m6A Target Gene Information
General Information of the m6A Target Gene (ID: M6ATAR00526)
Full List of m6A Methylation Regulator of This Target Gene and Corresponding Disease/Drug Response(s)
FGFR4
can be regulated by the following regulator(s), and cause disease/drug response(s). You can browse detail information of regulator(s) or disease/drug response(s).
Browse Regulator
Browse Disease
Browse Drug
Methyltransferase-like 14 (METTL14) [WRITER]
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL14 | ||
| Cell Line | HepG2 cell line | Homo sapiens |
|
Treatment: shMETTL14 HepG2 cells
Control: shCtrl HepG2 cells
|
GSE121949 | |
| Regulation |
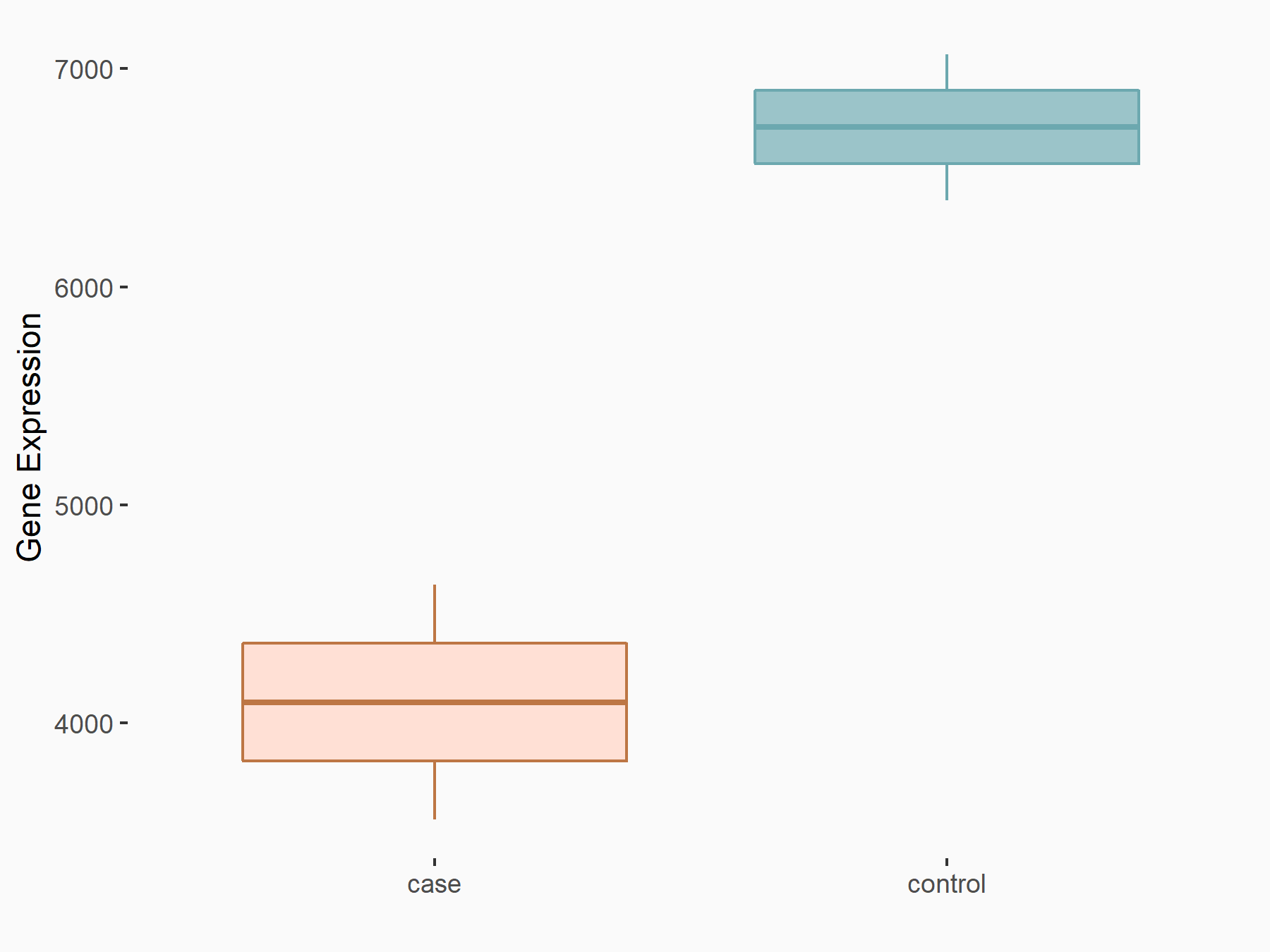 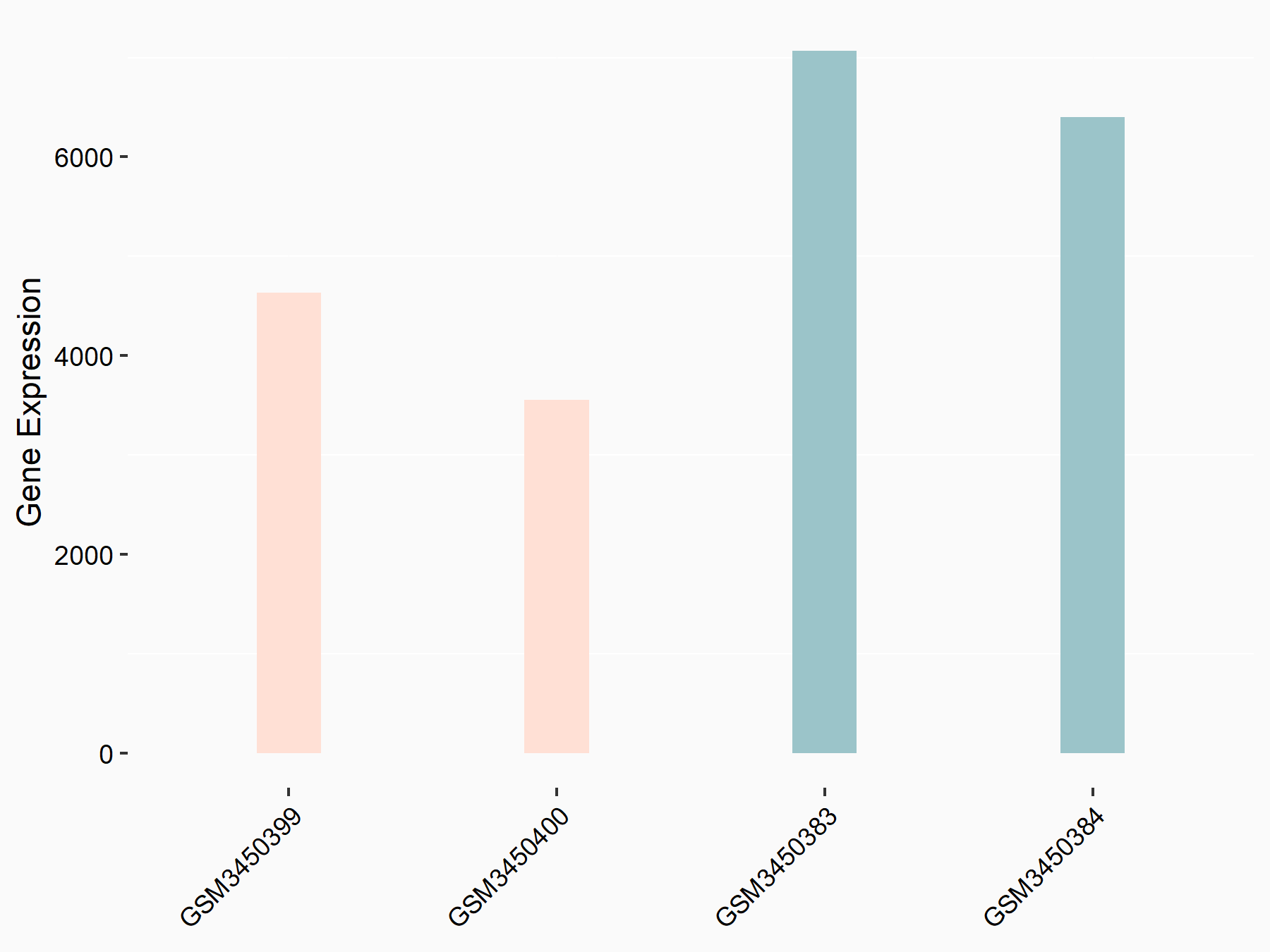 |
logFC: -7.17E-01 p-value: 3.71E-06 |
| More Results | Click to View More RNA-seq Results | |
| In total 3 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [1] | |||
| Response Summary | m6A-hypomethylation regulated Fibroblast growth factor receptor 4 (FGFR4) phosphorylates GSK-3beta and activates beta-catenin/TCF4-SLC7A11/FPN1 signaling to drive anti-HER2 resistance. Knockdown of METTL14 significantly increased the expression level of FGFR4 in HER2-positive breast cancer cells. FGFR4 reduced the sensitivity of HER2-positive breast cancer to trastuzumab plus pertuzumab or tucatinib. These results pinpoint a mechanism of anti-HER2 resistance and provide a strategy for overcoming resistance via FGFR4 inhibition in recalcitrant HER2-positive breast cancer. | |||
| Target Regulation | Down regulation | |||
| Responsed Disease | Breast cancer | ICD-11: 2C60 | ||
| Responsed Drug | Pertuzumab | Approved | ||
| Pathway Response | Wnt signaling pathway | hsa04310 | ||
| Cell Process | Glutathione synthesis | |||
| In-vitro Model | ZR-75-1 | Invasive breast carcinoma | Homo sapiens | CVCL_0588 |
| T-47D | Invasive breast carcinoma | Homo sapiens | CVCL_0553 | |
| SUM-159 (A mesenchymal triple-negative breast cancer cell line) | ||||
| SK-BR-3 | Breast adenocarcinoma | Homo sapiens | CVCL_0033 | |
| MDA-MB-468 | Breast adenocarcinoma | Homo sapiens | CVCL_0419 | |
| MDA-MB-453 | Breast adenocarcinoma | Homo sapiens | CVCL_0418 | |
| MDA-MB-361 | Breast adenocarcinoma | Homo sapiens | CVCL_0620 | |
| MDA-MB-231 | Breast adenocarcinoma | Homo sapiens | CVCL_0062 | |
| MCF-7 | Invasive breast carcinoma | Homo sapiens | CVCL_0031 | |
| MCF-10A | Normal | Homo sapiens | CVCL_0598 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| BT-549 | Invasive breast carcinoma | Homo sapiens | CVCL_1092 | |
| BT-474 | Invasive breast carcinoma | Homo sapiens | CVCL_0179 | |
| AU565 | Breast adenocarcinoma | Homo sapiens | CVCL_1074 | |
| In-vivo Model | Luciferase-labeled rSKBR3 and MDA-MB-361 cells (1 × 107 cells) mixed with 1:1 Matrigel (Corning, 356237) were subcutaneously injected into the fat pads of mice. After a tumor was palpable, the mice were randomized into four groups (five mice per group), and they were treated with vehicle, trastuzumab (20 mg/kg, intraperitoneal administration), roblitinib (30 mg/kg, oral administration), or a combination of both drugs. | |||
| Experiment 2 Reporting the m6A Methylation Regulator of This Target Gene | [1] | |||
| Response Summary | m6A-hypomethylation regulated Fibroblast growth factor receptor 4 (FGFR4) phosphorylates GSK-3beta and activates beta-catenin/TCF4-SLC7A11/FPN1 signaling to drive anti-HER2 resistance. Knockdown of METTL14 significantly increased the expression level of FGFR4 in HER2-positive breast cancer cells. FGFR4 reduced the sensitivity of HER2-positive breast cancer to trastuzumab plus pertuzumab or tucatinib. These results pinpoint a mechanism of anti-HER2 resistance and provide a strategy for overcoming resistance via FGFR4 inhibition in recalcitrant HER2-positive breast cancer. | |||
| Target Regulation | Down regulation | |||
| Responsed Disease | Breast cancer | ICD-11: 2C60 | ||
| Responsed Drug | Trastuzumab | Approved | ||
| Pathway Response | Wnt signaling pathway | hsa04310 | ||
| Cell Process | Glutathione synthesis | |||
| In-vitro Model | ZR-75-1 | Invasive breast carcinoma | Homo sapiens | CVCL_0588 |
| T-47D | Invasive breast carcinoma | Homo sapiens | CVCL_0553 | |
| SUM-159 (A mesenchymal triple-negative breast cancer cell line) | ||||
| SK-BR-3 | Breast adenocarcinoma | Homo sapiens | CVCL_0033 | |
| MDA-MB-468 | Breast adenocarcinoma | Homo sapiens | CVCL_0419 | |
| MDA-MB-453 | Breast adenocarcinoma | Homo sapiens | CVCL_0418 | |
| MDA-MB-361 | Breast adenocarcinoma | Homo sapiens | CVCL_0620 | |
| MDA-MB-231 | Breast adenocarcinoma | Homo sapiens | CVCL_0062 | |
| MCF-7 | Invasive breast carcinoma | Homo sapiens | CVCL_0031 | |
| MCF-10A | Normal | Homo sapiens | CVCL_0598 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| BT-549 | Invasive breast carcinoma | Homo sapiens | CVCL_1092 | |
| BT-474 | Invasive breast carcinoma | Homo sapiens | CVCL_0179 | |
| AU565 | Breast adenocarcinoma | Homo sapiens | CVCL_1074 | |
| In-vivo Model | Luciferase-labeled rSKBR3 and MDA-MB-361 cells (1 × 107 cells) mixed with 1:1 Matrigel (Corning, 356237) were subcutaneously injected into the fat pads of mice. After a tumor was palpable, the mice were randomized into four groups (five mice per group), and they were treated with vehicle, trastuzumab (20 mg/kg, intraperitoneal administration), roblitinib (30 mg/kg, oral administration), or a combination of both drugs. | |||
| Experiment 3 Reporting the m6A Methylation Regulator of This Target Gene | [1] | |||
| Response Summary | m6A-hypomethylation regulated Fibroblast growth factor receptor 4 (FGFR4) phosphorylates GSK-3beta and activates beta-catenin/TCF4-SLC7A11/FPN1 signaling to drive anti-HER2 resistance. Knockdown of METTL14 significantly increased the expression level of FGFR4 in HER2-positive breast cancer cells. FGFR4 reduced the sensitivity of HER2-positive breast cancer to trastuzumab plus pertuzumab or tucatinib. These results pinpoint a mechanism of anti-HER2 resistance and provide a strategy for overcoming resistance via FGFR4 inhibition in recalcitrant HER2-positive breast cancer. | |||
| Target Regulation | Down regulation | |||
| Responsed Disease | Breast cancer | ICD-11: 2C60 | ||
| Responsed Drug | Tucatinib | Approved | ||
| Pathway Response | Wnt signaling pathway | hsa04310 | ||
| Cell Process | Glutathione synthesis | |||
| In-vitro Model | ZR-75-1 | Invasive breast carcinoma | Homo sapiens | CVCL_0588 |
| T-47D | Invasive breast carcinoma | Homo sapiens | CVCL_0553 | |
| SUM-159 (A mesenchymal triple-negative breast cancer cell line) | ||||
| SK-BR-3 | Breast adenocarcinoma | Homo sapiens | CVCL_0033 | |
| MDA-MB-468 | Breast adenocarcinoma | Homo sapiens | CVCL_0419 | |
| MDA-MB-453 | Breast adenocarcinoma | Homo sapiens | CVCL_0418 | |
| MDA-MB-361 | Breast adenocarcinoma | Homo sapiens | CVCL_0620 | |
| MDA-MB-231 | Breast adenocarcinoma | Homo sapiens | CVCL_0062 | |
| MCF-7 | Invasive breast carcinoma | Homo sapiens | CVCL_0031 | |
| MCF-10A | Normal | Homo sapiens | CVCL_0598 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| BT-549 | Invasive breast carcinoma | Homo sapiens | CVCL_1092 | |
| BT-474 | Invasive breast carcinoma | Homo sapiens | CVCL_0179 | |
| AU565 | Breast adenocarcinoma | Homo sapiens | CVCL_1074 | |
| In-vivo Model | Luciferase-labeled rSKBR3 and MDA-MB-361 cells (1 × 107 cells) mixed with 1:1 Matrigel (Corning, 356237) were subcutaneously injected into the fat pads of mice. After a tumor was palpable, the mice were randomized into four groups (five mice per group), and they were treated with vehicle, trastuzumab (20 mg/kg, intraperitoneal administration), roblitinib (30 mg/kg, oral administration), or a combination of both drugs. | |||
Breast cancer [ICD-11: 2C60]
| In total 3 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [1] | |||
| Response Summary | m6A-hypomethylation regulated Fibroblast growth factor receptor 4 (FGFR4) phosphorylates GSK-3beta and activates beta-catenin/TCF4-SLC7A11/FPN1 signaling to drive anti-HER2 resistance. Knockdown of METTL14 significantly increased the expression level of FGFR4 in HER2-positive breast cancer cells. FGFR4 reduced the sensitivity of HER2-positive breast cancer to trastuzumab plus pertuzumab or tucatinib. These results pinpoint a mechanism of anti-HER2 resistance and provide a strategy for overcoming resistance via FGFR4 inhibition in recalcitrant HER2-positive breast cancer. | |||
| Responsed Disease | Breast cancer [ICD-11: 2C60] | |||
| Target Regulator | Methyltransferase-like 14 (METTL14) | WRITER | ||
| Target Regulation | Down regulation | |||
| Responsed Drug | Pertuzumab | Approved | ||
| Pathway Response | Wnt signaling pathway | hsa04310 | ||
| Cell Process | Glutathione synthesis | |||
| In-vitro Model | ZR-75-1 | Invasive breast carcinoma | Homo sapiens | CVCL_0588 |
| T-47D | Invasive breast carcinoma | Homo sapiens | CVCL_0553 | |
| SUM-159 (A mesenchymal triple-negative breast cancer cell line) | ||||
| SK-BR-3 | Breast adenocarcinoma | Homo sapiens | CVCL_0033 | |
| MDA-MB-468 | Breast adenocarcinoma | Homo sapiens | CVCL_0419 | |
| MDA-MB-453 | Breast adenocarcinoma | Homo sapiens | CVCL_0418 | |
| MDA-MB-361 | Breast adenocarcinoma | Homo sapiens | CVCL_0620 | |
| MDA-MB-231 | Breast adenocarcinoma | Homo sapiens | CVCL_0062 | |
| MCF-7 | Invasive breast carcinoma | Homo sapiens | CVCL_0031 | |
| MCF-10A | Normal | Homo sapiens | CVCL_0598 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| BT-549 | Invasive breast carcinoma | Homo sapiens | CVCL_1092 | |
| BT-474 | Invasive breast carcinoma | Homo sapiens | CVCL_0179 | |
| AU565 | Breast adenocarcinoma | Homo sapiens | CVCL_1074 | |
| In-vivo Model | Luciferase-labeled rSKBR3 and MDA-MB-361 cells (1 × 107 cells) mixed with 1:1 Matrigel (Corning, 356237) were subcutaneously injected into the fat pads of mice. After a tumor was palpable, the mice were randomized into four groups (five mice per group), and they were treated with vehicle, trastuzumab (20 mg/kg, intraperitoneal administration), roblitinib (30 mg/kg, oral administration), or a combination of both drugs. | |||
| Experiment 2 Reporting the m6A-centered Disease Response | [1] | |||
| Response Summary | m6A-hypomethylation regulated Fibroblast growth factor receptor 4 (FGFR4) phosphorylates GSK-3beta and activates beta-catenin/TCF4-SLC7A11/FPN1 signaling to drive anti-HER2 resistance. Knockdown of METTL14 significantly increased the expression level of FGFR4 in HER2-positive breast cancer cells. FGFR4 reduced the sensitivity of HER2-positive breast cancer to trastuzumab plus pertuzumab or tucatinib. These results pinpoint a mechanism of anti-HER2 resistance and provide a strategy for overcoming resistance via FGFR4 inhibition in recalcitrant HER2-positive breast cancer. | |||
| Responsed Disease | Breast cancer [ICD-11: 2C60] | |||
| Target Regulator | Methyltransferase-like 14 (METTL14) | WRITER | ||
| Target Regulation | Down regulation | |||
| Responsed Drug | Trastuzumab | Approved | ||
| Pathway Response | Wnt signaling pathway | hsa04310 | ||
| Cell Process | Glutathione synthesis | |||
| In-vitro Model | ZR-75-1 | Invasive breast carcinoma | Homo sapiens | CVCL_0588 |
| T-47D | Invasive breast carcinoma | Homo sapiens | CVCL_0553 | |
| SUM-159 (A mesenchymal triple-negative breast cancer cell line) | ||||
| SK-BR-3 | Breast adenocarcinoma | Homo sapiens | CVCL_0033 | |
| MDA-MB-468 | Breast adenocarcinoma | Homo sapiens | CVCL_0419 | |
| MDA-MB-453 | Breast adenocarcinoma | Homo sapiens | CVCL_0418 | |
| MDA-MB-361 | Breast adenocarcinoma | Homo sapiens | CVCL_0620 | |
| MDA-MB-231 | Breast adenocarcinoma | Homo sapiens | CVCL_0062 | |
| MCF-7 | Invasive breast carcinoma | Homo sapiens | CVCL_0031 | |
| MCF-10A | Normal | Homo sapiens | CVCL_0598 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| BT-549 | Invasive breast carcinoma | Homo sapiens | CVCL_1092 | |
| BT-474 | Invasive breast carcinoma | Homo sapiens | CVCL_0179 | |
| AU565 | Breast adenocarcinoma | Homo sapiens | CVCL_1074 | |
| In-vivo Model | Luciferase-labeled rSKBR3 and MDA-MB-361 cells (1 × 107 cells) mixed with 1:1 Matrigel (Corning, 356237) were subcutaneously injected into the fat pads of mice. After a tumor was palpable, the mice were randomized into four groups (five mice per group), and they were treated with vehicle, trastuzumab (20 mg/kg, intraperitoneal administration), roblitinib (30 mg/kg, oral administration), or a combination of both drugs. | |||
| Experiment 3 Reporting the m6A-centered Disease Response | [1] | |||
| Response Summary | m6A-hypomethylation regulated Fibroblast growth factor receptor 4 (FGFR4) phosphorylates GSK-3beta and activates beta-catenin/TCF4-SLC7A11/FPN1 signaling to drive anti-HER2 resistance. Knockdown of METTL14 significantly increased the expression level of FGFR4 in HER2-positive breast cancer cells. FGFR4 reduced the sensitivity of HER2-positive breast cancer to trastuzumab plus pertuzumab or tucatinib. These results pinpoint a mechanism of anti-HER2 resistance and provide a strategy for overcoming resistance via FGFR4 inhibition in recalcitrant HER2-positive breast cancer. | |||
| Responsed Disease | Breast cancer [ICD-11: 2C60] | |||
| Target Regulator | Methyltransferase-like 14 (METTL14) | WRITER | ||
| Target Regulation | Down regulation | |||
| Responsed Drug | Tucatinib | Approved | ||
| Pathway Response | Wnt signaling pathway | hsa04310 | ||
| Cell Process | Glutathione synthesis | |||
| In-vitro Model | ZR-75-1 | Invasive breast carcinoma | Homo sapiens | CVCL_0588 |
| T-47D | Invasive breast carcinoma | Homo sapiens | CVCL_0553 | |
| SUM-159 (A mesenchymal triple-negative breast cancer cell line) | ||||
| SK-BR-3 | Breast adenocarcinoma | Homo sapiens | CVCL_0033 | |
| MDA-MB-468 | Breast adenocarcinoma | Homo sapiens | CVCL_0419 | |
| MDA-MB-453 | Breast adenocarcinoma | Homo sapiens | CVCL_0418 | |
| MDA-MB-361 | Breast adenocarcinoma | Homo sapiens | CVCL_0620 | |
| MDA-MB-231 | Breast adenocarcinoma | Homo sapiens | CVCL_0062 | |
| MCF-7 | Invasive breast carcinoma | Homo sapiens | CVCL_0031 | |
| MCF-10A | Normal | Homo sapiens | CVCL_0598 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| BT-549 | Invasive breast carcinoma | Homo sapiens | CVCL_1092 | |
| BT-474 | Invasive breast carcinoma | Homo sapiens | CVCL_0179 | |
| AU565 | Breast adenocarcinoma | Homo sapiens | CVCL_1074 | |
| In-vivo Model | Luciferase-labeled rSKBR3 and MDA-MB-361 cells (1 × 107 cells) mixed with 1:1 Matrigel (Corning, 356237) were subcutaneously injected into the fat pads of mice. After a tumor was palpable, the mice were randomized into four groups (five mice per group), and they were treated with vehicle, trastuzumab (20 mg/kg, intraperitoneal administration), roblitinib (30 mg/kg, oral administration), or a combination of both drugs. | |||
Pertuzumab
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response | [1] | |||
| Response Summary | m6A-hypomethylation regulated Fibroblast growth factor receptor 4 (FGFR4) phosphorylates GSK-3beta and activates beta-catenin/TCF4-SLC7A11/FPN1 signaling to drive anti-HER2 resistance. Knockdown of METTL14 significantly increased the expression level of FGFR4 in HER2-positive breast cancer cells. FGFR4 reduced the sensitivity of HER2-positive breast cancer to trastuzumab plus pertuzumab or tucatinib. These results pinpoint a mechanism of anti-HER2 resistance and provide a strategy for overcoming resistance via FGFR4 inhibition in recalcitrant HER2-positive breast cancer. | |||
| Target Regulator | Methyltransferase-like 14 (METTL14) | WRITER | ||
| Target Regulation | Down regulation | |||
| Responsed Disease | Breast cancer | ICD-11: 2C60 | ||
| Pathway Response | Wnt signaling pathway | hsa04310 | ||
| Cell Process | Glutathione synthesis | |||
| In-vitro Model | ZR-75-1 | Invasive breast carcinoma | Homo sapiens | CVCL_0588 |
| T-47D | Invasive breast carcinoma | Homo sapiens | CVCL_0553 | |
| SUM-159 (A mesenchymal triple-negative breast cancer cell line) | ||||
| SK-BR-3 | Breast adenocarcinoma | Homo sapiens | CVCL_0033 | |
| MDA-MB-468 | Breast adenocarcinoma | Homo sapiens | CVCL_0419 | |
| MDA-MB-453 | Breast adenocarcinoma | Homo sapiens | CVCL_0418 | |
| MDA-MB-361 | Breast adenocarcinoma | Homo sapiens | CVCL_0620 | |
| MDA-MB-231 | Breast adenocarcinoma | Homo sapiens | CVCL_0062 | |
| MCF-7 | Invasive breast carcinoma | Homo sapiens | CVCL_0031 | |
| MCF-10A | Normal | Homo sapiens | CVCL_0598 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| BT-549 | Invasive breast carcinoma | Homo sapiens | CVCL_1092 | |
| BT-474 | Invasive breast carcinoma | Homo sapiens | CVCL_0179 | |
| AU565 | Breast adenocarcinoma | Homo sapiens | CVCL_1074 | |
| In-vivo Model | Luciferase-labeled rSKBR3 and MDA-MB-361 cells (1 × 107 cells) mixed with 1:1 Matrigel (Corning, 356237) were subcutaneously injected into the fat pads of mice. After a tumor was palpable, the mice were randomized into four groups (five mice per group), and they were treated with vehicle, trastuzumab (20 mg/kg, intraperitoneal administration), roblitinib (30 mg/kg, oral administration), or a combination of both drugs. | |||
Trastuzumab
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response | [1] | |||
| Response Summary | m6A-hypomethylation regulated Fibroblast growth factor receptor 4 (FGFR4) phosphorylates GSK-3beta and activates beta-catenin/TCF4-SLC7A11/FPN1 signaling to drive anti-HER2 resistance. Knockdown of METTL14 significantly increased the expression level of FGFR4 in HER2-positive breast cancer cells. FGFR4 reduced the sensitivity of HER2-positive breast cancer to trastuzumab plus pertuzumab or tucatinib. These results pinpoint a mechanism of anti-HER2 resistance and provide a strategy for overcoming resistance via FGFR4 inhibition in recalcitrant HER2-positive breast cancer. | |||
| Target Regulator | Methyltransferase-like 14 (METTL14) | WRITER | ||
| Target Regulation | Down regulation | |||
| Responsed Disease | Breast cancer | ICD-11: 2C60 | ||
| Pathway Response | Wnt signaling pathway | hsa04310 | ||
| Cell Process | Glutathione synthesis | |||
| In-vitro Model | ZR-75-1 | Invasive breast carcinoma | Homo sapiens | CVCL_0588 |
| T-47D | Invasive breast carcinoma | Homo sapiens | CVCL_0553 | |
| SUM-159 (A mesenchymal triple-negative breast cancer cell line) | ||||
| SK-BR-3 | Breast adenocarcinoma | Homo sapiens | CVCL_0033 | |
| MDA-MB-468 | Breast adenocarcinoma | Homo sapiens | CVCL_0419 | |
| MDA-MB-453 | Breast adenocarcinoma | Homo sapiens | CVCL_0418 | |
| MDA-MB-361 | Breast adenocarcinoma | Homo sapiens | CVCL_0620 | |
| MDA-MB-231 | Breast adenocarcinoma | Homo sapiens | CVCL_0062 | |
| MCF-7 | Invasive breast carcinoma | Homo sapiens | CVCL_0031 | |
| MCF-10A | Normal | Homo sapiens | CVCL_0598 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| BT-549 | Invasive breast carcinoma | Homo sapiens | CVCL_1092 | |
| BT-474 | Invasive breast carcinoma | Homo sapiens | CVCL_0179 | |
| AU565 | Breast adenocarcinoma | Homo sapiens | CVCL_1074 | |
| In-vivo Model | Luciferase-labeled rSKBR3 and MDA-MB-361 cells (1 × 107 cells) mixed with 1:1 Matrigel (Corning, 356237) were subcutaneously injected into the fat pads of mice. After a tumor was palpable, the mice were randomized into four groups (five mice per group), and they were treated with vehicle, trastuzumab (20 mg/kg, intraperitoneal administration), roblitinib (30 mg/kg, oral administration), or a combination of both drugs. | |||
Tucatinib
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response | [1] | |||
| Response Summary | m6A-hypomethylation regulated Fibroblast growth factor receptor 4 (FGFR4) phosphorylates GSK-3beta and activates beta-catenin/TCF4-SLC7A11/FPN1 signaling to drive anti-HER2 resistance. Knockdown of METTL14 significantly increased the expression level of FGFR4 in HER2-positive breast cancer cells. FGFR4 reduced the sensitivity of HER2-positive breast cancer to trastuzumab plus pertuzumab or tucatinib. These results pinpoint a mechanism of anti-HER2 resistance and provide a strategy for overcoming resistance via FGFR4 inhibition in recalcitrant HER2-positive breast cancer. | |||
| Target Regulator | Methyltransferase-like 14 (METTL14) | WRITER | ||
| Target Regulation | Down regulation | |||
| Responsed Disease | Breast cancer | ICD-11: 2C60 | ||
| Pathway Response | Wnt signaling pathway | hsa04310 | ||
| Cell Process | Glutathione synthesis | |||
| In-vitro Model | ZR-75-1 | Invasive breast carcinoma | Homo sapiens | CVCL_0588 |
| T-47D | Invasive breast carcinoma | Homo sapiens | CVCL_0553 | |
| SUM-159 (A mesenchymal triple-negative breast cancer cell line) | ||||
| SK-BR-3 | Breast adenocarcinoma | Homo sapiens | CVCL_0033 | |
| MDA-MB-468 | Breast adenocarcinoma | Homo sapiens | CVCL_0419 | |
| MDA-MB-453 | Breast adenocarcinoma | Homo sapiens | CVCL_0418 | |
| MDA-MB-361 | Breast adenocarcinoma | Homo sapiens | CVCL_0620 | |
| MDA-MB-231 | Breast adenocarcinoma | Homo sapiens | CVCL_0062 | |
| MCF-7 | Invasive breast carcinoma | Homo sapiens | CVCL_0031 | |
| MCF-10A | Normal | Homo sapiens | CVCL_0598 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| BT-549 | Invasive breast carcinoma | Homo sapiens | CVCL_1092 | |
| BT-474 | Invasive breast carcinoma | Homo sapiens | CVCL_0179 | |
| AU565 | Breast adenocarcinoma | Homo sapiens | CVCL_1074 | |
| In-vivo Model | Luciferase-labeled rSKBR3 and MDA-MB-361 cells (1 × 107 cells) mixed with 1:1 Matrigel (Corning, 356237) were subcutaneously injected into the fat pads of mice. After a tumor was palpable, the mice were randomized into four groups (five mice per group), and they were treated with vehicle, trastuzumab (20 mg/kg, intraperitoneal administration), roblitinib (30 mg/kg, oral administration), or a combination of both drugs. | |||
Full List of Crosstalk(s) between m6A Modification and Epigenetic Regulation Related to This Regulator
DNA modification
m6A Regulator: Methyltransferase-like 14 (METTL14)
| In total 9 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT02209 | ||
| Epigenetic Regulator | DNA (cytosine-5)-methyltransferase 3B (DNMT3B) | |
| Regulated Target | Methyltransferase-like protein 14 (METTL14) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Breast cancer | |
| Drug | Pertuzumab | |
| Crosstalk ID: M6ACROT02215 | ||
| Epigenetic Regulator | DNA (cytosine-5)-methyltransferase 3B (DNMT3B) | |
| Regulated Target | Methyltransferase-like protein 14 (METTL14) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Breast cancer | |
| Drug | Tucatinib | |
| Crosstalk ID: M6ACROT02222 | ||
| Epigenetic Regulator | DNA (cytosine-5)-methyltransferase 3B (DNMT3B) | |
| Regulated Target | Methyltransferase-like protein 14 (METTL14) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Breast cancer | |
| Drug | Trastuzumab | |
| Crosstalk ID: M6ACROT02233 | ||
| Epigenetic Regulator | Cysteine methyltransferase DNMT3A (DNMT3A) | |
| Regulated Target | Methyltransferase-like protein 14 (METTL14) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Breast cancer | |
| Drug | Pertuzumab | |
| Crosstalk ID: M6ACROT02239 | ||
| Epigenetic Regulator | Cysteine methyltransferase DNMT3A (DNMT3A) | |
| Regulated Target | Methyltransferase-like protein 14 (METTL14) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Breast cancer | |
| Drug | Tucatinib | |
| Crosstalk ID: M6ACROT02246 | ||
| Epigenetic Regulator | Cysteine methyltransferase DNMT3A (DNMT3A) | |
| Regulated Target | Methyltransferase-like protein 14 (METTL14) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Breast cancer | |
| Drug | Trastuzumab | |
| Crosstalk ID: M6ACROT02257 | ||
| Epigenetic Regulator | DNA (cytosine-5)-methyltransferase 1 (DNMT1) | |
| Regulated Target | Methyltransferase-like protein 14 (METTL14) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Breast cancer | |
| Drug | Pertuzumab | |
| Crosstalk ID: M6ACROT02263 | ||
| Epigenetic Regulator | DNA (cytosine-5)-methyltransferase 1 (DNMT1) | |
| Regulated Target | Methyltransferase-like protein 14 (METTL14) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Breast cancer | |
| Drug | Tucatinib | |
| Crosstalk ID: M6ACROT02270 | ||
| Epigenetic Regulator | DNA (cytosine-5)-methyltransferase 1 (DNMT1) | |
| Regulated Target | Methyltransferase-like protein 14 (METTL14) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Breast cancer | |
| Drug | Trastuzumab | |
RNA Modification Sequencing Data Associated with the Target (ID: M6ATAR00526)
| In total 3 m6A sequence/site(s) in this target gene | |||
| mod ID: M5CSITE003698 | Click to Show/Hide the Full List | ||
| mod site | chr5:177089902-177089903:+ | [2] | |
| Sequence | GCGAGAGAGGACTGGCCTTGCAGGGCGCAGGGCCCTAAGCT | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000509511.5; ENST00000292408.9; ENST00000513166.1; ENST00000503708.5; ENST00000502906.5; ENST00000430285.5; ENST00000507708.1; ENST00000393637.5; ENST00000393648.6; ENST00000426612.5; ENST00000510911.5; ENST00000514472.1 | ||
| External Link | RMBase: m5C_site_36448 | ||
| mod ID: M5CSITE003699 | Click to Show/Hide the Full List | ||
| mod site | chr5:177093250-177093251:+ | [3] | |
| Sequence | TGCTGCTGGCCGGGCTGTATCGAGGGCAGGCGCTCCACGGC | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000393637.5; ENST00000393648.6; ENST00000511076.1; ENST00000502906.5; ENST00000508139.1; ENST00000292408.9 | ||
| External Link | RMBase: m5C_site_36449 | ||
| mod ID: M5CSITE003700 | Click to Show/Hide the Full List | ||
| mod site | chr5:177097801-177097802:+ | [3] | |
| Sequence | CAGAGTTGCTGTGCCGTGTCCAAGGGCCGTGCCCTTGCCCT | ||
| Cell/Tissue List | heart; HEK293T; liver; testis; HeLa; T24 | ||
| Seq Type List | Bisulfite-seq; m5C-RIP-seq | ||
| Transcript ID List | ENST00000393648.6; ENST00000292408.9 | ||
| External Link | RMBase: m5C_site_36450 | ||
N6-methyladenosine (m6A)
| In total 44 m6A sequence/site(s) in this target gene | |||
| mod ID: M6ASITE072875 | Click to Show/Hide the Full List | ||
| mod site | chr5:177086956-177086957:+ | [4] | |
| Sequence | AGCGCTCGGGCTGTCTGCGGACCCTGCCGCGTGCAGGGGTC | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; HepG2; U2OS; H1A; H1B; A549; Jurkat; HEK293T; iSLK; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000510911.5; ENST00000507708.1; ENST00000502906.5; ENST00000393648.6; ENST00000292408.9; ENST00000503708.5; ENST00000514472.1 | ||
| External Link | RMBase: m6A_site_699487 | ||
| mod ID: M6ASITE072876 | Click to Show/Hide the Full List | ||
| mod site | chr5:177087042-177087043:+ | [4] | |
| Sequence | GGAGGAGCCAGGTGAGCAGGACCCTGTGCTGGGCGCGGAGT | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; HepG2; U2OS; H1A; H1B; A549; Jurkat; HEK293T; iSLK; endometrial; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000514472.1; ENST00000507708.1; ENST00000292408.9; ENST00000510911.5; ENST00000503708.5; ENST00000393648.6; ENST00000502906.5 | ||
| External Link | RMBase: m6A_site_699488 | ||
| mod ID: M6ASITE072877 | Click to Show/Hide the Full List | ||
| mod site | chr5:177087445-177087446:+ | [4] | |
| Sequence | GCCGGCGGCAGAGATGAGGGACCTGAGGCCCCGAAAAGTTC | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; HepG2; U2OS; H1A; H1B; A549; HEK293T; iSLK; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000503708.5; ENST00000513166.1; ENST00000292408.9; ENST00000514472.1; ENST00000393648.6; ENST00000507708.1; ENST00000502906.5; ENST00000510911.5 | ||
| External Link | RMBase: m6A_site_699489 | ||
| mod ID: M6ASITE072878 | Click to Show/Hide the Full List | ||
| mod site | chr5:177087518-177087519:+ | [4] | |
| Sequence | GTGCGGGAGGCTGAAGGAGAACCCAGGACTGTCTGATGCCT | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | HeLa; HepG2 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000502906.5; ENST00000514472.1; ENST00000503708.5; ENST00000510911.5; ENST00000393648.6; ENST00000292408.9; ENST00000513166.1; ENST00000507708.1 | ||
| External Link | RMBase: m6A_site_699490 | ||
| mod ID: M6ASITE072879 | Click to Show/Hide the Full List | ||
| mod site | chr5:177087525-177087526:+ | [4] | |
| Sequence | AGGCTGAAGGAGAACCCAGGACTGTCTGATGCCTAAGGCAG | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa; HepG2 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000514472.1; ENST00000503708.5; ENST00000507708.1; ENST00000393648.6; ENST00000292408.9; ENST00000513166.1; ENST00000502906.5; ENST00000510911.5 | ||
| External Link | RMBase: m6A_site_699491 | ||
| mod ID: M6ASITE072880 | Click to Show/Hide the Full List | ||
| mod site | chr5:177087611-177087612:+ | [5] | |
| Sequence | CATGGGTGACTCGACTAAGGACTCTGATATCAGGGCAGCCT | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HepG2 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000514472.1; ENST00000503708.5; ENST00000502906.5; ENST00000510911.5; ENST00000507708.1; ENST00000513166.1; ENST00000292408.9; ENST00000393648.6 | ||
| External Link | RMBase: m6A_site_699492 | ||
| mod ID: M6ASITE072881 | Click to Show/Hide the Full List | ||
| mod site | chr5:177088478-177088479:+ | [6] | |
| Sequence | GGGCAGGGGAAGCCAGATGGACTGGAAGTGGAGTGGCAGTG | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HepG2; HeLa; H1A; Huh7; HEC-1-A | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000510911.5; ENST00000292408.9; ENST00000507708.1; ENST00000502906.5; ENST00000513166.1; ENST00000514472.1; ENST00000503708.5; ENST00000393648.6 | ||
| External Link | RMBase: m6A_site_699493 | ||
| mod ID: M6ASITE072882 | Click to Show/Hide the Full List | ||
| mod site | chr5:177089892-177089893:+ | [4] | |
| Sequence | CACTGTGTCTGCGAGAGAGGACTGGCCTTGCAGGGCGCAGG | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa; HepG2; Huh7 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000510911.5; ENST00000502906.5; ENST00000426612.5; ENST00000503708.5; ENST00000430285.5; ENST00000509511.5; ENST00000513166.1; ENST00000393637.5; ENST00000393648.6; ENST00000514472.1; ENST00000292408.9; ENST00000507708.1 | ||
| External Link | RMBase: m6A_site_699494 | ||
| mod ID: M6ASITE072883 | Click to Show/Hide the Full List | ||
| mod site | chr5:177090646-177090647:+ | [7] | |
| Sequence | TGCAGAATCTCACCTTGATTACAGGTGGTAAGAGACTCTAG | ||
| Motif Score | 2.07285119 | ||
| Cell/Tissue List | kidney | ||
| Seq Type List | m6A-REF-seq | ||
| Transcript ID List | ENST00000503708.5; ENST00000430285.5; ENST00000502906.5; ENST00000393637.5; ENST00000509511.5; ENST00000393648.6; ENST00000426612.5; ENST00000292408.9 | ||
| External Link | RMBase: m6A_site_699495 | ||
| mod ID: M6ASITE072884 | Click to Show/Hide the Full List | ||
| mod site | chr5:177090774-177090775:+ | [8] | |
| Sequence | CTCCAGCAACGATGATGAGGACCCCAAGTCCCATAGGGACC | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | Huh7 | ||
| Seq Type List | MeRIP-seq | ||
| Transcript ID List | ENST00000509511.5; ENST00000502906.5; ENST00000292408.9; ENST00000503708.5; ENST00000430285.5; ENST00000393648.6; ENST00000426612.5; ENST00000393637.5 | ||
| External Link | RMBase: m6A_site_699496 | ||
| mod ID: M6ASITE072885 | Click to Show/Hide the Full List | ||
| mod site | chr5:177090792-177090793:+ | [8] | |
| Sequence | GGACCCCAAGTCCCATAGGGACCCCTCGAATAGGCACAGTT | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | Huh7 | ||
| Seq Type List | MeRIP-seq | ||
| Transcript ID List | ENST00000393648.6; ENST00000503708.5; ENST00000393637.5; ENST00000430285.5; ENST00000292408.9; ENST00000502906.5; ENST00000426612.5; ENST00000509511.5 | ||
| External Link | RMBase: m6A_site_699497 | ||
| mod ID: M6ASITE072886 | Click to Show/Hide the Full List | ||
| mod site | chr5:177090843-177090844:+ | [8] | |
| Sequence | AGGTCAGTAGGTCTCCAAGGACTTGTGTCCCCGCTGCTGCT | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | Huh7 | ||
| Seq Type List | MeRIP-seq | ||
| Transcript ID List | ENST00000502906.5; ENST00000503708.5; ENST00000292408.9; ENST00000430285.5; ENST00000393648.6; ENST00000426612.5; ENST00000393637.5; ENST00000509511.5 | ||
| External Link | RMBase: m6A_site_699498 | ||
| mod ID: M6ASITE072887 | Click to Show/Hide the Full List | ||
| mod site | chr5:177090997-177090998:+ | [9] | |
| Sequence | GCATGCAGTACCTGCGGGGAACACCGTCAAGTTCCGCTGTC | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000509511.5; ENST00000503708.5; ENST00000393648.6; ENST00000430285.5; ENST00000292408.9; ENST00000426612.5; ENST00000502906.5; ENST00000393637.5 | ||
| External Link | RMBase: m6A_site_699499 | ||
| mod ID: M6ASITE072888 | Click to Show/Hide the Full List | ||
| mod site | chr5:177091741-177091742:+ | [9] | |
| Sequence | TGGTGCCCTCGGACCGCGGCACATACACCTGCCTGGTAGAG | ||
| Motif Score | 2.830589286 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000292408.9; ENST00000502906.5; ENST00000393648.6; ENST00000393637.5; ENST00000509511.5 | ||
| External Link | RMBase: m6A_site_699500 | ||
| mod ID: M6ASITE072889 | Click to Show/Hide the Full List | ||
| mod site | chr5:177091745-177091746:+ | [9] | |
| Sequence | GCCCTCGGACCGCGGCACATACACCTGCCTGGTAGAGAACG | ||
| Motif Score | 2.110482143 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000509511.5; ENST00000393648.6; ENST00000502906.5; ENST00000292408.9; ENST00000393637.5 | ||
| External Link | RMBase: m6A_site_699501 | ||
| mod ID: M6ASITE072890 | Click to Show/Hide the Full List | ||
| mod site | chr5:177092365-177092366:+ | [9] | |
| Sequence | GCAGGCCGGGCTCCCGGCCAACACCACAGCCGTGGTGGGCA | ||
| Motif Score | 2.173910714 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000508139.1; ENST00000393648.6; ENST00000502906.5; ENST00000509511.5; ENST00000393637.5; ENST00000292408.9 | ||
| External Link | RMBase: m6A_site_699502 | ||
| mod ID: M6ASITE072891 | Click to Show/Hide the Full List | ||
| mod site | chr5:177092449-177092450:+ | [7] | |
| Sequence | CCACATCCAGTGGCTGAAGCACATCGTCATCAACGGCAGCA | ||
| Motif Score | 2.830589286 | ||
| Cell/Tissue List | liver | ||
| Seq Type List | m6A-REF-seq | ||
| Transcript ID List | ENST00000509511.5; ENST00000502906.5; ENST00000393637.5; ENST00000292408.9; ENST00000508139.1; ENST00000393648.6 | ||
| External Link | RMBase: m6A_site_699503 | ||
| mod ID: M6ASITE072892 | Click to Show/Hide the Full List | ||
| mod site | chr5:177092652-177092653:+ | [6] | |
| Sequence | TCCCCCACCCCAGACTGCAGACATCAATAGCTCAGAGGTGG | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HepG2; hESC-HEK293T | ||
| Seq Type List | m6A-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000508139.1; ENST00000509511.5; ENST00000502906.5; ENST00000292408.9; ENST00000393637.5; ENST00000393648.6 | ||
| External Link | RMBase: m6A_site_699504 | ||
| mod ID: M6ASITE072893 | Click to Show/Hide the Full List | ||
| mod site | chr5:177092718-177092719:+ | [7] | |
| Sequence | AGCCGAGGACGCAGGCGAGTACACCTGCCTCGCAGGCAATT | ||
| Motif Score | 2.856142857 | ||
| Cell/Tissue List | kidney; hESC-HEK293T | ||
| Seq Type List | m6A-REF-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000509511.5; ENST00000508139.1; ENST00000292408.9; ENST00000393637.5; ENST00000393648.6; ENST00000502906.5 | ||
| External Link | RMBase: m6A_site_699505 | ||
| mod ID: M6ASITE072894 | Click to Show/Hide the Full List | ||
| mod site | chr5:177093148-177093149:+ | [9] | |
| Sequence | CGAGGGCAGAGGAGGACCCCACATGGACCGCAGCAGCGCCC | ||
| Motif Score | 2.053113095 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000292408.9; ENST00000502906.5; ENST00000508139.1; ENST00000393648.6; ENST00000393637.5 | ||
| External Link | RMBase: m6A_site_699506 | ||
| mod ID: M6ASITE072895 | Click to Show/Hide the Full List | ||
| mod site | chr5:177093384-177093385:+ | [5] | |
| Sequence | GCCTGACTCCAGCAGGCAGAACCAAGTCTCCCACTTTGCAG | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | HepG2 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000508139.1; ENST00000393648.6; ENST00000292408.9; ENST00000502906.5; ENST00000511076.1; ENST00000393637.5 | ||
| External Link | RMBase: m6A_site_699507 | ||
| mod ID: M6ASITE072896 | Click to Show/Hide the Full List | ||
| mod site | chr5:177093547-177093548:+ | [4] | |
| Sequence | ACTATGGGAGTTCCCCCGGGACAGGTGCGCTGAGCTGTGTG | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; kidney | ||
| Seq Type List | m6A-seq; m6A-REF-seq | ||
| Transcript ID List | ENST00000502906.5; ENST00000393648.6; ENST00000292408.9; ENST00000393637.5; ENST00000511076.1 | ||
| External Link | RMBase: m6A_site_699508 | ||
| mod ID: M6ASITE072897 | Click to Show/Hide the Full List | ||
| mod site | chr5:177093724-177093725:+ | [4] | |
| Sequence | TGCAGAGGCCTTTGGCATGGACCCTGCCCGGCCTGACCAAG | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000502906.5; ENST00000393648.6; ENST00000393637.5; ENST00000511076.1; ENST00000292408.9 | ||
| External Link | RMBase: m6A_site_699509 | ||
| mod ID: M6ASITE072898 | Click to Show/Hide the Full List | ||
| mod site | chr5:177095329-177095330:+ | [5] | |
| Sequence | CCCGTCTGCTGCCCTTACAGACAACGCCTCTGACAAGGACC | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HepG2 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000393648.6; ENST00000483872.2; ENST00000292408.9; ENST00000393637.5; ENST00000502906.5; ENST00000511076.1 | ||
| External Link | RMBase: m6A_site_699510 | ||
| mod ID: M6ASITE072899 | Click to Show/Hide the Full List | ||
| mod site | chr5:177095341-177095342:+ | [9] | |
| Sequence | CCTTACAGACAACGCCTCTGACAAGGACCTGGCCGACCTGG | ||
| Motif Score | 2.859755952 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000292408.9; ENST00000483872.2; ENST00000393637.5; ENST00000502906.5; ENST00000511076.1; ENST00000393648.6 | ||
| External Link | RMBase: m6A_site_699511 | ||
| mod ID: M6ASITE072900 | Click to Show/Hide the Full List | ||
| mod site | chr5:177095347-177095348:+ | [5] | |
| Sequence | AGACAACGCCTCTGACAAGGACCTGGCCGACCTGGTCTCGG | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HepG2 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000502906.5; ENST00000483872.2; ENST00000511076.1; ENST00000393637.5; ENST00000393648.6; ENST00000292408.9 | ||
| External Link | RMBase: m6A_site_699512 | ||
| mod ID: M6ASITE072901 | Click to Show/Hide the Full List | ||
| mod site | chr5:177095396-177095397:+ | [9] | |
| Sequence | GTGATGAAGCTGATCGGCCGACACAAGAACATCATCAACCT | ||
| Motif Score | 2.865571429 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000393648.6; ENST00000502906.5; ENST00000511076.1; ENST00000292408.9; ENST00000393637.5; ENST00000483872.2 | ||
| External Link | RMBase: m6A_site_699513 | ||
| mod ID: M6ASITE072902 | Click to Show/Hide the Full List | ||
| mod site | chr5:177095404-177095405:+ | [6] | |
| Sequence | GCTGATCGGCCGACACAAGAACATCATCAACCTGCTTGGTG | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HepG2; hESC-HEK293T | ||
| Seq Type List | m6A-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000393637.5; ENST00000393648.6; ENST00000511076.1; ENST00000292408.9; ENST00000483872.2; ENST00000502906.5 | ||
| External Link | RMBase: m6A_site_699514 | ||
| mod ID: M6ASITE072903 | Click to Show/Hide the Full List | ||
| mod site | chr5:177095571-177095572:+ | [5] | |
| Sequence | GGAGTGCGCCGCCAAGGGAAACCTGCGGGAGTTCCTGCGGG | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | HepG2 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000393648.6; ENST00000483872.2; ENST00000502906.5; ENST00000393637.5; ENST00000511076.1; ENST00000292408.9 | ||
| External Link | RMBase: m6A_site_699515 | ||
| mod ID: M6ASITE072904 | Click to Show/Hide the Full List | ||
| mod site | chr5:177096069-177096070:+ | [5] | |
| Sequence | CCTGCAGTGTATCCACCGGGACCTGGCTGCCCGCAATGTGC | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HepG2 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000393637.5; ENST00000511076.1; ENST00000393648.6; ENST00000292408.9; ENST00000502906.5 | ||
| External Link | RMBase: m6A_site_699516 | ||
| mod ID: M6ASITE072905 | Click to Show/Hide the Full List | ||
| mod site | chr5:177096102-177096103:+ | [6] | |
| Sequence | CAATGTGCTGGTGACTGAGGACAATGTGATGAAGATTGCTG | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HepG2; liver | ||
| Seq Type List | m6A-seq; m6A-REF-seq | ||
| Transcript ID List | ENST00000393637.5; ENST00000502906.5; ENST00000393648.6; ENST00000292408.9; ENST00000511076.1 | ||
| External Link | RMBase: m6A_site_699517 | ||
| mod ID: M6ASITE072906 | Click to Show/Hide the Full List | ||
| mod site | chr5:177096702-177096703:+ | [4] | |
| Sequence | TTCTCGCTGCTGCGGGAGGGACATCGGATGGACCGACCCCC | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000292408.9; ENST00000393648.6; ENST00000393637.5; ENST00000502906.5 | ||
| External Link | RMBase: m6A_site_699518 | ||
| mod ID: M6ASITE072907 | Click to Show/Hide the Full List | ||
| mod site | chr5:177096713-177096714:+ | [4] | |
| Sequence | GCGGGAGGGACATCGGATGGACCGACCCCCACACTGCCCCC | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000393648.6; ENST00000292408.9; ENST00000502906.5; ENST00000393637.5 | ||
| External Link | RMBase: m6A_site_699519 | ||
| mod ID: M6ASITE072908 | Click to Show/Hide the Full List | ||
| mod site | chr5:177096723-177096724:+ | [9] | |
| Sequence | CATCGGATGGACCGACCCCCACACTGCCCCCCAGAGCTGTG | ||
| Motif Score | 2.053113095 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000393648.6; ENST00000393637.5; ENST00000502906.5; ENST00000292408.9 | ||
| External Link | RMBase: m6A_site_699520 | ||
| mod ID: M6ASITE072909 | Click to Show/Hide the Full List | ||
| mod site | chr5:177097192-177097193:+ | [4] | |
| Sequence | ACCCTCCTATCCCTCATCAAACTCCCCACCAAACTCCTCCC | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa; HepG2 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000292408.9; ENST00000513423.1; ENST00000393648.6; ENST00000393637.5; ENST00000502906.5 | ||
| External Link | RMBase: m6A_site_699521 | ||
| mod ID: M6ASITE072910 | Click to Show/Hide the Full List | ||
| mod site | chr5:177097204-177097205:+ | [4] | |
| Sequence | CTCATCAAACTCCCCACCAAACTCCTCCCCACCCAGAGAAC | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa; HepG2 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000393648.6; ENST00000292408.9; ENST00000502906.5; ENST00000393637.5; ENST00000513423.1 | ||
| External Link | RMBase: m6A_site_699522 | ||
| mod ID: M6ASITE072911 | Click to Show/Hide the Full List | ||
| mod site | chr5:177097223-177097224:+ | [4] | |
| Sequence | AACTCCTCCCCACCCAGAGAACCCCCGGTCCTCCCCTTCCT | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | HeLa; HepG2 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000393648.6; ENST00000513423.1; ENST00000292408.9; ENST00000393637.5; ENST00000502906.5 | ||
| External Link | RMBase: m6A_site_699523 | ||
| mod ID: M6ASITE072912 | Click to Show/Hide the Full List | ||
| mod site | chr5:177097368-177097369:+ | [6] | |
| Sequence | GCAGCTGGTGGAGGCGCTGGACAAGGTCCTGCTGGCCGTCT | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HepG2; hESC-HEK293T | ||
| Seq Type List | m6A-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000502906.5; ENST00000513423.1; ENST00000393648.6; ENST00000393637.5; ENST00000292408.9 | ||
| External Link | RMBase: m6A_site_699524 | ||
| mod ID: M6ASITE072913 | Click to Show/Hide the Full List | ||
| mod site | chr5:177097552-177097553:+ | [5] | |
| Sequence | GACCTCCGCCTGACCTTCGGACCCTATTCCCCCTCTGGTGG | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HepG2 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000292408.9; ENST00000393648.6; ENST00000502906.5; ENST00000393637.5; ENST00000513423.1 | ||
| External Link | RMBase: m6A_site_699525 | ||
| mod ID: M6ASITE072914 | Click to Show/Hide the Full List | ||
| mod site | chr5:177097670-177097671:+ | [6] | |
| Sequence | CCTTCGGGTCTGGGGTGCAGACATGAGCAAGGCTCAAGGCT | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HepG2; hESC-HEK293T | ||
| Seq Type List | m6A-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000292408.9; ENST00000513423.1; ENST00000393637.5; ENST00000502906.5; ENST00000393648.6 | ||
| External Link | RMBase: m6A_site_699526 | ||
| mod ID: M6ASITE072915 | Click to Show/Hide the Full List | ||
| mod site | chr5:177097882-177097883:+ | [10] | |
| Sequence | GGTTCTGCTCGGCTTCTTGGACCTTGGCGCTTAGTCCCCAT | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HepG2; Huh7 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000393648.6; ENST00000292408.9 | ||
| External Link | RMBase: m6A_site_699527 | ||
| mod ID: M6ASITE072916 | Click to Show/Hide the Full List | ||
| mod site | chr5:177097944-177097945:+ | [10] | |
| Sequence | CTGGAGAGCTGCTATGCTAAACCTCCTGCCTCCCAATACCA | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | HepG2; Huh7 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000393648.6; ENST00000292408.9 | ||
| External Link | RMBase: m6A_site_699528 | ||
| mod ID: M6ASITE072917 | Click to Show/Hide the Full List | ||
| mod site | chr5:177097987-177097988:+ | [8] | |
| Sequence | AGGAGGTTCTGGGCCTCTGAACCCCCTTTCCCCACACCTCC | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | Huh7 | ||
| Seq Type List | MeRIP-seq | ||
| Transcript ID List | ENST00000393648.6; ENST00000292408.9 | ||
| External Link | RMBase: m6A_site_699529 | ||
| mod ID: M6ASITE072918 | Click to Show/Hide the Full List | ||
| mod site | chr5:177098083-177098084:+ | [8] | |
| Sequence | AAGCTGGAAGCCTGCCGAAAACAGGAGCAAATGGCGTTTTA | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | Huh7 | ||
| Seq Type List | MeRIP-seq | ||
| Transcript ID List | ENST00000292408.9 | ||
| External Link | RMBase: m6A_site_699530 | ||
References