m6A Target Gene Information
General Information of the m6A Target Gene (ID: M6ATAR00209)
Full List of m6A Methylation Regulator of This Target Gene and Corresponding Disease/Drug Response(s)
CDK1
can be regulated by the following regulator(s), and cause disease/drug response(s). You can browse detail information of regulator(s) or disease/drug response(s).
Browse Regulator
Browse Disease
Browse Drug
Methyltransferase-like 3 (METTL3) [WRITER]
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | Liver | Mus musculus |
|
Treatment: Mettl3-deficient liver
Control: Wild type liver cells
|
GSE197800 | |
| Regulation |
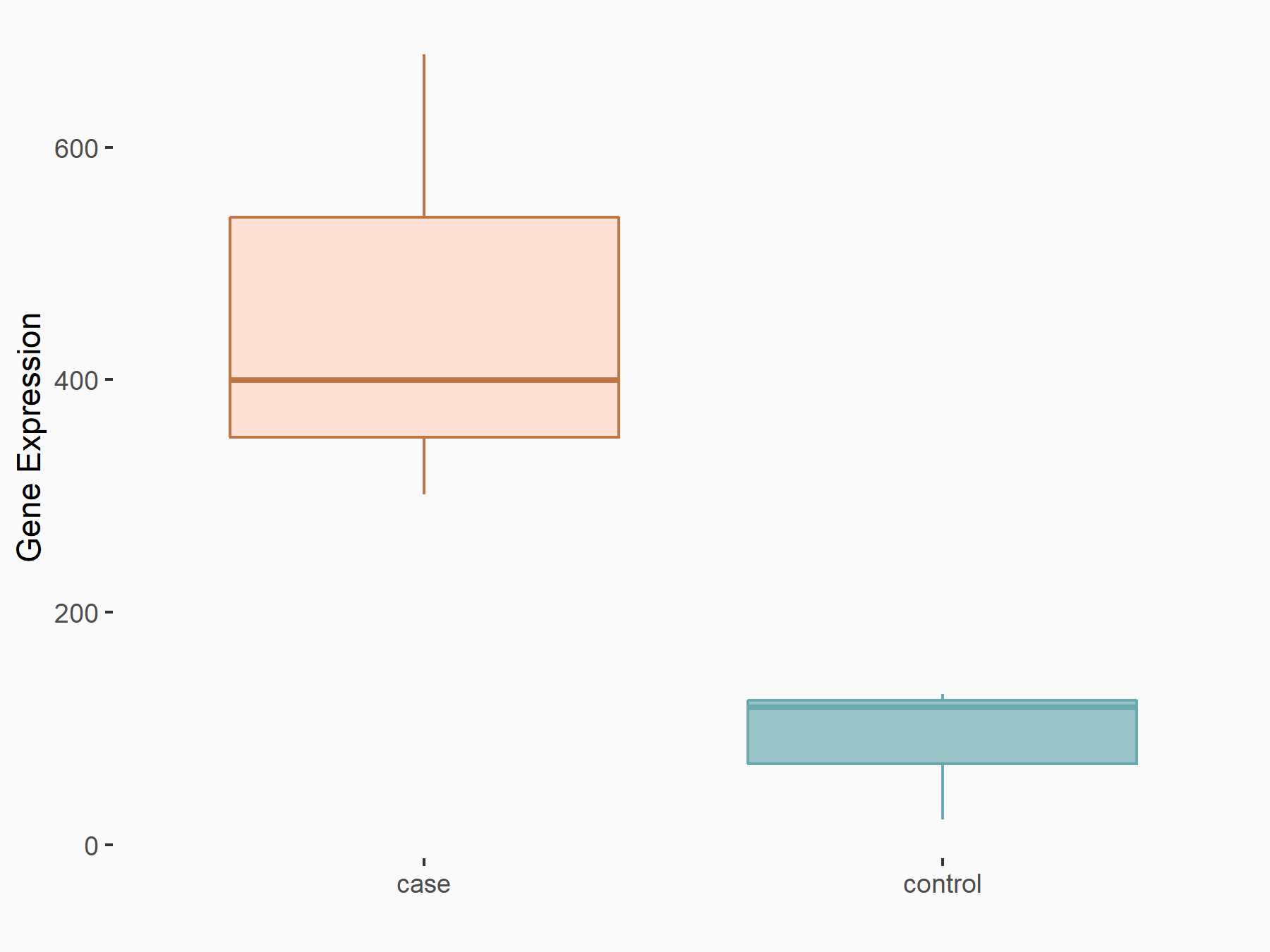  |
logFC: 2.36E+00 p-value: 3.11E-05 |
| More Results | Click to View More RNA-seq Results | |
| In total 1 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [1] | |||
| Response Summary | CircMETTL3 promotes breast cancer progression through circMETTL3/miR-31-5p/Cyclin-dependent kinase 1 (CDK1) axis. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Breast cancer | ICD-11: 2C60 | ||
| Pathway Response | Cell cycle | hsa04110 | ||
| Cell Process | Cell cycle | |||
| In-vitro Model | BT-474 | Invasive breast carcinoma | Homo sapiens | CVCL_0179 |
| HCC1806 | Breast squamous cell carcinoma | Homo sapiens | CVCL_1258 | |
| MCF-10A | Normal | Homo sapiens | CVCL_0598 | |
| MCF-7 | Invasive breast carcinoma | Homo sapiens | CVCL_0031 | |
| MDA-MB-231 | Breast adenocarcinoma | Homo sapiens | CVCL_0062 | |
| MDA-MB-453 | Breast adenocarcinoma | Homo sapiens | CVCL_0418 | |
| ZR-75-1 | Invasive breast carcinoma | Homo sapiens | CVCL_0588 | |
| In-vivo Model | Twelve female BALB/c nude mice (aged 4 weeks, 18-22g) were randomly divided into 2 groups. Stable circMETTL3-expression SUM1315 cells or control cells (1×106 cells in 0.1 mL PBS) was subcutaneously injected into mammary fat pads of the mice and the growth of tumors was followed up every week. Tumor volume was measured every week using a caliper, calculated as (length × width2)/2. After 4 weeks, mice were sacrificed and checked for final tumor weight. | |||
Protein virilizer homolog (VIRMA) [WRITER]
| Representative RNA-seq result indicating the expression of this target gene regulated by VIRMA | ||
| Cell Line | Human umbilical vein endothelial cells | Homo sapiens |
|
Treatment: siVIRMA HUVECs
Control: siControl HUVECs
|
GSE167067 | |
| Regulation |
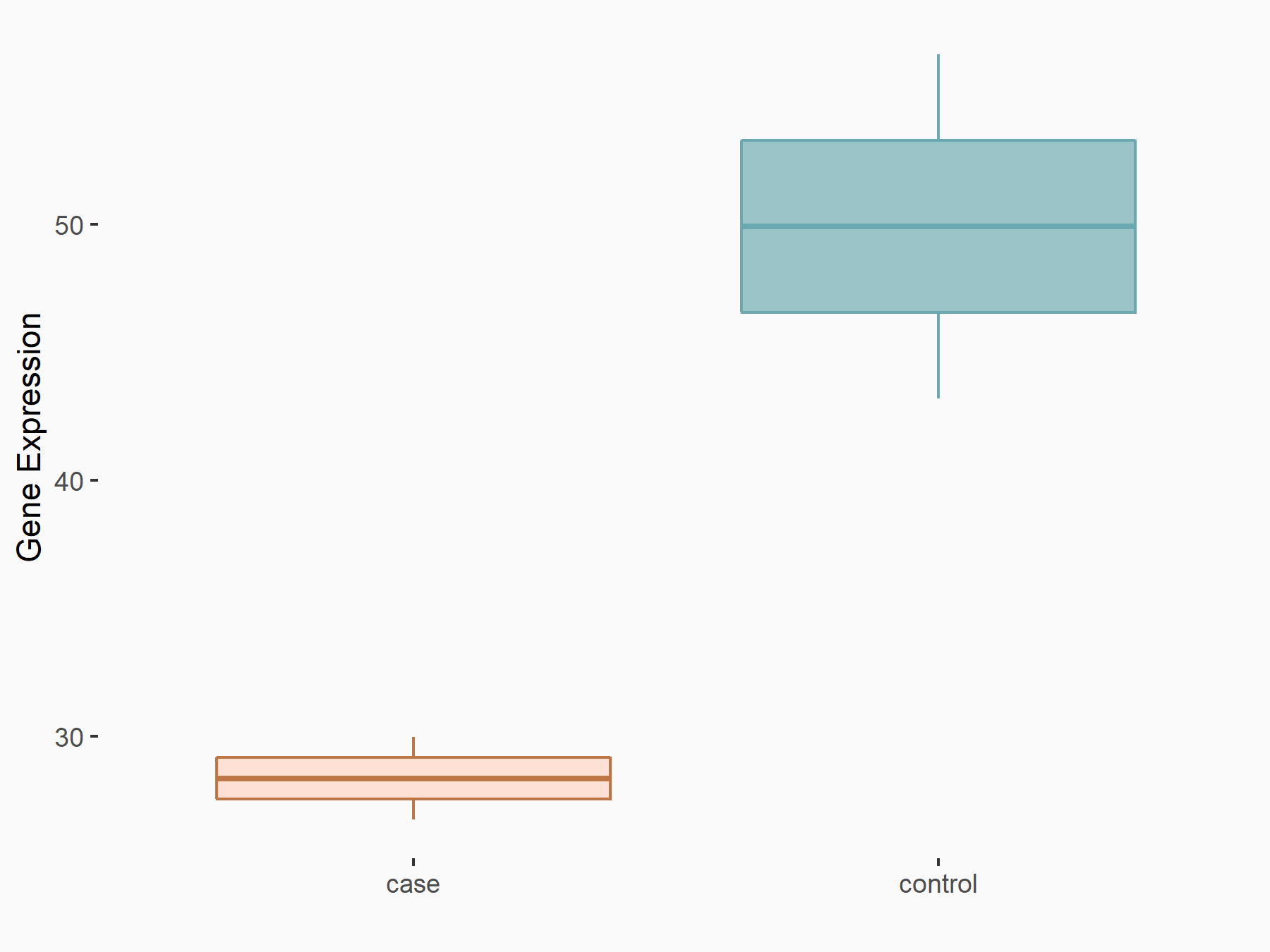  |
logFC: -7.84E-01 p-value: 3.08E-02 |
| More Results | Click to View More RNA-seq Results | |
| In total 1 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [2] | |||
| Response Summary | KIAA1429 acts as an oncogenic factor in breast cancer by regulating CDK1 in an N6-methyladenosine-independent manner.5'-fluorouracil was found to be very effective in reducing the expression of KIAA1429 and Cyclin-dependent kinase 1 (CDK1) in breast cancer. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Breast cancer | ICD-11: 2C60 | ||
| Responsed Drug | Fluorouracil | Approved | ||
| Pathway Response | Cell cycle | hsa04110 | ||
| Cell Process | Cell proliferation and metastasis | |||
Breast cancer [ICD-11: 2C60]
| In total 2 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [1] | |||
| Response Summary | CircMETTL3 promotes breast cancer progression through circMETTL3/miR-31-5p/Cyclin-dependent kinase 1 (CDK1) axis. | |||
| Responsed Disease | Breast cancer [ICD-11: 2C60] | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Cell cycle | hsa04110 | ||
| Cell Process | Cell cycle | |||
| In-vitro Model | BT-474 | Invasive breast carcinoma | Homo sapiens | CVCL_0179 |
| HCC1806 | Breast squamous cell carcinoma | Homo sapiens | CVCL_1258 | |
| MCF-10A | Normal | Homo sapiens | CVCL_0598 | |
| MCF-7 | Invasive breast carcinoma | Homo sapiens | CVCL_0031 | |
| MDA-MB-231 | Breast adenocarcinoma | Homo sapiens | CVCL_0062 | |
| MDA-MB-453 | Breast adenocarcinoma | Homo sapiens | CVCL_0418 | |
| ZR-75-1 | Invasive breast carcinoma | Homo sapiens | CVCL_0588 | |
| In-vivo Model | Twelve female BALB/c nude mice (aged 4 weeks, 18-22g) were randomly divided into 2 groups. Stable circMETTL3-expression SUM1315 cells or control cells (1×106 cells in 0.1 mL PBS) was subcutaneously injected into mammary fat pads of the mice and the growth of tumors was followed up every week. Tumor volume was measured every week using a caliper, calculated as (length × width2)/2. After 4 weeks, mice were sacrificed and checked for final tumor weight. | |||
| Experiment 2 Reporting the m6A-centered Disease Response | [2] | |||
| Response Summary | KIAA1429 acts as an oncogenic factor in breast cancer by regulating CDK1 in an N6-methyladenosine-independent manner.5'-fluorouracil was found to be very effective in reducing the expression of KIAA1429 and Cyclin-dependent kinase 1 (CDK1) in breast cancer. | |||
| Responsed Disease | Breast cancer [ICD-11: 2C60] | |||
| Target Regulator | Protein virilizer homolog (VIRMA) | WRITER | ||
| Target Regulation | Up regulation | |||
| Responsed Drug | Fluorouracil | Approved | ||
| Pathway Response | Cell cycle | hsa04110 | ||
| Cell Process | Cell proliferation and metastasis | |||
Fluorouracil
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response | [2] | |||
| Response Summary | KIAA1429 acts as an oncogenic factor in breast cancer by regulating CDK1 in an N6-methyladenosine-independent manner.5'-fluorouracil was found to be very effective in reducing the expression of KIAA1429 and Cyclin-dependent kinase 1 (CDK1) in breast cancer. | |||
| Target Regulator | Protein virilizer homolog (VIRMA) | WRITER | ||
| Target Regulation | Up regulation | |||
| Responsed Disease | Breast cancer | ICD-11: 2C60 | ||
| Pathway Response | Cell cycle | hsa04110 | ||
| Cell Process | Cell proliferation and metastasis | |||
RNA Modification Sequencing Data Associated with the Target (ID: M6ATAR00209)
| In total 1 m6A sequence/site(s) in this target gene | |||
| mod ID: 2OMSITE000570 | Click to Show/Hide the Full List | ||
| mod site | chr10:60793960-60793961:+ | [3] | |
| Sequence | TTAATGATTTGGACAATCAGATTAAGAAGATGTAGCTTTCT | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | Nm-seq | ||
| Transcript ID List | ENST00000373809.2; ENST00000395284.7; ENST00000614696.4; ENST00000448257.6; ENST00000316629.8 | ||
| External Link | RMBase: Nm_site_735 | ||
5-methylcytidine (m5C)
| In total 1 m6A sequence/site(s) in this target gene | |||
| mod ID: M5CSITE004693 | Click to Show/Hide the Full List | ||
| mod site | chr10:60794404-60794405:+ | [4] | |
| Sequence | GTTTCTTGCCATGTTGTTAACTATACAACCTGGCTAAAGAT | ||
| Cell/Tissue List | T24 | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000316629.8; ENST00000448257.6; ENST00000614696.4; ENST00000395284.7; ENST00000373809.2 | ||
| External Link | RMBase: m5C_site_5295 | ||
N6-methyladenosine (m6A)
| In total 50 m6A sequence/site(s) in this target gene | |||
| mod ID: M6ASITE000058 | Click to Show/Hide the Full List | ||
| mod site | chr10:60778470-60778471:+ | [5] | |
| Sequence | TTTAGCGCGGTGAGTTTGAAACTGCTCGCACTTGGCTTCAA | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | A549 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000519760.1; ENST00000475504.6; ENST00000519078.6; ENST00000395284.7; ENST00000316629.8 | ||
| External Link | RMBase: m6A_site_101998 | ||
| mod ID: M6ASITE000059 | Click to Show/Hide the Full List | ||
| mod site | chr10:60778661-60778662:+ | [6] | |
| Sequence | GGGTCCTCTAAGAAGGCCGGACTGGAGGTCAGGGATCTGCG | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | Huh7 | ||
| Seq Type List | MeRIP-seq | ||
| Transcript ID List | ENST00000519078.6; ENST00000395284.7; ENST00000519760.1; ENST00000475504.6; ENST00000316629.8; ENST00000614696.4; ENST00000448257.6 | ||
| External Link | RMBase: m6A_site_101999 | ||
| mod ID: M6ASITE000060 | Click to Show/Hide the Full List | ||
| mod site | chr10:60783497-60783498:+ | [6] | |
| Sequence | ATATTGCTACTTTCAGAGAAACTCATAAAAGCTATAGAAGA | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | Huh7 | ||
| Seq Type List | MeRIP-seq | ||
| Transcript ID List | ENST00000519078.6; ENST00000395284.7; ENST00000448257.6; ENST00000614696.4; ENST00000316629.8; ENST00000519760.1; ENST00000475504.6; ENST00000373809.2 | ||
| External Link | RMBase: m6A_site_102000 | ||
| mod ID: M6ASITE000061 | Click to Show/Hide the Full List | ||
| mod site | chr10:60784732-60784733:+ | [7] | |
| Sequence | GGAGTTGTGTATAAGGGTAGACACAAAACTACAGGTCAAGT | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000519078.6; ENST00000395284.7; ENST00000316629.8; ENST00000614696.4; ENST00000448257.6; ENST00000475504.6; ENST00000373809.2 | ||
| External Link | RMBase: m6A_site_102001 | ||
| mod ID: M6ASITE000062 | Click to Show/Hide the Full List | ||
| mod site | chr10:60785764-60785765:+ | [7] | |
| Sequence | TTCTATCCCTCCTGGTCAGTACATGGATTCTTCACTTGTTA | ||
| Motif Score | 2.856142857 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000448257.6; ENST00000373809.2; ENST00000395284.7; ENST00000316629.8; ENST00000614696.4; ENST00000475504.6; ENST00000519078.6 | ||
| External Link | RMBase: m6A_site_102002 | ||
| mod ID: M6ASITE000063 | Click to Show/Hide the Full List | ||
| mod site | chr10:60785838-60785839:+ | [8] | |
| Sequence | TGCACTGTGGATATAAAGGGACTATATATAGAAGTCCCTGC | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000448257.6; ENST00000373809.2; ENST00000614696.4; ENST00000395284.7; ENST00000519078.6; ENST00000475504.6; ENST00000316629.8 | ||
| External Link | RMBase: m6A_site_102003 | ||
| mod ID: M6ASITE000064 | Click to Show/Hide the Full List | ||
| mod site | chr10:60786134-60786135:+ | [8] | |
| Sequence | GTGTTTTATTCCAGTTTTGAACATGTTTTGGTCAATTTATT | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000316629.8; ENST00000448257.6; ENST00000519078.6; ENST00000373809.2; ENST00000614696.4; ENST00000395284.7; ENST00000475504.6 | ||
| External Link | RMBase: m6A_site_102004 | ||
| mod ID: M6ASITE000065 | Click to Show/Hide the Full List | ||
| mod site | chr10:60786159-60786160:+ | [8] | |
| Sequence | TTTTGGTCAATTTATTGTAGACATTTATTATATTTCAGGTA | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000614696.4; ENST00000448257.6; ENST00000475504.6; ENST00000316629.8; ENST00000395284.7; ENST00000373809.2; ENST00000519078.6 | ||
| External Link | RMBase: m6A_site_102005 | ||
| mod ID: M6ASITE000066 | Click to Show/Hide the Full List | ||
| mod site | chr10:60787775-60787776:+ | [8] | |
| Sequence | AAGAGGCTTCTGTGTAGAAAACCAATTTGTAATTTAAGGAT | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000395284.7; ENST00000614696.4; ENST00000373809.2; ENST00000487784.1; ENST00000316629.8; ENST00000448257.6; ENST00000519078.6 | ||
| External Link | RMBase: m6A_site_102006 | ||
| mod ID: M6ASITE000067 | Click to Show/Hide the Full List | ||
| mod site | chr10:60787801-60787802:+ | [8] | |
| Sequence | TTGTAATTTAAGGATCGGGGACCAAAAATGTTTAAAGACTA | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000373809.2; ENST00000487784.1; ENST00000395284.7; ENST00000519078.6; ENST00000316629.8; ENST00000448257.6; ENST00000614696.4 | ||
| External Link | RMBase: m6A_site_102007 | ||
| mod ID: M6ASITE000068 | Click to Show/Hide the Full List | ||
| mod site | chr10:60787818-60787819:+ | [8] | |
| Sequence | GGGACCAAAAATGTTTAAAGACTAAATGTGTTCACATTACT | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000316629.8; ENST00000614696.4; ENST00000519078.6; ENST00000395284.7; ENST00000487784.1; ENST00000373809.2; ENST00000448257.6 | ||
| External Link | RMBase: m6A_site_102008 | ||
| mod ID: M6ASITE000069 | Click to Show/Hide the Full List | ||
| mod site | chr10:60788123-60788124:+ | [8] | |
| Sequence | TAGAAGAGTTCTTCACAGAGACTTAAAACCTCAAAATCTCT | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000487784.1; ENST00000448257.6; ENST00000614696.4; ENST00000395284.7; ENST00000316629.8; ENST00000519078.6; ENST00000373809.2 | ||
| External Link | RMBase: m6A_site_102009 | ||
| mod ID: M6ASITE000070 | Click to Show/Hide the Full List | ||
| mod site | chr10:60788130-60788131:+ | [8] | |
| Sequence | GTTCTTCACAGAGACTTAAAACCTCAAAATCTCTTGATTGA | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000373809.2; ENST00000316629.8; ENST00000448257.6; ENST00000519078.6; ENST00000395284.7; ENST00000487784.1; ENST00000614696.4 | ||
| External Link | RMBase: m6A_site_102010 | ||
| mod ID: M6ASITE000071 | Click to Show/Hide the Full List | ||
| mod site | chr10:60788153-60788154:+ | [9] | |
| Sequence | TCAAAATCTCTTGATTGATGACAAAGGAACAATTAAACTGG | ||
| Motif Score | 2.859755952 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000395284.7; ENST00000316629.8; ENST00000487784.1; ENST00000614696.4; ENST00000448257.6; ENST00000373809.2; ENST00000519078.6 | ||
| External Link | RMBase: m6A_site_102011 | ||
| mod ID: M6ASITE000072 | Click to Show/Hide the Full List | ||
| mod site | chr10:60788161-60788162:+ | [8] | |
| Sequence | TCTTGATTGATGACAAAGGAACAATTAAACTGGCTGATTTT | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa; hESC-HEK293T | ||
| Seq Type List | m6A-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000614696.4; ENST00000519078.6; ENST00000487784.1; ENST00000316629.8; ENST00000395284.7; ENST00000373809.2; ENST00000448257.6 | ||
| External Link | RMBase: m6A_site_102012 | ||
| mod ID: M6ASITE000073 | Click to Show/Hide the Full List | ||
| mod site | chr10:60788169-60788170:+ | [8] | |
| Sequence | GATGACAAAGGAACAATTAAACTGGCTGATTTTGGCCTTGC | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000395284.7; ENST00000448257.6; ENST00000373809.2; ENST00000487784.1; ENST00000614696.4; ENST00000519078.6; ENST00000316629.8 | ||
| External Link | RMBase: m6A_site_102013 | ||
| mod ID: M6ASITE000074 | Click to Show/Hide the Full List | ||
| mod site | chr10:60788221-60788222:+ | [7] | |
| Sequence | GAATACCTATCAGAGTATATACACATGAGGCAAGTGGAATA | ||
| Motif Score | 2.110482143 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000614696.4; ENST00000373809.2; ENST00000395284.7; ENST00000448257.6; ENST00000519078.6; ENST00000487784.1; ENST00000316629.8 | ||
| External Link | RMBase: m6A_site_102014 | ||
| mod ID: M6ASITE000075 | Click to Show/Hide the Full List | ||
| mod site | chr10:60791956-60791957:+ | [9] | |
| Sequence | TCGTTACTCAACTCCAGTTGACATTTGGAGTATAGGCACCA | ||
| Motif Score | 2.859755952 | ||
| Cell/Tissue List | HEK293T; hESC-HEK293T | ||
| Seq Type List | DART-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000519078.6; ENST00000614696.4; ENST00000316629.8; ENST00000373809.2; ENST00000395284.7; ENST00000448257.6 | ||
| External Link | RMBase: m6A_site_102015 | ||
| mod ID: M6ASITE000076 | Click to Show/Hide the Full List | ||
| mod site | chr10:60791987-60791988:+ | [8] | |
| Sequence | ATAGGCACCATATTTGCTGAACTAGCAACTAAGAAACCACT | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa; HEK293T | ||
| Seq Type List | m6A-seq; DART-seq | ||
| Transcript ID List | ENST00000373809.2; ENST00000316629.8; ENST00000448257.6; ENST00000614696.4; ENST00000395284.7 | ||
| External Link | RMBase: m6A_site_102016 | ||
| mod ID: M6ASITE000077 | Click to Show/Hide the Full List | ||
| mod site | chr10:60792002-60792003:+ | [8] | |
| Sequence | GCTGAACTAGCAACTAAGAAACCACTTTTCCATGGGGATTC | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000614696.4; ENST00000395284.7; ENST00000448257.6; ENST00000316629.8; ENST00000373809.2 | ||
| External Link | RMBase: m6A_site_102017 | ||
| mod ID: M6ASITE000078 | Click to Show/Hide the Full List | ||
| mod site | chr10:60792035-60792036:+ | [9] | |
| Sequence | GGGGATTCAGAAATTGATCAACTCTTCAGGATTTTCAGGTA | ||
| Motif Score | 2.595904762 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000448257.6; ENST00000373809.2; ENST00000395284.7; ENST00000316629.8; ENST00000614696.4 | ||
| External Link | RMBase: m6A_site_102018 | ||
| mod ID: M6ASITE000079 | Click to Show/Hide the Full List | ||
| mod site | chr10:60792200-60792201:+ | [8] | |
| Sequence | AGAAGTGGAATCTTTACAGGACTATAAGAATACATTTCCCA | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa; AML | ||
| Seq Type List | m6A-seq; miCLIP | ||
| Transcript ID List | ENST00000316629.8; ENST00000448257.6; ENST00000614696.4; ENST00000395284.7; ENST00000373809.2 | ||
| External Link | RMBase: m6A_site_102019 | ||
| mod ID: M6ASITE000080 | Click to Show/Hide the Full List | ||
| mod site | chr10:60792211-60792212:+ | [9] | |
| Sequence | CTTTACAGGACTATAAGAATACATTTCCCAAATGGAAACCA | ||
| Motif Score | 2.110482143 | ||
| Cell/Tissue List | HEK293T; hESC-HEK293T | ||
| Seq Type List | DART-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000373809.2; ENST00000395284.7; ENST00000614696.4; ENST00000448257.6; ENST00000316629.8 | ||
| External Link | RMBase: m6A_site_102020 | ||
| mod ID: M6ASITE000081 | Click to Show/Hide the Full List | ||
| mod site | chr10:60792228-60792229:+ | [8] | |
| Sequence | AATACATTTCCCAAATGGAAACCAGGAAGCCTAGCATCCCA | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000395284.7; ENST00000614696.4; ENST00000316629.8; ENST00000448257.6; ENST00000373809.2 | ||
| External Link | RMBase: m6A_site_102021 | ||
| mod ID: M6ASITE000082 | Click to Show/Hide the Full List | ||
| mod site | chr10:60792257-60792258:+ | [8] | |
| Sequence | CCTAGCATCCCATGTCAAAAACTTGGATGAAAATGGCTTGG | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000316629.8; ENST00000614696.4; ENST00000448257.6; ENST00000395284.7; ENST00000373809.2 | ||
| External Link | RMBase: m6A_site_102022 | ||
| mod ID: M6ASITE000083 | Click to Show/Hide the Full List | ||
| mod site | chr10:60793952-60793953:+ | [8] | |
| Sequence | TCCATATTTTAATGATTTGGACAATCAGATTAAGAAGATGT | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; hESC-HEK293T | ||
| Seq Type List | m6A-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000316629.8; ENST00000614696.4; ENST00000448257.6; ENST00000395284.7; ENST00000373809.2 | ||
| External Link | RMBase: m6A_site_102023 | ||
| mod ID: M6ASITE000084 | Click to Show/Hide the Full List | ||
| mod site | chr10:60793982-60793983:+ | [9] | |
| Sequence | TAAGAAGATGTAGCTTTCTGACAAAAAGTTTCCATATGTTA | ||
| Motif Score | 2.859755952 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000614696.4; ENST00000395284.7; ENST00000316629.8; ENST00000448257.6; ENST00000373809.2 | ||
| External Link | RMBase: m6A_site_102024 | ||
| mod ID: M6ASITE000085 | Click to Show/Hide the Full List | ||
| mod site | chr10:60794008-60794009:+ | [9] | |
| Sequence | AGTTTCCATATGTTATATCAACAGATAGTTGTGTTTTTATT | ||
| Motif Score | 2.173910714 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000395284.7; ENST00000316629.8; ENST00000373809.2; ENST00000614696.4; ENST00000448257.6 | ||
| External Link | RMBase: m6A_site_102025 | ||
| mod ID: M6ASITE000086 | Click to Show/Hide the Full List | ||
| mod site | chr10:60794033-60794034:+ | [9] | |
| Sequence | TAGTTGTGTTTTTATTGTTAACTCTTGTCTATTTTTGTCTT | ||
| Motif Score | 2.590089286 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000614696.4; ENST00000395284.7; ENST00000316629.8; ENST00000373809.2; ENST00000448257.6 | ||
| External Link | RMBase: m6A_site_102026 | ||
| mod ID: M6ASITE000087 | Click to Show/Hide the Full List | ||
| mod site | chr10:60794076-60794077:+ | [9] | |
| Sequence | ATATATTTCTTTGTTATCAAACTTCAGCTGTACTTCGTCTT | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000448257.6; ENST00000395284.7; ENST00000614696.4; ENST00000316629.8; ENST00000373809.2 | ||
| External Link | RMBase: m6A_site_102027 | ||
| mod ID: M6ASITE000088 | Click to Show/Hide the Full List | ||
| mod site | chr10:60794087-60794088:+ | [9] | |
| Sequence | TGTTATCAAACTTCAGCTGTACTTCGTCTTCTAATTTCAAA | ||
| Motif Score | 3.278136905 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000614696.4; ENST00000316629.8; ENST00000373809.2; ENST00000395284.7; ENST00000448257.6 | ||
| External Link | RMBase: m6A_site_102028 | ||
| mod ID: M6ASITE000089 | Click to Show/Hide the Full List | ||
| mod site | chr10:60794114-60794115:+ | [9] | |
| Sequence | CTTCTAATTTCAAAAATATAACTTAAAAATGTAAATATTCT | ||
| Motif Score | 2.590089286 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000373809.2; ENST00000448257.6; ENST00000395284.7; ENST00000614696.4; ENST00000316629.8 | ||
| External Link | RMBase: m6A_site_102029 | ||
| mod ID: M6ASITE000090 | Click to Show/Hide the Full List | ||
| mod site | chr10:60794182-60794183:+ | [7] | |
| Sequence | TGTGTGTAGGTCTCACTGTAACAACTATTTGTTACTATAAT | ||
| Motif Score | 2.168095238 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000448257.6; ENST00000316629.8; ENST00000395284.7; ENST00000373809.2; ENST00000614696.4 | ||
| External Link | RMBase: m6A_site_102030 | ||
| mod ID: M6ASITE000091 | Click to Show/Hide the Full List | ||
| mod site | chr10:60794300-60794301:+ | [9] | |
| Sequence | ATTTTCCTGTAGTTGGAACTACTAAAATTTAGGAAAATGCT | ||
| Motif Score | 2.500660714 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000373809.2; ENST00000316629.8; ENST00000448257.6; ENST00000614696.4; ENST00000395284.7 | ||
| External Link | RMBase: m6A_site_102031 | ||
| mod ID: M6ASITE000092 | Click to Show/Hide the Full List | ||
| mod site | chr10:60794403-60794404:+ | [9] | |
| Sequence | AGTTTCTTGCCATGTTGTTAACTATACAACCTGGCTAAAGA | ||
| Motif Score | 2.590089286 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000614696.4; ENST00000448257.6; ENST00000316629.8; ENST00000373809.2; ENST00000395284.7 | ||
| External Link | RMBase: m6A_site_102032 | ||
| mod ID: M6ASITE000093 | Click to Show/Hide the Full List | ||
| mod site | chr10:60794408-60794409:+ | [7] | |
| Sequence | CTTGCCATGTTGTTAACTATACAACCTGGCTAAAGATGAAT | ||
| Motif Score | 2.110482143 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000316629.8; ENST00000614696.4; ENST00000448257.6; ENST00000373809.2; ENST00000395284.7 | ||
| External Link | RMBase: m6A_site_102033 | ||
| mod ID: M6ASITE000094 | Click to Show/Hide the Full List | ||
| mod site | chr10:60794456-60794457:+ | [9] | |
| Sequence | TACTGGTATTTTAATTTTTGACCTAAATGTTTAAGCATTCG | ||
| Motif Score | 2.839113095 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000448257.6; ENST00000614696.4; ENST00000373809.2; ENST00000395284.7; ENST00000316629.8 | ||
| External Link | RMBase: m6A_site_102034 | ||
| mod ID: M6ASITE000095 | Click to Show/Hide the Full List | ||
| mod site | chr10:60794487-60794488:+ | [8] | |
| Sequence | TAAGCATTCGGAATGAGAAAACTATACAGATTTGAGAAATG | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000316629.8; ENST00000448257.6; ENST00000373809.2; ENST00000614696.4; ENST00000395284.7 | ||
| External Link | RMBase: m6A_site_102035 | ||
| mod ID: M6ASITE000096 | Click to Show/Hide the Full List | ||
| mod site | chr10:60794548-60794549:+ | [7] | |
| Sequence | TTCAGTAACTTAAAAAGCTAACATGAGAGCATGCCAAAATT | ||
| Motif Score | 2.168095238 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000373809.2; ENST00000316629.8; ENST00000614696.4; ENST00000448257.6; ENST00000395284.7 | ||
| External Link | RMBase: m6A_site_102036 | ||
| mod ID: M6ASITE000097 | Click to Show/Hide the Full List | ||
| mod site | chr10:60794580-60794581:+ | [9] | |
| Sequence | GCCAAAATTTGCTAAGTCTTACAAAGATCAAGGGCTGTCCG | ||
| Motif Score | 2.07285119 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000373809.2; ENST00000614696.4; ENST00000316629.8; ENST00000448257.6; ENST00000395284.7 | ||
| External Link | RMBase: m6A_site_102037 | ||
| mod ID: M6ASITE000098 | Click to Show/Hide the Full List | ||
| mod site | chr10:60794613-60794614:+ | [8] | |
| Sequence | GCTGTCCGCAACAGGGAAGAACAGTTTTGAAAATTTATGAA | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000316629.8; ENST00000373809.2; ENST00000448257.6; ENST00000614696.4; ENST00000395284.7 | ||
| External Link | RMBase: m6A_site_102038 | ||
| mod ID: M6ASITE000099 | Click to Show/Hide the Full List | ||
| mod site | chr10:60794633-60794634:+ | [8] | |
| Sequence | ACAGTTTTGAAAATTTATGAACTATCTTATTTTTAGGTAGG | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa; HEK293T | ||
| Seq Type List | m6A-seq; DART-seq | ||
| Transcript ID List | ENST00000448257.6; ENST00000373809.2; ENST00000395284.7; ENST00000614696.4; ENST00000316629.8 | ||
| External Link | RMBase: m6A_site_102039 | ||
| mod ID: M6ASITE000100 | Click to Show/Hide the Full List | ||
| mod site | chr10:60794705-60794706:+ | [9] | |
| Sequence | ATGCCTTGGTCAGAGTAATAACTGAAGGAGTTGCTTATCTT | ||
| Motif Score | 2.590089286 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000373809.2; ENST00000316629.8; ENST00000395284.7; ENST00000614696.4; ENST00000448257.6 | ||
| External Link | RMBase: m6A_site_102040 | ||
| mod ID: M6ASITE000101 | Click to Show/Hide the Full List | ||
| mod site | chr10:60794748-60794749:+ | [8] | |
| Sequence | CTTTCGAGTCTGAGTTTAAAACTACACATTTTGACATAGTG | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa; HEK293T | ||
| Seq Type List | m6A-seq; DART-seq | ||
| Transcript ID List | ENST00000395284.7; ENST00000448257.6; ENST00000373809.2; ENST00000614696.4; ENST00000316629.8 | ||
| External Link | RMBase: m6A_site_102041 | ||
| mod ID: M6ASITE000102 | Click to Show/Hide the Full List | ||
| mod site | chr10:60794751-60794752:+ | [7] | |
| Sequence | TCGAGTCTGAGTTTAAAACTACACATTTTGACATAGTGTTT | ||
| Motif Score | 2.078666667 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000448257.6; ENST00000316629.8; ENST00000614696.4; ENST00000395284.7; ENST00000373809.2 | ||
| External Link | RMBase: m6A_site_102042 | ||
| mod ID: M6ASITE000103 | Click to Show/Hide the Full List | ||
| mod site | chr10:60794761-60794762:+ | [9] | |
| Sequence | GTTTAAAACTACACATTTTGACATAGTGTTTATTAGCAGCC | ||
| Motif Score | 2.859755952 | ||
| Cell/Tissue List | HEK293T; hESC-HEK293T | ||
| Seq Type List | DART-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000316629.8; ENST00000614696.4; ENST00000395284.7; ENST00000448257.6; ENST00000373809.2 | ||
| External Link | RMBase: m6A_site_102043 | ||
| mod ID: M6ASITE000104 | Click to Show/Hide the Full List | ||
| mod site | chr10:60794809-60794810:+ | [9] | |
| Sequence | AAGGCTCTAATGTATATTTAACTAAAATTACTAGCTTTGGG | ||
| Motif Score | 2.590089286 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000448257.6; ENST00000614696.4; ENST00000395284.7; ENST00000316629.8; ENST00000373809.2 | ||
| External Link | RMBase: m6A_site_102044 | ||
| mod ID: M6ASITE000105 | Click to Show/Hide the Full List | ||
| mod site | chr10:60794818-60794819:+ | [9] | |
| Sequence | ATGTATATTTAACTAAAATTACTAGCTTTGGGAATTAAACT | ||
| Motif Score | 2.494845238 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000373809.2; ENST00000316629.8; ENST00000614696.4; ENST00000448257.6; ENST00000395284.7 | ||
| External Link | RMBase: m6A_site_102045 | ||
| mod ID: M6ASITE000106 | Click to Show/Hide the Full List | ||
| mod site | chr10:60794836-60794837:+ | [8] | |
| Sequence | TTACTAGCTTTGGGAATTAAACTGTTTAACAAATAATGCTG | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000614696.4; ENST00000316629.8; ENST00000373809.2; ENST00000395284.7 | ||
| External Link | RMBase: m6A_site_102046 | ||
| mod ID: M6ASITE000107 | Click to Show/Hide the Full List | ||
| mod site | chr10:60794844-60794845:+ | [7] | |
| Sequence | TTTGGGAATTAAACTGTTTAACAAATAATGCTGCTCATTGT | ||
| Motif Score | 2.168095238 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000373809.2; ENST00000395284.7 | ||
| External Link | RMBase: m6A_site_102047 | ||
References