m6A Target Gene Information
General Information of the m6A Target Gene (ID: M6ATAR00457)
Full List of m6A Methylation Regulator of This Target Gene and Corresponding Disease/Drug Response(s)
ATP2A2
can be regulated by the following regulator(s), and cause disease/drug response(s). You can browse detail information of regulator(s) or disease/drug response(s).
Browse Regulator
Browse Disease
Fat mass and obesity-associated protein (FTO) [ERASER]
| Representative RNA-seq result indicating the expression of this target gene regulated by FTO | ||
| Cell Line | NB4 cell line | Homo sapiens |
|
Treatment: shFTO NB4 cells
Control: shNS NB4 cells
|
GSE103494 | |
| Regulation |
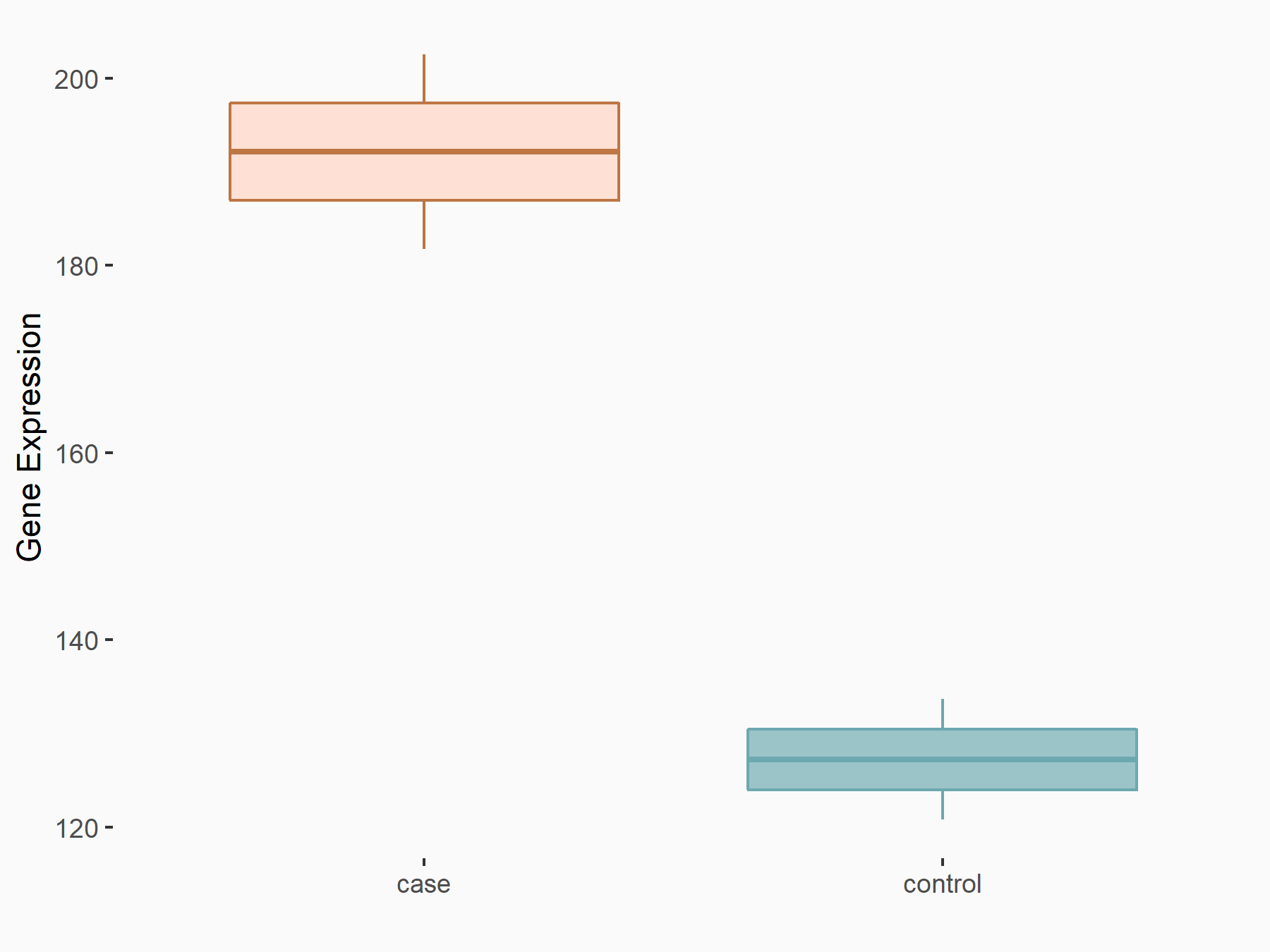  |
logFC: 5.91E-01 p-value: 1.58E-02 |
| More Results | Click to View More RNA-seq Results | |
| In total 1 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [1] | |||
| Response Summary | FTO modification of N6 -methyladenosine is associated with myocardial cell energy metabolism disorder. FTO reduced the m6A level of Sarcoplasmic/endoplasmic reticulum calcium ATPase 2 (SERCA2a/ATP2A2) mRNA through demethylation, thus promoting SERCA2a expression, maintaining calcium homeostasis, and improving energy metabolism of H/R cardiomyocytes. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Energy metabolism disorder | ICD-11: 5C53 | ||
| Cell Process | Energy metabolism | |||
| Cell apoptosis | ||||
| In-vitro Model | AC16 [Human hybrid cardiomyocyte] | Normal | Homo sapiens | CVCL_4U18 |
Energy metabolism disorder [ICD-11: 5C53]
| In total 1 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [1] | |||
| Response Summary | FTO modification of N6 -methyladenosine is associated with myocardial cell energy metabolism disorder. FTO reduced the m6A level of Sarcoplasmic/endoplasmic reticulum calcium ATPase 2 (SERCA2a/ATP2A2) mRNA through demethylation, thus promoting SERCA2a expression, maintaining calcium homeostasis, and improving energy metabolism of H/R cardiomyocytes. | |||
| Responsed Disease | Energy metabolism disorder [ICD-11: 5C53] | |||
| Target Regulator | Fat mass and obesity-associated protein (FTO) | ERASER | ||
| Target Regulation | Up regulation | |||
| Cell Process | Energy metabolism | |||
| Cell apoptosis | ||||
| In-vitro Model | AC16 [Human hybrid cardiomyocyte] | Normal | Homo sapiens | CVCL_4U18 |
Full List of Crosstalk(s) between m6A Modification and Epigenetic Regulation Related to This Regulator
Non-coding RNA
m6A Regulator: Fat mass and obesity-associated protein (FTO)
| In total 1 item(s) under this m6A regulator | ||
| Crosstalk ID: M6ACROT05226 | ||
| Epigenetic Regulator | Cardiac conduction regulatory RNA (CCRR) | |
| Regulated Target | FTO alpha-ketoglutarate dependent dioxygenase (FTO) | |
| Crosstalk relationship | ncRNA → m6A | |
| Disease | Acute myocardial infarction | |
RNA Modification Sequencing Data Associated with the Target (ID: M6ATAR00457)
| In total 1 m6A sequence/site(s) in this target gene | |||
| mod ID: AC4SITE000182 | Click to Show/Hide the Full List | ||
| mod site | chr12:110334064-110334065:+ | [3] | |
| Sequence | ACAGAGACTGCTCTCACTTGCCTAGTAGAGAAGATGAATGT | ||
| Cell/Tissue List | H1 | ||
| Seq Type List | ac4C-seq | ||
| Transcript ID List | ENST00000548169.2; ENST00000539276.7; ENST00000377685.9; ENST00000550262.1; ENST00000308664.10 | ||
| External Link | RMBase: ac4C_site_462 | ||
Adenosine-to-Inosine editing (A-to-I)
| In total 32 m6A sequence/site(s) in this target gene | |||
| mod ID: A2ISITE005763 | Click to Show/Hide the Full List | ||
| mod site | chr12:110287075-110287076:+ | [4] | |
| Sequence | CAGCCTGGCCAACATGGTGAAACCCCACCTGTACTAAAAAT | ||
| Transcript ID List | rmsk_3900604; ENST00000552636.1; ENST00000377685.9; ENST00000308664.10; ENST00000539276.7 | ||
| External Link | RMBase: RNA-editing_site_31800 | ||
| mod ID: A2ISITE005764 | Click to Show/Hide the Full List | ||
| mod site | chr12:110292291-110292292:+ | [4] | |
| Sequence | AATTAATTGTTTTCTGACACAGTCTCACTCTGTTGCCCAGG | ||
| Transcript ID List | ENST00000539276.7; ENST00000377685.9; ENST00000552636.1; ENST00000308664.10; ENST00000548169.2 | ||
| External Link | RMBase: RNA-editing_site_31801 | ||
| mod ID: A2ISITE005765 | Click to Show/Hide the Full List | ||
| mod site | chr12:110292340-110292341:+ | [4] | |
| Sequence | CAATGGTGTTGTCTCGGCTCACTGCAACCTCTTCCTCCCAG | ||
| Transcript ID List | ENST00000539276.7; ENST00000377685.9; ENST00000552636.1; ENST00000308664.10; ENST00000548169.2 | ||
| External Link | RMBase: RNA-editing_site_31802 | ||
| mod ID: A2ISITE005766 | Click to Show/Hide the Full List | ||
| mod site | chr12:110294225-110294226:+ | [5] | |
| Sequence | GCTAATTTTTTGTACTTTTAATAGAGACGGGGTTTCACCGT | ||
| Transcript ID List | ENST00000308664.10; ENST00000548169.2; ENST00000552636.1; ENST00000539276.7; ENST00000377685.9 | ||
| External Link | RMBase: RNA-editing_site_31803 | ||
| mod ID: A2ISITE005767 | Click to Show/Hide the Full List | ||
| mod site | chr12:110294227-110294228:+ | [5] | |
| Sequence | TAATTTTTTGTACTTTTAATAGAGACGGGGTTTCACCGTGT | ||
| Transcript ID List | ENST00000377685.9; ENST00000552636.1; ENST00000308664.10; ENST00000539276.7; ENST00000548169.2 | ||
| External Link | RMBase: RNA-editing_site_31804 | ||
| mod ID: A2ISITE005768 | Click to Show/Hide the Full List | ||
| mod site | chr12:110294241-110294242:+ | [5] | |
| Sequence | TTTAATAGAGACGGGGTTTCACCGTGTTAGCCAGGATGGTC | ||
| Transcript ID List | ENST00000548169.2; ENST00000539276.7; ENST00000377685.9; ENST00000308664.10; ENST00000552636.1 | ||
| External Link | RMBase: RNA-editing_site_31805 | ||
| mod ID: A2ISITE005769 | Click to Show/Hide the Full List | ||
| mod site | chr12:110294249-110294250:+ | [5] | |
| Sequence | AGACGGGGTTTCACCGTGTTAGCCAGGATGGTCTCGATCTC | ||
| Transcript ID List | ENST00000552636.1; ENST00000548169.2; ENST00000377685.9; ENST00000539276.7; ENST00000308664.10 | ||
| External Link | RMBase: RNA-editing_site_31806 | ||
| mod ID: A2ISITE005770 | Click to Show/Hide the Full List | ||
| mod site | chr12:110294304-110294305:+ | [5] | |
| Sequence | CGCCTGCCTCGGCCTCCCAAAGTGCCGGGATTACAAGTGTG | ||
| Transcript ID List | ENST00000552636.1; ENST00000539276.7; ENST00000548169.2; ENST00000308664.10; ENST00000377685.9 | ||
| External Link | RMBase: RNA-editing_site_31807 | ||
| mod ID: A2ISITE005771 | Click to Show/Hide the Full List | ||
| mod site | chr12:110294316-110294317:+ | [5] | |
| Sequence | CCTCCCAAAGTGCCGGGATTACAAGTGTGAGCCACTGTGCC | ||
| Transcript ID List | ENST00000539276.7; ENST00000548169.2; ENST00000308664.10; ENST00000552636.1; ENST00000377685.9 | ||
| External Link | RMBase: RNA-editing_site_31808 | ||
| mod ID: A2ISITE005772 | Click to Show/Hide the Full List | ||
| mod site | chr12:110294319-110294320:+ | [5] | |
| Sequence | CCCAAAGTGCCGGGATTACAAGTGTGAGCCACTGTGCCTGG | ||
| Transcript ID List | ENST00000308664.10; ENST00000539276.7; ENST00000548169.2; ENST00000377685.9; ENST00000552636.1 | ||
| External Link | RMBase: RNA-editing_site_31809 | ||
| mod ID: A2ISITE005773 | Click to Show/Hide the Full List | ||
| mod site | chr12:110294390-110294391:+ | [5] | |
| Sequence | ATATTTGGTCAGGGCATGGTAGCTCATGCCTGTAGTATCAG | ||
| Transcript ID List | ENST00000308664.10; ENST00000539276.7; ENST00000377685.9; rmsk_3900621; ENST00000548169.2; ENST00000552636.1 | ||
| External Link | RMBase: RNA-editing_site_31810 | ||
| mod ID: A2ISITE005774 | Click to Show/Hide the Full List | ||
| mod site | chr12:110294403-110294404:+ | [5] | |
| Sequence | GCATGGTAGCTCATGCCTGTAGTATCAGCAGTTTGGGAAGC | ||
| Transcript ID List | ENST00000539276.7; ENST00000308664.10; ENST00000548169.2; ENST00000377685.9; rmsk_3900621; ENST00000552636.1 | ||
| External Link | RMBase: RNA-editing_site_31811 | ||
| mod ID: A2ISITE005775 | Click to Show/Hide the Full List | ||
| mod site | chr12:110294409-110294410:+ | [5] | |
| Sequence | TAGCTCATGCCTGTAGTATCAGCAGTTTGGGAAGCTGAGGC | ||
| Transcript ID List | ENST00000377685.9; ENST00000552636.1; ENST00000308664.10; ENST00000548169.2; ENST00000539276.7; rmsk_3900621 | ||
| External Link | RMBase: RNA-editing_site_31812 | ||
| mod ID: A2ISITE005776 | Click to Show/Hide the Full List | ||
| mod site | chr12:110294421-110294422:+ | [5] | |
| Sequence | GTAGTATCAGCAGTTTGGGAAGCTGAGGCGGGTGGATCAGT | ||
| Transcript ID List | ENST00000539276.7; ENST00000552636.1; ENST00000377685.9; ENST00000308664.10; rmsk_3900621; ENST00000548169.2 | ||
| External Link | RMBase: RNA-editing_site_31813 | ||
| mod ID: A2ISITE005777 | Click to Show/Hide the Full List | ||
| mod site | chr12:110294444-110294445:+ | [5] | |
| Sequence | TGAGGCGGGTGGATCAGTTAAGGTCAGGAGGTCGAGACCAG | ||
| Transcript ID List | ENST00000377685.9; ENST00000548169.2; ENST00000539276.7; ENST00000552636.1; rmsk_3900621; ENST00000308664.10 | ||
| External Link | RMBase: RNA-editing_site_31814 | ||
| mod ID: A2ISITE005778 | Click to Show/Hide the Full List | ||
| mod site | chr12:110294449-110294450:+ | [5] | |
| Sequence | CGGGTGGATCAGTTAAGGTCAGGAGGTCGAGACCAGCCTGG | ||
| Transcript ID List | ENST00000308664.10; ENST00000548169.2; ENST00000377685.9; rmsk_3900621; ENST00000539276.7; ENST00000552636.1 | ||
| External Link | RMBase: RNA-editing_site_31815 | ||
| mod ID: A2ISITE005779 | Click to Show/Hide the Full List | ||
| mod site | chr12:110294472-110294473:+ | [5] | |
| Sequence | AGGTCGAGACCAGCCTGGCCAACATGGCAAGTCCCCGTCGC | ||
| Transcript ID List | ENST00000377685.9; ENST00000548169.2; ENST00000308664.10; ENST00000552636.1; ENST00000539276.7; rmsk_3900621 | ||
| External Link | RMBase: RNA-editing_site_31816 | ||
| mod ID: A2ISITE005780 | Click to Show/Hide the Full List | ||
| mod site | chr12:110294473-110294474:+ | [5] | |
| Sequence | GGTCGAGACCAGCCTGGCCAACATGGCAAGTCCCCGTCGCT | ||
| Transcript ID List | ENST00000377685.9; ENST00000552636.1; ENST00000308664.10; ENST00000539276.7; ENST00000548169.2; rmsk_3900621 | ||
| External Link | RMBase: RNA-editing_site_31817 | ||
| mod ID: A2ISITE005781 | Click to Show/Hide the Full List | ||
| mod site | chr12:110294480-110294481:+ | [5] | |
| Sequence | ACCAGCCTGGCCAACATGGCAAGTCCCCGTCGCTACTAAAA | ||
| Transcript ID List | ENST00000552636.1; ENST00000539276.7; rmsk_3900621; ENST00000548169.2; ENST00000308664.10; ENST00000377685.9 | ||
| External Link | RMBase: RNA-editing_site_31818 | ||
| mod ID: A2ISITE005782 | Click to Show/Hide the Full List | ||
| mod site | chr12:110294481-110294482:+ | [5] | |
| Sequence | CCAGCCTGGCCAACATGGCAAGTCCCCGTCGCTACTAAAAA | ||
| Transcript ID List | ENST00000552636.1; rmsk_3900621; ENST00000377685.9; ENST00000548169.2; ENST00000539276.7; ENST00000308664.10 | ||
| External Link | RMBase: RNA-editing_site_31819 | ||
| mod ID: A2ISITE005783 | Click to Show/Hide the Full List | ||
| mod site | chr12:110297228-110297229:+ | [6] | |
| Sequence | GCAGGCAGATCACGAGGTCAAGAGATTGAGACCACCCTGGC | ||
| Transcript ID List | ENST00000539276.7; ENST00000308664.10; ENST00000548169.2; ENST00000550248.2; ENST00000377685.9; rmsk_3900627 | ||
| External Link | RMBase: RNA-editing_site_31820 | ||
| mod ID: A2ISITE005784 | Click to Show/Hide the Full List | ||
| mod site | chr12:110299746-110299747:+ | [4] | |
| Sequence | CTCCCACCTCAGTCTCCTGAAAAACTGGGATCACAGGCATG | ||
| Transcript ID List | ENST00000377685.9; ENST00000308664.10; ENST00000548169.2; ENST00000539276.7; ENST00000550248.2 | ||
| External Link | RMBase: RNA-editing_site_31821 | ||
| mod ID: A2ISITE005785 | Click to Show/Hide the Full List | ||
| mod site | chr12:110309723-110309724:+ | [7] | |
| Sequence | GAGACCAGCCTGGGCAACATAGTAAAACCCCATCCCTACAA | ||
| Transcript ID List | ENST00000548169.2; rmsk_3900662; ENST00000308664.10; ENST00000377685.9; ENST00000550248.2; ENST00000539276.7 | ||
| External Link | RMBase: RNA-editing_site_31822 | ||
| mod ID: A2ISITE005786 | Click to Show/Hide the Full List | ||
| mod site | chr12:110313451-110313452:+ | [4] | |
| Sequence | CCTCCCCGTTAGCTGTGACTACAGGCACCCACCACAATGTC | ||
| Transcript ID List | ENST00000548169.2; ENST00000377685.9; ENST00000308664.10; ENST00000550248.2; ENST00000539276.7 | ||
| External Link | RMBase: RNA-editing_site_31823 | ||
| mod ID: A2ISITE005787 | Click to Show/Hide the Full List | ||
| mod site | chr12:110313849-110313850:+ | [7] | |
| Sequence | CTTTCCCATTAGCTGGAATTACAGGTGTGCACCACCCCACC | ||
| Transcript ID List | ENST00000550248.2; ENST00000308664.10; ENST00000377685.9; ENST00000548169.2; ENST00000539276.7 | ||
| External Link | RMBase: RNA-editing_site_31824 | ||
| mod ID: A2ISITE005788 | Click to Show/Hide the Full List | ||
| mod site | chr12:110319123-110319124:+ | [4] | |
| Sequence | GAGGCTGAGGTGGGAAGATGACTTGAGCCCAGGAGTTCAAG | ||
| Transcript ID List | ENST00000308664.10; ENST00000377685.9; ENST00000548169.2; ENST00000539276.7; ENST00000550248.2; rmsk_3900685 | ||
| External Link | RMBase: RNA-editing_site_31825 | ||
| mod ID: A2ISITE005789 | Click to Show/Hide the Full List | ||
| mod site | chr12:110319133-110319134:+ | [4] | |
| Sequence | TGGGAAGATGACTTGAGCCCAGGAGTTCAAGGCTGTAGTTC | ||
| Transcript ID List | ENST00000550248.2; ENST00000308664.10; ENST00000377685.9; ENST00000548169.2; rmsk_3900685; ENST00000539276.7 | ||
| External Link | RMBase: RNA-editing_site_31826 | ||
| mod ID: A2ISITE005790 | Click to Show/Hide the Full List | ||
| mod site | chr12:110322314-110322315:+ | [4] | |
| Sequence | AACTTGGAAATAAACATTTTATTTTATTTTACTTTTTGAGA | ||
| Transcript ID List | ENST00000308664.10; ENST00000548169.2; ENST00000550248.2; ENST00000377685.9; ENST00000539276.7 | ||
| External Link | RMBase: RNA-editing_site_31827 | ||
| mod ID: A2ISITE005791 | Click to Show/Hide the Full List | ||
| mod site | chr12:110322319-110322320:+ | [4] | |
| Sequence | GGAAATAAACATTTTATTTTATTTTACTTTTTGAGATGGAG | ||
| Transcript ID List | ENST00000548169.2; ENST00000539276.7; ENST00000550248.2; ENST00000377685.9; ENST00000308664.10 | ||
| External Link | RMBase: RNA-editing_site_31828 | ||
| mod ID: A2ISITE005792 | Click to Show/Hide the Full List | ||
| mod site | chr12:110323220-110323221:+ | [4] | |
| Sequence | TAGTGCTTTAGTCTCGCTCCATTGCCCAGGCTGGAGTGCAG | ||
| Transcript ID List | ENST00000308664.10; ENST00000539276.7; ENST00000377685.9; ENST00000548169.2 | ||
| External Link | RMBase: RNA-editing_site_31829 | ||
| mod ID: A2ISITE005793 | Click to Show/Hide the Full List | ||
| mod site | chr12:110328375-110328376:+ | [7] | |
| Sequence | AGGTCCGAAGTTTGAGACCAACCTGACCAAAATGGTGAAAC | ||
| Transcript ID List | ENST00000308664.10; ENST00000539276.7; rmsk_3900700; ENST00000548169.2; ENST00000547050.1; ENST00000549840.1; ENST00000377685.9 | ||
| External Link | RMBase: RNA-editing_site_31830 | ||
| mod ID: A2ISITE005794 | Click to Show/Hide the Full List | ||
| mod site | chr12:110350880-110350881:+ | [7] | |
| Sequence | TTTCGCACTGTATTAAATGTAACTTATTTAATGAAATCAGA | ||
| Transcript ID List | ENST00000313432.5; ENST00000308664.10; ENST00000539276.7; ENST00000377685.9 | ||
| External Link | RMBase: RNA-editing_site_31831 | ||
2'-O-Methylation (2'-O-Me)
| In total 1 m6A sequence/site(s) in this target gene | |||
| mod ID: 2OMSITE000075 | Click to Show/Hide the Full List | ||
| mod site | chr12:110332613-110332614:+ | [8] | |
| Sequence | CAGATGTTCATTCTGGACAGAGTGGAAGGTGATACTTGTTC | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | Nm-seq | ||
| Transcript ID List | ENST00000548169.2; ENST00000539276.7; ENST00000377685.9; ENST00000547050.1; ENST00000308664.10; ENST00000550262.1 | ||
| External Link | RMBase: Nm_site_1493 | ||
5-methylcytidine (m5C)
| In total 2 m6A sequence/site(s) in this target gene | |||
| mod ID: M5CSITE000096 | Click to Show/Hide the Full List | ||
| mod site | chr12:110281715-110281716:+ | [9] | |
| Sequence | GCTCCCGGGGTGGCACGAGCCCGCGGCCGGAGTGCGAGGCG | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000552636.1; ENST00000539276.7; ENST00000377685.9; ENST00000308664.10 | ||
| External Link | RMBase: m5C_site_10949 | ||
| mod ID: M5CSITE000097 | Click to Show/Hide the Full List | ||
| mod site | chr12:110347039-110347040:+ | ||
| Sequence | CCCCACCCCACCCCACCTCTCCCCACCTTACCCCCGCCCCG | ||
| Cell/Tissue List | T24 | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000539276.7; ENST00000308664.10; ENST00000313432.5; ENST00000553144.1; ENST00000377685.9; rmsk_3900724 | ||
| External Link | RMBase: m5C_site_10950 | ||
N6-methyladenosine (m6A)
| In total 167 m6A sequence/site(s) in this target gene | |||
| mod ID: M6ASITE014985 | Click to Show/Hide the Full List | ||
| mod site | chr12:110280868-110280869:+ | [10] | |
| Sequence | CCTCGAGACGCCCCGCGTGGACCGCGCTCCCAGCTCCTCGG | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | GSC-11 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000552636.1 | ||
| External Link | RMBase: m6A_site_208084 | ||
| mod ID: M6ASITE014986 | Click to Show/Hide the Full List | ||
| mod site | chr12:110281339-110281340:+ | [11] | |
| Sequence | CGGGCGTGCGGCGCTGAGGGACCCGGGCGAGCGCGCCGCGC | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; HepG2; A549; U2OS; hESCs; HEK293T; fibroblasts; GSC-11; HEK293A-TOA; MSC; TIME; TREX; iSLK | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000308664.10; ENST00000539276.7; ENST00000552636.1; ENST00000377685.9 | ||
| External Link | RMBase: m6A_site_208085 | ||
| mod ID: M6ASITE014987 | Click to Show/Hide the Full List | ||
| mod site | chr12:110281802-110281803:+ | [12] | |
| Sequence | CGAAGCCATGGAGAACGCGCACACCAAGACGGTGGAGGAGG | ||
| Motif Score | 2.830589286 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000539276.7; ENST00000377685.9; ENST00000308664.10; ENST00000552636.1 | ||
| External Link | RMBase: m6A_site_208086 | ||
| mod ID: M6ASITE014988 | Click to Show/Hide the Full List | ||
| mod site | chr12:110281869-110281870:+ | [11] | |
| Sequence | AGTACGGGGCTGAGCCTGGAACAGGTCAAGAAGCTTAAGGA | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; U2OS; hNPCs; fibroblasts; A549; LCLs; MT4; H1299; MM6; Huh7; Jurkat; CD4T; peripheral-blood; GSC-11; MSC; iSLK; TIME; endometrial; HEC-1-A; GSCs; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000539276.7; ENST00000377685.9; ENST00000308664.10; ENST00000552636.1 | ||
| External Link | RMBase: m6A_site_208087 | ||
| mod ID: M6ASITE014989 | Click to Show/Hide the Full List | ||
| mod site | chr12:110282717-110282718:+ | [11] | |
| Sequence | GTTTGTTTCTTACAGGAAAAACCTTGCTGGAACTTGTGATT | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | HeLa; HepG2; U2OS; hNPCs; fibroblasts; A549; LCLs; MM6; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293T; MSC; iSLK; endometrial; HEC-1-A; GSCs; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000308664.10; ENST00000552636.1; ENST00000377685.9; ENST00000539276.7 | ||
| External Link | RMBase: m6A_site_208088 | ||
| mod ID: M6ASITE014990 | Click to Show/Hide the Full List | ||
| mod site | chr12:110282728-110282729:+ | [11] | |
| Sequence | ACAGGAAAAACCTTGCTGGAACTTGTGATTGAGCAGTTTGA | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa; HepG2; U2OS; hNPCs; fibroblasts; A549; LCLs; MM6; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293T; MSC; iSLK; endometrial; HEC-1-A; GSCs; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000539276.7; ENST00000552636.1; ENST00000308664.10; ENST00000377685.9 | ||
| External Link | RMBase: m6A_site_208089 | ||
| mod ID: M6ASITE014991 | Click to Show/Hide the Full List | ||
| mod site | chr12:110282751-110282752:+ | [11] | |
| Sequence | TGTGATTGAGCAGTTTGAAGACTTGCTAGTTAGGATTTTAT | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa; HepG2; hNPCs; fibroblasts; A549; LCLs; Jurkat; CD4T; iSLK; MSC; endometrial; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000552636.1; ENST00000539276.7; ENST00000308664.10; ENST00000377685.9 | ||
| External Link | RMBase: m6A_site_208090 | ||
| mod ID: M6ASITE014992 | Click to Show/Hide the Full List | ||
| mod site | chr12:110292049-110292050:+ | [11] | |
| Sequence | GGTTTGAAGAAGGTGAAGAAACAATTACAGCCTTTGTAGAA | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa; hESC-HEK293T | ||
| Seq Type List | m6A-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000552636.1; ENST00000539276.7; ENST00000548169.2; ENST00000308664.10; ENST00000377685.9 | ||
| External Link | RMBase: m6A_site_208091 | ||
| mod ID: M6ASITE014993 | Click to Show/Hide the Full List | ||
| mod site | chr12:110292069-110292070:+ | [11] | |
| Sequence | ACAATTACAGCCTTTGTAGAACCTTTTGTAATTTTACTCAT | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000308664.10; ENST00000552636.1; ENST00000548169.2; ENST00000539276.7; ENST00000377685.9 | ||
| External Link | RMBase: m6A_site_208092 | ||
| mod ID: M6ASITE014994 | Click to Show/Hide the Full List | ||
| mod site | chr12:110296002-110296003:+ | [11] | |
| Sequence | TTAGTGTTGTCATTTTATAGACAGTAAACTTGTGTGTTGTG | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000548169.2; ENST00000550248.2; ENST00000552636.1; ENST00000377685.9; ENST00000539276.7; ENST00000308664.10 | ||
| External Link | RMBase: m6A_site_208093 | ||
| mod ID: M6ASITE014995 | Click to Show/Hide the Full List | ||
| mod site | chr12:110296009-110296010:+ | [11] | |
| Sequence | TGTCATTTTATAGACAGTAAACTTGTGTGTTGTGTAACCTC | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000308664.10; ENST00000550248.2; ENST00000552636.1; ENST00000539276.7; ENST00000548169.2; ENST00000377685.9 | ||
| External Link | RMBase: m6A_site_208094 | ||
| mod ID: M6ASITE014996 | Click to Show/Hide the Full List | ||
| mod site | chr12:110296065-110296066:+ | [11] | |
| Sequence | CTGTTAAGAGCCTGTATTGAACCTGGGCAGCTTAGCCCAGA | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000377685.9; ENST00000548169.2; ENST00000550248.2; ENST00000552636.1; ENST00000308664.10; ENST00000539276.7 | ||
| External Link | RMBase: m6A_site_208095 | ||
| mod ID: M6ASITE014997 | Click to Show/Hide the Full List | ||
| mod site | chr12:110296275-110296276:+ | [11] | |
| Sequence | GCTGTACTGCCTCTCATGAGACAGAATTGATTTTAAATGGC | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000550248.2; ENST00000308664.10; ENST00000548169.2; ENST00000377685.9; ENST00000552636.1; ENST00000539276.7 | ||
| External Link | RMBase: m6A_site_208096 | ||
| mod ID: M6ASITE014998 | Click to Show/Hide the Full List | ||
| mod site | chr12:110296671-110296672:+ | [11] | |
| Sequence | GGGCAAAGTGTATCGACAGGACAGAAAGAGTGTGCAGCGGA | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; hNPCs | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000550248.2; ENST00000377685.9; ENST00000548169.2; ENST00000308664.10; ENST00000539276.7; ENST00000552636.1 | ||
| External Link | RMBase: m6A_site_208097 | ||
| mod ID: M6ASITE014999 | Click to Show/Hide the Full List | ||
| mod site | chr12:110296704-110296705:+ | [11] | |
| Sequence | GCAGCGGATTAAAGCTAAAGACATAGTTCCTGGTGATATTG | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000308664.10; ENST00000548169.2; ENST00000539276.7; ENST00000550248.2; ENST00000377685.9; ENST00000552636.1 | ||
| External Link | RMBase: m6A_site_208098 | ||
| mod ID: M6ASITE015000 | Click to Show/Hide the Full List | ||
| mod site | chr12:110322997-110322998:+ | [12] | |
| Sequence | TTTCTCCTAATTAGTTGGTGACAAAGTTCCTGCTGATATAA | ||
| Motif Score | 2.859755952 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000550248.2; ENST00000548169.2; ENST00000308664.10; ENST00000377685.9; ENST00000539276.7 | ||
| External Link | RMBase: m6A_site_208099 | ||
| mod ID: M6ASITE015001 | Click to Show/Hide the Full List | ||
| mod site | chr12:110323041-110323042:+ | [12] | |
| Sequence | TAACTTCCATCAAATCTACCACACTAAGAGTTGACCAGTCA | ||
| Motif Score | 2.053113095 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000377685.9; ENST00000539276.7; ENST00000548169.2; ENST00000308664.10; ENST00000550248.2 | ||
| External Link | RMBase: m6A_site_208100 | ||
| mod ID: M6ASITE015002 | Click to Show/Hide the Full List | ||
| mod site | chr12:110325807-110325808:+ | [11] | |
| Sequence | ACCCTGGGCAACATGGTAAAACCTCATCTCTACAAAAAAAT | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000539276.7; ENST00000548169.2; rmsk_3900698; ENST00000308664.10; ENST00000377685.9; ENST00000547050.1 | ||
| External Link | RMBase: m6A_site_208101 | ||
| mod ID: M6ASITE015003 | Click to Show/Hide the Full List | ||
| mod site | chr12:110326413-110326414:+ | [12] | |
| Sequence | ATCTGTCTCTGTCATCAAGCACACTGATCCCGTCCCTGACC | ||
| Motif Score | 2.830589286 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000547050.1; ENST00000548169.2; ENST00000539276.7; ENST00000377685.9; ENST00000308664.10 | ||
| External Link | RMBase: m6A_site_208102 | ||
| mod ID: M6ASITE015004 | Click to Show/Hide the Full List | ||
| mod site | chr12:110326461-110326462:+ | [11] | |
| Sequence | TGTCAACCAAGATAAAAAGAACATGCTGTTTTCTGTAAGTA | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa; hESC-HEK293T | ||
| Seq Type List | m6A-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000547050.1; ENST00000377685.9; ENST00000539276.7; ENST00000308664.10; ENST00000548169.2 | ||
| External Link | RMBase: m6A_site_208103 | ||
| mod ID: M6ASITE015005 | Click to Show/Hide the Full List | ||
| mod site | chr12:110327559-110327560:+ | [11] | |
| Sequence | ACTTCTGTCCTAGGGTACAAACATTGCTGCTGGGAAAGCTA | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa; hESC-HEK293T; endometrial | ||
| Seq Type List | m6A-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000308664.10; ENST00000377685.9; ENST00000548169.2; ENST00000539276.7; ENST00000547050.1 | ||
| External Link | RMBase: m6A_site_208104 | ||
| mod ID: M6ASITE015006 | Click to Show/Hide the Full List | ||
| mod site | chr12:110327607-110327608:+ | [12] | |
| Sequence | GGTGGTAGCAACTGGAGTTAACACCGAAATTGGCAAGATCC | ||
| Motif Score | 2.168095238 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000539276.7; ENST00000548169.2; ENST00000308664.10; ENST00000547050.1; ENST00000377685.9 | ||
| External Link | RMBase: m6A_site_208105 | ||
| mod ID: M6ASITE015007 | Click to Show/Hide the Full List | ||
| mod site | chr12:110327645-110327646:+ | [12] | |
| Sequence | TCCGGGATGAAATGGTGGCAACAGAACAGGAGAGAACACCC | ||
| Motif Score | 2.173910714 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000539276.7; ENST00000308664.10; ENST00000547050.1; ENST00000548169.2; ENST00000377685.9 | ||
| External Link | RMBase: m6A_site_208106 | ||
| mod ID: M6ASITE015008 | Click to Show/Hide the Full List | ||
| mod site | chr12:110327650-110327651:+ | [11] | |
| Sequence | GATGAAATGGTGGCAACAGAACAGGAGAGAACACCCCTTCA | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa; hESC-HEK293T; hNPCs; fibroblasts; A549; endometrial | ||
| Seq Type List | m6A-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000308664.10; ENST00000377685.9; ENST00000547050.1; ENST00000548169.2; ENST00000539276.7 | ||
| External Link | RMBase: m6A_site_208107 | ||
| mod ID: M6ASITE015009 | Click to Show/Hide the Full List | ||
| mod site | chr12:110327660-110327661:+ | [11] | |
| Sequence | TGGCAACAGAACAGGAGAGAACACCCCTTCAGCAAAAACTA | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa; hESC-HEK293T; hNPCs; fibroblasts; A549; endometrial | ||
| Seq Type List | m6A-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000548169.2; ENST00000547050.1; ENST00000539276.7; ENST00000308664.10; ENST00000377685.9 | ||
| External Link | RMBase: m6A_site_208108 | ||
| mod ID: M6ASITE015010 | Click to Show/Hide the Full List | ||
| mod site | chr12:110327677-110327678:+ | [11] | |
| Sequence | AGAACACCCCTTCAGCAAAAACTAGATGAATTTGGGGAACA | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa; hNPCs; fibroblasts; A549; endometrial | ||
| Seq Type List | m6A-seq; m6A-CLIP/IP | ||
| Transcript ID List | ENST00000539276.7; ENST00000548169.2; ENST00000308664.10; ENST00000547050.1; ENST00000377685.9 | ||
| External Link | RMBase: m6A_site_208109 | ||
| mod ID: M6ASITE015011 | Click to Show/Hide the Full List | ||
| mod site | chr12:110327695-110327696:+ | [11] | |
| Sequence | AAACTAGATGAATTTGGGGAACAGCTTTCCAAAGTCATCTC | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa; hNPCs; fibroblasts; A549; endometrial | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000539276.7; ENST00000547050.1; ENST00000549840.1; ENST00000377685.9; ENST00000308664.10; ENST00000548169.2 | ||
| External Link | RMBase: m6A_site_208110 | ||
| mod ID: M6ASITE015012 | Click to Show/Hide the Full List | ||
| mod site | chr12:110327805-110327806:+ | [13] | |
| Sequence | GATCAGAGGTGCTATTTACTACTTTAAAATTGCAGTGGCCC | ||
| Motif Score | 2.500660714 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000548169.2; ENST00000539276.7; ENST00000547050.1; ENST00000549840.1; ENST00000377685.9; ENST00000308664.10 | ||
| External Link | RMBase: m6A_site_208111 | ||
| mod ID: M6ASITE015013 | Click to Show/Hide the Full List | ||
| mod site | chr12:110327888-110327889:+ | [11] | |
| Sequence | CCACCTGCCTGGCTCTTGGAACTCGCAGAATGGCAAAGAAA | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa; HepG2; A549; AML | ||
| Seq Type List | m6A-seq; miCLIP | ||
| Transcript ID List | ENST00000377685.9; ENST00000539276.7; ENST00000548169.2; ENST00000547050.1; ENST00000308664.10; ENST00000549840.1 | ||
| External Link | RMBase: m6A_site_208112 | ||
| mod ID: M6ASITE015014 | Click to Show/Hide the Full List | ||
| mod site | chr12:110327942-110327943:+ | [11] | |
| Sequence | GAAGCCTCCCGTCTGTGGAAACCCTTGGTTGTACTTCTGTT | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | HeLa; HepG2; A549 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000308664.10; ENST00000548169.2; ENST00000539276.7; ENST00000377685.9; ENST00000547050.1; ENST00000549840.1 | ||
| External Link | RMBase: m6A_site_208113 | ||
| mod ID: M6ASITE015015 | Click to Show/Hide the Full List | ||
| mod site | chr12:110327973-110327974:+ | [11] | |
| Sequence | TACTTCTGTTATCTGCTCAGACAAGACTGGTACACTTACAA | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa; HepG2 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000547050.1; ENST00000539276.7; ENST00000377685.9; ENST00000308664.10; ENST00000548169.2; ENST00000549840.1 | ||
| External Link | RMBase: m6A_site_208114 | ||
| mod ID: M6ASITE015016 | Click to Show/Hide the Full List | ||
| mod site | chr12:110327978-110327979:+ | [11] | |
| Sequence | CTGTTATCTGCTCAGACAAGACTGGTACACTTACAACAAAC | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa; HepG2 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000377685.9; ENST00000547050.1; ENST00000549840.1; ENST00000539276.7; ENST00000308664.10; ENST00000548169.2 | ||
| External Link | RMBase: m6A_site_208115 | ||
| mod ID: M6ASITE015017 | Click to Show/Hide the Full List | ||
| mod site | chr12:110327997-110327998:+ | [11] | |
| Sequence | GACTGGTACACTTACAACAAACCAGATGTCAGTCTGCAGGG | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | HeLa; HepG2 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000547050.1; ENST00000539276.7; ENST00000549840.1; ENST00000377685.9; ENST00000308664.10; ENST00000548169.2 | ||
| External Link | RMBase: m6A_site_208116 | ||
| mod ID: M6ASITE015018 | Click to Show/Hide the Full List | ||
| mod site | chr12:110332609-110332610:+ | [11] | |
| Sequence | CCTACAGATGTTCATTCTGGACAGAGTGGAAGGTGATACTT | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000548169.2; ENST00000550262.1; ENST00000308664.10; ENST00000539276.7; ENST00000377685.9; ENST00000547050.1 | ||
| External Link | RMBase: m6A_site_208117 | ||
| mod ID: M6ASITE015019 | Click to Show/Hide the Full List | ||
| mod site | chr12:110333195-110333196:+ | [11] | |
| Sequence | GGCAGGCATAAAGATGATAAACCAGTGAATTGTCACCAGTA | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | HeLa; Huh7 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000548169.2; ENST00000550262.1; ENST00000308664.10; ENST00000377685.9; ENST00000539276.7 | ||
| External Link | RMBase: m6A_site_208118 | ||
| mod ID: M6ASITE015020 | Click to Show/Hide the Full List | ||
| mod site | chr12:110333238-110333239:+ | [12] | |
| Sequence | ATGGTCTGGTAGAATTAGCAACAATTTGTGCTCTTTGTAAT | ||
| Motif Score | 2.173910714 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000377685.9; ENST00000539276.7; ENST00000548169.2; ENST00000550262.1; ENST00000308664.10 | ||
| External Link | RMBase: m6A_site_208119 | ||
| mod ID: M6ASITE015021 | Click to Show/Hide the Full List | ||
| mod site | chr12:110333260-110333261:+ | [13] | |
| Sequence | AATTTGTGCTCTTTGTAATGACTCTGCTTTGGATTACAATG | ||
| Motif Score | 3.28175 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000308664.10; ENST00000548169.2; ENST00000539276.7; ENST00000377685.9; ENST00000550262.1 | ||
| External Link | RMBase: m6A_site_208120 | ||
| mod ID: M6ASITE015022 | Click to Show/Hide the Full List | ||
| mod site | chr12:110333275-110333276:+ | [12] | |
| Sequence | TAATGACTCTGCTTTGGATTACAATGAGGTAAGTCTCTTCA | ||
| Motif Score | 2.07285119 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000548169.2; ENST00000550262.1; ENST00000539276.7; ENST00000377685.9; ENST00000308664.10 | ||
| External Link | RMBase: m6A_site_208121 | ||
| mod ID: M6ASITE015023 | Click to Show/Hide the Full List | ||
| mod site | chr12:110334044-110334045:+ | [14] | |
| Sequence | ATGAAAAAGTTGGAGAAGCTACAGAGACTGCTCTCACTTGC | ||
| Motif Score | 2.078666667 | ||
| Cell/Tissue List | kidney; liver | ||
| Seq Type List | m6A-REF-seq | ||
| Transcript ID List | ENST00000548169.2; ENST00000308664.10; ENST00000377685.9; ENST00000550262.1; ENST00000539276.7 | ||
| External Link | RMBase: m6A_site_208122 | ||
| mod ID: M6ASITE015024 | Click to Show/Hide the Full List | ||
| mod site | chr12:110334050-110334051:+ | [11] | |
| Sequence | AAGTTGGAGAAGCTACAGAGACTGCTCTCACTTGCCTAGTA | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000548169.2; ENST00000308664.10; ENST00000377685.9; ENST00000539276.7; ENST00000550262.1 | ||
| External Link | RMBase: m6A_site_208123 | ||
| mod ID: M6ASITE015025 | Click to Show/Hide the Full List | ||
| mod site | chr12:110334155-110334156:+ | [11] | |
| Sequence | GCAACTCAGTGAGTATTTGAACCTGGTTTATTGTTCATGCT | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000548169.2; ENST00000308664.10; ENST00000550262.1; ENST00000539276.7; ENST00000377685.9 | ||
| External Link | RMBase: m6A_site_208124 | ||
| mod ID: M6ASITE015026 | Click to Show/Hide the Full List | ||
| mod site | chr12:110334232-110334233:+ | [11] | |
| Sequence | CTGTCCTACTCTCTTAGAAAACTAATTATACCTTATCTCCT | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000377685.9; ENST00000539276.7; ENST00000548169.2; rmsk_3900708; ENST00000308664.10; ENST00000550262.1 | ||
| External Link | RMBase: m6A_site_208125 | ||
| mod ID: M6ASITE015027 | Click to Show/Hide the Full List | ||
| mod site | chr12:110334256-110334257:+ | [11] | |
| Sequence | ATTATACCTTATCTCCTCAAACTTACAGTATTTCCTTCCCC | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000550262.1; ENST00000308664.10; ENST00000539276.7; rmsk_3900708; ENST00000377685.9; ENST00000548169.2 | ||
| External Link | RMBase: m6A_site_208126 | ||
| mod ID: M6ASITE015028 | Click to Show/Hide the Full List | ||
| mod site | chr12:110339288-110339289:+ | [11] | |
| Sequence | TTCCCTTTGTAGGTCATTAAACAGCTGATGAAAAAGGAATT | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000377685.9; ENST00000308664.10; ENST00000539276.7; ENST00000548169.2 | ||
| External Link | RMBase: m6A_site_208127 | ||
| mod ID: M6ASITE015029 | Click to Show/Hide the Full List | ||
| mod site | chr12:110339366-110339367:+ | [11] | |
| Sequence | GTTTACTGTACACCAAATAAACCAAGCAGGACATCAATGAG | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000539276.7; ENST00000308664.10; ENST00000548169.2; ENST00000377685.9 | ||
| External Link | RMBase: m6A_site_208128 | ||
| mod ID: M6ASITE015030 | Click to Show/Hide the Full List | ||
| mod site | chr12:110339376-110339377:+ | [11] | |
| Sequence | CACCAAATAAACCAAGCAGGACATCAATGAGCAAGATGTTT | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; hESC-HEK293T | ||
| Seq Type List | m6A-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000308664.10; ENST00000377685.9; ENST00000539276.7; ENST00000548169.2 | ||
| External Link | RMBase: m6A_site_208129 | ||
| mod ID: M6ASITE015031 | Click to Show/Hide the Full List | ||
| mod site | chr12:110339524-110339525:+ | [14] | |
| Sequence | TGCTCCTGAAGGTGTCATTGACAGGTGCACCCACATTCGAG | ||
| Motif Score | 2.859755952 | ||
| Cell/Tissue List | kidney | ||
| Seq Type List | m6A-REF-seq | ||
| Transcript ID List | ENST00000308664.10; ENST00000548169.2; ENST00000539276.7; ENST00000377685.9 | ||
| External Link | RMBase: m6A_site_208130 | ||
| mod ID: M6ASITE015032 | Click to Show/Hide the Full List | ||
| mod site | chr12:110339536-110339537:+ | [14] | |
| Sequence | TGTCATTGACAGGTGCACCCACATTCGAGTTGGAAGTACTA | ||
| Motif Score | 2.053113095 | ||
| Cell/Tissue List | kidney; hESC-HEK293T | ||
| Seq Type List | m6A-REF-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000539276.7; ENST00000308664.10; ENST00000377685.9; ENST00000548169.2 | ||
| External Link | RMBase: m6A_site_208131 | ||
| mod ID: M6ASITE015033 | Click to Show/Hide the Full List | ||
| mod site | chr12:110339582-110339583:+ | [11] | |
| Sequence | CCTATGACCTCTGGAGTCAAACAGAAGATCATGTCTGTCAT | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa; HepG2 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000548169.2; ENST00000308664.10; ENST00000539276.7; ENST00000377685.9 | ||
| External Link | RMBase: m6A_site_208132 | ||
| mod ID: M6ASITE015034 | Click to Show/Hide the Full List | ||
| mod site | chr12:110339626-110339627:+ | [12] | |
| Sequence | AGAGTGGGGTAGTGGCAGCGACACACTGCGATGCCTGGCCC | ||
| Motif Score | 2.865571429 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000548169.2; ENST00000377685.9; ENST00000539276.7; ENST00000308664.10 | ||
| External Link | RMBase: m6A_site_208133 | ||
| mod ID: M6ASITE015035 | Click to Show/Hide the Full List | ||
| mod site | chr12:110339659-110339660:+ | [12] | |
| Sequence | CCTGGCCCTGGCCACTCATGACAACCCACTGAGAAGAGAAG | ||
| Motif Score | 2.859755952 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000377685.9; ENST00000539276.7; ENST00000308664.10; ENST00000548169.2 | ||
| External Link | RMBase: m6A_site_208134 | ||
| mod ID: M6ASITE015036 | Click to Show/Hide the Full List | ||
| mod site | chr12:110339695-110339696:+ | [11] | |
| Sequence | AGAAGAAATGCACCTTGAGGACTCTGCCAACTTTATTAAAT | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa; HepG2 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000548169.2; ENST00000377685.9; ENST00000308664.10; ENST00000539276.7 | ||
| External Link | RMBase: m6A_site_208135 | ||
| mod ID: M6ASITE015037 | Click to Show/Hide the Full List | ||
| mod site | chr12:110340773-110340774:+ | [11] | |
| Sequence | GGTCATCATGATCACTGGGGACAACAAGGGCACTGCTGTGG | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; CD34; HepG2; hESC-HEK293T | ||
| Seq Type List | m6A-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000539276.7; ENST00000308664.10; ENST00000548169.2; ENST00000377685.9 | ||
| External Link | RMBase: m6A_site_208136 | ||
| mod ID: M6ASITE015038 | Click to Show/Hide the Full List | ||
| mod site | chr12:110340776-110340777:+ | [12] | |
| Sequence | CATCATGATCACTGGGGACAACAAGGGCACTGCTGTGGCCA | ||
| Motif Score | 2.173910714 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000377685.9; ENST00000539276.7; ENST00000308664.10; ENST00000548169.2 | ||
| External Link | RMBase: m6A_site_208137 | ||
| mod ID: M6ASITE015039 | Click to Show/Hide the Full List | ||
| mod site | chr12:110340873-110340874:+ | [11] | |
| Sequence | ACAGGCCGGGAGTTTGATGAACTCAACCCCTCCGCCCAGCG | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa; CD34; HepG2; endometrial; AML | ||
| Seq Type List | m6A-seq; miCLIP | ||
| Transcript ID List | ENST00000548169.2; ENST00000539276.7; ENST00000308664.10; ENST00000377685.9 | ||
| External Link | RMBase: m6A_site_208138 | ||
| mod ID: M6ASITE015040 | Click to Show/Hide the Full List | ||
| mod site | chr12:110340933-110340934:+ | [11] | |
| Sequence | CGCTGTTTTGCTCGAGTTGAACCCTCCCACAAGTCTAAAAT | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | HeLa; HepG2 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000308664.10; ENST00000548169.2; ENST00000377685.9; ENST00000539276.7 | ||
| External Link | RMBase: m6A_site_208139 | ||
| mod ID: M6ASITE015041 | Click to Show/Hide the Full List | ||
| mod site | chr12:110340941-110340942:+ | [12] | |
| Sequence | TGCTCGAGTTGAACCCTCCCACAAGTCTAAAATCGTAGAAT | ||
| Motif Score | 2.053113095 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000548169.2; ENST00000539276.7; ENST00000308664.10; ENST00000377685.9 | ||
| External Link | RMBase: m6A_site_208140 | ||
| mod ID: M6ASITE015042 | Click to Show/Hide the Full List | ||
| mod site | chr12:110340985-110340986:+ | [14] | |
| Sequence | TTCAGTCTTTTGATGAGATTACAGCTATGGTGAGCATGTTT | ||
| Motif Score | 2.07285119 | ||
| Cell/Tissue List | liver | ||
| Seq Type List | m6A-REF-seq | ||
| Transcript ID List | ENST00000548169.2; ENST00000539276.7; ENST00000308664.10; ENST00000377685.9 | ||
| External Link | RMBase: m6A_site_208141 | ||
| mod ID: M6ASITE015043 | Click to Show/Hide the Full List | ||
| mod site | chr12:110342311-110342312:+ | [11] | |
| Sequence | CTGGCACTGCGGTGGCTAAAACCGCCTCTGAGATGGTCCTG | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000539276.7; ENST00000377685.9; ENST00000308664.10; ENST00000548169.2 | ||
| External Link | RMBase: m6A_site_208142 | ||
| mod ID: M6ASITE015044 | Click to Show/Hide the Full List | ||
| mod site | chr12:110342339-110342340:+ | [12] | |
| Sequence | TGAGATGGTCCTGGCGGATGACAACTTCTCCACCATTGTGG | ||
| Motif Score | 2.859755952 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000539276.7; ENST00000548169.2; ENST00000308664.10; ENST00000377685.9 | ||
| External Link | RMBase: m6A_site_208143 | ||
| mod ID: M6ASITE015045 | Click to Show/Hide the Full List | ||
| mod site | chr12:110342387-110342388:+ | [12] | |
| Sequence | TGAGGAGGGGCGGGCAATCTACAACAACATGAAACAGTTCA | ||
| Motif Score | 2.078666667 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000308664.10; ENST00000377685.9; ENST00000539276.7; ENST00000548169.2 | ||
| External Link | RMBase: m6A_site_208144 | ||
| mod ID: M6ASITE015046 | Click to Show/Hide the Full List | ||
| mod site | chr12:110342400-110342401:+ | [11] | |
| Sequence | GCAATCTACAACAACATGAAACAGTTCATCCGCTACCTCAT | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000377685.9; ENST00000539276.7; ENST00000308664.10; ENST00000548169.2 | ||
| External Link | RMBase: m6A_site_208145 | ||
| mod ID: M6ASITE015047 | Click to Show/Hide the Full List | ||
| mod site | chr12:110343353-110343354:+ | [11] | |
| Sequence | GTTCAACCCTCCTGATCTGGACATCATGAATAAACCTCCCC | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; hESC-HEK293T | ||
| Seq Type List | m6A-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000548169.2; ENST00000547792.1; ENST00000308664.10; ENST00000377685.9; ENST00000539276.7 | ||
| External Link | RMBase: m6A_site_208146 | ||
| mod ID: M6ASITE015048 | Click to Show/Hide the Full List | ||
| mod site | chr12:110343366-110343367:+ | [11] | |
| Sequence | GATCTGGACATCATGAATAAACCTCCCCGGAACCCAAAGGA | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000548169.2; ENST00000547792.1; ENST00000539276.7; ENST00000308664.10; ENST00000377685.9 | ||
| External Link | RMBase: m6A_site_208147 | ||
| mod ID: M6ASITE015049 | Click to Show/Hide the Full List | ||
| mod site | chr12:110343377-110343378:+ | [11] | |
| Sequence | CATGAATAAACCTCCCCGGAACCCAAAGGAACCATTGATCA | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000377685.9; ENST00000539276.7; ENST00000308664.10; ENST00000548169.2; ENST00000547792.1 | ||
| External Link | RMBase: m6A_site_208148 | ||
| mod ID: M6ASITE015050 | Click to Show/Hide the Full List | ||
| mod site | chr12:110343387-110343388:+ | [11] | |
| Sequence | CCTCCCCGGAACCCAAAGGAACCATTGATCAGCGGGTGGCT | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | HeLa; HEK293T | ||
| Seq Type List | m6A-seq; DART-seq | ||
| Transcript ID List | ENST00000377685.9; ENST00000548169.2; ENST00000308664.10; ENST00000547792.1; ENST00000539276.7 | ||
| External Link | RMBase: m6A_site_208149 | ||
| mod ID: M6ASITE015051 | Click to Show/Hide the Full List | ||
| mod site | chr12:110345273-110345274:+ | [12] | |
| Sequence | TTTCCTACAGTGTAAAGAGGACAACCCGGACTTTGAAGGCG | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000308664.10; ENST00000548169.2; ENST00000377685.9; ENST00000539276.7 | ||
| External Link | RMBase: m6A_site_208150 | ||
| mod ID: M6ASITE015052 | Click to Show/Hide the Full List | ||
| mod site | chr12:110345329-110345330:+ | [13] | |
| Sequence | TTGAATCCCCATACCCGATGACAATGGCGCTCTCTGTTCTA | ||
| Motif Score | 2.859755952 | ||
| Cell/Tissue List | HEK293T; hESC-HEK293T | ||
| Seq Type List | DART-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000548169.2; ENST00000539276.7; ENST00000377685.9; ENST00000308664.10 | ||
| External Link | RMBase: m6A_site_208151 | ||
| mod ID: M6ASITE015053 | Click to Show/Hide the Full List | ||
| mod site | chr12:110346011-110346012:+ | [11] | |
| Sequence | GCTCTGCAGCTTGTCCGAAAACCAGTCCTTGCTGAGGATGC | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | HeLa; Huh7 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000377685.9; ENST00000548169.2; ENST00000308664.10; ENST00000313432.5; ENST00000539276.7 | ||
| External Link | RMBase: m6A_site_208152 | ||
| mod ID: M6ASITE015054 | Click to Show/Hide the Full List | ||
| mod site | chr12:110346044-110346045:+ | [11] | |
| Sequence | GAGGATGCCCCCCTGGGAGAACATCTGGCTCGTGGGCTCCA | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa; hESC-HEK293T; Huh7 | ||
| Seq Type List | m6A-seq; MAZTER-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000548169.2; ENST00000308664.10; ENST00000539276.7; ENST00000313432.5; ENST00000377685.9 | ||
| External Link | RMBase: m6A_site_208153 | ||
| mod ID: M6ASITE015055 | Click to Show/Hide the Full List | ||
| mod site | chr12:110346108-110346109:+ | [11] | |
| Sequence | TTCCTGATCCTCTATGTCGAACCCTTGCCAGTAAGTGGTTG | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | HeLa; HepG2; Huh7 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000377685.9; ENST00000548169.2; ENST00000313432.5; ENST00000308664.10; ENST00000539276.7 | ||
| External Link | RMBase: m6A_site_208154 | ||
| mod ID: M6ASITE015056 | Click to Show/Hide the Full List | ||
| mod site | chr12:110346215-110346216:+ | [12] | |
| Sequence | GTCAGCTCATCTTCCAGATCACACCGCTGAACGTGACCCAG | ||
| Motif Score | 2.047297619 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000539276.7; ENST00000548169.2; ENST00000553144.1; ENST00000313432.5; ENST00000377685.9; ENST00000308664.10 | ||
| External Link | RMBase: m6A_site_208155 | ||
| mod ID: M6ASITE015057 | Click to Show/Hide the Full List | ||
| mod site | chr12:110346316-110346317:+ | [11] | |
| Sequence | GTGGCCCGCAACTACCTGGAACCTGGTAAAGAGTGTGTGCA | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | HeLa; HepG2; endometrial | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000308664.10; ENST00000548169.2; ENST00000553144.1; ENST00000313432.5; ENST00000539276.7; ENST00000377685.9 | ||
| External Link | RMBase: m6A_site_208156 | ||
| mod ID: M6ASITE015058 | Click to Show/Hide the Full List | ||
| mod site | chr12:110346438-110346439:+ | [11] | |
| Sequence | GATCTGGGTCTATAGCACAGACACTAACTTTAGCGATATGT | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa; CD34; hESC-HEK293T; HepG2; fibroblasts; A549; TIME; endometrial | ||
| Seq Type List | m6A-seq; MAZTER-seq; MeRIP-seq; m6A-CLIP/IP | ||
| Transcript ID List | ENST00000548169.2; ENST00000377685.9; ENST00000308664.10; ENST00000539276.7; ENST00000553144.1; ENST00000313432.5 | ||
| External Link | RMBase: m6A_site_208157 | ||
| mod ID: M6ASITE015059 | Click to Show/Hide the Full List | ||
| mod site | chr12:110346444-110346445:+ | [15] | |
| Sequence | GGTCTATAGCACAGACACTAACTTTAGCGATATGTTCTGGT | ||
| Motif Score | 2.590089286 | ||
| Cell/Tissue List | A549 | ||
| Seq Type List | m6A-CLIP/IP | ||
| Transcript ID List | ENST00000308664.10; ENST00000553144.1; ENST00000539276.7; ENST00000313432.5; ENST00000377685.9; ENST00000548169.2 | ||
| External Link | RMBase: m6A_site_208158 | ||
| mod ID: M6ASITE015060 | Click to Show/Hide the Full List | ||
| mod site | chr12:110346469-110346470:+ | [13] | |
| Sequence | AGCGATATGTTCTGGTCTTGACTGACAGTTTTCCATAAAGA | ||
| Motif Score | 3.28175 | ||
| Cell/Tissue List | HEK293T; AML | ||
| Seq Type List | DART-seq; miCLIP | ||
| Transcript ID List | ENST00000539276.7; ENST00000548169.2; ENST00000308664.10; ENST00000553144.1; ENST00000313432.5; ENST00000377685.9 | ||
| External Link | RMBase: m6A_site_208159 | ||
| mod ID: M6ASITE015061 | Click to Show/Hide the Full List | ||
| mod site | chr12:110346473-110346474:+ | [14] | |
| Sequence | ATATGTTCTGGTCTTGACTGACAGTTTTCCATAAAGAAGAT | ||
| Motif Score | 2.859755952 | ||
| Cell/Tissue List | kidney; liver; HEK293T | ||
| Seq Type List | m6A-REF-seq; DART-seq | ||
| Transcript ID List | ENST00000308664.10; ENST00000553144.1; ENST00000377685.9; ENST00000313432.5; ENST00000539276.7; ENST00000548169.2 | ||
| External Link | RMBase: m6A_site_208160 | ||
| mod ID: M6ASITE015062 | Click to Show/Hide the Full List | ||
| mod site | chr12:110346499-110346500:+ | [13] | |
| Sequence | TTCCATAAAGAAGATGTTTAACTTAATCAATTAATTTTTTT | ||
| Motif Score | 2.590089286 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000553144.1; ENST00000548169.2; ENST00000313432.5; ENST00000308664.10; ENST00000377685.9; ENST00000539276.7 | ||
| External Link | RMBase: m6A_site_208161 | ||
| mod ID: M6ASITE015063 | Click to Show/Hide the Full List | ||
| mod site | chr12:110346533-110346534:+ | [13] | |
| Sequence | TTTTTTTATTGTTTAAAGCAACTGTCTATTTCTGCTGAATT | ||
| Motif Score | 2.595904762 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000539276.7; ENST00000553144.1; ENST00000313432.5; ENST00000548169.2; ENST00000308664.10; ENST00000377685.9 | ||
| External Link | RMBase: m6A_site_208162 | ||
| mod ID: M6ASITE015064 | Click to Show/Hide the Full List | ||
| mod site | chr12:110346557-110346558:+ | [13] | |
| Sequence | TCTATTTCTGCTGAATTTTCACATGAACATACTGGCTGGTG | ||
| Motif Score | 2.047297619 | ||
| Cell/Tissue List | HEK293T; hESC-HEK293T | ||
| Seq Type List | DART-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000539276.7; ENST00000377685.9; ENST00000313432.5; ENST00000308664.10; ENST00000553144.1; ENST00000548169.2 | ||
| External Link | RMBase: m6A_site_208163 | ||
| mod ID: M6ASITE015065 | Click to Show/Hide the Full List | ||
| mod site | chr12:110346563-110346564:+ | [11] | |
| Sequence | TCTGCTGAATTTTCACATGAACATACTGGCTGGTGATGGAG | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa; hESC-HEK293T; fibroblasts; A549; iSLK | ||
| Seq Type List | m6A-seq; MAZTER-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000313432.5; ENST00000548169.2; ENST00000539276.7; ENST00000553144.1; ENST00000308664.10; ENST00000377685.9 | ||
| External Link | RMBase: m6A_site_208164 | ||
| mod ID: M6ASITE015066 | Click to Show/Hide the Full List | ||
| mod site | chr12:110346591-110346592:+ | [13] | |
| Sequence | GCTGGTGATGGAGGTTTCATACTCTAGATTTTGTTTTGCTT | ||
| Motif Score | 2.53247619 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000313432.5; ENST00000377685.9; ENST00000548169.2; ENST00000308664.10; ENST00000553144.1; ENST00000539276.7 | ||
| External Link | RMBase: m6A_site_208165 | ||
| mod ID: M6ASITE015067 | Click to Show/Hide the Full List | ||
| mod site | chr12:110346618-110346619:+ | [13] | |
| Sequence | ATTTTGTTTTGCTTTTTCTGACTCCAGTGGGGCAAGATTTT | ||
| Motif Score | 3.28175 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000313432.5; ENST00000553144.1; ENST00000377685.9; ENST00000308664.10; ENST00000539276.7; ENST00000548169.2 | ||
| External Link | RMBase: m6A_site_208166 | ||
| mod ID: M6ASITE015068 | Click to Show/Hide the Full List | ||
| mod site | chr12:110346649-110346650:+ | [12] | |
| Sequence | GCAAGATTTTCCTTTTTTATACACATAATTAAAGTGTCCAT | ||
| Motif Score | 2.110482143 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000308664.10; ENST00000539276.7; ENST00000377685.9; ENST00000313432.5; ENST00000553144.1 | ||
| External Link | RMBase: m6A_site_208167 | ||
| mod ID: M6ASITE015069 | Click to Show/Hide the Full List | ||
| mod site | chr12:110346651-110346652:+ | [13] | |
| Sequence | AAGATTTTCCTTTTTTATACACATAATTAAAGTGTCCATTG | ||
| Motif Score | 2.084928571 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000377685.9; ENST00000539276.7; ENST00000553144.1; ENST00000313432.5; ENST00000308664.10 | ||
| External Link | RMBase: m6A_site_208168 | ||
| mod ID: M6ASITE015070 | Click to Show/Hide the Full List | ||
| mod site | chr12:110346672-110346673:+ | [13] | |
| Sequence | CATAATTAAAGTGTCCATTGACATGTACAGAGAACTAACAC | ||
| Motif Score | 2.859755952 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000539276.7; ENST00000313432.5; ENST00000377685.9; ENST00000553144.1; ENST00000308664.10 | ||
| External Link | RMBase: m6A_site_208169 | ||
| mod ID: M6ASITE015071 | Click to Show/Hide the Full List | ||
| mod site | chr12:110346685-110346686:+ | [11] | |
| Sequence | TCCATTGACATGTACAGAGAACTAACACTATTTTATGCAAA | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa; iSLK | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000377685.9; ENST00000313432.5; ENST00000553144.1; ENST00000308664.10; ENST00000539276.7 | ||
| External Link | RMBase: m6A_site_208170 | ||
| mod ID: M6ASITE015072 | Click to Show/Hide the Full List | ||
| mod site | chr12:110346733-110346734:+ | [13] | |
| Sequence | TTGTAGATGAAAAAGCATGTACAGTGTTCTGTTTAATACTC | ||
| Motif Score | 2.856142857 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000377685.9; ENST00000313432.5; ENST00000539276.7; ENST00000308664.10; ENST00000553144.1 | ||
| External Link | RMBase: m6A_site_208171 | ||
| mod ID: M6ASITE015073 | Click to Show/Hide the Full List | ||
| mod site | chr12:110346750-110346751:+ | [13] | |
| Sequence | TGTACAGTGTTCTGTTTAATACTCATCCTTGTATAAAAAAA | ||
| Motif Score | 2.53247619 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000553144.1; ENST00000308664.10; ENST00000539276.7; ENST00000313432.5; ENST00000377685.9 | ||
| External Link | RMBase: m6A_site_208172 | ||
| mod ID: M6ASITE015074 | Click to Show/Hide the Full List | ||
| mod site | chr12:110346787-110346788:+ | [11] | |
| Sequence | AAAAATAGTTGAGCCAGCAGACATTGTCAGCAAATTAATTG | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa; HEK293T; hESC-HEK293T | ||
| Seq Type List | m6A-seq; DART-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000553144.1; ENST00000539276.7; ENST00000308664.10; ENST00000313432.5; ENST00000377685.9 | ||
| External Link | RMBase: m6A_site_208173 | ||
| mod ID: M6ASITE015075 | Click to Show/Hide the Full List | ||
| mod site | chr12:110346852-110346853:+ | [11] | |
| Sequence | TGTGTGGTTTTTTTTCTAAAACTAAATAGCATGTATTGTGT | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa; HEK293T | ||
| Seq Type List | m6A-seq; DART-seq | ||
| Transcript ID List | ENST00000377685.9; ENST00000539276.7; ENST00000313432.5; ENST00000553144.1; ENST00000308664.10 | ||
| External Link | RMBase: m6A_site_208174 | ||
| mod ID: M6ASITE015076 | Click to Show/Hide the Full List | ||
| mod site | chr12:110346907-110346908:+ | [13] | |
| Sequence | TCCGGATTTAATTTGATATCACAGTCTAATTTTTATTCATA | ||
| Motif Score | 2.047297619 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000553144.1; ENST00000539276.7; ENST00000377685.9; ENST00000308664.10; ENST00000313432.5 | ||
| External Link | RMBase: m6A_site_208175 | ||
| mod ID: M6ASITE015077 | Click to Show/Hide the Full List | ||
| mod site | chr12:110346989-110346990:+ | [13] | |
| Sequence | CAGTATTGTTTTGGTTTGCTACTTCCCTCACCCACTTTGGC | ||
| Motif Score | 2.500660714 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000539276.7; ENST00000308664.10; ENST00000377685.9; ENST00000553144.1; ENST00000313432.5 | ||
| External Link | RMBase: m6A_site_208176 | ||
| mod ID: M6ASITE015078 | Click to Show/Hide the Full List | ||
| mod site | chr12:110347100-110347101:+ | [12] | |
| Sequence | TGTGATGGTTCGTTCTGTTTACATCAGTTTTAACGAGAGGT | ||
| Motif Score | 2.07285119 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000553144.1; ENST00000308664.10; ENST00000377685.9; ENST00000539276.7; ENST00000313432.5 | ||
| External Link | RMBase: m6A_site_208177 | ||
| mod ID: M6ASITE015079 | Click to Show/Hide the Full List | ||
| mod site | chr12:110347112-110347113:+ | [13] | |
| Sequence | TTCTGTTTACATCAGTTTTAACGAGAGGTATGCCTGTACTC | ||
| Motif Score | 2.142029762 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000313432.5; ENST00000539276.7; ENST00000553144.1; ENST00000308664.10; ENST00000377685.9 | ||
| External Link | RMBase: m6A_site_208178 | ||
| mod ID: M6ASITE015080 | Click to Show/Hide the Full List | ||
| mod site | chr12:110347146-110347147:+ | [12] | |
| Sequence | TGTACTCGCTTGTGCAGAAAACATTGTTCCAGATTCAATCG | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000553144.1; ENST00000308664.10; ENST00000313432.5; ENST00000539276.7; ENST00000377685.9 | ||
| External Link | RMBase: m6A_site_208179 | ||
| mod ID: M6ASITE015081 | Click to Show/Hide the Full List | ||
| mod site | chr12:110347167-110347168:+ | [13] | |
| Sequence | CATTGTTCCAGATTCAATCGACTGGGTTTATGTCCCTTCAC | ||
| Motif Score | 3.287565476 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000553144.1; ENST00000313432.5; ENST00000539276.7; ENST00000377685.9; ENST00000308664.10 | ||
| External Link | RMBase: m6A_site_208180 | ||
| mod ID: M6ASITE015082 | Click to Show/Hide the Full List | ||
| mod site | chr12:110347186-110347187:+ | [13] | |
| Sequence | GACTGGGTTTATGTCCCTTCACATAGTTTTTAAGGTTATTT | ||
| Motif Score | 2.047297619 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000308664.10; ENST00000539276.7; ENST00000377685.9; ENST00000553144.1; ENST00000313432.5 | ||
| External Link | RMBase: m6A_site_208181 | ||
| mod ID: M6ASITE015083 | Click to Show/Hide the Full List | ||
| mod site | chr12:110347235-110347236:+ | [13] | |
| Sequence | GTCTAATGTATTTTATTGTAACAGACATTGTTTTGCCAACA | ||
| Motif Score | 2.168095238 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000308664.10; ENST00000539276.7; ENST00000553144.1; ENST00000377685.9; ENST00000313432.5 | ||
| External Link | RMBase: m6A_site_208182 | ||
| mod ID: M6ASITE015084 | Click to Show/Hide the Full List | ||
| mod site | chr12:110347239-110347240:+ | [11] | |
| Sequence | AATGTATTTTATTGTAACAGACATTGTTTTGCCAACATTGC | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa; HEK293T | ||
| Seq Type List | m6A-seq; DART-seq | ||
| Transcript ID List | ENST00000553144.1; ENST00000377685.9; ENST00000539276.7; ENST00000313432.5; ENST00000308664.10 | ||
| External Link | RMBase: m6A_site_208183 | ||
| mod ID: M6ASITE015085 | Click to Show/Hide the Full List | ||
| mod site | chr12:110347253-110347254:+ | [13] | |
| Sequence | TAACAGACATTGTTTTGCCAACATTGCCTATTTCAGTGGCA | ||
| Motif Score | 2.173910714 | ||
| Cell/Tissue List | HEK293T; hESC-HEK293T | ||
| Seq Type List | DART-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000553144.1; ENST00000377685.9; ENST00000539276.7; ENST00000313432.5; ENST00000308664.10 | ||
| External Link | RMBase: m6A_site_208184 | ||
| mod ID: M6ASITE015086 | Click to Show/Hide the Full List | ||
| mod site | chr12:110347299-110347300:+ | [11] | |
| Sequence | TCTAGTTTTAAAAAAATAAAACATTTTAAATGGACAGAGAA | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa; hESC-HEK293T | ||
| Seq Type List | m6A-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000377685.9; ENST00000553144.1; ENST00000308664.10; ENST00000539276.7; ENST00000313432.5 | ||
| External Link | RMBase: m6A_site_208185 | ||
| mod ID: M6ASITE015087 | Click to Show/Hide the Full List | ||
| mod site | chr12:110347312-110347313:+ | [11] | |
| Sequence | AAATAAAACATTTTAAATGGACAGAGAAAAATAACTGTCTT | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000308664.10; ENST00000377685.9; ENST00000553144.1; ENST00000539276.7; ENST00000313432.5 | ||
| External Link | RMBase: m6A_site_208186 | ||
| mod ID: M6ASITE015088 | Click to Show/Hide the Full List | ||
| mod site | chr12:110347362-110347363:+ | [11] | |
| Sequence | TCGTAAGTGGCTTACCTGGGACTAACAGACATGTCCAACTT | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000308664.10; ENST00000377685.9; ENST00000553144.1; ENST00000539276.7; ENST00000313432.5 | ||
| External Link | RMBase: m6A_site_208187 | ||
| mod ID: M6ASITE015089 | Click to Show/Hide the Full List | ||
| mod site | chr12:110347370-110347371:+ | [11] | |
| Sequence | GGCTTACCTGGGACTAACAGACATGTCCAACTTTCTCTCCA | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa; hESC-HEK293T | ||
| Seq Type List | m6A-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000539276.7; ENST00000308664.10; ENST00000313432.5; ENST00000553144.1; ENST00000377685.9 | ||
| External Link | RMBase: m6A_site_208188 | ||
| mod ID: M6ASITE015090 | Click to Show/Hide the Full List | ||
| mod site | chr12:110347416-110347417:+ | [13] | |
| Sequence | TAGCTCAGAACTTTAGTTGTACTCTGCTTGAGGGGAAGAAG | ||
| Motif Score | 3.278136905 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000313432.5; ENST00000553144.1; ENST00000539276.7; ENST00000377685.9; ENST00000308664.10 | ||
| External Link | RMBase: m6A_site_208189 | ||
| mod ID: M6ASITE015091 | Click to Show/Hide the Full List | ||
| mod site | chr12:110347549-110347550:+ | [11] | |
| Sequence | CTGCTGGCCTGGTATAGAGAACATAAGGGCAAGTGTGTATG | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa; hESC-HEK293T | ||
| Seq Type List | m6A-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000553144.1; ENST00000377685.9; ENST00000313432.5; ENST00000308664.10; ENST00000539276.7 | ||
| External Link | RMBase: m6A_site_208190 | ||
| mod ID: M6ASITE015092 | Click to Show/Hide the Full List | ||
| mod site | chr12:110347608-110347609:+ | [12] | |
| Sequence | TTGTAAAATCTGTAAATAGCACATGACCAAATGAACATATT | ||
| Motif Score | 2.830589286 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000553144.1; ENST00000377685.9; ENST00000539276.7; ENST00000308664.10; ENST00000313432.5 | ||
| External Link | RMBase: m6A_site_208191 | ||
| mod ID: M6ASITE015093 | Click to Show/Hide the Full List | ||
| mod site | chr12:110347613-110347614:+ | [13] | |
| Sequence | AAATCTGTAAATAGCACATGACCAAATGAACATATTGTATA | ||
| Motif Score | 2.839113095 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000308664.10; ENST00000377685.9; ENST00000539276.7; ENST00000553144.1; ENST00000313432.5 | ||
| External Link | RMBase: m6A_site_208192 | ||
| mod ID: M6ASITE015094 | Click to Show/Hide the Full List | ||
| mod site | chr12:110347622-110347623:+ | [11] | |
| Sequence | AATAGCACATGACCAAATGAACATATTGTATAGAACTATTT | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa; HEK293T | ||
| Seq Type List | m6A-seq; DART-seq | ||
| Transcript ID List | ENST00000377685.9; ENST00000308664.10; ENST00000553144.1; ENST00000539276.7; ENST00000313432.5 | ||
| External Link | RMBase: m6A_site_208193 | ||
| mod ID: M6ASITE015095 | Click to Show/Hide the Full List | ||
| mod site | chr12:110347636-110347637:+ | [11] | |
| Sequence | AAATGAACATATTGTATAGAACTATTTTTATTTGAATGTGG | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000313432.5; ENST00000539276.7; ENST00000308664.10; ENST00000377685.9; ENST00000553144.1 | ||
| External Link | RMBase: m6A_site_208194 | ||
| mod ID: M6ASITE015096 | Click to Show/Hide the Full List | ||
| mod site | chr12:110347785-110347786:+ | [13] | |
| Sequence | ATGCCTTCCAACATCCATCAACTAACGTGAGTATTTTCTTC | ||
| Motif Score | 2.595904762 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000539276.7; ENST00000308664.10; ENST00000553144.1; ENST00000377685.9; ENST00000313432.5 | ||
| External Link | RMBase: m6A_site_208195 | ||
| mod ID: M6ASITE015097 | Click to Show/Hide the Full List | ||
| mod site | chr12:110347997-110347998:+ | [16] | |
| Sequence | TGGAGGTGTGGCTTTCATAGACCTCCACAGGCTGCCTAGGA | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | CD34; endometrial | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000553144.1; ENST00000377685.9; ENST00000308664.10; ENST00000313432.5; ENST00000539276.7 | ||
| External Link | RMBase: m6A_site_208196 | ||
| mod ID: M6ASITE015098 | Click to Show/Hide the Full List | ||
| mod site | chr12:110348003-110348004:+ | [14] | |
| Sequence | TGTGGCTTTCATAGACCTCCACAGGCTGCCTAGGACAAGAT | ||
| Motif Score | 2.053113095 | ||
| Cell/Tissue List | kidney | ||
| Seq Type List | m6A-REF-seq | ||
| Transcript ID List | ENST00000553144.1; ENST00000313432.5; ENST00000377685.9; ENST00000539276.7; ENST00000308664.10 | ||
| External Link | RMBase: m6A_site_208197 | ||
| mod ID: M6ASITE015099 | Click to Show/Hide the Full List | ||
| mod site | chr12:110348017-110348018:+ | [17] | |
| Sequence | ACCTCCACAGGCTGCCTAGGACAAGATGACCAGGAGGGCCC | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | endometrial | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000308664.10; ENST00000539276.7; ENST00000313432.5; ENST00000553144.1; ENST00000377685.9 | ||
| External Link | RMBase: m6A_site_208198 | ||
| mod ID: M6ASITE015100 | Click to Show/Hide the Full List | ||
| mod site | chr12:110348045-110348046:+ | [12] | |
| Sequence | ACCAGGAGGGCCCAAGCCAGACAAAAGCCGGAGTGGGGAGA | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | hESC-HEK293T; endometrial | ||
| Seq Type List | MAZTER-seq; m6A-seq | ||
| Transcript ID List | ENST00000553144.1; ENST00000308664.10; ENST00000377685.9; ENST00000539276.7; ENST00000313432.5 | ||
| External Link | RMBase: m6A_site_208199 | ||
| mod ID: M6ASITE015101 | Click to Show/Hide the Full List | ||
| mod site | chr12:110348082-110348083:+ | [17] | |
| Sequence | GAGAGAGGCATTTCAGCCAGACCAACAGGCTGAAGGAGCTG | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | endometrial | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000553144.1; ENST00000377685.9; ENST00000313432.5; ENST00000539276.7; ENST00000308664.10 | ||
| External Link | RMBase: m6A_site_208200 | ||
| mod ID: M6ASITE015102 | Click to Show/Hide the Full List | ||
| mod site | chr12:110348086-110348087:+ | [14] | |
| Sequence | GAGGCATTTCAGCCAGACCAACAGGCTGAAGGAGCTGTGCA | ||
| Motif Score | 2.173910714 | ||
| Cell/Tissue List | kidney | ||
| Seq Type List | m6A-REF-seq | ||
| Transcript ID List | ENST00000313432.5; ENST00000539276.7; ENST00000377685.9; ENST00000553144.1; ENST00000308664.10 | ||
| External Link | RMBase: m6A_site_208201 | ||
| mod ID: M6ASITE015103 | Click to Show/Hide the Full List | ||
| mod site | chr12:110348108-110348109:+ | [17] | |
| Sequence | AGGCTGAAGGAGCTGTGCAGACTATTGCTAAAATGAGGGTT | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | endometrial | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000313432.5; ENST00000377685.9; ENST00000308664.10; ENST00000553144.1; ENST00000539276.7 | ||
| External Link | RMBase: m6A_site_208202 | ||
| mod ID: M6ASITE015104 | Click to Show/Hide the Full List | ||
| mod site | chr12:110348154-110348155:+ | [12] | |
| Sequence | CTGCCAGGAGTCATCCCAGAACATTGCTACTTATTTATTAA | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | hESC-HEK293T; endometrial | ||
| Seq Type List | MAZTER-seq; m6A-seq | ||
| Transcript ID List | ENST00000313432.5; ENST00000553144.1; ENST00000308664.10; ENST00000377685.9; ENST00000539276.7 | ||
| External Link | RMBase: m6A_site_208203 | ||
| mod ID: M6ASITE015105 | Click to Show/Hide the Full List | ||
| mod site | chr12:110348207-110348208:+ | [11] | |
| Sequence | TAGTGAAAGCAAGTAACTGAACCAGTGAACTCTGGGTATCG | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000308664.10; ENST00000553144.1; ENST00000313432.5; ENST00000377685.9; ENST00000539276.7 | ||
| External Link | RMBase: m6A_site_208204 | ||
| mod ID: M6ASITE015106 | Click to Show/Hide the Full List | ||
| mod site | chr12:110348215-110348216:+ | [11] | |
| Sequence | GCAAGTAACTGAACCAGTGAACTCTGGGTATCGATAGGTTC | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000313432.5; ENST00000308664.10; ENST00000377685.9; ENST00000553144.1; ENST00000539276.7 | ||
| External Link | RMBase: m6A_site_208205 | ||
| mod ID: M6ASITE015107 | Click to Show/Hide the Full List | ||
| mod site | chr12:110348302-110348303:+ | [11] | |
| Sequence | TCTTGTCCTTGGTGGCTAAGACTTAGCTCTGCAGGGGATGT | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000539276.7; ENST00000553144.1; ENST00000377685.9; ENST00000308664.10; ENST00000313432.5 | ||
| External Link | RMBase: m6A_site_208206 | ||
| mod ID: M6ASITE015108 | Click to Show/Hide the Full List | ||
| mod site | chr12:110348353-110348354:+ | [14] | |
| Sequence | TTAGTAGGACGTGGCTCTGCACAGCCCAAAAACCAGCTTAC | ||
| Motif Score | 2.830589286 | ||
| Cell/Tissue List | brain; kidney; liver | ||
| Seq Type List | m6A-REF-seq | ||
| Transcript ID List | ENST00000553144.1; ENST00000308664.10; ENST00000313432.5; ENST00000377685.9; ENST00000539276.7 | ||
| External Link | RMBase: m6A_site_208207 | ||
| mod ID: M6ASITE015109 | Click to Show/Hide the Full List | ||
| mod site | chr12:110348364-110348365:+ | [11] | |
| Sequence | TGGCTCTGCACAGCCCAAAAACCAGCTTACTCCTTAGCCAG | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000539276.7; ENST00000377685.9; ENST00000313432.5; ENST00000308664.10; ENST00000553144.1 | ||
| External Link | RMBase: m6A_site_208208 | ||
| mod ID: M6ASITE015110 | Click to Show/Hide the Full List | ||
| mod site | chr12:110348670-110348671:+ | [11] | |
| Sequence | GCTTGAGCCCAGGTACTCAAACCAGCCTGGGCAACAGAGTG | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000539276.7; ENST00000308664.10; ENST00000377685.9; ENST00000313432.5; ENST00000553144.1; rmsk_3900726 | ||
| External Link | RMBase: m6A_site_208209 | ||
| mod ID: M6ASITE015111 | Click to Show/Hide the Full List | ||
| mod site | chr12:110348791-110348792:+ | [11] | |
| Sequence | GCTGCTTTGCCCAGCAGGGAACATTTGGGGCAGGGGGTAAA | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa; hESC-HEK293T | ||
| Seq Type List | m6A-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000313432.5; ENST00000539276.7; ENST00000553144.1; ENST00000377685.9; ENST00000308664.10 | ||
| External Link | RMBase: m6A_site_208210 | ||
| mod ID: M6ASITE015112 | Click to Show/Hide the Full List | ||
| mod site | chr12:110348841-110348842:+ | [12] | |
| Sequence | TTTGAGCATCATGAGGTGTAACAAGAAATGGGTTGAATGGG | ||
| Motif Score | 2.168095238 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000313432.5; ENST00000539276.7; ENST00000308664.10; ENST00000377685.9; ENST00000553144.1 | ||
| External Link | RMBase: m6A_site_208211 | ||
| mod ID: M6ASITE015113 | Click to Show/Hide the Full List | ||
| mod site | chr12:110348894-110348895:+ | [11] | |
| Sequence | AGTGCATCTCTGGGCTGCAAACTGACTTGAGTGCTGCACTA | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000377685.9; ENST00000313432.5; ENST00000553144.1; ENST00000539276.7; ENST00000308664.10 | ||
| External Link | RMBase: m6A_site_208212 | ||
| mod ID: M6ASITE015114 | Click to Show/Hide the Full List | ||
| mod site | chr12:110348931-110348932:+ | [11] | |
| Sequence | ACTATTGCTATTCCGTGCAAACAAAACTCAGCTTTTCCTGA | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa; hESC-HEK293T | ||
| Seq Type List | m6A-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000308664.10; ENST00000313432.5; ENST00000539276.7; ENST00000553144.1; ENST00000377685.9 | ||
| External Link | RMBase: m6A_site_208213 | ||
| mod ID: M6ASITE015115 | Click to Show/Hide the Full List | ||
| mod site | chr12:110348936-110348937:+ | [11] | |
| Sequence | TGCTATTCCGTGCAAACAAAACTCAGCTTTTCCTGACTCAG | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000308664.10; ENST00000539276.7; ENST00000553144.1; ENST00000377685.9; ENST00000313432.5 | ||
| External Link | RMBase: m6A_site_208214 | ||
| mod ID: M6ASITE015116 | Click to Show/Hide the Full List | ||
| mod site | chr12:110348978-110348979:+ | [12] | |
| Sequence | TCCTTGACTTAGTGGCCTTTACAAAAAAAGTTGAGTAGTGT | ||
| Motif Score | 2.07285119 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000539276.7; ENST00000377685.9; ENST00000553144.1; ENST00000308664.10; ENST00000313432.5 | ||
| External Link | RMBase: m6A_site_208215 | ||
| mod ID: M6ASITE015117 | Click to Show/Hide the Full List | ||
| mod site | chr12:110349051-110349052:+ | [11] | |
| Sequence | TCATGGTTTCTCAGCTTCAGACCCCTCCAGCCCACAGAGGA | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000308664.10; ENST00000377685.9; ENST00000313432.5; ENST00000553144.1; ENST00000539276.7 | ||
| External Link | RMBase: m6A_site_208216 | ||
| mod ID: M6ASITE015118 | Click to Show/Hide the Full List | ||
| mod site | chr12:110349084-110349085:+ | [11] | |
| Sequence | ACAGAGGAGCCCATGGAGGGACCCACTTCCCTTGGTCCAGA | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000313432.5; ENST00000553144.1; ENST00000308664.10; ENST00000539276.7; ENST00000377685.9 | ||
| External Link | RMBase: m6A_site_208217 | ||
| mod ID: M6ASITE015119 | Click to Show/Hide the Full List | ||
| mod site | chr12:110349104-110349105:+ | [11] | |
| Sequence | ACCCACTTCCCTTGGTCCAGACAGCTGGGAGTGGGTTAGGC | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000539276.7; ENST00000377685.9; ENST00000313432.5; ENST00000308664.10; ENST00000553144.1 | ||
| External Link | RMBase: m6A_site_208218 | ||
| mod ID: M6ASITE015120 | Click to Show/Hide the Full List | ||
| mod site | chr12:110349127-110349128:+ | [13] | |
| Sequence | GCTGGGAGTGGGTTAGGCCCACTGCTGTTTTGAGCAGGGCC | ||
| Motif Score | 2.475107143 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000553144.1; ENST00000313432.5; ENST00000308664.10; ENST00000539276.7; ENST00000377685.9 | ||
| External Link | RMBase: m6A_site_208219 | ||
| mod ID: M6ASITE015121 | Click to Show/Hide the Full List | ||
| mod site | chr12:110349185-110349186:+ | [11] | |
| Sequence | GAAGGCTTTGCTGGGTGAAAACACTTCAGCATCTCCTCCTC | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa; hESC-HEK293T | ||
| Seq Type List | m6A-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000553144.1; ENST00000313432.5; ENST00000308664.10; ENST00000377685.9; ENST00000539276.7 | ||
| External Link | RMBase: m6A_site_208220 | ||
| mod ID: M6ASITE015122 | Click to Show/Hide the Full List | ||
| mod site | chr12:110349222-110349223:+ | [11] | |
| Sequence | CCTCAGGTCAACCCATAAAGACCAGGTCCAGCACCGTGGTC | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000539276.7; ENST00000553144.1; ENST00000377685.9; ENST00000308664.10; ENST00000313432.5 | ||
| External Link | RMBase: m6A_site_208221 | ||
| mod ID: M6ASITE015123 | Click to Show/Hide the Full List | ||
| mod site | chr12:110349248-110349249:+ | [12] | |
| Sequence | TCCAGCACCGTGGTCTTGGCACATCCCTGGCCTCAGGCCCT | ||
| Motif Score | 2.830589286 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000313432.5; ENST00000539276.7; ENST00000308664.10; ENST00000377685.9; ENST00000553144.1 | ||
| External Link | RMBase: m6A_site_208222 | ||
| mod ID: M6ASITE015124 | Click to Show/Hide the Full List | ||
| mod site | chr12:110349337-110349338:+ | [11] | |
| Sequence | TCAGGCAGCTCTTGCCTGAAACTTACTTCCACATTCTTTCC | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000377685.9; ENST00000553144.1; ENST00000308664.10; ENST00000539276.7; ENST00000313432.5 | ||
| External Link | RMBase: m6A_site_208223 | ||
| mod ID: M6ASITE015125 | Click to Show/Hide the Full List | ||
| mod site | chr12:110349347-110349348:+ | [12] | |
| Sequence | CTTGCCTGAAACTTACTTCCACATTCTTTCCTGATGGGCAG | ||
| Motif Score | 2.053113095 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000539276.7; ENST00000308664.10; ENST00000377685.9; ENST00000313432.5; ENST00000553144.1 | ||
| External Link | RMBase: m6A_site_208224 | ||
| mod ID: M6ASITE015126 | Click to Show/Hide the Full List | ||
| mod site | chr12:110349450-110349451:+ | [12] | |
| Sequence | GAGCTTTGCCCAGAAGACCAACAATCATACATACCCTAACT | ||
| Motif Score | 2.173910714 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000553144.1; ENST00000539276.7; ENST00000377685.9; ENST00000308664.10; ENST00000313432.5 | ||
| External Link | RMBase: m6A_site_208225 | ||
| mod ID: M6ASITE015127 | Click to Show/Hide the Full List | ||
| mod site | chr12:110349474-110349475:+ | [12] | |
| Sequence | TCATACATACCCTAACTGGGACACCACTCTGCAGAATGCAG | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000313432.5; ENST00000377685.9; ENST00000553144.1; ENST00000539276.7; ENST00000308664.10 | ||
| External Link | RMBase: m6A_site_208226 | ||
| mod ID: M6ASITE015128 | Click to Show/Hide the Full List | ||
| mod site | chr12:110349572-110349573:+ | [12] | |
| Sequence | GCATCTGGCCGACTTCCCTCACAACAGCTGCTCCCACATCC | ||
| Motif Score | 2.047297619 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000377685.9; ENST00000539276.7; ENST00000308664.10; ENST00000313432.5; ENST00000553144.1 | ||
| External Link | RMBase: m6A_site_208227 | ||
| mod ID: M6ASITE015129 | Click to Show/Hide the Full List | ||
| mod site | chr12:110349587-110349588:+ | [12] | |
| Sequence | CCCTCACAACAGCTGCTCCCACATCCCCTCGGACTGGAGCT | ||
| Motif Score | 2.053113095 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000377685.9; ENST00000308664.10; ENST00000553144.1; ENST00000539276.7; ENST00000313432.5 | ||
| External Link | RMBase: m6A_site_208228 | ||
| mod ID: M6ASITE015130 | Click to Show/Hide the Full List | ||
| mod site | chr12:110349648-110349649:+ | [14] | |
| Sequence | CAGACCTAAGACCTGAGACCACAAGATTAGCTCAGTGTCTA | ||
| Motif Score | 2.053113095 | ||
| Cell/Tissue List | brain; hESC-HEK293T | ||
| Seq Type List | m6A-REF-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000313432.5; ENST00000308664.10; ENST00000539276.7; ENST00000553144.1; ENST00000377685.9 | ||
| External Link | RMBase: m6A_site_208229 | ||
| mod ID: M6ASITE015131 | Click to Show/Hide the Full List | ||
| mod site | chr12:110349771-110349772:+ | [12] | |
| Sequence | GTAACCACCAAATTGTCCAAACACCCGCTGCAGTTAGCAAG | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000313432.5; ENST00000539276.7; ENST00000377685.9; ENST00000308664.10; ENST00000553144.1 | ||
| External Link | RMBase: m6A_site_208230 | ||
| mod ID: M6ASITE015132 | Click to Show/Hide the Full List | ||
| mod site | chr12:110349902-110349903:+ | [11] | |
| Sequence | CGGGTGCCCACAGGGCAGGGACAGGAAGGCTGTGCTGCTAC | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000553144.1; ENST00000377685.9; ENST00000313432.5; ENST00000539276.7; ENST00000308664.10 | ||
| External Link | RMBase: m6A_site_208231 | ||
| mod ID: M6ASITE015133 | Click to Show/Hide the Full List | ||
| mod site | chr12:110349971-110349972:+ | [11] | |
| Sequence | CCTCTGCACCACTAACCAAGACCTTGTCCTCTTGCCTGTCT | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000308664.10; ENST00000539276.7; ENST00000313432.5; ENST00000553144.1; ENST00000377685.9 | ||
| External Link | RMBase: m6A_site_208232 | ||
| mod ID: M6ASITE015134 | Click to Show/Hide the Full List | ||
| mod site | chr12:110350002-110350003:+ | [14] | |
| Sequence | TTGCCTGTCTCGCTGCTTTCACAGCTGCAACGATTGTGTCT | ||
| Motif Score | 2.047297619 | ||
| Cell/Tissue List | kidney; liver | ||
| Seq Type List | m6A-REF-seq | ||
| Transcript ID List | ENST00000377685.9; ENST00000308664.10; ENST00000553144.1; ENST00000313432.5; ENST00000539276.7 | ||
| External Link | RMBase: m6A_site_208233 | ||
| mod ID: M6ASITE015135 | Click to Show/Hide the Full List | ||
| mod site | chr12:110350135-110350136:+ | [13] | |
| Sequence | GTGTGGGCGGCACCTCAGGGACAGTAAATCAGAAATGCTGG | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000539276.7; ENST00000308664.10; ENST00000313432.5; ENST00000553144.1; ENST00000377685.9 | ||
| External Link | RMBase: m6A_site_208234 | ||
| mod ID: M6ASITE015136 | Click to Show/Hide the Full List | ||
| mod site | chr12:110350163-110350164:+ | [13] | |
| Sequence | TCAGAAATGCTGGTCTTGAAACCTTGAAAAGATCAAGCTGA | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000539276.7; ENST00000553144.1; ENST00000308664.10; ENST00000313432.5; ENST00000377685.9 | ||
| External Link | RMBase: m6A_site_208235 | ||
| mod ID: M6ASITE015137 | Click to Show/Hide the Full List | ||
| mod site | chr12:110350316-110350317:+ | [13] | |
| Sequence | CTGGAGTAACCGCTTCCTAAACCATTTTGCAGAAATGTAAG | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000539276.7; ENST00000553144.1; ENST00000377685.9; ENST00000313432.5; ENST00000308664.10 | ||
| External Link | RMBase: m6A_site_208236 | ||
| mod ID: M6ASITE015138 | Click to Show/Hide the Full List | ||
| mod site | chr12:110350394-110350395:+ | [13] | |
| Sequence | ATCTACCAACCCTGTGCATGACTGATGTTGGGGAAAAAGAA | ||
| Motif Score | 3.28175 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000539276.7; ENST00000308664.10; ENST00000313432.5; ENST00000377685.9 | ||
| External Link | RMBase: m6A_site_208237 | ||
| mod ID: M6ASITE015139 | Click to Show/Hide the Full List | ||
| mod site | chr12:110350466-110350467:+ | [13] | |
| Sequence | ATGTGGAGGAAATGTGTATTACCAATGGGGTTGTTAGCTTT | ||
| Motif Score | 2.052208333 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000539276.7; ENST00000308664.10; ENST00000313432.5; ENST00000377685.9 | ||
| External Link | RMBase: m6A_site_208238 | ||
| mod ID: M6ASITE015140 | Click to Show/Hide the Full List | ||
| mod site | chr12:110350498-110350499:+ | [13] | |
| Sequence | GTTAGCTTTTAAATCAAAATACTGATTACAGATGTACAATT | ||
| Motif Score | 2.53247619 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000539276.7; ENST00000377685.9; ENST00000308664.10; ENST00000313432.5 | ||
| External Link | RMBase: m6A_site_208239 | ||
| mod ID: M6ASITE015141 | Click to Show/Hide the Full List | ||
| mod site | chr12:110350505-110350506:+ | [13] | |
| Sequence | TTTAAATCAAAATACTGATTACAGATGTACAATTTAGCTTA | ||
| Motif Score | 2.07285119 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000313432.5; ENST00000308664.10; ENST00000539276.7; ENST00000377685.9 | ||
| External Link | RMBase: m6A_site_208240 | ||
| mod ID: M6ASITE015142 | Click to Show/Hide the Full List | ||
| mod site | chr12:110350513-110350514:+ | [13] | |
| Sequence | AAAATACTGATTACAGATGTACAATTTAGCTTAATCAGAAA | ||
| Motif Score | 2.856142857 | ||
| Cell/Tissue List | HEK293T; hESC-HEK293T | ||
| Seq Type List | DART-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000313432.5; ENST00000308664.10; ENST00000539276.7; ENST00000377685.9 | ||
| External Link | RMBase: m6A_site_208241 | ||
| mod ID: M6ASITE015143 | Click to Show/Hide the Full List | ||
| mod site | chr12:110350625-110350626:+ | [12] | |
| Sequence | AAGCCGTCAGCCAAGTCGCCACATTTCTCTGCAAAATGTCA | ||
| Motif Score | 2.053113095 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000313432.5; ENST00000539276.7; ENST00000308664.10; ENST00000377685.9 | ||
| External Link | RMBase: m6A_site_208242 | ||
| mod ID: M6ASITE015144 | Click to Show/Hide the Full List | ||
| mod site | chr12:110350720-110350721:+ | [13] | |
| Sequence | AGCCAGATCCCTCTCCAGTGACATTGGAACATGCTACTTTT | ||
| Motif Score | 2.859755952 | ||
| Cell/Tissue List | HEK293T; hESC-HEK293T | ||
| Seq Type List | DART-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000308664.10; ENST00000539276.7; ENST00000377685.9; ENST00000313432.5 | ||
| External Link | RMBase: m6A_site_208243 | ||
| mod ID: M6ASITE015145 | Click to Show/Hide the Full List | ||
| mod site | chr12:110350754-110350755:+ | [14] | |
| Sequence | TACTTTTTAATTGGCCCTGTACAGTTTGCTTATTTATAAAT | ||
| Motif Score | 2.856142857 | ||
| Cell/Tissue List | brain | ||
| Seq Type List | m6A-REF-seq | ||
| Transcript ID List | ENST00000539276.7; ENST00000308664.10; ENST00000313432.5; ENST00000377685.9 | ||
| External Link | RMBase: m6A_site_208244 | ||
| mod ID: M6ASITE015146 | Click to Show/Hide the Full List | ||
| mod site | chr12:110350853-110350854:+ | [14] | |
| Sequence | GTTATTTTCTGAGTGTGCAGACAGCTATTTCGCACTGTATT | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | kidney | ||
| Seq Type List | m6A-REF-seq | ||
| Transcript ID List | ENST00000539276.7; ENST00000313432.5; ENST00000308664.10; ENST00000377685.9 | ||
| External Link | RMBase: m6A_site_208245 | ||
| mod ID: M6ASITE015147 | Click to Show/Hide the Full List | ||
| mod site | chr12:110350909-110350910:+ | [11] | |
| Sequence | AATGAAATCAGAAGCAGTAGACAGATGTTGGTGCAATACAA | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa; HEK293T | ||
| Seq Type List | m6A-seq; DART-seq | ||
| Transcript ID List | ENST00000308664.10; ENST00000313432.5; ENST00000377685.9; ENST00000539276.7 | ||
| External Link | RMBase: m6A_site_208246 | ||
| mod ID: M6ASITE015148 | Click to Show/Hide the Full List | ||
| mod site | chr12:110350926-110350927:+ | [13] | |
| Sequence | TAGACAGATGTTGGTGCAATACAAATATTGTGATGCATTTA | ||
| Motif Score | 2.110482143 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000308664.10; ENST00000539276.7; ENST00000377685.9; ENST00000313432.5 | ||
| External Link | RMBase: m6A_site_208247 | ||
| mod ID: M6ASITE015149 | Click to Show/Hide the Full List | ||
| mod site | chr12:110350991-110350992:+ | [13] | |
| Sequence | TTTATCACTGCGCATGTTTGACTTTAGACTGTAAATAGAGA | ||
| Motif Score | 3.28175 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000313432.5; ENST00000308664.10; ENST00000377685.9; ENST00000539276.7 | ||
| External Link | RMBase: m6A_site_208248 | ||
| mod ID: M6ASITE015150 | Click to Show/Hide the Full List | ||
| mod site | chr12:110350998-110350999:+ | [13] | |
| Sequence | CTGCGCATGTTTGACTTTAGACTGTAAATAGAGATCAGTTT | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000377685.9; ENST00000539276.7; ENST00000308664.10; ENST00000313432.5 | ||
| External Link | RMBase: m6A_site_208249 | ||
| mod ID: M6ASITE015151 | Click to Show/Hide the Full List | ||
| mod site | chr12:110351038-110351039:+ | [13] | |
| Sequence | TGTTTCTTTCTGTGCTGGTAACAATGAGCGTCGCACAGACA | ||
| Motif Score | 2.168095238 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000377685.9; ENST00000313432.5; ENST00000308664.10; ENST00000539276.7 | ||
| External Link | RMBase: m6A_site_208250 | ||
References