m6A Target Gene Information
General Information of the m6A Target Gene (ID: M6ATAR00503)
Full List of m6A Methylation Regulator of This Target Gene and Corresponding Disease/Drug Response(s)
LXRA
can be regulated by the following regulator(s), and cause disease/drug response(s). You can browse detail information of regulator(s) or disease/drug response(s).
Browse Regulator
Browse Disease
YTH domain-containing family protein 2 (YTHDF2) [READER]
| Representative RNA-seq result indicating the expression of this target gene regulated by YTHDF2 | ||
| Cell Line | GSC11 cell line | Homo sapiens |
|
Treatment: siYTHDF2 GSC11 cells
Control: siControl GSC11 cells
|
GSE142825 | |
| Regulation |
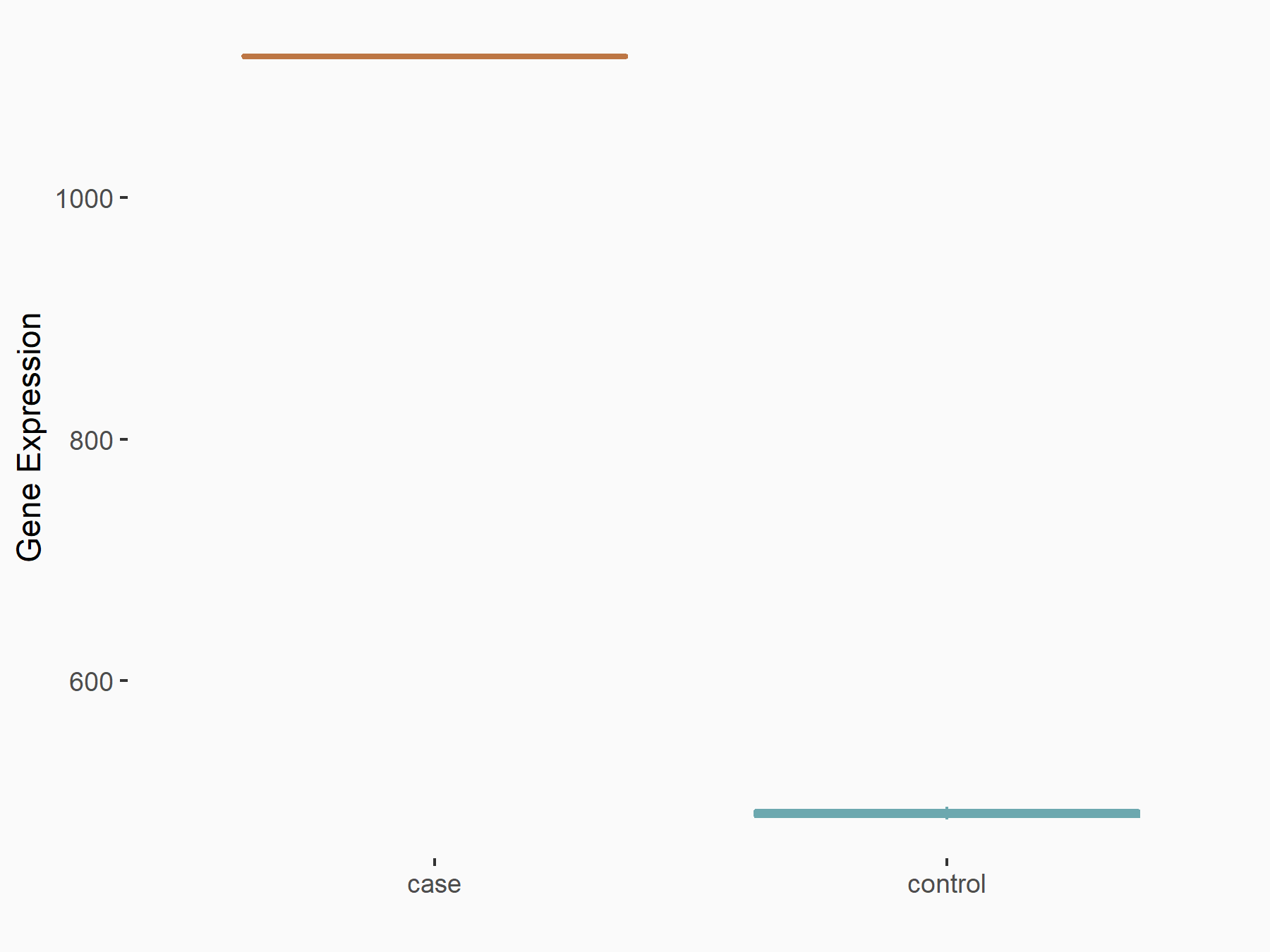  |
logFC: 1.19E+00 p-value: 2.27E-13 |
| More Results | Click to View More RNA-seq Results | |
| In total 1 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [1] | |||
| Response Summary | YTHDF2 facilitates m6A-dependent mRNA decay of Oxysterols receptor LXR-alpha (LXRA) and HIVEP2, which impacts the glioma patient survival. YTHDF2 promotes tumorigenesis of GBM cells, largely through the downregulation of LXRA and HIVEP2. | |||
| Target Regulation | Down regulation | |||
| Responsed Disease | Glioblastoma | ICD-11: 2A00.00 | ||
| Pathway Response | mRNA surveillance pathway | hsa03015), RNA degradation | ||
| Cell Process | RNA stability | |||
| In-vitro Model | U-87MG ATCC | Glioblastoma | Homo sapiens | CVCL_0022 |
| U-251MG | Astrocytoma | Homo sapiens | CVCL_0021 | |
| T98G | Glioblastoma | Homo sapiens | CVCL_0556 | |
| SW1783 | Anaplastic astrocytoma | Homo sapiens | CVCL_1722 | |
| LN-229 | Glioblastoma | Homo sapiens | CVCL_0393 | |
| Hs 683 | Oligodendroglioma | Homo sapiens | CVCL_0844 | |
| GSC7-2 (GSC7-2 were obtained from fresh surgical specimens of human primary and recurrent glioma) | ||||
| GSC6-27 (GSC6-27 were obtained from fresh surgical specimens of human primary and recurrent glioma) | ||||
| GSC23 | Glioblastoma | Homo sapiens | CVCL_DR59 | |
| GSC20 (GSC20 were obtained from fresh surgical specimens of human primary and recurrent glioma) | ||||
| GSC17 | Glioblastoma | Homo sapiens | CVCL_DR57 | |
| GSC11 | Glioblastoma | Homo sapiens | CVCL_DR55 | |
| In-vivo Model | For the studies of investigating mice survival, mice were intracranially injected with 10,000 GSC11, 10,000 GSC7-2, or 500,000 LN229 cells. | |||
Brain cancer [ICD-11: 2A00]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [1] | |||
| Response Summary | YTHDF2 facilitates m6A-dependent mRNA decay of Oxysterols receptor LXR-alpha (LXRA) and HIVEP2, which impacts the glioma patient survival. YTHDF2 promotes tumorigenesis of GBM cells, largely through the downregulation of LXRA and HIVEP2. | |||
| Responsed Disease | Glioblastoma [ICD-11: 2A00.00] | |||
| Target Regulator | YTH domain-containing family protein 2 (YTHDF2) | READER | ||
| Target Regulation | Down regulation | |||
| Pathway Response | mRNA surveillance pathway | hsa03015), RNA degradation | ||
| Cell Process | RNA stability | |||
| In-vitro Model | U-87MG ATCC | Glioblastoma | Homo sapiens | CVCL_0022 |
| U-251MG | Astrocytoma | Homo sapiens | CVCL_0021 | |
| T98G | Glioblastoma | Homo sapiens | CVCL_0556 | |
| SW1783 | Anaplastic astrocytoma | Homo sapiens | CVCL_1722 | |
| LN-229 | Glioblastoma | Homo sapiens | CVCL_0393 | |
| Hs 683 | Oligodendroglioma | Homo sapiens | CVCL_0844 | |
| GSC7-2 (GSC7-2 were obtained from fresh surgical specimens of human primary and recurrent glioma) | ||||
| GSC6-27 (GSC6-27 were obtained from fresh surgical specimens of human primary and recurrent glioma) | ||||
| GSC23 | Glioblastoma | Homo sapiens | CVCL_DR59 | |
| GSC20 (GSC20 were obtained from fresh surgical specimens of human primary and recurrent glioma) | ||||
| GSC17 | Glioblastoma | Homo sapiens | CVCL_DR57 | |
| GSC11 | Glioblastoma | Homo sapiens | CVCL_DR55 | |
| In-vivo Model | For the studies of investigating mice survival, mice were intracranially injected with 10,000 GSC11, 10,000 GSC7-2, or 500,000 LN229 cells. | |||
RNA Modification Sequencing Data Associated with the Target (ID: M6ATAR00503)
| In total 3 m6A sequence/site(s) in this target gene | |||
| mod ID: A2ISITE004714 | Click to Show/Hide the Full List | ||
| mod site | chr11:47256067-47256068:+ | [3] | |
| Sequence | TTTCTTTTTTTTATTTTGAGATGGAGTCTCAGTCTGTCACC | ||
| Transcript ID List | ENST00000531660.5; ENST00000395397.7; ENST00000616973.4; ENST00000527464.5; ENST00000487913.5; ENST00000407404.5; ENST00000481889.6; ENST00000529540.5; ENST00000436778.5; ENST00000481020.5; ENST00000532630.1; ENST00000405576.5; ENST00000495866.5 | ||
| External Link | RMBase: RNA-editing_site_21534 | ||
| mod ID: A2ISITE004715 | Click to Show/Hide the Full List | ||
| mod site | chr11:47262710-47262711:+ | [3] | |
| Sequence | AGTAGAGACAGGGTTTTACCATGTTGACCAGGCTGGTCTTG | ||
| Transcript ID List | ENST00000616973.4; ENST00000527949.1; ENST00000481020.5; ENST00000532630.1; ENST00000441012.7; ENST00000405576.5; ENST00000481889.6; ENST00000531660.5; ENST00000405853.7; ENST00000529540.5; ENST00000407404.5; ENST00000467728.5; ENST00000395397.7 | ||
| External Link | RMBase: RNA-editing_site_21535 | ||
| mod ID: A2ISITE004716 | Click to Show/Hide the Full List | ||
| mod site | chr11:47264211-47264212:+ | [3] | |
| Sequence | AGCAGGAAACCAAGTAGCTTAACGACTGGATGAGTGGGGCT | ||
| Transcript ID List | ENST00000395397.7; ENST00000405853.7; ENST00000527949.1; ENST00000531660.5; ENST00000467728.5; ENST00000529540.5; ENST00000441012.7; ENST00000481889.6; ENST00000616973.4; ENST00000407404.5; ENST00000405576.5; ENST00000481020.5; ENST00000532630.1 | ||
| External Link | RMBase: RNA-editing_site_21536 | ||
5-methylcytidine (m5C)
N6-methyladenosine (m6A)
| In total 48 m6A sequence/site(s) in this target gene | |||
| mod ID: M6ASITE005400 | Click to Show/Hide the Full List | ||
| mod site | chr11:47248331-47248332:+ | [4] | |
| Sequence | GCTAAGCAATGCTACTGGAGACCATAGGCAAAGCCAAGGTA | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000616973.4; ENST00000495866.5; ENST00000527464.5; ENST00000529540.5 | ||
| External Link | RMBase: m6A_site_139852 | ||
| mod ID: M6ASITE005401 | Click to Show/Hide the Full List | ||
| mod site | chr11:47248693-47248694:+ | [4] | |
| Sequence | ACCAAGGTAACGAAGCGCAGACTCCGGGCCCGGGTGGGCGG | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa; A549; GSC-11; iSLK; MSC; TIME | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000495866.5; ENST00000616973.4; ENST00000527464.5; ENST00000529540.5 | ||
| External Link | RMBase: m6A_site_139854 | ||
| mod ID: M6ASITE005402 | Click to Show/Hide the Full List | ||
| mod site | chr11:47248773-47248774:+ | [4] | |
| Sequence | AGCCGCCCGGCTCCAGCCGGACCGCTTGCCCGCCATCACCG | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; fibroblasts; A549; MSC; TIME; iSLK | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000529540.5; ENST00000616973.4; ENST00000527464.5; ENST00000495866.5 | ||
| External Link | RMBase: m6A_site_139856 | ||
| mod ID: M6ASITE005403 | Click to Show/Hide the Full List | ||
| mod site | chr11:47248812-47248813:+ | [4] | |
| Sequence | CGTTGTAATCTATGCAGCAAACAAGCTGGAACCCGCTGGGT | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa; A549; TIME | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000529540.5; ENST00000616973.4; ENST00000495866.5; ENST00000527464.5 | ||
| External Link | RMBase: m6A_site_139858 | ||
| mod ID: M6ASITE005404 | Click to Show/Hide the Full List | ||
| mod site | chr11:47248822-47248823:+ | [4] | |
| Sequence | TATGCAGCAAACAAGCTGGAACCCGCTGGGTGGCACCTGCA | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | HeLa; A549; TIME | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000495866.5; ENST00000527464.5; ENST00000529540.5; ENST00000616973.4 | ||
| External Link | RMBase: m6A_site_139859 | ||
| mod ID: M6ASITE005405 | Click to Show/Hide the Full List | ||
| mod site | chr11:47250505-47250506:+ | [4] | |
| Sequence | ATATATACTTCAGGATTTAAACTTTGCTTCCAGAGTTGTCA | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa; Huh7; Jurkat; HEK293A-TOA; MSC; TIME | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000405576.5; ENST00000487913.5; ENST00000407404.5; ENST00000481889.6; ENST00000532630.1; ENST00000481020.5; ENST00000395397.7; ENST00000495866.5; ENST00000529540.5; ENST00000531660.5; ENST00000436778.5; ENST00000616973.4; ENST00000527464.5 | ||
| External Link | RMBase: m6A_site_139860 | ||
| mod ID: M6ASITE005406 | Click to Show/Hide the Full List | ||
| mod site | chr11:47257619-47257620:+ | [4] | |
| Sequence | CAATTCCGGGCCGTGCTGGGACCTTTGCTCCACGAGGTGCC | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; HEK293A-TOA | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000527464.5; ENST00000405576.5; ENST00000444396.5; ENST00000481889.6; ENST00000495866.5; ENST00000481020.5; ENST00000487913.5; ENST00000532630.1; ENST00000529540.5; ENST00000436778.5; ENST00000407404.5; ENST00000395397.7; ENST00000616973.4; ENST00000531660.5 | ||
| External Link | RMBase: m6A_site_139861 | ||
| mod ID: M6ASITE005407 | Click to Show/Hide the Full List | ||
| mod site | chr11:47257655-47257656:+ | [4] | |
| Sequence | GTGCCTATGGAGGGGAGGGAACACGATTCTGGAGGCTGCTG | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000487913.5; ENST00000444396.5; ENST00000481889.6; ENST00000495866.5; ENST00000407404.5; ENST00000457932.5; ENST00000527464.5; ENST00000436778.5; ENST00000532630.1; ENST00000405576.5; ENST00000395397.7; ENST00000481020.5; ENST00000616973.4; ENST00000531660.5; ENST00000529540.5 | ||
| External Link | RMBase: m6A_site_139862 | ||
| mod ID: M6ASITE005408 | Click to Show/Hide the Full List | ||
| mod site | chr11:47258038-47258039:+ | [4] | |
| Sequence | CTCACTGGCTGGCCACCGAGACTTCTGGACAGGAAACTGCA | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa; GSC-11; iSLK; endometrial | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000405853.7; ENST00000436778.5; ENST00000444396.5; ENST00000481020.5; ENST00000527464.5; ENST00000486991.1; ENST00000476086.6; ENST00000405576.5; ENST00000532630.1; ENST00000437276.1; ENST00000487913.5; ENST00000529540.5; ENST00000412937.5; ENST00000467728.5; ENST00000420369.5; ENST00000495866.5; ENST00000616973.4; ENST00000461778.5; ENST00000449369.5; ENST00000395397.7; ENST00000419652.5; ENST00000457932.5; ENST00000531660.5; ENST00000436029.5; ENST00000525441.5; ENST00000481889.6; ENST00000441012.7; ENST00000407404.5 | ||
| External Link | RMBase: m6A_site_139863 | ||
| mod ID: M6ASITE005409 | Click to Show/Hide the Full List | ||
| mod site | chr11:47258046-47258047:+ | [4] | |
| Sequence | CTGGCCACCGAGACTTCTGGACAGGAAACTGCACCATCCTC | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; GSC-11; iSLK; endometrial | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000486991.1; ENST00000529540.5; ENST00000461778.5; ENST00000467728.5; ENST00000407404.5; ENST00000444396.5; ENST00000531660.5; ENST00000532630.1; ENST00000476086.6; ENST00000457932.5; ENST00000481889.6; ENST00000412937.5; ENST00000487913.5; ENST00000525441.5; ENST00000436029.5; ENST00000395397.7; ENST00000616973.4; ENST00000481020.5; ENST00000405576.5; ENST00000437276.1; ENST00000495866.5; ENST00000419652.5; ENST00000436778.5; ENST00000420369.5; ENST00000405853.7; ENST00000449369.5; ENST00000527464.5; ENST00000441012.7 | ||
| External Link | RMBase: m6A_site_139864 | ||
| mod ID: M6ASITE005410 | Click to Show/Hide the Full List | ||
| mod site | chr11:47258053-47258054:+ | [4] | |
| Sequence | CCGAGACTTCTGGACAGGAAACTGCACCATCCTCTTCTCCC | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa; GSC-11; iSLK; endometrial | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000461778.5; ENST00000412937.5; ENST00000441012.7; ENST00000481020.5; ENST00000437276.1; ENST00000532630.1; ENST00000444396.5; ENST00000486991.1; ENST00000395397.7; ENST00000531660.5; ENST00000481889.6; ENST00000405853.7; ENST00000407404.5; ENST00000436778.5; ENST00000420369.5; ENST00000529540.5; ENST00000495866.5; ENST00000419652.5; ENST00000476086.6; ENST00000457932.5; ENST00000527464.5; ENST00000436029.5; ENST00000616973.4; ENST00000405576.5; ENST00000525441.5; ENST00000467728.5; ENST00000487913.5; ENST00000449369.5 | ||
| External Link | RMBase: m6A_site_139865 | ||
| mod ID: M6ASITE005411 | Click to Show/Hide the Full List | ||
| mod site | chr11:47258092-47258093:+ | [4] | |
| Sequence | CCAGCAAGGGGGCTCCAGAGACTGCCCACCCAGGAAGTCTG | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa; GSC-11; iSLK; endometrial | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000531660.5; ENST00000420369.5; ENST00000457932.5; ENST00000395397.7; ENST00000481020.5; ENST00000405576.5; ENST00000481889.6; ENST00000527464.5; ENST00000407404.5; ENST00000437276.1; ENST00000486991.1; ENST00000436778.5; ENST00000461778.5; ENST00000616973.4; ENST00000476086.6; ENST00000444396.5; ENST00000529540.5; ENST00000449369.5; ENST00000525441.5; ENST00000532630.1; ENST00000441012.7; ENST00000419652.5; ENST00000467728.5; ENST00000412937.5; ENST00000495866.5; ENST00000436029.5; ENST00000405853.7; ENST00000487913.5 | ||
| External Link | RMBase: m6A_site_139866 | ||
| mod ID: M6ASITE005412 | Click to Show/Hide the Full List | ||
| mod site | chr11:47258258-47258259:+ | [4] | |
| Sequence | GATGGTGATGGAAGCTTGGGACAGGAGCAGGACTCTGGGTC | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000487913.5; ENST00000495866.5; ENST00000405853.7; ENST00000395397.7; ENST00000616973.4; ENST00000532630.1; ENST00000449369.5; ENST00000525441.5; ENST00000481020.5; ENST00000441012.7; ENST00000476086.6; ENST00000457932.5; ENST00000437276.1; ENST00000530310.1; ENST00000407404.5; ENST00000436778.5; ENST00000481889.6; ENST00000419652.5; ENST00000486991.1; ENST00000405576.5; ENST00000444396.5; ENST00000529540.5; ENST00000461778.5; ENST00000527464.5; ENST00000467728.5; ENST00000412937.5; ENST00000436029.5; ENST00000531660.5; ENST00000420369.5 | ||
| External Link | RMBase: m6A_site_139867 | ||
| mod ID: M6ASITE005413 | Click to Show/Hide the Full List | ||
| mod site | chr11:47258269-47258270:+ | [4] | |
| Sequence | AAGCTTGGGACAGGAGCAGGACTCTGGGTCCCAGAATAACT | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000436029.5; ENST00000476086.6; ENST00000467728.5; ENST00000481889.6; ENST00000495866.5; ENST00000616973.4; ENST00000461778.5; ENST00000527464.5; ENST00000407404.5; ENST00000420369.5; ENST00000525441.5; ENST00000530310.1; ENST00000531660.5; ENST00000405576.5; ENST00000419652.5; ENST00000457932.5; ENST00000444396.5; ENST00000529540.5; ENST00000412937.5; ENST00000441012.7; ENST00000449369.5; ENST00000487913.5; ENST00000437276.1; ENST00000486991.1; ENST00000395397.7; ENST00000405853.7; ENST00000436778.5; ENST00000481020.5; ENST00000532630.1 | ||
| External Link | RMBase: m6A_site_139868 | ||
| mod ID: M6ASITE005414 | Click to Show/Hide the Full List | ||
| mod site | chr11:47258344-47258345:+ | [4] | |
| Sequence | AATTGGGGAGATTCTTGGAGACAGGTTTGTGCTGGGACCCT | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000461778.5; ENST00000407404.5; ENST00000436778.5; ENST00000444396.5; ENST00000457932.5; ENST00000481889.6; ENST00000525441.5; ENST00000419652.5; ENST00000437276.1; ENST00000532630.1; ENST00000405576.5; ENST00000467728.5; ENST00000476086.6; ENST00000531660.5; ENST00000441012.7; ENST00000530310.1; ENST00000527464.5; ENST00000405853.7; ENST00000495866.5; ENST00000395397.7; ENST00000420369.5; ENST00000529540.5; ENST00000412937.5; ENST00000436029.5; ENST00000616973.4; ENST00000481020.5; ENST00000487913.5; ENST00000486991.1; ENST00000449369.5 | ||
| External Link | RMBase: m6A_site_139869 | ||
| mod ID: M6ASITE005415 | Click to Show/Hide the Full List | ||
| mod site | chr11:47258360-47258361:+ | [4] | |
| Sequence | GGAGACAGGTTTGTGCTGGGACCCTGGAAAGTGGCACGAGC | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000531660.5; ENST00000437276.1; ENST00000616973.4; ENST00000486991.1; ENST00000419652.5; ENST00000495866.5; ENST00000407404.5; ENST00000525441.5; ENST00000481020.5; ENST00000436778.5; ENST00000457932.5; ENST00000467728.5; ENST00000395397.7; ENST00000441012.7; ENST00000420369.5; ENST00000527464.5; ENST00000461778.5; ENST00000444396.5; ENST00000436029.5; ENST00000405576.5; ENST00000412937.5; ENST00000530310.1; ENST00000532630.1; ENST00000481889.6; ENST00000487913.5; ENST00000449369.5; ENST00000405853.7; ENST00000476086.6; ENST00000529540.5 | ||
| External Link | RMBase: m6A_site_139870 | ||
| mod ID: M6ASITE005416 | Click to Show/Hide the Full List | ||
| mod site | chr11:47258484-47258485:+ | [4] | |
| Sequence | AGCAGGGCAGATGCACCCAAACATTCCCTGCTGGCCTGGAG | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000457932.5; ENST00000395397.7; ENST00000405576.5; ENST00000532630.1; ENST00000495866.5; ENST00000487913.5; ENST00000436029.5; ENST00000412937.5; ENST00000437276.1; ENST00000527464.5; ENST00000476086.6; ENST00000420369.5; ENST00000436778.5; ENST00000530310.1; ENST00000441012.7; ENST00000616973.4; ENST00000419652.5; ENST00000486991.1; ENST00000467728.5; ENST00000481889.6; ENST00000407404.5; ENST00000525441.5; ENST00000531660.5; ENST00000481020.5; ENST00000405853.7; ENST00000449369.5; ENST00000461778.5; ENST00000529540.5; ENST00000444396.5 | ||
| External Link | RMBase: m6A_site_139871 | ||
| mod ID: M6ASITE005417 | Click to Show/Hide the Full List | ||
| mod site | chr11:47258508-47258509:+ | [4] | |
| Sequence | TCCCTGCTGGCCTGGAGGGGACTTTGGGGGAGAAAAGCCGG | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000405576.5; ENST00000461778.5; ENST00000531660.5; ENST00000436029.5; ENST00000419652.5; ENST00000527464.5; ENST00000412937.5; ENST00000529540.5; ENST00000481889.6; ENST00000476086.6; ENST00000436778.5; ENST00000525441.5; ENST00000449369.5; ENST00000405853.7; ENST00000395397.7; ENST00000437276.1; ENST00000487913.5; ENST00000495866.5; ENST00000420369.5; ENST00000616973.4; ENST00000457932.5; ENST00000481020.5; ENST00000407404.5; ENST00000441012.7; ENST00000532630.1; ENST00000467728.5; ENST00000444396.5; ENST00000530310.1 | ||
| External Link | RMBase: m6A_site_139872 | ||
| mod ID: M6ASITE005418 | Click to Show/Hide the Full List | ||
| mod site | chr11:47258549-47258550:+ | [4] | |
| Sequence | CACGGTGGCTGGGAGCCCAGACTTTGGAGTCAGAGGAGGGT | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000405576.5; ENST00000436029.5; ENST00000449369.5; ENST00000437276.1; rmsk_3543188; ENST00000436778.5; ENST00000457932.5; ENST00000481020.5; ENST00000412937.5; ENST00000476086.6; ENST00000495866.5; ENST00000419652.5; ENST00000616973.4; ENST00000487913.5; ENST00000532630.1; ENST00000525441.5; ENST00000444396.5; ENST00000395397.7; ENST00000420369.5; ENST00000530310.1; ENST00000405853.7; ENST00000441012.7; ENST00000407404.5; ENST00000461778.5; ENST00000527464.5; ENST00000467728.5; ENST00000481889.6; ENST00000531660.5; ENST00000529540.5 | ||
| External Link | RMBase: m6A_site_139873 | ||
| mod ID: M6ASITE005419 | Click to Show/Hide the Full List | ||
| mod site | chr11:47259760-47259761:+ | [5] | |
| Sequence | GAGCCGGGATGGGGCCTGAGACCCCCTTGTGCCTCTCTTTT | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | endometrial | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000616973.4; ENST00000527464.5; ENST00000395397.7; ENST00000437276.1; ENST00000473222.1; ENST00000530310.1; ENST00000498548.1; ENST00000467728.5; ENST00000487913.5; ENST00000525441.5; ENST00000532630.1; ENST00000449369.5; ENST00000457932.5; ENST00000441012.7; ENST00000531660.5; ENST00000461778.5; ENST00000407404.5; ENST00000405853.7; ENST00000483882.1; ENST00000481889.6; ENST00000436029.5; ENST00000529540.5; ENST00000495866.5; ENST00000419652.5; ENST00000476086.6; ENST00000444396.5; ENST00000405576.5; ENST00000481020.5; ENST00000412937.5; ENST00000436778.5; ENST00000420369.5 | ||
| External Link | RMBase: m6A_site_139874 | ||
| mod ID: M6ASITE005420 | Click to Show/Hide the Full List | ||
| mod site | chr11:47259790-47259791:+ | [4] | |
| Sequence | GCCTCTCTTTTGGAGCTCAGACTCTGCGGTGGAGCTGTGGA | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa; HepG2; CD4T; endometrial | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000407404.5; ENST00000437276.1; ENST00000457932.5; ENST00000444396.5; ENST00000405853.7; ENST00000529540.5; ENST00000461778.5; ENST00000441012.7; ENST00000395397.7; ENST00000481020.5; ENST00000436029.5; ENST00000487913.5; ENST00000527464.5; ENST00000531660.5; ENST00000419652.5; ENST00000483882.1; ENST00000473222.1; ENST00000525441.5; ENST00000498548.1; ENST00000530310.1; ENST00000481889.6; ENST00000467728.5; ENST00000476086.6; ENST00000420369.5; ENST00000532630.1; ENST00000412937.5; ENST00000436778.5; ENST00000616973.4; ENST00000405576.5; ENST00000449369.5; ENST00000495866.5 | ||
| External Link | RMBase: m6A_site_139875 | ||
| mod ID: M6ASITE005421 | Click to Show/Hide the Full List | ||
| mod site | chr11:47259971-47259972:+ | [4] | |
| Sequence | AGGGCAGAGCCCCCTTCAGAACCCACAGGTGAGGAGCTTCT | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | HeLa; HepG2; A549; CD4T; peripheral-blood; GSC-11; HEK293T; endometrial; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000420369.5; ENST00000457932.5; ENST00000395397.7; ENST00000529540.5; ENST00000525441.5; ENST00000531660.5; ENST00000495866.5; ENST00000436029.5; ENST00000407404.5; ENST00000481020.5; ENST00000436778.5; ENST00000461778.5; ENST00000527464.5; ENST00000481889.6; ENST00000532630.1; ENST00000530310.1; ENST00000444396.5; ENST00000405853.7; ENST00000616973.4; ENST00000473222.1; ENST00000487913.5; ENST00000449369.5; ENST00000412937.5; ENST00000476086.6; ENST00000441012.7; ENST00000467728.5; ENST00000483882.1; ENST00000405576.5 | ||
| External Link | RMBase: m6A_site_139876 | ||
| mod ID: M6ASITE005422 | Click to Show/Hide the Full List | ||
| mod site | chr11:47260418-47260419:+ | [6] | |
| Sequence | CTGTATCCAGAGATCCGTCCACAAAAGCGGAAAAAGGGGCC | ||
| Motif Score | 2.053113095 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000412937.5; ENST00000487913.5; ENST00000461778.5; ENST00000407404.5; ENST00000441012.7; ENST00000405853.7; ENST00000527464.5; ENST00000481020.5; ENST00000395397.7; ENST00000405576.5; ENST00000531660.5; ENST00000473222.1; ENST00000530310.1; ENST00000444396.5; ENST00000525441.5; ENST00000527949.1; ENST00000529540.5; ENST00000467728.5; ENST00000483882.1; ENST00000495866.5; ENST00000436029.5; ENST00000476086.6; ENST00000481889.6; ENST00000420369.5; ENST00000436778.5; ENST00000532630.1; ENST00000616973.4; ENST00000449369.5 | ||
| External Link | RMBase: m6A_site_139877 | ||
| mod ID: M6ASITE005423 | Click to Show/Hide the Full List | ||
| mod site | chr11:47260483-47260484:+ | [4] | |
| Sequence | GCTATGCAGCGTGTGTGGGGACAAGGCCTCGGGCTTCCACT | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; HepG2; hESC-HEK293T; CD4T; peripheral-blood; GSC-11; endometrial; NB4 | ||
| Seq Type List | m6A-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000420369.5; ENST00000481889.6; ENST00000527949.1; ENST00000476086.6; ENST00000405576.5; ENST00000532630.1; ENST00000407404.5; ENST00000412937.5; ENST00000436778.5; ENST00000495866.5; ENST00000481020.5; ENST00000530310.1; ENST00000444396.5; ENST00000483882.1; ENST00000531660.5; ENST00000616973.4; ENST00000405853.7; ENST00000461778.5; ENST00000395397.7; ENST00000529540.5; ENST00000473222.1; ENST00000436029.5; ENST00000441012.7; ENST00000487913.5; ENST00000525441.5; ENST00000449369.5; ENST00000467728.5; ENST00000527464.5 | ||
| External Link | RMBase: m6A_site_139878 | ||
| mod ID: M6ASITE005424 | Click to Show/Hide the Full List | ||
| mod site | chr11:47260504-47260505:+ | [6] | |
| Sequence | CAAGGCCTCGGGCTTCCACTACAATGTTCTGAGCTGCGAGG | ||
| Motif Score | 2.078666667 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000473222.1; ENST00000476086.6; ENST00000531660.5; ENST00000616973.4; ENST00000444396.5; ENST00000525441.5; ENST00000467728.5; ENST00000483882.1; ENST00000495866.5; ENST00000481889.6; ENST00000529540.5; ENST00000527949.1; ENST00000449369.5; ENST00000481020.5; ENST00000412937.5; ENST00000461778.5; ENST00000487913.5; ENST00000407404.5; ENST00000420369.5; ENST00000405853.7; ENST00000405576.5; ENST00000395397.7; ENST00000527464.5; ENST00000436778.5; ENST00000436029.5; ENST00000441012.7; ENST00000532630.1; ENST00000530310.1 | ||
| External Link | RMBase: m6A_site_139879 | ||
| mod ID: M6ASITE005425 | Click to Show/Hide the Full List | ||
| mod site | chr11:47260570-47260571:+ | [6] | |
| Sequence | CGTCATCAAGGGAGCGCACTACATCTGCCACAGTGGCGGCC | ||
| Motif Score | 2.078666667 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000436778.5; ENST00000487913.5; ENST00000532630.1; ENST00000449369.5; ENST00000436029.5; ENST00000481020.5; ENST00000483882.1; ENST00000441012.7; ENST00000525441.5; ENST00000481889.6; ENST00000461778.5; ENST00000405853.7; ENST00000420369.5; ENST00000444396.5; ENST00000527949.1; ENST00000531660.5; ENST00000412937.5; ENST00000476086.6; ENST00000616973.4; ENST00000530310.1; ENST00000473222.1; ENST00000395397.7; ENST00000407404.5; ENST00000527464.5; ENST00000467728.5; ENST00000529540.5; ENST00000405576.5 | ||
| External Link | RMBase: m6A_site_139880 | ||
| mod ID: M6ASITE005426 | Click to Show/Hide the Full List | ||
| mod site | chr11:47260603-47260604:+ | [4] | |
| Sequence | TGGCGGCCACTGCCCCATGGACACCTACATGCGTCGCAAGT | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; HepG2; MM6; GSC-11; endometrial; NB4 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000481889.6; ENST00000527949.1; ENST00000487913.5; ENST00000467728.5; ENST00000405576.5; ENST00000436029.5; ENST00000444396.5; ENST00000481020.5; ENST00000420369.5; ENST00000616973.4; ENST00000441012.7; ENST00000525441.5; ENST00000532630.1; ENST00000531660.5; ENST00000395397.7; ENST00000473222.1; ENST00000527464.5; ENST00000407404.5; ENST00000449369.5; ENST00000476086.6; ENST00000530310.1; ENST00000436778.5; ENST00000529540.5; ENST00000405853.7; ENST00000461778.5; ENST00000412937.5 | ||
| External Link | RMBase: m6A_site_139881 | ||
| mod ID: M6ASITE005427 | Click to Show/Hide the Full List | ||
| mod site | chr11:47260609-47260610:+ | [6] | |
| Sequence | CCACTGCCCCATGGACACCTACATGCGTCGCAAGTGCCAGG | ||
| Motif Score | 2.078666667 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000473222.1; ENST00000525441.5; ENST00000449369.5; ENST00000527949.1; ENST00000487913.5; ENST00000436778.5; ENST00000441012.7; ENST00000444396.5; ENST00000436029.5; ENST00000481889.6; ENST00000616973.4; ENST00000527464.5; ENST00000405576.5; ENST00000476086.6; ENST00000467728.5; ENST00000405853.7; ENST00000532630.1; ENST00000461778.5; ENST00000420369.5; ENST00000395397.7; ENST00000531660.5; ENST00000529540.5; ENST00000530310.1; ENST00000481020.5; ENST00000412937.5; ENST00000407404.5 | ||
| External Link | RMBase: m6A_site_139882 | ||
| mod ID: M6ASITE005428 | Click to Show/Hide the Full List | ||
| mod site | chr11:47261256-47261257:+ | [4] | |
| Sequence | TTAGGTGTCCTGTCAGAAGAACAGATCCGCCTGAAGAAACT | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa; HepG2; MM6; Huh7; CD4T; GSC-11; HEK293T; endometrial; HEC-1-A; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000405853.7; ENST00000449369.5; ENST00000395397.7; ENST00000441012.7; ENST00000407404.5; ENST00000405576.5; ENST00000461778.5; ENST00000532630.1; ENST00000436029.5; ENST00000487913.5; ENST00000527949.1; ENST00000412937.5; ENST00000420369.5; ENST00000481889.6; ENST00000436778.5; ENST00000525441.5; ENST00000527464.5; ENST00000467728.5; ENST00000531660.5; ENST00000444396.5; ENST00000616973.4; ENST00000481020.5; ENST00000529540.5 | ||
| External Link | RMBase: m6A_site_139883 | ||
| mod ID: M6ASITE005429 | Click to Show/Hide the Full List | ||
| mod site | chr11:47261274-47261275:+ | [4] | |
| Sequence | GAACAGATCCGCCTGAAGAAACTGAAGCGGCAAGAGGAGGA | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa; HepG2; MM6; Huh7; CD4T; GSC-11; HEK293T; endometrial; HEC-1-A; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000441012.7; ENST00000529540.5; ENST00000407404.5; ENST00000481020.5; ENST00000461778.5; ENST00000531660.5; ENST00000436029.5; ENST00000532630.1; ENST00000436778.5; ENST00000405853.7; ENST00000467728.5; ENST00000444396.5; ENST00000487913.5; ENST00000527949.1; ENST00000412937.5; ENST00000405576.5; ENST00000449369.5; ENST00000527464.5; ENST00000395397.7; ENST00000616973.4; ENST00000525441.5; ENST00000481889.6; ENST00000420369.5 | ||
| External Link | RMBase: m6A_site_139884 | ||
| mod ID: M6ASITE005430 | Click to Show/Hide the Full List | ||
| mod site | chr11:47261295-47261296:+ | [4] | |
| Sequence | CTGAAGCGGCAAGAGGAGGAACAGGCTCATGCCACATCCTT | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa; HepG2; kidney; MM6; Huh7; CD4T; GSC-11; HEK293T; endometrial; HEC-1-A; NB4 | ||
| Seq Type List | m6A-seq; m6A-REF-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000481889.6; ENST00000407404.5; ENST00000436778.5; ENST00000529540.5; ENST00000405853.7; ENST00000527949.1; ENST00000525441.5; ENST00000405576.5; ENST00000420369.5; ENST00000395397.7; ENST00000481020.5; ENST00000527464.5; ENST00000532630.1; ENST00000444396.5; ENST00000467728.5; ENST00000531660.5; ENST00000441012.7; ENST00000449369.5; ENST00000616973.4; ENST00000436029.5; ENST00000487913.5 | ||
| External Link | RMBase: m6A_site_139885 | ||
| mod ID: M6ASITE005431 | Click to Show/Hide the Full List | ||
| mod site | chr11:47261367-47261368:+ | [4] | |
| Sequence | CTGCCCCAGCTCAGCCCGGAACAACTGGGCATGATCGAGAA | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa; HepG2; MM6; Huh7; peripheral-blood; GSC-11; HEK293T; TIME; endometrial; HEC-1-A; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000532630.1; ENST00000529540.5; ENST00000467728.5; ENST00000527949.1; ENST00000481020.5; ENST00000436029.5; ENST00000531660.5; ENST00000405576.5; ENST00000481889.6; ENST00000395397.7; ENST00000527464.5; ENST00000405853.7; ENST00000616973.4; ENST00000441012.7; ENST00000525441.5; ENST00000407404.5; ENST00000487913.5 | ||
| External Link | RMBase: m6A_site_139886 | ||
| mod ID: M6ASITE005432 | Click to Show/Hide the Full List | ||
| mod site | chr11:47261656-47261657:+ | [4] | |
| Sequence | GAGATAGTTGACTTTGCTAAACAGCTACCCGGCTTCCTGCA | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; U2OS; H1A; H1B; A549; MM6; peripheral-blood; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; TREX; endometrial; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000487913.5; ENST00000531660.5; ENST00000481020.5; ENST00000441012.7; ENST00000405853.7; ENST00000527464.5; ENST00000405576.5; ENST00000532630.1; ENST00000527949.1; ENST00000467728.5; ENST00000407404.5; ENST00000529540.5; ENST00000395397.7; ENST00000616973.4; ENST00000481889.6 | ||
| External Link | RMBase: m6A_site_139887 | ||
| mod ID: M6ASITE005433 | Click to Show/Hide the Full List | ||
| mod site | chr11:47261691-47261692:+ | [4] | |
| Sequence | CCTGCAGCTCAGCCGGGAGGACCAGATTGCCCTGCTGAAGA | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; H1A; H1B; A549; MM6; CD4T; peripheral-blood; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; TREX; endometrial; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000467728.5; ENST00000487913.5; ENST00000531660.5; ENST00000395397.7; ENST00000481889.6; ENST00000616973.4; ENST00000481020.5; ENST00000532630.1; ENST00000527949.1; ENST00000405853.7; ENST00000441012.7; ENST00000407404.5; ENST00000527464.5; ENST00000405576.5; ENST00000529540.5 | ||
| External Link | RMBase: m6A_site_139888 | ||
| mod ID: M6ASITE005434 | Click to Show/Hide the Full List | ||
| mod site | chr11:47261711-47261712:+ | [4] | |
| Sequence | ACCAGATTGCCCTGCTGAAGACCTCTGCGATCGAGGTGGCT | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; H1A; H1B; A549; GM12878; MM6; CD4T; peripheral-blood; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; TREX; endometrial; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000616973.4; ENST00000405576.5; ENST00000405853.7; ENST00000467728.5; ENST00000531660.5; ENST00000407404.5; ENST00000481020.5; ENST00000481889.6; ENST00000441012.7; ENST00000527464.5; ENST00000529540.5; ENST00000395397.7; ENST00000487913.5; ENST00000527949.1; ENST00000532630.1 | ||
| External Link | RMBase: m6A_site_139889 | ||
| mod ID: M6ASITE005435 | Click to Show/Hide the Full List | ||
| mod site | chr11:47261793-47261794:+ | [4] | |
| Sequence | TCTGTGGGAGGGGCCTCCAGACATCGAGCTGGGAGAGCCAA | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; H1A; H1B; GM12878; MM6; Jurkat; peripheral-blood; GSC-11; HEK293A-TOA; iSLK; MSC; TREX; endometrial; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000532630.1; ENST00000531660.5; ENST00000405576.5; ENST00000395397.7; ENST00000441012.7; ENST00000487913.5; ENST00000405853.7; ENST00000407404.5; ENST00000616973.4; ENST00000467728.5; ENST00000481889.6; ENST00000527464.5; ENST00000527949.1; ENST00000529540.5; ENST00000481020.5 | ||
| External Link | RMBase: m6A_site_139890 | ||
| mod ID: M6ASITE005436 | Click to Show/Hide the Full List | ||
| mod site | chr11:47261873-47261874:+ | [4] | |
| Sequence | GGAAGAGGCCATGCTCCAAGACCAGCCCTCCTAGTCCCCGT | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa; H1A; MM6; Huh7; GSC-11; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000529540.5; ENST00000405853.7; ENST00000467728.5; ENST00000532630.1; ENST00000407404.5; ENST00000395397.7; ENST00000527949.1; ENST00000531660.5; ENST00000527464.5; ENST00000616973.4; ENST00000441012.7; ENST00000487913.5; ENST00000405576.5; ENST00000481020.5; ENST00000481889.6 | ||
| External Link | RMBase: m6A_site_139891 | ||
| mod ID: M6ASITE005437 | Click to Show/Hide the Full List | ||
| mod site | chr11:47261933-47261934:+ | [7] | |
| Sequence | TGCAGGTGATGCTTCTGGAGACATCTCGGAGGTACAACCCT | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | MM6; Huh7; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000527949.1; ENST00000531660.5; ENST00000532630.1; ENST00000405576.5; ENST00000481020.5; ENST00000441012.7; ENST00000487913.5; ENST00000467728.5; ENST00000407404.5; ENST00000481889.6; ENST00000616973.4; ENST00000405853.7; ENST00000395397.7; ENST00000527464.5; ENST00000529540.5 | ||
| External Link | RMBase: m6A_site_139892 | ||
| mod ID: M6ASITE005438 | Click to Show/Hide the Full List | ||
| mod site | chr11:47262003-47262004:+ | [8] | |
| Sequence | TTTCAGTTATAACCGGGAAGACTTTGCCAAAGCAGGTGAGA | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | Huh7; NB4 | ||
| Seq Type List | MeRIP-seq; m6A-seq | ||
| Transcript ID List | ENST00000467728.5; ENST00000487913.5; ENST00000616973.4; ENST00000531660.5; ENST00000407404.5; ENST00000441012.7; ENST00000481889.6; ENST00000405576.5; ENST00000405853.7; ENST00000527949.1; ENST00000481020.5; ENST00000532630.1; ENST00000529540.5; ENST00000395397.7 | ||
| External Link | RMBase: m6A_site_139893 | ||
| mod ID: M6ASITE005439 | Click to Show/Hide the Full List | ||
| mod site | chr11:47268260-47268261:+ | [4] | |
| Sequence | ACTCTCTTGGTGACCTATAGACCGGCCCAACGTGCAGGACC | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa; Huh7 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000441012.7; ENST00000395397.7; ENST00000494018.1; ENST00000527949.1; ENST00000405853.7; ENST00000529540.5; ENST00000407404.5; ENST00000405576.5; ENST00000616973.4; ENST00000532630.1; ENST00000462051.5; ENST00000467728.5; ENST00000481889.6; ENST00000481020.5 | ||
| External Link | RMBase: m6A_site_139894 | ||
| mod ID: M6ASITE005440 | Click to Show/Hide the Full List | ||
| mod site | chr11:47268278-47268279:+ | [4] | |
| Sequence | AGACCGGCCCAACGTGCAGGACCAGCTCCAGGTAGAGAGGC | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; Huh7 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000462051.5; ENST00000616973.4; ENST00000405853.7; ENST00000529540.5; ENST00000494018.1; ENST00000395397.7; ENST00000441012.7; ENST00000467728.5; ENST00000405576.5; ENST00000532630.1; ENST00000481020.5; ENST00000527949.1; ENST00000481889.6; ENST00000407404.5 | ||
| External Link | RMBase: m6A_site_139895 | ||
| mod ID: M6ASITE005441 | Click to Show/Hide the Full List | ||
| mod site | chr11:47268332-47268333:+ | [9] | |
| Sequence | TGTGGAAGCCCTGCATGCCTACGTCTCCATCCACCATCCCC | ||
| Motif Score | 2.05260119 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000407404.5; ENST00000481889.6; ENST00000467728.5; ENST00000527949.1; ENST00000462051.5; ENST00000529540.5; ENST00000481020.5; ENST00000494018.1; ENST00000405853.7; ENST00000395397.7; ENST00000441012.7; ENST00000532630.1; ENST00000405576.5; ENST00000616973.4 | ||
| External Link | RMBase: m6A_site_139896 | ||
| mod ID: M6ASITE005442 | Click to Show/Hide the Full List | ||
| mod site | chr11:47268581-47268582:+ | [4] | |
| Sequence | TTCCCACGGATGCTAATGAAACTGGTGAGCCTCCGGACCCT | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa; HEK293T; Huh7; HEK293A-TOA | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000529540.5; ENST00000532630.1; ENST00000441012.7; ENST00000395397.7; ENST00000467728.5; ENST00000616973.4; ENST00000527949.1; ENST00000405576.5; ENST00000481020.5; ENST00000481889.6; ENST00000407404.5; ENST00000405853.7; ENST00000494018.1; ENST00000462051.5 | ||
| External Link | RMBase: m6A_site_139897 | ||
| mod ID: M6ASITE005443 | Click to Show/Hide the Full List | ||
| mod site | chr11:47268597-47268598:+ | [4] | |
| Sequence | TGAAACTGGTGAGCCTCCGGACCCTGAGCAGCGTCCACTCA | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; HEK293T; A549; MM6; Huh7; HEK293A-TOA | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000616973.4; ENST00000405853.7; ENST00000462051.5; ENST00000529540.5; ENST00000395397.7; ENST00000481889.6; ENST00000527949.1; ENST00000481020.5; ENST00000532630.1; ENST00000405576.5; ENST00000441012.7; ENST00000467728.5; ENST00000407404.5; ENST00000494018.1 | ||
| External Link | RMBase: m6A_site_139898 | ||
| mod ID: M6ASITE005444 | Click to Show/Hide the Full List | ||
| mod site | chr11:47268646-47268647:+ | [4] | |
| Sequence | GTTTGCACTGCGTCTGCAGGACAAAAAGCTCCCACCGCTGC | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; A549; MM6; HEK293A-TOA; iSLK; MSC; TREX | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000481020.5; ENST00000405853.7; ENST00000407404.5; ENST00000462051.5; ENST00000529540.5; ENST00000395397.7; ENST00000494018.1; ENST00000481889.6; ENST00000616973.4; ENST00000527949.1; ENST00000532630.1; ENST00000441012.7; ENST00000467728.5; ENST00000405576.5 | ||
| External Link | RMBase: m6A_site_139899 | ||
| mod ID: M6ASITE005445 | Click to Show/Hide the Full List | ||
| mod site | chr11:47268768-47268769:+ | [4] | |
| Sequence | GGCTGCCTCCTAGAAGTGGAACAGACTGAGAAGGGCAAACA | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; A549; H1A; H1B; hNPCs; hESCs; GM12878; MM6; Huh7; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; TREX; endometrial; HEC-1-A; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000467728.5; ENST00000407404.5; ENST00000532630.1; ENST00000529540.5; ENST00000462051.5; ENST00000441012.7; ENST00000616973.4; ENST00000405853.7; ENST00000481889.6; ENST00000395397.7; ENST00000405576.5; ENST00000494018.1; ENST00000481020.5; ENST00000527949.1 | ||
| External Link | RMBase: m6A_site_139900 | ||
| mod ID: M6ASITE005446 | Click to Show/Hide the Full List | ||
| mod site | chr11:47268772-47268773:+ | [10] | |
| Sequence | GCCTCCTAGAAGTGGAACAGACTGAGAAGGGCAAACATTCC | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | A549 | ||
| Seq Type List | m6A-CLIP/IP | ||
| Transcript ID List | ENST00000494018.1; ENST00000481889.6; ENST00000616973.4; ENST00000405853.7; ENST00000462051.5; ENST00000529540.5; ENST00000467728.5; ENST00000407404.5; ENST00000441012.7; ENST00000532630.1; ENST00000395397.7; ENST00000405576.5; ENST00000481020.5; ENST00000527949.1 | ||
| External Link | RMBase: m6A_site_139901 | ||
| mod ID: M6ASITE005447 | Click to Show/Hide the Full List | ||
| mod site | chr11:47268786-47268787:+ | [4] | |
| Sequence | GAACAGACTGAGAAGGGCAAACATTCCTGGGAGCTGGGCAA | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; A549; H1A; H1B; hNPCs; hESCs; GM12878; MM6; Huh7; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; TREX; endometrial; HEC-1-A; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000532630.1; ENST00000467728.5; ENST00000616973.4; ENST00000529540.5; ENST00000494018.1; ENST00000405853.7; ENST00000441012.7; ENST00000462051.5; ENST00000481020.5; ENST00000527949.1; ENST00000395397.7; ENST00000405576.5; ENST00000481889.6 | ||
| External Link | RMBase: m6A_site_139902 | ||
References