m6A Target Gene Information
General Information of the m6A Target Gene (ID: M6ATAR00560)
Full List of m6A Methylation Regulator of This Target Gene and Corresponding Disease/Drug Response(s)
E2F8
can be regulated by the following regulator(s), and cause disease/drug response(s). You can browse detail information of regulator(s) or disease/drug response(s).
Browse Regulator
Browse Disease
Browse Drug
YTH domain-containing family protein 1 (YTHDF1) [READER]
| Representative RNA-seq result indicating the expression of this target gene regulated by YTHDF1 | ||
| Cell Line | AGS cell line | Homo sapiens |
|
Treatment: shYTHDF1 AGS
Control: shControl AGS
|
GSE159425 | |
| Regulation |
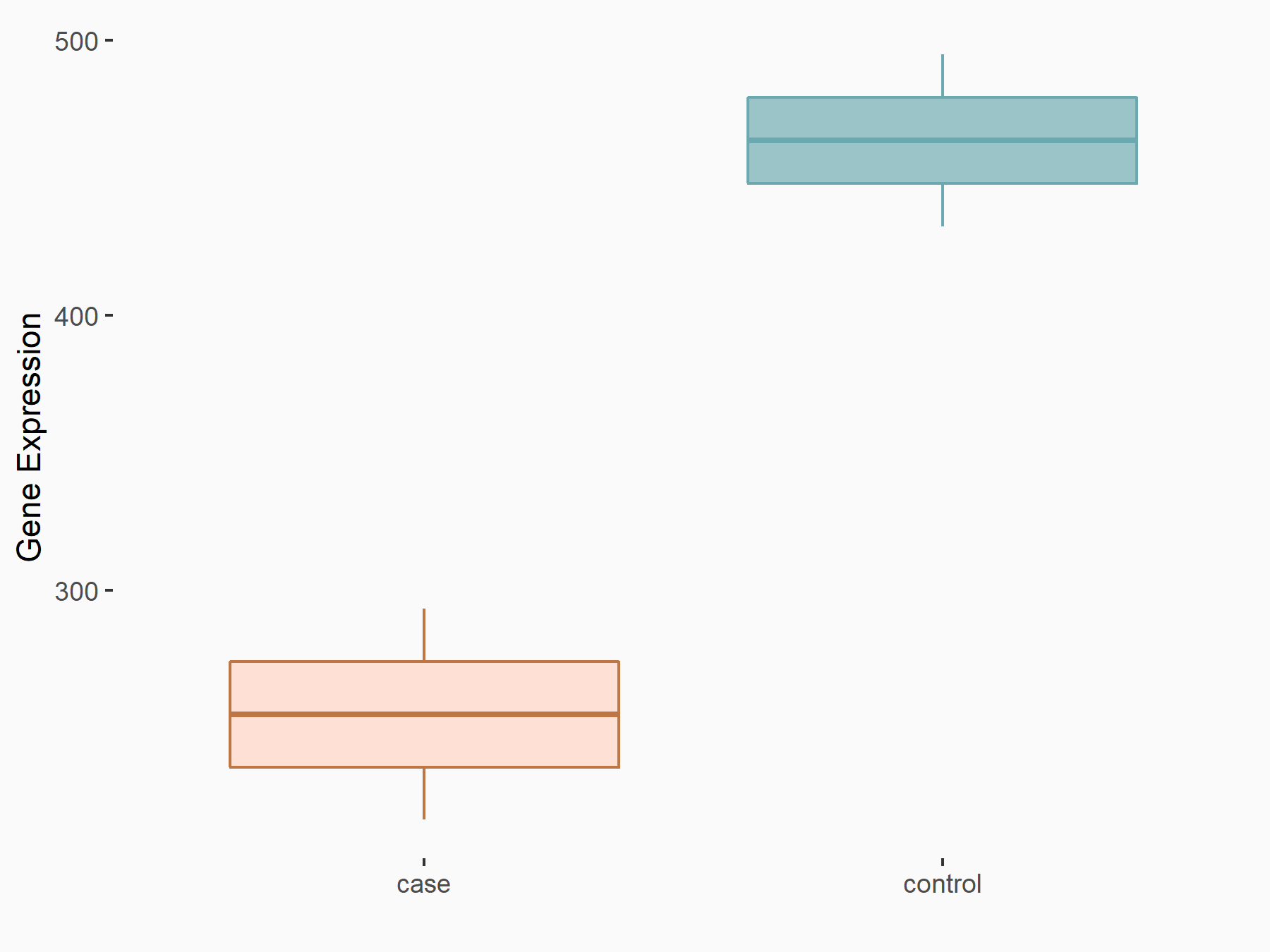  |
logFC: -8.62E-01 p-value: 7.08E-04 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between E2F8 and the regulator | ||
| Cell Line | Hela | Homo sapiens |
| Regulation | logFC: 1.18E+00 | GSE63591 |
| In total 3 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [1] | |||
| Response Summary | In breast cancer, accordingly YTHDF1 knockdown sensitizes breast cancer cells to Adriamycin and Cisplatin as well as Olaparib, a PARP inhibitor. Transcription factor E2F8 (E2F8) is a target molecule by YTHDF1 which modulates E2F8 mRNA stability and DNA damage repair in a METTL14-dependent manner. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Breast cancer | ICD-11: 2C60 | ||
| Responsed Drug | Cisplatin | Approved | ||
| Pathway Response | Nucleotide excision repair | hsa03420 | ||
| Cell Process | RNA stability | |||
| In-vitro Model | MDA-MB-231 | Breast adenocarcinoma | Homo sapiens | CVCL_0062 |
| MCF-7 | Invasive breast carcinoma | Homo sapiens | CVCL_0031 | |
| Hs 578T | Invasive breast carcinoma | Homo sapiens | CVCL_0332 | |
| In-vivo Model | 1×106 MDA-MB-231 cells were resuspended in 100 uL PBS with 50% Matrigel (Corning Costar, USA), and injected into the mammary fat pad of the mice. | |||
| Experiment 2 Reporting the m6A Methylation Regulator of This Target Gene | [1] | |||
| Response Summary | In breast cancer, accordingly YTHDF1 knockdown sensitizes breast cancer cells to Adriamycin and Cisplatin as well as Olaparib, a PARP inhibitor. Transcription factor E2F8 (E2F8) is a target molecule by YTHDF1 which modulates E2F8 mRNA stability and DNA damage repair in a METTL14-dependent manner. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Breast cancer | ICD-11: 2C60 | ||
| Responsed Drug | Doxil | Approved | ||
| Pathway Response | Nucleotide excision repair | hsa03420 | ||
| Cell Process | RNA stability | |||
| In-vitro Model | MDA-MB-231 | Breast adenocarcinoma | Homo sapiens | CVCL_0062 |
| MCF-7 | Invasive breast carcinoma | Homo sapiens | CVCL_0031 | |
| Hs 578T | Invasive breast carcinoma | Homo sapiens | CVCL_0332 | |
| In-vivo Model | 1×106 MDA-MB-231 cells were resuspended in 100 uL PBS with 50% Matrigel (Corning Costar, USA), and injected into the mammary fat pad of the mice. | |||
| Experiment 3 Reporting the m6A Methylation Regulator of This Target Gene | [1] | |||
| Response Summary | In breast cancer, accordingly YTHDF1 knockdown sensitizes breast cancer cells to Adriamycin and Cisplatin as well as Olaparib, a PARP inhibitor. Transcription factor E2F8 (E2F8) is a target molecule by YTHDF1 which modulates E2F8 mRNA stability and DNA damage repair in a METTL14-dependent manner. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Breast cancer | ICD-11: 2C60 | ||
| Responsed Drug | Olaparib | Approved | ||
| Pathway Response | Nucleotide excision repair | hsa03420 | ||
| Cell Process | RNA stability | |||
| In-vitro Model | MDA-MB-231 | Breast adenocarcinoma | Homo sapiens | CVCL_0062 |
| MCF-7 | Invasive breast carcinoma | Homo sapiens | CVCL_0031 | |
| Hs 578T | Invasive breast carcinoma | Homo sapiens | CVCL_0332 | |
| In-vivo Model | 1×106 MDA-MB-231 cells were resuspended in 100 uL PBS with 50% Matrigel (Corning Costar, USA), and injected into the mammary fat pad of the mice. | |||
Methyltransferase-like 14 (METTL14) [WRITER]
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL14 | ||
| Cell Line | MDA-MB-231 | Homo sapiens |
|
Treatment: siMETTL14 MDA-MB-231 cells
Control: MDA-MB-231 cells
|
GSE81164 | |
| Regulation |
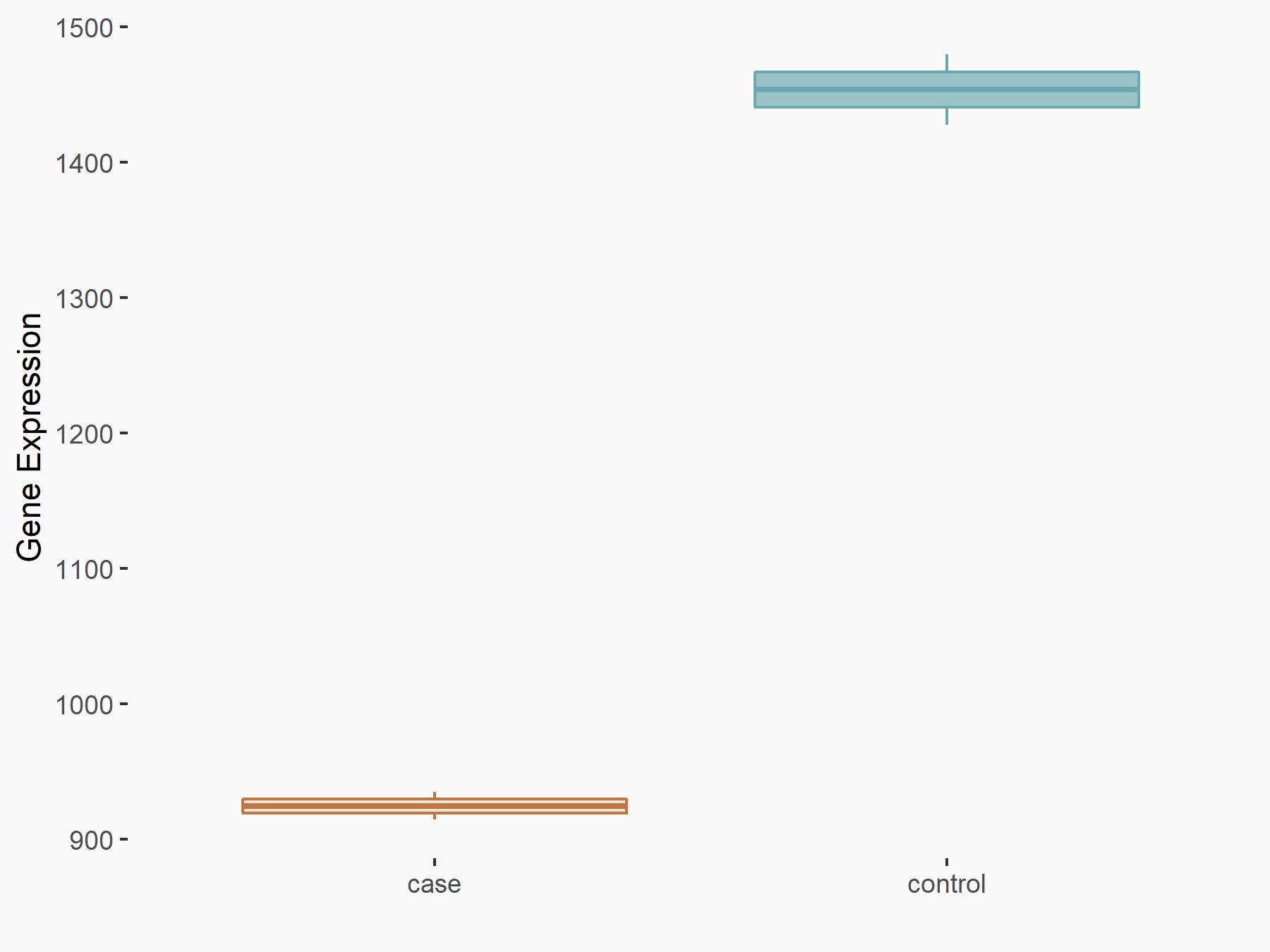  |
logFC: -6.53E-01 p-value: 4.62E-05 |
| More Results | Click to View More RNA-seq Results | |
| In total 3 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [1] | |||
| Response Summary | In breast cancer, accordingly YTHDF1 knockdown sensitizes breast cancer cells to Adriamycin and Cisplatin as well as Olaparib, a PARP inhibitor. Transcription factor E2F8 (E2F8) is a target molecule by YTHDF1 which modulates E2F8 mRNA stability and DNA damage repair in a METTL14-dependent manner. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Breast cancer | ICD-11: 2C60 | ||
| Responsed Drug | Cisplatin | Approved | ||
| Pathway Response | Nucleotide excision repair | hsa03420 | ||
| Cell Process | RNA stability | |||
| In-vitro Model | MDA-MB-231 | Breast adenocarcinoma | Homo sapiens | CVCL_0062 |
| MCF-7 | Invasive breast carcinoma | Homo sapiens | CVCL_0031 | |
| Hs 578T | Invasive breast carcinoma | Homo sapiens | CVCL_0332 | |
| In-vivo Model | 1×106 MDA-MB-231 cells were resuspended in 100 uL PBS with 50% Matrigel (Corning Costar, USA), and injected into the mammary fat pad of the mice. | |||
| Experiment 2 Reporting the m6A Methylation Regulator of This Target Gene | [1] | |||
| Response Summary | In breast cancer, accordingly YTHDF1 knockdown sensitizes breast cancer cells to Adriamycin and Cisplatin as well as Olaparib, a PARP inhibitor. Transcription factor E2F8 (E2F8) is a target molecule by YTHDF1 which modulates E2F8 mRNA stability and DNA damage repair in a METTL14-dependent manner. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Breast cancer | ICD-11: 2C60 | ||
| Responsed Drug | Doxil | Approved | ||
| Pathway Response | Nucleotide excision repair | hsa03420 | ||
| Cell Process | RNA stability | |||
| In-vitro Model | MDA-MB-231 | Breast adenocarcinoma | Homo sapiens | CVCL_0062 |
| MCF-7 | Invasive breast carcinoma | Homo sapiens | CVCL_0031 | |
| Hs 578T | Invasive breast carcinoma | Homo sapiens | CVCL_0332 | |
| In-vivo Model | 1×106 MDA-MB-231 cells were resuspended in 100 uL PBS with 50% Matrigel (Corning Costar, USA), and injected into the mammary fat pad of the mice. | |||
| Experiment 3 Reporting the m6A Methylation Regulator of This Target Gene | [1] | |||
| Response Summary | In breast cancer, accordingly YTHDF1 knockdown sensitizes breast cancer cells to Adriamycin and Cisplatin as well as Olaparib, a PARP inhibitor. Transcription factor E2F8 (E2F8) is a target molecule by YTHDF1 which modulates E2F8 mRNA stability and DNA damage repair in a METTL14-dependent manner. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Breast cancer | ICD-11: 2C60 | ||
| Responsed Drug | Olaparib | Approved | ||
| Pathway Response | Nucleotide excision repair | hsa03420 | ||
| Cell Process | RNA stability | |||
| In-vitro Model | MDA-MB-231 | Breast adenocarcinoma | Homo sapiens | CVCL_0062 |
| MCF-7 | Invasive breast carcinoma | Homo sapiens | CVCL_0031 | |
| Hs 578T | Invasive breast carcinoma | Homo sapiens | CVCL_0332 | |
| In-vivo Model | 1×106 MDA-MB-231 cells were resuspended in 100 uL PBS with 50% Matrigel (Corning Costar, USA), and injected into the mammary fat pad of the mice. | |||
Breast cancer [ICD-11: 2C60]
| In total 6 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [1] | |||
| Response Summary | In breast cancer, accordingly YTHDF1 knockdown sensitizes breast cancer cells to Adriamycin and Cisplatin as well as Olaparib, a PARP inhibitor. Transcription factor E2F8 (E2F8) is a target molecule by YTHDF1 which modulates E2F8 mRNA stability and DNA damage repair in a METTL14-dependent manner. | |||
| Responsed Disease | Breast cancer [ICD-11: 2C60] | |||
| Target Regulator | Methyltransferase-like 14 (METTL14) | WRITER | ||
| Target Regulation | Up regulation | |||
| Responsed Drug | Cisplatin | Approved | ||
| Pathway Response | Nucleotide excision repair | hsa03420 | ||
| Cell Process | RNA stability | |||
| In-vitro Model | MDA-MB-231 | Breast adenocarcinoma | Homo sapiens | CVCL_0062 |
| MCF-7 | Invasive breast carcinoma | Homo sapiens | CVCL_0031 | |
| Hs 578T | Invasive breast carcinoma | Homo sapiens | CVCL_0332 | |
| In-vivo Model | 1×106 MDA-MB-231 cells were resuspended in 100 uL PBS with 50% Matrigel (Corning Costar, USA), and injected into the mammary fat pad of the mice. | |||
| Experiment 2 Reporting the m6A-centered Disease Response | [1] | |||
| Response Summary | In breast cancer, accordingly YTHDF1 knockdown sensitizes breast cancer cells to Adriamycin and Cisplatin as well as Olaparib, a PARP inhibitor. Transcription factor E2F8 (E2F8) is a target molecule by YTHDF1 which modulates E2F8 mRNA stability and DNA damage repair in a METTL14-dependent manner. | |||
| Responsed Disease | Breast cancer [ICD-11: 2C60] | |||
| Target Regulator | Methyltransferase-like 14 (METTL14) | WRITER | ||
| Target Regulation | Up regulation | |||
| Responsed Drug | Doxil | Approved | ||
| Pathway Response | Nucleotide excision repair | hsa03420 | ||
| Cell Process | RNA stability | |||
| In-vitro Model | MDA-MB-231 | Breast adenocarcinoma | Homo sapiens | CVCL_0062 |
| MCF-7 | Invasive breast carcinoma | Homo sapiens | CVCL_0031 | |
| Hs 578T | Invasive breast carcinoma | Homo sapiens | CVCL_0332 | |
| In-vivo Model | 1×106 MDA-MB-231 cells were resuspended in 100 uL PBS with 50% Matrigel (Corning Costar, USA), and injected into the mammary fat pad of the mice. | |||
| Experiment 3 Reporting the m6A-centered Disease Response | [1] | |||
| Response Summary | In breast cancer, accordingly YTHDF1 knockdown sensitizes breast cancer cells to Adriamycin and Cisplatin as well as Olaparib, a PARP inhibitor. Transcription factor E2F8 (E2F8) is a target molecule by YTHDF1 which modulates E2F8 mRNA stability and DNA damage repair in a METTL14-dependent manner. | |||
| Responsed Disease | Breast cancer [ICD-11: 2C60] | |||
| Target Regulator | Methyltransferase-like 14 (METTL14) | WRITER | ||
| Target Regulation | Up regulation | |||
| Responsed Drug | Olaparib | Approved | ||
| Pathway Response | Nucleotide excision repair | hsa03420 | ||
| Cell Process | RNA stability | |||
| In-vitro Model | MDA-MB-231 | Breast adenocarcinoma | Homo sapiens | CVCL_0062 |
| MCF-7 | Invasive breast carcinoma | Homo sapiens | CVCL_0031 | |
| Hs 578T | Invasive breast carcinoma | Homo sapiens | CVCL_0332 | |
| In-vivo Model | 1×106 MDA-MB-231 cells were resuspended in 100 uL PBS with 50% Matrigel (Corning Costar, USA), and injected into the mammary fat pad of the mice. | |||
| Experiment 4 Reporting the m6A-centered Disease Response | [1] | |||
| Response Summary | In breast cancer, accordingly YTHDF1 knockdown sensitizes breast cancer cells to Adriamycin and Cisplatin as well as Olaparib, a PARP inhibitor. Transcription factor E2F8 (E2F8) is a target molecule by YTHDF1 which modulates E2F8 mRNA stability and DNA damage repair in a METTL14-dependent manner. | |||
| Responsed Disease | Breast cancer [ICD-11: 2C60] | |||
| Target Regulator | YTH domain-containing family protein 1 (YTHDF1) | READER | ||
| Target Regulation | Up regulation | |||
| Responsed Drug | Cisplatin | Approved | ||
| Pathway Response | Nucleotide excision repair | hsa03420 | ||
| Cell Process | RNA stability | |||
| In-vitro Model | MDA-MB-231 | Breast adenocarcinoma | Homo sapiens | CVCL_0062 |
| MCF-7 | Invasive breast carcinoma | Homo sapiens | CVCL_0031 | |
| Hs 578T | Invasive breast carcinoma | Homo sapiens | CVCL_0332 | |
| In-vivo Model | 1×106 MDA-MB-231 cells were resuspended in 100 uL PBS with 50% Matrigel (Corning Costar, USA), and injected into the mammary fat pad of the mice. | |||
| Experiment 5 Reporting the m6A-centered Disease Response | [1] | |||
| Response Summary | In breast cancer, accordingly YTHDF1 knockdown sensitizes breast cancer cells to Adriamycin and Cisplatin as well as Olaparib, a PARP inhibitor. Transcription factor E2F8 (E2F8) is a target molecule by YTHDF1 which modulates E2F8 mRNA stability and DNA damage repair in a METTL14-dependent manner. | |||
| Responsed Disease | Breast cancer [ICD-11: 2C60] | |||
| Target Regulator | YTH domain-containing family protein 1 (YTHDF1) | READER | ||
| Target Regulation | Up regulation | |||
| Responsed Drug | Doxil | Approved | ||
| Pathway Response | Nucleotide excision repair | hsa03420 | ||
| Cell Process | RNA stability | |||
| In-vitro Model | MDA-MB-231 | Breast adenocarcinoma | Homo sapiens | CVCL_0062 |
| MCF-7 | Invasive breast carcinoma | Homo sapiens | CVCL_0031 | |
| Hs 578T | Invasive breast carcinoma | Homo sapiens | CVCL_0332 | |
| In-vivo Model | 1×106 MDA-MB-231 cells were resuspended in 100 uL PBS with 50% Matrigel (Corning Costar, USA), and injected into the mammary fat pad of the mice. | |||
| Experiment 6 Reporting the m6A-centered Disease Response | [1] | |||
| Response Summary | In breast cancer, accordingly YTHDF1 knockdown sensitizes breast cancer cells to Adriamycin and Cisplatin as well as Olaparib, a PARP inhibitor. Transcription factor E2F8 (E2F8) is a target molecule by YTHDF1 which modulates E2F8 mRNA stability and DNA damage repair in a METTL14-dependent manner. | |||
| Responsed Disease | Breast cancer [ICD-11: 2C60] | |||
| Target Regulator | YTH domain-containing family protein 1 (YTHDF1) | READER | ||
| Target Regulation | Up regulation | |||
| Responsed Drug | Olaparib | Approved | ||
| Pathway Response | Nucleotide excision repair | hsa03420 | ||
| Cell Process | RNA stability | |||
| In-vitro Model | MDA-MB-231 | Breast adenocarcinoma | Homo sapiens | CVCL_0062 |
| MCF-7 | Invasive breast carcinoma | Homo sapiens | CVCL_0031 | |
| Hs 578T | Invasive breast carcinoma | Homo sapiens | CVCL_0332 | |
| In-vivo Model | 1×106 MDA-MB-231 cells were resuspended in 100 uL PBS with 50% Matrigel (Corning Costar, USA), and injected into the mammary fat pad of the mice. | |||
Cisplatin
[Approved]
| In total 2 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response | [1] | |||
| Response Summary | In breast cancer, accordingly YTHDF1 knockdown sensitizes breast cancer cells to Adriamycin and Cisplatin as well as Olaparib, a PARP inhibitor. Transcription factor E2F8 (E2F8) is a target molecule by YTHDF1 which modulates E2F8 mRNA stability and DNA damage repair in a METTL14-dependent manner. | |||
| Target Regulator | Methyltransferase-like 14 (METTL14) | WRITER | ||
| Target Regulation | Up regulation | |||
| Responsed Disease | Breast cancer | ICD-11: 2C60 | ||
| Pathway Response | Nucleotide excision repair | hsa03420 | ||
| Cell Process | RNA stability | |||
| In-vitro Model | MDA-MB-231 | Breast adenocarcinoma | Homo sapiens | CVCL_0062 |
| MCF-7 | Invasive breast carcinoma | Homo sapiens | CVCL_0031 | |
| Hs 578T | Invasive breast carcinoma | Homo sapiens | CVCL_0332 | |
| In-vivo Model | 1×106 MDA-MB-231 cells were resuspended in 100 uL PBS with 50% Matrigel (Corning Costar, USA), and injected into the mammary fat pad of the mice. | |||
| Experiment 2 Reporting the m6A-centered Drug Response | [1] | |||
| Response Summary | In breast cancer, accordingly YTHDF1 knockdown sensitizes breast cancer cells to Adriamycin and Cisplatin as well as Olaparib, a PARP inhibitor. Transcription factor E2F8 (E2F8) is a target molecule by YTHDF1 which modulates E2F8 mRNA stability and DNA damage repair in a METTL14-dependent manner. | |||
| Target Regulator | YTH domain-containing family protein 1 (YTHDF1) | READER | ||
| Target Regulation | Up regulation | |||
| Responsed Disease | Breast cancer | ICD-11: 2C60 | ||
| Pathway Response | Nucleotide excision repair | hsa03420 | ||
| Cell Process | RNA stability | |||
| In-vitro Model | MDA-MB-231 | Breast adenocarcinoma | Homo sapiens | CVCL_0062 |
| MCF-7 | Invasive breast carcinoma | Homo sapiens | CVCL_0031 | |
| Hs 578T | Invasive breast carcinoma | Homo sapiens | CVCL_0332 | |
| In-vivo Model | 1×106 MDA-MB-231 cells were resuspended in 100 uL PBS with 50% Matrigel (Corning Costar, USA), and injected into the mammary fat pad of the mice. | |||
Doxil
[Approved]
| In total 2 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response | [1] | |||
| Response Summary | In breast cancer, accordingly YTHDF1 knockdown sensitizes breast cancer cells to Adriamycin and Cisplatin as well as Olaparib, a PARP inhibitor. Transcription factor E2F8 (E2F8) is a target molecule by YTHDF1 which modulates E2F8 mRNA stability and DNA damage repair in a METTL14-dependent manner. | |||
| Target Regulator | Methyltransferase-like 14 (METTL14) | WRITER | ||
| Target Regulation | Up regulation | |||
| Responsed Disease | Breast cancer | ICD-11: 2C60 | ||
| Pathway Response | Nucleotide excision repair | hsa03420 | ||
| Cell Process | RNA stability | |||
| In-vitro Model | MDA-MB-231 | Breast adenocarcinoma | Homo sapiens | CVCL_0062 |
| MCF-7 | Invasive breast carcinoma | Homo sapiens | CVCL_0031 | |
| Hs 578T | Invasive breast carcinoma | Homo sapiens | CVCL_0332 | |
| In-vivo Model | 1×106 MDA-MB-231 cells were resuspended in 100 uL PBS with 50% Matrigel (Corning Costar, USA), and injected into the mammary fat pad of the mice. | |||
| Experiment 2 Reporting the m6A-centered Drug Response | [1] | |||
| Response Summary | In breast cancer, accordingly YTHDF1 knockdown sensitizes breast cancer cells to Adriamycin and Cisplatin as well as Olaparib, a PARP inhibitor. Transcription factor E2F8 (E2F8) is a target molecule by YTHDF1 which modulates E2F8 mRNA stability and DNA damage repair in a METTL14-dependent manner. | |||
| Target Regulator | YTH domain-containing family protein 1 (YTHDF1) | READER | ||
| Target Regulation | Up regulation | |||
| Responsed Disease | Breast cancer | ICD-11: 2C60 | ||
| Pathway Response | Nucleotide excision repair | hsa03420 | ||
| Cell Process | RNA stability | |||
| In-vitro Model | MDA-MB-231 | Breast adenocarcinoma | Homo sapiens | CVCL_0062 |
| MCF-7 | Invasive breast carcinoma | Homo sapiens | CVCL_0031 | |
| Hs 578T | Invasive breast carcinoma | Homo sapiens | CVCL_0332 | |
| In-vivo Model | 1×106 MDA-MB-231 cells were resuspended in 100 uL PBS with 50% Matrigel (Corning Costar, USA), and injected into the mammary fat pad of the mice. | |||
Olaparib
[Approved]
| In total 2 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response | [1] | |||
| Response Summary | In breast cancer, accordingly YTHDF1 knockdown sensitizes breast cancer cells to Adriamycin and Cisplatin as well as Olaparib, a PARP inhibitor. Transcription factor E2F8 (E2F8) is a target molecule by YTHDF1 which modulates E2F8 mRNA stability and DNA damage repair in a METTL14-dependent manner. | |||
| Target Regulator | Methyltransferase-like 14 (METTL14) | WRITER | ||
| Target Regulation | Up regulation | |||
| Responsed Disease | Breast cancer | ICD-11: 2C60 | ||
| Pathway Response | Nucleotide excision repair | hsa03420 | ||
| Cell Process | RNA stability | |||
| In-vitro Model | MDA-MB-231 | Breast adenocarcinoma | Homo sapiens | CVCL_0062 |
| MCF-7 | Invasive breast carcinoma | Homo sapiens | CVCL_0031 | |
| Hs 578T | Invasive breast carcinoma | Homo sapiens | CVCL_0332 | |
| In-vivo Model | 1×106 MDA-MB-231 cells were resuspended in 100 uL PBS with 50% Matrigel (Corning Costar, USA), and injected into the mammary fat pad of the mice. | |||
| Experiment 2 Reporting the m6A-centered Drug Response | [1] | |||
| Response Summary | In breast cancer, accordingly YTHDF1 knockdown sensitizes breast cancer cells to Adriamycin and Cisplatin as well as Olaparib, a PARP inhibitor. Transcription factor E2F8 (E2F8) is a target molecule by YTHDF1 which modulates E2F8 mRNA stability and DNA damage repair in a METTL14-dependent manner. | |||
| Target Regulator | YTH domain-containing family protein 1 (YTHDF1) | READER | ||
| Target Regulation | Up regulation | |||
| Responsed Disease | Breast cancer | ICD-11: 2C60 | ||
| Pathway Response | Nucleotide excision repair | hsa03420 | ||
| Cell Process | RNA stability | |||
| In-vitro Model | MDA-MB-231 | Breast adenocarcinoma | Homo sapiens | CVCL_0062 |
| MCF-7 | Invasive breast carcinoma | Homo sapiens | CVCL_0031 | |
| Hs 578T | Invasive breast carcinoma | Homo sapiens | CVCL_0332 | |
| In-vivo Model | 1×106 MDA-MB-231 cells were resuspended in 100 uL PBS with 50% Matrigel (Corning Costar, USA), and injected into the mammary fat pad of the mice. | |||
Full List of Crosstalk(s) between m6A Modification and Epigenetic Regulation Related to This Regulator
DNA modification
m6A Regulator: Methyltransferase-like 14 (METTL14)
| In total 9 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT02214 | ||
| Epigenetic Regulator | DNA (cytosine-5)-methyltransferase 3B (DNMT3B) | |
| Regulated Target | Methyltransferase-like protein 14 (METTL14) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Breast cancer | |
| Drug | Olaparib | |
| Crosstalk ID: M6ACROT02216 | ||
| Epigenetic Regulator | DNA (cytosine-5)-methyltransferase 3B (DNMT3B) | |
| Regulated Target | Methyltransferase-like protein 14 (METTL14) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Breast cancer | |
| Drug | Cisplatin | |
| Crosstalk ID: M6ACROT02217 | ||
| Epigenetic Regulator | DNA (cytosine-5)-methyltransferase 3B (DNMT3B) | |
| Regulated Target | Methyltransferase-like protein 14 (METTL14) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Breast cancer | |
| Drug | Adriamycin | |
| Crosstalk ID: M6ACROT02238 | ||
| Epigenetic Regulator | Cysteine methyltransferase DNMT3A (DNMT3A) | |
| Regulated Target | Methyltransferase-like protein 14 (METTL14) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Breast cancer | |
| Drug | Olaparib | |
| Crosstalk ID: M6ACROT02240 | ||
| Epigenetic Regulator | Cysteine methyltransferase DNMT3A (DNMT3A) | |
| Regulated Target | Methyltransferase-like protein 14 (METTL14) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Breast cancer | |
| Drug | Cisplatin | |
| Crosstalk ID: M6ACROT02241 | ||
| Epigenetic Regulator | Cysteine methyltransferase DNMT3A (DNMT3A) | |
| Regulated Target | Methyltransferase-like protein 14 (METTL14) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Breast cancer | |
| Drug | Adriamycin | |
| Crosstalk ID: M6ACROT02262 | ||
| Epigenetic Regulator | DNA (cytosine-5)-methyltransferase 1 (DNMT1) | |
| Regulated Target | Methyltransferase-like protein 14 (METTL14) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Breast cancer | |
| Drug | Olaparib | |
| Crosstalk ID: M6ACROT02264 | ||
| Epigenetic Regulator | DNA (cytosine-5)-methyltransferase 1 (DNMT1) | |
| Regulated Target | Methyltransferase-like protein 14 (METTL14) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Breast cancer | |
| Drug | Cisplatin | |
| Crosstalk ID: M6ACROT02265 | ||
| Epigenetic Regulator | DNA (cytosine-5)-methyltransferase 1 (DNMT1) | |
| Regulated Target | Methyltransferase-like protein 14 (METTL14) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Breast cancer | |
| Drug | Adriamycin | |
RNA Modification Sequencing Data Associated with the Target (ID: M6ATAR00560)
| In total 3 m6A sequence/site(s) in this target gene | |||
| mod ID: A2ISITE004580 | Click to Show/Hide the Full List | ||
| mod site | chr11:19233625-19233626:- | [2] | |
| Sequence | GCGTGGTGGCGTGCACCTGTAGTCCCAGTTACTCAGGAGGC | ||
| Transcript ID List | ENST00000527884.5; rmsk_3491527; ENST00000250024.9; ENST00000620009.4 | ||
| External Link | RMBase: RNA-editing_site_20802 | ||
| mod ID: A2ISITE004581 | Click to Show/Hide the Full List | ||
| mod site | chr11:19238845-19238846:- | [3] | |
| Sequence | CCATTACTTTCAGTAGCAAAAGCCACAATTACTTTTGCACC | ||
| Transcript ID List | ENST00000250024.9; ENST00000527884.5; ENST00000620009.4; ENST00000532666.1; rmsk_3491535 | ||
| External Link | RMBase: RNA-editing_site_20803 | ||
| mod ID: A2ISITE004582 | Click to Show/Hide the Full List | ||
| mod site | chr11:19239677-19239678:- | [3] | |
| Sequence | CCCCTCTTCAAAAATTCGTTAGTTTCATTACAACATTTCTG | ||
| Transcript ID List | ENST00000620009.4; ENST00000527884.5; ENST00000532666.1; ENST00000250024.9 | ||
| External Link | RMBase: RNA-editing_site_20804 | ||
5-methylcytidine (m5C)
| In total 1 m6A sequence/site(s) in this target gene | |||
| mod ID: M5CSITE005112 | Click to Show/Hide the Full List | ||
| mod site | chr11:19237389-19237390:- | [4] | |
| Sequence | GGATTATTGTGTCATAAGTTCTTAGCACGATATCCTAATTA | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000532666.1; ENST00000250024.9; ENST00000527884.5; ENST00000620009.4 | ||
| External Link | RMBase: m5C_site_7086 | ||
N6-methyladenosine (m6A)
| In total 69 m6A sequence/site(s) in this target gene | |||
| mod ID: M6ASITE004312 | Click to Show/Hide the Full List | ||
| mod site | chr11:19224149-19224150:- | [5] | |
| Sequence | ATCTTTTTGTAAAGTTTGCAACAATCCTCAATCAAGTCTAT | ||
| Motif Score | 2.173910714 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000250024.9; ENST00000620009.4; ENST00000527884.5 | ||
| External Link | RMBase: m6A_site_134371 | ||
| mod ID: M6ASITE004313 | Click to Show/Hide the Full List | ||
| mod site | chr11:19224262-19224263:- | [6] | |
| Sequence | CATTCCCTTAAACATTTTGCACAAAGAAAATGCTGTGTGAT | ||
| Motif Score | 2.830589286 | ||
| Cell/Tissue List | HEK293T; hESC-HEK293T | ||
| Seq Type List | DART-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000620009.4; ENST00000527884.5; ENST00000250024.9 | ||
| External Link | RMBase: m6A_site_134372 | ||
| mod ID: M6ASITE004314 | Click to Show/Hide the Full List | ||
| mod site | chr11:19224297-19224298:- | [6] | |
| Sequence | CCTCTAAGCAAATATGCTTGACATGCCTAACACAGCATTCC | ||
| Motif Score | 2.859755952 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000250024.9; ENST00000620009.4; ENST00000527884.5 | ||
| External Link | RMBase: m6A_site_134373 | ||
| mod ID: M6ASITE004315 | Click to Show/Hide the Full List | ||
| mod site | chr11:19224322-19224323:- | [6] | |
| Sequence | AAGTAACTGTATTAAAGTTTACTTCCCTCTAAGCAAATATG | ||
| Motif Score | 2.494845238 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000527884.5; ENST00000250024.9; ENST00000620009.4 | ||
| External Link | RMBase: m6A_site_134374 | ||
| mod ID: M6ASITE004316 | Click to Show/Hide the Full List | ||
| mod site | chr11:19224409-19224410:- | [6] | |
| Sequence | AAATAAAAGTAAAATGTTGAACTCTAAGATATATTAACTTC | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HEK293T; Huh7 | ||
| Seq Type List | DART-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000527884.5; ENST00000250024.9; ENST00000620009.4 | ||
| External Link | RMBase: m6A_site_134375 | ||
| mod ID: M6ASITE004317 | Click to Show/Hide the Full List | ||
| mod site | chr11:19224432-19224433:- | [6] | |
| Sequence | TAAGTTTAGCTTTCAATCCTACAAAATAAAAGTAAAATGTT | ||
| Motif Score | 2.078666667 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000250024.9; ENST00000620009.4; ENST00000527884.5 | ||
| External Link | RMBase: m6A_site_134376 | ||
| mod ID: M6ASITE004318 | Click to Show/Hide the Full List | ||
| mod site | chr11:19224581-19224582:- | [7] | |
| Sequence | TACACAGACTGATTTGGAGAACACATTCTCTGAAAATACTG | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HEK293T; A549; Huh7; HEK293A-TOA; TREX; TIME | ||
| Seq Type List | MeRIP-seq; m6A-seq | ||
| Transcript ID List | ENST00000527884.5; ENST00000250024.9; ENST00000620009.4 | ||
| External Link | RMBase: m6A_site_134377 | ||
| mod ID: M6ASITE004319 | Click to Show/Hide the Full List | ||
| mod site | chr11:19224594-19224595:- | [7] | |
| Sequence | AATAGAGAAAATGTACACAGACTGATTTGGAGAACACATTC | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HEK293T; A549; Huh7; HEK293A-TOA; TREX; TIME | ||
| Seq Type List | MeRIP-seq; m6A-seq; m6A-CLIP/IP | ||
| Transcript ID List | ENST00000527884.5; ENST00000620009.4; ENST00000250024.9 | ||
| External Link | RMBase: m6A_site_134378 | ||
| mod ID: M6ASITE004320 | Click to Show/Hide the Full List | ||
| mod site | chr11:19224600-19224601:- | [6] | |
| Sequence | TTGTTTAATAGAGAAAATGTACACAGACTGATTTGGAGAAC | ||
| Motif Score | 2.856142857 | ||
| Cell/Tissue List | HEK293T; hESC-HEK293T | ||
| Seq Type List | DART-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000620009.4; ENST00000250024.9; ENST00000527884.5 | ||
| External Link | RMBase: m6A_site_134379 | ||
| mod ID: M6ASITE004321 | Click to Show/Hide the Full List | ||
| mod site | chr11:19224653-19224654:- | [6] | |
| Sequence | CAGAGGATGTCCATTAATCAACAGATGTTGGCTTAGTTTAA | ||
| Motif Score | 2.173910714 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000620009.4; ENST00000527884.5; ENST00000250024.9 | ||
| External Link | RMBase: m6A_site_134380 | ||
| mod ID: M6ASITE004322 | Click to Show/Hide the Full List | ||
| mod site | chr11:19224687-19224688:- | [8] | |
| Sequence | CTCTTTGTCCCACAGCGAAAACTGGAAGTCTCAACAGAGGA | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa; HEK293T; A549; HEK293A-TOA; TREX | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000620009.4; ENST00000527884.5; ENST00000250024.9 | ||
| External Link | RMBase: m6A_site_134381 | ||
| mod ID: M6ASITE004323 | Click to Show/Hide the Full List | ||
| mod site | chr11:19224710-19224711:- | [8] | |
| Sequence | CTAATAAAACCTCCTTAGGAACTCTCTTTGTCCCACAGCGA | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa; HEK293T; A549; HEK293A-TOA | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000250024.9; ENST00000527884.5; ENST00000620009.4 | ||
| External Link | RMBase: m6A_site_134382 | ||
| mod ID: M6ASITE004324 | Click to Show/Hide the Full List | ||
| mod site | chr11:19224722-19224723:- | [8] | |
| Sequence | ATTTTGAGGGTGCTAATAAAACCTCCTTAGGAACTCTCTTT | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | HeLa; HEK293T; A549; HEK293A-TOA | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000620009.4; ENST00000527884.5; ENST00000250024.9 | ||
| External Link | RMBase: m6A_site_134383 | ||
| mod ID: M6ASITE004325 | Click to Show/Hide the Full List | ||
| mod site | chr11:19224774-19224775:- | [8] | |
| Sequence | TTCTTCCGTACCCCAGGTGGACCCACCAAGCCAACCAGCTC | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; HEK293T; A549; Huh7; HEK293A-TOA | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000527884.5; ENST00000620009.4; ENST00000250024.9 | ||
| External Link | RMBase: m6A_site_134384 | ||
| mod ID: M6ASITE004326 | Click to Show/Hide the Full List | ||
| mod site | chr11:19225555-19225556:- | [5] | |
| Sequence | ATTTTCCCTCTTTTCATGTAACACCGTTGAAGCTAATGGTC | ||
| Motif Score | 2.168095238 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000527884.5; ENST00000529188.1; ENST00000620009.4; ENST00000250024.9 | ||
| External Link | RMBase: m6A_site_134385 | ||
| mod ID: M6ASITE004327 | Click to Show/Hide the Full List | ||
| mod site | chr11:19225589-19225590:- | [6] | |
| Sequence | ATTCCAGTGACATCATCTGAACTCACTGCTGTTAATTTTCC | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000529188.1; ENST00000527884.5; ENST00000250024.9; ENST00000620009.4 | ||
| External Link | RMBase: m6A_site_134386 | ||
| mod ID: M6ASITE004328 | Click to Show/Hide the Full List | ||
| mod site | chr11:19225600-19225601:- | [5] | |
| Sequence | TTCCAGGTGTTATTCCAGTGACATCATCTGAACTCACTGCT | ||
| Motif Score | 2.859755952 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000620009.4; ENST00000250024.9; ENST00000529188.1; ENST00000527884.5 | ||
| External Link | RMBase: m6A_site_134387 | ||
| mod ID: M6ASITE004329 | Click to Show/Hide the Full List | ||
| mod site | chr11:19225748-19225749:- | [6] | |
| Sequence | TTCCCCAAACCACAGGATTTACAGCTCCCCAATTGCAGGTG | ||
| Motif Score | 2.07285119 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000529188.1; ENST00000250024.9; ENST00000620009.4; ENST00000527884.5 | ||
| External Link | RMBase: m6A_site_134388 | ||
| mod ID: M6ASITE004330 | Click to Show/Hide the Full List | ||
| mod site | chr11:19225863-19225864:- | [8] | |
| Sequence | CCTTCCTCCTTGTTTTCTAGACCTTGTTCCCATCAGGATAC | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa; peripheral-blood | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000529188.1; ENST00000620009.4; ENST00000527884.5; ENST00000250024.9 | ||
| External Link | RMBase: m6A_site_134389 | ||
| mod ID: M6ASITE004331 | Click to Show/Hide the Full List | ||
| mod site | chr11:19225885-19225886:- | [8] | |
| Sequence | TGTATTTGTGTGTAAAATAAACCCTTCCTCCTTGTTTTCTA | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | HeLa; peripheral-blood | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000620009.4; ENST00000527884.5; ENST00000529188.1; ENST00000250024.9 | ||
| External Link | RMBase: m6A_site_134390 | ||
| mod ID: M6ASITE004332 | Click to Show/Hide the Full List | ||
| mod site | chr11:19226074-19226075:- | [8] | |
| Sequence | CCACAGGTGGCATTTAAGAAACATGTTCTGCATTATTCGGG | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa; HepG2; A549; H1A; H1B; hESCs; GM12878; Jurkat; peripheral-blood; GSC-11; TIME; HEC-1-A; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000250024.9; ENST00000527884.5; ENST00000620009.4; ENST00000529188.1 | ||
| External Link | RMBase: m6A_site_134391 | ||
| mod ID: M6ASITE004333 | Click to Show/Hide the Full List | ||
| mod site | chr11:19229471-19229472:- | [8] | |
| Sequence | TTTAAAGAGGACCTAAAAGGACTTGAAAATGTCTCCGCAGT | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa; HepG2; A549; HEK293T; H1A; H1B; hESCs; GM12878; MM6; Jurkat; peripheral-blood; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; TREX; endometrial; HEC-1-A; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000527884.5; ENST00000250024.9; ENST00000620009.4 | ||
| External Link | RMBase: m6A_site_134392 | ||
| mod ID: M6ASITE004334 | Click to Show/Hide the Full List | ||
| mod site | chr11:19229481-19229482:- | [8] | |
| Sequence | CAAAAAGAAATTTAAAGAGGACCTAAAAGGACTTGAAAATG | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; HepG2; A549; HEK293T; H1A; H1B; hESCs; GM12878; MM6; Jurkat; peripheral-blood; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; TREX; endometrial; HEC-1-A; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000250024.9; ENST00000620009.4; ENST00000527884.5 | ||
| External Link | RMBase: m6A_site_134393 | ||
| mod ID: M6ASITE004335 | Click to Show/Hide the Full List | ||
| mod site | chr11:19229511-19229512:- | [8] | |
| Sequence | GAGGGCAAGCATGCTCGAGGACAGTGGTTCCAAAAAGAAAT | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; A549; H1A; H1B; hESCs; GM12878; MM6; Jurkat; peripheral-blood; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; TREX; endometrial; HEC-1-A; GSCs; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000527884.5; ENST00000250024.9; ENST00000620009.4 | ||
| External Link | RMBase: m6A_site_134394 | ||
| mod ID: M6ASITE004336 | Click to Show/Hide the Full List | ||
| mod site | chr11:19229563-19229564:- | [8] | |
| Sequence | GGCAAGGGGCAAAGAGCCGAACCAGGGAGCCAGCTGGAGAA | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; A549; H1A; H1B; hESCs; GM12878; MM6; Huh7; Jurkat; peripheral-blood; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; TREX; endometrial; HEC-1-A; GSCs; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000527884.5; ENST00000620009.4; ENST00000250024.9 | ||
| External Link | RMBase: m6A_site_134395 | ||
| mod ID: M6ASITE004337 | Click to Show/Hide the Full List | ||
| mod site | chr11:19229659-19229660:- | [6] | |
| Sequence | AAGACTCCACAGATGCCACCACTGAGAAGGCAGCCAATGAT | ||
| Motif Score | 2.475107143 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000250024.9; ENST00000527884.5; ENST00000620009.4 | ||
| External Link | RMBase: m6A_site_134396 | ||
| mod ID: M6ASITE004338 | Click to Show/Hide the Full List | ||
| mod site | chr11:19229676-19229677:- | [8] | |
| Sequence | TAAAGCTACTGGCTCAAAAGACTCCACAGATGCCACCACTG | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa; HEK293T; A549; GM12878; Huh7; HEK293A-TOA; MSC; TIME; TREX; iSLK | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000527884.5; ENST00000250024.9; ENST00000620009.4 | ||
| External Link | RMBase: m6A_site_134397 | ||
| mod ID: M6ASITE004339 | Click to Show/Hide the Full List | ||
| mod site | chr11:19229919-19229920:- | [8] | |
| Sequence | CAAACCAGTAGCCCCTCTGGACCCCCCAGTGAATGCTGAGA | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000620009.4; ENST00000527884.5; ENST00000250024.9 | ||
| External Link | RMBase: m6A_site_134398 | ||
| mod ID: M6ASITE004340 | Click to Show/Hide the Full List | ||
| mod site | chr11:19229936-19229937:- | [8] | |
| Sequence | GCAAGATCTGGACCCTGCAAACCAGTAGCCCCTCTGGACCC | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000250024.9; ENST00000620009.4; ENST00000527884.5 | ||
| External Link | RMBase: m6A_site_134399 | ||
| mod ID: M6ASITE004341 | Click to Show/Hide the Full List | ||
| mod site | chr11:19230635-19230636:- | [5] | |
| Sequence | CAGTAGCCCTATCAAGACCAACAAAGGTATGTTTTCAGGAA | ||
| Motif Score | 2.173910714 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000620009.4; ENST00000527884.5; ENST00000250024.9 | ||
| External Link | RMBase: m6A_site_134400 | ||
| mod ID: M6ASITE004342 | Click to Show/Hide the Full List | ||
| mod site | chr11:19230721-19230722:- | [5] | |
| Sequence | GGGAAACCAAACTTTACTCGACACCCATCTCTTATCAAATT | ||
| Motif Score | 2.865571429 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000250024.9; ENST00000527884.5; ENST00000620009.4 | ||
| External Link | RMBase: m6A_site_134401 | ||
| mod ID: M6ASITE004343 | Click to Show/Hide the Full List | ||
| mod site | chr11:19230745-19230746:- | [6] | |
| Sequence | GCCAAAAACCTCTTTTCCACACGTGGGAAACCAAACTTTAC | ||
| Motif Score | 2.058863095 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000527884.5; ENST00000620009.4; ENST00000250024.9 | ||
| External Link | RMBase: m6A_site_134402 | ||
| mod ID: M6ASITE004344 | Click to Show/Hide the Full List | ||
| mod site | chr11:19234359-19234360:- | [5] | |
| Sequence | TGGATAAAAGCAAGTTTAAAAGTAAGTGTCATGACTGCCAT | ||
| Motif Score | 1.756488095 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000620009.4; ENST00000250024.9; ENST00000527884.5 | ||
| External Link | RMBase: m6A_site_134403 | ||
| mod ID: M6ASITE004345 | Click to Show/Hide the Full List | ||
| mod site | chr11:19234499-19234500:- | [8] | |
| Sequence | TTCTGTAAACAGCCGCAAAGACAAGTCTTTAAGGGTAATGA | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa; hESC-HEK293T | ||
| Seq Type List | m6A-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000527884.5; ENST00000620009.4; ENST00000250024.9 | ||
| External Link | RMBase: m6A_site_134404 | ||
| mod ID: M6ASITE004346 | Click to Show/Hide the Full List | ||
| mod site | chr11:19234511-19234512:- | [8] | |
| Sequence | CTTCTCTAAAGCTTCTGTAAACAGCCGCAAAGACAAGTCTT | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000620009.4; ENST00000527884.5; ENST00000250024.9 | ||
| External Link | RMBase: m6A_site_134405 | ||
| mod ID: M6ASITE004347 | Click to Show/Hide the Full List | ||
| mod site | chr11:19234768-19234769:- | [8] | |
| Sequence | CCAGACATGTGTTTTGTGGAACTCCCTGGAGTGGAATTTCG | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa; HEK293T | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000527884.5; ENST00000620009.4; ENST00000250024.9 | ||
| External Link | RMBase: m6A_site_134406 | ||
| mod ID: M6ASITE004348 | Click to Show/Hide the Full List | ||
| mod site | chr11:19234784-19234785:- | [8] | |
| Sequence | TGGCCCAAATGGACACCCAGACATGTGTTTTGTGGAACTCC | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa; HEK293T; hESC-HEK293T | ||
| Seq Type List | m6A-seq; MeRIP-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000250024.9; ENST00000527884.5; ENST00000620009.4 | ||
| External Link | RMBase: m6A_site_134407 | ||
| mod ID: M6ASITE004349 | Click to Show/Hide the Full List | ||
| mod site | chr11:19234792-19234793:- | [8] | |
| Sequence | TCAAACACTGGCCCAAATGGACACCCAGACATGTGTTTTGT | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; HEK293T | ||
| Seq Type List | m6A-seq; MeRIP-seq; DART-seq | ||
| Transcript ID List | ENST00000620009.4; ENST00000527884.5; ENST00000250024.9 | ||
| External Link | RMBase: m6A_site_134408 | ||
| mod ID: M6ASITE004350 | Click to Show/Hide the Full List | ||
| mod site | chr11:19234808-19234809:- | [8] | |
| Sequence | GGATCATATCATCAAATCAAACACTGGCCCAAATGGACACC | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa; HEK293T | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000620009.4; ENST00000250024.9; ENST00000527884.5 | ||
| External Link | RMBase: m6A_site_134409 | ||
| mod ID: M6ASITE004351 | Click to Show/Hide the Full List | ||
| mod site | chr11:19234944-19234945:- | [6] | |
| Sequence | GGCGACACAATCTCAACAAAACCCTTGGCACCTTGAAGAGC | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | MeRIP-seq; DART-seq | ||
| Transcript ID List | ENST00000620009.4; ENST00000250024.9; ENST00000527884.5 | ||
| External Link | RMBase: m6A_site_134410 | ||
| mod ID: M6ASITE004352 | Click to Show/Hide the Full List | ||
| mod site | chr11:19234974-19234975:- | [6] | |
| Sequence | GCCTCGCCAAAAACAGGTACACTTGGCACGGGCGACACAAT | ||
| Motif Score | 2.506922619 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000250024.9; ENST00000527884.5; ENST00000620009.4 | ||
| External Link | RMBase: m6A_site_134411 | ||
| mod ID: M6ASITE004353 | Click to Show/Hide the Full List | ||
| mod site | chr11:19234982-19234983:- | ||
| Sequence | GGTGAGCCGCCTCGCCAAAAACAGGTACACTTGGCACGGGC | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | MeRIP-seq | ||
| Transcript ID List | ENST00000250024.9; ENST00000620009.4; ENST00000527884.5 | ||
| External Link | RMBase: m6A_site_134412 | ||
| mod ID: M6ASITE004354 | Click to Show/Hide the Full List | ||
| mod site | chr11:19237333-19237334:- | [6] | |
| Sequence | GAATAATGACATCTGCCTTGACGAAGTGGCAGAGGAACTTA | ||
| Motif Score | 2.833690476 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000250024.9; ENST00000620009.4; ENST00000527884.5 | ||
| External Link | RMBase: m6A_site_134413 | ||
| mod ID: M6ASITE004355 | Click to Show/Hide the Full List | ||
| mod site | chr11:19237345-19237346:- | [6] | |
| Sequence | CAACCCTGCTGTGAATAATGACATCTGCCTTGACGAAGTGG | ||
| Motif Score | 2.859755952 | ||
| Cell/Tissue List | HEK293T; hESC-HEK293T | ||
| Seq Type List | DART-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000250024.9; ENST00000620009.4; ENST00000527884.5 | ||
| External Link | RMBase: m6A_site_134414 | ||
| mod ID: M6ASITE004356 | Click to Show/Hide the Full List | ||
| mod site | chr11:19237467-19237468:- | [5] | |
| Sequence | TTACCTTTTCCTTTGTAGGAACACTTATCTGGAGATGAATT | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000532666.1; ENST00000620009.4; ENST00000250024.9; ENST00000527884.5 | ||
| External Link | RMBase: m6A_site_134415 | ||
| mod ID: M6ASITE004357 | Click to Show/Hide the Full List | ||
| mod site | chr11:19237920-19237921:- | [6] | |
| Sequence | TGTGAGCCCTGAGATCCGCAACAGAGATCAGAAAAGGGGTT | ||
| Motif Score | 2.173910714 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000527884.5; ENST00000250024.9; ENST00000620009.4; ENST00000532666.1 | ||
| External Link | RMBase: m6A_site_134416 | ||
| mod ID: M6ASITE004358 | Click to Show/Hide the Full List | ||
| mod site | chr11:19237969-19237970:- | [6] | |
| Sequence | AGGGAGAGCCGTGGACACCGACAGCCAACCTGAAAATGCTC | ||
| Motif Score | 2.865571429 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000620009.4; ENST00000527884.5; ENST00000250024.9; ENST00000532666.1 | ||
| External Link | RMBase: m6A_site_134417 | ||
| mod ID: M6ASITE004359 | Click to Show/Hide the Full List | ||
| mod site | chr11:19237975-19237976:- | [8] | |
| Sequence | GCTCTCAGGGAGAGCCGTGGACACCGACAGCCAACCTGAAA | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; HEK293T | ||
| Seq Type List | m6A-seq; DART-seq | ||
| Transcript ID List | ENST00000250024.9; ENST00000527884.5; ENST00000532666.1; ENST00000620009.4 | ||
| External Link | RMBase: m6A_site_134418 | ||
| mod ID: M6ASITE004360 | Click to Show/Hide the Full List | ||
| mod site | chr11:19238017-19238018:- | [5] | |
| Sequence | CTGACTTTGGCCCTTTAACCACACCTACCAAGCCCAAGGAA | ||
| Motif Score | 2.053113095 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000620009.4; ENST00000527884.5; ENST00000532666.1; ENST00000250024.9 | ||
| External Link | RMBase: m6A_site_134419 | ||
| mod ID: M6ASITE004361 | Click to Show/Hide the Full List | ||
| mod site | chr11:19238089-19238090:- | [8] | |
| Sequence | ATAAAAGGGGACTAATGAAAACACCTCTGAAAGAATCCACC | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000250024.9; ENST00000527884.5; ENST00000532666.1; ENST00000620009.4 | ||
| External Link | RMBase: m6A_site_134420 | ||
| mod ID: M6ASITE004362 | Click to Show/Hide the Full List | ||
| mod site | chr11:19238099-19238100:- | [8] | |
| Sequence | TGTGAGCCACATAAAAGGGGACTAATGAAAACACCTCTGAA | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000527884.5; ENST00000532666.1; ENST00000250024.9; ENST00000620009.4 | ||
| External Link | RMBase: m6A_site_134421 | ||
| mod ID: M6ASITE004363 | Click to Show/Hide the Full List | ||
| mod site | chr11:19238111-19238112:- | [5] | |
| Sequence | GAAAATCTCTTTTGTGAGCCACATAAAAGGGGACTAATGAA | ||
| Motif Score | 2.053113095 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000250024.9; ENST00000532666.1; ENST00000527884.5; ENST00000620009.4 | ||
| External Link | RMBase: m6A_site_134422 | ||
| mod ID: M6ASITE004364 | Click to Show/Hide the Full List | ||
| mod site | chr11:19240125-19240126:- | [6] | |
| Sequence | TCTAAATGTATGAGGAATTTACAGAATGGAGAACGAAAAGG | ||
| Motif Score | 2.07285119 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000527884.5; ENST00000250024.9; ENST00000532666.1; ENST00000620009.4 | ||
| External Link | RMBase: m6A_site_134423 | ||
| mod ID: M6ASITE004365 | Click to Show/Hide the Full List | ||
| mod site | chr11:19240190-19240191:- | [8] | |
| Sequence | CAATTGATTGGACTACTTGAACCATCGGGATTTGGGGAGGA | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000532666.1; ENST00000620009.4; ENST00000527884.5; ENST00000250024.9 | ||
| External Link | RMBase: m6A_site_134424 | ||
| mod ID: M6ASITE004366 | Click to Show/Hide the Full List | ||
| mod site | chr11:19240199-19240200:- | [8] | |
| Sequence | CTTTTAGTACAATTGATTGGACTACTTGAACCATCGGGATT | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000527884.5; ENST00000620009.4; ENST00000250024.9; ENST00000532666.1 | ||
| External Link | RMBase: m6A_site_134425 | ||
| mod ID: M6ASITE004367 | Click to Show/Hide the Full List | ||
| mod site | chr11:19240220-19240221:- | [8] | |
| Sequence | TTTTCTTTAAGGATATTAAAACTTTTAGTACAATTGATTGG | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000527884.5; ENST00000250024.9; ENST00000620009.4; ENST00000532666.1 | ||
| External Link | RMBase: m6A_site_134426 | ||
| mod ID: M6ASITE004368 | Click to Show/Hide the Full List | ||
| mod site | chr11:19240550-19240551:- | [8] | |
| Sequence | TCCGGAGCCCTGATCTGCGAACAGGTGGGTGCTTCCTAAAG | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000620009.4; ENST00000250024.9; ENST00000527884.5; ENST00000532666.1 | ||
| External Link | RMBase: m6A_site_134427 | ||
| mod ID: M6ASITE004369 | Click to Show/Hide the Full List | ||
| mod site | chr11:19240590-19240591:- | [8] | |
| Sequence | GTCGCGTGCGCTCAGCTGGGACCTGGGCTCGTGCGCTTAGT | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000620009.4; ENST00000527884.5; ENST00000250024.9; ENST00000532666.1 | ||
| External Link | RMBase: m6A_site_134428 | ||
| mod ID: M6ASITE004370 | Click to Show/Hide the Full List | ||
| mod site | chr11:19240631-19240632:- | [8] | |
| Sequence | ATCCAGGCATCTAGATGCAGACTTGTACCCAGTTACTTGGG | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000620009.4; ENST00000250024.9; ENST00000532666.1; ENST00000527884.5 | ||
| External Link | RMBase: m6A_site_134429 | ||
| mod ID: M6ASITE004371 | Click to Show/Hide the Full List | ||
| mod site | chr11:19240692-19240693:- | [6] | |
| Sequence | CTGTCAAGCGACCTCCCACGACTTTACTGCTGAGCCTGTGC | ||
| Motif Score | 3.287565476 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000527884.5; ENST00000250024.9; ENST00000620009.4; ENST00000532666.1 | ||
| External Link | RMBase: m6A_site_134430 | ||
| mod ID: M6ASITE004372 | Click to Show/Hide the Full List | ||
| mod site | chr11:19240718-19240719:- | [8] | |
| Sequence | GCTTTGGGACTGAATTTGGAACTTTCCTGTCAAGCGACCTC | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000250024.9; ENST00000620009.4; ENST00000527884.5; ENST00000532666.1 | ||
| External Link | RMBase: m6A_site_134431 | ||
| mod ID: M6ASITE004373 | Click to Show/Hide the Full List | ||
| mod site | chr11:19240730-19240731:- | [8] | |
| Sequence | TGCATACTTGGAGCTTTGGGACTGAATTTGGAACTTTCCTG | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000527884.5; ENST00000620009.4; ENST00000250024.9; ENST00000532666.1 | ||
| External Link | RMBase: m6A_site_134432 | ||
| mod ID: M6ASITE004374 | Click to Show/Hide the Full List | ||
| mod site | chr11:19240745-19240746:- | [6] | |
| Sequence | CTTTAAATGCCCAACTGCATACTTGGAGCTTTGGGACTGAA | ||
| Motif Score | 2.53247619 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000532666.1; ENST00000250024.9; ENST00000620009.4; ENST00000527884.5 | ||
| External Link | RMBase: m6A_site_134433 | ||
| mod ID: M6ASITE004375 | Click to Show/Hide the Full List | ||
| mod site | chr11:19240768-19240769:- | [8] | |
| Sequence | AAAAAAGTCTTAATTATAAGACTCTTTAAATGCCCAACTGC | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa; A549 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000532666.1; ENST00000527884.5; ENST00000620009.4; ENST00000250024.9 | ||
| External Link | RMBase: m6A_site_134434 | ||
| mod ID: M6ASITE004376 | Click to Show/Hide the Full List | ||
| mod site | chr11:19240842-19240843:- | [8] | |
| Sequence | GGTGTGAACTGGCCACCCGAACAGGGAGTAAATAGTGCTTT | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa; A549 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000620009.4; ENST00000250024.9; ENST00000532666.1; ENST00000527884.5 | ||
| External Link | RMBase: m6A_site_134435 | ||
| mod ID: M6ASITE004377 | Click to Show/Hide the Full List | ||
| mod site | chr11:19240855-19240856:- | [8] | |
| Sequence | AGTCTAACGCCGTGGTGTGAACTGGCCACCCGAACAGGGAG | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa; A549 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000620009.4; ENST00000532666.1; ENST00000250024.9; ENST00000527884.5 | ||
| External Link | RMBase: m6A_site_134436 | ||
| mod ID: M6ASITE004378 | Click to Show/Hide the Full List | ||
| mod site | chr11:19240921-19240922:- | [9] | |
| Sequence | GAGCGAGTGGCGGCCGGGGGACAGTACCTGCTTGCCTTATT | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | Jurkat | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000250024.9; ENST00000532666.1; ENST00000620009.4; ENST00000527884.5 | ||
| External Link | RMBase: m6A_site_134437 | ||
| mod ID: M6ASITE004379 | Click to Show/Hide the Full List | ||
| mod site | chr11:19241413-19241414:- | [9] | |
| Sequence | CGGCTCCCGCCGTGAGGTGGACTGACAGGTCAGCGGGACGC | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | Jurkat | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000527884.5; ENST00000620009.4; ENST00000532666.1 | ||
| External Link | RMBase: m6A_site_134438 | ||
| mod ID: M6ASITE004380 | Click to Show/Hide the Full List | ||
| mod site | chr11:19241507-19241508:- | [9] | |
| Sequence | CACCTCGGCGCTGCGGAAAGACTGACTGCCAGGTACGCGGG | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | Jurkat; HEK293T | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000527884.5; ENST00000620009.4; ENST00000532666.1 | ||
| External Link | RMBase: m6A_site_134439 | ||
References