m6A Target Gene Information
General Information of the m6A Target Gene (ID: M6ATAR00684)
Full List of m6A Methylation Regulator of This Target Gene and Corresponding Disease/Drug Response(s)
PRMT5
can be regulated by the following regulator(s), and cause disease/drug response(s). You can browse detail information of regulator(s) or disease/drug response(s).
Browse Regulator
Browse Disease
Methyltransferase-like 3 (METTL3) [WRITER]
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | Pancreatic islets | Mus musculus |
|
Treatment: Mettl3 knockout mice
Control: Mettl3 flox/flox mice
|
GSE155612 | |
| Regulation |
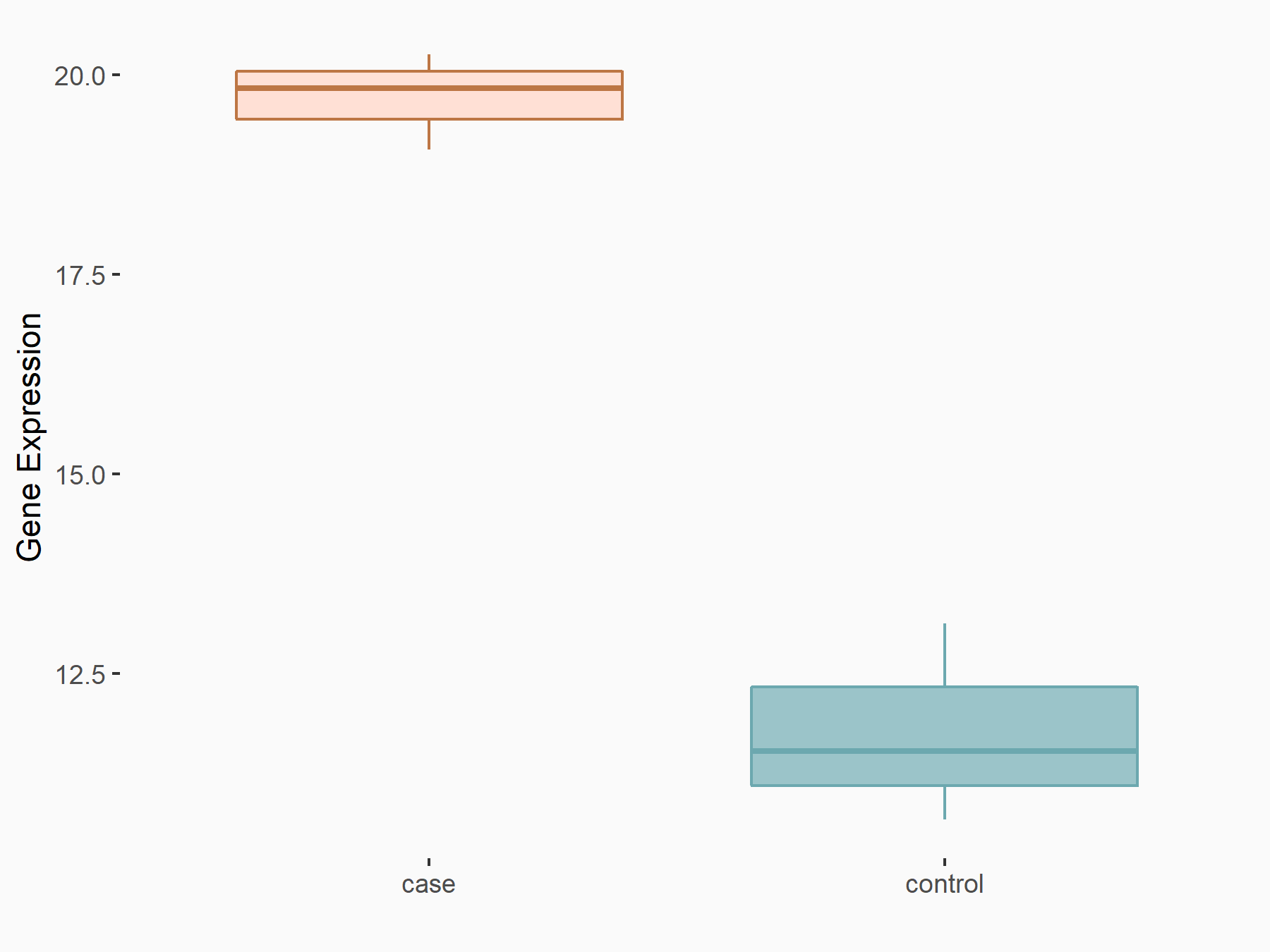  |
logFC: 7.02E-01 p-value: 6.96E-04 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between PRMT5 and the regulator | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
| Regulation | logFC: 5.94E+00 | GSE60213 |
| In total 1 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [1] | |||
| Response Summary | METTL3 intensified the metastasis and proliferation of OSCC by modulating the m6A amounts of Protein arginine N-methyltransferase 5 (PRMT5) and PD-L1. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Oral squamous cell carcinoma | ICD-11: 2B6E.0 | ||
| Pathway Response | PD-L1 expression and PD-1 checkpoint pathway in cancer | hsa05235 | ||
| In-vitro Model | SCC-9 | Tongue squamous cell carcinoma | Homo sapiens | CVCL_1685 |
| SCC-4 | Tongue squamous cell carcinoma | Homo sapiens | CVCL_1684 | |
| SCC-25 | Tongue squamous cell carcinoma | Homo sapiens | CVCL_1682 | |
| CAL-27 | Tongue squamous cell carcinoma | Homo sapiens | CVCL_1107 | |
| In-vivo Model | Six-week-old nude mice were randomly divided into two groups (three mice per group) and cultured with continuous access to sterile food and water in pathogen-free sterile conditions. To establish the OSCC xenograft model, we subcutaneously injected 5 × 106 SCC-9 cells stably transfected with METTL3 shRNA or sh-NC vectors into nude mice. | |||
Head and neck squamous carcinoma [ICD-11: 2B6E]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [1] | |||
| Response Summary | METTL3 intensified the metastasis and proliferation of OSCC by modulating the m6A amounts of Protein arginine N-methyltransferase 5 (PRMT5) and PD-L1. | |||
| Responsed Disease | Oral squamous cell carcinoma [ICD-11: 2B6E.0] | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Target Regulation | Up regulation | |||
| Pathway Response | PD-L1 expression and PD-1 checkpoint pathway in cancer | hsa05235 | ||
| In-vitro Model | SCC-9 | Tongue squamous cell carcinoma | Homo sapiens | CVCL_1685 |
| SCC-4 | Tongue squamous cell carcinoma | Homo sapiens | CVCL_1684 | |
| SCC-25 | Tongue squamous cell carcinoma | Homo sapiens | CVCL_1682 | |
| CAL-27 | Tongue squamous cell carcinoma | Homo sapiens | CVCL_1107 | |
| In-vivo Model | Six-week-old nude mice were randomly divided into two groups (three mice per group) and cultured with continuous access to sterile food and water in pathogen-free sterile conditions. To establish the OSCC xenograft model, we subcutaneously injected 5 × 106 SCC-9 cells stably transfected with METTL3 shRNA or sh-NC vectors into nude mice. | |||
RNA Modification Sequencing Data Associated with the Target (ID: M6ATAR00684)
| In total 1 m6A sequence/site(s) in this target gene | |||
| mod ID: M1ASITE000031 | Click to Show/Hide the Full List | ||
| mod site | chr14:22924484-22924485:- | [2] | |
| Sequence | AGAAGAGGAGAAGGATACCAATGTCCAGTGAGTGCTTTCCC | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | m1A-seq | ||
| Transcript ID List | ENST00000216350.12; ENST00000324366.13; ENST00000555530.5; ENST00000397441.6; ENST00000553915.5; ENST00000397440.8; ENST00000553897.5; ENST00000553502.5 | ||
| External Link | RMBase: m1A_site_300 | ||
5-methylcytidine (m5C)
| In total 3 m6A sequence/site(s) in this target gene | |||
| mod ID: M5CSITE000214 | Click to Show/Hide the Full List | ||
| mod site | chr14:22926236-22926237:- | [3] | |
| Sequence | TTGATCGCTGGCTTGGGGAGCCCATCAAAGCAGCCATTCTC | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000554867.5; ENST00000216350.12; ENST00000553550.5; ENST00000556043.5; ENST00000553787.5; ENST00000553641.5; ENST00000324366.13; ENST00000397441.6; ENST00000397440.8; ENST00000554716.5; ENST00000553897.5; ENST00000555530.5; ENST00000553915.5 | ||
| External Link | RMBase: m5C_site_12459 | ||
| mod ID: M5CSITE000215 | Click to Show/Hide the Full List | ||
| mod site | chr14:22928184-22928185:- | [3] | |
| Sequence | ATACGCTAATTGTGGGAAAGCTTTCTCCATGGATTCGTCCA | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000397441.6; ENST00000553787.5; ENST00000554910.5; ENST00000553641.5; ENST00000556032.1; ENST00000554867.5; ENST00000216350.12; ENST00000421938.2; ENST00000557415.5; ENST00000553915.5; ENST00000553550.5; ENST00000557015.1; ENST00000397440.8; ENST00000554716.5; ENST00000324366.13; ENST00000627278.1; ENST00000553897.5; ENST00000556616.5 | ||
| External Link | RMBase: m5C_site_12460 | ||
| mod ID: M5CSITE000216 | Click to Show/Hide the Full List | ||
| mod site | chr14:22929371-22929372:- | [3] | |
| Sequence | GAATCCCGGCGTGGACAGCGCGAGGAGAAAGATGGCGGCGA | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000554867.5; ENST00000556616.5; ENST00000324366.13; ENST00000553897.5; ENST00000557415.5; ENST00000553550.5; ENST00000553787.5; ENST00000557015.1; ENST00000556426.1; ENST00000397441.6; ENST00000216350.12 | ||
| External Link | RMBase: m5C_site_12461 | ||
N6-methyladenosine (m6A)
| In total 57 m6A sequence/site(s) in this target gene | |||
| mod ID: M6ASITE018854 | Click to Show/Hide the Full List | ||
| mod site | chr14:22920555-22920556:- | [4] | |
| Sequence | GGTTTTGTGTAAGAGGAAATACAAATAAAGTTATAGCCCTT | ||
| Motif Score | 2.110482143 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000397440.8; ENST00000216350.12; ENST00000324366.13; ENST00000557443.5; ENST00000397441.6; ENST00000476175.6 | ||
| External Link | RMBase: m6A_site_240002 | ||
| mod ID: M6ASITE018855 | Click to Show/Hide the Full List | ||
| mod site | chr14:22920623-22920624:- | [5] | |
| Sequence | TCCCTGGAATATCCCTGAGAACATGGGGTTTGAACGGATTT | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa; HEK293T; hESC-HEK293T; A549; MM6; AML | ||
| Seq Type List | m6A-seq; MeRIP-seq; MAZTER-seq; miCLIP | ||
| Transcript ID List | ENST00000397440.8; ENST00000557443.5; ENST00000216350.12; ENST00000324366.13; ENST00000476175.6; ENST00000397441.6 | ||
| External Link | RMBase: m6A_site_240007 | ||
| mod ID: M6ASITE018856 | Click to Show/Hide the Full List | ||
| mod site | chr14:22920645-22920646:- | [5] | |
| Sequence | CTACGCACTCAGCCTCAAGAACTCCCTGGAATATCCCTGAG | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa; HEK293T; fibroblasts; A549 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000476175.6; ENST00000557443.5; ENST00000397440.8; ENST00000324366.13; ENST00000397441.6; ENST00000216350.12 | ||
| External Link | RMBase: m6A_site_240008 | ||
| mod ID: M6ASITE018857 | Click to Show/Hide the Full List | ||
| mod site | chr14:22920706-22920707:- | [6] | |
| Sequence | CAAGCCACCAATCTATGAAGACCTCAGGCCAGGGGGTGAGG | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HEK293T; A549 | ||
| Seq Type List | MeRIP-seq; m6A-seq | ||
| Transcript ID List | ENST00000216350.12; ENST00000397441.6; ENST00000324366.13; ENST00000397440.8; ENST00000476175.6; ENST00000557443.5 | ||
| External Link | RMBase: m6A_site_240009 | ||
| mod ID: M6ASITE018858 | Click to Show/Hide the Full List | ||
| mod site | chr14:22920766-22920767:- | [4] | |
| Sequence | TCAATCTGCTTTCCTGCCTTACATCAAGGTGGGCAAGGGAT | ||
| Motif Score | 2.07285119 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000397441.6; ENST00000397440.8; ENST00000476175.6; ENST00000557443.5; ENST00000324366.13; ENST00000216350.12 | ||
| External Link | RMBase: m6A_site_240010 | ||
| mod ID: M6ASITE018859 | Click to Show/Hide the Full List | ||
| mod site | chr14:22920831-22920832:- | [4] | |
| Sequence | TGTAGTACAGAAGGTGCAGTACATCTATGGGCTGTGATTCC | ||
| Motif Score | 2.856142857 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000397441.6; ENST00000454731.6; ENST00000557443.5; ENST00000397440.8; ENST00000476175.6; ENST00000216350.12; ENST00000324366.13; ENST00000555454.5 | ||
| External Link | RMBase: m6A_site_240011 | ||
| mod ID: M6ASITE018860 | Click to Show/Hide the Full List | ||
| mod site | chr14:22920845-22920846:- | [7] | |
| Sequence | TCAGGTTCTGCTCCTGTAGTACAGAAGGTGCAGTACATCTA | ||
| Motif Score | 2.856142857 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000397441.6; ENST00000454731.6; ENST00000324366.13; ENST00000555454.5; ENST00000397440.8; ENST00000216350.12; ENST00000557443.5; ENST00000476175.6 | ||
| External Link | RMBase: m6A_site_240012 | ||
| mod ID: M6ASITE018861 | Click to Show/Hide the Full List | ||
| mod site | chr14:22920960-22920961:- | [7] | |
| Sequence | TATGAGTGGGCTGTGACAGCACCAGTCTGTTCTGCTATTCA | ||
| Motif Score | 2.809946429 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000553915.5; ENST00000476175.6; ENST00000557443.5; ENST00000555454.5; ENST00000553897.5; ENST00000397441.6; ENST00000324366.13; ENST00000216350.12; ENST00000454731.6; ENST00000397440.8 | ||
| External Link | RMBase: m6A_site_240013 | ||
| mod ID: M6ASITE018862 | Click to Show/Hide the Full List | ||
| mod site | chr14:22920965-22920966:- | [4] | |
| Sequence | TGTGGTATGAGTGGGCTGTGACAGCACCAGTCTGTTCTGCT | ||
| Motif Score | 2.859755952 | ||
| Cell/Tissue List | hESC-HEK293T; AML | ||
| Seq Type List | MAZTER-seq; miCLIP | ||
| Transcript ID List | ENST00000553897.5; ENST00000555454.5; ENST00000216350.12; ENST00000324366.13; ENST00000476175.6; ENST00000397441.6; ENST00000557443.5; ENST00000553915.5; ENST00000397440.8; ENST00000454731.6 | ||
| External Link | RMBase: m6A_site_240014 | ||
| mod ID: M6ASITE018863 | Click to Show/Hide the Full List | ||
| mod site | chr14:22922453-22922454:- | [5] | |
| Sequence | TGAGACTGTGCTTTATCAGGACATCACTCTGAGTGAGTGTC | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; hESC-HEK293T | ||
| Seq Type List | m6A-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000553915.5; ENST00000454731.6; ENST00000553897.5; ENST00000397441.6; ENST00000397440.8; ENST00000324366.13; ENST00000216350.12; ENST00000557758.1; ENST00000476175.6; ENST00000555454.5 | ||
| External Link | RMBase: m6A_site_240015 | ||
| mod ID: M6ASITE018864 | Click to Show/Hide the Full List | ||
| mod site | chr14:22922469-22922470:- | [5] | |
| Sequence | GCTTTGCCGGCTACTTTGAGACTGTGCTTTATCAGGACATC | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000553915.5; ENST00000216350.12; ENST00000555454.5; ENST00000557758.1; ENST00000553897.5; ENST00000324366.13; ENST00000397440.8; ENST00000476175.6; ENST00000397441.6; ENST00000454731.6 | ||
| External Link | RMBase: m6A_site_240016 | ||
| mod ID: M6ASITE018865 | Click to Show/Hide the Full List | ||
| mod site | chr14:22922494-22922495:- | [4] | |
| Sequence | GTGGAGGTGAACACAGTACTACATGGCTTTGCCGGCTACTT | ||
| Motif Score | 2.078666667 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000553897.5; ENST00000216350.12; ENST00000557758.1; ENST00000476175.6; ENST00000397441.6; ENST00000397440.8; ENST00000553915.5; ENST00000324366.13; ENST00000555454.5; ENST00000454731.6 | ||
| External Link | RMBase: m6A_site_240017 | ||
| mod ID: M6ASITE018866 | Click to Show/Hide the Full List | ||
| mod site | chr14:22922504-22922505:- | [5] | |
| Sequence | GGAATTTCCTGTGGAGGTGAACACAGTACTACATGGCTTTG | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa; Huh7 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000454731.6; ENST00000397441.6; ENST00000553897.5; ENST00000557758.1; ENST00000553915.5; ENST00000324366.13; ENST00000216350.12; ENST00000397440.8; ENST00000555454.5; ENST00000476175.6 | ||
| External Link | RMBase: m6A_site_240018 | ||
| mod ID: M6ASITE018867 | Click to Show/Hide the Full List | ||
| mod site | chr14:22922800-22922801:- | [4] | |
| Sequence | GCCTTATGTGGTACGGCTGCACAACTTCCACCAGCTCTCTG | ||
| Motif Score | 2.830589286 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000557758.1; ENST00000476175.6; ENST00000555454.5; ENST00000553897.5; ENST00000553915.5; ENST00000216350.12; ENST00000397441.6; ENST00000324366.13; ENST00000454731.6; ENST00000397440.8 | ||
| External Link | RMBase: m6A_site_240019 | ||
| mod ID: M6ASITE018868 | Click to Show/Hide the Full List | ||
| mod site | chr14:22923063-22923064:- | [5] | |
| Sequence | CCGAGCCTGTAGGGAGAAGGACCGTGACCCTGAGGTAAAGA | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000397441.6; ENST00000553897.5; ENST00000324366.13; ENST00000555454.5; ENST00000553915.5; ENST00000216350.12; ENST00000454731.6; ENST00000397440.8 | ||
| External Link | RMBase: m6A_site_240020 | ||
| mod ID: M6ASITE018869 | Click to Show/Hide the Full List | ||
| mod site | chr14:22923093-22923094:- | [4] | |
| Sequence | CATCTCTTCCTCCAAGCTGTACAATGAGGTCCGAGCCTGTA | ||
| Motif Score | 2.856142857 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000397440.8; ENST00000324366.13; ENST00000397441.6; ENST00000555454.5; ENST00000216350.12; ENST00000553897.5; ENST00000553915.5; ENST00000454731.6 | ||
| External Link | RMBase: m6A_site_240021 | ||
| mod ID: M6ASITE018870 | Click to Show/Hide the Full List | ||
| mod site | chr14:22923132-22923133:- | [4] | |
| Sequence | TGTGAGCATCCCCGGGGAGTACACTTCCTTTCTGGCTCCCA | ||
| Motif Score | 2.856142857 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000553915.5; ENST00000553897.5; ENST00000397441.6; ENST00000216350.12; ENST00000454731.6; ENST00000555454.5; ENST00000397440.8; ENST00000324366.13 | ||
| External Link | RMBase: m6A_site_240022 | ||
| mod ID: M6ASITE018871 | Click to Show/Hide the Full List | ||
| mod site | chr14:22924057-22924058:- | [4] | |
| Sequence | GCTTCTGGGCTCATTTGCTGACAATGAATTGTCGCCTGAGT | ||
| Motif Score | 2.859755952 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000555454.5; ENST00000553897.5; ENST00000324366.13; ENST00000397440.8; ENST00000553915.5; ENST00000397441.6; ENST00000216350.12 | ||
| External Link | RMBase: m6A_site_240025 | ||
| mod ID: M6ASITE018872 | Click to Show/Hide the Full List | ||
| mod site | chr14:22924093-22924094:- | [5] | |
| Sequence | GGTGGCTCCAGAGAAAGCAGACATCATTGTCAGTGAGCTTC | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000553897.5; ENST00000216350.12; ENST00000397440.8; ENST00000397441.6; ENST00000553915.5; ENST00000324366.13; ENST00000555454.5 | ||
| External Link | RMBase: m6A_site_240026 | ||
| mod ID: M6ASITE018873 | Click to Show/Hide the Full List | ||
| mod site | chr14:22924126-22924127:- | [5] | |
| Sequence | AGTGACCGTAGTCTCATCAGACATGAGGGAATGGGTGGCTC | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa; kidney; hESC-HEK293T | ||
| Seq Type List | m6A-seq; m6A-REF-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000553897.5; ENST00000324366.13; ENST00000216350.12; ENST00000553915.5; ENST00000397441.6; ENST00000555454.5; ENST00000397440.8 | ||
| External Link | RMBase: m6A_site_240027 | ||
| mod ID: M6ASITE018874 | Click to Show/Hide the Full List | ||
| mod site | chr14:22924174-22924175:- | [5] | |
| Sequence | TCTCTGTTTCAGGCTAGAGAACTGGCAGTTTGAAGAATGGG | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa; endometrial | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000324366.13; ENST00000553502.5; ENST00000553915.5; ENST00000555454.5; ENST00000553897.5; ENST00000397441.6; ENST00000397440.8; ENST00000216350.12 | ||
| External Link | RMBase: m6A_site_240028 | ||
| mod ID: M6ASITE018875 | Click to Show/Hide the Full List | ||
| mod site | chr14:22924287-22924288:- | [5] | |
| Sequence | GCTGTATGCTGTGGAGAAAAACCCAAATGCCGTGGTGACGT | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | HeLa; endometrial | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000216350.12; ENST00000553897.5; ENST00000553915.5; ENST00000397441.6; ENST00000553502.5; ENST00000324366.13; ENST00000397440.8 | ||
| External Link | RMBase: m6A_site_240029 | ||
| mod ID: M6ASITE018876 | Click to Show/Hide the Full List | ||
| mod site | chr14:22924361-22924362:- | [5] | |
| Sequence | GTGCTGGGAGCAGGACGGGGACCCCTGGTGAACGCTTCCCT | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000553502.5; ENST00000555530.5; ENST00000553897.5; ENST00000216350.12; ENST00000397441.6; ENST00000553915.5; ENST00000324366.13; ENST00000397440.8 | ||
| External Link | RMBase: m6A_site_240030 | ||
| mod ID: M6ASITE018877 | Click to Show/Hide the Full List | ||
| mod site | chr14:22924514-22924515:- | [5] | |
| Sequence | CATCTATAAATGTCTGCTAGACCGAGTACCAGAAGAGGAGA | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000556043.5; ENST00000555530.5; ENST00000553897.5; ENST00000397441.6; ENST00000553502.5; ENST00000324366.13; ENST00000397440.8; ENST00000216350.12; ENST00000553915.5 | ||
| External Link | RMBase: m6A_site_240031 | ||
| mod ID: M6ASITE018878 | Click to Show/Hide the Full List | ||
| mod site | chr14:22924659-22924660:- | [5] | |
| Sequence | ATATGAAGTGTTTGAAAAGGACCCCATCAAATACTCTCAGT | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000553502.5; ENST00000553915.5; ENST00000553897.5; ENST00000324366.13; ENST00000397441.6; ENST00000553550.5; ENST00000397440.8; ENST00000216350.12; ENST00000555530.5; ENST00000556043.5 | ||
| External Link | RMBase: m6A_site_240032 | ||
| mod ID: M6ASITE018879 | Click to Show/Hide the Full List | ||
| mod site | chr14:22924681-22924682:- | [5] | |
| Sequence | TGGACAATCTGGAATCTCAGACATATGAAGTGTTTGAAAAG | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa; hESC-HEK293T | ||
| Seq Type List | m6A-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000553915.5; ENST00000553502.5; ENST00000216350.12; ENST00000397441.6; ENST00000556043.5; ENST00000553550.5; ENST00000555530.5; ENST00000554716.5; ENST00000397440.8; ENST00000553897.5; ENST00000324366.13 | ||
| External Link | RMBase: m6A_site_240033 | ||
| mod ID: M6ASITE018880 | Click to Show/Hide the Full List | ||
| mod site | chr14:22924698-22924699:- | [5] | |
| Sequence | CATTCATCAGCCACTGATGGACAATCTGGAATCTCAGACAT | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; hESC-HEK293T | ||
| Seq Type List | m6A-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000553915.5; ENST00000553550.5; ENST00000554716.5; ENST00000216350.12; ENST00000397441.6; ENST00000555530.5; ENST00000324366.13; ENST00000397440.8; ENST00000553897.5; ENST00000553502.5; ENST00000556043.5 | ||
| External Link | RMBase: m6A_site_240034 | ||
| mod ID: M6ASITE018881 | Click to Show/Hide the Full List | ||
| mod site | chr14:22924900-22924901:- | [5] | |
| Sequence | CTTTGCCAAGGGCTATGAAGACTATCTGCAGTCCCCGCTTC | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000556043.5; ENST00000553550.5; ENST00000553915.5; ENST00000324366.13; ENST00000553502.5; ENST00000216350.12; ENST00000397440.8; ENST00000553897.5; ENST00000397441.6; ENST00000555530.5; ENST00000554716.5 | ||
| External Link | RMBase: m6A_site_240035 | ||
| mod ID: M6ASITE018882 | Click to Show/Hide the Full List | ||
| mod site | chr14:22924923-22924924:- | [5] | |
| Sequence | CCTCCACCTAATGCCTATGAACTCTTTGCCAAGGGCTATGA | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000555530.5; ENST00000324366.13; ENST00000553502.5; ENST00000216350.12; ENST00000397440.8; ENST00000397441.6; ENST00000554716.5; ENST00000553915.5; ENST00000556043.5; ENST00000553550.5; ENST00000553897.5 | ||
| External Link | RMBase: m6A_site_240036 | ||
| mod ID: M6ASITE018883 | Click to Show/Hide the Full List | ||
| mod site | chr14:22924948-22924949:- | [5] | |
| Sequence | CCTGGAATACTTAAGCCAGAACCGTCCTCCACCTAATGCCT | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000553502.5; ENST00000397441.6; ENST00000216350.12; ENST00000553915.5; ENST00000555530.5; ENST00000397440.8; ENST00000553550.5; ENST00000556043.5; ENST00000553897.5; ENST00000554716.5; ENST00000324366.13 | ||
| External Link | RMBase: m6A_site_240037 | ||
| mod ID: M6ASITE018884 | Click to Show/Hide the Full List | ||
| mod site | chr14:22926371-22926372:- | [5] | |
| Sequence | GCTTTCTCTTAAAGAAGTAAACAGATTTTGCGCAAAGGAGC | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000554910.5; ENST00000556043.5; ENST00000555530.5; ENST00000397440.8; ENST00000553897.5; ENST00000553550.5; ENST00000216350.12; ENST00000553915.5; ENST00000554867.5; ENST00000397441.6; ENST00000553787.5; ENST00000556616.5; ENST00000553641.5; ENST00000324366.13; ENST00000557415.5; ENST00000554716.5 | ||
| External Link | RMBase: m6A_site_240038 | ||
| mod ID: M6ASITE018885 | Click to Show/Hide the Full List | ||
| mod site | chr14:22926395-22926396:- | [5] | |
| Sequence | CTACTTCCCCTCTGTCTCAGACTGGCTTTCTCTTAAAGAAG | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000553550.5; ENST00000554716.5; ENST00000556616.5; ENST00000324366.13; ENST00000553915.5; ENST00000556043.5; ENST00000397440.8; ENST00000557415.5; ENST00000553897.5; ENST00000553641.5; ENST00000397441.6; ENST00000554910.5; ENST00000555530.5; ENST00000554867.5; ENST00000216350.12; ENST00000553787.5 | ||
| External Link | RMBase: m6A_site_240039 | ||
| mod ID: M6ASITE018886 | Click to Show/Hide the Full List | ||
| mod site | chr14:22926538-22926539:- | [5] | |
| Sequence | ACAGGTGGCACAACTTCCGGACTTTGTGTGACTATAGTAAG | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000557415.5; ENST00000553417.1; ENST00000553641.5; ENST00000553787.5; ENST00000554910.5; ENST00000397441.6; ENST00000555530.5; ENST00000324366.13; ENST00000556616.5; ENST00000554867.5; ENST00000397440.8; ENST00000553897.5; ENST00000553550.5; ENST00000553915.5; ENST00000554716.5; ENST00000216350.12 | ||
| External Link | RMBase: m6A_site_240040 | ||
| mod ID: M6ASITE018887 | Click to Show/Hide the Full List | ||
| mod site | chr14:22926549-22926550:- | [4] | |
| Sequence | TATTTGACTGAACAGGTGGCACAACTTCCGGACTTTGTGTG | ||
| Motif Score | 2.830589286 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000324366.13; ENST00000553897.5; ENST00000397441.6; ENST00000555530.5; ENST00000397440.8; ENST00000553641.5; ENST00000554867.5; ENST00000553417.1; ENST00000553787.5; ENST00000553915.5; ENST00000554716.5; ENST00000553550.5; ENST00000556616.5; ENST00000216350.12; ENST00000554910.5; ENST00000557415.5 | ||
| External Link | RMBase: m6A_site_240041 | ||
| mod ID: M6ASITE018888 | Click to Show/Hide the Full List | ||
| mod site | chr14:22926744-22926745:- | [4] | |
| Sequence | TAATTGAGAATGCACCAACTACACACACAGAGGAGTACAGT | ||
| Motif Score | 2.078666667 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000557415.5; ENST00000556616.5; ENST00000553641.5; ENST00000554867.5; ENST00000216350.12; ENST00000554910.5; ENST00000397440.8; ENST00000553787.5; ENST00000324366.13; ENST00000554716.5; ENST00000421938.2; ENST00000553550.5; ENST00000555530.5; ENST00000397441.6; ENST00000553417.1; ENST00000553897.5; ENST00000553915.5 | ||
| External Link | RMBase: m6A_site_240045 | ||
| mod ID: M6ASITE018889 | Click to Show/Hide the Full List | ||
| mod site | chr14:22926747-22926748:- | [8] | |
| Sequence | ATATAATTGAGAATGCACCAACTACACACACAGAGGAGTAC | ||
| Motif Score | 2.595904762 | ||
| Cell/Tissue List | A549 | ||
| Seq Type List | m6A-CLIP/IP | ||
| Transcript ID List | ENST00000555530.5; ENST00000557415.5; ENST00000553787.5; ENST00000421938.2; ENST00000554867.5; ENST00000397441.6; ENST00000324366.13; ENST00000397440.8; ENST00000554716.5; ENST00000554910.5; ENST00000553915.5; ENST00000553550.5; ENST00000553897.5; ENST00000556616.5; ENST00000553641.5; ENST00000216350.12; ENST00000553417.1 | ||
| External Link | RMBase: m6A_site_240046 | ||
| mod ID: M6ASITE018890 | Click to Show/Hide the Full List | ||
| mod site | chr14:22926779-22926780:- | [5] | |
| Sequence | ACCCTTGGTGGCACCAGAGGACCTGAGAGATGATATAATTG | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000324366.13; ENST00000556616.5; ENST00000554867.5; ENST00000216350.12; ENST00000557415.5; ENST00000421938.2; ENST00000554910.5; ENST00000553897.5; ENST00000553417.1; ENST00000553915.5; ENST00000553550.5; ENST00000555530.5; ENST00000397441.6; ENST00000554716.5; ENST00000553641.5; ENST00000553787.5; ENST00000397440.8 | ||
| External Link | RMBase: m6A_site_240047 | ||
| mod ID: M6ASITE018891 | Click to Show/Hide the Full List | ||
| mod site | chr14:22927547-22927548:- | [4] | |
| Sequence | AGTTTTGACCAACCACATCCACACTGGCCATCACTCTTCCA | ||
| Motif Score | 2.053113095 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000553915.5; ENST00000554867.5; ENST00000553641.5; ENST00000557415.5; ENST00000397441.6; ENST00000397440.8; ENST00000421938.2; ENST00000324366.13; ENST00000556032.1; ENST00000553787.5; ENST00000555530.5; ENST00000554910.5; ENST00000553417.1; ENST00000553550.5; ENST00000556616.5; ENST00000216350.12; ENST00000553897.5; ENST00000554716.5 | ||
| External Link | RMBase: m6A_site_240053 | ||
| mod ID: M6ASITE018892 | Click to Show/Hide the Full List | ||
| mod site | chr14:22927583-22927584:- | [4] | |
| Sequence | GCCCCTTAATCAGGAAGATAACACCAACCTGGCCAGAGTTT | ||
| Motif Score | 2.168095238 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000555530.5; ENST00000554716.5; ENST00000554867.5; ENST00000553417.1; ENST00000553641.5; ENST00000216350.12; ENST00000553787.5; ENST00000397441.6; ENST00000421938.2; ENST00000554910.5; ENST00000553897.5; ENST00000557415.5; ENST00000397440.8; ENST00000324366.13; ENST00000556032.1; ENST00000556616.5; ENST00000553550.5; ENST00000553915.5 | ||
| External Link | RMBase: m6A_site_240054 | ||
| mod ID: M6ASITE018893 | Click to Show/Hide the Full List | ||
| mod site | chr14:22927651-22927652:- | [9] | |
| Sequence | TCTCCGTTCCAGGCCATGTTACAGGAGCTGAATTTTGGTGC | ||
| Motif Score | 2.07285119 | ||
| Cell/Tissue List | brain | ||
| Seq Type List | m6A-REF-seq | ||
| Transcript ID List | ENST00000554716.5; ENST00000421938.2; ENST00000553897.5; ENST00000557415.5; ENST00000627278.1; ENST00000554867.5; ENST00000556032.1; ENST00000216350.12; ENST00000324366.13; ENST00000397440.8; ENST00000553641.5; ENST00000555530.5; ENST00000553550.5; ENST00000554910.5; ENST00000553787.5; ENST00000556616.5; ENST00000553915.5; ENST00000397441.6 | ||
| External Link | RMBase: m6A_site_240055 | ||
| mod ID: M6ASITE018894 | Click to Show/Hide the Full List | ||
| mod site | chr14:22928135-22928136:- | [5] | |
| Sequence | AGTGGAGAAGATTCGCAGGAACTCCGAGGCGGTAAGACTGT | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa; peripheral-blood | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000553787.5; ENST00000554867.5; ENST00000397440.8; ENST00000554716.5; ENST00000554910.5; ENST00000553897.5; ENST00000216350.12; ENST00000421938.2; ENST00000555530.5; ENST00000553641.5; ENST00000324366.13; ENST00000627278.1; ENST00000557415.5; ENST00000553550.5; ENST00000553915.5; ENST00000557015.1; ENST00000397441.6; ENST00000556032.1; ENST00000556616.5 | ||
| External Link | RMBase: m6A_site_240056 | ||
| mod ID: M6ASITE018895 | Click to Show/Hide the Full List | ||
| mod site | chr14:22928162-22928163:- | [5] | |
| Sequence | TTCTCCATGGATTCGTCCAGACTCAAAAGTGGAGAAGATTC | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa; peripheral-blood | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000553915.5; ENST00000627278.1; ENST00000557415.5; ENST00000553787.5; ENST00000397441.6; ENST00000556616.5; ENST00000554716.5; ENST00000216350.12; ENST00000324366.13; ENST00000421938.2; ENST00000553641.5; ENST00000397440.8; ENST00000554910.5; ENST00000556032.1; ENST00000553550.5; ENST00000553897.5; ENST00000554867.5; ENST00000557015.1 | ||
| External Link | RMBase: m6A_site_240057 | ||
| mod ID: M6ASITE018896 | Click to Show/Hide the Full List | ||
| mod site | chr14:22928210-22928211:- | [5] | |
| Sequence | TCTTGGGTGGGGGAGTGCAGACTGGAATACGCTAATTGTGG | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa; HepG2; A549; peripheral-blood | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000554910.5; ENST00000397440.8; ENST00000556032.1; ENST00000553915.5; ENST00000556616.5; ENST00000324366.13; ENST00000397441.6; ENST00000554716.5; ENST00000557015.1; ENST00000553897.5; ENST00000553787.5; ENST00000554867.5; ENST00000627278.1; ENST00000553550.5; ENST00000557415.5; ENST00000553641.5; ENST00000421938.2; ENST00000216350.12 | ||
| External Link | RMBase: m6A_site_240058 | ||
| mod ID: M6ASITE018897 | Click to Show/Hide the Full List | ||
| mod site | chr14:22928251-22928252:- | [5] | |
| Sequence | TTGAACAAGTAAAGTGCTGAACCTTATTCAGTATTTGTCTT | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | HeLa; HepG2; A549; peripheral-blood | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000216350.12; ENST00000553915.5; ENST00000553641.5; ENST00000397441.6; ENST00000553550.5; ENST00000554910.5; ENST00000554867.5; ENST00000556616.5; ENST00000553897.5; ENST00000627278.1; ENST00000397440.8; ENST00000324366.13; ENST00000557015.1; ENST00000554716.5; ENST00000556032.1; ENST00000557415.5; ENST00000553787.5; ENST00000421938.2 | ||
| External Link | RMBase: m6A_site_240059 | ||
| mod ID: M6ASITE018898 | Click to Show/Hide the Full List | ||
| mod site | chr14:22928267-22928268:- | [5] | |
| Sequence | TTCCCAAAACTAAAAATTGAACAAGTAAAGTGCTGAACCTT | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa; HepG2; A549; peripheral-blood | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000553897.5; ENST00000216350.12; ENST00000556032.1; ENST00000553915.5; ENST00000324366.13; ENST00000557015.1; ENST00000554910.5; ENST00000557415.5; ENST00000556616.5; ENST00000397440.8; ENST00000556426.1; ENST00000553787.5; ENST00000421938.2; ENST00000553550.5; ENST00000554867.5; ENST00000554716.5; ENST00000397441.6; ENST00000553641.5; ENST00000627278.1 | ||
| External Link | RMBase: m6A_site_240060 | ||
| mod ID: M6ASITE018899 | Click to Show/Hide the Full List | ||
| mod site | chr14:22928279-22928280:- | [5] | |
| Sequence | TTCTACTCTTGATTCCCAAAACTAAAAATTGAACAAGTAAA | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa; HepG2; A549; peripheral-blood | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000324366.13; ENST00000554716.5; ENST00000216350.12; ENST00000556032.1; ENST00000557015.1; ENST00000553915.5; ENST00000554910.5; ENST00000556426.1; ENST00000397440.8; ENST00000421938.2; ENST00000627278.1; ENST00000554867.5; ENST00000397441.6; ENST00000553641.5; ENST00000557415.5; ENST00000553787.5; ENST00000553550.5; ENST00000553897.5; ENST00000556616.5 | ||
| External Link | RMBase: m6A_site_240061 | ||
| mod ID: M6ASITE018900 | Click to Show/Hide the Full List | ||
| mod site | chr14:22928489-22928490:- | [5] | |
| Sequence | GCTGTCAGGAAGGGGTAGGGACCATCCCATATGCCCCTTAA | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; HepG2; A549; Jurkat; CD4T; peripheral-blood; GSC-11; iSLK; endometrial; HEC-1-A; GSCs; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000553897.5; ENST00000556426.1; ENST00000557015.1; ENST00000397440.8; ENST00000553550.5; ENST00000554867.5; ENST00000556616.5; ENST00000216350.12; ENST00000553641.5; ENST00000553787.5; ENST00000397441.6; ENST00000557415.5; ENST00000553915.5; ENST00000554716.5; ENST00000324366.13; ENST00000421938.2; ENST00000554910.5; ENST00000627278.1; ENST00000556032.1 | ||
| External Link | RMBase: m6A_site_240062 | ||
| mod ID: M6ASITE018901 | Click to Show/Hide the Full List | ||
| mod site | chr14:22928516-22928517:- | [5] | |
| Sequence | CGGTCCCCAGACACGATCAGACCTACTGCTGTCAGGAAGGG | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa; HepG2; A549; Jurkat; CD4T; peripheral-blood; GSC-11; iSLK; MSC; endometrial; HEC-1-A; GSCs; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000554910.5; ENST00000397440.8; ENST00000556032.1; ENST00000553550.5; ENST00000421938.2; ENST00000553897.5; ENST00000554716.5; ENST00000324366.13; ENST00000397441.6; ENST00000627278.1; ENST00000557415.5; ENST00000553641.5; ENST00000553787.5; ENST00000553915.5; ENST00000554867.5; ENST00000556616.5; ENST00000556426.1; ENST00000216350.12; ENST00000557015.1 | ||
| External Link | RMBase: m6A_site_240063 | ||
| mod ID: M6ASITE018902 | Click to Show/Hide the Full List | ||
| mod site | chr14:22928526-22928527:- | [5] | |
| Sequence | AGAATCGGCCCGGTCCCCAGACACGATCAGACCTACTGCTG | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa; HepG2; A549; hESC-HEK293T; Jurkat; CD4T; peripheral-blood; GSC-11; iSLK; MSC; endometrial; HEC-1-A; GSCs; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000216350.12; ENST00000553915.5; ENST00000627278.1; ENST00000556426.1; ENST00000397441.6; ENST00000556616.5; ENST00000421938.2; ENST00000557015.1; ENST00000553550.5; ENST00000397440.8; ENST00000553641.5; ENST00000556032.1; ENST00000324366.13; ENST00000554910.5; ENST00000553897.5; ENST00000557415.5; ENST00000553787.5; ENST00000554867.5; ENST00000554716.5 | ||
| External Link | RMBase: m6A_site_240064 | ||
| mod ID: M6ASITE018903 | Click to Show/Hide the Full List | ||
| mod site | chr14:22928554-22928555:- | [5] | |
| Sequence | AAGAGGGAGTTCATTCAGGAACCTGCTAAGAATCGGCCCGG | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | HeLa; HepG2; A549; Jurkat; CD4T; peripheral-blood; GSC-11; iSLK; MSC; endometrial; HEC-1-A; GSCs; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000627278.1; ENST00000553897.5; ENST00000397441.6; ENST00000557415.5; ENST00000421938.2; ENST00000557015.1; ENST00000556032.1; ENST00000553787.5; ENST00000324366.13; ENST00000397440.8; ENST00000556616.5; ENST00000553915.5; ENST00000216350.12; ENST00000553550.5; ENST00000554910.5; ENST00000554716.5; ENST00000554867.5; ENST00000553641.5; ENST00000556426.1 | ||
| External Link | RMBase: m6A_site_240065 | ||
| mod ID: M6ASITE018904 | Click to Show/Hide the Full List | ||
| mod site | chr14:22929189-22929190:- | [5] | |
| Sequence | TCGGGGACGGAGAAGGGCAGACTAGTCATCCCGGAGAAGCA | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa; HepG2; A549; H1B; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293A-TOA; iSLK; MSC; TREX; TIME; endometrial; HEC-1-A; GSCs; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000627278.1; ENST00000216350.12; ENST00000554867.5; ENST00000554716.5; ENST00000556426.1; ENST00000553641.5; ENST00000557015.1; ENST00000556616.5; ENST00000397440.8; ENST00000324366.13; ENST00000553787.5; ENST00000421938.2; ENST00000397441.6; ENST00000553550.5; ENST00000557415.5; ENST00000556032.1; ENST00000553915.5; ENST00000553897.5 | ||
| External Link | RMBase: m6A_site_240066 | ||
| mod ID: M6ASITE018905 | Click to Show/Hide the Full List | ||
| mod site | chr14:22929211-22929212:- | [5] | |
| Sequence | GAGCGGAATGCGGGGTCCGAACTCGGGGACGGAGAAGGGCA | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa; HepG2; A549; HEK293T; H1B; H1A; fibroblasts; MM6; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293A-TOA; iSLK; MSC; TREX; TIME; endometrial; HEC-1-A; GSCs; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000554716.5; ENST00000553641.5; ENST00000397440.8; ENST00000554867.5; ENST00000627278.1; ENST00000397441.6; ENST00000553897.5; ENST00000556616.5; ENST00000553915.5; ENST00000553787.5; ENST00000557015.1; ENST00000557415.5; ENST00000324366.13; ENST00000556032.1; ENST00000556426.1; ENST00000421938.2; ENST00000553550.5; ENST00000216350.12 | ||
| External Link | RMBase: m6A_site_240067 | ||
| mod ID: M6ASITE018906 | Click to Show/Hide the Full List | ||
| mod site | chr14:22929233-22929234:- | [7] | |
| Sequence | GGGGTGAGGGCCGGACCTCCACGAGCGGAATGCGGGGTCCG | ||
| Motif Score | 2.027047619 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000553787.5; ENST00000553641.5; ENST00000627278.1; ENST00000397440.8; ENST00000556616.5; ENST00000556032.1; ENST00000557015.1; ENST00000397441.6; ENST00000553915.5; ENST00000554716.5; ENST00000554867.5; ENST00000556426.1; ENST00000421938.2; ENST00000553550.5; ENST00000553897.5; ENST00000216350.12; ENST00000557415.5; ENST00000324366.13 | ||
| External Link | RMBase: m6A_site_240068 | ||
| mod ID: M6ASITE018907 | Click to Show/Hide the Full List | ||
| mod site | chr14:22929239-22929240:- | [5] | |
| Sequence | CAAGCAGGGGTGAGGGCCGGACCTCCACGAGCGGAATGCGG | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; HepG2; A549; HEK293T; H1B; H1A; hESCs; fibroblasts; GM12878; MM6; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; TREX; endometrial; HEC-1-A; GSCs; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000553550.5; ENST00000556426.1; ENST00000553897.5; ENST00000553787.5; ENST00000397440.8; ENST00000554867.5; ENST00000556616.5; ENST00000556032.1; ENST00000554716.5; ENST00000553641.5; ENST00000553915.5; ENST00000397441.6; ENST00000216350.12; ENST00000557415.5; ENST00000324366.13; ENST00000557015.1; ENST00000627278.1; ENST00000421938.2 | ||
| External Link | RMBase: m6A_site_240069 | ||
| mod ID: M6ASITE018908 | Click to Show/Hide the Full List | ||
| mod site | chr14:22929278-22929279:- | [4] | |
| Sequence | TTGCGTCCCCGAAATAGCTGACACACTAGGGGCTGTGGCCA | ||
| Motif Score | 2.859755952 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000553787.5; ENST00000216350.12; ENST00000421938.2; ENST00000553897.5; ENST00000556032.1; ENST00000627278.1; ENST00000324366.13; ENST00000397441.6; ENST00000553641.5; ENST00000557415.5; ENST00000553915.5; ENST00000554716.5; ENST00000556616.5; ENST00000556426.1; ENST00000397440.8; ENST00000554867.5; ENST00000557015.1; ENST00000553550.5 | ||
| External Link | RMBase: m6A_site_240070 | ||
| mod ID: M6ASITE018909 | Click to Show/Hide the Full List | ||
| mod site | chr14:22929305-22929306:- | [5] | |
| Sequence | CCGCGTGTCCAGCGGGAGGGACCTGAATTGCGTCCCCGAAA | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; HepG2; A549; HEK293T; H1A; H1B; hESCs; fibroblasts; GM12878; MM6; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; TREX; endometrial; HEC-1-A; GSCs; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000557415.5; ENST00000556032.1; ENST00000553897.5; ENST00000627278.1; ENST00000216350.12; ENST00000554716.5; ENST00000324366.13; ENST00000554867.5; ENST00000557015.1; ENST00000397441.6; ENST00000553915.5; ENST00000553550.5; ENST00000556426.1; ENST00000556616.5; ENST00000397440.8; ENST00000421938.2; ENST00000553641.5; ENST00000553787.5 | ||
| External Link | RMBase: m6A_site_240071 | ||
| mod ID: M6ASITE018910 | Click to Show/Hide the Full List | ||
| mod site | chr14:22929377-22929378:- | [5] | |
| Sequence | ATCAAGGAATCCCGGCGTGGACAGCGCGAGGAGAAAGATGG | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; hESC-HEK293T; HepG2; H1A; H1B; hESCs; fibroblasts; A549; GM12878; HEK293A-TOA; iSLK; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq; MAZTER-seq; m6A-CLIP/IP | ||
| Transcript ID List | ENST00000553550.5; ENST00000397441.6; ENST00000557415.5; ENST00000216350.12; ENST00000557015.1; ENST00000553897.5 | ||
| External Link | RMBase: m6A_site_240072 | ||
References