m6A Target Gene Information
General Information of the m6A Target Gene (ID: M6ATAR00342)
Full List of m6A Methylation Regulator of This Target Gene and Corresponding Disease/Drug Response(s)
MYD88
can be regulated by the following regulator(s), and cause disease/drug response(s). You can browse detail information of regulator(s) or disease/drug response(s).
Browse Regulator
Browse Disease
Methyltransferase-like 3 (METTL3) [WRITER]
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | MOLM-13 cell line | Homo sapiens |
|
Treatment: shMETTL3 MOLM13 cells
Control: MOLM13 cells
|
GSE98623 | |
| Regulation |
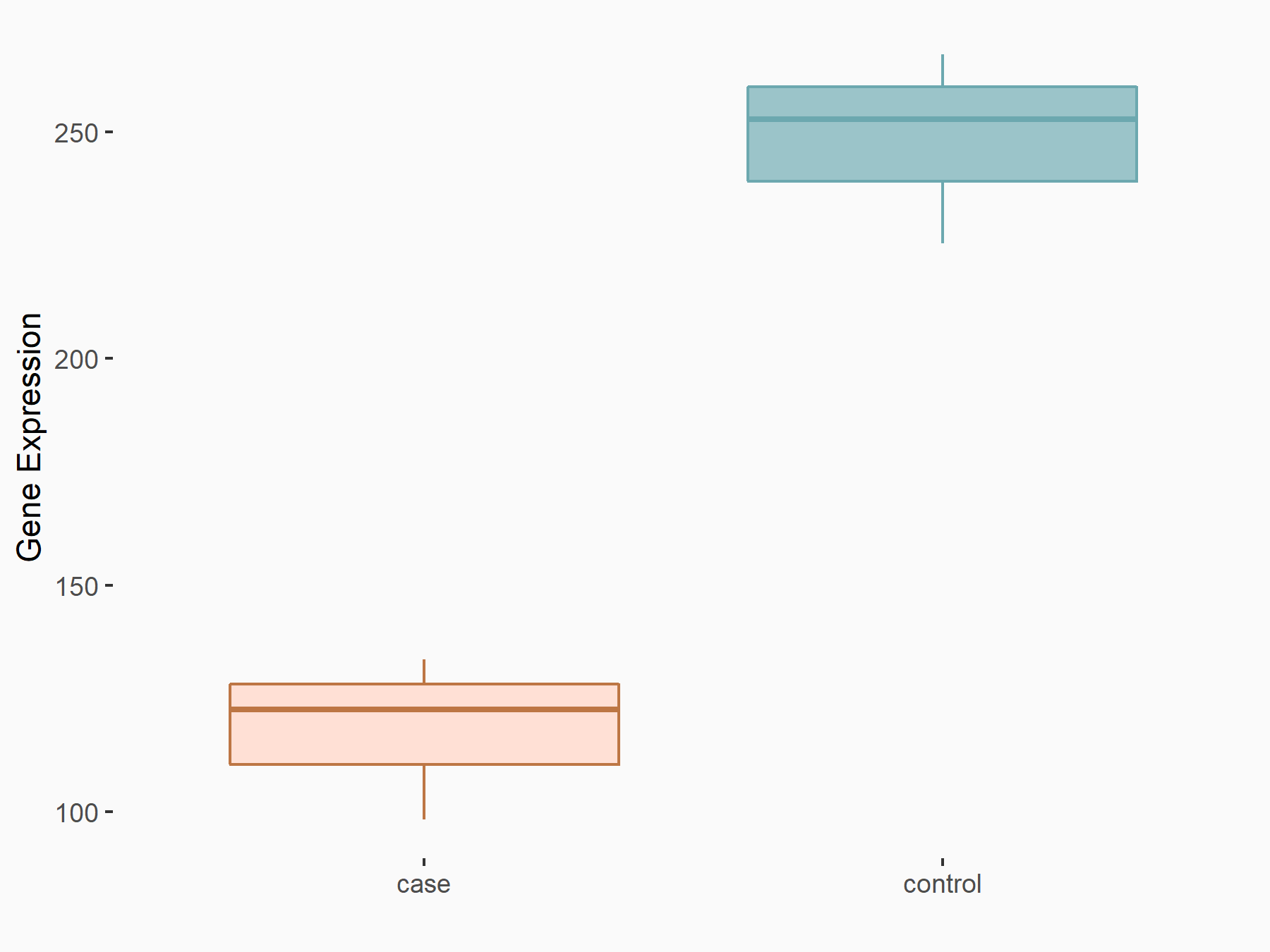  |
logFC: -1.07E+00 p-value: 1.83E-13 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between MYD88 and the regulator | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 1.16E+00 | GSE60213 |
| In total 2 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [1] | |||
| Response Summary | Knocked down METTL3 and demonstrated that METTL3 depletion decreased the expression of inflammatory cytokines and the phosphorylation of IKK-alpha/beta, p65 and IKappa-B-alpha in the NF-Kappa-B signalling pathway as well as p38, ERK and JNK in the MAPK signalling pathway in LPS-induced HDPCs. METTL3 inhibits the LPS-induced inflammatory response of HDPCs by regulating alternative splicing of Myeloid differentiation primary response protein MyD88 (MYD88). | |||
| Target Regulation | Down regulation | |||
| Responsed Disease | Pulpitis | ICD-11: DA09 | ||
| Pathway Response | MAPK signaling pathway | hsa04010 | ||
| Cell Process | Alternative splicing | |||
| Experiment 2 Reporting the m6A Methylation Regulator of This Target Gene | [2] | |||
| Response Summary | METTL3 positively regulates expression of Myeloid differentiation primary response protein MyD88 (MYD88), a critical upstream regulator of NF-Kappa-B signaling, by facilitating m6A methylation modification to MYD88-RNA, subsequently inducing the activation of NF-Kappa-B which is widely regarded as a repressor of osteogenesis and therefore suppressing osteogenic progression. The METTL3-mediated m6A methylation is found to be dynamically reversed by the demethylase ALKBH5. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Skeletal anomaly | ICD-11: LD24 | ||
| Pathway Response | Central carbon metabolism in cancer | hsa05230 | ||
| Cell Process | Glucose metabolism | |||
| In-vitro Model | Mesenchymal stem cell line (NP tissues were used to isolate NP cells) | |||
RNA demethylase ALKBH5 (ALKBH5) [ERASER]
| Representative RNA-seq result indicating the expression of this target gene regulated by ALKBH5 | ||
| Cell Line | 143B cell line | Homo sapiens |
|
Treatment: siALKBH5 transfected 143B cells
Control: siControl 143B cells
|
GSE154528 | |
| Regulation |
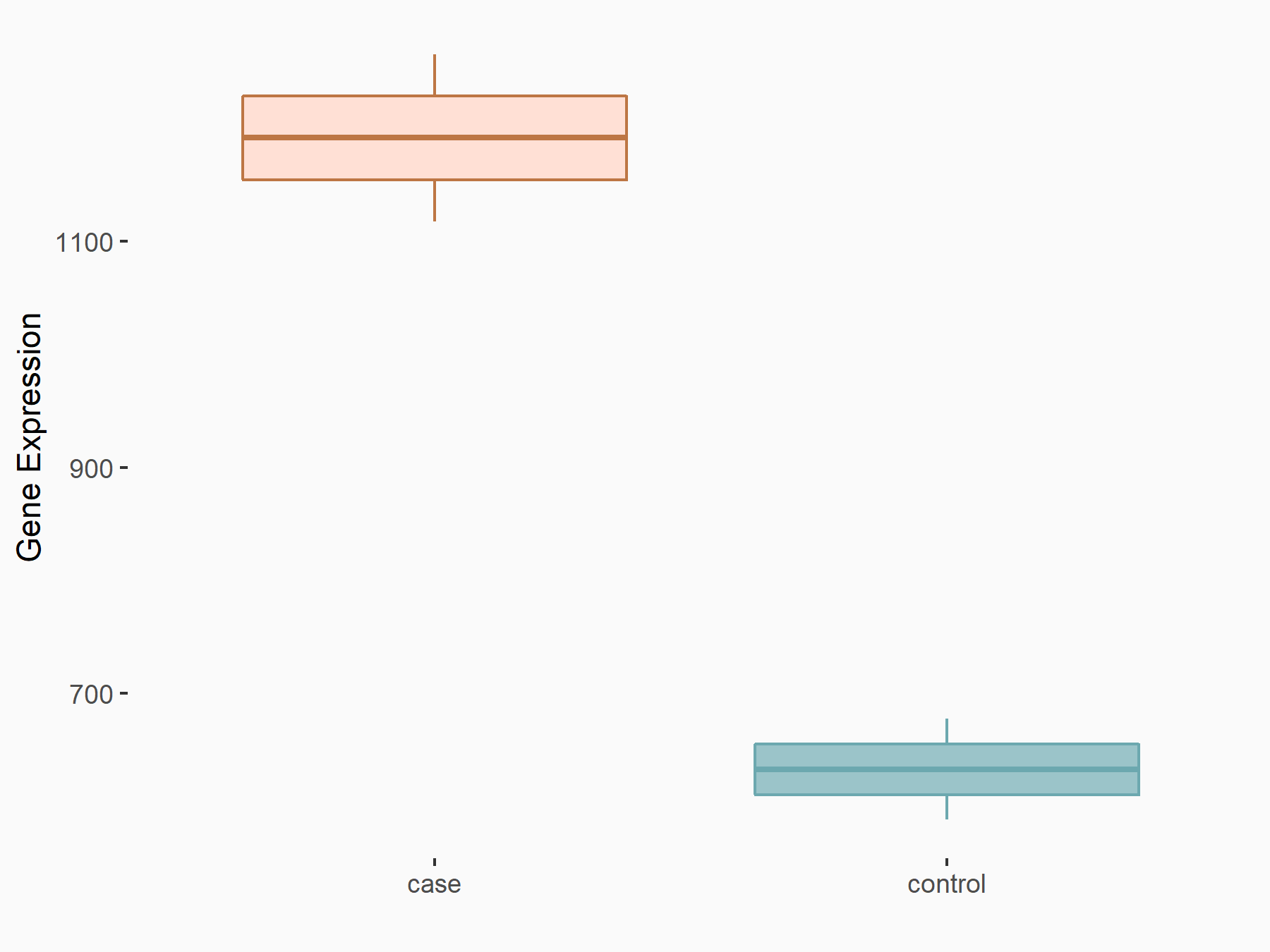  |
logFC: 9.11E-01 p-value: 2.76E-05 |
| More Results | Click to View More RNA-seq Results | |
| In total 1 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [2] | |||
| Response Summary | METTL3 positively regulates expression of Myeloid differentiation primary response protein MyD88 (MYD88), a critical upstream regulator of NF-Kappa-B signaling, by facilitating m6A methylation modification to MYD88-RNA, subsequently inducing the activation of NF-Kappa-B which is widely regarded as a repressor of osteogenesis and therefore suppressing osteogenic progression. The METTL3-mediated m6A methylation is found to be dynamically reversed by the demethylase ALKBH5. | |||
| Target Regulation | Down regulation | |||
| Responsed Disease | Skeletal anomaly | ICD-11: LD24 | ||
| Pathway Response | Central carbon metabolism in cancer | hsa05230 | ||
| Cell Process | Glucose metabolism | |||
| In-vitro Model | Mesenchymal stem cell line (NP tissues were used to isolate NP cells) | |||
RNA-binding motif protein 15 (RBM15) [WRITER]
| Representative RNA-seq result indicating the expression of this target gene regulated by RBM15 | ||
| Cell Line | Human BJ Fibroblast | Homo sapiens |
|
Treatment: siRBM15 BJ fibroblast
Control: siControl BJ fibroblast
|
GSE154148 | |
| Regulation |
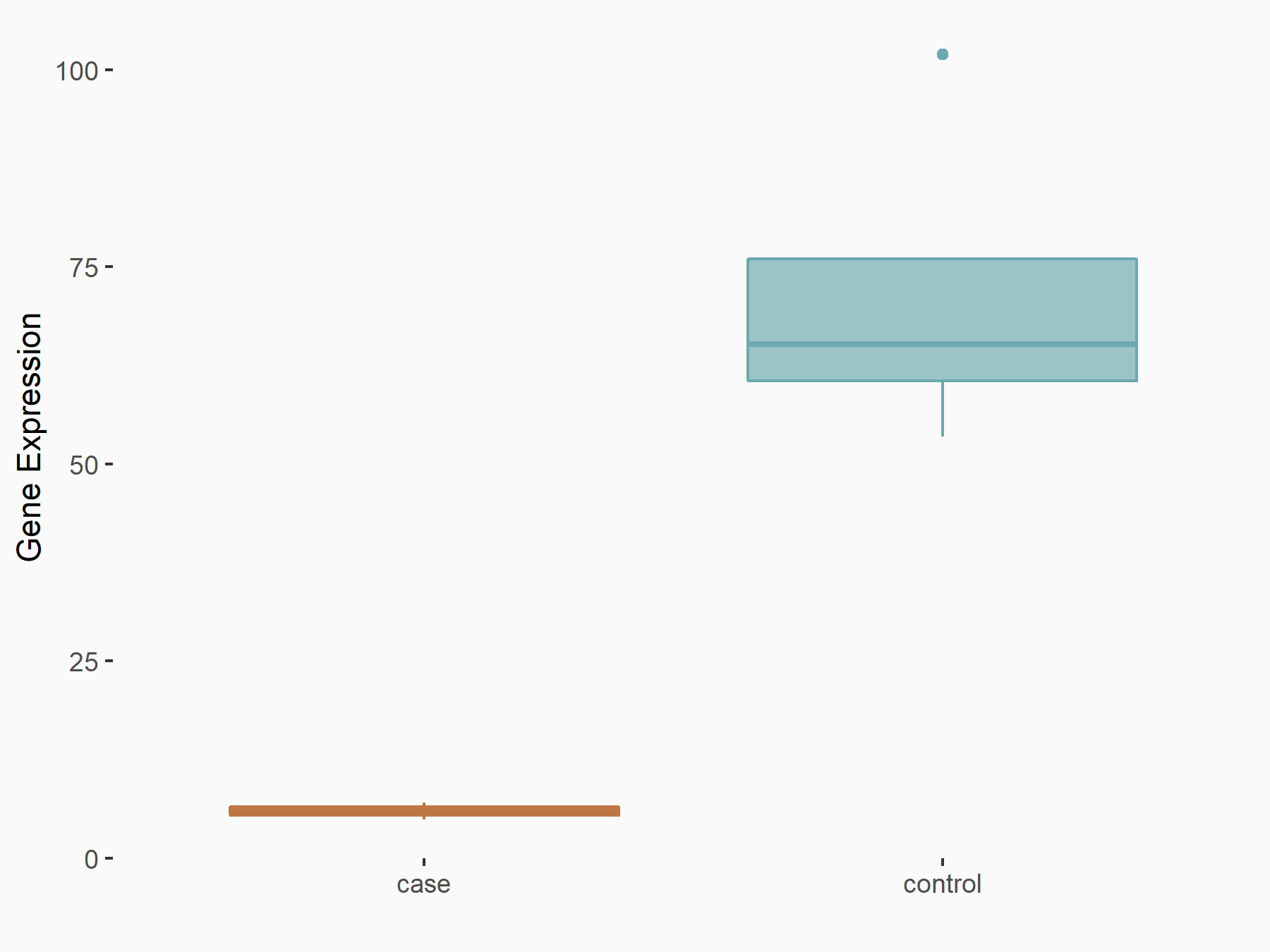 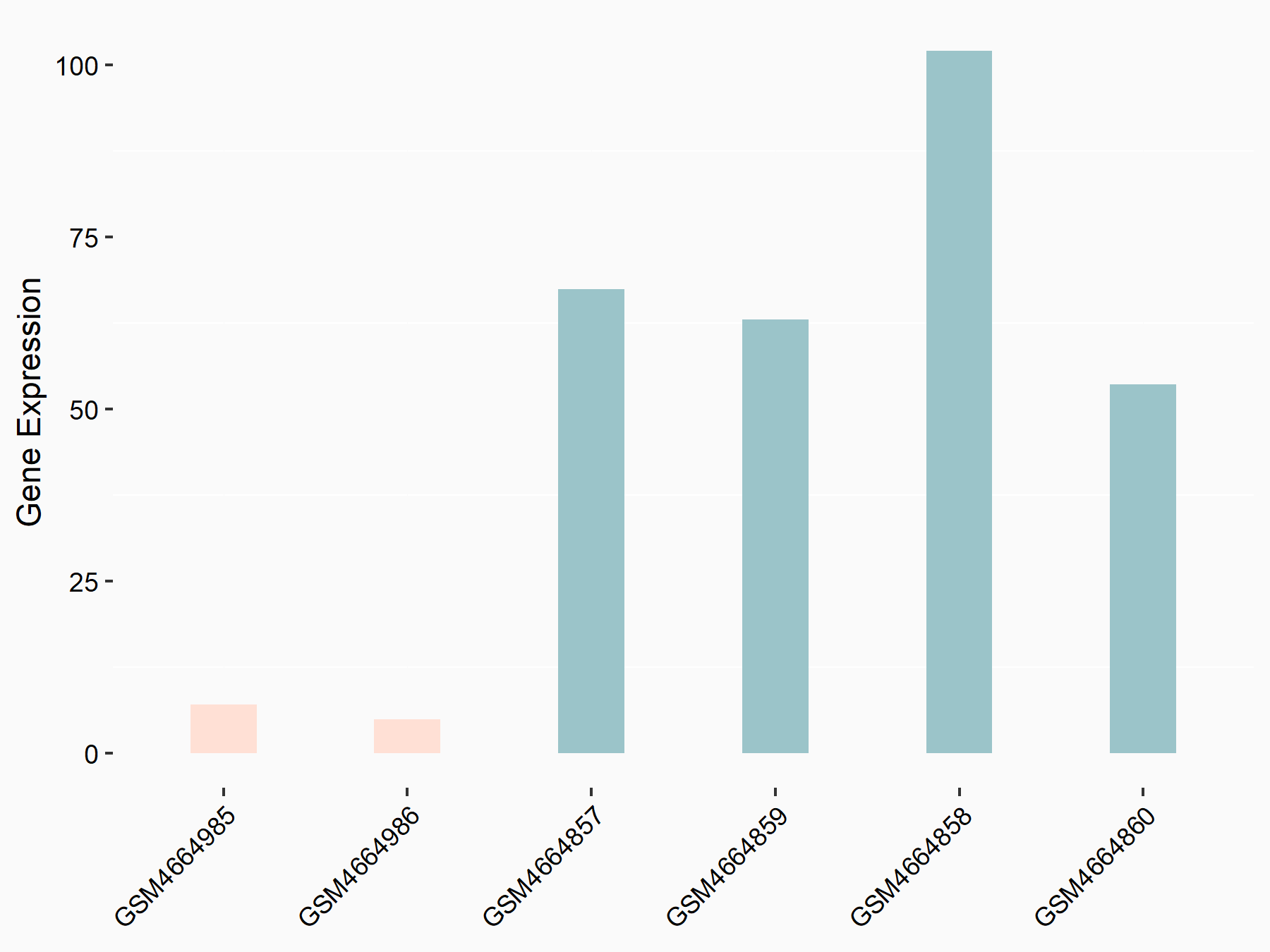 |
logFC: -3.36E+00 p-value: 3.10E-04 |
| More Results | Click to View More RNA-seq Results | |
| In total 1 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [3] | |||
| Response Summary | RBM15 silencing inhibited the CRC growth and metastasis in vitro and in vivo. RBM15 mediated m6A methylation modification of Myeloid differentiation primary response protein MyD88 (MYD88) mRNA in colorectal cancer cells. | |||
| Target Regulation | Down regulation | |||
| Responsed Disease | Colorectal cancer | ICD-11: 2B91 | ||
| Cell Process | Cell proliferative | |||
| Cell invasive | ||||
| In-vitro Model | HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 |
| SW480 | Colon adenocarcinoma | Homo sapiens | CVCL_0546 | |
Colorectal cancer [ICD-11: 2B91]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [3] | |||
| Response Summary | RBM15 silencing inhibited the CRC growth and metastasis in vitro and in vivo. RBM15 mediated m6A methylation modification of Myeloid differentiation primary response protein MyD88 (MYD88) mRNA in colorectal cancer cells. | |||
| Responsed Disease | Colorectal cancer [ICD-11: 2B91] | |||
| Target Regulator | RNA-binding motif protein 15 (RBM15) | WRITER | ||
| Target Regulation | Down regulation | |||
| Cell Process | Cell proliferative | |||
| Cell invasive | ||||
| In-vitro Model | HCT 116 | Colon carcinoma | Homo sapiens | CVCL_0291 |
| SW480 | Colon adenocarcinoma | Homo sapiens | CVCL_0546 | |
Pulpitis [ICD-11: DA09]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [1] | |||
| Response Summary | Knocked down METTL3 and demonstrated that METTL3 depletion decreased the expression of inflammatory cytokines and the phosphorylation of IKK-alpha/beta, p65 and IKappa-B-alpha in the NF-Kappa-B signalling pathway as well as p38, ERK and JNK in the MAPK signalling pathway in LPS-induced HDPCs. METTL3 inhibits the LPS-induced inflammatory response of HDPCs by regulating alternative splicing of Myeloid differentiation primary response protein MyD88 (MYD88). | |||
| Responsed Disease | Pulpitis [ICD-11: DA09] | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Target Regulation | Down regulation | |||
| Pathway Response | MAPK signaling pathway | hsa04010 | ||
| Cell Process | Alternative splicing | |||
Skeletal anomaly [ICD-11: LD24]
| In total 2 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [2] | |||
| Response Summary | METTL3 positively regulates expression of Myeloid differentiation primary response protein MyD88 (MYD88), a critical upstream regulator of NF-Kappa-B signaling, by facilitating m6A methylation modification to MYD88-RNA, subsequently inducing the activation of NF-Kappa-B which is widely regarded as a repressor of osteogenesis and therefore suppressing osteogenic progression. The METTL3-mediated m6A methylation is found to be dynamically reversed by the demethylase ALKBH5. | |||
| Responsed Disease | Skeletal anomaly [ICD-11: LD24] | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Central carbon metabolism in cancer | hsa05230 | ||
| Cell Process | Glucose metabolism | |||
| In-vitro Model | Mesenchymal stem cell line (NP tissues were used to isolate NP cells) | |||
| Experiment 2 Reporting the m6A-centered Disease Response | [2] | |||
| Response Summary | METTL3 positively regulates expression of Myeloid differentiation primary response protein MyD88 (MYD88), a critical upstream regulator of NF-Kappa-B signaling, by facilitating m6A methylation modification to MYD88-RNA, subsequently inducing the activation of NF-Kappa-B which is widely regarded as a repressor of osteogenesis and therefore suppressing osteogenic progression. The METTL3-mediated m6A methylation is found to be dynamically reversed by the demethylase ALKBH5. | |||
| Responsed Disease | Skeletal anomaly [ICD-11: LD24] | |||
| Target Regulator | RNA demethylase ALKBH5 (ALKBH5) | ERASER | ||
| Target Regulation | Down regulation | |||
| Pathway Response | Central carbon metabolism in cancer | hsa05230 | ||
| Cell Process | Glucose metabolism | |||
| In-vitro Model | Mesenchymal stem cell line (NP tissues were used to isolate NP cells) | |||
RNA Modification Sequencing Data Associated with the Target (ID: M6ATAR00342)
| In total 1 m6A sequence/site(s) in this target gene | |||
| mod ID: A2ISITE011663 | Click to Show/Hide the Full List | ||
| mod site | chr3:38141149-38141150:+ | [4] | |
| Sequence | AGGTGCCCATCAGAAGCGACTGATCCCCATCAAGTACAAGG | ||
| Transcript ID List | ENST00000463956.1; ENST00000484513.1; ENST00000652590.1; ENST00000417037.7; ENST00000652213.1; ENST00000421516.3; ENST00000443433.7; ENST00000648963.1; ENST00000424893.6; ENST00000650112.1; ENST00000481122.5; ENST00000650905.1; ENST00000652534.1; ENST00000495303.6; ENST00000651800.1; ENST00000416282.3; ENST00000396334.8 | ||
| External Link | RMBase: RNA-editing_site_95811 | ||
N6-methyladenosine (m6A)
| In total 41 m6A sequence/site(s) in this target gene | |||
| mod ID: M6ASITE060053 | Click to Show/Hide the Full List | ||
| mod site | chr3:38138658-38138659:+ | [5] | |
| Sequence | GAAAGAGGAAGCGCTGGCAGACAATGCGACCCGACCGCGCT | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa; fibroblasts; A549; Jurkat; GSC-11; MSC | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000651800.1; ENST00000396334.8; ENST00000650112.1; ENST00000495303.6; ENST00000424893.6; ENST00000417037.7; ENST00000652534.1; ENST00000652213.1; ENST00000460295.1; ENST00000416282.3 | ||
| External Link | RMBase: m6A_site_583356 | ||
| mod ID: M6ASITE060054 | Click to Show/Hide the Full List | ||
| mod site | chr3:38138690-38138691:+ | [5] | |
| Sequence | GACCGCGCTGAGGCTCCAGGACCGCCCGCCATGGCTGCAGG | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; HepG2; A549; H1A; H1B; fibroblasts; MM6; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293T; iSLK; MSC; TIME; TREX; endometrial; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000650112.1; ENST00000417037.7; ENST00000443433.7; ENST00000652534.1; ENST00000652213.1; ENST00000396334.8; ENST00000650905.1; ENST00000495303.6; ENST00000424893.6; ENST00000460295.1; ENST00000416282.3; ENST00000421516.3; ENST00000651800.1; ENST00000652590.1; ENST00000648963.1 | ||
| External Link | RMBase: m6A_site_583357 | ||
| mod ID: M6ASITE060055 | Click to Show/Hide the Full List | ||
| mod site | chr3:38138820-38138821:+ | [5] | |
| Sequence | CTCTGTTCTTGAACGTGCGGACACAGGTGGCGGCCGACTGG | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; HepG2; A549; H1B; MM6; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293T; MSC; TIME; TREX; iSLK; endometrial; HEC-1-A; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000650112.1; ENST00000652213.1; ENST00000460295.1; ENST00000421516.3; ENST00000650905.1; ENST00000651800.1; ENST00000424893.6; ENST00000416282.3; ENST00000648963.1; ENST00000652590.1; ENST00000495303.6; ENST00000396334.8; ENST00000443433.7; ENST00000652534.1; ENST00000417037.7 | ||
| External Link | RMBase: m6A_site_583358 | ||
| mod ID: M6ASITE060056 | Click to Show/Hide the Full List | ||
| mod site | chr3:38138841-38138842:+ | [5] | |
| Sequence | CACAGGTGGCGGCCGACTGGACCGCGCTGGCGGAGGAGATG | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; HepG2; A549; MM6; Jurkat; CD4T; peripheral-blood; GSC-11; MSC; TIME; TREX; iSLK; endometrial; HEC-1-A; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000652213.1; ENST00000651800.1; ENST00000417037.7; ENST00000416282.3; ENST00000396334.8; ENST00000652590.1; ENST00000650112.1; ENST00000424893.6; ENST00000460295.1; ENST00000421516.3; ENST00000648963.1; ENST00000652534.1; ENST00000495303.6; ENST00000650905.1; ENST00000443433.7 | ||
| External Link | RMBase: m6A_site_583359 | ||
| mod ID: M6ASITE060057 | Click to Show/Hide the Full List | ||
| mod site | chr3:38138863-38138864:+ | [5] | |
| Sequence | CGCGCTGGCGGAGGAGATGGACTTTGAGTACTTGGAGATCC | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa; HepG2; A549; MM6; Jurkat; CD4T; peripheral-blood; GSC-11; MSC; TIME; TREX; iSLK; endometrial; HEC-1-A; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq; m6A-CLIP/IP | ||
| Transcript ID List | ENST00000652590.1; ENST00000416282.3; ENST00000417037.7; ENST00000396334.8; ENST00000651800.1; ENST00000421516.3; ENST00000495303.6; ENST00000648963.1; ENST00000650112.1; ENST00000650905.1; ENST00000652213.1; ENST00000424893.6; ENST00000460295.1; ENST00000443433.7; ENST00000652534.1 | ||
| External Link | RMBase: m6A_site_583360 | ||
| mod ID: M6ASITE060058 | Click to Show/Hide the Full List | ||
| mod site | chr3:38138895-38138896:+ | [5] | |
| Sequence | TGGAGATCCGGCAACTGGAGACACAAGCGGACCCCACTGGC | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa; HepG2; A549; MM6; Jurkat; CD4T; peripheral-blood; GSC-11; TIME; iSLK; endometrial; HEC-1-A; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000648963.1; ENST00000416282.3; ENST00000443433.7; ENST00000650905.1; ENST00000652534.1; ENST00000495303.6; ENST00000652213.1; ENST00000460295.1; ENST00000651800.1; ENST00000421516.3; ENST00000417037.7; ENST00000650112.1; ENST00000396334.8; ENST00000652590.1; ENST00000424893.6 | ||
| External Link | RMBase: m6A_site_583361 | ||
| mod ID: M6ASITE060059 | Click to Show/Hide the Full List | ||
| mod site | chr3:38138905-38138906:+ | [5] | |
| Sequence | GCAACTGGAGACACAAGCGGACCCCACTGGCAGGCTGCTGG | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; HepG2; A549; MM6; Jurkat; CD4T; peripheral-blood; GSC-11; iSLK; endometrial; HEC-1-A; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000648963.1; ENST00000421516.3; ENST00000652590.1; ENST00000650112.1; ENST00000652534.1; ENST00000443433.7; ENST00000460295.1; ENST00000651800.1; ENST00000424893.6; ENST00000416282.3; ENST00000652213.1; ENST00000495303.6; ENST00000417037.7; ENST00000650905.1; ENST00000396334.8 | ||
| External Link | RMBase: m6A_site_583362 | ||
| mod ID: M6ASITE060060 | Click to Show/Hide the Full List | ||
| mod site | chr3:38139017-38139018:+ | [6] | |
| Sequence | GACGTGCTGCTGGAGCTGGGACCCAGCATTGGTGAGGACGT | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HepG2; A549; MM6; Jurkat; CD4T; peripheral-blood; HEC-1-A; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000652213.1; ENST00000650905.1; ENST00000648963.1; ENST00000652590.1; ENST00000424893.6; ENST00000416282.3; ENST00000443433.7; ENST00000421516.3; ENST00000417037.7; ENST00000495303.6; ENST00000396334.8; ENST00000651800.1; ENST00000460295.1; ENST00000650112.1; ENST00000652534.1 | ||
| External Link | RMBase: m6A_site_583363 | ||
| mod ID: M6ASITE060061 | Click to Show/Hide the Full List | ||
| mod site | chr3:38139160-38139161:+ | [6] | |
| Sequence | AAGAAGCCTGCAGAGGGAGAACCATGCGGGTCCCGTTCCTT | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | HepG2; A549; MM6; Jurkat; CD4T; peripheral-blood; HEC-1-A; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000652213.1; ENST00000650112.1; ENST00000443433.7; ENST00000424893.6; ENST00000651800.1; ENST00000396334.8; ENST00000481122.5; ENST00000495303.6; ENST00000652590.1; ENST00000416282.3; ENST00000650905.1; ENST00000421516.3; ENST00000652534.1; ENST00000648963.1; ENST00000460295.1; ENST00000417037.7 | ||
| External Link | RMBase: m6A_site_583364 | ||
| mod ID: M6ASITE060062 | Click to Show/Hide the Full List | ||
| mod site | chr3:38139211-38139212:+ | [7] | |
| Sequence | GGTCGCGGTTATTAAGAAGGACTGGAGAAAGGTCCGGATAG | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HepG2; peripheral-blood; HEC-1-A; NB4; MM6 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000651800.1; ENST00000652213.1; ENST00000650905.1; ENST00000416282.3; ENST00000396334.8; ENST00000648963.1; ENST00000460295.1; ENST00000495303.6; ENST00000417037.7; ENST00000424893.6; ENST00000481122.5; ENST00000650112.1; ENST00000443433.7; ENST00000421516.3; ENST00000652534.1; ENST00000652590.1 | ||
| External Link | RMBase: m6A_site_583365 | ||
| mod ID: M6ASITE060063 | Click to Show/Hide the Full List | ||
| mod site | chr3:38140472-38140473:+ | [8] | |
| Sequence | GAGATGATCCGGCAACTGGAACAGACAAACTATCGACTGAA | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | peripheral-blood | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000650905.1; ENST00000481122.5; ENST00000417037.7; ENST00000484513.1; ENST00000652590.1; ENST00000443433.7; ENST00000421516.3; ENST00000495303.6; ENST00000416282.3; ENST00000460295.1; ENST00000396334.8; ENST00000652213.1; ENST00000424893.6; ENST00000651800.1; ENST00000650112.1; ENST00000463956.1; ENST00000652534.1; ENST00000648963.1 | ||
| External Link | RMBase: m6A_site_583366 | ||
| mod ID: M6ASITE060064 | Click to Show/Hide the Full List | ||
| mod site | chr3:38140480-38140481:+ | [8] | |
| Sequence | CCGGCAACTGGAACAGACAAACTATCGACTGAAGTTGTGTG | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | peripheral-blood | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000443433.7; ENST00000650905.1; ENST00000651800.1; ENST00000652213.1; ENST00000495303.6; ENST00000652590.1; ENST00000481122.5; ENST00000421516.3; ENST00000463956.1; ENST00000417037.7; ENST00000416282.3; ENST00000648963.1; ENST00000396334.8; ENST00000484513.1; ENST00000460295.1; ENST00000652534.1; ENST00000650112.1; ENST00000424893.6 | ||
| External Link | RMBase: m6A_site_583367 | ||
| mod ID: M6ASITE060065 | Click to Show/Hide the Full List | ||
| mod site | chr3:38140820-38140821:+ | [9] | |
| Sequence | GCAAGGAATGTGACTTCCAGACCAAATTTGCACTCAGCCTC | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HEC-1-A | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000443433.7; ENST00000396334.8; ENST00000651800.1; ENST00000421516.3; ENST00000417037.7; ENST00000481122.5; ENST00000424893.6; ENST00000463956.1; ENST00000495303.6; ENST00000416282.3; ENST00000652534.1; ENST00000650905.1; ENST00000484513.1; ENST00000652590.1; ENST00000650112.1; ENST00000652213.1; ENST00000648963.1 | ||
| External Link | RMBase: m6A_site_583368 | ||
| mod ID: M6ASITE060066 | Click to Show/Hide the Full List | ||
| mod site | chr3:38141253-38141254:+ | [7] | |
| Sequence | GCACCAAATCTTGGTTCTGGACTCGCCTTGCCAAGGCCTTG | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HepG2; U2OS; H1A; H1B; hESCs; fibroblasts; A549; GM12878; MM6; Huh7; HEK293A-TOA; iSLK; MSC; TIME; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000650112.1; ENST00000463956.1; ENST00000650905.1; ENST00000443433.7; ENST00000648963.1; ENST00000495303.6; ENST00000417037.7; ENST00000651800.1; ENST00000652213.1; ENST00000484513.1; ENST00000424893.6; ENST00000416282.3; ENST00000396334.8; ENST00000481122.5; ENST00000421516.3; ENST00000652534.1; ENST00000652590.1 | ||
| External Link | RMBase: m6A_site_583369 | ||
| mod ID: M6ASITE060067 | Click to Show/Hide the Full List | ||
| mod site | chr3:38141288-38141289:+ | [6] | |
| Sequence | GCCTTGTCCCTGCCCTGAAGACTGTTCTGAGGCCCTGGGTG | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HepG2; A549; U2OS; H1B; H1A; hESCs; fibroblasts; GM12878; LCLs; MM6; Huh7; Jurkat; HEK293A-TOA; iSLK; MSC; TIME; TREX; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000650905.1; ENST00000484513.1; ENST00000652213.1; ENST00000650112.1; ENST00000495303.6; ENST00000421516.3; ENST00000443433.7; ENST00000396334.8; ENST00000651800.1; ENST00000652534.1; ENST00000424893.6; ENST00000481122.5; ENST00000416282.3; ENST00000652590.1; ENST00000417037.7 | ||
| External Link | RMBase: m6A_site_583370 | ||
| mod ID: M6ASITE060068 | Click to Show/Hide the Full List | ||
| mod site | chr3:38141420-38141421:+ | [6] | |
| Sequence | TGGAGATGCCAACTTCACAGACACGTCTGCAGCAGCTGGAC | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HepG2; HEK293T; A549; U2OS; H1B; hESCs; fibroblasts; GM12878; LCLs; MM6; Huh7; Jurkat; HEK293A-TOA; iSLK; MSC; TIME; TREX; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000396334.8; ENST00000651800.1; ENST00000443433.7; ENST00000416282.3; ENST00000495303.6; ENST00000424893.6; ENST00000652590.1; ENST00000652534.1; ENST00000650112.1; ENST00000650905.1; ENST00000652213.1; ENST00000421516.3; ENST00000417037.7; ENST00000484513.1 | ||
| External Link | RMBase: m6A_site_583371 | ||
| mod ID: M6ASITE060069 | Click to Show/Hide the Full List | ||
| mod site | chr3:38141439-38141440:+ | [6] | |
| Sequence | GACACGTCTGCAGCAGCTGGACATCACATTTCATGTCCTGC | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HepG2; HEK293T; A549; U2OS; H1B; hNPCs; hESCs; fibroblasts; GM12878; LCLs; MM6; Huh7; Jurkat; HEK293A-TOA; iSLK; MSC; TIME; TREX; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000396334.8; ENST00000652213.1; ENST00000650905.1; ENST00000495303.6; ENST00000652590.1; ENST00000650112.1; ENST00000421516.3; ENST00000416282.3; ENST00000417037.7; ENST00000484513.1; ENST00000443433.7; ENST00000424893.6; ENST00000652534.1; ENST00000651800.1 | ||
| External Link | RMBase: m6A_site_583372 | ||
| mod ID: M6ASITE060070 | Click to Show/Hide the Full List | ||
| mod site | chr3:38141465-38141466:+ | [6] | |
| Sequence | CATTTCATGTCCTGCATGGAACCAGTGGCTGTGAGTGGCAT | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | HepG2; HEK293T; A549; U2OS; H1B; H1A; hNPCs; hESCs; fibroblasts; GM12878; LCLs; CD8T; MM6; Huh7; Jurkat; CD4T; HEK293A-TOA; iSLK; MSC; TIME; TREX; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq; m6A-CLIP/IP | ||
| Transcript ID List | ENST00000652590.1; ENST00000651800.1; ENST00000417037.7; ENST00000421516.3; ENST00000424893.6; ENST00000652213.1; ENST00000650112.1; ENST00000396334.8; ENST00000652534.1; ENST00000416282.3; ENST00000650905.1; ENST00000495303.6; ENST00000484513.1 | ||
| External Link | RMBase: m6A_site_583373 | ||
| mod ID: M6ASITE060071 | Click to Show/Hide the Full List | ||
| mod site | chr3:38141512-38141513:+ | [6] | |
| Sequence | TTGCTGGATTATCAGCCAGGACACTATAGAACAGGACCAGC | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HepG2; HEK293T; kidney; A549; hESC-HEK293T; U2OS; H1B; H1A; hNPCs; hESCs; fibroblasts; GM12878; LCLs; CD8T; MM6; Huh7; Jurkat; CD4T; HEK293A-TOA; iSLK; MSC; TIME; TREX; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq; m6A-REF-seq; MAZTER-seq; m6A-CLIP/IP | ||
| Transcript ID List | ENST00000484513.1; ENST00000652213.1; ENST00000651800.1; ENST00000421516.3; ENST00000417037.7; ENST00000396334.8; ENST00000652590.1; ENST00000424893.6; ENST00000652534.1; ENST00000650112.1; ENST00000495303.6; ENST00000416282.3; ENST00000650905.1 | ||
| External Link | RMBase: m6A_site_583374 | ||
| mod ID: M6ASITE060072 | Click to Show/Hide the Full List | ||
| mod site | chr3:38141522-38141523:+ | [6] | |
| Sequence | ATCAGCCAGGACACTATAGAACAGGACCAGCTGAGACTAAG | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HepG2; HEK293T; A549; U2OS; H1B; H1A; hNPCs; hESCs; fibroblasts; GM12878; LCLs; MM6; Huh7; Jurkat; CD4T; HEK293A-TOA; iSLK; MSC; TIME; TREX; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000424893.6; ENST00000416282.3; ENST00000650112.1; ENST00000495303.6; ENST00000652213.1; ENST00000417037.7; ENST00000651800.1; ENST00000652590.1; ENST00000396334.8; ENST00000421516.3; ENST00000650905.1; ENST00000652534.1; ENST00000484513.1 | ||
| External Link | RMBase: m6A_site_583375 | ||
| mod ID: M6ASITE060073 | Click to Show/Hide the Full List | ||
| mod site | chr3:38141527-38141528:+ | [6] | |
| Sequence | CCAGGACACTATAGAACAGGACCAGCTGAGACTAAGAAGGA | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HepG2; HEK293T; A549; U2OS; H1A; H1B; hNPCs; hESCs; fibroblasts; GM12878; LCLs; MM6; Huh7; Jurkat; CD4T; HEK293A-TOA; iSLK; MSC; TIME; TREX; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000417037.7; ENST00000652590.1; ENST00000651800.1; ENST00000396334.8; ENST00000495303.6; ENST00000650905.1; ENST00000652213.1; ENST00000421516.3; ENST00000416282.3; ENST00000484513.1; ENST00000424893.6; ENST00000650112.1; ENST00000652534.1 | ||
| External Link | RMBase: m6A_site_583376 | ||
| mod ID: M6ASITE060074 | Click to Show/Hide the Full List | ||
| mod site | chr3:38141537-38141538:+ | [6] | |
| Sequence | ATAGAACAGGACCAGCTGAGACTAAGAAGGACCAGCAGAGC | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HepG2; HEK293T; A549; U2OS; H1A; H1B; hNPCs; hESCs; fibroblasts; GM12878; LCLs; MM6; Huh7; Jurkat; CD4T; HEK293A-TOA; iSLK; MSC; TIME; TREX; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq; m6A-CLIP/IP | ||
| Transcript ID List | ENST00000396334.8; ENST00000652590.1; ENST00000652213.1; ENST00000651800.1; ENST00000650112.1; ENST00000417037.7; ENST00000652534.1; ENST00000495303.6; ENST00000484513.1; ENST00000424893.6; ENST00000421516.3; ENST00000416282.3; ENST00000650905.1 | ||
| External Link | RMBase: m6A_site_583377 | ||
| mod ID: M6ASITE060075 | Click to Show/Hide the Full List | ||
| mod site | chr3:38141547-38141548:+ | [6] | |
| Sequence | ACCAGCTGAGACTAAGAAGGACCAGCAGAGCCAGCTCAGCT | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HepG2; HEK293T; A549; U2OS; H1A; H1B; hNPCs; hESCs; fibroblasts; GM12878; LCLs; MM6; Huh7; Jurkat; CD4T; HEK293A-TOA; iSLK; MSC; TIME; TREX; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000424893.6; ENST00000396334.8; ENST00000652590.1; ENST00000421516.3; ENST00000416282.3; ENST00000650112.1; ENST00000650905.1; ENST00000652534.1; ENST00000652213.1; ENST00000651800.1; ENST00000484513.1; ENST00000417037.7; ENST00000495303.6 | ||
| External Link | RMBase: m6A_site_583378 | ||
| mod ID: M6ASITE060076 | Click to Show/Hide the Full List | ||
| mod site | chr3:38141579-38141580:+ | [10] | |
| Sequence | AGCTCAGCTCTGAGCCATTCACACATCTTCACCCTCAGTTT | ||
| Motif Score | 2.047297619 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000424893.6; ENST00000416282.3; ENST00000650905.1; ENST00000651800.1; ENST00000652213.1; ENST00000484513.1; ENST00000396334.8; ENST00000417037.7; ENST00000421516.3; ENST00000652590.1; ENST00000650112.1; ENST00000652534.1; ENST00000495303.6 | ||
| External Link | RMBase: m6A_site_583379 | ||
| mod ID: M6ASITE060077 | Click to Show/Hide the Full List | ||
| mod site | chr3:38141627-38141628:+ | [5] | |
| Sequence | TGAGGAGTGGGATGGGGAGAACAGAGAGTAGCTGTGTTTGA | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; A549; U2OS; H1A; H1B; hESCs; fibroblasts; GM12878; LCLs; MM6; Huh7; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; TREX; endometrial; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000650112.1; ENST00000652534.1; ENST00000651800.1; ENST00000650905.1; ENST00000421516.3; ENST00000417037.7; ENST00000484513.1; ENST00000652213.1; ENST00000424893.6; ENST00000652590.1; ENST00000495303.6; ENST00000416282.3; ENST00000396334.8 | ||
| External Link | RMBase: m6A_site_583380 | ||
| mod ID: M6ASITE060078 | Click to Show/Hide the Full List | ||
| mod site | chr3:38141695-38141696:+ | [5] | |
| Sequence | CTCTGGGTCTCCTGGGGGAGACCAGGCTTGGCTGCGGGAGA | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa; HepG2; A549; U2OS; H1A; H1B; hESCs; HEK293T; fibroblasts; GM12878; LCLs; MM6; Huh7; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; TREX; endometrial; HEC-1-A; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000396334.8; ENST00000417037.7; ENST00000652534.1; ENST00000650905.1; ENST00000651800.1; ENST00000421516.3; ENST00000484513.1; ENST00000650112.1; ENST00000416282.3; ENST00000652213.1; ENST00000652590.1 | ||
| External Link | RMBase: m6A_site_583381 | ||
| mod ID: M6ASITE060079 | Click to Show/Hide the Full List | ||
| mod site | chr3:38141731-38141732:+ | [5] | |
| Sequence | GGAGAGCTGGCTGTTGCTGGACTACATGCTGGCCACTGCTG | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa; HepG2; A549; U2OS; H1A; H1B; hESCs; HEK293T; fibroblasts; GM12878; LCLs; MM6; Huh7; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; TREX; endometrial; HEC-1-A; NB4; AML | ||
| Seq Type List | m6A-seq; MeRIP-seq; miCLIP | ||
| Transcript ID List | ENST00000421516.3; ENST00000650112.1; ENST00000652590.1; ENST00000652213.1; ENST00000650905.1; ENST00000396334.8; ENST00000416282.3; ENST00000651800.1; ENST00000484513.1; ENST00000417037.7; ENST00000652534.1 | ||
| External Link | RMBase: m6A_site_583382 | ||
| mod ID: M6ASITE060080 | Click to Show/Hide the Full List | ||
| mod site | chr3:38141886-38141887:+ | [11] | |
| Sequence | GTTTGGCCCAGCCCAAGGAGACCCCACCTTGAGCCTTATTT | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | A549; U2OS; GM12878; LCLs; Huh7; peripheral-blood; iSLK; MSC; TIME; TREX; HEC-1-A; NB4; MM6 | ||
| Seq Type List | MeRIP-seq; m6A-seq | ||
| Transcript ID List | ENST00000652213.1; ENST00000396334.8; ENST00000417037.7; ENST00000650112.1; ENST00000652534.1; ENST00000651800.1; ENST00000652590.1; ENST00000650905.1; ENST00000421516.3; ENST00000416282.3; ENST00000484513.1 | ||
| External Link | RMBase: m6A_site_583383 | ||
| mod ID: M6ASITE060081 | Click to Show/Hide the Full List | ||
| mod site | chr3:38141988-38141989:+ | [11] | |
| Sequence | CAGTGACAAGTCCCCAAGAGACTCGCCTGAGCAGCTTGGGC | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | A549; H1A; GM12878; LCLs; Huh7; HEK293A-TOA; TREX; iSLK; HEC-1-A; NB4; MM6 | ||
| Seq Type List | MeRIP-seq; m6A-seq | ||
| Transcript ID List | ENST00000417037.7; ENST00000652213.1; ENST00000652590.1; ENST00000421516.3; ENST00000651800.1; ENST00000416282.3; ENST00000652534.1; ENST00000484513.1; ENST00000650905.1; ENST00000650112.1; ENST00000396334.8 | ||
| External Link | RMBase: m6A_site_583384 | ||
| mod ID: M6ASITE060082 | Click to Show/Hide the Full List | ||
| mod site | chr3:38142227-38142228:+ | [12] | |
| Sequence | ACAAAGTTATTTGTTTACAAACAGCGACCATATAAAAGCCT | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | CD34; MT4; Huh7 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000652590.1; ENST00000417037.7; ENST00000650905.1; ENST00000421516.3; ENST00000484513.1; ENST00000652213.1; ENST00000396334.8; ENST00000651800.1; ENST00000650112.1; ENST00000652534.1; ENST00000416282.3 | ||
| External Link | RMBase: m6A_site_583385 | ||
| mod ID: M6ASITE060083 | Click to Show/Hide the Full List | ||
| mod site | chr3:38142285-38142286:+ | [12] | |
| Sequence | GGGCACATGGGCACATACAGACTCACATACAGACACACACA | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | CD34; MT4; Huh7 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000484513.1; ENST00000416282.3; ENST00000650112.1; ENST00000396334.8; rmsk_936837; ENST00000650905.1; ENST00000417037.7; ENST00000651800.1; ENST00000652213.1; ENST00000652590.1; ENST00000421516.3; ENST00000652534.1 | ||
| External Link | RMBase: m6A_site_583386 | ||
| mod ID: M6ASITE060084 | Click to Show/Hide the Full List | ||
| mod site | chr3:38142297-38142298:+ | [12] | |
| Sequence | ACATACAGACTCACATACAGACACACACATATATGTACAGA | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | CD34; MT4; Huh7 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000416282.3; ENST00000421516.3; ENST00000652590.1; ENST00000417037.7; ENST00000650905.1; ENST00000652213.1; rmsk_936837; ENST00000484513.1; ENST00000650112.1; ENST00000651800.1; ENST00000652534.1; ENST00000396334.8 | ||
| External Link | RMBase: m6A_site_583387 | ||
| mod ID: M6ASITE060085 | Click to Show/Hide the Full List | ||
| mod site | chr3:38142317-38142318:+ | [13] | |
| Sequence | ACACACACATATATGTACAGACATGTACTCTCACACACACA | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | MT4; Huh7 | ||
| Seq Type List | MeRIP-seq | ||
| Transcript ID List | ENST00000651800.1; ENST00000396334.8; ENST00000416282.3; ENST00000652213.1; ENST00000484513.1; ENST00000652534.1; ENST00000650112.1; ENST00000652590.1; rmsk_936837; ENST00000421516.3; ENST00000650905.1; ENST00000417037.7 | ||
| External Link | RMBase: m6A_site_583388 | ||
| mod ID: M6ASITE060086 | Click to Show/Hide the Full List | ||
| mod site | chr3:38142380-38142381:+ | [14] | |
| Sequence | TCTAGGTACAGCTCCCAGGAACAGCTAGGTGGGAAAGTCCC | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | liver; A549; MT4; Huh7 | ||
| Seq Type List | m6A-REF-seq; m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000652213.1; ENST00000650112.1; ENST00000652534.1; ENST00000650905.1; ENST00000651800.1; ENST00000652590.1; ENST00000421516.3; ENST00000416282.3; ENST00000417037.7; ENST00000484513.1; ENST00000396334.8 | ||
| External Link | RMBase: m6A_site_583389 | ||
| mod ID: M6ASITE060087 | Click to Show/Hide the Full List | ||
| mod site | chr3:38142431-38142432:+ | [10] | |
| Sequence | GAGCCTAACCATGTCCCTGAACAAAAATTGGGCACTCATCT | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | hESC-HEK293T; LCLs; Huh7 | ||
| Seq Type List | MAZTER-seq; m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000652590.1; ENST00000652534.1; ENST00000651800.1; ENST00000652213.1; ENST00000396334.8; ENST00000650112.1; ENST00000416282.3; ENST00000650905.1; ENST00000417037.7; ENST00000421516.3; ENST00000484513.1 | ||
| External Link | RMBase: m6A_site_583390 | ||
| mod ID: M6ASITE060088 | Click to Show/Hide the Full List | ||
| mod site | chr3:38142484-38142485:+ | [15] | |
| Sequence | TTGTGTCCCTACTCATTGAAACCAAACTCTGGAAAGGACCC | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | A549; LCLs; Huh7 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000650112.1; ENST00000396334.8; ENST00000417037.7; ENST00000484513.1; ENST00000652213.1; ENST00000421516.3; ENST00000651800.1; ENST00000652534.1; ENST00000652590.1; ENST00000416282.3; ENST00000650905.1 | ||
| External Link | RMBase: m6A_site_583391 | ||
| mod ID: M6ASITE060089 | Click to Show/Hide the Full List | ||
| mod site | chr3:38142489-38142490:+ | [15] | |
| Sequence | TCCCTACTCATTGAAACCAAACTCTGGAAAGGACCCAATGT | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | A549; LCLs; Huh7 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000421516.3; ENST00000650905.1; ENST00000652213.1; ENST00000416282.3; ENST00000396334.8; ENST00000417037.7; ENST00000651800.1; ENST00000484513.1; ENST00000650112.1; ENST00000652590.1; ENST00000652534.1 | ||
| External Link | RMBase: m6A_site_583392 | ||
| mod ID: M6ASITE060090 | Click to Show/Hide the Full List | ||
| mod site | chr3:38142501-38142502:+ | [15] | |
| Sequence | GAAACCAAACTCTGGAAAGGACCCAATGTACCAGTATTTAT | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | A549; LCLs; Huh7 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000650905.1; ENST00000651800.1; ENST00000652213.1; ENST00000417037.7; ENST00000484513.1; ENST00000421516.3; ENST00000650112.1; ENST00000416282.3; ENST00000652534.1; ENST00000396334.8; ENST00000652590.1 | ||
| External Link | RMBase: m6A_site_583393 | ||
| mod ID: M6ASITE060091 | Click to Show/Hide the Full List | ||
| mod site | chr3:38142562-38142563:+ | [15] | |
| Sequence | AGAGGAAGAGAGCTGCTTAAACTCACACAACAATGAACTGC | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | hESCs; A549; HEC-1-A | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000417037.7; ENST00000416282.3; ENST00000484513.1; ENST00000650112.1; ENST00000652590.1; ENST00000396334.8; ENST00000651800.1; ENST00000652534.1; ENST00000421516.3; ENST00000652213.1; ENST00000650905.1 | ||
| External Link | RMBase: m6A_site_583394 | ||
| mod ID: M6ASITE060092 | Click to Show/Hide the Full List | ||
| mod site | chr3:38142578-38142579:+ | [15] | |
| Sequence | TTAAACTCACACAACAATGAACTGCAGACACAGCTGTTCTC | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | hESCs; A549; HEC-1-A | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000396334.8; ENST00000417037.7; ENST00000416282.3; ENST00000650905.1; ENST00000652213.1; ENST00000421516.3; ENST00000650112.1; ENST00000652534.1; ENST00000652590.1; ENST00000651800.1; ENST00000484513.1 | ||
| External Link | RMBase: m6A_site_583395 | ||
| mod ID: M6ASITE060093 | Click to Show/Hide the Full List | ||
| mod site | chr3:38142585-38142586:+ | [15] | |
| Sequence | CACACAACAATGAACTGCAGACACAGCTGTTCTCTCCCTCT | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | hESCs; A549; HEC-1-A | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000650112.1; ENST00000396334.8; ENST00000417037.7; ENST00000416282.3; ENST00000652590.1; ENST00000652534.1; ENST00000650905.1; ENST00000484513.1; ENST00000652213.1; ENST00000421516.3; ENST00000651800.1 | ||
| External Link | RMBase: m6A_site_583396 | ||
References