m6A-centered Crosstalk Information
Mechanism of Crosstalk between m6A Modification and Epigenetic Regulation
| Crosstalk ID |
M6ACROT05829
|
[1] | |||
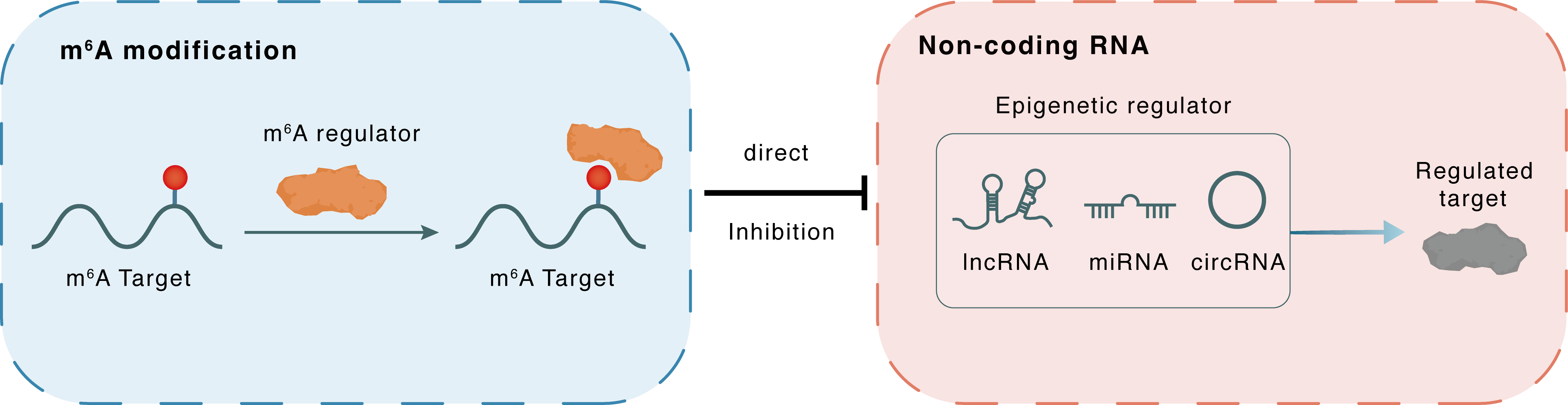 m6A modification
pre-miR-665
pre-miR-665
YTHDF1
m6A modification
pre-miR-665
pre-miR-665
YTHDF1
 : m6A sites
Direct
Inhibition
Non-coding RNA
miR-665
DLX3
lncRNA miRNA circRNA : m6A sites
Direct
Inhibition
Non-coding RNA
miR-665
DLX3
lncRNA miRNA circRNA
|
|||||
| m6A Modification: | |||||
|---|---|---|---|---|---|
| m6A Regulator | YTH domain-containing family protein 1 (YTHDF1) | READER | |||
| m6A Target | pre-miR-665 | ||||
| Epigenetic Regulation that have Cross-talk with This m6A Modification: | |||||
| Epigenetic Regulation Type | Non-coding RNA (ncRNA) | ||||
| Epigenetic Regulator | hsa-miR-665 | microRNA | View Details | ||
| Regulated Target | Homeobox protein DLX-3 (DLX3) | View Details | |||
| Crosstalk Relationship | m6A → ncRNA | Inhibition | |||
| Crosstalk Mechanism | m6A regulators directly modulate the functionality of ncRNAs through specific targeting ncRNA | ||||
| Crosstalk Summary | The targeting of Homeobox protein DLX-3 (DLX3) by hsa-miR-665 and the potential direct regulation of DLX3 expression by METTL3, mediated by the "reader" protein YTHDF1, were demonstrated. Overall, the METTL3/pre-miR-665/DLX3 pathway might provide a new target for SCAP-based tooth root/maxillofacial bone tissue regeneration. | ||||
| Responsed Disease | Structural developmental anomalies of teeth and periodontal tissues | ICD-11: LA30.0 | |||
| Cell Process | Cell differentiation | ||||
In-vitro Model |
HEK293T | Normal | Homo sapiens | CVCL_0063 | |