m6A Target Gene Information
General Information of the m6A Target Gene (ID: M6ATAR00228)
Full List of m6A Methylation Regulator of This Target Gene and Corresponding Disease/Drug Response(s)
DGCR8
can be regulated by the following regulator(s), and cause disease/drug response(s). You can browse detail information of regulator(s) or disease/drug response(s).
Browse Regulator
Browse Disease
Methyltransferase-like 3 (METTL3) [WRITER]
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | Liver | Mus musculus |
|
Treatment: Mettl3 knockout liver
Control: Wild type liver cells
|
GSE198513 | |
| Regulation |
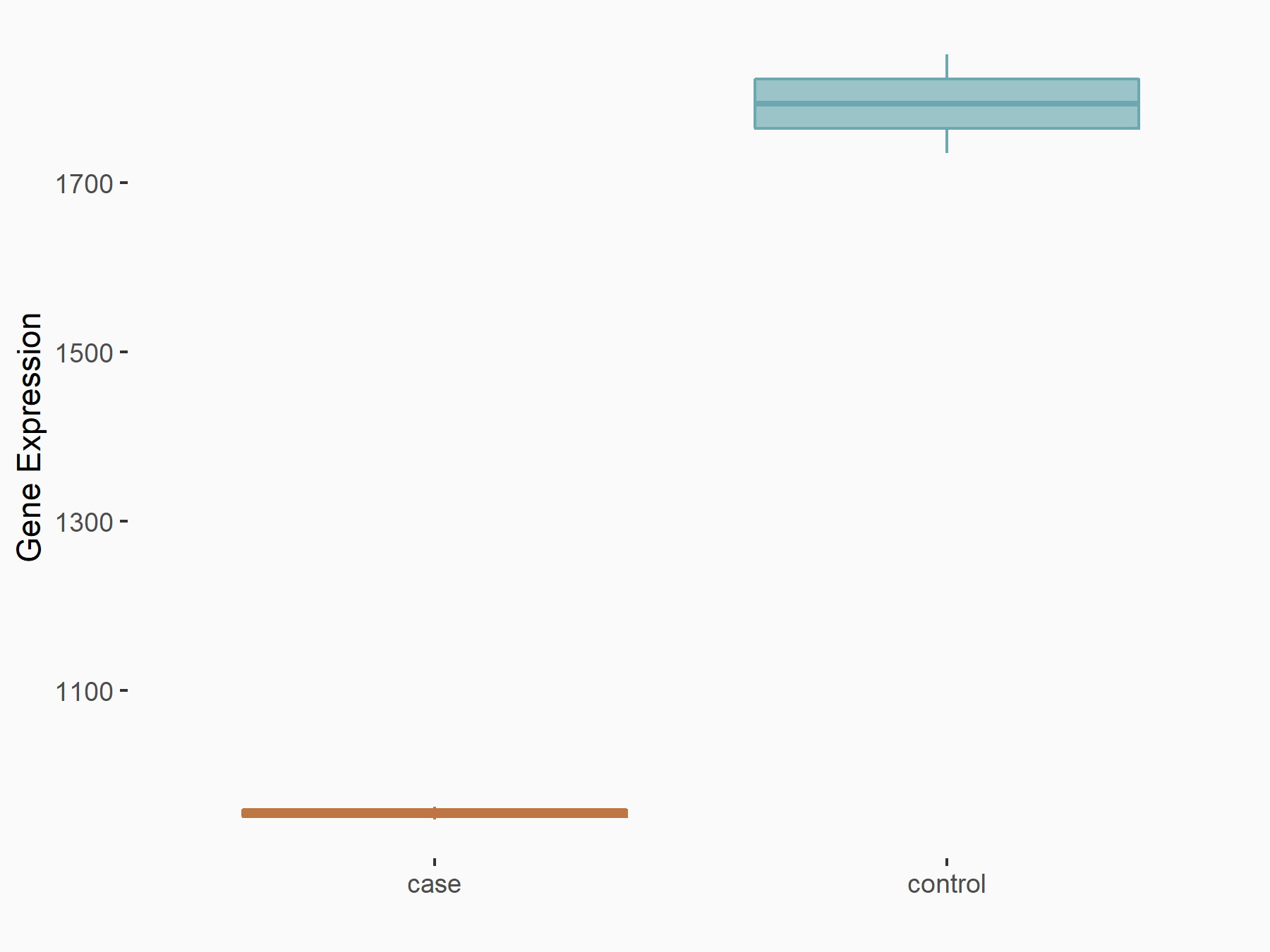 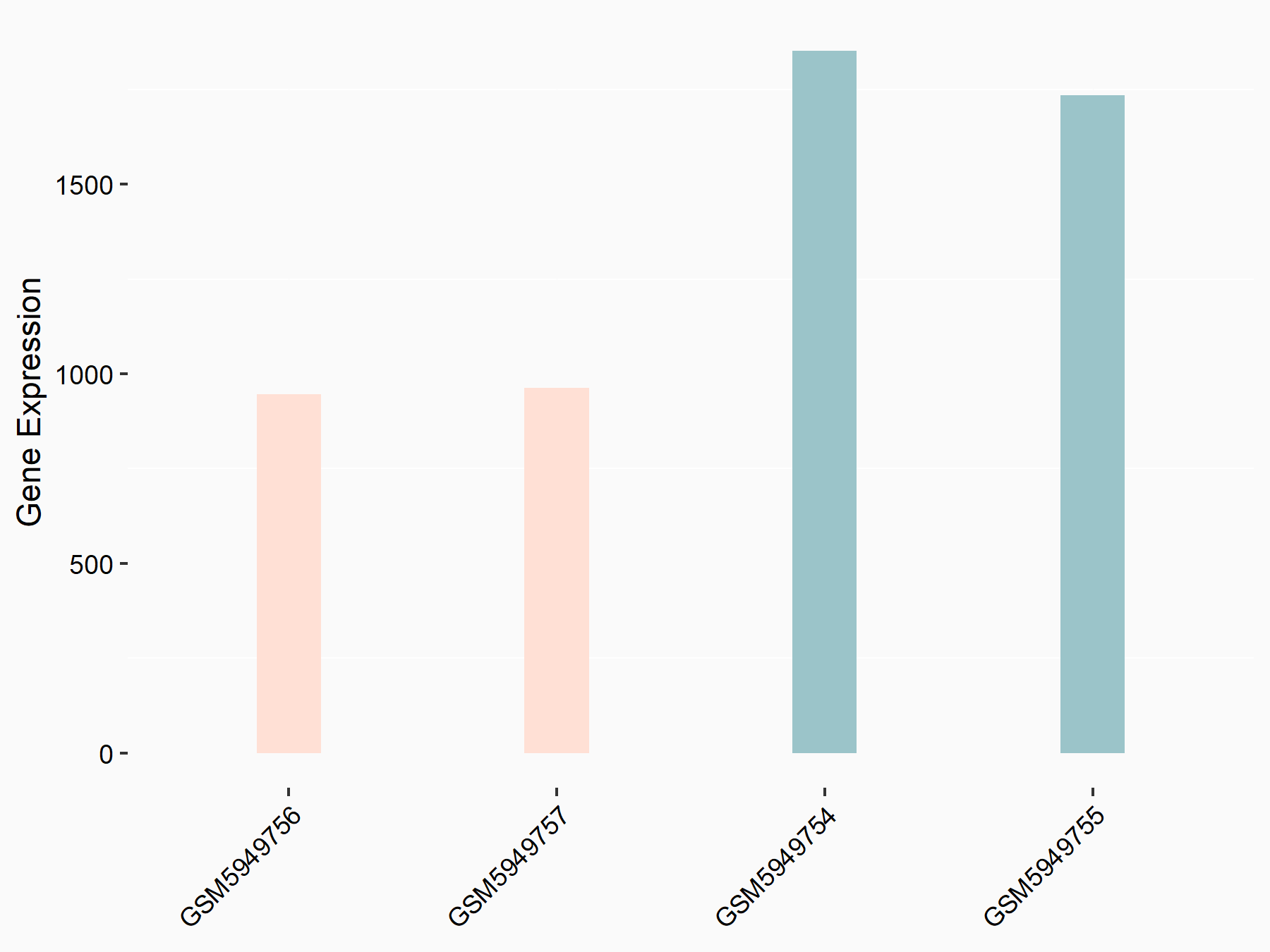 |
logFC: -9.09E-01 p-value: 2.99E-21 |
| More Results | Click to View More RNA-seq Results | |
| In total 4 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [1] | |||
| Response Summary | The m(6)A mark acts as a key post-transcriptional modification that promotes the initiation of miRNA biogenesis. METTL3 depletion reduced the binding of Microprocessor complex subunit DGCR8 (DGCR8) to pri-miRNAs and resulted in the global reduction of mature miRNAs and concomitant accumulation of unprocessed pri-miRNAs. | |||
| Target Regulation | Up regulation | |||
| Cell Process | miRNA maturation | |||
| MicroRNAs in cancer (hsa05206) | ||||
| In-vitro Model | HEK293T | Normal | Homo sapiens | CVCL_0063 |
| HeLa | Endocervical adenocarcinoma | Homo sapiens | CVCL_0030 | |
| HUVEC-C | Normal | Homo sapiens | CVCL_2959 | |
| MDA-MB-231 | Breast adenocarcinoma | Homo sapiens | CVCL_0062 | |
| Experiment 2 Reporting the m6A Methylation Regulator of This Target Gene | [2] | |||
| Response Summary | METTL3 could interact with Microprocessor complex subunit DGCR8 (DGCR8) protein and positively modulate pri-miR-320b maturation process in an N6-methyladenosine (m6A)-dependent manner. Therefore, our findings uncover a VEGF-C-independent mechanism of exosomal and intracellular miR-320b-mediated LN metastasis and identify miR-320b as a novel predictive marker and therapeutic target for LN metastasis in ESCC. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Esophageal squamous cell carcinoma | ICD-11: 2B70.1 | ||
| Pathway Response | PI3K-Akt signaling pathway | hsa04151 | ||
| Cell Process | Cell proliferation | |||
| Cell migration | ||||
| Cell invasion | ||||
| Epithelial-mesenchymal transition | ||||
| In-vitro Model | TE-1 | Esophageal squamous cell carcinoma | Homo sapiens | CVCL_1759 |
| KYSE-30 | Esophageal squamous cell carcinoma | Homo sapiens | CVCL_1351 | |
| KYSE-150 | Esophageal squamous cell carcinoma | Homo sapiens | CVCL_1348 | |
| HET-1A | Normal | Homo sapiens | CVCL_3702 | |
| CVCL_E307 | Esophageal squamous cell carcinoma | Homo sapiens | CVCL_E307 | |
| Eca-109 | Esophageal squamous cell carcinoma | Homo sapiens | CVCL_6898 | |
| In-vivo Model | Luciferase-labeled KYSE150 cells (5 × 106) were inoculated into the footpads of BALB/c nude mice (4-5 weeks old, 18-20 g) to establish the popliteal lymphatic metastasis model. | |||
| Experiment 3 Reporting the m6A Methylation Regulator of This Target Gene | [3] | |||
| Response Summary | METTL3 promoted cell proliferation, migration, invasion and tumorigenesis in PCa. METTL3 upregulating the level of m6A, and interacted with Microprocessor complex subunit DGCR8 (DGCR8) to recognize the m6A modification of pre-miR-182 to regulate its splicing and maturation and promote the high expression of miRNA. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Prostate cancer | ICD-11: 2C82 | ||
| In-vitro Model | WPMY-1 | Normal | Homo sapiens | CVCL_3814 |
| VCaP | Prostate carcinoma | Homo sapiens | CVCL_2235 | |
| PC-3 | Prostate carcinoma | Homo sapiens | CVCL_0035 | |
| LNCaP | Prostate carcinoma | Homo sapiens | CVCL_0395 | |
| DU145 | Prostate carcinoma | Homo sapiens | CVCL_0105 | |
| Experiment 4 Reporting the m6A Methylation Regulator of This Target Gene | [4] | |||
| Response Summary | Interleukin 1-beta (IL-1-beta) is an important inducer of cartilage degeneration that can induce an inflammatory cascade reaction in chondrocytes and inhibit the normal biological function of cells. METTL3 could regulate miR-126-5p maturation, we first confirmed that METTL3 can bind the key protein underlying pri-miRNA processing, Microprocessor complex subunit DGCR8 (DGCR8). Additionally, when METTL3 expression was inhibited, the miR-126-5p maturation process was blocked. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Chondropathies | ICD-11: FB82 | ||
| Cell Process | RNA mature | |||
| In-vitro Model | Cartilage cells (From the cartilage tissue samples from patients) | |||
Methyltransferase-like 14 (METTL14) [WRITER]
| In total 1 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [6] | |||
| Response Summary | METTL14 interacts with the microprocessor protein Microprocessor complex subunit DGCR8 (DGCR8) and positively modulates the primary microRNA 126 process in an m6 A-dependent manner. microRNA 126 inhibits the repressing effect of METTL14 in Hepatocellular carcinoma metastasis. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Hepatocellular carcinoma | ICD-11: 2C12.02 | ||
| Cell Process | Tumor metastasis | |||
| In-vitro Model | HCC-1664 cell line (Primary HCC cells) | |||
| Hep 3B2.1-7 | Childhood hepatocellular carcinoma | Homo sapiens | CVCL_0326 | |
| Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 | |
| SMMC-7721 | Endocervical adenocarcinoma | Homo sapiens | CVCL_0534 | |
| SMMC-7721 | Endocervical adenocarcinoma | Homo sapiens | CVCL_0534 | |
| In-vivo Model | Male athymic BALB/c nude mice (5 weeks old) were used. Subcutaneous tumor growth assays, liver metastasis model, and tail vein injection model were performed as described.Metastases were detected using the IVIS@Lumina II system (Caliper Life Sciences, Hopkinton, MA) 10 minutes after intraperitoneal injection of 4.0 mg luciferin (Gold Biotech) in 50 uL of saline. | |||
Esophageal cancer [ICD-11: 2B70]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [2] | |||
| Response Summary | METTL3 could interact with Microprocessor complex subunit DGCR8 (DGCR8) protein and positively modulate pri-miR-320b maturation process in an N6-methyladenosine (m6A)-dependent manner. Therefore, our findings uncover a VEGF-C-independent mechanism of exosomal and intracellular miR-320b-mediated LN metastasis and identify miR-320b as a novel predictive marker and therapeutic target for LN metastasis in ESCC. | |||
| Responsed Disease | Esophageal squamous cell carcinoma [ICD-11: 2B70.1] | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Target Regulation | Up regulation | |||
| Pathway Response | PI3K-Akt signaling pathway | hsa04151 | ||
| Cell Process | Cell proliferation | |||
| Cell migration | ||||
| Cell invasion | ||||
| Epithelial-mesenchymal transition | ||||
| In-vitro Model | TE-1 | Esophageal squamous cell carcinoma | Homo sapiens | CVCL_1759 |
| KYSE-30 | Esophageal squamous cell carcinoma | Homo sapiens | CVCL_1351 | |
| KYSE-150 | Esophageal squamous cell carcinoma | Homo sapiens | CVCL_1348 | |
| HET-1A | Normal | Homo sapiens | CVCL_3702 | |
| CVCL_E307 | Esophageal squamous cell carcinoma | Homo sapiens | CVCL_E307 | |
| Eca-109 | Esophageal squamous cell carcinoma | Homo sapiens | CVCL_6898 | |
| In-vivo Model | Luciferase-labeled KYSE150 cells (5 × 106) were inoculated into the footpads of BALB/c nude mice (4-5 weeks old, 18-20 g) to establish the popliteal lymphatic metastasis model. | |||
Liver cancer [ICD-11: 2C12]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [6] | |||
| Response Summary | METTL14 interacts with the microprocessor protein Microprocessor complex subunit DGCR8 (DGCR8) and positively modulates the primary microRNA 126 process in an m6 A-dependent manner. microRNA 126 inhibits the repressing effect of METTL14 in Hepatocellular carcinoma metastasis. | |||
| Responsed Disease | Hepatocellular carcinoma [ICD-11: 2C12.02] | |||
| Target Regulator | Methyltransferase-like 14 (METTL14) | WRITER | ||
| Target Regulation | Up regulation | |||
| Cell Process | Tumor metastasis | |||
| In-vitro Model | HCC-1664 cell line (Primary HCC cells) | |||
| Hep 3B2.1-7 | Childhood hepatocellular carcinoma | Homo sapiens | CVCL_0326 | |
| Hep-G2 | Hepatoblastoma | Homo sapiens | CVCL_0027 | |
| SMMC-7721 | Endocervical adenocarcinoma | Homo sapiens | CVCL_0534 | |
| SMMC-7721 | Endocervical adenocarcinoma | Homo sapiens | CVCL_0534 | |
| In-vivo Model | Male athymic BALB/c nude mice (5 weeks old) were used. Subcutaneous tumor growth assays, liver metastasis model, and tail vein injection model were performed as described.Metastases were detected using the IVIS@Lumina II system (Caliper Life Sciences, Hopkinton, MA) 10 minutes after intraperitoneal injection of 4.0 mg luciferin (Gold Biotech) in 50 uL of saline. | |||
Prostate cancer [ICD-11: 2C82]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [3] | |||
| Response Summary | METTL3 promoted cell proliferation, migration, invasion and tumorigenesis in PCa. METTL3 upregulating the level of m6A, and interacted with Microprocessor complex subunit DGCR8 (DGCR8) to recognize the m6A modification of pre-miR-182 to regulate its splicing and maturation and promote the high expression of miRNA. | |||
| Responsed Disease | Prostate cancer [ICD-11: 2C82] | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Target Regulation | Up regulation | |||
| In-vitro Model | WPMY-1 | Normal | Homo sapiens | CVCL_3814 |
| VCaP | Prostate carcinoma | Homo sapiens | CVCL_2235 | |
| PC-3 | Prostate carcinoma | Homo sapiens | CVCL_0035 | |
| LNCaP | Prostate carcinoma | Homo sapiens | CVCL_0395 | |
| DU145 | Prostate carcinoma | Homo sapiens | CVCL_0105 | |
Chondropathies [ICD-11: FB82]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [4] | |||
| Response Summary | Interleukin 1-beta (IL-1-beta) is an important inducer of cartilage degeneration that can induce an inflammatory cascade reaction in chondrocytes and inhibit the normal biological function of cells. METTL3 could regulate miR-126-5p maturation, we first confirmed that METTL3 can bind the key protein underlying pri-miRNA processing, Microprocessor complex subunit DGCR8 (DGCR8). Additionally, when METTL3 expression was inhibited, the miR-126-5p maturation process was blocked. | |||
| Responsed Disease | Chondropathies [ICD-11: FB82] | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Target Regulation | Up regulation | |||
| Cell Process | RNA mature | |||
| In-vitro Model | Cartilage cells (From the cartilage tissue samples from patients) | |||
Full List of Crosstalk(s) between m6A Modification and Epigenetic Regulation Related to This Regulator
Histone modification
m6A Regulator: Methyltransferase-like 3 (METTL3)
| In total 1 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT03374 | ||
| Regulated Target | Histone H3 lysine 18 lactylation (H3K18la) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Esophageal Squamous Cell Carcinoma | |
RNA Modification Sequencing Data Associated with the Target (ID: M6ATAR00228)
| In total 1 m6A sequence/site(s) in this target gene | |||
| mod ID: AC4SITE000057 | Click to Show/Hide the Full List | ||
| mod site | chr22:20107303-20107304:+ | [8] | |
| Sequence | AGTTGGAAAGCAGTTAGCCTCACAGAAGATCCTTCAGCTGC | ||
| Cell/Tissue List | H1 | ||
| Seq Type List | ac4C-seq | ||
| Transcript ID List | ENST00000495826.5; ENST00000485802.1; ENST00000383024.6; ENST00000495351.1; ENST00000498171.5; ENST00000407755.1; ENST00000351989.8 | ||
| External Link | RMBase: ac4C_site_1264 | ||
Adenosine-to-Inosine editing (A-to-I)
| In total 2 m6A sequence/site(s) in this target gene | |||
| mod ID: A2ISITE011030 | Click to Show/Hide the Full List | ||
| mod site | chr22:20085806-20085807:+ | [9] | |
| Sequence | TTCTAAAAGCTGTCTACATTAATGAAAAGAGCAATGTGGCC | ||
| Transcript ID List | ENST00000457069.1; ENST00000580330.1; MIMAT0017998; ENST00000383024.6; ENST00000351989.8 | ||
| External Link | RMBase: RNA-editing_site_90842 | ||
| mod ID: A2ISITE011031 | Click to Show/Hide the Full List | ||
| mod site | chr22:20101364-20101365:+ | [9] | |
| Sequence | GGCGGGCAGATTACGAGGTAAGGACATCGAGACCATCCTGG | ||
| Transcript ID List | ENST00000407755.1; rmsk_5248496; ENST00000491892.1; ENST00000495826.5; ENST00000498171.5; ENST00000383024.6; ENST00000351989.8 | ||
| External Link | RMBase: RNA-editing_site_90843 | ||
5-methylcytidine (m5C)
N6-methyladenosine (m6A)
| In total 76 m6A sequence/site(s) in this target gene | |||
| mod ID: M6ASITE056414 | Click to Show/Hide the Full List | ||
| mod site | chr22:20080289-20080290:+ | [10] | |
| Sequence | CCGGCGACCGGAGAGCCTGGACAGGCTTTCCAGATGGCTGC | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000383024.6; ENST00000351989.8 | ||
| External Link | RMBase: m6A_site_555351 | ||
| mod ID: M6ASITE056415 | Click to Show/Hide the Full List | ||
| mod site | chr22:20085882-20085883:+ | [10] | |
| Sequence | ACGCGCCCGGGTCTCAGCGGACTTGTGCATGTTAGCTGTGT | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa; HEK293T; A549; HepG2; hNPCs; hESCs; fibroblasts; GM12878; CD8T; MM6; HEK293A-TOA; iSLK; MSC; TIME; TREX | ||
| Seq Type List | m6A-seq; MeRIP-seq; m6A-CLIP/IP | ||
| Transcript ID List | ENST00000495826.5; ENST00000457069.1; ENST00000407755.1; ENST00000351989.8; ENST00000383024.6 | ||
| External Link | RMBase: m6A_site_555352 | ||
| mod ID: M6ASITE056416 | Click to Show/Hide the Full List | ||
| mod site | chr22:20085926-20085927:+ | [10] | |
| Sequence | TTTATGTGAGGGCTTGTAAAACTCTGGTCTTGTAAACTAGT | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa; HEK293T; A549; HepG2; U2OS; hESCs; fibroblasts; GM12878; H1299; MM6; HEK293A-TOA; iSLK; MSC; TIME; TREX; AML | ||
| Seq Type List | m6A-seq; MeRIP-seq; m6A-CLIP/IP; miCLIP | ||
| Transcript ID List | ENST00000407755.1; ENST00000383024.6; ENST00000495826.5; ENST00000457069.1; ENST00000351989.8 | ||
| External Link | RMBase: m6A_site_555353 | ||
| mod ID: M6ASITE056417 | Click to Show/Hide the Full List | ||
| mod site | chr22:20085941-20085942:+ | [10] | |
| Sequence | GTAAAACTCTGGTCTTGTAAACTAGTCTTAAGCGCTTTTAA | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa; HEK293T; A549; HepG2; U2OS; H1B; hESCs; fibroblasts; GM12878; H1299; MM6; Huh7; HEK293A-TOA; iSLK; MSC; TIME; TREX | ||
| Seq Type List | m6A-seq; MeRIP-seq; DART-seq | ||
| Transcript ID List | ENST00000351989.8; ENST00000457069.1; ENST00000383024.6; ENST00000495826.5; ENST00000407755.1 | ||
| External Link | RMBase: m6A_site_555354 | ||
| mod ID: M6ASITE056418 | Click to Show/Hide the Full List | ||
| mod site | chr22:20085969-20085970:+ | [10] | |
| Sequence | TAAGCGCTTTTAATATGGAGACAGATGAGAGCCCCTCTCCG | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa; HEK293T; A549; HepG2; U2OS; H1A; H1B; hESCs; fibroblasts; GM12878; H1299; MM6; Huh7; HEK293A-TOA; iSLK; MSC; TIME; TREX | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000383024.6; ENST00000457069.1; ENST00000351989.8; ENST00000495826.5; ENST00000407755.1 | ||
| External Link | RMBase: m6A_site_555355 | ||
| mod ID: M6ASITE056419 | Click to Show/Hide the Full List | ||
| mod site | chr22:20086133-20086134:+ | [10] | |
| Sequence | GTTGGCTCTGGTGGTGATGGACAGTCCGAACTCCCTGCTGA | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; A549; hESC-HEK293T; U2OS; H1A; H1B; hESCs; fibroblasts; GM12878; MM6; Huh7; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; TREX; endometrial; NB4; AML | ||
| Seq Type List | m6A-seq; MeRIP-seq; MAZTER-seq; m6A-CLIP/IP; miCLIP | ||
| Transcript ID List | ENST00000457069.1; ENST00000408439.3; ENST00000383024.6; ENST00000351989.8; ENST00000495826.5; ENST00000407755.1 | ||
| External Link | RMBase: m6A_site_555356 | ||
| mod ID: M6ASITE056420 | Click to Show/Hide the Full List | ||
| mod site | chr22:20086142-20086143:+ | [10] | |
| Sequence | GGTGGTGATGGACAGTCCGAACTCCCTGCTGAGGACCCCTT | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; A549; U2OS; H1A; H1B; hESCs; fibroblasts; GM12878; MM6; Huh7; CD4T; peripheral-blood; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; TREX; endometrial; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000407755.1; ENST00000351989.8; ENST00000457069.1; ENST00000495826.5; ENST00000383024.6 | ||
| External Link | RMBase: m6A_site_555357 | ||
| mod ID: M6ASITE056421 | Click to Show/Hide the Full List | ||
| mod site | chr22:20086156-20086157:+ | [10] | |
| Sequence | GTCCGAACTCCCTGCTGAGGACCCCTTCAACTTCTACGGAG | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; A549; U2OS; H1A; H1B; hESCs; fibroblasts; GM12878; MM6; Huh7; CD4T; peripheral-blood; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; TREX; endometrial; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000407755.1; ENST00000495826.5; ENST00000383024.6; ENST00000457069.1; ENST00000351989.8 | ||
| External Link | RMBase: m6A_site_555358 | ||
| mod ID: M6ASITE056422 | Click to Show/Hide the Full List | ||
| mod site | chr22:20086225-20086226:+ | [10] | |
| Sequence | TAAGGGCCGCCTCCTCATAGACCCGAACTGTAGTGGCCACA | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; A549; U2OS; H1A; H1B; hESCs; fibroblasts; GM12878; LCLs; H1299; MM6; Huh7; peripheral-blood; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; TREX; endometrial; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000383024.6; ENST00000457069.1; ENST00000351989.8; ENST00000495826.5; ENST00000407755.1 | ||
| External Link | RMBase: m6A_site_555359 | ||
| mod ID: M6ASITE056424 | Click to Show/Hide the Full List | ||
| mod site | chr22:20086231-20086232:+ | [10] | |
| Sequence | CCGCCTCCTCATAGACCCGAACTGTAGTGGCCACAGCCCGC | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; A549; U2OS; H1A; H1B; hESCs; fibroblasts; GM12878; LCLs; H1299; MM6; Huh7; peripheral-blood; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; TREX; endometrial; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000407755.1; ENST00000351989.8; ENST00000383024.6; ENST00000457069.1; ENST00000495826.5 | ||
| External Link | RMBase: m6A_site_555360 | ||
| mod ID: M6ASITE056425 | Click to Show/Hide the Full List | ||
| mod site | chr22:20086357-20086358:+ | [10] | |
| Sequence | CGAGAGCTGCAGGAGTAAGGACAGGAAGGTGCTGTACACAG | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; HEK293T; A549; HepG2; U2OS; hESCs; GM12878; MM6; peripheral-blood; HEK293A-TOA; iSLK; MSC; TIME; TREX | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000407755.1; ENST00000351989.8; ENST00000383024.6; ENST00000495826.5 | ||
| External Link | RMBase: m6A_site_555361 | ||
| mod ID: M6ASITE056426 | Click to Show/Hide the Full List | ||
| mod site | chr22:20086471-20086472:+ | [11] | |
| Sequence | CTTTGGCGGGAGTGTTGGTGACGGGGTAGGCATAGGGGGTG | ||
| Motif Score | 2.833690476 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000383024.6; ENST00000351989.8; ENST00000495826.5; ENST00000407755.1 | ||
| External Link | RMBase: m6A_site_555362 | ||
| mod ID: M6ASITE056427 | Click to Show/Hide the Full List | ||
| mod site | chr22:20086582-20086583:+ | [12] | |
| Sequence | TGAGTTAGAAGATTTTACTGACAATTTGGAGCTAGATGAAG | ||
| Motif Score | 2.859755952 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000383024.6; ENST00000351989.8; ENST00000407755.1; ENST00000495826.5 | ||
| External Link | RMBase: m6A_site_555363 | ||
| mod ID: M6ASITE056428 | Click to Show/Hide the Full List | ||
| mod site | chr22:20086645-20086646:+ | [10] | |
| Sequence | TAAAGCAATCGTTCAGAGAGACAGAGTGGATGAAGAGGCCT | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa; Huh7; peripheral-blood | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000495826.5; ENST00000383024.6; ENST00000407755.1; ENST00000351989.8 | ||
| External Link | RMBase: m6A_site_555364 | ||
| mod ID: M6ASITE056429 | Click to Show/Hide the Full List | ||
| mod site | chr22:20086722-20086723:+ | [10] | |
| Sequence | TCTAGAGGGCTCTTAGCAAAACCCAAAGAGAGATTTGGGAA | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000495826.5; ENST00000383024.6; ENST00000351989.8; ENST00000407755.1 | ||
| External Link | RMBase: m6A_site_555365 | ||
| mod ID: M6ASITE056430 | Click to Show/Hide the Full List | ||
| mod site | chr22:20086785-20086786:+ | [10] | |
| Sequence | GGAAATTAAAAAAAAAAAAAACAAAAACCAAAATCCCCTCT | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | rmsk_5248472; ENST00000351989.8; ENST00000383024.6; ENST00000495826.5; ENST00000407755.1 | ||
| External Link | RMBase: m6A_site_555366 | ||
| mod ID: M6ASITE056431 | Click to Show/Hide the Full List | ||
| mod site | chr22:20086791-20086792:+ | [10] | |
| Sequence | TAAAAAAAAAAAAAACAAAAACCAAAATCCCCTCTGAGGTG | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000495826.5; ENST00000383024.6; ENST00000351989.8; ENST00000407755.1; rmsk_5248472 | ||
| External Link | RMBase: m6A_site_555367 | ||
| mod ID: M6ASITE056432 | Click to Show/Hide the Full List | ||
| mod site | chr22:20087145-20087146:+ | [10] | |
| Sequence | CCAGGGGCTCTGGTGTCTGAACAGCGTGTTTTGCAGGATGA | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000407755.1; ENST00000495826.5; ENST00000383024.6; ENST00000351989.8; ENST00000498171.5 | ||
| External Link | RMBase: m6A_site_555368 | ||
| mod ID: M6ASITE056433 | Click to Show/Hide the Full List | ||
| mod site | chr22:20087171-20087172:+ | [12] | |
| Sequence | TGTTTTGCAGGATGACTTTGACAACGATGTGGATGCTCTGC | ||
| Motif Score | 2.859755952 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000383024.6; ENST00000407755.1; ENST00000498171.5; ENST00000495826.5; ENST00000351989.8 | ||
| External Link | RMBase: m6A_site_555369 | ||
| mod ID: M6ASITE056435 | Click to Show/Hide the Full List | ||
| mod site | chr22:20087227-20087228:+ | [10] | |
| Sequence | GTGCCCCCAAAAAGAGGCGAACAGAGGAAAAATATGGCGGA | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa; MT4; Huh7; peripheral-blood | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000383024.6; ENST00000407755.1; ENST00000351989.8; ENST00000498171.5; ENST00000495826.5 | ||
| External Link | RMBase: m6A_site_555370 | ||
| mod ID: M6ASITE056436 | Click to Show/Hide the Full List | ||
| mod site | chr22:20087249-20087250:+ | [10] | |
| Sequence | AGAGGAAAAATATGGCGGAGACAGCGACCATCCGTCCGATG | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa; HEK293; MT4; Huh7; peripheral-blood | ||
| Seq Type List | m6A-seq; m6A-REF-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000498171.5; ENST00000351989.8; ENST00000383024.6; ENST00000495826.5; ENST00000407755.1 | ||
| External Link | RMBase: m6A_site_555371 | ||
| mod ID: M6ASITE056437 | Click to Show/Hide the Full List | ||
| mod site | chr22:20087275-20087276:+ | [10] | |
| Sequence | ACCATCCGTCCGATGGAGAGACAAGTGTGCAGCCGATGATG | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa; MT4; Huh7 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000407755.1; ENST00000351989.8; ENST00000495826.5; ENST00000498171.5; ENST00000383024.6 | ||
| External Link | RMBase: m6A_site_555372 | ||
| mod ID: M6ASITE056438 | Click to Show/Hide the Full List | ||
| mod site | chr22:20087308-20087309:+ | [10] | |
| Sequence | CGATGATGACCAAGATTAAAACAGTGCTCAAAAGTACGTGT | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa; MT4; Huh7 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000407755.1; ENST00000495826.5; ENST00000498171.5; ENST00000351989.8; ENST00000383024.6 | ||
| External Link | RMBase: m6A_site_555373 | ||
| mod ID: M6ASITE056439 | Click to Show/Hide the Full List | ||
| mod site | chr22:20089977-20089978:+ | [12] | |
| Sequence | CAATTTCACTATCCACAGAAACACGACCCTCCTCTGAGTAG | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000407755.1; ENST00000498171.5; ENST00000495826.5; ENST00000383024.6; ENST00000351989.8 | ||
| External Link | RMBase: m6A_site_555374 | ||
| mod ID: M6ASITE056440 | Click to Show/Hide the Full List | ||
| mod site | chr22:20090030-20090031:+ | [10] | |
| Sequence | GCATTATAAGAAAATGAAGGACAACGAGGAACGGGAGCAAA | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; hESC-HEK293T; endometrial | ||
| Seq Type List | m6A-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000498171.5; ENST00000407755.1; ENST00000383024.6; ENST00000351989.8; ENST00000495826.5 | ||
| External Link | RMBase: m6A_site_555375 | ||
| mod ID: M6ASITE056441 | Click to Show/Hide the Full List | ||
| mod site | chr22:20090177-20090178:+ | [10] | |
| Sequence | GGGGCCCCCGGACGAGAAAGACCCACTAGGGGCTGAGGCAG | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa; peripheral-blood; HEK293T; endometrial; NB4 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000498171.5; ENST00000407755.1; ENST00000495826.5; ENST00000351989.8; ENST00000383024.6 | ||
| External Link | RMBase: m6A_site_555376 | ||
| mod ID: M6ASITE056442 | Click to Show/Hide the Full List | ||
| mod site | chr22:20091505-20091506:+ | [10] | |
| Sequence | TTACTGTGAAAAAATTCAGGACTTGGGCTGAGCGGCGGCAA | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa; peripheral-blood; HEK293T; endometrial; NB4 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000351989.8; ENST00000383024.6; ENST00000495826.5; ENST00000498171.5; ENST00000407755.1 | ||
| External Link | RMBase: m6A_site_555377 | ||
| mod ID: M6ASITE056443 | Click to Show/Hide the Full List | ||
| mod site | chr22:20091622-20091623:+ | [12] | |
| Sequence | TATCAGTGCAAGATGCACCCACAAAGAAAGGTATAAGCCTC | ||
| Motif Score | 2.053113095 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000498171.5; ENST00000495826.5; ENST00000407755.1; ENST00000383024.6; ENST00000351989.8 | ||
| External Link | RMBase: m6A_site_555378 | ||
| mod ID: M6ASITE056444 | Click to Show/Hide the Full List | ||
| mod site | chr22:20091919-20091920:+ | [12] | |
| Sequence | GGTCTGCATCCTGCACGAGTACATGCAGCGTGTCCTCAAGG | ||
| Motif Score | 2.856142857 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000407755.1; ENST00000498171.5; ENST00000495826.5; ENST00000351989.8; ENST00000383024.6 | ||
| External Link | RMBase: m6A_site_555379 | ||
| mod ID: M6ASITE056446 | Click to Show/Hide the Full List | ||
| mod site | chr22:20094720-20094721:+ | [12] | |
| Sequence | TTTTTTTCATAGCCCGAGCTACACTGGAAATCCTCATCCCT | ||
| Motif Score | 2.078666667 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000351989.8; ENST00000495826.5; ENST00000407755.1; ENST00000383024.6; ENST00000498171.5; ENST00000491892.1 | ||
| External Link | RMBase: m6A_site_555380 | ||
| mod ID: M6ASITE056447 | Click to Show/Hide the Full List | ||
| mod site | chr22:20094778-20094779:+ | [13] | |
| Sequence | CTCTGAAGAGAAGCCCAAAGACAGTGAAGAACTCGAGGTGA | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | kidney | ||
| Seq Type List | m6A-REF-seq | ||
| Transcript ID List | ENST00000407755.1; ENST00000491892.1; ENST00000383024.6; ENST00000498171.5; ENST00000351989.8; ENST00000495826.5 | ||
| External Link | RMBase: m6A_site_555381 | ||
| mod ID: M6ASITE056448 | Click to Show/Hide the Full List | ||
| mod site | chr22:20106186-20106187:+ | [12] | |
| Sequence | TGTGTTGTAGTATTTTAACCACATCAGCATCGAGGACTCGC | ||
| Motif Score | 2.053113095 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000495826.5; ENST00000407755.1; ENST00000498171.5; ENST00000491892.1; ENST00000351989.8; ENST00000495351.1; ENST00000383024.6 | ||
| External Link | RMBase: m6A_site_555382 | ||
| mod ID: M6ASITE056449 | Click to Show/Hide the Full List | ||
| mod site | chr22:20106608-20106609:+ | [12] | |
| Sequence | AAGAAACCATGGGATGGGTGACACGTCTATCAAGTTTGAAG | ||
| Motif Score | 2.859755952 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000498171.5; ENST00000495351.1; ENST00000383024.6; ENST00000351989.8; ENST00000495826.5; ENST00000485802.1; ENST00000491892.1; ENST00000407755.1 | ||
| External Link | RMBase: m6A_site_555383 | ||
| mod ID: M6ASITE056450 | Click to Show/Hide the Full List | ||
| mod site | chr22:20106961-20106962:+ | [10] | |
| Sequence | GGAAAGGCCCTCCTCCTCGAACAGCCAGCAGAATCCTGCTG | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000485802.1; ENST00000383024.6; ENST00000491892.1; ENST00000498171.5; ENST00000495351.1; ENST00000407755.1; ENST00000495826.5; ENST00000351989.8 | ||
| External Link | RMBase: m6A_site_555384 | ||
| mod ID: M6ASITE056451 | Click to Show/Hide the Full List | ||
| mod site | chr22:20107276-20107277:+ | [14] | |
| Sequence | CTTTCTCTGTGTAGGTAAGAACAAGAGAGTTGGAAAGCAGT | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | Huh7 | ||
| Seq Type List | MeRIP-seq | ||
| Transcript ID List | ENST00000407755.1; ENST00000485802.1; ENST00000495826.5; ENST00000351989.8; ENST00000498171.5; ENST00000495351.1; ENST00000383024.6 | ||
| External Link | RMBase: m6A_site_555385 | ||
| mod ID: M6ASITE056452 | Click to Show/Hide the Full List | ||
| mod site | chr22:20107331-20107332:+ | [13] | |
| Sequence | ATCCTTCAGCTGCTGCACCCACATGTCAAGAACTGGGGGTC | ||
| Motif Score | 2.053113095 | ||
| Cell/Tissue List | HEK293; hESC-HEK293T | ||
| Seq Type List | m6A-REF-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000485802.1; ENST00000495826.5; ENST00000383024.6; ENST00000351989.8; ENST00000495351.1; ENST00000407755.1; ENST00000498171.5 | ||
| External Link | RMBase: m6A_site_555386 | ||
| mod ID: M6ASITE056453 | Click to Show/Hide the Full List | ||
| mod site | chr22:20107342-20107343:+ | [15] | |
| Sequence | GCTGCACCCACATGTCAAGAACTGGGGGTCTTTACTGCGCA | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HepG2; Huh7 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000498171.5; ENST00000495351.1; ENST00000383024.6; ENST00000351989.8; ENST00000495826.5; ENST00000485802.1; ENST00000407755.1 | ||
| External Link | RMBase: m6A_site_555387 | ||
| mod ID: M6ASITE056454 | Click to Show/Hide the Full List | ||
| mod site | chr22:20108408-20108409:+ | [16] | |
| Sequence | GAGCTCTGGCATCCCGAGGAACCACAGCACCCTTGGGGCTG | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | HEK293T; endometrial | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000351989.8; ENST00000475941.1; ENST00000498171.5; ENST00000485802.1; ENST00000495826.5; ENST00000407755.1; ENST00000383024.6 | ||
| External Link | RMBase: m6A_site_555388 | ||
| mod ID: M6ASITE056455 | Click to Show/Hide the Full List | ||
| mod site | chr22:20108675-20108676:+ | [17] | |
| Sequence | GAAAGGCCTGGCAGCTATGGACTGCCCCTCGCTCTCTGCTG | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HEK293A-TOA | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000475941.1; ENST00000485802.1; ENST00000407755.1; ENST00000498171.5; ENST00000351989.8; ENST00000495826.5; ENST00000383024.6 | ||
| External Link | RMBase: m6A_site_555389 | ||
| mod ID: M6ASITE056457 | Click to Show/Hide the Full List | ||
| mod site | chr22:20108892-20108893:+ | [10] | |
| Sequence | GGTGCTATACTTCCCAGGAGACATCGGACAAGAGTGTGATT | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa; hESC-HEK293T; U2OS; MT4; Huh7 | ||
| Seq Type List | m6A-seq; MAZTER-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000351989.8; ENST00000485802.1; ENST00000495826.5; ENST00000475941.1; ENST00000407755.1; ENST00000383024.6; ENST00000498171.5 | ||
| External Link | RMBase: m6A_site_555390 | ||
| mod ID: M6ASITE056458 | Click to Show/Hide the Full List | ||
| mod site | chr22:20108899-20108900:+ | [10] | |
| Sequence | TACTTCCCAGGAGACATCGGACAAGAGTGTGATTGAGCTGC | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; U2OS; MT4; Huh7 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000383024.6; ENST00000495826.5; ENST00000485802.1; ENST00000475941.1; ENST00000407755.1; ENST00000351989.8; ENST00000498171.5 | ||
| External Link | RMBase: m6A_site_555391 | ||
| mod ID: M6ASITE056459 | Click to Show/Hide the Full List | ||
| mod site | chr22:20108938-20108939:+ | [10] | |
| Sequence | GCAGCAGTATGCCAAGAAGAACAAGCCCAACCTGCACATCC | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa; U2OS; MT4; Huh7; CD4T | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000475941.1; ENST00000383024.6; ENST00000498171.5; ENST00000495826.5; ENST00000485802.1; ENST00000407755.1; ENST00000351989.8 | ||
| External Link | RMBase: m6A_site_555392 | ||
| mod ID: M6ASITE056460 | Click to Show/Hide the Full List | ||
| mod site | chr22:20109066-20109067:+ | [10] | |
| Sequence | CCTGTGGCACCTGTGGAGGGACAGCAGACCCCAGGACCCGT | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; Huh7; CD4T; peripheral-blood; endometrial | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000498171.5; ENST00000351989.8; ENST00000485802.1; ENST00000495826.5; ENST00000383024.6; ENST00000475941.1; ENST00000407755.1 | ||
| External Link | RMBase: m6A_site_555393 | ||
| mod ID: M6ASITE056461 | Click to Show/Hide the Full List | ||
| mod site | chr22:20109073-20109074:+ | [10] | |
| Sequence | CACCTGTGGAGGGACAGCAGACCCCAGGACCCGTGCCTGAG | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa; Huh7; CD4T; peripheral-blood; endometrial | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000485802.1; ENST00000498171.5; ENST00000383024.6; ENST00000475941.1; ENST00000495826.5; ENST00000407755.1; ENST00000351989.8 | ||
| External Link | RMBase: m6A_site_555394 | ||
| mod ID: M6ASITE056462 | Click to Show/Hide the Full List | ||
| mod site | chr22:20109081-20109082:+ | [10] | |
| Sequence | GAGGGACAGCAGACCCCAGGACCCGTGCCTGAGCTGACACC | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; Huh7; CD4T; peripheral-blood; endometrial | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000383024.6; ENST00000495826.5; ENST00000351989.8; ENST00000485802.1; ENST00000407755.1; ENST00000498171.5; ENST00000475941.1 | ||
| External Link | RMBase: m6A_site_555395 | ||
| mod ID: M6ASITE056463 | Click to Show/Hide the Full List | ||
| mod site | chr22:20110030-20110031:+ | [10] | |
| Sequence | TGTTGTTCTTTCAGGAGGAGACTCGAAAGAAGCCCAAGATG | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa; HepG2; A549; U2OS; H1A; H1B; MM6; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293T; iSLK; endometrial; HEC-1-A; GSCs; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000383024.6; ENST00000475941.1; ENST00000498171.5; ENST00000407755.1; ENST00000495826.5; ENST00000351989.8 | ||
| External Link | RMBase: m6A_site_555396 | ||
| mod ID: M6ASITE056464 | Click to Show/Hide the Full List | ||
| mod site | chr22:20110160-20110161:+ | [10] | |
| Sequence | CCAGCCGCACTTCTGAGGAGACCAGCAGTCATGCATCGTGC | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; A549; U2OS; H1B; H1A; hESCs; GM12878; H1299; MM6; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293A-TOA; iSLK; TIME; TREX; MSC; endometrial; HEC-1-A; GSCs; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000407755.1; ENST00000475941.1; ENST00000495826.5; ENST00000498171.5; ENST00000351989.8; ENST00000383024.6 | ||
| External Link | RMBase: m6A_site_555397 | ||
| mod ID: M6ASITE056465 | Click to Show/Hide the Full List | ||
| mod site | chr22:20110268-20110269:+ | [10] | |
| Sequence | GGAGGCTTTAGTGTACAGGGACAGCCATGGCCACACAGCAC | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; A549; hESC-HEK293T; U2OS; H1A; H1B; hESCs; fibroblasts; GM12878; CD8T; H1299; MM6; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; TREX; endometrial; HEC-1-A; GSCs; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq; MAZTER-seq; m6A-CLIP/IP | ||
| Transcript ID List | ENST00000498171.5; ENST00000351989.8; ENST00000383024.6; ENST00000407755.1; ENST00000495826.5; ENST00000475941.1 | ||
| External Link | RMBase: m6A_site_555398 | ||
| mod ID: M6ASITE056466 | Click to Show/Hide the Full List | ||
| mod site | chr22:20110339-20110340:+ | [10] | |
| Sequence | CTCCAGGCCTGAATGGATGGACTCAGCGACTGCACCAGTGG | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; A549; U2OS; H1A; H1B; hNPCs; hESCs; fibroblasts; GM12878; LCLs; H1299; MM6; Huh7; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; TREX; endometrial; HEC-1-A; GSCs; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000351989.8; ENST00000475941.1; ENST00000383024.6; ENST00000495826.5; ENST00000498171.5; ENST00000407755.1 | ||
| External Link | RMBase: m6A_site_555399 | ||
| mod ID: M6ASITE056468 | Click to Show/Hide the Full List | ||
| mod site | chr22:20110376-20110377:+ | [10] | |
| Sequence | GTGGCAGCTGGTGACTGTGGACAGTGGTGGACCCTGCTTCT | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; A549; hESC-HEK293T; U2OS; H1A; H1B; hNPCs; hESCs; fibroblasts; GM12878; LCLs; H1299; MM6; Huh7; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; TREX; endometrial; HEC-1-A; GSCs; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq; DART-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000495826.5; ENST00000407755.1; ENST00000498171.5; ENST00000475941.1; ENST00000351989.8; ENST00000383024.6 | ||
| External Link | RMBase: m6A_site_555400 | ||
| mod ID: M6ASITE056469 | Click to Show/Hide the Full List | ||
| mod site | chr22:20110386-20110387:+ | [10] | |
| Sequence | GTGACTGTGGACAGTGGTGGACCCTGCTTCTGTGCACCTGC | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; A549; U2OS; H1A; H1B; hNPCs; hESCs; fibroblasts; GM12878; LCLs; H1299; MM6; Huh7; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; TREX; endometrial; HEC-1-A; GSCs; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000498171.5; ENST00000475941.1; ENST00000383024.6; ENST00000351989.8; ENST00000495826.5; ENST00000407755.1 | ||
| External Link | RMBase: m6A_site_555401 | ||
| mod ID: M6ASITE056470 | Click to Show/Hide the Full List | ||
| mod site | chr22:20110452-20110453:+ | [11] | |
| Sequence | CATGAATTTTAGTATGTAATACGCACTGACGACACATGATG | ||
| Motif Score | 2.084416667 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000383024.6; ENST00000407755.1; ENST00000498171.5; ENST00000351989.8; ENST00000495826.5; ENST00000475941.1 | ||
| External Link | RMBase: m6A_site_555402 | ||
| mod ID: M6ASITE056471 | Click to Show/Hide the Full List | ||
| mod site | chr22:20110570-20110571:+ | [10] | |
| Sequence | CCTGCAGGGCTGCTACAGGGACCTGGTCAGGAGGTGCACAT | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; A549; H1A; H1B; hESCs; fibroblasts; GM12878; H1299; MM6; Huh7; peripheral-blood; HEK293A-TOA; iSLK; MSC; TIME; TREX | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000475941.1; ENST00000351989.8; ENST00000383024.6; ENST00000407755.1; ENST00000495826.5; ENST00000498171.5 | ||
| External Link | RMBase: m6A_site_555403 | ||
| mod ID: M6ASITE056472 | Click to Show/Hide the Full List | ||
| mod site | chr22:20110587-20110588:+ | [12] | |
| Sequence | GGGACCTGGTCAGGAGGTGCACATGGTGCCCTGCCCTCACC | ||
| Motif Score | 2.830589286 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000475941.1; ENST00000407755.1; ENST00000383024.6; ENST00000498171.5; ENST00000351989.8; ENST00000495826.5 | ||
| External Link | RMBase: m6A_site_555404 | ||
| mod ID: M6ASITE056473 | Click to Show/Hide the Full List | ||
| mod site | chr22:20110662-20110663:+ | [18] | |
| Sequence | AAGAGAAAGGAATATTTTAAACCTTTTTGGCTTAAACAGAA | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | HeLa; A549; HEK293T; HepG2; H1A; H1B; GM12878; peripheral-blood; MSC | ||
| Seq Type List | MeRIP-seq; m6A-seq | ||
| Transcript ID List | ENST00000495826.5; ENST00000498171.5; ENST00000383024.6; ENST00000475941.1; ENST00000351989.8; ENST00000407755.1 | ||
| External Link | RMBase: m6A_site_555405 | ||
| mod ID: M6ASITE056474 | Click to Show/Hide the Full List | ||
| mod site | chr22:20110677-20110678:+ | [15] | |
| Sequence | TTTAAACCTTTTTGGCTTAAACAGAATTTTAGCATCAGAAC | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HepG2 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000383024.6; ENST00000475941.1; ENST00000495826.5; ENST00000407755.1; ENST00000498171.5; ENST00000351989.8 | ||
| External Link | RMBase: m6A_site_555406 | ||
| mod ID: M6ASITE056475 | Click to Show/Hide the Full List | ||
| mod site | chr22:20110696-20110697:+ | [15] | |
| Sequence | AACAGAATTTTAGCATCAGAACTAGCTTTCTGGGATTGGAG | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HepG2 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000383024.6; ENST00000475941.1; ENST00000407755.1; ENST00000495826.5; ENST00000498171.5; ENST00000351989.8 | ||
| External Link | RMBase: m6A_site_555407 | ||
| mod ID: M6ASITE056476 | Click to Show/Hide the Full List | ||
| mod site | chr22:20110721-20110722:+ | [15] | |
| Sequence | CTTTCTGGGATTGGAGGCAAACCATCAAGGTGGTCCCTCTC | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | HepG2; HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000475941.1; ENST00000495826.5; ENST00000498171.5; ENST00000351989.8; ENST00000407755.1; ENST00000383024.6 | ||
| External Link | RMBase: m6A_site_555408 | ||
| mod ID: M6ASITE056477 | Click to Show/Hide the Full List | ||
| mod site | chr22:20110750-20110751:+ | [12] | |
| Sequence | GTGGTCCCTCTCCAGTCTGGACACGATGCCAGCAAGGATGA | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | hESC-HEK293T; HepG2; HeLa | ||
| Seq Type List | MAZTER-seq; m6A-seq | ||
| Transcript ID List | ENST00000383024.6; ENST00000495826.5; ENST00000475941.1; ENST00000498171.5; ENST00000351989.8; ENST00000407755.1 | ||
| External Link | RMBase: m6A_site_555409 | ||
| mod ID: M6ASITE056479 | Click to Show/Hide the Full List | ||
| mod site | chr22:20110752-20110753:+ | [11] | |
| Sequence | GGTCCCTCTCCAGTCTGGACACGATGCCAGCAAGGATGACG | ||
| Motif Score | 2.058863095 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000351989.8; ENST00000498171.5; ENST00000407755.1; ENST00000383024.6; ENST00000475941.1; ENST00000495826.5 | ||
| External Link | RMBase: m6A_site_555410 | ||
| mod ID: M6ASITE056480 | Click to Show/Hide the Full List | ||
| mod site | chr22:20110856-20110857:+ | [11] | |
| Sequence | CCCTAAAAGCGCCTCTTTGGACACTGAGGCCCTCTCTGCCT | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HEK293T; hESC-HEK293T; HeLa | ||
| Seq Type List | DART-seq; MAZTER-seq; MeRIP-seq; m6A-seq | ||
| Transcript ID List | ENST00000407755.1; ENST00000351989.8; ENST00000495826.5; ENST00000498171.5; ENST00000383024.6; ENST00000475941.1 | ||
| External Link | RMBase: m6A_site_555412 | ||
| mod ID: M6ASITE056481 | Click to Show/Hide the Full List | ||
| mod site | chr22:20110903-20110904:+ | [11] | |
| Sequence | GCCTCCGGCAACAGTTTTTTACAAAGATTTTTTGCAGTCGA | ||
| Motif Score | 2.07285119 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000351989.8; ENST00000475941.1; ENST00000407755.1; ENST00000498171.5; ENST00000495826.5; ENST00000383024.6 | ||
| External Link | RMBase: m6A_site_555414 | ||
| mod ID: M6ASITE056482 | Click to Show/Hide the Full List | ||
| mod site | chr22:20110982-20110983:+ | [18] | |
| Sequence | ATATTTTAGCATTTTGAAAGACTTTCACAGTGAGAGTAGAA | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HEK293T; HeLa; HepG2; hNPCs; hESCs; fibroblasts; A549; GM12878; HEK293A-TOA; iSLK; MSC; TIME; TREX | ||
| Seq Type List | MeRIP-seq; m6A-seq | ||
| Transcript ID List | ENST00000351989.8; ENST00000495826.5; ENST00000383024.6; ENST00000407755.1; ENST00000475941.1; ENST00000498171.5 | ||
| External Link | RMBase: m6A_site_555417 | ||
| mod ID: M6ASITE056483 | Click to Show/Hide the Full List | ||
| mod site | chr22:20111036-20111037:+ | [18] | |
| Sequence | TCATGCATTTTAGCAAGTGGACTTGTTGAAACAGGAAGCAA | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HEK293T; HeLa; HepG2; hESCs; fibroblasts; A549; GM12878; HEK293A-TOA; iSLK; MSC; TIME; TREX | ||
| Seq Type List | MeRIP-seq; m6A-seq | ||
| Transcript ID List | ENST00000475941.1; ENST00000383024.6; ENST00000498171.5; ENST00000495826.5; ENST00000351989.8; ENST00000407755.1 | ||
| External Link | RMBase: m6A_site_555418 | ||
| mod ID: M6ASITE056484 | Click to Show/Hide the Full List | ||
| mod site | chr22:20111046-20111047:+ | [18] | |
| Sequence | TAGCAAGTGGACTTGTTGAAACAGGAAGCAAGGGCCTTCAG | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HEK293T; HeLa; HepG2; hESCs; fibroblasts; A549; GM12878; HEK293A-TOA; iSLK; MSC; TIME; TREX | ||
| Seq Type List | MeRIP-seq; m6A-seq | ||
| Transcript ID List | ENST00000351989.8; ENST00000383024.6; ENST00000495826.5; ENST00000475941.1; ENST00000407755.1; ENST00000498171.5 | ||
| External Link | RMBase: m6A_site_555419 | ||
| mod ID: M6ASITE056486 | Click to Show/Hide the Full List | ||
| mod site | chr22:20111251-20111252:+ | [14] | |
| Sequence | AAGGCTGGCCCTGGTGGTGGACTGGCACCTGTGCAGAGTGC | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | Huh7; peripheral-blood | ||
| Seq Type List | MeRIP-seq; m6A-seq | ||
| Transcript ID List | ENST00000383024.6; ENST00000498171.5; ENST00000495826.5; ENST00000407755.1; ENST00000475941.1; ENST00000351989.8 | ||
| External Link | RMBase: m6A_site_555423 | ||
| mod ID: M6ASITE056487 | Click to Show/Hide the Full List | ||
| mod site | chr22:20111353-20111354:+ | [11] | |
| Sequence | CCATGCCCCCTACAGGCGGTACTGATGGCGCTTTTTTTTTT | ||
| Motif Score | 3.278136905 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000351989.8; ENST00000498171.5; ENST00000383024.6; ENST00000407755.1; ENST00000495826.5 | ||
| External Link | RMBase: m6A_site_555424 | ||
| mod ID: M6ASITE056488 | Click to Show/Hide the Full List | ||
| mod site | chr22:20111389-20111390:+ | [14] | |
| Sequence | TTTTTTTTTCTGTCAGGAAAACAATGTTGGCCTGTGGGCCG | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | Huh7 | ||
| Seq Type List | MeRIP-seq | ||
| Transcript ID List | ENST00000383024.6; ENST00000495826.5; ENST00000407755.1; ENST00000498171.5; ENST00000351989.8 | ||
| External Link | RMBase: m6A_site_555425 | ||
| mod ID: M6ASITE056489 | Click to Show/Hide the Full List | ||
| mod site | chr22:20111416-20111417:+ | [12] | |
| Sequence | TGGCCTGTGGGCCGCCCACAACATATCCTTCCCTCACTACC | ||
| Motif Score | 2.173910714 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000407755.1; ENST00000495826.5; ENST00000498171.5; ENST00000383024.6; ENST00000351989.8 | ||
| External Link | RMBase: m6A_site_555426 | ||
| mod ID: M6ASITE056490 | Click to Show/Hide the Full List | ||
| mod site | chr22:20111505-20111506:+ | [12] | |
| Sequence | ACTCAAATTGCTATAATTAGACACTTGCTTCTGTCTTGCCT | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000407755.1; ENST00000498171.5; ENST00000351989.8; ENST00000383024.6; ENST00000495826.5 | ||
| External Link | RMBase: m6A_site_555427 | ||
| mod ID: M6ASITE056491 | Click to Show/Hide the Full List | ||
| mod site | chr22:20111594-20111595:+ | [10] | |
| Sequence | CCAGCCAGATGCGCCTGTGAACCAAAGCTTCGTGCACATGT | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | HeLa; HEK293A-TOA | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000351989.8; ENST00000498171.5; ENST00000495826.5; ENST00000407755.1; ENST00000383024.6 | ||
| External Link | RMBase: m6A_site_555428 | ||
| mod ID: M6ASITE056492 | Click to Show/Hide the Full List | ||
| mod site | chr22:20111609-20111610:+ | [12] | |
| Sequence | TGTGAACCAAAGCTTCGTGCACATGTGTTCCCCTAAAGGTT | ||
| Motif Score | 2.830589286 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000495826.5; ENST00000351989.8; ENST00000383024.6; ENST00000498171.5; ENST00000407755.1 | ||
| External Link | RMBase: m6A_site_555429 | ||
| mod ID: M6ASITE056494 | Click to Show/Hide the Full List | ||
| mod site | chr22:20111686-20111687:+ | [11] | |
| Sequence | ACCACAGCAGGTGCTGCCATACTCTTGTGGTCTCTGTGCGC | ||
| Motif Score | 2.53247619 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000498171.5; ENST00000407755.1; ENST00000351989.8; ENST00000495826.5; ENST00000383024.6 | ||
| External Link | RMBase: m6A_site_555430 | ||
| mod ID: M6ASITE056495 | Click to Show/Hide the Full List | ||
| mod site | chr22:20111754-20111755:+ | [12] | |
| Sequence | AGCATGGGTATGAATCGTGCACACAGCCATGCTTCAAGGCC | ||
| Motif Score | 2.830589286 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000407755.1; ENST00000351989.8; ENST00000383024.6; ENST00000498171.5; ENST00000495826.5 | ||
| External Link | RMBase: m6A_site_555431 | ||
| mod ID: M6ASITE056496 | Click to Show/Hide the Full List | ||
| mod site | chr22:20111848-20111849:+ | [12] | |
| Sequence | GGCGAGCCTTGATTGTCTGAACACATAAAGCAAACTGTCCA | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | hESC-HEK293T; Huh7 | ||
| Seq Type List | MAZTER-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000351989.8; ENST00000407755.1; ENST00000495826.5; ENST00000498171.5; ENST00000383024.6 | ||
| External Link | RMBase: m6A_site_555432 | ||
| mod ID: M6ASITE056497 | Click to Show/Hide the Full List | ||
| mod site | chr22:20111861-20111862:+ | [14] | |
| Sequence | TGTCTGAACACATAAAGCAAACTGTCCAGAAGGGAATGGCT | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | Huh7 | ||
| Seq Type List | MeRIP-seq | ||
| Transcript ID List | ENST00000498171.5; ENST00000383024.6; ENST00000495826.5; ENST00000407755.1; ENST00000351989.8 | ||
| External Link | RMBase: m6A_site_555433 | ||
Pseudouridine (Pseudo)
| In total 1 m6A sequence/site(s) in this target gene | |||
| mod ID: PSESITE000174 | Click to Show/Hide the Full List | ||
| mod site | chr22:20110703-20110704:+ | [19] | |
| Sequence | TTTTAGCATCAGAACTAGCTTTCTGGGATTGGAGGCAAACC | ||
| Transcript ID List | ENST00000383024.6; ENST00000475941.1; ENST00000498171.5; ENST00000407755.1; ENST00000351989.8; ENST00000495826.5 | ||
| External Link | RMBase: Pseudo_site_3396 | ||
References