m6A Target Gene Information
General Information of the m6A Target Gene (ID: M6ATAR00800)
Full List of m6A Methylation Regulator of This Target Gene and Corresponding Disease/Drug Response(s)
SENP1
can be regulated by the following regulator(s), and cause disease/drug response(s). You can browse detail information of regulator(s) or disease/drug response(s).
Browse Regulator
Browse Disease
RNA demethylase ALKBH5 (ALKBH5) [ERASER]
| Representative RNA-seq result indicating the expression of this target gene regulated by ALKBH5 | ||
| Cell Line | NOMO-1 cell line | Homo sapiens |
|
Treatment: shALKBH5 NOMO-1 cells
Control: shNS NOMO-1 cells
|
GSE144968 | |
| Regulation |
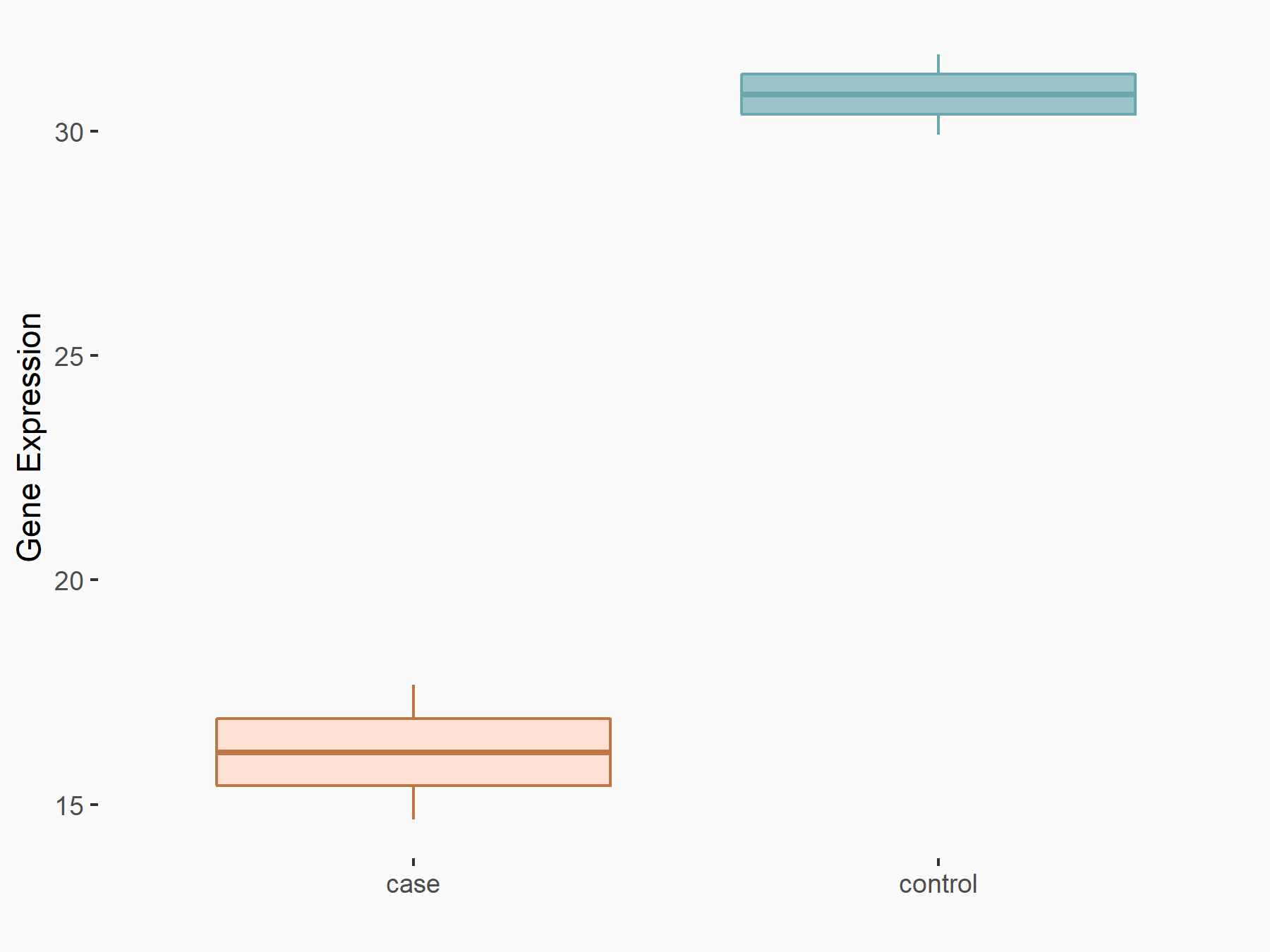  |
logFC: -8.95E-01 p-value: 1.12E-02 |
| More Results | Click to View More RNA-seq Results | |
| In total 1 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [1] | |||
| Response Summary | ROS promotes ALKBH5 SUMOylation through activating ERK/EPHB2/JNK signaling, leading to inhibition of ALKBH5 m6A demethylase activity by blocking substrate accessibility. Post-translational modification of ALKBH5 regulates ROS-induced DNA damage response. ROS specifically promotes ALKBH5 but not FTO, METTL3 and METTL14 SUMOylation by enhancing the interaction of ALKBH5 and UBC9 and inhibiting the association between ALKBH5 and SUMO specific peptidase 1 (SENP1). | |||
| Target Regulation | Down regulation | |||
| Responsed Disease | Diseases of the circulatory system | ICD-11: BE2Z | ||
| Pathway Response | Apoptosis | hsa04210 | ||
| Chemical carcinogenesis - reactive oxygen species | hsa05208 | |||
| Cell Process | Oxygen species(ROS)-induced stress | |||
| In-vitro Model | HEK293T | Normal | Homo sapiens | CVCL_0063 |
| In-vivo Model | For the ROS-induced DNA damage analysis, the indicated cell lines were treated with or without 100 uM hydrogen peroxide (H2O2), or 80 uM Carbonyl cyanide m-chlorophenylhydrazone (CCCP) for 6 hours. For the in vivo ROS study, DMSO and 5 mg/kg CCCP was intraperitoneally injected in to three pairs of mice. | |||
Diseases of the circulatory system [ICD-11: BE2Z]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [1] | |||
| Response Summary | ROS promotes ALKBH5 SUMOylation through activating ERK/EPHB2/JNK signaling, leading to inhibition of ALKBH5 m6A demethylase activity by blocking substrate accessibility. Post-translational modification of ALKBH5 regulates ROS-induced DNA damage response. ROS specifically promotes ALKBH5 but not FTO, METTL3 and METTL14 SUMOylation by enhancing the interaction of ALKBH5 and UBC9 and inhibiting the association between ALKBH5 and SUMO specific peptidase 1 (SENP1). | |||
| Responsed Disease | Diseases of the circulatory system [ICD-11: BE2Z] | |||
| Target Regulator | RNA demethylase ALKBH5 (ALKBH5) | ERASER | ||
| Target Regulation | Down regulation | |||
| Pathway Response | Apoptosis | hsa04210 | ||
| Chemical carcinogenesis - reactive oxygen species | hsa05208 | |||
| Cell Process | Oxygen species(ROS)-induced stress | |||
| In-vitro Model | HEK293T | Normal | Homo sapiens | CVCL_0063 |
| In-vivo Model | For the ROS-induced DNA damage analysis, the indicated cell lines were treated with or without 100 uM hydrogen peroxide (H2O2), or 80 uM Carbonyl cyanide m-chlorophenylhydrazone (CCCP) for 6 hours. For the in vivo ROS study, DMSO and 5 mg/kg CCCP was intraperitoneally injected in to three pairs of mice. | |||
Full List of Crosstalk(s) between m6A Modification and Epigenetic Regulation Related to This Regulator
Histone modification
m6A Regulator: Insulin-like growth factor 2 mRNA-binding protein 3 (IGF2BP3)
| In total 1 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT03115 | ||
| Epigenetic Regulator | Histone deacetylase 2 (HDAC2) | |
| Regulated Target | Epidermal growth factor receptor (EGFR) | |
| Crosstalk relationship | m6A → Histone modification | |
| Disease | Acute myeloid leukaemia | |
RNA Modification Sequencing Data Associated with the Target (ID: M6ATAR00800)
| In total 2 m6A sequence/site(s) in this target gene | |||
| mod ID: A2ISITE005518 | Click to Show/Hide the Full List | ||
| mod site | chr12:48054530-48054531:- | [3] | |
| Sequence | TCCCAAAGTGCTGGGATTACAGATGTGAGCCACTGCGCCTG | ||
| Transcript ID List | ENST00000549518.6; ENST00000448372.5; ENST00000549595.5; ENST00000552189.5 | ||
| External Link | RMBase: RNA-editing_site_28023 | ||
| mod ID: A2ISITE005519 | Click to Show/Hide the Full List | ||
| mod site | chr12:48085638-48085639:- | [3] | |
| Sequence | ACAGGGAAGAGGTAGTGGAGAGAGATGGTCAGGCTTCCTGT | ||
| Transcript ID List | ENST00000552189.5; ENST00000551592.5; ENST00000448372.5; ENST00000551798.1; ENST00000549595.5; ENST00000549518.6; ENST00000547886.5 | ||
| External Link | RMBase: RNA-editing_site_28024 | ||
N6-methyladenosine (m6A)
| In total 71 m6A sequence/site(s) in this target gene | |||
| mod ID: M6ASITE011629 | Click to Show/Hide the Full List | ||
| mod site | chr12:48043420-48043421:- | [4] | |
| Sequence | GCAATATGGTGTCGATTTGGACTATGAATCAAAAGACCTTT | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | A549 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000549518.6; rmsk_3781433; ENST00000552189.5; ENST00000448372.5 | ||
| External Link | RMBase: m6A_site_185264 | ||
| mod ID: M6ASITE011630 | Click to Show/Hide the Full List | ||
| mod site | chr12:48043746-48043747:- | [5] | |
| Sequence | TCCATTGTTAACTGAATGTTACTGTTTCCACTCCAGCAACT | ||
| Motif Score | 2.494845238 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000549518.6; ENST00000552189.5; ENST00000448372.5 | ||
| External Link | RMBase: m6A_site_185265 | ||
| mod ID: M6ASITE011631 | Click to Show/Hide the Full List | ||
| mod site | chr12:48043904-48043905:- | [5] | |
| Sequence | CTTTTCTCCTCTCCCGTTGTACCGCATTCTTTCCAGCATTG | ||
| Motif Score | 2.8355 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000448372.5; ENST00000549518.6; ENST00000552189.5 | ||
| External Link | RMBase: m6A_site_185266 | ||
| mod ID: M6ASITE011632 | Click to Show/Hide the Full List | ||
| mod site | chr12:48044089-48044090:- | [6] | |
| Sequence | GATCAACTTGAGATTTAAAAACTGCTATTCCTTTTGTTCTT | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa; hNPCs | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000552189.5; ENST00000549518.6; ENST00000448372.5 | ||
| External Link | RMBase: m6A_site_185267 | ||
| mod ID: M6ASITE011633 | Click to Show/Hide the Full List | ||
| mod site | chr12:48044112-48044113:- | [6] | |
| Sequence | AAAAAACGGGTGGAGTTTGGACTGATCAACTTGAGATTTAA | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa; hNPCs; AML | ||
| Seq Type List | m6A-seq; miCLIP | ||
| Transcript ID List | ENST00000549518.6; ENST00000448372.5; ENST00000552189.5 | ||
| External Link | RMBase: m6A_site_185268 | ||
| mod ID: M6ASITE011634 | Click to Show/Hide the Full List | ||
| mod site | chr12:48044301-48044302:- | [5] | |
| Sequence | TTTAAAGACTGAATGTGTTCACCATTTAGCCTGTAGATTTA | ||
| Motif Score | 2.026654762 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000549518.6; ENST00000552189.5; ENST00000448372.5 | ||
| External Link | RMBase: m6A_site_185269 | ||
| mod ID: M6ASITE011635 | Click to Show/Hide the Full List | ||
| mod site | chr12:48044500-48044501:- | [5] | |
| Sequence | TCTGACCAAGAAGAAAAAGGACCTTAGTATTTAGCATAAAA | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000552189.5; ENST00000549518.6; ENST00000448372.5 | ||
| External Link | RMBase: m6A_site_185270 | ||
| mod ID: M6ASITE011636 | Click to Show/Hide the Full List | ||
| mod site | chr12:48044549-48044550:- | [5] | |
| Sequence | GGAGCGGTTTCAGTGGCGCAACAAAGCACCACTTTTACTGT | ||
| Motif Score | 2.173910714 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000448372.5; ENST00000549518.6; ENST00000552189.5 | ||
| External Link | RMBase: m6A_site_185271 | ||
| mod ID: M6ASITE011637 | Click to Show/Hide the Full List | ||
| mod site | chr12:48044579-48044580:- | [6] | |
| Sequence | CAACACATGATACGGGGAAGACCCACCCAGGGAGCGGTTTC | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa; Huh7 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000552189.5; ENST00000549518.6; ENST00000448372.5 | ||
| External Link | RMBase: m6A_site_185272 | ||
| mod ID: M6ASITE011638 | Click to Show/Hide the Full List | ||
| mod site | chr12:48044601-48044602:- | [5] | |
| Sequence | AGCAGGGGTTTGCATGTGCTACCAACACATGATACGGGGAA | ||
| Motif Score | 2.05802381 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000448372.5; ENST00000552189.5; ENST00000549518.6 | ||
| External Link | RMBase: m6A_site_185273 | ||
| mod ID: M6ASITE011639 | Click to Show/Hide the Full List | ||
| mod site | chr12:48044657-48044658:- | [6] | |
| Sequence | GCACAGCACATGAGCCCTGGACAGGTCCCAAAGTTCCAAGC | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000552189.5; ENST00000448372.5; ENST00000549518.6 | ||
| External Link | RMBase: m6A_site_185274 | ||
| mod ID: M6ASITE011640 | Click to Show/Hide the Full List | ||
| mod site | chr12:48044764-48044765:- | [6] | |
| Sequence | CATAAGTCCCTTCCTCCAGGACCTTTCCTATTTATATGTCC | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000552189.5; ENST00000549518.6; ENST00000448372.5 | ||
| External Link | RMBase: m6A_site_185275 | ||
| mod ID: M6ASITE011641 | Click to Show/Hide the Full List | ||
| mod site | chr12:48044799-48044800:- | [6] | |
| Sequence | GGAATGTGACTGTAAACTGGACTTTGGGGCCCAGGCATAAG | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa; Huh7 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000448372.5; ENST00000549518.6; ENST00000552189.5 | ||
| External Link | RMBase: m6A_site_185276 | ||
| mod ID: M6ASITE011642 | Click to Show/Hide the Full List | ||
| mod site | chr12:48044804-48044805:- | [6] | |
| Sequence | GGTCAGGAATGTGACTGTAAACTGGACTTTGGGGCCCAGGC | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa; HEK293T; Huh7 | ||
| Seq Type List | m6A-seq; DART-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000552189.5; ENST00000448372.5; ENST00000549518.6 | ||
| External Link | RMBase: m6A_site_185277 | ||
| mod ID: M6ASITE011643 | Click to Show/Hide the Full List | ||
| mod site | chr12:48044856-48044857:- | [6] | |
| Sequence | ACACACACCCACACACACAAACACACACACACACACAATTT | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa; HEK293T; MT4; Huh7 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000552189.5; ENST00000549518.6; ENST00000448372.5 | ||
| External Link | RMBase: m6A_site_185278 | ||
| mod ID: M6ASITE011644 | Click to Show/Hide the Full List | ||
| mod site | chr12:48044882-48044883:- | [6] | |
| Sequence | AACCACCAGCATATACATGGACATACACACACACCCACACA | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; HEK293T; MT4; Huh7 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000448372.5; ENST00000549518.6; ENST00000552189.5 | ||
| External Link | RMBase: m6A_site_185279 | ||
| mod ID: M6ASITE011645 | Click to Show/Hide the Full List | ||
| mod site | chr12:48044901-48044902:- | [6] | |
| Sequence | GGCAAAAGGTAGGGATAAAAACCACCAGCATATACATGGAC | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | HeLa; HEK293T; MT4; Huh7 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000549518.6; ENST00000552189.5; ENST00000448372.5 | ||
| External Link | RMBase: m6A_site_185280 | ||
| mod ID: M6ASITE011646 | Click to Show/Hide the Full List | ||
| mod site | chr12:48044944-48044945:- | [5] | |
| Sequence | AATTCTTGTTGTTAGGCCTTACTAAGAAGTAGGAAAGGGCA | ||
| Motif Score | 2.494845238 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000448372.5; ENST00000549518.6; ENST00000552189.5 | ||
| External Link | RMBase: m6A_site_185281 | ||
| mod ID: M6ASITE011647 | Click to Show/Hide the Full List | ||
| mod site | chr12:48045001-48045002:- | [6] | |
| Sequence | ACTCAGTCCCTAATTATGGGACTGAGAAAAGCTTGGAAAGA | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa; CD34; HEK293T; U2OS; H1A; H1B; hESCs; fibroblasts; A549; GM12878; Huh7; iSLK; TIME; MSC | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000448372.5; ENST00000549518.6; ENST00000552189.5 | ||
| External Link | RMBase: m6A_site_185282 | ||
| mod ID: M6ASITE011648 | Click to Show/Hide the Full List | ||
| mod site | chr12:48045075-48045076:- | [6] | |
| Sequence | TCCCTAAGCTGAAGAGAGAGACTGCTTTTCACTTCTTCAGT | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; A549; U2OS; H1A; H1B; hESCs; fibroblasts; GM12878; LCLs; H1299; Huh7; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; TREX; NB4; MM6 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000448372.5; ENST00000552189.5; ENST00000549518.6 | ||
| External Link | RMBase: m6A_site_185283 | ||
| mod ID: M6ASITE011649 | Click to Show/Hide the Full List | ||
| mod site | chr12:48045098-48045099:- | [6] | |
| Sequence | TGCTGTGAAAGGGGTGAGGGACATCCCTAAGCTGAAGAGAG | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; A549; U2OS; H1A; H1B; hESCs; fibroblasts; GM12878; LCLs; Huh7; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; TREX; NB4; MM6 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000549518.6; ENST00000448372.5; ENST00000552189.5 | ||
| External Link | RMBase: m6A_site_185284 | ||
| mod ID: M6ASITE011650 | Click to Show/Hide the Full List | ||
| mod site | chr12:48045125-48045126:- | [6] | |
| Sequence | GATACTATTTTTGCAAAGAAACTTTGGTGCTGTGAAAGGGG | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; A549; U2OS; H1A; H1B; hESCs; fibroblasts; GM12878; Huh7; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; TREX; endometrial; NB4; MM6 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000549518.6; ENST00000448372.5; ENST00000552189.5 | ||
| External Link | RMBase: m6A_site_185285 | ||
| mod ID: M6ASITE011651 | Click to Show/Hide the Full List | ||
| mod site | chr12:48045177-48045178:- | [5] | |
| Sequence | GGCCCTGGCGAGATGCATTCACAAGCACATCTGCCTTTCCT | ||
| Motif Score | 2.047297619 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000552189.5; ENST00000549518.6; ENST00000448372.5 | ||
| External Link | RMBase: m6A_site_185286 | ||
| mod ID: M6ASITE011652 | Click to Show/Hide the Full List | ||
| mod site | chr12:48045201-48045202:- | [6] | |
| Sequence | TGTAACACCCTTGATCCTGGACCAGGCCCTGGCGAGATGCA | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; A549; U2OS; H1A; H1B; hESCs; fibroblasts; GM12878; MM6; Huh7; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; TREX; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000549518.6; ENST00000552189.5; ENST00000448372.5 | ||
| External Link | RMBase: m6A_site_185287 | ||
| mod ID: M6ASITE011653 | Click to Show/Hide the Full List | ||
| mod site | chr12:48045246-48045247:- | [6] | |
| Sequence | TACAGCCAGAGACCTTGGAAACAGCTGCTCCCAGCCCTCTG | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; A549; U2OS; fibroblasts; GM12878; Huh7; peripheral-blood; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; MM6 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000552189.5; ENST00000549518.6; ENST00000448372.5 | ||
| External Link | RMBase: m6A_site_185288 | ||
| mod ID: M6ASITE011654 | Click to Show/Hide the Full List | ||
| mod site | chr12:48045255-48045256:- | [6] | |
| Sequence | TTTGTTGTCTACAGCCAGAGACCTTGGAAACAGCTGCTCCC | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; A549; U2OS; fibroblasts; GM12878; Huh7; peripheral-blood; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; MM6 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000552189.5; ENST00000549518.6; ENST00000448372.5 | ||
| External Link | RMBase: m6A_site_185289 | ||
| mod ID: M6ASITE011655 | Click to Show/Hide the Full List | ||
| mod site | chr12:48045283-48045284:- | [6] | |
| Sequence | AGACCTTGACCATGTGGGGGACCAGCTCTTTGTTGTCTACA | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; A549; U2OS; fibroblasts; GM12878; Huh7; peripheral-blood; HEK293A-TOA; iSLK | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000448372.5; ENST00000552189.5; ENST00000549518.6 | ||
| External Link | RMBase: m6A_site_185290 | ||
| mod ID: M6ASITE011656 | Click to Show/Hide the Full List | ||
| mod site | chr12:48045301-48045302:- | [6] | |
| Sequence | AAGACTGTCTCACTTAGCAGACCTTGACCATGTGGGGGACC | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa; HEK293T; HepG2; U2OS; fibroblasts; A549; GM12878; Huh7; HEK293A-TOA; TREX | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000448372.5; ENST00000552189.5; ENST00000549518.6 | ||
| External Link | RMBase: m6A_site_185291 | ||
| mod ID: M6ASITE011657 | Click to Show/Hide the Full List | ||
| mod site | chr12:48045318-48045319:- | [6] | |
| Sequence | CACCGAAAACTCTTGTGAAGACTGTCTCACTTAGCAGACCT | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa; HEK293T; HepG2; U2OS; fibroblasts; A549; GM12878; Huh7; HEK293A-TOA; TREX | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000448372.5; ENST00000552189.5; ENST00000549518.6 | ||
| External Link | RMBase: m6A_site_185292 | ||
| mod ID: M6ASITE011658 | Click to Show/Hide the Full List | ||
| mod site | chr12:48045330-48045331:- | [6] | |
| Sequence | TGGGAGATCCTCCACCGAAAACTCTTGTGAAGACTGTCTCA | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa; HEK293T; HepG2; U2OS; fibroblasts; A549; HEK293A-TOA | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000549518.6; ENST00000549595.5; ENST00000552189.5; ENST00000448372.5 | ||
| External Link | RMBase: m6A_site_185293 | ||
| mod ID: M6ASITE011659 | Click to Show/Hide the Full List | ||
| mod site | chr12:48046377-48046378:- | [4] | |
| Sequence | TGCTGACTGTATTACCAAAGACAGACCAATCAACTTCACAC | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HEK293T; A549 | ||
| Seq Type List | MeRIP-seq; m6A-seq | ||
| Transcript ID List | ENST00000549518.6; ENST00000549595.5; ENST00000552189.5; ENST00000448372.5 | ||
| External Link | RMBase: m6A_site_185294 | ||
| mod ID: M6ASITE011660 | Click to Show/Hide the Full List | ||
| mod site | chr12:48048052-48048053:- | [5] | |
| Sequence | ACTTTAGAAAGAAGAATATTACCTATTACGACTCCATGGGT | ||
| Motif Score | 2.052208333 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000448372.5; ENST00000552189.5; ENST00000549518.6; ENST00000549595.5 | ||
| External Link | RMBase: m6A_site_185295 | ||
| mod ID: M6ASITE011661 | Click to Show/Hide the Full List | ||
| mod site | chr12:48048974-48048975:- | [5] | |
| Sequence | AGTAGATGTATTTTCTGTTGACATTCTTTTGGTGCCCATTC | ||
| Motif Score | 2.859755952 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000552189.5; ENST00000549518.6; ENST00000448372.5; ENST00000549595.5 | ||
| External Link | RMBase: m6A_site_185296 | ||
| mod ID: M6ASITE011662 | Click to Show/Hide the Full List | ||
| mod site | chr12:48054906-48054907:- | [7] | |
| Sequence | TAAAAATGTATATATTTTAAACATATATATGTTTATATGTA | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | GM12878; Huh7 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000549595.5; ENST00000468202.1; ENST00000448372.5; ENST00000549518.6; ENST00000552189.5 | ||
| External Link | RMBase: m6A_site_185297 | ||
| mod ID: M6ASITE011663 | Click to Show/Hide the Full List | ||
| mod site | chr12:48055017-48055018:- | [6] | |
| Sequence | GGCATGGATTCTTTGCAGGAACAACACCTACACCCAAGAGG | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa; HEK293T; A549; HepG2; GM12878; Huh7; HEK293A-TOA; iSLK; TIME; TREX | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000552189.5; ENST00000549595.5; ENST00000463559.1; ENST00000448372.5; ENST00000468202.1; ENST00000549518.6 | ||
| External Link | RMBase: m6A_site_185298 | ||
| mod ID: M6ASITE011664 | Click to Show/Hide the Full List | ||
| mod site | chr12:48055058-48055059:- | [6] | |
| Sequence | TGGTTGAATGAAGCACCTAAACATTGTATACTGCAGATTTA | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa; HEK293T; A549; HepG2; GM12878; Huh7; HEK293A-TOA; iSLK; TIME; TREX | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000549595.5; ENST00000463559.1; ENST00000549518.6; ENST00000552189.5; ENST00000468202.1; ENST00000448372.5 | ||
| External Link | RMBase: m6A_site_185299 | ||
| mod ID: M6ASITE011665 | Click to Show/Hide the Full List | ||
| mod site | chr12:48055111-48055112:- | [6] | |
| Sequence | GCAAGAGAAAGTGTAACTGGACTGCCACGGCTAAAAGAAAA | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa; HEK293T; HepG2; GM12878; Huh7; HEK293A-TOA; iSLK; TIME; TREX | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000549595.5; ENST00000552189.5; ENST00000549518.6; ENST00000463559.1; ENST00000448372.5; ENST00000468202.1 | ||
| External Link | RMBase: m6A_site_185300 | ||
| mod ID: M6ASITE011666 | Click to Show/Hide the Full List | ||
| mod site | chr12:48055116-48055117:- | [8] | |
| Sequence | CAAGCGCAAGAGAAAGTGTAACTGGACTGCCACGGCTAAAA | ||
| Motif Score | 2.590089286 | ||
| Cell/Tissue List | CD8T | ||
| Seq Type List | m6A-CLIP/IP | ||
| Transcript ID List | ENST00000549518.6; ENST00000448372.5; ENST00000463559.1; ENST00000468202.1; ENST00000552189.5; ENST00000549595.5 | ||
| External Link | RMBase: m6A_site_185301 | ||
| mod ID: M6ASITE011667 | Click to Show/Hide the Full List | ||
| mod site | chr12:48055246-48055247:- | [6] | |
| Sequence | AAGATAATAAGATGAAGGGAACATCGCCGTTTGGAAAGTGT | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa; HEK293T; HepG2; Huh7; HEK293A-TOA | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000549595.5; ENST00000552189.5; ENST00000448372.5; ENST00000468202.1; ENST00000549518.6; ENST00000463559.1 | ||
| External Link | RMBase: m6A_site_185302 | ||
| mod ID: M6ASITE011668 | Click to Show/Hide the Full List | ||
| mod site | chr12:48055442-48055443:- | [6] | |
| Sequence | TGCTGACTGATGCCTGGGGGACCAAGCCCATAATTGTGGAG | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; HEK293T; HepG2; GM12878; HEK293A-TOA | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000468202.1; ENST00000549518.6; ENST00000549595.5; ENST00000448372.5; ENST00000552189.5 | ||
| External Link | RMBase: m6A_site_185303 | ||
| mod ID: M6ASITE011669 | Click to Show/Hide the Full List | ||
| mod site | chr12:48055580-48055581:- | [6] | |
| Sequence | TACTAAACAGTATAGGGTAAACATCCAATTAAAAGGCAGAT | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000552189.5; ENST00000468202.1; ENST00000549595.5; ENST00000448372.5; ENST00000549518.6 | ||
| External Link | RMBase: m6A_site_185304 | ||
| mod ID: M6ASITE011670 | Click to Show/Hide the Full List | ||
| mod site | chr12:48058621-48058622:- | [8] | |
| Sequence | ATCACAGCACAAAGAAGGAGACAAAATGGACAAATATACTG | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | CD8T | ||
| Seq Type List | m6A-CLIP/IP | ||
| Transcript ID List | rmsk_3781455; ENST00000549595.5; ENST00000549518.6; ENST00000448372.5; ENST00000552189.5 | ||
| External Link | RMBase: m6A_site_185305 | ||
| mod ID: M6ASITE011671 | Click to Show/Hide the Full List | ||
| mod site | chr12:48058700-48058701:- | [8] | |
| Sequence | ATACCTCTAGAAGACAGGTGACTAAGGCAGAACTAAAACAA | ||
| Motif Score | 3.28175 | ||
| Cell/Tissue List | CD8T | ||
| Seq Type List | m6A-CLIP/IP | ||
| Transcript ID List | rmsk_3781455; ENST00000552189.5; ENST00000448372.5; ENST00000549595.5; ENST00000549518.6 | ||
| External Link | RMBase: m6A_site_185306 | ||
| mod ID: M6ASITE011672 | Click to Show/Hide the Full List | ||
| mod site | chr12:48058707-48058708:- | [8] | |
| Sequence | TTTTTTCATACCTCTAGAAGACAGGTGACTAAGGCAGAACT | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | CD8T | ||
| Seq Type List | m6A-CLIP/IP | ||
| Transcript ID List | rmsk_3781455; ENST00000552189.5; ENST00000549595.5; ENST00000448372.5; ENST00000549518.6 | ||
| External Link | RMBase: m6A_site_185307 | ||
| mod ID: M6ASITE011673 | Click to Show/Hide the Full List | ||
| mod site | chr12:48063673-48063674:- | [6] | |
| Sequence | TTTTTACATACAAATAGTAAACATTAAGTGGCAGTTAAATT | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000549518.6; ENST00000549595.5; ENST00000551358.1; ENST00000448372.5; ENST00000552189.5 | ||
| External Link | RMBase: m6A_site_185308 | ||
| mod ID: M6ASITE011674 | Click to Show/Hide the Full List | ||
| mod site | chr12:48065069-48065070:- | [9] | |
| Sequence | AAGATGAATTTCCTGAAATTACAGAGGTAAGTTACATACAC | ||
| Motif Score | 2.07285119 | ||
| Cell/Tissue List | HEK293 | ||
| Seq Type List | m6A-REF-seq | ||
| Transcript ID List | ENST00000551358.1; ENST00000552189.5; ENST00000549595.5; ENST00000448372.5; ENST00000549518.6 | ||
| External Link | RMBase: m6A_site_185309 | ||
| mod ID: M6ASITE011675 | Click to Show/Hide the Full List | ||
| mod site | chr12:48071679-48071680:- | [6] | |
| Sequence | TGAAAGTGAAAGATTCCCAGACTCCAACTCCCAGGTAAACT | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000552189.5; ENST00000549518.6; ENST00000448372.5; ENST00000549595.5 | ||
| External Link | RMBase: m6A_site_185310 | ||
| mod ID: M6ASITE011676 | Click to Show/Hide the Full List | ||
| mod site | chr12:48071714-48071715:- | [6] | |
| Sequence | TTGTGTTTCTTTAGGATCAGACTCTGTGATTTTACTGAAAG | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000448372.5; ENST00000549595.5; ENST00000549518.6; ENST00000552189.5 | ||
| External Link | RMBase: m6A_site_185311 | ||
| mod ID: M6ASITE011677 | Click to Show/Hide the Full List | ||
| mod site | chr12:48074474-48074475:- | [6] | |
| Sequence | AGCATTTTAACTAACCAGGAACAGCTGTCCCACAGTGTATA | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa; Huh7 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000549595.5; ENST00000549518.6; ENST00000552189.5; ENST00000448372.5 | ||
| External Link | RMBase: m6A_site_185312 | ||
| mod ID: M6ASITE011678 | Click to Show/Hide the Full List | ||
| mod site | chr12:48074547-48074548:- | [6] | |
| Sequence | CTCACTGTTTAAAAATGGAAACTCTTGTGCATCTCAGATCA | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000552189.5; ENST00000549595.5; ENST00000448372.5; ENST00000549518.6 | ||
| External Link | RMBase: m6A_site_185313 | ||
| mod ID: M6ASITE011679 | Click to Show/Hide the Full List | ||
| mod site | chr12:48074568-48074569:- | [6] | |
| Sequence | CAGTAAAAATACTTTGAAAGACTCACTGTTTAAAAATGGAA | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000448372.5; ENST00000549595.5; ENST00000552189.5; ENST00000549518.6 | ||
| External Link | RMBase: m6A_site_185314 | ||
| mod ID: M6ASITE011680 | Click to Show/Hide the Full List | ||
| mod site | chr12:48074715-48074716:- | [6] | |
| Sequence | AAACAGTTTACTATAGCCAAACCCACCACACATTTTCCTTT | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | HeLa; fibroblasts | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000549595.5; ENST00000552189.5; ENST00000448372.5; ENST00000549518.6 | ||
| External Link | RMBase: m6A_site_185315 | ||
| mod ID: M6ASITE011681 | Click to Show/Hide the Full List | ||
| mod site | chr12:48074733-48074734:- | [6] | |
| Sequence | CTACAGATGGTCACAGGGAAACAGTTTACTATAGCCAAACC | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa; fibroblasts | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000549595.5; ENST00000549518.6; ENST00000552189.5; ENST00000448372.5 | ||
| External Link | RMBase: m6A_site_185316 | ||
| mod ID: M6ASITE011682 | Click to Show/Hide the Full List | ||
| mod site | chr12:48074760-48074761:- | [6] | |
| Sequence | GAAGAAAGAGAGATTTACAGACAGCTGCTACAGATGGTCAC | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000552189.5; ENST00000549518.6; ENST00000448372.5; ENST00000549595.5 | ||
| External Link | RMBase: m6A_site_185317 | ||
| mod ID: M6ASITE011683 | Click to Show/Hide the Full List | ||
| mod site | chr12:48083683-48083684:- | [6] | |
| Sequence | GAAAAATCTTTTCCTATTAAACCTGTTCCAAGTCCATCTTG | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000448372.5; ENST00000547886.5; ENST00000549518.6; ENST00000551592.5; ENST00000552189.5; ENST00000549595.5 | ||
| External Link | RMBase: m6A_site_185318 | ||
| mod ID: M6ASITE011684 | Click to Show/Hide the Full List | ||
| mod site | chr12:48083729-48083730:- | [6] | |
| Sequence | CAGTTTTGCGGGAAAGTCAAACCATCACTGCCATGTATCTG | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000551798.1; ENST00000552189.5; ENST00000549595.5; ENST00000551592.5; ENST00000549518.6; ENST00000547886.5; ENST00000448372.5 | ||
| External Link | RMBase: m6A_site_185319 | ||
| mod ID: M6ASITE011685 | Click to Show/Hide the Full List | ||
| mod site | chr12:48083750-48083751:- | [6] | |
| Sequence | TTTTTTTAGTGGATTATCAAACAGTTTTGCGGGAAAGTCAA | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000549595.5; ENST00000551592.5; ENST00000551798.1; ENST00000552189.5; ENST00000547886.5; ENST00000549518.6; ENST00000448372.5 | ||
| External Link | RMBase: m6A_site_185320 | ||
| mod ID: M6ASITE011686 | Click to Show/Hide the Full List | ||
| mod site | chr12:48096398-48096399:- | [6] | |
| Sequence | TTCCAGGCAAGGACATTTGGACCGATCTTTTACATGTTCCA | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000549518.6; ENST00000551798.1; ENST00000547886.5; ENST00000448372.5; ENST00000552189.5; ENST00000549595.5; ENST00000547181.5; ENST00000551592.5 | ||
| External Link | RMBase: m6A_site_185322 | ||
| mod ID: M6ASITE011687 | Click to Show/Hide the Full List | ||
| mod site | chr12:48096406-48096407:- | [6] | |
| Sequence | ATTTTATCTTCCAGGCAAGGACATTTGGACCGATCTTTTAC | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000549518.6; ENST00000551798.1; ENST00000547181.5; ENST00000448372.5; ENST00000547886.5; ENST00000549595.5; ENST00000551592.5; ENST00000552189.5 | ||
| External Link | RMBase: m6A_site_185323 | ||
| mod ID: M6ASITE011688 | Click to Show/Hide the Full List | ||
| mod site | chr12:48097765-48097766:- | [6] | |
| Sequence | AGAACAGTACGTTTTAGAAAACTCATTGTTCCTCTGAATTT | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000552189.5; ENST00000549595.5; ENST00000448372.5; ENST00000547181.5; ENST00000549882.1; ENST00000549518.6; ENST00000551592.5; ENST00000551798.1 | ||
| External Link | RMBase: m6A_site_185324 | ||
| mod ID: M6ASITE011689 | Click to Show/Hide the Full List | ||
| mod site | chr12:48097782-48097783:- | [6] | |
| Sequence | TATGTATGAGTTCTTTGAGAACAGTACGTTTTAGAAAACTC | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000551798.1; ENST00000448372.5; ENST00000549518.6; ENST00000549595.5; ENST00000552189.5; ENST00000551592.5; ENST00000549882.1; ENST00000547181.5 | ||
| External Link | RMBase: m6A_site_185325 | ||
| mod ID: M6ASITE011690 | Click to Show/Hide the Full List | ||
| mod site | chr12:48097813-48097814:- | [6] | |
| Sequence | ATTAAGACAAGGACGTTAAGACAAAATAGGCTATGTATGAG | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000549595.5; ENST00000551592.5; ENST00000448372.5; ENST00000551798.1; ENST00000549518.6; ENST00000552189.5; ENST00000547181.5; ENST00000549882.1 | ||
| External Link | RMBase: m6A_site_185326 | ||
| mod ID: M6ASITE011691 | Click to Show/Hide the Full List | ||
| mod site | chr12:48098018-48098019:- | [6] | |
| Sequence | ACAAACAGGTTTTCCAGAGGACCAGCTTTCGCTTTCTGACC | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000547181.5; ENST00000549595.5; ENST00000549882.1; ENST00000552189.5; ENST00000448372.5; ENST00000551592.5; ENST00000551798.1; ENST00000549518.6 | ||
| External Link | RMBase: m6A_site_185327 | ||
| mod ID: M6ASITE011692 | Click to Show/Hide the Full List | ||
| mod site | chr12:48098034-48098035:- | [6] | |
| Sequence | AAACCCACCTCCTGCCACAAACAGGTTTTCCAGAGGACCAG | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000551798.1; ENST00000547181.5; ENST00000549882.1; ENST00000552189.5; ENST00000448372.5; ENST00000549518.6; ENST00000549595.5 | ||
| External Link | RMBase: m6A_site_185328 | ||
| mod ID: M6ASITE011693 | Click to Show/Hide the Full List | ||
| mod site | chr12:48098052-48098053:- | [6] | |
| Sequence | ACCACAACTCCGTATTCAAAACCCACCTCCTGCCACAAACA | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000552189.5; ENST00000448372.5; ENST00000549518.6; ENST00000551798.1; ENST00000549882.1; ENST00000547181.5; ENST00000549595.5 | ||
| External Link | RMBase: m6A_site_185329 | ||
| mod ID: M6ASITE011694 | Click to Show/Hide the Full List | ||
| mod site | chr12:48098072-48098073:- | [6] | |
| Sequence | TGGAGAAGTGACTTTAGTGAACCACAACTCCGTATTCAAAA | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000547181.5; ENST00000549595.5; ENST00000549518.6; ENST00000549882.1; ENST00000552189.5; ENST00000448372.5; ENST00000551798.1 | ||
| External Link | RMBase: m6A_site_185330 | ||
| mod ID: M6ASITE011695 | Click to Show/Hide the Full List | ||
| mod site | chr12:48105469-48105470:- | [6] | |
| Sequence | CATCAGAAAGAAACGTCTGGACCCTCCTGCTCAGGTTATGA | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000547181.5; ENST00000552189.5; ENST00000448372.5; ENST00000549518.6; ENST00000549882.1; ENST00000551798.1 | ||
| External Link | RMBase: m6A_site_185331 | ||
| mod ID: M6ASITE011696 | Click to Show/Hide the Full List | ||
| mod site | chr12:48105550-48105551:- | [6] | |
| Sequence | TACTTCTGCGTCTCACGTAAACATTTCTCCAACTCTCCTAC | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000551798.1; ENST00000547181.5; ENST00000549518.6; ENST00000549882.1; ENST00000448372.5; ENST00000552189.5 | ||
| External Link | RMBase: m6A_site_185332 | ||
| mod ID: M6ASITE011697 | Click to Show/Hide the Full List | ||
| mod site | chr12:48105591-48105592:- | [6] | |
| Sequence | CATCTCCCGCCTTCCTTGAGACCCGAGTGATATTTCTTGAC | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000549882.1; ENST00000448372.5; ENST00000552189.5; ENST00000551798.1; ENST00000549518.6; ENST00000547181.5 | ||
| External Link | RMBase: m6A_site_185333 | ||
| mod ID: M6ASITE011700 | Click to Show/Hide the Full List | ||
| mod site | chr12:48105765-48105766:- | [10] | |
| Sequence | CTCTCCTGGGCCGCGGAGAGACCGTCTCCCTGCCGTTACAG | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000549882.1; ENST00000549518.6; ENST00000551798.1; ENST00000552189.5; ENST00000547181.5; ENST00000448372.5 | ||
| External Link | RMBase: m6A_site_185336 | ||
| mod ID: M6ASITE011705 | Click to Show/Hide the Full List | ||
| mod site | chr12:48106057-48106058:- | [6] | |
| Sequence | CGTTCCGCGCCCGGCCGGAAACCCCTTCGCATGGCAGCCGG | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | HeLa; HEK293T | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000549882.1; ENST00000549518.6 | ||
| External Link | RMBase: m6A_site_185341 | ||
References