m6A Target Gene Information
General Information of the m6A Target Gene (ID: M6ATAR00665)
Full List of m6A Methylation Regulator of This Target Gene and Corresponding Disease/Drug Response(s)
PDGFC
can be regulated by the following regulator(s), and cause disease/drug response(s). You can browse detail information of regulator(s) or disease/drug response(s).
Browse Regulator
Browse Disease
YTH domain-containing family protein 2 (YTHDF2) [READER]
| Representative RNA-seq result indicating the expression of this target gene regulated by YTHDF2 | ||
| Cell Line | GSC11 cell line | Homo sapiens |
|
Treatment: siYTHDF2 GSC11 cells
Control: siControl GSC11 cells
|
GSE142825 | |
| Regulation |
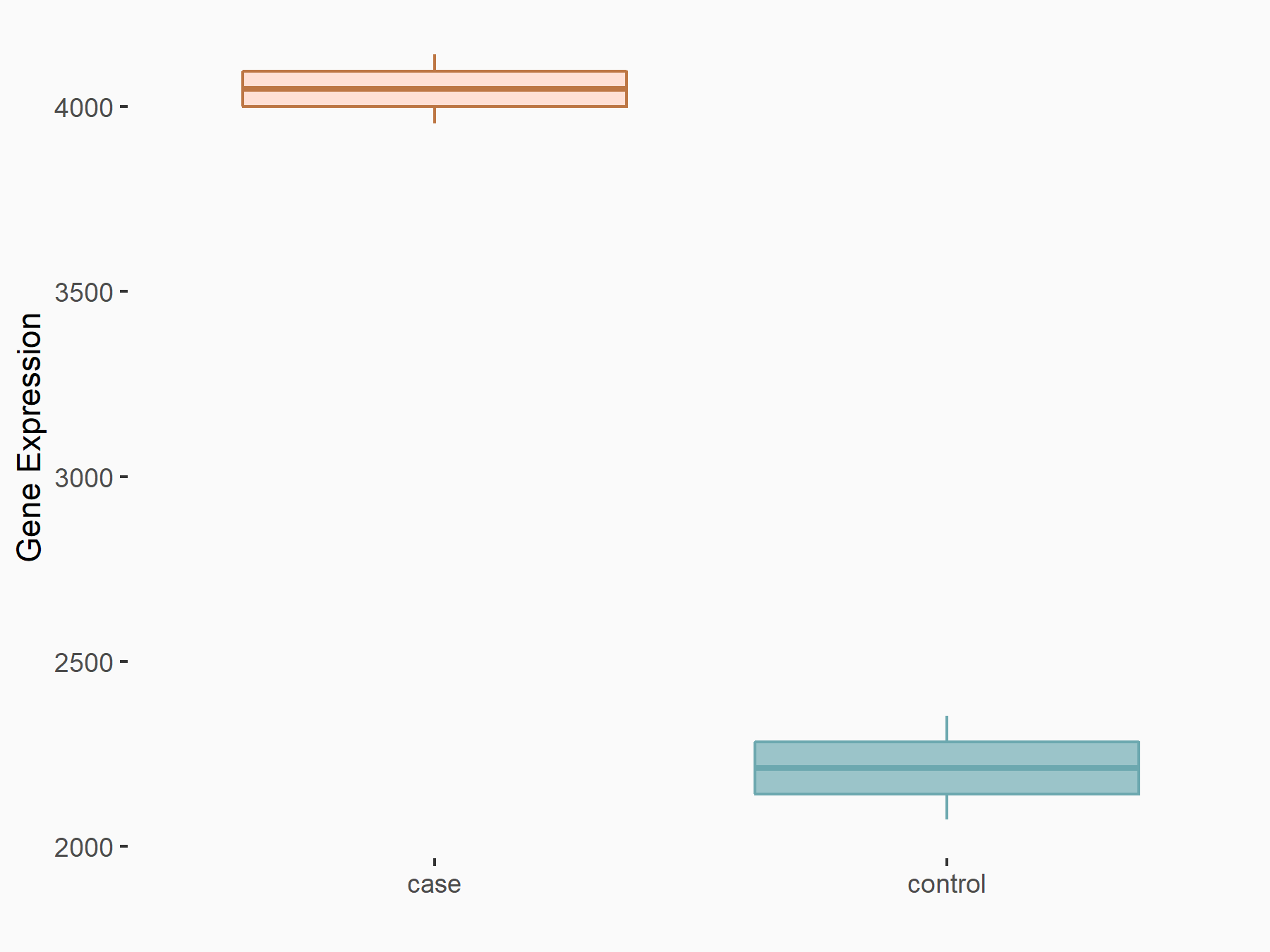  |
logFC: 8.71E-01 p-value: 1.24E-12 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between PDGFC and the regulator | ||
| Cell Line | Hela | Homo sapiens |
| Regulation | logFC: 1.41E+00 | GSE49339 |
| In total 1 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [1] | |||
| Response Summary | FTO downregulation leads to increased m6A modifications in the 3' UTR of Platelet-derived growth factor C (PDGFC) and then modulates the degradation of its transcriptional level in an m6A-YTHDF2-dependent manner, highlighting a potential therapeutic target for PDAC treatment and prognostic prediction. | |||
| Target Regulation | Down regulation | |||
| Responsed Disease | Pancreatic cancer | ICD-11: 2C10 | ||
| Pathway Response | PI3K-Akt signaling pathway | hsa04151 | ||
| In-vitro Model | SW1990 | Pancreatic adenocarcinoma | Homo sapiens | CVCL_1723 |
| PANC-1 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0480 | |
| MIA PaCa-2 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0428 | |
| HPDE | Normal | Homo sapiens | CVCL_4376 | |
| CFPAC-1 | Cystic fibrosis | Homo sapiens | CVCL_1119 | |
| Capan-1 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0237 | |
| BxPC-3 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0186 | |
| AsPC-1 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0152 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| In-vivo Model | The right flanks of mice were injected subcutaneously with 2 × 106 MiaPaCa-2 cells stably expressing shFTO and a scrambled shRNA in 100 uL PBS. Tumors were measured using an external caliper once per week, and tumor volume was calculated with the formula: (length × width2)/2. | |||
Fat mass and obesity-associated protein (FTO) [ERASER]
| Representative RNA-seq result indicating the expression of this target gene regulated by FTO | ||
| Cell Line | 253J cell line | Homo sapiens |
|
Treatment: siFTO 253J cells
Control: 253J cells
|
GSE150239 | |
| Regulation |
  |
logFC: 8.37E+00 p-value: 3.21E-02 |
| More Results | Click to View More RNA-seq Results | |
| In total 1 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [1] | |||
| Response Summary | FTO downregulation leads to increased m6A modifications in the 3' UTR of Platelet-derived growth factor C (PDGFC) and then modulates the degradation of its transcriptional level in an m6A-YTHDF2-dependent manner, highlighting a potential therapeutic target for PDAC treatment and prognostic prediction. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Pancreatic cancer | ICD-11: 2C10 | ||
| Pathway Response | PI3K-Akt signaling pathway | hsa04151 | ||
| In-vitro Model | SW1990 | Pancreatic adenocarcinoma | Homo sapiens | CVCL_1723 |
| PANC-1 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0480 | |
| MIA PaCa-2 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0428 | |
| HPDE | Normal | Homo sapiens | CVCL_4376 | |
| CFPAC-1 | Cystic fibrosis | Homo sapiens | CVCL_1119 | |
| Capan-1 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0237 | |
| BxPC-3 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0186 | |
| AsPC-1 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0152 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| In-vivo Model | The right flanks of mice were injected subcutaneously with 2 × 106 MiaPaCa-2 cells stably expressing shFTO and a scrambled shRNA in 100 uL PBS. Tumors were measured using an external caliper once per week, and tumor volume was calculated with the formula: (length × width2)/2. | |||
Pancreatic cancer [ICD-11: 2C10]
| In total 2 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [1] | |||
| Response Summary | FTO downregulation leads to increased m6A modifications in the 3' UTR of Platelet-derived growth factor C (PDGFC) and then modulates the degradation of its transcriptional level in an m6A-YTHDF2-dependent manner, highlighting a potential therapeutic target for PDAC treatment and prognostic prediction. | |||
| Responsed Disease | Pancreatic cancer [ICD-11: 2C10] | |||
| Target Regulator | Fat mass and obesity-associated protein (FTO) | ERASER | ||
| Target Regulation | Up regulation | |||
| Pathway Response | PI3K-Akt signaling pathway | hsa04151 | ||
| In-vitro Model | SW1990 | Pancreatic adenocarcinoma | Homo sapiens | CVCL_1723 |
| PANC-1 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0480 | |
| MIA PaCa-2 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0428 | |
| HPDE | Normal | Homo sapiens | CVCL_4376 | |
| CFPAC-1 | Cystic fibrosis | Homo sapiens | CVCL_1119 | |
| Capan-1 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0237 | |
| BxPC-3 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0186 | |
| AsPC-1 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0152 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| In-vivo Model | The right flanks of mice were injected subcutaneously with 2 × 106 MiaPaCa-2 cells stably expressing shFTO and a scrambled shRNA in 100 uL PBS. Tumors were measured using an external caliper once per week, and tumor volume was calculated with the formula: (length × width2)/2. | |||
| Experiment 2 Reporting the m6A-centered Disease Response | [1] | |||
| Response Summary | FTO downregulation leads to increased m6A modifications in the 3' UTR of Platelet-derived growth factor C (PDGFC) and then modulates the degradation of its transcriptional level in an m6A-YTHDF2-dependent manner, highlighting a potential therapeutic target for PDAC treatment and prognostic prediction. | |||
| Responsed Disease | Pancreatic cancer [ICD-11: 2C10] | |||
| Target Regulator | YTH domain-containing family protein 2 (YTHDF2) | READER | ||
| Target Regulation | Down regulation | |||
| Pathway Response | PI3K-Akt signaling pathway | hsa04151 | ||
| In-vitro Model | SW1990 | Pancreatic adenocarcinoma | Homo sapiens | CVCL_1723 |
| PANC-1 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0480 | |
| MIA PaCa-2 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0428 | |
| HPDE | Normal | Homo sapiens | CVCL_4376 | |
| CFPAC-1 | Cystic fibrosis | Homo sapiens | CVCL_1119 | |
| Capan-1 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0237 | |
| BxPC-3 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0186 | |
| AsPC-1 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0152 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| In-vivo Model | The right flanks of mice were injected subcutaneously with 2 × 106 MiaPaCa-2 cells stably expressing shFTO and a scrambled shRNA in 100 uL PBS. Tumors were measured using an external caliper once per week, and tumor volume was calculated with the formula: (length × width2)/2. | |||
RNA Modification Sequencing Data Associated with the Target (ID: M6ATAR00665)
| In total 7 m6A sequence/site(s) in this target gene | |||
| mod ID: A2ISITE000628 | Click to Show/Hide the Full List | ||
| mod site | chr4:156766529-156766530:- | [2] | |
| Sequence | TTTTGTAAAATTTAAAGTTGACTTTTCAAGAAGGATGGTTT | ||
| Transcript ID List | ENST00000504672.5; ENST00000506880.5; ENST00000274071.6; ENST00000502773.6; ENST00000422544.2 | ||
| External Link | RMBase: RNA-editing_site_107124 | ||
| mod ID: A2ISITE000629 | Click to Show/Hide the Full List | ||
| mod site | chr4:156766535-156766536:- | [2] | |
| Sequence | TAAAGGTTTTGTAAAATTTAAAGTTGACTTTTCAAGAAGGA | ||
| Transcript ID List | ENST00000502773.6; ENST00000506880.5; ENST00000422544.2; ENST00000504672.5; ENST00000274071.6 | ||
| External Link | RMBase: RNA-editing_site_107125 | ||
| mod ID: A2ISITE000630 | Click to Show/Hide the Full List | ||
| mod site | chr4:156766542-156766543:- | [2] | |
| Sequence | TGTAATTTAAAGGTTTTGTAAAATTTAAAGTTGACTTTTCA | ||
| Transcript ID List | ENST00000502773.6; ENST00000274071.6; ENST00000506880.5; ENST00000422544.2; ENST00000504672.5 | ||
| External Link | RMBase: RNA-editing_site_107126 | ||
| mod ID: A2ISITE000631 | Click to Show/Hide the Full List | ||
| mod site | chr4:156766543-156766544:- | [2] | |
| Sequence | GTGTAATTTAAAGGTTTTGTAAAATTTAAAGTTGACTTTTC | ||
| Transcript ID List | ENST00000502773.6; ENST00000422544.2; ENST00000506880.5; ENST00000504672.5; ENST00000274071.6 | ||
| External Link | RMBase: RNA-editing_site_107127 | ||
| mod ID: A2ISITE000632 | Click to Show/Hide the Full List | ||
| mod site | chr4:156767025-156767026:- | [2] | |
| Sequence | TTCTACAAAACATTAACATTAGACCAAGTCAAAATCTGCCC | ||
| Transcript ID List | ENST00000274071.6; ENST00000504672.5; ENST00000422544.2; ENST00000502773.6; ENST00000506880.5 | ||
| External Link | RMBase: RNA-editing_site_107128 | ||
| mod ID: A2ISITE000633 | Click to Show/Hide the Full List | ||
| mod site | chr4:156767124-156767125:- | [2] | |
| Sequence | GTGATGGAGTCTCCCAATTAAATCCCAACACCTCCCCCAGC | ||
| Transcript ID List | ENST00000274071.6; ENST00000502773.6; ENST00000422544.2; ENST00000506880.5; ENST00000504672.5 | ||
| External Link | RMBase: RNA-editing_site_107129 | ||
| mod ID: A2ISITE000634 | Click to Show/Hide the Full List | ||
| mod site | chr4:156767125-156767126:- | [2] | |
| Sequence | AGTGATGGAGTCTCCCAATTAAATCCCAACACCTCCCCCAG | ||
| Transcript ID List | ENST00000506880.5; ENST00000504672.5; ENST00000274071.6; ENST00000502773.6; ENST00000422544.2 | ||
| External Link | RMBase: RNA-editing_site_107130 | ||
5-methylcytidine (m5C)
| In total 5 m6A sequence/site(s) in this target gene | |||
| mod ID: M5CSITE003351 | Click to Show/Hide the Full List | ||
| mod site | chr4:156971508-156971509:- | [3] | |
| Sequence | CGCCCGGGCCGCCGTCGGCCCGGAGCCCCCCGGCCCCGCTC | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000502773.6 | ||
| External Link | RMBase: m5C_site_34420 | ||
| mod ID: M5CSITE003352 | Click to Show/Hide the Full List | ||
| mod site | chr4:156971509-156971510:- | [3] | |
| Sequence | GCGCCCGGGCCGCCGTCGGCCCGGAGCCCCCCGGCCCCGCT | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000502773.6 | ||
| External Link | RMBase: m5C_site_34421 | ||
| mod ID: M5CSITE003353 | Click to Show/Hide the Full List | ||
| mod site | chr4:156971510-156971511:- | [3] | |
| Sequence | TGCGCCCGGGCCGCCGTCGGCCCGGAGCCCCCCGGCCCCGC | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000502773.6 | ||
| External Link | RMBase: m5C_site_34422 | ||
| mod ID: M5CSITE003354 | Click to Show/Hide the Full List | ||
| mod site | chr4:156971519-156971520:- | [3] | |
| Sequence | CTCTCCGCCTGCGCCCGGGCCGCCGTCGGCCCGGAGCCCCC | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000502773.6 | ||
| External Link | RMBase: m5C_site_34423 | ||
| mod ID: M5CSITE003355 | Click to Show/Hide the Full List | ||
| mod site | chr4:156971520-156971521:- | [3] | |
| Sequence | TCTCTCCGCCTGCGCCCGGGCCGCCGTCGGCCCGGAGCCCC | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000502773.6 | ||
| External Link | RMBase: m5C_site_34424 | ||
N6-methyladenosine (m6A)
| In total 46 m6A sequence/site(s) in this target gene | |||
| mod ID: M6ASITE068553 | Click to Show/Hide the Full List | ||
| mod site | chr4:156761578-156761579:- | [4] | |
| Sequence | GCTCAAGATTTATTTCTTGGACTCTGAGAAAATGAAAGATA | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | Huh7 | ||
| Seq Type List | MeRIP-seq | ||
| Transcript ID List | ENST00000502773.6 | ||
| External Link | RMBase: m6A_site_655240 | ||
| mod ID: M6ASITE068554 | Click to Show/Hide the Full List | ||
| mod site | chr4:156761874-156761875:- | [5] | |
| Sequence | TGCTGTGCTGTGCAGTAGGAACACATCCTATTTATTGTGAT | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000502773.6; ENST00000274071.6 | ||
| External Link | RMBase: m6A_site_655241 | ||
| mod ID: M6ASITE068555 | Click to Show/Hide the Full List | ||
| mod site | chr4:156761939-156761940:- | [5] | |
| Sequence | TCTGAGCCTAGCTCAGAAAAACATAAAGCACCTTGAAAAAG | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000502773.6; ENST00000274071.6 | ||
| External Link | RMBase: m6A_site_655242 | ||
| mod ID: M6ASITE068556 | Click to Show/Hide the Full List | ||
| mod site | chr4:156762275-156762276:- | [5] | |
| Sequence | ATTTAATATTCTTTTTTATGACAACTTAGATCAACTATTTT | ||
| Motif Score | 2.859755952 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000502773.6; ENST00000274071.6 | ||
| External Link | RMBase: m6A_site_655243 | ||
| mod ID: M6ASITE068557 | Click to Show/Hide the Full List | ||
| mod site | chr4:156762352-156762353:- | [5] | |
| Sequence | ATTTTTATATTCTCCTTTTGACATTATAACTGTTGGCTTTT | ||
| Motif Score | 2.859755952 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000502773.6; ENST00000274071.6 | ||
| External Link | RMBase: m6A_site_655244 | ||
| mod ID: M6ASITE068558 | Click to Show/Hide the Full List | ||
| mod site | chr4:156762374-156762375:- | [5] | |
| Sequence | GTTTATTTGTTTCATTGTGTACATTTTTATATTCTCCTTTT | ||
| Motif Score | 2.856142857 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000274071.6; ENST00000502773.6 | ||
| External Link | RMBase: m6A_site_655245 | ||
| mod ID: M6ASITE068559 | Click to Show/Hide the Full List | ||
| mod site | chr4:156762439-156762440:- | [6] | |
| Sequence | TCATGGTTTGGAAGAGATAAACCTGAAAAGAAGAGTGGCCT | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | HEK293T; hNPCs; fibroblasts; Huh7; HEK293A-TOA; iSLK; MSC; TIME; TREX | ||
| Seq Type List | MeRIP-seq; m6A-seq | ||
| Transcript ID List | ENST00000274071.6; ENST00000502773.6 | ||
| External Link | RMBase: m6A_site_655246 | ||
| mod ID: M6ASITE068560 | Click to Show/Hide the Full List | ||
| mod site | chr4:156762463-156762464:- | [5] | |
| Sequence | CCATTTAGAAGAAGAGAACTACATTCATGGTTTGGAAGAGA | ||
| Motif Score | 2.078666667 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000274071.6; ENST00000502773.6 | ||
| External Link | RMBase: m6A_site_655247 | ||
| mod ID: M6ASITE068561 | Click to Show/Hide the Full List | ||
| mod site | chr4:156762466-156762467:- | [6] | |
| Sequence | CTGCCATTTAGAAGAAGAGAACTACATTCATGGTTTGGAAG | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HEK293T; U2OS; hNPCs; fibroblasts; A549; Huh7; HEK293A-TOA; iSLK; MSC; TIME; TREX | ||
| Seq Type List | MeRIP-seq; m6A-seq | ||
| Transcript ID List | ENST00000502773.6; ENST00000274071.6 | ||
| External Link | RMBase: m6A_site_655248 | ||
| mod ID: M6ASITE068562 | Click to Show/Hide the Full List | ||
| mod site | chr4:156762521-156762522:- | [6] | |
| Sequence | ACTTGTGTCGTGCTGATAGGACAGACTGGATTTTTCATATT | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HEK293T; U2OS; hNPCs; fibroblasts; A549; Huh7; HEK293A-TOA; iSLK; MSC; TIME; TREX | ||
| Seq Type List | MeRIP-seq; m6A-seq | ||
| Transcript ID List | ENST00000502773.6; ENST00000274071.6 | ||
| External Link | RMBase: m6A_site_655249 | ||
| mod ID: M6ASITE068563 | Click to Show/Hide the Full List | ||
| mod site | chr4:156762541-156762542:- | [6] | |
| Sequence | ACTATGTTGCTATGAATTAAACTTGTGTCGTGCTGATAGGA | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HEK293T; U2OS; hNPCs; fibroblasts; A549; Huh7; HEK293A-TOA; iSLK; MSC; TIME; TREX | ||
| Seq Type List | MeRIP-seq; m6A-seq | ||
| Transcript ID List | ENST00000502773.6; ENST00000274071.6 | ||
| External Link | RMBase: m6A_site_655250 | ||
| mod ID: M6ASITE068564 | Click to Show/Hide the Full List | ||
| mod site | chr4:156762561-156762562:- | [6] | |
| Sequence | AACCTGGTTTTTAAAAAGGAACTATGTTGCTATGAATTAAA | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HEK293T; U2OS; hNPCs; fibroblasts; A549; Huh7; HEK293A-TOA; iSLK; MSC; TIME; TREX | ||
| Seq Type List | MeRIP-seq; m6A-seq | ||
| Transcript ID List | ENST00000502773.6; ENST00000274071.6 | ||
| External Link | RMBase: m6A_site_655251 | ||
| mod ID: M6ASITE068565 | Click to Show/Hide the Full List | ||
| mod site | chr4:156762580-156762581:- | [6] | |
| Sequence | GAACATTCTATGTACTACAAACCTGGTTTTTAAAAAGGAAC | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | HEK293T; U2OS; hNPCs; fibroblasts; A549; Huh7; HEK293A-TOA; iSLK; MSC; TIME; TREX | ||
| Seq Type List | MeRIP-seq; m6A-seq | ||
| Transcript ID List | ENST00000274071.6; ENST00000502773.6 | ||
| External Link | RMBase: m6A_site_655252 | ||
| mod ID: M6ASITE068566 | Click to Show/Hide the Full List | ||
| mod site | chr4:156762584-156762585:- | [5] | |
| Sequence | ACCAGAACATTCTATGTACTACAAACCTGGTTTTTAAAAAG | ||
| Motif Score | 2.078666667 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000274071.6; ENST00000502773.6 | ||
| External Link | RMBase: m6A_site_655253 | ||
| mod ID: M6ASITE068567 | Click to Show/Hide the Full List | ||
| mod site | chr4:156762598-156762599:- | [6] | |
| Sequence | ATTCACATATGTAAACCAGAACATTCTATGTACTACAAACC | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HEK293T; U2OS; fibroblasts; A549; Huh7; HEK293A-TOA; iSLK; MSC; TIME; TREX | ||
| Seq Type List | MeRIP-seq; m6A-seq | ||
| Transcript ID List | ENST00000274071.6; ENST00000502773.6 | ||
| External Link | RMBase: m6A_site_655254 | ||
| mod ID: M6ASITE068568 | Click to Show/Hide the Full List | ||
| mod site | chr4:156762604-156762605:- | [6] | |
| Sequence | GCTCATATTCACATATGTAAACCAGAACATTCTATGTACTA | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | HEK293T; U2OS; fibroblasts; A549; Huh7; HEK293A-TOA; iSLK; MSC; TIME; TREX | ||
| Seq Type List | MeRIP-seq; m6A-seq | ||
| Transcript ID List | ENST00000502773.6; ENST00000274071.6 | ||
| External Link | RMBase: m6A_site_655255 | ||
| mod ID: M6ASITE068569 | Click to Show/Hide the Full List | ||
| mod site | chr4:156762614-156762615:- | [5] | |
| Sequence | TTTTTTTTTTGCTCATATTCACATATGTAAACCAGAACATT | ||
| Motif Score | 2.047297619 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000502773.6; ENST00000274071.6 | ||
| External Link | RMBase: m6A_site_655256 | ||
| mod ID: M6ASITE068570 | Click to Show/Hide the Full List | ||
| mod site | chr4:156762725-156762726:- | [7] | |
| Sequence | AATGTCAGTACAGGAAAAAAACTGTGCAAGTGAGCACCTGA | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HepG2; HEK293T; U2OS; fibroblasts; A549; Huh7; HEK293A-TOA; iSLK; MSC; TIME; TREX; AML | ||
| Seq Type List | m6A-seq; MeRIP-seq; miCLIP | ||
| Transcript ID List | ENST00000274071.6; ENST00000502773.6 | ||
| External Link | RMBase: m6A_site_655257 | ||
| mod ID: M6ASITE068571 | Click to Show/Hide the Full List | ||
| mod site | chr4:156762882-156762883:- | [7] | |
| Sequence | TTTGAGAGGAGGCCTAAAGGACAGGAGAAAAGGTCTTCAAT | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HepG2; HEK293T; U2OS; hNPCs; fibroblasts; A549; MM6; Huh7; HEK293A-TOA; iSLK; MSC; TIME; TREX; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000504672.5; ENST00000502773.6; ENST00000274071.6 | ||
| External Link | RMBase: m6A_site_655258 | ||
| mod ID: M6ASITE068572 | Click to Show/Hide the Full List | ||
| mod site | chr4:156762930-156762931:- | [7] | |
| Sequence | TCTGAAAGAGGAGACATCAAACAGAATTAGGAGTTGTGCAA | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HepG2; HEK293T; U2OS; hNPCs; fibroblasts; A549; MM6; Huh7; HEK293A-TOA; iSLK; MSC; TIME; TREX; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000504672.5; ENST00000274071.6; ENST00000502773.6 | ||
| External Link | RMBase: m6A_site_655259 | ||
| mod ID: M6ASITE068573 | Click to Show/Hide the Full List | ||
| mod site | chr4:156762937-156762938:- | [7] | |
| Sequence | AGTGCATTCTGAAAGAGGAGACATCAAACAGAATTAGGAGT | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HepG2; HEK293T; U2OS; hNPCs; fibroblasts; A549; MM6; Huh7; HEK293A-TOA; iSLK; MSC; TIME; TREX; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000502773.6; ENST00000504672.5; ENST00000274071.6 | ||
| External Link | RMBase: m6A_site_655260 | ||
| mod ID: M6ASITE068574 | Click to Show/Hide the Full List | ||
| mod site | chr4:156762959-156762960:- | [8] | |
| Sequence | ACCTTTCATCTTCAGGATTTACAGTGCATTCTGAAAGAGGA | ||
| Motif Score | 2.07285119 | ||
| Cell/Tissue List | brain; liver | ||
| Seq Type List | m6A-REF-seq | ||
| Transcript ID List | ENST00000274071.6; ENST00000506880.5; ENST00000502773.6; ENST00000504672.5 | ||
| External Link | RMBase: m6A_site_655261 | ||
| mod ID: M6ASITE068575 | Click to Show/Hide the Full List | ||
| mod site | chr4:156762979-156762980:- | [7] | |
| Sequence | CTCAGTTGTTTGCTTCAAGGACCTTTCATCTTCAGGATTTA | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HepG2; HEK293T; U2OS; hNPCs; fibroblasts; A549; MM6; Huh7; HEK293A-TOA; iSLK; MSC; TIME; TREX; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000506880.5; ENST00000504672.5; ENST00000502773.6; ENST00000274071.6 | ||
| External Link | RMBase: m6A_site_655262 | ||
| mod ID: M6ASITE068576 | Click to Show/Hide the Full List | ||
| mod site | chr4:156763100-156763101:- | [8] | |
| Sequence | ACTGTGTGTGCAGAGGGAGCACAGGAGGATAGCCGCATCAC | ||
| Motif Score | 2.830589286 | ||
| Cell/Tissue List | kidney | ||
| Seq Type List | m6A-REF-seq | ||
| Transcript ID List | ENST00000504672.5; ENST00000422544.2; ENST00000506880.5; ENST00000274071.6; ENST00000502773.6 | ||
| External Link | RMBase: m6A_site_655263 | ||
| mod ID: M6ASITE068577 | Click to Show/Hide the Full List | ||
| mod site | chr4:156763184-156763185:- | [9] | |
| Sequence | TCCTTCAGTTGAGACCAAAGACCGGTGTCAGGGGATTGCAC | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HEK293T; U2OS; hNPCs; endometrial | ||
| Seq Type List | MeRIP-seq; m6A-seq | ||
| Transcript ID List | ENST00000422544.2; ENST00000274071.6; ENST00000502773.6; ENST00000506880.5; ENST00000504672.5 | ||
| External Link | RMBase: m6A_site_655264 | ||
| mod ID: M6ASITE068578 | Click to Show/Hide the Full List | ||
| mod site | chr4:156763191-156763192:- | [9] | |
| Sequence | TTTCAGGTCCTTCAGTTGAGACCAAAGACCGGTGTCAGGGG | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HEK293T; U2OS; hNPCs; endometrial | ||
| Seq Type List | MeRIP-seq; m6A-seq | ||
| Transcript ID List | ENST00000274071.6; ENST00000502773.6; ENST00000422544.2; ENST00000504672.5; ENST00000506880.5 | ||
| External Link | RMBase: m6A_site_655265 | ||
| mod ID: M6ASITE068579 | Click to Show/Hide the Full List | ||
| mod site | chr4:156850276-156850277:- | [5] | |
| Sequence | GTAGAGGAAAATGTATGGATACAACTTACGTTTGATGAAAG | ||
| Motif Score | 2.110482143 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000422544.2; ENST00000502773.6; ENST00000274071.6; ENST00000512711.1 | ||
| External Link | RMBase: m6A_site_655266 | ||
| mod ID: M6ASITE068580 | Click to Show/Hide the Full List | ||
| mod site | chr4:156850411-156850412:- | [5] | |
| Sequence | ATGTTTTTTGCTTTAGGAGTACAAGATCCTCAGCATGAGAG | ||
| Motif Score | 2.856142857 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000274071.6; ENST00000422544.2; ENST00000512711.1; ENST00000502773.6 | ||
| External Link | RMBase: m6A_site_655267 | ||
| mod ID: M6ASITE068581 | Click to Show/Hide the Full List | ||
| mod site | chr4:156970792-156970793:- | [10] | |
| Sequence | CAGTTTTCCAGCAACAAGGAACAGAACGGTGAGTCTTTCCC | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | iSLK; MSC; TIME; endometrial | ||
| Seq Type List | MeRIP-seq; m6A-seq | ||
| Transcript ID List | ENST00000422544.2; ENST00000502773.6; ENST00000274071.6; ENST00000513664.1 | ||
| External Link | RMBase: m6A_site_655268 | ||
| mod ID: M6ASITE068582 | Click to Show/Hide the Full List | ||
| mod site | chr4:156970845-156970846:- | [11] | |
| Sequence | TGGCCGGCCAGAGACAGGGGACTCAGGCGGAATCCAACCTG | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa; CD34; HepG2; HEK293T; Jurkat; peripheral-blood; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; TREX; endometrial; HEC-1-A; GSCs; MM6 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000513664.1; ENST00000502773.6; ENST00000422544.2; ENST00000274071.6 | ||
| External Link | RMBase: m6A_site_655269 | ||
| mod ID: M6ASITE068583 | Click to Show/Hide the Full List | ||
| mod site | chr4:156970852-156970853:- | [11] | |
| Sequence | TCTGCCCTGGCCGGCCAGAGACAGGGGACTCAGGCGGAATC | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa; CD34; HepG2; HEK293T; Jurkat; peripheral-blood; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; TREX; endometrial; HEC-1-A; GSCs; MM6 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000274071.6; ENST00000502773.6; ENST00000513664.1; ENST00000422544.2 | ||
| External Link | RMBase: m6A_site_655270 | ||
| mod ID: M6ASITE068584 | Click to Show/Hide the Full List | ||
| mod site | chr4:156970963-156970964:- | [11] | |
| Sequence | CCCTGGCGGTGGTGAAAGAGACTCGGGAGTCGCTGCTTCCA | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa; CD34; HepG2; HEK293T; A549; H1A; H1B; fibroblasts; MM6; Huh7; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; TREX; endometrial; HEC-1-A; GSCs | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000502773.6; ENST00000513664.1; ENST00000274071.6 | ||
| External Link | RMBase: m6A_site_655271 | ||
| mod ID: M6ASITE068585 | Click to Show/Hide the Full List | ||
| mod site | chr4:156971044-156971045:- | [11] | |
| Sequence | TTGGATGGGATTATGTGGAAACTACCCTGCGATTCTCTGCT | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa; CD34; HepG2; HEK293T; A549; H1A; H1B; fibroblasts; MM6; Huh7; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; TREX; endometrial; HEC-1-A; GSCs | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000274071.6; ENST00000502773.6 | ||
| External Link | RMBase: m6A_site_655272 | ||
| mod ID: M6ASITE068586 | Click to Show/Hide the Full List | ||
| mod site | chr4:156971092-156971093:- | [11] | |
| Sequence | ATCCTTTTCAAAAACTGGAGACACAGAAGAGGGCTCTAGGA | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa; CD34; HepG2; HEK293T; H1A; H1B; fibroblasts; A549; MM6; Huh7; Jurkat; peripheral-blood; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; TREX; endometrial; HEC-1-A; GSCs | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000274071.6; ENST00000502773.6 | ||
| External Link | RMBase: m6A_site_655273 | ||
| mod ID: M6ASITE068587 | Click to Show/Hide the Full List | ||
| mod site | chr4:156971099-156971100:- | [11] | |
| Sequence | TGCTTTGATCCTTTTCAAAAACTGGAGACACAGAAGAGGGC | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa; CD34; HepG2; HEK293T; H1A; H1B; fibroblasts; A549; MM6; Huh7; Jurkat; peripheral-blood; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; TREX; endometrial; HEC-1-A; GSCs | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000502773.6; ENST00000274071.6 | ||
| External Link | RMBase: m6A_site_655274 | ||
| mod ID: M6ASITE068588 | Click to Show/Hide the Full List | ||
| mod site | chr4:156971139-156971140:- | [11] | |
| Sequence | ATTTGTTTAAACCTTGGGAAACTGGTTCAGGTCCAGGTTTT | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa; CD34; HepG2; HEK293T; H1A; H1B; fibroblasts; A549; MM6; Huh7; Jurkat; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; TREX; endometrial; HEC-1-A; GSCs | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000274071.6; ENST00000502773.6 | ||
| External Link | RMBase: m6A_site_655275 | ||
| mod ID: M6ASITE068589 | Click to Show/Hide the Full List | ||
| mod site | chr4:156971149-156971150:- | [11] | |
| Sequence | ACAGCTCAGGATTTGTTTAAACCTTGGGAAACTGGTTCAGG | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | HeLa; CD34; HepG2; HEK293T; H1A; H1B; fibroblasts; A549; MM6; Huh7; Jurkat; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; TREX; endometrial; HEC-1-A; GSCs | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000274071.6; ENST00000502773.6 | ||
| External Link | RMBase: m6A_site_655276 | ||
| mod ID: M6ASITE068590 | Click to Show/Hide the Full List | ||
| mod site | chr4:156971243-156971244:- | [12] | |
| Sequence | CTCCCAGCAACTTGCTGGGGACTTCTCGCCGCTCCCCCGCG | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | CD34; HepG2; HEK293T; fibroblasts; A549; MM6; Huh7; Jurkat; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; TREX; endometrial | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000502773.6; ENST00000274071.6 | ||
| External Link | RMBase: m6A_site_655277 | ||
| mod ID: M6ASITE068591 | Click to Show/Hide the Full List | ||
| mod site | chr4:156971274-156971275:- | [12] | |
| Sequence | GCCTCTGTCCTGTCCCCCGAACTGACAGGTGCTCCCAGCAA | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | CD34; HepG2; HEK293T; fibroblasts; A549; MM6; Huh7; Jurkat; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; TREX; endometrial | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000274071.6; ENST00000502773.6 | ||
| External Link | RMBase: m6A_site_655278 | ||
| mod ID: M6ASITE068592 | Click to Show/Hide the Full List | ||
| mod site | chr4:156971349-156971350:- | [12] | |
| Sequence | AGCTTTGTTATTGATCAGAAACTGCTCGCCGCCGACTTGGC | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | CD34; HEK293T; HEK293A-TOA; iSLK; MSC | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000502773.6 | ||
| External Link | RMBase: m6A_site_655279 | ||
| mod ID: M6ASITE068593 | Click to Show/Hide the Full List | ||
| mod site | chr4:156971468-156971469:- | [13] | |
| Sequence | CCGGCGCCCCGGCCCGTGGGACCCGCCGCGGCGCCCTCGCC | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HEK293T; GSC-11; HEK293A-TOA; iSLK; MSC | ||
| Seq Type List | MeRIP-seq; m6A-seq | ||
| Transcript ID List | ENST00000502773.6 | ||
| External Link | RMBase: m6A_site_655280 | ||
| mod ID: M6ASITE068594 | Click to Show/Hide the Full List | ||
| mod site | chr4:156971588-156971589:- | [11] | |
| Sequence | TCCGTGCTGCGGCGCTCCGAACCCGGGGCCAACTGCGCTCC | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; fibroblasts; MM6; Jurkat; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; TREX; endometrial; HEC-1-A; GSCs | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000502773.6 | ||
| External Link | RMBase: m6A_site_655281 | ||
| mod ID: M6ASITE068595 | Click to Show/Hide the Full List | ||
| mod site | chr4:156971628-156971629:- | [11] | |
| Sequence | GTGAACTTTGGTCGCGGAAAACTCAAGCCAGGTGCCGGAAT | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa; CD34; HepG2; HEK293T; fibroblasts; A549; H1299; MM6; Jurkat; peripheral-blood; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; TREX; endometrial; HEC-1-A; GSCs | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000502773.6 | ||
| External Link | RMBase: m6A_site_655282 | ||
| mod ID: M6ASITE068596 | Click to Show/Hide the Full List | ||
| mod site | chr4:156971644-156971645:- | [11] | |
| Sequence | GACGAGCCCGAACAAGGTGAACTTTGGTCGCGGAAAACTCA | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa; CD34; HepG2; HEK293T; fibroblasts; A549; H1299; MM6; Jurkat; peripheral-blood; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; TREX; endometrial; HEC-1-A; GSCs | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000502773.6 | ||
| External Link | RMBase: m6A_site_655283 | ||
| mod ID: M6ASITE068597 | Click to Show/Hide the Full List | ||
| mod site | chr4:156971653-156971654:- | [11] | |
| Sequence | GGAGCGGGGGACGAGCCCGAACAAGGTGAACTTTGGTCGCG | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa; CD34; HepG2; A549; H1A; H1B; HEK293T; fibroblasts; H1299; MM6; Jurkat; peripheral-blood; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; TREX; endometrial; HEC-1-A; GSCs | ||
| Seq Type List | m6A-seq; MeRIP-seq; m6A-CLIP/IP | ||
| Transcript ID List | ENST00000502773.6 | ||
| External Link | RMBase: m6A_site_655284 | ||
| mod ID: M6ASITE068598 | Click to Show/Hide the Full List | ||
| mod site | chr4:156971725-156971726:- | [11] | |
| Sequence | GTGGGGGGAGGGGACGGGGAACAACCGGAGCCCCGCGGTGA | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa; HepG2; hNPCs; fibroblasts; A549; MM6; Jurkat; GSC-11; HEK293T; HEK293A-TOA; iSLK; MSC; TIME; TREX; endometrial; HEC-1-A; GSCs | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000502773.6 | ||
| External Link | RMBase: m6A_site_655285 | ||
References