m6A-centered Crosstalk Information
Mechanism of Crosstalk between m6A Modification and Epigenetic Regulation
| Crosstalk ID |
M6ACROT03142
|
[1] | |||
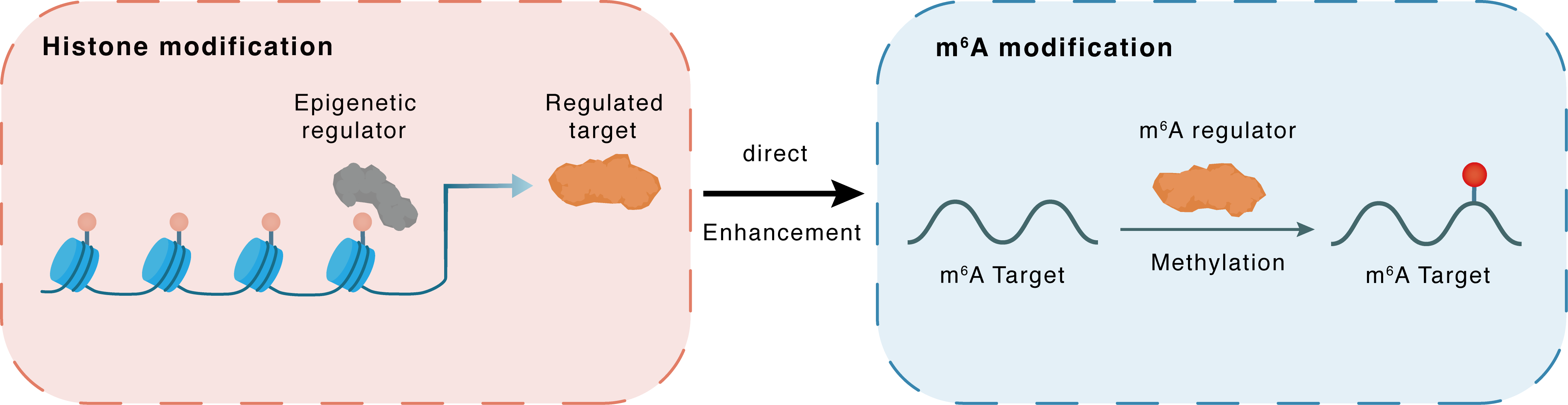 Histone modification
H3K36me1
SMYD2
METTL3
Direct
Enhancement
m6A modification
m6A Target
m6A Target
METTL3
Methylation
Histone modification
H3K36me1
SMYD2
METTL3
Direct
Enhancement
m6A modification
m6A Target
m6A Target
METTL3
Methylation
 : m6A sites : m6A sites
|
|||||
| m6A Modification: | |||||
|---|---|---|---|---|---|
| m6A Regulator | Methyltransferase-like 3 (METTL3) | WRITER | |||
| Epigenetic Regulation that have Cross-talk with This m6A Modification: | |||||
| Epigenetic Regulation Type | Histone modification (HistMod) | ||||
| Epigenetic Regulator | N-lysine methyltransferase SMYD2 (SMYD2) | WRITER | View Details | ||
| Regulated Target | Histone H3 lysine 36 monomethylation (H3K36me1) | View Details | |||
| Downstream Gene | METTL3 | View Details | |||
| Crosstalk Relationship | Histone modification → m6A | Enhancement | |||
| Crosstalk Mechanism | Histone modification directly impacts m6A modification through modulating the level of m6A regulator | ||||
| Crosstalk Summary | Histone H3 lysine 36 monomethylation (H3K36me1) plays an important role in HSCR, confirming that the methyltransferase SMYD2 can affect m6A methylation levels and intestinal nervous system development by regulating METTL3 expression. | ||||
| Responsed Disease | Structural developmental anomalies of large intestine | ICD-11: LB16.1 | |||
| Cell Process | Cell proliferation | ||||
| Cell migration | |||||
In-vitro Model |
SH-SY5Y | Neuroblastoma | Homo sapiens | CVCL_0019 | |
| HEK293T | Normal | Homo sapiens | CVCL_0063 | ||
Full List of Potential Compound(s) Related to This m6A-centered Crosstalk
| N-lysine methyltransferase SMYD2 (SMYD2) | 6 Compound(s) Regulating the Target | Click to Show/Hide the Full List | ||
| AZ505 | Preclinical | [2] | ||
| Synonyms |
AZ 505; AZ-505
Click to Show/Hide
|
|||
| MOA | Inhibitor | |||
| Activity | IC50 = 120 nM | |||
| External Link | ||||
| LLY-507 | Preclinical | [3] | ||
| Synonyms |
1793053-37-8; LLY507; CHEMBL3414623; 5-Cyano-2'-{4-[2-(3-Methyl-1h-Indol-1-Yl)ethyl]piperazin-1-Yl}-N-[3-(Pyrrolidin-1-Yl)propyl]biphenyl-3-Carboxamide; 3-cyano-5-(2-{4-[2-(3-methyl-1H-indol-1-yl)ethyl]piperazin-1-yl}phenyl)-N-[3-(pyrrolidin-1-yl)propyl]benzamide; 3-Cyano-5-[2-[4-[2-(3-methyl-1H-indol-1-yl)ethyl]piperazin-1-yl]phenyl]-N-[3-(pyrrolidin-1-yl)propyl]benzamide; GTPL8239; SCHEMBL19760400; EX-A899; LLY 507; MolPort-042-624-530; BCP17114; s7575; BDBM50075102; ZINC231558920; AKOS027470175; CS-5126
Click to Show/Hide
|
|||
| MOA | Inhibitor | |||
| Activity | IC50 < 15 nM | |||
| External Link | ||||
| EPZ032597 | Preclinical | [4] | ||
| MOA | Inhibitor | |||
| External Link | ||||
| EPZ033294 | Preclinical | [4] | ||
| MOA | Inhibitor | |||
| External Link | ||||
| A-893 | Preclinical | [5] | ||
| Synonyms |
1868232-32-9; (R)-N-cyclohexyl-3-((3,4-dichlorophenethyl)amino)-N-(2-((2-hydroxy-2-(3-oxo-3,4-dihydro-2H-benzo[b][1,4]oxazin-8-yl)ethyl)amino)ethyl)propanamide; CHEMBL3590526; N-Cyclohexyl-N~3~-[2-(3,4-Dichlorophenyl)ethyl]-N-(2-{[(2r)-2-Hydroxy-2-(3-Oxo-3,4-Dihydro-2h-1,4-Benzoxazin-8-Yl)ethyl]amino}ethyl)-Beta-Alaninamide; SCHEMBL17476248; EX-A2769; BDBM50095537; AKOS030235552; ZINC230499113; ACN-037539; AC-29886; HY-19563; CS-0015655; Q27454706; 4GQ; N-Cyclohexyl-3-[2-(3,4-dichlorophenyl)ethylamino]-N-[2-[[(2R)-2-hydroxy-2-(3-oxo-4H-1,4-benzoxazin-8-yl)ethyl]amino]ethyl]propanamide
Click to Show/Hide
|
|||
| MOA | Inhibitor | |||
| External Link | ||||
| BAY 598 | Preclinical | [6] | ||
| Synonyms |
BAY-598; 1906919-67-2; CHEMBL3818617; 1906919-67-2 (S-isomer); (S,E)-N-(1-(N'-cyano-N-(3-(difluoromethoxy)phenyl)carbamimidoyl)-3-(3,4-dichlorophenyl)-4,5-dihydro-1H-pyrazol-4-yl)-N-ethyl-2-hydroxyacetamide; BAY 598 - Bio-X; BAY598; GTPL8953; EX-A1835; BDBM50180955; ZINC504786915; AC-31567; BS-16389; HY-19546; CS-0015642; J3.601.000B; Q27074893; (S)-N-(1-(N'-Cyano-N-(3-(difluoromethoxy)phenyl)carbamimidoyl)-3-(3,4-dichlorophenyl)-4,5-dihydro-1H-pyrazol-4-yl)-N-ethyl-2-hydroxyacetamide; (S,E)-N-(1-(N-cyano-N'-(3-(difluoromethoxy)phenyl)carbamimidoyl)-3-(3,4-dichlorophenyl)-4,5-dihydro-1H-pyrazol-4-yl)-N-ethyl-2-hydroxyacetamide; (S,Z)-N-(1-(N-cyano-N'-(3-(difluoromethoxy)phenyl)carbamimidoyl)-3-(3,4-dichlorophenyl)-4,5-dihydro-1H-pyrazol-4-yl)-N-ethyl-2-hydroxyacetamide; N-[(4S)-1-[(Cyanoamino)[[3-(difluoromethoxy)phenyl]imino]methyl]-3-(3,4-dichlorophenyl)-4,5-dihydro-1H-pyrazol-4-yl]-N-ethyl-2-hydroxyacetamide; N-[(4S)-1-[(Z)-N'-cyano-N-[3-(difluoromethoxy)phenyl]carbamimidoyl]-3-(3,4-dichlorophenyl)-4,5-dihydro-1H-pyrazol-4-yl]-N-ethyl-2-hydroxyacetamide
Click to Show/Hide
|
|||
| MOA | Inhibitor | |||
| External Link | ||||
References