m6A-centered Crosstalk Information
Mechanism of Crosstalk between m6A Modification and Epigenetic Regulation
| Crosstalk ID |
M6ACROT05734
|
[1] | |||
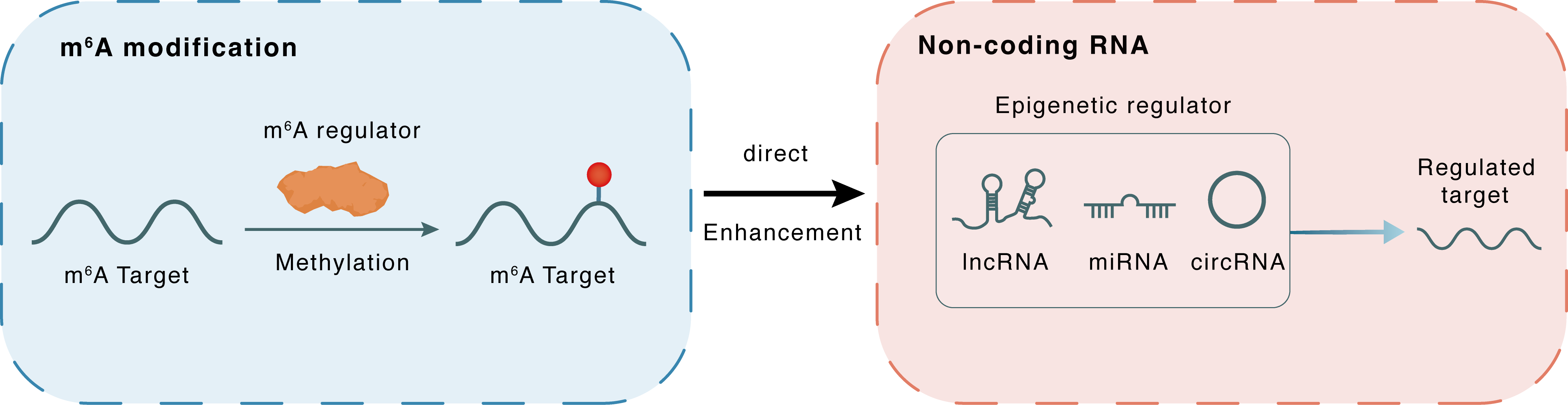 m6A modification
miR-6858
miR-6858
METTL14
Methylation
m6A modification
miR-6858
miR-6858
METTL14
Methylation
 : m6A sites
Direct
Enhancement
Non-coding RNA
miR-6858
GSDMC
lncRNA miRNA circRNA : m6A sites
Direct
Enhancement
Non-coding RNA
miR-6858
GSDMC
lncRNA miRNA circRNA
|
|||||
| m6A Modification: | |||||
|---|---|---|---|---|---|
| m6A Regulator | Methyltransferase-like 14 (METTL14) | WRITER | |||
| m6A Target | miR-6858 | ||||
| Epigenetic Regulation that have Cross-talk with This m6A Modification: | |||||
| Epigenetic Regulation Type | Non-coding RNA (ncRNA) | ||||
| Epigenetic Regulator | miR-6858 | microRNA | View Details | ||
| Regulated Target | Gasdermin C (GSDMC) | View Details | |||
| Crosstalk Relationship | m6A → ncRNA | Enhancement | |||
| Crosstalk Mechanism | m6A regulators directly modulate the functionality of ncRNAs through specific targeting ncRNA | ||||
| Crosstalk Summary | METTL14-upregulated miR-6858 triggers cell apoptosis in keratinocytes of oral lichen planus through decreasing Gasdermin C (GSDMC) | ||||
| Responsed Disease | Lichen planus | ICD-11: EA91.4 | |||
| Cell Process | mRNA degradation | ||||
| Cell apoptosis | |||||