m6A Target Gene Information
General Information of the m6A Target Gene (ID: M6ATAR00674)
Full List of m6A Methylation Regulator of This Target Gene and Corresponding Disease/Drug Response(s)
TRAF4
can be regulated by the following regulator(s), and cause disease/drug response(s). You can browse detail information of regulator(s) or disease/drug response(s).
Browse Regulator
Browse Disease
YTH domain-containing family protein 1 (YTHDF1) [READER]
| Representative RNA-seq result indicating the expression of this target gene regulated by YTHDF1 | ||
| Cell Line | AGS cell line | Homo sapiens |
|
Treatment: shYTHDF1 AGS
Control: shControl AGS
|
GSE159425 | |
| Regulation |
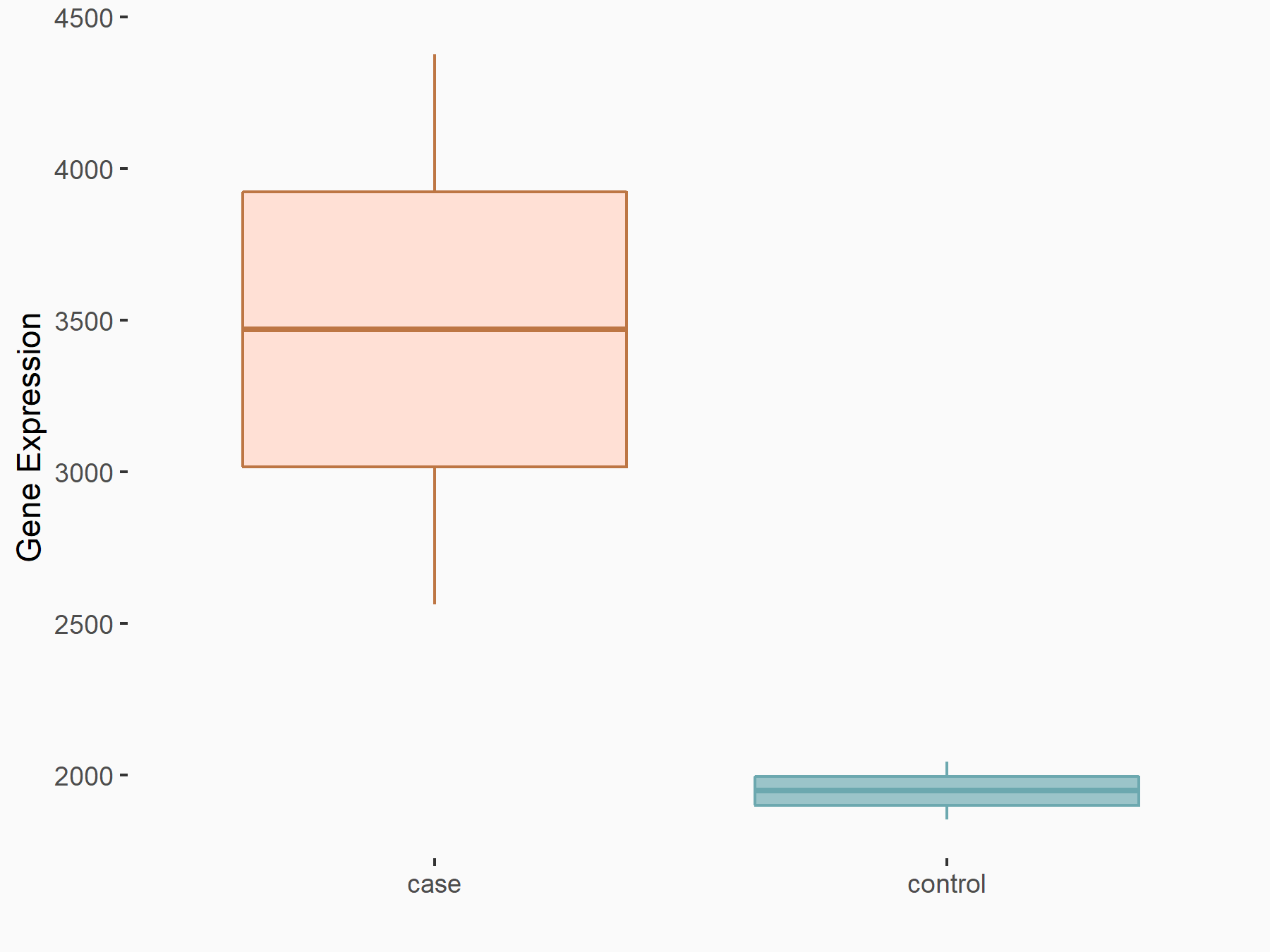 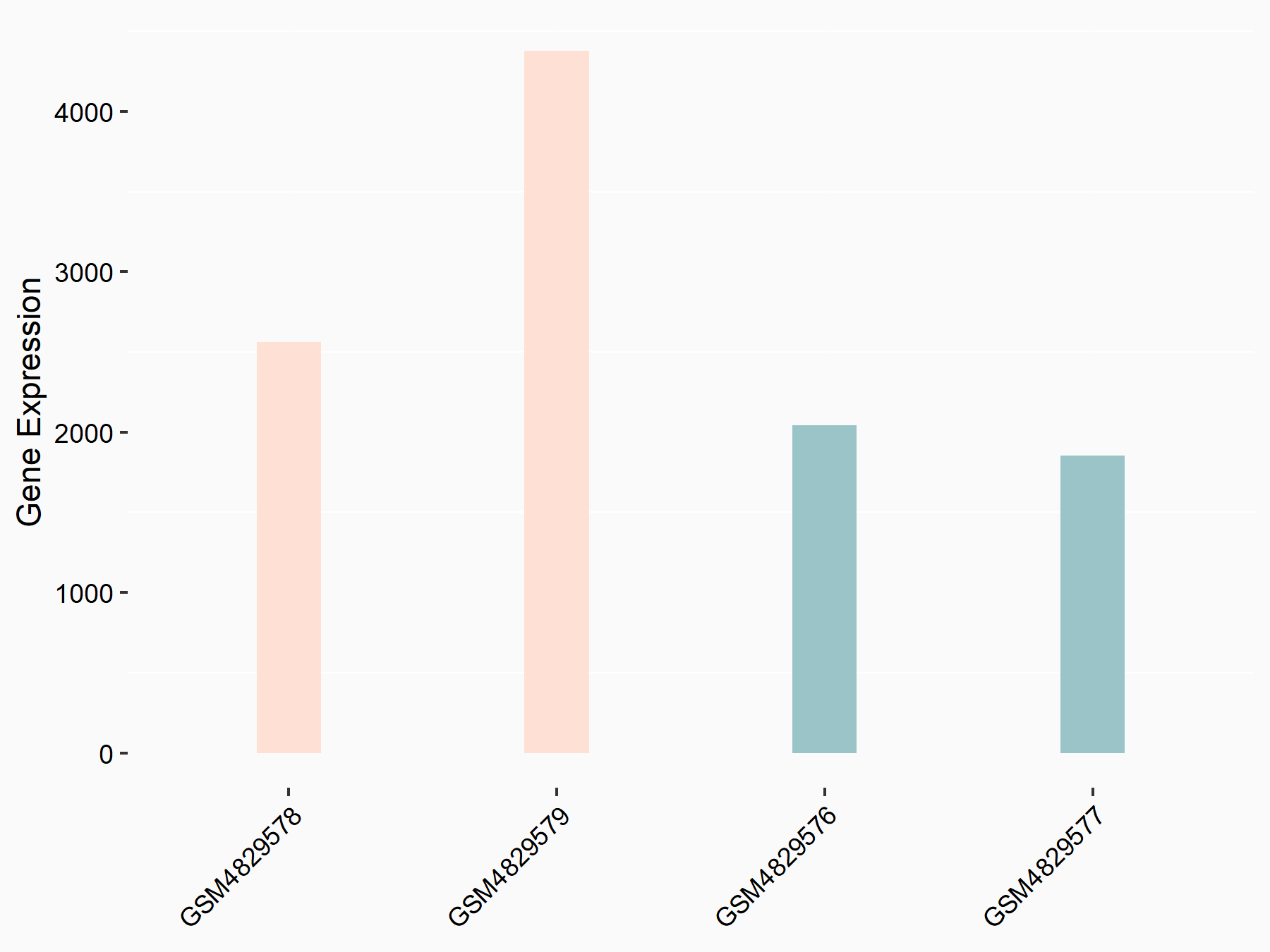 |
logFC: 8.33E-01 p-value: 9.45E-04 |
| More Results | Click to View More RNA-seq Results | |
| Representative RIP-seq result supporting the interaction between TRAF4 and the regulator | ||
| Cell Line | Hela | Homo sapiens |
| Regulation | logFC: 1.38E+00 | GSE63591 |
| In total 1 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [1] | |||
| Response Summary | m6A-dependent TNF receptor-associated factor 4 (TRAF4) expression upregulation by ALKBH5 and YTHDF1 contributes to curcumin-induced obesity prevention. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Obesity | ICD-11: 5B81 | ||
| Pathway Response | Ubiquitin mediated proteolysis | hsa04120), Proteasome | ||
| Cell Process | Ubiquitination degradation | |||
| In-vitro Model | SVF (Stromal vascular cell fraction (SVF) was isolated from minced inguinal adipose tissue of male C57BL/6J mice (3-4 weeks old)) | |||
| 3T3-L1 | Normal | Mus musculus | CVCL_0123 | |
| In-vivo Model | Before the tests, animals were fasted for 8 h. l glucose tolerance test (GTT) was conducted during week 11 on the diet. The mice were challenged with 2 g/kg body weight d-glucose (Sigma-Aldrich, USA). Insulin tolerance test (ITT) was conducted during week 12 on the diet. For insulin tolerance test, mice were injected intraperitoneally with 0.75 U/kg body weight insulin (Sigma-Aldrich, USA). For both tests, blood samples were taken from the tail vein and glucose levels were measured at indicated time points after administration using an AlphaTRAK glucometer. | |||
RNA demethylase ALKBH5 (ALKBH5) [ERASER]
| Representative RNA-seq result indicating the expression of this target gene regulated by ALKBH5 | ||
| Cell Line | 143B cell line | Homo sapiens |
|
Treatment: siALKBH5 transfected 143B cells
Control: siControl 143B cells
|
GSE154528 | |
| Regulation |
  |
logFC: -6.18E-01 p-value: 8.54E-03 |
| More Results | Click to View More RNA-seq Results | |
| In total 1 item(s) under this regulator | ||||
| Experiment 1 Reporting the m6A Methylation Regulator of This Target Gene | [1] | |||
| Response Summary | m6A-dependent TNF receptor-associated factor 4 (TRAF4) expression upregulation by ALKBH5 and YTHDF1 contributes to curcumin-induced obesity prevention. | |||
| Target Regulation | Up regulation | |||
| Responsed Disease | Obesity | ICD-11: 5B81 | ||
| Pathway Response | Ubiquitin mediated proteolysis | hsa04120), Proteasome | ||
| Cell Process | Ubiquitination degradation | |||
| In-vitro Model | SVF (Stromal vascular cell fraction (SVF) was isolated from minced inguinal adipose tissue of male C57BL/6J mice (3-4 weeks old)) | |||
| 3T3-L1 | Normal | Mus musculus | CVCL_0123 | |
| In-vivo Model | Before the tests, animals were fasted for 8 h. l glucose tolerance test (GTT) was conducted during week 11 on the diet. The mice were challenged with 2 g/kg body weight d-glucose (Sigma-Aldrich, USA). Insulin tolerance test (ITT) was conducted during week 12 on the diet. For insulin tolerance test, mice were injected intraperitoneally with 0.75 U/kg body weight insulin (Sigma-Aldrich, USA). For both tests, blood samples were taken from the tail vein and glucose levels were measured at indicated time points after administration using an AlphaTRAK glucometer. | |||
Obesity [ICD-11: 5B81]
| In total 2 item(s) under this disease | ||||
| Experiment 1 Reporting the m6A-centered Disease Response | [1] | |||
| Response Summary | m6A-dependent TNF receptor-associated factor 4 (TRAF4) expression upregulation by ALKBH5 and YTHDF1 contributes to curcumin-induced obesity prevention. | |||
| Responsed Disease | Obesity [ICD-11: 5B81] | |||
| Target Regulator | RNA demethylase ALKBH5 (ALKBH5) | ERASER | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Ubiquitin mediated proteolysis | hsa04120), Proteasome | ||
| Cell Process | Ubiquitination degradation | |||
| In-vitro Model | SVF (Stromal vascular cell fraction (SVF) was isolated from minced inguinal adipose tissue of male C57BL/6J mice (3-4 weeks old)) | |||
| 3T3-L1 | Normal | Mus musculus | CVCL_0123 | |
| In-vivo Model | Before the tests, animals were fasted for 8 h. l glucose tolerance test (GTT) was conducted during week 11 on the diet. The mice were challenged with 2 g/kg body weight d-glucose (Sigma-Aldrich, USA). Insulin tolerance test (ITT) was conducted during week 12 on the diet. For insulin tolerance test, mice were injected intraperitoneally with 0.75 U/kg body weight insulin (Sigma-Aldrich, USA). For both tests, blood samples were taken from the tail vein and glucose levels were measured at indicated time points after administration using an AlphaTRAK glucometer. | |||
| Experiment 2 Reporting the m6A-centered Disease Response | [1] | |||
| Response Summary | m6A-dependent TNF receptor-associated factor 4 (TRAF4) expression upregulation by ALKBH5 and YTHDF1 contributes to curcumin-induced obesity prevention. | |||
| Responsed Disease | Obesity [ICD-11: 5B81] | |||
| Target Regulator | YTH domain-containing family protein 1 (YTHDF1) | READER | ||
| Target Regulation | Up regulation | |||
| Pathway Response | Ubiquitin mediated proteolysis | hsa04120), Proteasome | ||
| Cell Process | Ubiquitination degradation | |||
| In-vitro Model | SVF (Stromal vascular cell fraction (SVF) was isolated from minced inguinal adipose tissue of male C57BL/6J mice (3-4 weeks old)) | |||
| 3T3-L1 | Normal | Mus musculus | CVCL_0123 | |
| In-vivo Model | Before the tests, animals were fasted for 8 h. l glucose tolerance test (GTT) was conducted during week 11 on the diet. The mice were challenged with 2 g/kg body weight d-glucose (Sigma-Aldrich, USA). Insulin tolerance test (ITT) was conducted during week 12 on the diet. For insulin tolerance test, mice were injected intraperitoneally with 0.75 U/kg body weight insulin (Sigma-Aldrich, USA). For both tests, blood samples were taken from the tail vein and glucose levels were measured at indicated time points after administration using an AlphaTRAK glucometer. | |||
RNA Modification Sequencing Data Associated with the Target (ID: M6ATAR00674)
| In total 3 m6A sequence/site(s) in this target gene | |||
| mod ID: A2ISITE007787 | Click to Show/Hide the Full List | ||
| mod site | chr17:28745817-28745818:+ | [2] | |
| Sequence | ACAGCCCTGGCTGGGCCCCAAGCTGCAGGCTAAGCTTCTGC | ||
| Transcript ID List | ENST00000618771.1; ENST00000461195.5; ENST00000586813.5; ENST00000262396.10; ENST00000475329.5; ENST00000262395.10; ENST00000498540.2; ENST00000394925.5; ENST00000422344.5; ENST00000444415.7 | ||
| External Link | RMBase: RNA-editing_site_56014 | ||
| mod ID: A2ISITE007788 | Click to Show/Hide the Full List | ||
| mod site | chr17:28745847-28745848:+ | [2] | |
| Sequence | TAAGCTTCTGCCTGAAGCCTAGGGACTTGCCTCTGTCCCCT | ||
| Transcript ID List | ENST00000618771.1; ENST00000422344.5; ENST00000498540.2; ENST00000262396.10; ENST00000586813.5; ENST00000475329.5; ENST00000444415.7; ENST00000262395.10; ENST00000394925.5; ENST00000461195.5 | ||
| External Link | RMBase: RNA-editing_site_56015 | ||
| mod ID: A2ISITE007789 | Click to Show/Hide the Full List | ||
| mod site | chr17:28746873-28746874:+ | [2] | |
| Sequence | CAGCGCAGCCAATGAAAACTAACCTAGGGGGAGGGGGTGAC | ||
| Transcript ID List | ENST00000475329.5; ENST00000262395.10; ENST00000262396.10; ENST00000394925.5; ENST00000584944.5; ENST00000498540.2; ENST00000422344.5; ENST00000444415.7; ENST00000461195.5; ENST00000586813.5; ENST00000618771.1 | ||
| External Link | RMBase: RNA-editing_site_56016 | ||
N1-methyladenosine (m1A)
| In total 2 m6A sequence/site(s) in this target gene | |||
| mod ID: M1ASITE000052 | Click to Show/Hide the Full List | ||
| mod site | chr17:28748367-28748368:+ | [3] | |
| Sequence | TGCCACCTCTGAGTGCCCCAAGCGCACTCAGCCCTGCACCT | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | m1A-MAP-seq; m1A-seq | ||
| Transcript ID List | ENST00000586813.5; ENST00000262396.10; ENST00000580073.1; ENST00000454852.6; ENST00000618771.1; ENST00000473421.1; ENST00000262395.10; ENST00000444415.7; ENST00000461195.5; ENST00000469529.6 | ||
| External Link | RMBase: m1A_site_444 | ||
| mod ID: M1ASITE000053 | Click to Show/Hide the Full List | ||
| mod site | chr17:28749735-28749736:+ | [4] | |
| Sequence | ACCACCCTCAGGTGCCTCCAATTGGTGCTTCAGCCCTGGCC | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | m1A-seq | ||
| Transcript ID List | ENST00000262395.10; ENST00000444415.7 | ||
| External Link | RMBase: m1A_site_445 | ||
5-methylcytidine (m5C)
| In total 12 m6A sequence/site(s) in this target gene | |||
| mod ID: M5CSITE001018 | Click to Show/Hide the Full List | ||
| mod site | chr17:28744013-28744014:+ | [5] | |
| Sequence | CCCTGGCCGCGACCGCCAGTCGGCGCCGCCCGGAGCCGGGA | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000262395.10; ENST00000618771.1 | ||
| External Link | RMBase: m5C_site_18361 | ||
| mod ID: M5CSITE001019 | Click to Show/Hide the Full List | ||
| mod site | chr17:28744016-28744017:+ | [5] | |
| Sequence | TGGCCGCGACCGCCAGTCGGCGCCGCCCGGAGCCGGGAGCG | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000618771.1; ENST00000262395.10 | ||
| External Link | RMBase: m5C_site_18362 | ||
| mod ID: M5CSITE001020 | Click to Show/Hide the Full List | ||
| mod site | chr17:28744018-28744019:+ | [5] | |
| Sequence | GCCGCGACCGCCAGTCGGCGCCGCCCGGAGCCGGGAGCGCC | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000262395.10; ENST00000461195.5; ENST00000618771.1; ENST00000478021.1 | ||
| External Link | RMBase: m5C_site_18363 | ||
| mod ID: M5CSITE001021 | Click to Show/Hide the Full List | ||
| mod site | chr17:28744065-28744066:+ | [6] | |
| Sequence | GCGAGGCGCGGGCTGTGGGGCCGCCGCGTGCCTGGCCCCGC | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000262395.10; ENST00000618771.1; ENST00000394925.5; ENST00000461195.5; ENST00000478021.1; ENST00000422344.5; ENST00000444415.7; ENST00000498540.2; ENST00000586813.5 | ||
| External Link | RMBase: m5C_site_18364 | ||
| mod ID: M5CSITE001022 | Click to Show/Hide the Full List | ||
| mod site | chr17:28744066-28744067:+ | [6] | |
| Sequence | CGAGGCGCGGGCTGTGGGGCCGCCGCGTGCCTGGCCCCGCT | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000262395.10; ENST00000586813.5; ENST00000394925.5; ENST00000478021.1; ENST00000444415.7; ENST00000618771.1; ENST00000498540.2; ENST00000461195.5; ENST00000422344.5 | ||
| External Link | RMBase: m5C_site_18365 | ||
| mod ID: M5CSITE001023 | Click to Show/Hide the Full List | ||
| mod site | chr17:28744080-28744081:+ | [6] | |
| Sequence | TGGGGCCGCCGCGTGCCTGGCCCCGCTCGCCCGTGCCGGCC | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000262395.10; ENST00000394925.5; ENST00000618771.1; ENST00000461195.5; ENST00000498540.2; ENST00000422344.5; ENST00000478021.1; ENST00000444415.7; ENST00000586813.5 | ||
| External Link | RMBase: m5C_site_18366 | ||
| mod ID: M5CSITE001024 | Click to Show/Hide the Full List | ||
| mod site | chr17:28744081-28744082:+ | [6] | |
| Sequence | GGGGCCGCCGCGTGCCTGGCCCCGCTCGCCCGTGCCGGCCG | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000461195.5; ENST00000422344.5; ENST00000394925.5; ENST00000444415.7; ENST00000478021.1; ENST00000586813.5; ENST00000262395.10; ENST00000618771.1; rmsk_4664083; ENST00000498540.2 | ||
| External Link | RMBase: m5C_site_18367 | ||
| mod ID: M5CSITE001025 | Click to Show/Hide the Full List | ||
| mod site | chr17:28744082-28744083:+ | [6] | |
| Sequence | GGGCCGCCGCGTGCCTGGCCCCGCTCGCCCGTGCCGGCCGC | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000262395.10; ENST00000478021.1; ENST00000394925.5; ENST00000461195.5; ENST00000618771.1; ENST00000498540.2; ENST00000422344.5; ENST00000586813.5; ENST00000444415.7; rmsk_4664083 | ||
| External Link | RMBase: m5C_site_18368 | ||
| mod ID: M5CSITE001026 | Click to Show/Hide the Full List | ||
| mod site | chr17:28744083-28744084:+ | [6] | |
| Sequence | GGCCGCCGCGTGCCTGGCCCCGCTCGCCCGTGCCGGCCGCT | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000478021.1; ENST00000394925.5; ENST00000586813.5; rmsk_4664083; ENST00000461195.5; ENST00000618771.1; ENST00000262395.10; ENST00000422344.5; ENST00000498540.2; ENST00000444415.7 | ||
| External Link | RMBase: m5C_site_18369 | ||
| mod ID: M5CSITE001027 | Click to Show/Hide the Full List | ||
| mod site | chr17:28745666-28745667:+ | [5] | |
| Sequence | CTCCACTGCACAGCCTGGAGCAGCCTCTGCCCTGCCTGTCC | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000444415.7; ENST00000262395.10; ENST00000475329.5; ENST00000618771.1; ENST00000262396.10; ENST00000461195.5; ENST00000422344.5; ENST00000394925.5; ENST00000586813.5; ENST00000498540.2 | ||
| External Link | RMBase: m5C_site_18370 | ||
| mod ID: M5CSITE001028 | Click to Show/Hide the Full List | ||
| mod site | chr17:28747240-28747241:+ | [5] | |
| Sequence | TCTTCAAGTGCCCTGAGGACCAGCTTCCTCTGGACTATGCC | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000498540.2; ENST00000586813.5; ENST00000422344.5; ENST00000444415.7; ENST00000262395.10; ENST00000473421.1; ENST00000618771.1; ENST00000394925.5; ENST00000584944.5; ENST00000262396.10; ENST00000454852.6; ENST00000461195.5; ENST00000475329.5 | ||
| External Link | RMBase: m5C_site_18371 | ||
| mod ID: M5CSITE001029 | Click to Show/Hide the Full List | ||
| mod site | chr17:28748938-28748939:+ | [5] | |
| Sequence | CCCATGGCCTGGGGCTTTGCCAACAGTGCCCTAAGCTGGCA | ||
| Seq Type List | Bisulfite-seq | ||
| Transcript ID List | ENST00000454852.6; ENST00000262395.10; ENST00000618771.1; ENST00000444415.7; ENST00000262396.10; ENST00000469529.6 | ||
| External Link | RMBase: m5C_site_18372 | ||
N6-methyladenosine (m6A)
| In total 48 m6A sequence/site(s) in this target gene | |||
| mod ID: M6ASITE031151 | Click to Show/Hide the Full List | ||
| mod site | chr17:28744128-28744129:+ | [7] | |
| Sequence | CGCCATGCCTGGCTTCGACTACAAGTTCCTGGAGAAGCCCA | ||
| Motif Score | 2.078666667 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000262396.10; ENST00000586813.5; ENST00000618771.1; ENST00000444415.7; ENST00000461195.5; ENST00000422344.5; ENST00000394925.5; ENST00000262395.10; ENST00000498540.2; ENST00000478021.1 | ||
| External Link | RMBase: m6A_site_354914 | ||
| mod ID: M6ASITE031152 | Click to Show/Hide the Full List | ||
| mod site | chr17:28745387-28745388:+ | [8] | |
| Sequence | ACACATTTACTCTCCTACAGACACAGCCCCATTGTCTCCGC | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa; MT4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000461195.5; ENST00000478021.1; ENST00000262395.10; ENST00000475329.5; ENST00000422344.5; ENST00000586813.5; ENST00000444415.7; ENST00000618771.1; ENST00000394925.5; ENST00000498540.2; ENST00000262396.10 | ||
| External Link | RMBase: m6A_site_354915 | ||
| mod ID: M6ASITE031153 | Click to Show/Hide the Full List | ||
| mod site | chr17:28746782-28746783:+ | [8] | |
| Sequence | GCCCTGGTTCTTGGCAGAGGACCTTCCTGGGGACTGGGCAG | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000475329.5; ENST00000444415.7; ENST00000262395.10; ENST00000618771.1; ENST00000584944.5; ENST00000422344.5; ENST00000586813.5; ENST00000394925.5; ENST00000461195.5; ENST00000262396.10; ENST00000498540.2 | ||
| External Link | RMBase: m6A_site_354916 | ||
| mod ID: M6ASITE031154 | Click to Show/Hide the Full List | ||
| mod site | chr17:28746794-28746795:+ | [8] | |
| Sequence | GGCAGAGGACCTTCCTGGGGACTGGGCAGCCTTCGCACTTA | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000394925.5; ENST00000586813.5; ENST00000584944.5; ENST00000618771.1; ENST00000262395.10; ENST00000262396.10; ENST00000422344.5; ENST00000444415.7; ENST00000461195.5; ENST00000498540.2; ENST00000475329.5 | ||
| External Link | RMBase: m6A_site_354917 | ||
| mod ID: M6ASITE031155 | Click to Show/Hide the Full List | ||
| mod site | chr17:28746870-28746871:+ | [8] | |
| Sequence | TTTCAGCGCAGCCAATGAAAACTAACCTAGGGGGAGGGGGT | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000586813.5; ENST00000618771.1; ENST00000262395.10; ENST00000444415.7; ENST00000262396.10; ENST00000422344.5; ENST00000584944.5; ENST00000461195.5; ENST00000498540.2; ENST00000475329.5; ENST00000394925.5 | ||
| External Link | RMBase: m6A_site_354918 | ||
| mod ID: M6ASITE031156 | Click to Show/Hide the Full List | ||
| mod site | chr17:28747536-28747537:+ | [9] | |
| Sequence | CCCTTCCGGTGGCAAGCCAGACCTGTGGCTGAGGGCCAGTA | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HepG2 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000262396.10; ENST00000473421.1; ENST00000444415.7; ENST00000461195.5; ENST00000262395.10; ENST00000394925.5; ENST00000498540.2; ENST00000454852.6; ENST00000422344.5; ENST00000618771.1; ENST00000584944.5; ENST00000586813.5; ENST00000475329.5 | ||
| External Link | RMBase: m6A_site_354923 | ||
| mod ID: M6ASITE031157 | Click to Show/Hide the Full List | ||
| mod site | chr17:28747593-28747594:+ | [9] | |
| Sequence | AAGGAAGCAGAGCCCCAGGGACAGCCTGGGATTGAGCAGGC | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HepG2 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000394925.5; ENST00000586813.5; ENST00000262395.10; ENST00000618771.1; ENST00000475329.5; ENST00000578917.1; ENST00000422344.5; ENST00000461195.5; ENST00000498540.2; ENST00000473421.1; ENST00000444415.7; ENST00000454852.6; ENST00000262396.10; ENST00000584944.5 | ||
| External Link | RMBase: m6A_site_354924 | ||
| mod ID: M6ASITE031158 | Click to Show/Hide the Full List | ||
| mod site | chr17:28747852-28747853:+ | [10] | |
| Sequence | CTACCCCCAGATCTACCCAGACCCGGAGCTGGAAGTACAAG | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa; MT4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000454852.6; ENST00000586813.5; ENST00000422344.5; ENST00000584944.5; ENST00000461195.5; ENST00000618771.1; ENST00000262396.10; ENST00000473421.1; ENST00000578917.1; ENST00000444415.7; ENST00000262395.10; ENST00000475329.5; ENST00000469529.6 | ||
| External Link | RMBase: m6A_site_354925 | ||
| mod ID: M6ASITE031159 | Click to Show/Hide the Full List | ||
| mod site | chr17:28748099-28748100:+ | [7] | |
| Sequence | AGCCGCCGTGATCTACCTGCACACTTGCAGCATGACTGCCC | ||
| Motif Score | 2.830589286 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000475329.5; ENST00000262396.10; ENST00000469529.6; ENST00000262395.10; ENST00000584944.5; ENST00000586813.5; ENST00000422344.5; ENST00000618771.1; ENST00000444415.7; ENST00000578917.1; ENST00000461195.5; ENST00000454852.6; ENST00000473421.1 | ||
| External Link | RMBase: m6A_site_354926 | ||
| mod ID: M6ASITE031160 | Click to Show/Hide the Full List | ||
| mod site | chr17:28748412-28748413:+ | [7] | |
| Sequence | CACTAAGGAGTTCGTCTTTGACACCATCCAGGTGAGGCCTT | ||
| Motif Score | 2.859755952 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000444415.7; ENST00000262395.10; ENST00000580073.1; ENST00000618771.1; ENST00000461195.5; ENST00000454852.6; ENST00000262396.10; ENST00000586813.5; ENST00000473421.1; ENST00000469529.6 | ||
| External Link | RMBase: m6A_site_354927 | ||
| mod ID: M6ASITE031161 | Click to Show/Hide the Full List | ||
| mod site | chr17:28748586-28748587:+ | [8] | |
| Sequence | GGGCACTGTGGCTCGGGAGGACCTGCCAGGCCATCTGAAGG | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; HepG2; peripheral-blood; HEK293A-TOA; endometrial; HEC-1-A; NB4 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000469529.6; ENST00000586813.5; ENST00000444415.7; ENST00000618771.1; ENST00000262395.10; ENST00000580073.1; ENST00000454852.6; ENST00000262396.10 | ||
| External Link | RMBase: m6A_site_354928 | ||
| mod ID: M6ASITE031162 | Click to Show/Hide the Full List | ||
| mod site | chr17:28748607-28748608:+ | [8] | |
| Sequence | CCTGCCAGGCCATCTGAAGGACAGCTGTAACACCGCCCTGG | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; HepG2; peripheral-blood; HEK293A-TOA; endometrial; HEC-1-A; NB4 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000444415.7; ENST00000454852.6; ENST00000618771.1; ENST00000262396.10; ENST00000580073.1; ENST00000262395.10; ENST00000469529.6 | ||
| External Link | RMBase: m6A_site_354929 | ||
| mod ID: M6ASITE031163 | Click to Show/Hide the Full List | ||
| mod site | chr17:28748646-28748647:+ | [8] | |
| Sequence | GGTGCTCTGCCCATTCAAAGACTCCGGCTGCAAGCACAGGG | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa; HepG2; A549; peripheral-blood; HEK293A-TOA; endometrial; HEC-1-A; NB4 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000444415.7; ENST00000469529.6; ENST00000454852.6; ENST00000262396.10; ENST00000618771.1; ENST00000262395.10; ENST00000580073.1 | ||
| External Link | RMBase: m6A_site_354930 | ||
| mod ID: M6ASITE031164 | Click to Show/Hide the Full List | ||
| mod site | chr17:28748661-28748662:+ | [11] | |
| Sequence | CAAAGACTCCGGCTGCAAGCACAGGGTGAGATGCCCCTTTT | ||
| Motif Score | 2.830589286 | ||
| Cell/Tissue List | kidney | ||
| Seq Type List | m6A-REF-seq | ||
| Transcript ID List | ENST00000580073.1; ENST00000469529.6; ENST00000444415.7; ENST00000262395.10; ENST00000454852.6; ENST00000618771.1; ENST00000262396.10 | ||
| External Link | RMBase: m6A_site_354931 | ||
| mod ID: M6ASITE031165 | Click to Show/Hide the Full List | ||
| mod site | chr17:28748715-28748716:+ | [8] | |
| Sequence | TTTTGCCCTTGAAGCCCTAGACAGAGGCTCAGCTTCTGATG | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa; HepG2; A549; peripheral-blood; HEK293T; HEK293A-TOA; endometrial; HEC-1-A | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000444415.7; ENST00000262395.10; ENST00000469529.6; ENST00000454852.6; ENST00000580073.1; ENST00000262396.10; ENST00000618771.1 | ||
| External Link | RMBase: m6A_site_354932 | ||
| mod ID: M6ASITE031166 | Click to Show/Hide the Full List | ||
| mod site | chr17:28749117-28749118:+ | [12] | |
| Sequence | TGGAAGATTGGCAGCTATGGACGGCGGCTACAGGAGGCCAA | ||
| Motif Score | 3.616982143 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000262396.10; ENST00000444415.7; ENST00000618771.1; ENST00000262395.10 | ||
| External Link | RMBase: m6A_site_354933 | ||
| mod ID: M6ASITE031167 | Click to Show/Hide the Full List | ||
| mod site | chr17:28749126-28749127:+ | [11] | |
| Sequence | GGCAGCTATGGACGGCGGCTACAGGAGGCCAAGGCCAAGCC | ||
| Motif Score | 2.078666667 | ||
| Cell/Tissue List | liver; HEK293T | ||
| Seq Type List | m6A-REF-seq; DART-seq | ||
| Transcript ID List | ENST00000618771.1; ENST00000444415.7; ENST00000262395.10; ENST00000262396.10 | ||
| External Link | RMBase: m6A_site_354934 | ||
| mod ID: M6ASITE031168 | Click to Show/Hide the Full List | ||
| mod site | chr17:28749149-28749150:+ | [12] | |
| Sequence | GGAGGCCAAGGCCAAGCCCAACCTTGAGTGCTTCAGCCCAG | ||
| Motif Score | 2.153267857 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000444415.7; ENST00000262395.10; ENST00000618771.1; ENST00000262396.10 | ||
| External Link | RMBase: m6A_site_354935 | ||
| mod ID: M6ASITE031169 | Click to Show/Hide the Full List | ||
| mod site | chr17:28749194-28749195:+ | [7] | |
| Sequence | CTACACACATAAGTATGGTTACAAGCTGCAGGTGTCTGCAT | ||
| Motif Score | 2.07285119 | ||
| Cell/Tissue List | hESC-HEK293T | ||
| Seq Type List | MAZTER-seq | ||
| Transcript ID List | ENST00000444415.7; ENST00000262396.10; ENST00000262395.10; ENST00000618771.1 | ||
| External Link | RMBase: m6A_site_354936 | ||
| mod ID: M6ASITE031170 | Click to Show/Hide the Full List | ||
| mod site | chr17:28749244-28749245:+ | [12] | |
| Sequence | GCAATGGCAGTGGTGAGGGCACACACCTCTCACTGTACATT | ||
| Motif Score | 2.830589286 | ||
| Cell/Tissue List | HEK293T; hESC-HEK293T | ||
| Seq Type List | DART-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000262396.10; ENST00000618771.1; ENST00000444415.7; ENST00000262395.10 | ||
| External Link | RMBase: m6A_site_354937 | ||
| mod ID: M6ASITE031171 | Click to Show/Hide the Full List | ||
| mod site | chr17:28749246-28749247:+ | [12] | |
| Sequence | AATGGCAGTGGTGAGGGCACACACCTCTCACTGTACATTCG | ||
| Motif Score | 2.084928571 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000618771.1; ENST00000262395.10; ENST00000262396.10; ENST00000444415.7 | ||
| External Link | RMBase: m6A_site_354938 | ||
| mod ID: M6ASITE031172 | Click to Show/Hide the Full List | ||
| mod site | chr17:28749287-28749288:+ | [11] | |
| Sequence | TGTGCTGCCTGGTGCCTTTGACAATCTCCTTGAGTGGCCCT | ||
| Motif Score | 2.859755952 | ||
| Cell/Tissue List | HEK293; kidney; liver; hESC-HEK293T | ||
| Seq Type List | m6A-REF-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000444415.7; ENST00000618771.1; ENST00000262396.10; ENST00000262395.10 | ||
| External Link | RMBase: m6A_site_354939 | ||
| mod ID: M6ASITE031173 | Click to Show/Hide the Full List | ||
| mod site | chr17:28749363-28749364:+ | [8] | |
| Sequence | AGCGACCCTGGGCTGGCTAAACCACAGCACGTCACTGAGAC | ||
| Motif Score | 2.185083333 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; A549; U2OS; H1A; H1B; hNPCs; hESCs; fibroblasts; GM12878; LCLs; MT4; MM6; Huh7; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; TREX; endometrial; HEC-1-A; GSCs; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000262396.10; ENST00000444415.7; ENST00000262395.10; ENST00000618771.1 | ||
| External Link | RMBase: m6A_site_354940 | ||
| mod ID: M6ASITE031174 | Click to Show/Hide the Full List | ||
| mod site | chr17:28749382-28749383:+ | [8] | |
| Sequence | AACCACAGCACGTCACTGAGACCTTCCACCCCGACCCAAAC | ||
| Motif Score | 2.876744048 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; A549; U2OS; H1A; H1B; hNPCs; hESCs; fibroblasts; GM12878; LCLs; MT4; H1299; MM6; Huh7; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; TREX; endometrial; HEC-1-A; GSCs; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000444415.7; ENST00000262395.10; ENST00000262396.10; ENST00000618771.1 | ||
| External Link | RMBase: m6A_site_354941 | ||
| mod ID: M6ASITE031175 | Click to Show/Hide the Full List | ||
| mod site | chr17:28749401-28749402:+ | [8] | |
| Sequence | GACCTTCCACCCCGACCCAAACTGGAAGAATTTCCAGAAGC | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; A549; U2OS; H1A; H1B; hNPCs; hESCs; fibroblasts; GM12878; LCLs; MT4; H1299; MM6; Huh7; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; TREX; endometrial; HEC-1-A; GSCs; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000262396.10; ENST00000262395.10; ENST00000444415.7; ENST00000618771.1 | ||
| External Link | RMBase: m6A_site_354942 | ||
| mod ID: M6ASITE031176 | Click to Show/Hide the Full List | ||
| mod site | chr17:28749494-28749495:+ | [8] | |
| Sequence | CAAGTTCATCTCCCACCAGGACATTCGAAAGCGAAACTATG | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; CD34; HepG2; HEK293T; brain; A549; hESC-HEK293T; U2OS; H1A; H1B; hNPCs; hESCs; fibroblasts; GM12878; LCLs; H1299; MM6; Huh7; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; TREX; endometrial; HEC-1-A; GSCs; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq; m6A-REF-seq; MAZTER-seq; m6A-CLIP/IP | ||
| Transcript ID List | ENST00000444415.7; ENST00000262395.10; ENST00000618771.1; ENST00000262396.10 | ||
| External Link | RMBase: m6A_site_354943 | ||
| mod ID: M6ASITE031177 | Click to Show/Hide the Full List | ||
| mod site | chr17:28749509-28749510:+ | [8] | |
| Sequence | CCAGGACATTCGAAAGCGAAACTATGTGCGGGATGATGCAG | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa; CD34; HepG2; HEK293T; A549; U2OS; H1A; H1B; hNPCs; hESCs; fibroblasts; GM12878; LCLs; CD8T; H1299; MM6; Huh7; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; TREX; endometrial; HEC-1-A; GSCs; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq; m6A-CLIP/IP | ||
| Transcript ID List | ENST00000262395.10; ENST00000618771.1; ENST00000444415.7; ENST00000262396.10 | ||
| External Link | RMBase: m6A_site_354944 | ||
| mod ID: M6ASITE031178 | Click to Show/Hide the Full List | ||
| mod site | chr17:28749552-28749553:+ | [8] | |
| Sequence | TTCATCCGTGCTGCTGTTGAACTGCCCCGGAAGATCCTCAG | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; A549; U2OS; H1A; H1B; hNPCs; hESCs; fibroblasts; GM12878; LCLs; H1299; MM6; Huh7; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; TREX; endometrial; HEC-1-A; GSCs; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq; m6A-CLIP/IP | ||
| Transcript ID List | ENST00000444415.7; ENST00000618771.1; ENST00000262396.10; ENST00000262395.10 | ||
| External Link | RMBase: m6A_site_354945 | ||
| mod ID: M6ASITE031179 | Click to Show/Hide the Full List | ||
| mod site | chr17:28749603-28749604:+ | [13] | |
| Sequence | GTGGGGTTCGAGGGGAAAGGACGATGGGGCATGACCTCAGT | ||
| Motif Score | 3.616982143 | ||
| Cell/Tissue List | A549 | ||
| Seq Type List | m6A-CLIP/IP | ||
| Transcript ID List | ENST00000444415.7; ENST00000262396.10; ENST00000262395.10 | ||
| External Link | RMBase: m6A_site_354946 | ||
| mod ID: M6ASITE031180 | Click to Show/Hide the Full List | ||
| mod site | chr17:28749638-28749639:+ | [8] | |
| Sequence | CTCAGTCAGGCACTGGCTGAACTTGGAGAGGGGGCCGGACC | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; A549; U2OS; H1A; H1B; hNPCs; hESCs; fibroblasts; GM12878; LCLs; H1299; MM6; Huh7; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; TREX; endometrial; HEC-1-A; GSCs; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000262395.10; ENST00000444415.7; ENST00000262396.10 | ||
| External Link | RMBase: m6A_site_354947 | ||
| mod ID: M6ASITE031181 | Click to Show/Hide the Full List | ||
| mod site | chr17:28749656-28749657:+ | [8] | |
| Sequence | GAACTTGGAGAGGGGGCCGGACCCCCGTCAGCTGCTTCTGC | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; A549; U2OS; H1A; H1B; hNPCs; hESCs; fibroblasts; GM12878; LCLs; H1299; MM6; Huh7; Jurkat; CD4T; peripheral-blood; GSC-11; HEK293A-TOA; iSLK; MSC; TIME; TREX; endometrial; HEC-1-A; GSCs; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000444415.7; ENST00000262395.10 | ||
| External Link | RMBase: m6A_site_354948 | ||
| mod ID: M6ASITE031182 | Click to Show/Hide the Full List | ||
| mod site | chr17:28749766-28749767:+ | [8] | |
| Sequence | AGCCCTGGCCCCTGTGGGGAACAGGTCTTGGGGTCATGAAG | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; A549; U2OS; H1A; H1B; LCLs; MM6; Jurkat; CD4T; peripheral-blood; GSC-11; iSLK; TREX; MSC; endometrial; HEC-1-A; GSCs; NB4 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000262395.10; ENST00000444415.7 | ||
| External Link | RMBase: m6A_site_354949 | ||
| mod ID: M6ASITE031183 | Click to Show/Hide the Full List | ||
| mod site | chr17:28749795-28749796:+ | [8] | |
| Sequence | GGGGTCATGAAGGGCTGGAAACAAGTGACCCCAGGGCCTGT | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; A549; hESC-HEK293T; U2OS; H1A; H1B; Jurkat; CD4T; peripheral-blood; GSC-11; iSLK; TREX; endometrial; HEC-1-A; NB4; MM6 | ||
| Seq Type List | m6A-seq; MeRIP-seq; MAZTER-seq | ||
| Transcript ID List | ENST00000262395.10; ENST00000444415.7 | ||
| External Link | RMBase: m6A_site_354950 | ||
| mod ID: M6ASITE031184 | Click to Show/Hide the Full List | ||
| mod site | chr17:28749837-28749838:+ | [8] | |
| Sequence | TCCCTTCTTGGGTAGGGCAGACATGCCTTGGTGCCGGTCAC | ||
| Motif Score | 2.897386905 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; A549; hESC-HEK293T; H1A; H1B; Jurkat; CD4T; peripheral-blood; GSC-11; TREX; endometrial; HEC-1-A; NB4; MM6 | ||
| Seq Type List | m6A-seq; MeRIP-seq; MAZTER-seq; m6A-CLIP/IP | ||
| Transcript ID List | ENST00000444415.7; ENST00000262395.10 | ||
| External Link | RMBase: m6A_site_354952 | ||
| mod ID: M6ASITE031185 | Click to Show/Hide the Full List | ||
| mod site | chr17:28749865-28749866:+ | [12] | |
| Sequence | TGGTGCCGGTCACACTCTACACGGACTGAGGTGCCTGCTCA | ||
| Motif Score | 2.058863095 | ||
| Cell/Tissue List | HEK293T | ||
| Seq Type List | DART-seq | ||
| Transcript ID List | ENST00000262395.10; ENST00000444415.7 | ||
| External Link | RMBase: m6A_site_354953 | ||
| mod ID: M6ASITE031186 | Click to Show/Hide the Full List | ||
| mod site | chr17:28749869-28749870:+ | [8] | |
| Sequence | GCCGGTCACACTCTACACGGACTGAGGTGCCTGCTCAGGTG | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa; HepG2; HEK293T; A549; H1A; H1B; peripheral-blood; GSC-11; endometrial; HEC-1-A; NB4; MM6 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000444415.7; ENST00000262395.10 | ||
| External Link | RMBase: m6A_site_354954 | ||
| mod ID: M6ASITE031187 | Click to Show/Hide the Full List | ||
| mod site | chr17:28750135-28750136:+ | [8] | |
| Sequence | TCCCTGGGAGGCTCAACCAAACTGAAGGCAAAGAAAGGACC | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000262395.10 | ||
| External Link | RMBase: m6A_site_354958 | ||
| mod ID: M6ASITE031188 | Click to Show/Hide the Full List | ||
| mod site | chr17:28750153-28750154:+ | [8] | |
| Sequence | AAACTGAAGGCAAAGAAAGGACCAGTCAGAGAAGGGCCGCT | ||
| Motif Score | 3.622404762 | ||
| Cell/Tissue List | HeLa; H1A; H1B; GSC-11; NB4 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000262395.10 | ||
| External Link | RMBase: m6A_site_354959 | ||
| mod ID: M6ASITE031189 | Click to Show/Hide the Full List | ||
| mod site | chr17:28750237-28750238:+ | [8] | |
| Sequence | CTCTGATGGACGCCGGGAAAACTGCATCGGGCTTTGTGTGG | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa; H1A; H1B; GSC-11; NB4 | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | ENST00000262395.10 | ||
| External Link | RMBase: m6A_site_354960 | ||
| mod ID: M6ASITE031190 | Click to Show/Hide the Full List | ||
| mod site | chr17:28750451-28750452:+ | [8] | |
| Sequence | TTATTATTTCATTGAAAAGAACTGTCAAGTGAAATACTTTT | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa; HEK293T; hESCs; GM12878; A549; Huh7 | ||
| Seq Type List | m6A-seq; MeRIP-seq; m6A-CLIP/IP | ||
| Transcript ID List | ENST00000262395.10 | ||
| External Link | RMBase: m6A_site_354961 | ||
| mod ID: M6ASITE031191 | Click to Show/Hide the Full List | ||
| mod site | chr17:28750572-28750573:+ | [8] | |
| Sequence | TGAATGCTTTCTAACTCCGAACCCCAGCCACATCCAGGGAC | ||
| Motif Score | 2.930744048 | ||
| Cell/Tissue List | HeLa; HEK293T; GM12878; Huh7; HEK293A-TOA; TREX | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000262395.10 | ||
| External Link | RMBase: m6A_site_354962 | ||
| mod ID: M6ASITE031192 | Click to Show/Hide the Full List | ||
| mod site | chr17:28750591-28750592:+ | [8] | |
| Sequence | AACCCCAGCCACATCCAGGGACTGGGTGTTGAGCAAAAGGG | ||
| Motif Score | 4.065041667 | ||
| Cell/Tissue List | HeLa; HEK293T; GM12878; Huh7; HEK293A-TOA; TREX | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000262395.10 | ||
| External Link | RMBase: m6A_site_354963 | ||
| mod ID: M6ASITE031193 | Click to Show/Hide the Full List | ||
| mod site | chr17:28750715-28750716:+ | [8] | |
| Sequence | ATCCACGTTTCACCTATGAAACATGAACAGAGGAGACTGAC | ||
| Motif Score | 2.20572619 | ||
| Cell/Tissue List | HeLa; Huh7; GSC-11 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000262395.10 | ||
| External Link | RMBase: m6A_site_354964 | ||
| mod ID: M6ASITE031194 | Click to Show/Hide the Full List | ||
| mod site | chr17:28750721-28750722:+ | [8] | |
| Sequence | GTTTCACCTATGAAACATGAACAGAGGAGACTGACTTATCA | ||
| Motif Score | 2.951386905 | ||
| Cell/Tissue List | HeLa; Huh7; GSC-11 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000262395.10 | ||
| External Link | RMBase: m6A_site_354965 | ||
| mod ID: M6ASITE031195 | Click to Show/Hide the Full List | ||
| mod site | chr17:28750730-28750731:+ | [8] | |
| Sequence | ATGAAACATGAACAGAGGAGACTGACTTATCAGTGATTCTT | ||
| Motif Score | 3.319380952 | ||
| Cell/Tissue List | HeLa; Huh7; GSC-11 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000262395.10 | ||
| External Link | RMBase: m6A_site_354966 | ||
| mod ID: M6ASITE031196 | Click to Show/Hide the Full List | ||
| mod site | chr17:28750763-28750764:+ | [8] | |
| Sequence | TGATTCTTCCGCGGGTTCGGACAGGGCCTCGATTCTGTTTT | ||
| Motif Score | 3.643047619 | ||
| Cell/Tissue List | HeLa; Huh7; GSC-11 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000262395.10 | ||
| External Link | RMBase: m6A_site_354967 | ||
| mod ID: M6ASITE031197 | Click to Show/Hide the Full List | ||
| mod site | chr17:28750786-28750787:+ | [8] | |
| Sequence | GGGCCTCGATTCTGTTTTAAACTCCAGTAGTCCCTAGAAAT | ||
| Motif Score | 2.627720238 | ||
| Cell/Tissue List | HeLa; Huh7; GSC-11 | ||
| Seq Type List | m6A-seq; MeRIP-seq | ||
| Transcript ID List | ENST00000262395.10 | ||
| External Link | RMBase: m6A_site_354968 | ||
| mod ID: M6ASITE031198 | Click to Show/Hide the Full List | ||
| mod site | chr17:28750945-28750946:+ | [8] | |
| Sequence | TTGTTGTAAAGATTAAATGAACTGGTCAGCACACAGTGCCT | ||
| Motif Score | 3.373380952 | ||
| Cell/Tissue List | HeLa | ||
| Seq Type List | m6A-seq | ||
| Transcript ID List | rmsk_4664087; ENST00000262395.10 | ||
| External Link | RMBase: m6A_site_354969 | ||
References