m6A Target Gene Information
General Information of the m6A Target Gene (ID: M6ATAR00369)
Full List of m6A Methylation Regulator of This Target Gene and Corresponding Disease/Drug Response(s)
PIK3CB
can be regulated by the following regulator(s), and cause disease/drug response(s). You can browse detail information of regulator(s) or disease/drug response(s).
Browse Regulator
Browse Disease
Browse Drug
Methyltransferase-like 14 (METTL14) [WRITER]
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL14 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
|
Treatment: siMETTL14 MDA-MB-231 cells
Control: MDA-MB-231 cells
|
GSE81164 | |
| Regulation |
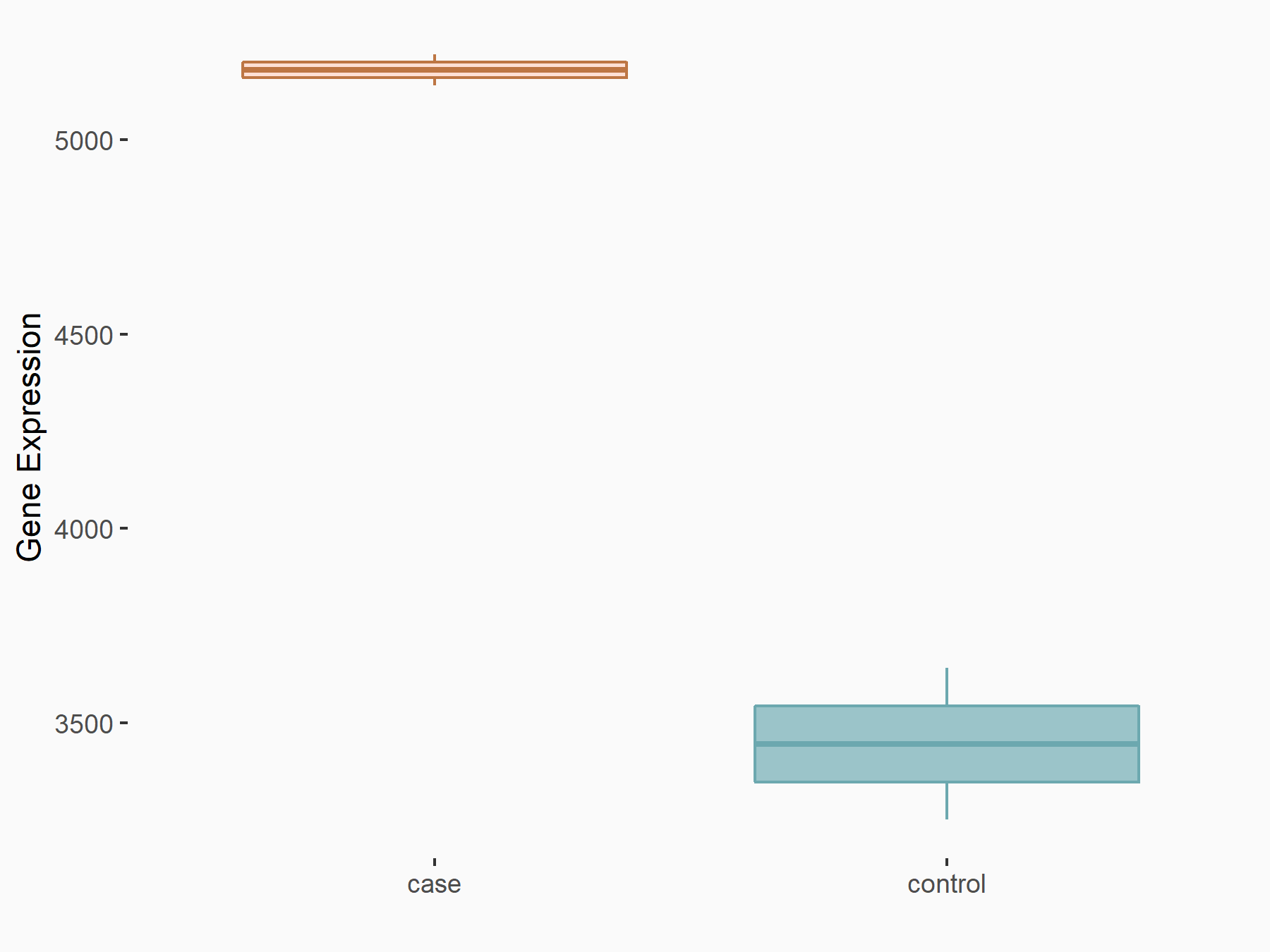 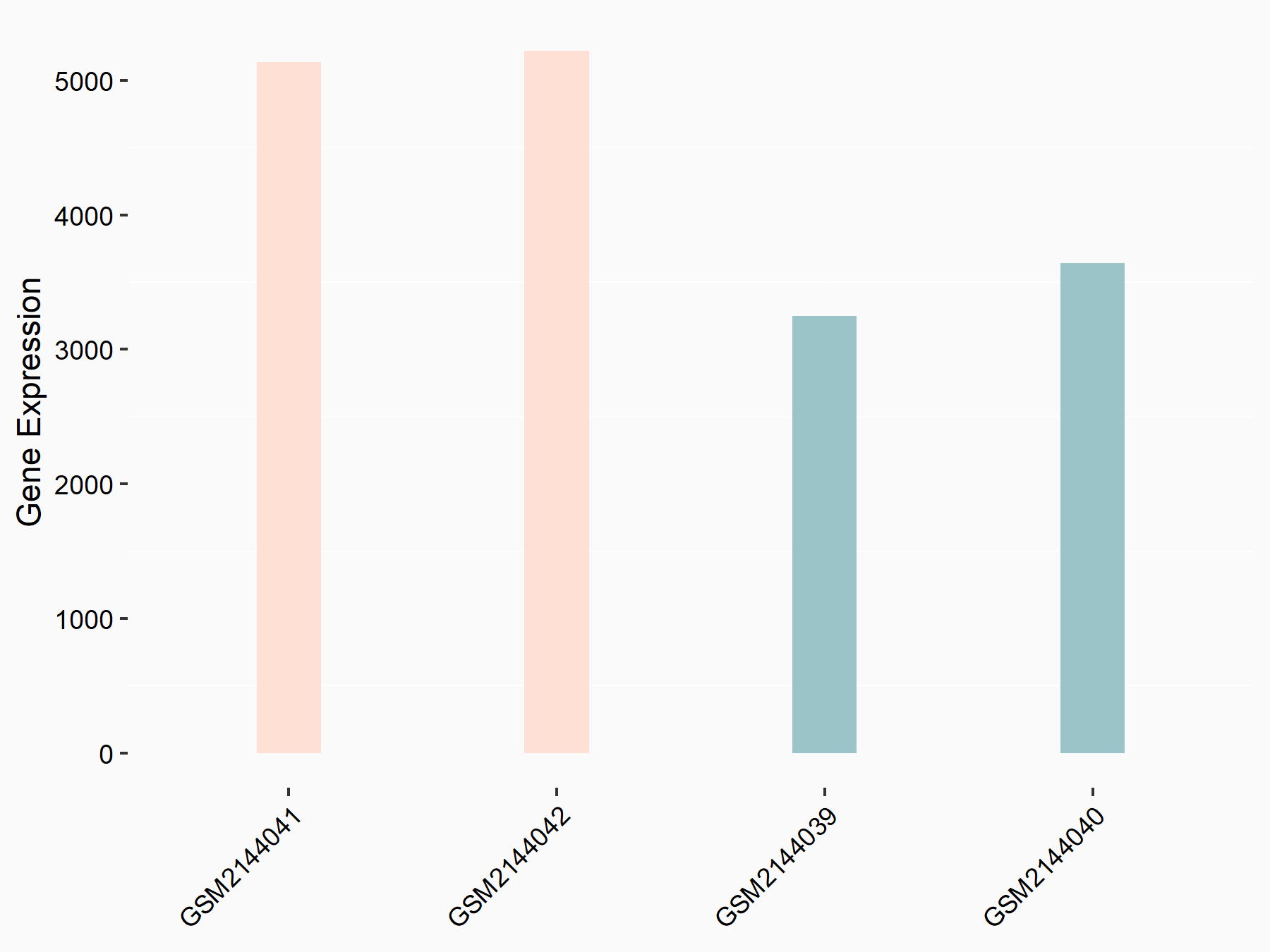 |
logFC: 5.88E-01 p-value: 1.08E-05 |
| More Results | Click to View More RNA-seq Results | |
| In total 1 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [1] | |||
| Response Summary | N6-methyladenosine mRNA methylation of PIK3CB regulates AKT signalling to promote PTEN-deficient pancreatic cancer progression. Rs142933486 is significantly associated with the overall survival of PDAC by reducing the PIK3CB m6A level, which facilitated its mRNA and protein expression levels mediated by the m6A 'writer' complex (METTL13/METTL14/WTAP) and the m6A 'reader' YTHDF2. KIN-193, a PI3-kinase subunit beta (PIK3CB)-selective inhibitor, is shown to serve as an effective anticancer agent for blocking PTEN-deficient PDAC. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Pancreatic cancer | ICD-11: 2C10 | ||
| Responsed Drug | AZD6482 | Terminated | ||
| Pathway Response | PI3K-Akt signaling pathway | hsa04151 | ||
| Glycolysis / Gluconeogenesis | hsa00010 | |||
| Cell Process | Glucose metabolism | |||
| In-vitro Model | BxPC-3 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0186 |
| PANC-1 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0480 | |
| In-vivo Model | Established cohorts of mice bearing tumour xenografts driven by PTEN-deficient BxPC-3 and PANC-1 cells with PIK3CB overexpression. When tumours grew to ~300 mm3, mice were grouped and administered with vehicle (DMSO) or KIN-193 via intraperitoneal injection (20 mg/kg) once daily. | |||
Wilms tumor 1-associating protein (WTAP) [WRITER]
| Representative RNA-seq result indicating the expression of this target gene regulated by WTAP | ||
| Cell Line | mice hepatocyte | Mus musculus |
|
Treatment: Wtap Hknockout mice hepatocyte
Control: Wtap flox/flox mice hepatocyte
|
GSE168850 | |
| Regulation |
  |
logFC: 5.88E-01 p-value: 1.20E-03 |
| More Results | Click to View More RNA-seq Results | |
| In total 1 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [1] | |||
| Response Summary | N6-methyladenosine mRNA methylation of PIK3CB regulates AKT signalling to promote PTEN-deficient pancreatic cancer progression. Rs142933486 is significantly associated with the overall survival of PDAC by reducing the PIK3CB m6A level, which facilitated its mRNA and protein expression levels mediated by the m6A 'writer' complex (METTL13/METTL14/WTAP) and the m6A 'reader' YTHDF2. KIN-193, a PI3-kinase subunit beta (PIK3CB)-selective inhibitor, is shown to serve as an effective anticancer agent for blocking PTEN-deficient PDAC. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Pancreatic cancer | ICD-11: 2C10 | ||
| Responsed Drug | AZD6482 | Terminated | ||
| Pathway Response | PI3K-Akt signaling pathway | hsa04151 | ||
| Glycolysis / Gluconeogenesis | hsa00010 | |||
| Cell Process | Glucose metabolism | |||
| In-vitro Model | BxPC-3 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0186 |
| PANC-1 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0480 | |
| In-vivo Model | Established cohorts of mice bearing tumour xenografts driven by PTEN-deficient BxPC-3 and PANC-1 cells with PIK3CB overexpression. When tumours grew to ~300 mm3, mice were grouped and administered with vehicle (DMSO) or KIN-193 via intraperitoneal injection (20 mg/kg) once daily. | |||
YTH domain-containing family protein 2 (YTHDF2) [READER]
| Representative RIP-seq result supporting the interaction between PIK3CB and the regulator | ||
| Cell Line | Hela | Homo sapiens |
| Regulation | logFC: 1.28E+00 | GSE49339 |
| In total 1 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [1] | |||
| Response Summary | N6-methyladenosine mRNA methylation of PIK3CB regulates AKT signalling to promote PTEN-deficient pancreatic cancer progression. Rs142933486 is significantly associated with the overall survival of PDAC by reducing the PIK3CB m6A level, which facilitated its mRNA and protein expression levels mediated by the m6A 'writer' complex (METTL13/METTL14/WTAP) and the m6A 'reader' YTHDF2. KIN-193, a PI3-kinase subunit beta (PIK3CB)-selective inhibitor, is shown to serve as an effective anticancer agent for blocking PTEN-deficient PDAC. | |||
| Target Regulation | Down regulation | |||
| Responsed Disease | Pancreatic cancer | ICD-11: 2C10 | ||
| Responsed Drug | AZD6482 | Terminated | ||
| Pathway Response | PI3K-Akt signaling pathway | hsa04151 | ||
| Glycolysis / Gluconeogenesis | hsa00010 | |||
| Cell Process | Glucose metabolism | |||
| In-vitro Model | BxPC-3 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0186 |
| PANC-1 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0480 | |
| In-vivo Model | Established cohorts of mice bearing tumour xenografts driven by PTEN-deficient BxPC-3 and PANC-1 cells with PIK3CB overexpression. When tumours grew to ~300 mm3, mice were grouped and administered with vehicle (DMSO) or KIN-193 via intraperitoneal injection (20 mg/kg) once daily. | |||
Methyltransferase-like 13 (METTL13) [WRITER]
| In total 1 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [1] | |||
| Response Summary | N6-methyladenosine mRNA methylation of PIK3CB regulates AKT signalling to promote PTEN-deficient pancreatic cancer progression. Rs142933486 is significantly associated with the overall survival of PDAC by reducing the PIK3CB m6A level, which facilitated its mRNA and protein expression levels mediated by the m6A 'writer' complex (METTL13/METTL14/WTAP) and the m6A 'reader' YTHDF2. KIN-193, a PI3-kinase subunit beta (PIK3CB)-selective inhibitor, is shown to serve as an effective anticancer agent for blocking PTEN-deficient PDAC. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Pancreatic cancer | ICD-11: 2C10 | ||
| Responsed Drug | AZD6482 | Terminated | ||
| Pathway Response | PI3K-Akt signaling pathway | hsa04151 | ||
| Glycolysis / Gluconeogenesis | hsa00010 | |||
| Cell Process | Glucose metabolism | |||
| In-vitro Model | BxPC-3 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0186 |
| PANC-1 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0480 | |
| In-vivo Model | Established cohorts of mice bearing tumour xenografts driven by PTEN-deficient BxPC-3 and PANC-1 cells with PIK3CB overexpression. When tumours grew to ~300 mm3, mice were grouped and administered with vehicle (DMSO) or KIN-193 via intraperitoneal injection (20 mg/kg) once daily. | |||
Pancreatic cancer [ICD-11: 2C10]
| In total 4 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [1] | |||
| Response Summary | N6-methyladenosine mRNA methylation of PIK3CB regulates AKT signalling to promote PTEN-deficient pancreatic cancer progression. Rs142933486 is significantly associated with the overall survival of PDAC by reducing the PIK3CB m6A level, which facilitated its mRNA and protein expression levels mediated by the m6A 'writer' complex (METTL13/METTL14/WTAP) and the m6A 'reader' YTHDF2. KIN-193, a PI3-kinase subunit beta (PIK3CB)-selective inhibitor, is shown to serve as an effective anticancer agent for blocking PTEN-deficient PDAC. | |||
| Responsed Disease | Pancreatic cancer [ICD-11: 2C10] | |||
| Target Regulator | Methyltransferase-like 13 (METTL13) | WRITER | ||
| Target Regulation | Up regulation | |||
| Responsed Drug | AZD6482 | Terminated | ||
| Pathway Response | PI3K-Akt signaling pathway | hsa04151 | ||
| Glycolysis / Gluconeogenesis | hsa00010 | |||
| Cell Process | Glucose metabolism | |||
| In-vitro Model | BxPC-3 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0186 |
| PANC-1 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0480 | |
| In-vivo Model | Established cohorts of mice bearing tumour xenografts driven by PTEN-deficient BxPC-3 and PANC-1 cells with PIK3CB overexpression. When tumours grew to ~300 mm3, mice were grouped and administered with vehicle (DMSO) or KIN-193 via intraperitoneal injection (20 mg/kg) once daily. | |||
| Experiment 2 Reporting the m6A-centered Disease Response | [1] | |||
| Response Summary | N6-methyladenosine mRNA methylation of PIK3CB regulates AKT signalling to promote PTEN-deficient pancreatic cancer progression. Rs142933486 is significantly associated with the overall survival of PDAC by reducing the PIK3CB m6A level, which facilitated its mRNA and protein expression levels mediated by the m6A 'writer' complex (METTL13/METTL14/WTAP) and the m6A 'reader' YTHDF2. KIN-193, a PI3-kinase subunit beta (PIK3CB)-selective inhibitor, is shown to serve as an effective anticancer agent for blocking PTEN-deficient PDAC. | |||
| Responsed Disease | Pancreatic cancer [ICD-11: 2C10] | |||
| Target Regulator | Methyltransferase-like 14 (METTL14) | WRITER | ||
| Target Regulation | Up regulation | |||
| Responsed Drug | AZD6482 | Terminated | ||
| Pathway Response | PI3K-Akt signaling pathway | hsa04151 | ||
| Glycolysis / Gluconeogenesis | hsa00010 | |||
| Cell Process | Glucose metabolism | |||
| In-vitro Model | BxPC-3 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0186 |
| PANC-1 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0480 | |
| In-vivo Model | Established cohorts of mice bearing tumour xenografts driven by PTEN-deficient BxPC-3 and PANC-1 cells with PIK3CB overexpression. When tumours grew to ~300 mm3, mice were grouped and administered with vehicle (DMSO) or KIN-193 via intraperitoneal injection (20 mg/kg) once daily. | |||
| Experiment 3 Reporting the m6A-centered Disease Response | [1] | |||
| Response Summary | N6-methyladenosine mRNA methylation of PIK3CB regulates AKT signalling to promote PTEN-deficient pancreatic cancer progression. Rs142933486 is significantly associated with the overall survival of PDAC by reducing the PIK3CB m6A level, which facilitated its mRNA and protein expression levels mediated by the m6A 'writer' complex (METTL13/METTL14/WTAP) and the m6A 'reader' YTHDF2. KIN-193, a PI3-kinase subunit beta (PIK3CB)-selective inhibitor, is shown to serve as an effective anticancer agent for blocking PTEN-deficient PDAC. | |||
| Responsed Disease | Pancreatic cancer [ICD-11: 2C10] | |||
| Target Regulator | Wilms tumor 1-associating protein (WTAP) | WRITER | ||
| Target Regulation | Up regulation | |||
| Responsed Drug | AZD6482 | Terminated | ||
| Pathway Response | PI3K-Akt signaling pathway | hsa04151 | ||
| Glycolysis / Gluconeogenesis | hsa00010 | |||
| Cell Process | Glucose metabolism | |||
| In-vitro Model | BxPC-3 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0186 |
| PANC-1 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0480 | |
| In-vivo Model | Established cohorts of mice bearing tumour xenografts driven by PTEN-deficient BxPC-3 and PANC-1 cells with PIK3CB overexpression. When tumours grew to ~300 mm3, mice were grouped and administered with vehicle (DMSO) or KIN-193 via intraperitoneal injection (20 mg/kg) once daily. | |||
| Experiment 4 Reporting the m6A-centered Disease Response | [1] | |||
| Response Summary | N6-methyladenosine mRNA methylation of PIK3CB regulates AKT signalling to promote PTEN-deficient pancreatic cancer progression. Rs142933486 is significantly associated with the overall survival of PDAC by reducing the PIK3CB m6A level, which facilitated its mRNA and protein expression levels mediated by the m6A 'writer' complex (METTL13/METTL14/WTAP) and the m6A 'reader' YTHDF2. KIN-193, a PI3-kinase subunit beta (PIK3CB)-selective inhibitor, is shown to serve as an effective anticancer agent for blocking PTEN-deficient PDAC. | |||
| Responsed Disease | Pancreatic cancer [ICD-11: 2C10] | |||
| Target Regulator | YTH domain-containing family protein 2 (YTHDF2) | READER | ||
| Target Regulation | Down regulation | |||
| Responsed Drug | AZD6482 | Terminated | ||
| Pathway Response | PI3K-Akt signaling pathway | hsa04151 | ||
| Glycolysis / Gluconeogenesis | hsa00010 | |||
| Cell Process | Glucose metabolism | |||
| In-vitro Model | BxPC-3 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0186 |
| PANC-1 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0480 | |
| In-vivo Model | Established cohorts of mice bearing tumour xenografts driven by PTEN-deficient BxPC-3 and PANC-1 cells with PIK3CB overexpression. When tumours grew to ~300 mm3, mice were grouped and administered with vehicle (DMSO) or KIN-193 via intraperitoneal injection (20 mg/kg) once daily. | |||
AZD6482
[Terminated]
| In total 4 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response | [1] | |||
| Response Summary | N6-methyladenosine mRNA methylation of PIK3CB regulates AKT signalling to promote PTEN-deficient pancreatic cancer progression. Rs142933486 is significantly associated with the overall survival of PDAC by reducing the PIK3CB m6A level, which facilitated its mRNA and protein expression levels mediated by the m6A 'writer' complex (METTL13/METTL14/WTAP) and the m6A 'reader' YTHDF2. KIN-193, a PI3-kinase subunit beta (PIK3CB)-selective inhibitor, is shown to serve as an effective anticancer agent for blocking PTEN-deficient PDAC. | |||
| Target Regulator | Methyltransferase-like 13 (METTL13) | WRITER | ||
| Target Regulation | Up regulation | |||
| Responsed Disease | Pancreatic cancer | ICD-11: 2C10 | ||
| Pathway Response | PI3K-Akt signaling pathway | hsa04151 | ||
| Glycolysis / Gluconeogenesis | hsa00010 | |||
| Cell Process | Glucose metabolism | |||
| In-vitro Model | BxPC-3 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0186 |
| PANC-1 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0480 | |
| In-vivo Model | Established cohorts of mice bearing tumour xenografts driven by PTEN-deficient BxPC-3 and PANC-1 cells with PIK3CB overexpression. When tumours grew to ~300 mm3, mice were grouped and administered with vehicle (DMSO) or KIN-193 via intraperitoneal injection (20 mg/kg) once daily. | |||
| Experiment 2 Reporting the m6A-centered Drug Response | [1] | |||
| Response Summary | N6-methyladenosine mRNA methylation of PIK3CB regulates AKT signalling to promote PTEN-deficient pancreatic cancer progression. Rs142933486 is significantly associated with the overall survival of PDAC by reducing the PIK3CB m6A level, which facilitated its mRNA and protein expression levels mediated by the m6A 'writer' complex (METTL13/METTL14/WTAP) and the m6A 'reader' YTHDF2. KIN-193, a PI3-kinase subunit beta (PIK3CB)-selective inhibitor, is shown to serve as an effective anticancer agent for blocking PTEN-deficient PDAC. | |||
| Target Regulator | Methyltransferase-like 14 (METTL14) | WRITER | ||
| Target Regulation | Up regulation | |||
| Responsed Disease | Pancreatic cancer | ICD-11: 2C10 | ||
| Pathway Response | PI3K-Akt signaling pathway | hsa04151 | ||
| Glycolysis / Gluconeogenesis | hsa00010 | |||
| Cell Process | Glucose metabolism | |||
| In-vitro Model | BxPC-3 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0186 |
| PANC-1 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0480 | |
| In-vivo Model | Established cohorts of mice bearing tumour xenografts driven by PTEN-deficient BxPC-3 and PANC-1 cells with PIK3CB overexpression. When tumours grew to ~300 mm3, mice were grouped and administered with vehicle (DMSO) or KIN-193 via intraperitoneal injection (20 mg/kg) once daily. | |||
| Experiment 3 Reporting the m6A-centered Drug Response | [1] | |||
| Response Summary | N6-methyladenosine mRNA methylation of PIK3CB regulates AKT signalling to promote PTEN-deficient pancreatic cancer progression. Rs142933486 is significantly associated with the overall survival of PDAC by reducing the PIK3CB m6A level, which facilitated its mRNA and protein expression levels mediated by the m6A 'writer' complex (METTL13/METTL14/WTAP) and the m6A 'reader' YTHDF2. KIN-193, a PI3-kinase subunit beta (PIK3CB)-selective inhibitor, is shown to serve as an effective anticancer agent for blocking PTEN-deficient PDAC. | |||
| Target Regulator | Wilms tumor 1-associating protein (WTAP) | WRITER | ||
| Target Regulation | Up regulation | |||
| Responsed Disease | Pancreatic cancer | ICD-11: 2C10 | ||
| Pathway Response | PI3K-Akt signaling pathway | hsa04151 | ||
| Glycolysis / Gluconeogenesis | hsa00010 | |||
| Cell Process | Glucose metabolism | |||
| In-vitro Model | BxPC-3 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0186 |
| PANC-1 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0480 | |
| In-vivo Model | Established cohorts of mice bearing tumour xenografts driven by PTEN-deficient BxPC-3 and PANC-1 cells with PIK3CB overexpression. When tumours grew to ~300 mm3, mice were grouped and administered with vehicle (DMSO) or KIN-193 via intraperitoneal injection (20 mg/kg) once daily. | |||
| Experiment 4 Reporting the m6A-centered Drug Response | [1] | |||
| Response Summary | N6-methyladenosine mRNA methylation of PIK3CB regulates AKT signalling to promote PTEN-deficient pancreatic cancer progression. Rs142933486 is significantly associated with the overall survival of PDAC by reducing the PIK3CB m6A level, which facilitated its mRNA and protein expression levels mediated by the m6A 'writer' complex (METTL13/METTL14/WTAP) and the m6A 'reader' YTHDF2. KIN-193, a PI3-kinase subunit beta (PIK3CB)-selective inhibitor, is shown to serve as an effective anticancer agent for blocking PTEN-deficient PDAC. | |||
| Target Regulator | YTH domain-containing family protein 2 (YTHDF2) | READER | ||
| Target Regulation | Down regulation | |||
| Responsed Disease | Pancreatic cancer | ICD-11: 2C10 | ||
| Pathway Response | PI3K-Akt signaling pathway | hsa04151 | ||
| Glycolysis / Gluconeogenesis | hsa00010 | |||
| Cell Process | Glucose metabolism | |||
| In-vitro Model | BxPC-3 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0186 |
| PANC-1 | Pancreatic ductal adenocarcinoma | Homo sapiens | CVCL_0480 | |
| In-vivo Model | Established cohorts of mice bearing tumour xenografts driven by PTEN-deficient BxPC-3 and PANC-1 cells with PIK3CB overexpression. When tumours grew to ~300 mm3, mice were grouped and administered with vehicle (DMSO) or KIN-193 via intraperitoneal injection (20 mg/kg) once daily. | |||
RNA Modification Sequencing Data Associated with the Target (ID: M6ATAR00369)
| In total 45 m6A sequence/site(s) in this target gene | |||
| mod ID: A2ISITE000011 | Click to Show/Hide the Full List | ||
| mod site | chr3:138665788-138665789:- | [2] | |
| Sequence | TCACTTGAACCCAGGAGTTCAAGAGCAGCCTGGGTAACATG | ||
| Transcript ID List | ENST00000469284.6; ENST00000477593.5; ENST00000462898.5; rmsk_1109893; ENST00000544716.5; ENST00000481749.5; ENST00000289153.6; ENST00000493568.5 | ||
| External Link | RMBase: RNA-editing_site_100125 | ||
| mod ID: A2ISITE000012 | Click to Show/Hide the Full List | ||
| mod site | chr3:138668083-138668084:- | [2] | |
| Sequence | AGAACTAGAAAAGTGCTTTTATTATTATTTTTTTCTGAGAT | ||
| Transcript ID List | ENST00000462898.5; ENST00000469284.6; ENST00000493568.5; ENST00000289153.6; ENST00000544716.5; ENST00000481749.5; ENST00000477593.5 | ||
| External Link | RMBase: RNA-editing_site_100126 | ||
| mod ID: A2ISITE000013 | Click to Show/Hide the Full List | ||
| mod site | chr3:138687518-138687519:- | [2] | |
| Sequence | TATGATTTGATTGTACCACTACACCCTAGCCTGGGCAACAG | ||
| Transcript ID List | ENST00000462898.5; ENST00000544716.5; ENST00000493568.5; ENST00000289153.6; ENST00000481749.5; rmsk_1109942; ENST00000469284.6; ENST00000477593.5; ENST00000473435.1 | ||
| External Link | RMBase: RNA-editing_site_100127 | ||
| mod ID: A2ISITE000014 | Click to Show/Hide the Full List | ||
| mod site | chr3:138712704-138712705:- | [2] | |
| Sequence | GTGTGGTGGCGGGTGCCTGTAATTCCAGCTACTCAGGAGGC | ||
| Transcript ID List | rmsk_1110007; ENST00000493568.5; ENST00000462898.5; ENST00000544716.5; ENST00000289153.6; ENST00000477593.5 | ||
| External Link | RMBase: RNA-editing_site_100128 | ||
| mod ID: A2ISITE000015 | Click to Show/Hide the Full List | ||
| mod site | chr3:138730809-138730810:- | [3] | |
| Sequence | CCTCCCAAGTAGCTAGGACTACAGGTATACACCATCACTTC | ||
| Transcript ID List | ENST00000477593.5; ENST00000462294.1; ENST00000462898.5; ENST00000289153.6 | ||
| External Link | RMBase: RNA-editing_site_100129 | ||
| mod ID: A2ISITE000016 | Click to Show/Hide the Full List | ||
| mod site | chr3:138731538-138731539:- | [2] | |
| Sequence | GCAAACAGGCTGGGCGTGGTAGCTCACACCTGTAATCCTAG | ||
| Transcript ID List | ENST00000462898.5; rmsk_1110039; ENST00000477593.5; ENST00000289153.6; ENST00000462294.1 | ||
| External Link | RMBase: RNA-editing_site_100130 | ||
| mod ID: A2ISITE000017 | Click to Show/Hide the Full List | ||
| mod site | chr3:138762765-138762766:- | [4] | |
| Sequence | AAGGTTTCACCATGTTGCCCAGGCTGGTCTCAAACTCCTGG | ||
| Transcript ID List | ENST00000465581.1; ENST00000477593.5; ENST00000483968.5; ENST00000461451.1; ENST00000462898.5 | ||
| External Link | RMBase: RNA-editing_site_100131 | ||
| mod ID: A2ISITE000018 | Click to Show/Hide the Full List | ||
| mod site | chr3:138762777-138762778:- | [4] | |
| Sequence | TTTAGTAGAGACAAGGTTTCACCATGTTGCCCAGGCTGGTC | ||
| Transcript ID List | ENST00000483968.5; ENST00000461451.1; ENST00000465581.1; ENST00000477593.5; ENST00000462898.5 | ||
| External Link | RMBase: RNA-editing_site_100132 | ||
| mod ID: A2ISITE000019 | Click to Show/Hide the Full List | ||
| mod site | chr3:138762784-138762785:- | [4] | |
| Sequence | TTGTATTTTTAGTAGAGACAAGGTTTCACCATGTTGCCCAG | ||
| Transcript ID List | ENST00000465581.1; ENST00000461451.1; ENST00000462898.5; ENST00000483968.5; ENST00000477593.5 | ||
| External Link | RMBase: RNA-editing_site_100133 | ||
| mod ID: A2ISITE000020 | Click to Show/Hide the Full List | ||
| mod site | chr3:138762794-138762795:- | [4] | |
| Sequence | GCTAATTTTTTTGTATTTTTAGTAGAGACAAGGTTTCACCA | ||
| Transcript ID List | ENST00000461451.1; ENST00000483968.5; ENST00000462898.5; ENST00000465581.1; ENST00000477593.5 | ||
| External Link | RMBase: RNA-editing_site_100134 | ||
| mod ID: A2ISITE000021 | Click to Show/Hide the Full List | ||
| mod site | chr3:138762811-138762812:- | [4] | |
| Sequence | GCCTGCCGCCATGCCTGGCTAATTTTTTTGTATTTTTAGTA | ||
| Transcript ID List | ENST00000483968.5; ENST00000465581.1; ENST00000477593.5; ENST00000461451.1; ENST00000462898.5 | ||
| External Link | RMBase: RNA-editing_site_100135 | ||
| mod ID: A2ISITE000022 | Click to Show/Hide the Full List | ||
| mod site | chr3:138762837-138762838:- | [4] | |
| Sequence | CCTCCCTGGTAGCTGGGATTACAGGTGCCTGCCGCCATGCC | ||
| Transcript ID List | ENST00000462898.5; ENST00000461451.1; ENST00000477593.5; ENST00000465581.1; ENST00000483968.5 | ||
| External Link | RMBase: RNA-editing_site_100136 | ||
| mod ID: A2ISITE000023 | Click to Show/Hide the Full List | ||
| mod site | chr3:138762872-138762873:- | [4] | |
| Sequence | CCACCCTCTGGGGTCAAGCAATTCTCCTGACTCAGCCTCCC | ||
| Transcript ID List | ENST00000465581.1; ENST00000483968.5; ENST00000461451.1; ENST00000477593.5; ENST00000462898.5 | ||
| External Link | RMBase: RNA-editing_site_100137 | ||
| mod ID: A2ISITE000024 | Click to Show/Hide the Full List | ||
| mod site | chr3:138762876-138762877:- | [4] | |
| Sequence | ACCTCCACCCTCTGGGGTCAAGCAATTCTCCTGACTCAGCC | ||
| Transcript ID List | ENST00000465581.1; ENST00000461451.1; ENST00000483968.5; ENST00000477593.5; ENST00000462898.5 | ||
| External Link | RMBase: RNA-editing_site_100138 | ||
| mod ID: A2ISITE000025 | Click to Show/Hide the Full List | ||
| mod site | chr3:138762890-138762891:- | [4] | |
| Sequence | CTTGGCCCACTGCAACCTCCACCCTCTGGGGTCAAGCAATT | ||
| Transcript ID List | ENST00000477593.5; ENST00000465581.1; ENST00000462898.5; ENST00000461451.1; ENST00000483968.5 | ||
| External Link | RMBase: RNA-editing_site_100139 | ||
| mod ID: A2ISITE000026 | Click to Show/Hide the Full List | ||
| mod site | chr3:138762897-138762898:- | [4] | |
| Sequence | GTGTAATCTTGGCCCACTGCAACCTCCACCCTCTGGGGTCA | ||
| Transcript ID List | ENST00000477593.5; ENST00000483968.5; ENST00000465581.1; ENST00000462898.5; ENST00000461451.1 | ||
| External Link | RMBase: RNA-editing_site_100140 | ||
| mod ID: A2ISITE000027 | Click to Show/Hide the Full List | ||
| mod site | chr3:138762913-138762914:- | [4] | |
| Sequence | AGGCTGGAGTGCAGTGGTGTAATCTTGGCCCACTGCAACCT | ||
| Transcript ID List | ENST00000461451.1; ENST00000462898.5; ENST00000477593.5; ENST00000483968.5; ENST00000465581.1 | ||
| External Link | RMBase: RNA-editing_site_100141 | ||
| mod ID: A2ISITE000028 | Click to Show/Hide the Full List | ||
| mod site | chr3:138763192-138763193:- | [4] | |
| Sequence | TGAGCTGAGATCATGCCACTACACTCCAGCCTGGGTGACAG | ||
| Transcript ID List | ENST00000462898.5; ENST00000483968.5; ENST00000461451.1; rmsk_1110130; ENST00000477593.5; ENST00000465581.1 | ||
| External Link | RMBase: RNA-editing_site_100142 | ||
| mod ID: A2ISITE000029 | Click to Show/Hide the Full List | ||
| mod site | chr3:138763220-138763221:- | [4] | |
| Sequence | GCTTGAACCTGGGAGGCAGAAGTTGCAGTGAGCTGAGATCA | ||
| Transcript ID List | ENST00000465581.1; rmsk_1110130; ENST00000477593.5; ENST00000483968.5; ENST00000461451.1; ENST00000462898.5 | ||
| External Link | RMBase: RNA-editing_site_100143 | ||
| mod ID: A2ISITE000030 | Click to Show/Hide the Full List | ||
| mod site | chr3:138763223-138763224:- | [4] | |
| Sequence | ATTGCTTGAACCTGGGAGGCAGAAGTTGCAGTGAGCTGAGA | ||
| Transcript ID List | ENST00000477593.5; rmsk_1110130; ENST00000483968.5; ENST00000462898.5; ENST00000461451.1; ENST00000465581.1 | ||
| External Link | RMBase: RNA-editing_site_100144 | ||
| mod ID: A2ISITE000031 | Click to Show/Hide the Full List | ||
| mod site | chr3:138763249-138763250:- | [4] | |
| Sequence | GCTACTCAGGAGGCTGAAGCAGGAGAATTGCTTGAACCTGG | ||
| Transcript ID List | ENST00000462898.5; ENST00000465581.1; ENST00000461451.1; rmsk_1110130; ENST00000483968.5; ENST00000477593.5 | ||
| External Link | RMBase: RNA-editing_site_100145 | ||
| mod ID: A2ISITE000032 | Click to Show/Hide the Full List | ||
| mod site | chr3:138763262-138763263:- | [4] | |
| Sequence | GCCTGTAATCCCAGCTACTCAGGAGGCTGAAGCAGGAGAAT | ||
| Transcript ID List | ENST00000465581.1; ENST00000462898.5; ENST00000483968.5; ENST00000461451.1; ENST00000477593.5; rmsk_1110130 | ||
| External Link | RMBase: RNA-editing_site_100146 | ||
| mod ID: A2ISITE000033 | Click to Show/Hide the Full List | ||
| mod site | chr3:138763266-138763267:- | [4] | |
| Sequence | GTGTGCCTGTAATCCCAGCTACTCAGGAGGCTGAAGCAGGA | ||
| Transcript ID List | ENST00000462898.5; ENST00000477593.5; ENST00000465581.1; rmsk_1110130; ENST00000483968.5; ENST00000461451.1 | ||
| External Link | RMBase: RNA-editing_site_100147 | ||
| mod ID: A2ISITE000034 | Click to Show/Hide the Full List | ||
| mod site | chr3:138763275-138763276:- | [4] | |
| Sequence | CATGATGGTGTGTGCCTGTAATCCCAGCTACTCAGGAGGCT | ||
| Transcript ID List | rmsk_1110130; ENST00000461451.1; ENST00000465581.1; ENST00000477593.5; ENST00000462898.5; ENST00000483968.5 | ||
| External Link | RMBase: RNA-editing_site_100148 | ||
| mod ID: A2ISITE000035 | Click to Show/Hide the Full List | ||
| mod site | chr3:138763307-138763308:- | [4] | |
| Sequence | CATCTCTACTAAAAATACAAAAATTAGCCGGGCATGATGGT | ||
| Transcript ID List | ENST00000483968.5; rmsk_1110130; ENST00000461451.1; ENST00000477593.5; ENST00000462898.5; ENST00000465581.1 | ||
| External Link | RMBase: RNA-editing_site_100149 | ||
| mod ID: A2ISITE000036 | Click to Show/Hide the Full List | ||
| mod site | chr3:138763331-138763332:- | [4] | |
| Sequence | AGCCTGACCAACATGGTGAAACCCCATCTCTACTAAAAATA | ||
| Transcript ID List | ENST00000461451.1; ENST00000483968.5; ENST00000477593.5; ENST00000462898.5; ENST00000465581.1; rmsk_1110130 | ||
| External Link | RMBase: RNA-editing_site_100150 | ||
| mod ID: A2ISITE000037 | Click to Show/Hide the Full List | ||
| mod site | chr3:138763341-138763342:- | [4] | |
| Sequence | GTTTGAGACTAGCCTGACCAACATGGTGAAACCCCATCTCT | ||
| Transcript ID List | ENST00000483968.5; ENST00000465581.1; ENST00000461451.1; ENST00000477593.5; rmsk_1110130; ENST00000462898.5 | ||
| External Link | RMBase: RNA-editing_site_100151 | ||
| mod ID: A2ISITE000038 | Click to Show/Hide the Full List | ||
| mod site | chr3:138763408-138763409:- | [4] | |
| Sequence | CACGGTGGCTCATGCCTGTAATCCCAGCACTTTGGGAGGCC | ||
| Transcript ID List | ENST00000477593.5; ENST00000461451.1; ENST00000465581.1; rmsk_1110130; ENST00000462898.5; ENST00000483968.5 | ||
| External Link | RMBase: RNA-editing_site_100152 | ||
| mod ID: A2ISITE000039 | Click to Show/Hide the Full List | ||
| mod site | chr3:138776441-138776442:- | [2] | |
| Sequence | CTGCCTCGCCCTCTCACAGTACTCGAATTACAGGCATAAGC | ||
| Transcript ID List | ENST00000483968.5; ENST00000461451.1; ENST00000462898.5; ENST00000477593.5 | ||
| External Link | RMBase: RNA-editing_site_100153 | ||
| mod ID: A2ISITE000040 | Click to Show/Hide the Full List | ||
| mod site | chr3:138781838-138781839:- | [5] | |
| Sequence | GCCTCGACTGCCTGGCGTCAAGTGATCCTCTTTTACTTCAG | ||
| Transcript ID List | ENST00000483968.5; ENST00000462898.5; ENST00000461451.1; ENST00000477593.5 | ||
| External Link | RMBase: RNA-editing_site_100154 | ||
| mod ID: A2ISITE000041 | Click to Show/Hide the Full List | ||
| mod site | chr3:138788880-138788881:- | [5] | |
| Sequence | ACCTCCGCCTCCCGGGTTCAAGCGATTCTCCTGCCTCAGCC | ||
| Transcript ID List | ENST00000462898.5; ENST00000461451.1; ENST00000483968.5; ENST00000477593.5 | ||
| External Link | RMBase: RNA-editing_site_100155 | ||
| mod ID: A2ISITE000042 | Click to Show/Hide the Full List | ||
| mod site | chr3:138789375-138789376:- | [2] | |
| Sequence | ACCTCCACCTCCTGTGCTCAAGTGATCCTCCCACCTCAGCC | ||
| Transcript ID List | ENST00000477593.5; ENST00000461451.1; ENST00000462898.5; ENST00000483968.5 | ||
| External Link | RMBase: RNA-editing_site_100156 | ||
| mod ID: A2ISITE000043 | Click to Show/Hide the Full List | ||
| mod site | chr3:138790361-138790362:- | [5] | |
| Sequence | CCTTGGCCTCACAAAATGCTAGGATTATAGGTGTGAGCCAC | ||
| Transcript ID List | ENST00000462898.5; ENST00000483968.5; ENST00000461451.1; ENST00000477593.5 | ||
| External Link | RMBase: RNA-editing_site_100157 | ||
| mod ID: A2ISITE000044 | Click to Show/Hide the Full List | ||
| mod site | chr3:138790366-138790367:- | [2] | |
| Sequence | GCCCACCTTGGCCTCACAAAATGCTAGGATTATAGGTGTGA | ||
| Transcript ID List | ENST00000462898.5; ENST00000461451.1; ENST00000477593.5; ENST00000483968.5 | ||
| External Link | RMBase: RNA-editing_site_100158 | ||
| mod ID: A2ISITE000045 | Click to Show/Hide the Full List | ||
| mod site | chr3:138792180-138792181:- | [5] | |
| Sequence | ATTTTTTGAAATGGAGTGTCACTTTGTTGCCCAGGCTGGAG | ||
| Transcript ID List | ENST00000483968.5; ENST00000461451.1; ENST00000462898.5; ENST00000477593.5 | ||
| External Link | RMBase: RNA-editing_site_100159 | ||
| mod ID: A2ISITE000046 | Click to Show/Hide the Full List | ||
| mod site | chr3:138793448-138793449:- | [5] | |
| Sequence | CATGCTCGACTGATTTTTGTATTTTTATTAGAGACGGGGTT | ||
| Transcript ID List | ENST00000477593.5; ENST00000461451.1; ENST00000483968.5; ENST00000462898.5 | ||
| External Link | RMBase: RNA-editing_site_100160 | ||
| mod ID: A2ISITE000047 | Click to Show/Hide the Full List | ||
| mod site | chr3:138819875-138819876:- | [5] | |
| Sequence | TCAAGCGATTCTCCTGCCTCAGCCACCCAAGTAGCCCGAGA | ||
| Transcript ID List | ENST00000461451.1; ENST00000477593.5; ENST00000462898.5; ENST00000483968.5 | ||
| External Link | RMBase: RNA-editing_site_100161 | ||
| mod ID: A2ISITE000048 | Click to Show/Hide the Full List | ||
| mod site | chr3:138820553-138820554:- | [5] | |
| Sequence | TTGCAGTGAGCTGAAATTGCACCACTGCACTCCAGCCTGGG | ||
| Transcript ID List | ENST00000461451.1; ENST00000462898.5; ENST00000483968.5; ENST00000477593.5; rmsk_1110292 | ||
| External Link | RMBase: RNA-editing_site_100162 | ||
| mod ID: A2ISITE000049 | Click to Show/Hide the Full List | ||
| mod site | chr3:138820591-138820592:- | [5] | |
| Sequence | CCGAGGCAGGAGAATTGCTTAAGCCCCGGAGGCAGTGGTTG | ||
| Transcript ID List | rmsk_1110292; ENST00000461451.1; ENST00000462898.5; ENST00000477593.5; ENST00000483968.5 | ||
| External Link | RMBase: RNA-editing_site_100163 | ||
| mod ID: A2ISITE000050 | Click to Show/Hide the Full List | ||
| mod site | chr3:138820661-138820662:- | [5] | |
| Sequence | GTCTCTAATAAAAGTACAAAAATTAGCCAGGTGTGGTGGTG | ||
| Transcript ID List | ENST00000461451.1; ENST00000483968.5; rmsk_1110292; ENST00000477593.5; ENST00000462898.5 | ||
| External Link | RMBase: RNA-editing_site_100164 | ||
| mod ID: A2ISITE000051 | Click to Show/Hide the Full List | ||
| mod site | chr3:138820768-138820769:- | [5] | |
| Sequence | AGGCACGGTGGCTCATGCCTATAATCCCAGCATTCTGAGAG | ||
| Transcript ID List | ENST00000477593.5; rmsk_1110292; ENST00000461451.1; ENST00000483968.5; ENST00000462898.5 | ||
| External Link | RMBase: RNA-editing_site_100165 | ||
| mod ID: A2ISITE000052 | Click to Show/Hide the Full List | ||
| mod site | chr3:138820788-138820789:- | [5] | |
| Sequence | CAGAATGAAGAAAGTGGACCAGGCACGGTGGCTCATGCCTA | ||
| Transcript ID List | ENST00000477593.5; ENST00000461451.1; ENST00000483968.5; ENST00000462898.5; rmsk_1110292 | ||
| External Link | RMBase: RNA-editing_site_100166 | ||
| mod ID: A2ISITE000053 | Click to Show/Hide the Full List | ||
| mod site | chr3:138827021-138827022:- | [5] | |
| Sequence | GGCTGGAGTGCAATGGTGCAATCTCGGTTCACTGCAACCTC | ||
| Transcript ID List | ENST00000477593.5; ENST00000483968.5; ENST00000462898.5; ENST00000461451.1 | ||
| External Link | RMBase: RNA-editing_site_100167 | ||
| mod ID: A2ISITE000054 | Click to Show/Hide the Full List | ||
| mod site | chr3:138827935-138827936:- | [5] | |
| Sequence | GAGTGCAGTGGCGCGATCTCAGTTTACTTCAAGCTCCGCCT | ||
| Transcript ID List | ENST00000462898.5; ENST00000477593.5; ENST00000483968.5; ENST00000461451.1 | ||
| External Link | RMBase: RNA-editing_site_100168 | ||
| mod ID: A2ISITE000055 | Click to Show/Hide the Full List | ||
| mod site | chr3:138833230-138833231:- | [2] | |
| Sequence | CTACCAAAAACACAAAAATTAGCAGGGCGTGGTGGCGGGCG | ||
| Transcript ID List | ENST00000462898.5; rmsk_1110331; ENST00000477593.5; ENST00000461451.1; ENST00000483968.5 | ||
| External Link | RMBase: RNA-editing_site_100169 | ||
N6-methyladenosine (m6A)
| In total 75 m6A sequence/site(s) in this target gene | |||
| mod ID: M6ASITE062456 | Click to Show/Hide the Full List | ||
| mod site | chr3:138654142-138654143:- | [6] | |
| Sequence | TCGGTGATATGGGAAAGAGAACTGAGTATTTGCCCTATGAC | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000477593.5; ENST00000289153.6; ENST00000469284.6; ENST00000544716.5; ENST00000462898.5 | ||
| External Link | RMBase: m6A_site_611932 | ||
| mod ID: M6ASITE062457 | Click to Show/Hide the Full List | ||
| mod site | chr3:138654204-138654205:- | [6] | |
| Sequence | TCCTGGCAACATCCAGCAAAACTACTGCTTATTCTCCAAAG | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000462898.5; ENST00000289153.6; ENST00000469284.6; ENST00000544716.5; ENST00000477593.5 | ||
| External Link | RMBase: m6A_site_611933 | ||
| mod ID: M6ASITE062458 | Click to Show/Hide the Full List | ||
| mod site | chr3:138654263-138654264:- | [6] | |
| Sequence | TGTTAATATGTGCAATAGGAACTACTGGTTTGAATGTGTAA | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000544716.5; ENST00000462898.5; ENST00000289153.6; ENST00000477593.5; ENST00000469284.6 | ||
| External Link | RMBase: m6A_site_611934 | ||
| mod ID: M6ASITE062459 | Click to Show/Hide the Full List | ||
| mod site | chr3:138654404-138654405:- | [6] | |
| Sequence | TTCTTACTGTATTTATTAAAACTTGTAATAATGTGATTTTT | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000289153.6; ENST00000477593.5; ENST00000462898.5; ENST00000544716.5; ENST00000469284.6 | ||
| External Link | RMBase: m6A_site_611935 | ||
| mod ID: M6ASITE062460 | Click to Show/Hide the Full List | ||
| mod site | chr3:138654466-138654467:- | [6] | |
| Sequence | GAAACCTATTTATGGAATAAACTCCAGATCTGAAATTCAGT | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa; HEK293T | ||
| Seq Type List | m6A-seq; DART-seq | ||
| Transcript ID List | ENST00000477593.5; ENST00000544716.5; ENST00000289153.6; ENST00000462898.5; ENST00000469284.6 | ||
| External Link | RMBase: m6A_site_611936 | ||
| mod ID: M6ASITE062461 | Click to Show/Hide the Full List | ||
| mod site | chr3:138654483-138654484:- | [6] | |
| Sequence | AAGCAAGCAAATGAAAAGAAACCTATTTATGGAATAAACTC | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000477593.5; ENST00000469284.6; ENST00000544716.5; ENST00000462898.5; ENST00000289153.6 | ||
| External Link | RMBase: m6A_site_611937 | ||
| mod ID: M6ASITE062462 | Click to Show/Hide the Full List | ||
| mod site | chr3:138655143-138655144:- | [7] | |
| Sequence | TGGATAATCATTTCCTGCTGACTTTGCACGCCAAGGAATGC | ||
| Motif Score | 3.28175 | ||
| Cell/Tissue List | A549 | ||
| Seq Type List | m6A-CLIP/IP | ||
| Transcript ID List | ENST00000544716.5; ENST00000462898.5; ENST00000469284.6; ENST00000481749.5; ENST00000289153.6; ENST00000477593.5 | ||
| External Link | RMBase: m6A_site_611938 | ||
| mod ID: M6ASITE062463 | Click to Show/Hide the Full List | ||
| mod site | chr3:138655171-138655172:- | [6] | |
| Sequence | TTCCAAGACTTATCATGAAAACTGTCAATGGATAATCATTT | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; A549; H1A; hNPCs; hESCs; fibroblasts; GM12878; MM6; Huh7; HEK293A-TOA; iSLK; MSC; TIME; TREX | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000481749.5; ENST00000477593.5; ENST00000289153.6; ENST00000544716.5; ENST00000469284.6; ENST00000462898.5; ENST00000493568.5 | ||
| External Link | RMBase: m6A_site_611939 | ||
| mod ID: M6ASITE062464 | Click to Show/Hide the Full List | ||
| mod site | chr3:138655184-138655185:- | [6] | |
| Sequence | GCTTTTTGAATGCTTCCAAGACTTATCATGAAAACTGTCAA | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; A549; H1A; hNPCs; hESCs; fibroblasts; GM12878; MM6; Huh7; HEK293A-TOA; iSLK; MSC; TIME; TREX | ||
| Seq Type List | m6A-seq; MeRIP-seq; m6A-CLIP/IP | ||
| Transcript ID List | ENST00000481749.5; ENST00000469284.6; ENST00000289153.6; ENST00000544716.5; ENST00000493568.5; ENST00000462898.5; ENST00000477593.5 | ||
| External Link | RMBase: m6A_site_611940 | ||
| mod ID: M6ASITE062465 | Click to Show/Hide the Full List | ||
| mod site | chr3:138655226-138655227:- | [6] | |
| Sequence | TCCCACAGAACCTAATCTGAACAATCCCCGATGATTCCCTC | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; A549; U2OS; H1A; hNPCs; hESCs; fibroblasts; GM12878; MM6; Huh7; HEK293A-TOA; iSLK; MSC; TIME; TREX | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000462898.5; ENST00000544716.5; ENST00000481749.5; ENST00000289153.6; ENST00000477593.5; ENST00000469284.6; ENST00000493568.5 | ||
| External Link | RMBase: m6A_site_611941 | ||
| mod ID: M6ASITE062466 | Click to Show/Hide the Full List | ||
| mod site | chr3:138655237-138655238:- | [6] | |
| Sequence | AGAACAAGGCATCCCACAGAACCTAATCTGAACAATCCCCG | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; A549; U2OS; H1A; hNPCs; hESCs; fibroblasts; GM12878; MM6; Huh7; HEK293A-TOA; iSLK; MSC; TIME; TREX | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000469284.6; ENST00000481749.5; ENST00000493568.5; ENST00000544716.5; ENST00000462898.5; ENST00000289153.6; ENST00000477593.5 | ||
| External Link | RMBase: m6A_site_611942 | ||
| mod ID: M6ASITE062467 | Click to Show/Hide the Full List | ||
| mod site | chr3:138655254-138655255:- | [6] | |
| Sequence | AACTCAGAGTTAAATTAAGAACAAGGCATCCCACAGAACCT | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; A549; U2OS; hNPCs; hESCs; fibroblasts; GM12878; CD8T; MM6; Huh7; HEK293A-TOA; iSLK; MSC; TIME; TREX | ||
| Seq Type List | m6A-seq; MeRIP-seq; m6A-CLIP/IP | ||
| Transcript ID List | ENST00000544716.5; ENST00000469284.6; ENST00000481749.5; ENST00000493568.5; ENST00000289153.6; ENST00000462898.5; ENST00000477593.5 | ||
| External Link | RMBase: m6A_site_611943 | ||
| mod ID: M6ASITE062468 | Click to Show/Hide the Full List | ||
| mod site | chr3:138655273-138655274:- | [6] | |
| Sequence | AAAGGGAATGAAATCCTGGAACTCAGAGTTAAATTAAGAAC | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; A549; U2OS; hNPCs; hESCs; fibroblasts; GM12878; MM6; Huh7; HEK293A-TOA; iSLK; MSC; TIME; TREX; AML | ||
| Seq Type List | m6A-seq; MeRIP-seq; miCLIP | ||
| Transcript ID List | ENST00000289153.6; ENST00000469284.6; ENST00000477593.5; ENST00000462898.5; ENST00000481749.5; ENST00000544716.5; ENST00000493568.5 | ||
| External Link | RMBase: m6A_site_611944 | ||
| mod ID: M6ASITE062469 | Click to Show/Hide the Full List | ||
| mod site | chr3:138655317-138655318:- | [6] | |
| Sequence | TTGCACTTGCACTAAATTGAACATGACCCTGTTAGAGATGT | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; A549; U2OS; hNPCs; hESCs; fibroblasts; GM12878; LCLs; MM6; Huh7; HEK293A-TOA; iSLK; MSC; TIME; TREX | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000481749.5; ENST00000462898.5; ENST00000289153.6; ENST00000469284.6; ENST00000493568.5; ENST00000477593.5; ENST00000544716.5 | ||
| External Link | RMBase: m6A_site_611945 | ||
| mod ID: M6ASITE062470 | Click to Show/Hide the Full List | ||
| mod site | chr3:138655401-138655402:- | [6] | |
| Sequence | GGCCCACACAGTTCGGAAAGACTACAGATCTTAACGATCAG | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; A549; U2OS; H1B; hNPCs; hESCs; fibroblasts; GM12878; MM6; Huh7; HEK293A-TOA; iSLK; MSC; TIME; AML | ||
| Seq Type List | m6A-seq; MeRIP-seq; miCLIP | ||
| Transcript ID List | ENST00000289153.6; ENST00000462898.5; ENST00000544716.5; ENST00000477593.5; ENST00000469284.6; ENST00000481749.5; ENST00000493568.5 | ||
| External Link | RMBase: m6A_site_611946 | ||
| mod ID: M6ASITE062471 | Click to Show/Hide the Full List | ||
| mod site | chr3:138655428-138655429:- | [6] | |
| Sequence | AAGCTGGACTACTAAAGTGAACTGGATGGCCCACACAGTTC | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa; HEK293T; A549; HepG2; H1B; hNPCs; hESCs; fibroblasts; GM12878; MM6; Huh7; HEK293A-TOA; iSLK; TIME | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000481749.5; ENST00000493568.5; ENST00000289153.6; ENST00000469284.6; ENST00000462898.5; ENST00000477593.5; ENST00000544716.5 | ||
| External Link | RMBase: m6A_site_611947 | ||
| mod ID: M6ASITE062472 | Click to Show/Hide the Full List | ||
| mod site | chr3:138655441-138655442:- | [6] | |
| Sequence | AGGCGCTCAGGGAAAGCTGGACTACTAAAGTGAACTGGATG | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa; HEK293T; A549; HepG2; H1B; hNPCs; hESCs; fibroblasts; GM12878; CD8T; MM6; Huh7; HEK293A-TOA; iSLK; TIME | ||
| Seq Type List | m6A-seq; MeRIP-seq; m6A-CLIP/IP | ||
| Transcript ID List | ENST00000477593.5; ENST00000469284.6; ENST00000544716.5; ENST00000462898.5; ENST00000289153.6; ENST00000493568.5; ENST00000481749.5 | ||
| External Link | RMBase: m6A_site_611948 | ||
| mod ID: M6ASITE062473 | Click to Show/Hide the Full List | ||
| mod site | chr3:138655484-138655485:- | [6] | |
| Sequence | AGTGAAGAAGAAGCACTCAAACAGTTTAAGCAAAAATTTGA | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa; HEK293T; HepG2; H1B; hNPCs; hESCs; fibroblasts; A549; GM12878; MM6; HEK293A-TOA; iSLK | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000544716.5; ENST00000477593.5; ENST00000462898.5; ENST00000481749.5; ENST00000469284.6; ENST00000289153.6; ENST00000493568.5 | ||
| External Link | RMBase: m6A_site_611949 | ||
| mod ID: M6ASITE062474 | Click to Show/Hide the Full List | ||
| mod site | chr3:138656174-138656175:- | [6] | |
| Sequence | TTGACTGCAGGGCTTCCTGAACTCACATCAGTCAAAGATAT | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa; HEK293T | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000477593.5; ENST00000544716.5; ENST00000481749.5; ENST00000462898.5; ENST00000493568.5; ENST00000469284.6; ENST00000289153.6 | ||
| External Link | RMBase: m6A_site_611950 | ||
| mod ID: M6ASITE062475 | Click to Show/Hide the Full List | ||
| mod site | chr3:138657714-138657715:- | [6] | |
| Sequence | ATGTCATTCAACAAGGAAAAACAGGAAATACAGAAAAGTTT | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000289153.6; ENST00000462898.5; ENST00000493568.5; ENST00000481749.5; ENST00000487552.1; ENST00000544716.5; ENST00000469284.6; ENST00000477593.5 | ||
| External Link | RMBase: m6A_site_611951 | ||
| mod ID: M6ASITE062476 | Click to Show/Hide the Full List | ||
| mod site | chr3:138657814-138657815:- | [6] | |
| Sequence | CTCTTCCACATTGACTTTGGACATATTCTTGGAAATTTCAA | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000469284.6; ENST00000462898.5; ENST00000544716.5; ENST00000289153.6; ENST00000481749.5; ENST00000487552.1; ENST00000477593.5; ENST00000493568.5 | ||
| External Link | RMBase: m6A_site_611952 | ||
| mod ID: M6ASITE062477 | Click to Show/Hide the Full List | ||
| mod site | chr3:138663913-138663914:- | [6] | |
| Sequence | ACAACATCATGGTCAAAAAAACTGGCCAGGTGAGCTGCTCC | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000477593.5; ENST00000462898.5; ENST00000289153.6; ENST00000469284.6; ENST00000487552.1; ENST00000544716.5; ENST00000493568.5; ENST00000481749.5 | ||
| External Link | RMBase: m6A_site_611953 | ||
| mod ID: M6ASITE062478 | Click to Show/Hide the Full List | ||
| mod site | chr3:138663941-138663942:- | [6] | |
| Sequence | GTCCTTGGGATTGGTGACAGACATAGTGACAACATCATGGT | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000289153.6; ENST00000493568.5; ENST00000477593.5; ENST00000469284.6; ENST00000481749.5; ENST00000462898.5; ENST00000544716.5 | ||
| External Link | RMBase: m6A_site_611954 | ||
| mod ID: M6ASITE062479 | Click to Show/Hide the Full List | ||
| mod site | chr3:138664017-138664018:- | [6] | |
| Sequence | CTTGTTCAGGGATGACCTGGACCGAGCCATTGAGGAATTTA | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000481749.5; ENST00000462898.5; ENST00000469284.6; ENST00000289153.6; ENST00000493568.5; ENST00000544716.5; ENST00000477593.5 | ||
| External Link | RMBase: m6A_site_611955 | ||
| mod ID: M6ASITE062480 | Click to Show/Hide the Full List | ||
| mod site | chr3:138665059-138665060:- | [6] | |
| Sequence | CAACAAAGATGCCCTTCTGAACTGGCTTAAAGAATACAACT | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000289153.6; ENST00000477593.5; ENST00000544716.5; ENST00000469284.6; ENST00000462898.5; ENST00000493568.5; ENST00000481749.5 | ||
| External Link | RMBase: m6A_site_611956 | ||
| mod ID: M6ASITE062481 | Click to Show/Hide the Full List | ||
| mod site | chr3:138665110-138665111:- | [6] | |
| Sequence | AATTGCTGACATTCAGCTGAACAGTAGCAATGTGGCTGCTG | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000481749.5; ENST00000469284.6; ENST00000477593.5; ENST00000462898.5; ENST00000493568.5; ENST00000289153.6; ENST00000544716.5 | ||
| External Link | RMBase: m6A_site_611957 | ||
| mod ID: M6ASITE062482 | Click to Show/Hide the Full List | ||
| mod site | chr3:138665132-138665133:- | [6] | |
| Sequence | AAGTTGTGAGCACCTCTGAAACAATTGCTGACATTCAGCTG | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000477593.5; ENST00000289153.6; ENST00000481749.5; ENST00000462898.5; ENST00000544716.5; ENST00000469284.6; ENST00000493568.5 | ||
| External Link | RMBase: m6A_site_611958 | ||
| mod ID: M6ASITE062483 | Click to Show/Hide the Full List | ||
| mod site | chr3:138684629-138684630:- | [6] | |
| Sequence | CCATGTGTTATCCTCTCAGAACTCTAGTAAGTGACTGTGCT | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000481749.5; ENST00000477593.5; ENST00000493568.5; ENST00000469284.6; ENST00000289153.6; ENST00000544716.5; ENST00000462898.5; ENST00000473435.1; ENST00000485060.1 | ||
| External Link | RMBase: m6A_site_611959 | ||
| mod ID: M6ASITE062484 | Click to Show/Hide the Full List | ||
| mod site | chr3:138684651-138684652:- | [6] | |
| Sequence | TGACCTGCAGTCACCCCTGAACCCATGTGTTATCCTCTCAG | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000462898.5; ENST00000477593.5; ENST00000544716.5; ENST00000493568.5; ENST00000289153.6; ENST00000473435.1; ENST00000481749.5; ENST00000485060.1; ENST00000469284.6 | ||
| External Link | RMBase: m6A_site_611960 | ||
| mod ID: M6ASITE062485 | Click to Show/Hide the Full List | ||
| mod site | chr3:138684698-138684699:- | [6] | |
| Sequence | GCCATGCATACCTGTTTAAAACAGAGTGCTTACCGGGAAGC | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000473435.1; ENST00000485060.1; ENST00000289153.6; ENST00000477593.5; ENST00000462898.5; ENST00000544716.5; ENST00000493568.5; ENST00000481749.5; ENST00000469284.6 | ||
| External Link | RMBase: m6A_site_611961 | ||
| mod ID: M6ASITE062486 | Click to Show/Hide the Full List | ||
| mod site | chr3:138684738-138684739:- | [6] | |
| Sequence | ACTGAATGCCGTGAAGTTAAACAGAGCCAAAGGGAAGGAGG | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000462898.5; ENST00000469284.6; ENST00000473435.1; ENST00000544716.5; ENST00000493568.5; ENST00000481749.5; ENST00000289153.6; ENST00000485060.1; ENST00000477593.5 | ||
| External Link | RMBase: m6A_site_611962 | ||
| mod ID: M6ASITE062487 | Click to Show/Hide the Full List | ||
| mod site | chr3:138684758-138684759:- | [6] | |
| Sequence | ACTTTAAATAGTTTAATCAAACTGAATGCCGTGAAGTTAAA | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000477593.5; ENST00000469284.6; ENST00000493568.5; ENST00000485060.1; ENST00000544716.5; ENST00000462898.5; ENST00000473435.1; ENST00000289153.6; ENST00000481749.5 | ||
| External Link | RMBase: m6A_site_611963 | ||
| mod ID: M6ASITE062488 | Click to Show/Hide the Full List | ||
| mod site | chr3:138684778-138684779:- | [6] | |
| Sequence | AAGCACTCAATAAGTTAAAAACTTTAAATAGTTTAATCAAA | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000481749.5; ENST00000289153.6; ENST00000477593.5; ENST00000462898.5; ENST00000485060.1; ENST00000493568.5; ENST00000473435.1; ENST00000469284.6; ENST00000544716.5 | ||
| External Link | RMBase: m6A_site_611964 | ||
| mod ID: M6ASITE062489 | Click to Show/Hide the Full List | ||
| mod site | chr3:138684815-138684816:- | [6] | |
| Sequence | ATGAATTTTTGGTGTGGGGAACTTATTTTTCAGGTTGAAGC | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000473435.1; ENST00000469284.6; ENST00000485060.1; ENST00000462898.5; ENST00000544716.5; ENST00000477593.5; ENST00000493568.5; ENST00000289153.6; ENST00000481749.5 | ||
| External Link | RMBase: m6A_site_611965 | ||
| mod ID: M6ASITE062490 | Click to Show/Hide the Full List | ||
| mod site | chr3:138694830-138694831:- | [6] | |
| Sequence | TCTGGATTTCAACTATCCAGACCAGTACGTTCGAGAATATG | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000493568.5; ENST00000289153.6; ENST00000473435.1; ENST00000477593.5; ENST00000544716.5; ENST00000462898.5; ENST00000481749.5; ENST00000469284.6 | ||
| External Link | RMBase: m6A_site_611966 | ||
| mod ID: M6ASITE062491 | Click to Show/Hide the Full List | ||
| mod site | chr3:138694877-138694878:- | [6] | |
| Sequence | CTGCTTCAGATTTGGCCTAAACTGCCCCCCCGGGAGGCCCT | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000469284.6; ENST00000493568.5; ENST00000481749.5; ENST00000462898.5; ENST00000289153.6; ENST00000473435.1; ENST00000477593.5; ENST00000544716.5 | ||
| External Link | RMBase: m6A_site_611967 | ||
| mod ID: M6ASITE062492 | Click to Show/Hide the Full List | ||
| mod site | chr3:138698924-138698925:- | [6] | |
| Sequence | CTGTCAATCAAGTGGAATAAACTTGAGGATGTTGCTCAGGT | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000477593.5; ENST00000473435.1; ENST00000493568.5; ENST00000462898.5; ENST00000469284.6; ENST00000544716.5; ENST00000289153.6; ENST00000481749.5 | ||
| External Link | RMBase: m6A_site_611968 | ||
| mod ID: M6ASITE062493 | Click to Show/Hide the Full List | ||
| mod site | chr3:138698985-138698986:- | [6] | |
| Sequence | TATTTGGACTTTGCGACAAGACTGCCGAGAGATTTTCCCAC | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000481749.5; ENST00000477593.5; ENST00000473435.1; ENST00000462898.5; ENST00000544716.5; ENST00000469284.6; ENST00000493568.5; ENST00000289153.6 | ||
| External Link | RMBase: m6A_site_611969 | ||
| mod ID: M6ASITE062494 | Click to Show/Hide the Full List | ||
| mod site | chr3:138698998-138698999:- | [6] | |
| Sequence | ATGAAATGGATCTTATTTGGACTTTGCGACAAGACTGCCGA | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000469284.6; ENST00000493568.5; ENST00000289153.6; ENST00000477593.5; ENST00000462898.5; ENST00000544716.5; ENST00000473435.1; ENST00000481749.5 | ||
| External Link | RMBase: m6A_site_611970 | ||
| mod ID: M6ASITE062495 | Click to Show/Hide the Full List | ||
| mod site | chr3:138699048-138699049:- | [6] | |
| Sequence | TGTATTGAAAGAAATCTTGGACAGGGATCCCTTGTCTCAAC | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000473435.1; ENST00000289153.6; ENST00000462898.5; ENST00000544716.5; ENST00000469284.6; ENST00000477593.5; ENST00000493568.5; ENST00000481749.5 | ||
| External Link | RMBase: m6A_site_611971 | ||
| mod ID: M6ASITE062496 | Click to Show/Hide the Full List | ||
| mod site | chr3:138707188-138707189:- | [6] | |
| Sequence | AAATTTCCAGAGAATAAAAAACAACCTTATTATTACCCTCC | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000544716.5; ENST00000481749.5; ENST00000477593.5; ENST00000469284.6; ENST00000493568.5; ENST00000473435.1; ENST00000289153.6; ENST00000462898.5 | ||
| External Link | RMBase: m6A_site_611972 | ||
| mod ID: M6ASITE062497 | Click to Show/Hide the Full List | ||
| mod site | chr3:138707247-138707248:- | [6] | |
| Sequence | ATCCAATGGGAACTGTTCAAACAAATCCATATACTGAAAAT | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000477593.5; ENST00000493568.5; ENST00000469284.6; ENST00000462898.5; ENST00000481749.5; ENST00000544716.5; ENST00000473435.1; ENST00000289153.6 | ||
| External Link | RMBase: m6A_site_611973 | ||
| mod ID: M6ASITE062498 | Click to Show/Hide the Full List | ||
| mod site | chr3:138707256-138707257:- | [6] | |
| Sequence | AAATGTTGAATCCAATGGGAACTGTTCAAACAAATCCATAT | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000473435.1; ENST00000481749.5; ENST00000493568.5; ENST00000469284.6; ENST00000477593.5; ENST00000544716.5; ENST00000289153.6; ENST00000462898.5 | ||
| External Link | RMBase: m6A_site_611974 | ||
| mod ID: M6ASITE062499 | Click to Show/Hide the Full List | ||
| mod site | chr3:138707284-138707285:- | [6] | |
| Sequence | TATTTGCATGTTTTAGATGAACTCGAAGAAATGTTGAATCC | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000477593.5; ENST00000493568.5; ENST00000462898.5; ENST00000469284.6; ENST00000289153.6; ENST00000473435.1; ENST00000481749.5; ENST00000544716.5 | ||
| External Link | RMBase: m6A_site_611975 | ||
| mod ID: M6ASITE062500 | Click to Show/Hide the Full List | ||
| mod site | chr3:138707312-138707313:- | [6] | |
| Sequence | TTTAAAGACATTTTTTAAGAACCAAAATTATTTGCATGTTT | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000469284.6; ENST00000481749.5; ENST00000462898.5; ENST00000289153.6; ENST00000477593.5; ENST00000493568.5; ENST00000473435.1; ENST00000544716.5 | ||
| External Link | RMBase: m6A_site_611976 | ||
| mod ID: M6ASITE062501 | Click to Show/Hide the Full List | ||
| mod site | chr3:138707325-138707326:- | [6] | |
| Sequence | TTTAAAGCCTAGTTTTAAAGACATTTTTTAAGAACCAAAAT | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000544716.5; ENST00000462898.5; ENST00000289153.6; ENST00000493568.5; ENST00000481749.5; ENST00000469284.6; ENST00000477593.5; ENST00000473435.1 | ||
| External Link | RMBase: m6A_site_611977 | ||
| mod ID: M6ASITE062502 | Click to Show/Hide the Full List | ||
| mod site | chr3:138712239-138712240:- | [6] | |
| Sequence | AGGACAATTGAGAACTGGAGACATAATATTACACAGCTGGT | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000473435.1; ENST00000493568.5; ENST00000544716.5; ENST00000477593.5; ENST00000462898.5; ENST00000289153.6; ENST00000469284.6 | ||
| External Link | RMBase: m6A_site_611978 | ||
| mod ID: M6ASITE062503 | Click to Show/Hide the Full List | ||
| mod site | chr3:138712246-138712247:- | [6] | |
| Sequence | ACTTTAAAGGACAATTGAGAACTGGAGACATAATATTACAC | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000469284.6; ENST00000473435.1; ENST00000477593.5; ENST00000544716.5; ENST00000289153.6; ENST00000493568.5; ENST00000462898.5 | ||
| External Link | RMBase: m6A_site_611979 | ||
| mod ID: M6ASITE062504 | Click to Show/Hide the Full List | ||
| mod site | chr3:138712256-138712257:- | [6] | |
| Sequence | ATGGTTTTTGACTTTAAAGGACAATTGAGAACTGGAGACAT | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000544716.5; ENST00000462898.5; ENST00000289153.6; ENST00000477593.5; ENST00000493568.5; ENST00000469284.6 | ||
| External Link | RMBase: m6A_site_611980 | ||
| mod ID: M6ASITE062505 | Click to Show/Hide the Full List | ||
| mod site | chr3:138714490-138714491:- | [6] | |
| Sequence | TTAATCCCTCTAAATATCAGACCATCAGGAAAGCTGGAAAA | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000477593.5; ENST00000493568.5; ENST00000462898.5; ENST00000289153.6 | ||
| External Link | RMBase: m6A_site_611981 | ||
| mod ID: M6ASITE062506 | Click to Show/Hide the Full List | ||
| mod site | chr3:138714514-138714515:- | [6] | |
| Sequence | AAACGAAGAAATCAACGAAAACTATTAATCCCTCTAAATAT | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000289153.6; ENST00000477593.5; ENST00000462898.5; ENST00000493568.5 | ||
| External Link | RMBase: m6A_site_611982 | ||
| mod ID: M6ASITE062507 | Click to Show/Hide the Full List | ||
| mod site | chr3:138714620-138714621:- | [6] | |
| Sequence | AATGATCATATTTGGAATGAACCACTGGAATTTGATATTAA | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000289153.6; ENST00000462898.5; ENST00000477593.5; ENST00000493568.5 | ||
| External Link | RMBase: m6A_site_611983 | ||
| mod ID: M6ASITE062508 | Click to Show/Hide the Full List | ||
| mod site | chr3:138714670-138714671:- | [6] | |
| Sequence | GTACTGAGCTCCTGTGTAAAACCATCGTAAGCTCAGAGGTA | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000462898.5; ENST00000289153.6; ENST00000462294.1; ENST00000477593.5; ENST00000493568.5 | ||
| External Link | RMBase: m6A_site_611984 | ||
| mod ID: M6ASITE062509 | Click to Show/Hide the Full List | ||
| mod site | chr3:138733368-138733369:- | [6] | |
| Sequence | ATAAACTTAACACAGAGGAAACTGTAAAAGTGAGTACCCAC | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000462294.1; ENST00000462898.5; ENST00000477593.5; ENST00000289153.6 | ||
| External Link | RMBase: m6A_site_611985 | ||
| mod ID: M6ASITE062510 | Click to Show/Hide the Full List | ||
| mod site | chr3:138733384-138733385:- | [6] | |
| Sequence | GTCTTGGTTAAGGGAAATAAACTTAACACAGAGGAAACTGT | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000462294.1; ENST00000477593.5; ENST00000462898.5; ENST00000289153.6 | ||
| External Link | RMBase: m6A_site_611986 | ||
| mod ID: M6ASITE062511 | Click to Show/Hide the Full List | ||
| mod site | chr3:138734647-138734648:- | [6] | |
| Sequence | TTCCATTACCACCAAAGAAAACACGAATTATTTCTGTAAGT | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000462294.1; ENST00000462898.5; ENST00000477593.5; ENST00000289153.6 | ||
| External Link | RMBase: m6A_site_611987 | ||
| mod ID: M6ASITE062512 | Click to Show/Hide the Full List | ||
| mod site | chr3:138734723-138734724:- | [6] | |
| Sequence | AAGATCAAGAAAATGTATGAACAAGAAATGATTGCCATAGA | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000289153.6; ENST00000462898.5; ENST00000477593.5; ENST00000462294.1 | ||
| External Link | RMBase: m6A_site_611988 | ||
| mod ID: M6ASITE062513 | Click to Show/Hide the Full List | ||
| mod site | chr3:138734781-138734782:- | [6] | |
| Sequence | TATCCGGAACTGTGTGATGAACAGAGCCCTGCCCCATTTTA | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000462294.1; ENST00000289153.6; ENST00000477593.5; ENST00000462898.5 | ||
| External Link | RMBase: m6A_site_611989 | ||
| mod ID: M6ASITE062514 | Click to Show/Hide the Full List | ||
| mod site | chr3:138734793-138734794:- | [6] | |
| Sequence | GCCCCTATAGTATATCCGGAACTGTGTGATGAACAGAGCCC | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000462294.1; ENST00000289153.6; ENST00000462898.5; ENST00000477593.5 | ||
| External Link | RMBase: m6A_site_611990 | ||
| mod ID: M6ASITE062515 | Click to Show/Hide the Full List | ||
| mod site | chr3:138742564-138742565:- | [6] | |
| Sequence | CGTAGCTGTTCATTTTGAAAACTGCCAGGTAGGATTATATT | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa; CD34 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000477593.5; ENST00000289153.6; ENST00000462294.1; ENST00000462898.5 | ||
| External Link | RMBase: m6A_site_611991 | ||
| mod ID: M6ASITE062516 | Click to Show/Hide the Full List | ||
| mod site | chr3:138742605-138742606:- | [6] | |
| Sequence | CCTGAAAACTTAGAAGATAAACTTTATGGGGGAAAGCTCAT | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa; CD34 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000462294.1; ENST00000289153.6; ENST00000477593.5; ENST00000462898.5 | ||
| External Link | RMBase: m6A_site_611992 | ||
| mod ID: M6ASITE062517 | Click to Show/Hide the Full List | ||
| mod site | chr3:138742618-138742619:- | [6] | |
| Sequence | TGAACCATCCATCCCTGAAAACTTAGAAGATAAACTTTATG | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000477593.5; ENST00000462898.5; ENST00000462294.1; ENST00000289153.6 | ||
| External Link | RMBase: m6A_site_611993 | ||
| mod ID: M6ASITE062518 | Click to Show/Hide the Full List | ||
| mod site | chr3:138742635-138742636:- | [6] | |
| Sequence | ACATATCCACCAGAGCATGAACCATCCATCCCTGAAAACTT | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000477593.5; ENST00000462294.1; ENST00000462898.5; ENST00000289153.6 | ||
| External Link | RMBase: m6A_site_611994 | ||
| mod ID: M6ASITE062519 | Click to Show/Hide the Full List | ||
| mod site | chr3:138742659-138742660:- | [6] | |
| Sequence | TCTTGGATGGACTGGCTAAAACAAACATATCCACCAGAGCA | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000462898.5; ENST00000289153.6; ENST00000477593.5; ENST00000462294.1 | ||
| External Link | RMBase: m6A_site_611995 | ||
| mod ID: M6ASITE062520 | Click to Show/Hide the Full List | ||
| mod site | chr3:138742669-138742670:- | [6] | |
| Sequence | TGTGGGATTGTCTTGGATGGACTGGCTAAAACAAACATATC | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000477593.5; ENST00000462898.5; ENST00000289153.6; ENST00000462294.1 | ||
| External Link | RMBase: m6A_site_611996 | ||
| mod ID: M6ASITE062521 | Click to Show/Hide the Full List | ||
| mod site | chr3:138755782-138755783:- | [6] | |
| Sequence | TGACCCAGGGGAAAAATTAGACTCAAAAATTGGAGTCCTTA | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000483968.5; ENST00000462898.5; ENST00000477593.5; ENST00000289153.6 | ||
| External Link | RMBase: m6A_site_611997 | ||
| mod ID: M6ASITE062522 | Click to Show/Hide the Full List | ||
| mod site | chr3:138755841-138755842:- | [6] | |
| Sequence | CGAAGACTCTGTGATGTCAGACCTTTTCTTCCAGTTCTCAA | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000483968.5; ENST00000462898.5; ENST00000477593.5; ENST00000289153.6 | ||
| External Link | RMBase: m6A_site_611998 | ||
| mod ID: M6ASITE062523 | Click to Show/Hide the Full List | ||
| mod site | chr3:138755856-138755857:- | [6] | |
| Sequence | CTTGAAGATGAAACACGAAGACTCTGTGATGTCAGACCTTT | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000289153.6; ENST00000477593.5; ENST00000461451.1; ENST00000462898.5; ENST00000483968.5 | ||
| External Link | RMBase: m6A_site_611999 | ||
| mod ID: M6ASITE062524 | Click to Show/Hide the Full List | ||
| mod site | chr3:138755864-138755865:- | [6] | |
| Sequence | ATGAGGAGCTTGAAGATGAAACACGAAGACTCTGTGATGTC | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000483968.5; ENST00000461451.1; ENST00000477593.5; ENST00000289153.6; ENST00000462898.5 | ||
| External Link | RMBase: m6A_site_612000 | ||
| mod ID: M6ASITE062525 | Click to Show/Hide the Full List | ||
| mod site | chr3:138755894-138755895:- | [6] | |
| Sequence | TGTTTGCATGTGTGAATCAGACTGCTGTATATGAGGAGCTT | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000289153.6; ENST00000462898.5; ENST00000483968.5; ENST00000461451.1; ENST00000477593.5 | ||
| External Link | RMBase: m6A_site_612001 | ||
| mod ID: M6ASITE062526 | Click to Show/Hide the Full List | ||
| mod site | chr3:138759305-138759306:- | [6] | |
| Sequence | AATGCCTCCTGCTATGGCAGACATCCTTGACATCTGGGCGG | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000462898.5; ENST00000483968.5; ENST00000465581.1; ENST00000477593.5; ENST00000461451.1; ENST00000289153.6 | ||
| External Link | RMBase: m6A_site_612002 | ||
| mod ID: M6ASITE062527 | Click to Show/Hide the Full List | ||
| mod site | chr3:138770071-138770072:- | [6] | |
| Sequence | AAACAGAAGAATGATACAAGACTGAGGATCTTGCCAGGCAA | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000483968.5; ENST00000461451.1; ENST00000477593.5; ENST00000465581.1; ENST00000462898.5 | ||
| External Link | RMBase: m6A_site_612003 | ||
| mod ID: M6ASITE062528 | Click to Show/Hide the Full List | ||
| mod site | chr3:138770089-138770090:- | [6] | |
| Sequence | TCTCTTGCTTTGATGCTCAAACAGAAGAATGATACAAGACT | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000461451.1; ENST00000477593.5; ENST00000462898.5; ENST00000483968.5; ENST00000465581.1 | ||
| External Link | RMBase: m6A_site_612004 | ||
| mod ID: M6ASITE062529 | Click to Show/Hide the Full List | ||
| mod site | chr3:138770133-138770134:- | [6] | |
| Sequence | AAAGTTTTCATAGTTTAAAAACTCTGCTTGTCCTTAGCCTT | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000477593.5; ENST00000483968.5; ENST00000462898.5; ENST00000465581.1; ENST00000461451.1 | ||
| External Link | RMBase: m6A_site_612005 | ||
| mod ID: M6ASITE062530 | Click to Show/Hide the Full List | ||
| mod site | chr3:138796487-138796488:- | [6] | |
| Sequence | GCCATCTGTTTTCCTAAAAGACTGAAATGGGGGCGACCATT | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000483968.5; ENST00000477593.5; ENST00000461451.1; ENST00000462898.5 | ||
| External Link | RMBase: m6A_site_612006 | ||
References