m6A-centered Crosstalk Information
Mechanism of Crosstalk between m6A Modification and Epigenetic Regulation
| Crosstalk ID |
M6ACROT05710
|
[1] | |||
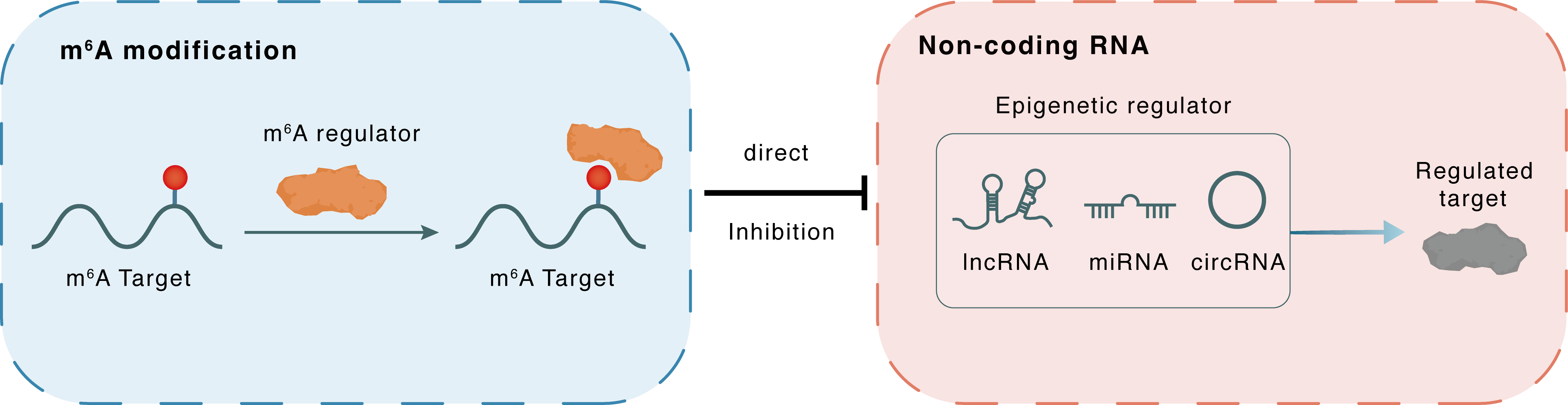 m6A modification
Circ_GPATCH2L
Circ_GPATCH2L
YTHDF2
m6A modification
Circ_GPATCH2L
Circ_GPATCH2L
YTHDF2
 : m6A sites
Direct
Inhibition
Non-coding RNA
circGPATCH2L
TRIM28
lncRNA miRNA circRNA : m6A sites
Direct
Inhibition
Non-coding RNA
circGPATCH2L
TRIM28
lncRNA miRNA circRNA
|
|||||
| m6A Modification: | |||||
|---|---|---|---|---|---|
| m6A Regulator | YTH domain-containing family protein 2 (YTHDF2) | READER | |||
| m6A Target | Circ_GPATCH2L | ||||
| Epigenetic Regulation that have Cross-talk with This m6A Modification: | |||||
| Epigenetic Regulation Type | Non-coding RNA (ncRNA) | ||||
| Epigenetic Regulator | Circ_GPATCH2L | circRNA | View Details | ||
| Regulated Target | Tripartite motif containing 28 (TRIM28) | View Details | |||
| Crosstalk Relationship | m6A → ncRNA | Inhibition | |||
| Crosstalk Mechanism | m6A regulators directly modulate the functionality of ncRNAs through specific targeting ncRNA | ||||
| Crosstalk Summary | m6A-methylated Circ_GPATCH2L is recognised and endoribonucleolytically cleaved by a YTHDF2-RPL10-RNase P/MRP complex to maintain the physiological state of nucleus pulposus cells, and then regulates DNA damage and apoptosis through Tripartite motif containing 28 (TRIM28) in intervertebral disc degeneration. | ||||
| Responsed Disease | Spinal pain | ICD-11: ME84.2 | |||
| Cell Process | Cell apoptosis | ||||
| In-vivo Model | Briefly, the spine was exposed through an anterior midline transperitoneal approach, and L4/5 were punctured by a 33-gauge needle (Hamilton, Reno, NV). A total of 2 ×108 viral genome particles per mouse of Adv-sh-Scramble (5 ×1011 pfu/ml) or Adv-sh-circGPATCH2L (1 ×1011 pfu/ml) were injected into the indicated discs. After the operation, the incision was closed in a layered fashion, and the animals were allowed to move freely around their cages. | ||||