m6A Target Gene Information
General Information of the m6A Target Gene (ID: M6ATAR00393)
Full List of m6A Methylation Regulator of This Target Gene and Corresponding Disease/Drug Response(s)
SIRT1
can be regulated by the following regulator(s), and cause disease/drug response(s). You can browse detail information of regulator(s) or disease/drug response(s).
Browse Regulator
Browse Disease
Browse Drug
Insulin-like growth factor 2 mRNA-binding protein 2 (IGF2BP2) [READER]
| Representative RNA-seq result indicating the expression of this target gene regulated by IGF2BP2 | ||
| Cell Line | Liver | Mus musculus |
|
Treatment: IMP2 -/- liver
Control: Wild type liver cells
|
GSE66440 | |
| Regulation |
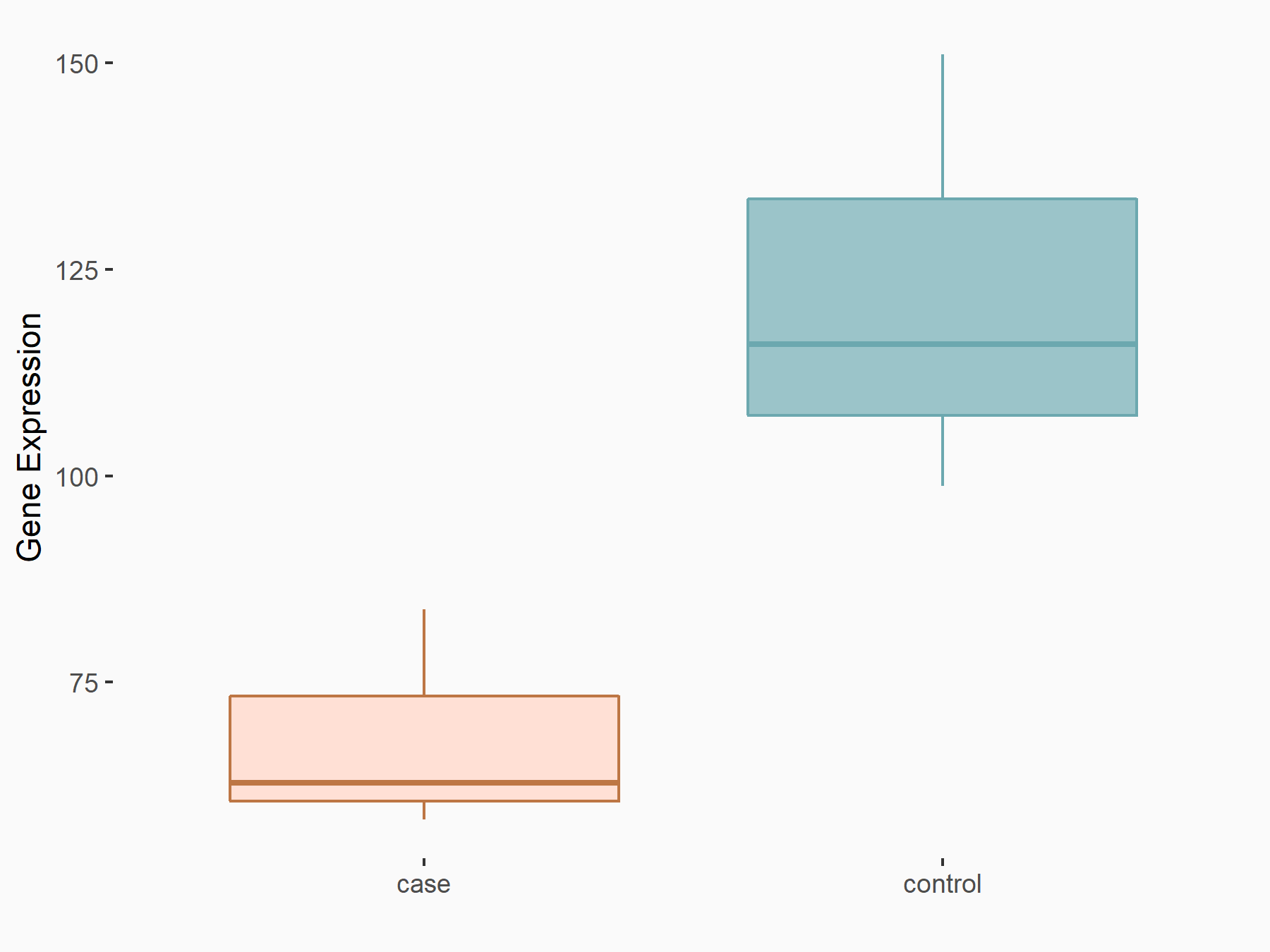 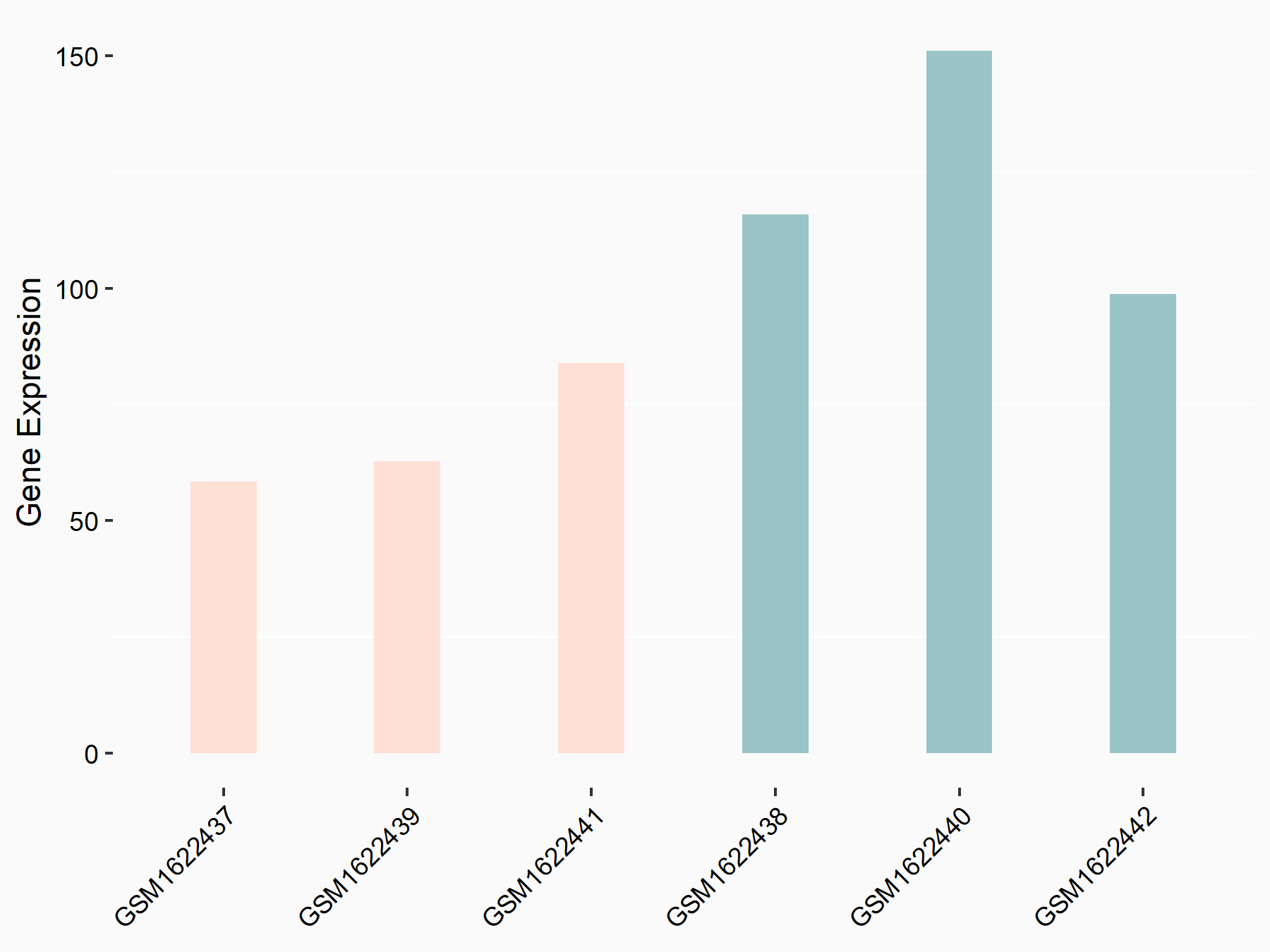 |
logFC: -8.22E-01 p-value: 2.08E-02 |
| More Results | Click to View More RNA-seq Results | |
| In total 1 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [1] | |||
| Response Summary | IGF2BP2 regulated GC the proliferation/migration through recognizing the m6A modification sites of NAD-dependent protein deacetylase sirtuin-1 (SIRT1) mRNA. | |||
| Responsed Disease | Gastric cancer | ICD-11: 2B72 | ||
| Cell Process | Cell proliferation | |||
| Cell migration | ||||
| In-vitro Model | SNU-216 | Gastric tubular adenocarcinoma | Homo sapiens | CVCL_3946 |
| MKN45 | Gastric adenocarcinoma | Homo sapiens | CVCL_0434 | |
| AGS | Gastric adenocarcinoma | Homo sapiens | CVCL_0139 | |
| GES-1 | Normal | Homo sapiens | CVCL_EQ22 | |
| In-vivo Model | About 5 × 106 MKN45 cells stably transfected with IGF2BP2 shRNA or sh-NC vectors were subcutaneously injected into flank of nude mice. | |||
Methyltransferase-like 14 (METTL14) [WRITER]
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL14 | ||
| Cell Line | HepG2 cell line | Homo sapiens |
|
Treatment: shMETTL14 HepG2 cells
Control: shCtrl HepG2 cells
|
GSE121949 | |
| Regulation |
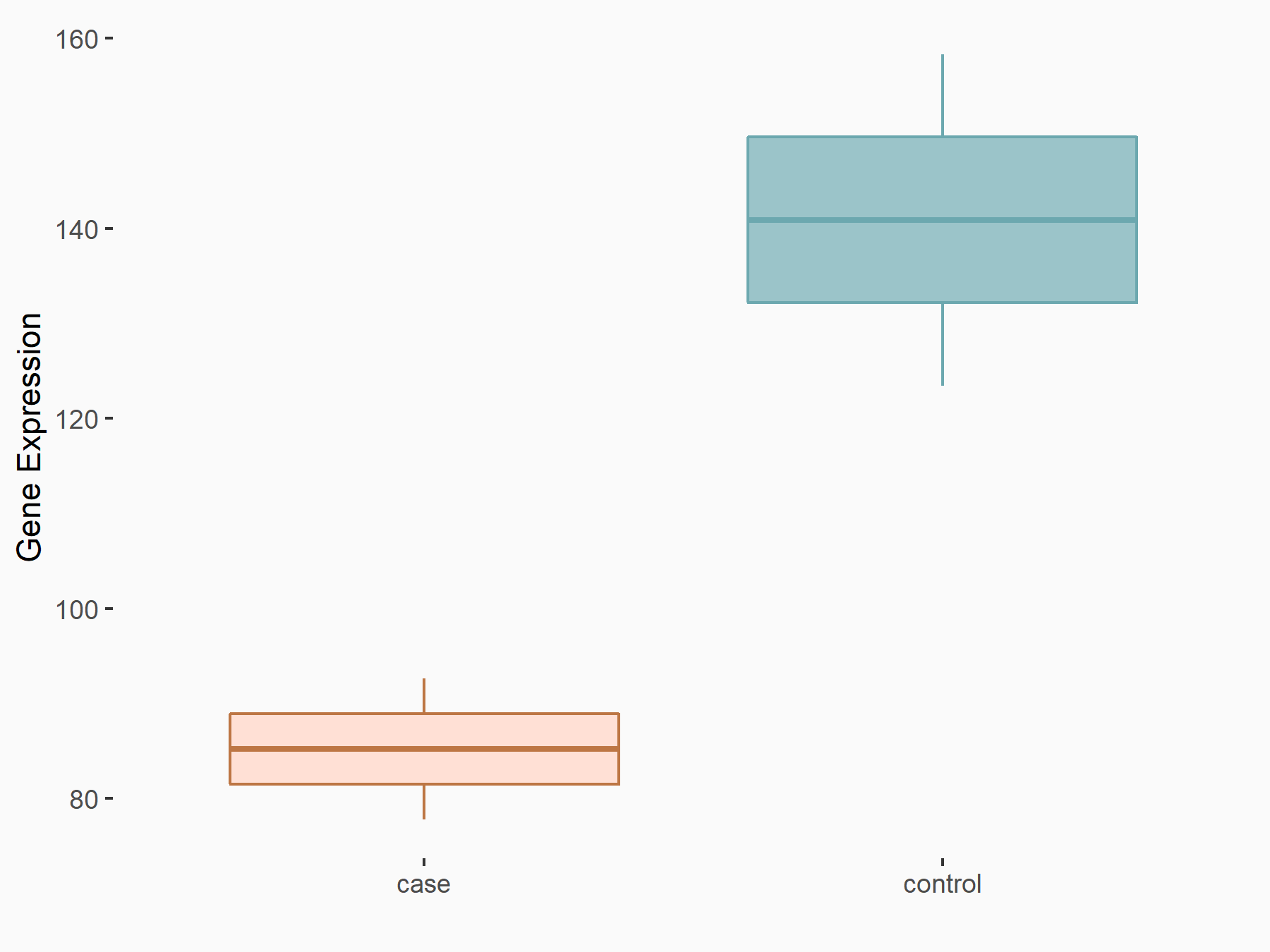 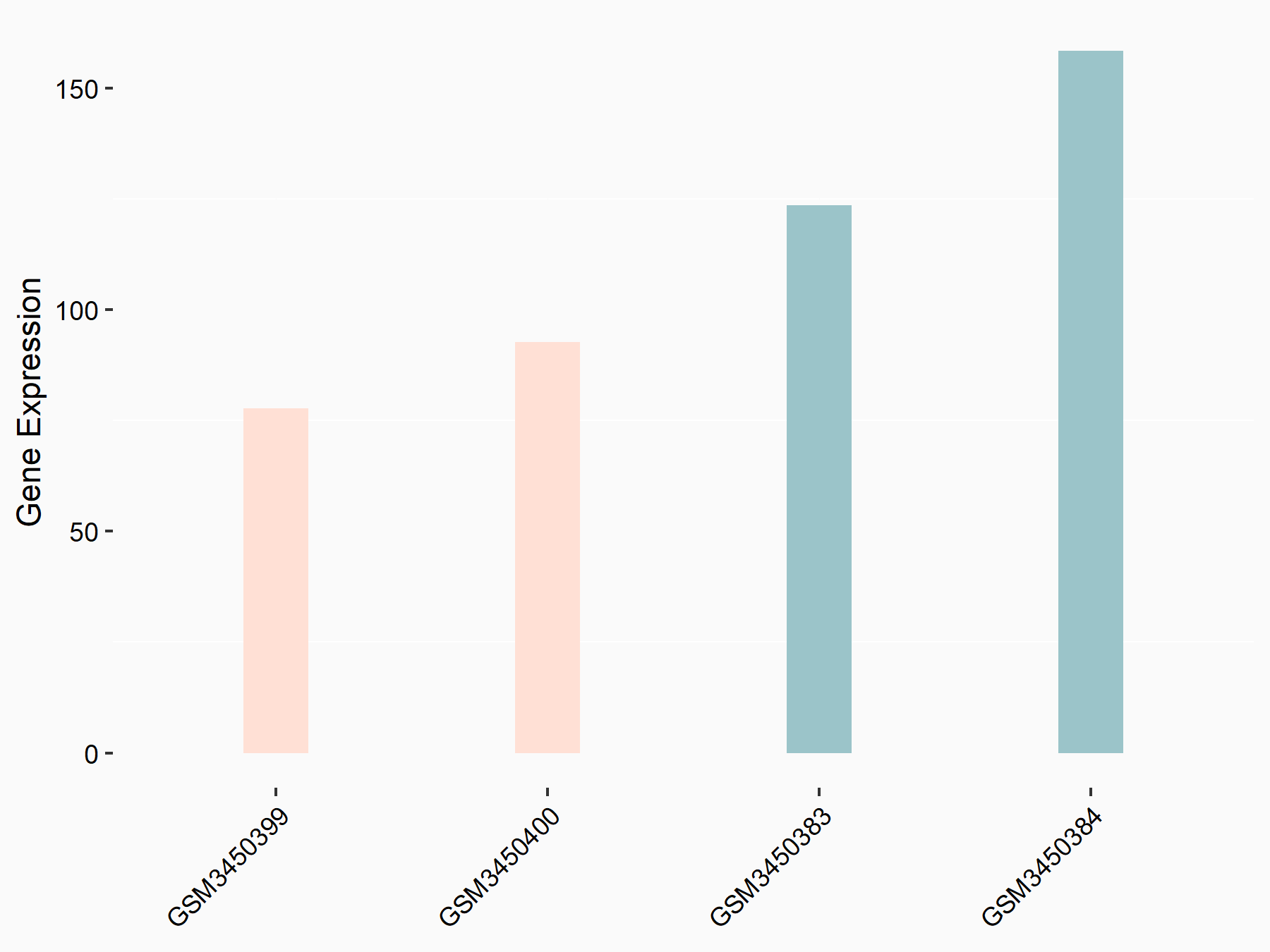 |
logFC: -7.26E-01 p-value: 1.60E-02 |
| More Results | Click to View More RNA-seq Results | |
| In total 1 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [2] | |||
| Response Summary | The elevated m6A RNA levels and the most upregulated METTL14 expression in kidneys of mice with adriamycin and diabetic nephropathy. METTL14-dependent RNA m6A modification contributes to podocyte injury through posttranscriptional regulation of NAD-dependent protein deacetylase sirtuin-1 (SIRT1) mRNA, which provide a potential approach for the diagnosis and treatment of podocytopathies. | |||
| Target Regulation | Down regulation | |||
| Responsed Disease | Chronic kidney disease | ICD-11: GB61.Z | ||
| Responsed Drug | Doxil | Approved | ||
| Cell Process | Cell apoptosis | |||
| In-vitro Model | Conditionally immortalized human podocytes (Podocytes) | |||
| In-vivo Model | To establish mice model with ADR nephropathy, adult male C57BL/6J mice (8-12 weeks of age) were purchased from Animal Center of Fudan University and injected with 19.5 mg/kg ADR (D1515, Sigma-Aldrich, St-Louis, MO, USA) intravenously via tail vein. | |||
Methyltransferase-like 3 (METTL3) [WRITER]
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | Embryonic stem cells | Mus musculus |
|
Treatment: METTL3 knockout mESCs
Control: Wild type mESCs
|
GSE156481 | |
| Regulation |
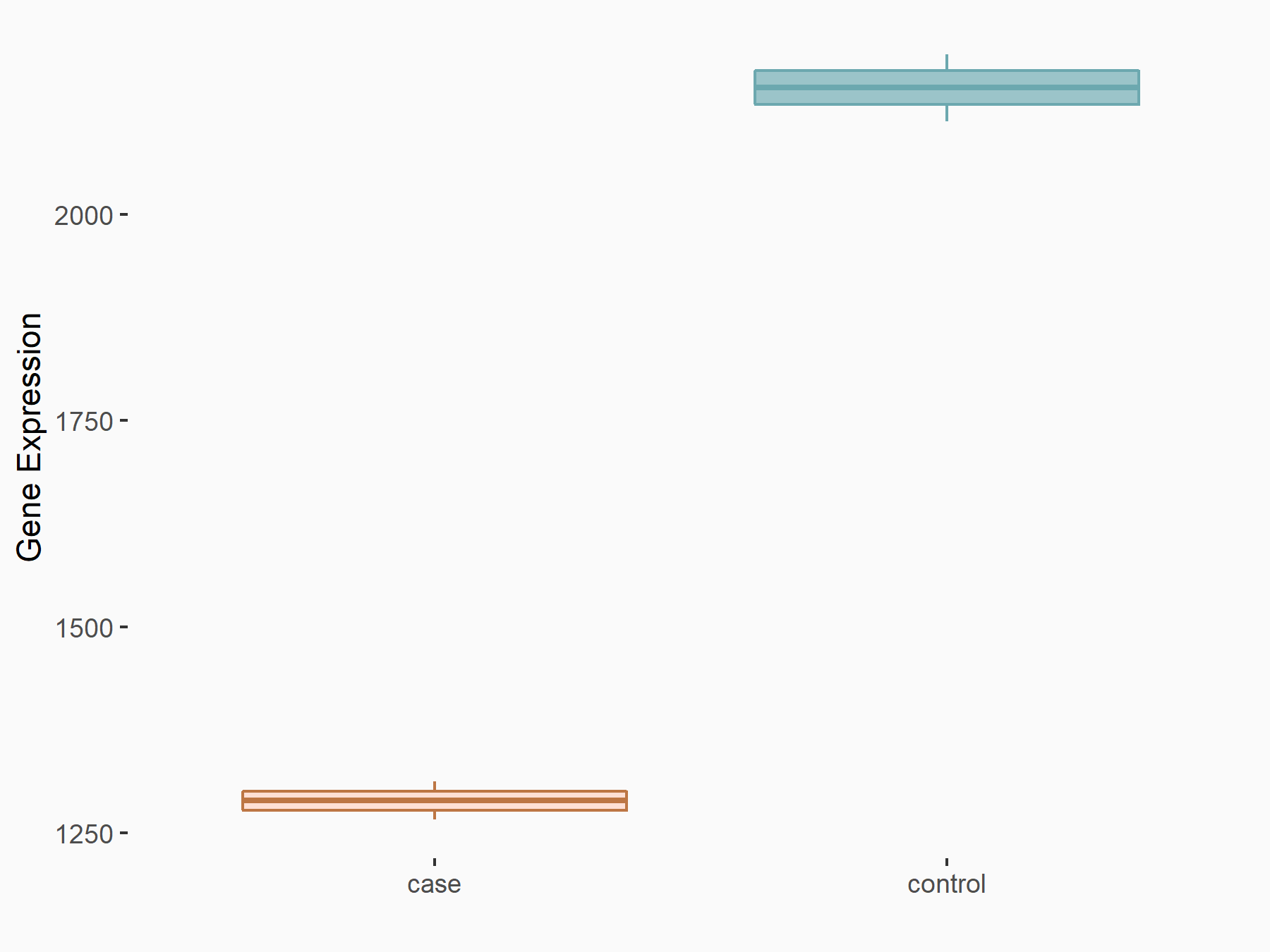  |
logFC: -7.40E-01 p-value: 1.00E-18 |
| More Results | Click to View More RNA-seq Results | |
| In total 2 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [3] | |||
| Response Summary | Type 2 diabetes (T2D) is characterized by lack of insulin, insulin resistance and high blood sugar. METTL3 silence decreased the m6A methylated and total mRNA level of Fatty acid synthase (Fasn), subsequently inhibited fatty acid metabolism. The expression of Acc1, Acly, Dgat2, Ehhadh, Fasn, Foxo, Pgc1a and NAD-dependent protein deacetylase sirtuin-1 (SIRT1), which are critical to the regulation of fatty acid synthesis and oxidation were dramatically decreased in livers of hepatocyte-specific METTL3 knockout mice. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Type 2 diabetes mellitus | ICD-11: 5A11 | ||
| Pathway Response | Insulin resistance | hsa04931 | ||
| Cell Process | Lipid metabolism | |||
| In-vitro Model | Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 |
| In-vivo Model | Hepatocyte-specific METTL3 knockout mice (TBG-Cre, METTL3 fl/fl) were generated by crossing mice with TBG-Cre Tg mice. METTL3 flox (METTL3 fl/fl) and hepatocyte-specific METTL3 knockout mice (TBG-Cre, METTL3 fl/fl) were used for experiments. | |||
| Experiment 2 Reporting the m6A Methylation Regulator of This Target Gene | [4] | |||
| Response Summary | m6A-mediated LINC00680 regulates the proliferation and ECM degradation of chondrocytes through LINC00680/m6A/NAD-dependent protein deacetylase sirtuin-1 (SIRT1) mRNA axis. METTL3-mediated LINC00680 accelerates osteoarthritis(OA) progression, which provides novel understanding of the role of m6A and lncRNA in OA. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Osteoarthritis | ICD-11: FA05 | ||
| In-vitro Model | Chondrocytes (Chondrocytes were isolated from human cartilage and cultured) | |||
Protein virilizer homolog (VIRMA) [WRITER]
| In total 1 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [10] | |||
| Response Summary | KIAA1429 increased the expression of NAD-dependent protein deacetylase sirtuin-1 (SIRT1) via regulating its mRNA stability in an m6A-dependent manner. More importantly, in vivo experiment showed that depletion of KIAA1429 significantly inhibited colorectal tumor growth. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Colorectal cancer | ICD-11: 2B91 | ||
| In-vitro Model | SW620 | Colon adenocarcinoma | Homo sapiens | CVCL_0547 |
| SW480 | Colon adenocarcinoma | Homo sapiens | CVCL_0546 | |
| NCM460 | Normal | Homo sapiens | CVCL_0460 | |
| LoVo | Colon adenocarcinoma | Homo sapiens | CVCL_0399 | |
| HT29 | Colon cancer | Mus musculus | CVCL_A8EZ | |
| HCT 8 | Colon adenocarcinoma | Homo sapiens | CVCL_2478 | |
| HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 | |
| In-vivo Model | 5.0 × 106 SW480 cells (infected with scr or KIAA1429 shRNA) that suspended in 50 ul PBS and mixed with an equal volume of matrigel were subcutaneously injected in a 6-weeks-old male NOD/SCID (The Jackson Laboratory, Stock No: 001303) mice flank. We started measuring tumor size at the indicated times one week after injection. | |||
Gastric cancer [ICD-11: 2B72]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [1] | |||
| Response Summary | IGF2BP2 regulated GC the proliferation/migration through recognizing the m6A modification sites of NAD-dependent protein deacetylase sirtuin-1 (SIRT1) mRNA. | |||
| Responsed Disease | Gastric cancer [ICD-11: 2B72] | |||
| Target Regulator | Insulin-like growth factor 2 mRNA-binding protein 2 (IGF2BP2) | READER | ||
| Cell Process | Cell proliferation | |||
| Cell migration | ||||
| In-vitro Model | SNU-216 | Gastric tubular adenocarcinoma | Homo sapiens | CVCL_3946 |
| MKN45 | Gastric adenocarcinoma | Homo sapiens | CVCL_0434 | |
| AGS | Gastric adenocarcinoma | Homo sapiens | CVCL_0139 | |
| GES-1 | Normal | Homo sapiens | CVCL_EQ22 | |
| In-vivo Model | About 5 × 106 MKN45 cells stably transfected with IGF2BP2 shRNA or sh-NC vectors were subcutaneously injected into flank of nude mice. | |||
Colorectal cancer [ICD-11: 2B91]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [10] | |||
| Response Summary | KIAA1429 increased the expression of NAD-dependent protein deacetylase sirtuin-1 (SIRT1) via regulating its mRNA stability in an m6A-dependent manner. More importantly, in vivo experiment showed that depletion of KIAA1429 significantly inhibited colorectal tumor growth. | |||
| Responsed Disease | Colorectal cancer [ICD-11: 2B91] | |||
| Target Regulator | Protein virilizer homolog (VIRMA) | WRITER | ||
| Target Regulation | Up regulation | |||
| In-vitro Model | SW620 | Colon adenocarcinoma | Homo sapiens | CVCL_0547 |
| SW480 | Colon adenocarcinoma | Homo sapiens | CVCL_0546 | |
| NCM460 | Normal | Homo sapiens | CVCL_0460 | |
| LoVo | Colon adenocarcinoma | Homo sapiens | CVCL_0399 | |
| HT29 | Colon cancer | Mus musculus | CVCL_A8EZ | |
| HCT 8 | Colon adenocarcinoma | Homo sapiens | CVCL_2478 | |
| HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 | |
| In-vivo Model | 5.0 × 106 SW480 cells (infected with scr or KIAA1429 shRNA) that suspended in 50 ul PBS and mixed with an equal volume of matrigel were subcutaneously injected in a 6-weeks-old male NOD/SCID (The Jackson Laboratory, Stock No: 001303) mice flank. We started measuring tumor size at the indicated times one week after injection. | |||
Type 2 diabetes mellitus [ICD-11: 5A11]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [3] | |||
| Response Summary | Type 2 diabetes (T2D) is characterized by lack of insulin, insulin resistance and high blood sugar. METTL3 silence decreased the m6A methylated and total mRNA level of Fatty acid synthase (Fasn), subsequently inhibited fatty acid metabolism. The expression of Acc1, Acly, Dgat2, Ehhadh, Fasn, Foxo, Pgc1a and NAD-dependent protein deacetylase sirtuin-1 (SIRT1), which are critical to the regulation of fatty acid synthesis and oxidation were dramatically decreased in livers of hepatocyte-specific METTL3 knockout mice. | |||
| Responsed Disease | Type 2 diabetes mellitus [ICD-11: 5A11] | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Insulin resistance | hsa04931 | ||
| Cell Process | Lipid metabolism | |||
| In-vitro Model | Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 |
| In-vivo Model | Hepatocyte-specific METTL3 knockout mice (TBG-Cre, METTL3 fl/fl) were generated by crossing mice with TBG-Cre Tg mice. METTL3 flox (METTL3 fl/fl) and hepatocyte-specific METTL3 knockout mice (TBG-Cre, METTL3 fl/fl) were used for experiments. | |||
Osteoarthritis [ICD-11: FA05]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [4] | |||
| Response Summary | m6A-mediated LINC00680 regulates the proliferation and ECM degradation of chondrocytes through LINC00680/m6A/NAD-dependent protein deacetylase sirtuin-1 (SIRT1) mRNA axis. METTL3-mediated LINC00680 accelerates osteoarthritis(OA) progression, which provides novel understanding of the role of m6A and lncRNA in OA. | |||
| Responsed Disease | Osteoarthritis [ICD-11: FA05] | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Target Regulation | Up regulation | |||
| In-vitro Model | Chondrocytes (Chondrocytes were isolated from human cartilage and cultured) | |||
Chronic kidney disease [ICD-11: GB61]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [2] | |||
| Response Summary | The elevated m6A RNA levels and the most upregulated METTL14 expression in kidneys of mice with adriamycin and diabetic nephropathy. METTL14-dependent RNA m6A modification contributes to podocyte injury through posttranscriptional regulation of NAD-dependent protein deacetylase sirtuin-1 (SIRT1) mRNA, which provide a potential approach for the diagnosis and treatment of podocytopathies. | |||
| Responsed Disease | Chronic kidney disease [ICD-11: GB61.Z] | |||
| Target Regulator | Methyltransferase-like 14 (METTL14) | WRITER | ||
| Target Regulation | Down regulation | |||
| Responsed Drug | Doxil | Approved | ||
| Cell Process | Cell apoptosis | |||
| In-vitro Model | Conditionally immortalized human podocytes (Podocytes) | |||
| In-vivo Model | To establish mice model with ADR nephropathy, adult male C57BL/6J mice (8-12 weeks of age) were purchased from Animal Center of Fudan University and injected with 19.5 mg/kg ADR (D1515, Sigma-Aldrich, St-Louis, MO, USA) intravenously via tail vein. | |||
Doxil
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response | [2] | |||
| Response Summary | The elevated m6A RNA levels and the most upregulated METTL14 expression in kidneys of mice with adriamycin and diabetic nephropathy. METTL14-dependent RNA m6A modification contributes to podocyte injury through posttranscriptional regulation of NAD-dependent protein deacetylase sirtuin-1 (SIRT1) mRNA, which provide a potential approach for the diagnosis and treatment of podocytopathies. | |||
| Target Regulator | Methyltransferase-like 14 (METTL14) | WRITER | ||
| Target Regulation | Down regulation | |||
| Responsed Disease | Chronic kidney disease | ICD-11: GB61.Z | ||
| Cell Process | Cell apoptosis | |||
| In-vitro Model | Conditionally immortalized human podocytes (Podocytes) | |||
| In-vivo Model | To establish mice model with ADR nephropathy, adult male C57BL/6J mice (8-12 weeks of age) were purchased from Animal Center of Fudan University and injected with 19.5 mg/kg ADR (D1515, Sigma-Aldrich, St-Louis, MO, USA) intravenously via tail vein. | |||
Full List of Crosstalk(s) between m6A Modification and Epigenetic Regulation Related to This Regulator
RNA modification
m6A Regulator: ELAV-like protein 1 (ELAVL1)
| In total 2 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT00478 | ||
| Epigenetic Regulator | Interferon-inducible protein 4 (ADAR1) | |
| Regulated Target | MicroRNA 155 (MIR155) | |
| Crosstalk relationship | A-to-I → m6A | |
| Crosstalk ID: M6ACROT00517 | ||
| Epigenetic Regulator | Double-stranded RNA-specific editase 1 (ADARB1) | |
| Regulated Target | MicroRNA 221 (MIR221) | |
| Crosstalk relationship | A-to-I → m6A | |
m6A Regulator: Fat mass and obesity-associated protein (FTO)
| In total 2 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT00479 | ||
| Epigenetic Regulator | Interferon-inducible protein 4 (ADAR1) | |
| Regulated Target | MicroRNA 155 (MIR155) | |
| Crosstalk relationship | A-to-I → m6A | |
| Crosstalk ID: M6ACROT00518 | ||
| Epigenetic Regulator | Double-stranded RNA-specific editase 1 (ADARB1) | |
| Regulated Target | MicroRNA 221 (MIR221) | |
| Crosstalk relationship | A-to-I → m6A | |
Non-coding RNA
m6A Regulator: Methyltransferase-like 14 (METTL14)
| In total 1 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT05148 | ||
| Epigenetic Regulator | hsa-miR-103-3p | |
| Regulated Target | Methyltransferase-like protein 14 (METTL14) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Osteoporosis | |
m6A Regulator: RNA demethylase ALKBH5 (ALKBH5)
| In total 1 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT05154 | ||
| Epigenetic Regulator | hsa-miR-193a-3p | |
| Regulated Target | RNA demethylase ALKBH5 (ALKBH5) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Injury of heart | |
| Drug | Suxiao Jiuxin Pill | |
RNA Modification Sequencing Data Associated with the Target (ID: M6ATAR00393)
| In total 27 m6A sequence/site(s) in this target gene | |||
| mod ID: A2ISITE004163 | Click to Show/Hide the Full List | ||
| mod site | chr10:67886294-67886295:+ | [12] | |
| Sequence | CTTGATAGGTGTTCATCTTTATCAATATTTGGGTCAGGCCG | ||
| Transcript ID List | ENST00000212015.11; ENST00000473922.1; ENST00000497639.5; ENST00000432464.5 | ||
| External Link | RMBase: RNA-editing_site_16063 | ||
| mod ID: A2ISITE004164 | Click to Show/Hide the Full List | ||
| mod site | chr10:67892451-67892452:+ | [13] | |
| Sequence | GCATGGTGGTGTGCACCTGTAGACCCATCCACTCAGTAGTC | ||
| Transcript ID List | ENST00000473922.1; ENST00000432464.5; ENST00000212015.11; ENST00000406900.5; rmsk_3332535 | ||
| External Link | RMBase: RNA-editing_site_16064 | ||
| mod ID: A2ISITE004165 | Click to Show/Hide the Full List | ||
| mod site | chr10:67895164-67895165:+ | [12] | |
| Sequence | CACAGTGGCTCACACCTGTAATCCCAGCACTTTGGGAGGCC | ||
| Transcript ID List | rmsk_3217008; ENST00000473922.1; ENST00000212015.11; ENST00000406900.5; ENST00000432464.5 | ||
| External Link | RMBase: RNA-editing_site_16065 | ||
| mod ID: A2ISITE004166 | Click to Show/Hide the Full List | ||
| mod site | chr10:67898050-67898051:+ | [14] | |
| Sequence | GAGGCCACAGCGGGCGGTCAAAAGTTTGAGACCAGCCTGAC | ||
| Transcript ID List | ENST00000473922.1; rmsk_3332555; ENST00000432464.5; ENST00000212015.11; ENST00000406900.5 | ||
| External Link | RMBase: RNA-editing_site_16066 | ||
| mod ID: A2ISITE004167 | Click to Show/Hide the Full List | ||
| mod site | chr10:67898051-67898052:+ | [14] | |
| Sequence | AGGCCACAGCGGGCGGTCAAAAGTTTGAGACCAGCCTGACC | ||
| Transcript ID List | rmsk_3332555; ENST00000212015.11; ENST00000406900.5; ENST00000432464.5; ENST00000473922.1 | ||
| External Link | RMBase: RNA-editing_site_16067 | ||
| mod ID: A2ISITE004168 | Click to Show/Hide the Full List | ||
| mod site | chr10:67898052-67898053:+ | [14] | |
| Sequence | GGCCACAGCGGGCGGTCAAAAGTTTGAGACCAGCCTGACCA | ||
| Transcript ID List | ENST00000212015.11; rmsk_3332555; ENST00000432464.5; ENST00000406900.5; ENST00000473922.1 | ||
| External Link | RMBase: RNA-editing_site_16068 | ||
| mod ID: A2ISITE004169 | Click to Show/Hide the Full List | ||
| mod site | chr10:67898073-67898074:+ | [14] | |
| Sequence | GTTTGAGACCAGCCTGACCAACATGGTGAAACCCTGTCTCT | ||
| Transcript ID List | ENST00000406900.5; rmsk_3332555; ENST00000212015.11; ENST00000432464.5; ENST00000473922.1 | ||
| External Link | RMBase: RNA-editing_site_16069 | ||
| mod ID: A2ISITE004170 | Click to Show/Hide the Full List | ||
| mod site | chr10:67898075-67898076:+ | [14] | |
| Sequence | TTGAGACCAGCCTGACCAACATGGTGAAACCCTGTCTCTAC | ||
| Transcript ID List | ENST00000406900.5; ENST00000473922.1; rmsk_3332555; ENST00000432464.5; ENST00000212015.11 | ||
| External Link | RMBase: RNA-editing_site_16070 | ||
| mod ID: A2ISITE004171 | Click to Show/Hide the Full List | ||
| mod site | chr10:67898081-67898082:+ | [14] | |
| Sequence | CCAGCCTGACCAACATGGTGAAACCCTGTCTCTACTAAAAA | ||
| Transcript ID List | rmsk_3332555; ENST00000473922.1; ENST00000212015.11; ENST00000432464.5; ENST00000406900.5 | ||
| External Link | RMBase: RNA-editing_site_16071 | ||
| mod ID: A2ISITE004172 | Click to Show/Hide the Full List | ||
| mod site | chr10:67898082-67898083:+ | [14] | |
| Sequence | CAGCCTGACCAACATGGTGAAACCCTGTCTCTACTAAAAAT | ||
| Transcript ID List | ENST00000473922.1; rmsk_3332555; ENST00000432464.5; ENST00000212015.11; ENST00000406900.5 | ||
| External Link | RMBase: RNA-editing_site_16072 | ||
| mod ID: A2ISITE004173 | Click to Show/Hide the Full List | ||
| mod site | chr10:67898083-67898084:+ | [14] | |
| Sequence | AGCCTGACCAACATGGTGAAACCCTGTCTCTACTAAAAATA | ||
| Transcript ID List | ENST00000432464.5; rmsk_3332555; ENST00000406900.5; ENST00000473922.1; ENST00000212015.11 | ||
| External Link | RMBase: RNA-editing_site_16073 | ||
| mod ID: A2ISITE004174 | Click to Show/Hide the Full List | ||
| mod site | chr10:67898100-67898101:+ | [14] | |
| Sequence | GAAACCCTGTCTCTACTAAAAATACAAAAATTAGATGGATG | ||
| Transcript ID List | ENST00000212015.11; ENST00000432464.5; rmsk_3332555; ENST00000406900.5; ENST00000473922.1 | ||
| External Link | RMBase: RNA-editing_site_16074 | ||
| mod ID: A2ISITE004175 | Click to Show/Hide the Full List | ||
| mod site | chr10:67898137-67898138:+ | [14] | |
| Sequence | GATGTGGTGGCGCATGCTGTAATCCCAGCTACTCAGGAGGC | ||
| Transcript ID List | ENST00000212015.11; ENST00000473922.1; rmsk_3332555; ENST00000432464.5; ENST00000406900.5 | ||
| External Link | RMBase: RNA-editing_site_16075 | ||
| mod ID: A2ISITE004176 | Click to Show/Hide the Full List | ||
| mod site | chr10:67898138-67898139:+ | [14] | |
| Sequence | ATGTGGTGGCGCATGCTGTAATCCCAGCTACTCAGGAGGCT | ||
| Transcript ID List | rmsk_3332555; ENST00000212015.11; ENST00000432464.5; ENST00000473922.1; ENST00000406900.5 | ||
| External Link | RMBase: RNA-editing_site_16076 | ||
| mod ID: A2ISITE004177 | Click to Show/Hide the Full List | ||
| mod site | chr10:67898161-67898162:+ | [14] | |
| Sequence | CCAGCTACTCAGGAGGCTGAAGCAGGAGAATCACTTGAACC | ||
| Transcript ID List | ENST00000473922.1; ENST00000432464.5; rmsk_3332555; ENST00000212015.11; ENST00000406900.5 | ||
| External Link | RMBase: RNA-editing_site_16077 | ||
| mod ID: A2ISITE004178 | Click to Show/Hide the Full List | ||
| mod site | chr10:67898164-67898165:+ | [14] | |
| Sequence | GCTACTCAGGAGGCTGAAGCAGGAGAATCACTTGAACCCAG | ||
| Transcript ID List | ENST00000473922.1; rmsk_3332555; ENST00000212015.11; ENST00000432464.5; ENST00000406900.5 | ||
| External Link | RMBase: RNA-editing_site_16078 | ||
| mod ID: A2ISITE004179 | Click to Show/Hide the Full List | ||
| mod site | chr10:67898173-67898174:+ | [14] | |
| Sequence | GAGGCTGAAGCAGGAGAATCACTTGAACCCAGGAGGCGGAG | ||
| Transcript ID List | rmsk_3332555; ENST00000406900.5; ENST00000212015.11; ENST00000432464.5; ENST00000473922.1 | ||
| External Link | RMBase: RNA-editing_site_16079 | ||
| mod ID: A2ISITE004180 | Click to Show/Hide the Full List | ||
| mod site | chr10:67898178-67898179:+ | [14] | |
| Sequence | TGAAGCAGGAGAATCACTTGAACCCAGGAGGCGGAGGTTGC | ||
| Transcript ID List | ENST00000432464.5; ENST00000212015.11; ENST00000406900.5; rmsk_3332555; ENST00000473922.1 | ||
| External Link | RMBase: RNA-editing_site_16080 | ||
| mod ID: A2ISITE004181 | Click to Show/Hide the Full List | ||
| mod site | chr10:67898183-67898184:+ | [14] | |
| Sequence | CAGGAGAATCACTTGAACCCAGGAGGCGGAGGTTGCAGTGA | ||
| Transcript ID List | ENST00000432464.5; ENST00000406900.5; ENST00000473922.1; rmsk_3332555; ENST00000212015.11 | ||
| External Link | RMBase: RNA-editing_site_16081 | ||
| mod ID: A2ISITE004182 | Click to Show/Hide the Full List | ||
| mod site | chr10:67898186-67898187:+ | [14] | |
| Sequence | GAGAATCACTTGAACCCAGGAGGCGGAGGTTGCAGTGAGCT | ||
| Transcript ID List | ENST00000432464.5; ENST00000406900.5; ENST00000473922.1; rmsk_3332555; ENST00000212015.11 | ||
| External Link | RMBase: RNA-editing_site_16082 | ||
| mod ID: A2ISITE004183 | Click to Show/Hide the Full List | ||
| mod site | chr10:67898203-67898204:+ | [14] | |
| Sequence | AGGAGGCGGAGGTTGCAGTGAGCTGAGATGGCACCACTGCA | ||
| Transcript ID List | ENST00000212015.11; rmsk_3332555; ENST00000473922.1; ENST00000406900.5; ENST00000432464.5 | ||
| External Link | RMBase: RNA-editing_site_16083 | ||
| mod ID: A2ISITE004184 | Click to Show/Hide the Full List | ||
| mod site | chr10:67898218-67898219:+ | [14] | |
| Sequence | CAGTGAGCTGAGATGGCACCACTGCACTCCAGCCTGGGCGA | ||
| Transcript ID List | rmsk_3332555; ENST00000406900.5; ENST00000212015.11; ENST00000473922.1; ENST00000432464.5 | ||
| External Link | RMBase: RNA-editing_site_16084 | ||
| mod ID: A2ISITE004185 | Click to Show/Hide the Full List | ||
| mod site | chr10:67900119-67900120:+ | [12] | |
| Sequence | TTAAATAAAATTATAAAAATAAAGCAGCACCCAGCTTTATT | ||
| Transcript ID List | ENST00000473922.1; ENST00000432464.5; ENST00000212015.11; ENST00000406900.5 | ||
| External Link | RMBase: RNA-editing_site_16085 | ||
| mod ID: A2ISITE004186 | Click to Show/Hide the Full List | ||
| mod site | chr10:67900120-67900121:+ | [12] | |
| Sequence | TAAATAAAATTATAAAAATAAAGCAGCACCCAGCTTTATTT | ||
| Transcript ID List | ENST00000473922.1; ENST00000406900.5; ENST00000432464.5; ENST00000212015.11 | ||
| External Link | RMBase: RNA-editing_site_16086 | ||
| mod ID: A2ISITE004187 | Click to Show/Hide the Full List | ||
| mod site | chr10:67901249-67901250:+ | [13] | |
| Sequence | TCAAGCGATCCTCCCACCTTAGCCTCCTGACTAGCTGTGAC | ||
| Transcript ID List | ENST00000432464.5; ENST00000212015.11; ENST00000473922.1; ENST00000406900.5 | ||
| External Link | RMBase: RNA-editing_site_16087 | ||
| mod ID: A2ISITE004188 | Click to Show/Hide the Full List | ||
| mod site | chr10:67903585-67903586:+ | [12] | |
| Sequence | TTTAGTAGAGACGGGGTTTCACCATCTTAGCCAGGATGGTC | ||
| Transcript ID List | ENST00000432464.5; ENST00000473922.1; ENST00000406900.5; ENST00000212015.11 | ||
| External Link | RMBase: RNA-editing_site_16088 | ||
| mod ID: A2ISITE004189 | Click to Show/Hide the Full List | ||
| mod site | chr10:67904371-67904372:+ | [12] | |
| Sequence | GTCTCGTACTCCTGGTCTCAAGTAACCTGCCCGCCTCGGCC | ||
| Transcript ID List | ENST00000473922.1; ENST00000406900.5; ENST00000212015.11; ENST00000432464.5 | ||
| External Link | RMBase: RNA-editing_site_16089 | ||
N6-methyladenosine (m6A)
| In total 76 m6A sequence/site(s) in this target gene | |||
| mod ID: M6ASITE000316 | Click to Show/Hide the Full List | ||
| mod site | chr10:67885025-67885026:+ | [15] | |
| Sequence | TGCGGCGGCTGGGGAAGGAGACAATGGGCCGGGCCTGCAGG | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa; CD34; HepG2; A549; hESC-HEK293T; U2OS; MT4; MM6; Jurkat; GSC-11; iSLK; endometrial | ||
| Seq Type List | m6A-seq; MeRIP-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000212015.11 | ||
| External Link | RMBase: m6A_site_102817 | ||
| mod ID: M6ASITE000317 | Click to Show/Hide the Full List | ||
| mod site | chr10:67885073-67885074:+ | [16] | |
| Sequence | TCGGGAGCCACCGCTGGCCGACAACTTGTACGACGAAGACG | ||
| Motif Score | 2.865571429 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000212015.11 | ||
| External Link | RMBase: m6A_site_102818 | ||
| mod ID: M6ASITE000318 | Click to Show/Hide the Full List | ||
| mod site | chr10:67887492-67887493:+ | [16] | |
| Sequence | GATGAGGAGGATAGAGCCTCACATGCAAGCTCTAGTGACTG | ||
| Motif Score | 2.047297619 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000473922.1; ENST00000212015.11; ENST00000497639.5; ENST00000432464.5 | ||
| External Link | RMBase: m6A_site_102819 | ||
| mod ID: M6ASITE000319 | Click to Show/Hide the Full List | ||
| mod site | chr10:67887514-67887515:+ | [15] | |
| Sequence | ATGCAAGCTCTAGTGACTGGACTCCAAGGCCACGGATAGGT | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000473922.1; ENST00000497639.5; ENST00000432464.5; ENST00000212015.11 | ||
| External Link | RMBase: m6A_site_102820 | ||
| mod ID: M6ASITE000320 | Click to Show/Hide the Full List | ||
| mod site | chr10:67888903-67888904:+ | [16] | |
| Sequence | CCATATACTTTTGTTCAGCAACATCTTATGATTGGCACAGA | ||
| Motif Score | 2.173910714 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000212015.11; ENST00000497639.5; ENST00000432464.5; ENST00000473922.1 | ||
| External Link | RMBase: m6A_site_102821 | ||
| mod ID: M6ASITE000321 | Click to Show/Hide the Full List | ||
| mod site | chr10:67888931-67888932:+ | [15] | |
| Sequence | TGATTGGCACAGATCCTCGAACAATTCTTAAAGATTTATTG | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000212015.11; ENST00000473922.1; ENST00000432464.5; ENST00000497639.5 | ||
| External Link | RMBase: m6A_site_102822 | ||
| mod ID: M6ASITE000322 | Click to Show/Hide the Full List | ||
| mod site | chr10:67888958-67888959:+ | [15] | |
| Sequence | TTAAAGATTTATTGCCGGAAACAATACCTCCACCTGAGTTG | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000473922.1; ENST00000212015.11; ENST00000497639.5; ENST00000432464.5 | ||
| External Link | RMBase: m6A_site_102823 | ||
| mod ID: M6ASITE000323 | Click to Show/Hide the Full List | ||
| mod site | chr10:67888988-67888989:+ | [16] | |
| Sequence | CACCTGAGTTGGATGATATGACACTGTGGCAGATTGTTATT | ||
| Motif Score | 2.859755952 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000497639.5; ENST00000212015.11; ENST00000432464.5; ENST00000473922.1 | ||
| External Link | RMBase: m6A_site_102824 | ||
| mod ID: M6ASITE000324 | Click to Show/Hide the Full List | ||
| mod site | chr10:67889023-67889024:+ | [15] | |
| Sequence | GTTATTAATATCCTTTCAGAACCACCAAAAAGGAAAAAAAG | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000432464.5; ENST00000473922.1; ENST00000212015.11; ENST00000497639.5 | ||
| External Link | RMBase: m6A_site_102825 | ||
| mod ID: M6ASITE000325 | Click to Show/Hide the Full List | ||
| mod site | chr10:67889057-67889058:+ | [16] | |
| Sequence | AAAAAAGAAAAGATATTAATACAATTGAAGATGCTGTGAAA | ||
| Motif Score | 2.110482143 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000432464.5; ENST00000497639.5; ENST00000473922.1; ENST00000212015.11 | ||
| External Link | RMBase: m6A_site_102826 | ||
| mod ID: M6ASITE000326 | Click to Show/Hide the Full List | ||
| mod site | chr10:67908081-67908082:+ | [16] | |
| Sequence | ATCTTGCCTGATTTGTAAATACAAAGTTGACTGTGAAGCTG | ||
| Motif Score | 2.110482143 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000406900.5; ENST00000432464.5; ENST00000212015.11; ENST00000403579.1 | ||
| External Link | RMBase: m6A_site_102827 | ||
| mod ID: M6ASITE000327 | Click to Show/Hide the Full List | ||
| mod site | chr10:67909290-67909291:+ | [15] | |
| Sequence | CCTAGGTGCCCAGCTGATGAACCGCTTGCTATCATGAAACC | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000432464.5; ENST00000403579.1; ENST00000212015.11; ENST00000406900.5 | ||
| External Link | RMBase: m6A_site_102828 | ||
| mod ID: M6ASITE000328 | Click to Show/Hide the Full List | ||
| mod site | chr10:67909308-67909309:+ | [15] | |
| Sequence | GAACCGCTTGCTATCATGAAACCAGAGATTGTGTTTTTTGG | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000406900.5; ENST00000212015.11; ENST00000403579.1; ENST00000432464.5 | ||
| External Link | RMBase: m6A_site_102829 | ||
| mod ID: M6ASITE000329 | Click to Show/Hide the Full List | ||
| mod site | chr10:67909344-67909345:+ | [15] | |
| Sequence | TTTGGTGAAAATTTACCAGAACAGTTTCATAGAGCCATGAA | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000432464.5; ENST00000406900.5; ENST00000403579.1; ENST00000212015.11 | ||
| External Link | RMBase: m6A_site_102830 | ||
| mod ID: M6ASITE000330 | Click to Show/Hide the Full List | ||
| mod site | chr10:67909422-67909423:+ | [15] | |
| Sequence | GGGTCTTCCCTCAAAGTAAGACCAGTAGCACTAATTCCAAG | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa; HEK293T | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000403579.1; ENST00000432464.5; ENST00000212015.11; ENST00000406900.5 | ||
| External Link | RMBase: m6A_site_102831 | ||
| mod ID: M6ASITE000331 | Click to Show/Hide the Full List | ||
| mod site | chr10:67912516-67912517:+ | [15] | |
| Sequence | CAGATATTAATTAATAGAGAACCTTTGCCTCATCTGCATTT | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | HeLa; HEK293T; hNPCs; hESCs; A549; HEK293A-TOA | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000432464.5; ENST00000403579.1; ENST00000406900.5; ENST00000212015.11 | ||
| External Link | RMBase: m6A_site_102832 | ||
| mod ID: M6ASITE000332 | Click to Show/Hide the Full List | ||
| mod site | chr10:67912557-67912558:+ | [15] | |
| Sequence | TGATGTAGAGCTTCTTGGAGACTGTGATGTCATAATTAATG | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa; HEK293T; HepG2; hNPCs; hESCs; fibroblasts; A549; CD8T; HEK293A-TOA | ||
| Seq Type List | m6A-seq; MeRIP-seq; m6A-CLIP/IP | ||
| Transcript ID List | ENST00000403579.1; ENST00000432464.5; ENST00000212015.11; ENST00000406900.5 | ||
| External Link | RMBase: m6A_site_102833 | ||
| mod ID: M6ASITE000333 | Click to Show/Hide the Full List | ||
| mod site | chr10:67912612-67912613:+ | [15] | |
| Sequence | TTAGGTGGTGAATATGCCAAACTTTGCTGTAACCCTGTAAA | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa; HEK293T; BGC823; HepG2; hNPCs; hESCs; fibroblasts; A549; GM12878; HEK293A-TOA; iSLK; MSC; TIME; TREX | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000432464.5; ENST00000406900.5; ENST00000403579.1; ENST00000212015.11 | ||
| External Link | RMBase: m6A_site_102834 | ||
| mod ID: M6ASITE000334 | Click to Show/Hide the Full List | ||
| mod site | chr10:67912654-67912655:+ | [15] | |
| Sequence | CTTTCAGAAATTACTGAAAAACCTCCACGAACACAAAAAGA | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | HeLa; HEK293T; BGC823; HepG2; hNPCs; hESCs; fibroblasts; A549; GM12878; Huh7; HEK293A-TOA; iSLK; MSC; TIME; TREX | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000403579.1; ENST00000432464.5; ENST00000212015.11; ENST00000406900.5 | ||
| External Link | RMBase: m6A_site_102835 | ||
| mod ID: M6ASITE000335 | Click to Show/Hide the Full List | ||
| mod site | chr10:67912664-67912665:+ | [15] | |
| Sequence | TTACTGAAAAACCTCCACGAACACAAAAAGAATTGGCTTAT | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa; HEK293T; hESC-HEK293T; BGC823; HepG2; hNPCs; hESCs; fibroblasts; A549; GM12878; Huh7; HEK293A-TOA; iSLK; MSC; TIME; TREX | ||
| Seq Type List | m6A-seq; MeRIP-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000212015.11; ENST00000406900.5; ENST00000432464.5; ENST00000403579.1 | ||
| External Link | RMBase: m6A_site_102836 | ||
| mod ID: M6ASITE000336 | Click to Show/Hide the Full List | ||
| mod site | chr10:67912703-67912704:+ | [16] | |
| Sequence | ATTTGTCAGAGTTGCCACCCACACCTCTTCATGTTTCAGAA | ||
| Motif Score | 2.053113095 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000212015.11; ENST00000432464.5; ENST00000406900.5; ENST00000403579.1 | ||
| External Link | RMBase: m6A_site_102837 | ||
| mod ID: M6ASITE000337 | Click to Show/Hide the Full List | ||
| mod site | chr10:67912725-67912726:+ | [15] | |
| Sequence | ACCTCTTCATGTTTCAGAAGACTCAAGTTCACCAGAAAGAA | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa; HEK293T; BGC823; hNPCs; hESCs; fibroblasts; A549; GM12878; Huh7; HEK293A-TOA; iSLK; MSC; TIME; TREX; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000432464.5; ENST00000403579.1; ENST00000406900.5; ENST00000212015.11 | ||
| External Link | RMBase: m6A_site_102838 | ||
| mod ID: M6ASITE000338 | Click to Show/Hide the Full List | ||
| mod site | chr10:67912745-67912746:+ | [15] | |
| Sequence | ACTCAAGTTCACCAGAAAGAACTTCACCACCAGATTCTTCA | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; BGC823; hNPCs; hESCs; fibroblasts; A549; GM12878; Huh7; Jurkat; peripheral-blood; HEK293A-TOA; iSLK; MSC; TIME; TREX; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000403579.1; ENST00000432464.5; ENST00000212015.11; ENST00000406900.5 | ||
| External Link | RMBase: m6A_site_102839 | ||
| mod ID: M6ASITE000339 | Click to Show/Hide the Full List | ||
| mod site | chr10:67912775-67912776:+ | [16] | |
| Sequence | CAGATTCTTCAGTGATTGTCACACTTTTAGACCAAGCAGCT | ||
| Motif Score | 2.047297619 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000403579.1; ENST00000212015.11; ENST00000432464.5; ENST00000406900.5 | ||
| External Link | RMBase: m6A_site_102840 | ||
| mod ID: M6ASITE000340 | Click to Show/Hide the Full List | ||
| mod site | chr10:67912785-67912786:+ | [15] | |
| Sequence | AGTGATTGTCACACTTTTAGACCAAGCAGCTAAGAGTAATG | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; BGC823; hNPCs; hESCs; fibroblasts; A549; GM12878; Huh7; Jurkat; peripheral-blood; HEK293A-TOA; iSLK; MSC; TIME; TREX; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000212015.11; ENST00000406900.5; ENST00000403579.1; ENST00000432464.5 | ||
| External Link | RMBase: m6A_site_102841 | ||
| mod ID: M6ASITE000341 | Click to Show/Hide the Full List | ||
| mod site | chr10:67912849-67912850:+ | [15] | |
| Sequence | AAAGGTTGTATGGAAGAAAAACCACAGGAAGTACAAACTTC | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; hNPCs; hESCs; fibroblasts; A549; GM12878; MM6; Huh7; Jurkat; peripheral-blood; HEK293A-TOA; iSLK; MSC; TREX; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000406900.5; ENST00000212015.11; ENST00000432464.5; ENST00000403579.1 | ||
| External Link | RMBase: m6A_site_102842 | ||
| mod ID: M6ASITE000342 | Click to Show/Hide the Full List | ||
| mod site | chr10:67912865-67912866:+ | [15] | |
| Sequence | AAAAACCACAGGAAGTACAAACTTCTAGGAATGTTGAAAGT | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; U2OS; hNPCs; hESCs; fibroblasts; A549; GM12878; MM6; Huh7; Jurkat; peripheral-blood; HEK293A-TOA; iSLK; MSC; TREX; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000403579.1; ENST00000212015.11; ENST00000432464.5; ENST00000406900.5 | ||
| External Link | RMBase: m6A_site_102843 | ||
| mod ID: M6ASITE000343 | Click to Show/Hide the Full List | ||
| mod site | chr10:67912894-67912895:+ | [15] | |
| Sequence | AATGTTGAAAGTATTGCTGAACAGATGGAAAATCCGGATTT | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; U2OS; hNPCs; hESCs; fibroblasts; A549; GM12878; MM6; Huh7; Jurkat; peripheral-blood; HEK293A-TOA; iSLK; MSC; TIME; TREX; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000432464.5; ENST00000212015.11; ENST00000406900.5; ENST00000403579.1 | ||
| External Link | RMBase: m6A_site_102844 | ||
| mod ID: M6ASITE000344 | Click to Show/Hide the Full List | ||
| mod site | chr10:67912955-67912956:+ | [15] | |
| Sequence | CTGGGGAGAAAAATGAAAGAACTTCAGTGGCTGGAACAGTG | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; A549; U2OS; hNPCs; hESCs; fibroblasts; GM12878; CD8T; MM6; Huh7; peripheral-blood; HEK293A-TOA; iSLK; MSC; TIME; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq; m6A-CLIP/IP | ||
| Transcript ID List | ENST00000432464.5; ENST00000403579.1; ENST00000406900.5; ENST00000212015.11 | ||
| External Link | RMBase: m6A_site_102845 | ||
| mod ID: M6ASITE000345 | Click to Show/Hide the Full List | ||
| mod site | chr10:67912970-67912971:+ | [15] | |
| Sequence | AAAGAACTTCAGTGGCTGGAACAGTGAGAAAATGCTGGCCT | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; A549; U2OS; hNPCs; hESCs; fibroblasts; GM12878; MM6; Huh7; peripheral-blood; HEK293A-TOA; iSLK; MSC; TIME; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000403579.1; ENST00000406900.5; ENST00000432464.5; ENST00000212015.11 | ||
| External Link | RMBase: m6A_site_102846 | ||
| mod ID: M6ASITE000346 | Click to Show/Hide the Full List | ||
| mod site | chr10:67916297-67916298:+ | [16] | |
| Sequence | GTTTTTGCCACCAAATCGTTACATTTTCCATGGCGCTGAGG | ||
| Motif Score | 2.07285119 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000432464.5; ENST00000403579.1; ENST00000212015.11; ENST00000406900.5 | ||
| External Link | RMBase: m6A_site_102847 | ||
| mod ID: M6ASITE000347 | Click to Show/Hide the Full List | ||
| mod site | chr10:67916327-67916328:+ | [15] | |
| Sequence | TGGCGCTGAGGTATATTCAGACTCTGAAGATGACGTCTTAT | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; A549; U2OS; hNPCs; hESCs; fibroblasts; GM12878; Huh7; HEK293A-TOA; iSLK; MSC; TIME | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000406900.5; ENST00000432464.5; ENST00000212015.11; ENST00000403579.1 | ||
| External Link | RMBase: m6A_site_102848 | ||
| mod ID: M6ASITE000348 | Click to Show/Hide the Full List | ||
| mod site | chr10:67916369-67916370:+ | [17] | |
| Sequence | CTCTAGTTCTTGTGGCAGTAACAGTGATAGTGGGACATGCC | ||
| Motif Score | 2.168095238 | ||
| Cell/Tissue List | kidney | ||
| Seq Type List | m6A-REF-seq | ||
| Transcript ID List | ENST00000212015.11; ENST00000406900.5; ENST00000432464.5; ENST00000403579.1 | ||
| External Link | RMBase: m6A_site_102849 | ||
| mod ID: M6ASITE000349 | Click to Show/Hide the Full List | ||
| mod site | chr10:67916383-67916384:+ | [15] | |
| Sequence | GCAGTAACAGTGATAGTGGGACATGCCAGAGTCCAAGTTTA | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; A549; hESC-HEK293T; U2OS; hNPCs; hESCs; fibroblasts; GM12878; Huh7; HEK293A-TOA; iSLK; MSC; TIME | ||
| Seq Type List | m6A-seq; MeRIP-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000403579.1; ENST00000212015.11; ENST00000432464.5; ENST00000406900.5 | ||
| External Link | RMBase: m6A_site_102850 | ||
| mod ID: M6ASITE000350 | Click to Show/Hide the Full List | ||
| mod site | chr10:67916409-67916410:+ | [15] | |
| Sequence | CAGAGTCCAAGTTTAGAAGAACCCATGGAGGATGAAAGTGA | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; A549; U2OS; hNPCs; hESCs; fibroblasts; GM12878; Huh7; HEK293A-TOA; iSLK; MSC; TREX | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000432464.5; ENST00000403579.1; ENST00000212015.11; ENST00000406900.5 | ||
| External Link | RMBase: m6A_site_102851 | ||
| mod ID: M6ASITE000351 | Click to Show/Hide the Full List | ||
| mod site | chr10:67916444-67916445:+ | [16] | |
| Sequence | AAGTGAAATTGAAGAATTCTACAATGGCTTAGAAGATGAGC | ||
| Motif Score | 2.078666667 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000403579.1; ENST00000406900.5; ENST00000432464.5; ENST00000212015.11 | ||
| External Link | RMBase: m6A_site_102852 | ||
| mod ID: M6ASITE000352 | Click to Show/Hide the Full List | ||
| mod site | chr10:67916503-67916504:+ | [15] | |
| Sequence | CTGGAGGAGCTGGATTTGGGACTGATGGAGATGATCAAGAG | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; A549; U2OS; hNPCs; hESCs; fibroblasts; GM12878; CD8T; MM6; Huh7; Jurkat; peripheral-blood; HEK293A-TOA; iSLK; MSC; TIME; TREX | ||
| Seq Type List | m6A-seq; MeRIP-seq; m6A-CLIP/IP | ||
| Transcript ID List | ENST00000212015.11; ENST00000406900.5; ENST00000403579.1; ENST00000432464.5 | ||
| External Link | RMBase: m6A_site_102853 | ||
| mod ID: M6ASITE000353 | Click to Show/Hide the Full List | ||
| mod site | chr10:67916550-67916551:+ | [15] | |
| Sequence | AATGAAGCTATATCTGTGAAACAGGAAGTAACAGACATGAA | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; A549; hESC-HEK293T; U2OS; hNPCs; hESCs; fibroblasts; GM12878; CD8T; MM6; Huh7; Jurkat; HEK293A-TOA; iSLK; MSC; TIME; TREX | ||
| Seq Type List | m6A-seq; MeRIP-seq; DART-seq; MAZTER-seq; m6A-CLIP/IP | ||
| Transcript ID List | ENST00000406900.5; ENST00000432464.5; ENST00000403579.1; ENST00000212015.11 | ||
| External Link | RMBase: m6A_site_102854 | ||
| mod ID: M6ASITE000354 | Click to Show/Hide the Full List | ||
| mod site | chr10:67916560-67916561:+ | [16] | |
| Sequence | TATCTGTGAAACAGGAAGTAACAGACATGAACTATCCATCA | ||
| Motif Score | 2.168095238 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000432464.5; ENST00000212015.11; ENST00000406900.5; ENST00000403579.1 | ||
| External Link | RMBase: m6A_site_102855 | ||
| mod ID: M6ASITE000355 | Click to Show/Hide the Full List | ||
| mod site | chr10:67916564-67916565:+ | [15] | |
| Sequence | TGTGAAACAGGAAGTAACAGACATGAACTATCCATCAAACA | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; A549; hESC-HEK293T; U2OS; hNPCs; hESCs; fibroblasts; GM12878; MM6; Huh7; Jurkat; HEK293A-TOA; iSLK; MSC; TIME; TREX | ||
| Seq Type List | m6A-seq; MeRIP-seq; DART-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000406900.5; ENST00000403579.1; ENST00000212015.11; ENST00000432464.5 | ||
| External Link | RMBase: m6A_site_102856 | ||
| mod ID: M6ASITE000356 | Click to Show/Hide the Full List | ||
| mod site | chr10:67916570-67916571:+ | [15] | |
| Sequence | ACAGGAAGTAACAGACATGAACTATCCATCAAACAAATCAT | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; A549; U2OS; hNPCs; hESCs; fibroblasts; GM12878; CD8T; MM6; Huh7; HEK293A-TOA; iSLK; MSC; TIME; TREX | ||
| Seq Type List | m6A-seq; MeRIP-seq; m6A-CLIP/IP | ||
| Transcript ID List | ENST00000212015.11; ENST00000406900.5; ENST00000403579.1; ENST00000432464.5 | ||
| External Link | RMBase: m6A_site_102857 | ||
| mod ID: M6ASITE000357 | Click to Show/Hide the Full List | ||
| mod site | chr10:67916582-67916583:+ | [15] | |
| Sequence | AGACATGAACTATCCATCAAACAAATCATAGTGTAATAATT | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; A549; U2OS; hNPCs; hESCs; fibroblasts; GM12878; MM6; Huh7; HEK293A-TOA; iSLK; MSC; TIME; TREX | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000432464.5; ENST00000406900.5; ENST00000403579.1; ENST00000212015.11 | ||
| External Link | RMBase: m6A_site_102858 | ||
| mod ID: M6ASITE000358 | Click to Show/Hide the Full List | ||
| mod site | chr10:67916611-67916612:+ | [18] | |
| Sequence | AGTGTAATAATTGTGCAGGTACAGGAATTGTTCCACCAGCA | ||
| Motif Score | 2.856142857 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000212015.11; ENST00000406900.5; ENST00000432464.5; ENST00000403579.1 | ||
| External Link | RMBase: m6A_site_102859 | ||
| mod ID: M6ASITE000359 | Click to Show/Hide the Full List | ||
| mod site | chr10:67916638-67916639:+ | [15] | |
| Sequence | TTGTTCCACCAGCATTAGGAACTTTAGCATGTCAAAATGAA | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; A549; U2OS; hNPCs; hESCs; fibroblasts; GM12878; Huh7; HEK293A-TOA; iSLK; MSC; TIME; TREX | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000212015.11; ENST00000403579.1; ENST00000406900.5; ENST00000432464.5 | ||
| External Link | RMBase: m6A_site_102860 | ||
| mod ID: M6ASITE000360 | Click to Show/Hide the Full List | ||
| mod site | chr10:67916672-67916673:+ | [15] | |
| Sequence | AAATGAATGTTTACTTGTGAACTCGATAGAGCAAGGAAACC | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; A549; U2OS; hNPCs; hESCs; fibroblasts; GM12878; Huh7; HEK293A-TOA; iSLK; MSC; TIME; TREX | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000403579.1; ENST00000432464.5; ENST00000212015.11; ENST00000406900.5 | ||
| External Link | RMBase: m6A_site_102861 | ||
| mod ID: M6ASITE000361 | Click to Show/Hide the Full List | ||
| mod site | chr10:67916690-67916691:+ | [15] | |
| Sequence | GAACTCGATAGAGCAAGGAAACCAGAAAGGTGTAATATTTA | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; A549; hNPCs; hESCs; fibroblasts; GM12878; Huh7; HEK293A-TOA; iSLK; MSC; TIME; TREX | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000432464.5; ENST00000403579.1; ENST00000212015.11; ENST00000406900.5 | ||
| External Link | RMBase: m6A_site_102862 | ||
| mod ID: M6ASITE000362 | Click to Show/Hide the Full List | ||
| mod site | chr10:67916751-67916752:+ | [18] | |
| Sequence | TTTTTCATGGATAATTTTTAACTTCATTATTTCTGTACTTG | ||
| Motif Score | 2.590089286 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000432464.5; ENST00000403579.1; ENST00000212015.11; ENST00000406900.5 | ||
| External Link | RMBase: m6A_site_102863 | ||
| mod ID: M6ASITE000363 | Click to Show/Hide the Full List | ||
| mod site | chr10:67916773-67916774:+ | [16] | |
| Sequence | TTCATTATTTCTGTACTTGTACAAACTCAACACTAACTTTT | ||
| Motif Score | 2.856142857 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000212015.11; ENST00000406900.5; ENST00000432464.5; ENST00000403579.1 | ||
| External Link | RMBase: m6A_site_102864 | ||
| mod ID: M6ASITE000364 | Click to Show/Hide the Full List | ||
| mod site | chr10:67916777-67916778:+ | [15] | |
| Sequence | TTATTTCTGTACTTGTACAAACTCAACACTAACTTTTTTTT | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa; HEK293T; Huh7; HEK293A-TOA; iSLK; MSC; TIME | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000432464.5; ENST00000403579.1; ENST00000212015.11; ENST00000406900.5 | ||
| External Link | RMBase: m6A_site_102865 | ||
| mod ID: M6ASITE000365 | Click to Show/Hide the Full List | ||
| mod site | chr10:67916930-67916931:+ | [16] | |
| Sequence | GTTTTGTCTAGTGAGTTTCAACATTTTTAAAGTTTTCAAAA | ||
| Motif Score | 2.173910714 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000403579.1; ENST00000212015.11; ENST00000432464.5; ENST00000406900.5 | ||
| External Link | RMBase: m6A_site_102866 | ||
| mod ID: M6ASITE000366 | Click to Show/Hide the Full List | ||
| mod site | chr10:67917028-67917029:+ | [16] | |
| Sequence | TATTTTCACTTTTCTTTGTAACATTGAATGGTTTGAAGTAC | ||
| Motif Score | 2.168095238 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000212015.11; ENST00000432464.5; ENST00000403579.1; ENST00000406900.5 | ||
| External Link | RMBase: m6A_site_102867 | ||
| mod ID: M6ASITE000367 | Click to Show/Hide the Full List | ||
| mod site | chr10:67917047-67917048:+ | [18] | |
| Sequence | AACATTGAATGGTTTGAAGTACTCAAAATCTGTTACGCTAA | ||
| Motif Score | 3.278136905 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000403579.1; ENST00000212015.11; ENST00000406900.5; ENST00000432464.5 | ||
| External Link | RMBase: m6A_site_102868 | ||
| mod ID: M6ASITE000368 | Click to Show/Hide the Full List | ||
| mod site | chr10:67917083-67917084:+ | [16] | |
| Sequence | GCTAAACTTTTGATTCTTTAACACAATTATTTTTAAACACT | ||
| Motif Score | 2.168095238 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000403579.1; ENST00000212015.11; ENST00000406900.5; ENST00000432464.5 | ||
| External Link | RMBase: m6A_site_102869 | ||
| mod ID: M6ASITE000369 | Click to Show/Hide the Full List | ||
| mod site | chr10:67917099-67917100:+ | [15] | |
| Sequence | TTTAACACAATTATTTTTAAACACTGGCATTTTCCAAAACT | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa; hESC-HEK293T | ||
| Seq Type List | m6A-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000403579.1; ENST00000406900.5; ENST00000212015.11; ENST00000432464.5 | ||
| External Link | RMBase: m6A_site_102870 | ||
| mod ID: M6ASITE000370 | Click to Show/Hide the Full List | ||
| mod site | chr10:67917117-67917118:+ | [15] | |
| Sequence | AAACACTGGCATTTTCCAAAACTGTGGCAGCTAACTTTTTA | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000432464.5; ENST00000212015.11; ENST00000403579.1; ENST00000406900.5 | ||
| External Link | RMBase: m6A_site_102871 | ||
| mod ID: M6ASITE000371 | Click to Show/Hide the Full List | ||
| mod site | chr10:67917150-67917151:+ | [16] | |
| Sequence | ACTTTTTAAAATCTCAAATGACATGCAGTGTGAGTAGAAGG | ||
| Motif Score | 2.859755952 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000212015.11; ENST00000432464.5; ENST00000406900.5; ENST00000403579.1 | ||
| External Link | RMBase: m6A_site_102872 | ||
| mod ID: M6ASITE000372 | Click to Show/Hide the Full List | ||
| mod site | chr10:67917177-67917178:+ | [16] | |
| Sequence | GTGTGAGTAGAAGGAAGTCAACAATATGTGGGGAGAGCACT | ||
| Motif Score | 2.173910714 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000212015.11; ENST00000432464.5; ENST00000406900.5; ENST00000403579.1 | ||
| External Link | RMBase: m6A_site_102873 | ||
| mod ID: M6ASITE000373 | Click to Show/Hide the Full List | ||
| mod site | chr10:67917258-67917259:+ | [18] | |
| Sequence | ATTTCAGGATTATTGTATTTACGTTCAAATGAAGATGGCTT | ||
| Motif Score | 2.046785714 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000403579.1; ENST00000406900.5; ENST00000212015.11; ENST00000432464.5 | ||
| External Link | RMBase: m6A_site_102874 | ||
| mod ID: M6ASITE000374 | Click to Show/Hide the Full List | ||
| mod site | chr10:67917294-67917295:+ | [16] | |
| Sequence | GGCTTTTGTACTTCCTGTGGACATGTAGCAATGTCTATATT | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000432464.5; ENST00000212015.11; ENST00000403579.1; ENST00000406900.5 | ||
| External Link | RMBase: m6A_site_102875 | ||
| mod ID: M6ASITE000375 | Click to Show/Hide the Full List | ||
| mod site | chr10:67917377-67917378:+ | [15] | |
| Sequence | ATGTTTCAGTTGCTTTAGAAACATTAGTGCCTGCCTGGATC | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa; HEK293T; hESC-HEK293T | ||
| Seq Type List | m6A-seq; DART-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000406900.5; ENST00000212015.11; ENST00000432464.5; ENST00000403579.1 | ||
| External Link | RMBase: m6A_site_102876 | ||
| mod ID: M6ASITE000376 | Click to Show/Hide the Full List | ||
| mod site | chr10:67917472-67917473:+ | [16] | |
| Sequence | AGCTTGCATTGATCTTTTCCACAAGTATTAAACTGCCAAAA | ||
| Motif Score | 2.053113095 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000432464.5; ENST00000212015.11; ENST00000406900.5; ENST00000403579.1 | ||
| External Link | RMBase: m6A_site_102877 | ||
| mod ID: M6ASITE000377 | Click to Show/Hide the Full List | ||
| mod site | chr10:67917653-67917654:+ | [18] | |
| Sequence | TTGTAATTCAGCCAGAAAGTACATGTCTCCCATTGGGAGGA | ||
| Motif Score | 2.856142857 | ||
| Cell/Tissue List | HEK293T; hESC-HEK293T | ||
| Seq Type List | DART-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000403579.1; ENST00000432464.5; ENST00000406900.5; ENST00000212015.11 | ||
| External Link | RMBase: m6A_site_102878 | ||
| mod ID: M6ASITE000378 | Click to Show/Hide the Full List | ||
| mod site | chr10:67917692-67917693:+ | [18] | |
| Sequence | GATTTGGTGTTAAATACCAAACTGCTAGCCCTAGTATTATG | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000432464.5; ENST00000212015.11; ENST00000406900.5; ENST00000403579.1 | ||
| External Link | RMBase: m6A_site_102879 | ||
| mod ID: M6ASITE000379 | Click to Show/Hide the Full List | ||
| mod site | chr10:67917720-67917721:+ | [15] | |
| Sequence | CCCTAGTATTATGGAGATGAACATGATGATGTAACTTGTAA | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa; HEK293T; hESC-HEK293T | ||
| Seq Type List | m6A-seq; DART-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000432464.5; ENST00000403579.1; ENST00000406900.5; ENST00000212015.11 | ||
| External Link | RMBase: m6A_site_102880 | ||
| mod ID: M6ASITE000380 | Click to Show/Hide the Full List | ||
| mod site | chr10:67917764-67917765:+ | [15] | |
| Sequence | CAGAATAGTTAATGAATGAAACTAGTTCTTATAATTTATCT | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa; HEK293T | ||
| Seq Type List | m6A-seq; DART-seq | ||
| Transcript ID List | ENST00000403579.1; ENST00000432464.5; ENST00000406900.5; ENST00000212015.11 | ||
| External Link | RMBase: m6A_site_102881 | ||
| mod ID: M6ASITE000381 | Click to Show/Hide the Full List | ||
| mod site | chr10:67917812-67917813:+ | [18] | |
| Sequence | AAAGCTTAGCCTGCCTTAAAACTAGAGATCAACTTTCTCAG | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000212015.11; ENST00000432464.5; ENST00000403579.1; ENST00000406900.5 | ||
| External Link | RMBase: m6A_site_102882 | ||
| mod ID: M6ASITE000382 | Click to Show/Hide the Full List | ||
| mod site | chr10:67917914-67917915:+ | [16] | |
| Sequence | ATTTTTCAGACCATTTTTGAACATCACTCCTAAATTAATAA | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000403579.1; ENST00000432464.5; ENST00000212015.11; ENST00000406900.5 | ||
| External Link | RMBase: m6A_site_102883 | ||
| mod ID: M6ASITE000383 | Click to Show/Hide the Full List | ||
| mod site | chr10:67917964-67917965:+ | [16] | |
| Sequence | CTGTTGCTTTAGTATTTATTACAATAAAAAGGGTTTGAAAT | ||
| Motif Score | 2.07285119 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000212015.11; ENST00000432464.5; ENST00000406900.5; ENST00000403579.1 | ||
| External Link | RMBase: m6A_site_102884 | ||
| mod ID: M6ASITE000384 | Click to Show/Hide the Full List | ||
| mod site | chr10:67918007-67918008:+ | [15] | |
| Sequence | AGCTGTTCTTTATGCATAAAACACCCAGCTAGGACCATTAC | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa; hESC-HEK293T | ||
| Seq Type List | m6A-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000212015.11; ENST00000403579.1; ENST00000432464.5; ENST00000406900.5 | ||
| External Link | RMBase: m6A_site_102885 | ||
| mod ID: M6ASITE000385 | Click to Show/Hide the Full List | ||
| mod site | chr10:67918020-67918021:+ | [15] | |
| Sequence | GCATAAAACACCCAGCTAGGACCATTACTGCCAGAGAAAAA | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000432464.5; ENST00000403579.1; ENST00000406900.5; ENST00000212015.11 | ||
| External Link | RMBase: m6A_site_102886 | ||
| mod ID: M6ASITE000386 | Click to Show/Hide the Full List | ||
| mod site | chr10:67918066-67918067:+ | [18] | |
| Sequence | TATTGAATGGCCATTTCCCTACTTATAAGATGTCTCAATCT | ||
| Motif Score | 2.500660714 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000406900.5; ENST00000403579.1; ENST00000432464.5; ENST00000212015.11 | ||
| External Link | RMBase: m6A_site_102887 | ||
| mod ID: M6ASITE000387 | Click to Show/Hide the Full List | ||
| mod site | chr10:67918101-67918102:+ | [16] | |
| Sequence | CAATCTGAATTTATTTGGCTACACTAAAGAATGCAGTATAT | ||
| Motif Score | 2.078666667 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000212015.11; ENST00000432464.5; ENST00000406900.5; ENST00000403579.1 | ||
| External Link | RMBase: m6A_site_102888 | ||
| mod ID: M6ASITE000388 | Click to Show/Hide the Full List | ||
| mod site | chr10:67918198-67918199:+ | [17] | |
| Sequence | GTTTGTTTTAAATTATTTTTACAGTGAAGACTGTTTTCAGC | ||
| Motif Score | 2.07285119 | ||
| Cell/Tissue List | brain; HEK293; HEK293T | ||
| Seq Type List | m6A-REF-seq; DART-seq | ||
| Transcript ID List | ENST00000432464.5; ENST00000403579.1; ENST00000212015.11; ENST00000406900.5 | ||
| External Link | RMBase: m6A_site_102889 | ||
| mod ID: M6ASITE000389 | Click to Show/Hide the Full List | ||
| mod site | chr10:67918207-67918208:+ | [18] | |
| Sequence | AAATTATTTTTACAGTGAAGACTGTTTTCAGCTCTTTTTAT | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000406900.5; ENST00000212015.11; ENST00000403579.1; ENST00000432464.5 | ||
| External Link | RMBase: m6A_site_102890 | ||
| mod ID: M6ASITE000390 | Click to Show/Hide the Full List | ||
| mod site | chr10:67918233-67918234:+ | [16] | |
| Sequence | TTCAGCTCTTTTTATATTGTACATAGTCTTTTATGTAATTT | ||
| Motif Score | 2.856142857 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000406900.5; ENST00000212015.11; ENST00000403579.1; ENST00000432464.5 | ||
| External Link | RMBase: m6A_site_102891 | ||
| mod ID: M6ASITE000391 | Click to Show/Hide the Full List | ||
| mod site | chr10:67918313-67918314:+ | [16] | |
| Sequence | CTTCCTTCAACTTTTGAAATACAAAACCAGTGTTTTTTACT | ||
| Motif Score | 2.110482143 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000406900.5; ENST00000403579.1; ENST00000212015.11; ENST00000432464.5 | ||
| External Link | RMBase: m6A_site_102892 | ||
References