m6A Target Gene Information
General Information of the m6A Target Gene (ID: M6ATAR00151)
Full List of m6A Methylation Regulator of This Target Gene and Corresponding Disease/Drug Response(s)
DLEU2
can be regulated by the following regulator(s), and cause disease/drug response(s). You can browse detail information of regulator(s) or disease/drug response(s).
Browse Regulator
Browse Disease
YTH domain-containing family protein 2 (YTHDF2) [READER]
| Representative RNA-seq result indicating the expression of this target gene regulated by YTHDF2 | ||
| Cell Line | Human umbilical cord blood CD34+ cells | Homo sapiens |
|
Treatment: YTHDF2 knockdown UCB CD34+ cells
Control: Wild type UCB CD34+ cells
|
GSE107956 | |
| Regulation |
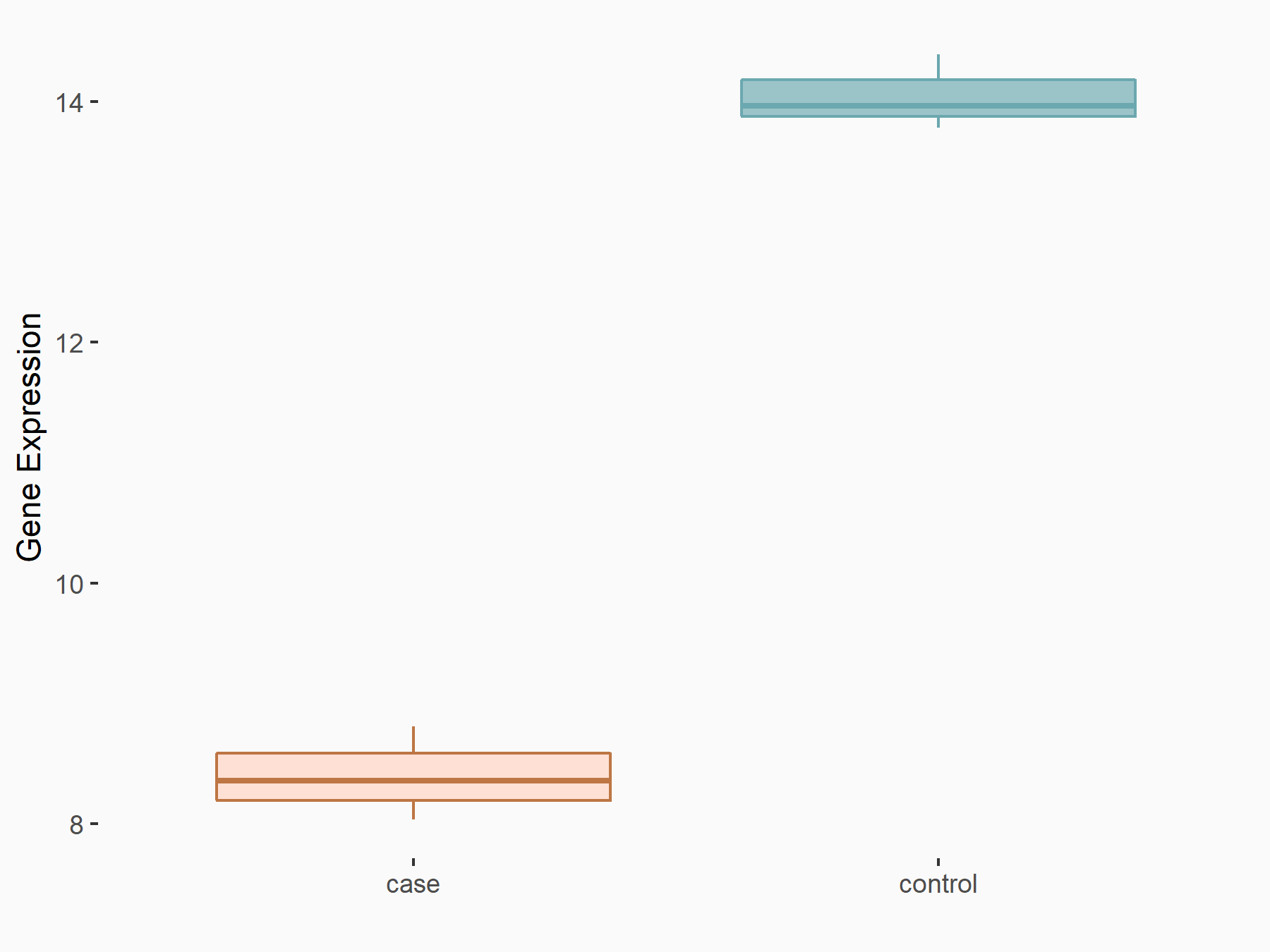 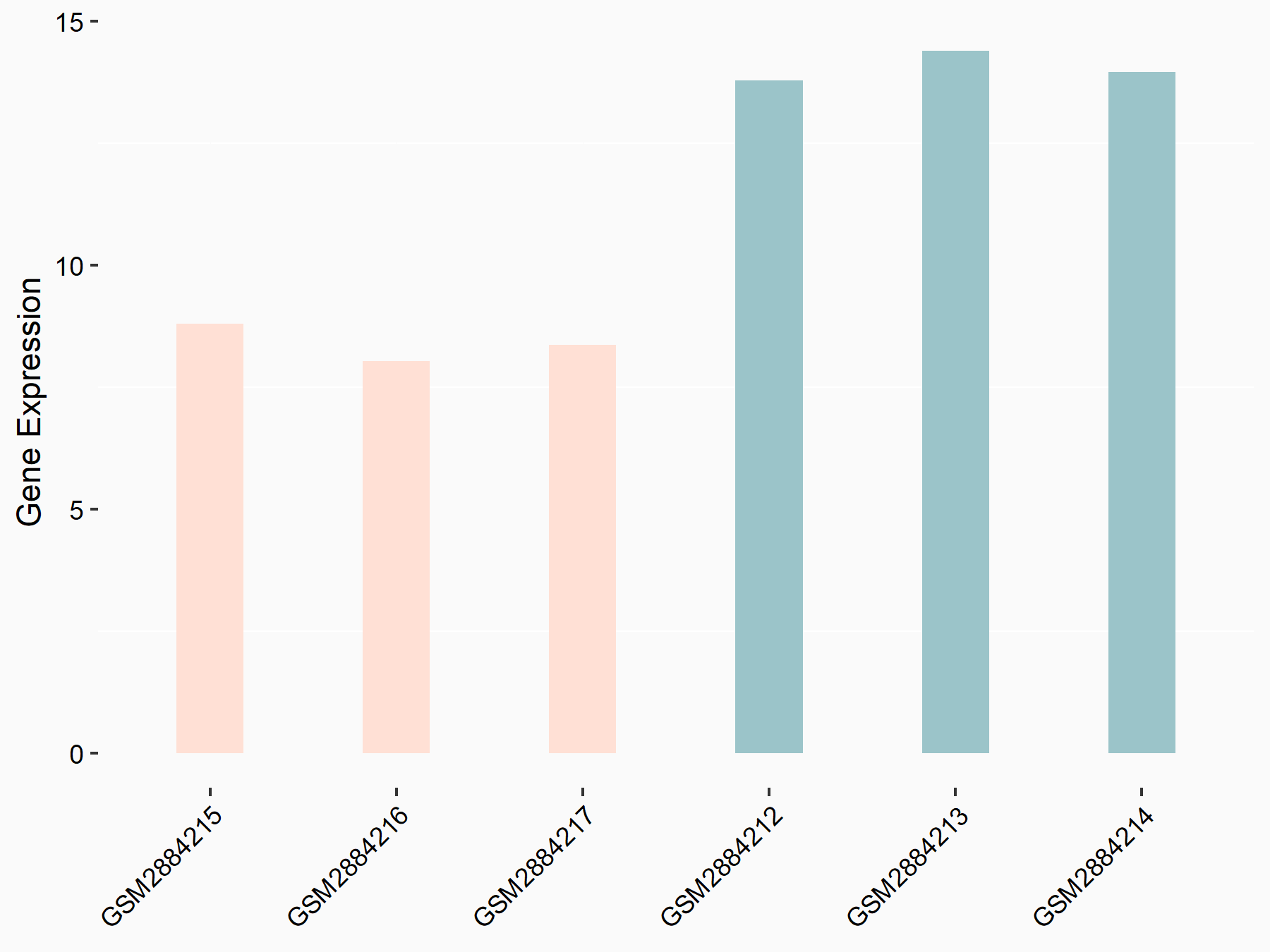 |
logFC: -6.80E-01 p-value: 2.37E-05 |
| More Results | Click to View More RNA-seq Results | |
| In total 1 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [1] | |||
| Response Summary | The elevated FTO in esophageal squamous cell carcinoma decreased m6A methylation of LINC00022 transcript, leading to the inhibition of Deleted in lymphocytic leukemia 2 (DLEU2/LINC00022) decay via the m6A reader YTHDF2. | |||
| Target Regulation | Down regulation | |||
| Responsed Disease | Esophageal squamous cell carcinoma | ICD-11: 2B70.1 | ||
| Pathway Response | Cell cycle | hsa04110 | ||
| Ubiquitin mediated proteolysis | hsa04120 | |||
| Cell Process | Ubiquitination degradation | |||
| Cell apoptosis | ||||
| Decreased G0/G1 phase | ||||
| In-vitro Model | HET-1A | Normal | Homo sapiens | CVCL_3702 |
| KYSE-150 | Esophageal squamous cell carcinoma | Homo sapiens | CVCL_1348 | |
| KYSE-450 | Esophageal squamous cell carcinoma | Homo sapiens | CVCL_1353 | |
| KYSE-70 | Esophageal squamous cell carcinoma | Homo sapiens | CVCL_1356 | |
| TE-1 | Esophageal squamous cell carcinoma | Homo sapiens | CVCL_1759 | |
| In-vivo Model | The number of cells inoculated in each mouse was 4 × 106, 1 × 106, 2 × 106 and 1 × 106, respectively. | |||
Fat mass and obesity-associated protein (FTO) [ERASER]
| In total 1 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [1] | |||
| Response Summary | The elevated FTO in esophageal squamous cell carcinoma decreased m6A methylation of Deleted in lymphocytic leukemia 2 (DLEU2/LINC00022) transcript, leading to the inhibition of LINC00022 decay via the m6A reader YTHDF2. | |||
| Target Regulation | Down regulation | |||
| Responsed Disease | Esophageal squamous cell carcinoma | ICD-11: 2B70.1 | ||
| Pathway Response | Cell cycle | hsa04110 | ||
| Ubiquitin mediated proteolysis | hsa04120 | |||
| Cell Process | Ubiquitination degradation | |||
| Cell apoptosis | ||||
| Decreased G0/G1 phase | ||||
| In-vitro Model | HET-1A | Normal | Homo sapiens | CVCL_3702 |
| KYSE-150 | Esophageal squamous cell carcinoma | Homo sapiens | CVCL_1348 | |
| KYSE-450 | Esophageal squamous cell carcinoma | Homo sapiens | CVCL_1353 | |
| KYSE-70 | Esophageal squamous cell carcinoma | Homo sapiens | CVCL_1356 | |
| TE-1 | Esophageal squamous cell carcinoma | Homo sapiens | CVCL_1759 | |
| In-vivo Model | The number of cells inoculated in each mouse was 4 × 106, 1 × 106, 2 × 106 and 1 × 106, respectively. | |||
Esophageal cancer [ICD-11: 2B70]
| In total 2 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [1] | |||
| Response Summary | The elevated FTO in esophageal squamous cell carcinoma decreased m6A methylation of Deleted in lymphocytic leukemia 2 (DLEU2/LINC00022) transcript, leading to the inhibition of LINC00022 decay via the m6A reader YTHDF2. | |||
| Responsed Disease | Esophageal squamous cell carcinoma [ICD-11: 2B70.1] | |||
| Target Regulator | Fat mass and obesity-associated protein (FTO) | ERASER | ||
| Target Regulation | Down regulation | |||
| Pathway Response | Cell cycle | hsa04110 | ||
| Ubiquitin mediated proteolysis | hsa04120 | |||
| Cell Process | Ubiquitination degradation | |||
| Cell apoptosis | ||||
| Decreased G0/G1 phase | ||||
| In-vitro Model | HET-1A | Normal | Homo sapiens | CVCL_3702 |
| KYSE-150 | Esophageal squamous cell carcinoma | Homo sapiens | CVCL_1348 | |
| KYSE-450 | Esophageal squamous cell carcinoma | Homo sapiens | CVCL_1353 | |
| KYSE-70 | Esophageal squamous cell carcinoma | Homo sapiens | CVCL_1356 | |
| TE-1 | Esophageal squamous cell carcinoma | Homo sapiens | CVCL_1759 | |
| In-vivo Model | The number of cells inoculated in each mouse was 4 × 106, 1 × 106, 2 × 106 and 1 × 106, respectively. | |||
| Experiment 2 Reporting the m6A-centered Disease Response | [1] | |||
| Response Summary | The elevated FTO in esophageal squamous cell carcinoma decreased m6A methylation of LINC00022 transcript, leading to the inhibition of Deleted in lymphocytic leukemia 2 (DLEU2/LINC00022) decay via the m6A reader YTHDF2. | |||
| Responsed Disease | Esophageal squamous cell carcinoma [ICD-11: 2B70.1] | |||
| Target Regulator | YTH domain-containing family protein 2 (YTHDF2) | READER | ||
| Target Regulation | Down regulation | |||
| Pathway Response | Cell cycle | hsa04110 | ||
| Ubiquitin mediated proteolysis | hsa04120 | |||
| Cell Process | Ubiquitination degradation | |||
| Cell apoptosis | ||||
| Decreased G0/G1 phase | ||||
| In-vitro Model | HET-1A | Normal | Homo sapiens | CVCL_3702 |
| KYSE-150 | Esophageal squamous cell carcinoma | Homo sapiens | CVCL_1348 | |
| KYSE-450 | Esophageal squamous cell carcinoma | Homo sapiens | CVCL_1353 | |
| KYSE-70 | Esophageal squamous cell carcinoma | Homo sapiens | CVCL_1356 | |
| TE-1 | Esophageal squamous cell carcinoma | Homo sapiens | CVCL_1759 | |
| In-vivo Model | The number of cells inoculated in each mouse was 4 × 106, 1 × 106, 2 × 106 and 1 × 106, respectively. | |||
Full List of Crosstalk(s) between m6A Modification and Epigenetic Regulation Related to This Regulator
Non-coding RNA
m6A Regulator: YTH domain-containing family protein 2 (YTHDF2)
| In total 1 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT05518 | ||
| Epigenetic Regulator | Deleted in lymphocytic leukemia 2 (DLEU2/LINC00022) | |
| Regulated Target | Cyclin-dependent kinase inhibitor 1 (CDKN1A) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Esophageal Squamous Cell Carcinoma | |
m6A Regulator: Fat mass and obesity-associated protein (FTO)
| In total 1 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT05519 | ||
| Epigenetic Regulator | Deleted in lymphocytic leukemia 2 (DLEU2/LINC00022) | |
| Regulated Target | Cyclin-dependent kinase inhibitor 1 (CDKN1A) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Esophageal Squamous Cell Carcinoma | |
RNA Modification Sequencing Data Associated with the Target (ID: M6ATAR00151)
| In total 55 m6A sequence/site(s) in this target gene | |||
| mod ID: A2ISITE006092 | Click to Show/Hide the Full List | ||
| mod site | chr13:49969078-49969079:- | [2] | |
| Sequence | AGGCTGGAGTGCAATGGCGCAATCTTGGCTCACTGCAAACT | ||
| Transcript ID List | ENST00000651713.1 | ||
| External Link | RMBase: RNA-editing_site_35462 | ||
| mod ID: A2ISITE006093 | Click to Show/Hide the Full List | ||
| mod site | chr13:49983152-49983153:- | [2] | |
| Sequence | GAGTGCAGTGGCACAATCTCAGCTCACTGCAACCTCTGACT | ||
| Transcript ID List | ENST00000651713.1; ENST00000621282.4 | ||
| External Link | RMBase: RNA-editing_site_35463 | ||
| mod ID: A2ISITE006094 | Click to Show/Hide the Full List | ||
| mod site | chr13:49988283-49988284:- | [2] | |
| Sequence | GAGTGCAGTGGTGAGATCTCAGCTCACTGCAGCCTCCACCT | ||
| Transcript ID List | ENST00000651713.1; ENST00000621282.4 | ||
| External Link | RMBase: RNA-editing_site_35464 | ||
| mod ID: A2ISITE006095 | Click to Show/Hide the Full List | ||
| mod site | chr13:49992034-49992035:- | [2] | |
| Sequence | TGAGGCAGGAGAATCACTTGAACCCAGGAGGCAGAGGTTGC | ||
| Transcript ID List | ENST00000651713.1; ENST00000621282.4; rmsk_4013622 | ||
| External Link | RMBase: RNA-editing_site_35465 | ||
| mod ID: A2ISITE006096 | Click to Show/Hide the Full List | ||
| mod site | chr13:49994789-49994790:- | [2] | |
| Sequence | TCAAGTGTTCCTCAGGCCTCAGCCTCCCAAAGTGCTGGGAT | ||
| Transcript ID List | ENST00000651713.1; ENST00000621282.4 | ||
| External Link | RMBase: RNA-editing_site_35466 | ||
| mod ID: A2ISITE006097 | Click to Show/Hide the Full List | ||
| mod site | chr13:49994857-49994858:- | [2] | |
| Sequence | GGCTAATTTTATTTTTTGTAAAGATAGAGTCTCACTATGTT | ||
| Transcript ID List | ENST00000651713.1; ENST00000621282.4 | ||
| External Link | RMBase: RNA-editing_site_35467 | ||
| mod ID: A2ISITE006098 | Click to Show/Hide the Full List | ||
| mod site | chr13:49995015-49995016:- | [2] | |
| Sequence | GTGTTGATTGATTTTGAGACAGGGTCTATCTCTGTCCCCCA | ||
| Transcript ID List | ENST00000651713.1; ENST00000621282.4 | ||
| External Link | RMBase: RNA-editing_site_35468 | ||
| mod ID: A2ISITE006099 | Click to Show/Hide the Full List | ||
| mod site | chr13:50021033-50021034:- | [2] | |
| Sequence | TAGGAGAATCACTTGTACCCAGGAGGTGGGGGTTGCAGTGA | ||
| Transcript ID List | ENST00000621282.4; rmsk_4013677 | ||
| External Link | RMBase: RNA-editing_site_35470 | ||
| mod ID: A2ISITE006100 | Click to Show/Hide the Full List | ||
| mod site | chr13:50021212-50021213:- | [3] | |
| Sequence | GCACGGTGGCTCACGCCTGTAATCTCAGCACTTTGGGAGGC | ||
| Transcript ID List | rmsk_4013677; ENST00000621282.4 | ||
| External Link | RMBase: RNA-editing_site_35471 | ||
| mod ID: A2ISITE006101 | Click to Show/Hide the Full List | ||
| mod site | chr13:50022703-50022704:- | [2] | |
| Sequence | ATCTCGAACTCCTGGGCTCAAGGATCCTTCCGCCTTGACCT | ||
| Transcript ID List | ENST00000621282.4 | ||
| External Link | RMBase: RNA-editing_site_35472 | ||
| mod ID: A2ISITE006102 | Click to Show/Hide the Full List | ||
| mod site | chr13:50024471-50024472:- | [2] | |
| Sequence | ACCTCTGCCTCCCAGATTCAAGCGATTCTCCTGCCTCAGCC | ||
| Transcript ID List | ENST00000621282.4 | ||
| External Link | RMBase: RNA-editing_site_35473 | ||
| mod ID: A2ISITE006103 | Click to Show/Hide the Full List | ||
| mod site | chr13:50024494-50024495:- | [2] | |
| Sequence | TGGTGCAATCTCAGCTTACTACAACCTCTGCCTCCCAGATT | ||
| Transcript ID List | ENST00000621282.4 | ||
| External Link | RMBase: RNA-editing_site_35474 | ||
| mod ID: A2ISITE006104 | Click to Show/Hide the Full List | ||
| mod site | chr13:50024515-50024516:- | [2] | |
| Sequence | TTTGCCCAGGCTGGAGGGCAATGGTGCAATCTCAGCTTACT | ||
| Transcript ID List | ENST00000621282.4 | ||
| External Link | RMBase: RNA-editing_site_35475 | ||
| mod ID: A2ISITE006105 | Click to Show/Hide the Full List | ||
| mod site | chr13:50027924-50027925:- | [3] | |
| Sequence | CTGCTGTAATTCTTTAAGGTAACTCTAAAATTAATGGAATG | ||
| Transcript ID List | ENST00000621282.4; ENST00000425586.5 | ||
| External Link | RMBase: RNA-editing_site_35476 | ||
| mod ID: A2ISITE006106 | Click to Show/Hide the Full List | ||
| mod site | chr13:50033091-50033092:- | [2] | |
| Sequence | CTAATTTTTGTACATTTAGTAGAGACGGGTTTCGCCATGTT | ||
| Transcript ID List | ENST00000425586.5; ENST00000621282.4 | ||
| External Link | RMBase: RNA-editing_site_35477 | ||
| mod ID: A2ISITE006107 | Click to Show/Hide the Full List | ||
| mod site | chr13:50033135-50033136:- | [2] | |
| Sequence | CCTCCCAAGTAGCTGGGATTACAGACACACACCACCGCGCC | ||
| Transcript ID List | ENST00000425586.5; ENST00000621282.4 | ||
| External Link | RMBase: RNA-editing_site_35478 | ||
| mod ID: A2ISITE006108 | Click to Show/Hide the Full List | ||
| mod site | chr13:50033171-50033172:- | [2] | |
| Sequence | TCTGCCTCCCGGGTTCAAACAATTCTCCTGCGTCAGCCTCC | ||
| Transcript ID List | ENST00000621282.4; ENST00000425586.5 | ||
| External Link | RMBase: RNA-editing_site_35479 | ||
| mod ID: A2ISITE006109 | Click to Show/Hide the Full List | ||
| mod site | chr13:50033174-50033175:- | [2] | |
| Sequence | ATCTCTGCCTCCCGGGTTCAAACAATTCTCCTGCGTCAGCC | ||
| Transcript ID List | ENST00000621282.4; ENST00000425586.5 | ||
| External Link | RMBase: RNA-editing_site_35480 | ||
| mod ID: A2ISITE006110 | Click to Show/Hide the Full List | ||
| mod site | chr13:50033219-50033220:- | [2] | |
| Sequence | TGTCACTCAGGCTAGAGTGCAGTGGCCTGATCTTGGCTCGC | ||
| Transcript ID List | ENST00000425586.5; ENST00000621282.4 | ||
| External Link | RMBase: RNA-editing_site_35481 | ||
| mod ID: A2ISITE006111 | Click to Show/Hide the Full List | ||
| mod site | chr13:50038366-50038367:- | [4] | |
| Sequence | CAAGGTGCTGAGATTACGGCATGAGGCACCATGCCTGGCCT | ||
| Transcript ID List | ENST00000425586.5; ENST00000621282.4 | ||
| External Link | RMBase: RNA-editing_site_35482 | ||
| mod ID: A2ISITE006112 | Click to Show/Hide the Full List | ||
| mod site | chr13:50038371-50038372:- | [4] | |
| Sequence | TCTCCCAAGGTGCTGAGATTACGGCATGAGGCACCATGCCT | ||
| Transcript ID List | ENST00000425586.5; ENST00000621282.4 | ||
| External Link | RMBase: RNA-editing_site_35483 | ||
| mod ID: A2ISITE006113 | Click to Show/Hide the Full List | ||
| mod site | chr13:50038393-50038394:- | [4] | |
| Sequence | TCAAGCAGTCCTCCCGCCTCAGTCTCCCAAGGTGCTGAGAT | ||
| Transcript ID List | ENST00000621282.4; ENST00000425586.5 | ||
| External Link | RMBase: RNA-editing_site_35484 | ||
| mod ID: A2ISITE006114 | Click to Show/Hide the Full List | ||
| mod site | chr13:50038407-50038408:- | [4] | |
| Sequence | GTCTGCTTCTGGGTTCAAGCAGTCCTCCCGCCTCAGTCTCC | ||
| Transcript ID List | ENST00000621282.4; ENST00000425586.5 | ||
| External Link | RMBase: RNA-editing_site_35485 | ||
| mod ID: A2ISITE006115 | Click to Show/Hide the Full List | ||
| mod site | chr13:50038411-50038412:- | [4] | |
| Sequence | GCATGTCTGCTTCTGGGTTCAAGCAGTCCTCCCGCCTCAGT | ||
| Transcript ID List | ENST00000621282.4; ENST00000425586.5 | ||
| External Link | RMBase: RNA-editing_site_35486 | ||
| mod ID: A2ISITE006116 | Click to Show/Hide the Full List | ||
| mod site | chr13:50038445-50038446:- | [4] | |
| Sequence | TTTGTTAGTGATATGGTCTCACTTTGTCGCCCAGGCATGTC | ||
| Transcript ID List | ENST00000621282.4; ENST00000425586.5 | ||
| External Link | RMBase: RNA-editing_site_35487 | ||
| mod ID: A2ISITE006117 | Click to Show/Hide the Full List | ||
| mod site | chr13:50038453-50038454:- | [4] | |
| Sequence | TAGAATTTTTTGTTAGTGATATGGTCTCACTTTGTCGCCCA | ||
| Transcript ID List | ENST00000621282.4; ENST00000425586.5 | ||
| External Link | RMBase: RNA-editing_site_35488 | ||
| mod ID: A2ISITE006118 | Click to Show/Hide the Full List | ||
| mod site | chr13:50038459-50038460:- | [4] | |
| Sequence | AATTTTTAGAATTTTTTGTTAGTGATATGGTCTCACTTTGT | ||
| Transcript ID List | ENST00000425586.5; ENST00000621282.4 | ||
| External Link | RMBase: RNA-editing_site_35489 | ||
| mod ID: A2ISITE006119 | Click to Show/Hide the Full List | ||
| mod site | chr13:50038470-50038471:- | [4] | |
| Sequence | CACACCCGGCTAATTTTTAGAATTTTTTGTTAGTGATATGG | ||
| Transcript ID List | ENST00000425586.5; ENST00000621282.4 | ||
| External Link | RMBase: RNA-editing_site_35490 | ||
| mod ID: A2ISITE006120 | Click to Show/Hide the Full List | ||
| mod site | chr13:50038506-50038507:- | [4] | |
| Sequence | CCTCCTGAGTAGTTGGGACTACAGGCATGCACCACCCACAC | ||
| Transcript ID List | ENST00000425586.5; ENST00000621282.4 | ||
| External Link | RMBase: RNA-editing_site_35491 | ||
| mod ID: A2ISITE006121 | Click to Show/Hide the Full List | ||
| mod site | chr13:50038516-50038517:- | [4] | |
| Sequence | CCCCACCCAGCCTCCTGAGTAGTTGGGACTACAGGCATGCA | ||
| Transcript ID List | ENST00000621282.4; ENST00000425586.5 | ||
| External Link | RMBase: RNA-editing_site_35492 | ||
| mod ID: A2ISITE006122 | Click to Show/Hide the Full List | ||
| mod site | chr13:50038545-50038546:- | [4] | |
| Sequence | GCCTCGATCTCCTGGGCTCAAGTGATTTTCCCCACCCAGCC | ||
| Transcript ID List | ENST00000425586.5; ENST00000621282.4 | ||
| External Link | RMBase: RNA-editing_site_35493 | ||
| mod ID: A2ISITE006123 | Click to Show/Hide the Full List | ||
| mod site | chr13:50038546-50038547:- | [4] | |
| Sequence | AGCCTCGATCTCCTGGGCTCAAGTGATTTTCCCCACCCAGC | ||
| Transcript ID List | ENST00000425586.5; ENST00000621282.4 | ||
| External Link | RMBase: RNA-editing_site_35494 | ||
| mod ID: A2ISITE006124 | Click to Show/Hide the Full List | ||
| mod site | chr13:50038566-50038567:- | [4] | |
| Sequence | GCACAATCATGGCTCACTACAGCCTCGATCTCCTGGGCTCA | ||
| Transcript ID List | ENST00000621282.4; ENST00000425586.5 | ||
| External Link | RMBase: RNA-editing_site_35495 | ||
| mod ID: A2ISITE006125 | Click to Show/Hide the Full List | ||
| mod site | chr13:50038568-50038569:- | [4] | |
| Sequence | TGGCACAATCATGGCTCACTACAGCCTCGATCTCCTGGGCT | ||
| Transcript ID List | ENST00000425586.5; ENST00000621282.4 | ||
| External Link | RMBase: RNA-editing_site_35496 | ||
| mod ID: A2ISITE006126 | Click to Show/Hide the Full List | ||
| mod site | chr13:50038590-50038591:- | [4] | |
| Sequence | TCTTGCTGTATTGGGAGTGCAGTGGCACAATCATGGCTCAC | ||
| Transcript ID List | ENST00000621282.4; ENST00000425586.5 | ||
| External Link | RMBase: RNA-editing_site_35497 | ||
| mod ID: A2ISITE006127 | Click to Show/Hide the Full List | ||
| mod site | chr13:50050058-50050059:- | [3] | |
| Sequence | GCCAGAAACAGGGAATAGGAAGATTAAGAGCTATGGGATTT | ||
| Transcript ID List | ENST00000235290.7; ENST00000421758.5; ENST00000458725.5; ENST00000425586.5; ENST00000443587.5; ENST00000449579.1; ENST00000433070.6; ENST00000621282.4 | ||
| External Link | RMBase: RNA-editing_site_35498 | ||
| mod ID: A2ISITE006128 | Click to Show/Hide the Full List | ||
| mod site | chr13:50050062-50050063:- | [3] | |
| Sequence | ATGGGCCAGAAACAGGGAATAGGAAGATTAAGAGCTATGGG | ||
| Transcript ID List | ENST00000449579.1; ENST00000421758.5; ENST00000621282.4; ENST00000443587.5; ENST00000458725.5; ENST00000425586.5; ENST00000433070.6; ENST00000235290.7 | ||
| External Link | RMBase: RNA-editing_site_35499 | ||
| mod ID: A2ISITE006129 | Click to Show/Hide the Full List | ||
| mod site | chr13:50050064-50050065:- | [2] | |
| Sequence | AAATGGGCCAGAAACAGGGAATAGGAAGATTAAGAGCTATG | ||
| Transcript ID List | ENST00000458725.5; ENST00000433070.6; ENST00000421758.5; ENST00000443587.5; ENST00000621282.4; ENST00000449579.1; ENST00000425586.5; ENST00000235290.7 | ||
| External Link | RMBase: RNA-editing_site_35500 | ||
| mod ID: A2ISITE006130 | Click to Show/Hide the Full List | ||
| mod site | chr13:50050075-50050076:- | [3] | |
| Sequence | CCTTACCTGGTAAATGGGCCAGAAACAGGGAATAGGAAGAT | ||
| Transcript ID List | ENST00000235290.7; ENST00000449579.1; ENST00000443587.5; ENST00000425586.5; ENST00000458725.5; ENST00000421758.5; ENST00000433070.6; ENST00000621282.4 | ||
| External Link | RMBase: RNA-editing_site_35501 | ||
| mod ID: A2ISITE006131 | Click to Show/Hide the Full List | ||
| mod site | chr13:50050091-50050092:- | [3] | |
| Sequence | AAACTAGGAGTTTCTTCCTTACCTGGTAAATGGGCCAGAAA | ||
| Transcript ID List | ENST00000421758.5; ENST00000621282.4; ENST00000433070.6; ENST00000458725.5; ENST00000443587.5; ENST00000425586.5; ENST00000449579.1; ENST00000235290.7 | ||
| External Link | RMBase: RNA-editing_site_35502 | ||
| mod ID: A2ISITE006132 | Click to Show/Hide the Full List | ||
| mod site | chr13:50050106-50050107:- | [2] | |
| Sequence | CATCTGAGAAAGTGGAAACTAGGAGTTTCTTCCTTACCTGG | ||
| Transcript ID List | ENST00000235290.7; ENST00000433070.6; ENST00000425586.5; ENST00000421758.5; ENST00000443587.5; ENST00000458725.5; ENST00000621282.4; ENST00000449579.1 | ||
| External Link | RMBase: RNA-editing_site_35503 | ||
| mod ID: A2ISITE006133 | Click to Show/Hide the Full List | ||
| mod site | chr13:50050142-50050143:- | [3] | |
| Sequence | TCTGATATCTTTTAGAGAAAAGGAATTTCATTTCTACATCT | ||
| Transcript ID List | ENST00000458725.5; ENST00000443587.5; ENST00000449579.1; ENST00000235290.7; ENST00000425586.5; ENST00000433070.6; ENST00000421758.5; ENST00000621282.4 | ||
| External Link | RMBase: RNA-editing_site_35504 | ||
| mod ID: A2ISITE006134 | Click to Show/Hide the Full List | ||
| mod site | chr13:50050143-50050144:- | [3] | |
| Sequence | TTCTGATATCTTTTAGAGAAAAGGAATTTCATTTCTACATC | ||
| Transcript ID List | ENST00000458725.5; ENST00000621282.4; ENST00000421758.5; ENST00000449579.1; ENST00000433070.6; ENST00000425586.5; ENST00000235290.7; ENST00000443587.5 | ||
| External Link | RMBase: RNA-editing_site_35505 | ||
| mod ID: A2ISITE006135 | Click to Show/Hide the Full List | ||
| mod site | chr13:50050149-50050150:- | [3] | |
| Sequence | GCAATTTTCTGATATCTTTTAGAGAAAAGGAATTTCATTTC | ||
| Transcript ID List | ENST00000425586.5; ENST00000621282.4; ENST00000458725.5; ENST00000235290.7; ENST00000421758.5; ENST00000449579.1; ENST00000433070.6; ENST00000443587.5 | ||
| External Link | RMBase: RNA-editing_site_35506 | ||
| mod ID: A2ISITE006136 | Click to Show/Hide the Full List | ||
| mod site | chr13:50050547-50050548:- | [3] | |
| Sequence | CTTACTCAGATGTAGAAATGAATTTCTTTTTCTCTAAAAGA | ||
| Transcript ID List | ENST00000621282.4; ENST00000433070.6; ENST00000458725.5; ENST00000421758.5; ENST00000443587.5; ENST00000449579.1; ENST00000425586.5; ENST00000235290.7 | ||
| External Link | RMBase: RNA-editing_site_35507 | ||
| mod ID: A2ISITE006137 | Click to Show/Hide the Full List | ||
| mod site | chr13:50050550-50050551:- | [3] | |
| Sequence | CCACTTACTCAGATGTAGAAATGAATTTCTTTTTCTCTAAA | ||
| Transcript ID List | ENST00000433070.6; ENST00000458725.5; ENST00000449579.1; ENST00000235290.7; ENST00000443587.5; ENST00000425586.5; ENST00000421758.5; ENST00000621282.4 | ||
| External Link | RMBase: RNA-editing_site_35508 | ||
| mod ID: A2ISITE006138 | Click to Show/Hide the Full List | ||
| mod site | chr13:50050649-50050650:- | [3] | |
| Sequence | TGAGATGAGAAGTAAAACAAATCCTGTAGCTCTTGATCTTC | ||
| Transcript ID List | ENST00000443587.5; ENST00000421758.5; ENST00000621282.4; ENST00000425586.5; ENST00000449579.1; ENST00000458725.5; ENST00000433070.6; ENST00000235290.7 | ||
| External Link | RMBase: RNA-editing_site_35509 | ||
| mod ID: A2ISITE006139 | Click to Show/Hide the Full List | ||
| mod site | chr13:50050650-50050651:- | [3] | |
| Sequence | CTGAGATGAGAAGTAAAACAAATCCTGTAGCTCTTGATCTT | ||
| Transcript ID List | ENST00000421758.5; ENST00000443587.5; ENST00000458725.5; ENST00000235290.7; ENST00000449579.1; ENST00000425586.5; ENST00000433070.6; ENST00000621282.4 | ||
| External Link | RMBase: RNA-editing_site_35510 | ||
| mod ID: A2ISITE006140 | Click to Show/Hide the Full List | ||
| mod site | chr13:50050653-50050654:- | [3] | |
| Sequence | ACCCTGAGATGAGAAGTAAAACAAATCCTGTAGCTCTTGAT | ||
| Transcript ID List | ENST00000458725.5; ENST00000443587.5; ENST00000449579.1; ENST00000235290.7; ENST00000421758.5; ENST00000425586.5; ENST00000433070.6; ENST00000621282.4 | ||
| External Link | RMBase: RNA-editing_site_35511 | ||
| mod ID: A2ISITE006141 | Click to Show/Hide the Full List | ||
| mod site | chr13:50050654-50050655:- | [3] | |
| Sequence | TACCCTGAGATGAGAAGTAAAACAAATCCTGTAGCTCTTGA | ||
| Transcript ID List | ENST00000425586.5; ENST00000421758.5; ENST00000621282.4; ENST00000458725.5; ENST00000433070.6; ENST00000449579.1; ENST00000235290.7; ENST00000443587.5 | ||
| External Link | RMBase: RNA-editing_site_35512 | ||
| mod ID: A2ISITE006142 | Click to Show/Hide the Full List | ||
| mod site | chr13:50050655-50050656:- | [3] | |
| Sequence | GTACCCTGAGATGAGAAGTAAAACAAATCCTGTAGCTCTTG | ||
| Transcript ID List | ENST00000443587.5; ENST00000433070.6; ENST00000621282.4; ENST00000425586.5; ENST00000449579.1; ENST00000458725.5; ENST00000235290.7; ENST00000421758.5 | ||
| External Link | RMBase: RNA-editing_site_35513 | ||
| mod ID: A2ISITE006143 | Click to Show/Hide the Full List | ||
| mod site | chr13:50057011-50057012:- | [5] | |
| Sequence | TCCCAGCACTTTGCGACACCAAGGCGGGAGGATCATTTGAG | ||
| Transcript ID List | ENST00000433070.6; rmsk_4013736; ENST00000235290.7; ENST00000458725.5; ENST00000443587.5; ENST00000449579.1; ENST00000421758.5; ENST00000425586.5; ENST00000621282.4 | ||
| External Link | RMBase: RNA-editing_site_35514 | ||
| mod ID: A2ISITE006144 | Click to Show/Hide the Full List | ||
| mod site | chr13:50065552-50065553:- | [2] | |
| Sequence | TTGCAGTGAACCGAGATCACACCACTGCACTCCAGCCTGGG | ||
| Transcript ID List | ENST00000443587.5; ENST00000621282.4; ENST00000433070.6; ENST00000425586.5; ENST00000449579.1; ENST00000235290.7; ENST00000421758.5; rmsk_4013753; ENST00000458725.5 | ||
| External Link | RMBase: RNA-editing_site_35515 | ||
| mod ID: A2ISITE006145 | Click to Show/Hide the Full List | ||
| mod site | chr13:50065568-50065569:- | [2] | |
| Sequence | ACCCGGAGGGCAGAGCTTGCAGTGAACCGAGATCACACCAC | ||
| Transcript ID List | ENST00000458725.5; ENST00000449579.1; ENST00000433070.6; ENST00000421758.5; ENST00000621282.4; rmsk_4013753; ENST00000443587.5; ENST00000235290.7; ENST00000425586.5 | ||
| External Link | RMBase: RNA-editing_site_35516 | ||
| mod ID: A2ISITE006146 | Click to Show/Hide the Full List | ||
| mod site | chr13:50073614-50073615:- | [3] | |
| Sequence | ATGTTGGAATTTGGGGGAAGAGATTTATTGTGTTAATTTTT | ||
| Transcript ID List | ENST00000443587.5; ENST00000421758.5; ENST00000433070.6; ENST00000458725.5; ENST00000449579.1; ENST00000235290.7; ENST00000425586.5; ENST00000621282.4 | ||
| External Link | RMBase: RNA-editing_site_35517 | ||
N6-methyladenosine (m6A)
| In total 52 m6A sequence/site(s) in this target gene | |||
| mod ID: M6ASITE017455 | Click to Show/Hide the Full List | ||
| mod site | chr13:49956816-49956817:- | [6] | |
| Sequence | GTTCCTGGACACTACAAGAAACCTGTGGTCCCAGCTACTCG | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000651713.1 | ||
| External Link | RMBase: m6A_site_228697 | ||
| mod ID: M6ASITE017456 | Click to Show/Hide the Full List | ||
| mod site | chr13:49956828-49956829:- | [6] | |
| Sequence | CATGAAGAGATGGTTCCTGGACACTACAAGAAACCTGTGGT | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000651713.1; rmsk_4013536 | ||
| External Link | RMBase: m6A_site_228698 | ||
| mod ID: M6ASITE017457 | Click to Show/Hide the Full List | ||
| mod site | chr13:49956849-49956850:- | [6] | |
| Sequence | TGATACATTGACAATAAAGAACATGAAGAGATGGTTCCTGG | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000651713.1; rmsk_4013536 | ||
| External Link | RMBase: m6A_site_228699 | ||
| mod ID: M6ASITE017458 | Click to Show/Hide the Full List | ||
| mod site | chr13:49956876-49956877:- | [6] | |
| Sequence | AGTGGGAGACATGAAACTAAACAGGAATGATACATTGACAA | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | rmsk_4013536; ENST00000651713.1 | ||
| External Link | RMBase: m6A_site_228700 | ||
| mod ID: M6ASITE017459 | Click to Show/Hide the Full List | ||
| mod site | chr13:49956881-49956882:- | [6] | |
| Sequence | CTCACAGTGGGAGACATGAAACTAAACAGGAATGATACATT | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000651713.1; rmsk_4013536 | ||
| External Link | RMBase: m6A_site_228701 | ||
| mod ID: M6ASITE017460 | Click to Show/Hide the Full List | ||
| mod site | chr13:49956888-49956889:- | [6] | |
| Sequence | CTTGTTTCTCACAGTGGGAGACATGAAACTAAACAGGAATG | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | rmsk_4013536; ENST00000651713.1 | ||
| External Link | RMBase: m6A_site_228702 | ||
| mod ID: M6ASITE017461 | Click to Show/Hide the Full List | ||
| mod site | chr13:49976368-49976369:- | [6] | |
| Sequence | AGAGTAGTCCTGACAGCCAGACCTCCAGGGTGGAAATGGGA | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000651713.1 | ||
| External Link | RMBase: m6A_site_228703 | ||
| mod ID: M6ASITE017462 | Click to Show/Hide the Full List | ||
| mod site | chr13:49993867-49993868:- | [7] | |
| Sequence | CCCTGGAAGAGCACAGTGGAACTAGATCCTAGTACAGATAG | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HEK293A-TOA | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000621282.4; ENST00000651713.1 | ||
| External Link | RMBase: m6A_site_228704 | ||
| mod ID: M6ASITE017463 | Click to Show/Hide the Full List | ||
| mod site | chr13:49993911-49993912:- | [7] | |
| Sequence | AAGATGACTGTCCAACTAAAACCTGCATACACAGCTACATC | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | HEK293A-TOA | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000651713.1; ENST00000621282.4 | ||
| External Link | RMBase: m6A_site_228705 | ||
| mod ID: M6ASITE017464 | Click to Show/Hide the Full List | ||
| mod site | chr13:49993945-49993946:- | [6] | |
| Sequence | CACAGAACAGCGACTGGGAAACTGCCACTAGAAAAAGATGA | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa; HEK293A-TOA | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000651713.1; ENST00000621282.4 | ||
| External Link | RMBase: m6A_site_228706 | ||
| mod ID: M6ASITE017465 | Click to Show/Hide the Full List | ||
| mod site | chr13:49997080-49997081:- | [6] | |
| Sequence | CCCGTCCTCCAACTACAGGGACCGCACGGTACGCGCCAAGC | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; HEK293A-TOA | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000651713.1; ENST00000621282.4 | ||
| External Link | RMBase: m6A_site_228707 | ||
| mod ID: M6ASITE017466 | Click to Show/Hide the Full List | ||
| mod site | chr13:49997180-49997181:- | [6] | |
| Sequence | CCCTCGCTGAAGGTTCGAGGACCACCCCGCTTTCCCGAGAG | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; Jurkat; peripheral-blood; GSC-11; HEK293T; iSLK; endometrial; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000621282.4 | ||
| External Link | RMBase: m6A_site_228708 | ||
| mod ID: M6ASITE017467 | Click to Show/Hide the Full List | ||
| mod site | chr13:49997271-49997272:- | [6] | |
| Sequence | GGTTTCTGCCGCGCTGCCGGACTGGAGCTCAGACGGCCTTC | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa; HEK293T; MM6; Jurkat; peripheral-blood; GSC-11; HEK293A-TOA; endometrial; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000621282.4 | ||
| External Link | RMBase: m6A_site_228710 | ||
| mod ID: M6ASITE017468 | Click to Show/Hide the Full List | ||
| mod site | chr13:49997352-49997353:- | [6] | |
| Sequence | GCAGGTTTTCTGGACCACAGACACACTGCCCCGGCGCCCTC | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa; HEK293T; MT4; peripheral-blood; GSC-11; HEK293A-TOA; endometrial; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000621282.4 | ||
| External Link | RMBase: m6A_site_228712 | ||
| mod ID: M6ASITE017469 | Click to Show/Hide the Full List | ||
| mod site | chr13:49997359-49997360:- | [6] | |
| Sequence | GGACGGAGCAGGTTTTCTGGACCACAGACACACTGCCCCGG | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; HEK293T; HEK293A-TOA; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000621282.4 | ||
| External Link | RMBase: m6A_site_228713 | ||
| mod ID: M6ASITE017470 | Click to Show/Hide the Full List | ||
| mod site | chr13:50014805-50014806:- | [8] | |
| Sequence | CTCAACCAAATTAGAGGAGGACTGAGGTAAAGAACTCTTAT | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | AML | ||
| Seq Type List | miCLIP | ||
| Transcript ID List | ENST00000621282.4 | ||
| External Link | RMBase: m6A_site_228754 | ||
| mod ID: M6ASITE017471 | Click to Show/Hide the Full List | ||
| mod site | chr13:50027174-50027175:- | [9] | |
| Sequence | GGGACTTACAGGAGAATTGAACAGTTGTTTCTGCTGACTCA | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | Huh7 | ||
| Seq Type List | MeRIP-seq | ||
| Transcript ID List | ENST00000621282.4; ENST00000425586.5 | ||
| External Link | RMBase: m6A_site_228775 | ||
| mod ID: M6ASITE017472 | Click to Show/Hide the Full List | ||
| mod site | chr13:50027191-50027192:- | [9] | |
| Sequence | AAAACTGTAAGTAACTGGGGACTTACAGGAGAATTGAACAG | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | Huh7 | ||
| Seq Type List | MeRIP-seq | ||
| Transcript ID List | ENST00000621282.4; ENST00000425586.5 | ||
| External Link | RMBase: m6A_site_228776 | ||
| mod ID: M6ASITE017473 | Click to Show/Hide the Full List | ||
| mod site | chr13:50027208-50027209:- | [9] | |
| Sequence | GATGATCACCTCAGCAGAAAACTGTAAGTAACTGGGGACTT | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | Huh7 | ||
| Seq Type List | MeRIP-seq | ||
| Transcript ID List | ENST00000425586.5; ENST00000621282.4 | ||
| External Link | RMBase: m6A_site_228777 | ||
| mod ID: M6ASITE017474 | Click to Show/Hide the Full List | ||
| mod site | chr13:50027296-50027297:- | [10] | |
| Sequence | AACTGAAAATGGAAACAGAAACACGTCTGTACAGAGCAGAG | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HEK293T; Huh7 | ||
| Seq Type List | MeRIP-seq | ||
| Transcript ID List | ENST00000425586.5; ENST00000621282.4 | ||
| External Link | RMBase: m6A_site_228778 | ||
| mod ID: M6ASITE017475 | Click to Show/Hide the Full List | ||
| mod site | chr13:50027302-50027303:- | [10] | |
| Sequence | AGAAGGAACTGAAAATGGAAACAGAAACACGTCTGTACAGA | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HEK293T; Huh7 | ||
| Seq Type List | MeRIP-seq | ||
| Transcript ID List | ENST00000621282.4; ENST00000425586.5 | ||
| External Link | RMBase: m6A_site_228779 | ||
| mod ID: M6ASITE017476 | Click to Show/Hide the Full List | ||
| mod site | chr13:50027315-50027316:- | [10] | |
| Sequence | TTAGACCATGTGGAGAAGGAACTGAAAATGGAAACAGAAAC | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HEK293T; Huh7 | ||
| Seq Type List | MeRIP-seq | ||
| Transcript ID List | ENST00000621282.4; ENST00000425586.5 | ||
| External Link | RMBase: m6A_site_228780 | ||
| mod ID: M6ASITE017477 | Click to Show/Hide the Full List | ||
| mod site | chr13:50027331-50027332:- | [10] | |
| Sequence | ACTAAACTACTGGTACTTAGACCATGTGGAGAAGGAACTGA | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HEK293T; Huh7 | ||
| Seq Type List | MeRIP-seq | ||
| Transcript ID List | ENST00000621282.4; ENST00000425586.5 | ||
| External Link | RMBase: m6A_site_228781 | ||
| mod ID: M6ASITE017478 | Click to Show/Hide the Full List | ||
| mod site | chr13:50027346-50027347:- | [10] | |
| Sequence | AAAAGGTGAGAACTGACTAAACTACTGGTACTTAGACCATG | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HEK293T; Huh7 | ||
| Seq Type List | MeRIP-seq | ||
| Transcript ID List | ENST00000621282.4; ENST00000425586.5 | ||
| External Link | RMBase: m6A_site_228782 | ||
| mod ID: M6ASITE017479 | Click to Show/Hide the Full List | ||
| mod site | chr13:50027355-50027356:- | [10] | |
| Sequence | TCCCCTCAAAAAAGGTGAGAACTGACTAAACTACTGGTACT | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HEK293T; Huh7 | ||
| Seq Type List | MeRIP-seq | ||
| Transcript ID List | ENST00000621282.4; ENST00000425586.5 | ||
| External Link | RMBase: m6A_site_228783 | ||
| mod ID: M6ASITE017480 | Click to Show/Hide the Full List | ||
| mod site | chr13:50029297-50029298:- | [10] | |
| Sequence | ACTTAAACTTATCAGAGAAGACTTATAAGAGGATATGAAAG | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HEK293T; Huh7 | ||
| Seq Type List | MeRIP-seq | ||
| Transcript ID List | ENST00000621282.4; ENST00000425586.5 | ||
| External Link | RMBase: m6A_site_228784 | ||
| mod ID: M6ASITE017481 | Click to Show/Hide the Full List | ||
| mod site | chr13:50029311-50029312:- | [10] | |
| Sequence | TGAAGGAAATGTGGACTTAAACTTATCAGAGAAGACTTATA | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HEK293T; Huh7 | ||
| Seq Type List | MeRIP-seq | ||
| Transcript ID List | ENST00000621282.4; ENST00000425586.5 | ||
| External Link | RMBase: m6A_site_228785 | ||
| mod ID: M6ASITE017482 | Click to Show/Hide the Full List | ||
| mod site | chr13:50029317-50029318:- | [10] | |
| Sequence | GAAGCTTGAAGGAAATGTGGACTTAAACTTATCAGAGAAGA | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HEK293T; Huh7 | ||
| Seq Type List | MeRIP-seq | ||
| Transcript ID List | ENST00000621282.4; ENST00000425586.5 | ||
| External Link | RMBase: m6A_site_228786 | ||
| mod ID: M6ASITE017483 | Click to Show/Hide the Full List | ||
| mod site | chr13:50029365-50029366:- | [10] | |
| Sequence | TAGAGAATAATGGAATGTAAACTCTGGCTTGCTAACTTAAG | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | MeRIP-seq | ||
| Transcript ID List | ENST00000425586.5; ENST00000621282.4 | ||
| External Link | RMBase: m6A_site_228787 | ||
| mod ID: M6ASITE017484 | Click to Show/Hide the Full List | ||
| mod site | chr13:50029390-50029391:- | [10] | |
| Sequence | ATTATACTAACATACAAGAAACAAGTAGAGAATAATGGAAT | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | MeRIP-seq | ||
| Transcript ID List | ENST00000621282.4; ENST00000425586.5 | ||
| External Link | RMBase: m6A_site_228788 | ||
| mod ID: M6ASITE017485 | Click to Show/Hide the Full List | ||
| mod site | chr13:50044043-50044044:- | [11] | |
| Sequence | AAAAATTTTAAGCCAGTTTTACACTAAATGTTCCTCAGTCT | ||
| Motif Score | 2.07285119 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000458725.5; ENST00000425586.5; ENST00000621282.4; ENST00000235290.7 | ||
| External Link | RMBase: m6A_site_228789 | ||
| mod ID: M6ASITE017486 | Click to Show/Hide the Full List | ||
| mod site | chr13:50044066-50044067:- | [11] | |
| Sequence | GATGTTGAGAAATTCTGCTGACAAAAAATTTTAAGCCAGTT | ||
| Motif Score | 2.859755952 | ||
| Cell/Tissue List | HEK293T; hESC-HEK293T | ||
| Seq Type List | DART-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000235290.7; ENST00000425586.5; ENST00000621282.4; ENST00000458725.5 | ||
| External Link | RMBase: m6A_site_228790 | ||
| mod ID: M6ASITE017487 | Click to Show/Hide the Full List | ||
| mod site | chr13:50044435-50044436:- | [11] | |
| Sequence | ACTTTGTTTACTTATAAAATACATCTCAGGTATTTCGGATG | ||
| Motif Score | 2.110482143 | ||
| Cell/Tissue List | HEK293T; hESC-HEK293T | ||
| Seq Type List | DART-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000235290.7; ENST00000621282.4; ENST00000425586.5; ENST00000458725.5; ENST00000433070.6 | ||
| External Link | RMBase: m6A_site_228791 | ||
| mod ID: M6ASITE017488 | Click to Show/Hide the Full List | ||
| mod site | chr13:50044446-50044447:- | [11] | |
| Sequence | TTTATGTTCTTACTTTGTTTACTTATAAAATACATCTCAGG | ||
| Motif Score | 2.494845238 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000235290.7; ENST00000433070.6; ENST00000458725.5; ENST00000621282.4; ENST00000425586.5 | ||
| External Link | RMBase: m6A_site_228792 | ||
| mod ID: M6ASITE017489 | Click to Show/Hide the Full List | ||
| mod site | chr13:50044508-50044509:- | [10] | |
| Sequence | TTTTATTACTAATAGCTTGAACCCTTTTAATGAAGACCTAA | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | MeRIP-seq | ||
| Transcript ID List | ENST00000235290.7; ENST00000433070.6; ENST00000425586.5; ENST00000621282.4; ENST00000458725.5 | ||
| External Link | RMBase: m6A_site_228793 | ||
| mod ID: M6ASITE017490 | Click to Show/Hide the Full List | ||
| mod site | chr13:50044535-50044536:- | [11] | |
| Sequence | ATGTCAAGGGATTACTTATAACTTCCATTTTATTACTAATA | ||
| Motif Score | 2.590089286 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000443587.5; ENST00000621282.4; ENST00000458725.5; ENST00000235290.7; ENST00000425586.5; ENST00000433070.6 | ||
| External Link | RMBase: m6A_site_228794 | ||
| mod ID: M6ASITE017491 | Click to Show/Hide the Full List | ||
| mod site | chr13:50044542-50044543:- | [11] | |
| Sequence | TTTACACATGTCAAGGGATTACTTATAACTTCCATTTTATT | ||
| Motif Score | 2.494845238 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000235290.7; ENST00000621282.4; ENST00000425586.5; ENST00000443587.5; ENST00000458725.5; ENST00000433070.6 | ||
| External Link | RMBase: m6A_site_228795 | ||
| mod ID: M6ASITE017492 | Click to Show/Hide the Full List | ||
| mod site | chr13:50044557-50044558:- | [11] | |
| Sequence | ACTAAAAAAATTTATTTTACACATGTCAAGGGATTACTTAT | ||
| Motif Score | 2.084928571 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000443587.5; ENST00000621282.4; ENST00000425586.5; ENST00000458725.5; ENST00000235290.7; ENST00000433070.6 | ||
| External Link | RMBase: m6A_site_228796 | ||
| mod ID: M6ASITE017493 | Click to Show/Hide the Full List | ||
| mod site | chr13:50044601-50044602:- | [10] | |
| Sequence | TCTGGGAGAAGAAATAAGAGACTATCATCAGTACATTCCCA | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HEK293T; A549; AML | ||
| Seq Type List | MeRIP-seq; m6A-seq; miCLIP | ||
| Transcript ID List | ENST00000425586.5; ENST00000621282.4; ENST00000458725.5; ENST00000443587.5; ENST00000433070.6; ENST00000235290.7 | ||
| External Link | RMBase: m6A_site_228797 | ||
| mod ID: M6ASITE017494 | Click to Show/Hide the Full List | ||
| mod site | chr13:50044673-50044674:- | [10] | |
| Sequence | AGGAAAAAAAAATGACCTAAACTTTTGAGATAGATTTGGCT | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HEK293T; A549 | ||
| Seq Type List | MeRIP-seq; m6A-seq | ||
| Transcript ID List | ENST00000433070.6; ENST00000621282.4; ENST00000235290.7; ENST00000425586.5; ENST00000443587.5; ENST00000458725.5 | ||
| External Link | RMBase: m6A_site_228798 | ||
| mod ID: M6ASITE017495 | Click to Show/Hide the Full List | ||
| mod site | chr13:50044679-50044680:- | [11] | |
| Sequence | TCAAAAAGGAAAAAAAAATGACCTAAACTTTTGAGATAGAT | ||
| Motif Score | 2.839113095 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000433070.6; ENST00000458725.5; ENST00000621282.4; ENST00000235290.7; ENST00000425586.5; ENST00000443587.5 | ||
| External Link | RMBase: m6A_site_228799 | ||
| mod ID: M6ASITE017496 | Click to Show/Hide the Full List | ||
| mod site | chr13:50049614-50049615:- | [12] | |
| Sequence | TGAAAGGTGTACTGCAAGGAACAAAATGTTTGTAAATTCTC | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000621282.4; ENST00000425586.5; ENST00000443587.5; ENST00000458725.5; ENST00000433070.6; ENST00000449579.1; ENST00000235290.7; ENST00000421758.5 | ||
| External Link | RMBase: m6A_site_228800 | ||
| mod ID: M6ASITE017497 | Click to Show/Hide the Full List | ||
| mod site | chr13:50054319-50054320:- | [8] | |
| Sequence | ACCCCGGGCAACAGAATGGGACTCCATCTCAAAAAAAAAAA | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | AML | ||
| Seq Type List | miCLIP | ||
| Transcript ID List | ENST00000433070.6; rmsk_4013733; ENST00000458725.5; ENST00000449579.1; ENST00000443587.5; ENST00000621282.4; ENST00000421758.5; ENST00000425586.5; ENST00000235290.7 | ||
| External Link | RMBase: m6A_site_228801 | ||
| mod ID: M6ASITE017498 | Click to Show/Hide the Full List | ||
| mod site | chr13:50054688-50054689:- | [8] | |
| Sequence | TACAAAACTCTTAGAAGAAAACAAAGGGCAAAAGCTTCAAG | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | AML | ||
| Seq Type List | miCLIP | ||
| Transcript ID List | rmsk_4013734; ENST00000443587.5; ENST00000449579.1; ENST00000425586.5; ENST00000235290.7; ENST00000421758.5; ENST00000458725.5; ENST00000433070.6; ENST00000621282.4 | ||
| External Link | RMBase: m6A_site_228802 | ||
| mod ID: M6ASITE017499 | Click to Show/Hide the Full List | ||
| mod site | chr13:50055052-50055053:- | [8] | |
| Sequence | TTCATATGGAATCTCAAAGGACATGAAATAACCAAAACAAA | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | AML | ||
| Seq Type List | miCLIP | ||
| Transcript ID List | ENST00000235290.7; ENST00000443587.5; ENST00000449579.1; ENST00000458725.5; ENST00000425586.5; ENST00000621282.4; ENST00000433070.6; ENST00000421758.5; rmsk_4013734 | ||
| External Link | RMBase: m6A_site_228803 | ||
| mod ID: M6ASITE017500 | Click to Show/Hide the Full List | ||
| mod site | chr13:50064003-50064004:- | [8] | |
| Sequence | ATAAAATGTACAAACGTAGGACTGACTGAAAATACTGCAAA | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | AML | ||
| Seq Type List | miCLIP | ||
| Transcript ID List | ENST00000449579.1; ENST00000433070.6; ENST00000621282.4; ENST00000235290.7; ENST00000421758.5; ENST00000443587.5; ENST00000425586.5; ENST00000458725.5 | ||
| External Link | RMBase: m6A_site_228804 | ||
| mod ID: M6ASITE017501 | Click to Show/Hide the Full List | ||
| mod site | chr13:50068137-50068138:- | [12] | |
| Sequence | ATAATGATAGAATGAAGACCACAATAAAAGAGACCTCTACT | ||
| Motif Score | 2.053113095 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000425586.5; ENST00000421758.5; ENST00000449579.1; ENST00000458725.5; ENST00000443587.5; ENST00000433070.6; ENST00000235290.7; ENST00000621282.4 | ||
| External Link | RMBase: m6A_site_228805 | ||
| mod ID: M6ASITE017502 | Click to Show/Hide the Full List | ||
| mod site | chr13:50071520-50071521:- | [9] | |
| Sequence | GCACCCATCCGTTTTTACAAACCCAGGTGGGAAGAGCCCAA | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | Huh7 | ||
| Seq Type List | MeRIP-seq | ||
| Transcript ID List | ENST00000235290.7; ENST00000421758.5; ENST00000621282.4; ENST00000443587.5; ENST00000425586.5; ENST00000449579.1; ENST00000458725.5; ENST00000433070.6 | ||
| External Link | RMBase: m6A_site_228807 | ||
| mod ID: M6ASITE017503 | Click to Show/Hide the Full List | ||
| mod site | chr13:50075618-50075619:- | [9] | |
| Sequence | CAGAGAACCAATTCTGGAGAACAGCCTCACTTCTTTGATTG | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | Huh7 | ||
| Seq Type List | MeRIP-seq | ||
| Transcript ID List | ENST00000449579.1; ENST00000425586.5; ENST00000421758.5; ENST00000443587.5; ENST00000433070.6; ENST00000235290.7; ENST00000621282.4; ENST00000458725.5 | ||
| External Link | RMBase: m6A_site_228808 | ||
| mod ID: M6ASITE017504 | Click to Show/Hide the Full List | ||
| mod site | chr13:50075632-50075633:- | [9] | |
| Sequence | GTTTTGACCTGTAGCAGAGAACCAATTCTGGAGAACAGCCT | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | Huh7 | ||
| Seq Type List | MeRIP-seq | ||
| Transcript ID List | ENST00000425586.5; ENST00000621282.4; ENST00000458725.5; ENST00000235290.7; ENST00000433070.6; ENST00000421758.5; ENST00000449579.1; ENST00000443587.5 | ||
| External Link | RMBase: m6A_site_228809 | ||
| mod ID: M6ASITE017505 | Click to Show/Hide the Full List | ||
| mod site | chr13:50080423-50080424:- | [8] | |
| Sequence | CGGTGACACGCTTCTTTAGGACTGCCTGCCTACAGAGTTTT | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | AML | ||
| Seq Type List | miCLIP | ||
| Transcript ID List | ENST00000421758.5; ENST00000425586.5; ENST00000458725.5; ENST00000433070.6; ENST00000449579.1; ENST00000235290.7; ENST00000621282.4; ENST00000443587.5 | ||
| External Link | RMBase: m6A_site_228810 | ||
| mod ID: M6ASITE017506 | Click to Show/Hide the Full List | ||
| mod site | chr13:50125417-50125418:- | [13] | |
| Sequence | GCGGGCCCCGCCGAGGGGGGACACCTGGCTGAGGCACAGCT | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000621282.4; ENST00000425586.5 | ||
| External Link | RMBase: m6A_site_228837 | ||
References