16] | |
| Sequence | CTTCTCCATGCAGATCTGGGACATGAGCGATGATGAGACCA | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000354956.9; ENST00000446450.6; ENST00000354449.7 | ||
| External Link | RMBase: m6A_site_577124 | ||
| mod ID: M6ASITE059537 | Click to Show/Hide the Full List | ||
| mod site | chr3:11554834-11554835:+ | [16] | |
| Sequence | GGGACATGAGCGATGATGAGACCATCTGAGATGGCCCCGCT | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa; Huh7 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000446450.6; ENST00000354449.7; ENST00000354956.9 | ||
| External Link | RMBase: m6A_site_577125 | ||
| mod ID: M6ASITE059538 | Click to Show/Hide the Full List | ||
| mod site | chr3:11554911-11554912:+ | [16] | |
| Sequence | CTCTCCATCGCCAGAGCAGGACTGCTGACCCCAGGCCTGGT | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa; MM6; endometrial | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000354956.9; ENST00000354449.7; ENST00000446450.6 | ||
| External Link | RMBase: m6A_site_577126 | ||
| mod ID: M6ASITE059539 | Click to Show/Hide the Full List | ||
| mod site | chr3:11555082-11555083:+ | [16] | |
| Sequence | GGCCTTGCTATTGACCTGGGACTTGGTCCTCCATGCAGTTT | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa; HEK293T; MM6; HEK293A-TOA; TREX | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000354449.7; ENST00000354956.9; rmsk_888046 | ||
| External Link | RMBase: m6A_site_577127 | ||
| mod ID: M6ASITE059540 | Click to Show/Hide the Full List | ||
| mod site | chr3:11555180-11555181:+ | [16] | |
| Sequence | GAGGGGGTGACCCAACACAGACCAAATGGGGAAATGAGCAA | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa; HEK293T; A549; iSLK; MSC; TIME; TREX | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000354449.7 | ||
| External Link | RMBase: m6A_site_577128 | ||
| mod ID: M6ASITE059541 | Click to Show/Hide the Full List | ||
| mod site | chr3:11555312-11555313:+ | [16] | |
| Sequence | TGGGCATGGGTGAGGGTGGGACCCCGTGAGCGCACTGCACC | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; A549; MM6; CD4T; peripheral-blood; HEK293T; HEK293A-TOA; TREX; iSLK; endometrial; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000354449.7 | ||
| External Link | RMBase: m6A_site_577129 | ||
| mod ID: M6ASITE059542 | Click to Show/Hide the Full List | ||
| mod site | chr3:11555381-11555382:+ | [16] | |
| Sequence | AGAGCCGAGCTGGGTACGAGACTAAAGGGCCCACATGACCC | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa; A549; H1A; MM6; CD4T; peripheral-blood; HEK293T; HEK293A-TOA; iSLK; TIME; TREX; endometrial; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000354449.7 | ||
| External Link | RMBase: m6A_site_577130 | ||
| mod ID: M6ASITE059543 | Click to Show/Hide the Full List | ||
| mod site | chr3:11555426-11555427:+ | [16] | |
| Sequence | ACGCCAGATTTCCACCAAGGACTGAGTGAGCTGCTCAGACA | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa; H1A; MM6; CD4T; peripheral-blood; GSC-11; HEK293T; HEK293A-TOA; iSLK; TIME; TREX; MSC; endometrial; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000354449.7 | ||
| External Link | RMBase: m6A_site_577131 | ||
| mod ID: M6ASITE059544 | Click to Show/Hide the Full List | ||
| mod site | chr3:11555444-11555445:+ | [16] | |
| Sequence | GGACTGAGTGAGCTGCTCAGACATGGCTTTCTGCCTCCCAG | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa; H1A; MM6; CD4T; peripheral-blood; GSC-11; HEK293T; HEK293A-TOA; iSLK; TIME; TREX; MSC; endometrial; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000354449.7 | ||
| External Link | RMBase: m6A_site_577132 | ||
| mod ID: M6ASITE059545 | Click to Show/Hide the Full List | ||
| mod site | chr3:11555995-11555996:+ | [16] | |
| Sequence | CTCATGGGAGCTTCATGGGGACACAGCCGGCACAGGTGCAG | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000354449.7 | ||
| External Link | RMBase: m6A_site_577133 | ||
| mod ID: M6ASITE059546 | Click to Show/Hide the Full List | ||
| mod site | chr3:11556248-11556249:+ | [16] | |
| Sequence | GCAGCGCGGCTCTGGGAAGAACTTCACGGAGCCCCTTCTTA | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa; HepG2; H1A; H1B; MM6; Jurkat; peripheral-blood; GSC-11; HEK293T; endometrial; HEC-1-A; NB4 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000354449.7 | ||
| External Link | RMBase: m6A_site_577136 | ||
| mod ID: M6ASITE059547 | Click to Show/Hide the Full List | ||
| mod site | chr3:11556377-11556378:+ | [16] | |
| Sequence | GATAGGTCAGTGCACCAGGGACCCGGCCGCCAGCACCGCCG | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; HepG2; H1A; H1B; MM6; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293T; endometrial; HEC-1-A; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000354449.7 | ||
| External Link | RMBase: m6A_site_577139 | ||
| mod ID: M6ASITE059549 | Click to Show/Hide the Full List | ||
| mod site | chr3:11556437-11556438:+ | [16] | |
| Sequence | CCTTGTTCACTGACAAAGAGACCTGTCCCAGGAGTGTCCTC | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa; HepG2; H1A; H1B; A549; MM6; Jurkat; endometrial | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000354449.7 | ||
| External Link | RMBase: m6A_site_577141 | ||
| mod ID: M6ASITE059550 | Click to Show/Hide the Full List | ||
| mod site | chr3:11556519-11556520:+ | [20] | |
| Sequence | CTCAGTAGCCTGTAGCAATAACAAACTCGTGGCTATGAATG | ||
| Motif Score | 2.168095238 | ||
| Cell/Tissue List | kidney; hESC-HEK293T | ||
| Seq Type List | m6A-REF-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000354449.7 | ||
| External Link | RMBase: m6A_site_577147 | ||
| mod ID: M6ASITE059551 | Click to Show/Hide the Full List | ||
| mod site | chr3:11556523-11556524:+ | [16] | |
| Sequence | GTAGCCTGTAGCAATAACAAACTCGTGGCTATGAATGCAGA | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa; A549 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000354449.7 | ||
| External Link | RMBase: m6A_site_577148 | ||
| mod ID: M6ASITE059552 | Click to Show/Hide the Full List | ||
| mod site | chr3:11556596-11556597:+ | [20] | |
| Sequence | CTTTTACAGACAAATCTACGACAAAAAAAAAGATCAACTTT | ||
| Motif Score | 2.865571429 | ||
| Cell/Tissue List | liver | ||
| Seq Type List | m6A-REF-seq | ||
| Transcript ID List | ENST00000354449.7 | ||
| External Link | RMBase: m6A_site_577152 | ||
| mod ID: M6ASITE059553 | Click to Show/Hide the Full List | ||
| mod site | chr3:11556689-11556690:+ | [19] | |
| Sequence | TAAATAGCTACATATCATTAACAAATTAATGTTCTTCAAAA | ||
| Motif Score | 2.168095238 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000354449.7 | ||
| External Link | RMBase: m6A_site_577153 | ||
| mod ID: M6ASITE059554 | Click to Show/Hide the Full List | ||
| mod site | chr3:11556932-11556933:+ | [20] | |
| Sequence | CCTGCAGCCACTCTGTGACTACAAGAGCCAGTCCTCCGACC | ||
| Motif Score | 2.078666667 | ||
| Cell/Tissue List | brain | ||
| Seq Type List | m6A-REF-seq | ||
| Transcript ID List | ENST00000354449.7 | ||
| External Link | RMBase: m6A_site_577158 | ||
| mod ID: M6ASITE059555 | Click to Show/Hide the Full List | ||
| mod site | chr3:11557257-11557258:+ | [20] | |
| Sequence | AAATAATAAATGCAGTAATAACAGTATAAAGTCAGAGGAAT | ||
| Motif Score | 2.168095238 | ||
| Cell/Tissue List | brain; HEK293 | ||
| Seq Type List | m6A-REF-seq | ||
| Transcript ID List | ENST00000354449.7 | ||
| External Link | RMBase: m6A_site_577166 | ||
References