m6A Target Gene Information
General Information of the m6A Target Gene (ID: M6ATAR00222)
Full List of m6A Methylation Regulator of This Target Gene and Corresponding Disease/Drug Response(s)
CTNNB1
can be regulated by the following regulator(s), and cause disease/drug response(s). You can browse detail information of regulator(s) or disease/drug response(s).
Browse Regulator
Browse Disease
Browse Drug
Methyltransferase-like 3 (METTL3) [WRITER]
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | Embryonic stem cells | Mus musculus |
|
Treatment: METTL3 knockout mESCs
Control: Wild type mESCs
|
GSE156481 | |
| Regulation |
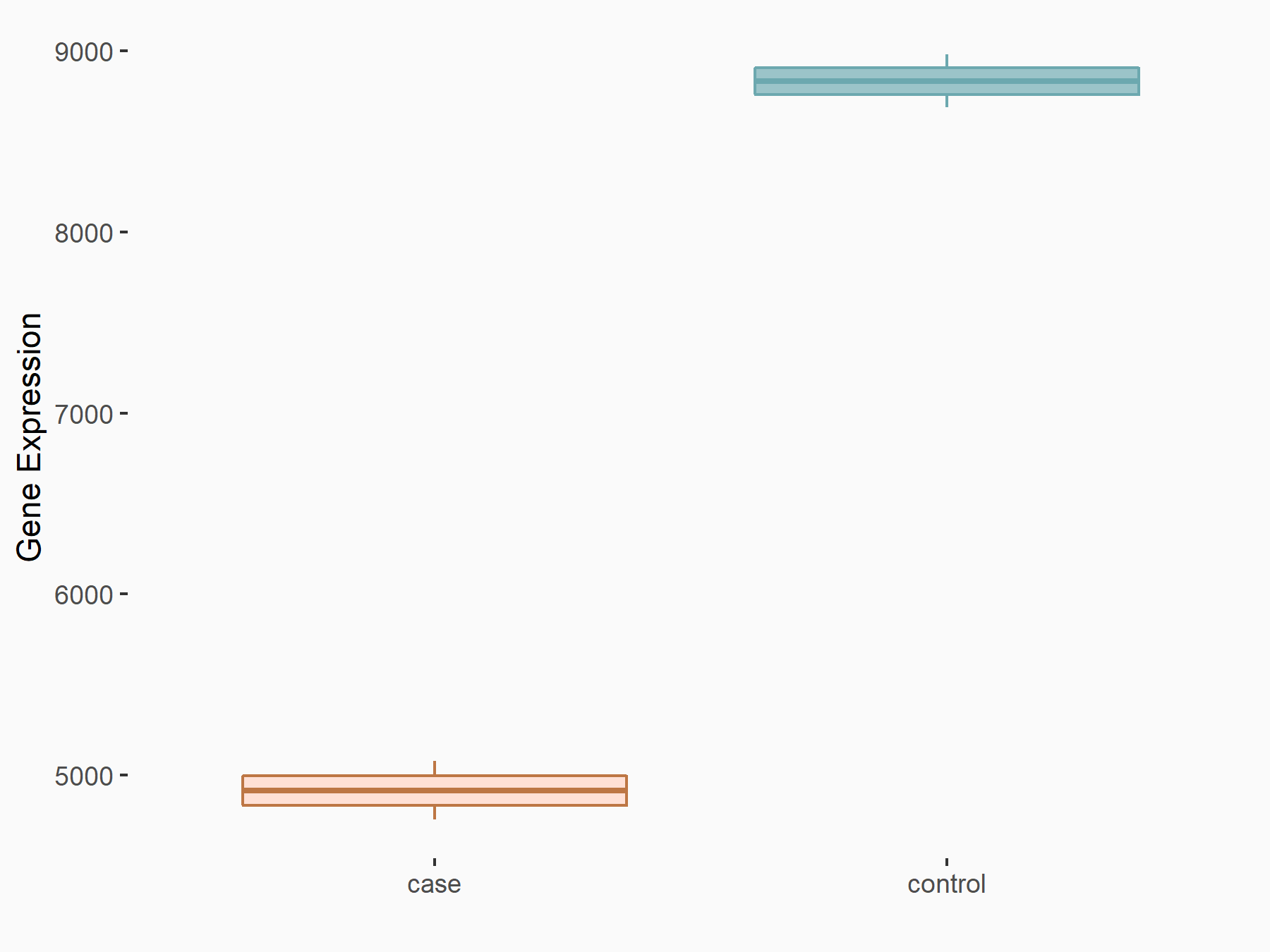  |
logFC: -8.46E-01 p-value: 2.97E-40 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between CTNNB1 and the regulator | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 1.37E+00 | GSE60213 |
| In total 3 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [1] | |||
| Response Summary | METTL3 activated the luciferase activity of TOPflash (a reporter for beta-catenin/TCF signaling), and downregulation of METTL3 inhibited the expression of Catenin beta-1 (CTNNB1/Beta-catenin)/TCF target genes vimentin and N-cadherin, which are two regulators of epithelial-mesenchymal transition. METTL3 silencing decreased the m6A methylation and total mRNA levels of Tankyrase, a negative regulator of axin. METTL3 is a therapeutic target for NPC. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Nasopharyngeal carcinoma | ICD-11: 2B6B | ||
| Pathway Response | Wnt signaling pathway | hsa04310 | ||
| Cell Process | Epithelial-mesenchymal transition | |||
| In-vitro Model | Neural progenitor cells (NPCs) (The progenitor cells of the CNS) | |||
| NP69 (A human immortalized nasopharyngeal epithelial) | ||||
| HNE-2 | Nasopharyngeal carcinoma | Homo sapiens | CVCL_FA07 | |
| HNE-1 | Nasopharyngeal carcinoma | Homo sapiens | CVCL_0308 | |
| CNE-2 | Nasopharyngeal carcinoma | Homo sapiens | CVCL_6889 | |
| CNE-1 | Normal | Homo sapiens | CVCL_6888 | |
| In-vivo Model | 1 × 105 HNE2 cells (with or without METTL3 knockdown) were labeled with luciferase gene and injected into the tail vein of the nude mice. | |||
| Experiment 2 Reporting the m6A Methylation Regulator of This Target Gene | [2] | |||
| Response Summary | METTL3 is significantly up-regulated in Hepatoblastoma(HB) and promotes HB development.m6A mRNA methylation contributes significantly to regulate the Wnt/beta-catenin pathway. Reduced m6A methylation can lead to a decrease in expression and stability of the Catenin beta-1 (CTNNB1/Beta-catenin). | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Hepatoblastoma | ICD-11: 2C12.01 | ||
| Pathway Response | Wnt signaling pathway | hsa04310 | ||
| Cell Process | RNA stability | |||
| In-vitro Model | HEK293T | Normal | Homo sapiens | CVCL_0063 |
| Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 | |
| L-02 | Endocervical adenocarcinoma | Homo sapiens | CVCL_6926 | |
| QSG-7701 | Endocervical adenocarcinoma | Homo sapiens | CVCL_6944 | |
| In-vivo Model | 5 × 106 cells were subcutaneously injected into the left or right flank of each mouse. | |||
| Experiment 3 Reporting the m6A Methylation Regulator of This Target Gene | [3] | |||
| Response Summary | SAM not only played a compensatory role, but also led to m6A modification changes in neural tube development and regulation. Ethionine affected m6A modification by reducing SAM metabolism. METTL3 is enriched in HT-22 cells, and METTL3 knockdown reduces cell proliferation and increases apoptosis through suppressing Wnt/Catenin beta-1 (CTNNB1/Beta-catenin) signaling pathway. Overexpression of ALKBH5 can only inhibit cell proliferation, but cannot promote cell apoptosis. | |||
| Target Regulation | Down regulation | |||
| Responsed Disease | Neural tube defect | ICD-11: LA02.Z | ||
| Pathway Response | Wnt signaling pathway | hsa04310 | ||
| Cell Process | Cell apoptosis | |||
| In-vitro Model | HT22 | Normal | Mus musculus | CVCL_0321 |
| In-vivo Model | The mice were maintained on a 12-h light/dark cycle (lights on from 8:00 a.m. to 8:00 p.m.). On day 7.5 of pregnancy (E7.5), ethionine (Sigma-Aldrich, USA) was intraperitoneally injected only once at a dose of 500 mg/kg to establish the NTDs embryo model. And SAM (MedChemExpress, USA) was intraperitoneally injected only once at a dose of 30 mg/kg. The same dose was intraperitoneally injected to the pregnant mice for control group. | |||
Methyltransferase-like 14 (METTL14) [WRITER]
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL14 | ||
| Cell Line | BMDM | Mus musculus |
|
Treatment: METTL14 knockout mice BMDM
Control: Wild type mice BMDM
|
GSE153512 | |
| Regulation |
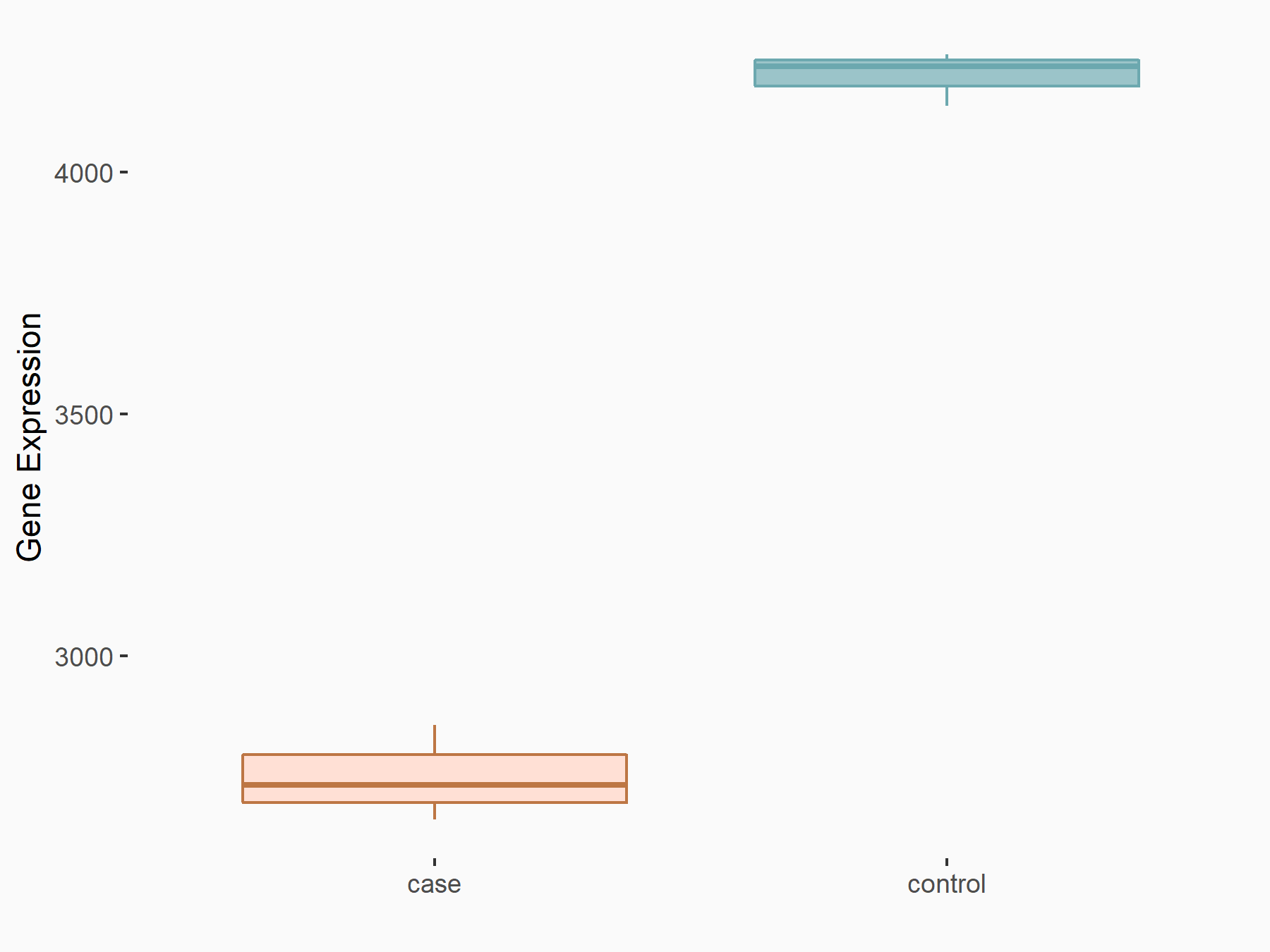  |
logFC: -6.10E-01 p-value: 2.52E-16 |
| More Results | Click to View More RNA-seq Results | |
| In total 4 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [4] | |||
| Response Summary | m6A-hypomethylation regulated FGFR4 phosphorylates GSK-3beta and activates Catenin beta-1 (CTNNB1/Beta-catenin)/TCF4-SLC7A11/FPN1 signaling to drive anti-HER2 resistance. Knockdown of METTL14 significantly increased the expression level of FGFR4 in HER2-positive breast cancer cells. FGFR4 reduced the sensitivity of HER2-positive breast cancer to trastuzumab plus pertuzumab or tucatinib. These results pinpoint a mechanism of anti-HER2 resistance and provide a strategy for overcoming resistance via FGFR4 inhibition in recalcitrant HER2-positive breast cancer. | |||
| Target Regulation | Down regulation | |||
| Responsed Disease | Breast cancer | ICD-11: 2C60 | ||
| Responsed Drug | Pertuzumab | Approved | ||
| Pathway Response | Wnt signaling pathway | hsa04310 | ||
| Cell Process | Glutathione synthesis | |||
| In-vitro Model | ZR-75-1 | Invasive breast carcinoma | Homo sapiens | CVCL_0588 |
| T-47D | Invasive breast carcinoma | Homo sapiens | CVCL_0553 | |
| SUM-159 (A mesenchymal triple-negative breast cancer cell line) | ||||
| SK-BR-3 | Breast adenocarcinoma | Homo sapiens | CVCL_0033 | |
| MDA-MB-468 | Breast adenocarcinoma | Homo sapiens | CVCL_0419 | |
| MDA-MB-453 | Breast adenocarcinoma | Homo sapiens | CVCL_0418 | |
| MDA-MB-361 | Breast adenocarcinoma | Homo sapiens | CVCL_0620 | |
| MDA-MB-231 | Breast adenocarcinoma | Homo sapiens | CVCL_0062 | |
| MCF-7 | Invasive breast carcinoma | Homo sapiens | CVCL_0031 | |
| MCF-10A | Normal | Homo sapiens | CVCL_0598 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| BT-549 | Invasive breast carcinoma | Homo sapiens | CVCL_1092 | |
| BT-474 | Invasive breast carcinoma | Homo sapiens | CVCL_0179 | |
| AU565 | Breast adenocarcinoma | Homo sapiens | CVCL_1074 | |
| In-vivo Model | Luciferase-labeled rSKBR3 and MDA-MB-361 cells (1 × 107 cells) mixed with 1:1 Matrigel (Corning, 356237) were subcutaneously injected into the fat pads of mice. After a tumor was palpable, the mice were randomized into four groups (five mice per group), and they were treated with vehicle, trastuzumab (20 mg/kg, intraperitoneal administration), roblitinib (30 mg/kg, oral administration), or a combination of both drugs. | |||
| Experiment 2 Reporting the m6A Methylation Regulator of This Target Gene | [4] | |||
| Response Summary | m6A-hypomethylation regulated FGFR4 phosphorylates GSK-3beta and activates Catenin beta-1 (CTNNB1/Beta-catenin)/TCF4-SLC7A11/FPN1 signaling to drive anti-HER2 resistance. Knockdown of METTL14 significantly increased the expression level of FGFR4 in HER2-positive breast cancer cells. FGFR4 reduced the sensitivity of HER2-positive breast cancer to trastuzumab plus pertuzumab or tucatinib. These results pinpoint a mechanism of anti-HER2 resistance and provide a strategy for overcoming resistance via FGFR4 inhibition in recalcitrant HER2-positive breast cancer. | |||
| Target Regulation | Down regulation | |||
| Responsed Disease | Breast cancer | ICD-11: 2C60 | ||
| Responsed Drug | Trastuzumab | Approved | ||
| Pathway Response | Wnt signaling pathway | hsa04310 | ||
| Cell Process | Glutathione synthesis | |||
| In-vitro Model | ZR-75-1 | Invasive breast carcinoma | Homo sapiens | CVCL_0588 |
| T-47D | Invasive breast carcinoma | Homo sapiens | CVCL_0553 | |
| SUM-159 (A mesenchymal triple-negative breast cancer cell line) | ||||
| SK-BR-3 | Breast adenocarcinoma | Homo sapiens | CVCL_0033 | |
| MDA-MB-468 | Breast adenocarcinoma | Homo sapiens | CVCL_0419 | |
| MDA-MB-453 | Breast adenocarcinoma | Homo sapiens | CVCL_0418 | |
| MDA-MB-361 | Breast adenocarcinoma | Homo sapiens | CVCL_0620 | |
| MDA-MB-231 | Breast adenocarcinoma | Homo sapiens | CVCL_0062 | |
| MCF-7 | Invasive breast carcinoma | Homo sapiens | CVCL_0031 | |
| MCF-10A | Normal | Homo sapiens | CVCL_0598 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| BT-549 | Invasive breast carcinoma | Homo sapiens | CVCL_1092 | |
| BT-474 | Invasive breast carcinoma | Homo sapiens | CVCL_0179 | |
| AU565 | Breast adenocarcinoma | Homo sapiens | CVCL_1074 | |
| In-vivo Model | Luciferase-labeled rSKBR3 and MDA-MB-361 cells (1 × 107 cells) mixed with 1:1 Matrigel (Corning, 356237) were subcutaneously injected into the fat pads of mice. After a tumor was palpable, the mice were randomized into four groups (five mice per group), and they were treated with vehicle, trastuzumab (20 mg/kg, intraperitoneal administration), roblitinib (30 mg/kg, oral administration), or a combination of both drugs. | |||
| Experiment 3 Reporting the m6A Methylation Regulator of This Target Gene | [4] | |||
| Response Summary | m6A-hypomethylation regulated FGFR4 phosphorylates GSK-3beta and activates Catenin beta-1 (CTNNB1/Beta-catenin)/TCF4-SLC7A11/FPN1 signaling to drive anti-HER2 resistance. Knockdown of METTL14 significantly increased the expression level of FGFR4 in HER2-positive breast cancer cells. FGFR4 reduced the sensitivity of HER2-positive breast cancer to trastuzumab plus pertuzumab or tucatinib. These results pinpoint a mechanism of anti-HER2 resistance and provide a strategy for overcoming resistance via FGFR4 inhibition in recalcitrant HER2-positive breast cancer. | |||
| Target Regulation | Down regulation | |||
| Responsed Disease | Breast cancer | ICD-11: 2C60 | ||
| Responsed Drug | Tucatinib | Approved | ||
| Pathway Response | Wnt signaling pathway | hsa04310 | ||
| Cell Process | Glutathione synthesis | |||
| In-vitro Model | ZR-75-1 | Invasive breast carcinoma | Homo sapiens | CVCL_0588 |
| T-47D | Invasive breast carcinoma | Homo sapiens | CVCL_0553 | |
| SUM-159 (A mesenchymal triple-negative breast cancer cell line) | ||||
| SK-BR-3 | Breast adenocarcinoma | Homo sapiens | CVCL_0033 | |
| MDA-MB-468 | Breast adenocarcinoma | Homo sapiens | CVCL_0419 | |
| MDA-MB-453 | Breast adenocarcinoma | Homo sapiens | CVCL_0418 | |
| MDA-MB-361 | Breast adenocarcinoma | Homo sapiens | CVCL_0620 | |
| MDA-MB-231 | Breast adenocarcinoma | Homo sapiens | CVCL_0062 | |
| MCF-7 | Invasive breast carcinoma | Homo sapiens | CVCL_0031 | |
| MCF-10A | Normal | Homo sapiens | CVCL_0598 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| BT-549 | Invasive breast carcinoma | Homo sapiens | CVCL_1092 | |
| BT-474 | Invasive breast carcinoma | Homo sapiens | CVCL_0179 | |
| AU565 | Breast adenocarcinoma | Homo sapiens | CVCL_1074 | |
| In-vivo Model | Luciferase-labeled rSKBR3 and MDA-MB-361 cells (1 × 107 cells) mixed with 1:1 Matrigel (Corning, 356237) were subcutaneously injected into the fat pads of mice. After a tumor was palpable, the mice were randomized into four groups (five mice per group), and they were treated with vehicle, trastuzumab (20 mg/kg, intraperitoneal administration), roblitinib (30 mg/kg, oral administration), or a combination of both drugs. | |||
| Experiment 4 Reporting the m6A Methylation Regulator of This Target Gene | [5] | |||
| Response Summary | Mettl14 resulted in enhanced levels of Wnt1 m6A modification and Wnt1 protein but not its transcript level.Furthermore, Mettl14 overexpression blocked I/R-induced downregulation of Wnt1 and Catenin beta-1 (CTNNB1/Beta-catenin) proteins, whereas Mettl14 hearts exhibited the opposite results. Mettl14 attenuates cardiac I/R injury by activating Wnt/Bete-catenin in an m6A-dependent manner, providing a novel therapeutic target for ischemic heart disease. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Ischemic heart disease | ICD-11: BA40-BA6Z | ||
| Pathway Response | Wnt signaling pathway | hsa04310 | ||
| In-vitro Model | Neonatal rat ventricular cardiomyocytes (Primary myocyte cells) | |||
| In-vivo Model | C57BL/6 mouse hearts were subjected to ischemia/reperfusion (I/R) in vivo as described previously (Bock-Marquette et al., 2004; Song et al., 2015; Brocard et al., 2017). I/R injury in mice was induced by 45-min ischemia, followed by 7-day and 4-week reperfusion in a loss-of-function study (Figure 1) and gain-of-function study (Figure 2), respectively. In brief, mice were anesthetized with 2% avertin (0.1 ml/10g body weight; Sigma-Aldrich Corporation, United States) through intraperitoneal injection. To generate I/R injury, the left anterior descending coronary artery (LAD) was ligated with 7-0 nylon for 45 min and then was removed. For the sham group, a suture was passed under the LAD but without ligation. According to the experimental requirements, at different time points of cardiac I/R, the mice were anesthetized for assessing heart function by echocardiographic measurement. All the mice survived during the process of I/R injury after the operation. | |||
RNA demethylase ALKBH5 (ALKBH5) [ERASER]
| Representative RNA-seq result indicating the expression of this target gene regulated by ALKBH5 | ||
| Cell Line | 143B cell line | Homo sapiens |
|
Treatment: siALKBH5 transfected 143B cells
Control: siControl 143B cells
|
GSE154528 | |
| Regulation |
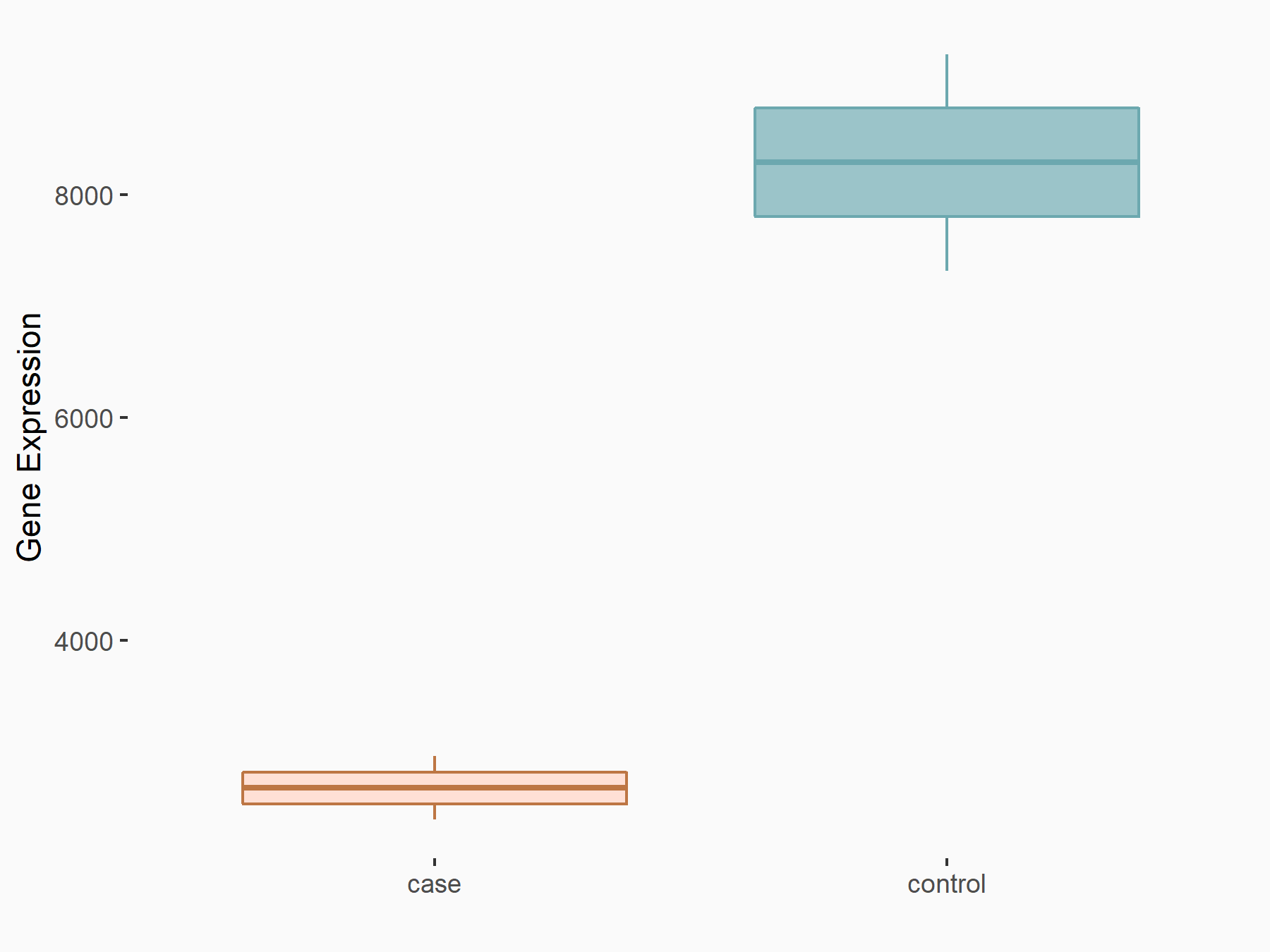 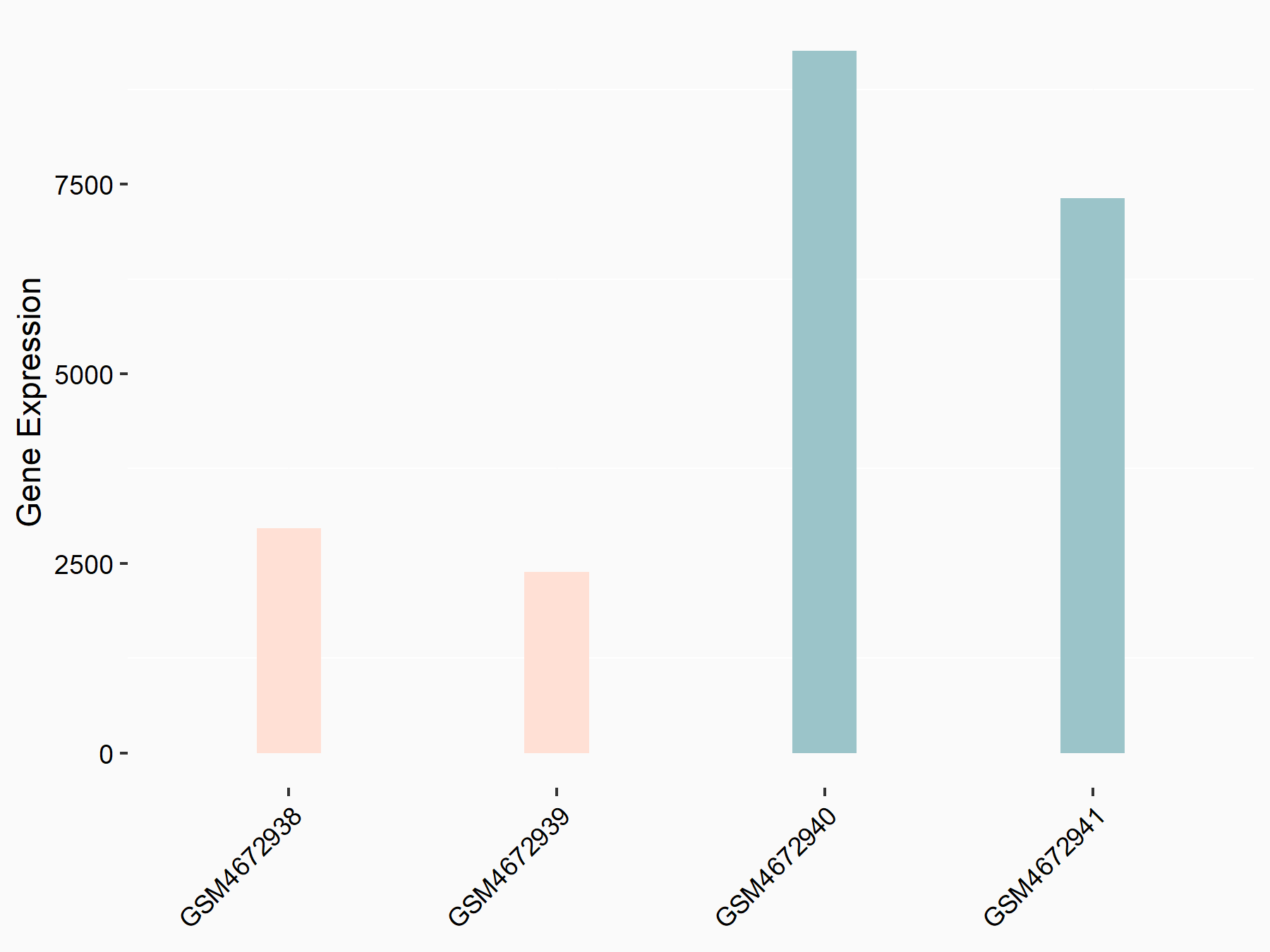 |
logFC: -1.63E+00 p-value: 8.42E-15 |
| More Results | Click to View More RNA-seq Results | |
| In total 1 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [3] | |||
| Response Summary | SAM not only played a compensatory role, but also led to m6A modification changes in neural tube development and regulation. Ethionine affected m6A modification by reducing SAM metabolism. METTL3 is enriched in HT-22 cells, and METTL3 knockdown reduces cell proliferation and increases apoptosis through suppressing Wnt/Catenin beta-1 (CTNNB1/Beta-catenin) signaling pathway. Overexpression of ALKBH5 can only inhibit cell proliferation, but cannot promote cell apoptosis. | |||
| Responsed Disease | Neural tube defect | ICD-11: LA02.Z | ||
| Pathway Response | Wnt signaling pathway | hsa04310 | ||
| Cell Process | Cell apoptosis | |||
| In-vitro Model | HT22 | Normal | Mus musculus | CVCL_0321 |
| In-vivo Model | The mice were maintained on a 12-h light/dark cycle (lights on from 8:00 a.m. to 8:00 p.m.). On day 7.5 of pregnancy (E7.5), ethionine (Sigma-Aldrich, USA) was intraperitoneally injected only once at a dose of 500 mg/kg to establish the NTDs embryo model. And SAM (MedChemExpress, USA) was intraperitoneally injected only once at a dose of 30 mg/kg. The same dose was intraperitoneally injected to the pregnant mice for control group. | |||
YTH domain-containing family protein 1 (YTHDF1) [READER]
| Representative RIP-seq result supporting the interaction between CTNNB1 and the regulator | ||
| Cell Line | Hela | Homo sapiens |
| Regulation | logFC: 2.05E+00 | GSE63591 |
| In total 2 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [6] | |||
| Response Summary | Silencing YTHDF1 significantly inhibited Wnt/Catenin beta-1 (CTNNB1/Beta-catenin) pathway activity in Colorectal cancer cells. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Colorectal cancer | ICD-11: 2B91 | ||
| Pathway Response | Wnt signaling pathway | hsa04310 | ||
| Signaling pathways regulating pluripotency of stem cells | hsa04550 | |||
| Cell Process | Cell Tumorigenicity | |||
| In-vitro Model | NCM460 | Normal | Homo sapiens | CVCL_0460 |
| SW480 | Colon adenocarcinoma | Homo sapiens | CVCL_0546 | |
| SW620 | Colon adenocarcinoma | Homo sapiens | CVCL_0547 | |
| In-vivo Model | 1 × 105 parental HT29-shNC cells, HT29-shYTHDF1 cells, HT29-shNC colonospheres, and HT29-shYTHDF1 colonospheres were inoculated subcutaneously into the left inguinal folds of the nude mice. | |||
| Experiment 2 Reporting the m6A Methylation Regulator of This Target Gene | [7] | |||
| Response Summary | ECs transmitted miR-376c into NSCLC cells through Evs, and inhibited the intracellular YTHDF1 expression and the Wnt/Catenin beta-1 (CTNNB1/Beta-catenin) pathway activation. YTHDF1 overexpression reversed the inhibitory role of miR-376c released by EC-Evs in non-small cell lung cancer cells. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Non-small-cell lung carcinoma | ICD-11: 2C25.Y | ||
| Cell Process | Cell proliferation | |||
| Cell invasion | ||||
| Cell migration | ||||
| Cell apoptosis | ||||
| In-vitro Model | A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 |
| NCI-1650 (Non-Small Cell Lung Cancer Cells) | ||||
| NCI-H1299 | Lung large cell carcinoma | Homo sapiens | CVCL_0060 | |
| NCI-H358 | Minimally invasive lung adenocarcinoma | Homo sapiens | CVCL_1559 | |
YTH domain-containing family protein 3 (YTHDF3) [READER]
| Representative RIP-seq result supporting the interaction between CTNNB1 and the regulator | ||
| Cell Line | Hela | Homo sapiens |
| Regulation | logFC: 1.43E+00 | GSE86214 |
| In total 1 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [8] | |||
| Response Summary | YTHDF3 enhances Catenin beta-1 (CTNNB1/Beta-catenin) translation through recognizing and binding the m6A peaks on CTNNB1 mRNA.m6A reading protein YTHDF3 promotes the translation of the target transcript CTNNB1, contributing to ocular melanoma propagation and migration through m6A methylation. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Melanoma of uvea | ICD-11: 2D0Y | ||
Fat mass and obesity-associated protein (FTO) [ERASER]
| In total 2 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [9] | |||
| Response Summary | FTO expression was significantly upregulated in HNSCC datasets and tissues. FTO expression was significantly correlated with Catenin beta-1 (CTNNB1/Beta-catenin) expression. Moreover, it exerted a tumorigenic effect by increasing CTNNB1 expression in an m6A-dependent manner. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Head and neck squamous carcinoma | ICD-11: 2B6E | ||
| Pathway Response | Adherens junction | hsa04520 | ||
| Cell Process | Cell proliferation and migration | |||
| In-vitro Model | CAL-27 | Tongue squamous cell carcinoma | Homo sapiens | CVCL_1107 |
| FaDu | Hypopharyngeal squamous cell carcinoma | Homo sapiens | CVCL_1218 | |
| Tu 686 | Laryngeal squamous cell carcinoma | Homo sapiens | CVCL_4916 | |
| HN-6 | Tongue squamous cell carcinoma | Homo sapiens | CVCL_8129 | |
| HEp-2 | Endocervical adenocarcinoma | Homo sapiens | CVCL_1906 | |
| Experiment 2 Reporting the m6A Methylation Regulator of This Target Gene | [10] | |||
| Response Summary | The mRNA level of FTO is elevated in cervical squamous cell carcinoma (CSCC) tissues when compared with respective adjacent normal tissues.FTO enhances the chemo-radiotherapy resistance both in vitro and in vivo through regulating expression of Catenin beta-1 (CTNNB1/Beta-catenin) by reducing m6A levels in its mRNA transcripts and in turn increases excision repair cross-complementation group 1 (ERCC1) activity. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Cervical squamous cell carcinoma | ICD-11: 2E66.2 | ||
| Cell Process | RNA decay | |||
| In-vitro Model | C-33 A | Cervical squamous cell carcinoma | Homo sapiens | CVCL_1094 |
| SiHa | Cervical squamous cell carcinoma | Homo sapiens | CVCL_0032 | |
| In-vivo Model | Either SiHa cells or FTO overexpressed SiHa cells were injected subcutaneously into the right flanks of 4- to 6-week-old female athymic nude mice (CAS, Beijing, China). The mice were divided into four groups (N = 6). After transplantation, tumor size was measured by caliper every other day. Tumor volume was calculated using the formula: volume = (length × width2)/2. When the tumor volumes reached 50 mm3, the animals were treated with intraperitoneal injection of cisplatin (3 mg/kg) every other day for seven times and local irradiation of 8 Gy one time. | |||
Nasopharyngeal carcinoma [ICD-11: 2B6B]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [1] | |||
| Response Summary | METTL3 activated the luciferase activity of TOPflash (a reporter for beta-catenin/TCF signaling), and downregulation of METTL3 inhibited the expression of Catenin beta-1 (CTNNB1/Beta-catenin)/TCF target genes vimentin and N-cadherin, which are two regulators of epithelial-mesenchymal transition. METTL3 silencing decreased the m6A methylation and total mRNA levels of Tankyrase, a negative regulator of axin. METTL3 is a therapeutic target for NPC. | |||
| Responsed Disease | Nasopharyngeal carcinoma [ICD-11: 2B6B] | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Wnt signaling pathway | hsa04310 | ||
| Cell Process | Epithelial-mesenchymal transition | |||
| In-vitro Model | Neural progenitor cells (NPCs) (The progenitor cells of the CNS) | |||
| NP69 (A human immortalized nasopharyngeal epithelial) | ||||
| HNE-2 | Nasopharyngeal carcinoma | Homo sapiens | CVCL_FA07 | |
| HNE-1 | Nasopharyngeal carcinoma | Homo sapiens | CVCL_0308 | |
| CNE-2 | Nasopharyngeal carcinoma | Homo sapiens | CVCL_6889 | |
| CNE-1 | Normal | Homo sapiens | CVCL_6888 | |
| In-vivo Model | 1 × 105 HNE2 cells (with or without METTL3 knockdown) were labeled with luciferase gene and injected into the tail vein of the nude mice. | |||
Head and neck squamous carcinoma [ICD-11: 2B6E]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [9] | |||
| Response Summary | FTO expression was significantly upregulated in HNSCC datasets and tissues. FTO expression was significantly correlated with Catenin beta-1 (CTNNB1/Beta-catenin) expression. Moreover, it exerted a tumorigenic effect by increasing CTNNB1 expression in an m6A-dependent manner. | |||
| Responsed Disease | Head and neck squamous carcinoma [ICD-11: 2B6E] | |||
| Target Regulator | Fat mass and obesity-associated protein (FTO) | ERASER | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Adherens junction | hsa04520 | ||
| Cell Process | Cell proliferation and migration | |||
| In-vitro Model | CAL-27 | Tongue squamous cell carcinoma | Homo sapiens | CVCL_1107 |
| FaDu | Hypopharyngeal squamous cell carcinoma | Homo sapiens | CVCL_1218 | |
| Tu 686 | Laryngeal squamous cell carcinoma | Homo sapiens | CVCL_4916 | |
| HN-6 | Tongue squamous cell carcinoma | Homo sapiens | CVCL_8129 | |
| HEp-2 | Endocervical adenocarcinoma | Homo sapiens | CVCL_1906 | |
Colorectal cancer [ICD-11: 2B91]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [6] | |||
| Response Summary | Silencing YTHDF1 significantly inhibited Wnt/Catenin beta-1 (CTNNB1/Beta-catenin) pathway activity in Colorectal cancer cells. | |||
| Responsed Disease | Colorectal cancer [ICD-11: 2B91] | |||
| Target Regulator | YTH domain-containing family protein 1 (YTHDF1) | READER | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Wnt signaling pathway | hsa04310 | ||
| Signaling pathways regulating pluripotency of stem cells | hsa04550 | |||
| Cell Process | Cell Tumorigenicity | |||
| In-vitro Model | NCM460 | Normal | Homo sapiens | CVCL_0460 |
| SW480 | Colon adenocarcinoma | Homo sapiens | CVCL_0546 | |
| SW620 | Colon adenocarcinoma | Homo sapiens | CVCL_0547 | |
| In-vivo Model | 1 × 105 parental HT29-shNC cells, HT29-shYTHDF1 cells, HT29-shNC colonospheres, and HT29-shYTHDF1 colonospheres were inoculated subcutaneously into the left inguinal folds of the nude mice. | |||
Liver cancer [ICD-11: 2C12]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [2] | |||
| Response Summary | METTL3 is significantly up-regulated in Hepatoblastoma(HB) and promotes HB development.m6A mRNA methylation contributes significantly to regulate the Wnt/beta-catenin pathway. Reduced m6A methylation can lead to a decrease in expression and stability of the Catenin beta-1 (CTNNB1/Beta-catenin). | |||
| Responsed Disease | Hepatoblastoma [ICD-11: 2C12.01] | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Wnt signaling pathway | hsa04310 | ||
| Cell Process | RNA stability | |||
| In-vitro Model | HEK293T | Normal | Homo sapiens | CVCL_0063 |
| Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 | |
| L-02 | Endocervical adenocarcinoma | Homo sapiens | CVCL_6926 | |
| QSG-7701 | Endocervical adenocarcinoma | Homo sapiens | CVCL_6944 | |
| In-vivo Model | 5 × 106 cells were subcutaneously injected into the left or right flank of each mouse. | |||
Lung cancer [ICD-11: 2C25]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [7] | |||
| Response Summary | ECs transmitted miR-376c into NSCLC cells through Evs, and inhibited the intracellular YTHDF1 expression and the Wnt/Catenin beta-1 (CTNNB1/Beta-catenin) pathway activation. YTHDF1 overexpression reversed the inhibitory role of miR-376c released by EC-Evs in non-small cell lung cancer cells. | |||
| Responsed Disease | Non-small-cell lung carcinoma [ICD-11: 2C25.Y] | |||
| Target Regulator | YTH domain-containing family protein 1 (YTHDF1) | READER | ||
| Target Regulation | Up regulation | |||
| Cell Process | Cell proliferation | |||
| Cell invasion | ||||
| Cell migration | ||||
| Cell apoptosis | ||||
| In-vitro Model | A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 |
| NCI-1650 (Non-Small Cell Lung Cancer Cells) | ||||
| NCI-H1299 | Lung large cell carcinoma | Homo sapiens | CVCL_0060 | |
| NCI-H358 | Minimally invasive lung adenocarcinoma | Homo sapiens | CVCL_1559 | |
Breast cancer [ICD-11: 2C60]
| In total 3 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [4] | |||
| Response Summary | m6A-hypomethylation regulated FGFR4 phosphorylates GSK-3beta and activates Catenin beta-1 (CTNNB1/Beta-catenin)/TCF4-SLC7A11/FPN1 signaling to drive anti-HER2 resistance. Knockdown of METTL14 significantly increased the expression level of FGFR4 in HER2-positive breast cancer cells. FGFR4 reduced the sensitivity of HER2-positive breast cancer to trastuzumab plus pertuzumab or tucatinib. These results pinpoint a mechanism of anti-HER2 resistance and provide a strategy for overcoming resistance via FGFR4 inhibition in recalcitrant HER2-positive breast cancer. | |||
| Responsed Disease | Breast cancer [ICD-11: 2C60] | |||
| Target Regulator | Methyltransferase-like 14 (METTL14) | WRITER | ||
| Target Regulation | Down regulation | |||
| Responsed Drug | Pertuzumab | Approved | ||
| Pathway Response | Wnt signaling pathway | hsa04310 | ||
| Cell Process | Glutathione synthesis | |||
| In-vitro Model | ZR-75-1 | Invasive breast carcinoma | Homo sapiens | CVCL_0588 |
| T-47D | Invasive breast carcinoma | Homo sapiens | CVCL_0553 | |
| SUM-159 (A mesenchymal triple-negative breast cancer cell line) | ||||
| SK-BR-3 | Breast adenocarcinoma | Homo sapiens | CVCL_0033 | |
| MDA-MB-468 | Breast adenocarcinoma | Homo sapiens | CVCL_0419 | |
| MDA-MB-453 | Breast adenocarcinoma | Homo sapiens | CVCL_0418 | |
| MDA-MB-361 | Breast adenocarcinoma | Homo sapiens | CVCL_0620 | |
| MDA-MB-231 | Breast adenocarcinoma | Homo sapiens | CVCL_0062 | |
| MCF-7 | Invasive breast carcinoma | Homo sapiens | CVCL_0031 | |
| MCF-10A | Normal | Homo sapiens | CVCL_0598 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| BT-549 | Invasive breast carcinoma | Homo sapiens | CVCL_1092 | |
| BT-474 | Invasive breast carcinoma | Homo sapiens | CVCL_0179 | |
| AU565 | Breast adenocarcinoma | Homo sapiens | CVCL_1074 | |
| In-vivo Model | Luciferase-labeled rSKBR3 and MDA-MB-361 cells (1 × 107 cells) mixed with 1:1 Matrigel (Corning, 356237) were subcutaneously injected into the fat pads of mice. After a tumor was palpable, the mice were randomized into four groups (five mice per group), and they were treated with vehicle, trastuzumab (20 mg/kg, intraperitoneal administration), roblitinib (30 mg/kg, oral administration), or a combination of both drugs. | |||
| Experiment 2 Reporting the m6A-centered Disease Response | [4] | |||
| Response Summary | m6A-hypomethylation regulated FGFR4 phosphorylates GSK-3beta and activates Catenin beta-1 (CTNNB1/Beta-catenin)/TCF4-SLC7A11/FPN1 signaling to drive anti-HER2 resistance. Knockdown of METTL14 significantly increased the expression level of FGFR4 in HER2-positive breast cancer cells. FGFR4 reduced the sensitivity of HER2-positive breast cancer to trastuzumab plus pertuzumab or tucatinib. These results pinpoint a mechanism of anti-HER2 resistance and provide a strategy for overcoming resistance via FGFR4 inhibition in recalcitrant HER2-positive breast cancer. | |||
| Responsed Disease | Breast cancer [ICD-11: 2C60] | |||
| Target Regulator | Methyltransferase-like 14 (METTL14) | WRITER | ||
| Target Regulation | Down regulation | |||
| Responsed Drug | Trastuzumab | Approved | ||
| Pathway Response | Wnt signaling pathway | hsa04310 | ||
| Cell Process | Glutathione synthesis | |||
| In-vitro Model | ZR-75-1 | Invasive breast carcinoma | Homo sapiens | CVCL_0588 |
| T-47D | Invasive breast carcinoma | Homo sapiens | CVCL_0553 | |
| SUM-159 (A mesenchymal triple-negative breast cancer cell line) | ||||
| SK-BR-3 | Breast adenocarcinoma | Homo sapiens | CVCL_0033 | |
| MDA-MB-468 | Breast adenocarcinoma | Homo sapiens | CVCL_0419 | |
| MDA-MB-453 | Breast adenocarcinoma | Homo sapiens | CVCL_0418 | |
| MDA-MB-361 | Breast adenocarcinoma | Homo sapiens | CVCL_0620 | |
| MDA-MB-231 | Breast adenocarcinoma | Homo sapiens | CVCL_0062 | |
| MCF-7 | Invasive breast carcinoma | Homo sapiens | CVCL_0031 | |
| MCF-10A | Normal | Homo sapiens | CVCL_0598 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| BT-549 | Invasive breast carcinoma | Homo sapiens | CVCL_1092 | |
| BT-474 | Invasive breast carcinoma | Homo sapiens | CVCL_0179 | |
| AU565 | Breast adenocarcinoma | Homo sapiens | CVCL_1074 | |
| In-vivo Model | Luciferase-labeled rSKBR3 and MDA-MB-361 cells (1 × 107 cells) mixed with 1:1 Matrigel (Corning, 356237) were subcutaneously injected into the fat pads of mice. After a tumor was palpable, the mice were randomized into four groups (five mice per group), and they were treated with vehicle, trastuzumab (20 mg/kg, intraperitoneal administration), roblitinib (30 mg/kg, oral administration), or a combination of both drugs. | |||
| Experiment 3 Reporting the m6A-centered Disease Response | [4] | |||
| Response Summary | m6A-hypomethylation regulated FGFR4 phosphorylates GSK-3beta and activates Catenin beta-1 (CTNNB1/Beta-catenin)/TCF4-SLC7A11/FPN1 signaling to drive anti-HER2 resistance. Knockdown of METTL14 significantly increased the expression level of FGFR4 in HER2-positive breast cancer cells. FGFR4 reduced the sensitivity of HER2-positive breast cancer to trastuzumab plus pertuzumab or tucatinib. These results pinpoint a mechanism of anti-HER2 resistance and provide a strategy for overcoming resistance via FGFR4 inhibition in recalcitrant HER2-positive breast cancer. | |||
| Responsed Disease | Breast cancer [ICD-11: 2C60] | |||
| Target Regulator | Methyltransferase-like 14 (METTL14) | WRITER | ||
| Target Regulation | Down regulation | |||
| Responsed Drug | Tucatinib | Approved | ||
| Pathway Response | Wnt signaling pathway | hsa04310 | ||
| Cell Process | Glutathione synthesis | |||
| In-vitro Model | ZR-75-1 | Invasive breast carcinoma | Homo sapiens | CVCL_0588 |
| T-47D | Invasive breast carcinoma | Homo sapiens | CVCL_0553 | |
| SUM-159 (A mesenchymal triple-negative breast cancer cell line) | ||||
| SK-BR-3 | Breast adenocarcinoma | Homo sapiens | CVCL_0033 | |
| MDA-MB-468 | Breast adenocarcinoma | Homo sapiens | CVCL_0419 | |
| MDA-MB-453 | Breast adenocarcinoma | Homo sapiens | CVCL_0418 | |
| MDA-MB-361 | Breast adenocarcinoma | Homo sapiens | CVCL_0620 | |
| MDA-MB-231 | Breast adenocarcinoma | Homo sapiens | CVCL_0062 | |
| MCF-7 | Invasive breast carcinoma | Homo sapiens | CVCL_0031 | |
| MCF-10A | Normal | Homo sapiens | CVCL_0598 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| BT-549 | Invasive breast carcinoma | Homo sapiens | CVCL_1092 | |
| BT-474 | Invasive breast carcinoma | Homo sapiens | CVCL_0179 | |
| AU565 | Breast adenocarcinoma | Homo sapiens | CVCL_1074 | |
| In-vivo Model | Luciferase-labeled rSKBR3 and MDA-MB-361 cells (1 × 107 cells) mixed with 1:1 Matrigel (Corning, 356237) were subcutaneously injected into the fat pads of mice. After a tumor was palpable, the mice were randomized into four groups (five mice per group), and they were treated with vehicle, trastuzumab (20 mg/kg, intraperitoneal administration), roblitinib (30 mg/kg, oral administration), or a combination of both drugs. | |||
Melanoma of uvea [ICD-11: 2D0Y]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [8] | |||
| Response Summary | YTHDF3 enhances Catenin beta-1 (CTNNB1/Beta-catenin) translation through recognizing and binding the m6A peaks on CTNNB1 mRNA.m6A reading protein YTHDF3 promotes the translation of the target transcript CTNNB1, contributing to ocular melanoma propagation and migration through m6A methylation. | |||
| Responsed Disease | Melanoma of uvea [ICD-11: 2D0Y] | |||
| Target Regulator | YTH domain-containing family protein 3 (YTHDF3) | READER | ||
| Target Regulation | Up regulation | |||
Cervical intraepithelial neoplasia [ICD-11: 2E66]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [10] | |||
| Response Summary | The mRNA level of FTO is elevated in cervical squamous cell carcinoma (CSCC) tissues when compared with respective adjacent normal tissues.FTO enhances the chemo-radiotherapy resistance both in vitro and in vivo through regulating expression of Catenin beta-1 (CTNNB1/Beta-catenin) by reducing m6A levels in its mRNA transcripts and in turn increases excision repair cross-complementation group 1 (ERCC1) activity. | |||
| Responsed Disease | Cervical squamous cell carcinoma [ICD-11: 2E66.2] | |||
| Target Regulator | Fat mass and obesity-associated protein (FTO) | ERASER | ||
| Target Regulation | Up regulation | |||
| Cell Process | RNA decay | |||
| In-vitro Model | C-33 A | Cervical squamous cell carcinoma | Homo sapiens | CVCL_1094 |
| SiHa | Cervical squamous cell carcinoma | Homo sapiens | CVCL_0032 | |
| In-vivo Model | Either SiHa cells or FTO overexpressed SiHa cells were injected subcutaneously into the right flanks of 4- to 6-week-old female athymic nude mice (CAS, Beijing, China). The mice were divided into four groups (N = 6). After transplantation, tumor size was measured by caliper every other day. Tumor volume was calculated using the formula: volume = (length × width2)/2. When the tumor volumes reached 50 mm3, the animals were treated with intraperitoneal injection of cisplatin (3 mg/kg) every other day for seven times and local irradiation of 8 Gy one time. | |||
Ischemic heart disease [ICD-11: BA40-BA6Z]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [5] | |||
| Response Summary | Mettl14 resulted in enhanced levels of Wnt1 m6A modification and Wnt1 protein but not its transcript level.Furthermore, Mettl14 overexpression blocked I/R-induced downregulation of Wnt1 and Catenin beta-1 (CTNNB1/Beta-catenin) proteins, whereas Mettl14 hearts exhibited the opposite results. Mettl14 attenuates cardiac I/R injury by activating Wnt/Bete-catenin in an m6A-dependent manner, providing a novel therapeutic target for ischemic heart disease. | |||
| Responsed Disease | Ischemic heart disease [ICD-11: BA40-BA6Z] | |||
| Target Regulator | Methyltransferase-like 14 (METTL14) | WRITER | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Wnt signaling pathway | hsa04310 | ||
| In-vitro Model | Neonatal rat ventricular cardiomyocytes (Primary myocyte cells) | |||
| In-vivo Model | C57BL/6 mouse hearts were subjected to ischemia/reperfusion (I/R) in vivo as described previously (Bock-Marquette et al., 2004; Song et al., 2015; Brocard et al., 2017). I/R injury in mice was induced by 45-min ischemia, followed by 7-day and 4-week reperfusion in a loss-of-function study (Figure 1) and gain-of-function study (Figure 2), respectively. In brief, mice were anesthetized with 2% avertin (0.1 ml/10g body weight; Sigma-Aldrich Corporation, United States) through intraperitoneal injection. To generate I/R injury, the left anterior descending coronary artery (LAD) was ligated with 7-0 nylon for 45 min and then was removed. For the sham group, a suture was passed under the LAD but without ligation. According to the experimental requirements, at different time points of cardiac I/R, the mice were anesthetized for assessing heart function by echocardiographic measurement. All the mice survived during the process of I/R injury after the operation. | |||
Spina bifida [ICD-11: LA02]
| In total 2 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [3] | |||
| Response Summary | SAM not only played a compensatory role, but also led to m6A modification changes in neural tube development and regulation. Ethionine affected m6A modification by reducing SAM metabolism. METTL3 is enriched in HT-22 cells, and METTL3 knockdown reduces cell proliferation and increases apoptosis through suppressing Wnt/Catenin beta-1 (CTNNB1/Beta-catenin) signaling pathway. Overexpression of ALKBH5 can only inhibit cell proliferation, but cannot promote cell apoptosis. | |||
| Responsed Disease | Neural tube defect [ICD-11: LA02.Z] | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Target Regulation | Down regulation | |||
| Pathway Response | Wnt signaling pathway | hsa04310 | ||
| Cell Process | Cell apoptosis | |||
| In-vitro Model | HT22 | Normal | Mus musculus | CVCL_0321 |
| In-vivo Model | The mice were maintained on a 12-h light/dark cycle (lights on from 8:00 a.m. to 8:00 p.m.). On day 7.5 of pregnancy (E7.5), ethionine (Sigma-Aldrich, USA) was intraperitoneally injected only once at a dose of 500 mg/kg to establish the NTDs embryo model. And SAM (MedChemExpress, USA) was intraperitoneally injected only once at a dose of 30 mg/kg. The same dose was intraperitoneally injected to the pregnant mice for control group. | |||
| Experiment 2 Reporting the m6A-centered Disease Response | [3] | |||
| Response Summary | SAM not only played a compensatory role, but also led to m6A modification changes in neural tube development and regulation. Ethionine affected m6A modification by reducing SAM metabolism. METTL3 is enriched in HT-22 cells, and METTL3 knockdown reduces cell proliferation and increases apoptosis through suppressing Wnt/Catenin beta-1 (CTNNB1/Beta-catenin) signaling pathway. Overexpression of ALKBH5 can only inhibit cell proliferation, but cannot promote cell apoptosis. | |||
| Responsed Disease | Neural tube defect [ICD-11: LA02.Z] | |||
| Target Regulator | RNA demethylase ALKBH5 (ALKBH5) | ERASER | ||
| Pathway Response | Wnt signaling pathway | hsa04310 | ||
| Cell Process | Cell apoptosis | |||
| In-vitro Model | HT22 | Normal | Mus musculus | CVCL_0321 |
| In-vivo Model | The mice were maintained on a 12-h light/dark cycle (lights on from 8:00 a.m. to 8:00 p.m.). On day 7.5 of pregnancy (E7.5), ethionine (Sigma-Aldrich, USA) was intraperitoneally injected only once at a dose of 500 mg/kg to establish the NTDs embryo model. And SAM (MedChemExpress, USA) was intraperitoneally injected only once at a dose of 30 mg/kg. The same dose was intraperitoneally injected to the pregnant mice for control group. | |||
Pertuzumab
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response | [4] | |||
| Response Summary | m6A-hypomethylation regulated FGFR4 phosphorylates GSK-3beta and activates Catenin beta-1 (CTNNB1/Beta-catenin)/TCF4-SLC7A11/FPN1 signaling to drive anti-HER2 resistance. Knockdown of METTL14 significantly increased the expression level of FGFR4 in HER2-positive breast cancer cells. FGFR4 reduced the sensitivity of HER2-positive breast cancer to trastuzumab plus pertuzumab or tucatinib. These results pinpoint a mechanism of anti-HER2 resistance and provide a strategy for overcoming resistance via FGFR4 inhibition in recalcitrant HER2-positive breast cancer. | |||
| Target Regulator | Methyltransferase-like 14 (METTL14) | WRITER | ||
| Target Regulation | Down regulation | |||
| Responsed Disease | Breast cancer | ICD-11: 2C60 | ||
| Pathway Response | Wnt signaling pathway | hsa04310 | ||
| Cell Process | Glutathione synthesis | |||
| In-vitro Model | ZR-75-1 | Invasive breast carcinoma | Homo sapiens | CVCL_0588 |
| T-47D | Invasive breast carcinoma | Homo sapiens | CVCL_0553 | |
| SUM-159 (A mesenchymal triple-negative breast cancer cell line) | ||||
| SK-BR-3 | Breast adenocarcinoma | Homo sapiens | CVCL_0033 | |
| MDA-MB-468 | Breast adenocarcinoma | Homo sapiens | CVCL_0419 | |
| MDA-MB-453 | Breast adenocarcinoma | Homo sapiens | CVCL_0418 | |
| MDA-MB-361 | Breast adenocarcinoma | Homo sapiens | CVCL_0620 | |
| MDA-MB-231 | Breast adenocarcinoma | Homo sapiens | CVCL_0062 | |
| MCF-7 | Invasive breast carcinoma | Homo sapiens | CVCL_0031 | |
| MCF-10A | Normal | Homo sapiens | CVCL_0598 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| BT-549 | Invasive breast carcinoma | Homo sapiens | CVCL_1092 | |
| BT-474 | Invasive breast carcinoma | Homo sapiens | CVCL_0179 | |
| AU565 | Breast adenocarcinoma | Homo sapiens | CVCL_1074 | |
| In-vivo Model | Luciferase-labeled rSKBR3 and MDA-MB-361 cells (1 × 107 cells) mixed with 1:1 Matrigel (Corning, 356237) were subcutaneously injected into the fat pads of mice. After a tumor was palpable, the mice were randomized into four groups (five mice per group), and they were treated with vehicle, trastuzumab (20 mg/kg, intraperitoneal administration), roblitinib (30 mg/kg, oral administration), or a combination of both drugs. | |||
Trastuzumab
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response | [4] | |||
| Response Summary | m6A-hypomethylation regulated FGFR4 phosphorylates GSK-3beta and activates Catenin beta-1 (CTNNB1/Beta-catenin)/TCF4-SLC7A11/FPN1 signaling to drive anti-HER2 resistance. Knockdown of METTL14 significantly increased the expression level of FGFR4 in HER2-positive breast cancer cells. FGFR4 reduced the sensitivity of HER2-positive breast cancer to trastuzumab plus pertuzumab or tucatinib. These results pinpoint a mechanism of anti-HER2 resistance and provide a strategy for overcoming resistance via FGFR4 inhibition in recalcitrant HER2-positive breast cancer. | |||
| Target Regulator | Methyltransferase-like 14 (METTL14) | WRITER | ||
| Target Regulation | Down regulation | |||
| Responsed Disease | Breast cancer | ICD-11: 2C60 | ||
| Pathway Response | Wnt signaling pathway | hsa04310 | ||
| Cell Process | Glutathione synthesis | |||
| In-vitro Model | ZR-75-1 | Invasive breast carcinoma | Homo sapiens | CVCL_0588 |
| T-47D | Invasive breast carcinoma | Homo sapiens | CVCL_0553 | |
| SUM-159 (A mesenchymal triple-negative breast cancer cell line) | ||||
| SK-BR-3 | Breast adenocarcinoma | Homo sapiens | CVCL_0033 | |
| MDA-MB-468 | Breast adenocarcinoma | Homo sapiens | CVCL_0419 | |
| MDA-MB-453 | Breast adenocarcinoma | Homo sapiens | CVCL_0418 | |
| MDA-MB-361 | Breast adenocarcinoma | Homo sapiens | CVCL_0620 | |
| MDA-MB-231 | Breast adenocarcinoma | Homo sapiens | CVCL_0062 | |
| MCF-7 | Invasive breast carcinoma | Homo sapiens | CVCL_0031 | |
| MCF-10A | Normal | Homo sapiens | CVCL_0598 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| BT-549 | Invasive breast carcinoma | Homo sapiens | CVCL_1092 | |
| BT-474 | Invasive breast carcinoma | Homo sapiens | CVCL_0179 | |
| AU565 | Breast adenocarcinoma | Homo sapiens | CVCL_1074 | |
| In-vivo Model | Luciferase-labeled rSKBR3 and MDA-MB-361 cells (1 × 107 cells) mixed with 1:1 Matrigel (Corning, 356237) were subcutaneously injected into the fat pads of mice. After a tumor was palpable, the mice were randomized into four groups (five mice per group), and they were treated with vehicle, trastuzumab (20 mg/kg, intraperitoneal administration), roblitinib (30 mg/kg, oral administration), or a combination of both drugs. | |||
Tucatinib
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response | [4] | |||
| Response Summary | m6A-hypomethylation regulated FGFR4 phosphorylates GSK-3beta and activates Catenin beta-1 (CTNNB1/Beta-catenin)/TCF4-SLC7A11/FPN1 signaling to drive anti-HER2 resistance. Knockdown of METTL14 significantly increased the expression level of FGFR4 in HER2-positive breast cancer cells. FGFR4 reduced the sensitivity of HER2-positive breast cancer to trastuzumab plus pertuzumab or tucatinib. These results pinpoint a mechanism of anti-HER2 resistance and provide a strategy for overcoming resistance via FGFR4 inhibition in recalcitrant HER2-positive breast cancer. | |||
| Target Regulator | Methyltransferase-like 14 (METTL14) | WRITER | ||
| Target Regulation | Down regulation | |||
| Responsed Disease | Breast cancer | ICD-11: 2C60 | ||
| Pathway Response | Wnt signaling pathway | hsa04310 | ||
| Cell Process | Glutathione synthesis | |||
| In-vitro Model | ZR-75-1 | Invasive breast carcinoma | Homo sapiens | CVCL_0588 |
| T-47D | Invasive breast carcinoma | Homo sapiens | CVCL_0553 | |
| SUM-159 (A mesenchymal triple-negative breast cancer cell line) | ||||
| SK-BR-3 | Breast adenocarcinoma | Homo sapiens | CVCL_0033 | |
| MDA-MB-468 | Breast adenocarcinoma | Homo sapiens | CVCL_0419 | |
| MDA-MB-453 | Breast adenocarcinoma | Homo sapiens | CVCL_0418 | |
| MDA-MB-361 | Breast adenocarcinoma | Homo sapiens | CVCL_0620 | |
| MDA-MB-231 | Breast adenocarcinoma | Homo sapiens | CVCL_0062 | |
| MCF-7 | Invasive breast carcinoma | Homo sapiens | CVCL_0031 | |
| MCF-10A | Normal | Homo sapiens | CVCL_0598 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| BT-549 | Invasive breast carcinoma | Homo sapiens | CVCL_1092 | |
| BT-474 | Invasive breast carcinoma | Homo sapiens | CVCL_0179 | |
| AU565 | Breast adenocarcinoma | Homo sapiens | CVCL_1074 | |
| In-vivo Model | Luciferase-labeled rSKBR3 and MDA-MB-361 cells (1 × 107 cells) mixed with 1:1 Matrigel (Corning, 356237) were subcutaneously injected into the fat pads of mice. After a tumor was palpable, the mice were randomized into four groups (five mice per group), and they were treated with vehicle, trastuzumab (20 mg/kg, intraperitoneal administration), roblitinib (30 mg/kg, oral administration), or a combination of both drugs. | |||
Full List of Crosstalk(s) between m6A Modification and Epigenetic Regulation Related to This Regulator
DNA modification
m6A Regulator: Methyltransferase-like 14 (METTL14)
| In total 9 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT02206 | ||
| Epigenetic Regulator | DNA (cytosine-5)-methyltransferase 3B (DNMT3B) | |
| Regulated Target | Methyltransferase-like protein 14 (METTL14) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Breast cancer | |
| Drug | Trastuzumab | |
| Crosstalk ID: M6ACROT02211 | ||
| Epigenetic Regulator | DNA (cytosine-5)-methyltransferase 3B (DNMT3B) | |
| Regulated Target | Methyltransferase-like protein 14 (METTL14) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Breast cancer | |
| Drug | Pertuzumab | |
| Crosstalk ID: M6ACROT02219 | ||
| Epigenetic Regulator | DNA (cytosine-5)-methyltransferase 3B (DNMT3B) | |
| Regulated Target | Methyltransferase-like protein 14 (METTL14) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Breast cancer | |
| Drug | Tucatinib | |
| Crosstalk ID: M6ACROT02230 | ||
| Epigenetic Regulator | Cysteine methyltransferase DNMT3A (DNMT3A) | |
| Regulated Target | Methyltransferase-like protein 14 (METTL14) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Breast cancer | |
| Drug | Trastuzumab | |
| Crosstalk ID: M6ACROT02235 | ||
| Epigenetic Regulator | Cysteine methyltransferase DNMT3A (DNMT3A) | |
| Regulated Target | Methyltransferase-like protein 14 (METTL14) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Breast cancer | |
| Drug | Pertuzumab | |
| Crosstalk ID: M6ACROT02243 | ||
| Epigenetic Regulator | Cysteine methyltransferase DNMT3A (DNMT3A) | |
| Regulated Target | Methyltransferase-like protein 14 (METTL14) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Breast cancer | |
| Drug | Tucatinib | |
| Crosstalk ID: M6ACROT02254 | ||
| Epigenetic Regulator | DNA (cytosine-5)-methyltransferase 1 (DNMT1) | |
| Regulated Target | Methyltransferase-like protein 14 (METTL14) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Breast cancer | |
| Drug | Trastuzumab | |
| Crosstalk ID: M6ACROT02259 | ||
| Epigenetic Regulator | DNA (cytosine-5)-methyltransferase 1 (DNMT1) | |
| Regulated Target | Methyltransferase-like protein 14 (METTL14) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Breast cancer | |
| Drug | Pertuzumab | |
| Crosstalk ID: M6ACROT02267 | ||
| Epigenetic Regulator | DNA (cytosine-5)-methyltransferase 1 (DNMT1) | |
| Regulated Target | Methyltransferase-like protein 14 (METTL14) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Breast cancer | |
| Drug | Tucatinib | |
Non-coding RNA
m6A Regulator: Methyltransferase-like 3 (METTL3)
| In total 1 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT05052 | ||
| Epigenetic Regulator | MicroRNA 186 (MIR186) | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Hepatoblastoma | |
m6A Regulator: YTH domain-containing family protein 1 (YTHDF1)
| In total 2 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT05267 | ||
| Epigenetic Regulator | hsa-miR-136-5p | |
| Regulated Target | YTH domain-containing family protein 1 (YTHDF1) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Colorectal cancer | |
| Crosstalk ID: M6ACROT05335 | ||
| Epigenetic Regulator | hsa-miR-376c | |
| Regulated Target | YTH domain-containing family protein 1 (YTHDF1) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Non-small cell lung cancer | |
RNA Modification Sequencing Data Associated with the Target (ID: M6ATAR00222)
| In total 2 m6A sequence/site(s) in this target gene | |||
| mod ID: A2ISITE011678 | Click to Show/Hide the Full List | ||
| mod site | chr3:41210928-41210929:+ | [12] | |
| Sequence | CCTCCCAAGTAGCTGGGACTACACCAGGCACCCCACCATGC | ||
| Transcript ID List | ENST00000644867.1; ENST00000647413.1; ENST00000643865.1; ENST00000646116.1; ENST00000642248.1; ENST00000643992.1; ENST00000645210.1; ENST00000349496.11; ENST00000642886.1; ENST00000646725.1; ENST00000488914.2; ENST00000433400.6; ENST00000642836.1; ENST00000647264.1; ENST00000643031.1; ENST00000642315.1; ENST00000645276.1; ENST00000643297.1; ENST00000645982.1; ENST00000642426.1; ENST00000426215.5; ENST00000644678.1; ENST00000643977.1; ENST00000642986.1; ENST00000453024.6; ENST00000450969.6; ENST00000646074.1; ENST00000405570.6; ENST00000645305.1; ENST00000645900.1; ENST00000646381.1; ENST00000643541.1; ENST00000646369.1; ENST00000396183.7; ENST00000646174.1; ENST00000642992.1; ENST00000644873.1; ENST00000647390.1; ENST00000644138.1; ENST00000643052.1; ENST00000431914.6; ENST00000644524.1; ENST00000396185.8; ENST00000645493.1 | ||
| External Link | RMBase: RNA-editing_site_96002 | ||
| mod ID: A2ISITE011679 | Click to Show/Hide the Full List | ||
| mod site | chr3:41221474-41221475:+ | [13] | |
| Sequence | CCTCAGCCTCCCCAGTGGCTAGGACTACAGGCACATACTAC | ||
| Transcript ID List | ENST00000645320.1; ENST00000644524.1; ENST00000643052.1; ENST00000645900.1; ENST00000433400.6; ENST00000643992.1; ENST00000646116.1; ENST00000645305.1; ENST00000643297.1; ENST00000396185.8; ENST00000642886.1; ENST00000642836.1; ENST00000453024.6; ENST00000647390.1; ENST00000642426.1; ENST00000643541.1; ENST00000645982.1; ENST00000643031.1; ENST00000644867.1; ENST00000396183.7; ENST00000647413.1; ENST00000405570.6; ENST00000644138.1; ENST00000643865.1; ENST00000426215.5; ENST00000642248.1; ENST00000643977.1; ENST00000646725.1; ENST00000431914.6; ENST00000441708.2; ENST00000488914.2; ENST00000349496.11; ENST00000644873.1; ENST00000645493.1; ENST00000450969.6; ENST00000646174.1; ENST00000645276.1; ENST00000642992.1; ENST00000646381.1; ENST00000644678.1; ENST00000647264.1; ENST00000646369.1; ENST00000642986.1; ENST00000646074.1; ENST00000645210.1; ENST00000642315.1 | ||
| External Link | RMBase: RNA-editing_site_96003 | ||
5-methylcytidine (m5C)
| In total 2 m6A sequence/site(s) in this target gene | |||
| mod ID: M5CSITE003064 | Click to Show/Hide the Full List | ||
| mod site | chr3:41225038-41225039:+ | [14] | |
| Sequence | TTAGATGAGGGCATGCAGATCCCATCTACACAGTTTGATGC | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000645276.1; ENST00000441708.2; ENST00000644873.1; ENST00000645210.1; ENST00000642315.1; ENST00000645900.1; ENST00000643052.1; ENST00000647264.1; ENST00000396183.7; ENST00000643992.1; ENST00000642836.1; ENST00000647413.1; ENST00000642426.1; ENST00000642886.1; ENST00000642992.1; ENST00000643865.1; ENST00000643031.1; ENST00000645982.1; ENST00000643977.1; ENST00000646074.1; ENST00000433400.6; ENST00000396185.8; ENST00000453024.6; ENST00000644867.1; ENST00000644138.1; ENST00000647021.1; ENST00000643297.1; ENST00000645320.1; ENST00000450969.6; ENST00000431914.6; ENST00000646116.1; ENST00000645493.1; ENST00000349496.11; ENST00000644524.1; ENST00000643541.1; ENST00000405570.6; ENST00000646381.1; ENST00000645305.1; ENST00000642248.1; ENST00000647390.1; ENST00000646174.1; ENST00000642986.1; ENST00000488914.2; ENST00000646725.1; ENST00000644678.1; ENST00000646369.1 | ||
| External Link | RMBase: m5C_site_31862 | ||
| mod ID: M5CSITE003065 | Click to Show/Hide the Full List | ||
| mod site | chr3:41236676-41236677:+ | [14] | |
| Sequence | CAGTTGAGCTGACCAGCTCTCTCTTCAGAACAGAGCCAATG | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000349496.11; ENST00000644867.1; ENST00000405570.6; ENST00000642886.1; ENST00000642836.1; ENST00000647390.1; ENST00000643052.1; ENST00000645982.1; ENST00000644524.1; ENST00000453024.6; ENST00000646074.1; ENST00000431914.6; ENST00000645320.1; ENST00000643297.1; ENST00000646174.1; ENST00000642986.1; ENST00000642315.1; ENST00000645927.1; ENST00000643031.1; ENST00000646369.1; ENST00000643992.1; ENST00000396183.7; ENST00000645276.1; ENST00000643865.1; ENST00000471014.2; ENST00000485265.2; ENST00000643977.1; ENST00000645763.1; ENST00000465552.2; ENST00000644678.1; ENST00000643541.1; ENST00000450969.6; ENST00000441708.2; ENST00000646725.1; ENST00000644873.1; ENST00000396185.8; ENST00000433400.6; ENST00000646116.1; ENST00000647413.1; ENST00000645900.1; ENST00000646381.1; ENST00000644138.1; ENST00000647264.1; ENST00000645305.1; ENST00000645210.1; ENST00000482042.1; ENST00000642426.1; ENST00000642248.1; ENST00000647021.1; ENST00000645493.1; ENST00000642992.1 | ||
| External Link | RMBase: m5C_site_31863 | ||
N6-methyladenosine (m6A)
| In total 98 m6A sequence/site(s) in this target gene | |||
| mod ID: M6ASITE060213 | Click to Show/Hide the Full List | ||
| mod site | chr3:41198766-41198767:+ | [15] | |
| Sequence | CAGGTCGAAATTCAAGCTGAACAGCCTGCTGAGAGGTGGGA | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | H1A; hESCs | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000642248.1; ENST00000405570.6; ENST00000642836.1; ENST00000431914.6; ENST00000642992.1; ENST00000433400.6; ENST00000426215.5; ENST00000646381.1; ENST00000645210.1; ENST00000643541.1 | ||
| External Link | RMBase: m6A_site_584440 | ||
| mod ID: M6ASITE060214 | Click to Show/Hide the Full List | ||
| mod site | chr3:41199637-41199638:+ | [16] | |
| Sequence | CGCAGGTCGAGGACGGTCGGACTCCCGCGGCGGGAGGAGCC | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa; CD34; HepG2; A549; H1B; H1A; fibroblasts; LCLs; MM6; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293T; HEK293A-TOA; iSLK; MSC; TIME; TREX; endometrial; HEC-1-A; GSCs; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000488914.2; ENST00000642248.1; ENST00000647264.1; ENST00000431914.6; ENST00000645276.1; ENST00000646074.1; ENST00000643052.1; ENST00000647390.1; ENST00000646381.1; ENST00000396183.7; ENST00000646116.1; ENST00000349496.11; ENST00000643031.1; ENST00000645900.1; ENST00000646725.1; ENST00000643992.1; ENST00000642426.1; ENST00000645982.1; ENST00000433400.6; ENST00000642836.1; ENST00000644873.1; ENST00000643541.1; ENST00000644867.1; ENST00000644678.1; ENST00000396185.8; ENST00000643865.1; ENST00000426215.5; ENST00000642992.1; ENST00000645210.1; ENST00000453024.6; ENST00000642986.1; ENST00000405570.6; ENST00000644138.1; ENST00000646174.1; ENST00000642886.1 | ||
| External Link | RMBase: m6A_site_584441 | ||
| mod ID: M6ASITE060215 | Click to Show/Hide the Full List | ||
| mod site | chr3:41199730-41199731:+ | [16] | |
| Sequence | CCGCTTCCTCTCGGAGCCAAACTTCGTAGCAGGCGCGCGGT | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa; HepG2; A549; MM6; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293T; iSLK; MSC; TIME; endometrial; HEC-1-A; GSCs; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000642426.1; ENST00000644867.1; ENST00000642886.1; ENST00000645982.1; ENST00000642836.1; ENST00000646174.1; ENST00000645900.1; ENST00000644138.1; ENST00000644873.1; ENST00000646116.1; ENST00000488914.2; ENST00000645210.1; ENST00000642986.1; ENST00000644678.1; ENST00000431914.6; ENST00000643541.1; ENST00000643992.1; ENST00000647264.1; ENST00000426215.5; ENST00000647390.1; ENST00000642992.1; ENST00000433400.6; ENST00000643865.1; ENST00000396183.7; ENST00000405570.6; ENST00000646074.1; ENST00000453024.6; ENST00000349496.11; ENST00000643031.1; ENST00000645493.1; ENST00000646725.1; ENST00000643052.1; ENST00000646369.1; ENST00000396185.8; ENST00000646381.1; ENST00000642248.1; ENST00000645276.1 | ||
| External Link | RMBase: m6A_site_584442 | ||
| mod ID: M6ASITE060216 | Click to Show/Hide the Full List | ||
| mod site | chr3:41224039-41224040:+ | [17] | |
| Sequence | GGGTATTTGAAGTATACCATACAACTGTTTTGAAAATCCAG | ||
| Motif Score | 2.110482143 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000349496.11; ENST00000642315.1; ENST00000642426.1; ENST00000645982.1; ENST00000646725.1; ENST00000643052.1; ENST00000647264.1; ENST00000645210.1; ENST00000643541.1; ENST00000646116.1; ENST00000645305.1; ENST00000643865.1; ENST00000433400.6; ENST00000642836.1; ENST00000647021.1; ENST00000642248.1; ENST00000644138.1; ENST00000645276.1; ENST00000643031.1; ENST00000642886.1; ENST00000450969.6; ENST00000431914.6; ENST00000396183.7; ENST00000647413.1; ENST00000426215.5; ENST00000453024.6; ENST00000642986.1; ENST00000642992.1; ENST00000441708.2; ENST00000643992.1; ENST00000646381.1; ENST00000646369.1; ENST00000488914.2; ENST00000643297.1; ENST00000646074.1; ENST00000644524.1; ENST00000646174.1; ENST00000644867.1; ENST00000643977.1; ENST00000645320.1; ENST00000647390.1; ENST00000645493.1; ENST00000644678.1; ENST00000645900.1; ENST00000396185.8; ENST00000644873.1; ENST00000405570.6 | ||
| External Link | RMBase: m6A_site_584443 | ||
| mod ID: M6ASITE060217 | Click to Show/Hide the Full List | ||
| mod site | chr3:41224065-41224066:+ | [16] | |
| Sequence | GTTTTGAAAATCCAGCGTGGACAATGGCTACTCAAGGTTTG | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; HepG2; A549; iSLK | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000644873.1; ENST00000642986.1; ENST00000646369.1; ENST00000647021.1; ENST00000643992.1; ENST00000645305.1; ENST00000647413.1; ENST00000644678.1; ENST00000647390.1; ENST00000431914.6; ENST00000642426.1; ENST00000643865.1; ENST00000488914.2; ENST00000642886.1; ENST00000646074.1; ENST00000645493.1; ENST00000396183.7; ENST00000645210.1; ENST00000642992.1; ENST00000441708.2; ENST00000642315.1; ENST00000643052.1; ENST00000643031.1; ENST00000396185.8; ENST00000450969.6; ENST00000647264.1; ENST00000405570.6; ENST00000644138.1; ENST00000643541.1; ENST00000426215.5; ENST00000453024.6; ENST00000646725.1; ENST00000646381.1; ENST00000645320.1; ENST00000642836.1; ENST00000645276.1; ENST00000433400.6; ENST00000644524.1; ENST00000645982.1; ENST00000349496.11; ENST00000643297.1; ENST00000646174.1; ENST00000643977.1; ENST00000646116.1; ENST00000645900.1; ENST00000642248.1; ENST00000644867.1 | ||
| External Link | RMBase: m6A_site_584444 | ||
| mod ID: M6ASITE060218 | Click to Show/Hide the Full List | ||
| mod site | chr3:41224543-41224544:+ | [16] | |
| Sequence | AGCTGATTTGATGGAGTTGGACATGGCCATGGAACCAGACA | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000642248.1; ENST00000349496.11; ENST00000431914.6; ENST00000646369.1; ENST00000646074.1; ENST00000645982.1; ENST00000647021.1; ENST00000643992.1; ENST00000645493.1; ENST00000441708.2; ENST00000642986.1; ENST00000450969.6; ENST00000645210.1; ENST00000642315.1; ENST00000647264.1; ENST00000642836.1; ENST00000644867.1; ENST00000646174.1; ENST00000642886.1; ENST00000647413.1; ENST00000426215.5; ENST00000645320.1; ENST00000405570.6; ENST00000396183.7; ENST00000642426.1; ENST00000645900.1; ENST00000643031.1; ENST00000453024.6; ENST00000433400.6; ENST00000646116.1; ENST00000645276.1; ENST00000396185.8; ENST00000643052.1; ENST00000643297.1; ENST00000646725.1; ENST00000646381.1; ENST00000642992.1; ENST00000488914.2; ENST00000644873.1; ENST00000645305.1; ENST00000643541.1; ENST00000643977.1; ENST00000644524.1; ENST00000647390.1; ENST00000644138.1; ENST00000643865.1; ENST00000644678.1 | ||
| External Link | RMBase: m6A_site_584445 | ||
| mod ID: M6ASITE060219 | Click to Show/Hide the Full List | ||
| mod site | chr3:41224556-41224557:+ | [16] | |
| Sequence | GAGTTGGACATGGCCATGGAACCAGACAGAAAAGCGGCTGT | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000453024.6; ENST00000642315.1; ENST00000645305.1; ENST00000647264.1; ENST00000642992.1; ENST00000396183.7; ENST00000646725.1; ENST00000644524.1; ENST00000646381.1; ENST00000646174.1; ENST00000643297.1; ENST00000644678.1; ENST00000647413.1; ENST00000644138.1; ENST00000643541.1; ENST00000642248.1; ENST00000441708.2; ENST00000644867.1; ENST00000642886.1; ENST00000349496.11; ENST00000642426.1; ENST00000642836.1; ENST00000645900.1; ENST00000645493.1; ENST00000488914.2; ENST00000643052.1; ENST00000645320.1; ENST00000433400.6; ENST00000426215.5; ENST00000450969.6; ENST00000645276.1; ENST00000643992.1; ENST00000642986.1; ENST00000643977.1; ENST00000645982.1; ENST00000645210.1; ENST00000647021.1; ENST00000647390.1; ENST00000405570.6; ENST00000646074.1; ENST00000643865.1; ENST00000646369.1; ENST00000396185.8; ENST00000431914.6; ENST00000644873.1; ENST00000643031.1; ENST00000646116.1 | ||
| External Link | RMBase: m6A_site_584446 | ||
| mod ID: M6ASITE060220 | Click to Show/Hide the Full List | ||
| mod site | chr3:41224561-41224562:+ | [16] | |
| Sequence | GGACATGGCCATGGAACCAGACAGAAAAGCGGCTGTTAGTC | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa; brain | ||
| Seq Type List | m6A-seq; m6A-REF-seq | ||
| Transcript ID List | ENST00000433400.6; ENST00000642986.1; ENST00000644678.1; ENST00000488914.2; ENST00000646381.1; ENST00000643992.1; ENST00000642886.1; ENST00000426215.5; ENST00000646074.1; ENST00000441708.2; ENST00000643052.1; ENST00000643297.1; ENST00000646725.1; ENST00000643865.1; ENST00000645305.1; ENST00000396185.8; ENST00000642315.1; ENST00000644524.1; ENST00000645900.1; ENST00000431914.6; ENST00000645276.1; ENST00000645982.1; ENST00000643977.1; ENST00000349496.11; ENST00000647264.1; ENST00000453024.6; ENST00000450969.6; ENST00000405570.6; ENST00000642426.1; ENST00000647390.1; ENST00000644867.1; ENST00000644138.1; ENST00000646174.1; ENST00000645493.1; ENST00000645320.1; ENST00000645210.1; ENST00000647021.1; ENST00000642836.1; ENST00000643541.1; ENST00000396183.7; ENST00000642992.1; ENST00000646369.1; ENST00000647413.1; ENST00000646116.1; ENST00000644873.1; ENST00000643031.1; ENST00000642248.1 | ||
| External Link | RMBase: m6A_site_584447 | ||
| mod ID: M6ASITE060221 | Click to Show/Hide the Full List | ||
| mod site | chr3:41224592-41224593:+ | [18] | |
| Sequence | GCTGTTAGTCACTGGCAGCAACAGTCTTACCTGGACTCTGG | ||
| Motif Score | 2.173910714 | ||
| Cell/Tissue List | brain | ||
| Seq Type List | m6A-REF-seq | ||
| Transcript ID List | ENST00000645982.1; ENST00000642248.1; ENST00000642992.1; ENST00000645305.1; ENST00000647390.1; ENST00000645210.1; ENST00000431914.6; ENST00000426215.5; ENST00000645900.1; ENST00000646074.1; ENST00000642836.1; ENST00000642426.1; ENST00000643865.1; ENST00000646725.1; ENST00000433400.6; ENST00000396183.7; ENST00000488914.2; ENST00000644873.1; ENST00000643541.1; ENST00000405570.6; ENST00000647264.1; ENST00000646174.1; ENST00000441708.2; ENST00000642315.1; ENST00000643992.1; ENST00000644867.1; ENST00000647413.1; ENST00000644678.1; ENST00000643297.1; ENST00000644138.1; ENST00000643031.1; ENST00000647021.1; ENST00000642886.1; ENST00000349496.11; ENST00000646381.1; ENST00000645276.1; ENST00000643052.1; ENST00000643977.1; ENST00000453024.6; ENST00000644524.1; ENST00000642986.1; ENST00000396185.8; ENST00000450969.6; ENST00000646116.1; ENST00000645493.1; ENST00000646369.1; ENST00000645320.1 | ||
| External Link | RMBase: m6A_site_584448 | ||
| mod ID: M6ASITE060222 | Click to Show/Hide the Full List | ||
| mod site | chr3:41224606-41224607:+ | [16] | |
| Sequence | GCAGCAACAGTCTTACCTGGACTCTGGAATCCATTCTGGTG | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa; peripheral-blood | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000644873.1; ENST00000450969.6; ENST00000646116.1; ENST00000453024.6; ENST00000644138.1; ENST00000642986.1; ENST00000646074.1; ENST00000646369.1; ENST00000488914.2; ENST00000396185.8; ENST00000645982.1; ENST00000647390.1; ENST00000643977.1; ENST00000642315.1; ENST00000643031.1; ENST00000642426.1; ENST00000643865.1; ENST00000645305.1; ENST00000349496.11; ENST00000647021.1; ENST00000642886.1; ENST00000645900.1; ENST00000645210.1; ENST00000645276.1; ENST00000642836.1; ENST00000642248.1; ENST00000645493.1; ENST00000647264.1; ENST00000643297.1; ENST00000405570.6; ENST00000643052.1; ENST00000396183.7; ENST00000646381.1; ENST00000642992.1; ENST00000644678.1; ENST00000433400.6; ENST00000647413.1; ENST00000644524.1; ENST00000643541.1; ENST00000646174.1; ENST00000441708.2; ENST00000644867.1; ENST00000643992.1; ENST00000426215.5; ENST00000431914.6; ENST00000645320.1; ENST00000646725.1 | ||
| External Link | RMBase: m6A_site_584449 | ||
| mod ID: M6ASITE060223 | Click to Show/Hide the Full List | ||
| mod site | chr3:41224712-41224713:+ | [16] | |
| Sequence | CAAGTCCTGTATGAGTGGGAACAGGGATTTTCTCAGTCCTT | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa; kidney; Huh7; peripheral-blood; endometrial | ||
| Seq Type List | m6A-seq; m6A-REF-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000644138.1; ENST00000643992.1; ENST00000643977.1; ENST00000645305.1; ENST00000453024.6; ENST00000644524.1; ENST00000645900.1; ENST00000644873.1; ENST00000645982.1; ENST00000646074.1; ENST00000647021.1; ENST00000643865.1; ENST00000647390.1; ENST00000643031.1; ENST00000646369.1; ENST00000642836.1; ENST00000646381.1; ENST00000645493.1; ENST00000645210.1; ENST00000646725.1; ENST00000431914.6; ENST00000644867.1; ENST00000396185.8; ENST00000450969.6; ENST00000644678.1; ENST00000646116.1; ENST00000441708.2; ENST00000642248.1; ENST00000642315.1; ENST00000643297.1; ENST00000488914.2; ENST00000349496.11; ENST00000396183.7; ENST00000647413.1; ENST00000642426.1; ENST00000645276.1; ENST00000642992.1; ENST00000643052.1; ENST00000645320.1; ENST00000643541.1; ENST00000405570.6; ENST00000642886.1; ENST00000642986.1; ENST00000433400.6; ENST00000647264.1; ENST00000646174.1 | ||
| External Link | RMBase: m6A_site_584450 | ||
| mod ID: M6ASITE060224 | Click to Show/Hide the Full List | ||
| mod site | chr3:41224742-41224743:+ | [16] | |
| Sequence | TCTCAGTCCTTCACTCAAGAACAAGTAGCTGGTAAGAGTAT | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa; Huh7; endometrial | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000643992.1; ENST00000645900.1; ENST00000646116.1; ENST00000645305.1; ENST00000642426.1; ENST00000646074.1; ENST00000647390.1; ENST00000643977.1; ENST00000642836.1; ENST00000645276.1; ENST00000645210.1; ENST00000644867.1; ENST00000642886.1; ENST00000643031.1; ENST00000349496.11; ENST00000643541.1; ENST00000405570.6; ENST00000647264.1; ENST00000644524.1; ENST00000642248.1; ENST00000643865.1; ENST00000645320.1; ENST00000642992.1; ENST00000643297.1; ENST00000644138.1; ENST00000453024.6; ENST00000396183.7; ENST00000646381.1; ENST00000441708.2; ENST00000647413.1; ENST00000645982.1; ENST00000450969.6; ENST00000646725.1; ENST00000431914.6; ENST00000642315.1; ENST00000646369.1; ENST00000646174.1; ENST00000643052.1; ENST00000642986.1; ENST00000645493.1; ENST00000647021.1; ENST00000488914.2; ENST00000644678.1; ENST00000396185.8; ENST00000644873.1; ENST00000433400.6 | ||
| External Link | RMBase: m6A_site_584451 | ||
| mod ID: M6ASITE060225 | Click to Show/Hide the Full List | ||
| mod site | chr3:41224963-41224964:+ | [16] | |
| Sequence | CCTTTTCCAGATATTGATGGACAGTATGCAATGACTCGAGC | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; Huh7; endometrial | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000643031.1; ENST00000453024.6; ENST00000645210.1; ENST00000645982.1; ENST00000646074.1; ENST00000644867.1; ENST00000433400.6; ENST00000349496.11; ENST00000647413.1; ENST00000642886.1; ENST00000642248.1; ENST00000642992.1; ENST00000643297.1; ENST00000646174.1; ENST00000396183.7; ENST00000642315.1; ENST00000645305.1; ENST00000646381.1; ENST00000646725.1; ENST00000643052.1; ENST00000644524.1; ENST00000643541.1; ENST00000643977.1; ENST00000396185.8; ENST00000644873.1; ENST00000645276.1; ENST00000405570.6; ENST00000645493.1; ENST00000642986.1; ENST00000431914.6; ENST00000450969.6; ENST00000642836.1; ENST00000643865.1; ENST00000644138.1; ENST00000644678.1; ENST00000642426.1; ENST00000645320.1; ENST00000647390.1; ENST00000647264.1; ENST00000647021.1; ENST00000441708.2; ENST00000645900.1; ENST00000643992.1; ENST00000646369.1; ENST00000488914.2; ENST00000646116.1 | ||
| External Link | RMBase: m6A_site_584452 | ||
| mod ID: M6ASITE060226 | Click to Show/Hide the Full List | ||
| mod site | chr3:41225015-41225016:+ | [16] | |
| Sequence | GAGCTGCTATGTTCCCTGAGACATTAGATGAGGGCATGCAG | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa; hESC-HEK293T; Huh7; endometrial | ||
| Seq Type List | m6A-seq; MAZTER-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000645210.1; ENST00000645493.1; ENST00000431914.6; ENST00000396185.8; ENST00000643031.1; ENST00000646116.1; ENST00000433400.6; ENST00000642836.1; ENST00000643977.1; ENST00000642886.1; ENST00000647390.1; ENST00000349496.11; ENST00000644867.1; ENST00000642992.1; ENST00000644678.1; ENST00000644873.1; ENST00000643541.1; ENST00000645982.1; ENST00000642986.1; ENST00000396183.7; ENST00000644138.1; ENST00000453024.6; ENST00000645305.1; ENST00000646074.1; ENST00000441708.2; ENST00000643297.1; ENST00000405570.6; ENST00000646369.1; ENST00000450969.6; ENST00000647264.1; ENST00000643865.1; ENST00000642248.1; ENST00000644524.1; ENST00000646381.1; ENST00000645900.1; ENST00000647021.1; ENST00000642426.1; ENST00000645320.1; ENST00000643992.1; ENST00000645276.1; ENST00000488914.2; ENST00000643052.1; ENST00000642315.1; ENST00000646174.1; ENST00000647413.1; ENST00000646725.1 | ||
| External Link | RMBase: m6A_site_584453 | ||
| mod ID: M6ASITE060227 | Click to Show/Hide the Full List | ||
| mod site | chr3:41225045-41225046:+ | [17] | |
| Sequence | AGGGCATGCAGATCCCATCTACACAGTTTGATGCTGCTCAT | ||
| Motif Score | 2.078666667 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000431914.6; ENST00000642315.1; ENST00000644867.1; ENST00000642248.1; ENST00000644138.1; ENST00000396183.7; ENST00000646369.1; ENST00000643977.1; ENST00000396185.8; ENST00000645900.1; ENST00000453024.6; ENST00000647264.1; ENST00000642836.1; ENST00000450969.6; ENST00000646725.1; ENST00000488914.2; ENST00000643297.1; ENST00000643865.1; ENST00000646174.1; ENST00000642992.1; ENST00000643031.1; ENST00000642986.1; ENST00000645210.1; ENST00000646074.1; ENST00000645493.1; ENST00000645320.1; ENST00000405570.6; ENST00000647390.1; ENST00000643541.1; ENST00000646116.1; ENST00000441708.2; ENST00000644524.1; ENST00000646381.1; ENST00000643992.1; ENST00000642886.1; ENST00000349496.11; ENST00000645276.1; ENST00000645982.1; ENST00000645305.1; ENST00000643052.1; ENST00000644873.1; ENST00000642426.1; ENST00000644678.1; ENST00000647021.1; ENST00000433400.6; ENST00000647413.1 | ||
| External Link | RMBase: m6A_site_584454 | ||
| mod ID: M6ASITE060228 | Click to Show/Hide the Full List | ||
| mod site | chr3:41225092-41225093:+ | [16] | |
| Sequence | AATGTCCAGCGTTTGGCTGAACCATCACAGATGCTGAAACA | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | HeLa; Huh7; endometrial | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000643992.1; ENST00000647390.1; ENST00000644138.1; ENST00000643052.1; ENST00000645493.1; ENST00000433400.6; ENST00000643977.1; ENST00000431914.6; ENST00000642836.1; ENST00000644873.1; ENST00000647021.1; ENST00000645982.1; ENST00000645276.1; ENST00000642886.1; ENST00000453024.6; ENST00000488914.2; ENST00000644867.1; ENST00000646116.1; ENST00000645305.1; ENST00000642248.1; ENST00000647264.1; ENST00000643031.1; ENST00000647413.1; ENST00000643541.1; ENST00000396185.8; ENST00000643865.1; ENST00000646725.1; ENST00000396183.7; ENST00000405570.6; ENST00000349496.11; ENST00000643297.1; ENST00000646369.1; ENST00000646174.1; ENST00000441708.2; ENST00000642986.1; ENST00000646381.1; ENST00000645900.1; ENST00000450969.6; ENST00000645320.1; ENST00000644678.1; ENST00000646074.1; ENST00000642426.1; ENST00000644524.1; ENST00000645210.1; ENST00000642315.1; ENST00000642992.1 | ||
| External Link | RMBase: m6A_site_584455 | ||
| mod ID: M6ASITE060229 | Click to Show/Hide the Full List | ||
| mod site | chr3:41225098-41225099:+ | [18] | |
| Sequence | CAGCGTTTGGCTGAACCATCACAGATGCTGAAACATGCAGT | ||
| Motif Score | 2.047297619 | ||
| Cell/Tissue List | kidney | ||
| Seq Type List | m6A-REF-seq | ||
| Transcript ID List | ENST00000642248.1; ENST00000643297.1; ENST00000644678.1; ENST00000643031.1; ENST00000647413.1; ENST00000645305.1; ENST00000645276.1; ENST00000642426.1; ENST00000441708.2; ENST00000643541.1; ENST00000431914.6; ENST00000405570.6; ENST00000646174.1; ENST00000642315.1; ENST00000488914.2; ENST00000643977.1; ENST00000646369.1; ENST00000642986.1; ENST00000642992.1; ENST00000643052.1; ENST00000645900.1; ENST00000644138.1; ENST00000646725.1; ENST00000644867.1; ENST00000646381.1; ENST00000642886.1; ENST00000644524.1; ENST00000450969.6; ENST00000396185.8; ENST00000643992.1; ENST00000643865.1; ENST00000647390.1; ENST00000642836.1; ENST00000646074.1; ENST00000645320.1; ENST00000645493.1; ENST00000645210.1; ENST00000644873.1; ENST00000645982.1; ENST00000396183.7; ENST00000646116.1; ENST00000647021.1; ENST00000453024.6; ENST00000433400.6; ENST00000647264.1; ENST00000349496.11 | ||
| External Link | RMBase: m6A_site_584456 | ||
| mod ID: M6ASITE060230 | Click to Show/Hide the Full List | ||
| mod site | chr3:41225110-41225111:+ | [16] | |
| Sequence | GAACCATCACAGATGCTGAAACATGCAGTTGTAAACTTGAT | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa; hESC-HEK293T; Huh7; endometrial | ||
| Seq Type List | m6A-seq; MAZTER-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000642886.1; ENST00000646074.1; ENST00000645276.1; ENST00000647390.1; ENST00000643992.1; ENST00000645982.1; ENST00000643297.1; ENST00000645900.1; ENST00000647021.1; ENST00000643977.1; ENST00000453024.6; ENST00000488914.2; ENST00000644138.1; ENST00000647264.1; ENST00000644867.1; ENST00000642315.1; ENST00000644873.1; ENST00000642986.1; ENST00000643865.1; ENST00000642248.1; ENST00000646116.1; ENST00000644524.1; ENST00000642992.1; ENST00000645210.1; ENST00000643031.1; ENST00000646725.1; ENST00000643541.1; ENST00000396185.8; ENST00000441708.2; ENST00000646381.1; ENST00000646174.1; ENST00000433400.6; ENST00000349496.11; ENST00000396183.7; ENST00000642426.1; ENST00000646369.1; ENST00000644678.1; ENST00000405570.6; ENST00000645493.1; ENST00000645305.1; ENST00000642836.1; ENST00000643052.1; ENST00000431914.6; ENST00000647413.1; ENST00000645320.1; ENST00000450969.6 | ||
| External Link | RMBase: m6A_site_584457 | ||
| mod ID: M6ASITE060231 | Click to Show/Hide the Full List | ||
| mod site | chr3:41225124-41225125:+ | [16] | |
| Sequence | GCTGAAACATGCAGTTGTAAACTTGATTAACTATCAAGATG | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa; Huh7 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000488914.2; ENST00000441708.2; ENST00000645982.1; ENST00000643297.1; ENST00000646725.1; ENST00000645305.1; ENST00000645276.1; ENST00000645210.1; ENST00000646074.1; ENST00000644138.1; ENST00000644867.1; ENST00000396183.7; ENST00000642248.1; ENST00000644524.1; ENST00000643992.1; ENST00000646116.1; ENST00000642315.1; ENST00000647264.1; ENST00000645320.1; ENST00000646369.1; ENST00000643541.1; ENST00000647413.1; ENST00000349496.11; ENST00000642836.1; ENST00000431914.6; ENST00000642426.1; ENST00000450969.6; ENST00000453024.6; ENST00000644873.1; ENST00000643031.1; ENST00000644678.1; ENST00000646381.1; ENST00000642886.1; ENST00000433400.6; ENST00000396185.8; ENST00000645900.1; ENST00000642986.1; ENST00000643052.1; ENST00000646174.1; ENST00000643977.1; ENST00000645493.1; ENST00000643865.1; ENST00000647390.1; ENST00000405570.6; ENST00000642992.1; ENST00000647021.1 | ||
| External Link | RMBase: m6A_site_584458 | ||
| mod ID: M6ASITE060232 | Click to Show/Hide the Full List | ||
| mod site | chr3:41225152-41225153:+ | [16] | |
| Sequence | AACTATCAAGATGATGCAGAACTTGCCACACGTGCAATCCC | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa; Huh7 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000647021.1; ENST00000643541.1; ENST00000645320.1; ENST00000450969.6; ENST00000646381.1; ENST00000646725.1; ENST00000646369.1; ENST00000396185.8; ENST00000644873.1; ENST00000453024.6; ENST00000431914.6; ENST00000645493.1; ENST00000645305.1; ENST00000405570.6; ENST00000647264.1; ENST00000645210.1; ENST00000644678.1; ENST00000642315.1; ENST00000642836.1; ENST00000646116.1; ENST00000433400.6; ENST00000642248.1; ENST00000642426.1; ENST00000643992.1; ENST00000645276.1; ENST00000644524.1; ENST00000646174.1; ENST00000647390.1; ENST00000643297.1; ENST00000644138.1; ENST00000643977.1; ENST00000647413.1; ENST00000643865.1; ENST00000642986.1; ENST00000643031.1; ENST00000644867.1; ENST00000642992.1; ENST00000643052.1; ENST00000488914.2; ENST00000645900.1; ENST00000349496.11; ENST00000441708.2; ENST00000645982.1; ENST00000646074.1; ENST00000642886.1; ENST00000396183.7 | ||
| External Link | RMBase: m6A_site_584459 | ||
| mod ID: M6ASITE060233 | Click to Show/Hide the Full List | ||
| mod site | chr3:41225159-41225160:+ | [17] | |
| Sequence | AAGATGATGCAGAACTTGCCACACGTGCAATCCCTGAACTG | ||
| Motif Score | 2.053113095 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000433400.6; ENST00000646174.1; ENST00000645305.1; ENST00000642248.1; ENST00000645493.1; ENST00000646725.1; ENST00000643865.1; ENST00000647390.1; ENST00000644678.1; ENST00000646369.1; ENST00000488914.2; ENST00000349496.11; ENST00000396183.7; ENST00000646074.1; ENST00000647264.1; ENST00000642315.1; ENST00000642886.1; ENST00000642992.1; ENST00000643031.1; ENST00000645320.1; ENST00000647413.1; ENST00000431914.6; ENST00000646116.1; ENST00000645276.1; ENST00000643977.1; ENST00000643297.1; ENST00000642986.1; ENST00000450969.6; ENST00000643992.1; ENST00000645982.1; ENST00000644867.1; ENST00000642426.1; ENST00000644138.1; ENST00000647021.1; ENST00000644524.1; ENST00000453024.6; ENST00000643541.1; ENST00000645210.1; ENST00000405570.6; ENST00000644873.1; ENST00000642836.1; ENST00000643052.1; ENST00000645900.1; ENST00000646381.1; ENST00000396185.8; ENST00000441708.2 | ||
| External Link | RMBase: m6A_site_584460 | ||
| mod ID: M6ASITE060234 | Click to Show/Hide the Full List | ||
| mod site | chr3:41225176-41225177:+ | [16] | |
| Sequence | GCCACACGTGCAATCCCTGAACTGACAAAACTGCTAAATGA | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa; Huh7 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000642315.1; ENST00000642426.1; ENST00000396185.8; ENST00000645210.1; ENST00000431914.6; ENST00000644138.1; ENST00000643992.1; ENST00000433400.6; ENST00000644524.1; ENST00000450969.6; ENST00000643052.1; ENST00000644678.1; ENST00000441708.2; ENST00000642836.1; ENST00000645982.1; ENST00000643541.1; ENST00000646381.1; ENST00000488914.2; ENST00000645276.1; ENST00000643977.1; ENST00000642992.1; ENST00000396183.7; ENST00000646174.1; ENST00000645320.1; ENST00000349496.11; ENST00000644873.1; ENST00000405570.6; ENST00000647413.1; ENST00000645305.1; ENST00000643031.1; ENST00000646116.1; ENST00000642986.1; ENST00000646074.1; ENST00000642248.1; ENST00000647021.1; ENST00000647390.1; ENST00000453024.6; ENST00000643865.1; ENST00000645493.1; ENST00000644867.1; ENST00000646725.1; ENST00000643297.1; ENST00000647264.1; ENST00000642886.1; ENST00000646369.1; ENST00000645900.1 | ||
| External Link | RMBase: m6A_site_584461 | ||
| mod ID: M6ASITE060235 | Click to Show/Hide the Full List | ||
| mod site | chr3:41225180-41225181:+ | [17] | |
| Sequence | CACGTGCAATCCCTGAACTGACAAAACTGCTAAATGACGAG | ||
| Motif Score | 2.859755952 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000643992.1; ENST00000647413.1; ENST00000647021.1; ENST00000647390.1; ENST00000644867.1; ENST00000433400.6; ENST00000405570.6; ENST00000645900.1; ENST00000396183.7; ENST00000642886.1; ENST00000643977.1; ENST00000450969.6; ENST00000644678.1; ENST00000643865.1; ENST00000396185.8; ENST00000431914.6; ENST00000642986.1; ENST00000646725.1; ENST00000643031.1; ENST00000646074.1; ENST00000646381.1; ENST00000349496.11; ENST00000642992.1; ENST00000643052.1; ENST00000644524.1; ENST00000642248.1; ENST00000642315.1; ENST00000645982.1; ENST00000644873.1; ENST00000644138.1; ENST00000642836.1; ENST00000646369.1; ENST00000488914.2; ENST00000643297.1; ENST00000645276.1; ENST00000645210.1; ENST00000643541.1; ENST00000645493.1; ENST00000642426.1; ENST00000645320.1; ENST00000646116.1; ENST00000647264.1; ENST00000441708.2; ENST00000453024.6; ENST00000645305.1; ENST00000646174.1 | ||
| External Link | RMBase: m6A_site_584462 | ||
| mod ID: M6ASITE060236 | Click to Show/Hide the Full List | ||
| mod site | chr3:41225185-41225186:+ | [16] | |
| Sequence | GCAATCCCTGAACTGACAAAACTGCTAAATGACGAGGACCA | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa; Huh7 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000646381.1; ENST00000646074.1; ENST00000645320.1; ENST00000645493.1; ENST00000433400.6; ENST00000647413.1; ENST00000441708.2; ENST00000646174.1; ENST00000644867.1; ENST00000396183.7; ENST00000647264.1; ENST00000450969.6; ENST00000643031.1; ENST00000645900.1; ENST00000643865.1; ENST00000645982.1; ENST00000645210.1; ENST00000642992.1; ENST00000644678.1; ENST00000642426.1; ENST00000643541.1; ENST00000646369.1; ENST00000645276.1; ENST00000642836.1; ENST00000643977.1; ENST00000396185.8; ENST00000405570.6; ENST00000644873.1; ENST00000647021.1; ENST00000646116.1; ENST00000642248.1; ENST00000453024.6; ENST00000643992.1; ENST00000642886.1; ENST00000349496.11; ENST00000645305.1; ENST00000642986.1; ENST00000643052.1; ENST00000647390.1; ENST00000431914.6; ENST00000488914.2; ENST00000646725.1; ENST00000642315.1; ENST00000644524.1; ENST00000644138.1; ENST00000643297.1 | ||
| External Link | RMBase: m6A_site_584463 | ||
| mod ID: M6ASITE060237 | Click to Show/Hide the Full List | ||
| mod site | chr3:41225202-41225203:+ | [16] | |
| Sequence | AAAACTGCTAAATGACGAGGACCAGGTAAGCAATGACATAG | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; Huh7 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000643031.1; ENST00000642248.1; ENST00000646074.1; ENST00000644678.1; ENST00000643865.1; ENST00000644873.1; ENST00000645493.1; ENST00000646116.1; ENST00000645982.1; ENST00000642426.1; ENST00000647413.1; ENST00000645320.1; ENST00000642886.1; ENST00000396185.8; ENST00000646725.1; ENST00000643052.1; ENST00000643297.1; ENST00000647264.1; ENST00000642986.1; ENST00000396183.7; ENST00000645305.1; ENST00000643541.1; ENST00000453024.6; ENST00000644524.1; ENST00000642315.1; ENST00000644867.1; ENST00000642992.1; ENST00000646174.1; ENST00000643977.1; ENST00000441708.2; ENST00000643992.1; ENST00000646381.1; ENST00000450969.6; ENST00000647021.1; ENST00000433400.6; ENST00000405570.6; ENST00000645900.1; ENST00000349496.11; ENST00000642836.1; ENST00000647390.1; ENST00000645276.1; ENST00000431914.6; ENST00000646369.1; ENST00000644138.1; ENST00000645210.1 | ||
| External Link | RMBase: m6A_site_584464 | ||
| mod ID: M6ASITE060238 | Click to Show/Hide the Full List | ||
| mod site | chr3:41225392-41225393:+ | [16] | |
| Sequence | TCTAAAAAGGAAGCTTCCAGACACGCTATCATGCGTTCTCC | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa; hESC-HEK293T | ||
| Seq Type List | m6A-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000645493.1; ENST00000642992.1; ENST00000642315.1; ENST00000433400.6; ENST00000643865.1; ENST00000642836.1; ENST00000644867.1; ENST00000642986.1; ENST00000642248.1; ENST00000646725.1; ENST00000431914.6; ENST00000645320.1; ENST00000646369.1; ENST00000644873.1; ENST00000646116.1; ENST00000642886.1; ENST00000647390.1; ENST00000643541.1; ENST00000643297.1; ENST00000645210.1; ENST00000644524.1; ENST00000645982.1; ENST00000643992.1; ENST00000441708.2; ENST00000647264.1; ENST00000643031.1; ENST00000450969.6; ENST00000644678.1; ENST00000645305.1; ENST00000644138.1; ENST00000396183.7; ENST00000642426.1; ENST00000647413.1; ENST00000646174.1; ENST00000643977.1; ENST00000396185.8; ENST00000405570.6; ENST00000647021.1; ENST00000349496.11; ENST00000643052.1; ENST00000646074.1; ENST00000645276.1; ENST00000645900.1; ENST00000646381.1; ENST00000453024.6 | ||
| External Link | RMBase: m6A_site_584465 | ||
| mod ID: M6ASITE060239 | Click to Show/Hide the Full List | ||
| mod site | chr3:41225450-41225451:+ | [17] | |
| Sequence | TTGTACGTACCATGCAGAATACAAATGATGTAGAAACAGCT | ||
| Motif Score | 2.110482143 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000349496.11; ENST00000647413.1; ENST00000644138.1; ENST00000646174.1; ENST00000642836.1; ENST00000441708.2; ENST00000645900.1; ENST00000396185.8; ENST00000645982.1; ENST00000643992.1; ENST00000646381.1; ENST00000431914.6; ENST00000433400.6; ENST00000644524.1; ENST00000642886.1; ENST00000643031.1; ENST00000647264.1; ENST00000396183.7; ENST00000644678.1; ENST00000645493.1; ENST00000644867.1; ENST00000646074.1; ENST00000643297.1; ENST00000645305.1; ENST00000647390.1; ENST00000645276.1; ENST00000646116.1; ENST00000642315.1; ENST00000644873.1; ENST00000643541.1; ENST00000647021.1; ENST00000642426.1; ENST00000645320.1; ENST00000642248.1; ENST00000643865.1; ENST00000405570.6; ENST00000642986.1; ENST00000450969.6; ENST00000453024.6; ENST00000646725.1; ENST00000645210.1; ENST00000646369.1; ENST00000642992.1; ENST00000643052.1; ENST00000643977.1 | ||
| External Link | RMBase: m6A_site_584466 | ||
| mod ID: M6ASITE060240 | Click to Show/Hide the Full List | ||
| mod site | chr3:41225465-41225466:+ | [16] | |
| Sequence | AGAATACAAATGATGTAGAAACAGCTCGTTGTACCGCTGGG | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000643865.1; ENST00000643052.1; ENST00000646381.1; ENST00000396183.7; ENST00000433400.6; ENST00000441708.2; ENST00000646725.1; ENST00000645305.1; ENST00000644678.1; ENST00000646116.1; ENST00000643977.1; ENST00000646369.1; ENST00000644138.1; ENST00000645982.1; ENST00000450969.6; ENST00000643541.1; ENST00000647413.1; ENST00000647264.1; ENST00000453024.6; ENST00000645900.1; ENST00000646074.1; ENST00000644524.1; ENST00000645276.1; ENST00000642836.1; ENST00000647390.1; ENST00000431914.6; ENST00000642426.1; ENST00000642315.1; ENST00000644867.1; ENST00000642886.1; ENST00000645210.1; ENST00000645320.1; ENST00000643992.1; ENST00000349496.11; ENST00000646174.1; ENST00000644873.1; ENST00000645493.1; ENST00000396185.8; ENST00000642986.1; ENST00000642248.1; ENST00000405570.6; ENST00000642992.1; ENST00000647021.1; ENST00000643031.1; ENST00000643297.1 | ||
| External Link | RMBase: m6A_site_584467 | ||
| mod ID: M6ASITE060241 | Click to Show/Hide the Full List | ||
| mod site | chr3:41225486-41225487:+ | [16] | |
| Sequence | CAGCTCGTTGTACCGCTGGGACCTTGCATAACCTTTCCCAT | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000647390.1; ENST00000644678.1; ENST00000643865.1; ENST00000645900.1; ENST00000645493.1; ENST00000643977.1; ENST00000644524.1; ENST00000645276.1; ENST00000646725.1; ENST00000642836.1; ENST00000396185.8; ENST00000441708.2; ENST00000644138.1; ENST00000646074.1; ENST00000645320.1; ENST00000644867.1; ENST00000643031.1; ENST00000645305.1; ENST00000396183.7; ENST00000646116.1; ENST00000433400.6; ENST00000646174.1; ENST00000643052.1; ENST00000453024.6; ENST00000643541.1; ENST00000349496.11; ENST00000642248.1; ENST00000642886.1; ENST00000642992.1; ENST00000647413.1; ENST00000647264.1; ENST00000642426.1; ENST00000431914.6; ENST00000642986.1; ENST00000643297.1; ENST00000450969.6; ENST00000644873.1; ENST00000647021.1; ENST00000642315.1; ENST00000643992.1; ENST00000645210.1; ENST00000646381.1; ENST00000646369.1; ENST00000405570.6; ENST00000645982.1 | ||
| External Link | RMBase: m6A_site_584468 | ||
| mod ID: M6ASITE060242 | Click to Show/Hide the Full List | ||
| mod site | chr3:41225693-41225694:+ | [17] | |
| Sequence | CTGTGTTGTTTTATGCCATTACAACTCTCCACAACCTTTTA | ||
| Motif Score | 2.07285119 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000645210.1; ENST00000642315.1; ENST00000645493.1; ENST00000643052.1; ENST00000646074.1; ENST00000643297.1; ENST00000644524.1; ENST00000645305.1; ENST00000646174.1; ENST00000646116.1; ENST00000450969.6; ENST00000396185.8; ENST00000645320.1; ENST00000349496.11; ENST00000642986.1; ENST00000644867.1; ENST00000643977.1; ENST00000642836.1; ENST00000644873.1; ENST00000647413.1; ENST00000433400.6; ENST00000643541.1; ENST00000644138.1; ENST00000431914.6; ENST00000643031.1; ENST00000642426.1; ENST00000646725.1; ENST00000646369.1; ENST00000646381.1; ENST00000642992.1; ENST00000645900.1; ENST00000405570.6; ENST00000441708.2; ENST00000647021.1; ENST00000642886.1; ENST00000644678.1; ENST00000647390.1; ENST00000642248.1; ENST00000453024.6; ENST00000643865.1; ENST00000643992.1; ENST00000645982.1; ENST00000396183.7; ENST00000645276.1; ENST00000647264.1 | ||
| External Link | RMBase: m6A_site_584469 | ||
| mod ID: M6ASITE060243 | Click to Show/Hide the Full List | ||
| mod site | chr3:41225716-41225717:+ | [19] | |
| Sequence | ACTCTCCACAACCTTTTATTACATCAAGAAGGAGCTAAAAT | ||
| Motif Score | 2.07285119 | ||
| Cell/Tissue List | HEK293T; hESC-HEK293T | ||
| Seq Type List | DART-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000450969.6; ENST00000644678.1; ENST00000645276.1; ENST00000645305.1; ENST00000642426.1; ENST00000642986.1; ENST00000646369.1; ENST00000644524.1; ENST00000646725.1; ENST00000647021.1; ENST00000642836.1; ENST00000647264.1; ENST00000643297.1; ENST00000396185.8; ENST00000647413.1; ENST00000642886.1; ENST00000647390.1; ENST00000642248.1; ENST00000642315.1; ENST00000644138.1; ENST00000349496.11; ENST00000643977.1; ENST00000645210.1; ENST00000645320.1; ENST00000645982.1; ENST00000646074.1; ENST00000643992.1; ENST00000642992.1; ENST00000644867.1; ENST00000646381.1; ENST00000643865.1; ENST00000643031.1; ENST00000441708.2; ENST00000453024.6; ENST00000643541.1; ENST00000405570.6; ENST00000396183.7; ENST00000646174.1; ENST00000643052.1; ENST00000645900.1; ENST00000431914.6; ENST00000645493.1; ENST00000646116.1; ENST00000433400.6; ENST00000644873.1 | ||
| External Link | RMBase: m6A_site_584470 | ||
| mod ID: M6ASITE060244 | Click to Show/Hide the Full List | ||
| mod site | chr3:41225784-41225785:+ | [17] | |
| Sequence | GAAAATGGTTGCCTTGCTCAACAAAACAAATGTTAAATTCT | ||
| Motif Score | 2.173910714 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000646725.1; ENST00000405570.6; ENST00000642886.1; ENST00000644524.1; ENST00000645900.1; ENST00000643297.1; ENST00000642836.1; ENST00000453024.6; ENST00000642248.1; ENST00000644678.1; ENST00000644873.1; ENST00000645493.1; ENST00000645210.1; ENST00000646074.1; ENST00000646116.1; ENST00000642426.1; ENST00000646369.1; ENST00000396185.8; ENST00000643031.1; ENST00000642992.1; ENST00000431914.6; ENST00000433400.6; ENST00000645982.1; ENST00000643977.1; ENST00000645276.1; ENST00000647413.1; ENST00000647264.1; ENST00000643992.1; ENST00000643541.1; ENST00000644867.1; ENST00000643052.1; ENST00000645305.1; ENST00000441708.2; ENST00000644138.1; ENST00000642986.1; ENST00000642315.1; ENST00000643865.1; ENST00000450969.6; ENST00000396183.7; ENST00000646381.1; ENST00000647021.1; ENST00000647390.1; ENST00000645320.1; ENST00000646174.1; ENST00000349496.11 | ||
| External Link | RMBase: m6A_site_584471 | ||
| mod ID: M6ASITE060245 | Click to Show/Hide the Full List | ||
| mod site | chr3:41225789-41225790:+ | [16] | |
| Sequence | TGGTTGCCTTGCTCAACAAAACAAATGTTAAATTCTTGGCT | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000644138.1; ENST00000642992.1; ENST00000405570.6; ENST00000643297.1; ENST00000441708.2; ENST00000431914.6; ENST00000645320.1; ENST00000396183.7; ENST00000642836.1; ENST00000643977.1; ENST00000643031.1; ENST00000643992.1; ENST00000396185.8; ENST00000647413.1; ENST00000645305.1; ENST00000646174.1; ENST00000646369.1; ENST00000645210.1; ENST00000646116.1; ENST00000647390.1; ENST00000642248.1; ENST00000644867.1; ENST00000647021.1; ENST00000647264.1; ENST00000453024.6; ENST00000643052.1; ENST00000646074.1; ENST00000645900.1; ENST00000645276.1; ENST00000349496.11; ENST00000645982.1; ENST00000642986.1; ENST00000645493.1; ENST00000644873.1; ENST00000450969.6; ENST00000646381.1; ENST00000646725.1; ENST00000642426.1; ENST00000644678.1; ENST00000433400.6; ENST00000642315.1; ENST00000643541.1; ENST00000644524.1; ENST00000642886.1; ENST00000643865.1 | ||
| External Link | RMBase: m6A_site_584472 | ||
| mod ID: M6ASITE060246 | Click to Show/Hide the Full List | ||
| mod site | chr3:41225816-41225817:+ | [18] | |
| Sequence | TTAAATTCTTGGCTATTACGACAGACTGCCTTCAAATTTTA | ||
| Motif Score | 2.865571429 | ||
| Cell/Tissue List | liver | ||
| Seq Type List | m6A-REF-seq | ||
| Transcript ID List | ENST00000646725.1; ENST00000349496.11; ENST00000645982.1; ENST00000642886.1; ENST00000645493.1; ENST00000396183.7; ENST00000642986.1; ENST00000645305.1; ENST00000642836.1; ENST00000643297.1; ENST00000642426.1; ENST00000441708.2; ENST00000643031.1; ENST00000646369.1; ENST00000644867.1; ENST00000647390.1; ENST00000645900.1; ENST00000643052.1; ENST00000646116.1; ENST00000642992.1; ENST00000405570.6; ENST00000646074.1; ENST00000646381.1; ENST00000453024.6; ENST00000450969.6; ENST00000647413.1; ENST00000645320.1; ENST00000644873.1; ENST00000396185.8; ENST00000644678.1; ENST00000643865.1; ENST00000643541.1; ENST00000645210.1; ENST00000646174.1; ENST00000431914.6; ENST00000642248.1; ENST00000643992.1; ENST00000643977.1; ENST00000644138.1; ENST00000645276.1; ENST00000647021.1; ENST00000644524.1; ENST00000642315.1; ENST00000433400.6; ENST00000647264.1 | ||
| External Link | RMBase: m6A_site_584473 | ||
| mod ID: M6ASITE060247 | Click to Show/Hide the Full List | ||
| mod site | chr3:41225820-41225821:+ | [16] | |
| Sequence | ATTCTTGGCTATTACGACAGACTGCCTTCAAATTTTAGCTT | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000644524.1; ENST00000645982.1; ENST00000453024.6; ENST00000405570.6; ENST00000646174.1; ENST00000644873.1; ENST00000433400.6; ENST00000643031.1; ENST00000646369.1; ENST00000645210.1; ENST00000643977.1; ENST00000642248.1; ENST00000643865.1; ENST00000643297.1; ENST00000647264.1; ENST00000642426.1; ENST00000642886.1; ENST00000396183.7; ENST00000646074.1; ENST00000644138.1; ENST00000441708.2; ENST00000643992.1; ENST00000642992.1; ENST00000646725.1; ENST00000645493.1; ENST00000646116.1; ENST00000642315.1; ENST00000644678.1; ENST00000643052.1; ENST00000646381.1; ENST00000645320.1; ENST00000642836.1; ENST00000647413.1; ENST00000645900.1; ENST00000644867.1; ENST00000643541.1; ENST00000647021.1; ENST00000450969.6; ENST00000396185.8; ENST00000642986.1; ENST00000647390.1; ENST00000645305.1; ENST00000349496.11; ENST00000645276.1; ENST00000431914.6 | ||
| External Link | RMBase: m6A_site_584474 | ||
| mod ID: M6ASITE060248 | Click to Show/Hide the Full List | ||
| mod site | chr3:41227230-41227231:+ | [16] | |
| Sequence | ATCATACTGGCTAGTGGTGGACCCCAAGCTTTAGTAAATAT | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000643541.1; ENST00000396185.8; ENST00000643031.1; ENST00000644873.1; ENST00000645982.1; ENST00000646074.1; ENST00000642248.1; ENST00000643992.1; ENST00000642836.1; ENST00000441708.2; ENST00000644678.1; ENST00000646116.1; ENST00000647390.1; ENST00000396183.7; ENST00000646174.1; ENST00000643865.1; ENST00000349496.11; ENST00000645276.1; ENST00000433400.6; ENST00000644524.1; ENST00000642426.1; ENST00000646369.1; ENST00000643297.1; ENST00000645320.1; ENST00000453024.6; ENST00000647021.1; ENST00000642992.1; ENST00000405570.6; ENST00000645305.1; ENST00000647413.1; ENST00000450969.6; ENST00000643052.1; ENST00000645900.1; ENST00000647264.1; ENST00000431914.6; ENST00000642315.1; ENST00000646381.1; ENST00000645210.1; ENST00000642986.1; ENST00000642886.1; ENST00000644138.1; ENST00000644867.1; ENST00000646725.1; ENST00000645493.1; ENST00000643977.1 | ||
| External Link | RMBase: m6A_site_584475 | ||
| mod ID: M6ASITE060249 | Click to Show/Hide the Full List | ||
| mod site | chr3:41227258-41227259:+ | [16] | |
| Sequence | CTTTAGTAAATATAATGAGGACCTATACTTACGAAAAACTA | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000642992.1; ENST00000644678.1; ENST00000431914.6; ENST00000405570.6; ENST00000645982.1; ENST00000643052.1; ENST00000643031.1; ENST00000453024.6; ENST00000645305.1; ENST00000644524.1; ENST00000647264.1; ENST00000645493.1; ENST00000450969.6; ENST00000645276.1; ENST00000433400.6; ENST00000642315.1; ENST00000647413.1; ENST00000396183.7; ENST00000643977.1; ENST00000643865.1; ENST00000441708.2; ENST00000644873.1; ENST00000647021.1; ENST00000642248.1; ENST00000642426.1; ENST00000644867.1; ENST00000646116.1; ENST00000643297.1; ENST00000642886.1; ENST00000644138.1; ENST00000646725.1; ENST00000646174.1; ENST00000396185.8; ENST00000645927.1; ENST00000645210.1; ENST00000646369.1; ENST00000643992.1; ENST00000647390.1; ENST00000645900.1; ENST00000643541.1; ENST00000646381.1; ENST00000642836.1; ENST00000646074.1; ENST00000349496.11; ENST00000642986.1; ENST00000645320.1 | ||
| External Link | RMBase: m6A_site_584476 | ||
| mod ID: M6ASITE060250 | Click to Show/Hide the Full List | ||
| mod site | chr3:41227275-41227276:+ | [16] | |
| Sequence | AGGACCTATACTTACGAAAAACTACTGTGGACCACAAGCAG | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000647264.1; ENST00000431914.6; ENST00000643541.1; ENST00000643992.1; ENST00000450969.6; ENST00000642248.1; ENST00000644867.1; ENST00000349496.11; ENST00000642986.1; ENST00000645493.1; ENST00000643977.1; ENST00000642836.1; ENST00000642426.1; ENST00000645305.1; ENST00000453024.6; ENST00000643865.1; ENST00000647390.1; ENST00000644873.1; ENST00000405570.6; ENST00000646381.1; ENST00000644524.1; ENST00000642886.1; ENST00000643297.1; ENST00000645210.1; ENST00000646725.1; ENST00000644138.1; ENST00000433400.6; ENST00000396185.8; ENST00000441708.2; ENST00000645276.1; ENST00000642992.1; ENST00000645900.1; ENST00000646174.1; ENST00000647021.1; ENST00000645320.1; ENST00000647413.1; ENST00000642315.1; ENST00000644678.1; ENST00000643052.1; ENST00000646074.1; ENST00000396183.7; ENST00000643031.1; ENST00000645927.1; ENST00000646369.1; ENST00000645982.1; ENST00000646116.1 | ||
| External Link | RMBase: m6A_site_584477 | ||
| mod ID: M6ASITE060251 | Click to Show/Hide the Full List | ||
| mod site | chr3:41227285-41227286:+ | [16] | |
| Sequence | CTTACGAAAAACTACTGTGGACCACAAGCAGAGTGCTGAAG | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; HEK293T | ||
| Seq Type List | m6A-seq; DART-seq | ||
| Transcript ID List | ENST00000643541.1; ENST00000450969.6; ENST00000644524.1; ENST00000647413.1; ENST00000645927.1; ENST00000643052.1; ENST00000433400.6; ENST00000396185.8; ENST00000396183.7; ENST00000646074.1; ENST00000647021.1; ENST00000643297.1; ENST00000642886.1; ENST00000642836.1; ENST00000645305.1; ENST00000642986.1; ENST00000647264.1; ENST00000644678.1; ENST00000642426.1; ENST00000642992.1; ENST00000645276.1; ENST00000441708.2; ENST00000645320.1; ENST00000645982.1; ENST00000643865.1; ENST00000431914.6; ENST00000642248.1; ENST00000646381.1; ENST00000645900.1; ENST00000453024.6; ENST00000643992.1; ENST00000405570.6; ENST00000644873.1; ENST00000647390.1; ENST00000645493.1; ENST00000646369.1; ENST00000646725.1; ENST00000644867.1; ENST00000643031.1; ENST00000644138.1; ENST00000643977.1; ENST00000646174.1; ENST00000645210.1; ENST00000642315.1; ENST00000646116.1; ENST00000349496.11 | ||
| External Link | RMBase: m6A_site_584478 | ||
| mod ID: M6ASITE060252 | Click to Show/Hide the Full List | ||
| mod site | chr3:41227288-41227289:+ | [17] | |
| Sequence | ACGAAAAACTACTGTGGACCACAAGCAGAGTGCTGAAGGTG | ||
| Motif Score | 2.053113095 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000642886.1; ENST00000644138.1; ENST00000643052.1; ENST00000396183.7; ENST00000405570.6; ENST00000646725.1; ENST00000647413.1; ENST00000644873.1; ENST00000642992.1; ENST00000396185.8; ENST00000645305.1; ENST00000645276.1; ENST00000644867.1; ENST00000643865.1; ENST00000433400.6; ENST00000643992.1; ENST00000644524.1; ENST00000645210.1; ENST00000642248.1; ENST00000642315.1; ENST00000645927.1; ENST00000644678.1; ENST00000645982.1; ENST00000646174.1; ENST00000643977.1; ENST00000646369.1; ENST00000645320.1; ENST00000642836.1; ENST00000646074.1; ENST00000441708.2; ENST00000349496.11; ENST00000643297.1; ENST00000643031.1; ENST00000431914.6; ENST00000647390.1; ENST00000643541.1; ENST00000647264.1; ENST00000453024.6; ENST00000647021.1; ENST00000642986.1; ENST00000645493.1; ENST00000646381.1; ENST00000646116.1; ENST00000642426.1; ENST00000645900.1; ENST00000450969.6 | ||
| External Link | RMBase: m6A_site_584479 | ||
| mod ID: M6ASITE060253 | Click to Show/Hide the Full List | ||
| mod site | chr3:41233234-41233235:+ | [16] | |
| Sequence | ACTTAGGTGAAAGGAAGTAAACTCTGGAAATACAGAAGGAC | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000644867.1; ENST00000642426.1; ENST00000643992.1; ENST00000646116.1; ENST00000645305.1; ENST00000646174.1; ENST00000647021.1; ENST00000645493.1; ENST00000645276.1; ENST00000647413.1; ENST00000433400.6; ENST00000642836.1; ENST00000643031.1; ENST00000646725.1; ENST00000396183.7; ENST00000643052.1; ENST00000643977.1; ENST00000642315.1; ENST00000441708.2; ENST00000642986.1; ENST00000643541.1; ENST00000645763.1; ENST00000646074.1; ENST00000431914.6; ENST00000644678.1; ENST00000396185.8; ENST00000645210.1; ENST00000643865.1; ENST00000453024.6; ENST00000644873.1; ENST00000646381.1; ENST00000645320.1; ENST00000644524.1; ENST00000647390.1; ENST00000642886.1; ENST00000645982.1; ENST00000643297.1; ENST00000645900.1; ENST00000646369.1; ENST00000642248.1; ENST00000642992.1; ENST00000349496.11; ENST00000450969.6; ENST00000645927.1; ENST00000405570.6; ENST00000647264.1; ENST00000644138.1 | ||
| External Link | RMBase: m6A_site_584480 | ||
| mod ID: M6ASITE060254 | Click to Show/Hide the Full List | ||
| mod site | chr3:41233253-41233254:+ | [16] | |
| Sequence | AACTCTGGAAATACAGAAGGACACCTCCTAAGGCTAGAACA | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000644524.1; ENST00000646174.1; ENST00000645763.1; ENST00000643992.1; ENST00000642992.1; ENST00000646369.1; ENST00000643031.1; ENST00000349496.11; ENST00000644867.1; ENST00000396185.8; ENST00000643977.1; ENST00000405570.6; ENST00000441708.2; ENST00000642426.1; ENST00000642248.1; ENST00000647413.1; ENST00000645927.1; ENST00000431914.6; ENST00000643297.1; ENST00000644873.1; ENST00000642836.1; ENST00000642886.1; ENST00000433400.6; ENST00000647390.1; ENST00000645320.1; ENST00000642315.1; ENST00000643052.1; ENST00000645210.1; ENST00000647264.1; ENST00000645982.1; ENST00000643865.1; ENST00000646381.1; ENST00000644678.1; ENST00000644138.1; ENST00000645305.1; ENST00000647021.1; ENST00000450969.6; ENST00000396183.7; ENST00000453024.6; ENST00000645493.1; ENST00000645276.1; ENST00000646725.1; ENST00000642986.1; ENST00000646074.1; ENST00000646116.1; ENST00000643541.1; ENST00000645900.1 | ||
| External Link | RMBase: m6A_site_584481 | ||
| mod ID: M6ASITE060255 | Click to Show/Hide the Full List | ||
| mod site | chr3:41233271-41233272:+ | [16] | |
| Sequence | GGACACCTCCTAAGGCTAGAACAGATATTTAGGATTGATAG | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000646074.1; ENST00000642248.1; ENST00000450969.6; ENST00000644678.1; ENST00000645900.1; ENST00000431914.6; ENST00000642986.1; ENST00000643865.1; ENST00000642992.1; ENST00000645982.1; ENST00000646369.1; ENST00000643031.1; ENST00000405570.6; ENST00000646725.1; ENST00000645493.1; ENST00000643297.1; ENST00000646381.1; ENST00000642886.1; ENST00000396185.8; ENST00000646116.1; ENST00000644867.1; ENST00000642315.1; ENST00000644873.1; ENST00000643977.1; ENST00000645210.1; ENST00000453024.6; ENST00000645763.1; ENST00000643052.1; ENST00000647021.1; ENST00000643992.1; ENST00000647264.1; ENST00000644138.1; ENST00000349496.11; ENST00000647413.1; ENST00000644524.1; ENST00000647390.1; ENST00000643541.1; ENST00000441708.2; ENST00000433400.6; ENST00000642836.1; ENST00000645305.1; ENST00000645927.1; ENST00000646174.1; ENST00000645276.1; ENST00000645320.1; ENST00000642426.1; ENST00000396183.7 | ||
| External Link | RMBase: m6A_site_584482 | ||
| mod ID: M6ASITE060256 | Click to Show/Hide the Full List | ||
| mod site | chr3:41233359-41233360:+ | [16] | |
| Sequence | GGTGGAATGCAAGCTTTAGGACTTCACCTGACAGATCCAAG | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000647390.1; ENST00000643977.1; ENST00000644138.1; ENST00000642426.1; ENST00000645210.1; ENST00000646369.1; ENST00000643031.1; ENST00000642836.1; ENST00000349496.11; ENST00000396183.7; ENST00000643865.1; ENST00000644524.1; ENST00000646174.1; ENST00000646725.1; ENST00000433400.6; ENST00000643297.1; ENST00000450969.6; ENST00000645982.1; ENST00000642248.1; ENST00000646074.1; ENST00000647413.1; ENST00000431914.6; ENST00000453024.6; ENST00000642992.1; ENST00000396185.8; ENST00000644873.1; ENST00000647264.1; ENST00000644678.1; ENST00000441708.2; ENST00000405570.6; ENST00000645927.1; ENST00000645320.1; ENST00000645493.1; ENST00000643992.1; ENST00000642886.1; ENST00000642986.1; ENST00000643052.1; ENST00000645900.1; ENST00000646381.1; ENST00000645763.1; ENST00000644867.1; ENST00000643541.1; ENST00000642315.1; ENST00000645276.1; ENST00000646116.1; ENST00000645305.1; ENST00000647021.1 | ||
| External Link | RMBase: m6A_site_584483 | ||
| mod ID: M6ASITE060257 | Click to Show/Hide the Full List | ||
| mod site | chr3:41233397-41233398:+ | [16] | |
| Sequence | AAGTCAACGTCTTGTTCAGAACTGTCTTTGGACTCTCAGGA | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000645493.1; ENST00000431914.6; ENST00000644138.1; ENST00000643297.1; ENST00000349496.11; ENST00000645305.1; ENST00000645320.1; ENST00000646174.1; ENST00000642315.1; ENST00000644867.1; ENST00000644524.1; ENST00000644678.1; ENST00000645763.1; ENST00000642986.1; ENST00000441708.2; ENST00000642248.1; ENST00000642992.1; ENST00000647413.1; ENST00000646381.1; ENST00000405570.6; ENST00000643052.1; ENST00000643977.1; ENST00000647264.1; ENST00000643992.1; ENST00000642886.1; ENST00000450969.6; ENST00000643865.1; ENST00000645210.1; ENST00000645927.1; ENST00000433400.6; ENST00000643541.1; ENST00000644873.1; ENST00000645900.1; ENST00000396185.8; ENST00000646369.1; ENST00000647021.1; ENST00000646074.1; ENST00000642836.1; ENST00000453024.6; ENST00000647390.1; ENST00000646116.1; ENST00000645982.1; ENST00000642426.1; ENST00000396183.7; ENST00000645276.1; ENST00000643031.1; ENST00000646725.1 | ||
| External Link | RMBase: m6A_site_584484 | ||
| mod ID: M6ASITE060258 | Click to Show/Hide the Full List | ||
| mod site | chr3:41233408-41233409:+ | [16] | |
| Sequence | TTGTTCAGAACTGTCTTTGGACTCTCAGGAATCTTTCAGAT | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000646381.1; ENST00000646074.1; ENST00000644138.1; ENST00000642248.1; ENST00000642986.1; ENST00000644867.1; ENST00000647390.1; ENST00000431914.6; ENST00000643052.1; ENST00000643992.1; ENST00000441708.2; ENST00000644524.1; ENST00000644873.1; ENST00000642886.1; ENST00000453024.6; ENST00000645763.1; ENST00000643977.1; ENST00000646369.1; ENST00000646725.1; ENST00000645900.1; ENST00000642426.1; ENST00000643297.1; ENST00000643865.1; ENST00000405570.6; ENST00000647264.1; ENST00000642992.1; ENST00000645276.1; ENST00000642836.1; ENST00000646174.1; ENST00000646116.1; ENST00000647413.1; ENST00000644678.1; ENST00000645305.1; ENST00000647021.1; ENST00000643031.1; ENST00000643541.1; ENST00000349496.11; ENST00000645320.1; ENST00000485265.2; ENST00000396183.7; ENST00000450969.6; ENST00000645927.1; ENST00000642315.1; ENST00000645982.1; ENST00000645493.1; ENST00000645210.1; ENST00000433400.6; ENST00000396185.8 | ||
| External Link | RMBase: m6A_site_584485 | ||
| mod ID: M6ASITE060259 | Click to Show/Hide the Full List | ||
| mod site | chr3:41233440-41233441:+ | [16] | |
| Sequence | CTTTCAGATGCTGCAACTAAACAGGTAAATTCTGAGTAAAC | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000441708.2; ENST00000349496.11; ENST00000642886.1; ENST00000450969.6; ENST00000642992.1; ENST00000644138.1; ENST00000645982.1; ENST00000646074.1; ENST00000433400.6; ENST00000405570.6; ENST00000642836.1; ENST00000645927.1; ENST00000643031.1; ENST00000643992.1; ENST00000644906.1; ENST00000642426.1; ENST00000647021.1; ENST00000485265.2; ENST00000644867.1; ENST00000643297.1; ENST00000396185.8; ENST00000645900.1; ENST00000453024.6; ENST00000646174.1; ENST00000642315.1; ENST00000644873.1; ENST00000645763.1; ENST00000647413.1; ENST00000645305.1; ENST00000645210.1; ENST00000646381.1; ENST00000644678.1; ENST00000647390.1; ENST00000643977.1; ENST00000646725.1; ENST00000646369.1; ENST00000643052.1; ENST00000647264.1; ENST00000643865.1; ENST00000644524.1; ENST00000645320.1; ENST00000642986.1; ENST00000396183.7; ENST00000645493.1; ENST00000642248.1; ENST00000645276.1; ENST00000646116.1; ENST00000431914.6; ENST00000643541.1 | ||
| External Link | RMBase: m6A_site_584486 | ||
| mod ID: M6ASITE060260 | Click to Show/Hide the Full List | ||
| mod site | chr3:41233459-41233460:+ | [16] | |
| Sequence | AACAGGTAAATTCTGAGTAAACTGGTGCCATGGGAATAGAG | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000644678.1; ENST00000433400.6; ENST00000646116.1; ENST00000644873.1; ENST00000396185.8; ENST00000647390.1; ENST00000645927.1; ENST00000643541.1; ENST00000645900.1; ENST00000647264.1; ENST00000643297.1; ENST00000645276.1; ENST00000645305.1; ENST00000642315.1; ENST00000646369.1; ENST00000646725.1; ENST00000646074.1; ENST00000647413.1; ENST00000642248.1; ENST00000644906.1; ENST00000643052.1; ENST00000642426.1; ENST00000643865.1; ENST00000645493.1; ENST00000642836.1; ENST00000453024.6; ENST00000643031.1; ENST00000647021.1; ENST00000450969.6; ENST00000405570.6; ENST00000642986.1; ENST00000441708.2; ENST00000645982.1; ENST00000396183.7; ENST00000646381.1; ENST00000645763.1; ENST00000645210.1; ENST00000431914.6; ENST00000349496.11; ENST00000643992.1; ENST00000644138.1; ENST00000642886.1; ENST00000643977.1; ENST00000645320.1; ENST00000485265.2; ENST00000646174.1; ENST00000642992.1; ENST00000644524.1; ENST00000644867.1 | ||
| External Link | RMBase: m6A_site_584487 | ||
| mod ID: M6ASITE060261 | Click to Show/Hide the Full List | ||
| mod site | chr3:41233552-41233553:+ | [16] | |
| Sequence | GGATGGAAGGTCTCCTTGGGACTCTTGTTCAGCTTCTGGGT | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000643992.1; ENST00000643031.1; ENST00000644524.1; ENST00000643865.1; ENST00000396185.8; ENST00000644867.1; ENST00000644873.1; ENST00000643297.1; ENST00000646074.1; ENST00000642426.1; ENST00000453024.6; ENST00000642248.1; ENST00000485265.2; ENST00000642992.1; ENST00000643541.1; ENST00000646381.1; ENST00000646116.1; ENST00000642315.1; ENST00000642836.1; ENST00000647390.1; ENST00000645927.1; ENST00000645276.1; ENST00000396183.7; ENST00000441708.2; ENST00000645982.1; ENST00000642986.1; ENST00000643977.1; ENST00000647264.1; ENST00000644906.1; ENST00000645210.1; ENST00000645493.1; ENST00000645320.1; ENST00000646174.1; ENST00000644678.1; ENST00000645763.1; ENST00000349496.11; ENST00000431914.6; ENST00000647021.1; ENST00000642886.1; ENST00000643052.1; ENST00000647413.1; ENST00000646369.1; ENST00000644138.1; ENST00000645900.1; ENST00000646725.1; ENST00000645305.1; ENST00000433400.6; ENST00000450969.6; ENST00000405570.6 | ||
| External Link | RMBase: m6A_site_584488 | ||
| mod ID: M6ASITE060262 | Click to Show/Hide the Full List | ||
| mod site | chr3:41233643-41233644:+ | [16] | |
| Sequence | CACTTGCAATAATTATAAGAACAAGATGATGGTCTGCCAAG | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa; CD34; hESC-HEK293T | ||
| Seq Type List | m6A-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000643052.1; ENST00000349496.11; ENST00000645900.1; ENST00000646116.1; ENST00000643977.1; ENST00000644138.1; ENST00000643865.1; ENST00000642886.1; ENST00000642315.1; ENST00000642986.1; ENST00000647264.1; ENST00000647413.1; ENST00000645320.1; ENST00000644873.1; ENST00000645493.1; ENST00000645276.1; ENST00000485265.2; ENST00000645305.1; ENST00000644678.1; ENST00000396183.7; ENST00000646381.1; ENST00000644906.1; ENST00000646725.1; ENST00000644524.1; ENST00000646074.1; ENST00000453024.6; ENST00000646369.1; ENST00000647021.1; ENST00000645927.1; ENST00000645210.1; ENST00000396185.8; ENST00000642426.1; ENST00000643031.1; ENST00000645763.1; ENST00000642836.1; ENST00000441708.2; ENST00000643992.1; ENST00000433400.6; ENST00000450969.6; ENST00000646174.1; ENST00000644867.1; ENST00000642992.1; ENST00000431914.6; ENST00000643541.1; ENST00000645982.1; ENST00000647390.1; ENST00000642248.1; ENST00000643297.1; ENST00000405570.6 | ||
| External Link | RMBase: m6A_site_584489 | ||
| mod ID: M6ASITE060263 | Click to Show/Hide the Full List | ||
| mod site | chr3:41233709-41233710:+ | [18] | |
| Sequence | TACTGTCCTTCGGGCTGGTGACAGGGAAGACATCACTGAGC | ||
| Motif Score | 2.859755952 | ||
| Cell/Tissue List | HEK293 | ||
| Seq Type List | m6A-REF-seq | ||
| Transcript ID List | ENST00000646116.1; ENST00000647413.1; ENST00000642315.1; ENST00000485265.2; ENST00000646381.1; ENST00000644867.1; ENST00000644873.1; ENST00000645276.1; ENST00000643031.1; ENST00000644524.1; ENST00000349496.11; ENST00000643977.1; ENST00000644138.1; ENST00000453024.6; ENST00000396183.7; ENST00000450969.6; ENST00000643992.1; ENST00000642836.1; ENST00000645927.1; ENST00000642886.1; ENST00000642426.1; ENST00000644906.1; ENST00000643865.1; ENST00000645493.1; ENST00000642992.1; ENST00000647021.1; ENST00000646174.1; ENST00000643297.1; ENST00000645210.1; ENST00000642248.1; ENST00000646725.1; ENST00000646074.1; ENST00000644678.1; ENST00000647264.1; ENST00000645305.1; ENST00000643541.1; ENST00000643052.1; ENST00000647390.1; ENST00000645982.1; ENST00000405570.6; ENST00000646369.1; ENST00000431914.6; ENST00000645763.1; ENST00000441708.2; ENST00000396185.8; ENST00000642986.1; ENST00000645320.1; ENST00000433400.6; ENST00000645900.1 | ||
| External Link | RMBase: m6A_site_584490 | ||
| mod ID: M6ASITE060264 | Click to Show/Hide the Full List | ||
| mod site | chr3:41233718-41233719:+ | [16] | |
| Sequence | TCGGGCTGGTGACAGGGAAGACATCACTGAGCCTGCCATCT | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa; CD34; liver; hESC-HEK293T; endometrial | ||
| Seq Type List | m6A-seq; m6A-REF-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000642992.1; ENST00000646174.1; ENST00000642986.1; ENST00000646074.1; ENST00000645982.1; ENST00000485265.2; ENST00000642315.1; ENST00000644678.1; ENST00000642836.1; ENST00000433400.6; ENST00000645276.1; ENST00000643031.1; ENST00000647413.1; ENST00000644524.1; ENST00000645763.1; ENST00000647021.1; ENST00000642886.1; ENST00000405570.6; ENST00000643865.1; ENST00000431914.6; ENST00000644867.1; ENST00000644906.1; ENST00000645927.1; ENST00000450969.6; ENST00000643977.1; ENST00000643052.1; ENST00000441708.2; ENST00000643992.1; ENST00000396183.7; ENST00000646381.1; ENST00000647264.1; ENST00000642426.1; ENST00000643297.1; ENST00000645900.1; ENST00000643541.1; ENST00000646725.1; ENST00000644873.1; ENST00000396185.8; ENST00000644138.1; ENST00000647390.1; ENST00000349496.11; ENST00000646369.1; ENST00000645320.1; ENST00000453024.6; ENST00000646116.1; ENST00000642248.1; ENST00000645305.1; ENST00000645210.1; ENST00000645493.1 | ||
| External Link | RMBase: m6A_site_584491 | ||
| mod ID: M6ASITE060265 | Click to Show/Hide the Full List | ||
| mod site | chr3:41233764-41233765:+ | [17] | |
| Sequence | CTTCGTCATCTGACCAGCCGACACCAAGAAGCAGAGATGGC | ||
| Motif Score | 2.865571429 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000647264.1; ENST00000646369.1; ENST00000644678.1; ENST00000646381.1; ENST00000642886.1; ENST00000645305.1; ENST00000642248.1; ENST00000646116.1; ENST00000645927.1; ENST00000433400.6; ENST00000349496.11; ENST00000396183.7; ENST00000396185.8; ENST00000645276.1; ENST00000646074.1; ENST00000644867.1; ENST00000441708.2; ENST00000643992.1; ENST00000642992.1; ENST00000643865.1; ENST00000642426.1; ENST00000643297.1; ENST00000645900.1; ENST00000485265.2; ENST00000643541.1; ENST00000642986.1; ENST00000645493.1; ENST00000644138.1; ENST00000644524.1; ENST00000647390.1; ENST00000645210.1; ENST00000642315.1; ENST00000646725.1; ENST00000645982.1; ENST00000647021.1; ENST00000645320.1; ENST00000405570.6; ENST00000450969.6; ENST00000646174.1; ENST00000647413.1; ENST00000431914.6; ENST00000644906.1; ENST00000453024.6; ENST00000642836.1; ENST00000644873.1; ENST00000645763.1; ENST00000643052.1; ENST00000643977.1; ENST00000643031.1 | ||
| External Link | RMBase: m6A_site_584492 | ||
| mod ID: M6ASITE060266 | Click to Show/Hide the Full List | ||
| mod site | chr3:41233812-41233813:+ | [16] | |
| Sequence | GCAGTTCGCCTTCACTATGGACTACCAGTTGTGGTTAAGCT | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa; CD34; endometrial | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000643031.1; ENST00000450969.6; ENST00000396185.8; ENST00000646381.1; ENST00000645276.1; ENST00000646116.1; ENST00000645305.1; ENST00000441708.2; ENST00000646725.1; ENST00000642248.1; ENST00000645320.1; ENST00000644678.1; ENST00000453024.6; ENST00000643297.1; ENST00000643992.1; ENST00000642315.1; ENST00000349496.11; ENST00000642992.1; ENST00000646369.1; ENST00000405570.6; ENST00000642426.1; ENST00000645900.1; ENST00000485265.2; ENST00000642886.1; ENST00000643052.1; ENST00000645210.1; ENST00000644873.1; ENST00000431914.6; ENST00000643865.1; ENST00000644906.1; ENST00000642986.1; ENST00000644867.1; ENST00000646074.1; ENST00000645982.1; ENST00000643977.1; ENST00000642836.1; ENST00000644524.1; ENST00000647413.1; ENST00000647390.1; ENST00000433400.6; ENST00000646174.1; ENST00000643541.1; ENST00000645763.1; ENST00000645493.1; ENST00000645927.1; ENST00000396183.7; ENST00000647264.1; ENST00000647021.1; ENST00000644138.1 | ||
| External Link | RMBase: m6A_site_584493 | ||
| mod ID: M6ASITE060267 | Click to Show/Hide the Full List | ||
| mod site | chr3:41233836-41233837:+ | [17] | |
| Sequence | CCAGTTGTGGTTAAGCTCTTACACCCACCATCCCACTGGCC | ||
| Motif Score | 2.07285119 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000441708.2; ENST00000642992.1; ENST00000643031.1; ENST00000642886.1; ENST00000645763.1; ENST00000645305.1; ENST00000405570.6; ENST00000642248.1; ENST00000647390.1; ENST00000643865.1; ENST00000645210.1; ENST00000643052.1; ENST00000645900.1; ENST00000646174.1; ENST00000647264.1; ENST00000644524.1; ENST00000485265.2; ENST00000644867.1; ENST00000645493.1; ENST00000647021.1; ENST00000643992.1; ENST00000646369.1; ENST00000645982.1; ENST00000644906.1; ENST00000349496.11; ENST00000646116.1; ENST00000646381.1; ENST00000644138.1; ENST00000642426.1; ENST00000645927.1; ENST00000450969.6; ENST00000644678.1; ENST00000642836.1; ENST00000642315.1; ENST00000644873.1; ENST00000643297.1; ENST00000645276.1; ENST00000396183.7; ENST00000643977.1; ENST00000453024.6; ENST00000431914.6; ENST00000646074.1; ENST00000646725.1; ENST00000433400.6; ENST00000647413.1; ENST00000645320.1; ENST00000396185.8; ENST00000643541.1; ENST00000642986.1 | ||
| External Link | RMBase: m6A_site_584494 | ||
| mod ID: M6ASITE060268 | Click to Show/Hide the Full List | ||
| mod site | chr3:41234242-41234243:+ | [17] | |
| Sequence | GTTCAGTTGCTTGTTCGTGCACATCAGGATACCCAGCGCCG | ||
| Motif Score | 2.830589286 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000643031.1; ENST00000433400.6; ENST00000441708.2; ENST00000643992.1; ENST00000644867.1; ENST00000646369.1; ENST00000642426.1; ENST00000450969.6; ENST00000642315.1; ENST00000645982.1; ENST00000645900.1; ENST00000647390.1; ENST00000396185.8; ENST00000645927.1; ENST00000643052.1; ENST00000645276.1; ENST00000647413.1; ENST00000396183.7; ENST00000643297.1; ENST00000642836.1; ENST00000644906.1; ENST00000642992.1; ENST00000405570.6; ENST00000643865.1; ENST00000646116.1; ENST00000644395.1; ENST00000645305.1; ENST00000431914.6; ENST00000644678.1; ENST00000645493.1; ENST00000485265.2; ENST00000646725.1; ENST00000646381.1; ENST00000645320.1; ENST00000643977.1; ENST00000644873.1; ENST00000645210.1; ENST00000349496.11; ENST00000647264.1; ENST00000647021.1; ENST00000465552.2; ENST00000453024.6; ENST00000644524.1; ENST00000646174.1; ENST00000645763.1; ENST00000642886.1; ENST00000646074.1; ENST00000644138.1; ENST00000642986.1; ENST00000643541.1; ENST00000642248.1 | ||
| External Link | RMBase: m6A_site_584495 | ||
| mod ID: M6ASITE060269 | Click to Show/Hide the Full List | ||
| mod site | chr3:41234279-41234280:+ | [16] | |
| Sequence | GCCGTACGTCCATGGGTGGGACACAGCAGCAATTTGTGGTA | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; hESC-HEK293T; HepG2; Huh7 | ||
| Seq Type List | m6A-seq; MAZTER-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000396183.7; ENST00000433400.6; ENST00000645900.1; ENST00000646725.1; ENST00000646381.1; ENST00000645982.1; ENST00000645493.1; ENST00000450969.6; ENST00000644678.1; ENST00000465552.2; ENST00000441708.2; ENST00000645210.1; ENST00000453024.6; ENST00000645305.1; ENST00000645927.1; ENST00000643992.1; ENST00000642886.1; ENST00000643541.1; ENST00000643297.1; ENST00000642248.1; ENST00000642426.1; ENST00000644138.1; ENST00000644867.1; ENST00000647021.1; ENST00000642986.1; ENST00000646074.1; ENST00000396185.8; ENST00000647264.1; ENST00000644906.1; ENST00000485265.2; ENST00000647390.1; ENST00000646174.1; ENST00000431914.6; ENST00000643052.1; ENST00000644873.1; ENST00000645320.1; ENST00000643977.1; ENST00000645763.1; ENST00000405570.6; ENST00000646369.1; ENST00000642836.1; ENST00000644395.1; ENST00000642315.1; ENST00000647413.1; ENST00000645276.1; ENST00000646116.1; ENST00000642992.1; ENST00000643865.1; ENST00000643031.1; ENST00000644524.1; ENST00000349496.11 | ||
| External Link | RMBase: m6A_site_584496 | ||
| mod ID: M6ASITE060270 | Click to Show/Hide the Full List | ||
| mod site | chr3:41234362-41234363:+ | [16] | |
| Sequence | ATAAATCCAAAGGATCCTGAACTTCTTTCTTTGGTCATTGG | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa; Huh7 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000645982.1; ENST00000465552.2; ENST00000644867.1; ENST00000643541.1; ENST00000396185.8; ENST00000450969.6; ENST00000647264.1; ENST00000645900.1; ENST00000642426.1; ENST00000643031.1; ENST00000647021.1; ENST00000644395.1; ENST00000645493.1; ENST00000431914.6; ENST00000349496.11; ENST00000643992.1; ENST00000441708.2; ENST00000647413.1; ENST00000645763.1; ENST00000642836.1; ENST00000433400.6; ENST00000642248.1; ENST00000644524.1; ENST00000645927.1; ENST00000645305.1; ENST00000645320.1; ENST00000644138.1; ENST00000642315.1; ENST00000645210.1; ENST00000643865.1; ENST00000644678.1; ENST00000643297.1; ENST00000485265.2; ENST00000646116.1; ENST00000642992.1; ENST00000647390.1; ENST00000643977.1; ENST00000642986.1; ENST00000396183.7; ENST00000643052.1; ENST00000646369.1; ENST00000646074.1; ENST00000645276.1; ENST00000646725.1; ENST00000642886.1; ENST00000646174.1; ENST00000453024.6; ENST00000644906.1; ENST00000644873.1; ENST00000646381.1; ENST00000405570.6 | ||
| External Link | RMBase: m6A_site_584497 | ||
| mod ID: M6ASITE060271 | Click to Show/Hide the Full List | ||
| mod site | chr3:41235772-41235773:+ | [17] | |
| Sequence | AGGTTGTACCGGAGCCCTTCACATCCTAGCTCGGGATGTTC | ||
| Motif Score | 2.047297619 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000647021.1; ENST00000642248.1; ENST00000646381.1; ENST00000642986.1; ENST00000644524.1; ENST00000645982.1; ENST00000643031.1; ENST00000647390.1; ENST00000642836.1; ENST00000441708.2; ENST00000644138.1; ENST00000646369.1; ENST00000643977.1; ENST00000642886.1; ENST00000396185.8; ENST00000645305.1; ENST00000642426.1; ENST00000465552.2; ENST00000646174.1; ENST00000645276.1; ENST00000646074.1; ENST00000645763.1; ENST00000642315.1; ENST00000405570.6; ENST00000644873.1; ENST00000646725.1; ENST00000433400.6; ENST00000644678.1; ENST00000645927.1; ENST00000647264.1; ENST00000642992.1; ENST00000647413.1; ENST00000482042.1; ENST00000643865.1; ENST00000643541.1; ENST00000645320.1; ENST00000643992.1; ENST00000450969.6; ENST00000645493.1; ENST00000453024.6; ENST00000645210.1; ENST00000644867.1; ENST00000644906.1; ENST00000643052.1; ENST00000646116.1; ENST00000485265.2; ENST00000431914.6; ENST00000645900.1; ENST00000396183.7; ENST00000349496.11; ENST00000643297.1 | ||
| External Link | RMBase: m6A_site_584498 | ||
| mod ID: M6ASITE060272 | Click to Show/Hide the Full List | ||
| mod site | chr3:41235793-41235794:+ | [17] | |
| Sequence | CATCCTAGCTCGGGATGTTCACAACCGAATTGTTATCAGAG | ||
| Motif Score | 2.047297619 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000441708.2; ENST00000431914.6; ENST00000643541.1; ENST00000645276.1; ENST00000646074.1; ENST00000645305.1; ENST00000646369.1; ENST00000396183.7; ENST00000646174.1; ENST00000647264.1; ENST00000646381.1; ENST00000647021.1; ENST00000482042.1; ENST00000647413.1; ENST00000643031.1; ENST00000644873.1; ENST00000642836.1; ENST00000644867.1; ENST00000642886.1; ENST00000645320.1; ENST00000644524.1; ENST00000642248.1; ENST00000642315.1; ENST00000645900.1; ENST00000642426.1; ENST00000645210.1; ENST00000646116.1; ENST00000645493.1; ENST00000465552.2; ENST00000645927.1; ENST00000453024.6; ENST00000433400.6; ENST00000643977.1; ENST00000643297.1; ENST00000450969.6; ENST00000644138.1; ENST00000645982.1; ENST00000642986.1; ENST00000396185.8; ENST00000644906.1; ENST00000642992.1; ENST00000646725.1; ENST00000643992.1; ENST00000405570.6; ENST00000349496.11; ENST00000485265.2; ENST00000647390.1; ENST00000645763.1; ENST00000643052.1; ENST00000643865.1; ENST00000644678.1 | ||
| External Link | RMBase: m6A_site_584499 | ||
| mod ID: M6ASITE060273 | Click to Show/Hide the Full List | ||
| mod site | chr3:41235815-41235816:+ | [16] | |
| Sequence | AACCGAATTGTTATCAGAGGACTAAATACCATTCCATTGTT | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000645320.1; ENST00000431914.6; ENST00000645210.1; ENST00000644678.1; ENST00000645927.1; ENST00000396185.8; ENST00000642836.1; ENST00000482042.1; ENST00000645276.1; ENST00000643977.1; ENST00000646381.1; ENST00000643052.1; ENST00000643541.1; ENST00000349496.11; ENST00000441708.2; ENST00000646174.1; ENST00000647413.1; ENST00000465552.2; ENST00000485265.2; ENST00000644138.1; ENST00000642986.1; ENST00000644873.1; ENST00000433400.6; ENST00000644867.1; ENST00000645900.1; ENST00000643992.1; ENST00000644524.1; ENST00000643031.1; ENST00000642886.1; ENST00000645493.1; ENST00000450969.6; ENST00000642248.1; ENST00000644906.1; ENST00000647021.1; ENST00000645305.1; ENST00000646369.1; ENST00000645763.1; ENST00000643297.1; ENST00000453024.6; ENST00000646116.1; ENST00000642992.1; ENST00000642426.1; ENST00000647390.1; ENST00000405570.6; ENST00000643865.1; ENST00000396183.7; ENST00000646725.1; ENST00000642315.1; ENST00000646074.1; ENST00000645982.1; ENST00000647264.1 | ||
| External Link | RMBase: m6A_site_584500 | ||
| mod ID: M6ASITE060274 | Click to Show/Hide the Full List | ||
| mod site | chr3:41235927-41235928:+ | [16] | |
| Sequence | AACATTTCAGAAAATTAGGGACCATAATAGGGTTACCAACA | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000642836.1; ENST00000644867.1; ENST00000645982.1; ENST00000485265.2; ENST00000644138.1; ENST00000645763.1; ENST00000647021.1; ENST00000644678.1; ENST00000647413.1; ENST00000647264.1; ENST00000644906.1; ENST00000646725.1; ENST00000645927.1; ENST00000405570.6; ENST00000396183.7; ENST00000643541.1; ENST00000482042.1; ENST00000642315.1; ENST00000642248.1; ENST00000642986.1; ENST00000645210.1; ENST00000645320.1; ENST00000646074.1; ENST00000349496.11; ENST00000644873.1; ENST00000645493.1; ENST00000433400.6; ENST00000642886.1; ENST00000646174.1; ENST00000646369.1; ENST00000645305.1; ENST00000643865.1; ENST00000642992.1; ENST00000646381.1; ENST00000643992.1; ENST00000450969.6; ENST00000396185.8; ENST00000644524.1; ENST00000643977.1; ENST00000453024.6; ENST00000643297.1; ENST00000441708.2; ENST00000643052.1; ENST00000647390.1; ENST00000645900.1; ENST00000646116.1; ENST00000643031.1; ENST00000465552.2; ENST00000642426.1; ENST00000645276.1; ENST00000431914.6 | ||
| External Link | RMBase: m6A_site_584501 | ||
| mod ID: M6ASITE060275 | Click to Show/Hide the Full List | ||
| mod site | chr3:41236370-41236371:+ | [16] | |
| Sequence | GCTTTATTCTCCCATTGAAAACATCCAAAGAGTAGCTGCAG | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa; hESC-HEK293T; MT4 | ||
| Seq Type List | m6A-seq; MAZTER-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000433400.6; ENST00000647021.1; ENST00000643992.1; ENST00000643865.1; ENST00000431914.6; ENST00000644873.1; ENST00000482042.1; ENST00000642986.1; ENST00000644524.1; ENST00000646074.1; ENST00000646725.1; ENST00000645900.1; ENST00000405570.6; ENST00000441708.2; ENST00000349496.11; ENST00000642992.1; ENST00000645210.1; ENST00000645982.1; ENST00000396183.7; ENST00000644906.1; ENST00000485265.2; ENST00000645320.1; ENST00000646174.1; ENST00000465552.2; ENST00000647413.1; ENST00000450969.6; ENST00000647390.1; ENST00000643977.1; ENST00000642426.1; ENST00000642248.1; ENST00000643541.1; ENST00000643297.1; ENST00000645763.1; ENST00000642886.1; ENST00000644138.1; ENST00000645276.1; ENST00000646381.1; ENST00000644867.1; ENST00000646369.1; ENST00000642315.1; ENST00000646116.1; ENST00000644678.1; ENST00000453024.6; ENST00000642836.1; ENST00000647264.1; ENST00000396185.8; ENST00000645493.1; ENST00000645305.1; ENST00000645927.1; ENST00000643052.1; ENST00000643031.1 | ||
| External Link | RMBase: m6A_site_584502 | ||
| mod ID: M6ASITE060276 | Click to Show/Hide the Full List | ||
| mod site | chr3:41236404-41236405:+ | [16] | |
| Sequence | GCTGCAGGGGTCCTCTGTGAACTTGCTCAGGACAAGGAAGC | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa; MT4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000642836.1; ENST00000453024.6; ENST00000643977.1; ENST00000645493.1; ENST00000646074.1; ENST00000644524.1; ENST00000645763.1; ENST00000644873.1; ENST00000645927.1; ENST00000642426.1; ENST00000642315.1; ENST00000645276.1; ENST00000642248.1; ENST00000647413.1; ENST00000643052.1; ENST00000646174.1; ENST00000450969.6; ENST00000644867.1; ENST00000405570.6; ENST00000485265.2; ENST00000349496.11; ENST00000643031.1; ENST00000482042.1; ENST00000647264.1; ENST00000642992.1; ENST00000644138.1; ENST00000642986.1; ENST00000396185.8; ENST00000645900.1; ENST00000646369.1; ENST00000645210.1; ENST00000642886.1; ENST00000646116.1; ENST00000643992.1; ENST00000645305.1; ENST00000643541.1; ENST00000646381.1; ENST00000433400.6; ENST00000431914.6; ENST00000645982.1; ENST00000643865.1; ENST00000646725.1; ENST00000645320.1; ENST00000647021.1; ENST00000647390.1; ENST00000396183.7; ENST00000465552.2; ENST00000644678.1; ENST00000643297.1; ENST00000441708.2 | ||
| External Link | RMBase: m6A_site_584503 | ||
| mod ID: M6ASITE060277 | Click to Show/Hide the Full List | ||
| mod site | chr3:41236415-41236416:+ | [16] | |
| Sequence | CCTCTGTGAACTTGCTCAGGACAAGGAAGCTGCAGAAGCTA | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; hESC-HEK293T; MT4 | ||
| Seq Type List | m6A-seq; MAZTER-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000642836.1; ENST00000644138.1; ENST00000643865.1; ENST00000643541.1; ENST00000643977.1; ENST00000482042.1; ENST00000645210.1; ENST00000643297.1; ENST00000642315.1; ENST00000396185.8; ENST00000645763.1; ENST00000647413.1; ENST00000645493.1; ENST00000441708.2; ENST00000642986.1; ENST00000643052.1; ENST00000450969.6; ENST00000646074.1; ENST00000645982.1; ENST00000645305.1; ENST00000642886.1; ENST00000647264.1; ENST00000642426.1; ENST00000431914.6; ENST00000645320.1; ENST00000642992.1; ENST00000646369.1; ENST00000433400.6; ENST00000453024.6; ENST00000645900.1; ENST00000647390.1; ENST00000644873.1; ENST00000644867.1; ENST00000643992.1; ENST00000405570.6; ENST00000646116.1; ENST00000644524.1; ENST00000485265.2; ENST00000645276.1; ENST00000643031.1; ENST00000646725.1; ENST00000642248.1; ENST00000647021.1; ENST00000646174.1; ENST00000644678.1; ENST00000349496.11; ENST00000646381.1; ENST00000465552.2; ENST00000396183.7; ENST00000645927.1 | ||
| External Link | RMBase: m6A_site_584504 | ||
| mod ID: M6ASITE060278 | Click to Show/Hide the Full List | ||
| mod site | chr3:41236513-41236514:+ | [16] | |
| Sequence | GTGTGGGTAAGTAAAAAGGAACCAAAGCCTTTAGCAGATGT | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000642992.1; ENST00000647021.1; ENST00000647390.1; ENST00000647264.1; ENST00000644873.1; ENST00000482042.1; ENST00000431914.6; ENST00000645982.1; ENST00000646074.1; ENST00000396185.8; ENST00000645900.1; ENST00000643052.1; ENST00000645493.1; ENST00000643992.1; ENST00000405570.6; ENST00000642315.1; ENST00000450969.6; ENST00000644678.1; ENST00000643541.1; ENST00000453024.6; ENST00000643977.1; ENST00000465552.2; ENST00000645210.1; ENST00000643031.1; ENST00000646369.1; ENST00000646174.1; ENST00000645320.1; ENST00000645927.1; ENST00000647413.1; ENST00000646381.1; ENST00000644867.1; ENST00000471014.2; ENST00000645276.1; ENST00000396183.7; ENST00000642886.1; ENST00000645305.1; ENST00000644138.1; ENST00000642248.1; ENST00000643297.1; ENST00000646725.1; ENST00000643865.1; ENST00000645763.1; ENST00000642426.1; ENST00000349496.11; ENST00000642836.1; ENST00000441708.2; ENST00000485265.2; ENST00000644524.1; ENST00000642986.1; ENST00000646116.1; ENST00000433400.6 | ||
| External Link | RMBase: m6A_site_584505 | ||
| mod ID: M6ASITE060279 | Click to Show/Hide the Full List | ||
| mod site | chr3:41236589-41236590:+ | [18] | |
| Sequence | TTTCTCCTTGTCTCTTAGCGACATATGCAGCTGCTGTTTTG | ||
| Motif Score | 2.865571429 | ||
| Cell/Tissue List | liver | ||
| Seq Type List | m6A-REF-seq | ||
| Transcript ID List | ENST00000647264.1; ENST00000453024.6; ENST00000349496.11; ENST00000646381.1; ENST00000646074.1; ENST00000643977.1; ENST00000471014.2; ENST00000642315.1; ENST00000642426.1; ENST00000642992.1; ENST00000482042.1; ENST00000644138.1; ENST00000647021.1; ENST00000646116.1; ENST00000405570.6; ENST00000433400.6; ENST00000644867.1; ENST00000396185.8; ENST00000465552.2; ENST00000643865.1; ENST00000643031.1; ENST00000396183.7; ENST00000646369.1; ENST00000645305.1; ENST00000642886.1; ENST00000643541.1; ENST00000644678.1; ENST00000643297.1; ENST00000645276.1; ENST00000441708.2; ENST00000644873.1; ENST00000644524.1; ENST00000645982.1; ENST00000645763.1; ENST00000645493.1; ENST00000646174.1; ENST00000642836.1; ENST00000646725.1; ENST00000642248.1; ENST00000642986.1; ENST00000485265.2; ENST00000647390.1; ENST00000645927.1; ENST00000647413.1; ENST00000450969.6; ENST00000645900.1; ENST00000645210.1; ENST00000643992.1; ENST00000431914.6; ENST00000645320.1; ENST00000643052.1 | ||
| External Link | RMBase: m6A_site_584506 | ||
| mod ID: M6ASITE060280 | Click to Show/Hide the Full List | ||
| mod site | chr3:41236641-41236642:+ | [17] | |
| Sequence | TGAGGACAAGCCACAAGATTACAAGAAACGGCTTTCAGTTG | ||
| Motif Score | 2.07285119 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000646074.1; ENST00000646116.1; ENST00000441708.2; ENST00000642886.1; ENST00000644524.1; ENST00000643865.1; ENST00000647390.1; ENST00000645305.1; ENST00000644873.1; ENST00000643541.1; ENST00000647264.1; ENST00000645320.1; ENST00000642315.1; ENST00000642426.1; ENST00000642836.1; ENST00000349496.11; ENST00000645982.1; ENST00000642992.1; ENST00000431914.6; ENST00000647021.1; ENST00000482042.1; ENST00000643977.1; ENST00000645276.1; ENST00000396185.8; ENST00000643031.1; ENST00000643052.1; ENST00000645900.1; ENST00000646369.1; ENST00000433400.6; ENST00000644867.1; ENST00000645763.1; ENST00000644678.1; ENST00000645210.1; ENST00000642248.1; ENST00000405570.6; ENST00000642986.1; ENST00000643297.1; ENST00000450969.6; ENST00000485265.2; ENST00000646725.1; ENST00000646381.1; ENST00000465552.2; ENST00000471014.2; ENST00000645927.1; ENST00000396183.7; ENST00000646174.1; ENST00000643992.1; ENST00000644138.1; ENST00000647413.1; ENST00000453024.6; ENST00000645493.1 | ||
| External Link | RMBase: m6A_site_584507 | ||
| mod ID: M6ASITE060281 | Click to Show/Hide the Full List | ||
| mod site | chr3:41236685-41236686:+ | [18] | |
| Sequence | TGACCAGCTCTCTCTTCAGAACAGAGCCAATGGCTTGGAAT | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | kidney | ||
| Seq Type List | m6A-REF-seq | ||
| Transcript ID List | ENST00000642426.1; ENST00000349496.11; ENST00000643992.1; ENST00000465552.2; ENST00000645900.1; ENST00000645320.1; ENST00000647413.1; ENST00000441708.2; ENST00000645305.1; ENST00000647021.1; ENST00000643977.1; ENST00000450969.6; ENST00000642886.1; ENST00000643865.1; ENST00000643541.1; ENST00000431914.6; ENST00000645493.1; ENST00000453024.6; ENST00000645927.1; ENST00000642986.1; ENST00000396183.7; ENST00000645210.1; ENST00000646381.1; ENST00000647264.1; ENST00000646369.1; ENST00000643031.1; ENST00000485265.2; ENST00000644524.1; ENST00000644873.1; ENST00000642248.1; ENST00000646725.1; ENST00000644138.1; ENST00000433400.6; ENST00000642315.1; ENST00000482042.1; ENST00000471014.2; ENST00000642992.1; ENST00000644867.1; ENST00000645276.1; ENST00000644678.1; ENST00000647390.1; ENST00000646074.1; ENST00000642836.1; ENST00000396185.8; ENST00000646116.1; ENST00000643052.1; ENST00000643297.1; ENST00000645763.1; ENST00000645982.1; ENST00000646174.1; ENST00000405570.6 | ||
| External Link | RMBase: m6A_site_584508 | ||
| mod ID: M6ASITE060282 | Click to Show/Hide the Full List | ||
| mod site | chr3:41239190-41239191:+ | [20] | |
| Sequence | CCAGGATGCCTTGGGTATGGACCCCATGATGGAACATGAGA | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HepG2 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000646174.1; ENST00000646369.1; ENST00000450969.6; ENST00000644867.1; ENST00000645210.1; ENST00000642992.1; ENST00000646116.1; ENST00000433400.6; ENST00000643297.1; ENST00000643977.1; ENST00000642426.1; ENST00000643541.1; ENST00000405570.6; ENST00000645900.1; ENST00000453024.6; ENST00000642248.1; ENST00000647264.1; ENST00000396185.8; ENST00000431914.6; ENST00000645305.1; ENST00000644501.1; ENST00000644873.1; ENST00000643031.1; ENST00000645320.1; ENST00000646074.1; ENST00000645982.1; ENST00000643992.1; ENST00000441708.2; ENST00000642836.1; ENST00000647413.1; ENST00000642315.1; ENST00000644524.1; ENST00000643052.1; ENST00000396183.7; ENST00000642886.1; ENST00000646725.1; ENST00000644678.1; ENST00000646381.1; ENST00000647390.1; ENST00000349496.11; ENST00000647021.1; ENST00000645493.1; ENST00000642986.1; ENST00000645276.1; ENST00000471014.2 | ||
| External Link | RMBase: m6A_site_584509 | ||
| mod ID: M6ASITE060283 | Click to Show/Hide the Full List | ||
| mod site | chr3:41239203-41239204:+ | [17] | |
| Sequence | GGTATGGACCCCATGATGGAACATGAGATGGGTGGCCACCA | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | hESC-HEK293T; HepG2 | ||
| Seq Type List | MAZTER-seq; m6A-seq | ||
| Transcript ID List | ENST00000396185.8; ENST00000645276.1; ENST00000645210.1; ENST00000643992.1; ENST00000647413.1; ENST00000396183.7; ENST00000441708.2; ENST00000646116.1; ENST00000644678.1; ENST00000642426.1; ENST00000450969.6; ENST00000349496.11; ENST00000646369.1; ENST00000647264.1; ENST00000453024.6; ENST00000646725.1; ENST00000642992.1; ENST00000642836.1; ENST00000644867.1; ENST00000643297.1; ENST00000646074.1; ENST00000644873.1; ENST00000405570.6; ENST00000471014.2; ENST00000642886.1; ENST00000642248.1; ENST00000431914.6; ENST00000646381.1; ENST00000644524.1; ENST00000642315.1; ENST00000643977.1; ENST00000645900.1; ENST00000643541.1; ENST00000643031.1; ENST00000642986.1; ENST00000647021.1; ENST00000645493.1; ENST00000645305.1; ENST00000645982.1; ENST00000647390.1; ENST00000643052.1; ENST00000646174.1; ENST00000433400.6; ENST00000645320.1; ENST00000644501.1 | ||
| External Link | RMBase: m6A_site_584510 | ||
| mod ID: M6ASITE060284 | Click to Show/Hide the Full List | ||
| mod site | chr3:41239277-41239278:+ | [21] | |
| Sequence | AGATCTGGGGCATGCCCAGGACCTCATGGATGGGCTGCCTC | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | CD34; HepG2; HeLa; A549 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000643052.1; ENST00000642248.1; ENST00000644524.1; ENST00000644678.1; ENST00000643031.1; ENST00000646381.1; ENST00000646174.1; ENST00000645210.1; ENST00000643977.1; ENST00000642992.1; ENST00000647390.1; ENST00000645493.1; ENST00000645320.1; ENST00000642886.1; ENST00000441708.2; ENST00000643992.1; ENST00000433400.6; ENST00000642986.1; ENST00000647413.1; ENST00000644867.1; ENST00000431914.6; ENST00000644873.1; ENST00000646725.1; ENST00000645982.1; ENST00000349496.11; ENST00000644501.1; ENST00000450969.6; ENST00000643297.1; ENST00000643541.1; ENST00000642315.1; ENST00000405570.6; ENST00000642836.1; ENST00000646369.1; ENST00000471014.2; ENST00000647264.1; ENST00000396185.8; ENST00000647021.1; ENST00000642426.1; ENST00000645305.1; ENST00000453024.6; ENST00000645900.1; ENST00000646074.1; ENST00000645276.1; ENST00000396183.7; ENST00000646116.1 | ||
| External Link | RMBase: m6A_site_584511 | ||
| mod ID: M6ASITE060285 | Click to Show/Hide the Full List | ||
| mod site | chr3:41239408-41239409:+ | [21] | |
| Sequence | ACTTTTACTCTGCCTACAGAACTTCAGAAAGACTTGGTTGG | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | CD34; HepG2; HeLa; hESCs; fibroblasts; A549 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000643992.1; ENST00000646074.1; ENST00000441708.2; ENST00000644678.1; ENST00000450969.6; ENST00000643052.1; ENST00000646725.1; ENST00000646381.1; ENST00000645305.1; ENST00000646116.1; ENST00000396183.7; ENST00000643297.1; ENST00000471014.2; ENST00000433400.6; ENST00000647021.1; ENST00000645982.1; ENST00000643977.1; ENST00000396185.8; ENST00000643031.1; ENST00000405570.6; ENST00000643541.1; ENST00000645210.1; ENST00000644524.1; ENST00000645320.1; ENST00000349496.11; ENST00000453024.6; ENST00000642315.1; ENST00000642836.1; ENST00000642986.1; ENST00000431914.6; ENST00000647264.1; ENST00000644501.1; ENST00000646174.1; ENST00000642992.1; ENST00000642426.1; ENST00000644873.1; ENST00000645493.1; ENST00000645900.1; ENST00000642248.1; ENST00000647390.1; ENST00000646369.1; ENST00000642886.1; ENST00000644867.1; ENST00000645276.1 | ||
| External Link | RMBase: m6A_site_584512 | ||
| mod ID: M6ASITE060286 | Click to Show/Hide the Full List | ||
| mod site | chr3:41239419-41239420:+ | [21] | |
| Sequence | GCCTACAGAACTTCAGAAAGACTTGGTTGGTAGGGTGGGAG | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | CD34; HepG2; HeLa; hESCs; A549 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000646116.1; ENST00000643031.1; ENST00000450969.6; ENST00000646725.1; ENST00000441708.2; ENST00000644501.1; ENST00000642886.1; ENST00000645276.1; ENST00000642836.1; ENST00000643992.1; ENST00000644867.1; ENST00000396185.8; ENST00000642315.1; ENST00000349496.11; ENST00000645493.1; ENST00000644524.1; ENST00000645210.1; ENST00000643977.1; ENST00000643052.1; ENST00000642248.1; ENST00000471014.2; ENST00000645305.1; ENST00000646174.1; ENST00000453024.6; ENST00000645900.1; ENST00000646369.1; ENST00000643297.1; ENST00000647390.1; ENST00000644678.1; ENST00000645320.1; ENST00000433400.6; ENST00000646381.1; ENST00000642426.1; ENST00000645982.1; ENST00000642986.1; ENST00000431914.6; ENST00000646074.1; ENST00000647264.1; ENST00000405570.6; ENST00000396183.7; ENST00000643541.1; ENST00000644873.1; ENST00000642992.1; ENST00000647021.1 | ||
| External Link | RMBase: m6A_site_584513 | ||
| mod ID: M6ASITE060287 | Click to Show/Hide the Full List | ||
| mod site | chr3:41239466-41239467:+ | [17] | |
| Sequence | AGGCTATTTGTAAATCTGCCACAAAAACAGGTATATACTTT | ||
| Motif Score | 2.053113095 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000405570.6; ENST00000645493.1; ENST00000646725.1; ENST00000644873.1; ENST00000646174.1; ENST00000644867.1; ENST00000643977.1; ENST00000646074.1; ENST00000644524.1; ENST00000645210.1; ENST00000642315.1; ENST00000433400.6; ENST00000453024.6; ENST00000645305.1; ENST00000643541.1; ENST00000441708.2; ENST00000646369.1; ENST00000396185.8; ENST00000471014.2; ENST00000349496.11; ENST00000643031.1; ENST00000646381.1; ENST00000645982.1; ENST00000645320.1; ENST00000645276.1; ENST00000647264.1; ENST00000642836.1; ENST00000644501.1; ENST00000643052.1; ENST00000431914.6; ENST00000396183.7; ENST00000642992.1; ENST00000645900.1; ENST00000450969.6; ENST00000642248.1; ENST00000642886.1; ENST00000643992.1; ENST00000644678.1; ENST00000647390.1; ENST00000646116.1; ENST00000647021.1; ENST00000643297.1; ENST00000642986.1; ENST00000642426.1 | ||
| External Link | RMBase: m6A_site_584514 | ||
| mod ID: M6ASITE060288 | Click to Show/Hide the Full List | ||
| mod site | chr3:41239472-41239473:+ | [22] | |
| Sequence | TTTGTAAATCTGCCACAAAAACAGGTATATACTTTGAAAGG | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa; hESCs; A549 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000645210.1; ENST00000647390.1; ENST00000645305.1; ENST00000642836.1; ENST00000643977.1; ENST00000644524.1; ENST00000396183.7; ENST00000644678.1; ENST00000646725.1; ENST00000645493.1; ENST00000450969.6; ENST00000471014.2; ENST00000647021.1; ENST00000643031.1; ENST00000643052.1; ENST00000645982.1; ENST00000642886.1; ENST00000644501.1; ENST00000433400.6; ENST00000642426.1; ENST00000644873.1; ENST00000646369.1; ENST00000396185.8; ENST00000643541.1; ENST00000643297.1; ENST00000642248.1; ENST00000642315.1; ENST00000644867.1; ENST00000645276.1; ENST00000642986.1; ENST00000453024.6; ENST00000646074.1; ENST00000441708.2; ENST00000431914.6; ENST00000646381.1; ENST00000642992.1; ENST00000645900.1; ENST00000643992.1; ENST00000647264.1; ENST00000646116.1; ENST00000349496.11; ENST00000646174.1; ENST00000645320.1; ENST00000405570.6 | ||
| External Link | RMBase: m6A_site_584515 | ||
| mod ID: M6ASITE060289 | Click to Show/Hide the Full List | ||
| mod site | chr3:41239505-41239506:+ | [17] | |
| Sequence | TTGAAAGGAGATGTCTTGGAACATTGGAATGTTCTCAGATT | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | hESC-HEK293T; hESCs; A549; iSLK | ||
| Seq Type List | MAZTER-seq; m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000645210.1; ENST00000646116.1; ENST00000643297.1; ENST00000642992.1; ENST00000642986.1; ENST00000645320.1; ENST00000396185.8; ENST00000433400.6; ENST00000646369.1; ENST00000645982.1; ENST00000642315.1; ENST00000643977.1; ENST00000450969.6; ENST00000441708.2; ENST00000643052.1; ENST00000642886.1; ENST00000405570.6; ENST00000645900.1; ENST00000644678.1; ENST00000643992.1; ENST00000642248.1; ENST00000453024.6; ENST00000643541.1; ENST00000645305.1; ENST00000645276.1; ENST00000644867.1; ENST00000646725.1; ENST00000645493.1; ENST00000646174.1; ENST00000431914.6; ENST00000471014.2; ENST00000646381.1; ENST00000647021.1; ENST00000644873.1; ENST00000643031.1; ENST00000647264.1; ENST00000642426.1; ENST00000349496.11; ENST00000646074.1; ENST00000642836.1; ENST00000647390.1; ENST00000644501.1; ENST00000644524.1; ENST00000396183.7 | ||
| External Link | RMBase: m6A_site_584516 | ||
| mod ID: M6ASITE060290 | Click to Show/Hide the Full List | ||
| mod site | chr3:41239579-41239580:+ | [19] | |
| Sequence | TAACTTTAATGTTTTTTGCCACAGCTTTTGCAACTTAATAC | ||
| Motif Score | 2.053113095 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000644867.1; ENST00000645493.1; ENST00000642986.1; ENST00000645305.1; ENST00000643992.1; ENST00000349496.11; ENST00000642426.1; ENST00000644501.1; ENST00000431914.6; ENST00000450969.6; ENST00000643052.1; ENST00000441708.2; ENST00000644524.1; ENST00000646381.1; ENST00000643297.1; ENST00000645982.1; ENST00000644873.1; ENST00000396183.7; ENST00000453024.6; ENST00000642836.1; ENST00000647264.1; ENST00000433400.6; ENST00000642248.1; ENST00000642315.1; ENST00000647021.1; ENST00000646725.1; ENST00000647390.1; ENST00000645210.1; ENST00000643031.1; ENST00000645320.1; ENST00000405570.6; ENST00000646116.1; ENST00000646174.1; ENST00000643541.1; ENST00000644678.1; ENST00000645276.1; ENST00000396185.8; ENST00000646369.1; ENST00000642886.1; ENST00000646074.1; ENST00000471014.2; ENST00000643977.1; ENST00000642992.1; ENST00000645900.1 | ||
| External Link | RMBase: m6A_site_584517 | ||
| mod ID: M6ASITE060291 | Click to Show/Hide the Full List | ||
| mod site | chr3:41239672-41239673:+ | [16] | |
| Sequence | TATACAGCTGTATTGTCTGAACTTGCATTGTGATTGGCCTG | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa; CD34; HEK293T; HepG2; U2OS; A549; TIME | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000644873.1; ENST00000349496.11; ENST00000646174.1; ENST00000643297.1; ENST00000642248.1; ENST00000645493.1; ENST00000450969.6; ENST00000647264.1; ENST00000642315.1; ENST00000642986.1; ENST00000646116.1; ENST00000396185.8; ENST00000642992.1; ENST00000644867.1; ENST00000645210.1; ENST00000433400.6; ENST00000642426.1; ENST00000453024.6; ENST00000644678.1; ENST00000646381.1; ENST00000643031.1; ENST00000644501.1; ENST00000471014.2; ENST00000646725.1; ENST00000643977.1; ENST00000645982.1; ENST00000431914.6; ENST00000642886.1; ENST00000645320.1; ENST00000643992.1; ENST00000647390.1; ENST00000645276.1; ENST00000642836.1; ENST00000646074.1; ENST00000441708.2; ENST00000396183.7; ENST00000644524.1; ENST00000645900.1; ENST00000645305.1; ENST00000646369.1; ENST00000643052.1; ENST00000405570.6; ENST00000643541.1; ENST00000647021.1 | ||
| External Link | RMBase: m6A_site_584518 | ||
| mod ID: M6ASITE060292 | Click to Show/Hide the Full List | ||
| mod site | chr3:41239745-41239746:+ | [17] | |
| Sequence | GGTATCTCAGAAAGTGCCTGACACACTAACCAAGCTGAGTT | ||
| Motif Score | 2.859755952 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000642315.1; ENST00000646381.1; ENST00000642836.1; ENST00000643052.1; ENST00000441708.2; ENST00000431914.6; ENST00000643541.1; ENST00000644678.1; ENST00000471014.2; ENST00000642426.1; ENST00000647021.1; ENST00000644867.1; ENST00000642986.1; ENST00000453024.6; ENST00000642992.1; ENST00000644873.1; ENST00000645982.1; ENST00000396185.8; ENST00000405570.6; ENST00000643977.1; ENST00000646116.1; ENST00000644501.1; ENST00000450969.6; ENST00000646725.1; ENST00000396183.7; ENST00000645276.1; ENST00000645493.1; ENST00000643031.1; ENST00000644524.1; ENST00000433400.6; ENST00000645320.1; ENST00000645900.1; ENST00000643992.1; ENST00000643297.1; ENST00000645210.1; ENST00000642886.1; ENST00000642248.1; ENST00000647390.1; ENST00000646369.1; ENST00000349496.11; ENST00000646074.1; ENST00000645305.1 | ||
| External Link | RMBase: m6A_site_584519 | ||
| mod ID: M6ASITE060293 | Click to Show/Hide the Full List | ||
| mod site | chr3:41239776-41239777:+ | [16] | |
| Sequence | AAGCTGAGTTTCCTATGGGAACAATTGAAGTAAACTTTTTG | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa; CD34; HepG2; HEK293T; A549; hESC-HEK293T; U2OS; hNPCs; hESCs; fibroblasts; GM12878; LCLs; MM6; Huh7; HEK293A-TOA; iSLK; MSC; TIME; TREX; endometrial | ||
| Seq Type List | m6A-seq; MeRIP-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000396185.8; ENST00000405570.6; ENST00000349496.11; ENST00000644873.1; ENST00000645320.1; ENST00000645982.1; ENST00000642315.1; ENST00000396183.7; ENST00000431914.6; ENST00000642986.1; ENST00000642886.1; ENST00000643297.1; ENST00000471014.2; ENST00000642836.1; ENST00000644678.1; ENST00000644501.1; ENST00000646725.1; ENST00000647021.1; ENST00000645493.1; ENST00000647390.1; ENST00000646074.1; ENST00000645305.1; ENST00000645900.1; ENST00000644867.1; ENST00000643992.1; ENST00000453024.6; ENST00000645276.1; ENST00000433400.6; ENST00000450969.6; ENST00000643031.1; ENST00000645210.1; ENST00000646369.1; ENST00000643977.1; ENST00000646116.1; ENST00000642248.1; ENST00000642426.1; ENST00000646381.1; ENST00000642992.1; ENST00000643052.1; ENST00000643541.1; ENST00000644524.1 | ||
| External Link | RMBase: m6A_site_584520 | ||
| mod ID: M6ASITE060294 | Click to Show/Hide the Full List | ||
| mod site | chr3:41239789-41239790:+ | [16] | |
| Sequence | TATGGGAACAATTGAAGTAAACTTTTTGTTCTGGTCCTTTT | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa; CD34; HepG2; HEK293T; A549; U2OS; H1A; H1B; hNPCs; hESCs; fibroblasts; GM12878; LCLs; CD8T; MM6; Huh7; Jurkat; HEK293A-TOA; iSLK; MSC; TIME; TREX; endometrial | ||
| Seq Type List | m6A-seq; MeRIP-seq; m6A-CLIP/IP | ||
| Transcript ID List | ENST00000643031.1; ENST00000642886.1; ENST00000642836.1; ENST00000450969.6; ENST00000646074.1; ENST00000645320.1; ENST00000645305.1; ENST00000647021.1; ENST00000645900.1; ENST00000642426.1; ENST00000453024.6; ENST00000471014.2; ENST00000646116.1; ENST00000642315.1; ENST00000643297.1; ENST00000644524.1; ENST00000396185.8; ENST00000431914.6; ENST00000644873.1; ENST00000643977.1; ENST00000644501.1; ENST00000396183.7; ENST00000642248.1; ENST00000646381.1; ENST00000644678.1; ENST00000645982.1; ENST00000647390.1; ENST00000349496.11; ENST00000433400.6; ENST00000642986.1; ENST00000643541.1; ENST00000645493.1; ENST00000643052.1; ENST00000405570.6; ENST00000646369.1; ENST00000645210.1; ENST00000645276.1; ENST00000642992.1; ENST00000643992.1; ENST00000646725.1; ENST00000644867.1 | ||
| External Link | RMBase: m6A_site_584521 | ||
| mod ID: M6ASITE060295 | Click to Show/Hide the Full List | ||
| mod site | chr3:41239823-41239824:+ | [17] | |
| Sequence | TCCTTTTTGGTCGAGGAGTAACAATACAAATGGATTTTGGG | ||
| Motif Score | 2.168095238 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000642315.1; ENST00000349496.11; ENST00000646074.1; ENST00000645320.1; ENST00000642986.1; ENST00000453024.6; ENST00000396185.8; ENST00000646725.1; ENST00000647390.1; ENST00000646369.1; ENST00000645493.1; ENST00000642992.1; ENST00000643992.1; ENST00000645305.1; ENST00000643031.1; ENST00000644501.1; ENST00000643297.1; ENST00000396183.7; ENST00000450969.6; ENST00000405570.6; ENST00000433400.6; ENST00000645900.1; ENST00000644867.1; ENST00000643052.1; ENST00000645276.1; ENST00000647021.1; ENST00000431914.6; ENST00000642886.1; ENST00000642836.1; ENST00000642426.1; ENST00000645210.1; ENST00000643541.1; ENST00000646116.1; ENST00000645982.1; ENST00000644524.1; ENST00000642248.1; ENST00000471014.2; ENST00000644873.1; ENST00000643977.1; ENST00000644678.1; ENST00000646381.1 | ||
| External Link | RMBase: m6A_site_584522 | ||
| mod ID: M6ASITE060296 | Click to Show/Hide the Full List | ||
| mod site | chr3:41239828-41239829:+ | [17] | |
| Sequence | TTTGGTCGAGGAGTAACAATACAAATGGATTTTGGGAGTGA | ||
| Motif Score | 2.110482143 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000453024.6; ENST00000643297.1; ENST00000645210.1; ENST00000642836.1; ENST00000471014.2; ENST00000644873.1; ENST00000643052.1; ENST00000645320.1; ENST00000645982.1; ENST00000433400.6; ENST00000645900.1; ENST00000642426.1; ENST00000646725.1; ENST00000646116.1; ENST00000643977.1; ENST00000642986.1; ENST00000647390.1; ENST00000645305.1; ENST00000405570.6; ENST00000644678.1; ENST00000647021.1; ENST00000643031.1; ENST00000396185.8; ENST00000643541.1; ENST00000645276.1; ENST00000642992.1; ENST00000646381.1; ENST00000642248.1; ENST00000450969.6; ENST00000396183.7; ENST00000642886.1; ENST00000643992.1; ENST00000644524.1; ENST00000642315.1; ENST00000349496.11; ENST00000644867.1; ENST00000644501.1; ENST00000646369.1; ENST00000646074.1; ENST00000645493.1; ENST00000431914.6 | ||
| External Link | RMBase: m6A_site_584523 | ||
| mod ID: M6ASITE060297 | Click to Show/Hide the Full List | ||
| mod site | chr3:41239868-41239869:+ | [19] | |
| Sequence | ACTCAAGAAGTGAAGAATGCACAAGAATGGATCACAAGATG | ||
| Motif Score | 2.830589286 | ||
| Cell/Tissue List | HEK293T; hESC-HEK293T | ||
| Seq Type List | DART-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000645305.1; ENST00000642426.1; ENST00000642886.1; ENST00000644678.1; ENST00000471014.2; ENST00000349496.11; ENST00000453024.6; ENST00000647390.1; ENST00000646116.1; ENST00000645276.1; ENST00000642315.1; ENST00000642992.1; ENST00000644873.1; ENST00000433400.6; ENST00000431914.6; ENST00000405570.6; ENST00000642986.1; ENST00000642248.1; ENST00000643297.1; ENST00000646369.1; ENST00000646725.1; ENST00000644867.1; ENST00000645210.1; ENST00000642836.1; ENST00000450969.6; ENST00000647021.1; ENST00000645320.1; ENST00000643541.1; ENST00000643992.1; ENST00000646381.1; ENST00000644524.1; ENST00000396185.8; ENST00000643052.1; ENST00000645900.1; ENST00000645493.1; ENST00000643977.1; ENST00000643031.1; ENST00000396183.7; ENST00000645982.1; ENST00000644501.1; ENST00000646074.1 | ||
| External Link | RMBase: m6A_site_584524 | ||
| mod ID: M6ASITE060298 | Click to Show/Hide the Full List | ||
| mod site | chr3:41239881-41239882:+ | [19] | |
| Sequence | AGAATGCACAAGAATGGATCACAAGATGGAATTTATCAAAC | ||
| Motif Score | 2.047297619 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000645900.1; ENST00000646116.1; ENST00000643031.1; ENST00000643992.1; ENST00000450969.6; ENST00000644867.1; ENST00000646074.1; ENST00000405570.6; ENST00000645320.1; ENST00000642836.1; ENST00000643052.1; ENST00000642315.1; ENST00000643541.1; ENST00000645493.1; ENST00000643977.1; ENST00000396183.7; ENST00000646381.1; ENST00000647021.1; ENST00000645210.1; ENST00000643297.1; ENST00000644873.1; ENST00000646369.1; ENST00000349496.11; ENST00000642992.1; ENST00000647390.1; ENST00000471014.2; ENST00000644678.1; ENST00000644524.1; ENST00000645982.1; ENST00000642248.1; ENST00000453024.6; ENST00000642426.1; ENST00000646725.1; ENST00000396185.8; ENST00000642886.1; ENST00000645276.1; ENST00000433400.6; ENST00000645305.1; ENST00000642986.1; ENST00000644501.1; ENST00000431914.6 | ||
| External Link | RMBase: m6A_site_584525 | ||
| mod ID: M6ASITE060299 | Click to Show/Hide the Full List | ||
| mod site | chr3:41239900-41239901:+ | [16] | |
| Sequence | CACAAGATGGAATTTATCAAACCCTAGCCTTGCTTGTTAAA | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; A549; U2OS; H1A; H1B; hNPCs; hESCs; fibroblasts; GM12878; LCLs; MM6; Huh7; Jurkat; HEK293A-TOA; iSLK; MSC; TIME; TREX; endometrial | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000645305.1; ENST00000396183.7; ENST00000643297.1; ENST00000433400.6; ENST00000642992.1; ENST00000471014.2; ENST00000647021.1; ENST00000646116.1; ENST00000643031.1; ENST00000642315.1; ENST00000396185.8; ENST00000644501.1; ENST00000645276.1; ENST00000642836.1; ENST00000349496.11; ENST00000645982.1; ENST00000453024.6; ENST00000645210.1; ENST00000644524.1; ENST00000646725.1; ENST00000645900.1; ENST00000644867.1; ENST00000642426.1; ENST00000642886.1; ENST00000644873.1; ENST00000405570.6; ENST00000646369.1; ENST00000646074.1; ENST00000642986.1; ENST00000643541.1; ENST00000645493.1; ENST00000450969.6; ENST00000643052.1 | ||
| External Link | RMBase: m6A_site_584526 | ||
| mod ID: M6ASITE060300 | Click to Show/Hide the Full List | ||
| mod site | chr3:41239962-41239963:+ | [19] | |
| Sequence | GAATATCTGTAATGGTACTGACTTTGCTTGCTTTGAAGTAG | ||
| Motif Score | 3.28175 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000349496.11; ENST00000642836.1; ENST00000396183.7; ENST00000645210.1; ENST00000642986.1; ENST00000643031.1; ENST00000396185.8; ENST00000471014.2; ENST00000433400.6 | ||
| External Link | RMBase: m6A_site_584527 | ||
| mod ID: M6ASITE060301 | Click to Show/Hide the Full List | ||
| mod site | chr3:41240058-41240059:+ | [19] | |
| Sequence | AGTGTTAAGTTATAGTGAATACTGCTACAGCAATTTCTAAT | ||
| Motif Score | 2.53247619 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000396183.7; ENST00000396185.8; ENST00000471014.2; ENST00000643031.1; ENST00000349496.11 | ||
| External Link | RMBase: m6A_site_584528 | ||
| mod ID: M6ASITE060302 | Click to Show/Hide the Full List | ||
| mod site | chr3:41240064-41240065:+ | [19] | |
| Sequence | AAGTTATAGTGAATACTGCTACAGCAATTTCTAATTTTTAA | ||
| Motif Score | 2.078666667 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000349496.11; ENST00000643031.1; ENST00000396183.7; ENST00000396185.8; ENST00000471014.2 | ||
| External Link | RMBase: m6A_site_584529 | ||
| mod ID: M6ASITE060303 | Click to Show/Hide the Full List | ||
| mod site | chr3:41240105-41240106:+ | [17] | |
| Sequence | GAATTGAGTAATGGTGTAGAACACTAATTCATAATCACTCT | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | hESC-HEK293T; Huh7 | ||
| Seq Type List | MAZTER-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000643031.1; ENST00000396185.8; ENST00000396183.7; ENST00000471014.2; ENST00000349496.11 | ||
| External Link | RMBase: m6A_site_584530 | ||
| mod ID: M6ASITE060304 | Click to Show/Hide the Full List | ||
| mod site | chr3:41240121-41240122:+ | [19] | |
| Sequence | TAGAACACTAATTCATAATCACTCTAATTAATTGTAATCTG | ||
| Motif Score | 2.469291667 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000396183.7; ENST00000396185.8; ENST00000471014.2; ENST00000643031.1; ENST00000349496.11 | ||
| External Link | RMBase: m6A_site_584531 | ||
| mod ID: M6ASITE060305 | Click to Show/Hide the Full List | ||
| mod site | chr3:41240153-41240154:+ | [19] | |
| Sequence | TGTAATCTGAATAAAGTGTAACAATTGTGTAGCCTTTTTGT | ||
| Motif Score | 2.168095238 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000349496.11; ENST00000471014.2; ENST00000643031.1; ENST00000396183.7; ENST00000396185.8 | ||
| External Link | RMBase: m6A_site_584532 | ||
| mod ID: M6ASITE060306 | Click to Show/Hide the Full List | ||
| mod site | chr3:41240183-41240184:+ | [16] | |
| Sequence | AGCCTTTTTGTATAAAATAGACAAATAGAAAATGGTCCAAT | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa; HEK293T; hESC-HEK293T; Huh7 | ||
| Seq Type List | m6A-seq; DART-seq; MAZTER-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000471014.2; ENST00000396183.7; ENST00000396185.8; ENST00000349496.11; ENST00000643031.1 | ||
| External Link | RMBase: m6A_site_584533 | ||
| mod ID: M6ASITE060307 | Click to Show/Hide the Full List | ||
| mod site | chr3:41240266-41240267:+ | [16] | |
| Sequence | TTTCATGTTTTTGATCAAAAACTATTTGGGATATGTATGGG | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa; HEK293T; Huh7 | ||
| Seq Type List | m6A-seq; DART-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000643031.1; ENST00000349496.11; ENST00000396183.7; ENST00000396185.8; ENST00000471014.2 | ||
| External Link | RMBase: m6A_site_584534 | ||
| mod ID: M6ASITE060308 | Click to Show/Hide the Full List | ||
| mod site | chr3:41240318-41240319:+ | [16] | |
| Sequence | AGTAAGAGGTGTTATTTGGAACCTTGTTTTGGACAGTTTAC | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | HeLa; Huh7 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000643031.1; ENST00000349496.11; ENST00000396185.8; ENST00000396183.7; ENST00000471014.2 | ||
| External Link | RMBase: m6A_site_584535 | ||
| mod ID: M6ASITE060309 | Click to Show/Hide the Full List | ||
| mod site | chr3:41240330-41240331:+ | [16] | |
| Sequence | TATTTGGAACCTTGTTTTGGACAGTTTACCAGTTGCCTTTT | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; Huh7 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000396183.7; ENST00000643031.1; ENST00000349496.11; ENST00000396185.8; ENST00000471014.2 | ||
| External Link | RMBase: m6A_site_584536 | ||
| mod ID: M6ASITE060310 | Click to Show/Hide the Full List | ||
| mod site | chr3:41240428-41240429:+ | [16] | |
| Sequence | AAAAAATGGTTCAGAATTAAACTTTTAATTCATTCGATTGT | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa; HEK293T; Huh7 | ||
| Seq Type List | m6A-seq; DART-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000396185.8; ENST00000349496.11; ENST00000396183.7; ENST00000471014.2; ENST00000643031.1 | ||
| External Link | RMBase: m6A_site_584537 | ||
References