m6A Regulator Information
General Information of the m6A Regulator (ID: REG00023)
| Regulator Name | YTH domain-containing protein 2 (YTHDC2) | ||||
|---|---|---|---|---|---|
| Synonyms |
3'-5' RNA helicase YTHDC2; hYTHDC2
Click to Show/Hide
|
||||
| Gene Name | YTHDC2 | ||||
| Sequence |
MSRPSSVSPRQPAPGGGGGGGPSPCGPGGGGRAKGLKDIRIDEEVKIAVNIALERFRYGD
QREMEFPSSLTSTERAFIHRLSQSLGLVSKSKGKGANRYLTVKKKDGSETAHAMMTCNLT HNTKHAVRSLIQRFPVTNKERTELLPKTERGNVFAVEAENREMSKTSGRLNNGIPQIPVK RGESEFDSFRQSLPVFEKQEEIVKIIKENKVVLIVGETGSGKTTQIPQFLLDDCFKNGIP CRIFCTQPRRLAAIAVAERVAAERRERIGQTIGYQIRLESRVSPKTLLTFCTNGVLLRTL MAGDSTLSTVTHVIVDEVHERDRFSDFLLTKLRDLLQKHPTLKLILSSAALDVNLFIRYF GSCPVIYIQGRPFEVKEMFLEDILRTTGYTNKEMLKYKKEKQQEEKQQTTLTEWYSAQEN SFKPESQRQRTVLNVTDEYDLLDDGGDAVFSQLTEKDVNCLEPWLIKEMDACLSDIWLHK DIDAFAQVFHLILTENVSVDYRHSETSATALMVAAGRGFASQVEQLISMGANVHSKASNG WMALDWAKHFGQTEIVDLLESYSATLEFGNLDESSLVQTNGSDLSAEDRELLKAYHHSFD DEKVDLDLIMHLLYNICHSCDAGAVLIFLPGYDEIVGLRDRILFDDKRFADSTHRYQVFM LHSNMQTSDQKKVLKNPPAGVRKIILSTNIAETSITVNDVVFVIDSGKVKEKSFDALNFV TMLKMVWISKASAIQRKGRAGRCRPGICFRLFSRLRFQNMLEFQTPELLRMPLQELCLHT KLLAPVNCPIADFLMKAPEPPPALIVRNAVQMLKTIDAMDTWEDLTELGYHLADLPVEPH LGKMVLCAVVLKCLDPILTIACTLAYRDPFVLPTQASQKRAAMLCRKRFTAGAFSDHMAL LRAFQAWQKARSDGWERAFCEKNFLSQATMEIIIGMRTQLLGQLRASGFVRARGGGDIRD VNTNSENWAVVKAALVAGMYPNLVHVDRENLVLTGPKEKKVRFHPASVLSQPQYKKIPPA NGQAAAIKALPTDWLIYDEMTRAHRIANIRCCSAVTPVTILVFCGPARLASNALQEPSSF RVDGIPNDSSDSEMEDKTTANLAALKLDEWLHFTLEPEAASLLLQLRQKWHSLFLRRMRA PSKPWSQVDEATIRAIIAVLSTEEQSAGLQQPSGIGQRPRPMSSEELPLASSWRSNNSRK SSADTEFSDECTTAERVLMKSPSPALHPPQKYKDRGILHPKRGTEDRSDQSSLKSTDSSS YPSPCASPSPPSSGKGSKSPSPRPNMPVRYFIMKSSNLRNLEISQQKGIWSTTPSNERKL NRAFWESSIVYLVFSVQGSGHFQGFSRMSSEIGREKSQDWGSAGLGGVFKVEWIRKESLP FQFAHHLLNPWNDNKKVQISRDGQELEPLVGEQLLQLWERLPLGEKNTTD Click to Show/Hide
|
||||
| Family | DEAD box helicase family; DEAH subfamily | ||||
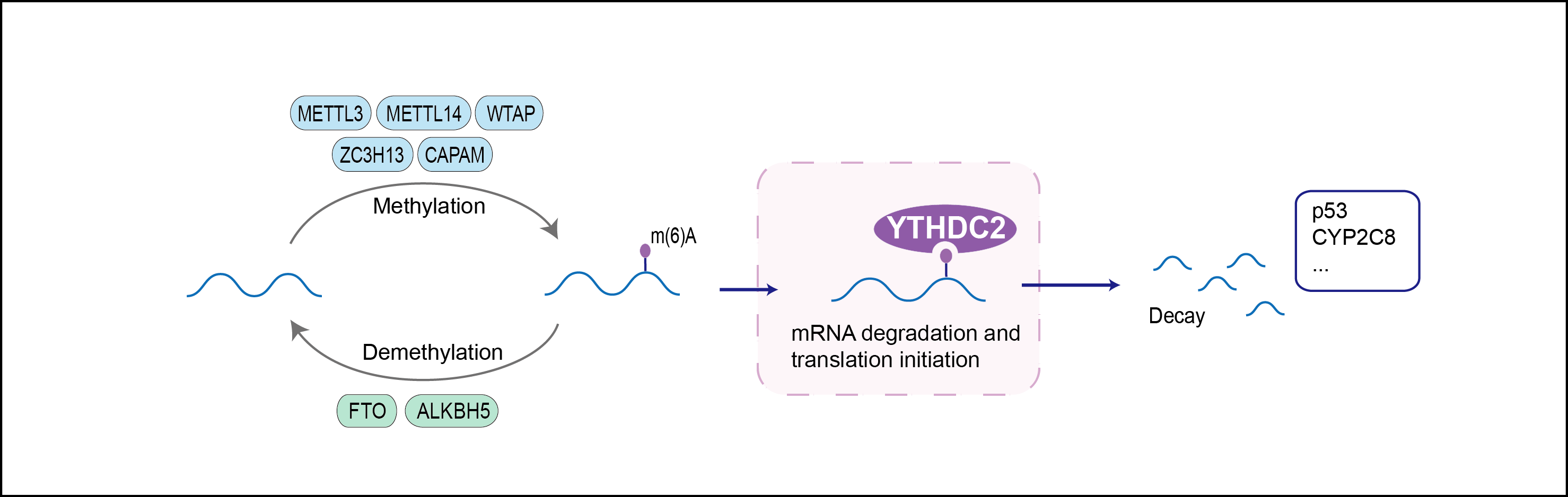
| Function |
3'-5' RNA helicase that plays a key role in the male and female germline by promoting transition from mitotic to meiotic divisions in stem cells. Specifically recognizes and binds N6-methyladenosine (m6A)-containing RNAs, a modification present at internal sites of mRNAs and some non-coding RNAs that plays a role in the efficiency of RNA processing and stability. Essential for ensuring a successful progression of the meiotic program in the germline by regulating the level of m6A-containing RNAs (By similarity). Acts by binding and promoting degradation of m6A-containing mRNAs: the 3'-5' RNA helicase activity is required for this process and RNA degradation may be mediated by XRN1 exoribonuclease. Required for both spermatogenesis and oogenesis (By similarity).
Click to Show/Hide
|
||||
| Gene ID | 64848 | ||||
| Uniprot ID | |||||
| Regulator Type | WRITER ERASER READER | ||||
| Mechanism Diagram | Click to View the Original Diagram | ||||
|
|||||
| Target Genes | Click to View Potential Target Genes of This Regulator | ||||
Full List of Target Gene(s) of This m6A Regulator and Corresponding Disease/Drug Response(s)
YTHDC2 can regulate the m6A methylation of following target genes, and result in corresponding disease/drug response(s). You can browse corresponding disease or drug response(s) resulted from the regulation of certain target gene.
Browse Target Gene related Disease
Browse Target Gene related Drug
40S ribosomal protein S6 (S6)
Nasopharyngeal carcinoma [ICD-11: 2B6B]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [1] | |||
| Responsed Disease | Nasopharyngeal carcinoma [ICD-11: 2B6B] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
HK1-IRR (HK1-IRR (HK1-ionizing radiation radioresistent cell line) was derived from HK1 after a prolonged exposure of irradiation.HK1, a generous gift from Prof. Ya Cao (Cancer Research Institute, Central South University), was established from a recurrent nasopharynx carcinoma of a Chinese 17-year-old male patient) | |||
| NPC/HK1 | Nasopharyngeal carcinoma | Homo sapiens | CVCL_7084 | |
| CNE2-IRR (CNE2-IRR (CNE2-ionizing radiation radioresistent cell line) was derived from CNE2 after a prolonged exposure of irradiation) | ||||
| CNE-2 | Nasopharyngeal carcinoma | Homo sapiens | CVCL_6889 | |
| In-vivo Model | 2 × 106 cells resuspended in 50 uL of Matrigel (Corning) were subcutaneously injected into 4-6 weeks old male nude mice. When tumor volumes reached 150-200 mm3, animals were divided into control group and radiotherapy group. In the radiotherapy group, tumors were treated with a single irradiation (4 Gy) when tumor volumes reached approximately 150-200 mm3. The tumor stopped growing in the next few days and then restarted growth. | |||
| Response Summary | YTHDC2 promotes radiotherapy resistance of NPC cells by activating the IGF1R/ATK/40S ribosomal protein S6 (S6) signaling axis and serves as a potential therapeutic target in radiosensitization of NPC cells. | |||
Acetyl-CoA carboxylase 1 (ACC1/ACACA)
Non-alcoholic fatty liver disease [ICD-11: DB92]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [2] | |||
| Responsed Disease | Non-alcoholic fatty liver disease [ICD-11: DB92] | |||
| Target Regulation | Down regulation | |||
| Pathway Response | RNA degradation | hsa03018 | ||
| Cell Process | RNA stability | |||
| In-vivo Model | All mice were housed at 21℃ ± 1℃ with a humidity of 55% ± 10% and a 12-hour light/dark cycle. The high-fat diets (HFDs), containing 60% kcal from fat, 20% kcal from carbohydrate, and 20% kcal from protein. | |||
| Response Summary | In nonalcoholic fatty liver disease, Ythdc2 could bind to mRNA of lipogenic genes, including sterol regulatory element-binding protein 1c, fatty acid synthase, stearoyl-CoA desaturase 1, and Acetyl-CoA carboxylase 1 (ACC1/ACACA), to decrease their mRNA stability and inhibit gene expression. | |||
Autophagy protein 5 (ATG5)
Liver cancer [ICD-11: 2C12]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [3] | |||
| Responsed Disease | Liver hepatocellular carcinoma [ICD-11: 2C12.02] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
Huh-7 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_0336 |
| Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 | |
| In-vivo Model | Four-week-old BALB/C-nu nude male mice were used for animal studies, and all animals were maintained in the specific pathogen-free (SPF) conditions at our institution. Huh-7 and stable YTHDC2 knockdown Huh-7 cells (approximately 1 × 107) resuspended with 50 μl of PBS and 50 μl of stromal gel were injected subcutaneously into the axilla of BALB/c nude mice to establish the subcutaneous xenograft model. When the volume of xenograft tumors up to 100 mm3, the mice were randomly divided into six groups, with five mice in each group. Erastin and sorafenib were dissolved in 10 % DMSO and 90 % corn oil and injected intraperitoneally into the mice every other day. After 28 days, mice were deeply anesthetized by intraperitoneal injection of sodium thiopental before decapitation, followed by tumors extraction, and stored in a -80 °C refrigerator for subsequent experiments. There were no occurrences of mouse mortality throughout the entire experimental period. | |||
Cyclin-dependent kinase inhibitor 1 (CDKN1A)
Esophageal cancer [ICD-11: 2B70]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [4] | |||
| Responsed Disease | Esophageal squamous cell carcinoma [ICD-11: 2B70.1] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | p53 signaling pathway | hsa04115 | ||
| NF-kappa B signaling pathway | hsa04064 | |||
| JAK-STAT signaling pathway | hsa04630 | |||
| Cell Process | Genetic variants | |||
In-vitro Model |
KYSE-150 | Esophageal squamous cell carcinoma | Homo sapiens | CVCL_1348 |
| KYSE-30 | Esophageal squamous cell carcinoma | Homo sapiens | CVCL_1351 | |
| Response Summary | Knockdown of YTHDC2 substantially promoted the proliferation rate of esophageal squamous cell carcinoma cells by affecting several cancer-related signaling pathways. | |||
Cystine/glutamate transporter (SLC7A11)
Lung cancer [ICD-11: 2C25]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [5] | |||
| Responsed Disease | Lung adenocarcinoma [ICD-11: 2C25.0] | |||
| Target Regulation | Down regulation | |||
In-vitro Model |
PC-9 | Lung adenocarcinoma | Homo sapiens | CVCL_B260 |
| NCI-H441 | Lung papillary adenocarcinoma | Homo sapiens | CVCL_1561 | |
| NCI-H292 | Lung mucoepidermoid carcinoma | Homo sapiens | CVCL_0455 | |
| NCI-H1975 | Lung adenocarcinoma | Homo sapiens | CVCL_1511 | |
| NCI-H1650 | Minimally invasive lung adenocarcinoma | Homo sapiens | CVCL_1483 | |
| NCI-H1299 | Lung large cell carcinoma | Homo sapiens | CVCL_0060 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| HCC827 | Lung adenocarcinoma | Homo sapiens | CVCL_2063 | |
| Calu-1 | Lung squamous cell carcinoma | Homo sapiens | CVCL_0608 | |
| BEAS-2B | Normal | Homo sapiens | CVCL_0168 | |
| A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 | |
| Response Summary | YTHDC2 destabilized Cystine/glutamate transporter (SLC7A11) mRNA in an m6A-dependent manner because YTHDC2 preferentially bound to m6A-modified SLC7A11 mRNA and thereafter promoted its decay. the promotion of cystine uptake via the suppression of YTHDC2 is critical for LUAD tumorigenesis. | |||
Cytochrome P450 2C8 (CYP2C8)
Liver cancer [ICD-11: 2C12]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [6] | |||
| Responsed Disease | Hepatocellular carcinoma [ICD-11: 2C12.02] | |||
| Target Regulation | Down regulation | |||
| Pathway Response | Drug metabolism - cytochrome P450 | hsa00982 | ||
| Cell Process | Drug-metabolizing | |||
In-vitro Model |
HepaRG | Hepatitis C infection | Homo sapiens | CVCL_9720 |
| Huh-7 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_0336 | |
| Response Summary | In the Hepatocellular carcinoma cells YTHDC2 promotes CYP2C8 mRNA degradation via recognizing the m6A in CYP2C8 mRNA, which is installed by METTL3/14 and removed by FTO. | |||
G1/S-specific cyclin-D2 (CCND2)
Esophageal cancer [ICD-11: 2B70]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [4] | |||
| Responsed Disease | Esophageal squamous cell carcinoma [ICD-11: 2B70.1] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | p53 signaling pathway | hsa04115 | ||
| NF-kappa B signaling pathway | hsa04064 | |||
| JAK-STAT signaling pathway | hsa04630 | |||
| Cell Process | Genetic variants | |||
In-vitro Model |
KYSE-150 | Esophageal squamous cell carcinoma | Homo sapiens | CVCL_1348 |
| KYSE-30 | Esophageal squamous cell carcinoma | Homo sapiens | CVCL_1351 | |
| Response Summary | Knockdown of YTHDC2 substantially promoted the proliferation rate of esophageal squamous cell carcinoma cells by affecting several cancer-related signaling pathways. | |||
G2/mitotic-specific cyclin-B2 (CCNB2)
Male infertility [ICD-11: GB04]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [7] | |||
| Responsed Disease | Male infertility [ICD-11: GB04] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | Cell cycle | hsa04110 | ||
| Cell Process | Block the G2/M phase | |||
In-vitro Model |
GC-1 spg | Normal | Mus musculus | CVCL_8872 |
| In-vivo Model | Mice in the control group received an i.p. injections of 0.9% NaCl. Mice in the low, medium, and high Mn groups received i.p. injections of 12.5, 25, and 50 mg/kg MnCl2. The volume of administration was 5 mL/kg body weight. The injection was given daily for 2 weeks. | |||
| Response Summary | Over-expression (OE) of YTHDC2 increased G2/mitotic-specific cyclin-B2 (CCNB2) levels, reduced cell cycle arrest, and improved reproductive toxicity after Mn exposure. | |||
Hypoxia-inducible factor 1-alpha (HIF-1-Alpha/HIF1A)
Colon cancer [ICD-11: 2B90]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [8] | |||
| Responsed Disease | Colon cancer [ICD-11: 2B90] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | HIF-1 signaling pathway | hsa04066 | ||
| Cell Process | Biological regulation | |||
In-vitro Model |
COS (From the African green monkey cell line (CV-1).) | |||
| HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 | |
| HT29 | Colon cancer | Mus musculus | CVCL_A8EZ | |
| In-vivo Model | HCT116, Y2KD-116 and con-116 cells were resuspended at 1 × 106 cells per 50 ul of PBS. Cells were injected into the exteriorized spleen after abdominal incision. | |||
| Response Summary | YTHDC2 contributes to colon tumor metastasis by promoting translation of Hypoxia-inducible factor 1-alpha (HIF-1-Alpha/HIF1A) and that YTHDC2 is potentially a diagnostic marker and target gene for treating colon cancer patients. | |||
Insulin-like growth factor 1 receptor (IGF1R)
Nasopharyngeal carcinoma [ICD-11: 2B6B]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [1] | |||
| Responsed Disease | Nasopharyngeal carcinoma [ICD-11: 2B6B] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
HK1-IRR (HK1-IRR (HK1-ionizing radiation radioresistent cell line) was derived from HK1 after a prolonged exposure of irradiation.HK1, a generous gift from Prof. Ya Cao (Cancer Research Institute, Central South University), was established from a recurrent nasopharynx carcinoma of a Chinese 17-year-old male patient) | |||
| NPC/HK1 | Nasopharyngeal carcinoma | Homo sapiens | CVCL_7084 | |
| CNE2-IRR (CNE2-IRR (CNE2-ionizing radiation radioresistent cell line) was derived from CNE2 after a prolonged exposure of irradiation) | ||||
| CNE-2 | Nasopharyngeal carcinoma | Homo sapiens | CVCL_6889 | |
| In-vivo Model | 2 × 106 cells resuspended in 50 uL of Matrigel (Corning) were subcutaneously injected into 4-6 weeks old male nude mice. When tumor volumes reached 150-200 mm3, animals were divided into control group and radiotherapy group. In the radiotherapy group, tumors were treated with a single irradiation (4 Gy) when tumor volumes reached approximately 150-200 mm3. The tumor stopped growing in the next few days and then restarted growth. | |||
| Response Summary | YTHDC2 promotes radiotherapy resistance of NPC cells by activating the Insulin-like growth factor 1 receptor (IGF1R)/ATK/S6 signaling axis and serves as a potential therapeutic target in radiosensitization of NPC cells. | |||
Interleukin-6 (IL-6)
Kaposi's sarcoma [ICD-11: 2B57]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [9] | |||
| Responsed Disease | Kaposi's sarcoma [ICD-11: 2B57] | |||
In-vitro Model |
iSLK.219 | Clear cell renal cell carcinoma | Homo sapiens | CVCL_B6YV |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| HEK293 | Normal | Homo sapiens | CVCL_0045 | |
| Response Summary | This modification recruits the m6A reader YTHDC2 and found that YTHDC2 is necessary for the escape of the IL-6 transcript. m6A modification is essential to confer SOX resistance to the Interleukin-6 (IL-6) mRNA. These results shed light on how the host cell has evolved to use RNA modifications to circumvent viral manipulation of RNA fate during KSHV infection Kaposi's sarcoma. | |||
Mutated in multiple advanced cancers 1 (PTEN)
Skin cancer [ICD-11: 2C3Z]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [10] | |||
| Responsed Disease | Skin cancer [ICD-11: 2C3Z] | |||
| Target Regulation | Down regulation | |||
In-vitro Model |
HaCaT | Normal | Homo sapiens | CVCL_0038 |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
Nuclear factor erythroid 2-related factor 2 (NFE2L2)
Male reproductive disorders [ICD-11: VV5Z]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [11] | |||
| Responsed Disease | Male reproductive disorders [ICD-11: VV5Z] | |||
| Cell Process | Oxidative stress | |||
| Cell apoptosis | ||||
| In-vivo Model | Exposed Sprague-Dawley rats to 0, 250, and 500 mg DEHP per kg body weight per day at the prepuberty stage from postnatal day 22 (PND 22) to PND 35 by oral gavage. | |||
| Response Summary | DEHP worsened testicular histology, decreased testosterone concentrations, downregulated expression of spermatogenesis inducers, enhanced oxidative stress, inhibited the Nuclear factor erythroid 2-related factor 2 (NFE2L2)-mediated antioxidant pathway, and increased apoptosis in testes. DEHP is a common environmental endocrine disrupting chemical that induces male reproductive disorders. Additionally, DEHP increased global levels of m6A RNA modification and altered the expression of two important RNA methylation modulator genes, FTO and YTHDC2. | |||
RAC-alpha serine/threonine-protein kinase (AKT1)
Nasopharyngeal carcinoma [ICD-11: 2B6B]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [1] | |||
| Responsed Disease | Nasopharyngeal carcinoma [ICD-11: 2B6B] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
HK1-IRR (HK1-IRR (HK1-ionizing radiation radioresistent cell line) was derived from HK1 after a prolonged exposure of irradiation.HK1, a generous gift from Prof. Ya Cao (Cancer Research Institute, Central South University), was established from a recurrent nasopharynx carcinoma of a Chinese 17-year-old male patient) | |||
| NPC/HK1 | Nasopharyngeal carcinoma | Homo sapiens | CVCL_7084 | |
| CNE2-IRR (CNE2-IRR (CNE2-ionizing radiation radioresistent cell line) was derived from CNE2 after a prolonged exposure of irradiation) | ||||
| CNE-2 | Nasopharyngeal carcinoma | Homo sapiens | CVCL_6889 | |
| In-vivo Model | 2 × 106 cells resuspended in 50 uL of Matrigel (Corning) were subcutaneously injected into 4-6 weeks old male nude mice. When tumor volumes reached 150-200 mm3, animals were divided into control group and radiotherapy group. In the radiotherapy group, tumors were treated with a single irradiation (4 Gy) when tumor volumes reached approximately 150-200 mm3. The tumor stopped growing in the next few days and then restarted growth. | |||
| Response Summary | YTHDC2 promotes radiotherapy resistance of NPC cells by activating the IGF1R/RAC-alpha serine/threonine-protein kinase (AKT1)/S6 signaling axis and serves as a potential therapeutic target in radiosensitization of NPC cells. | |||
Stearoyl-CoA desaturase (SCD)
Non-alcoholic fatty liver disease [ICD-11: DB92]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [2] | |||
| Responsed Disease | Non-alcoholic fatty liver disease [ICD-11: DB92] | |||
| Target Regulation | Down regulation | |||
| Pathway Response | RNA degradation | hsa03018 | ||
| Cell Process | RNA stability | |||
| In-vivo Model | All mice were housed at 21℃ ± 1℃ with a humidity of 55% ± 10% and a 12-hour light/dark cycle. The high-fat diets (HFDs), containing 60% kcal from fat, 20% kcal from carbohydrate, and 20% kcal from protein. | |||
| Response Summary | In nonalcoholic fatty liver disease, Ythdc2 could bind to mRNA of lipogenic genes, including sterol regulatory element-binding protein 1c, fatty acid synthase, Stearoyl-CoA desaturase (SCD), and acetyl-CoA carboxylase 1, to decrease their mRNA stability and inhibit gene expression. | |||
Papillary Thyroid Cancer [ICD-11: XH1ND9]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [12] | |||
| Responsed Disease | Papillary Thyroid Cancer [ICD-11: XH1ND9] | |||
| Responsed Drug | Simvastatin | Approved | ||
| Target Regulation | Down regulation | |||
In-vitro Model |
B-CPAP | Thyroid gland carcinoma | Homo sapiens | CVCL_0153 |
| TPC-1 | Thyroid gland papillary carcinoma | Homo sapiens | CVCL_6298 | |
| K1 | Thyroid gland papillary carcinoma | Homo sapiens | CVCL_2537 | |
| In-vivo Model | These mice were provided with unrestricted access to both water and food, following a 12-h light and 12-h dark cycle. Subcutaneous injections of stably transfected K1 cells (2 × 106 cells per mouse) were administered in the armpits of the mice. Once the tumors became palpable, their volume was measured every three days and calculated using the formula: length × width2 × 0.5. The tumor weight was detected after the mice were sacrificed. | |||
Sterol regulatory element-binding protein 1 (SREBF1)
Non-alcoholic fatty liver disease [ICD-11: DB92]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [2] | |||
| Responsed Disease | Non-alcoholic fatty liver disease [ICD-11: DB92] | |||
| Target Regulation | Down regulation | |||
| Pathway Response | RNA degradation | hsa03018 | ||
| Cell Process | RNA stability | |||
| In-vivo Model | All mice were housed at 21℃ ± 1℃ with a humidity of 55% ± 10% and a 12-hour light/dark cycle. The high-fat diets (HFDs), containing 60% kcal from fat, 20% kcal from carbohydrate, and 20% kcal from protein. | |||
| Response Summary | In nonalcoholic fatty liver disease, Ythdc2 could bind to mRNA of lipogenic genes, including Sterol regulatory element-binding protein 1 (SREBF1), fatty acid synthase, stearoyl-CoA desaturase 1, and acetyl-CoA carboxylase 1, to decrease their mRNA stability and inhibit gene expression. | |||
Transcriptional coactivator YAP1 (YAP1)
Gastric cancer [ICD-11: 2B72]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [13] | |||
| Responsed Disease | Gastric cancer [ICD-11: 2B72] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
HGC-27 | Gastric carcinoma | Homo sapiens | CVCL_1279 |
| AGS | Gastric adenocarcinoma | Homo sapiens | CVCL_0139 | |
| In-vivo Model | They were subcutaneously and caudal vein injected with YTHDC2 knockout AGS cells, respectively. After 7 weeks, the mice were sacrificed and tumor size and lung metastasis nodules were recorded. | |||
| Response Summary | High YTHDC2 was strongly positively correlated with high Transcriptional coactivator YAP1 (YAP1) in clinical GC tissues, YTHDC2 is a novel oncogene in GC, which provides the theoretical basis for the strategy of targeting YTHDC2 for GC patients. | |||
Transforming growth factor beta-2 proprotein (TGFB2)
Esophageal cancer [ICD-11: 2B70]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [4] | |||
| Responsed Disease | Esophageal squamous cell carcinoma [ICD-11: 2B70.1] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | p53 signaling pathway | hsa04115 | ||
| NF-kappa B signaling pathway | hsa04064 | |||
| JAK-STAT signaling pathway | hsa04630 | |||
| Cell Process | Genetic variants | |||
In-vitro Model |
KYSE-150 | Esophageal squamous cell carcinoma | Homo sapiens | CVCL_1348 |
| KYSE-30 | Esophageal squamous cell carcinoma | Homo sapiens | CVCL_1351 | |
| Response Summary | Knockdown of YTHDC2 substantially promoted the proliferation rate of esophageal squamous cell carcinoma cells by affecting several cancer-related signaling pathways. | |||
Tumor necrosis factor receptor superfamily member 6 (FAS)
Non-alcoholic fatty liver disease [ICD-11: DB92]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [2] | |||
| Responsed Disease | Non-alcoholic fatty liver disease [ICD-11: DB92] | |||
| Target Regulation | Down regulation | |||
| Pathway Response | RNA degradation | hsa03018 | ||
| Cell Process | RNA stability | |||
| In-vivo Model | All mice were housed at 21℃ ± 1℃ with a humidity of 55% ± 10% and a 12-hour light/dark cycle. The high-fat diets (HFDs), containing 60% kcal from fat, 20% kcal from carbohydrate, and 20% kcal from protein. | |||
| Response Summary | In nonalcoholic fatty liver disease, Ythdc2 could bind to mRNA of lipogenic genes, including sterol regulatory element-binding protein 1c, Tumor necrosis factor receptor superfamily member 6 (FAS), stearoyl-CoA desaturase 1, and acetyl-CoA carboxylase 1, to decrease their mRNA stability and inhibit gene expression. | |||
Ubiquitin carboxyl-terminal hydrolase CYLD (CYLD)
Lung cancer [ICD-11: 2C25]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [14] | |||
| Responsed Disease | Lung cancer [ICD-11: 2C25] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | NF-kappa B signaling pathway | hsa04064 | ||
In-vitro Model |
NCI-H1299 | Lung large cell carcinoma | Homo sapiens | CVCL_0060 |
| BEAS-2B | Normal | Homo sapiens | CVCL_0168 | |
| In-vivo Model | Approximately 5×106 normal H1299 cells or stable YTHDC2-overexpressing H1299 cells were implanted subcutaneously into the right flank of the animals (n=8 mice per group). Animals were euthanized by cervical dislocation ~30 days after implantation, and tumors were collected and photographed. | |||
| Response Summary | Smoking-related downregulation of YTHDC2 was associated with enhanced proliferation and migration in lung cancer cells, YTHDC2 functions as a tumor suppressor through the Ubiquitin carboxyl-terminal hydrolase CYLD (CYLD)/NF-Kappa-B signaling pathway, which is mediated by m6A modification. | |||
Adipogenesis regulatory factor (ADIRF)
Lung cancer [ICD-11: 2C25]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [15] | |||
| Responsed Disease | Lung adenocarcinoma [ICD-11: 2C25.0] | |||
| Target Regulation | Down regulation | |||
In-vitro Model |
A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 |
| H157 | Buccal mucosa squamous cell carcinoma | Homo sapiens | CVCL_2458 | |
| NCI-H460 | Lung large cell carcinoma | Homo sapiens | CVCL_0459 | |
| NCI-H1299 | Lung large cell carcinoma | Homo sapiens | CVCL_0060 | |
| PC-9 | Lung adenocarcinoma | Homo sapiens | CVCL_B260 | |
| BEAS-2B | Normal | Homo sapiens | CVCL_0168 | |
CALML3 antisense RNA 1 (CALML3-AS1)
Lung cancer [ICD-11: 2C25]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [16] | |||
| Responsed Disease | Non-small cell lung cancer [ICD-11: 2C25.Y] | |||
| Target Regulation | Down regulation | |||
Cell division cycle-associated protein 4 (CDCA4)
Lung cancer [ICD-11: 2C25]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [17] | |||
| Responsed Disease | Lung adenocarcinoma [ICD-11: 2C25.0] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
BEAS-2B | Normal | Homo sapiens | CVCL_0168 |
| NCI-H1299 | Lung large cell carcinoma | Homo sapiens | CVCL_0060 | |
| NCI-H157 | Lung squamous cell carcinoma | Homo sapiens | CVCL_0463 | |
| NCI-H1975 | Lung adenocarcinoma | Homo sapiens | CVCL_1511 | |
| Calu-3 | Lung adenocarcinoma | Homo sapiens | CVCL_0609 | |
| In-vivo Model | For the action of CDCA4, mice were set into Control, sh-NC, sh-CDCA4 groups, 6 mice/group. For the action of ALKBH5 in CDCA4, mice were set into oe-NC, oe-CDCA4, oe-CDCA4 + sh-NC, oe-CDCA4 + sh-ALKBH5 groups, 6 mice/group. In brief, 1 × 106/100 μL LLC cells were injected subcutaneously into the right flank of mice to establish a tumor model. LLC cells stably transfected with oe-NC, oe-CDCA4, oe-CDCA4 + sh-NC and oe-CDCA4 + sh-ALKBH5 were injected into each group of mice . | |||
Circ_YTHDC2
Hematological disorders [ICD-11: 3C0Z]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [18] | |||
| Responsed Disease | Hematological disorders [ICD-11: 3C0Z] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
A7r5 | Normal | Rattus norvegicus | CVCL_0137 |
| Response Summary | YTHDC2/Circ_YTHDC2/TET2 pathway is an important target of metformin in preventing the progression of VSMCs dysfunction under high glucose. | |||
DNA-binding protein inhibitor ID-3 (ID3)
Lung cancer [ICD-11: 2C25]
| In total 2 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [19] | |||
| Responsed Disease | Non-small cell lung cancer [ICD-11: 2C25.Y] | |||
| Responsed Drug | Cisplatin | Approved | ||
| Target Regulation | Up regulation | |||
In-vitro Model |
A549/DDP | Lung adenocarcinoma | Homo sapiens | CVCL_C0W4 |
| Experiment 2 Reporting the m6A-centered Disease Response of This Target Gene | [19] | |||
| Responsed Disease | Non-small cell lung cancer [ICD-11: 2C25.Y] | |||
| Responsed Drug | 3-deazidenosine | Investigative | ||
| Target Regulation | Up regulation | |||
In-vitro Model |
A549/DDP | Lung adenocarcinoma | Homo sapiens | CVCL_C0W4 |
Histone-lysine N-methyltransferase ASH1L (ASH1L)
Uveitis [ICD-11: 9A96]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [20] | |||
| Responsed Disease | Uveitis [ICD-11: 9A96] | |||
| Target Regulation | Up regulation | |||
Large ribosomal subunit protein bL12m (MRPL12)
Lung cancer [ICD-11: 2C25]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [21] | |||
| Responsed Disease | Lung adenocarcinoma [ICD-11: 2C25.0] | |||
| Target Regulation | Down regulation | |||
In-vitro Model |
A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 |
| NCI-H1573 | Lung adenocarcinoma | Homo sapiens | CVCL_1478 | |
LIM domain kinase 1 (LIMK1)
Colorectal cancer [ICD-11: 2B91]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [22] | |||
| Responsed Disease | Colorectal cancer [ICD-11: 2B91] | |||
| Responsed Drug | Fluorouracil | Approved | ||
| Target Regulation | Down regulation | |||
In-vitro Model |
LoVo | Colon adenocarcinoma | Homo sapiens | CVCL_0399 |
| HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 | |
| HT29 | Colon cancer | Mus musculus | CVCL_A8EZ | |
| Caco-2 | Colon adenocarcinoma | Homo sapiens | CVCL_0025 | |
| RKO | Colon carcinoma | Homo sapiens | CVCL_0504 | |
| SW480 | Colon adenocarcinoma | Homo sapiens | CVCL_0546 | |
| NCM460 | Normal | Homo sapiens | CVCL_0460 | |
| FHC | Normal | Homo sapiens | CVCL_3688 | |
| In-vivo Model | A total of 5 × 106 SW480 cells/100 μL of PBS were subcutaneously injected into the back of mice to establish a xenograft model, and tumor growth was measured with a vernier caliper every 3 days. 72 h after tumor inoculation, mice were injected intraperitoneally with 5-FU (6 mg/kg; Sigma, German) twice weekly for 21 days (6 doses in total). | |||
Lysine-specific demethylase 5B (KDM5B)
Chronic kidney disease [ICD-11: GB61]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [23] | |||
| Responsed Disease | Diabetic nephropathy [ICD-11: GB61.Z] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
RSC96
|
N.A. | Rattus norvegicus | CVCL_4694 |
| In-vivo Model | Male type 2 diabetic mice (db/db) [18-week-old] were purchased from SLAC (Shanghai, China) (n = 8). Corresponding 18-week-old heterozygotes mice (db/m) were used as controls. All mice were killed, and sciatic nerves were collected and stored at - 80 °C. | |||
NUTM2B antisense RNA 1 (NUTM2B-AS1)
Liver cancer [ICD-11: 2C12]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [24] | |||
| Responsed Disease | Liver cancer [ICD-11: 2C12] | |||
| Target Regulation | Up regulation | |||
pri-miR-17
Colorectal cancer [ICD-11: 2B91]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [25] | |||
| Responsed Disease | Colorectal cancer [ICD-11: 2B91] | |||
| Responsed Drug | Fluorouracil | Approved | ||
| Target Regulation | Down regulation | |||
In-vitro Model |
HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 |
| LoVo | Colon adenocarcinoma | Homo sapiens | CVCL_0399 | |
| SW480 | Colon adenocarcinoma | Homo sapiens | CVCL_0546 | |
| SW620 | Colon adenocarcinoma | Homo sapiens | CVCL_0547 | |
| Caco-2 | Colon adenocarcinoma | Homo sapiens | CVCL_0025 | |
| HT29 | Colon cancer | Mus musculus | CVCL_A8EZ | |
| RKO | Colon carcinoma | Homo sapiens | CVCL_0504 | |
| FHC | Normal | Homo sapiens | CVCL_3688 | |
| NCM460 | Normal | Homo sapiens | CVCL_0460 | |
Protein MROH8 (MROH8)
Pancreatic cancer [ICD-11: 2C10]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [26] | |||
| Responsed Disease | Pancreatic cancer [ICD-11: 2C10] | |||
In-vitro Model |
AsPC-1 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0152 |
| Capan-1 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0237 | |
| Capan-2 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0026 | |
| CFPAC-1 | Cystic fibrosis | Homo sapiens | CVCL_1119 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| HPDE6c7 | Normal | Homo sapiens | CVCL_0P38 | |
Rod cGMP-specific 3',5'-cyclic phosphodiesterase subunit beta (PDE6B)
Inherited retinal dystrophies [ICD-11: 9B70]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [28] | |||
| Responsed Disease | Inherited retinal dystrophies [ICD-11: 9B70] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
HEK293T | Normal | Homo sapiens | CVCL_0063 |
Serine/threonine-protein phosphatase with EF-hands 2 (PPEF2)
Inherited retinal dystrophies [ICD-11: 9B70]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [28] | |||
| Responsed Disease | Inherited retinal dystrophies [ICD-11: 9B70] | |||
| Target Regulation | Up regulation | |||
In-vitro Model |
HEK293T | Normal | Homo sapiens | CVCL_0063 |
SOX
Kaposi's sarcoma [ICD-11: 2B57]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [9] | |||
| Responsed Disease | Kaposi's sarcoma [ICD-11: 2B57] | |||
In-vitro Model |
iSLK.219 | Clear cell renal cell carcinoma | Homo sapiens | CVCL_B6YV |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| HEK293 | Normal | Homo sapiens | CVCL_0045 | |
| Response Summary | This modification recruits the m6A reader YTHDC2 and found that YTHDC2 is necessary for the escape of the IL-6 transcript. m6A modification is essential to confer SOX (SOX) resistance to the IL-6 mRNA. These results shed light on how the host cell has evolved to use RNA modifications to circumvent viral manipulation of RNA fate during KSHV infection Kaposi's sarcoma. | |||
Unspecific Target Gene
Head and neck squamous carcinoma [ICD-11: 2B6E]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [29] | |||
| Responsed Disease | Head and neck squamous carcinoma [ICD-11: 2B6E] | |||
| Pathway Response | Ubiquitin mediated proteolysis | hsa04120 | ||
| Cell Process | Ubiquitin-mediated proteolysis | |||
| Cell apoptosis | ||||
| Response Summary | In head and neck squamous cell carcinoma patients, a majority of highly expressed m6A regulatory genes is associated with poor OS, in particular ALKBH5, whereas YTHDC2 was associated with better prognosis. | |||
Rectum cancer [ICD-11: 2B92]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [30] | |||
| Responsed Disease | Rectum cancer [ICD-11: 2B92] | |||
| Target Regulation | Down regulation | |||
| Pathway Response | Nucleotide excision repair | hsa03420 | ||
| Cell Process | DNA repair | |||
| Epithelial-mesenchymal transition | ||||
| Response Summary | The m6A RNA methylation regulators, specifically YTHDC2 and METTL14, were significantly down-regulated and were potential prognostic biomarkers in rectal cancer. | |||
Coronary artery disease [ICD-11: BA8Z]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [31] | |||
| Responsed Disease | Coronary artery disease [ICD-11: BA8Z] | |||
Diseases of the musculoskeletal system [ICD-11: FC0Z]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [32] | |||
| Responsed Disease | Diseases of the musculoskeletal system [ICD-11: FC0Z] | |||
| Pathway Response | Ribosome biogenesis in eukaryotes | hsa03008), mRNA surveillance pathway | ||
| Cell Process | Adipogenic differentiation | |||
| Response Summary | YTHDC2 knockdown can promote the osteogenic differentiation of hBMSCs and inhibit the adipogenic differentiation. YTHDC2 knockdown cause changes in ribosome function. | |||
Menopausal disorder [ICD-11: GA30]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [33] | |||
| Responsed Disease | Premature ovarian failure [ICD-11: GA30.6] | |||
| Pathway Response | Oocyte meiosis | hsa04114 | ||
| Cell Process | Meiosis | |||
In-vitro Model |
HeLa | Endocervical adenocarcinoma | Homo sapiens | CVCL_0030 |
| Response Summary | YTHDC2 is a key regulator of meiosis in humans and pathogenic variants within this gene are associated with POI. | |||
Male infertility [ICD-11: GB04]
| In total 2 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response of This Target Gene | [34] | |||
| Responsed Disease | Male infertility [ICD-11: GB04] | |||
| Responsed Drug | Ethyl ester form of meclofenamic acid | Approved | ||
| Cell Process | Cell cycle | |||
| Cell proliferation | ||||
In-vitro Model |
GC-1 spg | Normal | Mus musculus | CVCL_8872 |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| In-vivo Model | Mouse GC-1 spg cells were treated with the ester form of meclofenamic acid (MA2) to inhibit the demethylase activity of FTO. | |||
| Response Summary | METTL3, METTL14, ALKBH5 and YTHDC2 are involved in the regulation of spermatogenesis and oogenesis. MA2 affected CDKs expression through the m6A-dependent mRNA degradation pathway, and thus repressed spermatogonial proliferation. Additionally, mutation of the predicted m6A sites in the Cdk2-3'UTR could mitigated the degradation of CDK2 mRNA after MA2 treatment. | |||
| Experiment 2 Reporting the m6A-centered Disease Response of This Target Gene | [35] | |||
| Responsed Disease | Male infertility [ICD-11: GB04] | |||
| Cell Process | RNA stability | |||
| RNA degradation (hsa03018) | ||||
In-vitro Model |
HeLa | Endocervical adenocarcinoma | Homo sapiens | CVCL_0030 |
| In-vivo Model | Mixture of Cas9 mRNA (20 ng/uL) and two sgRNAs (5 ng/uL each) were injected into the cytoplasm and male pronucleus of the zygote, obtained by CBF1 mating. | |||
| Response Summary | Ythdc2 knockout mice are infertile; males have significantly smaller testes and females have significantly smaller ovaries compared to those of littermates. | |||
Stearoyl-CoA desaturase (SCD)
Simvastatin
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [12] | |||
| Responsed Disease | Papillary Thyroid Cancer | ICD-11: XH1ND9 | ||
| Target Regulation | Down regulation | |||
| In-vitro Model | B-CPAP | Thyroid gland carcinoma | Homo sapiens | CVCL_0153 |
| TPC-1 | Thyroid gland papillary carcinoma | Homo sapiens | CVCL_6298 | |
| K1 | Thyroid gland papillary carcinoma | Homo sapiens | CVCL_2537 | |
| In-vivo Model | These mice were provided with unrestricted access to both water and food, following a 12-h light and 12-h dark cycle. Subcutaneous injections of stably transfected K1 cells (2 × 106 cells per mouse) were administered in the armpits of the mice. Once the tumors became palpable, their volume was measured every three days and calculated using the formula: length × width2 × 0.5. The tumor weight was detected after the mice were sacrificed. | |||
DNA-binding protein inhibitor ID-3 (ID3)
Cisplatin
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [19] | |||
| Responsed Disease | Non-small cell lung cancer | ICD-11: 2C25.Y | ||
| Target Regulation | Up regulation | |||
| In-vitro Model | A549/DDP | Lung adenocarcinoma | Homo sapiens | CVCL_C0W4 |
3-deazidenosine
[Investigative]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [19] | |||
| Responsed Disease | Non-small cell lung cancer | ICD-11: 2C25.Y | ||
| Target Regulation | Up regulation | |||
| In-vitro Model | A549/DDP | Lung adenocarcinoma | Homo sapiens | CVCL_C0W4 |
LIM domain kinase 1 (LIMK1)
Fluorouracil
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [22] | |||
| Responsed Disease | Colorectal cancer | ICD-11: 2B91 | ||
| Target Regulation | Down regulation | |||
| In-vitro Model | LoVo | Colon adenocarcinoma | Homo sapiens | CVCL_0399 |
| HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 | |
| HT29 | Colon cancer | Mus musculus | CVCL_A8EZ | |
| Caco-2 | Colon adenocarcinoma | Homo sapiens | CVCL_0025 | |
| RKO | Colon carcinoma | Homo sapiens | CVCL_0504 | |
| SW480 | Colon adenocarcinoma | Homo sapiens | CVCL_0546 | |
| NCM460 | Normal | Homo sapiens | CVCL_0460 | |
| FHC | Normal | Homo sapiens | CVCL_3688 | |
| In-vivo Model | A total of 5 × 106 SW480 cells/100 μL of PBS were subcutaneously injected into the back of mice to establish a xenograft model, and tumor growth was measured with a vernier caliper every 3 days. 72 h after tumor inoculation, mice were injected intraperitoneally with 5-FU (6 mg/kg; Sigma, German) twice weekly for 21 days (6 doses in total). | |||
pri-miR-17
Fluorouracil
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [25] | |||
| Responsed Disease | Colorectal cancer | ICD-11: 2B91 | ||
| Target Regulation | Down regulation | |||
| In-vitro Model | HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 |
| LoVo | Colon adenocarcinoma | Homo sapiens | CVCL_0399 | |
| SW480 | Colon adenocarcinoma | Homo sapiens | CVCL_0546 | |
| SW620 | Colon adenocarcinoma | Homo sapiens | CVCL_0547 | |
| Caco-2 | Colon adenocarcinoma | Homo sapiens | CVCL_0025 | |
| HT29 | Colon cancer | Mus musculus | CVCL_A8EZ | |
| RKO | Colon carcinoma | Homo sapiens | CVCL_0504 | |
| FHC | Normal | Homo sapiens | CVCL_3688 | |
| NCM460 | Normal | Homo sapiens | CVCL_0460 | |
RNA component of 7SK nuclear ribonucleoprotein (RN7SK)
Etoposide
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [27] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | Wnt signaling pathway | hsa04310 | ||
| In-vitro Model | BEAS-2B | Normal | Homo sapiens | CVCL_0168 |
| NCI-H1299 | Lung large cell carcinoma | Homo sapiens | CVCL_0060 | |
| NCI-H1975 | Lung adenocarcinoma | Homo sapiens | CVCL_1511 | |
| A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 | |
| PC-9 | Lung adenocarcinoma | Homo sapiens | CVCL_B260 | |
| BEL-7404 | Endocervical adenocarcinoma | Homo sapiens | CVCL_6568 | |
| BEL-7402 | Endocervical adenocarcinoma | Homo sapiens | CVCL_5492 | |
| Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 | |
| SW1990 | Pancreatic adenocarcinoma | Homo sapiens | CVCL_1723 | |
| PANC-1 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0480 | |
| MKN45 | Gastric adenocarcinoma | Homo sapiens | CVCL_0434 | |
| MGC-803 | Gastric mucinous adenocarcinoma | Homo sapiens | CVCL_5334 | |
| HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 | |
| HT29 | Colon cancer | Mus musculus | CVCL_A8EZ | |
| In-vivo Model | For the generation of PDX mouse models, fresh LUSC specimens (2-3 mm3), which were from Shanghai Chest Hospital, were implanted into 4- to 6-week-old athymic nude mice (Jiesijie, Shanghai, China). After successful tumor growth was confirmed, the tumor tissues were passaged and implanted into the next generation of mice. The third to fifth generations of PDX-bearing mice were used for drug administration. When tumors reached approximately 200 mm3, mice were injected daily with DMSO (no. ST038, Beyotime Biotechnology, Shanghai, China) with or without MIT (no. S2485, 5 mg/kg, Selleck, Houston, TX) or HYD (no. S1896, 20 mg/kg, Selleck) (n = 5 mice/group). Tumor growth was monitored, and sizes were calculated by 0.5 × L × W2 (L indicating length and W indicating width). The mice were euthanized at day 28 after implantation. All animal experiments were approved by the institutional ethics committee of Shanghai Chest Hospital (approval number KS(Y)21382). | |||
Hydroxyurea
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [27] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | Wnt signaling pathway | hsa04310 | ||
| In-vitro Model | BEAS-2B | Normal | Homo sapiens | CVCL_0168 |
| NCI-H1299 | Lung large cell carcinoma | Homo sapiens | CVCL_0060 | |
| NCI-H1975 | Lung adenocarcinoma | Homo sapiens | CVCL_1511 | |
| A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 | |
| PC-9 | Lung adenocarcinoma | Homo sapiens | CVCL_B260 | |
| BEL-7404 | Endocervical adenocarcinoma | Homo sapiens | CVCL_6568 | |
| BEL-7402 | Endocervical adenocarcinoma | Homo sapiens | CVCL_5492 | |
| Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 | |
| SW1990 | Pancreatic adenocarcinoma | Homo sapiens | CVCL_1723 | |
| PANC-1 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0480 | |
| MKN45 | Gastric adenocarcinoma | Homo sapiens | CVCL_0434 | |
| MGC-803 | Gastric mucinous adenocarcinoma | Homo sapiens | CVCL_5334 | |
| HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 | |
| HT29 | Colon cancer | Mus musculus | CVCL_A8EZ | |
| In-vivo Model | For the generation of PDX mouse models, fresh LUSC specimens (2-3 mm3), which were from Shanghai Chest Hospital, were implanted into 4- to 6-week-old athymic nude mice (Jiesijie, Shanghai, China). After successful tumor growth was confirmed, the tumor tissues were passaged and implanted into the next generation of mice. The third to fifth generations of PDX-bearing mice were used for drug administration. When tumors reached approximately 200 mm3, mice were injected daily with DMSO (no. ST038, Beyotime Biotechnology, Shanghai, China) with or without MIT (no. S2485, 5 mg/kg, Selleck, Houston, TX) or HYD (no. S1896, 20 mg/kg, Selleck) (n = 5 mice/group). Tumor growth was monitored, and sizes were calculated by 0.5 × L × W2 (L indicating length and W indicating width). The mice were euthanized at day 28 after implantation. All animal experiments were approved by the institutional ethics committee of Shanghai Chest Hospital (approval number KS(Y)21382). | |||
Mitoxantrone
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [27] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | Wnt signaling pathway | hsa04310 | ||
| In-vitro Model | BEAS-2B | Normal | Homo sapiens | CVCL_0168 |
| NCI-H1299 | Lung large cell carcinoma | Homo sapiens | CVCL_0060 | |
| NCI-H1975 | Lung adenocarcinoma | Homo sapiens | CVCL_1511 | |
| A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 | |
| PC-9 | Lung adenocarcinoma | Homo sapiens | CVCL_B260 | |
| BEL-7404 | Endocervical adenocarcinoma | Homo sapiens | CVCL_6568 | |
| BEL-7402 | Endocervical adenocarcinoma | Homo sapiens | CVCL_5492 | |
| Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 | |
| SW1990 | Pancreatic adenocarcinoma | Homo sapiens | CVCL_1723 | |
| PANC-1 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0480 | |
| MKN45 | Gastric adenocarcinoma | Homo sapiens | CVCL_0434 | |
| MGC-803 | Gastric mucinous adenocarcinoma | Homo sapiens | CVCL_5334 | |
| HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 | |
| HT29 | Colon cancer | Mus musculus | CVCL_A8EZ | |
| In-vivo Model | For the generation of PDX mouse models, fresh LUSC specimens (2-3 mm3), which were from Shanghai Chest Hospital, were implanted into 4- to 6-week-old athymic nude mice (Jiesijie, Shanghai, China). After successful tumor growth was confirmed, the tumor tissues were passaged and implanted into the next generation of mice. The third to fifth generations of PDX-bearing mice were used for drug administration. When tumors reached approximately 200 mm3, mice were injected daily with DMSO (no. ST038, Beyotime Biotechnology, Shanghai, China) with or without MIT (no. S2485, 5 mg/kg, Selleck, Houston, TX) or HYD (no. S1896, 20 mg/kg, Selleck) (n = 5 mice/group). Tumor growth was monitored, and sizes were calculated by 0.5 × L × W2 (L indicating length and W indicating width). The mice were euthanized at day 28 after implantation. All animal experiments were approved by the institutional ethics committee of Shanghai Chest Hospital (approval number KS(Y)21382). | |||
Oxaliplatin
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [27] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | Wnt signaling pathway | hsa04310 | ||
| In-vitro Model | BEAS-2B | Normal | Homo sapiens | CVCL_0168 |
| NCI-H1299 | Lung large cell carcinoma | Homo sapiens | CVCL_0060 | |
| NCI-H1975 | Lung adenocarcinoma | Homo sapiens | CVCL_1511 | |
| A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 | |
| PC-9 | Lung adenocarcinoma | Homo sapiens | CVCL_B260 | |
| BEL-7404 | Endocervical adenocarcinoma | Homo sapiens | CVCL_6568 | |
| BEL-7402 | Endocervical adenocarcinoma | Homo sapiens | CVCL_5492 | |
| Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 | |
| SW1990 | Pancreatic adenocarcinoma | Homo sapiens | CVCL_1723 | |
| PANC-1 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0480 | |
| MKN45 | Gastric adenocarcinoma | Homo sapiens | CVCL_0434 | |
| MGC-803 | Gastric mucinous adenocarcinoma | Homo sapiens | CVCL_5334 | |
| HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 | |
| HT29 | Colon cancer | Mus musculus | CVCL_A8EZ | |
| In-vivo Model | For the generation of PDX mouse models, fresh LUSC specimens (2-3 mm3), which were from Shanghai Chest Hospital, were implanted into 4- to 6-week-old athymic nude mice (Jiesijie, Shanghai, China). After successful tumor growth was confirmed, the tumor tissues were passaged and implanted into the next generation of mice. The third to fifth generations of PDX-bearing mice were used for drug administration. When tumors reached approximately 200 mm3, mice were injected daily with DMSO (no. ST038, Beyotime Biotechnology, Shanghai, China) with or without MIT (no. S2485, 5 mg/kg, Selleck, Houston, TX) or HYD (no. S1896, 20 mg/kg, Selleck) (n = 5 mice/group). Tumor growth was monitored, and sizes were calculated by 0.5 × L × W2 (L indicating length and W indicating width). The mice were euthanized at day 28 after implantation. All animal experiments were approved by the institutional ethics committee of Shanghai Chest Hospital (approval number KS(Y)21382). | |||
Raltitrexed
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [27] | |||
| Target Regulation | Up regulation | |||
| Pathway Response | Wnt signaling pathway | hsa04310 | ||
| In-vitro Model | BEAS-2B | Normal | Homo sapiens | CVCL_0168 |
| NCI-H1299 | Lung large cell carcinoma | Homo sapiens | CVCL_0060 | |
| NCI-H1975 | Lung adenocarcinoma | Homo sapiens | CVCL_1511 | |
| A-549 | Lung adenocarcinoma | Homo sapiens | CVCL_0023 | |
| PC-9 | Lung adenocarcinoma | Homo sapiens | CVCL_B260 | |
| BEL-7404 | Endocervical adenocarcinoma | Homo sapiens | CVCL_6568 | |
| BEL-7402 | Endocervical adenocarcinoma | Homo sapiens | CVCL_5492 | |
| Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 | |
| SW1990 | Pancreatic adenocarcinoma | Homo sapiens | CVCL_1723 | |
| PANC-1 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0480 | |
| MKN45 | Gastric adenocarcinoma | Homo sapiens | CVCL_0434 | |
| MGC-803 | Gastric mucinous adenocarcinoma | Homo sapiens | CVCL_5334 | |
| HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 | |
| HT29 | Colon cancer | Mus musculus | CVCL_A8EZ | |
| In-vivo Model | For the generation of PDX mouse models, fresh LUSC specimens (2-3 mm3), which were from Shanghai Chest Hospital, were implanted into 4- to 6-week-old athymic nude mice (Jiesijie, Shanghai, China). After successful tumor growth was confirmed, the tumor tissues were passaged and implanted into the next generation of mice. The third to fifth generations of PDX-bearing mice were used for drug administration. When tumors reached approximately 200 mm3, mice were injected daily with DMSO (no. ST038, Beyotime Biotechnology, Shanghai, China) with or without MIT (no. S2485, 5 mg/kg, Selleck, Houston, TX) or HYD (no. S1896, 20 mg/kg, Selleck) (n = 5 mice/group). Tumor growth was monitored, and sizes were calculated by 0.5 × L × W2 (L indicating length and W indicating width). The mice were euthanized at day 28 after implantation. All animal experiments were approved by the institutional ethics committee of Shanghai Chest Hospital (approval number KS(Y)21382). | |||
Unspecific Target Gene
Ethyl ester form of meclofenamic acid
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response of This Target Gene | [34] | |||
| Responsed Disease | Male infertility | ICD-11: GB04 | ||
| Cell Process | Cell cycle | |||
| Cell proliferation | ||||
| In-vitro Model | GC-1 spg | Normal | Mus musculus | CVCL_8872 |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| In-vivo Model | Mouse GC-1 spg cells were treated with the ester form of meclofenamic acid (MA2) to inhibit the demethylase activity of FTO. | |||
| Response Summary | METTL3, METTL14, ALKBH5 and YTHDC2 are involved in the regulation of spermatogenesis and oogenesis. MA2 affected CDKs expression through the m6A-dependent mRNA degradation pathway, and thus repressed spermatogonial proliferation. Additionally, mutation of the predicted m6A sites in the Cdk2-3'UTR could mitigated the degradation of CDK2 mRNA after MA2 treatment. | |||
Full List of Crosstalk(s) between m6A Modification and Epigenetic Regulation Related to This Regulator
DNA modification
m6A Target: ANKRD13C divergent transcript (ANKRD13C-DT)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT02006 | ||
| Epigenetic Regulator | Methylcytosine dioxygenase TET1 (TET1) | |
| Regulated Target | Long Terminal Repeat 7 (LTR7) | |
| Crosstalk relationship | m6A → DNA modification | |
m6A Target: Circ_YTHDC2
| In total 4 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT02027 | ||
| Epigenetic Regulator | Methylcytosine dioxygenase TET2 (TET2) | |
| Regulated Target | Myocardin (MYOCD) | |
| Crosstalk relationship | m6A → DNA modification | |
| Disease | Hematological disorders | |
| Drug | Metformin | |
| Crosstalk ID: M6ACROT02028 | ||
| Epigenetic Regulator | Methylcytosine dioxygenase TET2 (TET2) | |
| Regulated Target | Serum response factor (SRF) | |
| Crosstalk relationship | m6A → DNA modification | |
| Disease | Hematological disorders | |
| Drug | Metformin | |
| Crosstalk ID: M6ACROT02029 | ||
| Epigenetic Regulator | Methylcytosine dioxygenase TET2 (TET2) | |
| Regulated Target | Krueppel-like factor 4 (KLF4) | |
| Crosstalk relationship | m6A → DNA modification | |
| Disease | Hematological disorders | |
| Drug | Metformin | |
| Crosstalk ID: M6ACROT02097 | ||
| Epigenetic Regulator | Methylcytosine dioxygenase TET2 (TET2) | |
| Crosstalk relationship | m6A → DNA modification | |
| Disease | Hematological disorders | |
m6A Target: Stearoyl-CoA desaturase (SCD)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT02051 | ||
| Epigenetic Regulator | DNA (cytosine-5)-methyltransferase 1 (DNMT1) | |
| Regulated Target | Methyltransferase 16, RNA N6-adenosine (METTL16) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Papillary Thyroid Cancer | |
| Drug | Simvastatin | |
Histone modification
m6A Target: Mutated in multiple advanced cancers 1 (PTEN)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03043 | ||
| Epigenetic Regulator | Polycomb Repressive Complex 2 (PRC2) | |
| Regulated Target | Histone H3 lysine 27 trimethylation (H3K27me3) | |
| Crosstalk relationship | m6A → Histone modification | |
| Disease | Skin cancer | |
m6A Target: Histone-lysine N-methyltransferase ASH1L (ASH1L)
| In total 2 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03205 | ||
| Epigenetic Regulator | Histone-lysine N-methyltransferase ASH1L (ASH1L) | |
| Regulated Target | Histone H3 lysine 4 trimethylation (H3K4me3) | |
| Crosstalk relationship | m6A → Histone modification | |
| Disease | Uveitis | |
| Crosstalk ID: M6ACROT05852 | ||
| Epigenetic Regulator | Histone-lysine N-methyltransferase ASH1L (ASH1L) | |
| Regulated Target | Histone H3 lysine 4 trimethylation (H3K4me3) | |
| Crosstalk relationship | m6A → Histone modification | |
| Disease | Uveitis | |
m6A Target: NUTM2B antisense RNA 1 (NUTM2B-AS1)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03250 | ||
| Epigenetic Regulator | Histone-lysine N-methyltransferase 2A (KMT2A) | |
| Regulated Target | Histone H3 lysine 4 trimethylation (H3K4me3) | |
| Crosstalk relationship | m6A → Histone modification | |
| Disease | Liver cancer | |
m6A Target: Lysine-specific demethylase 5B (KDM5B)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT03255 | ||
| Epigenetic Regulator | Lysine-specific demethylase 5B (KDM5B) | |
| Regulated Target | Histone H3 lysine 4 trimethylation (H3K4me3) | |
| Crosstalk relationship | m6A → Histone modification | |
| Disease | Diabetic nephropathy | |
m6A Target: seRNA
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05868 | ||
| Epigenetic Regulator | Histone-lysine N-methyltransferase 2A (KMT2A) | |
| Regulated Target | Histone H3 lysine 4 trimethylation (H3K4me3) | |
| Crosstalk relationship | m6A → Histone modification | |
| Disease | Pancreatic ductal adenocarcinoma | |
Non-coding RNA
m6A Target: Dual specificity mitogen-activated protein kinase kinase 7 (MAP2K7)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05063 | ||
| Epigenetic Regulator | Lnc-AK311120 | |
| Regulated Target | YTH N6-methyladenosine RNA binding protein C2 (YTHDC2) | |
| Crosstalk relationship | ncRNA → m6A | |
m6A Target: Circ_YTHDC2
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05527 | ||
| Epigenetic Regulator | Circ_YTHDC2 | |
| Regulated Target | Tet methylcytosine dioxygenase 2 (TET2) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Hematological disorders | |
m6A Target: RNA component of 7SK nuclear ribonucleoprotein (RN7SK)
| In total 5 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05661 | ||
| Epigenetic Regulator | RNA component of 7SK nuclear ribonucleoprotein (RN7SK) | |
| Regulated Target | Cullin 1 (CUL1) | |
| Crosstalk relationship | m6A → ncRNA | |
| Drug | mitoxantrone (MIT) | |
| Crosstalk ID: M6ACROT06031 | ||
| Epigenetic Regulator | RNA component of 7SK nuclear ribonucleoprotein (RN7SK) | |
| Regulated Target | Cullin 1 (CUL1) | |
| Crosstalk relationship | m6A → ncRNA | |
| Drug | hydroxyurea (HYD) | |
| Crosstalk ID: M6ACROT06034 | ||
| Epigenetic Regulator | RNA component of 7SK nuclear ribonucleoprotein (RN7SK) | |
| Regulated Target | Cullin 1 (CUL1) | |
| Crosstalk relationship | m6A → ncRNA | |
| Drug | raltitrexed (RAL) | |
| Crosstalk ID: M6ACROT06037 | ||
| Epigenetic Regulator | RNA component of 7SK nuclear ribonucleoprotein (RN7SK) | |
| Regulated Target | Cullin 1 (CUL1) | |
| Crosstalk relationship | m6A → ncRNA | |
| Drug | oxaliplatin (OXA) | |
| Crosstalk ID: M6ACROT06040 | ||
| Epigenetic Regulator | RNA component of 7SK nuclear ribonucleoprotein (RN7SK) | |
| Regulated Target | Cullin 1 (CUL1) | |
| Crosstalk relationship | m6A → ncRNA | |
| Drug | etoposide (ETO) | |
m6A Target: CALML3 antisense RNA 1 (CALML3-AS1)
| In total 1 item(s) under this m6A target | ||
| Crosstalk ID: M6ACROT05759 | ||
| Epigenetic Regulator | CALML3 antisense RNA 1 (CALML3-AS1) | |
| Regulated Target | Histone-lysine N-methyltransferase EZH2 (EZH2) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Non-small cell lung cancer | |
Xenobiotics Compound(s) Regulating the m6A Methylation Regulator
| Compound Name | DEHP | Investigative |
|---|---|---|
| Synonyms |
DEHP; 117-81-7; BIS(2-ETHYLHEXYL)PHTHALATE; Di(2-ethylhexyl)phthalate; Di(2-ethylhexyl) phthalate; Diethylhexyl phthalate; 2-Ethylhexyl phthalate; Di-sec-octyl phthalate; Octyl phthalate; Fleximel; Octoil; Ethylhexyl phthalate; Palatinol AH; Celluflex DOP; Vestinol AH; Bisoflex DOP; Kodaflex DOP; Staflex DOP; Truflex DOP; Flexol DOP; Vinicizer 80; Bisoflex 81; Eviplast 80; Eviplast 81; Hercoflex 260; RC plasticizer DOP; Compound 889; Witcizer 312; Platinol dop; Di-2-ethylhexyl phthalate; Nuoplaz dop; Platinol ah; Hatcol dop; Reomol dop; Pittsburgh PX-138; Sansocizer DOP; Ergoplast FDO; Monocizer DOP; Plasthall DOP; Flexol plasticizer DOP; Mollan O; Jayflex DOP; Sicol 150; Ergoplast FDO-S; Di(2-ethylhexyl)orthophthalate; Good-rite gp 264; Reomol D 79P; Bis(2-ethylhexyl) benzene-1,2-dicarboxylate; Di(ethylhexyl) phthalate; Bis(ethylhexyl) phthalate; Rcra waste number U028; Px-138; Phthalic acid dioctyl ester; NCI-C52733; Di(2-ethylhexyl) o-phthalate; Phthalic acid di(2-ethylhexyl) ester; 1,2-Benzenedicarboxylic acid, bis(2-ethylhexyl) ester; DOP; Bis(2-ethylhexyl) 1,2-benzenedicarboxylate; UNII-C42K0PH13C; Bis(2-ethylhexyl) o-phthalate; Phthalic acid, bis(2-ethylhexyl) ester; CHEBI:17747; 1,2-Benzenedicarboxylic acid bis(2-ethylhexyl) ester; Benzenedicarboxylic acid, bis(2-ethylhexyl) ester; Phthalic Acid Bis(2-ethylhexyl) Ester; Bis-(2-ethylhexyl)ester kyseliny ftalove; C42K0PH13C; 1,2-Benzenedicarboxylic acid, 1,2-bis(2-ethylhexyl) ester; DTXSID5020607; Etalon; Bis-(2-ethylhexyl)ester kyseliny ftalove [Czech]; Di-(2-ethylhexyl) phthalate; BIS-(2-ETHYLHEXYL) PHTHALATE; NCGC00091499-05; Sconamoll DOP; Diacizer DOP; Kodaflex DEHP; 15495-94-0; Etalon (plasticizer); Sansocizer R 8000; Caswell No. 392K; Dioctylphthalate; Behp; Di-2-ethylhexylphthalate; Diplast O; ESBO-D 82; Ergoplast FDO; Ergoplast FDO-S; Etalon; Phthalic acid, bis-2-ethylhexyl ester; DOF [Russian plasticizer]; SMR000777878; CCRIS 237; Ethyl hexyl phthalate; HSDB 339; Di(2-ethylhexyl) orthophthalate; Bis-(2-ethylhexyl)ester kyseliny ftalove (czech); EINECS 204-211-0; NSC 17069; Diethylhexylphthalate (Bis-(2-ethylhexyl) Phthalate); RCRA waste no. U028; Union carbide flexol 380; EPA Pesticide Chemical Code 295200; BRN 1890696; AI3-04273; DAF 68; Palatinol DOP; Polycizer DOP; Merrol DOP; Palatinol AH-L; Hatco DOP; Vinycizer 80; Di(2-ethylhexyl)phthalate (DEHP); N-Dioctyl phthalate; MFCD00009493; Corflex 400; Dioctyl phthalate, 99%; DSSTox_CID_607; 1, bis(ethylhexyl) ester; Epitope ID:140107; EC 204-211-0; WLN: 8OVR BVO8; Di(2-Ethylhexyl phthalate); DSSTox_RID_75688; DSSTox_GSID_20607; SCHEMBL20271; 14C -DEHP; 8033-53-2; MLS001333173; MLS001333174; MLS002454397; Dioctyl phthalate, >=99.5%; 1,2-Benzenedicarboxylic acid, bis-(1-ethylhexyl) ester; CHEMBL1242017; SCHEMBL21733281; HMS2233C15; HMS3374J09; AMY40790; HY-B1945; NSC17069; Tox21_400084; Bis(2-ethylhexyl)ester phthalic acid; NSC-17069; s3360; AKOS024318875; Bis(2-ethylhexyl) phthalate-[13C6]; Phthalic acid bis(2-ethylhexyl ester); MCULE-4692716107; NCGC00091499-01; NCGC00091499-02; NCGC00091499-04; NCGC00091499-06; NCGC00091499-07; CAS-117-81-7; I887; Bis(2-ethylhexyl) 1, 2-benzenedicarboxylate; CS-0014050; FT-0624576; FT-0663286; P0297; WLN: 4Y2 & 1OVR BVO1Y4 & 2; Bis(2-ethylhexyl) phthalate, Selectophore(TM); C03690; A937603; Q418492; 1,2-Benzenedicarboxylic acid bis-(1-ethylhexyl) ester; benzene-1,2-dicarboxylic acid bis(2-ethylhexyl) ester; BRD-A89471977-001-05-2; Bis(2-ethylhexyl) phthalate 100 microg/mL in Methanol; Bis(2-ethylhexyl) phthalate 5000 microg/mL in Methanol; F0001-0292; Bis(2-ethylhexyl) phthalate, SAJ first grade, >=98.0%; Bis(2-ethylhexyl) phthalate, PESTANAL(R), analytical standard; Phthalic acid, bis-2-ethylhexyl ester 10 microg/mL in Cyclohexane; Plastic additive 01, European Pharmacopoeia (EP) Reference Standard; Bis(2-ethylhexyl) phthalate, certified reference material, TraceCERT(R); Plastic additive 14, United States Pharmacopeia (USP) Reference Standard; 1,2-Benzenedicarboxylic acid, bis(2-ethylhexyl) ester, labeled with carbon-14; 50885-87-5; 82208-43-3
Click to Show/Hide
|
|
| External link | ||
| Description |
DEHP worsened testicular histology, decreased testosterone concentrations, downregulated expression of spermatogenesis inducers, enhanced oxidative stress, inhibited theNrf2-mediated antioxidant pathway, and increased apoptosis in testes. DEHP is a common environmental endocrine disrupting chemical that inducesmale reproductive disorders. Additionally, DEHP increased global levels of m6A RNA modification and altered the expression of two important RNA methylation modulator genes, FTO and YTHDC2.
|
[11] |
| Compound Name | Trichostatin A | Investigative |
| Synonyms |
trichostatin A; 58880-19-6; Trichostatin; TSA; Trichostatin A (TSA); (2E,4E,6R)-7-[4-(Dimethylamino)phenyl]-N-hydroxy-4,6-dimethyl-7-oxohepta-2,4-dienamide; UNII-3X2S926L3Z; Antibiotic A-300; CHEBI:46024; C17H22N2O3; GNF-PF-1011; 3X2S926L3Z; 58880-19-6 (R-isomer); 2,4-Heptadienamide, 7-(4-(dimethylamino)phenyl)-N-hydroxy-4,6-dimethyl-7-oxo-; 7-(4-(Dimethylamino)phenyl)-N-hydroxy-4,6-dimethyl-7-oxo-2,4-heptadienamide; MFCD03848392; (R,2E,4E)-7-(4-(dimethylamino)phenyl)-N-hydroxy-4,6-dimethyl-7-oxohepta-2,4-dienamide; A-300-I; (2E,4E,6R)-7-(4-(dimethylamino)phenyl)-N-hydroxy-4,6-dimethyl-7-oxo-2,4-heptadienamide; [R-(E,E)]-7-[4-(Dimethylamino)phenyl]-N-hydroxy-4,6-dimethyl-7-oxo-2,4-heptadienamide; 2,4-Heptadienamide, 7-[4-(dimethylamino)phenyl]-N-hydroxy-4,6-dimethyl-7-oxo-, (2E,4E,6R)-; Trichostatin-A; Tricostatin A; 7-[4-(DIMETHYLAMINO)PHENYL]-N-HYDROXY-4,6-DIMETHYL-7-OXO-2,4-HEPTADIENAMIDE; 2,4-Heptadienamide, 7-(4-(dimethylamino)phenyl)-N-hydroxy-4,6-dimethyl-7-oxo-, (2E,4E,6R)-; TSN; Trichostatina; trichostatine a; Trichlostatin A; Trichostatin(s); (2E,4E,6R)-7-(4-dimethylaminophenyl)-4,6-dimethyl-7-oxo-hepta-2,4-dienehydroxamic acid; (2E,4E,6R)-7-(4-Dimethylaminophenyl)-N-hydroxy-4,6-dimethyl-7-oxo-hepta-2,4-dienamide; Trichostatin A,TSA; (R)-Trichostatin A; NCGC_TSA; 1c3r; 3f0r; Trichostatin-A - TSA; SCHEMBL19886; MLS006011095; SGCTO-002; SCHEMBL675951; GTPL7005; DTXSID6037063; CHEBI:93196; BCPP000035; HMS1362L09; HMS1792L09; HMS1990L09; HMS3403L09; HMS3649O20; BCP01776; EX-A1665; Trichostatin A, Ready Made Solution; BDBM50005711; LMPK01000055; s1045; Trichostatin A from Streptomyces sp.; AKOS015899840; ZINC100014731; CCG-208142; CCG-208681; CS-0499; DB04297; NSC 311042; NCGC00162453-01; NCGC00162453-02; NCGC00162453-03; NCGC00162453-04; NCGC00162453-05; NCGC00162453-15; 3C10; AS-74315; HY-15144; M984; SMR004702883; A8183; SW219664-1; T2477; A25618; M02571; 880T196; Q425894; SR-05000013796; Q-201864; SR-05000013796-3; BRD-K68202742-001-04-1; BRD-K68202742-001-05-8; Trichostatin A, >=98% (HPLC), from Streptomyces sp.; Trichostatin A, Streptomyces sp. - CAS 58880-19-6; UNII-30RHG284Z4 component RTKIYFITIVXBLE-QEQCGCAPSA-N; Trichostatin A??, Vetec(TM) reagent grade, from Streptomyces sp., >=98%; (2E,4E,6R)-7-(4-(Dimethylamino)phen yl)-N-hydroxy-4,6-dimethyl-7-oxo-2,4-heptadienamid e; (6R)-N-Hydroxy-4,6-dimethyl-7-oxo-7-[4-(dimethylamino)phenyl]-2,4-heptadienamide; 7-[4-(Dimethylamino)phenyl]-N-hydroxy-4,6R-dimethyl-7-oxo-2E,4E-heptadienamide; 2,4-Heptadienamide, 7-[4-(dimethylamino)phenyl]-N-hydroxy-4,6-dimethyl-7-oxo-, (2E,4E,6R)- (9CI); 2,4-Heptadienamide, 7-[4-(dimethylamino)phenyl]-N-hydroxy-4,6-dimethyl-7-oxo-, [R-(E,E)]-; 2,4-Heptadienamide,7-[4-(dimethylamino)phenyl]-N-hydroxy-4,6-dimethyl-7-oxo-, (2E,4E,6R)-
Click to Show/Hide
|
|
| External link | ||
| Description |
Treatment with the HDAC inhibitor, trichostatin A (TSA), reduces YTHDC2 expression in Huh7 and in TNF-Alpha-stimulated hepatocytes.
|
[36] |
References