m6A Target Gene Information
General Information of the m6A Target Gene (ID: M6ATAR00268)
Full List of m6A Methylation Regulator of This Target Gene and Corresponding Disease/Drug Response(s)
GSK3B
can be regulated by the following regulator(s), and cause disease/drug response(s). You can browse detail information of regulator(s) or disease/drug response(s).
Browse Regulator
Browse Disease
Browse Drug
Fat mass and obesity-associated protein (FTO) [ERASER]
| Representative RNA-seq result indicating the expression of this target gene regulated by FTO | ||
| Cell Line | B16-OVA cell line | Mus musculus |
|
Treatment: shFTO B16-OVA cells
Control: shNC B16-OVA cells
|
GSE154952 | |
| Regulation |
  |
logFC: 6.35E-01 p-value: 1.84E-09 |
| More Results | Click to View More RNA-seq Results | |
| In total 1 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [1] | |||
| Response Summary | FTO expression significantly contributes to the phenotype conversion of VSMCs and the aortic dissecting aneurysm by the demethylation function (m6A), thereby providing a novel therapeutic target. Knockdown of FTO suppresses the Glycogen synthase kinase-3 beta (GSK3Beta/GSK3B) levels and Klf5 expression regardless of AngII treatment. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Aortic aneurysm or dissection | ICD-11: BD50 | ||
| Pathway Response | Nucleotide excision repair | hsa03420 | ||
| Cell Process | DNA repair | |||
| Cell proliferation and migration | ||||
| In-vitro Model | VSMC (Human aortic vascular smooth muscle cells) | |||
Methyltransferase-like 14 (METTL14) [WRITER]
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL14 | ||
| Cell Line | BMDM | Mus musculus |
|
Treatment: METTL14 knockout mice BMDM
Control: Wild type mice BMDM
|
GSE153512 | |
| Regulation |
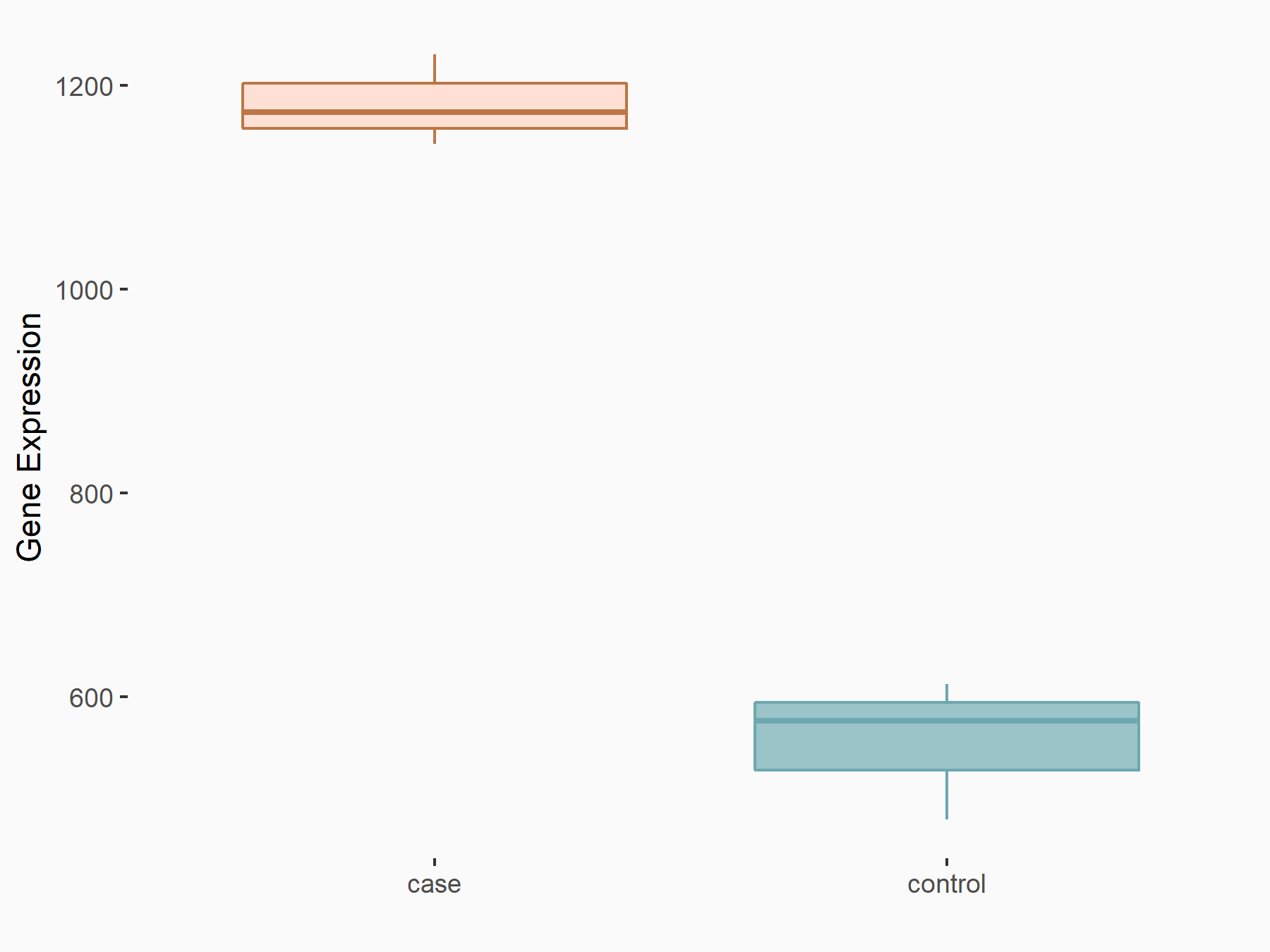 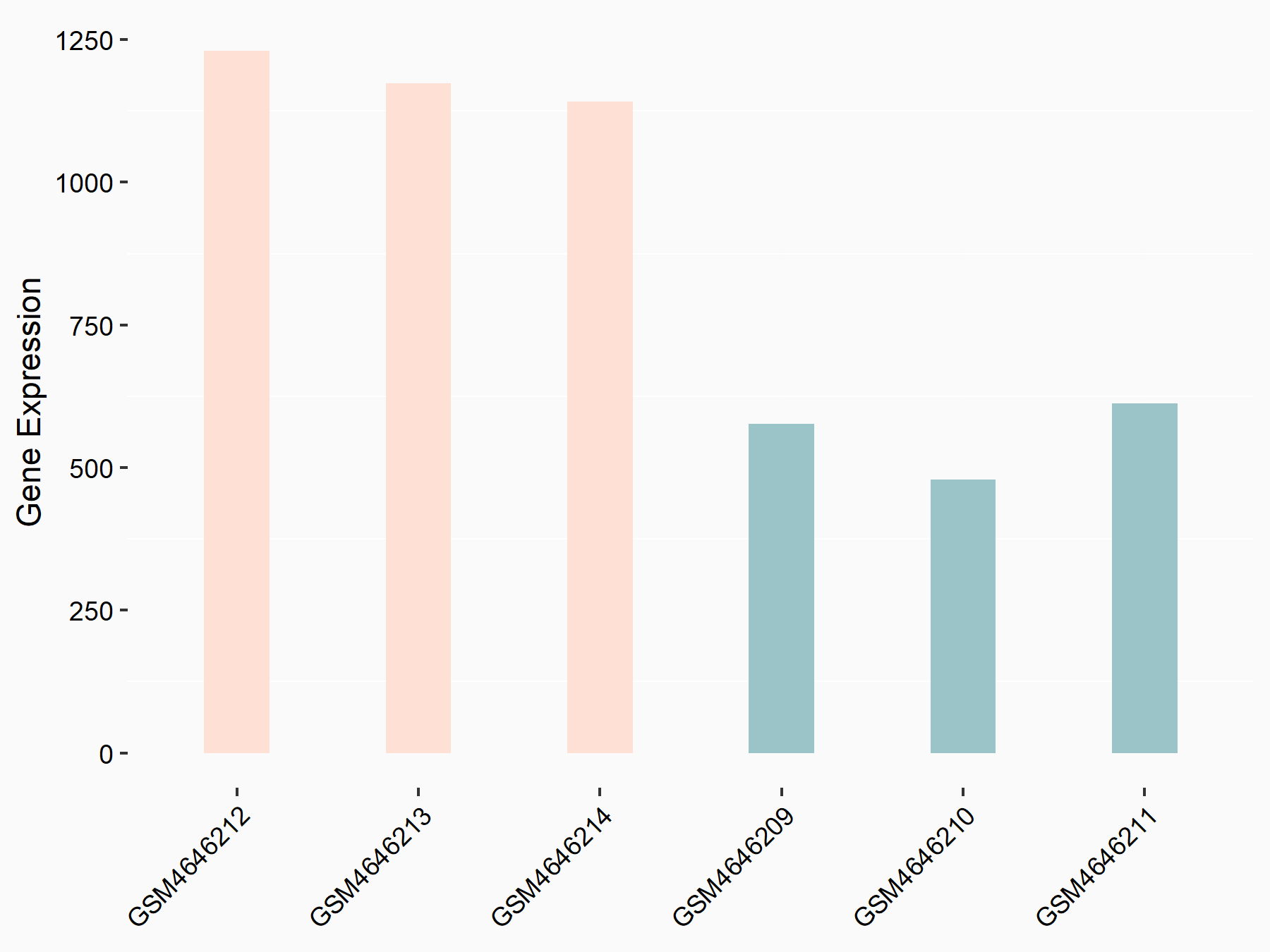 |
logFC: 1.08E+00 p-value: 1.69E-24 |
| More Results | Click to View More RNA-seq Results | |
| In total 3 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [2] | |||
| Response Summary | m6A-hypomethylation regulated FGFR4 phosphorylates Glycogen synthase kinase-3 beta (GSK3Beta/GSK3B) and activates beta-catenin/TCF4-SLC7A11/FPN1 signaling to drive anti-HER2 resistance. Knockdown of METTL14 significantly increased the expression level of FGFR4 in HER2-positive breast cancer cells. FGFR4 reduced the sensitivity of HER2-positive breast cancer to trastuzumab plus pertuzumab or tucatinib. These results pinpoint a mechanism of anti-HER2 resistance and provide a strategy for overcoming resistance via FGFR4 inhibition in recalcitrant HER2-positive breast cancer. | |||
| Target Regulation | Down regulation | |||
| Responsed Disease | Breast cancer | ICD-11: 2C60 | ||
| Responsed Drug | Pertuzumab | Approved | ||
| Pathway Response | Wnt signaling pathway | hsa04310 | ||
| Cell Process | Glutathione synthesis | |||
| In-vitro Model | ZR-75-1 | Invasive breast carcinoma | Homo sapiens | CVCL_0588 |
| T-47D | Invasive breast carcinoma | Homo sapiens | CVCL_0553 | |
| SUM-159 (A mesenchymal triple-negative breast cancer cell line) | ||||
| SK-BR-3 | Breast adenocarcinoma | Homo sapiens | CVCL_0033 | |
| MDA-MB-468 | Breast adenocarcinoma | Homo sapiens | CVCL_0419 | |
| MDA-MB-453 | Breast adenocarcinoma | Homo sapiens | CVCL_0418 | |
| MDA-MB-361 | Breast adenocarcinoma | Homo sapiens | CVCL_0620 | |
| MDA-MB-231 | Breast adenocarcinoma | Homo sapiens | CVCL_0062 | |
| MCF-7 | Invasive breast carcinoma | Homo sapiens | CVCL_0031 | |
| MCF-10A | Normal | Homo sapiens | CVCL_0598 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| BT-549 | Invasive breast carcinoma | Homo sapiens | CVCL_1092 | |
| BT-474 | Invasive breast carcinoma | Homo sapiens | CVCL_0179 | |
| AU565 | Breast adenocarcinoma | Homo sapiens | CVCL_1074 | |
| In-vivo Model | Luciferase-labeled rSKBR3 and MDA-MB-361 cells (1 × 107 cells) mixed with 1:1 Matrigel (Corning, 356237) were subcutaneously injected into the fat pads of mice. After a tumor was palpable, the mice were randomized into four groups (five mice per group), and they were treated with vehicle, trastuzumab (20 mg/kg, intraperitoneal administration), roblitinib (30 mg/kg, oral administration), or a combination of both drugs. | |||
| Experiment 2 Reporting the m6A Methylation Regulator of This Target Gene | [2] | |||
| Response Summary | m6A-hypomethylation regulated FGFR4 phosphorylates Glycogen synthase kinase-3 beta (GSK3Beta/GSK3B) and activates beta-catenin/TCF4-SLC7A11/FPN1 signaling to drive anti-HER2 resistance. Knockdown of METTL14 significantly increased the expression level of FGFR4 in HER2-positive breast cancer cells. FGFR4 reduced the sensitivity of HER2-positive breast cancer to trastuzumab plus pertuzumab or tucatinib. These results pinpoint a mechanism of anti-HER2 resistance and provide a strategy for overcoming resistance via FGFR4 inhibition in recalcitrant HER2-positive breast cancer. | |||
| Target Regulation | Down regulation | |||
| Responsed Disease | Breast cancer | ICD-11: 2C60 | ||
| Responsed Drug | Trastuzumab | Approved | ||
| Pathway Response | Wnt signaling pathway | hsa04310 | ||
| Cell Process | Glutathione synthesis | |||
| In-vitro Model | ZR-75-1 | Invasive breast carcinoma | Homo sapiens | CVCL_0588 |
| T-47D | Invasive breast carcinoma | Homo sapiens | CVCL_0553 | |
| SUM-159 (A mesenchymal triple-negative breast cancer cell line) | ||||
| SK-BR-3 | Breast adenocarcinoma | Homo sapiens | CVCL_0033 | |
| MDA-MB-468 | Breast adenocarcinoma | Homo sapiens | CVCL_0419 | |
| MDA-MB-453 | Breast adenocarcinoma | Homo sapiens | CVCL_0418 | |
| MDA-MB-361 | Breast adenocarcinoma | Homo sapiens | CVCL_0620 | |
| MDA-MB-231 | Breast adenocarcinoma | Homo sapiens | CVCL_0062 | |
| MCF-7 | Invasive breast carcinoma | Homo sapiens | CVCL_0031 | |
| MCF-10A | Normal | Homo sapiens | CVCL_0598 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| BT-549 | Invasive breast carcinoma | Homo sapiens | CVCL_1092 | |
| BT-474 | Invasive breast carcinoma | Homo sapiens | CVCL_0179 | |
| AU565 | Breast adenocarcinoma | Homo sapiens | CVCL_1074 | |
| In-vivo Model | Luciferase-labeled rSKBR3 and MDA-MB-361 cells (1 × 107 cells) mixed with 1:1 Matrigel (Corning, 356237) were subcutaneously injected into the fat pads of mice. After a tumor was palpable, the mice were randomized into four groups (five mice per group), and they were treated with vehicle, trastuzumab (20 mg/kg, intraperitoneal administration), roblitinib (30 mg/kg, oral administration), or a combination of both drugs. | |||
| Experiment 3 Reporting the m6A Methylation Regulator of This Target Gene | [2] | |||
| Response Summary | m6A-hypomethylation regulated FGFR4 phosphorylates Glycogen synthase kinase-3 beta (GSK3Beta/GSK3B) and activates beta-catenin/TCF4-SLC7A11/FPN1 signaling to drive anti-HER2 resistance. Knockdown of METTL14 significantly increased the expression level of FGFR4 in HER2-positive breast cancer cells. FGFR4 reduced the sensitivity of HER2-positive breast cancer to trastuzumab plus pertuzumab or tucatinib. These results pinpoint a mechanism of anti-HER2 resistance and provide a strategy for overcoming resistance via FGFR4 inhibition in recalcitrant HER2-positive breast cancer. | |||
| Target Regulation | Down regulation | |||
| Responsed Disease | Breast cancer | ICD-11: 2C60 | ||
| Responsed Drug | Tucatinib | Approved | ||
| Pathway Response | Wnt signaling pathway | hsa04310 | ||
| Cell Process | Glutathione synthesis | |||
| In-vitro Model | ZR-75-1 | Invasive breast carcinoma | Homo sapiens | CVCL_0588 |
| T-47D | Invasive breast carcinoma | Homo sapiens | CVCL_0553 | |
| SUM-159 (A mesenchymal triple-negative breast cancer cell line) | ||||
| SK-BR-3 | Breast adenocarcinoma | Homo sapiens | CVCL_0033 | |
| MDA-MB-468 | Breast adenocarcinoma | Homo sapiens | CVCL_0419 | |
| MDA-MB-453 | Breast adenocarcinoma | Homo sapiens | CVCL_0418 | |
| MDA-MB-361 | Breast adenocarcinoma | Homo sapiens | CVCL_0620 | |
| MDA-MB-231 | Breast adenocarcinoma | Homo sapiens | CVCL_0062 | |
| MCF-7 | Invasive breast carcinoma | Homo sapiens | CVCL_0031 | |
| MCF-10A | Normal | Homo sapiens | CVCL_0598 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| BT-549 | Invasive breast carcinoma | Homo sapiens | CVCL_1092 | |
| BT-474 | Invasive breast carcinoma | Homo sapiens | CVCL_0179 | |
| AU565 | Breast adenocarcinoma | Homo sapiens | CVCL_1074 | |
| In-vivo Model | Luciferase-labeled rSKBR3 and MDA-MB-361 cells (1 × 107 cells) mixed with 1:1 Matrigel (Corning, 356237) were subcutaneously injected into the fat pads of mice. After a tumor was palpable, the mice were randomized into four groups (five mice per group), and they were treated with vehicle, trastuzumab (20 mg/kg, intraperitoneal administration), roblitinib (30 mg/kg, oral administration), or a combination of both drugs. | |||
Breast cancer [ICD-11: 2C60]
| In total 3 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [2] | |||
| Response Summary | m6A-hypomethylation regulated FGFR4 phosphorylates Glycogen synthase kinase-3 beta (GSK3Beta/GSK3B) and activates beta-catenin/TCF4-SLC7A11/FPN1 signaling to drive anti-HER2 resistance. Knockdown of METTL14 significantly increased the expression level of FGFR4 in HER2-positive breast cancer cells. FGFR4 reduced the sensitivity of HER2-positive breast cancer to trastuzumab plus pertuzumab or tucatinib. These results pinpoint a mechanism of anti-HER2 resistance and provide a strategy for overcoming resistance via FGFR4 inhibition in recalcitrant HER2-positive breast cancer. | |||
| Responsed Disease | Breast cancer [ICD-11: 2C60] | |||
| Target Regulator | Methyltransferase-like 14 (METTL14) | WRITER | ||
| Target Regulation | Down regulation | |||
| Responsed Drug | Pertuzumab | Approved | ||
| Pathway Response | Wnt signaling pathway | hsa04310 | ||
| Cell Process | Glutathione synthesis | |||
| In-vitro Model | ZR-75-1 | Invasive breast carcinoma | Homo sapiens | CVCL_0588 |
| T-47D | Invasive breast carcinoma | Homo sapiens | CVCL_0553 | |
| SUM-159 (A mesenchymal triple-negative breast cancer cell line) | ||||
| SK-BR-3 | Breast adenocarcinoma | Homo sapiens | CVCL_0033 | |
| MDA-MB-468 | Breast adenocarcinoma | Homo sapiens | CVCL_0419 | |
| MDA-MB-453 | Breast adenocarcinoma | Homo sapiens | CVCL_0418 | |
| MDA-MB-361 | Breast adenocarcinoma | Homo sapiens | CVCL_0620 | |
| MDA-MB-231 | Breast adenocarcinoma | Homo sapiens | CVCL_0062 | |
| MCF-7 | Invasive breast carcinoma | Homo sapiens | CVCL_0031 | |
| MCF-10A | Normal | Homo sapiens | CVCL_0598 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| BT-549 | Invasive breast carcinoma | Homo sapiens | CVCL_1092 | |
| BT-474 | Invasive breast carcinoma | Homo sapiens | CVCL_0179 | |
| AU565 | Breast adenocarcinoma | Homo sapiens | CVCL_1074 | |
| In-vivo Model | Luciferase-labeled rSKBR3 and MDA-MB-361 cells (1 × 107 cells) mixed with 1:1 Matrigel (Corning, 356237) were subcutaneously injected into the fat pads of mice. After a tumor was palpable, the mice were randomized into four groups (five mice per group), and they were treated with vehicle, trastuzumab (20 mg/kg, intraperitoneal administration), roblitinib (30 mg/kg, oral administration), or a combination of both drugs. | |||
| Experiment 2 Reporting the m6A-centered Disease Response | [2] | |||
| Response Summary | m6A-hypomethylation regulated FGFR4 phosphorylates Glycogen synthase kinase-3 beta (GSK3Beta/GSK3B) and activates beta-catenin/TCF4-SLC7A11/FPN1 signaling to drive anti-HER2 resistance. Knockdown of METTL14 significantly increased the expression level of FGFR4 in HER2-positive breast cancer cells. FGFR4 reduced the sensitivity of HER2-positive breast cancer to trastuzumab plus pertuzumab or tucatinib. These results pinpoint a mechanism of anti-HER2 resistance and provide a strategy for overcoming resistance via FGFR4 inhibition in recalcitrant HER2-positive breast cancer. | |||
| Responsed Disease | Breast cancer [ICD-11: 2C60] | |||
| Target Regulator | Methyltransferase-like 14 (METTL14) | WRITER | ||
| Target Regulation | Down regulation | |||
| Responsed Drug | Trastuzumab | Approved | ||
| Pathway Response | Wnt signaling pathway | hsa04310 | ||
| Cell Process | Glutathione synthesis | |||
| In-vitro Model | ZR-75-1 | Invasive breast carcinoma | Homo sapiens | CVCL_0588 |
| T-47D | Invasive breast carcinoma | Homo sapiens | CVCL_0553 | |
| SUM-159 (A mesenchymal triple-negative breast cancer cell line) | ||||
| SK-BR-3 | Breast adenocarcinoma | Homo sapiens | CVCL_0033 | |
| MDA-MB-468 | Breast adenocarcinoma | Homo sapiens | CVCL_0419 | |
| MDA-MB-453 | Breast adenocarcinoma | Homo sapiens | CVCL_0418 | |
| MDA-MB-361 | Breast adenocarcinoma | Homo sapiens | CVCL_0620 | |
| MDA-MB-231 | Breast adenocarcinoma | Homo sapiens | CVCL_0062 | |
| MCF-7 | Invasive breast carcinoma | Homo sapiens | CVCL_0031 | |
| MCF-10A | Normal | Homo sapiens | CVCL_0598 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| BT-549 | Invasive breast carcinoma | Homo sapiens | CVCL_1092 | |
| BT-474 | Invasive breast carcinoma | Homo sapiens | CVCL_0179 | |
| AU565 | Breast adenocarcinoma | Homo sapiens | CVCL_1074 | |
| In-vivo Model | Luciferase-labeled rSKBR3 and MDA-MB-361 cells (1 × 107 cells) mixed with 1:1 Matrigel (Corning, 356237) were subcutaneously injected into the fat pads of mice. After a tumor was palpable, the mice were randomized into four groups (five mice per group), and they were treated with vehicle, trastuzumab (20 mg/kg, intraperitoneal administration), roblitinib (30 mg/kg, oral administration), or a combination of both drugs. | |||
| Experiment 3 Reporting the m6A-centered Disease Response | [2] | |||
| Response Summary | m6A-hypomethylation regulated FGFR4 phosphorylates Glycogen synthase kinase-3 beta (GSK3Beta/GSK3B) and activates beta-catenin/TCF4-SLC7A11/FPN1 signaling to drive anti-HER2 resistance. Knockdown of METTL14 significantly increased the expression level of FGFR4 in HER2-positive breast cancer cells. FGFR4 reduced the sensitivity of HER2-positive breast cancer to trastuzumab plus pertuzumab or tucatinib. These results pinpoint a mechanism of anti-HER2 resistance and provide a strategy for overcoming resistance via FGFR4 inhibition in recalcitrant HER2-positive breast cancer. | |||
| Responsed Disease | Breast cancer [ICD-11: 2C60] | |||
| Target Regulator | Methyltransferase-like 14 (METTL14) | WRITER | ||
| Target Regulation | Down regulation | |||
| Responsed Drug | Tucatinib | Approved | ||
| Pathway Response | Wnt signaling pathway | hsa04310 | ||
| Cell Process | Glutathione synthesis | |||
| In-vitro Model | ZR-75-1 | Invasive breast carcinoma | Homo sapiens | CVCL_0588 |
| T-47D | Invasive breast carcinoma | Homo sapiens | CVCL_0553 | |
| SUM-159 (A mesenchymal triple-negative breast cancer cell line) | ||||
| SK-BR-3 | Breast adenocarcinoma | Homo sapiens | CVCL_0033 | |
| MDA-MB-468 | Breast adenocarcinoma | Homo sapiens | CVCL_0419 | |
| MDA-MB-453 | Breast adenocarcinoma | Homo sapiens | CVCL_0418 | |
| MDA-MB-361 | Breast adenocarcinoma | Homo sapiens | CVCL_0620 | |
| MDA-MB-231 | Breast adenocarcinoma | Homo sapiens | CVCL_0062 | |
| MCF-7 | Invasive breast carcinoma | Homo sapiens | CVCL_0031 | |
| MCF-10A | Normal | Homo sapiens | CVCL_0598 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| BT-549 | Invasive breast carcinoma | Homo sapiens | CVCL_1092 | |
| BT-474 | Invasive breast carcinoma | Homo sapiens | CVCL_0179 | |
| AU565 | Breast adenocarcinoma | Homo sapiens | CVCL_1074 | |
| In-vivo Model | Luciferase-labeled rSKBR3 and MDA-MB-361 cells (1 × 107 cells) mixed with 1:1 Matrigel (Corning, 356237) were subcutaneously injected into the fat pads of mice. After a tumor was palpable, the mice were randomized into four groups (five mice per group), and they were treated with vehicle, trastuzumab (20 mg/kg, intraperitoneal administration), roblitinib (30 mg/kg, oral administration), or a combination of both drugs. | |||
Aortic aneurysm or dissection [ICD-11: BD50]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [1] | |||
| Response Summary | FTO expression significantly contributes to the phenotype conversion of VSMCs and the aortic dissecting aneurysm by the demethylation function (m6A), thereby providing a novel therapeutic target. Knockdown of FTO suppresses the Glycogen synthase kinase-3 beta (GSK3Beta/GSK3B) levels and Klf5 expression regardless of AngII treatment. | |||
| Responsed Disease | Aortic aneurysm or dissection [ICD-11: BD50] | |||
| Target Regulator | Fat mass and obesity-associated protein (FTO) | ERASER | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Nucleotide excision repair | hsa03420 | ||
| Cell Process | DNA repair | |||
| Cell proliferation and migration | ||||
| In-vitro Model | VSMC (Human aortic vascular smooth muscle cells) | |||
Pertuzumab
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response | [2] | |||
| Response Summary | m6A-hypomethylation regulated FGFR4 phosphorylates Glycogen synthase kinase-3 beta (GSK3Beta/GSK3B) and activates beta-catenin/TCF4-SLC7A11/FPN1 signaling to drive anti-HER2 resistance. Knockdown of METTL14 significantly increased the expression level of FGFR4 in HER2-positive breast cancer cells. FGFR4 reduced the sensitivity of HER2-positive breast cancer to trastuzumab plus pertuzumab or tucatinib. These results pinpoint a mechanism of anti-HER2 resistance and provide a strategy for overcoming resistance via FGFR4 inhibition in recalcitrant HER2-positive breast cancer. | |||
| Target Regulator | Methyltransferase-like 14 (METTL14) | WRITER | ||
| Target Regulation | Down regulation | |||
| Responsed Disease | Breast cancer | ICD-11: 2C60 | ||
| Pathway Response | Wnt signaling pathway | hsa04310 | ||
| Cell Process | Glutathione synthesis | |||
| In-vitro Model | ZR-75-1 | Invasive breast carcinoma | Homo sapiens | CVCL_0588 |
| T-47D | Invasive breast carcinoma | Homo sapiens | CVCL_0553 | |
| SUM-159 (A mesenchymal triple-negative breast cancer cell line) | ||||
| SK-BR-3 | Breast adenocarcinoma | Homo sapiens | CVCL_0033 | |
| MDA-MB-468 | Breast adenocarcinoma | Homo sapiens | CVCL_0419 | |
| MDA-MB-453 | Breast adenocarcinoma | Homo sapiens | CVCL_0418 | |
| MDA-MB-361 | Breast adenocarcinoma | Homo sapiens | CVCL_0620 | |
| MDA-MB-231 | Breast adenocarcinoma | Homo sapiens | CVCL_0062 | |
| MCF-7 | Invasive breast carcinoma | Homo sapiens | CVCL_0031 | |
| MCF-10A | Normal | Homo sapiens | CVCL_0598 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| BT-549 | Invasive breast carcinoma | Homo sapiens | CVCL_1092 | |
| BT-474 | Invasive breast carcinoma | Homo sapiens | CVCL_0179 | |
| AU565 | Breast adenocarcinoma | Homo sapiens | CVCL_1074 | |
| In-vivo Model | Luciferase-labeled rSKBR3 and MDA-MB-361 cells (1 × 107 cells) mixed with 1:1 Matrigel (Corning, 356237) were subcutaneously injected into the fat pads of mice. After a tumor was palpable, the mice were randomized into four groups (five mice per group), and they were treated with vehicle, trastuzumab (20 mg/kg, intraperitoneal administration), roblitinib (30 mg/kg, oral administration), or a combination of both drugs. | |||
Trastuzumab
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response | [2] | |||
| Response Summary | m6A-hypomethylation regulated FGFR4 phosphorylates Glycogen synthase kinase-3 beta (GSK3Beta/GSK3B) and activates beta-catenin/TCF4-SLC7A11/FPN1 signaling to drive anti-HER2 resistance. Knockdown of METTL14 significantly increased the expression level of FGFR4 in HER2-positive breast cancer cells. FGFR4 reduced the sensitivity of HER2-positive breast cancer to trastuzumab plus pertuzumab or tucatinib. These results pinpoint a mechanism of anti-HER2 resistance and provide a strategy for overcoming resistance via FGFR4 inhibition in recalcitrant HER2-positive breast cancer. | |||
| Target Regulator | Methyltransferase-like 14 (METTL14) | WRITER | ||
| Target Regulation | Down regulation | |||
| Responsed Disease | Breast cancer | ICD-11: 2C60 | ||
| Pathway Response | Wnt signaling pathway | hsa04310 | ||
| Cell Process | Glutathione synthesis | |||
| In-vitro Model | ZR-75-1 | Invasive breast carcinoma | Homo sapiens | CVCL_0588 |
| T-47D | Invasive breast carcinoma | Homo sapiens | CVCL_0553 | |
| SUM-159 (A mesenchymal triple-negative breast cancer cell line) | ||||
| SK-BR-3 | Breast adenocarcinoma | Homo sapiens | CVCL_0033 | |
| MDA-MB-468 | Breast adenocarcinoma | Homo sapiens | CVCL_0419 | |
| MDA-MB-453 | Breast adenocarcinoma | Homo sapiens | CVCL_0418 | |
| MDA-MB-361 | Breast adenocarcinoma | Homo sapiens | CVCL_0620 | |
| MDA-MB-231 | Breast adenocarcinoma | Homo sapiens | CVCL_0062 | |
| MCF-7 | Invasive breast carcinoma | Homo sapiens | CVCL_0031 | |
| MCF-10A | Normal | Homo sapiens | CVCL_0598 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| BT-549 | Invasive breast carcinoma | Homo sapiens | CVCL_1092 | |
| BT-474 | Invasive breast carcinoma | Homo sapiens | CVCL_0179 | |
| AU565 | Breast adenocarcinoma | Homo sapiens | CVCL_1074 | |
| In-vivo Model | Luciferase-labeled rSKBR3 and MDA-MB-361 cells (1 × 107 cells) mixed with 1:1 Matrigel (Corning, 356237) were subcutaneously injected into the fat pads of mice. After a tumor was palpable, the mice were randomized into four groups (five mice per group), and they were treated with vehicle, trastuzumab (20 mg/kg, intraperitoneal administration), roblitinib (30 mg/kg, oral administration), or a combination of both drugs. | |||
Tucatinib
[Approved]
| In total 1 item(s) under this drug | ||||
| Experiment 1 Reporting the m6A-centered Drug Response | [2] | |||
| Response Summary | m6A-hypomethylation regulated FGFR4 phosphorylates Glycogen synthase kinase-3 beta (GSK3Beta/GSK3B) and activates beta-catenin/TCF4-SLC7A11/FPN1 signaling to drive anti-HER2 resistance. Knockdown of METTL14 significantly increased the expression level of FGFR4 in HER2-positive breast cancer cells. FGFR4 reduced the sensitivity of HER2-positive breast cancer to trastuzumab plus pertuzumab or tucatinib. These results pinpoint a mechanism of anti-HER2 resistance and provide a strategy for overcoming resistance via FGFR4 inhibition in recalcitrant HER2-positive breast cancer. | |||
| Target Regulator | Methyltransferase-like 14 (METTL14) | WRITER | ||
| Target Regulation | Down regulation | |||
| Responsed Disease | Breast cancer | ICD-11: 2C60 | ||
| Pathway Response | Wnt signaling pathway | hsa04310 | ||
| Cell Process | Glutathione synthesis | |||
| In-vitro Model | ZR-75-1 | Invasive breast carcinoma | Homo sapiens | CVCL_0588 |
| T-47D | Invasive breast carcinoma | Homo sapiens | CVCL_0553 | |
| SUM-159 (A mesenchymal triple-negative breast cancer cell line) | ||||
| SK-BR-3 | Breast adenocarcinoma | Homo sapiens | CVCL_0033 | |
| MDA-MB-468 | Breast adenocarcinoma | Homo sapiens | CVCL_0419 | |
| MDA-MB-453 | Breast adenocarcinoma | Homo sapiens | CVCL_0418 | |
| MDA-MB-361 | Breast adenocarcinoma | Homo sapiens | CVCL_0620 | |
| MDA-MB-231 | Breast adenocarcinoma | Homo sapiens | CVCL_0062 | |
| MCF-7 | Invasive breast carcinoma | Homo sapiens | CVCL_0031 | |
| MCF-10A | Normal | Homo sapiens | CVCL_0598 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | |
| BT-549 | Invasive breast carcinoma | Homo sapiens | CVCL_1092 | |
| BT-474 | Invasive breast carcinoma | Homo sapiens | CVCL_0179 | |
| AU565 | Breast adenocarcinoma | Homo sapiens | CVCL_1074 | |
| In-vivo Model | Luciferase-labeled rSKBR3 and MDA-MB-361 cells (1 × 107 cells) mixed with 1:1 Matrigel (Corning, 356237) were subcutaneously injected into the fat pads of mice. After a tumor was palpable, the mice were randomized into four groups (five mice per group), and they were treated with vehicle, trastuzumab (20 mg/kg, intraperitoneal administration), roblitinib (30 mg/kg, oral administration), or a combination of both drugs. | |||
Full List of Crosstalk(s) between m6A Modification and Epigenetic Regulation Related to This Regulator
DNA modification
m6A Regulator: Methyltransferase-like 14 (METTL14)
| In total 9 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT02210 | ||
| Epigenetic Regulator | DNA (cytosine-5)-methyltransferase 3B (DNMT3B) | |
| Regulated Target | Methyltransferase-like protein 14 (METTL14) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Breast cancer | |
| Drug | Pertuzumab | |
| Crosstalk ID: M6ACROT02218 | ||
| Epigenetic Regulator | DNA (cytosine-5)-methyltransferase 3B (DNMT3B) | |
| Regulated Target | Methyltransferase-like protein 14 (METTL14) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Breast cancer | |
| Drug | Tucatinib | |
| Crosstalk ID: M6ACROT02223 | ||
| Epigenetic Regulator | DNA (cytosine-5)-methyltransferase 3B (DNMT3B) | |
| Regulated Target | Methyltransferase-like protein 14 (METTL14) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Breast cancer | |
| Drug | Trastuzumab | |
| Crosstalk ID: M6ACROT02234 | ||
| Epigenetic Regulator | Cysteine methyltransferase DNMT3A (DNMT3A) | |
| Regulated Target | Methyltransferase-like protein 14 (METTL14) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Breast cancer | |
| Drug | Pertuzumab | |
| Crosstalk ID: M6ACROT02242 | ||
| Epigenetic Regulator | Cysteine methyltransferase DNMT3A (DNMT3A) | |
| Regulated Target | Methyltransferase-like protein 14 (METTL14) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Breast cancer | |
| Drug | Tucatinib | |
| Crosstalk ID: M6ACROT02247 | ||
| Epigenetic Regulator | Cysteine methyltransferase DNMT3A (DNMT3A) | |
| Regulated Target | Methyltransferase-like protein 14 (METTL14) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Breast cancer | |
| Drug | Trastuzumab | |
| Crosstalk ID: M6ACROT02258 | ||
| Epigenetic Regulator | DNA (cytosine-5)-methyltransferase 1 (DNMT1) | |
| Regulated Target | Methyltransferase-like protein 14 (METTL14) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Breast cancer | |
| Drug | Pertuzumab | |
| Crosstalk ID: M6ACROT02266 | ||
| Epigenetic Regulator | DNA (cytosine-5)-methyltransferase 1 (DNMT1) | |
| Regulated Target | Methyltransferase-like protein 14 (METTL14) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Breast cancer | |
| Drug | Tucatinib | |
| Crosstalk ID: M6ACROT02271 | ||
| Epigenetic Regulator | DNA (cytosine-5)-methyltransferase 1 (DNMT1) | |
| Regulated Target | Methyltransferase-like protein 14 (METTL14) | |
| Crosstalk relationship | DNA modification → m6A | |
| Disease | Breast cancer | |
| Drug | Trastuzumab | |
Non-coding RNA
m6A Regulator: YTH domain-containing family protein 2 (YTHDF2)
| In total 1 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT05278 | ||
| Epigenetic Regulator | hsa-miR-6125 | |
| Regulated Target | YTH domain-containing family protein 2 (YTHDF2) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Colorectal cancer | |
RNA Modification Sequencing Data Associated with the Target (ID: M6ATAR00268)
| In total 1 m6A sequence/site(s) in this target gene | |||
| mod ID: AC4SITE000073 | Click to Show/Hide the Full List | ||
| mod site | chr3:119863466-119863467:- | [5] | |
| Sequence | TACGGGACCCAAATGTCAAACTACCAAATGGGCGAGACACA | ||
| Cell/Tissue List | H1 | ||
| Seq Type List | ac4C-seq | ||
| Transcript ID List | ENST00000650344.1; ENST00000316626.5; ENST00000264235.12; ENST00000473886.1 | ||
| External Link | RMBase: ac4C_site_1391 | ||
Adenosine-to-Inosine editing (A-to-I)
| In total 40 m6A sequence/site(s) in this target gene | |||
| mod ID: A2ISITE012015 | Click to Show/Hide the Full List | ||
| mod site | chr3:119824205-119824206:- | [6] | |
| Sequence | AATAAAAAGCTCCTCACCAAATTCAAGCTTGTACATTATAT | ||
| Transcript ID List | ENST00000264235.12 | ||
| External Link | RMBase: RNA-editing_site_98909 | ||
| mod ID: A2ISITE012016 | Click to Show/Hide the Full List | ||
| mod site | chr3:119826351-119826352:- | [7] | |
| Sequence | CTAGTGTGAGACTTTGGTATAGTGCACAGCTTGAAATTGGT | ||
| Transcript ID List | ENST00000264235.12 | ||
| External Link | RMBase: RNA-editing_site_98910 | ||
| mod ID: A2ISITE012017 | Click to Show/Hide the Full List | ||
| mod site | chr3:119843010-119843011:- | [8] | |
| Sequence | AGACAGGAGAATTGCTTGAAACCGGGAGGCGGAGGTTGCAT | ||
| Transcript ID List | ENST00000316626.5; ENST00000650344.1; ENST00000474830.1; ENST00000264235.12; rmsk_1074251; ENST00000473886.1 | ||
| External Link | RMBase: RNA-editing_site_98911 | ||
| mod ID: A2ISITE012018 | Click to Show/Hide the Full List | ||
| mod site | chr3:119843011-119843012:- | [8] | |
| Sequence | GAGACAGGAGAATTGCTTGAAACCGGGAGGCGGAGGTTGCA | ||
| Transcript ID List | ENST00000264235.12; rmsk_1074251; ENST00000473886.1; ENST00000474830.1; ENST00000316626.5; ENST00000650344.1 | ||
| External Link | RMBase: RNA-editing_site_98912 | ||
| mod ID: A2ISITE012019 | Click to Show/Hide the Full List | ||
| mod site | chr3:119843052-119843053:- | [8] | |
| Sequence | TGTGGTGGCAGGTGCCTGTAATCCCAGCTTCTGGGGAGGCT | ||
| Transcript ID List | ENST00000474830.1; ENST00000264235.12; ENST00000650344.1; ENST00000316626.5; rmsk_1074251; ENST00000473886.1 | ||
| External Link | RMBase: RNA-editing_site_98913 | ||
| mod ID: A2ISITE012020 | Click to Show/Hide the Full List | ||
| mod site | chr3:119843063-119843064:- | [8] | |
| Sequence | ACTTAGCCAGGTGTGGTGGCAGGTGCCTGTAATCCCAGCTT | ||
| Transcript ID List | ENST00000474830.1; rmsk_1074251; ENST00000650344.1; ENST00000264235.12; ENST00000473886.1; ENST00000316626.5 | ||
| External Link | RMBase: RNA-editing_site_98914 | ||
| mod ID: A2ISITE012021 | Click to Show/Hide the Full List | ||
| mod site | chr3:119843075-119843076:- | [8] | |
| Sequence | CTAAAAATAAAAACTTAGCCAGGTGTGGTGGCAGGTGCCTG | ||
| Transcript ID List | ENST00000473886.1; ENST00000650344.1; rmsk_1074251; ENST00000264235.12; ENST00000474830.1; ENST00000316626.5 | ||
| External Link | RMBase: RNA-editing_site_98915 | ||
| mod ID: A2ISITE012022 | Click to Show/Hide the Full List | ||
| mod site | chr3:119843079-119843080:- | [8] | |
| Sequence | TCTACTAAAAATAAAAACTTAGCCAGGTGTGGTGGCAGGTG | ||
| Transcript ID List | ENST00000473886.1; ENST00000650344.1; ENST00000316626.5; rmsk_1074251; ENST00000474830.1; ENST00000264235.12 | ||
| External Link | RMBase: RNA-editing_site_98916 | ||
| mod ID: A2ISITE012023 | Click to Show/Hide the Full List | ||
| mod site | chr3:119843089-119843090:- | [8] | |
| Sequence | AAACCTTGTCTCTACTAAAAATAAAAACTTAGCCAGGTGTG | ||
| Transcript ID List | rmsk_1074251; ENST00000650344.1; ENST00000264235.12; ENST00000474830.1; ENST00000316626.5; ENST00000473886.1 | ||
| External Link | RMBase: RNA-editing_site_98917 | ||
| mod ID: A2ISITE012024 | Click to Show/Hide the Full List | ||
| mod site | chr3:119843107-119843108:- | [8] | |
| Sequence | AGCCTGACCAACAAGGTGAAACCTTGTCTCTACTAAAAATA | ||
| Transcript ID List | ENST00000474830.1; ENST00000316626.5; ENST00000650344.1; ENST00000473886.1; ENST00000264235.12; rmsk_1074251 | ||
| External Link | RMBase: RNA-editing_site_98918 | ||
| mod ID: A2ISITE012025 | Click to Show/Hide the Full List | ||
| mod site | chr3:119843114-119843115:- | [8] | |
| Sequence | TGAGACCAGCCTGACCAACAAGGTGAAACCTTGTCTCTACT | ||
| Transcript ID List | ENST00000473886.1; ENST00000264235.12; ENST00000474830.1; ENST00000316626.5; rmsk_1074251; ENST00000650344.1 | ||
| External Link | RMBase: RNA-editing_site_98919 | ||
| mod ID: A2ISITE012026 | Click to Show/Hide the Full List | ||
| mod site | chr3:119843118-119843119:- | [8] | |
| Sequence | AGTTTGAGACCAGCCTGACCAACAAGGTGAAACCTTGTCTC | ||
| Transcript ID List | ENST00000650344.1; ENST00000264235.12; ENST00000474830.1; ENST00000473886.1; rmsk_1074251; ENST00000316626.5 | ||
| External Link | RMBase: RNA-editing_site_98920 | ||
| mod ID: A2ISITE012027 | Click to Show/Hide the Full List | ||
| mod site | chr3:119843121-119843122:- | [8] | |
| Sequence | AGGAGTTTGAGACCAGCCTGACCAACAAGGTGAAACCTTGT | ||
| Transcript ID List | ENST00000650344.1; ENST00000316626.5; ENST00000473886.1; ENST00000474830.1; ENST00000264235.12; rmsk_1074251 | ||
| External Link | RMBase: RNA-editing_site_98921 | ||
| mod ID: A2ISITE012028 | Click to Show/Hide the Full List | ||
| mod site | chr3:119843127-119843128:- | [8] | |
| Sequence | GAGGTCAGGAGTTTGAGACCAGCCTGACCAACAAGGTGAAA | ||
| Transcript ID List | ENST00000650344.1; ENST00000316626.5; ENST00000264235.12; ENST00000474830.1; rmsk_1074251; ENST00000473886.1 | ||
| External Link | RMBase: RNA-editing_site_98922 | ||
| mod ID: A2ISITE012029 | Click to Show/Hide the Full List | ||
| mod site | chr3:119843141-119843142:- | [8] | |
| Sequence | TGGGTGGATCACCTGAGGTCAGGAGTTTGAGACCAGCCTGA | ||
| Transcript ID List | ENST00000264235.12; ENST00000474830.1; ENST00000473886.1; rmsk_1074251; ENST00000650344.1; ENST00000316626.5 | ||
| External Link | RMBase: RNA-editing_site_98923 | ||
| mod ID: A2ISITE012030 | Click to Show/Hide the Full List | ||
| mod site | chr3:119843178-119843179:- | [8] | |
| Sequence | CTCACACCTGTAATCCCAGCACTTTGGGAGGCCGAGGTGGG | ||
| Transcript ID List | ENST00000650344.1; ENST00000474830.1; ENST00000264235.12; rmsk_1074251; ENST00000473886.1; ENST00000316626.5 | ||
| External Link | RMBase: RNA-editing_site_98924 | ||
| mod ID: A2ISITE012031 | Click to Show/Hide the Full List | ||
| mod site | chr3:119843181-119843182:- | [8] | |
| Sequence | TGGCTCACACCTGTAATCCCAGCACTTTGGGAGGCCGAGGT | ||
| Transcript ID List | ENST00000316626.5; ENST00000474830.1; rmsk_1074251; ENST00000650344.1; ENST00000264235.12; ENST00000473886.1 | ||
| External Link | RMBase: RNA-editing_site_98925 | ||
| mod ID: A2ISITE012032 | Click to Show/Hide the Full List | ||
| mod site | chr3:119843193-119843194:- | [8] | |
| Sequence | GGCTGGGCATGATGGCTCACACCTGTAATCCCAGCACTTTG | ||
| Transcript ID List | ENST00000264235.12; ENST00000316626.5; ENST00000650344.1; ENST00000473886.1; ENST00000474830.1; rmsk_1074251 | ||
| External Link | RMBase: RNA-editing_site_98926 | ||
| mod ID: A2ISITE012033 | Click to Show/Hide the Full List | ||
| mod site | chr3:119843202-119843203:- | [8] | |
| Sequence | CATATTCTGGGCTGGGCATGATGGCTCACACCTGTAATCCC | ||
| Transcript ID List | ENST00000650344.1; ENST00000264235.12; rmsk_1074251; ENST00000473886.1; ENST00000316626.5; ENST00000474830.1 | ||
| External Link | RMBase: RNA-editing_site_98927 | ||
| mod ID: A2ISITE012034 | Click to Show/Hide the Full List | ||
| mod site | chr3:119843271-119843272:- | [8] | |
| Sequence | AGCTGCTTCAACCCCCACAAATGCCACAGCAGCGTCAGGTA | ||
| Transcript ID List | ENST00000473886.1; ENST00000316626.5; ENST00000474830.1; ENST00000650344.1; ENST00000264235.12 | ||
| External Link | RMBase: RNA-editing_site_98928 | ||
| mod ID: A2ISITE012035 | Click to Show/Hide the Full List | ||
| mod site | chr3:119843272-119843273:- | [8] | |
| Sequence | CAGCTGCTTCAACCCCCACAAATGCCACAGCAGCGTCAGGT | ||
| Transcript ID List | ENST00000473886.1; ENST00000474830.1; ENST00000316626.5; ENST00000650344.1; ENST00000264235.12 | ||
| External Link | RMBase: RNA-editing_site_98929 | ||
| mod ID: A2ISITE012036 | Click to Show/Hide the Full List | ||
| mod site | chr3:119878396-119878397:- | [7] | |
| Sequence | GGTGGTATCTTGTTTTAATTAGCATTTCCCTAATGATTAAT | ||
| Transcript ID List | ENST00000650344.1; ENST00000316626.5; ENST00000264235.12 | ||
| External Link | RMBase: RNA-editing_site_98930 | ||
| mod ID: A2ISITE012037 | Click to Show/Hide the Full List | ||
| mod site | chr3:119878400-119878401:- | [7] | |
| Sequence | ATATGGTGGTATCTTGTTTTAATTAGCATTTCCCTAATGAT | ||
| Transcript ID List | ENST00000264235.12; ENST00000650344.1; ENST00000316626.5 | ||
| External Link | RMBase: RNA-editing_site_98931 | ||
| mod ID: A2ISITE012038 | Click to Show/Hide the Full List | ||
| mod site | chr3:119878533-119878534:- | [7] | |
| Sequence | AACTTCCAAACTGTTTTCCAAGGTGCTATATACCATTTCAC | ||
| Transcript ID List | ENST00000264235.12; ENST00000316626.5; ENST00000650344.1 | ||
| External Link | RMBase: RNA-editing_site_98932 | ||
| mod ID: A2ISITE012039 | Click to Show/Hide the Full List | ||
| mod site | chr3:119880225-119880226:- | [7] | |
| Sequence | AGGTATATGAGAGGTGCTCAACATCATTGATCATCAGAGAA | ||
| Transcript ID List | rmsk_1074303; ENST00000316626.5; ENST00000264235.12; ENST00000650344.1 | ||
| External Link | RMBase: RNA-editing_site_98933 | ||
| mod ID: A2ISITE012040 | Click to Show/Hide the Full List | ||
| mod site | chr3:119959576-119959577:- | [8] | |
| Sequence | GGTTGCAGTGAGCTGAGATCATGCCACTGCACTCCAGCCTG | ||
| Transcript ID List | ENST00000650344.1; ENST00000316626.5; rmsk_1074450; ENST00000264235.12 | ||
| External Link | RMBase: RNA-editing_site_98934 | ||
| mod ID: A2ISITE012041 | Click to Show/Hide the Full List | ||
| mod site | chr3:119959610-119959611:- | [8] | |
| Sequence | GAGGCAGGAGAATCACTTGAACCTGGGAGACAGAGGTTGCA | ||
| Transcript ID List | rmsk_1074450; ENST00000264235.12; ENST00000650344.1; ENST00000316626.5 | ||
| External Link | RMBase: RNA-editing_site_98935 | ||
| mod ID: A2ISITE012042 | Click to Show/Hide the Full List | ||
| mod site | chr3:119959611-119959612:- | [8] | |
| Sequence | TGAGGCAGGAGAATCACTTGAACCTGGGAGACAGAGGTTGC | ||
| Transcript ID List | ENST00000650344.1; ENST00000264235.12; rmsk_1074450; ENST00000316626.5 | ||
| External Link | RMBase: RNA-editing_site_98936 | ||
| mod ID: A2ISITE012043 | Click to Show/Hide the Full List | ||
| mod site | chr3:119959625-119959626:- | [8] | |
| Sequence | GCTACTCAGGAGGCTGAGGCAGGAGAATCACTTGAACCTGG | ||
| Transcript ID List | rmsk_1074450; ENST00000316626.5; ENST00000650344.1; ENST00000264235.12 | ||
| External Link | RMBase: RNA-editing_site_98937 | ||
| mod ID: A2ISITE012044 | Click to Show/Hide the Full List | ||
| mod site | chr3:119959638-119959639:- | [8] | |
| Sequence | GCGTGTAATCCCAGCTACTCAGGAGGCTGAGGCAGGAGAAT | ||
| Transcript ID List | rmsk_1074450; ENST00000650344.1; ENST00000264235.12; ENST00000316626.5 | ||
| External Link | RMBase: RNA-editing_site_98938 | ||
| mod ID: A2ISITE012045 | Click to Show/Hide the Full List | ||
| mod site | chr3:119959642-119959643:- | [8] | |
| Sequence | ACACGCGTGTAATCCCAGCTACTCAGGAGGCTGAGGCAGGA | ||
| Transcript ID List | ENST00000650344.1; rmsk_1074450; ENST00000316626.5; ENST00000264235.12 | ||
| External Link | RMBase: RNA-editing_site_98939 | ||
| mod ID: A2ISITE012046 | Click to Show/Hide the Full List | ||
| mod site | chr3:119959646-119959647:- | [8] | |
| Sequence | TGGTACACGCGTGTAATCCCAGCTACTCAGGAGGCTGAGGC | ||
| Transcript ID List | ENST00000316626.5; rmsk_1074450; ENST00000264235.12; ENST00000650344.1 | ||
| External Link | RMBase: RNA-editing_site_98940 | ||
| mod ID: A2ISITE012047 | Click to Show/Hide the Full List | ||
| mod site | chr3:119959652-119959653:- | [8] | |
| Sequence | GCATGGTGGTACACGCGTGTAATCCCAGCTACTCAGGAGGC | ||
| Transcript ID List | ENST00000316626.5; ENST00000650344.1; ENST00000264235.12; rmsk_1074450 | ||
| External Link | RMBase: RNA-editing_site_98941 | ||
| mod ID: A2ISITE012048 | Click to Show/Hide the Full List | ||
| mod site | chr3:119959685-119959686:- | [8] | |
| Sequence | CATCTCTACTAAAAATACAAAAAAATTAGCTGGGCATGGTG | ||
| Transcript ID List | ENST00000264235.12; ENST00000650344.1; rmsk_1074450; ENST00000316626.5 | ||
| External Link | RMBase: RNA-editing_site_98942 | ||
| mod ID: A2ISITE012049 | Click to Show/Hide the Full List | ||
| mod site | chr3:119959694-119959695:- | [8] | |
| Sequence | GTGAAACCCCATCTCTACTAAAAATACAAAAAAATTAGCTG | ||
| Transcript ID List | rmsk_1074450; ENST00000316626.5; ENST00000264235.12; ENST00000650344.1 | ||
| External Link | RMBase: RNA-editing_site_98943 | ||
| mod ID: A2ISITE012050 | Click to Show/Hide the Full List | ||
| mod site | chr3:119959695-119959696:- | [8] | |
| Sequence | GGTGAAACCCCATCTCTACTAAAAATACAAAAAAATTAGCT | ||
| Transcript ID List | ENST00000264235.12; ENST00000650344.1; ENST00000316626.5; rmsk_1074450 | ||
| External Link | RMBase: RNA-editing_site_98944 | ||
| mod ID: A2ISITE012051 | Click to Show/Hide the Full List | ||
| mod site | chr3:119971144-119971145:- | [7] | |
| Sequence | CCTAGATTGCTTCAACTTCAATCCATTTGGGGAATTTTGGA | ||
| Transcript ID List | ENST00000650344.1; ENST00000316626.5; ENST00000264235.12 | ||
| External Link | RMBase: RNA-editing_site_98945 | ||
| mod ID: A2ISITE012052 | Click to Show/Hide the Full List | ||
| mod site | chr3:120027293-120027294:- | [7] | |
| Sequence | ACCTCCACCTCCTGGGTTCAAGTGATTCTCCTGCCTCAGCC | ||
| Transcript ID List | ENST00000264235.12; ENST00000650344.1; ENST00000316626.5 | ||
| External Link | RMBase: RNA-editing_site_98946 | ||
| mod ID: A2ISITE012053 | Click to Show/Hide the Full List | ||
| mod site | chr3:120029082-120029083:- | [9] | |
| Sequence | TGATTCATATTGCCAAATAGTAGCTGATCCAAATGCACAGC | ||
| Transcript ID List | ENST00000264235.12; ENST00000316626.5; ENST00000650344.1 | ||
| External Link | RMBase: RNA-editing_site_98947 | ||
| mod ID: A2ISITE012054 | Click to Show/Hide the Full List | ||
| mod site | chr3:120074059-120074060:- | [7] | |
| Sequence | TATCCACCTCGGCCTCCCATAGTGCTGGGATTACAGTCGTG | ||
| Transcript ID List | ENST00000650344.1; ENST00000316626.5; ENST00000264235.12 | ||
| External Link | RMBase: RNA-editing_site_98948 | ||
5-methylcytidine (m5C)
| In total 1 m6A sequence/site(s) in this target gene | |||
| mod ID: M5CSITE003140 | Click to Show/Hide the Full List | ||
| mod site | chr3:119876478-119876479:- | [10] | |
| Sequence | ACTCCAACAAGGGAGCAAATCAGAGAAATGAACCCAAACTA | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000264235.12; ENST00000316626.5; ENST00000473886.1; ENST00000650344.1 | ||
| External Link | RMBase: m5C_site_32712 | ||
N6-methyladenosine (m6A)
| In total 116 m6A sequence/site(s) in this target gene | |||
| mod ID: M6ASITE062167 | Click to Show/Hide the Full List | ||
| mod site | chr3:119821819-119821820:- | [11] | |
| Sequence | ATTAATCTTTGTTATACAGAACAAGATATCAATACCACTTC | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000264235.12 | ||
| External Link | RMBase: m6A_site_604622 | ||
| mod ID: M6ASITE062168 | Click to Show/Hide the Full List | ||
| mod site | chr3:119821841-119821842:- | [11] | |
| Sequence | CCATTCCTCTAATTAGGGAAACATTAATCTTTGTTATACAG | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000264235.12 | ||
| External Link | RMBase: m6A_site_604623 | ||
| mod ID: M6ASITE062169 | Click to Show/Hide the Full List | ||
| mod site | chr3:119822046-119822047:- | [12] | |
| Sequence | TTCCCCATTCCCTTGTAAATACATTTTGTTCTATGTGACTT | ||
| Motif Score | 2.110482143 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000264235.12 | ||
| External Link | RMBase: m6A_site_604624 | ||
| mod ID: M6ASITE062170 | Click to Show/Hide the Full List | ||
| mod site | chr3:119822161-119822162:- | [12] | |
| Sequence | GGAAAAGCAAAATCTGTATAACATTATTACTACTTGAATGC | ||
| Motif Score | 2.168095238 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000264235.12 | ||
| External Link | RMBase: m6A_site_604625 | ||
| mod ID: M6ASITE062171 | Click to Show/Hide the Full List | ||
| mod site | chr3:119822210-119822211:- | [12] | |
| Sequence | TTGTATACAAACTATTGCAAACACTTGTGCAAATCTGTCTT | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000264235.12 | ||
| External Link | RMBase: m6A_site_604626 | ||
| mod ID: M6ASITE062172 | Click to Show/Hide the Full List | ||
| mod site | chr3:119822290-119822291:- | [11] | |
| Sequence | TGACCACTTCCATCTTAAAAACAAACCTAAAAAACAAAATT | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000264235.12 | ||
| External Link | RMBase: m6A_site_604627 | ||
| mod ID: M6ASITE062173 | Click to Show/Hide the Full List | ||
| mod site | chr3:119822327-119822328:- | [13] | |
| Sequence | GATGGATCACTTGGGCCTGTACACATACCAATTAGCGTGAC | ||
| Motif Score | 2.856142857 | ||
| Cell/Tissue List | HEK293T; hESC-HEK293T | ||
| Seq Type List | DART-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000264235.12 | ||
| External Link | RMBase: m6A_site_604628 | ||
| mod ID: M6ASITE062174 | Click to Show/Hide the Full List | ||
| mod site | chr3:119822475-119822476:- | [12] | |
| Sequence | GTACATAAGGGGTTAGCTTGACAAAGTAGACTTCCTTGTGT | ||
| Motif Score | 2.859755952 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000264235.12 | ||
| External Link | RMBase: m6A_site_604629 | ||
| mod ID: M6ASITE062175 | Click to Show/Hide the Full List | ||
| mod site | chr3:119822502-119822503:- | [12] | |
| Sequence | TCTGCAATAAAAGCAAAATGACAACCAGTACATAAGGGGTT | ||
| Motif Score | 2.859755952 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000264235.12 | ||
| External Link | RMBase: m6A_site_604630 | ||
| mod ID: M6ASITE062176 | Click to Show/Hide the Full List | ||
| mod site | chr3:119822569-119822570:- | [12] | |
| Sequence | CTGGGGTTTGGATTATTTTCACAAGGGTTATGCCGTTTTAT | ||
| Motif Score | 2.047297619 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000264235.12 | ||
| External Link | RMBase: m6A_site_604631 | ||
| mod ID: M6ASITE062177 | Click to Show/Hide the Full List | ||
| mod site | chr3:119822626-119822627:- | [11] | |
| Sequence | GAAGCACAGTAGAGTCCCAGACTGAGATCTACCTTTGAGAG | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa; HEK293T | ||
| Seq Type List | m6A-seq; DART-seq | ||
| Transcript ID List | ENST00000264235.12 | ||
| External Link | RMBase: m6A_site_604632 | ||
| mod ID: M6ASITE062178 | Click to Show/Hide the Full List | ||
| mod site | chr3:119822653-119822654:- | [11] | |
| Sequence | AGCCACTGAAAATAAAAGAGACAACTAGAAGCACAGTAGAG | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa; HEK293T; hESC-HEK293T | ||
| Seq Type List | m6A-seq; DART-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000264235.12 | ||
| External Link | RMBase: m6A_site_604633 | ||
| mod ID: M6ASITE062179 | Click to Show/Hide the Full List | ||
| mod site | chr3:119822678-119822679:- | [11] | |
| Sequence | ATCTGTCTTTTCTGGTAGAAACCTGAGCCACTGAAAATAAA | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000264235.12 | ||
| External Link | RMBase: m6A_site_604634 | ||
| mod ID: M6ASITE062180 | Click to Show/Hide the Full List | ||
| mod site | chr3:119822706-119822707:- | [12] | |
| Sequence | TGGAAAAGTAGGTATGCTTTACAATAAAATCTGTCTTTTCT | ||
| Motif Score | 2.07285119 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000264235.12 | ||
| External Link | RMBase: m6A_site_604635 | ||
| mod ID: M6ASITE062181 | Click to Show/Hide the Full List | ||
| mod site | chr3:119822744-119822745:- | [11] | |
| Sequence | TCATCTAATCACAGAGCCAAACACTTTTCTCCCCTGTGTGG | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000264235.12 | ||
| External Link | RMBase: m6A_site_604636 | ||
| mod ID: M6ASITE062182 | Click to Show/Hide the Full List | ||
| mod site | chr3:119822781-119822782:- | [13] | |
| Sequence | TGAAATTGTATCTGATTCCTACTGTTCATGTTAGTGATCAT | ||
| Motif Score | 2.500660714 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000264235.12 | ||
| External Link | RMBase: m6A_site_604637 | ||
| mod ID: M6ASITE062183 | Click to Show/Hide the Full List | ||
| mod site | chr3:119822803-119822804:- | [13] | |
| Sequence | GGGGGTTTTCTTTCTTTTCAACTGAAATTGTATCTGATTCC | ||
| Motif Score | 2.595904762 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000264235.12 | ||
| External Link | RMBase: m6A_site_604638 | ||
| mod ID: M6ASITE062184 | Click to Show/Hide the Full List | ||
| mod site | chr3:119822922-119822923:- | [13] | |
| Sequence | CTATCCCTGTGGATATGAAGACACTGGCATTTCATCTATTT | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000264235.12 | ||
| External Link | RMBase: m6A_site_604639 | ||
| mod ID: M6ASITE062185 | Click to Show/Hide the Full List | ||
| mod site | chr3:119823029-119823030:- | [11] | |
| Sequence | TAAGTCCTCTTAATTTTTAGACACATTTTTGGTTTATGTTT | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa; hESC-HEK293T | ||
| Seq Type List | m6A-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000264235.12 | ||
| External Link | RMBase: m6A_site_604640 | ||
| mod ID: M6ASITE062186 | Click to Show/Hide the Full List | ||
| mod site | chr3:119823069-119823070:- | [12] | |
| Sequence | ACTGCACCTTCTTTCCAGTGACATGCTGTGTCATTTTTTTT | ||
| Motif Score | 2.859755952 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000264235.12 | ||
| External Link | RMBase: m6A_site_604641 | ||
| mod ID: M6ASITE062187 | Click to Show/Hide the Full List | ||
| mod site | chr3:119823089-119823090:- | [11] | |
| Sequence | TCATTCCAGCAAGGCAGAAGACTGCACCTTCTTTCCAGTGA | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000264235.12 | ||
| External Link | RMBase: m6A_site_604642 | ||
| mod ID: M6ASITE062188 | Click to Show/Hide the Full List | ||
| mod site | chr3:119823120-119823121:- | [11] | |
| Sequence | CAAAGGTTACTATTTTTGAAACATCGTGTGTTCATTCCAGC | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa; hESC-HEK293T | ||
| Seq Type List | m6A-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000264235.12 | ||
| External Link | RMBase: m6A_site_604643 | ||
| mod ID: M6ASITE062189 | Click to Show/Hide the Full List | ||
| mod site | chr3:119823142-119823143:- | [11] | |
| Sequence | ATAGAAGAAAAAACTACCAAACCAAAGGTTACTATTTTTGA | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000264235.12 | ||
| External Link | RMBase: m6A_site_604644 | ||
| mod ID: M6ASITE062190 | Click to Show/Hide the Full List | ||
| mod site | chr3:119823150-119823151:- | [11] | |
| Sequence | TGTCTTGAATAGAAGAAAAAACTACCAAACCAAAGGTTACT | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000264235.12 | ||
| External Link | RMBase: m6A_site_604645 | ||
| mod ID: M6ASITE062191 | Click to Show/Hide the Full List | ||
| mod site | chr3:119823231-119823232:- | [12] | |
| Sequence | TAATTCTTACTGTGTGCCCAACACGAAGGCCTTTTTTGAAA | ||
| Motif Score | 2.173910714 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000264235.12 | ||
| External Link | RMBase: m6A_site_604646 | ||
| mod ID: M6ASITE062192 | Click to Show/Hide the Full List | ||
| mod site | chr3:119823335-119823336:- | [11] | |
| Sequence | ACTGTTGTTTTAACAAGGGAACTCTTAGCCCATTTCCTCCC | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000264235.12 | ||
| External Link | RMBase: m6A_site_604647 | ||
| mod ID: M6ASITE062193 | Click to Show/Hide the Full List | ||
| mod site | chr3:119823355-119823356:- | [11] | |
| Sequence | TGCCACCGTGCCAATAGAGGACTGTTGTTTTAACAAGGGAA | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000264235.12 | ||
| External Link | RMBase: m6A_site_604648 | ||
| mod ID: M6ASITE062194 | Click to Show/Hide the Full List | ||
| mod site | chr3:119823556-119823557:- | [11] | |
| Sequence | CCTAACCATGGGTCTTAAAAACAGCAGATTCTGGGAGCCTT | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000264235.12 | ||
| External Link | RMBase: m6A_site_604649 | ||
| mod ID: M6ASITE062195 | Click to Show/Hide the Full List | ||
| mod site | chr3:119823652-119823653:- | [11] | |
| Sequence | CATACTTAGTCCATTCAGGGACTTAGTGTAGCACCAGGGAG | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000264235.12 | ||
| External Link | RMBase: m6A_site_604650 | ||
| mod ID: M6ASITE062196 | Click to Show/Hide the Full List | ||
| mod site | chr3:119823685-119823686:- | [11] | |
| Sequence | TCCTCCTCGGTAGCAGTGAGACCGGTTTCATTTCATACTTA | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000264235.12 | ||
| External Link | RMBase: m6A_site_604651 | ||
| mod ID: M6ASITE062197 | Click to Show/Hide the Full List | ||
| mod site | chr3:119823990-119823991:- | [12] | |
| Sequence | TACCGGCCTGTATTATTGTAACAATAACTCTAGCAATGTAT | ||
| Motif Score | 2.168095238 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000264235.12 | ||
| External Link | RMBase: m6A_site_604652 | ||
| mod ID: M6ASITE062198 | Click to Show/Hide the Full List | ||
| mod site | chr3:119824018-119824019:- | [13] | |
| Sequence | CCATATAGTTTTAACTCTGTACAGTAGGTACCGGCCTGTAT | ||
| Motif Score | 2.856142857 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000264235.12 | ||
| External Link | RMBase: m6A_site_604653 | ||
| mod ID: M6ASITE062199 | Click to Show/Hide the Full List | ||
| mod site | chr3:119824064-119824065:- | [11] | |
| Sequence | TGATTCTGTTTAATAATCAGACAAAATGTAGACGAGCTTTT | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa; hESC-HEK293T | ||
| Seq Type List | m6A-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000264235.12 | ||
| External Link | RMBase: m6A_site_604654 | ||
| mod ID: M6ASITE062200 | Click to Show/Hide the Full List | ||
| mod site | chr3:119824122-119824123:- | [11] | |
| Sequence | GTAAATATGTGGACCCAGGAACTGTTATTAATGAGCAAAAA | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa; HEK293T | ||
| Seq Type List | m6A-seq; DART-seq | ||
| Transcript ID List | ENST00000264235.12 | ||
| External Link | RMBase: m6A_site_604655 | ||
| mod ID: M6ASITE062201 | Click to Show/Hide the Full List | ||
| mod site | chr3:119824130-119824131:- | [11] | |
| Sequence | TTTTGTATGTAAATATGTGGACCCAGGAACTGTTATTAATG | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; HEK293T | ||
| Seq Type List | m6A-seq; DART-seq | ||
| Transcript ID List | ENST00000264235.12 | ||
| External Link | RMBase: m6A_site_604656 | ||
| mod ID: M6ASITE062202 | Click to Show/Hide the Full List | ||
| mod site | chr3:119824193-119824194:- | [12] | |
| Sequence | CTCACCAAATTCAAGCTTGTACATTATATTTTCTTTCTGTG | ||
| Motif Score | 2.856142857 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000264235.12 | ||
| External Link | RMBase: m6A_site_604657 | ||
| mod ID: M6ASITE062203 | Click to Show/Hide the Full List | ||
| mod site | chr3:119824270-119824271:- | [12] | |
| Sequence | GGATCACAGAACTCTTTTATACAAGTGAGATCCAGGTCTCT | ||
| Motif Score | 2.110482143 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000264235.12 | ||
| External Link | RMBase: m6A_site_604658 | ||
| mod ID: M6ASITE062204 | Click to Show/Hide the Full List | ||
| mod site | chr3:119824280-119824281:- | [11] | |
| Sequence | GATCATGGTGGGATCACAGAACTCTTTTATACAAGTGAGAT | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000264235.12 | ||
| External Link | RMBase: m6A_site_604659 | ||
| mod ID: M6ASITE062205 | Click to Show/Hide the Full List | ||
| mod site | chr3:119824432-119824433:- | [11] | |
| Sequence | ATCAGTGCTTATTTTAGGAAACAGGTTGCCCCCCACAACTG | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000264235.12 | ||
| External Link | RMBase: m6A_site_604660 | ||
| mod ID: M6ASITE062206 | Click to Show/Hide the Full List | ||
| mod site | chr3:119824493-119824494:- | [11] | |
| Sequence | CAGCAAGCGTTTGGATGCAGACACTGCTCTGGACGTGGTAT | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000264235.12 | ||
| External Link | RMBase: m6A_site_604661 | ||
| mod ID: M6ASITE062207 | Click to Show/Hide the Full List | ||
| mod site | chr3:119824536-119824537:- | [11] | |
| Sequence | GGTCTGTGATAGAGAAATGGACCTGCATTCAGATCCAACTG | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000264235.12 | ||
| External Link | RMBase: m6A_site_604662 | ||
| mod ID: M6ASITE062208 | Click to Show/Hide the Full List | ||
| mod site | chr3:119824585-119824586:- | [11] | |
| Sequence | CGTTGATAGACCATTTTCAGACAGAATTTATAAAGAATCTG | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000264235.12 | ||
| External Link | RMBase: m6A_site_604663 | ||
| mod ID: M6ASITE062209 | Click to Show/Hide the Full List | ||
| mod site | chr3:119824596-119824597:- | [11] | |
| Sequence | AAAGGTAGTGGCGTTGATAGACCATTTTCAGACAGAATTTA | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000264235.12 | ||
| External Link | RMBase: m6A_site_604664 | ||
| mod ID: M6ASITE062210 | Click to Show/Hide the Full List | ||
| mod site | chr3:119824623-119824624:- | [13] | |
| Sequence | TCAAAGTATTTGGCAGCATAACTCCTTAAAGGTAGTGGCGT | ||
| Motif Score | 2.590089286 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000264235.12 | ||
| External Link | RMBase: m6A_site_604665 | ||
| mod ID: M6ASITE062211 | Click to Show/Hide the Full List | ||
| mod site | chr3:119824706-119824707:- | [14] | |
| Sequence | GCTCAGTCAAACCAAATCAAACAAATTGAATTTTATGTTTT | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | peripheral-blood | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000264235.12 | ||
| External Link | RMBase: m6A_site_604666 | ||
| mod ID: M6ASITE062212 | Click to Show/Hide the Full List | ||
| mod site | chr3:119824716-119824717:- | [14] | |
| Sequence | AAGGAAACATGCTCAGTCAAACCAAATCAAACAAATTGAAT | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | peripheral-blood | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000264235.12 | ||
| External Link | RMBase: m6A_site_604667 | ||
| mod ID: M6ASITE062213 | Click to Show/Hide the Full List | ||
| mod site | chr3:119824730-119824731:- | [14] | |
| Sequence | CAGAGTTTATGAGAAAGGAAACATGCTCAGTCAAACCAAAT | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | peripheral-blood | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000264235.12 | ||
| External Link | RMBase: m6A_site_604668 | ||
| mod ID: M6ASITE062214 | Click to Show/Hide the Full List | ||
| mod site | chr3:119824764-119824765:- | [14] | |
| Sequence | CCACAAAAGGGGTTTCACAAACCTCTGGATATATCAGAGTT | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | peripheral-blood | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000264235.12 | ||
| External Link | RMBase: m6A_site_604669 | ||
| mod ID: M6ASITE062215 | Click to Show/Hide the Full List | ||
| mod site | chr3:119825212-119825213:- | [11] | |
| Sequence | CCTTATATACTCTAAAATGAACTTTAGTCACCTTGGTGCTT | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa; HEK293T | ||
| Seq Type List | m6A-seq; DART-seq | ||
| Transcript ID List | ENST00000264235.12 | ||
| External Link | RMBase: m6A_site_604670 | ||
| mod ID: M6ASITE062216 | Click to Show/Hide the Full List | ||
| mod site | chr3:119825456-119825457:- | [13] | |
| Sequence | TGTCTACAGTATCAAAATTGACTGGTTGAAGCATGAGAAGA | ||
| Motif Score | 3.28175 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000264235.12 | ||
| External Link | RMBase: m6A_site_604671 | ||
| mod ID: M6ASITE062217 | Click to Show/Hide the Full List | ||
| mod site | chr3:119825587-119825588:- | [12] | |
| Sequence | TGGTAGAAATATTAAAAACTACACATCAGAATGATACAGTC | ||
| Motif Score | 2.078666667 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000264235.12 | ||
| External Link | RMBase: m6A_site_604672 | ||
| mod ID: M6ASITE062218 | Click to Show/Hide the Full List | ||
| mod site | chr3:119825633-119825634:- | [12] | |
| Sequence | TTTTTAACCTCATGCTTTTTACATTTATTTATTGATGCATG | ||
| Motif Score | 2.07285119 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000264235.12 | ||
| External Link | RMBase: m6A_site_604673 | ||
| mod ID: M6ASITE062219 | Click to Show/Hide the Full List | ||
| mod site | chr3:119825801-119825802:- | [11] | |
| Sequence | GGAAAGATACACTTGAGAGGACATTGTAGTTAAATAATGTG | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000264235.12 | ||
| External Link | RMBase: m6A_site_604674 | ||
| mod ID: M6ASITE062220 | Click to Show/Hide the Full List | ||
| mod site | chr3:119825890-119825891:- | [11] | |
| Sequence | GATGAGCCCAGAGGAAGGGGACAGGTCAGGGATACATCTCA | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000264235.12 | ||
| External Link | RMBase: m6A_site_604675 | ||
| mod ID: M6ASITE062221 | Click to Show/Hide the Full List | ||
| mod site | chr3:119825977-119825978:- | [11] | |
| Sequence | AAATCTTAGCACAGTTTCAAACTAGTGACCTGGGAGGAGAT | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000264235.12 | ||
| External Link | RMBase: m6A_site_604676 | ||
| mod ID: M6ASITE062222 | Click to Show/Hide the Full List | ||
| mod site | chr3:119826046-119826047:- | [12] | |
| Sequence | GCAGCAAGCAAGACTCAGGCACACATGCTCTACAGGTGGCT | ||
| Motif Score | 2.830589286 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000264235.12 | ||
| External Link | RMBase: m6A_site_604677 | ||
| mod ID: M6ASITE062223 | Click to Show/Hide the Full List | ||
| mod site | chr3:119826054-119826055:- | [11] | |
| Sequence | GAGGGGAAGCAGCAAGCAAGACTCAGGCACACATGCTCTAC | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000264235.12 | ||
| External Link | RMBase: m6A_site_604678 | ||
| mod ID: M6ASITE062224 | Click to Show/Hide the Full List | ||
| mod site | chr3:119826088-119826089:- | [15] | |
| Sequence | CTAGTGATGTAGAGGCTTGTACAGGAGGCTGCCAGAGGGGA | ||
| Motif Score | 2.856142857 | ||
| Cell/Tissue List | brain | ||
| Seq Type List | m6A-REF-seq | ||
| Transcript ID List | ENST00000264235.12 | ||
| External Link | RMBase: m6A_site_604679 | ||
| mod ID: M6ASITE062225 | Click to Show/Hide the Full List | ||
| mod site | chr3:119826167-119826168:- | [11] | |
| Sequence | GGAGGTCAATAAAGTTTGGAACTCTACAGGGAAGATTCTTA | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000264235.12 | ||
| External Link | RMBase: m6A_site_604680 | ||
| mod ID: M6ASITE062226 | Click to Show/Hide the Full List | ||
| mod site | chr3:119826217-119826218:- | [11] | |
| Sequence | GTTGGTGGCAGGTGTGGCAGACAAAGAAATGTGTATCATTC | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa; hESC-HEK293T | ||
| Seq Type List | m6A-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000264235.12 | ||
| External Link | RMBase: m6A_site_604681 | ||
| mod ID: M6ASITE062227 | Click to Show/Hide the Full List | ||
| mod site | chr3:119826252-119826253:- | [11] | |
| Sequence | ATATCTTCGGGTATTTATAGACTTGCCTTTGGCATGTTGGT | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa; HEK293T | ||
| Seq Type List | m6A-seq; DART-seq | ||
| Transcript ID List | ENST00000264235.12 | ||
| External Link | RMBase: m6A_site_604682 | ||
| mod ID: M6ASITE062228 | Click to Show/Hide the Full List | ||
| mod site | chr3:119826300-119826301:- | [11] | |
| Sequence | GCAGGTATAACTCAACGGGGACTTAAATGTCACTTGTAAAA | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000264235.12 | ||
| External Link | RMBase: m6A_site_604683 | ||
| mod ID: M6ASITE062229 | Click to Show/Hide the Full List | ||
| mod site | chr3:119826361-119826362:- | [11] | |
| Sequence | CTTTTAAAGTCTAGTGTGAGACTTTGGTATAGTGCACAGCT | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000264235.12 | ||
| External Link | RMBase: m6A_site_604684 | ||
| mod ID: M6ASITE062230 | Click to Show/Hide the Full List | ||
| mod site | chr3:119826391-119826392:- | [11] | |
| Sequence | GTGCTGATGGGTTTTTTTGAACTTTGTTTTCTTTTAAAGTC | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000264235.12 | ||
| External Link | RMBase: m6A_site_604685 | ||
| mod ID: M6ASITE062231 | Click to Show/Hide the Full List | ||
| mod site | chr3:119826421-119826422:- | [11] | |
| Sequence | CCCTTCCACAAAAGAAGAAAACCTTTTTCTGTGCTGATGGG | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000264235.12 | ||
| External Link | RMBase: m6A_site_604686 | ||
| mod ID: M6ASITE062232 | Click to Show/Hide the Full List | ||
| mod site | chr3:119826456-119826457:- | [11] | |
| Sequence | TTGTATACTTTAAAAACAGGACTCCTGCCTCATGCCCCTTC | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000264235.12 | ||
| External Link | RMBase: m6A_site_604687 | ||
| mod ID: M6ASITE062233 | Click to Show/Hide the Full List | ||
| mod site | chr3:119826461-119826462:- | [11] | |
| Sequence | TGCTGTTGTATACTTTAAAAACAGGACTCCTGCCTCATGCC | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000264235.12 | ||
| External Link | RMBase: m6A_site_604688 | ||
| mod ID: M6ASITE062234 | Click to Show/Hide the Full List | ||
| mod site | chr3:119826504-119826505:- | [12] | |
| Sequence | CATACAATCTCTCTCCCACGACAATCTTTTTTTATTAAAAG | ||
| Motif Score | 2.865571429 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000264235.12 | ||
| External Link | RMBase: m6A_site_604689 | ||
| mod ID: M6ASITE062235 | Click to Show/Hide the Full List | ||
| mod site | chr3:119826606-119826607:- | [13] | |
| Sequence | TAAAATATCTATAAATACAAACCAATTTCATTGTATTCTCA | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000264235.12 | ||
| External Link | RMBase: m6A_site_604690 | ||
| mod ID: M6ASITE062236 | Click to Show/Hide the Full List | ||
| mod site | chr3:119826610-119826611:- | [13] | |
| Sequence | CTTGTAAAATATCTATAAATACAAACCAATTTCATTGTATT | ||
| Motif Score | 2.110482143 | ||
| Cell/Tissue List | HEK293T; hESC-HEK293T | ||
| Seq Type List | DART-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000264235.12 | ||
| External Link | RMBase: m6A_site_604691 | ||
| mod ID: M6ASITE062237 | Click to Show/Hide the Full List | ||
| mod site | chr3:119826726-119826727:- | [12] | |
| Sequence | TACTTGAGTGTCACTCAGCAACACTGGTCACGTTTGGAAAG | ||
| Motif Score | 2.173910714 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000650344.1; ENST00000316626.5; ENST00000264235.12; ENST00000474830.1; ENST00000473886.1 | ||
| External Link | RMBase: m6A_site_604692 | ||
| mod ID: M6ASITE062238 | Click to Show/Hide the Full List | ||
| mod site | chr3:119826755-119826756:- | [11] | |
| Sequence | CAGCCAGCTGCACAGGAAAAACCACCAGTTACTTGAGTGTC | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000474830.1; ENST00000650344.1; ENST00000264235.12; ENST00000316626.5; ENST00000473886.1 | ||
| External Link | RMBase: m6A_site_604693 | ||
| mod ID: M6ASITE062239 | Click to Show/Hide the Full List | ||
| mod site | chr3:119826786-119826787:- | [11] | |
| Sequence | CAGCTTCCAACTCCACCTGAACAGTCCCGAGCAGCCAGCTG | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000473886.1; ENST00000650344.1; ENST00000316626.5; ENST00000474830.1; ENST00000264235.12 | ||
| External Link | RMBase: m6A_site_604694 | ||
| mod ID: M6ASITE062240 | Click to Show/Hide the Full List | ||
| mod site | chr3:119826832-119826833:- | [11] | |
| Sequence | GCTAATACTGGAGACCGTGGACAGACCAATAATGCTGCTTC | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000264235.12; ENST00000650344.1; ENST00000316626.5; ENST00000474830.1; ENST00000473886.1 | ||
| External Link | RMBase: m6A_site_604695 | ||
| mod ID: M6ASITE062241 | Click to Show/Hide the Full List | ||
| mod site | chr3:119843275-119843276:- | [12] | |
| Sequence | AAGCAGCTGCTTCAACCCCCACAAATGCCACAGCAGCGTCA | ||
| Motif Score | 2.053113095 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000316626.5; ENST00000473886.1; ENST00000650344.1; ENST00000474830.1; ENST00000264235.12 | ||
| External Link | RMBase: m6A_site_604696 | ||
| mod ID: M6ASITE062242 | Click to Show/Hide the Full List | ||
| mod site | chr3:119843351-119843352:- | [16] | |
| Sequence | GTGTCTTAACCTTCTATAGAACTGTCAAGTAATCCACCTCT | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | Huh7 | ||
| Seq Type List | MeRIP-seq | ||
| Transcript ID List | ENST00000316626.5; ENST00000474830.1; ENST00000473886.1; ENST00000650344.1; ENST00000264235.12 | ||
| External Link | RMBase: m6A_site_604697 | ||
| mod ID: M6ASITE062243 | Click to Show/Hide the Full List | ||
| mod site | chr3:119843383-119843384:- | [16] | |
| Sequence | GCATACAAAGTTAAGAGTGGACTCTAACATTTGTGTCTTAA | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | Huh7 | ||
| Seq Type List | MeRIP-seq | ||
| Transcript ID List | ENST00000264235.12; ENST00000474830.1; ENST00000650344.1; ENST00000316626.5; ENST00000473886.1 | ||
| External Link | RMBase: m6A_site_604698 | ||
| mod ID: M6ASITE062244 | Click to Show/Hide the Full List | ||
| mod site | chr3:119863450-119863451:- | [11] | |
| Sequence | CAAACTACCAAATGGGCGAGACACACCTGCACTCTTCAACT | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa; MT4; Huh7 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000473886.1; ENST00000264235.12; ENST00000650344.1; ENST00000316626.5 | ||
| External Link | RMBase: m6A_site_604699 | ||
| mod ID: M6ASITE062245 | Click to Show/Hide the Full List | ||
| mod site | chr3:119863467-119863468:- | [11] | |
| Sequence | TTACGGGACCCAAATGTCAAACTACCAAATGGGCGAGACAC | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa; Huh7 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000316626.5; ENST00000473886.1; ENST00000264235.12; ENST00000650344.1 | ||
| External Link | RMBase: m6A_site_604700 | ||
| mod ID: M6ASITE062246 | Click to Show/Hide the Full List | ||
| mod site | chr3:119863480-119863481:- | [11] | |
| Sequence | ATTTTTTGATGAATTACGGGACCCAAATGTCAAACTACCAA | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; Huh7 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000473886.1; ENST00000264235.12; ENST00000650344.1; ENST00000316626.5 | ||
| External Link | RMBase: m6A_site_604701 | ||
| mod ID: M6ASITE062247 | Click to Show/Hide the Full List | ||
| mod site | chr3:119863530-119863531:- | [13] | |
| Sequence | GAGTATACACCAACTGCCCGACTAACACCACTGGAAGCTTG | ||
| Motif Score | 3.287565476 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000473886.1; ENST00000264235.12; ENST00000650344.1; ENST00000316626.5 | ||
| External Link | RMBase: m6A_site_604702 | ||
| mod ID: M6ASITE062248 | Click to Show/Hide the Full List | ||
| mod site | chr3:119863544-119863545:- | [12] | |
| Sequence | GTAGCCGTCTGCTGGAGTATACACCAACTGCCCGACTAACA | ||
| Motif Score | 2.110482143 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000264235.12; ENST00000650344.1; ENST00000316626.5; ENST00000473886.1 | ||
| External Link | RMBase: m6A_site_604703 | ||
| mod ID: M6ASITE062249 | Click to Show/Hide the Full List | ||
| mod site | chr3:119863569-119863570:- | [13] | |
| Sequence | ACTCCACCGGAGGCAATTGCACTGTGTAGCCGTCTGCTGGA | ||
| Motif Score | 3.252583333 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000264235.12; ENST00000650344.1; ENST00000473886.1; ENST00000316626.5 | ||
| External Link | RMBase: m6A_site_604704 | ||
| mod ID: M6ASITE062250 | Click to Show/Hide the Full List | ||
| mod site | chr3:119863589-119863590:- | [11] | |
| Sequence | TGCAGGTCTTCCGACCCCGAACTCCACCGGAGGCAATTGCA | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa; Huh7 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000473886.1; ENST00000650344.1; ENST00000316626.5; ENST00000264235.12 | ||
| External Link | RMBase: m6A_site_604705 | ||
| mod ID: M6ASITE062251 | Click to Show/Hide the Full List | ||
| mod site | chr3:119866610-119866611:- | [11] | |
| Sequence | TAGGATTCGTCAGGAACAGGACATTTCACCTCAGGAGTGCG | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; hESC-HEK293T; Huh7 | ||
| Seq Type List | m6A-seq; MAZTER-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000264235.12; ENST00000473886.1; ENST00000650344.1; ENST00000316626.5 | ||
| External Link | RMBase: m6A_site_604706 | ||
| mod ID: M6ASITE062252 | Click to Show/Hide the Full List | ||
| mod site | chr3:119866615-119866616:- | [11] | |
| Sequence | ATCTTTAGGATTCGTCAGGAACAGGACATTTCACCTCAGGA | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa; Huh7 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000316626.5; ENST00000473886.1; ENST00000264235.12; ENST00000650344.1 | ||
| External Link | RMBase: m6A_site_604707 | ||
| mod ID: M6ASITE062253 | Click to Show/Hide the Full List | ||
| mod site | chr3:119876417-119876418:- | [16] | |
| Sequence | AAATTAAGGCACATCCTTGGACTAAGGTGAGTTAGATTTTT | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | Huh7 | ||
| Seq Type List | MeRIP-seq | ||
| Transcript ID List | ENST00000316626.5; ENST00000650344.1; ENST00000473886.1; ENST00000264235.12 | ||
| External Link | RMBase: m6A_site_604708 | ||
| mod ID: M6ASITE062254 | Click to Show/Hide the Full List | ||
| mod site | chr3:119876456-119876457:- | [13] | |
| Sequence | GAGAAATGAACCCAAACTACACAGAATTTAAATTCCCTCAA | ||
| Motif Score | 2.084928571 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000264235.12; ENST00000316626.5; ENST00000473886.1; ENST00000650344.1 | ||
| External Link | RMBase: m6A_site_604709 | ||
| mod ID: M6ASITE062255 | Click to Show/Hide the Full List | ||
| mod site | chr3:119876467-119876468:- | [13] | |
| Sequence | GGAGCAAATCAGAGAAATGAACCCAAACTACACAGAATTTA | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000316626.5; ENST00000264235.12; ENST00000650344.1; ENST00000473886.1 | ||
| External Link | RMBase: m6A_site_604710 | ||
| mod ID: M6ASITE062256 | Click to Show/Hide the Full List | ||
| mod site | chr3:119876492-119876493:- | [12] | |
| Sequence | TCTAGGTCCTGGGAACTCCAACAAGGGAGCAAATCAGAGAA | ||
| Motif Score | 2.173910714 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000650344.1; ENST00000316626.5; ENST00000473886.1; ENST00000264235.12 | ||
| External Link | RMBase: m6A_site_604711 | ||
| mod ID: M6ASITE062257 | Click to Show/Hide the Full List | ||
| mod site | chr3:119905808-119905809:- | [13] | |
| Sequence | TTGGCTGAGCTGTTACTAGGACAACCAATATTTCCAGGGGA | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HEK293T; hESC-HEK293T | ||
| Seq Type List | DART-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000264235.12; ENST00000316626.5; ENST00000650344.1 | ||
| External Link | RMBase: m6A_site_604712 | ||
| mod ID: M6ASITE062258 | Click to Show/Hide the Full List | ||
| mod site | chr3:119912756-119912757:- | [13] | |
| Sequence | TTCGTATATCTGTTCTCGGTACTATAGGGCACCAGAGTTGA | ||
| Motif Score | 3.278136905 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000264235.12; ENST00000650344.1; ENST00000316626.5 | ||
| External Link | RMBase: m6A_site_604713 | ||
| mod ID: M6ASITE062259 | Click to Show/Hide the Full List | ||
| mod site | chr3:119916052-119916053:- | [13] | |
| Sequence | TGCTGTATTAAAACTCTGTGACTTTGGAAGGTAAGCTACTG | ||
| Motif Score | 3.28175 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000650344.1; ENST00000264235.12; ENST00000316626.5 | ||
| External Link | RMBase: m6A_site_604714 | ||
| mod ID: M6ASITE062260 | Click to Show/Hide the Full List | ||
| mod site | chr3:119916060-119916061:- | [11] | |
| Sequence | CCTGATACTGCTGTATTAAAACTCTGTGACTTTGGAAGGTA | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000264235.12; ENST00000316626.5; ENST00000650344.1 | ||
| External Link | RMBase: m6A_site_604715 | ||
| mod ID: M6ASITE062261 | Click to Show/Hide the Full List | ||
| mod site | chr3:119916094-119916095:- | [11] | |
| Sequence | TCGGGATATTAAACCGCAGAACCTCTTGTTGGATCCTGATA | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000264235.12; ENST00000650344.1; ENST00000316626.5 | ||
| External Link | RMBase: m6A_site_604716 | ||
| mod ID: M6ASITE062262 | Click to Show/Hide the Full List | ||
| mod site | chr3:119916102-119916103:- | [11] | |
| Sequence | ATCTGCCATCGGGATATTAAACCGCAGAACCTCTTGTTGGA | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000316626.5; ENST00000650344.1; ENST00000264235.12 | ||
| External Link | RMBase: m6A_site_604717 | ||
| mod ID: M6ASITE062263 | Click to Show/Hide the Full List | ||
| mod site | chr3:119923399-119923400:- | [11] | |
| Sequence | AGACACTATAGTCGAGCCAAACAGACGCTCCCTGTGATTTA | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000650344.1; ENST00000316626.5; ENST00000264235.12 | ||
| External Link | RMBase: m6A_site_604718 | ||
| mod ID: M6ASITE062264 | Click to Show/Hide the Full List | ||
| mod site | chr3:119923417-119923418:- | [11] | |
| Sequence | ACAGTATACAGAGTTGCCAGACACTATAGTCGAGCCAAACA | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000650344.1; ENST00000264235.12; ENST00000316626.5 | ||
| External Link | RMBase: m6A_site_604719 | ||
| mod ID: M6ASITE062265 | Click to Show/Hide the Full List | ||
| mod site | chr3:119923437-119923438:- | [11] | |
| Sequence | TGCTGGACTATGTTCCGGAAACAGTATACAGAGTTGCCAGA | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000650344.1; ENST00000316626.5; ENST00000264235.12 | ||
| External Link | RMBase: m6A_site_604720 | ||
| mod ID: M6ASITE062266 | Click to Show/Hide the Full List | ||
| mod site | chr3:119923451-119923452:- | [11] | |
| Sequence | CTATCTTAATCTGGTGCTGGACTATGTTCCGGAAACAGTAT | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000264235.12; ENST00000316626.5; ENST00000650344.1 | ||
| External Link | RMBase: m6A_site_604721 | ||
| mod ID: M6ASITE062267 | Click to Show/Hide the Full List | ||
| mod site | chr3:119947310-119947311:- | [12] | |
| Sequence | GAGAAAGCTAGATCACTGTAACATAGTCCGATTGCGTTATT | ||
| Motif Score | 2.168095238 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000316626.5; ENST00000264235.12; ENST00000650344.1 | ||
| External Link | RMBase: m6A_site_604722 | ||
| mod ID: M6ASITE062268 | Click to Show/Hide the Full List | ||
| mod site | chr3:120002058-120002059:- | [11] | |
| Sequence | CATCAAGAAAGTATTGCAGGACAAGAGATTTAAGGTAAAAT | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; hESC-HEK293T | ||
| Seq Type List | m6A-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000650344.1; ENST00000316626.5; ENST00000264235.12 | ||
| External Link | RMBase: m6A_site_604723 | ||
| mod ID: M6ASITE062269 | Click to Show/Hide the Full List | ||
| mod site | chr3:120002087-120002088:- | [11] | |
| Sequence | AAACTTTGTGATTCAGGAGAACTGGTCGCCATCAAGAAAGT | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000316626.5; ENST00000264235.12; ENST00000650344.1 | ||
| External Link | RMBase: m6A_site_604724 | ||
| mod ID: M6ASITE062270 | Click to Show/Hide the Full List | ||
| mod site | chr3:120002105-120002106:- | [11] | |
| Sequence | GGTGTGGTATATCAAGCCAAACTTTGTGATTCAGGAGAACT | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000316626.5; ENST00000650344.1; ENST00000264235.12 | ||
| External Link | RMBase: m6A_site_604725 | ||
| mod ID: M6ASITE062271 | Click to Show/Hide the Full List | ||
| mod site | chr3:120002154-120002155:- | [11] | |
| Sequence | ACAAGAAGTCAGCTATACAGACACTAAAGTGATTGGAAATG | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa; hESC-HEK293T; MM6; Jurkat; peripheral-blood; endometrial; NB4 | ||
| Seq Type List | m6A-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000316626.5; ENST00000650344.1; ENST00000264235.12 | ||
| External Link | RMBase: m6A_site_604726 | ||
| mod ID: M6ASITE062272 | Click to Show/Hide the Full List | ||
| mod site | chr3:120002181-120002182:- | [11] | |
| Sequence | AACTCCTGGGCAGGGTCCAGACAGGCCACAAGAAGTCAGCT | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa; HepG2; MM6; Jurkat; CD4T; peripheral-blood; endometrial; HEC-1-A; NB4 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000264235.12; ENST00000650344.1; ENST00000316626.5 | ||
| External Link | RMBase: m6A_site_604727 | ||
| mod ID: M6ASITE062273 | Click to Show/Hide the Full List | ||
| mod site | chr3:120002235-120002236:- | [11] | |
| Sequence | TTCCTTTCTTCCTTTAGGAGACAAGGACGGCAGCAAGGTGA | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa; HepG2; hESC-HEK293T; LCLs; MM6; Jurkat; CD4T; peripheral-blood; endometrial; HEC-1-A; NB4 | ||
| Seq Type List | m6A-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000264235.12; ENST00000650344.1; ENST00000316626.5 | ||
| External Link | RMBase: m6A_site_604728 | ||
| mod ID: M6ASITE062274 | Click to Show/Hide the Full List | ||
| mod site | chr3:120093415-120093416:- | [11] | |
| Sequence | TCATGTCAGGGCGGCCCAGAACCACCTCCTTTGCGGAGAGC | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; hNPCs; fibroblasts; LCLs; MM6; Jurkat; CD4T; peripheral-blood; endometrial; HEC-1-A; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000316626.5; ENST00000650344.1; ENST00000264235.12 | ||
| External Link | RMBase: m6A_site_604730 | ||
| mod ID: M6ASITE062275 | Click to Show/Hide the Full List | ||
| mod site | chr3:120093488-120093489:- | [11] | |
| Sequence | CCTAACACCCCAACATAAAGACAAAAGGAAGAAAAGGAGGA | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa; HEK293T; hESC-HEK293T; hNPCs; fibroblasts; A549; LCLs; MT4; MM6; Jurkat; iSLK | ||
| Seq Type List | m6A-seq; MeRIP-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000316626.5; ENST00000264235.12; ENST00000650344.1 | ||
| External Link | RMBase: m6A_site_604731 | ||
| mod ID: M6ASITE062276 | Click to Show/Hide the Full List | ||
| mod site | chr3:120093504-120093505:- | [12] | |
| Sequence | CTGCATTTATCGTTAACCTAACACCCCAACATAAAGACAAA | ||
| Motif Score | 2.168095238 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000316626.5; ENST00000650344.1; ENST00000264235.12 | ||
| External Link | RMBase: m6A_site_604732 | ||
| mod ID: M6ASITE062277 | Click to Show/Hide the Full List | ||
| mod site | chr3:120093532-120093533:- | [11] | |
| Sequence | TCTTTTTCTTCTGTGGGAGAACTTAATGCTGCATTTATCGT | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa; HEK293T; hNPCs; fibroblasts; A549; LCLs; MT4; iSLK | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000650344.1; ENST00000264235.12; ENST00000316626.5 | ||
| External Link | RMBase: m6A_site_604733 | ||
| mod ID: M6ASITE062278 | Click to Show/Hide the Full List | ||
| mod site | chr3:120093589-120093590:- | [12] | |
| Sequence | TGTTTTGTTTTTTATAGTATACAAAAGGAGTGAAAAGCCAA | ||
| Motif Score | 2.110482143 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000264235.12; ENST00000650344.1; ENST00000316626.5 | ||
| External Link | RMBase: m6A_site_604734 | ||
| mod ID: M6ASITE062279 | Click to Show/Hide the Full List | ||
| mod site | chr3:120093831-120093832:- | [17] | |
| Sequence | GCCGCTTCCCTTCTTCATTGACTCGGAAAAAAAATCCCCGA | ||
| Motif Score | 3.28175 | ||
| Cell/Tissue List | CD8T | ||
| Seq Type List | m6A-CLIP/IP | ||
| Transcript ID List | ENST00000264235.12; ENST00000650344.1 | ||
| External Link | RMBase: m6A_site_604735 | ||
| mod ID: M6ASITE062280 | Click to Show/Hide the Full List | ||
| mod site | chr3:120093866-120093867:- | [11] | |
| Sequence | CCTTTCTCTTCAAGCGTGAGACTCGTGATCCTTCCGCCGCT | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; A549; U2OS; H1B; hESCs; fibroblasts; GM12878; LCLs; MM6; Huh7; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; TREX; HEC-1-A; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000264235.12; ENST00000650344.1 | ||
| External Link | RMBase: m6A_site_604736 | ||
| mod ID: M6ASITE062281 | Click to Show/Hide the Full List | ||
| mod site | chr3:120094168-120094169:- | [11] | |
| Sequence | ATGTCCCCGGCGAATGGGGAACAGTCGAGGAGCCGCTGCCT | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa; CD34; HepG2; A549; HEK293T; U2OS; H1A; H1B; H1299; MM6; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293A-TOA; iSLK; TREX; MSC; endometrial; HEC-1-A; GSCs; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000264235.12 | ||
| External Link | RMBase: m6A_site_604737 | ||
| mod ID: M6ASITE062282 | Click to Show/Hide the Full List | ||
| mod site | chr3:120094297-120094298:- | [11] | |
| Sequence | TGGGCTGGGGCAAAGCGCGGACACTTCCTGAGCGGGCACCG | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; CD34; HepG2; A549; HEK293T; U2OS; H1A; H1B; hNPCs; hESCs; fibroblasts; MT4; H1299; MM6; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; TREX; endometrial; HEC-1-A; GSCs; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000264235.12 | ||
| External Link | RMBase: m6A_site_604738 | ||
Pseudouridine (Pseudo)
| In total 1 m6A sequence/site(s) in this target gene | |||
| mod ID: PSESITE000189 | Click to Show/Hide the Full List | ||
| mod site | chr3:120093530-120093531:- | [18] | |
| Sequence | TTTTTCTTCTGTGGGAGAACTTAATGCTGCATTTATCGTTA | ||
| Transcript ID List | ENST00000650344.1; ENST00000264235.12; ENST00000316626.5 | ||
| External Link | RMBase: Pseudo_site_3581 | ||
References