m6A Regulator Information
General Information of the m6A Regulator (ID: REG00025)
| Regulator Name | YTH domain-containing family protein 3 (YTHDF3) | ||||
|---|---|---|---|---|---|
| Synonyms |
DF3
Click to Show/Hide
|
||||
| Gene Name | YTHDF3 | ||||
| Sequence |
MSATSVDQRPKGQGNKVSVQNGSIHQKDAVNDDDFEPYLSSQTNQSNSYPPMSDPYMPSY
YAPSIGFPYSLGEAAWSTAGDQPMPYLTTYGQMSNGEHHYIPDGVFSQPGALGNTPPFLG QHGFNFFPGNADFSTWGTSGSQGQSTQSSAYSSSYGYPPSSLGRAITDGQAGFGNDTLSK VPGISSIEQGMTGLKIGGDLTAAVTKTVGTALSSSGMTSIATNSVPPVSSAAPKPTSWAA IARKPAKPQPKLKPKGNVGIGGSAVPPPPIKHNMNIGTWDEKGSVVKAPPTQPVLPPQTI IQQPQPLIQPPPLVQSQLPQQQPQPPQPQQQQGPQPQAQPHQVQPQQQQLQNRWVAPRNR GAGFNQNNGAGSENFGLGVVPVSASPSSVEVHPVLEKLKAINNYNPKDFDWNLKNGRVFI IKSYSEDDIHRSIKYSIWCSTEHGNKRLDAAYRSLNGKGPLYLLFSVNGSGHFCGVAEMK SVVDYNAYAGVWSQDKWKGKFEVKWIFVKDVPNNQLRHIRLENNDNKPVTNSRDTQEVPL EKAKQVLKIIATFKHTTSIFDDFAHYEKRQEEEEAMRRERNRNKQ Click to Show/Hide
|
||||
| Family | YTHDF family; YTHDF3 subfamily | ||||
| Function |
Specifically recognizes and binds N6-methyladenosine (m6A)-containing RNAs, and regulates their stability. M6A is a modification present at internal sites of mRNAs and some non-coding RNAs and plays a role in mRNA stability and processing. Acts as a regulator of mRNA stability by promoting degradation of m6A-containing mRNAs via interaction with the CCR4-NOT complex or PAN3. The YTHDF paralogs (YTHDF1, YTHDF2 and YTHDF3) share m6A-containing mRNAs targets and act redundantly to mediate mRNA degradation and cellular differentiation. Acts as a negative regulator of type I interferon response by down-regulating interferon-stimulated genes (ISGs) expression: acts by binding to FOXO3 mRNAs (By similarity). Binds to FOXO3 mRNAs independently of METTL3-mediated m6A modification (By similarity). Can also act as a regulator of mRNA stability in cooperation with YTHDF2 by binding to m6A-containing mRNA and promoting their degradation. Recognizes and binds m6A-containing circular RNAs (circRNAs); circRNAs are generated through back-splicing of pre-mRNAs, a non-canonical splicing process promoted by dsRNA structures across circularizing exons. Promotes formation of phase-separated membraneless compartments, such as P-bodies or stress granules, by undergoing liquid-liquid phase separation upon binding to mRNAs containing multiple m6A-modified residues: polymethylated mRNAs act as a multivalent scaffold for the binding of YTHDF proteins, juxtaposing their disordered regions and thereby leading to phase separation. The resulting mRNA-YTHDF complexes then partition into different endogenous phase-separated membraneless compartments, such as P-bodies, stress granules or neuronal RNA granules. May also recognize and bind N1-methyladenosine (m1A)-containing mRNAs: inhibits trophoblast invasion by binding to m1A-methylated transcripts of IGF1R, promoting their degradation; Has some antiviral activity against HIV-1 virus: incorporated into HIV-1 particles in a nucleocapsid-dependent manner and reduces viral infectivity in the next cycle of infection . May interfere with this early step of the viral life cycle by binding to N6-methyladenosine (m6A) modified sites on the HIV-1 RNA genome.
Click to Show/Hide
|
||||
| Gene ID | 253943 | ||||
| Uniprot ID | |||||
| Regulator Type | WRITER ERASER READER | ||||
| Mechanism Diagram | Click to View the Original Diagram | ||||
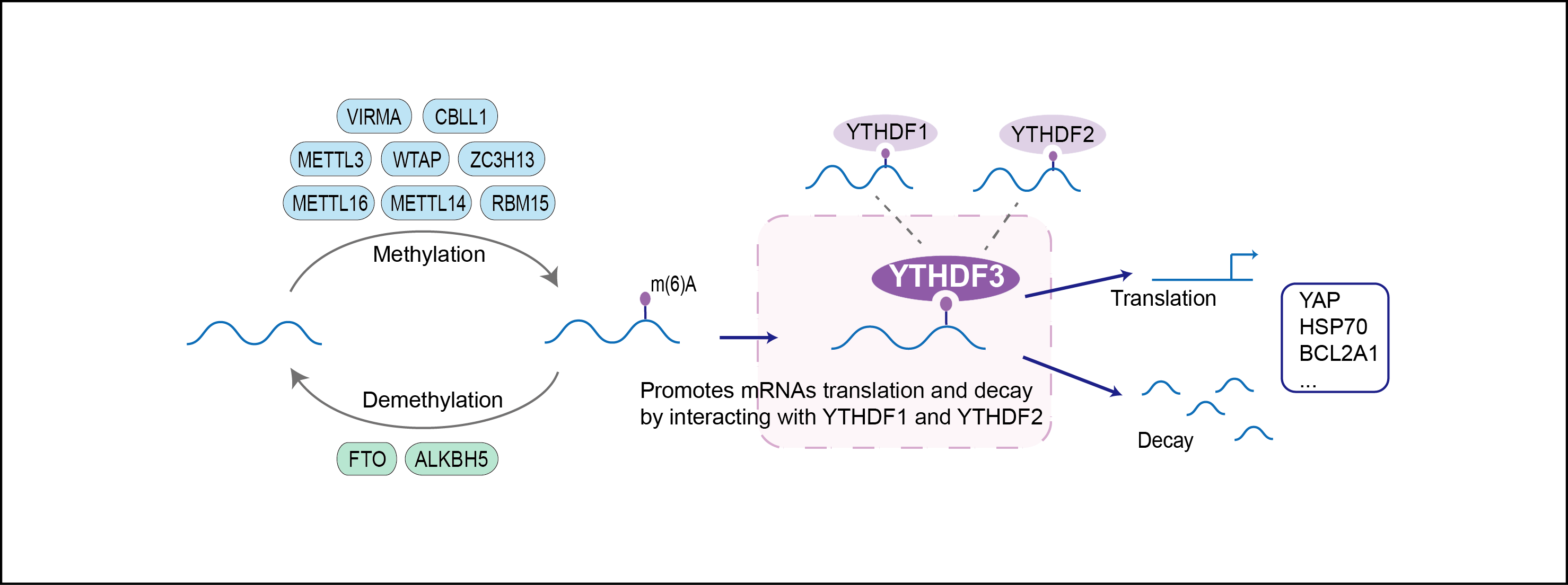
|
|||||
| Target Genes | Click to View Potential Target Genes of This Regulator | ||||
Full List of Target Gene(s) of This m6A Regulator and Corresponding Disease/Drug Response(s)
YTHDF3 can regulate the m6A methylation of following target genes, and result in corresponding disease/drug response(s). You can browse corresponding disease or drug response(s) resulted from the regulation of certain target gene.
Browse Target Gene related Disease
Browse Target Gene related Drug
Gap junction alpha-1 protein (GJA1)
| Representative RNA-seq result indicating the expression of this target gene regulated by YTHDF3 | ||
| Cell Line | Mouse embryonic fibroblasts | Mus musculus |
|
Treatment: shYthdf3 embryonic fibroblasts
Control: shLuc embryonic fibroblasts
|
GSE156437 | |
| Regulation |
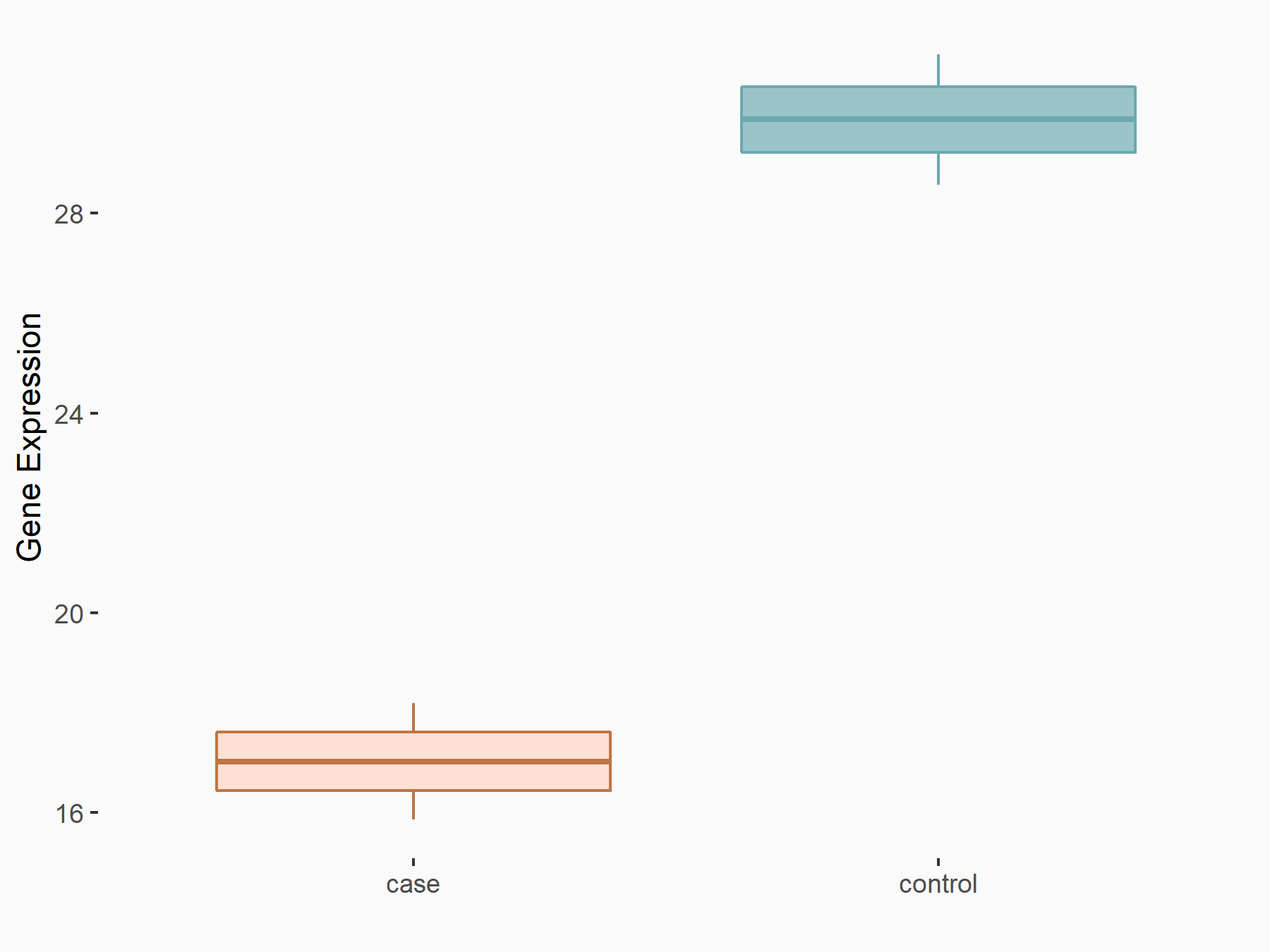  |
logFC: -7.79E-01 p-value: 7.79E-03 |
| More Results | Click to View More RNA-seq Results | |
Breast cancer [ICD-11: 2C60]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [1] | |||
| Responsed Disease | Breast cancer [ICD-11: 2C60] | |||
| Target Regulation | Up regulation | |||
| Cell Process | Cell metastasis | |||
In-vitro Model |
MDA-MB-231Br (After brain metastases of MDA-MB-361 breast adenocarcinoma cells) | |||
| MDA-MB-231 | Breast adenocarcinoma | Homo sapiens | CVCL_0062 | |
| MDA-IBC-3 | Breast inflammatory carcinoma | Homo sapiens | CVCL_HC47 | |
| JIMT-1Br3 (After brain metastases of JIMT-1 breast cancer cells) | ||||
| JIMT-1 | Breast ductal carcinoma | Homo sapiens | CVCL_2077 | |
| HEK293-FT | Normal | Homo sapiens | CVCL_6911 | |
| HCC1954Br (After brain metastases of HCC1954 breast cancer cells) | ||||
| HCC1954 | Breast ductal carcinoma | Homo sapiens | CVCL_1259 | |
| bEnd.3 | Cerebrovascular endothelioma cells from mice | Mus musculus | CVCL_0170 | |
| BEAS-2B | Normal | Homo sapiens | CVCL_0168 | |
| 4T1Br (After brain metastases of 4T1 mouse breast cancer cells) | ||||
| 4T1 | Normal | Mus musculus | CVCL_0125 | |
| In-vivo Model | For the in vivo brain and bone extravasation and seeding assays, cancer cells labeled with CMFDA C2925 (Thermo fisher scientific) or GFP were injected intracardially into the nude mice. Cell number and injection procedure were described in "Animal Experiments". For the in vivo lung extravasation and seeding assays, cancer cells labeled with GFP (2.5 × 105 cells/mouse) were injected into the tail vein of nude mice. At 24 or 48 hrs later, the mice were sacrificed. | |||
| Response Summary | Mechanistically, YTHDF3 enhances the translation of m6A-enriched transcripts for ST6GALNAC5, Gap junction alpha-1 protein (GJA1), and EGFR, all associated with breast cancer brain metastasis. This work uncovers an essential role of YTHDF3 in controlling the interaction between cancer cells and brain microenvironment, thereby inducing brain metastatic competence. | |||
Catenin beta-1 (CTNNB1/Beta-catenin)
| Representative RIP-seq result supporting the interaction between the target gene and YTHDF3 | ||
| Cell Line | Hela | Homo sapiens |
| Regulation | logFC: 1.43E+00 | GSE86214 |
Melanoma of uvea [ICD-11: 2D0Y]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [2] | |||
| Responsed Disease | Melanoma of uvea [ICD-11: 2D0Y] | |||
| Target Regulation | Up regulation | |||
| Response Summary | YTHDF3 enhances Catenin beta-1 (CTNNB1/Beta-catenin) translation through recognizing and binding the m6A peaks on CTNNB1 mRNA.m6A reading protein YTHDF3 promotes the translation of the target transcript CTNNB1, contributing to ocular melanoma propagation and migration through m6A methylation. | |||
Epidermal growth factor receptor (EGFR)
| Representative RIP-seq result supporting the interaction between the target gene and YTHDF3 | ||
| Cell Line | Hela | Homo sapiens |
| Regulation | logFC: 2.94E+00 | GSE86214 |
Breast cancer [ICD-11: 2C60]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [1] | |||
| Responsed Disease | Breast cancer [ICD-11: 2C60] | |||
| Target Regulation | Up regulation | |||
| Cell Process | Cell metastasis | |||
In-vitro Model |
MDA-MB-231Br (After brain metastases of MDA-MB-361 breast adenocarcinoma cells) | |||
| MDA-MB-231 | Breast adenocarcinoma | Homo sapiens | CVCL_0062 | |
| MDA-IBC-3 | Breast inflammatory carcinoma | Homo sapiens | CVCL_HC47 | |
| JIMT-1Br3 (After brain metastases of JIMT-1 breast cancer cells) | ||||
| JIMT-1 | Breast ductal carcinoma | Homo sapiens | CVCL_2077 | |
| HEK293-FT | Normal | Homo sapiens | CVCL_6911 | |
| HCC1954Br (After brain metastases of HCC1954 breast cancer cells) | ||||
| HCC1954 | Breast ductal carcinoma | Homo sapiens | CVCL_1259 | |
| bEnd.3 | Cerebrovascular endothelioma cells from mice | Mus musculus | CVCL_0170 | |
| BEAS-2B | Normal | Homo sapiens | CVCL_0168 | |
| 4T1Br (After brain metastases of 4T1 mouse breast cancer cells) | ||||
| 4T1 | Normal | Mus musculus | CVCL_0125 | |
| In-vivo Model | For the in vivo brain and bone extravasation and seeding assays, cancer cells labeled with CMFDA C2925 (Thermo fisher scientific) or GFP were injected intracardially into the nude mice. Cell number and injection procedure were described in "Animal Experiments". For the in vivo lung extravasation and seeding assays, cancer cells labeled with GFP (2.5 × 105 cells/mouse) were injected into the tail vein of nude mice. At 24 or 48 hrs later, the mice were sacrificed. | |||
| Response Summary | Mechanistically, YTHDF3 enhances the translation of m6A-enriched transcripts for ST6GALNAC5, GJA1, and Epidermal growth factor receptor (EGFR), all associated with breast cancer brain metastasis. This work uncovers an essential role of YTHDF3 in controlling the interaction between cancer cells and brain microenvironment, thereby inducing brain metastatic competence. | |||
Liver cancer [ICD-11: 2C12]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [11] | |||
| Responsed Disease | Liver hepatocellular carcinoma [ICD-11: 2C12.02] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
Hep 3B2.1-7 | Childhood hepatocellular carcinoma | Homo sapiens | CVCL_0326 |
| MHCC97-H | Adult hepatocellular carcinoma | Homo sapiens | CVCL_4972 | |
| BEL-7402 | Endocervical adenocarcinoma | Homo sapiens | CVCL_5492 | |
| Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 | |
| Huh-7 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_0336 | |
| HCCLM3 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_6832 | |
| SMMC-7721 | Endocervical adenocarcinoma | Homo sapiens | CVCL_0534 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| In-vivo Model | For mice in each group, 1 × 107 cells in 200 μL of PBS were injected into the subcutaneous tissue on the right flank. After injection, tumor parameters were measured every 2 days, and the tumor volume was calculated as (length × width × width)/2. | |||
Forkhead box protein M1 (FOXM1)
| Representative RIP-seq result supporting the interaction between the target gene and YTHDF3 | ||
| Cell Line | Hela | Homo sapiens |
| Regulation | logFC: 1.19E+00 | GSE86214 |
Neoplasms of haematopoietic or lymphoid tissues [ICD-11: 2B3Z]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [3] | |||
| Responsed Disease | Neoplasms of haematopoietic or lymphoid tissues [ICD-11: 2B3Z] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | mRNA surveillance pathway | hsa03015), RNA degradation | ||
| Cell Process | RNA stability | |||
| Response Summary | m6A reader Ythdf3 protects hematopoietic stem cell integrity under stress by promoting the translation of Forkhead box protein M1 (FOXM1) and Asxl1 transcripts. | |||
Forkhead box protein O3 (FOXO3)
| Representative RIP-seq result supporting the interaction between the target gene and YTHDF3 | ||
| Cell Line | Hela | Homo sapiens |
| Regulation | logFC: 2.93E+00 | GSE86214 |
Inflammatory response [ICD-11: MG46]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [4] | |||
| Responsed Disease | Inflammatory response [ICD-11: MG46] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | FoxO signaling pathway | hsa04068 | ||
| In-vivo Model | YTHDF3-/- mice were generated using the CRISPR-Cas9 system. | |||
| Response Summary | YTHDF3 as a negative regulator of antiviral immunity through the translational promotion of Forkhead box protein O3 (FOXO3) mRNA under homeostatic conditions, adding insight into the networks of RNA-binding protein-RNA interactions in homeostatically maintaining host antiviral immune function and preventing inflammatory response. | |||
Integrin alpha-6 (ITGA6)
| Representative RIP-seq result supporting the interaction between the target gene and YTHDF3 | ||
| Cell Line | Hela | Homo sapiens |
| Regulation | logFC: 1.67E+00 | GSE86214 |
Liver cancer [ICD-11: 2C12]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [5] | |||
| Responsed Disease | Hepatocellular carcinoma [ICD-11: 2C12.02] | |||
| Cell Process | Cell proliferation | |||
| Cell migration | ||||
| Cell invasion | ||||
| Cell apoptosis | ||||
In-vitro Model |
HEK293T | Normal | Homo sapiens | CVCL_0063 |
| Hep 3B2.1-7 | Childhood hepatocellular carcinoma | Homo sapiens | CVCL_0326 | |
| SMMC-7721 | Endocervical adenocarcinoma | Homo sapiens | CVCL_0534 | |
| In-vivo Model | HCC cells (5 × 106 cells/mouse) that had been transfected with oe-NC + sh-NC, oe-KDM5B + sh-NC, or oe-KDM5B + sh-ITGA6, or treated with NS, GSK-467 (a selective inhibitor of KDM5B) + oe-NC or GSK-467 + oe-ITGA6 were then subcutaneously implanted into the back of mice. | |||
| Response Summary | KDM5B regulates the YTHDF3/ITGA6 axis by inhibiting the expression of miR-448 to promote the occurrence of hepatocellular carcinoma. miR-448 could target YTHDF3 and inhibit the YTHDF3/Integrin alpha-6 (ITGA6) axis, thereby inhibiting the occurrence of HCC. | |||
Bladder cancer [ICD-11: 2C94]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [6] | |||
| Responsed Disease | Bladder cancer [ICD-11: 2C94] | |||
| Pathway Response | Cell adhesion molecules | hsa04514 | ||
| Cell Process | Cell adhesion | |||
| Cell migration | ||||
| Cell invasion | ||||
In-vitro Model |
5637 | Bladder carcinoma | Homo sapiens | CVCL_0126 |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| J82 | Bladder carcinoma | Homo sapiens | CVCL_0359 | |
| SV-HUC-1 | Normal | Homo sapiens | CVCL_3798 | |
| T24 | Bladder carcinoma | Homo sapiens | CVCL_0554 | |
| UM-UC-3 | Bladder carcinoma | Homo sapiens | CVCL_1783 | |
| In-vivo Model | For the subcutaneous implantation model, 1 × 107 cells were subcutaneously implanted into 5-week-old BALB/cJNju-Foxn1nu/Nju nude mice. | |||
| Response Summary | m6A writer METTL3 and eraser ALKBH5 altered cell adhesion by regulating Integrin alpha-6 (ITGA6) expression in bladder cancer cells. m6A is highly enriched within the ITGA6 transcripts, and increased m6A methylations of the ITGA6 mRNA 3'UTR promotes the translation of ITGA6 mRNA via binding of the m6A readers YTHDF1 and YTHDF3. Inhibition of ITGA6 results in decreased growth and progression of bladder cancer cells in vitro and in vivo. | |||
Polycomb group protein ASXL1 (Asxl1)
| Representative RIP-seq result supporting the interaction between the target gene and YTHDF3 | ||
| Cell Line | Hela | Homo sapiens |
| Regulation | logFC: 2.55E+00 | GSE86214 |
Neoplasms of haematopoietic or lymphoid tissues [ICD-11: 2B3Z]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [3] | |||
| Responsed Disease | Neoplasms of haematopoietic or lymphoid tissues [ICD-11: 2B3Z] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | mRNA surveillance pathway | hsa03015), RNA degradation | ||
| Cell Process | RNA stability | |||
| Response Summary | m6A reader Ythdf3 protects hematopoietic stem cell integrity under stress by promoting the translation of Foxm1 and Polycomb group protein ASXL1 (Asxl1) transcripts. | |||
Transcriptional coactivator YAP1 (YAP1)
| Representative RIP-seq result supporting the interaction between the target gene and YTHDF3 | ||
| Cell Line | Hela | Homo sapiens |
| Regulation | logFC: 1.33E+00 | GSE86214 |
Lung cancer [ICD-11: 2C25]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [7] | |||
| Responsed Disease | Non-small-cell lung carcinoma [ICD-11: 2C25.Y] | |||
| Responsed Drug | Cisplatin | Approved | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Hippo signaling pathway | hsa04390 | ||
| Cell Process | Metabolic | |||
In-vitro Model |
A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 |
| Calu-6 | Lung adenocarcinoma | Homo sapiens | CVCL_0236 | |
| NCI-H1299 | Lung large cell carcinoma | Homo sapiens | CVCL_0060 | |
| NCI-H520 | Lung squamous cell carcinoma | Homo sapiens | CVCL_1566 | |
| In-vivo Model | Mice were injected with 5 × 106 lung cancer cells with stably expression of relevant plasmids and randomly divided into two groups (five mice per group) after the diameter of the xenografted tumors had reached approximately 5 mm in diameter. Xenografted mice were then administrated with PBS or DDP (3 mg/kg per day) for three times a week, and tumor volume were measured every second day. | |||
| Response Summary | METTL3, YTHDF3, YTHDF1, and eIF3b directly promoted YAP translation through an interaction with the translation initiation machinery. METTL3 knockdown inhibits tumor growth and enhances sensitivity to DDP in vivo.m6A mRNA methylation initiated by METTL3 directly promotes YAP translation and increases YAP activity by regulating the MALAT1-miR-1914-3p-Transcriptional coactivator YAP1 (YAP1) axis to induce Non-small cell lung cancer drug resistance and metastasis. | |||
Zinc finger E-box-binding homeobox 1 (ZEB1)
| Representative RIP-seq result supporting the interaction between the target gene and YTHDF3 | ||
| Cell Line | Hela | Homo sapiens |
| Regulation | logFC: 1.92E+00 | GSE86214 |
Liver cancer [ICD-11: 2C12]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [8] | |||
| Responsed Disease | Hepatocellular carcinoma [ICD-11: 2C12.02] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | RNA degradation | hsa03018 | ||
| Cell Process | RNA stability | |||
In-vitro Model |
BEL-7402 | Endocervical adenocarcinoma | Homo sapiens | CVCL_5492 |
| BEL-7404 | Endocervical adenocarcinoma | Homo sapiens | CVCL_6568 | |
| Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 | |
| Huh-7 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_0336 | |
| L-02 | Endocervical adenocarcinoma | Homo sapiens | CVCL_6926 | |
| SMMC-7721 | Endocervical adenocarcinoma | Homo sapiens | CVCL_0534 | |
| In-vivo Model | 1 × 107 Bel-7404 cells in 200 uL PBS were injected into the right flank of nude mice. | |||
| Response Summary | Circ_KIAA1429 could accelerate HCC advancement, maintained the expression of Zeb1 through the mechanism of m6A-YTHDF3-Zinc finger E-box-binding homeobox 1 (ZEB1) in hepatocellular carcinoma. | |||
Breast cancer [ICD-11: 2C60]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [9] | |||
| Responsed Disease | Breast cancer [ICD-11: 2C60] | |||
| Target Regulation | Up regulation | |||
| Cell Process | Cell migration | |||
| Cell invasion | ||||
| Epithelial-mesenchymal transition | ||||
In-vitro Model |
MDA-MB-231 | Breast adenocarcinoma | Homo sapiens | CVCL_0062 |
| Hs 578T | Invasive breast carcinoma | Homo sapiens | CVCL_0332 | |
| HCC1937 | Breast ductal carcinoma | Homo sapiens | CVCL_0290 | |
| BT-549 | Invasive breast carcinoma | Homo sapiens | CVCL_1092 | |
| Response Summary | YTHDF3 positively regulated cell migration, invasion, and EMT in triple-negative breast cancer cells. Moreover, Zinc finger E-box-binding homeobox 1 (ZEB1) was identified as a key downstream target for YTHDF3 and YTHDF3 could enhance ZEB1 mRNA stability in an m6A-dependent manner. | |||
Metastasis associated lung adenocarcinoma transcript 1 (MALAT1)
| Representative RIP-seq result supporting the interaction between the target gene and YTHDF3 | ||
| Cell Line | Hela | Homo sapiens |
| Regulation | logFC: 1.66E+00 | GSE86214 |
Lung cancer [ICD-11: 2C25]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [7] | |||
| Responsed Disease | Non-small-cell lung carcinoma [ICD-11: 2C25.Y] | |||
| Responsed Drug | Cisplatin | Approved | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Hippo signaling pathway | hsa04390 | ||
| Cell Process | Metabolic | |||
In-vitro Model |
A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 |
| Calu-6 | Lung adenocarcinoma | Homo sapiens | CVCL_0236 | |
| NCI-H1299 | Lung large cell carcinoma | Homo sapiens | CVCL_0060 | |
| NCI-H520 | Lung squamous cell carcinoma | Homo sapiens | CVCL_1566 | |
| In-vivo Model | Mice were injected with 5 × 106 lung cancer cells with stably expression of relevant plasmids and randomly divided into two groups (five mice per group) after the diameter of the xenografted tumors had reached approximately 5 mm in diameter. Xenografted mice were then administrated with PBS or DDP (3 mg/kg per day) for three times a week, and tumor volume were measured every second day. | |||
| Response Summary | METTL3, YTHDF3, YTHDF1, and eIF3b directly promoted YAP translation through an interaction with the translation initiation machinery. METTL3 knockdown inhibits tumor growth and enhances sensitivity to DDP in vivo.m6A mRNA methylation initiated by METTL3 directly promotes YAP translation and increases YAP activity by regulating the Metastasis associated lung adenocarcinoma transcript 1 (MALAT1)-miR-1914-3p-YAP axis to induce Non-small cell lung cancer drug resistance and metastasis. | |||
Apoptosis regulator Bcl-2 (BCL2)
Pancreatic cancer [ICD-11: 2C10]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [10] | |||
| Responsed Disease | Pancreatic cancer [ICD-11: 2C10] | |||
| Responsed Drug | Celastrol | Preclinical | ||
| Target Regulation | Up regulation | |||
In-vitro Model |
PANC-1 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0480 |
| SW1990 | Pancreatic adenocarcinoma | Homo sapiens | CVCL_1723 | |
| AsPC-1 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0152 | |
| BxPC-3 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0186 | |
| In-vivo Model | AsPC-1 cells suspended in 100 μl of PBS (2 × 106 cells/100 μl) were injected subcutaneously into the lateral flank of the mice, which were randomly divided into solvent group (n = 6), 1.0 mg/kg of celastrol group (n = 6) and 3.0 mg/kg of celastrol group (n = 6). The administration of celastrol was performed by intraperitoneal injection into tumor-bearing mice every 2 day after 10 day inoculation. The tumor sizes were monitored with calipers every 5 days, and the tumor volume was calculated with the formula: Volume (mm3) = 1/2 × length × width2. | |||
GD1 alpha synthase (ST6GALNAC5)
Breast cancer [ICD-11: 2C60]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [1] | |||
| Responsed Disease | Breast cancer [ICD-11: 2C60] | |||
| Target Regulation | Up regulation | |||
| Cell Process | Cell metastasis | |||
In-vitro Model |
MDA-MB-231Br (After brain metastases of MDA-MB-361 breast adenocarcinoma cells) | |||
| MDA-MB-231 | Breast adenocarcinoma | Homo sapiens | CVCL_0062 | |
| MDA-IBC-3 | Breast inflammatory carcinoma | Homo sapiens | CVCL_HC47 | |
| JIMT-1Br3 (After brain metastases of JIMT-1 breast cancer cells) | ||||
| JIMT-1 | Breast ductal carcinoma | Homo sapiens | CVCL_2077 | |
| HEK293-FT | Normal | Homo sapiens | CVCL_6911 | |
| HCC1954Br (After brain metastases of HCC1954 breast cancer cells) | ||||
| HCC1954 | Breast ductal carcinoma | Homo sapiens | CVCL_1259 | |
| bEnd.3 | Cerebrovascular endothelioma cells from mice | Mus musculus | CVCL_0170 | |
| BEAS-2B | Normal | Homo sapiens | CVCL_0168 | |
| 4T1Br (After brain metastases of 4T1 mouse breast cancer cells) | ||||
| 4T1 | Normal | Mus musculus | CVCL_0125 | |
| In-vivo Model | For the in vivo brain and bone extravasation and seeding assays, cancer cells labeled with CMFDA C2925 (Thermo fisher scientific) or GFP were injected intracardially into the nude mice. Cell number and injection procedure were described in "Animal Experiments". For the in vivo lung extravasation and seeding assays, cancer cells labeled with GFP (2.5 × 105 cells/mouse) were injected into the tail vein of nude mice. At 24 or 48 hrs later, the mice were sacrificed. | |||
| Response Summary | Mechanistically, YTHDF3 enhances the translation of m6A-enriched transcripts for GD1 alpha synthase (ST6GALNAC5), GJA1, and EGFR, all associated with breast cancer brain metastasis. This work uncovers an essential role of YTHDF3 in controlling the interaction between cancer cells and brain microenvironment, thereby inducing brain metastatic competence. | |||
Interleukin-1 beta (IL1B)
Gangrene or necrosis of lung [ICD-11: CA43]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [13] | |||
| Responsed Disease | Gangrene or necrosis of lung [ICD-11: CA43] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | MAPK signaling pathway | hsa04010 | ||
| PI3K-Akt signaling pathway | hsa04151 | |||
| Cell Process | Biological regulation | |||
| Cell apoptosis | ||||
In-vitro Model |
BEAS-2B | Normal | Homo sapiens | CVCL_0168 |
| In-vivo Model | After being anesthetized with urethane (i.p.), SD rats were endotracheally intubated and ventilated using an animal ventilator under the conditions: respiratory rate of 70 breaths/min, tidal volume of 20 ml/kg, and inspiratory/expiratory ratio of 1:1. | |||
| Response Summary | N6-methyladenosine (m6A) methylation modification is implicated in the pathogenesis of lung ischemia-reperfusion injury. YTHDF3 or IGF2BP2 knockdown inhibited hypoxia/reoxygenation-activated p38, ERK1/2, AKT, and NF-Kappa-B pathways in BEAS-2B cells, and inhibited p-p65, Interleukin-1 beta (IL1B) and TNF-alpha secretion. | |||
Mitogen-activated protein kinase 1 (MAPK/ERK2/MAPK1)
Gangrene or necrosis of lung [ICD-11: CA43]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [13] | |||
| Responsed Disease | Gangrene or necrosis of lung [ICD-11: CA43] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | MAPK signaling pathway | hsa04010 | ||
| PI3K-Akt signaling pathway | hsa04151 | |||
| Apoptosis | hsa04210 | |||
| Cell Process | Biological regulation | |||
| Cell apoptosis | ||||
In-vitro Model |
BEAS-2B | Normal | Homo sapiens | CVCL_0168 |
| In-vivo Model | After being anesthetized with urethane (i.p.), SD rats were endotracheally intubated and ventilated using an animal ventilator under the conditions: respiratory rate of 70 breaths/min, tidal volume of 20 ml/kg, and inspiratory/expiratory ratio of 1:1. | |||
| Response Summary | N6-methyladenosine (m6A) methylation modification is implicated in the pathogenesis of lung ischemia-reperfusion injury. YTHDF3 or IGF2BP2 knockdown inhibited hypoxia/reoxygenation-activated p38, Mitogen-activated protein kinase 1 (MAPK/ERK2/MAPK1), AKT, and NF-Kappa-B pathways in BEAS-2B cells, and inhibited p-p65, IL-1-beta and TNF-alpha secretion. | |||
Mitogen-activated protein kinase 14 (p38/MAPK14)
Gangrene or necrosis of lung [ICD-11: CA43]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [13] | |||
| Responsed Disease | Gangrene or necrosis of lung [ICD-11: CA43] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | MAPK signaling pathway | hsa04010 | ||
| PI3K-Akt signaling pathway | hsa04151 | |||
| Cell Process | Biological regulation | |||
| Cell apoptosis | ||||
In-vitro Model |
BEAS-2B | Normal | Homo sapiens | CVCL_0168 |
| In-vivo Model | After being anesthetized with urethane (i.p.), SD rats were endotracheally intubated and ventilated using an animal ventilator under the conditions: respiratory rate of 70 breaths/min, tidal volume of 20 ml/kg, and inspiratory/expiratory ratio of 1:1. | |||
| Response Summary | N6-methyladenosine (m6A) methylation modification is implicated in the pathogenesis of lung ischemia-reperfusion injury. YTHDF3 or IGF2BP2 knockdown inhibited hypoxia/reoxygenation-activated Mitogen-activated protein kinase 14 (p38/MAPK14), ERK1/2, AKT, and NF-Kappa-B pathways in BEAS-2B cells, and inhibited p-p65, IL-1-beta and TNF-alpha secretion. | |||
RAC-alpha serine/threonine-protein kinase (AKT1)
Gangrene or necrosis of lung [ICD-11: CA43]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [13] | |||
| Responsed Disease | Gangrene or necrosis of lung [ICD-11: CA43] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | MAPK signaling pathway | hsa04010 | ||
| PI3K-Akt signaling pathway | hsa04151 | |||
| Apoptosis | hsa04210 | |||
| Cell Process | Biological regulation | |||
| Cell apoptosis | ||||
In-vitro Model |
BEAS-2B | Normal | Homo sapiens | CVCL_0168 |
| In-vivo Model | After being anesthetized with urethane (i.p.), SD rats were endotracheally intubated and ventilated using an animal ventilator under the conditions: respiratory rate of 70 breaths/min, tidal volume of 20 ml/kg, and inspiratory/expiratory ratio of 1:1. | |||
| Response Summary | N6-methyladenosine (m6A) methylation modification is implicated in the pathogenesis of lung ischemia-reperfusion injury. YTHDF3 or IGF2BP2 knockdown inhibited hypoxia/reoxygenation-activated p38, ERK1/2, RAC-alpha serine/threonine-protein kinase (AKT1), and NF-Kappa-B pathways in BEAS-2B cells, and inhibited p-p65, IL-1-beta and TNF-alpha secretion. | |||
Transcription factor p65 (RELA)
Gangrene or necrosis of lung [ICD-11: CA43]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [13] | |||
| Responsed Disease | Gangrene or necrosis of lung [ICD-11: CA43] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | MAPK signaling pathway | hsa04010 | ||
| PI3K-Akt signaling pathway | hsa04151 | |||
| Apoptosis | hsa04210 | |||
| Cell Process | Biological regulation | |||
| Cell apoptosis | ||||
In-vitro Model |
BEAS-2B | Normal | Homo sapiens | CVCL_0168 |
| In-vivo Model | After being anesthetized with urethane (i.p.), SD rats were endotracheally intubated and ventilated using an animal ventilator under the conditions: respiratory rate of 70 breaths/min, tidal volume of 20 ml/kg, and inspiratory/expiratory ratio of 1:1. | |||
| Response Summary | N6-methyladenosine (m6A) methylation modification is implicated in the pathogenesis of lung ischemia-reperfusion injury. YTHDF3 or IGF2BP2 knockdown inhibited hypoxia/reoxygenation-activated p38, ERK1/2, AKT, and NF-Kappa-B pathways in BEAS-2B cells, and inhibited Transcription factor p65 (RELA), IL-1-beta and TNF-alpha secretion. | |||
Tumor necrosis factor (TNF/TNF-alpha)
Gangrene or necrosis of lung [ICD-11: CA43]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [13] | |||
| Responsed Disease | Gangrene or necrosis of lung [ICD-11: CA43] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | MAPK signaling pathway | hsa04010 | ||
| PI3K-Akt signaling pathway | hsa04151 | |||
| Apoptosis | hsa04210 | |||
| Cell Process | Biological regulation | |||
| Cell apoptosis | ||||
In-vitro Model |
BEAS-2B | Normal | Homo sapiens | CVCL_0168 |
| In-vivo Model | After being anesthetized with urethane (i.p.), SD rats were endotracheally intubated and ventilated using an animal ventilator under the conditions: respiratory rate of 70 breaths/min, tidal volume of 20 ml/kg, and inspiratory/expiratory ratio of 1:1. | |||
| Response Summary | N6-methyladenosine (m6A) methylation modification is implicated in the pathogenesis of lung ischemia-reperfusion injury. YTHDF3 or IGF2BP2 knockdown inhibited hypoxia/reoxygenation-activated p38, ERK1/2, AKT, and NF-Kappa-B pathways in BEAS-2B cells, and inhibited p-p65, IL-1-beta and Tumor necrosis factor (TNF/TNF-alpha) secretion. | |||
Tyrosine-protein kinase EIF2AK2 (eIF2AK2/p68)
Colorectal cancer [ICD-11: 2B91]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [14] | |||
| Responsed Disease | Colorectal cancer [ICD-11: 2B91] | |||
In-vitro Model |
HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 |
| HCT 8 | Colon adenocarcinoma | Homo sapiens | CVCL_2478 | |
| Caco-2 | Colon adenocarcinoma | Homo sapiens | CVCL_0025 | |
| RKO | Colon carcinoma | Homo sapiens | CVCL_0504 | |
| HCoEpiC (Healthy colon epithelial HCoEpiC cells) | ||||
| FHC | Normal | Homo sapiens | CVCL_3688 | |
| Response Summary | YTHDF3 was highly expressed in oxaliplatin-resistant (OXAR) CRC tissues and cells. YTHDF3 as a novel hallmark and revealed the molecular mechanism of YTHDF3 on gene translation via coordination with Tyrosine-protein kinase EIF2AK2 (eIF2AK2/p68) in OXAR CRC cells. | |||
Growth arrest specific 5 (GAS5)
Colorectal cancer [ICD-11: 2B91]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [15] | |||
| Responsed Disease | Colorectal cancer [ICD-11: 2B91] | |||
| Target Regulation | Down regulation | |||
| Pathway Response | Hippo signaling pathway | hsa04390 | ||
| Cell Process | Ubiquitination degradation | |||
In-vitro Model |
DLD-1 | Colon adenocarcinoma | Homo sapiens | CVCL_0248 |
| HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 | |
| HT29 | Colon cancer | Mus musculus | CVCL_A8EZ | |
| LoVo | Colon adenocarcinoma | Homo sapiens | CVCL_0399 | |
| LS174T | Colon adenocarcinoma | Homo sapiens | CVCL_1384 | |
| RKO | Colon carcinoma | Homo sapiens | CVCL_0504 | |
| SW480 | Colon adenocarcinoma | Homo sapiens | CVCL_0546 | |
| SW620 | Colon adenocarcinoma | Homo sapiens | CVCL_0547 | |
| Response Summary | A new mechanism for m6A-induced decay of Growth arrest specific 5 (GAS5) on YAP signaling in progression of Colorectal cancer which offers a promising approach for CRC treatment. LncRNA GAS5 expressions is negatively correlated with YAP and YTHDF3 protein levels in tumors from CRC patients. | |||
hsa-miR-1914-3p
Lung cancer [ICD-11: 2C25]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [7] | |||
| Responsed Disease | Non-small-cell lung carcinoma [ICD-11: 2C25.Y] | |||
| Responsed Drug | Cisplatin | Approved | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Hippo signaling pathway | hsa04390 | ||
| Cell Process | Metabolic | |||
In-vitro Model |
A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 |
| Calu-6 | Lung adenocarcinoma | Homo sapiens | CVCL_0236 | |
| NCI-H1299 | Lung large cell carcinoma | Homo sapiens | CVCL_0060 | |
| NCI-H520 | Lung squamous cell carcinoma | Homo sapiens | CVCL_1566 | |
| In-vivo Model | Mice were injected with 5 × 106 lung cancer cells with stably expression of relevant plasmids and randomly divided into two groups (five mice per group) after the diameter of the xenografted tumors had reached approximately 5 mm in diameter. Xenografted mice were then administrated with PBS or DDP (3 mg/kg per day) for three times a week, and tumor volume were measured every second day. | |||
| Response Summary | METTL3, YTHDF3, YTHDF1, and eIF3b directly promoted YAP translation through an interaction with the translation initiation machinery. METTL3 knockdown inhibits tumor growth and enhances sensitivity to DDP in vivo.m6A mRNA methylation initiated by METTL3 directly promotes YAP translation and increases YAP activity by regulating the MALAT1-hsa-miR-1914-3p-YAP axis to induce Non-small cell lung cancer drug resistance and metastasis. | |||
hsa-miR-5586-5p
Pancreatic cancer [ICD-11: 2C10]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [16] | |||
| Responsed Disease | Pancreatic cancer [ICD-11: 2C10] | |||
| Pathway Response | Glycolysis / Gluconeogenesis | hsa00010 | ||
| Cell Process | Glycolysis | |||
In-vitro Model |
SW1990 | Pancreatic adenocarcinoma | Homo sapiens | CVCL_1723 |
| PANC-1 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0480 | |
| HPDE | Normal | Homo sapiens | CVCL_4376 | |
| BxPC-3 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0186 | |
| AsPC-1 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0152 | |
| Response Summary | Theses results implicate a negative feedback of m6A reader YTHDF3 and glycolytic lncRNA DICER1-AS1 is involved in glycolysis and tumorigenesis of pancreatic cancer. YTHDF3 and lncRNA DICER1-AS1 promotes glycolysis of pancreatic cancer through inhibiting maturation of hsa-miR-5586-5p. | |||
2,4-dienoyl-CoA reductase [ (DECR1)
Liver cancer [ICD-11: 2C12]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [17] | |||
| Responsed Disease | Liver cancer [ICD-11: 2C12] | |||
| Responsed Drug | Sorafenib | Approved | ||
| Target Regulation | Down regulation | |||
In-vitro Model |
Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 |
| MHCC97-H | Adult hepatocellular carcinoma | Homo sapiens | CVCL_4972 | |
| SNU-449 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_0454 | |
| Huh-7 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_0336 | |
|
MIHA
|
N.A. | Homo sapiens | CVCL_SA11 | |
| In-vivo Model | For subcutaneous HCC mice xenograft model, the cell suspension was injected into the right flank of the mice (n = 5 per group). | |||
Chromobox protein homolog 1 (CBX1)
Nasopharyngeal carcinoma [ICD-11: 2B6B]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [18] | |||
| Responsed Disease | Nasopharyngeal carcinoma [ICD-11: 2B6B] | |||
| Target Regulation | Down regulation | |||
Claspin (CLSPN)
Pancreatic cancer [ICD-11: 2C10]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [10] | |||
| Responsed Disease | Pancreatic cancer [ICD-11: 2C10] | |||
| Responsed Drug | Celastrol | Preclinical | ||
| Target Regulation | Up regulation | |||
In-vitro Model |
PANC-1 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0480 |
| SW1990 | Pancreatic adenocarcinoma | Homo sapiens | CVCL_1723 | |
| AsPC-1 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0152 | |
| BxPC-3 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0186 | |
| In-vivo Model | AsPC-1 cells suspended in 100 μl of PBS (2 × 106 cells/100 μl) were injected subcutaneously into the lateral flank of the mice, which were randomly divided into solvent group (n = 6), 1.0 mg/kg of celastrol group (n = 6) and 3.0 mg/kg of celastrol group (n = 6). The administration of celastrol was performed by intraperitoneal injection into tumor-bearing mice every 2 day after 10 day inoculation. The tumor sizes were monitored with calipers every 5 days, and the tumor volume was calculated with the formula: Volume (mm3) = 1/2 × length × width2. | |||
Cytochrome c oxidase subunit 4 isoform 1, mitochondrial (COX4I1)
Asthma [ICD-11: CA23]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [19] | |||
| Responsed Disease | Asthma [ICD-11: CA23] | |||
| Target Regulation | Down regulation | |||
DICER1 antisense RNA 1 (DICER1-AS1)
Pancreatic cancer [ICD-11: 2C10]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [16] | |||
| Responsed Disease | Pancreatic cancer [ICD-11: 2C10] | |||
| Target Regulation | Down regulation | |||
In-vitro Model |
SW1990 | Pancreatic adenocarcinoma | Homo sapiens | CVCL_1723 |
| AsPC-1 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0152 | |
| PANC-1 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0480 | |
| BxPC-3 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0186 | |
| HPDE | Normal | Homo sapiens | CVCL_4376 | |
DNA repair protein RAD51 homolog 4 (RAD51D)
Cervical cancer [ICD-11: 2C77]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [20] | |||
| Responsed Disease | Cervical cancer [ICD-11: 2C77] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
HeLa | Endocervical adenocarcinoma | Homo sapiens | CVCL_0030 |
| ME-180 | Human papillomavirus-related cervical squamous cell carcinoma | Homo sapiens | CVCL_1401 | |
| SiHa | Cervical squamous cell carcinoma | Homo sapiens | CVCL_0032 | |
| MS751 | Human papillomavirus-related cervical squamous cell carcinoma | Homo sapiens | CVCL_4996 | |
| In-vivo Model | 1 × 107 ME180 cells with stable overexpression of HNF1alpha or negative control ME180 cells were resuspended in 100 μL of phosphate-buffered saline and mixed with 100 μL of Matrigel. | |||
Forkhead box protein O6 (FOXO6)
Chronic glomerulonephritis [ICD-11: GB40]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [21] | |||
| Responsed Disease | Chronic glomerulonephritis [ICD-11: GB40] | |||
| Target Regulation | Down regulation | |||
Glypican-4 (GPC4)
Uveitis [ICD-11: 9A96]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [22] | |||
| Responsed Disease | Uveitis [ICD-11: 9A96] | |||
| Responsed Drug | FB23-2 | Investigative | ||
| Target Regulation | Down regulation | |||
In-vitro Model |
HMC3
|
N.A. | Homo sapiens | CVCL_II76 |
Heat shock 70 kDa protein 13 (HSPA13)
Renal cell carcinoma [ICD-11: 2C90]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [23] | |||
| Responsed Disease | Renal cell carcinoma [ICD-11: 2C90] | |||
NEDD4 family-interacting protein 1 (NDFIP1)
Liver cancer [ICD-11: 2C12]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [24] | |||
| Responsed Disease | Intrahepatic cholangiocarcinoma [ICD-11: 2C12.10] | |||
In-vitro Model |
HIBEpic (Human intrahepatic bile duct epithelial cells) | |||
| HuCC-T1 | Intrahepatic cholangiocarcinoma | Homo sapiens | CVCL_0324 | |
| RBE | Intrahepatic cholangiocarcinoma | Homo sapiens | CVCL_4896 | |
| HCCC-9810 | Intrahepatic cholangiocarcinoma | Homo sapiens | CVCL_6908 | |
| In-vivo Model | Mice were housed in a specific pathogen-free animal facility with a 12-h light-dark cycle. Lentivirus-infected HUCCT1 or HCCC-9810 cells (4 × 106) were suspended in 0.1 mL of phosphate-buffered saline and were subcutaneously injected into the right flank of nude mice. Tumor size was monitored every 4 days for a total of 28 days, and tumor volume was calculated as length × width2/2. After 4 weeks, the mice were euthanized by intraperitoneal injection of 200 mg/kg sodium pentobarbital. Then, tumor tissues were excised and weighed for further analysis. | |||
Phosphoglycerate kinase 1 (PGK1)
Osteosarcoma [ICD-11: 2B51]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [25] | |||
| Responsed Disease | Osteosarcoma [ICD-11: 2B51] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
hFOB 1.19 | Normal | Homo sapiens | CVCL_3708 |
| MG-63 | Osteosarcoma | Homo sapiens | CVCL_0426 | |
| SaOS-2 | Osteosarcoma | Homo sapiens | CVCL_0548 | |
| In-vivo Model | Twelve 6-week-old BALB/c male nude mice (Vitalriver, Beijing, China) were randomly divided to two groups. One group nude mice were subcutaneously injected with MG63 cells of YTHDF3 knockdown (sh-YTHDF3) or controls (sh-NC) at the right flank. The mice xenografts had been approved by the Laboratory Animal Welfare & Ethics Committee of Xi'an Jiao Tong University. All these animal procedures and handling care were performed in accordance with the Health guide for the care National Institutes and Laboratory animals. | |||
Pituitary homeobox 3 (PTX3)
Asthma [ICD-11: CA23]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [26] | |||
| Responsed Disease | Allergic asthma [ICD-11: CA23.0] | |||
| Target Regulation | Down regulation | |||
In-vitro Model |
THP-1 | Childhood acute monocytic leukemia | Homo sapiens | CVCL_0006 |
| In-vivo Model | Briefly, the Mettl3 targeting vector was designed to flank exon 2-3 with LoxP sites and electroporated into C57BL/6 embryonic stem cells (ES). Positive ES clones were confirmed by PCR and sequencing, and injected into C57BL/6 blastocysts to generate chimeric offspring. Chimeric mice were mated with C57BL/6 mice to obtain Mettl3flox/flox mice. These Mettl3flox/flox mice were crossed with Lyz2-Cre mice (The Jackson Laboratory) to generate Mettl3fl/fl (WT) and Mettl3fl/flLyz2Cre/+ (KO) mice. | |||
Prolyl 4-hydroxylase subunit alpha-2 (P4HA2)
Thyroid Cancer [ICD-11: 2D10]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [27] | |||
| Responsed Disease | Papillary thyroid cancer [ICD-11: 2D10.1] | |||
In-vitro Model |
Nthy-ori 3-1 | Normal | Homo sapiens | CVCL_2659 |
| TPC-1 | Thyroid gland papillary carcinoma | Homo sapiens | CVCL_6298 | |
| K1 | Thyroid gland papillary carcinoma | Homo sapiens | CVCL_2537 | |
| IHH-4 | Thyroid gland papillary carcinoma | Homo sapiens | CVCL_2960 | |
| B-CPAP | Thyroid gland carcinoma | Homo sapiens | CVCL_0153 | |
Pseudorabies Virus (PRV)
Rabies [ICD-11: 1C82]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [28] | |||
| Responsed Disease | Rabies [ICD-11: 1C82] | |||
| Responsed Drug | 3-deazidenosine | Investigative | ||
| Target Regulation | Up regulation | |||
In-vitro Model |
PK-15
|
N.A. | Sus scrofa | CVCL_2160 |
Receptor-interacting serine/threonine-protein kinase 3 (RIP3)
Aortic aneurysm or dissection [ICD-11: BD50]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [29] | |||
| Responsed Disease | Abdominal aortic aneurysm [ICD-11: BD50.4] | |||
| Target Regulation | Up regulation | |||
Thioredoxin-dependent peroxide reductase, mitochondrial (PRDX3)
Cognitive impairment [ICD-11: MB21]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [30] | |||
| Responsed Disease | Cognitive impairment [ICD-11: MB21] | |||
| Target Regulation | Up regulation | |||
Unspecific Target Gene
Testicular cancer [ICD-11: 2C80]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [31] | |||
| Responsed Disease | Testicular cancer [ICD-11: 2C80] | |||
| Cell Process | Transcription | |||
| Response Summary | Abundance of m6A and expression of VIRMA/YTHDF3 were different among Testicular Germ Cell Tumors subtypes, with higher levels in SEs, suggesting a contribution to SE phenotype maintenance. | |||
Acute ischemic stroke [ICD-11: 8B11]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [32] | |||
| Responsed Disease | Acute ischemic stroke [ICD-11: 8B11] | |||
| In-vivo Model | 36 male C57BL/6J mice (20-22 g, 2-month-old) were purchased from SPF (Beijing) biotechnology co., Ltd (Beijing, China) and maintained in the specific pathogen-free (SPF) animal laboratory with a 12/12 h light/dark cycle with free access to food and water. The mice were randomly assigned into six groups (n = 6 per group): (1) sham-operated group (Sham), (2) MCAO 6 h, (3) MCAO 12 h, (4) MCAO 1 d, (5) MCAO 3 d, and (6) MCAO 7 d. | |||
Aortic aneurysm or dissection [ICD-11: BD50]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [33] | |||
| Responsed Disease | Abdominal aortic aneurysm [ICD-11: BD50.4] | |||
| Cell Process | Inflammatory infiltrates | |||
| Neovascularization | ||||
In-vitro Model |
PBMCs (Human peripheral blood mononuclear cells (PBMCs) are isolated from peripheral blood and identified as any blood cell with a round nucleus) | |||
| SMCs (Aneurysmal smooth muscle cells) | ||||
| Response Summary | YTHDF3 represented an even greater risk of rupture. Regarding the cellular location, METTL14 seemed to be associated with inflammatory infiltrates and neovascularization. Furthermore, a strong correlation was seen between FTO and aneurysmal smooth muscle cells (SMCs), YTHDF3, and macrophage infiltrate. The results also reveal the important roles of m6A modulators, including YTHDF3, FTO, and METTL14, in the pathogenesis of human human abdominal aortic aneurysm(AAA) and provide a new view on m6A modification in AAA. | |||
Asthma [ICD-11: CA23]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [34] | |||
| Responsed Disease | Asthma [ICD-11: CA23] | |||
| In-vivo Model | Modelled two groups of female 6-week-old BALB/C mice: severe asthma group and blank control group (n = 3 per group). They had the same feeding conditions and growth environment. Immunization solution: Dissolve 20 mg ovalbumin (OVA) in 1 ml normal saline (NS), after OVA is completely dissolved, dilute 0.4-10 ml and mix well, then it was mixed with the same volume of liquid aluminium adjuvant and placed on a shaking table at 4℃ for 30 min. Challenge solution: Add 0.5 g OVA into 10 ml NS, fully dissolve it, and shake it on a shaking table at 4℃ for 30 min. Immunization: Mice were injected intraperitoneally on days 0 and 12, each with 0.2 ml; the control group was treated with equal volume of normal saline. Challenge: On days 18-23, the mice were atomized by ultrasound in a closed container at a dose of 10 ml once a day for 20 min. Lung tissue was taken 24 h after the last atomization and immediately stored in liquid nitrogen. | |||
| Response Summary | m6A(YTHDF3 and YTHDC1) modification plays a key role in severe asthma, and is able to guide the future strategy of immunotherapy. | |||
Liver disease [ICD-11: DB9Z]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [35] | |||
| Responsed Disease | Liver disease [ICD-11: DB9Z] | |||
| Pathway Response | PPAR signaling pathway | hsa03320 | ||
| Cell Process | Fatty degeneration | |||
| In-vivo Model | A total of 24 male mice were randomly allocated to LFD (low-fat diet), LFDR (low-fat diet + resveratrol), HFD (high-fat diet), and HFDR (high-fat diet + resveratrol) groups for 12 weeks (n = 6/group). | |||
| Response Summary | The beneficial effect of resveratrol on lipid metabolism disorder under HFD is due to a decrease of m6A RNA methylation and an increase of PPARalpha mRNA, providing mechanistic insights into the function of resveratrol in alleviating the disturbance of lipid metabolism in mice. The resveratrol in HFD increased the transcript levels of methyltransferase like 3 (METTL3), alkB homolog 5 (ALKBH5), fat mass and obesity associated protein (FTO), and YTH domain family 2 (YTHDF2), whereas it decreased the level of YTH domain family 3 (YTHDF3) and m6A abundance in mice liver. | |||
Transcriptional coactivator YAP1 (YAP1)
| Representative RIP-seq result supporting the interaction between the target gene and YTHDF3 | ||
| Cell Line | Hela | Homo sapiens |
| Regulation | logFC: 1.33E+00 | GSE86214 |
Cisplatin
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [7] | |||
| Responsed Disease | Non-small-cell lung carcinoma | ICD-11: 2C25.Y | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Hippo signaling pathway | hsa04390 | ||
| Cell Process | Metabolic | |||
| In-vitro Model | A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 |
| Calu-6 | Lung adenocarcinoma | Homo sapiens | CVCL_0236 | |
| NCI-H1299 | Lung large cell carcinoma | Homo sapiens | CVCL_0060 | |
| NCI-H520 | Lung squamous cell carcinoma | Homo sapiens | CVCL_1566 | |
| In-vivo Model | Mice were injected with 5 × 106 lung cancer cells with stably expression of relevant plasmids and randomly divided into two groups (five mice per group) after the diameter of the xenografted tumors had reached approximately 5 mm in diameter. Xenografted mice were then administrated with PBS or DDP (3 mg/kg per day) for three times a week, and tumor volume were measured every second day. | |||
| Response Summary | METTL3, YTHDF3, YTHDF1, and eIF3b directly promoted YAP translation through an interaction with the translation initiation machinery. METTL3 knockdown inhibits tumor growth and enhances sensitivity to DDP in vivo.m6A mRNA methylation initiated by METTL3 directly promotes YAP translation and increases YAP activity by regulating the MALAT1-miR-1914-3p-Transcriptional coactivator YAP1 (YAP1) axis to induce Non-small cell lung cancer drug resistance and metastasis. | |||
Metastasis associated lung adenocarcinoma transcript 1 (MALAT1)
| Representative RIP-seq result supporting the interaction between the target gene and YTHDF3 | ||
| Cell Line | Hela | Homo sapiens |
| Regulation | logFC: 1.66E+00 | GSE86214 |
Cisplatin
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [7] | |||
| Responsed Disease | Non-small-cell lung carcinoma | ICD-11: 2C25.Y | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Hippo signaling pathway | hsa04390 | ||
| Cell Process | Metabolic | |||
| In-vitro Model | A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 |
| Calu-6 | Lung adenocarcinoma | Homo sapiens | CVCL_0236 | |
| NCI-H1299 | Lung large cell carcinoma | Homo sapiens | CVCL_0060 | |
| NCI-H520 | Lung squamous cell carcinoma | Homo sapiens | CVCL_1566 | |
| In-vivo Model | Mice were injected with 5 × 106 lung cancer cells with stably expression of relevant plasmids and randomly divided into two groups (five mice per group) after the diameter of the xenografted tumors had reached approximately 5 mm in diameter. Xenografted mice were then administrated with PBS or DDP (3 mg/kg per day) for three times a week, and tumor volume were measured every second day. | |||
| Response Summary | METTL3, YTHDF3, YTHDF1, and eIF3b directly promoted YAP translation through an interaction with the translation initiation machinery. METTL3 knockdown inhibits tumor growth and enhances sensitivity to DDP in vivo.m6A mRNA methylation initiated by METTL3 directly promotes YAP translation and increases YAP activity by regulating the Metastasis associated lung adenocarcinoma transcript 1 (MALAT1)-miR-1914-3p-YAP axis to induce Non-small cell lung cancer drug resistance and metastasis. | |||
Apoptosis regulator Bcl-2 (BCL2)
Celastrol
[Preclinical]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [10] | |||
| Responsed Disease | Pancreatic cancer | ICD-11: 2C10 | ||
| Target Regulation | Up regulation | |||
| In-vitro Model | PANC-1 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0480 |
| SW1990 | Pancreatic adenocarcinoma | Homo sapiens | CVCL_1723 | |
| AsPC-1 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0152 | |
| BxPC-3 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0186 | |
| In-vivo Model | AsPC-1 cells suspended in 100 μl of PBS (2 × 106 cells/100 μl) were injected subcutaneously into the lateral flank of the mice, which were randomly divided into solvent group (n = 6), 1.0 mg/kg of celastrol group (n = 6) and 3.0 mg/kg of celastrol group (n = 6). The administration of celastrol was performed by intraperitoneal injection into tumor-bearing mice every 2 day after 10 day inoculation. The tumor sizes were monitored with calipers every 5 days, and the tumor volume was calculated with the formula: Volume (mm3) = 1/2 × length × width2. | |||
hsa-miR-1914-3p
Cisplatin
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [7] | |||
| Responsed Disease | Non-small-cell lung carcinoma | ICD-11: 2C25.Y | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Hippo signaling pathway | hsa04390 | ||
| Cell Process | Metabolic | |||
| In-vitro Model | A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 |
| Calu-6 | Lung adenocarcinoma | Homo sapiens | CVCL_0236 | |
| NCI-H1299 | Lung large cell carcinoma | Homo sapiens | CVCL_0060 | |
| NCI-H520 | Lung squamous cell carcinoma | Homo sapiens | CVCL_1566 | |
| In-vivo Model | Mice were injected with 5 × 106 lung cancer cells with stably expression of relevant plasmids and randomly divided into two groups (five mice per group) after the diameter of the xenografted tumors had reached approximately 5 mm in diameter. Xenografted mice were then administrated with PBS or DDP (3 mg/kg per day) for three times a week, and tumor volume were measured every second day. | |||
| Response Summary | METTL3, YTHDF3, YTHDF1, and eIF3b directly promoted YAP translation through an interaction with the translation initiation machinery. METTL3 knockdown inhibits tumor growth and enhances sensitivity to DDP in vivo.m6A mRNA methylation initiated by METTL3 directly promotes YAP translation and increases YAP activity by regulating the MALAT1-hsa-miR-1914-3p-YAP axis to induce Non-small cell lung cancer drug resistance and metastasis. | |||
2,4-dienoyl-CoA reductase [ (DECR1)
Sorafenib
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [17] | |||
| Responsed Disease | Liver cancer | ICD-11: 2C12 | ||
| Target Regulation | Down regulation | |||
| In-vitro Model | Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 |
| MHCC97-H | Adult hepatocellular carcinoma | Homo sapiens | CVCL_4972 | |
| SNU-449 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_0454 | |
| Huh-7 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_0336 | |
|
MIHA
|
N.A. | Homo sapiens | CVCL_SA11 | |
| In-vivo Model | For subcutaneous HCC mice xenograft model, the cell suspension was injected into the right flank of the mice (n = 5 per group). | |||
Claspin (CLSPN)
Celastrol
[Preclinical]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [10] | |||
| Responsed Disease | Pancreatic cancer | ICD-11: 2C10 | ||
| Target Regulation | Up regulation | |||
| In-vitro Model | PANC-1 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0480 |
| SW1990 | Pancreatic adenocarcinoma | Homo sapiens | CVCL_1723 | |
| AsPC-1 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0152 | |
| BxPC-3 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0186 | |
| In-vivo Model | AsPC-1 cells suspended in 100 μl of PBS (2 × 106 cells/100 μl) were injected subcutaneously into the lateral flank of the mice, which were randomly divided into solvent group (n = 6), 1.0 mg/kg of celastrol group (n = 6) and 3.0 mg/kg of celastrol group (n = 6). The administration of celastrol was performed by intraperitoneal injection into tumor-bearing mice every 2 day after 10 day inoculation. The tumor sizes were monitored with calipers every 5 days, and the tumor volume was calculated with the formula: Volume (mm3) = 1/2 × length × width2. | |||
Glypican-4 (GPC4)
FB23-2
[Investigative]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [22] | |||
| Responsed Disease | Uveitis | ICD-11: 9A96 | ||
| Target Regulation | Down regulation | |||
| In-vitro Model |
HMC3
|
N.A. | Homo sapiens | CVCL_II76 |
Pseudorabies Virus (PRV)
3-deazidenosine
[Investigative]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [28] | |||
| Responsed Disease | Rabies | ICD-11: 1C82 | ||
| Target Regulation | Up regulation | |||
| In-vitro Model |
PK-15
|
N.A. | Sus scrofa | CVCL_2160 |
Full List of Crosstalk(s) between m6A Modification and Epigenetic Regulation Related to This Regulator
RNA modification
m6A Target: Growth arrest specific 5 (GAS5)
| In total 6 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT00407 | ||
| Epigenetic Regulator | Double-stranded RNA-specific editase 1 (ADARB1) | |
| Regulated Target | MicroRNA 21 (MIR21) | |
| Crosstalk relationship | m6A → A-to-I | |
| Crosstalk ID: M6ACROT00408 | ||
| Epigenetic Regulator | Interferon-inducible protein 4 (ADAR1) | |
| Regulated Target | MicroRNA 21 (MIR21) | |
| Crosstalk relationship | m6A → A-to-I | |
| Crosstalk ID: M6ACROT00477 | ||
| Epigenetic Regulator | Interferon-inducible protein 4 (ADAR1) | |
| Regulated Target | MicroRNA 155 (MIR155) | |
| Crosstalk relationship | m6A → A-to-I | |
| Crosstalk ID: M6ACROT00509 | ||
| Epigenetic Regulator | Interferon-inducible protein 4 (ADAR1) | |
| Regulated Target | hsa-mir-18a | |
| Crosstalk relationship | m6A → A-to-I | |
| Crosstalk ID: M6ACROT00516 | ||
| Epigenetic Regulator | Double-stranded RNA-specific editase 1 (ADARB1) | |
| Regulated Target | MicroRNA 221 (MIR221) | |
| Crosstalk relationship | m6A → A-to-I | |
| Crosstalk ID: M6ACROT00564 | ||
| Epigenetic Regulator | RNA cytosine C(5)-methyltransferase NSUN2 (NSUN2) | |
| Regulated Target | Cyclin-dependent kinase inhibitor 1C (CDKN1C) | |
| Crosstalk relationship | m6A → m5C | |
m6A Target: Epidermal growth factor receptor (EGFR)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT00443 | ||
| Epigenetic Regulator | Double-stranded RNA-specific editase 1 (ADARB1) | |
| Regulated Target | MicroRNA 214 (MIR214) | |
| Crosstalk relationship | A-to-I → m6A | |
Histone modification
m6A Target: Thioredoxin-dependent peroxide reductase, mitochondrial (PRDX3)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03138 | ||
| Epigenetic Regulator | Histone acetyltransferase p300 (P300) | |
| Regulated Target | Histone H3 lysine 18 lactylation (H3K18la) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Cognitive impairment | |
m6A Target: Chromobox protein homolog 1 (CBX1)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03186 | ||
| Epigenetic Regulator | Chromobox protein homolog 1 (CBX1) | |
| Regulated Target | Histone H3 lysine 9 trimethylation (H3K9me3) | |
| Crosstalk relationship | m6A → Histone modification | |
| Disease | Nasopharyngeal carcinoma | |
m6A Target: Integrin alpha-6 (ITGA6)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03213 | ||
| Epigenetic Regulator | Lysine-specific demethylase 5B (KDM5B) | |
| Regulated Target | Histone H3 lysine 4 trimethylation (H3K4me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Liver cancer | |
m6A Target: Zinc finger E-box-binding homeobox 1 (ZEB1)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03537 | ||
| Epigenetic Regulator | Lysine-specific demethylase 5B (KDM5B) | |
| Regulated Target | Histone H3 lysine 4 trimethylation (H3K4me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Liver cancer | |
m6A Target: Epidermal growth factor receptor (EGFR)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03538 | ||
| Epigenetic Regulator | Lysine-specific demethylase 5B (KDM5B) | |
| Regulated Target | Histone H3 lysine 4 trimethylation (H3K4me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Liver cancer | |
Non-coding RNA
m6A Target: Microtubule-associated protein tau (TAU)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05088 | ||
| Epigenetic Regulator | DPPA2 upstream binding RNA (DUBR) | |
| Regulated Target | YTH N6-methyladenosine RNA binding protein F3 (YTHDF3) | |
| Crosstalk relationship | ncRNA → m6A | |
m6A Target: Calmodulin-1 (CALM1)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05090 | ||
| Epigenetic Regulator | DPPA2 upstream binding RNA (DUBR) | |
| Regulated Target | YTH N6-methyladenosine RNA binding protein F3 (YTHDF3) | |
| Crosstalk relationship | ncRNA → m6A | |
m6A Target: Cytochrome c oxidase subunit 4 isoform 1, mitochondrial (COX4I1)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05096 | ||
| Epigenetic Regulator | PM2.5-associated exosomal transcript PAET | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Asthma | |
m6A Target: Integrin alpha-6 (ITGA6)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05113 | ||
| Epigenetic Regulator | hsa-miR-502-3p | |
| Regulated Target | YTH N6-methyladenosine RNA binding protein F3 (YTHDF3) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Liver hepatocellular carcinoma | |
| Crosstalk ID: M6ACROT05114 | ||
| Epigenetic Regulator | Small nucleolar RNA host gene 3 (SNHG3) | |
| Regulated Target | hsa-miR-502-3p | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Liver hepatocellular carcinoma | |
m6A Target: 2,4-dienoyl-CoA reductase [ (DECR1)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05228 | ||
| Epigenetic Regulator | HNF4A antisense RNA 1 (HNF4A-AS1) | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Liver cancer | |
| Drug | Sorafenib | |
m6A Target: DICER1 antisense RNA 1 (DICER1-AS1)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05281 | ||
| Epigenetic Regulator | hsa-miR-5586-5p | |
| Regulated Target | YTH N6-methyladenosine RNA binding protein F3 (YTHDF3) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Pancreatic cancer | |
m6A Target: Growth arrest specific 5 (GAS5)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05400 | ||
| Epigenetic Regulator | Growth arrest specific 5 (GAS5) | |
| Regulated Target | YTH N6-methyladenosine RNA binding protein F3 (YTHDF3) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Colorectal cancer | |
m6A Target: Metastasis associated lung adenocarcinoma transcript 1 (MALAT1)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05403 | ||
| Epigenetic Regulator | Metastasis associated lung adenocarcinoma transcript 1 (MALAT1) | |
| Regulated Target | Transcriptional coactivator YAP1 (YAP1) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Breast cancer | |
| Drug | Tamoxifen | |
m6A Target: hsa-miR-1914-3p
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05407 | ||
| Epigenetic Regulator | hsa-miR-1914-3p | |
| Regulated Target | Transcriptional coactivator YAP1 (YAP1) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Non-small cell lung cancer | |
| Drug | Cisplatin | |
Xenobiotics Compound(s) Regulating the m6A Methylation Regulator
| Compound Name | Cholecalciferol | Investigative |
|---|---|---|
| Synonyms |
Vitamin D3; cholecalciferol; 67-97-0; Calciol; Colecalciferol; Oleovitamin D3; Ricketon; Arachitol; Trivitan; Deparal; Delsterol; Vigorsan; Ebivit; Activated 7-dehydrocholesterol; vitamin d-3; Colecalcipherol; Quintox; Colecalciferolum; Cholecalciferolum; (+)-Vitamin D3; D3-Vicotrat; D3-Vigantol; Vi-de-3-hydrosol; NEO Dohyfral D3; Vitinc Dan-Dee-3; 1406-16-2; Cholecalciferol, D3; Vi-De3; Provitina; Duphafral D3 1000; FeraCol; Delta-D; CC; CHEBI:28940; MFCD00078131; (1S,3Z)-3-[(2E)-2-[(1R,3aS,7aR)-7a-methyl-1-[(2R)-6-methylheptan-2-yl]-2,3,3a,5,6,7-hexahydro-1H-inden-4-ylidene]ethylidene]-4-methylidenecyclohexan-1-ol; NSC 375571; 9,10-Secocholesta-5,7,10(19)-trien-3-beta-ol; 7-Dehydrocholesterol activated; Micro-dee; 7-Dehydrocholesterol, Activated; VidDe-3-hydrosol; Vitamin D3 solution; NSC-375571; Colecalciferol (INN); Colecalciferol [INN]; NCGC00159331-02; Rampage; (3beta,5Z,7E)-9,10-secocholesta-5,7,10(19)-trien-3-ol; (5Z,7E)-(3S)-9,10-seco-5,7,10(19)-cholestatrien-3-ol; (5Z,7E)-(3S)-9,10-secocholesta-5,7,10(19)-trien-3-ol; DSSTox_CID_6294; DSSTox_RID_78090; DSSTox_GSID_26294; Vitamin D3 (Cholecalciferol); UNII-1C6V77QF41; 9,10-Seco(5Z,7E)-5,7,10(19)-cholestatrien-3beta-ol; Colecalciferolo; (3S,5Z,7E)-9,10-secocholesta-5,7,10(19)-trien-3-ol; Colecalciferolo [DCIT]; 9,10-Secocholesta-5(Z),7(E),10(19)-trien-3(.beta.)-ol; Vigantol Oil; (5e)-cholecalciferol; 22350-41-0; Colecalciferol D3; (1S,3Z)-3-[(2E)-2-[(1R,3aS,7aR)-1-[(1R)-1,5-dimethylhexyl]-7a-methyl-2,3,3a,5,6,7-hexahydro-1H-inden-4-ylidene]ethylidene]-4-methylene-cyclohexanol; (S,Z)-3-(2-((1R,3aS,7aR,E)-7a-methyl-1-((R)-6-methylheptan-2-yl)octahydro-4H-inden-4-ylidene)ethylidene)-4-methylenecyclohexan-1-ol; Cyclohexanol, 3-[(2E)-2-[(1R,3aS,7aR)-1-[(1R)-1,5-dimethylhexyl]octahydro-7a-methyl-4H-inden-4-ylidene]ethylidene]-4-methylene-, (1S,3Z)-; Colecalciferolum [INN-Latin]; Vitamin D 3; 7-Dehydrocholestrol, activated; Irradiated 7-dehydrocholesterol; CCRIS 5813; CCRIS 6286; HSDB 820; 7-Dehydrocholesterol, irradiated; Vitamin D3 emulsifiable; EINECS 200-673-2; EINECS 215-797-2; EPA Pesticide Chemical Code 202901; Vitamin D3; Cholecalciferol; 1C6V77QF41; Devaron; Videkhol; NSC375571; Granuvit D3; DP-R206; CAS-67-97-0; Prestwick_63; Cholecalciferol D3; Cyclohexanol, 3-((2E)-2-((1R,3aS,7aR)-1-((1R)-1,5-dimethylhexyl)octahydro-7a-methyl-4H-inden-4-ylidene)ethylidene)-4-methylene-, (1S,3Z)-; Cholecalciferol [USP:BAN:JAN:ISO]; ()-Vitamin D3; 9,10-Seco(5Z,7E)-5,7,10(19)-cholestatrien-3-ol; Delta-D (TN); 9,10-Secocholesta-5,7,10(19)-trien-3-ol, (3beta,5Z,7E)-; Prestwick3_000429; bmse000507; UPCMLD-DP152; SCHEMBL3126; Vitamin d (cholecalciferol); CHEMBL1042; BSPBio_000418; Cholecalciferol; 67-97-0; Cholecalciferol (JP17/USP); BPBio1_000460; MEGxm0_000458; DTXSID6026294; UPCMLD-DP152:001; ACon1_001997; Vitamin d3 (as cholecalciferol); HMS2096E20; Vitamin d assay system suitability; Cholecalciferol, >=98% (HPLC); 9,10-Secocholestra-5,7,10(19)-trien-3-ol, (3beta,5Z,7E)-; Cholecalciferol, analytical standard; ZINC4474460; Tox21_111578; Tox21_202546; BDBM50030475; LMST03020001; s4063; AKOS015950641; AC-8884; CCG-268466; CS-1179; DB00169; SMP1_000068; AK R215 COMPONENT COLECALCIFEROL; AK-R215 COMPONENT COLECALCIFEROL; NCGC00091072-01; NCGC00159331-04; NCGC00260095-01; BS-42465; HY-15398; K119; Vitamin D3 10 microg/mL in Acetonitrile; 9,10-secocholesta-5,7,10-trien-3-ol; Cholecalciferol (D3), analytical standard; C05443; D00188; 9,10-Secocholesta-5,7,10(19)-trien-3-ol; Cholecalciferol, meets USP testing specifications; 078V131; 9,10-Secocholesta-5,7,10(19)-trien-3?-ol; Q139347; (5E,7E)-9,10-Secocholesta-5,7,10-trien-3-ol; Q-201931; 3-beta,Z,7E-9,10-Secocholestr-5,7,10(19)-trien-3-ol; Vitamin D3 solution, 100 mug/mL in ethanol, 97% (CP); (3beta,Z,7E)-9,10-Secocholesta-5,7,10(19)-trien-3-ol; 9,10-Secocholesta-5,7,10(19)-trien-3-ol, (3b,5Z,7E)-; Cholecalciferol, European Pharmacopoeia (EP) Reference Standard; Colecalciferol, British Pharmacopoeia (BP) Reference Standard; Cholecalciferol, United States Pharmacopeia (USP) Reference Standard; Cholecalciferol for system suitability, European Pharmacopoeia (EP) Reference Standard; Vitamin D3 solution, 1 mg/mL in ethanol, ampule of 1 mL, certified reference material; (1S,3Z)-3-[(2E)-2-[7a-Methyl-1-(6-methylheptan-2-yl)-2,3,3a,5,6,7-hexahydro-1H-inden-4-ylidene]ethylidene]-4-methylidenecyclohexan-1-ol; Cholecalciferol (Vitamin D3), Pharmaceutical Secondary Standard; Certified Reference Material
Click to Show/Hide
|
|
| External link | ||
| Description |
vitamin D3 protects vascular endothelial cells from HCMV-induced apoptosis by reducing the elevated translation of MCU induced by HCMV through METTL3- and YTHDF3-dependent mechanisms via VDR/AMPK/METTL3 pathway.
|
[36] |
References