m6A Regulator Information
General Information of the m6A Regulator (ID: REG00024)
| Regulator Name | YTH domain-containing family protein 1 (YTHDF1) | ||||
|---|---|---|---|---|---|
| Synonyms |
DF1; Dermatomyositis associated with cancer putative autoantigen 1; DACA-1; C20orf21
Click to Show/Hide
|
||||
| Gene Name | YTHDF1 | ||||
| Sequence |
MSATSVDTQRTKGQDNKVQNGSLHQKDTVHDNDFEPYLTGQSNQSNSYPSMSDPYLSSYY
PPSIGFPYSLNEAPWSTAGDPPIPYLTTYGQLSNGDHHFMHDAVFGQPGGLGNNIYQHRF NFFPENPAFSAWGTSGSQGQQTQSSAYGSSYTYPPSSLGGTVVDGQPGFHSDTLSKAPGM NSLEQGMVGLKIGDVSSSAVKTVGSVVSSVALTGVLSGNGGTNVNMPVSKPTSWAAIASK PAKPQPKMKTKSGPVMGGGLPPPPIKHNMDIGTWDNKGPVPKAPVPQQAPSPQAAPQPQQ VAQPLPAQPPALAQPQYQSPQQPPQTRWVAPRNRNAAFGQSGGAGSDSNSPGNVQPNSAP SVESHPVLEKLKAAHSYNPKEFEWNLKSGRVFIIKSYSEDDIHRSIKYSIWCSTEHGNKR LDSAFRCMSSKGPVYLLFSVNGSGHFCGVAEMKSPVDYGTSAGVWSQDKWKGKFDVQWIF VKDVPNNQLRHIRLENNDNKPVTNSRDTQEVPLEKAKQVLKIISSYKHTTSIFDDFAHYE KRQEEEEVVRKERQSRNKQ Click to Show/Hide
|
||||
| Family | YTHDF family; YTHDF1 subfamily | ||||
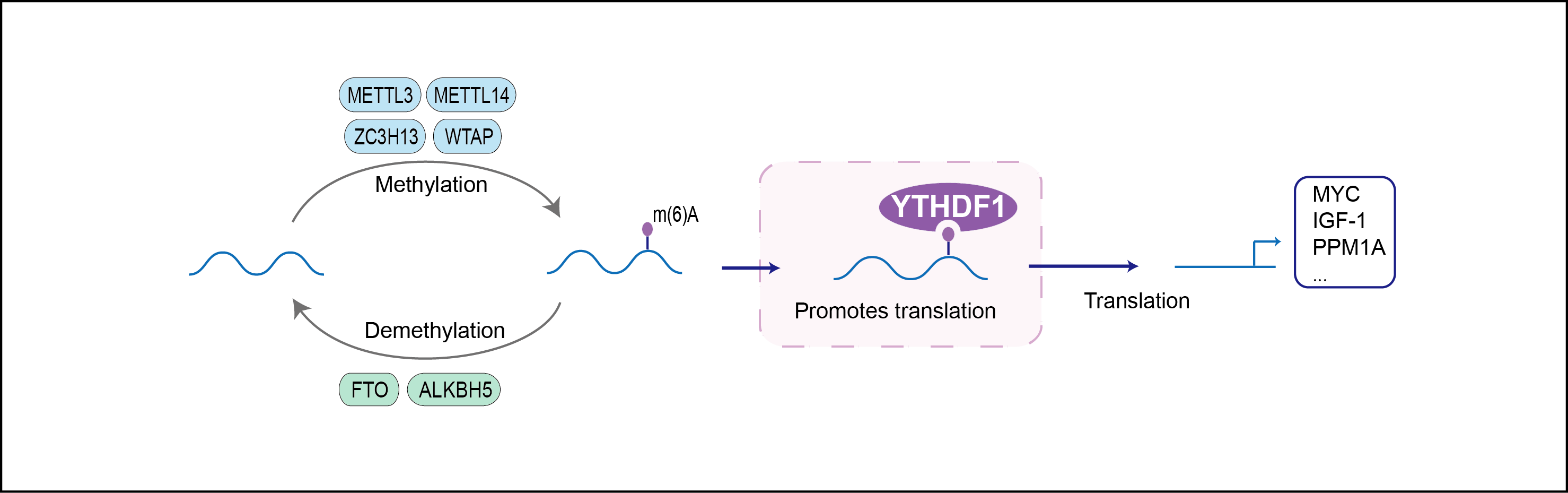
| Function |
Specifically recognizes and binds N6-methyladenosine (m6A)-containing mRNAs, and regulates their stability. M6A is a modification present at internal sites of mRNAs and some non-coding RNAs and plays a role in mRNA stability and processing. Acts as a regulator of mRNA stability by promoting degradation of m6A-containing mRNAs via interaction with the CCR4-NOT complex. The YTHDF paralogs (YTHDF1, YTHDF2 and YTHDF3) shares m6A-containing mRNAs targets and act redundantly to mediate mRNA degradation and cellular differentiation. Required to facilitate learning and memory formation in the hippocampus by binding to m6A-containing neuronal mRNAs (By similarity). Acts as a regulator of axon guidance by binding to m6A-containing ROBO3 transcripts (By similarity). Acts as a negative regulator of antigen cross-presentation in myeloid dendritic cells (By similarity). In the context of tumorigenesis, negative regulation of antigen cross-presentation limits the anti-tumor response by reducing efficiency of tumor-antigen cross-presentation (By similarity). Promotes formation of phase-separated membraneless compartments, such as P-bodies or stress granules, by undergoing liquid-liquid phase separation upon binding to mRNAs containing multiple m6A-modified residues: polymethylated mRNAs act as a multivalent scaffold for the binding of YTHDF proteins, juxtaposing their disordered regions and thereby leading to phase separation. The resulting mRNA-YTHDF complexes then partition into different endogenous phase-separated membraneless compartments, such as P-bodies, stress granules or neuronal RNA granules.
Click to Show/Hide
|
||||
| Gene ID | 54915 | ||||
| Uniprot ID | |||||
| Regulator Type | WRITER ERASER READER | ||||
| Mechanism Diagram | Click to View the Original Diagram | ||||
|
|||||
| Target Genes | Click to View Potential Target Genes of This Regulator | ||||
Full List of Target Gene(s) of This m6A Regulator and Corresponding Disease/Drug Response(s)
YTHDF1 can regulate the m6A methylation of following target genes, and result in corresponding disease/drug response(s). You can browse corresponding disease or drug response(s) resulted from the regulation of certain target gene.
Browse Target Gene related Disease
Browse Target Gene related Drug
Ephrin type-B receptor 2 (ERK/EPHB2)
| Representative RNA-seq result indicating the expression of this target gene regulated by YTHDF1 | ||
| Cell Line | AGS cell line | Homo sapiens |
|
Treatment: shYTHDF1 AGS
Control: shControl AGS
|
GSE159425 | |
| Regulation |
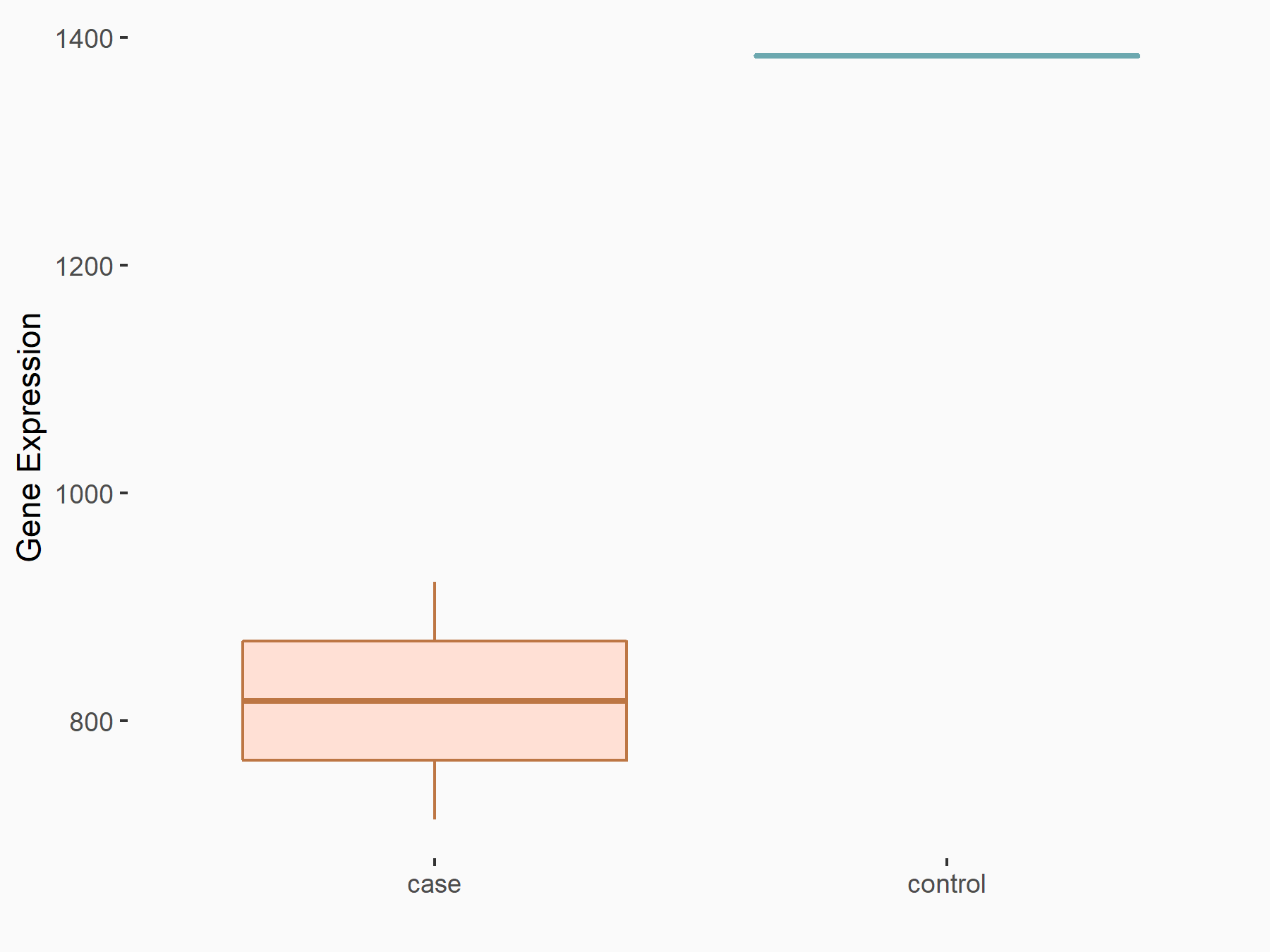  |
logFC: -7.59E-01 p-value: 2.20E-04 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and YTHDF1 | ||
| Cell Line | Hela | Homo sapiens |
| Regulation | logFC: 1.91E+00 | GSE63591 |
Muscular dystrophies [ICD-11: 8C70]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [1] | |||
| Responsed Disease | Muscular dystrophies [ICD-11: 8C70] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | MAPK signaling pathway | hsa04010 | ||
In-vitro Model |
HEK293T | Normal | Homo sapiens | CVCL_0063 |
| C2C12 | Normal | Mus musculus | CVCL_0188 | |
| In-vivo Model | For mouse muscle injury and regeneration experiment, tibialis anterior (TA) muscles of 6-week-old male mice were injected with 25 uL of 10 uM cardiotoxin (CTX, Merck Millipore, 217503), 0.9% normal saline (Saline) were used as control. The regenerated muscles were collected at day 1, 3, 5, and 10 post-injection. TA muscles were isolated for Hematoxylin and eosin staining or frozen in liquid nitrogen for RNA and protein extraction. | |||
| Response Summary | m6A writers METTL3/METTL14 and the m6A reader YTHDF1 orchestrate MNK2 expression posttranscriptionally and thus control Ephrin type-B receptor 2 (ERK/EPHB2) signaling, which is required for the maintenance of muscle myogenesis and contribute to regeneration. | |||
Epidermal growth factor receptor (EGFR)
| Representative RNA-seq result indicating the expression of this target gene regulated by YTHDF1 | ||
| Cell Line | AGS cell line | Homo sapiens |
|
Treatment: shYTHDF1 AGS
Control: shControl AGS
|
GSE159425 | |
| Regulation |
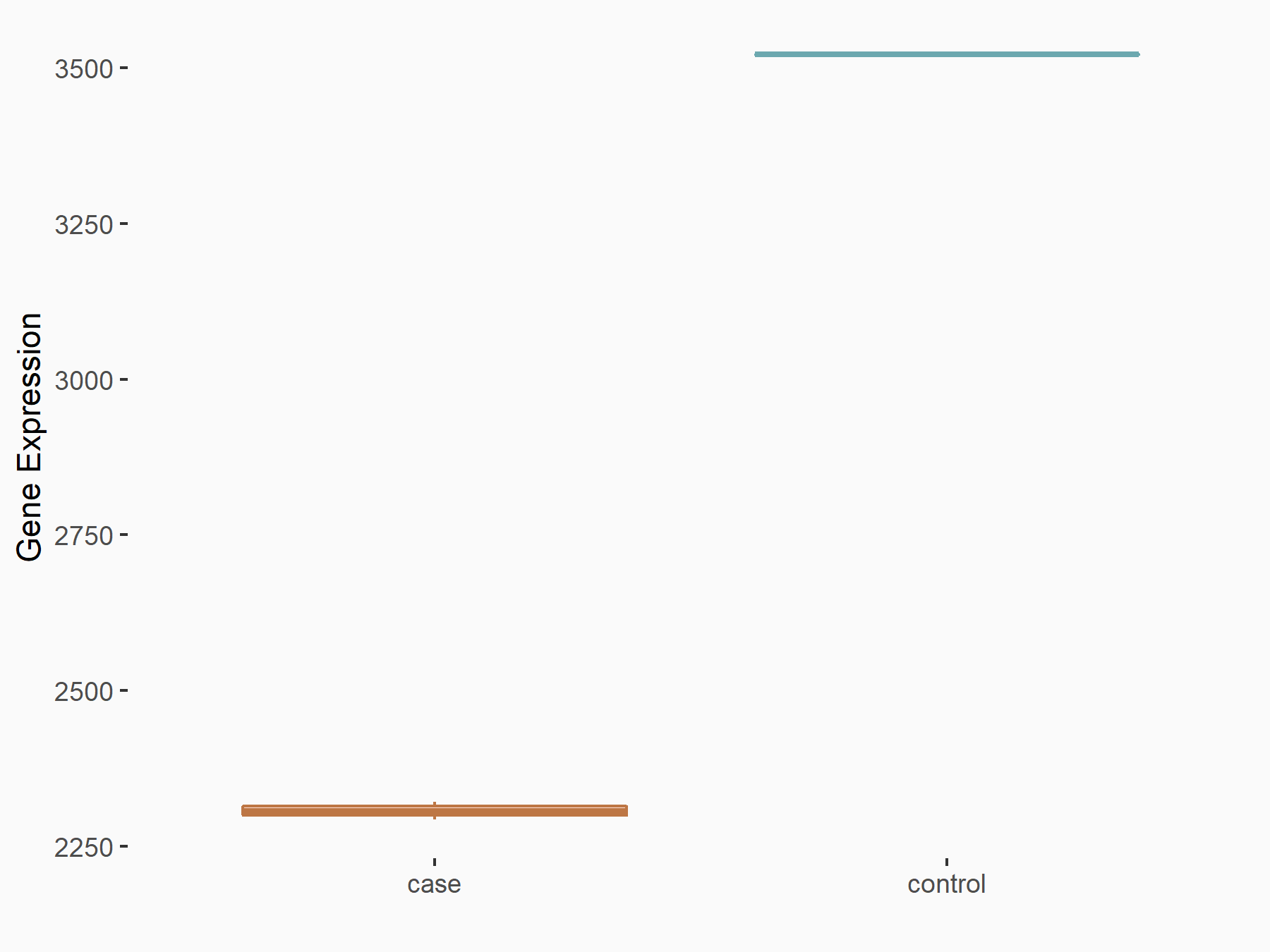 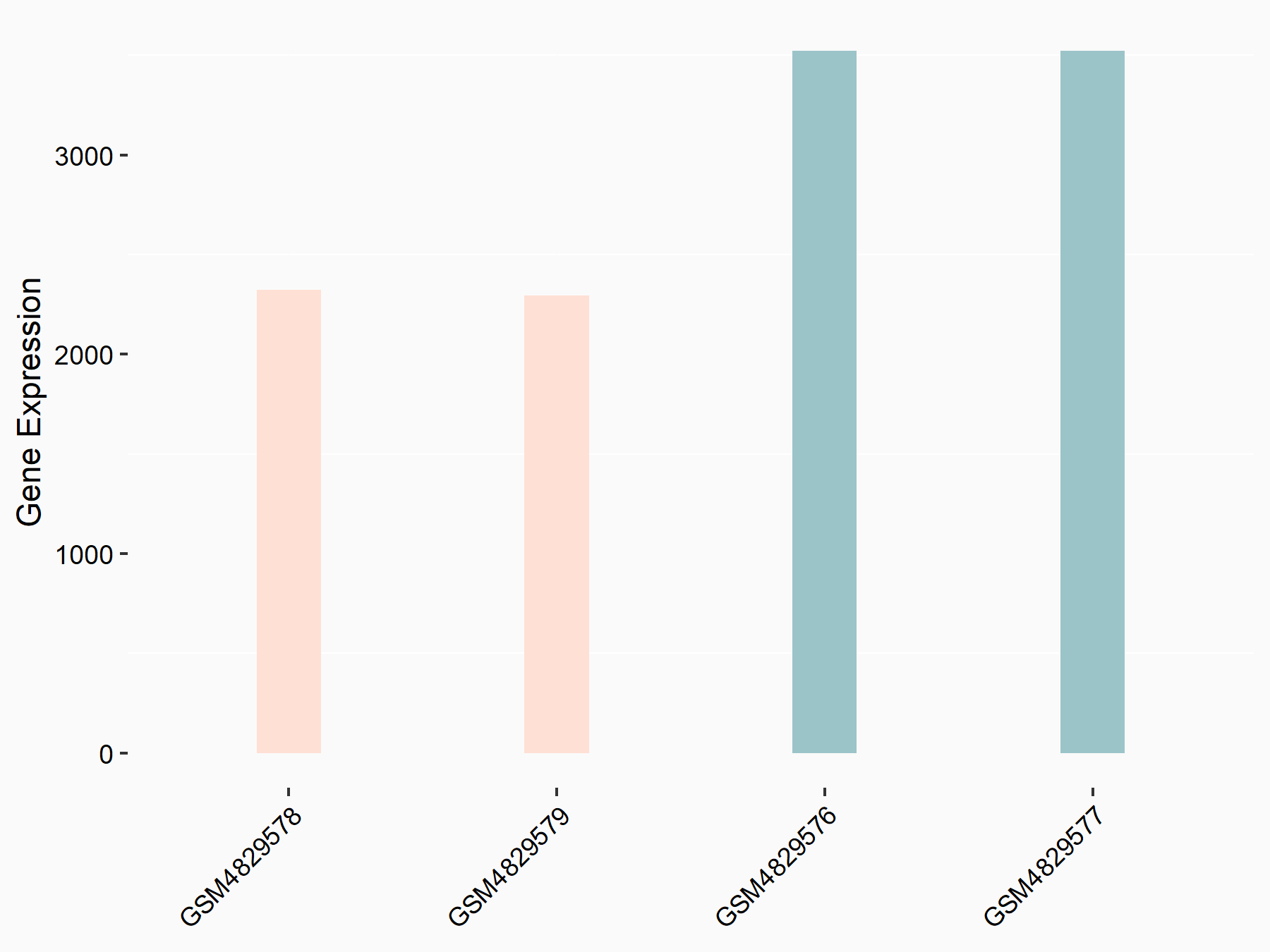 |
logFC: -6.10E-01 p-value: 2.21E-04 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and YTHDF1 | ||
| Cell Line | Hela | Homo sapiens |
| Regulation | logFC: 2.78E+00 | GSE63591 |
Liver cancer [ICD-11: 2C12]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [2] | |||
| Responsed Disease | Intrahepatic cholangiocarcinoma [ICD-11: 2C12.10] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | PI3K-Akt signaling pathway | hsa04151 | ||
| Cell Process | Cell proliferation | |||
| Cell migration | ||||
| Cell nvasion | ||||
In-vitro Model |
HuCC-T1 | Intrahepatic cholangiocarcinoma | Homo sapiens | CVCL_0324 |
| RBE | Intrahepatic cholangiocarcinoma | Homo sapiens | CVCL_4896 | |
| In-vivo Model | For the subcutaneous implantation ICC mouse model, 6-week-old male NCG mice (Jiangsu, China) were randomly enrolled into shNC group and shYTHDF1 group (n = 9); 1 × 106 HuCCT1 cells in 0.1-mL PBS transfected with shNC or shYTHDF1 were subcutaneously inoculated in the right flanks of the mice. For AKT/YapS127A-induced orthotopic ICC mouse model, 16 mice were divided into two groups randomly. For the control group, 20-ug AKT, 30-ug Yap, and 2-ug pCMV/SB plasmids plus 20-ug vector plasmids as control were diluted in 2-mL saline and then were injected into the lateral tail vein within 7 s. For the YTHDF1-overexpressed group, mice were injected with additional 20-ug YTHDF1 plasmids under the same conditions. Mice were sacrificed at 4 weeks after injection, and liver tissues were harvested for analysis. | |||
| Response Summary | YTHDF1 regulates the translation of Epidermal growth factor receptor (EGFR) mRNA via binding m6 A sites in the 3'-UTR of EGFR transcript. YTHDF1 is upregulated in ICC and associated with shorter survival of ICC patients. | |||
Forkhead box protein O3 (FOXO3)
| Representative RNA-seq result indicating the expression of this target gene regulated by YTHDF1 | ||
| Cell Line | AGS cell line | Homo sapiens |
|
Treatment: shYTHDF1 AGS
Control: shNC AGS
|
GSE166972 | |
| Regulation |
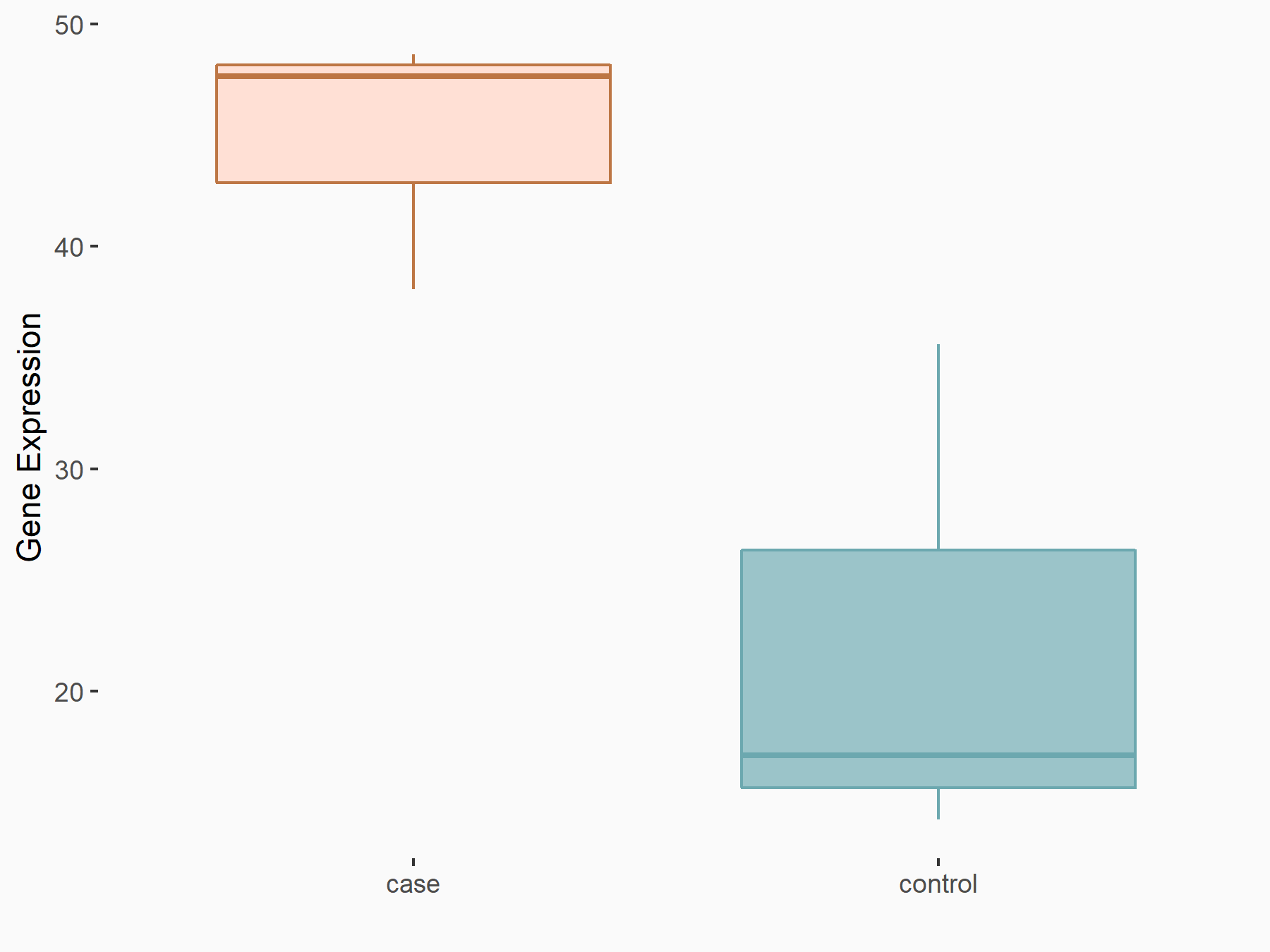 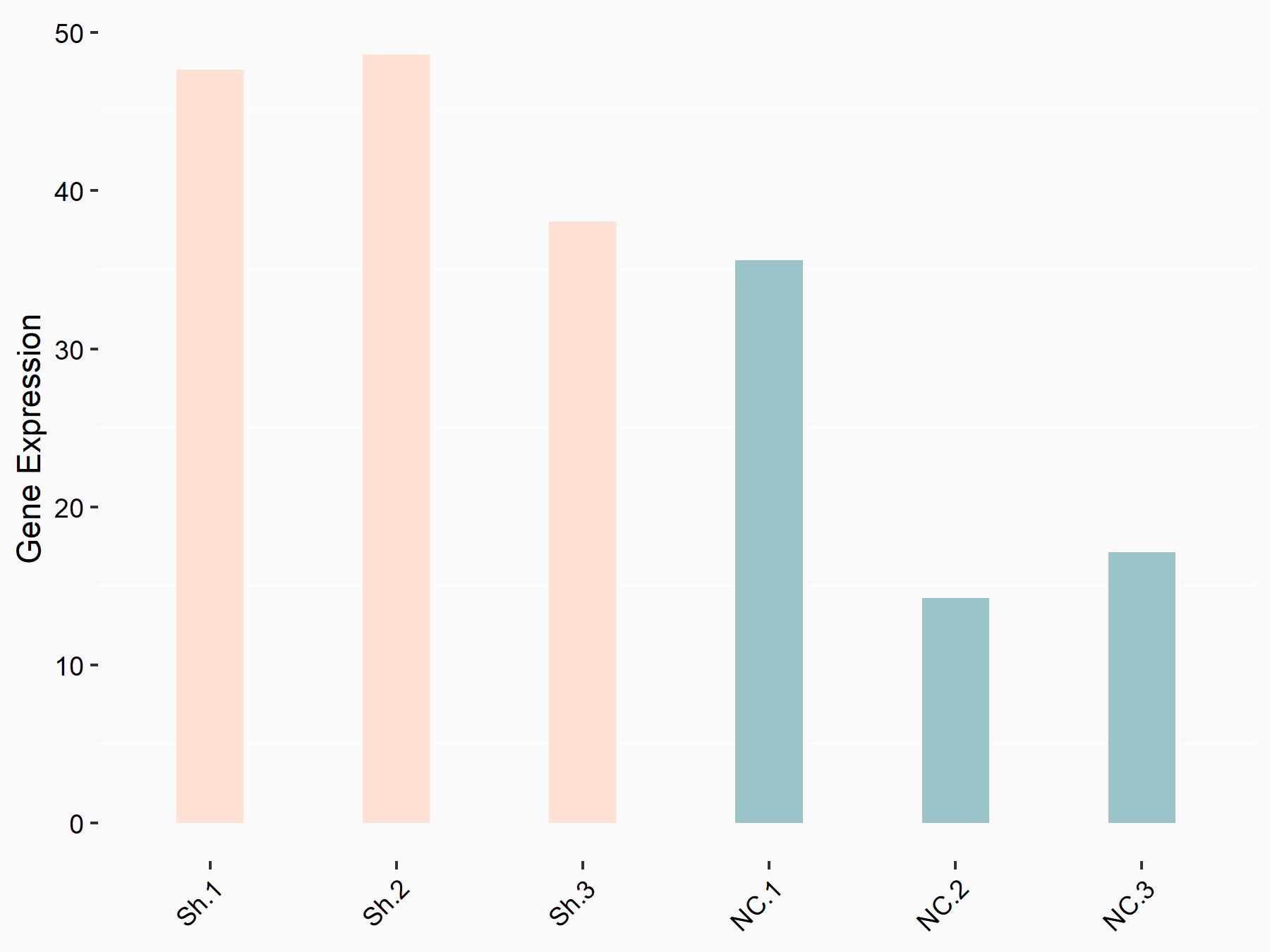 |
logFC: 1.07E+00 p-value: 3.17E-02 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and YTHDF1 | ||
| Cell Line | Hela | Homo sapiens |
| Regulation | logFC: 2.44E+00 | GSE63591 |
Liver cancer [ICD-11: 2C12]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [4] | |||
| Responsed Disease | Hepatocellular carcinoma [ICD-11: 2C12.02] | |||
| Responsed Drug | Sorafenib | Approved | ||
| Target Regulation | Up regulation | |||
| Pathway Response | FoxO signaling pathway | hsa04068 | ||
| Cell Process | Cell autophagy | |||
| Response Summary | METTL3 can sensitise hepatocellular carcinoma cells to sorafenib through stabilising Forkhead box protein O3 (FOXO3) in an m6A-dependent manner and translated by YTHDF1, thereby inhibiting the transcription of autophagy-related genes, including ATG3, ATG5, ATG7, ATG12, and ATG16L1. | |||
Injury of heart [ICD-11: NB31]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [48] | |||
| Responsed Disease | Myocardial injury [ICD-11: NB31.Z] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
H9c2(2-1) | Normal | Rattus norvegicus | CVCL_0286 |
| In-vivo Model | For exercise-trained rats, SD rats were treadmill trained 5 days per week continuing six weeks. In brief, exercise was carried out on a motor-driven treadmill, set at a 10.5% incline, 5 days per week for 6 weeks in an adjoining room maintained at 20 °C. Running duration and speed were gradually increased more than 22 days to 60 min at 30 m/min, corresponding to about 80% VO2 max, and maintained for 2-3 weeks. | |||
Hexokinase-2 (HK2)
| Representative RNA-seq result indicating the expression of this target gene regulated by YTHDF1 | ||
| Cell Line | AGS cell line | Homo sapiens |
|
Treatment: shYTHDF1 AGS
Control: shControl AGS
|
GSE159425 | |
| Regulation |
 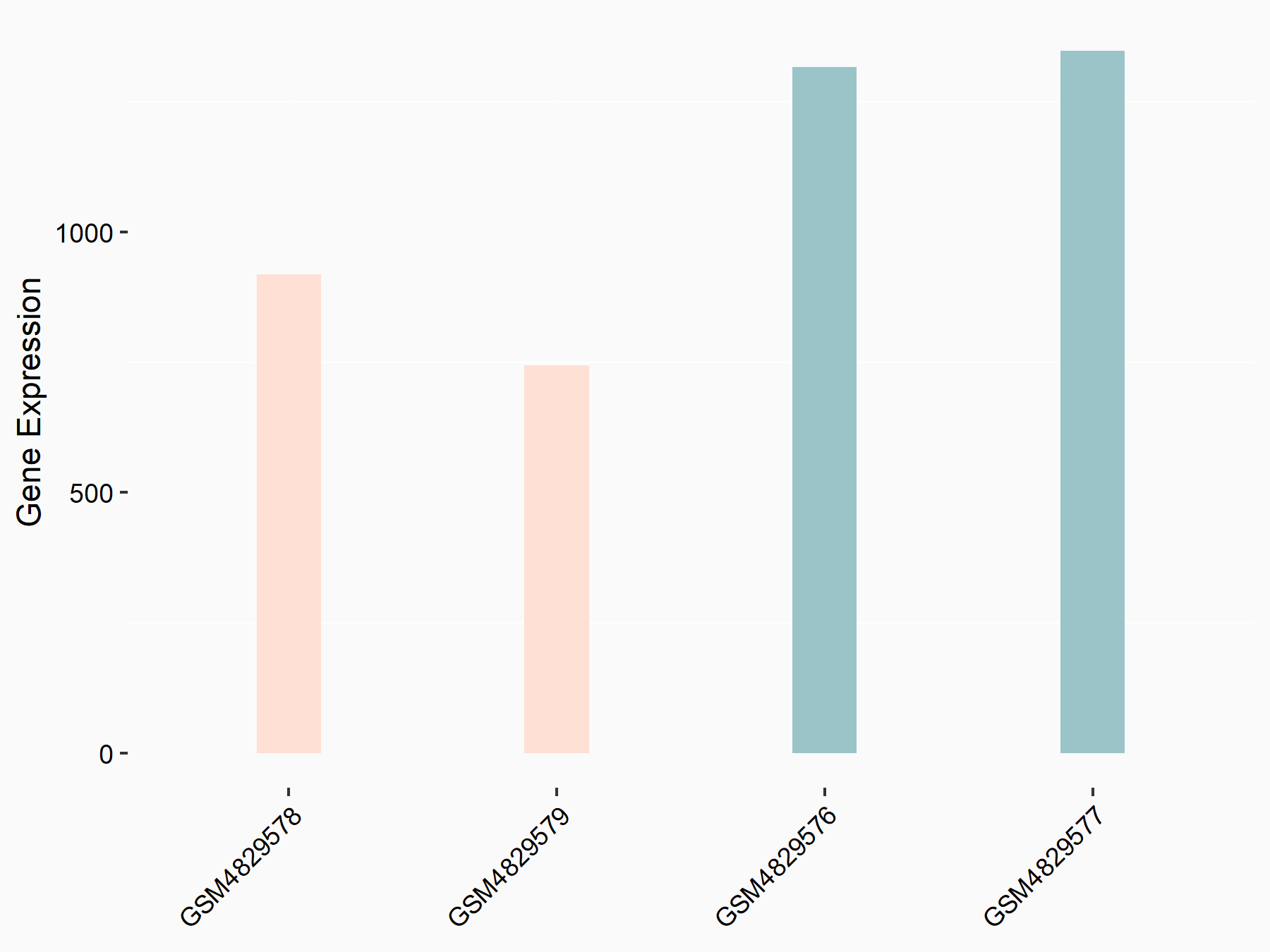 |
logFC: -6.80E-01 p-value: 6.28E-04 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and YTHDF1 | ||
| Cell Line | Hela | Homo sapiens |
| Regulation | logFC: 1.46E+00 | GSE63591 |
Cervical cancer [ICD-11: 2C77]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [5] | |||
| Responsed Disease | Cervical cancer [ICD-11: 2C77] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | Glycolysis / Gluconeogenesis | hsa00010), Central carbon metabolism in cancer | ||
| Cell Process | Glycolysis | |||
In-vitro Model |
SiHa | Cervical squamous cell carcinoma | Homo sapiens | CVCL_0032 |
| HT-3 | Cervical carcinoma | Homo sapiens | CVCL_1293 | |
| Ca Ski | Cervical squamous cell carcinoma | Homo sapiens | CVCL_1100 | |
| C-33 A | Cervical squamous cell carcinoma | Homo sapiens | CVCL_1094 | |
| In-vivo Model | Five-week-old male nude BALB/C mice were applied for this animal studies and fed with certified standard diet and tap water ad libitum in a light/dark cycle of 12 h on/12 h off.The assay was performed in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals. Stable transfection of METTL3 knockdown (sh-METTL3) or negative control (sh-blank) in SiHa cells (5 × 106 cells per 0.1 mL) were injected into the flank of mice. | |||
| Response Summary | METTL3 enhanced the Hexokinase-2 (HK2) stability through YTHDF1-mediated m6A modification, thereby promoting the Warburg effect of CC, which promotes a novel insight for the CC treatment. | |||
Esophageal cancer [ICD-11: 2B70]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [51] | |||
| Responsed Disease | Esophageal Squamous Cell Carcinoma [ICD-11: 2B70.1] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
HET-1A | Normal | Homo sapiens | CVCL_3702 |
| Eca-109 | Esophageal squamous cell carcinoma | Homo sapiens | CVCL_6898 | |
| TE-10 | Esophageal squamous cell carcinoma | Homo sapiens | CVCL_1760 | |
| In-vivo Model | Six-week-old male nude BALB/C mice were randomly divided into two groups, NC and sh-HCP5#1 groups. 2 × 106 EC109 cells were subcutaneously injected into nude mice and grown for 5 weeks. | |||
Integrin alpha-6 (ITGA6)
| Representative RNA-seq result indicating the expression of this target gene regulated by YTHDF1 | ||
| Cell Line | AGS cell line | Homo sapiens |
|
Treatment: shYTHDF1 AGS
Control: shControl AGS
|
GSE159425 | |
| Regulation |
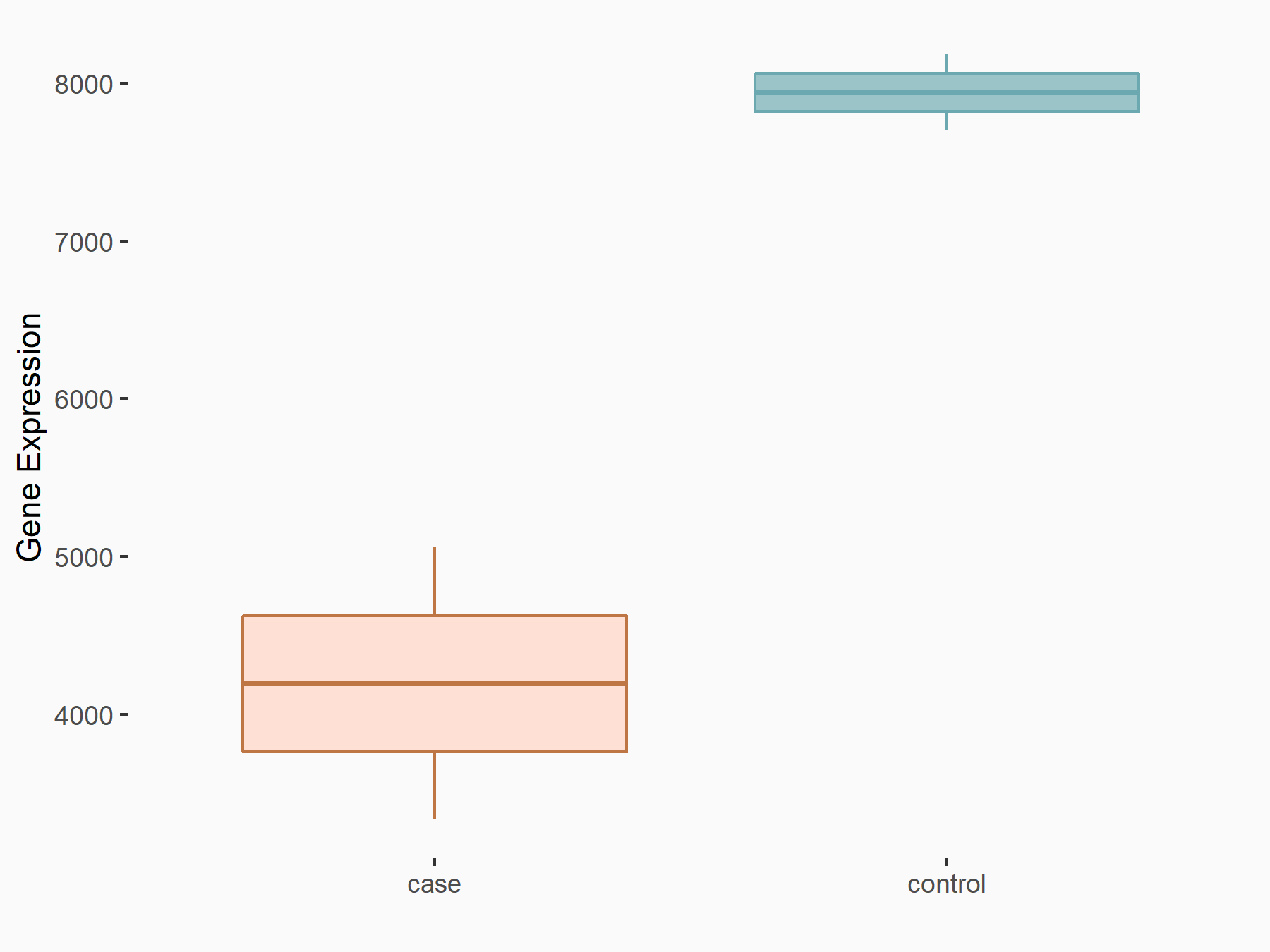 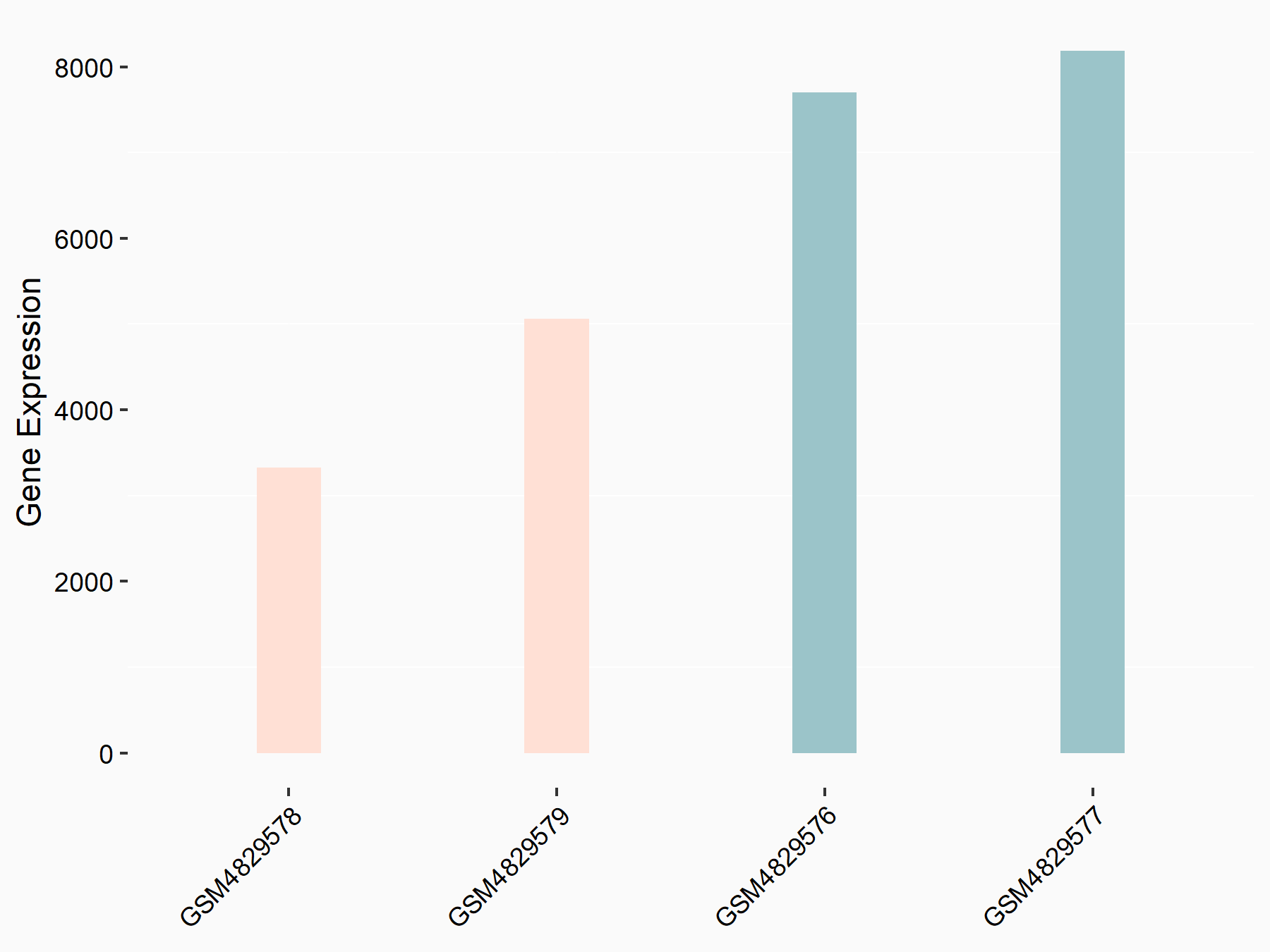 |
logFC: -9.21E-01 p-value: 3.27E-05 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and YTHDF1 | ||
| Cell Line | Hela | Homo sapiens |
| Regulation | logFC: 2.14E+00 | GSE63591 |
Bladder cancer [ICD-11: 2C94]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [6] | |||
| Responsed Disease | Bladder cancer [ICD-11: 2C94] | |||
| Pathway Response | Cell adhesion molecules | hsa04514 | ||
| Cell Process | Cell adhesion | |||
| Cell migration | ||||
| Cell invasion | ||||
In-vitro Model |
5637 | Bladder carcinoma | Homo sapiens | CVCL_0126 |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| J82 | Bladder carcinoma | Homo sapiens | CVCL_0359 | |
| SV-HUC-1 | Normal | Homo sapiens | CVCL_3798 | |
| T24 | Bladder carcinoma | Homo sapiens | CVCL_0554 | |
| UM-UC-3 | Bladder carcinoma | Homo sapiens | CVCL_1783 | |
| In-vivo Model | For the subcutaneous implantation model, 1 × 107 cells were subcutaneously implanted into 5-week-old BALB/cJNju-Foxn1nu/Nju nude mice. | |||
| Response Summary | m6A writer METTL3 and eraser ALKBH5 altered cell adhesion by regulating Integrin alpha-6 (ITGA6) expression in bladder cancer cells. m6A is highly enriched within the ITGA6 transcripts, and increased m6A methylations of the ITGA6 mRNA 3'UTR promotes the translation of ITGA6 mRNA via binding of the m6A readers YTHDF1 and YTHDF3. Inhibition of ITGA6 results in decreased growth and progression of bladder cancer cells in vitro and in vivo. | |||
Kelch-like ECH-associated protein 1 (KEAP1)
| Representative RNA-seq result indicating the expression of this target gene regulated by YTHDF1 | ||
| Cell Line | AGS cell line | Homo sapiens |
|
Treatment: shYTHDF1 AGS
Control: shControl AGS
|
GSE159425 | |
| Regulation |
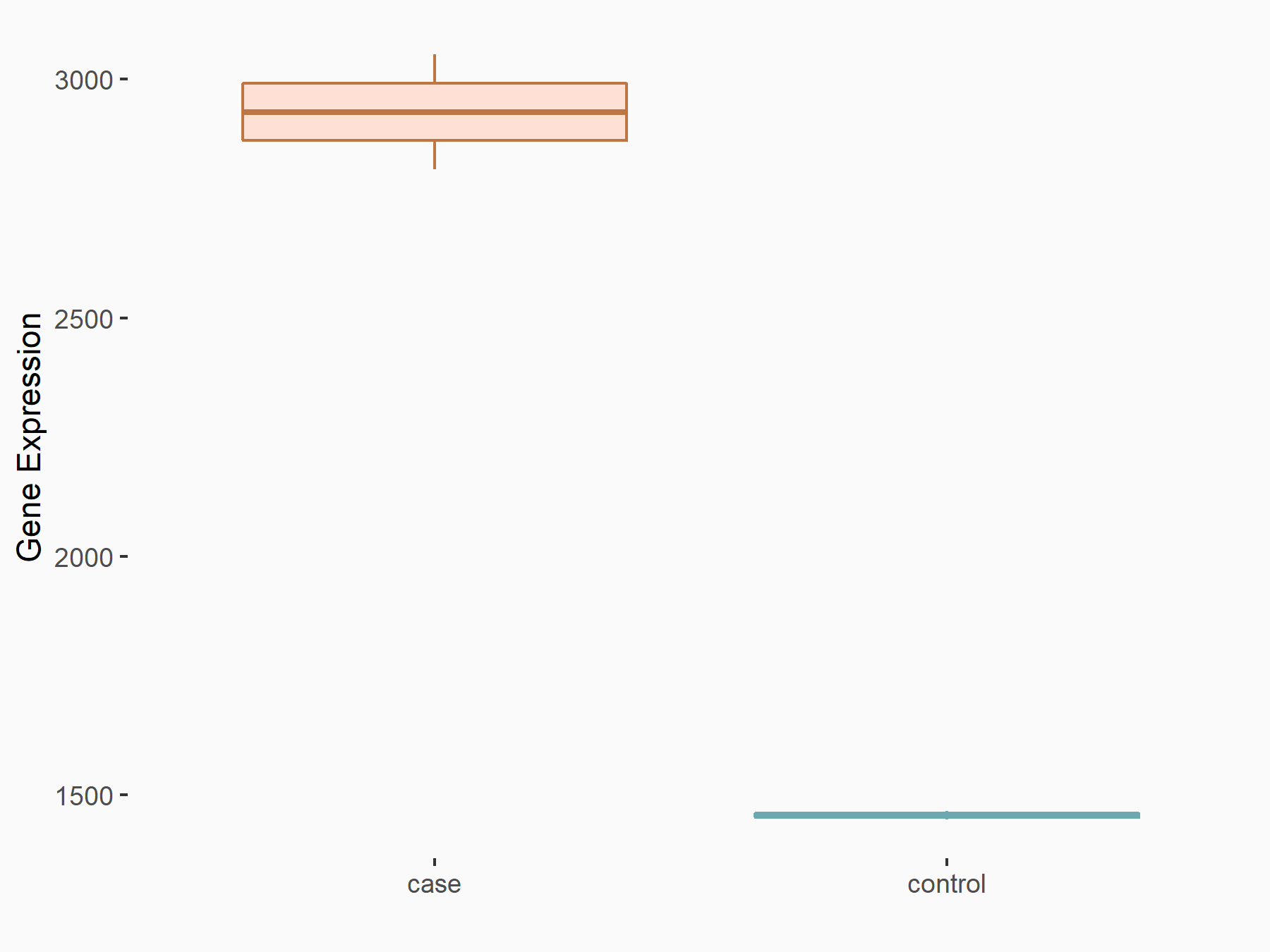 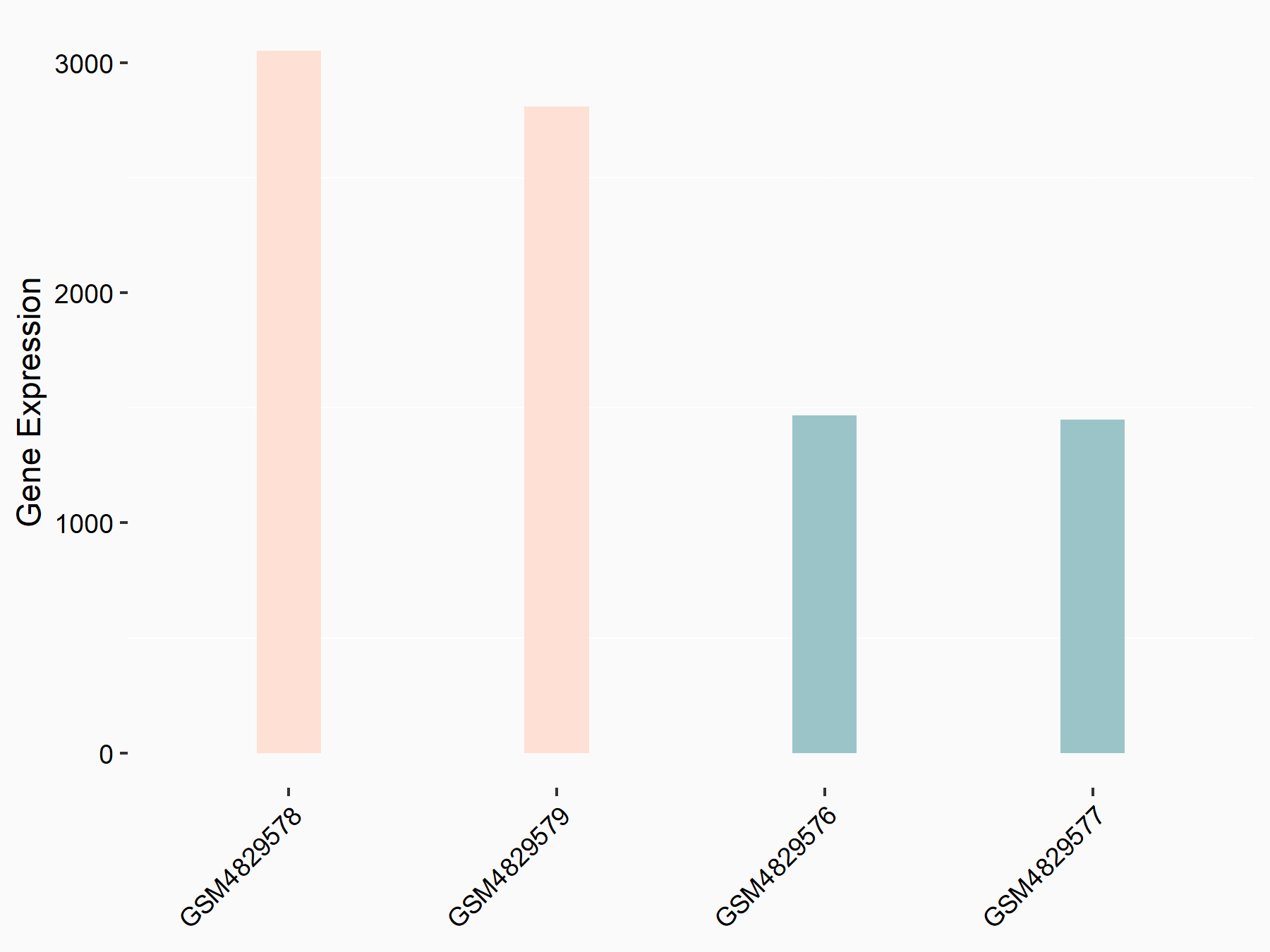 |
logFC: 1.01E+00 p-value: 4.73E-09 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and YTHDF1 | ||
| Cell Line | Hela | Homo sapiens |
| Regulation | logFC: 1.61E+00 | GSE63591 |
Lung cancer [ICD-11: 2C25]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [7] | |||
| Responsed Disease | Non-small-cell lung carcinoma [ICD-11: 2C25.Y] | |||
| Responsed Drug | Cisplatin | Approved | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Chemical carcinogenesis - reactive oxygen species | hsa05208 | ||
| Cell cycle | hsa04110 | |||
| Cell Process | Biological regulation | |||
In-vitro Model |
A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 |
| A549-DDP (Human lung adenocarcinoma is resistant to cisplatin) | ||||
| GLC-82 | Endocervical adenocarcinoma | Homo sapiens | CVCL_3371 | |
| NCI-H1299 | Lung large cell carcinoma | Homo sapiens | CVCL_0060 | |
| NCI-H1975 | Lung adenocarcinoma | Homo sapiens | CVCL_1511 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| NCI-H1650 | Minimally invasive lung adenocarcinoma | Homo sapiens | CVCL_1483 | |
| NCI-H838 | Lung adenocarcinoma | Homo sapiens | CVCL_1594 | |
| SPC-A1 | Endocervical adenocarcinoma | Homo sapiens | CVCL_6955 | |
| In-vivo Model | Mice were treated via nasal inhalation of adenovirus carrying Cre recombinase (5 × 106 p.f.u for Ad-Cre, Biowit Inc., Shenzhen, Guangdong), and were then killed at indicated times for gross inspection and histopathological examination. | |||
| Response Summary | YTHDF1 deficiency inhibits Non-small cell lung cancer cell proliferation and xenograft tumor formation through regulating the translational efficiency of CDK2, CDK4, p27, and cyclin D1, and that YTHDF1 depletion restrains de novo lung adenocarcinomas (ADC) progression. Mechanistic studies identified the Kelch-like ECH-associated protein 1 (KEAP1)-Nrf2-AKR1C1 axis as the downstream mediator of YTHDF1. YTHDF1 high expression correlates with better clinical outcome, with its depletion rendering cancerous cells resistant to cisplatin (DDP) treatment. | |||
Myc proto-oncogene protein (MYC)
| Representative RNA-seq result indicating the expression of this target gene regulated by YTHDF1 | ||
| Cell Line | Embryonic stem cells | Mus musculus |
|
Treatment: YTHDF1-/- mESCs
Control: Wild type ESCs
|
GSE147849 | |
| Regulation |
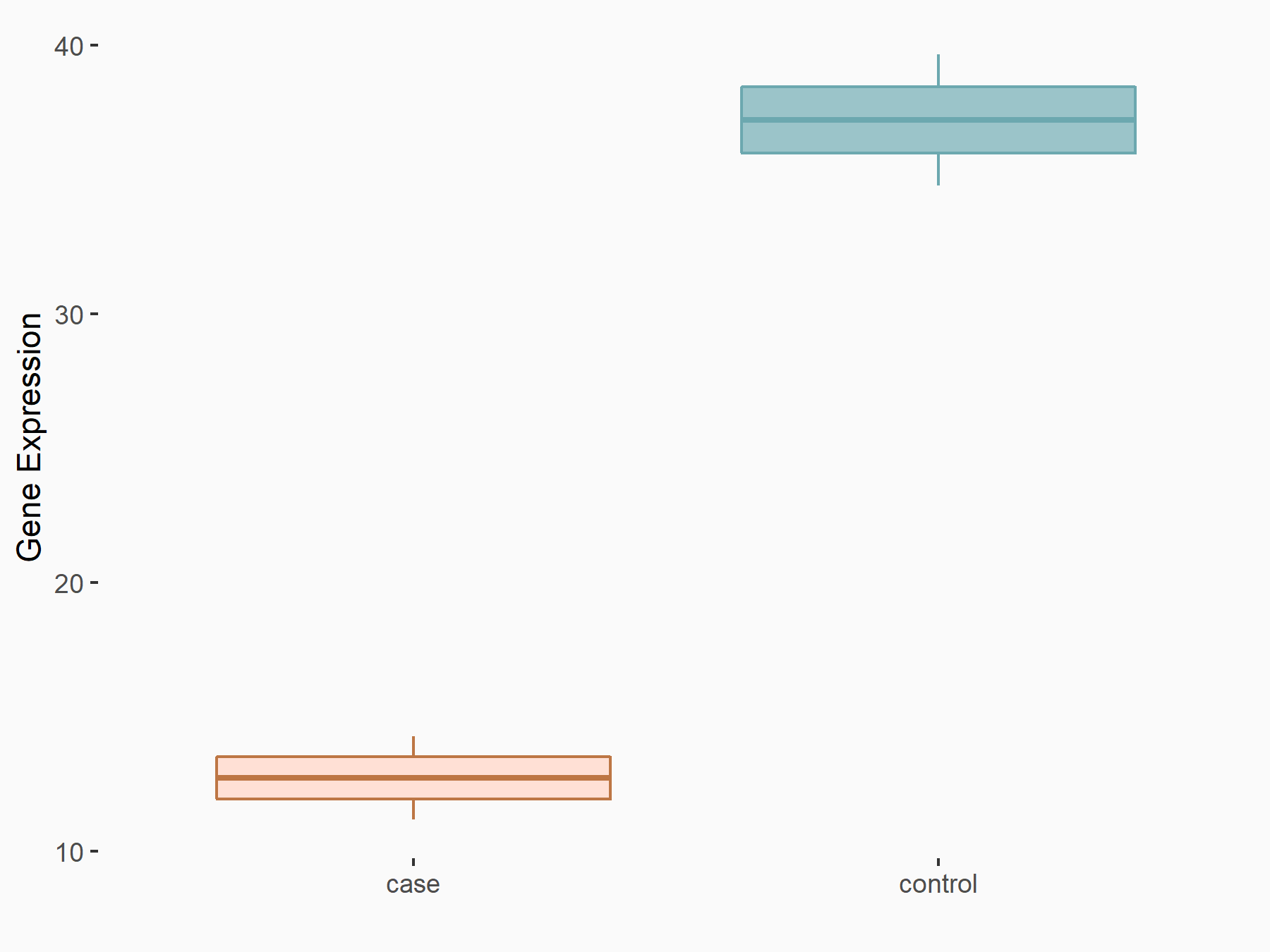 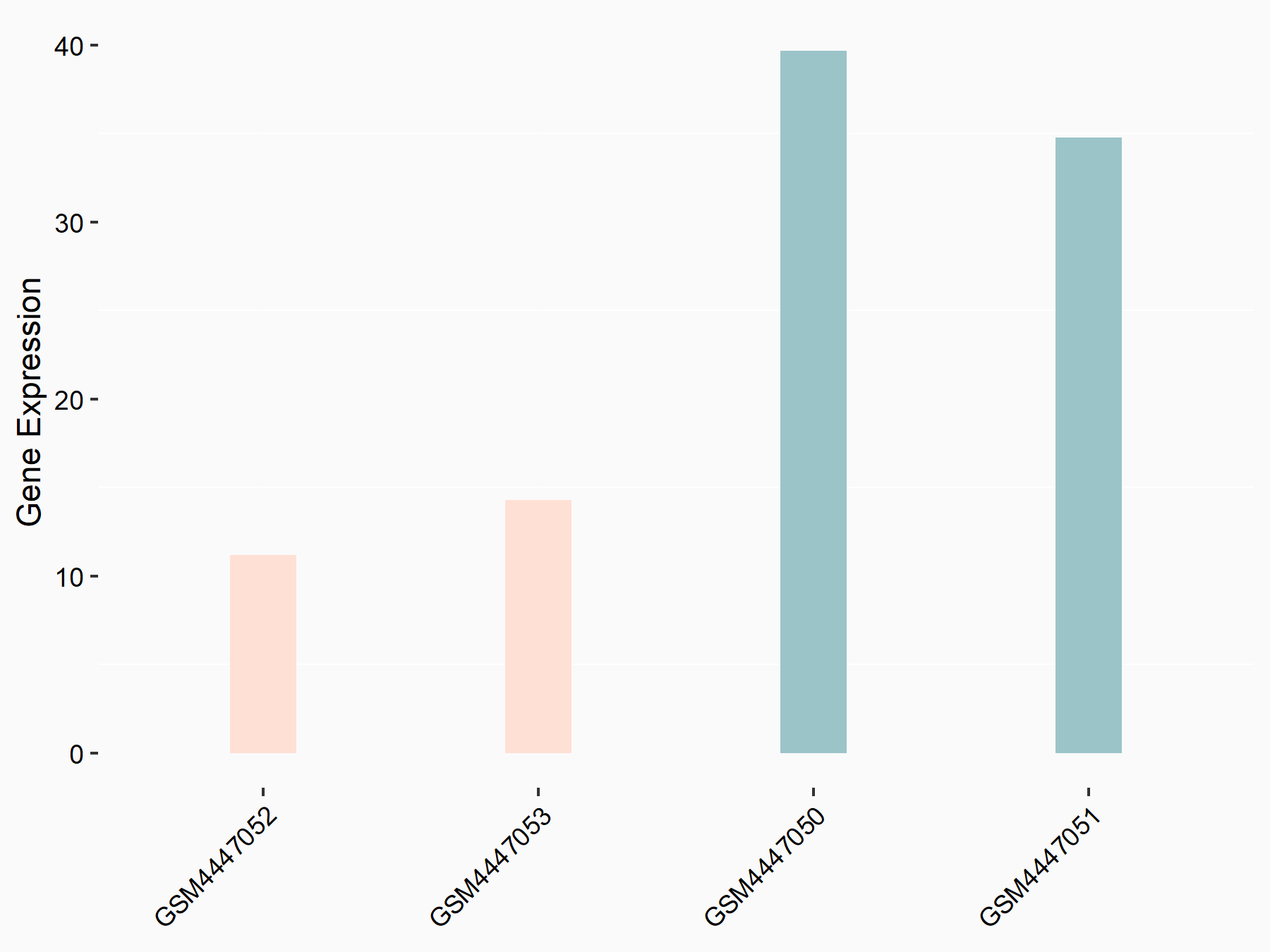 |
logFC: -1.55E+00 p-value: 2.76E-03 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and YTHDF1 | ||
| Cell Line | Hela | Homo sapiens |
| Regulation | logFC: 1.21E+00 | GSE63591 |
Head and neck squamous carcinoma [ICD-11: 2B6E]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [8] | |||
| Responsed Disease | Oral squamous cell carcinoma [ICD-11: 2B6E.0] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | RNA degradation | hsa03018 | ||
| Cell Process | RNA stability | |||
In-vitro Model |
CAL-27 | Tongue squamous cell carcinoma | Homo sapiens | CVCL_1107 |
| NHOK (Normal oral keratinocytes) | ||||
| SCC-15 | Tongue squamous cell carcinoma | Homo sapiens | CVCL_1681 | |
| SCC-25 | Tongue squamous cell carcinoma | Homo sapiens | CVCL_1682 | |
| TSCCa | Endocervical adenocarcinoma | Homo sapiens | CVCL_VL15 | |
| In-vivo Model | The stable transfection of SCC25 cells (1 × 107 cells in 0.1 mL) with lenti-sh-METTL3 or blank vectors was injected subcutaneously into BALB/c nude mice. | |||
| Response Summary | In oral squamous cell carcinoma, YTH N6-methyladenosine RNA binding protein 1 (YTH domain family, member 1 [YTHDF1]) mediated the m6A-increased stability of Myc proto-oncogene protein (MYC) mRNA catalyzed by METTL3. | |||
Lung cancer [ICD-11: 2C25]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [9] | |||
| Responsed Disease | Non-small-cell lung carcinoma [ICD-11: 2C25.Y] | |||
| Pathway Response | p53 signaling pathway | hsa04115 | ||
| Central carbon metabolism in cancer | hsa05230 | |||
| PD-L1 expression and PD-1 checkpoint pathway in cancer | hsa05235 | |||
| Response Summary | This study revealed that m6A methylation is closely related to the poor prognosis of non-small cell lung cancer patients via interference with the TIME, which suggests that m6A plays a role in optimizing individualized immunotherapy management and improving prognosis. The expression levels of METTL3, FTO and YTHDF1 in non-small cell lung cancer were changed. Patients in Cluster 1 had lower immunoscores, higher programmed death-ligand 1 (PD-L1) expression, and shorter overall survival compared to patients in Cluster 2. The Myc proto-oncogene protein (MYC) targets, E2 transcription Factor (E2F) targets were significantly enriched. | |||
PR domain zinc finger protein 2 (PRDM2)
| Representative RNA-seq result indicating the expression of this target gene regulated by YTHDF1 | ||
| Cell Line | AGS cell line | Homo sapiens |
|
Treatment: shYTHDF1 AGS
Control: shNC AGS
|
GSE166972 | |
| Regulation |
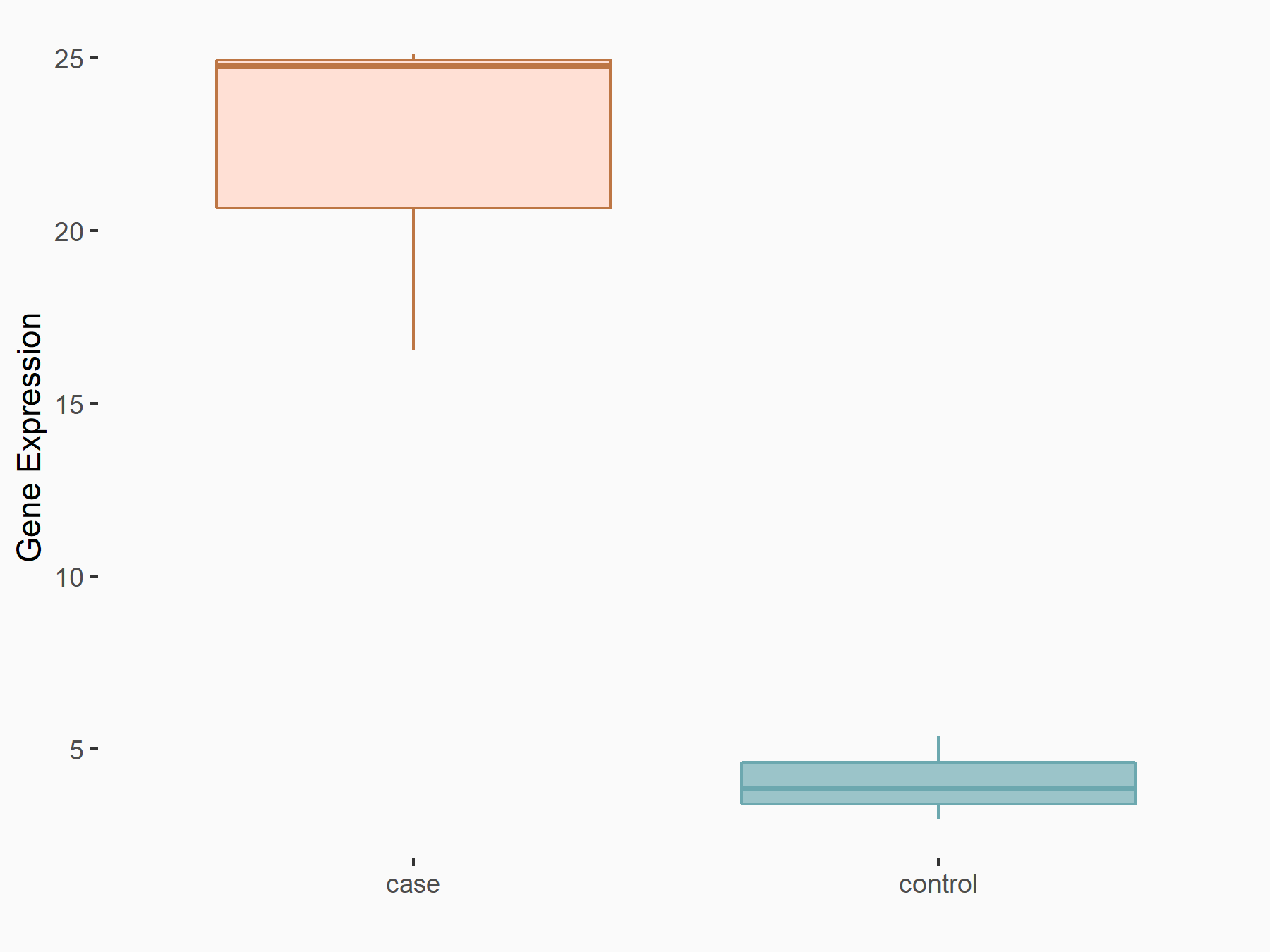 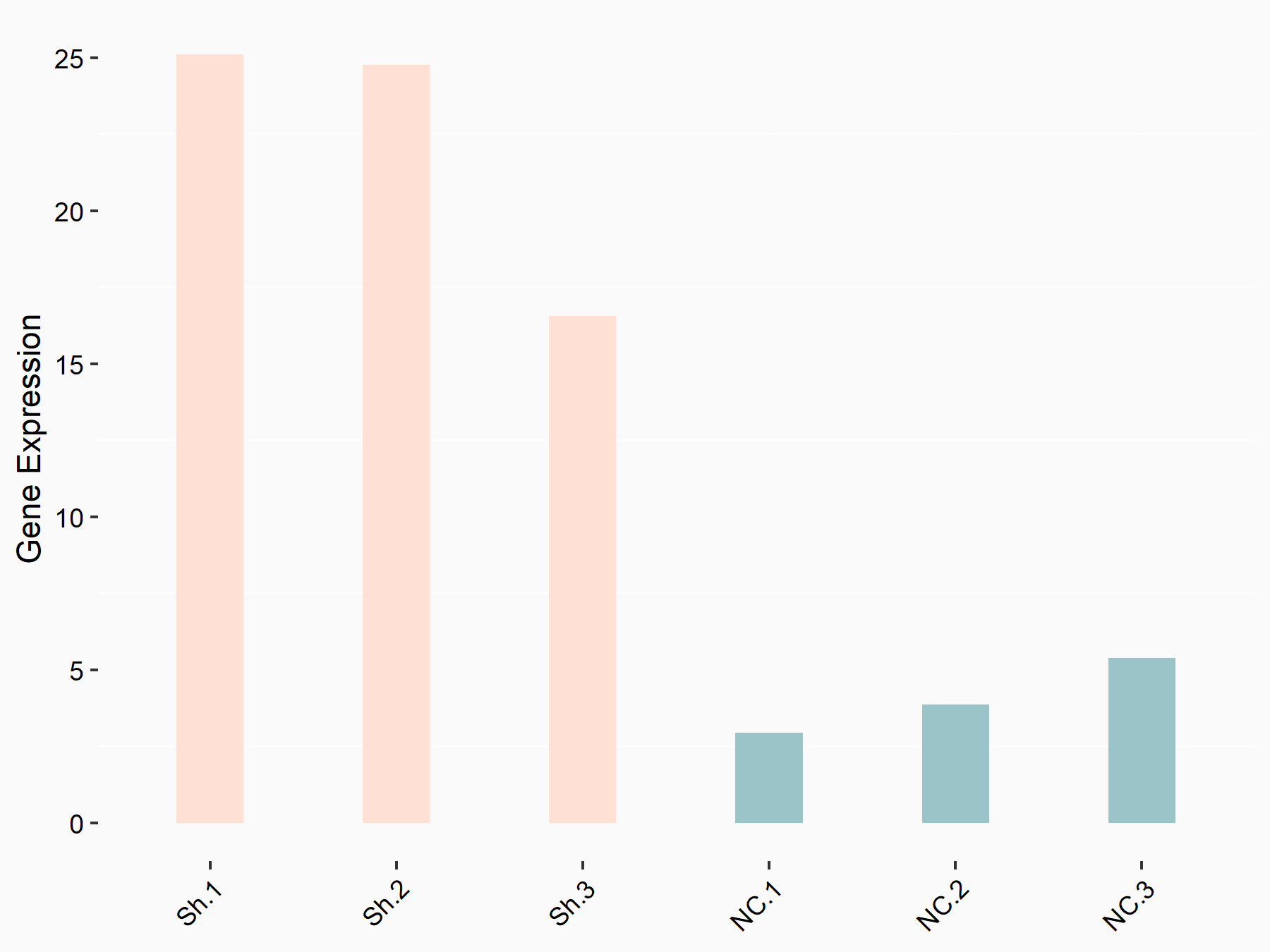 |
logFC: 2.20E+00 p-value: 3.42E-04 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and YTHDF1 | ||
| Cell Line | Hela | Homo sapiens |
| Regulation | logFC: 1.84E+00 | GSE63591 |
Solid tumour/cancer [ICD-11: 2A00-2F9Z]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [10] | |||
| Responsed Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | |||
| Responsed Drug | Arsenite | Phase 2 | ||
| Target Regulation | Down regulation | |||
| Pathway Response | p53 signaling pathway | hsa04115 | ||
In-vitro Model |
HaCaT | Normal | Homo sapiens | CVCL_0038 |
| Response Summary | METTL3 significantly decreased m6A level, restoring p53 activation and inhibiting cellular transformation phenotypes in the arsenite-transformed cells. m6A downregulated the expression of the positive p53 regulator, PR domain zinc finger protein 2 (PRDM2), through the YTHDF2-promoted decay of PRDM2 mRNAs. m6A upregulated the expression of the negative p53 regulator, YY1 and MDM2 through YTHDF1-stimulated translation of YY1 and MDM2 mRNA. This study further sheds light on the mechanisms of arsenic carcinogenesis via RNA epigenetics. | |||
TNF receptor-associated factor 4 (TRAF4)
| Representative RNA-seq result indicating the expression of this target gene regulated by YTHDF1 | ||
| Cell Line | AGS cell line | Homo sapiens |
|
Treatment: shYTHDF1 AGS
Control: shControl AGS
|
GSE159425 | |
| Regulation |
 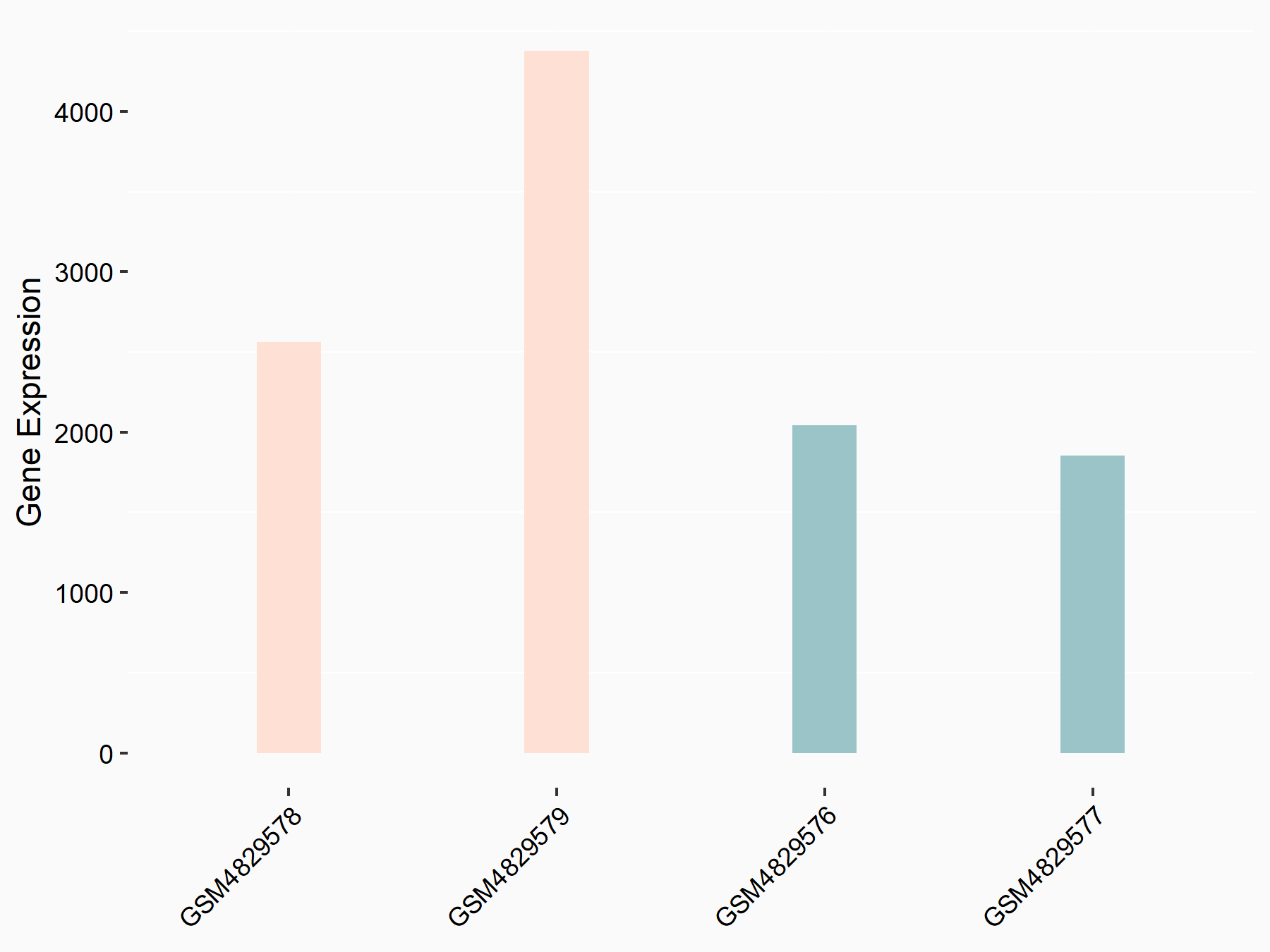 |
logFC: 8.33E-01 p-value: 9.45E-04 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and YTHDF1 | ||
| Cell Line | Hela | Homo sapiens |
| Regulation | logFC: 1.38E+00 | GSE63591 |
Obesity [ICD-11: 5B81]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [11] | |||
| Responsed Disease | Obesity [ICD-11: 5B81] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | Ubiquitin mediated proteolysis | hsa04120), Proteasome | ||
| Cell Process | Ubiquitination degradation | |||
In-vitro Model |
SVF (Stromal vascular cell fraction (SVF) was isolated from minced inguinal adipose tissue of male C57BL/6J mice (3-4 weeks old)) | |||
| 3T3-L1 | Normal | Mus musculus | CVCL_0123 | |
| In-vivo Model | Before the tests, animals were fasted for 8 h. l glucose tolerance test (GTT) was conducted during week 11 on the diet. The mice were challenged with 2 g/kg body weight d-glucose (Sigma-Aldrich, USA). Insulin tolerance test (ITT) was conducted during week 12 on the diet. For insulin tolerance test, mice were injected intraperitoneally with 0.75 U/kg body weight insulin (Sigma-Aldrich, USA). For both tests, blood samples were taken from the tail vein and glucose levels were measured at indicated time points after administration using an AlphaTRAK glucometer. | |||
| Response Summary | m6A-dependent TNF receptor-associated factor 4 (TRAF4) expression upregulation by ALKBH5 and YTHDF1 contributes to curcumin-induced obesity prevention. | |||
Transcription factor E2F8 (E2F8)
| Representative RNA-seq result indicating the expression of this target gene regulated by YTHDF1 | ||
| Cell Line | AGS cell line | Homo sapiens |
|
Treatment: shYTHDF1 AGS
Control: shControl AGS
|
GSE159425 | |
| Regulation |
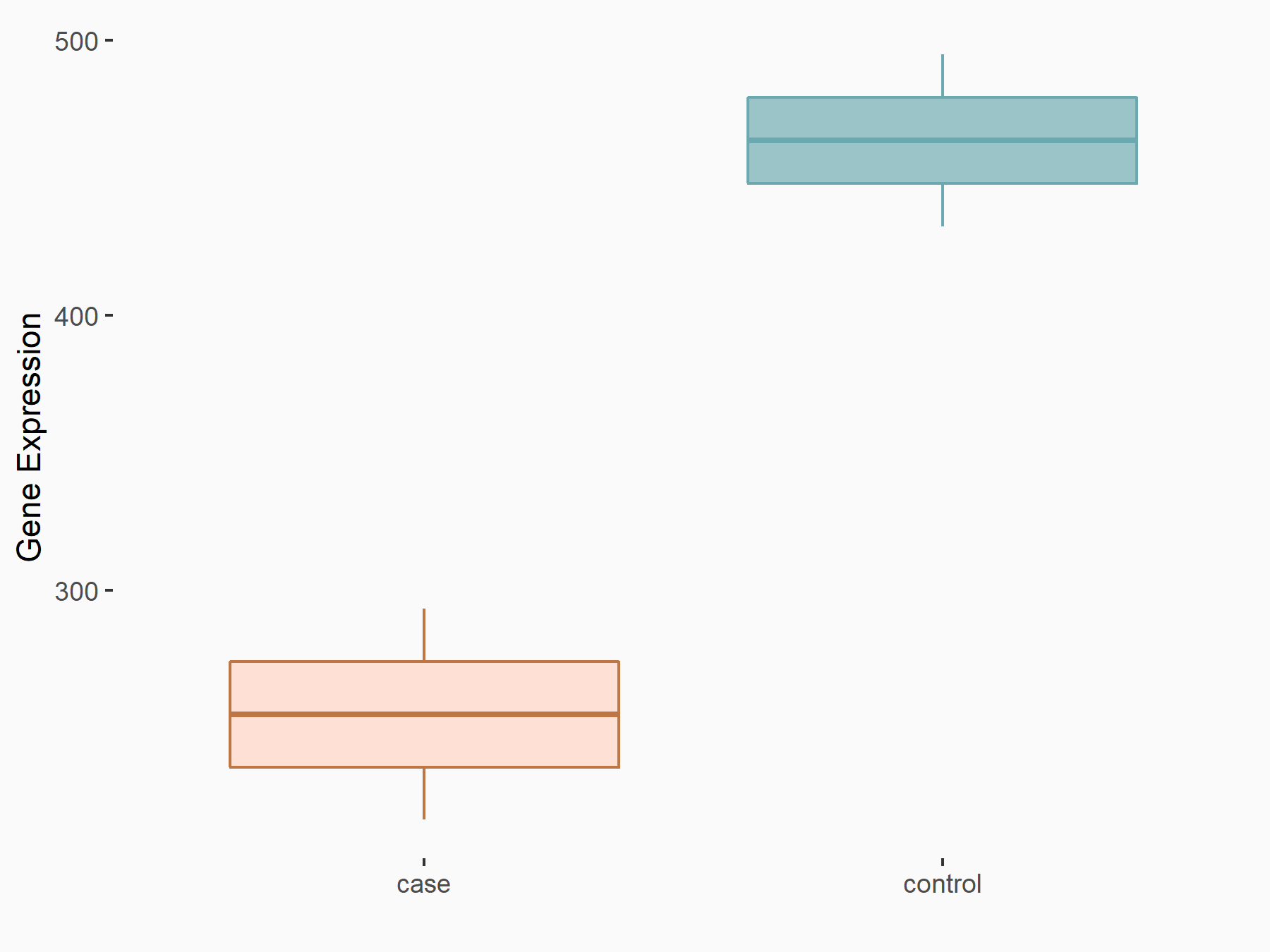 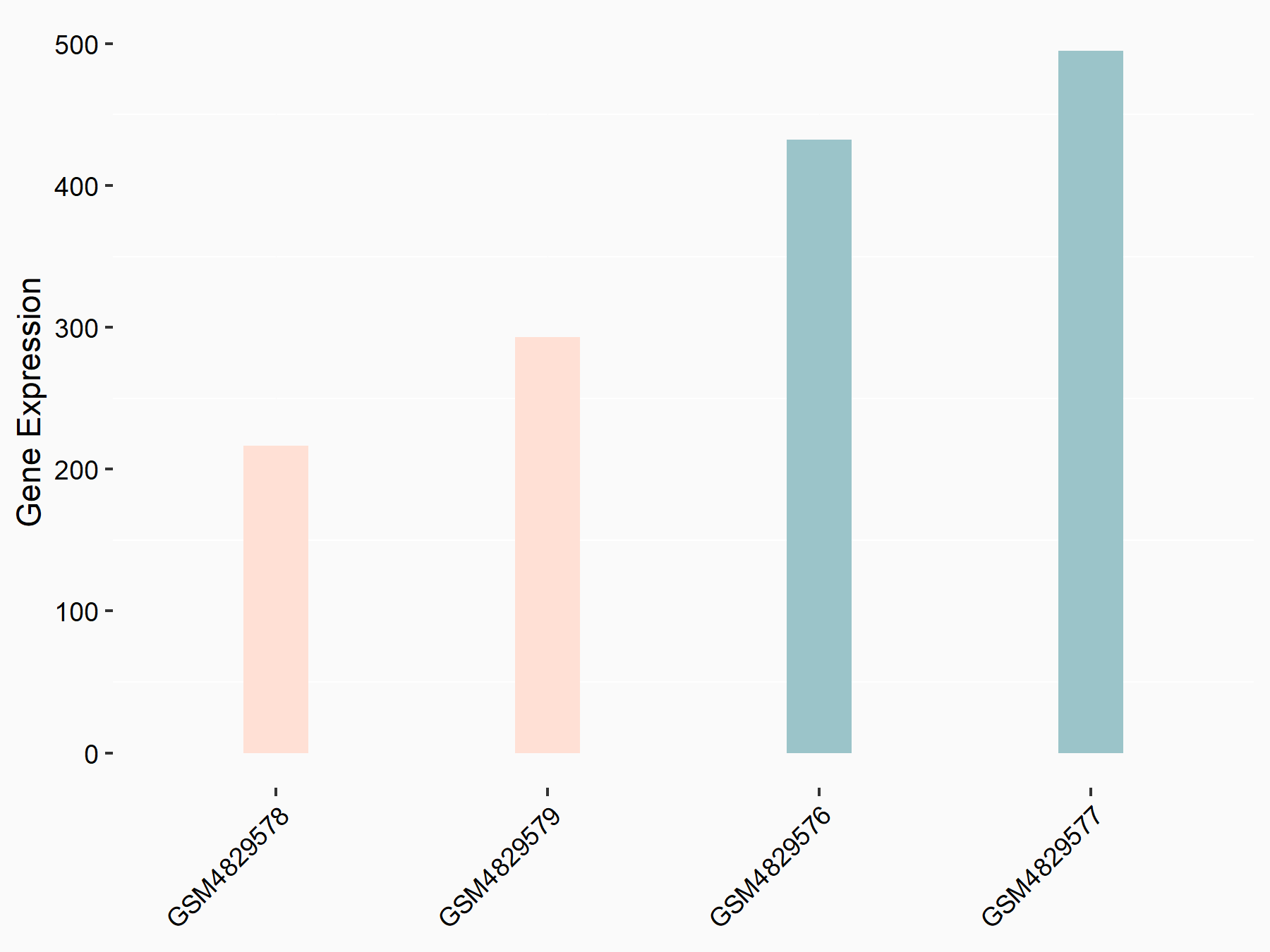 |
logFC: -8.62E-01 p-value: 7.08E-04 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and YTHDF1 | ||
| Cell Line | Hela | Homo sapiens |
| Regulation | logFC: 1.18E+00 | GSE63591 |
Breast cancer [ICD-11: 2C60]
| In total 3 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [12] | |||
| Responsed Disease | Breast cancer [ICD-11: 2C60] | |||
| Responsed Drug | Cisplatin | Approved | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Nucleotide excision repair | hsa03420 | ||
| Cell Process | RNA stability | |||
In-vitro Model |
MDA-MB-231 | Breast adenocarcinoma | Homo sapiens | CVCL_0062 |
| MCF-7 | Invasive breast carcinoma | Homo sapiens | CVCL_0031 | |
| Hs 578T | Invasive breast carcinoma | Homo sapiens | CVCL_0332 | |
| In-vivo Model | 1×106 MDA-MB-231 cells were resuspended in 100 uL PBS with 50% Matrigel (Corning Costar, USA), and injected into the mammary fat pad of the mice. | |||
| Response Summary | In breast cancer, accordingly YTHDF1 knockdown sensitizes breast cancer cells to Adriamycin and Cisplatin as well as Olaparib, a PARP inhibitor. Transcription factor E2F8 (E2F8) is a target molecule by YTHDF1 which modulates E2F8 mRNA stability and DNA damage repair in a METTL14-dependent manner. | |||
| Experiment 2 Reporting the m6A-centered Disease Response of This Target Gene | [12] | |||
| Responsed Disease | Breast cancer [ICD-11: 2C60] | |||
| Responsed Drug | Doxil | Approved | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Nucleotide excision repair | hsa03420 | ||
| Cell Process | RNA stability | |||
In-vitro Model |
MDA-MB-231 | Breast adenocarcinoma | Homo sapiens | CVCL_0062 |
| MCF-7 | Invasive breast carcinoma | Homo sapiens | CVCL_0031 | |
| Hs 578T | Invasive breast carcinoma | Homo sapiens | CVCL_0332 | |
| In-vivo Model | 1×106 MDA-MB-231 cells were resuspended in 100 uL PBS with 50% Matrigel (Corning Costar, USA), and injected into the mammary fat pad of the mice. | |||
| Response Summary | In breast cancer, accordingly YTHDF1 knockdown sensitizes breast cancer cells to Adriamycin and Cisplatin as well as Olaparib, a PARP inhibitor. Transcription factor E2F8 (E2F8) is a target molecule by YTHDF1 which modulates E2F8 mRNA stability and DNA damage repair in a METTL14-dependent manner. | |||
| Experiment 3 Reporting the m6A-centered Disease Response of This Target Gene | [12] | |||
| Responsed Disease | Breast cancer [ICD-11: 2C60] | |||
| Responsed Drug | Olaparib | Approved | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Nucleotide excision repair | hsa03420 | ||
| Cell Process | RNA stability | |||
In-vitro Model |
MDA-MB-231 | Breast adenocarcinoma | Homo sapiens | CVCL_0062 |
| MCF-7 | Invasive breast carcinoma | Homo sapiens | CVCL_0031 | |
| Hs 578T | Invasive breast carcinoma | Homo sapiens | CVCL_0332 | |
| In-vivo Model | 1×106 MDA-MB-231 cells were resuspended in 100 uL PBS with 50% Matrigel (Corning Costar, USA), and injected into the mammary fat pad of the mice. | |||
| Response Summary | In breast cancer, accordingly YTHDF1 knockdown sensitizes breast cancer cells to Adriamycin and Cisplatin as well as Olaparib, a PARP inhibitor. Transcription factor E2F8 (E2F8) is a target molecule by YTHDF1 which modulates E2F8 mRNA stability and DNA damage repair in a METTL14-dependent manner. | |||
Transcriptional coactivator YAP1 (YAP1)
| Representative RNA-seq result indicating the expression of this target gene regulated by YTHDF1 | ||
| Cell Line | AGS cell line | Homo sapiens |
|
Treatment: shYTHDF1 AGS
Control: shNC AGS
|
GSE166972 | |
| Regulation |
 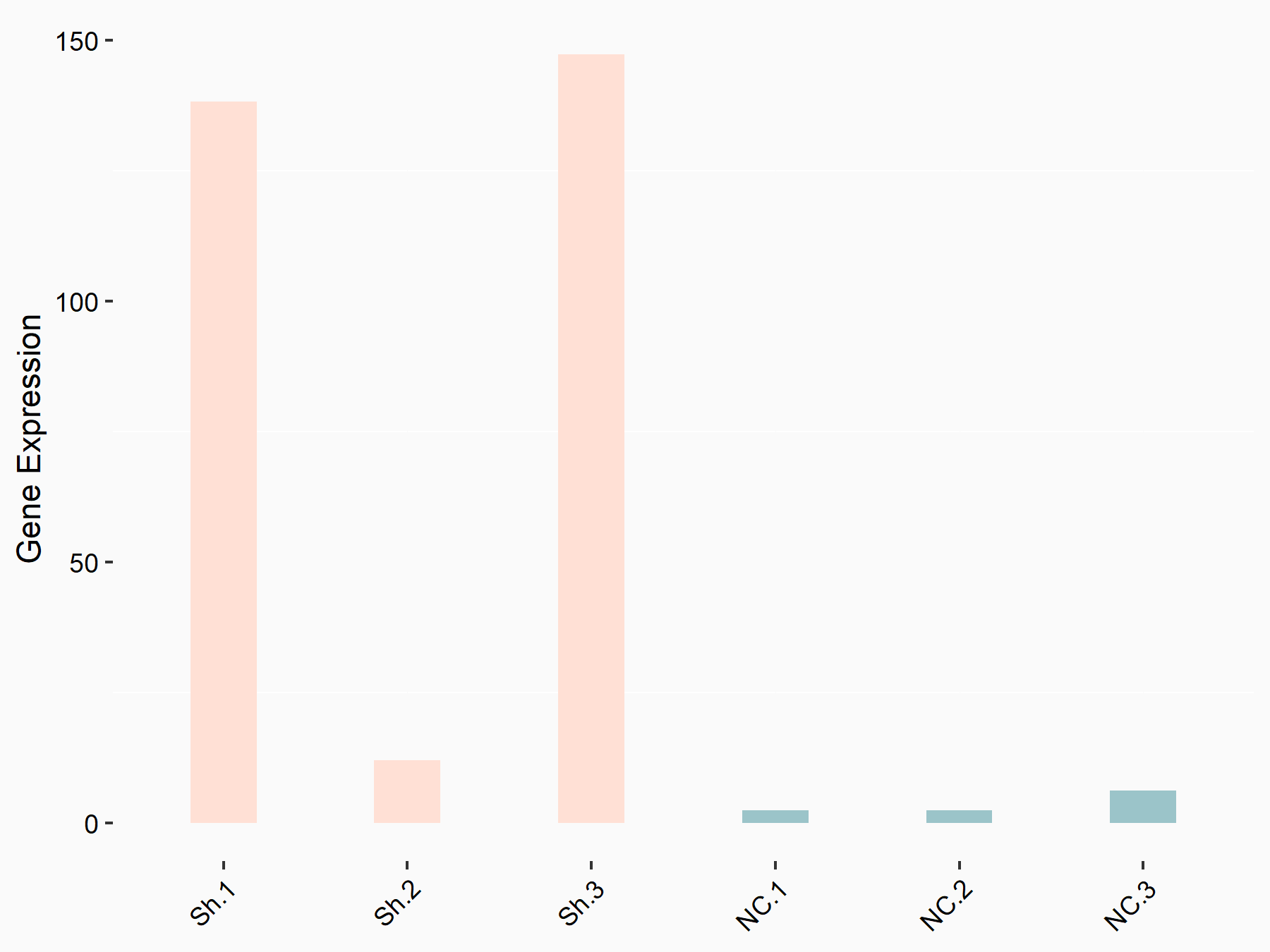 |
logFC: 3.89E+00 p-value: 1.09E-02 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and YTHDF1 | ||
| Cell Line | Hela | Homo sapiens |
| Regulation | logFC: 1.35E+00 | GSE63591 |
Osteosarcoma [ICD-11: 2B51]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [13] | |||
| Responsed Disease | Osteosarcoma [ICD-11: 2B51] | |||
| Target Regulation | Up regulation | |||
| Cell Process | Cell growth | |||
| Cell migration | ||||
| Cell invasion | ||||
| Cell apoptosis | ||||
In-vitro Model |
U2OS | Osteosarcoma | Homo sapiens | CVCL_0042 |
| In-vivo Model | Three-week-old BABL/c female nude mice were randomized into three groups. 5 × 106 143B cells were subcutaneously injected in mice, and the tumor volume was assessed every 2 weeks. Eight weeks after injection, the animals were killed. The xenograft tumors were harvested and the tumor volumes were calculated by the standard formula: length × width2/2. | |||
| Response Summary | ALKBH5 is an anti-tumor factor or a pro-apoptotic factor, acting at least partially by suppressing Transcriptional coactivator YAP1 (YAP1) expression through dual mechanisms with direct m6A methylation of YAP and indirect downregulation of YAP level due to methylation of pre-miR-181b-1. Further results revealed that m6A methylated pre-miR-181b-1 was subsequently recognized by m6A-binding protein YTHDF2 to mediate RNA degradation. However, methylated YAP transcripts were recognized by YTHDF1 to promote its translation. ALKBH5 overexpression was considered a new approach of replacement therapy for osteosarcoma treatment. | |||
Lung cancer [ICD-11: 2C25]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [14] | |||
| Responsed Disease | Non-small-cell lung carcinoma [ICD-11: 2C25.Y] | |||
| Responsed Drug | Cisplatin | Approved | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Hippo signaling pathway | hsa04390 | ||
| Cell Process | Metabolic | |||
In-vitro Model |
A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 |
| Calu-6 | Lung adenocarcinoma | Homo sapiens | CVCL_0236 | |
| NCI-H1299 | Lung large cell carcinoma | Homo sapiens | CVCL_0060 | |
| NCI-H520 | Lung squamous cell carcinoma | Homo sapiens | CVCL_1566 | |
| In-vivo Model | Mice were injected with 5 × 106 lung cancer cells with stably expression of relevant plasmids and randomly divided into two groups (five mice per group) after the diameter of the xenografted tumors had reached approximately 5 mm in diameter. Xenografted mice were then administrated with PBS or DDP (3 mg/kg per day) for three times a week, and tumor volume were measured every second day. | |||
| Response Summary | METTL3, YTHDF3, YTHDF1, and eIF3b directly promoted YAP translation through an interaction with the translation initiation machinery. METTL3 knockdown inhibits tumor growth and enhances sensitivity to DDP in vivo.m6A mRNA methylation initiated by METTL3 directly promotes YAP translation and increases YAP activity by regulating the MALAT1-miR-1914-3p-Transcriptional coactivator YAP1 (YAP1) axis to induce Non-small cell lung cancer drug resistance and metastasis. | |||
Kidney disorders [ICD-11: GB90]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [15] | |||
| Responsed Disease | Kidney disorders [ICD-11: GB90] | |||
| Target Regulation | Up regulation | |||
| Response Summary | YTHDF1 knockdown alleviated the progression of renal fibrosis both in cultured cells induced by transforming growth factor-beta administration and in the UUO mouse model. Transcriptional coactivator YAP1 (YAP1) was accordingly down-regulated when YTHDF1 was inhibited. | |||
Adenosine 5'-monophosphoramidase HINT2 (HINT2)
| Representative RNA-seq result indicating the expression of this target gene regulated by YTHDF1 | ||
| Cell Line | AGS cell line | Homo sapiens |
|
Treatment: shYTHDF1 AGS
Control: shControl AGS
|
GSE159425 | |
| Regulation |
 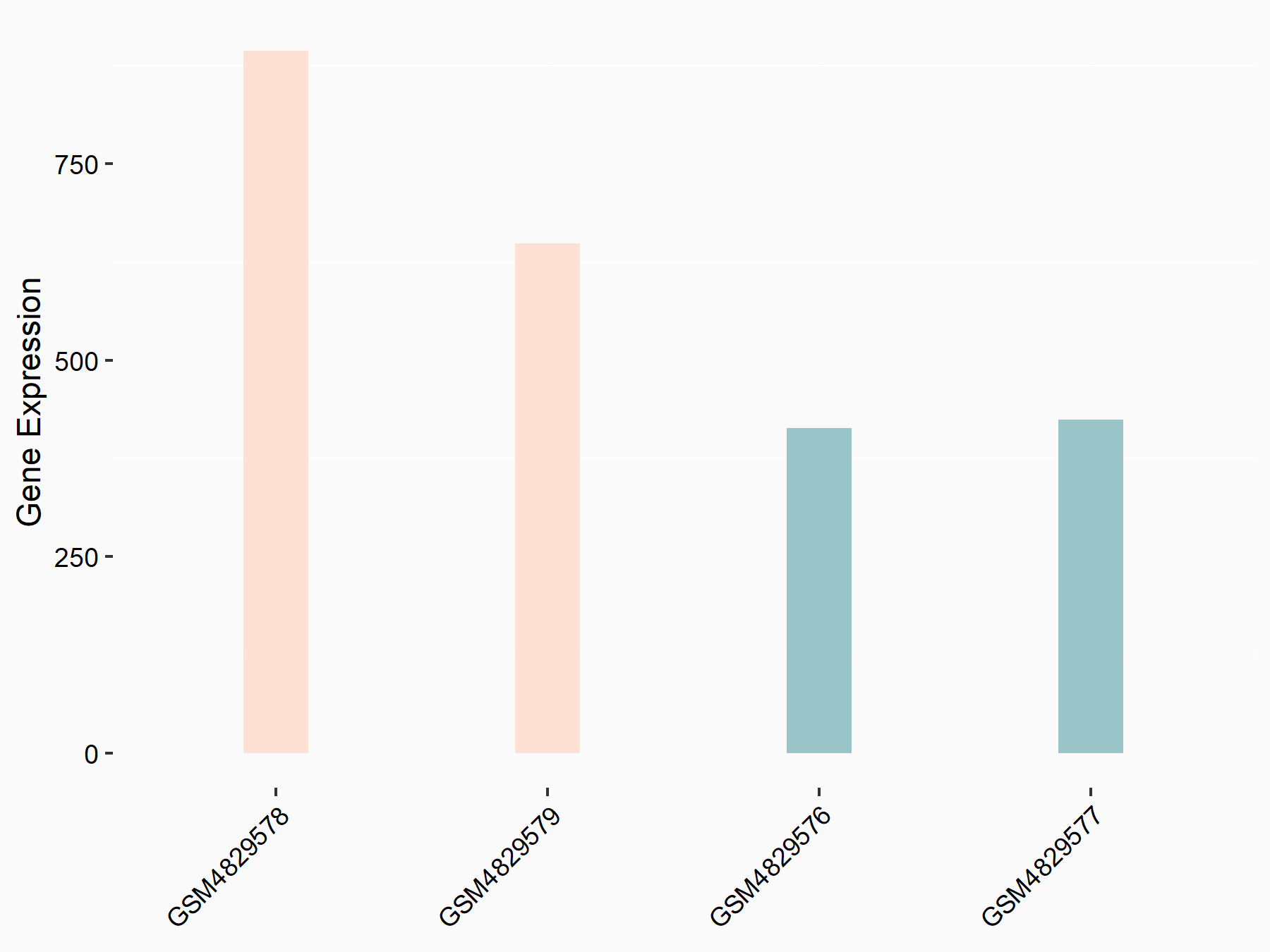 |
logFC: 8.82E-01 p-value: 1.64E-04 |
| More Results | Click to View More RNA-seq Results | |
Melanoma [ICD-11: 2C30]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [16] | |||
| Responsed Disease | Melanoma [ICD-11: 2C30] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
CM2005.1 | Conjunctival melanoma | Homo sapiens | CVCL_M592 |
| CRMM-1 | Conjunctival melanoma | Homo sapiens | CVCL_M593 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| OCM-1 | Amelanotic melanoma | Homo sapiens | CVCL_6934 | |
| OCM-1A | Amelanotic melanoma | Homo sapiens | CVCL_6935 | |
| OM431 | Uveal melanoma | Homo sapiens | CVCL_J308 | |
| PIG1 | Normal | Homo sapiens | CVCL_S410 | |
| In-vivo Model | Approximately 1 × 106 melanoma cells from each group were injected subcutaneously into the right side of the abdomen of BALB/c nude mice (male, 6 weeks old). | |||
| Response Summary | YTHDF1 promoted the translation of methylated Adenosine 5'-monophosphoramidase HINT2 (HINT2) mRNA, a tumor suppressor in ocular melanoma. | |||
Autophagy-related protein 16-1 (ATG16L1)
| Representative RNA-seq result indicating the expression of this target gene regulated by YTHDF1 | ||
| Cell Line | AGS cell line | Homo sapiens |
|
Treatment: shYTHDF1 AGS
Control: shNC AGS
|
GSE166972 | |
| Regulation |
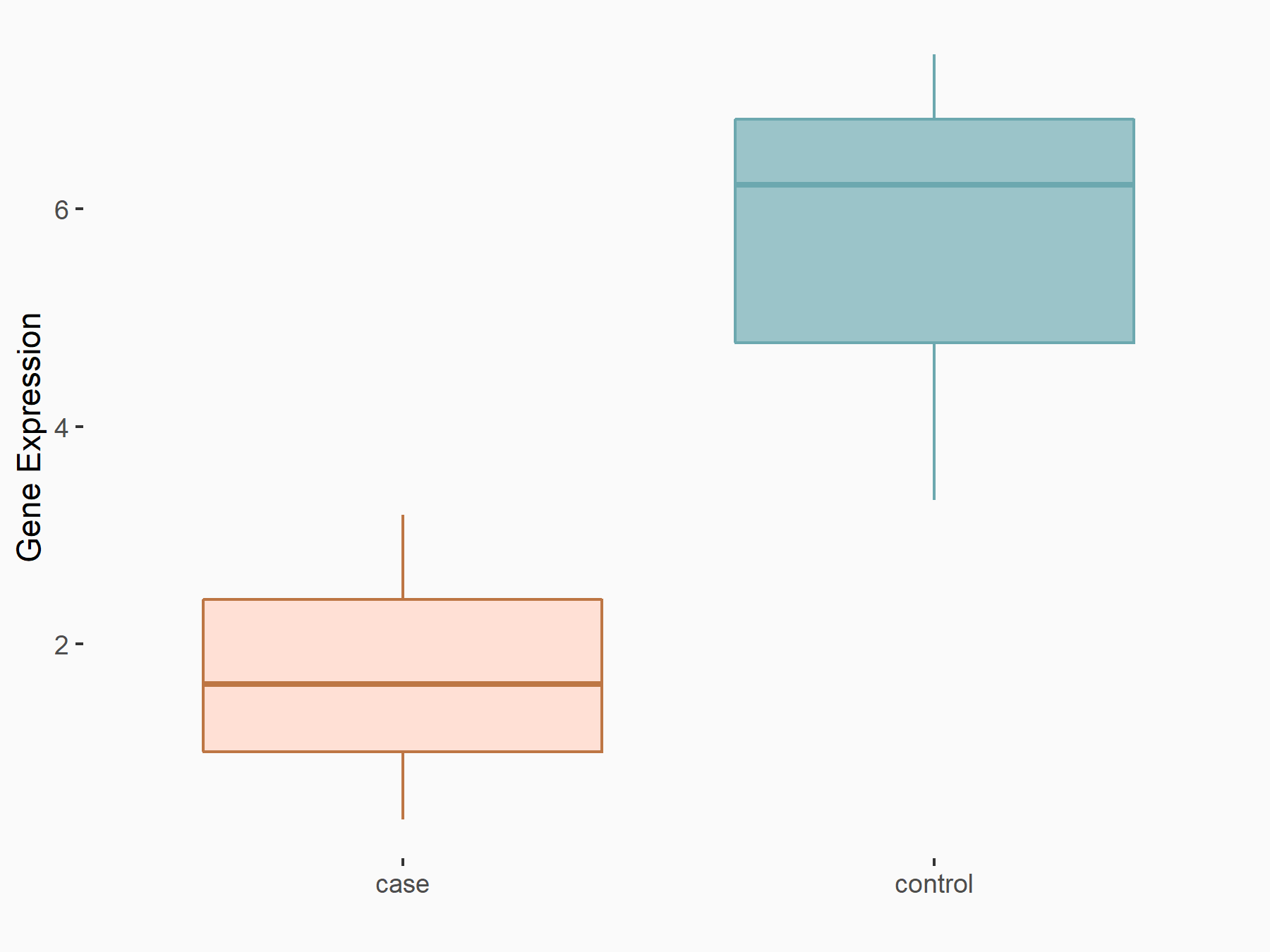 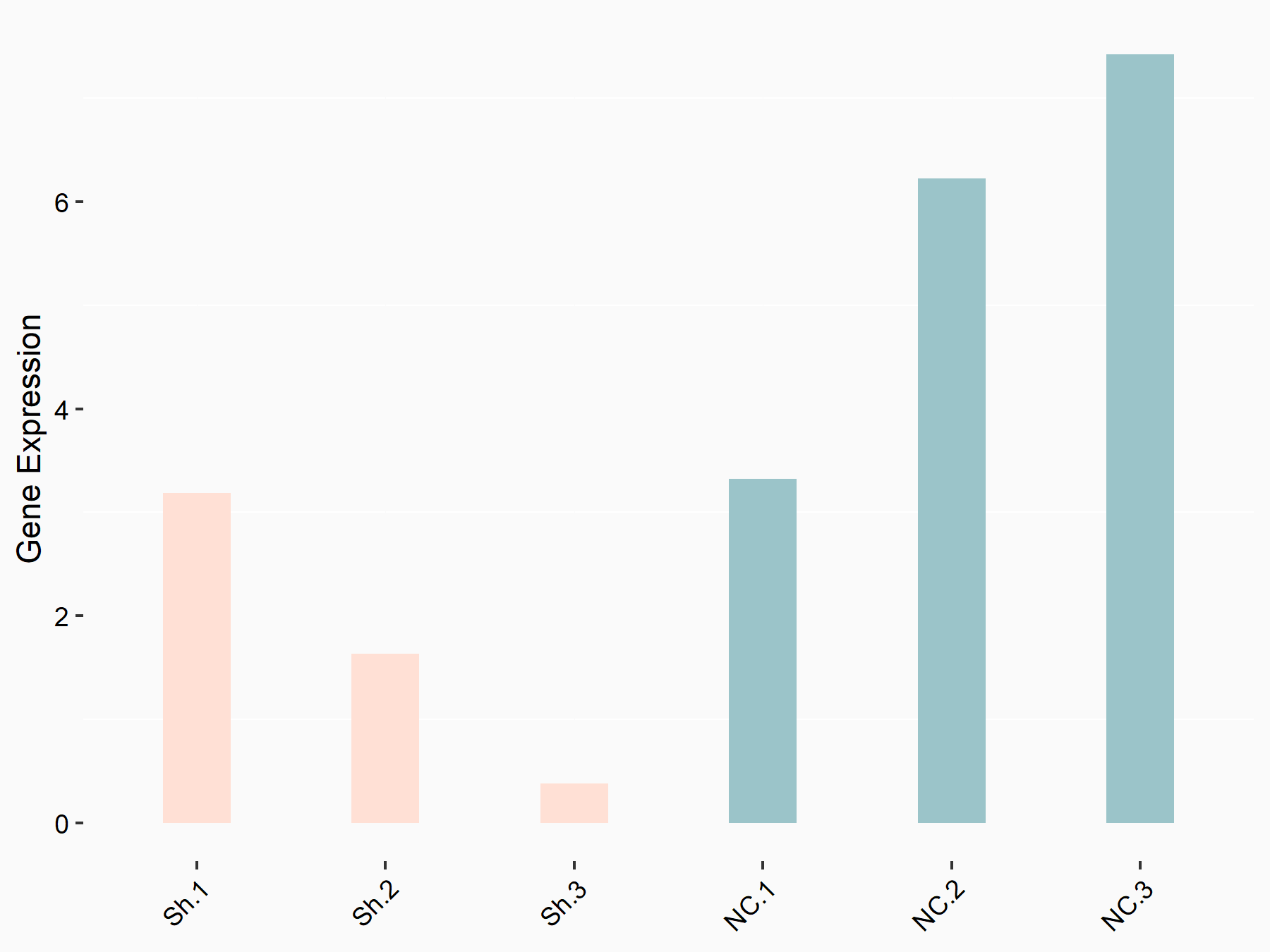 |
logFC: -1.37E+00 p-value: 3.41E-02 |
| More Results | Click to View More RNA-seq Results | |
Liver cancer [ICD-11: 2C12]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [4] | |||
| Responsed Disease | Hepatocellular carcinoma [ICD-11: 2C12.02] | |||
| Responsed Drug | Sorafenib | Approved | ||
| Target Regulation | Up regulation | |||
| Pathway Response | FoxO signaling pathway | hsa04068 | ||
| Autophagy | hsa04140 | |||
| Cell Process | Cell autophagy | |||
| Response Summary | METTL3 can sensitise hepatocellular carcinoma cells to sorafenib through stabilising forkhead box class O3 (FOXO3) in an m6A-dependent manner and translated by YTHDF1, thereby inhibiting the transcription of autophagy-related genes, including ATG3, ATG5, ATG7, ATG12, and Autophagy-related protein 16-1 (ATG16L1). | |||
Beclin-1 (BECN1)
| Representative RNA-seq result indicating the expression of this target gene regulated by YTHDF1 | ||
| Cell Line | AGS cell line | Homo sapiens |
|
Treatment: shYTHDF1 AGS
Control: shNC AGS
|
GSE166972 | |
| Regulation |
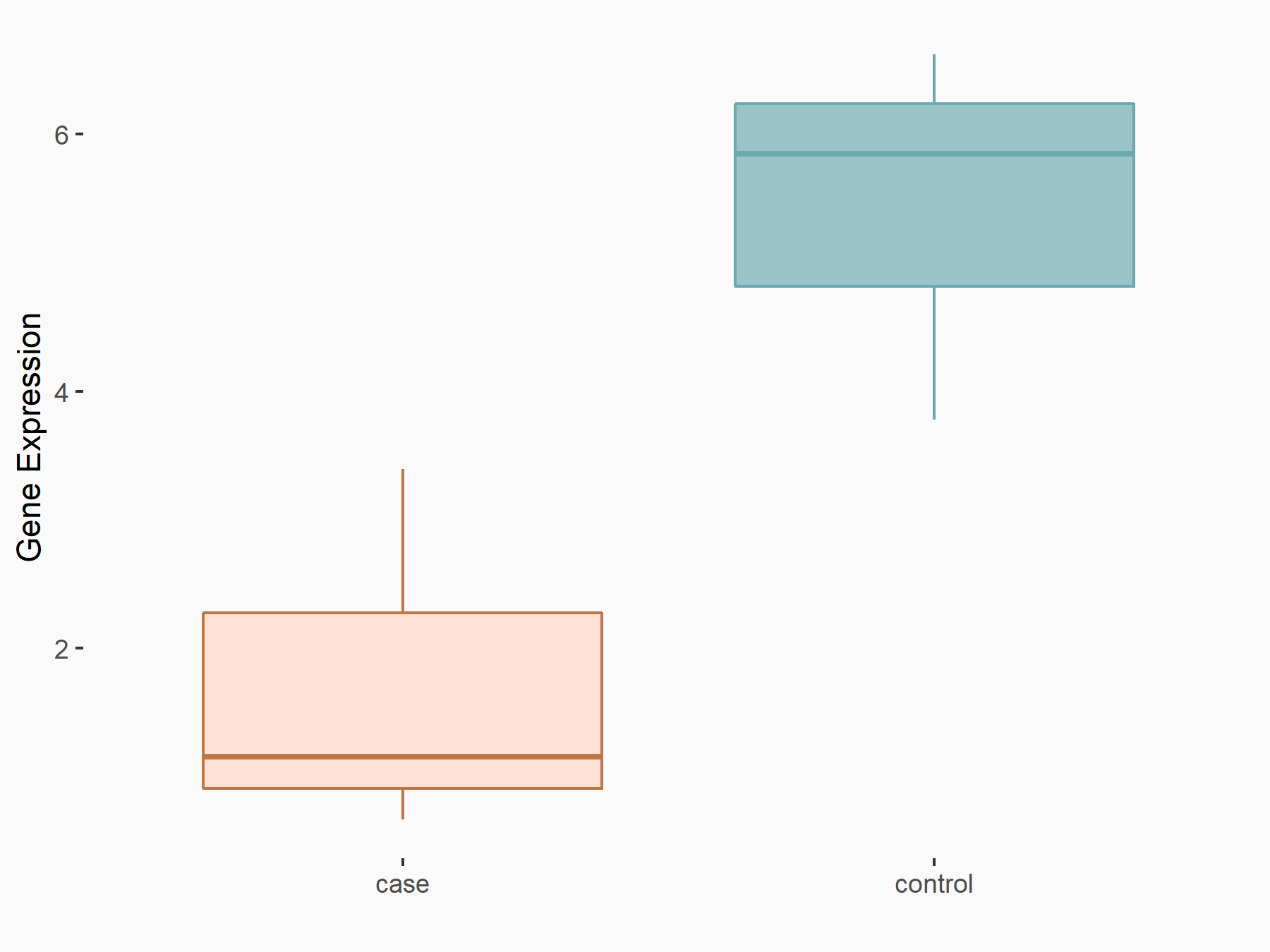 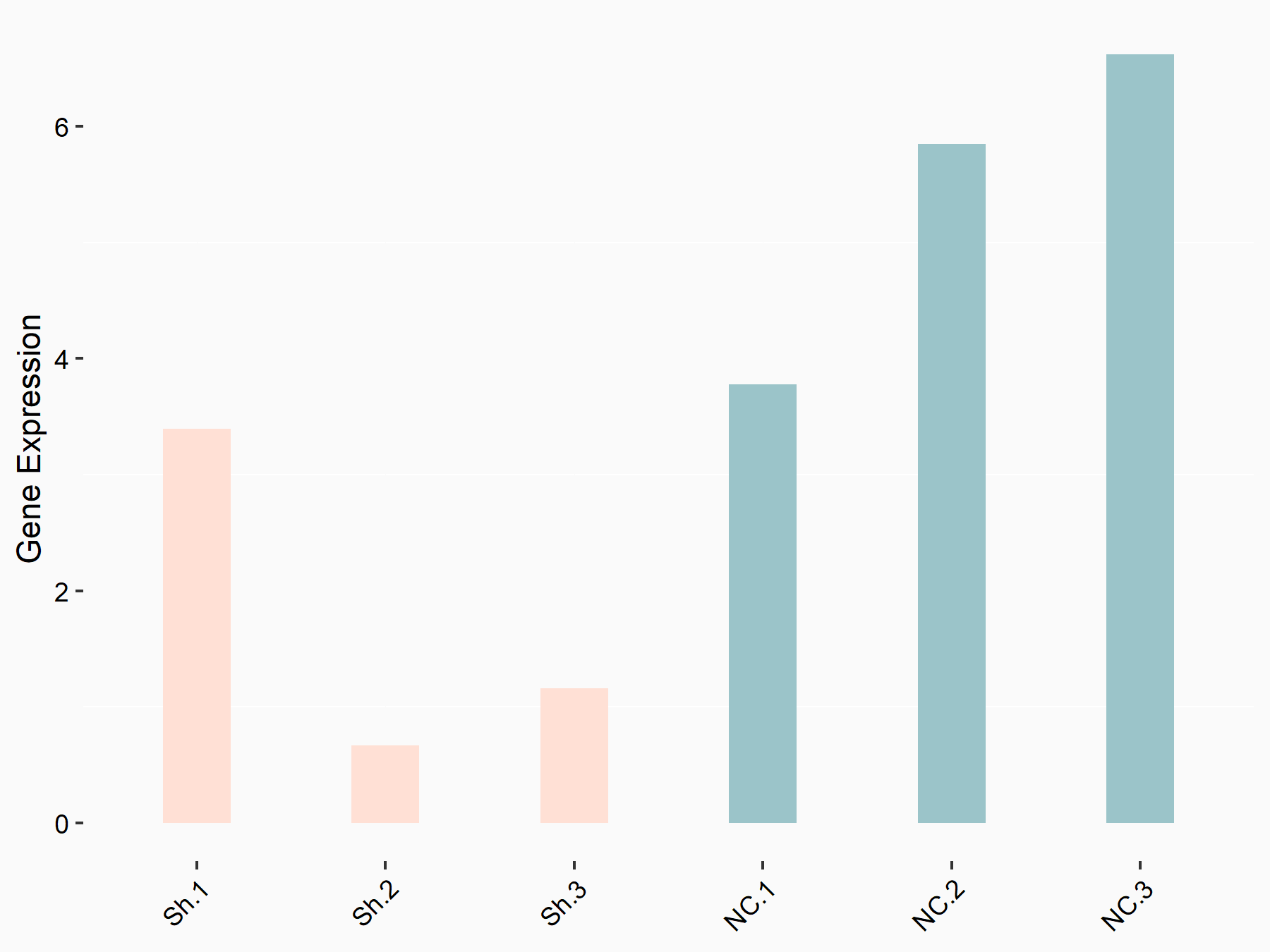 |
logFC: -1.33E+00 p-value: 2.29E-02 |
| More Results | Click to View More RNA-seq Results | |
Liver cancer [ICD-11: 2C12]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [17] | |||
| Responsed Disease | Hepatocellular carcinoma [ICD-11: 2C12.02] | |||
| Responsed Drug | Sorafenib | Approved | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Ferroptosis | hsa04216 | ||
| Autophagy | hsa04140 | |||
| Cell Process | Ferroptosis | |||
| Cell autophagy | ||||
In-vitro Model |
HSC (Hematopoietic stem cell) | |||
| In-vivo Model | VA-Lip-Mettl4-shRNA, VA-Lip-Fto-Plasmid and VA-Lip-Ythdf1-shRNA (0.75 mg/kg) were injected intravenously 3 times a week. | |||
| Response Summary | Analyzed the effect of sorafenib on HSC ferroptosis and m6A modification in advanced fibrotic patients with hepatocellular carcinoma receiving sorafenib monotherapy. YTHDF1 promotes Beclin-1 (BECN1) mRNA stability and autophagy activation via recognizing the m6A binding site within BECN1 coding regions. | |||
Cyclin-dependent kinase 2 (CDK2)
| Representative RNA-seq result indicating the expression of this target gene regulated by YTHDF1 | ||
| Cell Line | AGS cell line | Homo sapiens |
|
Treatment: shYTHDF1 AGS
Control: shControl AGS
|
GSE159425 | |
| Regulation |
 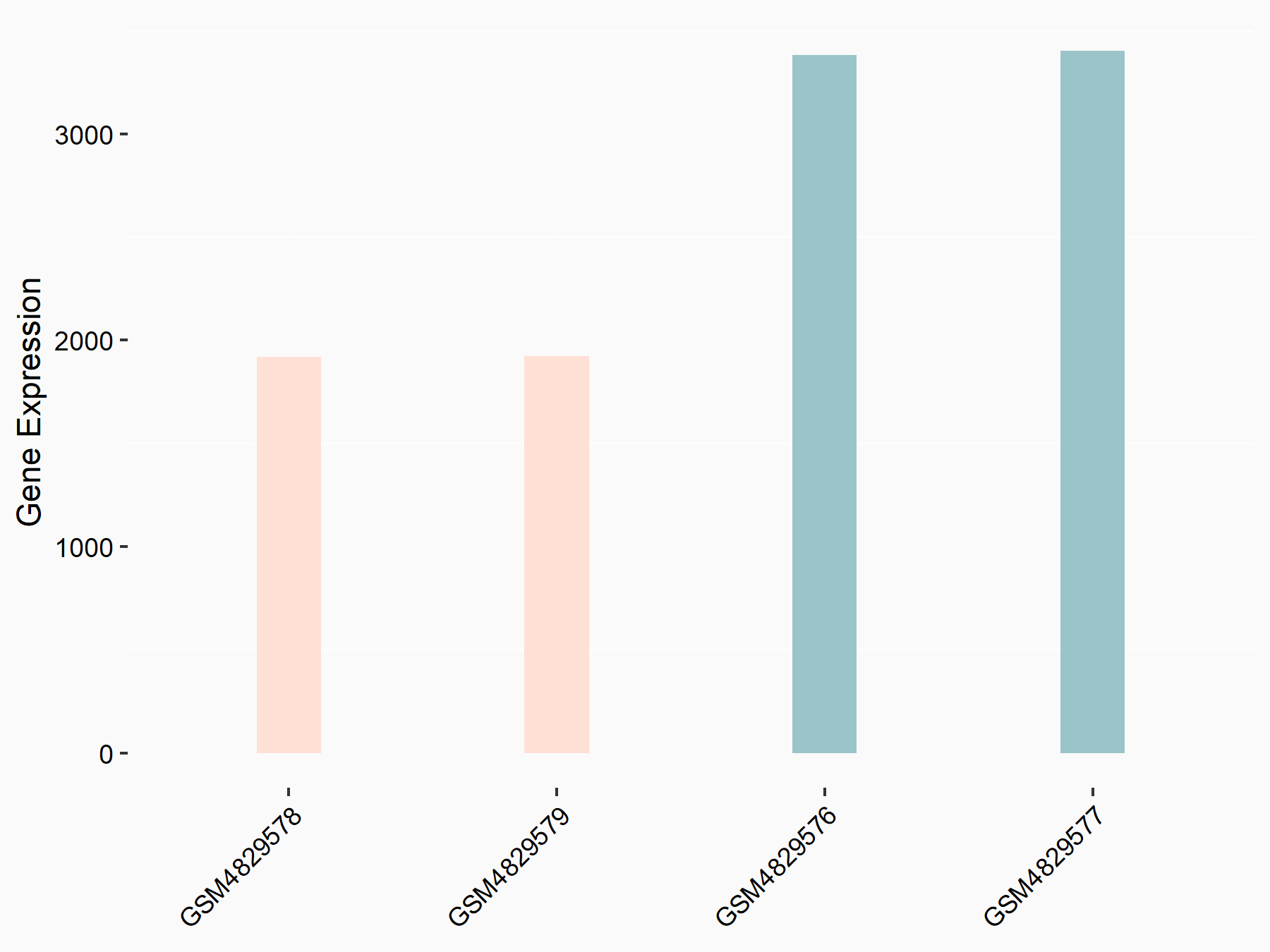 |
logFC: -8.20E-01 p-value: 7.96E-07 |
| More Results | Click to View More RNA-seq Results | |
Lung cancer [ICD-11: 2C25]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [7] | |||
| Responsed Disease | Non-small-cell lung carcinoma [ICD-11: 2C25.Y] | |||
| Responsed Drug | Cisplatin | Approved | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Chemical carcinogenesis - reactive oxygen species | hsa05208 | ||
| Cell cycle | hsa04110 | |||
| Cell Process | Biological regulation | |||
In-vitro Model |
A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 |
| A549-DDP (Human lung adenocarcinoma is resistant to cisplatin) | ||||
| GLC-82 | Endocervical adenocarcinoma | Homo sapiens | CVCL_3371 | |
| NCI-H1299 | Lung large cell carcinoma | Homo sapiens | CVCL_0060 | |
| NCI-H1975 | Lung adenocarcinoma | Homo sapiens | CVCL_1511 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| NCI-H1650 | Minimally invasive lung adenocarcinoma | Homo sapiens | CVCL_1483 | |
| NCI-H838 | Lung adenocarcinoma | Homo sapiens | CVCL_1594 | |
| SPC-A1 | Endocervical adenocarcinoma | Homo sapiens | CVCL_6955 | |
| In-vivo Model | Mice were treated via nasal inhalation of adenovirus carrying Cre recombinase (5 × 106 p.f.u for Ad-Cre, Biowit Inc., Shenzhen, Guangdong), and were then killed at indicated times for gross inspection and histopathological examination. | |||
| Response Summary | YTHDF1 deficiency inhibits Non-small cell lung cancer cell proliferation and xenograft tumor formation through regulating the translational efficiency of Cyclin-dependent kinase 2 (CDK2), CDK4, p27, and cyclin D1, and that YTHDF1 depletion restrains de novo lung adenocarcinomas (ADC) progression. Mechanistic studies identified the Keap1-Nrf2-AKR1C1 axis as the downstream mediator of YTHDF1. YTHDF1 high expression correlates with better clinical outcome, with its depletion rendering cancerous cells resistant to cisplatin (DDP) treatment. | |||
Forkhead box protein M1 (FOXM1)
| Representative RNA-seq result indicating the expression of this target gene regulated by YTHDF1 | ||
| Cell Line | AGS cell line | Homo sapiens |
|
Treatment: shYTHDF1 AGS
Control: shNC AGS
|
GSE166972 | |
| Regulation |
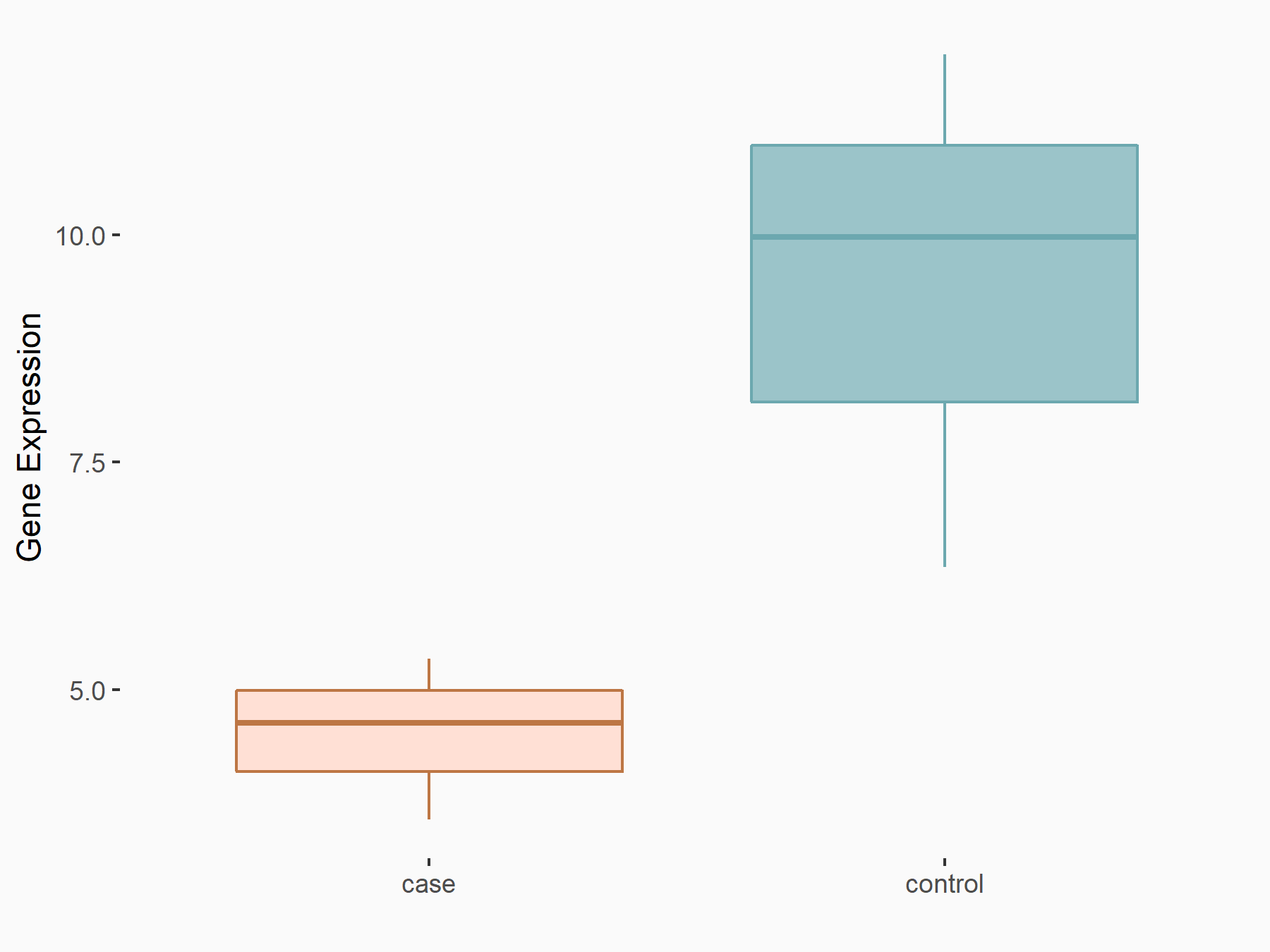 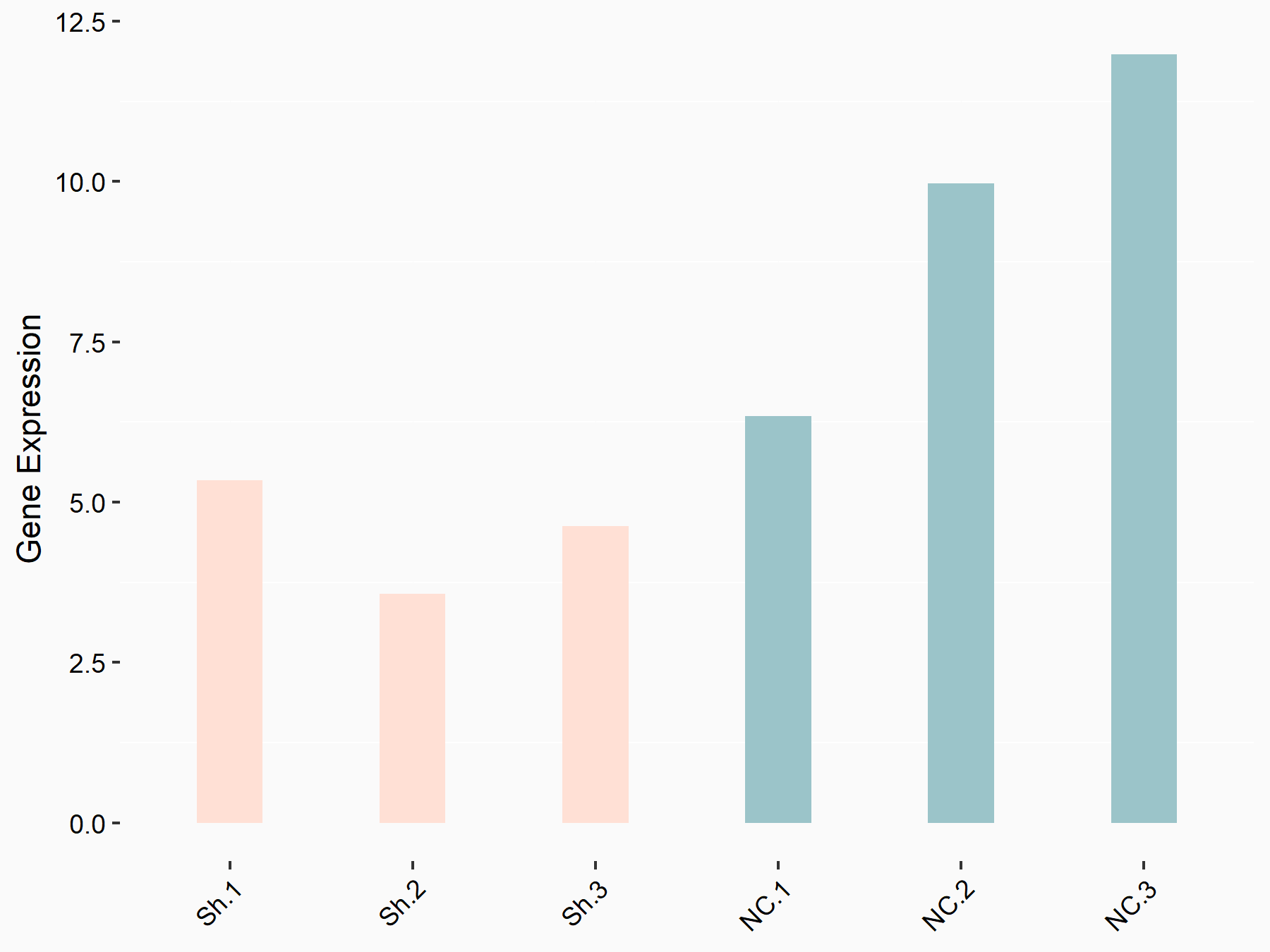 |
logFC: -8.94E-01 p-value: 2.29E-02 |
| More Results | Click to View More RNA-seq Results | |
Breast cancer [ICD-11: 2C60]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [18] | |||
| Responsed Disease | Breast cancer [ICD-11: 2C60] | |||
| Target Regulation | Up regulation | |||
| Cell Process | Epithelial-mesenchymal transformation | |||
| Cell proliferation | ||||
| Cell invasion | ||||
In-vitro Model |
T-47D | Invasive breast carcinoma | Homo sapiens | CVCL_0553 |
| SK-BR-3 | Breast adenocarcinoma | Homo sapiens | CVCL_0033 | |
| MDA-MB-231 | Breast adenocarcinoma | Homo sapiens | CVCL_0062 | |
| MCF-7 | Invasive breast carcinoma | Homo sapiens | CVCL_0031 | |
| MCF-10A | Normal | Homo sapiens | CVCL_0598 | |
| BT-549 | Invasive breast carcinoma | Homo sapiens | CVCL_1092 | |
| In-vivo Model | NOD/SCID immune-deficient mice were purchased from Shanghai Experimental Animal Center. 2 * 106 MCF-7 cells transduced with sh-NC or sh-YTHDF1-were subcutaneously injected into the mice (5/group). Tumor width and length were measured every 7 days. Tumor volume = (length * width2)/2. After 7 weeks, mice were sacrificed, and the weight of tumors was detected. Xenografts were collected for HE staining, immunohistochemistry staining and western blot analysis.For spontaneous lung metastasis assay, 4 * 106 sh-NC or sh-YTHDF1#2 transduced MCF-7 cells were injected into the mammary fat pads of the NOD/SCID mice (5/group). The primary tumor was removed when its volume reached 150 mm3. The mice were sacrificed, and lung metastasis nodules were counted 12 weeks after the removal. | |||
| Response Summary | Forkhead box protein M1 (FOXM1) is a target of YTHDF1. Through recognizing and binding to the m6A-modified mRNA of FOXM1, YTHDF1 accelerated the translation process of FOXM1 and promoted breast cancer metastasis. | |||
Multiple myeloma [ICD-11: 2A83]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [46] | |||
| Responsed Disease | Multiple myeloma [ICD-11: 2A83.1] | |||
In-vitro Model |
NCI-H929 | Plasma cell myeloma | Homo sapiens | CVCL_1600 |
| MM1.S | Plasma cell myeloma | Homo sapiens | CVCL_8792 | |
| U266B1 | Plasma cell myeloma | Homo sapiens | CVCL_0566 | |
| CAG | Plasma cell myeloma | Homo sapiens | CVCL_D569 | |
Pulmonary hypertension [ICD-11: BB01]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [47] | |||
| Responsed Disease | Pulmonary arterial hypertension [ICD-11: BB01.0] | |||
| Target Regulation | Up regulation | |||
| In-vivo Model | Twenty-four C57BL/6 mice, 6-8-week-old, weighing 20-25 g, were obtained from the animal center of Nanchang University. All the animals were raised in specific-pathogen-free (SPF) conditions in a 12-h light-dark cycle and supplied with free food and water. The animal experiments were conducted following the standard guidelines and permitted by the ethics committee of The First Affiliated Hospital of Nanchang University. The mice were randomly assigned into 4 groups (n = 6 per group). Mice in the control (normoxia) group were housed at 21% O2 (room air) for 4 weeks. Mice in the PAH (hypoxia) group were housed in a chamber with 10% O2 and 90% N2 for 4 weeks. The chamber has an adsorption-type oxygen concentrator to control the flow rates of compressed air and nitrogen (Teijin, Tokyo, Japan). Mice in the PAH + si-YTHDF1 group received a tail vein injection of adenovirus containing si-YTHDF1 (Ad-si-YTHDF1). Mice in the PAH + si-NC group received a tail vein injection of Ad-si-NC. The adenovirus was immediately injected when hypoxia subjection and the adenovirus dose was 1.0 × 109 PFU with 100 μL saline for each mouse. | |||
Insulin-like growth factor I (IGF1)
| Representative RNA-seq result indicating the expression of this target gene regulated by YTHDF1 | ||
| Cell Line | AGS cell line | Homo sapiens |
|
Treatment: shYTHDF1 AGS
Control: shNC AGS
|
GSE166972 | |
| Regulation |
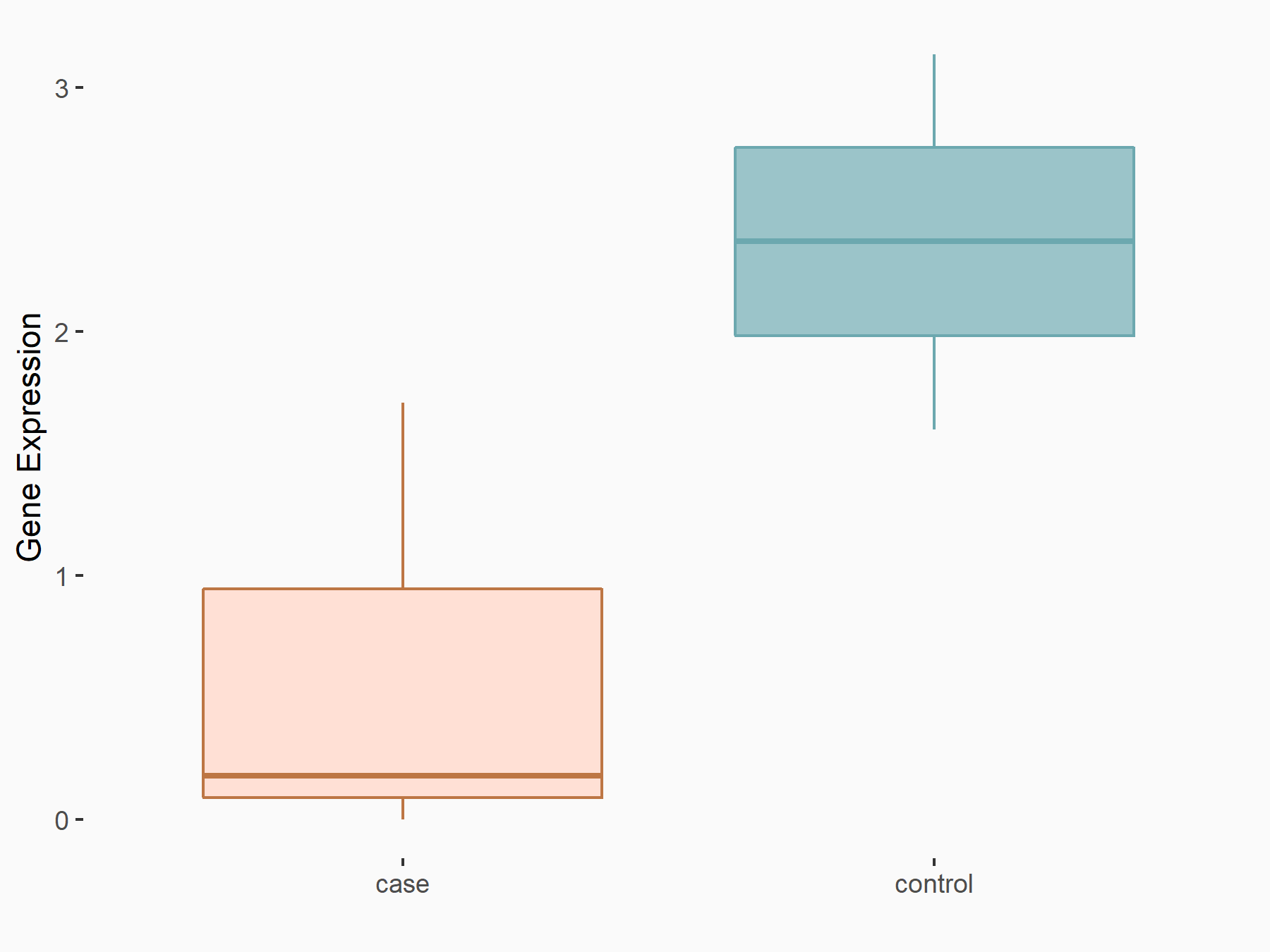 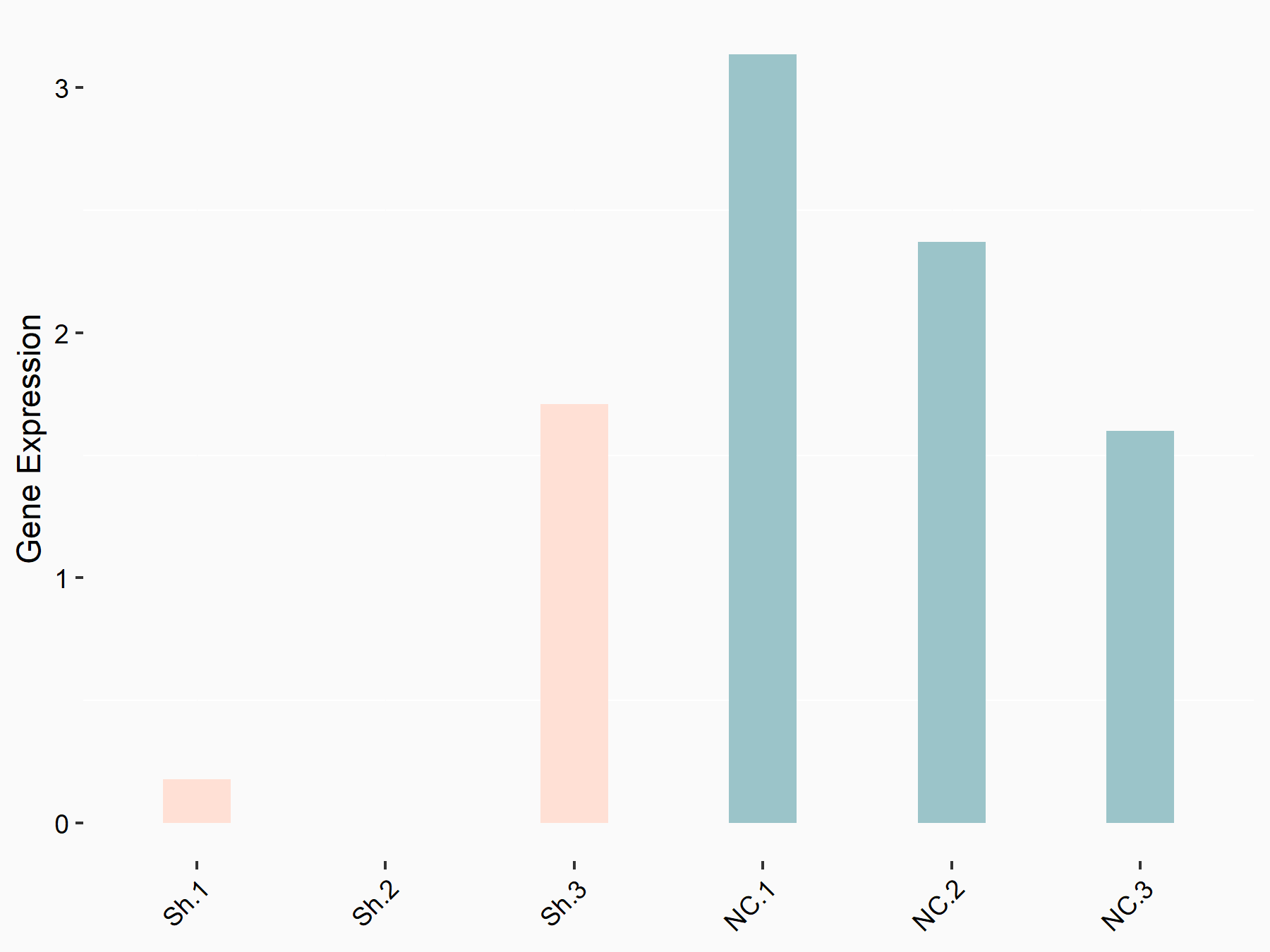 |
logFC: -1.17E+00 p-value: 4.12E-02 |
| More Results | Click to View More RNA-seq Results | |
Ageing-related disease [ICD-11: 9B10-9B60]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [19] | |||
| Responsed Disease | Ageing-related disease [ICD-11: 9B10-9B60] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | Nucleotide excision repair | hsa03420 | ||
| Cell Process | DNA repair and mitochondrial stress | |||
In-vitro Model |
Mouse fibroblasts (Major cellular components of loose connective tissue) | |||
| Response Summary | The long-lived endocrine mutants - Snell dwarf, growth hormone receptor deletion and pregnancy-associated plasma protein-A knockout - all show increases in the N6-adenosine-methyltransferases (METTL3/14) that catalyze 6-methylation of adenosine (m6A) in the 5' UTR region of select mRNAs. In addition, these mice have elevated levels of YTHDF1, which recognizes m6A and promotes translation by a cap-independent mechanism. Augmented translation by cap-independent pathways facilitated by m6A modifications contribute to the stress resistance and increased healthy longevity of mice with diminished GH and Insulin-like growth factor I (IGF1) signals. | |||
Nucleolar protein 3 (ARC)
| Representative RNA-seq result indicating the expression of this target gene regulated by YTHDF1 | ||
| Cell Line | AGS cell line | Homo sapiens |
|
Treatment: shYTHDF1 AGS
Control: shControl AGS
|
GSE159425 | |
| Regulation |
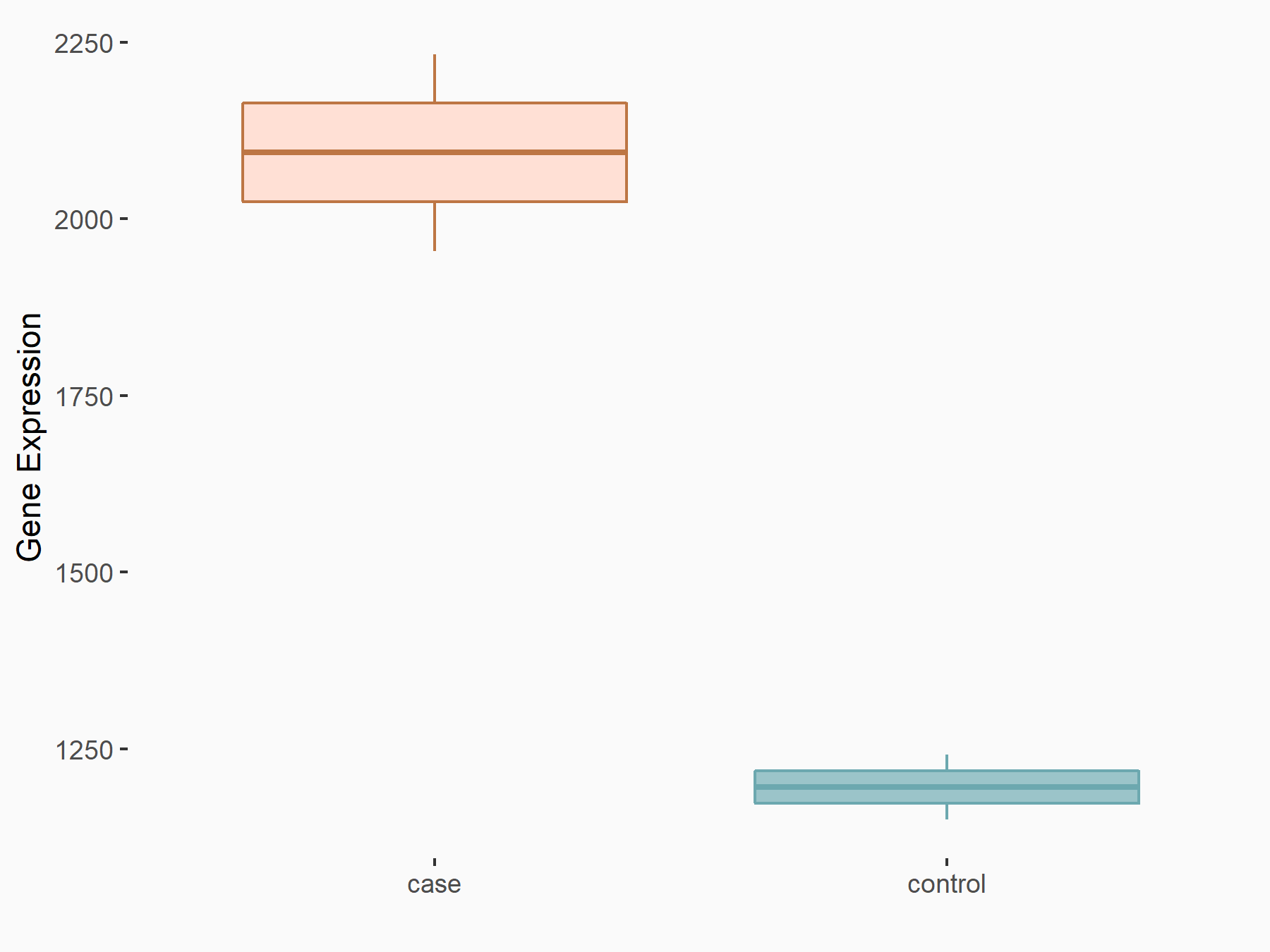 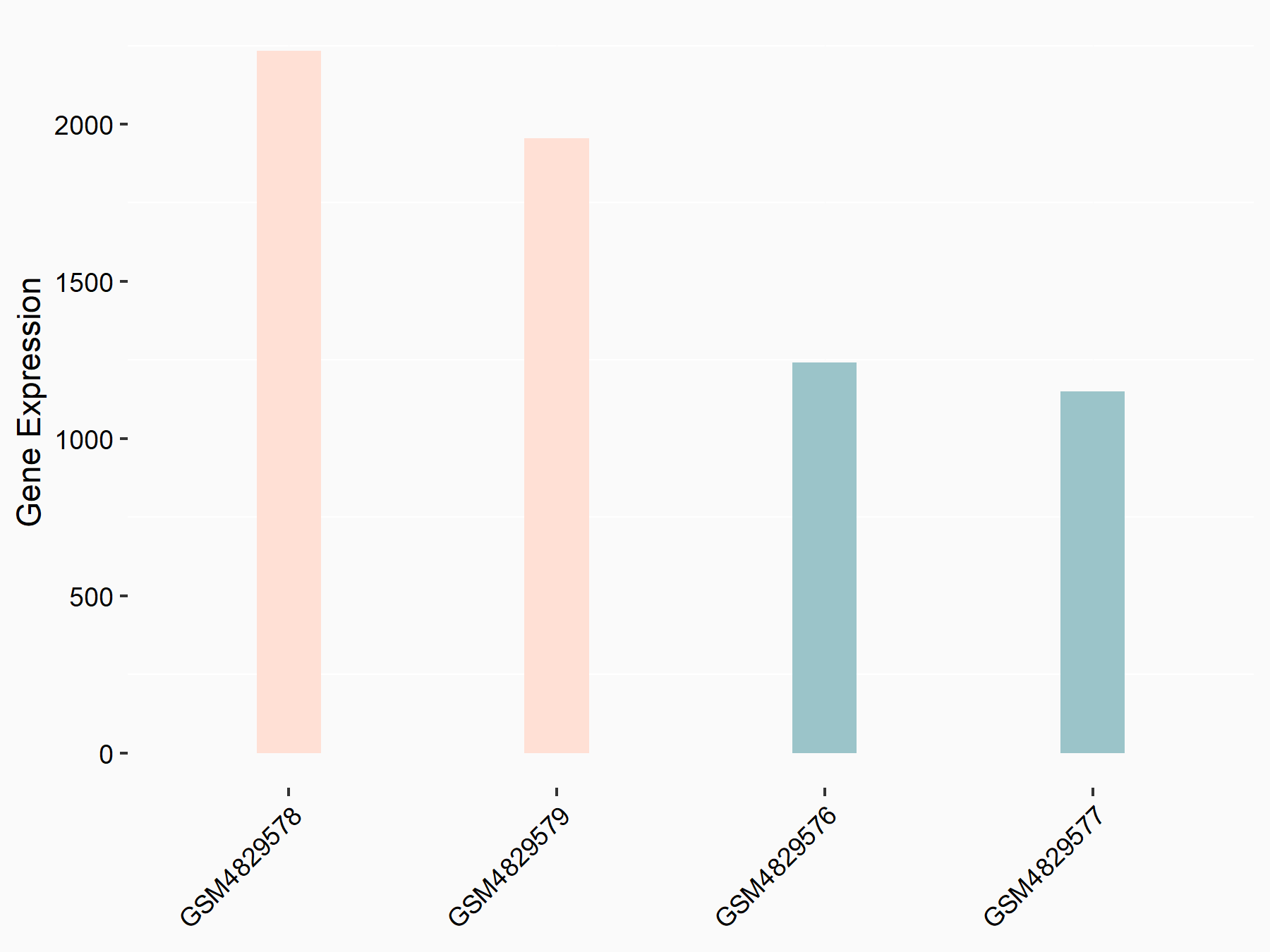 |
logFC: 8.08E-01 p-value: 1.08E-05 |
| More Results | Click to View More RNA-seq Results | |
Alzheimer disease [ICD-11: 8A20]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [20] | |||
| Responsed Disease | Alzheimer disease [ICD-11: 8A20] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
SH-SY5Y | Neuroblastoma | Homo sapiens | CVCL_0019 |
| Response Summary | METTL3 rescues the A-Bete-induced reduction of Nucleolar protein 3 (ARC) expression via YTHDF1-Dependent m6A modification, which suggests an important mechanism of epigenetic alteration in AD. | |||
Pyruvate kinase PKM (PKM2/PKM)
| Representative RNA-seq result indicating the expression of this target gene regulated by YTHDF1 | ||
| Cell Line | AGS cell line | Homo sapiens |
|
Treatment: shYTHDF1 AGS
Control: shControl AGS
|
GSE159425 | |
| Regulation |
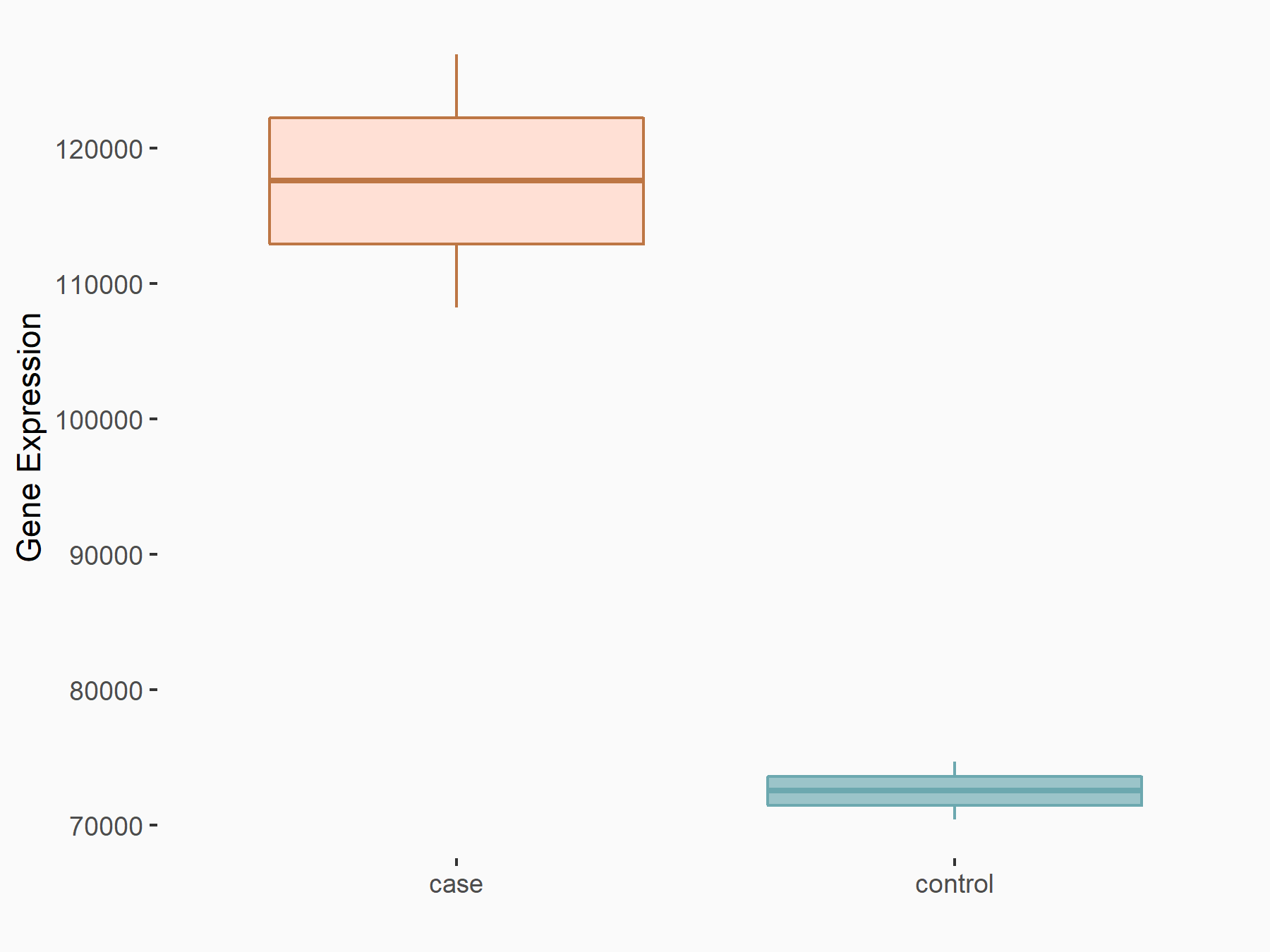 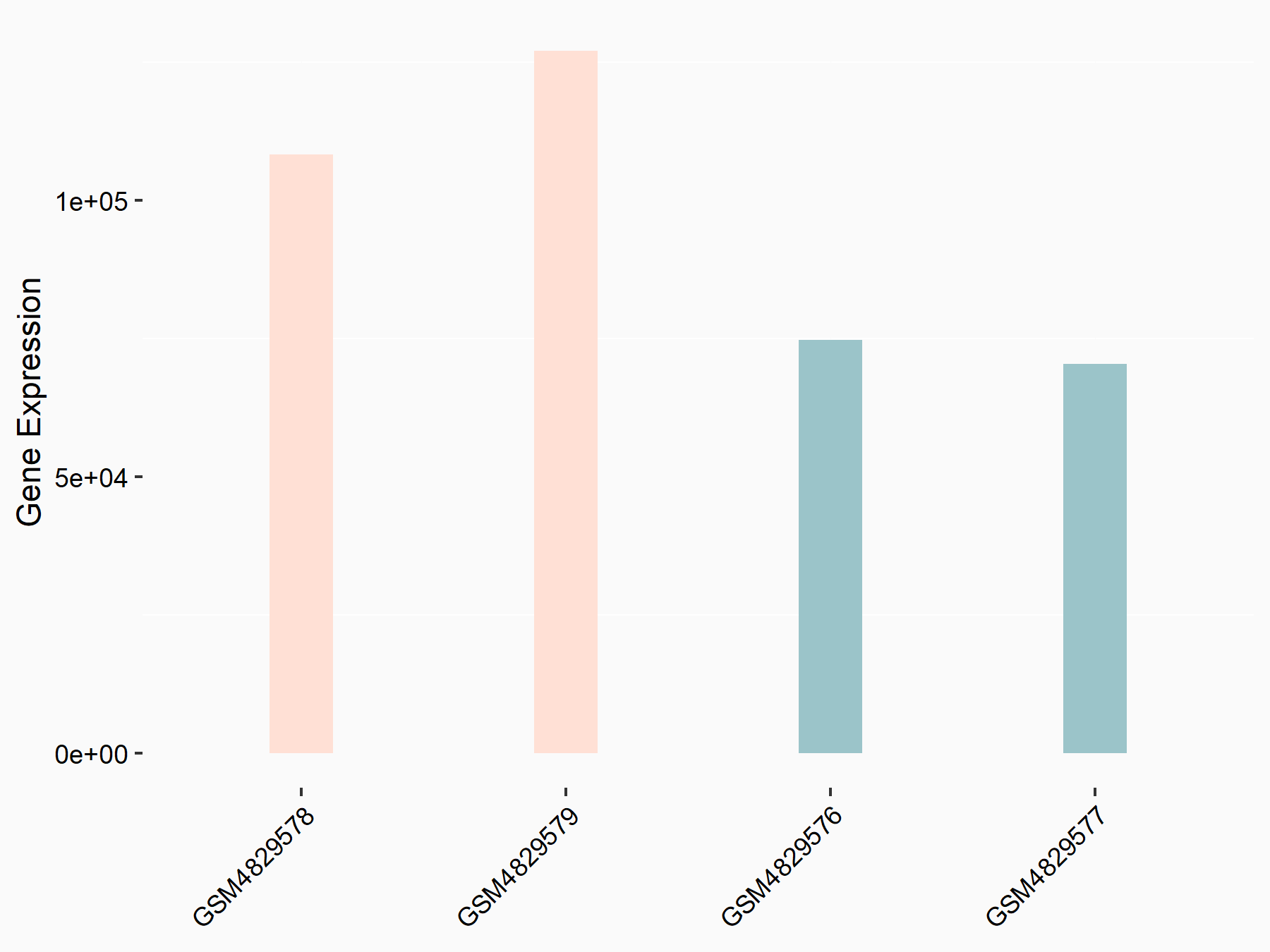 |
logFC: 6.97E-01 p-value: 4.17E-05 |
| More Results | Click to View More RNA-seq Results | |
Breast cancer [ICD-11: 2C60]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [21] | |||
| Responsed Disease | Breast cancer [ICD-11: 2C60] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | HIF-1 signaling pathway | hsa04066 | ||
| Cell Process | Glycolysis | |||
In-vitro Model |
T-47D | Invasive breast carcinoma | Homo sapiens | CVCL_0553 |
| MDA-MB-231 | Breast adenocarcinoma | Homo sapiens | CVCL_0062 | |
| MCF-7 | Invasive breast carcinoma | Homo sapiens | CVCL_0031 | |
| MCF-10A | Normal | Homo sapiens | CVCL_0598 | |
| BT-474 | Invasive breast carcinoma | Homo sapiens | CVCL_0179 | |
| In-vivo Model | Each mouse was injected subcutaneously with 5 × 106 units of tumor cells to construct the tumor model. | |||
| Response Summary | Tumor hypoxia can transcriptionally induce HIF1alpha and post-transcriptionally inhibit the expression of miR-16-5p to promote YTHDF1 expression, which could sequentially enhance tumor glycolysis by upregulating Pyruvate kinase PKM (PKM2/PKM) and eventually increase the tumorigenesis and metastasis potential of breast cancer cells. | |||
RAC-alpha serine/threonine-protein kinase (AKT1)
| Representative RNA-seq result indicating the expression of this target gene regulated by YTHDF1 | ||
| Cell Line | AGS cell line | Homo sapiens |
|
Treatment: shYTHDF1 AGS
Control: shNC AGS
|
GSE166972 | |
| Regulation |
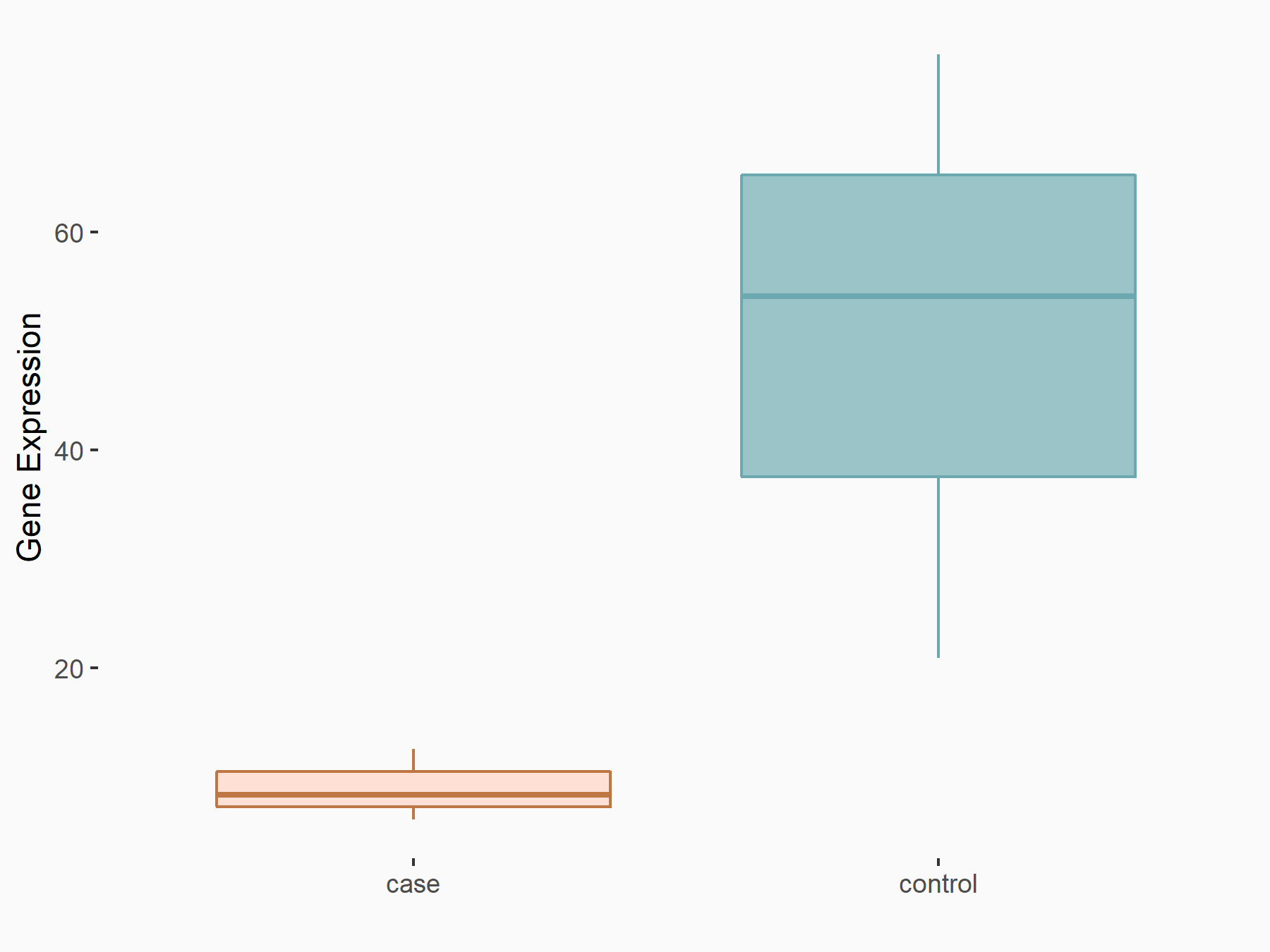 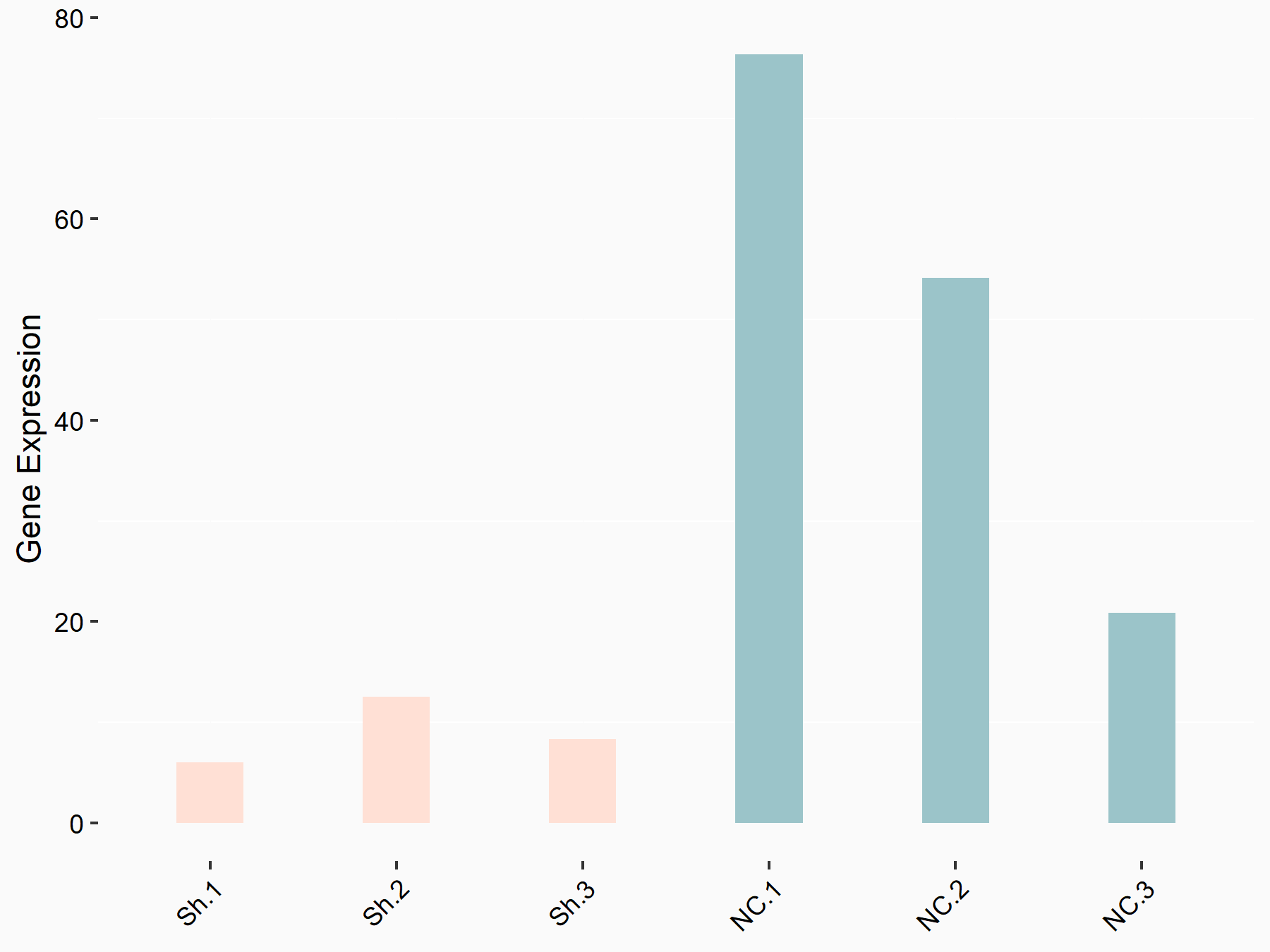 |
logFC: -2.24E+00 p-value: 7.14E-03 |
| More Results | Click to View More RNA-seq Results | |
Liver cancer [ICD-11: 2C12]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [22] | |||
| Responsed Disease | Hepatocellular carcinoma [ICD-11: 2C12.02] | |||
| Target Regulation | Down regulation | |||
| Pathway Response | PI3K-Akt signaling pathway | hsa04151 | ||
| mTOR signaling pathway | hsa04150 | |||
| Cell Process | Epithelial-mesenchymal transition | |||
| Cell migration | ||||
| Cell invasion | ||||
| Cell proliferation | ||||
In-vitro Model |
SNU-398 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_0077 |
| SK-HEP-1 | Liver and intrahepatic bile duct epithelial neoplasm | Homo sapiens | CVCL_0525 | |
| PLC/PRF/5 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_0485 | |
| L-02 | Endocervical adenocarcinoma | Homo sapiens | CVCL_6926 | |
| Huh-7 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_0336 | |
| Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 | |
| Hep 3B2.1-7 | Childhood hepatocellular carcinoma | Homo sapiens | CVCL_0326 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| HCCLM3 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_6832 | |
| In-vivo Model | Ten four-week-old BALB/c male nude mice (GemPharmatech, Jiangsu, China) were subcutaneously injected with control Huh7 cells 2 × 106 (left-back) and stable knockdown of YTHDF1 Huh7 cells 2 × 106 (right-back). These cells were respectively premixed with 50 ul Matrigel (Corning, 354,234) in 100 ul PBS. | |||
| Response Summary | YTHDF1 contributes to the progression of HCC by activating PI3K/RAC-alpha serine/threonine-protein kinase (AKT1)/mTOR signaling pathway and inducing EMT. | |||
Transcription factor E2F1 (E2F1)
| Representative RNA-seq result indicating the expression of this target gene regulated by YTHDF1 | ||
| Cell Line | AGS cell line | Homo sapiens |
|
Treatment: shYTHDF1 AGS
Control: shNC AGS
|
GSE166972 | |
| Regulation |
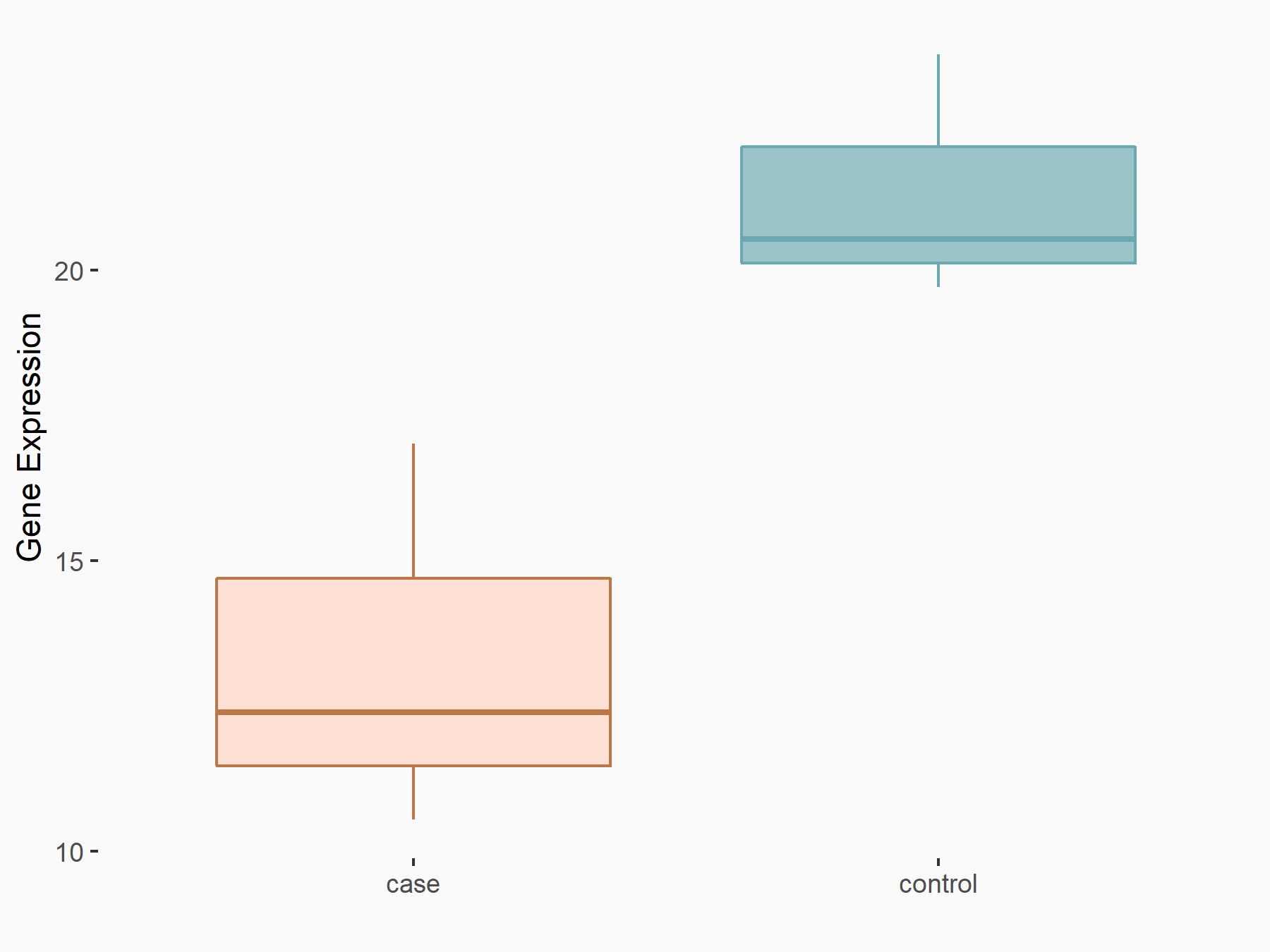 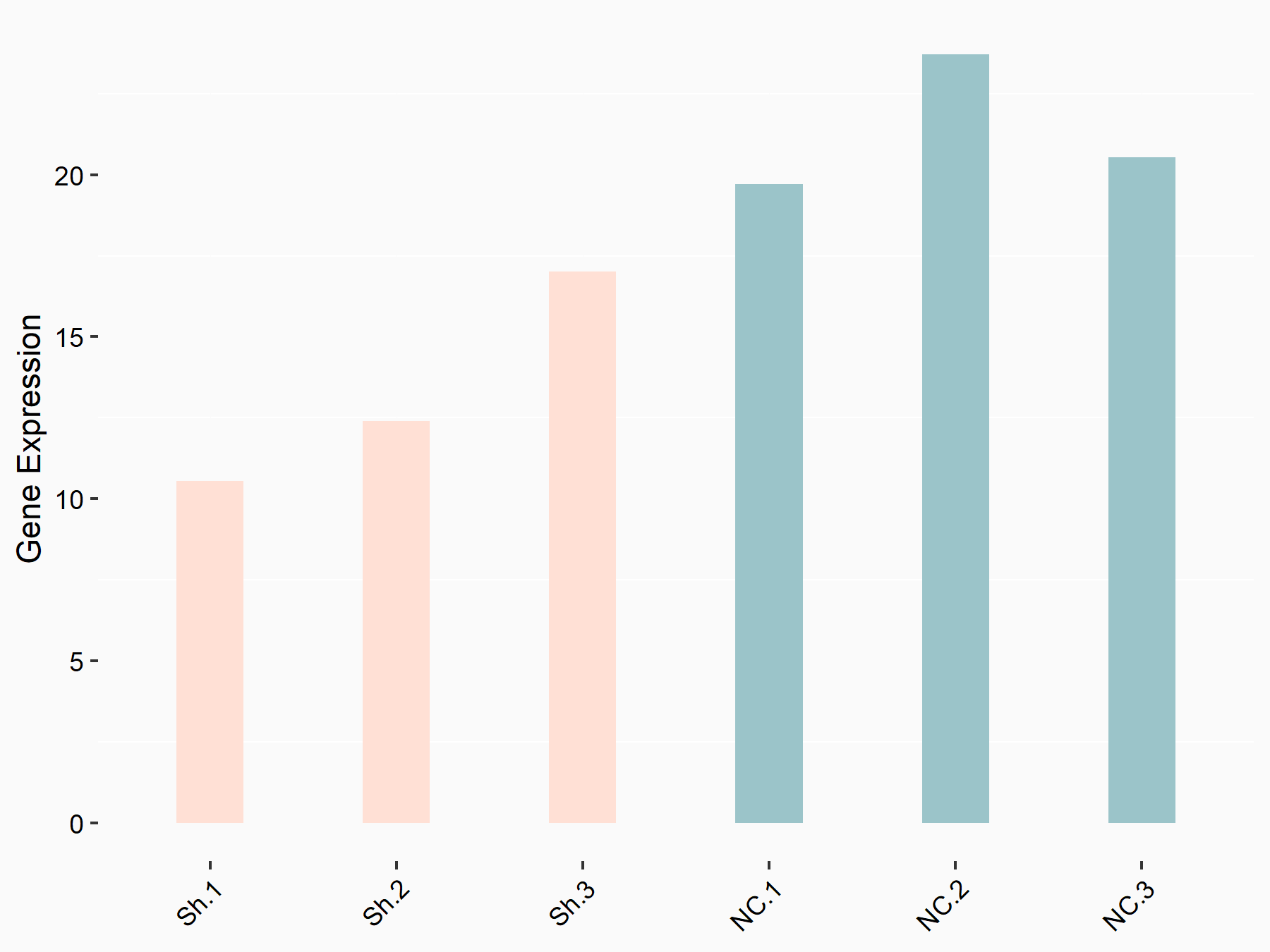 |
logFC: -6.62E-01 p-value: 3.39E-02 |
| More Results | Click to View More RNA-seq Results | |
Lung cancer [ICD-11: 2C25]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [9] | |||
| Responsed Disease | Non-small-cell lung carcinoma [ICD-11: 2C25.Y] | |||
| Pathway Response | p53 signaling pathway | hsa04115 | ||
| Central carbon metabolism in cancer | hsa05230 | |||
| PD-L1 expression and PD-1 checkpoint pathway in cancer | hsa05235 | |||
| Response Summary | This study revealed that m6A methylation is closely related to the poor prognosis of non-small cell lung cancer patients via interference with the TIME, which suggests that m6A plays a role in optimizing individualized immunotherapy management and improving prognosis. The expression levels of METTL3, FTO and YTHDF1 in non-small cell lung cancer were changed. Patients in Cluster 1 had lower immunoscores, higher programmed death-ligand 1 (PD-L1) expression, and shorter overall survival compared to patients in Cluster 2. The hallmarks of the Myelocytomatosis viral oncogene (MYC) targets, Transcription factor E2F1 (E2F1) targets were significantly enriched. | |||
Tripartite motif-containing protein 29 (TRIM29)
| Representative RNA-seq result indicating the expression of this target gene regulated by YTHDF1 | ||
| Cell Line | AGS cell line | Homo sapiens |
|
Treatment: shYTHDF1 AGS
Control: shNC AGS
|
GSE166972 | |
| Regulation |
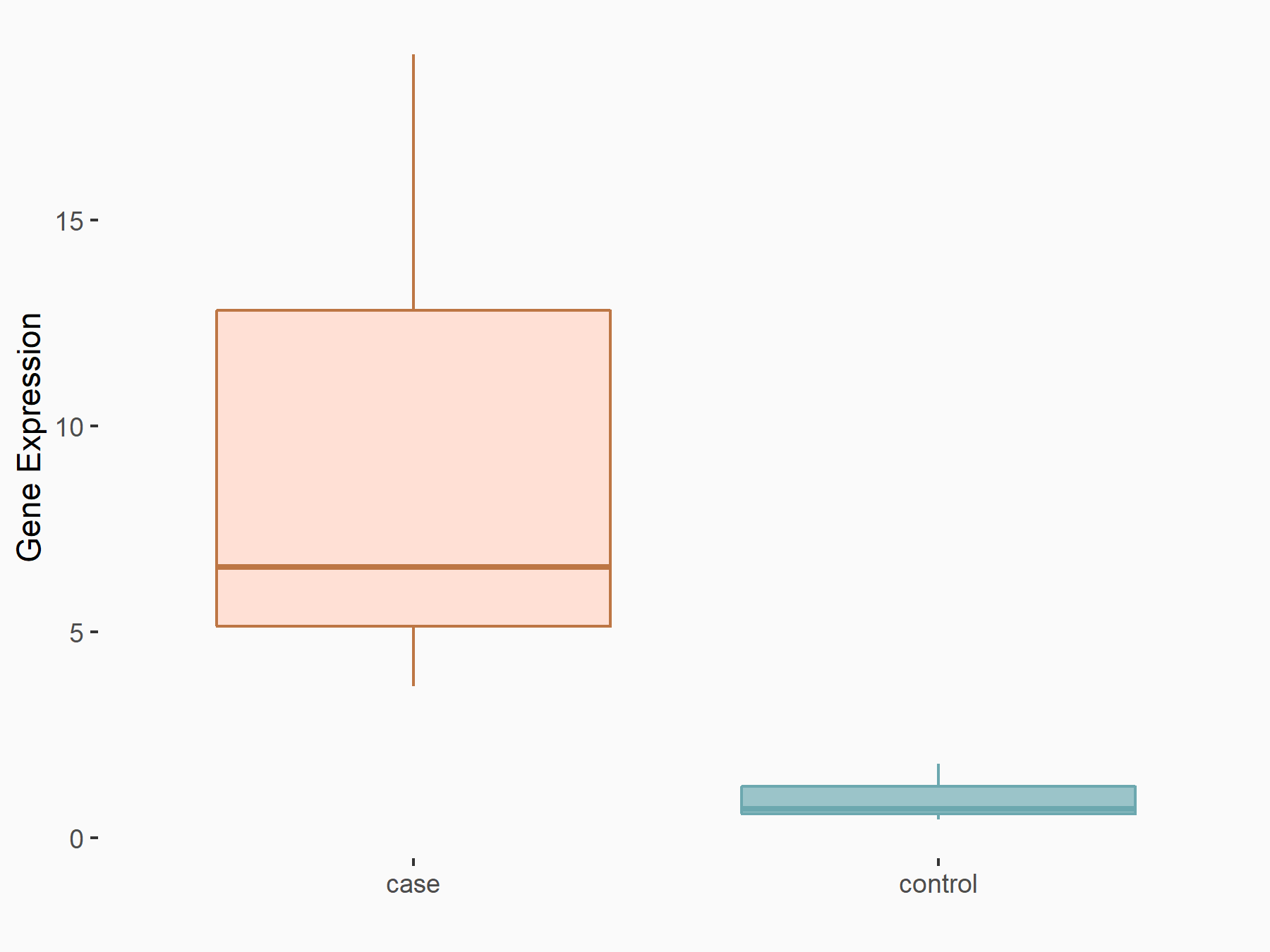 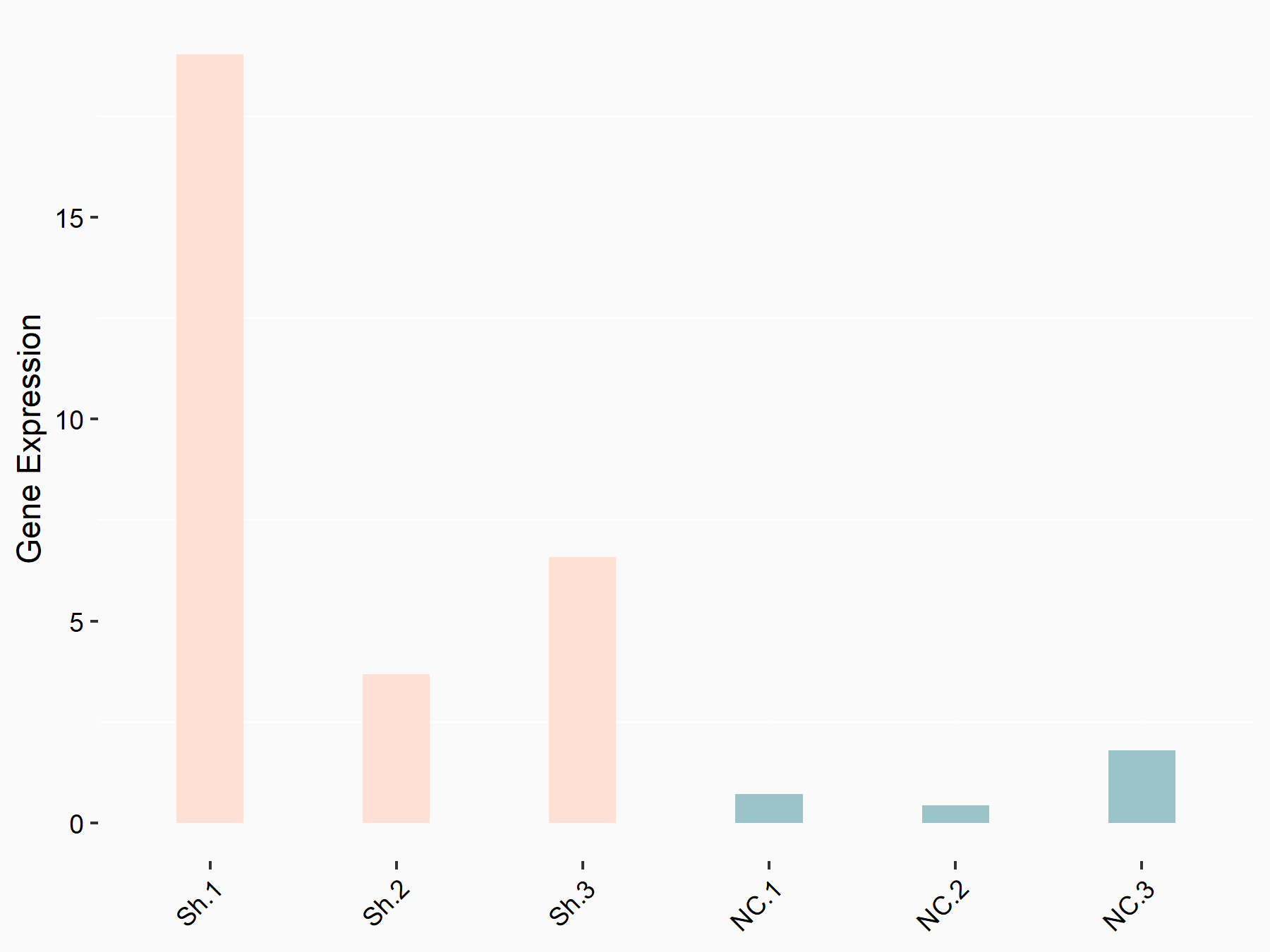 |
logFC: 2.23E+00 p-value: 1.11E-02 |
| More Results | Click to View More RNA-seq Results | |
Ovarian cancer [ICD-11: 2C73]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [23] | |||
| Responsed Disease | Ovarian cancer [ICD-11: 2C73] | |||
| Responsed Drug | Cisplatin | Approved | ||
| Target Regulation | Up regulation | |||
| Cell Process | Ectopic expression | |||
In-vitro Model |
A2780 | Ovarian endometrioid adenocarcinoma | Homo sapiens | CVCL_0134 |
| SK-OV-3 | Ovarian serous cystadenocarcinoma | Homo sapiens | CVCL_0532 | |
| In-vivo Model | The specified number of viable SKOV3/DDP cells and SKOV3/DDP cells with TRIM29 knock down were resuspended in 100 uL PBS, injected subcutaneously under the left and right back of 4-week old nude mice respectively (n = 3 per group). | |||
| Response Summary | m6A-YTHDF1-mediated Tripartite motif-containing protein 29 (TRIM29) upregulation facilitates the stem cell-like phenotype of cisplatin-resistant ovarian cancer cells. TRIM29 acts as an oncogene to promote the CSC-like features of cisplatin-resistant ovarian cancer in an m6A-YTHDF1-dependent manner. | |||
Ubiquitin carboxyl-terminal hydrolase 14 (USP14)
| Representative RNA-seq result indicating the expression of this target gene regulated by YTHDF1 | ||
| Cell Line | AGS cell line | Homo sapiens |
|
Treatment: shYTHDF1 AGS
Control: shNC AGS
|
GSE166972 | |
| Regulation |
 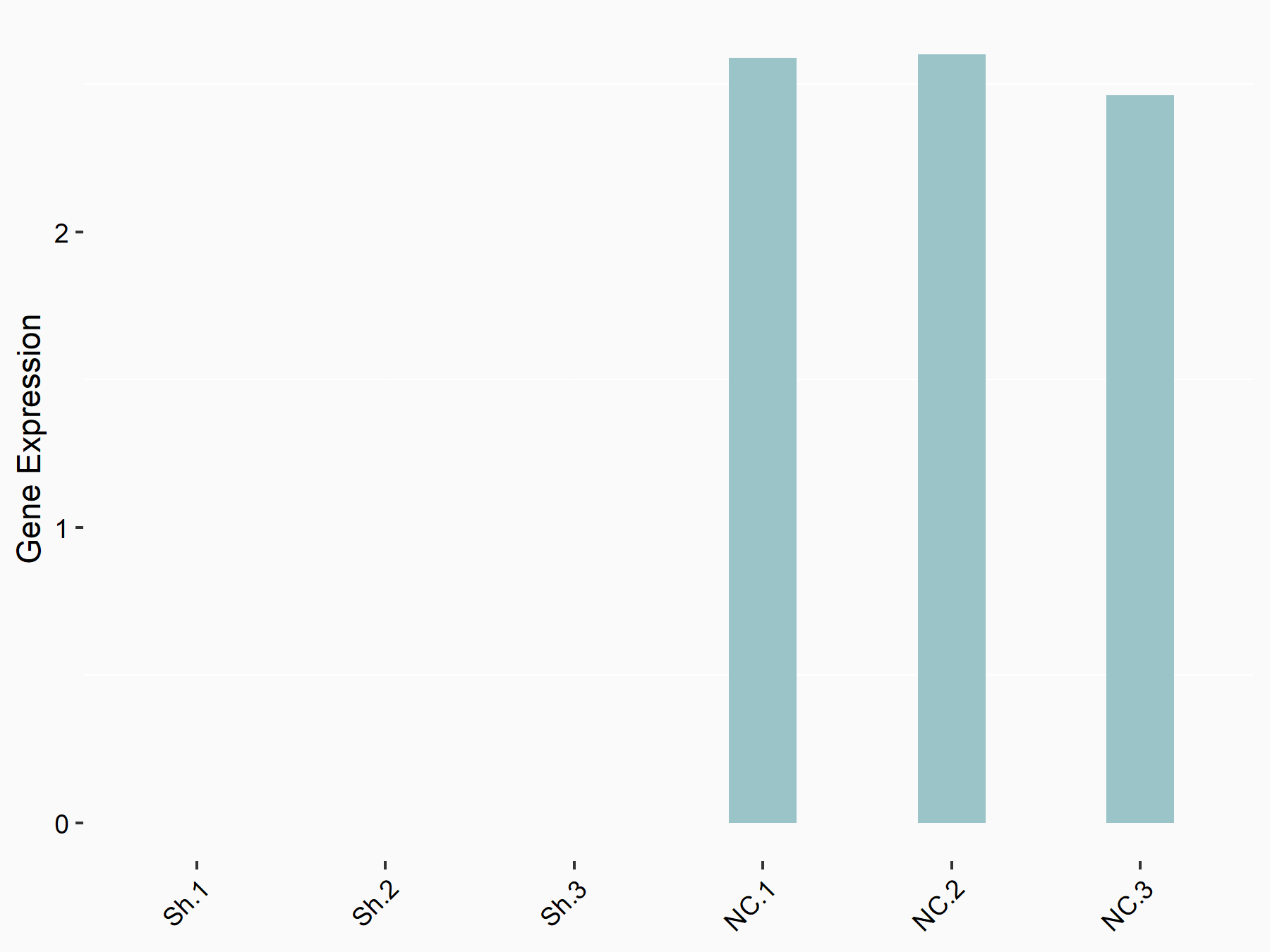 |
logFC: -1.83E+00 p-value: 4.80E-05 |
| More Results | Click to View More RNA-seq Results | |
Gastric cancer [ICD-11: 2B72]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [24] | |||
| Responsed Disease | Gastric cancer [ICD-11: 2B72] | |||
| Target Regulation | Up regulation | |||
| Cell Process | Cell proliferation and invasion | |||
| Cell apoptosis | ||||
In-vitro Model |
AGS | Gastric adenocarcinoma | Homo sapiens | CVCL_0139 |
| BGC-823 | Gastric carcinoma | Homo sapiens | CVCL_3360 | |
| GES-1 | Normal | Homo sapiens | CVCL_EQ22 | |
| HGC-27 | Gastric carcinoma | Homo sapiens | CVCL_1279 | |
| MKN28 | Gastric tubular adenocarcinoma | Homo sapiens | CVCL_1416 | |
| SGC-7901 | Gastric carcinoma | Homo sapiens | CVCL_0520 | |
| In-vivo Model | BGC-823 cells (5 × 106) with shRNAs targeting YTHDF1 (sh-YTHDF1) or shRNAs targeting control (sh-NC) were trypsinized and suspended in 0.1 mL PBS and injected subcutaneously into the BALB/c mice (n = 5 mice per group). | |||
| Response Summary | YTHDF1 promoted Ubiquitin carboxyl-terminal hydrolase 14 (USP14) protein translation in an m6A-dependent manner. USP14 upregulation was positively correlated with YTHDF1 expression and indicated a poor prognosis in gastric cancer. | |||
Apoptosis regulator Bcl-2 (BCL2)
| Representative RIP-seq result supporting the interaction between the target gene and YTHDF1 | ||
| Cell Line | Hela | Homo sapiens |
| Regulation | logFC: 2.25E+00 | GSE63591 |
B-cell lymphomas [ICD-11: 2A86]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [25] | |||
| Responsed Disease | B-cell lymphomas [ICD-11: 2A86] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | Apoptosis | hsa04210 | ||
| Autophagy | hsa04140 | |||
| Cell Process | Cell proliferation and metastasis | |||
| Cell apoptosis | ||||
In-vitro Model |
ATDC-5 | Mouse teratocarcinoma | Mus musculus | CVCL_3894 |
| In-vivo Model | For MIA + SAH control, S-adenosylhomocysteine (SAH), Mettl3 inhibitor (10 mg/kg) (MCE, NJ, USA) was injected intraperitoneally before MIA injection and maintained twice a week until mice were sacrificed. | |||
| Response Summary | Mettl3 inhibitor, S-adenosylhomocysteine promoted the apoptosis and autophagy of chondrocytes with inflammation in vitro and aggravated the degeneration of chondrocytes and subchondral bone in monosodium iodoacetate (MIA) induced temporomandibular joint osteoarthritis mice in vivo. Bcl2 protein interacted with Beclin1 protein in chondrocytes induced by TNF-alpha stimulation. Mettl3 inhibits the apoptosis and autophagy of chondrocytes in inflammation through m6A/Ythdf1/Apoptosis regulator Bcl-2 (BCL2) signal axis which provides promising therapeutic strategy for temporomandibular joint osteoarthritis. | |||
Dentofacial anomalies [ICD-11: DA0E]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [25] | |||
| Responsed Disease | Temporomandibular joint disorders [ICD-11: DA0E.8] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | Apoptosis | hsa04210 | ||
| Autophagy | hsa04140 | |||
| Cell Process | Cell proliferation and metastasis | |||
| Cell apoptosis | ||||
In-vitro Model |
ATDC-5 | Mouse teratocarcinoma | Mus musculus | CVCL_3894 |
| In-vivo Model | For MIA + SAH control, S-adenosylhomocysteine (SAH), Mettl3 inhibitor (10 mg/kg) (MCE, NJ, USA) was injected intraperitoneally before MIA injection and maintained twice a week until mice were sacrificed. | |||
| Response Summary | Mettl3 inhibitor, S-adenosylhomocysteine promoted the apoptosis and autophagy of chondrocytes with inflammation in vitro and aggravated the degeneration of chondrocytes and subchondral bone in monosodium iodoacetate (MIA) induced temporomandibular joint osteoarthritis mice in vivo. Bcl2 protein interacted with Beclin1 protein in chondrocytes induced by TNF-alpha stimulation. Mettl3 inhibits the apoptosis and autophagy of chondrocytes in inflammation through m6A/Ythdf1/Apoptosis regulator Bcl-2 (BCL2) signal axis which provides promising therapeutic strategy for temporomandibular joint osteoarthritis. | |||
Autophagy-related protein 2 homolog A (ATG2A)
| Representative RIP-seq result supporting the interaction between the target gene and YTHDF1 | ||
| Cell Line | Hela | Homo sapiens |
| Regulation | logFC: 2.31E+00 | GSE63591 |
Liver cancer [ICD-11: 2C12]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [26] | |||
| Responsed Disease | Hepatocellular carcinoma [ICD-11: 2C12.02] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | Autophagy | hsa04140 | ||
| HIF-1 signaling pathway | hsa04066 | |||
| Cell Process | Cell proliferation | |||
| Cell migration | ||||
| Cell invasion | ||||
| Cell autophagy | ||||
In-vitro Model |
Hep 3B2.1-7 | Childhood hepatocellular carcinoma | Homo sapiens | CVCL_0326 |
| Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 | |
| Huh-7 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_0336 | |
| SMMC-7721 | Endocervical adenocarcinoma | Homo sapiens | CVCL_0534 | |
| In-vivo Model | HCC cells (1 × 106/100 uL PBS) were administered to 4-week-old female BALB/c nude mice by subcutaneous injection (n = 6). | |||
| Response Summary | HIF-1-alpha-induced YTHDF1 expression was associated with hypoxia-induced autophagy and autophagy-related hepatocellular carcinoma progression via promoting translation of autophagy-related genes Autophagy-related protein 2 homolog A (ATG2A) and ATG14 in a m6A-dependent manner. | |||
Beclin-1 associated RUN domain containing protein (RUBCN/Rubicon)
| Representative RIP-seq result supporting the interaction between the target gene and YTHDF1 | ||
| Cell Line | Hela | Homo sapiens |
| Regulation | logFC: 1.78E+00 | GSE63591 |
Non-alcoholic fatty liver disease [ICD-11: DB92]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [27] | |||
| Responsed Disease | Non-alcoholic fatty liver disease [ICD-11: DB92] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | Autophagy | hsa04140 | ||
| Cell Process | Cell autophagy | |||
In-vitro Model |
AML12 | Normal | Mus musculus | CVCL_0140 |
| Hepa 1-6 | Hepatocellular carcinoma of the mouse | Mus musculus | CVCL_0327 | |
| Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 | |
| In-vivo Model | All mice were housed in specific pathogen-free conditions in the animal facility with constant temperature and humidity under a 12-h light/12-h dark cycle, with free access to water and food. After a week of adaptation, they were then divided into two groups randomly and fed with a HFD (60 kcal% fat, D12492, Research Diets) or standard normal chow diet (CD) for 16 weeks, respectively. | |||
| Response Summary | The upregulation of METTL3 and YTHDF1 induced by lipotoxicity contributes to the elevated expression level of Beclin-1 associated RUN domain containing protein (RUBCN/Rubicon) in an m6A-dependent manner, which inhibits the fusion of autophagosomes and lysosomes and further suppresses the clearance of LDs via lysosomes in nonalcoholic fatty liver disease. | |||
Catenin beta-1 (CTNNB1/Beta-catenin)
| Representative RIP-seq result supporting the interaction between the target gene and YTHDF1 | ||
| Cell Line | Hela | Homo sapiens |
| Regulation | logFC: 2.05E+00 | GSE63591 |
Colorectal cancer [ICD-11: 2B91]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [28] | |||
| Responsed Disease | Colorectal cancer [ICD-11: 2B91] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | Wnt signaling pathway | hsa04310 | ||
| Signaling pathways regulating pluripotency of stem cells | hsa04550 | |||
| Cell Process | Cell Tumorigenicity | |||
In-vitro Model |
NCM460 | Normal | Homo sapiens | CVCL_0460 |
| SW480 | Colon adenocarcinoma | Homo sapiens | CVCL_0546 | |
| SW620 | Colon adenocarcinoma | Homo sapiens | CVCL_0547 | |
| In-vivo Model | 1 × 105 parental HT29-shNC cells, HT29-shYTHDF1 cells, HT29-shNC colonospheres, and HT29-shYTHDF1 colonospheres were inoculated subcutaneously into the left inguinal folds of the nude mice. | |||
| Response Summary | Silencing YTHDF1 significantly inhibited Wnt/Catenin beta-1 (CTNNB1/Beta-catenin) pathway activity in Colorectal cancer cells. | |||
Lung cancer [ICD-11: 2C25]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [29] | |||
| Responsed Disease | Non-small-cell lung carcinoma [ICD-11: 2C25.Y] | |||
| Target Regulation | Up regulation | |||
| Cell Process | Cell proliferation | |||
| Cell invasion | ||||
| Cell migration | ||||
| Cell apoptosis | ||||
In-vitro Model |
A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 |
| NCI-1650 (Non-Small Cell Lung Cancer Cells) | ||||
| NCI-H1299 | Lung large cell carcinoma | Homo sapiens | CVCL_0060 | |
| NCI-H358 | Minimally invasive lung adenocarcinoma | Homo sapiens | CVCL_1559 | |
| Response Summary | ECs transmitted miR-376c into NSCLC cells through Evs, and inhibited the intracellular YTHDF1 expression and the Wnt/Catenin beta-1 (CTNNB1/Beta-catenin) pathway activation. YTHDF1 overexpression reversed the inhibitory role of miR-376c released by EC-Evs in non-small cell lung cancer cells. | |||
E3 SUMO-protein ligase RanBP2 (RANBP2)
| Representative RIP-seq result supporting the interaction between the target gene and YTHDF1 | ||
| Cell Line | Hela | Homo sapiens |
| Regulation | logFC: 2.70E+00 | GSE63591 |
Cervical cancer [ICD-11: 2C77]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [30] | |||
| Responsed Disease | Cervical cancer [ICD-11: 2C77] | |||
| Target Regulation | Up regulation | |||
| Cell Process | Cell growth | |||
| Cell migration | ||||
| Cell invasion | ||||
In-vitro Model |
HEK293T | Normal | Homo sapiens | CVCL_0063 |
| HeLa | Endocervical adenocarcinoma | Homo sapiens | CVCL_0030 | |
| SiHa | Cervical squamous cell carcinoma | Homo sapiens | CVCL_0032 | |
| In-vivo Model | Balb/c female nude mice at 4-6 weeks old were injected with 4 × 106 cells subcutaneously on the back. | |||
| Response Summary | The oncogenic role of YTHDF1 in cervical cancer by regulating E3 SUMO-protein ligase RanBP2 (RANBP2) expression and YTHDF1 represents a potential target for cervical cancer therapy. | |||
Frizzled-7 (FZD7)
| Representative RIP-seq result supporting the interaction between the target gene and YTHDF1 | ||
| Cell Line | Hela | Homo sapiens |
| Regulation | logFC: 1.73E+00 | GSE63591 |
Gastric cancer [ICD-11: 2B72]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [31] | |||
| Responsed Disease | Gastric cancer [ICD-11: 2B72] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | Wnt signaling pathway | hsa04310 | ||
In-vitro Model |
MGC-803 | Gastric mucinous adenocarcinoma | Homo sapiens | CVCL_5334 |
| HGC-27 | Gastric carcinoma | Homo sapiens | CVCL_1279 | |
| In-vivo Model | Stable short hairpin (shRNA)-expressing MGC-803 cells (3 × 106) were suspended in 0.1 mL PBS and injected into the flanks of BALB/c mice (n = 8 mice/group) at 5-6 weeks of age. For the flanks injected mice, tumor growth was examined every 3 days. After 5 weeks, mice were sacrificed by cervical dislocation, and weight of xenografts was tested.BALB/c mice were randomly divided into three groups (n = 4 mice/group). A total of 3 × 106 stable shRNA-expressing MGC-803 cells were resuspended in 0.1 mL PBS and injected into the abdominal cavity. After 4 weeks, mice were sacrificed by cervical dislocation, abdominal cavities were opened, and the numbers of implantation metastasis were counted.For the pulmonary metastasis model, NOD/SCID mice were randomly divided into three groups (n = 4 mice/group). A total of 1 × 105 stable shRNA-expressing MGC-803 cells were resuspended in 0.1 mL PBS and injected into the lateral tail vein. After 7 weeks, mice were sacrificed by cervical dislocation, and lungs were extracted and fixed 4% paraformaldehyde in PBS. Paraffin embedding, sectioning, and staining with hematoxylin and eosin were performed. Visible lung metastases were measured and counted using a microscope. | |||
| Response Summary | Mutated YTHDF1 enhanced expression of Frizzled-7 (FZD7), leading to hyperactivation of the Wnt/Bete-catenin pathway and promotion of gastric cancer carcinogenesis. | |||
Poly [ADP-ribose] polymerase 1 (PARP1)
| Representative RIP-seq result supporting the interaction between the target gene and YTHDF1 | ||
| Cell Line | Hela | Homo sapiens |
| Regulation | logFC: 1.06E+00 | GSE63591 |
Gastric cancer [ICD-11: 2B72]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [32] | |||
| Responsed Disease | Gastric cancer [ICD-11: 2B72] | |||
| Responsed Drug | Oxaliplatin | Approved | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Nucleotide excision repair | hsa03420 | ||
| Signaling pathways regulating pluripotency of stem cells | hsa04550 | |||
| Cell Process | RNA stability | |||
| Excision repair | ||||
In-vitro Model |
SNU-719 | Gastric tubular adenocarcinoma | Homo sapiens | CVCL_5086 |
| MKN74 | Gastric tubular adenocarcinoma | Homo sapiens | CVCL_2791 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| AGS | Gastric adenocarcinoma | Homo sapiens | CVCL_0139 | |
| In-vivo Model | 100,000 pLKO and PARP1-sh1 (PT1 and PT2) cells were mixed with matrix gel and inoculate into BALB/C nude mice, respectively. After 25 days, 6 organoid transplanted tumor mice were treated with oxaliplatin (Sellekchem, s1224) twice a week for 4 weeks at a dose of 5 mg/kg. | |||
| Response Summary | m6A methyltransferase METTL3 facilitates oxaliplatin resistance in CD133+ gastric cancer stem cells by Promoting PARP1 mRNA stability which increases base excision repair pathway activity. METTTL3 enhances the stability of PARP1 by recruiting Poly [ADP-ribose] polymerase 1 (PARP1) to target the 3'-untranslated Region (3'-UTR) of PARP1 mRNA. | |||
Protein phosphatase 1A (PPM1A)
| Representative RIP-seq result supporting the interaction between the target gene and YTHDF1 | ||
| Cell Line | Hela | Homo sapiens |
| Regulation | logFC: 1.22E+00 | GSE63591 |
Male infertility [ICD-11: GB04]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [33] | |||
| Responsed Disease | Azoospermia [ICD-11: GB04.0] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | Autophagy | hsa04140 | ||
| Cell Process | RNA stability | |||
| Cell autophagy | ||||
In-vitro Model |
TM3 | Normal | Mus musculus | CVCL_4326 |
| In-vivo Model | Male SPF BALB/c mice (qls02-0202) were purchased from Qinglongshan animal breeding farm. Mice were sacrificed by CO2 asphyxiation and testes were obtained for following histopathological analyses. | |||
| Response Summary | m6A modification promoted translation of Protein phosphatase 1A (PPM1A) (protein phosphatase 1A, magnesium dependent, alpha isoform), a negative AMP-activated protein kinase (AMPK) regulator, but decreased expression of CAMKK2 (calcium/calmodulin-dependent protein kinase kinase 2, beta), a positive AMPK regulator, by reducing its RNA stability. Similar regulation of METTL14, ALKBH5, and m6A was also observed in LCs upon treatment with human chorionic gonadotropin (HsCG). Knock down of YTHDF1 failed to change the expression of CAMKK2 Providing insight into novel therapeutic strategies by exploiting m6A RNA methylation as targets for treating azoospermatism and oligospermatism patients with reduction in serum testosterone. Rapamycin could effectively inhibit phosphorylation of RPS6KB1. | |||
Rho guanine nucleotide exchange factor 2 (ARHGEF2)
| Representative RIP-seq result supporting the interaction between the target gene and YTHDF1 | ||
| Cell Line | Hela | Homo sapiens |
| Regulation | logFC: 1.80E+00 | GSE63591 |
Colorectal cancer [ICD-11: 2B91]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [34] | |||
| Responsed Disease | Colorectal cancer [ICD-11: 2B91] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
SW1116 | Colon adenocarcinoma | Homo sapiens | CVCL_0544 |
| RKO | Colon carcinoma | Homo sapiens | CVCL_0504 | |
| LS180 | Colon adenocarcinoma | Homo sapiens | CVCL_0397 | |
| LoVo | Colon adenocarcinoma | Homo sapiens | CVCL_0399 | |
| HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 | |
| DLD-1 | Colon adenocarcinoma | Homo sapiens | CVCL_0248 | |
| Response Summary | YTHDF1 promotes cell growth in CRC cell lines and primary organoids and lung and liver metastasis in vivo. YTHDF1 binds to m6A sites of Rho guanine nucleotide exchange factor 2 (ARHGEF2) messenger RNA, resulting in enhanced translation of ARHGEF2. | |||
Serine/threonine-protein kinase mTOR (MTOR)
| Representative RIP-seq result supporting the interaction between the target gene and YTHDF1 | ||
| Cell Line | Hela | Homo sapiens |
| Regulation | logFC: 1.73E+00 | GSE63591 |
Liver cancer [ICD-11: 2C12]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [22] | |||
| Responsed Disease | Hepatocellular carcinoma [ICD-11: 2C12.02] | |||
| Target Regulation | Down regulation | |||
| Pathway Response | PI3K-Akt signaling pathway | hsa04151), mTOR signaling pathway | ||
| Cell Process | Epithelial-mesenchymal transition | |||
| Cell migration | ||||
| Cell invasion | ||||
| Cell proliferation | ||||
In-vitro Model |
SNU-398 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_0077 |
| SK-HEP-1 | Liver and intrahepatic bile duct epithelial neoplasm | Homo sapiens | CVCL_0525 | |
| PLC/PRF/5 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_0485 | |
| L-02 | Endocervical adenocarcinoma | Homo sapiens | CVCL_6926 | |
| Huh-7 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_0336 | |
| Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 | |
| Hep 3B2.1-7 | Childhood hepatocellular carcinoma | Homo sapiens | CVCL_0326 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| HCCLM3 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_6832 | |
| In-vivo Model | Ten four-week-old BALB/c male nude mice (GemPharmatech, Jiangsu, China) were subcutaneously injected with control Huh7 cells 2 × 106 (left-back) and stable knockdown of YTHDF1 Huh7 cells 2 × 106 (right-back). These cells were respectively premixed with 50 ul Matrigel (Corning, 354,234) in 100 ul PBS. | |||
| Response Summary | YTHDF1 contributes to the progression of HCC by activating PI3K/AKT/Serine/threonine-protein kinase mTOR (MTOR) signaling pathway and inducing EMT. | |||
Signal transducer and activator of transcription 2 (STAT2)
| Representative RIP-seq result supporting the interaction between the target gene and YTHDF1 | ||
| Cell Line | Hela | Homo sapiens |
| Regulation | logFC: 1.30E+00 | GSE63591 |
Congenital pneumonia [ICD-11: KB24]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [35] | |||
| Responsed Disease | Congenital pneumonia [ICD-11: KB24] | |||
| Target Regulation | Down regulation | |||
| Cell Process | Cell apoptosis | |||
In-vitro Model |
WI-38 | Normal | Homo sapiens | CVCL_0579 |
| In-vivo Model | All mice were housed under a 12 h light/dark cycle with constant temperature about 25 ℃ and relative humidity approximating 55 %. The mice had free access to food and water for 10 days prior to the experiment. Forty mice were randomly selected and divided into four groups of 10 mice each. After 10 days, mice received an intraperitoneal injection of 22 mg / mL sodium pentobarbital (diluted in saline) followed by 167 uM LPS (60 uL) Saline solution was instilled into the oral cavity through the posterior pharyngeal wall. Pinch the nares quickly and hold for 30 s, model is successful when all fluid is absorbed into the nasal cavity, and slight tracheal rales appear. Lentiviral vectors containing pcDNA-SNHG4 (150 uM) or pcDNA-3.1 were intratracheally injected into mice. Twenty-one days after establishing the model, mice were intraperitoneally injected with 3% sodium pentobarbital and euthanized by overdose anesthesia at a dose of 90 mL/Kg, and organs and tissues were removed for follow-up studies. | |||
| Response Summary | SNHG4 was downregulated in the neonatal pneumonia patient serum and its overexpression could inhibit LPS induced inflammatory injury in human lung fibroblasts and mouse lung tissue. The molecular mechanism underlying this protective effect was achieved by suppression of METTL3-mediated m6A modification levels of YTHDF1-dependent Signal transducer and activator of transcription 2 (STAT2) mRNA. | |||
Transferrin receptor protein 1 (TFRC)
| Representative RIP-seq result supporting the interaction between the target gene and YTHDF1 | ||
| Cell Line | Hela | Homo sapiens |
| Regulation | logFC: 2.40E+00 | GSE63591 |
Hypopharyngeal cancer [ICD-11: 2B6D]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [36] | |||
| Responsed Disease | Hypopharyngeal squamous cell carcinoma [ICD-11: 2B6D.0] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | Ferroptosis | hsa04216 | ||
| Cell Process | Ferroptosis | |||
In-vitro Model |
YCU-MS861 | Maxillary sinus squamous cell carcinoma | Homo sapiens | CVCL_8021 |
| YCU-MS861 | Maxillary sinus squamous cell carcinoma | Homo sapiens | CVCL_8021 | |
| FaDu | Hypopharyngeal squamous cell carcinoma | Homo sapiens | CVCL_1218 | |
| Detroit 562 | Pharyngeal squamous cell carcinoma | Homo sapiens | CVCL_1171 | |
| Response Summary | YTHDF1 enhanced Transferrin receptor protein 1 (TFRC) expression in HPSCC through an m6A-dependent mechanism. | |||
Metastasis associated lung adenocarcinoma transcript 1 (MALAT1)
| Representative RIP-seq result supporting the interaction between the target gene and YTHDF1 | ||
| Cell Line | Hela | Homo sapiens |
| Regulation | logFC: 2.44E+00 | GSE63591 |
Lung cancer [ICD-11: 2C25]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [14] | |||
| Responsed Disease | Non-small-cell lung carcinoma [ICD-11: 2C25.Y] | |||
| Responsed Drug | Cisplatin | Approved | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Hippo signaling pathway | hsa04390 | ||
| Cell Process | Metabolic | |||
In-vitro Model |
A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 |
| Calu-6 | Lung adenocarcinoma | Homo sapiens | CVCL_0236 | |
| NCI-H1299 | Lung large cell carcinoma | Homo sapiens | CVCL_0060 | |
| NCI-H520 | Lung squamous cell carcinoma | Homo sapiens | CVCL_1566 | |
| In-vivo Model | Mice were injected with 5 × 106 lung cancer cells with stably expression of relevant plasmids and randomly divided into two groups (five mice per group) after the diameter of the xenografted tumors had reached approximately 5 mm in diameter. Xenografted mice were then administrated with PBS or DDP (3 mg/kg per day) for three times a week, and tumor volume were measured every second day. | |||
| Response Summary | METTL3, YTHDF3, YTHDF1, and eIF3b directly promoted YAP translation through an interaction with the translation initiation machinery. METTL3 knockdown inhibits tumor growth and enhances sensitivity to DDP in vivo.m6A mRNA methylation initiated by METTL3 directly promotes YAP translation and increases YAP activity by regulating the Metastasis associated lung adenocarcinoma transcript 1 (MALAT1)-miR-1914-3p-YAP axis to induce Non-small cell lung cancer drug resistance and metastasis. | |||
Aldo-keto reductase family 1 member C1 (AKR1C1)
Lung cancer [ICD-11: 2C25]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [7] | |||
| Responsed Disease | Non-small-cell lung carcinoma [ICD-11: 2C25.Y] | |||
| Responsed Drug | Cisplatin | Approved | ||
| Target Regulation | Down regulation | |||
| Pathway Response | Chemical carcinogenesis - reactive oxygen species | hsa05208 | ||
| Cell cycle | hsa04110 | |||
| Cell Process | Biological regulation | |||
In-vitro Model |
A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 |
| A549-DDP (Human lung adenocarcinoma is resistant to cisplatin) | ||||
| GLC-82 | Endocervical adenocarcinoma | Homo sapiens | CVCL_3371 | |
| NCI-H1299 | Lung large cell carcinoma | Homo sapiens | CVCL_0060 | |
| NCI-H1975 | Lung adenocarcinoma | Homo sapiens | CVCL_1511 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| NCI-H1650 | Minimally invasive lung adenocarcinoma | Homo sapiens | CVCL_1483 | |
| NCI-H838 | Lung adenocarcinoma | Homo sapiens | CVCL_1594 | |
| SPC-A1 | Endocervical adenocarcinoma | Homo sapiens | CVCL_6955 | |
| In-vivo Model | Mice were treated via nasal inhalation of adenovirus carrying Cre recombinase (5 × 106 p.f.u for Ad-Cre, Biowit Inc., Shenzhen, Guangdong), and were then killed at indicated times for gross inspection and histopathological examination. | |||
| Response Summary | YTHDF1 deficiency inhibits Non-small cell lung cancer cell proliferation and xenograft tumor formation through regulating the translational efficiency of CDK2, CDK4, p27, and cyclin D1, and that YTHDF1 depletion restrains de novo lung adenocarcinomas (ADC) progression. Mechanistic studies identified the Keap1-Nrf2-Aldo-keto reductase family 1 member C1 (AKR1C1) axis as the downstream mediator of YTHDF1. YTHDF1 high expression correlates with better clinical outcome, with its depletion rendering cancerous cells resistant to cisplatin (DDP) treatment. | |||
Alpha-enolase (ENO1)
Bladder cancer [ICD-11: 2C94]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [37] | |||
| Responsed Disease | Bladder cancer [ICD-11: 2C94] | |||
In-vitro Model |
T24 | Bladder carcinoma | Homo sapiens | CVCL_0554 |
| 5637 | Bladder carcinoma | Homo sapiens | CVCL_0126 | |
| UM-UC-3 | Bladder carcinoma | Homo sapiens | CVCL_1783 | |
| RT-4 | Bladder carcinoma | Homo sapiens | CVCL_0036 | |
| J82 | Bladder carcinoma | Homo sapiens | CVCL_0359 | |
| SV-HUC-1 | Normal | Homo sapiens | CVCL_3798 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| In-vivo Model | For the subcutaneous implantation model, 5 × 106 cells (n = 6 per group) were subcutaneously injected into the flank regions of 4-5 weeks female BALB/c nude mice. Tumor volume was calculated as 0.5 × W2 × L (where W and L represent a tumor's width and length, respectively). After xenografts were generated, DMSOand SB431542 (10 mg/kg, #301836-41-9, SANTA CRUZ BIOTECHNOLOGY), or ENOblock (#1177827-73-4, 10 mg/kg, MCE) were administered daily. All animal experiments were approved by The Affiliated Hospital of Qingdao University Committee on Animal Care. | |||
Autophagy protein 5 (ATG5)
Liver cancer [ICD-11: 2C12]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [4] | |||
| Responsed Disease | Hepatocellular carcinoma [ICD-11: 2C12.02] | |||
| Responsed Drug | Sorafenib | Approved | ||
| Target Regulation | Up regulation | |||
| Pathway Response | FoxO signaling pathway | hsa04068 | ||
| Autophagy | hsa04140 | |||
| Cell Process | Cell autophagy | |||
| Response Summary | METTL3 can sensitise hepatocellular carcinoma cells to sorafenib through stabilising forkhead box class O3 (FOXO3) in an m6A-dependent manner and translated by YTHDF1, thereby inhibiting the transcription of autophagy-related genes, including ATG3, Autophagy protein 5 (ATG5), ATG7, ATG12, and ATG16L1. | |||
Beclin 1-associated autophagy-related key regulator (ATG14)
Liver cancer [ICD-11: 2C12]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [26] | |||
| Responsed Disease | Hepatocellular carcinoma [ICD-11: 2C12.02] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | Autophagy | hsa04140 | ||
| HIF-1 signaling pathway | hsa04066 | |||
| Cell Process | Cell proliferation | |||
| Cell migration | ||||
| Cell invasion | ||||
| Cell autophagy | ||||
In-vitro Model |
Hep 3B2.1-7 | Childhood hepatocellular carcinoma | Homo sapiens | CVCL_0326 |
| Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 | |
| Huh-7 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_0336 | |
| SMMC-7721 | Endocervical adenocarcinoma | Homo sapiens | CVCL_0534 | |
| In-vivo Model | HCC cells (1 × 106/100 uL PBS) were administered to 4-week-old female BALB/c nude mice by subcutaneous injection (n = 6). | |||
| Response Summary | HIF-1-alpha-induced YTHDF1 expression was associated with hypoxia-induced autophagy and autophagy-related hepatocellular carcinoma progression via promoting translation of autophagy-related genes ATG2A and Beclin 1-associated autophagy-related key regulator (ATG14) in a m6A-dependent manner. | |||
Caveolin-1 (CAV1)
Vascular Calcification [ICD-11: BE2Y]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [38] | |||
| Responsed Disease | Vascular Calcification [ICD-11: BE2Y] | |||
In-vitro Model |
CHO-S/H9C2
|
N.A. | Cricetulus griseus | CVCL_A0TS |
| In-vivo Model | 2-month-old mice were given doses of tamoxifen at 40 mg/kg body weight/ day for 5 consecutive days by intraperitoneal injection. Littermate mice with wild-type levels of Ythdf1, either expressing Cre recombinase only or containing the Ythdf1 flox/flox-only, were used as controls (Ctrl). All mice received tamoxifen injections. All in vivo experiments and analyses were performed 1 month after the first tamoxifen injection. | |||
CCR4-NOT transcription complex subunit 7 (CNOT7)
Osteosarcoma [ICD-11: 2B51]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [39] | |||
| Responsed Disease | Osteosarcoma [ICD-11: 2B51] | |||
| Target Regulation | Up regulation | |||
| Cell Process | Cell proliferation | |||
| Cell migration | ||||
| Cell invasion | ||||
In-vitro Model |
U2OS | Osteosarcoma | Homo sapiens | CVCL_0042 |
| SaOS-2 | Osteosarcoma | Homo sapiens | CVCL_0548 | |
| MG-63 | Osteosarcoma | Homo sapiens | CVCL_0426 | |
| HOS | Osteosarcoma | Homo sapiens | CVCL_0312 | |
| hFOB 1.19 | Normal | Homo sapiens | CVCL_3708 | |
| In-vivo Model | The mice were divided into two groups with three mice in each group and their flank was subcutaneously injected with 1 × 107 OS cells. 28 days following subcutaneous injection, the mice were sacrificed through carbon dioxide euthanasia (30%/min) to obtain tumor weight and volume measurements. | |||
| Response Summary | YTHDF1 could recognize the m6A sites of CCR4-NOT transcription complex subunit 7 (CNOT7) and promote its expression in an m6A manner. Inhibition of YTHDF1 could suppress the proliferation, migration and invasion of the OS cells. | |||
Cellular tumor antigen p53 (TP53/p53)
Solid tumour/cancer [ICD-11: 2A00-2F9Z]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [10] | |||
| Responsed Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | |||
| Responsed Drug | Arsenite | Phase 2 | ||
| Target Regulation | Down regulation | |||
| Pathway Response | p53 signaling pathway | hsa04115 | ||
In-vitro Model |
HaCaT | Normal | Homo sapiens | CVCL_0038 |
| Response Summary | METTL3 significantly decreased m6A level, restoring Cellular tumor antigen p53 (TP53/p53) activation and inhibiting cellular transformation phenotypes in the arsenite-transformed cells. m6A downregulated the expression of the positive p53 regulator, PRDM2, through the YTHDF2-promoted decay of PRDM2 mRNAs. m6A upregulated the expression of the negative p53 regulator, YY1 and MDM2 through YTHDF1-stimulated translation of YY1 and MDM2 mRNA. This study further sheds light on the mechanisms of arsenic carcinogenesis via RNA epigenetics. | |||
Collagen alpha-1 (III) chain (COL3A1)
Hepatic fibrosis/cirrhosis [ICD-11: DB93]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [40] | |||
| Responsed Disease | Hepatic fibrosis/cirrhosis [ICD-11: DB93] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
JS1
|
N.A. | Mus musculus | CVCL_C7PL |
| In-vivo Model | Following 1 week of adaptive feeding, the mice were randomly divided into CCl4 + siCON and CCl4 + siYTHDF1 groups. Both groups were injected intraperitoneally with 5 ml/kg of 10% carbon tetrachloride (CCl4) twice a week for 4 weeks, to establish liver fibrosis. | |||
Cyclin-dependent kinase 4 (CDK4)
Lung cancer [ICD-11: 2C25]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [7] | |||
| Responsed Disease | Non-small-cell lung carcinoma [ICD-11: 2C25.Y] | |||
| Responsed Drug | Cisplatin | Approved | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Chemical carcinogenesis - reactive oxygen species | hsa05208 | ||
| Cell cycle | hsa04110 | |||
| Cell Process | Biological regulation | |||
In-vitro Model |
A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 |
| A549-DDP (Human lung adenocarcinoma is resistant to cisplatin) | ||||
| GLC-82 | Endocervical adenocarcinoma | Homo sapiens | CVCL_3371 | |
| NCI-H1299 | Lung large cell carcinoma | Homo sapiens | CVCL_0060 | |
| NCI-H1975 | Lung adenocarcinoma | Homo sapiens | CVCL_1511 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| NCI-H1650 | Minimally invasive lung adenocarcinoma | Homo sapiens | CVCL_1483 | |
| NCI-H838 | Lung adenocarcinoma | Homo sapiens | CVCL_1594 | |
| SPC-A1 | Endocervical adenocarcinoma | Homo sapiens | CVCL_6955 | |
| In-vivo Model | Mice were treated via nasal inhalation of adenovirus carrying Cre recombinase (5 × 106 p.f.u for Ad-Cre, Biowit Inc., Shenzhen, Guangdong), and were then killed at indicated times for gross inspection and histopathological examination. | |||
| Response Summary | YTHDF1 deficiency inhibits Non-small cell lung cancer cell proliferation and xenograft tumor formation through regulating the translational efficiency of CDK2, Cyclin-dependent kinase 4 (CDK4), p27, and cyclin D1, and that YTHDF1 depletion restrains de novo lung adenocarcinomas (ADC) progression. Mechanistic studies identified the Keap1-Nrf2-AKR1C1 axis as the downstream mediator of YTHDF1. YTHDF1 high expression correlates with better clinical outcome, with its depletion rendering cancerous cells resistant to cisplatin (DDP) treatment. | |||
Cyclin-dependent kinase inhibitor 1B (CDKN1B/p27)
Lung cancer [ICD-11: 2C25]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [7] | |||
| Responsed Disease | Non-small-cell lung carcinoma [ICD-11: 2C25.Y] | |||
| Responsed Drug | Cisplatin | Approved | ||
| Target Regulation | Down regulation | |||
| Pathway Response | Chemical carcinogenesis - reactive oxygen species | hsa05208 | ||
| Cell cycle | hsa04110 | |||
| Cell Process | Biological regulation | |||
In-vitro Model |
A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 |
| A549-DDP (Human lung adenocarcinoma is resistant to cisplatin) | ||||
| GLC-82 | Endocervical adenocarcinoma | Homo sapiens | CVCL_3371 | |
| NCI-H1299 | Lung large cell carcinoma | Homo sapiens | CVCL_0060 | |
| NCI-H1975 | Lung adenocarcinoma | Homo sapiens | CVCL_1511 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| NCI-H1650 | Minimally invasive lung adenocarcinoma | Homo sapiens | CVCL_1483 | |
| NCI-H838 | Lung adenocarcinoma | Homo sapiens | CVCL_1594 | |
| SPC-A1 | Endocervical adenocarcinoma | Homo sapiens | CVCL_6955 | |
| In-vivo Model | Mice were treated via nasal inhalation of adenovirus carrying Cre recombinase (5 × 106 p.f.u for Ad-Cre, Biowit Inc., Shenzhen, Guangdong), and were then killed at indicated times for gross inspection and histopathological examination. | |||
| Response Summary | YTHDF1 deficiency inhibits Non-small cell lung cancer cell proliferation and xenograft tumor formation through regulating the translational efficiency of CDK2, CDK4, Cyclin-dependent kinase inhibitor 1B (CDKN1B/p27), and cyclin D1, and that YTHDF1 depletion restrains de novo lung adenocarcinomas (ADC) progression. Mechanistic studies identified the Keap1-Nrf2-AKR1C1 axis as the downstream mediator of YTHDF1. YTHDF1 high expression correlates with better clinical outcome, with its depletion rendering cancerous cells resistant to cisplatin (DDP) treatment. | |||
DNA (cytosine-5)-methyltransferase 3B (DNMT3B)
Gastric cancer [ICD-11: 2B72]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [41] | |||
| Responsed Disease | Gastric cancer [ICD-11: 2B72] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
AGS | Gastric adenocarcinoma | Homo sapiens | CVCL_0139 |
| BGC-823 | Gastric carcinoma | Homo sapiens | CVCL_3360 | |
| HGC-27 | Gastric carcinoma | Homo sapiens | CVCL_1279 | |
E3 ubiquitin-protein ligase Mdm2 (Mdm2)
Acute kidney failure [ICD-11: GB60]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [42] | |||
| Responsed Disease | Acute kidney failure [ICD-11: GB60] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
HK-2 [Human kidney] | Normal | Homo sapiens | CVCL_0302 |
Eukaryotic translation initiation factor 2 subunit 1 (EIF2S1/eIF2-Alpha)
Pulmonary hypertension [ICD-11: BB01]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [43] | |||
| Responsed Disease | Pulmonary arterial hypertension [ICD-11: BB01.0] | |||
| Target Regulation | Up regulation | |||
| In-vivo Model | Adult male Sprague-Dawley (150-200 g in body weight) rats were randomly divided into control or MCT/vehicle rats. Experimental rats were administered an intraperitoneal injection of monocrotaline (60 mg/kg, Sigma, 315-22-0), and their littermates were injected with saline. For GSK2606414 treatment, rats that underwent monocrotaline treatment were treated either with vehicle or GSK2606414 (10 mg/kg, Sigma, 1,337,531-89-1) by intraperitoneal injection per day. GSK2606414 was dissolved in a mixture of dimethyl sulfoxide (DMSO): polyethylene glycol (PEG) 400: distilled water (1: 4: 5). After 4 weeks, RV systolic blood pressure (RVSP) was measured with pressure transducers under anesthesia. The RV hypertrophy was analyzed as a ratio of RV to left ventricular and septal weight. Left lung tissues were fixed in 4% paraformaldehyde solution for the following histology staining; right lung tissues and pulmonary arteries were excised and immediately frozen in liquid nitrogen for other experiments. | |||
Eukaryotic translation initiation factor 3 subunit A (EIF3A/EIF3)
Merkel cell carcinoma [ICD-11: 2C34]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [44] | |||
| Responsed Disease | Merkel cell carcinoma [ICD-11: 2C34] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | mRNA surveillance pathway | hsa03015 | ||
| Cell Process | Translation | |||
In-vitro Model |
MCC13 | Merkel cell carcinoma | Homo sapiens | CVCL_2583 |
| MCC26 | Merkel cell carcinoma | Homo sapiens | CVCL_2585 | |
| MKL-1 | Merkel cell carcinoma | Homo sapiens | CVCL_2600 | |
| MKL-2 | Merkel cell carcinoma | Homo sapiens | CVCL_D027 | |
| PeTa | Merkel cell carcinoma | Homo sapiens | CVCL_LC73 | |
| UISO-MCC-1 | Merkel cell carcinoma | Homo sapiens | CVCL_E996 | |
| WaGa | Merkel cell carcinoma | Homo sapiens | CVCL_E998 | |
| Response Summary | Knockdown of YTHDF1 in Merkel cell carcinoma (MCC) cell lines negatively affected the translation initiation factor Eukaryotic translation initiation factor 3 subunit A (EIF3A/EIF3) and reduced proliferation and clonogenic capacity in vitro. | |||
Eukaryotic translation initiation factor 3 subunit C (EIF3C)
Ovarian cancer [ICD-11: 2C73]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [45] | |||
| Responsed Disease | Ovarian cancer [ICD-11: 2C73] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | mRNA surveillance pathway | hsa03015 | ||
| Cell Process | Tumorigenesis and metastasis | |||
In-vitro Model |
A2780 | Ovarian endometrioid adenocarcinoma | Homo sapiens | CVCL_0134 |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| SK-OV-3 | Ovarian serous cystadenocarcinoma | Homo sapiens | CVCL_0532 | |
| In-vivo Model | 5 × 106 cells in PBS were injected subcutaneously into one side of the posterior flanks of Balb/C nude mice at 6-8 weeks old. | |||
| Response Summary | YTHDF1 augments the translation of Eukaryotic translation initiation factor 3 subunit C (EIF3C) in an m6A-dependent manner by binding to m6A-modified EIF3C mRNA and concomitantly promotes the overall translational output, thereby facilitating tumorigenesis and metastasis of ovarian cancer. | |||
G1/S-specific cyclin-D1 (CCND1)
Lung cancer [ICD-11: 2C25]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [7] | |||
| Responsed Disease | Non-small-cell lung carcinoma [ICD-11: 2C25.Y] | |||
| Responsed Drug | Cisplatin | Approved | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Chemical carcinogenesis - reactive oxygen species | hsa05208 | ||
| Cell cycle | hsa04110 | |||
| Cell Process | Biological regulation | |||
In-vitro Model |
A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 |
| A549-DDP (Human lung adenocarcinoma is resistant to cisplatin) | ||||
| GLC-82 | Endocervical adenocarcinoma | Homo sapiens | CVCL_3371 | |
| NCI-H1299 | Lung large cell carcinoma | Homo sapiens | CVCL_0060 | |
| NCI-H1975 | Lung adenocarcinoma | Homo sapiens | CVCL_1511 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| NCI-H1650 | Minimally invasive lung adenocarcinoma | Homo sapiens | CVCL_1483 | |
| NCI-H838 | Lung adenocarcinoma | Homo sapiens | CVCL_1594 | |
| SPC-A1 | Endocervical adenocarcinoma | Homo sapiens | CVCL_6955 | |
| In-vivo Model | Mice were treated via nasal inhalation of adenovirus carrying Cre recombinase (5 × 106 p.f.u for Ad-Cre, Biowit Inc., Shenzhen, Guangdong), and were then killed at indicated times for gross inspection and histopathological examination. | |||
| Response Summary | YTHDF1 deficiency inhibits Non-small cell lung cancer cell proliferation and xenograft tumor formation through regulating the translational efficiency of CDK2, CDK4, p27, and G1/S-specific cyclin-D1 (CCND1), and that YTHDF1 depletion restrains de novo lung adenocarcinomas (ADC) progression. Mechanistic studies identified the Keap1-Nrf2-AKR1C1 axis as the downstream mediator of YTHDF1. YTHDF1 high expression correlates with better clinical outcome, with its depletion rendering cancerous cells resistant to cisplatin (DDP) treatment. | |||
Asthma [ICD-11: CA23]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [49] | |||
| Responsed Disease | Asthma [ICD-11: CA23] | |||
| Target Regulation | Up regulation | |||
Heat shock factor protein 1 (HSF1)
Colorectal cancer [ICD-11: 2B91]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [50] | |||
| Responsed Disease | Colorectal cancer [ICD-11: 2B91] | |||
In-vitro Model |
DLD-1 | Colon adenocarcinoma | Homo sapiens | CVCL_0248 |
| SW480 | Colon adenocarcinoma | Homo sapiens | CVCL_0546 | |
Interferon gamma receptor 1 (IFNGR1)
Gastric cancer [ICD-11: 2B72]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [52] | |||
| Responsed Disease | Gastric cancer [ICD-11: 2B72] | |||
| Target Regulation | Down regulation | |||
| Pathway Response | JAK-STAT signaling pathway | hsa04630 | ||
| Cell Process | Immunity | |||
In-vitro Model |
YTN16 (Mouse gastric cancer cell line (YTN16)) | |||
| MKN74 | Gastric tubular adenocarcinoma | Homo sapiens | CVCL_2791 | |
| BGC-823 | Gastric carcinoma | Homo sapiens | CVCL_3360 | |
| AGS | Gastric adenocarcinoma | Homo sapiens | CVCL_0139 | |
| In-vivo Model | MKN74 cells (5×106/tumor) expressing shNC, shYTHDF1-1, or shYTHDF1-2 were suspended in ice-cold 100 ul PBS:Matrigel gel (1:1, v/v) (Corning, USA), and subcutaneously implanted into the right dorsal flank of 4-week-old NOD. | |||
| Response Summary | Loss of YTHDF1 mediated the overexpression of Interferon gamma receptor 1 (IFNGR1) and JAK/STAT1 signaling pathway in tumor cells, which contributes to restored sensitivity to antitumor immunity. YTHDF1 is overexpressed in GC and promotes GC by inducing cell proliferation and repression of DCs-mediated antitumor immune response. | |||
MAP kinase signal-integrating kinase 2 (MNK2)
Muscular dystrophies [ICD-11: 8C70]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [1] | |||
| Responsed Disease | Muscular dystrophies [ICD-11: 8C70] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | MAPK signaling pathway | hsa04010 | ||
In-vitro Model |
HEK293T | Normal | Homo sapiens | CVCL_0063 |
| C2C12 | Normal | Mus musculus | CVCL_0188 | |
| In-vivo Model | For mouse muscle injury and regeneration experiment, tibialis anterior (TA) muscles of 6-week-old male mice were injected with 25 uL of 10 uM cardiotoxin (CTX, Merck Millipore, 217503), 0.9% normal saline (Saline) were used as control. The regenerated muscles were collected at day 1, 3, 5, and 10 post-injection. TA muscles were isolated for Hematoxylin and eosin staining or frozen in liquid nitrogen for RNA and protein extraction. | |||
| Response Summary | m6A writers METTL3/METTL14 and the m6A reader YTHDF1 orchestrate MAP kinase signal-integrating kinase 2 (MNK2) expression posttranscriptionally and thus control ERK signaling, which is required for the maintenance of muscle myogenesis contributes to regeneration. | |||
Melanoma-associated antigen D1 (MAGED1)
Pulmonary hypertension [ICD-11: BB01]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [53] | |||
| Responsed Disease | Pulmonary hypertension [ICD-11: BB01] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
PASMC cell line (Pulmonary artery smooth muscle cell) | |||
| In-vivo Model | SMC-specific TWIST1-deficient mice were generated by crossing TWIST1flox/flox mice with animals expressing Sm22-Cre (smooth muscle protein 22-Cre). Deletion of TWIST1 in SMCs was confirmed by histology and Western blot. | |||
| Response Summary | YTHDF1 promotes PASMC proliferation and pulmonary hypertension by enhancing Melanoma-associated antigen D1 (MAGED1) translation. | |||
Mutated in multiple advanced cancers 1 (PTEN)
Renal cell carcinoma [ICD-11: 2C90]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [54] | |||
| Responsed Disease | Renal cell carcinoma of kidney [ICD-11: 2C90.0] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | PI3K-Akt signaling pathway | hsa04151 | ||
| Response Summary | Upregulation of METTL14 inhibited ccRCC cells proliferation and migration in vitro. Overexpression of METTL14 increased the m6A enrichment of Mutated in multiple advanced cancers 1 (PTEN), and promoted Pten expression. METTL14-enhanced Pten mRNA stability was dependent on YTHDF1. | |||
Acute ischemic stroke [ICD-11: 8B11]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [55] | |||
| Responsed Disease | Acute ischemic stroke [ICD-11: 8B11] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
PC12 | Rat adrenal gland pheochromocytoma | Rattus norvegicus | CVCL_0481 |
Nuclear factor erythroid 2-related factor 2 (NFE2L2)
Lung cancer [ICD-11: 2C25]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [7] | |||
| Responsed Disease | Non-small-cell lung carcinoma [ICD-11: 2C25.Y] | |||
| Responsed Drug | Cisplatin | Approved | ||
| Target Regulation | Down regulation | |||
| Pathway Response | Chemical carcinogenesis - reactive oxygen species | hsa05208 | ||
| Cell cycle | hsa04110 | |||
| Cell Process | Biological regulation | |||
In-vitro Model |
A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 |
| A549-DDP (Human lung adenocarcinoma is resistant to cisplatin) | ||||
| GLC-82 | Endocervical adenocarcinoma | Homo sapiens | CVCL_3371 | |
| NCI-H1299 | Lung large cell carcinoma | Homo sapiens | CVCL_0060 | |
| NCI-H1975 | Lung adenocarcinoma | Homo sapiens | CVCL_1511 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| NCI-H1650 | Minimally invasive lung adenocarcinoma | Homo sapiens | CVCL_1483 | |
| NCI-H838 | Lung adenocarcinoma | Homo sapiens | CVCL_1594 | |
| SPC-A1 | Endocervical adenocarcinoma | Homo sapiens | CVCL_6955 | |
| In-vivo Model | Mice were treated via nasal inhalation of adenovirus carrying Cre recombinase (5 × 106 p.f.u for Ad-Cre, Biowit Inc., Shenzhen, Guangdong), and were then killed at indicated times for gross inspection and histopathological examination. | |||
| Response Summary | YTHDF1 deficiency inhibits Non-small cell lung cancer cell proliferation and xenograft tumor formation through regulating the translational efficiency of CDK2, CDK4, p27, and cyclin D1, and that YTHDF1 depletion restrains de novo lung adenocarcinomas (ADC) progression. Mechanistic studies identified the Keap1-Nuclear factor erythroid 2-related factor 2 (NFE2L2)-AKR1C1 axis as the downstream mediator of YTHDF1. YTHDF1 high expression correlates with better clinical outcome, with its depletion rendering cancerous cells resistant to cisplatin (DDP) treatment. | |||
Bladder cancer [ICD-11: 2C94]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [57] | |||
| Responsed Disease | Bladder cancer [ICD-11: 2C94] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
SV-HUC-1 | Normal | Homo sapiens | CVCL_3798 |
| J82 | Bladder carcinoma | Homo sapiens | CVCL_0359 | |
| UM-UC-3 | Bladder carcinoma | Homo sapiens | CVCL_1783 | |
Potassium voltage-gated channel subfamily H member 6 (KCNH6)
Idiopathic interstitial pneumonitis [ICD-11: CB03]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [58] | |||
| Responsed Disease | Idiopathic pulmonary fibrosis [ICD-11: CB03.4] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
WI-38 | Normal | Homo sapiens | CVCL_0579 |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| In-vivo Model | Animals were bred and housed in the pathogen-free facility of the Laboratory Animal Center of Shanghai General Hospital (Shanghai, China). All lungs were collected 4 weeks after BLM treatment for histology and further study. Lung microsections (5 uM) were applied to Masson's trichrome and Sirius red staining to visualize fibrotic lesions. | |||
| Response Summary | Lowering m6A levels through silencing METTL3 suppresses the FMT process in vitro and in vivo. m6A modification regulates EMT by modulating the translation of Potassium voltage-gated channel subfamily H member 6 (KCNH6) mRNA in a YTHDF1-dependent manner. Manipulation of m6A modification through targeting METTL3 becomes a promising strategy for the treatment of idiopathic pulmonary fibrosis. | |||
Programmed cell death 1 ligand 1 (CD274/PD-L1)
Lung cancer [ICD-11: 2C25]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [9] | |||
| Responsed Disease | Non-small-cell lung carcinoma [ICD-11: 2C25.Y] | |||
| Pathway Response | p53 signaling pathway | hsa04115 | ||
| Central carbon metabolism in cancer | hsa05230 | |||
| PD-L1 expression and PD-1 checkpoint pathway in cancer | hsa05235 | |||
| Response Summary | This study revealed that m6A methylation is closely related to the poor prognosis of non-small cell lung cancer patients via interference with the TIME, which suggests that m6A plays a role in optimizing individualized immunotherapy management and improving prognosis. The expression levels of METTL3, FTO and YTHDF1 in non-small cell lung cancer were changed. Patients in Cluster 1 had lower immunoscores, higher Programmed cell death 1 ligand 1 (CD274/PD-L1) expression, and shorter overall survival compared to patients in Cluster 2. The hallmarks of the Myelocytomatosis viral oncogene (MYC) targets, E2 transcription Factor (E2F) targets were significantly enriched. | |||
Protocadherin Fat 4 (FAT4)
Melanoma of uvea [ICD-11: 2D0Y]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [59] | |||
| Responsed Disease | Melanoma of uvea [ICD-11: 2D0Y] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
Mel290 | Uveal melanoma | Homo sapiens | CVCL_C304 |
| OMM2.3 | Uveal melanoma | Homo sapiens | CVCL_C306 | |
| OMM-1 | Uveal melanoma | Homo sapiens | CVCL_6939 | |
| CRMM-1 | Conjunctival melanoma | Homo sapiens | CVCL_M593 | |
| CRMM-2 | Conjunctival melanoma | Homo sapiens | CVCL_M594 | |
| CM2005.1 | Conjunctival melanoma | Homo sapiens | CVCL_M592 | |
| MuM-2B | Uveal melanoma | Homo sapiens | CVCL_3447 | |
| ARPE-19 | Normal | Homo sapiens | CVCL_0145 | |
| A-375 | Amelanotic melanoma | Homo sapiens | CVCL_0132 | |
| SK-MEL-28 | Cutaneous melanoma | Homo sapiens | CVCL_0526 | |
| PIG1 | Normal | Homo sapiens | CVCL_S410 | |
Rho GTPase activating protein 5 (ARHGAP5)
Breast cancer [ICD-11: 2C60]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [60] | |||
| Responsed Disease | Breast cancer [ICD-11: 2C60] | |||
| Cell Process | Cell proliferation | |||
| Cell migration | ||||
| Cell invasion | ||||
In-vitro Model |
MDA-MB-231 | Breast adenocarcinoma | Homo sapiens | CVCL_0062 |
| T-47D | Invasive breast carcinoma | Homo sapiens | CVCL_0553 | |
| In-vivo Model | Roughly 1 × 107 of MCF-7 or MDA-MB-231 cells stably transfected with ADAR1 sgRNA or Ctrl sgRNA (as described above) were injected subcutaneously into the thighs of the female athymic nude mice (5 weeks old, 17-18 g, three mice per group). Tumor growth was measured by measuring the width (W) and length (L) with calipers, and the volume (V) of the tumor was figured using the criterion V = (W2 × L)/2. At 43 days after injection, the mice were euthanized and tumors were removed and weighed. The tumor samples were further analyzed with IHC and Western blot assay. The animal studies were performed according to the institutional ethics guidelines for animal experiments and approved by the Chongqing Medical University Animal Care and Use Committee. | |||
Signal transducer and activator of transcription 5A (STAT5A)
Acute ischemic stroke [ICD-11: 8B11]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [61] | |||
| Responsed Disease | Acute ischemic stroke [ICD-11: 8B11] | |||
| Target Regulation | Down regulation | |||
In-vitro Model |
HT22 | Normal | Mus musculus | CVCL_0321 |
SRSF protein kinase 2 (SRPK2)
Gastric cancer [ICD-11: 2B72]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [62] | |||
| Responsed Disease | Gastric cancer [ICD-11: 2B72] | |||
Staphylococcal nuclease domain-containing protein 1 (SND1)
Nasal-type natural killer/T-cell lymphoma [ICD-11: XH3400]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [63] | |||
| Responsed Disease | Nasal-type natural killer/T-cell lymphoma [ICD-11: XH3400] | |||
| Responsed Drug | Cisplatin | Approved | ||
| Target Regulation | Up regulation | |||
In-vitro Model |
NK-92 | Natural killer cell lymphoblastic leukemia/lymphoma | Homo sapiens | CVCL_2142 |
| YTS | Lymphoblastic leukemia/lymphoma | Homo sapiens | CVCL_D324 | |
| SNT-8 | Nasal type extranodal NK/T-cell lymphoma | Homo sapiens | CVCL_A677 | |
| SNK-6 | Nasal type extranodal NK/T-cell lymphoma | Homo sapiens | CVCL_A673 | |
| In-vivo Model | A total of 32 nude mice were randomly divided into four groups with eight mice in each group. Nude mice were subcutaneously injected with 2 × 106 cells (suspended in 100 μl of PBS) transfected with NC shRNA or SND1 shRNA into the back flank of mice. In DDP administration group, DDP (3 mg/kg per week for 2 weeks) was intraperitoneally injected into mice every 3 days. The control group received 200 μl of 0.1% DMSO. The tumor volume was measured every week for a total of 4 weeks. The mice were sacrificed after 4 weeks, then the tumors were harvested for weight measurement and other analysis. The tumor volume (mm3) was estimated with the formula (0.5 × length × width2). | |||
Stearoyl-CoA desaturase (SCD)
Non-alcoholic fatty liver disease [ICD-11: DB92]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [64] | |||
| Responsed Disease | Nonalcoholic fatty liver disease [ICD-11: DB92.Z] | |||
| Target Regulation | Up regulation | |||
Thrombospondin-1 (THBS1)
Acute pericarditis [ICD-11: BB20]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [65] | |||
| Responsed Disease | Acute pericarditis [ICD-11: BB20.2] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
MC3T3-E1 | Normal | Mus musculus | CVCL_0409 |
Tissue factor pathway inhibitor 2 (TFPI-2)
Pancreatic cancer [ICD-11: 2C10]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [66] | |||
| Responsed Disease | Pancreatic cancer [ICD-11: 2C10] | |||
| Target Regulation | Up regulation | |||
| Cell Process | Cell growth | |||
| cell migration | ||||
| cell invasion | ||||
| Response Summary | Knockdown of FTO increases m6A methylation of Tissue factor pathway inhibitor 2 (TFPI-2) mRNA in PC cells, thereby increasing mRNA stability via the m6A reader YTHDF1. | |||
Transcription factor p65 (RELA)
Acute ischemic stroke [ICD-11: 8B11]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [67] | |||
| Responsed Disease | Acute ischemic stroke [ICD-11: 8B11] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
BV-2 | Normal | Mus musculus | CVCL_0182 |
Transcriptional repressor protein YY1 (YY1)
Gastric cancer [ICD-11: 2B72]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [68] | |||
| Responsed Disease | Gastric cancer [ICD-11: 2B72] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
MGC-803 | Gastric mucinous adenocarcinoma | Homo sapiens | CVCL_5334 |
| AGS | Gastric adenocarcinoma | Homo sapiens | CVCL_0139 | |
| MKN45 | Gastric adenocarcinoma | Homo sapiens | CVCL_0434 | |
| BGC-823 | Gastric carcinoma | Homo sapiens | CVCL_3360 | |
| GES-1 | Normal | Homo sapiens | CVCL_EQ22 | |
| In-vivo Model | Five-week-old male BALB/c nude mice were used for tumor growth studies in vivo. Briefly, AGS cells were subcutaneously injected into the dorsal side of mice blindly and randomly (n = 5 per group). | |||
Tumor necrosis factor (TNF/TNF-alpha)
B-cell lymphomas [ICD-11: 2A86]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [25] | |||
| Responsed Disease | B-cell lymphomas [ICD-11: 2A86] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | Apoptosis | hsa04210 | ||
| Cell Process | Cell proliferation and metastasis | |||
| Cell apoptosis | ||||
In-vitro Model |
ATDC-5 | Mouse teratocarcinoma | Mus musculus | CVCL_3894 |
| In-vivo Model | For MIA + SAH control, S-adenosylhomocysteine (SAH), Mettl3 inhibitor (10 mg/kg) (MCE, NJ, USA) was injected intraperitoneally before MIA injection and maintained twice a week until mice were sacrificed. | |||
| Response Summary | Mettl3 inhibitor, S-adenosylhomocysteine promoted the apoptosis and autophagy of chondrocytes with inflammation in vitro and aggravated the degeneration of chondrocytes and subchondral bone in monosodium iodoacetate (MIA) induced temporomandibular joint osteoarthritis mice in vivo. Bcl2 protein interacted with Beclin1 protein in chondrocytes induced by Tumor necrosis factor (TNF/TNF-alpha) stimulation. Mettl3 inhibits the apoptosis and autophagy of chondrocytes in inflammation through m6A/Ythdf1/Bcl2 signal axis which provides promising therapeutic strategy for temporomandibular joint osteoarthritis. | |||
Dentofacial anomalies [ICD-11: DA0E]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [25] | |||
| Responsed Disease | Temporomandibular joint disorders [ICD-11: DA0E.8] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | Apoptosis | hsa04210 | ||
| Cell Process | Cell proliferation and metastasis | |||
| Cell apoptosis | ||||
In-vitro Model |
ATDC-5 | Mouse teratocarcinoma | Mus musculus | CVCL_3894 |
| In-vivo Model | For MIA + SAH control, S-adenosylhomocysteine (SAH), Mettl3 inhibitor (10 mg/kg) (MCE, NJ, USA) was injected intraperitoneally before MIA injection and maintained twice a week until mice were sacrificed. | |||
| Response Summary | Mettl3 inhibitor, S-adenosylhomocysteine promoted the apoptosis and autophagy of chondrocytes with inflammation in vitro and aggravated the degeneration of chondrocytes and subchondral bone in monosodium iodoacetate (MIA) induced temporomandibular joint osteoarthritis mice in vivo. Bcl2 protein interacted with Beclin1 protein in chondrocytes induced by Tumor necrosis factor (TNF/TNF-alpha) stimulation. Mettl3 inhibits the apoptosis and autophagy of chondrocytes in inflammation through m6A/Ythdf1/Bcl2 signal axis which provides promising therapeutic strategy for temporomandibular joint osteoarthritis. | |||
Ubiquitin-like modifier-activating enzyme ATG7 (ATG7)
Liver cancer [ICD-11: 2C12]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [4] | |||
| Responsed Disease | Hepatocellular carcinoma [ICD-11: 2C12.02] | |||
| Responsed Drug | Sorafenib | Approved | ||
| Target Regulation | Up regulation | |||
| Pathway Response | FoxO signaling pathway | hsa04068 | ||
| Autophagy | hsa04140 | |||
| Cell Process | Cell autophagy | |||
| Response Summary | METTL3 can sensitise hepatocellular carcinoma cells to sorafenib through stabilising forkhead box class O3 (FOXO3) in an m6A-dependent manner and translated by YTHDF1, thereby inhibiting the transcription of autophagy-related genes, including ATG3, ATG5, Ubiquitin-like modifier-activating enzyme ATG7 (ATG7), ATG12, and ATG16L1. | |||
Ubiquitin-like protein ATG12 (ATG12)
Liver cancer [ICD-11: 2C12]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [4] | |||
| Responsed Disease | Hepatocellular carcinoma [ICD-11: 2C12.02] | |||
| Responsed Drug | Sorafenib | Approved | ||
| Target Regulation | Up regulation | |||
| Pathway Response | FoxO signaling pathway | hsa04068 | ||
| Autophagy | hsa04140 | |||
| Cell Process | Cell autophagy | |||
| Response Summary | METTL3 can sensitise hepatocellular carcinoma cells to sorafenib through stabilising forkhead box class O3 (FOXO3) in an m6A-dependent manner and translated by YTHDF1, thereby inhibiting the transcription of autophagy-related genes, including ATG3, ATG5, ATG7, Ubiquitin-like protein ATG12 (ATG12), and ATG16L1. | |||
Ubiquitin-like-conjugating enzyme ATG3 (ATG3)
Liver cancer [ICD-11: 2C12]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [4] | |||
| Responsed Disease | Hepatocellular carcinoma [ICD-11: 2C12.02] | |||
| Responsed Drug | Sorafenib | Approved | ||
| Target Regulation | Up regulation | |||
| Pathway Response | FoxO signaling pathway | hsa04068 | ||
| Autophagy | hsa04140 | |||
| Cell Process | Cell autophagy | |||
| Response Summary | METTL3 can sensitise hepatocellular carcinoma cells to sorafenib through stabilising forkhead box class O3 (FOXO3) in an m6A-dependent manner and translated by YTHDF1, thereby inhibiting the transcription of autophagy-related genes, including Ubiquitin-like-conjugating enzyme ATG3 (ATG3), ATG5, ATG7, ATG12, and ATG16L1. | |||
Zinc finger protein SNAI1 (SNAI1)
Gastric cancer [ICD-11: 2B72]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [69] | |||
| Responsed Disease | Gastric cancer [ICD-11: 2B72] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
AGS | Gastric adenocarcinoma | Homo sapiens | CVCL_0139 |
| MKN45 | Gastric adenocarcinoma | Homo sapiens | CVCL_0434 | |
| MKN7 | Gastric tubular adenocarcinoma | Homo sapiens | CVCL_1417 | |
| HGC-27 | Gastric carcinoma | Homo sapiens | CVCL_1279 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| GES-1 | Normal | Homo sapiens | CVCL_EQ22 | |
| In-vivo Model | To establish a murine subcutaneous tumor model, sh-PLAGL2 or sh-PLAGL2 + oe-Snail HGC-27 stable transfected HGC-27 cells (5 × 106 cells/kg) subcutaneously to the right of the dorsal midline of mice (n = 6 for each). | |||
Liver cancer [ICD-11: 2C12]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [70] | |||
| Responsed Disease | Hepatocellular carcinoma [ICD-11: 2C12.02] | |||
| Target Regulation | Up regulation | |||
| Cell Process | Epithelial-mesenchymal transition | |||
In-vitro Model |
A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 |
| HeLa | Endocervical adenocarcinoma | Homo sapiens | CVCL_0030 | |
| Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 | |
| Huh-7 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_0336 | |
| In-vivo Model | All animal experiments complied with Zhongshan School of Medicine Policy on Care and Use of Laboratory Animals. For subcutaneous transplanted model, sh-control and sh-METTL3 Huh7 cells (5 × 106 per mouse, n = 5 for each group) were diluted in 200 uL of PBS + 200 uL Matrigel (BD Biosciences) and subcutaneously injected into immunodeficient female mice to investigate tumor growth. | |||
| Response Summary | The upregulation of METTL3 and YTHDF1 act as adverse prognosis factors for overall survival (OS) rate of liver cancer patients. m6A-sequencing and functional studies confirm that Zinc finger protein SNAI1 (SNAI1), a key transcription factor of EMT, is involved in m6A-regulated EMT. | |||
hsa-miR-1914-3p
Lung cancer [ICD-11: 2C25]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [14] | |||
| Responsed Disease | Non-small-cell lung carcinoma [ICD-11: 2C25.Y] | |||
| Responsed Drug | Cisplatin | Approved | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Hippo signaling pathway | hsa04390 | ||
| Cell Process | Metabolic | |||
In-vitro Model |
A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 |
| Calu-6 | Lung adenocarcinoma | Homo sapiens | CVCL_0236 | |
| NCI-H1299 | Lung large cell carcinoma | Homo sapiens | CVCL_0060 | |
| NCI-H520 | Lung squamous cell carcinoma | Homo sapiens | CVCL_1566 | |
| In-vivo Model | Mice were injected with 5 × 106 lung cancer cells with stably expression of relevant plasmids and randomly divided into two groups (five mice per group) after the diameter of the xenografted tumors had reached approximately 5 mm in diameter. Xenografted mice were then administrated with PBS or DDP (3 mg/kg per day) for three times a week, and tumor volume were measured every second day. | |||
| Response Summary | METTL3, YTHDF3, YTHDF1, and eIF3b directly promoted YAP translation through an interaction with the translation initiation machinery. METTL3 knockdown inhibits tumor growth and enhances sensitivity to DDP in vivo.m6A mRNA methylation initiated by METTL3 directly promotes YAP translation and increases YAP activity by regulating the MALAT1-hsa-miR-1914-3p-YAP axis to induce Non-small cell lung cancer drug resistance and metastasis. | |||
Anillin (ANLN)
Liver cancer [ICD-11: 2C12]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [71] | |||
| Responsed Disease | Liver hepatocellular carcinoma [ICD-11: 2C12.02] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | mTOR signaling pathway | hsa04150 | ||
In-vitro Model |
Huh-7 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_0336 |
| HCCLM3 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_6832 | |
| PLC/PRF/5 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_0485 | |
| Hep 3B2.1-7 | Childhood hepatocellular carcinoma | Homo sapiens | CVCL_0326 | |
| BEL-7402 | Endocervical adenocarcinoma | Homo sapiens | CVCL_5492 | |
| HUVEC-C | Normal | Homo sapiens | CVCL_2959 | |
| RAW 264.7 | Mouse leukemia | Mus musculus | CVCL_0493 | |
| HeLa | Endocervical adenocarcinoma | Homo sapiens | CVCL_0030 | |
| In-vivo Model | Using a 100 μl Hamilton Microliter syringe, all the groups of 1 × 107 luciferase-labeled Huh-7 cells in 50 μl 1 x phosphate-buffered saline (PBS) was injected intracardially in the left ventricle of nu/nu, female 4-6-week-old nude mice. | |||
Bone morphogenetic protein 8B (BMP8B)
Obesity [ICD-11: 5B81]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [72] | |||
| Responsed Disease | Obesity [ICD-11: 5B81] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
3T3-L1 | Normal | Mus musculus | CVCL_0123 |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| In-vivo Model | Mice were intraperitoneally injected with CL316,243(Sigma) at 1 mg/kg/day for seven consecutive days. To exclude the BAT function, interscapular BAT was surgically removed37,38. The OCR of adipose tissues was performed using Clark-type oxygen electrodes (Strathkelvin Instruments). | |||
cAMP-responsive element modulator (CREM)
Cognitive disorders [ICD-11: 6E0Z]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [73] | |||
| Responsed Disease | Cognitive disorders [ICD-11: 6E0Z] | |||
| Target Regulation | Up regulation | |||
| In-vivo Model | 40 male Sprague Dawley (SD) rats (7-week-old) were obtained from SJA LABORATORY Animal Co, Ltd. (Hunan, China). After acclimation for 1 week, rats were randomly divided into four groups: the sham group, the sevoflurane group, the sevoflurane + Ad-NC group and the sevoflurane + Ad-YTHDF1 group, with ten animals in each group. Rats in sevoflurane groups were inhaled with 2.2% sevoflurane in the anesthetizing chamber at a flow rate of 1 L per minute for 6 h, and rats in the sham group were inhaled with 40% oxygen. The Ad-NC and Ad-YTHDF1 were constructed by GenePharma (Shanghai, China). Rats in the sevoflurane + Ad-NC group and the sevoflurane + Ad-YTHDF1 group were intracerebroventricularly injected with Ad-NC or Ad-YTHDF1 plasmids (100 μL; 1 × 109 PFU) by a Hamilton microsurgically gauged syringe into the left lateral cerebral ventricles 1 h before sevoflurane anesthesia. | |||
Collagen alpha-1 (COL1A1)
Hepatic fibrosis/cirrhosis [ICD-11: DB93]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [40] | |||
| Responsed Disease | Hepatic fibrosis/cirrhosis [ICD-11: DB93] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
JS1
|
N.A. | Mus musculus | CVCL_C7PL |
| In-vivo Model | Following 1 week of adaptive feeding, the mice were randomly divided into CCl4 + siCON and CCl4 + siYTHDF1 groups. Both groups were injected intraperitoneally with 5 ml/kg of 10% carbon tetrachloride (CCl4) twice a week for 4 weeks, to establish liver fibrosis. | |||
Keloid [ICD-11: EE60]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [74] | |||
| Responsed Disease | Keloid [ICD-11: EE60] | |||
| Target Regulation | Up regulation | |||
| Cell Process | mRNA stability | |||
In-vitro Model |
CHSE-EC
|
N.A. | Oncorhynchus tshawytscha | CVCL_DG46 |
| In-vivo Model | Six- to eight-week-old C57BL/6 mice were purchased from Cyagen Biosciences, Inc. (Cyagen, Suzhou, China). Alkbh3-/- C57BL/6 mice were generated via the CRISPR/Cas9 system. Single-guide RNAs (sgRNAs) were designed to target exons 3 to 5 of Alkbh3 and were coinjected with Cas9 into the zygotes. The pups obtained were genotyped by PCR. After genotyping, the F0 mice were subjected to serial mating to generate homozygous mutant offspring. | |||
Collagen alpha-2(I) chain (COL1A2)
Hepatic fibrosis/cirrhosis [ICD-11: DB93]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [40] | |||
| Responsed Disease | Hepatic fibrosis/cirrhosis [ICD-11: DB93] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
JS1
|
N.A. | Mus musculus | CVCL_C7PL |
| In-vivo Model | Following 1 week of adaptive feeding, the mice were randomly divided into CCl4 + siCON and CCl4 + siYTHDF1 groups. Both groups were injected intraperitoneally with 5 ml/kg of 10% carbon tetrachloride (CCl4) twice a week for 4 weeks, to establish liver fibrosis. | |||
Collagen alpha-2(V) chain (COL5A2)
Hepatic fibrosis/cirrhosis [ICD-11: DB93]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [40] | |||
| Responsed Disease | Hepatic fibrosis/cirrhosis [ICD-11: DB93] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
JS1
|
N.A. | Mus musculus | CVCL_C7PL |
| In-vivo Model | Following 1 week of adaptive feeding, the mice were randomly divided into CCl4 + siCON and CCl4 + siYTHDF1 groups. Both groups were injected intraperitoneally with 5 ml/kg of 10% carbon tetrachloride (CCl4) twice a week for 4 weeks, to establish liver fibrosis. | |||
Collagen alpha-2(VI) chain (COL6A2)
Hepatic fibrosis/cirrhosis [ICD-11: DB93]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [40] | |||
| Responsed Disease | Hepatic fibrosis/cirrhosis [ICD-11: DB93] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
JS1
|
N.A. | Mus musculus | CVCL_C7PL |
| In-vivo Model | Following 1 week of adaptive feeding, the mice were randomly divided into CCl4 + siCON and CCl4 + siYTHDF1 groups. Both groups were injected intraperitoneally with 5 ml/kg of 10% carbon tetrachloride (CCl4) twice a week for 4 weeks, to establish liver fibrosis. | |||
Cysteine methyltransferase DNMT3A (DNMT3A)
Alzheimer disease [ICD-11: 8A20]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [76] | |||
| Responsed Disease | Alzheimer disease [ICD-11: 8A20] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
HEK293T | Normal | Homo sapiens | CVCL_0063 |
Dickkopf-related protein 3 (DKK3)
Chronic kidney disease [ICD-11: GB61]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [77] | |||
| Responsed Disease | Diabetic nephropathy [ICD-11: GB61.Z] | |||
In-vitro Model |
HK-2 [Human kidney] | Normal | Homo sapiens | CVCL_0302 |
| In-vivo Model | Female mice (8 weeks old, 20-25 g) on a C57BL/6J background were fasted for 12 h but were allowed to drink water freely. They were then injected intraperitoneally with 50 mg/kg body weight of freshly dissolved STZ in sterile PBS for four consecutive days. Mice were given sterile PBS alone in the same way as an untreated control. The mice's blood glucose levels were assessed two weeks after their most recent treatment. Mice that exhibited glucose levels greater than 202 mg/dL were classified as successful hyperglycemic models and were utilized in subsequent studies. Five months after the final dose of STZ, the mice were euthanized, and the renal tissues were collected for pathological examination to confirm the successful establishment of the model of diabetes nephropathy induced by hyperglycemia. All animal experiments were performed according to the guidelines of the Institutional Animal Care and Use Committee at the China Pharmaceutical University. Isoflurane was used to anesthetize mice. | |||
Discoidin domain-containing receptor 2 (DDR2)
Ovarian cancer [ICD-11: 2C73]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [78] | |||
| Responsed Disease | Ovarian cancer [ICD-11: 2C73] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
SK-OV-3 | Ovarian serous cystadenocarcinoma | Homo sapiens | CVCL_0532 |
| In-vivo Model | BALB nude mice were purchased from Beijing Vital River Laboratory Animal Technology Co., Ltd. SKOV3 cells were transfected with sh-METTL3, sh-DDR2, DDR2 over-expression plasmid (oe-DDR2) or negative control. About one week later, the above SKOV3 cells were digested into single cells and subcutaneously injected to nude mice in different groups. After a duration of 4 weeks, the mice were euthanized by intraperitoneal injection of pentobarbital at a dosage 120 mg/kg. Finally, the subcutaneous tumors were removed to measure the size and volume (calculated according to volume = L (longest diameter) x W2 (shorter diameter)/2). | |||
Dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit 2 (RPN2)
Bladder cancer [ICD-11: 2C94]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [79] | |||
| Responsed Disease | Bladder cancer [ICD-11: 2C94] | |||
| Responsed Drug | Cisplatin | Approved | ||
| Pathway Response | PI3K-Akt signaling pathway | hsa04151 | ||
| mTOR signaling pathway | hsa04150 | |||
In-vitro Model |
5637 | Bladder carcinoma | Homo sapiens | CVCL_0126 |
| T24 | Bladder carcinoma | Homo sapiens | CVCL_0554 | |
| UM-UC-3 | Bladder carcinoma | Homo sapiens | CVCL_1783 | |
E3 ubiquitin-protein ligase TRIM68 (TRIM68)
Prostate cancer [ICD-11: 2C82]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [80] | |||
| Responsed Disease | Prostate cancer [ICD-11: 2C82] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
RWPE-2
|
N.A. | Homo sapiens | CVCL_3792 |
| LNCaP | Prostate carcinoma | Homo sapiens | CVCL_0395 | |
| DU145 | Prostate carcinoma | Homo sapiens | CVCL_0105 | |
| PC-3 | Prostate carcinoma | Homo sapiens | CVCL_0035 | |
| In-vivo Model | Cells (si-NC group, si-YTHDF1 group, si-TRIM68 group, si-YTHDF1 + OE-TRIM68 group) were digested with 0.25% trypsin, and fetal bovine serum was added to prevent excessive digestion. After centrifugation at 1000 rpm for 5 min, cells were collected and suspended with culture medium without serum, and cell density was adjusted to 5 × 107 cells/ml. SPF male BALB/c-nu nude mice (n = 20) were used in this study. Tumor cells (100 μl) were inoculated subcutaneously in the axils of the right forelimbs of mice. After the injection, the mental state, activity, diet, urine and feces of the nude mice were observed regularly every day. The body weight was measured weekly with an electronic balance, and the diameters of subcutaneous graft were measured weekly with vernier caliper. After the tumor grew to about 40 mm3, the nude mice were given flutamine (100 mg/kg; continuous administration for 1 week). The subcutaneous graft tumor volume V (mm3) = longest tumor diameter (mm) × shortest tumor diameter (mm) × 0.5. According to the tumor volume obtained, the growth curve of transplanted tumor was plotted. Five weeks later, the mice were weighed, and sacrificed by cervical dislocation method. The tumor was separated and rinsed with sterile PBS, and then weighed. After weighing, whole blood was taken to separate serum, and tumor tissue was taken. One part was fixed in 4% paraformaldehyde solution, and the other part was stored at -80 °C. | |||
Fibronectin (FN1)
Keloid [ICD-11: EE60]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [74] | |||
| Responsed Disease | Keloid [ICD-11: EE60] | |||
| Target Regulation | Up regulation | |||
| Cell Process | mRNA stability | |||
In-vitro Model |
CHSE-EC
|
N.A. | Oncorhynchus tshawytscha | CVCL_DG46 |
| In-vivo Model | Six- to eight-week-old C57BL/6 mice were purchased from Cyagen Biosciences, Inc. (Cyagen, Suzhou, China). Alkbh3-/- C57BL/6 mice were generated via the CRISPR/Cas9 system. Single-guide RNAs (sgRNAs) were designed to target exons 3 to 5 of Alkbh3 and were coinjected with Cas9 into the zygotes. The pups obtained were genotyped by PCR. After genotyping, the F0 mice were subjected to serial mating to generate homozygous mutant offspring. | |||
G1/S-specific cyclin-E2 (CCNE2)
Acute myeloid leukaemia [ICD-11: 2A60]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [81] | |||
| Responsed Disease | Acute myeloid leukaemia [ICD-11: 2A60] | |||
| Responsed Drug | Tegaserod | Approved | ||
| Target Regulation | Up regulation | |||
In-vitro Model |
THP-1 | Childhood acute monocytic leukemia | Homo sapiens | CVCL_0006 |
| MV4-11 | Childhood acute monocytic leukemia | Homo sapiens | CVCL_0064 | |
Glucose-6-phosphate exchanger SLC37A2 (SLC37A2)
Inflammatory bowel disease [ICD-11: DD7Z]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [82] | |||
| Responsed Disease | Inflammatory bowel disease [ICD-11: DD7Z] | |||
In-vitro Model |
RAW 264.7 | Mouse leukemia | Mus musculus | CVCL_0493 |
| THP-1 | Childhood acute monocytic leukemia | Homo sapiens | CVCL_0006 | |
| In-vivo Model | Mice were randomly allocated into three groups (n = 5/group): the negative control (NEG) group, the DSS-induced colitis (DSS) group, and the hucMSC-Ex-treated colitis (hucMSC-Ex) group. Mice in the NEG group were fed autoclaved water during the experiment, while mice in the DSS and hucMSC-Ex groups were fed autoclaved water containing 3% DSS (MP Biomedicals, USA). A total of 1 mg of hucMSC-Ex was administered by caudal vein to mice in the hucMSC-Ex group on the 3rd, 6th, and 9th day of modeling, while mice in other groups were injected with PBS. All mice were sacrificed when severe hematochezia and weight loss were observed in the DSS group. The disease progression of mice in the different groups was evaluated by weight change, DAI score, colorectal length, and weight ratio. After the mice were sacrificed, the general view of the spleen and colorectum of the mice was observed. The colorectal mucosa and spleen tissues of the mice in each group were separated, and RNA and protein of the tissues were extracted for subsequent experiments. | |||
Glucose-induced degradation protein 8 homolog (GID8)
Colorectal cancer [ICD-11: 2B91]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [83] | |||
| Responsed Disease | Colorectal cancer [ICD-11: 2B91] | |||
In-vitro Model |
SW620 | Colon adenocarcinoma | Homo sapiens | CVCL_0547 |
| HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 | |
| NCM460 | Normal | Homo sapiens | CVCL_0460 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| In-vivo Model | Sh-GID8 stable cells (2 × 106) were inoculated subcutaneously into the left axilla of athymic nude mice (n = 5 mice/group). Every three days, the tumor's size was measured with a digital caliper, and its volume was computed using the following formula: tumor volume (mm3) = length × width × width × 0.52. When the subcutaneous tumor reached approximately 50-100 mm3, glutamine (75 mg/kg) [13] or PBS was injected intratumorally every day. Tumors were collected for TUNEL staining or other analysis when tumor volumes reached the humane endpoint. For the metastasis model, the constructed HCT116 cells (2 × 106) with stable YTHDF1 or GID8 knockdown were injected into nude mice through the tail vein (n = 4 or n = 5 mice/group). Two months later, all the mice were sacrificed to observe tumor metastasis in the lungs. | |||
Glycogen phosphorylase, brain form (PYGB)
Pancreatic cancer [ICD-11: 2C10]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [84] | |||
| Responsed Disease | Pancreatic ductal adenocarcinoma [ICD-11: 2C10.0] | |||
In-vitro Model |
BxPC-3 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0186 |
| PANC-1 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0480 | |
| MIA PaCa-2 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0428 | |
| HPDE | Normal | Homo sapiens | CVCL_4376 | |
| In-vivo Model | Six BALB/c nude mice (male, 4 weeks) were randomly divided into 2 groups and inoculated with BXPC-3 cells (1 × 106) transfected with shPYGB or shNC by subcutaneous injection. Tumor volume was recorded every 7 days and mice were sacrificed after 6 weeks. To evaluate metastasis in vivo, transfected cells (5 × 106) were injected into nude mice through the tail vein. | |||
GTPase HRas (HRAS)
Soft tissue sarcoma [ICD-11: XH4UM7]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [85] | |||
| Responsed Disease | Soft tissue sarcoma [ICD-11: XH4UM7] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
5637 | Bladder carcinoma | Homo sapiens | CVCL_0126 |
| Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 | |
| T24 | Bladder carcinoma | Homo sapiens | CVCL_0554 | |
| 769-P | Renal cell carcinoma | Homo sapiens | CVCL_1050 | |
| AsPC-1 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0152 | |
| HCC827 | Lung adenocarcinoma | Homo sapiens | CVCL_2063 | |
| HeLa | Endocervical adenocarcinoma | Homo sapiens | CVCL_0030 | |
| MCF-7 | Invasive breast carcinoma | Homo sapiens | CVCL_0031 | |
| PC-3 | Prostate carcinoma | Homo sapiens | CVCL_0035 | |
| SW480 | Colon adenocarcinoma | Homo sapiens | CVCL_0546 | |
| HGC-27 | Gastric carcinoma | Homo sapiens | CVCL_1279 | |
| In-vivo Model | A total of 5637 cells with ectopic expression of dm6ACRISPR systems were injected subcutaneously (1 × 107 cells/inoculum) into the flanks of 5-wk-old nude mice (Shanghai Model Organisms Center). Tumor formation/growth was assessed until the experimental endpoint, and tumor volume was calculated by the formula: (width)2 × length/2. For tail vein injection, 5637 cells with ectopic expression of dm6ACRISPR systems (5 × 106 cells/0.1 mL PBS) were injected into the lateral tail vein of 5-wk-old nude mice. | |||
Histone-lysine N-methyltransferase NSD2 (NSD2)
Chronic kidney disease [ICD-11: GB61]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [86] | |||
| Responsed Disease | Diabetic nephropathy [ICD-11: GB61.Z] | |||
| Target Regulation | Up regulation | |||
Homeobox protein DLX-3 (DLX3)
Structural developmental anomalies of teeth and periodontal tissues [ICD-11: LA30]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [87] | |||
| Responsed Disease | Structural developmental anomalies of teeth and periodontal tissues [ICD-11: LA30.0] | |||
Insulin-like growth factor-binding protein 4 (IGFBP4)
Endometrial cancer [ICD-11: 2C76]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [88] | |||
| Responsed Disease | Endometrial cancer [ICD-11: 2C76] | |||
| Pathway Response | PI3K-Akt signaling pathway | hsa04151 | ||
| NF-kappa B signaling pathway | hsa04064 | |||
In-vitro Model |
HEC-1-A | Endometrial adenocarcinoma | Homo sapiens | CVCL_0293 |
| In-vivo Model | Ten specific pathogen-free (SPF) BALB/c nude mice (4-6 weeks old, 16 ± 2 g) were purchased from Shanghai Experimental Animal Center, Chinese Academy of Sciences. All experimental animals were kept in a SPF-level aseptic layer at 22-26 °C and 55 ± 5% humidity. Nude mice were randomly divided into sh-NC and sh-METTL3 groups, and these mice were anesthetized by intraperitoneal injection of pentobarbital sodium (60 mg/kg). After anesthesia, the right back skin of the nude mice was disinfected with conventional iodine. The mice in the sh-NC group were injected with 0.2 mL HEC-1-A cell suspension (1 × 106), and those in the sh-METTL3 group were injected with an equal volume of HEC-1-A cells that were stably transfected with sh-METTL3. The injector was slowly withdrawn, and the injection site was pressed for about 10 s to prevent an outflow of cell suspension. Three weeks later, the nude mice were euthanized by cervical dislocation, and subcutaneous tumors were separated and weighted. Tumor volume was measured as follows: tumor volume = 1/2 × length × width2. | |||
Lactadherin (MFGE8)
Acute liver failure [ICD-11: DB91]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [89] | |||
| Responsed Disease | Acute liver failure [ICD-11: DB91] | |||
| Target Regulation | Up regulation | |||
Long intergenic non-protein coding RNA 901 (LINC00901)
Pancreatic cancer [ICD-11: 2C10]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [90] | |||
| Responsed Disease | Pancreatic cancer [ICD-11: 2C10] | |||
| Target Regulation | Down regulation | |||
In-vitro Model |
MIA PaCa-2 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0428 |
| In-vivo Model | Nude (nu/nu) mice (4-5 weeks old) were purchased from Harlan Laboratories (Indianapolis, IN, USA). All animal studies were conducted in accordance with NIH animal use guidelines and a protocol approved by the UMMC Animal Care Committee. MIA PaCa-2 cells with LINC00901 overexpression or vector control (pCDH); LINC00901 KO or vector control (LCV2-m) at the exponential stage were harvested and mixed with 50% matrigel, and then injected into mice (2.5 million cells/spot) s.c. as described previously.21 Tumor growth was measured every 4 days, starting 2 weeks after injection of tumor cells. | |||
M-phase inducer phosphatase 1 (CDC25A)
Traumatic brain injury induced by controlled cortical impact injury [ICD-11: NA07]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [91] | |||
| Responsed Disease | Traumatic brain injury induced by controlled cortical impact injury [ICD-11: NA07] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
HT22 | Normal | Mus musculus | CVCL_0321 |
| In-vivo Model | A total of twenty-seven adult specific-pathogen-free C57BL/6 mice (male, 10 weeks old, 22-25 g) were obtained from the Animal Center of Xi'an JiaoTong University (Xi'an, China). Mice were housed in a standard environment of 22 ± 0.5°C, 70% humidity with a 12-h light/dark cycle and free access to food and water. | |||
Malignant T-cell-amplified sequence 1 (MCT1)
Cervical cancer [ICD-11: 2C77]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [92] | |||
| Responsed Disease | Cervical cancer [ICD-11: 2C77] | |||
In-vitro Model |
C-33 A | Cervical squamous cell carcinoma | Homo sapiens | CVCL_1094 |
| HeLa | Endocervical adenocarcinoma | Homo sapiens | CVCL_0030 | |
| Ca Ski | Cervical squamous cell carcinoma | Homo sapiens | CVCL_1100 | |
| SiHa | Cervical squamous cell carcinoma | Homo sapiens | CVCL_0032 | |
| HaCaT | Normal | Homo sapiens | CVCL_0038 | |
Methyl-CpG-binding protein 2 (MECP2)
Vascular disorders of the liver [ICD-11: DB98]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [93] | |||
| Responsed Disease | Vascular disorders of the liver [ICD-11: DB98.8] | |||
| Target Regulation | Up regulation | |||
Neuronal regeneration-related protein (NREP)
Idiopathic interstitial pneumonitis [ICD-11: CB03]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [94] | |||
| Responsed Disease | Pulmonary Fibrosis [ICD-11: CB03.4] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
MLE-12
|
N.A. | Mus musculus | CVCL_3751 |
| In-vivo Model | C57BL/6J mice (6-8 weeks old), purchased from Shanghai Slac Laboratory Animal Co., Ltd (Shanghai, China), were exposed to 0, 10, or 20 ppm NaAsO2 (Sigma, Japan) in drinking water for 6 months [23]. The arsenite concentration in high arsenic groundwater of Northern China is approximately 2 ppm, and the residents are mainly exposed to arsenite via drinking the water from artesian wells [51]. The dose of 20 ppm used in the present study was calculated according to factor (10) for animal-to-human extrapolation. Therefore, the arsenite concentrations used in our animal experiments are related to the human exposure level. As a positive control for pulmonary fibrosis, C57BL/6J mice were injected intraperitoneally with bleomycin (35 mg/kg) (Solarbio, China) twice weekly for 8 weeks. To downregulate, specifically, the levels of YTHDF1 in lung tissues, As-IPF AAV9-SP-Cp-shYTHDF1/AAV9-NC (Shanghai Genechem Co., Ltd, China) was intravenously injected at the 5th month of exposure of 20 ppm NaAsO2. Mice were housed on a 12-hour/12-hour light/dark cycle with free access to normal food. | |||
Oxidized low-density lipoprotein receptor 1 (OLR1)
Brain cancer [ICD-11: 2A00]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [95] | |||
| Responsed Disease | Glioma [ICD-11: 2A00.0] | |||
| Responsed Drug | Temozolomide | Approved | ||
| Target Regulation | Up regulation | |||
In-vitro Model |
U-343MG | Glioblastoma | Homo sapiens | CVCL_S471 |
| U-251MG | Astrocytoma | Homo sapiens | CVCL_0021 | |
Pappalysin-1 (PAPPA)
Endometrial cancer [ICD-11: 2C76]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [88] | |||
| Responsed Disease | Endometrial cancer [ICD-11: 2C76] | |||
| Pathway Response | PI3K-Akt signaling pathway | hsa04151 | ||
| NF-kappa B signaling pathway | hsa04064 | |||
In-vitro Model |
HEC-1-A | Endometrial adenocarcinoma | Homo sapiens | CVCL_0293 |
| In-vivo Model | Ten specific pathogen-free (SPF) BALB/c nude mice (4-6 weeks old, 16 ± 2 g) were purchased from Shanghai Experimental Animal Center, Chinese Academy of Sciences. All experimental animals were kept in a SPF-level aseptic layer at 22-26 °C and 55 ± 5% humidity. Nude mice were randomly divided into sh-NC and sh-METTL3 groups, and these mice were anesthetized by intraperitoneal injection of pentobarbital sodium (60 mg/kg). After anesthesia, the right back skin of the nude mice was disinfected with conventional iodine. The mice in the sh-NC group were injected with 0.2 mL HEC-1-A cell suspension (1 × 106), and those in the sh-METTL3 group were injected with an equal volume of HEC-1-A cells that were stably transfected with sh-METTL3. The injector was slowly withdrawn, and the injection site was pressed for about 10 s to prevent an outflow of cell suspension. Three weeks later, the nude mice were euthanized by cervical dislocation, and subcutaneous tumors were separated and weighted. Tumor volume was measured as follows: tumor volume = 1/2 × length × width2. | |||
Phosphoglycerate kinase 1 (PGK1)
Head and neck squamous carcinoma [ICD-11: 2B6E]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [96] | |||
| Responsed Disease | Oral squamous cell carcinoma [ICD-11: 2B6E.0] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
HOK | Normal | Hexagrammos otakii | CVCL_YE19 |
| SCC-9 | Tongue squamous cell carcinoma | Homo sapiens | CVCL_1685 | |
| CAL-27 | Tongue squamous cell carcinoma | Homo sapiens | CVCL_1107 | |
PR domain zinc finger protein 15 (PRDM15)
Liver cancer [ICD-11: 2C12]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [97] | |||
| Responsed Disease | Intrahepatic cholangiocarcinoma [ICD-11: 2C12.10] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
CC-LP-1 | Intrahepatic cholangiocarcinoma | Homo sapiens | CVCL_0205 |
| HuCC-T1 | Intrahepatic cholangiocarcinoma | Homo sapiens | CVCL_0324 | |
| In-vivo Model | For subcutaneous inoculation, 1 × 106 control or METTL16-depleted CCA cells suspended in 100 μL PBS mixed with matrix gel (BD, 356,234) at 1:1 ratio was injected into the mice's flanks. Tumor sizes were measured at the relevant time intervals. At the end of the study, the mice were euthanized, and the tumors were dissected and weighed. For intrahepatic inoculation, 1 × 106 control or METTL16-depleted CCLP1 cells suspended in 10 μL PBS mixed with matrix gel (BD, 356,234) at a 1:1 ratio was implanted into the livers of NOD-SCID mice. The mice were sacrificed 8 weeks after injection, their livers were surgically dissected, and the liver/body weight ratios were evaluated. | |||
pre-miR-665
Structural developmental anomalies of teeth and periodontal tissues [ICD-11: LA30]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [87] | |||
| Responsed Disease | Structural developmental anomalies of teeth and periodontal tissues [ICD-11: LA30.0] | |||
Proliferating cell nuclear antigen (PCNA)
Prostate cancer [ICD-11: 2C82]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [98] | |||
| Responsed Disease | Prostate cancer [ICD-11: 2C82] | |||
| Target Regulation | Down regulation | |||
In-vitro Model |
PC-3 | Prostate carcinoma | Homo sapiens | CVCL_0035 |
| DU145 | Prostate carcinoma | Homo sapiens | CVCL_0105 | |
| 22Rv1 | Prostate carcinoma | Homo sapiens | CVCL_1045 | |
|
RWPE-2
|
N.A. | Homo sapiens | CVCL_3792 | |
| In-vivo Model | The mice were kept under standard conditions and provided with care in accordance with established protocols. For the experiment, 1 × 106 DU145 cells with reduced RBM15B expression and corresponding control cells were injected subcutaneously into the right flank of each nude mouse. Tumor length (L) and width (W) were measured weekly, and tumor volume was calculated using the formula: Volume = (W2 × L) / 2. Four weeks following the injections, mice were euthanized by intraperitoneal administration of sodium pentobarbital at a dose of 151 mg/kg. Tumors were then surgically removed and weighed. | |||
Ras-related protein Rab-27B (RAB27B)
Malignant haematopoietic neoplasm [ICD-11: 2B33]
| In total 2 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [99] | |||
| Responsed Disease | Chronic myeloid leukaemia [ICD-11: 2B33.2] | |||
| Responsed Drug | Rucaparib | Approved | ||
| Target Regulation | Up regulation | |||
In-vitro Model |
K-562 | Chronic myelogenous leukemia | Homo sapiens | CVCL_0004 |
| KCL-22 | Chronic myelogenous leukemia | Homo sapiens | CVCL_2091 | |
| In-vivo Model | K562 cells (1 × 106) were suspended in 100 μL of normal saline and the suspension was mixed with an equal volume of Matrigel. This mixture was subcutaneously injected into the right armpit of 4-week-old mice. Tumor size measurements were initiated on the day of inoculation. The tumor size was calculated using the formula: 0.5 × (long diameter) × (short diameter).2 When the tumor volume reached 100 mm3 (± 20%), rucaparib was given via intragastric administration at 50 mg/kg/d. | |||
| Experiment 2 Reporting the m6A-centered Disease Response of This Target Gene | [99] | |||
| Responsed Disease | Chronic myeloid leukaemia [ICD-11: 2B33.2] | |||
| Responsed Drug | Imatinib | Approved | ||
| Target Regulation | Up regulation | |||
In-vitro Model |
K-562 | Chronic myelogenous leukemia | Homo sapiens | CVCL_0004 |
| KCL-22 | Chronic myelogenous leukemia | Homo sapiens | CVCL_2091 | |
| In-vivo Model | K562 cells (1 × 106) were suspended in 100 μL of normal saline and the suspension was mixed with an equal volume of Matrigel. This mixture was subcutaneously injected into the right armpit of 4-week-old mice. Tumor size measurements were initiated on the day of inoculation. The tumor size was calculated using the formula: 0.5 × (long diameter) × (short diameter).2 When the tumor volume reached 100 mm3 (± 20%), rucaparib was given via intragastric administration at 50 mg/kg/d. | |||
Receptor-interacting serine/threonine-protein kinase 4 (RIPK4)
Malignant mixed epithelial mesenchymal tumour [ICD-11: 2B5D]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [100] | |||
| Responsed Disease | Malignant mixed epithelial mesenchymal tumour of ovary [ICD-11: 2B5D.0] | |||
| Responsed Drug | Cisplatin | Approved | ||
| Target Regulation | Up regulation | |||
| Pathway Response | NF-kappa B signaling pathway | hsa04064 | ||
In-vitro Model |
SK-OV-3 | Ovarian serous cystadenocarcinoma | Homo sapiens | CVCL_0532 |
| A2780 | Ovarian endometrioid adenocarcinoma | Homo sapiens | CVCL_0134 | |
| In-vivo Model | Animals were purchased from Changzhou Cavens Laboratory Animal Company (Changzhou, China) and maintained under specific pathogen-free (SPF) conditions. All animal experiments were performed in accordance with the guidelines of the Laboratory Animal Health Committee of Jiangsu University. Approximately 1 × 107 cells were injected subcutaneously into the dorsal surface of female BALB/c nude mice (n = 5 for each group, 4 weeks old, 15 ± 2 g). When the xenograft volume reached approximately 60 mm3 (after 1 week), DDP was intraperitoneally injected three times per week for 3 weeks. On Day 28, the mice were sacrificed and the tumors were frozen at -80 °C for follow-up experiments. | |||
Scavenger receptor class B member 1 (SCARB1)
Gastric cancer [ICD-11: 2B72]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [101] | |||
| Responsed Disease | Gastric cancer [ICD-11: 2B72] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
BGC-823 | Gastric carcinoma | Homo sapiens | CVCL_3360 |
| AGS | Gastric adenocarcinoma | Homo sapiens | CVCL_0139 | |
| SGC-7901 | Gastric carcinoma | Homo sapiens | CVCL_0520 | |
| MKN45 | Gastric adenocarcinoma | Homo sapiens | CVCL_0434 | |
| NCI-N87 | Gastric tubular adenocarcinoma | Homo sapiens | CVCL_1603 | |
| In-vivo Model | ShRNA control (empty vector) and sh-H19 were transfected into BGC-823 cells for 24 h. The cells were collected after digested by trypsin. The cells were washed using phosphate buffered saline (PBS) (Gibco, USA) twice and resuspended with PBS before being subcutaneously injected into nude mice. | |||
Serine/threonine-protein kinase B-raf (BRAF)
Atherosclerosis [ICD-11: BD40]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [102] | |||
| Responsed Disease | Atherosclerosis [ICD-11: BD40.Z] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
RAW 264.7 | Mouse leukemia | Mus musculus | CVCL_0493 |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
SH3 domain-binding protein 5 (SH3BP5)
Lung cancer [ICD-11: 2C25]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [103] | |||
| Responsed Disease | Lung cancer [ICD-11: 2C25] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
NCI-H522 | Lung adenocarcinoma | Homo sapiens | CVCL_1567 |
| A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 | |
| WI-38 | Normal | Homo sapiens | CVCL_0579 | |
| IMR-90 | Normal | Homo sapiens | CVCL_0347 | |
Solute carrier family 2, facilitated glucose transporter member 3 (SLC2A3)
Low bone mass disorder [ICD-11: FB83]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [104] | |||
| Responsed Disease | Osteoporosis [ICD-11: FB83.1] | |||
In-vitro Model |
MC3T3-E1 | Normal | Mus musculus | CVCL_0409 |
| In-vivo Model | Female C57BL/6 mice, aged eight weeks, were selected for the study. The mice were divided into three groups with six mice per group. After intramuscular anesthesia (xylazine 10 mg/kg, ketamine 100 mg/kg), two groups underwent bilateral ovariectomy (OVX), while the remaining group had a sham operation. Ovariectomized mice were administered weekly intratibial injections of Mettl14 overexpression adenovirus containing at a concentration of 109 plaque-forming unit per injection, for a duration of 8 weeks. The mice were humanely euthanized one week after the final injection, and their tibia were gathered for further analysis. This study was approved by the ethics committee of Tongji University (NO: TJBH00823101). All the experimental methods were carried out in accordance with the approved guidelines. All experimental procedures involving mice were carried out in strict accordance with the recommendations in the ARRIVE guidelines (Animal Research: Reporting of In Vivo Experiments). | |||
Toll-like receptor 2 (TLR2)
Hypopharyngeal cancer [ICD-11: 2B6D]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [105] | |||
| Responsed Disease | Hypopharyngeal squamous cell carcinoma [ICD-11: 2B6D.0] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
FaDu | Hypopharyngeal squamous cell carcinoma | Homo sapiens | CVCL_1218 |
| Detroit 562 | Pharyngeal squamous cell carcinoma | Homo sapiens | CVCL_1171 | |
| In-vivo Model | HPSCC FaDu or Detroit 562 cells (1 × 106 cells) expressing vector control and construct lentiviruses were subcutaneously injected into the right flanks of 4-week-old male nude mice. Tumor diameters and body weight were recorded every 3 days for 1-6 weeks. Detroit 562 and FaDu cells (1 × 106 cells) were subcutaneously injected into the right flanks of 4-week-old male nude mice. | |||
Tubulointerstitial nephritis antigen-like (TINAGL1)
Esophageal cancer [ICD-11: 2B70]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [106] | |||
| Responsed Disease | Esophageal cancer [ICD-11: 2B70] | |||
In-vitro Model |
TE-1 | Esophageal squamous cell carcinoma | Homo sapiens | CVCL_1759 |
| Eca-109 | Esophageal squamous cell carcinoma | Homo sapiens | CVCL_6898 | |
| In-vivo Model | 1 × 107 cells were injected into the right axillary region of nude mice. After 28 days, the mice were sacrificed and tumors were removed. ESCA cells (2 × 107 cells) were injected into nude mice via the tail vein. After 6 weeks, the mice were sacrificed and the lungs were obtained and fixed with 4% paraformaldehyde. The number of metastatic nodules was counted following H&E staining. | |||
Zinc finger protein RFP (TRIM27)
Colorectal cancer [ICD-11: 2B91]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [107] | |||
| Responsed Disease | Colorectal cancer [ICD-11: 2B91] | |||
| Responsed Drug | Cisplatin | Approved | ||
| Target Regulation | Up regulation | |||
In-vitro Model |
HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 |
| SW620 | Colon adenocarcinoma | Homo sapiens | CVCL_0547 | |
Unspecific Target Gene
Brain cancer [ICD-11: 2A00]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [108] | |||
| Responsed Disease | Glioma [ICD-11: 2A00.0] | |||
In-vitro Model |
SHG-44 | Astrocytoma | Homo sapiens | CVCL_6728 |
| U87 (A primary glioblastoma cell line) | ||||
| In-vivo Model | About 5 × 106 cells transfected with pc-YTHDF1 in SHG-44 cell (si-YTHDF1 in U87 cell) were suspended in 0.1 mL of PBS and injected subcutaneously into nude mice. | |||
| Response Summary | m6A RNA methylation regulators play a potential role in the progression of gliomas. Predicted one microRNA, microRNA 346 that regulated and binded to 3'UTR of YTHDF1, which was confirmed by our fluorescent enzyme reporter gene experiment. | |||
Colon cancer [ICD-11: 2B90]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [109] | |||
| Responsed Disease | Colon cancer [ICD-11: 2B90] | |||
| Responsed Drug | Cisplatin | Approved | ||
| Pathway Response | Metabolic pathways | hsa01100 | ||
| Cell Process | Glutamine metabolism | |||
In-vitro Model |
LoVo | Colon adenocarcinoma | Homo sapiens | CVCL_0399 |
| HT-29 | Colon adenocarcinoma | Homo sapiens | CVCL_0320 | |
| DLD-1 | Colon adenocarcinoma | Homo sapiens | CVCL_0248 | |
| HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 | |
| SW480 | Colon adenocarcinoma | Homo sapiens | CVCL_0546 | |
| CRL-1790 (The normal colon epithelial cell line CRL-1790, were purchased from the American Type Culture Collection (ATCC).) | ||||
| In-vivo Model | Mice were injected subcutaneously with LoVo (1 × 106) cells, which were stably transfected with the control shRNA or YTHDF1 shRNA. | |||
| Response Summary | YTHDF1-promoted cisplatin resistance, contributing to overcoming chemoresistant colon cancers. | |||
Colorectal cancer [ICD-11: 2B91]
| In total 2 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [110] | |||
| Responsed Disease | Colorectal cancer [ICD-11: 2B91] | |||
| Responsed Drug | Fluorouracil | Approved | ||
| Cell Process | Cells proliferation | |||
| Cells migration | ||||
| Cells invasion | ||||
In-vitro Model |
Caco-2 | Colon adenocarcinoma | Homo sapiens | CVCL_0025 |
| DLD-1 | Colon adenocarcinoma | Homo sapiens | CVCL_0248 | |
| HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 | |
| HT29 | Colon cancer | Mus musculus | CVCL_A8EZ | |
| KM12-SM | Colon carcinoma | Homo sapiens | CVCL_9548 | |
| LoVo | Colon adenocarcinoma | Homo sapiens | CVCL_0399 | |
| RKO | Colon carcinoma | Homo sapiens | CVCL_0504 | |
| SW480 | Colon adenocarcinoma | Homo sapiens | CVCL_0546 | |
| Response Summary | The knockdown of YTHDF1 resulted in the suppression of cancer proliferation and sensitization to the exposure of anticancer drugs such as fluorouracil and oxaliplatin. m6A reader Ythdf1 plays a significant role in colorectal cancer progression. An oncogenic transcription factor MYC was associated with YTHDF1 in both expression and chromatin immunoprecipitation data. | |||
| Experiment 2 Reporting the m6A-centered Disease Response of This Target Gene | [110] | |||
| Responsed Disease | Colorectal cancer [ICD-11: 2B91] | |||
| Responsed Drug | Oxaliplatin | Approved | ||
| Cell Process | Cells proliferation | |||
| Cells migration | ||||
| Cells invasion | ||||
In-vitro Model |
Caco-2 | Colon adenocarcinoma | Homo sapiens | CVCL_0025 |
| DLD-1 | Colon adenocarcinoma | Homo sapiens | CVCL_0248 | |
| HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 | |
| HT29 | Colon cancer | Mus musculus | CVCL_A8EZ | |
| KM12-SM | Colon carcinoma | Homo sapiens | CVCL_9548 | |
| LoVo | Colon adenocarcinoma | Homo sapiens | CVCL_0399 | |
| RKO | Colon carcinoma | Homo sapiens | CVCL_0504 | |
| SW480 | Colon adenocarcinoma | Homo sapiens | CVCL_0546 | |
| Response Summary | The knockdown of YTHDF1 resulted in the suppression of cancer proliferation and sensitization to the exposure of anticancer drugs such as fluorouracil and oxaliplatin. m6A reader Ythdf1 plays a significant role in colorectal cancer progression. An oncogenic transcription factor MYC was associated with YTHDF1 in both expression and chromatin immunoprecipitation data. | |||
Pancreatic cancer [ICD-11: 2C10]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [111] | |||
| Responsed Disease | Pancreatic cancer [ICD-11: 2C10] | |||
| Responsed Drug | Gemcitabine | Approved | ||
| Pathway Response | Adipocytokine signaling pathway | hsa04920 | ||
| Cell Process | Epithelial-mesenchymal transition | |||
In-vitro Model |
BxPC-3 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0186 |
| HDE-CT cell line (A normal human pancreatic cell line) | ||||
| MIA PaCa-2 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0428 | |
| Response Summary | Lasso regression identified a six-m6A-regulator-signature prognostic model (KIAA1429, HNRNPC, METTL3, YTHDF1, IGF2BP2, and IGF2BP3). Gene set enrichment analysis revealed m6A regulators (KIAA1429, HNRNPC, and IGF2BP2) were related to multiple biological behaviors in pancreatic cancer, including adipocytokine signaling, the well vs. poorly differentiated tumor pathway, tumor metastasis pathway, epithelial mesenchymal transition pathway, gemcitabine resistance pathway, and stemness pathway. | |||
Liver cancer [ICD-11: 2C12]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [112] | |||
| Responsed Disease | Hepatocellular carcinoma [ICD-11: 2C12.02] | |||
| Target Regulation | Up regulation | |||
| Cell Process | Epithelial-mesenchymal transition | |||
| Cell apoptosis | ||||
In-vitro Model |
BEL-7404 | Endocervical adenocarcinoma | Homo sapiens | CVCL_6568 |
| HCCLM3 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_6832 | |
| Hep 3B2.1-7 | Childhood hepatocellular carcinoma | Homo sapiens | CVCL_0326 | |
| Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 | |
| MHCC97-H | Adult hepatocellular carcinoma | Homo sapiens | CVCL_4972 | |
| MHCC97-L | Adult hepatocellular carcinoma | Homo sapiens | CVCL_4973 | |
| SMMC-7721 | Endocervical adenocarcinoma | Homo sapiens | CVCL_0534 | |
| Response Summary | YTHDF1 promotes the aggressive phenotypes by facilitating epithelial-mesenchymal transition (EMT) and activating AKT/glycogen synthase kinase (GSK)-3-beta/beta-catenin signaling in hepatocellular carcinoma. | |||
Metabolic dysfunction-associated steatotic liver disease [ICD-11: 5C90]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [115] | |||
| Responsed Disease | Metabolic dysfunction-associated steatotic liver disease [ICD-11: 5C90] | |||
In-vitro Model |
AML12 | Normal | Mus musculus | CVCL_0140 |
Alzheimer disease [ICD-11: 8A20]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [116] | |||
| Responsed Disease | Alzheimer disease [ICD-11: 8A20] | |||
| Cell Process | Oxidative stress | |||
| Cell apoptosis | ||||
| Mitochondrial dysfunction | ||||
In-vitro Model |
CCF-STTG1 | Astrocytoma | Homo sapiens | CVCL_1118 |
| Response Summary | Perturbed m6A signaling can be contributing to Alzheimer's disease pathogenesis, likely by compromising astrocyte bioenergetics. MO-I-500, a novel pharmacological inhibitor of FTO, can strongly reduce the adverse effects of STZ. STZ-treated astrocytes expressed significantly higher levels of m6A demethylase FTO and m6A reader YTHDF1. | |||
Acute ischemic stroke [ICD-11: 8B11]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [117] | |||
| Responsed Disease | Acute ischemic stroke [ICD-11: 8B11] | |||
| In-vivo Model | 36 male C57BL/6J mice (20-22 g, 2-month-old) were purchased from SPF (Beijing) biotechnology co., Ltd (Beijing, China) and maintained in the specific pathogen-free (SPF) animal laboratory with a 12/12 h light/dark cycle with free access to food and water. The mice were randomly assigned into six groups (n = 6 per group): (1) sham-operated group (Sham), (2) MCAO 6 h, (3) MCAO 12 h, (4) MCAO 1 d, (5) MCAO 3 d, and (6) MCAO 7 d. | |||
Myasthenia gravis [ICD-11: 8C60]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [118] | |||
| Responsed Disease | Myasthenia gravis [ICD-11: 8C60] | |||
| Pathway Response | Wnt signaling pathway | hsa04310 | ||
Forkhead box protein O3 (FOXO3)
| Representative RNA-seq result indicating the expression of this target gene regulated by YTHDF1 | ||
| Cell Line | AGS cell line | Homo sapiens |
|
Treatment: shYTHDF1 AGS
Control: shNC AGS
|
GSE166972 | |
| Regulation |
  |
logFC: 1.07E+00 p-value: 3.17E-02 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and YTHDF1 | ||
| Cell Line | Hela | Homo sapiens |
| Regulation | logFC: 2.44E+00 | GSE63591 |
Sorafenib
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [4] | |||
| Responsed Disease | Hepatocellular carcinoma | ICD-11: 2C12.02 | ||
| Target Regulation | Up regulation | |||
| Pathway Response | FoxO signaling pathway | hsa04068 | ||
| Cell Process | Cell autophagy | |||
| Response Summary | METTL3 can sensitise hepatocellular carcinoma cells to sorafenib through stabilising Forkhead box protein O3 (FOXO3) in an m6A-dependent manner and translated by YTHDF1, thereby inhibiting the transcription of autophagy-related genes, including ATG3, ATG5, ATG7, ATG12, and ATG16L1. | |||
Kelch-like ECH-associated protein 1 (KEAP1)
| Representative RNA-seq result indicating the expression of this target gene regulated by YTHDF1 | ||
| Cell Line | AGS cell line | Homo sapiens |
|
Treatment: shYTHDF1 AGS
Control: shControl AGS
|
GSE159425 | |
| Regulation |
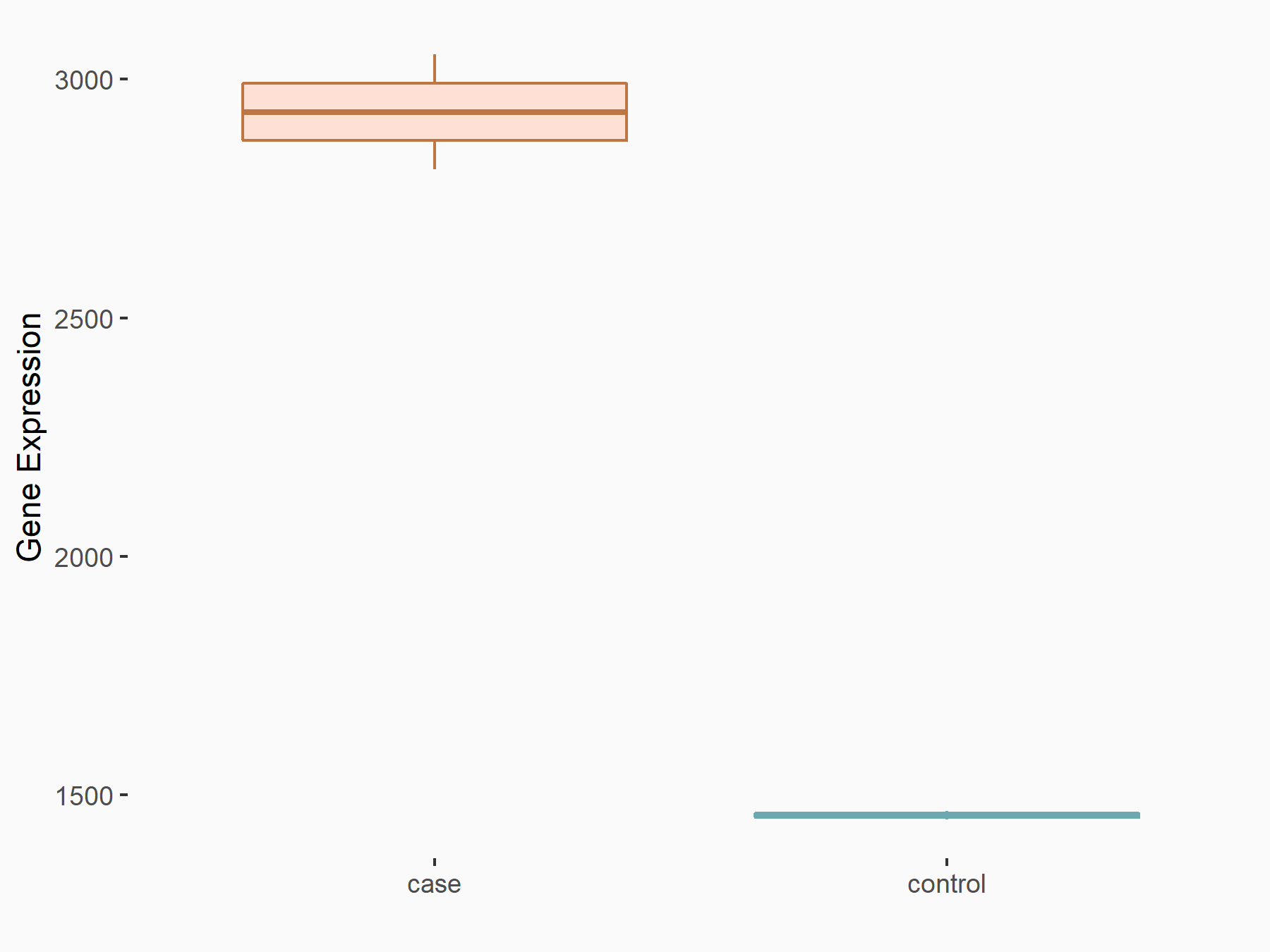 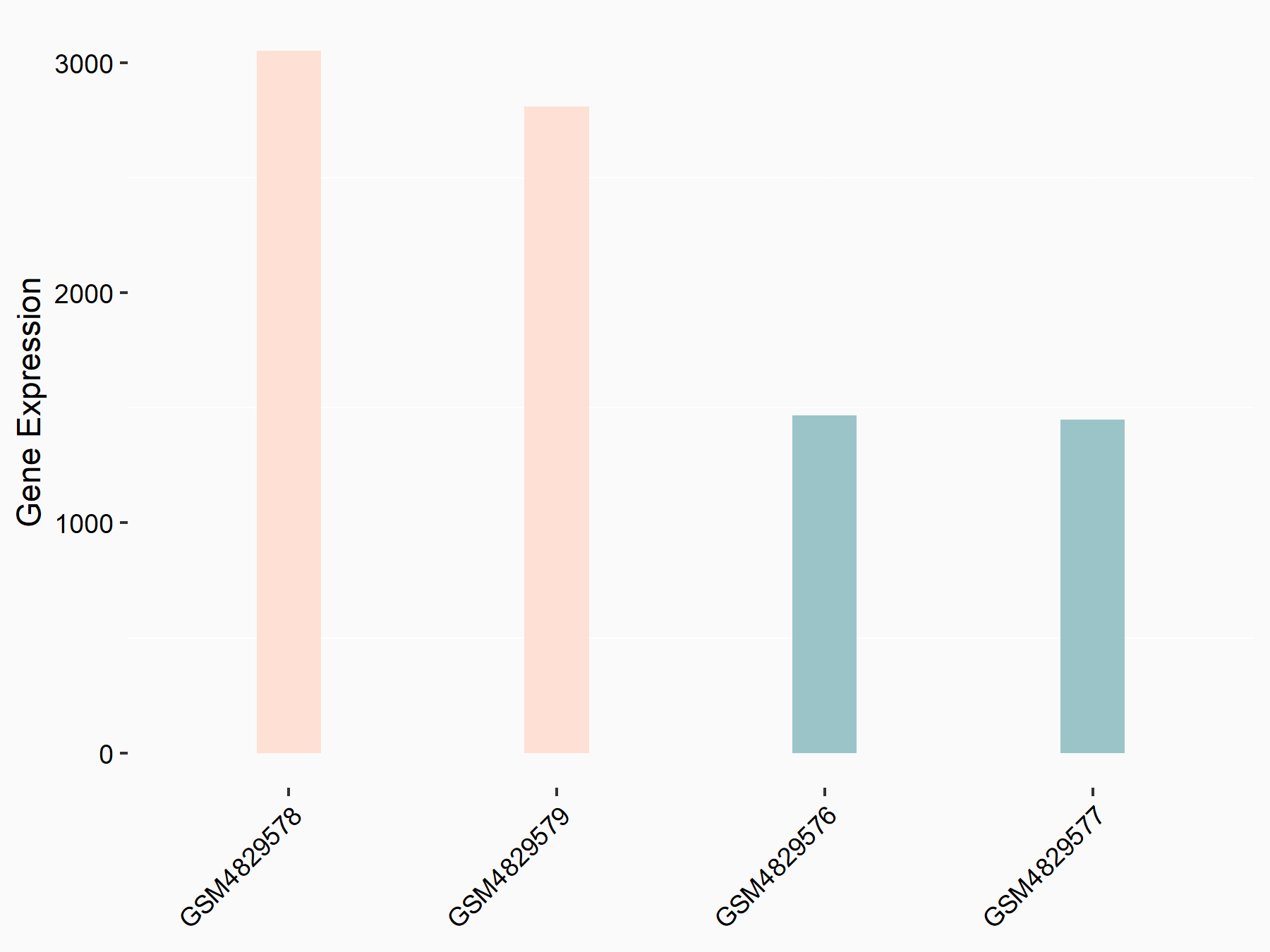 |
logFC: 1.01E+00 p-value: 4.73E-09 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and YTHDF1 | ||
| Cell Line | Hela | Homo sapiens |
| Regulation | logFC: 1.61E+00 | GSE63591 |
Cisplatin
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [7] | |||
| Responsed Disease | Non-small-cell lung carcinoma | ICD-11: 2C25.Y | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Chemical carcinogenesis - reactive oxygen species | hsa05208 | ||
| Cell cycle | hsa04110 | |||
| Cell Process | Biological regulation | |||
| In-vitro Model | A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 |
| A549-DDP (Human lung adenocarcinoma is resistant to cisplatin) | ||||
| GLC-82 | Endocervical adenocarcinoma | Homo sapiens | CVCL_3371 | |
| NCI-H1299 | Lung large cell carcinoma | Homo sapiens | CVCL_0060 | |
| NCI-H1975 | Lung adenocarcinoma | Homo sapiens | CVCL_1511 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| NCI-H1650 | Minimally invasive lung adenocarcinoma | Homo sapiens | CVCL_1483 | |
| NCI-H838 | Lung adenocarcinoma | Homo sapiens | CVCL_1594 | |
| SPC-A1 | Endocervical adenocarcinoma | Homo sapiens | CVCL_6955 | |
| In-vivo Model | Mice were treated via nasal inhalation of adenovirus carrying Cre recombinase (5 × 106 p.f.u for Ad-Cre, Biowit Inc., Shenzhen, Guangdong), and were then killed at indicated times for gross inspection and histopathological examination. | |||
| Response Summary | YTHDF1 deficiency inhibits Non-small cell lung cancer cell proliferation and xenograft tumor formation through regulating the translational efficiency of CDK2, CDK4, p27, and cyclin D1, and that YTHDF1 depletion restrains de novo lung adenocarcinomas (ADC) progression. Mechanistic studies identified the Kelch-like ECH-associated protein 1 (KEAP1)-Nrf2-AKR1C1 axis as the downstream mediator of YTHDF1. YTHDF1 high expression correlates with better clinical outcome, with its depletion rendering cancerous cells resistant to cisplatin (DDP) treatment. | |||
PR domain zinc finger protein 2 (PRDM2)
| Representative RNA-seq result indicating the expression of this target gene regulated by YTHDF1 | ||
| Cell Line | AGS cell line | Homo sapiens |
|
Treatment: shYTHDF1 AGS
Control: shNC AGS
|
GSE166972 | |
| Regulation |
 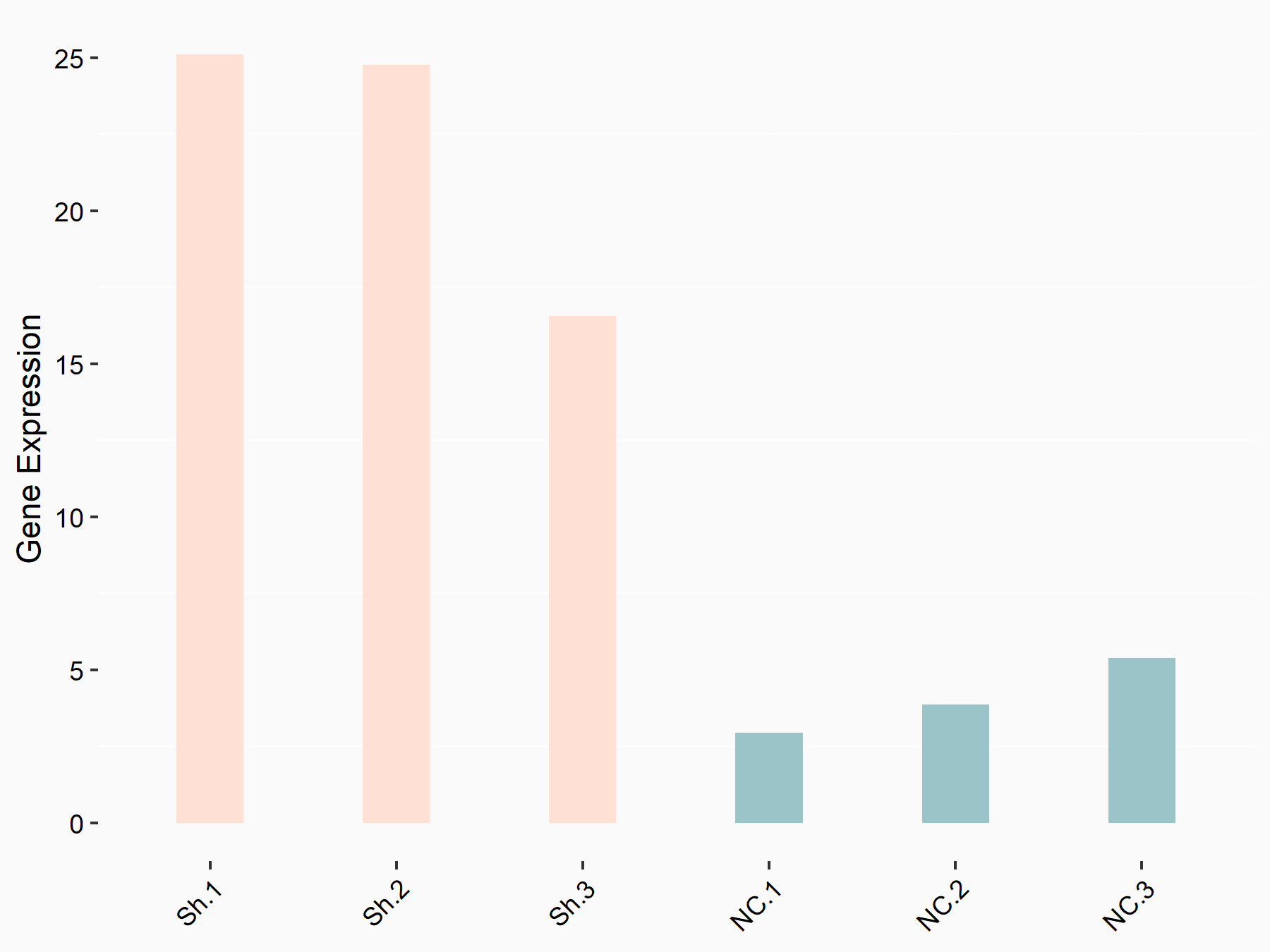 |
logFC: 2.20E+00 p-value: 3.42E-04 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and YTHDF1 | ||
| Cell Line | Hela | Homo sapiens |
| Regulation | logFC: 1.84E+00 | GSE63591 |
Arsenite
[Phase 2]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [10] | |||
| Responsed Disease | Solid tumour/cancer | ICD-11: 2A00-2F9Z | ||
| Target Regulation | Down regulation | |||
| Pathway Response | p53 signaling pathway | hsa04115 | ||
| In-vitro Model | HaCaT | Normal | Homo sapiens | CVCL_0038 |
| Response Summary | METTL3 significantly decreased m6A level, restoring p53 activation and inhibiting cellular transformation phenotypes in the arsenite-transformed cells. m6A downregulated the expression of the positive p53 regulator, PR domain zinc finger protein 2 (PRDM2), through the YTHDF2-promoted decay of PRDM2 mRNAs. m6A upregulated the expression of the negative p53 regulator, YY1 and MDM2 through YTHDF1-stimulated translation of YY1 and MDM2 mRNA. This study further sheds light on the mechanisms of arsenic carcinogenesis via RNA epigenetics. | |||
Transcription factor E2F8 (E2F8)
| Representative RNA-seq result indicating the expression of this target gene regulated by YTHDF1 | ||
| Cell Line | AGS cell line | Homo sapiens |
|
Treatment: shYTHDF1 AGS
Control: shControl AGS
|
GSE159425 | |
| Regulation |
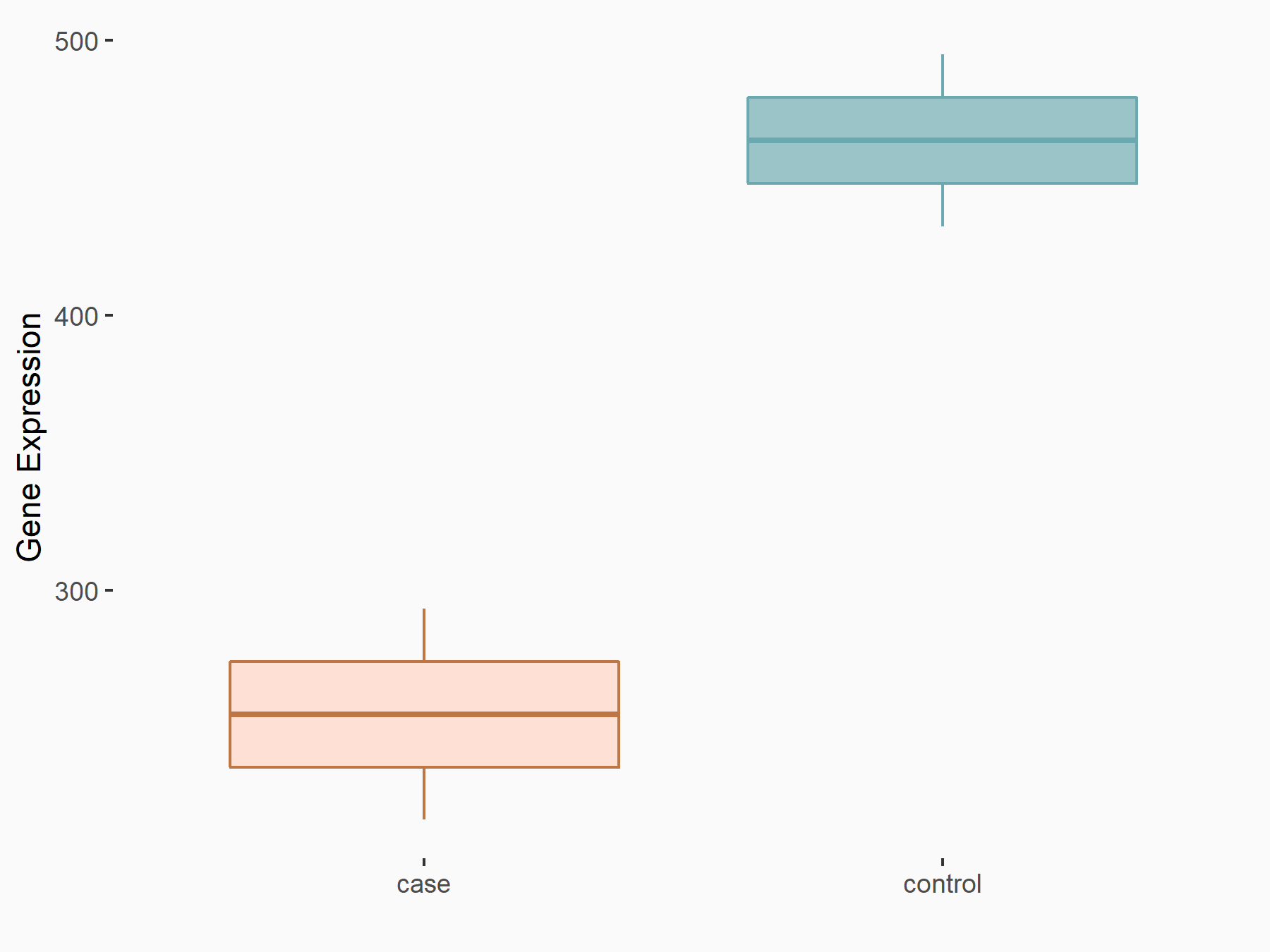 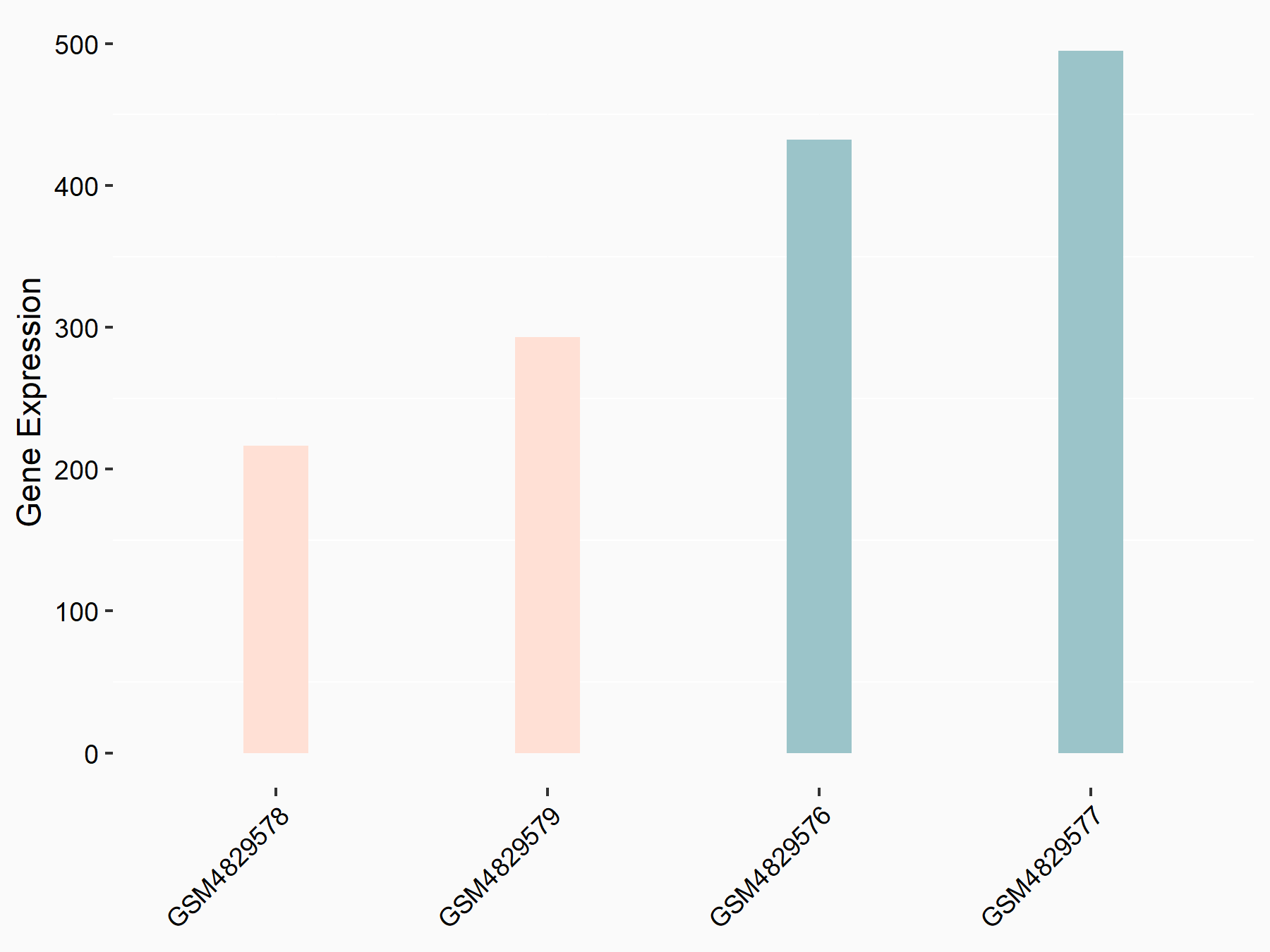 |
logFC: -8.62E-01 p-value: 7.08E-04 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and YTHDF1 | ||
| Cell Line | Hela | Homo sapiens |
| Regulation | logFC: 1.18E+00 | GSE63591 |
Cisplatin
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [12] | |||
| Responsed Disease | Breast cancer | ICD-11: 2C60 | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Nucleotide excision repair | hsa03420 | ||
| Cell Process | RNA stability | |||
| In-vitro Model | MDA-MB-231 | Breast adenocarcinoma | Homo sapiens | CVCL_0062 |
| MCF-7 | Invasive breast carcinoma | Homo sapiens | CVCL_0031 | |
| Hs 578T | Invasive breast carcinoma | Homo sapiens | CVCL_0332 | |
| In-vivo Model | 1×106 MDA-MB-231 cells were resuspended in 100 uL PBS with 50% Matrigel (Corning Costar, USA), and injected into the mammary fat pad of the mice. | |||
| Response Summary | In breast cancer, accordingly YTHDF1 knockdown sensitizes breast cancer cells to Adriamycin and Cisplatin as well as Olaparib, a PARP inhibitor. Transcription factor E2F8 (E2F8) is a target molecule by YTHDF1 which modulates E2F8 mRNA stability and DNA damage repair in a METTL14-dependent manner. | |||
Doxil
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [12] | |||
| Responsed Disease | Breast cancer | ICD-11: 2C60 | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Nucleotide excision repair | hsa03420 | ||
| Cell Process | RNA stability | |||
| In-vitro Model | MDA-MB-231 | Breast adenocarcinoma | Homo sapiens | CVCL_0062 |
| MCF-7 | Invasive breast carcinoma | Homo sapiens | CVCL_0031 | |
| Hs 578T | Invasive breast carcinoma | Homo sapiens | CVCL_0332 | |
| In-vivo Model | 1×106 MDA-MB-231 cells were resuspended in 100 uL PBS with 50% Matrigel (Corning Costar, USA), and injected into the mammary fat pad of the mice. | |||
| Response Summary | In breast cancer, accordingly YTHDF1 knockdown sensitizes breast cancer cells to Adriamycin and Cisplatin as well as Olaparib, a PARP inhibitor. Transcription factor E2F8 (E2F8) is a target molecule by YTHDF1 which modulates E2F8 mRNA stability and DNA damage repair in a METTL14-dependent manner. | |||
Olaparib
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [12] | |||
| Responsed Disease | Breast cancer | ICD-11: 2C60 | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Nucleotide excision repair | hsa03420 | ||
| Cell Process | RNA stability | |||
| In-vitro Model | MDA-MB-231 | Breast adenocarcinoma | Homo sapiens | CVCL_0062 |
| MCF-7 | Invasive breast carcinoma | Homo sapiens | CVCL_0031 | |
| Hs 578T | Invasive breast carcinoma | Homo sapiens | CVCL_0332 | |
| In-vivo Model | 1×106 MDA-MB-231 cells were resuspended in 100 uL PBS with 50% Matrigel (Corning Costar, USA), and injected into the mammary fat pad of the mice. | |||
| Response Summary | In breast cancer, accordingly YTHDF1 knockdown sensitizes breast cancer cells to Adriamycin and Cisplatin as well as Olaparib, a PARP inhibitor. Transcription factor E2F8 (E2F8) is a target molecule by YTHDF1 which modulates E2F8 mRNA stability and DNA damage repair in a METTL14-dependent manner. | |||
Transcriptional coactivator YAP1 (YAP1)
| Representative RNA-seq result indicating the expression of this target gene regulated by YTHDF1 | ||
| Cell Line | AGS cell line | Homo sapiens |
|
Treatment: shYTHDF1 AGS
Control: shNC AGS
|
GSE166972 | |
| Regulation |
 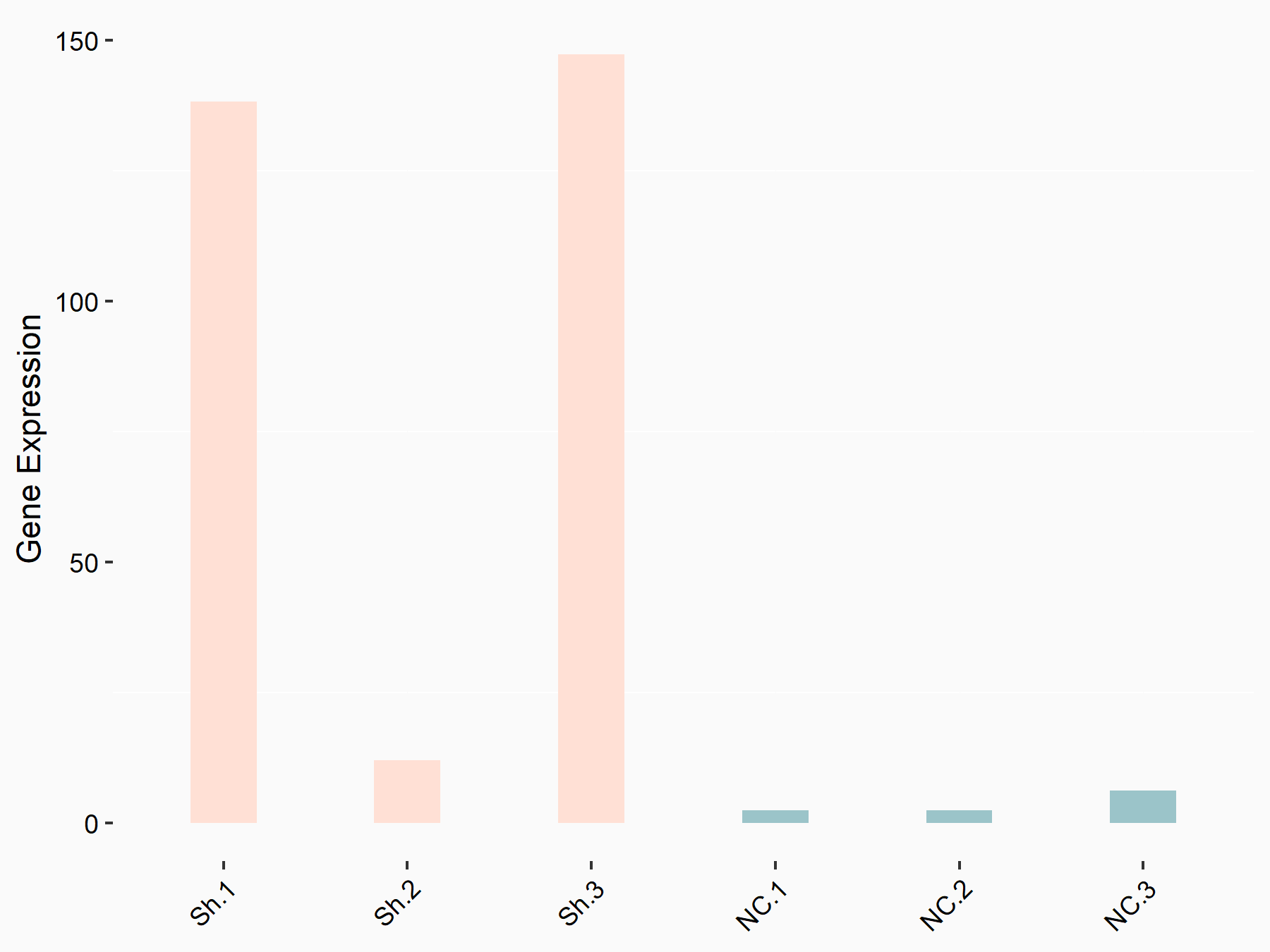 |
logFC: 3.89E+00 p-value: 1.09E-02 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between the target gene and YTHDF1 | ||
| Cell Line | Hela | Homo sapiens |
| Regulation | logFC: 1.35E+00 | GSE63591 |
Cisplatin
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [14] | |||
| Responsed Disease | Non-small-cell lung carcinoma | ICD-11: 2C25.Y | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Hippo signaling pathway | hsa04390 | ||
| Cell Process | Metabolic | |||
| In-vitro Model | A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 |
| Calu-6 | Lung adenocarcinoma | Homo sapiens | CVCL_0236 | |
| NCI-H1299 | Lung large cell carcinoma | Homo sapiens | CVCL_0060 | |
| NCI-H520 | Lung squamous cell carcinoma | Homo sapiens | CVCL_1566 | |
| In-vivo Model | Mice were injected with 5 × 106 lung cancer cells with stably expression of relevant plasmids and randomly divided into two groups (five mice per group) after the diameter of the xenografted tumors had reached approximately 5 mm in diameter. Xenografted mice were then administrated with PBS or DDP (3 mg/kg per day) for three times a week, and tumor volume were measured every second day. | |||
| Response Summary | METTL3, YTHDF3, YTHDF1, and eIF3b directly promoted YAP translation through an interaction with the translation initiation machinery. METTL3 knockdown inhibits tumor growth and enhances sensitivity to DDP in vivo.m6A mRNA methylation initiated by METTL3 directly promotes YAP translation and increases YAP activity by regulating the MALAT1-miR-1914-3p-Transcriptional coactivator YAP1 (YAP1) axis to induce Non-small cell lung cancer drug resistance and metastasis. | |||
Autophagy-related protein 16-1 (ATG16L1)
| Representative RNA-seq result indicating the expression of this target gene regulated by YTHDF1 | ||
| Cell Line | AGS cell line | Homo sapiens |
|
Treatment: shYTHDF1 AGS
Control: shNC AGS
|
GSE166972 | |
| Regulation |
 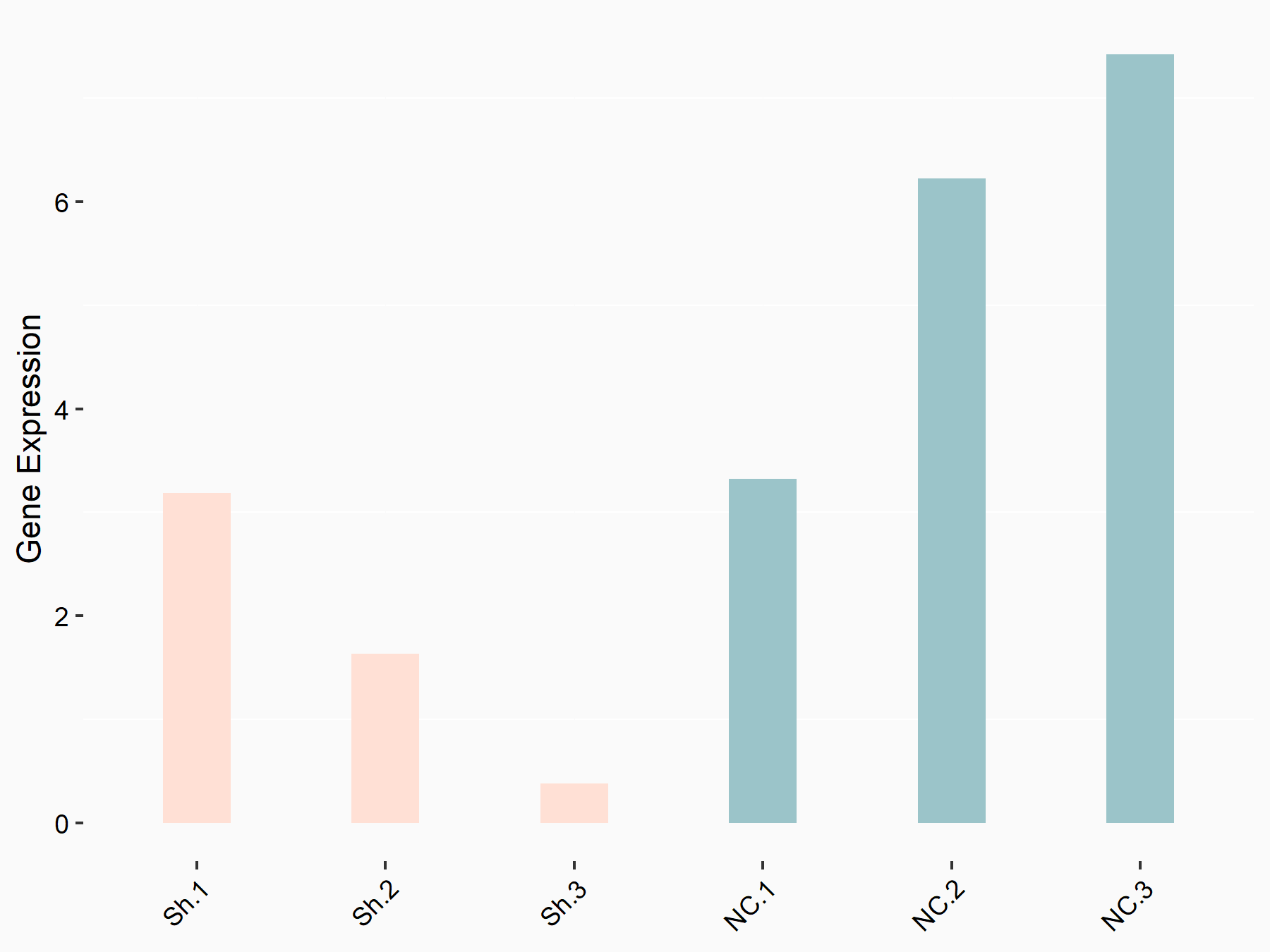 |
logFC: -1.37E+00 p-value: 3.41E-02 |
| More Results | Click to View More RNA-seq Results | |
Sorafenib
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [4] | |||
| Responsed Disease | Hepatocellular carcinoma | ICD-11: 2C12.02 | ||
| Target Regulation | Up regulation | |||
| Pathway Response | FoxO signaling pathway | hsa04068 | ||
| Autophagy | hsa04140 | |||
| Cell Process | Cell autophagy | |||
| Response Summary | METTL3 can sensitise hepatocellular carcinoma cells to sorafenib through stabilising forkhead box class O3 (FOXO3) in an m6A-dependent manner and translated by YTHDF1, thereby inhibiting the transcription of autophagy-related genes, including ATG3, ATG5, ATG7, ATG12, and Autophagy-related protein 16-1 (ATG16L1). | |||
Beclin-1 (BECN1)
| Representative RNA-seq result indicating the expression of this target gene regulated by YTHDF1 | ||
| Cell Line | AGS cell line | Homo sapiens |
|
Treatment: shYTHDF1 AGS
Control: shNC AGS
|
GSE166972 | |
| Regulation |
 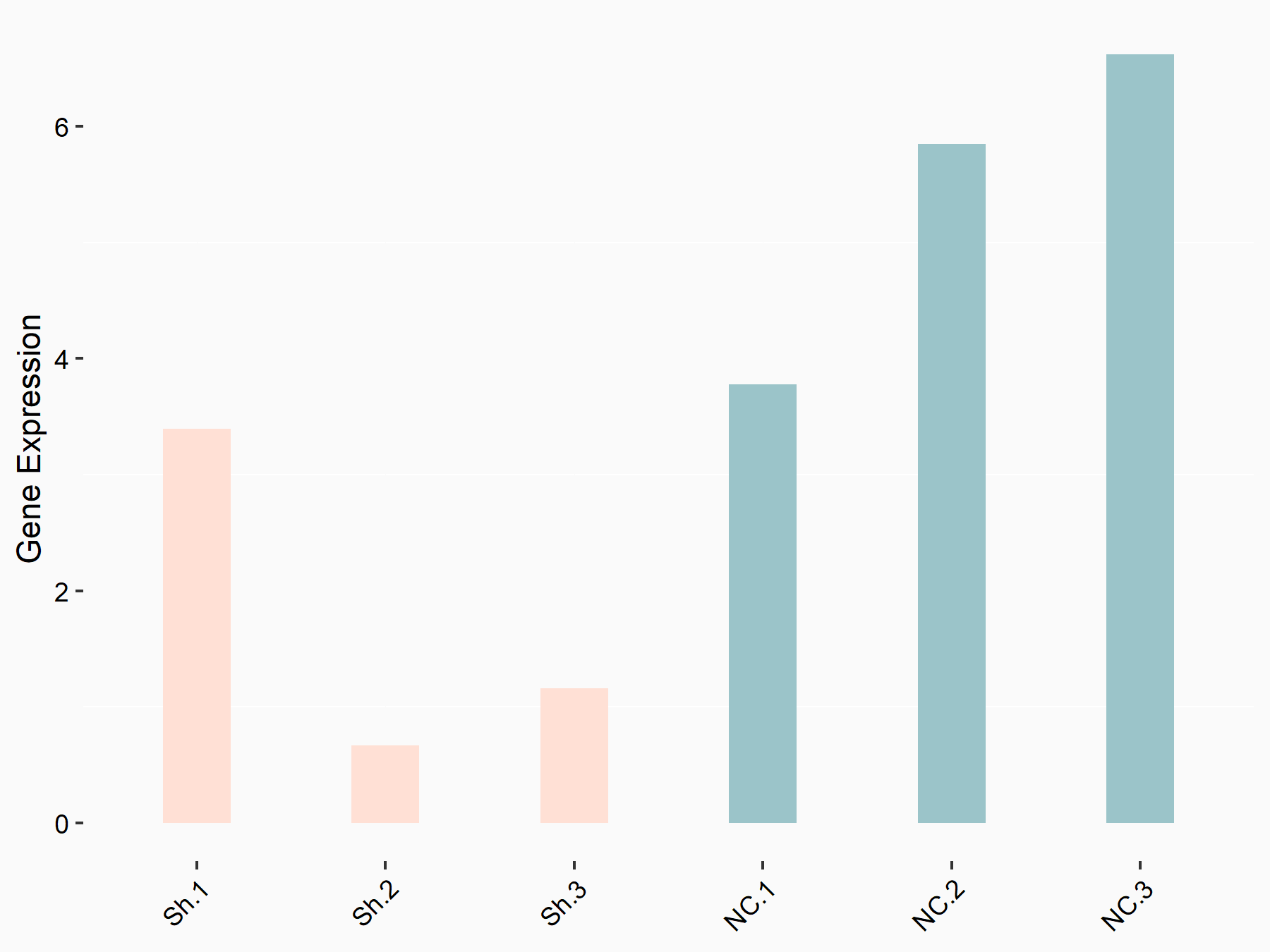 |
logFC: -1.33E+00 p-value: 2.29E-02 |
| More Results | Click to View More RNA-seq Results | |
Sorafenib
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [17] | |||
| Responsed Disease | Hepatocellular carcinoma | ICD-11: 2C12.02 | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Ferroptosis | hsa04216 | ||
| Autophagy | hsa04140 | |||
| Cell Process | Ferroptosis | |||
| Cell autophagy | ||||
| In-vitro Model | HSC (Hematopoietic stem cell) | |||
| In-vivo Model | VA-Lip-Mettl4-shRNA, VA-Lip-Fto-Plasmid and VA-Lip-Ythdf1-shRNA (0.75 mg/kg) were injected intravenously 3 times a week. | |||
| Response Summary | Analyzed the effect of sorafenib on HSC ferroptosis and m6A modification in advanced fibrotic patients with hepatocellular carcinoma receiving sorafenib monotherapy. YTHDF1 promotes Beclin-1 (BECN1) mRNA stability and autophagy activation via recognizing the m6A binding site within BECN1 coding regions. | |||
Cyclin-dependent kinase 2 (CDK2)
| Representative RNA-seq result indicating the expression of this target gene regulated by YTHDF1 | ||
| Cell Line | AGS cell line | Homo sapiens |
|
Treatment: shYTHDF1 AGS
Control: shControl AGS
|
GSE159425 | |
| Regulation |
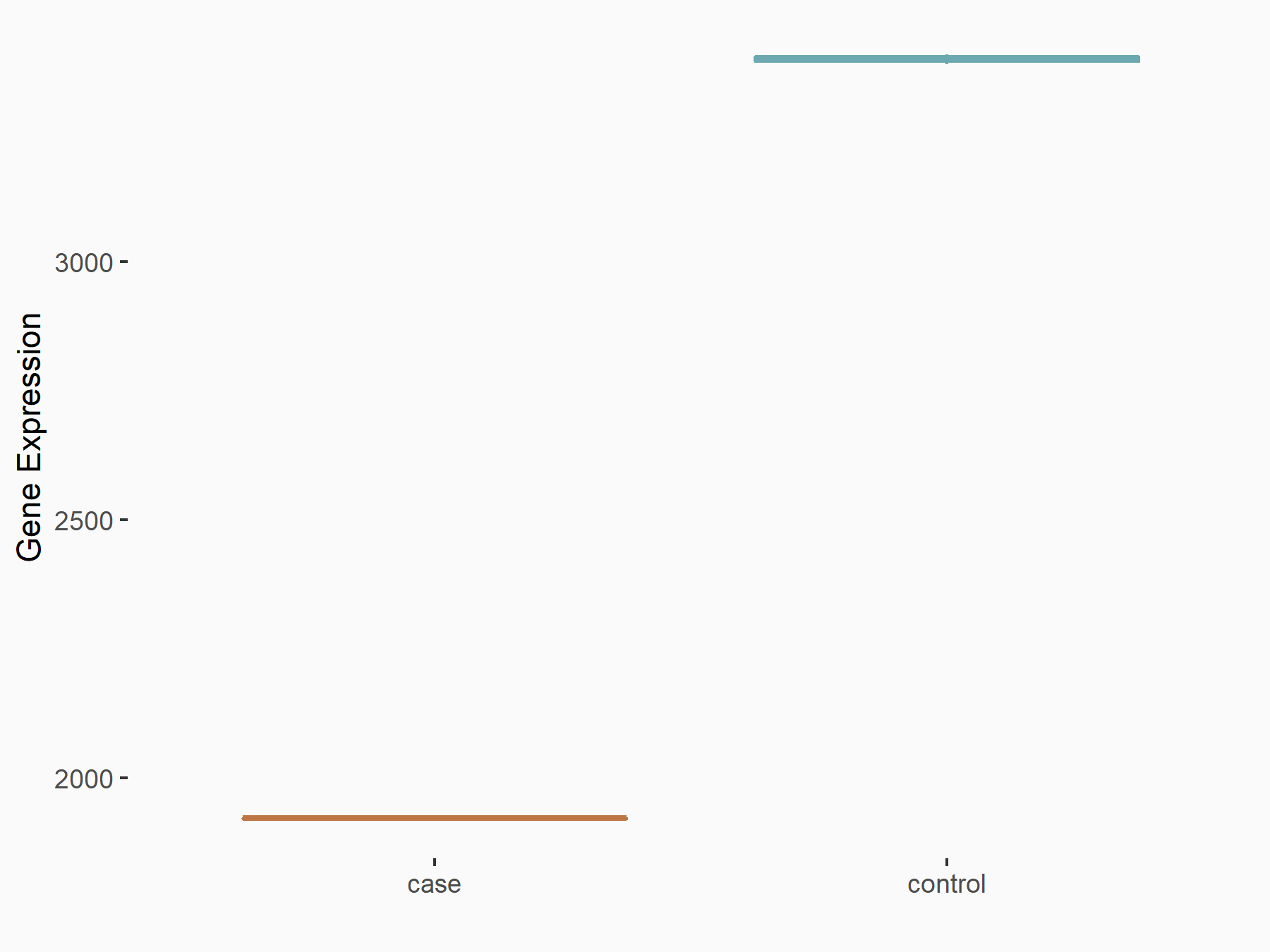 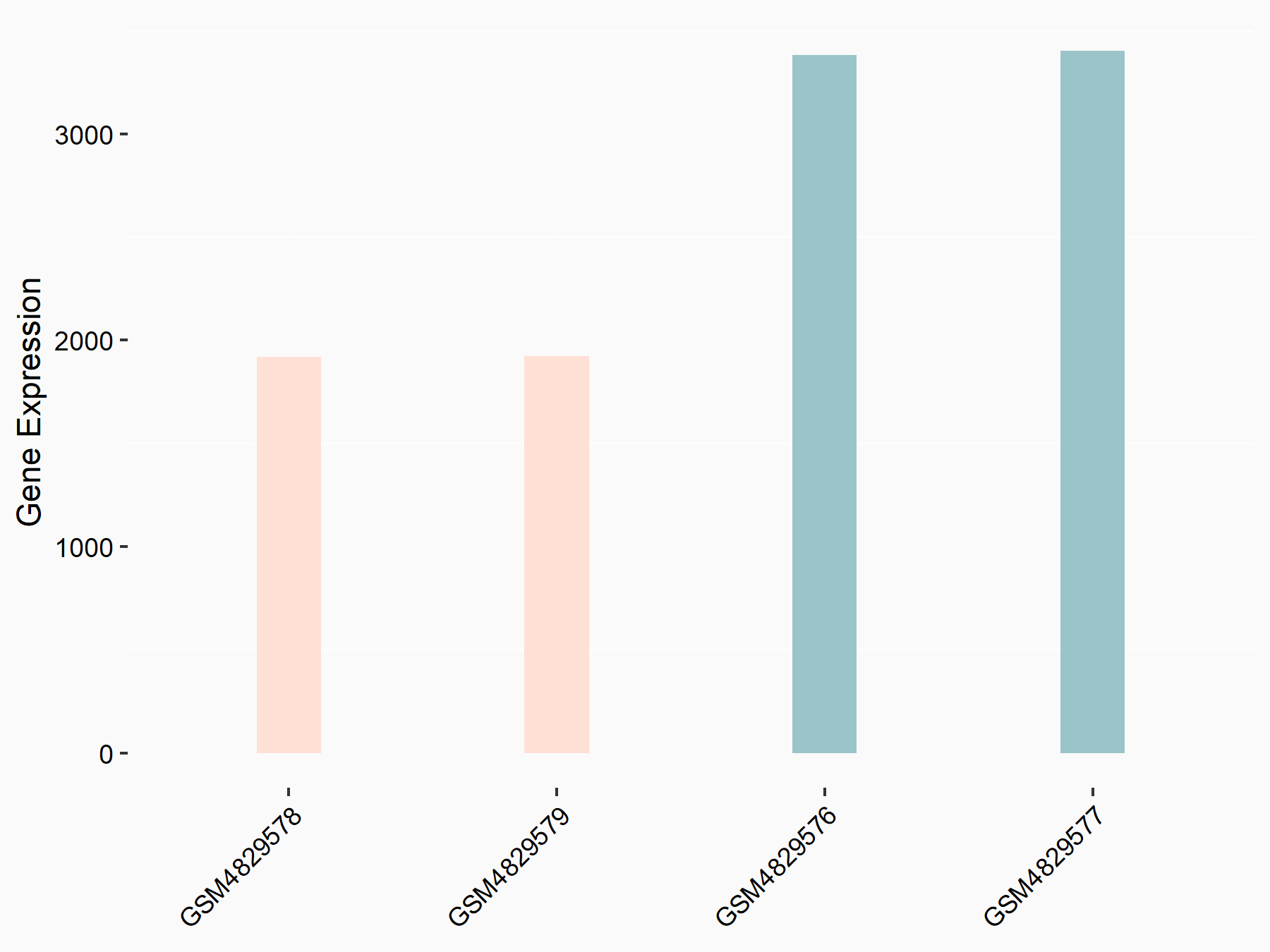 |
logFC: -8.20E-01 p-value: 7.96E-07 |
| More Results | Click to View More RNA-seq Results | |
Cisplatin
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [7] | |||
| Responsed Disease | Non-small-cell lung carcinoma | ICD-11: 2C25.Y | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Chemical carcinogenesis - reactive oxygen species | hsa05208 | ||
| Cell cycle | hsa04110 | |||
| Cell Process | Biological regulation | |||
| In-vitro Model | A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 |
| A549-DDP (Human lung adenocarcinoma is resistant to cisplatin) | ||||
| GLC-82 | Endocervical adenocarcinoma | Homo sapiens | CVCL_3371 | |
| NCI-H1299 | Lung large cell carcinoma | Homo sapiens | CVCL_0060 | |
| NCI-H1975 | Lung adenocarcinoma | Homo sapiens | CVCL_1511 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| NCI-H1650 | Minimally invasive lung adenocarcinoma | Homo sapiens | CVCL_1483 | |
| NCI-H838 | Lung adenocarcinoma | Homo sapiens | CVCL_1594 | |
| SPC-A1 | Endocervical adenocarcinoma | Homo sapiens | CVCL_6955 | |
| In-vivo Model | Mice were treated via nasal inhalation of adenovirus carrying Cre recombinase (5 × 106 p.f.u for Ad-Cre, Biowit Inc., Shenzhen, Guangdong), and were then killed at indicated times for gross inspection and histopathological examination. | |||
| Response Summary | YTHDF1 deficiency inhibits Non-small cell lung cancer cell proliferation and xenograft tumor formation through regulating the translational efficiency of Cyclin-dependent kinase 2 (CDK2), CDK4, p27, and cyclin D1, and that YTHDF1 depletion restrains de novo lung adenocarcinomas (ADC) progression. Mechanistic studies identified the Keap1-Nrf2-AKR1C1 axis as the downstream mediator of YTHDF1. YTHDF1 high expression correlates with better clinical outcome, with its depletion rendering cancerous cells resistant to cisplatin (DDP) treatment. | |||
Tripartite motif-containing protein 29 (TRIM29)
| Representative RNA-seq result indicating the expression of this target gene regulated by YTHDF1 | ||
| Cell Line | AGS cell line | Homo sapiens |
|
Treatment: shYTHDF1 AGS
Control: shNC AGS
|
GSE166972 | |
| Regulation |
 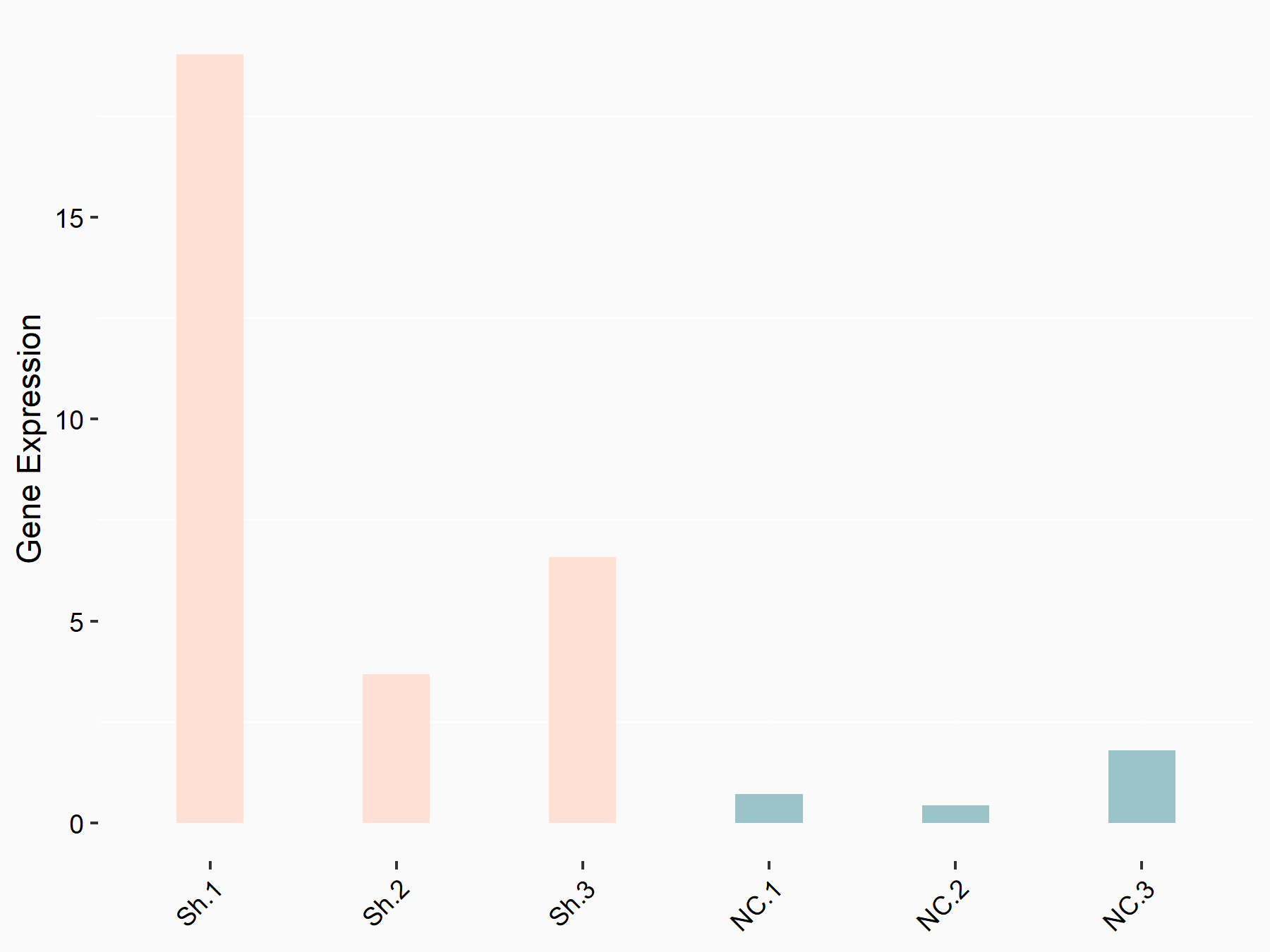 |
logFC: 2.23E+00 p-value: 1.11E-02 |
| More Results | Click to View More RNA-seq Results | |
Cisplatin
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [23] | |||
| Responsed Disease | Ovarian cancer | ICD-11: 2C73 | ||
| Target Regulation | Up regulation | |||
| Cell Process | Ectopic expression | |||
| In-vitro Model | A2780 | Ovarian endometrioid adenocarcinoma | Homo sapiens | CVCL_0134 |
| SK-OV-3 | Ovarian serous cystadenocarcinoma | Homo sapiens | CVCL_0532 | |
| In-vivo Model | The specified number of viable SKOV3/DDP cells and SKOV3/DDP cells with TRIM29 knock down were resuspended in 100 uL PBS, injected subcutaneously under the left and right back of 4-week old nude mice respectively (n = 3 per group). | |||
| Response Summary | m6A-YTHDF1-mediated Tripartite motif-containing protein 29 (TRIM29) upregulation facilitates the stem cell-like phenotype of cisplatin-resistant ovarian cancer cells. TRIM29 acts as an oncogene to promote the CSC-like features of cisplatin-resistant ovarian cancer in an m6A-YTHDF1-dependent manner. | |||
Poly [ADP-ribose] polymerase 1 (PARP1)
| Representative RIP-seq result supporting the interaction between the target gene and YTHDF1 | ||
| Cell Line | Hela | Homo sapiens |
| Regulation | logFC: 1.06E+00 | GSE63591 |
Oxaliplatin
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [32] | |||
| Responsed Disease | Gastric cancer | ICD-11: 2B72 | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Nucleotide excision repair | hsa03420 | ||
| Signaling pathways regulating pluripotency of stem cells | hsa04550 | |||
| Cell Process | RNA stability | |||
| Excision repair | ||||
| In-vitro Model | SNU-719 | Gastric tubular adenocarcinoma | Homo sapiens | CVCL_5086 |
| MKN74 | Gastric tubular adenocarcinoma | Homo sapiens | CVCL_2791 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| AGS | Gastric adenocarcinoma | Homo sapiens | CVCL_0139 | |
| In-vivo Model | 100,000 pLKO and PARP1-sh1 (PT1 and PT2) cells were mixed with matrix gel and inoculate into BALB/C nude mice, respectively. After 25 days, 6 organoid transplanted tumor mice were treated with oxaliplatin (Sellekchem, s1224) twice a week for 4 weeks at a dose of 5 mg/kg. | |||
| Response Summary | m6A methyltransferase METTL3 facilitates oxaliplatin resistance in CD133+ gastric cancer stem cells by Promoting PARP1 mRNA stability which increases base excision repair pathway activity. METTTL3 enhances the stability of PARP1 by recruiting Poly [ADP-ribose] polymerase 1 (PARP1) to target the 3'-untranslated Region (3'-UTR) of PARP1 mRNA. | |||
Metastasis associated lung adenocarcinoma transcript 1 (MALAT1)
| Representative RIP-seq result supporting the interaction between the target gene and YTHDF1 | ||
| Cell Line | Hela | Homo sapiens |
| Regulation | logFC: 2.44E+00 | GSE63591 |
Cisplatin
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [14] | |||
| Responsed Disease | Non-small-cell lung carcinoma | ICD-11: 2C25.Y | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Hippo signaling pathway | hsa04390 | ||
| Cell Process | Metabolic | |||
| In-vitro Model | A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 |
| Calu-6 | Lung adenocarcinoma | Homo sapiens | CVCL_0236 | |
| NCI-H1299 | Lung large cell carcinoma | Homo sapiens | CVCL_0060 | |
| NCI-H520 | Lung squamous cell carcinoma | Homo sapiens | CVCL_1566 | |
| In-vivo Model | Mice were injected with 5 × 106 lung cancer cells with stably expression of relevant plasmids and randomly divided into two groups (five mice per group) after the diameter of the xenografted tumors had reached approximately 5 mm in diameter. Xenografted mice were then administrated with PBS or DDP (3 mg/kg per day) for three times a week, and tumor volume were measured every second day. | |||
| Response Summary | METTL3, YTHDF3, YTHDF1, and eIF3b directly promoted YAP translation through an interaction with the translation initiation machinery. METTL3 knockdown inhibits tumor growth and enhances sensitivity to DDP in vivo.m6A mRNA methylation initiated by METTL3 directly promotes YAP translation and increases YAP activity by regulating the Metastasis associated lung adenocarcinoma transcript 1 (MALAT1)-miR-1914-3p-YAP axis to induce Non-small cell lung cancer drug resistance and metastasis. | |||
Aldo-keto reductase family 1 member C1 (AKR1C1)
Cisplatin
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [7] | |||
| Responsed Disease | Non-small-cell lung carcinoma | ICD-11: 2C25.Y | ||
| Target Regulation | Down regulation | |||
| Pathway Response | Chemical carcinogenesis - reactive oxygen species | hsa05208 | ||
| Cell cycle | hsa04110 | |||
| Cell Process | Biological regulation | |||
| In-vitro Model | A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 |
| A549-DDP (Human lung adenocarcinoma is resistant to cisplatin) | ||||
| GLC-82 | Endocervical adenocarcinoma | Homo sapiens | CVCL_3371 | |
| NCI-H1299 | Lung large cell carcinoma | Homo sapiens | CVCL_0060 | |
| NCI-H1975 | Lung adenocarcinoma | Homo sapiens | CVCL_1511 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| NCI-H1650 | Minimally invasive lung adenocarcinoma | Homo sapiens | CVCL_1483 | |
| NCI-H838 | Lung adenocarcinoma | Homo sapiens | CVCL_1594 | |
| SPC-A1 | Endocervical adenocarcinoma | Homo sapiens | CVCL_6955 | |
| In-vivo Model | Mice were treated via nasal inhalation of adenovirus carrying Cre recombinase (5 × 106 p.f.u for Ad-Cre, Biowit Inc., Shenzhen, Guangdong), and were then killed at indicated times for gross inspection and histopathological examination. | |||
| Response Summary | YTHDF1 deficiency inhibits Non-small cell lung cancer cell proliferation and xenograft tumor formation through regulating the translational efficiency of CDK2, CDK4, p27, and cyclin D1, and that YTHDF1 depletion restrains de novo lung adenocarcinomas (ADC) progression. Mechanistic studies identified the Keap1-Nrf2-Aldo-keto reductase family 1 member C1 (AKR1C1) axis as the downstream mediator of YTHDF1. YTHDF1 high expression correlates with better clinical outcome, with its depletion rendering cancerous cells resistant to cisplatin (DDP) treatment. | |||
Autophagy protein 5 (ATG5)
Sorafenib
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [4] | |||
| Responsed Disease | Hepatocellular carcinoma | ICD-11: 2C12.02 | ||
| Target Regulation | Up regulation | |||
| Pathway Response | FoxO signaling pathway | hsa04068 | ||
| Autophagy | hsa04140 | |||
| Cell Process | Cell autophagy | |||
| Response Summary | METTL3 can sensitise hepatocellular carcinoma cells to sorafenib through stabilising forkhead box class O3 (FOXO3) in an m6A-dependent manner and translated by YTHDF1, thereby inhibiting the transcription of autophagy-related genes, including ATG3, Autophagy protein 5 (ATG5), ATG7, ATG12, and ATG16L1. | |||
Cellular tumor antigen p53 (TP53/p53)
Arsenite
[Phase 2]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [10] | |||
| Responsed Disease | Solid tumour/cancer | ICD-11: 2A00-2F9Z | ||
| Target Regulation | Down regulation | |||
| Pathway Response | p53 signaling pathway | hsa04115 | ||
| In-vitro Model | HaCaT | Normal | Homo sapiens | CVCL_0038 |
| Response Summary | METTL3 significantly decreased m6A level, restoring Cellular tumor antigen p53 (TP53/p53) activation and inhibiting cellular transformation phenotypes in the arsenite-transformed cells. m6A downregulated the expression of the positive p53 regulator, PRDM2, through the YTHDF2-promoted decay of PRDM2 mRNAs. m6A upregulated the expression of the negative p53 regulator, YY1 and MDM2 through YTHDF1-stimulated translation of YY1 and MDM2 mRNA. This study further sheds light on the mechanisms of arsenic carcinogenesis via RNA epigenetics. | |||
Cyclin-dependent kinase 4 (CDK4)
Cisplatin
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [7] | |||
| Responsed Disease | Non-small-cell lung carcinoma | ICD-11: 2C25.Y | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Chemical carcinogenesis - reactive oxygen species | hsa05208 | ||
| Cell cycle | hsa04110 | |||
| Cell Process | Biological regulation | |||
| In-vitro Model | A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 |
| A549-DDP (Human lung adenocarcinoma is resistant to cisplatin) | ||||
| GLC-82 | Endocervical adenocarcinoma | Homo sapiens | CVCL_3371 | |
| NCI-H1299 | Lung large cell carcinoma | Homo sapiens | CVCL_0060 | |
| NCI-H1975 | Lung adenocarcinoma | Homo sapiens | CVCL_1511 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| NCI-H1650 | Minimally invasive lung adenocarcinoma | Homo sapiens | CVCL_1483 | |
| NCI-H838 | Lung adenocarcinoma | Homo sapiens | CVCL_1594 | |
| SPC-A1 | Endocervical adenocarcinoma | Homo sapiens | CVCL_6955 | |
| In-vivo Model | Mice were treated via nasal inhalation of adenovirus carrying Cre recombinase (5 × 106 p.f.u for Ad-Cre, Biowit Inc., Shenzhen, Guangdong), and were then killed at indicated times for gross inspection and histopathological examination. | |||
| Response Summary | YTHDF1 deficiency inhibits Non-small cell lung cancer cell proliferation and xenograft tumor formation through regulating the translational efficiency of CDK2, Cyclin-dependent kinase 4 (CDK4), p27, and cyclin D1, and that YTHDF1 depletion restrains de novo lung adenocarcinomas (ADC) progression. Mechanistic studies identified the Keap1-Nrf2-AKR1C1 axis as the downstream mediator of YTHDF1. YTHDF1 high expression correlates with better clinical outcome, with its depletion rendering cancerous cells resistant to cisplatin (DDP) treatment. | |||
Cyclin-dependent kinase inhibitor 1B (CDKN1B/p27)
Cisplatin
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [7] | |||
| Responsed Disease | Non-small-cell lung carcinoma | ICD-11: 2C25.Y | ||
| Target Regulation | Down regulation | |||
| Pathway Response | Chemical carcinogenesis - reactive oxygen species | hsa05208 | ||
| Cell cycle | hsa04110 | |||
| Cell Process | Biological regulation | |||
| In-vitro Model | A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 |
| A549-DDP (Human lung adenocarcinoma is resistant to cisplatin) | ||||
| GLC-82 | Endocervical adenocarcinoma | Homo sapiens | CVCL_3371 | |
| NCI-H1299 | Lung large cell carcinoma | Homo sapiens | CVCL_0060 | |
| NCI-H1975 | Lung adenocarcinoma | Homo sapiens | CVCL_1511 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| NCI-H1650 | Minimally invasive lung adenocarcinoma | Homo sapiens | CVCL_1483 | |
| NCI-H838 | Lung adenocarcinoma | Homo sapiens | CVCL_1594 | |
| SPC-A1 | Endocervical adenocarcinoma | Homo sapiens | CVCL_6955 | |
| In-vivo Model | Mice were treated via nasal inhalation of adenovirus carrying Cre recombinase (5 × 106 p.f.u for Ad-Cre, Biowit Inc., Shenzhen, Guangdong), and were then killed at indicated times for gross inspection and histopathological examination. | |||
| Response Summary | YTHDF1 deficiency inhibits Non-small cell lung cancer cell proliferation and xenograft tumor formation through regulating the translational efficiency of CDK2, CDK4, Cyclin-dependent kinase inhibitor 1B (CDKN1B/p27), and cyclin D1, and that YTHDF1 depletion restrains de novo lung adenocarcinomas (ADC) progression. Mechanistic studies identified the Keap1-Nrf2-AKR1C1 axis as the downstream mediator of YTHDF1. YTHDF1 high expression correlates with better clinical outcome, with its depletion rendering cancerous cells resistant to cisplatin (DDP) treatment. | |||
G1/S-specific cyclin-D1 (CCND1)
Cisplatin
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [7] | |||
| Responsed Disease | Non-small-cell lung carcinoma | ICD-11: 2C25.Y | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Chemical carcinogenesis - reactive oxygen species | hsa05208 | ||
| Cell cycle | hsa04110 | |||
| Cell Process | Biological regulation | |||
| In-vitro Model | A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 |
| A549-DDP (Human lung adenocarcinoma is resistant to cisplatin) | ||||
| GLC-82 | Endocervical adenocarcinoma | Homo sapiens | CVCL_3371 | |
| NCI-H1299 | Lung large cell carcinoma | Homo sapiens | CVCL_0060 | |
| NCI-H1975 | Lung adenocarcinoma | Homo sapiens | CVCL_1511 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| NCI-H1650 | Minimally invasive lung adenocarcinoma | Homo sapiens | CVCL_1483 | |
| NCI-H838 | Lung adenocarcinoma | Homo sapiens | CVCL_1594 | |
| SPC-A1 | Endocervical adenocarcinoma | Homo sapiens | CVCL_6955 | |
| In-vivo Model | Mice were treated via nasal inhalation of adenovirus carrying Cre recombinase (5 × 106 p.f.u for Ad-Cre, Biowit Inc., Shenzhen, Guangdong), and were then killed at indicated times for gross inspection and histopathological examination. | |||
| Response Summary | YTHDF1 deficiency inhibits Non-small cell lung cancer cell proliferation and xenograft tumor formation through regulating the translational efficiency of CDK2, CDK4, p27, and G1/S-specific cyclin-D1 (CCND1), and that YTHDF1 depletion restrains de novo lung adenocarcinomas (ADC) progression. Mechanistic studies identified the Keap1-Nrf2-AKR1C1 axis as the downstream mediator of YTHDF1. YTHDF1 high expression correlates with better clinical outcome, with its depletion rendering cancerous cells resistant to cisplatin (DDP) treatment. | |||
Nuclear factor erythroid 2-related factor 2 (NFE2L2)
Cisplatin
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [7] | |||
| Responsed Disease | Non-small-cell lung carcinoma | ICD-11: 2C25.Y | ||
| Target Regulation | Down regulation | |||
| Pathway Response | Chemical carcinogenesis - reactive oxygen species | hsa05208 | ||
| Cell cycle | hsa04110 | |||
| Cell Process | Biological regulation | |||
| In-vitro Model | A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 |
| A549-DDP (Human lung adenocarcinoma is resistant to cisplatin) | ||||
| GLC-82 | Endocervical adenocarcinoma | Homo sapiens | CVCL_3371 | |
| NCI-H1299 | Lung large cell carcinoma | Homo sapiens | CVCL_0060 | |
| NCI-H1975 | Lung adenocarcinoma | Homo sapiens | CVCL_1511 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| NCI-H1650 | Minimally invasive lung adenocarcinoma | Homo sapiens | CVCL_1483 | |
| NCI-H838 | Lung adenocarcinoma | Homo sapiens | CVCL_1594 | |
| SPC-A1 | Endocervical adenocarcinoma | Homo sapiens | CVCL_6955 | |
| In-vivo Model | Mice were treated via nasal inhalation of adenovirus carrying Cre recombinase (5 × 106 p.f.u for Ad-Cre, Biowit Inc., Shenzhen, Guangdong), and were then killed at indicated times for gross inspection and histopathological examination. | |||
| Response Summary | YTHDF1 deficiency inhibits Non-small cell lung cancer cell proliferation and xenograft tumor formation through regulating the translational efficiency of CDK2, CDK4, p27, and cyclin D1, and that YTHDF1 depletion restrains de novo lung adenocarcinomas (ADC) progression. Mechanistic studies identified the Keap1-Nuclear factor erythroid 2-related factor 2 (NFE2L2)-AKR1C1 axis as the downstream mediator of YTHDF1. YTHDF1 high expression correlates with better clinical outcome, with its depletion rendering cancerous cells resistant to cisplatin (DDP) treatment. | |||
Staphylococcal nuclease domain-containing protein 1 (SND1)
Cisplatin
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [63] | |||
| Responsed Disease | Nasal-type natural killer/T-cell lymphoma | ICD-11: XH3400 | ||
| Target Regulation | Up regulation | |||
| In-vitro Model | NK-92 | Natural killer cell lymphoblastic leukemia/lymphoma | Homo sapiens | CVCL_2142 |
| YTS | Lymphoblastic leukemia/lymphoma | Homo sapiens | CVCL_D324 | |
| SNT-8 | Nasal type extranodal NK/T-cell lymphoma | Homo sapiens | CVCL_A677 | |
| SNK-6 | Nasal type extranodal NK/T-cell lymphoma | Homo sapiens | CVCL_A673 | |
| In-vivo Model | A total of 32 nude mice were randomly divided into four groups with eight mice in each group. Nude mice were subcutaneously injected with 2 × 106 cells (suspended in 100 μl of PBS) transfected with NC shRNA or SND1 shRNA into the back flank of mice. In DDP administration group, DDP (3 mg/kg per week for 2 weeks) was intraperitoneally injected into mice every 3 days. The control group received 200 μl of 0.1% DMSO. The tumor volume was measured every week for a total of 4 weeks. The mice were sacrificed after 4 weeks, then the tumors were harvested for weight measurement and other analysis. The tumor volume (mm3) was estimated with the formula (0.5 × length × width2). | |||
Ubiquitin-like modifier-activating enzyme ATG7 (ATG7)
Sorafenib
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [4] | |||
| Responsed Disease | Hepatocellular carcinoma | ICD-11: 2C12.02 | ||
| Target Regulation | Up regulation | |||
| Pathway Response | FoxO signaling pathway | hsa04068 | ||
| Autophagy | hsa04140 | |||
| Cell Process | Cell autophagy | |||
| Response Summary | METTL3 can sensitise hepatocellular carcinoma cells to sorafenib through stabilising forkhead box class O3 (FOXO3) in an m6A-dependent manner and translated by YTHDF1, thereby inhibiting the transcription of autophagy-related genes, including ATG3, ATG5, Ubiquitin-like modifier-activating enzyme ATG7 (ATG7), ATG12, and ATG16L1. | |||
Ubiquitin-like protein ATG12 (ATG12)
Sorafenib
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [4] | |||
| Responsed Disease | Hepatocellular carcinoma | ICD-11: 2C12.02 | ||
| Target Regulation | Up regulation | |||
| Pathway Response | FoxO signaling pathway | hsa04068 | ||
| Autophagy | hsa04140 | |||
| Cell Process | Cell autophagy | |||
| Response Summary | METTL3 can sensitise hepatocellular carcinoma cells to sorafenib through stabilising forkhead box class O3 (FOXO3) in an m6A-dependent manner and translated by YTHDF1, thereby inhibiting the transcription of autophagy-related genes, including ATG3, ATG5, ATG7, Ubiquitin-like protein ATG12 (ATG12), and ATG16L1. | |||
Ubiquitin-like-conjugating enzyme ATG3 (ATG3)
Sorafenib
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [4] | |||
| Responsed Disease | Hepatocellular carcinoma | ICD-11: 2C12.02 | ||
| Target Regulation | Up regulation | |||
| Pathway Response | FoxO signaling pathway | hsa04068 | ||
| Autophagy | hsa04140 | |||
| Cell Process | Cell autophagy | |||
| Response Summary | METTL3 can sensitise hepatocellular carcinoma cells to sorafenib through stabilising forkhead box class O3 (FOXO3) in an m6A-dependent manner and translated by YTHDF1, thereby inhibiting the transcription of autophagy-related genes, including Ubiquitin-like-conjugating enzyme ATG3 (ATG3), ATG5, ATG7, ATG12, and ATG16L1. | |||
hsa-miR-1914-3p
Cisplatin
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [14] | |||
| Responsed Disease | Non-small-cell lung carcinoma | ICD-11: 2C25.Y | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Hippo signaling pathway | hsa04390 | ||
| Cell Process | Metabolic | |||
| In-vitro Model | A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 |
| Calu-6 | Lung adenocarcinoma | Homo sapiens | CVCL_0236 | |
| NCI-H1299 | Lung large cell carcinoma | Homo sapiens | CVCL_0060 | |
| NCI-H520 | Lung squamous cell carcinoma | Homo sapiens | CVCL_1566 | |
| In-vivo Model | Mice were injected with 5 × 106 lung cancer cells with stably expression of relevant plasmids and randomly divided into two groups (five mice per group) after the diameter of the xenografted tumors had reached approximately 5 mm in diameter. Xenografted mice were then administrated with PBS or DDP (3 mg/kg per day) for three times a week, and tumor volume were measured every second day. | |||
| Response Summary | METTL3, YTHDF3, YTHDF1, and eIF3b directly promoted YAP translation through an interaction with the translation initiation machinery. METTL3 knockdown inhibits tumor growth and enhances sensitivity to DDP in vivo.m6A mRNA methylation initiated by METTL3 directly promotes YAP translation and increases YAP activity by regulating the MALAT1-hsa-miR-1914-3p-YAP axis to induce Non-small cell lung cancer drug resistance and metastasis. | |||
CREB-binding protein (CREBBP)
PT2385
[Investigative]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [75] | |||
| Target Regulation | Up regulation | |||
Dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit 2 (RPN2)
Cisplatin
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [79] | |||
| Responsed Disease | Bladder cancer | ICD-11: 2C94 | ||
| Pathway Response | PI3K-Akt signaling pathway | hsa04151 | ||
| mTOR signaling pathway | hsa04150 | |||
| In-vitro Model | 5637 | Bladder carcinoma | Homo sapiens | CVCL_0126 |
| T24 | Bladder carcinoma | Homo sapiens | CVCL_0554 | |
| UM-UC-3 | Bladder carcinoma | Homo sapiens | CVCL_1783 | |
G1/S-specific cyclin-E2 (CCNE2)
Tegaserod
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [81] | |||
| Responsed Disease | Acute myeloid leukaemia | ICD-11: 2A60 | ||
| Target Regulation | Up regulation | |||
| In-vitro Model | THP-1 | Childhood acute monocytic leukemia | Homo sapiens | CVCL_0006 |
| MV4-11 | Childhood acute monocytic leukemia | Homo sapiens | CVCL_0064 | |
Oxidized low-density lipoprotein receptor 1 (OLR1)
Temozolomide
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [95] | |||
| Responsed Disease | Glioma | ICD-11: 2A00.0 | ||
| Target Regulation | Up regulation | |||
| In-vitro Model | U-343MG | Glioblastoma | Homo sapiens | CVCL_S471 |
| U-251MG | Astrocytoma | Homo sapiens | CVCL_0021 | |
Ras-related protein Rab-27B (RAB27B)
Rucaparib
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [99] | |||
| Responsed Disease | Chronic myeloid leukaemia | ICD-11: 2B33.2 | ||
| Target Regulation | Up regulation | |||
| In-vitro Model | K-562 | Chronic myelogenous leukemia | Homo sapiens | CVCL_0004 |
| KCL-22 | Chronic myelogenous leukemia | Homo sapiens | CVCL_2091 | |
| In-vivo Model | K562 cells (1 × 106) were suspended in 100 μL of normal saline and the suspension was mixed with an equal volume of Matrigel. This mixture was subcutaneously injected into the right armpit of 4-week-old mice. Tumor size measurements were initiated on the day of inoculation. The tumor size was calculated using the formula: 0.5 × (long diameter) × (short diameter).2 When the tumor volume reached 100 mm3 (± 20%), rucaparib was given via intragastric administration at 50 mg/kg/d. | |||
Imatinib
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [99] | |||
| Responsed Disease | Chronic myeloid leukaemia | ICD-11: 2B33.2 | ||
| Target Regulation | Up regulation | |||
| In-vitro Model | K-562 | Chronic myelogenous leukemia | Homo sapiens | CVCL_0004 |
| KCL-22 | Chronic myelogenous leukemia | Homo sapiens | CVCL_2091 | |
| In-vivo Model | K562 cells (1 × 106) were suspended in 100 μL of normal saline and the suspension was mixed with an equal volume of Matrigel. This mixture was subcutaneously injected into the right armpit of 4-week-old mice. Tumor size measurements were initiated on the day of inoculation. The tumor size was calculated using the formula: 0.5 × (long diameter) × (short diameter).2 When the tumor volume reached 100 mm3 (± 20%), rucaparib was given via intragastric administration at 50 mg/kg/d. | |||
Receptor-interacting serine/threonine-protein kinase 4 (RIPK4)
Cisplatin
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [100] | |||
| Responsed Disease | Malignant mixed epithelial mesenchymal tumour of ovary | ICD-11: 2B5D.0 | ||
| Target Regulation | Up regulation | |||
| Pathway Response | NF-kappa B signaling pathway | hsa04064 | ||
| In-vitro Model | SK-OV-3 | Ovarian serous cystadenocarcinoma | Homo sapiens | CVCL_0532 |
| A2780 | Ovarian endometrioid adenocarcinoma | Homo sapiens | CVCL_0134 | |
| In-vivo Model | Animals were purchased from Changzhou Cavens Laboratory Animal Company (Changzhou, China) and maintained under specific pathogen-free (SPF) conditions. All animal experiments were performed in accordance with the guidelines of the Laboratory Animal Health Committee of Jiangsu University. Approximately 1 × 107 cells were injected subcutaneously into the dorsal surface of female BALB/c nude mice (n = 5 for each group, 4 weeks old, 15 ± 2 g). When the xenograft volume reached approximately 60 mm3 (after 1 week), DDP was intraperitoneally injected three times per week for 3 weeks. On Day 28, the mice were sacrificed and the tumors were frozen at -80 °C for follow-up experiments. | |||
Zinc finger protein RFP (TRIM27)
Cisplatin
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [107] | |||
| Responsed Disease | Colorectal cancer | ICD-11: 2B91 | ||
| Target Regulation | Up regulation | |||
| In-vitro Model | HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 |
| SW620 | Colon adenocarcinoma | Homo sapiens | CVCL_0547 | |
Unspecific Target Gene
Cisplatin
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [109] | |||
| Responsed Disease | Colon cancer | ICD-11: 2B90 | ||
| Pathway Response | Metabolic pathways | hsa01100 | ||
| Cell Process | Glutamine metabolism | |||
| In-vitro Model | LoVo | Colon adenocarcinoma | Homo sapiens | CVCL_0399 |
| HT-29 | Colon adenocarcinoma | Homo sapiens | CVCL_0320 | |
| DLD-1 | Colon adenocarcinoma | Homo sapiens | CVCL_0248 | |
| HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 | |
| SW480 | Colon adenocarcinoma | Homo sapiens | CVCL_0546 | |
| CRL-1790 (The normal colon epithelial cell line CRL-1790, were purchased from the American Type Culture Collection (ATCC).) | ||||
| In-vivo Model | Mice were injected subcutaneously with LoVo (1 × 106) cells, which were stably transfected with the control shRNA or YTHDF1 shRNA. | |||
| Response Summary | YTHDF1-promoted cisplatin resistance, contributing to overcoming chemoresistant colon cancers. | |||
Fluorouracil
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [110] | |||
| Responsed Disease | Colorectal cancer | ICD-11: 2B91 | ||
| Cell Process | Cells proliferation | |||
| Cells migration | ||||
| Cells invasion | ||||
| In-vitro Model | Caco-2 | Colon adenocarcinoma | Homo sapiens | CVCL_0025 |
| DLD-1 | Colon adenocarcinoma | Homo sapiens | CVCL_0248 | |
| HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 | |
| HT29 | Colon cancer | Mus musculus | CVCL_A8EZ | |
| KM12-SM | Colon carcinoma | Homo sapiens | CVCL_9548 | |
| LoVo | Colon adenocarcinoma | Homo sapiens | CVCL_0399 | |
| RKO | Colon carcinoma | Homo sapiens | CVCL_0504 | |
| SW480 | Colon adenocarcinoma | Homo sapiens | CVCL_0546 | |
| Response Summary | The knockdown of YTHDF1 resulted in the suppression of cancer proliferation and sensitization to the exposure of anticancer drugs such as fluorouracil and oxaliplatin. m6A reader Ythdf1 plays a significant role in colorectal cancer progression. An oncogenic transcription factor MYC was associated with YTHDF1 in both expression and chromatin immunoprecipitation data. | |||
Oxaliplatin
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [110] | |||
| Responsed Disease | Colorectal cancer | ICD-11: 2B91 | ||
| Cell Process | Cells proliferation | |||
| Cells migration | ||||
| Cells invasion | ||||
| In-vitro Model | Caco-2 | Colon adenocarcinoma | Homo sapiens | CVCL_0025 |
| DLD-1 | Colon adenocarcinoma | Homo sapiens | CVCL_0248 | |
| HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 | |
| HT29 | Colon cancer | Mus musculus | CVCL_A8EZ | |
| KM12-SM | Colon carcinoma | Homo sapiens | CVCL_9548 | |
| LoVo | Colon adenocarcinoma | Homo sapiens | CVCL_0399 | |
| RKO | Colon carcinoma | Homo sapiens | CVCL_0504 | |
| SW480 | Colon adenocarcinoma | Homo sapiens | CVCL_0546 | |
| Response Summary | The knockdown of YTHDF1 resulted in the suppression of cancer proliferation and sensitization to the exposure of anticancer drugs such as fluorouracil and oxaliplatin. m6A reader Ythdf1 plays a significant role in colorectal cancer progression. An oncogenic transcription factor MYC was associated with YTHDF1 in both expression and chromatin immunoprecipitation data. | |||
Gemcitabine
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [111] | |||
| Responsed Disease | Pancreatic cancer | ICD-11: 2C10 | ||
| Pathway Response | Adipocytokine signaling pathway | hsa04920 | ||
| Cell Process | Epithelial-mesenchymal transition | |||
| In-vitro Model | BxPC-3 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0186 |
| HDE-CT cell line (A normal human pancreatic cell line) | ||||
| MIA PaCa-2 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0428 | |
| Response Summary | Lasso regression identified a six-m6A-regulator-signature prognostic model (KIAA1429, HNRNPC, METTL3, YTHDF1, IGF2BP2, and IGF2BP3). Gene set enrichment analysis revealed m6A regulators (KIAA1429, HNRNPC, and IGF2BP2) were related to multiple biological behaviors in pancreatic cancer, including adipocytokine signaling, the well vs. poorly differentiated tumor pathway, tumor metastasis pathway, epithelial mesenchymal transition pathway, gemcitabine resistance pathway, and stemness pathway. | |||
Full List of Crosstalk(s) between m6A Modification and Epigenetic Regulation Related to This Regulator
RNA modification
m6A Target: Collagen alpha-1 (COL1A1)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT00054 | ||
| Epigenetic Regulator | Alpha-ketoglutarate-dependent dioxygenase alkB homolog 3 (ALKBH3) | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | m1A → m6A | |
| Disease | Keloid | |
| Crosstalk ID: M6ACROT00056 | ||
| Epigenetic Regulator | YTH domain-containing family protein 2 (YTHDF2) | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | m1A → m6A | |
| Disease | Keloid | |
m6A Target: Fibronectin (FN1)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT00055 | ||
| Epigenetic Regulator | Alpha-ketoglutarate-dependent dioxygenase alkB homolog 3 (ALKBH3) | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | m1A → m6A | |
| Disease | Keloid | |
| Drug | Ocriplasmin | |
| Crosstalk ID: M6ACROT00057 | ||
| Epigenetic Regulator | YTH domain-containing family protein 2 (YTHDF2) | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | m1A → m6A | |
| Disease | Keloid | |
| Drug | Ocriplasmin | |
m6A Target: Interferon-inducible protein 4 (ADAR1)
| In total 4 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT00060 | ||
| Epigenetic Regulator | Interferon-inducible protein 4 (ADAR1) | |
| Regulated Target | Tankyrase (TNKS) | |
| Crosstalk relationship | m6A → A-to-I | |
| Drug | PMID27841036-Compound-37 | |
| Crosstalk ID: M6ACROT00061 | ||
| Epigenetic Regulator | Interferon-inducible protein 4 (ADAR1) | |
| Regulated Target | Tyrosine-protein kinase EIF2AK2 (eIF2AK2/p68) | |
| Crosstalk relationship | m6A → A-to-I | |
| Drug | US9650366, 12 | |
| Crosstalk ID: M6ACROT00062 | ||
| Epigenetic Regulator | Interferon-inducible protein 4 (ADAR1) | |
| Regulated Target | Proteasome 20S subunit beta 2 (PSMB2) | |
| Crosstalk relationship | m6A → A-to-I | |
| Crosstalk ID: M6ACROT00064 | ||
| Epigenetic Regulator | Interferon-inducible protein 4 (ADAR1) | |
| Regulated Target | Cyclin-dependent kinase 2 (CDK2) | |
| Crosstalk relationship | m6A → A-to-I | |
| Disease | Glioblastoma | |
| Drug | PHA848125 | |
m6A Target: Rho GTPase activating protein 5 (ARHGAP5)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT00066 | ||
| Epigenetic Regulator | Interferon-inducible protein 4 (ADAR1) | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | A-to-I → m6A | |
| Disease | Breast cancer | |
m6A Target: Epidermal growth factor receptor (EGFR)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT00445 | ||
| Epigenetic Regulator | Double-stranded RNA-specific editase 1 (ADARB1) | |
| Regulated Target | MicroRNA 214 (MIR214) | |
| Crosstalk relationship | A-to-I → m6A | |
m6A Target: Transferrin receptor protein 1 (TFRC)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT00452 | ||
| Epigenetic Regulator | Double-stranded RNA-specific editase 1 (ADARB1) | |
| Regulated Target | MicroRNA 214 (MIR214) | |
| Crosstalk relationship | A-to-I → m6A | |
DNA modification
m6A Target: Collagen alpha-1 (COL1A1)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT02001 | ||
| Epigenetic Regulator | Cysteine methyltransferase DNMT3A (DNMT3A) | |
| Regulated Target | Suppressor of cytokine signaling 3 (SOCS3) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Hepatic fibrosis/cirrhosis | |
m6A Target: Collagen alpha-2(I) chain (COL1A2)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT02002 | ||
| Epigenetic Regulator | Cysteine methyltransferase DNMT3A (DNMT3A) | |
| Regulated Target | Suppressor of cytokine signaling 3 (SOCS3) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Hepatic fibrosis/cirrhosis | |
m6A Target: Collagen alpha-1 (III) chain (COL3A1)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT02003 | ||
| Epigenetic Regulator | Cysteine methyltransferase DNMT3A (DNMT3A) | |
| Regulated Target | Suppressor of cytokine signaling 3 (SOCS3) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Hepatic fibrosis/cirrhosis | |
m6A Target: Collagen alpha-2(V) chain (COL5A2)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT02004 | ||
| Epigenetic Regulator | Cysteine methyltransferase DNMT3A (DNMT3A) | |
| Regulated Target | Suppressor of cytokine signaling 3 (SOCS3) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Hepatic fibrosis/cirrhosis | |
m6A Target: Collagen alpha-2(VI) chain (COL6A2)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT02005 | ||
| Epigenetic Regulator | Cysteine methyltransferase DNMT3A (DNMT3A) | |
| Regulated Target | Suppressor of cytokine signaling 3 (SOCS3) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Hepatic fibrosis/cirrhosis | |
m6A Target: DNA (cytosine-5)-methyltransferase 3B (DNMT3B)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT02074 | ||
| Epigenetic Regulator | DNA (cytosine-5)-methyltransferase 3B (DNMT3B) | |
| Crosstalk relationship | m6A → DNA modification | |
| Disease | Gastric cancer | |
m6A Target: Cysteine methyltransferase DNMT3A (DNMT3A)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT02077 | ||
| Epigenetic Regulator | Cysteine methyltransferase DNMT3A (DNMT3A) | |
| Regulated Target | Alpha tubulin acetyltransferase 1 (ATAT1) | |
| Crosstalk relationship | m6A → DNA modification | |
| Disease | Alzheimer disease | |
m6A Target: Methyl-CpG-binding protein 2 (MECP2)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT02108 | ||
| Epigenetic Regulator | Methyl-CpG-binding protein 2 (MECP2) | |
| Regulated Target | Solute carrier family 31 member 1 (SLC31A1) | |
| Crosstalk relationship | m6A → DNA modification | |
| Disease | Vascular disorders of the liver | |
Histone modification
m6A Target: Protocadherin Fat 4 (FAT4)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03053 | ||
| Epigenetic Regulator | Histone Deacetylase (HDAC) | |
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Melanoma of uvea | |
| Crosstalk ID: M6ACROT03054 | ||
| Epigenetic Regulator | Histone Deacetylase (HDAC) | |
| Regulated Target | Histone H3 lysine 9 acetylation (H3K9Ac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Melanoma of uvea | |
m6A Target: PR domain zinc finger protein 15 (PRDM15)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03070 | ||
| Epigenetic Regulator | Histone acetyltransferase p300 (P300) | |
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Intrahepatic cholangiocarcinoma | |
m6A Target: Stearoyl-CoA desaturase (SCD)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03135 | ||
| Epigenetic Regulator | Lactate dehydrogenase A (LDHA) | |
| Regulated Target | Histone H3 lysine 18 lactylation (H3K18lac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Nonalcoholic fatty liver disease | |
m6A Target: Histone-lysine N-methyltransferase NSD2 (NSD2)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03210 | ||
| Epigenetic Regulator | Histone-lysine N-methyltransferase NSD2 (NSD2) | |
| Regulated Target | Histone H3 lysine 36 dimethylation (H3K36me2) | |
| Crosstalk relationship | m6A → Histone modification | |
| Disease | Diabetic nephropathy | |
| Crosstalk ID: M6ACROT03212 | ||
| Epigenetic Regulator | Histone-lysine N-methyltransferase NSD2 (NSD2) | |
| Regulated Target | Histone H3 lysine 36 trimethylation (H3K36me3) | |
| Crosstalk relationship | m6A → Histone modification | |
| Disease | Diabetic nephropathy | |
m6A Target: CREB-binding protein (CREBBP)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03218 | ||
| Epigenetic Regulator | CREB-binding protein (CREBBP) | |
| Crosstalk relationship | m6A → Histone modification | |
| Drug | BHBA | |
m6A Target: E3 ubiquitin-protein ligase RING2 (RNF2)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03220 | ||
| Epigenetic Regulator | E3 ubiquitin-protein ligase RING2 (RNF2) | |
| Regulated Target | Histone H2A lysine 119 ubiquitination (H2AK119ub) | |
| Crosstalk relationship | m6A → Histone modification | |
m6A Target: Dickkopf-related protein 3 (DKK3)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03354 | ||
| Epigenetic Regulator | Histone acetyltransferase p300 (P300) | |
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Diabetic nephropathy | |
| Drug | SETDB1-TTD-IN-1 | |
Non-coding RNA
m6A Target: Microtubule-associated protein tau (TAU)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05087 | ||
| Epigenetic Regulator | DPPA2 upstream binding RNA (DUBR) | |
| Regulated Target | YTH domain-containing family protein 1 (YTHDF1) | |
| Crosstalk relationship | ncRNA → m6A | |
m6A Target: Calmodulin-1 (CALM1)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05089 | ||
| Epigenetic Regulator | DPPA2 upstream binding RNA (DUBR) | |
| Regulated Target | YTH domain-containing family protein 1 (YTHDF1) | |
| Crosstalk relationship | ncRNA → m6A | |
m6A Target: Myc proto-oncogene protein (MYC)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05115 | ||
| Epigenetic Regulator | DLGAP1 antisense RNA 2 (DLGAP1-AS2) | |
| Regulated Target | YTH domain-containing family protein 1 (YTHDF1) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Non-small cell lung cancer | |
m6A Target: Hexokinase-2 (HK2)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05116 | ||
| Epigenetic Regulator | HLA complex P5 (HCP5) | |
| Regulated Target | YTH domain-containing family protein 1 (YTHDF1) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Esophageal Squamous Cell Carcinoma | |
m6A Target: Scavenger receptor class B member 1 (SCARB1)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05117 | ||
| Epigenetic Regulator | H19 imprinted maternally expressed transcript (H19) | |
| Regulated Target | YTH domain-containing family protein 1 (YTHDF1) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Gastric cancer | |
m6A Target: Interleukin-18 (IL18)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05191 | ||
| Epigenetic Regulator | hsa-miR-381 | |
| Regulated Target | YTH domain-containing family protein 1 (YTHDF1) | |
| Crosstalk relationship | ncRNA → m6A | |
m6A Target: Rho GTPase activating protein 5 (ARHGAP5)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05219 | ||
| Epigenetic Regulator | hsa-miR-532-5p | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Breast cancer | |
m6A Target: Zinc finger protein SNAI1 (SNAI1)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05230 | ||
| Epigenetic Regulator | hsa-miR-145-5p | |
| Regulated Target | YTH domain-containing family protein 1 (YTHDF1) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Gastric cancer | |
m6A Target: Transcription factor p65 (RELA)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05263 | ||
| Epigenetic Regulator | hsa-miR-421-3p | |
| Regulated Target | YTH domain-containing family protein 1 (YTHDF1) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Acute ischemic stroke | |
m6A Target: Catenin beta-1 (CTNNB1/Beta-catenin)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05267 | ||
| Epigenetic Regulator | hsa-miR-136-5p | |
| Regulated Target | YTH domain-containing family protein 1 (YTHDF1) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Colorectal cancer | |
| Crosstalk ID: M6ACROT05335 | ||
| Epigenetic Regulator | hsa-miR-376c | |
| Regulated Target | YTH domain-containing family protein 1 (YTHDF1) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Non-small cell lung cancer | |
m6A Target: Rho guanine nucleotide exchange factor 2 (ARHGEF2)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05268 | ||
| Epigenetic Regulator | hsa-miR-136-5p | |
| Regulated Target | YTH domain-containing family protein 1 (YTHDF1) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Colorectal cancer | |
m6A Target: Zinc finger protein RFP (TRIM27)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05269 | ||
| Epigenetic Regulator | hsa-miR-136-5p | |
| Regulated Target | YTH domain-containing family protein 1 (YTHDF1) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Colorectal cancer | |
| Drug | Cisplatin | |
m6A Target: Glucose-induced degradation protein 8 homolog (GID8)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05270 | ||
| Epigenetic Regulator | hsa-miR-136-5p | |
| Regulated Target | YTH domain-containing family protein 1 (YTHDF1) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Colorectal cancer | |
m6A Target: Pyruvate kinase PKM (PKM2/PKM)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05322 | ||
| Epigenetic Regulator | hsa-miR-16-5p | |
| Regulated Target | YTH domain-containing family protein 1 (YTHDF1) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Breast cancer | |
m6A Target: Oxidized low-density lipoprotein receptor 1 (OLR1)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05359 | ||
| Epigenetic Regulator | Circ_TTLL13 | |
| Regulated Target | YTH domain-containing family protein 1 (YTHDF1) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Brain cancer | |
| Drug | Temozolomide | |
m6A Target: Metastasis associated lung adenocarcinoma transcript 1 (MALAT1)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05405 | ||
| Epigenetic Regulator | Metastasis associated lung adenocarcinoma transcript 1 (MALAT1) | |
| Regulated Target | Transcriptional coactivator YAP1 (YAP1) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Non-small cell lung cancer | |
| Drug | Cisplatin | |
m6A Target: hsa-miR-1914-3p
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05408 | ||
| Epigenetic Regulator | hsa-miR-1914-3p | |
| Regulated Target | Transcriptional coactivator YAP1 (YAP1) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Non-small cell lung cancer | |
| Drug | Cisplatin | |
m6A Target: Testis associated oncogenic lncRNA (THORLNC)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05443 | ||
| Epigenetic Regulator | Testis associated oncogenic lncRNA (THORLNC) | |
| Crosstalk relationship | m6A → ncRNA | |
m6A Target: Long intergenic non-protein coding RNA 901 (LINC00901)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05672 | ||
| Epigenetic Regulator | Long intergenic non-protein coding RNA 901 (LINC00901) | |
| Regulated Target | Insulin like growth factor 2 mRNA binding protein 2 (IGF2BP2) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Pancreatic cancer | |
m6A Target: pre-miR-665
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05829 | ||
| Epigenetic Regulator | hsa-miR-665 | |
| Regulated Target | Homeobox protein DLX-3 (DLX3) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Structural developmental anomalies of teeth and periodontal tissues | |
Xenobiotics Compound(s) Regulating the m6A Methylation Regulator
| Compound Name | MO-I-500 | Investigative |
|---|---|---|
| Description |
Perturbed m6A signaling can be contributing toAlzheimer's disease pathogenesis, likely by compromising astrocyte bioenergetics. MO-I-500, a novel pharmacological inhibitor of FTO, can strongly reduce the adverse effects of STZ. STZ-treated astrocytes expressed significantly higher levels of m6A demethylase FTO and m6A reader YTHDF1.
|
[116] |
| Compound Name | AdoHcy | Investigative |
| Synonyms |
S-Adenosyl-L-homocysteine; S-adenosylhomocysteine; S-adenosyl-L-homocysteine; 979-92-0; AdoHcy; S-(5'-adenosyl)-L-homocysteine; adenosylhomocysteine; Formycinylhomocysteine; Adenosyl-L-homocysteine; S-(5'-deoxyadenosin-5'-yl)-L-homocysteine; 2-S-adenosyl-L-homocysteine; 5'-Deoxy-S-adenosyl-L-homocysteine; S-adenosyl-homocysteine; S-Adenosyl Homocysteine; L-S-Adenosylhomocysteine; L-Homocysteine, S-(5'-deoxyadenosin-5'-yl)-; adenosylhomo-cys; adenosyl-homo-cys; UNII-8K31Q2S66S; (S)-5'-(S)-(3-Amino-3-carboxypropyl)-5'-thioadenosine; BRN 5166233; SAH; S-Adenosylhomocysteine; S-Adenosyl-L-Homocysteine
Click to Show/Hide
|
|
| External link | ||
| Description |
Mettl3 inhibitor, S-adenosylhomocysteine promoted the apoptosis and autophagy of chondrocytes with inflammation in vitro and aggravated the degeneration of chondrocytes and subchondral bone in monosodium iodoacetate (MIA) inducedtemporomandibular joint osteoarthritis mice in vivo. Bcl2 protein interacted with Beclin1 protein in chondrocytes induced by TNF-alpha stimulation. Mettl3 inhibits the apoptosis and autophagy of chondrocytes in inflammation through m6A/Ythdf1/Bcl2 signal axis which provides promising therapeutic strategy fortemporomandibular joint osteoarthritis.
|
[25] |
| Compound Name | Olean-28,13Beta-lactam(B28) | Investigative |
| Description |
B28 disrupts YTDFH1-GLS1 axis to induce ROS-dependent cell bioenergetics crisis and cell death which finally suppress PAAD cell growth, indicating that this synthesized olean-28,13Beta-lactam maybe a potent agent for PAAD intervention.
|
[119] |
References