m6A Target Gene Information
General Information of the m6A Target Gene (ID: M6ATAR00573)
Full List of m6A Methylation Regulator of This Target Gene and Corresponding Disease/Drug Response(s)
LGALS9
can be regulated by the following regulator(s), and cause disease/drug response(s). You can browse detail information of regulator(s) or disease/drug response(s).
Browse Regulator
Browse Disease
Heterogeneous nuclear ribonucleoproteins C1/C2 (HNRNPC) [READER]
| Representative RNA-seq result indicating the expression of this target gene regulated by HNRNPC | ||
| Cell Line | MG63 cell line | Homo sapiens |
|
Treatment: HNRNPC knockdown MG63 cells
Control: Wild type MG63 cells
|
GSE63086 | |
| Regulation |
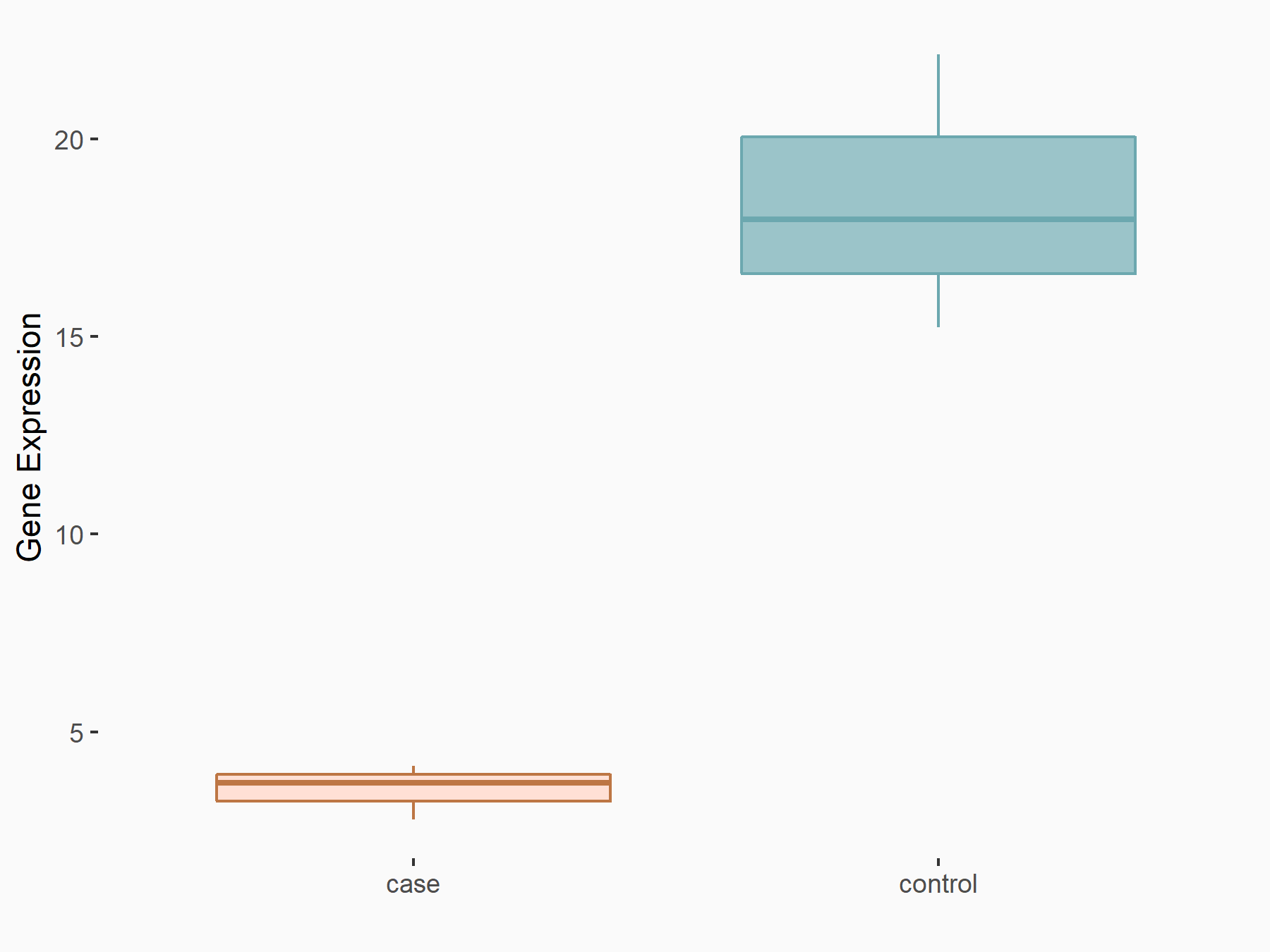  |
logFC: -2.10E+00 p-value: 1.66E-04 |
| More Results | Click to View More RNA-seq Results | |
| In total 1 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [1] | |||
| Response Summary | HNRNPA2B1 and HNRNPC were extensively expressed in the Glioblastoma multiforme(GBM) microenvironment. m6A regulators promoted the stemness state in GBM cancer cells. Cell communication analysis identified genes in the GALECTIN signaling network in GBM samples, and expression of these genes (Galectin-9 (LGALS9), CD44, CD45, and HAVCR2) correlated with that of m6A regulators. | |||
| Responsed Disease | Glioblastoma | ICD-11: 2A00.00 | ||
| In-vitro Model | U-87MG ATCC | Glioblastoma | Homo sapiens | CVCL_0022 |
| THP-1 | Childhood acute monocytic leukemia | Homo sapiens | CVCL_0006 | |
Heterogeneous nuclear ribonucleoproteins A2/B1 (HNRNPA2B1) [READER]
| In total 1 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [1] | |||
| Response Summary | HNRNPA2B1 and HNRNPC were extensively expressed in the Glioblastoma multiforme(GBM) microenvironment.m6A regulators promoted the stemness state in GBM cancer cells. Cell communication analysis identified genes in the GALECTIN signaling network in GBM samples, and expression of these genes (Galectin-9 (LGALS9), CD44, CD45, and HAVCR2) correlated with that of m6A regulators. | |||
| Responsed Disease | Glioblastoma | ICD-11: 2A00.00 | ||
| In-vitro Model | U-87MG ATCC | Glioblastoma | Homo sapiens | CVCL_0022 |
| THP-1 | Childhood acute monocytic leukemia | Homo sapiens | CVCL_0006 | |
Brain cancer [ICD-11: 2A00]
| In total 2 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [1] | |||
| Response Summary | HNRNPA2B1 and HNRNPC were extensively expressed in the Glioblastoma multiforme(GBM) microenvironment.m6A regulators promoted the stemness state in GBM cancer cells. Cell communication analysis identified genes in the GALECTIN signaling network in GBM samples, and expression of these genes (Galectin-9 (LGALS9), CD44, CD45, and HAVCR2) correlated with that of m6A regulators. | |||
| Responsed Disease | Glioblastoma [ICD-11: 2A00.00] | |||
| Target Regulator | Heterogeneous nuclear ribonucleoproteins A2/B1 (HNRNPA2B1) | READER | ||
| In-vitro Model | U-87MG ATCC | Glioblastoma | Homo sapiens | CVCL_0022 |
| THP-1 | Childhood acute monocytic leukemia | Homo sapiens | CVCL_0006 | |
| Experiment 2 Reporting the m6A-centered Disease Response | [1] | |||
| Response Summary | HNRNPA2B1 and HNRNPC were extensively expressed in the Glioblastoma multiforme(GBM) microenvironment. m6A regulators promoted the stemness state in GBM cancer cells. Cell communication analysis identified genes in the GALECTIN signaling network in GBM samples, and expression of these genes (Galectin-9 (LGALS9), CD44, CD45, and HAVCR2) correlated with that of m6A regulators. | |||
| Responsed Disease | Glioblastoma [ICD-11: 2A00.00] | |||
| Target Regulator | Heterogeneous nuclear ribonucleoproteins C1/C2 (HNRNPC) | READER | ||
| In-vitro Model | U-87MG ATCC | Glioblastoma | Homo sapiens | CVCL_0022 |
| THP-1 | Childhood acute monocytic leukemia | Homo sapiens | CVCL_0006 | |
RNA Modification Sequencing Data Associated with the Target (ID: M6ATAR00573)
| In total 1 m6A sequence/site(s) in this target gene | |||
| mod ID: A2ISITE007786 | Click to Show/Hide the Full List | ||
| mod site | chr17:27637344-27637345:+ | [2] | |
| Sequence | GTGAAACACCTTCCTATTTTACAGATGAGGAAACTGAGGCA | ||
| Transcript ID List | ENST00000467111.5; ENST00000448970.6; ENST00000579290.1; ENST00000584386.5; ENST00000580779.5; ENST00000577392.5; ENST00000395473.7; ENST00000584605.5; ENST00000584661.5; ENST00000302228.9; ENST00000579402.1; ENST00000313648.10; ENST00000579930.5 | ||
| External Link | RMBase: RNA-editing_site_55857 | ||
5-methylcytidine (m5C)
N6-methyladenosine (m6A)
| In total 12 m6A sequence/site(s) in this target gene | |||
| mod ID: M6ASITE031091 | Click to Show/Hide the Full List | ||
| mod site | chr17:27631215-27631216:+ | [3] | |
| Sequence | AGTCGTTCCCTCTACAAAGGACTTCCTAGTGGGTGTGAAAG | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | endometrial | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000584605.5; ENST00000467111.5; ENST00000395473.7; ENST00000448970.6; ENST00000579930.5; ENST00000579290.1; ENST00000579402.1; ENST00000580779.5; ENST00000313648.10; ENST00000584661.5; ENST00000577392.5; ENST00000302228.9 | ||
| External Link | RMBase: m6A_site_353620 | ||
| mod ID: M6ASITE031092 | Click to Show/Hide the Full List | ||
| mod site | chr17:27638280-27638281:+ | [4] | |
| Sequence | AGGCTGTCCCCTTTTCTGGGACTATTCAAGGAGGTCTCCAG | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; A549; H1A; H1B; hNPCs; hESCs; fibroblasts; GM12878; LCLs; Huh7; GSC-11; HEK293A-TOA; MSC; TIME; TREX; endometrial; GSCs | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000584661.5; ENST00000448970.6; ENST00000584605.5; ENST00000579290.1; ENST00000580779.5; ENST00000579930.5; ENST00000579402.1; ENST00000467111.5; ENST00000302228.9; ENST00000577392.5; ENST00000313648.10; ENST00000395473.7; ENST00000584386.5 | ||
| External Link | RMBase: m6A_site_353621 | ||
| mod ID: M6ASITE031093 | Click to Show/Hide the Full List | ||
| mod site | chr17:27638306-27638307:+ | [4] | |
| Sequence | CAAGGAGGTCTCCAGGACGGACTTCAGATCACTGTCAATGG | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; A549; H1A; H1B; hESCs; fibroblasts; GM12878; Huh7; GSC-11; HEK293A-TOA; MSC; TIME; TREX; endometrial; HEC-1-A; GSCs | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000448970.6; ENST00000580779.5; ENST00000302228.9; ENST00000313648.10; ENST00000579402.1; ENST00000584661.5; ENST00000584386.5; ENST00000579930.5; ENST00000584605.5; ENST00000467111.5; ENST00000577392.5; ENST00000579290.1; ENST00000395473.7 | ||
| External Link | RMBase: m6A_site_353622 | ||
| mod ID: M6ASITE031094 | Click to Show/Hide the Full List | ||
| mod site | chr17:27638328-27638329:+ | [4] | |
| Sequence | TTCAGATCACTGTCAATGGGACCGTTCTCAGCTCCAGTGGA | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; A549; H1A; H1B; HEK293A-TOA; TIME; endometrial; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000580779.5; ENST00000467111.5; ENST00000584386.5; ENST00000448970.6; ENST00000584605.5; ENST00000577392.5; ENST00000579930.5; ENST00000302228.9; ENST00000579290.1; ENST00000584661.5; ENST00000395473.7; ENST00000313648.10; ENST00000579402.1 | ||
| External Link | RMBase: m6A_site_353623 | ||
| mod ID: M6ASITE031095 | Click to Show/Hide the Full List | ||
| mod site | chr17:27638349-27638350:+ | [4] | |
| Sequence | CCGTTCTCAGCTCCAGTGGAACCAGGTGTGTGTGTATATGG | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | HeLa; CD34; HepG2; HEK293T; A549; H1A; H1B; HEK293A-TOA; TIME; endometrial; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000579402.1; ENST00000584661.5; ENST00000395473.7; ENST00000584605.5; ENST00000467111.5; ENST00000448970.6; ENST00000584386.5; ENST00000580779.5; ENST00000579290.1; ENST00000577392.5; ENST00000313648.10; ENST00000579930.5; ENST00000302228.9 | ||
| External Link | RMBase: m6A_site_353624 | ||
| mod ID: M6ASITE031096 | Click to Show/Hide the Full List | ||
| mod site | chr17:27638406-27638407:+ | [5] | |
| Sequence | CCAGCCTCGTCCCTTAGTAAACCACTGTGCCTGTGAGCCTG | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | CD34; endometrial | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000584661.5; ENST00000584605.5; ENST00000579290.1; ENST00000395473.7; ENST00000579402.1; ENST00000577392.5; ENST00000584386.5; ENST00000467111.5; ENST00000448970.6; ENST00000580779.5; ENST00000313648.10; ENST00000302228.9; ENST00000579930.5 | ||
| External Link | RMBase: m6A_site_353625 | ||
| mod ID: M6ASITE031097 | Click to Show/Hide the Full List | ||
| mod site | chr17:27640582-27640583:+ | [4] | |
| Sequence | TGGGCCTAGGTTTGCTGTGAACTTTCAGACTGGCTTCAGTG | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa; HEK293A-TOA; endometrial; HEC-1-A | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000395473.7; ENST00000577392.5; ENST00000302228.9; ENST00000583671.1; ENST00000579290.1; ENST00000448970.6; ENST00000584605.5; ENST00000580779.5; ENST00000313648.10; ENST00000584661.5; ENST00000579930.5; ENST00000584386.5; ENST00000467111.5 | ||
| External Link | RMBase: m6A_site_353626 | ||
| mod ID: M6ASITE031098 | Click to Show/Hide the Full List | ||
| mod site | chr17:27640590-27640591:+ | [4] | |
| Sequence | GGTTTGCTGTGAACTTTCAGACTGGCTTCAGTGGAAATGAC | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa; HEK293A-TOA; endometrial; HEC-1-A | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000580779.5; ENST00000395473.7; ENST00000583671.1; ENST00000584386.5; ENST00000577392.5; ENST00000579290.1; ENST00000302228.9; ENST00000448970.6; ENST00000584661.5; ENST00000313648.10; ENST00000467111.5; ENST00000579930.5; ENST00000584605.5 | ||
| External Link | RMBase: m6A_site_353627 | ||
| mod ID: M6ASITE031099 | Click to Show/Hide the Full List | ||
| mod site | chr17:27640704-27640705:+ | [6] | |
| Sequence | GGGGGCCCGAGGAGAGGAAGACACACATGCCTTTCCAGAAG | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | peripheral-blood; endometrial; HEC-1-A; NB4 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000584386.5; ENST00000448970.6; ENST00000577392.5; ENST00000584605.5; ENST00000395473.7; ENST00000302228.9; ENST00000584661.5; ENST00000583671.1; ENST00000313648.10; ENST00000579930.5; ENST00000467111.5; ENST00000580779.5 | ||
| External Link | RMBase: m6A_site_353628 | ||
| mod ID: M6ASITE031100 | Click to Show/Hide the Full List | ||
| mod site | chr17:27640999-27641000:+ | [6] | |
| Sequence | CTTTTACTCTGAAAATAAGAACTAGAGGAGTCATCACGTTG | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | peripheral-blood; endometrial | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000302228.9; ENST00000584386.5; ENST00000448970.6; ENST00000395473.7; ENST00000581710.5; ENST00000580779.5; ENST00000583671.1; ENST00000313648.10; ENST00000467111.5; ENST00000584661.5; ENST00000579930.5; ENST00000577392.5 | ||
| External Link | RMBase: m6A_site_353629 | ||
| mod ID: M6ASITE031102 | Click to Show/Hide the Full List | ||
| mod site | chr17:27647298-27647299:+ | [7] | |
| Sequence | CAACCTGTGCTCTGGGAACCACATCGCCTTCCACCTGAACC | ||
| Motif Score | 2.053113095 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000580779.5; ENST00000302228.9; ENST00000581710.5; ENST00000578944.1; ENST00000395473.7; ENST00000486774.1; ENST00000313648.10; ENST00000481514.5; ENST00000467111.5 | ||
| External Link | RMBase: m6A_site_353630 | ||
| mod ID: M6ASITE031103 | Click to Show/Hide the Full List | ||
| mod site | chr17:27649500-27649501:+ | [8] | |
| Sequence | CAGAGTGTTCTCTTCAGAGGACTGGCTCCTTTCCCAGTGTC | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | LCLs | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000467111.5; ENST00000302228.9; ENST00000395473.7; ENST00000481514.5; ENST00000486774.1 | ||
| External Link | RMBase: m6A_site_353631 | ||
References