m6A-centered Crosstalk Information
Mechanism of Crosstalk between m6A Modification and Epigenetic Regulation
| Crosstalk ID |
M6ACROT05399
|
[1] | |||
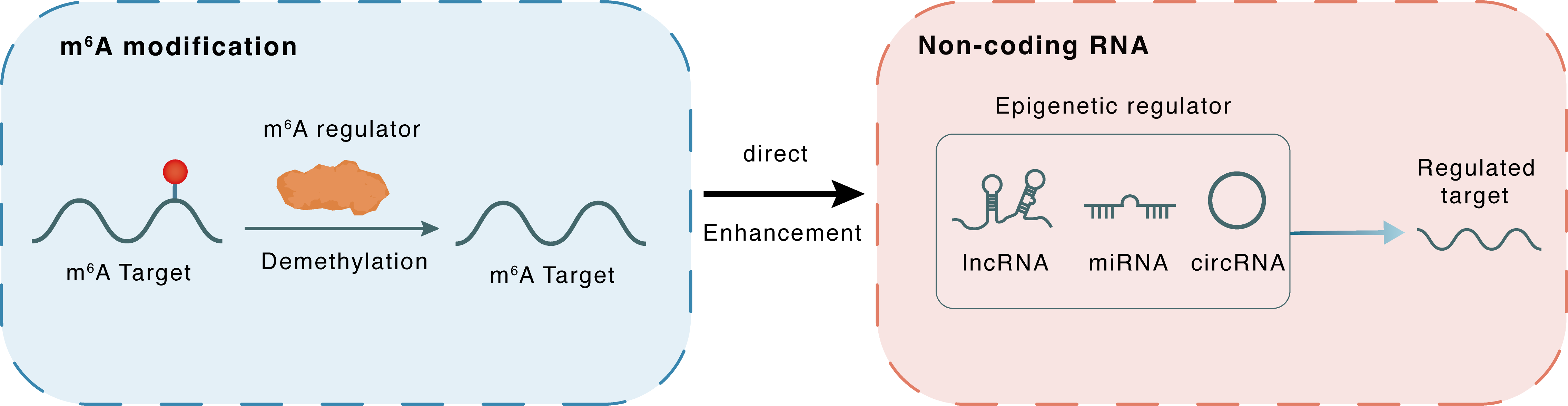 m6A modification
GAS5
GAS5
ALKBH5
Demethylation
m6A modification
GAS5
GAS5
ALKBH5
Demethylation
 : m6A sites
Direct
Enhancement
Non-coding RNA
GAS5
GAS5-AS1
lncRNA miRNA circRNA : m6A sites
Direct
Enhancement
Non-coding RNA
GAS5
GAS5-AS1
lncRNA miRNA circRNA
|
|||||
| m6A Modification: | |||||
|---|---|---|---|---|---|
| m6A Regulator | RNA demethylase ALKBH5 (ALKBH5) | ERASER | |||
| m6A Target | Growth arrest specific 5 (GAS5) | ||||
| Epigenetic Regulation that have Cross-talk with This m6A Modification: | |||||
| Epigenetic Regulation Type | Non-coding RNA (ncRNA) | ||||
| Epigenetic Regulator | Growth arrest specific 5 (GAS5) | LncRNA | View Details | ||
| Regulated Target | GAS5 antisense RNA 1 (GAS5-AS1) | View Details | |||
| Crosstalk Relationship | m6A → ncRNA | Enhancement | |||
| Crosstalk Mechanism | m6A regulators directly modulate the functionality of ncRNAs through specific targeting ncRNA | ||||
| Crosstalk Summary | The GAS5 antisense RNA 1 (GAS5-AS1) expression in cervical cancer tissues was markedly decreased when compared with that in the adjacent normal tissues. GAS5-AS1 interacted with the tumor suppressor Growth arrest specific 5 (GAS5), and increased its stability by interacting with RNA demethylase ALKBH5 and decreasing GAS5 N6-methyladenosine (m6A) modification. m6A-mediated GAS5 RNA degradation relied on the m6A reader protein YTHDF2-dependent pathway. | ||||
| Responsed Disease | Cervical cancer | ICD-11: 2C77 | |||
| Cell Process | Cell proliferation and metastasis | ||||
In-vitro Model |
Ca Ski | Cervical squamous cell carcinoma | Homo sapiens | CVCL_1100 | |
| SiHa | Cervical squamous cell carcinoma | Homo sapiens | CVCL_0032 | ||
| C-33 A | Cervical squamous cell carcinoma | Homo sapiens | CVCL_1094 | ||
| HeLa | Endocervical adenocarcinoma | Homo sapiens | CVCL_0030 | ||
| In-vivo Model | 200 uL PBS containing 1× 107 cells of stable cells were subcutaneously injected into male BALB/c athymic nude mice (6-week old, 18-20 g). | ||||