m6A Target Gene Information
General Information of the m6A Target Gene (ID: M6ATAR00312)
Full List of m6A Methylation Regulator of This Target Gene and Corresponding Disease/Drug Response(s)
PKM
can be regulated by the following regulator(s), and cause disease/drug response(s). You can browse detail information of regulator(s) or disease/drug response(s).
Browse Regulator
Browse Disease
Browse Drug
Fat mass and obesity-associated protein (FTO) [ERASER]
| Representative RNA-seq result indicating the expression of this target gene regulated by FTO | ||
| Cell Line | NB4 cell line | Homo sapiens |
|
Treatment: shFTO NB4 cells
Control: shNS NB4 cells
|
GSE103494 | |
| Regulation |
  |
logFC: 1.52E+00 p-value: 5.79E-04 |
| More Results | Click to View More RNA-seq Results | |
| In total 1 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [1] | |||
| Response Summary | The overexpression of demethylase FTO in the HCC tissue and cells. FTO could regulate the demethylation of Pyruvate kinase PKM (PKM2/PKM) in the hepatocellular carcinoma. | |||
| Target Regulation | Down regulation | |||
| Responsed Disease | Hepatocellular carcinoma | ICD-11: 2C12.02 | ||
| Pathway Response | Central carbon metabolism in cancer | hsa05230 | ||
| Cell Process | Glucose metabolism | |||
| In-vitro Model | Hep 3B2.1-7 | Childhood hepatocellular carcinoma | Homo sapiens | CVCL_0326 |
| Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 | |
| Huh-7 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_0336 | |
| L-02 | Endocervical adenocarcinoma | Homo sapiens | CVCL_6926 | |
| SMMC-7721 | Endocervical adenocarcinoma | Homo sapiens | CVCL_0534 | |
| In-vivo Model | The transfected cells (2×106) were directly subcutaneously injected in to flank of mice. The width and length were measured every six days. After three weeks, the mice were killed and the necropsies were weighted. | |||
YTH domain-containing family protein 1 (YTHDF1) [READER]
| Representative RNA-seq result indicating the expression of this target gene regulated by YTHDF1 | ||
| Cell Line | AGS cell line | Homo sapiens |
|
Treatment: shYTHDF1 AGS
Control: shControl AGS
|
GSE159425 | |
| Regulation |
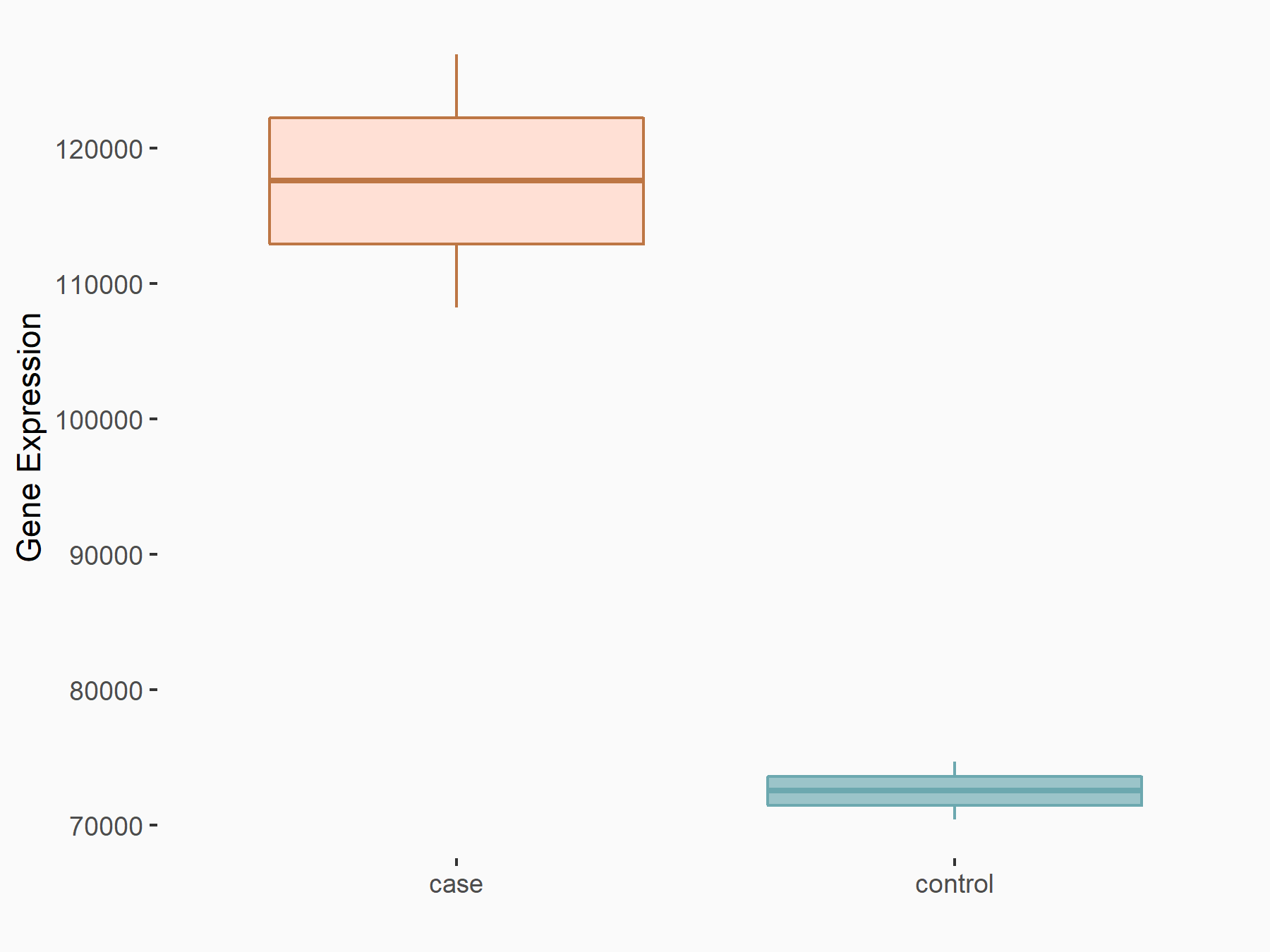  |
logFC: 6.97E-01 p-value: 4.17E-05 |
| More Results | Click to View More RNA-seq Results | |
| In total 1 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [2] | |||
| Response Summary | Tumor hypoxia can transcriptionally induce HIF1alpha and post-transcriptionally inhibit the expression of miR-16-5p to promote YTHDF1 expression, which could sequentially enhance tumor glycolysis by upregulating Pyruvate kinase PKM (PKM2/PKM) and eventually increase the tumorigenesis and metastasis potential of breast cancer cells. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Breast cancer | ICD-11: 2C60 | ||
| Pathway Response | HIF-1 signaling pathway | hsa04066 | ||
| Cell Process | Glycolysis | |||
| In-vitro Model | T-47D | Invasive breast carcinoma | Homo sapiens | CVCL_0553 |
| MDA-MB-231 | Breast adenocarcinoma | Homo sapiens | CVCL_0062 | |
| MCF-7 | Invasive breast carcinoma | Homo sapiens | CVCL_0031 | |
| MCF-10A | Normal | Homo sapiens | CVCL_0598 | |
| BT-474 | Invasive breast carcinoma | Homo sapiens | CVCL_0179 | |
| In-vivo Model | Each mouse was injected subcutaneously with 5 × 106 units of tumor cells to construct the tumor model. | |||
Zinc finger CCCH domain-containing protein 13 (ZC3H13) [WRITER]
| Representative RNA-seq result indicating the expression of this target gene regulated by ZC3H13 | ||
| Cell Line | Human umbilical vein endothelial cells | Homo sapiens |
|
Treatment: siKIAA0853 HUVECs
Control: siControl HUVECs
|
GSE167067 | |
| Regulation |
  |
logFC: -9.34E-01 p-value: 1.41E-02 |
| More Results | Click to View More RNA-seq Results | |
| In total 1 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [3] | |||
| Response Summary | ZC3H13 overexpression sensitized to cisplatin and weakened metabolism reprogramming of HCC cells, ZC3H13-induced m6A modified patterns substantially abolished Pyruvate kinase PKM (PKM2/PKM) mRNA stability. | |||
| Target Regulation | Down regulation | |||
| Responsed Disease | Hepatocellular carcinoma | ICD-11: 2C12.02 | ||
| Responsed Drug | Cisplatin | Approved | ||
| Pathway Response | Central carbon metabolism in cancer | hsa05230 | ||
| Glycolysis / Gluconeogenesis | hsa00010 | |||
| Cell Process | Glycolysis | |||
| In-vitro Model | SMMC-7721 | Endocervical adenocarcinoma | Homo sapiens | CVCL_0534 |
| Huh-7 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_0336 | |
| Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 | |
| Hep 3B2.1-7 | Childhood hepatocellular carcinoma | Homo sapiens | CVCL_0326 | |
Liver cancer [ICD-11: 2C12]
| In total 2 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [1] | |||
| Response Summary | The overexpression of demethylase FTO in the HCC tissue and cells. FTO could regulate the demethylation of Pyruvate kinase PKM (PKM2/PKM) in the hepatocellular carcinoma. | |||
| Responsed Disease | Hepatocellular carcinoma [ICD-11: 2C12.02] | |||
| Target Regulator | Fat mass and obesity-associated protein (FTO) | ERASER | ||
| Target Regulation | Down regulation | |||
| Pathway Response | Central carbon metabolism in cancer | hsa05230 | ||
| Cell Process | Glucose metabolism | |||
| In-vitro Model | Hep 3B2.1-7 | Childhood hepatocellular carcinoma | Homo sapiens | CVCL_0326 |
| Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 | |
| Huh-7 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_0336 | |
| L-02 | Endocervical adenocarcinoma | Homo sapiens | CVCL_6926 | |
| SMMC-7721 | Endocervical adenocarcinoma | Homo sapiens | CVCL_0534 | |
| In-vivo Model | The transfected cells (2×106) were directly subcutaneously injected in to flank of mice. The width and length were measured every six days. After three weeks, the mice were killed and the necropsies were weighted. | |||
| Experiment 2 Reporting the m6A-centered Disease Response | [3] | |||
| Response Summary | ZC3H13 overexpression sensitized to cisplatin and weakened metabolism reprogramming of HCC cells, ZC3H13-induced m6A modified patterns substantially abolished Pyruvate kinase PKM (PKM2/PKM) mRNA stability. | |||
| Responsed Disease | Hepatocellular carcinoma [ICD-11: 2C12.02] | |||
| Target Regulator | Zinc finger CCCH domain-containing protein 13 (ZC3H13) | WRITER | ||
| Target Regulation | Down regulation | |||
| Responsed Drug | Cisplatin | Approved | ||
| Pathway Response | Central carbon metabolism in cancer | hsa05230 | ||
| Glycolysis / Gluconeogenesis | hsa00010 | |||
| Cell Process | Glycolysis | |||
| In-vitro Model | SMMC-7721 | Endocervical adenocarcinoma | Homo sapiens | CVCL_0534 |
| Huh-7 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_0336 | |
| Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 | |
| Hep 3B2.1-7 | Childhood hepatocellular carcinoma | Homo sapiens | CVCL_0326 | |
Breast cancer [ICD-11: 2C60]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [2] | |||
| Response Summary | Tumor hypoxia can transcriptionally induce HIF1alpha and post-transcriptionally inhibit the expression of miR-16-5p to promote YTHDF1 expression, which could sequentially enhance tumor glycolysis by upregulating Pyruvate kinase PKM (PKM2/PKM) and eventually increase the tumorigenesis and metastasis potential of breast cancer cells. | |||
| Responsed Disease | Breast cancer [ICD-11: 2C60] | |||
| Target Regulator | YTH domain-containing family protein 1 (YTHDF1) | READER | ||
| Target Regulation | Up regulation | |||
| Pathway Response | HIF-1 signaling pathway | hsa04066 | ||
| Cell Process | Glycolysis | |||
| In-vitro Model | T-47D | Invasive breast carcinoma | Homo sapiens | CVCL_0553 |
| MDA-MB-231 | Breast adenocarcinoma | Homo sapiens | CVCL_0062 | |
| MCF-7 | Invasive breast carcinoma | Homo sapiens | CVCL_0031 | |
| MCF-10A | Normal | Homo sapiens | CVCL_0598 | |
| BT-474 | Invasive breast carcinoma | Homo sapiens | CVCL_0179 | |
| In-vivo Model | Each mouse was injected subcutaneously with 5 × 106 units of tumor cells to construct the tumor model. | |||
Cisplatin
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response | [3] | |||
| Response Summary | ZC3H13 overexpression sensitized to cisplatin and weakened metabolism reprogramming of HCC cells, ZC3H13-induced m6A modified patterns substantially abolished Pyruvate kinase PKM (PKM2/PKM) mRNA stability. | |||
| Target Regulator | Zinc finger CCCH domain-containing protein 13 (ZC3H13) | WRITER | ||
| Target Regulation | Down regulation | |||
| Responsed Disease | Hepatocellular carcinoma | ICD-11: 2C12.02 | ||
| Pathway Response | Central carbon metabolism in cancer | hsa05230 | ||
| Glycolysis / Gluconeogenesis | hsa00010 | |||
| Cell Process | Glycolysis | |||
| In-vitro Model | SMMC-7721 | Endocervical adenocarcinoma | Homo sapiens | CVCL_0534 |
| Huh-7 | Adult hepatocellular carcinoma | Homo sapiens | CVCL_0336 | |
| Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 | |
| Hep 3B2.1-7 | Childhood hepatocellular carcinoma | Homo sapiens | CVCL_0326 | |
Full List of Crosstalk(s) between m6A Modification and Epigenetic Regulation Related to This Regulator
Histone modification
m6A Regulator: YTH domain-containing family protein 2 (YTHDF2)
| In total 2 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT03418 | ||
| Regulated Target | Histone H3 lysine 4 trimethylation (H3K4me3) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Liver cancer | |
| Crosstalk ID: M6ACROT03429 | ||
| Regulated Target | Histone H3 lysine 27 acetylation (H3K27ac) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Liver cancer | |
Non-coding RNA
m6A Regulator: YTH domain-containing family protein 2 (YTHDF2)
| In total 2 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT05046 | ||
| Epigenetic Regulator | MicroRNA 145 (MIR145) | |
| Regulated Target | YTH domain-containing family protein 2 (YTHDF2) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Liver cancer | |
| Crosstalk ID: M6ACROT05316 | ||
| Epigenetic Regulator | FTO intronic transcript 1 (FTO-IT1) | |
| Regulated Target | FTO alpha-ketoglutarate dependent dioxygenase (FTO) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Liver cancer | |
m6A Regulator: Fat mass and obesity-associated protein (FTO)
| In total 1 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT05314 | ||
| Epigenetic Regulator | FTO intronic transcript 1 (FTO-IT1) | |
| Regulated Target | FTO alpha-ketoglutarate dependent dioxygenase (FTO) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Liver cancer | |
m6A Regulator: YTH domain-containing family protein 1 (YTHDF1)
| In total 1 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT05322 | ||
| Epigenetic Regulator | hsa-miR-16-5p | |
| Regulated Target | YTH domain-containing family protein 1 (YTHDF1) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Breast cancer | |
RNA Modification Sequencing Data Associated with the Target (ID: M6ATAR00312)
| In total 6 m6A sequence/site(s) in this target gene | |||
| mod ID: A2ISITE007040 | Click to Show/Hide the Full List | ||
| mod site | chr15:72202815-72202816:- | [6] | |
| Sequence | ATGTCACAGGCACTTGGTGAAGGACTGGTTTCTGTGGAGTC | ||
| Transcript ID List | ENST00000563986.1; ENST00000565184.5; ENST00000319622.10; ENST00000568459.5; ENST00000564440.5; ENST00000568883.5; ENST00000389093.7; ENST00000449901.6; ENST00000569857.5; ENST00000567118.5; ENST00000335181.10; ENST00000565154.5; ENST00000570166.1; ENST00000561609.5 | ||
| External Link | RMBase: RNA-editing_site_44978 | ||
| mod ID: A2ISITE007041 | Click to Show/Hide the Full List | ||
| mod site | chr15:72203341-72203342:- | [6] | |
| Sequence | AGTTGGACTTTCATCACCCTACCTCTGCATCTGCCTGAAGG | ||
| Transcript ID List | ENST00000569857.5; ENST00000570166.1; ENST00000567118.5; ENST00000565154.5; ENST00000561609.5; ENST00000335181.10; ENST00000568883.5; ENST00000449901.6; ENST00000563986.1; ENST00000568459.5; ENST00000319622.10; ENST00000565184.5; ENST00000564440.5; ENST00000389093.7 | ||
| External Link | RMBase: RNA-editing_site_44979 | ||
| mod ID: A2ISITE007042 | Click to Show/Hide the Full List | ||
| mod site | chr15:72213143-72213144:- | [6] | |
| Sequence | TCCATGACCAAGTAGATTTTATTCTAGGAGAACAAAATTCT | ||
| Transcript ID List | ENST00000562997.5; ENST00000565184.5; ENST00000568883.5; ENST00000567118.5; ENST00000562784.1; ENST00000564276.1; ENST00000335181.10; ENST00000565154.5; ENST00000319622.10; ENST00000567087.5; ENST00000569857.5; ENST00000561609.5; ENST00000389093.7; ENST00000449901.6; ENST00000568459.5; ENST00000564178.5; rmsk_4378892 | ||
| External Link | RMBase: RNA-editing_site_44980 | ||
| mod ID: A2ISITE007043 | Click to Show/Hide the Full List | ||
| mod site | chr15:72224677-72224678:- | [7] | |
| Sequence | TAGTGATGGTGTTTCACCATATTAGCCAGGCTGGTCTCGAA | ||
| Transcript ID List | ENST00000567118.5; ENST00000565184.5; ENST00000449901.6; ENST00000564276.1; ENST00000567087.5; ENST00000335181.10; ENST00000569050.1; ENST00000568459.5; ENST00000561609.5; ENST00000565154.5; ENST00000566809.1; ENST00000568883.5; ENST00000389093.7; ENST00000569857.5; ENST00000319622.10 | ||
| External Link | RMBase: RNA-editing_site_44981 | ||
| mod ID: A2ISITE007044 | Click to Show/Hide the Full List | ||
| mod site | chr15:72224679-72224680:- | [7] | |
| Sequence | AGTAGTGATGGTGTTTCACCATATTAGCCAGGCTGGTCTCG | ||
| Transcript ID List | ENST00000565184.5; ENST00000389093.7; ENST00000568459.5; ENST00000568883.5; ENST00000449901.6; ENST00000319622.10; ENST00000567118.5; ENST00000565154.5; ENST00000569857.5; ENST00000567087.5; ENST00000566809.1; ENST00000335181.10; ENST00000561609.5; ENST00000564276.1; ENST00000569050.1 | ||
| External Link | RMBase: RNA-editing_site_44982 | ||
| mod ID: A2ISITE007045 | Click to Show/Hide the Full List | ||
| mod site | chr15:72224796-72224797:- | [6] | |
| Sequence | CAGTGGTGCGATCTCCGTTCACTGCAGCCTCCACTTCCTGG | ||
| Transcript ID List | ENST00000561609.5; ENST00000389093.7; ENST00000564276.1; ENST00000335181.10; ENST00000569050.1; ENST00000319622.10; ENST00000449901.6; ENST00000568459.5; ENST00000566809.1; ENST00000568883.5; ENST00000569857.5; ENST00000567118.5; ENST00000567087.5; ENST00000565184.5; ENST00000565154.5 | ||
| External Link | RMBase: RNA-editing_site_44983 | ||
5-methylcytidine (m5C)
| In total 9 m6A sequence/site(s) in this target gene | |||
| mod ID: M5CSITE000475 | Click to Show/Hide the Full List | ||
| mod site | chr15:72199594-72199595:- | [8] | |
| Sequence | ACCCCCTTCCCCCAGCCCATCCATTAGGCCAGCAACGCTTG | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000564440.5; ENST00000335181.10; ENST00000449901.6; ENST00000565154.5; ENST00000565184.5; ENST00000389093.7; ENST00000319622.10; ENST00000565143.1; ENST00000567118.5; ENST00000568883.5 | ||
| External Link | RMBase: m5C_site_14290 | ||
| mod ID: M5CSITE000476 | Click to Show/Hide the Full List | ||
| mod site | chr15:72202624-72202625:- | [8] | |
| Sequence | CTCTGGACGGATGTTGCTCCCCTAGATTGCCCGTGAGGCAG | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000567118.5; ENST00000335181.10; ENST00000568459.5; ENST00000564440.5; ENST00000565184.5; ENST00000569857.5; ENST00000568883.5; ENST00000389093.7; ENST00000319622.10; ENST00000561609.5; ENST00000449901.6; ENST00000563986.1; ENST00000565154.5 | ||
| External Link | RMBase: m5C_site_14291 | ||
| mod ID: M5CSITE000477 | Click to Show/Hide the Full List | ||
| mod site | chr15:72202626-72202627:- | [8] | |
| Sequence | CCCTCTGGACGGATGTTGCTCCCCTAGATTGCCCGTGAGGC | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000569857.5; ENST00000564440.5; ENST00000565154.5; ENST00000567118.5; ENST00000335181.10; ENST00000389093.7; ENST00000449901.6; ENST00000319622.10; ENST00000568459.5; ENST00000561609.5; ENST00000563986.1; ENST00000565184.5; ENST00000568883.5 | ||
| External Link | RMBase: m5C_site_14292 | ||
| mod ID: M5CSITE000478 | Click to Show/Hide the Full List | ||
| mod site | chr15:72204236-72204237:- | ||
| Sequence | ACACTGGCCAACAGTTCCTTCCATTAACTGTTCACTGCTTT | ||
| Cell/Tissue List | T24 | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000570166.1; ENST00000568459.5; ENST00000563986.1; ENST00000449901.6; ENST00000389093.7; ENST00000565184.5; ENST00000561609.5; ENST00000564440.5; ENST00000319622.10; ENST00000565154.5; ENST00000335181.10; ENST00000569857.5; ENST00000567118.5; ENST00000568883.5 | ||
| External Link | RMBase: m5C_site_14293 | ||
| mod ID: M5CSITE000479 | Click to Show/Hide the Full List | ||
| mod site | chr15:72206131-72206132:- | [8] | |
| Sequence | GCTGTGCCATTGGGGTACAGCTGCTGCTCTTACTCTAGACC | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000319622.10; ENST00000389093.7; ENST00000565184.5; ENST00000335181.10; ENST00000567118.5; ENST00000561609.5; ENST00000449901.6; ENST00000568459.5; ENST00000563986.1; ENST00000570166.1; ENST00000569857.5; ENST00000568883.5; ENST00000565154.5 | ||
| External Link | RMBase: m5C_site_14294 | ||
| mod ID: M5CSITE000480 | Click to Show/Hide the Full List | ||
| mod site | chr15:72206886-72206887:- | [8] | |
| Sequence | TCTCACTAATGCTCTGTCCCCTCCCAGATGCTGGAGAGCAT | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000319622.10; ENST00000389093.7; ENST00000568883.5; ENST00000561609.5; ENST00000335181.10; ENST00000564993.1; ENST00000563275.1; ENST00000569857.5; ENST00000449901.6; ENST00000565184.5; ENST00000565154.5; ENST00000568459.5; ENST00000567118.5 | ||
| External Link | RMBase: m5C_site_14295 | ||
| mod ID: M5CSITE000481 | Click to Show/Hide the Full List | ||
| mod site | chr15:72209862-72209863:- | [8] | |
| Sequence | TCTTGCTTACTCTCTTGTCCCTAGAGCGGCACTGCAGAGGT | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000562997.5; ENST00000568743.1; ENST00000319622.10; ENST00000564178.5; ENST00000565154.5; ENST00000562784.1; ENST00000449901.6; ENST00000335181.10; ENST00000565184.5; ENST00000567118.5; ENST00000568883.5; ENST00000569857.5; ENST00000561609.5; ENST00000389093.7; ENST00000568459.5; ENST00000567087.5 | ||
| External Link | RMBase: m5C_site_14296 | ||
| mod ID: M5CSITE000482 | Click to Show/Hide the Full List | ||
| mod site | chr15:72219113-72219114:- | [8] | |
| Sequence | CTCTTTCAATTTGTCTCGACCCAGGACCTCAGCAGCCATGT | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000564178.5; ENST00000335181.10; ENST00000567118.5; ENST00000567087.5; ENST00000561609.5; ENST00000568459.5; ENST00000565154.5; ENST00000389093.7; ENST00000566809.1; ENST00000449901.6; ENST00000569050.1; ENST00000565184.5; ENST00000562997.5; ENST00000569857.5; ENST00000319622.10; ENST00000568883.5; ENST00000564276.1 | ||
| External Link | RMBase: m5C_site_14297 | ||
| mod ID: M5CSITE000483 | Click to Show/Hide the Full List | ||
| mod site | chr15:72219117-72219118:- | [8] | |
| Sequence | CACTCTCTTTCAATTTGTCTCGACCCAGGACCTCAGCAGCC | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000335181.10; ENST00000566809.1; ENST00000568459.5; ENST00000449901.6; ENST00000565154.5; ENST00000561609.5; ENST00000565184.5; ENST00000567087.5; ENST00000567118.5; ENST00000568883.5; ENST00000564178.5; ENST00000569857.5; ENST00000562997.5; ENST00000569050.1; ENST00000319622.10; ENST00000564276.1; ENST00000389093.7 | ||
| External Link | RMBase: m5C_site_14298 | ||
N6-methyladenosine (m6A)
| In total 62 m6A sequence/site(s) in this target gene | |||
| mod ID: M6ASITE024058 | Click to Show/Hide the Full List | ||
| mod site | chr15:72199044-72199045:- | [9] | |
| Sequence | GCCTACCTGTATGTCAATAAACAACAGCTGAAGCACCTGTT | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa; HEK293T; hESC-HEK293T; HepG2; H1299; Huh7; TREX | ||
| Seq Type List | m6A-seq; MeRIP-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000568883.5; ENST00000565143.1; ENST00000565184.5; ENST00000389093.7; ENST00000335181.10; ENST00000319622.10; ENST00000564440.5; ENST00000449901.6 | ||
| External Link | RMBase: m6A_site_286174 | ||
| mod ID: M6ASITE024059 | Click to Show/Hide the Full List | ||
| mod site | chr15:72199095-72199096:- | [10] | |
| Sequence | ACCTTCCATTTTCCCCCACTACTGCAGCACCTCCAGGCCTG | ||
| Motif Score | 2.500660714 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000565184.5; ENST00000565143.1; ENST00000335181.10; ENST00000564440.5; ENST00000568883.5; ENST00000449901.6; ENST00000389093.7; ENST00000319622.10 | ||
| External Link | RMBase: m6A_site_286175 | ||
| mod ID: M6ASITE024060 | Click to Show/Hide the Full List | ||
| mod site | chr15:72199098-72199099:- | [10] | |
| Sequence | TCCACCTTCCATTTTCCCCCACTACTGCAGCACCTCCAGGC | ||
| Motif Score | 2.475107143 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000565143.1; ENST00000449901.6; ENST00000564440.5; ENST00000319622.10; ENST00000389093.7; ENST00000565184.5; ENST00000335181.10; ENST00000568883.5 | ||
| External Link | RMBase: m6A_site_286176 | ||
| mod ID: M6ASITE024061 | Click to Show/Hide the Full List | ||
| mod site | chr15:72199129-72199130:- | [9] | |
| Sequence | ACTCAGCTGTCCTGCAGCAAACACTCCACCCTCCACCTTCC | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa; HEK293T; hESC-HEK293T; HepG2; A549; H1299; Huh7; TREX; endometrial | ||
| Seq Type List | m6A-seq; MeRIP-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000319622.10; ENST00000335181.10; ENST00000564440.5; ENST00000567118.5; ENST00000568883.5; ENST00000389093.7; ENST00000565184.5; ENST00000565143.1; ENST00000449901.6 | ||
| External Link | RMBase: m6A_site_286177 | ||
| mod ID: M6ASITE024062 | Click to Show/Hide the Full List | ||
| mod site | chr15:72199221-72199222:- | [10] | |
| Sequence | CTGTTGTTCCATTGAAGCCGACTCTGGCCCTGGCCCTTACT | ||
| Motif Score | 3.287565476 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000335181.10; ENST00000389093.7; ENST00000565143.1; ENST00000567118.5; ENST00000564440.5; ENST00000565184.5; ENST00000568883.5; ENST00000319622.10; ENST00000449901.6 | ||
| External Link | RMBase: m6A_site_286178 | ||
| mod ID: M6ASITE024063 | Click to Show/Hide the Full List | ||
| mod site | chr15:72199277-72199278:- | [9] | |
| Sequence | CCCTGGCTTGGGGTCAAGAAACAGCCAGCAAGAGTTAGGGG | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; liver; H1A; H1B; A549; H1299; MM6; Huh7; peripheral-blood; TREX; endometrial; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq; m6A-REF-seq | ||
| Transcript ID List | ENST00000567118.5; ENST00000319622.10; ENST00000564440.5; ENST00000449901.6; ENST00000335181.10; ENST00000389093.7; ENST00000568883.5; ENST00000565184.5; ENST00000565143.1 | ||
| External Link | RMBase: m6A_site_286179 | ||
| mod ID: M6ASITE024064 | Click to Show/Hide the Full List | ||
| mod site | chr15:72199305-72199306:- | [9] | |
| Sequence | AAAAAATGGATGCCCAGAGGACTCCCAACCCTGGCTTGGGG | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; H1A; H1B; A549; H1299; MM6; Huh7; peripheral-blood; TREX; endometrial; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000335181.10; ENST00000564440.5; ENST00000565143.1; ENST00000568883.5; ENST00000449901.6; ENST00000319622.10; ENST00000565184.5; ENST00000567118.5; ENST00000389093.7 | ||
| External Link | RMBase: m6A_site_286180 | ||
| mod ID: M6ASITE024065 | Click to Show/Hide the Full List | ||
| mod site | chr15:72199345-72199346:- | [10] | |
| Sequence | CAGCTTCCTTTCCTGTGTGTACTCTGTCCAGTTCCTTTAGA | ||
| Motif Score | 3.278136905 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000449901.6; ENST00000565184.5; ENST00000568883.5; ENST00000335181.10; ENST00000319622.10; ENST00000564440.5; ENST00000389093.7; ENST00000565143.1; ENST00000567118.5 | ||
| External Link | RMBase: m6A_site_286181 | ||
| mod ID: M6ASITE024066 | Click to Show/Hide the Full List | ||
| mod site | chr15:72199366-72199367:- | [11] | |
| Sequence | AGCCAGATGGCAAGAGGGTGACAGCTTCCTTTCCTGTGTGT | ||
| Motif Score | 2.859755952 | ||
| Cell/Tissue List | liver; HEK293T | ||
| Seq Type List | m6A-REF-seq; DART-seq | ||
| Transcript ID List | ENST00000389093.7; ENST00000565143.1; ENST00000564440.5; ENST00000568883.5; ENST00000319622.10; ENST00000449901.6; ENST00000567118.5; ENST00000565184.5; ENST00000335181.10 | ||
| External Link | RMBase: m6A_site_286182 | ||
| mod ID: M6ASITE024067 | Click to Show/Hide the Full List | ||
| mod site | chr15:72199401-72199402:- | [9] | |
| Sequence | GGAAGAAGGAGGAATGCTGGACTGGAGGCCCCTGGAGCCAG | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; H1A; H1B; A549; H1299; MM6; peripheral-blood; TREX; endometrial; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq; m6A-CLIP/IP | ||
| Transcript ID List | ENST00000319622.10; ENST00000389093.7; ENST00000449901.6; ENST00000564440.5; ENST00000567118.5; ENST00000565143.1; ENST00000565184.5; ENST00000568883.5; ENST00000335181.10 | ||
| External Link | RMBase: m6A_site_286183 | ||
| mod ID: M6ASITE024068 | Click to Show/Hide the Full List | ||
| mod site | chr15:72199467-72199468:- | [9] | |
| Sequence | TTGCAGCCTGCTCTAGTGGGACAGCCCAGAGCCTGGCTGCC | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; hESC-HEK293T; H1A; H1B; A549; H1299; MM6; Huh7; peripheral-blood; TREX; endometrial; HEC-1-A; AML | ||
| Seq Type List | m6A-seq; MeRIP-seq; MAZTER-seq; miCLIP | ||
| Transcript ID List | ENST00000568883.5; ENST00000319622.10; ENST00000389093.7; ENST00000565184.5; ENST00000567118.5; ENST00000449901.6; ENST00000565143.1; ENST00000564440.5; ENST00000335181.10 | ||
| External Link | RMBase: m6A_site_286184 | ||
| mod ID: M6ASITE024069 | Click to Show/Hide the Full List | ||
| mod site | chr15:72199499-72199500:- | [9] | |
| Sequence | AAGATCAACGCCTCACTGAAACATGGCTGTGTTTGCAGCCT | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; hESC-HEK293T; H1A; H1B; A549; H1299; MM6; Huh7; peripheral-blood; TREX; endometrial; AML | ||
| Seq Type List | m6A-seq; MeRIP-seq; MAZTER-seq; miCLIP | ||
| Transcript ID List | ENST00000564440.5; ENST00000565143.1; ENST00000389093.7; ENST00000319622.10; ENST00000568883.5; ENST00000335181.10; ENST00000449901.6; ENST00000567118.5; ENST00000565184.5 | ||
| External Link | RMBase: m6A_site_286185 | ||
| mod ID: M6ASITE024070 | Click to Show/Hide the Full List | ||
| mod site | chr15:72199531-72199532:- | [9] | |
| Sequence | CGTGGCACTGGTAGGTTGGGACACCAGGGAAGAAGATCAAC | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; hESC-HEK293T; H1A; H1B; A549; H1299; MM6; Huh7; peripheral-blood; TREX; endometrial | ||
| Seq Type List | m6A-seq; MeRIP-seq; m6A-REF-seq; MAZTER-seq; m6A-CLIP/IP | ||
| Transcript ID List | ENST00000449901.6; ENST00000567118.5; ENST00000568883.5; ENST00000389093.7; ENST00000564440.5; ENST00000319622.10; ENST00000565143.1; ENST00000565184.5; ENST00000335181.10 | ||
| External Link | RMBase: m6A_site_286186 | ||
| mod ID: M6ASITE024071 | Click to Show/Hide the Full List | ||
| mod site | chr15:72199552-72199553:- | [12] | |
| Sequence | AGAACTCACTCTGGGCTGTAACGTGGCACTGGTAGGTTGGG | ||
| Motif Score | 2.142029762 | ||
| Cell/Tissue List | A549 | ||
| Seq Type List | m6A-CLIP/IP | ||
| Transcript ID List | ENST00000449901.6; ENST00000335181.10; ENST00000319622.10; ENST00000389093.7; ENST00000567118.5; ENST00000565143.1; ENST00000564440.5; ENST00000568883.5; ENST00000565184.5 | ||
| External Link | RMBase: m6A_site_286187 | ||
| mod ID: M6ASITE024072 | Click to Show/Hide the Full List | ||
| mod site | chr15:72199565-72199566:- | [10] | |
| Sequence | CAGCAACGCTTGTAGAACTCACTCTGGGCTGTAACGTGGCA | ||
| Motif Score | 2.469291667 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000389093.7; ENST00000567118.5; ENST00000565184.5; ENST00000319622.10; ENST00000449901.6; ENST00000335181.10; ENST00000568883.5; ENST00000564440.5; ENST00000565143.1 | ||
| External Link | RMBase: m6A_site_286188 | ||
| mod ID: M6ASITE024073 | Click to Show/Hide the Full List | ||
| mod site | chr15:72199569-72199570:- | [9] | |
| Sequence | AGGCCAGCAACGCTTGTAGAACTCACTCTGGGCTGTAACGT | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; CD8T; A549; H1299; MM6; Huh7; peripheral-blood; TREX; endometrial; AML | ||
| Seq Type List | m6A-seq; MeRIP-seq; m6A-CLIP/IP; miCLIP | ||
| Transcript ID List | ENST00000335181.10; ENST00000449901.6; ENST00000565143.1; ENST00000567118.5; ENST00000319622.10; ENST00000568883.5; ENST00000389093.7; ENST00000564440.5; ENST00000565184.5 | ||
| External Link | RMBase: m6A_site_286189 | ||
| mod ID: M6ASITE024074 | Click to Show/Hide the Full List | ||
| mod site | chr15:72199580-72199581:- | [12] | |
| Sequence | GCCCATCCATTAGGCCAGCAACGCTTGTAGAACTCACTCTG | ||
| Motif Score | 2.147845238 | ||
| Cell/Tissue List | A549 | ||
| Seq Type List | m6A-CLIP/IP | ||
| Transcript ID List | ENST00000449901.6; ENST00000319622.10; ENST00000568883.5; ENST00000564440.5; ENST00000565184.5; ENST00000389093.7; ENST00000567118.5; ENST00000335181.10; ENST00000565143.1 | ||
| External Link | RMBase: m6A_site_286190 | ||
| mod ID: M6ASITE024075 | Click to Show/Hide the Full List | ||
| mod site | chr15:72199645-72199646:- | [13] | |
| Sequence | TTGTTCCTGTGCCGTGATGGACCCCAGAGCCCCTCCTCCAG | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HepG2; A549; MM6; Huh7; peripheral-blood; endometrial | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000568883.5; ENST00000565154.5; ENST00000389093.7; ENST00000567118.5; ENST00000565143.1; ENST00000564440.5; ENST00000568459.5; ENST00000565184.5; ENST00000319622.10; ENST00000449901.6; ENST00000335181.10 | ||
| External Link | RMBase: m6A_site_286191 | ||
| mod ID: M6ASITE024076 | Click to Show/Hide the Full List | ||
| mod site | chr15:72199677-72199678:- | [14] | |
| Sequence | CCCTGGCTCCGGCTTCACCAACACCATGCGTGTTGTTCCTG | ||
| Motif Score | 2.173910714 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000565184.5; ENST00000335181.10; ENST00000565143.1; ENST00000564440.5; ENST00000568883.5; ENST00000319622.10; ENST00000568459.5; ENST00000389093.7; ENST00000565154.5; ENST00000449901.6; ENST00000567118.5 | ||
| External Link | RMBase: m6A_site_286192 | ||
| mod ID: M6ASITE024077 | Click to Show/Hide the Full List | ||
| mod site | chr15:72200490-72200491:- | [15] | |
| Sequence | GGACGTGGACCTCCGGGTGAACTTTGCCATGAATGTTGGTA | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | CD34; HepG2; peripheral-blood; endometrial | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000335181.10; ENST00000568883.5; ENST00000567118.5; ENST00000565154.5; ENST00000319622.10; ENST00000389093.7; ENST00000565143.1; ENST00000568459.5; ENST00000565184.5; ENST00000564440.5; ENST00000449901.6 | ||
| External Link | RMBase: m6A_site_286193 | ||
| mod ID: M6ASITE024078 | Click to Show/Hide the Full List | ||
| mod site | chr15:72200502-72200503:- | [13] | |
| Sequence | GGCCTGGGCTGAGGACGTGGACCTCCGGGTGAACTTTGCCA | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HepG2; peripheral-blood; endometrial | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000389093.7; ENST00000564440.5; ENST00000565154.5; ENST00000568883.5; ENST00000565184.5; ENST00000568459.5; ENST00000335181.10; ENST00000567118.5; ENST00000565143.1; ENST00000319622.10; ENST00000449901.6 | ||
| External Link | RMBase: m6A_site_286194 | ||
| mod ID: M6ASITE024079 | Click to Show/Hide the Full List | ||
| mod site | chr15:72200535-72200536:- | [13] | |
| Sequence | CTTCCCTGTGCTGTGCAAGGACCCAGTCCAGGAGGCCTGGG | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HepG2; MT4; peripheral-blood | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000568459.5; ENST00000335181.10; ENST00000449901.6; ENST00000565143.1; ENST00000565184.5; ENST00000565154.5; ENST00000567118.5; ENST00000389093.7; ENST00000564440.5; ENST00000561609.5; ENST00000319622.10; ENST00000568883.5 | ||
| External Link | RMBase: m6A_site_286195 | ||
| mod ID: M6ASITE024080 | Click to Show/Hide the Full List | ||
| mod site | chr15:72200587-72200588:- | [13] | |
| Sequence | CTGTGACCCGGAATCCCCAGACAGCTCGTCAGGCCCACCTG | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HepG2; MT4; peripheral-blood | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000449901.6; ENST00000565184.5; ENST00000335181.10; ENST00000568883.5; ENST00000568459.5; ENST00000561609.5; ENST00000564440.5; ENST00000319622.10; ENST00000389093.7; ENST00000565154.5; ENST00000567118.5; ENST00000565143.1 | ||
| External Link | RMBase: m6A_site_286196 | ||
| mod ID: M6ASITE024081 | Click to Show/Hide the Full List | ||
| mod site | chr15:72201409-72201410:- | [9] | |
| Sequence | GCCCCTCCCCAGCTCCTGAGACACAGCTGCAGCTACAAGTG | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000561609.5; ENST00000449901.6; ENST00000568883.5; ENST00000564440.5; ENST00000389093.7; ENST00000335181.10; ENST00000565143.1; ENST00000567118.5; ENST00000565154.5; ENST00000319622.10; ENST00000565184.5; ENST00000568459.5 | ||
| External Link | RMBase: m6A_site_286197 | ||
| mod ID: M6ASITE024082 | Click to Show/Hide the Full List | ||
| mod site | chr15:72202184-72202185:- | [10] | |
| Sequence | ACATAACAGTGCACAGGCTGACTACAAATGGTTATTTGATA | ||
| Motif Score | 3.28175 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000565184.5; ENST00000561609.5; ENST00000563986.1; ENST00000567118.5; ENST00000568459.5; ENST00000319622.10; ENST00000564440.5; ENST00000449901.6; ENST00000565154.5; ENST00000389093.7; ENST00000335181.10; ENST00000568883.5 | ||
| External Link | RMBase: m6A_site_286198 | ||
| mod ID: M6ASITE024083 | Click to Show/Hide the Full List | ||
| mod site | chr15:72202569-72202570:- | [9] | |
| Sequence | CACTTGCAATTATTTGAGGAACTCCGCCGCCTGGCGCCCAT | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa; HepG2; Huh7; endometrial | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000561609.5; ENST00000568459.5; ENST00000565154.5; ENST00000335181.10; ENST00000567118.5; ENST00000568883.5; ENST00000389093.7; ENST00000563986.1; ENST00000565184.5; ENST00000319622.10; ENST00000449901.6; ENST00000569857.5; ENST00000564440.5 | ||
| External Link | RMBase: m6A_site_286199 | ||
| mod ID: M6ASITE024084 | Click to Show/Hide the Full List | ||
| mod site | chr15:72202732-72202733:- | [9] | |
| Sequence | CTTGGCCTTTGTTCCCAGGAACATGTTCCTCACCAGCTGTC | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000568459.5; ENST00000335181.10; ENST00000319622.10; ENST00000389093.7; ENST00000564440.5; ENST00000570166.1; ENST00000569857.5; ENST00000568883.5; ENST00000563986.1; ENST00000561609.5; ENST00000565154.5; ENST00000449901.6; ENST00000565184.5; ENST00000567118.5 | ||
| External Link | RMBase: m6A_site_286200 | ||
| mod ID: M6ASITE024085 | Click to Show/Hide the Full List | ||
| mod site | chr15:72203112-72203113:- | [11] | |
| Sequence | TGCGAGCCTCAAGTCACTCCACAGACCTCATGGAAGCCATG | ||
| Motif Score | 2.053113095 | ||
| Cell/Tissue List | brain; HEK293 | ||
| Seq Type List | m6A-REF-seq | ||
| Transcript ID List | ENST00000563986.1; ENST00000319622.10; ENST00000565154.5; ENST00000389093.7; ENST00000449901.6; ENST00000570166.1; ENST00000567118.5; ENST00000335181.10; ENST00000568459.5; ENST00000564440.5; ENST00000565184.5; ENST00000561609.5; ENST00000569857.5; ENST00000568883.5 | ||
| External Link | RMBase: m6A_site_286201 | ||
| mod ID: M6ASITE024086 | Click to Show/Hide the Full List | ||
| mod site | chr15:72203320-72203321:- | [9] | |
| Sequence | CCTCTGCATCTGCCTGAAGGACAGATTTAGCCAATTAACCT | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000389093.7; ENST00000563986.1; ENST00000561609.5; ENST00000568459.5; ENST00000564440.5; ENST00000570166.1; ENST00000567118.5; ENST00000335181.10; ENST00000449901.6; ENST00000565154.5; ENST00000568883.5; ENST00000565184.5; ENST00000569857.5; ENST00000319622.10 | ||
| External Link | RMBase: m6A_site_286202 | ||
| mod ID: M6ASITE024087 | Click to Show/Hide the Full List | ||
| mod site | chr15:72203355-72203356:- | [9] | |
| Sequence | GCACCTCCTGAAGCAGTTGGACTTTCATCACCCTACCTCTG | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000563986.1; ENST00000449901.6; ENST00000570166.1; ENST00000561609.5; ENST00000567118.5; ENST00000319622.10; ENST00000569857.5; ENST00000565154.5; ENST00000389093.7; ENST00000565184.5; ENST00000335181.10; ENST00000568883.5; ENST00000568459.5; ENST00000564440.5 | ||
| External Link | RMBase: m6A_site_286203 | ||
| mod ID: M6ASITE024088 | Click to Show/Hide the Full List | ||
| mod site | chr15:72206761-72206762:- | [9] | |
| Sequence | TGGAGAAACAGCCAAAGGGGACTATCCTCTGGAGGCTGTGC | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa; CD34; HepG2; peripheral-blood; HEK293T; endometrial; NB4; MM6 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000570166.1; ENST00000564993.1; ENST00000389093.7; ENST00000569857.5; ENST00000565184.5; ENST00000567118.5; ENST00000565154.5; ENST00000568883.5; ENST00000568459.5; ENST00000561609.5; ENST00000335181.10; ENST00000563986.1; ENST00000563275.1; ENST00000319622.10; ENST00000449901.6 | ||
| External Link | RMBase: m6A_site_286204 | ||
| mod ID: M6ASITE024089 | Click to Show/Hide the Full List | ||
| mod site | chr15:72206774-72206775:- | [9] | |
| Sequence | GCATCATGCTGTCTGGAGAAACAGCCAAAGGGGACTATCCT | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa; CD34; HepG2; peripheral-blood; HEK293T; endometrial; NB4; MM6 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000565184.5; ENST00000568883.5; ENST00000568459.5; ENST00000563275.1; ENST00000319622.10; ENST00000570166.1; ENST00000449901.6; ENST00000567118.5; ENST00000564993.1; ENST00000561609.5; ENST00000335181.10; ENST00000389093.7; ENST00000569857.5; ENST00000565154.5; ENST00000563986.1 | ||
| External Link | RMBase: m6A_site_286205 | ||
| mod ID: M6ASITE024090 | Click to Show/Hide the Full List | ||
| mod site | chr15:72208665-72208666:- | [9] | |
| Sequence | CCTGGGAGAGAAGGGAAAGAACATCAAGATTATCAGCAAAA | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa; hESC-HEK293T; HepG2; peripheral-blood; endometrial; NB4; MM6 | ||
| Seq Type List | m6A-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000568883.5; ENST00000568459.5; ENST00000449901.6; ENST00000565154.5; ENST00000561609.5; ENST00000569857.5; ENST00000567118.5; ENST00000389093.7; ENST00000319622.10; ENST00000565184.5; ENST00000335181.10; ENST00000568743.1; ENST00000563275.1 | ||
| External Link | RMBase: m6A_site_286206 | ||
| mod ID: M6ASITE024091 | Click to Show/Hide the Full List | ||
| mod site | chr15:72208782-72208783:- | [9] | |
| Sequence | GCCTGCTGTGTCGGAGAAGGACATCCAGGATCTGAAGTTTG | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; hESC-HEK293T; HepG2; peripheral-blood; endometrial; NB4; MM6 | ||
| Seq Type List | m6A-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000568883.5; ENST00000319622.10; ENST00000335181.10; ENST00000563275.1; ENST00000569857.5; ENST00000567118.5; ENST00000565154.5; ENST00000568743.1; ENST00000568459.5; ENST00000449901.6; ENST00000561609.5; ENST00000565184.5; ENST00000389093.7 | ||
| External Link | RMBase: m6A_site_286207 | ||
| mod ID: M6ASITE024092 | Click to Show/Hide the Full List | ||
| mod site | chr15:72208806-72208807:- | [9] | |
| Sequence | CCTTCCTGGGGCTGCTGTGGACTTGCCTGCTGTGTCGGAGA | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa; HepG2; peripheral-blood; endometrial; NB4; MM6; AML | ||
| Seq Type List | m6A-seq; miCLIP | ||
| Transcript ID List | ENST00000568459.5; ENST00000565154.5; ENST00000449901.6; ENST00000565184.5; ENST00000563275.1; ENST00000561609.5; ENST00000335181.10; ENST00000569857.5; ENST00000568883.5; ENST00000319622.10; ENST00000567118.5; ENST00000568743.1; ENST00000389093.7 | ||
| External Link | RMBase: m6A_site_286208 | ||
| mod ID: M6ASITE024093 | Click to Show/Hide the Full List | ||
| mod site | chr15:72208827-72208828:- | [9] | |
| Sequence | GGGCAGCAAGAAGGGTGTGAACCTTCCTGGGGCTGCTGTGG | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | HeLa; HepG2; peripheral-blood; endometrial; NB4; MM6 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000563275.1; ENST00000567118.5; ENST00000568883.5; ENST00000389093.7; ENST00000335181.10; ENST00000565154.5; ENST00000568743.1; ENST00000319622.10; ENST00000568459.5; ENST00000565184.5; ENST00000561609.5; ENST00000569857.5; ENST00000449901.6 | ||
| External Link | RMBase: m6A_site_286209 | ||
| mod ID: M6ASITE024094 | Click to Show/Hide the Full List | ||
| mod site | chr15:72209620-72209621:- | [9] | |
| Sequence | AAACAGTCTTTTGTCTCTAAACTTCCTTGACACAAGGAAGA | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa; HepG2 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000389093.7; ENST00000569857.5; ENST00000565184.5; ENST00000335181.10; ENST00000449901.6; ENST00000319622.10; ENST00000568743.1; ENST00000565154.5; ENST00000567118.5; ENST00000568459.5; ENST00000568883.5; ENST00000562784.1; ENST00000561609.5 | ||
| External Link | RMBase: m6A_site_286210 | ||
| mod ID: M6ASITE024095 | Click to Show/Hide the Full List | ||
| mod site | chr15:72209638-72209639:- | [9] | |
| Sequence | CTGGAGTCCAGTTGTCTAAAACAGTCTTTTGTCTCTAAACT | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa; HepG2; peripheral-blood; MM6 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000567118.5; ENST00000569857.5; ENST00000568743.1; ENST00000449901.6; ENST00000568459.5; ENST00000335181.10; ENST00000389093.7; ENST00000565154.5; ENST00000561609.5; ENST00000565184.5; ENST00000568883.5; ENST00000319622.10; ENST00000562784.1 | ||
| External Link | RMBase: m6A_site_286211 | ||
| mod ID: M6ASITE024096 | Click to Show/Hide the Full List | ||
| mod site | chr15:72209749-72209750:- | [9] | |
| Sequence | CCTGTGGCTGGACTACAAGAACATCTGCAAGGTGGTGGAAG | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa; hESC-HEK293T; HepG2; peripheral-blood; endometrial; NB4; MM6 | ||
| Seq Type List | m6A-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000389093.7; ENST00000568459.5; ENST00000335181.10; ENST00000568743.1; ENST00000567118.5; ENST00000319622.10; ENST00000562784.1; ENST00000565184.5; ENST00000561609.5; ENST00000449901.6; ENST00000569857.5; ENST00000565154.5; ENST00000568883.5; ENST00000564178.5 | ||
| External Link | RMBase: m6A_site_286212 | ||
| mod ID: M6ASITE024097 | Click to Show/Hide the Full List | ||
| mod site | chr15:72209758-72209759:- | [9] | |
| Sequence | CGAGAACATCCTGTGGCTGGACTACAAGAACATCTGCAAGG | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa; HepG2; peripheral-blood; HEK293T; endometrial; NB4; MM6 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000568743.1; ENST00000335181.10; ENST00000449901.6; ENST00000319622.10; ENST00000568459.5; ENST00000561609.5; ENST00000562784.1; ENST00000567118.5; ENST00000565184.5; ENST00000565154.5; ENST00000564178.5; ENST00000568883.5; ENST00000562997.5; ENST00000569857.5; ENST00000389093.7 | ||
| External Link | RMBase: m6A_site_286213 | ||
| mod ID: M6ASITE024098 | Click to Show/Hide the Full List | ||
| mod site | chr15:72209773-72209774:- | [9] | |
| Sequence | CATGGAAAAGTGTGACGAGAACATCCTGTGGCTGGACTACA | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa; kidney; liver; HepG2; peripheral-blood; HEK293T; endometrial; NB4; MM6 | ||
| Seq Type List | m6A-seq; m6A-REF-seq | ||
| Transcript ID List | ENST00000568743.1; ENST00000565184.5; ENST00000568459.5; ENST00000562784.1; ENST00000568883.5; ENST00000561609.5; ENST00000319622.10; ENST00000567118.5; ENST00000449901.6; ENST00000564178.5; ENST00000335181.10; ENST00000389093.7; ENST00000569857.5; ENST00000562997.5; ENST00000565154.5 | ||
| External Link | RMBase: m6A_site_286214 | ||
| mod ID: M6ASITE024099 | Click to Show/Hide the Full List | ||
| mod site | chr15:72209794-72209795:- | [11] | |
| Sequence | AATCACGCTGGATAACGCCTACATGGAAAAGTGTGACGAGA | ||
| Motif Score | 2.078666667 | ||
| Cell/Tissue List | kidney; hESC-HEK293T | ||
| Seq Type List | m6A-REF-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000389093.7; ENST00000562784.1; ENST00000561609.5; ENST00000568883.5; ENST00000569857.5; ENST00000335181.10; ENST00000562997.5; ENST00000567118.5; ENST00000567087.5; ENST00000319622.10; ENST00000568459.5; ENST00000564178.5; ENST00000565154.5; ENST00000568743.1; ENST00000449901.6; ENST00000565184.5 | ||
| External Link | RMBase: m6A_site_286215 | ||
| mod ID: M6ASITE024100 | Click to Show/Hide the Full List | ||
| mod site | chr15:72209884-72209885:- | [9] | |
| Sequence | TACTGGTGTCTCCAGTTTGGACTCTTGCTTACTCTCTTGTC | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa; HepG2; peripheral-blood; HEK293T; endometrial; NB4; MM6 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000562784.1; ENST00000568883.5; ENST00000567087.5; ENST00000567118.5; ENST00000568459.5; ENST00000449901.6; ENST00000564178.5; ENST00000561609.5; ENST00000565184.5; ENST00000335181.10; ENST00000319622.10; ENST00000565154.5; ENST00000568743.1; ENST00000389093.7; ENST00000569857.5; ENST00000562997.5 | ||
| External Link | RMBase: m6A_site_286216 | ||
| mod ID: M6ASITE024101 | Click to Show/Hide the Full List | ||
| mod site | chr15:72209983-72209984:- | [9] | |
| Sequence | TTAGAGAATAAGAGCAAAAAACTGTATTACTTTTAGCAGTG | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000561609.5; ENST00000565184.5; ENST00000567087.5; ENST00000449901.6; ENST00000335181.10; ENST00000568883.5; ENST00000319622.10; ENST00000389093.7; ENST00000564178.5; ENST00000568459.5; ENST00000562784.1; ENST00000562997.5; ENST00000567118.5; ENST00000569857.5; ENST00000565154.5; ENST00000568743.1 | ||
| External Link | RMBase: m6A_site_286217 | ||
| mod ID: M6ASITE024102 | Click to Show/Hide the Full List | ||
| mod site | chr15:72210291-72210292:- | [16] | |
| Sequence | GTAGGCAATATGCCCCAGAGACATGTCCTCCAAAGCGCTGG | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HepG2 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000568459.5; ENST00000319622.10; ENST00000562997.5; ENST00000561609.5; ENST00000564276.1; ENST00000562784.1; ENST00000389093.7; ENST00000565154.5; ENST00000564178.5; ENST00000567087.5; ENST00000568883.5; ENST00000569857.5; ENST00000335181.10; ENST00000449901.6; ENST00000565184.5; ENST00000567118.5 | ||
| External Link | RMBase: m6A_site_286218 | ||
| mod ID: M6ASITE024103 | Click to Show/Hide the Full List | ||
| mod site | chr15:72210363-72210364:- | [9] | |
| Sequence | CTAAAGGACCTGAGATCCGAACTGGGCTCATCAAGGGCGTG | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa; HepG2; peripheral-blood; endometrial | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000564276.1; ENST00000567118.5; ENST00000564178.5; ENST00000568459.5; ENST00000561609.5; ENST00000562784.1; ENST00000449901.6; ENST00000335181.10; ENST00000562997.5; ENST00000568883.5; ENST00000319622.10; ENST00000569857.5; ENST00000565154.5; ENST00000565184.5; ENST00000389093.7; ENST00000567087.5 | ||
| External Link | RMBase: m6A_site_286219 | ||
| mod ID: M6ASITE024104 | Click to Show/Hide the Full List | ||
| mod site | chr15:72210376-72210377:- | [9] | |
| Sequence | GTGGCTCTAGACACTAAAGGACCTGAGATCCGAACTGGGCT | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; HepG2; peripheral-blood; endometrial | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000568459.5; ENST00000564178.5; ENST00000565154.5; ENST00000564276.1; ENST00000569857.5; ENST00000389093.7; ENST00000449901.6; ENST00000561609.5; ENST00000565184.5; ENST00000335181.10; ENST00000567118.5; ENST00000568883.5; ENST00000562784.1; ENST00000319622.10; ENST00000567087.5; ENST00000562997.5 | ||
| External Link | RMBase: m6A_site_286220 | ||
| mod ID: M6ASITE024105 | Click to Show/Hide the Full List | ||
| mod site | chr15:72210386-72210387:- | [9] | |
| Sequence | GCCCGTTGCTGTGGCTCTAGACACTAAAGGACCTGAGATCC | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa; hESC-HEK293T; HepG2; peripheral-blood; endometrial | ||
| Seq Type List | m6A-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000562997.5; ENST00000389093.7; ENST00000562784.1; ENST00000449901.6; ENST00000564276.1; ENST00000565154.5; ENST00000569857.5; ENST00000568459.5; ENST00000565184.5; ENST00000567087.5; ENST00000568883.5; ENST00000561609.5; ENST00000319622.10; ENST00000567118.5; ENST00000335181.10; ENST00000564178.5 | ||
| External Link | RMBase: m6A_site_286221 | ||
| mod ID: M6ASITE024106 | Click to Show/Hide the Full List | ||
| mod site | chr15:72210465-72210466:- | [9] | |
| Sequence | TCCCCCAGTACCATGCGGAGACCATCAAGAATGTGCGCACA | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa; HepG2; peripheral-blood; endometrial; AML | ||
| Seq Type List | m6A-seq; miCLIP | ||
| Transcript ID List | ENST00000449901.6; ENST00000569857.5; ENST00000335181.10; ENST00000389093.7; ENST00000319622.10; ENST00000568883.5; ENST00000564178.5; ENST00000562997.5; ENST00000564276.1; ENST00000567118.5; ENST00000565184.5; ENST00000567087.5; ENST00000565154.5; ENST00000562784.1; ENST00000561609.5; ENST00000568459.5 | ||
| External Link | RMBase: m6A_site_286222 | ||
| mod ID: M6ASITE024107 | Click to Show/Hide the Full List | ||
| mod site | chr15:72217416-72217417:- | [9] | |
| Sequence | GTCTGAACTTCTCTCATGGAACTCATGAGGTGAGCTGTGGC | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa; HepG2; peripheral-blood; endometrial | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000566809.1; ENST00000562784.1; ENST00000567087.5; ENST00000319622.10; ENST00000561609.5; ENST00000335181.10; ENST00000564178.5; ENST00000569857.5; ENST00000565154.5; ENST00000568883.5; ENST00000564276.1; ENST00000389093.7; ENST00000449901.6; ENST00000568459.5; ENST00000567118.5; ENST00000562997.5; ENST00000565184.5 | ||
| External Link | RMBase: m6A_site_286223 | ||
| mod ID: M6ASITE024108 | Click to Show/Hide the Full List | ||
| mod site | chr15:72217430-72217431:- | [9] | |
| Sequence | AATGAATGTGGCTCGTCTGAACTTCTCTCATGGAACTCATG | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa; HepG2; peripheral-blood; endometrial; MM6 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000567087.5; ENST00000567118.5; ENST00000565154.5; ENST00000565184.5; ENST00000449901.6; ENST00000561609.5; ENST00000562997.5; ENST00000335181.10; ENST00000562784.1; ENST00000389093.7; ENST00000568883.5; ENST00000569857.5; ENST00000564178.5; ENST00000566809.1; ENST00000564276.1; ENST00000319622.10; ENST00000568459.5 | ||
| External Link | RMBase: m6A_site_286224 | ||
| mod ID: M6ASITE024109 | Click to Show/Hide the Full List | ||
| mod site | chr15:72218966-72218967:- | [9] | |
| Sequence | ACCACCCATCACAGCCCGGAACACTGGCATCATCTGTACCA | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa; HepG2; hESC-HEK293T; MT4; CD4T; peripheral-blood; endometrial; MM6; AML | ||
| Seq Type List | m6A-seq; MAZTER-seq; MeRIP-seq; miCLIP | ||
| Transcript ID List | ENST00000568459.5; ENST00000567118.5; ENST00000319622.10; ENST00000389093.7; ENST00000565154.5; ENST00000564276.1; ENST00000449901.6; ENST00000562997.5; ENST00000566809.1; ENST00000567087.5; ENST00000335181.10; ENST00000565184.5; ENST00000561609.5; ENST00000569050.1; ENST00000569857.5; ENST00000564178.5; ENST00000568883.5 | ||
| External Link | RMBase: m6A_site_286225 | ||
| mod ID: M6ASITE024110 | Click to Show/Hide the Full List | ||
| mod site | chr15:72218996-72218997:- | [9] | |
| Sequence | GGAGCACATGTGCCGCCTGGACATTGATTCACCACCCATCA | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; HepG2; MT4; CD4T; peripheral-blood; HEK293T; endometrial; MM6 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000319622.10; ENST00000335181.10; ENST00000569857.5; ENST00000567118.5; ENST00000564178.5; ENST00000449901.6; ENST00000565154.5; ENST00000564276.1; ENST00000568459.5; ENST00000566809.1; ENST00000389093.7; ENST00000567087.5; ENST00000569050.1; ENST00000562997.5; ENST00000561609.5; ENST00000565184.5; ENST00000568883.5 | ||
| External Link | RMBase: m6A_site_286226 | ||
| mod ID: M6ASITE024111 | Click to Show/Hide the Full List | ||
| mod site | chr15:72219026-72219027:- | [14] | |
| Sequence | GCTGCACGCAGCCATGGCTGACACATTCCTGGAGCACATGT | ||
| Motif Score | 2.859755952 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000568883.5; ENST00000567087.5; ENST00000569050.1; ENST00000319622.10; ENST00000564276.1; ENST00000567118.5; ENST00000569857.5; ENST00000568459.5; ENST00000565154.5; ENST00000335181.10; ENST00000561609.5; ENST00000562997.5; ENST00000449901.6; ENST00000565184.5; ENST00000389093.7; ENST00000564178.5; ENST00000566809.1 | ||
| External Link | RMBase: m6A_site_286227 | ||
| mod ID: M6ASITE024112 | Click to Show/Hide the Full List | ||
| mod site | chr15:72219054-72219055:- | [9] | |
| Sequence | CCGGGACTGCCTTCATTCAGACCCAGCAGCTGCACGCAGCC | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa; HepG2; MT4; Huh7; CD4T; peripheral-blood; HEK293T; endometrial; HEC-1-A; MM6 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000564178.5; ENST00000565184.5; ENST00000389093.7; ENST00000335181.10; ENST00000561609.5; ENST00000568459.5; ENST00000564276.1; ENST00000569050.1; ENST00000562997.5; ENST00000569857.5; ENST00000567087.5; ENST00000319622.10; ENST00000565154.5; ENST00000566809.1; ENST00000449901.6; ENST00000567118.5; ENST00000568883.5 | ||
| External Link | RMBase: m6A_site_286228 | ||
| mod ID: M6ASITE024113 | Click to Show/Hide the Full List | ||
| mod site | chr15:72219069-72219070:- | [9] | |
| Sequence | AGCCCCATAGTGAAGCCGGGACTGCCTTCATTCAGACCCAG | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa; HepG2; MT4; Huh7; CD4T; peripheral-blood; HEK293T; endometrial; HEC-1-A; MM6 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000568883.5; ENST00000389093.7; ENST00000565184.5; ENST00000569050.1; ENST00000567118.5; ENST00000569857.5; ENST00000564276.1; ENST00000449901.6; ENST00000335181.10; ENST00000562997.5; ENST00000568459.5; ENST00000565154.5; ENST00000566809.1; ENST00000564178.5; ENST00000319622.10; ENST00000561609.5; ENST00000567087.5 | ||
| External Link | RMBase: m6A_site_286229 | ||
| mod ID: M6ASITE024114 | Click to Show/Hide the Full List | ||
| mod site | chr15:72221173-72221174:- | [9] | |
| Sequence | CGGAAGCCCAACCCCAGAGAACCAAAGGTATGACCTGATAT | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | HeLa; HepG2; MT4; A549; MM6; CD4T; peripheral-blood; HEK293T; endometrial; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000564276.1; ENST00000568883.5; ENST00000567118.5; ENST00000568459.5; ENST00000389093.7; ENST00000335181.10; ENST00000562997.5; ENST00000569050.1; ENST00000566809.1; ENST00000565184.5; ENST00000565154.5; ENST00000449901.6; ENST00000319622.10; ENST00000561609.5; ENST00000569857.5; ENST00000567087.5 | ||
| External Link | RMBase: m6A_site_286230 | ||
| mod ID: M6ASITE024115 | Click to Show/Hide the Full List | ||
| mod site | chr15:72231551-72231552:- | [9] | |
| Sequence | GGGCCGAGAGCCAAGAAAAGACACCCCATCTGGCAGCCCAA | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa; HEK293T; H1A; H1B; GM12878; HEK293A-TOA; iSLK; MSC; TREX; TIME | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000566809.1; ENST00000569050.1; ENST00000568883.5; ENST00000567087.5 | ||
| External Link | RMBase: m6A_site_286231 | ||
| mod ID: M6ASITE024116 | Click to Show/Hide the Full List | ||
| mod site | chr15:72231587-72231588:- | [9] | |
| Sequence | CCCTGCCCAGGTAGGCCGGGACAGCTGGGGTGGCCTGGGCC | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; HEK293T; H1A; H1B; GM12878; HEK293A-TOA; iSLK; MSC; TREX; TIME | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000568883.5; ENST00000569050.1; ENST00000567087.5; ENST00000566809.1 | ||
| External Link | RMBase: m6A_site_286232 | ||
| mod ID: M6ASITE024117 | Click to Show/Hide the Full List | ||
| mod site | chr15:72231622-72231623:- | [9] | |
| Sequence | CGGGAGAATGCTGCCCCGGAACCCATAAATCTGGGCCCTGC | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | HeLa; HEK293T; H1A; H1B; GM12878; HEK293A-TOA; iSLK; MSC; TREX; TIME | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000566809.1; ENST00000569050.1; ENST00000567087.5; ENST00000568883.5 | ||
| External Link | RMBase: m6A_site_286233 | ||
| mod ID: M6ASITE024118 | Click to Show/Hide the Full List | ||
| mod site | chr15:72231676-72231677:- | [9] | |
| Sequence | CCGAGGGTGCGCCAGAGCAGACACCCGGAGGAGTGGGGAGT | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa; HEK293T; H1A; H1B; GM12878; MM6; CD4T; HEK293A-TOA; MSC; TIME; TREX; iSLK | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000569050.1; ENST00000567087.5; ENST00000568883.5 | ||
| External Link | RMBase: m6A_site_286234 | ||
| mod ID: M6ASITE024119 | Click to Show/Hide the Full List | ||
| mod site | chr15:72231752-72231753:- | [9] | |
| Sequence | CCCCTCCGAAGGCGGCCAGGACCTCCAACCACGCACAAGTC | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; HEK293T; HepG2; H1A; H1B; hNPCs; GM12878; MM6; CD4T; peripheral-blood; HEK293A-TOA; iSLK; MSC; TIME; TREX | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000568883.5; ENST00000569050.1 | ||
| External Link | RMBase: m6A_site_286235 | ||
2'-O-Methylation (2'-O-Me)
| In total 3 m6A sequence/site(s) in this target gene | |||
| mod ID: 2OMSITE000124 | Click to Show/Hide the Full List | ||
| mod site | chr15:72199333-72199334:- | [17] | |
| Sequence | CTGTGTGTACTCTGTCCAGTTCCTTTAGAAAAAATGGATGC | ||
| Cell/Tissue List | HEK293 | ||
| Seq Type List | Nm-seq | ||
| Transcript ID List | ENST00000389093.7; ENST00000319622.10; ENST00000568883.5; ENST00000565184.5; ENST00000449901.6; ENST00000564440.5; ENST00000567118.5; ENST00000335181.10; ENST00000565143.1 | ||
| External Link | RMBase: Nm_site_1966 | ||
| mod ID: 2OMSITE000125 | Click to Show/Hide the Full List | ||
| mod site | chr15:72207129-72207130:- | [17] | |
| Sequence | AAGCCTGTCATCTGTGCTACTCAGGCATGTGCCCACCCTTC | ||
| Cell/Tissue List | HEK293 | ||
| Seq Type List | Nm-seq | ||
| Transcript ID List | ENST00000567118.5; ENST00000563275.1; ENST00000389093.7; ENST00000564993.1; ENST00000335181.10; ENST00000569857.5; ENST00000561609.5; ENST00000568459.5; ENST00000449901.6; ENST00000565184.5; ENST00000319622.10; ENST00000565154.5; ENST00000568883.5 | ||
| External Link | RMBase: Nm_site_1967 | ||
| mod ID: 2OMSITE000126 | Click to Show/Hide the Full List | ||
| mod site | chr15:72210477-72210478:- | [17] | |
| Sequence | TCTTCCTTCTTTTCCCCCAGTACCATGCGGAGACCATCAAG | ||
| Cell/Tissue List | HEK293; HeLa | ||
| Seq Type List | Nm-seq | ||
| Transcript ID List | ENST00000389093.7; ENST00000568459.5; ENST00000567087.5; ENST00000564276.1; ENST00000565184.5; ENST00000564178.5; ENST00000568883.5; ENST00000567118.5; ENST00000561609.5; ENST00000562784.1; ENST00000449901.6; ENST00000335181.10; ENST00000565154.5; ENST00000319622.10; ENST00000569857.5; ENST00000562997.5 | ||
| External Link | RMBase: Nm_site_1968 | ||
Pseudouridine (Pseudo)
| In total 1 m6A sequence/site(s) in this target gene | |||
| mod ID: PSESITE000064 | Click to Show/Hide the Full List | ||
| mod site | chr15:72200295-72200296:- | [18] | |
| Sequence | GTTCATTCTGAATCTCTCATTCTCCGAAGTCCTAAGCTGAG | ||
| Transcript ID List | ENST00000565184.5; ENST00000568883.5; ENST00000565154.5; ENST00000449901.6; ENST00000335181.10; ENST00000319622.10; ENST00000568459.5; ENST00000565143.1; ENST00000389093.7; ENST00000564440.5; ENST00000567118.5 | ||
| External Link | RMBase: Pseudo_site_1665 | ||
References