m6A Regulator Information
General Information of the m6A Regulator (ID: REG00013)
| Regulator Name | Insulin-like growth factor 2 mRNA-binding protein 2 (IGF2BP2) | ||||
|---|---|---|---|---|---|
| Synonyms |
IGF2 mRNA-binding protein 2; IMP-2; Hepatocellular carcinoma autoantigen p62; IGF-II mRNA-binding protein 2; VICKZ family member 2; IMP2; VICKZ2
Click to Show/Hide
|
||||
| Gene Name | IGF2BP2 | ||||
| Sequence |
MMNKLYIGNLSPAVTADDLRQLFGDRKLPLAGQVLLKSGYAFVDYPDQNWAIRAIETLSG
KVELHGKIMEVDYSVSKKLRSRKIQIRNIPPHLQWEVLDGLLAQYGTVENVEQVNTDTET AVVNVTYATREEAKIAMEKLSGHQFENYSFKISYIPDEEVSSPSPPQRAQRGDHSSREQG HAPGGTSQARQIDFPLRILVPTQFVGAIIGKEGLTIKNITKQTQSRVDIHRKENSGAAEK PVTIHATPEGTSEACRMILEIMQKEADETKLAEEIPLKILAHNGLVGRLIGKEGRNLKKI EHETGTKITISSLQDLSIYNPERTITVKGTVEACASAEIEIMKKLREAFENDMLAVNQQA NLIPGLNLSALGIFSTGLSVLSPPAGPRGAPPAAPYHPFTTHSGYFSSLYPHHQFGPFPH HHSYPEQEIVNLFIPTQAVGAIIGKKGAHIKQLARFAGASIKIAPAEGPDVSERMVIITG PPEAQFKAQGRIFGKLKEENFFNPKEEVKLEAHIRVPSSTAGRVIGKGGKTVNELQNLTS AEVIVPRDQTPDENEEVIVRIIGHFFASQTAQRKIREIVQQVKQQEQKYPQGVASQRSK Click to Show/Hide
|
||||
| Family | RRM IMP/VICKZ family | ||||
| Function |
RNA-binding factor that recruits target transcripts to cytoplasmic protein-RNA complexes (mRNPs). This transcript 'caging' into mRNPs allows mRNA transport and transient storage. It also modulates the rate and location at which target transcripts encounter the translational apparatus and shields them from endonuclease attacks or microRNA-mediated degradation (By similarity). Binds to the 5'-UTR of the insulin-like growth factor 2 (IGF2) mRNAs. Binding is isoform-specific. Binds to beta-actin/ACTB and MYC transcripts.
Click to Show/Hide
|
||||
| Gene ID | 10644 | ||||
| Uniprot ID | |||||
| Regulator Type | WRITER ERASER READER | ||||
| Mechanism Diagram | Click to View the Original Diagram | ||||
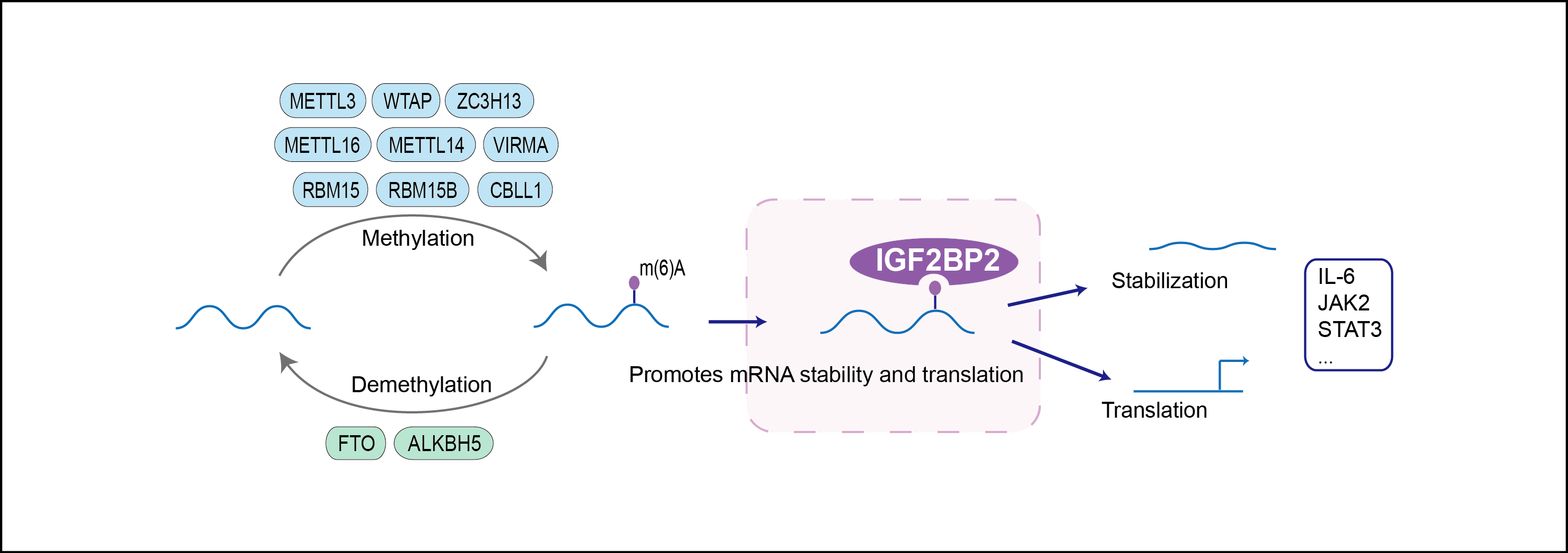
|
|||||
| Target Genes | Click to View Potential Target Genes of This Regulator | ||||
Full List of Target Gene(s) of This m6A Regulator and Corresponding Disease/Drug Response(s)
IGF2BP2 can regulate the m6A methylation of following target genes, and result in corresponding disease/drug response(s). You can browse corresponding disease or drug response(s) resulted from the regulation of certain target gene.
Browse Target Gene related Disease
Browse Target Gene related Drug
Interleukin-1 beta (IL1B)
| Representative RNA-seq result indicating the expression of this target gene regulated by IGF2BP2 | ||
| Cell Line | ES-2 cell line | Homo sapiens |
|
Treatment: siIGF2BP2 ES-2 cells
Control: siControl ES-2 cells
|
GSE109604 | |
| Regulation |
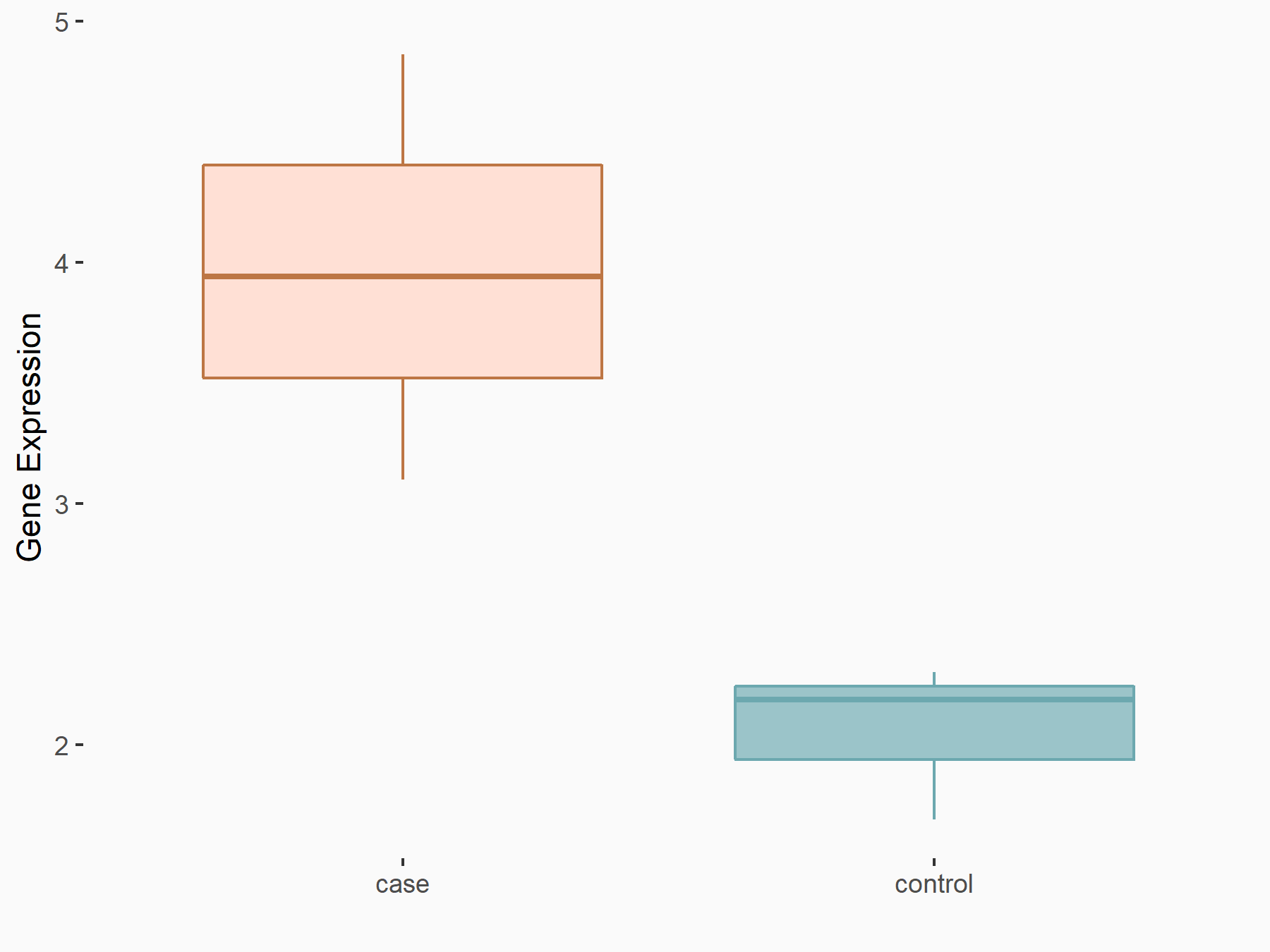  |
logFC: 6.90E-01 p-value: 1.24E-02 |
| More Results | Click to View More RNA-seq Results | |
Gangrene or necrosis of lung [ICD-11: CA43]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [1] | |||
| Responsed Disease | Gangrene or necrosis of lung [ICD-11: CA43] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | MAPK signaling pathway | hsa04010 | ||
| PI3K-Akt signaling pathway | hsa04151 | |||
| Cell Process | Biological regulation | |||
| Cell apoptosis | ||||
In-vitro Model |
BEAS-2B | Normal | Homo sapiens | CVCL_0168 |
| In-vivo Model | After being anesthetized with urethane (i.p.), SD rats were endotracheally intubated and ventilated using an animal ventilator under the conditions: respiratory rate of 70 breaths/min, tidal volume of 20 ml/kg, and inspiratory/expiratory ratio of 1:1. | |||
| Response Summary | N6-methyladenosine (m6A) methylation modification is implicated in the pathogenesis of lung ischemia-reperfusion injury. YTHDF3 or IGF2BP2 knockdown inhibited hypoxia/reoxygenation-activated p38, ERK1/2, AKT, and NF-Kappa-B pathways in BEAS-2B cells, and inhibited p-p65, Interleukin-1 beta (IL1B) and TNF-alpha secretion. | |||
Krueppel-like factor 12 (KLF12)
| Representative RNA-seq result indicating the expression of this target gene regulated by IGF2BP2 | ||
| Cell Line | Liver | Mus musculus |
|
Treatment: IMP2 -/- liver
Control: Wild type liver cells
|
GSE66440 | |
| Regulation |
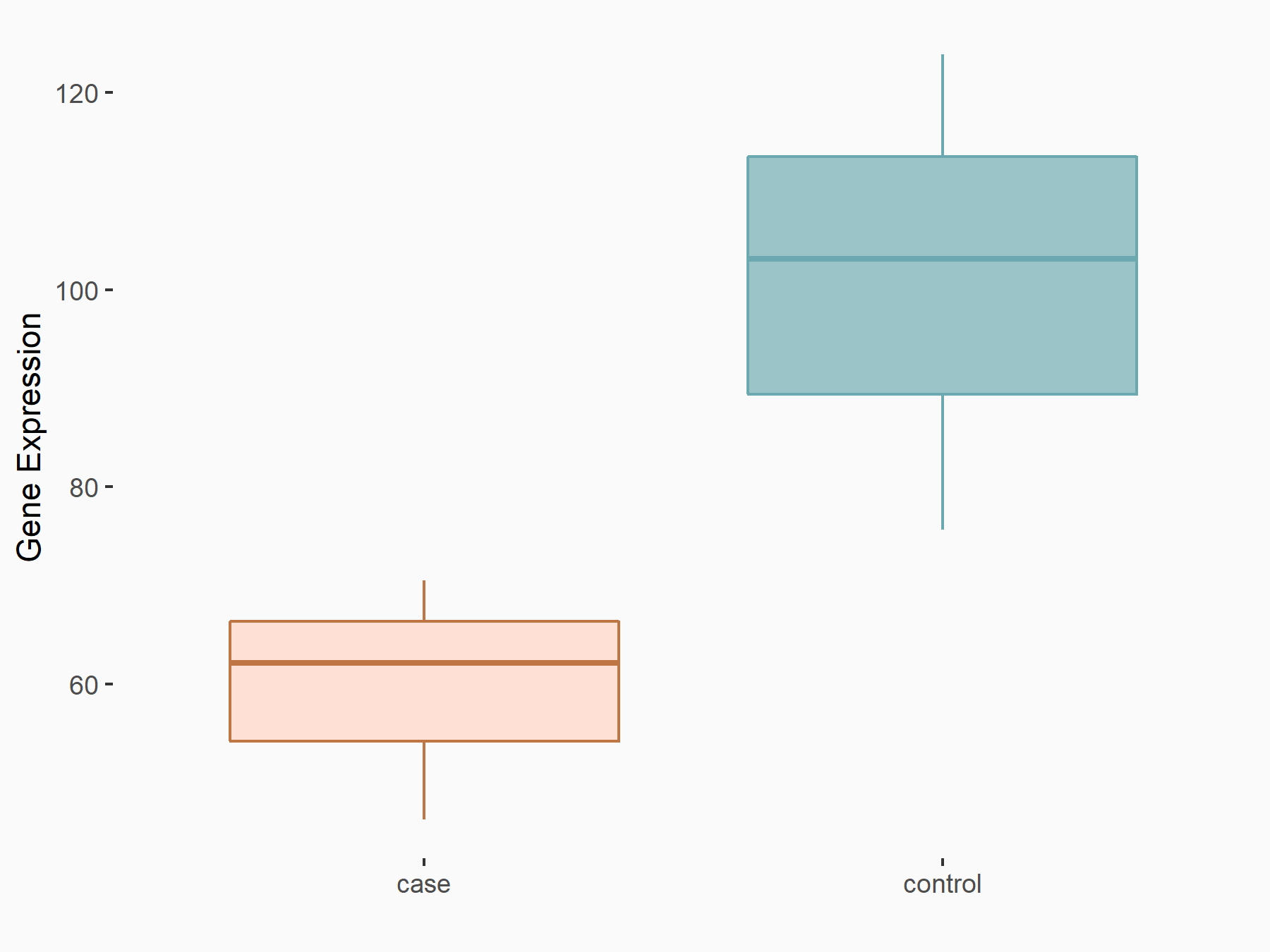 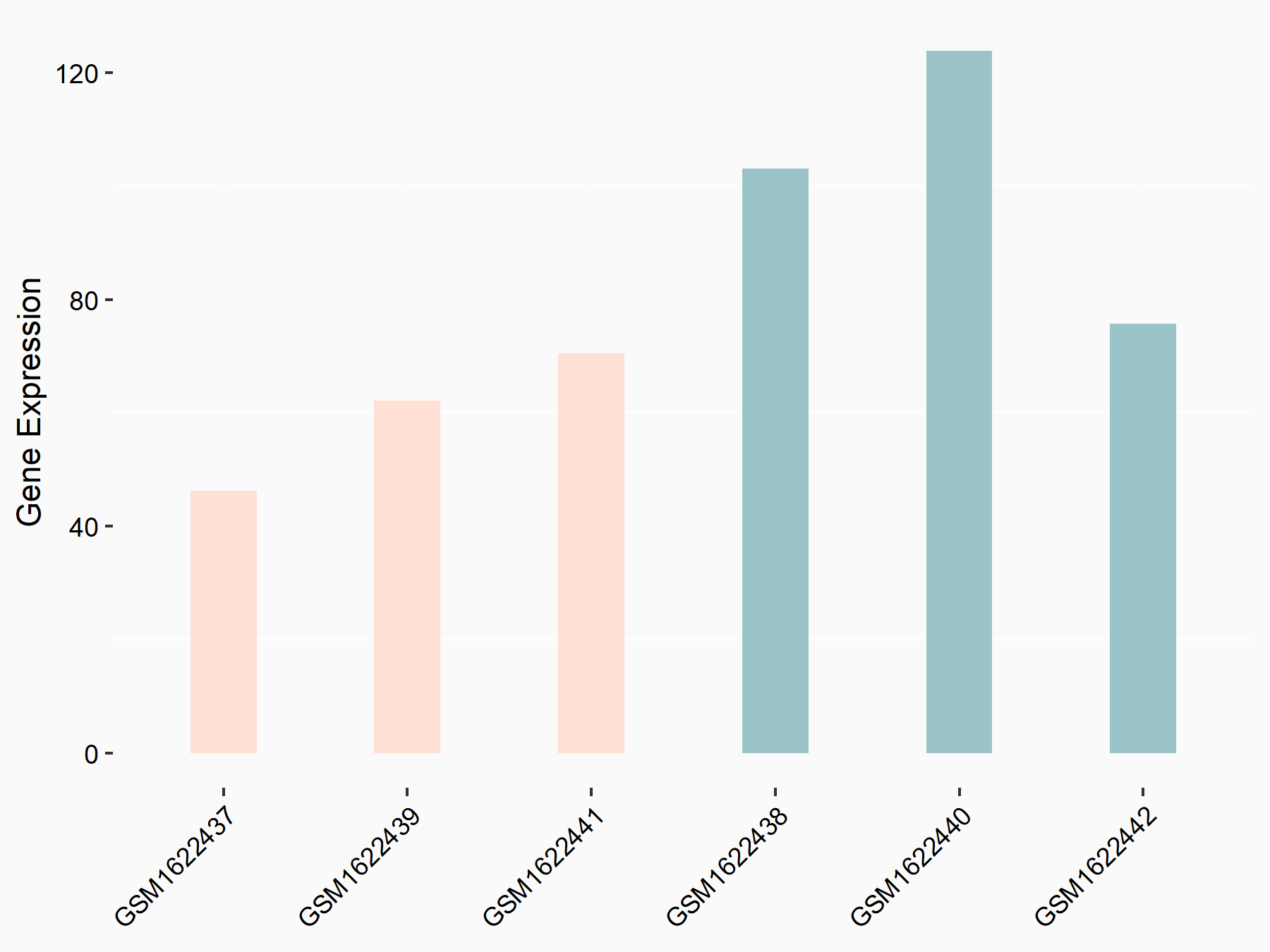 |
logFC: -7.41E-01 p-value: 4.43E-02 |
| More Results | Click to View More RNA-seq Results | |
Pancreatic cancer [ICD-11: 2C10]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [2] | |||
| Responsed Disease | Pancreatic ductal adenocarcinoma [ICD-11: 2C10.0] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | mRNA surveillance pathway | hsa03015 | ||
| RNA degradation | hsa03018 | |||
| Cell Process | RNA stability | |||
In-vitro Model |
THP-1 | Childhood acute monocytic leukemia | Homo sapiens | CVCL_0006 |
| PATU-8988 (Human pancreatic adenocarcinoma cell) | ||||
| PANC-1 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0480 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| 37 (Pancreatic cancer cell) | ||||
| In-vivo Model | BALB/c nude mice which were co-injected with THP-1 cells and PATU-8988 cells subcutaneously. | |||
| Response Summary | LncRNA-PACERR which bound to IGF2BP2 acts as an m6A-dependent manner to enhance the stability of Krueppel-like factor 12 (KLF12) and c-myc in cytoplasm. This study found that LncRNA-PACERR functions as key regulator of TAMs in PDAC microenvironment and revealed the novel mechanisms in cytoplasm and in nucleus. | |||
NAD-dependent protein deacetylase sirtuin-1 (SIRT1)
| Representative RNA-seq result indicating the expression of this target gene regulated by IGF2BP2 | ||
| Cell Line | Liver | Mus musculus |
|
Treatment: IMP2 -/- liver
Control: Wild type liver cells
|
GSE66440 | |
| Regulation |
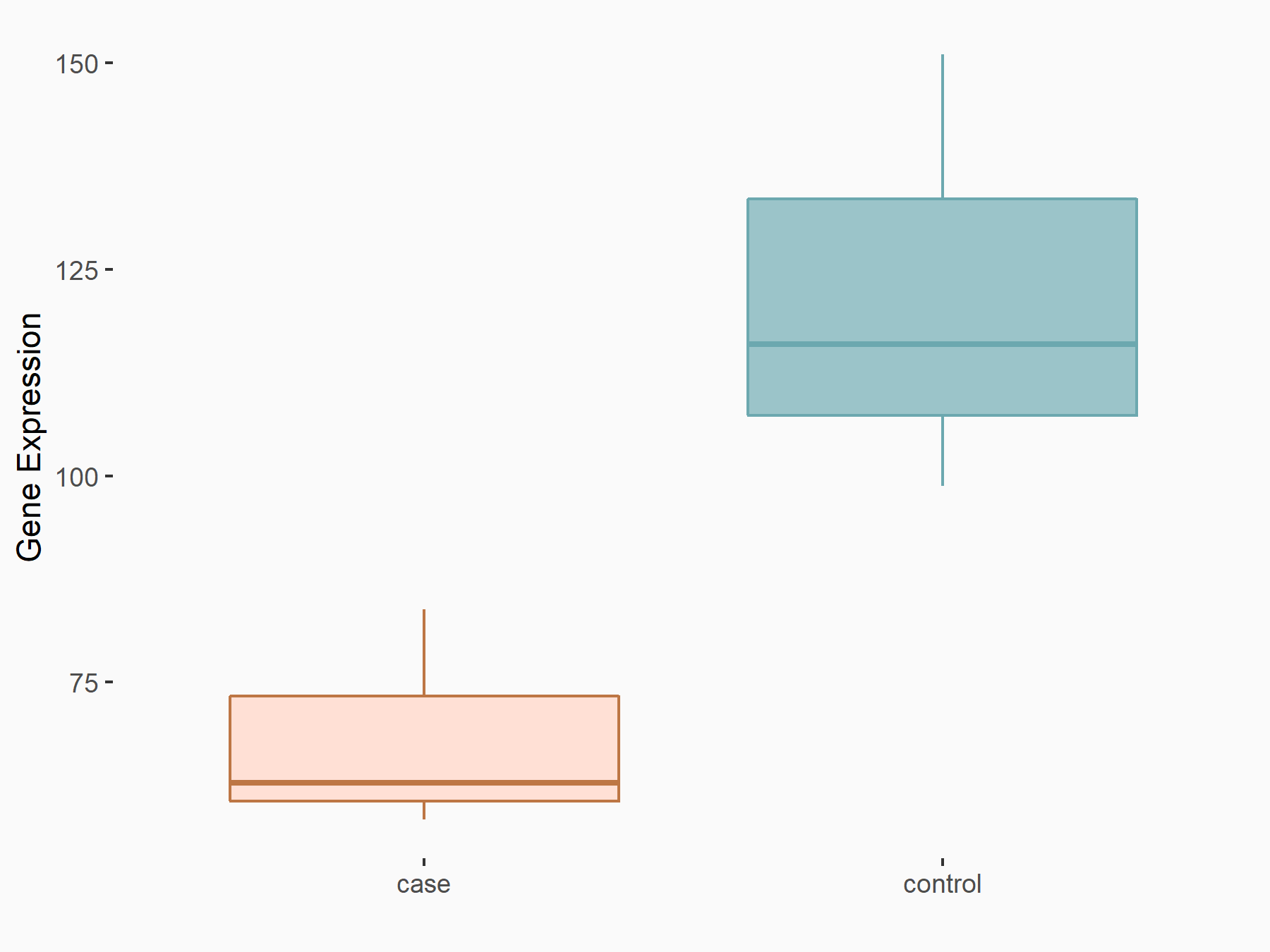 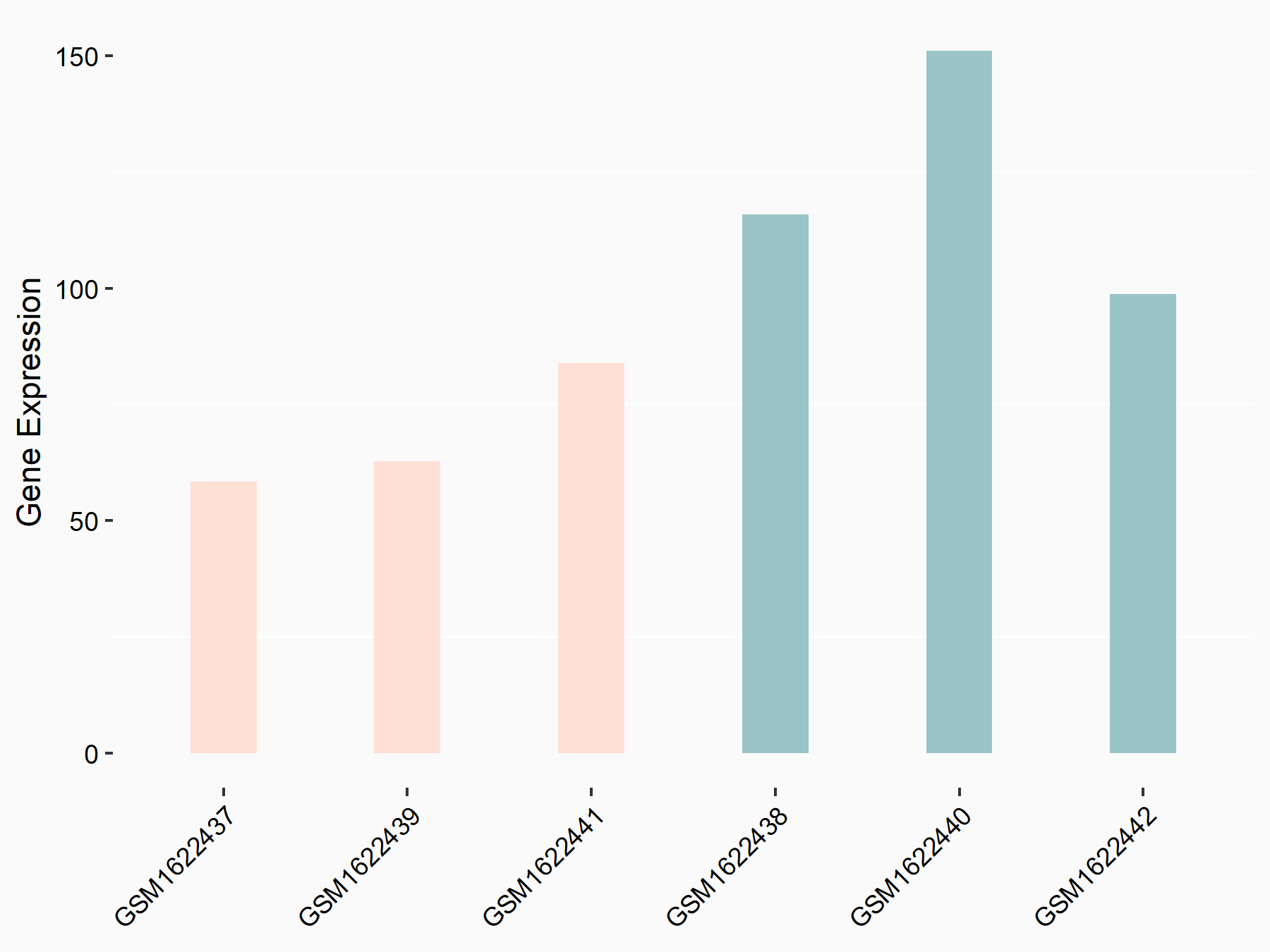 |
logFC: -8.22E-01 p-value: 2.08E-02 |
| More Results | Click to View More RNA-seq Results | |
Gastric cancer [ICD-11: 2B72]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [3] | |||
| Responsed Disease | Gastric cancer [ICD-11: 2B72] | |||
| Cell Process | Cell proliferation | |||
| Cell migration | ||||
In-vitro Model |
SNU-216 | Gastric tubular adenocarcinoma | Homo sapiens | CVCL_3946 |
| MKN45 | Gastric adenocarcinoma | Homo sapiens | CVCL_0434 | |
| AGS | Gastric adenocarcinoma | Homo sapiens | CVCL_0139 | |
| GES-1 | Normal | Homo sapiens | CVCL_EQ22 | |
| In-vivo Model | About 5 × 106 MKN45 cells stably transfected with IGF2BP2 shRNA or sh-NC vectors were subcutaneously injected into flank of nude mice. | |||
| Response Summary | IGF2BP2 regulated GC the proliferation/migration through recognizing the m6A modification sites of NAD-dependent protein deacetylase sirtuin-1 (SIRT1) mRNA. | |||
Vascular endothelial growth factor A (VEGFA)
| Representative RNA-seq result indicating the expression of this target gene regulated by IGF2BP2 | ||
| Cell Line | ES-2 cell line | Homo sapiens |
|
Treatment: siIGF2BP2 ES-2 cells
Control: siControl ES-2 cells
|
GSE109604 | |
| Regulation |
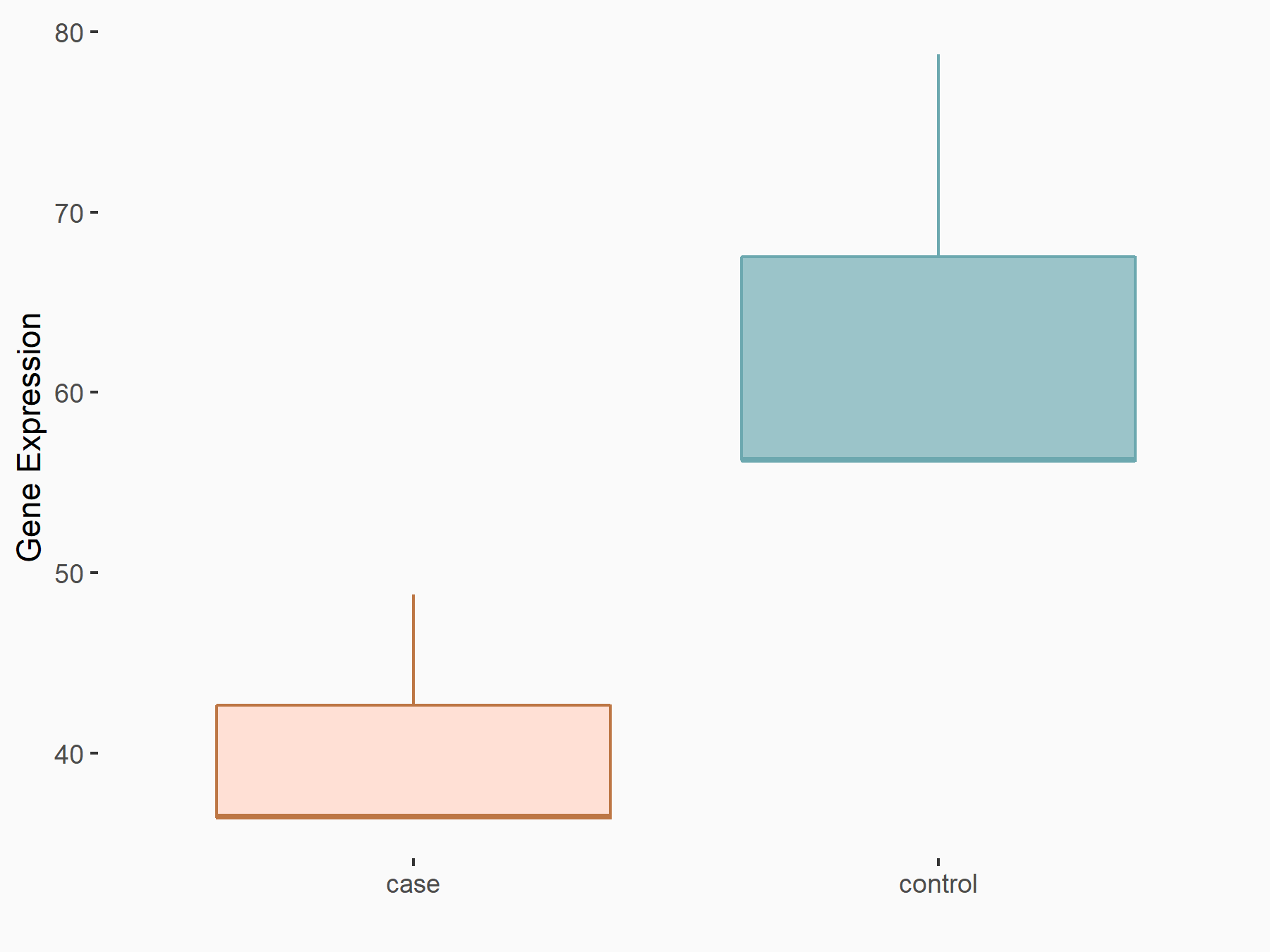 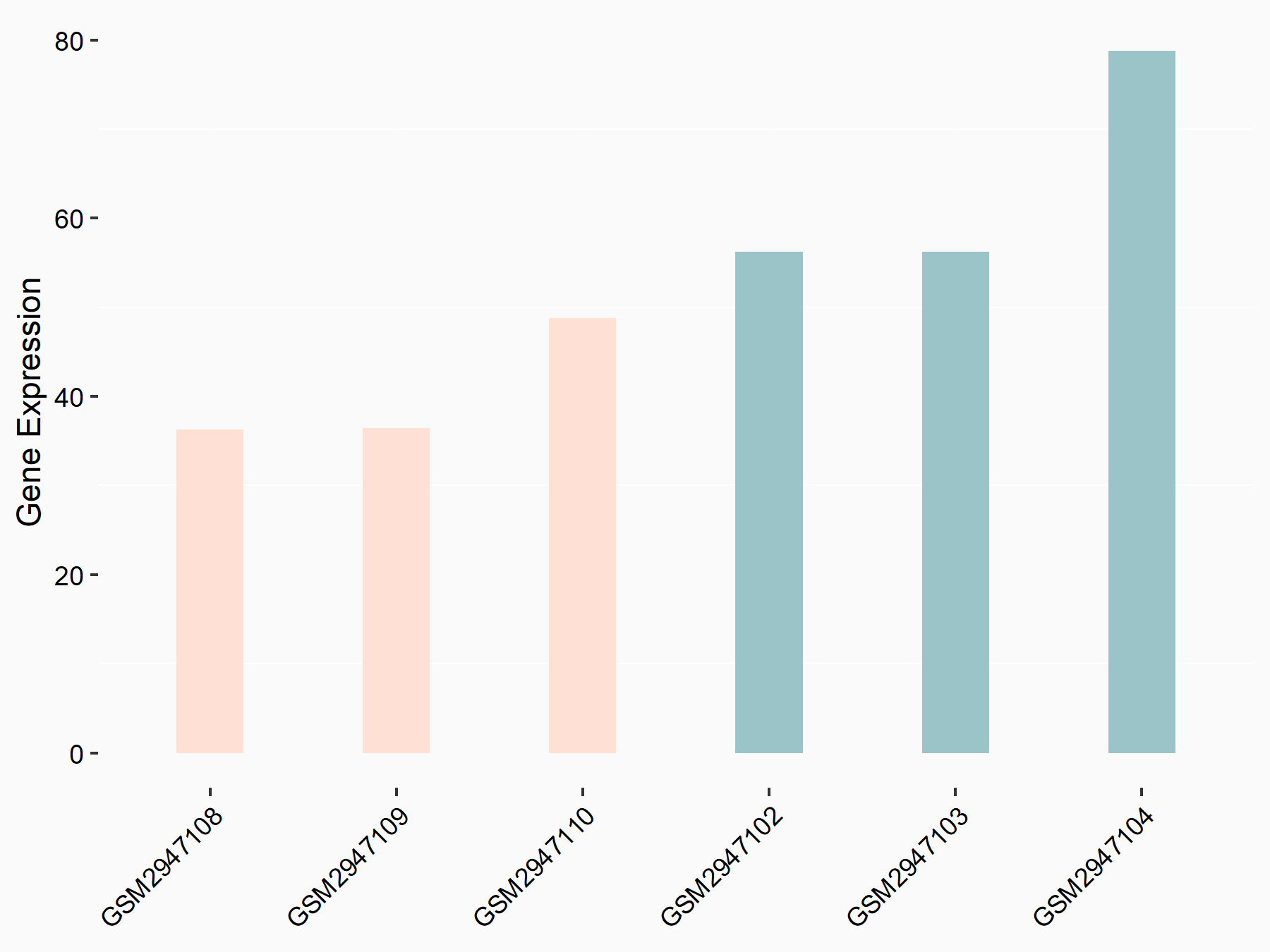 |
logFC: -6.36E-01 p-value: 3.11E-02 |
| More Results | Click to View More RNA-seq Results | |
Colorectal cancer [ICD-11: 2B91]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [4] | |||
| Responsed Disease | Colorectal cancer [ICD-11: 2B91] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | PI3K-Akt signaling pathway | hsa04151 | ||
In-vitro Model |
SW620 | Colon adenocarcinoma | Homo sapiens | CVCL_0547 |
| SW480 | Colon adenocarcinoma | Homo sapiens | CVCL_0546 | |
| NCM460 | Normal | Homo sapiens | CVCL_0460 | |
| LoVo | Colon adenocarcinoma | Homo sapiens | CVCL_0399 | |
| HT29 | Colon cancer | Mus musculus | CVCL_A8EZ | |
| HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 | |
| DLD-1 | Colon adenocarcinoma | Homo sapiens | CVCL_0248 | |
| In-vivo Model | A total of 8 × 106 wild-type (WT) or METTL3-knockdown cells were injected into the dorsal flanks of 6-week-old nude mice. Seven mice were randomly selected to calculate the volume according to the following formula: V = (width2 × length)/2. Mice were euthanized three weeks after injection and tumors removed, weighed, fixed, and embedded for immunohistochemical analysis. | |||
| Response Summary | EphA2 and Vascular endothelial growth factor A (VEGFA) targeted by METTL3 via different IGF2BP2-dependent mechanisms were found to promote vasculogenic mimicry (VM) formation via PI3K/AKT/mTOR and ERK1/2 signaling in CRC. | |||
High mobility group protein HMG-I/HMG-Y (HMGA1)
| Representative RIP-seq result supporting the interaction between the target gene and IGF2BP2 | ||
| Cell Line | HEK293T | Homo sapiens |
| Regulation | logFC: 1.10E+00 | GSE90639 |
Colorectal cancer [ICD-11: 2B91]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [5] | |||
| Responsed Disease | Colorectal cancer [ICD-11: 2B91] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | mRNA surveillance pathway | hsa03015 | ||
| Cell Process | mRNA stability | |||
| Epithelial-mesenchymal transition | ||||
In-vitro Model |
DLD-1 | Colon adenocarcinoma | Homo sapiens | CVCL_0248 |
| FHC | Normal | Homo sapiens | CVCL_3688 | |
| HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| HT29 | Colon cancer | Mus musculus | CVCL_A8EZ | |
| LoVo | Colon adenocarcinoma | Homo sapiens | CVCL_0399 | |
| SW480 | Colon adenocarcinoma | Homo sapiens | CVCL_0546 | |
| SW620 | Colon adenocarcinoma | Homo sapiens | CVCL_0547 | |
| In-vivo Model | Groups of HCT116-Luc-shCtrl, HCT116-Luc-shLINC00460, and HCT116-Luc-shLINC00460 + HMGA1 cells (5 × 106) were injected subcutaneously into the flanks of mice correspondingly. | |||
| Response Summary | LINC00460 is a novel oncogene of colorectal cancer through interacting with IGF2BP2 and DHX9 and bind to the m6A modified High mobility group protein HMG-I/HMG-Y (HMGA1) mRNA to enhance the HMGA1 mRNA stability. The N6-methyladenosine (m6A) modification of HMGA1 mRNA by METTL3 enhanced HMGA1 expression in CRC. | |||
MARCKS-related protein (MARCKSL1)
| Representative RIP-seq result supporting the interaction between the target gene and IGF2BP2 | ||
| Cell Line | HEK293T | Homo sapiens |
| Regulation | logFC: 1.20E+00 | GSE90639 |
Liver cancer [ICD-11: 2C12]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [6] | |||
| Responsed Disease | Hepatocellular carcinoma [ICD-11: 2C12.02] | |||
| Cell Process | RNA decay | |||
In-vitro Model |
Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| HeLa | Endocervical adenocarcinoma | Homo sapiens | CVCL_0030 | |
| Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 | |
| Response Summary | In contrast to the mRNA-decay-promoting function of YTH domain-containing family protein 2, IGF2BPs promote the stability and storage of their target mRNAs (for example, MYC) in an m6A-dependent manner under normal and stress conditions and therefore affect gene expression output. Four representative high confidence targets, including MYC, FSCN1, TK1, and MARCKS-related protein (MARCKSL1), exhibit strong binding with IGF2BPs around their m6A motifs in control cells. Knocking down of each individual IGF2BPs in Hela (cervical cancer) and HepG2 (liver cancer) cells significantly repressed MYC expression. | |||
Cervical cancer [ICD-11: 2C77]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [6] | |||
| Responsed Disease | Cervical cancer [ICD-11: 2C77] | |||
| Cell Process | RNA decay | |||
In-vitro Model |
Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| HeLa | Endocervical adenocarcinoma | Homo sapiens | CVCL_0030 | |
| Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 | |
| Response Summary | In contrast to the mRNA-decay-promoting function of YTH domain-containing family protein 2, IGF2BPs promote the stability and storage of their target mRNAs (for example, MYC) in an m6A-dependent manner under normal and stress conditions and therefore affect gene expression output. Four representative high confidence targets, including MYC, FSCN1, TK1, and MARCKS-related protein (MARCKSL1), exhibit strong binding with IGF2BPs around their m6A motifs in control cells. Knocking down of each individual IGF2BPs in Hela (cervical cancer) and HepG2 (liver cancer) cells significantly repressed MYC expression. | |||
Serine/arginine-rich splicing factor 7 (SRSF7)
| Representative RIP-seq result supporting the interaction between the target gene and IGF2BP2 | ||
| Cell Line | HEK293T | Homo sapiens |
| Regulation | logFC: 1.26E+00 | GSE90639 |
Brain cancer [ICD-11: 2A00]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [7] | |||
| Responsed Disease | Glioblastoma [ICD-11: 2A00.00] | |||
| Pathway Response | Spliceosome | hsa03040 | ||
In-vitro Model |
HEK293T | Normal | Homo sapiens | CVCL_0063 |
| U87MG (Astroblastoma cells from human brain) | ||||
| LN-229 | Glioblastoma | Homo sapiens | CVCL_0393 | |
| A-172 | Glioblastoma | Homo sapiens | CVCL_0131 | |
| LN-18 | Glioblastoma | Homo sapiens | CVCL_0392 | |
| LN-428 | Glioblastoma | Homo sapiens | CVCL_3959 | |
| LN-443 | Glioblastoma | Homo sapiens | CVCL_3960 | |
| SNB-19 | Astrocytoma | Homo sapiens | CVCL_0535 | |
| T98G | Glioblastoma | Homo sapiens | CVCL_0556 | |
| U-118MG | Astrocytoma | Homo sapiens | CVCL_0633 | |
| U251 (Fibroblasts or fibroblast like cells) | ||||
| U-138MG | Astrocytoma | Homo sapiens | CVCL_0020 | |
| Response Summary | The gene expression of Serine/arginine-rich splicing factor 7 (SRSF7) is positively correlated with glioblastoma (GBM) cell-specific m6A methylation. The two m6A sites on PDZ-binding kinase (PBK) are regulated by SRSF7 and partially mediate the effects of SRSF7 in GBM cells through recognition by IGF2BP2. | |||
Y-box-binding protein 1 (YBX1)
| Representative RIP-seq result supporting the interaction between the target gene and IGF2BP2 | ||
| Cell Line | HEK293T | Homo sapiens |
| Regulation | logFC: 1.14E+00 | GSE90639 |
Malignant haematopoietic neoplasm [ICD-11: 2B33]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [8] | |||
| Responsed Disease | Myeloid leukaemia [ICD-11: 2B33.1] | |||
| Target Regulation | Up regulation | |||
| Cell Process | Cell apoptosis | |||
In-vitro Model |
Leukemia stem cell line (Leukemia stem cell line) | |||
| Kasumi-1 | Myeloid leukemia with maturation | Homo sapiens | CVCL_0589 | |
| MOLM-13 | Adult acute myeloid leukemia | Homo sapiens | CVCL_2119 | |
| THP-1 | Childhood acute monocytic leukemia | Homo sapiens | CVCL_0006 | |
| MV4-11 | Childhood acute monocytic leukemia | Homo sapiens | CVCL_0064 | |
| BV-173 | Chronic myelogenous leukemia | Homo sapiens | CVCL_0181 | |
| NOMO-1 | Adult acute monocytic leukemia | Homo sapiens | CVCL_1609 | |
| K-562 | Chronic myelogenous leukemia | Homo sapiens | CVCL_0004 | |
| KG-1a | Adult acute myeloid leukemia | Homo sapiens | CVCL_1824 | |
| Response Summary | Y-box-binding protein 1 (YBX1) selectively functions in regulating survival of myeloid leukemia cells. YBX1 interacts with insulin-like growth factor 2 messenger RNA (mRNA)-binding proteins (IGF2BPs) and stabilizes m6A-tagged RNA. YBX1 deficiency dysregulates the expression of apoptosis-related genes and promotes mRNA decay of MYC and BCL2 in an m6A-dependent manner, which contributes to the defective survival that results from deletion of YBX1. | |||
Metastasis associated lung adenocarcinoma transcript 1 (MALAT1)
| Representative RIP-seq result supporting the interaction between the target gene and IGF2BP2 | ||
| Cell Line | HEK293T | Homo sapiens |
| Regulation | logFC: 1.31E+00 | GSE90639 |
Lung cancer [ICD-11: 2C25]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [9] | |||
| Responsed Disease | Non-small-cell lung carcinoma [ICD-11: 2C25.Y] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | Lysosome | hsa04142 | ||
| Cell Process | Cell autophagy | |||
In-vitro Model |
NCI-H157 | Lung squamous cell carcinoma | Homo sapiens | CVCL_0463 |
| NCI-H460 | Lung large cell carcinoma | Homo sapiens | CVCL_0459 | |
| NCI-H1975 | Lung adenocarcinoma | Homo sapiens | CVCL_1511 | |
| NCI-H1703 | Lung squamous cell carcinoma | Homo sapiens | CVCL_1490 | |
| NCI-H1299 | Lung large cell carcinoma | Homo sapiens | CVCL_0060 | |
| BEAS-2B | Normal | Homo sapiens | CVCL_0168 | |
| A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 | |
| In-vivo Model | Mice (male and 6 weeks old) were subcutaneously injected with NSCLC cells (1.0*106 cells/200 uL). The mice were terminated after 4 weeks of induction, and the tumor volume and tumor weight were measured. | |||
| Response Summary | IGF2BP2 promotes Metastasis associated lung adenocarcinoma transcript 1 (MALAT1) stability in an m6A-dependent mechanism, thus promoting its downstream target autophagy-related (ATG)12 expression and NSCLC proliferation. | |||
Apoptosis regulator Bcl-2 (BCL2)
Malignant haematopoietic neoplasm [ICD-11: 2B33]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [8] | |||
| Responsed Disease | Myeloid leukaemia [ICD-11: 2B33.1] | |||
| Target Regulation | Up regulation | |||
| Cell Process | Cell apoptosis | |||
In-vitro Model |
Leukemia stem cell line (Leukemia stem cell line) | |||
| Kasumi-1 | Myeloid leukemia with maturation | Homo sapiens | CVCL_0589 | |
| MOLM-13 | Adult acute myeloid leukemia | Homo sapiens | CVCL_2119 | |
| THP-1 | Childhood acute monocytic leukemia | Homo sapiens | CVCL_0006 | |
| MV4-11 | Childhood acute monocytic leukemia | Homo sapiens | CVCL_0064 | |
| BV-173 | Chronic myelogenous leukemia | Homo sapiens | CVCL_0181 | |
| NOMO-1 | Adult acute monocytic leukemia | Homo sapiens | CVCL_1609 | |
| K-562 | Chronic myelogenous leukemia | Homo sapiens | CVCL_0004 | |
| KG-1a | Adult acute myeloid leukemia | Homo sapiens | CVCL_1824 | |
| Response Summary | YBX1 selectively functions in regulating survival of myeloid leukemia cells. YBX1 interacts with insulin-like growth factor 2 messenger RNA (mRNA)-binding proteins (IGF2BPs) and stabilizes m6A-tagged RNA. YBX1 deficiency dysregulates the expression of apoptosis-related genes and promotes mRNA decay of MYC and Apoptosis regulator Bcl-2 (BCL2) in an m6A-dependent manner, which contributes to the defective survival that results from deletion of YBX1. | |||
ATP-dependent translocase ABCB1 (ABCB1)
Breast cancer [ICD-11: 2C60]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [10] | |||
| Responsed Disease | Breast cancer [ICD-11: 2C60] | |||
| Responsed Drug | Adriamycin | Phase 3 | ||
| Target Regulation | Up regulation | |||
In-vitro Model |
MCF-7 | Invasive breast carcinoma | Homo sapiens | CVCL_0031 |
| MDA-MB-231 | Breast adenocarcinoma | Homo sapiens | CVCL_0062 | |
| In-vivo Model | The xenograft mouse models were established by injecting MCF-7/ADR cells (1 × 107 in 100 μL RPMI 1640 medium) into the mouse right flank. Tumor size was monitored every week. When the average tumor size reached approximately 100 mm3, 5.0 mg/kg adriamycin were subsequently subjected through tail vein every other day. Mice were sacrificed after 4 weeks, and tumors were excised. | |||
Cellular tumor antigen p53 (TP53/p53)
Vascular disorders of the liver [ICD-11: DB98]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [11] | |||
| Responsed Disease | Vascular disorders of the liver [ICD-11: DB98.8] | |||
| Target Regulation | Up regulation | |||
Complex I-AGGG (NDUFB2)
Lung cancer [ICD-11: 2C25]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [12] | |||
| Responsed Disease | Non-small-cell lung carcinoma [ICD-11: 2C25.Y] | |||
| Target Regulation | Down regulation | |||
| Pathway Response | Ubiquitin mediated proteolysis | hsa04120 | ||
| Cell Process | Tumour immunology | |||
| Ubiquitination degradation | ||||
In-vitro Model |
A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 |
| BEAS-2B | Normal | Homo sapiens | CVCL_0168 | |
| NCI-H1299 | Lung large cell carcinoma | Homo sapiens | CVCL_0060 | |
| NCI-H1650 | Minimally invasive lung adenocarcinoma | Homo sapiens | CVCL_1483 | |
| NCI-H1703 | Lung squamous cell carcinoma | Homo sapiens | CVCL_1490 | |
| NCI-H1975 | Lung adenocarcinoma | Homo sapiens | CVCL_1511 | |
| NCI-H460 | Lung large cell carcinoma | Homo sapiens | CVCL_0459 | |
| HCC827 | Lung adenocarcinoma | Homo sapiens | CVCL_2063 | |
| LL/2 (LLC1) | Malignant tumors | Mus musculus | CVCL_4358 | |
| In-vivo Model | A549 cells were transfected with the pZW1-FCS-circNDUFB2 plasmid or pZW1-FCS-Vector plasmid, and selected with G418 (800 ug/ml) for 4 weeks, and then 2 × 106 A549 cells were subcutaneously injected into the right flank of each mouse. | |||
| Response Summary | Complex I-AGGG (NDUFB2) interacts with IGF2BP1/2/3 in NSCLC cells. circNDUFB2 participates in the degradation of IGF2BPs and activation of anti-tumor immunity during NSCLC progression via the modulation of both protein ubiquitination and degradation, as well as cellular immune responses. | |||
Cyclin-dependent kinase 6 (CDK6)
Laryngeal cancer [ICD-11: 2C23]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [13] | |||
| Responsed Disease | Laryngeal squamous cell carcinoma [ICD-11: 2C23.Z] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
16HBE14o- | Normal | Homo sapiens | CVCL_0112 |
| In-vivo Model | Six-week-old male BALB/c nude mice were randomly assigned to three groups (n = 6 in each group): Lv-sh-NC, Lv-sh-IGF2BP2#1, and Lv-sh-IGF2BP2#2. Mice in each group received the subcutaneous injection of 1 × 106 cells (0.1 mL) into the upper right flanks of mice. Cells were pre-transduced with sh-NC, sh-IGF2BP2#1, or sh-IGF2BP2#2 for 48 h before the injection. Starting from day 10 of the injection, the tumor volume was measured every 3 days. Mice were anesthetized and euthanized at day 25 of the injection, tumors were removed and collected, tumor weight was determined in a blind manner. | |||
Cyclin-dependent kinase inhibitor 2A (CDKN2A)
Mature T-cell lymphoma [ICD-11: 2A90]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [14] | |||
| Responsed Disease | Mature T-cell lymphoma [ICD-11: 2A90] | |||
| Target Regulation | Up regulation | |||
| Cell Process | Cell proliferation | |||
| Cell migration | ||||
| Response Summary | The decline in METTL3 levels was responsible for CTCL cell proliferation and migration,Cyclin-dependent kinase inhibitor 2A (CDKN2A) was a key regulator during this process in vitro and in vivo, and insufficient methylation modification blocked the interaction between CDKN2A and m6A reader IGF2BP2, resulting in mRNA degradation. | |||
Differentiation antagonizing non-protein coding RNA (DANCR)
Pancreatic cancer [ICD-11: 2C10]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [15] | |||
| Responsed Disease | Pancreatic cancer [ICD-11: 2C10] | |||
| Target Regulation | Up regulation | |||
| Cell Process | Cell proliferation | |||
In-vitro Model |
BxPC-3 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0186 |
| SW1990 | Pancreatic adenocarcinoma | Homo sapiens | CVCL_1723 | |
| In-vivo Model | DANCR KO or empty vector control were harvested and then mixed with matrigel (1:1) (BD Biosciences). Three different numbers of cells (1 × 104, 1 × 105, and 5 × 105 cells) were subcutaneously injected into nude mice, five animals per group. | |||
| Response Summary | IGF2BP2 functions in partnerships with Putative uncharacterized protein DANCR (DANCR) to regulate its stability. In tumor cells, IGF2BP2 is upregulated, which increases the chance of IGF2PB2 to interact with and stabilize DANCR.DANCR is a novel target for IGF2BP2 through m6A modification, and IGF2BP2 and DANCR work together to promote cancer stemness-like properties and pancreatic cancer pathogenesis. | |||
Ephrin type-A receptor 2 (EphA2)
Colorectal cancer [ICD-11: 2B91]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [4] | |||
| Responsed Disease | Colorectal cancer [ICD-11: 2B91] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | PI3K-Akt signaling pathway | hsa04151 | ||
In-vitro Model |
SW620 | Colon adenocarcinoma | Homo sapiens | CVCL_0547 |
| SW480 | Colon adenocarcinoma | Homo sapiens | CVCL_0546 | |
| NCM460 | Normal | Homo sapiens | CVCL_0460 | |
| LoVo | Colon adenocarcinoma | Homo sapiens | CVCL_0399 | |
| HT29 | Colon cancer | Mus musculus | CVCL_A8EZ | |
| HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 | |
| DLD-1 | Colon adenocarcinoma | Homo sapiens | CVCL_0248 | |
| In-vivo Model | A total of 8 × 106 wild-type (WT) or METTL3-knockdown cells were injected into the dorsal flanks of 6-week-old nude mice. Seven mice were randomly selected to calculate the volume according to the following formula: V = (width2 × length)/2. Mice were euthanized three weeks after injection and tumors removed, weighed, fixed, and embedded for immunohistochemical analysis. | |||
| Response Summary | Ephrin type-A receptor 2 (EphA2) and VEGFA targeted by METTL3 via different IGF2BP2-dependent mechanisms were found to promote vasculogenic mimicry (VM) formation via PI3K/AKT/mTOR and ERK1/2 signaling in CRC. | |||
Fascin (FSCN1)
Liver cancer [ICD-11: 2C12]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [6] | |||
| Responsed Disease | Hepatocellular carcinoma [ICD-11: 2C12.02] | |||
| Cell Process | RNA decay | |||
In-vitro Model |
Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| HeLa | Endocervical adenocarcinoma | Homo sapiens | CVCL_0030 | |
| Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 | |
| Response Summary | In contrast to the mRNA-decay-promoting function of YTH domain-containing family protein 2, IGF2BPs promote the stability and storage of their target mRNAs (for example, MYC) in an m6A-dependent manner under normal and stress conditions and therefore affect gene expression output. Four representative high confidence targets, including MYC, Fascin (FSCN1), TK1, and MARCKSL1, exhibit strong binding with IGF2BPs around their m6A motifs in control cells. Knocking down of each individual IGF2BPs in Hela (cervical cancer) and HepG2 (liver cancer) cells significantly repressed MYC expression. | |||
Cervical cancer [ICD-11: 2C77]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [6] | |||
| Responsed Disease | Cervical cancer [ICD-11: 2C77] | |||
| Cell Process | RNA decay | |||
In-vitro Model |
Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| HeLa | Endocervical adenocarcinoma | Homo sapiens | CVCL_0030 | |
| Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 | |
| Response Summary | In contrast to the mRNA-decay-promoting function of YTH domain-containing family protein 2, IGF2BPs promote the stability and storage of their target mRNAs (for example, MYC) in an m6A-dependent manner under normal and stress conditions and therefore affect gene expression output. Four representative high confidence targets, including MYC, Fascin (FSCN1), TK1, and MARCKSL1, exhibit strong binding with IGF2BPs around their m6A motifs in control cells. Knocking down of each individual IGF2BPs in Hela (cervical cancer) and HepG2 (liver cancer) cells significantly repressed MYC expression. | |||
Flap endonuclease 1 (FEN1)
Liver cancer [ICD-11: 2C12]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [16] | |||
| Responsed Disease | Hepatocellular carcinoma [ICD-11: 2C12.02] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
PLC/PRF/5 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_0485 |
| MHCC97-L | Adult hepatocellular carcinoma | Homo sapiens | CVCL_4973 | |
| Huh-7 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_0336 | |
| Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 | |
| In-vivo Model | A total of 40 BALB/c nude mice were chosen and assigned to two groups: shCtrl group (injected with HepG2 cells) and shIGF2BP2 group (injected with HepG2 cells with IGF2BP2 knockdown). 200 ul of the above cell suspension containing 2 × 105 cells was injected into the left or right back of each mice. | |||
| Response Summary | IGF2BP2 overexpression promoted HCC proliferation in vitro and in vivo, IGF2BP2 directly recognized and bound to the m6A site on FEN1 mRNA and enhanced Flap endonuclease 1 (FEN1) mRNA stability. | |||
Forkhead box protein M1 (FOXM1)
Colorectal cancer [ICD-11: 2B91]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [17] | |||
| Responsed Disease | Colorectal cancer [ICD-11: 2B91] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
HCoEpiC (Healthy colon epithelial HCoEpiC cells) | |||
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 | |
| SW480 | Colon adenocarcinoma | Homo sapiens | CVCL_0546 | |
| SW620 | Colon adenocarcinoma | Homo sapiens | CVCL_0547 | |
| DLD-1 | Colon adenocarcinoma | Homo sapiens | CVCL_0248 | |
| LoVo | Colon adenocarcinoma | Homo sapiens | CVCL_0399 | |
| HT-29 | Colon adenocarcinoma | Homo sapiens | CVCL_0320 | |
| In-vivo Model | Harvested cells were resuspended in PBS and each side of mouse was injected about 1 × 106 cells. Tumor volume was estimated every four days and calculated as 0.5 × length × width2. | |||
G1/S-specific cyclin-D1 (CCND1)
Colorectal cancer [ICD-11: 2B91]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [18] | |||
| Responsed Disease | Colorectal cancer [ICD-11: 2B91] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 |
| HT-29 | Colon adenocarcinoma | Homo sapiens | CVCL_0320 | |
| SW480 | Colon adenocarcinoma | Homo sapiens | CVCL_0546 | |
| SW620 | Colon adenocarcinoma | Homo sapiens | CVCL_0547 | |
| DLD-1 | Colon adenocarcinoma | Homo sapiens | CVCL_0248 | |
| LoVo | Colon adenocarcinoma | Homo sapiens | CVCL_0399 | |
| HUVEC-C | Normal | Homo sapiens | CVCL_2959 | |
| HCoEpiC (Healthy colon epithelial HCoEpiC cells) | ||||
| In-vivo Model | Ten BALB/C nude mice (4 weeks old, female) were injected in 1 × 106 HCT116 cells in 100 uL PBS at each side. The tumor size was detected every four days after the injection of cells and calculated according to the formula. | |||
Glucose transporter type 1 (GLUT1)
Colorectal cancer [ICD-11: 2B91]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [19] | |||
| Responsed Disease | Colorectal cancer [ICD-11: 2B91] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | Glycolysis / Gluconeogenesis | hsa00010 | ||
| Cell Process | Glucose metabolism | |||
| Response Summary | METTL3 stabilizes HK2 and Glucose transporter type 1 (SLC2A1) (GLUT1) expression in colorectal cancer through an m6A-IGF2BP2/3- dependent mechanism. | |||
Liver cancer [ICD-11: 2C12]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [21] | |||
| Responsed Disease | Liver cancer [ICD-11: 2C12] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
Hep 3B2.1-7 | Childhood hepatocellular carcinoma | Homo sapiens | CVCL_0326 |
| BEL-7402 | Endocervical adenocarcinoma | Homo sapiens | CVCL_5492 | |
| Huh-7 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_0336 | |
| SMMC-7721 | Endocervical adenocarcinoma | Homo sapiens | CVCL_0534 | |
| In-vivo Model | BEL-7404 cells or BEL-7404 miR4458HG-KO cells were infused in the right flank of randomly selected 4-week-old male BALB/c mice. | |||
Hepatocyte nuclear factor 1-alpha (HNF1A/TCF1)
Thyroid Cancer [ICD-11: 2D10]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [22] | |||
| Responsed Disease | Thyroid Cancer [ICD-11: 2D10] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | Wnt signaling pathway | hsa04310 | ||
| Cell Process | Cell migratory | |||
In-vitro Model |
B-CPAP | Thyroid gland carcinoma | Homo sapiens | CVCL_0153 |
| Nthy-ori 3-1 | Normal | Homo sapiens | CVCL_2659 | |
| TPC-1 | Thyroid gland papillary carcinoma | Homo sapiens | CVCL_6298 | |
| Response Summary | Silence of METTL3 inhibited migratory ability and Wnt activity in TPC-1 cells. METTL3 positively regulated the enrichment abundance of Hepatocyte nuclear factor 1-alpha (HNF1A/TCF1) in anti-IGF2BP2. TCF1 was responsible for METTL3-regulated thyroid carcinoma progression via the m6A methylation. | |||
Hexokinase-2 (HK2)
Colorectal cancer [ICD-11: 2B91]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [19] | |||
| Responsed Disease | Colorectal cancer [ICD-11: 2B91] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | Glycolysis / Gluconeogenesis | hsa00010 | ||
| Cell Process | Glucose metabolism | |||
| Response Summary | METTL3 stabilizes Hexokinase-2 (HK2) and SLC2A1 (GLUT1) expression in colorectal cancer through an m6A-IGF2BP2/3- dependent mechanism. | |||
Liver cancer [ICD-11: 2C12]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [21] | |||
| Responsed Disease | Liver cancer [ICD-11: 2C12] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
Hep 3B2.1-7 | Childhood hepatocellular carcinoma | Homo sapiens | CVCL_0326 |
| BEL-7402 | Endocervical adenocarcinoma | Homo sapiens | CVCL_5492 | |
| Huh-7 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_0336 | |
| SMMC-7721 | Endocervical adenocarcinoma | Homo sapiens | CVCL_0534 | |
| In-vivo Model | BEL-7404 cells or BEL-7404 miR4458HG-KO cells were infused in the right flank of randomly selected 4-week-old male BALB/c mice. | |||
High mobility group protein HMGI-C (HMGA2)
Colorectal cancer [ICD-11: 2B91]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [5] | |||
| Responsed Disease | Colorectal cancer [ICD-11: 2B91] | |||
| Target Regulation | Up regulation | |||
Hypoxia-inducible factor 1-alpha (HIF-1-Alpha/HIF1A)
Injury of other or unspecified intrathoracic organs [ICD-11: NB32]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [25] | |||
| Responsed Disease | Injury of other or unspecified intrathoracic organs [ICD-11: NB32.3] | |||
| Target Regulation | Up regulation | |||
Insulin-like growth factor 1 receptor (IGF1R)
Gastric cancer [ICD-11: 2B72]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [26] | |||
| Responsed Disease | Gastric cancer [ICD-11: 2B72] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
SGC-7901 | Gastric carcinoma | Homo sapiens | CVCL_0520 |
| MKN45 | Gastric adenocarcinoma | Homo sapiens | CVCL_0434 | |
| MKN1 | Gastric adenosquamous carcinoma | Homo sapiens | CVCL_1415 | |
| MGC-803 | Gastric mucinous adenocarcinoma | Homo sapiens | CVCL_5334 | |
| GES-1 | Normal | Homo sapiens | CVCL_EQ22 | |
| In-vivo Model | A total of 30 BALB/c nude mice were chosen and assigned to three groups: (1) control (injected with 0.2 mL PBS), (2) si-NC (injected with si-NC transfected SGC7901 cells) and (3) si-IGF2BP2 (injected with si-IGF2BP2 transfected SGC7901 cells (n = 5 per group). 2 × 106 SGC7901 cells were injected into the left right back of each mouse through subcutaneous injection. Tumor sizes were recorded once per week. After 28 days, the mice were euthanized, and tumor tissues were weighted. | |||
| Response Summary | IGF2BP2, as a m6A reader, was proved to increase the expression of Insulin-like growth factor 1 receptor (IGF1R) by identifying m6A methylation modification sites in IGF1R mRNA, thus activating RhoA-ROCK pathway. The oncogenic role of IGF2BP2 in gastric cancer carcinogenesis and confirmed its activation is partly due to the activation of IGF1R-RhoA-ROCK signaling pathway. | |||
Prostate cancer [ICD-11: 2C82]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [27] | |||
| Responsed Disease | Prostate cancer [ICD-11: 2C82] | |||
| Target Regulation | Up regulation | |||
| Cell Process | RNA stability | |||
In-vitro Model |
PC-3 | Prostate carcinoma | Homo sapiens | CVCL_0035 |
| LNCaP C4-2B | Prostate carcinoma | Homo sapiens | CVCL_4784 | |
| In-vivo Model | At 1 week post-injection with PC-3 cells, mice were randomly assigned to three groups (n = 8 per group): the ASO-NC group (injection with ASO negative control targeting unknown sequence, 5 nmol in 100 uL PBS for each mouse), the ASO-L group (injection with low-dose ASO targeting PCAT6, 5 nmol in 100 uL PBS for each mouse), and the ASO-H group (injection with high-dose ASO targeting PCAT6, 10 nmol in 100 uL PBS for each mouse). | |||
| Response Summary | METTL3-mediated m6A modification contributed to PCAT6 upregulation in an IGF2BP2-dependent manner. Furthermore, PCAT6 upregulated Insulin-like growth factor 1 receptor (IGF1R) expression by enhancing IGF1R mRNA stability through the PCAT6/IGF2BP2/IGF1R RNA-protein three-dimensional complex. The m6 A-induced PCAT6/IGF2BP2/IGF1R axis promotes PCa bone metastasis and tumor growth, suggesting that PCAT6 serves as a promising prognostic marker and therapeutic target against bone-metastatic PCa. | |||
LIM and SH3 domain protein 1 (LASP1)
Nasopharyngeal carcinoma [ICD-11: 2B6B]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [28] | |||
| Responsed Disease | Nasopharyngeal carcinoma [ICD-11: 2B6B] | |||
| Target Regulation | Up regulation | |||
| Response Summary | WTAP-mediated m6A modification of LIM and SH3 domain protein 1 (LASP1) enhanced its stability relying on the m6A reader IGF2BP2-dependent pathway. Furthermore, DIAPH1-AS1 acted as a molecular adaptor that promoted MTDH-LASP1 complex formation and upregulated LASP1 expression, ultimately facilitating NPC growth and metastasis. | |||
Metastasis-associated protein MTA1 (MTA1)
Colorectal cancer [ICD-11: 2B91]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [29] | |||
| Responsed Disease | Colorectal cancer [ICD-11: 2B91] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | mRNA surveillance pathway | hsa03015 | ||
| RNA degradation | hsa03018 | |||
| Cell Process | RNA stability | |||
In-vitro Model |
SW620 | Colon adenocarcinoma | Homo sapiens | CVCL_0547 |
| SW480 | Colon adenocarcinoma | Homo sapiens | CVCL_0546 | |
| RKO | Colon carcinoma | Homo sapiens | CVCL_0504 | |
| LS174T | Colon adenocarcinoma | Homo sapiens | CVCL_1384 | |
| LoVo | Colon adenocarcinoma | Homo sapiens | CVCL_0399 | |
| HT29 | Colon cancer | Mus musculus | CVCL_A8EZ | |
| HCT 8 | Colon adenocarcinoma | Homo sapiens | CVCL_2478 | |
| HCT 15 | Colon adenocarcinoma | Homo sapiens | CVCL_0292 | |
| HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 | |
| DLD-1 | Colon adenocarcinoma | Homo sapiens | CVCL_0248 | |
| CW-2 | Colon adenocarcinoma | Homo sapiens | CVCL_1151 | |
| In-vivo Model | FTO-overexpressing and control cells (2 × 106 suspended in 100 ul PBS) were subcutaneously injected into each mouse. | |||
| Response Summary | FTO inhibited CRC metastasis both in vitro and in vivo. FTO exerted a tumor suppressive role by inhibiting Metastasis-associated protein MTA1 (MTA1) expression in an m6A-dependent manner. Methylated MTA1 transcripts were recognized by an m6A "reader", insulin-like growth factor 2 mRNA binding protein 2 (IGF2BP2), which then stabilized its mRNA. | |||
Mitogen-activated protein kinase 1 (MAPK/ERK2/MAPK1)
Gangrene or necrosis of lung [ICD-11: CA43]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [1] | |||
| Responsed Disease | Gangrene or necrosis of lung [ICD-11: CA43] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | MAPK signaling pathway | hsa04010 | ||
| PI3K-Akt signaling pathway | hsa04151 | |||
| Apoptosis | hsa04210 | |||
| Cell Process | Biological regulation | |||
| Cell apoptosis | ||||
In-vitro Model |
BEAS-2B | Normal | Homo sapiens | CVCL_0168 |
| In-vivo Model | After being anesthetized with urethane (i.p.), SD rats were endotracheally intubated and ventilated using an animal ventilator under the conditions: respiratory rate of 70 breaths/min, tidal volume of 20 ml/kg, and inspiratory/expiratory ratio of 1:1. | |||
| Response Summary | N6-methyladenosine (m6A) methylation modification is implicated in the pathogenesis of lung ischemia-reperfusion injury. YTHDF3 or IGF2BP2 knockdown inhibited hypoxia/reoxygenation-activated p38, Mitogen-activated protein kinase 1 (MAPK/ERK2/MAPK1), AKT, and NF-Kappa-B pathways in BEAS-2B cells, and inhibited p-p65, IL-1-beta and TNF-alpha secretion. | |||
Mitogen-activated protein kinase 14 (p38/MAPK14)
Gangrene or necrosis of lung [ICD-11: CA43]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [1] | |||
| Responsed Disease | Gangrene or necrosis of lung [ICD-11: CA43] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | MAPK signaling pathway | hsa04010 | ||
| PI3K-Akt signaling pathway | hsa04151 | |||
| Cell Process | Biological regulation | |||
| Cell apoptosis | ||||
In-vitro Model |
BEAS-2B | Normal | Homo sapiens | CVCL_0168 |
| In-vivo Model | After being anesthetized with urethane (i.p.), SD rats were endotracheally intubated and ventilated using an animal ventilator under the conditions: respiratory rate of 70 breaths/min, tidal volume of 20 ml/kg, and inspiratory/expiratory ratio of 1:1. | |||
| Response Summary | N6-methyladenosine (m6A) methylation modification is implicated in the pathogenesis of lung ischemia-reperfusion injury. YTHDF3 or IGF2BP2 knockdown inhibited hypoxia/reoxygenation-activated Mitogen-activated protein kinase 14 (p38/MAPK14), ERK1/2, AKT, and NF-Kappa-B pathways in BEAS-2B cells, and inhibited p-p65, IL-1-beta and TNF-alpha secretion. | |||
Mitogen-activated protein kinase 3 (ERK1/MAPK3)
Gangrene or necrosis of lung [ICD-11: CA43]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [1] | |||
| Responsed Disease | Gangrene or necrosis of lung [ICD-11: CA43] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | MAPK signaling pathway | hsa04010 | ||
| PI3K-Akt signaling pathway | hsa04151 | |||
| Apoptosis | hsa04210 | |||
| Cell Process | Biological regulation | |||
| Cell apoptosis | ||||
In-vitro Model |
BEAS-2B | Normal | Homo sapiens | CVCL_0168 |
| In-vivo Model | After being anesthetized with urethane (i.p.), SD rats were endotracheally intubated and ventilated using an animal ventilator under the conditions: respiratory rate of 70 breaths/min, tidal volume of 20 ml/kg, and inspiratory/expiratory ratio of 1:1. | |||
| Response Summary | N6-methyladenosine (m6A) methylation modification is implicated in the pathogenesis of lung ischemia-reperfusion injury. YTHDF3 or IGF2BP2 knockdown inhibited hypoxia/reoxygenation-activated p38, Mitogen-activated protein kinase 3 (ERK1/MAPK3), AKT, and NF-Kappa-B pathways in BEAS-2B cells, and inhibited p-p65, IL-1-beta and TNF-alpha secretion. | |||
Myc proto-oncogene protein (MYC)
Malignant haematopoietic neoplasm [ICD-11: 2B33]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [8] | |||
| Responsed Disease | Myeloid leukaemia [ICD-11: 2B33.1] | |||
| Target Regulation | Up regulation | |||
| Cell Process | Cell apoptosis | |||
In-vitro Model |
Leukemia stem cell line (Leukemia stem cell line) | |||
| Kasumi-1 | Myeloid leukemia with maturation | Homo sapiens | CVCL_0589 | |
| MOLM-13 | Adult acute myeloid leukemia | Homo sapiens | CVCL_2119 | |
| THP-1 | Childhood acute monocytic leukemia | Homo sapiens | CVCL_0006 | |
| MV4-11 | Childhood acute monocytic leukemia | Homo sapiens | CVCL_0064 | |
| BV-173 | Chronic myelogenous leukemia | Homo sapiens | CVCL_0181 | |
| NOMO-1 | Adult acute monocytic leukemia | Homo sapiens | CVCL_1609 | |
| K-562 | Chronic myelogenous leukemia | Homo sapiens | CVCL_0004 | |
| KG-1a | Adult acute myeloid leukemia | Homo sapiens | CVCL_1824 | |
| Response Summary | YBX1 selectively functions in regulating survival of myeloid leukemia cells. YBX1 interacts with insulin-like growth factor 2 messenger RNA (mRNA)-binding proteins (IGF2BPs) and stabilizes m6A-tagged RNA. YBX1 deficiency dysregulates the expression of apoptosis-related genes and promotes mRNA decay of Myc proto-oncogene protein (MYC) and BCL2 in an m6A-dependent manner, which contributes to the defective survival that results from deletion of YBX1. | |||
Esophageal cancer [ICD-11: 2B70]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [30] | |||
| Responsed Disease | Esophageal cancer [ICD-11: 2B70] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
TE-1 | Esophageal squamous cell carcinoma | Homo sapiens | CVCL_1759 |
| Eca-109 | Esophageal squamous cell carcinoma | Homo sapiens | CVCL_6898 | |
| KYSE-150 | Esophageal squamous cell carcinoma | Homo sapiens | CVCL_1348 | |
| TE-10 | Esophageal squamous cell carcinoma | Homo sapiens | CVCL_1760 | |
| In-vivo Model | The mice were acclimatized and fed for one week. Then, they were randomly divided into two groups: sh-NC group and sh-SHMT2 group. Cells transfected with sh-NC or sh-SHMT2 were subsequently cultured routinely. Next, cells with logarithmic growth phase (1 ×106) were taken and injected to the right axilla of nude mice. After subcutaneous inoculation, the mice were observed for their mental status, activity, and tumor formation. The tumor volume was monitored every 4 days, and the mice were euthanized after 28 days. Tumor tissues were separated and weighed from nude mice. A portion of the dissected tumor tissue was fixed overnight in 4% paraformaldehyde, embedded in paraffin blocks and sectioned. | |||
Colorectal cancer [ICD-11: 2B91]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [31] | |||
| Responsed Disease | Colorectal cancer [ICD-11: 2B91] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | Ubiquitin mediated proteolysis | hsa04120 | ||
| Glycolysis / Gluconeogenesis | hsa00010 | |||
| Cell Process | Autophagy-lysosome pathway | |||
| Ubiquitination | ||||
| Glycolysis | ||||
In-vitro Model |
DLD-1 | Colon adenocarcinoma | Homo sapiens | CVCL_0248 |
| HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 | |
| In-vivo Model | For the orthotopic models, 2 × 106 cells with negative control (NC, sh-NC), sh-1 or sh-2 in 0.5 mL of PBS were subcutaneously injected into the dorsal flank of 2 mice respectively. Then 15 mice were separated into 3 groups (sh-NC, sh-1 and sh-2), of which the tumor pieces were tied to the base of the ceca. The growth of the tumors was monitored every 2 weeks after intraperitoneal injection of D-luciferin with a Xenogen IVIS 100 Bioluminescent Imaging System. | |||
| Response Summary | LINRIS blocked K139 ubiquitination of IGF2BP2, maintaining its stability. This process prevented the degradation of IGF2BP2 through the autophagy-lysosome pathway (ALP). The LINRIS-IGF2BP2-Myc proto-oncogene protein (MYC) axis promotes the progression of Colorectal cancer and is a promising therapeutic target. MYC-mediated glycolysis was influenced by the interaction between LINRIS and IGF2BP2. | |||
Pancreatic cancer [ICD-11: 2C10]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [2] | |||
| Responsed Disease | Pancreatic ductal adenocarcinoma [ICD-11: 2C10.0] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | mRNA surveillance pathway | hsa03015 | ||
| RNA degradation | hsa03018 | |||
| Cell Process | RNA stability | |||
In-vitro Model |
THP-1 | Childhood acute monocytic leukemia | Homo sapiens | CVCL_0006 |
| PATU-8988 (Human pancreatic adenocarcinoma cell) | ||||
| PANC-1 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0480 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| 37 (Pancreatic cancer cell) | ||||
| In-vivo Model | BALB/c nude mice which were co-injected with THP-1 cells and PATU-8988 cells subcutaneously. | |||
| Response Summary | LncRNA-PACERR which bound to IGF2BP2 acts as an m6A-dependent manner to enhance the stability of KLF12 and Myc proto-oncogene protein (MYC) in cytoplasm. This study found that LncRNA-PACERR functions as key regulator of TAMs in PDAC microenvironment and revealed the novel mechanisms in cytoplasm and in nucleus. | |||
Liver cancer [ICD-11: 2C12]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [6] | |||
| Responsed Disease | Hepatocellular carcinoma [ICD-11: 2C12.02] | |||
| Cell Process | RNA decay | |||
In-vitro Model |
Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| HeLa | Endocervical adenocarcinoma | Homo sapiens | CVCL_0030 | |
| Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 | |
| Response Summary | In contrast to the mRNA-decay-promoting function of YTH domain-containing family protein 2, IGF2BPs promote the stability and storage of their target mRNAs (for example, MYC) in an m6A-dependent manner under normal and stress conditions and therefore affect gene expression output. Four representative high confidence targets, including Myc proto-oncogene protein (MYC), FSCN1, TK1, and MARCKSL1, exhibit strong binding with IGF2BPs around their m6A motifs in control cells. Knocking down of each individual IGF2BPs in Hela (cervical cancer) and HepG2 (liver cancer) cells significantly repressed MYC expression. | |||
Cervical cancer [ICD-11: 2C77]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [6] | |||
| Responsed Disease | Cervical cancer [ICD-11: 2C77] | |||
| Cell Process | RNA decay | |||
In-vitro Model |
Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| HeLa | Endocervical adenocarcinoma | Homo sapiens | CVCL_0030 | |
| Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 | |
| Response Summary | In contrast to the mRNA-decay-promoting function of YTH domain-containing family protein 2, IGF2BPs promote the stability and storage of their target mRNAs (for example, MYC) in an m6A-dependent manner under normal and stress conditions and therefore affect gene expression output. Four representative high confidence targets, including Myc proto-oncogene protein (MYC), FSCN1, TK1, and MARCKSL1, exhibit strong binding with IGF2BPs around their m6A motifs in control cells. Knocking down of each individual IGF2BPs in Hela (cervical cancer) and HepG2 (liver cancer) cells significantly repressed MYC expression. | |||
Obg-like ATPase 1 (OLA1)
Colorectal cancer [ICD-11: 2B91]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [34] | |||
| Responsed Disease | Colorectal cancer [ICD-11: 2B91] | |||
| Target Regulation | Up regulation | |||
| Cell Process | Warburg effect | |||
| Mitochondrial energy metabolism | ||||
In-vitro Model |
Caco-2 | Colon adenocarcinoma | Homo sapiens | CVCL_0025 |
| HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 | |
| HIEC (Normal intestinal epithelial cells) | ||||
| HT29 | Colon cancer | Mus musculus | CVCL_A8EZ | |
| LoVo | Colon adenocarcinoma | Homo sapiens | CVCL_0399 | |
| RKO | Colon carcinoma | Homo sapiens | CVCL_0504 | |
| SW480 | Colon adenocarcinoma | Homo sapiens | CVCL_0546 | |
| SW480 | Colon adenocarcinoma | Homo sapiens | CVCL_0546 | |
| SW620 | Colon adenocarcinoma | Homo sapiens | CVCL_0547 | |
| Response Summary | The critical modulation network underlying m6A readers stabilizes lncRNAs, and they jointly promote mitochondrial energy metabolism in the pathogenesis of colorectal cancer. N6-methyladenosine reader stabilizes the ZFAS1/OLA1 axis. Thus, direct interaction between the KH3-4 domain of IMP2 and ZFAS1 where IMP2 serves as a reader for m6A-modified ZFAS1 and promotes the RNA stability of ZFAS1 is critical for CRC development. | |||
POU domain, class 5, transcription factor 1 (POU5F1)
Esophageal cancer [ICD-11: 2B70]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [35] | |||
| Responsed Disease | Esophageal Squamous Cell Carcinoma [ICD-11: 2B70.1] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
YES-2 | Esophageal squamous cell carcinoma | Homo sapiens | CVCL_E322 |
| KYSE-30 | Esophageal squamous cell carcinoma | Homo sapiens | CVCL_1351 | |
| KYSE-70 | Esophageal squamous cell carcinoma | Homo sapiens | CVCL_1356 | |
| KYSE-140 | Esophageal squamous cell carcinoma | Homo sapiens | CVCL_1347 | |
| KYSE-150 | Esophageal squamous cell carcinoma | Homo sapiens | CVCL_1348 | |
| KYSE-180 | Esophageal squamous cell carcinoma | Homo sapiens | CVCL_1349 | |
| KYSE-410 | Esophageal squamous cell carcinoma | Homo sapiens | CVCL_1352 | |
| KYSE-450 | Esophageal squamous cell carcinoma | Homo sapiens | CVCL_1353 | |
| KYSE-510 | Esophageal squamous cell carcinoma | Homo sapiens | CVCL_1354 | |
| COLO 680N | Esophageal squamous cell carcinoma | Homo sapiens | CVCL_1131 | |
|
Ne2/1b4.14
|
N.A. | Mus musculus | CVCL_0E04 | |
Programmed cell death 1 ligand 1 (CD274/PD-L1)
Pancreatic cancer [ICD-11: 2C10]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [36] | |||
| Responsed Disease | Pancreatic ductal adenocarcinoma [ICD-11: 2C10.0] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
PANC-1 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0480 |
| Capan-2 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0026 | |
| BxPC-3 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0186 | |
| CFPAC-1 | Cystic fibrosis | Homo sapiens | CVCL_1119 | |
| In-vivo Model | Male BALB/c nude mice (5-6 weeks) were obtained from Slac Laboratory Animal Center (Shanghai, China) and maintained under pathogen-free conditions. PANC-1 cells (2 × 106 cells suspended in 100 μl PBS) transfected with circMYO1C knockdown (sh-circMYO1C) or controls (sh-NC) were subcutaneously injected into the flank of nude mice. One week later, the tumor size was measured every three days. | |||
Putative pituitary tumor-transforming gene 3 protein (PTTG3P)
Colorectal cancer [ICD-11: 2B91]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [37] | |||
| Responsed Disease | Colorectal cancer [ICD-11: 2B91] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | Hippo signaling pathway | hsa04390 | ||
| Cell Process | Cell proliferation and suppression of apoptosis | |||
In-vitro Model |
FHC | Normal | Homo sapiens | CVCL_3688 |
| HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 | |
| HCT 8 | Colon adenocarcinoma | Homo sapiens | CVCL_2478 | |
| HT29 | Colon cancer | Mus musculus | CVCL_A8EZ | |
| NCM460 | Normal | Homo sapiens | CVCL_0460 | |
| SW480 | Colon adenocarcinoma | Homo sapiens | CVCL_0546 | |
| SW620 | Colon adenocarcinoma | Homo sapiens | CVCL_0547 | |
| In-vivo Model | Indicated cells (1 × 107) were subcutaneously injected into 4-week-old male nude mice. Tumor volume was measured every 5 days. | |||
| Response Summary | In colorectal cancer, n6-methyladenosine (m6A) subunit METTL3 increased PTTG3P expression by influencing its stability, while insulin-like growth factor 2 mRNA binding protein 2 (IGF2BP2) could identify Putative pituitary tumor-transforming gene 3 protein (PTTG3P) m6A methylation status and bind to it. | |||
RAC-alpha serine/threonine-protein kinase (AKT1)
Gangrene or necrosis of lung [ICD-11: CA43]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [1] | |||
| Responsed Disease | Gangrene or necrosis of lung [ICD-11: CA43] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | MAPK signaling pathway | hsa04010 | ||
| PI3K-Akt signaling pathway | hsa04151 | |||
| Apoptosis | hsa04210 | |||
| Cell Process | Biological regulation | |||
| Cell apoptosis | ||||
In-vitro Model |
BEAS-2B | Normal | Homo sapiens | CVCL_0168 |
| In-vivo Model | After being anesthetized with urethane (i.p.), SD rats were endotracheally intubated and ventilated using an animal ventilator under the conditions: respiratory rate of 70 breaths/min, tidal volume of 20 ml/kg, and inspiratory/expiratory ratio of 1:1. | |||
| Response Summary | N6-methyladenosine (m6A) methylation modification is implicated in the pathogenesis of lung ischemia-reperfusion injury. YTHDF3 or IGF2BP2 knockdown inhibited hypoxia/reoxygenation-activated p38, ERK1/2, RAC-alpha serine/threonine-protein kinase (AKT1), and NF-Kappa-B pathways in BEAS-2B cells, and inhibited p-p65, IL-1-beta and TNF-alpha secretion. | |||
RB1-inducible coiled-coil protein 1 (RB1CC1/FIP200)
Head and neck squamous carcinoma [ICD-11: 2B6E]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [38] | |||
| Responsed Disease | Oral squamous cell carcinoma [ICD-11: 2B6E.0] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
CAL-33 | Tongue squamous cell carcinoma | Homo sapiens | CVCL_1108 |
| In-vivo Model | Thirty-six specific pathogen-free male BALB/c-nude mice (age, 5-6 weeks) were randomly assigned to the groups: CAL33/shMETTL14#2, CAL33/shMETTL14#3, CAL33/shNC and HSC3/shMETTL14#2, HSC3/shMETTL14#3, HSC3/shNC (n = 6 per group). Fifty microliters of PBS buffer containing approximately 1 × 106 cells was injected into the left tongue under 2% pentobarbital sodium intraperitoneal injection anesthesia to establish a tumor xenograft. The weight of the mice was measured every 3 days after one week until they lost more than 15% of their body weight in a short period of time. | |||
Receptor tyrosine-protein kinase erbB-2 (ERBB2)
Colorectal cancer [ICD-11: 2B91]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [39] | |||
| Responsed Disease | Colorectal cancer [ICD-11: 2B91] | |||
| Responsed Drug | Temozolomide | Approved | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Hippo signaling pathway | hsa04390 | ||
| Cell Process | Cell apoptosis | |||
In-vitro Model |
HCT 8 | Colon adenocarcinoma | Homo sapiens | CVCL_2478 |
| SW480 | Colon adenocarcinoma | Homo sapiens | CVCL_0546 | |
| SW620 | Colon adenocarcinoma | Homo sapiens | CVCL_0547 | |
| In-vivo Model | IGF2BP2 activates the expression of ErbB2 by recognizing the m6A of YAP, thus affecting the cell cycle of CRC, inhibiting cell apoptosis, and promoting proliferation. | |||
| Response Summary | IGF2BP2 activates the expression of Receptor tyrosine-protein kinase erbB-2 (ERBB2) by recognizing the m6A of YAP, thus affecting the cell cycle of colorectal cancer, inhibiting cell apoptosis, and promoting proliferation. | |||
RNA cytosine C(5)-methyltransferase NSUN2 (NSUN2)
Colorectal cancer [ICD-11: 2B91]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [40] | |||
| Responsed Disease | Colorectal cancer [ICD-11: 2B91] | |||
| Target Regulation | Up regulation | |||
| Cell Process | Cells invasion | |||
In-vitro Model |
HEK293T | Normal | Homo sapiens | CVCL_0063 |
| DLD-1 | Colon adenocarcinoma | Homo sapiens | CVCL_0248 | |
| HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 | |
| LoVo | Colon adenocarcinoma | Homo sapiens | CVCL_0399 | |
| SW1116 | Colon adenocarcinoma | Homo sapiens | CVCL_0544 | |
| SW620 | Colon adenocarcinoma | Homo sapiens | CVCL_0547 | |
| In-vivo Model | 2 × 106 cells suspended in 40 uL PBS were injected into the inferior hemispleen into each 6-week-old BALB/c nude mouse. | |||
| Response Summary | N6-methyladenosine modification of RNA cytosine C(5)-methyltransferase NSUN2 (NSUN2) modulates cytoplasmic export and stabilizes HMGA2 to promote Colorectal carcinoma LM. By forming a circNSUN2/IGF2BP2/HMGA2 RNA-protein ternary complex in the cytoplasm, circNSUN2 enhances the stability of HMGA2 mRNA to promote CRC metastasis progression. | |||
Stimulator of interferon genes protein (STING1)
Colorectal cancer [ICD-11: 2B91]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [41] | |||
| Responsed Disease | Colorectal cancer [ICD-11: 2B91] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
FHC | Normal | Homo sapiens | CVCL_3688 |
| LoVo | Colon adenocarcinoma | Homo sapiens | CVCL_0399 | |
| HT-29 | Colon adenocarcinoma | Homo sapiens | CVCL_0320 | |
| SW480 | Colon adenocarcinoma | Homo sapiens | CVCL_0546 | |
| SW620 | Colon adenocarcinoma | Homo sapiens | CVCL_0547 | |
| THP-1 | Childhood acute monocytic leukemia | Homo sapiens | CVCL_0006 | |
Thymidine kinase, cytosolic (TK1)
Liver cancer [ICD-11: 2C12]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [6] | |||
| Responsed Disease | Hepatocellular carcinoma [ICD-11: 2C12.02] | |||
| Cell Process | RNA decay | |||
In-vitro Model |
Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| HeLa | Endocervical adenocarcinoma | Homo sapiens | CVCL_0030 | |
| Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 | |
| Response Summary | In contrast to the mRNA-decay-promoting function of YTH domain-containing family protein 2, IGF2BPs promote the stability and storage of their target mRNAs (for example, MYC) in an m6A-dependent manner under normal and stress conditions and therefore affect gene expression output. Four representative high confidence targets, including MYC, FSCN1, Thymidine kinase, cytosolic (TK1), and MARCKSL1, exhibit strong binding with IGF2BPs around their m6A motifs in control cells. Knocking down of each individual IGF2BPs in Hela (cervical cancer) and HepG2 (liver cancer) cells significantly repressed MYC expression. | |||
Lung cancer [ICD-11: 2C25]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [42] | |||
| Responsed Disease | Lung cancer [ICD-11: 2C25] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
NHBE (Normal bronchial epithelial cells) | |||
| NCI-H460 | Lung large cell carcinoma | Homo sapiens | CVCL_0459 | |
| NCI-H1299 | Lung large cell carcinoma | Homo sapiens | CVCL_0060 | |
| A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 | |
| In-vivo Model | Suspension of H1299 cells (5.0 × 105) was subcutaneously injected into the right flanks of the mice. | |||
| Response Summary | In lung cancer, IGF2BP2 modified m6A to increase the expression of Thymidine kinase, cytosolic (TK1), thus promoting angiogenesis. | |||
Cervical cancer [ICD-11: 2C77]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [6] | |||
| Responsed Disease | Cervical cancer [ICD-11: 2C77] | |||
| Cell Process | RNA decay | |||
In-vitro Model |
Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| HeLa | Endocervical adenocarcinoma | Homo sapiens | CVCL_0030 | |
| Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 | |
| Response Summary | In contrast to the mRNA-decay-promoting function of YTH domain-containing family protein 2, IGF2BPs promote the stability and storage of their target mRNAs (for example, MYC) in an m6A-dependent manner under normal and stress conditions and therefore affect gene expression output. Four representative high confidence targets, including MYC, FSCN1, Thymidine kinase, cytosolic (TK1), and MARCKSL1, exhibit strong binding with IGF2BPs around their m6A motifs in control cells. Knocking down of each individual IGF2BPs in Hela (cervical cancer) and HepG2 (liver cancer) cells significantly repressed MYC expression. | |||
Transcription factor E2F3 (E2F3)
Liver cancer [ICD-11: 2C12]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [43] | |||
| Responsed Disease | Liver cancer [ICD-11: 2C12] | |||
| Target Regulation | Up regulation | |||
Transcription factor p65 (RELA)
Gangrene or necrosis of lung [ICD-11: CA43]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [1] | |||
| Responsed Disease | Gangrene or necrosis of lung [ICD-11: CA43] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | MAPK signaling pathway | hsa04010 | ||
| PI3K-Akt signaling pathway | hsa04151 | |||
| Cell Process | Biological regulation | |||
| Cell apoptosis | ||||
In-vitro Model |
BEAS-2B | Normal | Homo sapiens | CVCL_0168 |
| In-vivo Model | After being anesthetized with urethane (i.p.), SD rats were endotracheally intubated and ventilated using an animal ventilator under the conditions: respiratory rate of 70 breaths/min, tidal volume of 20 ml/kg, and inspiratory/expiratory ratio of 1:1. | |||
| Response Summary | N6-methyladenosine (m6A) methylation modification is implicated in the pathogenesis of lung ischemia-reperfusion injury. YTHDF3 or IGF2BP2 knockdown inhibited hypoxia/reoxygenation-activated p38, ERK1/2, AKT, and NF-Kappa-B pathways in BEAS-2B cells, and inhibited Transcription factor p65 (RELA), IL-1-beta and TNF-alpha secretion. | |||
Transcription factor SOX-2 (SOX2)
Brain cancer [ICD-11: 2A00]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [44] | |||
| Responsed Disease | Glioma [ICD-11: 2A00.0] | |||
| Target Regulation | Up regulation | |||
Colorectal cancer [ICD-11: 2B91]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [45] | |||
| Responsed Disease | Colorectal cancer [ICD-11: 2B91] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | Signaling pathways regulating pluripotency of stem cells | hsa04550 | ||
| Cell Process | Cell self-renewal | |||
| Stem cell frequency | ||||
| Cell migration | ||||
In-vitro Model |
CCD-112CoN | Normal | Homo sapiens | CVCL_6382 |
| DLD-1 | Colon adenocarcinoma | Homo sapiens | CVCL_0248 | |
| HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 | |
| HCT 15 | Colon adenocarcinoma | Homo sapiens | CVCL_0292 | |
| HCT 8 | Colon adenocarcinoma | Homo sapiens | CVCL_2478 | |
| LS174T | Colon adenocarcinoma | Homo sapiens | CVCL_1384 | |
| RKO | Colon carcinoma | Homo sapiens | CVCL_0504 | |
| SW480 | Colon adenocarcinoma | Homo sapiens | CVCL_0546 | |
| SW620 | Colon adenocarcinoma | Homo sapiens | CVCL_0547 | |
| Response Summary | METTL3, acting as an oncogene, maintained Transcription factor SOX-2 (SOX2) expression through an m6A-IGF2BP2-dependent mechanism in CRC cells, and indicated a potential biomarker panel for prognostic prediction in Colorectal carcinoma. | |||
Transcriptional coactivator YAP1 (YAP1)
Colorectal cancer [ICD-11: 2B91]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [39] | |||
| Responsed Disease | Colorectal cancer [ICD-11: 2B91] | |||
| Responsed Drug | Temozolomide | Approved | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Hippo signaling pathway | hsa04390 | ||
| Cell Process | Cell apoptosis | |||
In-vitro Model |
HCT 8 | Colon adenocarcinoma | Homo sapiens | CVCL_2478 |
| SW480 | Colon adenocarcinoma | Homo sapiens | CVCL_0546 | |
| SW620 | Colon adenocarcinoma | Homo sapiens | CVCL_0547 | |
| In-vivo Model | IGF2BP2 activates the expression of ErbB2 by recognizing the m6A of YAP, thus affecting the cell cycle of CRC, inhibiting cell apoptosis, and promoting proliferation. | |||
| Response Summary | IGF2BP2 activates the expression of ErbB2 by recognizing the m6A of Transcriptional coactivator YAP1 (YAP1), thus affecting the cell cycle of colorectal cancer, inhibiting cell apoptosis, and promoting proliferation. | |||
Lung cancer [ICD-11: 2C25]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [46] | |||
| Responsed Disease | Non-small cell lung cancer [ICD-11: 2C25.Y] | |||
| Responsed Drug | Trans-3,5,4'-trimethoxystilbene | Investigative | ||
| Target Regulation | Up regulation | |||
In-vitro Model |
16HBE14o- | Normal | Homo sapiens | CVCL_0112 |
| PC-9 | Lung adenocarcinoma | Homo sapiens | CVCL_B260 | |
| NCI-H460 | Lung large cell carcinoma | Homo sapiens | CVCL_0459 | |
| NCI-H1299 | Lung large cell carcinoma | Homo sapiens | CVCL_0060 | |
| NCI-H1975 | Lung adenocarcinoma | Homo sapiens | CVCL_1511 | |
| A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 | |
Triple-negative breast cancer [ICD-11: 2C6Z]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [47] | |||
| Responsed Disease | Triple-negative breast cancer [ICD-11: 2C6Z] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
U2OS | Osteosarcoma | Homo sapiens | CVCL_0042 |
|
WI-38 VA13 subline 2RA
|
N.A. | Homo sapiens | CVCL_2759 | |
| CAL-27 | Tongue squamous cell carcinoma | Homo sapiens | CVCL_1107 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
Injuries of spine or trunk [ICD-11: ND51]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [48] | |||
| Responsed Disease | Spinal cord injury [ICD-11: ND51.2] | |||
| Target Regulation | Up regulation | |||
Tumor necrosis factor (TNF/TNF-alpha)
Gangrene or necrosis of lung [ICD-11: CA43]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [1] | |||
| Responsed Disease | Gangrene or necrosis of lung [ICD-11: CA43] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | MAPK signaling pathway | hsa04010 | ||
| PI3K-Akt signaling pathway | hsa04151 | |||
| Apoptosis | hsa04210 | |||
| Cell Process | Biological regulation | |||
| Cell apoptosis | ||||
In-vitro Model |
BEAS-2B | Normal | Homo sapiens | CVCL_0168 |
| In-vivo Model | After being anesthetized with urethane (i.p.), SD rats were endotracheally intubated and ventilated using an animal ventilator under the conditions: respiratory rate of 70 breaths/min, tidal volume of 20 ml/kg, and inspiratory/expiratory ratio of 1:1. | |||
| Response Summary | N6-methyladenosine (m6A) methylation modification is implicated in the pathogenesis of lung ischemia-reperfusion injury. YTHDF3 or IGF2BP2 knockdown inhibited hypoxia/reoxygenation-activated p38, ERK1/2, AKT, and NF-Kappa-B pathways in BEAS-2B cells, and inhibited p-p65, IL-1-beta and Tumor necrosis factor (TNF/TNF-alpha) secretion. | |||
Vang-like protein 1 (VANGL1)
Lung cancer [ICD-11: 2C25]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [49] | |||
| Responsed Disease | Lung adenocarcinoma [ICD-11: 2C25.0] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | Nucleotide excision repair | hsa03420 | ||
| Cell Process | DNA repair | |||
In-vitro Model |
A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 |
| NCI-H1299 | Lung large cell carcinoma | Homo sapiens | CVCL_0060 | |
| In-vivo Model | Two hundred milliliters of A549 cells (1 × 106) were injected into the left flank of the back of each mouse. | |||
| Response Summary | Up-regulation of Vang-like protein 1 (VANGL1) by IGF2BPs and miR-29b-3p attenuates the detrimental effect of irradiation on lung adenocarcinoma. Increased m6A level of VANGL1 and reduced miR-29b-3p took the responsibility of VANGL1 overexpression upon irradiation. | |||
Zinc finger protein SNAI1 (SNAI1)
Nasopharyngeal carcinoma [ICD-11: 2B6B]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [50] | |||
| Responsed Disease | Nasopharyngeal carcinoma [ICD-11: 2B6B] | |||
| Target Regulation | Up regulation | |||
| Cell Process | Epithelial-mesenchymal transition | |||
In-vitro Model |
SUNE1 | Nasopharyngeal carcinoma | Homo sapiens | CVCL_6946 |
| Response Summary | Overexpression of Zinc finger protein SNAI1 (SNAI1) partially reversed the regulatory effects of METTL3 on EMT-related gene expressions and metastatic abilities in nasopharyngeal carcinoma. Knockdown of METTL3 decreased the enrichment abundance of Snail in anti-IGF2BP2. | |||
Zinc finger protein SNAI2 (Slug)
Head and neck squamous carcinoma [ICD-11: 2B6E]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [51] | |||
| Responsed Disease | Head and neck squamous carcinoma [ICD-11: 2B6E] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | Adherens junction | hsa04520 | ||
| Cell Process | Epithelial-mesenchymal transition | |||
In-vitro Model |
SCC-15 | Tongue squamous cell carcinoma | Homo sapiens | CVCL_1681 |
| FaDu | Hypopharyngeal squamous cell carcinoma | Homo sapiens | CVCL_1218 | |
| In-vivo Model | To construct the metastasis model, 5 × 106 FaDu cells were transfected with sh-IGF2BP2-luc and sh-NC-luc, suspended in 60 uL PBS, and then injected into the footpads of the mice. Six weeks after injection, mice were subjected to bioluminescence imaging to evaluate lymphatic metastasis. For bioluminescence imaging, mice were anesthetized by inhaling 2% isoflurane for approximately 5 min, injected intraperitoneally with D-Luciferin potassium salt (200 uL, 150 ug/ml, ST196, Beyotime, Shanghai, China), and imaged with a bioluminescence system (NightOwl II LB983, Berthold Technologies, Germany). | |||
| Response Summary | Mechanistic investigations revealed that Zinc finger protein SNAI2 (Slug), a key EMT-related transcriptional factor, is the direct target of IGF2BP2, and essential for IGF2BP2-regulated EMT and metastasis in HNSCC. | |||
Cancer susceptibility 9 (CASC9)
Brain cancer [ICD-11: 2A00]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [52] | |||
| Responsed Disease | Glioblastoma [ICD-11: 2A00.00] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | Glycolysis / Gluconeogenesis | hsa00010 | ||
| Cell Process | Aerobic glycolysis | |||
In-vitro Model |
NHA (Normal human astrocytes) | |||
| U251 (Fibroblasts or fibroblast like cells) | ||||
| U-87MG ATCC | Glioblastoma | Homo sapiens | CVCL_0022 | |
| Response Summary | Cancer susceptibility 9 (CASC9)/IGF2BP2/HK2 axis promotes the aerobic glycolysis of glioblastoma multiforme. | |||
DIAPH1 antisense RNA 1 (DIAPH1-AS1)
Nasopharyngeal carcinoma [ICD-11: 2B6B]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [28] | |||
| Responsed Disease | Nasopharyngeal carcinoma [ICD-11: 2B6B] | |||
| Target Regulation | Up regulation | |||
| Response Summary | WTAP-mediated m6A modification of DIAPH1 antisense RNA 1 (DIAPH1-AS1) enhanced its stability relying on the m6A reader IGF2BP2-dependent pathway. Furthermore, DIAPH1-AS1 acted as a molecular adaptor that promoted MTDH-LASP1 complex formation and upregulated LASP1 expression, ultimately facilitating NPC growth and metastasis. | |||
HLA complex group 11 (HCG11)
Lung cancer [ICD-11: 2C25]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [53] | |||
| Responsed Disease | Lung adenocarcinoma [ICD-11: 2C25.0] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | Nucleotide excision repair | hsa03420 | ||
| Cell Process | RNA stabilization | |||
In-vitro Model |
A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 |
| HBE (Human bronchial epithelial cell line) | ||||
| NCI-H1975 | Lung adenocarcinoma | Homo sapiens | CVCL_1511 | |
| NCI-H522 | Lung adenocarcinoma | Homo sapiens | CVCL_1567 | |
| PC-9 | Lung adenocarcinoma | Homo sapiens | CVCL_B260 | |
| In-vivo Model | LUAD cells stably HCG11 and/or LATS1 overexpressed or silenced were subcutaneously injected into the flank of the BALB/c nude mice (male, 4 weeks old). | |||
| Response Summary | HLA complex group 11 (HCG11) mediated by METTL14 inhibited the growth of lung adenocarcinoma via IGF2BP2/LATS1. The m6A modification of HCG11 promoted its nuclear exportation and binding by Insulin Like Growth Factor 2 MRNA Binding Protein 2 (IGF2BP2), resulting in increased stability. | |||
HOXD antisense growth-associated long non-coding RNA (HAGLR)
Thyroid Cancer [ICD-11: 2D10]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [54] | |||
| Responsed Disease | Thyroid Cancer [ICD-11: 2D10] | |||
| Target Regulation | Up regulation | |||
| Cell Process | Cell proliferation | |||
| Cell migration | ||||
| Cell invasion | ||||
| Cell apoptosis | ||||
| Cell cycle progression | ||||
In-vitro Model |
TPC-1 | Thyroid gland papillary carcinoma | Homo sapiens | CVCL_6298 |
| Response Summary | IGF2BP2 loss inhibited cell proliferation, migration and invasion, and induced cell apoptosis and cell cycle arrest by down-regulating HOXD antisense growth-associated long non-coding RNA (HAGLR) expression in an m6A-dependent manner in thyroid cancer cells. | |||
Retinopathy [ICD-11: 9B71]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [55] | |||
| Responsed Disease | Diabetic retinopathy [ICD-11: 9B71.0] | |||
| Target Regulation | Up regulation | |||
Prostate cancer associated transcript 6 (PCAT6)
Prostate cancer [ICD-11: 2C82]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [27] | |||
| Responsed Disease | Prostate cancer [ICD-11: 2C82] | |||
| Target Regulation | Up regulation | |||
| Cell Process | RNA stability | |||
In-vitro Model |
PC-3 | Prostate carcinoma | Homo sapiens | CVCL_0035 |
| LNCaP C4-2B | Prostate carcinoma | Homo sapiens | CVCL_4784 | |
| In-vivo Model | At 1 week post-injection with PC-3 cells, mice were randomly assigned to three groups (n = 8 per group): the ASO-NC group (injection with ASO negative control targeting unknown sequence, 5 nmol in 100 uL PBS for each mouse), the ASO-L group (injection with low-dose ASO targeting PCAT6, 5 nmol in 100 uL PBS for each mouse), and the ASO-H group (injection with high-dose ASO targeting PCAT6, 10 nmol in 100 uL PBS for each mouse). | |||
| Response Summary | METTL3-mediated m6A modification contributed to Prostate cancer associated transcript 6 (PCAT6) upregulation in an IGF2BP2-dependent manner. Furthermore, PCAT6 upregulated IGF1R expression by enhancing IGF1R mRNA stability through the PCAT6/IGF2BP2/IGF1R RNA-protein three-dimensional complex. The m6 A-induced PCAT6/IGF2BP2/IGF1R axis promotes PCa bone metastasis and tumor growth, suggesting that PCAT6 serves as a promising prognostic marker and therapeutic target against bone-metastatic PCa. | |||
PTGS2 antisense RNA 1 (PACERR)
Pancreatic cancer [ICD-11: 2C10]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [2] | |||
| Responsed Disease | Pancreatic ductal adenocarcinoma [ICD-11: 2C10.0] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | mRNA surveillance pathway | hsa03015 | ||
| RNA degradation | hsa03018 | |||
| Cell Process | RNA stability | |||
In-vitro Model |
THP-1 | Childhood acute monocytic leukemia | Homo sapiens | CVCL_0006 |
| PATU-8988 (Human pancreatic adenocarcinoma cell) | ||||
| PANC-1 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0480 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| 37 (Pancreatic cancer cell) | ||||
| In-vivo Model | BALB/c nude mice which were co-injected with THP-1 cells and PATU-8988 cells subcutaneously. | |||
| Response Summary | PTGS2 antisense RNA 1 (PACERR) which bound to IGF2BP2 acts as an m6A-dependent manner to enhance the stability of KLF12 and c-myc in cytoplasm. This study found that LncRNA-PACERR functions as key regulator of TAMs in PDAC microenvironment and revealed the novel mechanisms in cytoplasm and in nucleus. | |||
ZNFX1 antisense RNA 1 (ZFAS1)
Colorectal cancer [ICD-11: 2B91]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [34] | |||
| Responsed Disease | Colorectal cancer [ICD-11: 2B91] | |||
| Target Regulation | Up regulation | |||
| Cell Process | Warburg effect | |||
| Mitochondrial energy metabolism | ||||
In-vitro Model |
Caco-2 | Colon adenocarcinoma | Homo sapiens | CVCL_0025 |
| HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 | |
| HIEC (Normal intestinal epithelial cells) | ||||
| HT29 | Colon cancer | Mus musculus | CVCL_A8EZ | |
| LoVo | Colon adenocarcinoma | Homo sapiens | CVCL_0399 | |
| RKO | Colon carcinoma | Homo sapiens | CVCL_0504 | |
| SW480 | Colon adenocarcinoma | Homo sapiens | CVCL_0546 | |
| SW480 | Colon adenocarcinoma | Homo sapiens | CVCL_0546 | |
| SW620 | Colon adenocarcinoma | Homo sapiens | CVCL_0547 | |
| Response Summary | The critical modulation network underlying m6A readers stabilizes lncRNAs, and they jointly promote mitochondrial energy metabolism in the pathogenesis of colorectal cancer. N6-methyladenosine reader stabilizes the ZFAS1/OLA1 axis. Thus, direct interaction between the KH3-4 domain of IMP2 and ZFAS1 where IMP2 serves as a reader for m6A-modified ZFAS1 and promotes the RNA stability of ZFAS1 is critical for CRC development. | |||
hsa-miR-133a-3p
Cardiomegaly [ICD-11: BC45]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [56] | |||
| Responsed Disease | Cardiomegaly [ICD-11: BC45] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
CM | Pancreatic insulinoma | Homo sapiens | CVCL_H246 |
| In-vivo Model | Neonatal mouse cardiomyocytes (CMs) were blinded to isolate from 7-10 cases of postnatal 1-day old wildtype C57/B mice and cultured using Pierce Primary Cardi Page 10.omyocyte Isolation Kit (Thermo Fisher Scientific) as the manufacturer's instructions. | |||
| Response Summary | IGF2BP2 physically interacted with AGO2 and increased more hsa-miR-133a-3p accumulation on its target site.m6A modification promoted the repression of specific miRNA during heart development and hypertrophy. | |||
hsa_circ_0000231 (circ_ARHGAP12)
Cervical cancer [ICD-11: 2C77]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [57] | |||
| Responsed Disease | Cervical cancer [ICD-11: 2C77] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | Cellular senescence | hsa04218 | ||
In-vitro Model |
C-33 A | Cervical squamous cell carcinoma | Homo sapiens | CVCL_1094 |
| Ca Ski | Cervical squamous cell carcinoma | Homo sapiens | CVCL_1100 | |
| HaCaT | Normal | Homo sapiens | CVCL_0038 | |
| HT-3 | Cervical carcinoma | Homo sapiens | CVCL_1293 | |
| SiHa | Cervical squamous cell carcinoma | Homo sapiens | CVCL_0032 | |
| In-vivo Model | CircARHGAP12-stable knockdown cervical cancer cells (100 uL PBS containing 5 × 106 cells) were subcutaneously injected into the lateral flank of BALB/c nude mice. | |||
| Response Summary | m6A-modified hsa_circ_0000231 (circARHGAP12) interacts with IGF2BP2 to enhance FOXM1 mRNA stability, forming circARHGAP12/IGF2BP2/FOXM1 complex, thereby promoting the proliferation and migration of cervical cancer cells. | |||
hsa_circ_0058493 (Circ_RHBDD1)
Gastric cancer [ICD-11: 2B72]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [58] | |||
| Responsed Disease | Gastric cancer [ICD-11: 2B72] | |||
In-vitro Model |
MKN28 | Gastric tubular adenocarcinoma | Homo sapiens | CVCL_1416 |
5-hydroxytryptamine receptor 3A (HTR3A)
Esophageal cancer [ICD-11: 2B70]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [59] | |||
| Responsed Disease | Esophageal Squamous Cell Carcinoma [ICD-11: 2B70.1] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
KYSE-180 | Esophageal squamous cell carcinoma | Homo sapiens | CVCL_1349 |
| TE-5 | Esophageal squamous cell carcinoma | Homo sapiens | CVCL_1764 | |
| KYSE-150 | Esophageal squamous cell carcinoma | Homo sapiens | CVCL_1348 | |
| KYSE-510 | Esophageal squamous cell carcinoma | Homo sapiens | CVCL_1354 | |
| KYSE-140 | Esophageal squamous cell carcinoma | Homo sapiens | CVCL_1347 | |
AC026356.1
Lung cancer [ICD-11: 2C25]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [60] | |||
| Responsed Disease | Lung adenocarcinoma [ICD-11: 2C25.0] | |||
| Target Regulation | Up regulation | |||
| In-vivo Model | Cells (2 × 104) were seeded onto 12-well plates and cultured in serum-free 1640 medium. Cell spheroids were documented and quantified using an inverted microscope (Olympus, Japan) after two weeks. | |||
Ankyrin repeat domain-containing protein 22 (ANKRD22)
Nasopharyngeal carcinoma [ICD-11: 2B6B]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [61] | |||
| Responsed Disease | Nasopharyngeal carcinoma [ICD-11: 2B6B] | |||
| Target Regulation | Up regulation | |||
ATP-dependent 6-phosphofructokinase, liver type (PFKL)
Lung cancer [ICD-11: 2C25]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [62] | |||
| Responsed Disease | Non-small cell lung cancer [ICD-11: 2C25.Y] | |||
In-vitro Model |
NCI-H1299 | Lung large cell carcinoma | Homo sapiens | CVCL_0060 |
| A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 | |
BACE1 antisense RNA (BACE1-AS)
Colorectal cancer [ICD-11: 2B91]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [63] | |||
| Responsed Disease | Colorectal cancer [ICD-11: 2B91] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | Wnt signaling pathway | hsa04310 | ||
In-vitro Model |
CCD-841CoN
|
N.A. | Homo sapiens | CVCL_2871 |
| SW480 | Colon adenocarcinoma | Homo sapiens | CVCL_0546 | |
| HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 | |
| SW620 | Colon adenocarcinoma | Homo sapiens | CVCL_0547 | |
| LoVo | Colon adenocarcinoma | Homo sapiens | CVCL_0399 | |
| In-vivo Model | BACE1-AS knockout SW620 or parental SW620 cells were injected into the inferior Hemi-spleen of the mice. The weights of mice were recorded every 3 days. After 7 weeks, the mice were euthanized. The whole livers of mice were resected and photographed to assess metastatic burden. . | |||
Beta-1,4-glucuronyltransferase 1 (B3GNT6)
Pancreatic cancer [ICD-11: 2C10]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [64] | |||
| Responsed Disease | Pancreatic carcinoma [ICD-11: 2C10.Y] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
CFPAC-1 | Cystic fibrosis | Homo sapiens | CVCL_1119 |
| PANC-1 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0480 | |
| In-vivo Model | To evaluate the effect of IGF2BP2 on the tumor formation ability in vivo, 2 × 106 cells were injected into the axilla of nude mice for subcutaneous tumor formation. After 9 days, modified IGF2BP2-si or si-NC was injected into the tumors every 3 days for 3 weeks. Tumor size was measured every 3 days. | |||
Cell division control protein 45 homolog (CDC45)
Liver cancer [ICD-11: 2C12]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [65] | |||
| Responsed Disease | Liver hepatocellular carcinoma [ICD-11: 2C12.02] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
Huh-7 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_0336 |
| SK-HEP-1 | Liver and intrahepatic bile duct epithelial neoplasm | Homo sapiens | CVCL_0525 | |
| HCCLM3 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_6832 | |
| Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 | |
| In-vivo Model | Following one week of adaptive feeding, the mice were haphazardly allocated to one of two groups: sh-NC or sh-CDC45. The former received subcutaneous injection of SK-Hep-1 cells transfected with sh-NC while the latter received injection of SK-Hep-1 cells transfected with sh-CDC45. Each mouse was injected with 3 × 10 [6] cells in a volume of 150 μl at the left armpit. Tumor volume was monitored biweekly following implantation. | |||
Cell division control protein 6 homolog (CDC6)
Lung cancer [ICD-11: 2C25]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [66] | |||
| Responsed Disease | Lung cancer [ICD-11: 2C25] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 |
| Calu-1 | Lung squamous cell carcinoma | Homo sapiens | CVCL_0608 | |
| HOP-62 | Lung adenocarcinoma | Homo sapiens | CVCL_1285 | |
| In-vivo Model | For the in vivo tumorigenicity assay, female BALB/c nude mice (ages 4-5 weeks) were randomly divided into two groups (n = 5 per group). A549 cells (2 × 106) that had been stably transfected with sh-LCAT1 or scramble were implanted subcutaneously into the nude mice. | |||
Cell division cycle and apoptosis regulator protein 1 (CCAR1)
Prostate cancer [ICD-11: 2C82]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [67] | |||
| Responsed Disease | Prostate cancer [ICD-11: 2C82] | |||
| Target Regulation | Up regulation | |||
Cohesin subunit SA-3 (STAG3)
Colorectal cancer [ICD-11: 2B91]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [68] | |||
| Responsed Disease | Colorectal cancer [ICD-11: 2B91] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 |
| HT29 | Colon cancer | Mus musculus | CVCL_A8EZ | |
| SW480 | Colon adenocarcinoma | Homo sapiens | CVCL_0546 | |
| LoVo | Colon adenocarcinoma | Homo sapiens | CVCL_0399 | |
| SW620 | Colon adenocarcinoma | Homo sapiens | CVCL_0547 | |
| NCM460 | Normal | Homo sapiens | CVCL_0460 | |
| In-vivo Model | Oe-STAG3 and sh-METTL3 were transfected into HCT116 cells. Trypsin digestion and cell counting were performed on each set of cells after they had reached around 80% confluence. Nude mice were subcutaneously injected with 200 μL cells (2 × 106) and randomly assigned to 5 groups (n = 6 in each group): control group (mice were injected with cells without transfection plasmid), oe-NC group (mice were injected with cells transfected with oe-NC), oe-STAG3 group (mice were injected with cells transfected with oe-STAG3), oe-STAG3 + sh-NC group (mice were injected with cells transfected with oe-STAG3 and sh-NC) and oe-STAG3 + sh-METTL3 group. | |||
Cyclic AMP-responsive element-binding protein 1 (CREB1)
Colorectal cancer [ICD-11: 2B91]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [69] | |||
| Responsed Disease | Colorectal cancer [ICD-11: 2B91] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
COLO205
|
N.A. | Homo sapiens | CVCL_F402 |
| HCT 15 | Colon adenocarcinoma | Homo sapiens | CVCL_0292 | |
| LoVo | Colon adenocarcinoma | Homo sapiens | CVCL_0399 | |
| SW620 | Colon adenocarcinoma | Homo sapiens | CVCL_0547 | |
| HT-29 | Colon adenocarcinoma | Homo sapiens | CVCL_0320 | |
| HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 | |
| In-vivo Model | To investigate tumor metastasis, approximately 1 × 106 HCT116 (Control or circEZH2 KD) cells suspended in 100 μL of 1 × PBS per mouse were injected into the tail vein of male BALB/c nude mice. | |||
Cytoskeleton-associated protein 2-like (CKAP2L)
Ovarian cancer [ICD-11: 2C73]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [70] | |||
| Responsed Disease | Ovarian cancer [ICD-11: 2C73] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
A2780 | Ovarian endometrioid adenocarcinoma | Homo sapiens | CVCL_0134 |
| OVCAR-3 | Ovarian serous adenocarcinoma | Homo sapiens | CVCL_0465 | |
| SK-OV-3 | Ovarian serous cystadenocarcinoma | Homo sapiens | CVCL_0532 | |
| ES-2 | Ovarian clear cell adenocarcinoma | Homo sapiens | CVCL_3509 | |
| 12Z | Endometriosis | Homo sapiens | CVCL_0Q73 | |
|
IOSE-80
|
N.A. | Homo sapiens | CVCL_5546 | |
| In-vivo Model | To establish an orthotopic xenograft tumor model, 12 healthy female Balb/c nude mice were chosen and assigned to two groups. Nude mice were anesthetized by isoflurane inhalation, and after disinfecting the skin with iodophor, a 5 mm incision was made in the skin and abdominal wall, parallel and ventral to the spine, midway, and between the last rib and the iliac crest. After pulling out the ovary, the cell suspensions (10 μL) containing 1 × 106 IGF2BP2-overexpressing or control SKOV3 cells were inoculated after inserting the needle at the junction between the bursa and the fat pad. The ovary was put back in place, and if no bleeding was noted, the incision on the muscle layer and body wall was closed separately. | |||
Dipeptidyl peptidase 4 (DPP4)
Thyroid Cancer [ICD-11: 2D10]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [71] | |||
| Responsed Disease | Papillary thyroid cancer [ICD-11: 2D10.1] | |||
| Target Regulation | Up regulation | |||
Ena/VASP-like protein (EVL)
Macroscopic changes of size of the kidney [ICD-11: MF54]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [72] | |||
| Responsed Disease | Macroscopic changes of size of the kidney [ICD-11: MF54.0] | |||
| Responsed Drug | Isoforsythiaside | Investigative | ||
| Target Regulation | Up regulation | |||
In-vitro Model |
HEK293 | Normal | Homo sapiens | CVCL_0045 |
| HK-2 [Human kidney] | Normal | Homo sapiens | CVCL_0302 | |
| In-vivo Model | For the unilateral ureteral obstruction (UUO)-induced ON model, mice were subjected to permanent ureteral ligation of the left kidney ureter under aseptic and anaesthetic conditions, whereas sham-operated mice merely lacked ligation under equivalent conditions. The mice were sacrificed under anaesthesia at 3, 7 and 14 days postoperatively. For the ischemia/reperfusion (I/R)-induced renal fibrosis model, mice were recovered with blood supply after 42 min of bilateral renal artery clamping under both aseptic and anaesthetic conditions. These mice were sacrificed under anaesthesia at 7 and 14 days postoperatively. For drug treatment, isoforsythiaside was injected intraperitoneally at concentrations of 10, 25 and 50 mg/kg per day when the model was established. Similarly, the sham-operated group received an equivalent volume of saline intraperitoneally as a control. The harvested kidneys have been primarily used for morphological and histopathological observations and molecular biology studies. | |||
F-box only protein 43 (FBXO43)
Liver cancer [ICD-11: 2C12]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [73] | |||
| Responsed Disease | Liver hepatocellular carcinoma [ICD-11: 2C12.02] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
HCCLM3 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_6832 |
| Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 | |
| Huh-7 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_0336 | |
| L-02 | Endocervical adenocarcinoma | Homo sapiens | CVCL_6926 | |
Fatty acid-binding protein 5 (FABP5)
Pancreatic cancer [ICD-11: 2C10]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [74] | |||
| Responsed Disease | Pancreatic neuroendocrine neoplasms [ICD-11: 2C10.1] | |||
| Target Regulation | Down regulation | |||
In-vitro Model |
QGP-1 | Pancreatic somatostatinoma | Homo sapiens | CVCL_3143 |
| In-vivo Model | For tumor xenograft models, QGP-1cells (5 × 106) with ALKBH5 over-expression, ALKBH5 over-expression with FABP5 knockdown, and negative control were subcutaneously injected into the right axilla of female BALB/c nude mice (4-6 weeks). After 4 weeks, the mice were sacrificed via a form of euthanasia. | |||
FBXL19 antisense RNA 1 (FBXL19-AS1)
Brain cancer [ICD-11: 2A00]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [75] | |||
| Responsed Disease | Glioma [ICD-11: 2A00.0] | |||
Fibroblast growth factor receptor 2 (FGFR2)
Breast cancer [ICD-11: 2C60]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [76] | |||
| Responsed Disease | Breast cancer [ICD-11: 2C60] | |||
| Target Regulation | Up regulation | |||
Flotillin-1 (FLOT1)
Brain cancer [ICD-11: 2A00]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [77] | |||
| Responsed Disease | Glioma [ICD-11: 2A00.0] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
U-251MG | Astrocytoma | Homo sapiens | CVCL_0021 |
| T98G | Glioblastoma | Homo sapiens | CVCL_0556 | |
| U-87MG ATCC | Glioblastoma | Homo sapiens | CVCL_0022 | |
| U-118MG | Astrocytoma | Homo sapiens | CVCL_0633 | |
Fructose-bisphosphate aldolase A (ALDOA)
Hepatic fibrosis/cirrhosis [ICD-11: DB93]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [78] | |||
| Responsed Disease | Hepatic fibrosis/cirrhosis [ICD-11: DB93] | |||
| Target Regulation | Up regulation | |||
glucosylceramidase beta 1 like, pseudogene (GBA1LP)
Liver cancer [ICD-11: 2C12]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [79] | |||
| Responsed Disease | Liver hepatocellular carcinoma [ICD-11: 2C12.02] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
L-02 | Endocervical adenocarcinoma | Homo sapiens | CVCL_6926 |
| Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 | |
| Huh-7 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_0336 | |
| Hep 3B2.1-7 | Childhood hepatocellular carcinoma | Homo sapiens | CVCL_0326 | |
| MHCC97-H | Adult hepatocellular carcinoma | Homo sapiens | CVCL_4972 | |
| SMMC-7721 | Endocervical adenocarcinoma | Homo sapiens | CVCL_0534 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| In-vivo Model | In vivo tumor growth assay, 1 × 107 Hep3B-GBAP1 or MHCC97H-shGBAP1 subclones and the corresponding control cells were transplanted into the body of 6-week-old BALB/c nude mice (Slac Laboratory Animal Center, Shanghai, China) via subcutaneous injection. Tumor size was measured every 3 days. Twenty-one days later, tumors were removed from mice in different groups. Tumor weight was calculated after dissection. Permission of conducting animal study was obtained from the Research Ethics Committee of Xi'an Jiaotong University. In vivo lung metastasis model, 1 × 106 cells were intravenously injected into the lateral tail vein of nude mice (n = 5 mice/group). | |||
Glutaredoxin-1 (GLRX)
Parkinson disease [ICD-11: 8A00]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [80] | |||
| Responsed Disease | Parkinson disease [ICD-11: 8A00] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
BV-2 | Normal | Mus musculus | CVCL_0182 |
GNAS antisense RNA 1 (GNAS-AS1)
Acute myeloid leukaemia [ICD-11: 2A60]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [81] | |||
| Responsed Disease | Acute myeloid leukaemia [ICD-11: 2A60] | |||
| Responsed Drug | Chidamide | Investigative | ||
In-vitro Model |
HL-60 | Adult acute myeloid leukemia | Homo sapiens | CVCL_0002 |
| HEL | Erythroleukemia | Homo sapiens | CVCL_0001 | |
| MOLM-13 | Adult acute myeloid leukemia | Homo sapiens | CVCL_2119 | |
| MV4-11 | Childhood acute monocytic leukemia | Homo sapiens | CVCL_0064 | |
| Kasumi-1 | Myeloid leukemia with maturation | Homo sapiens | CVCL_0589 | |
|
HS-5
|
N.A. | Homo sapiens | CVCL_3720 | |
| In-vivo Model | Subcutaneous tumorigenesis was induced in mice by subcutaneous transplantation of 1 × 106 HL-60 cells. To stably overexpress GNAS-AS1, lentivirus-packaged OE-GNAS-AS1 (1 × 108 plaque-forming units). When the tumor volume reached 150-200 mm3, mice in the control group were orally treated with 1% DMSO (containing 0.2% carboxymethylcellulose and 0.1% Tween 80). Mice in the Chidamide, Chidamide + OE-NC, and Chidamide + OE-GNAS-AS1 groups were orally treated with Chidamide (25 mg/kg body weight, formulated with 1% DMSO containing 0.2% carboxymethylcellulose and 0.1% Tween 80). All treatments were repeated three times a week and for two weeks. Tumor volume was recorded every 5 days. | |||
Granulocyte-macrophage colony-stimulating factor (CSF2)
Gastric cancer [ICD-11: 2B72]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [82] | |||
| Responsed Disease | Gastric cancer [ICD-11: 2B72] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
HGC-27 | Gastric carcinoma | Homo sapiens | CVCL_1279 |
| In-vivo Model | 48 male BALB/c nu/nu mice (Slake Experimental Animal Center in Shanghai) at the age of 4 weeks were randomly divided into 8 groups (n = 6/group). All groups received subcutaneous injections of HGC-27 (2 × 106 cells in 200 μL PBS), Additionally, MSCs transfected with or without CSF2 plasmid, P-MSCs transfected with or without CSF2/IGF2BP2 inhibitor, and GC-MSCs transfected with or without CSF2/IGF2BP2 inhibitor were co-injected with HGC-27 cells in a 1:1 ratio. The mice were monitored every 2 days. Tumor volumes were calculated using the modified ellipsoidal formula: V = 1/2 (length × width2). | |||
Histone-lysine N-methyltransferase SUV39H2 (SUV39H2)
Gastric cancer [ICD-11: 2B72]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [83] | |||
| Responsed Disease | Gastric cancer [ICD-11: 2B72] | |||
| Target Regulation | Up regulation | |||
| In-vivo Model | The nude mice were inoculated in the inguinal region subcutaneously with 1 × 106 stably transfected GC cells which were suspended in 100 μL PBS. After one week, preestablished tumor xenografts were treated with DMSO or OTS186935 (50 mg/kg, 2 × /wk × 3). | |||
Homeobox protein Hox-C6 (HOXC6)
Prostate cancer [ICD-11: 2C82]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [84] | |||
| Responsed Disease | Prostate cancer [ICD-11: 2C82] | |||
In-vitro Model |
VCaP | Prostate carcinoma | Homo sapiens | CVCL_2235 |
| DU145 | Prostate carcinoma | Homo sapiens | CVCL_0105 | |
| HNC PC3 | Retromolar trigone squamous cell carcinoma | Homo sapiens | CVCL_C8XA | |
Homeobox protein MSX-1 (MSX1)
Colorectal cancer [ICD-11: 2B91]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [85] | |||
| Responsed Disease | Colorectal cancer [ICD-11: 2B91] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 |
| SW620 | Colon adenocarcinoma | Homo sapiens | CVCL_0547 | |
| SW480 | Colon adenocarcinoma | Homo sapiens | CVCL_0546 | |
| LoVo | Colon adenocarcinoma | Homo sapiens | CVCL_0399 | |
| HT29 | Colon cancer | Mus musculus | CVCL_A8EZ | |
JmjC domain-containing protein 8 (JMJD8)
Colorectal cancer [ICD-11: 2B91]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [86] | |||
| Responsed Disease | Colorectal cancer [ICD-11: 2B91] | |||
In-vitro Model |
SW480 | Colon adenocarcinoma | Homo sapiens | CVCL_0546 |
| SW620 | Colon adenocarcinoma | Homo sapiens | CVCL_0547 | |
Kremen protein 1 (KRM1)
Calcific aortic valve disease [ICD-11: BB71 ]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [87] | |||
| Responsed Disease | Calcific aortic valve disease [ICD-11: BB71 ] | |||
| Target Regulation | Up regulation | |||
Krueppel-like factor 6 (KLF6)
Injury of heart [ICD-11: NB31]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [88] | |||
| Responsed Disease | Myocardial injury [ICD-11: NB31.Z] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
HL-1 | Normal | Mus musculus | CVCL_0303 |
Large neutral amino acids transporter small subunit 1 (SLC7A5)
Lung cancer [ICD-11: 2C25]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [89] | |||
| Responsed Disease | Lung cancer [ICD-11: 2C25] | |||
| Target Regulation | Up regulation | |||
long intergenic non-protein coding RNA 941 (LINC00941)
Pancreatic cancer [ICD-11: 2C10]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [90] | |||
| Responsed Disease | Pancreatic ductal adenocarcinoma [ICD-11: 2C10.0] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
PANC-1 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0480 |
| MIA PaCa-2 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0428 | |
| SW1990 | Pancreatic adenocarcinoma | Homo sapiens | CVCL_1723 | |
| AsPC-1 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0152 | |
| BxPC-3 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0186 | |
| HPDE6c7 | Normal | Homo sapiens | CVCL_0P38 | |
| In-vivo Model | PANC-1 cells were stably transfected with LV-METTL14, LV-NC, sh-METTL14, sh-NC, or sh-LINC00941. A mixture of 2×106 cells and 100 μL PBS was injected into the spleen of every BALB/c nude mouse. After two months of housing in a sterile environment, the mice were sacrificed. Their liver tissues were removed for hematoxylin and eosin (HE) staining. | |||
Macrophage colony-stimulating factor 1 (CSF1)
Pancreatic cancer [ICD-11: 2C10]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [33] | |||
| Responsed Disease | Pancreatic cancer [ICD-11: 2C10] | |||
| Target Regulation | Up regulation | |||
Nuclear transcription factor Y subunit alpha (NFYA)
Ovarian cancer [ICD-11: 2C73]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [91] | |||
| Responsed Disease | Ovarian cancer [ICD-11: 2C73] | |||
| Target Regulation | Up regulation | |||
| Cell Process | mRNA stability | |||
Phospholipid hydroperoxide glutathione peroxidase GPX4 (GPX4)
Lung cancer [ICD-11: 2C25]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [92] | |||
| Responsed Disease | Small cell lung cancer [ICD-11: 2C25.1] | |||
In-vitro Model |
A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 |
| PC-9 | Lung adenocarcinoma | Homo sapiens | CVCL_B260 | |
| In-vivo Model | For xenograft establishment, six-week-old female athymic nude mice (n = 6 per group) were used, following ethical guidelines approved by the Institutional Animal Care and Use Committee. Each mouse received subcutaneous injections in the flank region with 1 × 10^6 cells suspended in 100 μL of serum-free medium, either sh-METTL3-transfected A549 cells or sh-NC-transfected controls. Tumour growth was monitored bi-dimensionally every week using callipers, with tumour volume calculated using the formula (length × width^2)/2. After 4 weeks, the mice were anaesthetised and euthanized by intraperitoneal injection of 3% pentobarbital sodium (100 mg/kg). The tumours were weighted and used for HE and Immunohistochemistry staining. | |||
Ulcerative colitis [ICD-11: DD71]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [93] | |||
| Responsed Disease | Ulcerative colitis [ICD-11: DD71] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
NCM460 | Normal | Homo sapiens | CVCL_0460 |
| In-vivo Model | All mice were randomly categorized into 6 groups (n = 6 / group): the control group (mice were fed with normal drinking water), the DSS group, DSS + Lv-oe-NC group, DSS + Lv-oe-IGF2BP2 group, DSS + Lv-oe-IGF2BP2 + Lv-sh-NC group, and DSS + Lv-oe-IGF2BP2 + Lv-sh-GPX4 group. The last four groups of mice underwent anaesthesia via intraperitoneal injection of 30 mg/kg pentobarbital sodium. Next, mice were intracolonically administrated with Lv-oe-NC alone, Lv-oe-IGF2BP2 alone, Lv-oe-IGF2BP2 plus Lv-sh-NC, or Lv-oe-IGF2BP2 plus Lv-sh-GPX4 at a dose of 1 × 109 IU in a final volume of 100 μL PBS [18]. After 48 h, except for the control group, the acute UC model in mice was established by feeding with drinking water containing 3 % DSS for 7 days [19]. On the 10th day, all mice underwent euthanasia via inhalation of overdose CO2, after which the colons of mice in each group were collected, measured using a ruler. | |||
Procathepsin L (CTSL)
Cervical cancer [ICD-11: 2C77]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [94] | |||
| Responsed Disease | Cervical cancer [ICD-11: 2C77] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
Ca Ski | Cervical squamous cell carcinoma | Homo sapiens | CVCL_1100 |
| SiHa | Cervical squamous cell carcinoma | Homo sapiens | CVCL_0032 | |
| HEK293-FT | Normal | Homo sapiens | CVCL_6911 | |
| In-vivo Model | Female BALB/c nude mice (4-5 weeks old) were purchased from the Center of Experimental Animals of Guangdong. To establish a tail vein metastasis model, 2 × 106 SiHa cells in 200 μl PBS were injected into the tail vein of each mouse (n = 6 for both METTL3-overexpressing and empty vector groups). The mice were killed at approximately 8 weeks, and lung tissues were isolated and embedded in paraffin. Hematoxylin and eosin staining was then used to determine the number of lung metastasis nodules. To establish the popliteal lymph node metastasis model, 1 × 106 SiHa cells in 50 μl PBS were injected subcutaneously into the footpad of each mouse (n = 6 for both groups). Cells from the experimental and control groups were inoculated under the right and left footpads of each mouse, respectively. After 8 weeks, the popliteal lymph nodes were excised. | |||
Procollagen-lysine,2-oxoglutarate 5-dioxygenase 2 (PLOD2)
Male infertility [ICD-11: GB04]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [95] | |||
| Responsed Disease | Male infertility [ICD-11: GB04] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
GC-2spd(ts)
|
N.A. | Mus musculus | CVCL_6633 |
| In-vivo Model | 20 adult male Wistar rats (180-200 g) were purchased from the Guangdong Provincial Center for Disease Control and Prevention. A 12 h/12 h light/dark cycle was maintained for the animals and they were fed standard food pellets and water as needed. | |||
Protein arginine N-methyltransferase 6 (PRMT6)
Acute myeloid leukaemia [ICD-11: 2A60]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [96] | |||
| Responsed Disease | Acute myeloid leukaemia [ICD-11: 2A60] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
MOLM-13 | Adult acute myeloid leukemia | Homo sapiens | CVCL_2119 |
| MV4-11 | Childhood acute monocytic leukemia | Homo sapiens | CVCL_0064 | |
Protein c-Fos (FOS)
Gastric cancer [ICD-11: 2B72]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [97] | |||
| Responsed Disease | Gastric cancer [ICD-11: 2B72] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
Akata | EBV-related Burkitt lymphoma | Homo sapiens | CVCL_0148 |
| In-vivo Model | To evaluate the tumorigenic effect of FTO, FTO knockdown or control AGS B95.8 cells (1 × 107 suspended in 150 μL of PBS) were subcutaneously injected into the flanks of B-NDG mice. The diameter and width of the tumours were measured every 4 days and used to estimate the tumour volumes by the standard formula: 0.5 × length × width2. At the end stage, the tumours were removed, imaged and weighed. To investigate the peritoneal dissemination ability of EBVaGC cells, intraperitoneal injection was performed. Briefly, 1 × 107 FTO silencing or control EBVaGC cells suspended in 0.4 mL of PBS were injected into the peritoneal cavity of each mouse. Eight weeks later, all the mice were sacrificed, and the abdominal and intestinal metastatic nodules were excised, counted, photographed and paraffin embedded. For the lung metastasis model, 200 μL of 1 × 106 luciferase-labelled EBVaGC cells from different groups were directly injected into the tail vein of B-NDG mice, and distant and lung metastasis were evaluated using bioluminescent imaging. After 6 or 8 weeks, the mice were euthanized, and the lungs were embedded in paraffin and subjected to haematoxylin and eosin (H&E) staining to record the micrometastatic nodules using a microscope. All animal experiments were approved by our Institutional Animal Care. | |||
Protein Jumonji (JARID2)
Colorectal cancer [ICD-11: 2B91]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [85] | |||
| Responsed Disease | Colorectal cancer [ICD-11: 2B91] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 |
| SW620 | Colon adenocarcinoma | Homo sapiens | CVCL_0547 | |
| SW480 | Colon adenocarcinoma | Homo sapiens | CVCL_0546 | |
| LoVo | Colon adenocarcinoma | Homo sapiens | CVCL_0399 | |
| HT29 | Colon cancer | Mus musculus | CVCL_A8EZ | |
ribonuclease P RNA component H1 (RPPH1)
Breast cancer [ICD-11: 2C60]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [76] | |||
| Responsed Disease | Breast cancer [ICD-11: 2C60] | |||
Semaphorin-3F (SEMA3F)
Prostate cancer [ICD-11: 2C82]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [98] | |||
| Responsed Disease | Prostate cancer [ICD-11: 2C82] | |||
In-vitro Model |
RWPE-1 | Normal | Homo sapiens | CVCL_3791 |
| DU145 | Prostate carcinoma | Homo sapiens | CVCL_0105 | |
| PC-3 | Prostate carcinoma | Homo sapiens | CVCL_0035 | |
| 22Rv1 | Prostate carcinoma | Homo sapiens | CVCL_1045 | |
| LNCaP | Prostate carcinoma | Homo sapiens | CVCL_0395 | |
| LNCaP C4-2B | Prostate carcinoma | Homo sapiens | CVCL_4784 | |
| VCaP | Prostate carcinoma | Homo sapiens | CVCL_2235 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| In-vivo Model | All the animal studies and protocols followed the institutional guidelines of the First Affiliated Hospital, School of Medicine, Zhejiang University. | |||
Serine/threonine-protein kinase Nek7 (NEK7)
Intracerebral hemorrhage [ICD-11: 8B00]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [99] | |||
| Responsed Disease | Intracerebral hemorrhage [ICD-11: 8B00] | |||
| Target Regulation | Down regulation | |||
In-vitro Model |
HT22 | Normal | Mus musculus | CVCL_0321 |
Serine/threonine-protein phosphatase CPPED1 (CPPED1)
Laryngeal cancer [ICD-11: 2C23]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [100] | |||
| Responsed Disease | Laryngeal squamous cell carcinoma [ICD-11: 2C23.Z] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
Tu 177 | Laryngeal squamous cell carcinoma | Homo sapiens | CVCL_4913 |
| AMC-HN-8 | Laryngeal squamous cell carcinoma | Homo sapiens | CVCL_5966 | |
|
NP69SV40T
|
N.A. | Homo sapiens | CVCL_F755 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
Syndecan-4 (SDC4)
Thyroid Cancer [ICD-11: 2D10]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [101] | |||
| Responsed Disease | Thyroid Cancer [ICD-11: 2D10] | |||
In-vitro Model |
BHP 10-3 | Thyroid gland papillary carcinoma | Homo sapiens | CVCL_6278 |
|
Ca
|
N.A. | Drosophila melanogaster | CVCL_Z839 | |
| K1 | Thyroid gland papillary carcinoma | Homo sapiens | CVCL_2537 | |
Toll-like receptor 2 (TLR2)
Hypopharyngeal cancer [ICD-11: 2B6D]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [102] | |||
| Responsed Disease | Hypopharyngeal squamous cell carcinoma [ICD-11: 2B6D.0] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
FaDu | Hypopharyngeal squamous cell carcinoma | Homo sapiens | CVCL_1218 |
| Detroit 562 | Pharyngeal squamous cell carcinoma | Homo sapiens | CVCL_1171 | |
| In-vivo Model | HPSCC FaDu or Detroit 562 cells (1 × 106 cells) expressing vector control and construct lentiviruses were subcutaneously injected into the right flanks of 4-week-old male nude mice. Tumor diameters and body weight were recorded every 3 days for 1-6 weeks. Detroit 562 and FaDu cells (1 × 106 cells) were subcutaneously injected into the right flanks of 4-week-old male nude mice. | |||
Transcription factor E2F6 (E2F6)
Liver cancer [ICD-11: 2C12]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [43] | |||
| Responsed Disease | Liver cancer [ICD-11: 2C12] | |||
| Target Regulation | Up regulation | |||
Transcription factor E2F7 (E2F7)
Nasopharyngeal carcinoma [ICD-11: 2B6B]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [103] | |||
| Responsed Disease | Nasopharyngeal carcinoma [ICD-11: 2B6B] | |||
| Target Regulation | Up regulation | |||
| In-vivo Model | For the tumor growth model, 1 × 106 SUNE-1 cells stably expressing scrambled or sh-VIRMA were injected subcutaneously into the axilla of mice, and the tumor size was measured every 4 days. After 32 days, the mice were sacrificed, and the tumors were retrieved. For the tumor inguinal lymph node metastasis model, 1 × 106 scrambled or sh-VIRMA SUNE-1 cells were injected into the footpads of mice. After 6 weeks, the mice were euthanized. The footpad tumors and inguinal lymph nodes were excised. Tumors and lymph nodes were subjected to subsequent in situ hybridization and immunohistochemistry analysis. | |||
Transcription factor SOX-6 (SOX6)
Liver cancer [ICD-11: 2C12]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [104] | |||
| Responsed Disease | Intrahepatic cholangiocarcinoma [ICD-11: 2C12.10] | |||
| Target Regulation | Up regulation | |||
Transcriptional enhancer factor TEF-1 (TEAD1)
Liver disease [ICD-11: DB9Z]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [105] | |||
| Responsed Disease | Liver disease [ICD-11: DB9Z] | |||
| Target Regulation | Up regulation | |||
Transient receptor potential cation channel subfamily M member 7 (TRPM7)
Lung cancer [ICD-11: 2C25]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [106] | |||
| Responsed Disease | Non-small cell lung cancer [ICD-11: 2C25.Y] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
NCI-H520 | Lung squamous cell carcinoma | Homo sapiens | CVCL_1566 |
| SK-MES-1 | Lung squamous cell carcinoma | Homo sapiens | CVCL_0630 | |
| Calu-1 | Lung squamous cell carcinoma | Homo sapiens | CVCL_0608 | |
| PC-9 | Lung adenocarcinoma | Homo sapiens | CVCL_B260 | |
| A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 | |
| BEAS-2B | Normal | Homo sapiens | CVCL_0168 | |
| In-vivo Model | The BALB/c nude mice (male, 4-6 weeks) were provided via Vital River Laboratory Animal Technology (Beijing, China). For xenograft tumor growth assay, 15 mice were randomly classified into 3 groups (n = 5/group), including control, sh-NC or sh-DGUOK-AS1 group. In sh-NC or sh-DGUOK-AS1 group, mice were subcutaneously injected with SK-MES-1 cells (3 × 106) stably transfected with sh-NC or sh-DGUOK-AS1; Moreover, mice were subcutaneously injected with non-transfected SK-MES-1 cells (3 × 106) as control group. The tumors were monitored every 7 days, and size was calculated by (length × width2)/2. Mice were euthanized after 4 weeks, and the tumors were collected and weighed, followed via collection for further analyses. Ki67 and CD31 levels were measured through immunohistochemistry staining using Ki67 (ab15580, 1:2000 dilution, Abcam) or CD31 (ab18298, 1:3000 dilution, Abcam) according to the protocols. For pulmonary metastatic assay, 3 × 106 SK-MES-1 cells stably transfected with sh-NC or sh-DGUOK-AS1 or non-transfected SK-MES-1 cells were intravenously injected into mice. Mice were sacrificed after 8 weeks, and lung specimens were collected, followed by hematoxylin-eosin (HE) staining assay for metastatic lesions in lung tissues. All experiments were approved via the Animal Ethics Committee of The Second Hospital of Shandong University. | |||
Ubiquitin carboxyl-terminal hydrolase 13 (USP13)
Gastrointestinal stromal tumor [ICD-11: 2B5B]
| In total 2 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [107] | |||
| Responsed Disease | Gastrointestinal stromal tumor [ICD-11: 2B5B] | |||
| Responsed Drug | 3-Methyladenine | Investigative | ||
| Target Regulation | Up regulation | |||
In-vitro Model |
GIST-T1 | Gastrointestinal stromal tumor | Homo sapiens | CVCL_4976 |
| GIST882 | Gastrointestinal stromal tumor | Homo sapiens | CVCL_7044 | |
| GIST882 | Gastrointestinal stromal tumor | Homo sapiens | CVCL_7044 | |
| GIST-T1 | Gastrointestinal stromal tumor | Homo sapiens | CVCL_4976 | |
| In-vivo Model | 6 × 106 logarithmically growing GIST cells resuspended in 100 μL PBS were injected subcutaneously into the flank of 6-week-old female nude mice for tumor growth assays. The following treatments were initiated when the tumor volume reached approximately 300 mm3. (1) IM-sensitive GIST cell lines and IM-resistant GIST cell lines were used to establish a subcutaneous xenograft model of GIST (n = 6 mice/group). When the tumors grew to the required size, mice were treated with IM via drinking water; (2) GIST cells were pretreated with USP13 lentivirus or USP13 control lentivirus. In addition, IM-resistant GIST cells were pretreated with USP13 inhibitors or control. These lentivirus-transfected cells were implanted into subcutaneous mice tumors to construct the USP13 positive, USP13-negative, and control groups (n = 6 mice/group). When the transplanted tumor grew to the required volume, mice in each group were treated with IM via drinking water. (3) IM-resistant GIST cells-xenografted mice and control mice were treated with USP13 inhibitors or with USP13 inhibitors and autophagy inhibitors in combination therapy to assess reversal of IM resistance (n = 6 mice/group). Mice were treated with IM (45 mg/kg/day) and with or without 3-MA (15 mg/kg/day) via drinking water when the tumors grew to the required size. The USP13 inhibitor Spautin-1 was used at 20 mg/kg/day when required. | |||
| Experiment 2 Reporting the m6A-centered Disease Response of This Target Gene | [107] | |||
| Responsed Disease | Gastrointestinal stromal tumor [ICD-11: 2B5B] | |||
| Responsed Drug | Spautin 1 | Preclinical | ||
| Target Regulation | Up regulation | |||
In-vitro Model |
GIST-T1 | Gastrointestinal stromal tumor | Homo sapiens | CVCL_4976 |
| GIST882 | Gastrointestinal stromal tumor | Homo sapiens | CVCL_7044 | |
| GIST882 | Gastrointestinal stromal tumor | Homo sapiens | CVCL_7044 | |
| GIST-T1 | Gastrointestinal stromal tumor | Homo sapiens | CVCL_4976 | |
| In-vivo Model | 6 × 106 logarithmically growing GIST cells resuspended in 100 μL PBS were injected subcutaneously into the flank of 6-week-old female nude mice for tumor growth assays. The following treatments were initiated when the tumor volume reached approximately 300 mm3. (1) IM-sensitive GIST cell lines and IM-resistant GIST cell lines were used to establish a subcutaneous xenograft model of GIST (n = 6 mice/group). When the tumors grew to the required size, mice were treated with IM via drinking water; (2) GIST cells were pretreated with USP13 lentivirus or USP13 control lentivirus. In addition, IM-resistant GIST cells were pretreated with USP13 inhibitors or control. These lentivirus-transfected cells were implanted into subcutaneous mice tumors to construct the USP13 positive, USP13-negative, and control groups (n = 6 mice/group). When the transplanted tumor grew to the required volume, mice in each group were treated with IM via drinking water. (3) IM-resistant GIST cells-xenografted mice and control mice were treated with USP13 inhibitors or with USP13 inhibitors and autophagy inhibitors in combination therapy to assess reversal of IM resistance (n = 6 mice/group). Mice were treated with IM (45 mg/kg/day) and with or without 3-MA (15 mg/kg/day) via drinking water when the tumors grew to the required size. The USP13 inhibitor Spautin-1 was used at 20 mg/kg/day when required. | |||
Zinc finger protein 281 (ZNF281)
Breast cancer [ICD-11: 2C60]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [108] | |||
| Responsed Disease | Breast cancer [ICD-11: 2C60] | |||
| Responsed Drug | RS-102895 | Investigative | ||
| Target Regulation | Up regulation | |||
In-vitro Model |
MDA-MB-231 | Breast adenocarcinoma | Homo sapiens | CVCL_0062 |
| In-vivo Model | MDA-MB-231 cells (1×106 cells/mouse) were mixed with an equivalent number of NFs/Ctrl, CAFs/shNC, CAFs/sh lncSNHG5, CAFs/shZNF281, CAFs/sh lncSNHG5/ZNF281, CAFs/RS102895, CAFs/Maraviroc or CAFs/Cenicriviroc in 200 μl PBS:Matrigel at a ratio of 1:1. | |||
Zinc finger protein RFP (TRIM27)
Psoriasis [ICD-11: EA90]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [109] | |||
| Responsed Disease | Psoriasis [ICD-11: EA90] | |||
In-vitro Model |
HaCaT | Normal | Homo sapiens | CVCL_0038 |
Unspecific Target Gene
Pancreatic cancer [ICD-11: 2C10]
| In total 2 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [110] | |||
| Responsed Disease | Pancreatic cancer [ICD-11: 2C10] | |||
| Responsed Drug | Gemcitabine | Approved | ||
| Pathway Response | Adipocytokine signaling pathway | hsa04920 | ||
| Cell Process | Epithelial-mesenchymal transition | |||
In-vitro Model |
BxPC-3 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0186 |
| HDE-CT cell line (A normal human pancreatic cell line) | ||||
| MIA PaCa-2 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0428 | |
| Response Summary | Lasso regression identified a six-m6A-regulator-signature prognostic model (KIAA1429, HNRNPC, METTL3, YTHDF1, IGF2BP2, and IGF2BP3). Gene set enrichment analysis revealed m6A regulators (KIAA1429, HNRNPC, and IGF2BP2) were related to multiple biological behaviors in pancreatic cancer, including adipocytokine signaling, the well vs. poorly differentiated tumor pathway, tumor metastasis pathway, epithelial mesenchymal transition pathway, gemcitabine resistance pathway, and stemness pathway. | |||
| Experiment 2 Reporting the m6A-centered Disease Response of This Target Gene | [111] | |||
| Responsed Disease | Pancreatic ductal adenocarcinoma [ICD-11: 2C10.0] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
PANC-1 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0480 |
| SW1990 | Pancreatic adenocarcinoma | Homo sapiens | CVCL_1723 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| In-vivo Model | 0.1 ml of cell suspension containing 2 × 106 luciferase-labeled cells was injected into tail veins. | |||
Liver cancer [ICD-11: 2C12]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [104] | |||
| Responsed Disease | Intrahepatic cholangiocarcinoma [ICD-11: 2C12.10] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
SK-ChA-1 | Cholangiocarcinoma | Homo sapiens | CVCL_6952 |
| RBE | Intrahepatic cholangiocarcinoma | Homo sapiens | CVCL_4896 | |
| Mz-ChA-1 | Gallbladder carcinoma | Homo sapiens | CVCL_6932 | |
| HEK293 | Normal | Homo sapiens | CVCL_0045 | |
| In-vivo Model | Six-week-old male BALB/c nude mice were maintained under specific pathogen-free conditions,Mice were randomly assigned to 2 groups (N = 6). In each group, lentiviral-transduced SK-Cha-1 cells (2.5 × 106) were subcutaneously injected into the dorsal right flanks of the mice, and the mice were monitored every 3 days for tumor growth. | |||
| Response Summary | The combination of m6A writers, IGF2BP2, and CTNNB1 distinguished CCA tissues from normal tissues. | |||
ATP-dependent translocase ABCB1 (ABCB1)
Adriamycin
[Phase 3]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [10] | |||
| Responsed Disease | Breast cancer | ICD-11: 2C60 | ||
| Target Regulation | Up regulation | |||
| In-vitro Model | MCF-7 | Invasive breast carcinoma | Homo sapiens | CVCL_0031 |
| MDA-MB-231 | Breast adenocarcinoma | Homo sapiens | CVCL_0062 | |
| In-vivo Model | The xenograft mouse models were established by injecting MCF-7/ADR cells (1 × 107 in 100 μL RPMI 1640 medium) into the mouse right flank. Tumor size was monitored every week. When the average tumor size reached approximately 100 mm3, 5.0 mg/kg adriamycin were subsequently subjected through tail vein every other day. Mice were sacrificed after 4 weeks, and tumors were excised. | |||
Receptor tyrosine-protein kinase erbB-2 (ERBB2)
Temozolomide
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [39] | |||
| Responsed Disease | Colorectal cancer | ICD-11: 2B91 | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Hippo signaling pathway | hsa04390 | ||
| Cell Process | Cell apoptosis | |||
| In-vitro Model | HCT 8 | Colon adenocarcinoma | Homo sapiens | CVCL_2478 |
| SW480 | Colon adenocarcinoma | Homo sapiens | CVCL_0546 | |
| SW620 | Colon adenocarcinoma | Homo sapiens | CVCL_0547 | |
| In-vivo Model | IGF2BP2 activates the expression of ErbB2 by recognizing the m6A of YAP, thus affecting the cell cycle of CRC, inhibiting cell apoptosis, and promoting proliferation. | |||
| Response Summary | IGF2BP2 activates the expression of Receptor tyrosine-protein kinase erbB-2 (ERBB2) by recognizing the m6A of YAP, thus affecting the cell cycle of colorectal cancer, inhibiting cell apoptosis, and promoting proliferation. | |||
Transcriptional coactivator YAP1 (YAP1)
Temozolomide
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [39] | |||
| Responsed Disease | Colorectal cancer | ICD-11: 2B91 | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Hippo signaling pathway | hsa04390 | ||
| Cell Process | Cell apoptosis | |||
| In-vitro Model | HCT 8 | Colon adenocarcinoma | Homo sapiens | CVCL_2478 |
| SW480 | Colon adenocarcinoma | Homo sapiens | CVCL_0546 | |
| SW620 | Colon adenocarcinoma | Homo sapiens | CVCL_0547 | |
| In-vivo Model | IGF2BP2 activates the expression of ErbB2 by recognizing the m6A of YAP, thus affecting the cell cycle of CRC, inhibiting cell apoptosis, and promoting proliferation. | |||
| Response Summary | IGF2BP2 activates the expression of ErbB2 by recognizing the m6A of Transcriptional coactivator YAP1 (YAP1), thus affecting the cell cycle of colorectal cancer, inhibiting cell apoptosis, and promoting proliferation. | |||
Trans-3,5,4'-trimethoxystilbene
[Investigative]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [46] | |||
| Responsed Disease | Non-small cell lung cancer | ICD-11: 2C25.Y | ||
| Target Regulation | Up regulation | |||
| In-vitro Model | 16HBE14o- | Normal | Homo sapiens | CVCL_0112 |
| PC-9 | Lung adenocarcinoma | Homo sapiens | CVCL_B260 | |
| NCI-H460 | Lung large cell carcinoma | Homo sapiens | CVCL_0459 | |
| NCI-H1299 | Lung large cell carcinoma | Homo sapiens | CVCL_0060 | |
| NCI-H1975 | Lung adenocarcinoma | Homo sapiens | CVCL_1511 | |
| A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 | |
Ena/VASP-like protein (EVL)
Isoforsythiaside
[Investigative]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [72] | |||
| Responsed Disease | Macroscopic changes of size of the kidney | ICD-11: MF54.0 | ||
| Target Regulation | Up regulation | |||
| In-vitro Model | HEK293 | Normal | Homo sapiens | CVCL_0045 |
| HK-2 [Human kidney] | Normal | Homo sapiens | CVCL_0302 | |
| In-vivo Model | For the unilateral ureteral obstruction (UUO)-induced ON model, mice were subjected to permanent ureteral ligation of the left kidney ureter under aseptic and anaesthetic conditions, whereas sham-operated mice merely lacked ligation under equivalent conditions. The mice were sacrificed under anaesthesia at 3, 7 and 14 days postoperatively. For the ischemia/reperfusion (I/R)-induced renal fibrosis model, mice were recovered with blood supply after 42 min of bilateral renal artery clamping under both aseptic and anaesthetic conditions. These mice were sacrificed under anaesthesia at 7 and 14 days postoperatively. For drug treatment, isoforsythiaside was injected intraperitoneally at concentrations of 10, 25 and 50 mg/kg per day when the model was established. Similarly, the sham-operated group received an equivalent volume of saline intraperitoneally as a control. The harvested kidneys have been primarily used for morphological and histopathological observations and molecular biology studies. | |||
GNAS antisense RNA 1 (GNAS-AS1)
Chidamide
[Investigative]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [81] | |||
| Responsed Disease | Acute myeloid leukaemia | ICD-11: 2A60 | ||
| In-vitro Model | HL-60 | Adult acute myeloid leukemia | Homo sapiens | CVCL_0002 |
| HEL | Erythroleukemia | Homo sapiens | CVCL_0001 | |
| MOLM-13 | Adult acute myeloid leukemia | Homo sapiens | CVCL_2119 | |
| MV4-11 | Childhood acute monocytic leukemia | Homo sapiens | CVCL_0064 | |
| Kasumi-1 | Myeloid leukemia with maturation | Homo sapiens | CVCL_0589 | |
|
HS-5
|
N.A. | Homo sapiens | CVCL_3720 | |
| In-vivo Model | Subcutaneous tumorigenesis was induced in mice by subcutaneous transplantation of 1 × 106 HL-60 cells. To stably overexpress GNAS-AS1, lentivirus-packaged OE-GNAS-AS1 (1 × 108 plaque-forming units). When the tumor volume reached 150-200 mm3, mice in the control group were orally treated with 1% DMSO (containing 0.2% carboxymethylcellulose and 0.1% Tween 80). Mice in the Chidamide, Chidamide + OE-NC, and Chidamide + OE-GNAS-AS1 groups were orally treated with Chidamide (25 mg/kg body weight, formulated with 1% DMSO containing 0.2% carboxymethylcellulose and 0.1% Tween 80). All treatments were repeated three times a week and for two weeks. Tumor volume was recorded every 5 days. | |||
Ubiquitin carboxyl-terminal hydrolase 13 (USP13)
3-Methyladenine
[Investigative]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [107] | |||
| Responsed Disease | Gastrointestinal stromal tumor | ICD-11: 2B5B | ||
| Target Regulation | Up regulation | |||
| In-vitro Model | GIST-T1 | Gastrointestinal stromal tumor | Homo sapiens | CVCL_4976 |
| GIST882 | Gastrointestinal stromal tumor | Homo sapiens | CVCL_7044 | |
| GIST882 | Gastrointestinal stromal tumor | Homo sapiens | CVCL_7044 | |
| GIST-T1 | Gastrointestinal stromal tumor | Homo sapiens | CVCL_4976 | |
| In-vivo Model | 6 × 106 logarithmically growing GIST cells resuspended in 100 μL PBS were injected subcutaneously into the flank of 6-week-old female nude mice for tumor growth assays. The following treatments were initiated when the tumor volume reached approximately 300 mm3. (1) IM-sensitive GIST cell lines and IM-resistant GIST cell lines were used to establish a subcutaneous xenograft model of GIST (n = 6 mice/group). When the tumors grew to the required size, mice were treated with IM via drinking water; (2) GIST cells were pretreated with USP13 lentivirus or USP13 control lentivirus. In addition, IM-resistant GIST cells were pretreated with USP13 inhibitors or control. These lentivirus-transfected cells were implanted into subcutaneous mice tumors to construct the USP13 positive, USP13-negative, and control groups (n = 6 mice/group). When the transplanted tumor grew to the required volume, mice in each group were treated with IM via drinking water. (3) IM-resistant GIST cells-xenografted mice and control mice were treated with USP13 inhibitors or with USP13 inhibitors and autophagy inhibitors in combination therapy to assess reversal of IM resistance (n = 6 mice/group). Mice were treated with IM (45 mg/kg/day) and with or without 3-MA (15 mg/kg/day) via drinking water when the tumors grew to the required size. The USP13 inhibitor Spautin-1 was used at 20 mg/kg/day when required. | |||
Spautin 1
[Preclinical]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [107] | |||
| Responsed Disease | Gastrointestinal stromal tumor | ICD-11: 2B5B | ||
| Target Regulation | Up regulation | |||
| In-vitro Model | GIST-T1 | Gastrointestinal stromal tumor | Homo sapiens | CVCL_4976 |
| GIST882 | Gastrointestinal stromal tumor | Homo sapiens | CVCL_7044 | |
| GIST882 | Gastrointestinal stromal tumor | Homo sapiens | CVCL_7044 | |
| GIST-T1 | Gastrointestinal stromal tumor | Homo sapiens | CVCL_4976 | |
| In-vivo Model | 6 × 106 logarithmically growing GIST cells resuspended in 100 μL PBS were injected subcutaneously into the flank of 6-week-old female nude mice for tumor growth assays. The following treatments were initiated when the tumor volume reached approximately 300 mm3. (1) IM-sensitive GIST cell lines and IM-resistant GIST cell lines were used to establish a subcutaneous xenograft model of GIST (n = 6 mice/group). When the tumors grew to the required size, mice were treated with IM via drinking water; (2) GIST cells were pretreated with USP13 lentivirus or USP13 control lentivirus. In addition, IM-resistant GIST cells were pretreated with USP13 inhibitors or control. These lentivirus-transfected cells were implanted into subcutaneous mice tumors to construct the USP13 positive, USP13-negative, and control groups (n = 6 mice/group). When the transplanted tumor grew to the required volume, mice in each group were treated with IM via drinking water. (3) IM-resistant GIST cells-xenografted mice and control mice were treated with USP13 inhibitors or with USP13 inhibitors and autophagy inhibitors in combination therapy to assess reversal of IM resistance (n = 6 mice/group). Mice were treated with IM (45 mg/kg/day) and with or without 3-MA (15 mg/kg/day) via drinking water when the tumors grew to the required size. The USP13 inhibitor Spautin-1 was used at 20 mg/kg/day when required. | |||
Zinc finger protein 281 (ZNF281)
RS-102895
[Investigative]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [108] | |||
| Responsed Disease | Breast cancer | ICD-11: 2C60 | ||
| Target Regulation | Up regulation | |||
| In-vitro Model | MDA-MB-231 | Breast adenocarcinoma | Homo sapiens | CVCL_0062 |
| In-vivo Model | MDA-MB-231 cells (1×106 cells/mouse) were mixed with an equivalent number of NFs/Ctrl, CAFs/shNC, CAFs/sh lncSNHG5, CAFs/shZNF281, CAFs/sh lncSNHG5/ZNF281, CAFs/RS102895, CAFs/Maraviroc or CAFs/Cenicriviroc in 200 μl PBS:Matrigel at a ratio of 1:1. | |||
Unspecific Target Gene
Gemcitabine
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [110] | |||
| Responsed Disease | Pancreatic cancer | ICD-11: 2C10 | ||
| Pathway Response | Adipocytokine signaling pathway | hsa04920 | ||
| Cell Process | Epithelial-mesenchymal transition | |||
| In-vitro Model | BxPC-3 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0186 |
| HDE-CT cell line (A normal human pancreatic cell line) | ||||
| MIA PaCa-2 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0428 | |
| Response Summary | Lasso regression identified a six-m6A-regulator-signature prognostic model (KIAA1429, HNRNPC, METTL3, YTHDF1, IGF2BP2, and IGF2BP3). Gene set enrichment analysis revealed m6A regulators (KIAA1429, HNRNPC, and IGF2BP2) were related to multiple biological behaviors in pancreatic cancer, including adipocytokine signaling, the well vs. poorly differentiated tumor pathway, tumor metastasis pathway, epithelial mesenchymal transition pathway, gemcitabine resistance pathway, and stemness pathway. | |||
Full List of Crosstalk(s) between m6A Modification and Epigenetic Regulation Related to This Regulator
RNA modification
m6A Target: Nuclear transcription factor Y subunit alpha (NFYA)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT00058 | ||
| Epigenetic Regulator | rRNA 2'-O-methyltransferase fibrillarin (FBL) | |
| Regulated Target | Insulin like growth factor 2 mRNA binding protein 2 (IGF2BP2) | |
| Crosstalk relationship | 2'-O-methylation → m6A | |
| Disease | Ovarian cancer | |
m6A Target: Glucose transporter type 1 (GLUT1)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT00489 | ||
| Epigenetic Regulator | Fat mass and obesity-associated protein (FTO) | |
| Regulated Target | hsa-mir-22 | |
| Crosstalk relationship | m6Am → m6A | |
m6A Target: Hypoxia-inducible factor 1-alpha (HIF-1-Alpha/HIF1A)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT00525 | ||
| Epigenetic Regulator | Double-stranded RNA-specific editase 1 (ADARB1) | |
| Regulated Target | MicroRNA 142 (MIR142) | |
| Crosstalk relationship | A-to-I → m6A | |
| Crosstalk ID: M6ACROT00529 | ||
| Epigenetic Regulator | Fat mass and obesity-associated protein (FTO) | |
| Regulated Target | MicroRNA 576 (MIR576) | |
| Crosstalk relationship | m6Am → m6A | |
m6A Target: Insulin-like growth factor 2 (IGF2)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT00539 | ||
| Epigenetic Regulator | Interferon-inducible protein 4 (ADAR1) | |
| Regulated Target | MicroRNA 432 (MIR432) | |
| Crosstalk relationship | A-to-I → m6A | |
m6A Target: Fascin (FSCN1)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT00546 | ||
| Epigenetic Regulator | Interferon-inducible protein 4 (ADAR1) | |
| Regulated Target | MicroRNA 200b (MIR200B) | |
| Crosstalk relationship | A-to-I → m6A | |
| Crosstalk ID: M6ACROT00547 | ||
| Epigenetic Regulator | Y-box-binding protein 1 (YBX1) | |
| Regulated Target | MicroRNA 200b (MIR200B) | |
| Crosstalk relationship | m5C → m6A | |
m6A Target: Transcription factor SOX-6 (SOX6)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT00552 | ||
| Epigenetic Regulator | Double-stranded RNA-specific editase 1 (ADARB1) | |
| Regulated Target | MicroRNA 122 (MIR122) | |
| Crosstalk relationship | A-to-I → m6A | |
m6A Target: Differentiation antagonizing non-protein coding RNA (DANCR)
| In total 3 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT00559 | ||
| Epigenetic Regulator | Interferon-inducible protein 4 (ADAR1) | |
| Regulated Target | Transcription factor SOX-2 (SOX2) | |
| Crosstalk relationship | m6A → A-to-I | |
| Crosstalk ID: M6ACROT00561 | ||
| Epigenetic Regulator | Fat mass and obesity-associated protein (FTO) | |
| Regulated Target | Transcription factor SOX-2 (SOX2) | |
| Crosstalk relationship | m6A → m6Am | |
| Crosstalk ID: M6ACROT00563 | ||
| Epigenetic Regulator | Methyltransferase-like protein 1 (METTL1) | |
| Regulated Target | Transcription factor SOX-2 (SOX2) | |
| Crosstalk relationship | m6A → m7G | |
DNA modification
m6A Target: Semaphorin-3F (SEMA3F)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT02016 | ||
| Epigenetic Regulator | DNA (cytosine-5)-methyltransferase 1 (DNMT1) | |
| Regulated Target | Semaphorin-3F (SEMA3F) | |
| Crosstalk relationship | m6A → DNA modification | |
| Disease | Prostate cancer | |
| Drug | Docetaxel | |
Histone modification
m6A Target: Fructose-bisphosphate aldolase A (ALDOA)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03079 | ||
| Regulated Target | Histone H3 lysine 18 lactylation (H3K18la) | |
| Crosstalk relationship | m6A → Histone modification | |
| Disease | Hepatic fibrosis/cirrhosis | |
m6A Target: Krueppel-like factor 12 (KLF12)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03080 | ||
| Epigenetic Regulator | Histone acetyltransferase p300 (P300) | |
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | m6A → Histone modification | |
| Disease | Pancreatic ductal adenocarcinoma | |
| Crosstalk ID: M6ACROT03475 | ||
| Epigenetic Regulator | Histone acetyltransferase p300 (P300) | |
| Regulated Target | Histone H3 lysine 18 lactylation (H3K18la) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Pancreatic cancer | |
m6A Target: JmjC domain-containing protein 8 (JMJD8)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03102 | ||
| Epigenetic Regulator | Histone deacetylase 2 (HDAC2) | |
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
m6A Target: Large neutral amino acids transporter small subunit 1 (SLC7A5)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03128 | ||
| Epigenetic Regulator | Histone-lysine N-methyltransferase SETD1A (SETD1A) | |
| Regulated Target | Histone H3 lysine 4 trimethylation (H3K4me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Lung cancer | |
m6A Target: Macrophage colony-stimulating factor 1 (CSF1)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03132 | ||
| Epigenetic Regulator | Histone acetyltransferase p300 (P300) | |
| Regulated Target | Histone H3 lysine 18 lactylation (H3K18la) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Pancreatic cancer | |
| Crosstalk ID: M6ACROT06041 | ||
| Epigenetic Regulator | Histone acetyltransferase p300 (P300) | |
| Regulated Target | Histone H3 lysine 18 lactylation (H3K18la) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Pancreatic cancer | |
| Drug | Gemcitabine | |
m6A Target: Myc proto-oncogene protein (MYC)
| In total 4 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03133 | ||
| Epigenetic Regulator | Histone acetyltransferase p300 (P300) | |
| Regulated Target | Histone H3 lysine 18 lactylation (H3K18la) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Pancreatic cancer | |
| Crosstalk ID: M6ACROT03476 | ||
| Epigenetic Regulator | Histone acetyltransferase p300 (P300) | |
| Regulated Target | Histone H3 lysine 18 lactylation (H3K18la) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Pancreatic cancer | |
| Crosstalk ID: M6ACROT03477 | ||
| Epigenetic Regulator | Histone acetyltransferase p300 (P300) | |
| Regulated Target | Histone H3 lysine 18 lactylation (H3K18la) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Pancreatic cancer | |
| Crosstalk ID: M6ACROT06042 | ||
| Epigenetic Regulator | Histone acetyltransferase p300 (P300) | |
| Regulated Target | Histone H3 lysine 18 lactylation (H3K18la) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Pancreatic cancer | |
| Drug | Gemcitabine | |
m6A Target: Ankyrin repeat domain-containing protein 22 (ANKRD22)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03153 | ||
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Nasopharyngeal carcinoma | |
m6A Target: Histone-lysine N-methyltransferase SUV39H2 (SUV39H2)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03191 | ||
| Epigenetic Regulator | Histone-lysine N-methyltransferase SUV39H2 (SUV39H2) | |
| Regulated Target | Histone H3 lysine 9 trimethylation (H3K9me3) | |
| Crosstalk relationship | m6A → Histone modification | |
| Disease | Gastric cancer | |
| Drug | Cisplatin | |
m6A Target: Hexokinase-2 (HK2)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03367 | ||
| Epigenetic Regulator | Histone deacetylase 2 (HDAC2) | |
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
| Crosstalk ID: M6ACROT03368 | ||
| Epigenetic Regulator | Histone deacetylase 2 (HDAC2) | |
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Colorectal cancer | |
m6A Target: Vang-like protein 1 (VANGL1)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03465 | ||
| Epigenetic Regulator | Histone-lysine N-methyltransferase SETD1A (SETD1A) | |
| Regulated Target | Histone H3 lysine 4 trimethylation (H3K4me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Lung cancer | |
m6A Target: HLA complex group 11 (HCG11)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03466 | ||
| Epigenetic Regulator | Histone-lysine N-methyltransferase SETD1A (SETD1A) | |
| Regulated Target | Histone H3 lysine 4 trimethylation (H3K4me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Lung cancer | |
m6A Target: Thymidine kinase, cytosolic (TK1)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03467 | ||
| Epigenetic Regulator | Histone-lysine N-methyltransferase SETD1A (SETD1A) | |
| Regulated Target | Histone H3 lysine 4 trimethylation (H3K4me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Lung cancer | |
m6A Target: AC026356.1
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03468 | ||
| Epigenetic Regulator | Histone-lysine N-methyltransferase SETD1A (SETD1A) | |
| Regulated Target | Histone H3 lysine 4 trimethylation (H3K4me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Lung cancer | |
m6A Target: Putative uncharacterized protein DANCR (DANCR)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03473 | ||
| Epigenetic Regulator | Histone acetyltransferase p300 (P300) | |
| Regulated Target | Histone H3 lysine 18 lactylation (H3K18la) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Pancreatic cancer | |
m6A Target: PTGS2 antisense RNA 1 (PACERR)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03474 | ||
| Epigenetic Regulator | Histone acetyltransferase p300 (P300) | |
| Regulated Target | Histone H3 lysine 18 lactylation (H3K18la) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Pancreatic cancer | |
m6A Target: Beta-1,4-glucuronyltransferase 1 (B3GNT6)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03478 | ||
| Epigenetic Regulator | Histone acetyltransferase p300 (P300) | |
| Regulated Target | Histone H3 lysine 18 lactylation (H3K18la) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Pancreatic cancer | |
m6A Target: Programmed cell death 1 ligand 1 (CD274/PD-L1)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03479 | ||
| Epigenetic Regulator | Histone acetyltransferase p300 (P300) | |
| Regulated Target | Histone H3 lysine 18 lactylation (H3K18la) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Pancreatic cancer | |
m6A Target: long intergenic non-protein coding RNA 941 (LINC00941)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03480 | ||
| Epigenetic Regulator | Histone acetyltransferase p300 (P300) | |
| Regulated Target | Histone H3 lysine 18 lactylation (H3K18la) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Pancreatic cancer | |
Non-coding RNA
m6A Target: 5-hydroxytryptamine receptor 3A (HTR3A)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05021 | ||
| Epigenetic Regulator | Long intergenic non-protein coding RNA 1305 (LINC01305) | |
| Regulated Target | Insulin like growth factor 2 mRNA binding protein 2 (IGF2BP2) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Esophageal Squamous Cell Carcinoma | |
m6A Target: Forkhead box protein M1 (FOXM1)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05027 | ||
| Epigenetic Regulator | LINC00035 (ABHD11-AS1) | |
| Regulated Target | Insulin like growth factor 2 mRNA binding protein 2 (IGF2BP2) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Colorectal cancer | |
m6A Target: Myc proto-oncogene protein (MYC)
| In total 3 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05055 | ||
| Epigenetic Regulator | Long intergenic non-protein coding RNA 920 (LINC00920) | |
| Regulated Target | Insulin like growth factor 2 mRNA binding protein 2 (IGF2BP2) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Colorectal cancer | |
| Crosstalk ID: M6ACROT05319 | ||
| Epigenetic Regulator | PTGS2 antisense RNA 1 (PACERR) | |
| Regulated Target | Insulin like growth factor 2 mRNA binding protein 2 (IGF2BP2) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Pancreatic ductal adenocarcinoma | |
| Crosstalk ID: M6ACROT05950 | ||
| Epigenetic Regulator | Long intergenic non-protein coding RNA 901 (LINC00901) | |
| Regulated Target | Insulin like growth factor 2 mRNA binding protein 2 (IGF2BP2) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | pancreatic cancer | |
m6A Target: Cellular tumor antigen p53 (TP53/p53)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05064 | ||
| Epigenetic Regulator | Antisense of IGF2R non-protein coding RNA (AIRN) | |
| Regulated Target | Insulin like growth factor 2 mRNA binding protein 2 (IGF2BP2) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Vascular disorders of the liver | |
m6A Target: Transcription factor E2F6 (E2F6)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05066 | ||
| Epigenetic Regulator | Read-through Circular RNA E2F | |
| Regulated Target | Insulin like growth factor 2 mRNA binding protein 2 (IGF2BP2) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Liver cancer | |
m6A Target: Transcription factor E2F3 (E2F3)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05067 | ||
| Epigenetic Regulator | Read-through Circular RNA E2F | |
| Regulated Target | Insulin like growth factor 2 mRNA binding protein 2 (IGF2BP2) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Liver cancer | |
m6A Target: Homeobox protein MSX-1 (MSX1)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05070 | ||
| Epigenetic Regulator | P53 upregulated regulator of p53 levels (PURPL) | |
| Regulated Target | Insulin like growth factor 2 mRNA binding protein 2 (IGF2BP2) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Colorectal cancer | |
m6A Target: Protein Jumonji (JARID2)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05071 | ||
| Epigenetic Regulator | P53 upregulated regulator of p53 levels (PURPL) | |
| Regulated Target | Insulin like growth factor 2 mRNA binding protein 2 (IGF2BP2) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Colorectal cancer | |
m6A Target: Transcriptional coactivator YAP1 (YAP1)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05091 | ||
| Epigenetic Regulator | piR-31115 | |
| Regulated Target | Methyltransferase-like protein 3 (METTL3) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Triple-negative breast cancer | |
| Crosstalk ID: M6ACROT05190 | ||
| Epigenetic Regulator | Circ_PACRGL | |
| Regulated Target | Insulin like growth factor 2 mRNA binding protein 2 (IGF2BP2) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Non-small cell lung cancer | |
| Drug | Trans-3,5,4'-trimethoxystilbene | |
m6A Target: G1/S-specific cyclin-D1 (CCND1)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05098 | ||
| Epigenetic Regulator | HNF1A antisense RNA 1 (HNF1A-AS1) | |
| Regulated Target | Insulin like growth factor 2 mRNA binding protein 2 (IGF2BP2) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Colorectal cancer | |
m6A Target: Cell division control protein 6 homolog (CDC6)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05109 | ||
| Epigenetic Regulator | Lysocardiolipin acyltransferase 1 (LCLAT1) | |
| Regulated Target | Insulin like growth factor 2 mRNA binding protein 2 (IGF2BP2) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Lung cancer | |
m6A Target: Hexokinase-2 (HK2)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05158 | ||
| Epigenetic Regulator | MIR4458 host gene (MIR4458HG) | |
| Regulated Target | Insulin like growth factor 2 mRNA binding protein 2 (IGF2BP2) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Liver cancer | |
m6A Target: Glucose transporter type 1 (SLC2A1)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05159 | ||
| Epigenetic Regulator | MIR4458 host gene (MIR4458HG) | |
| Regulated Target | Insulin like growth factor 2 mRNA binding protein 2 (IGF2BP2) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Liver cancer | |
m6A Target: Serine/threonine-protein phosphatase CPPED1 (CPPED1)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05176 | ||
| Epigenetic Regulator | Circ_CDK1 | |
| Regulated Target | Insulin like growth factor 2 mRNA binding protein 2 (IGF2BP2) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Laryngeal squamous cell carcinoma | |
m6A Target: Zinc finger protein 281 (ZNF281)
| In total 3 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05197 | ||
| Epigenetic Regulator | Small nucleolar RNA host gene 5 (SNHG5) | |
| Regulated Target | Insulin like growth factor 2 mRNA binding protein 2 (IGF2BP2) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Breast cancer | |
| Drug | RS102895 | |
| Crosstalk ID: M6ACROT05993 | ||
| Epigenetic Regulator | Small nucleolar RNA host gene 5 (SNHG5) | |
| Regulated Target | Insulin like growth factor 2 mRNA binding protein 2 (IGF2BP2) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Breast cancer | |
| Drug | Marasviroc | |
| Crosstalk ID: M6ACROT05994 | ||
| Epigenetic Regulator | Small nucleolar RNA host gene 5 (SNHG5) | |
| Regulated Target | Insulin like growth factor 2 mRNA binding protein 2 (IGF2BP2) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Breast cancer | |
| Drug | Cenicriviroc | |
m6A Target: Cyclic AMP-responsive element-binding protein 1 (CREB1)
| In total 3 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05198 | ||
| Epigenetic Regulator | hsa_circ_0006357 (Circ_EZH2) | |
| Regulated Target | Insulin like growth factor 2 mRNA binding protein 2 (IGF2BP2) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Colorectal cancer | |
| Crosstalk ID: M6ACROT05199 | ||
| Epigenetic Regulator | hsa-miR-133b | |
| Regulated Target | Insulin like growth factor 2 mRNA binding protein 2 (IGF2BP2) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Colorectal cancer | |
| Crosstalk ID: M6ACROT05200 | ||
| Epigenetic Regulator | hsa_circ_0006357 (Circ_EZH2) | |
| Regulated Target | hsa-miR-133b | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Colorectal cancer | |
m6A Target: Kremen protein 1 (KRM1)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05206 | ||
| Epigenetic Regulator | Circ_HIPK3 | |
| Regulated Target | Insulin like growth factor 2 mRNA binding protein 2 (IGF2BP2) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Calcific aortic valve disease | |
m6A Target: Stimulator of interferon genes protein (STING1)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05222 | ||
| Epigenetic Regulator | Circ_ASPH | |
| Regulated Target | Insulin like growth factor 2 mRNA binding protein 2 (IGF2BP2) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Colorectal cancer | |
m6A Target: High mobility group protein HMG-I/HMG-Y (HMGA1)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05274 | ||
| Epigenetic Regulator | Long intergenic non-protein coding RNA 460 (LINC00460) | |
| Regulated Target | Insulin like growth factor 2 mRNA binding protein 2 (IGF2BP2) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Colorectal cancer | |
| Crosstalk ID: M6ACROT05372 | ||
| Epigenetic Regulator | Long intergenic non-protein coding RNA 460 (LINC00460) | |
| Regulated Target | Insulin like growth factor 2 mRNA binding protein 2 (IGF2BP2) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Colorectal cancer | |
m6A Target: GNAS antisense RNA 1 (GNAS-AS1)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05289 | ||
| Epigenetic Regulator | hsa-miR-34a-5p | |
| Regulated Target | Insulin like growth factor 2 mRNA binding protein 2 (IGF2BP2) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Breast cancer | |
| Drug | Tamoxifen | |
| Crosstalk ID: M6ACROT05291 | ||
| Epigenetic Regulator | GNAS antisense RNA 1 (GNAS-AS1) | |
| Regulated Target | hsa-miR-34a-5p | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Breast cancer | |
| Drug | Tamoxifen | |
m6A Target: Protein arginine N-methyltransferase 6 (PRMT6)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05290 | ||
| Epigenetic Regulator | hsa-miR-34a-5p | |
| Regulated Target | Insulin like growth factor 2 mRNA binding protein 2 (IGF2BP2) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Acute myeloid leukaemia | |
| Crosstalk ID: M6ACROT05292 | ||
| Epigenetic Regulator | GNAS antisense RNA 1 (GNAS-AS1) | |
| Regulated Target | hsa-miR-34a-5p | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Acute myeloid leukaemia | |
m6A Target: Insulin-like growth factor 1 receptor (IGF1R)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05294 | ||
| Epigenetic Regulator | Prostate cancer associated transcript 6 (PCAT6) | |
| Regulated Target | Insulin like growth factor 2 mRNA binding protein 2 (IGF2BP2) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Prostate cancer | |
m6A Target: ATP-dependent translocase ABCB1 (ABCB1)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05307 | ||
| Epigenetic Regulator | A1BG antisense RNA 1 (A1BG-AS1) | |
| Regulated Target | Insulin like growth factor 2 mRNA binding protein 2 (IGF2BP2) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Breast cancer | |
| Drug | Adriamycin | |
m6A Target: Krueppel-like factor 12 (KLF12)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05318 | ||
| Epigenetic Regulator | PTGS2 antisense RNA 1 (PACERR) | |
| Regulated Target | Insulin like growth factor 2 mRNA binding protein 2 (IGF2BP2) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Pancreatic ductal adenocarcinoma | |
m6A Target: Fibroblast growth factor receptor 2 (FGFR2)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05362 | ||
| Epigenetic Regulator | Ribonuclease P RNA component H1 (RPPH1) | |
| Regulated Target | Insulin like growth factor 2 mRNA binding protein 2 (IGF2BP2) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Breast cancer | |
m6A Target: Thymidine kinase, cytosolic (TK1)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05367 | ||
| Epigenetic Regulator | hsa-miR-320b | |
| Regulated Target | Insulin like growth factor 2 mRNA binding protein 2 (IGF2BP2) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Lung cancer | |
m6A Target: miR-133a
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05495 | ||
| Epigenetic Regulator | hsa-miR-133a-3p | |
| Regulated Target | Protein argonaute-2 (AGO2) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Cardiomegaly | |
m6A Target: HOXD antisense growth-associated long non-coding RNA (HAGLR)
| In total 3 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05501 | ||
| Epigenetic Regulator | HOXD antisense growth-associated long non-coding RNA (HAGLR) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Thyroid Cancer | |
| Crosstalk ID: M6ACROT05768 | ||
| Epigenetic Regulator | HOXD antisense growth-associated long non-coding RNA (HAGLR) | |
| Regulated Target | hsa-miR-106b-5p | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Diabetic retinopathy | |
| Crosstalk ID: M6ACROT05895 | ||
| Epigenetic Regulator | hsa-miR-106b-5p | |
| Regulated Target | Mutated in multiple advanced cancers 1 (PTEN) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Diabetic retinopathy | |
m6A Target: hsa_circ_0000231 (circ_ARHGAP12)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05505 | ||
| Epigenetic Regulator | hsa_circ_0000231 (Circ_ARHGAP12) | |
| Regulated Target | Forkhead box protein M1 (FOXM1) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Cervical cancer | |
m6A Target: HLA complex group 11 (HCG11)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05524 | ||
| Epigenetic Regulator | HLA complex group 11 (HCG11) | |
| Regulated Target | Serine/threonine-protein kinase LATS1 (LATS1) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Lung cancer | |
m6A Target: Cancer susceptibility 9 (CASC9)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05525 | ||
| Epigenetic Regulator | Cancer susceptibility 9 (CASC9) | |
| Regulated Target | Hexokinase-2 (HK2) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Brain cancer | |
m6A Target: ZNFX1 antisense RNA 1 (ZFAS1)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05537 | ||
| Epigenetic Regulator | ZNFX1 antisense RNA 1 (ZFAS1) | |
| Regulated Target | Obg-like ATPase 1 (OLA1) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Colorectal cancer | |
m6A Target: Prostate cancer associated transcript 6 (PCAT6)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05572 | ||
| Epigenetic Regulator | Prostate cancer associated transcript 6 (PCAT6) | |
| Regulated Target | Insulin like growth factor 2 mRNA binding protein 2 (IGF2BP2) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Prostate cancer | |
m6A Target: PTGS2 antisense RNA 1 (PACERR)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05574 | ||
| Epigenetic Regulator | PTGS2 antisense RNA 1 (PACERR) | |
| Regulated Target | Krueppel-like factor 12 (KLF12) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Pancreatic ductal adenocarcinoma | |
m6A Target: DIAPH1 antisense RNA 1 (DIAPH1-AS1)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05602 | ||
| Epigenetic Regulator | DIAPH1 antisense RNA 1 (DIAPH1-AS1) | |
| Regulated Target | LIM and SH3 domain protein 1 (LASP1) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Nasopharyngeal carcinoma | |
m6A Target: Metastasis associated lung adenocarcinoma transcript 1 (MALAT1)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05631 | ||
| Epigenetic Regulator | Metastasis associated lung adenocarcinoma transcript 1 (MALAT1) | |
| Regulated Target | Ubiquitin-like protein ATG12 (ATG12) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Non-small cell lung cancer | |
m6A Target: BACE1 antisense RNA (BACE1-AS)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05641 | ||
| Epigenetic Regulator | BACE1 antisense RNA (BACE1-AS) | |
| Regulated Target | hsa-miR-214-3p | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Colorectal cancer | |
m6A Target: Long intergenic non-protein coding RNA 941 (LINC00941)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05736 | ||
| Epigenetic Regulator | Long intergenic non-protein coding RNA 941 (LINC00941) | |
| Regulated Target | Insulin like growth factor 2 mRNA binding protein 2 (IGF2BP2) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Pancreatic cancer | |
m6A Target: AC026356.1
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05774 | ||
| Epigenetic Regulator | AC026356.1 | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Lung cancer | |
m6A Target: FBXL19 antisense RNA 1 (FBXL19-AS1)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05802 | ||
| Epigenetic Regulator | FBXL19 antisense RNA 1 (FBXL19-AS1) | |
| Regulated Target | Zinc finger protein 765 (ZNF765) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Glioma | |
| Drug | DOX | |
| Crosstalk ID: M6ACROT05885 | ||
| Epigenetic Regulator | FBXL19 antisense RNA 1 (FBXL19-AS1) | |
| Regulated Target | Zinc finger protein 765 (ZNF765) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Glioma | |
| Drug | DOX | |
m6A Target: OIP5 antisense RNA 1 (OIP5-AS1)
| In total 3 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05812 | ||
| Epigenetic Regulator | OIP5 antisense RNA 1 (OIP5-AS1) | |
| Regulated Target | hsa-miR-495-3p | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Glioma | |
| Crosstalk ID: M6ACROT05875 | ||
| Epigenetic Regulator | hsa-miR-495-3p | |
| Regulated Target | Hypoxia-inducible factor 1-alpha (HIF-1-Alpha/HIF1A) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Glioma | |
| Crosstalk ID: M6ACROT05876 | ||
| Epigenetic Regulator | hsa-miR-495-3p | |
| Regulated Target | Matrix metalloproteinase-14 (MMP14) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Glioma | |
m6A Target: Urothelial cancer associated 1 (UCA1)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05817 | ||
| Epigenetic Regulator | Urothelial cancer associated 1 (UCA1) | |
| Crosstalk relationship | m6A → ncRNA | |
m6A Target: Cell division cycle and apoptosis regulator protein 1 (CCAR1)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05918 | ||
| Epigenetic Regulator | Circ_ABCC4 | |
| Regulated Target | Insulin like growth factor 2 mRNA binding protein 2 (IGF2BP2) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Prostate cancer | |
m6A Target: Transcription factor SOX-2 (SOX2)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05921 | ||
| Epigenetic Regulator | Circ_EPHB4 | |
| Regulated Target | Insulin like growth factor 2 mRNA binding protein 2 (IGF2BP2) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Brain cancer | |
m6A Target: Programmed cell death 1 ligand 1 (CD274/PD-L1)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05923 | ||
| Epigenetic Regulator | hsa_circ_0058493 (Circ_RHBDD1) | |
| Regulated Target | Insulin like growth factor 2 mRNA binding protein 2 (IGF2BP2) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Gastric cancer | |
Xenobiotics Compound(s) Regulating the m6A Methylation Regulator
| Compound Name | Lapatinib | Investigative |
|---|---|---|
| Synonyms |
Lapatinib; 231277-92-2; Lapatinib Ditosylate; Tykerb; GW572016; Lapatinib base; GW 572016; Tyverb; 388082-78-8; Lapatinib free base; UNII-0VUA21238F; Lapatinib (free base); 231277-92-2 (free base); GSK572016; MFCD09264194; FMM; N-(3-chloro-4-((3-fluorobenzyl)oxy)phenyl)-6-(5-(((2-(methylsulfonyl)ethyl)amino)methyl)furan-2-yl)quinazolin-4-amine; N-{3-CHLORO-4-[(3-FLUOROBENZYL)OXY]PHENYL}-6-[5-({[2-(METHYLSULFONYL)ETHYL]AMINO}METHYL)-2-FURYL]-4-QUINAZOLINAMINE; N-{3-chloro-4-[(3-fluorophenyl)methoxy]phenyl}-6-(5-{[(2-methanesulfonylethyl)amino]methyl}furan-2-yl)quinazolin-4-amine; CHEMBL554; N-[3-chloro-4-[(3-fluorophenyl)methoxy]phenyl]-6-[5-[(2-methylsulfonylethylamino)methyl]furan-2-yl]quinazolin-4-amine; CHEBI:49603; 0VUA21238F; NSC745750; GW-572016; NCGC00167507-01; C29H26ClFN4O4S; DSSTox_CID_26675; DSSTox_RID_81812; DSSTox_GSID_46675; 1210608-87-9; Lapatinib (INN); 4-Quinazolinamine, N-(3-chloro-4-((3-fluorophenyl)methoxy)phenyl)-6-(5-(((2-(methylsulfonyl)ethyl)amino)methyl)-2-furanyl)-; 4-Quinazolinamine, N-[3-chloro-4-[(3-fluorophenyl)methoxy]phenyl]-6-[5-[[[2-(methylsulfonyl)ethyl]amino]methyl]-2-furanyl]-; N-(3-chloro-4-((3-fluorophenyl)methoxy)phenyl)-6-(5-(((2-(methylsulfonyl)ethyl)amino)methyl)-2-furanyl)-4-quinazolinamine; N-(3-chloro-4-(3-fluorobenzyloxy)phenyl)-6-(5-((2-(methylsulfonyl)ethylamino)methyl)furan-2-yl)quinazolin-4-amine; n-[3-chloro-4-[(3-fluorophenyl)methoxy]phenyl]-6-[5-[[[2-(methylsulfonyl)ethyl]amino]methyl]-2-furanyl]-4-quinazolinamine; N-{3-chloro-4-[(3-fluorobenzyl)oxy]phenyl}-6-[5-({[2-(methylsulfonyl)ethyl]amino}methyl)furan-2-yl]quinazolin-4-amine; GSK 572016; CAS-231277-92-2; GW-2016; Lapatinib [INN:BAN]; GW 282974X; HSDB 8209; 1xkk; N-[3-Chloro-4-[(3-fluorobenzyl)oxy]phenyl]-6-[5-({[2-(methanesulphonyl)ethyl]amino}methyl)-2-furyl]-4-quinazolinamine; N-{3-Chloro-4-[(3-fluorobenzyl)oxy]phenyl}-6-[5-({[2-(methanesulphonyl)ethyl]amino}methyl)-2-furyl]-4-quinazolinamine; Lapatinib, Free base; Lapatinib 13C-D7; nchembio866-comp20; Kinome_3684; Kinome_3685; Lapatinib base- Bio-X; SCHEMBL8100; Lapatinib (GW572016); BDBM5445; cid_208908; GTPL5692; QCR-63; DTXSID7046675; AOB5254; EX-A402; SYN1052; BCPP000188; BCPP000189; HMS2089H10; HMS3244N06; HMS3244N10; HMS3244N14; HMS3744K11; Tykerb (TN) (Glaxo Smith Kline); BCP01874; ZINC1550477; Tox21_112505; NSC800780; AKOS005145766; Tox21_112505_1; AC-1314; BCP9000837; BCP9000838; CCG-270133; CM14421; DB01259; GSK-572016; GW-572016X; NSC-745750; NSC-800780; SB16918; NCGC00167507-02; NCGC00167507-03; NCGC00167507-04; NCGC00167507-09; 913989-15-8; AS-14065; BC164610; HY-50898; N-(3-Chloro-4-((3-fluorophenyl)methoxy)phenyl)-6-(5-((2-methylsulfonylethylamino)methyl)-2-furyl)quinazolin-4-amine; N-(3-Chloro-4-{[(3-fluorophenyl)methyl]oxy}phenyl)-6-[5-({[2-(methylsulfonyl)ethyl]amino}methyl)-2-furanyl]-4-quinazolinamine; Q570; AM20090641; FT-0659650; SW199101-5; A25184; D08108; J90020; AB01273965-01; AB01273965-02; AB01273965-03; AB01273965_04; AB01273965_05; 277L922; Q420323; Q-101353; SR-05000001472-1; BRD-K19687926-001-01-7; BRD-K19687926-379-02-5; GW572016;GW-572016;GW 572016; 1092929-10-6; GW-2016;N-(3-chloro-4-((3-fluorobenzyl)oxy)phenyl)-6-(5-(((2-(methylsulfonyl)ethyl)amino)methyl)furan-2-yl)quinazolin-4-amine;4-[[3-Chloro-4-(3-fluorobenzyloxy)phenyl]amino]-6-[5-[[(2-methanesulfonylethyl)amino]methyl]furan-2-yl]quinazoline; N-[3-chloro-4-(3-fluorobenzyloxy)phenyl]-6-[5-({[2-(methanesulfonyl)ethyl]amino}methyl)furan-2-yl]quinazolin-4-amine; N-[3-chloro-4-[(3-fluorophenyl)methoxy]phenyl]-6-[5-[(2-methylsulfonylethylamino)methyl]-2-furyl] quinazolin-4-amine; N-{3-chloro-4-[(3-fluoro-benzyl)oxy]phenyl}-6-[5-({2-(methylsulfonyl)ethyl]amino}methyl)-2-furyl]-4-quinazolinamine; N-{3-Chloro-4-[(3-fluorobenzyl)oxy]phenyl}-6-[5-({[2-(methane sulphonyl)ethyl]amino}methyl)-2-furyl]-4-quinazolinamine; N-{3-Chloro-4[(3-fluorobenzyl)oxy]phenyl}-6-[5-({[2-(methane sulphonyl)ethyl]amino}methyl)-2-furyl]-4-quinazolinamine; N3-Chloro-4-(3-fluorophenyl)methoxyphenyl-6-5-(2-methylsulfonylethylamino)methyl-2-furylquinazolin-4-amine
Click to Show/Hide
|
|
| External link | ||
| Activity |
IC50 = 9.95uM (TPC1SR cell)
|
[112] |
References