m6A Target Gene Information
General Information of the m6A Target Gene (ID: M6ATAR00410)
Full List of m6A Methylation Regulator of This Target Gene and Corresponding Disease/Drug Response(s)
SQSTM1
can be regulated by the following regulator(s), and cause disease/drug response(s). You can browse detail information of regulator(s) or disease/drug response(s).
Browse Regulator
Browse Disease
Browse Drug
Methyltransferase-like 3 (METTL3) [WRITER]
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | HUVEC cell line | Homo sapiens |
|
Treatment: shMETTL3 HUVEC cells
Control: shScramble HUVEC cells
|
GSE157544 | |
| Regulation |
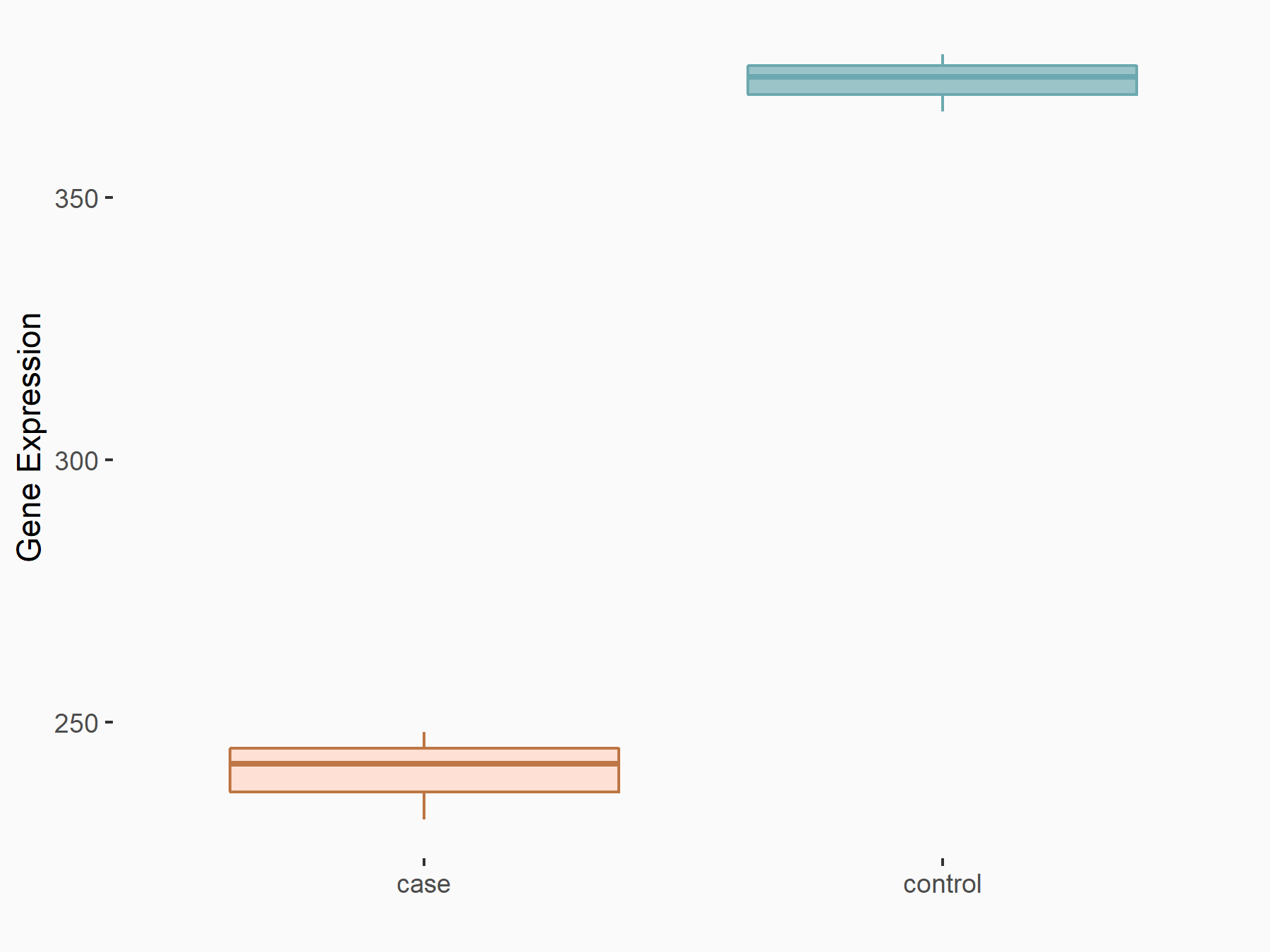  |
logFC: -6.28E-01 p-value: 1.94E-05 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between SQSTM1 and the regulator | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 2.03E+00 | GSE60213 |
| In total 3 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [1] | |||
| Response Summary | METTL3 could positively regulate the autophagy by targeting the autophagy-related genes such as ATG5, ATG7, LC3B, and Sequestosome-1 (SQSTM1). beta-elemene inhibited the autophagy flux by preventing autophagic lysosome acidification, resulting in increasing expression of SQSTM1 and LC3B-II. beta-elemene could reverse gefitinib resistance in non-small cell lung cancer cells by inhibiting cell autophagy process in a manner of chloroquine. METTL3-mediated autophagy in reversing gefitinib resistance of NSCLC cells by beta-elemene, which shed light on providing potential molecular-therapy target and clinical-treatment method in NSCLC patients with gefitinib resistance. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Non-small-cell lung carcinoma | ICD-11: 2C25.Y | ||
| Responsed Drug | Chloroquine | Approved | ||
| Pathway Response | Autophagy | hsa04140 | ||
| Cell Process | Autophagic lysosome acidification | |||
| In-vitro Model | Gefitinib-resistant cell line HCC827GR (Gefitinib-resistant HCC827 cell line) | |||
| Gefitinib-resistant cell line PC9GR (Gefitinib-resistant PC9 cell line) | ||||
| HCC827 | Lung adenocarcinoma | Homo sapiens | CVCL_2063 | |
| PC-9 | Lung adenocarcinoma | Homo sapiens | CVCL_B260 | |
| In-vivo Model | NSCLC gefitinib-resistant cells (5 × 106 cells in 100 uL PBS) were injected subcutaneously into the lateral surface of the left abdomen of 6-week-old female BALB/c nude mice (at least five mice per group to ensure accuracy). | |||
| Experiment 2 Reporting the m6A Methylation Regulator of This Target Gene | [1] | |||
| Response Summary | METTL3 could positively regulate the autophagy by targeting the autophagy-related genes such as ATG5, ATG7, LC3B, and Sequestosome-1 (SQSTM1). beta-elemene inhibited the autophagy flux by preventing autophagic lysosome acidification, resulting in increasing expression of SQSTM1 and LC3B-II. beta-elemene could reverse gefitinib resistance in non-small cell lung cancer cells by inhibiting cell autophagy process in a manner of chloroquine. METTL3-mediated autophagy in reversing gefitinib resistance of NSCLC cells by beta-elemene, which shed light on providing potential molecular-therapy target and clinical-treatment method in NSCLC patients with gefitinib resistance. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Non-small-cell lung carcinoma | ICD-11: 2C25.Y | ||
| Responsed Drug | Gefitinib | Approved | ||
| Pathway Response | Autophagy | hsa04140 | ||
| Cell Process | Autophagic lysosome acidification | |||
| In-vitro Model | Gefitinib-resistant cell line HCC827GR (Gefitinib-resistant HCC827 cell line) | |||
| Gefitinib-resistant cell line PC9GR (Gefitinib-resistant PC9 cell line) | ||||
| HCC827 | Lung adenocarcinoma | Homo sapiens | CVCL_2063 | |
| PC-9 | Lung adenocarcinoma | Homo sapiens | CVCL_B260 | |
| In-vivo Model | NSCLC gefitinib-resistant cells (5 × 106 cells in 100 uL PBS) were injected subcutaneously into the lateral surface of the left abdomen of 6-week-old female BALB/c nude mice (at least five mice per group to ensure accuracy). | |||
| Experiment 3 Reporting the m6A Methylation Regulator of This Target Gene | [1] | |||
| Response Summary | METTL3 could positively regulate the autophagy by targeting the autophagy-related genes such as ATG5, ATG7, LC3B, and Sequestosome-1 (SQSTM1). beta-elemene inhibited the autophagy flux by preventing autophagic lysosome acidification, resulting in increasing expression of SQSTM1 and LC3B-II. beta-elemene could reverse gefitinib resistance in non-small cell lung cancer cells by inhibiting cell autophagy process in a manner of chloroquine. METTL3-mediated autophagy in reversing gefitinib resistance of NSCLC cells by beta-elemene, which shed light on providing potential molecular-therapy target and clinical-treatment method in NSCLC patients with gefitinib resistance. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Non-small-cell lung carcinoma | ICD-11: 2C25.Y | ||
| Responsed Drug | Beta-Elemen | Phase 3 | ||
| Pathway Response | Autophagy | hsa04140 | ||
| Cell Process | Autophagic lysosome acidification | |||
| In-vitro Model | Gefitinib-resistant cell line HCC827GR (Gefitinib-resistant HCC827 cell line) | |||
| Gefitinib-resistant cell line PC9GR (Gefitinib-resistant PC9 cell line) | ||||
| HCC827 | Lung adenocarcinoma | Homo sapiens | CVCL_2063 | |
| PC-9 | Lung adenocarcinoma | Homo sapiens | CVCL_B260 | |
| In-vivo Model | NSCLC gefitinib-resistant cells (5 × 106 cells in 100 uL PBS) were injected subcutaneously into the lateral surface of the left abdomen of 6-week-old female BALB/c nude mice (at least five mice per group to ensure accuracy). | |||
Fat mass and obesity-associated protein (FTO) [ERASER]
| Representative RNA-seq result indicating the expression of this target gene regulated by FTO | ||
| Cell Line | 253J cell line | Homo sapiens |
|
Treatment: siFTO 253J cells
Control: 253J cells
|
GSE150239 | |
| Regulation |
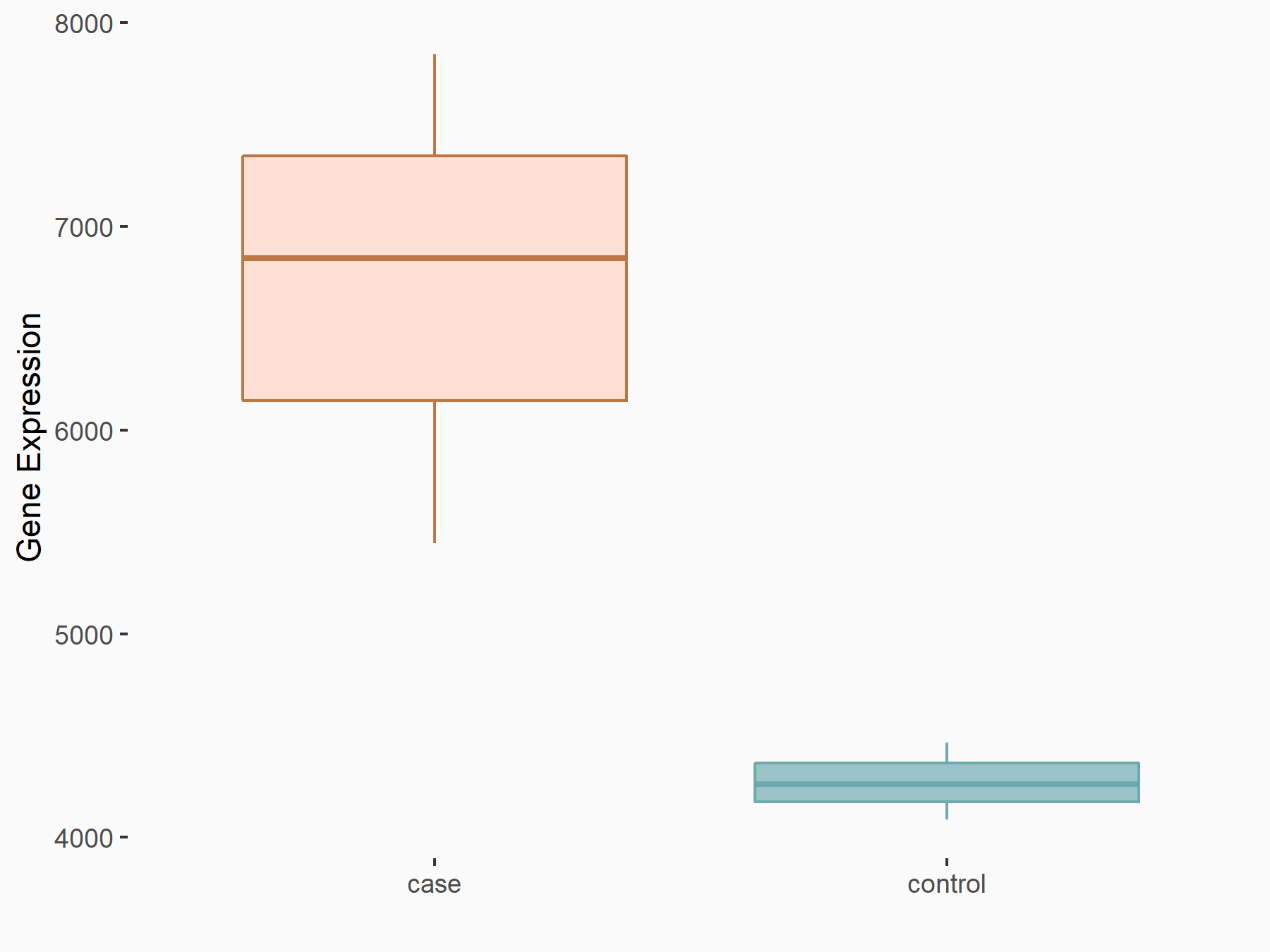  |
logFC: 6.53E-01 p-value: 1.38E-07 |
| More Results | Click to View More RNA-seq Results | |
| In total 3 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [2] | |||
| Response Summary | The m6A changes caused by FTO influence the stability of ULK1 transcripts, likely through a YTHDF2-dependent manner.Under both basal and rapamycin-induced autophagy conditions, depletion of FTO significantly reduced the formation of GFP-LC3B puncta. The level of Sequestosome-1 (SQSTM1)/SQSTM1 (an autophagy substrate) was higher in FTO-knockdown cells than that in control cells. FTO specifically upregulates the ULK1 protein abundance. ULK1 mRNA undergoes m6A modification in the 3'-UTR and the m6A-marked ULK1 transcripts can further be targeted for degradation by YTHDF2. | |||
| Target Regulation | Down regulation | |||
| Responsed Drug | Rapamycin | Approved | ||
| Pathway Response | Autophagy | hsa04140 | ||
| Cell Process | RNA stability | |||
| Cell autophagy | ||||
| In-vitro Model | HEK293T | Normal | Homo sapiens | CVCL_0063 |
| HeLa | Endocervical adenocarcinoma | Homo sapiens | CVCL_0030 | |
| Experiment 2 Reporting the m6A Methylation Regulator of This Target Gene | [3] | |||
| Response Summary | FTO deletion inhibited arsenic-induced tumorigenesis. Epidermis-specific FTO deletion prevented skin tumorigenesis induced by arsenic and UVB irradiation. NEDD4L was identified as the m6A-modified gene target of FTO. Arsenic stabilizes FTO protein through inhibiting Sequestosome-1 (SQSTM1)-mediated selective autophagy. FTO-mediated dysregulation of mRNA m6A methylation as an epitranscriptomic mechanism to promote arsenic tumorigenicity. Arsenic suppresses p62 expression by downregulating the NF-kappaB pathway to upregulate FTO. | |||
| Target Regulation | Down regulation | |||
| Responsed Disease | Solid tumour/cancer | ICD-11: 2A00-2F9Z | ||
| Pathway Response | Autophagy | hsa04140 | ||
| Cell Process | Cellular Processes | |||
| Transport and catabolism | ||||
| Cell autophagy | ||||
| In-vitro Model | HaCaT | Normal | Homo sapiens | CVCL_0038 |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| HeLa | Endocervical adenocarcinoma | Homo sapiens | CVCL_0030 | |
| MEF (Mouse embryonic fibroblasts) | ||||
| In-vivo Model | As cells (5 million) in Matrigel or As-T (1 million) cells in PBS with or without gene manipulations were injected subcutaneously into the right flanks of female mice (6-8 weeks of age). For treatment with CS1 or CS2, As-T cells (1 million) in PBS were injected subcutaneously into the right flanks of 6-week-old female nude mice. | |||
| Experiment 3 Reporting the m6A Methylation Regulator of This Target Gene | [3] | |||
| Response Summary | FTO deletion inhibited arsenic-induced tumorigenesis. Epidermis-specific FTO deletion prevented skin tumorigenesis induced by arsenic and UVB irradiation. NEDD4L was identified as the m6A-modified gene target of FTO. Arsenic stabilizes FTO protein through inhibiting Sequestosome-1 (SQSTM1)-mediated selective autophagy. FTO-mediated dysregulation of mRNA m6A methylation as an epitranscriptomic mechanism to promote arsenic tumorigenicity. Arsenic suppresses p62 expression by downregulating the NF-kappaB pathway to upregulate FTO. | |||
| Target Regulation | Down regulation | |||
| Responsed Disease | Human skin lesions | ICD-11: ME60 | ||
| Pathway Response | Autophagy | hsa04140 | ||
| Cell Process | Cellular Processes | |||
| Transport and catabolism | ||||
| Cell autophagy | ||||
| In-vitro Model | HaCaT | Normal | Homo sapiens | CVCL_0038 |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| HeLa | Endocervical adenocarcinoma | Homo sapiens | CVCL_0030 | |
| MEF (Mouse embryonic fibroblasts) | ||||
| In-vivo Model | As cells (5 million) in Matrigel or As-T (1 million) cells in PBS with or without gene manipulations were injected subcutaneously into the right flanks of female mice (6-8 weeks of age). For treatment with CS1 or CS2, As-T cells (1 million) in PBS were injected subcutaneously into the right flanks of 6-week-old female nude mice. | |||
YTH domain-containing protein 1 (YTHDC1) [READER]
| Representative RNA-seq result indicating the expression of this target gene regulated by YTHDC1 | ||
| Cell Line | MOLM-13 cell line | Homo sapiens |
|
Treatment: shYTHDC1 MOLM13 cells
Control: shControl MOLM13 cells
|
GSE168565 | |
| Regulation |
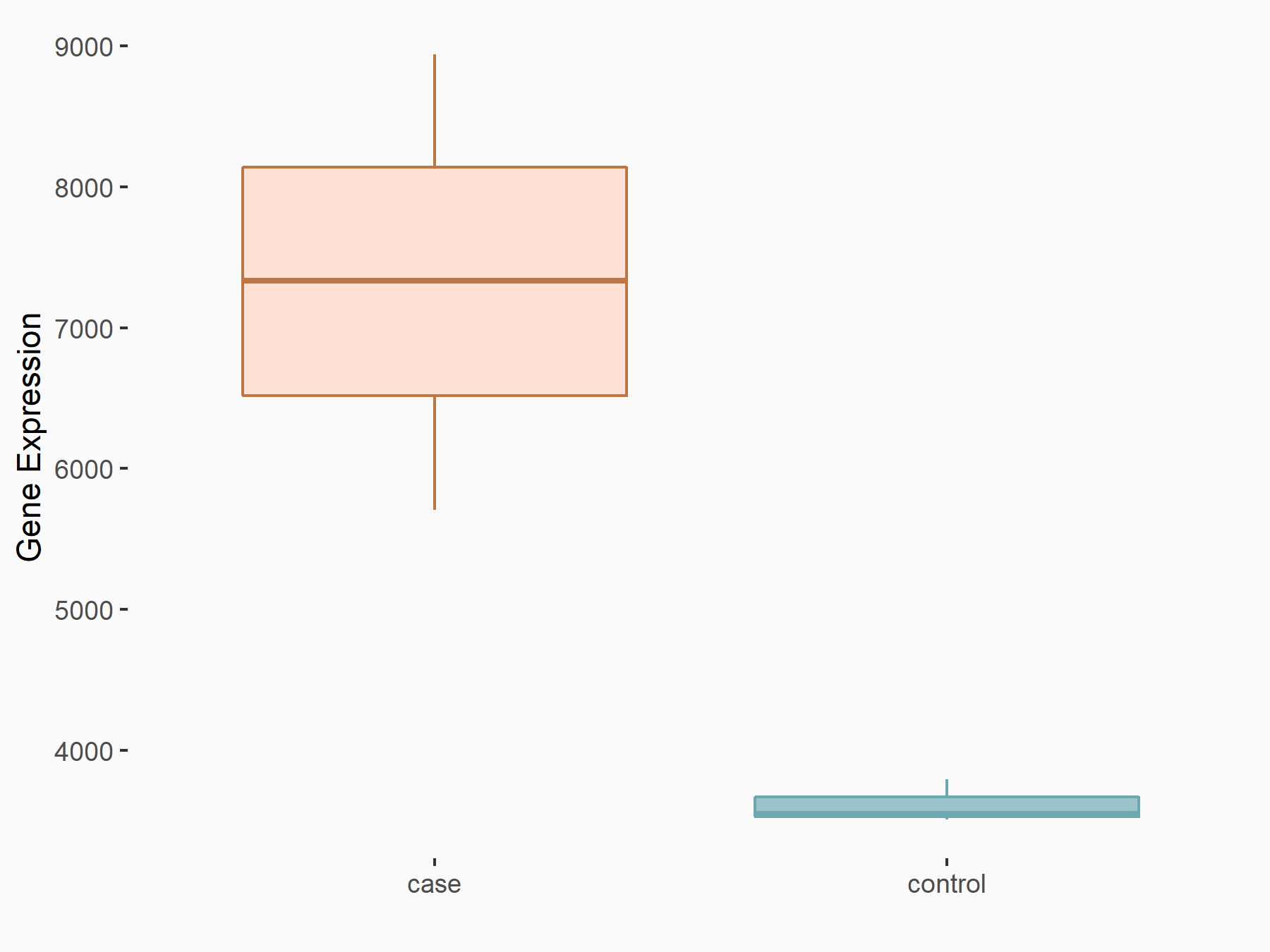  |
logFC: 1.02E+00 p-value: 3.33E-09 |
| More Results | Click to View More RNA-seq Results | |
| In total 2 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [4] | |||
| Response Summary | In diabetes/diabetic skin, YTHDC1 interacted and cooperated with ELAVL1/HuR (ELAV like RNA binding protein 1) in modulating the expression of Sequestosome-1 (SQSTM1). | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Diabetes | ICD-11: 5A10-5A14 | ||
| Pathway Response | Autophagy | hsa04140 | ||
| Cell Process | Cellular Processes | |||
| Cellular Transport | ||||
| Cellular catabolism | ||||
| Cell apoptosis | ||||
| Cell autophagy | ||||
| In-vitro Model | HaCaT | Normal | Homo sapiens | CVCL_0038 |
| NHEK (Normal human epithelial keratinocytes) | ||||
| In-vivo Model | The WT-si-NC, WT-si-Ythdc1 and WT-si-Sqstm1 groups were intracutaneously injected with corresponding siRNAs (si-NC, si-Ythdc1, or si-Sqstm1, 2.5 nmol) on the circle. | |||
| Experiment 2 Reporting the m6A Methylation Regulator of This Target Gene | [4] | |||
| Response Summary | In diabetes/diabetic skin, YTHDC1 interacted and cooperated with ELAVL1/HuR (ELAV like RNA binding protein 1) in modulating the expression of Sequestosome-1 (SQSTM1). | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Diabetic skin lesions | ICD-11: EB90.0 | ||
| Pathway Response | Autophagy | hsa04140 | ||
| Cell Process | Cellular Processes | |||
| Cellular Transport | ||||
| Cellular catabolism | ||||
| Cell apoptosis | ||||
| Cell autophagy | ||||
| In-vitro Model | HaCaT | Normal | Homo sapiens | CVCL_0038 |
| NHEK (Normal human epithelial keratinocytes) | ||||
| In-vivo Model | The WT-si-NC, WT-si-Ythdc1 and WT-si-Sqstm1 groups were intracutaneously injected with corresponding siRNAs (si-NC, si-Ythdc1, or si-Sqstm1, 2.5 nmol) on the circle. | |||
Solid tumour/cancer [ICD-11: 2A00-2F9Z]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [3] | |||
| Response Summary | FTO deletion inhibited arsenic-induced tumorigenesis. Epidermis-specific FTO deletion prevented skin tumorigenesis induced by arsenic and UVB irradiation. NEDD4L was identified as the m6A-modified gene target of FTO. Arsenic stabilizes FTO protein through inhibiting Sequestosome-1 (SQSTM1)-mediated selective autophagy. FTO-mediated dysregulation of mRNA m6A methylation as an epitranscriptomic mechanism to promote arsenic tumorigenicity. Arsenic suppresses p62 expression by downregulating the NF-kappaB pathway to upregulate FTO. | |||
| Responsed Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | |||
| Target Regulator | Fat mass and obesity-associated protein (FTO) | ERASER | ||
| Target Regulation | Down regulation | |||
| Pathway Response | Autophagy | hsa04140 | ||
| Cell Process | Cellular Processes | |||
| Transport and catabolism | ||||
| Cell autophagy | ||||
| In-vitro Model | HaCaT | Normal | Homo sapiens | CVCL_0038 |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| HeLa | Endocervical adenocarcinoma | Homo sapiens | CVCL_0030 | |
| MEF (Mouse embryonic fibroblasts) | ||||
| In-vivo Model | As cells (5 million) in Matrigel or As-T (1 million) cells in PBS with or without gene manipulations were injected subcutaneously into the right flanks of female mice (6-8 weeks of age). For treatment with CS1 or CS2, As-T cells (1 million) in PBS were injected subcutaneously into the right flanks of 6-week-old female nude mice. | |||
Lung cancer [ICD-11: 2C25]
| In total 3 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [1] | |||
| Response Summary | METTL3 could positively regulate the autophagy by targeting the autophagy-related genes such as ATG5, ATG7, LC3B, and Sequestosome-1 (SQSTM1). beta-elemene inhibited the autophagy flux by preventing autophagic lysosome acidification, resulting in increasing expression of SQSTM1 and LC3B-II. beta-elemene could reverse gefitinib resistance in non-small cell lung cancer cells by inhibiting cell autophagy process in a manner of chloroquine. METTL3-mediated autophagy in reversing gefitinib resistance of NSCLC cells by beta-elemene, which shed light on providing potential molecular-therapy target and clinical-treatment method in NSCLC patients with gefitinib resistance. | |||
| Responsed Disease | Non-small-cell lung carcinoma [ICD-11: 2C25.Y] | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Target Regulation | Up regulation | |||
| Responsed Drug | Chloroquine | Approved | ||
| Pathway Response | Autophagy | hsa04140 | ||
| Cell Process | Autophagic lysosome acidification | |||
| In-vitro Model | Gefitinib-resistant cell line HCC827GR (Gefitinib-resistant HCC827 cell line) | |||
| Gefitinib-resistant cell line PC9GR (Gefitinib-resistant PC9 cell line) | ||||
| HCC827 | Lung adenocarcinoma | Homo sapiens | CVCL_2063 | |
| PC-9 | Lung adenocarcinoma | Homo sapiens | CVCL_B260 | |
| In-vivo Model | NSCLC gefitinib-resistant cells (5 × 106 cells in 100 uL PBS) were injected subcutaneously into the lateral surface of the left abdomen of 6-week-old female BALB/c nude mice (at least five mice per group to ensure accuracy). | |||
| Experiment 2 Reporting the m6A-centered Disease Response | [1] | |||
| Response Summary | METTL3 could positively regulate the autophagy by targeting the autophagy-related genes such as ATG5, ATG7, LC3B, and Sequestosome-1 (SQSTM1). beta-elemene inhibited the autophagy flux by preventing autophagic lysosome acidification, resulting in increasing expression of SQSTM1 and LC3B-II. beta-elemene could reverse gefitinib resistance in non-small cell lung cancer cells by inhibiting cell autophagy process in a manner of chloroquine. METTL3-mediated autophagy in reversing gefitinib resistance of NSCLC cells by beta-elemene, which shed light on providing potential molecular-therapy target and clinical-treatment method in NSCLC patients with gefitinib resistance. | |||
| Responsed Disease | Non-small-cell lung carcinoma [ICD-11: 2C25.Y] | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Target Regulation | Up regulation | |||
| Responsed Drug | Gefitinib | Approved | ||
| Pathway Response | Autophagy | hsa04140 | ||
| Cell Process | Autophagic lysosome acidification | |||
| In-vitro Model | Gefitinib-resistant cell line HCC827GR (Gefitinib-resistant HCC827 cell line) | |||
| Gefitinib-resistant cell line PC9GR (Gefitinib-resistant PC9 cell line) | ||||
| HCC827 | Lung adenocarcinoma | Homo sapiens | CVCL_2063 | |
| PC-9 | Lung adenocarcinoma | Homo sapiens | CVCL_B260 | |
| In-vivo Model | NSCLC gefitinib-resistant cells (5 × 106 cells in 100 uL PBS) were injected subcutaneously into the lateral surface of the left abdomen of 6-week-old female BALB/c nude mice (at least five mice per group to ensure accuracy). | |||
| Experiment 3 Reporting the m6A-centered Disease Response | [1] | |||
| Response Summary | METTL3 could positively regulate the autophagy by targeting the autophagy-related genes such as ATG5, ATG7, LC3B, and Sequestosome-1 (SQSTM1). beta-elemene inhibited the autophagy flux by preventing autophagic lysosome acidification, resulting in increasing expression of SQSTM1 and LC3B-II. beta-elemene could reverse gefitinib resistance in non-small cell lung cancer cells by inhibiting cell autophagy process in a manner of chloroquine. METTL3-mediated autophagy in reversing gefitinib resistance of NSCLC cells by beta-elemene, which shed light on providing potential molecular-therapy target and clinical-treatment method in NSCLC patients with gefitinib resistance. | |||
| Responsed Disease | Non-small-cell lung carcinoma [ICD-11: 2C25.Y] | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Target Regulation | Up regulation | |||
| Responsed Drug | Beta-Elemen | Phase 3 | ||
| Pathway Response | Autophagy | hsa04140 | ||
| Cell Process | Autophagic lysosome acidification | |||
| In-vitro Model | Gefitinib-resistant cell line HCC827GR (Gefitinib-resistant HCC827 cell line) | |||
| Gefitinib-resistant cell line PC9GR (Gefitinib-resistant PC9 cell line) | ||||
| HCC827 | Lung adenocarcinoma | Homo sapiens | CVCL_2063 | |
| PC-9 | Lung adenocarcinoma | Homo sapiens | CVCL_B260 | |
| In-vivo Model | NSCLC gefitinib-resistant cells (5 × 106 cells in 100 uL PBS) were injected subcutaneously into the lateral surface of the left abdomen of 6-week-old female BALB/c nude mice (at least five mice per group to ensure accuracy). | |||
Diabetes [ICD-11: 5A10-5A14]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [4] | |||
| Response Summary | In diabetes/diabetic skin, YTHDC1 interacted and cooperated with ELAVL1/HuR (ELAV like RNA binding protein 1) in modulating the expression of Sequestosome-1 (SQSTM1). | |||
| Responsed Disease | Diabetes [ICD-11: 5A10-5A14] | |||
| Target Regulator | YTH domain-containing protein 1 (YTHDC1) | READER | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Autophagy | hsa04140 | ||
| Cell Process | Cellular Processes | |||
| Cellular Transport | ||||
| Cellular catabolism | ||||
| Cell apoptosis | ||||
| Cell autophagy | ||||
| In-vitro Model | HaCaT | Normal | Homo sapiens | CVCL_0038 |
| NHEK (Normal human epithelial keratinocytes) | ||||
| In-vivo Model | The WT-si-NC, WT-si-Ythdc1 and WT-si-Sqstm1 groups were intracutaneously injected with corresponding siRNAs (si-NC, si-Ythdc1, or si-Sqstm1, 2.5 nmol) on the circle. | |||
Diabetic skin lesions [ICD-11: EB90]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [4] | |||
| Response Summary | In diabetes/diabetic skin, YTHDC1 interacted and cooperated with ELAVL1/HuR (ELAV like RNA binding protein 1) in modulating the expression of Sequestosome-1 (SQSTM1). | |||
| Responsed Disease | Diabetic skin lesions [ICD-11: EB90.0] | |||
| Target Regulator | YTH domain-containing protein 1 (YTHDC1) | READER | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Autophagy | hsa04140 | ||
| Cell Process | Cellular Processes | |||
| Cellular Transport | ||||
| Cellular catabolism | ||||
| Cell apoptosis | ||||
| Cell autophagy | ||||
| In-vitro Model | HaCaT | Normal | Homo sapiens | CVCL_0038 |
| NHEK (Normal human epithelial keratinocytes) | ||||
| In-vivo Model | The WT-si-NC, WT-si-Ythdc1 and WT-si-Sqstm1 groups were intracutaneously injected with corresponding siRNAs (si-NC, si-Ythdc1, or si-Sqstm1, 2.5 nmol) on the circle. | |||
Human skin lesions [ICD-11: ME60]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [3] | |||
| Response Summary | FTO deletion inhibited arsenic-induced tumorigenesis. Epidermis-specific FTO deletion prevented skin tumorigenesis induced by arsenic and UVB irradiation. NEDD4L was identified as the m6A-modified gene target of FTO. Arsenic stabilizes FTO protein through inhibiting Sequestosome-1 (SQSTM1)-mediated selective autophagy. FTO-mediated dysregulation of mRNA m6A methylation as an epitranscriptomic mechanism to promote arsenic tumorigenicity. Arsenic suppresses p62 expression by downregulating the NF-kappaB pathway to upregulate FTO. | |||
| Responsed Disease | Human skin lesions [ICD-11: ME60] | |||
| Target Regulator | Fat mass and obesity-associated protein (FTO) | ERASER | ||
| Target Regulation | Down regulation | |||
| Pathway Response | Autophagy | hsa04140 | ||
| Cell Process | Cellular Processes | |||
| Transport and catabolism | ||||
| Cell autophagy | ||||
| In-vitro Model | HaCaT | Normal | Homo sapiens | CVCL_0038 |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| HeLa | Endocervical adenocarcinoma | Homo sapiens | CVCL_0030 | |
| MEF (Mouse embryonic fibroblasts) | ||||
| In-vivo Model | As cells (5 million) in Matrigel or As-T (1 million) cells in PBS with or without gene manipulations were injected subcutaneously into the right flanks of female mice (6-8 weeks of age). For treatment with CS1 or CS2, As-T cells (1 million) in PBS were injected subcutaneously into the right flanks of 6-week-old female nude mice. | |||
Chloroquine
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response | [1] | |||
| Response Summary | METTL3 could positively regulate the autophagy by targeting the autophagy-related genes such as ATG5, ATG7, LC3B, and Sequestosome-1 (SQSTM1). beta-elemene inhibited the autophagy flux by preventing autophagic lysosome acidification, resulting in increasing expression of SQSTM1 and LC3B-II. beta-elemene could reverse gefitinib resistance in non-small cell lung cancer cells by inhibiting cell autophagy process in a manner of chloroquine. METTL3-mediated autophagy in reversing gefitinib resistance of NSCLC cells by beta-elemene, which shed light on providing potential molecular-therapy target and clinical-treatment method in NSCLC patients with gefitinib resistance. | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Target Regulation | Up regulation | |||
| Responsed Disease | Non-small-cell lung carcinoma | ICD-11: 2C25.Y | ||
| Pathway Response | Autophagy | hsa04140 | ||
| Cell Process | Autophagic lysosome acidification | |||
| In-vitro Model | Gefitinib-resistant cell line HCC827GR (Gefitinib-resistant HCC827 cell line) | |||
| Gefitinib-resistant cell line PC9GR (Gefitinib-resistant PC9 cell line) | ||||
| HCC827 | Lung adenocarcinoma | Homo sapiens | CVCL_2063 | |
| PC-9 | Lung adenocarcinoma | Homo sapiens | CVCL_B260 | |
| In-vivo Model | NSCLC gefitinib-resistant cells (5 × 106 cells in 100 uL PBS) were injected subcutaneously into the lateral surface of the left abdomen of 6-week-old female BALB/c nude mice (at least five mice per group to ensure accuracy). | |||
Gefitinib
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response | [1] | |||
| Response Summary | METTL3 could positively regulate the autophagy by targeting the autophagy-related genes such as ATG5, ATG7, LC3B, and Sequestosome-1 (SQSTM1). beta-elemene inhibited the autophagy flux by preventing autophagic lysosome acidification, resulting in increasing expression of SQSTM1 and LC3B-II. beta-elemene could reverse gefitinib resistance in non-small cell lung cancer cells by inhibiting cell autophagy process in a manner of chloroquine. METTL3-mediated autophagy in reversing gefitinib resistance of NSCLC cells by beta-elemene, which shed light on providing potential molecular-therapy target and clinical-treatment method in NSCLC patients with gefitinib resistance. | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Target Regulation | Up regulation | |||
| Responsed Disease | Non-small-cell lung carcinoma | ICD-11: 2C25.Y | ||
| Pathway Response | Autophagy | hsa04140 | ||
| Cell Process | Autophagic lysosome acidification | |||
| In-vitro Model | Gefitinib-resistant cell line HCC827GR (Gefitinib-resistant HCC827 cell line) | |||
| Gefitinib-resistant cell line PC9GR (Gefitinib-resistant PC9 cell line) | ||||
| HCC827 | Lung adenocarcinoma | Homo sapiens | CVCL_2063 | |
| PC-9 | Lung adenocarcinoma | Homo sapiens | CVCL_B260 | |
| In-vivo Model | NSCLC gefitinib-resistant cells (5 × 106 cells in 100 uL PBS) were injected subcutaneously into the lateral surface of the left abdomen of 6-week-old female BALB/c nude mice (at least five mice per group to ensure accuracy). | |||
Rapamycin
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response | [2] | |||
| Response Summary | The m6A changes caused by FTO influence the stability of ULK1 transcripts, likely through a YTHDF2-dependent manner.Under both basal and rapamycin-induced autophagy conditions, depletion of FTO significantly reduced the formation of GFP-LC3B puncta. The level of Sequestosome-1 (SQSTM1)/SQSTM1 (an autophagy substrate) was higher in FTO-knockdown cells than that in control cells. FTO specifically upregulates the ULK1 protein abundance. ULK1 mRNA undergoes m6A modification in the 3'-UTR and the m6A-marked ULK1 transcripts can further be targeted for degradation by YTHDF2. | |||
| Target Regulator | Fat mass and obesity-associated protein (FTO) | ERASER | ||
| Target Regulation | Down regulation | |||
| Pathway Response | Autophagy | hsa04140 | ||
| Cell Process | RNA stability | |||
| Cell autophagy | ||||
| In-vitro Model | HEK293T | Normal | Homo sapiens | CVCL_0063 |
| HeLa | Endocervical adenocarcinoma | Homo sapiens | CVCL_0030 | |
Beta-Elemen
[Phase 3]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response | [1] | |||
| Response Summary | METTL3 could positively regulate the autophagy by targeting the autophagy-related genes such as ATG5, ATG7, LC3B, and Sequestosome-1 (SQSTM1). beta-elemene inhibited the autophagy flux by preventing autophagic lysosome acidification, resulting in increasing expression of SQSTM1 and LC3B-II. beta-elemene could reverse gefitinib resistance in non-small cell lung cancer cells by inhibiting cell autophagy process in a manner of chloroquine. METTL3-mediated autophagy in reversing gefitinib resistance of NSCLC cells by beta-elemene, which shed light on providing potential molecular-therapy target and clinical-treatment method in NSCLC patients with gefitinib resistance. | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Target Regulation | Up regulation | |||
| Responsed Disease | Non-small-cell lung carcinoma | ICD-11: 2C25.Y | ||
| Pathway Response | Autophagy | hsa04140 | ||
| Cell Process | Autophagic lysosome acidification | |||
| In-vitro Model | Gefitinib-resistant cell line HCC827GR (Gefitinib-resistant HCC827 cell line) | |||
| Gefitinib-resistant cell line PC9GR (Gefitinib-resistant PC9 cell line) | ||||
| HCC827 | Lung adenocarcinoma | Homo sapiens | CVCL_2063 | |
| PC-9 | Lung adenocarcinoma | Homo sapiens | CVCL_B260 | |
| In-vivo Model | NSCLC gefitinib-resistant cells (5 × 106 cells in 100 uL PBS) were injected subcutaneously into the lateral surface of the left abdomen of 6-week-old female BALB/c nude mice (at least five mice per group to ensure accuracy). | |||
Full List of Crosstalk(s) between m6A Modification and Epigenetic Regulation Related to This Regulator
DNA modification
m6A Regulator: Methyltransferase-like 14 (METTL14)
| In total 6 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT02226 | ||
| Epigenetic Regulator | DNA (cytosine-5)-methyltransferase 3B (DNMT3B) | |
| Regulated Target | Methyltransferase-like protein 14 (METTL14) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Breast cancer | |
| Drug | Paclitaxel | |
| Crosstalk ID: M6ACROT02250 | ||
| Epigenetic Regulator | Cysteine methyltransferase DNMT3A (DNMT3A) | |
| Regulated Target | Methyltransferase-like protein 14 (METTL14) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Breast cancer | |
| Drug | Paclitaxel | |
| Crosstalk ID: M6ACROT02274 | ||
| Epigenetic Regulator | DNA (cytosine-5)-methyltransferase 1 (DNMT1) | |
| Regulated Target | Methyltransferase-like protein 14 (METTL14) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Breast cancer | |
| Drug | Paclitaxel | |
| Crosstalk ID: M6ACROT05977 | ||
| Epigenetic Regulator | DNA (cytosine-5)-methyltransferase 3B (DNMT3B) | |
| Regulated Target | Methyltransferase-like protein 14 (METTL14) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Breast cancer | |
| Drug | STM2457 | |
| Crosstalk ID: M6ACROT05979 | ||
| Epigenetic Regulator | Cysteine methyltransferase DNMT3A (DNMT3A) | |
| Regulated Target | Methyltransferase-like protein 14 (METTL14) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Breast cancer | |
| Drug | STM2457 | |
| Crosstalk ID: M6ACROT05981 | ||
| Epigenetic Regulator | DNA (cytosine-5)-methyltransferase 1 (DNMT1) | |
| Regulated Target | Methyltransferase-like protein 14 (METTL14) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Breast cancer | |
| Drug | STM2457 | |
RNA Modification Sequencing Data Associated with the Target (ID: M6ATAR00410)
| In total 1 m6A sequence/site(s) in this target gene | |||
| mod ID: AC4SITE000098 | Click to Show/Hide the Full List | ||
| mod site | chr5:179833104-179833105:+ | [9] | |
| Sequence | CCCGTCTCTCCAGAGAGTTCCAGCACAGAGGAGAAGAGCAG | ||
| Cell/Tissue List | H1 | ||
| Seq Type List | ac4C-seq | ||
| Transcript ID List | ENST00000466342.1; ENST00000360718.5; ENST00000510187.5; ENST00000389805.9 | ||
| External Link | RMBase: ac4C_site_1536 | ||
Adenosine-to-Inosine editing (A-to-I)
| In total 8 m6A sequence/site(s) in this target gene | |||
| mod ID: A2ISITE001325 | Click to Show/Hide the Full List | ||
| mod site | chr5:179808629-179808630:+ | [10] | |
| Sequence | GATCGAGACCATCCTGGCTAACACGGTGAAACCCCGTCTCT | ||
| Transcript ID List | rmsk_1845595; ENST00000506690.1; ENST00000506042.5; ENST00000514093.5 | ||
| External Link | RMBase: RNA-editing_site_114142 | ||
| mod ID: A2ISITE001326 | Click to Show/Hide the Full List | ||
| mod site | chr5:179811146-179811147:+ | [10] | |
| Sequence | GGAGGCTGAGGCAGGAGAATAGCTTCAACCTGTGAGGCGGA | ||
| Transcript ID List | ENST00000506042.5; ENST00000514093.5; rmsk_1845604; ENST00000506690.1 | ||
| External Link | RMBase: RNA-editing_site_114143 | ||
| mod ID: A2ISITE001327 | Click to Show/Hide the Full List | ||
| mod site | chr5:179818924-179818925:+ | [10] | |
| Sequence | GACTGCATCGGGAAAGGGGAAGGAGTCAGAGGCGAGAAGGG | ||
| Transcript ID List | ENST00000422245.5; ENST00000514093.5 | ||
| External Link | RMBase: RNA-editing_site_114144 | ||
| mod ID: A2ISITE001328 | Click to Show/Hide the Full List | ||
| mod site | chr5:179829944-179829945:+ | [11] | |
| Sequence | ATCCCCCACTTCCCGTGTCTACAAACACTATGAAAATTAGC | ||
| Transcript ID List | ENST00000466342.1; rmsk_1845634; ENST00000360718.5; ENST00000389805.9; ENST00000510187.5 | ||
| External Link | RMBase: RNA-editing_site_114145 | ||
| mod ID: A2ISITE001329 | Click to Show/Hide the Full List | ||
| mod site | chr5:179829986-179829987:+ | [11] | |
| Sequence | AGGCATGGAGTTGTGTACTTATGGCCCCAGCTACGTGGGAG | ||
| Transcript ID List | ENST00000360718.5; rmsk_1845634; ENST00000389805.9; ENST00000466342.1; ENST00000510187.5 | ||
| External Link | RMBase: RNA-editing_site_114146 | ||
| mod ID: A2ISITE001330 | Click to Show/Hide the Full List | ||
| mod site | chr5:179830644-179830645:+ | [11] | |
| Sequence | CTCAGCTCACTGCAGTTTCCACCTCCTGGGTTCAAGCAATT | ||
| Transcript ID List | ENST00000389805.9; ENST00000466342.1; ENST00000510187.5; ENST00000360718.5 | ||
| External Link | RMBase: RNA-editing_site_114147 | ||
| mod ID: A2ISITE001331 | Click to Show/Hide the Full List | ||
| mod site | chr5:179830658-179830659:+ | [11] | |
| Sequence | GTTTCCACCTCCTGGGTTCAAGCAATTTTCTGCCTCAGCCT | ||
| Transcript ID List | ENST00000389805.9; ENST00000510187.5; ENST00000466342.1; ENST00000360718.5 | ||
| External Link | RMBase: RNA-editing_site_114148 | ||
| mod ID: A2ISITE001332 | Click to Show/Hide the Full List | ||
| mod site | chr5:179830817-179830818:+ | [10] | |
| Sequence | TACCTGCCTTCGTCTCCCAAAGTGCTGGGATTACAGGCATG | ||
| Transcript ID List | ENST00000466342.1; ENST00000360718.5; ENST00000510187.5; ENST00000389805.9 | ||
| External Link | RMBase: RNA-editing_site_114149 | ||
N1-methyladenosine (m1A)
| In total 1 m6A sequence/site(s) in this target gene | |||
| mod ID: M1ASITE000097 | Click to Show/Hide the Full List | ||
| mod site | chr5:179820904-179820905:+ | [12] | |
| Sequence | GCGCGGCGGCTGCGACCGGGACGGCCCGTTTTCCGCCAGCT | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | m1A-MAP-seq | ||
| Transcript ID List | ENST00000626660.1; ENST00000453046.5; ENST00000422245.5; ENST00000514093.5; ENST00000508284.5; ENST00000504627.1; ENST00000464493.5; ENST00000481335.5; ENST00000389805.9 | ||
| External Link | RMBase: m1A_site_753 | ||
5-methylcytidine (m5C)
| In total 19 m6A sequence/site(s) in this target gene | |||
| mod ID: M5CSITE003701 | Click to Show/Hide the Full List | ||
| mod site | chr5:179808617-179808618:+ | ||
| Sequence | ACGAGGTCAGGAGATCGAGACCATCCTGGCTAACACGGTGA | ||
| Cell/Tissue List | T24 | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | rmsk_1845595; ENST00000506690.1; ENST00000506042.5; ENST00000514093.5 | ||
| External Link | RMBase: m5C_site_36586 | ||
| mod ID: M5CSITE003702 | Click to Show/Hide the Full List | ||
| mod site | chr5:179808618-179808619:+ | ||
| Sequence | CGAGGTCAGGAGATCGAGACCATCCTGGCTAACACGGTGAA | ||
| Cell/Tissue List | T24 | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000506042.5; ENST00000514093.5; rmsk_1845595; ENST00000506690.1 | ||
| External Link | RMBase: m5C_site_36587 | ||
| mod ID: M5CSITE003703 | Click to Show/Hide the Full List | ||
| mod site | chr5:179808621-179808622:+ | ||
| Sequence | GGTCAGGAGATCGAGACCATCCTGGCTAACACGGTGAAACC | ||
| Cell/Tissue List | T24 | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | rmsk_1845595; ENST00000514093.5; ENST00000506042.5; ENST00000506690.1 | ||
| External Link | RMBase: m5C_site_36588 | ||
| mod ID: M5CSITE003704 | Click to Show/Hide the Full List | ||
| mod site | chr5:179808622-179808623:+ | ||
| Sequence | GTCAGGAGATCGAGACCATCCTGGCTAACACGGTGAAACCC | ||
| Cell/Tissue List | T24 | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | rmsk_1845595; ENST00000506042.5; ENST00000506690.1; ENST00000514093.5 | ||
| External Link | RMBase: m5C_site_36589 | ||
| mod ID: M5CSITE003705 | Click to Show/Hide the Full List | ||
| mod site | chr5:179808626-179808627:+ | ||
| Sequence | GGAGATCGAGACCATCCTGGCTAACACGGTGAAACCCCGTC | ||
| Cell/Tissue List | T24 | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000514093.5; ENST00000506042.5; ENST00000506690.1; rmsk_1845595 | ||
| External Link | RMBase: m5C_site_36590 | ||
| mod ID: M5CSITE003706 | Click to Show/Hide the Full List | ||
| mod site | chr5:179808640-179808641:+ | ||
| Sequence | TCCTGGCTAACACGGTGAAACCCCGTCTCTACTAAAAAATA | ||
| Cell/Tissue List | T24 | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000506042.5; rmsk_1845595; ENST00000506690.1; ENST00000514093.5 | ||
| External Link | RMBase: m5C_site_36591 | ||
| mod ID: M5CSITE003707 | Click to Show/Hide the Full List | ||
| mod site | chr5:179808641-179808642:+ | ||
| Sequence | CCTGGCTAACACGGTGAAACCCCGTCTCTACTAAAAAATAC | ||
| Cell/Tissue List | T24 | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | rmsk_1845595; ENST00000506042.5; ENST00000506690.1; ENST00000514093.5 | ||
| External Link | RMBase: m5C_site_36592 | ||
| mod ID: M5CSITE003708 | Click to Show/Hide the Full List | ||
| mod site | chr5:179808642-179808643:+ | ||
| Sequence | CTGGCTAACACGGTGAAACCCCGTCTCTACTAAAAAATACA | ||
| Cell/Tissue List | T24 | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000514093.5; ENST00000506690.1; ENST00000506042.5; rmsk_1845595 | ||
| External Link | RMBase: m5C_site_36593 | ||
| mod ID: M5CSITE003709 | Click to Show/Hide the Full List | ||
| mod site | chr5:179808643-179808644:+ | ||
| Sequence | TGGCTAACACGGTGAAACCCCGTCTCTACTAAAAAATACAA | ||
| Cell/Tissue List | T24 | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000506690.1; ENST00000506042.5; ENST00000514093.5; rmsk_1845595 | ||
| External Link | RMBase: m5C_site_36594 | ||
| mod ID: M5CSITE003710 | Click to Show/Hide the Full List | ||
| mod site | chr5:179811579-179811580:+ | ||
| Sequence | CTCACTCTTCAAAGTGTCTGCGAGATTAATCTCTCATGGCC | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m5C-RIP-seq | ||
| Transcript ID List | ENST00000514093.5; ENST00000506690.1; ENST00000506042.5; rmsk_1845606 | ||
| External Link | RMBase: m5C_site_36595 | ||
| mod ID: M5CSITE003711 | Click to Show/Hide the Full List | ||
| mod site | chr5:179820905-179820906:+ | [13] | |
| Sequence | CGCGGCGGCTGCGACCGGGACGGCCCGTTTTCCGCCAGCTC | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000626660.1; ENST00000464493.5; ENST00000504627.1; ENST00000514093.5; ENST00000389805.9; ENST00000481335.5; ENST00000453046.5; ENST00000422245.5; ENST00000508284.5 | ||
| External Link | RMBase: m5C_site_36596 | ||
| mod ID: M5CSITE003712 | Click to Show/Hide the Full List | ||
| mod site | chr5:179820909-179820910:+ | [13] | |
| Sequence | GCGGCTGCGACCGGGACGGCCCGTTTTCCGCCAGCTCGCCG | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000504627.1; ENST00000626660.1; ENST00000481335.5; ENST00000389805.9; ENST00000514093.5; ENST00000453046.5; ENST00000508284.5; ENST00000464493.5; ENST00000422245.5 | ||
| External Link | RMBase: m5C_site_36597 | ||
| mod ID: M5CSITE003713 | Click to Show/Hide the Full List | ||
| mod site | chr5:179820910-179820911:+ | [13] | |
| Sequence | CGGCTGCGACCGGGACGGCCCGTTTTCCGCCAGCTCGCCGC | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000508284.5; ENST00000626660.1; ENST00000481335.5; ENST00000514093.5; ENST00000464493.5; ENST00000422245.5; ENST00000453046.5; ENST00000504627.1; ENST00000389805.9 | ||
| External Link | RMBase: m5C_site_36598 | ||
| mod ID: M5CSITE003714 | Click to Show/Hide the Full List | ||
| mod site | chr5:179824092-179824093:+ | [13] | |
| Sequence | GGGCACCTGTCTGAGGTGAGCAGGCCCTCTGTGCAGGCCTG | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000466342.1; ENST00000514093.5; ENST00000485412.1; ENST00000510187.5; ENST00000422245.5; ENST00000389805.9; ENST00000464493.5; ENST00000508284.5; ENST00000360718.5 | ||
| External Link | RMBase: m5C_site_36599 | ||
| mod ID: M5CSITE003715 | Click to Show/Hide the Full List | ||
| mod site | chr5:179831401-179831402:+ | ||
| Sequence | CTGGGTGCAGTGGCTCATGCCTGTAATCCAAGCACTTTGGG | ||
| Cell/Tissue List | T24 | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000510187.5; ENST00000466342.1; ENST00000389805.9; rmsk_1845641; ENST00000360718.5 | ||
| External Link | RMBase: m5C_site_36600 | ||
| mod ID: M5CSITE003716 | Click to Show/Hide the Full List | ||
| mod site | chr5:179831415-179831416:+ | ||
| Sequence | TCATGCCTGTAATCCAAGCACTTTGGGAGGCCAAGGCAGGC | ||
| Cell/Tissue List | T24 | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | rmsk_1845641; ENST00000510187.5; ENST00000466342.1; ENST00000360718.5; ENST00000389805.9 | ||
| External Link | RMBase: m5C_site_36601 | ||
| mod ID: M5CSITE003717 | Click to Show/Hide the Full List | ||
| mod site | chr5:179833492-179833493:+ | [13] | |
| Sequence | CTTAACTGCACGTGTGCATGCGTGCTCCCCGACTGTCTGCC | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000360718.5; ENST00000389805.9; ENST00000510187.5 | ||
| External Link | RMBase: m5C_site_36602 | ||
| mod ID: M5CSITE003718 | Click to Show/Hide the Full List | ||
| mod site | chr5:179836691-179836692:+ | [13] | |
| Sequence | GTACGGGCCAGTTTCTCTGCCTTCTTCCAGGATCAGGGGTT | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000510187.5; ENST00000389805.9; ENST00000360718.5 | ||
| External Link | RMBase: m5C_site_36604 | ||
| mod ID: M5CSITE003719 | Click to Show/Hide the Full List | ||
| mod site | chr5:179836868-179836869:+ | [13] | |
| Sequence | AGCGCGCTCCTGACCCCTCCCTGCAGGGGCTACGTTAGCAG | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000389805.9; ENST00000510187.5; ENST00000360718.5 | ||
| External Link | RMBase: m5C_site_36605 | ||
N6-methyladenosine (m6A)
| In total 34 m6A sequence/site(s) in this target gene | |||
| mod ID: M6ASITE072983 | Click to Show/Hide the Full List | ||
| mod site | chr5:179806516-179806517:+ | [14] | |
| Sequence | GCTGAGTGCCGCGTACCAGGACAGCGAGAGGAAGGCGCACA | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | brain | ||
| Seq Type List | m6A-REF-seq | ||
| Transcript ID List | ENST00000514093.5; ENST00000506042.5 | ||
| External Link | RMBase: m6A_site_701557 | ||
| mod ID: M6ASITE072984 | Click to Show/Hide the Full List | ||
| mod site | chr5:179812842-179812843:+ | [15] | |
| Sequence | TTCCAAGGTTGAGGGTTGGAACTCCACAGCCTCAGCCTCCC | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000514093.5; ENST00000512234.1; ENST00000506042.5; ENST00000506690.1 | ||
| External Link | RMBase: m6A_site_701559 | ||
| mod ID: M6ASITE072985 | Click to Show/Hide the Full List | ||
| mod site | chr5:179820904-179820905:+ | [16] | |
| Sequence | GCGCGGCGGCTGCGACCGGGACGGCCCGTTTTCCGCCAGCT | ||
| Motif Score | 3.616982143 | ||
| Cell/Tissue List | CD8T | ||
| Seq Type List | m6A-CLIP/IP | ||
| Transcript ID List | ENST00000508284.5; ENST00000514093.5; ENST00000453046.5; ENST00000626660.1; ENST00000389805.9; ENST00000422245.5; ENST00000481335.5; ENST00000504627.1; ENST00000464493.5 | ||
| External Link | RMBase: m6A_site_701560 | ||
| mod ID: M6ASITE072986 | Click to Show/Hide the Full List | ||
| mod site | chr5:179821061-179821062:+ | [15] | |
| Sequence | GAGGCTGCGGCGGGTCCGGGACCCTGCGAGCGGCTGCTGAG | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000508284.5; ENST00000453046.5; ENST00000481335.5; ENST00000510187.5; ENST00000504627.1; ENST00000422245.5; ENST00000626660.1; ENST00000514093.5; ENST00000464493.5; ENST00000389805.9 | ||
| External Link | RMBase: m6A_site_701561 | ||
| mod ID: M6ASITE072987 | Click to Show/Hide the Full List | ||
| mod site | chr5:179821641-179821642:+ | [15] | |
| Sequence | CAAGCGCGGGGGTGCGGGGGACTCGCGAGCGCCGCGACAGC | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000508284.5; ENST00000481335.5; ENST00000514093.5; ENST00000464493.5; ENST00000453046.5; ENST00000422245.5; ENST00000510187.5; ENST00000504627.1; ENST00000389805.9 | ||
| External Link | RMBase: m6A_site_701562 | ||
| mod ID: M6ASITE072988 | Click to Show/Hide the Full List | ||
| mod site | chr5:179823044-179823045:+ | [17] | |
| Sequence | GGATGACATCTTCCGAATCTACATTAAAGGTAAGGGGCTGC | ||
| Motif Score | 2.078666667 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000481335.5; ENST00000504627.1; ENST00000453046.5; ENST00000389805.9; ENST00000510187.5; ENST00000508284.5; ENST00000514093.5; ENST00000360718.5; ENST00000464493.5; ENST00000422245.5 | ||
| External Link | RMBase: m6A_site_701563 | ||
| mod ID: M6ASITE072989 | Click to Show/Hide the Full List | ||
| mod site | chr5:179823914-179823915:+ | [17] | |
| Sequence | TGCTCAGGAGGCGCCCCGCAACATGGTGCACCCCAATGTGA | ||
| Motif Score | 2.173910714 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000514093.5; ENST00000481335.5; ENST00000464493.5; ENST00000485412.1; ENST00000508284.5; ENST00000360718.5; ENST00000504627.1; ENST00000453046.5; ENST00000466342.1; ENST00000389805.9; ENST00000422245.5; ENST00000510187.5 | ||
| External Link | RMBase: m6A_site_701564 | ||
| mod ID: M6ASITE072990 | Click to Show/Hide the Full List | ||
| mod site | chr5:179823974-179823975:+ | [14] | |
| Sequence | GCCTGTGGTAGGAACCCGCTACAAGTGCAGCGTCTGCCCAG | ||
| Motif Score | 2.078666667 | ||
| Cell/Tissue List | brain; hESC-HEK293T | ||
| Seq Type List | m6A-REF-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000485412.1; ENST00000464493.5; ENST00000466342.1; ENST00000389805.9; ENST00000504627.1; ENST00000510187.5; ENST00000508284.5; ENST00000514093.5; ENST00000422245.5; ENST00000360718.5 | ||
| External Link | RMBase: m6A_site_701565 | ||
| mod ID: M6ASITE072991 | Click to Show/Hide the Full List | ||
| mod site | chr5:179824043-179824044:+ | [14] | |
| Sequence | AAAGGGCTTGCACCGGGGGCACACCAAGCTCGCATTCCCCA | ||
| Motif Score | 2.830589286 | ||
| Cell/Tissue List | brain; hESC-HEK293T | ||
| Seq Type List | m6A-REF-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000389805.9; ENST00000466342.1; ENST00000464493.5; ENST00000514093.5; ENST00000485412.1; ENST00000504627.1; ENST00000510187.5; ENST00000360718.5; ENST00000422245.5; ENST00000508284.5 | ||
| External Link | RMBase: m6A_site_701566 | ||
| mod ID: M6ASITE072992 | Click to Show/Hide the Full List | ||
| mod site | chr5:179824222-179824223:+ | [17] | |
| Sequence | CTCCGGAAGGTGAAACACGGACACTTCGGGTGGCCAGGATG | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000360718.5; ENST00000514093.5; ENST00000508284.5; ENST00000422245.5; ENST00000464493.5; ENST00000389805.9; ENST00000510187.5; ENST00000485412.1; ENST00000466342.1 | ||
| External Link | RMBase: m6A_site_701567 | ||
| mod ID: M6ASITE072993 | Click to Show/Hide the Full List | ||
| mod site | chr5:179833108-179833109:+ | [14] | |
| Sequence | TCTCTCCAGAGAGTTCCAGCACAGAGGAGAAGAGCAGCTCA | ||
| Motif Score | 2.830589286 | ||
| Cell/Tissue List | HEK293; kidney | ||
| Seq Type List | m6A-REF-seq | ||
| Transcript ID List | ENST00000360718.5; ENST00000510187.5; ENST00000389805.9; ENST00000466342.1 | ||
| External Link | RMBase: m6A_site_701568 | ||
| mod ID: M6ASITE072994 | Click to Show/Hide the Full List | ||
| mod site | chr5:179833275-179833276:+ | [18] | |
| Sequence | TGCCCCTCCCGCCACCTGGGACCACGGCCAGCCTAGTGATC | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HepG2; HeLa; LCLs; H1299; MM6; Huh7; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293T; endometrial; HEC-1-A; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000466342.1; ENST00000510187.5; ENST00000360718.5; ENST00000389805.9 | ||
| External Link | RMBase: m6A_site_701569 | ||
| mod ID: M6ASITE072995 | Click to Show/Hide the Full List | ||
| mod site | chr5:179833348-179833349:+ | [19] | |
| Sequence | CTCTGCAGCCCCACTTACAAACCCGAGGGAGCTGCTGCTGC | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | H1299 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000510187.5; ENST00000466342.1; ENST00000360718.5; ENST00000389805.9 | ||
| External Link | RMBase: m6A_site_701570 | ||
| mod ID: M6ASITE072996 | Click to Show/Hide the Full List | ||
| mod site | chr5:179833415-179833416:+ | [16] | |
| Sequence | CACGCTGGGAACCTGCTAGAACTTTGTAGTTACTTGGTCTT | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | A549 | ||
| Seq Type List | m6A-CLIP/IP | ||
| Transcript ID List | ENST00000360718.5; ENST00000389805.9; ENST00000510187.5 | ||
| External Link | RMBase: m6A_site_701571 | ||
| mod ID: M6ASITE072997 | Click to Show/Hide the Full List | ||
| mod site | chr5:179833664-179833665:+ | [14] | |
| Sequence | CAAAAGAAGTGGACCCGTCTACAGGTGAACTCCAGTCCCTA | ||
| Motif Score | 2.078666667 | ||
| Cell/Tissue List | brain | ||
| Seq Type List | m6A-REF-seq | ||
| Transcript ID List | ENST00000389805.9; ENST00000510187.5; ENST00000360718.5 | ||
| External Link | RMBase: m6A_site_701572 | ||
| mod ID: M6ASITE072998 | Click to Show/Hide the Full List | ||
| mod site | chr5:179833672-179833673:+ | [18] | |
| Sequence | GTGGACCCGTCTACAGGTGAACTCCAGTCCCTACAGATGCC | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HepG2; HeLa; endometrial | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000360718.5; ENST00000389805.9; ENST00000510187.5 | ||
| External Link | RMBase: m6A_site_701573 | ||
| mod ID: M6ASITE072999 | Click to Show/Hide the Full List | ||
| mod site | chr5:179833684-179833685:+ | [14] | |
| Sequence | ACAGGTGAACTCCAGTCCCTACAGATGCCAGAATCCGAAGG | ||
| Motif Score | 2.078666667 | ||
| Cell/Tissue List | brain; kidney | ||
| Seq Type List | m6A-REF-seq | ||
| Transcript ID List | ENST00000510187.5; ENST00000360718.5; ENST00000389805.9 | ||
| External Link | RMBase: m6A_site_701574 | ||
| mod ID: M6ASITE073000 | Click to Show/Hide the Full List | ||
| mod site | chr5:179833719-179833720:+ | [15] | |
| Sequence | CGAAGGGCCAAGCTCTCTGGACCCCTCCCAGGAGGGACCCA | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; HepG2; peripheral-blood; endometrial; HEC-1-A | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000510187.5; ENST00000389805.9; ENST00000360718.5 | ||
| External Link | RMBase: m6A_site_701575 | ||
| mod ID: M6ASITE073001 | Click to Show/Hide the Full List | ||
| mod site | chr5:179833735-179833736:+ | [15] | |
| Sequence | CTGGACCCCTCCCAGGAGGGACCCACAGGGCTGAAGGAAGC | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; HepG2; peripheral-blood; HEK293T; endometrial; HEC-1-A | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000389805.9; ENST00000360718.5; ENST00000510187.5 | ||
| External Link | RMBase: m6A_site_701576 | ||
| mod ID: M6ASITE073002 | Click to Show/Hide the Full List | ||
| mod site | chr5:179833768-179833769:+ | [17] | |
| Sequence | AAGGAAGCTGCCTTGTACCCACATCTCCCGCCAGGCAAGTG | ||
| Motif Score | 2.053113095 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000389805.9; ENST00000360718.5; ENST00000510187.5 | ||
| External Link | RMBase: m6A_site_701577 | ||
| mod ID: M6ASITE073003 | Click to Show/Hide the Full List | ||
| mod site | chr5:179836524-179836525:+ | [15] | |
| Sequence | GGCTCACCAGGCTCCTGCAGACCAAGAACTATGACATCGGA | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa; HepG2; peripheral-blood; endometrial; HEC-1-A | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000360718.5; ENST00000389805.9; ENST00000510187.5 | ||
| External Link | RMBase: m6A_site_701583 | ||
| mod ID: M6ASITE073004 | Click to Show/Hide the Full List | ||
| mod site | chr5:179836531-179836532:+ | [15] | |
| Sequence | CAGGCTCCTGCAGACCAAGAACTATGACATCGGAGCGGCTC | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa; HepG2; peripheral-blood; endometrial; HEC-1-A | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000360718.5; ENST00000389805.9; ENST00000510187.5 | ||
| External Link | RMBase: m6A_site_701584 | ||
| mod ID: M6ASITE073005 | Click to Show/Hide the Full List | ||
| mod site | chr5:179836537-179836538:+ | [14] | |
| Sequence | CCTGCAGACCAAGAACTATGACATCGGAGCGGCTCTGGACA | ||
| Motif Score | 2.859755952 | ||
| Cell/Tissue List | kidney; hESC-HEK293T | ||
| Seq Type List | m6A-REF-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000510187.5; ENST00000360718.5; ENST00000389805.9 | ||
| External Link | RMBase: m6A_site_701585 | ||
| mod ID: M6ASITE073006 | Click to Show/Hide the Full List | ||
| mod site | chr5:179836555-179836556:+ | [15] | |
| Sequence | TGACATCGGAGCGGCTCTGGACACCATCCAGTATTCAAAGC | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; hESC-HEK293T | ||
| Seq Type List | m6A-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000389805.9; ENST00000360718.5; ENST00000510187.5 | ||
| External Link | RMBase: m6A_site_701586 | ||
| mod ID: M6ASITE073007 | Click to Show/Hide the Full List | ||
| mod site | chr5:179836742-179836743:+ | [15] | |
| Sequence | AAGCCATTTAGGGCAGCAAAACAAGTGACATGAAGGGAGGG | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa; hESC-HEK293T | ||
| Seq Type List | m6A-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000510187.5; ENST00000360718.5; ENST00000389805.9 | ||
| External Link | RMBase: m6A_site_701590 | ||
| mod ID: M6ASITE073008 | Click to Show/Hide the Full List | ||
| mod site | chr5:179836749-179836750:+ | [17] | |
| Sequence | TTAGGGCAGCAAAACAAGTGACATGAAGGGAGGGTCCCTGT | ||
| Motif Score | 2.859755952 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000389805.9; ENST00000360718.5; ENST00000510187.5 | ||
| External Link | RMBase: m6A_site_701591 | ||
| mod ID: M6ASITE073009 | Click to Show/Hide the Full List | ||
| mod site | chr5:179836832-179836833:+ | [15] | |
| Sequence | AGCAGGGCTGGGCCTGCGAGACCCAAGGCTCACTGCAGCGC | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000389805.9; ENST00000510187.5; ENST00000360718.5 | ||
| External Link | RMBase: m6A_site_701594 | ||
| mod ID: M6ASITE073010 | Click to Show/Hide the Full List | ||
| mod site | chr5:179836895-179836896:+ | [17] | |
| Sequence | GGCTACGTTAGCAGCCCAGCACATAGCTTGCCTAATGGCTT | ||
| Motif Score | 2.830589286 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000510187.5; ENST00000389805.9; ENST00000360718.5 | ||
| External Link | RMBase: m6A_site_701595 | ||
| mod ID: M6ASITE073011 | Click to Show/Hide the Full List | ||
| mod site | chr5:179836940-179836941:+ | [20] | |
| Sequence | TTTCTCTTTTGTTTTAAATGACTCATAGGTCCCTGACATTT | ||
| Motif Score | 3.28175 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000510187.5; ENST00000360718.5; ENST00000389805.9 | ||
| External Link | RMBase: m6A_site_701596 | ||
| mod ID: M6ASITE073012 | Click to Show/Hide the Full List | ||
| mod site | chr5:179836955-179836956:+ | [20] | |
| Sequence | AAATGACTCATAGGTCCCTGACATTTAGTTGATTATTTTCT | ||
| Motif Score | 2.859755952 | ||
| Cell/Tissue List | HEK293T; hESC-HEK293T | ||
| Seq Type List | DART-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000360718.5; ENST00000510187.5; ENST00000389805.9 | ||
| External Link | RMBase: m6A_site_701597 | ||
| mod ID: M6ASITE073013 | Click to Show/Hide the Full List | ||
| mod site | chr5:179836990-179836991:+ | [17] | |
| Sequence | TTTTCTGCTACAGACCTGGTACACTCTGATTTTAGATAAAG | ||
| Motif Score | 2.856142857 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000389805.9; ENST00000360718.5; ENST00000510187.5 | ||
| External Link | RMBase: m6A_site_701598 | ||
| mod ID: M6ASITE073014 | Click to Show/Hide the Full List | ||
| mod site | chr5:179837219-179837220:+ | [17] | |
| Sequence | CCATCCTGTTAAATTTGTAAACAATCTAATTAAATGGCATC | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000360718.5; ENST00000389805.9 | ||
| External Link | RMBase: m6A_site_701599 | ||
| mod ID: M6ASITE073015 | Click to Show/Hide the Full List | ||
| mod site | chr5:179837697-179837698:+ | [14] | |
| Sequence | ACCTTGGCTGCTCACTGTCCACATGTGAACTTTTTCTAGGT | ||
| Motif Score | 2.053113095 | ||
| Cell/Tissue List | kidney | ||
| Seq Type List | m6A-REF-seq | ||
| Transcript ID List | ENST00000389805.9 | ||
| External Link | RMBase: m6A_site_701607 | ||
| mod ID: M6ASITE073016 | Click to Show/Hide the Full List | ||
| mod site | chr5:179837922-179837923:+ | [17] | |
| Sequence | CATGCCCTCCATGTGTAAGAACAATGCCAGGGCCCAGGAGG | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000389805.9 | ||
| External Link | RMBase: m6A_site_701612 | ||
2'-O-Methylation (2'-O-Me)
| In total 4 m6A sequence/site(s) in this target gene | |||
| mod ID: 2OMSITE000388 | Click to Show/Hide the Full List | ||
| mod site | chr5:179825170-179825171:+ | [21] | |
| Sequence | GGTCCATCGGAGGATCCGAGTGTGAATTTCCTGAAGAACGT | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | Nm-seq | ||
| Transcript ID List | ENST00000510187.5; ENST00000466342.1; ENST00000514093.5; ENST00000389805.9; ENST00000360718.5 | ||
| External Link | RMBase: Nm_site_5630 | ||
| mod ID: 2OMSITE000389 | Click to Show/Hide the Full List | ||
| mod site | chr5:179825172-179825173:+ | [21] | |
| Sequence | TCCATCGGAGGATCCGAGTGTGAATTTCCTGAAGAACGTTG | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | Nm-seq | ||
| Transcript ID List | ENST00000389805.9; ENST00000360718.5; ENST00000514093.5; ENST00000466342.1; ENST00000510187.5 | ||
| External Link | RMBase: Nm_site_5631 | ||
| mod ID: 2OMSITE000390 | Click to Show/Hide the Full List | ||
| mod site | chr5:179833609-179833610:+ | [21] | |
| Sequence | CAGATGGAGTCGGATAACTGTTCAGGAGGAGATGATGACTG | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | Nm-seq | ||
| Transcript ID List | ENST00000360718.5; ENST00000510187.5; ENST00000389805.9 | ||
| External Link | RMBase: Nm_site_5632 | ||
| mod ID: 2OMSITE000391 | Click to Show/Hide the Full List | ||
| mod site | chr5:179833724-179833725:+ | [21] | |
| Sequence | GGCCAAGCTCTCTGGACCCCTCCCAGGAGGGACCCACAGGG | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | Nm-seq | ||
| Transcript ID List | ENST00000510187.5; ENST00000360718.5; ENST00000389805.9 | ||
| External Link | RMBase: Nm_site_5633 | ||
References