m6A Target Gene Information
General Information of the m6A Target Gene (ID: M6ATAR00168)
Full List of m6A Methylation Regulator of This Target Gene and Corresponding Disease/Drug Response(s)
ACACA
can be regulated by the following regulator(s), and cause disease/drug response(s). You can browse detail information of regulator(s) or disease/drug response(s).
Browse Regulator
Browse Disease
Methyltransferase-like 3 (METTL3) [WRITER]
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
|
Treatment: METTL3 knockdown MDA-MB-231 cells
Control: MDA-MB-231 cells
|
GSE70061 | |
| Regulation |
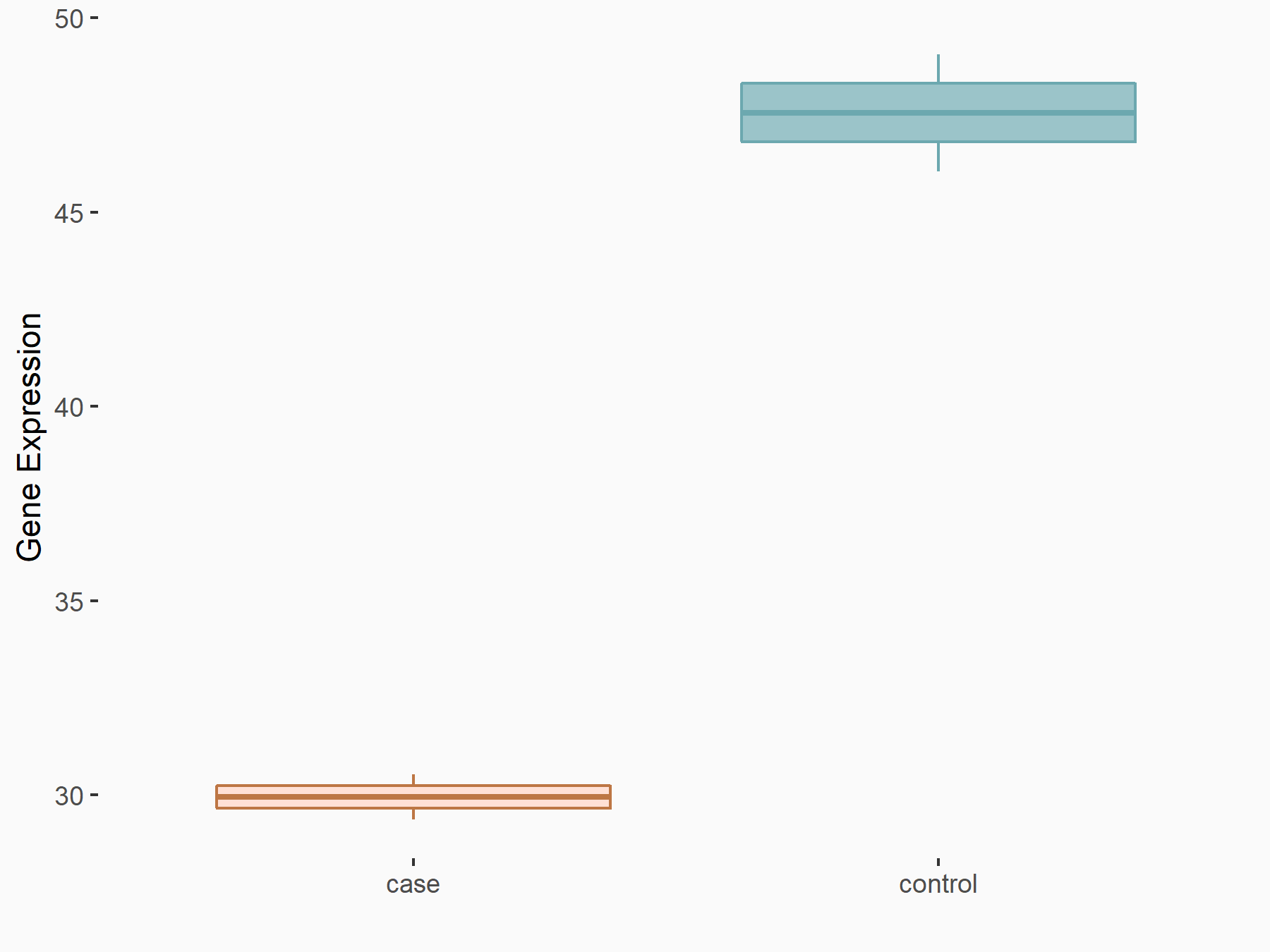  |
logFC: -6.50E-01 p-value: 2.31E-03 |
| More Results | Click to View More RNA-seq Results | |
| In total 1 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [1] | |||
| Response Summary | Type 2 diabetes (T2D) is characterized by lack of insulin, insulin resistance and high blood sugar. METTL3 silence decreased the m6A methylated and total mRNA level of Fatty acid synthase (Fasn), subsequently inhibited fatty acid metabolism. The expression of Acetyl-CoA carboxylase 1 (ACC1/ACACA), Acly, Dgat2, Ehhadh, Fasn, Foxo, Pgc1a and Sirt1, which are critical to the regulation of fatty acid synthesis and oxidation were dramatically decreased in livers of hepatocyte-specific METTL3 knockout mice. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Type 2 diabetes mellitus | ICD-11: 5A11 | ||
| Pathway Response | Insulin resistance | hsa04931 | ||
| Cell Process | Lipid metabolism | |||
| In-vitro Model | Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 |
| In-vivo Model | Hepatocyte-specific METTL3 knockout mice (TBG-Cre, METTL3 fl/fl) were generated by crossing mice with TBG-Cre Tg mice. METTL3 flox (METTL3 fl/fl) and hepatocyte-specific METTL3 knockout mice (TBG-Cre, METTL3 fl/fl) were used for experiments. | |||
Heterogeneous nuclear ribonucleoproteins A2/B1 (HNRNPA2B1) [READER]
| In total 1 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [2] | |||
| Response Summary | The levels of m6A and its regulator HNRNPA2B1 were significantly increased in cancerous tissues of esophageal cancer(ESCA), and overexpression of HNRNPA2B1 promotes ESCA progression via up-regulation of de novo fatty acid synthetic enzymes ACLY and Acetyl-CoA carboxylase 1 (ACC1/ACACA). | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Esophageal cancer | ICD-11: 2B70 | ||
| Pathway Response | Metabolic pathways | hsa01100 | ||
| Cell Process | Fatty acid synthesis | |||
| In-vitro Model | TE-10 | Esophageal squamous cell carcinoma | Homo sapiens | CVCL_1760 |
| Eca-109 | Esophageal squamous cell carcinoma | Homo sapiens | CVCL_6898 | |
YTH domain-containing protein 2 (YTHDC2) [READER]
| In total 1 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [3] | |||
| Response Summary | In nonalcoholic fatty liver disease, Ythdc2 could bind to mRNA of lipogenic genes, including sterol regulatory element-binding protein 1c, fatty acid synthase, stearoyl-CoA desaturase 1, and Acetyl-CoA carboxylase 1 (ACC1/ACACA), to decrease their mRNA stability and inhibit gene expression. | |||
| Target Regulation | Down regulation | |||
| Responsed Disease | Non-alcoholic fatty liver disease | ICD-11: DB92 | ||
| Pathway Response | RNA degradation | hsa03018 | ||
| Cell Process | RNA stability | |||
| In-vivo Model | All mice were housed at 21℃ ± 1℃ with a humidity of 55% ± 10% and a 12-hour light/dark cycle. The high-fat diets (HFDs), containing 60% kcal from fat, 20% kcal from carbohydrate, and 20% kcal from protein. | |||
Esophageal cancer [ICD-11: 2B70]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [2] | |||
| Response Summary | The levels of m6A and its regulator HNRNPA2B1 were significantly increased in cancerous tissues of esophageal cancer(ESCA), and overexpression of HNRNPA2B1 promotes ESCA progression via up-regulation of de novo fatty acid synthetic enzymes ACLY and Acetyl-CoA carboxylase 1 (ACC1/ACACA). | |||
| Responsed Disease | Esophageal cancer [ICD-11: 2B70] | |||
| Target Regulator | Heterogeneous nuclear ribonucleoproteins A2/B1 (HNRNPA2B1) | READER | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Metabolic pathways | hsa01100 | ||
| Cell Process | Fatty acid synthesis | |||
| In-vitro Model | TE-10 | Esophageal squamous cell carcinoma | Homo sapiens | CVCL_1760 |
| Eca-109 | Esophageal squamous cell carcinoma | Homo sapiens | CVCL_6898 | |
Type 2 diabetes mellitus [ICD-11: 5A11]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [1] | |||
| Response Summary | Type 2 diabetes (T2D) is characterized by lack of insulin, insulin resistance and high blood sugar. METTL3 silence decreased the m6A methylated and total mRNA level of Fatty acid synthase (Fasn), subsequently inhibited fatty acid metabolism. The expression of Acetyl-CoA carboxylase 1 (ACC1/ACACA), Acly, Dgat2, Ehhadh, Fasn, Foxo, Pgc1a and Sirt1, which are critical to the regulation of fatty acid synthesis and oxidation were dramatically decreased in livers of hepatocyte-specific METTL3 knockout mice. | |||
| Responsed Disease | Type 2 diabetes mellitus [ICD-11: 5A11] | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Insulin resistance | hsa04931 | ||
| Cell Process | Lipid metabolism | |||
| In-vitro Model | Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 |
| In-vivo Model | Hepatocyte-specific METTL3 knockout mice (TBG-Cre, METTL3 fl/fl) were generated by crossing mice with TBG-Cre Tg mice. METTL3 flox (METTL3 fl/fl) and hepatocyte-specific METTL3 knockout mice (TBG-Cre, METTL3 fl/fl) were used for experiments. | |||
Non-alcoholic fatty liver disease [ICD-11: DB92]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [3] | |||
| Response Summary | In nonalcoholic fatty liver disease, Ythdc2 could bind to mRNA of lipogenic genes, including sterol regulatory element-binding protein 1c, fatty acid synthase, stearoyl-CoA desaturase 1, and Acetyl-CoA carboxylase 1 (ACC1/ACACA), to decrease their mRNA stability and inhibit gene expression. | |||
| Responsed Disease | Non-alcoholic fatty liver disease [ICD-11: DB92] | |||
| Target Regulator | YTH domain-containing protein 2 (YTHDC2) | READER | ||
| Target Regulation | Down regulation | |||
| Pathway Response | RNA degradation | hsa03018 | ||
| Cell Process | RNA stability | |||
| In-vivo Model | All mice were housed at 21℃ ± 1℃ with a humidity of 55% ± 10% and a 12-hour light/dark cycle. The high-fat diets (HFDs), containing 60% kcal from fat, 20% kcal from carbohydrate, and 20% kcal from protein. | |||
RNA Modification Sequencing Data Associated with the Target (ID: M6ATAR00168)
| In total 38 m6A sequence/site(s) in this target gene | |||
| mod ID: A2ISITE007924 | Click to Show/Hide the Full List | ||
| mod site | chr17:37092216-37092217:- | [4] | |
| Sequence | GATGACTCACTTTCAGCCTCAACCTCCTGGGCTCATGTGAT | ||
| Transcript ID List | ENST00000619546.4; ENST00000614428.4; ENST00000614482.4; ENST00000612895.4; ENST00000617649.4; ENST00000613776.1; ENST00000616317.5 | ||
| External Link | RMBase: RNA-editing_site_56976 | ||
| mod ID: A2ISITE007925 | Click to Show/Hide the Full List | ||
| mod site | chr17:37092222-37092223:- | [4] | |
| Sequence | AGGTATGATGACTCACTTTCAGCCTCAACCTCCTGGGCTCA | ||
| Transcript ID List | ENST00000612895.4; ENST00000614482.4; ENST00000613776.1; ENST00000619546.4; ENST00000617649.4; ENST00000616317.5; ENST00000614428.4 | ||
| External Link | RMBase: RNA-editing_site_56977 | ||
| mod ID: A2ISITE007926 | Click to Show/Hide the Full List | ||
| mod site | chr17:37158947-37158948:- | [5] | |
| Sequence | TGGCCTCAAGTGATCCTCCCACCTTGGCCTCCCCAAGTGCT | ||
| Transcript ID List | ENST00000619546.4; ENST00000614482.4; ENST00000616317.5; ENST00000612895.4; ENST00000617649.4; ENST00000614428.4 | ||
| External Link | RMBase: RNA-editing_site_56978 | ||
| mod ID: A2ISITE007927 | Click to Show/Hide the Full List | ||
| mod site | chr17:37184092-37184093:- | [5] | |
| Sequence | TTGGGAGGCCGAGGTGGGCAAATCAGCTGAGGTTAGGAGTT | ||
| Transcript ID List | rmsk_4683021; ENST00000612895.4; ENST00000619546.4; ENST00000617649.4; ENST00000618575.1; ENST00000616317.5; ENST00000614428.4 | ||
| External Link | RMBase: RNA-editing_site_56979 | ||
| mod ID: A2ISITE007928 | Click to Show/Hide the Full List | ||
| mod site | chr17:37184249-37184250:- | [5] | |
| Sequence | GAGTAGGAGAGAGTAAACTAAGACCACATGCTTGTGATGAT | ||
| Transcript ID List | ENST00000616317.5; ENST00000617649.4; ENST00000619546.4; ENST00000614428.4; ENST00000618575.1; ENST00000612895.4 | ||
| External Link | RMBase: RNA-editing_site_56980 | ||
| mod ID: A2ISITE007929 | Click to Show/Hide the Full List | ||
| mod site | chr17:37199914-37199915:- | [4] | |
| Sequence | AGATGAGCCTGGGCAACATAATGAGACCCAGTCCTCTATTA | ||
| Transcript ID List | ENST00000612895.4; rmsk_4683040; ENST00000619546.4; ENST00000616317.5; ENST00000617649.4; ENST00000614428.4 | ||
| External Link | RMBase: RNA-editing_site_56981 | ||
| mod ID: A2ISITE007930 | Click to Show/Hide the Full List | ||
| mod site | chr17:37199915-37199916:- | [4] | |
| Sequence | GAGATGAGCCTGGGCAACATAATGAGACCCAGTCCTCTATT | ||
| Transcript ID List | ENST00000617649.4; ENST00000616317.5; ENST00000612895.4; ENST00000619546.4; ENST00000614428.4; rmsk_4683040 | ||
| External Link | RMBase: RNA-editing_site_56982 | ||
| mod ID: A2ISITE007931 | Click to Show/Hide the Full List | ||
| mod site | chr17:37199962-37199963:- | [4] | |
| Sequence | GCACTTTGGGAGGCTGAGTCAGGAAAATTGCTTGAGCCCAC | ||
| Transcript ID List | rmsk_4683040; ENST00000614428.4; ENST00000619546.4; ENST00000616317.5; ENST00000612895.4; ENST00000617649.4 | ||
| External Link | RMBase: RNA-editing_site_56983 | ||
| mod ID: A2ISITE007932 | Click to Show/Hide the Full List | ||
| mod site | chr17:37201328-37201329:- | [5] | |
| Sequence | CATGCCTCATTCTCTCAAGTAGCTGGGATTACAGGAATGTG | ||
| Transcript ID List | ENST00000614428.4; ENST00000616317.5; ENST00000617649.4; ENST00000612895.4; ENST00000614450.4; ENST00000619546.4 | ||
| External Link | RMBase: RNA-editing_site_56984 | ||
| mod ID: A2ISITE007933 | Click to Show/Hide the Full List | ||
| mod site | chr17:37212785-37212786:- | [4] | |
| Sequence | ATGAAAAATTTGTAGTGGCCAGGTTTGCTGGCTCATGCCTA | ||
| Transcript ID List | ENST00000612120.4; rmsk_4683056; ENST00000614450.4; ENST00000616317.5; ENST00000614428.4; ENST00000612895.4; ENST00000617649.4 | ||
| External Link | RMBase: RNA-editing_site_56985 | ||
| mod ID: A2ISITE007934 | Click to Show/Hide the Full List | ||
| mod site | chr17:37242951-37242952:- | [5] | |
| Sequence | ACCCCTGCCTCCCGAGTTCAAGCAATTCTCCTGCCTCAGCC | ||
| Transcript ID List | ENST00000616317.5; ENST00000614428.4; ENST00000617649.4; ENST00000613146.4; ENST00000612895.4 | ||
| External Link | RMBase: RNA-editing_site_56986 | ||
| mod ID: A2ISITE007935 | Click to Show/Hide the Full List | ||
| mod site | chr17:37302229-37302230:- | [6] | |
| Sequence | AGCCTGGGTCACAAGAGCGAAACCCTGTCTCAAAAAAAAAA | ||
| Transcript ID List | ENST00000617548.1; ENST00000614789.4; ENST00000613146.4; ENST00000616317.5; rmsk_4683242; ENST00000614428.4; ENST00000621960.1; ENST00000616352.4; ENST00000615229.4 | ||
| External Link | RMBase: RNA-editing_site_56987 | ||
| mod ID: A2ISITE007936 | Click to Show/Hide the Full List | ||
| mod site | chr17:37302236-37302237:- | [6] | |
| Sequence | GCACTCCAGCCTGGGTCACAAGAGCGAAACCCTGTCTCAAA | ||
| Transcript ID List | ENST00000616317.5; ENST00000614428.4; ENST00000617548.1; ENST00000616352.4; ENST00000615229.4; ENST00000614789.4; ENST00000613146.4; rmsk_4683242; ENST00000621960.1 | ||
| External Link | RMBase: RNA-editing_site_56988 | ||
| mod ID: A2ISITE007937 | Click to Show/Hide the Full List | ||
| mod site | chr17:37302249-37302250:- | [6] | |
| Sequence | AGATCGCGCCATTGCACTCCAGCCTGGGTCACAAGAGCGAA | ||
| Transcript ID List | ENST00000621960.1; ENST00000616352.4; ENST00000617548.1; rmsk_4683242; ENST00000615229.4; ENST00000614428.4; ENST00000616317.5; ENST00000614789.4; ENST00000613146.4 | ||
| External Link | RMBase: RNA-editing_site_56989 | ||
| mod ID: A2ISITE007938 | Click to Show/Hide the Full List | ||
| mod site | chr17:37302269-37302270:- | [6] | |
| Sequence | TTGCAGATTGCAGTGAGCCGAGATCGCGCCATTGCACTCCA | ||
| Transcript ID List | ENST00000614428.4; ENST00000614789.4; ENST00000617548.1; ENST00000616317.5; rmsk_4683242; ENST00000616352.4; ENST00000615229.4; ENST00000621960.1; ENST00000613146.4 | ||
| External Link | RMBase: RNA-editing_site_56990 | ||
| mod ID: A2ISITE007939 | Click to Show/Hide the Full List | ||
| mod site | chr17:37302274-37302275:- | [6] | |
| Sequence | GGAGGTTGCAGATTGCAGTGAGCCGAGATCGCGCCATTGCA | ||
| Transcript ID List | ENST00000621960.1; ENST00000617548.1; rmsk_4683242; ENST00000613146.4; ENST00000614428.4; ENST00000615229.4; ENST00000616352.4; ENST00000614789.4; ENST00000616317.5 | ||
| External Link | RMBase: RNA-editing_site_56991 | ||
| mod ID: A2ISITE007940 | Click to Show/Hide the Full List | ||
| mod site | chr17:37302314-37302315:- | [6] | |
| Sequence | CGGGAGGCTGAGGCGGGAGAATCGCTTGAACCCGGGAGGTG | ||
| Transcript ID List | ENST00000614428.4; ENST00000617548.1; ENST00000614789.4; ENST00000613146.4; ENST00000616352.4; ENST00000616317.5; ENST00000621960.1; ENST00000615229.4; rmsk_4683242 | ||
| External Link | RMBase: RNA-editing_site_56992 | ||
| mod ID: A2ISITE007941 | Click to Show/Hide the Full List | ||
| mod site | chr17:37302324-37302325:- | [6] | |
| Sequence | CCCAGCTACTCGGGAGGCTGAGGCGGGAGAATCGCTTGAAC | ||
| Transcript ID List | ENST00000617548.1; ENST00000616352.4; ENST00000616317.5; rmsk_4683242; ENST00000614789.4; ENST00000621960.1; ENST00000615229.4; ENST00000613146.4; ENST00000614428.4 | ||
| External Link | RMBase: RNA-editing_site_56993 | ||
| mod ID: A2ISITE007942 | Click to Show/Hide the Full List | ||
| mod site | chr17:37302337-37302338:- | [6] | |
| Sequence | AGTCACCTGTAATCCCAGCTACTCGGGAGGCTGAGGCGGGA | ||
| Transcript ID List | ENST00000621960.1; rmsk_4683242; ENST00000616317.5; ENST00000614428.4; ENST00000615229.4; ENST00000613146.4; ENST00000614789.4; ENST00000617548.1; ENST00000616352.4 | ||
| External Link | RMBase: RNA-editing_site_56994 | ||
| mod ID: A2ISITE007943 | Click to Show/Hide the Full List | ||
| mod site | chr17:37302341-37302342:- | [6] | |
| Sequence | TGGCAGTCACCTGTAATCCCAGCTACTCGGGAGGCTGAGGC | ||
| Transcript ID List | ENST00000616352.4; ENST00000617548.1; ENST00000614428.4; ENST00000621960.1; ENST00000615229.4; ENST00000613146.4; rmsk_4683242; ENST00000614789.4; ENST00000616317.5 | ||
| External Link | RMBase: RNA-editing_site_56995 | ||
| mod ID: A2ISITE007944 | Click to Show/Hide the Full List | ||
| mod site | chr17:37302346-37302347:- | [6] | |
| Sequence | CATGGTGGCAGTCACCTGTAATCCCAGCTACTCGGGAGGCT | ||
| Transcript ID List | ENST00000613146.4; ENST00000616317.5; ENST00000616352.4; ENST00000615229.4; ENST00000621960.1; rmsk_4683242; ENST00000614789.4; ENST00000614428.4; ENST00000617548.1 | ||
| External Link | RMBase: RNA-editing_site_56996 | ||
| mod ID: A2ISITE007945 | Click to Show/Hide the Full List | ||
| mod site | chr17:37302357-37302358:- | [6] | |
| Sequence | TATTAGCCTGGCATGGTGGCAGTCACCTGTAATCCCAGCTA | ||
| Transcript ID List | rmsk_4683242; ENST00000616352.4; ENST00000613146.4; ENST00000617548.1; ENST00000621960.1; ENST00000614789.4; ENST00000615229.4; ENST00000614428.4; ENST00000616317.5 | ||
| External Link | RMBase: RNA-editing_site_56997 | ||
| mod ID: A2ISITE007946 | Click to Show/Hide the Full List | ||
| mod site | chr17:37302373-37302374:- | [6] | |
| Sequence | CTACTAAAAATACAAATATTAGCCTGGCATGGTGGCAGTCA | ||
| Transcript ID List | ENST00000614789.4; ENST00000614428.4; rmsk_4683242; ENST00000616317.5; ENST00000617548.1; ENST00000621960.1; ENST00000613146.4; ENST00000616352.4; ENST00000615229.4 | ||
| External Link | RMBase: RNA-editing_site_56998 | ||
| mod ID: A2ISITE007947 | Click to Show/Hide the Full List | ||
| mod site | chr17:37302379-37302380:- | [6] | |
| Sequence | CCATCTCTACTAAAAATACAAATATTAGCCTGGCATGGTGG | ||
| Transcript ID List | ENST00000613146.4; ENST00000614428.4; rmsk_4683242; ENST00000616352.4; ENST00000615229.4; ENST00000614789.4; ENST00000616317.5; ENST00000617548.1; ENST00000621960.1 | ||
| External Link | RMBase: RNA-editing_site_56999 | ||
| mod ID: A2ISITE007948 | Click to Show/Hide the Full List | ||
| mod site | chr17:37302382-37302383:- | [6] | |
| Sequence | ACCCCATCTCTACTAAAAATACAAATATTAGCCTGGCATGG | ||
| Transcript ID List | ENST00000614789.4; ENST00000617548.1; ENST00000613146.4; ENST00000621960.1; ENST00000616317.5; rmsk_4683242; ENST00000616352.4; ENST00000614428.4; ENST00000615229.4 | ||
| External Link | RMBase: RNA-editing_site_57000 | ||
| mod ID: A2ISITE007949 | Click to Show/Hide the Full List | ||
| mod site | chr17:37302384-37302385:- | [6] | |
| Sequence | AAACCCCATCTCTACTAAAAATACAAATATTAGCCTGGCAT | ||
| Transcript ID List | ENST00000616317.5; ENST00000617548.1; ENST00000616352.4; ENST00000615229.4; rmsk_4683242; ENST00000614428.4; ENST00000613146.4; ENST00000614789.4; ENST00000621960.1 | ||
| External Link | RMBase: RNA-editing_site_57001 | ||
| mod ID: A2ISITE007950 | Click to Show/Hide the Full List | ||
| mod site | chr17:37302388-37302389:- | [6] | |
| Sequence | GGTTAAACCCCATCTCTACTAAAAATACAAATATTAGCCTG | ||
| Transcript ID List | ENST00000617548.1; rmsk_4683242; ENST00000614428.4; ENST00000616317.5; ENST00000613146.4; ENST00000614789.4; ENST00000616352.4; ENST00000621960.1; ENST00000615229.4 | ||
| External Link | RMBase: RNA-editing_site_57002 | ||
| mod ID: A2ISITE007951 | Click to Show/Hide the Full List | ||
| mod site | chr17:37302410-37302411:- | [6] | |
| Sequence | TCGAGACCAGTCTGGCCAACATGGTTAAACCCCATCTCTAC | ||
| Transcript ID List | ENST00000615229.4; ENST00000616317.5; ENST00000617548.1; ENST00000616352.4; ENST00000614789.4; ENST00000621960.1; ENST00000613146.4; rmsk_4683242; ENST00000614428.4 | ||
| External Link | RMBase: RNA-editing_site_57003 | ||
| mod ID: A2ISITE007952 | Click to Show/Hide the Full List | ||
| mod site | chr17:37302412-37302413:- | [6] | |
| Sequence | GTTCGAGACCAGTCTGGCCAACATGGTTAAACCCCATCTCT | ||
| Transcript ID List | ENST00000613146.4; ENST00000617548.1; ENST00000614428.4; ENST00000615229.4; rmsk_4683242; ENST00000614789.4; ENST00000616352.4; ENST00000621960.1; ENST00000616317.5 | ||
| External Link | RMBase: RNA-editing_site_57004 | ||
| mod ID: A2ISITE007953 | Click to Show/Hide the Full List | ||
| mod site | chr17:37302436-37302437:- | [6] | |
| Sequence | TGGGTGGATCACCTGAGGTCAGGAGTTCGAGACCAGTCTGG | ||
| Transcript ID List | ENST00000615229.4; ENST00000614789.4; ENST00000613146.4; ENST00000616352.4; ENST00000617548.1; ENST00000621960.1; rmsk_4683242; ENST00000616317.5; ENST00000614428.4 | ||
| External Link | RMBase: RNA-editing_site_57005 | ||
| mod ID: A2ISITE007954 | Click to Show/Hide the Full List | ||
| mod site | chr17:37302441-37302442:- | [6] | |
| Sequence | TGAGGTGGGTGGATCACCTGAGGTCAGGAGTTCGAGACCAG | ||
| Transcript ID List | ENST00000615229.4; ENST00000616352.4; ENST00000614789.4; ENST00000617548.1; ENST00000616317.5; ENST00000614428.4; ENST00000621960.1; rmsk_4683242; ENST00000613146.4 | ||
| External Link | RMBase: RNA-editing_site_57006 | ||
| mod ID: A2ISITE007955 | Click to Show/Hide the Full List | ||
| mod site | chr17:37302446-37302447:- | [6] | |
| Sequence | GAGGCTGAGGTGGGTGGATCACCTGAGGTCAGGAGTTCGAG | ||
| Transcript ID List | rmsk_4683242; ENST00000616352.4; ENST00000617548.1; ENST00000616317.5; ENST00000613146.4; ENST00000615229.4; ENST00000621960.1; ENST00000614428.4; ENST00000614789.4 | ||
| External Link | RMBase: RNA-editing_site_57007 | ||
| mod ID: A2ISITE007956 | Click to Show/Hide the Full List | ||
| mod site | chr17:37302465-37302466:- | [6] | |
| Sequence | TGTAATCCCAGCACTTTGGGAGGCTGAGGTGGGTGGATCAC | ||
| Transcript ID List | ENST00000617548.1; ENST00000614789.4; rmsk_4683242; ENST00000616352.4; ENST00000613146.4; ENST00000614428.4; ENST00000621960.1; ENST00000615229.4; ENST00000616317.5 | ||
| External Link | RMBase: RNA-editing_site_57008 | ||
| mod ID: A2ISITE007957 | Click to Show/Hide the Full List | ||
| mod site | chr17:37302473-37302474:- | [6] | |
| Sequence | CTCACGCCTGTAATCCCAGCACTTTGGGAGGCTGAGGTGGG | ||
| Transcript ID List | ENST00000617548.1; ENST00000616317.5; ENST00000615229.4; ENST00000614428.4; ENST00000613146.4; rmsk_4683242; ENST00000621960.1; ENST00000614789.4; ENST00000616352.4 | ||
| External Link | RMBase: RNA-editing_site_57009 | ||
| mod ID: A2ISITE007958 | Click to Show/Hide the Full List | ||
| mod site | chr17:37302481-37302482:- | [6] | |
| Sequence | TGTGGTGGCTCACGCCTGTAATCCCAGCACTTTGGGAGGCT | ||
| Transcript ID List | ENST00000616352.4; ENST00000616317.5; ENST00000617548.1; ENST00000614428.4; ENST00000614789.4; ENST00000613146.4; rmsk_4683242; ENST00000621960.1; ENST00000615229.4 | ||
| External Link | RMBase: RNA-editing_site_57010 | ||
| mod ID: A2ISITE007959 | Click to Show/Hide the Full List | ||
| mod site | chr17:37302490-37302491:- | [6] | |
| Sequence | CCGGCCGGGTGTGGTGGCTCACGCCTGTAATCCCAGCACTT | ||
| Transcript ID List | rmsk_4683242; ENST00000616352.4; ENST00000614789.4; ENST00000613146.4; ENST00000616317.5; ENST00000614428.4; ENST00000617548.1; ENST00000621960.1; ENST00000615229.4 | ||
| External Link | RMBase: RNA-editing_site_57011 | ||
| mod ID: A2ISITE007960 | Click to Show/Hide the Full List | ||
| mod site | chr17:37320761-37320762:- | [5] | |
| Sequence | TAGAGACAGGGTTTCACCATATTGCTCAGGCTGGTCTCGAA | ||
| Transcript ID List | ENST00000616352.4; ENST00000617548.1; ENST00000616317.5; ENST00000615229.4; ENST00000621960.1; ENST00000614428.4; ENST00000614789.4; ENST00000613146.4 | ||
| External Link | RMBase: RNA-editing_site_57012 | ||
| mod ID: A2ISITE007961 | Click to Show/Hide the Full List | ||
| mod site | chr17:37337907-37337908:- | [5] | |
| Sequence | AGATGGGGTTTCACCATGTTAGCCAGGATGGTCTTGATCTC | ||
| Transcript ID List | ENST00000616352.4; ENST00000621960.1; ENST00000616317.5; ENST00000614789.4; ENST00000617548.1; ENST00000612007.1; ENST00000615229.4; ENST00000613146.4; ENST00000614428.4 | ||
| External Link | RMBase: RNA-editing_site_57013 | ||
2'-O-Methylation (2'-O-Me)
| In total 1 m6A sequence/site(s) in this target gene | |||
| mod ID: 2OMSITE000150 | Click to Show/Hide the Full List | ||
| mod site | chr17:37116520-37116521:- | [7] | |
| Sequence | TTTCCTACTCACAGTCTTGGGGCCGAAAAATACTTGTTTGG | ||
| Cell/Tissue List | HEK293 | ||
| Seq Type List | RiboMeth-seq | ||
| Transcript ID List | ENST00000614428.4; ENST00000612895.4; ENST00000613776.1; ENST00000616317.5; ENST00000614482.4; ENST00000617649.4; ENST00000619546.4 | ||
| External Link | RMBase: Nm_site_2307 | ||
5-methylcytidine (m5C)
| In total 3 m6A sequence/site(s) in this target gene | |||
| mod ID: M5CSITE001092 | Click to Show/Hide the Full List | ||
| mod site | chr17:37085749-37085750:- | [8] | |
| Sequence | TCCCCTTCTTCAGGAGGAATCTGTGCGGATAGATTGGCTGG | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000619546.4; ENST00000612895.4; ENST00000616317.5; ENST00000614428.4 | ||
| External Link | RMBase: m5C_site_18591 | ||
| mod ID: M5CSITE001093 | Click to Show/Hide the Full List | ||
| mod site | chr17:37179333-37179334:- | [8] | |
| Sequence | CCCCTCTGCCTTCTGACATGCTGACTTACACTGAACTGGTA | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000617649.4; ENST00000618575.1; ENST00000616317.5; ENST00000614428.4; ENST00000619546.4; ENST00000612895.4 | ||
| External Link | RMBase: m5C_site_18592 | ||
| mod ID: M5CSITE001094 | Click to Show/Hide the Full List | ||
| mod site | chr17:37291233-37291234:- | ||
| Sequence | GCGGTGAGCTGAGATTGCACCATTGTACTCCAGCCTGGGCA | ||
| Cell/Tissue List | T24 | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000616352.4; ENST00000612895.4; ENST00000617649.4; ENST00000615229.4; rmsk_4683216; ENST00000613146.4; ENST00000621960.1; ENST00000614789.4; ENST00000616317.5; ENST00000614428.4; ENST00000617548.1 | ||
| External Link | RMBase: m5C_site_18594 | ||
N6-methyladenosine (m6A)
| In total 115 m6A sequence/site(s) in this target gene | |||
| mod ID: M6ASITE031998 | Click to Show/Hide the Full List | ||
| mod site | chr17:37085029-37085030:- | [9] | |
| Sequence | GAGCTTCAGATGCACTCTTTACACATTTTGTTGAAATAAAC | ||
| Motif Score | 2.07285119 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000619546.4; ENST00000612895.4; ENST00000614428.4; ENST00000616317.5 | ||
| External Link | RMBase: m6A_site_359473 | ||
| mod ID: M6ASITE031999 | Click to Show/Hide the Full List | ||
| mod site | chr17:37085104-37085105:- | [9] | |
| Sequence | ATTATCTGTTCTCTTCGTTCACAAAACAGCCTTCACTTGTC | ||
| Motif Score | 2.047297619 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000612895.4; ENST00000614428.4; ENST00000619546.4; ENST00000616317.5 | ||
| External Link | RMBase: m6A_site_359474 | ||
| mod ID: M6ASITE032000 | Click to Show/Hide the Full List | ||
| mod site | chr17:37085200-37085201:- | [10] | |
| Sequence | ACATTCCCACAGCCCCTCAAACATGCAAGGCTACTAAGGCA | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HEK293T; hESC-HEK293T | ||
| Seq Type List | DART-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000619546.4; ENST00000614428.4; ENST00000616317.5; ENST00000612895.4 | ||
| External Link | RMBase: m6A_site_359475 | ||
| mod ID: M6ASITE032001 | Click to Show/Hide the Full List | ||
| mod site | chr17:37085220-37085221:- | [10] | |
| Sequence | CACAGAACATGAGCAATCTGACATTCCCACAGCCCCTCAAA | ||
| Motif Score | 2.859755952 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000616317.5; ENST00000614428.4; ENST00000619546.4; ENST00000612895.4 | ||
| External Link | RMBase: m6A_site_359476 | ||
| mod ID: M6ASITE032002 | Click to Show/Hide the Full List | ||
| mod site | chr17:37085264-37085265:- | [9] | |
| Sequence | TACTGTAGGTCTTACCCTGGACAAGGATTTTTTCAAGTTGG | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000619546.4; ENST00000612895.4; ENST00000614428.4; ENST00000616317.5 | ||
| External Link | RMBase: m6A_site_359477 | ||
| mod ID: M6ASITE032003 | Click to Show/Hide the Full List | ||
| mod site | chr17:37085283-37085284:- | [10] | |
| Sequence | CCCCATCCCCAGAGCTCCTTACTGTAGGTCTTACCCTGGAC | ||
| Motif Score | 2.494845238 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000612895.4; ENST00000619546.4; ENST00000614428.4; ENST00000616317.5 | ||
| External Link | RMBase: m6A_site_359478 | ||
| mod ID: M6ASITE032004 | Click to Show/Hide the Full List | ||
| mod site | chr17:37085349-37085350:- | [10] | |
| Sequence | CCTCTCCCCACCAAATTATGACACAAACGAGTATGTTTCCT | ||
| Motif Score | 2.859755952 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000619546.4; ENST00000614428.4; ENST00000616317.5; ENST00000612895.4 | ||
| External Link | RMBase: m6A_site_359479 | ||
| mod ID: M6ASITE032005 | Click to Show/Hide the Full List | ||
| mod site | chr17:37085383-37085384:- | [10] | |
| Sequence | TCAGAGGCAGGGTGGGTTACACTCATTTACCTCCCCTCTCC | ||
| Motif Score | 2.506922619 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000614428.4; ENST00000619546.4; ENST00000612895.4; ENST00000616317.5 | ||
| External Link | RMBase: m6A_site_359480 | ||
| mod ID: M6ASITE032006 | Click to Show/Hide the Full List | ||
| mod site | chr17:37085385-37085386:- | [9] | |
| Sequence | CTTCAGAGGCAGGGTGGGTTACACTCATTTACCTCCCCTCT | ||
| Motif Score | 2.07285119 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000612895.4; ENST00000614428.4; ENST00000616317.5; ENST00000619546.4 | ||
| External Link | RMBase: m6A_site_359481 | ||
| mod ID: M6ASITE032007 | Click to Show/Hide the Full List | ||
| mod site | chr17:37085454-37085455:- | [9] | |
| Sequence | AAATCAAATCCCACATATCTACAAGTGGTATGAAGTCCTGC | ||
| Motif Score | 2.078666667 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000616317.5; ENST00000614428.4; ENST00000612895.4; ENST00000619546.4 | ||
| External Link | RMBase: m6A_site_359482 | ||
| mod ID: M6ASITE032008 | Click to Show/Hide the Full List | ||
| mod site | chr17:37085462-37085463:- | [9] | |
| Sequence | TTTGCAATAAATCAAATCCCACATATCTACAAGTGGTATGA | ||
| Motif Score | 2.053113095 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000619546.4; ENST00000612895.4; ENST00000616317.5; ENST00000614428.4 | ||
| External Link | RMBase: m6A_site_359483 | ||
| mod ID: M6ASITE032009 | Click to Show/Hide the Full List | ||
| mod site | chr17:37085505-37085506:- | [10] | |
| Sequence | AAGAATTGGATTCATTTTTGACCAGATTATTCTTCTATGCT | ||
| Motif Score | 2.839113095 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000619546.4; ENST00000614428.4; ENST00000616317.5; ENST00000612895.4 | ||
| External Link | RMBase: m6A_site_359484 | ||
| mod ID: M6ASITE032010 | Click to Show/Hide the Full List | ||
| mod site | chr17:37085532-37085533:- | [10] | |
| Sequence | CCATTGCTGGCTATTTGCTTACCTAGTAAGAATTGGATTCA | ||
| Motif Score | 2.052208333 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000619546.4; ENST00000612895.4; ENST00000614428.4; ENST00000616317.5 | ||
| External Link | RMBase: m6A_site_359485 | ||
| mod ID: M6ASITE032011 | Click to Show/Hide the Full List | ||
| mod site | chr17:37085581-37085582:- | [9] | |
| Sequence | AGGGCATAAAGTGGTGGGGGACAATTCTCAGTCCAGGAAAA | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | hESC-HEK293T; AML | ||
| Seq Type List | MAZTER-seq; miCLIP | ||
| Transcript ID List | ENST00000614428.4; ENST00000619546.4; ENST00000612895.4; ENST00000616317.5 | ||
| External Link | RMBase: m6A_site_359486 | ||
| mod ID: M6ASITE032012 | Click to Show/Hide the Full List | ||
| mod site | chr17:37085673-37085674:- | [11] | |
| Sequence | TATGGCTGGGTATGGAGCGGACAGCCCCCAGGAGTCAGAGC | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; liver | ||
| Seq Type List | m6A-seq; m6A-REF-seq | ||
| Transcript ID List | ENST00000614428.4; ENST00000612895.4; ENST00000616317.5; ENST00000619546.4 | ||
| External Link | RMBase: m6A_site_359487 | ||
| mod ID: M6ASITE032013 | Click to Show/Hide the Full List | ||
| mod site | chr17:37085728-37085729:- | [11] | |
| Sequence | TGTGCGGATAGATTGGCTGGACTTTTCAATGGTTCTGGGTT | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa; A549 | ||
| Seq Type List | m6A-seq; m6A-CLIP/IP | ||
| Transcript ID List | ENST00000616317.5; ENST00000619546.4; ENST00000614428.4; ENST00000612895.4 | ||
| External Link | RMBase: m6A_site_359488 | ||
| mod ID: M6ASITE032014 | Click to Show/Hide the Full List | ||
| mod site | chr17:37085859-37085860:- | [10] | |
| Sequence | CCAAGGAGGAAATCACTAAGACATTTGAGAAGCAGTGGTAT | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HEK293T; hESC-HEK293T | ||
| Seq Type List | DART-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000614428.4; ENST00000612895.4; ENST00000616317.5; ENST00000619546.4 | ||
| External Link | RMBase: m6A_site_359489 | ||
| mod ID: M6ASITE032015 | Click to Show/Hide the Full List | ||
| mod site | chr17:37086004-37086005:- | [10] | |
| Sequence | GTGGAGAGAGTTTACTTTCAACAACTAGTTTATTCAAGAAA | ||
| Motif Score | 2.173910714 | ||
| Cell/Tissue List | HEK293T; hESC-HEK293T | ||
| Seq Type List | DART-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000612895.4; ENST00000619546.4; ENST00000616317.5; ENST00000614428.4 | ||
| External Link | RMBase: m6A_site_359490 | ||
| mod ID: M6ASITE032016 | Click to Show/Hide the Full List | ||
| mod site | chr17:37086050-37086051:- | [10] | |
| Sequence | AGGCTTACTGGCCCCTCCTGACTGGCATCAGGGGCTTCCTC | ||
| Motif Score | 3.28175 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000619546.4; ENST00000616317.5; ENST00000614428.4; ENST00000612895.4 | ||
| External Link | RMBase: m6A_site_359491 | ||
| mod ID: M6ASITE032017 | Click to Show/Hide the Full List | ||
| mod site | chr17:37086099-37086100:- | [9] | |
| Sequence | ACATGACCTTAAGATTATTTACAACTCAGTCATGGTGCTGC | ||
| Motif Score | 2.07285119 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000616317.5; ENST00000612895.4; ENST00000619546.4; ENST00000614428.4 | ||
| External Link | RMBase: m6A_site_359492 | ||
| mod ID: M6ASITE032018 | Click to Show/Hide the Full List | ||
| mod site | chr17:37086130-37086131:- | [9] | |
| Sequence | ATGAGGGAGGCTTCCAGGAAACACAGAATCCACATGACCTT | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000619546.4; ENST00000616317.5; ENST00000612895.4; ENST00000614428.4 | ||
| External Link | RMBase: m6A_site_359493 | ||
| mod ID: M6ASITE032019 | Click to Show/Hide the Full List | ||
| mod site | chr17:37086181-37086182:- | [9] | |
| Sequence | AAAGGACCCTGGTGACAGCTACACCCTCAGCACCAGGAGCT | ||
| Motif Score | 2.078666667 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000612895.4; ENST00000616317.5; ENST00000614428.4; ENST00000619546.4 | ||
| External Link | RMBase: m6A_site_359494 | ||
| mod ID: M6ASITE032020 | Click to Show/Hide the Full List | ||
| mod site | chr17:37086302-37086303:- | [9] | |
| Sequence | GAGAGGTTAATGGTGCCTCCACACTCACTCTTCCCTAGTTC | ||
| Motif Score | 2.053113095 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000616317.5; ENST00000614428.4; ENST00000612895.4; ENST00000619546.4 | ||
| External Link | RMBase: m6A_site_359495 | ||
| mod ID: M6ASITE032021 | Click to Show/Hide the Full List | ||
| mod site | chr17:37086609-37086610:- | [9] | |
| Sequence | CTTGGGTTCAGTCTCTGAGGACAAAGGGAAAGGTAGTTGTT | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000614428.4; ENST00000616317.5; ENST00000619546.4; ENST00000612895.4 | ||
| External Link | RMBase: m6A_site_359496 | ||
| mod ID: M6ASITE032022 | Click to Show/Hide the Full List | ||
| mod site | chr17:37086631-37086632:- | [10] | |
| Sequence | TCTTGATTTAGGGGACCTCAACCTTGGGTTCAGTCTCTGAG | ||
| Motif Score | 2.153267857 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000612895.4; ENST00000619546.4; ENST00000616317.5; ENST00000614428.4 | ||
| External Link | RMBase: m6A_site_359497 | ||
| mod ID: M6ASITE032023 | Click to Show/Hide the Full List | ||
| mod site | chr17:37086727-37086728:- | [9] | |
| Sequence | ATTAGAGCATAAAATCTACTACAAGCTCCATAGGAACTCAA | ||
| Motif Score | 2.078666667 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000616317.5; ENST00000612895.4; ENST00000614428.4; ENST00000619546.4 | ||
| External Link | RMBase: m6A_site_359498 | ||
| mod ID: M6ASITE032024 | Click to Show/Hide the Full List | ||
| mod site | chr17:37086730-37086731:- | [10] | |
| Sequence | AAAATTAGAGCATAAAATCTACTACAAGCTCCATAGGAACT | ||
| Motif Score | 2.500660714 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000614428.4; ENST00000612895.4; ENST00000619546.4; ENST00000616317.5 | ||
| External Link | RMBase: m6A_site_359499 | ||
| mod ID: M6ASITE032025 | Click to Show/Hide the Full List | ||
| mod site | chr17:37086774-37086775:- | [9] | |
| Sequence | GAATGATTCCCAGTTTAACCACACTACGGTACCTTTTATGA | ||
| Motif Score | 2.053113095 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000612895.4; ENST00000614428.4; ENST00000619546.4; ENST00000616317.5 | ||
| External Link | RMBase: m6A_site_359500 | ||
| mod ID: M6ASITE032026 | Click to Show/Hide the Full List | ||
| mod site | chr17:37087056-37087057:- | [12] | |
| Sequence | TCCTCCACTCCCTGCACAGGACTGAGAAGGCAATGAAAGGT | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | A549 | ||
| Seq Type List | m6A-CLIP/IP | ||
| Transcript ID List | ENST00000616317.5; ENST00000612895.4; ENST00000614482.4; ENST00000619546.4; ENST00000617649.4; ENST00000614428.4; ENST00000618053.1 | ||
| External Link | RMBase: m6A_site_359501 | ||
| mod ID: M6ASITE032027 | Click to Show/Hide the Full List | ||
| mod site | chr17:37087118-37087119:- | [9] | |
| Sequence | CCCCCTCCAGCCCACCAGTCACACCTACCCCATTCAGTATT | ||
| Motif Score | 2.047297619 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000617649.4; ENST00000618053.1; ENST00000614428.4; ENST00000614482.4; ENST00000616317.5; ENST00000619546.4; ENST00000612895.4 | ||
| External Link | RMBase: m6A_site_359502 | ||
| mod ID: M6ASITE032028 | Click to Show/Hide the Full List | ||
| mod site | chr17:37087145-37087146:- | [13] | |
| Sequence | ACATCCTGGGATGTAAGATCACAGAATCCCCCTCCAGCCCA | ||
| Motif Score | 2.047297619 | ||
| Cell/Tissue List | HEK293 | ||
| Seq Type List | m6A-REF-seq | ||
| Transcript ID List | ENST00000614482.4; ENST00000614428.4; ENST00000618053.1; ENST00000617649.4; ENST00000616317.5; ENST00000619546.4; ENST00000612895.4 | ||
| External Link | RMBase: m6A_site_359503 | ||
| mod ID: M6ASITE032029 | Click to Show/Hide the Full List | ||
| mod site | chr17:37087165-37087166:- | [9] | |
| Sequence | CTCGTTTCAGGTTATGCATGACATCCTGGGATGTAAGATCA | ||
| Motif Score | 2.859755952 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000618053.1; ENST00000617649.4; ENST00000614482.4; ENST00000619546.4; ENST00000612895.4; ENST00000616317.5; ENST00000614428.4 | ||
| External Link | RMBase: m6A_site_359504 | ||
| mod ID: M6ASITE032030 | Click to Show/Hide the Full List | ||
| mod site | chr17:37087255-37087256:- | [9] | |
| Sequence | AAGGGCTAGAGCTGCCTTTTACAACTGTAACCACTGTAATG | ||
| Motif Score | 2.07285119 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000619546.4; ENST00000612895.4; ENST00000614482.4; ENST00000617649.4; ENST00000618053.1; ENST00000613776.1; ENST00000616317.5; ENST00000614428.4 | ||
| External Link | RMBase: m6A_site_359505 | ||
| mod ID: M6ASITE032031 | Click to Show/Hide the Full List | ||
| mod site | chr17:37087338-37087339:- | [9] | |
| Sequence | AAGTCATACGGATCCTCTCCACAATGGATTCCCCTTCCACG | ||
| Motif Score | 2.053113095 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000619546.4; ENST00000612895.4; ENST00000617649.4; ENST00000618053.1; ENST00000613776.1; ENST00000614482.4; ENST00000616317.5; ENST00000614428.4 | ||
| External Link | RMBase: m6A_site_359506 | ||
| mod ID: M6ASITE032032 | Click to Show/Hide the Full List | ||
| mod site | chr17:37087382-37087383:- | [9] | |
| Sequence | CATCATCCATATGACGCAGCACATATCACCCACTCAGCGAG | ||
| Motif Score | 2.830589286 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000614482.4; ENST00000612895.4; ENST00000613776.1; ENST00000616317.5; ENST00000619546.4; ENST00000618053.1; ENST00000614428.4; ENST00000617649.4 | ||
| External Link | RMBase: m6A_site_359507 | ||
| mod ID: M6ASITE032033 | Click to Show/Hide the Full List | ||
| mod site | chr17:37088982-37088983:- | [9] | |
| Sequence | TCACTCGGTAATAGAGGAAAACATCAAATGCATCAGCAGAG | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000616317.5; ENST00000619546.4; ENST00000617649.4; ENST00000613776.1; ENST00000618053.1; ENST00000614428.4; ENST00000614482.4; ENST00000612895.4 | ||
| External Link | RMBase: m6A_site_359508 | ||
| mod ID: M6ASITE032034 | Click to Show/Hide the Full List | ||
| mod site | chr17:37097077-37097078:- | [9] | |
| Sequence | CCTGGTCAAGAAGAAAATCCACAATGCCAACCCTGAGCTGA | ||
| Motif Score | 2.053113095 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000612895.4; ENST00000614428.4; ENST00000616317.5; ENST00000614482.4; ENST00000619546.4; ENST00000613776.1; ENST00000617649.4 | ||
| External Link | RMBase: m6A_site_359509 | ||
| mod ID: M6ASITE032035 | Click to Show/Hide the Full List | ||
| mod site | chr17:37097147-37097148:- | [9] | |
| Sequence | AGGATATCCTGGATTGGAAAACATCCCGTACCTTCTTCTAC | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000616317.5; ENST00000614482.4; ENST00000613776.1; ENST00000614428.4; ENST00000617649.4; ENST00000619546.4; ENST00000612895.4 | ||
| External Link | RMBase: m6A_site_359510 | ||
| mod ID: M6ASITE032036 | Click to Show/Hide the Full List | ||
| mod site | chr17:37097866-37097867:- | [9] | |
| Sequence | GCAGTTTGCTGACTTGCACGACACACCAGGCCGGATGCAGG | ||
| Motif Score | 2.865571429 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000613776.1; ENST00000612895.4; ENST00000616317.5; ENST00000617649.4; ENST00000619546.4; ENST00000614428.4; ENST00000614482.4 | ||
| External Link | RMBase: m6A_site_359511 | ||
| mod ID: M6ASITE032037 | Click to Show/Hide the Full List | ||
| mod site | chr17:37097941-37097942:- | [9] | |
| Sequence | TGAGCGGAAGGAGTTGGAGAACAAGTTGAAGGAGCGGGAGG | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000617649.4; ENST00000614482.4; ENST00000613776.1; ENST00000616317.5; ENST00000612895.4; ENST00000619546.4; ENST00000614428.4 | ||
| External Link | RMBase: m6A_site_359512 | ||
| mod ID: M6ASITE032038 | Click to Show/Hide the Full List | ||
| mod site | chr17:37097966-37097967:- | [13] | |
| Sequence | CAGGGACCCCAGAGCTAAGCACAGCTGAGCGGAAGGAGTTG | ||
| Motif Score | 2.830589286 | ||
| Cell/Tissue List | HEK293 | ||
| Seq Type List | m6A-REF-seq | ||
| Transcript ID List | ENST00000617649.4; ENST00000614428.4; ENST00000614482.4; ENST00000616317.5; ENST00000619546.4; ENST00000613776.1; ENST00000612895.4 | ||
| External Link | RMBase: m6A_site_359513 | ||
| mod ID: M6ASITE032039 | Click to Show/Hide the Full List | ||
| mod site | chr17:37111553-37111554:- | [9] | |
| Sequence | GCGTCGGGTGGACCCAGTCTACATCCACTTGGCTGAGCGAT | ||
| Motif Score | 2.078666667 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000614482.4; ENST00000619546.4; ENST00000612895.4; ENST00000617649.4; ENST00000616317.5; ENST00000613776.1; ENST00000614428.4 | ||
| External Link | RMBase: m6A_site_359514 | ||
| mod ID: M6ASITE032040 | Click to Show/Hide the Full List | ||
| mod site | chr17:37111617-37111618:- | [13] | |
| Sequence | CTGTTCTGGAGCCAGAAGGGACAGTAGAAATCAAATTCCGC | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | brain | ||
| Seq Type List | m6A-REF-seq | ||
| Transcript ID List | ENST00000619546.4; ENST00000617649.4; ENST00000614482.4; ENST00000612895.4; ENST00000614428.4; ENST00000616317.5; ENST00000613776.1 | ||
| External Link | RMBase: m6A_site_359515 | ||
| mod ID: M6ASITE032041 | Click to Show/Hide the Full List | ||
| mod site | chr17:37113117-37113118:- | [9] | |
| Sequence | CTCCTCCATCAACCCCCGGCACATGGAGATGTATGCTGACC | ||
| Motif Score | 2.830589286 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000614428.4; ENST00000613776.1; ENST00000617649.4; ENST00000612895.4; ENST00000619546.4; ENST00000616317.5; ENST00000614482.4 | ||
| External Link | RMBase: m6A_site_359516 | ||
| mod ID: M6ASITE032042 | Click to Show/Hide the Full List | ||
| mod site | chr17:37113186-37113187:- | [9] | |
| Sequence | CTGCCAGCCTGTGCTGGTTTACATTCCTCCCCAGGCTGAGC | ||
| Motif Score | 2.07285119 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000616317.5; ENST00000612895.4; ENST00000614482.4; ENST00000614428.4; ENST00000619546.4; ENST00000617649.4; ENST00000613776.1 | ||
| External Link | RMBase: m6A_site_359517 | ||
| mod ID: M6ASITE032043 | Click to Show/Hide the Full List | ||
| mod site | chr17:37113231-37113232:- | [9] | |
| Sequence | AGTGCTGAAGTTTGGTGCTTACATTGTGGATGGCTTGAGGG | ||
| Motif Score | 2.07285119 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000614428.4; ENST00000616317.5; ENST00000613776.1; ENST00000614482.4; ENST00000619546.4; ENST00000612895.4; ENST00000617649.4 | ||
| External Link | RMBase: m6A_site_359518 | ||
| mod ID: M6ASITE032044 | Click to Show/Hide the Full List | ||
| mod site | chr17:37129415-37129416:- | [9] | |
| Sequence | GAATCATCGAGTTTGTTCCCACAAAGACCCCATACGATCCT | ||
| Motif Score | 2.053113095 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000617649.4; ENST00000619546.4; ENST00000612895.4; ENST00000616317.5; ENST00000614428.4; ENST00000614482.4 | ||
| External Link | RMBase: m6A_site_359519 | ||
| mod ID: M6ASITE032045 | Click to Show/Hide the Full List | ||
| mod site | chr17:37130084-37130085:- | [9] | |
| Sequence | TGTCCTGCACTGGCTGTCTTACATGCCCAAGGTGAGCCCTT | ||
| Motif Score | 2.07285119 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000619546.4; ENST00000617649.4; ENST00000614482.4; ENST00000614428.4; ENST00000616317.5; ENST00000612895.4 | ||
| External Link | RMBase: m6A_site_359520 | ||
| mod ID: M6ASITE032046 | Click to Show/Hide the Full List | ||
| mod site | chr17:37130156-37130157:- | [9] | |
| Sequence | GGGCATCCAGATTATGCACAACAATGGGGTGACCCACTGCA | ||
| Motif Score | 2.173910714 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000612895.4; ENST00000614482.4; ENST00000616317.5; ENST00000619546.4; ENST00000614428.4; ENST00000617649.4 | ||
| External Link | RMBase: m6A_site_359521 | ||
| mod ID: M6ASITE032047 | Click to Show/Hide the Full List | ||
| mod site | chr17:37143616-37143617:- | [10] | |
| Sequence | AACCCCCCTGTATACAACTCACTCCTTTTAAAGAAAAAAAT | ||
| Motif Score | 2.469291667 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000612895.4; ENST00000615653.1; ENST00000616317.5; ENST00000617649.4; ENST00000619546.4; ENST00000614428.4; ENST00000614482.4 | ||
| External Link | RMBase: m6A_site_359522 | ||
| mod ID: M6ASITE032048 | Click to Show/Hide the Full List | ||
| mod site | chr17:37143620-37143621:- | [10] | |
| Sequence | ATTTAACCCCCCTGTATACAACTCACTCCTTTTAAAGAAAA | ||
| Motif Score | 2.595904762 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000612895.4; ENST00000616317.5; ENST00000615653.1; ENST00000617649.4; ENST00000614482.4; ENST00000614428.4; ENST00000619546.4 | ||
| External Link | RMBase: m6A_site_359523 | ||
| mod ID: M6ASITE032049 | Click to Show/Hide the Full List | ||
| mod site | chr17:37143623-37143624:- | [10] | |
| Sequence | AGCATTTAACCCCCCTGTATACAACTCACTCCTTTTAAAGA | ||
| Motif Score | 2.110482143 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000614428.4; ENST00000614482.4; ENST00000612895.4; ENST00000616317.5; ENST00000617649.4; ENST00000619546.4; ENST00000615653.1 | ||
| External Link | RMBase: m6A_site_359524 | ||
| mod ID: M6ASITE032050 | Click to Show/Hide the Full List | ||
| mod site | chr17:37155708-37155709:- | [9] | |
| Sequence | CTCAACTCTGTCCATTGTGAACACGTGGAAGATGAAGGAGA | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000616317.5; ENST00000614482.4; ENST00000614428.4; ENST00000617649.4; ENST00000619546.4; ENST00000612895.4 | ||
| External Link | RMBase: m6A_site_359525 | ||
| mod ID: M6ASITE032051 | Click to Show/Hide the Full List | ||
| mod site | chr17:37161784-37161785:- | [9] | |
| Sequence | GGTAGATCCTGAGGATCCTTACAAGGTACACACTAAGAGCA | ||
| Motif Score | 2.07285119 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000614482.4; ENST00000619546.4; ENST00000618575.1; ENST00000612895.4; ENST00000614428.4; ENST00000616317.5; ENST00000617649.4 | ||
| External Link | RMBase: m6A_site_359526 | ||
| mod ID: M6ASITE032052 | Click to Show/Hide the Full List | ||
| mod site | chr17:37161965-37161966:- | [9] | |
| Sequence | TTGTTATTGGCAATGACATCACATACCGAATTGGGTCCTTT | ||
| Motif Score | 2.047297619 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000619546.4; ENST00000617649.4; ENST00000614428.4; ENST00000618575.1; ENST00000612895.4; ENST00000616317.5 | ||
| External Link | RMBase: m6A_site_359527 | ||
| mod ID: M6ASITE032053 | Click to Show/Hide the Full List | ||
| mod site | chr17:37161970-37161971:- | [9] | |
| Sequence | TATCATTGTTATTGGCAATGACATCACATACCGAATTGGGT | ||
| Motif Score | 2.859755952 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000614428.4; ENST00000618575.1; ENST00000612895.4; ENST00000617649.4; ENST00000616317.5; ENST00000619546.4 | ||
| External Link | RMBase: m6A_site_359528 | ||
| mod ID: M6ASITE032054 | Click to Show/Hide the Full List | ||
| mod site | chr17:37179338-37179339:- | [9] | |
| Sequence | ATCTCCCCCTCTGCCTTCTGACATGCTGACTTACACTGAAC | ||
| Motif Score | 2.859755952 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000618575.1; ENST00000616317.5; ENST00000619546.4; ENST00000612895.4; ENST00000617649.4; ENST00000614428.4 | ||
| External Link | RMBase: m6A_site_359529 | ||
| mod ID: M6ASITE032055 | Click to Show/Hide the Full List | ||
| mod site | chr17:37181231-37181232:- | [9] | |
| Sequence | ACAATCCTTAGGGACAACATACATATATGATATCCCAGAGA | ||
| Motif Score | 2.110482143 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000617649.4; ENST00000619546.4; ENST00000614428.4; ENST00000616317.5; ENST00000618575.1; ENST00000612895.4 | ||
| External Link | RMBase: m6A_site_359530 | ||
| mod ID: M6ASITE032056 | Click to Show/Hide the Full List | ||
| mod site | chr17:37188344-37188345:- | [9] | |
| Sequence | TTCCCATCCGCCTCTTCCTGACAAACGAGTCTGGCTATTAC | ||
| Motif Score | 2.859755952 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000617649.4; ENST00000619245.1; ENST00000619546.4; ENST00000614428.4; ENST00000612895.4; ENST00000616317.5 | ||
| External Link | RMBase: m6A_site_359531 | ||
| mod ID: M6ASITE032057 | Click to Show/Hide the Full List | ||
| mod site | chr17:37188394-37188395:- | [9] | |
| Sequence | CCAGGCAGAACTGAAAATCAACATTCGCCTGACGCCAACTG | ||
| Motif Score | 2.173910714 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000616317.5; ENST00000614428.4; ENST00000612895.4; ENST00000619245.1; ENST00000617649.4; ENST00000619546.4 | ||
| External Link | RMBase: m6A_site_359532 | ||
| mod ID: M6ASITE032058 | Click to Show/Hide the Full List | ||
| mod site | chr17:37191165-37191166:- | [9] | |
| Sequence | TGTCCGCACTGACTGTAACCACATCTTCCTCAACTTTGTGC | ||
| Motif Score | 2.053113095 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000619245.1; ENST00000617649.4; ENST00000612895.4; ENST00000619546.4; ENST00000616317.5; ENST00000614428.4 | ||
| External Link | RMBase: m6A_site_359533 | ||
| mod ID: M6ASITE032059 | Click to Show/Hide the Full List | ||
| mod site | chr17:37191195-37191196:- | [9] | |
| Sequence | TGAGTTGGAAGTTGCTTTTAACAATACAAATGTCCGCACTG | ||
| Motif Score | 2.168095238 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000619245.1; ENST00000617649.4; ENST00000619546.4; ENST00000614428.4; ENST00000616317.5; ENST00000612895.4 | ||
| External Link | RMBase: m6A_site_359534 | ||
| mod ID: M6ASITE032060 | Click to Show/Hide the Full List | ||
| mod site | chr17:37192198-37192199:- | [13] | |
| Sequence | TGCCATTCCATGTGCTAATCACAAGATGCACCTGTATCTCG | ||
| Motif Score | 2.047297619 | ||
| Cell/Tissue List | brain; hESC-HEK293T | ||
| Seq Type List | m6A-REF-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000619546.4; ENST00000619245.1; ENST00000614428.4; ENST00000617649.4; ENST00000616317.5; ENST00000612895.4 | ||
| External Link | RMBase: m6A_site_359535 | ||
| mod ID: M6ASITE032061 | Click to Show/Hide the Full List | ||
| mod site | chr17:37192243-37192244:- | [11] | |
| Sequence | GGCTTTCCAGTTAGAGCTGAACCGGATGAGAAATTTTGACC | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000617649.4; ENST00000619245.1; ENST00000614428.4; ENST00000619546.4; ENST00000616317.5; ENST00000612895.4 | ||
| External Link | RMBase: m6A_site_359536 | ||
| mod ID: M6ASITE032062 | Click to Show/Hide the Full List | ||
| mod site | chr17:37193393-37193394:- | [9] | |
| Sequence | GAGAATTCCCTAAATTTTTTACATTCCGAGCAAGGGATAAG | ||
| Motif Score | 2.07285119 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000614428.4; ENST00000616317.5; ENST00000612895.4; ENST00000617649.4; ENST00000619245.1; ENST00000619546.4 | ||
| External Link | RMBase: m6A_site_359537 | ||
| mod ID: M6ASITE032063 | Click to Show/Hide the Full List | ||
| mod site | chr17:37200432-37200433:- | [9] | |
| Sequence | CGCCTTACTTTCCTGGTTGCACAAAAGGTACTGACATGTGA | ||
| Motif Score | 2.830589286 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000612895.4; ENST00000617649.4; ENST00000614450.4; ENST00000619546.4; ENST00000614428.4; ENST00000616317.5 | ||
| External Link | RMBase: m6A_site_359538 | ||
| mod ID: M6ASITE032064 | Click to Show/Hide the Full List | ||
| mod site | chr17:37205849-37205850:- | [9] | |
| Sequence | TCCCAGGGATGAACCAATTCACATTCTCAATGTGGCTATCA | ||
| Motif Score | 2.047297619 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000612895.4; ENST00000617649.4; ENST00000614450.4; ENST00000614428.4; ENST00000616317.5 | ||
| External Link | RMBase: m6A_site_359539 | ||
| mod ID: M6ASITE032065 | Click to Show/Hide the Full List | ||
| mod site | chr17:37206807-37206808:- | [9] | |
| Sequence | CACATTCCCTGAGGCAGGTCACACGTCTCTTTATGATGAGG | ||
| Motif Score | 2.047297619 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000616317.5; ENST00000614450.4; ENST00000612895.4; ENST00000614428.4; ENST00000617649.4 | ||
| External Link | RMBase: m6A_site_359540 | ||
| mod ID: M6ASITE032066 | Click to Show/Hide the Full List | ||
| mod site | chr17:37206826-37206827:- | [13] | |
| Sequence | ACTCCCCACCCCAGAGTCCCACATTCCCTGAGGCAGGTCAC | ||
| Motif Score | 2.053113095 | ||
| Cell/Tissue List | brain; hESC-HEK293T | ||
| Seq Type List | m6A-REF-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000614428.4; ENST00000617649.4; ENST00000612895.4; ENST00000614450.4; ENST00000616317.5 | ||
| External Link | RMBase: m6A_site_359541 | ||
| mod ID: M6ASITE032067 | Click to Show/Hide the Full List | ||
| mod site | chr17:37207728-37207729:- | [9] | |
| Sequence | TGTCAGCGATGTACTGTTGGACAACTCATTCACTCCACCTT | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000614450.4; ENST00000612895.4; ENST00000616317.5; ENST00000617649.4; ENST00000614428.4 | ||
| External Link | RMBase: m6A_site_359542 | ||
| mod ID: M6ASITE032068 | Click to Show/Hide the Full List | ||
| mod site | chr17:37223582-37223583:- | [9] | |
| Sequence | AGAAACTCATCCTATCAGAAACATCTATTTTTGATGTCCTA | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000616317.5; ENST00000614428.4; ENST00000612895.4; ENST00000617649.4; ENST00000613146.4; ENST00000612120.4 | ||
| External Link | RMBase: m6A_site_359543 | ||
| mod ID: M6ASITE032069 | Click to Show/Hide the Full List | ||
| mod site | chr17:37225018-37225019:- | [9] | |
| Sequence | TCAGCTATTGACATGTATGGACATCAATTTTGCATTGAGAA | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000612895.4; ENST00000612120.4; ENST00000614428.4; ENST00000617649.4; ENST00000613146.4; ENST00000616317.5 | ||
| External Link | RMBase: m6A_site_359544 | ||
| mod ID: M6ASITE032070 | Click to Show/Hide the Full List | ||
| mod site | chr17:37234985-37234986:- | [9] | |
| Sequence | CCAAGAAGAATCTTCTGGTCACAATGCTTATTGTGAGTACC | ||
| Motif Score | 2.047297619 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000617649.4; ENST00000614428.4; ENST00000613146.4; ENST00000616317.5; ENST00000612895.4 | ||
| External Link | RMBase: m6A_site_359545 | ||
| mod ID: M6ASITE032071 | Click to Show/Hide the Full List | ||
| mod site | chr17:37235029-37235030:- | [9] | |
| Sequence | CATGAACACTGTACTGAACTACATCTTCTCTCACGCTCAAG | ||
| Motif Score | 2.078666667 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000612895.4; ENST00000614428.4; ENST00000616317.5; ENST00000613146.4; ENST00000617649.4 | ||
| External Link | RMBase: m6A_site_359546 | ||
| mod ID: M6ASITE032072 | Click to Show/Hide the Full List | ||
| mod site | chr17:37240490-37240491:- | [11] | |
| Sequence | GGCAGTACCTGCGAGTAGAGACACAATTCCAGAATGGTAAG | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa; hESC-HEK293T | ||
| Seq Type List | m6A-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000616317.5; ENST00000614428.4; ENST00000612895.4; ENST00000617649.4; ENST00000613146.4 | ||
| External Link | RMBase: m6A_site_359547 | ||
| mod ID: M6ASITE032073 | Click to Show/Hide the Full List | ||
| mod site | chr17:37240540-37240541:- | [9] | |
| Sequence | CCGAAGTGGCATCCGAGGCCACATGAAGGCTGTGGTGATGG | ||
| Motif Score | 2.053113095 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000613146.4; ENST00000614428.4; ENST00000612895.4; ENST00000617649.4; ENST00000616317.5 | ||
| External Link | RMBase: m6A_site_359548 | ||
| mod ID: M6ASITE032074 | Click to Show/Hide the Full List | ||
| mod site | chr17:37242022-37242023:- | [9] | |
| Sequence | TCCTAGATAGCCATGCAGCTACATTGAACCGGAAATCTGAA | ||
| Motif Score | 2.078666667 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000616317.5; ENST00000617649.4; ENST00000613146.4; ENST00000614428.4; ENST00000612895.4 | ||
| External Link | RMBase: m6A_site_359549 | ||
| mod ID: M6ASITE032075 | Click to Show/Hide the Full List | ||
| mod site | chr17:37242045-37242046:- | [9] | |
| Sequence | TTTTATACCTTAGATTGCAAACATCCTAGATAGCCATGCAG | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000614428.4; ENST00000616317.5; ENST00000612895.4; ENST00000617649.4; ENST00000613146.4 | ||
| External Link | RMBase: m6A_site_359550 | ||
| mod ID: M6ASITE032076 | Click to Show/Hide the Full List | ||
| mod site | chr17:37243402-37243403:- | [9] | |
| Sequence | CTCAGTATGCTAGCAACATCACATCAGTCCTCTGTCAGTTT | ||
| Motif Score | 2.047297619 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000617649.4; ENST00000614428.4; ENST00000616317.5; ENST00000613146.4; ENST00000612895.4 | ||
| External Link | RMBase: m6A_site_359551 | ||
| mod ID: M6ASITE032077 | Click to Show/Hide the Full List | ||
| mod site | chr17:37245101-37245102:- | [9] | |
| Sequence | ACTAGCCAAAATGCAACTGGACAACCCCAGCAAGGTTCAGC | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000616317.5; ENST00000613146.4; ENST00000614428.4; ENST00000612895.4; ENST00000617649.4 | ||
| External Link | RMBase: m6A_site_359552 | ||
| mod ID: M6ASITE032078 | Click to Show/Hide the Full List | ||
| mod site | chr17:37246880-37246881:- | [9] | |
| Sequence | TGCTGGGAAGTTAATCCAGTACATTGTAGAAGATGGAGGTC | ||
| Motif Score | 2.856142857 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000613146.4; ENST00000616317.5; ENST00000612895.4; ENST00000617649.4; ENST00000614428.4 | ||
| External Link | RMBase: m6A_site_359553 | ||
| mod ID: M6ASITE032079 | Click to Show/Hide the Full List | ||
| mod site | chr17:37246965-37246966:- | [9] | |
| Sequence | TGTCTCCTAGATATCGCATCACAATTGGCAATAAAACCTGT | ||
| Motif Score | 2.047297619 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000612895.4; ENST00000613146.4; ENST00000616317.5; ENST00000614428.4; ENST00000617649.4 | ||
| External Link | RMBase: m6A_site_359554 | ||
| mod ID: M6ASITE032080 | Click to Show/Hide the Full List | ||
| mod site | chr17:37248090-37248091:- | [9] | |
| Sequence | TCATGTGTAGAAGTAGATGTACATCGGCTGAGTGACGGTGG | ||
| Motif Score | 2.856142857 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000616317.5; ENST00000612926.1; ENST00000614428.4; ENST00000617649.4; ENST00000612895.4; ENST00000613146.4 | ||
| External Link | RMBase: m6A_site_359555 | ||
| mod ID: M6ASITE032081 | Click to Show/Hide the Full List | ||
| mod site | chr17:37248598-37248599:- | [10] | |
| Sequence | TATGAGGGAGTCAAGTATGTACTTAAGGTATGCAGAACCCC | ||
| Motif Score | 3.278136905 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000612895.4; ENST00000616317.5; ENST00000612926.1; ENST00000617649.4; ENST00000613146.4; ENST00000614428.4 | ||
| External Link | RMBase: m6A_site_359556 | ||
| mod ID: M6ASITE032082 | Click to Show/Hide the Full List | ||
| mod site | chr17:37248651-37248652:- | [9] | |
| Sequence | GTCAAGTCCTTCCTGCTCATACACTTCTGAATACAGTAGAT | ||
| Motif Score | 2.110482143 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000612895.4; ENST00000617649.4; ENST00000613146.4; ENST00000612926.1; ENST00000614428.4; ENST00000616317.5 | ||
| External Link | RMBase: m6A_site_359557 | ||
| mod ID: M6ASITE032083 | Click to Show/Hide the Full List | ||
| mod site | chr17:37252094-37252095:- | [9] | |
| Sequence | CTTTCAGGCTGAGCGACCTGACACCATGTTGGGGGTTGTGT | ||
| Motif Score | 2.859755952 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000613146.4; ENST00000612895.4; ENST00000614428.4; ENST00000617649.4; ENST00000616317.5 | ||
| External Link | RMBase: m6A_site_359558 | ||
| mod ID: M6ASITE032084 | Click to Show/Hide the Full List | ||
| mod site | chr17:37252888-37252889:- | [13] | |
| Sequence | AGACTGATAGCAGAAAAAGTACAGGTGTGTATTTCTCCACA | ||
| Motif Score | 2.856142857 | ||
| Cell/Tissue List | brain | ||
| Seq Type List | m6A-REF-seq | ||
| Transcript ID List | ENST00000613146.4; ENST00000612895.4; ENST00000617649.4; ENST00000616317.5; ENST00000614428.4 | ||
| External Link | RMBase: m6A_site_359559 | ||
| mod ID: M6ASITE032085 | Click to Show/Hide the Full List | ||
| mod site | chr17:37253033-37253034:- | [9] | |
| Sequence | TGGCTTCTGTTCTCCCAGAAACATGGTGGTGGCTTTGAAGG | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000617649.4; ENST00000613146.4; ENST00000616317.5; ENST00000614428.4; ENST00000612895.4 | ||
| External Link | RMBase: m6A_site_359560 | ||
| mod ID: M6ASITE032086 | Click to Show/Hide the Full List | ||
| mod site | chr17:37258274-37258275:- | [9] | |
| Sequence | ATTGATTTTGAAGATTCTGCACACGTTCCTTGTCCAAGGGG | ||
| Motif Score | 2.830589286 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000616317.5; ENST00000612895.4; ENST00000613146.4; ENST00000617649.4; ENST00000614428.4 | ||
| External Link | RMBase: m6A_site_359561 | ||
| mod ID: M6ASITE032087 | Click to Show/Hide the Full List | ||
| mod site | chr17:37259484-37259485:- | [10] | |
| Sequence | TGGGTTATGTGAGTGCTGGGACTGTGGAATACCTGTACAGC | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000616317.5; ENST00000617649.4; ENST00000613146.4; ENST00000612895.4; ENST00000614428.4 | ||
| External Link | RMBase: m6A_site_359562 | ||
| mod ID: M6ASITE032088 | Click to Show/Hide the Full List | ||
| mod site | chr17:37263696-37263697:- | [9] | |
| Sequence | GCTACTCCAGCAGTATTTGAACACATGGAACAGGTACATAT | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000613146.4; ENST00000616317.5; ENST00000612895.4; ENST00000614428.4; ENST00000617649.4 | ||
| External Link | RMBase: m6A_site_359563 | ||
| mod ID: M6ASITE032089 | Click to Show/Hide the Full List | ||
| mod site | chr17:37263762-37263763:- | [9] | |
| Sequence | TTTGGTCGTGATTGCTCTGTACAACGCAGGCATCAGAAGAT | ||
| Motif Score | 2.856142857 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000612895.4; ENST00000614428.4; ENST00000613146.4; ENST00000616317.5; ENST00000617649.4 | ||
| External Link | RMBase: m6A_site_359564 | ||
| mod ID: M6ASITE032090 | Click to Show/Hide the Full List | ||
| mod site | chr17:37263843-37263844:- | [9] | |
| Sequence | TTTGTGATGAGACTAGCCAAACAATCTCGTCATCTGGAGGT | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000614428.4; ENST00000613146.4; ENST00000617649.4; ENST00000616317.5; ENST00000612895.4 | ||
| External Link | RMBase: m6A_site_359565 | ||
| mod ID: M6ASITE032091 | Click to Show/Hide the Full List | ||
| mod site | chr17:37270784-37270785:- | [9] | |
| Sequence | GAAGGGAATTAGAAAAGTCAACAATGCAGATGACTTCCCTA | ||
| Motif Score | 2.173910714 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000617649.4; ENST00000612895.4; ENST00000616317.5; ENST00000613146.4; ENST00000614428.4 | ||
| External Link | RMBase: m6A_site_359566 | ||
| mod ID: M6ASITE032092 | Click to Show/Hide the Full List | ||
| mod site | chr17:37277956-37277957:- | [9] | |
| Sequence | GCCAGTGCCTGGAGGACCAAACAACAACAACTATGCAAATG | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000615229.4; ENST00000617649.4; ENST00000612895.4; ENST00000614428.4; ENST00000616317.5; ENST00000613146.4 | ||
| External Link | RMBase: m6A_site_359567 | ||
| mod ID: M6ASITE032093 | Click to Show/Hide the Full List | ||
| mod site | chr17:37278001-37278002:- | [9] | |
| Sequence | TTCGCTTGCATTTTCAGAATACATTAAGATGGCAGATCACT | ||
| Motif Score | 2.110482143 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000614428.4; ENST00000613146.4; ENST00000616317.5; ENST00000617649.4; ENST00000615229.4; ENST00000612895.4; ENST00000621960.1 | ||
| External Link | RMBase: m6A_site_359568 | ||
| mod ID: M6ASITE032094 | Click to Show/Hide the Full List | ||
| mod site | chr17:37283293-37283294:- | [9] | |
| Sequence | TTAGATTCGTTGTCATGGTCACACCTGAAGACCTTAAAGCC | ||
| Motif Score | 2.047297619 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000613146.4; ENST00000614428.4; ENST00000612895.4; ENST00000615229.4; ENST00000621960.1; ENST00000616317.5; ENST00000617649.4 | ||
| External Link | RMBase: m6A_site_359569 | ||
| mod ID: M6ASITE032095 | Click to Show/Hide the Full List | ||
| mod site | chr17:37283391-37283392:- | [9] | |
| Sequence | CTCCCAGGTTCTTATTGCTAACAATGGCATTGCAGCAGTGA | ||
| Motif Score | 2.168095238 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000621960.1; ENST00000614428.4; ENST00000617649.4; ENST00000613146.4; ENST00000615229.4; ENST00000616317.5; ENST00000612895.4 | ||
| External Link | RMBase: m6A_site_359570 | ||
| mod ID: M6ASITE032096 | Click to Show/Hide the Full List | ||
| mod site | chr17:37284896-37284897:- | [10] | |
| Sequence | TAGATTCTCAACGAGATTTCACTGTGGCTTCTCCAGCAGAA | ||
| Motif Score | 2.469291667 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000617548.1; ENST00000616352.4; ENST00000621960.1; ENST00000612895.4; ENST00000614789.4; ENST00000616317.5; ENST00000617649.4; ENST00000614428.4; ENST00000613146.4; ENST00000615229.4 | ||
| External Link | RMBase: m6A_site_359571 | ||
| mod ID: M6ASITE032097 | Click to Show/Hide the Full List | ||
| mod site | chr17:37330390-37330391:- | [11] | |
| Sequence | TTTGGAGGAATAATGGATGAACCATCTCCCTTGGCCCAACC | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000614789.4; ENST00000617548.1; ENST00000615229.4; ENST00000613146.4; ENST00000612007.1; ENST00000616317.5; ENST00000614438.1; ENST00000621960.1; ENST00000614428.4; ENST00000616352.4 | ||
| External Link | RMBase: m6A_site_359572 | ||
| mod ID: M6ASITE032098 | Click to Show/Hide the Full List | ||
| mod site | chr17:37339821-37339822:- | [11] | |
| Sequence | GGAAGTGGATATCTACTCAGACAGTAAGAATTATAAGAGGT | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000615229.4; ENST00000612007.1; ENST00000616352.4; ENST00000617548.1; ENST00000616317.5; ENST00000621960.1; ENST00000614789.4; ENST00000613146.4; ENST00000614428.4 | ||
| External Link | RMBase: m6A_site_359573 | ||
| mod ID: M6ASITE032099 | Click to Show/Hide the Full List | ||
| mod site | chr17:37406351-37406352:- | [11] | |
| Sequence | AATTGTCTTCTTTGGAAAGAACCATCCCCTCTTTGGGCTTC | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | HeLa; HEK293A-TOA; TREX; MSC; TIME | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000618351.1; ENST00000616317.5; ENST00000616352.4; ENST00000617548.1; ENST00000615229.4 | ||
| External Link | RMBase: m6A_site_359574 | ||
| mod ID: M6ASITE032100 | Click to Show/Hide the Full List | ||
| mod site | chr17:37406427-37406428:- | [11] | |
| Sequence | GCTGCTCGTGGATGAACCAGACTGATTTTAAGGGGTGAAGA | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa; HEK293T; H1A; H1B; HEK293A-TOA; MSC; TIME; TREX | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000617548.1; ENST00000615229.4; ENST00000616317.5; ENST00000618351.1; ENST00000616352.4 | ||
| External Link | RMBase: m6A_site_359575 | ||
| mod ID: M6ASITE032101 | Click to Show/Hide the Full List | ||
| mod site | chr17:37406432-37406433:- | [11] | |
| Sequence | GAAGGGCTGCTCGTGGATGAACCAGACTGATTTTAAGGGGT | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | HeLa; HEK293T; H1A; H1B; A549; HEK293A-TOA; MSC; TIME; TREX | ||
| Seq Type List | m6A-seq; MeRIP-seq; m6A-CLIP/IP | ||
| Transcript ID List | ENST00000617548.1; ENST00000616317.5; ENST00000616352.4; ENST00000615229.4; ENST00000618351.1 | ||
| External Link | RMBase: m6A_site_359576 | ||
| mod ID: M6ASITE032102 | Click to Show/Hide the Full List | ||
| mod site | chr17:37406500-37406501:- | [11] | |
| Sequence | GGGAAATTGAGGCTGAGGGAACTGGGCCCAGGGACGGCGAG | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa; HEK293T; H1A; H1B; HEK293A-TOA; MSC; TIME; TREX | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000616352.4; ENST00000615229.4; ENST00000617548.1; ENST00000616317.5; ENST00000618351.1 | ||
| External Link | RMBase: m6A_site_359577 | ||
| mod ID: M6ASITE032103 | Click to Show/Hide the Full List | ||
| mod site | chr17:37406570-37406571:- | [11] | |
| Sequence | GTGAGGCGGGCCGAGCCGGGACTGCCTGGGTTTGGGGATAA | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa; HEK293A-TOA | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000616317.5; ENST00000618351.1; ENST00000616352.4; ENST00000615229.4; ENST00000617548.1 | ||
| External Link | RMBase: m6A_site_359578 | ||
| mod ID: M6ASITE094655 | Click to Show/Hide the Full List | ||
| mod site | KI270857.1:1566285-1566286:- | [11] | |
| Sequence | CAGCAACCTGGTGAAGTTGGACCTACTGGAGGAGAAGGAGG | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000633325.1; ENST00000613687.4; ENST00000634140.1; ENST00000632486.1; ENST00000633050.1; ENST00000632550.1; ENST00000615918.2; ENST00000632723.1; ENST00000619487.4; ENST00000632361.1 | ||
| External Link | RMBase: m6A_site_881257 | ||
| mod ID: M6ASITE094656 | Click to Show/Hide the Full List | ||
| mod site | KI270857.1:1566363-1566364:- | [11] | |
| Sequence | GGCCCAACCTCTGGAGCTGAACCAGCACTCTCGATTCATAA | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000632486.1; ENST00000632361.1; ENST00000633325.1; ENST00000633050.1; ENST00000619487.4; ENST00000632723.1; ENST00000615918.2; ENST00000632550.1; ENST00000613687.4; ENST00000634140.1 | ||
| External Link | RMBase: m6A_site_881258 | ||
| mod ID: M6ASITE094657 | Click to Show/Hide the Full List | ||
| mod site | KI270857.1:1566395-1566396:- | [11] | |
| Sequence | TTTGGAGGAATAATGGATGAACCATCTCCCTTGGCCCAACC | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000613687.4; ENST00000632486.1; ENST00000632361.1; ENST00000632723.1; ENST00000615918.2; ENST00000632550.1; ENST00000634140.1; ENST00000633325.1; ENST00000633050.1; ENST00000619487.4 | ||
| External Link | RMBase: m6A_site_881259 | ||
| mod ID: M6ASITE094658 | Click to Show/Hide the Full List | ||
| mod site | KI270857.1:1575846-1575847:- | [11] | |
| Sequence | GGAAGTGGATATCTACTCAGACAGTAAGAATTATAAGAGGT | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000633050.1; ENST00000632486.1; ENST00000632361.1; ENST00000632550.1; ENST00000619487.4; ENST00000615918.2; ENST00000633325.1; ENST00000613687.4; ENST00000632723.1 | ||
| External Link | RMBase: m6A_site_881260 | ||
| mod ID: M6ASITE094659 | Click to Show/Hide the Full List | ||
| mod site | KI270857.1:1645506-1645507:- | [11] | |
| Sequence | AATTGTCTTCTTTGGAAAGAACCATCCCCTCTTTGGGCTTC | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | HeLa; HEK293T; A549; peripheral-blood; TIME; iSLK; TREX; MSC; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000632361.1; ENST00000615918.2; ENST00000631769.1; ENST00000619487.4; ENST00000633050.1 | ||
| External Link | RMBase: m6A_site_881261 | ||
| mod ID: M6ASITE094660 | Click to Show/Hide the Full List | ||
| mod site | KI270857.1:1645582-1645583:- | [11] | |
| Sequence | GCTGCTCGTGGATGAACCAGACTGATTTTAAGGGGTGAAGA | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; A549; H1A; H1B; MM6; peripheral-blood; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; TREX; endometrial; HEC-1-A; GSCs; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000633050.1; ENST00000615918.2; ENST00000631769.1; ENST00000619487.4; ENST00000632361.1 | ||
| External Link | RMBase: m6A_site_881262 | ||
| mod ID: M6ASITE094661 | Click to Show/Hide the Full List | ||
| mod site | KI270857.1:1645587-1645588:- | [11] | |
| Sequence | GAAGGGCTGCTCGTGGATGAACCAGACTGATTTTAAGGGGT | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; A549; H1A; H1B; MM6; peripheral-blood; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; TREX; endometrial; HEC-1-A; GSCs; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000619487.4; ENST00000632361.1; ENST00000631769.1; ENST00000615918.2; ENST00000633050.1 | ||
| External Link | RMBase: m6A_site_881263 | ||
| mod ID: M6ASITE094662 | Click to Show/Hide the Full List | ||
| mod site | KI270857.1:1645656-1645657:- | [11] | |
| Sequence | GGGAAATTGAGGCTGAGGGAACTGGGCCCAGGGACGGCGAG | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; A549; H1A; H1B; peripheral-blood; GSC-11; HEK293A-TOA; TREX; iSLK; TIME; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000615918.2; ENST00000619487.4; ENST00000632361.1; ENST00000631769.1; ENST00000633050.1 | ||
| External Link | RMBase: m6A_site_881264 | ||
| mod ID: M6ASITE094663 | Click to Show/Hide the Full List | ||
| mod site | KI270857.1:1645726-1645727:- | [11] | |
| Sequence | GTGAGGCGGGCCGAGCCGGGACTGCCTGGGTTTGGGGATAA | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa; HEK293T; HEK293A-TOA | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000619487.4; ENST00000633050.1; ENST00000631769.1; ENST00000632361.1; ENST00000615918.2 | ||
| External Link | RMBase: m6A_site_881265 | ||
References