m6A Target Gene Information
General Information of the m6A Target Gene (ID: M6ATAR00119)
Full List of m6A Methylation Regulator of This Target Gene and Corresponding Disease/Drug Response(s)
MIR370
can be regulated by the following regulator(s), and cause disease/drug response(s). You can browse detail information of regulator(s) or disease/drug response(s).
Browse Regulator
Browse Disease
Methyltransferase-like 3 (METTL3) [WRITER]
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | Embryonic stem cells | Mus musculus |
|
Treatment: METTL3 knockout mESCs
Control: Wild type mESCs
|
GSE156481 | |
| Regulation |
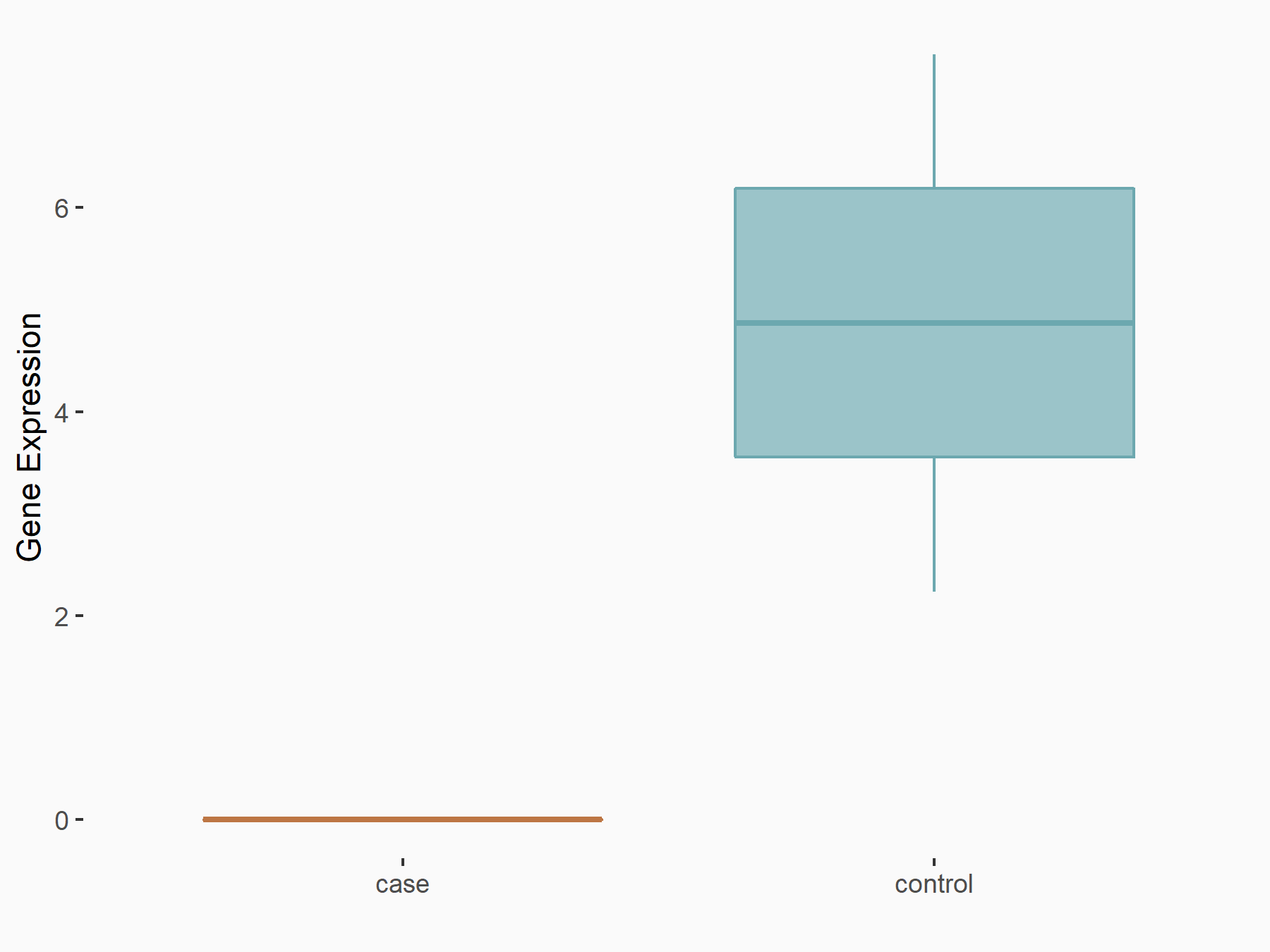 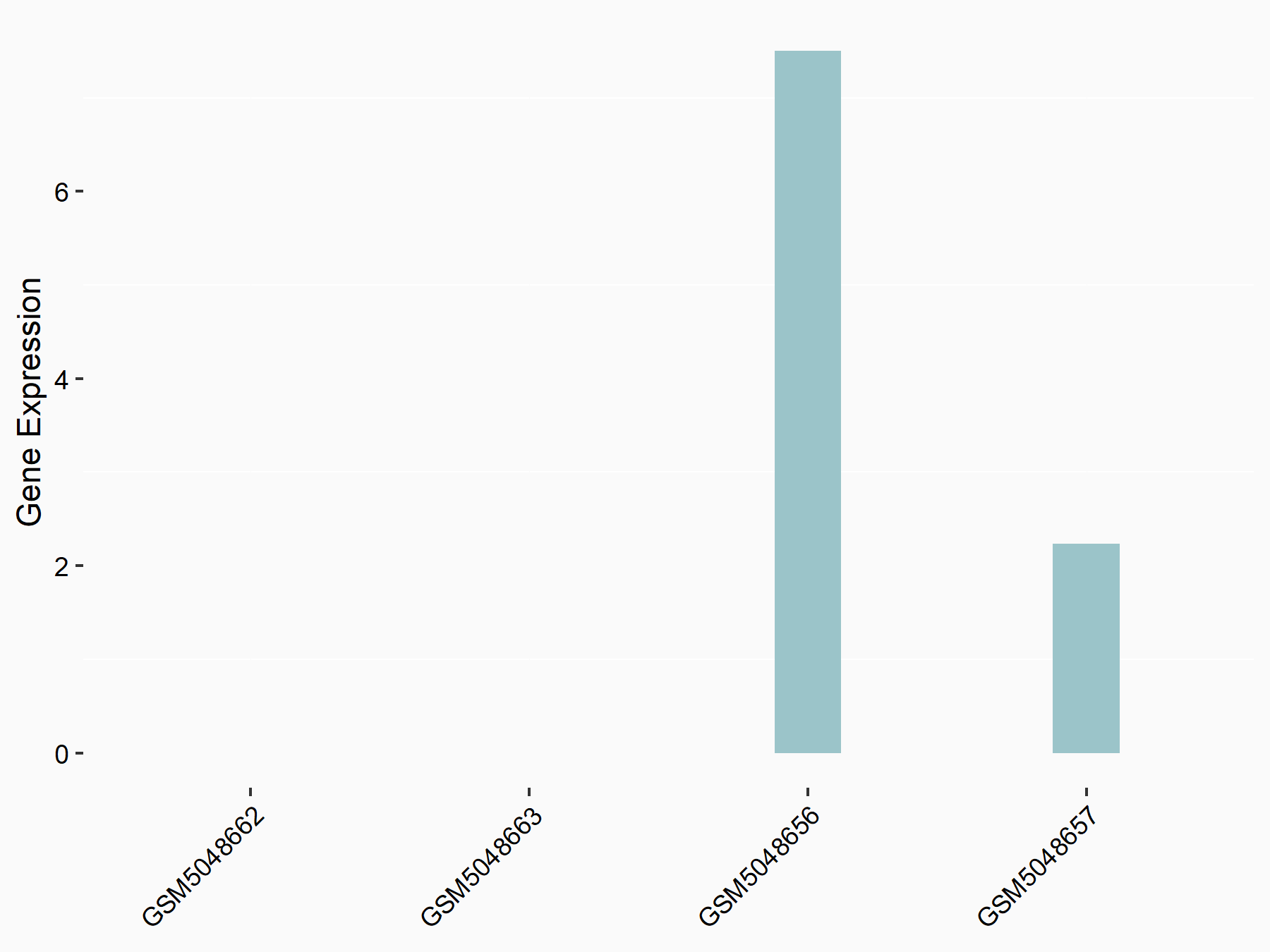 |
logFC: -4.78E+00 p-value: 3.06E-02 |
| More Results | Click to View More RNA-seq Results | |
| In total 1 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [1] | |||
| Response Summary | In hepatocellular carcinoma, METTL3 could direct the formation of circHPS5, and specific m6A controlled the accumulation of circHPS5. YTHDC1 facilitated the cytoplasmic output of circHPS5 under m6A modification. CircHPS5 can act as a microRNA 370 (MIR370) sponge to regulate the expression of HMGA2 and further accelerate hepatocellular carcinoma cell tumorigenesis. | |||
| Responsed Disease | Hepatocellular carcinoma | ICD-11: 2C12.02 | ||
| Pathway Response | Transcriptional misregulation in cancer | hsa05202 | ||
| Cell Process | Epithelial-mesenchymal transition | |||
| Cell autophagy | ||||
| In-vitro Model | Hep 3B2.1-7 | Childhood hepatocellular carcinoma | Homo sapiens | CVCL_0326 |
| L-02 | Endocervical adenocarcinoma | Homo sapiens | CVCL_6926 | |
| In-vivo Model | To create the xenograft neoplasm system, 40 male BALB/c nude mice aged 5 weeks were randomly separated into sh-NC, sh-circHPS5, sh-circHPS5+CTRL, and sh-circHPS5+SAH groups (n = 5 for each group). HCC cells were subcutaneously injected into the axilla of the nude mice. | |||
YTH domain-containing protein 1 (YTHDC1) [READER]
| In total 1 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [1] | |||
| Response Summary | In hepatocellular carcinoma, METTL3 could direct the formation of circHPS5, and specific m6A controlled the accumulation of circHPS5. YTHDC1 facilitated the cytoplasmic output of circHPS5 under m6A modification. CircHPS5 can act as a microRNA 370 (MIR370) sponge to regulate the expression of HMGA2 and further accelerate hepatocellular carcinoma cell tumorigenesis. | |||
| Responsed Disease | Hepatocellular carcinoma | ICD-11: 2C12.02 | ||
| Pathway Response | Transcriptional misregulation in cancer | hsa05202 | ||
| Cell Process | Epithelial-mesenchymal transition | |||
| Cell autophagy | ||||
| In-vitro Model | Hep 3B2.1-7 | Childhood hepatocellular carcinoma | Homo sapiens | CVCL_0326 |
| L-02 | Endocervical adenocarcinoma | Homo sapiens | CVCL_6926 | |
| In-vivo Model | To create the xenograft neoplasm system, 40 male BALB/c nude mice aged 5 weeks were randomly separated into sh-NC, sh-circHPS5, sh-circHPS5+CTRL, and sh-circHPS5+SAH groups (n = 5 for each group). HCC cells were subcutaneously injected into the axilla of the nude mice. | |||
Liver cancer [ICD-11: 2C12]
| In total 2 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [1] | |||
| Response Summary | In hepatocellular carcinoma, METTL3 could direct the formation of circHPS5, and specific m6A controlled the accumulation of circHPS5. YTHDC1 facilitated the cytoplasmic output of circHPS5 under m6A modification. CircHPS5 can act as a microRNA 370 (MIR370) sponge to regulate the expression of HMGA2 and further accelerate hepatocellular carcinoma cell tumorigenesis. | |||
| Responsed Disease | Hepatocellular carcinoma [ICD-11: 2C12.02] | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Pathway Response | Transcriptional misregulation in cancer | hsa05202 | ||
| Cell Process | Epithelial-mesenchymal transition | |||
| Cell autophagy | ||||
| In-vitro Model | Hep 3B2.1-7 | Childhood hepatocellular carcinoma | Homo sapiens | CVCL_0326 |
| L-02 | Endocervical adenocarcinoma | Homo sapiens | CVCL_6926 | |
| In-vivo Model | To create the xenograft neoplasm system, 40 male BALB/c nude mice aged 5 weeks were randomly separated into sh-NC, sh-circHPS5, sh-circHPS5+CTRL, and sh-circHPS5+SAH groups (n = 5 for each group). HCC cells were subcutaneously injected into the axilla of the nude mice. | |||
| Experiment 2 Reporting the m6A-centered Disease Response | [1] | |||
| Response Summary | In hepatocellular carcinoma, METTL3 could direct the formation of circHPS5, and specific m6A controlled the accumulation of circHPS5. YTHDC1 facilitated the cytoplasmic output of circHPS5 under m6A modification. CircHPS5 can act as a microRNA 370 (MIR370) sponge to regulate the expression of HMGA2 and further accelerate hepatocellular carcinoma cell tumorigenesis. | |||
| Responsed Disease | Hepatocellular carcinoma [ICD-11: 2C12.02] | |||
| Target Regulator | YTH domain-containing protein 1 (YTHDC1) | READER | ||
| Pathway Response | Transcriptional misregulation in cancer | hsa05202 | ||
| Cell Process | Epithelial-mesenchymal transition | |||
| Cell autophagy | ||||
| In-vitro Model | Hep 3B2.1-7 | Childhood hepatocellular carcinoma | Homo sapiens | CVCL_0326 |
| L-02 | Endocervical adenocarcinoma | Homo sapiens | CVCL_6926 | |
| In-vivo Model | To create the xenograft neoplasm system, 40 male BALB/c nude mice aged 5 weeks were randomly separated into sh-NC, sh-circHPS5, sh-circHPS5+CTRL, and sh-circHPS5+SAH groups (n = 5 for each group). HCC cells were subcutaneously injected into the axilla of the nude mice. | |||
Full List of Crosstalk(s) between m6A Modification and Epigenetic Regulation Related to This Regulator
Histone modification
m6A Regulator: YTH domain-containing protein 1 (YTHDC1)
| In total 1 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT03484 | ||
| Epigenetic Regulator | Histone acetyltransferase p300 (P300) | |
| Regulated Target | Histone H3 lysine 18 lactylation (H3K18la) | |
| Crosstalk relationship | Histone modification → m6A | |
| Disease | Liver cancer | |
Non-coding RNA
m6A Regulator: Methyltransferase-like 3 (METTL3)
| In total 1 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT05534 | ||
| Epigenetic Regulator | MicroRNA 370 (MIR370) | |
| Regulated Target | High mobility group protein HMGI-C (HMGA2) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Liver cancer | |
m6A Regulator: YTH domain-containing protein 1 (YTHDC1)
| In total 1 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT05535 | ||
| Epigenetic Regulator | MicroRNA 370 (MIR370) | |
| Regulated Target | High mobility group protein HMGI-C (HMGA2) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Liver cancer | |