m6A Target Gene Information
General Information of the m6A Target Gene (ID: M6ATAR00276)
Full List of m6A Methylation Regulator of This Target Gene and Corresponding Disease/Drug Response(s)
HMGA2
can be regulated by the following regulator(s), and cause disease/drug response(s). You can browse detail information of regulator(s) or disease/drug response(s).
Browse Regulator
Browse Disease
Methyltransferase-like 3 (METTL3) [WRITER]
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | LX2 cell line | Homo sapiens |
|
Treatment: shMETTL3 LX2 cells
Control: shLuc LX2 cells
|
GSE207909 | |
| Regulation |
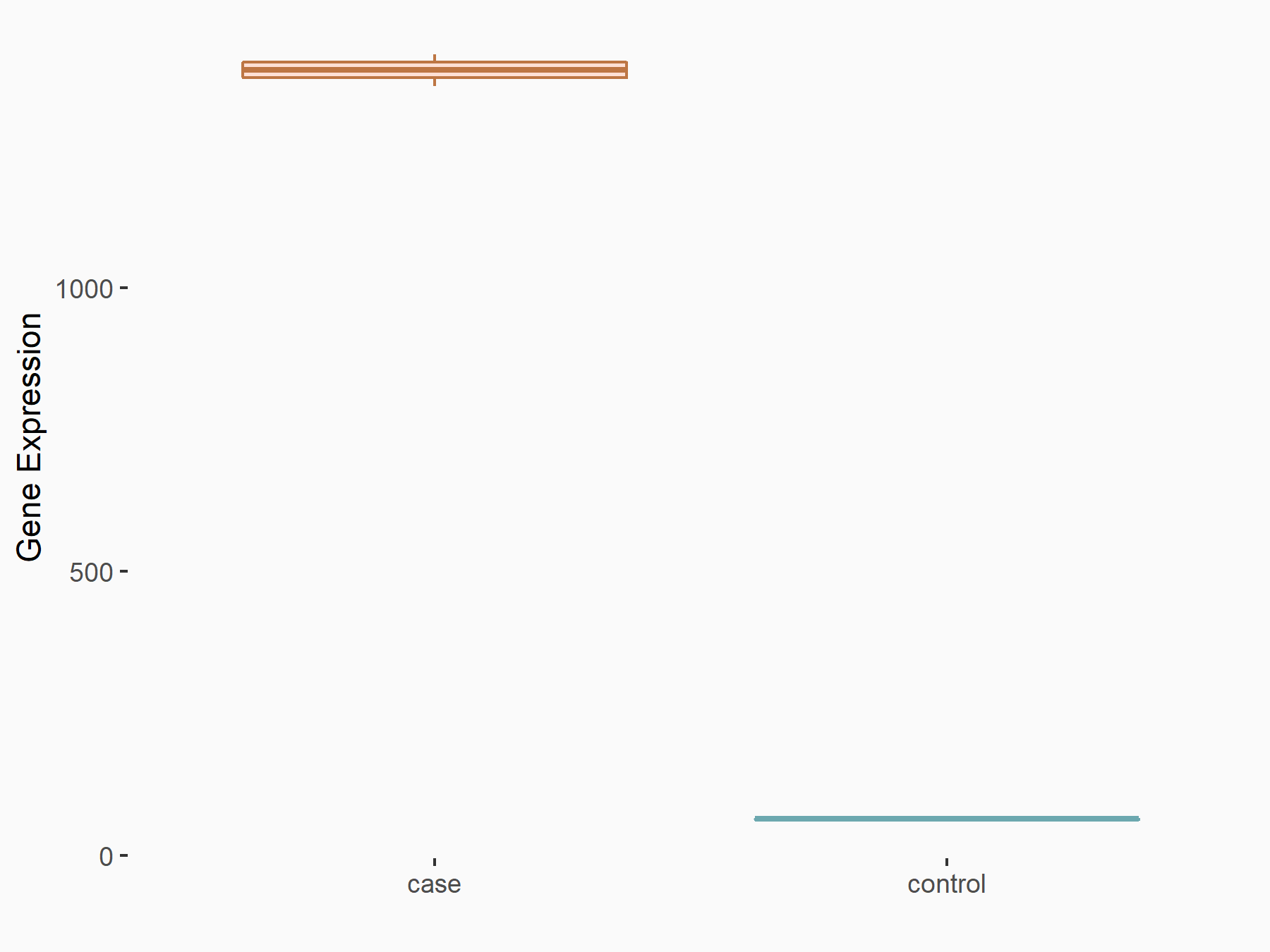 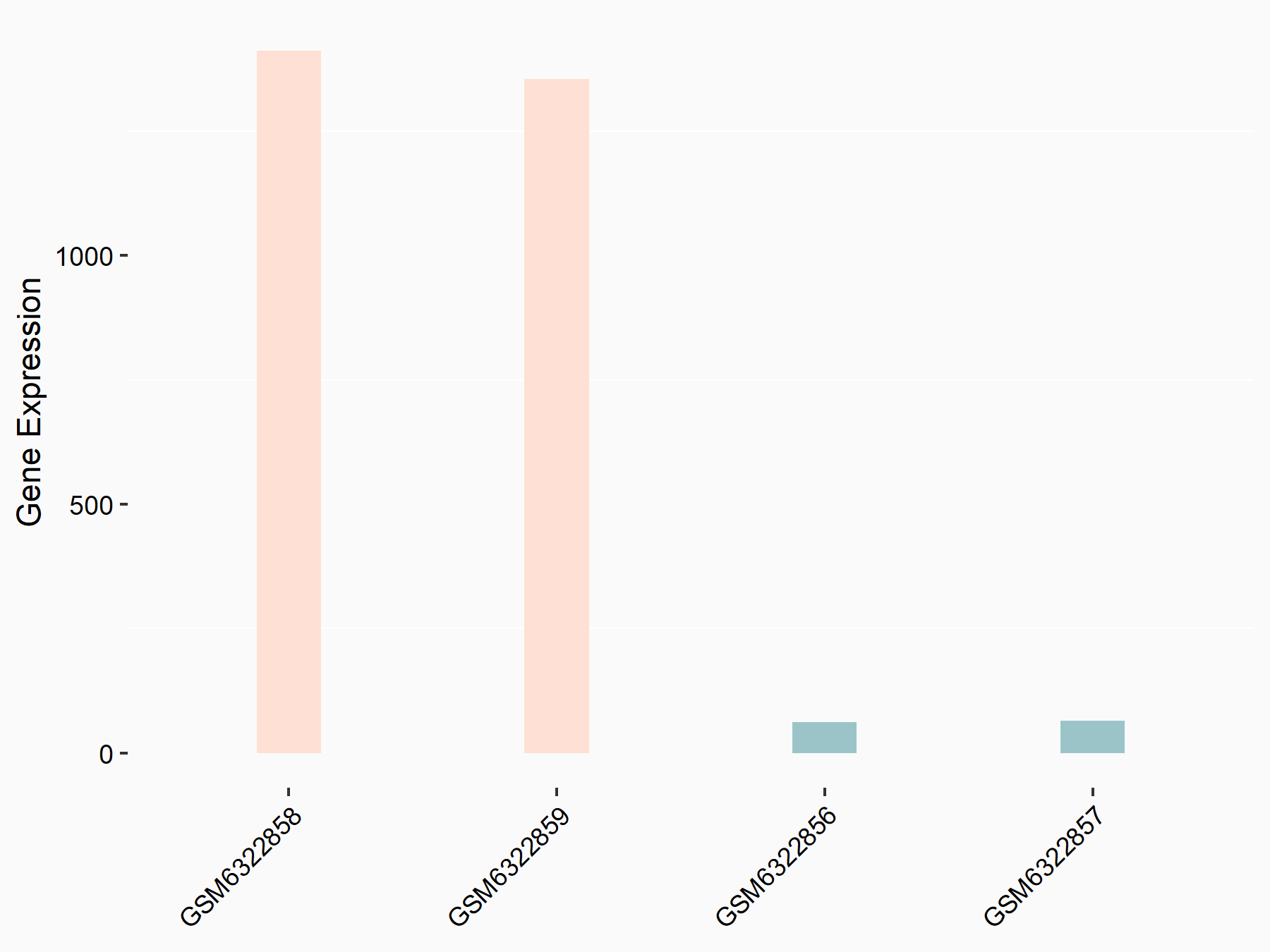 |
logFC: 4.44E+00 p-value: 5.85E-138 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between HMGA2 and the regulator | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 1.68E+00 | GSE60213 |
| In total 2 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [1] | |||
| Response Summary | In hepatocellular carcinoma, METTL3 could direct the formation of circHPS5, and specific m6A controlled the accumulation of circHPS5. YTHDC1 facilitated the cytoplasmic output of circHPS5 under m6A modification. CircHPS5 can act as a miR-370 sponge to regulate the expression of High mobility group protein HMGI-C (HMGA2) and further accelerate hepatocellular carcinoma cell tumorigenesis. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Hepatocellular carcinoma | ICD-11: 2C12.02 | ||
| Pathway Response | Transcriptional misregulation in cancer | hsa05202 | ||
| Cell Process | Epithelial-mesenchymal transition | |||
| Cell autophagy | ||||
| In-vitro Model | Hep 3B2.1-7 | Childhood hepatocellular carcinoma | Homo sapiens | CVCL_0326 |
| L-02 | Endocervical adenocarcinoma | Homo sapiens | CVCL_6926 | |
| In-vivo Model | To create the xenograft neoplasm system, 40 male BALB/c nude mice aged 5 weeks were randomly separated into sh-NC, sh-circHPS5, sh-circHPS5+CTRL, and sh-circHPS5+SAH groups (n = 5 for each group). HCC cells were subcutaneously injected into the axilla of the nude mice. | |||
| Experiment 2 Reporting the m6A Methylation Regulator of This Target Gene | [2] | |||
| Response Summary | Silencing METTL3 down-regulate MALAT1 and High mobility group protein HMGI-C (HMGA2) by sponging miR-26b, and finally inhibit EMT, migration and invasion in BC, providing a theoretical basis for clinical treatment of BC. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Breast cancer | ICD-11: 2C60 | ||
| Cell Process | Epithelial-mesenchymal transition | |||
| In-vitro Model | MDA-MB-468 | Breast adenocarcinoma | Homo sapiens | CVCL_0419 |
| MDA-MB-231 | Breast adenocarcinoma | Homo sapiens | CVCL_0062 | |
| MCF-7 | Invasive breast carcinoma | Homo sapiens | CVCL_0031 | |
| MCF-10A | Normal | Homo sapiens | CVCL_0598 | |
| In-vivo Model | Eighteen BALB/C female nude mice aged 4-5 weeks and weighing 15-18 g were randomly assigned into three groups of six mice. The MCF-7 cell lines stably transfected with sh-NC + oe-NC, sh-METTL3 + oe-NC and sh-METTL3 + oe-HMGA2 were selected for subcutaneous establishment of the BC cell line MCF-7 as xenografts in the nude mice. For this purpose, MCF-7 cell lines in the logarithmic growth stage were prepared into a suspension with a concentration of about 1 × 107 cells/ml. The prepared cell suspension was injected into the left armpit of the mice, and the subsequent tumor growth was recorded. | |||
YTH domain-containing protein 1 (YTHDC1) [READER]
| In total 1 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [1] | |||
| Response Summary | In hepatocellular carcinoma, METTL3 could direct the formation of circHPS5, and specific m6A controlled the accumulation of circHPS5. YTHDC1 facilitated the cytoplasmic output of circHPS5 under m6A modification. CircHPS5 can act as a miR-370 sponge to regulate the expression of High mobility group protein HMGI-C (HMGA2) and further accelerate hepatocellular carcinoma cell tumorigenesis. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Hepatocellular carcinoma | ICD-11: 2C12.02 | ||
| Pathway Response | Transcriptional misregulation in cancer | hsa05202 | ||
| Cell Process | Epithelial-mesenchymal transition | |||
| Cell autophagy | ||||
| In-vitro Model | Hep 3B2.1-7 | Childhood hepatocellular carcinoma | Homo sapiens | CVCL_0326 |
| L-02 | Endocervical adenocarcinoma | Homo sapiens | CVCL_6926 | |
| In-vivo Model | To create the xenograft neoplasm system, 40 male BALB/c nude mice aged 5 weeks were randomly separated into sh-NC, sh-circHPS5, sh-circHPS5+CTRL, and sh-circHPS5+SAH groups (n = 5 for each group). HCC cells were subcutaneously injected into the axilla of the nude mice. | |||
Liver cancer [ICD-11: 2C12]
| In total 2 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [1] | |||
| Response Summary | In hepatocellular carcinoma, METTL3 could direct the formation of circHPS5, and specific m6A controlled the accumulation of circHPS5. YTHDC1 facilitated the cytoplasmic output of circHPS5 under m6A modification. CircHPS5 can act as a miR-370 sponge to regulate the expression of High mobility group protein HMGI-C (HMGA2) and further accelerate hepatocellular carcinoma cell tumorigenesis. | |||
| Responsed Disease | Hepatocellular carcinoma [ICD-11: 2C12.02] | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Transcriptional misregulation in cancer | hsa05202 | ||
| Cell Process | Epithelial-mesenchymal transition | |||
| Cell autophagy | ||||
| In-vitro Model | Hep 3B2.1-7 | Childhood hepatocellular carcinoma | Homo sapiens | CVCL_0326 |
| L-02 | Endocervical adenocarcinoma | Homo sapiens | CVCL_6926 | |
| In-vivo Model | To create the xenograft neoplasm system, 40 male BALB/c nude mice aged 5 weeks were randomly separated into sh-NC, sh-circHPS5, sh-circHPS5+CTRL, and sh-circHPS5+SAH groups (n = 5 for each group). HCC cells were subcutaneously injected into the axilla of the nude mice. | |||
| Experiment 2 Reporting the m6A-centered Disease Response | [1] | |||
| Response Summary | In hepatocellular carcinoma, METTL3 could direct the formation of circHPS5, and specific m6A controlled the accumulation of circHPS5. YTHDC1 facilitated the cytoplasmic output of circHPS5 under m6A modification. CircHPS5 can act as a miR-370 sponge to regulate the expression of High mobility group protein HMGI-C (HMGA2) and further accelerate hepatocellular carcinoma cell tumorigenesis. | |||
| Responsed Disease | Hepatocellular carcinoma [ICD-11: 2C12.02] | |||
| Target Regulator | YTH domain-containing protein 1 (YTHDC1) | READER | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Transcriptional misregulation in cancer | hsa05202 | ||
| Cell Process | Epithelial-mesenchymal transition | |||
| Cell autophagy | ||||
| In-vitro Model | Hep 3B2.1-7 | Childhood hepatocellular carcinoma | Homo sapiens | CVCL_0326 |
| L-02 | Endocervical adenocarcinoma | Homo sapiens | CVCL_6926 | |
| In-vivo Model | To create the xenograft neoplasm system, 40 male BALB/c nude mice aged 5 weeks were randomly separated into sh-NC, sh-circHPS5, sh-circHPS5+CTRL, and sh-circHPS5+SAH groups (n = 5 for each group). HCC cells were subcutaneously injected into the axilla of the nude mice. | |||
Breast cancer [ICD-11: 2C60]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [2] | |||
| Response Summary | Silencing METTL3 down-regulate MALAT1 and High mobility group protein HMGI-C (HMGA2) by sponging miR-26b, and finally inhibit EMT, migration and invasion in BC, providing a theoretical basis for clinical treatment of BC. | |||
| Responsed Disease | Breast cancer [ICD-11: 2C60] | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Target Regulation | Up regulation | |||
| Cell Process | Epithelial-mesenchymal transition | |||
| In-vitro Model | MDA-MB-468 | Breast adenocarcinoma | Homo sapiens | CVCL_0419 |
| MDA-MB-231 | Breast adenocarcinoma | Homo sapiens | CVCL_0062 | |
| MCF-7 | Invasive breast carcinoma | Homo sapiens | CVCL_0031 | |
| MCF-10A | Normal | Homo sapiens | CVCL_0598 | |
| In-vivo Model | Eighteen BALB/C female nude mice aged 4-5 weeks and weighing 15-18 g were randomly assigned into three groups of six mice. The MCF-7 cell lines stably transfected with sh-NC + oe-NC, sh-METTL3 + oe-NC and sh-METTL3 + oe-HMGA2 were selected for subcutaneous establishment of the BC cell line MCF-7 as xenografts in the nude mice. For this purpose, MCF-7 cell lines in the logarithmic growth stage were prepared into a suspension with a concentration of about 1 × 107 cells/ml. The prepared cell suspension was injected into the left armpit of the mice, and the subsequent tumor growth was recorded. | |||
Full List of Crosstalk(s) between m6A Modification and Epigenetic Regulation Related to This Regulator
RNA modification
m6A Regulator: ELAV-like protein 1 (ELAVL1)
| In total 2 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT00465 | ||
| Epigenetic Regulator | Interferon-inducible protein 4 (ADAR1) | |
| Regulated Target | MicroRNA 149 (MIR149) | |
| Crosstalk relationship | A-to-I → m6A | |
| Crosstalk ID: M6ACROT00466 | ||
| Epigenetic Regulator | Methyltransferase-like protein 1 (METTL1) | |
| Regulated Target | MicroRNA 149 (MIR149) | |
| Crosstalk relationship | m7G → m6A | |
m6A Regulator: Methyltransferase-like 14 (METTL14)
| In total 2 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT00467 | ||
| Epigenetic Regulator | Interferon-inducible protein 4 (ADAR1) | |
| Regulated Target | MicroRNA 149 (MIR149) | |
| Crosstalk relationship | A-to-I → m6A | |
| Crosstalk ID: M6ACROT00468 | ||
| Epigenetic Regulator | Methyltransferase-like protein 1 (METTL1) | |
| Regulated Target | MicroRNA 149 (MIR149) | |
| Crosstalk relationship | m7G → m6A | |
Histone modification
m6A Regulator: YTH domain-containing protein 1 (YTHDC1)
| In total 1 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT03485 | ||
| Epigenetic Regulator | Histone acetyltransferase p300 (P300) | |
| Regulated Target | Histone H3 lysine 18 lactylation (H3K18la) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Liver cancer | |
RNA Modification Sequencing Data Associated with the Target (ID: M6ATAR00276)
| In total 63 m6A sequence/site(s) in this target gene | |||
| mod ID: M6ASITE013755 | Click to Show/Hide the Full List | ||
| mod site | chr12:65824478-65824479:+ | [6] | |
| Sequence | ACTTGAATCTTGGGGCAGGAACTCAGAAAACTTCCAGCCCG | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HepG2; HEK293T; U2OS; H1A; H1B; hNPCs; hESCs; fibroblasts; A549; H1299; Huh7; GSC-11; HEK293A-TOA; iSLK; MSC; TIME | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000537275.5; ENST00000403681.6; ENST00000537429.5; ENST00000393578.7; ENST00000536545.5; ENST00000354636.7; ENST00000425208.6 | ||
| External Link | RMBase: m6A_site_197400 | ||
| mod ID: M6ASITE013756 | Click to Show/Hide the Full List | ||
| mod site | chr12:65824487-65824488:+ | [6] | |
| Sequence | TTGGGGCAGGAACTCAGAAAACTTCCAGCCCGGGCAGCGCG | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HepG2; HEK293T; U2OS; H1A; H1B; hNPCs; hESCs; fibroblasts; A549; H1299; Huh7; GSC-11; HEK293A-TOA; iSLK; MSC; TIME | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000536545.5; ENST00000403681.6; ENST00000537275.5; ENST00000393578.7; ENST00000537429.5; ENST00000354636.7; ENST00000425208.6 | ||
| External Link | RMBase: m6A_site_197401 | ||
| mod ID: M6ASITE013757 | Click to Show/Hide the Full List | ||
| mod site | chr12:65824521-65824522:+ | [7] | |
| Sequence | CAGCGCGCGCTTGGTGCAAGACTCAGGAGCTAGCAGCCCGT | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; U2OS; H1B; hESCs; fibroblasts; A549; H1299; Huh7; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000536545.5; ENST00000537275.5; ENST00000425208.6; ENST00000403681.6; ENST00000354636.7; ENST00000537429.5; ENST00000393578.7 | ||
| External Link | RMBase: m6A_site_197402 | ||
| mod ID: M6ASITE013758 | Click to Show/Hide the Full List | ||
| mod site | chr12:65824823-65824824:+ | [7] | |
| Sequence | CCCGCCTAACATTTCAAGGGACACAATTCACTCCAAGTCTC | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; CD34; HepG2; HEK293T; U2OS; H1A; fibroblasts; Huh7; GSC-11; iSLK; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000403681.6; ENST00000354636.7; ENST00000545998.1; ENST00000536545.5; ENST00000537275.5; ENST00000393578.7; ENST00000425208.6; ENST00000537429.5 | ||
| External Link | RMBase: m6A_site_197403 | ||
| mod ID: M6ASITE013759 | Click to Show/Hide the Full List | ||
| mod site | chr12:65825320-65825321:+ | [7] | |
| Sequence | CCGTCCACTTCAGCCCAGGGACAACCTGCCGCCCCAGCGCC | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; HepG2; hESC-HEK293T; U2OS; H1299; GSC-11; iSLK; MSC; HEC-1-A; GSCs; NB4 | ||
| Seq Type List | m6A-seq; MAZTER-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000545998.1; ENST00000537429.5; ENST00000393577.7; ENST00000537275.5; ENST00000425208.6; ENST00000393578.7; ENST00000541363.5; ENST00000536545.5; ENST00000354636.7; ENST00000403681.6 | ||
| External Link | RMBase: m6A_site_197404 | ||
| mod ID: M6ASITE013760 | Click to Show/Hide the Full List | ||
| mod site | chr12:65828002-65828003:+ | [7] | |
| Sequence | TTTTTCCCTCACAATTAGGAACCAACCGGTGAGCCCTCTCC | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | HeLa; HepG2; U2OS; hNPCs; fibroblasts; H1299; GSC-11; HEK293T; iSLK; MSC; endometrial; HEC-1-A; GSCs; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000425208.6; ENST00000354636.7; ENST00000545998.1; ENST00000541363.5; ENST00000403681.6; ENST00000537275.5; ENST00000539662.1; ENST00000537429.5; ENST00000536545.5; ENST00000393577.7; ENST00000393578.7 | ||
| External Link | RMBase: m6A_site_197406 | ||
| mod ID: M6ASITE013761 | Click to Show/Hide the Full List | ||
| mod site | chr12:65828029-65828030:+ | [7] | |
| Sequence | GGTGAGCCCTCTCCTAAGAGACCCAGGGGAAGACCCAAAGG | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa; HepG2; U2OS; hNPCs; fibroblasts; H1299; Huh7; GSC-11; HEK293T; iSLK; endometrial; HEC-1-A; GSCs; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000536545.5; ENST00000537275.5; ENST00000545998.1; ENST00000541363.5; ENST00000403681.6; ENST00000393577.7; ENST00000539662.1; ENST00000393578.7; ENST00000537429.5; ENST00000354636.7; ENST00000425208.6 | ||
| External Link | RMBase: m6A_site_197407 | ||
| mod ID: M6ASITE013762 | Click to Show/Hide the Full List | ||
| mod site | chr12:65828041-65828042:+ | [7] | |
| Sequence | CCTAAGAGACCCAGGGGAAGACCCAAAGGCAGCAAAAACAA | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa; HepG2; U2OS; hNPCs; fibroblasts; H1299; Huh7; GSC-11; HEK293T; iSLK; endometrial; HEC-1-A; GSCs; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000403681.6; ENST00000541363.5; ENST00000536545.5; ENST00000425208.6; ENST00000537429.5; ENST00000539662.1; ENST00000354636.7; ENST00000537275.5; ENST00000393578.7; ENST00000393577.7; ENST00000545998.1 | ||
| External Link | RMBase: m6A_site_197408 | ||
| mod ID: M6ASITE013763 | Click to Show/Hide the Full List | ||
| mod site | chr12:65828058-65828059:+ | [7] | |
| Sequence | AAGACCCAAAGGCAGCAAAAACAAGAGTCCCTCTAAAGCAG | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa; HepG2; hESC-HEK293T; U2OS; hNPCs; fibroblasts; H1299; Huh7; GSC-11; iSLK; endometrial; HEC-1-A; GSCs; NB4 | ||
| Seq Type List | m6A-seq; MAZTER-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000541363.5; ENST00000536545.5; ENST00000393578.7; ENST00000537429.5; ENST00000425208.6; ENST00000354636.7; ENST00000403681.6; ENST00000539662.1; ENST00000537275.5; ENST00000545998.1; ENST00000393577.7 | ||
| External Link | RMBase: m6A_site_197409 | ||
| mod ID: M6ASITE013764 | Click to Show/Hide the Full List | ||
| mod site | chr12:65828205-65828206:+ | [7] | |
| Sequence | TCCCAACTGGGTAACACAAGACTCATTTCTTACATTTAACA | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa; U2OS; H1299 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000537429.5; ENST00000545998.1; ENST00000536545.5; ENST00000537275.5; ENST00000393577.7; ENST00000541363.5; ENST00000403681.6; ENST00000354636.7; ENST00000539662.1; ENST00000393578.7; ENST00000425208.6 | ||
| External Link | RMBase: m6A_site_197410 | ||
| mod ID: M6ASITE013765 | Click to Show/Hide the Full List | ||
| mod site | chr12:65830919-65830920:+ | [8] | |
| Sequence | CTACTTAATATTCCATTAAAACTAATTACTATGCATGACCA | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | Huh7 | ||
| Seq Type List | MeRIP-seq | ||
| Transcript ID List | ENST00000615897.1; ENST00000537275.5; ENST00000393577.7; ENST00000537429.5; ENST00000536545.5; ENST00000425208.6; ENST00000354636.7; ENST00000393578.7; ENST00000403681.6; ENST00000539662.1; ENST00000541363.5 | ||
| External Link | RMBase: m6A_site_197411 | ||
| mod ID: M6ASITE013766 | Click to Show/Hide the Full List | ||
| mod site | chr12:65830969-65830970:+ | [8] | |
| Sequence | GATTCTAGTCATTCCTCAGAACTCCAATCTGAGTGCTCAAA | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | Huh7 | ||
| Seq Type List | MeRIP-seq | ||
| Transcript ID List | ENST00000393577.7; ENST00000537429.5; ENST00000393578.7; ENST00000539662.1; ENST00000354636.7; ENST00000403681.6; ENST00000425208.6; ENST00000536545.5; ENST00000541363.5; ENST00000615897.1; ENST00000537275.5 | ||
| External Link | RMBase: m6A_site_197412 | ||
| mod ID: M6ASITE013767 | Click to Show/Hide the Full List | ||
| mod site | chr12:65838556-65838557:+ | [9] | |
| Sequence | GAAAAACGGCCAAGAGGCAGACCTAGGAAATGGGTGAGTAA | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | H1A; hNPCs; H1299; Huh7; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000354636.7; ENST00000403681.6; ENST00000393578.7; ENST00000541363.5; ENST00000537429.5; ENST00000536545.5; ENST00000537275.5; ENST00000539662.1; ENST00000425208.6; ENST00000393577.7 | ||
| External Link | RMBase: m6A_site_197413 | ||
| mod ID: M6ASITE013768 | Click to Show/Hide the Full List | ||
| mod site | chr12:65867784-65867785:+ | [10] | |
| Sequence | ACATAAAGCTTGTCATCTGAACAGGACTATAAGCATCTCGA | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | iSLK | ||
| Seq Type List | MeRIP-seq | ||
| Transcript ID List | ENST00000403681.6; ENST00000393577.7; ENST00000537429.5; ENST00000541363.5; ENST00000354636.7; ENST00000536545.5; ENST00000425208.6; ENST00000539662.1 | ||
| External Link | RMBase: m6A_site_197439 | ||
| mod ID: M6ASITE013769 | Click to Show/Hide the Full List | ||
| mod site | chr12:65867789-65867790:+ | [10] | |
| Sequence | AAGCTTGTCATCTGAACAGGACTATAAGCATCTCGAGGGTA | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | iSLK | ||
| Seq Type List | MeRIP-seq | ||
| Transcript ID List | ENST00000403681.6; ENST00000541363.5; ENST00000393577.7; ENST00000354636.7; ENST00000425208.6; ENST00000536545.5; ENST00000537429.5; ENST00000539662.1 | ||
| External Link | RMBase: m6A_site_197440 | ||
| mod ID: M6ASITE013770 | Click to Show/Hide the Full List | ||
| mod site | chr12:65881793-65881794:+ | [11] | |
| Sequence | GGAAGCCTGCTGATCCCGGAACTGGCCTTGCGCGCAAGTGG | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | fibroblasts; iSLK; TIME | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000425208.6; ENST00000393577.7; ENST00000537429.5; ENST00000354636.7; ENST00000539662.1; ENST00000536545.5; ENST00000403681.6; ENST00000541363.5 | ||
| External Link | RMBase: m6A_site_197446 | ||
| mod ID: M6ASITE013771 | Click to Show/Hide the Full List | ||
| mod site | chr12:65881825-65881826:+ | [11] | |
| Sequence | CGCAAGTGGTGGGATGACAGACACATTCCCTTCCTTGAACT | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | fibroblasts; GSC-11; iSLK; TIME; TREX; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000539662.1; ENST00000541363.5; ENST00000393577.7; ENST00000536545.5; ENST00000537429.5; ENST00000354636.7; ENST00000425208.6; ENST00000403681.6 | ||
| External Link | RMBase: m6A_site_197447 | ||
| mod ID: M6ASITE013772 | Click to Show/Hide the Full List | ||
| mod site | chr12:65881843-65881844:+ | [11] | |
| Sequence | AGACACATTCCCTTCCTTGAACTGCACTCTTTTCCCTTGCA | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | fibroblasts; peripheral-blood; GSC-11; iSLK; TIME; TREX; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000537429.5; ENST00000354636.7; ENST00000393577.7; ENST00000403681.6; ENST00000536545.5; ENST00000425208.6; ENST00000541363.5; ENST00000539662.1 | ||
| External Link | RMBase: m6A_site_197448 | ||
| mod ID: M6ASITE013773 | Click to Show/Hide the Full List | ||
| mod site | chr12:65951384-65951385:+ | [12] | |
| Sequence | TTTTCTTTTCCTCCTTAGCCACAACAAGTTGTTCAGAAGAA | ||
| Motif Score | 2.053113095 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000393577.7; ENST00000541363.5; ENST00000403681.6; ENST00000539662.1 | ||
| External Link | RMBase: m6A_site_197459 | ||
| mod ID: M6ASITE013774 | Click to Show/Hide the Full List | ||
| mod site | chr12:65951387-65951388:+ | [12] | |
| Sequence | TCTTTTCCTCCTTAGCCACAACAAGTTGTTCAGAAGAAGCC | ||
| Motif Score | 2.173910714 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000541363.5; ENST00000539662.1; ENST00000393577.7; ENST00000403681.6 | ||
| External Link | RMBase: m6A_site_197460 | ||
| mod ID: M6ASITE013775 | Click to Show/Hide the Full List | ||
| mod site | chr12:65963250-65963251:+ | [13] | |
| Sequence | TTTGTGTGTTCCAGGAGGAAACTGAAGAGACATCCTCACAA | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HEK293T; HEC-1-A; NB4 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000539662.1; ENST00000403681.6 | ||
| External Link | RMBase: m6A_site_197461 | ||
| mod ID: M6ASITE013776 | Click to Show/Hide the Full List | ||
| mod site | chr12:65963259-65963260:+ | [12] | |
| Sequence | TCCAGGAGGAAACTGAAGAGACATCCTCACAAGAGTCTGCC | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | hESC-HEK293T; HEC-1-A; NB4 | ||
| Seq Type List | MAZTER-seq; m6A-seq | ||
| Transcript ID List | ENST00000539662.1; ENST00000403681.6 | ||
| External Link | RMBase: m6A_site_197462 | ||
| mod ID: M6ASITE013777 | Click to Show/Hide the Full List | ||
| mod site | chr12:65963267-65963268:+ | [12] | |
| Sequence | GAAACTGAAGAGACATCCTCACAAGAGTCTGCCGAAGAGGA | ||
| Motif Score | 2.047297619 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000403681.6; ENST00000539662.1 | ||
| External Link | RMBase: m6A_site_197463 | ||
| mod ID: M6ASITE013778 | Click to Show/Hide the Full List | ||
| mod site | chr12:65963287-65963288:+ | [13] | |
| Sequence | ACAAGAGTCTGCCGAAGAGGACTAGGGGGCGCCAACGTTCG | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HEK293T; HEC-1-A; NB4 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000403681.6; ENST00000539662.1 | ||
| External Link | RMBase: m6A_site_197464 | ||
| mod ID: M6ASITE013779 | Click to Show/Hide the Full List | ||
| mod site | chr12:65963349-65963350:+ | [12] | |
| Sequence | GGATCTTTTGAAGGGAGAAGACACTGCAGTGACCACTTATT | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | hESC-HEK293T; Huh7; HEC-1-A; NB4 | ||
| Seq Type List | MAZTER-seq; MeRIP-seq; m6A-seq | ||
| Transcript ID List | ENST00000539662.1; ENST00000403681.6 | ||
| External Link | RMBase: m6A_site_197465 | ||
| mod ID: M6ASITE013780 | Click to Show/Hide the Full List | ||
| mod site | chr12:65963452-65963453:+ | [12] | |
| Sequence | GGGTGGGGTGGGGAGAAATCACATAACCTTAAAAAGGACTA | ||
| Motif Score | 2.047297619 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000403681.6 | ||
| External Link | RMBase: m6A_site_197466 | ||
| mod ID: M6ASITE013781 | Click to Show/Hide the Full List | ||
| mod site | chr12:65963469-65963470:+ | [8] | |
| Sequence | ATCACATAACCTTAAAAAGGACTATATTAATCACCTTCTTT | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | Huh7 | ||
| Seq Type List | MeRIP-seq | ||
| Transcript ID List | ENST00000403681.6 | ||
| External Link | RMBase: m6A_site_197467 | ||
| mod ID: M6ASITE013782 | Click to Show/Hide the Full List | ||
| mod site | chr12:65963523-65963524:+ | [8] | |
| Sequence | AGTCCCAGGTTTAGTGAAAAACTGCTGTAAACACAGGGGAC | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | Huh7 | ||
| Seq Type List | MeRIP-seq | ||
| Transcript ID List | ENST00000403681.6 | ||
| External Link | RMBase: m6A_site_197468 | ||
| mod ID: M6ASITE013783 | Click to Show/Hide the Full List | ||
| mod site | chr12:65963533-65963534:+ | [8] | |
| Sequence | TTAGTGAAAAACTGCTGTAAACACAGGGGACACAGCTTAAC | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | Huh7 | ||
| Seq Type List | MeRIP-seq | ||
| Transcript ID List | ENST00000403681.6 | ||
| External Link | RMBase: m6A_site_197469 | ||
| mod ID: M6ASITE013784 | Click to Show/Hide the Full List | ||
| mod site | chr12:65963552-65963553:+ | [12] | |
| Sequence | AACACAGGGGACACAGCTTAACAATGCAACTTTTAATTACT | ||
| Motif Score | 2.168095238 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000403681.6 | ||
| External Link | RMBase: m6A_site_197470 | ||
| mod ID: M6ASITE013785 | Click to Show/Hide the Full List | ||
| mod site | chr12:65963678-65963679:+ | [12] | |
| Sequence | AATCACTGCACTGCATGCAAACAAGAAACGTGTCACACTTG | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000403681.6 | ||
| External Link | RMBase: m6A_site_197471 | ||
| mod ID: M6ASITE013786 | Click to Show/Hide the Full List | ||
| mod site | chr12:65963692-65963693:+ | [12] | |
| Sequence | ATGCAAACAAGAAACGTGTCACACTTGTGACGTCGGGCATT | ||
| Motif Score | 2.047297619 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000403681.6 | ||
| External Link | RMBase: m6A_site_197472 | ||
| mod ID: M6ASITE013787 | Click to Show/Hide the Full List | ||
| mod site | chr12:65963737-65963738:+ | [12] | |
| Sequence | TAGGAAGAACGCGGTGTGTAACACTGTGTACACCTCAAATA | ||
| Motif Score | 2.168095238 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000403681.6 | ||
| External Link | RMBase: m6A_site_197473 | ||
| mod ID: M6ASITE013788 | Click to Show/Hide the Full List | ||
| mod site | chr12:65963746-65963747:+ | [12] | |
| Sequence | CGCGGTGTGTAACACTGTGTACACCTCAAATACCACCCCAA | ||
| Motif Score | 2.856142857 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000403681.6 | ||
| External Link | RMBase: m6A_site_197474 | ||
| mod ID: M6ASITE013789 | Click to Show/Hide the Full List | ||
| mod site | chr12:65963798-65963799:+ | [12] | |
| Sequence | TAGTGAATCCTCTGTTTAGAACACCAAAGATAAGGACTAGA | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000403681.6 | ||
| External Link | RMBase: m6A_site_197475 | ||
| mod ID: M6ASITE013790 | Click to Show/Hide the Full List | ||
| mod site | chr12:65963813-65963814:+ | [14] | |
| Sequence | TTAGAACACCAAAGATAAGGACTAGATACTACTTTCTCTTT | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000403681.6 | ||
| External Link | RMBase: m6A_site_197476 | ||
| mod ID: M6ASITE013791 | Click to Show/Hide the Full List | ||
| mod site | chr12:65963851-65963852:+ | [12] | |
| Sequence | TTTTTCGTATAATCTTGTAGACACTTACTTGATGATTTTTA | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000403681.6 | ||
| External Link | RMBase: m6A_site_197477 | ||
| mod ID: M6ASITE013792 | Click to Show/Hide the Full List | ||
| mod site | chr12:65963857-65963858:+ | [14] | |
| Sequence | GTATAATCTTGTAGACACTTACTTGATGATTTTTAACTTTT | ||
| Motif Score | 2.494845238 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000403681.6 | ||
| External Link | RMBase: m6A_site_197478 | ||
| mod ID: M6ASITE013793 | Click to Show/Hide the Full List | ||
| mod site | chr12:65963924-65963925:+ | [12] | |
| Sequence | TGTATCCTTTCATTCAGCTAACAAACTAGAAAAGGTTATGT | ||
| Motif Score | 2.168095238 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000403681.6 | ||
| External Link | RMBase: m6A_site_197479 | ||
| mod ID: M6ASITE013794 | Click to Show/Hide the Full List | ||
| mod site | chr12:65963972-65963973:+ | [12] | |
| Sequence | TCAAAAAGGGAAGTAAGCAAACAAATATTGCCAACTCTTCT | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000403681.6 | ||
| External Link | RMBase: m6A_site_197480 | ||
| mod ID: M6ASITE013795 | Click to Show/Hide the Full List | ||
| mod site | chr12:65964271-65964272:+ | [12] | |
| Sequence | ATTTCTTTTGCTATAGTTATACATCAATTTAAAAAGCAAAA | ||
| Motif Score | 2.110482143 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000403681.6 | ||
| External Link | RMBase: m6A_site_197481 | ||
| mod ID: M6ASITE013796 | Click to Show/Hide the Full List | ||
| mod site | chr12:65964442-65964443:+ | [12] | |
| Sequence | ACTTCAAGTAATACGTTTTGACATAAGATGGTTGACCAAGG | ||
| Motif Score | 2.859755952 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000403681.6 | ||
| External Link | RMBase: m6A_site_197482 | ||
| mod ID: M6ASITE013797 | Click to Show/Hide the Full List | ||
| mod site | chr12:65964562-65964563:+ | [12] | |
| Sequence | CAATACTACCTCTGAATGTTACAACGAATTTACAGTCTAGT | ||
| Motif Score | 2.07285119 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000403681.6 | ||
| External Link | RMBase: m6A_site_197483 | ||
| mod ID: M6ASITE013798 | Click to Show/Hide the Full List | ||
| mod site | chr12:65964667-65964668:+ | [12] | |
| Sequence | TCATCTGCAGTTCTTTTTGTACACTTAATTACAGTTAAAGA | ||
| Motif Score | 2.856142857 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000403681.6 | ||
| External Link | RMBase: m6A_site_197484 | ||
| mod ID: M6ASITE013799 | Click to Show/Hide the Full List | ||
| mod site | chr12:65964716-65964717:+ | [14] | |
| Sequence | CCTTACTGTGTTTCAGCATGACTATGTATTTTTCTATGTTT | ||
| Motif Score | 3.28175 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000403681.6 | ||
| External Link | RMBase: m6A_site_197485 | ||
| mod ID: M6ASITE013800 | Click to Show/Hide the Full List | ||
| mod site | chr12:65964789-65964790:+ | [12] | |
| Sequence | AGCTTCTCTGCTAGATTTCTACATTAACTTGAAAATTTTTT | ||
| Motif Score | 2.078666667 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000403681.6 | ||
| External Link | RMBase: m6A_site_197486 | ||
| mod ID: M6ASITE013801 | Click to Show/Hide the Full List | ||
| mod site | chr12:65964853-65964854:+ | [12] | |
| Sequence | AAGGATAATTTTCCTCAATCACACTACACATCACACAAGAT | ||
| Motif Score | 2.047297619 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000403681.6 | ||
| External Link | RMBase: m6A_site_197487 | ||
| mod ID: M6ASITE013802 | Click to Show/Hide the Full List | ||
| mod site | chr12:65964985-65964986:+ | [14] | |
| Sequence | CCAGACTTCAAAAGGAATGAACTTGTTACTTGCAGCATTCA | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000403681.6 | ||
| External Link | RMBase: m6A_site_197488 | ||
| mod ID: M6ASITE013803 | Click to Show/Hide the Full List | ||
| mod site | chr12:65965058-65965059:+ | [14] | |
| Sequence | TGCAGCTAACCCTAGTCAAAACTATTTTTGTAAAAGACATT | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000403681.6 | ||
| External Link | RMBase: m6A_site_197489 | ||
| mod ID: M6ASITE013804 | Click to Show/Hide the Full List | ||
| mod site | chr12:65965074-65965075:+ | [14] | |
| Sequence | CAAAACTATTTTTGTAAAAGACATTTGATAGAAAGGAACAC | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000403681.6 | ||
| External Link | RMBase: m6A_site_197490 | ||
| mod ID: M6ASITE013805 | Click to Show/Hide the Full List | ||
| mod site | chr12:65965158-65965159:+ | [14] | |
| Sequence | AAAAGCCAACCTTCAAAGAAACTTGAAGCTTTGTAGGTGAG | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000403681.6 | ||
| External Link | RMBase: m6A_site_197491 | ||
| mod ID: M6ASITE013806 | Click to Show/Hide the Full List | ||
| mod site | chr12:65965184-65965185:+ | [12] | |
| Sequence | AGCTTTGTAGGTGAGATGCAACAAGCCCTGCTTTTGCATAA | ||
| Motif Score | 2.173910714 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000403681.6 | ||
| External Link | RMBase: m6A_site_197492 | ||
| mod ID: M6ASITE013807 | Click to Show/Hide the Full List | ||
| mod site | chr12:65965257-65965258:+ | [14] | |
| Sequence | TGAATATAAGAAAATGCTTGACAAATATTTTCATGTATTTT | ||
| Motif Score | 2.859755952 | ||
| Cell/Tissue List | HEK293T; hESC-HEK293T | ||
| Seq Type List | DART-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000403681.6 | ||
| External Link | RMBase: m6A_site_197493 | ||
| mod ID: M6ASITE013808 | Click to Show/Hide the Full List | ||
| mod site | chr12:65965374-65965375:+ | [14] | |
| Sequence | CTCTCCTAGTGAGTGTGCTGACTTTTTAACATGGTATTATC | ||
| Motif Score | 3.28175 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000403681.6 | ||
| External Link | RMBase: m6A_site_197494 | ||
| mod ID: M6ASITE013809 | Click to Show/Hide the Full List | ||
| mod site | chr12:65965382-65965383:+ | [12] | |
| Sequence | GTGAGTGTGCTGACTTTTTAACATGGTATTATCAACTGGGC | ||
| Motif Score | 2.168095238 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000403681.6 | ||
| External Link | RMBase: m6A_site_197495 | ||
| mod ID: M6ASITE013810 | Click to Show/Hide the Full List | ||
| mod site | chr12:65965516-65965517:+ | [12] | |
| Sequence | GGGAGGTAGTTTCAAAGGCCACATACCTCTCTGAGACTGGC | ||
| Motif Score | 2.053113095 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000403681.6 | ||
| External Link | RMBase: m6A_site_197496 | ||
| mod ID: M6ASITE013811 | Click to Show/Hide the Full List | ||
| mod site | chr12:65965591-65965592:+ | [12] | |
| Sequence | ATGGAGAGAATTAAAACTCAACATTACTGTTAACTGTGCGT | ||
| Motif Score | 2.173910714 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000403681.6 | ||
| External Link | RMBase: m6A_site_197497 | ||
| mod ID: M6ASITE013812 | Click to Show/Hide the Full List | ||
| mod site | chr12:65965733-65965734:+ | [12] | |
| Sequence | CAATTGCTCATGTTGGCCAAACATGGTGCACCGAGTGATTT | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000403681.6 | ||
| External Link | RMBase: m6A_site_197498 | ||
| mod ID: M6ASITE013813 | Click to Show/Hide the Full List | ||
| mod site | chr12:65965795-65965796:+ | [14] | |
| Sequence | TTTTATTTCCTGTATGTTGTACAATCAAAACACACTACTAC | ||
| Motif Score | 2.856142857 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000403681.6 | ||
| External Link | RMBase: m6A_site_197499 | ||
| mod ID: M6ASITE013814 | Click to Show/Hide the Full List | ||
| mod site | chr12:65965885-65965886:+ | [12] | |
| Sequence | TGGCCAATGGAACAGTAAGAACATCATAAAATTTTTATATA | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000403681.6 | ||
| External Link | RMBase: m6A_site_197500 | ||
| mod ID: M6ASITE013815 | Click to Show/Hide the Full List | ||
| mod site | chr12:65965970-65965971:+ | [12] | |
| Sequence | TGCTGTTGTTGGTCGCAGCTACATAAGACTGGACATTTAAC | ||
| Motif Score | 2.078666667 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000403681.6 | ||
| External Link | RMBase: m6A_site_197501 | ||
| mod ID: M6ASITE013816 | Click to Show/Hide the Full List | ||
| mod site | chr12:65966134-65966135:+ | [14] | |
| Sequence | GAATTGCTGTCTATGATTGTACTTTGAATCGCTTGCTTGTT | ||
| Motif Score | 3.278136905 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000403681.6 | ||
| External Link | RMBase: m6A_site_197502 | ||
| mod ID: M6ASITE013817 | Click to Show/Hide the Full List | ||
| mod site | chr12:65966194-65966195:+ | [14] | |
| Sequence | ATTATCACTGTCTGTTCTGCACAATAAACATAACAGCCTCT | ||
| Motif Score | 2.830589286 | ||
| Cell/Tissue List | HEK293T; hESC-HEK293T | ||
| Seq Type List | DART-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000403681.6 | ||
| External Link | RMBase: m6A_site_197503 | ||
Pseudouridine (Pseudo)
| In total 1 m6A sequence/site(s) in this target gene | |||
| mod ID: PSESITE000022 | Click to Show/Hide the Full List | ||
| mod site | chr12:65964467-65964468:+ | [15] | |
| Sequence | AGATGGTTGACCAAGGTGCTTTTCTTCGGCTTGAGTTCACC | ||
| Transcript ID List | ENST00000403681.6 | ||
| External Link | RMBase: Pseudo_site_1170 | ||
References