m6A Target Gene Information
General Information of the m6A Target Gene (ID: M6ATAR00653)
Full List of m6A Methylation Regulator of This Target Gene and Corresponding Disease/Drug Response(s)
hsa-miR-26b
can be regulated by the following regulator(s), and cause disease/drug response(s). You can browse detail information of regulator(s) or disease/drug response(s).
Browse Regulator
Browse Disease
Methyltransferase-like 3 (METTL3) [WRITER]
| Representative RNA-seq result indicating the expression of this target gene regulated by METTL3 | ||
| Cell Line | Mouse testis | Mus musculus |
|
Treatment: Mettl3 knockout mouse testis
Control: Mouse testis
|
GSE99771 | |
| Regulation |
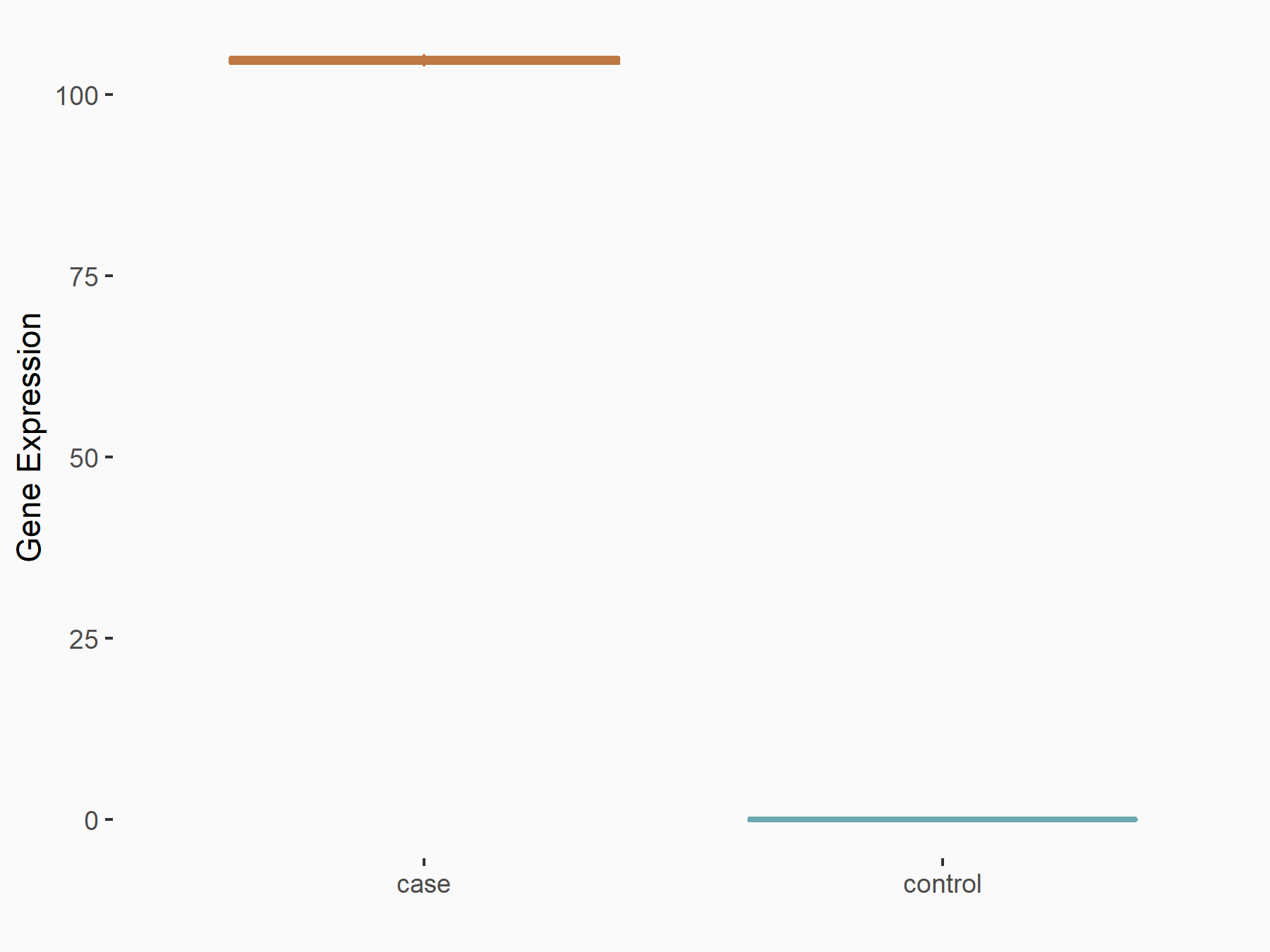 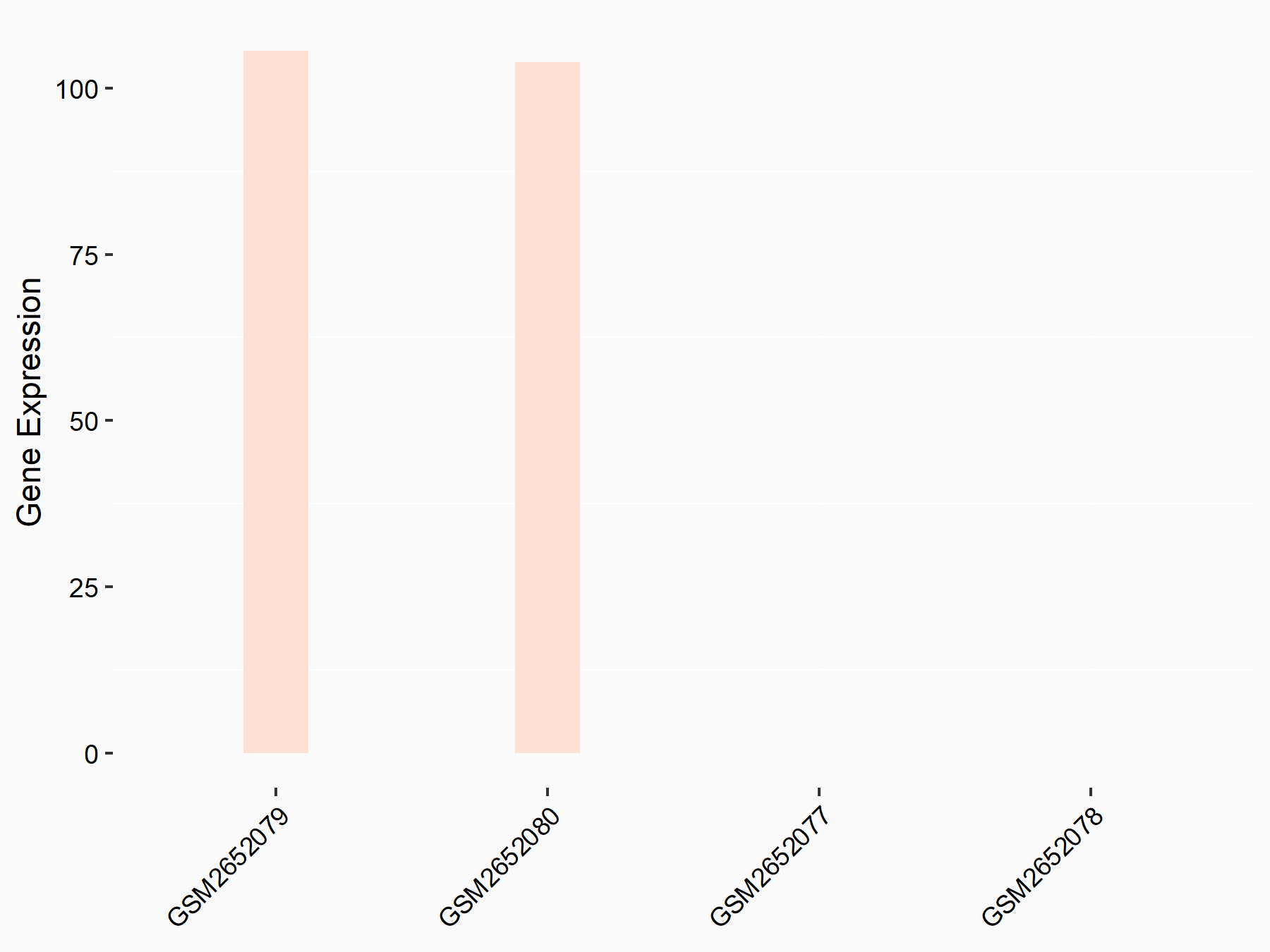 |
logFC: 6.72E+00 p-value: 1.75E-06 |
| More Results | Click to View More RNA-seq Results | |
| In total 1 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [1] | |||
| Response Summary | Silencing METTL3 down-regulate MALAT1 and HMGA2 by sponging hsa-miR-26b, and finally inhibit EMT, migration and invasion in BC, providing a theoretical basis for clinical treatment of BC. | |||
| Target Regulation | Down regulation | |||
| Responsed Disease | Breast cancer | ICD-11: 2C60 | ||
| Cell Process | Epithelial-mesenchymal transition | |||
| In-vitro Model | MDA-MB-468 | Breast adenocarcinoma | Homo sapiens | CVCL_0419 |
| MDA-MB-231 | Breast adenocarcinoma | Homo sapiens | CVCL_0062 | |
| MCF-7 | Invasive breast carcinoma | Homo sapiens | CVCL_0031 | |
| MCF-10A | Normal | Homo sapiens | CVCL_0598 | |
| In-vivo Model | Eighteen BALB/C female nude mice aged 4-5 weeks and weighing 15-18 g were randomly assigned into three groups of six mice. The MCF-7 cell lines stably transfected with sh-NC + oe-NC, sh-METTL3 + oe-NC and sh-METTL3 + oe-HMGA2 were selected for subcutaneous establishment of the BC cell line MCF-7 as xenografts in the nude mice. For this purpose, MCF-7 cell lines in the logarithmic growth stage were prepared into a suspension with a concentration of about 1 × 107 cells/ml. The prepared cell suspension was injected into the left armpit of the mice, and the subsequent tumor growth was recorded. | |||
Breast cancer [ICD-11: 2C60]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [1] | |||
| Response Summary | Silencing METTL3 down-regulate MALAT1 and HMGA2 by sponging hsa-miR-26b, and finally inhibit EMT, migration and invasion in BC, providing a theoretical basis for clinical treatment of BC. | |||
| Responsed Disease | Breast cancer [ICD-11: 2C60] | |||
| Target Regulator | Methyltransferase-like 3 (METTL3) | WRITER | ||
| Target Regulation | Down regulation | |||
| Cell Process | Epithelial-mesenchymal transition | |||
| In-vitro Model | MDA-MB-468 | Breast adenocarcinoma | Homo sapiens | CVCL_0419 |
| MDA-MB-231 | Breast adenocarcinoma | Homo sapiens | CVCL_0062 | |
| MCF-7 | Invasive breast carcinoma | Homo sapiens | CVCL_0031 | |
| MCF-10A | Normal | Homo sapiens | CVCL_0598 | |
| In-vivo Model | Eighteen BALB/C female nude mice aged 4-5 weeks and weighing 15-18 g were randomly assigned into three groups of six mice. The MCF-7 cell lines stably transfected with sh-NC + oe-NC, sh-METTL3 + oe-NC and sh-METTL3 + oe-HMGA2 were selected for subcutaneous establishment of the BC cell line MCF-7 as xenografts in the nude mice. For this purpose, MCF-7 cell lines in the logarithmic growth stage were prepared into a suspension with a concentration of about 1 × 107 cells/ml. The prepared cell suspension was injected into the left armpit of the mice, and the subsequent tumor growth was recorded. | |||
Full List of Crosstalk(s) between m6A Modification and Epigenetic Regulation Related to This Regulator
Non-coding RNA
m6A Regulator: Methyltransferase-like 3 (METTL3)
| In total 1 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT05610 | ||
| Epigenetic Regulator | hsa-miR-26b | |
| Regulated Target | High mobility group protein HMGI-C (HMGA2) | |
| Crosstalk relationship | m6A → ncRNA | |
| Disease | Breast cancer | |