m6A Target Gene Information
General Information of the m6A Target Gene (ID: M6ATAR00199)
Full List of m6A Methylation Regulator of This Target Gene and Corresponding Disease/Drug Response(s)
ARNTL
can be regulated by the following regulator(s), and cause disease/drug response(s). You can browse detail information of regulator(s) or disease/drug response(s).
Browse Regulator
Browse Disease
Methyltransferase-like 3 (METTL3) [WRITER]
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | mouse embryonic stem cells | Mus musculus |
|
Treatment: METTL3-/- ESCs
Control: Wild type ESCs
|
GSE145309 | |
| Regulation |
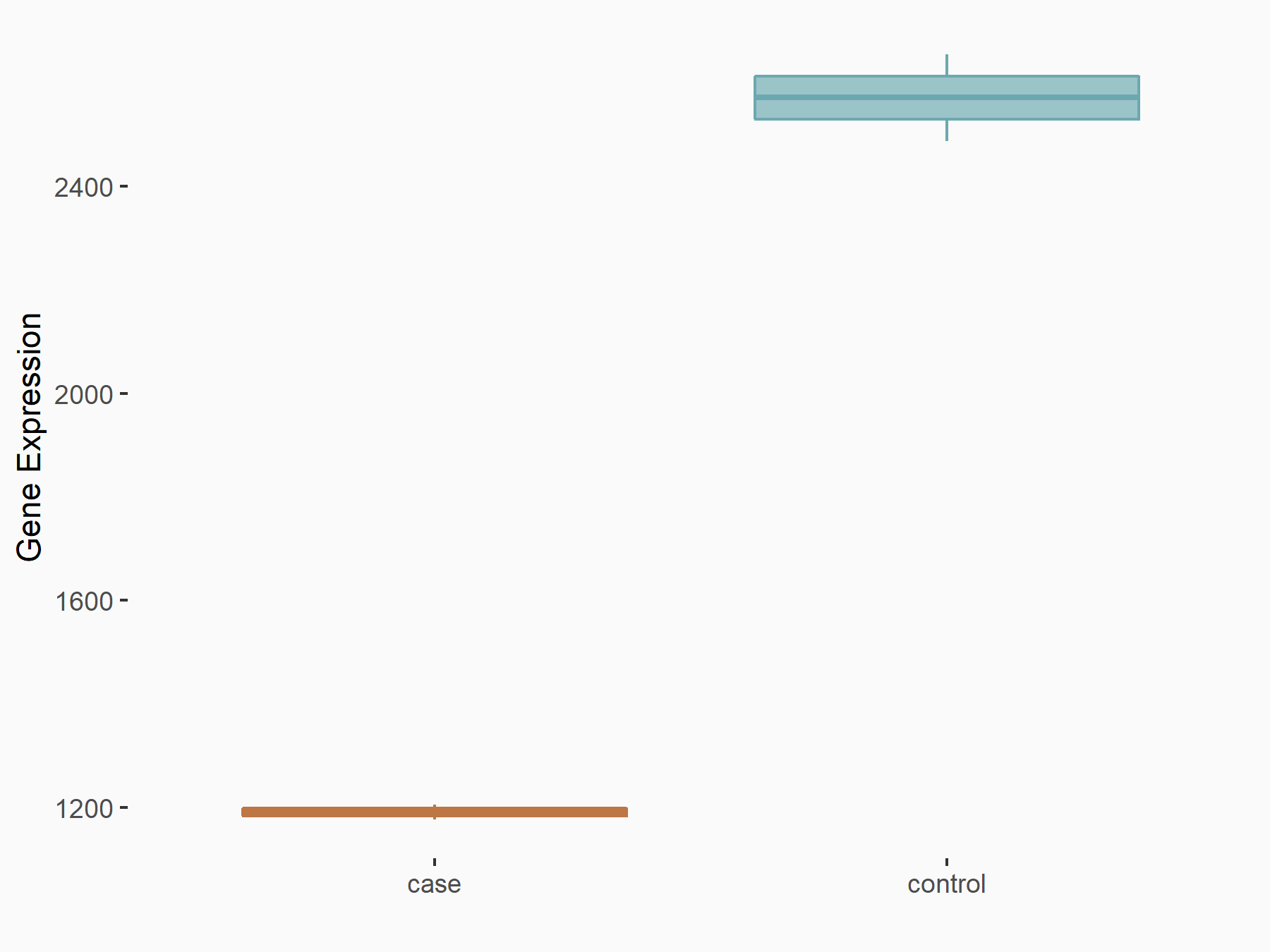 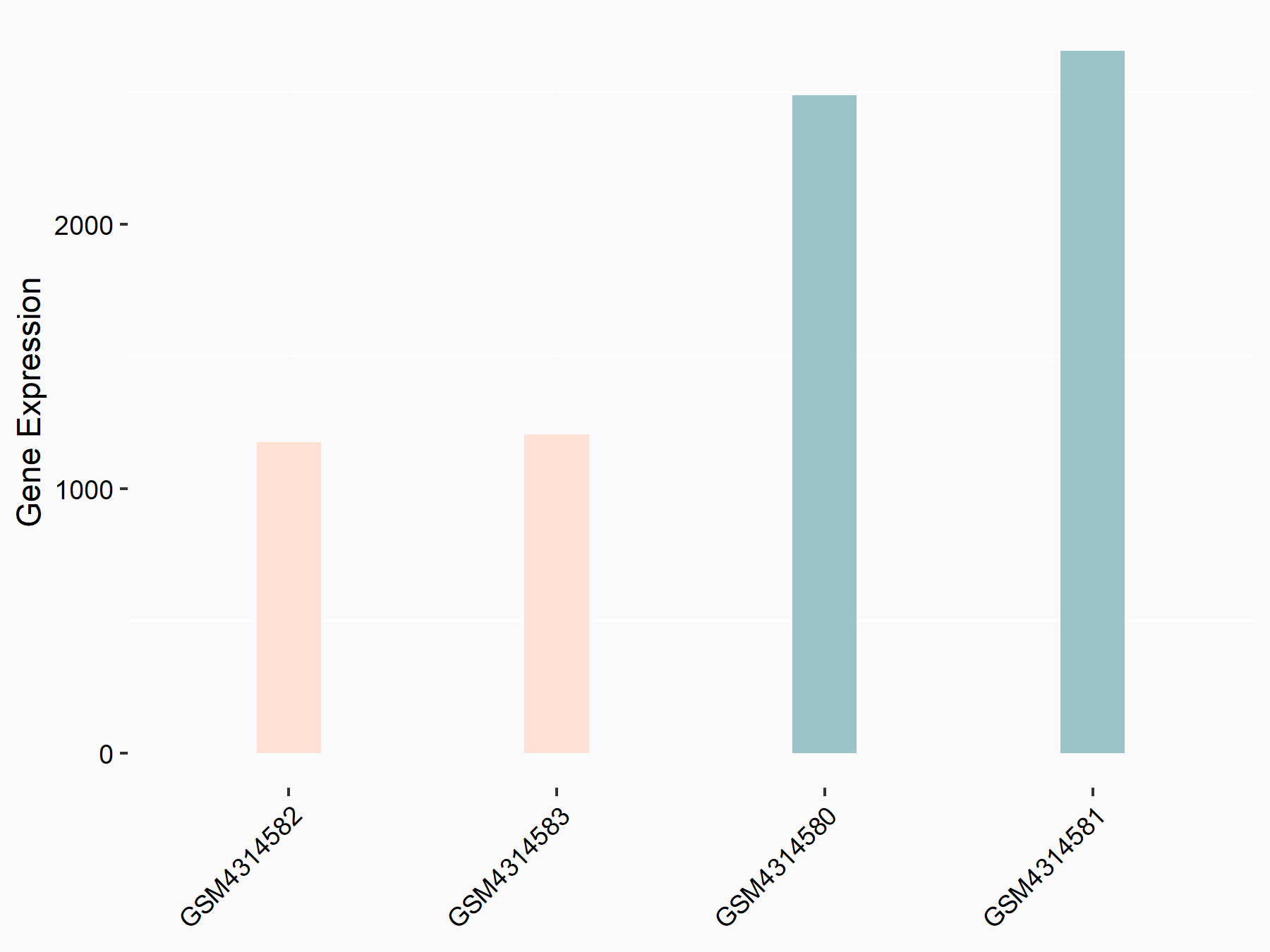 |
logFC: -1.11E+00 p-value: 1.53E-46 |
| More Results | Click to View More RNA-seq Results | |
| In total 2 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [1] | |||
| Response Summary | PPaRalpha to mediate its mRNA stability to regulate lipid metabolism. Hepatic deletion of Brain and muscle ARNT-like 1 (Bmal1/ARNTL) increases m6A mRNA methylation, particularly of PPaRalpha. Inhibition of m6A methylation via knockdown of m6A methyltransferase METTL3 decreases PPaR-Alpha m6A abundance and increases PPaRalpha mRNA lifetime and expression, reducing lipid accumulation in cells in vitro. YTHDF2 binds to PPaRalpha to mediate its mRNA stability to regulate lipid metabolism. Transcriptional regulation of circadian rhythms is essential for lipid metabolic homeostasis, disruptions of which can lead to metabolic diseases. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Metabolic disorders | ICD-11: 5D2Z | ||
| Pathway Response | Adipocytokine signaling pathway | hsa04920 | ||
| Cell Process | Llipid metabolism | |||
| In-vitro Model | Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 |
| Hepa 1-6 | Hepatocellular carcinoma of the mouse | Mus musculus | CVCL_0327 | |
| In-vivo Model | Liver-specific Bmal1f/f-AlbCre-knockout mice were purchased from Jackson Laboratory. C57BI/6J or Bmal1f/f-AlbCre-knockout male mice were maintained under a 12 hr light/12 hr dark (LD) cycle (ZT0 = 6 AM) and fed ad libitum with normal rodent chow (2018 Global 18% Protein diet, Envigo) and water. At 10-14 weeks of age, 10 male mice per group were sacrificed via CO2 asphyxiation at Zeitgeber Time (ZT) 0,2,6,10,12,14,18,22. In order to induce high levels of ROS in the liver, WT male mice were fasted 12 h and followed by intraperitoneal injection with 300 mg/kg APAP dissolved in PBS and re-fed. | |||
| Experiment 2 Reporting the m6A Methylation Regulator of This Target Gene | [2] | |||
| Response Summary | Liver-specific Mettl3 knockout mice exhibited global decrease in m6A on polyadenylated RNAs and pathologic features associated with nonalcoholic fatty liver disease. Studies in the M3LKO model indicated that METTL3 exhibits pleotropic function to maintain liver homeostasis by deregulating m6A profile and expression of the liver transcriptome. A significant decrease in total Brain and muscle ARNT-like 1 (Bmal1/ARNTL) and Clock mRNAs but an increase in their nuclear levels were observed in M3LKO livers, suggesting impaired nuclear export. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Non-alcoholic fatty liver disease | ICD-11: DB92 | ||
| In-vivo Model | M3LKO (Mettl3fl/fl; Alb-Cre) mice were generated by crossing Mettl3fl/fl mice (provided by Dr. Jacob Hanna) with albumin-Cre mice (Jackson Laboratories, Bar Harbor, ME), and genotypes were confirmed by tail-DNA PCR using primers as previously described. | |||
YTH domain-containing family protein 2 (YTHDF2) [READER]
| Representative RNA-seq result indicating the expression of this target gene regulated by YTHDF2 | ||
| Cell Line | B18-hi B cell line | Mus musculus |
|
Treatment: YTHDF2 knockout B18-hi B cells
Control: Wild type B18-hi B cells
|
GSE189819 | |
| Regulation |
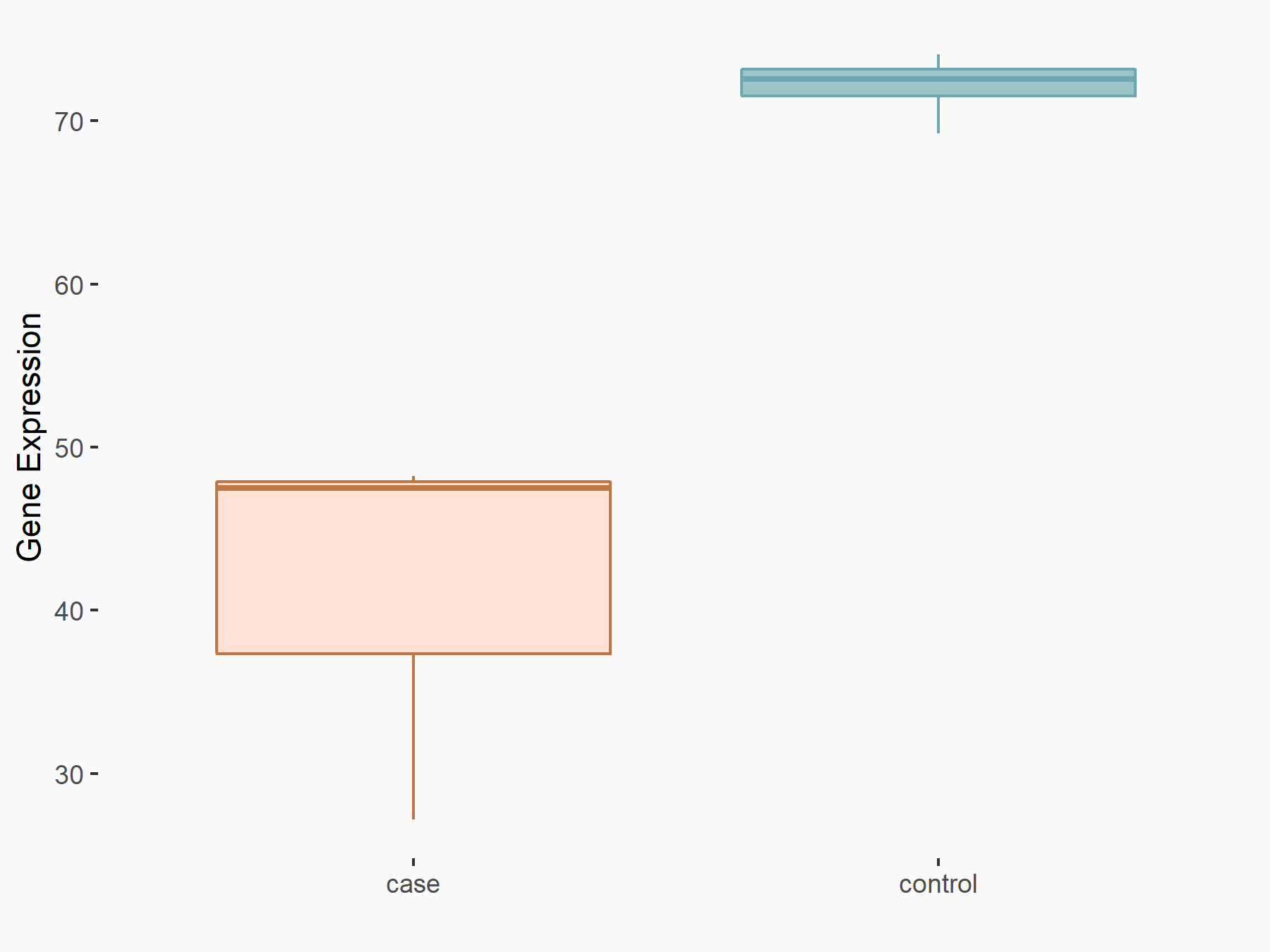 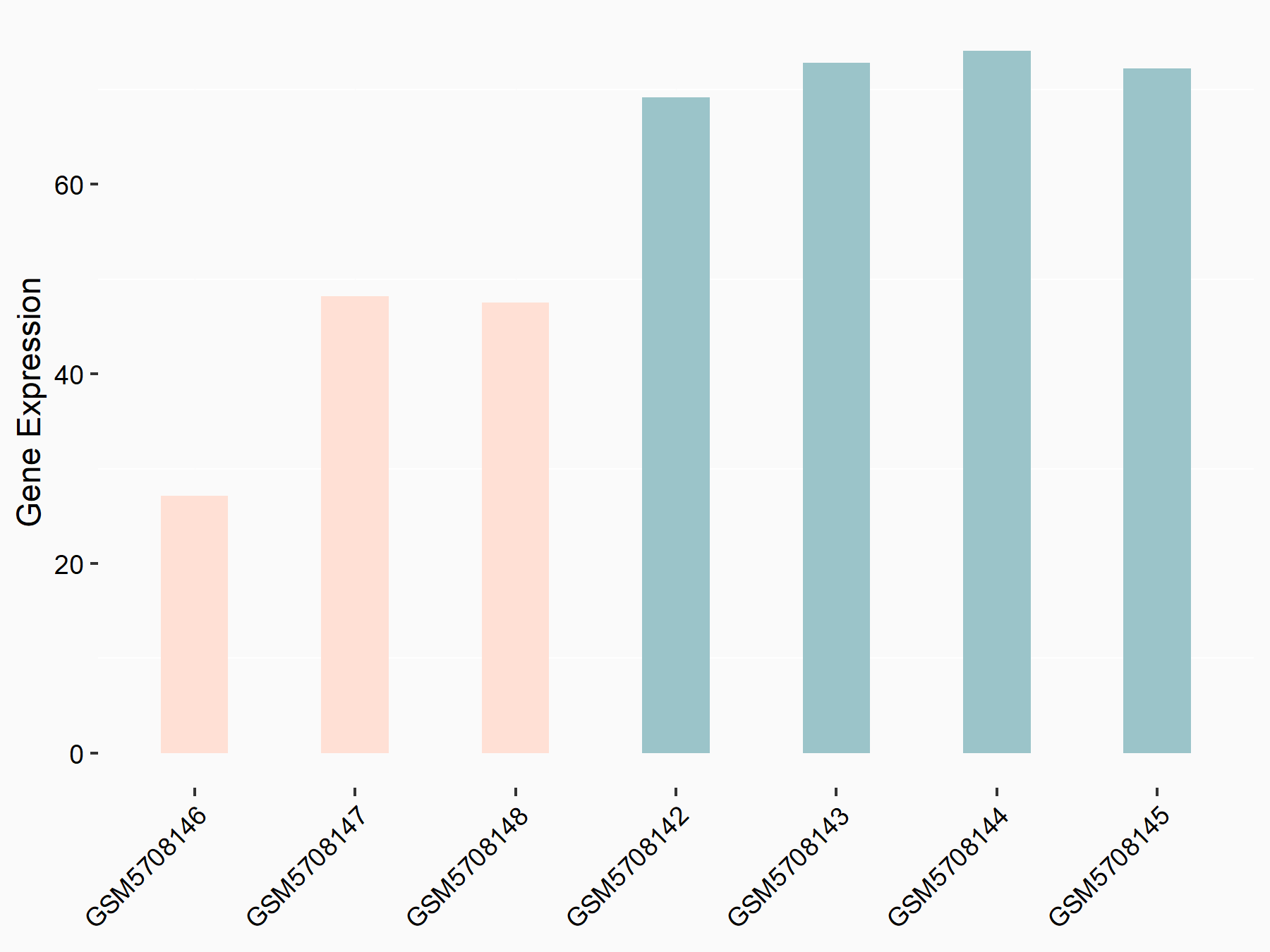 |
logFC: -7.97E-01 p-value: 5.55E-03 |
| More Results | Click to View More RNA-seq Results | |
| In total 1 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [1] | |||
| Response Summary | PPaRalpha to mediate its mRNA stability to regulate lipid metabolism. Hepatic deletion of Brain and muscle ARNT-like 1 (Bmal1/ARNTL) increases m6A mRNA methylation, particularly of PPaRalpha. Inhibition of m6A methylation via knockdown of m6A methyltransferase METTL3 decreases PPaR-Alpha m6A abundance and increases PPaRalpha mRNA lifetime and expression, reducing lipid accumulation in cells in vitro. YTHDF2 binds to PPaRalpha to mediate its mRNA stability to regulate lipid metabolism. Transcriptional regulation of circadian rhythms is essential for lipid metabolic homeostasis, disruptions of which can lead to metabolic diseases. | |||
| Target Regulation | Down regulation | |||
| Responsed Disease | Metabolic disorders | ICD-11: 5D2Z | ||
| Pathway Response | PPAR signaling pathway | hsa03320 | ||
| Adipocytokine signaling pathway | hsa04920 | |||
| Cell Process | Llipid metabolism | |||
| In-vitro Model | Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 |
| Hepa 1-6 | Hepatocellular carcinoma of the mouse | Mus musculus | CVCL_0327 | |
| In-vivo Model | Liver-specific Bmal1f/f-AlbCre-knockout mice were purchased from Jackson Laboratory. C57BI/6J or Bmal1f/f-AlbCre-knockout male mice were maintained under a 12 hr light/12 hr dark (LD) cycle (ZT0 = 6 AM) and fed ad libitum with normal rodent chow (2018 Global 18% Protein diet, Envigo) and water. At 10-14 weeks of age, 10 male mice per group were sacrificed via CO2 asphyxiation at Zeitgeber Time (ZT) 0,2,6,10,12,14,18,22. In order to induce high levels of ROS in the liver, WT male mice were fasted 12 h and followed by intraperitoneal injection with 300 mg/kg APAP dissolved in PBS and re-fed. | |||
Metabolic disorders [ICD-11: 5D2Z]
| In total 2 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [1] | |||
| Response Summary | PPaRalpha to mediate its mRNA stability to regulate lipid metabolism. Hepatic deletion of Brain and muscle ARNT-like 1 (Bmal1/ARNTL) increases m6A mRNA methylation, particularly of PPaRalpha. Inhibition of m6A methylation via knockdown of m6A methyltransferase METTL3 decreases PPaR-Alpha m6A abundance and increases PPaRalpha mRNA lifetime and expression, reducing lipid accumulation in cells in vitro. YTHDF2 binds to PPaRalpha to mediate its mRNA stability to regulate lipid metabolism. Transcriptional regulation of circadian rhythms is essential for lipid metabolic homeostasis, disruptions of which can lead to metabolic diseases. | |||
| Responsed Disease | Metabolic disorders [ICD-11: 5D2Z] | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Adipocytokine signaling pathway | hsa04920 | ||
| Cell Process | Llipid metabolism | |||
| In-vitro Model | Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 |
| Hepa 1-6 | Hepatocellular carcinoma of the mouse | Mus musculus | CVCL_0327 | |
| In-vivo Model | Liver-specific Bmal1f/f-AlbCre-knockout mice were purchased from Jackson Laboratory. C57BI/6J or Bmal1f/f-AlbCre-knockout male mice were maintained under a 12 hr light/12 hr dark (LD) cycle (ZT0 = 6 AM) and fed ad libitum with normal rodent chow (2018 Global 18% Protein diet, Envigo) and water. At 10-14 weeks of age, 10 male mice per group were sacrificed via CO2 asphyxiation at Zeitgeber Time (ZT) 0,2,6,10,12,14,18,22. In order to induce high levels of ROS in the liver, WT male mice were fasted 12 h and followed by intraperitoneal injection with 300 mg/kg APAP dissolved in PBS and re-fed. | |||
| Experiment 2 Reporting the m6A-centered Disease Response | [1] | |||
| Response Summary | PPaRalpha to mediate its mRNA stability to regulate lipid metabolism. Hepatic deletion of Brain and muscle ARNT-like 1 (Bmal1/ARNTL) increases m6A mRNA methylation, particularly of PPaRalpha. Inhibition of m6A methylation via knockdown of m6A methyltransferase METTL3 decreases PPaR-Alpha m6A abundance and increases PPaRalpha mRNA lifetime and expression, reducing lipid accumulation in cells in vitro. YTHDF2 binds to PPaRalpha to mediate its mRNA stability to regulate lipid metabolism. Transcriptional regulation of circadian rhythms is essential for lipid metabolic homeostasis, disruptions of which can lead to metabolic diseases. | |||
| Responsed Disease | Metabolic disorders [ICD-11: 5D2Z] | |||
| Target Regulator | YTH domain-containing family protein 2 (YTHDF2) | READER | ||
| Target Regulation | Down regulation | |||
| Pathway Response | PPAR signaling pathway | hsa03320 | ||
| Adipocytokine signaling pathway | hsa04920 | |||
| Cell Process | Llipid metabolism | |||
| In-vitro Model | Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 |
| Hepa 1-6 | Hepatocellular carcinoma of the mouse | Mus musculus | CVCL_0327 | |
| In-vivo Model | Liver-specific Bmal1f/f-AlbCre-knockout mice were purchased from Jackson Laboratory. C57BI/6J or Bmal1f/f-AlbCre-knockout male mice were maintained under a 12 hr light/12 hr dark (LD) cycle (ZT0 = 6 AM) and fed ad libitum with normal rodent chow (2018 Global 18% Protein diet, Envigo) and water. At 10-14 weeks of age, 10 male mice per group were sacrificed via CO2 asphyxiation at Zeitgeber Time (ZT) 0,2,6,10,12,14,18,22. In order to induce high levels of ROS in the liver, WT male mice were fasted 12 h and followed by intraperitoneal injection with 300 mg/kg APAP dissolved in PBS and re-fed. | |||
Non-alcoholic fatty liver disease [ICD-11: DB92]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [2] | |||
| Response Summary | Liver-specific Mettl3 knockout mice exhibited global decrease in m6A on polyadenylated RNAs and pathologic features associated with nonalcoholic fatty liver disease. Studies in the M3LKO model indicated that METTL3 exhibits pleotropic function to maintain liver homeostasis by deregulating m6A profile and expression of the liver transcriptome. A significant decrease in total Brain and muscle ARNT-like 1 (Bmal1/ARNTL) and Clock mRNAs but an increase in their nuclear levels were observed in M3LKO livers, suggesting impaired nuclear export. | |||
| Responsed Disease | Non-alcoholic fatty liver disease [ICD-11: DB92] | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Target Regulation | Up regulation | |||
| In-vivo Model | M3LKO (Mettl3fl/fl; Alb-Cre) mice were generated by crossing Mettl3fl/fl mice (provided by Dr. Jacob Hanna) with albumin-Cre mice (Jackson Laboratories, Bar Harbor, ME), and genotypes were confirmed by tail-DNA PCR using primers as previously described. | |||
RNA Modification Sequencing Data Associated with the Target (ID: M6ATAR00199)
| In total 22 m6A sequence/site(s) in this target gene | |||
| mod ID: M6ASITE004206 | Click to Show/Hide the Full List | ||
| mod site | chr11:13277856-13277857:+ | [3] | |
| Sequence | TGCCTGTTTACCCGCGCCGGACTCACCGCCGCCGCCGCCGC | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | U2OS; HEK293A-TOA; TIME | ||
| Seq Type List | MeRIP-seq; m6A-seq | ||
| Transcript ID List | ENST00000529825.5; ENST00000403510.7; ENST00000533520.5; ENST00000534544.5; ENST00000389707.8; ENST00000529050.5; ENST00000527998.5; ENST00000529388.5; ENST00000403290.5; ENST00000480685.5; ENST00000401424.5; ENST00000530357.5 | ||
| External Link | RMBase: m6A_site_132518 | ||
| mod ID: M6ASITE004207 | Click to Show/Hide the Full List | ||
| mod site | chr11:13310015-13310016:+ | [3] | |
| Sequence | GCTAGAGTGTATACGTTTGGACCCAAGCTTAACTTTTCCAA | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | U2OS; iSLK; TIME | ||
| Seq Type List | MeRIP-seq | ||
| Transcript ID List | ENST00000529825.5; ENST00000530357.5; ENST00000403290.5; ENST00000389707.8; ENST00000531665.5; ENST00000529388.5; ENST00000401424.5; ENST00000534544.5; ENST00000533520.5; ENST00000527998.5; ENST00000480685.5; ENST00000529050.5; ENST00000482049.5; ENST00000403510.7 | ||
| External Link | RMBase: m6A_site_132519 | ||
| mod ID: M6ASITE004208 | Click to Show/Hide the Full List | ||
| mod site | chr11:13349984-13349985:+ | [4] | |
| Sequence | GCCTGAAAAGAAATTATAAAACATGAAAATCGCTTTGAGGT | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000529825.5; ENST00000403290.5; ENST00000527998.5; ENST00000389707.8; ENST00000529050.5; ENST00000401424.5; ENST00000534544.5; ENST00000403510.7; ENST00000529388.5; ENST00000533520.5; ENST00000485918.2; ENST00000482049.5; ENST00000531665.5; ENST00000530357.5; ENST00000480685.5 | ||
| External Link | RMBase: m6A_site_132520 | ||
| mod ID: M6ASITE004209 | Click to Show/Hide the Full List | ||
| mod site | chr11:13358581-13358582:+ | [5] | |
| Sequence | TGGCTGTTCAGCACATGAAAACATTAAGAGGTGAGACCCTG | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | Huh7 | ||
| Seq Type List | MeRIP-seq | ||
| Transcript ID List | ENST00000401424.5; ENST00000389707.8; ENST00000403482.7; ENST00000403290.5; ENST00000497429.5; ENST00000403510.7; ENST00000530357.5; ENST00000529388.5 | ||
| External Link | RMBase: m6A_site_132521 | ||
| mod ID: M6ASITE004210 | Click to Show/Hide the Full List | ||
| mod site | chr11:13360371-13360372:+ | [5] | |
| Sequence | CAATCCATACACAGAAGCAAACTACAAACCAACTTTTCTAT | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | Huh7 | ||
| Seq Type List | MeRIP-seq | ||
| Transcript ID List | ENST00000529388.5; ENST00000401424.5; ENST00000403482.7; ENST00000530357.5; ENST00000389707.8; ENST00000403290.5; ENST00000497429.5; ENST00000403510.7 | ||
| External Link | RMBase: m6A_site_132522 | ||
| mod ID: M6ASITE004211 | Click to Show/Hide the Full List | ||
| mod site | chr11:13360378-13360379:+ | [5] | |
| Sequence | TACACAGAAGCAAACTACAAACCAACTTTTCTATCAGACGA | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | Huh7 | ||
| Seq Type List | MeRIP-seq | ||
| Transcript ID List | ENST00000403482.7; ENST00000403290.5; ENST00000530357.5; ENST00000389707.8; ENST00000497429.5; ENST00000401424.5; ENST00000403510.7 | ||
| External Link | RMBase: m6A_site_132523 | ||
| mod ID: M6ASITE004212 | Click to Show/Hide the Full List | ||
| mod site | chr11:13372217-13372218:+ | [6] | |
| Sequence | GGCTATTTGAAAAGCTGGCCACCCACAAAGATGGGGCTGGA | ||
| Motif Score | 2.032470238 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000389707.8; ENST00000403510.7; ENST00000403482.7; ENST00000401424.5; ENST00000529390.1; ENST00000403290.5; ENST00000497429.5 | ||
| External Link | RMBase: m6A_site_132524 | ||
| mod ID: M6ASITE004213 | Click to Show/Hide the Full List | ||
| mod site | chr11:13372243-13372244:+ | [6] | |
| Sequence | AAAGATGGGGCTGGATGAAGACAACGAACCAGACAATGAGG | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HEK293T; MT4 | ||
| Seq Type List | DART-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000401424.5; ENST00000403290.5; ENST00000529390.1; ENST00000389707.8; ENST00000403510.7; ENST00000497429.5; ENST00000403482.7 | ||
| External Link | RMBase: m6A_site_132525 | ||
| mod ID: M6ASITE004214 | Click to Show/Hide the Full List | ||
| mod site | chr11:13372246-13372247:+ | [6] | |
| Sequence | GATGGGGCTGGATGAAGACAACGAACCAGACAATGAGGGGT | ||
| Motif Score | 2.147845238 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000403290.5; ENST00000389707.8; ENST00000403482.7; ENST00000401424.5; ENST00000529390.1; ENST00000497429.5; ENST00000403510.7 | ||
| External Link | RMBase: m6A_site_132526 | ||
| mod ID: M6ASITE004215 | Click to Show/Hide the Full List | ||
| mod site | chr11:13372250-13372251:+ | [7] | |
| Sequence | GGGCTGGATGAAGACAACGAACCAGACAATGAGGGGTGTAA | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | MT4 | ||
| Seq Type List | MeRIP-seq | ||
| Transcript ID List | ENST00000529390.1; ENST00000497429.5; ENST00000403510.7; ENST00000403290.5; ENST00000403482.7; ENST00000401424.5; ENST00000389707.8 | ||
| External Link | RMBase: m6A_site_132527 | ||
| mod ID: M6ASITE004216 | Click to Show/Hide the Full List | ||
| mod site | chr11:13372255-13372256:+ | [4] | |
| Sequence | GGATGAAGACAACGAACCAGACAATGAGGGGTGTAACCTCA | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000403290.5; ENST00000403482.7; ENST00000389707.8; ENST00000403510.7; ENST00000401424.5; ENST00000497429.5; ENST00000529390.1 | ||
| External Link | RMBase: m6A_site_132528 | ||
| mod ID: M6ASITE004217 | Click to Show/Hide the Full List | ||
| mod site | chr11:13372319-13372320:+ | [4] | |
| Sequence | CTGCATTCTCATGTAGTTCCACAACCAGTGAACGGGGAAAT | ||
| Motif Score | 2.053113095 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000403510.7; ENST00000529390.1; ENST00000389707.8; ENST00000497429.5; ENST00000401424.5; ENST00000403482.7; ENST00000403290.5 | ||
| External Link | RMBase: m6A_site_132529 | ||
| mod ID: M6ASITE004218 | Click to Show/Hide the Full List | ||
| mod site | chr11:13374137-13374138:+ | [8] | |
| Sequence | TACCACAAGAACTTCTAGGCACATCGTGTTATGAATATTTT | ||
| Motif Score | 2.830589286 | ||
| Cell/Tissue List | brain | ||
| Seq Type List | m6A-REF-seq | ||
| Transcript ID List | ENST00000389707.8; ENST00000524392.5; ENST00000401424.5; ENST00000529390.1; ENST00000403290.5; ENST00000403482.7; ENST00000403510.7 | ||
| External Link | RMBase: m6A_site_132530 | ||
| mod ID: M6ASITE004219 | Click to Show/Hide the Full List | ||
| mod site | chr11:13374168-13374169:+ | [6] | |
| Sequence | TGAATATTTTCACCAAGATGACATAGGACATCTTGCAGAAT | ||
| Motif Score | 2.859755952 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000401424.5; ENST00000389707.8; ENST00000524392.5; ENST00000403482.7; ENST00000403290.5; ENST00000529390.1; ENST00000403510.7 | ||
| External Link | RMBase: m6A_site_132531 | ||
| mod ID: M6ASITE004220 | Click to Show/Hide the Full List | ||
| mod site | chr11:13375679-13375680:+ | [4] | |
| Sequence | TCAAAGATGGTTCTTTTATCACACTACGGAGTCGATGGTTC | ||
| Motif Score | 2.047297619 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000472842.1; ENST00000524392.5; ENST00000403290.5; ENST00000403482.7; ENST00000403510.7; ENST00000389707.8; ENST00000529390.1; ENST00000401424.5 | ||
| External Link | RMBase: m6A_site_132532 | ||
| mod ID: M6ASITE004221 | Click to Show/Hide the Full List | ||
| mod site | chr11:13375749-13375750:+ | [4] | |
| Sequence | AGAATATATTGTCTCAACTAACACTGTTGTTTTGTAAGTAC | ||
| Motif Score | 2.168095238 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000403290.5; ENST00000403482.7; ENST00000401424.5; ENST00000529390.1; ENST00000524392.5; ENST00000472842.1; ENST00000389707.8; ENST00000403510.7 | ||
| External Link | RMBase: m6A_site_132533 | ||
| mod ID: M6ASITE004222 | Click to Show/Hide the Full List | ||
| mod site | chr11:13376704-13376705:+ | [6] | |
| Sequence | AGCATCCCCCCACAGCATGGACAGCATGCTGCCCTCTGGAG | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000403510.7; ENST00000403290.5; ENST00000472842.1; ENST00000524392.5; ENST00000401424.5; ENST00000529390.1; ENST00000389707.8; ENST00000403482.7 | ||
| External Link | RMBase: m6A_site_132534 | ||
| mod ID: M6ASITE004223 | Click to Show/Hide the Full List | ||
| mod site | chr11:13386710-13386711:+ | [9] | |
| Sequence | CTCTTGGAAGCAGATGCTGGACTGGGTGGCCCTGTTGACTT | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa; HepG2; A549; HEK293A-TOA; TIME | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000524392.5; ENST00000389707.8; ENST00000403510.7; ENST00000401424.5; ENST00000472842.1; ENST00000403482.7; ENST00000403290.5 | ||
| External Link | RMBase: m6A_site_132535 | ||
| mod ID: M6ASITE004224 | Click to Show/Hide the Full List | ||
| mod site | chr11:13386736-13386737:+ | [6] | |
| Sequence | TGGCCCTGTTGACTTTAGTGACTTGCCATGGCCGCTGTAAA | ||
| Motif Score | 3.28175 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000524392.5; ENST00000401424.5; ENST00000389707.8; ENST00000403510.7; ENST00000403290.5; ENST00000472842.1; ENST00000403482.7 | ||
| External Link | RMBase: m6A_site_132536 | ||
| mod ID: M6ASITE004225 | Click to Show/Hide the Full List | ||
| mod site | chr11:13386756-13386757:+ | [9] | |
| Sequence | ACTTGCCATGGCCGCTGTAAACACTACATGTTGCTTTGGCA | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa; HEK293T; HepG2; A549; GM12878; HEK293A-TOA; MSC; TIME; iSLK | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000403482.7; ENST00000403510.7; ENST00000401424.5; ENST00000389707.8; ENST00000524392.5; ENST00000472842.1; ENST00000403290.5 | ||
| External Link | RMBase: m6A_site_132537 | ||
| mod ID: M6ASITE004226 | Click to Show/Hide the Full List | ||
| mod site | chr11:13387107-13387108:+ | [4] | |
| Sequence | TTCAGATGTATGGTGTTTTTACACTACAAAGAAGTCCCCCA | ||
| Motif Score | 2.07285119 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000389707.8; ENST00000401424.5; ENST00000403510.7; ENST00000524392.5; ENST00000472842.1; ENST00000403482.7; ENST00000403290.5 | ||
| External Link | RMBase: m6A_site_132538 | ||
| mod ID: M6ASITE004227 | Click to Show/Hide the Full List | ||
| mod site | chr11:13387112-13387113:+ | [4] | |
| Sequence | ATGTATGGTGTTTTTACACTACAAAGAAGTCCCCCATGTGG | ||
| Motif Score | 2.078666667 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000403482.7; ENST00000403510.7; ENST00000401424.5; ENST00000389707.8; ENST00000403290.5; ENST00000524392.5; ENST00000472842.1 | ||
| External Link | RMBase: m6A_site_132539 | ||
References